Abstract
The multi-domain protein Scd2 acts as a scaffold upon which the small GTPase Cdc42 (cell division cycle 42), its nucleotide-exchange factor Scd1 and the p21-activated kinase Shk1 assemble to regulate cell polarity and the mating response in fission yeast. In the present study, we show using isothermal titration calorimetry that Scd2 binds two molecules of active GTP-bound Cdc42 simultaneously, but independently of one another. The two binding sites have significantly different affinities, 21 nM and 3 μM, suggesting that they play distinct roles in the Shk1 signalling network. Each of the Cdc42-binding sites includes one of the SH3 (Src homology 3) domains of Scd2. Our data indicate that complex formation does not occur in a conventional manner via the conserved SH3 domain ligand-binding surface. Neither of the isolated SH3 domains is sufficient to interact with the GTPase, and they both require adjacent regions to either stabilize their conformations or contribute to the formation of the Cdc42-binding surface. Furthermore, we show that there is no evidence for an intramolecular PX–SH3 domain interaction, which could interfere with SH3 domain function. This work suggests that SH3 domains might contribute directly to signalling through small GTPases and thereby adds another aspect to the diverse nature of SH3 domains as protein–protein-interaction modules.
Keywords: cell polarity, GTPase signalling, phox homology domain (PX domain), protein–protein interaction, Src homology 3 domain (SH3 domain)
Abbreviations: Cdc42, cell division cycle 42; CB, Cdc42-binding; CRIB, Cdc42 and Rac-interactive binding; DTT, dithiothreitol; GAP, GTPase-activating protein; GEF, guanine nucleotide-exchange factor; GST, glutathione S-transferase; ITC, isothermal titration calorimetry; PAK, p21-activated serine/threonine kinase; PB1, Phox and Bem1; PX, phox homology; Scd2C, C-terminal portion of Scd2; Scd2N, N-terminal portion of Scd2; SH3, Src homology 3
INTRODUCTION
Cdc42 (cell division cycle 42) is a member of the Rho family of small GTPases that regulates a wide range of cellular processes, including actin organization, gene transcription and cell-cycle progression in eukaryotic cells [1–3]. Like other members of the Ras superfamily of small GTPases, Cdc42 acts as a molecular switch, cycling between an inactive (GDP-bound), and an active (GTP-bound) state [4]. The switch is tightly controlled by regulatory proteins: GEFs (guanine nucleotide-exchange factors), which catalyse the dissociation of GDP and allow association with GTP, and GAPs (GTPase-activating proteins), which act as potent down-regulators by increasing the rate of GTP hydrolysis by several orders of magnitude [5,6]. In the active GTP-bound state, Rho-family GTPases are able to bind to a variety of downstream effectors and activate specific signalling pathways. These pathways are often not linear, but involve the formation of signalling complexes that contain regulators, GTPases and effectors, such as the Par polarity complex, the Ste5/Pbs2-containing MAPK (mitogen-activated protein kinase) pathways in yeast or the PAK (p21-activated serine/threonine kinase) signalling network [7–11]. The formation of such signalling complexes has the advantage of allowing an increased level of control by combining positive and negative regulators, thereby creating sophisticated feedback loops. Adaptor proteins are often central to the formation of such signalling complexes providing a scaffold upon which the signalling components can assemble [12].
The establishment of cell polarity and the mating response in the fission yeast Schizosaccharomyces pombe are regulated by a signalling complex that involves the scaffold protein Scd2, the GTPase Cdc42, its GEF Scd1, the effector kinase Shk1 and a number of other proteins that activate or inhibit Shk1 kinase activity [13–18]. Shk1 is a member of the family of PAKs that is highly conserved amongst eukaryotes, and contains an N-terminal regulatory domain and a C-terminal kinase domain, which interact to form an autoinhibited conformation that represses kinase activity [8,19,20]. Kinase activation follows interaction with the small GTPases Cdc42 or Rac, which bind to a CRIB (Cdc42 and Rac-interactive binding) motif within the regulatory domain [21,22]. This causes a conformational change that relieves the auto-inhibited conformation [20], enabling the kinase to phosphorylate downstream effectors to induce a specific cellular response, such as regulation of microtubule dynamics or cytoskeletal remodelling.
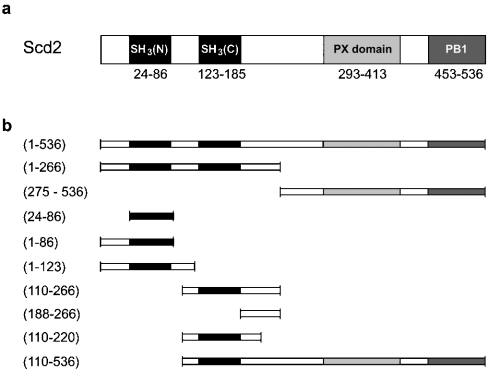
The underlying mechanisms by which the scaffold protein Scd2 contributes to the regulation of these processes are currently unknown. Although genetic analysis, co-precipitation assays and yeast two-hybrid analysis have provided an insight into the organizing/scaffolding activities of Scd2 [15,16,23], details of its interactions at the molecular level have not been examined. Scd2 is a multi-domain adaptor protein that acts as a positive regulator of Shk1 kinase activity and consists of two SH3 (Src homology 3) domains (SH3A and SH3B), a PX (phox homology) domain and a PB1 (Phox and Bem1) domain (Figure 1a). PB1 domains mediate protein–protein interactions, and structural studies have explained how this domain allows the formation of homodimers, as well as heterodimers and multimers [24,25]. Within the Shk1 signalling complex, the PB1 domain of Scd2 is believed to interact with a PB1 domain in the C-terminal portion of Scd1. SH3 domains are protein–protein-interaction modules that recognize proline-rich sequences in their target molecules that often contain a consensus PxxP (Pro-Xaa-Xaa-Pro) motif [26,27]. PX domains are membrane-targeting modules that recognize phosphoinositides with varying specificities [28–30]. In addition, some PX domains contain a consensus PxxP motif and have been proposed to bind to SH3 domains. In fact, Hiroaki et al. [31] have shown that the isolated PX domain of the NADPH oxidase component p47phox is capable of binding to its C-terminal SH3 domain with an affinity of approx. 50 μM, leading to the suggestion that an intramolecular PX–SH3 domain interaction might negatively regulate lipid binding of the PX domain. In support of this model, lipid binding to p47phox is inhibited in the context of the full-length protein, but is restored in an SH3B mutant [32,33]. In contrast, the recent crystal structure of the autoinhibited core of p47phox revealed that the conserved ligand-binding surface of SH3B is not available for binding to the PX domain because it is already engaged in an intramolecular interaction with a non-consensus binding motif in the C-terminal portion of the protein, indicating that communication between the PX and SH3 domains must happen by a different mechanism [34]. Endo et al. [23] have shown recently by GST (glutathione S-transferase) pull-down assays that Scd2 contains two binding sites for active Cdc42, but it was not investigated in their study whether these binding events happen independently or whether there is co-operation between the two sites. In addition, it was suggested in this work that an intramolecular interaction between the PX domain of Scd2 and its second SH3 domain occurs, which in this case is believed to inhibit binding of Cdc42 [23].
Figure 1. Schematic diagram illustrating the domain structure of Scd2.
(a) The positions of the N-terminal tandem SH3 domains and the C-terminal PX and PB1 domains are indicated as determined by PROSITE (http://us.expasy.org/prosite/). (b) Representation of the constructs used in the present study.
In the present study, we have used ITC (isothermal titration calorimetry) to investigate the molecular details of this novel Cdc42–effector/scaffold interaction, and to determine whether the PX domain can modulate complex formation between Scd2 and the GTPase. We have shown that Scd2 binds two molecules of Cdc42 simultaneously in a nucleotide-dependent fashion, but independently of the absence or presence of the PX domain. The two GTPase-binding sites in Scd2 have significantly different affinities, suggesting that they play distinct roles in Shk1-regulated signalling pathways. Furthermore, each of the two binding sites includes one of the SH3 domains, pointing to a novel function of SH3 domains in intracellular signalling through small GTPases.
EXPERIMENTAL
Protein expression and purification
The genes encoding Scd2 and Cdc42, and fragments, were cloned into pGEX-4T1 (Amersham Biosciences). The point mutations Scd2Trp62→Arg, Scd2Trp160→Arg and Cdc42Gln61→Leu were introduced using the QuikChange Site-Directed Mutagenesis kit from Stratagene. All DNA sequences were verified by nucleotide sequencing.
GST-fusion proteins were expressed in Escherichia coli strain BL21 (Novagen), purified on glutathione–Sepharose 4B (Amersham Biosciences) and cleaved on-column with human α-thrombin protease. Cleaved protein was purified further by gel filtration on Superdex 75, for Cdc42 and fragments of Scd2, or Superdex 200 (Amersham Biosciences), for full-length Scd2. All proteins were concentrated in VivaSpin concentrators (Viva-Science) to between 500 and 1000 μM, and were stored at −70 °C after shock freezing in liquid nitrogen. Purity and identity of the proteins was confirmed by SDS/PAGE analysis (12 or 15% gels) and electrospray MS. Protein concentrations were determined spectrophotometrically at 280 nm, using molar absorption coefficients calculated based on amino acid sequence. The molar absorption coefficient for Cdc42 was corrected for absorption of the guanine nucleotide. Figure 1(b) shows a schematic of the Scd2 constructs used in the present study.
Nucleotide analysis
The nucleotide content of purified wild-type Cdc42 (hereafter referred to as Cdc42.GDP) and the constitutively active Cdc42 mutant Cdc42Gln61→Leu (hereafter referred to as Cdc42.GTP) was analysed by reverse-phase HPLC [35]. Samples (1 nmol) of Cdc42 were made up to 44 μl in 50 mM Hepes, pH 7.0, 50 mM NaCl and 2 mM DTT (dithiothreitol), and the protein was denatured by the addition of 2.5 μl of 10% perchloric acid, followed by the addition of 3.5 μl of 2 M sodium acetate, pH 4.0. Precipitated protein was removed by centrifugation at 16000 g in a benchtop centrifuge for 2 min, and the supernatant was added to an equal volume of 100 mM KH2PO4/K2HPO4, pH 6.5, and 10 mM tetrabutylammonium bromide. Samples were analysed by reverse-phase HPLC on a Zorbax SB-C18 column (4.6 mm×250 mm; Hichrom), run isocratically in 100 mM KH2PO4/K2HPO4, pH 6.5, 10 mM tetrabutylammonium bromide and 8.5% acetonitrile at a flow rate of 1 ml/min, and the nucleotide absorbance was measured at 254 nm. Nucleotides present were compared with standards of GDP and GTP run separately.
ITC measurements
Complex formation between Scd2 and Cdc42 was measured by ITC using a MicroCal VP-ITC microcalorimeter (MicroCal, Northampton, MA, U.S.A.) [36]. A NAP 5 column was used to exchange all proteins into ITC buffer (25 mM Hepes, pH 7.5, 2 mM MgCl2, 50 mM NaCl and 2 mM DTT). Experiments were performed at 18 °C. In general, the sample cell contained Cdc42.GDP or Cdc42.GTP at concentrations in the range 25–50 μM, and constructs of Scd2 at 300–500 μM were injected in 10 μl samples from a total of 290 μl. Heats of dilution, determined by titrating Scd2 into buffer alone, were subtracted from the raw titration data before analysis. Data were fitted by least-squares procedures using the evaluation software, Microcal Origin version 7.0 provided by the manufacturer. All titrations using full-length Scd2 or constructs containing two Cdc42-binding sites were fitted using the ‘ligand in the cell’ assumption. All measurements were repeated two to five times.
Surface plasmon resonance
The interaction between Scd2-(110–220) and Cdc42.GTP or Cdc42.GDP and Scd2-(275–536) was analysed using a BIAcore 2000 instrument (BIAcore). Scd2-(110–220) was prepared in coupling buffer (25 mM Hepes, pH 6.8, 50 mM NaCl and 2 mM MgCl2) and immobilized on a CM5 sensor chip (BIAcore), with EDC [N-ethyl-N′-(dimethylaminopropyl)carbodi-imide] and NHS (N-hydroxysuccinimide). A flow cell without immobilized protein was used as a control. All other proteins were diluted to the appropriate concentrations using running buffer [10 mM Hepes, pH 7.4, 150 mM NaCl and 0.005% (v/v) P20 surfactant]. Each protein was injected on to the channels at a flow rate of 5 μl/min for 10 min, followed by a wash with running buffer for 150 s. For analysis, the reference flow cell sensorgram was subtracted from the corresponding sensorgrams to abolish baseline drift, bulk and non-specific interaction contributions.
Gel-filtration HPLC
Complex formation between Scd2-(1–536) and Cdc42 was monitored by analytical gel filtration on a Superdex 200 column. Scd2 (20 μM) was mixed with various concentrations of Cdc42.GDP or Cdc42.GTP (20 μM or 40 μM) in a total volume of 250 μl. A sample (100 μl) was loaded on to the column, which had been equilibrated in 50 mM Tris/HCl, pH 7.9, 100 mM NaCl and 2 mM MgCl2. The column was run at 0.4 ml/min, and 0.5 ml fractions were collected for analysis by SDS/PAGE (15% gels). Complex formation between Scd2-(1–266), Scd2-(275–536) or Scd2-(110–536), and Ccd42.GDP or Cdc42.GTP was analysed in a similar fashion, except that each protein was used at 100 μM.
RESULTS
Scd2 interacts with two molecules of Cdc42 simultaneously
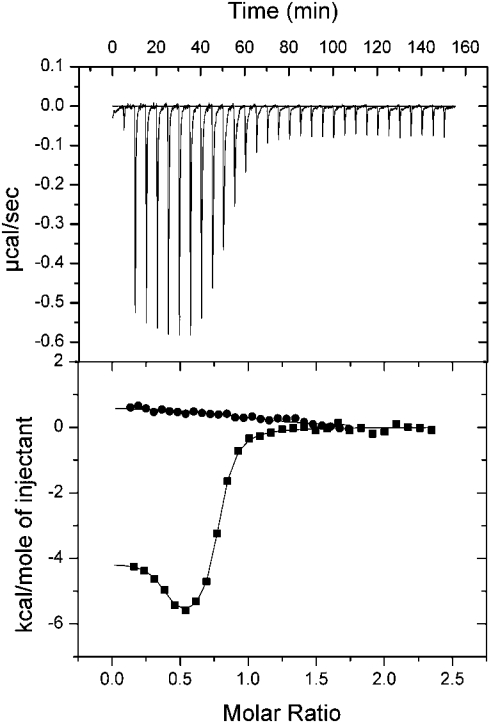
ITC allows the direct measurement of the equilibrium binding constant Ka (Kd=1/Ka), the enthalpy of complex formation (ΔH) and complex stoichiometry of a protein–protein interaction without the need for modification of the proteins under investigation. Titration of full-length Scd2 with active GTP-bound Cdc42 resulted in a binding isotherm, which could not be fitted to a 1:1 binding model. Fitting of the data to a two-site binding model resulted in a good fit with a stoichiometry of 1:1 for each site, indicating that two molecules of Cdc42 can interact simultaneously with Scd2 (Figure 2). The two Cdc42-binding sites in Scd2 are significantly different and include a high-affinity site, Kd1=21 nM, that is exothermic, and a lower-affinity site, Kd2=3.0 μM, binding to which is entirely entropy driven (Table 1). To examine whether both binding events occurred in a nucleotide-dependent manner, we titrated Cdc42.GDP with Scd2. No significant heat change was observed, suggesting that the proteins either do not or only so weakly interact that it was undetectable under the experimental conditions or that ΔH is zero at the experimental temperature (Figure 2).
Figure 2. Characterization of the interaction between Scd2 and Cdc42.GTP or Cdc42.GDP by ITC.
Full-length Scd2 was titrated into Cdc42 in 25 mM Hepes, pH 7.5, 50 mM NaCl, 2 mM MgCl2 and 2 mM DTT at 18 °C. The upper plot shows the raw calorimetric data for the Cdc42.GTP–Scd2 interaction. The lower plot shows the isotherm of binding after subtraction of the heat of dilution. The data for Cdc42.GTP binding to Scd2 have been fitted to a two-site binding model. Scd2 titration into Cdc42.GTP (■); Scd2 titrated into Cdc42.GDP (●).
Table 1. ITC measurements for complex formation between Scd2 constructs and Cdc42.GTP or Cdc42.GDP.
All measurements were performed using 25 mM Hepes, pH 7.5, 50 mM NaCl, 2 mM MgCl2 and 2 mM DTT at 18 °C. Scd2 constructs were used at concentrations in the range 300–500 μM, and were titrated into Cdc42.GTP, Cdc42.GDP or Cdc42.GTPΔ14 at concentrations between 30 and 50 μM. Kd is given in units of 10−6 M, ΔH and TΔS are given in kcal/mol (1 kcal/mol≡4.184 kJ/mol). The stoichiometry of complex formation for each binding site is N=1.0±0.1 for titrations with Cdc42.GDP and N=1.0±0.25 for titrations with Cdc42.GTP (Cdc42Gln61→Leu generally contained 75–90% GTP, as determined by reverse-phase HPLC).
| Kd1 | ΔH1 | TΔS1 | Kd2 | ΔH2 | TΔS2 | |
|---|---|---|---|---|---|---|
| Titrated into Cdc42.GTP | ||||||
| Scd2-(1–536) | 0.021±0.009 | −3.99±0.06 | 6.30±0.34 | 3.01±1.11 | 0.84±0.15 | 8.23±0.12 |
| Scd2-(1–266) | 0.028±0.003 | −3.87±0.02 | 6.20±0.09 | 2.71±0.41 | 0.83±0.04 | 8.25±0.04 |
| Scd2-(275–536) | No binding | |||||
| Scd2-(1–86) | 3.00±1.01 | −2.35±0.23 | 5.03±0.03 | |||
| Scd2-(110–266) | 2.93±0.40 | 1.91±0.01 | 9.28±0.09 | |||
| Scd2-(110–536) | 4.06±0.57 | 2.67±0.54 | 9.85±0.63 | |||
| Titrated into Cdc42.GDP | ||||||
| Scd2-(1–536) | No binding | |||||
| Scd2-(1–266) | No binding | |||||
| Titrated into Cdc42.GTPΔ14 | ||||||
| Scd2-(1–536) | 0.021±0.01 | −5.24±0.04 | 4.97±0.15 | 6.21±0.32 | 1.04±0.19 | 7.97±0.3 |
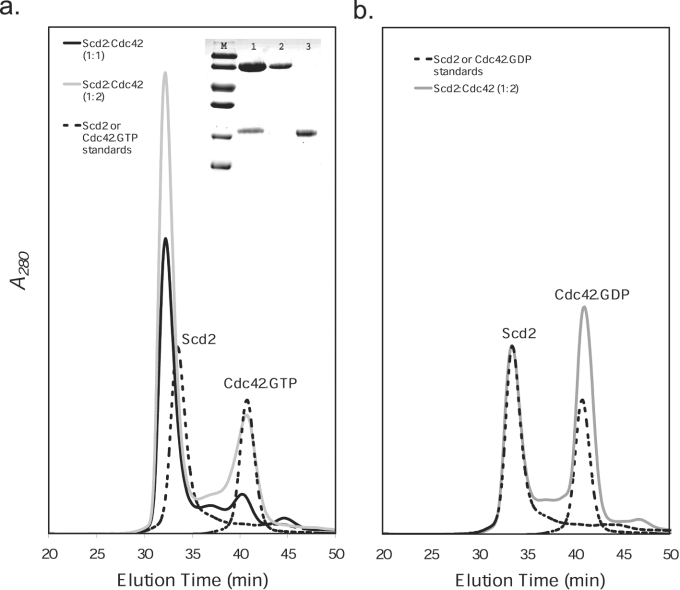
To investigate complex formation further, we carried out analytical gel-filtration analysis. Mixtures of Cdc42.GTP or Cdc42.GDP with Scd2 at different stoichiometries were prepared, and their elution profiles were compared with those of the individual proteins, which elute as single symmetrical peaks from an S200 column (Figures 3a and 3b). Under the same experimental conditions, mixtures at molar ratios of 1:1 and 2:1 Cdc42.GTP/Scd2 were applied to the column. Figure 3(a) shows that a 1:1 mixture elutes as a single peak with a shorter retention time than the isolated proteins, indicating the formation of a complex between the two proteins. SDS/PAGE analysis (15% gels) of the peak fraction confirmed this by showing that both Cdc42 and Scd2 are present. Furthermore, when comparing the elution profiles of the 1:1 and 2:1 Cdc42.GTP/Scd2 mixtures (Figure 3a), the peak relating to the complex is seen to increase with addition of Cdc42.GTP, indicating that Scd2 is not saturated at 1:1 stoichiometry. These experiments were repeated with mixtures of Cdc42.GDP and Scd2 at stoichiometries of 1:1 (results not shown) and 2:1 (Figure 3b). In each case, the mixtures elute as two peaks, each peak having retention times identical with the separate protein standards without formation of a higher-molecular-mass complex.
Figure 3. Analytical gel-filtration analysis of complex formation between Scd2 and Cdc42.
Mixtures of Cdc42 and Scd2 were run on Superdex 200 in a buffer of 50 mM Tris/HCl, pH 7.9, 100 mM NaCl and 2 mM MgCl2 at a flow rate of 0.4 ml/min. (a) Analysis of mixtures of Cdc42.GTP and Scd2 at 20 μM. Scd2–Cdc42.GTP (1:1) (solid black line), Scd2–Cdc42.GTP (1:2) (solid grey line), Scd2 and Cdc42 run individually (broken line). The Cdc42 used in the present study was 75–90% GTP-bound as determined by reverse-phase HPLC. Inset, SDS/PAGE (15% gel) analysis of protein fractions. Lane M, molecular-mass markers (97.0, 66.0, 45.0, 30.0, 20.1 and 14.4 kDa); lane 1, fraction at 31.5 min, containing Scd2 and Cdc42; lane 2, Scd2; lane 3, Cdc42. (b) Analysis of mixtures of Cdc42.GDP and Scd2 at 20 μM. Scd2–Cdc42.GDP (1:2) (solid grey line), Scd2 and Cdc42 run individually (broken line).
The binding sites for Cdc42 are located in the N-terminal tandem SH3-domain-containing region of Scd2
Scd2 contains no CRIB motif or sequence homology with previously identified Cdc42-binding motifs. Using GST pull-down assays, Endo et al. [23] identified two regions in the N-terminal portion of Scd2 that interact with Cdc42.GTP, which were termed CB1 (Cdc42-binding 1; amino acids 1–86) and CB2 (Cdc42-binding 2; amino acids 110–266) regions respectively. Each of these regions contains one of the SH3 domains. To test whether each of these regions is sufficient for high-affinity complex formation and to investigate whether the C-terminal portion of Scd2 might contribute to complex formation or possibly reduce GTPase binding due to an intramolecular PX–SH3 domain interaction, we measured binding of Cdc42 to the N-terminal portion of Scd2 (Scd2N; amino acids 1–266), containing the two SH3 domains, and to the C-terminal portion (Scd2C; amino acids 275–536), containing the PX and PB1 domains. Titration of Scd2N into Cdc42.GTP resulted in a binding isotherm that was almost indistinguishable from that of the full-length protein, and accordingly could only be fitted to a two-site binding model (Table 1). The dissociation constants (Kd1=28 nM and Kd2=2.7 μM) and binding enthalpies (ΔH1=−3.87 kcal/mol and ΔH2=0.83 kcal/mol; 1 kcal/mol≡4.184 kJ/mol) of each interaction were in good agreement with those determined for the full-length protein, indicating that Scd2N is sufficient to confer high-affinity binding to Cdc42.GTP. As for the full-length protein, there was no detectable interaction with Cdc42.GDP, confirming the nucleotide-dependence of both binding events. Furthermore, no interaction was detected between Scd2C and Cdc42.GTP or Scd2.GDP (results not shown). Taken together, these data clearly indicate that the N-terminal half of Scd2 contains both binding sites for Cdc42, and that interaction with the GTPase is neither positively nor negatively influenced by the remainder of Scd2.
Identification of minimal Cdc42-binding domains
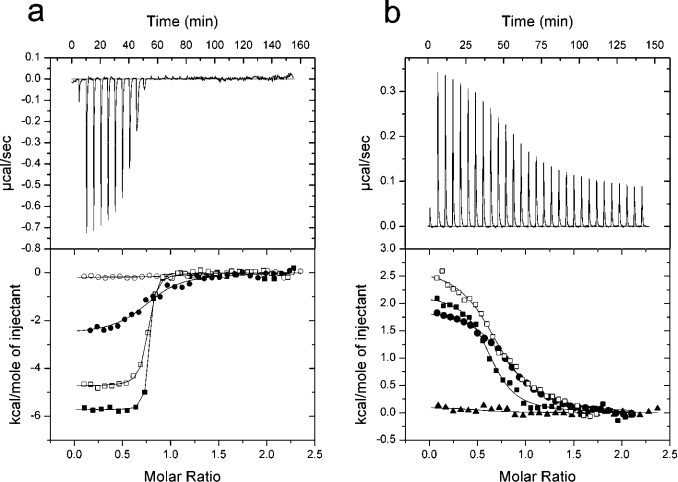
Titration of Cdc42.GTP with Scd2 fragments corresponding to CB1 and CB2 confirmed that each region binds one molecule of Cdc42 in a nucleotide-dependent fashion (Tables 1–3, and Figures 4a and 4b). Binding of Cdc42.GTP to CB2, which contains SH3B, was endothermic, with an affinity of 2.93 μM, suggesting that it represents the low-affinity binding site identified in full-length Scd2. In contrast, binding of CB1 to Cdc42.GTP was almost 150-fold weaker than the high-affinity binding site within full-length Scd2 or Scd2N (Kd=3.0 μM, ΔH=−2.35 kcal/mol), indicating either that CB1 may be lacking part of the Cdc42-interaction surface or alternatively that both sites act synergistically to bind Cdc42.
Figure 4. ITC describing the two Cdc42-binding regions in Scd2.
Each Scd2 construct was titrated into Cdc42 in 25 mM Hepes, pH 7.5, 50 mM NaCl, 2 mM MgCl2 and 2 mM DTT at 18 °C. The upper plot shows the raw calorimetric data. The lower plot represents the binding isotherm after subtraction of the heat of dilution. Data for each binding event is fitted to a one-site binding model. (a) Titration of Scd2-(1–86) into Cdc42.GTP (●), titration of Scd2-(1–86)Trp62→Arg into Cdc42.GTP (○), titration of Scd2-(1–123) into Cdc42.GTP (■), and Scd2-(1–123)Trp62→Arg into Cdc42.GTP (□). (b) Titration of Scd2-(188–266) into Cdc42.GTP (▲), titration of Scd2-(110–220) into Cdc42.GTP (■), titration of Scd2-(110–220)Trp160→Arg into Cdc42.GTP (□) and Scd2-(110–266) into Cdc42.GTP (●).
To characterize further the high-affinity binding site in Scd2, and, in particular, to address the role of SH3A in complex formation, we studied the binding of a construct that contains only the predicted SH3 domain (amino acids 24–86) to Cdc42.GTP (Table 2). No binding was detected, indicating that residues 1–23 are important for complex formation with Cdc42.GTP, either because they form part of the Cdc42-binding site or because they contribute to the stabilization of SH3A. Furthermore, we extended CB1 to include residues 87–123 that connect the two SH3 domains to test whether this region may contribute to complex formation with Cdc42.GTP. Indeed, titration of Cdc42.GTP with Scd2-(1–123) produced a binding isotherm that could be fitted to a one-site binding model, with a Kd of 17 nM and a ΔH of −5.72 kcal/mol (Figure 4a). These values are comparable with those of the high-affinity binding site of the full-length Scd2–Cdc42.GTP complex, indicating that there is no synergy between CB1 and CB2, but that the region around 87–123 contributes significantly to complex formation. Taken together, these results suggest that the regions N- and C-terminal to SH3A are both important to form a high-affinity binding surface for Cdc42. SH3 domains contain a number of highly conserved amino acids that are vital for the interaction with a consensus PxxP motif. One of these is a tryptophan residue that stacks against a proline residue and mutation of which generally disrupts binding to proline-rich ligands. Titration of the corresponding Scd2-(1–123)Trp62→Arg with Cdc42.GTP shows that such a point mutation in SH3A has only a weak effect, and reduces the affinity of this interaction 4-fold when compared with wild-type Scd2-(1–123) (Table 2 and Figure 4a). In contrast, the same mutation within the Scd2-(1–86) construct completely abolished complex formation with Cdc42.GTP.
Table 2. ITC measurements to delineate the first Cdc42.GTP-binding region of Scd2.
Experimental conditions and units are as in Table 1.
| Kd | ΔH | TΔS | |
|---|---|---|---|
| Scd2-(1–86) | 3.00±1.01 | −2.35±0.23 | 5.03±0.03 |
| Scd2-(1–86) Trp62→Arg | No binding | ||
| Scd2-(24–86) | No binding | ||
| Scd2-(1–123) | 0.017±0.002 | −5.72±0.01 | 4.65±0.07 |
| Scd2-(1–123) Trp62→Arg | 0.074±0.002 | −4.88±0.13 | 4.63±0.15 |
Using a similar strategy, we analysed complex formation between Cdc42.GTP and the CB2-binding region of Scd2 that contains the low-affinity GTPase-binding site. In this case, the isolated SH3B domain, Scd2-(123–188), was insoluble, and we were not able to test whether it was sufficient for complex formation with Cdc42.GTP. The smallest construct that contained SH3B and was still soluble was Scd2-(110–220). As shown in Figure 4(b) and Table 3, this construct bound Cdc42.GTP with a similar affinity and binding energetics as did Scd2-(110–266) (ΔH=2.55 kcal/mol, Kd=1.71 μM), indicating that residues 221–266 of CB2 are not required for complex formation with Cdc42.GTP. To characterize this interaction further, we mutated the conserved tryptophan residue in the ligand-binding site of SH3B. This mutation, Trp160→Arg, did not weaken the interaction (Figure 4b), suggesting either that the SH3 domain is not required for complex formation or at least that this interaction does not occur via the conserved proline residue binding surface. On the other hand, a construct containing residues Scd2-(186–266) that lacks SH3B was not sufficient to allow complex formation with Cdc42.GTP either, indicating that at least some features of the SH3 domain are required to form a binding surface for Cdc42.
Table 3. ITC measurements to delineate the second Cdc42.GTP-binding region of Scd2.
Experimental conditions and units are as in Table 1.
| Kd | ΔH | TΔS | |
|---|---|---|---|
| Scd2-(110–266) | 2.93±0.40 | 1.91±0.01 | 9.28±0.09 |
| Scd2-(188–266) | No binding | ||
| Scd2-(110–220) | 1.71±0.15 | 2.55±0.38 | 10.14±0.42 |
| Scd2-(110–220) Trp160→Arg | 2.52±0.85 | 3.07±0.37 | 10.55±0.16 |
The reduced binding of Scd2-(1–123)Trp62→Arg to Cdc42.GTP suggested that the conserved ligand-binding surface of SH3A may be at least partially involved in the interaction with Cdc42.GTP. Cdc42 is one of a handful of GTPases that contain a PxxP motif at their C-termini. The placental form of human Cdc42, R-Ras and TC21 all contain PxxP motifs, although only the PxxP motif of R-Ras has been previously reported to bind to a SH3 domain. However, this interaction between R-Ras and the scaffold protein Nck was not nucleotide-specific [37]. In order to assess whether the C-terminal PxxP motif was important for binding of Cdc42.GTP to Scd2, we produced a Cdc42 mutant lacking this region (Cdc42.Gln61→Leu.Δ14) and investigated its interaction with full-length Scd2 by ITC. The resulting binding isotherm was very similar to that produced by full-length Cdc42.GTP, and indicated formation of a Cdc42–Scd2 complex with a 2:1 stoichiometry (Table 1).
The PX and SH3 domains of Scd2 do not interact
In a previous report, it was suggested that an intramolecular interaction between the PX and SH3B domains of Scd2 kept Scd2 in an autoinhibited conformation, thereby preventing binding of Cdc42 to the CB2 region [23]. This model was based on a GST pull-down study, showing that Scd2-(110–536), which contains SH3B, the PX and PB1 domains, was not able to bind Ccd42.GTP. However, our binding studies show unambiguously that two molecules of Cdc42.GTP bind to full-length Scd2, arguing against an intramolecular autoinhibitory interaction. This observation is corroborated further by the titration of Cdc42.GTP with Scd2-(110–536), which produced a similar ITC profile to that of Scd2-(110–266) (results not shown). The interaction is endothermic with a ΔH of 2.67 kcal/mol and a Kd of 4.06 μM (Table 1). Furthermore, analytical gel-filtration experiments showed that a 1:1 molar ratio of Scd2-(110–536) and Cdc42.GTP eluted as a single peak with a shorter retention time than either of the individual proteins. The interaction was nucleotide-specific, since no complex was formed between Scd2-(110–536) and Cdc42.GDP under identical conditions (results not shown).
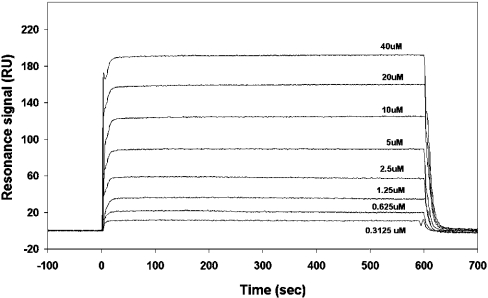
To probe this discrepancy with previous results, we investigated whether Scd2N containing the tandem SH3 domains was able to interact with Scd2C containing the PX and PB1 domains in an intermolecular fashion. Titration of Scd2C into Scd2N showed no heat change, nor did analytical gel-filtration experiments of the two proteins at high concentrations (100 μM) provide any evidence for complex formation (results not shown). To support these findings, we analysed binding of Scd2-(110–220), containing SH3B, to Scd2-(275–536), containing the PX domain, by surface plasmon resonance. Scd2-(110–220) was immobilized on a CM5 sensor chip, and in order to validate the immobilization procedure, Cdc42.GTP and Cdc42.GDP were applied to the chip as positive and negative controls respectively. Binding was determined using equilibrium analysis. A serial dilution of Cdc42.GTP (or Cdc42.GDP) was injected across the surface of the chip at a flow rate of 5 μl/min for 10 min at 25 °C. Analysis was performed on sensorgrams generated by dilutions ranging from 0.3125 μM to 40 μM (Figure 5) and 0.3125 μM to 100 μM Cdc42.GDP. The Cdc42.GTP data were fitted to a 1:1 binding model, and yielded a Kd of 7.07±0.08 μM. As expected, Cdc42.GDP showed no increase in resonance units when injected over the chip surface, indicating that no binding to Scd2-(110–220) takes place. Next, Scd2-(275–536) containing the PX domain was passed over immobilized Scd2-(110–220) at concentrations between 6.25 and 800 μM. No change in resonance units was detected in this experiment, even at very high protein concentrations, supporting further the notion that there is no intramolecular SH3B–PX domain interaction.
Figure 5. Sensorgram depicting the binding of Cdc42.GTP to Scd2-(110–220).
Various concentrations of Cdc42.GTP in 10 mM Hepes, pH 7.4, 150 mM NaCl and 0.005% P20 surfactant were passed over immobilized Scd2-(110–220) at a flow rate of 5 μl/min, and binding was measured by surface plasmon resonance.
DISCUSSION
The adaptor protein Scd2 is an important component of the PAK Shk1-dependent signalling network, regulating the establishment of cell polarity and the mating response in fission yeast [15,16,38]. It acts as a protein scaffold, upon which the GTPase Cdc42, its GEF Scd1, the downstream effector kinase Shk1 and a number of other proteins assemble [15,16,23]. We have conducted a thermodynamic study of the interaction between Scd2 and Cdc42, and have shown that the adapter protein Scd2 binds two molecules of Cdc42 simultaneously, but independently, in a nucleotide-specific manner. This supports and significantly extends previous work, which identified an interaction between Cdc42 and Scd2 using yeast two-hybrid analysis [16] and in vitro co-precipitation assays [23], and confirms that Scd2 is a bona fide effector of Cdc42, acting downstream of the GTPase in the signalling pathway.
ITC studies using Cdc42 and various Scd2 deletions located both binding sites in the N-terminal half of Scd2, as suggested previously [23]. Importantly, our experiments establish that the two sites are non-equivalent, and that the N-terminal site binds Cdc42.GTP with high affinity (21 nM) with a favourable enthalpic contribution, while the C-terminal site is over 140-fold weaker, and the interaction is entirely entropy driven. Each of the two binding sites includes one of the SH3 domains, pointing to a novel function of SH3 domains as downstream effectors of small GTPases. The isolated SH3A domain is not capable of forming a complex with Cdc42.GTP and regions N- as well as C-terminal to the SH3 domain are required to allow the formation of a high-affinity binding site. This is in contrast with previous results, which suggested that Scd2-(1–86) is sufficient for complex formation, but can be rationalized by the non-quantitative nature of that study, which might have underestimated the importance of additional sequences [23].
Interestingly, mutation of a conserved tryptophan residue in the conserved ligand-binding surface of SH3A led to a complete loss of binding in the context of the Scd2-(1–86) construct, while the same mutation within Scd2-(1–123) only weakens the affinity for Cdc42.GTP 4-fold. This suggests that the conserved ligand-binding pocket of SH3A contributes only marginally to complex formation with Cdc42, while the regions N-terminal (amino acids 1–23) and C-terminal (amino acids 87–123) to the SH3 domain seem to be vital for the full biological interaction. Given the proximity of the N- and C-termini of SH3 domains, one may speculate that they interact to form the Cdc42 ligand-binding surface, which might partially overlap with the binding surface for canonical PxxP-containing ligands. Although this would constitute a novel interaction between a GTPase and an SH3 domain, the importance of residues flanking SH3 domains has been highlighted previously in the interaction of the SH3 domain of PLCγ1 (phosphoinositide-specific phospholipase Cγ1) with the adapter protein Cbl [39]. Here, the interaction between the core SH3 domain and the Cbl ligand is rather weak, and inclusion of amino acids flanking the core SH3 domain substantially increases the affinity of this interaction. Just as the high-affinity Cdc42-binding site in Scd2 is formed, in part, by SH3A, formation of the low-affinity binding site requires the presence of SH3 domain B. Due to the low solubility of SH3B, it was not possible to assess the interaction of the isolated domain with Cdc42 directly. The smallest soluble fragment that includes SH3B was Scd2-(110–220), which contains an extra 35 residues C-terminal to SH3B. This fragment bound Cdc42 with essentially the same affinity as the full-length protein. This is in contrast with construct Scd2-(188–266) that contains only the region C-terminal to SH3B and which is not able to interact with Cdc42, highlighting the importance of SH3B. However, the observation that Scd2-(110–220)Trp160→Arg bound Cdc42 with the same affinity as the wild-type protein suggests that complex formation does not occur via the conserved PxxP-motif-binding surface.
SH3 domains are ubiquitous protein–protein-interaction modules that are approx. 60 amino acids in length and share a conserved β-barrel structure. Variations upon the canonical PxxP-containing ligand-binding mode are known, and a number of SH3 domain ligands have been identified that do not contain a consensus PxxP motif, but still bind SH3 domains via their conserved ligand-binding surfaces [40–42]. Furthermore, recent studies by a number of groups have revealed that SH3 domains can use other surfaces, distinct from those that recognize proline-rich ligands, to bind target proteins [43–45]. These interactions may take place simultaneously to those with proline-rich ligands, thereby extending the repertoire of SH3 domains as binary protein-interaction modules to mediate the formation of ternary complexes. An example for such a novel mode of SH3-domain-mediated interaction is peroxisomal Pex13 that interacts with a consensus PxxP motif in Pex14, while simultaneously binding to Pex5 via a concave surface composed of β-strands, β1 and β2 [45]. Given that Scd2 SH3B is known to bind to a PxxP motif in the N-terminal portion of Shk1 via its consensus ligand-binding surface [15,23], it is tempting to speculate that the SH3B domain of Scd2 might be another example of a bifunctional SH3 domain that interacts with two different proteins simultaneously. Further studies are now required to establish whether this is the case or whether both binding events are mutually exclusive.
PX domains have received a lot of attention recently due to the possibility that they may act as bifunctional protein-interaction modules that couple phospholipid binding to protein–protein interactions through intramolecular PX–SH3 domain interactions. Such an intramolecular autoinhibitory interaction that is believed to interfere with Cdc42 binding has been proposed for Scd2 as well as its Saccharomyces cerevisiae homologue Bem1 [23,46]. However, the data of the present study do not support this model, but clearly show, at least in the case of Scd2, that the presence of the PX domain does not influence binding to Cdc42, as illustrated by the nearly identical affinities of Cdc42 for full-length Scd2 and an Scd2 fragment in which the PX domain has been deleted. Furthermore, we have found no evidence for an interaction between the SH3-domain-containing N-terminal and PX-domain-containing C-terminal portion of Scd2 using a variety of techniques. These data imply that care has to be taken in the interpretation of binding studies involving PX- and SH3-domain-containing proteins, and highlight the requirement for further structural and biochemical studies in order to understand the molecular details of a potential interplay between PX and SH3 domains.
Combining our findings with genetic data from Schiz. pombe and the homologous pathway in S. cerevisiae, we propose a model of how Scd2 may play a central role in providing a scaffold to co-localize the essential cell polarity GTPase, Cdc42, its activator Scd1 and downstream effector, Shk1. In budding yeast, the scaffold protein Bem1 is localized to the site of polarization by activated Cdc42 [47]. We suggest that the N-terminal high-affinity binding site provides a region with which Scd2 anchors to the cell membrane via an interaction with Cdc42.GTP. Localization of Scd2 could then enable further recruitment and, more importantly, maintenance of active Scd1 at this site, possibly by preventing the reassociation of an autoinhibited conformation of the GEF, as suggested for Cdc24 in S. cerevisiae [47–49]. This may produce a self-propagating mechanism whereby activation of more Cdc42.GTP and further recruitment of Scd2 magnifies the initial signal at the plasma membrane [46]. Increased local concen-trations of Cdc42.GTP could then allow binding to the low-affinity site of Scd2. Assuming that this interaction could occur simultaneously with that of Shk1 and SH3B, this may induce co-localization of Cdc42 and Shk1, thereby promoting activation of the kinase, which in turn could allow the polarized assembly of the cytoskeleton.
The present paper adds to the growing literature that highlights the diverse nature of SH3 domains. Originally identified as protein modules that utilize a conserved hydrophobic pocket to bind to PxxP motifs, their protein-recognition repertoire has been extended to include atypical motifs in target ligands or utilize novel surfaces on the SH3 domain for ligand interactions. Our data suggest that there may be a novel function of SH3 domains, in which they act as downstream effectors for small GTPases. Structural studies are now required to unravel the molecular details of these interactions and enable us to predict similar interactions in other systems.
Acknowledgments
The genes encoding Scd2 and Cdc42 were gifts from Stevan Marcus (University of Texas) and John Armstrong (University of Sussex) respectively. We are grateful to Steve Howell and Lesley Haire (NIMR) for mass spectrometry, Susan Smith for help with protein purification and Yvonne Groemping for helpful discussions.
References
- 1.Johnson D. I. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature (London) 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 3.Etienne-Manneville S. Cdc42 – the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 4.Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature (London) 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 5.Sprang S. R. G protein mechanisms: insights from structural analysis. Annu. Rev. Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 6.Vetter I. R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 7.Elion E. A. The Ste5p scaffold. J. Cell Sci. 2001;114:3967–3978. doi: 10.1242/jcs.114.22.3967. [DOI] [PubMed] [Google Scholar]
- 8.Bokoch G. M. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 9.Pawson T., Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 10.Macara I. G. Parsing the polarity code. Nat. Rev. Mol. Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- 11.Henrique D., Schweisguth F. Cell polarity: the ups and downs of the Par6/aPKC complex. Curr. Opin. Genet. Dev. 2003;13:341–350. doi: 10.1016/s0959-437x(03)00077-7. [DOI] [PubMed] [Google Scholar]
- 12.Pawson T., Scott J. D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 13.Miller P. J., Johnson D. I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbreth M., Yang P., Wang D., Frost J., Polverino A., Cobb M. H., Marcus S. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc. Natl. Acad. Sci. U.S.A. 1996;93:13802–13807. doi: 10.1073/pnas.93.24.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang E., Bartholomeusz G., Pimental R., Chen J., Lai H., Wang L., Yang P., Marcus S. Direct binding and in vivo regulation of the fission yeast p21-activated kinase shk1 by the SH3 domain protein scd2. Mol. Cell. Biol. 1999;19:8066–8074. doi: 10.1128/mcb.19.12.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang E. C., Barr M., Wang Y., Jung V., Xu H. P., Wigler M. H. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 17.Yang P., Pimental R., Lai H., Marcus S. Direct activation of the fission yeast PAK Shk1 by the novel SH3 domain protein, Skb5. J. Biol. Chem. 1999;274:36052–36057. doi: 10.1074/jbc.274.51.36052. [DOI] [PubMed] [Google Scholar]
- 18.Kim H. W., Yang P., Qyang Y., Lai H., Du H., Henkel J. S., Kumar K., Bao S., Liu M., Marcus S. Genetic and molecular characterization of Skb15, a highly conserved inhibitor of the fission yeast PAK, Shk1. Mol. Cell. 2001;7:1095–1101. doi: 10.1016/s1097-2765(01)00248-9. [DOI] [PubMed] [Google Scholar]
- 19.Tu H., Wigler M. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol. Cell. Biol. 1999;19:602–611. doi: 10.1128/mcb.19.1.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell. 2000;102:387–397. doi: 10.1016/s0092-8674(00)00043-x. [DOI] [PubMed] [Google Scholar]
- 21.Burbelo P. D., Drechsel D., Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J. Biol. Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 22.Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., Lowe P. N., Laue E. D. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- 23.Endo M., Shirouzu M., Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- 24.Ito T., Matsui Y., Ago T., Ota K., Sumimoto H. Novel modular domain PB1 recognizes PC motif to mediate functional protein–protein interactions. EMBO J. 2001;20:3938–3946. doi: 10.1093/emboj/20.15.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson M. I., Gill D. J., Perisic O., Quinn M. T., Williams R. L. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell. 2003;12:39–50. doi: 10.1016/s1097-2765(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 26.Mayer B. J. SH3 domains: complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 27.Musacchio A. How SH3 domains recognize proline. Adv. Protein Chem. 2002;61:211–268. doi: 10.1016/s0065-3233(02)61006-x. [DOI] [PubMed] [Google Scholar]
- 28.Ellson C. D., Gobert-Gosse S., Anderson K. E., Davidson K., Erdjument-Bromage H., Tempst P., Thuring J. W., Cooper M. A., Lim Z. Y., Holmes A. B., et al. PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat. Cell Biol. 2001;3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Hortsman H., Seet L., Wong S. H., Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- 30.Kanai F., Liu H., Field S. J., Akbary H., Matsuo T., Brown G. E., Cantley L. C., Yaffe M. B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 31.Hiroaki H., Ago T., Ito T., Sumimoto H., Kohda D. Solution structure of the PX domain, a target of the SH3 domain. Nat. Struct. Biol. 2001;8:526–530. doi: 10.1038/88591. [DOI] [PubMed] [Google Scholar]
- 32.Karathanassis D., Stahelin R. V., Bravo J., Perisic O., Pacold C. M., Cho W., Williams R. L. Binding of the PX domain of p47phox to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J. 2002;21:5057–5068. doi: 10.1093/emboj/cdf519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ago T., Kuribayashi F., Hiroaki H., Takeya R., Ito T., Kohda D., Sumimoto H. Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxidase activation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4474–4479. doi: 10.1073/pnas.0735712100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groemping Y., Lapouge K., Smerdon S. J., Rittinger K. Molecular basis of phosphorylation-induced activation of the NADPH oxidase. Cell. 2003;113:343–355. doi: 10.1016/s0092-8674(03)00314-3. [DOI] [PubMed] [Google Scholar]
- 35.Smith S. J., Rittinger K. Preparation of GTPases for structural and biophysical analysis. Methods Mol. Biol. 2002;189:13–24. doi: 10.1385/1-59259-281-3:013. [DOI] [PubMed] [Google Scholar]
- 36.Wiseman T., Williston S., Brandts J. F., Lin L. N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 37.Wang B., Zou J. X., Ek-Rylander B., Ruoslahti E. R-Ras contains a proline-rich site that binds to SH3 domains and is required for integrin activation by R-Ras. J. Biol. Chem. 2000;275:5222–5227. doi: 10.1074/jbc.275.7.5222. [DOI] [PubMed] [Google Scholar]
- 38.Sawin K. E., Nurse P. Regulation of cell polarity by microtubules in fission yeast. J. Cell Biol. 1998;142:457–471. doi: 10.1083/jcb.142.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham L. J., Stoica B. A., Shapiro M., DeBell K. E., Rellahan B., Laborda J., Bonvini E. Sequences surrounding the Src-homology 3 domain of phospholipase Cγ-1 increase the domain's association with Cbl. Biochem. Biophys. Res. Commun. 1998;249:537–541. doi: 10.1006/bbrc.1998.9177. [DOI] [PubMed] [Google Scholar]
- 40.Liu Q., Berry D., Nash P., Pawson T., McGlade C. J., Li S. S. Structural basis for specific binding of the Gads SH3 domain to an RxxK motif-containing SLP-76 peptide: a novel mode of peptide recognition. Mol. Cell. 2003;11:471–481. doi: 10.1016/s1097-2765(03)00046-7. [DOI] [PubMed] [Google Scholar]
- 41.Harkiolaki M., Lewitzky M., Gilbert R. J., Jones E. Y., Bourette R. P., Mouchiroud G., Sondermann H., Moarefi I., Feller S. M. Structural basis for SH3 domain-mediated high-affinity binding between Mona/Gads and SLP-76. EMBO J. 2003;22:2571–2582. doi: 10.1093/emboj/cdg258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mongiovi A. M., Romano P. R., Panni S., Mendoza M., Wong W. T., Musacchio A., Cesareni G., Di Fiore P. P. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kami K., Takeya R., Sumimoto H., Kohda D. Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67phox, Grb2 and Pex13p. EMBO J. 2002;21:4268–4276. doi: 10.1093/emboj/cdf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishida M., Nagata K., Hachimori Y., Horiuchi M., Ogura K., Mandiyan V., Schlessinger J., Inagaki F. Novel recognition mode between Vav and Grb2 SH3 domains. EMBO J. 2001;20:2995–3007. doi: 10.1093/emboj/20.12.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douangamath A., Filipp F. V., Klein A. T., Barnett P., Zou P., Voorn-Brouwer T., Vega M. C., Mayans O. M., Sattler M., Distel B., Wilmanns M. Topography for independent binding of α-helical and PPII-helical ligands to a peroxisomal SH3 domain. Mol. Cell. 2002;10:1007–1017. doi: 10.1016/s1097-2765(02)00749-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irazoqui J. E., Gladfelter A. S., Lew D. J. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 47.Butty A. C., Perrinjaquet N., Petit A., Jaquenoud M., Segall J. E., Hofmann K., Zwahlen C., Peter M. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gulli M. P., Jaquenoud M., Shimada Y., Niederhauser G., Wiget P., Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 49.Shimada Y., Wiget P., Gulli M. P., Bi E., Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]