Abstract
Seed production generally requires the mating of opposite sex gametes. Apomixis, an asexual mode of reproduction, avoids both meiotic reduction and egg fertilization. The essential feature of apomixis is that an embryo is formed autonomously by parthenogenesis from an unreduced egg of an embryo sac generated through apomeiosis. If apomixis were well understood and harnessed, it could be exploited to indefinitely propagate superior hybrids or specific genotypes bearing complex gene sets. A more profound knowledge of the mechanisms that regulate reproductive events would contribute fundamentally to understanding the genetic control of the apomictic pathway. In Poa pratensis, we isolated and characterized two genes, PpSERK (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE) and APOSTART. These full-length genes were recovered by rapid amplification of cDNA ends and their temporal and spatial expression patterns were assessed by reverse transcription-polymerase chain reaction and in situ hybridization, respectively. The expression of PpSERK and APOSTART differed in apomictic and sexual genotypes. Their putative role in cell-signaling transduction cascades and trafficking events required during sporogenesis, gametogenesis, and embryogenesis in plants is reported and discussed. We propose that, in nucellar cells of apomictic genotypes, PpSERK is the switch that channels embryo sac development and that it may also redirect signaling gene products to compartments other than their typical ones. The involvement of APOSTART in meiosis and programmed cell death is also discussed.
Sex is the queen of problems in evolutionary biology. Perhaps no other natural phenomenon has aroused so much interest; certainly none has caused as much confusion (Bell, 1982). One of the major questions is as follows: What is the meaning and the rationale of sexuality, which leads to its maintenance under natural selection in biological populations? The easiest answer is that sex, through genetic recombination at meiosis and genomic fusion by fertilization, allows genotype rearrangement, diversification, and adaptation. But even the most dogmatic interpretation of the meaning of sex can be challenged. For example, the existence of apomixis, which is reproduction through seeds but without sex, can be an argument that questions the necessity of sex. In apomictic plants, the embryo is fertilization independent and develops inside an embryo sac formed from either a megaspore mother cell (MMC; diplospory) or a somatic cell of the nucellus (apospory) and retains the maternal genotype (Savidan, 2000; Grimanelli et al., 2001; Spillane et al., 2001). Fertilization may be required for endosperm formation, which can either retain the 2:1 maternal:paternal genome ratio or completely lack a paternal genome equivalent (Nogler, 1984; Spillane et al., 2001; Tucker et al., 2003).
Defining the nature and genetic control of apomixis may be crucial for both understanding the trait itself and better illustrating the meaning of sexuality. Although many years of descriptive studies have provided a solid documentation of the types of apomictic processes that occur in a wide variety of plant species, molecular studies aimed at understanding the basis of apomixis have failed to shed more than a dim and wavering light on its central mystery, partly because the majority of apomicts do not constitute agriculturally important crops and, with few exceptions (e.g. Tripsacum and maize [Zea mays]), do not have agriculturally important relatives (Bicknell and Koltunow, 2004). Furthermore, the polyploidy of most apomicts hinders genetic mapping studies and the building up of populations for reverse genetics. Even though apomixis may be variously influenced by environmental factors (Mazzucato et al., 1996), at least for facultative apomicts, it is generally accepted that apomictic behavior is under strong genetic control (Nogler, 1984; Savidan, 1990; Asker and Jerling, 1992; Savidan, 2000). An early theory regarding genetic control of apomixis proposed that the trait is regulated by a delicate gene balance (Müntzing, 1940) of recessive genes and that this might be disturbed after crosses. Today, basic regulation is usually considered to depend on a few dominant or codominant genes (Asker and Jerling, 1992; Koltunow et al., 1995), which allow a somatic nucellar cell to form an embryo sac without meiosis and an embryo to develop from an egg cell without fertilization. Once apomictic genes initiate nucellar embryo development and the initial cell forms and divides, the genes controlling embryo cell formation, structure, and embryo pattern formation are probably the same as those required for sexual embryo development. Whether the product of apomictic genes are proteins not produced in sexually reproducing plants (gain of function) or proteins that normally function to initiate events in sexual reproduction, but have an altered activity or spatial and temporal distribution during development (loss of function), is still not well understood. Today, a number of researchers support the hypothesis that zygotic embryogenesis and apomictic parthenogenesis follow similar pathways during embryo and seed production (Albertini et al., 2004; Bicknell and Koltunow, 2004). Specific genes are activated, modulated, or silenced in the primary steps of plant reproduction to ensure that functioning embryo sacs develop from meiotic spores and/or apomeiotic cells. As additional genes may be specifically or differentially expressed in sexually and apomictically reproducing plants and operate during embryo development, we would be better equipped to understand apomixis if the genes responsible for controlling the specific and differential expression in embryo sac and embryo formation were to be detected.
Chaudhury and Peacock (1993) hypothesized that genes isolated in model species such as Arabidopsis (Arabidopsis thaliana) would be important for the study of apomixis. Analysis of meiotic mutations resembling the apomictic process in Arabidopsis has led to the isolation of a number of genes involved in early male and female sporogenesis (SPL, NZZ; Schiefthaler et al., 1999; Yang et al., 1999), in the acquisition of embryogenic competence from somatic cells (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE [SERK]; Schmidt et al., 1997; Hecht et al., 2001) or spontaneous induction of embryo production when overexpressed (LEC1, LEC2) or repressed (PKL; Ogas et al., 1999). Endosperm formation is required for viable seed production in both sexually and apomictically reproducing species. While endosperm development in sexual plants requires fertilization, in apomicts it may proceed autonomously or require fertilization (pseudogamy), depending on the species. Three genes, FIS1 (or MEA), FIS2, and FIS3 (or FIE), whose proteins repress endosperm development in sexual plants in the absence of fertilization, have been identified (Ohad et al., 1996; Grossniklaus and Schneitz, 1998; Kiyosue et al., 1999; Luo et al., 1999; Vielle-Calzada et al., 1999). Mutations in any of the three genes allow partial endosperm development in the absence of fertilization. Mutations that block expression of FIS-class genes, or their downstream targets, might, therefore, contribute to autonomous endosperm development in apomictic plants. Analysis of genes differentially expressed in apomictically and sexually reproducing genotypes should, therefore, reveal key differences in their gene expression programs (Bicknell and Koltunow, 2004).
Comparative gene expression studies have been carried out during the early stages of apomictic and sexual embryo sac development in Panicum maximum (Chen et al., 1999), Brachiaria sp. (Leblanc et al., 1997; Dusi, 2001; Rodrigues et al., 2003), Pennisetum sp. (Vielle-Calzada et al., 1996; Jessup et al., 2003), Paspalum sp. (Pessino et al., 2001), as well as in apomeiotic mutants of Medicago falcata (Barcaccia et al., 2001). However, most of them were based on subtractive hybridization techniques and isolated only a few genes to which, disappointingly, no clear function could be assigned. Hybridization-based studies, even if negative in context, add support to the proposal that sexual and apomictic developmental pathways differ primarily in their ability to regulate common elements (Bicknell and Koltunow, 2004). Tucker et al. (2003) confirmed that apospory and the components of autonomous endosperm development share gene expression and regulatory components with sexual reproduction through the expression patterns of AtSPL:GUS, AtSERK1:GUS, and various AtFIS-class:GUS chimeric constructs in sexual and apomictic Hieracium sp. Their results indicate that aspects of meiosis are avoided because direction onto a mitotic embryo sac formation pathway is combined with fertilization-independent embryo and endosperm development in the aposporic initial cell that differentiates from the nucellus. They also hypothesize that apomixis is manifested by induction of a sexual program deregulated in both time and space that leads to changes in cell fate and the omission of critical steps in the sexual process.
Since natural apomixis does not, on the basis of available information, seem to result from the failure of a single reproductive pathway gene, but rather from an epistatic, possibly silencing, action exerted on the normal sexual reproductive pathway by a set of genes inherited as a unit and evolved in polyploid plants (Ozias-Akins et al., 1998), studying apomixis in polyploid species with a relatively large genome may be an obligate strategy.
The high polyploidy and contrasting mode of reproduction of Poa pratensis should make it a model species for investigating apomixis. Sexually, P. pratensis reproduces through out-crossing or selfing, whereas, apomictically, it is a pseudogamous aposporic parthenogenetic species. The combination of a pollen recognition system and the aposporic nature of apomixis confers a strong ability to hybridize with, and retain, alien genomes (Wedin and Huff, 1996) and so determine high polyploidy levels and unusual chromosome numbers (x = 7, 2n = 28–147; Speckmann and van Dijk, 1972). Despite the difficulties mapping such complex systems might present, their highly flexible reproductive mode should permit genotypes recombinant for features of apomixis to be isolated (Matzk et al., 2005). Such recombinants should prove of immense value in gene expression studies in accurately assigning a specific gene to one of its apomictic features rather than to the apomictic process as a whole. Overall results in P. pratensis indicate that two distinct genetic factors, or systems, control apospory and parthenogenesis, which may be developmentally uncoupled (Albertini et al., 2001). Recombinant genotypes (aposporic but nonparthenogenetic) were retrieved within a segregating population of P. pratensis by Albertini et al. (2001).
We (Albertini et al., 2004) previously used the cDNA-amplified fragment-length polymorphism (AFLP) transcriptional profiling technique to isolate messengers from developmentally staged inflorescences. More than 2,000 transcript-derived fragments were visualized and 179 of them were differentially expressed in apomictic and sexual genotypes. A major finding was that most of the genes expressed in florets displayed similar patterns in sexual and apomictic pathways. Of the approximately 8% of mRNAs that were differentially expressed in apomictic and sexual genotypes, the vast majority were attributable to genes whose temporal expression differed during flowering. As only 1.6% sexual or apomictic genotype-specific mRNAs were found, the developmental program seems to be highly conserved during zygotic embryogenesis and apomeiotic parthenogenesis (Albertini et al., 2004).
We now report the isolation and characterization of two genes, starting from the expressed sequence tag (EST) clones previously isolated in P. pratensis. Their genomic organization and characterization through temporal and spatial expression analysis of transcripts in reproductive tissues are reported. The putative involvement of these two genes in the process of ovule development and somatic embryo induction is also reported and discussed.
RESULTS
Cloning of Differentially Expressed Genes in Apomictically and Sexually Reproducing P. pratensis Genotypes
A differential display of mRNAs that combines cDNA-AFLP and developmentally staged inflorescences (Fig. 1) allowed Albertini et al. (2004) to select 2,248 ESTs, of which about 60% were specific to floral organs and/or involved in megasporogenesis, embryo, and seed development. In particular, EST number 6 was 181 nucleotides in length and showed highly significant amino acid sequence similarity (E-value 1e-35, 93% identity, 95% positivity) with the putative SERK1 gene of rice (Oryza sativa; accession no. BAD27594) and as much (E-value 5e-30, 83% identity, 93% positivity) with a SERK family member (accession no. AAL91629) located on chromosome 3 of Arabidopsis (locus At3g25560). Since SERK is believed to play a fundamental role not only in somatic embryogenesis but also in apomixis, this cDNA fragment was chosen for further molecular characterization and was named PpSERK.
Figure 1.
Morphological appearance of inflorescences (top) at the developmental stages collected as verified by cytohistologial investigation (bottom): premeiotic (MMC formed), meiotic (dyads or triads), postmeiotic (functional and degenerating megaspores for sexual genotypes and aposporous initials and degenerating megaspores for apomictic individuals), and anthesis (one or more embryo sacs formed). oi, Outer integument; ow, ovary wall; ae, aposporous embryo sac; ai, aposporous initial after the first mitotic division; c, calaza; dm, degenerated megaspore; dy, dyad; es, sexual embryo sac; fm, functional megaspore; ii, inner integument; mi, micropyle; mmc, megaspore mother cell; pn, polar nuclei.
Clone number 64 was 200 nucleotides in length and showed high similarity (E-value 9e-29, 83% identity) with an EST (accession no. BM084049) isolated from a pistil-specific cDNA library of apomictic Pennisetum ciliare (Jessup et al., 2003). It also showed high similarity (E-value 2e-44, 89% identity, 93% positivity) with the lipid-binding START domain-containing protein from Arabidopsis (accession no. NP193639). This P. pratensis cDNA fragment was named APOSTART and was chosen for further molecular characterization since its homolog of Arabidopsis (locus At4g19040) is located next to an unknown gene (locus At4g19050) that is strongly similar to a MOB1-like gene shown to be putatively involved in 2n egg formation in an apomeiotic mutant of M. falcata (Barcaccia et al., 2001). In Arabidopsis, both APOSTART-like and MOB1-like genes are also located on chromosome 5 in inverted position (loci At5g45560 and At5g45550, respectively).
Genomic Structure and Organization of PpSERK-Like and APOSTART Members
Forward and reverse primers were designed for both 5′- and 3′-RACE. Several RACE experiments were required to obtain the entire 5′-end of both PpSERK and APOSTART. RACE identified two members of PpSERK and two of APOSTART. On the basis of full lengths, amplification of cDNA and genomic DNA samples with PpSERK1, PpSERK2, APOSTART1, and APOSTART2 specific primers enabled us to perform the end-to-end PCR and obtain complete cDNA and DNA clones of each allele. Allele specificity was confirmed in replicated experiments by directly sequencing the amplified products.
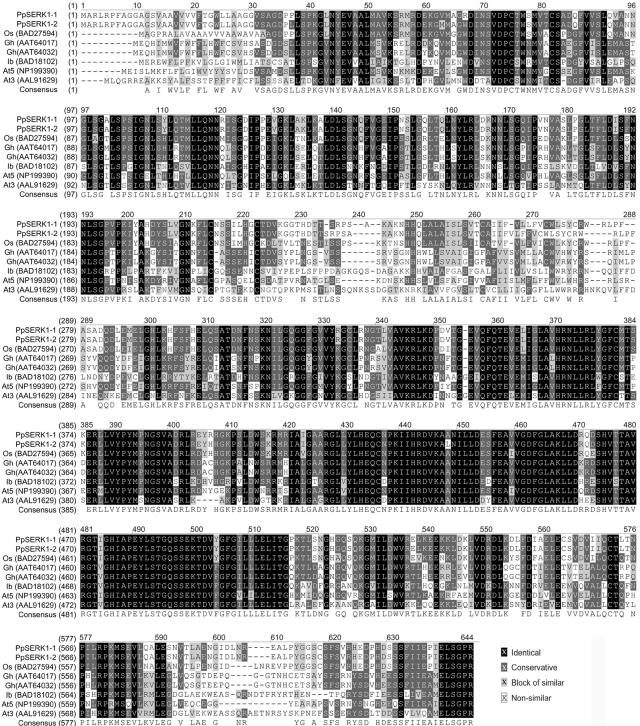
PpSERK1 mRNA sequence is 2,206 nucleotides in length and contains an open reading frame of 1,890 nucleotides, which encodes for a protein of 629 amino acids, and a 3′-untranslated region (UTR) of 316 nucleotides (accession no. AJ841698). Genomic sequencing of PpSERK1 showed that it is 5,188 nucleotides in length and contains 11 exons and 10 introns with a conserved exon/intron structure (supplemental data) identified in other SERK genes (Nolan et al., 2003). PpSERK2 is 2,246 nucleotides in length. It contains a 1,890-nucleotide coding sequence (CDS; accession no. AJ841697), which encodes for a putative protein of 629 amino acids, and a 3′-UTR region of 365 nucleotides. The genomic clone is of 5,290 nucleotides and contains 11 exons and 10 introns as well as PpSERK1.
PpSERK1 and PpSERK2 amino acid sequence comparison performed with Vector AlignX software resulted in 97.0% identity and 98.1% positivity. In particular, 12 residues with different, and seven with similar, properties were scored. The overall nucleotide alignment scored 92.1% identity. The CDSs differed for 49 residues (97.4% identity), whereas the 3′-UTRs showed only 66.1% identity due not only to single residues but also to three indel regions.
The final length of the genomic clone for APOSTART1 was 5,475 nucleotides and its sequence alignment with the relative full-length cDNA disclosed 21 introns (accession no. AJ786392). The 22 exons of APOSTART1 gave rise to a 2,187-nucleotide CDS encoding for a 728-amino acid protein (accession no. AJ786392). The APOSTART2 genomic clone was 5,441 nucleotides and, like APOSTART1, contained 21 introns and 22 exons for a 2,187-nucleotide CDS codifying for a 728-amino acid protein (accession no. AJ786393). The intron/exon structure showed to be conserved between species when compared with Arabidopsis and rice (see supplemental data).
APOSTART1 and APOSTART2 amino acid sequence alignment resulted in 98.2% identity and 98.5% positivity. In particular, 10 residues with different, and three with similar, properties were scored along the 728-amino acid alignment.
Bioinformatic Characterization of PpSERK and APOSTART Members
Bioinformatic characterization of isolated cDNAs was carried out using the predicted proteins obtained from the full-length sequences. Using PpSERK as query enabled SERK-like proteins to be detected not only in Arabidopsis, rice, Medicago truncatula, and maize, but also in other less-known plant genomes like Daucus carota, Citrus unshiu, Ipomoea batata, and Gossypium hirsutum. In particular, PpSERK1 protein alignments showed it was very similar (E-value 0.0, 88% identity, 93% positivity) to the putative SERK1 protein from rice (accession no. BAD27594). It also showed 83% identity and 77% positivity (E-value of 0.0) with the Leu-rich repeat (LRR) transmembrane protein kinase of Arabidopsis (accession no. NP199390). Moreover, amino acid identity was 64% with GhSERK1-like, 46.1% with ZmSERK1, 45.4% with MtSERK1, 45.3% with AtSERK1, and 40.4% with DcSERK (accession nos. AAT64017, CAC37638, AAN64293, CAB42254, and AAB61708, respectively). It is worth noting that up to seven structurally distinct members were found in Arabidopsis. Two completely differentiated members were also found in rice. Therefore, plant genomes appear to contain multiple types of SERK genes. BLAST analysis revealed high similarity (E-value 0.0, 88% identity, 93% positivity) between PpSERK2 and the putative SERK1 protein from rice (accession no. BAD27594). It also showed 53% identity and 70% positivity (E-value of e-180) with the putative SERK located on chromosome 3 (locus At3g25560) of Arabidopsis (accession no. AAL91629). The alignment between PpSERK2 and SERK1-like proteins from other species resulted in values comparable to those obtained for PpSERK1.
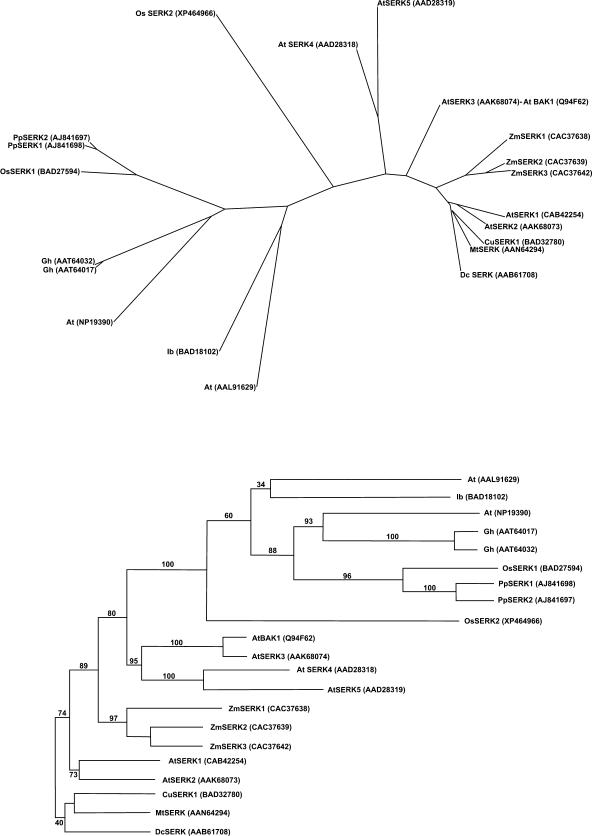
Multiple sequence alignments of a plant SERK protein subset were used to perform a phylogenetic analysis and obtain the consensus tree from the substitution matrix (Fig. 2). Differences in amino acid composition were consistent, mainly due to single substitutions and multiple residue gaps. Ordination analysis of both characterized SERK and SERK-like proteins according to their sequence similarity distinguished two main subgroups for plants. Figure 3 shows the amino acid alignment of PpSERK-like proteins mostly similar to PpSERK1 and PpSERK2.
Figure 2.
Homology dendrogram and phylogenetic tree of both characterized SERK and SERK-like proteins. PpSERK clustered together with SERK1 from rice and other SERK-like proteins, which, in addition to the characterized SERK proteins, included uncharacterized Arabidopsis members.
Figure 3.
Multiple alignment of amino acid sequences deduced from plant SERK-like genes recorded in public genomic databases mostly similar to PpSERK along with the consensus sequence. The SERK-like members of P. pratensis (PpSERK) share between 97.5% and 95.5% amino acid sequence similarity (and between 95.5 and 50.0% amino acid sequence identity) with proteins deduced from O. sativa (Os), G. hirsutum (Gh), I. batatas (Ib), and A. thaliana (At).
Alignment of both PpSERK1 and PpSERK2 revealed the LRR domain (E-value 1e-07, score 52.3 bits, 25.6% aligned) and the Tyr kinase, catalytic (TyrKc) domain (E-value 9e-41, score 162 bits, 74.7% aligned). LRR domain-containing proteins are short sequences involved in protein-protein interactions, but the function of the LRR domain is still unknown, whereas the Tyr kinase is a phosphotransferase of the Tyr-specific kinase subfamily. Enzymes with TyrKc domains belong to an extensive family of proteins that share a conserved catalytic core common to both Ser/Thr and Tyr protein kinases.
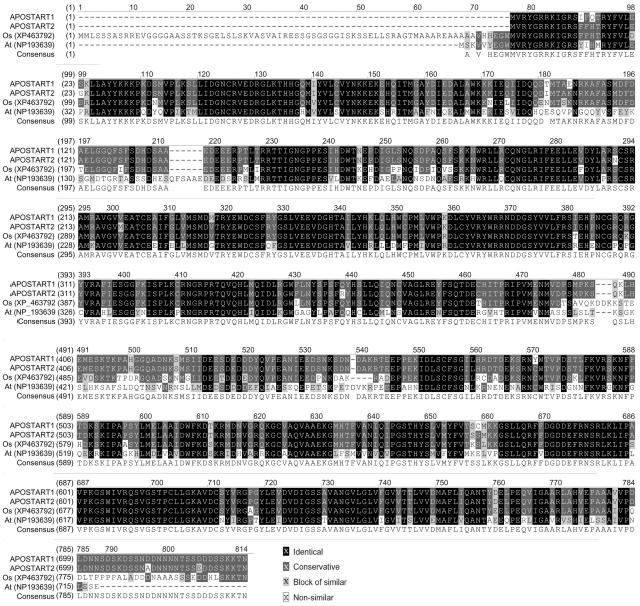
Database searches revealed a strong similarity (E-value 0.0, 86% identity, 90% positivity) between APOSTART1 and an unknown protein from rice (accession no. XP463792). It also showed that APOSTART1 was very similar (E-value 0.0, 65% identity, 79% positivity) to the pleckstrin homology (PH) domain-containing protein/lipid-binding START domain-containing protein (accession no. NP193639) from Arabidopsis (Fig. 4). Alignment revealed the presence of three conserved domains (see supplemental data): PH (E-value 4e-04, score 41.3 bits, 91.3% aligned), START (E-value 3e-28, score 121 bits, 87.7% aligned), and DUF1336 (E-value 2e-56, score 214 bits, 100% aligned).
Figure 4.
Multiple alignment of amino acid sequences deduced from plant APOSTART-like genes recorded in public genomic databases and APOSTART along with the consensus sequence. The APOSTART members of P. pratensis (APOSTART) share 90.0% and 79.0% amino acid sequence similarity (and 86.0 and 65.0% amino acid sequence identity) with protein deduced from O. sativa (Os) and A. thaliana (At), respectively.
The PH domain is commonly found in eukaryotic signaling proteins. The domain family possesses multiple functions, including the ability to bind inositol phosphates and various proteins. PH domains have been found to possess inserted domains (such as in phospholipase C-γ syntrophins) and to be inserted within other domains. START domains are 200 to 210 amino acids in length and occur in proteins involved in lipid transport (phosphatidylcholine) and metabolism, signal transduction, and transcriptional regulation. The most striking feature of the START domain structure is a predominantly hydrophobic tunnel that extends for nearly the entire length of the protein and is used for binding a single molecule of large lipophilic compounds, for example, cholesterol. The DUF1336 domain is the C terminus of approximately 250 residues in a number of hypothetical plant proteins of unknown function.
Although APOSTART2 and APOSTART1 are the same length, BLAST analysis revealed only two domains for APOSTART2: START (E-value 3e-27, score 118 bits, 87.7% aligned) and DUF1336 (E-value 1e-56, score 215 bits, 100% aligned). Even though the best amino acid alignment for APOSTART2 protein was detected in the same proteins of APOSTART1 (E-value 4e-74, 28% identity, 46% similarity), APOSTART2 also shared features with both an unknown protein (locus At5g35180) of Arabidopsis (accession no. BAA98203; E-value 1e-82, 31% identity, 48% similarity) and with an unknown protein of rice (accession no. BAD09877). This is important because, like APOSTART2, both the Arabidopsis and rice proteins (accessions BAA98203 and BAD09877) lack the PH domain.
PSORT (version 6.4) analysis for plant proteins predicted the putative localization of both APOSTART1 (87.4% of probability) and APOSTART2 (86.6% probability) to the inner membrane of the mitochondrion.
P. pratensis Contains at Least Eight Members of SERK and Nine Members of APOSTART Genes
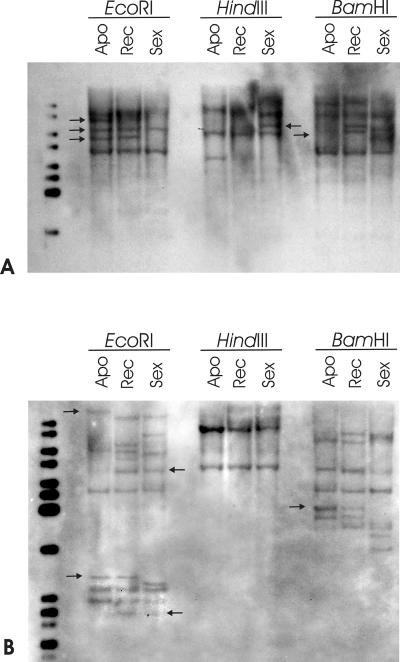
Southern-blot hybridization analysis was performed to investigate the genomic organization of PpSERK and APOSTART. Total genomic DNAs were extracted from apomictic, sexual, and recombinant genotypes and digested with EcoRI, HindIII, and BamHI restriction enzymes. A 250-bp-long probe was obtained from genomic DNA amplification with specific primers corresponding to the C-terminal domain of PpSERK. The hybridization on genomic DNA digested with EcoRI resulted in six bands for the apomictic, six for the recombinant, and four for the sexual genotypes (Fig. 5A). Five hybridization products were found for genomic DNA of apomictic, four for the recombinant, and six for the sexual genotype when digested with HindIII. As high as eight hybridization products were found for the apomictic and recombinant genotypes, whereas only seven fragments were retrieved for sexual genotypes when genomic DNAs were digested with BamHI. Intriguingly, all three enzymes revealed that apomictic, sexual, and recombinant genotypes differed for several polymorphisms (Fig. 5A).
Figure 5.
Southern analysis of PpSERK (A) and APOSTART (B) in apomictic (Apo), recombinant (Rec), and sexual (Sex) P. pratensis genotypes. Genomic DNA was digested with EcoRI, HindIII, and BamHI and probed with a 250-bp SERK-specific and a 280-bp APOSTART-specific probe. Band patterns revealed the presence of at least six alleles/members for PpSERK and at least nine alleles/members for APOSTART. Intriguingly several polymorphisms between apomictic, sexual, and recombinant genotypes were detected with all three restriction enzymes for both genes as indicated by arrows.
Specific primers for APOSTART were designed and produced a genomic fragment of 280 bp, which was used as a probe. As shown in Figure 5B, there were several hybridizing products in each digestion. APOSTART hybridization patterns suggest the presence of at least nine alleles/members in apomictic genotypes (HindIII digestion), eight in recombinant genotypes (HindIII digestion), and seven for sexual genotypes (EcoRI and HindIII digestion). As for PpSERK1, all three enzymes revealed that apomictic, sexual, and recombinant genotypes differed (Fig. 5B). The presence of multiple copies of APOSTART and SERK genes in the P. pratensis genome agrees with the finding of multiple members documented in genomes of other species.
PpSERK and APOSTART Are Differentially Expressed in Genotypes with Contrasting Modes of Reproduction
Expression of PpSERK and APOSTART was assayed in genotypes with different modes of reproduction by using allele-specific primers in the reverse transcription (RT)-PCR analyses. Allele specificity was verified by directly sequencing an aliquot of the amplified products of each experiment. Reactions were performed in triplicate on independently isolated and retrotranscribed mRNAs from three apomictic, three sexual, and two recombinant genotypes and differences in quantitative data were tested for significance using ANOVA. The expression of a β-tubulin allele was used as a reference for data standardization. Figure 6 shows the results of only one apomictic, one sexual, and one recombinant genotype since results for each group of genotypes were not statistically different (the mean-square ratios from one-way ANOVA never reached the critical value for significance at P = 0.05; see supplemental data).
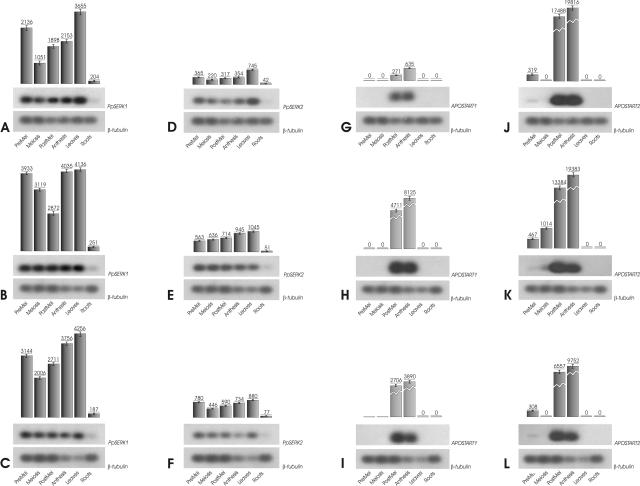
Figure 6.
Expression patterns and level of mRNA encoded by PpSERK1 (A–C), PpSERK2 (D–F), APOSTART1 (G–I), and APOSTART2 (L–N) in apomictic (A, D, G, and L), sexual (B, E, H, and M), and recombinant (C, F, I, and N) genotypes of P. pratensis. The PCR products were blotted to a nylon membrane and hybridized. Hybridized blots were exposed to x-ray films (figures) and also used for the analysis of bound radioactivity. The background-corrected gross counts (absolute gross counts subtracted by the background levels from corresponding lanes) for each PCR product were used for relative mRNA quantification with InstantImager electronic autoradiography software (Packard). Gross counts were performed after 22 cycles of RT-PCR for PpSERK1, 24 cycles for PpSERK2, and 30 cycles for APOSTART1 and APOSTART2. To evaluate the temporal changes in relative levels of mRNAs, the corrected gross counts for the target gene were normalized against those of the housekeeping β-tubulin mRNA. Values are expressed in gross counts. Each experiment was performed in triplicate. The sd is shown for each sample. Figures show the results of only one apomictic, one sexual, and one recombinant genotype since quantitative differences between genotypes with the same mode of reproduction were not statistically significant (see supplemental data).
Both PpSERK1 and PpSERK2 expression were relatively high in leaves and low in roots (Fig. 6, A–F). The expression pattern of two alleles was almost identical in apomictic and recombinant genotypes, but differed markedly in the sexual genotypes. Whereas in sexual genotypes PpSERK1 expression was high during premeiosis and decreased during meiosis and postmeiosis (Fig. 6B), it was modulated in the apomictic and recombinant genotypes (Fig. 6, A and C). In the last two, it was high in premeiosis, dropped during meiosis, then rose again to a level comparable to that recorded by sexual genotypes (Fig. 6, A–C). PpSERK2 expression rose from premeiosis to anthesis (Fig. 6E) in sexual genotypes, while in the apomictic and recombinant genotypes expression in inflorescences was higher in premeiosis than at any other stage (Fig. 6, D and F). In particular, expression dropped to a low level during meiosis, then, although it increased during postmeiosis and anthesis, it did not reach the premeiosis level (Fig. 6, D and F).
APOSTART transcripts were visualized in inflorescences of apomictic, sexual, and recombinant genotypes, but no expression was detected in leaves or root tissues (Fig. 6, G–L). The APOSTART1 profile failed to differentiate between genotypes with different reproductive behavior. However, closer inspection of the quantitative data showed that APOSTART1 expression was higher in sexual and recombinant genotypes (17-fold and 10-fold, respectively) than in apomictic genotypes (Fig. 6, G–I). APOSTART2 was expressed at a low level in sexual genotypes during premeiosis and meiosis, but strongly during postmeiosis and anthesis (Fig. 6K). The expression pathway of the apomictic and recombinant genotypes was similar (Fig. 6, J and L). However, although the gene was expressed at a low level during premeiosis, it was not detected during meiosis, even when the number of amplification cycles was high (Fig. 6, J and L).
In Situ Hybridization Studies
The spatial distribution of the PpSERK and APOSTART transcripts within reproducing organs of apomictic and sexual genotypes of P. pratensis was determined by a high number of independent in situ hybridization (ISH) experiments using digoxigenin (DIG)-labeled probes (see supplemental data). It should, however, be noted that, because of the very high sequence similarities between PpSERK1 and PpSERK2 and between APOSTART1 and APOSTART2, the probes used for ISH experiments could not distinguish alleles/members.
In order to identify the putative role of PpSERK members in apomictic reproduction, analysis of their transcripts was limited to the first stages of ovule development. A partial PpSERK cDNA fragment containing a portion of the LRR and of the TyrKc domains and the entire region comprised between them was used as a PpSERK-specific probe. Results of the ISH experiments are given in Figure 7. Expression of PpSERK was first detected in the whole ovule primordium but not in other flower tissues (data not shown). A strong signal was detected during MMC differentiation in sexual genotypes (Fig. 7A). Interestingly, PpSERK transcripts were not detected in the MMC of apomictic flowers, whereas strong signals were found in some neighboring nucellar cells in all analyzed sections (Fig. 7B; see supplemental data).
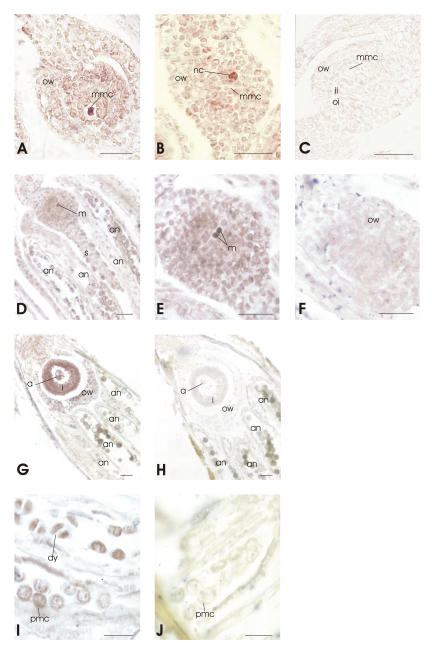
Figure 7.
PpSERK (A–C) and APOSTART (D–J) expression in longitudinal sections of flowers studied by ISH (bars = 40 μm). Sections were probed with DIG-labeled antisense (A, B, D, E, G, and I) or sense (C, F, H, and J) RNAs and viewed under a microscope bright field that gives a purple label. No signals were detected in the sections hybridized with the sense probes. A, Longitudinal section of ovule of a sexual genotype containing the MMC, which presents a strong hybridization signal. B, An apomictic genotype ovule where the hybridization signal was not detected in the MMC, but was present in two neighbor nucellar cells. D and E, Ovule during megasporogenesis, at different magnifications, when a strong signal was present in the megaspores (m), but was not detected in the other portions of the ovule. G, Flower during megagametogenesis; a strong hybridization signal is visualized in antipodal cells of the embryo sac and in nucellar cells. I and J, Anthers during microsporogenesis. Signals were observed in pollen mother cells (pmc) and in the dyads. an, Anthers; dy, dyads; fm, functional megaspore; I, integuments; m, megaspore; mmc, megaspore mother cell; nc, nucellar cell; ii, inner integument; oi, outer integument; ow, ovary wall; pmc, pollen mother cell; pn, polar nuclei; s, stigma.
Because APOSTART is an uncharacterized gene, its cellular expression was investigated in both anthers and ovaries. Ovaries were checked from the ovule primordium stage to mature embryo sac. Hybridization signals were recorded in both male and female reproductive cells for APOSTART (Fig. 7, D, E, G, and I), but no signals were detected in the sense slides (Fig. 7, F, H, and J). In particular, strong signals were detected in the megaspores during meiosis (Fig. 7, D and E) in all nucellar cells, as well as in the embryo sac at anthesis (Fig. 7G) in both apomictic and sexual genotypes, although the expression was higher in apomictic than sexual ones. Moreover, the signal was strong within anthers from pollen mother cells to the tetrad stage (Fig. 7I).
DISCUSSION
The Expression Pattern of PpSERK Is Compatible with Its Having a Role in the Specification of Aposporous Initials
The amino acid sequence of one of the 179 fragments (EST no. 6) was very like that of the SERK gene in Arabidopsis (At3g25560). This gene was named PpSERK. SERK is the only gene known to be implicated in the acquisition of embryogenic competence in plant cells. It was first isolated in carrot (Schmidt et al., 1997), where it was shown to be characteristic of embryogenic cell cultures and somatic embryos. AtSERK1 is expressed during megagametogenesis. Its mRNA level increases at the time of functional megaspore differentiation, persists during embryo sac development, but decreases to a very low level after fertilization (Hecht et al., 2001). SERK is a small gene family with at least five elements in Arabidopsis (Hecht et al., 2001), five in M. truncatula (Nolan et al., 2003), four in Helianthus annuus (Thomas et al., 2004) and three in maize (Baudino et al., 2001).
We have cloned two SERK genes (PpSERK1 and PpSERK2) from the eight putative members derived from Southern hybridization analysis; both were expressed in sexual, apomictic, and recombinant genotypes, but their expression and/or activation/inactivation timing differed.
In apospory, a cell of the nucellus becomes an aposporous initial and then develops into a nonreduced embryo sac which, through parthenogenesis, gives rise to a viable embryo. How and why somatic cells of the ovule change their developmental fate and gain embryogenic potency is not known (Fehér et al., 2003). Whereas the zygote, formed as a consequence of egg cell fertilization, is clearly predetermined to follow the embryogenic cell fate, in other forms of plant embryogenesis, including apomixis, there is a transition phase during which competent and embryogenic cell types are formed (Fehér et al., 2003). The transition phase is clearly very complex, but an understanding of its underlying mechanisms should lead to a deeper appreciation of the developmental strategy adopted by apomictic plants. The changing fate/acquisition of embryogenic competence mainly relies on dedifferentiation, a process whereby existing transcriptional and translational profiles are erased or altered so that the cell can set a new developmental program. Schmidt et al. (1997) hypothesized that, while most elements concerned with the origin and target processes of cell-to-cell communication in early plant embryogenesis are lacking, the SERK gene may be a significant component in the mechanism essential for formation of plant cells destined to become embryos. Tucker et al. (2003) studied the expression of the AtSERK1 homolog gene in Hieracium and found the pattern between sexual and apomictic plants to be conserved.
Our ISH data revealed that PpSERK is expressed in the MMC of sexual genotypes, but not in that of apomictic plants. In contrast, the strong signals detected in nucellar cells neighboring MMC suggest that PpSERK is involved in embryo sac development from nucellar cells.
APOSTART Is Involved in Sporogenesis and Programmed Cell Death
EST number 64 was almost identical to an EST isolated from a pistil-specific cDNA library of apomictic P. ciliare (Jessup et al., 2003). Because EST number 64 showed many features in common with a series of START-domain-containing genes, we decided to characterize it further. One of the highest scores was with an Arabidopsis gene located on chromosome 4 (At4g19040). This gene is closely linked with a gene (At4g19050) whose homolog seems to be involved in the production of 2n eggs in an apomeiotic mutant of M. falcata (Barcaccia et al., 2001). Because of its START domain and putative involvement in apomixis, we named this gene APOSTART. START was named after the discovery of the StAR gene involved in human congenital lipoid adrenal hyperplasia, which is characterized by a marked impairment in the biosynthesis of adrenal and gonadal steroid hormones. The clinical phenotype of this disease includes the onset of profound adrenocortical insufficiency shortly after birth, hyperpigmentation due to increased production of propiomelanocortin, elevated plasma renin activity as a consequence of reduced aldosterone synthesis, and male pseudohermaphroditism resulting from deficient fetal testicular testosterone synthesis (Christenson and Strauss, 2001). The START domain was initially identified in multicellular eukaryotes (plants and animals) as a widespread lipid-binding domain that had a regulatory role in signal transduction (Lakshminarayan et al., 2001). The presence of START domains in such evolutionarily distinct species suggests a conserved mechanism for the interaction of proteins with lipids/sterols (Ponting and Aravind, 1999; Schrick et al., 2004). Due to cell proliferation during evolution, the START domain acquired two distinct functions; the first appears to be generally related to the stress response (Osmark et al., 1998; Gamas et al., 1998), the second to signaling mediated by lipid binding. In the last, the START domain underwent the lineage-specific fusions to other effector domains that are typical of multidomain eukaryotic signaling proteins (Ponting and Aravind, 1999). At least 60 START-containing multidomain proteins have been found in Arabidopsis and rice. Most contain a homeodomain, while a lesser number contain both a PH and a recently identified domain of unknown function (DUF1336). Because PH domains are characterized by their ability to bind phosphoinositides, they influence membrane and/or protein interactions (Lemmon and Ferguson, 2001). A single START-DUF1336 protein of about the same size, but lacking the strong sequence similarity with PH at its amino terminus, is present in both Arabidopsis and rice. Strikingly, the START domain sequence correlates with the type of START protein. It would therefore seem that domain shuffling led to duplication and subsequent evolution of START domains after the initial manifestation of the novel protein structure (Schrick et al., 2004). APOSTART1 and APOSTART2 were expressed exclusively in inflorescences of P. pratensis; none was detected in leaves or roots. Overall expression of APOSTART2 was greater than that of APOSTART1. APOSTART1 expression was also lower in apomictic than in sexual genotypes. We propose that the APOSTART1 gene is involved in sporogenesis and that this is confirmed by its almost complete lack of expression in apomictic genotypes. We have preliminary data in Arabidopsis that indicate that when APOSTART is disrupted by T-DNA insertion, both mega- and microsporogenesis are adversely affected (E. Albertini and L. Colombo, unpublished data). The low APOSTART1 expression recorded in apomictic genotypes may be due to the facultative nature of apomixis in P. pratensis. The average apospory ratio in genotypes used for RT-PCR was 92% (Albertini et al., 2001). The recombinant genotype expression was about 10-fold that of apomictic genotypes, but lower than that of strictly sexual genotypes, which could also be due to the apospory ratio, which in these genotypes was 62% (Albertini et al., 2001).
The finding that APOSTART2 lacks the PH domain agrees with evidence that there is a single START-DUF1336 gene in both Arabidopsis and rice. Despite BLAST showing that APOSTART2 was most similar to the PH-START-DUF1336 gene of Arabidopsis, differences in the N-terminal amino acids typical of the PH domain were the same as those found in the members of Arabidopsis (At5g35180) and rice that lacked PH. Whether the lack of APOSTART2 during meiosis in both apomictic and recombinant genotypes has any effect on apomictic reproduction requires more extensive investigation.
ISH analysis revealed that APOSTART members are expressed during both male and female meiosis in all micro- and megaspores. Moreover, strong signals were recorded up to the mature embryo sac stage.
Overall these data suggest that, in P. pratensis, APOSTART expression may also be related to the programmed cell death that is involved in the nonfunctional megaspore and nucellar cell degeneration events that permit enlargement of maturing embryo sacs (Wu and Cheung, 2000). The microarray data for Arabidopsis support this interpretation. The Genevestigator database (Zimmermann et al., 2004) investigation of the expression of the APOSTART homolog of Arabidopsis (At4g19040) showed that this gene is exclusively up-regulated during senescence. The expression in “programmed cell death: senescence” was almost double that of any other hormone-treated or stress-induced case. This datum recalls the picture seen in Arabidopsis for the MMD1 (MALE MEIOCYTE DEATH1) gene (Yang et al., 2003). The MMD1 gene was shown to be involved in the regulation of gene expression during meiosis and mmd1 mutation triggers cell death in male meiocytes (Yang et al., 2003). Our data could also resemble that seen in nonplant organisms where genes involved in meiosis have been related to apoptosis (Xu et al., 1996; Takanami et al., 1998; Gartner et al., 2000; Roeder and Bailis 2000). The predicted localization of APOSTART in the mitochondrial membrane and its putative role in regulating mitochondrial membrane permeabilization makes APOSTART a prime candidate for apoptosis (Kroemer et al., 1995; Zamzami et al., 1995; Christenson and Strauss, 2001; Schrick et al., 2004).
Role of PpSERK and APOSTART in Apomixis
The two genes isolated in this study, as well as others reported previously (Albertini et al., 2004), are involved in cell-to-cell interaction of both the signaling pathway and hormone stimulation/production. Hecht et al. (2001) proposed that AtSERK is a component of an embryogenesis-signaling pathway. Competent cells may contain an inactive receptor that is activated by the presence of a proper ligand to switch on the embryogenesis program. According to Hecht et al. (2001), the acquisition of embryogenic competence in tissue cultures requires the presence of nonembryogenic cells that produce and secrete molecules that can be perceived by other cells, which, in turn, express their competence and develop into embryos (Pennell et al., 1992; de Jong et al., 1993). It is thought that there is a point at which ovule nucellar cells act as a sink for all sources of nutrients and shunt the regular nutrient traffic from the dividing megaspore MMC to itself. We have previously hypothesized (Albertini et al., 2004) that some genes that are differentially expressed between apomictic and sexual P. pratensis genotypes are differently timed and have diverse localization of transcripts. We propose that PpSERK activation in nucellar cells of apomictic genotypes is the switch that channels embryo sac development and that it could redirect signaling gene products to compartments other than their typical ones. The SERK-mediated signaling pathway may well, at some point, interact with the auxin/hormonal pathway controlled by APOSTART, but PpSERK is certainly not an integral part of it.
A detailed analysis of these genes and their specific ligands/substrates could provide us with a tool that would enable us to crack the code of apomixis.
MATERIALS AND METHODS
Plant Material
A segregating F1 population of 68 plants was produced by crossing a completely sexual clone (S1/1-7, nonaposporic and nonparthenogenetic) with a highly apomictic genotype (RS7-3, aposporic and parthenogenetic). The sexual clone was derived from a cross between two completely sexual genotypes selected from German cultivars (Matzk, 1991; Barcaccia et al., 1998), while the apomictic clone was selected from a natural Italian population (Mazzucato, 1995).
Cytological investigations showed that the chromosome number of parental genotypes was 2n = 36 for S1/1-7 and 2n = 64 for RS7-3. All progeny plants investigated were sired by agamospermous pollen and had a complement of 50 chromosomes (Porceddu et al., 2002). Plants were investigated for their genetic capacity for apospory and parthenogenesis over a period of 2 years. This allowed us to classify F1 genotypes into three classes (Albertini et al., 2001): apomictic (aposporic and parthenogenetic), sexual (nonaposporic and nonparthenogenetic), and recombinant (aposporic but nonparthenogenetic).
RNA Isolation and cDNA Synthesis
Florets of an apomictic, a sexual, and a recombinant genotype of Poa pratensis, were harvested and classified into four developmental stages (premeiosis, meiosis, postmeiosis, and anthesis), according to cytohistological investigations. Nucleic acids were isolated from about 0.5 g of fresh tissue (inflorescences at different developmental stages, leaves, and roots) using the GenElute mammalian total RNA miniprep kit (Sigma, St. Louis), according to the manufacturer's instructions, with some modifications to adapt it to plants. Total RNA was purified from residual genomic DNA by using the DNA-free (Ambion, Austin, TX) and the mRNA poly(A)+ purified by using the GenElute mRNA miniprep kit (Sigma). RT and second-strand synthesis was carried out with 1 μg of mRNA polyA+ and the standard procedure followed (Sambrook and Russell, 2001).
Sequence Data Analysis
Similarities for all cDNA-amplified fragments were searched in the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov) database using the 2.2.9 release with BLASTN (est_others database), BLASTX, and BLASTP (nr database) applications (Altschul et al., 1990, 1997; Altschul and Lipman, 1990) to compare nucleotides, translated sequences, and deduced proteins, respectively. Either the BLOSUM62 or the PAM30 substitution matrices were used for scoring protein alignments. The most significant amino acid sequence homologies were used for multiple sequence alignments with the VECTOR NTI Suite 8 AlignX software (InforMax/Invitrogen, Carlsbad, CA) to highlight conserved and variable regions with respect to the cDNA sequence adopted as query. The substitution matrix obtained from the multiple alignment of a plant SERK-like protein subset was used to perform a phylogenetic analysis using the FastME method (Desper and Gascuel, 2002) and bootstrap as a resampling procedure. The majority-rule consensus tree was generated from 1,000 datasets using the DAMBE program (Xia and Xie, 2001).
Rapid Amplification of cDNA Ends Analysis
Clone-specific primers were used for performing both 5′- and 3′-RACE to obtain the full-length genes. The SMART RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA) was applied to the mRNA poly(A)+ of the stage where the cDNA was scored, according to the manufacturer's instructions. Eight colonies for each RACE experiment were sequenced and full-length cDNA sequences were reconstructed from RACE fragments using VECTOR NTI Suite 8 Contig Express (InforMax).
Cloning of a Full-Length Genomic Gene
Clone-specific primers were used for performing end-to-end amplifications of genomic DNA to obtain the entire transcriptional unit. An 0.7-μL aliquot of PCR-derived products was sticky-end ligated into a pCR4-TOPO vector using the TOPO TA cloning kit for sequencing (Invitrogen). The plasmid DNA was purified from 5 mL of an overnight culture on Luria-Bertani medium of Escherichia coli using the GenElute plasmid miniprep kit (Sigma). After a first confirmation sequence performed using M13 forward and reverse as primers, plasmids were used with the GeneJumper primer insertion kit for sequencing (Invitrogen). Sequences were then performed using the transposon-specific primers included in the kit. Full-length sequences were then obtained using Contig Express software (Invitrogen). Alignment between the full-length cDNAs and genomic clones disclosed the intron/exon structures of the genes.
Southern-Blot Hybridization
For Southern analysis, 12 μg of P. pratensis genomic DNA (isolated with the Sigma GenElute plant genomic DNA kit) were digested with 60 units of EcoRI, HindIII, and BamHI, respectively (New England Biolabs, Beverly, MA). Restriction fragments were resolved by electrophoresis in a 0.8% agarose gel, blotted by capillary transfer, and linked at 80°C for 2 h to a Nitran N nylon membrane (Schleicher & Schull, Keene, NH). About 4 ng of each selected DNA or cDNA probe were labeled by PCR, using specific primers, in the presence of 32P-dCTP. Prehybridization, hybridization, and posthybridization washing of DNA membranes were performed according to Sambrook and Russell (2001).
RT-PCR Analysis
First-strand cDNA was synthesized from 1 μg of P. pratensis total RNA in a 50-μL volume (Sambrook and Russell, 2001). RT-PCR reactions were performed in a total volume of 50 μL containing 1 μL of first-strand cDNA, 1 μm of each primer, 1× PCR buffer, 1.5 mm MgCl2, 0.2 μm dNTPs, and 2 units of Taq DNA polymerase recombinant (Invitrogen). The reaction was denatured at 94°C for 1 min and then subjected to 15 to 30 cycles of 94°C denaturation for 1 min, melting temperature (specific for each primer pair) for 1 min, and 72°C for 1 min, plus a final extension at 72°C for 10 min. PCR products were collected after 18, 20, 22, 24, 26, 28, and 30 cycles to determine the linearity of the PCR. The P. pratensis β-tubulin gene was cloned using degenerate primers designed on the most conserved region of this gene in grass species, sequenced, and then specific primers (5′-GTGGAGTGGATCCCCAACAA-3′ and 5′-AAAGCCTTCCTCCTGAACATGG-3′) were designed. The Ppβ-tubulin gene was used as control in all experiments. PCR products of the Ppβ-tubulin gene were collected after 14, 16, 18, and 20 cycles. Within each experiment and for each gene analyzed, the complete set of samples was processed in parallel in a single PCR using aliquot of the same master mix. RT-PCR experiments were carried out on three apomictic, three sexual, and two recombinant genotypes. Each set of determinations was performed in triplicate. RT-PCR patterns were reported by using the following terminology: up-regulated or down-regulated when the expression of a gene increased or decreased, respectively, from premeiosis to postmeiosis. The term modulated was used when the expression increased from premeiosis to meiosis, then decreased from meiosis to postmeiosis, or vice versa.
Analysis of Amplification Products
The amplified PCR-generated products were directly sequenced to verify the allele specificity of primer pairs used and then separated on 2% agarose gel, blotted by capillary transfer, and linked at 80°C for 2 h to a Nitran N nylon membrane (Schleicher & Schull). The 32P-labeled probes were synthesized by PCR from purified PCR fragments. Filters were hybridized as in Vicient et al., (1999). All blots of the same gene were hybridized together so the probe concentration was identical for all filters. Hybridized filters were washed successively with 2× SSC, 0.1% SDS, 0.1× SSC, and 0.1% SDS at 55°C. The hybridized blots were first exposed with Kodak AR films (Eastman-Kodak, Rochester, NY) and then used for the analysis of bound radioactivity with the InstantImager (Packard, Meriden, CT). The background-corrected gross counts (absolute gross counts subtracted by the background levels from corresponding lanes) for each PCR product were quantified using InstantImager electronic autoradiography software (Packard). To evaluate the temporal changes in relative levels of mRNAs, the corrected gross counts for the target gene were normalized against those of the housekeeping β-tubulin mRNA. sds referred to replicated experiments. Values were expressed in gross counts.
ISH
During inflorescence development, in sexual and apomictic P. pratensis genotypes, we distinguished four different stages (premeiosis, meiosis, postmeiosis, and anthesis). At each stage, single spikelets were collected, fixed in ethanol-formaldehyde-acetic acid, embedded in paraffin, and used for ISH experiments. Tissue preparation and hybridization conditions were the same as described by Angenent et al. (1995).
Sense and antisense probes were obtained by in vitro transcription using cloned PCR-derived fragments of PpSERK1 and APOSTART as templates. In particular, PpSERK1 riboprobes were synthesized from a 1,110-bp fragment obtained using internal primers designed in a way so that the fragment contained small parts of the LRR and of the TyrKc domains and the entire region comprised between them. Moreover, APOSTART riboprobes were obtained from a single cDNA fragment of 1,060 bp, which comprised only a small part of the START and DUF1336 domains and the sequence between the two domains. Both PpSERK and APOSTART riboprobes did not discriminate between alleles due to the very small differences in sequences.
DIG-UTP sense and antisense riboprobes were synthesized by the T3 and T7 RNA polymerase. Transcripts were partially hydrolyzed by incubation at 60°C in 0.2 m Na2CO3/NaHCO3 buffer, pH 10.2, for about 35 min. Immunological detection was performed as described by Cañas et al. (1994).
The genomic DNA and mRNA nucleotide sequences and the deduced amino acid sequences of the isolated PpSERK and APOSTART genes have been recorded. Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AJ841698, AJ841697, AJ786392, and AJ786393.
Supplementary Material
Acknowledgments
We wish to thank Prof. Chris Gehring for providing Genevestigator data, Dr. Stefano Capomaccio for helping to name APOSTART and giving advice on the artwork, Dr. Luigi Russi and Prof. Peggy Ozias-Akins for critical reading of the manuscript, both Dr. Luca Pallottini and Dr. Lorenzo Raggi for their technical help, and Judy Etherington for her invaluable help with the English editing of the manuscript.
This work was supported by the Ministry of University and Research (project “Genetic aspects of seed production: an integrated approach towards the understanding of apomixis”).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062059.
References
- Albertini E, Marconi G, Barcaccia G, Raggi L, Falcinelli M (2004) Isolation of candidate genes in Poa pratensis L. Plant Mol Biol 56: 879–894 [DOI] [PubMed] [Google Scholar]
- Albertini E, Porceddu A, Ferranti F, Reale L, Barcaccia G, Romano B, Falcinelli M (2001) Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sex Plant Reprod 14: 213–217 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Lipman DJ (1990) Protein database searches for multiple alignments. Proc Natl Acad Sci USA 87: 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJM, van Tunen AJ (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7: 1569–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker SE, Jerling L (1992) Apomixis in Plants. CRC Press, London
- Barcaccia G, Mazzucato A, Albertini E, Zethof J, Gerats T, Pezzotti M, Falcinelli M (1998) Inheritance of parthenogenesis in Poa pratensis L.: auxin test and AFLP linkage analysis support monogenic control. Theor Appl Genet 97: 74–82 [Google Scholar]
- Barcaccia G, Varotto S, Meneghetti S, Albertini E, Porceddu A, Parrini P, Lucchin M (2001) Analysis of gene expression during flowering in apomeiotic mutants of Medicago spp: cloning of ESTs and candidate genes for 2n eggs. Sex Plant Reprod 14: 233–238 [DOI] [PubMed] [Google Scholar]
- Baudino S, Hansen S, Brettschneider R, Hecht VF, Dresselhaus T, Lorz H, Dumas C, Rogowsky PM (2001) Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 213: 1–10 [DOI] [PubMed] [Google Scholar]
- Bell G (1982) The Masterpiece of Nature. The Evolution and Genetics of Sexuality. University of California Press, Berkeley and Los Angeles, CA
- Bicknell RA, Koltunow AM (2004) Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16: 228–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas LA, Busscher M, Angenent GC, Beltrán J, van Tunen AJ (1994) Nuclear localization of the petunia MADS box protein FBP1. Plant J 6: 597–604 [Google Scholar]
- Chaudhury AM, Peacock JW (1993) Approaches to isolating apomictic mutants in Arabidopsis thaliana: Prospects and progress. In GS Khush, ed, Apomixis: Exploiting Hybrid Vigor in Rice. International Rice Research Institute, Manila, The Philippines, pp 66–71
- Chen LZ, Miyazaki C, Kojima A, Saito A, Adachi T (1999) Isolation and characterization of a gene expressed during early embryo sac development in apomictic guinea grass (Panicum maximum). J Plant Physiol 154: 55–62 [Google Scholar]
- Christenson LK, Strauss JF (2001) Steroidogenic acute regulatory protein: an update on its regulation and mechanism of action. Arch Med Res 32: 576–586 [DOI] [PubMed] [Google Scholar]
- Desper R, Gascuel O (2002) Fast and accurate phylogeny reconstruction algorithms based on the minimum-evolution principle. J Comput Biol 9: 687–705 [DOI] [PubMed] [Google Scholar]
- Dusi DMA (2001) Apomixis in Brachiaria decumbens Stapf. PhD Thesis, University of Wageningen, The Netherlands
- Fehér A, Pasternak TP, Dudits D (2003) Transition of somatic cells to an embryogenic state. Plant Cell Tissue Organ Cult 74: 201–228 [Google Scholar]
- Gamas P, de Billy F, Truchet G (1998) Symbiosis-specific expression of two Medicago truncatula nodulin genes, MtN1 and MtN13, encoding products homologous to plant defense proteins. Mol Plant Microbe Interact 11: 393–403 [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5: 435–443 [DOI] [PubMed] [Google Scholar]
- Grimanelli D, Leblanc O, Perotti E, Grossniklaus U (2001) Developmental genetics of gametophytic apomixis. Trends Genet 17: 597–604 [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Schneitz K (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9: 227–238 [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803–816 [PMC free article] [PubMed] [Google Scholar]
- Jessup RW, Burson BL, Burow G, Wang YW, Chang C, Li Z, Paterson AH, Hussey MA (2003) Segmental allotetraploidy and allelic interactions in buffelgrass (Pennisetum ciliare (L) Link syn Cenchrus ciliaris L) as revealed by genome mapping. Genome 46: 304–313 [DOI] [PubMed] [Google Scholar]
- de Jong AJ, Schmidt EDL, de Vries SC (1993) Early events in higher-plant embryogenesis. Plant Mol Biol 22: 367–377 [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Ohad N, Yadegari R, Hannon M, Dinneny J, Wells D, Katz A, Margossian L, Harada JJ, Goldberg RB, et al (1999) Control of fertilization-independent endosperm development by the MEDEA polycomb gene Arabidopsis. Proc Natl Acad Sci USA 96: 4186–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow AM, Bicknell RA, Chaudhury AM (1995) Apomixis: molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiol 108: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Petit P, Zamzami N, Vayssiere JL, Mignotte B (1995) The biochemistry of programmed cell death. FASEB J 9: 1277–1287 [DOI] [PubMed] [Google Scholar]
- Lakshminarayan MI, Koonin EV, Aravind L (2001) Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins Struct Funct Genet 43: 134–144 [DOI] [PubMed] [Google Scholar]
- Leblanc O, Armstead I, Pessino S, Ortiz JP, Evans C, doValle C, Hayward MD (1997) Non-radioactive mRNA fingerprinting to visualise gene expression in mature ovaries of Brachiaria hybrids derived from B. brizantha, an apomictic tropical forage. Plant Sci 126: 49–58 [Google Scholar]
- Lemmon MA, Ferguson KM (2001) Molecular determinants in pleckstrin homology domains that allow specific recognition of phosphoinositides. Biochem Soc Trans 29: 377–384 [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM (1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 296–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzk F (1991) New efforts to overcome apomixis in Poa pratensis L. Euphytica 55: 65–72 [Google Scholar]
- Matzk F, Prodanovic S, Baumlein H, Schubert I (2005) The inheritance of apomixis in Poa pratensis confirms a five locus model with differences in gene expressivity and penetrance. Plant Cell 17: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucato A (1995) Italian germplasm of Poa pratensis L. II. Isozyme progeny test to characterize genotype for their mode of reproduction. J Genet Breed 49: 119–126 [Google Scholar]
- Mazzucato A, Veronesi F, Falcinelli M (1996) Evolution and adaptedness in facultatively apomictic grass Poa pratensis L. Euphytica 92: 13–19 [Google Scholar]
- Müntzing A (1940) Further studies on apomixis and sexuality in Poa. Hereditas 26: 115–190 [Google Scholar]
- Nogler GA (1984) Gametophytic apomixis. In BM Johri, ed, Embriology of Angiosperms. Springer-Verlag, New York, pp 475–518
- Nolan KE, Irwanto RR, Rose RJ (2003) Auxin up-regulates MtSERK1 expression in both Medicago truncatula root-forming and embryogenic cultures. Plant Physiol 133: 218–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Margossian L, Hsu YC, Williams C, Repetti P, Fischer RL (1996) A mutation that allows endosperm development without fertilization. Proc Natl Acad Sci USA 93: 5319–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmark P, Boyle B, Brisson N (1998) Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol Biol 38: 1243–1246 [DOI] [PubMed] [Google Scholar]
- Ozias-Akins P, Roche D, Hanna WW (1998) Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc Natl Acad Sci USA 95: 5127–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Scofield GN, Booij H, de Vries SC, Roberts K (1992) Identification of a transitional cell state in the developmental pathway to carrot somatic embryogenesis. J Cell Biol 119: 1371–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessino SC, Espinoza F, Martinez EJ, Ortiz JPA, Valle EM, Quarin CL (2001) Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas 134: 35–42 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1999) START: a lipid-binding domain in StAR, HD-ZIP and signaling proteins. Trends Biochem Sci 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Porceddu A, Albertini E, Barcaccia G, Falistocco E, Falcinelli M (2002) Linkage mapping in apomictic and sexual Kentucky bluegrass (Poa pratensis L.) genotypes using a two way pseudo-testcross strategy based on AFLP and SAMPL markers. Theor Appl Genet 104: 273–280 [DOI] [PubMed] [Google Scholar]
- Rodrigues JCM, Cabral GB, Dusi DMA, de Mello LV, Rigden DJ, Carneiro VTC (2003) Identification of differentially expressed cDNA sequences in ovaries of sexual and apomictic plants of Brachiaria brizantha. Plant Mol Biol 53: 745–757 [DOI] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM (2000) The pachytene checkpoint. Trends Genet 16: 395–403 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Savidan Y (1990) The genetic control of apomixis. Apomixis Newsl 2: 24–26 [Google Scholar]
- Savidan Y (2000) Apomixis: genetics and breeding. In J Janick, ed, Plant Breeding Review, Vol 18. John Wiley & Sons, New York, pp 13–86
- Schiefthaler U, Balasubramanian S, Sieber P, Chevalier D, Wisman E, Schneitz K (1999) Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt ED, Guzzo F, Toonen MA, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049–2062 [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KFX (2004) START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann GJ, van Dijk GE (1972) Chromosome numbers and plant morphology in some ecotypes of Poa pratensis L. Euphytica 21: 171–180 [Google Scholar]
- Spillane C, Steimer A, Grossniklaus U (2001) Apomixis in agriculture: the quest for clonal seeds. Sex Plant Reprod 14: 179–187 [DOI] [PubMed] [Google Scholar]
- Takanami T, Sato S, Ishihara T, Katsura I, Takahashi H, Higashitani A (1998) Characterization of a Caenorhabditis elegans recA-like gene Ce-rdh-1 involved in meiotic recombination. DNA Res 5: 373–377 [DOI] [PubMed] [Google Scholar]
- Thomas C, Meyer D, Himber C, Steinmetz A (2004) Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol Biochem 42: 35–42 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Araujo ACG, Paech NA, Hecht V, Schmidt EDL, Rossell JB, de Vries SC, Koltunow AMG (2003) Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15: 1524–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon Bare-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11: 1769–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Nuccio ML, Budiman MA, Thomas TL, Burson BL, Hussey MA, Wing RA (1996) Comparative gene expression in sexual and apomictic ovaries of Pennisetum ciliare (L) Link. Plant Mol Biol 32: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Vielle-Calzada JP, Thomas J, Spillane C, Coluccio A, Hoeppner MA, Grossniklaus U (1999) Maintenance of genomic imprinting at the Arabidopsis medea locus requires zygotic DDM1 activity. Genes Dev 13: 2971–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedin WJ, Huff DR (1996) Bluegrass. In LE Moser, ed, Cool-Season Forage Grasses, Vol 34. American Society of Agronomy Monograph Series, ASA-CSSA-SSSA, Madison, WI, pp 665–691
- Wu H, Cheung AY (2000) Programmed cell death in plant reproduction. Plant Mol Biol 44: 267–281 [DOI] [PubMed] [Google Scholar]
- Xia X, Xie Z (2001) DAMBE: Data analysis in molecular biology and evolution. J Hered 92: 371–373 [DOI] [PubMed] [Google Scholar]
- Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D (1996) Targeted disruption of ATMleads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev 10: 2411–2422 [DOI] [PubMed] [Google Scholar]
- Yang WC, Ye D, Xu J, Sundaresan V (1999) The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Makaroff CA, Ma H (2003) The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15: 1281–1295 [PMC free article] [PubMed] [Google Scholar]
- Zamzami N, Marchetti P, Castedo M, Decaudin D, Macho A, Hirsch T, Susin SA, Petit PX, Mignotte B, Kroemer G (1995) Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J Exp Med 182: 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Henning L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 134: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.