Abstract
The transcription factor dMyc is the sole Drosophila ortholog of the vertebrate c-myc protooncogenes and a central regulator of growth and cell-cycle progression during normal development. We have investigated the molecular basis of dMyc function by analyzing its interaction with the putative transcriptional cofactors Tip48/Reptin (Rept) and Tip49/Pontin (Pont). We demonstrate that Rept and Pont have conserved their ability to bind to Myc during evolution. All three proteins are required for tissue growth in vivo, because mitotic clones mutant for either dmyc, pont,or rept suffer from cell competition. Most importantly, pont shows a strong dominant genetic interaction with dmyc that is manifested in the duration of development, rates of survival and size of the adult animal and, in particular, of the eye. The molecular basis for these effects may be found in the repression of certain target genes, such as mfas, by dMyc:Pont complexes. These findings indicate that dMyc:Pont complexes play an essential role in the control of cellular growth and proliferation during normal development.
Keywords: repression, transcription
Myc proteins are essential regulators of growth, proliferation, and apoptosis in metazoans (1–3). These proteins act as transcription factors to control the expression of numerous target genes involved in growth, metabolism, and other processes (4–7). Less is known about the molecular mechanism that allows Myc to control the expression of these targets. In recent years, different modes of gene activation by Myc have been proposed, notably recruitment of chromatin remodelers (8), histone acetylases (e.g., ref. 9), or RNA pol II kinases (10), but the physiological relevance of these different factors for Myc-dependent biological functions needs to be demonstrated. We therefore set out to study the mechanisms of Myc-controlled growth and proliferation during normal development by using Drosophila as a model system. Initially, we focused on the interaction of Myc with two specific components of cofactor complexes, Tip48 and Tip49, because of the availability of null mutations in the corresponding genes [called reptin (rept) and pontin (pont) in flies, respectively].
Tip48 and Tip49 are closely related proteins that show a high similarity to the bacterial ATP-dependent AAA+ super family DNA helicase RuvB. Orthologs of Tip48 and Tip49 have been identified in plants, yeast, and animals (e.g., refs. 11 and 12). Different observations strongly suggest that one major function of the Tip proteins resides in the control of transcription. Initially, vertebrate Tip49 was found to be a Tata-binding protein-interacting protein (13–16); later Tip48 and Tip49 were also shown to interact physically with the different transcription factors β-catenin (11, 14), c-Myc (12), E2F1 (only Tip49) (17), and ATF2 (only Tip48) (18), raising the possibility that the Tip proteins could bridge basic transcription machinery and sequence-specific activators. Both proteins were also purified as part of several multiprotein complexes involved in transcriptional regulation: the Ino80 chromatin remodeling complex in yeast (19, 20), Polycomb repressive complex 1 in Drosophila (only Tip48) (21), the Tip60 HAT complex in vertebrates (22), and the Uri complex regulating nutrition-dependent gene expression in yeast and in vertebrates (23). Interestingly, three other proteins that were found to bind the N terminus of c-Myc share residence with Tip48 and Tip49 in the Ino80 (BAF53 and β-actin) (24) or Tip60 complex (transformation/transcription-domain-associated protein, BAF53, and β-actin) (24, 25). Further support for an involvement of Tip48 and Tip49 in transcription is provided by the observations that both proteins colocalize with c-Myc on the nucleolin promoter (26) and that elimination of Tip48 or Tip49 function in yeast rapidly affects the expression of a large number of targets (16, 27, 28). Such a transcriptional role is also consistent with the described genetic interactions between a tip48 mutation and β-catenin in zebrafish and interactions of tip48 and tip49 with a β-catenin-reporter system in Drosophila (11, 29); in both of these in vivo interactions, Tip48 behaved as a negative component and Tip49 behaved as a positive component of the Wg signaling cascade. Similar opposing activities were also documented in a human cell line by assaying the ability of the β-catenin–T cell factor complex to activate a reporter gene (11). A potential role for Tip49 in Myc-dependent functions was addressed in a recent study that examined the consequences of coexpressing wild-type or putative dominant-negative forms of Tip49 with c-Myc. Neither form had any effect on control cells, but both enhanced the apoptosis caused by overexpressed c-Myc, and they reduced the ability of c-Myc in combination with activated Ras to transform rat embryo fibroblasts, which indicates that, upon forced overexpression, Myc might require Tip49 activity (17).
In the present study we show that the physical interaction between Myc and Pont/Rept is conserved in flies, that pont/rept are essential for tissue growth in vivo, and that dmyc and pont show a strong genetic interaction. We further identify the gene mfas as a transcriptional target that is repressed by dMyc:Pont complexes. These studies provide the first evidence that Pont and Rept are essential cofactors for the normal functions of Myc in vivo.
Materials and Methods
Fly Lines and Clonal Analysis. Fly stocks were obtained from the Bloomington Stock Center (Indiana University), with the exception of dmP0/FM7 (30), pont5.1/TM3, Ser (putative null allele, referred to as pont in the text) (11), rept35/TM3 (putative null allele, referred to as rept in the text) (11), and dmPL35/FM7 (31); the revertant lines pontrev5 (used as control for pont5.1) and reptrevΔ23 (used as control for rept35) were generated in parallel with the null alleles pont5.1 and rept35 (with which they are isogenic) by precise excision of the P-element insertions 0229/05 and P1706 (11), respectively.
Mitotic recombination was induced by using the FLP/FRT method (32). Females y w hs-flp122; rept06945 FRT2A/TM6B were crossed with males w;P(ubi-GFP.nls)3L1 P(ubi-GFP.nls)3L2 FRT2A or w;M(3)i55 hs-nGFP FRT(2A)/TM6B (33). Females y w hs-flp122;FRT82B pont5.1/TM6B were crossed with males w;FRT82B P(Ubi-GFP(S65T)nls)3R or w;FRT82B P(w+, c-myc)87E Sb63 M3(96C) (34). Larvae were heat-shocked at 37°C for 1 h.
ey>dmPL35 = y w dmPL35 tub>FRT>(dmyc-cDNA) >FRT> GAL4/Y; +/CyO, ey-Flp; ey>dmP0 = w dmP0 tub>FRT>(dmyc-cDNA)>FRT>GAL4 ey-Flp/Y; and ey>dm+ = y w dm+ tub>FRT>(dmyc-cDNA)>FRT>GAL4 ey-Flp/Y. These different genotypes express a rescuing dmyc cDNA (35) in all parts of the body except for the head capsule, where Flp-mediated recombination eliminates the cDNA and allows expression of GAL4 instead.
Molecular Biology. The sequences for pGEX-dMyc (expressing GST fused to amino acids 46–507 of dMyc), pCasper-hsp-HA-dMyc [expressing three copies of the hemagglutinin (HA)-epitope tag fused to full-length dMyc, H-dMyc], pUAS-HA-dMyc, pUAS-AU1-Rept (expressing one copy of the AU1-epitope tag fused to full-length Rept, A-Rept), and pUAS-9E10-Pont (expressing three copies of the c-Myc epitope tag recognized by monoclonal antibody 9E10, M-Pont) are available upon request.
Cell Culture and Transfection. Drosophila Schneider S2 cells were grown at 25°C with Schneider medium (GIBCO) supplemented with 10% heat-inactivated FCS and 100 units of penicillin/streptomycin (GIBCO). For stable transfection, 2 × 105 cells per ml were seeded in a medium flask. The day after, the medium was removed and 10 μg of pCasper-hsp-HA-dMyc and 4 μg of pcP4 plasmids (36) were mixed with the Cellfectin reagent (GIBCO) in serum-free medium and added to the cells. After 16 h, complete medium was added, and pools of clones were selected for 2 weeks in α-amanitin (200 μg/ml).
For transient transfections, 106 S2 cells per ml were plated in a medium flask. One day later, 10 μg of plasmid was transfected with the Cellfectin reagent as described above. After 48 h, transfected cells were subjected to a heat shock at 37°C for 1 h. After 2 h, cells were harvested and lysates were prepared for Western blot analysis.
Western Blot and Antibodies. Transfected S2 cells were washed in PBS and lysed with a buffer containing 50 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 0.5% Tween 20, and protease inhibitors. Untransfected cells (5 × 107 cells per immunoprecipitation reaction) were lysed in 50 mM Hepes, pH 7.5, containing 0.1% Triton X-100, 250 mM NaCl, and protease inhibitors. Fifty yw larvae were homogenized and lysed (by shredding and sonication) in the same buffer. The different lysates were subjected to sonication for 30 sec on ice. After measurement of the protein concentration using the Bio-Rad kit, lysates were immunoprecipitated by using specific antibodies bound to Protein G-Sepharose (CL-4B, Amersham Pharmacia Biotech). After immunoprecipitation, proteins were separated by SDS/PAGE, electrotransferred to a nitrocellulose membrane, subjected to Western blot analysis with specific antibodies, and visualized by enhanced chemiluminescence (ECL-Plus, Amersham Pharmacia Biotech). Primary antibodies were rat anti-HA monoclonal antibody (Roche), mouse anti-AU1 monoclonal antibody (Covance, Richmond, CA), mouse anti-9E10 monoclonal antibody (Covance), rabbit anti-9E10 polyclonal antiserum (Santa Cruz Biotechnology), rabbit anti-Pont antiserum, guinea pig anti-Rept antiserum, and nonimmune hybridoma supernatant for control.
In Vitro Binding Assays. GST and GST–dMyc fusion proteins were produced in bacteria (BL21) after induction with 0.1 mM isopropyl β-d-thiogalactoside for 5 h at 37°C. Cells were washed, resuspended in STE buffer (100 mM NaCl/10 mM Tris·HCl, pH 8.0/1 mM EDTA) and sonicated for 2 min on ice. After centrifugation, the supernatants were incubated for 2 h with GST agarose beads (Amersham Pharmacia Biotech). After extensive washes with STE buffer, the GST proteins immobilized on the beads were separated by SDS/PAGE, and protein concentration was estimated by staining with Coomassie blue. Equal amount of GST and GST–dMyc proteins were used for in vitro binding assays with the different cell lysates. Binding was performed for 3 h at 4°C. After washing, the bound material was resuspended in Laemmli sample buffer and separated by SDS/PAGE. The proteins were then visualized by Western blot analysis.
Analysis of Adult Flies. Flies were reared under identical growth condition and were age-matched (3-day-old males) before weighing with a precision scale (range 0.001–10 mg; Mettler ME30). To determine ommatidial number, all ommatidia were counted from scanning electron micrographs of the indicated number of eyes from different animals. From the same photographs, the size of the ommatidia was calculated by measuring the area of 20 ommatidia located in the center of the eye using photoshop (Adobe Systems, San Jose, CA).
Results
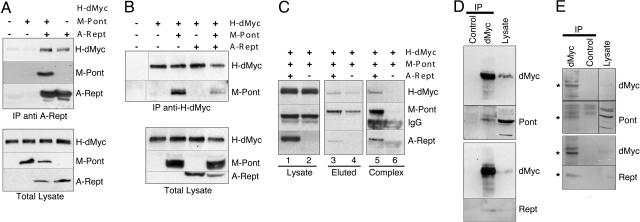
Physical Interaction Between dMyc and Pont/Rept. The interaction with human Tip48 and Tip49 requires a short sequence element in the N terminus of c-Myc called Myc Box 2. This functionally important Myc Box 2 is conserved in dMyc (37), suggesting that dMyc also has the potential to interact with Pont and Rept. Indeed, we could observe a specific interaction between the N terminus of dMyc and Pont or Rept in vitro (Fig. 4, which is published as supporting information on the PNAS web site). To demonstrate these interactions in cells, we coexpressed H-dMyc with M-Pont and/or A-Rept in Drosophila S2 cells and probed the lysates for the existence of different protein complexes. As shown in Fig. 1 A and B, all binary complexes could be observed. By using sequential immunoprecipitation with a 9E10 antibody, elution with 9E10 peptide, and reprecipitation with anti-AU1 antibodies, we could further demonstrate the existence of a ternary complex containing H-dMyc, M-Pont, and A-Rept (Fig. 1C). Although these experiments involved overexpression of the different proteins, we could also detect the interaction of endogenous dMyc with endogenous Pont or Rept (albeit more weakly), both from nontransfected S2 cells (Fig. 1D) and from third-instar larvae (Fig. 1E). These data demonstrate that dMyc forms complexes with Pont and Rept during normal development and that dMyc, Pont, and Rept can form a ternary complex in Drosophila cells.
Fig. 1.
Rept and Pont associate with dMyc in vivo. (A and B) M-Pont and/or A-Rept were transiently transfected into S2 cells stably expressing H-dMyc (as indicated). (Upper) Whole-cell lysates were immunoprecipitated with anti-AU1 antibodies for A-Rept (A) or anti-HA antibodies recognizing H-dMyc (B), followed by immunoblotting with anti-tag antibodies to detect the proteins indicated on the right. (Lower) Immunoblots of whole-cell lysates to reveal the relative expression levels of the indicated proteins. Positions of H-dMyc, M-Pont, and A-Rept, respectively, are indicated. The first lane in B Upper contains lysate of nontransfected S2 cells. (C) M-Pont and A-Rept (lanes 1, 3, and 5) or M-Pont alone (lanes 2, 4, and 6) were transiently transfected into S2 cells stably expressing H-dMyc. Cell lysates were incubated with 9E10 antibodies, and the immunoprecipitate was eluted with the 9E10 peptide (lanes 3 and 4). The eluate was then reimmunoprecipitated with anti-AU1 antibodies (lane 5 and 6), and the immunoprecipitate was analyzed by immunoblotting. (D and E) S2 cell lysates (D) or third-instar larval extracts (E) were incubated with anti-dMyc antibodies or control hybridoma supernatant. Immunoprecipitates were blotted with anti-dMyc antibodies, anti-Pont, or anti-Rept antisera as indicated. The rightmost lanes show immunoblots of whole-cell lysates. Asterisks indicate the migration of the endogenous proteins.
In the course of these experiments, we also noticed that, upon strong overexpression, M-Pont can compete with A-Rept for binding to dMyc in vitro and that the M-Pont:dMyc complex is more resistant to increasing NaCl concentrations than the A-Rept:dMyc interaction (Fig. 4). These observations suggest that Pont can also interact with dMyc in the absence of any bound Rept, although the physiological significance of such a complex is currently unclear.
Function of pont and rept in Vivo. If the physical interaction with Pont and Rept is important for dMyc function, we would expect mutations in pont and rept to affect some of the processes that are controlled by dMyc. However, currently there is only limited information available concerning the function of Tip48 and Tip49 in metazoans: Flies carrying null mutations in pont or rept were reported to die during early larval stages (11), whereas overexpression of putative dominant-negative versions of Tip49 in vertebrate cells showed no defects (12, 17). A closer inspection of larvae mutant for pont or rept further indicated that both genes might also control growth and proliferation. Homozygous pont-/- or rept-/- larvae hatch at normal rates, showing that the zygotic product of neither gene is essential for embryogenesis. The mutant larvae live for up to 7 days but grow only minimally in size (Fig. 2A); this phenotype is similar to that of mutants in growth-controlling genes (e.g., ref. 38) and to dmyc-null mutants (data not shown) (39). The defects of pont-/-rept-/- double mutants are very similar to the single mutants, although these larvae don't survive for longer than 5 days after egg deposition (Fig. 2A). Clones of pont-/- or rept-/- cells located within phenotypically wild-type imaginal discs grow poorly, indicating that the observed defects are cell-autonomous. Furthermore, these clones suffer from a phenomenon called “cell competition”: When surrounded by phenotypically wild-type cells, only a few small pont-/- or rept-/- clones survive at 72 h after the induction of the clones (Fig. 2B), and none can be observed at 120 h (data not shown). In contrast, when the cells surrounding the pont or rept clones are growth-impaired (because they carry a dominantly acting mutation in a gene coding for a ribosomal protein, a so-called Minute mutation), the mutant clones thrive even at 120 h after induction (Fig. 2C). Only few growth-controlling genes have been demonstrated to suffer from cell competition, notably the Minute loci and dmyc (30, 35, 40). This similarity of mutant phenotypes is consistent with the possibility that dMyc, Pont, and Rept control similar biological processes, as is the observation that all three genes are expressed in very similar patterns during embryogenesis (dynamically regulated in the mesoderm and midgut) and larval development (ubiquitously) (11, 30, 37). It should be noted that pont and dmyc also show some differences in their mutant phenotypes. Unlike dmyc mutant cells, pont-/- cells are not smaller than wild-type control cells, which is consistent with an involvement of Pont in additional processes that are less affected by dmyc mutations (e.g., in the control of cell-cycle progression).
Fig. 2.
Consequences of pont and/or rept inactivation in vivo. (A) The sizes of pont-/-, rept-/-, or rept-/-pont-/- homozygous mutant larvae are shown in comparison with those of wild-type larvae. Larvae of the indicated genotypes were reared at 25°C and photographed at the indicated times after egg deposition (in days). No rept pont double mutant larvae are alive after day 5. (B and C) Analysis of mitotic pont (B)or rept (C) clones in third-instar imaginal wing discs. Seventy-two hours after their induction, clones of cells with a homozygous mutation for pont (B) or rept (data not shown) are consistently smaller than their corresponding twin clones or even completely absent. (C)If the clones are induced in animals heterozygous for a Minute mutation, their growth is less disadvantaged and their size is still important 120 h after induction in wing imaginal discs, as shown for clones with a homozygous mutant for rept. (B Inset) A pont mutant clone is shown in higher magnification (outlined with white dots in B, the corresponding twin spot is marked by bright color and outlined with a solid line in B Inset). (C) The rept-/- clones are marked by the absence of color; the corresponding twin spots do not survive.
Genetic Interaction of dmyc with pont/rept in Vivo. The following sections provide direct evidence for a genetic interaction between dmyc and pont (and to a lesser extent rept) in vivo. Because the only available pont and rept alleles are recessive lethal, we tested for dominant interactions between pont and/or rept and hypomorphic dmyc alleles (called dm). The dmP0 allele is caused by a P-element insertion in the dmyc promoter, which results in a reduced dmyc expression; dmP0/Y mutant males are characterized by a slightly delayed development, thin bristles, and a weak reduction in body size and viability (Table 1) (30). The severity of this phenotype is dramatically enhanced when such dmP0/Y mutant flies also carry one mutant allele of pont: The viability drops from 54% to 12%, and the surviving flies take 2.4 days longer for their development and, despite the extended period, ultimately eclose with a significantly lower weight (Table 1). None of these defects are seen in control crosses with dm+ flies. rept does not show any dominant interaction with dmP0, and the pont rept double-mutant chromosome behaves like a pont single mutant. In addition, dmP0/Y; pont-/+ flies (but none of the other genotypes) occasionally show notches in the posterior wing margin (data not shown). A closer inspection reveals a significant decrease in the total wing area of dmP0/Y; pont-/+ flies compared with control, consistent with the functions of dMyc and Pont in growth control. A large part of this size difference can be attributed to a decrease in cell size (Table 3, which is published as supporting information on the PNAS web site).
Table 1. Phenotypes of male y w and dmP0 mutant flies carrying a pont and/or rept mutation.
| Genotype | Viability, % | Development, days | Weight, μg |
|---|---|---|---|
| y w/Y;+/+ | 98 (339) | 10.3 ± 0.6 (339) | 863 ± 56 (47) |
| y w/Y;pont–/+ | 108 (281) | 10.1 ± 0.6 (281) | 854 ± 37 (27) |
| y w/Y;rept–/+ | 92 (487) | 10.0 ± 0.5 (487) | 828 ± 44 (44) |
| y w/Y;rept–/+pont–/+ | 108 (363) | 10.1 ± 0.4 (363) | 852 ± 46 (33) |
| dmP0/Y;+/+ | 54 (287) | 10.6 ± 0.4 (287) | 772 ± 53 (46) |
| dmP0/Y;pont–/+ | 12 (55) | 13.0 ± 0.8 (55) | 693 ± 65 (21) |
| dmP0/Y;rept–/+ | 60 (301) | 10.8 ± 0.4 (301) | 774 ± 33 (29) |
| dmP0/Y;rept–/+pont–/+ | 20 (81) | 12.4 ± 0.7 (81) | 702 ± 70 (25) |
Viability is calculated as the percentage of the total of the expected genotype. Development indicates the time from egg deposition until eclosion. Values differing significantly (P < 0.01) from their control (dmP0/Y;+/+) are shown in bold. Standard deviations were calculated based on the total number of animals reported (indicated in parentheses). None of the y w/Y genotypes differed significantly from y w/Y;+/+. Note that relevant comparisons can only be made within the dm+(y w) and the dmP0 flies, because the two lines are not isogenic.
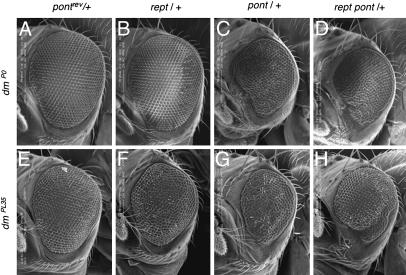
The Interaction of dmyc with pont and rept Is Essential for Eye Development. The most striking manifestation of the genetic interaction between dmyc and pont is seen in the eye. Approximately 90% of dmP0/Y; pont-/+ flies have small, irregularly shaped, and slightly rough eyes. Penetrance and expressivity of the phenotype are variable, but the anterior and ventral portions of the eye are always most defective (Fig. 3C). Interestingly, the same defect can also be observed in dmP0/Y flies, albeit at a very low frequency (ref. 30 and unpublished data), but not in dm+/Y flies that are heterozygous for pont. A quantitative analysis of adult eyes revealed a significant reduction in ommatidial size in all dmP0/Y mutant flies, which is significantly exacerbated by heterozygosity for pont (Table 2). In addition, dmP0/Y;pont-/+ eyes contain a reduced number of ommatidia, indicating an essential role of the dmyc/pont interaction for proliferation and growth during development.
Fig. 3.
Heterozygosity for pont or rept results in eye defects in hypomorphic dmyc mutants. Representative scanning electron micrographs of adult eyes. The genotypes shown are dmP0/Y; +/+ (A), dmP0/Y; rept-/+ (B), dmP0/Y; pont-/+ (C), dmP0/Y; rept-/+ pont-/+ (D), ey>dmPL35/Y; ±/+ (E), ey>dmPL35/Y; rept-/+ (F), ey>dmPL35/Y; pont+/+ (G), ey>dmPL35/Y; rept-/+ pont-/+ (H). All pictures are shown at the same magnification; anterior is to the left.
Table 2. Effect of the pont:dmyc interaction on eye and ommatidial size.
| Genotype | Ommatidia, n | Ommatidial size, μm2 | Area eye discs, μm2 × 103 |
|---|---|---|---|
| y w/Y;+/+ | 735 ± 18 (5) | 246 ± 5 (5) | 1.86 ± 0.26 (33) |
| y w/Y;pont–/+ | 748 ± 12 (5) | 241 ± 6 (5) | 1.74 ± 0.31 (51) |
| y w/Y;rept–/+ | 727 ± 12 (5) | 244 ± 5 (5) | 1.73 ± 0.19 (20) |
| y w/Y;rept–/+pont–/+ | 726 ± 12 (5) | 240 ± 9 (5) | nd |
| dmP0/Y;+/+ | 722 ± 18 (5) | 217 ± 8 (5) | 1.44 ± 0.15 (33) |
| dmP0/Y;pont–/+ | 561 ± 89 (7)* | 157 ± 16 (7)* | 0.90 ± 0.17 (51) |
| dmP0/Y;rept–/+ | 695 ± 33 (5) | 200 ± 5 (5)** | 1.51 ± 0.24 (17) |
| dmP0/Y;rept–/+pont–/+ | 532 ± 82 (7)* | 162 ± 7 (7)* | nd |
| ey>dmPL35/Y;+/+ | 685 ± 35 (5) | 194 ± 10 (5) | nd |
| ey>dmPL35/Y;pont–/+ | 591 ± 39 (5)* | 174 ± 7 (5)** | nd |
| ey>dmPL35/Y;rept–/+ | 637 ± 10 (5)** | 192 ± 12 (5) | nd |
| ey>dmPL35/Y;rept–/+pont–/+ | 583 ± 72 (5)** | 156 ± 9 (5)* | nd |
| ey>dm+;+/+ | 729 ± 10 (5) | 244 ± 8 (4) | nd |
| ey>dm+;UAS-H-dMyc/+ | 615 ± 17 (5)* | 329 ± 27 (5)* | nd |
| ey>dm+;UAS-M-Pont/+ | 737 ± 14 (5) | 234 ± 7 (5) | nd |
| ey>dm+;UAS-A-Rept/+ | 562 ± 88 (5)* | 226 ± 13 (5) | nd |
| ey>dmP0/Y;UAS-lacZ/+ | 738 ± 40 (10) | 208 ± 16 (10) | nd |
| ey>dmP0/Y;UAS-dMyc/+ | 615 ± 90 (10)* | 295 ± 11 (10)* | nd |
| ey>dmP0/Y;UAS-M-Pont/+ | 754 ± 37 (10) | 216 ± 7 (10) | nd |
| ey>dmP0/Y;UAS-A-Rept/+ | 535 ± 108 (10)* | 201 ± 26 (10) | nd |
| ey>dmP0/Y;pont–/+ | 514 ± 34 (4) | 169 ± 19 (4) | nd |
| ey>dmP0/Y;pont–/+ UAS-H-dMyc | 661 ± 31 (3)* | 312 ± 4 (3)* | nd |
| ey>dmP0/Y;pont–/+ UAS-M-Pont | 729 ± 46 (3)* | 227 ± 5 (3)* | nd |
| ey>dmP0/Y;pont–/+ UAS-A-Rept | 198 ± 95 (3)* | 167 ± 20 (3) | nd |
Standard deviations were calculated based on the total number of animals (reported in parentheses). Values differing significantly from their controls (lines 1, 5, 9, 13, 17, and 21 for samples 2–4, 6–8, 10– 12, 14–16, 18–20, and 22–24, respectively) are marked with asterisks. *, P < 0.01; **, P < 0.05. The genotypes are explained in the text. nd, not determined.
Similar genetic interactions were also observed with a second independent dmyc allele. ey>dmPL35 flies express the allele dmPL35 specifically in the eye (see Materials and Methods); these eyes are only moderately affected in their appearance, but heterozygosity for pont induces similar defects as described above (Table 2 and Fig. 5, which is published as supporting information on the PNAS web site). In addition, heterozygosity for rept also results in a weak reduction of ommatidial number in ey>dmPL35eyes, providing evidence that Rept might also act as a positive cofactor for dMyc.
As a further demonstration of specificity, the ectopic expression of M-Pont in the eyes of ey>dmP0;pont-/+ flies (Materials and Methods) fully rescued their defects (Table 2 and Fig. 5). Ectopic expression of H-dMyc also rescued the morphology defect of ey>dmP0;pont-/+ eyes but in addition produced gain-of-function phenotypes, such as an increase in ommatidial size. These gain-of-function effects are more pronounced in an ey>dmP0 or ey>dm+ background (Table 2 and Fig. 5), and they illustrate the documented abilities of dMyc to promote growth and apoptosis (ref. 30 and L. Montero and P.G., unpublished data).
In contrast, ectopic expression of A-Rept in ey>dmP0;pont-/+ flies enhanced penetrance and expressivity of their eye defects (Table 2 and Fig. 5). Even when overexpressed in an ey>dmP0 or an ey>dm+ background, A-Rept induced a rough eye appearance and a slight reduction in ommatidial number (Table 2 and data not shown); qualitatively similar results were obtained with two independent transgenes in ey>dm+ flies. These results indicate that A-Rept cannot substitute for Pont; instead, overexpression of A-Rept seems to have a dominant-negative effect on ommatidial number in an ey>dm+;pont+/+ background and on ommatidial size and number in an ey>dmP0;pont-/+ background. The basis for this effect is not known, but it is possible that the imbalance of Rept to Pont levels alters the composition of the multiprotein complexes that mediate the functions of these two proteins and thereby interferes with their function. It is further conceivable that endogenous Pont levels are relatively lower than Rept levels, which would explain why mild Pont overexpression does not cause any dominant-negative effects and why heterozygosity for pont results in a stronger genetic interaction with dmyc than heterozygosity for rept.
Cellular Basis of the Eye Defect in dmyc/pont Mutant Flies. A comparison of control, dmP0/Y and dmP0/Y;pont-/+ third-instar larval eye imaginal discs revealed no obvious defects in any of the three genotypes with respect to overall shape and the pattern of differentiating ommatidia (Fig. 6, which is published as supporting information on the PNAS web site). However, all dmP0/Y mutant discs are significantly smaller than the control discs, and the pont mutation (but not the rept mutation) further reduces their size; these data are in excellent agreement with the effects of the different genotypes on adult eye size (Table 2 and data not shown). This size reduction presumably reflects a reduction in cell size in the dmP0/Y eye discs as compared with control discs. Heterozygosity for pont further reduces this cell size and additionally reduces the number of cells.
This effect on cell number could be caused by a decreased rate of cell division and/or an increased rate of apoptosis. The different dmP0 genotypes contain similar numbers of mitotic cells (Fig. 6 and data not shown), suggesting at most minor differences in cell-cycle progression rates. However, a difference of only 5% in cell doubling time would be sufficient to explain the differences in cell number between the genotype with the fewest cells (dmP0/Y;pont-/+) and the genotype with the most cells (y w), and such a difference would have escaped our detection. In contrast, we did not find any indications for an involvement of apoptosis in this phenotype, because dmP0/Y;pont-/+ and dmP0/Y;pont+/+ discs do not show any difference in the number of apoptotic cells; in addition, expression of the viral caspase inhibitor p35 did not suppress the morphological defects in ey>dmP0/Y;pont-/+ eyes, nor did it affect the number of ommatidia in these eyes (Fig. 7, which is published as supporting information on the PNAS web site). Thus, we consider a proliferation defect in dmP0/Y;pont-/+ flies the most likely explanation for the observed reduction of ommatidia in the adult eyes.
Molecular Basis of the dMyc:Pont Interaction. An explanation for the observed genetic interaction might be found in the genes whose expression is controlled by dMyc:Pont complexes. To identify such genes, we combined microarrays with an RNA interference (RNAi) approach in S2 cells (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). As shown in Fig. 8, which is published as supporting information on the PNAS web site, Pont and Rept were found to control largely similar genes, but there is surprisingly little overlap with the dMyc targets; in particular, the genes activated by dMyc through canonical Myc binding sites called E-boxes (7, 41) were not significantly affected by RNAi against pont or rept. However, 11 genes were significantly and in the same direction affected by RNAi against all three proteins (8 up-regulated and 3 down-regulated) (Table 4, which is published as supporting information on the PNAS web site). Although we deem it unlikely that the misregulation of these genes explains all of the observed defects in the dm/Y;pont-/+ mutants, these genes may serve as examples for a larger class of genes that are controlled by dMyc:Pont[:Rept] complexes. To confirm these microarray results, we focused on one model target, midline fasciclin (mfas). Mfas is involved in cell–cell adhesion; its levels are up-regulated upon RNAi against dmyc, pont, or rept and rapidly down-regulated upon dMyc overexpression in vivo (7). Quantitative real-time PCR experiments showed that mfas levels are increased in dmP0/Y;pont-/+ double-mutant eye discs but not in single-mutant or wild-type control discs, confirming that mfas is also a target of dMyc:Pont complexes in vivo (Fig. 9, which is published as supporting information on the PNAS web site). To determine whether this effect is direct, we overexpressed H-dMyc in S2 cells and carried out chromatin immunoprecipitation experiments. As shown in Fig. 10 and Table 5, which are published as supporting information on the PNAS web site, under these conditions H-dMyc and endogenous Pont were found to be associated with a broad region of the mfas promoter (which does not contain any recognizable dMyc binding sites). These data suggest that dMyc:Pont complexes bind to certain target promoters such as mfas through as yet unidentified sequence motifs and mediate the transcriptional repression of these genes. Impairment of this process (through a combination of mutations in dmyc and pont) may then contribute to a reduction in cellular growth and proliferation.
Discussion
This study provides evidence that Tip49/Pont (and possibly Tip48/Rept) is an essential partner for Myc during normal development and that it plays an important role in the control of Myc-dependent transcription, growth, and proliferation. These conclusions are supported by four lines of evidence. First, we show that dMyc physically interacts with Rept and Pont in vitro, in cells, and in larvae. Although ternary complexes containing dMyc, Rept, and Pont can exist, we also provide evidence that dMyc can associate with Pont in the absence of Rept, although it is unclear whether such complexes lacking Rept have any physiological role in vivo. The stronger genetic interaction with pont raises the possibility that some of dMyc's functions might be mediated by such complexes, but the large degree of overlap between the targets of Pont and Rept and the fact that in most biochemically purified complexes Tip48 is accompanied by Tip49 suggest that most often these two proteins function together.
Second, flies lacking zygotic pont or rept gene products arrest their growth early during larval development, and mitotic clones homozygous mutant for pont or rept suffer from the same type of cell competition as do dmyc clones. These characteristics indicate a requirement for Pont/Rept for cellular proliferation and growth, which is consistent with their functioning as cofactors for dMyc.
Third, pont shows a strong dominant genetic interaction with dmyc. The causes for this interaction are likely to be defects in cellular growth and proliferation. The control of growth is most sensitive to variations in dMyc levels, because the moderate reduction of dMyc activity achieved in hypomorphic dmyc alleles already results in a decrease in cell size but not cell numbers. Removal of one copy of the pont gene exacerbates the growth defect and results in a reduction of cell numbers. We did not find any indication that apoptosis contributes to this reduction in cell number and, therefore, conclude that the cell number defects primarily reflect a proliferation defect. It is important to stress that none of these defects are seen in flies that are heterozygous for pont but wild-type for dmyc, arguing strongly against purely additive effect of the pont and dmyc mutations. Although we cannot strictly rule out the possibility that Pont and dMyc act in parallel growth-controlling pathways, such a dominant genetic interaction is indicative of close functional connections. We have not observed a dominant effect of the pont mutation on dMyc overexpression phenotypes (data not shown), suggesting that Pont is not limiting in situations of mildly increased dMyc levels. However, by using a vertebrate tissue culture system (Rat1 cells), Wood and colleagues (12) demonstrated that dominant-negative Pont/Tip49 inhibits the ability of human c-Myc to transform Rat1 fibroblasts in conjunction with activated Ras. Overexpression of a dominant-negative protein mutant potentially allows a stronger reduction of Pont/Tip49 activity than can be obtained in a heterozygous pont-/+ situation, and, thus, these experiments further reinforce our observation of a genetic interaction between myc and pont.
Fourth, we have shown that the expression of several genes, including mfas, is increased upon down-regulation of dmyc, pont, or rept in S2 cells and in dmyc/pont double-mutant eye imaginal discs in vivo. Chromatin immunoprecipitation experiments further suggest that mfas is a direct transcriptional target of Pont and dMyc.
Taken together, these data strongly argue that dMyc:Pont complexes are essential regulators of proliferation and growth in vivo and that they act at least partly by repressing the expression of target genes, such as mfas. A similar repressive function has recently also been found for Xenopus Pont and Rept (42); it was proposed that the well characterized repression of the transactivator Miz1 by c-Myc is mediated by Pont and Rept. Although it is tempting to speculate that Drosophila Pont functions analogously, no fly homolog of Miz1 has been identified. In addition, we currently do not know which of the reported Pont-containing complexes (see the Introduction) is responsible for the observed effect.
The function of Rept is less clear, because a rept mutant shows only a weak interaction and only with one dmyc allele. In contrast, overexpression of Rept strongly enhances the dmyc/pont mutant phenotypes. This observation could indicate that Rept also acts as antagonist of Pont and of dMyc:Pont complexes, analogously to what has been proposed for the interaction between Rept/Pont and β-Catenin (11). Alternatively, overexpression of Rept functions in a dominant-negative fashion, possibly by titrating Pont and/or other factors away from the multiprotein complexes in which they normally reside; in addition, Rept might be relatively more abundant than Pont such that heterozygosity for rept does not show any effects in most situations. Although we currently cannot rule out either explanation, our identification of mfas as a common target for dMyc, Pont, and Rept is more consistent with the latter possibility.
In conclusion, we have shown here that Pont, and possibly Rept, assists dMyc in the control of cellular proliferation and growth during normal development, presumably in part by repressing the expression of certain target genes.
Supplementary Material
Acknowledgments
We thank Lilian Montero for helpful discussions; Nadine Müller, Michael Furrer, Dominik Steiger, and Marco Ciro for assistance; and Konrad Basler and Andreas Trumpp for critical reading of the manuscript. This work was supported by a Marie-Heim-Vögtlein Fellowship (to P.B.) and grants from the Universität Zürich (to P.B.), the Swiss National Science Foundation and Fonds zur Förderung des Akademischen Nachwuchses/Schwyzer-Stiftung (to P.G.), the Centre National de la Recherche Scientifique, and La Ligue Contre le Cancer (to J.P.).
Author contributions: P.B. and P.G. designed research; P.B., T.H., S.B.D., F.U., D.A., and P.G. performed research; P.B., J.P., and D.A. contributed new reagents/analytic tools; P.B. and P.G. analyzed data; and P.G. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Pont, Pontin; Rept, Reptin; H-dMyc, HA-dMyc; M-Pont, 9E10-Pont; A-Rept, AU1-Rept.
References
- 1.Henriksson, M. & Luscher, B. (1996) Adv. Cancer Res. 68, 109-182. [DOI] [PubMed] [Google Scholar]
- 2.Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. (2002) Adv. Cancer Res. 84, 81-154. [DOI] [PubMed] [Google Scholar]
- 3.Green, D. R. & Evan, G. I. (2002) Cancer Cell 1, 19-30. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez, P. C., Frank, S. R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A. & Amati, B. (2003) Genes Dev. 17, 1115-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeller, K. I., Jegga, A. G., Aronow, B. J., O'Donnell, K. A. & Dang, C. V. (2003) Genome Biol. 4, R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel, J. H., Loboda, A. P., Showe, M. K., Showe, L. C. & McMahon, S. B. (2004) Nat. Rev. Cancer 4, 562-568. [DOI] [PubMed] [Google Scholar]
- 7.Orian, A., Van Steensel, B., Delrow, J., Bussemaker, H. J., Li, L., Sawado, T., Williams, E., Loo, L. W., Cowley, S. M., Yost, C., et al. (2003) Genes Dev. 17, 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, S. W., Davies, K. P., Yung, E., Beltran, R. J., Yu, J. & Kalpana, G. V. (1999) Nat. Genet. 22, 102-105. [DOI] [PubMed] [Google Scholar]
- 9.Park, J., Kunjibettu, S., McMahon, S. B. & Cole, M. D. (2001) Genes Dev. 15, 1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhardy, S. R. & Farnham, P. J. (2001) J. Biol. Chem. 276, 48562-48571. [DOI] [PubMed] [Google Scholar]
- 11.Bauer, A., Chauvet, S., Huber, O., Usseglio, F., Rothbacher, U., Aragnol, D., Kemler, R. & Pradel, J. (2000) EMBO J. 19, 6121-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood, M. A., McMahon, S. B. & Cole, M. D. (2000) Mol. Cell 5, 321-330. [DOI] [PubMed] [Google Scholar]
- 13.Kanemaki, M., Makino, Y., Yoshida, T., Kishimoto, T., Koga, A., Yamamoto, K., Yamamoto, M., Moncollin, V., Egly, J. M., Muramatsu, M. & Tamura, T. (1997) Biochem. Biophys. Res. Commun. 235, 64-68. [DOI] [PubMed] [Google Scholar]
- 14.Bauer, A., Huber, O. & Kemler, R. (1998) Proc. Natl. Acad. Sci. USA 95, 14787-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanemaki, M., Kurokawa, Y., Matsu-ura, T., Makino, Y., Masani, A., Okazaki, K., Morishita, T. & Tamura, T. A. (1999) J. Biol. Chem. 274, 22437-22444. [DOI] [PubMed] [Google Scholar]
- 16.Ohdate, H., Lim, C. R., Kokubo, T., Matsubara, K., Kimata, Y. & Kohno, K. (2003) J. Biol. Chem. 278, 14647-14656. [DOI] [PubMed] [Google Scholar]
- 17.Dugan, K. A., Wood, M. A. & Cole, M. D. (2002) Oncogene 21, 5835-5843. [DOI] [PubMed] [Google Scholar]
- 18.Cho, S. G., Bhoumik, A., Broday, L., Ivanov, V., Rosenstein, B. & Ronai, Z. (2001) Mol. Cell. Biol. 21, 8398-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen, X., Mizuguchi, G., Hamiche, A. & Wu, C. (2000) Nature 406, 541-544. [DOI] [PubMed] [Google Scholar]
- 20.Jonsson, Z. O., Jha, S., Wohlschlegel, J. A. & Dutta, A. (2004) Mol. Cell 16, 465-477. [DOI] [PubMed] [Google Scholar]
- 21.Saurin, A. J., Shao, Z., Erdjument-Bromage, H., Tempst, P. & Kingston, R. E. (2001) Nature 412, 655-660. [DOI] [PubMed] [Google Scholar]
- 22.Ikura, T., Ogryzko, V. V., Grigoriev, M., Groisman, R., Wang, J., Horikoshi, M., Scully, R., Qin, J. & Nakatani, Y. (2000) Cell 102, 463-473. [DOI] [PubMed] [Google Scholar]
- 23.Gstaiger, M., Luke, B., Hess, D., Oakeley, E. J., Wirbelauer, C., Blondel, M., Vigneron, M., Peter, M. & Krek, W. (2003) Science 302, 1208-1212. [DOI] [PubMed] [Google Scholar]
- 24.Park, J., Wood, M. A. & Cole, M. D. (2002) Mol. Cell. Biol. 22, 1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon, S. B., Van, B. H., Dugan, K. A., Copeland, T. D. & Cole, M. D. (1998) Cell 94, 363-374. [DOI] [PubMed] [Google Scholar]
- 26.Frank, S. R., Parisi, T., Taubert, S., Fernandez, P., Fuchs, M., Chan, H. M., Livingston, D. M. & Amati, B. (2003) EMBO Rep. 4, 575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson, Z. O., Dhar, S. K., Narlikar, G. J., Auty, R., Wagle, N., Pellman, D., Pratt, R. E., Kingston, R. & Dutta, A. (2001) J. Biol. Chem. 276, 16279-16288. [DOI] [PubMed] [Google Scholar]
- 28.Lim, C. R., Kimata, Y., Ohdate, H., Kokubo, T., Kikuchi, N., Horigome, T. & Kohno, K. (2000) J. Biol. Chem. 275, 22409-22417. [DOI] [PubMed] [Google Scholar]
- 29.Rottbauer, W., Saurin, A. J., Lickert, H., Shen, X., Burns, C. G., Wo, Z. G., Kemler, R., Kingston, R., Wu, C. & Fishman, M. (2002) Cell 111, 661-672. [DOI] [PubMed] [Google Scholar]
- 30.Johnston, L. A., Prober, D. A., Edgar, B. A., Eisenman, R. N. & Gallant, P. (1999) Cell 98, 779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourbon, H. M., Gonzy-Treboul, G., Peronnet, F., Alin, M. F., Ardourel, C., Benassayag, C., Cribbs, D., Deutsch, J., Ferrer, P., Haenlin, M., Lepesant, J. A., Noselli, S. & Vincent, A. (2002) Mech. Dev. 110, 71-83. [DOI] [PubMed] [Google Scholar]
- 32.Xu, T. & Rubin, G. M. (1993) Development (Cambridge, U.K.) 117, 1223-1237. [DOI] [PubMed] [Google Scholar]
- 33.Beuchle, D., Struhl, G. & Muller, J. (2001) Development (Cambridge, U.K.) 128, 993-1004. [DOI] [PubMed] [Google Scholar]
- 34.Zaffran, S., Chartier, A., Gallant, P., Astier, M., Arquier, N., Doherty, D., Gratecos, D. & Semeriva, M. (1998) Development (Cambridge, U.K.) 125, 3571-3584. [DOI] [PubMed] [Google Scholar]
- 35.De La Cova, C., Abril, M., Bellosta, P., Gallant, P. & Johnston, L. A. (2004) Cell 117, 107-116. [DOI] [PubMed] [Google Scholar]
- 36.Bellosta, P., Costa, M., Lin, D. A. & Basilico, C. (1995) Mol. Cell. Biol. 15, 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallant, P., Shiio, Y., Cheng, P. F., Parkhurst, S. M. & Eisenman, R. N. (1996) Science 274, 1523-1527. [DOI] [PubMed] [Google Scholar]
- 38.Galloni, M. & Edgar, B. A. (1999) Development (Cambridge, U.K.) 126, 2365-2375. [DOI] [PubMed] [Google Scholar]
- 39.Pierce, S. B., Yost, C., Britton, J. S., Loo, L. W., Flynn, E. M., Edgar, B. A. & Eisenman, R. N. (2004) Development (Cambridge, U.K.) 131, 2317-2327. [DOI] [PubMed] [Google Scholar]
- 40.Moreno, E. & Basler, K. (2004) Cell 117, 117-129. [DOI] [PubMed] [Google Scholar]
- 41.Hulf, T., Bellosta, P., Furrer, M., Steiger, D., Svensson, D., Barbour, A. & Gallant, P. (2005) Mol. Cell. Biol. 25, 3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etard, C., Gradl, D., Kunz, M., Eilers, M. & Wedlich, D. (2005) Mech. Dev. 122, 545-556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.