Abstract
Virus-induced cytoplasmic inclusion bodies (referred to as virus replication complexes [VRCs]) consisting of virus and host components are observed in plant cells infected with tobacco mosaic virus, but the components that modulate their form and function are not fully understood. Here, we show that the tobacco mosaic virus 126-kD protein fused with green fluorescent protein formed cytoplasmic bodies (126-bodies) in the absence of other viral components. Using mutant 126-kD:green fluorescent fusion proteins and viral constructs expressing the corresponding mutant 126-kD proteins, it was determined that the size of the 126-bodies and the corresponding VRCs changed in synchrony for each 126-kD protein mutation tested. Through colabeling experiments, we observed the coalignment and intracellular trafficking of 126-bodies and, regardless of size, VRCs, along microfilaments (MFs). Disruption of MFs with MF-depolymerizing agents or through virus-induced gene silencing compromised the intracellular trafficking of the 126-bodies and VRCs and virus cell-to-cell movement, but did not decrease virus accumulation to levels that would affect virus movement or prevent VRC formation. Our results indicate that (1) the 126-kD protein modulates VRC size and traffics along MFs in cells; (2) VRCs traffic along MFs in cells, possibly through an interaction with the 126-kD protein, and the negative effect of MF antagonists on 126-body and VRC intracellular movement and virus cell-to-cell movement correlates with the disruption of this association; and (3) virus movement was not correlated with VRC size.
Many plant viruses form cytoplasmic structures during the infection process. The content and intracellular movement of these structures is now under intensive study to better understand their function during virus infection and host response. Irregularly shaped cytoplasmic inclusion bodies, termed X-bodies, viroplasms, or virus replication complexes (VRCs), are observed in tobacco mosaic virus (TMV)-infected plant cells (Shalla, 1964; Beachy and Zaitlin, 1975; Hills et al., 1987; Heinlein et al., 1998; Más and Beachy, 1999; Szécsi et al., 1999; Asurmendi et al., 2004; Kawakami et al., 2004). These inclusion bodies contain ribosomes, tubule-like structures, endoplasmic reticulum, viral movement protein (MP), viral 126- and 183-kD proteins, and viral RNA (vRNA; Shalla, 1964; Martelli and Russo, 1977; Hills et al., 1987; Saito et al., 1987; Heinlein et al., 1998; Más and Beachy, 1999). In addition, the inclusion bodies are dynamic entities whose protein content and subcellular location change over time (Heinlein et al., 1998; Más and Beachy, 1999; Szécsi et al., 1999). The full content and function of these TMV-induced bodies, however, remains unclear. The inclusions have been suggested to represent (1) “viral factories” where viral translation and replication occur in the same subcellular site (Heinlein et al., 1998; Más and Beachy, 1999), (2) locations where an overexpressed viral protein(s) is accumulated before degradation (Padgett et al., 1996), and (3) cell-to-cell movement complexes (Szécsi et al., 1999; Asurmendi et al., 2004; Kawakami et al., 2004).
In regard to TMV-encoded proteins and virus cell-to-cell movement, studies have focused on the MP and its interactions with host factors (for review, see Haywood et al., 2002; Lazarowitz and Beachy, 1999; Roberts and Oparka, 2003; Heinlein and Epel, 2004; Oparka, 2004; Waigmann et al., 2004). However, recently it was shown conclusively through studies with mutant viruses that the 126- and/or the amino-coterminal 183-kD protein, independent of any involvement in virus replication, are/is necessary for the cell-to-cell movement of this virus (Hirashima and Watanabe, 2001, 2003). The mechanism by which the 126-kD protein and/or the 183-kD protein in VRCs support(s) the cell-to-cell movement of the virus is unknown.
In regard to host factors, cytoskeletal elements have been implicated in assisting the movement of some plant viruses (Reichel and Beachy, 1999; Aaziz et al., 2001; Heinlein, 2002; Roberts and Oparka, 2003; Heinlein and Epel, 2004; Oparka, 2004; Waigmann et al., 2004; Nelson, 2005). During TMV infection, microtubules (MTs) and the viral MP colocalize in infected cells (McLean et al., 1995; Heinlein et al., 1995, 1998; Boyko et al., 2000b). This association is disrupted by treatment with a pharmacological agent that depolymerizes MTs (Heinlein et al., 1995; Más and Beachy, 1999, 2000). Results from several studies using TMV mutants expressing modified MPs or cells treated to disrupt MT arrays support a role of MTs in viral cell-to-cell movement (Heinlein et al., 1995, 1998; Más and Beachy, 1999, 2000; Boyko et al., 2000a, 2002). However, results from a recent study where the MT array was disrupted in situ with pharmacological agents or by silencing of the α-tubulin gene indicated that disruption of the MTs did not alter TMV cell-to-cell spread (Gillespie et al., 2002). These authors suggested that MTs function to transport TMV MP to a subcellular site for degradation. This conclusion is further supported by the finding that a host protein, MPB2C, which binds to the MP and colocalizes with it at MTs, interferes with MP cell-to-cell movement (Kragler et al., 2003).
Less is known about the role of other cytoskeletal elements during TMV accumulation and movement. Actin and myosin were detected in plasmodesmata (PD) through immunocytological methods (White et al., 1994; Radford and White, 1998). In addition, inhibitors of actin activity increased the size exclusion limit of PD in treated tissue (Ding et al., 1996). Microfilaments (MFs) coaligned with or bound to TMV MP, respectively, in Bright Yellow-2 (BY-2) protoplast cells or in vitro (McLean et al., 1995). Although an MF antagonist, latrunculin B (LatB), blocks VRC cell-to-cell trafficking (Kawakami et al., 2004), it remains to be determined how this pharmacological treatment functions within the cell to prevent this movement (i.e. does it block movement indirectly by modifying the size exclusion limit of PD or directly by interfering with the interaction between VRCs and MFs).
In this study, we provide additional insights on the importance of the 126-kD protein for the formation of cytoplasmic inclusion bodies when expressed both in the absence (126-bodies) and presence (VRCs) of other viral components. Furthermore, we show by double-labeling the association of the 126-bodies and VRCs with the MFs, the movement of these bodies along MFs, and the effect of MF antagonists on this colocalization and intracellular movement. We also investigated the effect of MF antagonists and VRC size on the cell-to-cell movement of the virus. Our results lend support for the consideration of a new model describing TMV movement to PD involving the 126-kD protein.
RESULTS
The 126-kD Protein Modulates VRC Size within the Plant Cell
The viral component(s) that modulates VRC size and structure during TMV infection has not been determined. To address this issue, variants of the 126-kD protein fused with green fluorescent protein (GFP) and infectious transcripts of TMV-MP:GFP containing the variant 126-kD protein open reading frames (ORFs) were used in transient expression and infection studies, respectively (Fig. 1). The 126-kD protein encoded by the MIC and U1 strains of TMV differ at eight amino acids (arbitrarily labeled 1–8 from the N to C terminus; Holt et al., 1990). Although these strains differ in the systemic symptoms they induce, they spread cell to cell at the same rate in inoculated leaves of Nicotiana tabacum (Nelson et al., 1993; Bao et al., 1996). Two mutants of MIC, MIC1,3 and MIC2 (amino acids 1 and 3 or 2 altered from the parental MIC-encoded to U1-encoded residue), were also used in this study since the two mutant 126-kD protein:GFP (126:GFP) fusions induced different sized fluorescent bodies when expressed in cells (Ding et al., 2004). In addition, MIC1,3 and MIC2 moved cell to cell at rates similar to MIC (Bao et al., 1996; Ding et al., 2004).
Figure 1.
Schematics of transient expression constructs (A–D) and virus clones (E and G) used to study the function of the TMV 126-kD protein in cytoplasmic body formation and virus intracellular and cell-to-cell movement. The schematics are not drawn to scale. Horizontal lines represent 5′ and 3′ untranslated regions, rectangular boxes represent open reading frames, and arrowed rectangles represent promoter sequences. Promoter-gene-terminator constructs for particle bombardment (D) or agroinfiltration (A, B, C, D) studies and TMV constructs for virus infection studies (E and G) are shown. The amino acid alterations and their positions within the 126-kD protein, the effects of which were subsequently studied in both transient expression and infection experiments, are shown within the TMV genome (F). Asterisks indicate no change from the parental MIC sequence. ▾, UGA leaky amber termination codon that is read through to produce the 183-kD protein. Domains I and II (I, II) are regions with less sequence similarity between sindbis-like plant viruses and that regulate suppressor activity (Kubota et al., 2003; Ding et al., 2004). MT, Methyltransferase core domain; HEL, helicase core domain; POL, RNA-dependent RNA polymerase domain; 2 × 35S, enhanced 35S promoter; 126, 126-kD protein; 183, 183-kD protein.
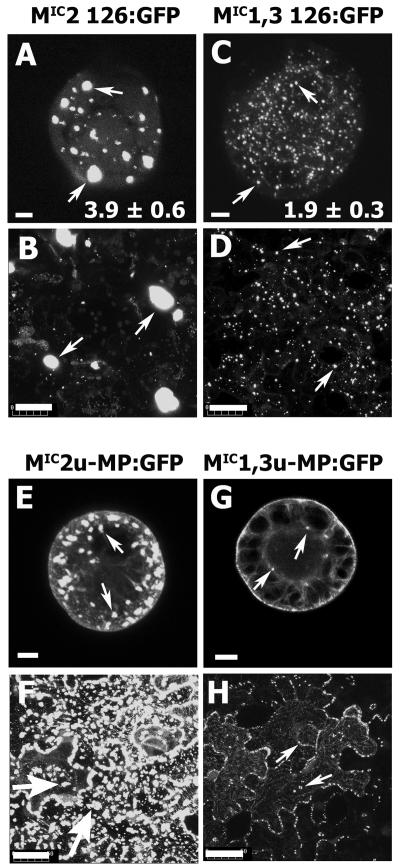
Expression of 126-kD mutant proteins fused with GFP in the absence of other viral components caused the formation of fluorescent cytoplasmic bodies (126-bodies) in BY-2 suspension cells and Nicotiana benthamiana leaf epidermal cells (Fig. 2, A–D). The 126-bodies formed by MIC2 126:GFP were larger than those formed by MIC1,3 126:GFP, in agreement with previous observations (Fig. 2, A–D; Ding et al., 2004). The decreased 126-body size corresponds to a decrease in RNA suppressor activity of the 126-kD protein when infections with viruses containing the corresponding mutant 126-kD protein were analyzed (Ding et al., 2004). Expression of free GFP displayed a diffuse fluorescence (data not shown).
Figure 2.
The effect of the 126-kD protein on cytoplasmic body formation in the absence (126-bodies) or presence (VRCs) of other viral factors. MIC2 126:GFP and MIC1,3 126:GFP fusion proteins were transiently expressed either in N. tabacum cv BY-2 suspension cells through particle bombardment (A and C) or in N. benthamiana leaves through agroinfiltration (B and D). Numbers in A and C indicate the mean diameters of the 126-bodies for three replicate means per treatment (each replicate mean consisted of 10 126-bodies per cell). The difference in mean values was significant at the 5% level of significance as determined by analysis of variance followed by lsd calculations (Gomez and Gomez, 1984). The same relative variation in 126-body and VRC diameter for each set of treatments was observed in the other horizontal panels (data not shown). The images were captured using confocal laser scanning microscopy either at 20 h postbombardment (A and C) or at 3 d postinfiltration (B and D). Example cytoplasmic bodies are indicated by arrows. Infectious viral transcripts containing altered 126-kD protein ORF were either electroporated into BY-2 protoplasts (E and G) or directly inoculated onto leaves of N. benthamiana (F and H). The VRCs were captured using confocal laser scanning microscopy at either 20 h postinoculation of protoplasts or at 4 dpi of leaves. Example VRCs are indicated by arrows. Bars = 10 μm (A, C, E and G) and 50 μm (B, D, F, and H).
Previous studies determined that the 126-kD protein and MP often colocalize within the cytoplasmic bodies during virus infection in cells (e.g. Heinlein et al., 1998; Más and Beachy, 1999; Szécsi et al., 1999). We used this information to study the effect of the 126-kD protein on VRC size during TMV infection through observation of a reporter MP:GFP fusion expressed from chimeric virus genomes, each expressing one of the 126-kD protein variants (U1-MP:GFP, MIC2u-MP:GFP, MICu-MP:GFP, and MIC1,3u-MP:GFP; see “Materials and Methods” for explanation of chimeric virus names). We determined that the 126-kD protein and the MP:GFP fusion colocalized in BY-2 protoplasts and N. benthamiana leaf cells during the phases of infection we utilized in our studies (Fig. 3, A–D). VRCs formed by MIC2u-MP:GFP and MIC1,3u-MP:GFP varied in size similarly to the 126-bodies formed by the respective variant 126:GFP fusion in protoplasts and leaves (Fig. 2, compare A–D and E–H). In addition, VRCs formed by U1-MP:GFP or MICu-MP:GFP were larger or intermediate in size, respectively, to those produced by MIC2u-MP:GFP and MIC1,3u-MP:GFP (Table I). Thus, through the expression of the mutant 126-kD protein during infection, it was apparent that altering the 126-kD protein modulated VRC size in BY-2 protoplasts and/or N. benthamiana leaves.
Figure 3.
The 126-kD protein colocalized with MP:GFP in VRCs during virus infection stages studied in both N. benthamiana leaves and BY-2 protoplasts. N. benthamiana leaf tissues infected mechanically or BY-2 protoplasts inoculated by electroporation with MIC2u-MP:GFP transcripts were collected at 4 dpi (A) or at 8 h postinoculation (B), 16 h postinoculation (C), or 24 h postinoculation (D), which represent early, mid, and late infection stages, respectively. The collected leaf tissues or protoplasts were fixed and labeled with antisera raised against 126-kD protein and rhodamine-conjugated secondary antibody. The fluorescent signals from MP:GFP and rhodamine-labeled 126-kD protein were scanned in separate channels through confocal laser scanning microscopy. Arrows indicated the colocalization of MP:GFP and 126-kD protein. Bars = 20 μm in A and 10 μm in B to D.
Table I.
VRC size of TMV strains and mutants
| TMV Strains and Mutants
|
VRC Diametersa
|
|---|---|
| N. benthamiana | |
| U1-MP:GFP | 6.9 ± 2.7 |
| MIC2u-MP:GFP | 5.9 ± 1.7 |
| MICu-MP:GFP | 4.2 ± 1.2 |
| MIC1,3u-MP:GFP | 3.2 ± 0.8 |
Mean diameters for three replicates (each replicate consisting of diameters from 10 VRCs per leaf) ± sd.
The 126-Bodies and the VRCs Coalign with and Traffic along MFs
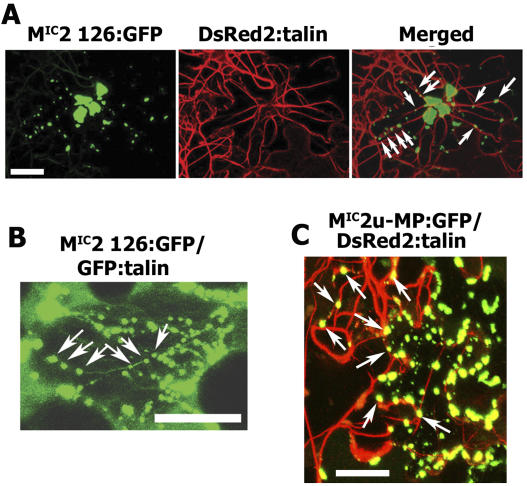
VRCs containing the 126-kD protein are observed on either side of PD at the infection front and move to and accumulate on either side of PD within cells at this location during virus spread (Szécsi et al., 1999; Kawakami et al., 2004). In a recent report, it was shown that LatB decreases cell-to-cell movement of VRCs, suggesting that TMV movement is dependent on MFs (Kawakami et al., 2004). However, an association between VRCs and MFs was not demonstrated. To clarify this issue, we analyzed the association of the 126-bodies and VRCs with MFs through dual-labeling experiments. A DsRed2:talin fusion was constructed as a reporter for MFs and coexpressed with 126:GFP fusions in leaf cells of N. benthamiana through agroinfiltration. In cells expressing both the 126-bodies, representing the MIC2 126:GFP fusion, and DsRed2:talin, the 126-bodies mostly coaligned with MFs (Fig. 4A). Unlike GFP, DsRed forms a tetramer in order to fluoresce (Baird et al., 2000). As a result, the MFs decorated by DsRed2:talin were thicker compared with the finer filaments characteristic of the actin network. To determine the authenticity of results obtained with DsRed2:talin, a GFP:talin fusion was used as a marker for MF position (Blancaflor, 2002). Despite the similar emission spectra for GFP:talin and MIC2 126:GFP, it was possible to distinguish between the filamentous structures decorated by GFP:talin and the bodies formed by the MIC2 126:GFP. In agreement with the DsRed2:talin results, the 126-bodies mostly coaligned with the MF marker (Fig. 4B). GFP:talin or DsRed2:talin in the absence of the 126-kD protein did not form cytoplasmic inclusion bodies (data not shown). The coalignment of the MIC2 126:GFP with MFs was further confirmed using a novel F-actin marker based on the actin binding domain 2 of fimbrin (ABD2:GFP; Wang et al., 2004). The ABD2:GFP was shown to decorate a more dynamic actin network compared with the more popular talin reporters (Wang et al., 2004). Coinfiltration of ABD2:GFP and MIC2 126:GFP also showed coalignment of the fused proteins (data not shown). Therefore, using several MF markers, we demonstrated that the 126:GFP fusion was coaligned with MFs.
Figure 4.
Coalignment of 126-bodies and VRCs with MFs. A, Expression of MIC2 126:GFP and DsRed2:talin in an N. benthamiana leaf epidermal cell after agroinfiltration. B, Expression of MIC2 126:GFP and GFP:talin in an N. benthamiana leaf epidermal cell after agroinfiltration. Images in A and B were captured at 1.5 d postinfiltration. Arrows indicate coalignment of MF markers with 126-bodies. Bars = 50 μm. C, Infection with MIC2u-MP:GFP and expression of DsRed2:talin in an N. benthamiana leaf epidermal cell. Image was taken at 4 dpi with MIC2u-MP:GFP and 2 d postinfiltration with DsRed2:talin. Arrows indicate coalignment of MF marker with VRCs. Bar = 50 μm.
To determine whether VRCs also coaligned with MFs, leaves from N. benthamiana were inoculated with infectious transcripts of MIC2u-MP:GFP, and, at 2 d postinoculation (dpi), Agrobacterium harboring the DsRed2:talin construct was infiltrated into the inoculated leaves. In cells that expressed both GFP and DsRed2, VRCs mostly coaligned with MFs (Fig. 4C). Similar results were obtained when GFP:talin was infiltrated into infected leaves (data not shown). Thus, both VRCs and 126-bodies coalign with MFs and raise the possibility that the association of the VRCs with MFs is via the 126-kD protein.
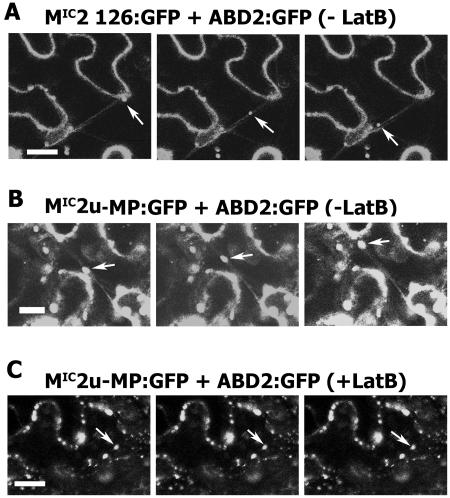
Although our double-labeling results showed the colocalization of 126-bodies and VRCs with MFs, it was not clear from static images whether this association was critical for intracellular movement of these complexes. We therefore utilized time-lapse imaging to determine the dynamic status of the 126-bodies and VRCs on MFs. We conducted experiments with both DsRed2:talin and ABD2:GFP to visualize MFs. Using either MF reporter, the 126-bodies trafficked along the decorated MFs (Fig. 5A; Supplemental Movies 1 and 2). VRCs also trafficked along ABD2:GFP-decorated MFs (Fig. 5B; Supplemental Movie 3). Average velocities were approximately 1 μm/s for 126-bodies and VRCs (max. ∼8 μm/s). These results show that the 126-bodies and VRCs not only align with MFs but also move intracellularly along these cytoskeletal elements.
Figure 5.
Trafficking of 126-bodies and VRCs along MFs and the effect of LatB on intracellular movement of VRCs. A, Expression of MIC2 126:GFP and ABD2:GFP in N. benthamiana leaf epidermal cells after agroinfiltration. At 2 d postinfiltration, the movement of 126-bodies along actin filaments was captured using time-lapse imaging. Images were taken every 3 s, and those shown represent three consecutive frames. Arrows indicate the position of a single 126-body over time. B, Expression of infectious transcripts of MIC2u-MP:GFP after inoculation onto an N. benthamiana leaf. At 1 dpi, Agrobacterium solution containing an expression vector for ADB2:GFP was infiltrated into the same leaf. Two days later, the movement of VRCs was captured using time-lapse imaging under conditions described above. The images represent three consecutive frames. Arrows indicate the positions of a single VRC over time. C, LatB (5 μm) was infiltrated into the abaxial surface of N. benthamiana leaves that were transiently expressing the same fusion proteins shown in B. The images were captured at 1 h post LatB application using the same conditions described in B. Arrows indicate the position of a single VRC over time. Bars = 25 μm.
MF Antagonists Allow 126-Body and VRC Formation But Disrupt MFs and 126-Body and VRC Intracellular Movement
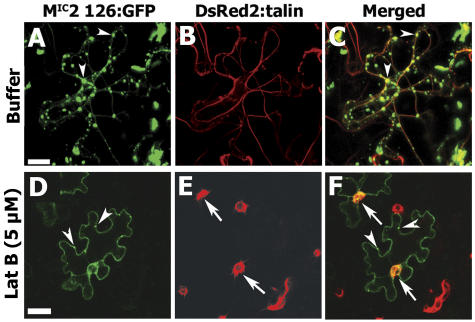
Since our double-labeling experiment showed the coalignment of 126-bodies with MFs, we wanted to know whether disrupting the MFs would alter their association. We therefore applied LatB, an inhibitor of actin polymerization, to leaves and MFs (visualized through fluorescence of DsRed2:talin), and 126-bodies were observed over time by confocal microscopy. When LatB was coinfiltrated with Agrobacterium expressing the MIC2 126:GFP fusion or DsRed2-talin, the MF strands were disrupted 1.5 d posttreatment, leading to the formation of ring-like structures (Fig. 6, compare B and E; see arrows). The 126-bodies were able to form, although not as large or as numerous as in untreated cells (Fig. 6, compare A and D; see arrowheads), indicating that MFs were necessary for the normal formation of these bodies. After LatB treatment, the 126-bodies did not always coalign with the shortened or ring-like MF structures (Fig. 6F, see arrows and arrowheads), indicating that their continued existence did not depend on intact, coaligned MFs.
Figure 6.
Effect of an MF antagonist on 126-body formation and 126-body alignment with MFs. LatB at 5 μm concentration or buffer was infiltrated into N. benthamiana leaves with a mixture of Agrobacterium containing vectors for expression of MIC2 126:GFP or DsRed2:talin. At 1.5 d postinfiltration, the infiltrated area was observed by confocal laser scanning microscopy. Arrows indicate disrupted MFs after LatB treatment. Arrowheads indicate 126-bodies formed in the presence or absence of LatB. The 126-bodies coaligned with MFs in the absence of LatB (A–C). The 126-bodies still formed after treatment with LatB (D–F), but were smaller than in the absence of LatB (A) and did not always coalign with collapsed MFs (F). Bars = 50 μm.
Because the 126-protein and/or the 183-kD protein are/is necessary for cell-to-cell movement (Hirashima and Watanabe, 2001, 2003), both proteins are components of VRCs, and VRC cell-to-cell movement is inhibited by treatment with an MF antagonist (Kawakami et al., 2004), we studied the effect of MF antagonists on VRC formation and intracellular movement. The effect of LatB and cytochalasin D (CD), another inhibitor of actin polymerization, on the formation of VRCs was analyzed in BY-2 protoplasts after inoculation with a transcript of MIC2u-MP:GFP. Application of LatB (5 μm) or CD (50 μg/mL) at the time of inoculation did not prevent the appearance of fluorescence from the virus-expressed GFP or the formation of the VRCs at the perinuclear region and the periphery of the cells (Fig. 7, LatB and CD). However, the number and size of the VRCs was decreased compared with those observed for the buffer treatment (Fig. 7). To investigate the effect of LatB on the intracellular movement of VRCs, N. benthamiana leaves were inoculated with transcripts from MIC2u-MP:GFP, and at 3 dpi LatB was infiltrated into the inoculated tissue. Time-lapse images were captured at 1 h postinfiltration. LatB treatment disrupted MFs and blocked intracellular trafficking of existing VRCs (Fig. 5C; Supplemental Movie 4). Trafficking of 126-bodies was also blocked with LatB treatment (data not shown). These results indicated that MF antagonists inhibit VRC intracellular movement, but do not lead to the destruction of existing VRCs within the cell or prevent the accumulation of the viral MP:GFP, represented by the diffuse fluorescence signal in cells treated with pharmacological agents (Fig. 7). Thus, the intracellular movement of existing VRCs relies on intact MFs and is separate from the effect of LatB on VRC formation.
Figure 7.
Effect of MF antagonists on VRC formation. A, N. tabacum cv BY-2 protoplasts were treated with buffer (buffer), LatB (5 μm), or CD (50 μg/mL) at the time of inoculation with MIC2u-MP:GFP and observed at 16 h postinoculation for VRC formation. VRCs were smaller and less numerous due to treatment with LatB and CD. Arrows indicate VRCs. Values for VRC number and size represent means ± sd for five individual cells. The experiment was repeated twice with similar results. Bars = 10 μm.
Virus-Induced Gene Silencing of Host Actin Transcript and MF Antagonists Inhibit the Cell-to-Cell Movement of TMV
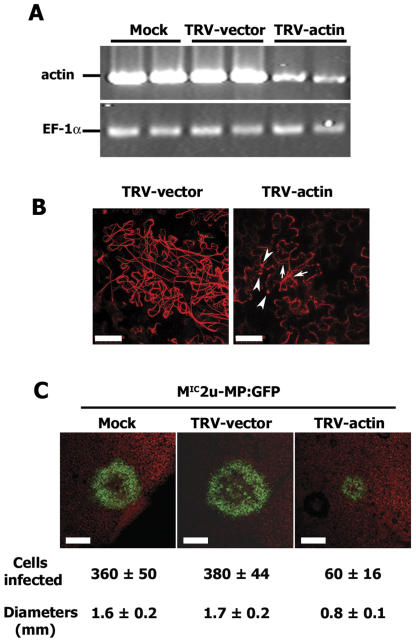
Virus-induced gene silencing (VIGS) was used as a genetic method to further study the role of MFs in the cell-to-cell spread of TMV. N. benthamiana plants were inoculated with a tobacco rattle virus (TRV) vector modified to express a region of an actin sequence from N. benthamiana leaves sharing high sequence identity with all known actin genes from N. tabacum (81%–92% identity; data not shown). By 8 dpi, the plants inoculated with TRV-actin showed a stunted growth pattern (small leaves and shortened internodes) compared with plants inoculated with TRV not containing an actin insert (TRV-vector); however, they continued to develop through the experimental period with further leaf formation, maintenance of green tissue, and flowering (data not shown). These results are similar to those reported for Arabidopsis and rye seedlings treated with LatB (Baluška et al., 2001). Those authors showed that the loss of MFs in cells prevented cell elongation but not continued plant development. At 12 dpi, the target actin transcript level in leaves of plants inoculated with TRV-actin was approximately 20% that observed for plants inoculated with the TRV-vector (Fig. 8A). As expected, the number of intact MFs was dramatically decreased at this time in the actin-silenced plants (Fig. 8B).
Figure 8.
Effect of silencing actin transcript expression on TMV cell-to-cell movement. Transcript of leaf-expressed actin 1 from N. benthamiana (accession no. AY594294) was silenced through expression of a fragment (876 nt) of the gene from within a TRV vector. TRV was inoculated to leaves through agroinfiltration. MIC2u-MP:GFP was inoculated to systemic leaves at 12 d postinfiltration, and spread of the virus was visualized by confocal laser scanning microscopy. A, Semiquantitative RT-PCR analysis of actin transcript levels in N. benthamiana leaves at 12 d postinfiltration with Agrobacterium containing (TRV-actin) or not containing (TRV-vector) the actin gene fragment in the TRV genome. Transcript levels also were determined for buffer-infiltrated leaves (Mock). Each lane represented a sample from an individual plant. Leaves analyzed for actin transcript (A) were representative of leaves then challenged with MIC2u-MP:GFP. B, Disruption of MFs in epidermal cells at 12 d postinfiltration with TRV-actin. Leaves were infiltrated with Agrobacterium containing a DsRed2:talin expression vector 2 d prior to image capture. Arrows and arrowheads, respectively, indicate intact and fragmented MFs in cells of plants infected with TRV-actin. Bars = 50μm. C, Fluorescence imaging of MP:GFP location, representing cell-to-cell spread of MIC2u-MP:GFP, in N. benthamiana leaves with (mock and TRV-vector) or without (TRV-actin) intact MFs. Images were taken with a 2.5× objective lens at 4 dpi with MIC2u-MP:GFP. The number of epidermal cells infected was calculated on the basis of the average size of the respective cells and the sizes of the fluorescent rings (estimated as circles). The diameters of the fluorescent rings were measured. Cell numbers and diameters represent the mean ± sd for five replicate infection loci per treatment. The experiment was repeated twice with similar results. Bars = 800 μm.
Actin-silenced plants at 12 dpi were challenged with MIC2u-MP:GFP to determine the effect of limited actin on the cell-to-cell movement of TMV. Individual green fluorescent rings representing cell-to-cell spread of the virus were frequently observed in leaves of plants initially inoculated with buffer or the TRV-vector (15–20 rings per inoculation), but were less frequent in leaves of plants initially inoculated with TRV-actin (3–5 rings per inoculation). The few fluorescent rings that did appear on plants inoculated with TRV-actin were smaller in diameter than those on plants inoculated with buffer or TRV-vector (Fig. 8C). The smaller ring was due to fewer infected cells and not due to the smaller size of the cells in the plants silenced for actin expression (Fig. 8C). Since VIGS often results in incomplete silencing of target transcripts (Ekengren et al., 2003), we speculate that the few fluorescent rings that did form were in areas where silencing was not complete.
Similar results were obtained using pharmacological agents to study TMV cell-to-cell movement. Mature N. benthamiana leaves were infiltrated with buffer containing or lacking LatB (5 μm) or CD (50 μg/mL) and with Agrobacterium containing an expression vector for DsRed2:talin. At 2 h postinfiltration, a period in which MFs were disrupted by LatB and CD in most leaf cells (data not shown), infectious TMV-MP-GFP-coat protein (CP) was inoculated to the infiltrated leaves. Infectious TMV-MP-GFP-CP was used in these experiments because it produced a stronger fluorescent signal from its reporter than did viruses expressing the MP:GFP fusion as a reporter. It was previously determined that the relative cell-to-cell spread of the green fluorescent lesion over time was not affected between TMV-MP-GFP-CP and those expressing MP:GFP fusions (Cheng et al., 2000; data not shown). At 3 dpi, green fluorescent spots representing cell-to-cell spread of virus were observed in leaves without LatB treatment (data not shown). However, far fewer (percentage of lesions compared with buffer treatment: 20%–30%, LatB; 30%–40%, CD) and smaller (diameters: 1.4 ± 0.2 mm, buffer; 0.8 ± 0.2 mm, LatB; 1.0 ± 0.2 mm, CD) green fluorescence spots were observed in leaves infiltrated with LatB or CD. These results from both VIGS and pharmacological treatments indicated that cell-to-cell movement of virus was inhibited by the disruption of MFs.
VRC Size and Virus Accumulation Rates Do Not Correlate with the Ability of the Virus to Move Cell to Cell
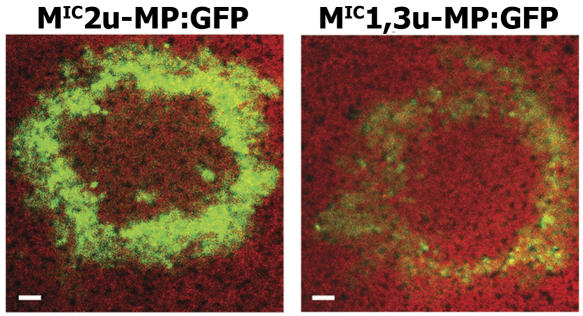
To study the effect of VRC size on virus cell-to-cell movement in intact plants, we compared the cell-to-cell movement of TMV mutants, which formed VRCs that varied in size. N. benthamiana leaves were inoculated with MIC2u-MP:GFP (large VRCs) or MIC1,3u-MP:GFP (small VRCs) and observed for the spread of green fluorescence. The two viruses spread equally well cell to cell, as determined by the size of the respective virus-induced green fluorescent rings (Fig. 9). Therefore, large VRCs were not necessary for virus cell-to-cell movement.
Figure 9.
Cell-to-cell movement of viruses that produce different size VRCs and accumulate different levels of MP:GFP. MIC2u-MP:GFP and MIC1,3u-MP:GFP transcripts were inoculated onto N. benthamiana leaves, and the size and intensity of the fluorescent rings were determined by confocal laser scanning microscopy and example images given. The mutations within the 126-kD protein ORF affected the accumulation, but not the spread, of MP:GFP, representing the location of the virus. Images were taken with the same settings and at the same focal distances. Mean values for ring diameters were 1.42 and 1.39 mm, respectively, for MIC2u-MP:GFP and MIC1,3u-MP:GFP for three replicates (independent rings) per virus (not significant; P > 0.5 by paired t test). The experiment was repeated once with similar results. Bar = 100 μm.
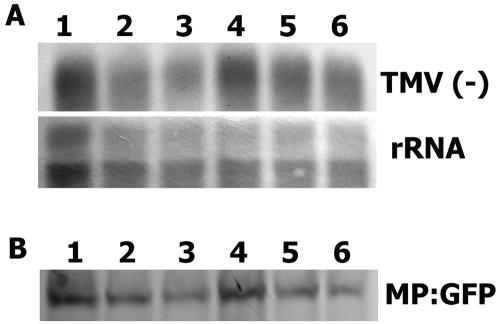
To determine if the diminished virus cell-to-cell movement observed in the leaves after LatB treatment was due to limited virus RNA or MP accumulation in cells, BY-2 protoplasts were inoculated with MIC2u-MP:GFP or MIC1,3u-MP:GFP transcripts and treated with LatB (5 μm). Protoplasts were analyzed for minus-strand vRNA [vRNA(−)] and MP:GFP accumulation (Fig. 10). Both vRNA(−) and MP:GFP accumulation were affected by LatB treatment (Fig. 10, A and B). However, the MP:GFP fusion protein accumulated to levels higher than would be expected if it was limiting virus movement (greater than 2%–5%; Arce-Johnson et al., 1995). Additionally, the accumulation of vRNA(−) and MP for MIC1,3u-MP:GFP was less than that observed for the LatB-treated infection by MIC2u-MP:GFP (Fig. 10). The low MP accumulation for MIC1,3u-MP:GFP in protoplasts is in agreement with the low level of MP:GFP observed in whole-plant infections with this virus (Fig. 9). Considering that MIC1,3u-MP:GFP moves normally between cells, these results indicated that the disrupted cell-to-cell movement in LatB-treated cells was due to the lack of intact MFs and not limited virus accumulation.
Figure 10.
RNA and MP accumulation levels for MIC2u-MP:GFP in the presence of LatB (5 μm) are comparable to that of MIC1,3u-MP:GFP in the absence of LatB in BY-2 protoplasts. BY-2 protoplasts inoculated with either MIC2u-MP:GFP (lanes 1 and 2, and 4 and 5) or MIC1,3u-MP:GFP (lanes 3 and 6) in the presence (lane 2 and 5) or absence (lanes 1 and 3, and 4 and 6) of LatB (5 μm) were analyzed for negative-strand viral RNA [TMV(−), A] and MP:GFP (B) accumulation. Protoplasts were sampled at 12 (lane 1–3) and 24 h postinoculation (lane 4–6) for the analysis. RNA (10 μg loaded per lane) was probed with a digoxigenin-labeled positive strand of a full-length fragment of TMV-MP:GFP. Ribosomal RNA (rRNA) on the blot was stained with methylene blue and then imaged. Viral MP:GFP (5 × 104 cells loaded per lane) was detected with anti-MP antibody followed by anti-rabbit IgG secondary antibody. The percentage of cells infected for each treatment was similar (all approximately 40%).
DISCUSSION
The 126-kD Protein Modulates the Size of the VRC
The 126-kD and 183-kD proteins are components of VRCs formed during TMV infection (e.g. Heinlein et al., 1998; Más and Beachy, 1999; Kawakami et al., 2004). In addition, the 126-kD protein functions as a heterodimer with the 183-kD protein during TMV replication and is produced in approximately 10-fold excess to the 183-kD protein (Nelson et al., 1993; Watanabe et al., 1999; Lewandowski and Dawson, 2000). The entire 126-kD protein, or portions thereof, can self-interact, form oligomers, or associate with membranes or membrane-associated host proteins (Yamanaka et al., 2000; Goregaoker et al., 2001; dos Reis Figueira et al., 2002; Goregaoker and Culver, 2003; Hagiwara et al., 2003; Tsujimoto et al., 2003). The 126-kD protein is not necessary for TMV replication but enhances its efficiency since a mutant virus expressing only the 183-kD protein replicated at only 10% of wild-type virus level (Ishikawa et al., 1986; Lewandowski and Dawson, 2000). The method by which the 126-kD protein functioned was not determined from these studies, although the quantity of the protein produced could be interpreted to indicate that at least a portion of its function was nonenzymatic. Here, we show through the expression of 126-kD protein variants alone or from within the genome of an infectious transcript that the 126-kD protein modulates the size of the VRCs (Fig. 2; Table I). Considering that portions of the 126-kD protein can form oligomers (Goregaoker and Culver, 2003), it is possible that the mutations made in the 126-kD protein prevent oligomers from forming efficiently, thereby resulting in smaller 126-bodies and VRCs. However, other activities of the 126-kD protein may have been affected to limit 126-body and VRC formation, such as its suppressor activity (Kubota et al., 2003; Ding et al., 2004), leading to less accumulation of viral mRNA and, therefore, less 126-kD protein and smaller bodies.
Previous reports showed that viral membrane-associated proteins, often associated with virus replication, form cytoplasmic bodies with VRC-like attributes (Schaad et al., 1997; Restrepo-Hartwig and Ahlquist, 1999; Carette et al., 2002). The 1a protein of brome mosaic virus, an apparent ortholog of the 126-kD protein of TMV, is among the best characterized of these proteins (for review, see Ahlquist et al., 2003). The 1a protein associates with the endoplasmic reticulum and forms homo-oligomers (Kao et al., 1992; O'Reilly et al., 1997; Restrepo-Hartwig and Ahlquist, 1999; den Boon et al., 2001). This protein also is responsible for the formation of cytoplasmic inclusion bodies (spherules) in yeast during virus accumulation in this host (Schwartz et al., 2002). Recently, it was determined that altering the ratio of 1a protein to 2a protein (polymerase) can substantially alter the structure of the spherules (Schwartz et al., 2004). The ability of the 126-kD protein to modulate the size of VRCs formed during virus infection further suggests the similar characteristics of these proteins. The intracellular and intercellular movement function of the 126-kD protein orthologs requires further investigation.
The Size of the VRCs Is Not Correlated with the Cell-to-Cell Movement Ability of the Mutant Viruses
The size of the VRCs was not correlated with the ability of TMV to move cell to cell (Figs. 2 and 9). Thus, we were able to separate the region of the 126-kD protein required for large body formation from the region required for cell-to-cell movement (e.g. the region required to associate with MFs or other host factors that associate with MFs).
Previous studies determined that the 126-kD protein and, in particular, the sequence between the methyltransferase and helicase core domains in TMV and in the closely related tomato mosaic virus control these virus's ability to suppress silencing (Kubota et al., 2003; Ding et al., 2004). Here, we determined that a virus, MIC1,3u-MP:GFP, expressing an attenuated suppressor (Ding et al., 2004) moved normally cell to cell (Fig. 9). Thus, the suppressor activity also can be unlinked from the cell-to-cell movement function of the 126-kD protein. Suppressor activity was also unlinked from the cell-to-cell movement function of the P15 protein of the pecluvirus peanut clump virus and the long-distance movement function of the helper component-proteinase (HC-Pro) of the potyvirus tobacco etch virus (Kasschau et al., 1997; Kasschau and Carrington, 2001; Donoyer et al., 2002).
The 126-Bodies and the VRCs Coalign with and Traffic along MFs
As reported here and in a recent study (Kawakami et al., 2004), treatment of leaf tissue with LatB, which disrupts MFs, inhibits the cell-to-cell movement of VRCs. However, the colocalization and intracellular movement of the VRCs along the MFs were not demonstrated. MF antagonists alter PD by increasing their size exclusion limits (Ding et al., 1996), possibly through an effect on the actin present in these structures (White et al., 1994). These antagonists therefore could affect VRC trafficking on MFs, MTs, or other structural components indirectly by disrupting cell communication. To differentiate between the various effects of MF antagonists on VRC movement, it was important to show that VRCs align with and move along MFs. We determined that both 126-bodies and VRCs align with and move along MFs within cells (Figs. 4–6; Supplemental Movies 1, 2, and 3). The velocity at which the VRCs move, approximately 1 μm/s, is similar to those previously described for VRCs (Kawakami et al., 2004) and for organelle trafficking (Nebenführ et al., 1999, and refs. therein). We determined that disruption of MFs did not decrease virus accumulation sufficiently to account for the reduced cell-to-cell spread of the virus (Fig. 10). Disruption of MFs did abolish the colocalization of MFs with 126-bodies and VRCs, the normal formation of the 126-bodies and VRCs, and intracellular movement of VRCs along MFs (Figs. 5–7; Supplemental Movie 4). These results now link the physical position of the VRCs with MFs and support the theory that this association is necessary for TMV intracellular and intercellular movement. The association of 126-bodies with MFs may explain the requirement of 126/183-kD protein for cell-to-cell movement of tomato mosaic virus (Hirashima and Watanabe, 2001, 2003) and brings into question whether the viral 126-kD protein or the viral MP is responsible for targeting the VRC to the cell wall area and PD. The MP has previously been implicated in this function (for review, see Waigmann et al., 2004). Recently, it was shown that the membrane-associated viral proteins, triple gene block proteins 2 and 3, necessary for cell-to-cell movement of a group 1 hordei-like virus, and triple gene block protein 2, likewise necessary for cell-to-cell movement of group 2 potato virus X, were associated with MFs (Haupt et al., 2005; Ju et al., 2005). The possibility that the 126-kD protein functions similarly to these proteins in supporting TMV movement requires further investigation.
A Model for TMV Movement
The information presented here allows for the presentation of a modified model for TMV intracellular and cell-to-cell movement (Fig. 11). In the model, the 126-kD protein has a role in forming VRCs that compartmentalize required components for TMV accumulation and protect progeny RNA and potentially other viral factors from host defense systems (Más and Beachy, 1999; Ding et al., 2004). The VRCs can be modified by the MP and CP, as suggested by others (Asurmendi et al., 2004). The VRCs coalign with and traffic along MFs, linking the physical location of the VRCs with the cytoskeletal element proposed to function in movement (Kawakami et al., 2004). Thus, the MFs likely have a direct effect on VRC movement rather than an indirect effect through an alteration of plasmodesmal structure. The MFs also are necessary for the normal formation of VRCs. Although it is possible that decreased VRC intracellular and intercellular movement was an indirect effect of the fewer VRCs available for movement after LatB treatment, the majority of the effect may be direct since intracellular movement of existing and maintained VRCs did not occur after LatB treatment. The 126-kD protein alone is able to align with MFs. As a result of these findings, it is now a question whether the 126-kD protein or the MP of TMV directs movement of the VRC to the cell wall area. The MP of TMV is required for cell-to-cell movement of the virus, binds RNA and MFs, alters the size exclusion limits of PD, and traffics itself cell to cell (for review, see Waigmann et al., 2004). The MP therefore has been considered to function in transporting TMV RNA between cells. However, the movement to the cell wall of an MP:vRNA complex in the absence of other viral factors has not been observed. Thus, it remains a possibility that the VRC could be transported to the cell wall through a direct or indirect interaction between the 126-kD protein and MFs, wherein the MP binds the vRNA and transports it through the PD either as part of a virus movement complex or alone (compare variations 1 and 2, Fig. 11; for references discussing TMV movement within PD, see Kawakami et al., 2004; Waigmann et al., 2004). Further experiments are clearly necessary to determine the roles of the MP and 126-kD protein in movement. If the 126-kD protein controls the intracellular movement of VRCs, the binding of the VRCs to MFs could be via membranes associated with the 126-kD protein (dos Reis Figueira et al., 2002), possibly through the association of the 126-kD protein with membrane-anchored proteins (Yamanaka et al., 2000; Hagiwara et al., 2003; Tsujimoto et al., 2003) or through the 126-kD protein itself. Future work will clarify the viral and/or host factors that bind the VRCs to MFs and the mechanism by which TMV RNA moves intracellularly and cell to cell.
Figure 11.
Model for TMV intracellular and intercellular movement. TMV VRCs align with and traffic along MFs within cells (this report; a and b) and may move along the MFs to locations next to PD (represented by hollow cylinder; Kawakami et al., 2004). There, viral elements move through the PD, as either viral movement complexes (VMCs; Kawakami et al., 2004) or viral RNA and MP complexes (Waigmann et al., 2004), to establish new VRCs in neighboring cells (c and d in variations 1 and 2). The 126-kD protein modulates VRC form (this report), is necessary for intracellular movement (this report), and may protect the VRC against host defense systems (e; Ding et al., 2004). The 126-kD protein and/or read-through 183-kD protein are/is necessary for virus cell-to-cell movement (Hirashima and Watanabe, 2001, 2003), possibly through the above mentioned functions. Thus, it is now unclear where the 126-kD protein and MP function to move the VRC to and through a plasmodesma. The 126-kD protein either directly interacts with the MFs (a) or indirectly interacts with the MFs (b) through its interaction with membranes (dos Reis Figueira et al., 2002) or with membrane-associated proteins (e.g. Yamanaka et al., 2000; Hagiwara et al., 2003; Tsujimoto et al., 2003). Small VRCs may merge to form large VRCs (f), but the large VRCs are not necessary for virus movement (this report). N, Nucleus.
MATERIALS AND METHODS
Virus Strains, Plasmids, and Plant Materials
The “masked” (M) and U1 strains of TMV were from described sources (Holt et al., 1990). MIC (IC, infectious clone) refers to the progeny of infectious transcript produced from a cDNA clone of the M strain (Holt et al., 1990). The MIC2 and MIC1,3 mutant strains were described previously (Shintaku et al., 1996; Ding et al., 2004). TMV-MP:GFP (previously TMV-M:GfusBr) and TMV-MP-GFP-CP (previously 4GD-GFP) were described previously (Casper and Holt, 1996; Oparka et al., 1997). pDsRed2:talin, pABD2:GFP, and pGFP:talin fusion constructs were made as described (Blancaflor, 2002; Wang et al., 2004). Nicotiana benthamiana and Nicotiana tabacum cv BY-2 suspension cells, a gift from Richard Cyr (Pennsylvania State University, College Park, PA), were used.
Construction of 126:GFP Fusions for Particle Bombardment and Agroinfiltration
Generation of p126-2:GFP and p126-1,3:GFP fusions, representing MIC2 or MIC1,3 126:GFP fusions in the pRTL2 vector (Restrepo et al., 1990), were described previously (Ding et al., 2004). These clones were used in particle bombardment transient expression studies.
Promoter-gene-terminator fragments from p126-2:GFP and p126-1,3:GFP fusions within the pRTL2 vector were introduced into the binary vector pGA482 (An et al., 1988) to generate pMIC2 126:GFP and pMIC1,3 126:GFP (Ding et al., 2004). The resulting plasmid was analyzed for sequence authenticity. The binary vectors containing the different 126:GFP fusions were transformed into ElectroMAX Agrobacterium tumefaciens LBA4404 competent cells by electroporation according to the manufacturer's protocol (Gibco BRL, Rockville, MD) and used in agroinfiltration transient expression studies.
Construction of TMV-MP:GFP Mutants
An approximately 2.9-kb nucleotide fragment from a plasmid containing TMV-MP:GFP (equivalent to TMV-M:GfusBr; Oparka et al., 1997) was removed after digestion with SphI and BamHI and replaced with the corresponding fragments from pMIC (previously L19; Shintaku et al., 1996), pMIC2, and pMIC1,3 (previously pMICm1,3; Shintaku et al., 1996). The final clones represented chimeric viruses with sequences between the SphI and BamHI sites from mutants of the M strain and sequences outside the restriction sites from the U1 strain of TMV. These clones are designated with a “u” (e.g. MIC2u) to indicate the presence of U1 sequence. TMV-MP:GFP moves cell to cell (Szécsi et al., 1999; Cheng et al., 2000).
Suspension Cell Culture and Particle Bombardment
N. tabacum cv BY-2 suspension cells were grown in 50 mL of culture media (4.3 g L−1 Murashige and Skoog salt, 100 mg L−1 myoinositol, 1 mg L−1 thiamine, 0.2 mg L−1 2,4-dichlorophenoxyacetic acid, 255 mg L−1 KH2PO4, 30 g L−1 Suc, pH 5.0) at 26°C with constant shaking at 150 rpm. The suspension cells were subcultured weekly. Plasmid DNA was introduced to BY-2 suspension cells by particle bombardment (model PDS-1000; DuPont, Wilmington, DE) as described (Marc et al., 1998).
Agroinfiltration
Agrobacterium containing the binary vectors were grown and infiltrated to leaves as described (Ding et al., 2004).
Inoculation of Protoplasts and Plant Leaves with Infectious Transcripts
Preparation of protoplasts from N. tabacum cv BY-2 suspension cells and inoculation of the protoplasts with infectious transcripts by electroporation were as described (Watanabe et al., 1987). After washing, protoplasts were resuspended in 4 mL of medium and cultured in the dark at 28°C. Plant leaves were inoculated mechanically by rubbing with infectious transcripts or purified viruses having equivalent infectivities as determined through bioassay (Nelson et al., 1993). After virus inoculation, plants were left in a greenhouse under previously described conditions (Nelson et al., 1993).
Treatment with MF Destabilizers and Visualization of MFs
Treatment of protoplasts or leaves with MF inhibitors, 5 μm LatB or 50 μg/mL CD (CalBiochem, La Jolla, CA), by addition to media or by infiltration, was as described (Heinlein et al., 1998; Gillespie et al., 2002). MFs in living tissue were visualized using a DsRed2:talin or GFP:talin fusion (Blancaflor, 2002; Mitra et al., 2003) or a fimbrin:GFP fusion (Wang et al., 2004).
Immunolabeling
Immunolabeling was performed with modification as described (Blancaflor and Hasenstein, 2000). The leaf tissues of N. benthamiana plants inoculated with infectious MIC2u-MP:GFP transcripts were fixed in 3.7% (v/v) formaldehyde and 5% (v/v) dimethyl sulfoxide in PHME buffer (60 mm PIPES, 25 mm HEPES, 5 mm EGTA, 2 mm MgCl2, pH 6.9) for 2 h. After fixation, the leaf tissues were digested with cell wall-degrading enzymes (1% Cellulase, 0.1% Pectolyase, and 0.1% bovine serum albumin in PHEM buffer) for 2 h and then incubated in 1% (v/v) Triton X-100 for 20 min, followed by treatment in ice-cold methanol for 10 min. Three washes with PHEM were required after each step. After an overnight incubation with rabbit anti-126-kD protein (1:100 dilution; Nelson et al., 1993) followed by a 2-h incubation with goat anti-rabbit IgG conjugated with rhodamine (Molecular Probes, Eugene, OR; 1:100 dilution), leaf tissues were mounted on slides in phosphate-buffered saline, pH 8.5, containing 20% (w/v) Mowiol 4-88 (CalBiochem) and 0.1% (w/v) phenylenediamine before observation with a confocal microscope. Protoplasts inoculated with infectious MIC2u-MP:GFP transcripts were fixed for 15 min in PHEM buffer containing 1.5% (v/v) formaldehyde and 5% (v/v) DMSO and then spun onto a cover slide and covered with a thin layer of 0.7% agar membrane. The remaining steps were the same as described for labeling in leaf tissues except for omission of the cell wall digestion step.
Synthesis and Expression of an N. benthamiana Actin Sequence from a TRV Vector for Virus-Induced Silencing
Total RNA isolated from N. benthamiana leaves was used to obtain an 876-bp nucleotide fragment from an actin gene (accession no. AY594294) through RT-PCR (Invitrogen, Carlsbad, CA). Primer names and sequences used in the reaction were 5′-attB1-Nb-actin1 (5′GGGGACAAGTTTGTACAAAAAAGCAGGCTGCAACTGGGATGATATGGAGAA3′) and 3′-attB2-Nb-actin1 (5′GGGGACCACTTTGTACAAGAAAGCTGGGTTCTGCCTTTGCAATCCACATCTG3′). The underlines indicate attB1 and attB2 sequences, respectively. The PCR product was recombined in vitro into the entry vector pDONR207 (Invitrogen) and subsequently into the pTRV2-attR2-attR1 high-throughput vector (Liu et al., 2002a, 2002b), all as described by the manufacturer (Invitrogen), to generate pTRV2-actin. For the VIGS assay, pTRV1, pTRV2, or pTRV2-actin were introduced into Agrobacterium strain GV3101 using standard protocols. Agrobacterium growth and infiltration were undertaken as described (Ding et al., 2004).
Western-Blot Analysis
For MP:GFP detection during virus infections, BY-2 protoplasts were harvested by centrifugation at 100g and resuspended 1:1 (v/v) with 2× SDS loading dye. Aliquots that corresponded to 1 × 104 protoplasts per lane were subjected to SDS-PAGE, and western-blot analyses were undertaken as described (Derrick et al., 1997). Source and purity of antibody against the MP were as described (Derrick et al., 1997). Antibody against MP was used at 1:6,000 dilution.
Northern-Blot Analysis
Total RNA was extracted from 5 × 104 protoplasts using TRIzol reagent (Invitrogen). RNA was denatured with glyoxal (Sambrook et al., 1989), and equal amounts of RNA (10 μg) were loaded per lane and separated in a 1.0% agarose gel containing 10 mm sodium phosphate, pH 7.0. The RNA was transferred to a nylon membrane (Amersham, Little Chalfont, UK) in 10× SSC. Before hybridization, the membrane was stained with methylene blue (Sambrook et al., 1989) to visualize RNA loads. Membranes were hybridized with digoxigenin-labeled positive strand of TMV-MP:GFP RNA probe. For probe labeling, TMV-MP:GFP construct (Oparka et al., 1997) was linearized with Acc65I and then labeled with a T7 digoxigenin-labeling kit according to the manufacturer's protocol (Roche Biochemicals, Mannheim, Germany). Prehybridization, hybridization, washing, and detection were also performed according to the manufacturer's protocol (Roche Biochemicals).
Semiquantitative RT-PCR
Total RNA was extracted from systemic leaves infected by TRV1 plus TRV2 or TRV1 plus TRV2-actin using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized using 1 μg of total RNA as template, an oligo-d(T) primer, and reverse transcriptase (Promega, Madison, WI). Semiquantitative reverse transcription (RT)-PCR was performed as described (Ding et al., 2004). Primers specific for Nb-actin and EF-1α, as an internal control, were used to amplify fragments. Primer sequences were as follows: 5′Nb-actin1, 5′GCAACTGGGATGATATGGAGAA3′; 3′Nb-actin1, 5′CTGCCTTTGCAATCCACATCTG3′; 5′EF-1α, 5′TGGTGTCCTCAAGCCTGGTATGGTTGT3′; and 3′EF-1α, 5′ACGCTTGAGATCCTTAACCGCAACATTCTT3′.
Confocal Microscopy
Images were captured with a confocal laser scanning system and fluorescence microscope as described (Cheng et al., 2000). GFP was detected by exciting tissue with the 488-nm line of the krypton/argon laser and emission captured at 522 nm. DsRed was detected by exciting tissue with the 568-nm line and emission captured at 598 nm. For time-lapse imaging, leaf tissue expressing the 126:GFP or MP:GFP after, respectively, agroinfiltration or mechanical inoculation (described earlier) was infiltrated with Agrobacterium expressing DsRed2:talin or ABD2:GFP 2 d post initial treatment as described (Ding et al., 2004). Images were obtained every 3 s for 2.25 min from a single optical plane using the confocal system. Time-lapse movies were generated using the Metamorph 4.5 program (Universal Imaging, West Chester, PA).
Resource Availability
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to signing of material transfer agreements between requestor and supplier and the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY594294.
Supplementary Material
Acknowledgments
We thank Dr. Roger Beachy for TMV-MP:GFP (TMV-M:GfusBr); Dr. Curtis A. Holt for TMV-MP-GFP-CP (4GD-GFP); Dr. S.P. Dinesh-Kumar for the TRV-silencing system; Shelly Carter for maintaining BY-2 suspension cells; Kirankumar Mysore, Ping Xu, and Anthony Cole for valuable comments on the manuscript; and Tom Wallace, Kristy Richerson, Janie Galloway, and Karen Flowers for maintenance of plants.
This work was supported by the Samuel Roberts Noble Foundation.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065722.
References
- Aaziz R, Dinant S, Epel BL (2001) The cytoskeleton and plasmodesmata: for better or for worse. Trends Plant Sci 6: 326–330 [DOI] [PubMed] [Google Scholar]
- Ahlquist PA, Noueiry O, Lee WM, Kushner DB, Dye BT (2003) Host factors in positive-strand RNA virus genome replication. J Virol 77: 8181–8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An GP, Ebert R, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, DPS Verma, eds, Plant Molecular Biology Manual, Vol 2. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3: 1–19
- Arce-Johnson P, Kahn TW, Reimann-Philipp U, Rivera-Bustamante R, Beachy RN (1995) The amount of movement protein produced in transgenic plants influences the establishment, local movement and systemic spread of infection by movement protein-deficient tobacco mosaic virus. Mol Plant Microbe Interact 8: 415–423 [Google Scholar]
- Asurmendi S, Berg RH, Koo JC, Beachy RN (2004) Coat protein regulates formation of replication complexes during tobacco mosaic virus infection. Proc Natl Acad Sci USA 101: 1415–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY (2000) Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci USA 97: 11984–11989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F, Jasik J, Edelmann HG, Salajová T, Volkmann D (2001) Latrunculin B-induced dwarfism: plant cell elongation is F-actin dependent. Dev Biol 231: 113–124 [DOI] [PubMed] [Google Scholar]
- Bao YM, Carter SA, Nelson RS (1996) The 126- and 183-kilodalton proteins of tobacco mosaic virus, and not their common nucleotide sequence, control mosaic symptom formation in tobacco. J Virol 70: 6378–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy RN, Zaitlin M (1975) Replication of tobacco mosaic virus. VI. Replicative intermediate and other viral related RNAs associated with polyribosomes. Virology 63: 84–97 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB (2002) The cytoskeleton and gravitropism in higher plants. J Plant Growth Regul 21: 120–136 [DOI] [PubMed] [Google Scholar]
- Blancaflor EB, Hasenstein KH (2000) Methods for detection and identification of F-actin in fixed and permeabilized plant tissues. In CJ Staiger, F Baluška, D Volkmann, PW Barlow, eds, Actin: A Dynamic Framework for Multiple Plant Cell Functions. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 601–618
- Boyko V, Ashby JA, Suslova E, Ferralli J, Sterthaus O, Deom CM, Heinlein M (2002) Intramolecular complementing mutations in tobacco mosaic virus movement protein confirm a role for microtubule association in viral RNA transport. J Virol 76: 3974–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M (2000. a) Function of microtubules in intercellular transport of plant virus RNA. Nat Cell Biol 2: 826–832 [DOI] [PubMed] [Google Scholar]
- Boyko V, Ferralli J, Heinlein M (2000. b) Intercellular movement of TMV RNA is temperature-dependent and corresponds to the association of movement protein with microtubules. Plant J 22: 315–325 [DOI] [PubMed] [Google Scholar]
- Carette JE, Lent JV, MacFarlane SA, Wellink J, van Kammen A (2002) Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J Virol 76: 6293–6301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper SJ, Holt CA (1996) Expression of the green fluorescent protein-encoding gene from a tobacco mosaic virus-based vector. Gene 173: 69–73 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Su CL, Carter SA, Nelson RS (2000) Vascular invasion routes and systemic accumulation patterns of tobacco mosaic virus in Nicotiana benthamiana. Plant J 23: 349–362 [DOI] [PubMed] [Google Scholar]
- den Boon J, Chen J, Ahlquist P (2001) Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J Virol 75: 12370–12381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrick PM, Carter SA, Nelson RS (1997) Mutation of the 126/183 kD proteins of tobacco mosaic tobamovirus: the relationship of phloem-dependent accumulation with viral protein accumulation. Mol Plant Microbe Interact 10: 589–596 [Google Scholar]
- Ding B, Kwon MO, Warnberg L (1996) Evidence that actin filaments are involved in controlling the permeability of plasmodesmata in tobacco mesophyll. Plant J 10: 157–164 [Google Scholar]
- Ding XS, Liu JZ, Cheng NH, Folimonov A, Hou YM, Bao YM, Katagi C, Carter SA, Nelson RS (2004) The Tobacco mosaic virus 126 kD protein associated with virus replication and movement suppresses RNA silencing. Mol Plant Microbe Interact 17: 583–592 [DOI] [PubMed] [Google Scholar]
- Donoyer P, Pfeffer S, Fritsch C, Hemmer O, Voinnet O, Richards KE (2002) Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J 29: 555–567 [DOI] [PubMed] [Google Scholar]
- dos Reis Figueira A, Golem S, Goregaoker SP, Culver JN (2002) A nuclear localization signal and a membrane association domain contribute to the cellular localization of the tobacco mosaic virus 126-kD replicase protein. Virology 301: 81–89 [DOI] [PubMed] [Google Scholar]
- Ekengren S, Liu Y, Schiff M, Dinesh-Kumar SP, Martin G (2003) Two MAPK cascades, NPR1, and TGA transcription factors play a role in Pto-mediated disease resistance in tomato. Plant J 36: 905–917 [DOI] [PubMed] [Google Scholar]
- Gillespie T, Boevink P, Haupt S, Roberts AG, Toth R, Valentine T, Chapman S, Oparka KJ (2002) Functional analysis of a DNA-shuffled movement protein reveals that microtubules are dispensable for the intercellular movement of Tobacco mosaic virus. Plant Cell 14: 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez KA, Gomez AA (1984) Statistical Procedures for Agricultural Research, Ed 2. John Wiley & Sons, New York
- Goregaoker SP, Culver JN (2003) Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J Virol 77: 3549–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goregaoker SP, Lewandowski DJ, Culver JN (2001) Identification and functional analysis of an interaction between domains of the 126/183-kD replicase-associated proteins of tobacco mosaic virus. Virology 282: 320–328 [DOI] [PubMed] [Google Scholar]
- Hagiwara Y, Komoda K, Yamanaka T, Tamai A, Meshi T, Funada R, Tsuchiya T, Naito S, Ishikawa M (2003) Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J 22: 344–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Cowan GH, Ziegler A, Roberts AG, Oparka KJ, Torrance L (2005) Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17: 164–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood V, Kragler F, Lucas WJ (2002) Plasmodesmata: pathways for protein and ribonucleoprotein signaling. Plant Cell (Suppl) 14: S303–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M (2002) Plasmodesmata: dynamic regulation and role in macromolecular cell-to-cell signaling. Curr Opin Plant Biol 5: 543–552 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL (2004) Macromolecular transport and signaling through plasmodesmata. Int Rev Cytol 235: 93–164 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Epel BL, Padgett HS, Beachy RN (1995) Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270: 1983–1985 [DOI] [PubMed] [Google Scholar]
- Heinlein M, Padgett HS, Gens JS, Pickard BG, Casper SJ, Epel BL, Beachy RN (1998) Changing patterns of localization of the tobacco mosaic virus movement protein and replicase to the endoplasmic reticulum and microtubules during infection. Plant Cell 10: 1107–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hills GJ, Plaskitt KA, Young ND, Dunigan DD, Watts JW, Wilson TMA, Zaitlin M (1987) Immunogold localization of the intracellular sites of structural and nonstructural tobacco mosaic virus proteins. Virology 161: 488–496 [DOI] [PubMed] [Google Scholar]
- Hirashima K, Watanabe Y (2001) Tobamovirus replicase coding region is involved in intercellular movement. J Virol 75: 8831–8836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirashima K, Watanabe Y (2003) RNA helicase domain of tobamovirus replicase executes intercellular movement possibly through collaboration with its nonconserved region. J Virol 77: 12357–12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CA, Hudgson RAJ, Coker FA, Beachy RN, Nelson RS (1990) Characterization of masked strain of tobacco mosaic virus: identification of region responsible for symptom attenuation by analysis of an infectious cDNA clone. Mol Plant Microbe Interact 3: 417–423 [DOI] [PubMed] [Google Scholar]
- Ishikawa M, Meshi T, Motoyoshi F, Takamatsu N, Okada Y (1986) In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res 14: 8291–8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H-J, Samuels TD, Wang Y-S, Blancaflor E, Payton M, Mitra R, Krishnamurthy K, Nelson RS, Verchot-Lubicz J (2005) The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol 138: 1877–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CC, Quadt R, Hershberger RP, Ahlquist P (1992) Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J Virol 66: 6322–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Carrington JC (2001) Long distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285: 71–81 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Cronin S, Carrington JC (1997) Genome application and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228: 251–262 [DOI] [PubMed] [Google Scholar]
- Kawakami S, Watanabe Y, Beachy RB (2004) Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc Natl Acad Sci USA 101: 6291–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragler F, Curin M, Trutnyeva K, Gansch A, Waigmann E (2003) MPB2C, a microtubule-associated plant protein binds to and interferes with cell-to-cell transport of tobacco mosaic virus movement protein. Plant Physiol 132: 1870–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Tsuda S, Tamai A, Meshi T (2003) Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J Virol 77: 11016–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Beachy RN (1999) Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski DJ, Dawson WO (2000) Function of the 126- and 183-kD proteins of tobacco mosaic virus. Virology 271: 90–98 [DOI] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Dinesh-Kumar SP (2002. a) Tobacco Rar1, EDS and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Dinesh-Kumar SP (2002. b) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao TH, McCubbin AG, Cyr RJ (1998) GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli GP, Russo M (1977) Plant virus inclusion bodies. Adv Virus Res 21: 175–266 [DOI] [PubMed] [Google Scholar]
- Más P, Beachy RN (1999) Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intercellular distribution of viral RNA. J Cell Biol 147: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Beachy RN (2000) Role of microtubules in the intracellular distribution of tobacco mosaic virus movement protein. Proc Natl Acad Sci USA 97: 12345–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean BG, Zupan J, Zambryski P (1995) Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco plants. Plant Cell 7: 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Krishnamurthy K, Blancaflor E, Payton M, Nelson RS, Verchot-Lubicz J (2003) The Potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral intercellular movement. Virology 312: 35–48 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA (1999) Stop-and-go movements of plant golgi stacks are mediated by the acto-myosin system. Plant Physiol 121: 1127–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RS (2005) Movement of viruses to and through plasmodesmata. In K Oparka, ed, Plasmodesmata. Blackwell Publishing, Oxford, pp 188–209
- Nelson RS, Li G, Hodgson RAJ, Beachy RN, Shintaku MH (1993) Impeded systemic accumulation of the masked strain of tobacco mosaic virus. Mol Plant Microbe Interact 6: 45–54 [Google Scholar]
- Oparka KJ (2004) Getting the message across: how do plant cells exchange macromolecular complexes. Trends Plant Sci 9: 33–41 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DAM, Santa Cruz S, Padgett HS, Beachy RN (1997) Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus. Plant J 12: 781–789 [DOI] [PubMed] [Google Scholar]
- O'Reilly EK, Paul JD, Kao CC (1997) Analysis of the interaction of viral RNA replication proteins by using the yeast two-hybrid assay. J Virol 71: 7526–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett HS, Epel BL, Heinlein MH, Watanabe Y, Beachy RN (1996) Distribution of tobamovirus movement protein in infected cells and implications for cell-to-cell spread of infection. Plant J 10: 1079–1099 [DOI] [PubMed] [Google Scholar]
- Radford JE, White RG (1998) Localization of a myosin-like protein to plasmodesmata. Plant J 19: 555–567 [DOI] [PubMed] [Google Scholar]
- Reichel C, Beachy RN (1999) The role of the ER and cytoskeleton in plant viral trafficking. Trends Plant Sci 4: 458–462 [DOI] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo-Hartwig M, Ahlquist P (1999) Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J Virol 73: 10303–10309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AG, Oparka KJ (2003) Plasmodesmata and the control of symplastic transport. Plant Cell Environ 26: 103–124 [Google Scholar]
- Saito T, Hosokawa D, Meshi T, Okada Y (1987) Immunocytochemical localization of the 130K and 180K proteins (putative replicase components) of tobacco mosaic virus. Virology 160: 477–481 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schaad MC, Jensen PE, Carrington JC (1997) Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J 16: 4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Chen JB, Janda M, Sullivan M, den Boon J, Ahlquist P (2002) A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9: 505–514 [DOI] [PubMed] [Google Scholar]
- Schwartz M, Chen JB, Lee WM, Janda M, Ahlquist P (2004) Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc Natl Acad Sci USA 101: 11263–11268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalla TA (1964) Assembly and aggregation of tobacco mosaic virus in tomato leaflets. J Cell Biol 21: 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintaku MH, Carter SA, Bao YM, Nelson RS (1996) Mapping nucleotides in the 126 kD protein gene that control the differential symptoms induced by two strains of tobacco mosaic virus. Virology 221: 218–225 [DOI] [PubMed] [Google Scholar]
- Szécsi J, Ding XS, Lim CO, Bendahmane M, Cho MJ, Nelson RS, Beachy RN (1999) Development of tobacco mosaic virus infection sites in Nicotiana benthamiana. Mol Plant Microbe Interact 12: 143–152 [Google Scholar]
- Tsujimoto Y, Numaga T, Ohshima K, Yano M, Ohsawa R, Goto D, Naito S, Ishikawa M (2003) Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J 22: 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waigmann E, Ueki S, Trutnyeva K, Citovsky V (2004) The ins and outs of nondestructive cell-to-cell and systemic movement of plant viruses. Crit Rev Plant Sci 23: 195–250 [Google Scholar]
- Wang SY, Motes CM, Mohamalawari DR, Blancaflor EB (2004) Green fluorescent protein fusions to Arabidopsis finbrin 1 for spatio-temporal imaging of F-actin dynamics in roots. Cell Motil Cytoskeleton 59: 79–93 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Honda A, Iwata A, Ueda S, Hibi T, Ishihama A (1999) Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J Virol 73: 2633–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Meshi T, Okada Y (1987) Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett 219: 65–69 [Google Scholar]
- White RG, Badelt K, Overall RL, Vesk M (1994) Actin associated with plasmodesmata. Protoplasma 180: 169–184 [Google Scholar]
- Yamanaka T, Ohta T, Takahashi M, Meshi M, Schmidt R, Dean C, Naito S, Ishikawa M (2000) TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci USA 97: 10107–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.