Abstract
Autophagy is an important mechanism for nonselective intracellular breakdown whereby cytosol and organelles are encapsulated in vesicles, which are then engulfed and digested by lytic vacuoles/lysosomes. In yeast, this encapsulation employs a set of autophagy (ATG) proteins that direct the conjugation of two ubiquitin-like protein tags, ATG8 and ATG12, to phosphatidylethanolamine and the ATG5 protein, respectively. Using an Arabidopsis (Arabidopsis thaliana) atg7 mutant unable to ligate either tag, we previously showed that the ATG8/12 conjugation system is important for survival under nitrogen-limiting growth conditions. By reverse-genetic analyses of the single Arabidopsis gene encoding ATG5, we show here that the subpathway that forms the ATG12-ATG5 conjugate also has an essential role in plant nutrient recycling. Similar to plants missing ATG7, those missing ATG5 display early senescence and are hypersensitive to either nitrogen or carbon starvation, which is accompanied by a more rapid loss of organellar and cytoplasmic proteins. Multiple ATG8 isoforms could be detected immunologically in seedling extracts. Their abundance was substantially elevated in both the atg5 and atg7 mutants, caused in part by an increase in abundance of several ATG8 mRNAs. Using a green fluorescent protein-ATG8a fusion in combination with concanamycin A, we also detected the accumulation of autophagic bodies inside the vacuole. This accumulation was substantially enhanced by starvation but blocked in the atg7 background. The use of this fusion in conjunction with atg mutants now provides an important marker to track autophagic vesicles in planta.
Protein turnover plays numerous essential roles in plants, including the precise removal of short-lived regulatory proteins, the elimination of abnormal proteins, the maintenance of amino acid pools needed for continual protein synthesis, and the recycling of carbon (C) and nitrogen (N) during senescence and apoptosis (Vierstra, 1996). Whereas much of regulatory protein turnover is accomplished by the highly selective ubiquitin (Ub)/26S proteasome pathway (Smalle and Vierstra, 2004), plants, like other eukaryotes, also use less-selective high-throughput mechanisms for degrading intracellular constituents in bulk. One of the most conspicuous in the cytoplasm is autophagy. Here, individual proteins, protein complexes, and entire organelles become engulfed in membrane-bound vesicles, which are delivered to the vacuole (yeast/plants) or lysosome (animals; Ohsumi, 2001; Klionsky, 2004; Thompson and Vierstra, 2005). These vesicles and their contents are then degraded by a wide range of vacuolar proteases, peptidases, lipases, and other hydrolytic enzymes.
In yeasts, autophagic engulfment and delivery to the vacuole occurs by two morphologically distinct but mechanistically overlapping routes, microautophagy and macroautophagy (Klionsky, 2004). Microautophagy involves the sequestration of cytosol by tubular invaginations of the tonoplast. These tubes then pinch off to release intravacuolar vesicles called autophagic bodies, whose cargo is then degraded by vacuolar hydrolases following breakdown of the limiting membrane. Macroautophagy, in contrast, involves the entrapment of cytosol by double membrane-bound vesicles called autophagosomes. These vesicles appear to arise from a tubular network called the preautophagosomal structure. It encircles portions of the cytoplasm in a cage-like structure, which then coalesces and seals to form an autophagosome. The outer membrane of the autophagosome fuses with the vacuole/lysosome to release the internal vesicle as an autophagic body. In yeast, a smaller version of the autophagosome is used to deliver functional protein complexes to the vacuole via a pathway called cytoplasm-to-vacuole targeting (Huang and Klionsky, 2002).
While microautophagy and macroautophagy have been extensively described at the cytological level, only recently have the underpinning mechanisms been revealed. Especially informative have been genetic screens in the yeasts Saccharomyces cerevisiae and Pichia pastoris that identified a diverse set of autophagy (ATG) proteins (also known as APG, AUT, CVT, and GSA; Klionsky et al., 2003). In particular, two Ub-like conjugation systems have emerged as essential for autophagosome formation during macroautophagy and invagination of the tonoplast during microautophagy (Ohsumi, 2001; Klionsky, 2004). These conjugation pathways employ two Ub-like peptide tags: ATG8 and ATG12. Through ATP-dependent conjugation cascades analogous to ubiquitination, these tags become activated by the common E1, ATG7, and then are transferred to the E2s, ATG3 and ATG10, which are specific for ATG8 and ATG12, respectively. Finally, ATG8 and ATG12 are covalently attached to their respective targets, the lipid phosphatidylethanolamine (PE) and the ATG5 protein. Linkage is via an amide bond between the C-terminal Gly carboxyl group of the tags and the free amino group in PE and the ɛ-amino group of a specific Lys in ATG5. Production of the ATG8-PE and ATG12-ATG5 conjugates appears to help promote the formation and closure of autophagic vesicles and their subsequent delivery to the vacuole (Ohsumi, 2001; Klionsky, 2004). The ATG8-PE conjugate becomes bound to the autophagic membrane presumably via its lipid moiety. The ATG12-ATG5 conjugate assembles with ATG16 to form a proposed hetero-octameric structure loosely associated with the vesicle. The dynamics of these conjugates in vivo suggest that formation of the ATG12-ATG5 adduct precedes formation of the ATG8-PE adduct and enhances this lipidation reaction via crosstalk between the two conjugation reactions (Suzuki et al., 2001).
Following the discovery of the yeast ATG system, sequence relatives were detected in other eukaryotes, suggesting that the autophagic process is strongly conserved (Mizushima et al., 2001; Thumm and Kadowaki, 2001; Tanida et al., 2002; Marino et al., 2003; Melendez et al., 2003; Otto et al., 2003). For plants in particular, we (Doelling et al., 2002; Thompson and Vierstra, 2005) and Ohsumi and colleagues (Hanaoka et al., 2002) retrieved a collection of ATG orthologs from the Arabidopsis (Arabidopsis thaliana) genomic database. In contrast to S. cerevisiae, where single ATG8 and ATG12 genes are present, Arabidopsis encodes nine ATG8 (AtATG8a–i) and two ATG12 (AtATG12a and AtATG12b) isoforms, suggesting that the autophagic process in plants is more complex. Arabidopsis mutants disrupted in the single loci encoding ATG7 and ATG9 (a transmembrane protein also needed for autophagosome assembly) and the two loci encoding the ATG4 protease (needed for maturation of ATG8) grow normally but display accelerated senescence and are hypersensitive to N-deficient conditions (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004). A similar phenotype was apparent in an Arabidopsis mutant missing the vesicular soluble n-ethylmaleimide-sensitive factor attachment protein receptor (v-SNARE) VTI12 (Vps ten interacting), which may be required for docking and fusion of autophagosomes to the tonoplast of the vacuole (Surpin et al., 2003). Therefore, while autophagy may not be essential to plants under N-rich growth conditions, it becomes important for survival when N is limiting.
Here, we report the continued description of autophagy in plants through a further analysis of the ATG8/12 conjugation system. From reverse-genetic analyses of the single gene that encodes ATG5, we show that disruption of the ATG12 conjugation subpathway is just as debilitating as an inability to conjugate both ATG8 and ATG12. Furthermore, we found that the ATG12-ATG5 conjugation is important not only for survival under N-limiting conditions but also during C starvation. Using a green fluorescent protein (GFP)-ATG8a fusion in combination with concanamycin A, we detected the accumulation of intravacuolar vesicles that may represent autophagic bodies awaiting catabolism. Both the expanding collection of atg mutants and the GFP-ATG8a reporter provide new tools to follow this previously enigmatic recycling process.
RESULTS
Expression Patterns of the ATG Genes
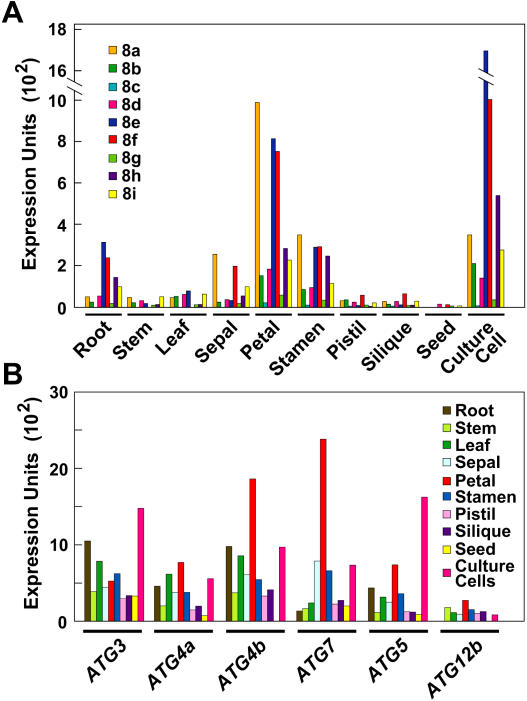
In previous studies, we and others discovered a collection of Arabidopsis genes related to the yeast ATG group, including small families that encode components of the ATG8/12 conjugation pathway (Doelling et al., 2002; Hanaoka et al., 2002). From an analysis of the Expressed Sequence Tag (EST; http://www.arabidopsis.org) and Massively Parallel Signature Sequences (http://mpss.udel.edu/at) databases and the DNA microarray results of Ma et al. (2005), it appears that all known Arabidopsis loci required for ATG8/12 conjugation are expressed (Fig. 1; data not shown). In fact, one or more ATG3, 4, 5, 7, 8, and 12 transcripts were detected in all tissues examined, indicating that this pathway is not restricted to a specific tissue type or developmental window (Fig. 1, A and B). High-level expression was observed in floral organs, including sepals, petals, and stamens (Fig. 1), implying that these ephemeral tissues use autophagy during their rapid senescence following fertilization. Comparably low-level expression was evident in germinating seeds, suggesting that autophagic events involving ATG proteins do not play a major role in seed metabolism.
Figure 1.
Expression levels of Arabidopsis ATG genes. Microarray expression levels of the nine-member ATG8 gene family (A) and other ATG genes (B) in various Arabidopsis tissues. Expression levels are graphed as normalized expression units × 102.
We also observed significant differences in expression among members of the ATG4 and ATG8 gene families, implying that tissue/cell-specific roles among their isoforms exist. Within the nine-member ATG8 family, ATG8a displayed the highest expression level in various flower parts, whereas ATG8e was highest in roots (Fig. 1A). In most cases, the microarray data for the family members were supported by analysis of the EST database. For example, the higher expression of ATG4b relative to ATG4a was paralleled by a 3-fold higher number of cDNAs in the EST collection. Within the ATG8 family, the three highly expressed isoforms ATG8a, e, and f had 21, 25, and 52 ESTs, respectively, whereas two less expressed isoforms, ATG8c and g, were represented by eight and five ESTs, respectively.
Isolation of Mutants Affecting ATG5
Previous studies with an Arabidopsis T-DNA disruption mutant affecting ATG7 showed that conjugation of both ATG8 and ATG12, while not essential, is important for survival under N-deficient growth conditions (Doelling et al., 2002). To help define the functions of the ATG12 conjugation pathway individually, we searched for mutants affecting its target protein ATG5. BLASTP searches of the near-complete genomes of Arabidopsis and rice (Oryza sativa) identified single loci encoding ATG5 in both species (At5g17290 and AP004084, respectively). By analysis of genomic and full-length cDNA sequences, the Arabidopsis ATG5 gene was predicted to encode a 337-amino acid protein with 66%, 31%, 42%, and 44% similarity to its rice, S. cerevisiae, Drosophila melanogaster, and human counterparts, respectively (Fig. 2). Among the numerous conserved residues present within the ATG5 collection is a Lys (position 128 in AtATG5) identified in the yeast version as the attachment site for ATG12.
Figure 2.
Sequence comparison of the ATG5 protein among eukaryotes. Amino acid sequence comparison of Arabidopsis (At) ATG5 (At5g17290) with the rice (Os; AP004084), yeast (S. cerevisiae, Sc; Kametaka et al., 1996), Drosophila (Dm; AAL39741), and human (Hs) orthologs (Hammond et al., 1998). Identical and similar amino acids are shown in reverse type and gray boxes, respectively. Dots denote gaps. Numbers at the end indicate the amino acid length of each protein. The asterisk marks the Lys (K128 in At) that provides the attachment site for ATG12. The arrowhead locates the T-DNA insertion site in the atg5-1 mutant.
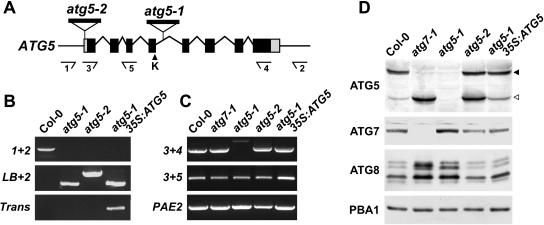
In a screen of the Syngenta Arabidopsis Insertion Library (SAIL) T-DNA insertion population in the wild-type Columbia-0 (Col-0) background (Sessions et al., 2002), we identified two mutant alleles of ATG5, designated atg5-1 and atg5-2 (Fig. 3A). The T-DNA of atg5-1 inserted within the fourth intron, just downstream of the exon bearing the codon for Lys-128, without altering the surrounding genomic sequence. For atg5-2, the T-DNA sequence interrupted the 5′ untranslated region (UTR), 40 nucleotides upstream of the translation start codon. Basta herbicide resistance conferred by the BAR gene within the T-DNA segregated in a 3:1 pattern for both mutant alleles, suggesting that each has a single insertion site. In populations of homozygous atg5-1 or atg5-2 plants, genomic PCR with 5′ and 3′ gene-specific primers (primers 1 + 2) confirmed the absence of an uninterrupted wild-type ATG5 gene, whereas PCR with a 5′ gene-specific primer and a T-DNA left-border (LB) primer (primers LB + 2) showed the presence of the introduced T-DNA sequences (Fig. 3B). Reverse transcription (RT)-PCR analysis demonstrated that the T-DNA prevented accumulation of the normal ATG5 mRNA in atg5-1 seedlings. Although a PCR product containing the region upstream of the T-DNA was generated from the mRNA (primers 3 + 5), a product encompassing the full-length transcript (primers 3 + 4) was not apparent (Fig. 3C). For atg5-2, however, RT-PCR showed that expression of the gene was not appreciably attenuated by the T-DNA. Both sets of primers were able to amplify the ATG5 mRNA, indicating that expression of the full-length protein was possible (Fig. 3C).
Figure 3.
Description of the Arabidopsis atg5-1 and atg5-2 mutants. A, Diagram of the Arabidopsis ATG5 gene. Lines indicate introns. Boxes indicate exons, with coding regions in black and the 5′ and 3′ UTRs in gray. The arrowhead locates the Lys (K-128) that provides the attachment site for ATG12. The locations of the T-DNA in the atg5-1 and atg5-2 mutants are shown. Arrows locate the priming sites used in B and C. B, Genomic PCR analysis of the atg5-1 and atg5-2 mutants. Genomic DNA isolated from wild-type Col-0, atg5-1, atg5-2, and atg5-1 complemented with 35S:ATG5 was subjected to PCR using either the number 1 and number 2 primer pairs (1 + 2), the number 2 primer with the LB of the T-DNA (LB + 2), or primers specific to the 35S:ATG5 transgene (Trans). C, RT-PCR analysis of the atg5-1 and atg5-2 mutants. Total RNA isolated from wild-type Col-0, atg7-1, atg5-1, atg5-2, and atg5-1 complemented with 35S:ATG5 was subjected to RT-PCR using either the number 3 and number 4 primer pair (3 + 4) or the number 3 and number 5 primer pair (3 + 5). A primer pair specific for PAE2 was used as an internal control. D, Immunoblot analysis of the atg5-1 and atg5-2 mutants. Crude protein extracts of wild-type Col-0, atg7-1, atg5-1, atg5-2, and atg5-1 complemented with 35S:ATG5 prepared from 10-d-old seedlings were subjected to SDS-PAGE and immunoblot analysis with anti-ATG7, anti-ATG5, and anti-ATG8 antibodies. Equal protein loads were confirmed by immunoblot analysis with anti-PBA1 antibodies. Black and white arrowheads identify the presumed ATG12-ATG5 conjugate (50 kD) and free ATG5 (40 kD), respectively.
To test if the atg5-1 and atg5-2 mutants affected accumulation of the ATG5 protein, we performed immunoblot analysis on crude extracts from homozygous seedling using antibodies generated against recombinant ATG5. As shown in Figure 3D, an abundant 50-kD protein and a minor 40-kD protein were detected in wild-type extracts. Both species (and smaller forms) were absent in the extracts from atg5-1 seedlings, demonstrating not only that the atg5-1 mutation likely represents a null allele but also that both species were derived from the ATG5 locus. The 40-kD species was close in apparent mass to that predicted for ATG5 (38,479 D), whereas the 50-kD species was close to the prediction for the ATG12-ATG5 conjugates (49,033 and 48,819 D for the ATG12a and b isoforms, respectively). To help confirm these assignments, we analyzed extracts from atg7-1 seedlings in the wild-type Wassilewskija (WS) background that fail to accumulate the E1 needed to conjugate ATG12 to ATG5 (Doelling et al., 2002). Here, the 50-kD species was completely absent whereas the 40-kD species was substantially more abundant (Fig. 3D), thus strongly supporting the proposal that the 50-kD protein is the ATG12-ATG5 conjugate while the 40-kD form is free ATG5.
Consistent with RT-PCR analyses showing that a full-length ATG5 transcript could be generated from the atg5-2 allele, we detected the corresponding ATG5 protein in homozygous atg5-2 seedlings. In contrast to wild-type seedlings, both free ATG5 and the ATG12-ATG5 conjugate were present in near equal amounts, indicating that the T-DNA may misregulate ATG5 expression (Fig. 3D). Since the atg5-2 mutation did not affect the level of the ATG12-ATG5 conjugate and was phenotypically silent, it was not characterized further.
Given the possibility that loss of ATG5 affects ATG8 conjugation in Arabidopsis as it does in yeast (Suzuki et al., 2001), we prepared antibodies against recombinant ATG8a to assay this protein family. This antibody also readily identified recombinant ATG8i, the most dissimilar isoform in this highly conserved family (69%–96% amino acid sequence identity within the family; Doelling et al., 2002), demonstrating that it should recognize all nine members of the ATG8 family (data not shown). In both wild-type Col-0 and WS seedling extracts, we detected several ATG8 proteins of 11 to 15 kD, indicating that multiple ATG isoforms accumulate (Fig. 3D). Their steady-state levels were substantially higher in the homozygous atg5-1 and atg7-1 backgrounds but not in the atg5-2 background, suggesting that defects in the entire ATG8/12 conjugation system and that for ATG12 specifically either enhance the synthesis of ATG8 proteins or interfere with their breakdown (see below).
Molecular Characterization of the N- and C-Limiting Phenotypes of atg5-1
Similar to Arabidopsis T-DNA mutants disrupting ATG7 (Doelling et al., 2002) and ATG9 (Hanaoka et al., 2002), homozygous atg5-1 seedlings germinated and developed normally, and flowered and set seed at the same rate as wild-type plants under a long-day photoperiod (16 h light/8 h dark; Fig. 4B; data not shown). However, under a short-day photoperiod (8 h light/16 h dark), the atg5-1 plants, like atg7-1 plants (Doelling et al., 2002), grew slower resulting in smaller plants, flowered later than wild type with reduced fecundity, and showed enhanced senescence of rosette leaves (data not shown; Fig. 9A).
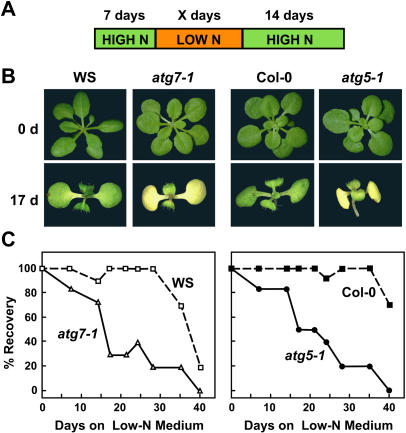
Figure 4.
Enhanced sensitivity of atg5-1 plants to N-deficient conditions. Plant lines include atg7-1 and atg5-1 and their respective wild-types WS and Col-0. A, Diagram of treatment. One-week-old seedlings were sown on N-rich agar medium and transferred to N-deficient agar medium for various lengths of time before transfer back to N-rich medium, all under a long-day photoperiod. B, Representative seedlings after 0 d and 17 d on N-deficient medium. C, The percentage of plants that survived growth for various days on N-deficient medium as determined by resumption of growth when transferred to N-rich medium. Each point represents the average of 10 seedlings.
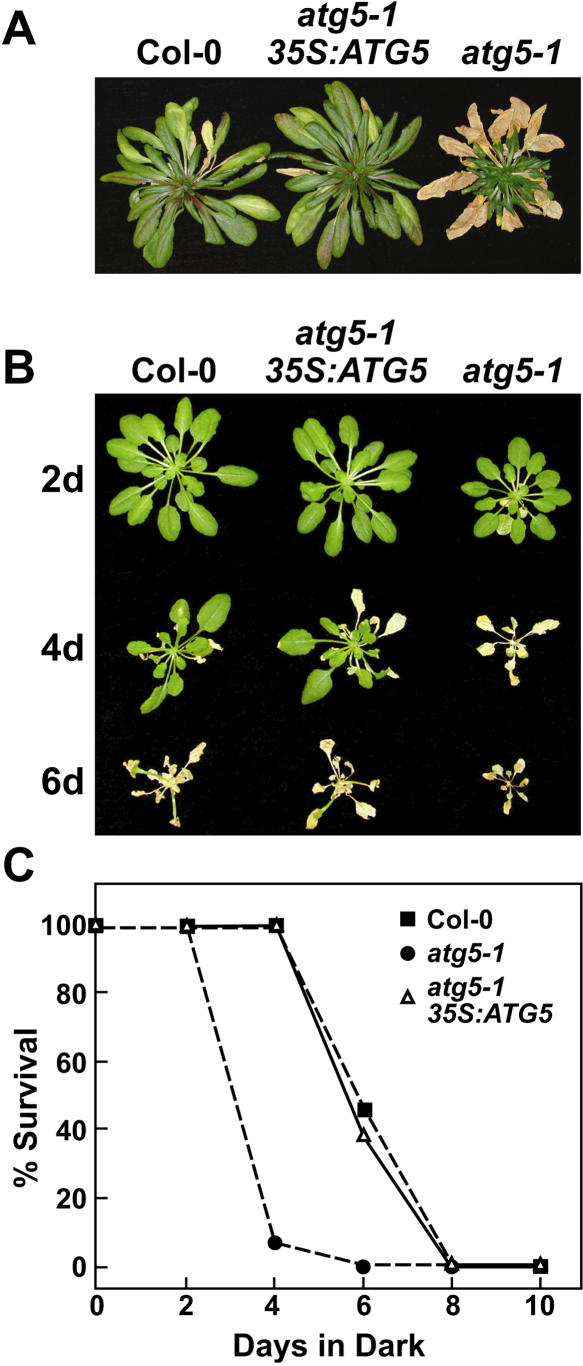
Figure 9.
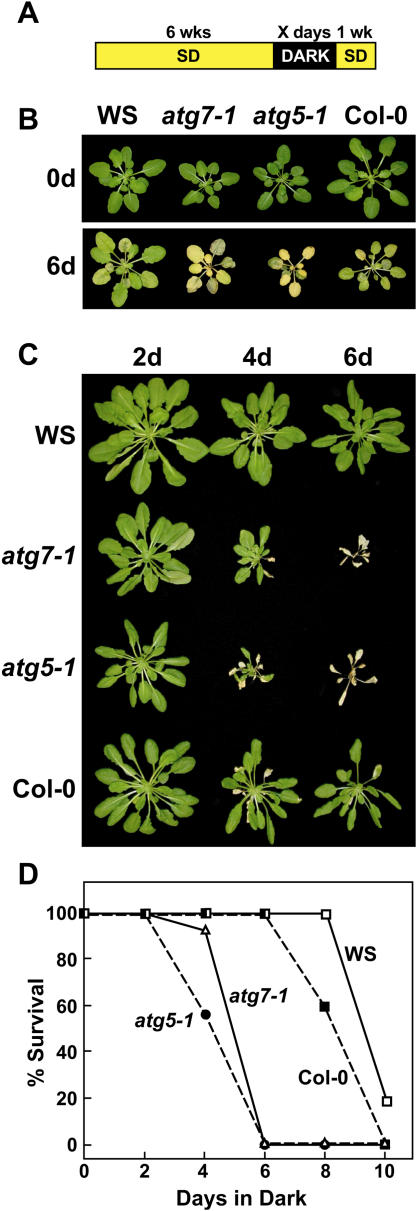
Complementation of the atg5-1 mutation with ATG5. Plant lines include wild type (Col-0), atg5-1, and atg5-1 complemented with a 35S:ATG5 transgene. A, Plants grown on soil for 3 months under a short-day photoperiod. B, Six-week-old plants grown under a short-day photoperiod, transferred to darkness for 2 to 6 d, and then transferred back to the short-day photoperiod. Images were taken after a 1-week recovery from the dark treatment. C, Percentage of plants that survived extended darkness as determined by resumption of growth is plotted versus days in the dark. Each point represents the average of 10 seedlings.
Prior growth studies showed that plants missing ATG7, and thus unable to conjugate both ATG8 and ATG12, are hypersensitive to N-deficient growth conditions (Doelling et al., 2002). A similar response was observed for homozygous atg5-1 seedlings when grown under a long-day photoperiod on a Suc-containing medium without N. Following 17 d on N-deficient medium, the atg5-1 seedlings, like atg7-1 seedlings, displayed enhanced chlorosis of the cotyledons and reduced true leaf formation as compared to wild-type seedlings (Fig. 4B). Enhanced chlorosis was also evident for detached leaves and intact plants grown in N-deficient liquid medium (data not shown). When the plants were exposed to various lengths of N starvation and then returned to N-rich medium, both the atg5-1 and atg7-1 mutants experienced a dramatic drop in the percentage of seedlings that recovered. While nearly 100% of wild type resumed growth after 28 d on N-deficient medium, only approximately 20% of the atg5-1 and atg7-1 plants recovered (Fig. 4C).
To test whether atg5-1 and atg7-1 plants also displayed slower growth and accelerated senescence under C-limiting conditions, we developed an assay to test the effects of C starvation specifically. Seedlings were first grown for 3 weeks on N/C-rich agar medium and transferred to soil for 3 more weeks in short days to maintain an adequate supply of photosynthate (Brouquisse et al., 1998; Matt et al., 1998). The seedlings were then placed in darkness for extended periods before a 1-week recovery under the short-day photoperiod. Previous studies showed that such prolonged darkness is a facile way to activate vacuole-specific proteolysis in corn, possibly via an autophagic mechanism (Brouquisse et al., 1998), and to accentuate autophagic defects in Arabidopsis (Hanaoka et al., 2002). As can be seen in Figure 5, both the wild-type WS and Col-0 parents could tolerate this dark incubation for up to 8 d and then resume growth in the light. At 10 d, all of the Col-0 plants and most of the WS plants died, thus defining an upper limit for tolerance to extended darkness. When the atg5-1 and atg7-1 seedlings were tested similarly, a striking hypersensitivity to darkness was apparent. Both mutants barely recovered after 4 d in the dark, whereas none survived after 6 d (Fig. 5, C and D).
Figure 5.
Enhanced sensitivity of atg7-1 and atg5-1 plants to C-limiting conditions induced by extended darkness. Plant lines include atg7-1 and atg5-1 and their respective wild types WS and Col-0. A, Diagram of treatment. Six-week-old plants were grown under a short-day photoperiod, transferred to darkness for various lengths of time, and then transferred back to the short-day photoperiod. B, Plants immediately before or after a 6-d dark treatment. C, Plants after 1-week recovery from 2-, 4-, and 6-d dark treatments. D, Percentage of plants that survived this regime as determined by resumption of growth is plotted versus days in the dark. Each point represents the average of 10 seedlings.
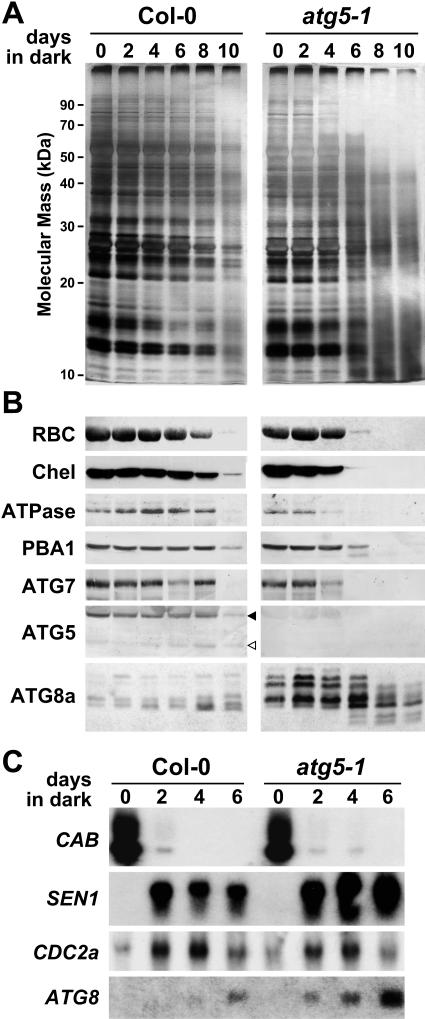
To examine the effects of C starvation induced by darkness at the molecular level, we collected wild-type and atg5-1 plants immediately after the dark treatment and examined both protein and mRNA levels. Plants left for progressively longer times in the dark became severely chlorotic and flaccid, a condition that was more pronounced in the atg5-1 mutant (Fig. 5B). Consistent with these problems, the atg5-1 mutant lost protein faster than its respective Col-0 wild type during the dark treatment, as seen by both silver staining of total protein and by immunoblot analyses for specific polypeptides (Fig. 6, A and B). Spectrophotometric analysis showed that chlorophyll was also degraded at an accelerated rate in the mutants. Despite the faster rate, the complement of proteins degraded in the atg5-1 background (as detected by silver staining) was indistinguishable from wild type, with little evidence that specific proteins were selectively retained or lost in the mutant (Fig. 6A). This comparable breakdown was supported by immunoblot analyses with a battery of antibodies against cytosolic and organellar proteins. As can be seen in Figure 6B, Rubisco and magnesium-protoporphyrin chelatase, which are markers for chloroplasts, a mitochondrial marker (the ATPase F1 subunit), and a cytosolic/nuclear marker (the 26S proteasome β1 subunit PBA1), all disappeared similarly during the dark treatment, with their loss accelerated in the atg5-1 seedlings.
Figure 6.
Protein and RNA analyses of atg5-1 plants exposed to C-limiting conditions induced by extended darkness. Tissue was collected from wild-type Col-0 and atg5-1 seedlings just after removing the plants from the indicated lengths of extended darkness (see Fig. 5B). A and B, SDS-PAGE analysis of total protein from an equal amount of tissue by fresh weight. A, Gels silver stained for total protein. B, Gels subjected to immunoblot analysis with antibodies against the large subunit of Rubisco, magnesium-protoporphyrin chelatase (Chel), the F1 subunit of mitochondrial ATPase (ATPase), β-subunit A of the 26S proteasome (PBA1), ATG7, ATG5, and ATG8. Black and white arrowheads identify the presumed ATG12-ATG5 conjugate (50 kD) and free ATG5 (40 kD), respectively. C, RNA gel-blot analysis of equal amounts of total RNA using probes for CAB, SEN1, CDC2a, and ATG8a. Equal loading of total RNA was confirmed by staining for rRNA with methylene blue (data not shown).
With respect to the ATG8/12 conjugation pathways, we found that the levels of ATG7 also decreased faster in the atg5-1 mutant background. While the free 40-kD form of ATG5 was barely detectable in wild-type plants before the dark treatment, this species became more abundant following prolonged incubation in the dark (Fig. 6B). Not surprisingly, this increase in free ATG5 in the Col-0 plants coincided with the drop in ATG7, which is necessary for its conjugation. Unlike ATG5 and ATG7 and other seedling proteins, the levels of the ATG8 protein family in wild-type seedlings did not drop during dark-induced C starvation but increased slightly despite the substantial loss of most other polypeptides (Fig. 6B). Similar to the observations in Figure 3D, the levels of the ATG8 proteins were substantially higher in the homozygous atg5-1 background, with several new species becoming predominant (Fig. 6B). Most of these species peaked in abundance between 2 to 6 d of darkness and declined thereafter. After more extended dark treatments, two lower molecular mass species of approximately 8 kD appeared (Fig. 6B). Although it remains possible that these species are ATG8-PE conjugates, their appearance at a time when ATG7 was nearly undetectable argued otherwise.
To confirm that the response of the plants to extended darkness was a senescence-related process, we examined the abundance of several transcripts (Fig. 6C), including that from SEN1, a gene specifically expressed during senescence (Weaver et al., 1998). Similar to previous studies with detached leaves (Doelling et al., 2002), CAB mRNA levels dropped rapidly following a dark incubation as short as 2 d, consistent with the instability of the CAB mRNA and the light requirement for continued CAB transcription (Giuliano et al., 1988). Conversely, transcripts from the cell-division regulator CDC2a and the senescence-associated SEN1 genes increased during the C-limiting treatment. Whereas the CDC2a mRNA levels fell after prolonged dark exposure, levels of the SEN1 transcript remain high especially in the atg5-1 background, supporting the view that this dark-induced chlorosis represents a form of senescence.
Previous studies found that the Arabidopsis ATG8 mRNAs are mildly increased during the Suc starvation of culture cells (Contento et al., 2004) and during the senescence of detached leaves (Doelling et al., 2002). To test if a similar increase could explain the increase in ATG8 protein during dark-induced C starvation in wild-type plants and the large increase in ATG8 protein in the atg5-1 background, we compared the abundance of the ATG8 transcripts and proteins. Similar to detached leaves (Doelling et al., 2002), the abundance of the ATG8 mRNA increased during dark-induced C starvation of intact wild-type plants (Fig. 6C). More ATG8 mRNA was evident in the atg5-1 background, suggesting that expression of these genes is up-regulated by this autophagic defect.
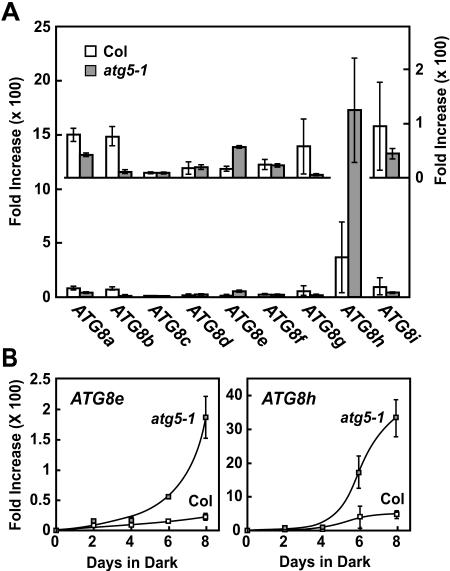
To help identify which of the nine ATG8 transcripts were responsible for this increase during C starvation and the further increase in the atg5-1 mutant, we analyzed the abundance of each during the dark treatment by real-time RT-PCR. We quantified ATG8a to ATG8i mRNA levels using the comparative threshold cycle (CT) method (Giulietti et al., 2001). Values were normalized to an internal control mRNA (histone H2A) and expressed relative to those before the dark treatment. As can be seen in Figure 7A, the relative abundance of most, if not all, ATG8 mRNAs was slightly or dramatically increased by a 6-d C starvation. Several were further increased in the atg5-1 background, especially ATG8e and ATG8h (Fig. 7B). A time course of this induction showed that both ATG8e and ATG8h transcripts rose dramatically in relative abundance beginning at day 6 and further increased at day 8 despite that fact that tissue became noticeably senescent with reduced total protein (Figs. 5B and 6, A and B). Collectively, the data show that ATG8 mRNA levels are up-regulated by C starvation with several responsive to the mutation in ATG5. These changes may in turn be responsible for the increase in ATG8 proteins in the atg5-1 seedlings during C starvation (Fig. 6B).
Figure 7.
Relative abundance of individual ATG8 mRNAs in Col-0 and atg5-1 plants exposed to C-limiting conditions induced by extended darkness. Relative abundance was obtained by real-time RT-PCR using the CT method and expressed as the average of triplicate technical repeats with se. All values are graphed as a fold increase in mRNA abundance relative to that at day 0 and were normalized against the abundance of histone H2A mRNA, used here as an internal control. A, Relative increases in mRNA abundance of each ATG8 transcript after 6 d of dark treatment as compared to abundance before treatment. Inset shows an expanded scale for the ATG8a to ATG8g and ATG8i transcripts. B, mRNA abundance for ATG8e and ATG8h over the course of an 8-d dark treatment relative to that before treatment.
Disruptions of ATG5 and ATG7 Affect the Profile of ATG8 Proteins
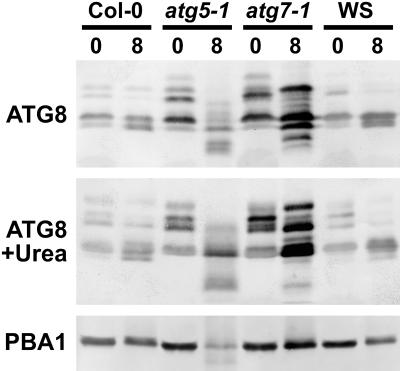
Recent studies by Yoshimoto et al. (2004) detected a new ATG8 species during N starvation that could represent one or more ATG8-PE adducts based on its faster migration rate. To further examine this possibility, we compared the profiles of ATG8 proteins in nonstarved and C-starved wild-type, atg5-1, and atg7-1 seedlings, using SDS-PAGE with and without 6 m urea. The inclusion of urea was based on previous studies with yeast ATG8 showing that the PE-conjugate migrates faster than free ATG8 under this electrophoretic condition (Kirisako et al., 1999). Unfortunately, while a number of ATG8 species were observed, none matched the expectations for the ATG8-PE adducts. As shown in Figure 8, several ATG8 isoforms were detected in wild-type Col-0 and WS seedlings grown in the light, with their abundance and complexity further enhanced by the atg5-1 and atg7-1 mutations. In wild-type plants, no new species (especially faster migrating forms) became evident (with or without urea) following a prolonged dark treatment of 8 d sufficient to impair survival. A faster migrating form was detected in C-starved atg5-1 plants in both the absence and presence of urea (Fig. 8). However, similarly sized species were also observed (albeit weaker) in atg7-1 plants, which should lack the capacity to form these lipidated adducts.
Figure 8.
Differential accumulation of ATG8 proteins under C-limiting growth conditions induced by extended darkness. Tissue was collected from wild types (Col-0 and WS), atg5-1, and atg7-1 immediately before or after 8 d in extended dark. ATG8 proteins were detected by immunoblot analysis with anti-ATG8 antibodies following SDS-PAGE with (bottom) or without (top) 6 m urea in the separating gel. Total protein from equivalent amounts of tissue fresh weight was used in each case. Intensities of PBA1 as detected with anti-PBA1 antibodies reflect the levels of total protein.
In an attempt to enrich for possible ATG8-PE adducts, we separated total cell lysates from C-starved and nonstarved seedlings by ultracentrifugation and assayed by immunoblotting the membrane-associated fraction for these species. Although we found the presumed ATG12-ATG5 conjugate of 50 kD in both the supernatant (soluble) and membrane fraction (pellet) consistent with a peripheral membrane association, none of the ATG8 species were in the pellet (data not shown). Collectively, the results suggest that the ATG8-PE conjugates, if produced, do not become a significant portion of the ATG8 pool under the conditions studied here.
Complementation of atg5-1
To confirm that the atg5-1 phenotypes were directly caused by a loss of ATG5 protein, we attempted to complement the mutation by ectopic expression of the wild-type ATG5 cDNA. atg5-1 plants transformed with an ATG5 transgene under the control of the cauliflower mosaic virus (CaMV) 35S promoter were selected by hygromycin resistance conferred by the T-DNA. T1 plants were self-fertilized, and progenies homozygous for the atg5-1 allele and 35S:ATG5 transgene were identified by PCR and hygromycin resistance, respectively (Fig. 3B). Both the ATG5 transcript and ATG5 proteins were detected in the complementation line, with the ratio of the 40- and 50-kD proteins similar to that found in wild-type seedlings (Fig. 3, C and D). Phenotypically, plants harboring the 35S:ATG5 transgene behaved like wild-type Col-0. Under the short-day photoperiod, they senesced similar to the wild type and much later than the atg5-1 mutant parent (Fig. 9A). The homozygous atg5-1/35S:ATG5 seedlings like wild type were also much less sensitive to C-limiting conditions induced by darkness than atg5-1 seedlings (Fig. 9, B and C). We observed no new phenotypes among complementation lines, suggesting that possible misexpression of ATG5 caused by the use of the CaMV 35S promoter does not interfere with autophagy in particular and overall growth in general.
Visualization of ATG8 Labeled Vesicles Is Dependent on ATG7
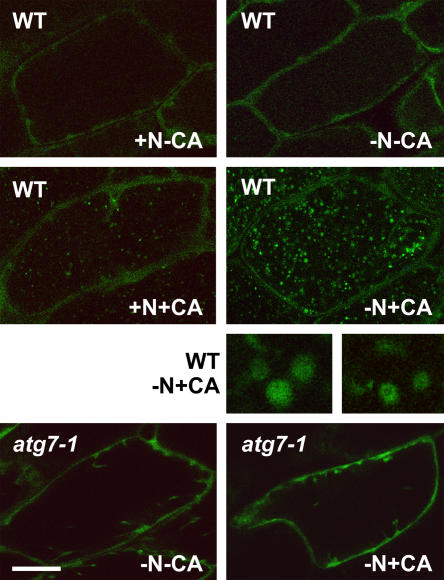
To help detect the various structures associated with autophagy, we examined the possibility of using GFP fusions to ATG8 proteins as cytological markers. Previous studies with both yeast and mammalian cells demonstrated that such fusions can be good markers for visualizing autophagic bodies in vivo (Suzuki et al., 2001; Mizushima et al., 2004). More recently, a similar GFP-ATG8 fusion was found to label autophagic body-like vesicles in Arabidopsis vacuoles (Yoshimoto et al., 2004). Their visualization in Arabidopsis required pretreatment with the vacuolar H+-ATPase inhibitor concanamycin A, which appears to block vacuolar hydrolysis by raising the internal pH above the optimum for many vacuolar enzymes (Tamura et al., 2003).
Here, we introduced a transgene expressing a GFP fusion to the N terminus of ATG8a into wild-type Col-0 and atg7-1 plants. (The atg5-1 mutant was not used since it does not a priori affect the conjugation of ATG8.) Immunoblot analysis with anti-ATG8a and anti-GFP antibodies detected a 40-kD protein in both plants expressing GFP-ATG8a, consistent with the expected size of the chimeric protein (data not shown). Using fluorescence confocal microscopy, we monitored hypocotyl cells of young seedlings after 2 d in N-rich and N-deficient media. As shown in the top sections of Figure 10, GFP-ATG8a is predominately cytoplasmic in wild-type cells with or without N starvation. Upon concanamycin A treatment, punctate structures accumulated in the central vacuole, with substantially more present upon N starvation (Fig. 10, middle sections). Upon closer examination, they resembled autophagic bodies, some appearing as hollow spherical vesicles with a diameter of 0.5 to 1.5 μm. Unlike transvacuolar strands/tubes that are highly dynamic and can appear as vesicles in cross section (Hoffmann and Nebenfuhr, 2004; Takatsuka et al., 2004), the vesicles labeled with GFP-ATG8a displayed a slow random motion, indicating that they were free-floating within the vacuolar lumen. The accumulation of these vesicles required the ATG8/12 conjugation system, further supporting the possibility that they represents autophagic bodies. Even after N starvation and treatment with concanamycin A, we failed to detect these vesicles in atg7-1 seedlings (Fig. 10, bottom sections). This pattern was also distinct from that observed with wild-type plants expressing GFP alone from the CaMV 35S promoter. Here, we observed diffuse staining of just the cytoplasm, which did not change upon N starvation with or without concanamycin A (data not shown).
Figure 10.
Use of a GFP-ATG8 fusion to detect possible autophagic vesicles inside the vacuole. Eight-day-old wild-type Col-0 and atg7-1 seedlings expressing a GFP-ATG8a fusion were grown in N-rich liquid medium for 6 d, transferred to N-rich (+N) or N-deficient (−N) liquid media for approximately 1.5 d, and then incubated for an additional 12 to 16 h with 0.5 μm concanamycin A (+CA) or an equal volume of dimethyl sulfoxide (−CA). Hypocotyls were visualized by fluorescence confocal microscopy of GFP. Size bars for images are equal to approximately 50 μm. Five-times magnifications of possible autophagic bodies in the vacuole of wild-type plants are included.
DISCUSSION
Although autophagy has been proposed to play important roles in nutrient recycling in plants, the underpinning mechanism(s) have remained elusive in the absence of molecular, cytological, and genetic markers to analyze the process (Moriyasu and Klionsky, 2004; Thompson and Vierstra, 2005). Clearly, significant advances are now possible with the discovery of orthologs to many components of the yeast ATG system in Arabidopsis, including the Ub-like conjugation system that attaches the peptide tags ATG8 and ATG12 to PE and the ATG5 protein, respectively (Doelling et al., 2002; Hanaoka et al., 2002; Menand et al., 2002; Surpin et al., 2003). Through reverse-genetic analyses of ATG7 (the E1 required for ATG8 and ATG12 attachment) and ATG5 (the protein target of ATG12), we show that the ATG system in particular and possibly autophagy in general, while not essential, is important for mobilizing nutrients during senescence and survival under N- and C-limiting conditions (Doelling et al., 2002; this article). Disruption of the single genes encoding either ATG5 or ATG7 generated plants that senesce earlier and are hypersensitive to N-deficient media and C starvation induced by extended darkness. However, other aspects of plant development that might have employed autophagic events occur normally in the mutants, including seed germination, pollen function, and various forms of apoptosis (e.g. vessel element and fiber maturation). Whether this insensitivity indicates that these processes do not require autophagy or use ATG-independent mechanism(s) is not yet known.
The phenotypes of the atg5-1 mutant grown under C- and N-rich and limiting conditions were indistinguishable from those of atg7-1, demonstrating that disruption of just the ATG12 conjugation pathway is as effective as simultaneously abrogating both the ATG8 and ATG12 conjugation pathways in Arabidopsis. Both the severity of the starvation phenotypes and the ability of the plants to recover were equally exacerbated in the atg5-1 and atg7-1 mutants. Similar phenotypes were evident for plants defective in ATG9, a scaffolding protein that functions upstream of ATG8/12 conjugation during autophagy (Hanaoka et al., 2002), and plants missing the v-SNARE VTI12, which likely functions downstream in vesicle docking with the tonoplast (Surpin et al., 2003). A recently published work by Yoshimoto et al. (2004) reported a similar set of phenotypes for a double mutant missing the ATG4a and ATG4b proteases required for maturation of seven of the nine ATG8s and their possible recycling from the PE adduct. Taken together, these mutant phenotypes suggest that disruption of the ATG system anywhere within its sequence cascade likely will induce the same developmental and growth defects. Given the central importance of the encoded proteins, the corresponding mutant phenotypes may represent the most severe abnormalities caused by inactivation of the ATG system.
Contrary to expectations, disruption of the ATG system and thus autophagy led to an acceleration of senescence and faster breakdown of both cytoplasmic and organellar proteins upon starvation. As judged by the loss of Rubisco and magnesium-protoporphyrin chelatase, chloroplasts were lost faster in the atg5-1 and atg7-1 mutants (Doelling et al., 2002; this article), despite the proposal that autophagic events are responsible for their removal (e.g. Moriyasu and Klionsky, 2004; Niwa et al., 2004). Clearly, this faster breakdown implies that an alternative route(s) became more prominent upon loss of the ATG system to maintain the supply of C and N needed for survival. Remarkably, these alternatives degrade cytosolic, chloroplastic, and mitochondrial proteins faster than normal but appear to compromise the viability of mutant plants. The nature of this alternative route(s) is unknown. The Ub/26S proteasome does not appear to represent one option given that the 26S proteasome (as judged by the PBA1 subunit) is not selectively retained during C starvation of the atg5-1 mutant.
Analysis of ATG5 further supports the expectations that the ATG8/12 conjugation pathway in plants is mechanistically similar to that in yeast. In wild-type Arabidopsis seedlings, most of the ATG5 pool exists as a higher molecular mass species. Both the apparent mass of this species and its absence in atg7-1 plants support its identity as the ATG12-ATG5 adduct. Surprisingly, little free ATG5 was available in wild-type plants even under nonstarvation conditions, implying that its conjugation to ATG12 is constitutively active. However, the free ATG5 pool did rise in wild-type plants during starvation. Its coincidence with the drop in ATG7 implies that the conjugation machinery may have become limiting. A significant portion of the ATG5 pool was also in the free form in homozygous atg5-2 plants. In this case, one possible scenario is that the level of ATG5 in these lines exceeded that of ATG12. Another is that this free form reflects misexpression of ATG5 proteins in cells/tissues not active in ATG12 conjugation. For atg5-2, such upregulation and/or misregulation could be induced by the insertion of the T-DNA within the 5′ UTR. Regardless of the reason, the rise in free ATG5 does not appear to be deleterious as the phenotype of the corresponding atg5-2 plants appeared indistinguishable from wild type.
With respect to ATG8, a number of distinct species were evident in both C-starved and nonstarved wild-type seedlings with their complexity further enhanced by the atg5-1 and atg7-1 mutations. Which, if any, of these species are the ATG8-PE adducts is unclear. SDS-PAGE analyses with or without urea of total seedling extracts failed to detect likely candidates even in fractions enriched for membrane proteins. The reason for this failure to detect the lipidated form is unknown. Certainly, the complex banding patterns of the ATG8 protein family could interfere with detection of the ATG8-PE conjugates. Another possibility is the ATG8-PE adducts are rapidly turned over in vivo and/or rapidly disassembled by cytosolic lipases in vitro before analysis. With regard to in vitro artifacts, even extraction of the tissue directly in SDS-PAGE sample buffer followed by rapid heat denaturation failed to detect new ATG8 species. Yoshimoto et al. (2004) recently identified several ATG8 species that could be ATG8-PE adducts, using cell fractionation coupled with an atg4a atg4b double mutant missing the ATG4 protease. However, given that only some of the ATG8 isoforms need this protease for maturation, it remains possible that these new species represent partial cleavage products of ATG8 and not the lipidated forms as proposed. Clearly, biochemical analyses of these species are needed to resolve this issue.
In accord with recent data, we found that the levels of most, if not all, Arabidopsis ATG8 mRNAs are increased by starvation. Whereas our data analyzed the effects of dark-induced C starvation, similar global up-regulation was evident during N and C starvation of whole seedlings (Yoshimoto et al., 2004; Buchanan-Wollaston et al., 2005) and Suc starvation of suspension culture cells (Contento et al., 2004; Rose et al., 2005). A further increase was discovered in the atg5-1 seedlings during C starvation (this article), suggesting that impairment of the autophagic process further activates expression of this gene family. In particular, the relative abundance of both ATG8e and ATG8h mRNAs was dramatically increased, implying that a subset of ATG8 genes is more responsive to this defect. Coincidently, these same two isoforms are also the ones identified by Contento et al. (2004) as most dramatically up-regulated by Suc starvation of cultured cells. The increased levels of ATG8 mRNAs in turn are likely responsible for the increased ATG8 protein levels seen in the atg5-1 and atg7-1 backgrounds. However, we cannot rule out the possibility that the defects in autophagy caused by these mutations also block the turnover of the ATG8 proteins. Current models for both microautophagy and macroautophagy propose that a portion of the ATG8 pool (possibly the ATG8-PE conjugate) decorates the preautophagosomal structure membranes that form autophagosomes during macroautophagy and the tonoplast invaginations that generate autophagic bodies during microautophagy (Ohsumi, 2001; Suzuki et al., 2001). During this process, a portion of the ATG8 pool would become trapped within the resulting vesicles and be degraded along with the vesicle in the vacuole. Our ability to tag possible autophagic bodies with GFP-ATG8a fusions is consistent with this scenario.
DNA microarray analyses indicate that most if not all components of the Arabidopsis ATG8/12 conjugation pathway are expressed in all organ types examined. Paralog-specific patterns were also evident among members of the ATG4 and ATG8 families, suggesting that tissue specificity exists among protein isoforms. Like previous studies with yeast and mammalian cells (Suzuki et al., 2001; Mizushima et al., 2004), we found that the ATG8 proteins may provide valuable markers for detecting autophagic events in plants. Using Arabidopsis expressing a GFP fusion to ATG8a, we detected by fluorescence confocal microscopy of hypocotyl cells ATG8a diffusively throughout the cytoplasm and concentrated in small spherical vesicles within the vacuolar lumen. These vesicles morphologically resembled autophagic bodies described in other eukaryotes, and, like autophagic bodies, their accumulation required ATG7 and was enhanced upon N starvation. In recent work by Yoshimoto et al. (2004), similar punctate structures were observed by fluorescence microscopy of root cells expressing GFP fusions to ATG8a, ATG8e, and ATG8i. Electron microscopy of these vesicles identified some cargo as organelles, strongly suggesting that they are autophagic bodies (Yoshimoto et al., 2004). Under our and their conditions, accumulation of these vesicles was dramatically aided by prior incubation of the seedlings with concanamycin A. By blocking the tonoplast H+-ATPase, this inhibitor raises the vacuolar pH, which in turn inactivates various vacuolar hydrolyases with acidic pH optima (Drose et al., 1993; Matsuoka et al., 1997). Presumably, this inactivation then stabilizes autophagic bodies from breakdown. The protease inhibitor E64c appears to have a similar effect on blocking autophagy-mediated events by inhibiting vacuolar Cys proteases (Moriyasu and Ohsumi, 1996; Takatsuka et al., 2004). By these mechanisms, both concanamycin A and E64c have also been shown to effectively stabilize GFP targeted to the vacuole by other transport pathways (Tamura et al., 2003).
One surprising feature of these possible autophagic bodies is their abundance. As can be seen in Figure 10, the vacuoles of N-starved hypocotyls became filled with hundreds of these vesicles during the 16-h incubation with concanamycin A. Such activity implies that autophagy can encapsulate substantial portions of the cytoplasm under nutrient-limiting conditions. Although much fewer vesicles were evident in nonstarved plants, their presence suggests that autophagy is active even under nonlimiting growth conditions. While it remains possible that autophagy can be accomplished by other routes in plants, the complete absence of these vesicles in atg7-1 cells argues that most if not all autophagic recycling requires the ATG system.
Certainly, the exploitation of the ATG system by both reverse genetics and cell biological approaches offers an excellent opportunity to dissect autophagy in plants. In particular, the discovery that GFP-ATG8 fusions appear to label autophagic bodies, which can be visualized in live cells by fluorescence confocal microscopy, now provides a marker to follow their formation in both cell- and environmental-specific manners. Labeled ATG8s may also enable the purification of autophagic bodies, whose contents can then be analyzed to define the types of cargo degraded by autophagic processes.
MATERIALS AND METHODS
Expression Analysis of ATG Genes
DNA microarray design and experimental data were extracted from Ma et al. (2005). The 70-mer oligonucleotide set of the Arabidopsis (Arabidopsis thaliana) Col-0 genome was designed and synthesized by Qiagen/Operon (http://oligos.qiagen.com/arrays/omad.php) based on the Arabidopsis genome information available on February 20, 2002. Roots from 6-d-old seedlings, stems from 4-week-old plants, rosette leaves from 3-week-old plants, floral organs at flowering, siliques 3 d after pollination, and seeds were collected from wild-type Col-0 Arabidopsis. Suspension culture cells were prepared from seeds and collected at the logarithmic growth phase. A negative control cutoff value for determining genes that were “expressed” versus “not expressed” for each hybridization was obtained by ranking the values of the negative control mismatch oligonucleotides from high to low and using the 90th percentile spot as the cutoff value (Ma et al., 2005). Expression data were included here only if the signal for the majority of the corresponding spots from multiple experiments exceeded this cutoff. The signal intensities of different organs were normalized by equalizing the median value of all individual gene intensities from each organ.
Sequence Analysis of ATG5 Proteins
ATG5 loci were identified in the Arabidopsis ecotype Col-0 (http://www.arabidopsis.org) and rice (Oryza sativa; http://www.tigr.org) genomic databases using yeast ATG5 as the query. Intron/exon positions of AtATG5 were determined by alignment with the full-length cDNA sequence from The Arabidopsis Information Resource (http://www.arabidopsis.org). Amino acid sequence comparisons were performed using MACBOXSHADE (Institute of Animal Health, Pirbright, UK). The GenBank accession numbers for the sequences described in this article are BAB10516 (AtATG5), AP004084 (OsATG5), NP_015176 (ScATG5), AAL39741 (DmATG5), and AAH02699 (HsATG5).
Isolation of atg5-1 and atg5-2 and Complementation of atg5-1
The atg5-1 and atg5-2 T-DNA insertion mutants generated with the Col-0 ecotype were obtained from the SAIL (Sessions et al., 2002). The mutations were tracked by PCR using the 3′ gene-specific primers CAATTCACAGATGGATTGTAACTGCAGAG (atg5-1) and CACAAAGGAGATCGAAAAGAACACCTGTTG (atg5-2) in combination with the LB T-DNA-specific primer GCCTTTTCAGAAATGGATAAATAGCCTTGCTT (Sessions et al., 2002), and by Basta resistance conferred by the T-DNAs. The mutants were backcrossed three times to the wild-type Col-0 to help remove extraneous mutations. Homozygous atg5-1 and atg5-2 mutants generated by self-pollination of heterozygous plants were verified by the presence of a T-DNA-specific product and by the absence of a full-length ATG5 gene product using PCR with the 5′ gene-specific primer ACGTTAGCCACCAACAGATTAAGCAGTGT in combination with the above 3′ gene-specific primers.
For complementation, the full-length coding region of the ATG5 cDNA was amplified by PCR using primers CTGCATATGGCGAAGGAAGCGGTCAAGTATGTA and TGTCTAGACAATTCACAGATGGATTGTAACTGCAGA designed to introduce an NdeI site at the 5′ end and an XbaI site at the 3′ end, respectively. A single Myc tag cassette was appended to the 5′ end of the ATG5 coding sequence, which was preceded by an XbaI site. The resulting product was digested with XbaI and inserted downstream of the CaMV 35S promoter into the binary vector pGSVE9, which was digested similarly (E. Babiychuk and S. Kushnir, unpublished data). The vector was introduced into Agrobacterium tumefaciens strain GV3101 and then transformed into homozygous atg5-1 plants by the floral dip method (Clough and Bent, 1998). The 35S:ATG5 transgene and the atg5-1 mutation were followed by hygromycin and Basta resistance, respectively. T2 plants homozygous for the atg5-1 mutation were confirmed to also contain the transgene by PCR. T3 seeds resulting from self-fertilization were used for phenotypic analyses.
Plant Growth Conditions
Arabidopsis seeds were vapor-phase sterilized, incubated in water at 4°C for 2 d, and grown on Gamborg's B5 (Sigma, St. Louis) agar or liquid medium with 1% Suc. The plates or liquid cultures were incubated at 21°C in 16-h-light/8-h-dark photoperiod for long days, 8-h-light/16-h-dark photoperiod for short days, or in continuous light. For plants exposed to a long-day photoperiod, plants were transferred to soil after 2 weeks.
For exposure to N-limiting conditions, 1-week-old seedlings grown in the long-day photoperiod were transferred to N-deficient agar medium: Murashige and Skoog micronutrient salts (Sigma), 3 mm CaCl2, 1.5 mm MgSO4, 1.25 mm KH2PO4, 5 mm KCl, 2 mm MES, pH 5.7. After various amounts of time on the N-deficient medium, seedlings were transferred back to Gamborg's B5 agar. For exposure to C-limiting conditions, seedlings grown in a short-day photoperiod for 3 weeks were transferred to soil and grown for 3 more weeks. The plants were then transferred to continuous darkness for various lengths of time, and either collected immediately or returned to the short-day photoperiod for a 1-week recovery. For confocal microscopy, seeds were germinated in Gamborg's B5 liquid. After 1 week, the seedlings were transferred to N-deficient medium for a 2-d incubation. Twelve to 16 h prior to examination, 0.5 μm concanamycin A (Sigma) was added to the growth medium. The seedlings were then incubated thereafter in the dark to enhance visualization of GFP in the vacuole (Tamura et al., 2003).
Protein Isolation and Immunoblot Analysis
Total protein was isolated from liquid-grown or soil-grown plants by homogenization in 2:1 (volume to fresh weight) SDS-PAGE sample buffer (125 mm Tris, pH 6.8, 5% SDS, 20% glycerol, and 10% 2-mercaptoethanol), and extracts were clarified by low-speed centrifugation. Proteins were subjected to SDS-PAGE with or without 6 m urea in the separating gel, and either stained with silver or transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA) for immunoblot analysis using alkaline phosphatase-labeled or peroxidase-labeled goat anti-mouse or goat anti-rabbit immunoglobulins (Kirkegaard & Perry Laboratories, Gaithersburg, MD) for detection. Sample sizes were adjusted to reflect either equal protein or equal fresh weight as indicated.
Antibodies against ATG5 and ATG8a were produced in rabbits (Polyclonal Antibody Service, Madison, WI) using recombinant proteins expressed with an N-terminal His-6 tag. The full-length coding regions were inserted as NdeI/SacI fragments into the NdeI/SacI site of pET28a (Stratagene, La Jolla, CA) and introduced into Escherichia coli BL21 Codon Plus cells (Novagen, Madison, WI). Following a 3-h induction of log-phase cultures by the addition of 1 mm isopropyl-β-d-thiogalactoside, soluble ATG5 and ATG8a proteins were purified on a NiNTA column (QIAGEN Sciences, Germantown, MD). For ATG5, the protein was further purified by SDS-PAGE and the gel fragments were injected directly. For ATG8a, the His-6 tag was removed by thrombin cleavage, and the protein was further purified by a second pass over the NiNTA column. Antibodies against the large subunit of spinach Rubisco and the CH-42 subunit of pea magnesium-protoporphyrin chelatase (Guo et al., 1998) were provided by Drs. Archie Portis and Tanya Falbel, respectively. Antibody against the F1 subunit of the mitochondrial ATPase was as described (Luethy et al., 1993). The anti-PBA1 and ATG7 antibodies were from Doelling et al. (2002) and Smalle et al. (2002).
DNA/RNA Isolation, RT-PCR, and Analysis of mRNA Abundance
Genomic DNA was isolated from 10-d -old seedlings, and PCR amplified for 35 cycles with Ex-Taq polymerase (TaKaRa, Shiga, Japan) and the ATG5 5′ gene-specific primer ACGTTAGCCACCAACAGATTAAGCAGTGT (primer 1; Fig. 3), the ATG5 3′ gene-specific primer CAATTCACAGATGGATTGTAACTGCAGAG (primer 2; Fig. 3), the LB T-DNA primer, or primers specific to the 35S:ATG5 transgene (GGAGAGGTACGTATTTTTACAA and TCACCTTTGAGGAGCTTT).
RNA was isolated from liquid-grown and soil-grown plants using Trizol reagent (Invitrogen, Carlsbad, CA). RNA for RT-PCR was treated with DNase RQI (Promega, Madison, WI) prior to the synthesis of first-strand cDNA by Moloney murine leukemia virus-reverse transcriptase (Promega). The first-strand synthesis primers were the ATG5 gene-specific primers TCACCTTTGAGGAGCTTT or CACAAAGGAGATCGAAAAGAACACCTGTTG (primers 4 and 5, respectively; Fig. 3), or the PAE2 3′ gene-specific primer CTCAACTCGATAAAATCCATTATCTG (control; Fig. 3). RT-PCR included 35 cycles with Ex-Taq polymerase (TaKaRa), the first-strand synthesis primer, and either the ATG5 5′ gene-specific primer ATGGCGAAGGAAGCGGTCA (primer 3; Fig. 3) or the PAE2 5′ gene-specific primer CTGACATTGAGGTTTATCTCAGATCG (control; Fig. 3).
For RNA gel-blot analysis, total RNA was isolated according to Smalle et al. (2002). 32P-labeled riboprobes were synthesized with T7, SP6, or T3 RNA polymerase using the linearized pGEMT (Promega) or pBluescript (Stratagene) cDNA constructions and the Riboprobe system (Promega). The CAB, CDC2a, SEN1, and ATG8a probes were from Doelling et al. (2002). Membranes were hybridized overnight at 68°C and washed according to Smalle et al. (2002) prior to autoradiography.
Abundance of individual ATG8 mRNAs was quantified using fluorescence-based real-time RT-PCR. DNase-treated mRNA (1 μg) from soil-grown tissue was used for first-strand cDNA synthesis with oligo(dT)18 and Moloney murine leukemia virus-reverse transcriptase. PCR reactions were performed according to manufacturer's recommendations, using an iCycler iQ Multi-Color Real-Time PCR detection system (Bio-Rad, Hercules, CA) and iQ SYBR Green Supermix (Bio-Rad). Primer pairs were engineered to generate approximately the same size PCR product (approximately 300 bp) and spanned an intron to ensure amplification from cDNA versus genomic DNA. Amplification conditions were 3 min at 95°C followed by 47 cycles of PCR (denaturing, 95°C for 10 s; annealing, 63°C for 30 s; extension, 72°C for 30 s). Fold increases in RNA levels were obtained using the CT method (Giulietti et al., 2001). ΔCT values of the samples were determined by subtracting the CT values for the target gene in dark-treated tissue from the CT values for the target gene in tissue before the dark treatment for the same background (Col-0 or atg5-1). To correct for sample variations, the relative mRNA abundances were normalized against that for the Histone H2A transcript. Triplicate values for fold increases were reported with se. Gene-specific primers used are as follows: ATG8a (At4g21980), TTCAAGATCTCTAACCCTCTCGAGGCA and AGACATCAATGCAGCAGTTGGAGGCAA; ATG8b (At4g04620), TCCTCTGGAGATGAGAATGGCTGAGTCTA and TAGAAAGATCCACCAAATGTGTTCTCTCC; ATG8c (At1g62040), TTCCAGTGATCGTAGAGAGAGCTGAAAGA and GTTCTCTCCACTGTAAGTCATGTAGAGAA; ATG8d (At2g05630), CTCTTGAAAAGAGACAAGCTGAAGCAG and ACCACTGTAACTCATGTATAGAAACCCGT; ATG8e (At2g45170), GCATCTTTAAGATGGACGACGATTTCGAA and ATGTGTTCTCGCCACTGTAAGTGATGTAA; ATG8f (At4g16520), GAATGGCAAAAAGCTCGTTCAAGCAAGAG and CATCATCCTTTTTCTCTTCGTACACAGAA; ATG8g (At3g60640), ATTTCGAGAAGAGGAAAGCTGAGGCTTTA and GTCTTCTTCCTTATTCTCATCGTAAATGG; ATG8h (At3g06420), AGTCTTTCAAGGATCAATTCTCCTCTGAT and AAAGTATTGTAGAGAGAGTCCATGCGACT; ATG8i (At3g15580), TGAAATCGTTCAAGGAACAATACACGTTG and TAAGATTCGTAGACCGAGTCCATCAGAGC; and H2A (At4g27230), CGATTTTTGAAAGCCGGTAAGTACGCCGA and GCAACTTGCTTAGCTCCTCATCATTCCTC. Sequence analysis of the PCR products confirmed the transcript specificity of each primer pair.
GFP-ATG8 Lines and Fluorescence Confocal Microscopy
The full-length coding region of the ATG8a cDNA was amplified by PCR using primers CTCAATTGATGGCTAAGAGTTCCTTC and TCGGATCCTCATCCAAAAGTGTTCTC designed to introduce a MunI site at the 5′ end and a BamHI site at the 3′ end, respectively. The resulting product was digested with these two enzymes and inserted downstream of the CaMV 35S promoter and GFP coding region into the binary vector pEGAD (Cutler et al., 2000), which was digested with EcoRI and BamHI. The vector was introduced into A. tumefaciens strain GV3101 and then transformed into wild-type Col-0 and homozygous atg7-1 plants by the floral dip method (Clough and Bent, 1998). The 35S:GFP-ATG8a transgene was followed by Basta resistance. T2 plants homozygous for the atg7-1 mutation were confirmed by PCR. T3 seeds resulting from self-fertilization were tested for GFP expression using a fluorescence dissecting microscope and by immunoblot analysis with anti-GFP antibodies. Antibodies against GFP were produced in rabbits (Polyclonal Antibody Service) using recombinant protein as described by Davis and Vierstra (1998).
Confocal fluorescence microscopy was conducted using a Bio-Rad MCR1024 laser scanning confocal microscope at 600× magnification (Richmond, CA). Excitation was at 480 nm and optical sections were collected with a GFP filter. Images were processed with NIH ImageJ (http://rsb.info.nih.gov/ij/).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers BAB10516 (AtATG5), AP004084 (OsATG5), NP_015176 (ScATG5), AAL39741 (DmATG5), and AAH02699 (HsATG5).
Acknowledgments
We thank Drs. Archie Portis, Tanya Falbel, and Thomas Elthon for the supply of various antibodies; Dr. Dave Ehrhardt for the pEGAD vector; Drs. Sean Carrol and Steve Paddock for help with the confocal microscopy; Dr. Patrick Krysan and Maria Cristina Suarez-Rodriguez for help with real-time RT-PCR; Dr. Xing-Wang Deng and Ligeng Ma for access to their microarray expression data in advance of publication; Dr. Jan Smalle for technical assistance; and Dr. Brian Downes and Joseph Walker for helpful discussions.
This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (grant no. 2002–01452 to R.D.V.), by the National Institutes of Health (Postdoctoral Fellowship to J.H.D.), by a Thailand Predoctoral Fellowship (to A.S.), and by the Wisconsin Alumni Research Foundation and Louis and Elsa Thomsen Wisconsin Distinguished Predoctoral Fellowships (to A.R.T.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.060673.
References
- Brouquisse R, Gaudillere JP, Raymond P (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol 117: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97: 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD (1998) Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol Biol 36: 521–528 [DOI] [PubMed] [Google Scholar]
- Doelling JH, Walker JM, Friedman EM, Thompson AR, Vierstra RD (2002) The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol Chem 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Drose S, Bindseil KU, Bowmama EJ, Siebers A, Zeeck A, Altendorf K (1993) Inhibitory effect of modified bafilomycins and concanamycins and P- and V-type adenosinetriphosphatases. Biochemistry 32: 3902–3906 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Hoffman NE, Ko K, Scolnik PA, Cashmore AR (1988) A light-entrained circadian clock controls transcription of several plant genes. EMBO J 7: 3635–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C (2001) An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25: 386–401 [DOI] [PubMed] [Google Scholar]
- Guo R, Luo M, Weinstein JD (1998) Magnesium-chelatase from developing pea leaves: characterization of a soluble extract from chloroplasts and resolution into three required protein fractions. Plant Physiol 116: 605–615 [PMC free article] [Google Scholar]
- Hammond EM, Brunet CL, Johnson GD, Parkhill J, Milner AE, Brady G, Gregory CD, Grand RJ (1998) Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett 425: 391–395 [DOI] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y (2002) Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Nebenfuhr A (2004) Dynamic rearrangement of transvacuolar strands in BY-2 cells imply a role of myosin in remodeling the plant actin cytoskeleton. Protoplasma 224: 201–210 [DOI] [PubMed] [Google Scholar]
- Huang WP, Klionsky DJ (2002) Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct 27: 409–420 [DOI] [PubMed] [Google Scholar]
- Kametaka S, Matsuura A, Wada Y, Ohsumi Y (1996) Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 178: 139–143 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2004) Autophagy. Eurekah.com/Landes Bioscience, Georgetown, TX
- Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, et al (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Luethy MH, Horak A, Elthon TE (1993) Monoclonal antibodies to the α- and β-subunits of the plant mitochondrial F1-ATPase. Plant Physiol 101: 931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Sun N, Liu X, Jiao Y, Zhao H, Deng X-W (2005) Organ-specific expression of Arabidopsis genome during development. Plant Physiol 138: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 278: 3671–3678 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Higuchi T, Maeshima M, Nakamura K (1997) A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt P, Schurr U, Klein D, Krapp A, Stitt M (1998) Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the NIA transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta 207: 27–41 [DOI] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B (2003) Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391 [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci USA 99: 6422–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 152: 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyasu Y, Klionsky DJ (2004) Autophagy in plants. In DJ Klionsky, ed, Autophagy. Eurekah.com/Landes Bioscience, Georgetown, TX, pp 208–215
- Moriyasu Y, Ohsumi Y (1996) Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol 111: 1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Kato T, Tabata S, Seki M, Kobayashi M, Shinozaki K, Moriyasu Y (2004) Disposal of chloroplasts with abnormal function into the vacuole in Arabidopsis thaliana cotyledon cells. Protoplasma 223: 229–232 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH (2003) Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 278: 17636–17645 [DOI] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F (2005) Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell doi/10.1042/BC20040516 [DOI] [PubMed]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD (2002) Cytokinin growth responses in Arabidopsis involve the 26S proteasome subunit RPN12. Plant Cell 14: 17–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15: 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y (2004) 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 45: 265–274 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi S, Watanabe M, Nishimura M, Hara-Nishimura I (2003) Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E (2002) Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 277: 13739–13744 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Vierstra RD (2005) Autophagic recycling: lessons from yeast help define the process in plants. Curr Opin Plant Biol 8: 165–173 [DOI] [PubMed] [Google Scholar]
- Thumm M, Kadowaki T (2001) The loss of Drosophila APG4/AUT2 function modifies the phenotypes of cut and Notch signaling pathway mutants. Mol Genet Genomics 266: 657–663 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32: 275–302 [DOI] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]