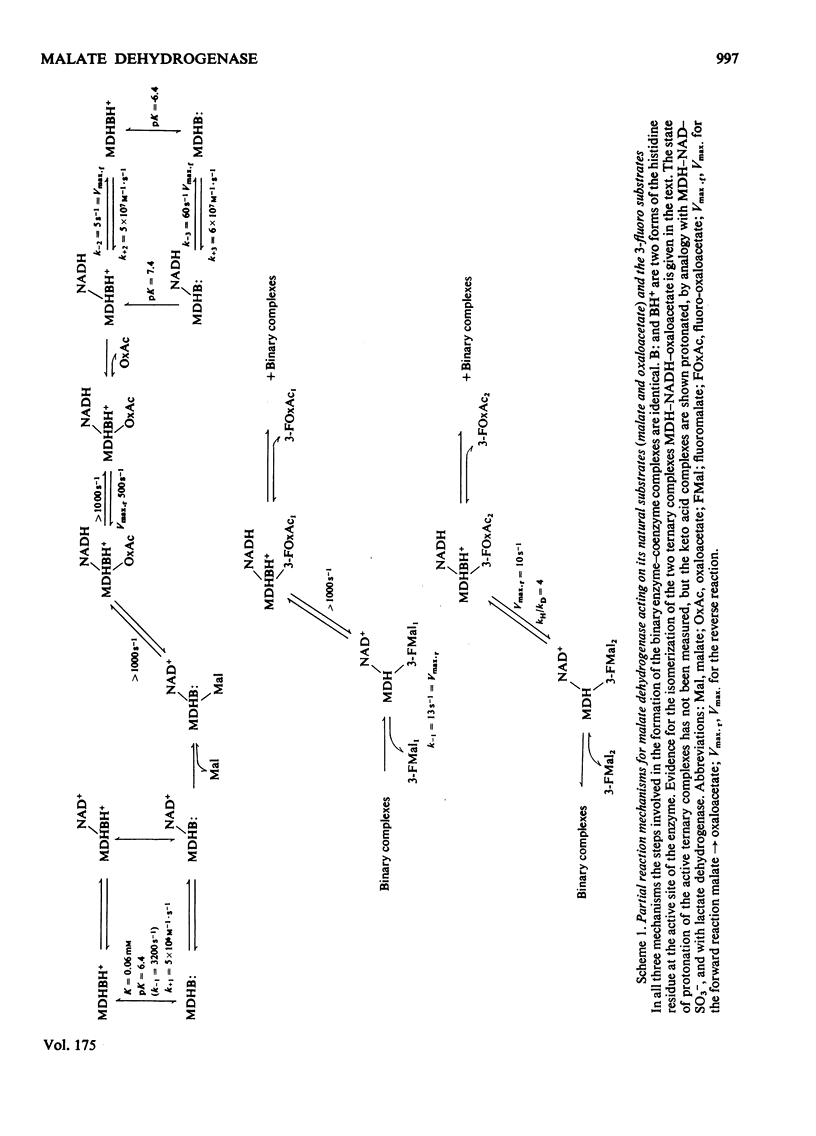
Abstract
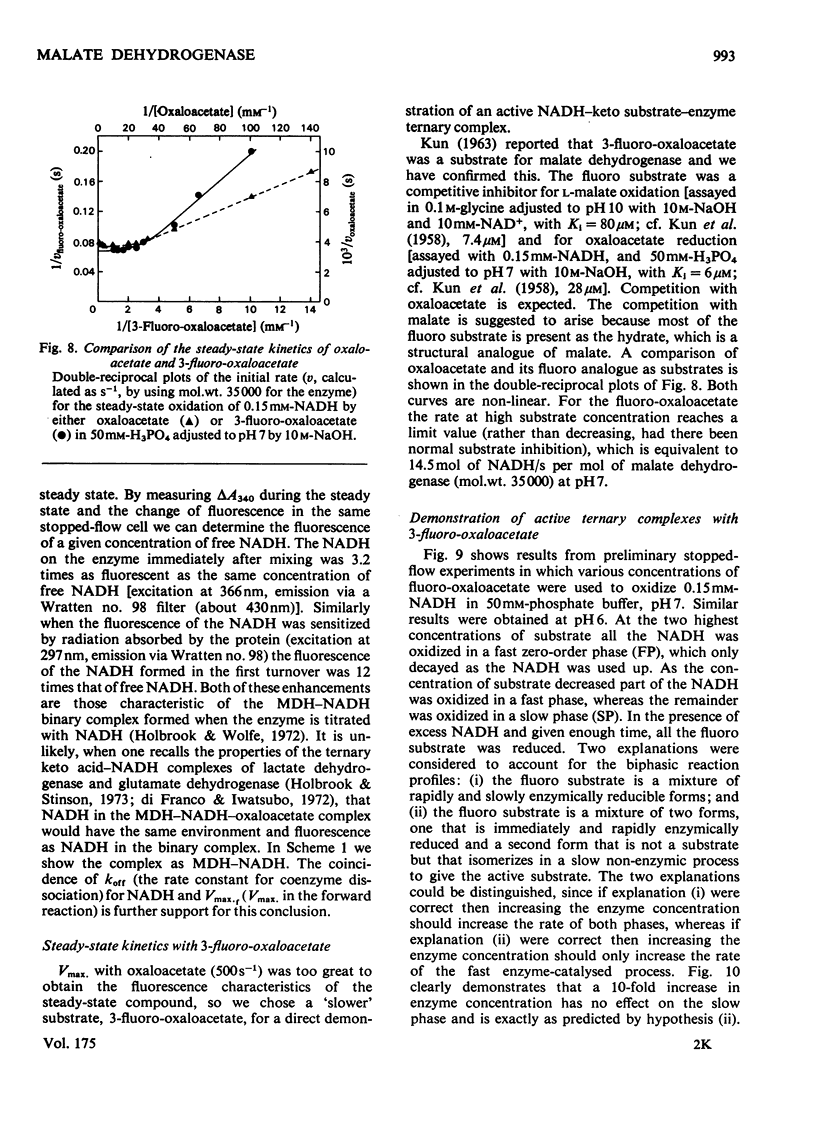
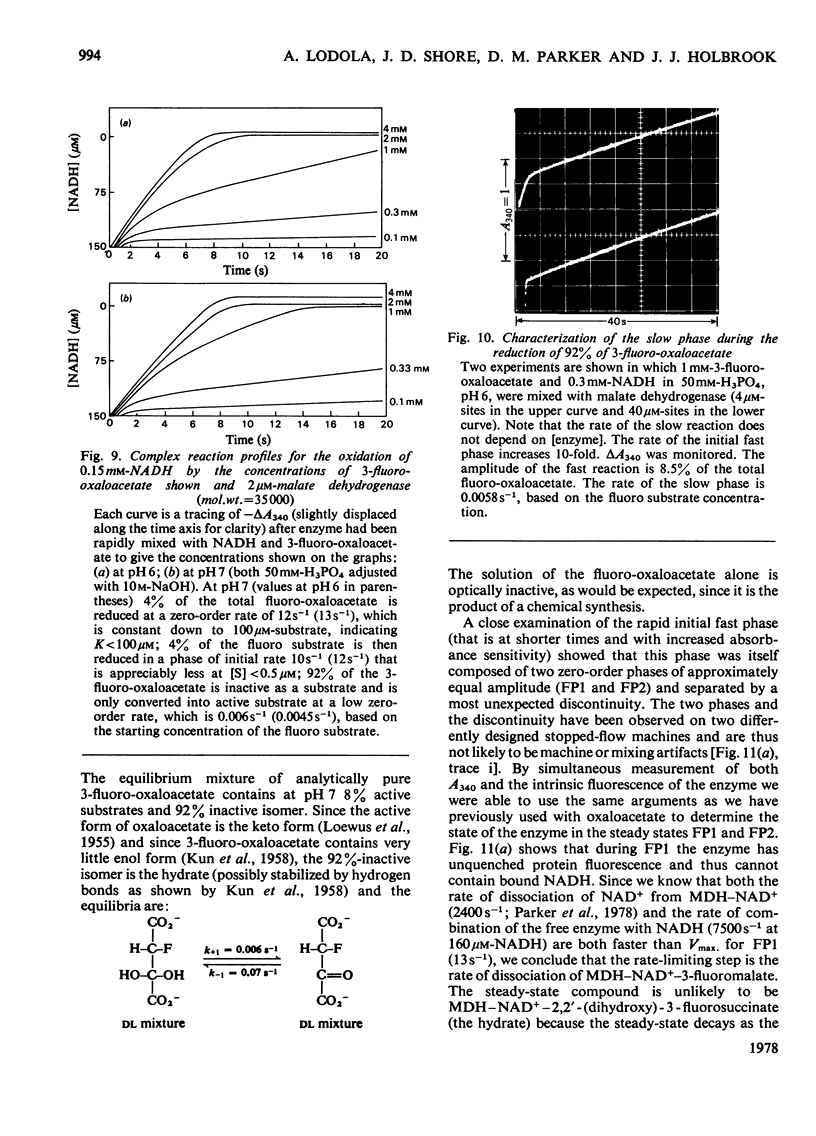
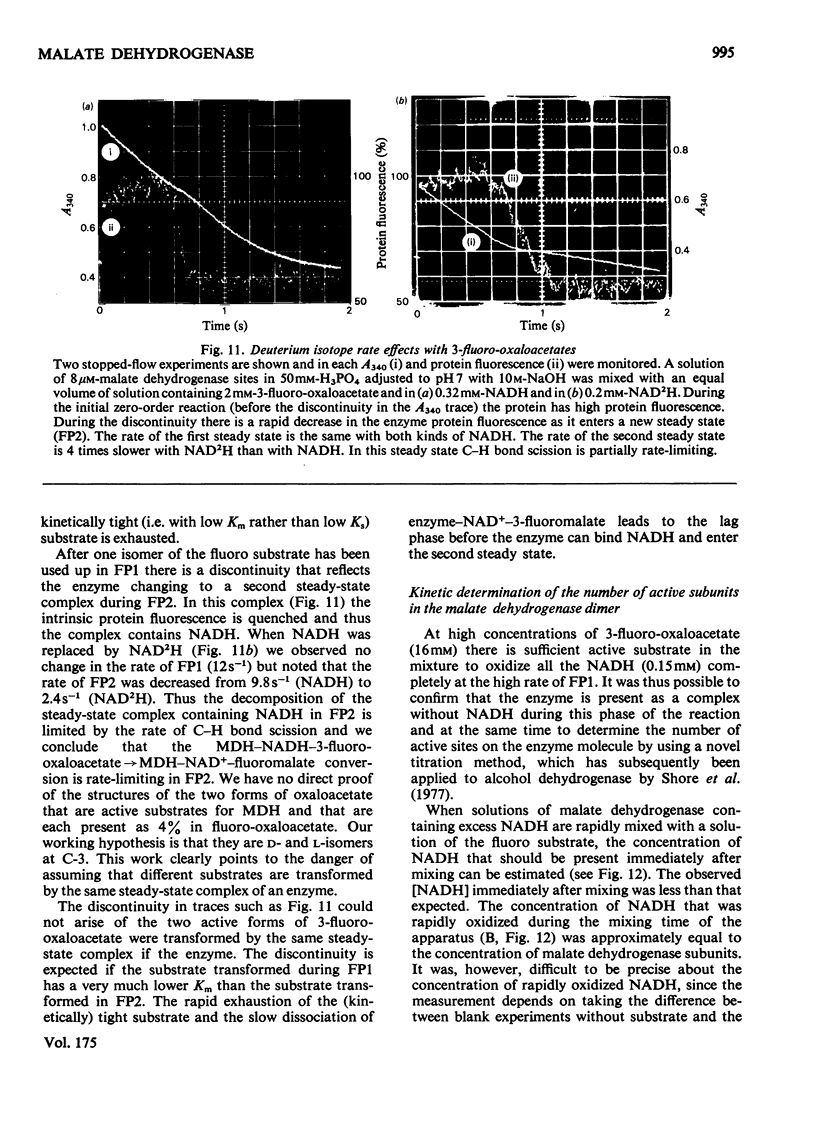
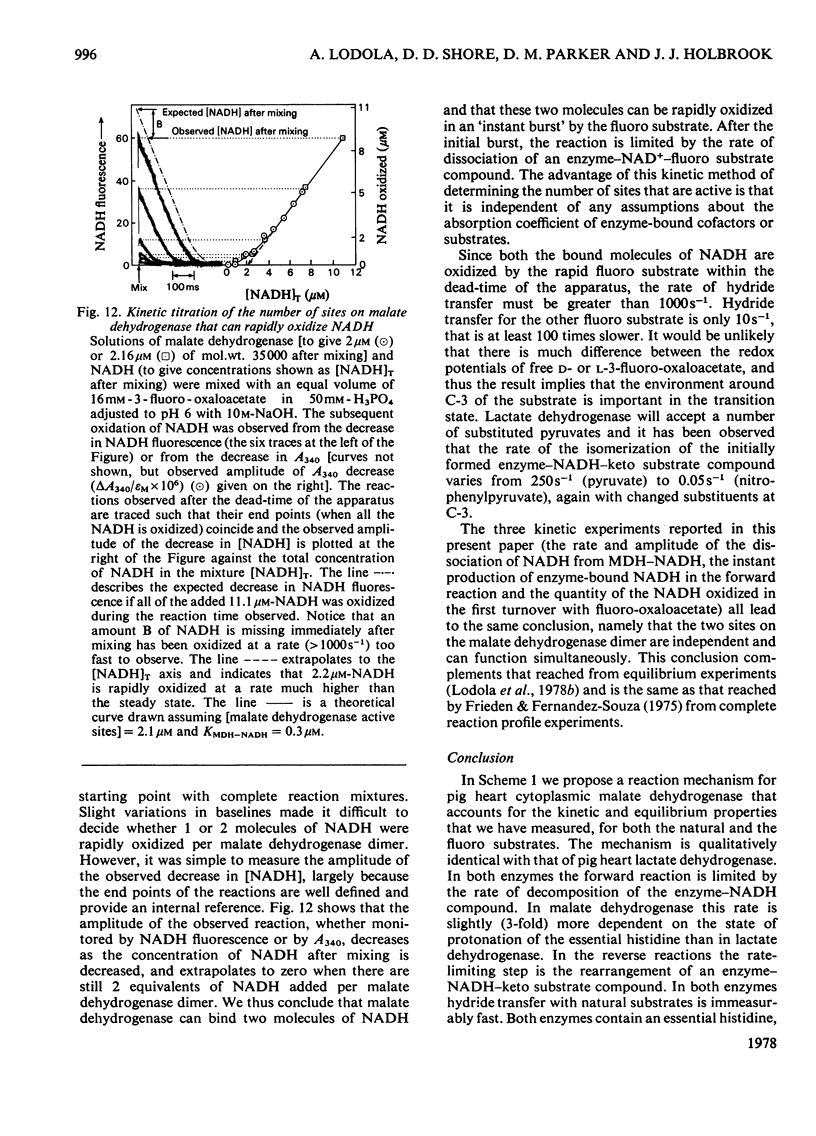
1. The mechanisms of the reduction of oxaloacetate and of 3-fluoro-oxaloacetate by NADH catalysed by cytoplasmic pig heart malate dehydrogenase (MDH) were investigated. 2. One mol of dimeric enzyme produces 1.7+/-0.4 mol of enzyme-bound NADH when mixed with saturating NAD+ and L-malate at a rate much higher than the subsequent turnover at pH 7.5. 3. Transient measurements of protein and nucleotide fluorescence show that the steady-state complex in the forward direction is MDH-NADH and in the reverse direction MDH-NADH-oxaloacetate. 4. The rate of dissociation of MDH-NADH was measured and is the same as Vmax. in the forward direction at pH 7.5. Both NADH-binding sites are kinetically equivalent. The rate of dissociation varies with pH, as does the equilibrium binding constant for NADH. 5. 3-Fluoro-oxaloacetate is composed of three forms (F1, F2 and S) of which F1 and F2 are immediately substrates for the enzyme. The third form, S, is not a substrate, but when the F forms are used up form S slowly and non-enzymically equilibrates to yield the active substrate forms. S is 2,2-dihydroxy-3-fluorosuccinate. 6. The steady-state compound during the reduction of form F1 is an enzyme form that does not contain NADH, probably MDH-NAD+-fluoromalate. The steady-state compound for form F2 is an enzyme form containing NADH, probably MDH-NADH-fluoro-oxaloacetate. 7. The rate-limiting reaction in the reduction of form F2 shows a deuterium isotope rate ratio of 4 when NADH is replaced by its deuterium analogue, and the rate-limiting reaction is concluded to be hydride transfer. 8. A novel titration was used to show that dimeric cytoplasmic malate dehydrogenase contains two sites that can rapidly reduce the F1 form of 3-fluoro-oxaloacetate. The enzyme shows 'all-of-the-sites' behaviour. 9. Partial mechanisms are proposed to explain the enzyme-catalysed transformations of the natural and the fluoro substrates. These mechanisms are similar to the mechanism of pig heart lactate dehydrogenase and this, and the structural results of others, can be explained if the two enzymes are a product of divergent evolution.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cassman M., Englard S. Beef heart malic dehydrogenases. V. A kinetic study of the reaction catalyzed by the supernatant enzyme. J Biol Chem. 1966 Feb 25;241(4):793–799. [PubMed] [Google Scholar]
- Di Franco A., Iwatsubo M. Reaction mechanism of L-glutamate dehydrogenase. Characterization of optical and kinetic properties of various enzyme-reduced-coenzyme complexes. Eur J Biochem. 1972 Nov 7;30(3):517–532. doi: 10.1111/j.1432-1033.1972.tb02123.x. [DOI] [PubMed] [Google Scholar]
- ENGLARD S., SIEGEL L., BREIGER H. H. Purification and properties of beef heart muscle "cytoplasmic" malic dehydrogenase. Biochem Biophys Res Commun. 1960 Sep;3:323–327. doi: 10.1016/0006-291x(60)90250-3. [DOI] [PubMed] [Google Scholar]
- Frieden C., Fernandez-Sousa J. Kinetic studies on pig heart cytoplasmic malate dehydrogenase. J Biol Chem. 1975 Mar 25;250(6):2106–2113. [PubMed] [Google Scholar]
- Hill E., Tsernoglou D., Webb L., Banaszak L. J. Polypeptide conformation of cytoplasmic malate dehydrogenase from an electron density map at 3.0 angstrom resolution. J Mol Biol. 1972 Dec 30;72(3):577–589. doi: 10.1016/0022-2836(72)90176-3. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Gutfreund H. Approaches to the study of enzyme mechanisms lactate dehydrogenase. FEBS Lett. 1973 Apr 15;31(2):157–169. doi: 10.1016/0014-5793(73)80095-x. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Hardman M. J., Parker D. M., Yates D. W., Gutfreund H. The putative isomerization of the lactate dehydrogenase--NADH complex. FEBS Lett. 1977;78(1):46–48. doi: 10.1016/0014-5793(77)80269-x. [DOI] [PubMed] [Google Scholar]
- Holbrook J. J., Stinson R. A. The use of ternary complexes to study ionizations and isomerizations during catalysis by lactate dehydrogenase. Biochem J. 1973 Apr;131(4):739–748. doi: 10.1042/bj1310739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook J. J., Wolfe R. G. Malate dehydrogenase. X. Fluorescence microtitration studies of D-malate, hydroxymalonate, nicotinamide dinucleotide, and dihydronicotinamide-adenine dinucleotide binding by mitochondrial and supernatant porcine heart enzymes. Biochemistry. 1972 Jun 20;11(13):2499–2502. doi: 10.1021/bi00763a018. [DOI] [PubMed] [Google Scholar]
- Kitto G. B., Lewis R. G. Purification and properties of tuna supernatant and mitochondrial malate dehydrogenases. Biochim Biophys Acta. 1967 May 16;139(1):1–15. doi: 10.1016/0005-2744(67)90107-6. [DOI] [PubMed] [Google Scholar]
- LOEWUS F. A., TCHEN T. T., VENNESLAND B. The enzymatic transfer of hydrogen. III. The reaction catalyzed by malic dehydrogenase. J Biol Chem. 1955 Feb;212(2):787–800. [PubMed] [Google Scholar]
- Lodola A., Parker D. M., Jeck R., Holbrook J. J. Malate dehydrogenase of the cytosol. Ionizations of the enzyme-reduced-coenzyme complex and a comparison with lactate dehydrogenase. Biochem J. 1978 Aug 1;173(2):597–605. doi: 10.1042/bj1730597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodola A., Spragg S. P., Holbrook J. J. Malate dehydrogenase of the cytosol. Preparation and reduced nicotinamide-adenine dinucleotide-binding studies. Biochem J. 1978 Mar 1;169(3):577–588. doi: 10.1042/bj1690577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D. M., Lodola A., Holbrook J. J. Use of the sulphite adduct of nicotinamide-adenine dinucleotide to study ionizations and the kinetics of lactate dehydrogenase and malate dehydrogenase. Biochem J. 1978 Sep 1;173(3):959–967. doi: 10.1042/bj1730959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAVAL D. N., WOLFE R. G. Malic dehydrogenase. II. Kinetic studies of the reaction mechanism. Biochemistry. 1962 Mar;1:263–269. doi: 10.1021/bi00908a012. [DOI] [PubMed] [Google Scholar]
- Rao S. T., Rossmann M. G. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241–256. doi: 10.1016/0022-2836(73)90388-4. [DOI] [PubMed] [Google Scholar]
- Silverstein E., Sulebele G. Catalytic mechanism of pig heart mitochondrial malate dehydrogenase studied by kinetics at equilibrium. Biochemistry. 1969 Jun;8(6):2543–2550. doi: 10.1021/bi00834a042. [DOI] [PubMed] [Google Scholar]
- Stinson R. A., Holbrook J. J. Equilibrium binding of nicotinamide nucleotides to lactate dehydrogenases. Biochem J. 1973 Apr;131(4):719–728. doi: 10.1042/bj1310719. [DOI] [PMC free article] [PubMed] [Google Scholar]


