Abstract
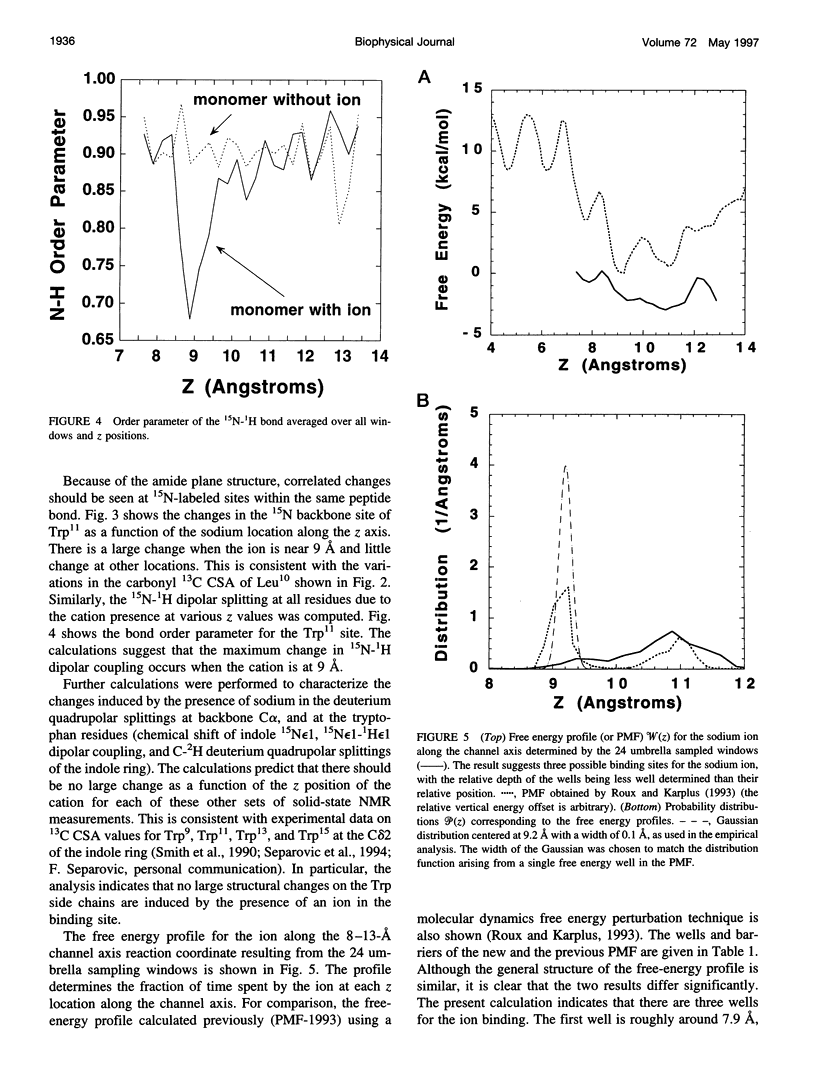
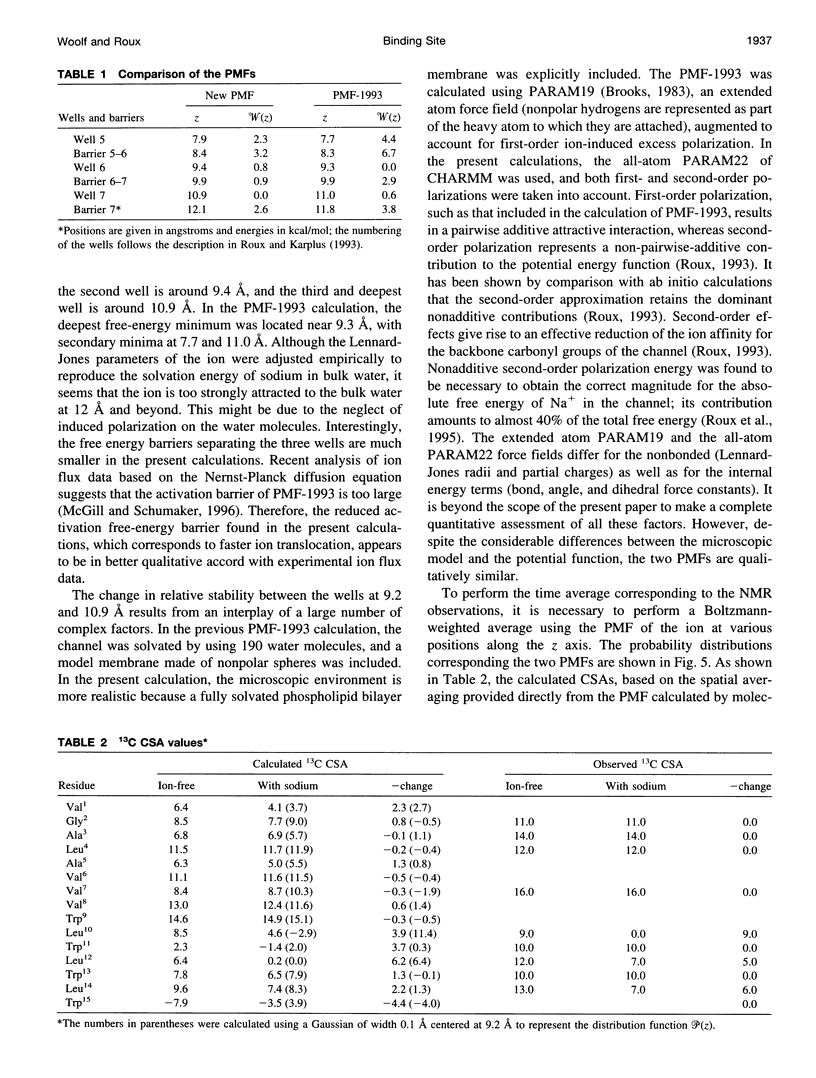
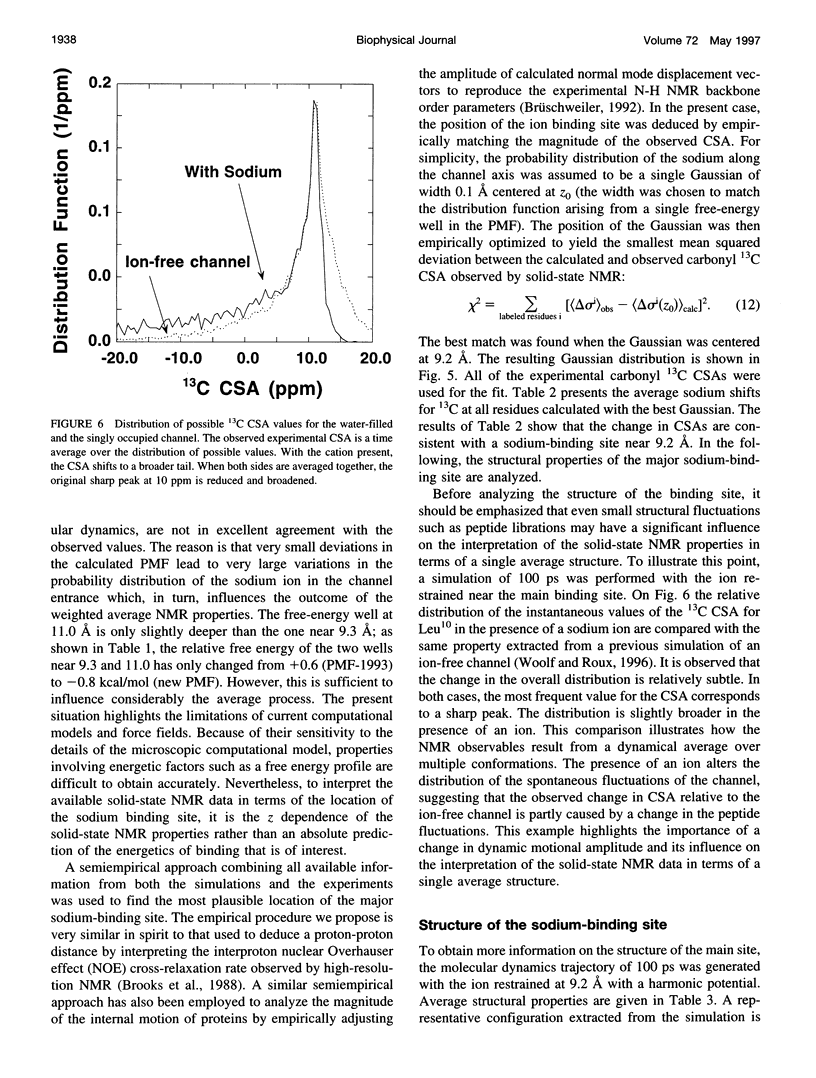
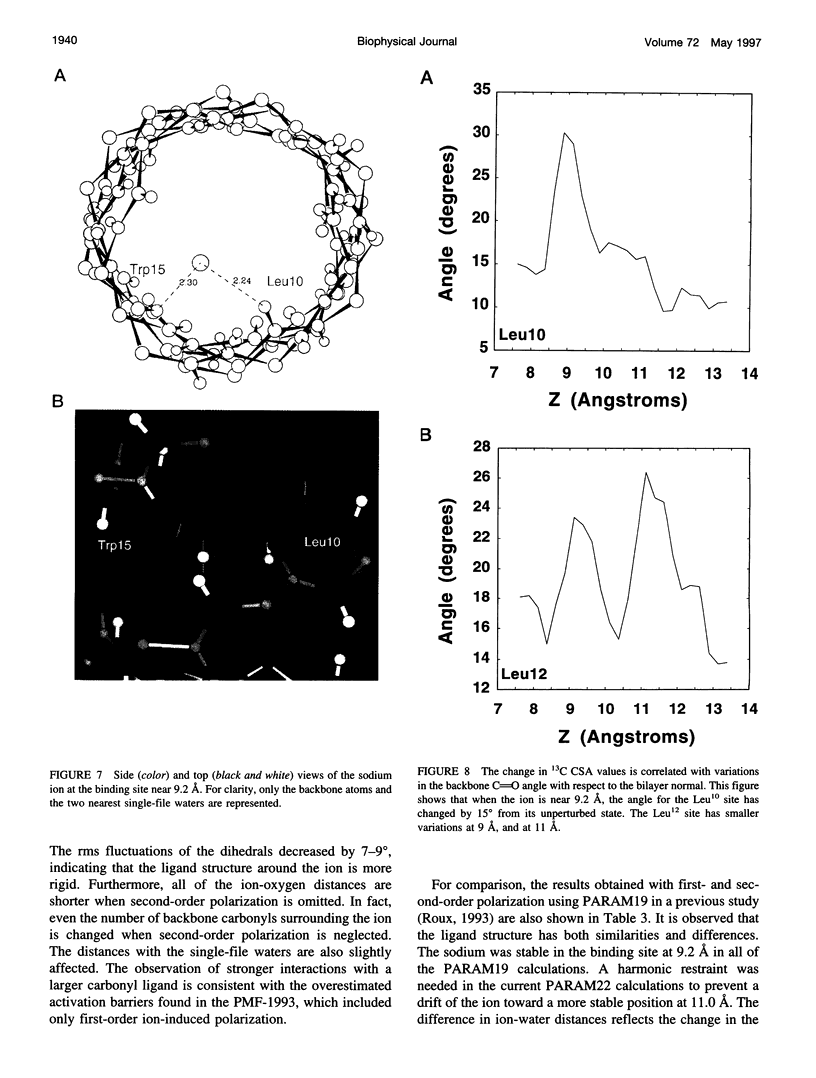
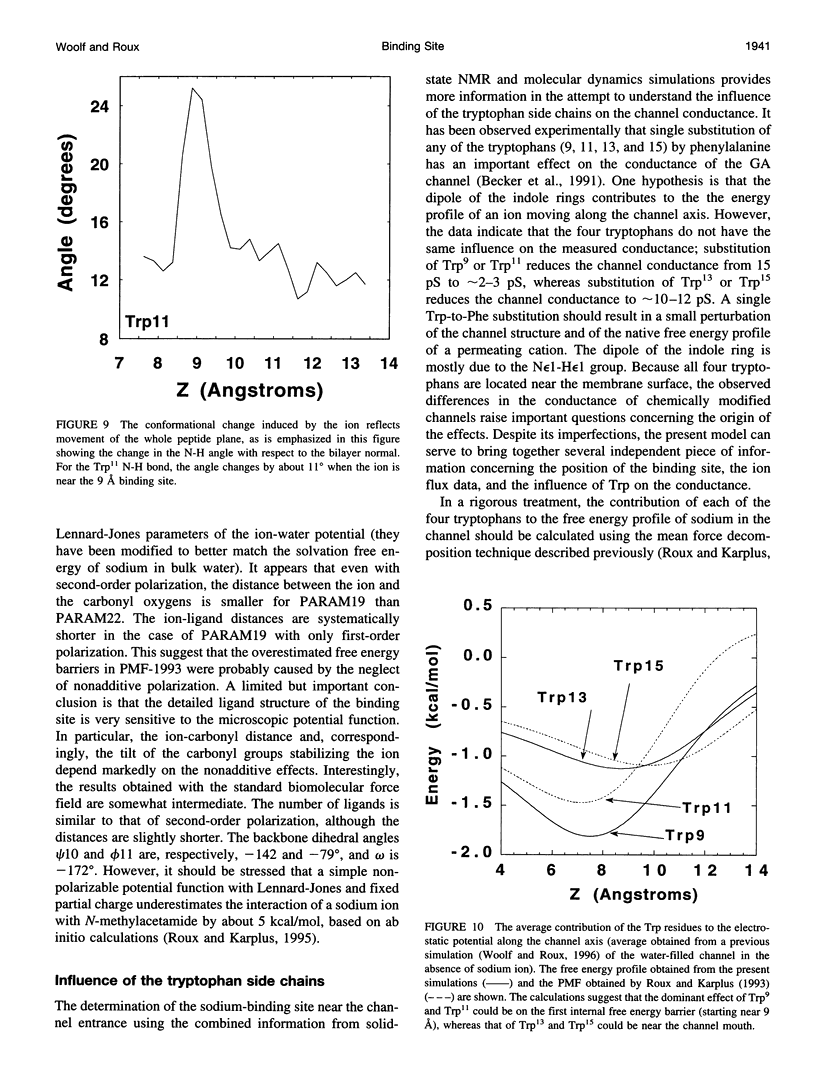
The location of the main binding site for sodium in the gramicidin A (GA) channel was investigated with molecular dynamics simulations, using an atomic model of the channel embedded in a fully hydrated dimyristoyl phosphatidycholine (DMPC) bilayer. Twenty-four separate simulations in which a sodium was restrained at different locations along the channel axis were generated. The results are compared with carbonyl 13C chemical shift anisotropy solid-state NMR experimental data previously obtained with oriented GA:DMPC samples. Predictions are made for other solid-state NMR properties that could be observed experimentally. The combined information from experiment and simulation strongly suggests that the main binding sites for sodium are near the channel's mouth, approximately 9.2 A from the center of the dimer channel. The 13C chemical shift anisotropy of Leu10 is the most affected by the presence of a sodium ion in the binding site. In the binding site, the sodium ion is lying off-axis, making contact with two carbonyl oxygens and two single-file water molecules. The main channel ligand is provided by the carbonyl group of the Leu10-Trp11 peptide linkage, which exhibits the largest deviation from the ion-free channel structure. Transient contacts with the carbonyl group of Val8 and Trp15 are also present. The influence of the tryptophan side chains on the channel conductance is examined based on the current information about the binding site.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen O. S., Koeppe R. E., 2nd Molecular determinants of channel function. Physiol Rev. 1992 Oct;72(4 Suppl):S89–158. doi: 10.1152/physrev.1992.72.suppl_4.S89. [DOI] [PubMed] [Google Scholar]
- Andersen O. S., Procopio J. Ion movement through gramicidin A channels. On the importance of the aqueous diffusion resistance and ion-water interactions. Acta Physiol Scand Suppl. 1980;481:27–35. [PubMed] [Google Scholar]
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Greathouse D. V., Koeppe R. E., 2nd, Andersen O. S. Amino acid sequence modulation of gramicidin channel function: effects of tryptophan-to-phenylalanine substitutions on the single-channel conductance and duration. Biochemistry. 1991 Sep 10;30(36):8830–8839. doi: 10.1021/bi00100a015. [DOI] [PubMed] [Google Scholar]
- Becker M. D., Koeppe R. E., 2nd, Andersen O. S. Amino acid substitutions and ion channel function. Model-dependent conclusions. Biophys J. 1992 Apr;62(1):25–27. doi: 10.1016/S0006-3495(92)81767-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busath D., Szabo G. Permeation characteristics of gramicidin conformers. Biophys J. 1988 May;53(5):697–707. doi: 10.1016/S0006-3495(88)83151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross T. A., Opella S. J. Protein structure by solid state nuclear magnetic resonance. Residues 40 to 45 of bacteriophage fd coat protein. J Mol Biol. 1985 Apr 5;182(3):367–381. doi: 10.1016/0022-2836(85)90197-4. [DOI] [PubMed] [Google Scholar]
- Eisenman G., Horn R. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J Membr Biol. 1983;76(3):197–225. doi: 10.1007/BF01870364. [DOI] [PubMed] [Google Scholar]
- Feller S. E., Pastor R. W. On simulating lipid bilayers with an applied surface tension: periodic boundary conditions and undulations. Biophys J. 1996 Sep;71(3):1350–1355. doi: 10.1016/S0006-3495(96)79337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B., Schwarz W. Potassium channels as multi-ion single-file pores. J Gen Physiol. 1978 Oct;72(4):409–442. doi: 10.1085/jgp.72.4.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Cross T. A. Tryptophan hydrogen bonding and electric dipole moments: functional roles in the gramicidin channel and implications for membrane proteins. Biochemistry. 1995 Oct 31;34(43):14147–14155. doi: 10.1021/bi00043a020. [DOI] [PubMed] [Google Scholar]
- Jing N., Prasad K. U., Urry D. W. The determination of binding constants of micellar-packaged gramicidin A by 13C-and 23Na-NMR. Biochim Biophys Acta. 1995 Aug 23;1238(1):1–11. doi: 10.1016/0005-2736(95)00095-k. [DOI] [PubMed] [Google Scholar]
- Jähnig F. What is the surface tension of a lipid bilayer membrane? Biophys J. 1996 Sep;71(3):1348–1349. doi: 10.1016/S0006-3495(96)79336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Koeppe R. E., 2nd, Killian J. A., Greathouse D. V. Orientations of the tryptophan 9 and 11 side chains of the gramicidin channel based on deuterium nuclear magnetic resonance spectroscopy. Biophys J. 1994 Jan;66(1):14–24. doi: 10.1016/S0006-3495(94)80748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs H., Mark A. E., Johansson J., van Gunsteren W. F. The effect of environment on the stability of an integral membrane helix: molecular dynamics simulations of surfactant protein C in chloroform, methanol and water. J Mol Biol. 1995 Apr 7;247(4):808–822. doi: 10.1016/s0022-2836(05)80156-1. [DOI] [PubMed] [Google Scholar]
- Läuger P. Ion transport through pores: a rate-theory analysis. Biochim Biophys Acta. 1973 Jul 6;311(3):423–441. doi: 10.1016/0005-2736(73)90323-4. [DOI] [PubMed] [Google Scholar]
- Nicholson L. K., Cross T. A. Gramicidin cation channel: an experimental determination of the right-handed helix sense and verification of beta-type hydrogen bonding. Biochemistry. 1989 Nov 28;28(24):9379–9385. doi: 10.1021/bi00450a019. [DOI] [PubMed] [Google Scholar]
- Olah G. A., Huang H. W., Liu W. H., Wu Y. L. Location of ion-binding sites in the gramicidin channel by X-ray diffraction. J Mol Biol. 1991 Apr 20;218(4):847–858. doi: 10.1016/0022-2836(91)90272-8. [DOI] [PubMed] [Google Scholar]
- Prosser R. S., Davis J. H., Dahlquist F. W., Lindorfer M. A. 2H nuclear magnetic resonance of the gramicidin A backbone in a phospholipid bilayer. Biochemistry. 1991 May 14;30(19):4687–4696. doi: 10.1021/bi00233a008. [DOI] [PubMed] [Google Scholar]
- Roux B., Karplus M. Ion transport in a model gramicidin channel. Structure and thermodynamics. Biophys J. 1991 May;59(5):961–981. doi: 10.1016/S0006-3495(91)82311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Karplus M. Molecular dynamics simulations of the gramicidin channel. Annu Rev Biophys Biomol Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- Roux B., Prod'hom B., Karplus M. Ion transport in the gramicidin channel: molecular dynamics study of single and double occupancy. Biophys J. 1995 Mar;68(3):876–892. doi: 10.1016/S0006-3495(95)80264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Separovic F., Gehrmann J., Milne T., Cornell B. A., Lin S. Y., Smith R. Sodium ion binding in the gramicidin A channel. Solid-state NMR studies of the tryptophan residues. Biophys J. 1994 Oct;67(4):1495–1500. doi: 10.1016/S0006-3495(94)80623-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R., Thomas D. E., Atkins A. R., Separovic F., Cornell B. A. Solid-state 13C-NMR studies of the effects of sodium ions on the gramicidin A ion channel. Biochim Biophys Acta. 1990 Jul 24;1026(2):161–166. doi: 10.1016/0005-2736(90)90059-w. [DOI] [PubMed] [Google Scholar]
- Smith R., Thomas D. E., Separovic F., Atkins A. R., Cornell B. A. Determination of the structure of a membrane-incorporated ion channel. Solid-state nuclear magnetic resonance studies of gramicidin A. Biophys J. 1989 Aug;56(2):307–314. doi: 10.1016/S0006-3495(89)82677-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Q., Nicholson L. K., Cross T. A. Experimental determination of torsion angles in the polypeptide backbone of the gramicidin A channel by solid state nuclear magnetic resonance. J Mol Biol. 1991 Apr 5;218(3):607–619. doi: 10.1016/0022-2836(91)90705-b. [DOI] [PubMed] [Google Scholar]
- Tian F., Lee K. C., Hu W., Cross T. A. Monovalent cation transport: lack of structural deformation upon cation binding. Biochemistry. 1996 Sep 17;35(37):11959–11966. doi: 10.1021/bi961170k. [DOI] [PubMed] [Google Scholar]
- Urry D. W., Prasad K. U., Trapane T. L. Location of monovalent cation binding sites in the gramicidin channel. Proc Natl Acad Sci U S A. 1982 Jan;79(2):390–394. doi: 10.1073/pnas.79.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W., Trapane T. L., Prasad K. U. Is the gramicidin a transmembrane channel single-stranded or double-stranded helix? A simple unequivocal determination. Science. 1983 Sep 9;221(4615):1064–1067. doi: 10.1126/science.221.4615.1064. [DOI] [PubMed] [Google Scholar]
- Urry D. W., Walker J. T., Trapane T. L. Ion interactions in (1-13C)D-Val8 and D-Leu14 analogs of gramicidin A, the helix sense of the channel and location of ion binding sites. J Membr Biol. 1982;69(3):225–231. doi: 10.1007/BF01870401. [DOI] [PubMed] [Google Scholar]
- Venable R. M., Zhang Y., Hardy B. J., Pastor R. W. Molecular dynamics simulations of a lipid bilayer and of hexadecane: an investigation of membrane fluidity. Science. 1993 Oct 8;262(5131):223–226. doi: 10.1126/science.8211140. [DOI] [PubMed] [Google Scholar]
- Woolf T. B., Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T. B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996 Jan;24(1):92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]