Abstract
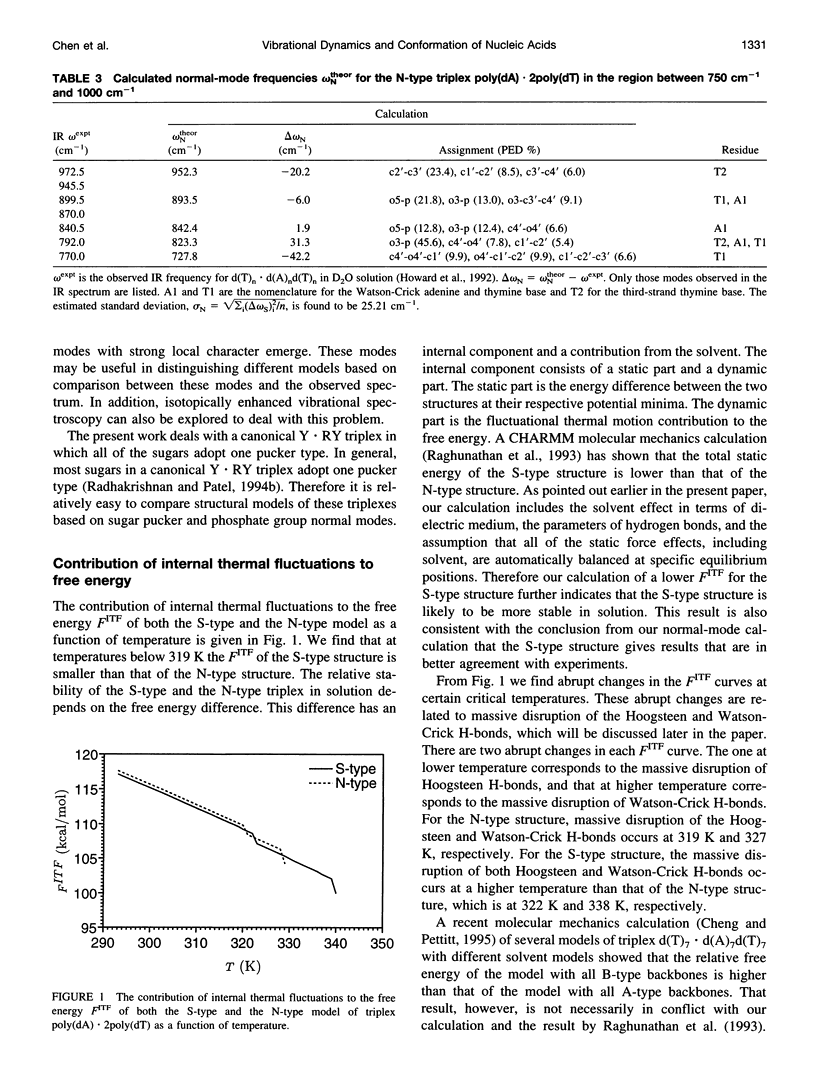
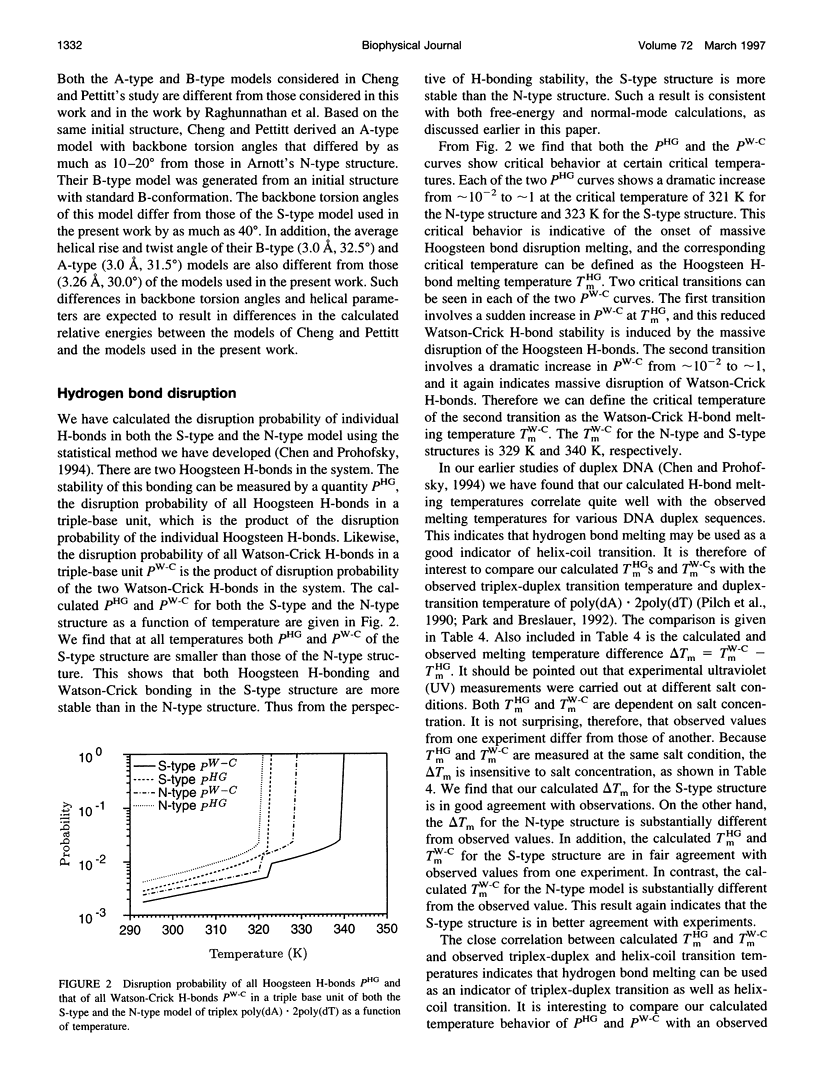
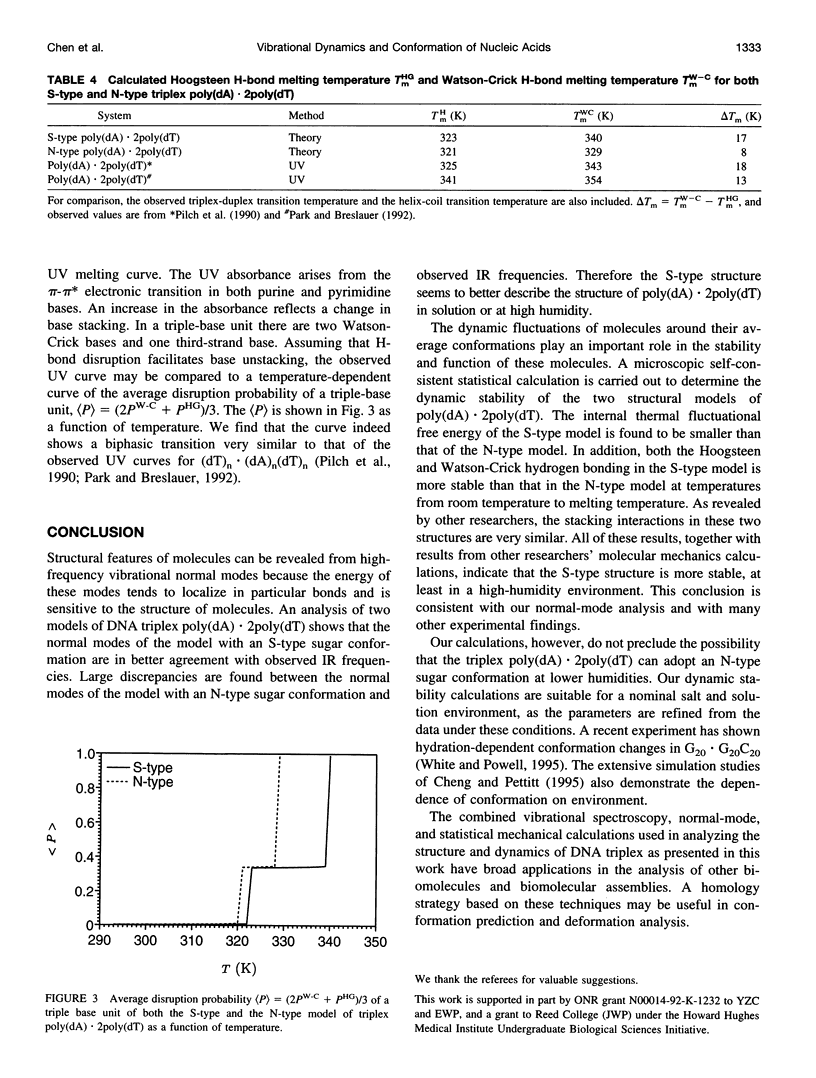
A normal-mode and statistical mechanical calculation was carried out to determine the vibrational normal modes, contribution of internal fluctuations to the free energy, and hydrogen bond disruption of DNA triplex poly(dA).2poly(dT). The calculation was performed on both the x-ray fiber diffraction model with a N-type sugar conformation, and a newly proposed model with a S-type sugar conformation. Our calculated normal modes for the S-type structure are in better agreement with observed IR spectra for samples in D2O solution. We also find that the contribution of internal fluctuations to free energy, premelting hydrogen bond disruption probability, and hydrogen bond melting temperatures for the Hoogsteen and Watson-Crick hydrogen bonds all show that the S-type structure is dynamically more stable than the N-type structure in a nominal solution environment. Therefore our calculation supports experimental findings that the triplex d(T)n.d(A)nd(T)n most likely adopts a S-type sugar conformation in solution or at high humidity. Our calculations, however, do not preclude the possibility of an N-type conformation at lower humidities.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Prohofsky E. W. Normal mode calculation of a netropsin-DNA complex: effect of structural deformation on vibrational spectrum. Biopolymers. 1995 Jun;35(6):657–666. doi: 10.1002/bip.360350611. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Prohofsky E. W. Sequence and temperature dependence of the interbase hydrogen-bond breathing modes in B-DNA polymers: comparison with low-frequency Raman peaks and their role in helix melting. Biopolymers. 1995 Jun;35(6):573–582. doi: 10.1002/bip.360350603. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Prohofsky EW. Near-neighbor effects in cooperative modified self-consistent phonon approximation melting in DNA. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 1994 Jan;49(1):873–881. doi: 10.1103/physreve.49.873. [DOI] [PubMed] [Google Scholar]
- Cheng Y. K., Pettitt B. M. Solvent effects on model d(CG.G)7 and d(TA.T)7 DNA triple helices. Biopolymers. 1995 May;35(5):457–473. doi: 10.1002/bip.360350505. [DOI] [PubMed] [Google Scholar]
- Dagneaux C., Liquier J., Taillandier E. Sugar conformations in DNA and RNA-DNA triple helices determined by FTIR spectroscopy: role of backbone composition. Biochemistry. 1995 Dec 26;34(51):16618–16623. doi: 10.1021/bi00051a009. [DOI] [PubMed] [Google Scholar]
- Durand M., Thuong N. T., Maurizot J. C. Binding of netropsin to a DNA triple helix. J Biol Chem. 1992 Dec 5;267(34):24394–24399. [PubMed] [Google Scholar]
- Howard F. B., Miles H. T., Liu K., Frazier J., Raghunathan G., Sasisekharan V. Structure of d(T)n.d(A)n.d(T)n: the DNA triple helix has B-form geometry with C2'-endo sugar pucker. Biochemistry. 1992 Nov 10;31(44):10671–10677. doi: 10.1021/bi00159a005. [DOI] [PubMed] [Google Scholar]
- Laughton C. A., Neidle S. Molecular dynamics simulation of the DNA triplex d(TC)5.d(GA)5.d(C+T)5. J Mol Biol. 1992 Jan 20;223(2):519–529. doi: 10.1016/0022-2836(92)90667-9. [DOI] [PubMed] [Google Scholar]
- Laughton C. A., Neidle S. Prediction of the structure of the Y+.R-.R(+)-type DNA triple helix by molecular modelling. Nucleic Acids Res. 1992 Dec 25;20(24):6535–6541. doi: 10.1093/nar/20.24.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letellier R., Ghomi M., Taillandier E. Interpretation of DNA vibration modes: I--The guanosine and cytidine residues involved in poly(dG-dC).poly(dG-dC) and d(CG)3.d(CG)3. J Biomol Struct Dyn. 1986 Feb;3(4):671–687. doi: 10.1080/07391102.1986.10508455. [DOI] [PubMed] [Google Scholar]
- Liquier J., Coffinier P., Firon M., Taillandier E. Triple helical polynucleotidic structures: sugar conformations determined by FTIR spectroscopy. J Biomol Struct Dyn. 1991 Dec;9(3):437–445. doi: 10.1080/07391102.1991.10507927. [DOI] [PubMed] [Google Scholar]
- Lu K. C., Prohofsky E. W., Van Zandt L. L. Vibrational modes of A-DNA, B-DNA, and A-RNA backbones: an application of a green-function refinement procedure. Biopolymers. 1977 Nov;16(11):2491–2506. doi: 10.1002/bip.1977.360161112. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Macaya R., Wang E., Schultze P., Sklenár V., Feigon J. Proton nuclear magnetic resonance assignments and structural characterization of an intramolecular DNA triplex. J Mol Biol. 1992 Jun 5;225(3):755–773. doi: 10.1016/0022-2836(92)90399-5. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. J. Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry. 1990 Sep 18;29(37):8820–8826. doi: 10.1021/bi00489a045. [DOI] [PubMed] [Google Scholar]
- Manzini G., Xodo L. E., Gasparotto D., Quadrifoglio F., van der Marel G. A., van Boom J. H. Triple helix formation by oligopurine-oligopyrimidine DNA fragments. Electrophoretic and thermodynamic behavior. J Mol Biol. 1990 Jun 20;213(4):833–843. doi: 10.1016/S0022-2836(05)80267-0. [DOI] [PubMed] [Google Scholar]
- Mergny J. L., Duval-Valentin G., Nguyen C. H., Perrouault L., Faucon B., Rougée M., Montenay-Garestier T., Bisagni E., Hélène C. Triple helix-specific ligands. Science. 1992 Jun 19;256(5064):1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- Ouali M., Letellier R., Adnet F., Liquier J., Sun J. S., Lavery R., Taillandier E. A possible family of B-like triple helix structures: comparison with the Arnott A-like triple helix. Biochemistry. 1993 Mar 2;32(8):2098–2103. doi: 10.1021/bi00059a030. [DOI] [PubMed] [Google Scholar]
- Park Y. W., Breslauer K. J. Drug binding to higher ordered DNA structures: netropsin complexation with a nucleic acid triple helix. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6653–6657. doi: 10.1073/pnas.89.14.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch D. S., Brousseau R., Shafer R. H. Thermodynamics of triple helix formation: spectrophotometric studies on the d(A)10.2d(T)10 and d(C+3T4C+3).d(G3A4G3).d(C3T4C3) triple helices. Nucleic Acids Res. 1990 Oct 11;18(19):5743–5750. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G. E., Park Y. W., Singleton S. F., Dervan P. B., Breslauer K. J. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu VV, Young L, Prohofsky EW. Hydrogen-bond melting in B-DNA copolymers in a mean-field self-consistent phonon approach. Phys Rev B Condens Matter. 1989 Mar 15;39(8):5436–5443. doi: 10.1103/physrevb.39.5436. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J. DNA triplexes: solution structures, hydration sites, energetics, interactions, and function. Biochemistry. 1994 Sep 27;33(38):11405–11416. doi: 10.1021/bi00204a001. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J., Gao X. Three-dimensional homonuclear NOESY-TOCSY of an intramolecular pyrimidine.purine.pyrimidine DNA triplex containing a central G.TA triple: nonexchangeable proton assignments and structural implications. Biochemistry. 1992 Mar 10;31(9):2514–2523. doi: 10.1021/bi00124a011. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J. Solution structure and hydration patterns of a pyrimidine.purine.pyrimidine DNA triplex containing a novel T.CG base-triple. J Mol Biol. 1994 Aug 26;241(4):600–619. doi: 10.1006/jmbi.1994.1534. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J. Solution structure of a pyrimidine.purine.pyrimidine DNA triplex containing T.AT, C+.GC and G.TA triples. Structure. 1994 Jan 15;2(1):17–32. doi: 10.1016/s0969-2126(00)00005-8. [DOI] [PubMed] [Google Scholar]
- Raghunathan G., Miles H. T., Sasisekharan V. Symmetry and molecular structure of a DNA triple helix: d(T)n.d(A)n.d(T)n. Biochemistry. 1993 Jan 19;32(2):455–462. doi: 10.1021/bi00053a009. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Singleton S. F., Dervan P. B. Equilibrium association constants for oligonucleotide-directed triple helix formation at single DNA sites: linkage to cation valence and concentration. Biochemistry. 1993 Dec 7;32(48):13171–13179. doi: 10.1021/bi00211a028. [DOI] [PubMed] [Google Scholar]
- Surewicz W. K., Mantsch H. H., Chapman D. Determination of protein secondary structure by Fourier transform infrared spectroscopy: a critical assessment. Biochemistry. 1993 Jan 19;32(2):389–394. doi: 10.1021/bi00053a001. [DOI] [PubMed] [Google Scholar]
- Van Meervelt L., Vlieghe D., Dautant A., Gallois B., Précigoux G., Kennard O. High-resolution structure of a DNA helix forming (C.G)*G base triplets. Nature. 1995 Apr 20;374(6524):742–744. doi: 10.1038/374742a0. [DOI] [PubMed] [Google Scholar]
- Wang E., Malek S., Feigon J. Structure of a G.T.A triplet in an intramolecular DNA triplex. Biochemistry. 1992 May 26;31(20):4838–4846. doi: 10.1021/bi00135a015. [DOI] [PubMed] [Google Scholar]
- White A. P., Powell J. W. Observation of the hydration-dependent conformation of the (dG)20.(dG)20(dC)20 oligonucleotide triplex using FTIR spectroscopy. Biochemistry. 1995 Jan 31;34(4):1137–1142. doi: 10.1021/bi00004a006. [DOI] [PubMed] [Google Scholar]


