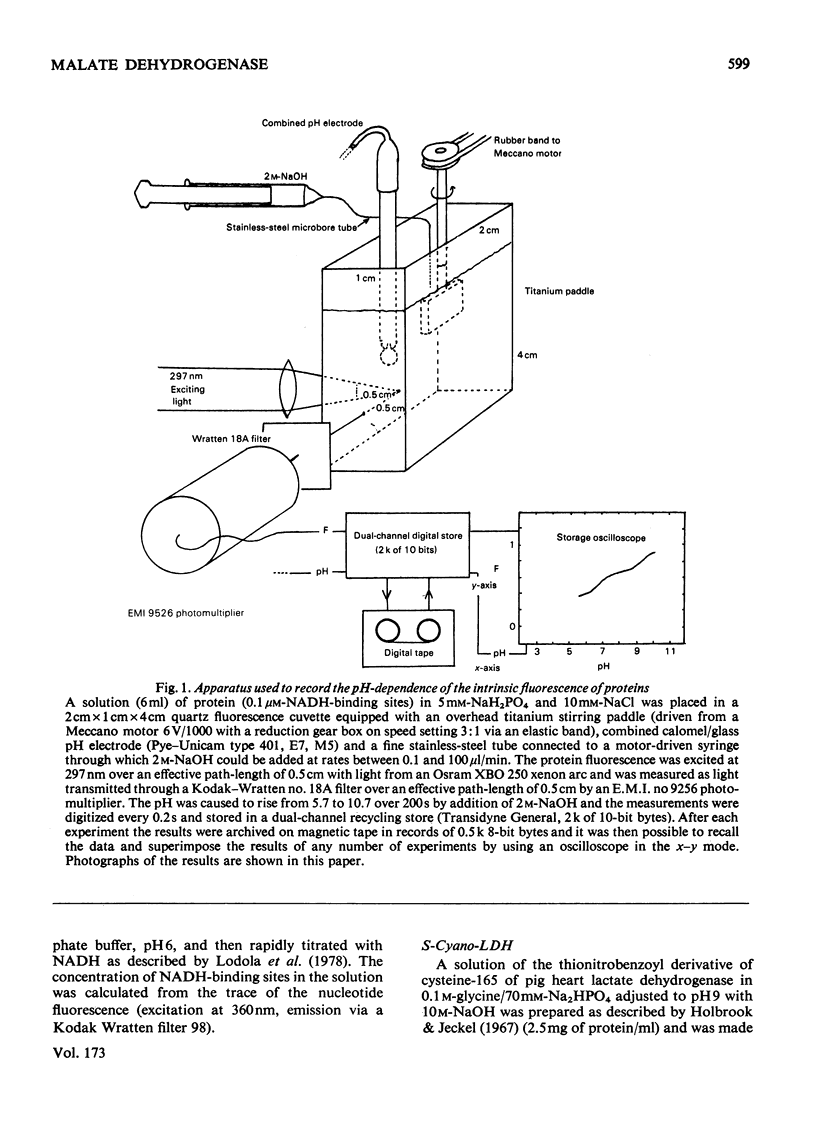
Abstract
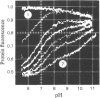
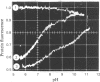
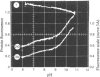
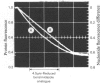
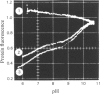
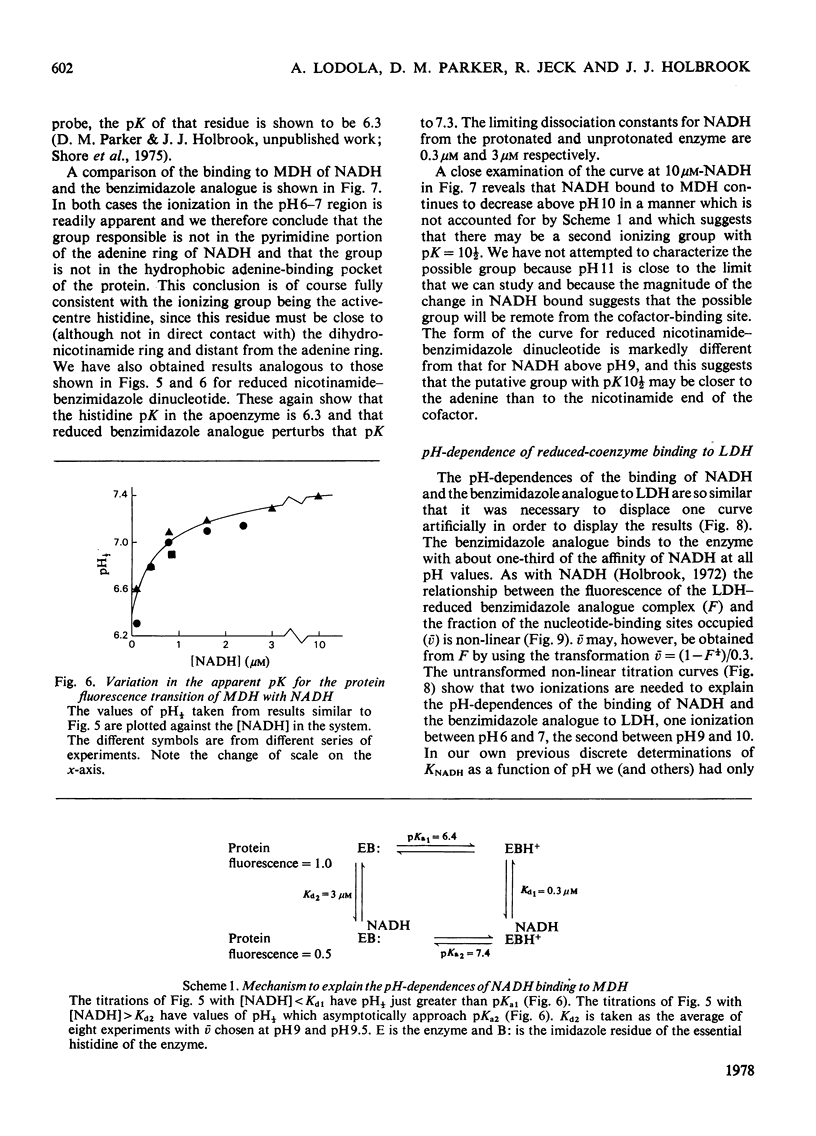
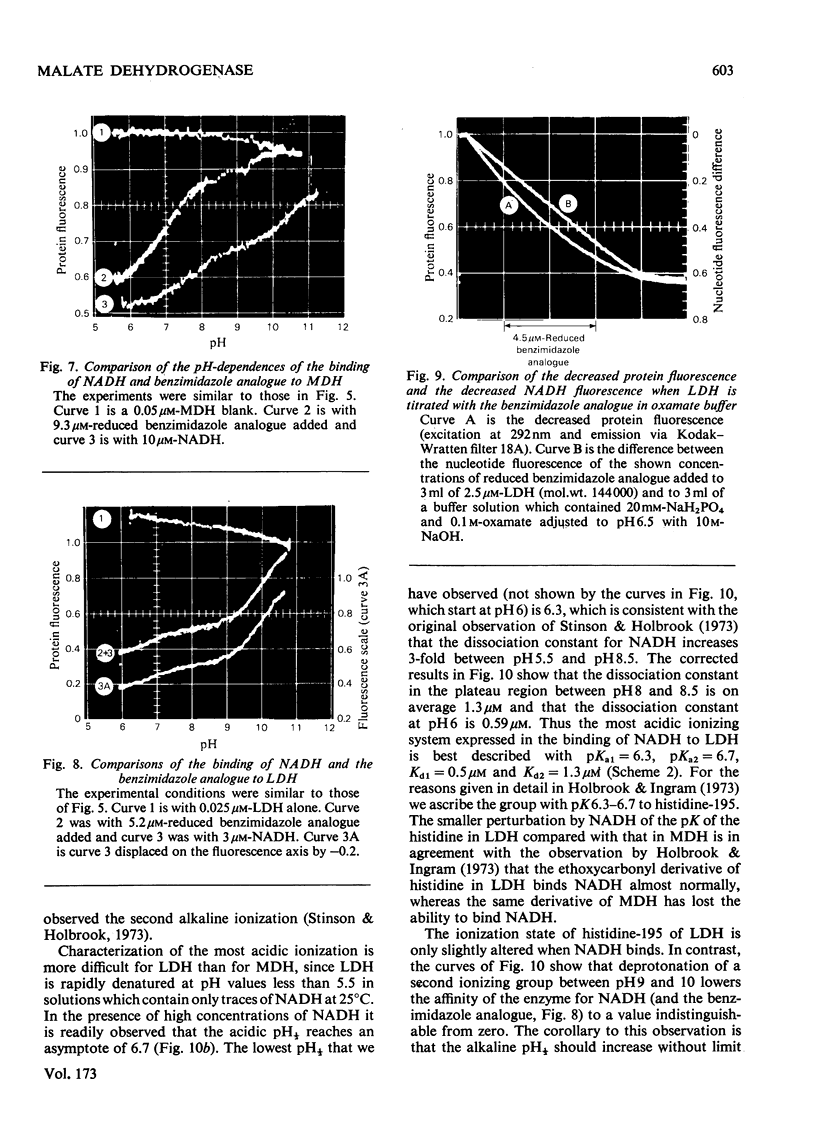
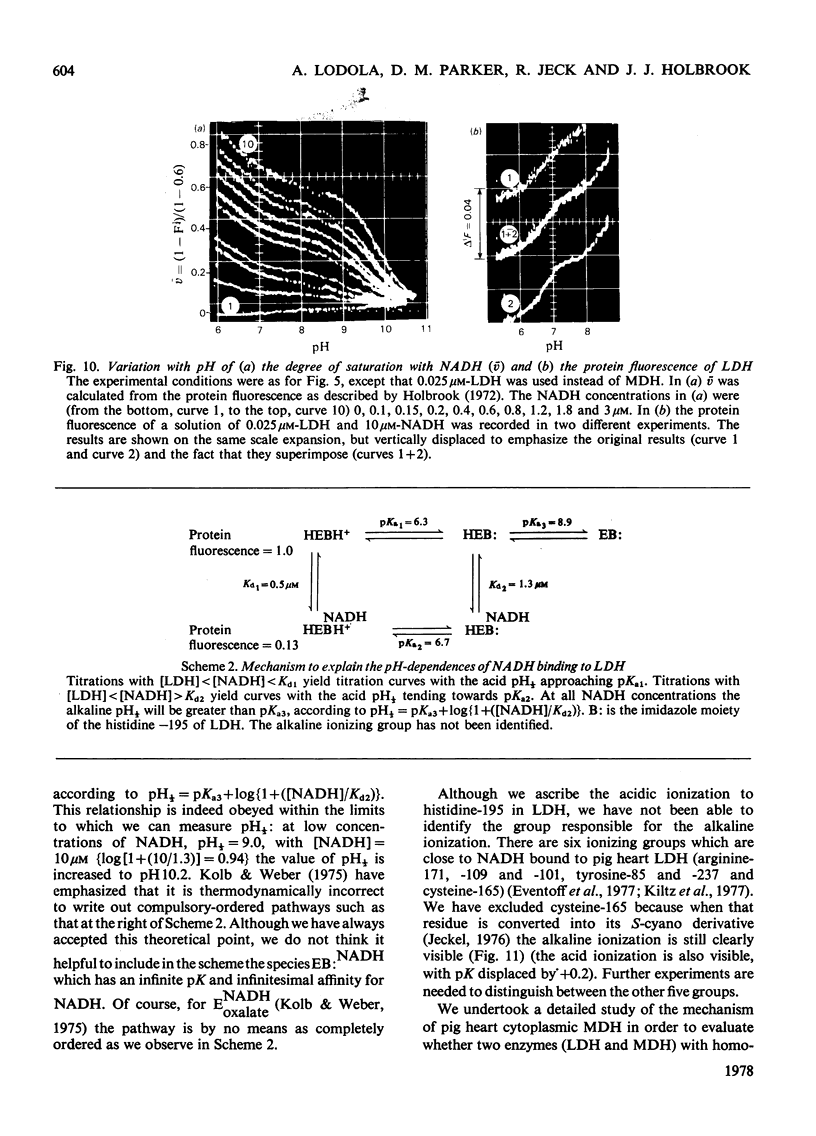
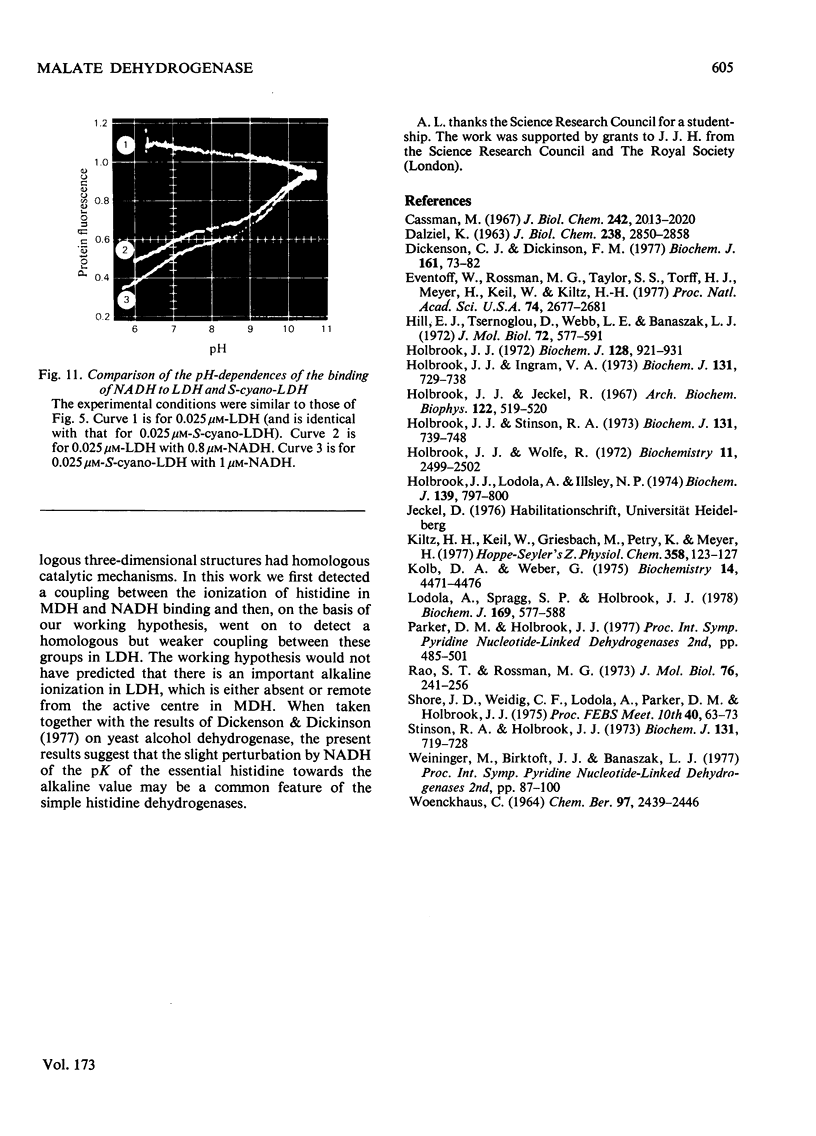
1. The pH-dependencies of the binding of NADH and reduced nicotinamide--benzimidazole dinucleotide to pig heart cytoplasmic malate dehydrogenase and lactate dehydrogenase are reported. 2. Two ionizing groups were observed in the binding of both reduced coenzymes to lactate dehydrogenase. One group, with pKa in the range 6.3--6.7, is the active-site histidine residue and its deprotonation weakens binding of reduced coenzyme 3-fold. Binding of both coenzymes is decreased to zero when a second group, of pKa 8.9, deprotonates. This group is not cysteine-165.3. Only one ionization is required to characterize the binding of the two reduced coenzymes to malate dehydrogenase. The group involved appears to be the active-site histidine residue, since its ethoxycarbonylation inhibits the enzyme and abolishes binding of reduced coenzyme. Binding of either reduced coenzyme increases the pKa of the group from 6.4 to 7.4, and deprotonation of the group is accompanied by a 10-fold weakening of coenzyme binding. 4. Two reactive histidine residues were detected per malate dehydrogenase dimer. 5. A mechanism which emphasizes the homology between the two enzymes is presented.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cassman M. Beef heart malic dehydrogenases. VI. A pH-dependent transition observed in the presence of reduced diphosphopyridine nucleotide. J Biol Chem. 1967 May 10;242(9):2013–2020. [PubMed] [Google Scholar]
- DALZIEL K. KINETIC STUDIES OF LIVER ALCOHOL DEHYDROGENASE AND PH EFFECTS WITH COENZYME PREPARATIONS OF HIGH PURITY. J Biol Chem. 1963 Aug;238:2850–2858. [PubMed] [Google Scholar]
- Dickenson C. J., Dickinson F. M. A study of the ionic properties of the essential histidine residue of yeast alcohol dehydrogenase in complexes of the enzyme with its coenzymes and substrates. Biochem J. 1977 Jan 1;161(1):73–82. doi: 10.1042/bj1610073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G., Taylor S. S., Torff H. J., Meyer H., Keil W., Kiltz H. H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E., Tsernoglou D., Webb L., Banaszak L. J. Polypeptide conformation of cytoplasmic malate dehydrogenase from an electron density map at 3.0 angstrom resolution. J Mol Biol. 1972 Dec 30;72(3):577–589. doi: 10.1016/0022-2836(72)90176-3. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Ingram V. A. Ionic properties of an essential histidine residue in pig heart lactate dehydrogenase. Biochem J. 1973 Apr;131(4):729–738. doi: 10.1042/bj1310729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Jeckel R. Thermal motion as the rate-limiting step in the superprecipitation of actomyosin gels. Arch Biochem Biophys. 1967 Nov;122(2):519–521. doi: 10.1016/0003-9861(67)90229-9. [DOI] [PubMed] [Google Scholar]
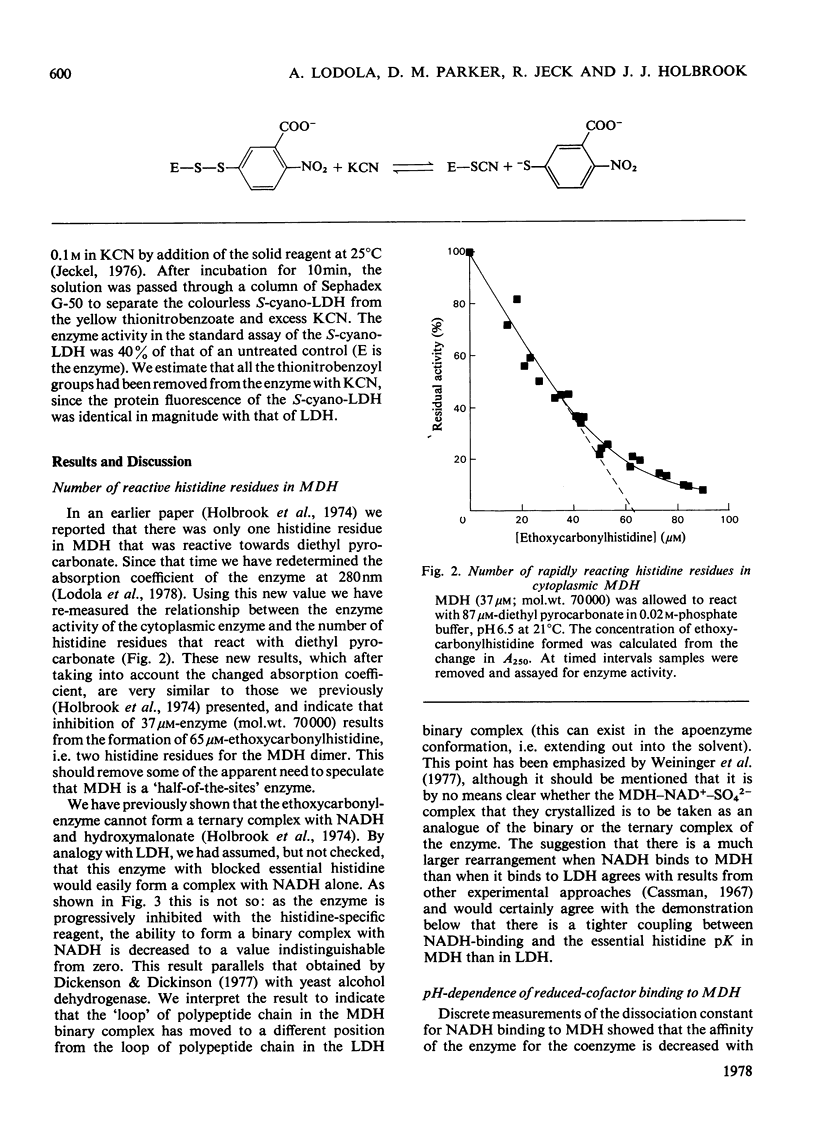
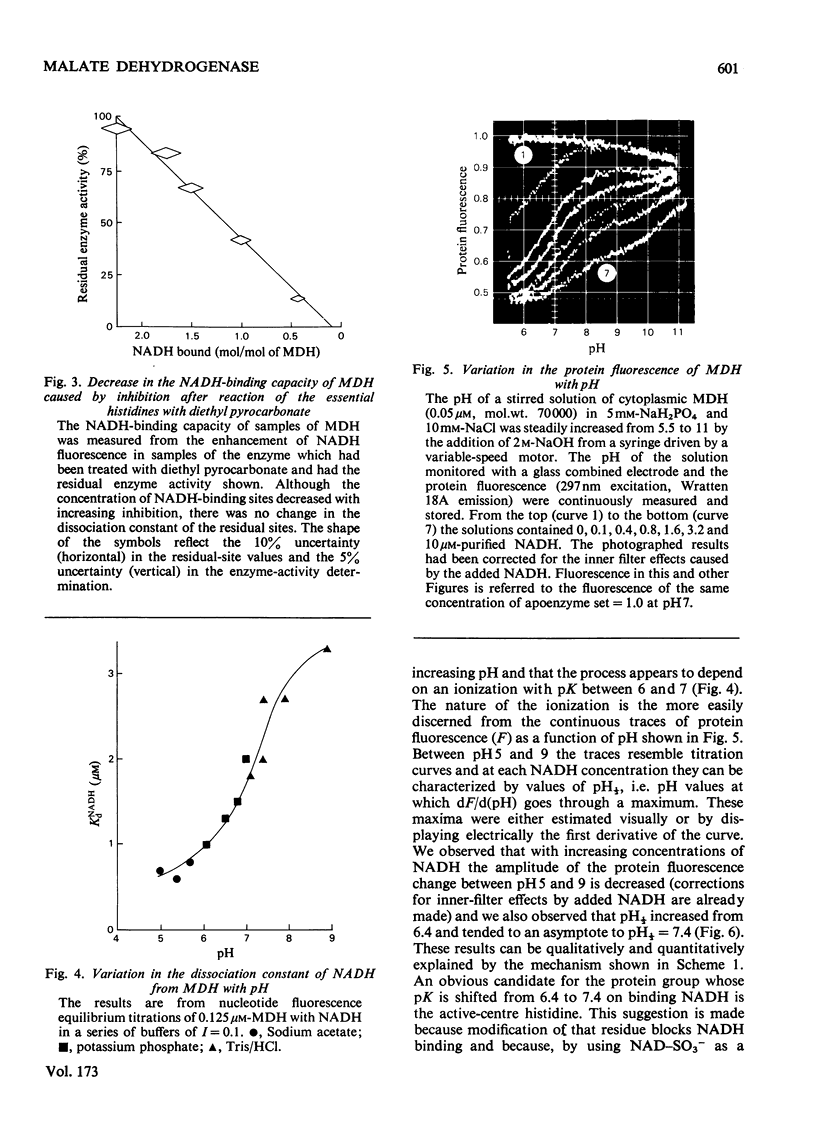
- Holbrook J. J., Lodola A., Illsley N. P. Histidine residues and the enzyme activity of pig heart supernatant malate dehydrogenase. Biochem J. 1974 Jun;139(3):797–800. doi: 10.1042/bj1390797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J. Protein fluorescence of lactate dehydrogenase. Biochem J. 1972 Jul;128(4):921–931. doi: 10.1042/bj1280921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Stinson R. A. The use of ternary complexes to study ionizations and isomerizations during catalysis by lactate dehydrogenase. Biochem J. 1973 Apr;131(4):739–748. doi: 10.1042/bj1310739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Wolfe R. G. Malate dehydrogenase. X. Fluorescence microtitration studies of D-malate, hydroxymalonate, nicotinamide dinucleotide, and dihydronicotinamide-adenine dinucleotide binding by mitochondrial and supernatant porcine heart enzymes. Biochemistry. 1972 Jun 20;11(13):2499–2502. doi: 10.1021/bi00763a018. [DOI] [PubMed] [Google Scholar]
- Kiltz H. H., Keil W., Griesbach M., Petry K., Meyer H. The primary structure of porcine lactate dehydrogenase: isoenzymes M4 and H4. Hoppe Seylers Z Physiol Chem. 1977 Jan;358(1):123–127. doi: 10.1515/bchm2.1977.358.1.123. [DOI] [PubMed] [Google Scholar]
- Kolb D. A., Weber G. Quantitative demonstration of the reciprocity of ligand effects in the ternary complex of chicken heart lactate dehydrogenase with nicotinamide adenine dinucleotide oxalate. Biochemistry. 1975 Oct 7;14(20):4471–4476. doi: 10.1021/bi00691a020. [DOI] [PubMed] [Google Scholar]
- Lodola A., Spragg S. P., Holbrook J. J. Malate dehydrogenase of the cytosol. Preparation and reduced nicotinamide-adenine dinucleotide-binding studies. Biochem J. 1978 Mar 1;169(3):577–588. doi: 10.1042/bj1690577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Stinson R. A., Holbrook J. J. Equilibrium binding of nicotinamide nucleotides to lactate dehydrogenases. Biochem J. 1973 Apr;131(4):719–728. doi: 10.1042/bj1310719. [DOI] [PMC free article] [PubMed] [Google Scholar]