Abstract
Cyclic nucleotide PDEs (phosphodiesterases) are important enzymes that regulate intracellular levels of cAMP and cGMP. In the present study, we identify and characterize novel PDEs in the genetic model, Drosophila melanogaster. The Drosophila genome encodes five novel PDE genes in addition to dunce. Predicted PDE sequences of Drosophila show highly conserved critical domains when compared with human PDEs. Thus PDE-encoding genes of D. melanogaster are CG14940-PDE1C, CG8279-PDE6β, CG5411-PDE8A, CG32648-PDE9 and CG10231-PDE11. Reverse transcriptase–PCRs of adult tissues reveal widespread expression of PDE genes. Drosophila Malpighian (renal) tubules express all the six PDEs: Drosophila PDE1, dunce (PDE4), PDE6, PDE8, PDE9 and PDE11. Antipeptide antibodies were raised against PDE1, PDE6, PDE9 and PDE11. Verification of antibody specificity by Western blotting of cloned and expressed PDE constructs allowed the immunoprecipitation studies of adult Drosophila lysates. Biochemical characterization of immunoprecipitated endogenous PDEs showed that PDE1 is a dual-specificity PDE (Michaelis constant Km for cGMP: 15.3±1 μM; Km cAMP: 20.5±1.5 μM), PDE6 is a cGMP-specific PDE (Km cGMP: 37±13 μM) and PDE11 is a dual-specificity PDE (Km cGMP: 6±2 μM; Km cAMP: 18.5±5.5 μM). Drosophila PDE1, PDE6 and PDE11 display sensitivity to vertebrate PDE inhibitors, zaprinast (IC50 was 71±39 μM for PDE1, 0.65±0.015 μM for PDE6 and 1.6±0.5 μM for PDE11) and sildenafil (IC50 was 1.3±0.9 μM for PDE1, 0.025±0.005 μM for PDE6 and 0.12±0.06 μM for PDE11). We provide the first characterization of a cGMP-specific PDE and two dual-specificity PDEs in Drosophila, and show a high degree of similarity in structure and function between human and Drosophila PDEs.
Keywords: cGMP-specific phosphodiesterase, Drosophila melanogaster, dunce, mammalian homologue, sildenafil, zaprinast
Abbreviations: cGK, cGMP-dependent protein kinase; PDE, phosphodiesterase; cA-PDE, cAMP-specific PDE; cG-PDE, cGMP-specific PDE; EST, expressed sequence tag; IP, immunoprecipitation; PAS, Per, ARNT, Sim; PKA, cAMP-dependent protein kinase; RT, reverse transcriptase; UTR, untranslated region
INTRODUCTION
cGMP signalling has been implicated in an increasing number of physiological processes. Recent works on several cell and tissue systems suggest that the enzymes that regulate the breakdown of cGMP, as opposed to its synthesis, are pivotal in maintaining the role of cGMP in cellular function [1–3].
In vertebrates, hydrolysis of cGMP is performed by cyclic nucleotide PDEs (phosphodiesterases), including PDE1, PDE5, PDE6, PDE9, PDE 10 and PDE 11 [4,5]. While some of these, notably PDE5, PDE6 and PDE9, are cG-PDEs (cGMP-specific PDEs), the others are dual-specificity enzymes that hydrolyse both cAMP and cGMP. PDEs are important drug targets, and as such, much is known about the pharmacology and biochemistry of these enzymes. For example, PDE5, the cellular target of sildenafil (Viagra), has been extensively characterized [5].
The use of genetic model organisms (Drosophila melanogaster and Mus musculus) has been a powerful tool in the demonstration of in vivo roles for PDEs [6–8]. However, these studies have focused on the cA-PDEs (cAMP-specific PDEs) and the Drosophila PDE, dunce, which is a member of the well-characterized PDE4 enzyme family [9]. In D. melanogaster, dunce mutants exhibit learning defects, while, in mammals, PDE4 selective inhibitors show anti-depressant and anti-inflammatory effects [10,11]. Much less, however, is known about cG-PDE function in an organotypic context, using model organisms. The elegant targeted expression systems available to Drosophila, e.g. the GAL4-UAS system (where UAS stands for upstream activating sequence) [12], allow for ectopic expression or disruption of genes of choice in particular cell types or tissues in the intact animal. Thus cell-specific signalling roles of cG-PDE can be assessed in intact tissues [13]. Indeed, we have evaluated cGMP signalling mechanisms in vivo, using the genetic model, the Drosophila Malpighian (renal) tubule [14]. By doing this, we have shown that cGMP signalling is an important modulator of renal function in Drosophila [2,13,15–18]. More specifically, we have shown that cG-PDE activity is critical for tubule function [2,13,17].
The discovery of the key role of dunce in learning and memory may have obscured the possibility that other important PDEs exist in the fly. As an important prelude to understand further the roles of cyclic nucleotide PDEs in vivo, we have identified and characterized five novel PDEs encoded by the Drosophila genome, and show that these are widely expressed in the fly. Specifically, we have identified the novel fly PDEs as PDE1, PDE6, PDE8, PDE9 and PDE11.
Antibodies were raised to PDE1, PDE6, PDE9 and PDE11, and biochemical characterization and inhibitor studies performed for PDE1, PDE6 and PDE11, thus providing the first such characterization of novel Drosophila cG-PDE function.
In the present study, we demonstrate conservation of both structure and function between vertebrate and Drosophila PDEs and suggest the widespread importance of PDEs in D. melanogaster.
EXPERIMENTAL
Drosophila stocks
Stocks were maintained on standard Drosophila diet at 25 °C and 55% humidity, during a 12 h:12 h photoperiod. The Drosophila line used in the present study was a standard wild-type strain (Oregon R), used at 7 days post-emergence in all experiments.
Bioinformatics
Polypeptide sequences of the 11 known mammalian PDE gene families were used as probes to obtain the annotated genome sequence of the D. melanogaster database. These were submitted to the FLYBLAST server and hits screened for signature HX3HX21–23D/E 3′-5′ cyclic nucleotide PDE motifs. Alignments of deduced Drosophila PDEs were made against relevant human PDEs using ClustalW (http://www.ebi.ac.uk/clustalw/), and drawn using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). In all alignments (Figure 1; also see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm), residues shaded black are identical, while grey shading indicates similar residues, using a PAM250 scoring matrix at 70% stringency.
Figure 1. Alignment of the catalytic domains of putative D. melanogaster PDEs with H. sapiens homologues.
The Berkeley Drosophila genome project database (now at http://flybase.net/) was searched with representative human PDE protein sequences, and putative Drosophila PDEs were identified. Sequence alignments using the H. sapiens homologues were performed using ClustalW and drawn using BioEdit. Comparisons of Drosophila and human PDEs are shown, where identical residues are shaded black; grey shading indicates residues with 70% similarity. Complete predicted amino acid sequences for Drosophila PDE1, PDE6, PDE8, PDE9 and PDE11 are contained in Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm.
RT (reverse transcriptase)–PCR
Expression of PDE genes was confirmed by performing RT–PCR on wild-type fly head and tubule cDNA preparations. Ten Drosophila heads or 50 tubules were dissected, poly(A)+ (polyadenylated) RNA was extracted (Dynal mRNA direct kit) and reverse-transcribed with MMLV (Moloney murine leukaemia virus) RNaseH-superscript reverse transcriptase (Invitrogen) as described previously [15]. Reverse transcription reaction mixture (1 μl) was used as a template for PCR containing PDE gene-specific primer pairs based on FlyBase sequences (Table 1). Additionally, to control against genomic contamination in cDNA preparations, primers were used which had been designed around intron/exon boundaries. Such primers verified the cDNA quality used in PCRs.
Table 1. Gene-specific primers for D. melanogaster PDE genes.
Gene-specific primer pairs based on FlyBase sequences of putative genes encoding PDEs (http://fly.ebi.ac.uk:7081/) were used for PCR of adult fly cDNA.
| Primer | Sequence 5′–3′ |
|---|---|
| CG8279F | CAAGATTCTGGTCAATGTCGGA |
| CG8279R | ACAAAAGGTCAAATAGCGGCG |
| CG10797F | GGAACCAGAAACACTGAGCGAC |
| CG10797R | TGCGGCTTGCGGAACTTTAG |
| CG14940F | AGACGCAGGAGAAGAAGAAAAGG |
| CG14940R | CCAGTTCATCAGACCCGTGTTG |
| CG10231F | ATGGTCTTCCGCATTCTCACCC |
| CG10231R | CCTCCACAAAGTTCTCGTCGTTC |
| CG32648F | GGACTCGTTCTCCTGCCACTATTC |
| CG32648R | GTTGCCAAAATGACAGCGATTG |
| CG5411F | CATTGCGAGAACCATTCGTCAC |
| CG5411R | TTGAAAATCAGGTGCGTGGGGG |
PCR cycle conditions were as follows: 94 °C (2 min), 30 cycles of [94 °C (15 s), 55 °C (30 s), 72 °C (3 min)], 72 °C (10 min). PCR products (400–800 bp) obtained from such RT–PCR experiments were cloned using the Invitrogen Topoisomerase (TOPO TA Cloning) system. Cloned plasmids were purified using Qiagen kits and sequenced to confirm their identity. The cloned PCR products shared 100% sequence identity with predicted PDE transcripts (results not shown).
Anti-PDE antibodies
Polyclonal rabbit anti-peptide antibodies were generated by Genosphere Technologies (Paris, France) against the following C-terminal epitopes for each putative cG-PDE as follows: PDE1, EQAVKDAEARALAT; PDE6, HGSEDSHTPEHQRS; PDE9, MDPDKVSKPGSQVR; and PDE11, PTSTQPSDDDNDAD. Antibodies were affinity-purified before use as described previously [19].
Cloning and expression of PDE1, PDE6 and PDE11 in Drosophila S2 cells
Cloning and expression studies were performed using ESTs (expressed sequence tags) as specified, obtained from BDGP (the Berkeley Drosophila Genome Project, http://www.fruitfly.org/EST/index.shtml).
PDE1: a full-length EST (RE56844) was available for this gene. All subsequent subcloning was performed using this EST as a template.
PDE6: one EST (GH27433) for PDE6 was available. However, the genome annotation predicted that this EST lacked 271 bp of the 5′-end of the open reading frame. Therefore an RT–PCR strategy was used to obtain the full-length open reading frame. A forward PCR primer (PDE6ORFF) was designed complementary to the 5′-end of the open reading frame. To establish whether or not the PDE6 transcript extended to the 5′-end of the predicted open reading frame, the reverse primer (PDE6R673) was designed to span the first intron, thus enabling the distinction to be made between amplification from genomic DNA versus cDNA. RT–PCR was then performed using gene-specific cDNA synthesized from total head RNA primed with the reverse primer as a template. This produced a fragment of the expected size, which was cloned and sequenced to confirm that it represented the 5′-end of the PDE6 transcript. The full-length open reading frame was then assembled using fusion PCR, in which the separate open reading frame moieties were amplified in two PCRs. The PCR products were purified and used in equal amounts as templates in a further PCR experiment using PDE6ORFF and PDE6ORFR primers. This gave a single product comprising the full-length open reading frame of PDE6, with an in-frame stop codon occurring at the 5′-end of 57 bp to the predicted ATG start codon. Northern blotting using a digoxigenin-labelled riboprobe complementary to the 3′-UTR (3′-untranslated region) of PDE6 showed that the full-length PDE6 transcript (∼7 kb) is detectable in both head and body total RNA (results not shown). The full-length open reading frame was sequenced to check for PCR errors and PCR artifacts, and the construct (pCR2.1PDE6ORF) was used as a template in all subsequent subcloning procedures.
PDE9: no ESTs were available for PDE9. However, as it is possible to characterize PDEs from cloned catalytic constructs alone [20], primers (PDE9catF and PDE9catDESR) were designed to encompass the catalytic domain of PDE9. Using the Malpighian (renal)-tubule-specific template cDNA, a fragment of the expected size was amplified and verified, and used for all subsequent cloning.
PDE11: initially, two incomplete ESTs coding for PDE11 were available from BDGP (RH43346 and LP04047). These encoded the 3′-UTR, including a poly(A)+ tail and approximately half of the coding sequence of CG10231. Library screening and 5′-rapid amplification of cDNA ends were used to obtain the full-length sequence for CG10231. However, a newly released EST, SD13096, was found to encode the full-length PDE11 (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). This was used for subsequent cloning; however, expression of PDE11 by this construct in S2 cells proved to be difficult (results not shown). Therefore to achieve the expression of PDE11 for validation of the anti-PDE11 antibody (Figure 4), a construct comprising the sequence for the PDE11 catalytic domain and the C-terminal region was used. This 2130 bp gene fragment was amplified using a forward primer 5′ to the catalytic domain (PDE11catF 5′-ATGGAGGCGTTCGCCATCTTCTGC-3′) and a reverse primer at the 3′-end of the open reading frame omitting the stop codon (PDE11V5R5′-TTTTTCAACCGCCATAGCGG-3′). The fragment was cloned into the S2 cell expression vector pMT V5 His TOPO (below) and the resultant constructs were screened for the correct orientation of the insert.
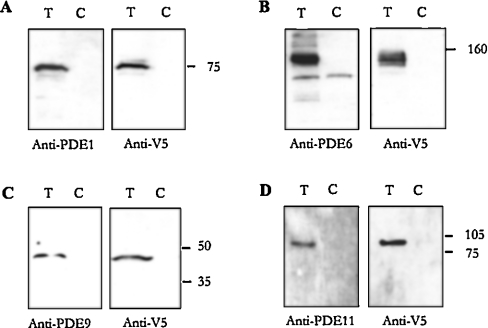
Figure 4. Anti-PDE antibodies recognize specific PDEs.
V5-tagged constructs for PDE1, PDE6, PDE9 and PDE11 were transfected into S2 cells, and cell extracts prepared from transfected cells (T) and untransfected controls (C). Western blotting was performed with cell extracts using polyclonal anti-PDE antibodies. Left panel: (A) PDE1, (B) PDE6, (C) PDE9 and (D) PDE11. Right panel: anti-V5 antibody [18] (A–D) PDE1–PDE11, same as left panel. Bands identified by both anti-PDE and anti-V5 antibodies in samples from transfected but not control cells were as follows: PDE1: 75 kDa; PDE6: 150 kDa; PDE9: 47 kDa; PDE11: 100 kDa, indicated by use of molecular mass standards (kDa).
All PDE constructs for cloning were sequence-verified and cloned into expression vector pMT/V5-HIS-TOPO (Invitrogen). This vector incorporates the C-terminal V5 epitope-tag [GKPIPNPLLGLDST, derived from a small epitope (Pk) present on the P and V proteins of the paramyxovirus of SV5 (simian virus 5)] and His6 tag. These plasmids were then used for the transient transfection of S2 cells under conditions of Cu2+-inducible expression. Cells were cultured according to standard methods described in [21]. Approximately 3×106 cells were used in each transfection, which was performed using calcium phosphate according to standard techniques (Invitrogen); transfection efficiencies were routinely 10%.
Western-blot analysis
Samples of 3×106 PDE1-, PDE6-, PDE9- and PDE11-transfected S2 cells were collected by centrifugation at 5000 g for 3 min. Cells were resuspended in PBS, spun down once more and resuspended in lysis buffer {50 mM Tris (pH 7.5), 150 mM NaCl, 1% (v/v) Igepal CA-630 [octylphenyl-poly(ethylene glycol); Sigma] and 1 μg/ml protease inhibitor cocktail with PMSF (Sigma)}. Cells were disrupted by sonication and centrifuged at 15000 g for 5 min, and the supernatants were collected. The protein content of each sample was estimated using the Bradford assay. Samples (10 μg of protein each) were loaded on to each lane for Western-blot analysis, performed according to standard methods described in [13] using the ECL® system (Amersham Biosciences). Blots were probed with anti-V5-epitope antibody (1/5000) (Invitrogen) or rabbit polyclonal anti-PDE antibodies (1/400). Horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) were used at a concentration of 1/5000. This procedure was also performed for untransfected (control) cells for blotting with anti-PDE antibodies.
IP (immunoprecipitation) assays and cG-PDE assays
IP assays were performed from heads of 7–9-day-old wild-type (Oregon R) adult Drosophila. For each IP sample, 20 heads were dissected and homogenized in 1 ml of lysis buffer [10%, v/v, glycerol, 1%, v/v, Triton X-100, 150 mM NaCl and 50 mM Hepes (pH 7.5), with 10 μl of protease inhibitor cocktail]. Samples were centrifuged at 15000 g at 4 °C for 5 min to remove insoluble material and the supernatant was removed to a fresh tube. Samples were precleared by an end-over-end incubation for 1 h at 4 °C with 25 μl of Protein A–Sepharose beads (Sigma) that had been prewashed with lysis buffer. Beads were pelleted by centrifugation at 5000 g at 4 °C for 1 min and the supernatant was removed to a fresh tube. Samples were then incubated with 5 μg of PDE-specific antibody or 5 μg of IgG control (Sigma) with an end-over-end mixing at 4 °C for 1 h. Protein was then precipitated by adding 10 μl of prewashed Protein A beads. After 30 min of incubation at 4 °C with mixing, beads were pelleted by centrifugation, washed three times with 0.5 ml of lysis buffer, followed by three washes with ice-cold KHEM buffer (50 mM KCl, 50 mM Hepes, pH 7.2, 10 mM EGTA and 1.92 mM MgCl2). Samples were assayed for PDE activity as described previously [22] in the presence of substrate and other appropriate reagents as described in the legends of Figures 5–7. Results are expressed as PDE activity: cGMP or cAMP hydrolysed·(IP assay)−1·min−1, mean±S.E.M., n=3–6. Where appropriate, statistically significant data are indicated by *P<0.05 (Student's t test, unpaired samples).
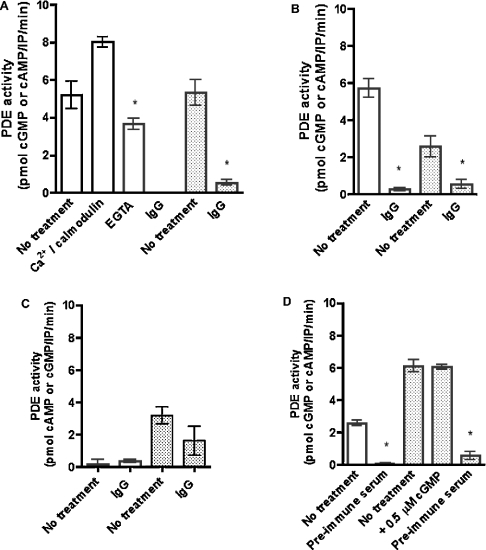
Figure 5. PDEs encoded by Drosophila PDE genes exhibit cG- and cA-PDE activity.
PDEs were immunoprecipitated from adult Drosophila head lysates using affinity-purified antibodies (PDE1, PDE6 and PDE9) and whole serum (PDE11). Control IPs were performed with IgG (preimmune serum for PDE11). Each immunoprecipitated sample was assayed for cGMP- and cAMP-specific PDE activity, where cG-PDE activity is indicated by unshaded bars; and cAMP-PDE activity is indicated by shaded bars. For each PDE, activities were assessed under the conditions described in the x-axis for each graph: (A) PDE1, (B) PDE6, (C) PDE9 and (D) PDE11. PDE activities were assayed at either 10 μM cGMP (for cG-PDE activity) or 10 μM cAMP (for cA-PDE activity). Ca2+/calmodulin-stimulated PDE1 activity was assayed at 0.2 mM Ca2+ and 0.4 mg/ml calmodulin. Data are represented as PDE activity [pmol of cGMP or cAMP·(IP assay)−1·min−1±S.E.M., n=6]. cG-PDE activity in the IgG-treated fraction from PDE1 samples (A) was negligible. *, Data statistically significant between antibody-specific and control IPs, assayed for cG- and cA-specific PDE activity, where P<0.05 (Student's unpaired t test). Additionally, in PDE1 panel, * indicates statistical significance (P<0.05, Student's unpaired t test) between EGTA-treated (0.1 mM EGTA) and untreated samples assayed for cG-PDE activity.
RESULTS
The D. melanogaster genome encodes five novel putative PDEs
Using the mammalian vertebrate sequences for comparison and the HX3HX21–23D/E protein cyclic nucleotide PDE motif as bait in an in silico screen, we identified five genes encoding putative novel PDEs in the Drosophila genome (Table 2). This was in addition to the previously characterized cAMP-PDE, dunce (Table 2). Homologues to mammalian PDE1, PDE6, PDE8, PDE9 and PDE11 were identified based on sequence similarity within the predicted catalytic domain, as well as by the presence of sequences proposed to be regulatory and post-translational modification sequences in the vertebrate enzymes [23]. Comparison of the Drosophila PDE predicted protein sequences with their mammalian homologues revealed high sequence identity (51–77%) within the conserved catalytic region (Table 2). Lower sequence identity (28–40%) was found over the complete protein sequence (Table 2).
Table 2. Assignment of D. melanogaster homologues of vertebrate PDEs.
Sequence information for novel PDE genes and deduced proteins identified from the D. melanogaster genome is listed, together with closest human homologues. Full-length ESTs for Drosophila PDE genes are also listed, where available (http://www.fruitfly.org/EST/index.shtml); except for GH27433*, which is not full-length. The percentage identities and similarities for each gene are calculated over the length of the shorter (human) homologue, and also in relation to the catalytic domain.
| Percentage amino acid identity (similarity) | |||||
|---|---|---|---|---|---|
| Gene | Human homologue | Human homologue | Catalytic domain | Predicted length of polypeptide (amino acids) | ESTs |
| CG14940 | PDE1 | 40 (56) | 63 (79) | 1818 | RE56844 |
| CG8279 | PDE6 | 28 (46) | 51 (69) | 1131 | GH27433* |
| CG5411 transcript A | PDE8 | 34 (52) | 60 (79) | 914 | SD18711 |
| CG5411 transcript B | PDE8 | 35 (53) | 60 (79) | 904 | RE31467 |
| CG5411 transcript C | PDE8 | 47 (66) | 60 (79) | 400 | GH21295 |
| CG5411 transcript D | PDE8 | 37 (57) | 60 (79) | 805 | RE35136 |
| CG5411 transcript E | PDE8 | 34 (52) | 60 (79) | 914 | RE07805 |
| CG5411 transcript F | PDE8 | 23 (9) | 60 (79) | 400 | LD46553 |
| CG32648 | PDE9 | 26 (34) | 63 (76) | 2080 | None |
| CG10231 | PDE11 | 38 (55) | 77 (96) | 1545 | SD13096 |
The five putative Drosophila PDEs contain domains and motifs homologous with their mammalian counterparts
An alignment was made of the catalytic domains of each Drosophila PDE with its human homologue (Figure 1), where dunce was included as a positive control. Inspection of the deduced protein sequences (Figure 1) reveals that each putative PDE encodes the signature HX3HX21–23D/E metal-dependent hydrolase motif [23]. Comparison of Drosophila with human PDEs confirms the high percentage of sequence identity within the catalytic domain (Table 2).
Drosophila PDE1
Although calcium (Ca2+)/calmodulin-sensitive dual-specificity PDE activity has been determined in Drosophila [24], its molecular identity was unknown. In the present study, we show that CG14940 encodes the closest Drosophila homologue of the mammalian Ca2+/calmodulin-sensitive PDE, PDE1. It shares 40% overall amino acid identity with mammalian PDE1C, but 63% identity with the conserved catalytic domain (Figure 1). The N-terminal autoinhibitory motif of mammalian PDE1A [25] appears to be conserved between mammals and flies; however, only one of the two mammalian calmodulin-binding sites appears to be present in Drosophila PDE1 (Figure 2). The EST sequence for CG14940 (RE56844) provides strong evidence that Drosophila PDE1 has an open reading frame of 1815 nucleotides, which encodes a polypeptide of 605 amino acids. Sequence comparisons between Drosophila PDE1 and Homo sapiens PDE1C revealed that the fly gene encodes an extra 13 amino acids at the N-terminus and is truncated by 133 amino acids at the C-terminus (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). Since no study has uncovered a role for the C-terminal region of PDE1, the significance of this latter finding is unknown.
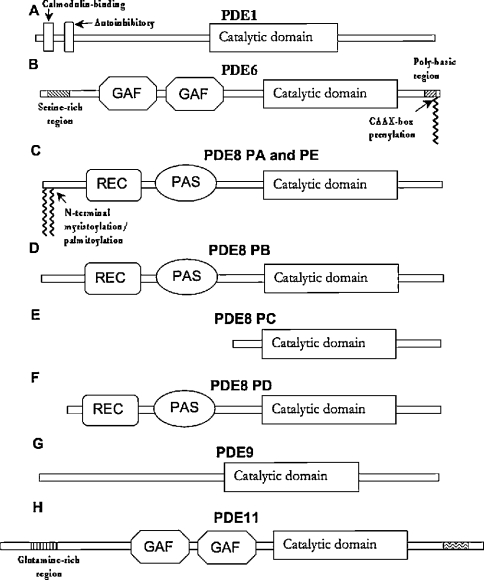
Figure 2. Schematic diagram of novel Drosophila PDEs.
Schematic representations of deduced protein sequences are derived from ClustalW alignments. These were made for each putative Drosophila PDE: (A) PDE1, (B) PDE6, (C–F) PDE8, (G) PDE9 and (H) PDE11 with their closest mammalian homologue, as ascertained by lowest BLAST expect score and closest overall sequence similarity.
Recent structural studies have illuminated the mechanism behind PDE substrate-specificity, where a conserved glutamine residue changes orientation with respect to the bound substrate (the ‘Q switch’ mechanism) [20]. The residues surrounding this glutamine residue, in particular critical histidine residues, confer this rotational freedom, which is supposed to form the basis of mechanism of action of dual-specificity PDEs. This glutamine residue (Gln-426 in PDE1C and Gln-439 in Drosophila PDE1), which co-ordinates the nucleoside purine, was identified from sequence alignments between the catalytic domains of the Drosophila and H. sapiens PDEs (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). Three other critical residues surrounding this glutamine residue, His-381, His-373 and Trp-496 in mammalian PDE1C, are conserved in Drosophila PDE1 (the corresponding residues are His-399, His-391 and Trp-539). It therefore appears that CG14940 has the structural correlates required to encode dual-specificity PDE1 activity in D. melanogaster.
Drosophila PDE6
The deduced protein encoded by CG8279 shares 28% overall sequence identity with mammalian PDE6β, but shows 51% identity within the catalytic domain. However, it appears to be more similar to mammalian PDE5 (58% identity within the catalytic domain, Table 2), which suggests that CG8279 may encode an ancestral form of mammalian PDE5. Interestingly, one of the most prominent features of the predicted protein sequence of CG8279 is the presence of a C-terminal CAAX-box prenylation motif (Figure 2, and see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). This and the presence of conserved catalytic-domain residues led to its designation as a PDE6 homologue. The DNA sequence predicted by the genome annotation comprises a 3393 bp open reading frame, coding for a polypeptide of 1131 amino acids. One EST, GH27433, comprising bp 271 of the predicted open reading frame to a poly(A)+ tail 1.6 kb downstream of the TGA stop codon, was available (BDGP). Further cloning and sequencing work (see the Experimental section) verified the Drosophila genome annotation for CG8279.
Drosophila PDE6 contains regions similar to the tandem cGMP-binding and dimerization mediating GAF domains [23,26] that are found within the N-terminal region of several mammalian PDEs (Figure 2). Proximal to the C-terminal CAAX-box prenylation motif lies a polybasic region comprising four lysine, one arginine and two serine residues, and also contains a consensus cGK (cGMP-dependent protein kinase) or PKA (cAMP-dependent protein kinase) phosphorylation motif (KKRS). Another cGK/PKA phosphorylation motif (KRPS) occurs at predicted amino acids 197–200. This second phosphorylation motif may be homologous with that of mammalian PDE5, which also lies N-terminal to the GAF domains and is supposed to modulate the binding of cGMP to these regulatory domains [23]. Comparison of Drosophila PDE6 polypeptide with H. sapiens PDE6β shows that the former has an extra 99 amino acids at the N-terminus and a 111-amino-acid insertion between the end of the catalytic domain and the prenylation motif (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm).
The canonical vertebrate cG-PDE, PDE5A, specifically hydrolyses cGMP. Structural studies have uncovered the main mechanism conferring cGMP-specificity on PDE5A [20]. This involves a key Gln-817 that is orientated to form a clamp by hydrogen-bonding interactions between Gln-817 and Gln-775 and also by hydrogen bonds between the carboxyl backbone of Ala-767 and Gln-775, and Trp-853 and Gln-775. Each of these residues is conserved in Drosophila PDE6 (Q935, Q893, A885 and W970) and indeed in mammalian PDE6β, another cG-PDE (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). This suggests that Drosophila PDE6 is a cG-PDE.
Drosophila PDE8
The predicted polypeptide sequence of the gene CG5411 bears close resemblance to human PDE8, sharing 30% overall amino acid sequence identity and 60% identity within the catalytic domain (Table 2 and Figure 1). At the N-terminus is a six-amino-acid consensus myristoylation/palmitoylation motif, MGCAP, which is almost identical with that contained in human PDE8A1 (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). N-myristoyltransferase strictly requires a glycine residue at position 2 and a preference for serine or threonine residue at position 6; therefore MGCAPS represents a strong consensus sequence for this enzyme [27]. Palmitoylated proteins fall into three categories; one group comprises transmembrane proteins that are acylated near the membrane; the second group of proteins, typified by the Ras family, is modified near the C-terminus; and the third group is modified at the N-terminus. PDE8 falls into the last category; the cysteine residue at position 3 is a prime candidate for post-translational palmitoylation. The close conservation between mammalian and fly PDE8 at the N-terminus suggests that lipid modification is important for the function of both proteins.
The predicted polypeptide sequence of CG5411 also possesses two conserved domains N-terminal to the catalytic domain. First, a REC (recA) domain, which shows 38% identity and 54% similarity with human PDE8A REC domain, was identified as a phosphate acceptor domain in the bacterial two-component system [28]. Secondly, there is a PAS (Per, ARNT, Sim) domain (34% identity and 53% similarity with human PDE8) that has been shown to serve several functions, including circadian cycling, dimerization and mediation of protein–protein interactions [29]. Taking into account all of these similarities, CG5411 was designated as the Drosophila PDE8 homologue.
The predicted nucleotide sequence of Drosophila PDE8 had 19 exons and covers over 13 kb of genomic sequence. To confirm the predicted nucleotide sequence, ESTs were obtained. Three ESTs, which have been sequenced at the 5′- and 3′-ends, were identified, which matched the PDE8 predicted nucleotide sequence. They were GH21295, LD46553 and SD18711 (Table 2). The sequence of GH21295 comprised a short 280 bp region immediately 5′ from the predicted 13th exon, whilst the poly(A)+ tail occurred 1.35 kb 3′ to the putative stop codon (Table 2). At 30 bp upstream from the poly(A)+ tail is a poly(A)+ signal sequence (ATAAA). EST LD46553 comprises the same transcription start site as GH21295 yet had 2.1 kb of sequence between the putative stop codon and the poly(A)+ tail (Table 2). SD18711 encoded the full predicted nucleotide sequence, in addition to 320 bp of 5′-UTR, resulting in an open reading frame of 3180 bp, which translates into a polypeptide of 1060 amino acids.
Since this analysis was performed, version 3 of the Drosophila genome annotation (http://flybase.net/annot/) was released. This has identified five transcripts encoding four different polypeptides for PDE8 (Figure 2), in which the open reading frames of transcripts A and E are identical. Analysis of LD46553 suggests that a further transcript exists, identical with transcript C with an 800 bp extension to the 3′-UTR, designated as transcript F. Transcripts B, C and D do not encode the N-terminal myristoylation/palmitoylation motif (Figure 2). Interestingly, all seven splice forms of H. sapiens PDE8A and PDE8B do encode this motif. Transcript C encodes neither the PAS nor REC domain, being truncated near the N-terminus of the catalytic domain (Figure 2). Transcripts A, B, C and D all encode unique N-terminal sequences. Since the N-terminal regions of several PDEs have been shown to account for their differential localization and choice of interacting partner, this was of particular interest. Therefore PSORT (http://psort.nibb.ac.jp/) predictions for the subcellular localization of each PDE8 isoform were made. Proteins A, C and D were predicted to be cytoplasmic, whereas Protein B was predicted to be confined to mitochondria.
Mammalian PDE8 is a high-affinity cAMP-specific PDE [30,31]. However, the crystal structure of this PDE has not been reported. Comparison of mammalian PDE8 with the structure of the cAMP-specific PDE4B suggests that two residues, the conserved Gln-369 and Asn-321, make contact with the substrate adenine. The conserved residue corresponding to Asn-321 in Drosophila PDE8 is Asn-729, which could possibly interact with the substrate in a similar way to PDE4B. Moreover, each of the four residues within the nucleoside-binding pocket is conserved between Drosophila PDE8 and human PDE8: Asn-803, Cys-811, Gln-851 and Trp-885 in Drosophila PDE8, corresponding to Cys-729, Asn-737, Gln-778 and Trp-812 in human PDE8A. This would suggest that Drosophila PDE8 could possess the same substrate-specificity as human PDE8A.
Drosophila PDE9
The catalytic domain of mammalian PDE9 shows low sequence identity (29–35%) with other mammalian PDEs [32]. However, the predicted polypeptide sequence encoded by Drosophila gene CG32648 shows 63% amino acid identity within the catalytic domain (Figure 1). Therefore CG32648 was designated as Drosophila PDE9. PDE9 shows no sequence similarity to Drosophila PDE9 outside of the catalytic domain. This may indicate that the sequence annotation does not represent the actual gene structure. However, as no ESTs were available for study, it is not possible at this stage to verify the PDE9 sequence.
Drosophila PDE11
Alignment of the predicted polypeptide sequence of CG10231 with mammalian PDE11A showed that these genes are closely related. Sequence identity within the catalytic domain is 77%, although the overall sequence identity is 38% (Figure 1 and Table 2). Like human PDE11, the predicted polypeptide sequence of CG10231 contains tandem GAF domains at the N-terminal region (Figure 2). Because of this close similarity to human PDE11A (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm), CG10231 was designated as Drosophila PDE11. Interestingly, PDE5, which is phosphorylated by cGK [33], contains one cGK phosphorylation motif in each subunit [34]. We have identified four such consensus motifs in PDE11 (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm). Although this does suggest that PDE11 may be a substrate for cGK in Drosophila, the significance of four putative phosphorylation sites is currently unknown.
When this study was initiated, two incomplete ESTs (LP04097 and RH43346) were available from BDGP. RH43346 provided the longest sequence, encoding the last six predicted exons, part of the seventh exon and a 3′-UTR of 1.6 kb with a poly(A)+ tail and poly(A)+ signal sequence.
However, a newly released EST, SD13096, was found to encode the full-length PDE11 (Supplementary Figure 1). Northern blotting of RNA from adult heads and bodies using a digoxigenin-labelled riboprobe, complementary to the 3′-UTR of SD13096, showed that the PDE11 transcript was approx. 5.8 kb long, the same length as SD13096 (results not shown), confirming that SD13096 encodes a full-length cDNA.
Widespread expression of PDE genes in adult tissue
Using RT–PCR, we investigated the expression of the putative PDE genes in tissues of interest, i.e. in adult head and Malpighian tubules. Results in Figures 3(A)–3(F) show widespread expression of all the PDEs. Interestingly, expression of PDE6 in Drosophila is widespread and is not confined to the eye tissue, as is mammalian PDE6.
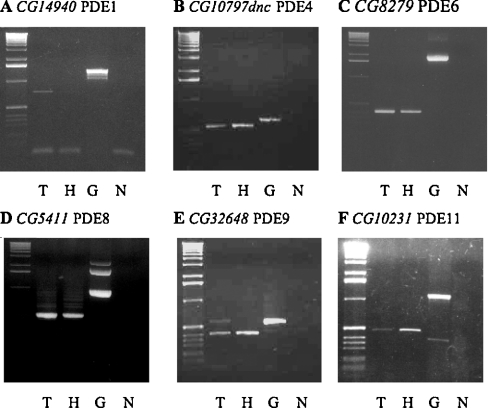
Figure 3. PDE gene expression in adult D. melanogaster.
RT–PCR using cDNA templates from tubule (T), head (H), and control genomic DNA (G) using intron-spanning gene-specific primers for putative PDE-encoding genes. (N) No template control. In all gels, the first lane is a 1 kb DNA ladder (Invitrogen). Expected PCR products indicated were obtained for all putative PDE genes: (A) CG14940 (PDE1C), genomic-1438 bp and cDNA-743 bp; (B) CG10797 (dnc) (PDE4), genomic-603 bp and cDNA-438 bp; (C) CG8279 (PDE6), genomic-1969 bp and cDNA-406 bp; (D) CG5411 (PDE8), genomic-998 bp and cDNA-506 bp; (E) CG32648 (PDE9), genomic-567 bp, cDNA-391 bp; (F) CG10231 (PDE11), genomic-1118 bp and cDNA-472 bp. PCR products were cloned and sequenced and found to have 100% identity with PDE genes (results not shown).
Although CG32648 expression in the head by RT–PCR using gene-specific primers (see the Experimental section) is not documented, this gene is expressed in a head cDNA library (GH library, Berkeley Drosophila genome project http://www.fruitfly.org/EST/; results not shown). Expression of PDE9 in adult Drosophila tissues is further confirmed by microarray analysis using Affymetrix arrays [35].
Microarray analysis of PDE gene expression in both tubules and in the rest of adult flies confirms the expression data using gene-specific primers (Figure 3). In particular, CG8279-PDE6, CG5411-PDE8 and CG10231-PDE11 are all significantly enriched in tubules compared with the rest of the fly (http://www.mblab.gla.ac.uk/%7Ejulian/arraysearch.cgi).
This confirms the importance of cyclic nucleotide signalling in Drosophila renal function. Finally, given that at least three of the expressed genes are putative cG-PDEs (CG8279-PDE6, CG14940-PDE1C and CG32648-PDE9), this suggests that regulation of cGMP signalling is necessarily complex, even in this simple epithelium.
Multiple cG-PDEs in D. melanogaster
To perform biochemical characterization of the PDEs encoded by the identified PDE genes, cloning and expression studies were attempted. Cloning of the full-length open reading frames of PDE1, PDE6, PDE8, PDE11 as well as the PDE9 catalytic domain was performed; although expression of all these constructs in Drosophila S2 cells was achieved, with the exception of PDE6 [36], obtaining active recombinant enzymes in S2 extracts from transfected cells proved to be highly problematic. We currently do not have a good explanation for this. The lack of activity associated with the PDE protein expressed in this model system may be associated with incorrect processing in Drosophila S2 cells, a macrophage-like line. Indeed, it could be due to the lack of appropriate chaperones or scaffolding partners in these cells that are required for folding, such as those identified for vertebrate PDEs, and needed to form a catalytically competent enzyme [20,37–39]. Therefore another approach was used to characterize the Drosophila PDEs, in particular putative cG-PDEs and dual-specificity PDEs. Polyclonal antibodies were raised to the unique C-termini of PDE1, PDE6, PDE9 and PDE11. Successful expression of recombinant protein for PDE1, PDE6, as well as the catalytic domain of PDE9 and catalytic domain plus C-terminal region of PDE11 in S2 cells, allowed antibody specificity to be verified by Western blotting (Figure 4). Expected sizes of polypeptides were obtained for each PDE construct.
Successful generation of PDE-specific antibodies allowed biochemical characterization of novel PDE1, PDE6 and PDE11 from immunoprecipitated samples of adult Drosophila head extract, subjected to PDE assays.
The lack of PDE activity from immunoprecipitated samples using control IgG fraction, compared with PDE activity obtained using anti-PDE antibodies (Figure 5), further confirms the specificity of these antibodies. Figure 5(A) demonstrates that PDE1 hydrolyses both cGMP and cAMP when assayed at a substrate concentration of 10 μM. As expected by sequence identity, PDE1 is a Ca2+/calmodulin-regulated enzyme. We show that cG-PDE activity is inhibited by the calcium chelator EGTA, but is stimulated by the addition of 0.2 mM Ca2+ and 0.4 mg/ml calmodulin.
PDE6 displays cG-PDE activity at 10 μM cGMP; however, significant cAMP-specific PDE activity was also found (Figure 5B). However, assay of PDE6 activity from transfected Drosophila S2 cells does not reveal cA-PDE activity [36].
PDE11 hydrolyses both cGMP and cAMP (Figure 5D). Interestingly, cAMP hydrolysis was not augmented by the addition of 0.5 μM cGMP to the assay mix.
PDE9 activity was not recovered successfully from immunoprecipitated samples using PDE9-specific antibody (Figure 5C). Although a small amount of cAMP-PDE activity was recovered, this activity was not statistically significant when compared with IgG controls. Thus all further characterization was performed for PDE1, PDE6 and PDE11 only.
Biochemical characteristics of novel Drosophila PDEs
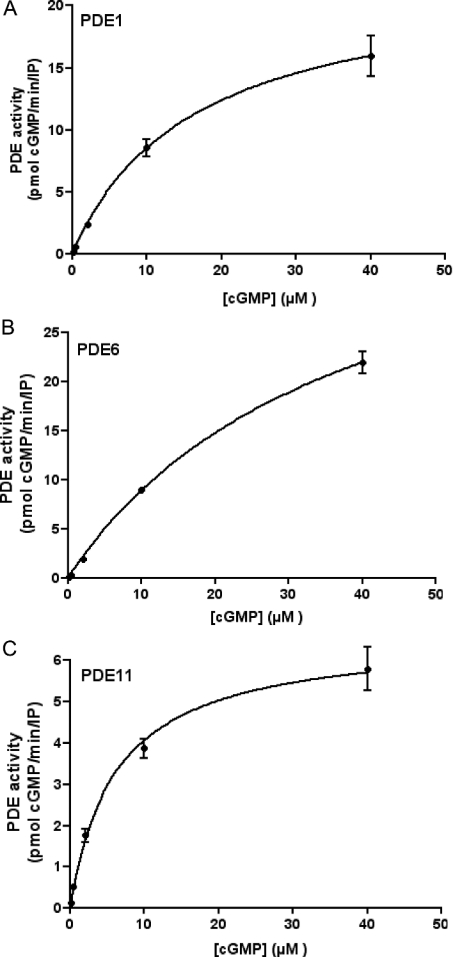
Figure 6 shows cGMP dependence of PDE1 (panel A), PDE6 (panel B) and PDE11 (panel C) activities. Values of the Michaelis constant Km were determined by non-linear regression plots (Figure 6) and confirmed by Lineweaver–Burk analysis (results not shown). PDE1 displayed a Km of 15.3±1 μM (15 μM by Line-weaver–Burk analysis) for cGMP; PDE6 showed a Km of 37±13 μM (50 μM by Lineweaver–Burk analysis), whereas PDE11 had a Km of 6±2 μM (4.6 μM by Lineweaver–Burk analysis). Although PDE6 activity could have been assayed at higher substrate concentrations, the approximate Km deduced from data shown in Figure 6(B) is in agreement with that obtained by analysis of PDE6 kinetics in PDE6-transfected S2 cell extracts [36].
Figure 6. Kinetic analysis of Drosophila cG-PDEs.
Kinetics of cG-PDE activity in immunoprecipitated samples from Drosophila head lysate using antibodies raised against three novel putative Drosophila PDEs. cG-PDE assays were performed at five substrate concentrations: 0.5, 2, 4, 10 and 40 μM cGMP. (A–C) Best-fit square-hyperbola non-linear regression plots. Km values calculated from analysis of the data (GraphPad Prism4) are shown in the text. (A) PDE1, (B) PDE6 and (C) PDE11. Results are expressed as cG-PDE activity [pmol of cGMP hydrolysed·(IP assay)−1·min−1±S.E.M., n=3].
Taken together, our kinetic data show that PDE11 is the Drosophila PDE with highest cGMP affinity among the PDEs analysed so far. These values also confirm the idea that Drosophila cG-PDEs are enzymes with high Km values, as these are in close agreement with previous data for cG-PDE activity assayed in Drosophila renal tubule extracts, which show a Km of 15.88±10.22 μM for cGMP [13].
Similar kinetic experiments performed for cAMP-dependent activity for PDE1 and PDE11 showed that PDE1 displayed a Km of 20.5±1.5 μM, whereas PDE11 has a Km of 18.5±1.5 μM for cAMP (results not shown).
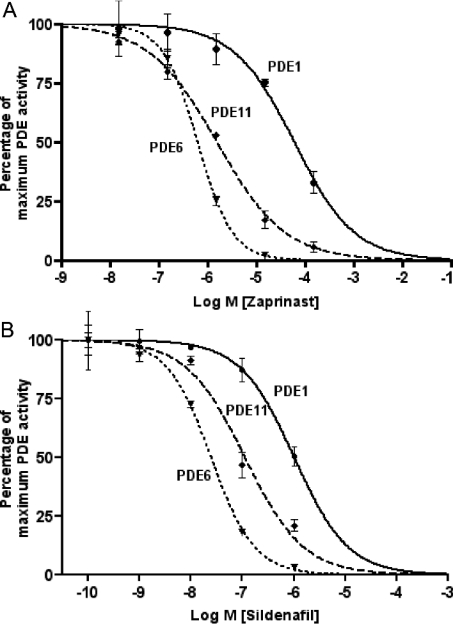
Previous work has shown that cG-PDE activity in Drosophila renal tissue is sensitive to inhibitors of vertebrate cG-PDEs, for example zaprinast [2] and the PDE5-specific inhibitor, sildenafil [13]. In the present study, we show that isolated cG-PDE activities are sensitive to both these inhibitors (Figures 7A and 7B).
Figure 7. Inhibition of Drosophila PDEs by zaprinast and sildenafil.
Immunoprecipitated samples from Drosophila head lysates using antibodies specific for PDE1, PDE6 and PDE11 were assayed for cG-PDE activity [pmol of cGMP hydrolysed·(IP assay)−1·min−1±S.E.M. at 10 μM substrate concentration] in the absence (control) or in the presence of 1.5×10−9, 1.5×10−8, 1.5×10−7, 1.5×10−6, 1.5×10−5 and 1.5×10−4 M zaprinast (A), and 10−10, 10−9, 10−8, 10−7 and 10−6 M sildenafil citrate (B). To aid comparison, results are expressed as percentage of maximum (control) PDE activity ±S.E.M., n=3. Control PDE activity [pmol of cGMP hydrolysed·(IP assay)−1·min−1 ±S.E.M.] for PDE1: 2.95±0.16; PDE6: 11.11±0.195; and PDE11: 3.65±0.15.
PDE1 is inhibited by zaprinast, with an IC50 value of 71±39 μM (Figure 7A), and by sildenafil with an IC50 value of 1.3±0.9 μM (Figure 7B). PDE6 is most sensitive to sildenafil, with an IC50 value of 25±5 nM (Figure 7B); for zaprinast, this value is 0.65±0.15 μM (Figure 7A). Interestingly, previous work has shown that Drosophila tubule cG-PDE activity exhibits nanomolar sensitivity to sildenafil [13]. It is possible, then, that PDE6 may be the main in vivo target for this drug in tubules. Finally, PDE11 shows sensitivity to both zaprinast and sildenafil, with IC50 values of 1.6±0.5 μM (Figure 7A) and 0.12±0.06 μM (Figure 7B) respectively.
DISCUSSION
Previous work has shown that cG-PDE activity is important for regulating fluid transport by the Drosophila Malpighian tubule. However, surprisingly, Drosophila PDEs have not been well characterized to date, beyond the work on dunce. Given this, we investigated candidate PDEs using D. melanogaster genome resources. We show that Drosophila encodes several novel PDEs. By homology, these include two cG-PDEs (PDE6 and PDE9), a cA-PDE (PDE8A) and two dual-specificity PDEs (PDE1 and PDE11). This, together with documented widespread expression of all PDE genes in the adult fly, suggests an important role for complex cyclic nucleotide regulation in the physiology of the fly.
Interestingly, the Drosophila genome does not encode homologues of mammalian PDE2, PDE3, PDE5, PDE7 and PDE10. This perhaps suggests that the Drosophila enzymes are products of ancestral genes, which in vertebrates evolved to encompass several other related PDEs that confer specialization of signalling processes in a necessarily more complicated body plan. For example, in D. melanogaster, PDE6 is the closest homologue of vertebrate PDE5. In mammals, PDE6 expression is confined to the eye and pineal gland, whereas in eye, it is an essential component of phototransduction [40]; recently, however, PDE6 expression has also been documented in Chinese hamster ovary and mouse F9 stem cells [41]. In the present study, we show that D. melanogaster PDE6 is widely expressed throughout the adult fly, suggesting multiple physiological roles for this PDE in vivo.
A striking observation from our work is the close similarity between the novel fly PDEs and their mammalian homologues. Not only is there a close sequence identity between the catalytic domains (59–77%), but the regulatory and post-translational modification domains and motifs are also very similar. However, in common with many proteins encoded by the Drosophila genome, the Drosophila PDEs have long insertions at the N- and C-termini. Although the significance of these is unknown, it may suggest that further regulation may exist for Drosophila PDE function. Another example of this is demonstrated by comparing Drosophila PDE6 with mammalian retinal PDE6. Whilst all three mammalian retinal PDE6 catalytic subunits contain the CAAX-box prenylation motif, none has the proximal polybasic region contained in the Drosophila enzyme. Thus sequence information raises interesting questions regarding regulation, expression and post-translational modification of these novel PDEs.
We also present the first detailed biochemical analysis of novel Drosophila PDEs. The present study allows assignments of these Drosophila PDEs with their mammalian counterparts, based on biochemical function, in addition to sequence similarity. From these studies, we show that D. melanogaster PDE1 is a dual-specificity, Ca2+/calmodulin-regulated enzyme. Thus, in spite of having structural differences from mammalian PDE1C, e.g. the Drosophila enzyme contains only one calmodulin-binding site and not two (Figure 2, and see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm), this enzyme performs as bona fide PDE1.
PDE6 is the most sensitive enzyme to cGMP, although low cAMP-hydrolysing activity is detected in IP samples of this enzyme. However, given that PDE6 is the closet homologue of vertebrate PDE5 and that residues deemed critical to the PDE5 mode of action [20] are conserved within the PDE6 catalytic domain (Q935, Q893, A885 and W970, see Supplementary Figure 1 at http://www.BiochemJ.org/bj/388/bj3880333add.htm) allowed us to assign PDE6 as a cG-PDE.
PDE11 is a dual-specificity PDE, as is its vertebrate homologue. However, compared with PDE1 and PDE6, in Drosophila, PDE11 possesses the highest affinity for cGMP.
All of the novel Drosophila PDEs characterized in the present study display sensitivity to known inhibitors of vertebrate PDEs, i.e. zaprinast and sildenafil (Figure 6 and Table 3). This correlates well with previous studies that demonstrated sensitivity of cG-PDE activity and Malpighian (renal) tubule function to zaprinast [2] and sildenafil [13]. PDE6, in particular, displays nanomolar sensitivity to sildenafil, as well as submicromolar sensitivity to zaprinast.
Table 3. Comparison of known Drosophila cG-PDEs with vertebrate cG-PDEs.
Data from Figures 5–7 are tabulated to aid comparison with vertebrate homologues of Drosophila PDEs. Values for Km (cGMP and cAMP) and IC50 values for zaprinast and sildenafil are listed, where known, with appropriate references. ND, not determined.
| Enzyme | Km cGMP (μM) | Km cAMP (μM) | IC50 zaprinast (μM) | IC50 sildenafil (μM) |
|---|---|---|---|---|
| Drosophila PDE1 | 15.3±1 | 20.5±1.5 | 71±39 | 1.3±0.9 |
| PDE1C | 2.2±0.1 (Mouse [43]) | 3.5±0.3 (Mouse [43]) | 3.5±0.6 (Human [44]) | 1.1±0.275 (Bovine aorta PDE1 [45]) |
| Drosophila PDE6 | 37±13 | ND | 0.65±0.15 | 0.025±0.005 |
| PDE6β | 17±7 (Bovine [46]) | ND | 0.18±0.01 {Human (Ki) [47]} | 0.02±0.001 (Human (Ki) [47]) |
| Drosophila PDE11 | 6±2 | 18.5±5.5 | 1.6±0.5 | 0.12±0.06 |
| PDE11A | 0.52±0.34 (Human PDE11A1 [48]) | 1.04±0.23 (Human PDE11A1 [48]) | 12 (Human PDE11A1 [48]) | 3.8±0.75 (Human PDE11A4 [49]) |
Thus, taken together, the Drosophila PDEs have reassuring similarities but intriguing differences, at both the gene and protein level, compared with vertebrate PDEs. For example, although dunce and PDE4 are close homologues, dunce-encoded PDE is not inhibited by the PDE4-specific inhibitor, rolipram [42]. Thus, given the value of PDEs as drug targets, our findings may allow rational drug design based on identified structure–function relationships between vertebrate and Drosophila enzymes and may also allow strategies for novel insecticide discovery.
Furthermore, following on from the characterization studies, analysis of PDE function in vivo, using transgenesis and mutagenesis in Drosophila, will allow discovery of fundamental physiological processes governed by cG-PDEs.
Acknowledgments
This work was supported by grants from the U.K. Biotechnology and Biological Sciences Research Council (to S.-A. D. and J. A. T. D.), the U.K. Medical Research Council (to M. D. H.) and a Wellcome Trust studentship (to J. P. D.). We are grateful to S. Francis (Vanderbilt School of Medicine, Nashville, TN, U.S.A.) for the gift of sildenafil citrate; to S. Francis, N. J. Pyne and K. E. Broderick for helpful discussion; and to M. R. MacPherson and A. C. Godfrey for help with experiments.
References
- 1.Mullershausen F., Friebe A., Feil R., Thompson W. J., Hofmann F., Koesling D. Direct activation of PDE5 by cGMP: long-term effects within NO/cGMP signaling. J. Cell Biol. 2003;160:719–727. doi: 10.1083/jcb.200211041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broderick K. E., MacPherson M. R., Regulski M., Tully T., Dow J. A. T., Davies S. A. Interactions between epithelial nitric oxide signaling and phosphodiesterase activity in Drosophila. Am. J. Physiol. Cell Physiol. 2003;285:C1207–C1218. doi: 10.1152/ajpcell.00123.2003. [DOI] [PubMed] [Google Scholar]
- 3.Murthy K. S. Activation of phosphodiesterase 5 and inhibition of guanylate cyclase by cGMP-dependent protein kinase in smooth muscle. Biochem. J. 2001;360:199–208. doi: 10.1042/0264-6021:3600199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehats C., Andersen C. B., Filopanti M., Jin S. L., Conti M. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol. Metab. 2002;13:29–35. [Google Scholar]
- 5.Francis S. H., Turko I. V., Corbin J. D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2001;65:1–52. doi: 10.1016/s0079-6603(00)65001-8. [DOI] [PubMed] [Google Scholar]
- 6.Davis R. L., Dauwalder B. The Drosophila dunce locus: learning and memory genes in the fly. Trends Genet. 1991;7:224–229. doi: 10.1016/0168-9525(91)90369-2. [DOI] [PubMed] [Google Scholar]
- 7.Hansen G., Jin S., Umetsu D. T., Conti M. Absence of muscarinic cholinergic airway responses in mice deficient in the cyclic nucleotide phosphodiesterase PDE4D. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6751–6756. doi: 10.1073/pnas.97.12.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houslay M. D. PDE4 cAMP-specific phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:249–315. doi: 10.1016/s0079-6603(01)69049-4. [DOI] [PubMed] [Google Scholar]
- 9.Houslay M. D., Adams D. R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell J. M., Zhang H. T. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol. Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Spina D. The potential of PDE4 inhibitors in respiratory disease. Curr. Drug Targets Inflamm. Allergy. 2004;3:231–236. doi: 10.2174/1568010043343822. [DOI] [PubMed] [Google Scholar]
- 12.Brand A. H., Perrimon N. Targetted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 13.Broderick K. E., Kean L., Dow J. A. T., Pyne N. J., Davies S. A. Ectopic expression of bovine type 5 phosphodiesterase confers a renal phenotype in Drosophila. J. Biol. Chem. 2004;279:8159–8168. doi: 10.1074/jbc.M304679200. [DOI] [PubMed] [Google Scholar]
- 14.Dow J. A. T., Davies S. A. Integrative physiology, functional genomics and epithelial function in a genetic model organism. Physiol. Rev. 2003;83:687–729. doi: 10.1152/physrev.00035.2002. [DOI] [PubMed] [Google Scholar]
- 15.Dow J. A. T., Maddrell S. H., Davies S. A., Skaer N. J., Kaiser K. A novel role for the nitric oxide-cGMP signaling pathway: the control of epithelial function in Drosophila. Am. J. Physiol. 1994;266:R1716–R1719. doi: 10.1152/ajpregu.1994.266.5.R1716. [DOI] [PubMed] [Google Scholar]
- 16.Davies S.-A. Nitric oxide signalling in insects. Insect. Biochem. Mol. Biol. 2000;30:1123–1138. doi: 10.1016/s0965-1748(00)00118-1. [DOI] [PubMed] [Google Scholar]
- 17.MacPherson M. R., Broderick K. E., Graham S., Day J. P., Houslay M. D., Dow J. A., Davies S. A. The dg2 (for) gene confers a renal phenotype in Drosophila by modulation of cGMP-specific phosphodiesterase. J. Exp. Biol. 2004;207:2769–2776. doi: 10.1242/jeb.01086. [DOI] [PubMed] [Google Scholar]
- 18.MacPherson M. R., Lohmann S. M., Davies S. A. Analysis of Drosophila cGMP-dependent protein kinases and assessment of their in vivo roles by targetted expression in a renal transporting epithelium. J. Biol. Chem. 2004;279:40026–40034. doi: 10.1074/jbc.M405619200. [DOI] [PubMed] [Google Scholar]
- 19.Torrie L. S., Radford J. C., Southall T. D., Kean L., Dinsmore A. J., Davies S. A., Dow J. A. Resolution of the insect ouabain paradox. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13689–13693. doi: 10.1073/pnas.0403087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K. Y., Card G. L., Suzuki Y., Artis D. R., Fong D., Gillette S., Hsieh D., Neiman J., West B. L., Zhang C., et al. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol. Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Radford J. C., Davies S. A., Dow J. A. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. J. Biol. Chem. 2002;277:38810–38817. doi: 10.1074/jbc.M203694200. [DOI] [PubMed] [Google Scholar]
- 22.Bolger G. B., McCahill A., Huston E., Cheung Y. F., McSorley T., Baillie G. S., Houslay M. D. The unique amino-terminal region of the PDE4D5 cAMP phosphodiesterase isoform confers preferential interaction with beta-arrestins. J. Biol. Chem. 2003;278:49230–49238. doi: 10.1074/jbc.M303772200. [DOI] [PubMed] [Google Scholar]
- 23.Charbonneau H., Prusti R. K., LeTrong H., Sonnenburg W. K., Mullaney P. J., Walsh K. A., Beavo J. A. Identification of a noncatalytic cGMP-binding domain conserved in both the cGMP-stimulated and photoreceptor cyclic nucleotide phosphodiesterases. Proc. Natl. Acad. Sci. U.S.A. 1990;87:288–292. doi: 10.1073/pnas.87.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter M. F., Kiger J. A., Jr The Dunce gene of Drosophila: roles of Ca2+ and calmodulin in adenosine 3′:5′-cyclic monophosphate-specific phosphodiesterase activity. J. Neurosci. 1984;4:495–501. doi: 10.1523/JNEUROSCI.04-02-00495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonnenburg W. K., Seger D., Kwak K. S., Huang J., Charbonneau H., Beavo J. A. Identification of inhibitory and calmodulin-binding domains of the PDE1A1 and PDE1A2 calmodulin-stimulated cyclic nucleotide phosphodiesterases. J. Biol. Chem. 1995;270:30989–31000. doi: 10.1074/jbc.270.52.30989. [DOI] [PubMed] [Google Scholar]
- 26.Muradov K. G., Boyd K. K., Martinez S. E., Beavo J. A., Artemyev N. O. The GAFa domains of rod cGMP-phosphodiesterase 6 determine the selectivity of the enzyme dimerization. J. Biol. Chem. 2003;278:10594–10601. doi: 10.1074/jbc.M208456200. [DOI] [PubMed] [Google Scholar]
- 27.Resh M. D. Membrane targeting of lipid modified signal transduction proteins. Subcell. Biochem. 2004;37:217–232. doi: 10.1007/978-1-4757-5806-1_6. [DOI] [PubMed] [Google Scholar]
- 28.Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J. Struct. Biol. 2004;148:259–267. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 29.Ponting C. P., Aravind L. PAS: a multifunctional domain family comes to light. Curr. Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi M., Matsushima K., Ohashi H., Tsunoda H., Murase S., Kawarada Y., Tanaka T. Molecular cloning and characterization of human PDE8B, a novel thyroid-specific isozyme of 3′,5′-cyclic nucleotide phosphodiesterase. Biochem. Biophys. Res. Commun. 1998;250:751–756. doi: 10.1006/bbrc.1998.9379. [DOI] [PubMed] [Google Scholar]
- 31.Soderling S. H., Bayuga S. J., Beavo J. A. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc. Natl. Acad. Sci. U.S.A. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soderling S. H., Bayuga S. J., Beavo J. A. Identification and characterization of a novel family of cyclic nucleotide phosphodiesterases. J. Biol. Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 33.Corbin J. D., Turko I. V., Beasley A., Francis S. H. Phosphorylation of phosphodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic and allosteric cGMP-binding activities. Eur. J. Biochem. 2000;267:2760–2767. doi: 10.1046/j.1432-1327.2000.01297.x. [DOI] [PubMed] [Google Scholar]
- 34.Thomas M. K., Francis S. H., Corbin J. D. Substrate- and kinase-directed regulation of phosphorylation of a cGMP-binding phosphodiesterase by cGMP. J. Biol. Chem. 1990;265:14971–14978. [PubMed] [Google Scholar]
- 35.Wang J., Kean L., Yang J., Allan A. K., Davies S. A., Herzyk P., Dow J. A. Function-informed transcriptome analysis of Drosophila renal tubule. Genome Biol. 2004;5:R69. doi: 10.1186/gb-2004-5-9-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day J. P., Houslay M. D., Davies S. A. Cloning and characterisation of a novel cGMP-specific phosphodiesterase from Drosophila melanogaster. BMC Meeting Abstracts: 1st International Conference on cGMP: NO/sGC Interaction and its Therapeutic Implications, 1:p0014. 2003.
- 37.Fink T. L., Francis S. H., Beasley A., Grimes K. A., Corbin J. D. Expression of an active, monomeric catalytic domain of the cGMP-binding cGMP-specific phosphodiesterase (PDE5) J. Biol. Chem. 1999;274:34613–34620. doi: 10.1074/jbc.274.49.34613. [DOI] [PubMed] [Google Scholar]
- 38.Xu R. X., Hassell A. M., Vanderwall D., Lambert M. H., Holmes W. D., Luther M. A., Rocque W. J., Milburn M. V., Zhao Y., Ke H., et al. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 39.Xu R. X., Rocque W. J., Lambert M. H., Vanderwall D. E., Luther M. A., Nolte R. T. Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J. Mol. Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 40.Yamazaki A., Moskvin O., Yamazaki R. K. Phosphorylation by cyclin-dependent protein kinase 5 of the regulatory subunit (Pgamma) of retinal cgmp phosphodiesterase (PDE6): its implications in phototransduction. Adv. Exp. Med. Biol. 2002;514:131–153. doi: 10.1007/978-1-4615-0121-3_9. [DOI] [PubMed] [Google Scholar]
- 41.Ahumada A., Slusarski D. C., Liu X., Moon R. T., Malbon C. C., Wang H. Y. Signaling of rat frizzled-2 through phosphodiesterase and cyclic GMP. Science. 2002;298:2006–2010. doi: 10.1126/science.1073776. [DOI] [PubMed] [Google Scholar]
- 42.Henkel-Tigges J., Davis R. L. Rat homologs of the Drosophila dunce gene code for cyclic AMP phosphodiesterases sensitive to rolipram and RO 20-1724. Mol. Pharmacol. 1990;37:7–10. [PubMed] [Google Scholar]
- 43.Yan C., Zhao A. Z., Bentley J. K., Beavo J. A. The calmodulin-dependent phosphodiesterase gene PDE1C encodes several functionally different splice variants in a tissue-specific manner. J. Biol. Chem. 1996;271:25699–25706. doi: 10.1074/jbc.271.41.25699. [DOI] [PubMed] [Google Scholar]
- 44.Loughney K., Martins T. J., Harris E. A., Sadhu K., Hicks J. B., Sonnenburg W. K., Beavo J. A., Ferguson K. Isolation and characterization of cDNAs corresponding to two human calcium, calmodulin-regulated, 3′,5′-cyclic nucleotide phosphodiesterases. J. Biol. Chem. 1996;271:796–806. doi: 10.1074/jbc.271.2.796. [DOI] [PubMed] [Google Scholar]
- 45.Daugan A., Grondin P., Ruault C., Le Monnier de Gouville A. C., Coste H., Linget J. M., Kirilovsky J., Hyafil F., Labaudiniere R. The discovery of tadalafil: a novel and highly selective PDE5 inhibitor. 2: 2,3,6,7,12,12a-hexahydropyrazino[1′,2′:1,6]pyrido[3,4-b]indole-1,4-dione analogues. J. Med. Chem. 2003;46:4533–4542. doi: 10.1021/jm0300577. [DOI] [PubMed] [Google Scholar]
- 46.Gillespie P. G., Beavo J. A. Characterization of a bovine cone photoreceptor phosphodiesterase purified by cyclic GMP-sepharose chromatography. J. Biol. Chem. 1988;263:8133–8141. [PubMed] [Google Scholar]
- 47.Zhang J., Kuvelkar R., Wu P., Egan R. W., Billah M. M., Wang P. Differential inhibitor sensitivity between human recombinant and native photoreceptor cGMP-phosphodiesterases (PDE6s) Biochem. Pharmacol. 2004;68:867–873. doi: 10.1016/j.bcp.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 48.Fawcett L., Baxendale R., Stacey P., McGrouther C., Harrow I., Soderling S., Hetman J., Beavo J. A., Phillips S. C. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc. Natl. Acad. Sci. U.S.A. 2000;97:3702–3707. doi: 10.1073/pnas.050585197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weeks J. L., Zoraghi R., Beasley A., Sekhar K. R., Francis S. H., Corbin J. D. High biochemical selectivity of tadalafil, sildenafil and vardenafil for human phosphodiesterase 5A1 (PDE5) over PDE11A4 suggests the absence of PDE11A4 cross-reaction in patients. Int. J. Impot. Res. 2004;17:5–9. doi: 10.1038/sj.ijir.3901283. [DOI] [PubMed] [Google Scholar]