Abstract
We report the cloning and functional characterization of an endo-β-1,3-glucanase from the pinewood nematode Bursaphelenchus xylophilus acquired by horizontal gene transfer from bacteria. This is the first gene of this type from any nematode species. We show that a similar cDNA is also present in another closely related species B. mucronatus, but that similar sequences are not present in any other nematode studied to date. The B. xylophilus gene is expressed solely in the oesophageal gland cells of the nematode and the protein is present in the nematode's secretions. The deduced amino acid sequence of the gene is very similar to glycosyl hydrolase family 16 proteins. The recombinant protein, expressed in Escherichia coli, preferentially hydrolysed the β-1,3-glucan laminarin, and had very low levels of activity on β-1,3-1,4-glucan, lichenan and barley β-glucan. Laminarin was degraded in an endoglucanase mode by the enzyme. The optimal temperature and pH for activity of the recombinant enzyme were 65 °C and pH 4.9. The protein is probably important in allowing the nematodes to feed on fungi. Sequence comparisons suggest that the gene encoding the endo-β-1,3-glucanase was acquired by horizontal gene transfer from bacteria. B. xylophilus therefore contains genes that have been acquired by this process from both bacteria and fungi. These findings support the idea that multiple independent horizontal gene transfer events have helped in shaping the evolution of several different life strategies in nematodes.
Keywords: β-1,3-endoglucanase; fungal feeding; horizontal gene transfer; laminarin; pinewood nematode (Bursaphelenchus xylophilus); secretion
Abbreviations: EST, expressed sequence tag; gDNA, genomic DNA; GHF, glycosyl hydrolase family; ORF, open reading frame
INTRODUCTION
β-1,3-Glucanases are widely distributed among bacteria, fungi and higher plants. They are classified into two groups: exo-β-1,3-glucanase (EC 3.2.1.58) and endo-β-1,3-glucanases (EC 3.2.1.6 and EC 3.2.1.39). β-1,3-Glucanases catalyse the hydrolysis of β-1,3-D-glucosidic linkages in β-1,3-D-glucan. This polymer is a major component of fungal cell walls and a major structural and storage polysaccharide (in the form of laminarin) in marine macroalgae [1]. The physiological functions of β-1,3-glucanases are distinct and depend on their source. In plants, the enzymes are considered to be involved in cell differentiation and defence against pathogenic fungi [2]. In bacteria, the enzymes are released to break down fungal cell walls to allow them to be used as a food source [3]. In fungi, β-1,3-glucanases may play roles in development and differentiation, β-glucan mobilization and interactions of plant pathogenic fungi with their hosts [4]. Although they have the same hydrolytic activity, the bacterial enzymes are classified into GHF16 (glycosyl hydrolase family 16), whereas most plant and fungal enzymes are grouped into GHF17, on the basis of differences in their amino acid sequences [5].
In the animal kingdom, functionally characterized β-1,3-glucanases are restricted to marine invertebrates. Genes encoding β-1,3-glucanases have been cloned from the purple sea urchin (Strongylocentrotus purpuratus) [6] and the bivalve mollusc Spisula sachalinensis (Sakhalin surf-clam) [7]. They are classified in GHF16 and thought to be involved in the digestion of algal food. In addition, many β-1,3-glucanase-like proteins have been isolated, and the encoding genes cloned, from insects [8–12] and other invertebrates [13–16]. Although these sequences contain regions that are very similar to the activation region of GHF16 β-1,3-glucanases, they have not been shown to exhibit glucanase activity. These proteins bind specifically to β-1,3-glucan, which is found in the cell surface of microbes, but is absent from the host, and they have been shown to play a role in the innate immune system by recognizing foreign material. For example, Manduca sexta (tobacco hornworm) β-1,3-glucan recognition protein [10], crayfish (Pacifastacus leniusculus) lipopolysaccharide- and β-1,3-glucan binding protein [15] and earthworm (Eisenia foetida) coelomic cytolytic factor-1 [14] have been shown to mediate activation of the prophenol oxidase activating system, which is supposed to be a recognition and defence system in many invertebrates [17].
Bursaphelenchus is a large group of nematodes that has a worldwide distribution. Most Bursaphelenchus species are solely fungal feeders and all species rely on fungi as a food source at some stage of their life cycle. Many Bursaphelenchus species feed on fungi colonizing dead trees. Bursaphelenchus xylophilus and a few other pathogenic species described to date are unique in their capacity to feed on live trees as well as feed on fungi. In the B. xylophilus pathogenic life cycle, the nematode is transmitted from trees killed by pine wilt to healthy pines by vector beetles. Once the nematodes enter the tree, they feed on plant cells in the tree, leading to disruption of pine tissues and lethal wilt. As the pine wilts and dies, the nematodes begin to feed on fungi that invade the dying tree. This pine wilt disease is the most serious forest disease in Japan and East Asia. Many Bursaphelenchus species, including B. xylophilus, therefore have a close association with fungi. Since β-1,3-glucan is the main structural component of fungal cell walls, it seems probable that β-1,3-glucanases may play an important role in the life cycle of these nematodes.
Plant-cell-wall-degrading enzymes produced by animals have been extensively studied in a variety of plant parasitic cyst and root-knot nematodes, including Heterodera, Globodera and Meloidogyne species. Several genes encoding cell-wall-degrading enzymes, such as cellulase (endo-β-1,4-glucanase) [18,19], pectate lyase [20,21] and polygalacturonase [22] have been identified in these species. These enzymes are produced within the oesophageal gland cells of the nematodes and are secreted through the nematode stylet into host tissues. Intriguingly, these genes appear to have been acquired by horizontal gene transfer from bacteria. These studies form part of an increasing body of literature on the molecular basis of plant–nematode interactions. Significant progress has been made in understanding the nature of the molecules produced by nematodes that allow them to parasitize plants (reviewed in [23,24]) and on the changes that are induced in the hosts as a result of nematode infection [25]. Most of this work has been performed on tylenchid (cyst-forming and root-knot nematodes) and, in contrast, little is known about the molecular basis of host–parasite interactions in Bursaphelenchus species.
In a previous study we identified and functionally characterized a family of cellulase (endo-β-1,4-glucanase) genes from B. xylophilus and showed that these genes were probably acquired by horizontal gene transfer, not from bacteria but from fungi [26]. It was suggested that these genes were acquired from fungi on which ancestors of the current nematode species fed. These cellulases may have a role in the parasitic process. In the present study, we present the cloning and biochemical characterization of the first nematode β-1,3-glucanase genes, identified from B. xylophilus and B. mucronatus during an EST (expressed sequence tag) project. The B. xylophilus gene is expressed in oesophageal gland cells and, like the previously characterized cellulases, the enzyme is secreted from the nematode stylet. However, in contrast with the cellulases, the glucanase is most similar to bacterial enzymes. The present study represents the first identification of this class of enzymes from any nematode species and includes a detailed analysis of the biochemical properties of the recombinant protein.
EXPERIMENTAL
Biological material
B. xylophilus (Ka-4 isolate) was cultured on the fungus Botrytis cinerea grown on autocleaved barley grains as described previously [26]. Nematodes for experiments were separated from B. cinerea hyphae on a Baermann funnel for 2 h at 25 °C. The nematodes were then washed five times in M9 buffer (42.3 mM Na2HPO4/22 mM KH2PO4/85.6 mM NaCl/1 mM MgSO4, pH 7.0) with centrifugation at 700 g for 2 min to remove any remaining B. cinerea mycelium.
Isolation of cDNA and gDNA (genomic DNA) clones
A cDNA library was constructed using mRNA derived from mixed-stage B. xylophilus that were vigorously growing on B. cinerea at 25 °C. The β-1,3-glucanase gene was identified during an EST project performed using this library (T. Kikuchi, unpublished work). One full-length cDNA (clone 03BK1-02-C05) encoding a β-1,3-glucanase, designated Bx-lam16A (see the Results section below) was identified during BLAST analysis of sequences generated in this project. The plasmid clone from which the sequence was obtained was identified and re-sequenced in both directions to obtain full-length cDNA sequences.
The Bx-lam16A genomic coding region was obtained by PCR amplification from B. xylophilus genomic DNA, using a gene-specific primer flanking each ORF (open reading frame). PCR products were cloned using the pGEM-T Easy vector (Promega, Chilworth, Southampton, U.K.) and sequenced.
Genomic Southern hybridization
A probe specific to Bx-lam16A was made by PCR amplification of the full-length cDNA from the original plasmid with primers b13-0s (5′-ATGAGAGTTGTCATTGCC-3′) and b13-0a (5′-CACCGAAAACTACAACGT-3′). This probe was used in Southern hybridizations of B. xylophilus and B. cinerea genomic DNA as described in [26].
Expression and purification of recombinant protein
The Bx-lam16A coding region without the putative signal sequence was amplified by PCR from the original plasmid using the primers bl3-1s (5′-GGAGTCATTTGGCAAGAGGAC-3′) and bl3-0a. The resulting PCR product was ligated directly into the pQE-30UA vector (Qiagen, Crawley, West Sussex, U.K.) and then transformed into Escherichia coli M15[pREP4] (Qiagen). Plasmids with the insert in the correct reading frame and which showed no sequence changes during the PCR process were selected after sequencing of the plasmid clones. Recombinant E. coli containing these constructs were grown at 25 °C in 500 ml of Luria–Bertani medium containing 100 μg/ml ampicillin and 25 μg/ml kanamycin until a D600 (attenuance) of 0.5 was reached. IPTG (isopropyl β-D-thiogalactoside) was added to a final concentration of 0.1 mM and incubation was continued for a further 10 h. The cells were harvested by centrifugation at 9000 g for 15 min and resuspended in 1 ml of lysis buffer (50 mM NaH2PO4/300 mM NaCl/10 mM imidazole, pH 8.0). The cells were sonicated twice for 1 min and cell debris was removed by centrifugation. The enzymes were purified from the supernatant using HisTrap HP in accordance with the manufacturer's instructions (Amersham Biosciences).
SDS/PAGE and Western-blot analysis
An antiserum was raised against BxLAM16A by injecting recombinant protein purified from E. coli into rabbits. For SDS/PAGE analysis, protein samples were prepared by boiling it for 5 min in an equal volume of 2×sample buffer [100 mM Tris (pH 6.8)/12% (v/v) 2-mercaptoethanol/4% SDS/20% (v/v) glycerol/0.01% Bromophenol Blue) and run on 12% (w/v) polyacrylamide gels prepared using standard methods. Proteins within the gels were either stained with Coomassie Blue or transferred on to PVDF membrane for Western-blot analysis. Membranes were blocked with 1% (w/v) blocking reagent (Roche, Lewes, East Sussex, U.K.) and 1% (v/v) normal goat serum in PBS with 0.2% Tween 20 for 16 h at 23 °C. Blots were then immunolabelled for 3 h at 23 °C with the antiserum against BxLAM16A diluted 1:1000 in PBS containing 0.2% Tween 20. Bound antiserum was detected with alkaline phosphatase-conjugated secondary antibodies and Nitro Blue Tetrazolium/5-bromo-4-chloroindol-3-yl phosphate colour reaction.
Total homogenate and secretions from B. xylophilus were prepared for Western-blot analysis as described previously [26], except that 0.1% laminarin was added to the M9 buffer.
Enzyme assays
Assays were performed in a reaction mixture containing 50 μl of 0.5% (w/v) laminarin (Sigma) (or other substrates where appropriate), 40 μl of citrate/phosphate buffer (0.1 M citric acid/0.2 M Na2HPO4, pH 4.9) and 10 μl of appropriately diluted enzyme solution. The pH value of the citrate/phosphate buffer was confirmed at room temperature (23 °C). After incubation at 40 °C for 10 min, the reducing power released was measured by the p-hydroxybenzoic acid hydrazide method [27], using D-glucose as the standard. One unit of β-glucanase activity was defined as the amount of the enzyme liberating 1 μmol of reducing sugar/min. Xyloglucan endotransferase activity was measured by the procedure of Nishitani and Tominaga [28], using 2-aminobenzamide-labelled xyloglucan oligomer [29] as an acceptor and tamarind (Tamarindus indica) xyloglucan as a donor molecule. A mixture of 11 nmol of a fluorescence labelled acceptor molecule and 250 μg of unlabelled high-Mr polymer was incubated with the enzyme preparation in 100 μl of 0.4×citrate/phosphate buffer, pH 4.9, at 40 °C for 10 min. The reaction product was chromatographed on Superdex 75HR (Amersham Biosciences) and detected by refractometry and fluorimetry. Protein concentrations were determined by the method of Lowry using the DC Protein Assay system (Bio-Rad Laboratories, Hemel Hempstead, Herts., U.K.), with BSA as the standard.
The substrates laminarin [from Laminaria digitata (fingered kelp)], pustulan [from Umbilicaria papulosa (toadskin lichen)] and carboxymethylcellulose were obtained from Nakalai Tesque (Kyoto, Japan) and glycol chitin from Seikagaku (Chuo-Ku, Tokyo, Japan). Lichenan [from the lichen Cetraria islandica (Iceland moss)], β-D-glucan [from barley (Hordeum vulgare) and baker's yeast (Saccharomyces cerevisiae)] and κ-carrageenan (from the red alga Eucheuma cottonii) were obtained from Sigma, whereas xylan [from birchwood (Betula alba)] was obtained from Fluka (Gillingham, Dorset, U.K.). Agarose was obtained from Wako (Tokyo, Japan) and tamarind xyloglucan (Glyloid 6c) was from Dainippon Pharmaceutical (Osaka, Japan). 2-Aminobenzamide-labelled xyloglucan oligomer was kindly provided by Dr Tadashi Ishii of the Forestry and Forest Products Research Institute.
Detection of hydrolytic products
The hydrolytic products of laminarin were determined by HPLC. Laminarin (0.25%, w/v) was digested in 0.4×citrate/phosphate buffer (pH 4.9) at 40 °C for various time intervals. The reaction was stopped by incubating at 98 °C for 5 min. The hydrolysis products were separated at 60 °C in water using tandem-connected SUGAR KS-802 columns fitted with a guard column (SUGAR KS-G; Shodex, Kawasaki, Kanagawa, Japan) and detected by refractometry.
Temperature and pH optima, and thermal stability
The optimum temperature was determined by running the standard assay at temperatures from 30 to 80 °C. The optimum pH of the enzyme was determined by running the standard assay at 40 °C using citrate/phosphate and 0.1 M KH2PO4/0.05 M Na2B4O7 buffers for pH ranges 3.0–7.4 and 6.4–8.9 respectively. Thermal stability was determined using diluted enzyme (0.65 mg/ml of purified enzyme was diluted 100 times in 0.4×citrate/phosphate buffer, pH 4.9), incubated at various temperatures for 60 min, with the residual activity determined using the standard assay at 40 °C.
In situ hybridization and immunolocalization
In situ hybridizations were performed as described previously [30]. Nematode sections were hybridized with digoxigenin-labelled sense or antisense probes generated using the b13-0s and b13-0a primers shown above. Specimens were examined with differential interference contrast microscopy.
Immunofluorescence labelling of B. xylophilus with antisera against BxLAM16A was performed essentially as described previously [31]. Rabbit antisera raised against BxLAM16A were diluted 1:100 in PBS containing 0.2% Tween 20 and 1% blocking reagent (Roche). The primary antiserum was detected using goat anti-rabbit antibodies conjugated to Alexa 488 (Molecular Probes) used at a final dilution of 1:500. Labelled nematodes were examined with a Nikon fluorescence microscope.
Phylogenetic analysis
The deduced amino acid sequence of B. xylophilus and B. mucronatus β-1,3-glucanases were compared with the sequences of GHF16 β-1,3-glucanases and β-1,3-glucanase-like proteins from bacteria, fungi and animals in a phylogenetic analysis. Protein sequences were aligned with Clustal X version 1.81 [32], using Blosum 30 as the protein weight matrix. Several values for both gap-opening and gap-extension penalties were tested, and a combination of 6 and 1 for these factors was finally adopted, as this gave rise to an alignment in which the conserved domains were aligned most accurately with few gaps. Minor corrections were then performed manually on the alignment based on the crystal structure of the endo-β-1,3-1,4-glucanases from Bacillus licheniformis and B. macerans [33,34]. Tentative phylogenetic trees for the alignment were calculated with neighbour joining and maximum parsimony methods using PAUP* v. 4.0b10 [35]. The tree topologies thus obtained were subjected to maximum likelihood) analysis using PROTML in the MOLPHY v.2.3b3 package [36] with the local rearrangement and the JTT-F options of amino acid substitution model. Local bootstrap probability was estimated using the resampling of estimated log-likelihood (RELL) method with the best tree.
RESULTS
Isolation of B. xylophilus β-1,3-glucanase gene
During an EST project performed on a B. xylophilus cDNA library, a clone with similarity to β-1,3-glucanases was identified. This cDNA was designated Bx-lam16A and the predicted protein BxLAM16A, in accordance with the terminology suggested in [37]. The putative full-length Bx-lam16A cDNA comprises 803 bp containing an ORF of 753 bp that ended with a TAA stop codon. The cDNA contained a 46 bp 3′-untranslated region, which contained a polyadenylation signal (AATAAA) 15 nt upstream of the poly(A) tail. This spacing is similar to that seen for many Caenorhabditis elegans genes. The predicted amino acid sequence includes a hydrophobic signal peptide, which is predicted by the SignalP program [38] to be cleaved between Ala15 and Gly16, giving rise to a mature peptide with a predicted molecular mass of 26441.73 Da and a predicted pI of 4.7.
A similarity search based on the BLASTP program showed that the deduced amino acid sequence BxLAM16A was most similar to GHF16 β-1,3-glucanases from bacteria. BxLAM16A shared 49–43% identity with β-1,3-glucanases from Xanthomonas campestris (cabbage-black-rot bacterium), X. axonopodis and Pseudomonas sp. Multiple alignments of the deduced amino acid sequence encoded by Bx-lam16A and other β-1,3-glucanases showed that conserved residues of the GHF16 glucanases were present in BxLAM16A (Figure 1). A cDNA that could encode a similar protein was found amongst ESTs generated from a related, but solely fungal feeding species, B. mucronatus. This cDNA, designated Bm-lam16A, encodes a product of 217 amino acid residues exhibiting 73% identity with BxLAM16A (Figure 1). The signal peptides of the Bursaphelenchus-predicted proteins had features as expected from eukaryotic signal peptides (a short hydrophobic region followed by a predicted cleavage site) rather than those of bacterial signals for secretion [38]. The Bursaphelenchus signal peptides are short (16 amino acids for BxLAM16A) whereas prokaryotic sequences tend to be longer. In addition, the sequence immediately following the peptide cleavage site does not contain alanine, aspartic or glutamic, serine or threonine residues. Such residues are usually present within five amino acids of the cleavage site of prokaryotic signal peptides. However, the BxLAM16A signal peptide does contain alanine residues at positions – 1 and – 3 relative to the predicted cleavage site, a feature almost always seen in prokaryotic signal peptides, although this character cannot be considered to be diagnostic.
Figure 1. Multiple alignment of the amino acid sequence of BxLAM16A and BmLAM16A with other GHF16 glucanases from bacteria and invertebrates.
The black shading indicates identical amino acids and the grey shading indicates conservative replacements. The numbers to the left indicate the amino acid position of the respective proteins. Residues identified as catalytic amino acids are labelled with asterisks. The position of the intron in the B. xylophilus sequence is indicated by an arrow above the alignment. BxLAMa, B. xylophilus sequence from the present study; BmLAMa, B. mucronatus sequence from the present study; X.axon, bacterium Xanthomonas axonopodis (AAM36156); P.sp, bacteria Pseudomonas sp. (BAC16331); C.cell, bacterium Cellulosimicrobium cellulans (AAC44371); T.neap, bacterium Thermotoga neapolitana (CAA88008); S.purp, sea urchin S. purpuratus (AAC47235); P.sach, bivalve mollusc Pseudocardium (=Spisula) sachalinensis (AAP74223).
Endogenous origin of the B. xylophilus β-1,3-glucanase
Analysis of genomic DNA was performed to confirm the nematode origin of Bx-lam16A. A Southern blot containing genomic DNA from B. xylophilus, and from the fungus on which the nematodes were reared (B. cinerea), as a negative control, was made and hybridized with a DNA probe generated from the Bx-lam16A cDNA. The probe hybridized specifically to fragments in both the EcoRI and HindIII digests of B. xylophilus DNA (Figure 2). No signal was obtained from B. cinerea genomic DNA. In addition, the entire coding region of the Bx-lam16A gene was amplified from B. xylophilus gDNA, and the resulting PCR product was cloned and sequenced. Analysis of this sequence showed that it was exactly the same as the cDNA sequence, except that one intron was present at the site indicated in Figure 1. This intron is 401 bp in length, is bordered by canonical cis-splicing sequences and is adenine/thymine-rich (59.7%). This sequence, and the band pattern observed on the Southern blot, suggested that the β-1,3-glucanase is present as a small gene family in B. xylophilus, since no EcoRI or HindIII recognition sites are present in the sequenced region of the gene recognized by the probe. Southern blotting of B. mucronatus genomic DNA suggested that a gene family of size similar to that in B. xylophilus is present in B. mucronatus (results not shown).
Figure 2. Southern-blot analysis of Bx-lam16A.
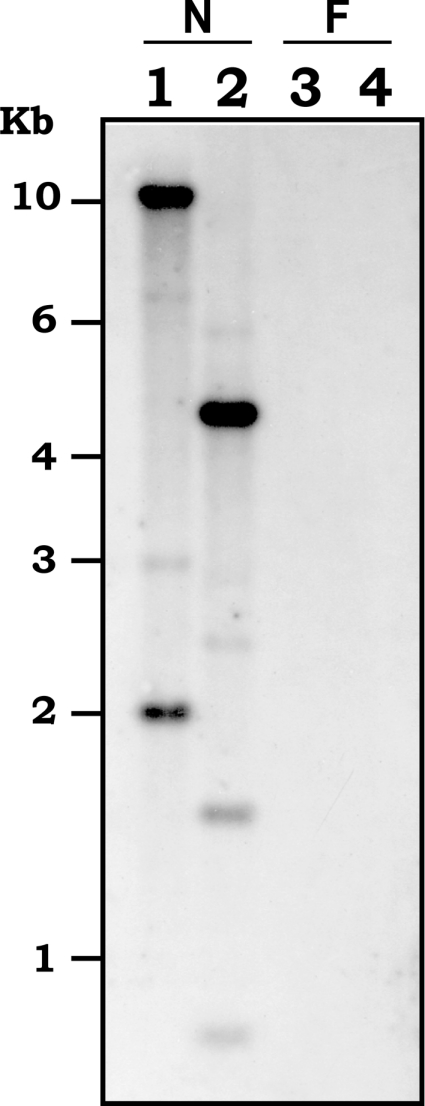
Genomic DNA from B. xylophilus (N) and B. cinerea (F) were digested with EcoRI (lanes 1 and 3) or HindIII (lanes 2 and 4). The blot was hybridized with a probe generated from Bx-lam16A cDNA.
Expression and purification of recombinant BxLAM16A
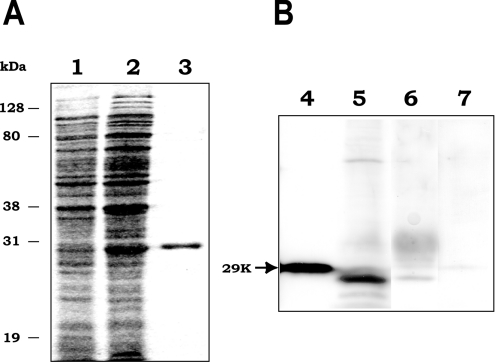
The BxLAM16A coding region without the putative signal sequence was amplified by PCR from the original plasmid, cloned into the expression vector and then expressed in E. coli as a polyhistidine-tagged fusion protein. The resulting recombinant protein had the expected molecular mass of 29 kDa, corresponding to 26.5 kDa from the Bx-lam16A ORF and 2.5 kDa encoded by the expression vector from the translational start codon to the cloning site, including a His6 tag. The recombinant protein was then purified using metal-affinity column chromatography to electrophoretic homogeneity (Figure 3A).
Figure 3. Detection of native and recombinant BxLAM16A.
(A) Coomassie Blue-stained SDS/PAGE gel showing expression and purification of recombinant BxLAM16A. Lanes 1 and 2, crude extract of E. coli containing the expression plasmid, non-induced (1) or induced (2) with IPTG; lane 3, purified recombinant BxLAM16A. (B) Westernblot analysis with antiserum raised against recombinant BxLAM16A. Lane 4, E. coli crude extract expressing BxLAM16A; lane 5, B. xylophilus homogenate; lane 6, B. xylophilus secretions; lane 7, crude extracts of B. cinerea as a negative control.
BxLAM16A was detected in nematode homogenate and secretions by Western-blot analysis using antisera raised against recombinant BxLAM16A (Figure 3B). The product of nematode appeared to be slightly smaller than that expressed in E. coli, which is in agreement with the expected sizes of these proteins. No signal was detected in crude cell extracts of B. cinerea on which the nematodes were reared or when the preimmune serum was used to probe Western blots (results not shown).
Biochemical analysis of recombinant BxLAM16A
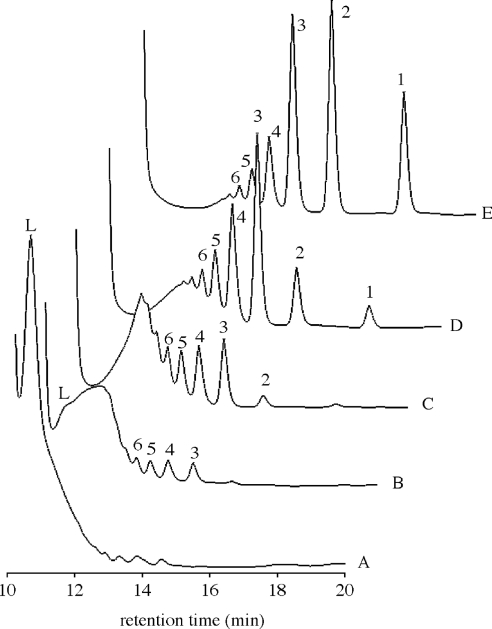
The activities of purified recombinant BxLAM16A with various substrates are summarized in Table 1. These results indicate that the enzyme had the highest specificity for the soluble β-1,3-glucan laminarin. Lower activity was found with β-1,3-glucan from baker's yeast, probably because of its low solubility. Very low levels of activity were observed on β-1,3-1,4-linked glucans, such as barley β-D-glucan and lichenan. The β-1,3-1,6-linked glucan (pustulan) and the β-1,4-linked substrates (carboxymethylcellulose, chitin and xylan) were not hydrolysed. Although some GHF16 proteins exhibit agarase, κ-carrageenase or xyloglucan endotransferase activity, BxLAM16A did not have these activities (Table 1). The mode of action of this enzyme was then examined by analysing the hydrolysis products by HPLC (Figure 4). Laminarin was immediately oligomerized and products larger than laminaribiose appeared. After prolonged incubation, the oligosaccharides gradually degraded into laminaribiose and glucose. These results confirm that BxLAM16A is an endo-β-1,3-glucanase (EC 3.2.1.39).
Table 1. Substrate specificity of purified recombinant BxLAM16A.
The enzyme reaction was performed at 40 °C for 10 min by incubating 0.065 μg of purified enzyme with 0.25% (w/v) substrate in a final 0.1 ml of buffer solution. The results are means for three independent experiments±S.D. No activity was detected in the case of pustulan, carboxymethylcellulose, glycol chitin, xylan, agarose, κ-carrageenan and xyloglucan. For xyloglucan, xyloglucan endotransferase activity was measured as described in the Experimental section.
| Substrate | Specific activity (units/mg) |
|---|---|
| Laminarin | 337±21 |
| Yeast glucan | 57±7 |
| Barley β-D-glucan | 10±4 |
| Lichenan | 7±3 |
Figure 4. HPLC analysis of BxLAM16A action on laminarin (0.25%, w/v).
Traces A, B, C, D and E, the enzyme (0.65 μg/ml) was incubated at 40 °C for 0, 3, 10, 210 min and overnight respectively. Samples were analysed on SUGAR KS-802 column ×2 (Shodex). Identified products are glucose (1), laminaribiose (2), laminari-oligomers (3–6) and laminarin (L).
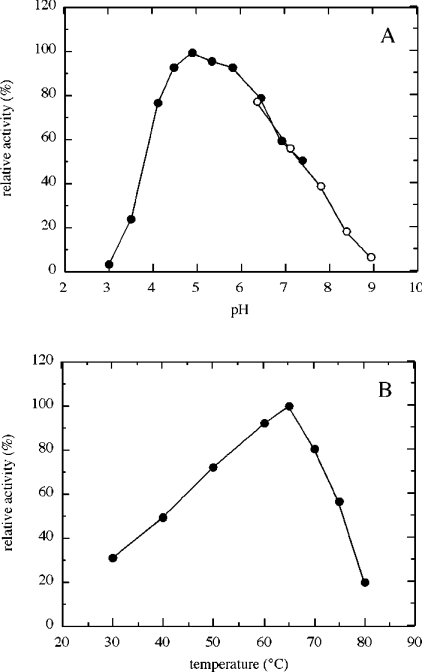
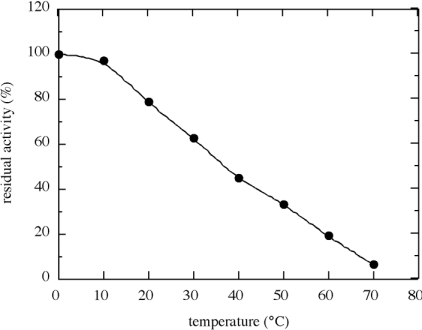
The laminarinase enzyme activity was investigated at different pH values and temperatures (Figure 5). The optimum pH was found to be 4.9 and the enzyme retained 90% of its activity between pH 4.5 and 5.8 (Figure 5A). There was a sharp decrease in activity below pH 4 and a gradual decrease in activity at increasingly alkaline pH values. The temperature at which maximum activity was observed was 65 °C (Figure 5B). Thermal stability was investigated by incubating the purified BxLAM16A at various temperatures for 60 min (Figure 6). Although the enzyme retained almost all of its activity at 10 °C, the residual activities of the enzyme gradually decreased as the incubation temperature increased.
Figure 5. Influence of pH and temperature on the activity of BxLAM16A.
(A) Enzyme activity was measured at 40 °C for 10 min, using laminarin as substrate, in citrate/phosphate buffer (●) at pH 3.0–7.4 and in KH2PO4/Na2B4O7 buffer (○) at pH 6.4–8.9. (B) Enzyme activity was measured in citrate/phosphate buffer, pH 4.9, for 10 min, using laminarin as substrate. Each assay was performed with 0.65 μg of protein/ml.
Figure 6. Thermal stability of BxLAM16A.
The enzyme (6.5 μg/ml) was incubated for 60 min at each temperature in citrate/phosphate buffer, pH 4.9. After the incubation, residual activity was measured at 40 °C.
Expression site and protein localization
To determine which nematode tissues express Bx-lam16A, in situ mRNA hybridization was performed. Digoxigenin-labelled antisense probes generated from Bx-lam16A, specifically hybridized with transcripts in the oesophageal gland cells of B. xylophilus (Figure 7A). No hybridization was observed in B. xylophilus with the control sense cDNA probes (Figure 7B). Hybridization signals were detected in female, male and propagative larvae of B. xylophilus. We could not determine precisely which gland cell or cells were the sites of Bx-lam16A expression. This is because it is difficult to distinguish each gland cell, as the three oesophageal gland cells of B. xylophilus are dorsally overlapping and all connect to similar positions in the large median oesophageal bulb [39].
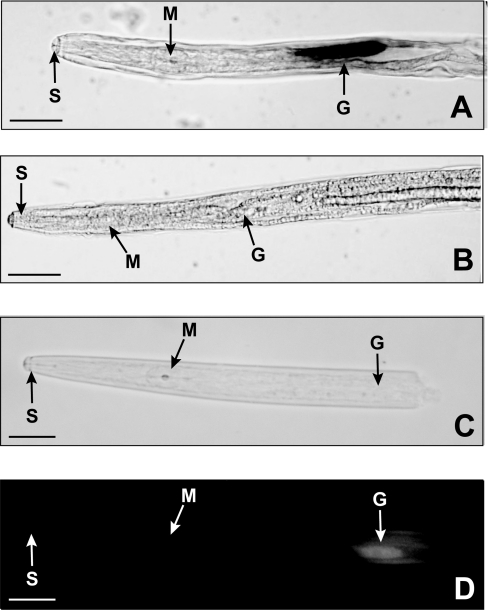
Figure 7. Localization of the BxLAM16A transcript and protein.
(A, B) Localization by in situ hybridization of Bx-lam16A transcripts in the oesophageal gland cells of B. xylophilus adult female with antisense (A) and sense (B) Bx-lam16A digoxigenin-labelled cDNA probes. Expression is restricted to oesophageal gland cells. (C, D) Immunofluorescence localization with antiserum against recombinant BxLAM16A, showing that the protein is present in the oesophageal gland cells of the nematode. (C) Illustrates the bright-field image, whereas (D) illustrates the same specimen viewed under fluorescence optics. G, oesophageal glands; S, stylet; M, metacarpus (scale bar, 20 μm).
Immunolocalization studies with antibodies raised against the recombinant BxLAM16A showed that the protein was present in the oesophageal glands of the nematodes (Figures 7C and 7D). No such binding was observed when using preimmune serum in place of the antiserum (results not shown).
Phylogenetic analysis
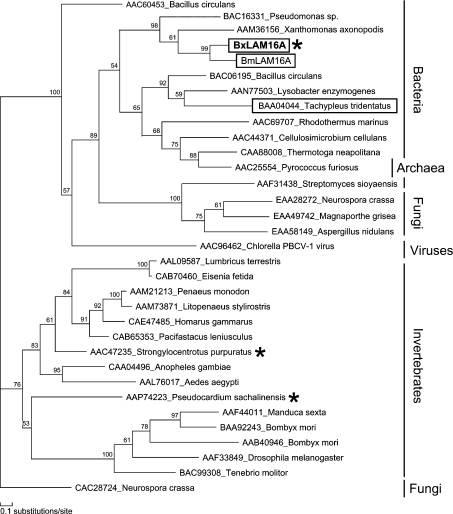
A phylogenetic tree generated using maximum likelihood analysis from an alignment of the BxLAM16A and BmLAM16A deduced proteins with selected GHF 16 proteins from bacteria, fungi and animals is shown in Figure 8. Analysis using neighbour-joining method generated trees with similar topology. This analysis showed that the BxLAM16A and BmLAM16A sequences are more closely related to bacterial sequences compared with animal sequences.
Figure 8. Unrooted phylogenetic tree of selected GHF16 β-1,3-glucanases and β-1,3-glucanase-like proteins generated using maximum-likelihood analysis.
Bootstrap probabilities are estimated for each node by the RELL method. Functionally characterized β-1,3-glucanases from animals are labelled with asterisks. Animal sequences that cluster in the bacterial branch are boxed. Scale bar, 10 substitutions/100 amino acid positions.
DISCUSSION
In the present paper, we describe the first β-1,3-glucanase gene from any nematode and confirmed its nematode origin.
The presence of a hydrophobic N-terminal sequence (signal peptide) in the deduced amino acid sequence of Bx-lam16A and its mRNA expression and protein localization in the oesophageal glands suggest that BxLAM16A is secreted from the stylet of the nematode. Western-blot analysis using nematode secretions supports this. Although B. xylophilus is a pathogen of trees, it can complete its life cycle feeding solely on fungi, including Botrytis, Alternaria and blue stain fungi such as Ophiostoma. β-1,3-Glucan is the main structural component of the cell walls of these fungi. It is probable therefore that BxLAM16A is secreted from the nematode stylet and weakens fungal cell walls to allow the nematode to feed more easily. Further evidence that the enzyme plays a role in fungal feeding rather than in parasitism of plants comes from the fact that a similar cDNA was identified in ESTs from a related nematode species, B. mucronatus, that feeds exclusively on fungi.
Other functions for the protein may be possible. β-1,3-Glucanase-like proteins, as well as lipopolysaccharide-binding proteins and peptidoglycan-binding proteins, have been suggested to play a role in invertebrate innate immunity by acting as pattern-recognition receptors [17]. They bind to β-1,3-glucan present in the cell surface of bacteria and fungi, triggering the activation of prophenol oxidase cascade, which results in the formation of cytotoxic and antimicrobial compounds and thus represents an important defence mechanism in a variety of invertebrates [17]. In recent years, C. elegans has begun to be used to study host–pathogen interactions and now is recognized as a powerful model system to study innate immunity [40]. However, many details, especially regarding pathogen recognition, are poorly understood and warrant further investigation [40]. Although no sequence similar to β-1,3-glucanase is present in C. elegans genome and it is unclear whether B. xylophilus β-1,3-glucanase has a role in the nematode immune system, the presence of the β-1,3-glucanase in at least one nematode species may help to understand the mechanism and evolution of nematode innate immune systems as proteins that play a role in microbe recognition in C. elegans become identified.
The biochemical properties of BxLAM16A were similar to those of bacterial β-1,3-glucanases from GHF16. BxLAM16A efficiently hydrolysed β-1,3-glucans and to a lesser degree lichenan. Similar substrate preferences have been found in bacterial β-1,3-glucanases, including those from Streptomyces sioyaensis [1], Thermotoga neapolitana [41] and Cellvibrio mixtus [42]. The optimum temperature of BxLAM16A is 65 °C and its optimum activity was observed at pH 4.9. Many bacterial β-1,3-glucanases show optimal activities between pH 4.0 and 6.0. BxLAM16A seems to be particularly active at temperatures that are much higher than those found in the normal environmental conditions that the nematodes would encounter. This is also the case for several non-thermophilic bacterial hydrolases including those from B. circulans [43] and S. sioyaensis [1]. Although the optimum temperature of BxLAM16A was high (65 °C), BxLAM16A was heat-labile, since the enzyme retained only 20% of its activity after incubation for 1 h at 60 °C. This might indicate that laminarin in the reaction solution contributes to the stabilization of the enzyme.
Many β-1,3-glucanases and β-1,3-glucanase-like proteins from a variety of invertebrates are present in GHF16 [5] (http://afmb.cnrs-mrs.fr/CAZY/). Phylogenetic analysis showed that these animal proteins were clustered in one clade with the exception of one protein, coagulation factor G, from the Japanese horseshoe crab (Tachypleus tridentatus). BxLAM16A was clustered together with glucanases from bacteria and this was supported by high bootstrap values, indicating that B. xylophilus β-1,3-glucanase is more closely related to those from bacteria than similar proteins from eukaryotes. In addition, no nematode sequences similar to the Bx-lam16A sequence described here are present in any publicly accessible database, despite the fact that genome sequences for C. elegans and C. briggsae and over 200000 EST sequences from other nematodes from across the phylum are now available [44]. It is therefore probable that the B. xylophilus endo-β-1,3-glucanase described in the present study was acquired by horizontal gene transfer from bacteria.
In plant parasitic cyst and root-knot nematode, a variety of genes considered to be involved in host–parasite interactions, including plant cell wall degrading enzymes [18–22] and chorismate mutase [45,46] are supposed to have been acquired by horizontal gene transfer from bacteria. It has been suggested that these transfers occurred from bacteria closely associated with an ancestor of these plant parasitic cyst and root-knot nematodes [47,48]. In a previous study, we suggested that cellulase (endo-β-1,4-glucanase) genes of B. xylophilus were acquired by horizontal gene transfer from fungi due to a close association of the ancestor of B. xylophilus with fungi rather than with bacteria [26]. The findings of this study support the independent horizontal-gene-transfer hypothesis that suggests that horizontal gene transfer has occurred on several independent occasions within the Nematoda and that it has been an important factor in the evolution of the ability to parasitize plants on each occasion. The results presented here suggest that similar horizontal-gene-transfer processes have enhanced the ability of a group of nematodes (Bursaphelenchus spp.) to feed on fungi. It is possible that these nematodes acquired β-1,3-glucanase genes from bacteria to obtain a fungal feeding ability, and a subgroup subsequently acquired cellulase genes from fungi, which permitted them to parasitize plants.
There are now a number of documented horizontal-gene-transfer events from both prokaryotes and eukaryotes to nematodes. At least three horizontal-gene-transfer events have occurred within tylenchid plant parasitic nematodes [48] and the present results show that at least two events have occurred within the Bursaphelenchus group. In addition, a large-scale analysis of the complete C. elegans genome sequence suggested that four alcohol dehydrogenase encoding genes were present in C. elegans and a closely related nematode that had been acquired by horizontal gene transfer from fungi [49]. Fewer studies have been performed on horizontal gene transfer in other phylogenetic groups, but there have been occasional reports of cell-wall-degrading enzymes in other invertebrates (e.g. [50]). As the number of genome sequencing and EST projects has increased, it has become clear that horizontal gene transfer to eukaryotes is a more widespread phenomenon than was thought previously. It is also clear that, on a limited number of occasions, it has allowed organisms to exploit niches that may have previously been unavailable to them.
It is intriguing to note that the genomic sequence of Bx-lam16A shows some features that are similar to some cyst and root-knot nematode genes acquired by horizontal gene transfer from bacteria. This is in spite of the fact that these genes were acquired independently and, presumably, from different sources. Whereas most nematode introns are small (the majority of C. elegans introns being less than 60 bp) the single intron in the Bx-lam16A gene is considerably larger at 401 bp. Some of the introns in plant parasitic-cyst- and root-knot-nematode genes considered to be acquired by horizontal gene transfer from bacteria are also considerably longer than standard nematode introns. In addition, although it is probable that the bacterial proteins would have been secreted and would therefore have had bacterial signal peptides at their N-termini, the signal peptides of the proteins reported in the present study and those encoded by other nematode genes acquired by horizontal gene transfer have the features of typical eukaryotic signal peptides. This may suggest that similar mechanisms have been used to modify these horizontally acquired genes on several occasions.
Acknowledgments
We thank all members of our working group for helpful advice and support. We also thank Dr T. Ishii for help with enzyme assays and K. Mishima for technical assistance. This work was supported in part by Research grant no. 200302 of the Forestry and Forest Products Research Institute (to T.K.) and by Scottish Executive Environment and Rural Affairs Department (project no. SCR/615/04 (to J.T.J.).
References
- 1.Hong T. Y., Cheng C. W., Huang J. W., Meng M. Isolation and biochemical characterization of an endo-1,3-β-glucanase from Streptomyces sioyaensis containing a C-terminal family 6 carbohydrate-binding module that binds to 1,3-β-glucan. Microbiology. 2002;148:1151–1159. doi: 10.1099/00221287-148-4-1151. [DOI] [PubMed] [Google Scholar]
- 2.Castresana C., de Carvalho F., Gheysen G., Habets M., Inze D., Van Montagu M. Tissue-specific and pathogen-induced regulation of a Nicotiana plumbaginifolia β-1,3-glucanase gene. Plant Cell. 1990;2:1131–1143. doi: 10.1105/tpc.2.12.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watanabe T., Kasahara N., Aida K., Tanaka H. Three N-terminal domains of β-1,3-glucanase A1 are involved in binding to insoluble β-1,3-glucan. J. Bacteriol. 1992;174:186–190. doi: 10.1128/jb.174.1.186-190.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Cruz J., Pintor-Toro J. A., Benitez T., Llobell A., Romero L. C. A novel endo-β-1,3-glucanase, BGN13.1, involved in the mycoparasitism of Trichoderma harzianum. J. Bacteriol. 1995;177:6937–6945. doi: 10.1128/jb.177.23.6937-6945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B., Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachman E. S., McClay D. R. Molecular cloning of the first metazoan β-1,3 glucanase from eggs of the sea urchin Strongylocentrotus purpuratus. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6808–6813. doi: 10.1073/pnas.93.13.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozhemyako V. B., Rebrikov D. V., Lukyanov S. A., Bogdanova E. A., Marin A., Mazur A. K., Kovalchuk S. N., Agafonova E. V., Sova V. V., Elyakova L. A., et al. Molecular cloning and characterization of an endo-1,3-β-D-glucanase from the mollusk Spisula sachalinensis. Comp. Biochem. Physiol. 2004;137:169–178. doi: 10.1016/j.cbpc.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Dimopoulos G., Richman A., Muller H. M., Kafatos F. C. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y. S., Ryu J. H., Han S. J., Choi K. H., Nam K. B., Jang I. H., Lemaitre B., Brey P. T., Lee W. J. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J. Biol. Chem. 2000;275:32721–32727. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 10.Ma C. C., Kanost M. R. A β 1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 2000;275:7505–7514. doi: 10.1074/jbc.275.11.7505. [DOI] [PubMed] [Google Scholar]
- 11.Ochiai M., Ashida M. A pattern-recognition protein for β-1,3-glucan – the binding domain and the cDNA cloning of β-1,3-glucan recognition protein from the silkworm, Bombyx mori. J. Biol. Chem. 2000;275:4995–5002. doi: 10.1074/jbc.275.7.4995. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R., Cho H. Y., Kim H. S., Ma Y. G., Osaki T., Kawabata S., Soderhall K., Lee B. L. Characterization and properties of a 1,3-β-D-glucan pattern recognition protein of Tenebrio molitor larvae that is specifically degraded by serine protease during prophenoloxidase activation. J. Biol. Chem. 2003;278:42072–42079. doi: 10.1074/jbc.M307475200. [DOI] [PubMed] [Google Scholar]
- 13.Seki N., Muta T., Oda T., Iwaki D., Kuma K., Miyata T., Iwanaga S. Horseshoe-crab (1,3)-β-D-glucan-sensitive coagulation factor-G – a serine-protease zymogen heterodimer with similarities to β-glucan-binding proteins. J. Biol. Chem. 1994;269:1370–1374. [PubMed] [Google Scholar]
- 14.Beschin A., Bilej M., Hanssens F., Raymakers J., Van Dyck E., Revets H., Brys L., Gomez J., De Baetselier P., Timmermans M. Identification and cloning of a glucan- and liopoplysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J. Biol. Chem. 1998;273:24948–24954. doi: 10.1074/jbc.273.38.24948. [DOI] [PubMed] [Google Scholar]
- 15.Lee S. Y., Wang R. G., Soderhall K. A lipopolysaccharide- and β-1,3-glucan-binding protein from hemocytes of the freshwater crayfish Pacifastacus leniusculus – purification, characterization, and cDNA cloning. J. Biol. Chem. 2000;275:1337–1343. doi: 10.1074/jbc.275.2.1337. [DOI] [PubMed] [Google Scholar]
- 16.Sritunyalucksana K., Lee S. Y., Soderhall K. A β-1,3-glucan binding protein from the black tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 2002;26:237–245. doi: 10.1016/s0145-305x(01)00074-x. [DOI] [PubMed] [Google Scholar]
- 17.Soderhall K., Cerenius L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998;10:23–28. doi: 10.1016/s0952-7915(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 18.Smant G., Stokkermans J. P., Yan Y., de Boer J. M., Baum T. J., Wang X., Hussey R. S., Gommers F. J., Henrissat B., Davis E. L., et al. Endogenous cellulases in animals: isolation of β-1,4-endoglucanase genes from two species of plant-parasitic cyst nematodes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4906–4911. doi: 10.1073/pnas.95.9.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosso M. N., Favery B., Piotte C., Arthaud L., de Boer J. M., Hussey R. S., Bakker J., Baum T. J., Abad P. Isolation of a cDNA encoding a β-1,4-endoglucanase in the root-knot nematode Meloidogyne incognita and expression analysis during plant parasitism. Mol. Plant Microbe Interact. 1999;12:585–591. doi: 10.1094/MPMI.1999.12.7.585. [DOI] [PubMed] [Google Scholar]
- 20.Popeijus H., Overmars H., Jones J., Blok V., Goverse A., Helder J., Schots A., Bakker J., Smant G. Degradation of plant cell walls by a nematode. Nature (London) 2000;406:36–37. doi: 10.1038/35017641. [DOI] [PubMed] [Google Scholar]
- 21.Doyle E. A., Lambert K. N. Cloning and characterization of an esophageal-gland-specific pectate lyase from the root-knot nematode Meloidogyne javanica. Mol. Plant Microbe Interact. 2002;15:549–556. doi: 10.1094/MPMI.2002.15.6.549. [DOI] [PubMed] [Google Scholar]
- 22.Jaubert S., Laffaire J. B., Abad P., Rosso M. N. A polygalacturonase of animal origin isolated from the root-knot nematode Meloidogyne incognita. FEBS Lett. 2002;522:109–112. doi: 10.1016/s0014-5793(02)02906-x. [DOI] [PubMed] [Google Scholar]
- 23.Williamson V. M., Gleason C. A. Plant-nematode interactions. Curr. Opin. Plant Biol. 2003;6:327–333. doi: 10.1016/s1369-5266(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 24.Vanholme B., De Meutter J., Tytgat T., Van Montagu M., Coomans A., Gheysen G. Secretions of plant-parasitic nematodes: a molecular update. Gene. 2004;332:13–27. doi: 10.1016/j.gene.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Gheysen G., Fenoll C. Gene expression in nematode feeding sites. Annu. Rev. Phytopathol. 2002;40:191–219. doi: 10.1146/annurev.phyto.40.121201.093719. [DOI] [PubMed] [Google Scholar]
- 26.Kikuchi T., Jones J. T., Aikawa T., Kosaka H., Ogura N. A family of glycosyl hydrolase family 45 cellulases from the pine wood nematode Bursaphelenchus xylophilus. FEBS Lett. 2004;572:201–205. doi: 10.1016/j.febslet.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 27.Lever M. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 28.Nishitani K., Tominaga R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992;267:21058–21064. [PubMed] [Google Scholar]
- 29.Ishii T., Ichita J., Matsue H., Ono H., Maeda I. Fluorescent labeling of pectic oligosaccharides with 2-aminobenzamide and enzyme assay for pectin. Carbohydr. Res. 2002;337:1023–1032. doi: 10.1016/s0008-6215(02)00087-3. [DOI] [PubMed] [Google Scholar]
- 30.de Boer J. M., Yan Y., Smant G., Davis E. L., Baum T. J. In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 1998;30:309–312. [PMC free article] [PubMed] [Google Scholar]
- 31.Goverse A., Davis E. L., Hussey R. S. Monoclonal-antibodies to the esophageal glands and stylet secretions of Heterodera glycines. J. Nematol. 1994;26:251–259. [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keitel T., Simon O., Borriss R., Heinemann U. Molecular and active-site structure of a Bacillus 1,3-1,4-β-glucanase. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5287–5291. doi: 10.1073/pnas.90.11.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn M., Olsen O., Politz O., Borriss R., Heinemann U. Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-β-glucanase. J. Biol. Chem. 1995;270:3081–3088. doi: 10.1074/jbc.270.7.3081. [DOI] [PubMed] [Google Scholar]
- 35.Swofford D. L. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other methods) Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 36.Adachi J., Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics Based on Maximum Likelihood. Tokyo: Institute of Statistical Mathematics; 1996. [Google Scholar]
- 37.Henrissat B., Teeri T. T., Warren R. A. J. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen H., Engelbrecht J., Brunak S., vonHeijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Nickle W. R., Golden A. M., Mamiya Y., Wergin W. P. On the taxonomy and morphology of the pine wood nematode, Bursaphelenchus xylophilus (Steiner & Buhrer 1934) Nickle 1970. J. Nematol. 1981;13:385–392. [PMC free article] [PubMed] [Google Scholar]
- 40.Millet A. C., Ewbank J. J. Immunity in Caenorhabditis elegans. Curr. Opin. Immunol. 2004;16:4–9. doi: 10.1016/j.coi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 41.Zverlov V. V., Volkov I. Y., Velikodvorskaya T. V., Schwarz W. H. Highly thermostable endo-1,3-β-glucanase (laminarinase) LamA from Thermotoga neapolitana: nucleotide sequence of the gene and characterization of the recombinant gene product. Microbiology. 1997;143:1701–1708. doi: 10.1099/00221287-143-5-1701. [DOI] [PubMed] [Google Scholar]
- 42.Sakellaris H., Pemberton J. M., Manners J. M. Genes from Cellvibrio mixtus encoding β-1,3 endoglucanase. Appl. Environ. Microbiol. 1990;56:3204–3208. doi: 10.1128/aem.56.10.3204-3208.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asano T., Taki J., Yamamoto M., Aono R. Cloning and structural analysis of bglM gene coding for the fungal cell wall-lytic β-1,3-glucan-hydrolase BglM of Bacillus circulans IAM1165. Biosci. Biotechnol. Biochem. 2002;66:1246–1255. doi: 10.1271/bbb.66.1246. [DOI] [PubMed] [Google Scholar]
- 44.Parkinson J., Mitreva M., Hall N., Blaxter M., McCarter J. P. 400000 nematode ESTs on the Net. Trends Parasitol. 2003;19:283–286. doi: 10.1016/s1471-4922(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 45.Bekal S., Niblack T. L., Lambert K. N. A chorismate mutase from the soybean cyst nematode Heterodera glycines shows polymorphisms that correlate with virulence. Mol. Plant Microbe Interact. 2003;16:439–446. doi: 10.1094/MPMI.2003.16.5.439. [DOI] [PubMed] [Google Scholar]
- 46.Jones J. T., Furlanetto C., Bakker E., Banks B., Blok V., Chen Q., Phillips M., Prior A. Characterization of a chorismate mutase from the potato cyst nematode Globodera pallida. Mol. Plant Pathol. 2003;4:43–50. doi: 10.1046/j.1364-3703.2003.00140.x. [DOI] [PubMed] [Google Scholar]
- 47.Bird D. M., Koltai H. Plant parasitic nematodes: habitats, hormones, and horizontally-acquired genes. J. Plant Growth Regul. 2000;19:183–194. doi: 10.1007/s003440000022. [DOI] [PubMed] [Google Scholar]
- 48.Scholl E. H., Thorne J. L., McCarter J. P., Bird D. M. Horizontally transferred genes in plant-parasitic nematodes: a high-throughput genomic approach. Genome Biol. 2003;4:R39. doi: 10.1186/gb-2003-4-6-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkinson J., Blaxter M. SimiTri – visualizing similarity relationships for groups of sequences. Bioinformatics. 2003;19:390–395. doi: 10.1093/bioinformatics/btf870. [DOI] [PubMed] [Google Scholar]
- 50.Sugimura M., Watanabe H., Lo N., Saito H. Purification, characterization, cDNA cloning and nucleotide sequencing of a cellulase from the yellow-spotted longicorn beetle, Psacothea hilaris. Eur. J. Biochem. 2003;270:3455–3460. doi: 10.1046/j.1432-1033.2003.03735.x. [DOI] [PubMed] [Google Scholar]