Abstract
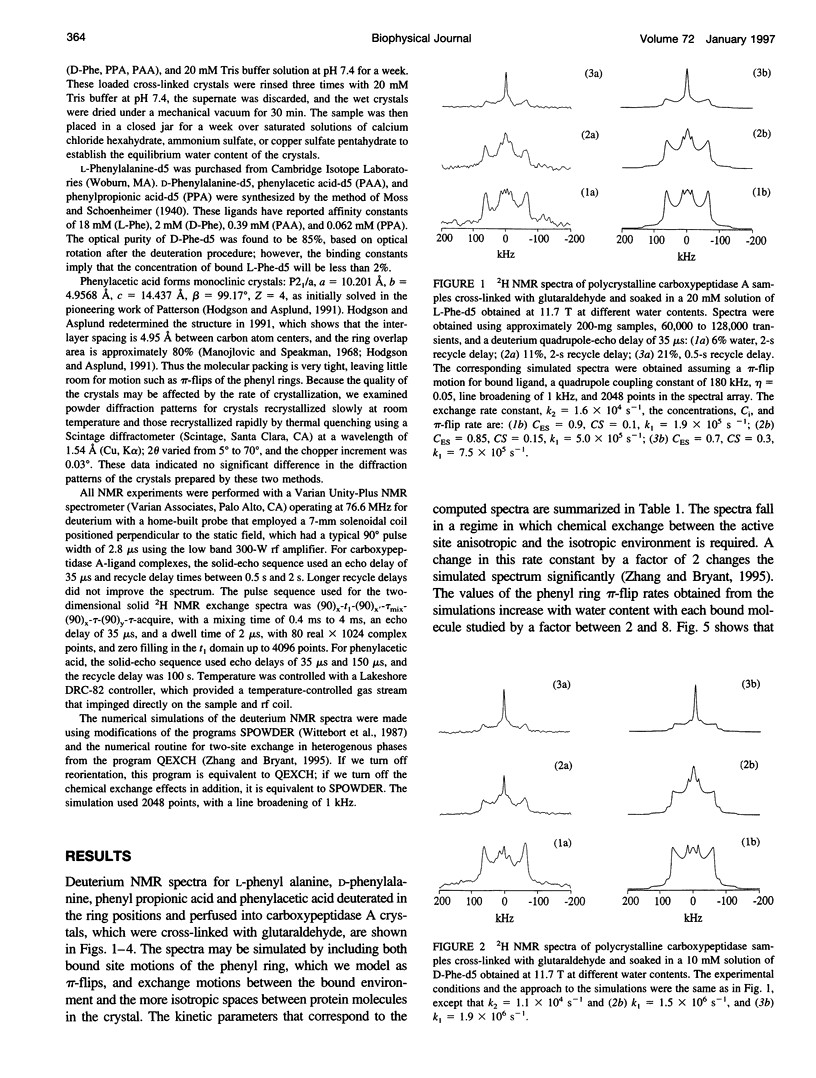
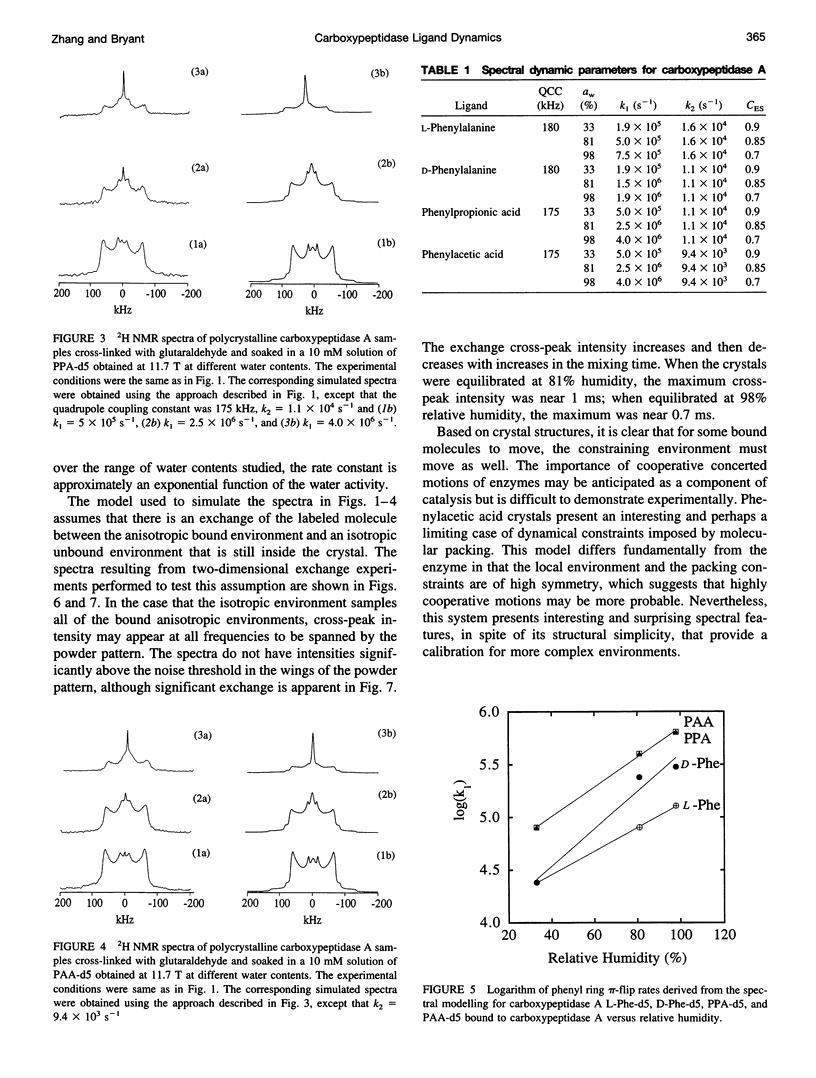
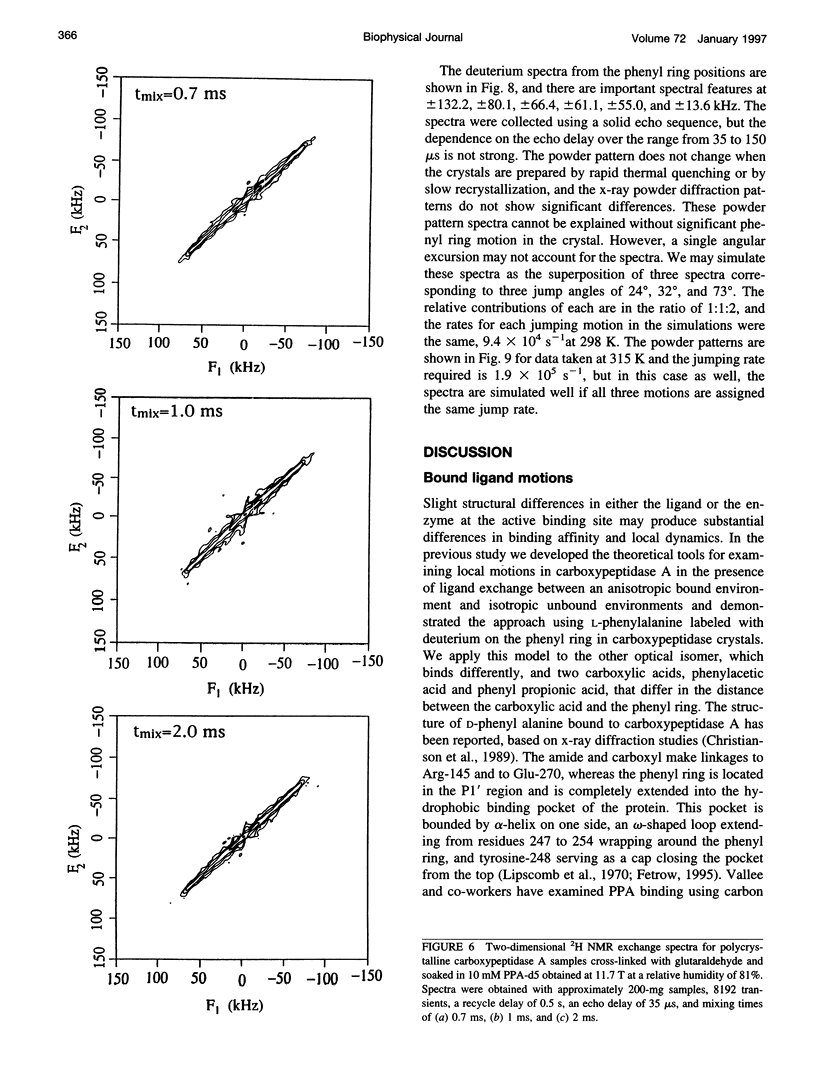
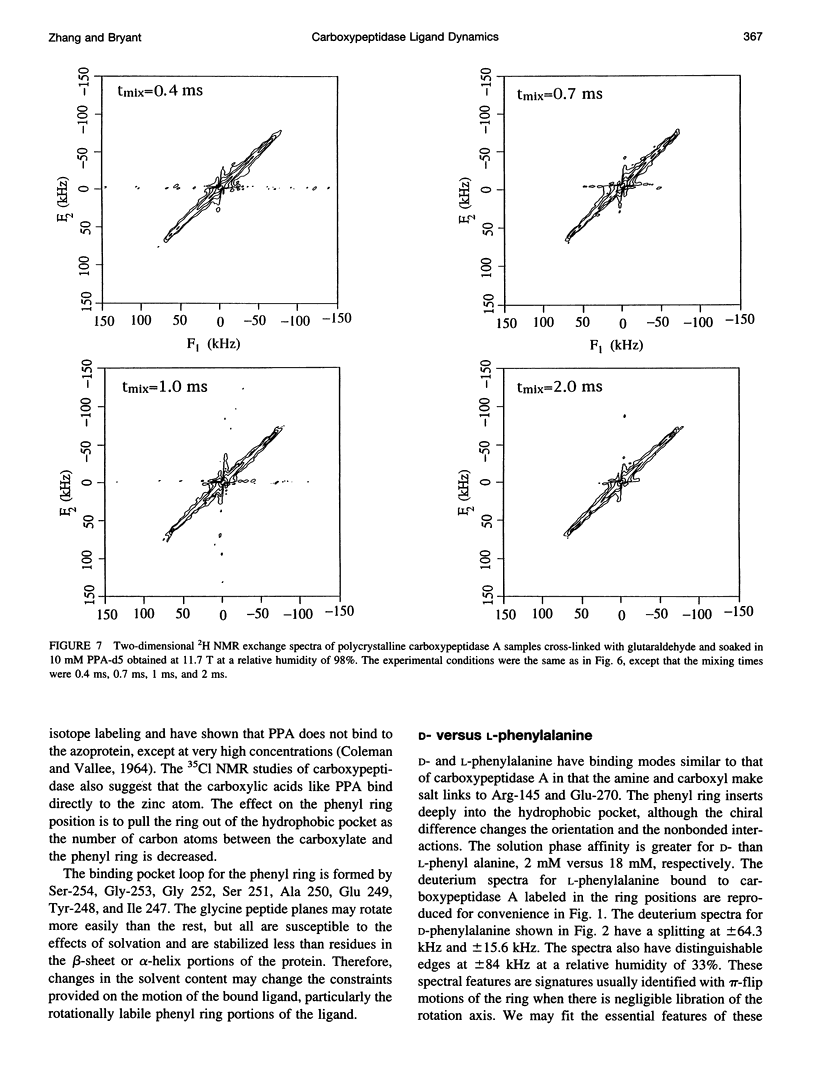
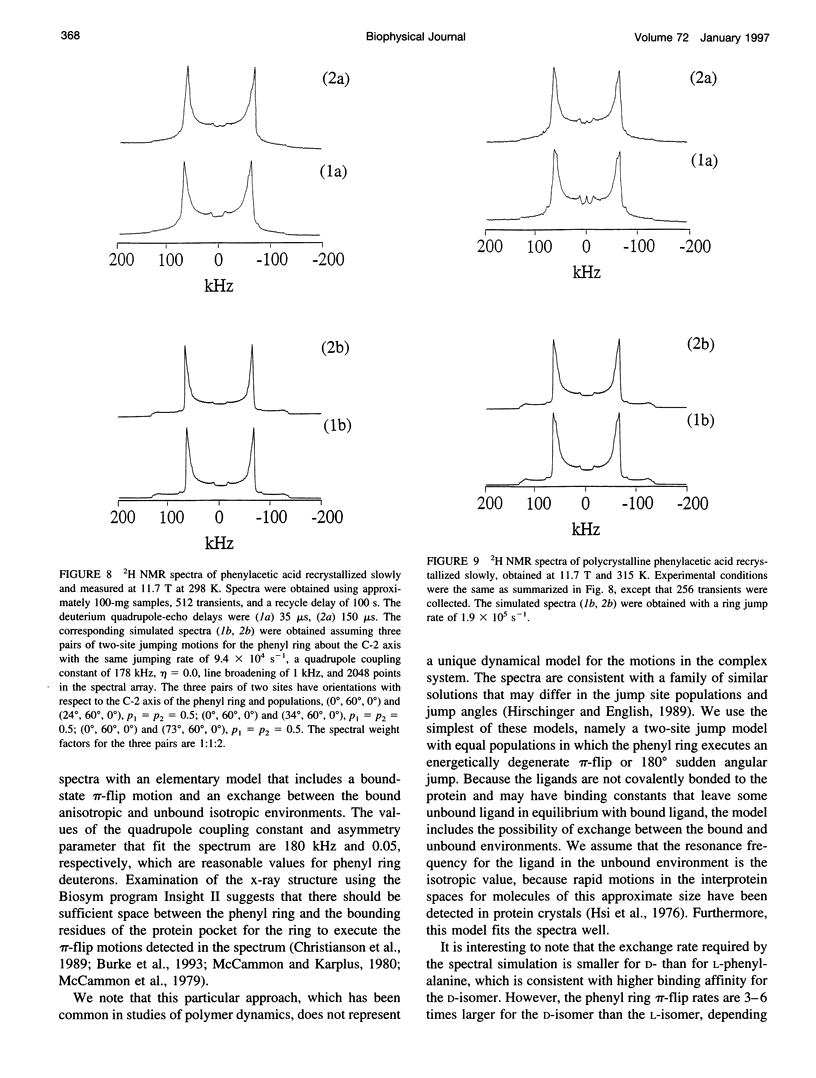
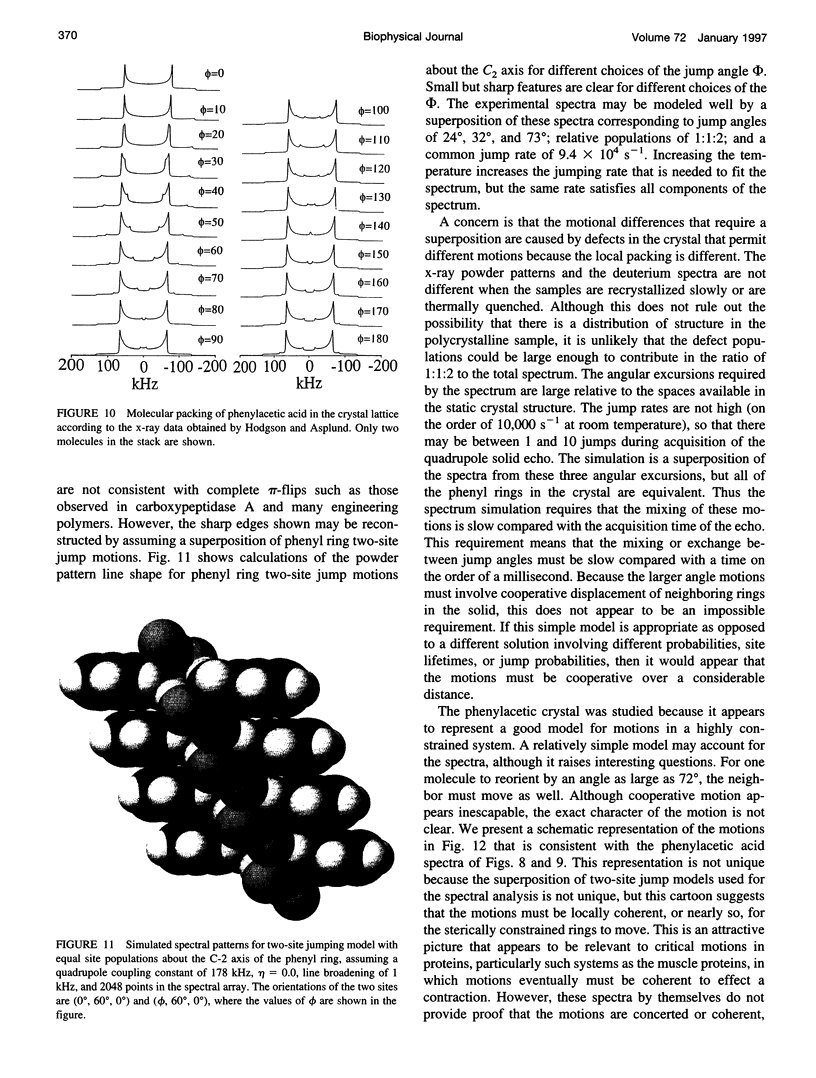
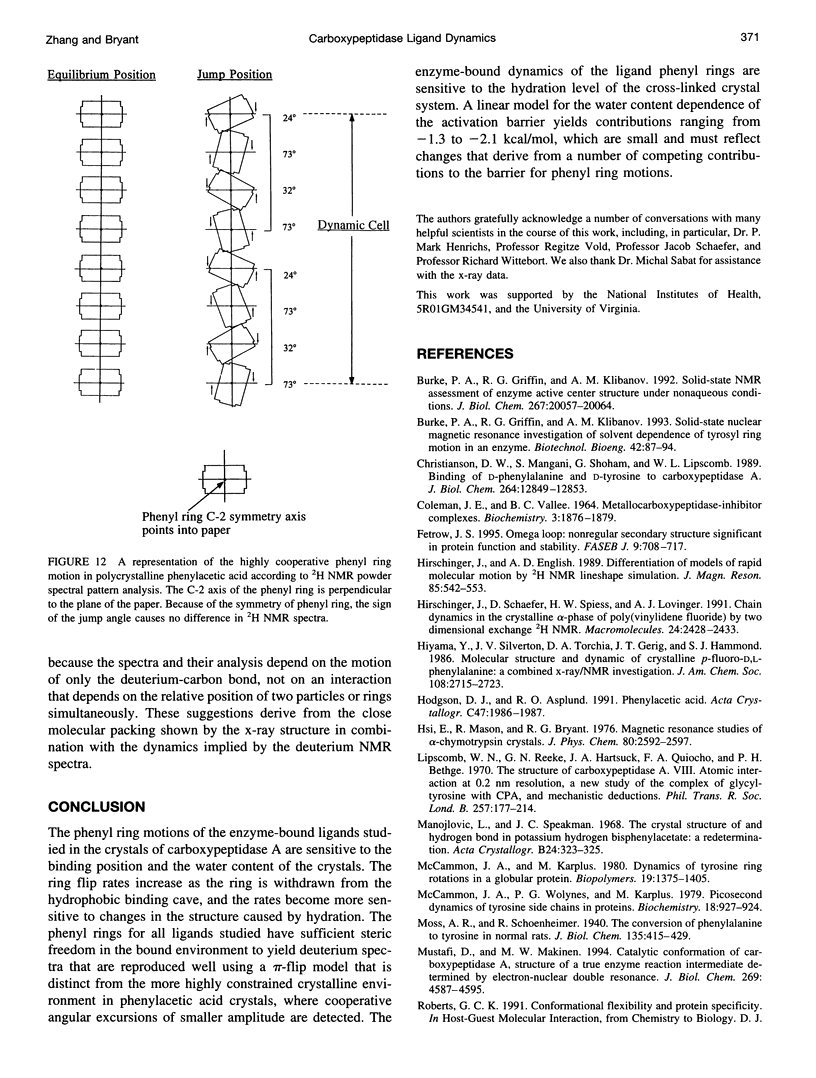
Deuterium NMR spectra for the phenyl ring deuterons have been obtained for D-phenylalanine, L-phenylalanine, phenylacetic acid, and phenyl propionic acid in randomly oriented crystals of carboxypeptidase A as a function of water content. The spectra are analyzed using a two-site jump model for phenyl ring pi-flips when the ligand is bound to the protein, and the model includes the possibility that the ligand may exchange with isotropic or unbound environments within the crystal. Although the binding pocket may impose local dynamical constraints, a complete pi-flip motion is consistent with the spectra of all ligands at all water contents. The rate constants for the pi-flip at 298 K are found to be 7.5 x 10(5) S-1, 1.9 x 10(6) S-1, 4.0 x 10(6) S-1, and 4.0 x 10(6) S-1 for L-phenylalanine, D-phenylalanine, phenyl propionic acid, and phenylacetic acid, respectively, at water activity of 0.98. The pi-flip rate for the ligand bound to the enzyme increases with water content. Assuming that the activation barrier may be written, delta G+2 = delta G+2o + baw, where aw is the water activity, and the value of b is -1.9 kcal/mol for phenylacetic acid and phenyl propionic acid, -1.3 kcal/mol for L-phenylalanine, and -2.1 kcal/mol for D-phenylalanine. Phenylacetic acid crystals were studied as an example of a phenyl ring motion that is highly constrained by a known and symmetrical packing environment. The deuterium spectra are complex and are not consistent with pi-flip motions, but they are consistent with a superposition of ring jump motions of 24 degrees, 34 degrees, and 72 degrees, with probabilities in the ratio of 1:1:2. Because of the limited space for motion imposed by the tight packing in the crystal, these motions must be highly cooperative and probably locally coherent; however, the spectra by themselves do not prove this intuitively reasonable hypothesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biering-Sørensen M., Biering-Sørensen K., Kaufmann B. Opfølgning i skolealderen af screening af 4-årige børn. Specielt med henblik på vurdering af sprogfunktionen. Ugeskr Laeger. 1989 May 15;151(20):1240–1244. [PubMed] [Google Scholar]
- Burke P. A., Griffin R. G., Klibanov A. M. Solid-state NMR assessment of enzyme active center structure under nonaqueous conditions. J Biol Chem. 1992 Oct 5;267(28):20057–20064. [PubMed] [Google Scholar]
- COLEMAN J. E., VALLEE B. L. METALLOCARBOXYPEPTIDASE-INHIBITOR COMPLEXES. Biochemistry. 1964 Dec;3:1874–1879. doi: 10.1021/bi00900a014. [DOI] [PubMed] [Google Scholar]
- Christianson D. W., Mangani S., Shoham G., Lipscomb W. N. Binding of D-phenylalanine and D-tyrosine to carboxypeptidase A. J Biol Chem. 1989 Aug 5;264(22):12849–12853. [PubMed] [Google Scholar]
- Fetrow J. S. Omega loops: nonregular secondary structures significant in protein function and stability. FASEB J. 1995 Jun;9(9):708–717. [PubMed] [Google Scholar]
- Lipscomb W. N., Reeke G. N., Jr, Hartsuck J. A., Quiocho F. A., Bethge P. H. The structure of carboxypeptidase A. 8. Atomic interpretation at 0.2 nm resolution, a new study of the complex of glycyl-L-tyrosine with CPA, and mechanistic deductions. Philos Trans R Soc Lond B Biol Sci. 1970 Feb 12;257(813):177–214. doi: 10.1098/rstb.1970.0020. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Wolynes P. G., Karplus M. Picosecond dynamics of tyrosine side chains in proteins. Biochemistry. 1979 Mar 20;18(6):927–942. doi: 10.1021/bi00573a001. [DOI] [PubMed] [Google Scholar]
- Mustafi D., Makinen M. W. Catalytic conformation of carboxypeptidase A. Structure of a true enzyme reaction intermediate determined by electron nuclear double resonance. J Biol Chem. 1994 Feb 11;269(6):4587–4595. [PubMed] [Google Scholar]
- Paupardin-Tritsch D., Hammond C., Gerschenfeld H. M. Serotonin and cyclic GMP both induce an increase of the calcium current in the same identified molluscan neurons. J Neurosci. 1986 Sep;6(9):2715–2723. doi: 10.1523/JNEUROSCI.06-09-02715.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994 Nov;20(3):216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- Rupley J. A., Careri G. Protein hydration and function. Adv Protein Chem. 1991;41:37–172. doi: 10.1016/s0065-3233(08)60197-7. [DOI] [PubMed] [Google Scholar]
- Smith R. L., Oldfield E. Dynamic structure of membranes by deuterium NMR. Science. 1984 Jul 20;225(4659):280–288. doi: 10.1126/science.6740310. [DOI] [PubMed] [Google Scholar]
- Steitz T. A., Ludwig M. L., Quiocho F. A., Lipscomb W. N. The structure of carboxypepidase A. V. Studies of enzyme-substrate and enzyme-inhibitor complexes at 6 A resolution. J Biol Chem. 1967 Oct 25;242(20):4662–4668. [PubMed] [Google Scholar]
- Zhang H., Bryant R. G. Characterization of enzyme-bound ligand dynamics by solid-state NMR in the presence of ligand exchange: L-phenylalanine on carboxypeptidase A. Biophys J. 1995 Jan;68(1):303–311. doi: 10.1016/S0006-3495(95)80188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]



