Abstract
The potential health benefits of dietary isoflavones have generated considerable interest in engineering the synthesis of these phytoestrogens into plants. Genistein glucoside production (up to 50 nmol g−1 fresh weight) was engineered in alfalfa (Medicago sativa) leaves by constitutive expression of isoflavone synthase from Medicago truncatula (MtIFS1). Glucosides of biochanin A (4′-O-methylgenistein) and pratensein (3′-hydroxybiochanin A) also accumulated. Although MtIFS1 was highly expressed in all organs examined, genistein accumulation was limited to leaves. MtIFS1-expressing lines accumulated several additional isoflavones, including formononetin and daidzein, in response to UV-B or Phoma medicaginis, whereas the chalcone and flavanone precursors of these compounds accumulated in control lines. Enhanced accumulation of the phytoalexin medicarpin was observed in P. medicaginis-infected leaves of MtIFS1-expressing plants. Microarray profiling indicated that MtIFS1 expression does not significantly alter global gene expression in the leaves. Our results highlight some of the challenges associated with metabolic engineering of plant natural products, including tissue-specific accumulation, potential for further modification by endogenous enzyme activities (hydroxylation, methylation, and glycosylation), and the differential response of engineered plants to environmental factors.
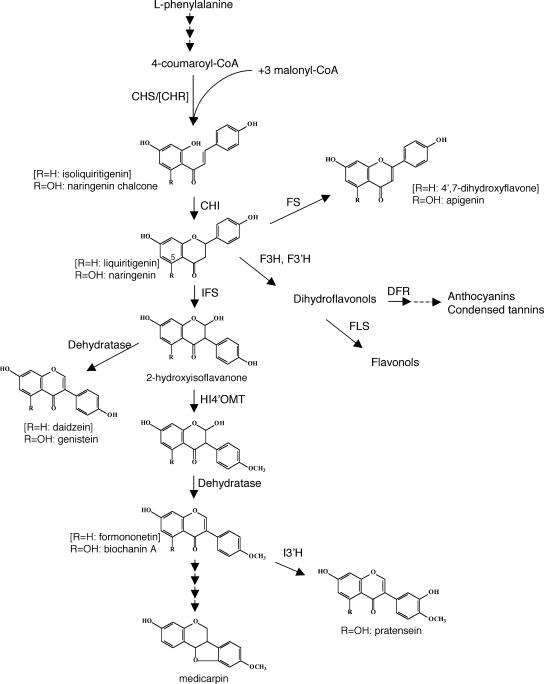
Isoflavonoids are a predominantly legume-specific subclass of flavonoid secondary metabolites, with roles in plant defense and nodulation (Dixon, 1999). The protective effects of the soy (Glycine max) isoflavones genistein and daidzein against hormone-dependent cancers, cardiovascular disease, osteoporosis, and menopausal symptoms (for review, see Cornwell et al., 2004; Dixon, 2004) have generated considerable interest in engineering these isoflavones into more commonly consumed food crops (Dixon and Steele, 1999). Isoflavones are synthesized from common flavanone intermediates (either liquiritigenin or naringenin; Fig. 1) by aryl migration catalyzed by the enzyme 2-hydroxyisoflavanone synthase (also referred to as isoflavone synthase or IFS). Cloning of IFS has allowed isoflavone synthesis to be engineered into plants that normally do not make isoflavones, as demonstrated in Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and cell cultures of maize (Zea mays; Jung et al., 2000a, 2000b; Yu et al., 2000; Liu et al., 2002). These studies have also highlighted some of the factors impacting isoflavone accumulation in plants. For example, in IFS-expressing Arabidopsis, accumulation of genistein was limited by competition from the flavonol pathway (Liu et al., 2002), whereas in IFS-expressing tobacco, it was correlated with the activity of the phenylpropanoid pathway leading to the accumulation of anthocyanins (Yu et al., 2000). In all cases, levels of genistein accumulating in transgenic plants were several orders of magnitude lower than levels of genistein in soybean.
Figure 1.
Schematic of biosynthetic pathways leading to flavonoid and isoflavonoid natural products. CHI, Chalcone isomerase; CHR, chalcone reductase; CHS, chalcone synthase; DFR, dihydroflavonol 4-reductase; F3H, flavanone 3-hydroxylase; FS, flavone synthase; F3′H, flavonoid 3′-hydroxylase; FLS, flavonol synthase; HI4′OMT, 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase; I3′H, isoflavone 3′-hydroxylase; IFS, 2-hydroxyisoflavanone synthase.
Engineering isoflavonoid biosynthesis in leguminous plants may provide enhanced levels of these compounds for plant-based dietary supplements and additional benefits related to plant defense and nodulation (Jung et al., 2003). There are only two examples of isoflavonoid engineering in legumes. A preliminary report of IFS-expressing soybean seeds showed that only two of 14 independent transgenic lines had significantly higher levels of seed isoflavones (maximal levels were 1.7-fold greater than controls), while in four lines isoflavone levels were reduced (Jung et al., 2003). In an alternative approach to increase isoflavone accumulation in soybean, the phenylpropanoid pathway-activating transcription factors C1 and R from maize were expressed in seed (Yu et al., 2003). The transgenic seed had a reduced genistein-to-daidzein ratio and only a modest (approximately 2-fold) increase in total isoflavones. Additional suppression of flavonol synthesis through down-regulation of flavanone 3-hydroxylase allowed for higher levels of isoflavone accumulation, as reported for IFS-expressing Arabidopsis (Liu et al., 2002).
In this study, we have generated transgenic alfalfa (Medicago sativa) expressing IFS from the model legume Medicago truncatula and examined the effect of constitutive IFS expression on the flavonoid composition of alfalfa, the metabolic responses of the transgenic plants to abiotic and biotic stress, and the impact of IFS expression on global gene expression patterns. These studies reveal differential tissue- and environment-specific effects on flavonoid metabolism as a result of IFS transgene expression. We also discuss the potential for using metabolic engineering to generate novel plant material for dietary studies designed to address the effect of isoflavones on animal health.
RESULTS
Generation of Transgenic Alfalfa with Constitutive Expression of MtIFS1
A cDNA encoding M. truncatula IFS (MtIFS1) was initially isolated by screening an M. truncatula root cDNA library with the soybean IFS CYP93C1v2 (Steele et al., 1999). This cDNA was later found to be identical to TC45136 present in The Institute for Genomic Research (TIGR) M. truncatula expressed sequence tag database (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=medicago). The coding region of MtIFS1 is 77.3% identical to CYP93C1v2 at the nucleotide level and 93.4% identical to a second MtIFS (accession no. AY167424), designated as MtIFS2. MtIFS1 is 94.8% identical to MtIFS2 and 80% identical to CYP93C1v2 and three M. sativa IFS sequences (Jung et al., 2000a) at the amino acid level. The activity of MtIFS1 was confirmed by assaying conversion of liquiritigenin and naringenin to the corresponding isoflavones daidzein and genistein, respectively, in microsomes from yeast (Saccharomyces cerevisiae) expressing MtIFS1 (data not shown). The in vitro properties of MtIFS1 were very similar to those of IFS from other plants (Akashi et al., 1999; Jung et al., 2000a).
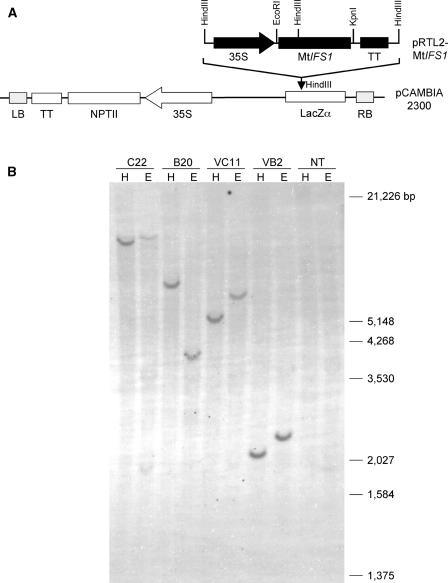
Alfalfa plants expressing MtIFS1 under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Fig. 2A) were generated. Because alfalfa contains multiple copies of the IFS gene (data not shown), plants were screened by PCR using primers specific to the NPTII gene; a subset of these lines was additionally confirmed to harbor the NPTII gene by DNA gel-blot analysis (Fig. 2B).
Figure 2.
Vector construct for alfalfa transformation and molecular characterization of transgenic lines. A, The MtIFS1 cDNA was subcloned into the pRTL2 vector and the expression cassette containing MtIFS1 flanked by CaMV 35S promoter and terminator sequences was cloned into the HindIII site of the binary vector pCAMBIA 2300. B, DNA gel-blot analysis of MtIFS1-expressing lines C22 and B20, vector control lines VC11 and VB2, and nontransgenic alfalfa cv Regen SY-4D (NT). Genomic DNA was digested with either HindIII (H) or EcoRI (E) restriction enzymes and transferred to a nylon membrane. The membrane was hybridized with a PCR fragment of the NPTII gene. Molecular mass markers are indicated.
Metabolite Analysis of MtIFS1-Expressing Alfalfa
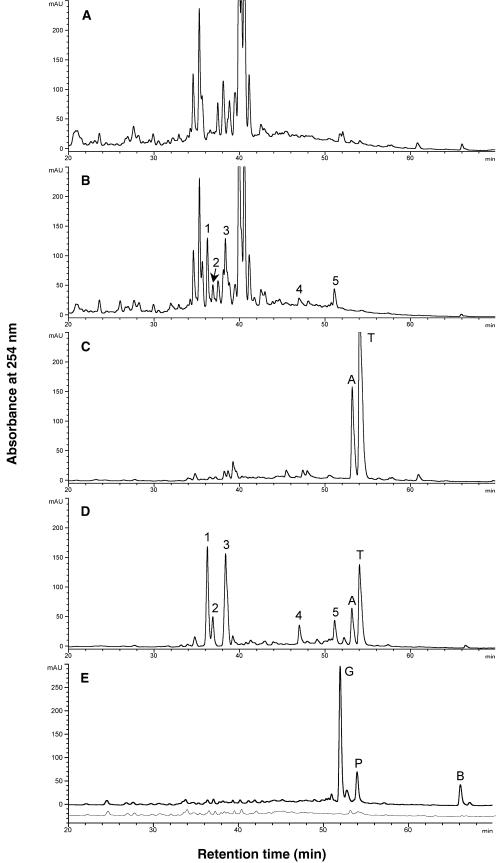
Alfalfa shoots accumulate GlcUA conjugates of the 5-hydroxy flavones apigenin, luteolin, tricin, and chrysoeriol (Stochmal et al., 2001a, 2001b). Comparison of HPLC traces of leaf extracts from vector control lines before (Fig. 3A) and after (Fig. 3C) digestion with β-glucuronidase confirmed the presence of flavone glucuronides. Unhydrolyzed leaf extracts showed several peaks with UV spectra indicative of flavones (Fig. 3A). These peaks were shifted to two peaks after digestion with β-glucuronidase (retention time [Rt] = 53.1 and 54.1 min; Fig. 3C). The aglycones were identified as the flavones apigenin (Rt = 53.1 min) and tricin (Rt = 54.1 min) by comparison of Rts and UV spectra with those of authentic standards.
Figure 3.
Effect of MtIFS1 expression on the isoflavonoid composition of alfalfa leaf extracts. HPLC traces of unhydrolyzed leaf extracts of vector control line VC11 (A) and MtIFS1-expressing line C22 (B). Peaks with UV spectra similar to genistein and not present in the control extracts are numbered 1 to 5. Peaks 1 and 4 were identified as genistin and sissotrin, respectively. Leaf extracts of control line VC11 (C) and MtIFS1-expressing line C22 (D) after digestion with β-glucuronidase. Apigenin and tricin aglycone were detected in both C and D, peaks A and T, respectively. E, HPLC traces of leaf extracts of line C22 (top trace) and VC11 (bottom trace, offset by 25 mAu) after digestion with purified β-glucosidase. Residues were resuspended in anhydrous MeOH to minimize solubilization of flavone glucuronides. Peaks labeled G, P, and B were identified as genistein, pratensein, and biochanin A, respectively.
Relative to controls, leaf extracts of MtIFS1-expressing lines contained five new peaks with UV spectra similar to genistein and related isoflavones (Fig. 3B). These peaks were not shifted after treatment with β-glucuronidase (Fig. 3D), but were shifted after digestion with purified β-glucosidase (Fig. 3E). The major peak present in β-glucosidase-treated extracts of MtIFS1-expressing lines was identified as genistein by comparison of Rt (51.8 min) and UV spectrum with those of an authentic standard, and by liquid chromatography (LC)-mass spectrometry (MS; molecular ion at m/z 269.0 [M-H]−). The other two peaks were identified as pratensein (5,7,3′-trihydroxy-4′-methoxyisoflavone, Rt = 53.9 min, molecular ion at m/z 299.0 [M-H]−) and biochanin A (5,7-dihydroxy-4′-methoxyisoflavone, Rt = 66.1 min, molecular ion at m/z 283.5 [M-H]−). Based on the results of the enzyme digestion, the five peaks present in unhydrolyzed MtIFS1-expressing leaf extracts likely represent Glc conjugates of these three isoflavones. Peaks 1 (Rt = 36.2 min) and 4 (Rt = 46.9 min) were further identified as genistin (7-O-glucosyl-genistein) and sissotrin (7-O-glucosyl-biochanin A), respectively, by comparison of Rts and UV spectra with those of authentic standards, and by LC-MS analysis.
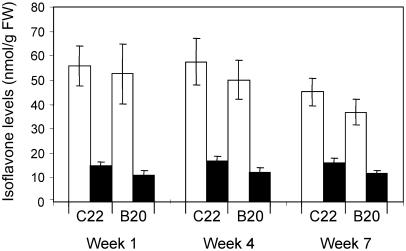
In preliminary experiments using plants grown under standard greenhouse conditions, the amount of genistein in β-glucosidase-treated leaf extracts of 42 independent transgenic lines varied from 0 (undetectable by the HPLC method) to approximately 100 nmol g−1 fresh weight (FW; approximately 27 μg g−1 FW; Supplemental Fig. 1), and was roughly correlated with the abundance of IFS transcripts detected by RNA gel-blot analysis (data not shown). Two lines, designated as C22 and B20, consistently accumulated the highest amount of genistein and were chosen for further analysis. DNA gel-blot analysis indicated that both lines harbored a single copy of the transgene (Fig. 2B). The level of genistein accumulating in these lines was found to vary depending on the growing conditions. However, under environmentally controlled greenhouse conditions (described in “Materials and Methods”), genistein levels in the leaves of these two lines were consistently around 50 nmol g−1 FW (Fig. 4), independent of the developmental stage of the plants. Levels of biochanin A were 3- to 4-fold lower than those of genistein (12 and 16 nmol g−1 FW for C22 and B20, respectively). Pratensein levels were not routinely quantified due to coelution with tricin.
Figure 4.
Isoflavone levels in leaves of MtIFS1-expressing lines C22 and B20. Leaves were harvested from plants grown in a greenhouse over the course of 7 weeks. Isoflavone levels in hydrolyzed leaf extracts were quantified by HPLC. White bars, Genistein; black bars, biochanin A. No isoflavones were detected in vector control lines.
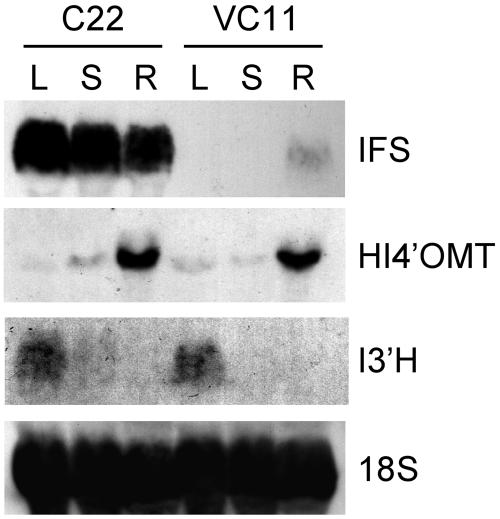
RNA gel-blot analysis demonstrated that MtIFS1 was highly expressed in leaf, stem, and root tissues of line C22, whereas in the vector control line VC11, IFS expression was only detected in the roots (Fig. 5). Surprisingly, no differences in flavonoid/isoflavonoid composition were observed in methanol extracts of stem, root, or flower tissues from lines C22 and B20 when compared to vector control lines (data not shown). Low levels of flavone conjugates were detected in stem extracts, similar to what was observed in leaf extracts. Root extracts of MtIFS1-expressing and vector control lines contained conjugates of the isoflavonoids formononetin, medicarpin, and coumestrol, but there was no significant difference in their levels in MtIFS1-expressing roots compared to controls. Flowers contained conjugates of the flavonols kaempferol and quercetin.
Figure 5.
Tissue-specific expression of IFS, HI4′OMT, and I3′H in MtIFS1-expressing line C22 and vector control line VC11. Total RNA from leaf (L), stem (S), and root (R) tissues was fractionated on a 1% formaldehyde-agarose gel and transferred to nylon membrane. Blots were hybridized with probes for IFS, HI4′OMT, I3′H, and 18S rRNA.
Leaves of MtIFS1-expressing and control plants contained low levels of transcripts encoding the enzymes 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase (HI4′OMT) and isoflavone 3′-hydroxylase (I3′H; Fig. 5), the activities of which are most likely responsible for the synthesis of biochanin A and pratensein in MtIFS1-expressing plants (Fig. 1). HI4′OMT has been shown to methylate the 2,7,4′-trihydroxyisoflavanone product of IFS on the 4′ position to form 2,7-dihydroxy-4′-methoxyisoflavanone, an intermediate in the synthesis of 5-deoxyisoflavonoids in legumes (Akashi et al., 2003). This enzyme is also predicted to methylate 5-hydroxyisoflavanones leading to the formation of biochanin A. I3′H from M. truncatula has been previously shown to hydroxylate biochanin A on the 3′ position in vitro to form pratensein (Liu et al., 2003). Expression levels of HI4′OMT and I3′H in the different tissues of MtIFS1-expressing line C22 and vector control line VC11 were similar, suggesting that MtIFS1 expression does not alter the normal expression level of these genes.
Response of MtIFS1-Expressing Plants to UV-B
The accumulation of phenylpropanoid compounds in response to UV-B light is well known, and UV-B induces isoflavonoids in legumes (Hadwiger and Schwochau, 1971; Bridge and Klarman, 1973; Beggs et al., 1985; Harborne and Williams, 2000). UV treatment of IFS-expressing Arabidopsis increased genistein levels by 2.5-fold; anthocyanin levels were also increased in these plants (Yu et al., 2000). To determine whether UV exposure would lead to a similar increase in isoflavone accumulation in MtIFS1-expressing alfalfa plants, leaf extracts from plants illuminated with UV-B for 6 h were analyzed by HPLC. In contrast to the situation in Arabidopsis and other plants, no anthocyanin was detected in any of the alfalfa plants after exposure to varying intensities of UV-B (data not shown). A lack of anthocyanin accumulation in alfalfa grown under other stressful environmental conditions (such as high light intensity, cold, outdoors in summer) has been reported (Ray et al., 2003) and may be related to the absence of flavanone 3-hydroxylase expression in the leaves (Charrier et al., 1995).
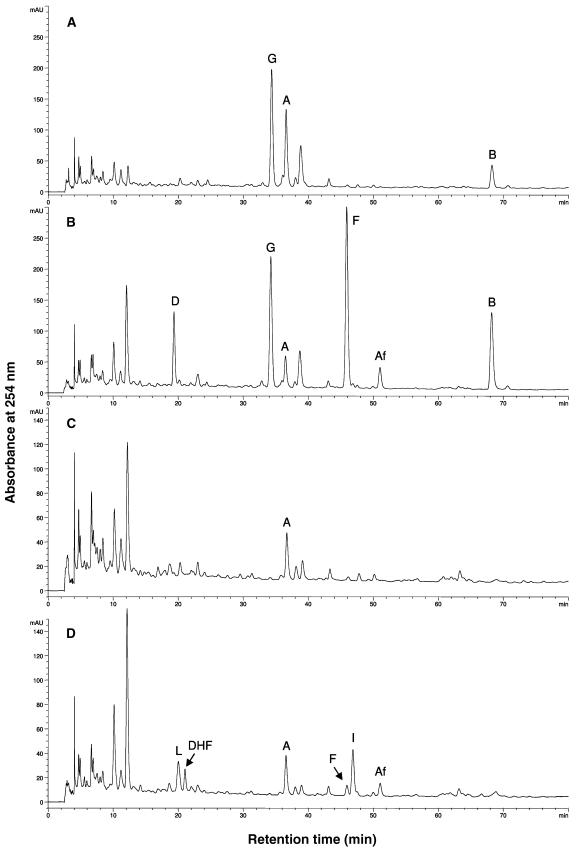
UV-B treatment led to the appearance of several peaks on HPLC chromatograms in both MtIFS1-expressing and vector control lines, consistent with induction of isoflavonoid biosynthesis (Fig. 6). Most notably, UV-B treatment induced two compounds of Rt = 19.6 and 46.2 min, identified as the 5-deoxyisoflavones daidzein and formononetin, respectively, in MtIFS1-expressing plants (Fig. 6B, peaks D and F, and Fig. 1). Formononetin is a known alfalfa phytoalexin (Paiva et al., 1994), whereas daidzein is not commonly reported to accumulate in alfalfa (Bisby et al., 1994) and was not detected in any of the vector control lines. Although a small peak of formononetin was detected in UV-B-treated vector control lines (Fig. 6D, peak F), these plants primarily accumulated the chalcone and flavanone precursors of formononetin (isoliquiritigenin and liquiritigenin, respectively; Fig. 1), as well as the related flavone 7,4′-dihydroxyflavone (Fig. 6D, peaks I, L, DHF). The isoflavone afromosin (7-hydroxy-6,4′-methoxyisoflavone) was also detected in UV-B-treated MtIFS1-expressing and vector control lines (Fig. 6, B and D, peak Af).
Figure 6.
Effect of UV-B on the flavonoid profile of MtIFS1-expressing and vector control plants. HPLC traces of hydrolyzed leaf extracts of MtIFS1-expressing line C22 (A and B) and vector control line VC11 (C and D) from control (A and C) and UV-B-treated (B and D) plants. Peaks labeled A, Af, B, D, DHF, F, G, I, and L were identified as apigenin, afromosin, biochanin A, daidzein, 7,4′-dihydroxyflavone, formononetin, genistein, isoliquiritigenin, and liquiritigenin, respectively. Peaks at 10 and 12 min represent unhydrolyzed conjugates of the flavone apigenin. Similar results were obtained for MtIFS1-expressing line B20 and vector control line VB2. Note that images A and B and images C and D are shown at different scales.
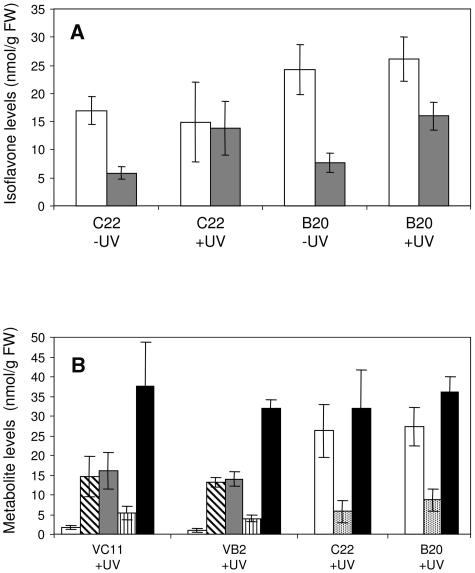
Although genistein levels were not significantly different after UV exposure, UV-B treatment more than doubled the level of biochanin A (Fig. 7A) and increased total isoflavone levels over 2-fold in MtIFS1-expressing lines C22 and B20 (from 22.8–60.8 nmol g−1 FW for C22 and from 31.9–78.1 nmol g−1 FW for B20), 40% to 50% of the increase in total isoflavones being attributable to the accumulation of formononetin. The plants used in this experiment were maintained in a growth chamber, and the lower levels of genistein and biochanin A in their leaves (when compared to levels in greenhouse-grown plants; Fig. 4) may be attributed to the different environmental conditions, in particular the much higher light intensity in the greenhouse (detailed in “Materials and Methods”). The amount of formononetin accumulated in the UV-B-treated MtIFS1-expressing lines was more than 15-fold higher than in the vector control lines (Fig. 7B, white bars). However, total levels of 5-deoxy compounds induced in response to UV-B (formononetin and daidzein in MtIFS1-expressing lines; isoliquiritigenin, liquiritigenin, and dihydroxyflavone in vector control lines) were similar (Fig. 7B, black bars).
Figure 7.
Isoflavone levels in MtIFS1-expressing and vector control lines in response to UV-B. A, Effect of UV-B on levels of genistein and biochanin A in MtIFS1-expressing lines C22 and B20. Plants were exposed to UV-B for 6 h and isoflavone levels in hydrolyzed leaf extracts were quantified by HPLC. White bars, Genistein; gray bars, biochanin A. B, Levels of 5-deoxyflavonoids in UV-B-treated MtIFS1-expressing (C22 and B20) and vector control (VC11 and VB2) lines. White bars, Formononetin; diagonally striped bars, liquiritigenin; gray bars, isoliquiritigenin; vertically striped bars, 7,4′-dihydroxyflavone; stipled bars, daidzein; black bars, total (summed) levels of 5-deoxyflavonoids.
Response of MtIFS1-Expressing Plants to Phoma medicaginis
Infection with Phoma medicaginis, a fungal pathogen responsible for spring black stem and leaf spot disease of alfalfa (Leath, 1990), induces the accumulation of isoflavonoids such as formononetin glucoside and the phytoalexin medicarpin in leaves of alfalfa (Paiva et al., 1994; He and Dixon, 2000). To determine whether MtIFS1 expression alters the biochemical response of alfalfa to this pathogen, MtIFS1-expressing and vector control lines were sprayed with a spore suspension of P. medicaginis and leaves were harvested 0, 24, 48, and 72 h after treatment. This experiment was of insufficient duration for the development of typical leaf spot symptoms, although subtle phenotypic changes, such as yellowing of the leaves, were routinely observed for infected plants. Flavonoids were extracted from leaves, hydrolyzed to aglycones, and analyzed by HPLC (Fig. 8).
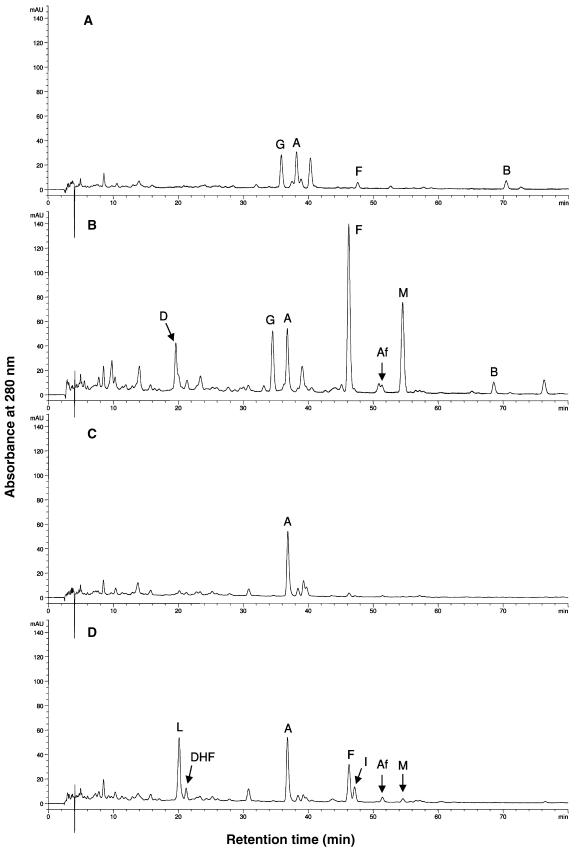
Figure 8.
Effect of P. medicaginis infection on the flavonoid profile of MtIFS1-expressing and vector control plants. HPLC traces of hydrolyzed leaf extracts of MtIFS1-expressing line C22 (A and B) and vector control line VC11 (C and D) from control (A and C) and spore-inoculated (B and D) plants at 24 h postinfection. Peaks labeled A, Af, B, D, DHF, F, G, I, L, and M were identified as apigenin, afromosin, biochanin A, daidzein, 7,4′-dihydroxyflavone, formononetin, genistein, isoliquiritigenin, liquiritigenin, and medicarpin, respectively. Similar results were obtained for MtIFS1-expressing line B20 and vector control line VB2. Note that A280 is shown due to the low absorbance of medicarpin at 254 nm.
There was no change in the flavonoid profile of vector control plants sprayed with a control solution (0.1% Tween 20) over the 72-h period (data not shown). However, MtIFS1-expressing plants sprayed with the same solution accumulated a small amount of formononetin at 24 to 72 h (Fig. 8A). Formononetin was not detected in any of the samples harvested at the start of the experiment (T = 0), and its synthesis may be in response to the cool, humid conditions used during the experiment.
Spore inoculation had a similar effect on the leaf flavonoid composition as did UV-B treatment, the major difference being the significant accumulation of medicarpin in P. medicaginis-infected tissues (Fig. 8, B and D). Only traces of medicarpin were detected in UV-B-treated leaves (data not shown), which suggests that its accumulation in response to P. medicaginis may be related to its antifungal activity (Blount et al., 1993). As observed in the UV-B experiment, formononetin, isoliquiritigenin, liquiritigenin, and 7,4′-dihydroxyflavone accumulated in leaves of vector control lines in response to P. medicaginis (Fig. 8D), whereas daidzein and formononetin accumulated in MtIFS1-expressing lines (Fig. 8B). Traces of afromosin were also detected in both lines (Fig. 8, B and D, peak Af). Genistein, pratensein, and biochanin A were still present in infected leaves of MtIFS1-expressing plants (Fig. 8B), and levels of genistein and biochanin A were not significantly changed due to infection, although a considerable amount of variation was observed (data not shown).
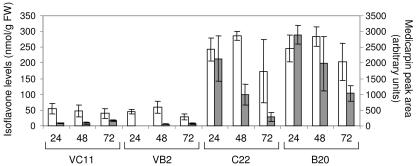
MtIFS1-expressing plants accumulated much higher concentrations of formononetin and medicarpin than the vector control lines 24 to 72 h after infection (Fig. 9). Levels of formononetin, which ranged from 173 to 286 nmol g−1 FW in infected leaves of C22 and B20, were 4- to 7-fold higher than levels accumulating in the vector control lines and more than 2-fold higher than those of genistein. In infected leaves of VC11 and VB2, medicarpin represented a minor peak (Fig. 8D), whereas much higher levels were found to accumulate in MtIFS1-expressing lines (Fig. 9). Maximal levels of medicarpin in leaves of MtIFS1-expressing lines were observed 24 h after infection and exceeded levels observed in the vector control lines by >25-fold (Fig. 9).
Figure 9.
Isoflavonoid phytoalexin levels in leaves of MtIFS1-expressing and vector control lines in response to P. medicaginis infection. Plants were sprayed with a spore suspension and leaves were harvested 0 to 72 h later. Isoflavone levels in hydrolyzed leaf extracts were quantified by HPLC. Formononetin and medicarpin were not detected at the start of the experiment (T = 0) in any of the transgenic lines (data not shown). Medicarpin concentration is presented as peak area (arbitrary units). White bars, Formononetin; gray bars, medicarpin.
Microarray Analysis of MtIFS1-Expressing Plants
Genistein is a known signaling molecule in soybean-rhizobium interactions (Pan et al., 1998) and is also a mammalian cell cycle inhibitor and inhibitor of Tyr kinase (Akiyama et al., 1987) and DNA topoisomerase (Okura et al., 1988; Markovits et al., 1989) activities. Because genistein is normally not synthesized in the leaves of alfalfa, we asked whether its accumulation caused any unexpected effect on gene expression. We used an M. truncatula 16K oligonucleotide array to profile gene expression in the leaves of MtIFS1-expressing plants. Due to the high degree of gene sequence conservation between alfalfa and M. truncatula, this array has been used to reliably profile gene expression in alfalfa (Aziz et al., 2005). RNA purified from leaves of MtIFS1-expressing lines C22 and B20 and vector control line VC11 was used to generate Cy5-labeled cDNA. RNA purified from a second vector control line, VB2, was used to generate Cy3-labeled reference cDNA. Three independently propagated cuttings of each line were used and a total of nine hybridizations were performed.
Pairwise comparisons of gene expression between lines C22 and VC11 and lines B20 and VC11 showed that, out of 16,086 spots, only 10 (C22 versus VC11) and 13 (B20 versus VC11) spots showed a statistically significant 2-fold or greater change in expression (P ≤ 0.05; Table I; Supplemental Table I). When the two MtIFS1-expressing lines were compared, 12 spots showed a 2-fold or greater change in expression (Supplemental Table II). As expected, the greatest change in gene expression was observed for spots predicted to hybridize to the introduced IFS transgene (TC45135 and TC45136, up-regulated 17- and 9-fold in line C22 and 5- and 4-fold in line B20, respectively; the 70-mer oligos for TC45135 and TC45136 differ at 8 and 9 nucleotides, respectively, compared to MtIFS1, explaining the differential hybridization levels). There was no change in gene expression for any enzymes of phenylpropanoid, flavonoid, or isoflavonoid biosynthesis, consistent with the results of the phytochemical and RNA gel-blot analyses (Figs. 4 and 5).
Table I.
Genes with statistically significant changes in gene expression identified from microarray analyses of MtIFS1-expressing line C22 and vector control line VC11
UTR, Untranslated region; EST, expressed sequence tag.
| TIGR Identifier | No. of ESTsa | Fold Change (C22/VC11) | P Value | Gene Annotation of Closest BLAST Homolog |
|---|---|---|---|---|
| TC45135b | 4 | 17.66 | 0.0015 | Isoflavone synthase |
| TC45136b | 36 | 8.86 | 0.0000 | Isoflavone synthase |
| TC46175b | 6 | 2.47 | 0.0000 | 3′ UTR of ubiquitin/ribosomal extension protein |
| TC58751 | 3 | 2.26 | 0.0000 | Transcription factor E2F (Chenopodium rubrum) |
| TC45391 | 8 | 2.16 | 0.0000 | No significant hit |
| TC45908 | 6 | −2.20 | 0.0497 | At1g04870/F13M7_12 (Arabidopsis) |
| TC49689 | 2 | −2.27 | 0.0051 | No significant hit |
| TC48121 | 3 | −2.75 | 0.0111 | At1g65020 (Arabidopsis) |
| TC50867 | 4 | −2.85 | 0.0000 | Putative thaumatin protein (Arabidopsis) |
| TC44464 | 27 | −3.10 | 0.0001 | Hypothetical protein (Lotus corniculatus) |
Number of ESTs in TIGR Medicago Gene Index Release 7.0 (May 2003).
TCs identified as up-regulated in both MtIFS1-expressing lines relative to vector control line VC11.
To address whether the experimental and/or biological variation was too great to identify differentially expressed genes, we analyzed a set of housekeeping genes that were spotted 17 times per array and found that the within-array coefficient of variation (range from 9%–29%) was similar, but often slightly higher than the between-array variation (range from 4%–31%) when identical spots were compared. This suggests that the analytical and biological variation, although quite high, would not limit the statistical resolution of the experiment.
We predicted that genes whose expression was regulated in response to genistein accumulation would be similarly up- or down-regulated in both MtIFS1-expressing lines when compared to vector control line VC11. Using this criterion, only one tentative consensus (TC) sequence (TC46175, up-regulated 2.5- and 2.7-fold) was identified with a statistically significant change in gene expression in lines C22 and B20 relative to the vector control line VC11 (Table I; Supplemental Tables I and II). However, we could not detect expression of this TC in any of the leaf samples used in the microarray experiment when examined by reverse transcription (RT)-PCR or northern gel-blot analysis. Furthermore, TC46175 could not be amplified from M. truncatula genomic DNA or leaf cDNA. TC46175 shares homology with the 3′-untranslated region of a ubiquitin/ribosomal extension protein, and it is not clear how expression of this TC may be related to isoflavone biosynthesis. From these results, we conclude that constitutive MtIFS1 expression does not significantly alter global gene expression in the leaves of alfalfa.
DISCUSSION
Factors Impacting Isoflavone Engineering in Plants
The numerous health benefits ascribed to dietary intake of soy isoflavones has generated significant interest in engineering these compounds in more widely consumed plants, despite conflicting evidence of their benefits to consumers (Dixon, 2004). This work, together with previous reports of isoflavone engineering in Arabidopsis, tobacco, and soybean (Jung et al., 2000a, 2000b, 2003; Liu et al., 2002; Yu et al., 2003), has additionally highlighted some of the factors impacting isoflavone engineering in plants. Genistein levels in leaves of MtIFS1-expressing alfalfa (approximately 50 nmol g−1 FW or approximately 13.5 μg g−1 FW) were severalfold higher than reported for IFS-expressing Arabidopsis (2 ng mg−1 FW [Jung et al., 2000a, 2000b], 7.4 nmol g−1 FW [Liu et al., 2002]) and tobacco (only detectable by gas chromatography-MS [Yu et al., 2000]). In each case, expression of IFS was driven by the CaMV 35S promoter, although different IFS homologs were used (soybean IFS in previous studies, M. truncatula IFS1 in this study). The high sequence identity between soybean IFS and MtIFS1 (approximately 77%), as well as the similar in vitro activity of yeast-expressed MtIFS1 and soybean IFS (data not shown), suggests that differences in genistein accumulation in transgenic plants are likely attributable to the different host plants. Because isoflavonoid synthesis is intrinsic to alfalfa, IFS may be better able to assimilate into the endogenous leaf flavonoid pathway. Furthermore, differences in activities of specific branches of the flavonoid pathway, leading primarily to flavonols in Arabidopsis and flavones in alfalfa leaves, may account for the differences in genistein accumulation. In Arabidopsis, competition between IFS and flavanone-3-hydroxylase (F3H) was reported to limit the accumulation of genistein in plants expressing IFS, presumably due to competition between IFS and F3H for the common flavanone substrate naringenin (Liu et al., 2002). Blocking flavonol synthesis increased the amount of genistein to levels (31–169 nmol g−1 FW) similar to those reported here for alfalfa (Liu et al., 2002). No expression of F3H was detected in alfalfa leaves (Charrier et al., 1995), and we could not detect any flavonols in the leaves of transgenic plants. The lack of correlation between levels of genistein and apigenin in the leaves of healthy MtIFS1-expressing plants (data not shown) initially suggested that competition between IFS and flavone synthase may not be a major factor limiting genistein synthesis in alfalfa. However, the disappearance of 7,4′-dihydroxyflavone in UV-B and Phoma-infected leaves of MtIFS1-expressing lines suggests that IFS may compete with flavone synthase for the common 5-deoxy flavone liquiritigenin under conditions of increased pathway flux arising from biotic or abiotic stress.
Although MtIFS1 was expressed in all tissues examined, changes in isoflavonoid/flavonoid composition were, surprisingly, only detected in the leaves. This was unexpected based on a previous report that tobacco plants expressing a soybean IFS accumulated a higher concentration of genistein in flowers than in leaves, presumably due to the anthocyanin pathway supplying naringenin in flowers (Yu et al., 2000). The similar flavone composition of alfalfa leaf and stem suggests that the restricted distribution of genistein in MtIFS1-expressing alfalfa was not necessarily correlated with a specific branch of the flavonoid pathway. In IFS-expressing Arabidopsis, whole shoots were analyzed and the distribution of genistein in different tissues was not reported (Jung et al., 2000a; Liu et al., 2002). However, from the available data for IFS-expressing alfalfa and tobacco (Yu et al., 2000), it is clear that additional factors may govern where engineered isoflavones accumulate in the plant. Although the level of IFS transcript was greater in roots of MtIFS1-expressing lines compared to control lines, the fact that this did not lead to increased levels of isoflavonoids suggests that IFS expression is not limiting for isoflavonoid accumulation in the roots. A similar result was reported for expression of IFS in soybean seeds, an organ with high endogenous IFS activity; only two out of 14 lines had significantly higher levels of seed isoflavones, and maximal isoflavone levels were only 1.7-fold greater than controls (Jung et al., 2003).
In all reports of metabolic engineering of isoflavones, genistein accumulated as glycosides (Yu et al., 2000; Liu et al., 2002). In Arabidopsis, engineered genistein was conjugated with Glc and rhamnose in a manner similar to the conjugation of the endogenous flavonols (Liu et al., 2002). In leaves of MtIFS1-expressing alfalfa, isoflavones accumulated as glucosides rather than as glucuronides, the expected conjugates based on the presence of glucuronidated flavones in the leaves. Genistein is likely conjugated by endogenous glycosyltransferases, which must be promiscuous in their choice of substrate given that they glycosylate compounds not normally produced in the plant. Metabolic engineering of isoflavone glucosides rather than aglycones may be advantageous because glycosylation may enhance the stability of these compounds in planta and facilitate their sequestration in the vacuole where they are less likely to interfere with metabolism. In alfalfa, endogenous O-methyltransferase and hydroxylase activities additionally metabolized a small fraction of the engineered 2-hydroxyisoflavanone to biochanin A and pratensein. No such modifications were reported in IFS-expressing Arabidopsis or tobacco, presumably because these plants lack enzymes for isoflavonoid biosynthesis.
Differences in the flavonoid composition of MtIFS1-expressing and vector control plants were even greater when these plants were subjected to UV-B or infection with P. medicaginis. In both cases, vector control and MtIFS1-expressing lines responded by accumulating the phytoalexins formononetin and medicarpin (the latter of which accumulated primarily in response to P. medicaginis), with levels of these isoflavonoids in MtIFS1-expressing plants greatly exceeding those in vector control lines (Figs. 7 and 9). In addition, MtIFS1-expressing plants accumulated daidzein, an isoflavone that was not detected in treated leaves of vector control plants and has not been reported to accumulate in alfalfa. The presence of daidzein in treated leaves of MtIFS1-expressing plants suggests that the activity of the 4′-O-methyltransferase immediately downstream of IFS in the pathway to formononetin and medicarpin (Fig. 1) may, under some conditions, be limiting. The instability of the 2-hydroxyisoflavanone product of IFS coupled with potential activity of a recently cloned 2-hydroxyisoflavanone dehydratase (Akashi et al., 2005), may have facilitated the formation of daidzein. The observation that this compound accumulated in MtIFS1-expressing plants only in response to stress emphasizes the importance of profiling transgenic plants under a range of environmental conditions.
Potential of Transgenic Alfalfa for Dietary Studies on Isoflavone Efficacy
The variable chemical composition of research material has been acknowledged as a major factor contributing to inconsistencies in the literature regarding the health effects of isoflavones (Messina et al., 2004). In this study, we have characterized plant lines that either accumulate (MtIFS1-expressing lines) or lack (vector control lines) isoflavones in their leaves and therefore have the potential to be used in studies to address the value of dietary delivery of isoflavones on animal health. The advantage of using metabolic engineering to generate material for dietary studies is that results are interpreted relative to control material lacking isoflavones, which is currently not possible with soy-derived materials. Alfalfa is an excellent model for these studies, since alfalfa pellets are widely used as a feed for laboratory animals. However, considering current recommended doses of isoflavones (Branca and Lorenzetti, 2005), additional genetic modification (such as suppression of the competing flavone pathway) may be required to enhance isoflavone levels in MtIFS1-expressing alfalfa.
Microarray analysis suggests that constitutive MtIFS1 expression in alfalfa leaves does not alter the expression of genes leading to new or altered levels of metabolites that might pose food safety concerns or limit the use of transgenic plants in feeding studies. The lack of a global alteration of gene expression in MtIFS1-expressing plants is favorable from the perspective of genetic engineering since it may provide support for substantial equivalence (Kuiper et al., 2001). The small number of genes (<1%) whose expression in independent MtIFS1 transgenic plants was significantly different from that in controls suggests that MtIFS1 expression and/or isoflavone accumulation in the leaves has no greater effect on gene expression than interplant variation. This is supported by the fact that the number of genes identified as having a statistically significant change in gene expression was similar when the two MtIFS1-expressing lines were compared to the vector control line or to each other. A similar result was recently reported for transgenic Arabidopsis plants engineered to accumulate the cyanogenic glucoside dhurrin, in which microarray analysis showed only marginal effects on the transcriptome (Kristensen et al., 2005). The observation that genistein as well as biochanin A and pratensein are glycosylated suggests that these isoflavones are stored in the vacuole and this sequestration may be sufficient to prevent any unintended effects on gene expression.
MATERIALS AND METHODS
Plant Material and Greenhouse Conditions
Plants were potted in Metro Mix 350 and grown in a controlled environment greenhouse with a 16-h photoperiod. The temperature was maintained at 20°C to 23°C during the day and 18°C to 20°C at night. Humidity was set at 50% during the day and 75% at night. During the course of these experiments, measured light intensity in the greenhouse ranged from 266 to 417 μmol m−2 s−1 photosynthetically activated radiation (PAR). Plants were watered daily with Peters 20:10:20 fertilizer (100 ppm N; Scotts, Marysville, OH) by flood irrigation. Independent transgenic lines were vegetatively propagated by cuttings as described (Hipskind and Paiva, 2000).
Vector Constructs and Plant Transformation
The MtIFS1 cDNA was subcloned into the EcoRI/KpnI sites of the pRTL2 vector (Restrepo et al., 1990) to create an expression cassette containing the MtIFS1 cDNA flanked by CaMV 35S promoter and terminator sequences. This 2,938-bp expression cassette was isolated following partial digestion with HindIII and cloned into the pCAMBIA 2300 binary vector (Hellens and Mullineaux, 2000; Fig. 2A). The pCAMBIA2300-MtIFS1 plasmid was transformed into Agrobacterium tumefaciens strain LBA44044 by electroporation. In preparation for transformation, an overnight culture of A. tumefaciens was grown to OD550 = 1.0 at 28°C in yeast extract peptone media (Trieu et al., 2000) supplemented with the appropriate antibiotics. Cells were harvested by centrifugation and resuspended in 2 volumes of modified basal medium (BM; described below) prior to incubation with leaf discs. Trifoliates from the alfalfa (Medicago sativa) cv Regen SY-4D (Bingham, 1991) were surface sterilized for 10 s in 70% (v/v) ethanol and 15 min in 10% (v/v) bleach containing 0.1% (v/v) Tween 20 followed by three washes in sterile distilled, deionized water. Leaf discs were incubated with the A. tumefaciens suspension for 10 to 60 min and blotted on sterile filter paper before transfer to solid media. The medium used for culture of explants was based on BM (Thomas and Rose, 1988), except that casamino acids were replaced with casein hydrolysate. Leaf explants were plated on BM containing 10 μm 2,4-D and 5 μm benzylaminopurine, as described (Thomas et al., 1990), with the additional inclusion of 100 μm acetosyringone. Callus induction and embryo development were essentially as described (Thomas et al., 1990), except that the kanamycin concentration was lowered to 25 μg/mL and cefotaxime was omitted. Rooted plantlets were transferred to BM containing 25 μg/mL kanamycin and 250 μg/mL carbenicillin in magenta boxes until plants were large enough to be transferred to the greenhouse.
DNA Extraction and Analysis
Genomic DNA was extracted from 100 mg of leaf tissue using plant DNAzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. PCR was performed with 250 ng of genomic DNA and NPT-II-specific primers (Trieu et al., 2000) using standard cycling parameters. For DNA gel-blot analysis, genomic DNA was purified from leaf tissue using the Plant DNeasy mini prep kit (Qiagen, Valencia, CA). Twenty micrograms of HindIII- or EcoRI-digested genomic DNA were fractionated on a 0.8% (w/v) Tris-acetate EDTA gel. Processing of gel, DNA transfer to nylon membrane, probe labeling, and hybridization were performed as described in the AlkPhos direct labeling kit (Amersham, Piscataway, NJ). Blots were hybridized with an NPT-II probe (Trieu et al., 2000) and developed using the CDP-Star detection reagent (Amersham).
RNA Extraction and Analysis
RNA was extracted from plant tissues using TRI reagent (Molecular Research Center, Cincinnati) according to the manufacturer's instructions. Twenty micrograms of RNA were fractionated on a 1% (w/v) formaldehyde-agarose gel and transferred to a nylon membrane by standard protocols (Ausubel et al., 1994). Probe labeling, blot hybridization, and signal detection were performed as described in the ECL direct labeling kit (Amersham). Blots were probed with a full-length MtIFS1 cDNA (TC45136) or M. truncatula cDNA corresponding to HI4′OMT (TC44044), I3′H (TC70025; Liu et al., 2003), or 18S rRNA (Maldonado-Mendoza et al., 2002) amplified from pBluescript using the T7 and T3 vector primers.
For RT-PCR, 2 μg of total RNA were transcribed into cDNA using ready-to-go RT-PCR beads (Amersham) and oligo(dT) primer. Two microliters of cDNA were used in each PCR reaction (50 μL total) with Ex-Taq PCR reagents (TaKaRa Shuzo, Shiga, Japan) and the following primers: actin, 5′-CAATTTCTCGCTCTGCTGAGGTGG-3′, 5′-GGCTGGATTTGCTGGAGATGATGC-3′; MtIFS1, 5′-CCCATGAAGCTACTTCC-3′, 5′-AGTCGTTCATGATAAGC-3′; TC54732, 5′-GTTCGTGCTTTACGGGTTGT-3′, 5′-AAAGCAGGGGGCAACATAGT-3′; TC76580, 5′-GTGGCGCTAAGAAGCGTAAG-3′, 5′-TCAAAACGCACAAAATTGTATTG-3′; TC56026, 5′-CGGTGTCGTGCTTTACCTTT-3′, 5′-ACCTACACAATGCCCCTCAA-3′; TC52595, 5′-GTTTTTGCACCAACTGCAAC-3′, 5′-GCCTCCACCCTTCTTCTTTT-3′; TC46499, 5′-TGCTAGTAAATGGTCTGCCAAT-3′, 5′-GCTAGGCAACGCTAGGACAT-3′; and TC44464, 5′-GACCGAGACGGAAATAGCAG-3′, 5′-ATGGAGCCGTTTTGTTCTTG-3′. PCR conditions were 94°C, 5 min, 25 to 32 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, followed by 72°C for 10 min. PCR products were resolved on a 1% (w/v) Tris-acetate EDTA-agarose gel, visualized with ethidium bromide, and band intensities were quantified using LabWorks software version 4.0 (UVP, Upland, CA). Band intensities were normalized relative to actin.
Metabolite Analysis and HPLC
Plant tissue was routinely harvested in the greenhouse, snap frozen in liquid N2, and stored at −80°C. Tissues were ground in liquid N2 and 0.5 to 1.0 g extracted two times in 10 volumes of 80% (v/v) MeOH. Extracts were combined and dried under a stream of N2. For analysis of isoflavone conjugates, residues were resuspended by vortexing in 80% or 100% MeOH, briefly sonicated, and clarified by centrifugation before loading on the HPLC. For digestion with β-glucosidase or β-glucuronidase, residues were resuspended in 0.6 volume citrate-phosphate buffer, pH 5.2. A crude β-glucosidase preparation from almonds (Sigma, St. Louis) was found to contain both β-glucosidase and β-glucuronidase activity and was used at 3 mg mL−1 for quantification of isoflavone aglycones. Additional enzymes used were purified β-glucosidase from almonds (Fig. 3E) or β-glucuronidase (Sigma); both were used at 25 units per reaction. Reactions were incubated overnight at 37°C and extracted two times with 1 volume of ethyl acetate. Ethyl acetate extracts were combined, dried under N2, and residues resuspended in 0.1 volume (relative to initial extraction volume) of MeOH, as described above. Samples (20 μL) were applied to an ODS2 reverse-phase column (5-μm particle size, 4.6 × 250 mm) and eluted in 1% (v/v) phosphoric acid with an increasing gradient of acetonitrile (0 to 5 min, 5%; 5 to 10 min, 5% to 10%; 10 to 25 min, 10% to 17%; 25 to 30 min, 17% to 23%; 30 to 65 min, 23% to 50%; 65 to 69 min, 50% to 100%) at a flow rate of 1 mL min−1. Isoflavone aglycones were identified by comparing Rts and UV spectra with those of authentic standards of genistein, biochanin A (both purchased from Indofine, Hillsborough, NJ), and pratensein (ChromaDex, Santa Ana, CA). Genistein and biochanin A levels were quantified from HPLC traces by calculating peak area and comparison to a standard curve. HPLC-MS was performed as previously described (Liu et al., 2003). For MS analysis of pratensein, β-glucosidase-treated extracts were further fractionated on Si250(F) silica gel thin-layer chromatography plates (J.T. Baker, Phillipsburg, NJ) using chloroform:methanol (96:4, v/v) as solvent to remove a contaminant that interfered with the analysis. Plates were visualized under UV illumination (254 nm) and the spot running at the same RF as the pratensein standard was scraped off the plate, eluted with methanol, and subjected to MS analysis.
UV-B Treatment
Cuttings rooted in 96-well flats were grown in a growth chamber under fluorescent lighting (approximately 150−255 μmol m−2 s−1 PAR). The chamber was set to a 16-h photoperiod, with a day/night temperature of 23°C/19°C, and 50% humidity. Plants (140- to 260-cm tall over the course of three independent experiments) were exposed to UV-B light (XX-15MR bench lamp, MR 302 nm, 115 V; UVP) for 6 h. The intensity of the light as measured with a 310-nm UV meter was 0.370 to 0.734 mW cm−2 from the top of the soil to the average maximal plant height. Prior to the start of the experiment, the fluorescent lighting in the chamber was lowered to half-intensity (approximately 80–130 μmol m−2 s−1 PAR). Control plants (non-UV treated) were kept in a separate growth chamber under identical conditions. In each experiment, leaves from three replicate plants were pooled and frozen in liquid N2 and the experiment was repeated three times. Flavonoid extraction and hydrolysis were performed as described above. Hydrolyzed isoflavones were further purified using C18 cartridges (Waters Sep-Pak C18 cartridges; Waters, Milford, MA) as follows. Extracts, dissolved in a small volume of MeOH, were diluted 1,000-fold with water and loaded on a C18 cartridge that had been preconditioned by washing with 10 volumes of MeOH followed by water. After washing with water, flavonoids were eluted with 6 volumes of MeOH. Eluates were dried under N2 and residues resuspended in 100 μL MeOH. Samples were loaded on the HPLC and eluted in 1% (v/v) phosphoric acid with an increasing gradient of acetonitrile (0 to 5 min, 25%; 5 to 80 min, 25% to 45%; 80 to 81 min, 45% to 100%) at a flow rate of 0.8 mL min−1.
Phoma medicaginis Infection
Phoma medicaginis Malbranche et Roumeguere was from the American Type Culture Collection (Manassas, VA). Conidial suspensions of P. medicaginis were plated on potato dextrose agar and incubated in the dark at 21°C until sporangia were visible (approximately 4–6 weeks). Spores were collected by flooding the surface of the plate with a sterile 0.1% (v/v) Tween 20 solution followed by gentle scraping with a spatula. Chunks of agar and mycelia were removed by filtration. Spore concentrations, as determined by hemacytometer counting, were routinely 1 to 2 × 108 spores mL−1. Plants were sprayed with the spore suspension or a control solution (0.1% Tween 20) in a growth chamber until dripping wet and bagged for 24 h. The chamber was set to a 12-h photoperiod with a day/night temperature of 21°C/16°C and 95% humidity, conditions reported to be optimal for growth chamber infection of Medicago sativa with P. medicaginis (Barbetti, 1987, 1991). Light intensity in the chamber was approximately 125 μmol m−2 s−1 PAR. Leaves were harvested at the given times and frozen in liquid N2 and the experiment was repeated three times. Flavonoid extraction and HPLC analysis were performed as described above.
Microarray Experiments
The Medicago Genome Oligo Set version 1.0 (Qiagen Operon, Alameda, CA) includes 70-mer oligonucleotides representing 16,086 M. truncatula TC sequences from TIGR Gene Index Database (www.tigr.org/tdb/tgi/mtgi) MtGI Release 5.0, as well as both positive and negative controls. Details of this oligo set are available at http://oligos.qiagen.com/arrays/omad.php. The oligos were spotted in 3× SSC onto Superamine (Telechem, Sunnyvale, CA) glass slides by the Galbraith lab at the University of Arizona (Tucson, AZ). A single array element was printed per oligo. After printing, slides were baked at 80°C for 1 h and stored at room temperature. Prior to use, slides were UV-cross-linked at 200 mJ and prehybridized in 5× SSC, 0.1% (w/v) SDS, and 1% (w/v) bovine serum albumin for 45 min at 42°C. Slides were then rinsed five times in distilled, deionized water, spun dry, and used immediately. Approximately 90% of spots on this array showed significant signal when hybridized with labeled alfalfa leaf cDNA.
PolyA RNA was isolated from 400 μg of total leaf RNA using the Poly(A) Purist MAG kit from Ambion (Austin, TX). Two micrograms of polyA were transcribed into cDNA and fluorescently labeled with Cy3 and Cy5 dyes using the SuperScript indirect cDNA labeling system (Invitrogen) according to the manufacturer's instructions. Labeled cDNA for experimental (Cy5) and reference (Cy3) samples were combined, ethanol precipitated, and resuspended in 10 μL of water. The cDNA was heated at 100°C for 3 min before the addition of 70 μL of SlideHyb glass array hybridization buffer number 1 (Ambion). The hybridization solution was pipetted onto slides covered with a gapped coverslip (LifterSlips; Erie Scientific Company, Portsmouth, NH). Slides were incubated overnight at 42°C in Corning hybridization chambers, then washed in 1× SSC, 0.1% (w/v) SDS for 5 min at 42°C, followed by 5-min washes in 0.1× SSC, 0.1% SDS (w/v), and 0.05× SSC at room temperature. Slides were scanned using the ScanArray 4000 microarray analysis system. Based on an initial low-resolution (50 μm) scan, laser power settings were manually adjusted to balance the intensities between the two channels and slides were rescanned at 10-μm resolution. Typical laser settings were Cy3 laser power = 100, photomultiplier tube = 80; Cy5 laser power = 85, photomultiplier tube = 75. GPR files were generated using GenePix Pro 4.1 software and the mean pixel intensities for each spot were used in subsequent data analysis. Data files were imported into GeneTraffic Duo version 2.6, and data for each hybridization were normalized using the Lowess SubGrid method. Spots flagged by the program under the default settings (spots with a raw pixel intensity <100 or lower than the average background and spots with an intensity/background intensity ratio <1) were removed from further analysis. Significance analysis was done in GeneTraffic by performing two class, unpaired t tests with the variance stabilization and Benjamini-Hochberg P-value correction options. Analysis was first performed on a dataset that excluded spots lacking one or more value over the three replicates. A second analysis was performed on a dataset that included spots missing one value and in which case the missing value was imputed by determining its k-nearest neighbor. This second analysis increased the number of spots analyzed by approximately 2,000 and increased the number of significant spots by 1 to 5, depending on the comparison.
Four TCs (TC54732, TC56026, TC57633, TC58892) that were initially identified with a statistically significant change in gene expression in either one or both MtIFS1-expressing lines were seen to be spotted in close proximity to the spot corresponding to IFS (TC45135) on the arrays used in this experiment. Expression of these spots could not be verified by RT-PCR or RNA gel-blot analysis, and the TCs were not identified as differentially expressed in a second experiment using a different array batch. Because of the possibility that the potential up-regulation of these TCs was artifactual, they were eliminated from our analysis.
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requester.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY939826 (MtIFS1). Gene expression data have been deposited with the GEO repository under accession number GSE2546.
Supplementary Material
Acknowledgments
We thank Jack Blount for help with HPLC analysis, Dr. Naveed Aziz for help with microarrays, David Huhman and Dr. Lloyd Sumner for assistance with LC-MS analysis, Yuanji Zhang for BLAST analysis, Corey Broeckling and members of the Dixon lab for helpful discussions, and Drs. Li Tian and Rujin Chen for critical review of the manuscript.
This work was supported by a grant from the Oklahoma Center for the Advancement of Science and Technology (project no. HR02–040R to R.A.D.) and by the Samuel Roberts Noble Foundation.
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.062539.
References
- Akashi T, Aoki T, Ayabe S (1999) Cloning and functional expression of a cytochrome P450 cDNA encoding 2-hydroxyisoflavanone synthase involved in biosynthesis of the isoflavonoid skeleton in licorice. Plant Physiol 121: 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Aoki T, Ayabe S (2005) Molecular and biochemical characterization of 2-hydroxyisoflavanone dehydratase. Involvement of carboxylesterase-like proteins in leguminous isoflavone biosynthesis. Plant Physiol 137: 882–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Sawada Y, Shimada H, Sakurai N, Aoki T, Ayabe S (2003) cDNA cloning and biochemical characterization of S-adenosyl-L-methionine: 2,7,4′-trihydroxyisoflavanone 4′-O-methyltransferase, a critical enzyme of the legume isoflavonoid phytoalexin pathway. Plant Cell Physiol 44: 103–112 [DOI] [PubMed] [Google Scholar]
- Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y (1987) Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem 262: 5592–5595 [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology. John Wiley and Sons, New York
- Aziz N, Paiva NL, May GD, Dixon RA (2005) Transcriptome analysis of alfalfa glandular trichomes. Planta 221: 28–38 [DOI] [PubMed] [Google Scholar]
- Barbetti MJ (1987) Effects of temperature and humidity on disease caused by Phoma medicaginis, resistance in some Medicago cultivars and the incidence of seed-borne inoculum. Aust J Exp Agric 27: 851–856 [Google Scholar]
- Barbetti MJ (1991) Effects of temperature and humidity on disease caused by Phoma medicaginis and Leptosphaerulina trifolii in lucerne (Medicago sativa). Plant Pathol 40: 296–301 [Google Scholar]
- Beggs CJ, Stolzer-Jehle A, Wellmann E (1985) Isoflavonoid formation as an indicator of UV stress in bean (Phaseolus vulgaris L.) leaves. Plant Physiol 79: 630–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham ET (1991) Registration of alfalfa hybrid Regen-SY germplasm for tissue culture and transformation research. Crop Sci 31: 1098 [Google Scholar]
- Bisby FA, Buckingham J, Harborne JB , eds (1994) Phytochemical Dictionary of the Leguminosae, Vol I. Plants and Their Constituents. Chapman and Hall, New York
- Blount JW, Dixon RA, Paiva NL (1993) Stress responses in alfalfa (Medicago sativa L.) XVI. Antifungal activity of medicarpin and its biosynthetic precursors; implications for the genetic manipulation of stress metabolites. Physiol Mol Plant Pathol 41: 333–349 [Google Scholar]
- Branca F, Lorenzetti S (2005) Health effects of phytoestrogens. Forum Nutr 57: 100–111 [DOI] [PubMed] [Google Scholar]
- Bridge MA, Klarman WL (1973) Soybean phytoalexin, hydroxyphaseollin, induced by ultraviolet irradiation. Phytopathology 63: 606–609 [Google Scholar]
- Charrier B, Coronado C, Kondorosi A, Ratet P (1995) Molecular characterization and expression of alfalfa (Medicago sativa L) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant Mol Biol 29: 773–786 [DOI] [PubMed] [Google Scholar]
- Cornwell T, Cohick W, Raskin I (2004) Dietary phytoestrogens and health. Phytochemistry 65: 995–1016 [DOI] [PubMed] [Google Scholar]
- Dixon RA (1999) Isoflavonoids: biochemistry, molecular biology and biological functions. In U Sankawa, ed, Comprehensive Natural Products Chemistry, Vol 1. Elsevier, Oxford, pp 773–823
- Dixon RA (2004) Phytoestrogens. Annu Rev Plant Biol 55: 225–261 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Steele CL (1999) Flavonoids and isoflavonoids—a gold mine for metabolic engineering. Trends Plant Sci 4: 394–400 [DOI] [PubMed] [Google Scholar]
- Hadwiger LA, Schwochau ME (1971) Ultraviolet light-induced formation of pisatin and phenylalanine ammonia lyase. Plant Physiol 47: 588–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55: 481–504 [DOI] [PubMed] [Google Scholar]
- He X-Z, Dixon RA (2000) Genetic manipulation of isoflavone 7-O-methyltransferase enhances the biosynthesis of 4′-O-methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell 12: 1689–170211006341 [Google Scholar]
- Hellens R, Mullineaux P (2000) A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446–451 [DOI] [PubMed] [Google Scholar]
- Hipskind JD, Paiva NL (2000) Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol Plant Microbe Interact 13: 551–562 [DOI] [PubMed] [Google Scholar]
- Jung W, Chung I-M, Heo H-Y (2003) Manipulating isoflavone levels in plants. Plant Biotechnol J 5: 149–155 [Google Scholar]
- Jung W, Yu O, Lau S-MC, O'Keefe DP, Odell J, Fader G, McGonigle B (2000. a) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18: 208–212 [DOI] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau S-MC, O'Keefe DP, Odell J, Fader G, McGonigle B (2000. b) Erratum. Nat Biotechnol 18: 559. [DOI] [PubMed] [Google Scholar]
- Kristensen C, Morant M, Olsen CE, Ekstrom CT, Galbraith DW, Moller BL, Bak S (2005) Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci USA 102: 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper HA, Kleter GA, Noteborn HPJM, Kok EJ (2001) Assessment of the food safety issues related to genetically modified foods. Plant J 27: 503–528 [DOI] [PubMed] [Google Scholar]
- Leath K (1990) Spring black stem and leaf spot. In D Stuteville, DC Erwin, eds, Compendium of Alfalfa Diseases, Ed 2. American Phytopathological Society, St. Paul, pp 16–17
- Liu C-J, Blount JW, Steele CL, Dixon RA (2002) Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc Natl Acad Sci USA 99: 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-J, Huhman D, Sumner LW, Dixon RA (2003) Regiospecific hydroxylation of isoflavones by cytochrome P450 81E enzymes from Medicago truncatula. Plant J 36: 471–484 [DOI] [PubMed] [Google Scholar]
- Maldonado-Mendoza IE, Dewbre GR, van Buuren ML, Vershaw WK, Harrison MJ (2002) Methods to estimate the proportion of plant and fungal RNA in an arbuscular mycorrhiza. Mycorrhiza 12: 67–74 [DOI] [PubMed] [Google Scholar]
- Markovits J, Linassier C, Fosse P, Couprie J, Pierre J, Jacquemin-Sablon A, Saucier JM, Le Pecq JB, Larsen AK (1989) Inhibitory effects of the tyrosine kinase inhibitor genistein on mammalian DNA topoisomerase II. Cancer Res 49: 5111–5117 [PubMed] [Google Scholar]
- Messina M, Erdman J, Setchell KDR (2004) Introduction to and perspectives from the fifth international symposium on the role of soy in preventing and treating chronic diseases. J Nutr 134: 1205S–1206S [DOI] [PubMed] [Google Scholar]
- Okura A, Arakawa H, Oka H, Yoshinari T, Monden Y (1988) Effect of genistein on topoisomerase activity and on the growth of [Val 12]Ha-ras-transformed NIH 3T3 cells. Biochem Biophys Res Commun 157: 183–189 [DOI] [PubMed] [Google Scholar]
- Paiva NL, Oommen A, Harrison MJ, Dixon RA (1994) Regulation of isoflavonoid metabolism in alfalfa. Plant Cell Tissue Organ Cult 38: 213–220 [Google Scholar]
- Pan B, Zhang F, Smith DL (1998) Genistein addition to the rooting medium of soybean at the onset of nitrogen fixation increases nodulation. J Plant Nutr 21: 1631–1639 [Google Scholar]
- Ray H, Yu M, Auser P, Blahut-Beatty L, McKersie B, Bowley S, Westcott N, Coulman B, Lloyd A, Gruber MY (2003) Expression of anthocyanins and proanthocyanins after transformation of alfalfa with maize Lc. Plant Physiol 132: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367: 147–150 [DOI] [PubMed] [Google Scholar]
- Stochmal A, Piacente S, Pizza C, DeRiccardis F, Leitz R, Oleszek W (2001. a) Alfalfa (Medicago sativa L.) flavonoids. 1. Apigenin and luteolin glycosides from aerial parts. J Agric Food Chem 49: 753–758 [DOI] [PubMed] [Google Scholar]
- Stochmal A, Simonet AM, Macias FA, Oleszek W (2001. b) Alfalfa (Medicago sativa L.) flavonoids. 2. Tricin and chrysoeriol glycosides from aerial parts. J Agric Food Chem 49: 5310–5314 [DOI] [PubMed] [Google Scholar]
- Thomas MR, Johnson LB, White FF (1990) Selection of interspecific somatic hybrids of Medicago by using Agrobacterium transformed tissue. Plant Sci 69: 189–198 [Google Scholar]
- Thomas MR, Rose RJ (1988) Enrichment for Nicotiana heterokaryons after protoplast fusion and subsequent growth in agarose microdrops. Planta 175: 396–402 [DOI] [PubMed] [Google Scholar]
- Trieu AT, Burleigh SH, Kardailsky IV, Maldonado-Mendoza IE, Versaw WK, Blaylock LA, Shin H, Choiu T-J, Katagi H, Dewbre GR, et al (2000) Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J 6: 531–542 [DOI] [PubMed] [Google Scholar]
- Yu O, Jung W, Shi J, Croes RA, Fader GM, McGonigle B, Odell JT (2000) Production of the isoflavones genistein and daidzein in non-legume dicot and monocot tissues. Plant Physiol 124: 781–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O, Shi J, Hession AO, Maxwell CA, McGonigle B, Odell JT (2003) Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63: 753–763 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.