Abstract
The capacity to synthesize vitamin C (ascorbate) is widespread in eukaryotes but is absent from humans. The last step in the biosynthetic pathway involves the conversion of an aldonolactone substrate to ascorbate, a reaction catalyzed by members of an FAD-dependent family of oxidoreductases. Here we demonstrate that both the African trypanosome, Trypanosoma brucei, and the American trypanosome, Trypanosoma cruzi, have the capacity to synthesize vitamin C and show that this reaction occurs in a unique single-membrane organelle, the glycosome. The corresponding T. brucei flavoprotein (TbALO) obeys Michaelis–Menten kinetics and can utilize both l-galactono-γ-lactone and d-arabinono-γ-lactone as substrate, properties characteristic of plant and fungal enzymes. We could detect no activity toward the mammalian enzyme substrate l-gulono-γ-lactone. TbALO null mutants (bloodstream form) were found to display a transient growth defect, a trait that was enhanced when they were cultured in medium in which the essential serum component had been pretreated with ascorbate oxidase to deplete vitamin C. It is implicit, therefore, that bloodstream-form trypanosomes also possess a capacity for ascorbate transport.
Keywords: ascorbic acid, FAD, oxidase, Trypanosoma brucei
African sleeping sickness (Trypanosoma brucei), American trypanosomiasis (Trypanosoma cruzi), and cutaneous/visceral leishamaniasis (Leishmania spp.) are caused by protozoan parasites belonging to the family Trypanosomatidae. Over 30 million people are infected by these pathogens. Development of improved chemotherapy is a priority, because current treatments are problematic. Many drugs have toxic side effects, fail to clear parasitaemia, and are cost-prohibitive.
The pathways used by trypanosomatids to remove reactive oxygen species are distinct from mammals and have features that may be therapeutically exploitable. For example, trypanosomes detoxify superoxide anions via the activity of Fe-superoxide dismutases, an enzyme normally associated with prokaryotes (1, 2), whereas hydroperoxide metabolism is centered on the parasite-specific thiol trypanothione (3). In concert with the NADPH-dependent flavoprotein trypanothione reductase, trypanothione can detoxify hydroperoxides by nonenzymatic and enzyme-mediated mechanisms. The reduced thiol drives a series of two-component oxidoreductase cascades (4–6). The initial interaction involves transfer of reducing equivalents from trypanothione to an intermediary molecule such as tryparedoxin, glutathione, or ascorbate, which in turn facilitates reduction of the appropriate peroxidase. Several trypanosomal peroxidases have now been characterized, including an endoplasmic reticulum (ER)-localized hemoperoxidase from T. cruzi that uses ascorbate as an electron donor and functions by depleting H2O2 (6).
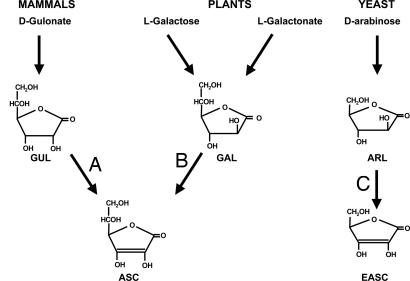
Ascorbate has been implicated in many biological processes. It is a cofactor for several enzymatic steps in the synthesis of collagen, neurotransmitters, peptide hormones, and carnitine (7, 8) and plays an important role in antioxidant defense at a number of levels. It can directly metabolize reactive oxygen species, acts to maintain α-tocopherol (vitamin E) in its reduced form, and mediates electron transfer to ascorbate-dependent peroxidases (8, 9). A number of pathways have evolved for synthesis of ascorbate (Fig. 1). Generally, animals use the uronic acid pathway (10), whereas several different mechanisms have been reported in plants (11–13). Yeasts are unusual in that they predominantly synthesize erythroascorbate, a five-carbon analogue of ascorbate that possesses all of its biochemical properties (14). The last step in all of the above pathways is common, involving the conversion of an aldonolactone substrate to ascorbate (or analogues), reactions catalyzed by a family of aldonolactone oxidase/dehydrogenases (Fig. 1). This group of flavoproteins can be divided into three classes, based on substrate specificity, electron acceptor, and subcellular location. Most mammals contain gulonolactone oxidase (GLO), a microsomal enzyme that synthesizes ascorbate from l-gulono-γ-lactone by using oxygen as electron acceptor (15, 16). In humans, the gene is defective, and the lack of GLO activity makes us susceptible to scurvy (17). The second class of enzyme is found in plants where several pathways generate l-galactono-γ-lactone (11, 12). This, in the presence of the electron acceptor cytochrome c, is converted to ascorbate by the activity of a mitochondrially located galactonolactone dehydrogenase (GALDH) (18). The third class of enzyme is typified by those in yeast mitochondria, where erythroascorbate is formed from d-arabinono-γ-lactone by the activity of arabinonolactone oxidase (ALO) by using oxygen as an electron acceptor (14, 19).
Fig. 1.
Ascorbate synthesis in mammals, plants, and yeast. The structures ofl-gulono-γ-lactone (GUL), l-galactono-γ-lactone (GAL), d-arabinono-γ-lactone (ARL), ascorbate (ASC), and erythroascorbate (EASC) are shown. The reactions catalysed by GLO (A), GALDH (B), and ALO (C) are indicated.
Trypanosomes contain significant levels of ascorbate (20), and regeneration from its oxidized dehydroascorbate form occurs by a nonenzymatic reaction involving trypanothione (6, 21). However, the role of ascorbate in trypanosomatid biology is poorly understood. Here we describe the properties and functional importance of an enzyme from T. brucei that catalyzes the last step in ascorbate synthesis. Furthermore, we demonstrate that this activity is localized to the glycosome, an unusual kinetoplastid-specific organelle.
Materials and Methods
Parasites. T. brucei [bloodstream form (BSF) (MITat 427 strain; clone 221a) and procyclic form (PCF) (strain 29–13)] (22–24) and T. cruzi (MHOM/BR/78/Silvio-X10/6) epimastigotes (6) were grown as described. DNA and RNA was extracted by using the DNeasy Tissue and RNeasy minikits (Qiagen, Valencia, CA), respectively.
Biochemical Properties. T. brucei ALO (TbALO) was amplified from genomic DNA with the primers TbALO4/TbALO5 (see Table 2, which is published as supporting information on the PNAS web site). The fragment was digested with BamHI/EcoRI, then cloned into the corresponding sites of the vector pTrcHis-C (Invitrogen). In this system, the expressed protein contains an N-terminal histidine tag and an epitope detectable with an anti-Xpress monoclonal antibody (Invitrogen). DNA was sequenced by using a dye terminator cycle sequencing kit (Applied Biosystems) and an Applied Biosystems Prism 377 sequencer. Escherichia coli BL-21+ was transformed with pTrcHis-TbALO and protein expression induced by isopropyl β-d-thiogalactoside. His-tagged TbALO was affinity purified on a Ni-NTA column (Qiagen). The cell lysis, column wash, and elution steps were carried out in the presence of protease inhibitors (Roche Applied Science, Indianapolis). Fractions were analyzed by SDS/PAGE and protein concentrations determined by the BCA protein assay system (Pierce).
TbALO activity was measured by following changes in absorbance at 550 nm because of the ascorbate-mediated reduction of cytochrome c (18). A standard 1-ml reaction containing 50 mM sodium phosphate, pH 7.5, 1 mM EDTA, 100 μM ferricytochrome c, and 1.2 μM TbALO was incubated at room temperature for 5 min. The background rate of cytochrome c reduction was determined, and the reaction initiated by the addition of d-arabinono-γ-lactone (Dextra Lab, Reading, U.K.), l-galactono-γ-lactone (Sigma), or l-gulono-γ-lactone (Acros, Geel, Belgium). Enzyme activity was calculated by using an ε value of 17.3 mM–1 and assumes that two molecules of ferricytochrome c are reduced per molecule ascorbate oxidized (18). Data were analyzed by fitting to a rectangular hyperbola (kinenort program, A. G. Clark, University of Wellington, Wellington, U.K.). The presence of flavin in TbALO was inferred from the spectral differences between the oxidized and reduced enzyme (25).
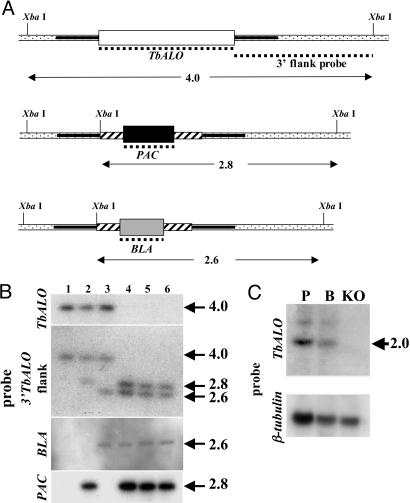
Construction of T. brucei Vectors and Parasite Transformation. The vectors used to delete TbALO, pKO-TbALO-PAC, and pKO-TbALO-BLA were generated by sequentially cloning TbALO flanking sequences either side of a puromycin or blasticidin resistance cassette (see Fig. 4A). The constructs were linearized with SacI/KpnI (pKO-ALO-PAC) or SacII/KpnI (pKO-ALO-BLA), then electroporated into BSF parasites (67). The vector used for localization was generated as follows: TbALO was amplified by using the primers TbALO12/TbALO7. The resultant fragment was digested with XbaI/BamHI and cloned into the tetracycline inducible vector pLEW-tagN (A. K. Ingram and D.H., unpublished data). Ligation was performed such that a 10-aa epitope (9E10) derived from human c-myc would be added to the N terminus of TbALO. The NotI linearized construct was electroporated into PCF parasites (26).
Fig. 4.
Targeted gene replacement of TbALO. (A) Diagram of the TbALO allele and the effects of gene replacement. The 5′ and 3′ flanking sequences (solid bars) were amplified and cloned sequentially either side of cassettes containing the selectable marker [puromycin (PAC) or blasticidin (BLA)] plus tubulin intergenic elements required for processing of mRNA (hashed boxes). The dotted lines correspond to probes used to check integration. The position of the predicted XbaI sites plus the band sizes (in kbp) obtained after hybridization are shown. (B) Autoradiographs of XbaI-digested genomic DNA from T. brucei 221 wild type (lane 1), ALO::PAC single knockout (lane 2), ALO::BLA single knockout (lane 3), and three independent ALO::PAC+BLA double TbALO knockout (lanes 4–6) cell lines. Blots were hybridized with labeled TbALO, 3′ TbALO flank, PAC, and BLA probes. Sizes given are in kbp. Integration was also verified by PCR (data not shown). (C) Blots containing 15 μgofT. brucei total RNA from procyclic form (P), bloodstream form (B), and ALO::PAC+BLA double knockout (KO) cell lines were hybridized with a labeled TbALO probe. Sizes given are in kb. RNA loading was judged by hybridization by using the T. brucei β-tubulin gene.
Phenotypic Analysis. To monitor parasite growth, dense cultures (>2 × 106 cells ml–1) of BSF trypanosomes (parental and null mutant) were seeded at 1 × 104 cells ml–1 and incubated at 37°C. Parasite growth was examined microscopically with subculturing to 1 × 104 cells ml–1 when necessary. We used modified Iscove's medium, where the serum had been pretreated with ascorbate oxidase, as indicated.
Localization. PCF T. brucei transformed with pLEW-tagN-TbALO were seeded at 5 × 105 cells ml–1 and incubated for 48 h with 1 μg ml–1 tetracycline. Parasites were fixed in 2% paraformaldehyde in PBS, then air dried onto microscope slides. Blocking using 50% horse serum/0.1% saponin in PBS was carried out before costaining with mouse anti-c-myc (9E10) antibody (1:200) (Santa Cruz Biotechnology) and rabbit anti-T. brucei GAPDH serum (1:400). Slides were washed extensively with 0.1% saponin in PBS, then incubated with Alexa-Fluor 488 goat anti-mouse and Alexa-Fluor 546 goat anti-rabbit serum (Molecular Probes) (both 1:400). DNA was stained with 200 pM DAPI (Sigma) in 50% glycerol in PBS, and slides were viewed by using a Zeiss LSM 510 confocal microscope. Fractionation studies were carried out on T. cruzi epimastigotes in the exponential phase of growth (6). Samples were collected and the activities of hexokinase, NADP+-dependent isocitrate dehydrogenase, trypanothione reductase, and ALO determined (5, 18).
Results
Isolation of a T. brucei ALO Gene. A sequence (AAX79383) in the T. brucei database was identified as containing an ORF related to GLO/ALO/GALDHs. Based on this, we amplified a 1,566-bp fragment with the potential to encode a 59-kDa protein (designated TbALO). The corresponding transcript is expressed in both BSF and PCF trypanosomes (see Fig. 4C). TbALO has highest identity (15–23%) with proteins that catalyze the final step in ascorbate synthesis (Fig. 2). Although there are significant differences among animal, plant, and fungal enzymes, one common feature is their use of FAD as a cofactor (27). There is an FAD-binding motif toward the N terminus of TbALO (residues 22–55) (pfam designation PF01565). This motif usually contains a histidine by which FAD is covalently attached via an 8-α-(N3-histidyl)-riboflavin linkage. However, TbALO lacks this histidine (residue 55), instead containing a lysine (Fig. 2). This is similar to plant GALDHs, where leucine is found at this site, leading to the suggestion that plant enzymes may interact noncovalently with FAD (28, 29). In the related flavoprotein vanillyl–alcohol oxidase (VAO), the equivalent histidine facilitates flavinylation at a second histidine residue near the C terminus (30). This second histidine is conserved at a corresponding position in all GLO/ALO/GALDH sequences reported (residue 462 in TbALO) (Fig. 2) and may perform a similar function as in VAO.
Fig. 2.
Sequence analysis of TbALO. Alignment of ALO sequences from T. brucei, T. cruzi (GenBank accession no. Tc00.1047053509179.100), and Saccharomyces cerevisiae (P54783), GALDH from Arabidopsis thaliana (T06690) and GLO from Rattus norvegicus (P10867). The residues common with the T. brucei sequence are highlighted in gray; dashes represent gaps in the sequence to optimize alignment. Amino acids that form the FAD-binding motif (red) (pfam designation PF01565) and the putative peroxisomal targeting signal (green) are highlighted. The second histidine (yellow) that may participate in flavinylation is shown.
TbALO has two other features of interest. First, sequences in the carboxyl region (residues 332–501) constitute a domain restricted to enzymes that synthesize ascorbate (Fig. 2) (pfam designation PF04030). There are no detailed reports on the mechanism by which these enzymes function, and the role of this region has not been elucidated. The second histidine alluded to above is found within this domain (Fig. 2). Another feature of interest is a putative targeting signal formed by the last three amino acids. This tripeptide (SHL) is reminiscent of a SKL-type signal that mediates glycosomal targeting (31). There are homologues of TbALO in T. cruzi, Leishmania major, and Leishmania infantum. They share 43–58% identity. As with TbALO, these enzymes contain a lysine in the FAD-binding motif, and all have the domain characteristic of ascorbate biosynthetic enzymes. The T. cruzi CL-Brener strain possesses two TbALO orthologues, whereas L. major and L. infantum have one. Both T. cruzi ALO enzymes have SKL-type signal at their C termini, whereas the Leishmania enzymes lack this targeting motif.
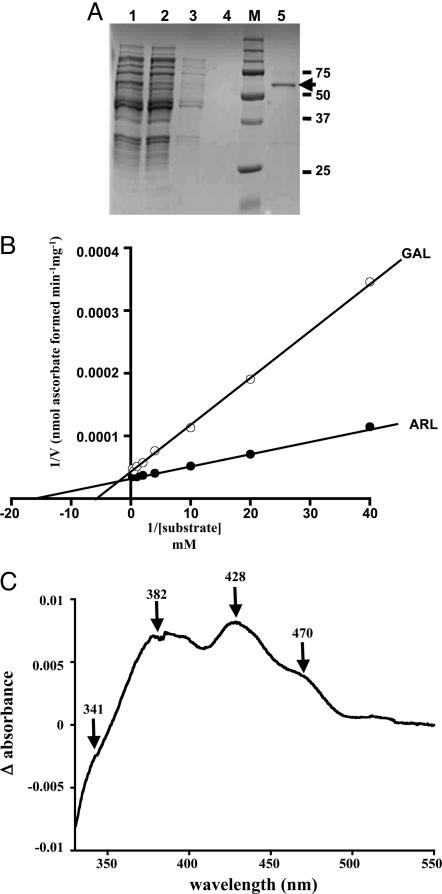
Biochemical Properties. To investigate TbALO activity, the gene was expressed in E. coli. A 60-kDa isopropyl β-d-thiogalactoside-inducible protein could be readily purified after one round of affinity chromatography (Fig. 3A). Fractions containing the recombinant protein were yellow, indicative of a flavoprotein. TbALO activity was assessed in the presence of various aldonolactone substrates. The parasite enzyme was shown to synthesize ascorbate only when d-arabinono-γ-lactone (a fungal metabolite) or l-galactono-γ-lactone (a plant metabolite) was used as substrate (Table 1). No significant activity was detected with l-gulono-γ-lactone, the preferred substrate for the mammalian enzyme (Table 1). Kinetic studies were carried out to investigate the interaction of TbALO with the fungal/plant metabolites. Double reciprocal plots of 1/TbALO activity against 1/[d-arabinono-γ-lactone] or 1/[l-galactono-γ-lactone] were linear (Fig. 3B) for substrate concentrations up to 4 mM. Extrapolation of the slopes allowed kinetic constants for both metabolites to be calculated (Table 1). The KM value toward l-galactono-γ-lactone is comparable to those of plant GALDHs (60–150 μM) (18, 28, 29). TbALO exhibited a higher affinity and activity toward d-arabinono-γ-lactone than to l-galactono-γ-lactone, as indicated by its higher apparent Vmax, turnover constant, catalytic efficiency, and lower apparent KM value. Interestingly, when TbALO activity was determined at substrate concentrations above 4 mM, inhibition was observed (data not shown), similar to that reported for sweet pea GALDH (18). For d-arabinono-γ-lactone, enzyme activity fell by 77% at 100 mM as compared with 2 mM. We also explored whether TbALO activity was inhibited by divalent metal ions, as observed with plant GALDH and yeast ALO activities (19, 29). Only copper (1 mM) appeared to show any degree of inhibition, reducing activity by 30% (data not shown), less than that observed for the plant/yeast enzymes.
Fig. 3.
Biochemical properties of TbALO. (A) Fractions obtained during the TbALO purification were resolved by SDS/PAGE (10% gel) and visualized after Coomassie staining. A clarified fraction of an E. coli (pTrcHis-TbALO) lysate after 6-h isopropyl β-d-thiogalactoside induction (lane 1) was loaded on a Ni-NTA column and the flow-through collected (lane 2). The column was washed extensively with 20 mM imidazole (lanes 3 and 4) and the recombinant protein eluted with 200 mM imidazole (lane 5). Markers (M) are in kilodaltons. The band of 60 kDa (indicated) corresponds to recombinant TbALO. (B) TbALO activity was assayed by following the ascorbate-dependent reduction of ferricytochrome c (18) in the presence of 1.2 μM TbALO and d-arabinono-γ-lactone (ARL) (0.025–4 mM) or l-galactono-γ-lactone (GAL) (0.025–4 mM) (Materials and Methods). (C) The absorbance spectrum (330–550 nm) of 1 μM TbALO was determined (oxidized). The sample was then saturated with argon and d-arabinono-γ-lactone (2 mM) added. After 2 h, the absorbance spectrum of TbALO (reduced) was examined. The difference between the oxidized and reduced spectra was then plotted.
Table 1. Kinetic parameters of vitamin C formation by TbALO.
| Substrate | Michaelis—Menten constant (KM), μM | Activity (Vmax), nmol ascorbate formed min-1mg-1 | Percent activity | Turnover constant (Kcat), s-1 | Catalytic efficiency (Kcat/KM), M-1s-1 |
|---|---|---|---|---|---|
| d-arabinono-γ-lactone | 55 ± 3 | 29,661 ± 366 | 100 | 27.2 | 4.9 × 105 |
| l-galactono-γ-lactone | 154 ± 24 | 22,786 ± 838 | 77 | 20.9 | 1.4 × 105 |
| l-gulono-γ-lactone | ND | 995 | 3 | ND | ND |
Enzyme activity (Vmax) was calculated using an ε value of 17.3 mM-1 (18). Kcat assumes one catalytic site per 60-kDa monomer. ND, not determined.
During purification of recombinant TbALO, yellow fractions were always associated with maximum enzyme activity. The spectral properties of oxidized TbALO were compared with the reduced enzyme to determine whether it displayed a flavin-binding signature (Fig. 3C). A series of peaks were observed, indicating the presence of a flavin cofactor (25). As with the yeast enzyme, the presence of a shoulder at 341 nm indicated covalent binding of the flavin group to the enzyme, despite the replacement of the histidine residue in the conserved FAD-binding motif by lysine (Fig. 2).
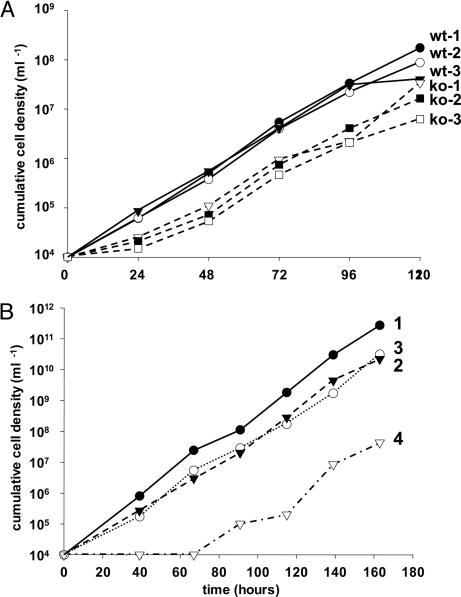
Properties of TbALO Null Mutants. To further assess the role of ascorbate in BSF T. brucei, both copies of the TbALO gene were deleted from the genome (Fig. 4 A and B). Northern blotting confirmed that the resulting cells no longer expressed TbALO mRNA (Fig. 4C). By inference, TbALO is nonessential to BSF parasites under normal culture conditions. We first investigated the effect on parasite growth. Over the initial 24-h period, null mutant cells consistently grew slower (70%) than wild-type parasites, but only when dense cultures (>2 × 106 cells ml–1) were used as the inoculum (Fig. 5); when using cultures with lower cell densities, this was not observed. After this initial lag period, the null mutants grew at the same rate as controls (≈10-fold in 24 h). Serum is an essential component of T. brucei growth medium and contains ≈40 μM ascorbate (32). In a second series of experiments, we used a medium where the serum had been pretreated with ascorbate oxidase, an enzyme that converts ascorbate to the oxidized unstable dehydroascorbate form (Fig. 5). Wild-type cells in this depleted medium also exhibited a 24-h lag period before growing at a rate comparable to that in normal medium. When TbALO null mutants were cultured in this pretreated medium, the lag period was extended to between 48 and 65 h. In all cases, the mutant clones then proceeded to grow out at a rate comparable to control cells. This suggests that depletion of ascorbate has an adverse effect on growth and further implies that normally parasites can scavenge ascorbate.
Fig. 5.
Growth of wild-type and TbALO mutants. (A) Cumulative density of wild-type (wt) and null mutant (KO) BSF parasites grown in standard medium over 5 days. (B) Cumulative density of BSF parasites grown in standard or ascorbate oxidase pretreated medium. 1, T. brucei wild type in standard medium; 2, T. brucei wild type in pretreated medium; 3, T. brucei null mutants grown in standard medium; 4, T. brucei null mutants in pretreated medium. A second null mutant clone displayed similar growth (data not shown).
A number of other phenotypic properties were also investigated. We examined whether the inability to synthesize ascorbate altered sensitivity to oxidants (H2O2), superoxide generating agents (phenazine methosulfate and paraquat), and trypanocidal drugs that are believed to mediate part of their activity by the production of reactive oxygen species (nifurtimox, benznidazole, and gentian violet). For all agents tested, null mutants seeded from exponentially growing cultures showed no significant difference when compared with wild-type controls (data not shown). The ability of mutants to differentiate from the BSF into the PCF was also assessed. This was achieved in vitro by subjecting BSF trypanosomes to a temperature drop to 27°C in medium containing citrate and cis-aconitate (33). Under these conditions, the null mutant could readily undergo differentiation. We also examined whether the TbALO mutants could infect mice. In these experiments, the mutant cell line produced a lethal infection in CD1 mice 3–4 days postinoculation, similar to wild type.
Localization of ALO in Trypanosomatids. A distinctive feature of TbALO is the presence of a C-terminal SKL-type glycosomal targeting sequence (SHL). To assess subcellular location, TbALO was cloned into an inducible expression vector downstream of the 9E10 epitope. The construct was then introduced into PCF cells that had been modified to support tetracycline-inducible expression (24). We could detect a 60-kDa band that was specific to extracts derived from induced cultures (Fig. 6A). Parasites expressing the recombinant protein were then fixed and stained for the 9E10 tag. This generated a highly punctate pattern throughout the cell body, suggestive of a glycosomal location. To confirm this, cells were costained with antiserum raised against glycosomal GADPH and the DNA dye DAPI (Fig. 6B). When the images were superimposed, the pattern of colocalization indicated that TbALO is glycosomal (Fig. 6B4).
Fig. 6.
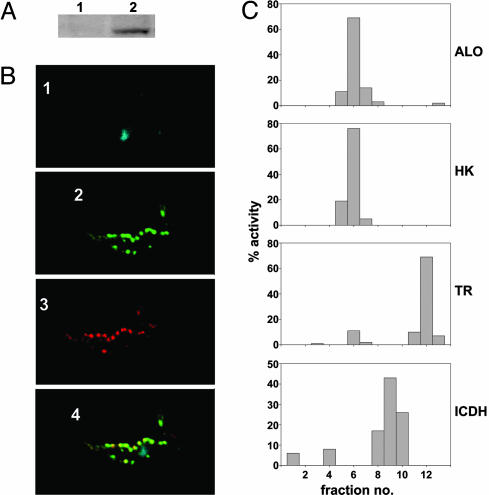
Localization of ALO to the glycosome of T. brucei and T. cruzi. (A) Blot containing lysates from 1 × 107 T. brucei PCF cells expressing epitope-tagged TbALO from uninduced (lane 1) and tetracycline-induced (lane 2) cultures probed with a c-myc (9E10) monoclonal antibody. Protein loading was judged by Coomassie staining. (B) Tetracycline-induced T. brucei PCF cells expressing epitope-tagged TbALO were stained with DAPI (blue; 1), anti-9E10 (green; 2), and antiglycosomal GAPDH (red; 3). When the pattern of TbALO and glycosomal GAPDH staining was superimposed, colocalization was observed (yellow; 4). (C) T. cruzi epimastigotes (1 × 1010) were homogenized by using silicon carbide and fractionated on a continuous sucrose gradient (0.4–2 M). Fractions (0.75 ml) were collected and assayed for ALO, hexokinase (HK), trypanothione reductase (TR), and isocitrate dehydrogenase (ICDH) activity (Materials and Methods). The percent activity collected in each sample was determined.
To investigate whether ALO has the same subcellular location in other trypanosome species, a homogenized T. cruzi extract was fractionated on a continuous sucrose gradient (0.4–2 M). Fractions were collected, then assayed for hexokinase (glycosomal marker) NADP+-dependent isocitrate dehydrogenase (mitochondrial marker) and trypanothione reductase (cytosolic marker) activities (Fig. 6C). In these gradients, hexokinase activity was found in fractions 5–7, whereas the predominant trypanothione reductase activity was associated with fractions 11–13. NADP+-dependent isocitrate dehydrogenase activity was predominantly in fractions between the glycosomal and cytosolic markers (fractions 8–10). Maximal ALO activity was found in fractions with the highest hexokinase activity. It can therefore be inferred that TcALO, whose C-terminal tripeptide is SHL, is also glycosomal.
Discussion
This study demonstrates that trypanosomes possess the capacity to synthesize vitamin C. The enzyme that catalyzes the final step in the biosynthetic pathway (TbALO) has a substrate specificity restricted to d-arabinono-γ-lactone and l-galactono-γ-lactone, compounds in which the hydroxyl groups on the ring structure are arranged in a trans-configuration (Fig. 1). This specificity is similar to the corresponding FAD-dependent oxidoreductases in plants and fungi (18, 19) but distinct from enzymes of animal origin, which can utilize aldonolactone substrates with both cis- and trans-configurations.
TbALO is FAD-dependent, displays Michaelis–Menten kinetics (Fig. 3B), and has a higher affinity toward d-arabinono-γ-lactone (Table 1). The yeast enzyme also exhibits this preference, with the result that erythroascorbate is the predominant form of vitamin C in this organism. If d-arabinono-γ-lactone is present in significant amounts in trypanosomes, vitamin C synthesis may therefore occur by a fungal route. Other fungal-like pathways have been reported in trypanosomes, notably those associated with lipid metabolism (34, 35). However, examination of the T. brucei genome has identified sequences that suggest that ascorbate synthesis proceeds via l-galactose and l-galactono-γ-lactone (ref. 11; S.R.W. and J.M.K., unpublished data), metabolites characteristic of the plant pathway (Fig. 1). Previously, we have noted that other components of enzyme-mediated oxidative defense in trypanosomes share surprising similarity with their plant counterparts, including an ascorbate-dependent hemoperoxidase from T. cruzi (6, 36). Genome analysis now strongly suggests that the presence of these and other “plant-like” enzymes, notably those involved in carbohydrate metabolism, represent evidence that trypanosomatids possessed a plastid at some point in their evolution (37). Despite subsequent loss of the organelle, many of the plastid genes have been retained and, interestingly, several appear to encode proteins that are targeted to the glycosome. It is tempting to speculate that components of the ascorbate synthetic pathway may have such an origin.
In both T. brucei and T. cruzi, we have shown that ALO is glycosomal, and that vitamin C synthesis therefore represents a previously uncharacterized role for this unique organelle. Glycosomes are single-membrane-bound structures related to peroxisomes and are the sites where glycolysis, β-oxidation of fatty acids, lipid ether synthesis, and purine salvage are compartmentalized (38). Generally, plant GALDH and yeast ALO are mitochondrial enzymes, whereas their mammalian counterparts are ER-localized (14, 16, 18). One reason that loss of ascorbate synthetic capacity may have undergone positive selection in humans and other primates is that it reduces the oxidative burden within the ER (39). GLO generates both a reductant (ascorbate) and an oxidant (H2O2) at equimolar levels. Glutathione-dependent peroxidases are the major H2O2-metabolizing enzymes in the ER (40), and loss of GLO activity could therefore act to maintain the level of reduced glutathione. This could be important, because the level of reduced glutathione in the ER is low compared with other subcellular compartments (41). The glycosomal oxidative defense system includes a nonselenium glutathione-dependent peroxidase that is essential for parasite survival (5, 42). One reason for sequestration of TbALO in the glycosome could be to maintain a close association with this peroxide-metabolizing system.
In humans, ascorbate deficiency results in scurvy, a disease caused by the inability to crosslink collagen. GLO-negative mice display severe vascular problems in the absence of dietary vitamin C (43). When ascorbate levels in plants are reduced, similar problems of structural maintenance are observed (44). This is accompanied by adverse effects on cell division and alterations in sensitivity toward ozone and photooxidative stress (44–46). In trypanosomes, TbALO null mutants were found to exhibit a lower initial growth rate compared with controls. This “lag” period was exacerbated when the essential serum component of the culture medium had been pretreated with ascorbate oxidase (Fig. 5). In plants, ascorbate may play a role in the cell cycle by controlling transition from G1 to S phase, although the mechanism(s) has yet to be fully explored (44). Our finding that the growth inhibition in T. brucei is transient suggests that ascorbate may have a role in initiating cell proliferation but thereafter is nonessential. Alternatively, the parasites may undergo a biochemical adaptation during this “lag” period so that another reductant is then able to compensate for lack of ascorbate, and normal growth can resume. It is also implicit from our data (Fig. 5) that trypanosomes can take up ascorbate from the culture medium, and that this capacity is able to complement the biosynthetic deficiency. Generally, vitamin C transport can occur by two mechanisms: uptake of dehydroascorbate, which is mediated by members of the glucose transporter family, and uptake of ascorbate, which is carried out by a sodium-dependent vitamin C transporter (47, 48). In human serum, 99% of vitamin C is in the reduced form (32). It is likely, therefore, that transport involves predominantly ascorbate, at least in the case of BSF parasites.
Surprisingly little is known about the role of vitamin C in trypanosome biology. As in other organisms, it is likely to be a significant free radical scavenger and to be involved in several other key physiological reactions. In T. cruzi, ascorbate acts as a shuttle molecule that transfers reducing equivalents from trypanothione to the ER-localized hemoperoxidase TcAPX (6). The functional relevance of this redox pathway has been demonstrated by the observation that overexpression of the enzyme confers increased resistance to exogenous H2O2. Genes related to TcAPX are also present in Leishmania species but not in T. brucei. This absence could reflect the host environment; T. cruzi and Leishmania are intracellular parasites, whereas African trypanosomes are extracellular. As a result, ascorbate-based antioxidant defense during infection may be more crucial in T. cruzi and Leishmania than in T. brucei. Further examination of these pathways is therefore warranted to establish whether aspects of ascorbate biochemistry in these important pathogens have potential as chemotherapeutic targets.
Supplementary Material
Acknowledgments
We thank Fred Opperdoes (Université Catholique de Louvain, Brussels) for the anti-GAPDH antibody and members of the T. brucei genome project for sequencing data. This study was supported by the Wellcome Trust.
Author contributions: S.R.W. and J.M.K. designed research; S.R.W., S.R.P., M.C.T., and D.H. performed research; S.R.W. and J.M.K. analyzed data; and S.R.W. and J.M.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office. C.C.W. is a guest editor invited by the Editorial Board.
Abbreviations: ER, endoplasmic reticulum; ALO, arabinonolactone oxidase; GLO, gulonolactone oxidase; GALDH, galactonolactone dehydrogenase; BSF, bloodstream form; PCF, procyclic form; TbALO, T. brucei ALO.
References
- 1.Temperton, N. J., Wilkinson, S. R. & Kelly, J. M. (1996) Mol. Biochem. Parasitol. 76, 339–343. [DOI] [PubMed] [Google Scholar]
- 2.Kabiri, M. & Steverding, D. (2001) Biochem. J. 360, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairlamb, A. H. & Cerami, A. (1992) Annu. Rev. Microbiol. 46, 695–729. [DOI] [PubMed] [Google Scholar]
- 4.Nogoceke, E., Gommel, D. U., Kiess, M., Kalisz, H. M. & Flohe, L. (1997) Biol. Chem. 378, 827–836. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson, S. R., Meyer, D. J., Taylor, M. C., Bromley, E. V., Miles, M. A. & Kelly, J. M. (2002) J. Biol. Chem. 277, 17062–17071. [DOI] [PubMed] [Google Scholar]
- 6.Wilkinson, S. R., Obado, S. O., Mauricio, I. L. & Kelly, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 13453–13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine, M., Rumsey, S. C., Daruwala, R., Park, J. B. & Wang, Y. (1999) J. Am. Med. Assoc. 281, 1415–1423. [DOI] [PubMed] [Google Scholar]
- 8.Halliwell, B. (2001) Mutat. Res. 475, 29–35. [DOI] [PubMed] [Google Scholar]
- 9.Asada, K. (1992) Physiol. Plant 85, 235–241. [Google Scholar]
- 10.Isherwood, F. A., Chen, Y. T. & Mapson, L. W. (1954) Biochem. J. 56, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler, G. L., Jones, M. A. & Smirnoff, N. (1998) Nature 393, 365–369. [DOI] [PubMed] [Google Scholar]
- 12.Lorence, A., Chevone, B. I., Mendes, P. & Nessler, C. L. (2004) Plant Physiol. 134, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agius, F., Gonzalez-Lamothe, R., Caballero, J. L., Munoz-Blanco, J., Botella, M. A. & Valpuesta, V. (2004) Nat. Biotechnol. 21, 177–181. [DOI] [PubMed] [Google Scholar]
- 14.Huh, W. K., Lee, B. H., Kim, S. T., Kim, Y. R., Rhie, G. E., Baek, Y. W., Hwang, C. S., Lee, J. S. & Kang, S. O. (1998) Mol. Microbiol. 30, 895–903. [DOI] [PubMed] [Google Scholar]
- 15.Brush, J. S. & May, H. E. (1966) J. Biol. Chem. 241, 2907–2912. [PubMed] [Google Scholar]
- 16.Puskas, F., Braun, L., Csala, M., Kardon, T., Marcolongo, P., Benedetti, A., Mandl, J. & Banhegyi, G. (1998) FEBS Lett. 430, 293–296. [DOI] [PubMed] [Google Scholar]
- 17.Sato, P. & Udenfriend, S. (1978) Arch. Biochem. Biophys. 187, 158–162. [DOI] [PubMed] [Google Scholar]
- 18.Oba, K., Ishikawa, S., Nishikawa, M., Mizuno, H. & Yamamoto, T. (1995) J. Biochem. (Tokyo) 117, 120–124. [DOI] [PubMed] [Google Scholar]
- 19.Huh, W. K., Kim, S. T., Yang, K. S., Seok, Y. J., Hah, Y. C. & Kang, S. O. (1994) Eur. J. Biochem. 225, 1073–1079. [DOI] [PubMed] [Google Scholar]
- 20.Clark, D., Albrecht, M. & Arevalo, J. (1994) Mol. Biochem. Parasitol. 66, 143–145. [DOI] [PubMed] [Google Scholar]
- 21.Krauth-Siegel, R. L. & Ludemann, H. (1996) Mol. Biochem. Parasitol. 80, 203–208. [DOI] [PubMed] [Google Scholar]
- 22.Brun, R. & Schonenberger, M. (1979) Acta Trop. 36, 289–292. [PubMed] [Google Scholar]
- 23.Hirumi, H. & Hirumi, K. (1989) J. Parasitol. 75, 985–989. [PubMed] [Google Scholar]
- 24.Wirtz, E., Leal, S., Ochatt, C. & Cross, G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101. [DOI] [PubMed] [Google Scholar]
- 25.Bleeg, H. S. & Christensen, F. (1982) Eur. J. Biochem. 127, 391–396. [DOI] [PubMed] [Google Scholar]
- 26.Ingram, A. K., Cross, G. A. & Horn, D. (2000) Mol. Biochem. Parasitol. 111, 309–317. [DOI] [PubMed] [Google Scholar]
- 27.Yagi, K. & Nishikimi, M. (1997) Methods Enzymol. 279, 24–29. [DOI] [PubMed] [Google Scholar]
- 28.Imai, T., Karita, S., Shiratori, G., Hattori, M., Nunome, T., Oba, K. & Hirai, M. (1998) Plant Cell Physiol. 39, 1350–1358. [DOI] [PubMed] [Google Scholar]
- 29.Yabuta, Y., Yoshimura, K., Takeda, T. & Shigeoka, S. (2000) Plant Cell Physiol. 41, 666–675. [DOI] [PubMed] [Google Scholar]
- 30.Fraaije, M. W., van den Heuvel, R. H., van Berkel, W. J. & Mattevi A. (1999) J. Biol. Chem. 274, 35514–35520. [DOI] [PubMed] [Google Scholar]
- 31.Sommer, J. M. & Wang, C. C. (1994) Annu. Rev. Microbiol. 48, 105–138. [DOI] [PubMed] [Google Scholar]
- 32.Lykkesfeldt, J., Loft, S. & Poulsen, H. E. (1995) Anal. Biochem. 229, 329–335. [DOI] [PubMed] [Google Scholar]
- 33.Overath, P., Czichos, J. & Haas, C. (1986) Eur. J. Biochem. 160, 175–182. [DOI] [PubMed] [Google Scholar]
- 34.Gelb, M. H., Van Voorhis, W. C., Buckner, F. S., Yokoyama, K., Eastman, R., Carpenter, E. P., Panethymitaki, C., Brown, K. A. & Smith, D. F. (2003) Mol. Biochem. Parasitol. 126, 155–163. [DOI] [PubMed] [Google Scholar]
- 35.Roberts, C. W., McLeod, R., Rice, D. W., Ginger, M., Chance, M. L. & Goad, L. J. (2003) Mol. Biochem. Parasitol. 126, 129–142. [DOI] [PubMed] [Google Scholar]
- 36.Wilkinson, S. R., Taylor, M. C., Touitha, S., Mauricio, I. L., Meyer, D. J. & Kelly, J. M. (2002) Biochem. J. 364, 787–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hannaert, V., Saavedra, E., Duffieux, F., Szikora, J. P., Rigden, D. J., Michels, P. A. & Opperdoes, F. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons, M. (2004) Mol. Microsc. 53, 717–724. [DOI] [PubMed] [Google Scholar]
- 39.Banhegyi, G., Braun, L., Csala, M., Puskas, F. & Mandl, J. (1997) Free Radic. Biol. Med. 23, 793–803. [DOI] [PubMed] [Google Scholar]
- 40.Marinho, H. S., Antunes, F. & Pinto, R. E. (1997) Free Radic. Biol. Med. 22, 871–883. [DOI] [PubMed] [Google Scholar]
- 41.Hwang, C., Sinskey, A. J. & Lodish, H. F. (1992) Science 257, 1496–1502. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson, S. R., Horn, D., Prathalingam, S. R. & Kelly, J. M. (2003) J. Biol. Chem. 278, 31640–31646. [DOI] [PubMed] [Google Scholar]
- 43.Maeda, N., Hagihara, H., Nakata, Y., Hiller, S., Wilder, J. & Reddick, R. (2000) Proc. Natl. Acad. Sci. USA 97, 841–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabata, K., Oba, K., Suzuki, K. & Esaka, M. (2001) Plant J. 27, 139–148. [DOI] [PubMed] [Google Scholar]
- 45.Conklin, P. L., Williams, E. H. & Last, R. L. (1996) Proc. Natl. Acad. Sci. USA 93, 9970–9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veljovic-Jovanovic, S. D., Pignocchi, C., Noctor, G. & Foyer, C. H. (2001) Plant Physiol. 127, 426–435. [PMC free article] [PubMed] [Google Scholar]
- 47.Welch, R. W., Wang, Y., Crossman, A., Jr., Park, J. B., Kirk, K. L. & Levine, M. (1995) J. Biol. Chem. 270, 12584–12592. [DOI] [PubMed] [Google Scholar]
- 48.Tsukaguchi, H., Tokui, T., Mackenzie, B., Berger, U. V., Chen, X. Z., Wang, Y., Brubaker, R. F. & Hediger, M. A. (1999) Nature 399, 70–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.