Abstract
Forkhead box O (Foxo) transcription factors are emerging as critical transcriptional integrators among pathways regulating differentiation, proliferation, and survival, yet the role of the distinct Foxo family members in angiogenic activity of endothelial cells and postnatal vessel formation has not been studied. Here, we show that Foxo1 and Foxo3a are the most abundant Foxo isoforms in mature endothelial cells and that overexpression of constitutively active Foxo1 or Foxo3a, but not Foxo4, significantly inhibits endothelial cell migration and tube formation in vitro. Silencing of either Foxo1 or Foxo3a gene expression led to a profound increase in the migratory and sprout-forming capacity of endothelial cells. Gene expression profiling showed that Foxo1 and Foxo3a specifically regulate a nonredundant but overlapping set of angiogenesis- and vascular remodeling–related genes. Whereas angiopoietin 2 (Ang2) was exclusively regulated by Foxo1, eNOS, which is essential for postnatal neovascularization, was regulated by Foxo1 and Foxo3a. Consistent with these findings, constitutively active Foxo1 and Foxo3a repressed eNOS protein expression and bound to the eNOS promoter. In vivo, Foxo3a deficiency increased eNOS expression and enhanced postnatal vessel formation and maturation. Thus, our data suggest an important role for Foxo transcription factors in the regulation of vessel formation in the adult.
Introduction
The forkhead box O (Foxo) subclass of transcription factors is evolutionary conserved and plays an important role in the control of cell and organismal growth, development, metabolism and longevity. These forkhead or winged helix proteins are structurally related transcriptional regulators, of which the mammalian Foxo members Foxo1 (Fkhr), Foxo3a (Fkhr-l1), and Foxo4 (Afx) represent a subfamily that is regulated by growth factor–dependent activation of the PI3K pathway (1–3). Foxo proteins are involved in various cellular processes ranging from programmed cell death and cell cycle progression to stress detoxification. At the organismal level, Foxo transcription factors play a crucial role in the regulation of tissue homeostasis in organs such as the pancreas and the ovaries and complex diseases such as diabetes and cancer (4–8). Activation of the PI3K pathway blocks the function of all 3 Foxo members by Akt-dependent phosphorylation of 3 conserved residues, which leads to inhibition of DNA binding, nuclear exclusion, and subsequent sequestration in the cytoplasm (9–11). Dephosphorylation of Foxo factors in turn stimulates nuclear entry, leading to the activation or repression of apoptosis- and cell cycle–related genes such as Bim, p27 Kip1, MnSOD, or GADD45 (12–15).
Despite redundant functions of Foxo proteins in vitro, their in vivo roles in development and physiology are diverse, and genetic loss of the distinct Foxo isoforms results in different phenotypes. For example, mice homozygous for a Foxo1–/– allele, but not Foxo3a–/– or Foxo4–/– mice, die during embryogenesis from defects in vascular development (16, 17). Although these studies suggest an essential role of Foxo1 in the formation and maturation of the nascent vasculature, relatively little is known about the function and significance of the distinct Foxo family members for the angiogenic activity of endothelial cells and postnatal vessel formation. In mature endothelial cells, inhibition of Foxo1 activity has been shown to be an important mechanism through which angiopoietin 1 (Ang1) modulates endothelial function (18). In addition, overexpression of Foxo3a enhances apoptosis via downregulation of the antiapoptotic protein flice-like inhibitory protein (19) and blocks growth factor–induced proliferation of endothelial cells (20). In endothelial progenitor cells, Foxo4 has been recently shown to play a pivotal role in the regulation of apoptosis by upregulating the proapoptotic gene Bim (21).
Here, we demonstrate that the Foxo transcription factors Foxo1 and Foxo3a, but not Foxo4, are critical regulators of endothelial sprout formation and migration in vitro. Gene expression profiling showed that endogenous Foxo1 and Foxo3a specifically regulate a nonredundant but overlapping set of angiogenesis- and vascular remodeling–related genes, including eNOS, which is essential for postnatal neovascularization. In vivo, Foxo3a deficiency increased eNOS expression and enhanced vessel formation and maturation in 2 models of postnatal neovascularization. Thus, our data suggest an important role for Foxo transcription factors in the regulation of vessel formation in the adult.
Results
Foxo1 and Foxo3a are the predominant Foxo factors in endothelial cells.
To investigate the role of Foxo transcription factors in the regulation of endothelial cell function, we first assessed the expression profile of the Foxo isoforms Foxo1, Foxo3a, and Foxo4 in endothelial cells. Microarray analysis of total RNA isolated from human umbilical vein endothelial cells (HUVECs) showed that Foxo1 and Foxo3a are the most abundant Foxo isoforms expressed, whereas Foxo4 expression was comparably low (Figure 1A). Western blotting of cell extracts confirmed the presence of Foxo1 and Foxo3a in endothelial cells (Figure 1B). Moreover, treatment of HUVECs with VEGF led to time-dependent phosphorylation of Foxo1 and Foxo3a; this was prevented by preincubation with the PI3K inhibitor LY294002, which suggests that Foxos are not only expressed but also regulated by physiologically important stimuli (Figure 1, C and D).
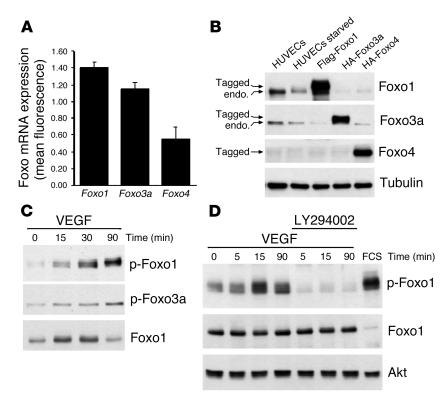
Figure 1.
Foxo1 and Foxo3a are the predominant Foxo transcription factors in endothelial cells. (A) Statistical summary of the Foxo expression profile as assessed in a microarray analysis of total RNA isolated from HUVECs. Data are presented as mean ± SEM; n = 4. The values on the y axis represent a ratio normalized to the mean fluorescent intensity of all genes on the chip. (B) Cell fractions were prepared from HUVECs that were either left untreated, serum starved for 24 hours, or transfected with Flag-Foxo1, HA-Foxo3a, or HA-Foxo4 and subjected to SDS-PAGE. Western blot analysis was performed using the indicated antibodies. Tagged, epitope-tagged; endo., endogenous. (C) HUVECs were serum-starved for 24 hours and then either left untreated or stimulated with VEGF (50 ng/ml) for the indicated times. Cell lysates were subjected to Western blot analysis with antibodies against phospho-Foxo1 (Thr 24), phospho-Foxo3a (Thr 32), or total Foxo1. (D) HUVECs were serum starved for 24 hours, then incubated with 10 μM LY294002 for 1 hour and stimulated with VEGF (50 ng/ml) for the indicated times. Cell lysates were analyzed by Western blot analysis using an antibody directed against phospho-Foxo1. The total level of protein was assayed by Western blot analysis using an anti-Foxo1 and anti-Akt antibody. FCS (20%) was used as a positive control.
Foxo1 and Foxo3a inhibit endothelial tube formation and migration.
To characterize the importance of Foxo1 and Foxo3a for endothelial cell function, we examined whether transfection of HUVECs with constitutively active constructs of Foxo1 and Foxo3a influences endothelial tube formation and migration. Therefore, we used the constitutively active Foxo constructs lacking the 3 Akt-dependent phosphorylation residues (Foxo A3), which are rendered resistant toward Akt-dependent inhibition (Figure 2A). Overexpression of constitutively active Foxo1 (Foxo1 A3) and Foxo3a (Foxo3a A3) led to a significant impairment of tube-forming activity (Figure 2, C–E) and VEGF-induced endothelial cell migration (Figure 2E). In contrast, transfection of constitutively active Foxo4 had no effect on either tube formation or endothelial cell migration (Figure 2, C and D), despite efficient expression (Figure 2A) and comparable activity on a forkhead responsive element–driven luciferase construct with 6 repeated binding sites (6xDBE) (Figure 2B).
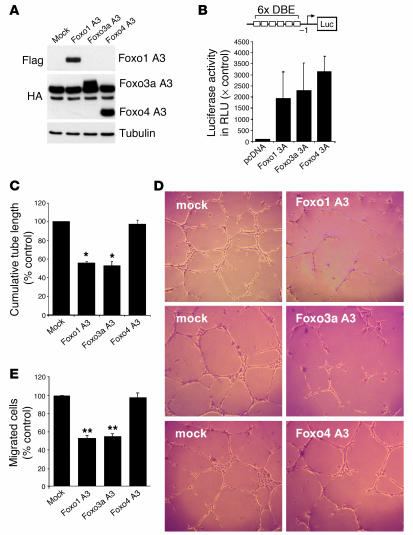
Figure 2.
Overexpression of a gain-of-function mutant of Foxo1 or Foxo3a inhibits endothelial sprout formation and migration. (A) HUVECs were transfected with constitutively active Foxo1, Foxo3a, Foxo4, or mock control. Twenty-four hours later, cells were lysed and subjected to Western blot analysis with antibodies against Flag and HA. An antibody directed against tubulin was used as loading control. (B) HUVECs were transfected with a forkhead-responsive element reporter construct (6xDBE) along with plasmids encoding either constitutively active Foxo1, Foxo3a, or Foxo4. A transfected empty vector (pcDNA) was used a control. At 24 hours after transfection, cells were lysed, and luciferase relative to renilla luciferase activity was measured. × Control, fold value relative to pcDNA-transfected cells. The statistical summary represents the mean ± SEM; n = 3. (C and D) Statistical summary and representative micrographs of the tube-forming activity. HUVECs were seeded on Matrigel Basement Membrane Matrix 18 hours after transfection with the indicated plasmids. The length of capillary-like structures was measured by light microscopy after 24 hours in a blinded fashion. Data are presented as mean ± SEM; n = 5 (Foxo1), n = 6 (Foxo3a), n = 5 (Foxo4). *P < 0.001 versus control. Magnification, ×50. (E) HUVECs were transfected with the constitutively active constructs of Foxo1, Foxo3a, and Foxo4 and were seeded in the upper chamber of a modified Boyden chamber 18 hours after transfection. Endothelial cell migration was assessed using VEGF (50 ng/ml) as chemoattractant after 24 hours of incubation. Data are presented as mean ± SEM. **P < 0.05 versus control; n = 3.
Conversely, silencing of endogenous Foxo1 or Foxo3a gene expression using small interfering RNA (siRNA) significantly increased endothelial migration and tube formation in the Matrigel assay (Figure 3, A–D). Western blotting confirmed the efficient and specific suppression of Foxo1 and Foxo3a by the respective siRNA oligonucleotides (Figure 3A). As a control for the specificity of this approach, a second Foxo1- and Foxo3a-specific siRNA was generated and gave identical results in terms of migration and tube formation in the Matrigel assay (Figure 3, A–C and data not shown).
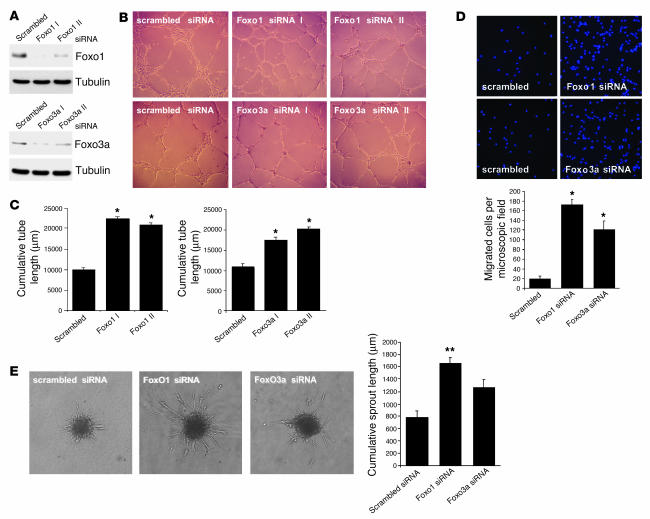
Figure 3.
Silencing Foxo1 or Foxo3a activity enhances the angiogenic activity of endothelial cells. (A) HUVECs were transfected with 2 different siRNAs targeted against either Foxo1 (Foxo1 I and Foxo1 II) or Foxo3a (Foxo3a I and Foxo3a II). A nonrelated scrambled siRNA was used a control. Cell lysates were subjected to Western blotting using antibodies against Foxo1 and Foxo3a. Tubulin was used a loading control. (B and C) HUVECs were transfected with 2 different siRNAs targeted against Foxo1, Foxo3a, or a scrambled oligonucleotide. After 18 hours, cells were seeded on a Growth Factor Reduced Matrigel in the presence of VEGF (50 ng/ml). Cumulative sprout length of capillary-like structures was measured by light microscopy after 24 hours. Representative micrographs and statistical summary are shown. Data are presented as mean ± SEM; n = 4 (Foxo1), n = 6 (Foxo3a). *P < 0.001 versus control. (D) Representative micrographs and statistical summary of endothelial cells transfected with siRNAs targeted against Foxo1, Foxo3a, or scrambled control. After 18 hours, cells were seeded in the upper chamber of modified Boyden chamber. Endothelial cell migration was assessed using VEGF (50 ng/ml) as chemoattractant. After 24 hours, nonmigrating cells on the upper side of the chamber were mechanically removed, and the remaining cells on the lower side were fixed and stained with DAPI. Data are presented as mean ± SEM; n = 5. *P < 0.001 versus control. (E) Three-dimensional in vitro angiogenesis with collagen gel–embedded spheroids of Foxo1, Foxo3a, or scrambled siRNA–transfected endothelial cells. Cumulative length of all sprouts originating from an individual spheroid was quantified after 24 hours. Statistical summary represents the mean ± SEM; n = 3. **P < 0.05 versus control. Magnification: ×50 (B); ×200 (D); ×100 (E).
To further substantiate a role of Foxo1 and Foxo3a in angiogenic activity of endothelial cells, we employed a 3-dimensional spheroidal system of endothelial differentiation and in vitro angiogenesis. Therefore, Foxo1 or Foxo3a siRNA–transfected endothelial cell spheroids were embedded in collagen gels, and outgrowth of capillary-like structures was assessed. Consistent with the enhanced tube formation in the Matrigel assay, silencing of Foxo1 or Foxo3a enhanced the sprouting activity of the endothelial spheroids (Figure 3E), which indicates that endogenous Foxo1 and Foxo3a are functionally important for the angiogenic activity of mature endothelial cells.
Transcriptome analysis of Foxo-regulated genes in endothelial cells.
In order to identify Foxo-regulated genes that might mediate the effect of endogenous Foxos on angiogenesis, we analyzed the expression profile of isolated mRNA in Foxo1- and Foxo3a-silenced endothelial cells by microarray technology. HUVECs were transfected with Foxo1- and Foxo3a-specific siRNAs or the respective scrambled control, and total RNA was isolated after 24 hours. In this set of experiments, knockdown of Foxo1 and Foxo3a expression resulted in changes in expression of known Foxo target genes that were in accordance with previously described functions of Foxo (Table 1 and Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI23126DS1).
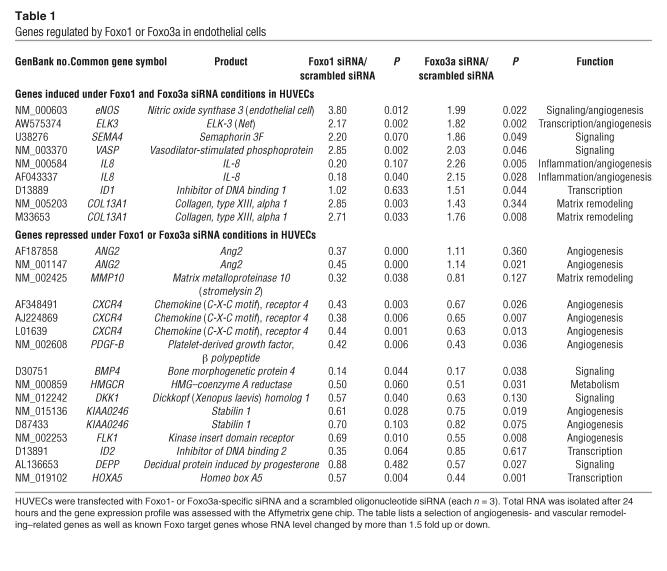
Table 1.
Genes regulated by Foxo1 or Foxo3a in endothelial cells
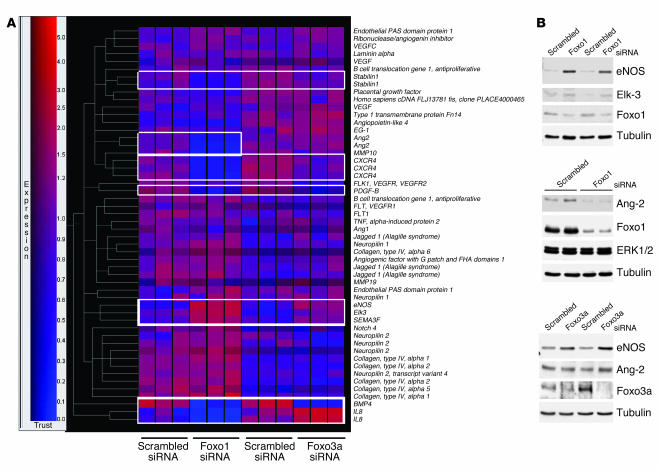
In addition to those of known Foxo target genes, we observed changes in expression of several angiogenesis- and vascular remodeling–related genes. In particular, Foxo1 and Foxo3a shared a subset of target genes with important vascular functions. Among these, we identified eNOS and the proangiogenic transcription factor ETS domain protein Elk-3 (ELK-3, also known as Net) as Foxo-regulated genes that were upregulated in Foxo-silenced endothelial cells (Table 1 and Figure 4, A and B). Silencing of Foxo1 or Foxo3a additionally downregulated signaling molecules, including bone morphogenic protein 4 (BMP4), chemokine (C-X-C motif) receptor 4 (CXCR4), stabilin-1, and PDGF, β polypeptide (PDGF-B) (Table 1).
Figure 4.
Transcriptome of endogenous Foxo1 and Foxo3a in endothelial cells. HUVECs were transfected with a Foxo1- and Foxo3a-specific siRNA or a scrambled oligonucleotide siRNA (each n = 3). Total RNA was isolated after 24 hours, and the gene expression profile was assessed with the Affymetrix gene chip expression assay. (A) Gene tree analysis of selected angiogenesis-related genes. Red indicates high expression. Genes regulated by Foxo1 and Foxo3a siRNA transfection are indicated by white boxes. (B) Validation of angiogenesis-related Foxo1- and Foxo3a-regulated genes. HUVECs were transfected with Foxo1- and Foxo3a-specific siRNAs and lysed 24 hours after transfection. Cell lysates were subjected to SDS-PAGE, and expression of eNOS, Elk-3, Ang2, Foxo1, and Foxo3a was determined by Western blot analysis. Tubulin and ERK1/2 served as loading controls.
Evidently, there were also apparent differences in the respective transcriptional profiles. Thus, Ang2 was significantly downregulated in Foxo1-silenced endothelial cells (18) but was unaffected in Foxo3a siRNA–transfected endothelial cells (Table 1). Likewise, several other angiogenesis- and vascular remodeling–related genes such as MMP10 or the transcription factor inhibitor of DNA binding 2 (ID2) were regulated by Foxo1 siRNA transfection but not in Foxo3a-silenced endothelial cells. This differential regulation also involved known Foxo target genes implicated in the well-established proapoptotic function of Foxo factors (22–25), which suggests a specific and unique role of each Foxo isoform in the endothelium. For example, knockdown of Foxo1 resulted in enhanced expression of the apoptosis inhibitor survivin (1.8-fold increase), whereas silencing of Foxo3a was associated with reduced expression of the proapoptotic genes Bim (0.44-fold reduction) and TNF-related apoptosis-inducing ligand (TRAIL) (0.6-fold reduction).
The changes in gene expression in response to Foxo siRNA transfection of the target genes eNOS, Ang2, and ELK-3, which might contribute to the antiangiogenic activity of Foxos in endothelial cells, was additionally confirmed by Western blotting (Figure 4B). Taken together, our data show that both Foxo transcription factors Foxo1 and Foxo3a regulate distinct but overlapping sets of angiogenesis- and vascular remodeling–related genes, which supports an important and unique role of Foxo factors in the vascular endothelium.
Foxo-dependent regulation of eNOS expression.
Since eNOS is a novel Foxo target that is essential for endothelial function and postnatal neovascularization (26–28), we further elucidated Foxo-dependent regulation of eNOS expression.
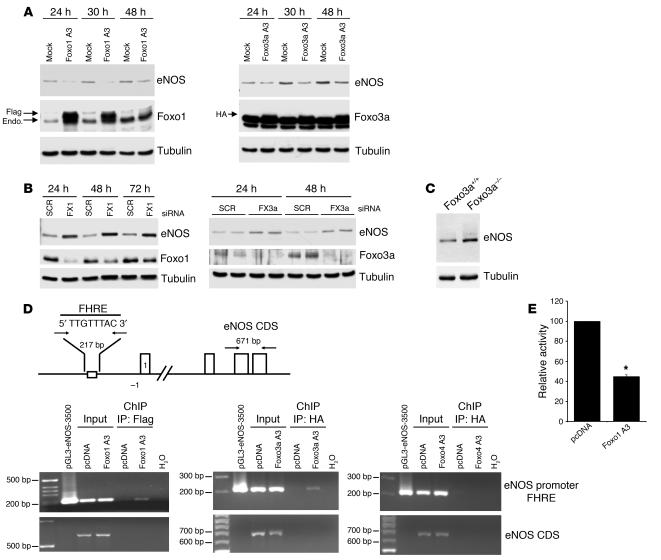
Overexpression of both Foxo1 A3 and Foxo3a A3 significantly decreased eNOS protein expression (Figure 5A). Consistent with these results, eNOS protein expression was significantly upregulated in the Foxo1 and Foxo3a siRNA–transfected endothelial cells (Figure 5B). Moreover, compared with that in wild-type mice, eNOS protein expression was enhanced in explanted aortas of Foxo3a–/– mice (Figure 5C), which suggests that eNOS is a physiologically relevant Foxo target gene in vivo.
Figure 5.
Foxo1 and Foxo3a are transcriptional repressors of eNOS. (A) eNOS expression in HUVECs that were transfected with constitutively active constructs of Flag–Foxo1 A3 and HA–Foxo3a A3. Cell lysates were prepared at the indicated time points, and expression of eNOS, Flag, HA, and tubulin was determined by Western blotting using the respective antibodies. (B) eNOS expression in HUVECs that were transfected with Foxo1- or Foxo3a-specific siRNA and lysed at the indicated time points. eNOS, Foxo1, and Foxo3a expression was determined by immunoblotting. SCR, scrambled siRNA; FX1, Foxo1 siRNA; FX3a, Foxo3a siRNA. (C) eNOS expression in the aorta of Foxo3a+/+ and Foxo3a–/– mice. After the aortas from the indicated groups of mice were removed, cell lysates were prepared, and expression of eNOS was determined by immunoblotting. Equal protein loading was confirmed with a tubulin antibody. (D) HUVECs were transfected with Foxo1 A3 (Flag-Foxo1 A3), Foxo3a A3 (HA–Foxo3a A3), Foxo4 A3 (HA–Foxo4 A3), or vector control. After 24 hours, chromatin-bound DNA was immunoprecipitated with an antibody against the Flag or HA epitope. Immunoprecipitated DNA was analyzed by PCR using a primer combination that encompassed the forkhead responsive element (FHRE). The pGL3-eNOS-3500 plasmid was used as a positive control for the PCR. eNOS primers for the eNOS coding sequence were used as a negative control to exclude nonspecific precipitated DNA. CDS, coding sequence. (E) HUVECs were cotransfected with pGL3-eNOS-3500 and either Foxo1 A3 or the empty vector pcDNA3 (pcDNA). Luciferase activity was measured 24 hours later. Values are expressed as the level of luciferase activity of Foxo1 A3 relative to that of pcDNA, which was set as 100%. Data are presented as mean ± SEM; n = 6. *P < 0.001.
Promoter analysis revealed that the eNOS promoter contains a conserved optimal forkhead-responsive element (FHRE) at position –2,753 relative to the start codon (TTGTTTAC) (29). Chromatin immunoprecipitation (ChIP) assays showed that Foxo1 and Foxo3a bind to the eNOS promoter region encompassing this DNA binding element (Figure 5D). However, no binding was detected when exon-specific primers were used (Figure 5D), which confirmed the specificity of the Foxo/eNOS promoter interaction. Additionally, overexpression of Foxo1 A3 inhibited the basal transcriptional activation of the eNOS promoter as assessed by a reporter gene assay using a 3,500-bp fragment of the human eNOS gene (Figure 5E).
Foxo3a is a modulator of postnatal neovascularization in vivo.
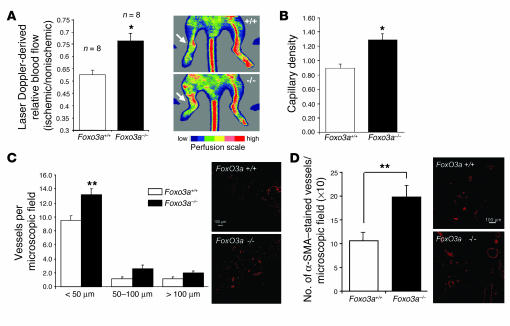
In order to determine the in vivo significance of these findings, we assessed the neovascularization capacity after inducing hind limb ischemia in genetically deficient mice. Since Foxo1–/– mice die during embryogenesis (16, 17), we used Foxo3a–/– mice. As shown in Figure 6, Foxo3a–/– mice had significantly increased limb perfusion 14 days after the induction of ischemia compared with wild-type Foxo3a (Foxo3a+/+) mice (Figure 6A). The enhanced recovery of blood flow was associated with an increased capillary density in Foxo3a–/– mice after hind limb ischemia (Figure 6B and Supplemental Figure 2). Consistent with these findings, the number of small-sized (<50 μm) actin-positive vessels was significantly increased in Foxo3a–/– versus wild-type mice (Figure 6C), which suggests that Foxo factors are involved not only in the formation but also in the maturation and stabilization of the nascent vasculature. To further validate the in vivo relevance of Foxo3a for postnatal neovascularization, we investigated the effect of Foxo3a deficiency in a Matrigel plug assay. Matrigel implants were administered subcutaneously to wild-type and Foxo3a–/– mice, and blood vessel infiltration of the implants was quantified. Compared with wild-type mice, Foxo3a–/– mice had a significantly elevated number of vessels infiltrating the Matrigel (Figure 6D). In summary, these data indicate that Foxo factors are important negative regulators of postnatal vessel formation and maturation in vivo and that a loss of Foxo function augments postnatal neovascularization capacity.
Figure 6.
Foxo3a modulates neovascularization capacity in vivo. (A) Foxo3a+/+ and Foxo3a–/– mice were subjected to hind limb ischemia, and perfusion was assessed 14 days after onset of ischemia using laser Doppler imaging. Low or no perfusion is shown as dark blue, whereas the highest perfusion level is shown as red. Arrows indicate the ischemic leg. Quantitative results are presented as mean ± SEM; n = 8. *P = 0.002. (B) Capillary density (ratio of the number of capillaries to the number of myocytes) was determined in 8-μm frozen sections of the adductor and semimembraneous muscles. Quantitative results are presented as mean ± SEM; n = 8 (Foxo3a+/+), n = 7 (Foxo3a–/–). (C) Conductance vessels in the adductor and semimembraneous muscles were identified by size (>20 μm) and smooth muscle actin staining using a Cy3-labeled mouse monoclonal antibody for smooth muscle actin. The number of small (<50 μm), medium (50–100 μm), and large vessels was determined separately. Data are presented as mean ± SEM; n = 6 (Foxo3a+/+), n = 5 (Foxo3a–/–). **P = 0.01. (D) Statistical summary and representative micrographs of blood vessel infiltration in Matrigel sections stained with a smooth muscle actin antibody in wild-type and Foxo3a–/– mice. Quantitative results are presented as mean ± SEM; n = 7 (Foxo3a+/+), n = 8 (Foxo3a–/–). Scale bars in C and D, 100 μm.
Discussion
Forkhead transcription factors of the Foxo family function as important transcriptional integrators of several signaling cascades, thereby serving as a transcriptional endpoint for pathways regulating differentiation, cell growth, and survival. However, the role of Foxo family members in the regulation of postnatal neovascularization has not been defined. This study identifies Foxo transcription factors as important modulators of vessel formation and maturation in the adult, thus providing evidence for a central function of Foxo factors in vascular homeostasis.
We show that Foxo1 and Foxo3a are the predominant Foxo isoforms expressed in mature endothelial cells and that overexpression of a gain-of-function mutant of Foxo1 or Foxo3a significantly inhibits endothelial cell migration and tube formation in vitro. Accordingly, knockdown of either Foxo1 or Foxo3a gene expression using siRNA led to a profound increase in endothelial sprout formation and migration. Notably, overexpression of a constitutively active Foxo4 had no effect on either migration or sprout formation, which indicated a specific regulatory effect of Foxo1 and Foxo3a as opposed to Foxo4.
The formation and maturation of the vasculature is a complex process involving multiple gene products all contributing to an integrated sequence of events. Consistent with an important role of Foxo transcription factors in these processes, our study identified a number of novel Foxo target genes involved at different stages of angiogenesis. Moreover, direct comparison of gene expression changes in Foxo1- and Foxo3a-silenced endothelial cells revealed that both transcription factors appear to regulate a common transcriptional program important for the angiogenic activity and maintenance of vascular homeostasis, thereby providing a mechanistic basis for the functional overlapping roles of Foxo1 and Foxo3a in this study. Among these genes, we identified eNOS as a novel Foxo target gene that was significantly upregulated in Foxo1- and Foxo3a- silenced endothelial cells. Several other genes with important vascular functions, such as PDGF-B, BMP4, and ELK-3 (Net), were also regulated by Foxo1 or Foxo3a gene silencing. However, there were also gene expression changes that do not support the proangiogenic phenotype of the Foxo-silenced endothelial cells, such as a reduction in mRNA levels of kinase insert domain receptor (KDR, or FLK-1) and CXCR4.
Transcriptional repression of eNOS by Foxo1 and Foxo3a, as shown by our data, might particularly contribute to the antiangiogenic effects of Foxo factors in endothelial cells. Moreover, that eNOS expression is enhanced in Foxo3a–/– mice indicates that eNOS is a Foxo-regulated gene in vivo, which adds to the proangiogenic phenotype of the Foxo3a–/– mice. Indeed, NO synthesized by the eNOS is essential for endothelial cell survival, migration (30, 31) and postnatal neovascularization (26–28). Although eNOS was initially defined as a constitutively expressed enzyme, recent studies highlight the contribution of both transcription and RNA stabilization to the expression level of eNOS mRNA (32, 33). Evidence for a direct effect of Foxo1 and Foxo3a on eNOS transcription was obtained by ChIP, which revealed that both Foxo factors bind to a conserved FHRE in the eNOS promoter located at –2,765 bp upstream of the start codon (29). Likewise, constitutively active Foxo1 significantly inhibited basal promoter activity of a reporter gene driven by the human eNOS promoter. Interestingly, Foxo4 neither bound to the eNOS promoter nor affected eNOS promoter activity (Figure 5D and data not shown), which supports the functional diversification of Foxo proteins in endothelial cells.
Our data show that, in addition to regulating eNOS, Foxo1 and Foxo3a also share several other target genes implicated in vessel formation and stabilization. For instance, ELK-3, which is an ETS ternary complex transcription factor, was significantly increased in Foxo1 and Foxo3a siRNA–treated endothelial cells. ELK-3 has recently been described to be expressed at sites of angiogenesis and vasculogenesis (34) and has been demonstrated to promote the angiogenic switch (35). Although further studies are necessary to confirm the direct transcriptional suppression of ELK-3 by Foxo factors, ELK-3 contains a conserved, inverse Foxo binding site (GTAAACAA) in the proximal promoter region that might be targeted by Foxos.
Despite these overlaps, there were also apparent differences in the respective transcriptional profiles of Foxo1 and Foxo3a. These included known apoptosis-related Foxo target genes such as Bim, TRAIL, and survivin as well as angiogenesis- and vascular remodeling–related genes such as Ang2 and MMP10. For example, Ang2, which has recently been described to be an important target gene of Foxo1 in endothelial cells (18), was significantly downregulated in Foxo1-silenced cells but remained essentially unchanged in Foxo3a siRNA–transfected endothelial cells. By binding but not activating Tie-2, Ang2 is thought to block a stabilizing signal provided by Ang1, thus contributing to vessel instability and postnatal remodeling (36–38). The downregulation of Ang2 in Foxo1 siRNA–treated endothelial cells might therefore affect vessel stabilization. Likewise, the changes in expression of several extracellular matrix proteins such as collagen and MMPs indicate that Foxos might be involved in regulating vessel remodeling as well.
A similar role for Foxo1 as a critical regulator of vascular maturation and remodeling has recently been shown by Daly and colleagues, who investigated the role of Foxo1 as transcriptional effector of the Ang1/Akt axis in endothelial cells (18). Likewise, some of our transcriptional profiling data are consistent with this study. Nevertheless, there were also significant differences in the respective data sets, which might be related to the different experimental strategies used in the 2 studies to identify Foxo1-regulated genes in endothelial cells. In fact, Daly and colleagues used a constitutively active Foxo1 adenovirus to analyze the transcriptional changes in endothelial cells, whereas our study used a gene-silencing approach. Indeed, many of the Foxo1-regulated genes described by Daly et al. are expressed at very low levels under basal cell culture conditions, which makes it difficult to detect any decrease in expression that results from siRNA-mediated transient downregulation. Likewise, knockdown of Foxo1 would be expected to primarily affect expression of target genes, whose basal expression level critically depends on Foxo1 activity. We found several Foxo1-regulated genes described by Daly and coworkers, including Ang2, ID2, and survivin, which suggests that functional Foxo activity is important for the basal expression of these genes (Supplemental Figure 1).
The physiological significance of Foxo factors in the vasculature is underscored by recent work showing that Foxo1 deficiency is embryonically lethal due to defects in vascular development (16, 17). In contrast, Foxo3a–/– mice are viable, which suggests that this isoform is not essential for embryonic vascular development (5). Nevertheless, Foxo3a–/– mice showed a significantly augmented neovascularization capacity in 2 models of postnatal vessel formation. This phenotype is reminiscent of that of the eNOS–/– mice, which do not show an embryonal developmental defect but have profoundly impaired adult neovascularization, which indicates that embryonic vascular development and postnatal vascularization are not necessarily mediated by the same signals (26–28). The question remains why Foxo1–/– mice exhibit a phenotype distinct from that of Foxo3a–/– mice, although both transcription factors show similar effects in mature endothelial cells. There may be a variety of reasons for the apparent differences in phenotype, such as spatiotemporal changes in Foxo isoform expression, which has been shown to occur during adipocyte progenitor differentiation (39). Additionally, given that transcriptional profiling experiments revealed important nonredundant functions of Foxos in regulation of target genes, it is tempting to speculate that such differential regulation of genes with crucial vascular functions might contribute to the differences in the phenotype of the knockout mice. Since all Foxo factors utilize the same forkhead responsive element (29), it will be therefore of interest to identify factors that determine the specific biological output of Foxo signaling in endothelial cells.
In view of the fact that vascular development of the Foxo1–/– mice is severely impaired, which indicates that disruption of Foxo1 leads to defective embryonic angiogenesis, the finding that Foxo1 has an entirely opposite effect on mature endothelial cells is intriguing (16, 17). Although the reasons underlying this seemingly opposing role of Foxo1 in embryonic versus adult vessel formation are unclear, one could presume that the molecular signals and mechanisms that govern the activity and specific transcriptional output of Foxo1 in embryonic vascular development are not necessarily functional during postnatal vessel formation. For instance, Ang2 is an important Foxo1 target gene in mature endothelial cells but was essentially unchanged in Foxo1–/– embryonic stem cell–derived endothelial cells (17, 18).
Taken together, these findings suggest that Foxo transcription factors play important and unique roles in the vascular endothelium by functioning as important transcriptional integrators of multiple downstream signaling molecules important for vessel formation and maturation. Since downregulation of Foxo3a enhanced neovascularization in vivo, the regulation of the expression and activity of Foxo transcription might evolve as a potential target for modulating therapeutic angiogenesis and revascularization after critical ischemia in the adult. In contrast, forced expression or activation of Foxos might limit unwanted neovascularization — as in the case of tumor angiogenesis — thereby revealing another aspect of the well-known antitumorigenic activity of Foxos (3).
Methods
Cell culture.
Pooled HUVECs were purchased from CellSystems and cultured in endothelial basal medium (EBM; Cambrex) supplemented with hydrocortisone (1 μg/ml), bovine brain extract (12 μg/ml), gentamicin (50 μg/ml), amphotericin B (50 ng/ml), epidermal growth factor (10 ng/ml), and 10% FCS (Invitrogen Corp.) until the third passage. After detachment with trypsin, cells were grown in 6-cm culture dishes for at least 18 hours.
Plasmid constructs and transfection.
HUVECs (3.5 × 105 cells/6-cm well) were grown to 60–70% confluence and then transfected with 4 μg of plasmids. Plasmids included constitutive active Foxo1 A3 (pcDNA3-Flag Foxo1 A3), constitutively active Foxo3a A3 (pECE-HA Foxo3a A3), and constitutively active Foxo4 A3 (pMT2-HA Foxo4 A3). The Foxo1 construct was described previously (12) and was kindly provided by William R. Sellers (Dana-Farber Cancer Institute). The Foxo3a plasmid and the pGL3-6xDBE reporter construct were from Paul Coffer (University Medical Center, Utrecht, Netherlands), and the Foxo4 construct was a gift from Boudewijn M.T. Burgering (University Medical Center, Utrecht, Netherlands) (10).
Transfection was performed using the GeneTrans II reagent (MoBiTec) according to the manufacture’s protocol. Cells were incubated with the DNA transfection reagent complexes at 37°C for 4 hours, and this was followed by recovery in the presence of 10% FCS. Transfection efficiency was approximately 40% as determined using green fluorescent protein, and maximal levels of protein expression were observed between 24 and 48 hours.
RNA interference.
To silence Foxo gene expression, we performed transfection of siRNA duplex using GeneTrans II (MoBiTec). Foxo1 and Foxo3a siRNAs were synthesized by Eurogentec. Foxo1 target sequences were 5′-GAGCGTGCCCTACTTCAAG-3′ (Foxo1 siRNA I) and 5′-TCTCCTAGGAGAAGAGCTG-3′ (Foxo1 siRNA II) and corresponded to nucleotides 576–594 as well as 745–763 relative to the first nucleotide of the start codon of the human Foxo1 coding sequence. The human Foxo3a siRNA target sequences were 5′-CAACCTGTCACTGCATAGT-3′ (Foxo3a siRNA I) and 5′-GAGCTCTTGGTGGATCATC-3′ (Foxo3a siRNA II) (8), which corresponded to 636–654 bp and 690–708 bp, respectively. A nonrelated, scrambled siRNA without any other match in the human genomic sequence was used as a control (5′-TTCTCCGAACGTGGCACGA-3′).
Microarray analysis.
Gene expression profiling was performed using the gene chip expression assay (HG-U133A). The protocol for sample preparation and microarray processing was carried out according to the standard Affymetrix GeneChip protocol (Affymetrix). Data were analyzed with GeneSpring software version 4.2 (Agilent Technologies) as previously described (40).
Western blot analysis.
For Western blot analysis, HUVECs were lysed with 150 μl lysis buffer (20 mmol/l Tris [pH 7.4], 150 mmol/l NaCl, 1 mmol/l EDTA, 1 mmol/l EGTA, 1% Triton, 2.5 mmol/l sodium pyrophosphate, 1 mmol/l β-glycerophosphate, 1 mmol Na3VO4, 1μg/ml leupeptin, and 1 mmol/l phenylmethylsulfonyl fluoride) for 15 minutes on ice. After centrifugation for 15 minutes at 20,000 g (4°C), the protein content of the samples was determined according to the Bradford method (31). Proteins (40 μg per lane) were loaded onto SDS-polyacrylamide gels and blotted onto nitrocellulose membranes. Western blots were performed by using antibodies directed against Foxo1 (FKHR, 1:1000; Cell Signaling Technology), Foxo3a (FKHR-L1, 1:500; Upstate), Foxo3a (FKHR-L1, 1:1,000; Cell Signaling Technology), Foxo4 (AFX, 1:1,000; Santa Cruz Biotechnology Inc.), phospho-Foxo1/Foxo3a (phospho-FKHR/FKHR-L1, 1:1000; Cell Signaling Technology), eNOS (1:2,500; BD Biosciences — Transduction Laboratories), Flag (M2 and M5, 1:1,000; Sigma-Aldrich), HA (1:1,000; 12CA5; Roche Diagnostics Corp.), Ang2 (1:1,000; R&D Systems), Elk-3 (Net A-20, 1:200; Santa Cruz Biotechnology Inc.), and tubulin (1:1,000; Labvision/Neomarkers). Enhanced chemiluminescence was performed according to the manufacturer’s instructions (Amersham Biosciences).
Migration assays.
To determine the migration of endothelial cells, HUVECs were detached with trypsin, harvested by centrifugation, resuspended in 500 μl EBM without supplements, counted, and placed in the upper chamber of a modified Boyden chamber (5 × 104 cells per chamber; pore size, 8 μm; BD Biosciences). The chamber was placed in a 24-well culture dish containing EBM with 10% FCS and growth factors. After incubation at 37°C, the nonmigrating cells on the upper side of the chamber were mechanically removed, and the remaining cells on the lower side fixed with 4% paraformaldehyde. For quantification, cell nuclei were stained with DAPI. Cells migrating into the lower chamber were counted manually in 3 random microscopic fields.
Tube formation assays.
HUVECs (1 × 105) were cultured in a 12-well plate (Greiner Bio-One) coated with 200 μl Matrigel Basement Membrane Matrix or Growth Factor Reduced Matrigel (BD Biosciences). Tube length was quantified after 24 hours by measuring the cumulative tube length in 3 random microscopic fields with a computer-assisted microscope using the program KS300 3.0 (Zeiss).
Spheroid-based angiogenesis assay.
Endothelial cell spheroids of defined cell number were generated as described previously (41). In brief, 12 hours after transfection, HUVECs were suspended in culture medium containing 0.20% (wt/vol) carboxymethylcellulose (Sigma-Aldrich) and seeded in nonadherent round-bottom 96-well plates (Greiner Bio-One). Under these conditions, all suspended cells contribute to the formation of a single spheroid per well of defined size and cell number (400 cells/spheroid). Spheroids were generated overnight, after which they were embedded into collagen gels. The spheroid-containing gel was rapidly transferred into prewarmed 24-well plates and allowed to polymerize (30 minutes), after which 100 μl EBM was added on top of the gel. After 24 hours, in vitro angiogenesis was quantified by measuring the cumulative length of the sprouts that had grown out of each spheroid using digital imaging software (AxioVision 3.1; Zeiss), with 10 spheroids analyzed per experimental group and experiment.
ChIP assay.
We cross-linked HUVECs (1 × 106) for 10 minutes by directly adding 1% formaldehyde to the culture medium. The fixed cells were lysed with lysis buffer (1% Triton X-100, 0.32 mol/l sucrose, 5 mmol/l EDTA, 1 mmol/l PMSF, 1 μg/ml leupeptin, 10 mmol/l Tris/HCl, pH 8.0) and sonified 5 times for 10 seconds with output 5 (Branson Sonifire 450; Branson). For ChIP, cell lysates were incubated with an antibody against Flag (M2; Sigma-Aldrich) or HA (12CA5; Roche Diagnostics Corp.). The following steps were performed according to the manufacturer’s instructions (Upstate). The isolated precipitated DNA was amplified by PCR with primers corresponding to an approximately 215-bp fragment of the human eNOS promoter (forward, 5′-CGGAGCAGGTGATAGAAGCTAGG-3′ and reverse, 5′-GCTTCCCTGGAGTCTTGTGTAGG-3′) and the eNOS coding sequence (forward, 5′-CCAGCTAGCCAAAGTCACCAT-3′ and reverse, 5′-GTCTCGGAGCCATACAGGATT-3′).
Luciferase assay.
Reporter gene constructs were generated using the vector plasmid pGL3-Basic (Promega), which contains the luciferase gene, as described previously (42). pGL3-eNOS-3500 contained a 3.5-kb human eNOS promoter fragment. HUVECs were transiently transfected with 2 μg of reporter gene and 2 μg of the indicated plasmid using the TransGene II transfection reagent (MoBiTec). After 24 hours of incubation, cells were lysed in passive lysis buffer (Promega), and luciferase activity was measured using the Luciferase Assay System (Promega) with a luminometer (Lumat LB 9501; Berthold).
Murine model of hind limb ischemia.
The role of Foxo3a in ischemia-induced neovascularization was investigated in a murine model of hind limb ischemia using Foxo3a+/+ and Foxo3a–/– mice (FVB/n), which have been described in ref. 5. The present study was performed with permission of the State of Hesse, Regierungspräsidium Darmstadt, according to section 8 of the German Law for the Protection of Animals, and conforms to the German Guide for the Care and Use of Laboratory Animals. The proximal portion of the femoral artery, including the superficial and the deep branch, and the distal portion of the saphenous artery as well as all arterial branches between the ligations were obliterated using an electrical coagulator. The overlying skin was closed using 3 surgical staples. Fourteen days after surgery, the ratio of ischemic (right) to normal (left) limb blood flow was measured using a laser Doppler blood flow meter (Laser Doppler Perfusion Imager System, moorLDI-Mark 2; Moor Instruments). Before scanning was initiated, mice were placed on a heating pad at 37°C to minimize variations in temperature. After laser Doppler color images were recorded, the perfusions of the ischemic and nonischemic limb were calculated on the basis of colored histogram pixels. To minimize variables, including ambient light and temperature, calculated perfusion was expressed as the ratio of ischemic to nonischemic hind limb perfusion.
Vascular histology.
Capillary density was determined in 8-μm frozen sections of the adductor and semimembraneous muscles. Endothelial cells were stained for CD31 (PE-labeled; BD Biosciences). Myocyte membranes were stained using an antibody to laminin (rabbit) followed by anti-rabbit Alexa 488. Conductance vessels in the adductor and semimembraneous muscles were identified by size (>20 μm) and staining using a Cy3-labeled mouse monoclonal antibody specific for smooth muscle actin (Sigma-Aldrich). The number of small (<50 μm), medium (50–100 μm), and large vessels was determined separately.
In vivo Matrigel plug assay.
This assay was carried out as described (43) in wild-type and Foxo3a–/– mice using 0.5 ml of Matrigel Basement Membrane Matrix (BD Biosciences) containing 40 units/ml of heparin (Sigma-Aldrich). At 14 days, blood vessel infiltration in Matrigel pellets was quantified by analysis of smooth muscle actin–stained (Cy3-labeled; Sigma-Aldrich) sections using a Zeiss confocal microscope (LSM 510).
Statistical analysis.
Data are expressed as mean ± SEM. Two treatment groups were compared by the unpaired Student’s t test; 1-way ANOVA was performed for serial analysis. P values less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The authors are indebted to B. Zimmermann and M. Muhly-Reinholz for expert technical assistance. We are thankful to U. Fiedler and H.G. Augustin (University of Freiburg) for assistance with the establishment of the spheroid-based angiogenesis assay. This study was supported by the Deutsche Forschungsgemeinschaft (Di 600/6-1 – FOR501) and a Young Investigator’s grant from the University of Frankfurt (to M. Potente). K. Sasaki was supported in part by the Japan Heart Foundation/Bayer Yakuhin Research Grant Abroad.
Footnotes
Nonstandard abbreviations used: Ang1, angiopoietin 1; ChIP, chromatin immunoprecipitation; DBE, DNA-binding element; EBM, endothelial basal medium; ELK-3, ETS domain protein Elk-3; Foxo, forkhead box O; HUVEC, human umbilical vein endothelial cell; siRNA, small interfering RNA.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Tran H, Brunet A, Griffith EC, Greenberg ME. The many forks in FOXO’s road [review] Sci STKE. 2003;2003:RE5. doi: 10.1126/stke.2003.172.re5. [DOI] [PubMed] [Google Scholar]
- 2.Burgering BM, Medema RH. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty [review] J. Leukoc. Biol. 2003;73:689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- 3.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 4.Kitamura T, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic β cell growth. J. Clin. Invest. 2002;110:1839–1847. doi:10.1172/JCI200216857. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 6.Nakae J, et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat. Genet. 2002;32:245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 7.Xia SJ, Pressey JG, Barr FG. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol. Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 8.Hu MC, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 9.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 10.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 11.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 12.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 13.Tran H, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 14.Kops GJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 15.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 16.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuyama T, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J. Biol. Chem. 2004;279:34741–34749. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 18.Daly C, et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1) Genes Dev. 2004;18:1060–1071. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skurk C, et al. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J. Biol. Chem. 2004;279:1513–1525. doi: 10.1074/jbc.M304736200. [DOI] [PubMed] [Google Scholar]
- 20.Potente M, Fisslthaler B, Busse R, Fleming I. 11,12-Epoxyeicosatrienoic acid-induced inhibition of FOXO factors promotes endothelial proliferation by down-regulating p27Kip1. J. Biol. Chem. 2003;278:29619–29625. doi: 10.1074/jbc.M305385200. [DOI] [PubMed] [Google Scholar]
- 21.Urbich C, et al. FOXO-dependent expression of the proapoptotic protein Bim: pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J. 2005;19:974–976. doi: 10.1096/fj.04-2727fje. [DOI] [PubMed] [Google Scholar]
- 22.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 23.Stahl M, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 24.Dijkers PF, et al. FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J. Cell Biol. 2002;156:531–542. doi: 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J. Biol. Chem. 2002;277:47928–47937. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 26.Murohara T, et al. Nitric oxide synthase modulates angiogenesis in response to tissue ischemia. J. Clin. Invest. 1998;101:2567–2578. doi: 10.1172/JCI1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papapetropoulos A, Garcia-Cardena G, Madri JA, Sessa WC. Nitric oxide production contributes to the angiogenic properties of vascular endothelial growth factor in human endothelial cells. J. Clin. Invest. 1997;100:3131–3139. doi: 10.1172/JCI119868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aicher A, et al. Essential role of endothelial nitric oxide synthase for mobilization of stem and progenitor cells. Nat. Med. 2003;9:1370–1376. doi: 10.1038/nm948. [DOI] [PubMed] [Google Scholar]
- 29.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ziche M, et al. Nitric oxide mediates angiogenesis in vivo and endothelial cell growth and migration in vitro promoted by substance P. J. Clin. Invest. 1994;94:2036–2044. doi: 10.1172/JCI117557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dimmeler S, Haendeler J, Nehls M, Zeiher AM. Suppression of apoptosis by nitric oxide via inhibition of interleukin-1beta-converting enzyme (ICE)-like and cysteine protease protein (CPP)-32-like proteases. J. Exp. Med. 1997;185:601–607. doi: 10.1084/jem.185.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tai SC, Robb GB, Marsden PA. Endothelial nitric oxide synthase: a new paradigm for gene regulation in the injured blood vessel [review] Arterioscler. Thromb. Vasc. Biol. 2004;24:405–412. doi: 10.1161/01.ATV.0000109171.50229.33. [DOI] [PubMed] [Google Scholar]
- 33.SenBanerjee S, et al. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayadi A, Suelves M, Dolle P, Wasylyk B. Net, an Ets ternary complex transcription factor, is expressed in sites of vasculogenesis, angiogenesis, and chondrogenesis during mouse development. Mech. Dev. 2001;102:205–208. doi: 10.1016/s0925-4773(01)00289-1. [DOI] [PubMed] [Google Scholar]
- 35.Zheng H, et al. The transcription factor Net regulates the angiogenic switch. Genes Dev. 2003;17:2283–2297. doi: 10.1101/gad.272503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holash J, Wiegand SJ, Yancopoulos GD. New model of tumor angiogenesis: dynamic balance between vessel regression and growth mediated by angiopoietins and VEGF [review] Oncogene. 1999;18:5356–5362. doi: 10.1038/sj.onc.1203035. [DOI] [PubMed] [Google Scholar]
- 37.Maisonpierre PC, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 38.Gale NW, et al. Angiopoietin-2 is required for postnatal angiogenesis and lymphatic patterning, and only the latter role is rescued by Angiopoietin-1. Dev. Cell. 2002;3:411–423. doi: 10.1016/s1534-5807(02)00217-4. [DOI] [PubMed] [Google Scholar]
- 39.Nakae J, et al. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 40.Hofmann WK, et al. Altered apoptosis pathways in mantle cell lymphoma detected by oligonucleotide microarray. Blood. 2001;98:787–794. doi: 10.1182/blood.v98.3.787. [DOI] [PubMed] [Google Scholar]
- 41.Korff T, Augustin HG. Integration of endothelial cells in multicellular spheroids prevents apoptosis and induces differentiation. J. Cell Biol. 1998;143:1341–1352. doi: 10.1083/jcb.143.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. Activation of protein kinase C alpha and/or epsilon enhances transcription of the human endothelial nitric oxide synthase gene. Mol. Pharmacol. 1998;53:630–637. doi: 10.1124/mol.53.4.630. [DOI] [PubMed] [Google Scholar]
- 43.Passaniti A, et al. A simple, quantitative method for assessing angiogenesis and antiangiogenic agents using reconstituted basement membrane, heparin, and fibroblast growth factor. Lab. Invest. 1992;67:519–528. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.