Abstract
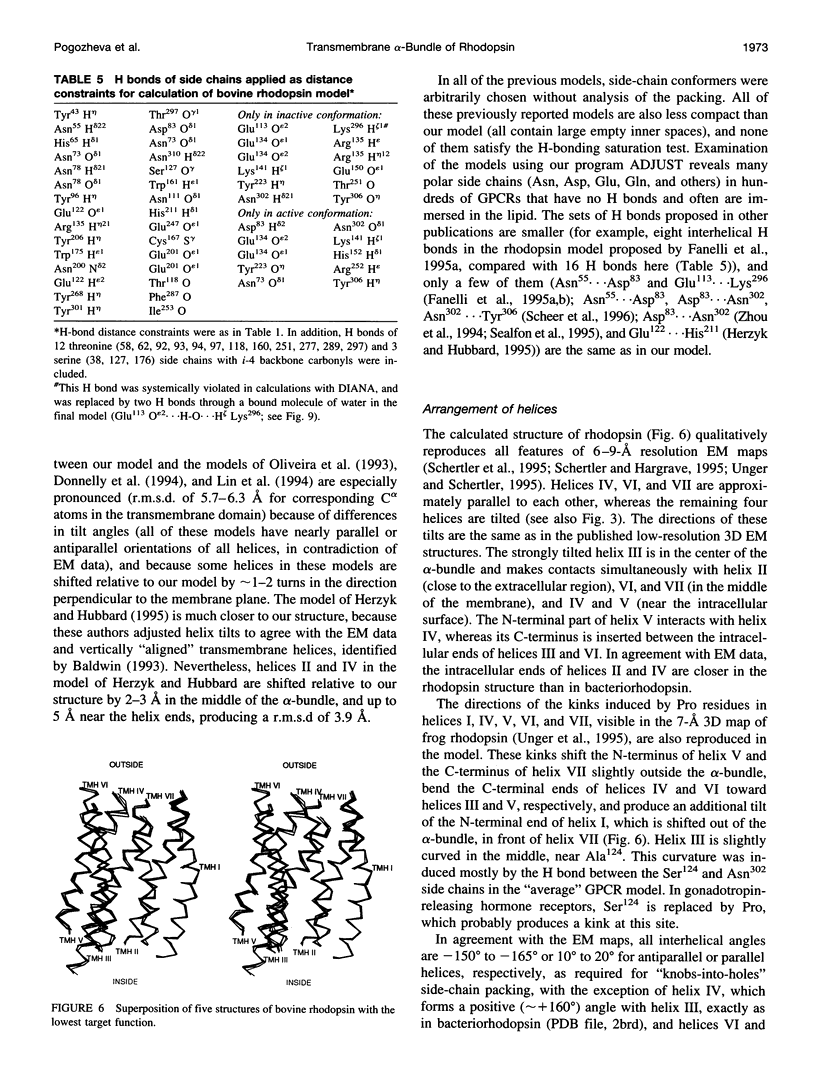
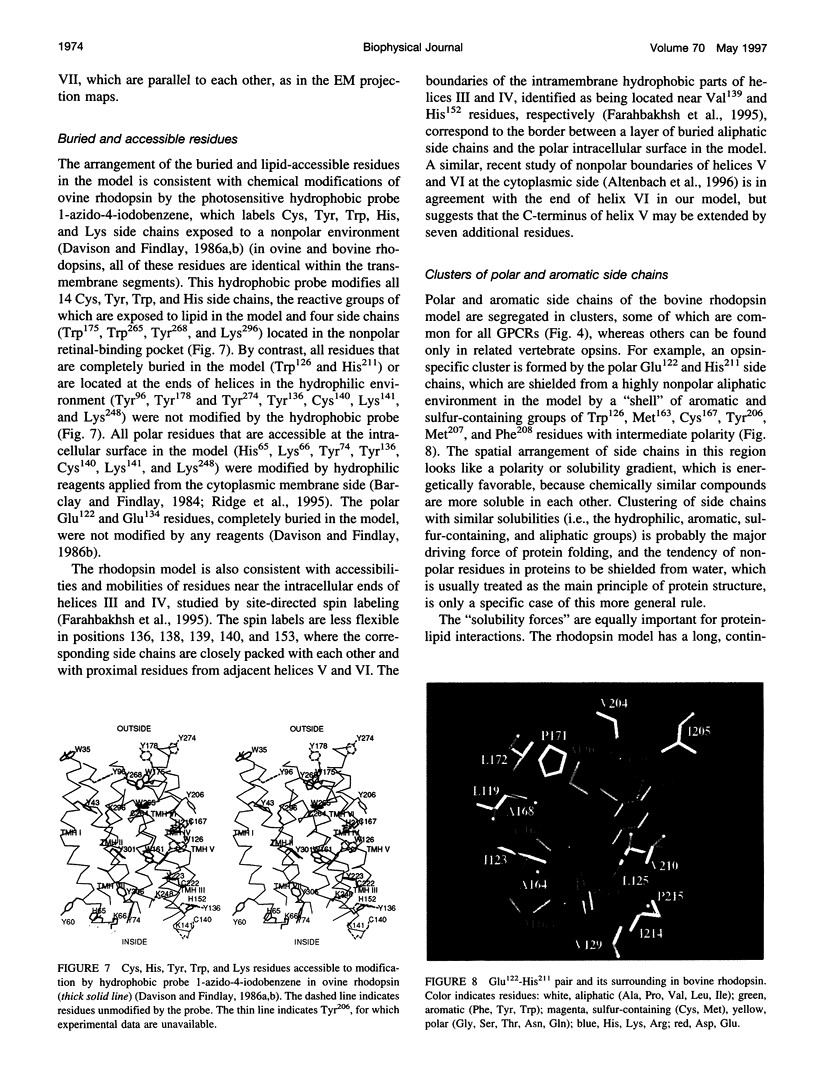
A 3D model of the transmembrane 7-alpha-bundle of rhodopsin-like G-protein-coupled receptors (GPCRs) was calculated using an iterative distance geometry refinement with an evolving system of hydrogen bonds, formed by intramembrane polar side chains in various proteins of the family and collectively applied as distance constraints. The alpha-bundle structure thus obtained provides H bonding of nearly all buried polar side chains simultaneously in the 410 GPCRs considered. Forty evolutionarily conserved GPCR residues form a single continuous domain, with an aliphatic "core" surrounded by six clusters of polar and aromatic side chains. The 7-alpha-bundle of a specific GPCR can be calculated using its own set of H bonds as distance constraints and the common "average" model to restrain positions of the helices. The bovine rhodopsin model thus determined is closely packed, but has a few small polar cavities, presumably filled by water, and has a binding pocket that is complementary to 11-cis (6-s-cis, 12-s-trans, C = N anti)-retinal or to all-trans-retinal, depending on conformations of the Lys296 and Trp265 side chains. A suggested mechanism of rhodopsin photoactivation, triggered by the cis-trans isomerization of retinal, involves rotations of Glu134, Tyr223, Trp265, Lys296, and Tyr306 side chains and rearrangement of their H bonds. The model is in agreement with published electron cryomicroscopy, mutagenesis, chemical modification, cross-linking, Fourier transform infrared spectroscopy, Raman spectroscopy, electron paramagnetic resonance spectroscopy, NMR, and optical spectroscopy data. The rhodopsin model and the published structure of bacteriorhodopsin have very similar retinal-binding pockets.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altenbach C., Yang K., Farrens D. L., Farahbakhsh Z. T., Khorana H. G., Hubbell W. L. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: a site-directed spin-labeling study. Biochemistry. 1996 Sep 24;35(38):12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- Altschuh D., Lesk A. M., Bloomer A. C., Klug A. Correlation of co-ordinated amino acid substitutions with function in viruses related to tobacco mosaic virus. J Mol Biol. 1987 Feb 20;193(4):693–707. doi: 10.1016/0022-2836(87)90352-4. [DOI] [PubMed] [Google Scholar]
- Arnis S., Fahmy K., Hofmann K. P., Sakmar T. P. A conserved carboxylic acid group mediates light-dependent proton uptake and signaling by rhodopsin. J Biol Chem. 1994 Sep 30;269(39):23879–23881. [PubMed] [Google Scholar]
- Asenjo A. B., Rim J., Oprian D. D. Molecular determinants of human red/green color discrimination. Neuron. 1994 May;12(5):1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- Bagley K. A., Balogh-Nair V., Croteau A. A., Dollinger G., Ebrey T. G., Eisenstein L., Hong M. K., Nakanishi K., Vittitow J. Fourier-transform infrared difference spectroscopy of rhodopsin and its photoproducts at low temperature. Biochemistry. 1985 Oct 22;24(22):6055–6071. doi: 10.1021/bi00343a006. [DOI] [PubMed] [Google Scholar]
- Baker E. N., Hubbard R. E. Hydrogen bonding in globular proteins. Prog Biophys Mol Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- Baldwin J. M. Structure and function of receptors coupled to G proteins. Curr Opin Cell Biol. 1994 Apr;6(2):180–190. doi: 10.1016/0955-0674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Baldwin J. M. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993 Apr;12(4):1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak L. S., Tiberi M., Freedman N. J., Kwatra M. M., Lefkowitz R. J., Caron M. G. A highly conserved tyrosine residue in G protein-coupled receptors is required for agonist-mediated beta 2-adrenergic receptor sequestration. J Biol Chem. 1994 Jan 28;269(4):2790–2795. [PubMed] [Google Scholar]
- Barclay P. L., Findlay J. B. Labelling of the cytoplasmic domains of ovine rhodopsin with hydrophilic chemical probes. Biochem J. 1984 May 15;220(1):75–84. doi: 10.1042/bj2200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. Helix geometry in proteins. J Mol Biol. 1988 Jun 5;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Blaber M., Zhang X. J., Lindstrom J. D., Pepiot S. D., Baase W. A., Matthews B. W. Determination of alpha-helix propensity within the context of a folded protein. Sites 44 and 131 in bacteriophage T4 lysozyme. J Mol Biol. 1994 Jan 14;235(2):600–624. doi: 10.1006/jmbi.1994.1016. [DOI] [PubMed] [Google Scholar]
- Borjigin J., Nathans J. Insertional mutagenesis as a probe of rhodopsin's topography, stability, and activity. J Biol Chem. 1994 May 20;269(20):14715–14722. [PubMed] [Google Scholar]
- Callender R. H., Doukas A., Crouch R., Nakanishi K. Molecular flow resonance Raman effect from retinal and rhodopsin. Biochemistry. 1976 Apr 20;15(8):1621–1629. doi: 10.1021/bi00653a005. [DOI] [PubMed] [Google Scholar]
- Ceresa B. P., Limbird L. E. Mutation of an aspartate residue highly conserved among G-protein-coupled receptors results in nonreciprocal disruption of alpha 2-adrenergic receptor-G-protein interactions. A negative charge at amino acid residue 79 forecasts alpha 2A-adrenergic receptor sensitivity to allosteric modulation by monovalent cations and fully effective receptor/G-protein coupling. J Biol Chem. 1994 Nov 25;269(47):29557–29564. [PubMed] [Google Scholar]
- Chabre M., Breton J. Orientation of aromatic residues in rhodopsin. Rotation of one tryptophan upon the meta I to meta II transition afer illumination. Photochem Photobiol. 1979 Aug;30(2):295–299. doi: 10.1111/j.1751-1097.1979.tb07150.x. [DOI] [PubMed] [Google Scholar]
- Chabre M., Breton J. The orientation of the chromophore of vertebrate rhodopsin in the "meta" intermediate states and the reversibility of the meta II-meta III transition. Vision Res. 1979;19(9):1005–1018. doi: 10.1016/0042-6989(79)90226-8. [DOI] [PubMed] [Google Scholar]
- Chan T., Lee M., Sakmar T. P. Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 1992 May 15;267(14):9478–9480. [PubMed] [Google Scholar]
- Chanda P. K., Minchin M. C., Davis A. R., Greenberg L., Reilly Y., McGregor W. H., Bhat R., Lubeck M. D., Mizutani S., Hung P. P. Identification of residues important for ligand binding to the human 5-hydroxytryptamine1A serotonin receptor. Mol Pharmacol. 1993 Apr;43(4):516–520. [PubMed] [Google Scholar]
- Chen J. G., Nakamura T., Ebrey T. G., Ok H., Konno K., Derguini F., Nakanishi K., Honig B. Wavelength regulation in iodopsin, a cone pigment. Biophys J. 1989 Apr;55(4):725–729. doi: 10.1016/S0006-3495(89)82871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Lesk A. M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986 Apr;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J., Henrick K., Fersht A. R. Disulfide mutants of barnase. I: Changes in stability and structure assessed by biophysical methods and X-ray crystallography. J Mol Biol. 1995 Oct 27;253(3):493–504. doi: 10.1006/jmbi.1995.0568. [DOI] [PubMed] [Google Scholar]
- Cohen G. B., Yang T., Robinson P. R., Oprian D. D. Constitutive activation of opsin: influence of charge at position 134 and size at position 296. Biochemistry. 1993 Jun 15;32(23):6111–6115. doi: 10.1021/bi00074a024. [DOI] [PubMed] [Google Scholar]
- Davies A., Schertler G. F., Gowen B. E., Saibil H. R. Projection structure of an invertebrate rhodopsin. J Struct Biol. 1996 Jul-Aug;117(1):36–44. doi: 10.1006/jsbi.1996.0067. [DOI] [PubMed] [Google Scholar]
- Davison M. D., Findlay J. B. Identification of the sites in opsin modified by photoactivated azido[125I]iodobenzene. Biochem J. 1986 Jun 1;236(2):389–395. doi: 10.1042/bj2360389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison M. D., Findlay J. B. Modification of ovine opsin with the photosensitive hydrophobic probe 1-azido-4-[125I]iodobenzene. Labelling of the chromophore-attachment domain. Biochem J. 1986 Mar 1;234(2):413–420. doi: 10.1042/bj2340413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenhofer J., Michel H. Structures of bacterial photosynthetic reaction centers. Annu Rev Cell Biol. 1991;7:1–23. doi: 10.1146/annurev.cb.07.110191.000245. [DOI] [PubMed] [Google Scholar]
- Deng H., Huang L., Callender R., Ebrey T. Evidence for a bound water molecule next to the retinal Schiff base in bacteriorhodopsin and rhodopsin: a resonance Raman study of the Schiff base hydrogen/deuterium exchange. Biophys J. 1994 Apr;66(4):1129–1136. doi: 10.1016/S0006-3495(94)80893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman H. G., Bouvier M., Benovic J. L., Caron M. G., Lefkowitz R. J. The multiple membrane spanning topography of the beta 2-adrenergic receptor. Localization of the sites of binding, glycosylation, and regulatory phosphorylation by limited proteolysis. J Biol Chem. 1987 Oct 15;262(29):14282–14288. [PubMed] [Google Scholar]
- Donnelly D., Findlay J. B., Blundell T. L. The evolution and structure of aminergic G protein-coupled receptors. Receptors Channels. 1994;2(1):61–78. [PubMed] [Google Scholar]
- Donnelly D., Overington J. P., Ruffle S. V., Nugent J. H., Blundell T. L. Modeling alpha-helical transmembrane domains: the calculation and use of substitution tables for lipid-facing residues. Protein Sci. 1993 Jan;2(1):55–70. doi: 10.1002/pro.5560020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elling C. E., Nielsen S. M., Schwartz T. W. Conversion of antagonist-binding site to metal-ion site in the tachykinin NK-1 receptor. Nature. 1995 Mar 2;374(6517):74–77. doi: 10.1038/374074a0. [DOI] [PubMed] [Google Scholar]
- Fahmy K., Jäger F., Beck M., Zvyaga T. A., Sakmar T. P., Siebert F. Protonation states of membrane-embedded carboxylic acid groups in rhodopsin and metarhodopsin II: a Fourier-transform infrared spectroscopy study of site-directed mutants. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanelli F., Menziani M. C., De Benedetti P. G. Computer simulations of signal transduction mechanism in alpha 1B-adrenergic and m3-muscarinic receptors. Protein Eng. 1995 Jun;8(6):557–564. doi: 10.1093/protein/8.6.557. [DOI] [PubMed] [Google Scholar]
- Farahbakhsh Z. T., Hideg K., Hubbell W. L. Photoactivated conformational changes in rhodopsin: a time-resolved spin label study. Science. 1993 Nov 26;262(5138):1416–1419. doi: 10.1126/science.8248781. [DOI] [PubMed] [Google Scholar]
- Farahbakhsh Z. T., Ridge K. D., Khorana H. G., Hubbell W. L. Mapping light-dependent structural changes in the cytoplasmic loop connecting helices C and D in rhodopsin: a site-directed spin labeling study. Biochemistry. 1995 Jul 11;34(27):8812–8819. doi: 10.1021/bi00027a033. [DOI] [PubMed] [Google Scholar]
- Farber G. K., Petsko G. A. The evolution of alpha/beta barrel enzymes. Trends Biochem Sci. 1990 Jun;15(6):228–234. doi: 10.1016/0968-0004(90)90035-a. [DOI] [PubMed] [Google Scholar]
- Fernandez L. M., Puett D. Identification of amino acid residues in transmembrane helices VI and VII of the lutropin/choriogonadotropin receptor involved in signaling. Biochemistry. 1996 Apr 2;35(13):3986–3993. doi: 10.1021/bi952421c. [DOI] [PubMed] [Google Scholar]
- Fernandez L. M., Puett D. Lys583 in the third extracellular loop of the lutropin/choriogonadotropin receptor is critical for signaling. J Biol Chem. 1996 Jan 12;271(2):925–930. doi: 10.1074/jbc.271.2.925. [DOI] [PubMed] [Google Scholar]
- Ganter U. M., Schmid E. D., Perez-Sala D., Rando R. R., Siebert F. Removal of the 9-methyl group of retinal inhibits signal transduction in the visual process. A Fourier transform infrared and biochemical investigation. Biochemistry. 1989 Jul 11;28(14):5954–5962. doi: 10.1021/bi00440a036. [DOI] [PubMed] [Google Scholar]
- Goldsmith T. H. Optimization, constraint, and history in the evolution of eyes. Q Rev Biol. 1990 Sep;65(3):281–322. doi: 10.1086/416840. [DOI] [PubMed] [Google Scholar]
- Grigorieff N., Ceska T. A., Downing K. H., Baldwin J. M., Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996 Jun 14;259(3):393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Gärtner W., Ullrich D., Vogt K. Quantum yield of CHAPSO-solubilized rhodopsin and 3-hydroxy retinal containing bovine opsin. Photochem Photobiol. 1991 Dec;54(6):1047–1055. doi: 10.1111/j.1751-1097.1991.tb02128.x. [DOI] [PubMed] [Google Scholar]
- Güntert P., Braun W., Wüthrich K. Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. J Mol Biol. 1991 Feb 5;217(3):517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- Han M., DeDecker B. S., Smith S. O. Localization of the retinal protonated Schiff base counterion in rhodopsin. Biophys J. 1993 Aug;65(2):899–906. doi: 10.1016/S0006-3495(93)81117-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M., Lin S. W., Minkova M., Smith S. O., Sakmar T. P. Functional interaction of transmembrane helices 3 and 6 in rhodopsin. Replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J Biol Chem. 1996 Dec 13;271(50):32337–32342. doi: 10.1074/jbc.271.50.32337. [DOI] [PubMed] [Google Scholar]
- Han M., Smith S. O. High-resolution structural studies of the retinal--Glu113 interaction in rhodopsin. Biophys Chem. 1995 Sep-Oct;56(1-2):23–29. doi: 10.1016/0301-4622(95)00011-l. [DOI] [PubMed] [Google Scholar]
- Han M., Smith S. O. NMR constraints on the location of the retinal chromophore in rhodopsin and bathorhodopsin. Biochemistry. 1995 Jan 31;34(4):1425–1432. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I., Kondo M., Nishimura M., Hara R., Hara T. Amino acid sequence surrounding the retinal-binding site in retinochrome of the squid, Todarodes pacificus. FEBS Lett. 1993 Nov 29;335(1):94–98. doi: 10.1016/0014-5793(93)80447-3. [DOI] [PubMed] [Google Scholar]
- Hargrave P. A., McDowell J. H., Feldmann R. J., Atkinson P. H., Rao J. K., Argos P. Rhodopsin's protein and carbohydrate structure: selected aspects. Vision Res. 1984;24(11):1487–1499. doi: 10.1016/0042-6989(84)90311-0. [DOI] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Herzyk P., Hubbard R. E. Automated method for modeling seven-helix transmembrane receptors from experimental data. Biophys J. 1995 Dec;69(6):2419–2442. doi: 10.1016/S0006-3495(95)80112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig B., Greenberg A. D., Dinur U., Ebrey T. G. Visual-pigment spectra: implications of the protonation of the retinal Schiff base. Biochemistry. 1976 Oct 19;15(21):4593–4599. doi: 10.1021/bi00666a008. [DOI] [PubMed] [Google Scholar]
- Horstman D. A., Brandon S., Wilson A. L., Guyer C. A., Cragoe E. J., Jr, Limbird L. E. An aspartate conserved among G-protein receptors confers allosteric regulation of alpha 2-adrenergic receptors by sodium. J Biol Chem. 1990 Dec 15;265(35):21590–21595. [PubMed] [Google Scholar]
- Hunyady L., Bor M., Balla T., Catt K. J. Critical role of a conserved intramembrane tyrosine residue in angiotensin II receptor activation. J Biol Chem. 1995 Apr 28;270(17):9702–9705. doi: 10.1074/jbc.270.17.9702. [DOI] [PubMed] [Google Scholar]
- Hänggi G., Braun W. Pattern recognition and self-correcting distance geometry calculations applied to myohemerythrin. FEBS Lett. 1994 May 16;344(2-3):147–153. doi: 10.1016/0014-5793(94)00366-1. [DOI] [PubMed] [Google Scholar]
- Irving C. S., Byers G. W., Leermakers P. A. Effect of solvent polarizability on the absorption spectrum of all-trans-retinylpyrrolidiniminium perchlorate. J Am Chem Soc. 1969 Apr 9;91(8):2141–2143. doi: 10.1021/ja01036a066. [DOI] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994 Mar 15;33(10):3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- Jäger F., Fahmy K., Sakmar T. P., Siebert F. Identification of glutamic acid 113 as the Schiff base proton acceptor in the metarhodopsin II photointermediate of rhodopsin. Biochemistry. 1994 Sep 13;33(36):10878–10882. doi: 10.1021/bi00202a005. [DOI] [PubMed] [Google Scholar]
- Jäger F., Jäger S., Krütle O., Friedman N., Sheves M., Hofmann K. P., Siebert F. Interactions of the beta-ionone ring with the protein in the visual pigment rhodopsin control the activation mechanism. An FTIR and fluorescence study on artificial vertebrate rhodopsins. Biochemistry. 1994 Jun 14;33(23):7389–7397. doi: 10.1021/bi00189a045. [DOI] [PubMed] [Google Scholar]
- Kandori H., Maeda A. FTIR spectroscopy reveals microscopic structural changes of the protein around the rhodopsin chromophore upon photoisomerization. Biochemistry. 1995 Oct 31;34(43):14220–14229. doi: 10.1021/bi00043a029. [DOI] [PubMed] [Google Scholar]
- Kaushal S., Khorana H. G. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994 May 24;33(20):6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- Kim J., Wess J., van Rhee A. M., Schöneberg T., Jacobson K. A. Site-directed mutagenesis identifies residues involved in ligand recognition in the human A2a adenosine receptor. J Biol Chem. 1995 Jun 9;270(23):13987–13997. doi: 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliger D. S., Milder S. J., Dratz E. A. Solvent effects on the spectra of retinal Schiff bases--I. models for the bathochromic shift of the chromophore spectrum in visual pigments. Photochem Photobiol. 1977 Mar;25(3):277–286. doi: 10.1111/j.1751-1097.1977.tb06911.x. [DOI] [PubMed] [Google Scholar]
- Kong H., Raynor K., Yasuda K., Bell G. I., Reisine T. Mutation of an aspartate at residue 89 in somatostatin receptor subtype 2 prevents Na+ regulation of agonist binding but does not alter receptor-G protein association. Mol Pharmacol. 1993 Aug;44(2):380–384. [PubMed] [Google Scholar]
- Kong H., Raynor K., Yasuda K., Moe S. T., Portoghese P. S., Bell G. I., Reisine T. A single residue, aspartic acid 95, in the delta opioid receptor specifies selective high affinity agonist binding. J Biol Chem. 1993 Nov 5;268(31):23055–23058. [PubMed] [Google Scholar]
- LIEBMAN P. A. In situ microspectrophotometric studies on the pigments of single retinal rods. Biophys J. 1962 Mar;2:161–178. doi: 10.1016/s0006-3495(62)86847-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba O. P., Borchman D., O'Brien P. J. Fourier transform infrared study of the rod outer segment disk and plasma membranes of vertebrate retina. Biochemistry. 1994 Feb 22;33(7):1704–1712. doi: 10.1021/bi00173a012. [DOI] [PubMed] [Google Scholar]
- Laue L., Chan W. Y., Hsueh A. J., Kudo M., Hsu S. Y., Wu S. M., Blomberg L., Cutler G. B., Jr Genetic heterogeneity of constitutively activating mutations of the human luteinizing hormone receptor in familial male-limited precocious puberty. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1906–1910. doi: 10.1073/pnas.92.6.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. W., Imamoto Y., Fukada Y., Shichida Y., Yoshizawa T., Mathies R. A. What makes red visual pigments red? A resonance Raman microprobe study of retinal chromophore structure in iodopsin. Biochemistry. 1994 Mar 1;33(8):2151–2160. doi: 10.1021/bi00174a023. [DOI] [PubMed] [Google Scholar]
- Lin S. W., Sakmar T. P., Franke R. R., Khorana H. G., Mathies R. A. Resonance Raman microprobe spectroscopy of rhodopsin mutants: effect of substitutions in the third transmembrane helix. Biochemistry. 1992 Jun 9;31(22):5105–5111. doi: 10.1021/bi00137a003. [DOI] [PubMed] [Google Scholar]
- Lin S. W., Sakmar T. P. Specific tryptophan UV-absorbance changes are probes of the transition of rhodopsin to its active state. Biochemistry. 1996 Aug 27;35(34):11149–11159. doi: 10.1021/bi960858u. [DOI] [PubMed] [Google Scholar]
- Liu J., Schöneberg T., van Rhee M., Wess J. Mutational analysis of the relative orientation of transmembrane helices I and VII in G protein-coupled receptors. J Biol Chem. 1995 Aug 18;270(33):19532–19539. doi: 10.1074/jbc.270.33.19532. [DOI] [PubMed] [Google Scholar]
- MATTHEWS R. G., HUBBARD R., BROWN P. K., WALD G. TAUTOMERIC FORMS OF METARHODOPSIN. J Gen Physiol. 1963 Nov;47:215–240. doi: 10.1085/jgp.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Ohkita Y. J., Sasaki J., Shichida Y., Yoshizawa T. Water structural changes in lumirhodopsin, metarhodopsin I, and metarhodopsin II upon photolysis of bovine rhodopsin: analysis by Fourier transform infrared spectroscopy. Biochemistry. 1993 Nov 16;32(45):12033–12038. doi: 10.1021/bi00096a013. [DOI] [PubMed] [Google Scholar]
- Maeda A., Shichida Y., Yoshizawa T. Formation of 7-cis- and 13-cis-retinal pigments by irradiating squid rhodopsin. Biochemistry. 1979 Apr 17;18(8):1449–1453. doi: 10.1021/bi00575a010. [DOI] [PubMed] [Google Scholar]
- Martynov V. I., Kostina M. B., Feigina M. Iu, Miroshnikov A. I. Issledovanie molekuliarnoi organizatsii zritel'nogo rodopsina v fotoretseptornykh membranakh metodom ogranichennogo proteinoliza. Bioorg Khim. 1983 Jun;9(6):734–745. [PubMed] [Google Scholar]
- Mathies R., Stryer L. Retinal has a highly dipolar vertically excited singlet state: implications for vision. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2169–2173. doi: 10.1073/pnas.73.7.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M., Matthews B. W. Stabilization of functional proteins by introduction of multiple disulfide bonds. Methods Enzymol. 1991;202:336–356. doi: 10.1016/0076-6879(91)02018-5. [DOI] [PubMed] [Google Scholar]
- McDonald I. K., Thornton J. M. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994 May 20;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Mizobe T., Maze M., Lam V., Suryanarayana S., Kobilka B. K. Arrangement of transmembrane domains in adrenergic receptors. Similarity to bacteriorhodopsin. J Biol Chem. 1996 Feb 2;271(5):2387–2389. doi: 10.1074/jbc.271.5.2387. [DOI] [PubMed] [Google Scholar]
- Mollevanger L. C., Kentgens A. P., Pardoen J. A., Courtin J. M., Veeman W. S., Lugtenburg J., de Grip W. J. High-resolution solid-state 13C-NMR study of carbons C-5 and C-12 of the chromophore of bovine rhodopsin. Evidence for a 6-S-cis conformation with negative-charge perturbation near C-12. Eur J Biochem. 1987 Feb 16;163(1):9–14. doi: 10.1111/j.1432-1033.1987.tb10729.x. [DOI] [PubMed] [Google Scholar]
- Monnot C., Bihoreau C., Conchon S., Curnow K. M., Corvol P., Clauser E. Polar residues in the transmembrane domains of the type 1 angiotensin II receptor are required for binding and coupling. Reconstitution of the binding site by co-expression of two deficient mutants. J Biol Chem. 1996 Jan 19;271(3):1507–1513. doi: 10.1074/jbc.271.3.1507. [DOI] [PubMed] [Google Scholar]
- Mumenthaler C., Braun W. Predicting the helix packing of globular proteins by self-correcting distance geometry. Protein Sci. 1995 May;4(5):863–871. doi: 10.1002/pro.5560040506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A. G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993 Mar;12(3):861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin A. G. Structural classification of proteins: new superfamilies. Curr Opin Struct Biol. 1996 Jun;6(3):386–394. doi: 10.1016/s0959-440x(96)80059-5. [DOI] [PubMed] [Google Scholar]
- Nakayama T. A., Khorana H. G. Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem. 1991 Mar 5;266(7):4269–4275. [PubMed] [Google Scholar]
- Nakayama T. A., Khorana H. G. Orientation of retinal in bovine rhodopsin determined by cross-linking using a photoactivatable analog of 11-cis-retinal. J Biol Chem. 1990 Sep 15;265(26):15762–15769. [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: identification of the retinylidene Schiff's base counterion in bovine rhodopsin. Biochemistry. 1990 Oct 16;29(41):9746–9752. doi: 10.1021/bi00493a034. [DOI] [PubMed] [Google Scholar]
- Nathans J. Determinants of visual pigment absorbance: role of charged amino acids in the putative transmembrane segments. Biochemistry. 1990 Jan 30;29(4):937–942. doi: 10.1021/bi00456a013. [DOI] [PubMed] [Google Scholar]
- Oprian D. D. The ligand-binding domain of rhodopsin and other G protein-linked receptors. J Bioenerg Biomembr. 1992 Apr;24(2):211–217. doi: 10.1007/BF00762679. [DOI] [PubMed] [Google Scholar]
- Oseroff A. R., Callender R. H. Resonance Raman spectroscopy of rhodopsin in retinal disk membranes. Biochemistry. 1974 Sep 24;13(20):4243–4248. doi: 10.1021/bi00717a027. [DOI] [PubMed] [Google Scholar]
- Palings I., Pardoen J. A., van den Berg E., Winkel C., Lugtenburg J., Mathies R. A. Assignment of fingerprint vibrations in the resonance Raman spectra of rhodopsin, isorhodopsin, and bathorhodopsin: implications for chromophore structure and environment. Biochemistry. 1987 May 5;26(9):2544–2556. doi: 10.1021/bi00383a021. [DOI] [PubMed] [Google Scholar]
- Persson B., Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994 Mar 25;237(2):182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Pittel Z., Wess J. Intramolecular interactions in muscarinic acetylcholine receptors studied with chimeric m2/m5 receptors. Mol Pharmacol. 1994 Jan;45(1):61–64. [PubMed] [Google Scholar]
- Quintana J., Wang H., Ascoli M. The regulation of the binding affinity of the luteinizing hormone/choriogonadotropin receptor by sodium ions is mediated by a highly conserved aspartate located in the second transmembrane domain of G protein-coupled receptors. Mol Endocrinol. 1993 Jun;7(6):767–775. doi: 10.1210/mend.7.6.8395653. [DOI] [PubMed] [Google Scholar]
- Rao V. R., Cohen G. B., Oprian D. D. Rhodopsin mutation G90D and a molecular mechanism for congenital night blindness. Nature. 1994 Feb 17;367(6464):639–642. doi: 10.1038/367639a0. [DOI] [PubMed] [Google Scholar]
- Rao V. R., Oprian D. D. Activating mutations of rhodopsin and other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- Rath P., DeCaluwé L. L., Bovee-Geurts P. H., DeGrip W. J., Rothschild K. J. Fourier transform infrared difference spectroscopy of rhodopsin mutants: light activation of rhodopsin causes hydrogen-bonding change in residue aspartic acid-83 during meta II formation. Biochemistry. 1993 Oct 5;32(39):10277–10282. doi: 10.1021/bi00090a001. [DOI] [PubMed] [Google Scholar]
- Resek J. F., Farahbakhsh Z. T., Hubbell W. L., Khorana H. G. Formation of the meta II photointermediate is accompanied by conformational changes in the cytoplasmic surface of rhodopsin. Biochemistry. 1993 Nov 16;32(45):12025–12032. doi: 10.1021/bi00096a012. [DOI] [PubMed] [Google Scholar]
- Ridge K. D., Zhang C., Khorana H. G. Mapping of the amino acids in the cytoplasmic loop connecting helices C and D in rhodopsin. Chemical reactivity in the dark state following single cysteine replacements. Biochemistry. 1995 Jul 11;34(27):8804–8811. doi: 10.1021/bi00027a032. [DOI] [PubMed] [Google Scholar]
- Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995 Mar;4(3):521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G. Glutamic acid-113 serves as the retinylidene Schiff base counterion in bovine rhodopsin. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar T. P., Franke R. R., Khorana H. G. The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength absorbance and Schiff base pKa. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3079–3083. doi: 10.1073/pnas.88.8.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer A., Fanelli F., Costa T., De Benedetti P. G., Cotecchia S. Constitutively active mutants of the alpha 1B-adrenergic receptor: role of highly conserved polar amino acids in receptor activation. EMBO J. 1996 Jul 15;15(14):3566–3578. [PMC free article] [PubMed] [Google Scholar]
- Schertler G. F., Hargrave P. A. Projection structure of frog rhodopsin in two crystal forms. Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11578–11582. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler G. F., Villa C., Henderson R. Projection structure of rhodopsin. Nature. 1993 Apr 22;362(6422):770–772. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Sealfon S. C., Chi L., Ebersole B. J., Rodic V., Zhang D., Ballesteros J. A., Weinstein H. Related contribution of specific helix 2 and 7 residues to conformational activation of the serotonin 5-HT2A receptor. J Biol Chem. 1995 Jul 14;270(28):16683–16688. doi: 10.1074/jbc.270.28.16683. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Palings I., Copié V., Raleigh D. P., Courtin J., Pardoen J. A., Lugtenburg J., Mathies R. A., Griffin R. G. Low-temperature solid-state 13C NMR studies of the retinal chromophore in rhodopsin. Biochemistry. 1987 Mar 24;26(6):1606–1611. doi: 10.1021/bi00380a018. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Palings I., Miley M. E., Courtin J., de Groot H., Lugtenburg J., Mathies R. A., Griffin R. G. Solid-state NMR studies of the mechanism of the opsin shift in the visual pigment rhodopsin. Biochemistry. 1990 Sep 4;29(35):8158–8164. doi: 10.1021/bi00487a025. [DOI] [PubMed] [Google Scholar]
- Smith S. O., de Groot H., Gebhard R., Lugtenburg J. Magic angle spinning NMR studies on the metarhodopsin II intermediate of bovine rhodopsin: evidence for an unprotonated Schiff base. Photochem Photobiol. 1992 Dec;56(6):1035–1039. doi: 10.1111/j.1751-1097.1992.tb09726.x. [DOI] [PubMed] [Google Scholar]
- Soppa J. Two hypotheses--one answer. Sequence comparison does not support an evolutionary link between halobacterial retinal proteins including bacteriorhodopsin and eukaryotic G-protein-coupled receptors. FEBS Lett. 1994 Mar 28;342(1):7–11. doi: 10.1016/0014-5793(94)80573-3. [DOI] [PubMed] [Google Scholar]
- Steinberg G., Ottolenghi M., Sheves M. pKa of the protonated Schiff base of bovine rhodopsin. A study with artificial pigments. Biophys J. 1993 May;64(5):1499–1502. doi: 10.1016/S0006-3495(93)81518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayana S., von Zastrow M., Kobilka B. K. Identification of intramolecular interactions in adrenergic receptors. J Biol Chem. 1992 Nov 5;267(31):21991–21994. [PubMed] [Google Scholar]
- Taylor W. R., Jones D. T., Green N. M. A method for alpha-helical integral membrane protein fold prediction. Proteins. 1994 Mar;18(3):281–294. doi: 10.1002/prot.340180309. [DOI] [PubMed] [Google Scholar]
- Thirstrup K., Elling C. E., Hjorth S. A., Schwartz T. W. Construction of a high affinity zinc switch in the kappa-opioid receptor. J Biol Chem. 1996 Apr 5;271(14):7875–7878. doi: 10.1074/jbc.271.14.7875. [DOI] [PubMed] [Google Scholar]
- Thomas D. D., Stryer L. Transverse location of the retinal chromophore of rhodopsin in rod outer segment disc membranes. J Mol Biol. 1982 Jan 5;154(1):145–157. doi: 10.1016/0022-2836(82)90422-3. [DOI] [PubMed] [Google Scholar]
- Unger V. M., Schertler G. F. Low resolution structure of bovine rhodopsin determined by electron cryo-microscopy. Biophys J. 1995 May;68(5):1776–1786. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D., Eisenhaber F., Argos P. Principles of helix-helix packing in proteins: the helical lattice superposition model. J Mol Biol. 1996 Jan 26;255(3):536–553. doi: 10.1006/jmbi.1996.0044. [DOI] [PubMed] [Google Scholar]
- Weitz C. J., Nathans J. Rhodopsin activation: effects on the metarhodopsin I-metarhodopsin II equilibrium of neutralization or introduction of charged amino acids within putative transmembrane segments. Biochemistry. 1993 Dec 28;32(51):14176–14182. doi: 10.1021/bi00214a016. [DOI] [PubMed] [Google Scholar]
- Yarden Y., Rodriguez H., Wong S. K., Brandt D. R., May D. C., Burnier J., Harkins R. N., Chen E. Y., Ramachandran J., Ullrich A. The avian beta-adrenergic receptor: primary structure and membrane topology. Proc Natl Acad Sci U S A. 1986 Sep;83(18):6795–6799. doi: 10.1073/pnas.83.18.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeagle P. L., Alderfer J. L., Albert A. D. Structure of the carboxy-terminal domain of bovine rhodopsin. Nat Struct Biol. 1995 Oct;2(10):832–834. doi: 10.1038/nsb1095-832. [DOI] [PubMed] [Google Scholar]
- Yeagle P. L., Alderfer J. L., Albert A. D. Structure of the third cytoplasmic loop of bovine rhodopsin. Biochemistry. 1995 Nov 14;34(45):14621–14625. doi: 10.1021/bi00045a002. [DOI] [PubMed] [Google Scholar]
- Yu H., Kono M., McKee T. D., Oprian D. D. A general method for mapping tertiary contacts between amino acid residues in membrane-embedded proteins. Biochemistry. 1995 Nov 21;34(46):14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]
- Zhou W., Flanagan C., Ballesteros J. A., Konvicka K., Davidson J. S., Weinstein H., Millar R. P., Sealfon S. C. A reciprocal mutation supports helix 2 and helix 7 proximity in the gonadotropin-releasing hormone receptor. Mol Pharmacol. 1994 Feb;45(2):165–170. [PubMed] [Google Scholar]
- Zhukovsky E. A., Oprian D. D. Effect of carboxylic acid side chains on the absorption maximum of visual pigments. Science. 1989 Nov 17;246(4932):928–930. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- Zhukovsky E. A., Robinson P. R., Oprian D. D. Changing the location of the Schiff base counterion in rhodopsin. Biochemistry. 1992 Oct 27;31(42):10400–10405. doi: 10.1021/bi00157a030. [DOI] [PubMed] [Google Scholar]
- Zvyaga T. A., Min K. C., Beck M., Sakmar T. P. Movement of the retinylidene Schiff base counterion in rhodopsin by one helix turn reverses the pH dependence of the metarhodopsin I to metarhodopsin II transition. J Biol Chem. 1993 Mar 5;268(7):4661–4667. [PubMed] [Google Scholar]