Abstract
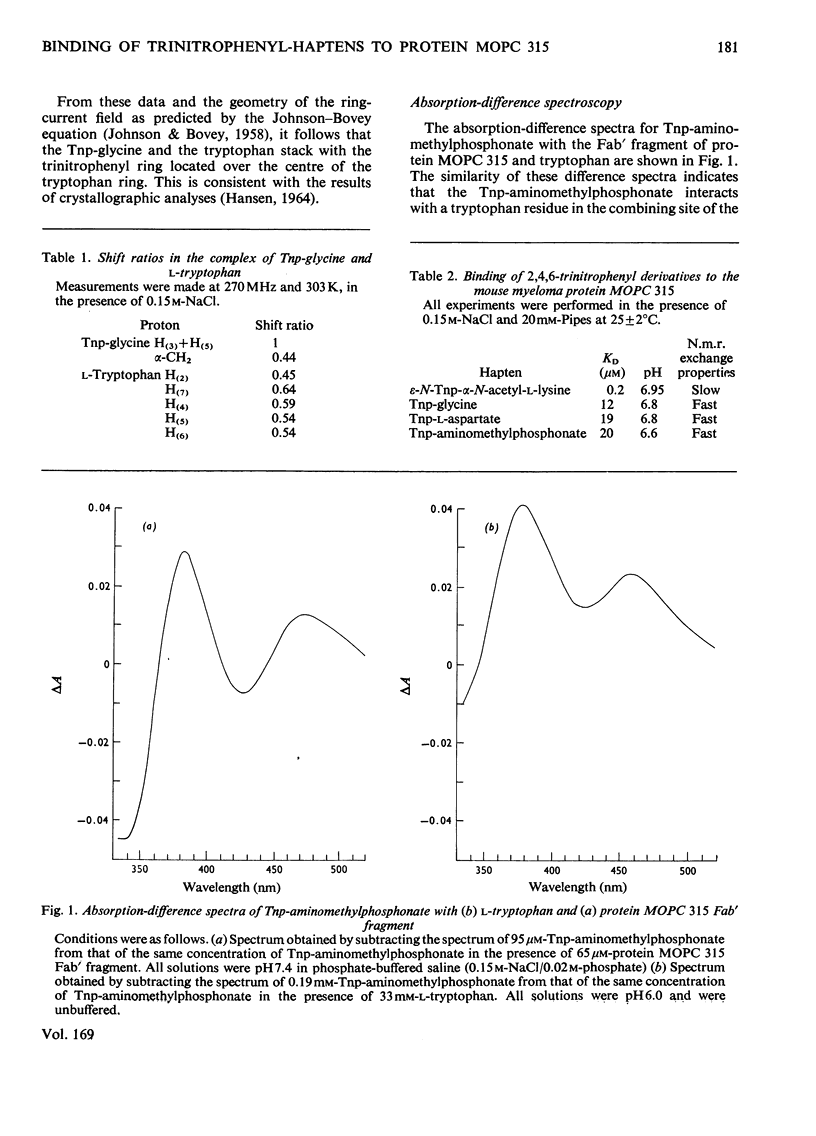
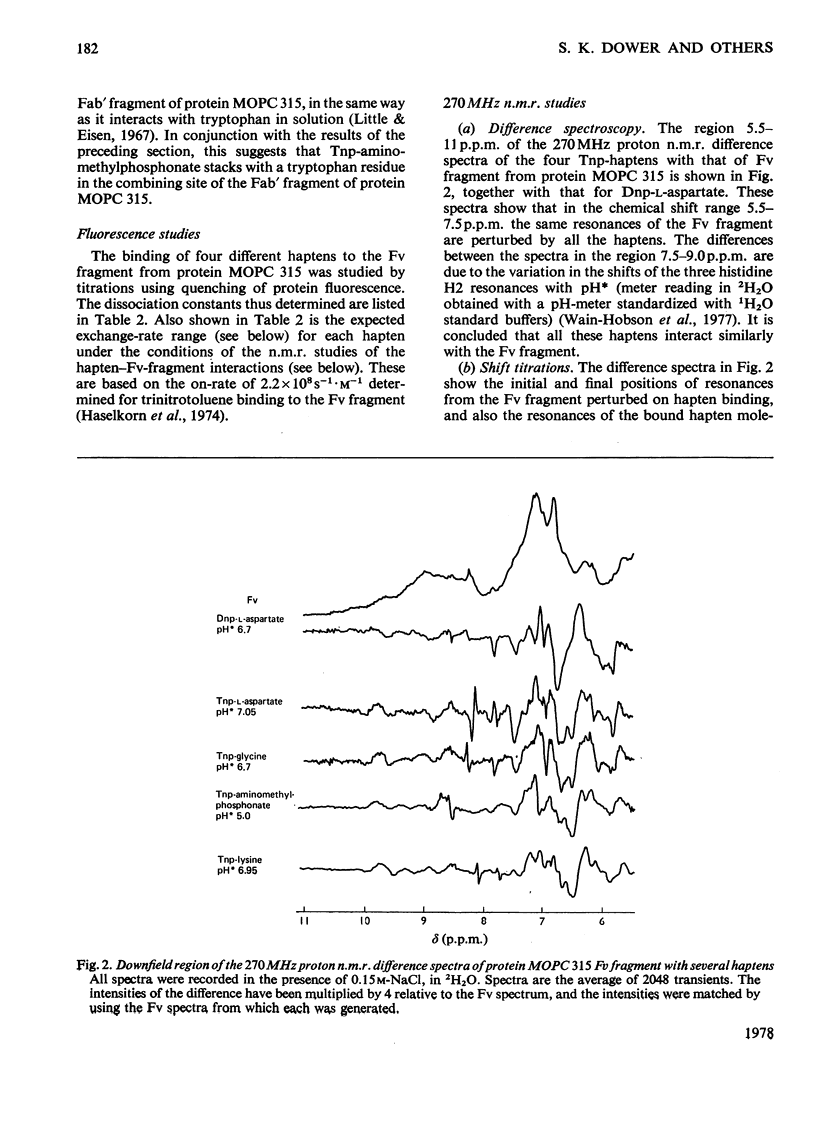
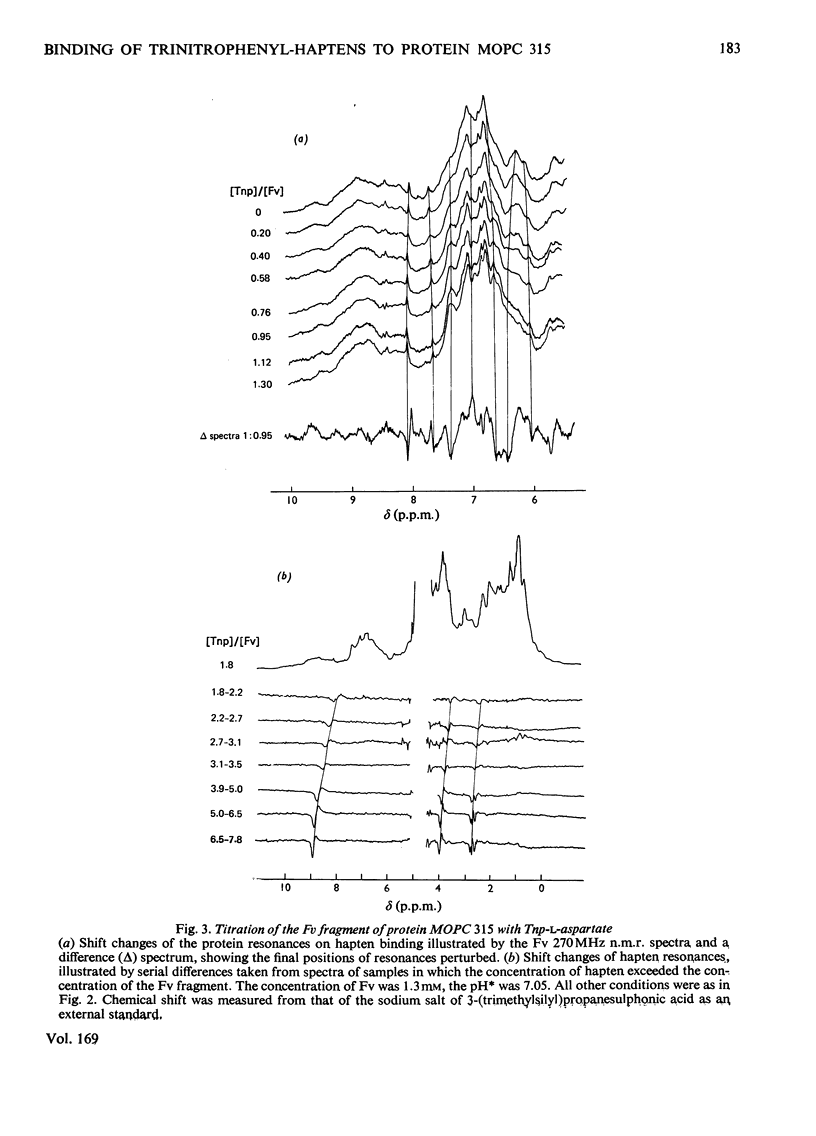
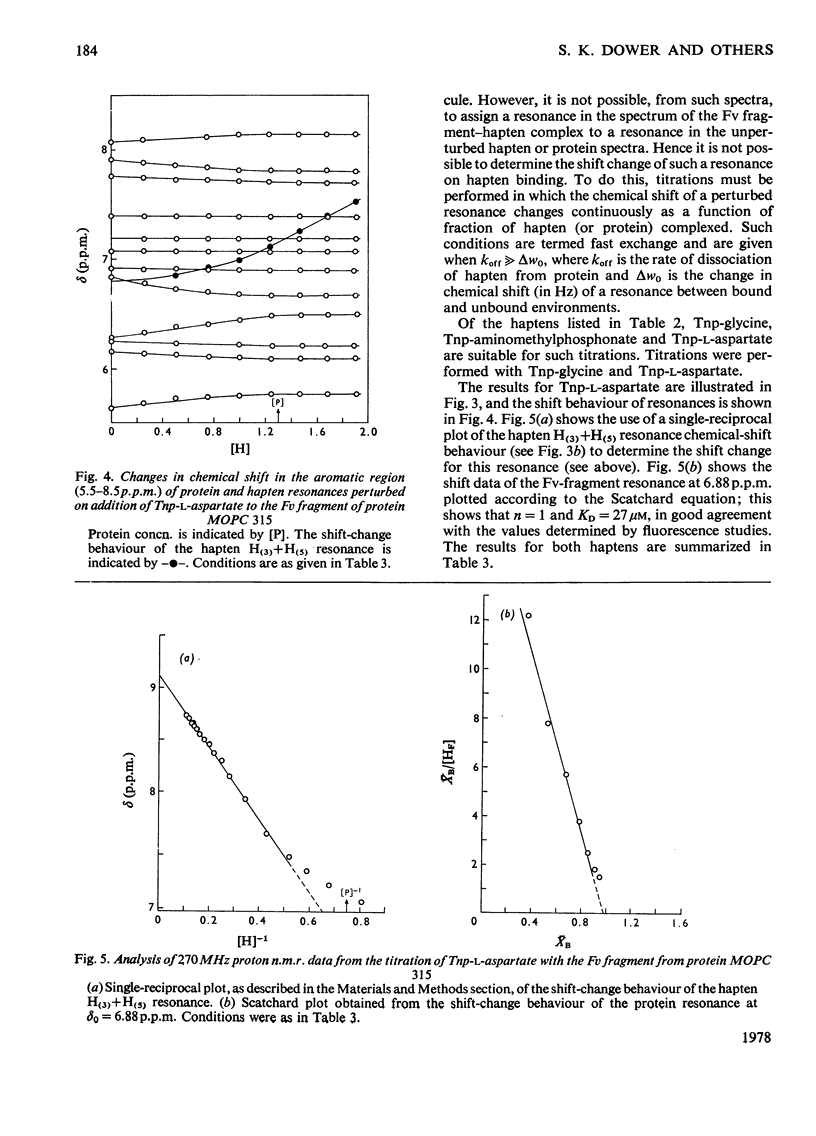
The binding of Tnp (2,4,6-trinitrophenyl) derivatives to the Fv fragment (variable region of heavy and light chains) of the mouse myeloma IgA protein MOPC 315 was investigated by 270MHz proton nuclear magnetic resonance. Two of the haptens, Tnp-glycine and Tnp-l-aspartate, are in fast exchange with the Fv fragment, and the changes in chemical shifts for both protein and hapten resonances were determined by titrations. For the tightly binding hapten ε-N-Tnp-α-N-acetyl-l-lysine, which is in slow exchange with the Fv fragment, the changes in chemical shifts for the hapten H3+H5 resonances were determined by cross-saturation. By using these data and the known structure of the combining site of protein MOPC 315 [Dwek, Wain-Hobson, Dower, Gettins, Sutton, Perkins & Givol (1977), Nature (London) 266, 31–37] the mode of binding of Tnp derivatives is deduced by ring-current calculations. The trinitrophenyl ring stacks with tryptophan-93L (light chain) in the `aromatic box' formed by tryptophan-93L, tyrosine-34L and phenyl-alanine-34H (heavy chain). Further evidence for the stacking interaction with a tryptophan residue is provided by the similarity of the optical-difference spectra observed with Tnp-aminomethylphosphonate in the presence of either the Fab fragment (light chain and N-terminal half of heavy chain) of protein MOPC 315 or tryptophan. These data show that the modes of binding of all the Tnp derivatives are very similar, despite a 100-fold range in their affinities. It is also concluded that the modes of binding of Dnp (2,4-dinitrophenyl) and Tnp derivatives to protein MOPC 315 are very similar, and that the structural basis for this is that the aromatic box is large enought to allow the trinitrophenyl ring to stack with tryptophan-93L while still forming hydrogen bonds to asparagine-36L and tyrosine-34L.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Davies D. R., Padlan E. A., Segal D. M. Three-dimensional structure of immunoglobulins. Annu Rev Biochem. 1975;44:639–667. doi: 10.1146/annurev.bi.44.070175.003231. [DOI] [PubMed] [Google Scholar]
- Dower S. K., Wain-Hobson S., Gettins P., Givol D., Jackson W. R., Perkins S. J., Sunderland C. A., Sutton B. J., Wright C. E., Dwek R. A. The combining site of the dinitrophenyl-binding immunoglobulin A myeloma protein MOPC 315. Biochem J. 1977 Aug 1;165(2):207–223. doi: 10.1042/bj1650207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek R. A., Givol D., Jones R., McLaughlin A. C., Wain-Hobson S., White A. I., Wright C. Interactions of the lanthanide- and hapten-binding sites in the Fv fragment from the myeloma protein MOPC 315. Biochem J. 1976 Apr 1;155(1):37–53. doi: 10.1042/bj1550037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwek R. A., Wain-Hobson S., Dower S., Gettins P., Sutton B., Perkins S. J., Givol D. Structure of an antibody combining site by magnetic resonance. Nature. 1977 Mar 3;266(5597):31–37. doi: 10.1038/266031a0. [DOI] [PubMed] [Google Scholar]
- Eisen H. N., Michaelides M. C., Underdown B. J., Schulenburg E. P., Simms E. S. Experimental approaches to homogenous antibody populations. Myeloma proteins with antihapten antibody activity. Fed Proc. 1970 Jan-Feb;29(1):78–84. [PubMed] [Google Scholar]
- Eisen H. N., Simms E. S., Potter M. Mouse myeloma proteins with antihapten antibody acitivity. The protein produced by plasma cell tumor MOPC-315. Biochemistry. 1968 Nov;7(11):4126–4134. doi: 10.1021/bi00851a048. [DOI] [PubMed] [Google Scholar]
- Haselkorn D., Friedman S., Givol D., Pecht I. Kinetic mapping of the antibody combining site by chemical relaxation spectrometry. Biochemistry. 1974 May 7;13(10):2210–2222. doi: 10.1021/bi00707a030. [DOI] [PubMed] [Google Scholar]
- Inbar D., Hochman J., Givol D. Localization of antibody-combining sites within the variable portions of heavy and light chains. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2659–2662. doi: 10.1073/pnas.69.9.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. F., Barisas B. G., Sturtevant J. M. Thermodynamics of hapten binding to MOPC 315 and MOPC 460 mouse myeloma proteins. Biochemistry. 1974 Jan 15;13(2):390–396. doi: 10.1021/bi00699a026. [DOI] [PubMed] [Google Scholar]
- Little J. R., Eisen H. N. Evidence for tryptophan in the active sites of antibodies to polynitrobenzenes. Biochemistry. 1967 Oct;6(10):3119–3125. doi: 10.1021/bi00862a020. [DOI] [PubMed] [Google Scholar]
- Little J. R., Eisen H. N. Specificity of the immune response to the 2,4-dinitrophenyl and 2,4,6-trinitrophenyl groups. Ligand binding and fluorescence properties of cross-reacting antibodies. J Exp Med. 1969 Feb 1;129(2):247–265. doi: 10.1084/jem.129.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padlan E. A., Davies D. R., Pecht I., Givol D., Wright C. Model-building studies of antigen-binding sites: the hapten-binding site of mopc-315. Cold Spring Harb Symp Quant Biol. 1977;41(Pt 2):627–637. doi: 10.1101/sqb.1977.041.01.072. [DOI] [PubMed] [Google Scholar]
- Wain-Hobson S., Dower S. K., Gettins P., Givol D., McLaughlin A. C., Pecht I., Sunderland C. A., Dwek R. A. Specificity of interactions of hapten side chains with the combining site of the myeloma protein MOPC 315. Biochem J. 1977 Aug 1;165(2):227–235. doi: 10.1042/bj1650227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T., Kabat E. A. An analysis of the sequences of the variable regions of Bence Jones proteins and myeloma light chains and their implications for antibody complementarity. J Exp Med. 1970 Aug 1;132(2):211–250. doi: 10.1084/jem.132.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]


