Abstract
Background: Gastric infection with Helicobacter pylori is the major cause of chronic active gastritis and is associated with the pathogenesis of peptic ulcer and gastric carcinoma. Gastric mucosal damage involves both host and H pylori dependent factors, such as the presence of the cag pathogenicity island and allelic variations of the vacA and iceA genes.
Aims: To evaluate the association of these virulence factors with the development of gastric malignancies, a retrospective study was performed on archived tissue routinely obtained for diagnostic histopathology.
Methods: DNA was extracted from formalin fixed, paraffin wax embedded gastric tissue sections of 93 patients with chronic active gastritis (n = 39), adenocarcinoma (n = 28), or mucosa associated lymphoid tissue (MALT) lymphoma (n = 24). The extracted DNA was used to perform a polymerase chain reaction based, simultaneous analysis of the following: (1) cagA status, (2) allelic variation of the iceA genes (iceA1, iceA2), allelic variation of the signal peptide (s1a, s1b, s2) and the midregion (m1, m1a, m2) of the vacA gene.
Results: The iceA1 gene showed a 3.6 fold and the vacA s1a variant a 4.2 fold higher prevalence in gastric adenocarcinoma than in gastritis. The combined presence of both the vacA s1a and iceA1 genes had a 5.6 fold higher frequency in adenocarcinoma. The vacA m2 allele was the predominant subtype in MALT lymphoma and the combination of the vacA m2 subtypes with the vacA s1 and the iceA1 variants occurred in MALT lymphoma nearly five times more often than in chronic active gastritis.
Conclusions: Certain H pylori subtype combinations possess a differentiating and predictive value for the development of gastric adenocarcinoma and MALT lymphoma.
Keywords: Helicobacter pylori, chronic active gastritis, gastric adenocarcinoma, mucosa associated lymphoid tissue lymphoma, virulence factors, cag pathogenicity island, cagA status, iceA, multiplex analysis
The human pathogen Helicobacter pylori is associated with the development of a variety of gastroduodenal diseases, such as chronic active gastritis (GA), peptic ulcer disease (PUD), gastric cancer (GC), and gastric non-Hodgkin’s lymphoma of mucosa associated lymphoid tissue (MALT lymphoma).1,2 Several prospective serological studies indicate that H pylori infection causes a three to sixfold higher risk for the development of GC.3,4 A seroepidemiological study with two large cohorts from different continents has also revealed a sixfold increased risk for the development of MALT lymphoma in patients with anti-H pylori antibodies compared with uninfected controls.5 However, in spite of the high prevalence of H pylori infections only a minority of infected individuals will develop one of the two malignancies. In addition to a variety of host and environmental factors, there are specific bacterial virulence genes that might determine the possible sequelae of infection by H pylori.6 In the past few years, many H pylori virulence factors have been described that contribute to the survival of H pylori in the acidic environment of the stomach and its colonisation of that organ. Other virulence factors associated with the development of disease, such as the vacA, cagA, and iceA genes are not present in all H pylori strains or show different allelic variations.6
The vacuolating cytotoxin (VacA) acts directly on the surrounding tissue, causing vacuolar degeneration of gastric epithelial cells by interfering with intracellular membrane fusion.6 The cytotoxin is encoded by the vacA gene, which shows sequence variations in two regions: the signal region and the midregion.7 The signal region has two different alleles (s1 and s2) and the midregion also has two (m1 and m2).8 The s1 region can be further subtyped into s1a or s1b.8 In Germany, another subtype of the vacA midregion (m1a) has been described as a variant of the m1 type.9
Although the vacA gene is present in every H pylori strain only about half of the strains produce the active cytotoxin.10,11 Studies of vacA diversity in different H pylori strains revealed that the production and activity of the cytotoxin is determined by the chimaeric combination of the signal region and midregion alleles of the gene.8 In vitro studies have shown that H pylori strains containing vacA s1/m1 produce large amounts of the cytotoxin, whereas vacA s1/m2 strains produce moderate amounts, and vacA s2/m2 strains little or no active vacuolating cytotoxin.8 In addition, vacA s2/m1 strains have recently been described, but are very rare.12
“Virulence factors associated with the development of disease, such as the vacA, cagA, and iceA genes are not present in all H pylori strains or show different allelic variations”
The cagA gene, which encodes an immunodominant antigen, is not present in all H pylori strains.13 It is part of the cag pathogenicity island, a 40 kb locus containing 31 genes, which are associated with the induction of interleukin 8 secretion by host epithelial cells.14 Some of the genes found within the cag pathogenicity island encode components of a type IV secretion machinery, which may be responsible for the export of H pylori effector proteins into the gastric target cells.15 The detection of the cagA gene indicates the presence of the entire cag pathogenicity island of the respective H pylori strain.14
Another important virulence determinant is the iceA gene, which can occur as an iceA1 or iceA2 variant.16 Both iceA alleles are flanked by the conserved genes, cysE and hpyIM, but they differ greatly in their genetic organisation and sequence.17 The expression of the iceA genes is induced by contact between H pylori and the epithelial cells of the stomach.16 The precise function of the IceA peptides is still unknown, although sequence homology suggests a role in endonuclease activity.17 Previous studies have shown that the presence of iceA1 is associated with increased mucosal interleukin 8 concentrations and a higher risk of developing PUD.16,18
Here, we describe a procedure for the simultaneous genotyping of the H pylori virulence determinants, vacA, cagA, and iceA, in formaldehyde fixed and paraffin wax embedded specimens. The polymerase chain reaction (PCR) assay opens up the possibility of testing for genomic H pylori subtypes without the need for microbiological cultures using tissue routinely obtained for diagnostic histopathology. Retrospective studies on wide panels of gastric tissue samples, taken from the files of the archives of pathology, can also be carried out using this method and might be useful for evaluating genomic H pylori risk factors in different diseases. In our present study, we compared the gene subtypes in gastric samples from patients with GA, GC, and low grade gastric MALT lymphoma. We show that certain combinations of different virulence determinants have a higher differentiating and predictive value for the development of gastric adenocarcinoma or MALT lymphoma when compared with single virulence factors alone.
MATERIAL AND METHODS
Tissue material and histology
Paraffin wax embedded specimens of gastric tissue were analysed from a total of 121 patients filed in the archives of the institutes for pathology of the University of Cologne (Germany) and the Johannes-Gutenberg-University of Mainz (Germany). Of these 121 patients, 91 were found to be H pylori positive by PCR and complete genotyping was carried out. The study population comprised 39 patients with GA, 28 patients with GC, and 24 patients with low grade MALT lymphoma. The mean ages were 55 years for the patients with GA, 60 years for those with GC, and 66 years for those with MALT lymphoma. Of the 28 patients with GC, 10 had a carcinoma of the intestinal type, 16 had a diffuse type carcinoma, and two were classified as mixed type carcinoma according to the Lauren classification. The patients with the low grade MALT lymphoma were all in clinical stage E1.
Histological analyses were performed independently by two experienced pathologists on 3 μm thick sections stained with haematoxylin and eosin and modified Giemsa. The presence of H pylori was assessed by histology and by single PCR amplification of the vacA and iceA alleles of H pylori. The PCR assays detecting different H pylori virulence target sequences were more sensitive than histology because in four cases of GC and five cases of MALT lymphoma H pylori infections were only detected by PCR.
DNA extraction from paraffin wax embedded gastric tissue
One to three 3 μm thick paraffin wax embedded sections were dewaxed in xylene by incubation at 65°C for 20 minute. The dewaxed sections were then washed in 500 μl 100% ethanol. After lysis in 200 μl proteinase K buffer (500 μg/ml proteinase K (Gibco, Gaithersburg, USA), 50mM Tris/HCl, pH 7.4, and 5mM EDTA, pH 8), nucleic acids were extracted by phenol/chloroform and subsequently precipitated with 300mM sodium acetate and isopropanol.
Each series of DNA extractions included sections taken from blocks of pure paraffin wax, which were cut in between each case. These sections were used as a contamination control because they were handled in between each specimen during all steps of the procedure. To assess whether a sufficient amount of DNA of adequate quality had been extracted we performed PCRs with primers for the human β globin locus (forward: ACA-CAA-CTG-TGT-TCA-CTA-GC and reverse: CAA-CTT-CAT-CCA-CGT-TCA-CC).
Primer and PCR design
For the detection of the H pylori vacA, cagA, and iceA genes in DNA extracted from the paraffin wax embedded tissue, we selected primers (table 1 ▶) based on the sequences of the vacA, cagA, and icA genes published in the Genbank database of the NCBI, controlled for their specificity by BLAST search, and synthesised by MWG-Biotech (Ebersberg, Germany).
Table 1.
Oligonucleotides used to detect the cagA gene, the iceA variants iceA1 and iceA2, and the signal regions s1 and s2 and mid regions m1 and m2 of the vacA gene
| Gene | Region | Primer | Primer sequence | Size (bp) PCR product |
| vacA | s1/ s2 | vacA1F | 5′-CTG-GT(C/T)-TAA-AGT-CGC-ACC-CTT-TGT-GC-3′ | s1: 120 |
| s2: 150 | ||||
| vacA1R | 5′-CAA-TGG-CTG-GAA-TGA-TCA-CGG-TTG-T(A/G)-3′ | |||
| vacA2F | 5′-CAA-ACA-CAC-CGC-AAA-ATC-AAT-CGC-CC-3′ | |||
| m1 | m1F1 | 5′-CAA-CAA-TCA-AGG-CAC-TAT-CAA-(C/T)TA-3′ | 301 | |
| m1R1 | 5′-CCG-CAT-GCT-TTA-ATG-TCA-TCA-G-3′ | |||
| m1F2 | 5′-TGG-TCC-GAG-GCG-GG(A/C)-AAG-T-3′ | |||
| m1R2 | 5′-TCA-TCA-GTA-TTT-CGC-ACC-ACA-C-3′ | |||
| m2 | m2F1 | 5′-TTT-GGA-GC(C/T)-CCA-GGA-AAC-ATT-G-3′ | 102 | |
| m2R1 | 5′-C(C/T)A-CAC-GCC-CAT-CTT-GGA-CAA-3′ | |||
| m2F2 | 5′-ACC-CTA-AA(C/T)-AGC-AAC-GCA-AGC-3′ | |||
| m2R2 | 5′-GAC-AAA-AAG-ATT-CAT-CGT-GCC-TT-3′ | |||
| cagA | cagA1F | 5′-CGT-TGA-TAA-GAA-(C/T)GA-TAG-GGA-TAA-C-3′ | 181 | |
| cagA1R | 5′-GAT-CCC-CAA-ATT-TCT-GAA-AGC-TCT-T-3′ | |||
| cagA2F | 5′-(C/T)GA-TAG-GGA-TAA-CAG-GCA-AGC-TT-3′ | |||
| cagA2R | 5′-CTG-AAA-GCT-CTT-TGT-GGA-AGA-TTC-3′ | |||
| iceA | iceA1 | ice1.1F | 5′-ATC-ATA-AAG-ACG-GCC-GCA-AAG-AT-3′ | 218 |
| ice1.1R | 5′-AT(A/G)-GGG-TCA-TAT-TGA-TAA-CA(A/G)-CC-3′ | |||
| ice1.2F | 5′-CCG-CAA-GGA-TGA-TT(C/T)-AAG-AGT-TTC-3′ | |||
| ice1.2R | 5′-GTC-ATA-TTG-ATA-ACA-(A/G)CC-CAC-ACA-3′ | |||
| iceA2 | ice2.1F | 5′-CGC-TGT-TTT-TCT-AGC-GGT-GTT-TTA-3′ | 247 | |
| ice2.1R | 5′-CAT-TGA-TCT-(A/G)TG-TTT-GTA-TGC-TTC-3′ | |||
| ice2.2F | 5′-CGG-TGT-TTT-AAT-GAG-(C/T)(A/G)G-TTG-CG-3′ | |||
| ice2.2R | 5′-ATG-CTT-CTT-TGA-AAA-TGG-TAT-GGC-3′ |
The primer sets were used in a nested PCR and generated amplification products with a maximum length of 301 bp (table 1 ▶). The combined use of primer sets in multiplex PCR was possible because each primer set required an annealing temperature of 58°C and all PCR products were distinguishable by size after their separation using submarine Tris/acetate/EDTA (TAE) electrophoresis in 3% agarose gels. The PCR reaction mix consisted of 5 μl extracted DNA, 5 μl dNTP (Roche, Mannheim, Germany; 1mM), 5 μl 10× PCR buffer (Roche; 25mM MgCl2, 100mM Tris/HCl, and 500mM KCl; pH 8.3 at 20°C), 1 μl 10μM sense primers, 1 μl antisense primers (10μM), and 0.2 μl Taq DNA polymerase (Roche; 5 units/μl) in a total volume of 50 μl. The samples were covered with mineral oil and PCR was performed in a thermal cycler (Biometra, Göttingen, Germany) starting with preincubation at 95°C for five minutes, then followed by 35 cycles of denaturation at 95°C for one minute, annealing at 58°C for one minute, and extension at 72°C for one minute. For the nested PCR amplification, 1 μl DNA template from the first PCR and 2 μl each of sense and antisense primers were used, and PCR was performed according to the PCR protocol described above. Aliquots of 10 μl of the amplified PCR products were resolved by electrophoresis in 3% agarose TAE gels.
The results of H pylori genotyping by multiplex PCR were validated previously by single PCR assays of each target sequence and subsequent sequencing of the respective amplicons, using sequence specific primers labelled with the IRD 800 fluorescence dye (MWG-Biotech). The sequences of the amplificates were analysed in an automated sequencer (LI-COR DNA analyser Gene Reader 4200; MWG-Biotech) and compared with the published data of the NCBI database by BLAST analysis.
Further analysis of the vacA s1 and m1 regions
The vacA signal regions s1 and s2 can be distinguished by the size of the amplified PCR product after gel electrophoresis. Further differentiation of vacA s1 into the subtypes s1a or s1b was performed by sequence analysis of the PCR vacA s1 amplificate and comparison with published s1a and s1b sequences (Genbank accession numbers Z26883, U05676, and AE001511).
The vacA m1a subtype, a variant of the m1 type described by Strobel et al,9 was distinguished from the m1 subtype by digestion of the m1 PCR product with the restriction enzymes Asp718 and RcaI (Roche), which produced fragments of 192 and 109 bp (m1, Asp718) or 100 and 201 bp (m1a, RcaI).
Statistical analysis
The correlations between the H pylori genotypes and GA, GC, or MALT lymphoma were analysed by the calculation of the odds ratio (OR).
RESULTS
Simultaneous detection of the genetic H pylori virulence subtypes
After extraction of DNA from the paraffin wax embedded gastric tissue of the 39 patients with GA, the 28 patients with GC, and the 24 patients with MALT lymphoma a single genotype of the vacA signal region and midregion, in addition to one of the two variants of the iceA gene were detected by PCR in all cases. The cagA gene was detected in 64 of the 91 cases.
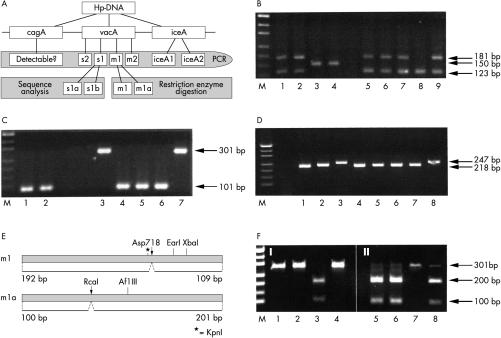
For the efficient genotyping of vacA, cagA, and iceA, oligonucleotide primers were designed in a way that allowed the simultaneous analysis of different subtypes by multiplex PCR. We selected primers that had the same annealing temperature (58°C) but produced PCR amplification products of different sizes (table 1 ▶), so that the primers specific for the different genomic virulence determinants could all be used in a single multiplex PCR assay and their products distinguished by gel electrophoresis. When this multiplex PCR amplification procedure was tested on a broad spectrum of diagnostic samples, we found that in some cases low amounts of amplificate gave PCR results that were difficult to interpret. However, the multiplex PCR assays (fig 1B–D ▶) with a limited number of primer sets produced reliable, distinct amplification products of all specimens, making it a potentially useful diagnostic tool for routine pathology testing. To this end, the seminested PCR for the detection of the vacA signal regions s1 and s2 was combined with the nested PCR for cagA showing products of 123 bp for vacA s1, 181 bp for cagA, and 150 bp for vacA s2 (fig 1B ▶). Simultaneous detection of the vacA midregions m1 (301 bp) and m2 (101 bp) or the iceA1 (218 bp) and iceA2 (247 bp) alleles was also routinely performed by combined PCR assays (fig 1C, D ▶).
Figure 1.
Multiplex analyses of the genomic Helicobacter pylori virulence determinants cagA, vacA, and ice A. (A) A schematic overview of the virulence genes and the method for their simultaneous detection. The cagA, vacA s1 or s2, vacA m1 or m2, and iceA 1 or 2 domains were amplified by the polymerase chain reaction (PCR) and detected by electrophoresis. The vacA s1 and m1 alleles were further analysed by sequencing or restriction enzyme digests. (B) Multiplex PCR assay of vacA signal regions s1 (123 bp) and s2 (150 bp), in addition to the cagA gene (181 bp). Samples in lanes 1, 2, 5, 6, 7, and 9 are vacA s1/cagA positive; those in lanes 3 and 4 are vacA s2 positive/cagA negative, and the sample in lane 8 is vacA s1 positive/cagA negative. (C) Combined PCR detection of vacA midregion m1 (101 bp) and m2 (301 bp). Helicobacter pylori in lanes 1, 2, and 4–6 are of the vacA m1 subtype and those in lanes 3 and 7 are of the vacA m2 subtype. (D) Simultaneous PCR analysis of the iceA1 (218 bp) and iceA2 (247 bp) alleles. All samples except lanes 3 and 8, which contain the iceA2 subtype, are iceA1 positive. (E) Schematic illustration of restriction enzyme polymorphisms within the m1 and m1a sequences. (F) Restriction enzyme digestion of vacA m1 PCR products with Asp718 (I) and RcaI (II). Lane 3 shows a vacA m1 subtype whereas lanes 5, 6, and 8 show m1a subtypes.
The vacA signal region s1 was further differentiated into the s1a or s1b subtype by sequence analysis. In Germany, another subtype of the vacA midregion, named m1a, has been described as a variant of the m1 type.9 To distinguish the m1a from the m1 subtype, we used a restriction enzyme polymorphism of the m1 subtypes, m1 and m1a. The restriction enzyme Asp718 cuts the m1 sequence, whereas RcaI cuts the m1a sequence only (fig 1E, F ▶). Before subtyping of vacA m1 by restriction enzyme digestion the vacA m1 PCR amplificate is called m1t (total: m1 and m1a).
Distribution of different H pylori genotypes in patients with GA, GC, or MALT lymphoma
Of the 91 patients analysed, 76 had the vacA signal region s1t (s1a and s1b), whereas the signal region s2 was found in only 15 patients. The prevalence of vacA s1t was highest in the GC group (93%), followed by the MALT lymphoma (83%), and GA (77%) groups. After differentiation of the s1t subtype into s1a and s1b by sequence analysis, vacA s1a was found to be the predominant type (70% of all patients), particularly in patients with GC (89%). The vacA s1b subtype was found in only 12% of studied tissues but in 25% of MALT lymphomas. One vacA s1 PCR product from a patient with MALT lymphoma contained sequence similarities of both vacA s1a and s1b; this isolate was finally considered to be untypable. The highest prevalence of vacA s2 was found among those patients with GA.
In general, vacA m2 was more prevalent than m1t (m1 and m1a). vacA m1t was found in 40% of the overall study population, being present in 61% of the patients with GC but only 17% of those with MALT lymphoma. In the MALT lymphoma group, 83% of patients were typed as vacA m2, whereas only 39% of those with GC were found to be vacA m2.
The cagA gene was frequently detected, and there was no significant difference between the three groups (63% of patients with MALT lymphoma, 69% of patients with GA, and 79% of those with GC).
In general, the iceA1 gene was more prevalent than iceA2. Patients with GA had the highest prevalence of iceA2 (44%).
Relation between the different combinations of H pylori subtypes and gastric diseases
To define risk factors for the development of GC or MALT lymphoma, in addition to single parameters (as described above), the different combinations of the various virulence genes were analysed for their correlation with the gastric malignancies investigated.
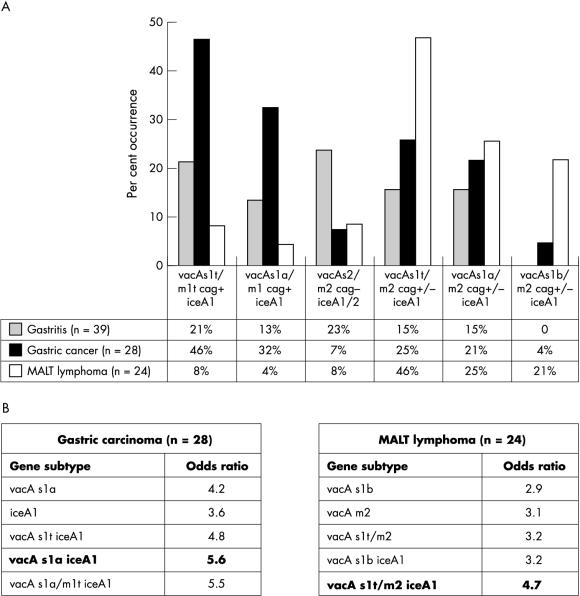
The vacA signal and midregions showed the most differential prevalence in GA, GC, and MALT lymphoma. The combined genotype vacA s1t/m1t cagA+ iceA1 occurred predominantly in patients with GC (46%) compared with 21% of those with GA and only 8% of those with MALT lymphoma. After further subdividing the signal and midregion of this gene subtype by sequence or restriction enzyme analysis, we found vacA s1a/m1 cagA+ iceA1 in 32% of the patients with GC, in 13% of those with GA, and in 4% of those with MALT lymphoma (fig 2A ▶).
Figure 2.
Helicobacter pylori genotypes in patients with chronic active gastritis (GA), gastric cancer (GC), and mucosa associated lymphoid tissue (MALT) lymphoma. (A) Distribution of vacA, cagA, and iceA subtype combinations in patients with GA, GC, and MALT lymphoma. The vacA s1a/m1 and iceA1 genes were more often detected in tissue from patients with GC than in tissue from patients with GA and MALT lymphoma. The allelic vacA s1t/m2 combination occurred primarily in patients with MALT lymphoma. The vacA s2/m2 cagA subtype combined with iceA1 or iceA2 was seen more frequently in patients with GA. (B) Odds ratios of the predominant H pylori genotypes in GC or MALT lymphoma.
Independent of the iceA status, vacA s2/m2 cagA(–) was relatively frequent in patients with GA (23%) but seldom found in patients with GC (7%) or MALT lymphoma (8%).
The presence of the vacA s1t/m2 subtype in combination with iceA1, regardless of the cagA status, predominated among the patients with MALT lymphoma (46%). The vacA s1b subtype combined with the vacA m2 and iceA1 gene was found in 21% of patients with MALT lymphoma, but only in 4% of those with GC and in none of the patients with GA.
The association of a certain subtype composition with GC or MALT lymphoma was clarified by comparison of the ORs of single virulence factors with their combinations found in gastric malignancies (fig 2B ▶). The iceA1 gene had a 3.6 fold higher prevalence and the vacA s1a a 4.2 fold higher prevalence in GC. However, the combination of both (the vacA s1a and iceA1 virulence determinants) in H pylori strains had a 5.6 fold higher prevalence in GC. The predominant single subtype in MALT lymphoma, vacA m2, had a 3.1 fold higher frequency than in GA, but the combination of vacA s1t/m2 and iceA1 (OR, 4.7) had a nearly fivefold higher frequency. Therefore, the combinations of the different H pylori virulence factor variants have to be taken into account to give a more reliable assessment of the risk of developing gastric malignancies.
DISCUSSION
The first aim of our study was to design a combined detection method for the H pylori virulence genes vacA, cagA, and iceA by means of PCR on paraffin wax embedded gastric tissue. The accessibility of paraffin wax embedded tissues allows the pathologist to genotype H pylori DNA using archival tissue specimens and also to compare H pylori genotypes with histology and clinical course.
“The vacA s1 positive H pylori genotype was the prominent subtype, being found in 93% of the patients with gastric cancer and 77% of those with chronic active gastritis”
When genotyping H pylori DNA extracted from cultured bacteria it is possible that specific H pylori strains may be favoured, whereas genotyping of DNA extracted from paraffin wax embedded tissue eliminates the pre-selection of strains or the modification of virulence genes by in vitro culture. However, to determine which H pylori genotypes are associated with different gastric diseases it is important to determine whether a given patient is infected with a single H pylori strain. Although it is possible that patients can be infected with more than one H pylori strain,19 we detected only single strain infections in a group of 23 patients, in whom H pylori genotyping was performed on specimens obtained from more than one site in the stomach, such as the antrum and corpus or cardia. These data correspond to the results of Miehlke et al, who showed by DNA fingerprinting that H pylori isolates collected from different stomach sites had identical genotypes.20
The vacA s1 positive H pylori genotype was the prominent subtype, being found in 93% of the patients with GC and 77% of those with GA. In patients with MALT lymphoma, vacA s1 was detected slightly more frequently than in patients with GA (table 2 ▶). Therefore, vacA s1 does not appear to be a high risk factor for the development of MALT lymphoma. However, the risk of developing GC is increased almost fourfold, particularly when the patient is infected with the s1a subtype (table 2 ▶). The vac s1t variant was frequently associated with the m1 allele (m1t), and H pylori strains were genotyped as vacA s1t/m1t in 61% of patients with GC compared with only 38% of those with GA. A correlation between vacA s1/m1 status and the development of GC has been demonstrated in studies from other European countries,21–23 whereas in most studies from Japan no correlation with gastric malignancies was found because of the almost exclusive prevalence of this subtype combination.24,25
Table 2.
Distribution of vacA and iceA subtypes and cagA positivity in biopsies from patients with gastritis, gastric carcinoma, or mucosal associated lymphoid tissue (MALT) lymphoma
| Gene subtype | Gastritis (n=39) | Gastric carcinoma (n=28) | MALT lymphoma (n=24) |
| vacA s1t | 30 (77%) | 26 (93%) | 20 (83%)* |
| vacA s1a | 26 (67%) | 25 (89%) | 13 (54%) |
| vacA s1b | 4 (10%) | 1 (4%) | 6 (25%) |
| vacA s2 | 9 (23%) | 2 (7%) | 4 (17%) |
| vacA m1t | 15 (38%) | 17 (61%) | 4 (17%) |
| vacA m1 | 11 (28%) | 11 (39%) | 1 (4%) |
| vacA m1a | 4 (10%) | 6 (21%) | 3 (13%) |
| vacAm2 | 24 (62%) | 11 (39%) | 20 (83%) |
| cagA positive | 27 (69%) | 22 (79%) | 15 (63%) |
| iceA1 | 22 (56%) | 23 (82%) | 18 (75%) |
| iceA2 | 17 (44%) | 5 (18%) | 6 (25%) |
vacA s1t is s1a and s1b; vacA m1t is m1 and m1a. *One case of MALT lymphoma was a mixed s1a/s1b subtype and was therefore classified as s1t, without subdivision into s1a or s1b.
The vacA midregion variant m1a has been described as a new m1 variant in Germany9 and was also detected in our study. It was more frequently detected among patients with GC than in those with GA, but differentiating the vacA m1t allele into m1 and m1a was not useful for predicting the risk of developing GC.
There was a strong association between the presence of cagA (as a marker gene for the cag pathogenicity island) and the vacA s1 subtype: none of the vacA s2 subtypes was found to be cagA positive and only 13% of all vacA s1 subtypes were cagA negative. This association of vacA s1 and cagA positivity has been shown previously.26 Although the presence of cagA has been reported to be a risk factor for GC in some European studies,21–23 we found the overall prevalence of cagA to be similar in patients with GA (69%), GC (79%), and MALT lymphoma (63%), and could find no correlation with a specific disease. The OR of 1.6 for cagA positivity indicates only a slightly higher risk of developing GC. Our data are in agreement with those of Louw et al, who found no correlation between cagA prevalence and GC risk.27 When compared with the other genes associated with H pylori virulence, cagA does not seem to be a specific risk factor for GC. The iceA1 allele, however, was detected predominantly in patients with GC (82%) and was associated with a 3.6 fold increased risk of developing GC. Two studies from South Africa also described GC as being associated with the presence of iceA1 positive H pylori.27,28 However, the association between iceA1 and GC could not be confirmed by studies from Japan,29 probably because of the regional differences of subtype occurrence.
In contrast to the data found in the patients with GC, there was a high prevalence of vacA m2 in tissues from the patients with MALT lymphoma: 83% of the patients with MALT lymphoma were infected by H pylori types vacA m2. In GA or in GC, however, only 62% and 39%, respectively, of H pylori strains were vacA m2 positive. Therefore, the presence of vacA m2 is associated with a threefold increased risk of developing MALT lymphoma.
Most H pylori strains in patients with MALT lymphoma were iceA1 positive, and the prevalence of iceA1 was similar in MALT lymphoma (75%) and GC (82%), but lower in patients with GA (56%). The presence of iceA1 was associated with a 2.3 fold increased risk of developing MALT lymphoma and Miehlke et al found a similar prevalence of iceA1 in patients with MALT lymphoma.30 In addition to iceA1, studies using serological detection methods have postulated that cagA is also a prognostic marker for the risk of developing MALT lymphoma.31–33 However, two reports using PCR based detection of cagA in cultured H pylori isolates found no correlation between cagA and MALT lymphoma.30,34 Although the in vitro culture of H pylori may lead to altered prevalence rates as a result of strain selection, the loss of genes, or a low culture rate,35 these data correspond to our findings, confirming that the cagA status is not a prognostic factor for the development of GC or of MALT lymphoma.
Take home messages.
We have developed a method for the simultaneous detection of several Helicobacter pylori virulence genes, which is convenient, economical, and applicable to formalin fixed, paraffin wax embedded tissue
The combined presence of vacA s1a and iceA1 appeared to be useful for identifying those patients at increased risk of developing gastric cancer
Helicobacter pylori strains harbouring the vacA s1t/m2 iceA1 genotype were associated with the highest risk for developing mucosa associated lymphoid tissue lymphoma
When evaluating the possible sequelae of gastric mucosal damage caused by H pylori infection, the composition of virulence subtypes should be taken into account
In our study, the main difference between the H pylori genotypes associated with GC or MALT lymphoma was in the occurrence of the vacA midregion and the vacA signal sequences, s1a and s1b. GC was frequently associated with the vacA s1a/m1 genotype, whereas MALT lymphomas were mainly associated with the vacA s1b/m2 genotype. In vitro analyses of toxic activities, caused by different H pylori strains in HeLa cells, have shown that toxic activity depends on the H pylori genotype.8 Helicobacter pylori strains containing vacA s1/m1 produce high amounts of toxic activity, whereas vacA s1/m2 strains produce moderate amounts, and vacA s2/m2 strains show no toxic activity.8 Recent studies using different epithelial cell types hosting H pylori suggest that the reduced H pylori toxicity caused by genetic vacA m2 variants in HeLa cells is a function of the host cell type used for the in vitro studies because in epithelial cells other than HeLa cells the H pylori vacA m2 strains were fully toxic.36 To understand the modulation of cytotoxin activity—for example, determined by the vacA midregion, further studies with different cell lines are required.
“The highest risk for developing mucosa associated lymphoid tissue lymphoma is thought to be associated with H pylori strains harbouring the vacA s1t/m2 iceA1 genotype”
Helicobacter pylori genotyping on archived gastric tissue revealed that H pylori strains with certain combinations of virulence subtypes are associated with GC or MALT lymphoma. Thus, the virulence subtype composition vacA s1a and iceA1 occurs mainly in GC (OR, 5.6), whereas the vacA s1t/m2 iceA1 combination (OR, 4.7) is found in MALT lymphoma, showing that the subtype composition has differentiating value. Therefore, when evaluating the possible sequelae of gastric mucosal damage caused by H pylori infection, the composition of virulence subtypes should be taken into account. The simultaneous detection of several virulence genes by the method described here is a convenient technique, with low costs, which is also applicable to formalin fixed, paraffin wax embedded tissue. Our present retrospective study suggests that the combined presence of vacA s1a and iceA1 is most effective for identifying those patients at increased risk of developing GC. However, the highest risk for developing MALT lymphoma is thought to be associated with H pylori strains harbouring the vacA s1t/m2 iceA1 genotype.
Acknowledgments
We thank D Lohfink for excellent technical support, and S Baldus and V Dries for support in histological analyses and critical discussions.
Abbreviations
GA, chronic active gastritis
GC, gastric cancer
MALT, mucosa associated lymphoid tissue
OR, odds ratio
PCR, polymerase chain reaction
PUD, peptic ulcer disease
TAE, Tris/acetate/EDTA
VacA, vacuolating cytotoxin
Footnotes
The first two authors contributed equally to this work.
REFERENCES
- 1.Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev 1997;10:720–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2002;2:28–37. [DOI] [PubMed] [Google Scholar]
- 3.Eurogast Study Group. An international association between Helicobacter pylori infection and gastric cancer. Lancet 1993;341:1359–62. [PubMed] [Google Scholar]
- 4.Hansson LE, Engstrand L, Nyren O, et al. Helicobacter pylori infection: independent risk indicator of gastric adenocarcinoma. Gastroenterology 1994;106:1398–400. [DOI] [PubMed] [Google Scholar]
- 5.Parsonnet J, Hansen S, Rodriguez L, et al. Helicobacter pylori infection and gastric lymphoma. N Engl J Med 1994;330:1267–71. [DOI] [PubMed] [Google Scholar]
- 6.Covacci A, Telford JL, Del Giudice G, et al. Helicobacter pylori virulence and genetic geography. Science 1999;284:1328–33. [DOI] [PubMed] [Google Scholar]
- 7.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol 1994;12:307–19. [DOI] [PubMed] [Google Scholar]
- 8.Atherton JC, Cao P, Peek RM, et al. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J Biol Chem 1995;270:17771–7. [DOI] [PubMed] [Google Scholar]
- 9.Strobel S, Bereswill S, Balig P, et al. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J Clin Microbiol 1998;36:1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leunk RD, Johnson PT, David BC, et al. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol 1988;26:93–9. [DOI] [PubMed] [Google Scholar]
- 11.Cover TL, Tummuru MKR, Cao P, et al. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem 1994;269:10566–73. [PubMed] [Google Scholar]
- 12.Letley DP, Lastovica A, Louw JA, et al. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J Clin Microbiol 1999;37:1203–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Covacci A, Censini S, Bugnoli M, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A 1993;90:5791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Censini S, Lange C, Xiang Z, et al. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A 1996;93:14648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odenbreit S, Püls J, Sedlmaier B, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 2000;287:1497–500. [DOI] [PubMed] [Google Scholar]
- 16.Peek RM, Jr, Thompson SA, Donahue JP, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Assoc Am Physicians 1998;110:531–44. [PubMed] [Google Scholar]
- 17.Figueiredo C, Quint WGV, Sanna R, et al. Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori. Gene 2000;246:59–68. [DOI] [PubMed] [Google Scholar]
- 18.van Doorn L-J, Figueiredo C, Sanna R, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 1998;115:58–66. [DOI] [PubMed] [Google Scholar]
- 19.Taylor NS, Fox JG, Akopyants NS, et al. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol 1995;33:918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miehlke S, Thomas R, Guiterrez O, et al. DNA fingerprinting of single colonies of Helicobacter pylori from gastric cancer patients suggests infection with a single predominant strain. J Clin Microbiol 1999;37:245–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basso D, Navaglia F, Brigato L, et al. Analysis of Helicobacter pylori vacA and cagA genotypes and serum antibody profile in benign and malignant diseases. Gut 1998;43:182–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miehlke S, Kirsch C, Agha-Amiri K, et al. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int J Cancer 2000;87:322–7. [PubMed] [Google Scholar]
- 23.van Doorn LJ, Figueiredo C, Rossau R, et al. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol 1998;36:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeda S, Ogura K, Yoshida H, et al. Major virulence factors, vacA and cagA, are commonly positive in Helicobacter pylori isolates in Japan. Gut 1998;42:338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito Y, Azuma T, Ito S, et al. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J Clin Microbiol 1997;35:1710–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudi J, Kuck D, Rudy A, et al. Helicobacter pylori vacA genotypes and cagA gene in a series of 383 H. pylori-positive patients. Z Gastroenterol 2000;38:559–64. [DOI] [PubMed] [Google Scholar]
- 27.Louw JA, Kidd MSG, Kummer AF, et al. The relationship between Helicobacter pylori infection, the virulence genotypes of the infecting strain and gastric cancer in the African setting. Helicobacter 2001;6:268–73. [DOI] [PubMed] [Google Scholar]
- 28.Kidd M, Peek RM, Lastovica AJ, et al. Analysis of iceA genotypes in South African Helicobacter pylori strains and relationship to clinically significant disease. Gut 2001;49:629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishiya D, Shimoyama T, Fukuda S, et al. Evaluation of the clinical relevance of the iceA1 gene in patients with Helicobacter pylori infection in Japan. Scand J Gastroenterol 2000;35:36–9. [DOI] [PubMed] [Google Scholar]
- 30.Miehlke S, Yu J, Schuppler M, et al. Helicobacter pylori vacA, iceA, and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease. Am J Gastroenterol 2001;96:1008–13. [DOI] [PubMed] [Google Scholar]
- 31.Eck M, Schmauβer B, Haas R, et al. MALT-type lymphoma of the stomach is associated with Helicobacter pylori strains expressing the CagA protein. Gastroenterology 1997;112:1482–6. [DOI] [PubMed] [Google Scholar]
- 32.Fischbach W, Jung T, Goebeler-Kolve M, et al. Comparative analysis of the Helicobacter pylori status in patients with gastric MALT-type lymphoma and their respective spouses. Z Gastroenterol 2000;38:627–30. [DOI] [PubMed] [Google Scholar]
- 33.Konturek PC, Konturek SJ, Starzyska T, et al. Helicobacter pylori–gastrin link in MALT lymphoma. Aliment Pharmacol Ther 2000;14:1311–18. [DOI] [PubMed] [Google Scholar]
- 34.De Jong D, van der Hulst RW, Pals G, et al. Gastric non-Hodgkin lymphomas of mucosa-associated lymphoid tissue are not associated with more aggressive Helicobacter pylori strains as identified by CagA. Am J Clin Pathol 1996;106:670–5. [DOI] [PubMed] [Google Scholar]
- 35.De Jong D, van Dijk WC, van der Hulst RWM. CagA+ H. pylori strains and gastric lymphoma. Gastroenterology 1997;113:2022–23. [PubMed] [Google Scholar]
- 36.Figura N, Vindigni C, Presenti L, et al. New acquisitions in Helicobacter pylori characteristics. Ital J Gastroenterol Hepatol 1998;30(suppl 3):254–8. [PubMed] [Google Scholar]