Abstract
Serpin superfamily proteins, most of which are serine protease inhibitors, share an unusual mechanism rooted in their conserved metastable tertiary structure. Although serpins have been identified in isolated members of archea, bacteria, and plants, a remarkable expansion is found in vertebrates. The chicken protein ovalbumin, a storage protein from egg white, lacking protease inhibitory activity, is an historical member of the superfamily and the founding member of the subgroup known as ov-serpins (ovalbumin-related serpins) or clade B serpins. In the human, ov-serpins include 13 proteins involved in the regulation of inflammation, apoptosis, angiogenesis, and embryogenesis. Here, a detailed analysis of the chicken (Gallus gallus) genome identified 10 clade B serpin genes that map to a single ≈150-kb locus and contain the signature protein sequence of serpins and the gene structure of ov-serpins, with either seven or eight exons. Orthologues of PAI-2 (SERPINB2), MNEI (SERPINB1), PI-6 (SERPINB6), and maspin (SERPINB5) are highly conserved. Comparison with human ov-serpins identified avian-specific and mammal-specific genes. Importantly, a unique model of mammalian ov-serpin evolution is revealed from the comparative analysis of the chicken and human loci. The presence of a subset of ov-serpin genes in zebrafish (Danio rerio) gives insight into the ancestral locus. This comparative genomic study provides a valuable perspective on the evolutionary pathway for the clade B serpins, allowing the identification of genes with functions that may have been conserved since the origin of vertebrates. In addition, it suggests that “newer” serpins, such as ovalbumin, have contributed to vertebrate adaptation.
Keywords: ov-serpin, multigene locus, maspin, MNEI, zebrafish
This year marks the 25th anniversary of a landmark study by Lois Hunt and Margaret Dayhoff (1), identifying the surprising phylogenetic relationship between the genes for the chicken-egg storage protein ovalbumin and two human-plasma protease inhibitors α1-antitrypsin and antithrombin III. That discovery, among the first to use protein sequence alignment to identify gene families, helped launch the modern field of Bioinformatics and provided the cornerstone for the superfamily now known as serpins. Although a few serpins have evolved functions distinct from protease inhibition, such as storage (ovalbumin), blood pressure regulation (angiotensinogen), or molecular chaperoning (heat shock protein 47), most serpins are serine protease inhibitors. Indeed, serpins participate in the precise regulation of key proteolytic events and cascades indispensable to multicellular organisms (2).
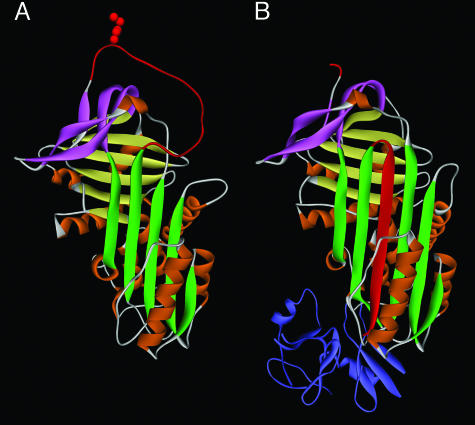
Serpin superfamily proteins share a tertiary structure, consisting of nine alpha helices and three beta sheets, with an exposed and mobile reactive center loop (RCL) that serves as bait for target proteases (3) (Fig. 1A). This highly ordered native structure is metastable (4), and it is the strain of this thermodynamically unstable structure that is the basis for the unique inhibitory mechanism, in which the serpin, after binding its protease, rapidly converts from the native stressed (S) conformation to a relaxed (R) hyperstable structure. As part of the S → R rearrangement, the cleaved RCL is inserted as a new strand in the center of the molecule, and the bound protease is irreversibly trapped and inactivated (5, 6) (Fig. 1B).
Fig. 1.
Protein structure of inhibitory serpins. (A) The native or stressed (S) conformation. The five-stranded β-sheet A is shown in green. The RCL shown at the top in red includes the specificity-determining bait “P1” residue (side chain is shown), which is exposed and free to interact with protease. In the inhibitory reaction, the protease approaches from the top, binds reversibly at the P1 site, and initiates proteolysis by forming a covalent acyl “intermediate” bond at P1 and cleaving after P1. Cleavage releases the S conformation, allowing opening of β-sheet A and rapid topological conversion to the relaxed (R) conformation. (B) The serpin–protease complex. The serpin is in the R conformation. Note that the cleaved RCL has been inserted in the center of the serpin as an additional strand in β-sheet A and has dragged the covalently bound protease (shown in blue) by >70 Å to the opposite end of the serpin. The mechanism, called “kinetic trapping,” irreversibly inactivates the protease by gross distortion of its catalytic center. Protein coordinates in A and B are from α1-antitrypsin (SERPINA1) in the native state (1QLP) and in complex with protease (1EZX), respectively. Sheet B is shown in yellow, sheet C in pink, and the nine helices in orange.
Not unexpectedly, a number of residues were found to be strictly conserved in distant species, because they occupy strategic buried positions that maintain the overall serpin structure (7, 8). Residues that contribute to the hinge, breach, and shutter regions (described in Results and Discussion), required for swift S → R rearrangement, are conserved in inhibitory serpins. In contrast, even a single missense mutation in the RCL can suffice to change specificity and, thereby, generate a new function in a duplicated gene. Serpins are found in scattered archea, bacteria, and unicellular eukaryotes, and the origin of the serpin fold is unresolved (9). The superfamily is dramatically expanded in multicellular organisms, likely because of the advantage of efficient regulation of endogenous and exogenous proteases (10) and, from an evolutionary perspective, because of the adaptability of the serpin structure and mechanism.
As the number of known serpins increased, a closer phylogenetic analysis verified the prediction of Hunt and Dayhoff (1), and a subgroup of ovalbumin-related serpins (ov-serpins) was identified (11). The ov-serpin family, now recognized as clade B serpins (2), comprises 13 genes in the human that have regulatory roles in inflammation, apoptosis, fibrinolysis, angiogenesis, and embryogenesis (reviewed in ref. 12). As described in Methods, we use the recently completed chicken genome sequence (13) to examine the structures and organization of clade B serpins in this species and to perform a comparative genomic analysis, which provides substantial insight into the evolutionary pathway of this family.
Methods
Sequences. Chicken (Gga) and zebrafish (Dre) serpins were searched by using the programs blast (14) and discontiguous mega blast with the human (Hsa) clade B serpin coding sequences (Build 35.1). The databases searched were the genomic Build 1.1 for both Gga and Dre and the nonRefSeq protein and EST databases of Gga and Dre (January 2005). blast searches were performed without filter and low stringency (expect = 10) to maximize matches. The protein sequences of matches were back-searched by using blastp to the Hsa nonredundant (nr) database. Identified clade B serpins of Gga and Dre were used in a second round of searches of their respective genome database and trace archives, as described above. The organization of the Gga ov-serpin locus was based on genome Build 1.1 by using the program mapviewer on the National Center for Biotechnology Information web site. Exon/intron boundaries were searched with the program genscan (15). Accession numbers, annotation, and proposed nomenclature of chicken and zebrafish genes, respectively, are shown in Tables 1 and 2, which are published as supporting information on the PNAS web site.
Alignments and Phylogenetic Analysis. The percentage of sequence identity was calculated by pairwise blast with the vector nti suite 8 for PC (Informax, Bethesda), with gaps included. Protein alignments were performed by using the program clustalw 1.8 (16), and manually optimized. Phylogenetic trees were inferred from the protein alignments by the neighbor-joining-distance-based method (observed divergence and global gap removal) (17) and bootstrapped 1,000 times by using the program phylowin 2.0 (18).
Results and Discussion
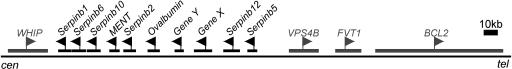
Identification of 10 Chicken Clade B Serpins. There are four known ov-serpin genes in chicken: ovalbumin, the related genes X and Y (19), and MENT (Mature Erythrocyte Nuclear Termination state-specific protein) (20). Using the sequences of the 13 human and 4 chicken ov-serpins, blast searches of publicly available protein and nucleic acid databases of chicken (Gallus gallus, Gga) yielded 10 candidate ov-serpin genes, all of which map to an ≈150-kb locus on chromosome 2q (Fig. 2). Multiple comparison methods: percentage of sequence identity, protein alignment, gene structure and position within the locus, and analysis of conserved and gene-specific residues collectively identified six of these as orthologues of human genes.
Fig. 2.
Chicken ov-serpin locus. The ten ov-serpins and flanking genes with their respective orientation on chicken chromosome 2q are drawn to scale. Assignment of gene names is by established convention in the case of ovalbumin, gene Y, gene X, and MENT or is based on orthology with human genes (presented in the text). An alternate name for MENT is serpinb10b. Accession numbers are shown in Table 1, which is published as supporting information on the PNAS web site. With the exception of chicken serpinb10, these genes have been found in EST databanks. The serpin cluster is flanked downstream by the Werner helicase-interacting protein (WHIP) and upstream by the vacuolar sorting protein 4b (VPS4B), follicular lymphoma variant translocation 1 (FVT1), and B cell leukemia/lymphoma 2 (BCL2).
Protein Identities and Alignment. The percentage of protein sequence identity was calculated for all pairs of chicken and human clade B serpins. Scores of the six pairs designated as orthologues, SERPINB1, -B5, -B6, -B2, -B10, and -B12, ranged from 69% to 46% (Fig. 3A). These pairs were also grouped in the neighbor-joining phylogenetic tree (Fig. 3B). Chicken serpinb10, the orthologue of human SERPINB10, and MENT (serpinb10b) clustered together in the phylogenetic tree and are designated as paralogues; they share 57% sequence identity with each other. Serpinb10 is 50% and MENT is 47% identical to human SERPINB10. Alignment of the 10 chicken and 13 human genes (see Fig. 9, which is published as supporting information on the PNAS web site) and the neighbor-joining phylogenetic tree (Fig. 3B) indicate that ovalbumin and the ovalbumin-like gene Y and gene X are paralogues and have no orthologue in humans. In accordance with the serpin nomenclature guidelines (2), ovalbumin, gene Y, and gene X become serpinb14, serpinb14b, and serpinb14c, respectively. Conversely, the chicken genome has no orthologue for human SERPINB8 or -B9, which cluster with SERPINB6, or for SERPINB3 (squamous-cell carcinoma antigen-1, SCCA-1), -B4 (SCCA-2), -B7 (megsin), -B11 or -B13 (headpin, hurpin), which cluster with SERPINB12 (Fig. 3 A and B), suggesting that these genes were either lost in the chicken or arose after the avian and mammalian lineages diverged. The finding that the chicken locus contains no pseudogenes favors the latter interpretation.
Fig. 3.
Comparative analyses of 10 chicken and 13 human clade B serpins. (A) Percentage of amino acid identity between chicken and human clade B serpins. Pairs with concordant highest-identity scores are highlighted. (B) Neighbor-joining tree of the chicken and human clade B serpins with human SERPINA1 (α1-antitrypsin) used as a root (348 sites compared). Code for bootstrap values is provided in the figure. (C) Gene structure of chicken clade B serpins. Boxes representing exons are drawn to scale with untranslated regions shown open, coding regions common to seven- and eight-exon genes shown shaded, and coding regions exclusive to the eight-exon genes shown black. Introns, which are shown as lines linking exons, are not drawn to scale. By using standard serpin numbering, based on α1-antitrypsin (7), the locations and phase of the introns are: 5′ untranslated region; amino acid (aa) 78, phase 0; aa 85, phase 0 (eight-exon genes only); aa 128, phase 0; aa 167, phase 1; aa 212, phase 0; and aa 262, phase 0.
Gene Structures. Serpin superfamily genes have extremely variable exon/intron organization patterns, and the analysis of intron locations has proved to be a valuable method for determining phylogenic relatedness within this superfamily (11, 21). Characterized clade B genes have either of two completely conserved patterns: a seven-exon/six-intron structure or an eight-exon structure, with identical placement of six of the introns. Each of the chicken genes has one of these structures, further supporting their classification as clade B serpins (Fig. 3C). Moreover, the chicken genes that are orthologues of seven-exon human genes, chicken serpinb1, -b6, and -b5, have the seven-exon structure, and the genes that are orthologues of eight-exon human genes, chicken serpinb10, -b10b/MENT, -b2, and -b12, have the eight-exon structure. In one of the earliest gene structure analyses, ovalbumin/serpinb14, gene Y/serpinb14b, and gene X/serpinb14c were shown to have the eight-exon structure (22).
Conserved and Variable Residues. Because three-dimensional crystal structures have been established for several serpins, conserved residues identified for the superfamily have been correlated with structural elements, including the breach, which forms the opening of β-sheet A, where the cleaved RCL is inserted, and the shutter region that lies below the point of entry; these elements are critical for the rapid S → R rearrangement required for efficient protease inhibition (7, 8) (Fig. 1). Analysis of 60 conserved positions distributed throughout the linear sequence (Fig. 9) shows that all but one of the chicken genes have ≥95% conservation (five or fewer variant residues), suggesting conservation of the overall serpin fold. The exception, chicken serpinb5, has 15 variant residues, many in the breach and shutter regions. Ten of the variant positions are shared with human SERPINB5, which is not a protease inhibitor and does not undergo the S → R rearrangement (23). The two SERPINB5 orthologues share high overall amino acid identity with each other but are the most divergent within clade B (Fig. 3 A and B).
We also analyzed the RCL, the N-terminal portion of which represents the hinge residues required for insertion of the loop in the inhibitory rearrangement reaction (24). Consensus hinge regions were found in chicken serpinb1, -b6, -b2, -b12, -b10, and -b10b (Fig. 4). The consensus hinge regions, together with the overall conservation of the serpin fold, make it likely that these genes encode protease inhibitors. The residues at the scissile bond, P1–P1′ in the canonical nomenclature, are the critical determinants of inhibitory specificity, and adjacent residues ≈P5–P4′ can influence the efficiency of inhibition. Therefore, orthologous genes with identical P1–P1′ and conservatively altered adjacent residues are likely to share function. These criteria are met for chicken serpinb1, the orthologue of MNEI (Monocyte Neutrophil Elastase Inhibitor) (SERPINB1), an efficient inhibitor of elastase-like and chymotrypsin-like serine proteases; serpinb6, the orthologue of PI-6 (SERPINB6), which inhibits thrombin, trypsin, cathepsin G, and chymotrypsin; serpinb2, the orthologue of plasminogen-activator inhibitor-2 (PAI-2/SERPINB2); and, possibly, for serpinb12, the orthologue of SERPINB12 but not for chicken serpinb10 or serpinb10b/MENT, which differ at the P1–P1′ positions from SERPINB10, an inhibitor of thrombin (Fig. 4).
Fig. 4.
Alignment of the RCL of chicken clade B serpins and human orthologues. Human SERPINB8 and -B9 are included for comparison. Shown at the top is the canonical numbering proximal or distal to the specificity-determining scissile bond (P1–P1′, boxed). The consensus hinge region of inhibitory serpins is E at P17, E/K/R at P16, G at P15, T/S at P14, small aliphatic residues (A/G/S) at P12-P9 (8), and T/SatP8 (25). Residues are colored according to type: polar uncharged (green), acidic (red), basic (blue), and nonpolar (yellow). Information on the inhibitory activity of human ov-serpins is reviewed in ref 12. Of note, chicken ovalbumin and human SERPINB5/maspin are not protease inhibitors.
The RCL of chicken ovalbumin/serpinb14 has multiple deviations in the hinge, including a bulky arginine residue at P14 and an alanine rather than a threonine at P8 (Fig. 4). Also, two of the variant structural residues, Ser-53 → Cys and Ser-56 → Ala, are shutter-region residues that function to receive the loop at P8 in the full RCL-insertion reaction (25). Although it is not an inhibitor, ovalbumin cleaved at P1–P1′ in the RCL undergoes slow and incomplete insertion of the loop (P15 to P10) with a modest increase in thermal stability (26). In its physiological role, ovalbumin, which is synthesized in the hen oviduct (27), is the major protein of egg white and is consumed as a nutrient supplemental to the yolk in developing avian embryos. During embryogenesis, ovalbumin changes to a more heat-stable conformation, while migrating from egg white to amniotic fluid, embryonic organs, and yolk (28). Whether this conformational change of ovalbumin is related (29) or unrelated (30) to classical serpin rearrangement is disputed. Relative to ovalbumin, the paralogues gene Y and gene X have hinge regions that deviate less from the consensus, and these genes, particularly gene Y, may encode inhibitory serpins.
Finally, the nearly identical RCL of chicken serpinb5 and human SERPINB5/maspin deviate from the consensus for inhibitors at multiple hinge positions and are four amino acids shorter than other serpins (Fig. 4). Despite the deviant sequence and lack of protease inhibition, this region is required for the inhibition of tumor-cell invasion by human SERPINB5 (maspin) (31). The recently obtained three-dimensional structure of human SERPINB5/maspin revealed an atypically rigid RCL that lies closer to the core of the molecule and is stabilized by interactions of specific loop residues with residues in β-sheet C (32), all of which are present also in the chicken orthologue, strongly suggesting shared function. Although the human protein was named for its link with mammary gland cancer, SERPINB5/maspin likely has important functions in embryogenesis, as suggested by the early embryonic lethality of the maspin knockout mice (33). These features indicate that serpinb5 of chicken would be a fruitful system for functional studies.
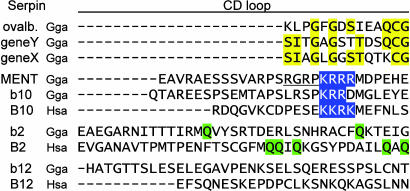
Interhelical CD Loop. The additional sequence in eight-exon ovserpins contributes to a variable-length loop between helices C and D, which, in structural models, does not perturb the serpin fold (7). Using the clusters established by the phylogenetic tree, we analyzed the CD loops for the possible presence of shared motifs. For the serpinb14 paralogues, ovalbumin, gene Y, and gene X, sequence identities are, indeed, found in the CD loop, which may indicate a shared function for this region or may simply reflect recent duplication (Fig. 5). In the second group, chicken serpinb10b/MENT and human SERPINB10/bomapin are both known to localize to the nucleus, and both have nuclear targeting signals within the CD loop: KKRK in SERPINB10 (34) and KRRR in MENT (20). The third member of this group, the previously undescribed chicken serpinb10, has a related motif (KRR) in a comparable position. However, the DNA-binding “AT hook,” which is required for chromatin condensation, is found only in MENT (Fig. 5), a protein characterized in chicken nucleated erythrocytes long before it was recognized as a serpin (20). The chromatin-condensation activity of MENT has yet to be described in another serpin, suggesting that this serpin function arose after birds and mammals split. Functional divergence has also been noted between SERPINB10 orthologues in humans (bomapin), rats (trespin), and even different strains of mice (35).
Fig. 5.
CD loops of chicken eight-exon clade B serpins. For the serpinb14 (ovalbumin) group, identical residues in the loop are highlighted in yellow. For the SERPINB10 group, nuclear-localization signals or related motif (chicken serpinb10) are indicated in blue. The AT-rich DNA-binding motif, present only in MENT, is underlined. For the SERPINB2 orthologues, glutamine residues required for crosslinking of human SERPINB2/PAI-2 are highlighted in green. In standard numbering, based on α1-antitrypsin, the CD loop replaces amino acids 84–87.
For SERPINB2 orthologues, the CD loops lack sequence homology but share greater length (37 aa), expected to support greater loop projection. SERPINB2/PAI-2 has additional functions that require the CD loop, including crosslinking to fibrin and other proteins (36, 37) and protecting cells from TNF-α-induced apoptosis (38). Glutamine residues required for crosslinking of human PAI-2 by transglutaminase (37), although fewer, are also found in the CD loop of chicken serpinb2 (Fig. 5). For the CD loops of SERPINB12 orthologues, there is no homology nor are there shared motifs or shared length, and no function has been reported for this region.
A Unique Model of Vertebrate Clade B Serpin Evolution. Unlike the chicken genome, with its single ov-serpin locus, the human genome has two loci, one at 6p25 and the other at 18q21 (12). In rodents, orthologues of the human genes are found on syntenic loci on mouse chromosomes 13A and 1D (39, 40) and on rat chromosomes 17p12 and 13p13, respectively (35), indicating that the two loci existed in the common ancestor of humans and rodents. One model explained the origin of the second locus by a duplication en masse of the chromosome 6p genes to chromosome 18 (41). A second model explained the process by two interchromosomal and multiple intrachromosomal duplications (42).
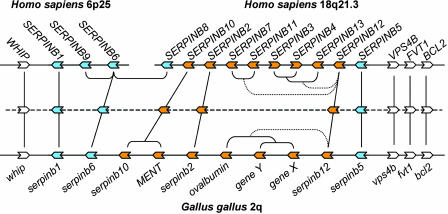
Here, we propose a radically different and more parsimonious evolution of mammalian ov-serpins, whereby the two loci were generated by the split of the ancestral single locus. This model is illustrated in Fig. 6 and is supported by the following points: (i) the relative position and orientation of the human and chicken orthologues are conserved;. (ii) the chicken locus is flanked downstream by the WHIP gene, the orthologue of which is downstream of SERPINB1 on human 6p25, mouse 13A, and rat 17p12; (iii) the three genes upstream of the chicken locus are the orthologues of VPS4B, FVT1, and BCL2, which are upstream of SERPINB5 on human 18q21, mouse 1D, and rat 13p13; (iv) the relative orientation of WHIP, VPS4B, FVT1, and BCL2 genes to the adjacent chicken and mammal ov-serpins is conserved; (v) genes with homology to WHIP on human 18q21.3 and to VPS4B, FVT1, and BCL2 on human 6p25 are absent, and they are also absent in the respective syntenic regions in rodents.
Fig. 6.
Comparison of human and chicken clade B serpin loci. The two human loci are shown at the top. We propose that these existed as a single locus that split, before the divergence of primates and rodents. The chicken locus is shown at the bottom. Serpin genes and their orientation are represented by blue (seven-exon genes) or orange (eight-exon genes) block arrows. Flanking genes are shown in white. Vertical lines link human–chicken orthologues. The deduced ancestor of chicken and mammals is shown in the middle, with phylogenetic relationships indicated. The loci are not drawn to scale: the chicken locus is ≈150 kb, whereas the human 6p25 and 18q23 are ≈250 kb and ≈600 kb, respectively.
The site of partition of the mammalian ancestor locus is depicted in the schematic of Fig. 6 as occurring between SERPINB6 and SERPINB8. In actuality, the human loci have two SERPINB8-like pseudogenes that are not depicted in the figure, one upstream of SERPINB6 on 6p25 and one downstream of SERPINB8 on 18q21.3 (42). So, more precisely, the mammalian ancestor locus split between the two pseudogenes, leaving SERPINB1, -B9, -B6, and a -B8-like pseudogene on one side and the other -B8-like pseudogene with SERPINB8, all of the eight-exon genes, and SERPINB5 on the other side. In the common ancestor of mammals and birds (represented in the middle in Fig. 6), the eight-exon genes were flanked downstream and upstream by seven-exon genes that were already highly divergent, suggesting that the eight-exon structure arose from a duplicated ancestral seven-exon gene.
After divergence of the two lineages, SERPINB6 duplicated twice in the mammalian ancestor to give SERPINB9 and SERPINB8 (Fig. 6), which, in turn, duplicated before the locus was split, giving rise to what are now two -B8-like pseudogenes (not shown). The three eight-exon genes in the ancestor locus have been maintained in chicken and mammals. SERPINB12 was duplicated independently in birds and mammals to give three new paralogues in chicken (ovalbumin, gene Y, and gene X) and five new paralogues in human (SERPINB7, -B11, -B3, -B4, and -B13) (Fig. 6). Further rounds of gene duplication and loss occurred in rat and mouse ov-serpins, resulting in complex loci, requiring detailed work to identify orthologues for comparative and gene-deletion studies (39, 40, 43).
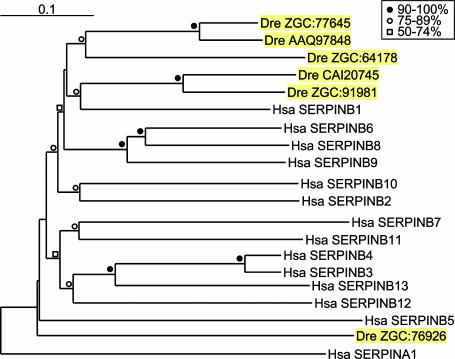
Ov-Serpins of Zebrafish. To further investigate the root of the clade B serpins in vertebrates, chicken and human nucleic acid and protein sequences were used to search the public databases of zebrafish (Danio rerio). Six ov-serpins were identified, and the phylogenetic analysis revealed that five of them clustered with some degree of certainty with SERPINB1 and that the remaining gene, ZGC:76926, diverged early from the ov-serpin ancestor (Fig. 7). Protein alignment and the percentage of sequence identity, respectively, are shown in Fig. 10 and Table 2, which are published as supporting information on the PNAS website. The five serpinb1-like genes have ≥95% structurally defined residues and conserved hinge regions, suggesting protease-inhibitory function. Serpin nomenclature is not proposed for the zebrafish genes, pending further mapping and sequencing. However, existing evidence is sufficient to identify ZGC:91981 as the orthologue of SERPINB1, including a strikingly conserved RCL. ZGC:91981 and CAI20745 are found within a few kilobases in the zebrafish genome, together with a serpinb1-like pseudogene, and they have greater protein sequence identity with each other (75%) than with human SERPINB1 (57% and 51%, respectively), indicating a relatively recent gene duplication within the fish lineage.
Fig. 7.
Neighbor-joining tree of zebrafish and human clade B serpins. Human SERPINA1 was used as a root (344 sites compared). The code for bootstrap values is included in the figure.
Zebrafish serpinb1-like genes have a seven-exon structure, except ZGC:77645, which has the eight-exon structure. This finding is interesting, because ZGC:77645 has a higher sequence identity with seven-exon genes than with the eight-exon genes of chickens and humans (Fig. 7) and is 75% identical to AAQ97848 (Table 2), indicative of recent duplication. The sixth zebrafish ov-serpin ZGC:76926 varies substantially at conserved positions (16 variations) and, in this respect, resembles human and chicken SERPINB5 (Fig. 10) but lacks the structural features identified in human and chicken SERPINB5/maspin that are essential for inhibition of tumor-cell invasion activity.
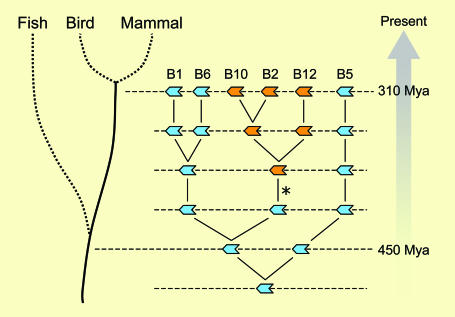
Root of Clade B Evolution. By including the zebrafish genes in the analysis, a more complete model of ov-serpin evolution and diversification emerges, in which a single seven-exon gene (Fig. 8, bottom line) evolved into the six-gene locus deduced for the last common ancestor of birds and mammals, 310 million years ago (Fig. 8, top line). The most parsimonious model involves a series of duplication events, the first of which generated the ancestor of SERPINB5, the most divergent ov-serpin. The resulting two-gene locus is proposed as the last common ancestor of fish, birds, and mammals, 450 million years ago, consistent with finding only serpinb1 and -b5-like genes in zebrafish. The events responsible for the multiple serpinb1-like genes in modern zebrafish may include an early duplication of the locus as part of the whole-genome duplication in the ray-finned fish lineage (44), in addition to the recent duplications noted above. In the tetrapod lineage, the SERPINB1 ancestor duplicated twice more, and a gene in a central position gained genetic material within the third exon, in a segment encoding the junction between helices C and D [Fig. 8, (*)], generating the ancestral tetrapod eight-exon ov-serpin and the first large CD loop. A similar independent insertional event likely explains the recent generation of an eight-exon structure in fish.
Fig. 8.
Evolution of the clade B serpins. The model shows serpin genes as blue (seven-exon genes) or orange (eight-exon genes) block arrows. Shown, starting at the bottom, is the ancestral locus with a single seven-exon gene, followed by subsequent stages to generate the ancestor locus of birds and mammals deduced in Fig. 6 and shown here at the top. Within the tetrapod lineage, the asterisk (*) represents an event in which additional sequence, including a CD loop, was added within a seven-exon gene, generating an eight-exon gene. Further details are in the text.
The absence of tertiary structure constraints on the amino acids linking helices C and D allowed the newly formed eight-exon genes to accumulate neutral mutations, in some cases, or to develop new functions, in other cases, as illustrated in chicken MENT and mammalian PAI-2 (Fig. 5). Because pressure to maintain original function would be decreased on duplicated genes, some “newer” genes may have lost the efficiency of their protease-inhibitory activity, while acquiring new functions. Conversely, ancient seven-exon genes retain a highly conserved efficient reactive center, indicative of a deeply rooted antiprotease role, such as that of SERPINB1.
Conclusion
The study of mammal, chicken, and zebrafish ov-serpins offers an insightful perspective on the evolution of these important proteins in vertebrates. High duplication rates, followed by acquisition of new functions or redundancy and gene loss, reflect the contemporary dynamic nature of the ov-serpins. The conserved protease-inhibitory activities of the orthologues of SERPINB1, SERPINB2, and SERPINB6 and their known roles in immunity suggest that their antiprotease activity is anchored with the regulation of the immune system of all vertebrates. Finally, and most provocatively, the link between ovalbumin as a bird-specific clade B serpin and its preponderant role in avian eggs suggest that the new function acquired by this ov-serpin may have played a role in adaptation of these vertebrates to new habitats and lifestyles.
Supplementary Material
Acknowledgments
We thank Alvin Davis, III, Margaret Collins, and Susanna Remold for critical reading of the manuscript and suggestions for data presentation. This work was supported by National Institutes of Health Grant HL66548 and a grant from the Cystic Fibrosis Foundation.
Author contributions: C.B. and E.R.-O. designed research; C.B. and E.R.-O. performed research; C.B. and E.R.-O. analyzed data; and C.B. and E.R.-O. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: RCL, reactive center loop.
References
- 1.Hunt, L. T. & Dayhoff, M. O. (1980) Biochem. Biophys. Res. Commun. 95, 864–871. [DOI] [PubMed] [Google Scholar]
- 2.Silverman, G. A., Bird, P. I., Carrell, R. W., Church, F. C., Coughlin, P. B., Gettins, P. G., Irving, J. A., Lomas, D. A., Luke, C. J., Moyer, R. W., et al. (2001) J. Biol. Chem. 276, 33293–33296. [DOI] [PubMed] [Google Scholar]
- 3.Stein, P. E., Leslie, A. G., Finch, J. T., Turnell, W. G., McLaughlin, P. J. & Carrell, R. W. (1990) Nature 347, 99–102. [DOI] [PubMed] [Google Scholar]
- 4.Carrell, R. W. & Owen, M. C. (1985) Nature 317, 730–732. [DOI] [PubMed] [Google Scholar]
- 5.Stratikos, E. & Gettins, P. G. (1999) Proc. Natl. Acad. Sci. USA 96, 4808–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huntington, J. A., Read, R. J. & Carrell, R. W. (2000) Nature 407, 923–926. [DOI] [PubMed] [Google Scholar]
- 7.Huber, R. & Carrell, R. W. (1989) Biochemistry 28, 8951–8966. [DOI] [PubMed] [Google Scholar]
- 8.Irving, J. A., Pike, R. N., Lesk, A. M. & Whisstock, J. C. (2000) Genome Res. 10, 1845–1864. [DOI] [PubMed] [Google Scholar]
- 9.Irving, J. A., Steenbakkers, P. J., Lesk, A. M., Op den Camp, H. J., Pike, R. N. & Whisstock, J. C. (2002) Mol. Biol. Evol. 19, 1881–1890. [DOI] [PubMed] [Google Scholar]
- 10.Hill, R. E. & Hastie, N. D. (1987) Nature 326, 96–99. [DOI] [PubMed] [Google Scholar]
- 11.Remold-O'Donnell, E. (1993) FEBS Lett. 315, 105–108. [DOI] [PubMed] [Google Scholar]
- 12.Silverman, G. A., Whisstock, J. C., Askew, D. J., Pak, S. C., Luke, C. J., Cataltepe, S., Irving, J. A. & Bird, P. I. (2004) Cell. Mol. Life Sci. 61, 301–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillier, L. W., Miller, W., Birney, E., Warren, W., Hardison, R. C., Ponting, C. P., Bork, P., Burt, D. W., Groenen, M. A., Delany, M. E., et al. (2004) Nature 432, 695–716.15592404 [Google Scholar]
- 14.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 16.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saitou, N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 18.Galtier, N., Gouy, M. & Gautier, C. (1996) Comput. Appl. Biosci. 12, 543–548. [DOI] [PubMed] [Google Scholar]
- 19.Heilig, R., Perrin, F., Gannon, F., Mandel, J. L. & Chambon, P. (1980) Cell 20, 625–637. [DOI] [PubMed] [Google Scholar]
- 20.Grigoryev, S. A., Bednar, J. & Woodcock, C. L. (1999) J. Biol. Chem. 274, 5626–5636. [DOI] [PubMed] [Google Scholar]
- 21.Ragg, H., Lokot, T., Kamp, P. B., Atchley, W. R. & Dress, A. (2001) Mol. Biol. Evol. 18, 577–584. [DOI] [PubMed] [Google Scholar]
- 22.Breathnach, R., Benoist, C., O'Hare, K., Gannon, F. & Chambon, P. (1978) Proc. Natl. Acad. Sci. USA 75, 4853–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pemberton, P. A., Wong, D. T., Gibson, H. L., Kiefer, M. C., Fitzpatrick, P. A., Sager, R. & Barr, P. J. (1995) J. Biol. Chem. 270, 15832–15837. [DOI] [PubMed] [Google Scholar]
- 24.Hopkins, P. C., Carrell, R. W. & Stone, S. R. (1993) Biochemistry 32, 7650–7657. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, A., Stein, P. E., Huntington, J. A. & Carrell, R. W. (2003) J. Biol. Chem. 278, 15116–15122. [DOI] [PubMed] [Google Scholar]
- 26.Huntington, J. A., Fan, B., Karlsson, K. E., Deinum, J., Lawrence, D. A. & Gettins, P. G. (1997) Biochemistry 36, 5432–5440. [DOI] [PubMed] [Google Scholar]
- 27.Palmiter, R. D., Mulvihill, E. R., Shepherd, J. H. & McKnight, G. S. (1981) J. Biol. Chem. 256, 7910–7916. [PubMed] [Google Scholar]
- 28.Sugimoto, Y., Sanuki, S., Ohsako, S., Higashimoto, Y., Kondo, M., Kurawaki, J., Ibrahim, H. R., Aoki, T., Kusakabe, T. & Koga, K. (1999) J. Biol. Chem. 274, 11030–11037. [DOI] [PubMed] [Google Scholar]
- 29.Huntington, J. A., Patston, P. A. & Gettins, P. G. (1995) Protein Sci. 4, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamasaki, M., Takahashi, N. & Hirose, M. (2003) J. Biol. Chem. 278, 35524–35530. [DOI] [PubMed] [Google Scholar]
- 31.Ngamkitidechakul, C., Warejcka, D. J., Burke, J. M., O'Brien, W. J. & Twining, S. S. (2003) J. Biol. Chem. 278, 31796–31806. [DOI] [PubMed] [Google Scholar]
- 32.Al-Ayyoubi, M., Gettins, P. G. & Volz, K. (2004) J. Biol. Chem. 279, 55540–55544. [DOI] [PubMed] [Google Scholar]
- 33.Gao, F., Shi, H. Y., Daughty, C., Cella, N. & Zhang, M. (2004) Development (Cambridge, U.K.) 131, 1479–1489. [DOI] [PubMed] [Google Scholar]
- 34.Chuang, T. L. & Schleef, R. R. (1999) J. Biol. Chem. 274, 11194–11198. [DOI] [PubMed] [Google Scholar]
- 35.Puente, X. S. & Lopez-Otin, C. (2004) Genome Res. 14, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie, H., Lawrie, L. C., Crombie, P. W., Mosesson, M. W. & Booth, N. A. (2000) J. Biol. Chem. 275, 24915–24920. [DOI] [PubMed] [Google Scholar]
- 37.Jensen, P. H., Schuler, E., Woodrow, G., Richardson, M., Goss, N., Hojrup, P., Petersen, T. E. & Rasmussen, L. K. (1994) J. Biol. Chem. 269, 15394–15398. [PubMed] [Google Scholar]
- 38.Dickinson, J. L., Norris, B. J., Jensen, P. H. & Antalis, T. M. (1998) Cell Death Differ. 5, 163–171. [DOI] [PubMed] [Google Scholar]
- 39.Kaiserman, D., Knaggs, S., Scarff, K. L., Gillard, A., Mirza, G., Cadman, M., McKeone, R., Denny, P., Cooley, J., Benarafa, C., et al. (2002) Genomics 79, 349–362. [DOI] [PubMed] [Google Scholar]
- 40.Askew, D. J., Askew, Y. S., Kato, Y., Turner, R. F., Dewar, K., Lehoczky, J. & Silverman, G. A. (2004) Genomics 84, 176–184. [DOI] [PubMed] [Google Scholar]
- 41.Bartuski, A. J., Kamachi, Y., Schick, C., Overhauser, J. & Silverman, G. A. (1997) Genomics 43, 321–328. [DOI] [PubMed] [Google Scholar]
- 42.Scott, F. L., Eyre, H. J., Lioumi, M., Ragoussis, J., Irving, J. A., Sutherland, G. A. & Bird, P. I. (1999) Genomics 62, 490–499. [DOI] [PubMed] [Google Scholar]
- 43.Benarafa, C., Cooley, J., Zeng, W., Bird, P. I. & Remold-O'Donnell, E. (2002) J. Biol. Chem. 277, 42028–42033. [DOI] [PubMed] [Google Scholar]
- 44.Jaillon, O., Aury, J. M., Brunet, F., Petit, J. L., Stange-Thomann, N., Mauceli, E., Bouneau, L., Fischer, C., Ozouf-Costaz, C., Bernot, A., et al. (2004) Nature 431, 946–957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.