Abstract
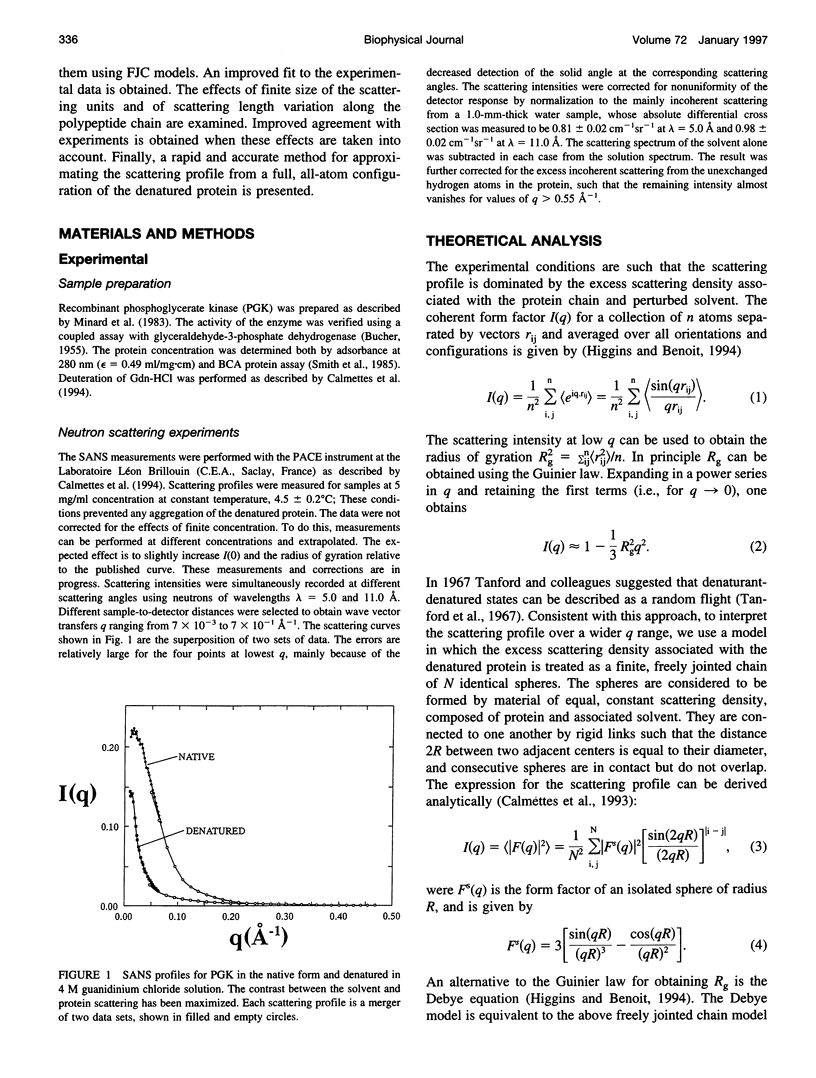
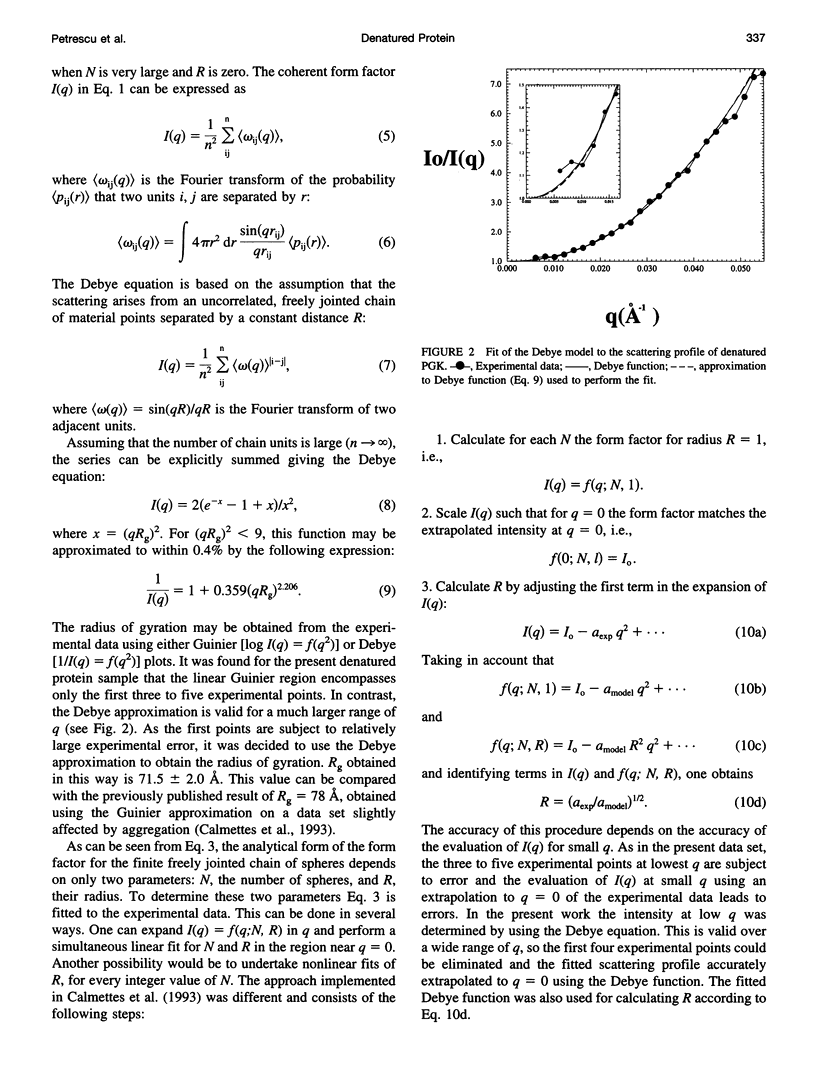
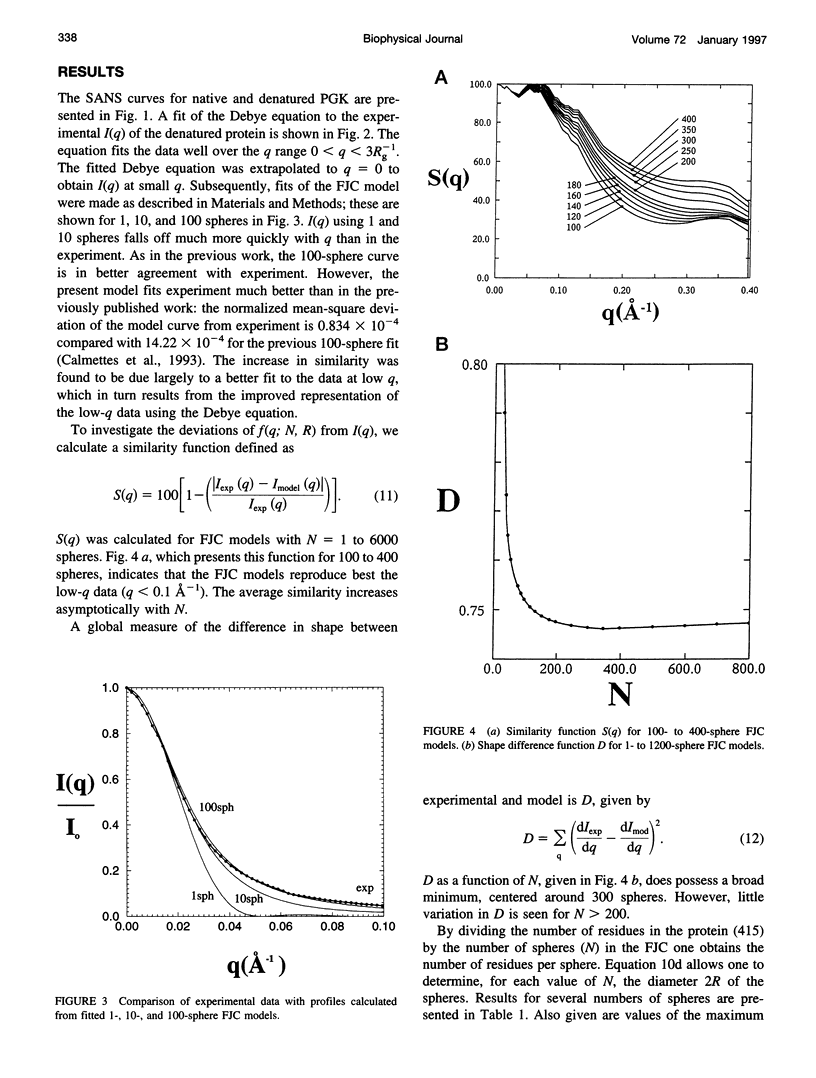
Small-angle neutron scattering profiles are presented from phosphoglycerate kinase, in the native form and strongly denatured in 4 M guanidinium chloride (GdnHCl) solution. The data are interpreted using a model in which the excess scattering density associated with the protein is represented as a finite freely jointed chain of spheres. The similarity of the model-derived scattering function to experiment increases asymptotically with the number of spheres. The improvement of the fit obtained with more than approximately 200 spheres (i.e., two residues per sphere) is insignificant. The effects of finite size of the scattering units and of scattering length variation along the polypeptide chain are examined. Improved agreement with experiment is obtained when these effects are taken into account. A method for rapid calculation of the scattering profile of a full, all-atom configuration is examined. It is found that a representation of the chain containing two scattering units per residue, placed at the backbone and side-chain scattering length centroids, reproduces the full, all-atom profile to within 2%.
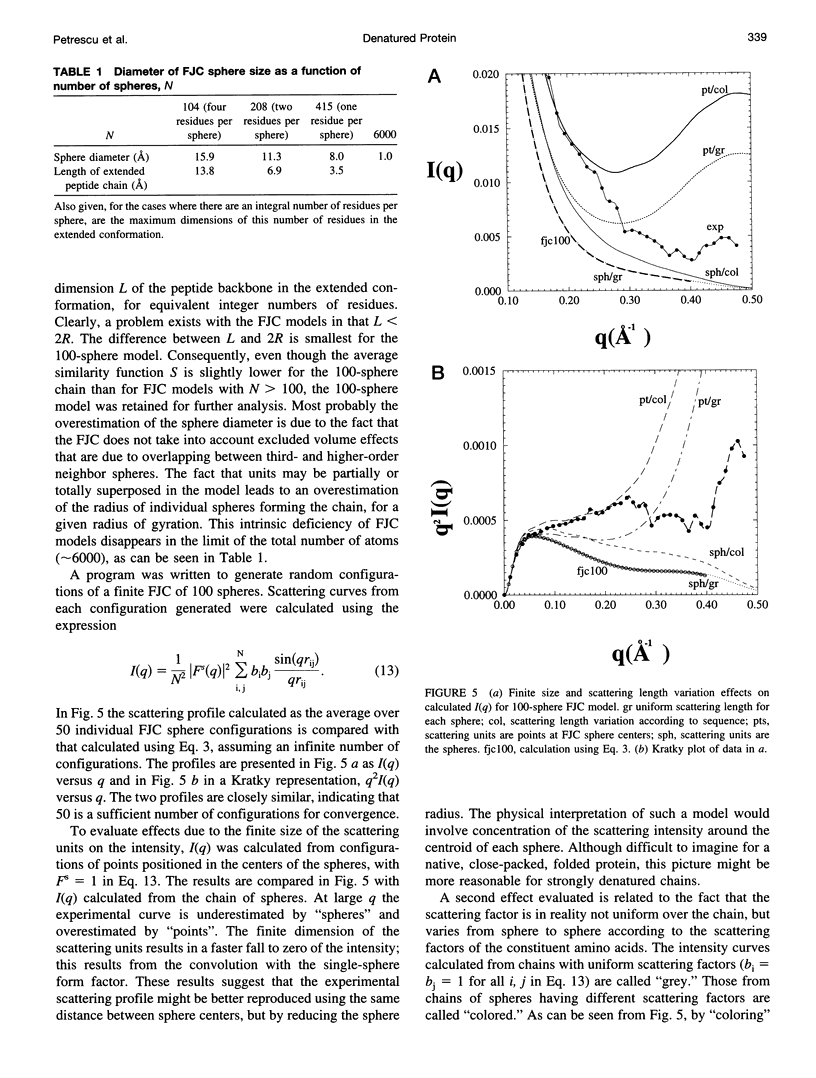
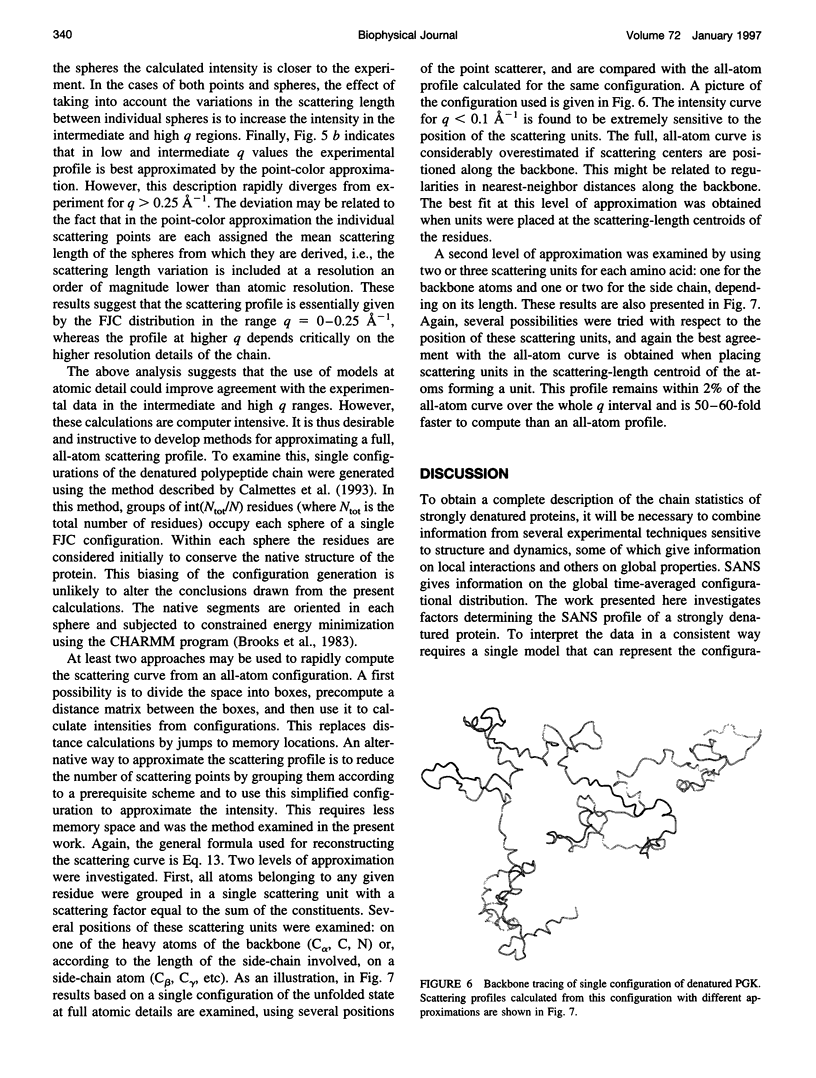
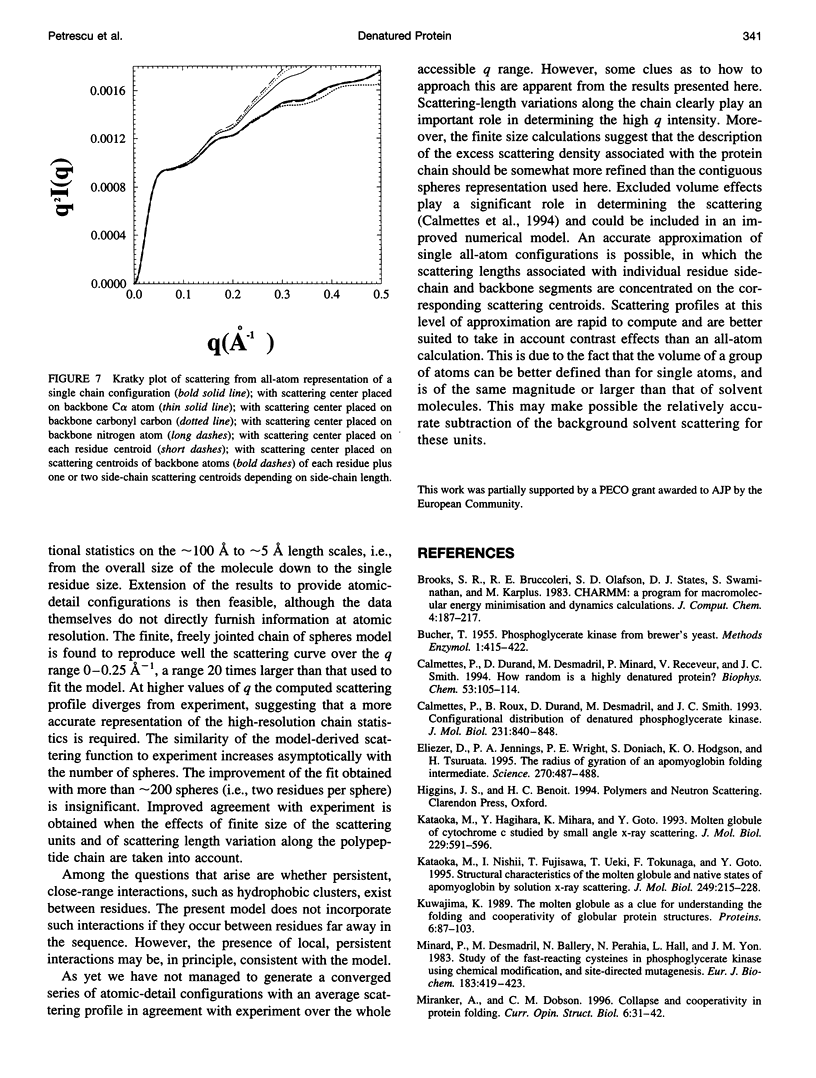
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Calmettes P., Durand D., Desmadril M., Minard P., Receveur V., Smith J. C. How random is a highly denatured protein? Biophys Chem. 1994 Dec;53(1-2):105–113. doi: 10.1016/0301-4622(94)00081-6. [DOI] [PubMed] [Google Scholar]
- Calmettes P., Roux B., Durand D., Desmadril M., Smith J. C. Configurational distribution of denatured phosphoglycerate kinase. J Mol Biol. 1993 Jun 5;231(3):840–848. doi: 10.1006/jmbi.1993.1330. [DOI] [PubMed] [Google Scholar]
- Eliezer D., Jennings P. A., Wright P. E., Doniach S., Hodgson K. O., Tsuruta H. The radius of gyration of an apomyoglobin folding intermediate. Science. 1995 Oct 20;270(5235):487–488. doi: 10.1126/science.270.5235.487. [DOI] [PubMed] [Google Scholar]
- Kataoka M., Hagihara Y., Mihara K., Goto Y. Molten globule of cytochrome c studied by small angle X-ray scattering. J Mol Biol. 1993 Feb 5;229(3):591–596. doi: 10.1006/jmbi.1993.1064. [DOI] [PubMed] [Google Scholar]
- Kataoka M., Nishii I., Fujisawa T., Ueki T., Tokunaga F., Goto Y. Structural characterization of the molten globule and native states of apomyoglobin by solution X-ray scattering. J Mol Biol. 1995 May 26;249(1):215–228. doi: 10.1006/jmbi.1995.0290. [DOI] [PubMed] [Google Scholar]
- Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6(2):87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- Miranker A. D., Dobson C. M. Collapse and cooperativity in protein folding. Curr Opin Struct Biol. 1996 Feb;6(1):31–42. doi: 10.1016/s0959-440x(96)80092-3. [DOI] [PubMed] [Google Scholar]
- Neri D., Billeter M., Wider G., Wüthrich K. NMR determination of residual structure in a urea-denatured protein, the 434-repressor. Science. 1992 Sep 11;257(5076):1559–1563. doi: 10.1126/science.1523410. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Dobson C. M., Evans P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992 Jul 23;358(6384):302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- Shortle D. R. Structural analysis of non-native states of proteins by NMR methods. Curr Opin Struct Biol. 1996 Feb;6(1):24–30. doi: 10.1016/s0959-440x(96)80091-1. [DOI] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]