Abstract
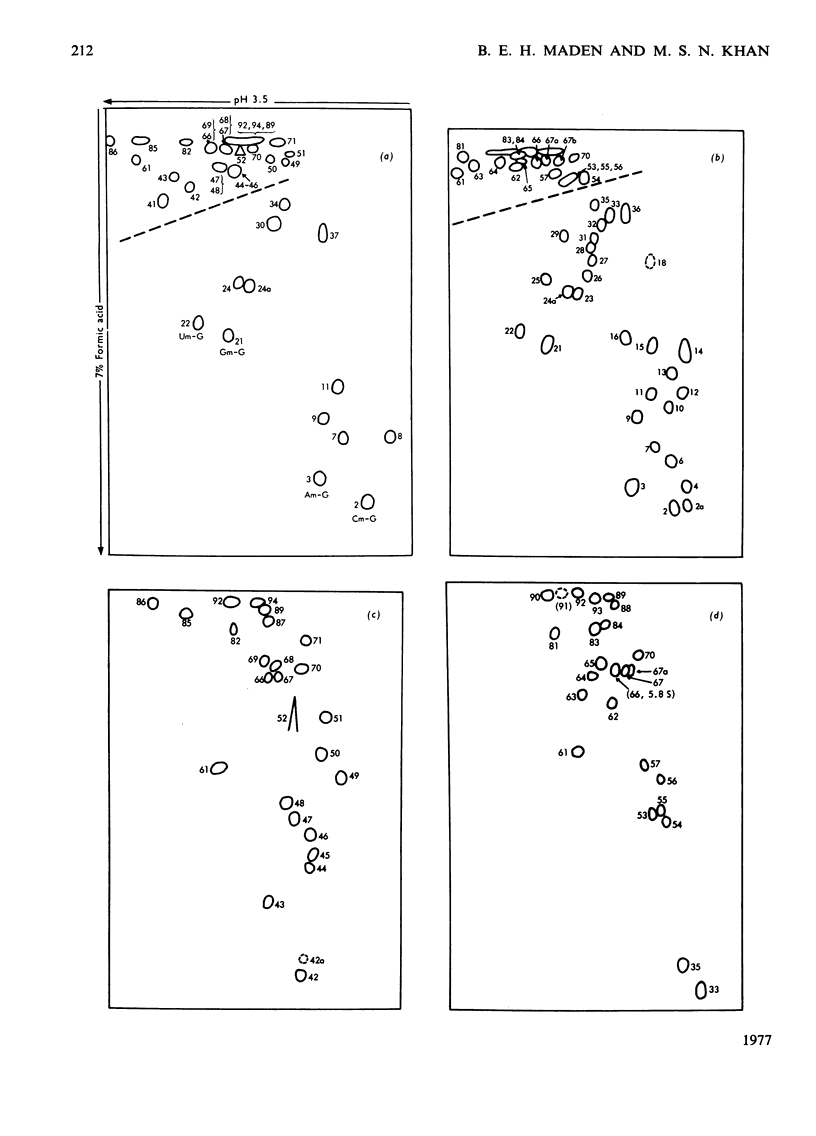
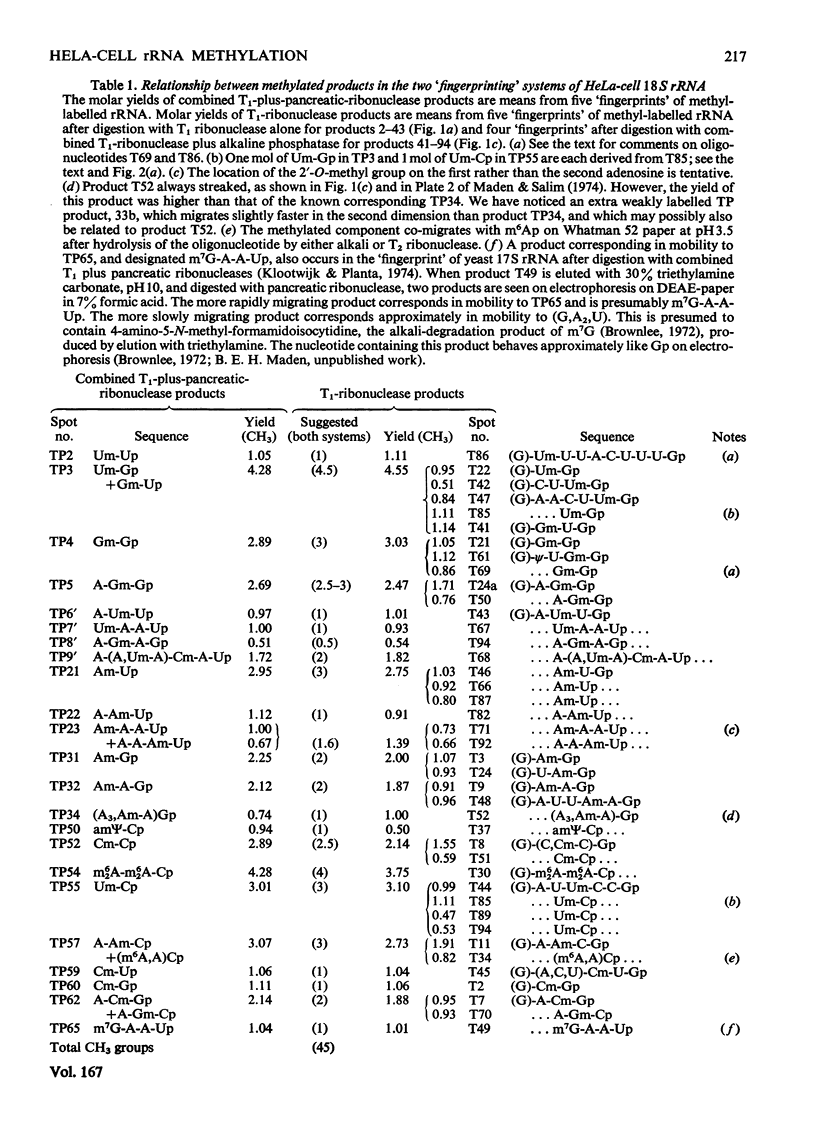
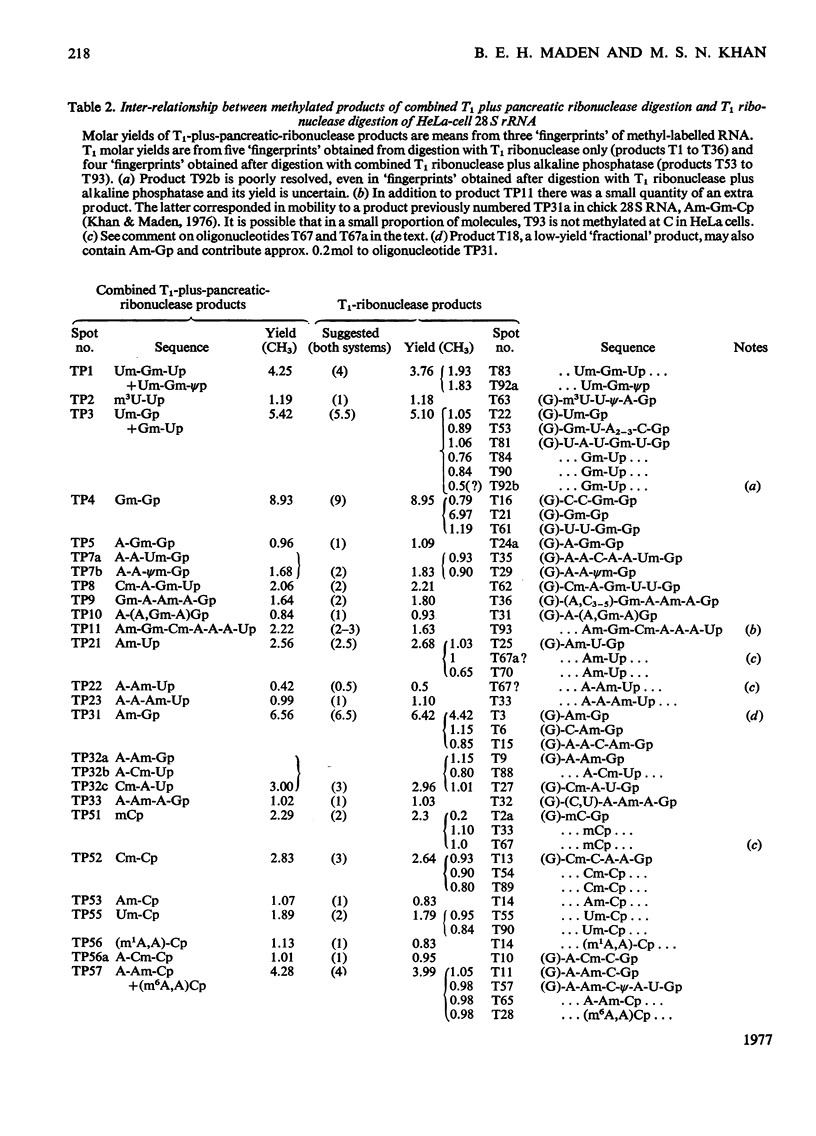
The methylated nucleotide sequences in HeLa-cell rRNA were previously characterized after enzymic digestion of the rRNA by T1 ribonuclease alone or by combined T1 plus pancreatic ribonucleases. For any methylated product occurring in a T1-ribonuclease digest there must be one or more corresponding products in a combined T1-plus-pancreatic-ribonuclease digest. Here we correlate fully the inter-relationship between the methylated products occurring in the two digestion systems. The analysis has led to the resolution of some previous uncertainties and has permitted an almost complete qualitative and quantitative description of the methylated components in HeLa-cell rRNA. The data are compared with those reported by other authors for HeLa-cell rRNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Iwanami Y., Brown G. M. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch Biochem Biophys. 1968 Jul;126(1):8–15. doi: 10.1016/0003-9861(68)90553-5. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequences within the ribosomal ribonucleic acids of HeLa cells, Xenopus laevis and chick embryo fibroblasts. J Mol Biol. 1976 Feb 25;101(2):235–254. doi: 10.1016/0022-2836(76)90375-2. [DOI] [PubMed] [Google Scholar]
- Klagsbrun M. An evolutionary study of the methylation of transfer and ribosomal ribonucleic acid in prokaryote and eukaryote organisms. J Biol Chem. 1973 Apr 10;248(7):2612–2620. [PubMed] [Google Scholar]
- Macon J. B., Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry. 1968 Oct;7(10):3453–3458. doi: 10.1021/bi00850a021. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Forbes J., de Jonge P., Klootwijk J. Presence of a hypermodified nucleotide in HeLa cell 18 S and Saccharomyces carlsbergensis 17 S ribosomal RNAs. FEBS Lett. 1975 Nov 1;59(1):60–63. doi: 10.1016/0014-5793(75)80341-3. [DOI] [PubMed] [Google Scholar]
- Maden B. E.H., Lees C. D., Salim M. Some methylated sequences and the numbers of methyl groups in HeLa cell rRNA. FEBS Lett. 1972 Dec 15;28(3):293–296. doi: 10.1016/0014-5793(72)80734-8. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Salim M. The methylated nucleotide sequences in HELA cell ribosomal RNA and its precursors. J Mol Biol. 1974 Sep 5;88(1):133–152. doi: 10.1016/0022-2836(74)90299-x. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O., Busch H. Tissue specific differences in the 2'-O-methylation of eukaryotic 5.8S ribosomal RNA. FEBS Lett. 1975 Nov 1;59(1):83–87. doi: 10.1016/0014-5793(75)80346-2. [DOI] [PubMed] [Google Scholar]
- Saponara A. G., Enger M. D. The isolation from ribonucleic acid of substituted uridines containing alpha-aminobutyrate moieties derived from methionine. Biochim Biophys Acta. 1974 Apr 27;349(1):61–77. doi: 10.1016/0005-2787(74)90009-4. [DOI] [PubMed] [Google Scholar]



