Abstract
The etiological origin of type 2 diabetes mellitus (T2DM) has long been controversial. The body of literature related to T2DM is vast and varied in focus, making a broad epidemiological perspective difficult, if not impossible. A data-mining approach was used to analyze all electronically available scientific literature, over 12 million Medline records, for “objects” such as genes, diseases, phenotypes, and chemical compounds linked to other objects within the T2DM literature but were not themselves within the T2DM literature. The goal of this analysis was to conduct a comprehensive survey to identify novel factors implicated in the pathology of T2DM by statistically evaluating mutually shared associations. Surprisingly, epigenetic factors were among the highest statistical scores in this analysis, strongly implicating epigenetic changes within the body as causal factors in the pathogenesis of T2DM. Further analysis implicates adipocytes as the potential tissue of origin, and cytokines or cytokine-like genes as the dysregulated factor(s) responsible for the T2DM phenotype. The analysis provides a wealth of literature supporting this hypothesis, which—if true—represents an important paradigm shift for researchers studying the pathogenesis of T2DM.
INTRODUCTION
The biomedical literature is vast and growing rapidly, with approximately 12.7 million records in Medline at the time of this writing and growing at a rate of 500 000 new records per year. Time and interest are natural limitations we all share in our awareness of past publications and in keeping up with current ones. As such, our perspective is limited and there is an increasing need for computational methods of literature analysis to gain a broader perspective [1]—particularly ones that lead researchers to reasonable hypotheses that they may not have been able to arrive at independently [2, 3].
Recently, a method was developed for a full-scale analysis of Medline records, with the intention of enabling software to take over some of this tedium so that a global analysis of literature trends could be conducted and statistically relevant associations brought to the attention of the user [4, 5]. This software package, entitled IRIDESCENT (Implicit Relationship IDEntification by in-Silico Construction of an Entity-based Network from Text), “reads” Medline to identify both the primary names (eg, “type 2 diabetes”) and synonyms (eg, “adult-onset diabetes”) of diseases, genes, phenotypes, and chemical compounds (collectively referred to as “objects”) as they appear together within Medline titles and abstracts. As objects appear together more frequently in the same Medline records, their relationship strength is deemed to increase. After processing all of Medline, this database reflects a historical record of research and a summary of what is known or, at least, published. Given an object of interest such as type 2 diabetes mellitus (T2DM), IRIDESCENT first identifies all related objects and then uses these objects to identify implied relationships (Figure 1). That is, it identifies new objects that are unrelated to T2DM yet share many relationships with T2DM. Each of these new objects shares a set of relationships with the original object of interest, but is not itself related. Each set of shared relationships is ranked against a random network model for its statistical relevance. These implicitly related objects identified by the system have the potential to offer insight into novel mechanisms of disease etiology and drug action, among other things. We have applied IRIDESCENT to the study of noninsulin-dependent diabetes mellitus (NIDDM), more commonly known as T2DM, in the hope of elucidating novel relationships pertinent to its pathogenesis.
Figure 1.

Identifying new relationships that are potentially implicit from known relationships. IRIDESCENT's method of analysis begins with a primary object of interest such as T2DM (black node) and then identifies all co-citations with other objects (gray nodes) observed within Medline. IRIDESCENT then examines all these directly related (gray) nodes in turn for their relationships with other objects (white nodes) that are not themselves related to the primary object. Once all of Medline is analyzed and all implicit relationships (white nodes) are identified, all relationships they share (gray nodes) with the primary node are individually scored for their statistical significance of co-occurrence and collectively scored for statistical significance against a random network model.
T2DM is an increasingly prevalent disease in the world, especially the United States, where the number of new patients grew 49% between 1991 and 2000. The economic cost of T2DM is staggering, with the Center for Disease Control estimating costs at $132 billion annually (May 2003 estimates) and affecting more than 6% of the population in the United States (http://www.cdc.gov/diabetes/pubs/estimates.htm). Many factors that correlate with the risk of developing T2DM have been identified, but causality has proven elusive. T2DM has consequently been termed a “complex” disorder [6], thought to be a result of a complex interaction between environmental influence and genetic background. A large number of studies so far have focused upon single-nucleotide polymorphisms (SNPs), the hypothesis being that while their individual associations are weak, they may be acting synergistically and possibly in concert with environmental influences. Table 1, for example, is a nonexhaustive list of 25 different SNPs reported to have “significant” associations with T2DM obtained from a PubMed query on the words “(type 2 diabetes or NIDDM) and polymorphism.” The odds any one person would inherit even a significant fraction of these 25, much less than all of them, are extremely low (allele frequencies aside, note their distributions across chromosomes). Thus far, little attention has been paid to the hypothesis that T2DM may be epigenetic in origin, but this is changing as researchers realize that traditional theories on complex disease etiology are lacking [7].
Table 1.
A nonexhaustive list of SNP studies that have reported finding a “significant” association between one or more SNPs and a cohort of T2DM patients. NCBI's sequence viewer was used to obtain flanking sequence. a denotes PMID = PubMed ID of paper publishing the SNP. b denotes that C allele was also present. c means that no dbSNP entry was found in exact map position as given, but the closest dbSNP entries had the same alleles as published, so these were used instead.
| Gene | Chr | Region | Mutation | Position | CpG? | PMIDa | Sequence | dbSNP |
| IDE | 10q23 | 3′ UTR | multiple | — | — | 12765971 | — | — |
| GFPT2 | 2p13 | 3′ UTR | multiple | — | — | 14764791 | — | — |
| IsI-1 | 5q11.2 | 5′ UTR | A→G | −47 | — | 11978668 | — | — |
| KIR6.2 | 11p15.1 | Coding | E23K | — | — | 12643262 | — | — |
| SUR1 | 11p15.1 | Coding | G→A | 3819 | — | 11030411 | — | — |
| PLA2G4A | 1q25 | Coding | F479L | — | — | 12765847 | — | — |
| PTP-1B | 20q13.1 | Coding | C→T | 981 | — | 11836311 | — | — |
| GFPT2 | 2p13 | Coding | I471V | — | — | 14764791 | — | — |
| PPARG2 | 3p25 | Coding | P12A | — | — | 12829658 | — | — |
| GLUT2 | 3q26.1 | Coding | A→G | 103 | — | 12017192 | — | — |
| PGC-1 | 4p15.1 | Coding | G482S | — | — | 12606537 | — | — |
| NR3C1 | 5q31 | Coding | A→G | 1220 | — | 12864802 | — | — |
| Syntaxin 1A | 7q11.23 | Coding | T→C | 204 | — | 11719842 | — | — |
| PRKCZ | 1p36.33 | Intron | G→A | — | — | 12970910 | — | rs436045 |
| CAPN10 | 2q37.3 | Intron | G→A | — | — | 14730479 | — | SNP43 |
| UCP2 | 11q13 | Promoter | G→A | −866 | Yb | 12915397 | CTGAGGCGT | rs659366 |
| Resistin | 19p13.3 | Promoter | C→G | −180 | Y | 12829623 | AAGACGGAG | rs1862513 |
| CRP | 1q21 | Promoter | C→T | −700 | Y | 12618085 | AACACGCGG | SNP133552 |
| PTGS2 | 1q25.2 | Promoter | C→G | −766 | Y | 12920574 | TCCCGCCTC | rs20417 |
| PCK1 | 20q13.31 | Promoter | C→G | −232 | Yc | 14764811 | CAACCTTGT | rs6025628 |
| GLUT2 | 3q26.2 | Promoter | A→C | −269 | N | 12017192 | AATCACATG | None |
| APM1 | 3q27 | Promoter | A→G | −11426 | Yc | 14749263 | TCTCGGCTC | rs12631446 |
| APM1 | 3q27 | Promoter | C→G | −11377 | Nc | 14749263 | ATTACAGGT | rs10937272 |
| TNF-alpha | 6p21.3 | Promoter | G→A | −308 | Nc | 12818408 | CATGGGGA | rs1800629 |
| IL-6 | 7p21 | Promoter | G→C | −174 | Y | 12589429 | TTGCCATGC | rs1800795 |
DNA methylation is a fundamentally important phenomenon within eukaryotes, serving as a means to distinguish host DNA from foreign DNA, to determine which strand of DNA is newly replicated [8] and to provide a signal for chromatin condensation such that transcriptional programs can be inactivated, a process especially important during normal development [9]. Loss of methylation in regulatory DNA regions has been an active research area in cancer, with a number of genes known to be dysregulated from a loss of methylation in certain tumors [10]. Erosion of normal DNA methylation patterns seems to occur in most tissues [11] and is time dependent [12, 13]. Although loss of DNA methylation can be induced chemically (eg, 5-aza-2'-deoxycytidine) or occur as a result of nutrient deficiency (eg, folate), neither of these cases likely apply to T2DM patients, so it is not clear whether other environmental factors could have either a protective or causative effect with regard to T2DM.
MATERIALS AND METHODS
At the time of this analysis, IRIDESCENT was capable of recognizing 33 534 unique objects within text. A total of 2105 of these were cataloged as being directly related to T2DM. IRIDESCENT then analyzed each of these 2105 related objects (schematically illustrated in Figure 1) for their relationships, removing those already in the list of direct relationships. The resulting list contained objects indirectly related to T2DM. That is to say, they shared a large set of relationships with T2DM but were not themselves related to it. At the time of analysis, none of these implicitly related objects were found comentioned with T2DM in the body of any Medline title or abstract. Each implicit relationship was then evaluated by IRIDESCENT based upon the number of relationships it shared with T2DM, relative strength of each relationship, quality of the relationships (statistical probability that each relationship is valid), and the probability that the two objects would share a similar set of relationships by chance, given the relative abundance of both objects and their shared intermediates within the network. A total of 1287 relationships were identified as being shared by the objects “methylation” and “T2DM.” Not all of these are necessarily causal, correlative, or even meaningful, but many are. Collectively, they provide evidence that a relationship does exist between epigenetic control and T2DM and enabled us to develop a more comprehensive theory regarding an epigenetic etiology and pathogenesis of T2DM. We will limit the discussion of shared relationships to those we believe are most pertinent (summarized in Figure 2).
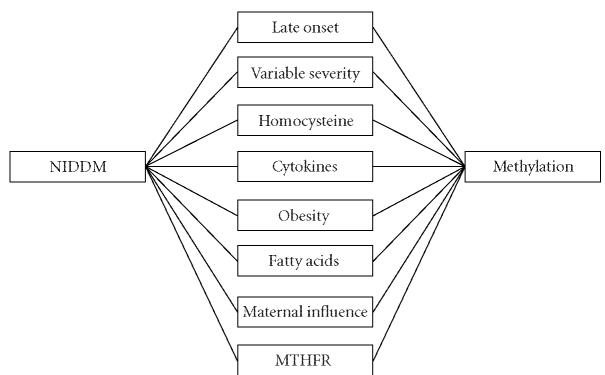
Figure 2.
Important relationships shared by methylation and T2DM (referred to as “NIDDM” in the system). A total of 1287 co-cited objects were identified between the two, about 959 of these reflect actual relationships of a nontrivial nature. Only relationships emphasized within this report are shown here. A full list is available online at http://innovation.swmed.edu/IRIDESCENT/NIDDM_theory.htm.
RESULTS
IRIDESCENT was used to identify and rank objects within Medline implicitly related to T2DM and identified “methylation” and “chromatin” as top scoring hits (Table 2). Methylation is a fundamentally important phenomenon within eukaryotes for the development and regulation of cellular processes, including the modification and regulation of proteins, lipids, and DNA [14]. Although the relationships linking T2DM and methylation may refer to more than just DNA methylation, much of the discussion here will be focused upon DNA methylation primarily because of the strong link to chromatin and because of the nature of the relationships explored that suggest a permanent change to cellular state is taking place in T2DM as opposed to something caused by a temporary equilibrium shift in molecular methylation capacity.
Table 2.
Objects linked to T2DM solely by virtue of relationships they share within Medline. At the time of analysis, none of these objects were documented in Medline to have a relationship with T2DM (shown at top as a positive control for the query). The nature of each of these implicit relationships varies and must be determined by examination of the intermediate connections. The expected value represents how many shared relationships would be expected given a randomly connected network of relationships with the same properties of the literature-derived one. The quality score represents a statistical weighting of co-mention frequency to reflect confidence that the relationship is not of a trivial nature. The observed to expected ratio (Obs/exp) provides a ranking of how statistically exceptional any given set of shared relationships is.
| Rank | Shared relationships | Implicit relationship | Quality | Expect | Obs/exp |
| — | 2105 | T2DM | 1421 | 329 | 4.32 |
| 1 | 1361 | Endotoxin | 1054 | 308 | 3.42 |
| 2 | 1312 | Hydrocortisone | 991 | 296 | 3.35 |
| 3 | 1301 | Neuroblastoma | 975 | 339 | 2.88 |
| 4 | 1287 | Methylation | 959 | 346 | 2.77 |
| 5 | 1256 | Chromatin | 938 | 339 | 2.77 |
IRIDESCENT identified a number of common phenotypes in the onset and pathology of T2DM that are also shared by diseases associated with a change in methylation state. These shared relationships offer a perspective on some of the puzzling properties of T2DM not easily explained by environmental or genetic mutation models. For example, the onset of T2DM varies, but usually occurs later in life and the probability of affliction generally increases with age. This pattern is also characteristic of epigenetic disorders such as aberrant expression of X-linked genes [15], onset of Huntington's disease [16], and the oncogenesis of tumors [17, 18]. Not all late-onset illnesses are caused by epigenetic changes, but most others share physical accumulations that are unique to the disease, such as the accumulation of amyloid precursor proteins in Alzheimer's disease [19] or Lewy bodies in Parkinsons [20]. T2DM is highly correlated with the presence of obesity and advanced glycosylation end products (AGEs), but neither is a requirement for its development nor unique to it as a disease. T2DM also varies in its severity, generally increasing over time. This is a phenotype shared with some tumors that have undergone methylation changes in promoter sequences, leading to higher gene expression and a more aggressive phenotype [17]. Another interesting observation about T2DM is the “maternal effect” in which T2DM patients report a higher frequency of maternal history of diabetes [21]. While this is not without controversy [22], such an effect could be explained if de novo methylation of DNA sequences during development was due to maternal influence. This type of phenomenon, in fact, has been observed in mice [23, 24].
METABOLIC CHANGES IN T2DM
IRIDESCENT also identified a number of metabolic alterations in the body's ability to methylate DNA that correlate with the existence of or predisposition to T2DM. For example, elevated levels of homocysteine have been found in T2DM patients, correlating with increased severity of the disease as defined by mortality [25]. Homocysteine is a critical metabolic intermediate responsible for carrying out methylation reactions, and the elevated serum levels of it are also correlated with DNA hypomethylation [26]. It has also been reported that sulfur-poor diets that force synthesis of cysteine from methionine predispose individuals to type 2 diabetes later in life [27, 28]. Since methionine affects S-adenosyl methionine (SAM), which is the methyl donor for the methylation of newly synthesized DNA, these individuals develop with an impaired ability to establish de novo DNA methylation patterns. Genetic factors that lead to deficiencies in the methylation pathway have also been shown to predispose individuals to develop T2DM. There is a well-known polymorphism (C677T) in the methylenetetrahydrofolate reductase (MTHFR) gene that decreases its efficiency, leading to a global hypomethylation of DNA [29]. Individuals with this mutation are also predisposed to develop T2DM and other complications of the metabolic syndrome [30]. It has also been found that lowering the amount of dietary methyl-providing compounds provided to mice during pregnancy leads to a global hypomethylation of DNA in their offspring and a corresponding modification of offspring hair coat color (increased yellow versus agouti). Expression of this yellow coat color is associated with increased risks of obesity, diabetes, and cancer [24]. And finally, it has also been shown that aberrant methylation patterns have been shown to induce diabetic symptoms in another form of diabetes, transient neonatal diabetes mellitus (TNDM), which is a result of genetic imprinting [31]. The same imprinted region responsible for TNDM, however, is not known to be responsible for T2DM [32].
A number of these metabolic correlations were very recently noted independently of our own analysis by a company proposing to do a genome-wide scan for alterations in DNA methylation patterns [33], giving further credence to the idea that changes in DNA methylation is a causative factor in the etiology of T2DM. While identifying an etiology is important, even in general terms, perhaps equally or more important is elucidating the causative factors in the pathogenesis of T2DM. If epigenetic alterations are responsible for T2DM, then at least three questions naturally arise: first, what secreted factors are responsible for the T2DM phenotype; second, what tissue type(s) is responsible for expressing the factors that induce the T2DM phenotype; and third, what environmental factors could lead to a loss of methylation and consequent dysregulation of the secreted factors.
CAUSAL FACTOR ANALYSIS
A possible answer for the first question above comes from the highest scoring object on IRIDESCENT's list of implicitly related objects (Table 2): endotoxins. While endotoxins are not known to be associated or causal in T2DM, they have been shown to induce obesity and insulin resistance [34]. Most of the relationships shared between T2DM and endotoxins are objects that either affect or are involved in the immune response, especially cytokines and inflammatory factors. Expression of acute-phase markers such as C-reactive protein (CRP), and proinflammatory cytokines such as IL-6 and TNF-alpha is highly correlated with the presence and severity of T2DM symptoms [35, 36]. These proinflammatory cytokines are also positively correlated with obesity [37]. Furthermore, TNF-alpha has been found to induce insulin resistance [38]. Indeed, there is a growing body of evidence that cytokines, more specifically the proinflammatory cytokines, could be responsible for the T2DM phenotype. It has been observed, for example, that a reversal of T2DM symptoms can be induced by disruption of the inflammatory pathway with high doses of aspirin [39]. Troglitazone, a widely used medication to treat T2DM, has also been found to have anti-inflammatory properties [40], and the lifestyle changes of exercise and dietary changes prescribed to T2DM patients that have been successful in reversing T2DM phenotypes have also been associated with decreases in inflammatory cytokines [41, 42].
If a gene(s) were to be dysregulated, it would likely happen via demethylation of its (or their) promoters. It is possible that some of the SNP association studies may have been linked to promoters with fewer methylatable sites. Within the 25 SNP studies that were surveyed earlier, 10 were found within the promoter region. None of them, however, made the association between the reported SNP and the possibility that the SNP would alter the number of CpGs in the promoter. So to examine this possibility, we used NCBI's sequence viewer to obtain the surrounding sequence for these SNPs. We found that, of the 10 promoter SNPs, 7 altered the number of CpGs present within the promoter (see Table 1). Of these 7, 3 were proinflammatory cytokines associated with T2DM-resistin [43], CRP [44], and IL-6 [45]. Interestingly, IL-6 controls both resistin [46] and CRP expression, with the -174 SNP in IL-6 having been linked to heritably high CRP levels [47]. No study has been done yet to see if this IL-6 polymorphism can be linked to resistin levels as well.
TISSUE OF ORIGIN ANALYSIS
Whereas proinflammatory cytokines have been implicated as a causal factor in T2DM, it is known that besides B cells and T cells, adipocytes and endothelial cells are the only other cell types known to normally produce cytokines. We see that within T cells, cytokine expression is determined by DNA methylation patterns [48] and can be altered by demethylating agents [49]. Neither T cells nor B cells seem a likely candidate since they are not very metabolically active in their naïve or memory forms, and their more active differentiated forms are relatively short lived. Adipocytes, however, are the primary repository for lipids and produce cytokines in proportion to factors such as their size and surrounding obesity [50]. Interestingly, a study by Benjamin and Jost demonstrated that short-chain fatty acids (SCFAs) can promote the demethylation of actively transcribed regions [51]. SCFAs can also affect chromatin structure by inhibiting HDAC, causing hyperacetylation of histones [52] and making regions of DNA more accessible to transcription factors. SCFAs are not normally present in high concentrations within adipocytes, but are normal metabolic byproducts of the long-chain fatty acids stored within. Since the rate of lipolysis within adipocytes is increased in T2DM [53], and can be induced by factors such as TNF-alpha already known to be elevated in T2DM [54], this would have an effect upon the relative concentrations of SCFAs within adipocytes. Higher amounts of SCFA metabolites within adipocytes might provide an environment in which loss of DNA methylation could occur and, coupled with active transcriptional activity, could lead to the hypomethylation and consequent dysregulation of cytokines or cytokine-like factors that lead to T2DM. We see suggestive evidence of this in a study by Laimer et al involving IL-6 and TNF-alpha levels in 20 women before and 1 year after gastric banding surgery. They found that the levels of other obesity markers such as CRP declined, while IL-6 and TNF-alpha did not [55].
Within the proposed model, the etiology of T2DM occurs within adipocytes, involving a gradual loss of DNA methylation around the promoters of cytokines and/or cytokine-like factors normally secreted by the adipocyte. This loss of methylation is favored under the conditions provided by obesity and is caused by transcriptional activity. The subsequent loss of methylation leads to dysregulation of these factors, resulting in a constitutive increase in the production of cytokines from adipocytes. Negative regulatory factors decrease the expression of these factors, enabling management of the T2DM phenotype, but only as long as they are present.
ETIOLOGICAL MODELS OF T2DM
We examine this new proposed model in the context of the three dominant models for the etiology and pathogenesis of T2DM: genetic, environmental, and a complex interaction of both factors. Genetic studies have shown that inheritance plays a role in determining an individual's risk of developing T2DM [56]. Linkage studies, while delineating a number of potential susceptibility regions, have yet to be successful in identifying a specific gene or set of genes responsible for the most popular form of T2DM, despite the large cohorts involved. The well-established correlation between obesity and T2DM also indicates that environmental variables affect the pathogenesis of T2DM. The prevailing “complex disease” theory is that the onset of T2DM is caused by one or more environmental variables acting upon a genetic background, of which there may be many contributing genes [6]. This theory explains how susceptibility to T2DM correlates with genetic background, such as race, as well as with environmental variables such as diet and exercise. But there are at least two observations about the nature of T2DM that the complex disease model does not explain while the epigenetic model does: time dependency and systemic memory.
Even when environmental variables are present on a susceptible genetic background, the onset of T2DM is still time dependent. That is to say, the risk of developing T2DM is positively correlated with age. The complex disease model does not easily explain this except to postulate an as-yet-unknown “trigger” event, such as an infection. Even if this were true, it would not explain the persistence of T2DM after onset. T2DM is diagnozed by the levels of insulin resistance and glucose intolerance experienced by a patient, levels which can be altered to prediabetic levels by sufficient changes in lifestyle. T2DM, however, cannot be reversed [57]. None of the existing models account for a mechanism by which the body can “remember” its state. The methylation status of genes, however, is intended to be a relatively persistent phenomenon, responsible for committing cells into their differentiated states [58]. Given that loss of DNA methylation is correlated with age [59], that the number of methylated sites in a genome is determined by inheritance, and that loss of methylation can be affected by environmental variables, the proposed epigenetic model merits serious consideration. An excellent review was recently published, contrasting the properties of epigenetic disorders with the other models of disease etiology discussed here [60].
DISCUSSION
Contrary to the mutation-centric model, which assumes alterations in function or activity based upon either somatic or inherited mutations in DNA, an epigenetic model implies dysregulation of a gene or set of genes. Thus, phenotypes resulting from the expression of such genes would make biological sense under other physiological conditions. Preventing energy influx into cells by inducing insulin resistance makes sense when considered within the context of the role of the immune system. Acquired immunity in the form of B-cell maturation and antibody production takes time during which pathogens are able to replicate. Part of the early immune response consists of an increase in the presence of proinflammatory cytokines within the circulating bloodstream [61, 62]. It would make sense that one role of these early responders would be to stem the influx of resources like glucose into cells to prevent their utilization by invading pathogens. Since adipocytes contain a large reservoir of energy, this makes them ideal targets for invading pathogens and could necessitate their taking a more active role in fighting infection beyond that of other somatic cells.
Identifying expression changes via microarray analysis and subsequently examining the methylation status of their promoters can obtain a candidate list for genes that have undergone epigenetic dysregulation. If this theory is ultimately shown to be correct, it will allow us the ability to diagnoze the current level of epigenetic progression towards T2DM in patients and offer hope for a T2DM cure that could not be easily provided in a mutation-centric model. It is not apparent how region-specific methylation could be reintroduced to affected regions, but since de novo methylation is a normal process during development and certain viruses can “shut off” the expression of immune-related genes by hypermethylation, it stands to reason that the mechanism to do so is already in place.
ACKNOWLEDGMENT
We would like to thank Dr Roger Unger for his very helpful review of the manuscript and suggestions. This work was funded in part by NSF-EPSCoR Grant no EPS-0132534, NIH/NCI Grant no R33 CA81656, and NIH/NHLBI Grant no P50 CA70907.
References
- 1.Yandell M.D, Majoros W.H. Genomics and natural language processing. Nat Rev Genet. 2002;3(8):601–610. doi: 10.1038/nrg861. [DOI] [PubMed] [Google Scholar]
- 2.Bray D. Reasoning for results. Nature. 2001;412(6850):863. doi: 10.1038/35091132. [DOI] [PubMed] [Google Scholar]
- 3.Blagosklonny M.V, Pardee A.B. Conceptual biology: unearthing the gems. Nature. 2002;416(6879):373. doi: 10.1038/416373a. [DOI] [PubMed] [Google Scholar]
- 4.Wren J.D, Garner H.R. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20(2):191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 5.Wren J.D, Bekeredjian R, Stewart J.A, Shohet R.V, Garner H.R. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20(3):389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 6.Marx J. Unraveling the causes of diabetes. Science. 2002;296(5568):686–689. doi: 10.1126/science.296.5568.686. [DOI] [PubMed] [Google Scholar]
- 7.Dennis C. Epigenetics and disease: altered states. Nature. 2003;421(6924):686–688. doi: 10.1038/421686a. [DOI] [PubMed] [Google Scholar]
- 8.Woodcock D.M, Simmons D.L, Crowther P.J, Cooper I.A, Trainor K.J, Morley A.A. Delayed DNA methylation is an integral feature of DNA replication in mammalian cells. Exp Cell Res. 1986;166(1):103–112. doi: 10.1016/0014-4827(86)90511-2. [DOI] [PubMed] [Google Scholar]
- 9.Attwood J.T, Yung R.L, Richardson B.C. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59(2):241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M, Herman J.G. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumors. J Pathol. 2002;196(1):1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 11.Cooney C.A. Are somatic cells inherently deficient in methylation metabolism? A proposed mechanism for DNA methylation loss, senescence and aging. Growth Dev Aging. 1993;57(4):261–273. [PubMed] [Google Scholar]
- 12.Wareham K.A, Lyon M.F, Glenister P.H, Williams E.D. Age related reactivation of an X-linked gene. Nature. 1987;327(6124):725–727. doi: 10.1038/327725a0. [DOI] [PubMed] [Google Scholar]
- 13.Fuke C, Shimabukuro M, Petronis A, et al. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study [published correction appears in Ann Hum Genet. 2005;69(pt 1):134] Ann Hum Genet. 2004;68(pt 3):196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 14.Hoffman R.M. Methioninase: a therapeutic for diseases related to altered methionine metabolism and transmethylation: cancer, heart disease, obesity, aging, and Parkinson's disease. Hum Cell. 1997;10(1):69–80. [PubMed] [Google Scholar]
- 15.Anderson C.L, Brown B.J. Variability of X chromosome inactivation: effect on levels of TIMP1 RNA and role of DNA methylation. Hum Genet. 2002;110(3):271–278. doi: 10.1007/s00439-002-0676-8. [DOI] [PubMed] [Google Scholar]
- 16.Reik W, Maher E.R, Morrison P.J, Harding A.E, Simpson S.A. Age at onset in Huntington's disease and methylation at D4S95. J Med Genet. 1993;30(3):185–188. doi: 10.1136/jmg.30.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y.I, Giuliano A, Hatch K.D, et al. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74(3):893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 18.Qu G, Dubeau L, Narayan A, Yu M.C, Ehrlich M. Satellite DNA hypomethylation vs overall genomic hypomethylation in ovarian epithelial tumors of different malignant potential. Mutat Res. 1999;423(1–2):91–101. doi: 10.1016/s0027-5107(98)00229-2. [DOI] [PubMed] [Google Scholar]
- 19.Hardy J. The Alzheimer family of diseases: many etiologies, one pathogenesis? Proc Natl Acad Sci USA. 1997;94(6):2095–2097. doi: 10.1073/pnas.94.6.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nussbaum R.L, Polymeropoulos M.H. Genetics of Parkinson's disease. Hum Mol Genet. 1997;6(10):1687–1691. doi: 10.1093/hmg/6.10.1687. [DOI] [PubMed] [Google Scholar]
- 21.Alcolado J.C, Laji K, Gill-Randall R. Maternal transmission of diabetes. Diabet Med. 2002;19(2):89–98. doi: 10.1046/j.1464-5491.2002.00675.x. [DOI] [PubMed] [Google Scholar]
- 22.Thorand B, Liese A.D, Metzger M.H, Reitmeir P, Schneider A, Lowel H. Can inaccuracy of reported parental history of diabetes explain the maternal transmission hypothesis for diabetes? Int J Epidemiol. 2001;30(5):1084–1089. doi: 10.1093/ije/30.5.1084. [DOI] [PubMed] [Google Scholar]
- 23.Pickard B, Dean W, Engemann S, et al. Epigenetic targeting in the mouse zygote marks DNA for later methylation: a mechanism for maternal effects in development. Mech Dev. 2001;103(1–2):35–47. doi: 10.1016/s0925-4773(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 24.Wolff G.L, Kodell R.L, Moore S.R, Cooney C.A. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957. [PubMed] [Google Scholar]
- 25.Stehouwer C.D, Gall M.A, Hougaard P, Jakobs C, Parving H.H. Plasma homocysteine concentration predicts mortality in non-insulin-dependent diabetic patients with and without albuminuria. Kidney Int. 1999;55(1):308–314. doi: 10.1046/j.1523-1755.1999.00256.x. [DOI] [PubMed] [Google Scholar]
- 26.Yi P, Melnyk S, Pogribna M, Pogribny I.P, Hine R.J, James S.J. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem. 2000;275(38):29318–29323. doi: 10.1074/jbc.M002725200. [DOI] [PubMed] [Google Scholar]
- 27.Rees W.D. Manipulating the sulfur amino acid content of the early diet and its implications for long-term health. Proc Nutr Soc. 2002;61(1):71–77. doi: 10.1079/pns2001137. [DOI] [PubMed] [Google Scholar]
- 28.Barker D.J, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1(8489):1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 29.Stern L.L, Mason J.B, Selhub J, Choi S.W. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9(8):849–853. [PubMed] [Google Scholar]
- 30.Benes P, Kankova K, Muzik J, et al. Methylenetetrahydrofolate reductase polymorphism, type II diabetes mellitus, coronary artery disease, and essential hypertension in the Czech population. Mol Genet Metab. 2001;73(2):188–195. doi: 10.1006/mgme.2001.3188. [DOI] [PubMed] [Google Scholar]
- 31.Temple I.K, Gardner R.J, Robinson D.O, et al. Further evidence for an imprinted gene for neonatal diabetes localised to chromosome 6q22-q23. Hum Mol Genet. 1996;5(8):1117–1121. doi: 10.1093/hmg/5.8.1117. [DOI] [PubMed] [Google Scholar]
- 32.Shield J, Owen K, Robinson D.O, et al. Maturity onset diabetes of the young (MODY) and early onset type II diabetes are not caused by loss of imprinting at the transient neonatal diabetes (TNDM) locus. Diabetologia. 2001;44(7):924. doi: 10.1007/s001250100557. [DOI] [PubMed] [Google Scholar]
- 33.Maier S, Olek A. Diabetes: a candidate disease for efficient DNA methylation profiling. J Nutr. 2002;132(suppl 8):2440S–2443S. doi: 10.1093/jn/132.8.2440S. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson C, Larsson B.M, Jennische E, et al. Maternal endotoxemia results in obesity and insulin resistance in adult male offspring. Endocrinology. 2001;142(6):2622–2630. doi: 10.1210/endo.142.6.8191. [DOI] [PubMed] [Google Scholar]
- 35.Pradhan A.D, Manson J.E, Rifai N, Buring J.E, Ridker P.M. C-reactive protein, interleukin 6, and risk of developing type 2 diabetes mellitus. JAMA. 2001;286(3):327–334. doi: 10.1001/jama.286.3.327. [DOI] [PubMed] [Google Scholar]
- 36.Kern P.A, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2001;280(5):E745–751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 37.Das U.N. Is obesity an inflammatory condition? Nutrition. 2001;17(11–12):953–966. doi: 10.1016/s0899-9007(01)00672-4. [DOI] [PubMed] [Google Scholar]
- 38.Moller D.E. Potential role of TNF-alpha in the pathogenesis of insulin resistance and type 2 diabetes. Trends Endocrinol Metab. 2000;11(6):212–217. doi: 10.1016/s1043-2760(00)00272-1. [DOI] [PubMed] [Google Scholar]
- 39.Yuan M, Konstantopoulos N, Lee J, et al. Reversal of obesity- and diet-induced insulin resistance with salicylates or targeted disruption of Ikkbeta [published correction appears in Science. 2002;295(5553):277] Science. 2001;293(5535):1673–1677. doi: 10.1126/science.1061620. [DOI] [PubMed] [Google Scholar]
- 40.Aljada A, Garg R, Ghanim H, et al. Nuclear factor-kappaB suppressive and inhibitor-kappaB stimulatory effects of troglitazone in obese patients with type 2 diabetes: evidence of an antiinflammatory action? J Clin Endocrinol Metab. 2001;86(7):3250–3256. doi: 10.1210/jcem.86.7.7564. [DOI] [PubMed] [Google Scholar]
- 41.Ziccardi P, Nappo F, Giugliano G, et al. Reduction of inflammatory cytokine concentrations and improvement of endothelial functions in obese women after weight loss over one year. Circulation. 2002;105(7):804–809. doi: 10.1161/hc0702.104279. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen B.K, Steensberg A, Fischer C, Keller C, Ostrowski K, Schjerling P. Exercise and cytokines with particular focus on muscle-derived IL-6. Exerc Immunol Rev. 2001;7:18–31. [PubMed] [Google Scholar]
- 43.Smith S.R, Bai F, Charbonneau C, Janderova L, Argyropoulos G. A promoter genotype and oxidative stress potentially link resistin to human insulin resistance. Diabetes. 2003;52(7):1611–1618. doi: 10.2337/diabetes.52.7.1611. [DOI] [PubMed] [Google Scholar]
- 44.Wolford J.K, Gruber J.D, Ossowski V.M, et al. A C-reactive protein promoter polymorphism is associated with type 2 diabetes mellitus in Pima Indians [published correction appears in Mol Genet Metab. 2003;79(3):231] Mol Genet Metab. 2003;78(2):136–144. doi: 10.1016/s1096-7192(02)00230-5. [DOI] [PubMed] [Google Scholar]
- 45.Vozarova B, Fernandez-Real J.M, Knowler W.C, et al. The interleukin-6 (-174) G/C promoter polymorphism is associated with type-2 diabetes mellitus in Native Americans and Aaucasians. Hum Genet. 2003;112(4):409–413. doi: 10.1007/s00439-003-0912-x. [DOI] [PubMed] [Google Scholar]
- 46.Kaser S, Kaser A, Sandhofer A, Ebenbichler C.F, Tilg H, Patsch J.R. Resistin messenger-RNA expression is increased by proinflammatory cytokines in vitro. Biochem Biophys Res Commun. 2003;309(2):286–290. doi: 10.1016/j.bbrc.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Vickers M.A, Green F.R, Terry C, et al. Genotype at a promoter polymorphism of the interleukin-6 gene is associated with baseline levels of plasma C-reactive protein. Cardiovasc Res. 2002;53(4):1029–1034. doi: 10.1016/s0008-6363(01)00534-x. [DOI] [PubMed] [Google Scholar]
- 48.Wilson C.B, Makar K.W, Perez-Melgosa M. Epigenetic regulation of T cell fate and function. J Infect Dis. 2002;185(suppl 1):S37–45. doi: 10.1086/338001. [DOI] [PubMed] [Google Scholar]
- 49.Rothenburg S, Koch-Nolte F, Thiele H.G, Haag F. DNA methylation contributes to tissue- and allele-specific expression of the T-cell differentiation marker RT6. Immunogenetics. 2001;52(3–4):231–241. doi: 10.1007/s002510000267. [DOI] [PubMed] [Google Scholar]
- 50.Coppack S.W. Pro-inflammatory cytokines and adipose tissue. Proc Nutr Soc. 2001;60(3):349–356. doi: 10.1079/pns2001110. [DOI] [PubMed] [Google Scholar]
- 51.Benjamin D, Jost J.P. Reversal of methylation-mediated repression with short-chain fatty acids: evidence for an additional mechanism to histone deacetylation. Nucleic Acids Res. 2001;29(17):3603–3610. doi: 10.1093/nar/29.17.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sealy L, Chalkley R. The effect of sodium butyrate on histone modification. Cell. 1978;14(1):115–121. doi: 10.1016/0092-8674(78)90306-9. [DOI] [PubMed] [Google Scholar]
- 53.Foley J.E, Kashiwagi A, Verso M.A, Reaven G, Andrews J. Improvement in in vitro insulin action after one month of insulin therapy in obese noninsulin-dependent diabetics. Measurements of glucose transport and metabolism, insulin binding, and lipolysis in isolated adipocytes. J Clin Invest. 1983;72(6):1901–1909. doi: 10.1172/JCI111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green A, Dobias S.B, Walters D.J, Brasier A.R. Tumor necrosis factor increases the rate of lipolysis in primary cultures of adipocytes without altering levels of hormone-sensitive lipase. Endocrinology. 1994;134(6):2581–2588. doi: 10.1210/endo.134.6.8194485. [DOI] [PubMed] [Google Scholar]
- 55.Laimer M, Ebenbichler C.F, Kaser S, et al. Markers of chronic inflammation and obesity: a prospective study on the reversibility of this association in middle-aged women undergoing weight loss by surgical intervention. Int J Obes Relat Metab Disord. 2002;26(5):659–662. doi: 10.1038/sj.ijo.0801970. [DOI] [PubMed] [Google Scholar]
- 56.Haffner S.M, Stern M.P, Hazuda H.P, Pugh J.A, Patterson J.K. Hyperinsulinemia in a population at high risk for non-insulin-dependent diabetes mellitus. N Engl J Med. 1986;315(4):220–224. doi: 10.1056/NEJM198607243150403. [DOI] [PubMed] [Google Scholar]
- 57.Nathan D.M. Prevention of long-term complications of non-insulin-dependent diabetes mellitus. Clin Invest Med. 1995;18(4):332–339. [PubMed] [Google Scholar]
- 58.Michalowsky L.A, Jones P.A. DNA methylation and differentiation. Environ Health Perspect. 1989;80:189–197. doi: 10.1289/ehp.8980189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Catania J, Fairweather D.S. DNA methylation and cellular ageing. Mutat Res. 1991;256(2–6):283–293. doi: 10.1016/0921-8734(91)90019-8. [DOI] [PubMed] [Google Scholar]
- 60.Bjornsson H.T, Fallin M.D, Feinberg A.P. An integrated epigenetic and genetic approach to common human disease. Trends Genet. 2004;20(8):350–358. doi: 10.1016/j.tig.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 61.Toossi Z. The inflammatory response in mycobacterium tuberculosis infection. Arch Immunol Ther Exp (Warsz) 2000;48(6):513–519. [PubMed] [Google Scholar]
- 62.Cooper M.A, Fehniger T.A, Ponnappan A, Mehta V, Wewers M.D, Caligiuri M.A. Interleukin-1beta costimulates interferon-gamma production by human natural killer cells. Eur J Immunol. 2001;31(3):792–801. doi: 10.1002/1521-4141(200103)31:3<792::aid-immu792>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]