Abstract
The axial body pattern of Arabidopsis is determined during embryogenesis by auxin signaling and differential gene expression. Here we demonstrate that another pathway, cell-to-cell communication through plasmodesmata (PD), is regulated during apical–basal pattern formation. The SHOOT MERISTEMLESS (STM) promoter was used to drive expression in the shoot apical meristem (SAM) and a subset of cells at the base of the hypocotyl of 1×,2×, and 3× soluble green fluorescent proteins (sGFPs), and the P30 movement protein of Tobacco mosaic virus (TMV) translationally fused to 1× and 2× sGFP. In the early heart stage, 2× sGFP (54 kDa) moves throughout the whole embryo, whereas 3× sGFP (81 kDa) shows more restricted movement. As the embryo develops, PD apertures are down regulated to form local subdomains allowing transport of different sized tracers. For example, movement of 2× sGFP to the cotyledon, and 3× sGFP to root tips, becomes restricted. Subdomains of cell-to-cell transport align with the apical–basal embryo body axis and correspond to the shoot apex, cotyledons, hypocotyl, and root. Studies with P30–GFP fusions reinforce the distinction between embryonic symplastic subdomains. Although P30 targets embryo cell walls as puncta (diagnostic for functional localization of P30 to PD in adult plants), P30 cannot dilate embryonic PD to overcome the barriers for transport between symplastic subdomains, suggesting that specific boundaries separate symplastic subdomains of the embryo. Thus, cell-to-cell communication via plasmodesmata conveys positional information critical to establish the axial body pattern during embryogenesis in Arabidopsis.
Keywords: GFP, symplast, Tobacco mosaic virus movement protein P30, STM
Arabidopsis seedlings show an apical–basal body pattern along the main axis comprised of structures such as the shoot apical meristem (SAM), cotyledons, hypocotyl, and root. Clonal analyses and histological techniques predict the contribution of each embryonic cell to the seedling body plan, and reflect the regular pattern of cell divisions in early embryogenesis (1). Although cell lineage plays an important role in early embryogenesis, when a regular pattern of cell division is disturbed, each cell differentiates according to its final position (2, 3). Overall, positional information prevails in the formation of the body pattern, and lineage-dependent cell fate specifies local patterning (reviewed in refs. 4 and 5). In the present report, we investigate the role of intercellular communication in conveying positional information to form basic axial patterning during late embryogenesis.
Plant cells are encased in rigid cell walls, and their cytoplasms are connected through dynamic intercellular channels termed plasmodesmata (PD) (6–9). The functional measure of PD is their size exclusion limit (SEL), the upper limit of the size of macromolecules that can diffuse from cell to cell. PD selectively allow movement of some transcription factors and RNAs critical in cell-fate determination (reviewed in ref. 10). Plant viral proteins exemplify exogenous proteins that interact specifically with PD to allow their own passage as well as that of other macromolecules (reviewed in ref. 11).
Embryos offer an advantageous system to examine the genetic and developmental controls of PD because they are enclosed by maternal tissues and are symplastically isolated from maternal input by the torpedo stage. Thus, the embryo is minimally affected by physiology and environment. PD in Arabidopsis embryos were first viewed ultrastructurally (12), and a few studies assessed PD function during embryogenesis (13–15).
Here we report extensive analyses on the size limits for protein transport during embryogenesis using the promoter of the SHOOT MERISTEMLESS (STM) gene (16) to drive transgenic expression of 1×, 2×, and 3× sGFP as well as Tobacco mosaic virus (TMV) P30 translationally fused to 1× and 2× sGFP in the SAM and a subset of cells in the hypocotyl. The subsequent movement of these tracers from their site of synthesis was monitored at three stages of embryogenesis. The results reveal that four subdomains of cell-to-cell transport are established by late embryogenesis.
Materials and Methods
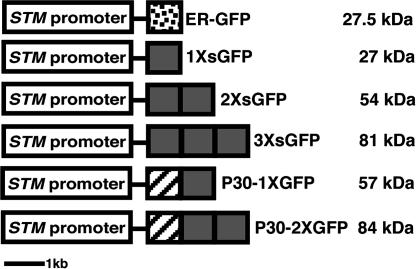
Construction of Plasmids and Transgenic Arabidopsis. STM coding sequences downstream of the 3.3-kb STM promoter in pJL8† were replaced with the coding sequences of different sized sGFPs (1×, 2×, and 3×) and TMV P30 from pRTL2 (17). The Tobacco etch virus (TEV) enhancer region (18) was inserted between the promoter and each coding sequence in all constructs except for 1× and 2× sGFP. Each plasmid was used to transform Arabidopsis thaliana, ecotype Columbia and Landsberg erecta, by floral dipping methods (19), and transgenic Columbia ecotype plants were used for the studies reported herein because of their higher expression of GFP. Thirty to 40 primary transgenic plants for each of the six constructs analyzed were screened by using epifluorescence microscopy, and six lines with strong and comparable expression of GFP were chosen for detailed analyses.
Microscopy. Embryos from transgenic Arabidopsis grown under greenhouse conditions were observed by conventional microscopy for β-glucuronidase (GUS) staining patterns and by confocal laser scanning microscopy for GFP expression/movement patterns as described (14).
Results
To study changes in PD aperture during embryogenesis requires the introduction of symplastic tracers and monitoring of their subsequent movement. Soluble GFP (sGFP) is an exceptionally useful symplastic tracer, because it is not membrane permeable and it remains in the hydrophilic nuclear and cytoplasmic compartments into which it is introduced. GFP is nontoxic and, because it is an exogenous protein, it does not bind to any cell components (20). We chose to induce the expression of sGFP by the SHOOT MERISTEMLESS (STM) promoter (Fig. 1) as the STM gene is transcribed throughout embryogenesis, and its mRNA is confined to a subset of cells at the shoot apical meristem (SAM) (16) (Fig. 2 A, G, and M).
Fig. 1.
Constructs transformed into Arabidopsis plants. White box, 3.3-kb STM promoter; dotted box, ER-tethered GFP with signal peptide and KDEL ER retention signal (18); gray box, soluble GFP; striped box, TMV P30.
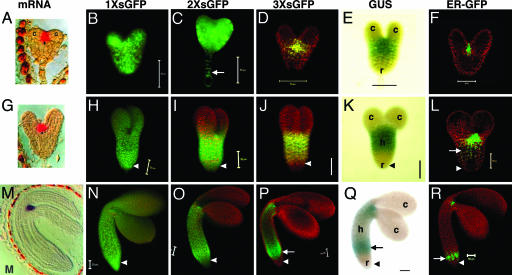
Fig. 2.
Movement pattern of soluble GFP during embryo development. Early heart (A–F), late heart (G–L), and midtorpedo (M–R) embryos for 1× sGFP (B, H, and N), 2× sGFP (C, I, and O), and 3× sGFP (D, J, and P) movement. STM promoter activity is shown by GUS (E, K, and Q) and ER-GFP (F, L, and R). Arrows indicate nucleus in suspensor cells (C), and ectopic expression of STM promoter in hypocotyls (L and P–R). Arrowheads indicate root. c, cotyledons; h, hypocotyl; r, root. (Scale bars, 50 μm.) [Images in A, G, and M are reprinted with permission from ref. 16 (Copyright 1998, Company of Biologists, Ltd.).]
To mark the initial site of protein synthesis and thus promoter activity, two cell-autonomous reporters, GUS and endoplasmic reticulum (ER)-tethered GFP (ER-GFP) (17) were transcriptionally fused to the same promoter. Three developmental stages were analyzed to highlight distinctive changes in the movement pattern of sGFP.
sGFP Movement in the Early Heart Embryo. In the early heart stage, the ER-GFP expression pattern (Fig. 2F) shows that the STM promoter is active in the SAM mimicking the localization of the STM transcript (Fig. 2 A) (16). [Here we define the early heart stage as when embryos are clearly heart shaped, but are small, representing an earlier stage than the one analyzed in ref. 15 where movement of 2× sGFP was limited]. The region defined by GUS activity is more extensive than ER-GFP expression (compare Fig. 2 E and F), which may result either from weak promoter activity in cells adjacent to the SAM (detected due to the enzymatically amplified GUS signal), or from diffusion of the GUS product from the SAM to adjacent cells (21). Nevertheless, cotyledons and the root were clearly devoid of GUS activity confirming that the SAM (and close neighboring cells) is the major site of STM promoter activity.
Both 1× sGFP (27 kDa) and 2× sGFP (54 kDa) move throughout the entire embryo and to the connected basal suspensor cells (Fig. 2 B and C, arrow). In a slightly later heart staged embryo (bigger in size without obvious hypocotyl elongation) (15), movement of 2× sGFP begins to be limited in the area of the cotyledons so that 2× sGFP shows a movement pattern similar to that shown for 3× sGFP in Fig. 2D. The 3× sGFP (81 kDa) shows limited movement around the SAM (Fig. 2D), and the cotyledons do not allow the transit of 3× sGFP from the SAM.
Formation of cotyledons at the early heart stage is a critical developmental time point in Arabidopsis embryogenesis, marking the morphogenetic transition from radial (in the prior globular stage) to bilateral symmetry (22). The localized down-regulation of symplastic transport into cotyledons (detected by 3× sGFP) may play a role in developmental signaling to direct the formation of cotyledons. See Fig. 7, which is published as supporting information on the PNAS web site, for images of only GFP signal without red autofluorescence.
sGFP Movement in the Late Heart Embryo. In late heart embryos (defined by an ≈1:1.2 ratio in length between cotyledons and hypocotyls), STM promoter activity is significantly broader than STM mRNA localization so that GUS and ER-GFP appear not only in the SAM, but also in a major portion of the hypocotyl (compare Fig. 2 G, K, and L, arrow). The 1× sGFP freely moves (Fig. 2H). Although both 2× and 3× sGFP appear among all cells in the SAM and hypocotyl, they exhibit restricted movement toward the cotyledons (Fig. 2 I and J). In higher magnification, 2× and 3× sGFP move from the SAM to several cell layers into the cotyledons but do not proceed further (data not shown, and see Fig. 3J), delimiting a boundary for a symplastic subdomain of the shoot apex (see below). Intriguingly, whereas 2× sGFP moves down to the end of the root tip, 3× sGFP does not (Fig. 2 I and J, arrowheads) delimiting another boundary for a symplastic subdomain of the root (see also Fig. 7 N and O). Note that, because the root is devoid either of GUS or ER-GFP (Fig. 2 K and L, arrowheads), the 1× and 2× sGFP found there must result from bone fide symplastic movement. The localized down-regulation in transport of 2× sGFP to the cotyledons, and of 3× sGFP to the root, becomes more evident from this stage onward and correlates with the development of more apparent tissue types. Note that cotyledon SEL is now lower than at the previous early heart stage, where 2× sGFP moves freely (compare Fig. 2 C and I).
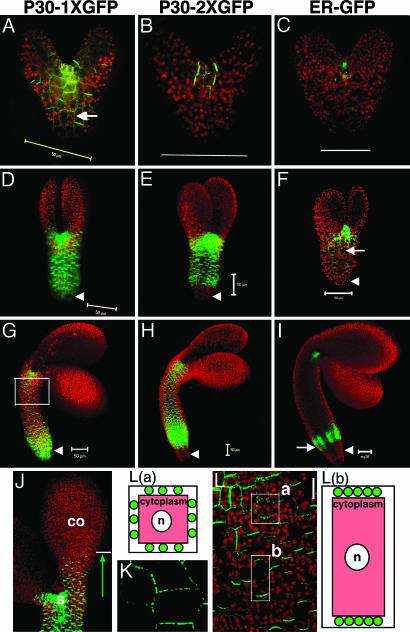
Fig. 3.
Movement pattern of TMV P30-GFP during embryo development. Early heart (A–C), early torpedo (D–F), and midtorpedo (G–I) embryos for TMV P30–1× GFP (A, D, G, and J), and TMV P30–2× GFP (B, E, and H) movement. ER-GFP shows the site of protein synthesis (C, F, and I). Arrows indicate P30–1× GFP localization to puncta in cell walls (A) and the ectopic expression of STM promoter in hypocotyls (F and I). Arrowheads indicate root. Cells in the box in G are shown in L.(J) P30–1× GFP in the SAM (s) moves several cell layers into cotyledons (co) but stops advancing further. (K) Higher magnification image of P30–1× GFP localizing to puncta in cell walls within SAM. (L) P30–1× GFP targets cell walls, evenly in four sides of cells within (close to) SAM (box a) but preferably goes to apical and basal sides in cells of hypocotyls (box b). Diagrams are individual cells; red represents cytoplasm; green circles represent TMV P30-GFP localized to cell walls as puncta. n, nuclei. (Scale bars, 50 μm.)
sGFP Movement in the Midtorpedo Embryo. The midtorpedo stage is referred to as “bent cotyledon” when embryos within their seed coats are observed in longitudinal sections (4); such embryos appear completely folded in an inverted “U” shape (Fig. 2M). When seed coats are removed and living embryos are exposed to a hydrating environment, embryos display only 90° bending of their cotyledons relative to the hypocotyl, due to removal of constraining seed coats.
In midtorpedo embryos, the STM promoter induces an unexpected ectopic expression at the base of the hypocotyl (Fig. 2 Q and R, arrows) in addition to the SAM shown by in situ mRNA localization (Fig. 2M). This finding reflects that regulatory elements of the STM gene required for its tight spatial localization to the SAM are not present in the 3.3-kb upstream promoter region used for the present studies. Additional regulatory elements located upstream, in introns, or downstream of the transcriptional unit may be required to regulate endogenous STM mRNA localization. Cis-regulatory elements, required for the correct spatiotemporal expression of genes, located far outside the transcriptional unit, have been reported in human genomes (23). However, unexpectedly and significantly (as described below), this ectopic expression provides an independent assay to identify the boundaries of symplastic subdomains between the hypocotyl and root, as well as between the shoot apex (including the SAM) and the hypocotyl.
In midtorpedo embryos, 1× sGFP sustains its free cell-to-cell transit pattern (Figs. 2N and 4K). Both 2× and 3× sGFP move in a similar pattern as in the previous late heart stage (Figs. 2 O and P and 4K). First, neither moves into the cotyledon. Second, both appear in the cytoplasm and nuclei throughout the length of the hypocotyl. Third, 3× sGFP does not move down to the root tip (Fig. 2P, arrowhead, and Fig. 4 A and F for higher magnification views), whereas 2× sGFP does (Fig. 2O, arrowhead, and Figs. 4K and 7 U and V). Fig. 7 shows the higher magnification view of the root tip for 1× sGFP and 2× sGFP movement at the midtorpedo stage; it is clear that 2× sGFP moves to the bottom cells of the root tip, but less extensively than 1× sGFP (compare Fig. 7 R and S with U and V), likely reflecting the decreased SEL of PD in this region. Taken together, the PD SEL for movement between subdomains remains the same as the previous late heart stage.
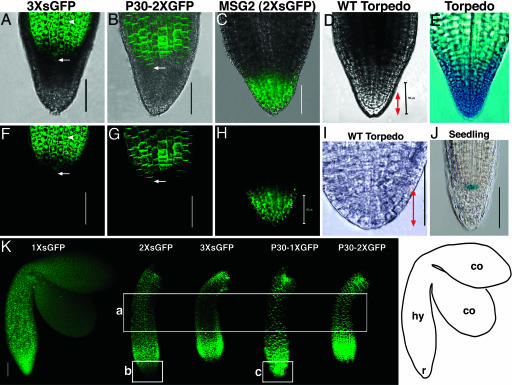
Fig. 4.
Movement of 3× sGFP and P30–2× GFP in roots of midtorpedo embryos. Overlapping bright field and green GFP fluorescence (A–C) and GFP fluorescence alone (F–H) are shown. (D and I) Wild-type embryo roots in bright field. (E) Torpedo embryo root showing root cap-specific GUS expression. (J) Seedling root showing quiescent center-specific GUS expression in line QC25 (29). (K) GFP signal in midtorpedo embryos; a, the mid region of hypocotyls; b and c, roots. Arrows indicate cells that mark the boundary between the hypocotyl (hy) and the root (r) in A, B, F, and G. Arrowheads shows nuclear localization of 3× sGFP (A and F). Bidirectional red arrows (D and I) indicate the central root cap. (Scale bars, 50 μm.) Image in I was adapted with permission from ref. 14. [Image in E reprinted with permission from ref. 26 (Copyright 1999, Company of Biologists, Ltd.).]
GUS activity appears more extended than ER-GFP expression so that only the center of hypocotyls (approximately onethird of total hypocotyl length) is free from the blue stain (compare Fig. 2 Q and R). Nevertheless, 3× sGFP is present in the middle region of the hypocotyl (where GUS is absent). This GFP signal intensity of 3× sGFP is weaker than that observed for 2× sGFP (compare Fig. 2 O and P). Nevertheless, nuclei of cells in the mid hypocotyl (in all tissues including protodermis, ground, and provascular tissues) are clearly loaded with 3× sGFP in high-magnification view (data not shown), and there is a faint green signal (above the red autofluorescence) indicative of movement compared to the ER-GFP control (compare Fig. 2 O, P, and R; see also box a in Fig. 4K).
Notably at this stage, there is a down-regulation within a subdomain, shown as the gradient in sGFP signal intensity in the mid hypocotyl that becomes more obvious as sGFP size increases (compare Fig. 2 N, O, and P; see also box a in Fig. 4K). Thus, this mid hypocotyl region is less open for transit of larger sized sGFP. In contrast, all sized sGFP signals move homogeneously within the hypocotyl of late heart embryos without a noticeable gradient (compare Fig. 2 H, I, and J). The ectopic expression in the bottom of the hypocotyl at this midtorpedo stage suggests that the lack of 3× sGFP movement into the root from the late heart stage onward is not simply due to the distance between the site of synthesis at the SAM and the root.
The results suggest that distinct subdomains of cell-to-cell transport arise during the course of embryogenesis. The first subdomain is detected when the embryo forms cotyledons at the early heart stage. As development proceeds, four subdomains become distinct: shoot apex including the SAM, cotyledon (embryonic leaf), hypocotyl (embryonic stem), and root. These subdomains, corresponding to the axial body pattern, are most evident at the midtorpedo stage defined here. Embryos begin dehydration and overall GFP expression level decreases beyond the midtorpedo stage (data not shown).
Note that the distinct PD SELs observed in different regions of the embryo confirm that the fusion proteins used in this study are stable. If 2× or 3× sGFP would break down to 1× sGFP, we would see sGFP signal in cotyledons (compare Fig. 2 N, O, and P). Similarly, if 3× sGFP would break down to 2× sGFP (or 1× sGFP), we would have seen signal in the root in 3× sGFP-transgenic lines (compare Fig. 2 N, O, and P, arrowheads; also see Fig. 4 A and F). To easily view both embryonic shape as well as sGFP movement, the fluorescent panels in Fig. 2 show green GFP fluorescence and red chlorophyll autofluorescence.
TMV P30-GFP Movement During Embryogenesis. The movement protein P30 of TMV and its GFP fusion derivatives dilate (gate) PD and thereby facilitate P30 movement beyond the limits imposed by native PD aperture in adult plants (17, 24, 25). To examine the movement function of TMV P30 during embryogenesis, we stably expressed TMV P30 translationally fused to 1× and 2× sGFP, respectively, under the control of the same STM promoter (Fig. 1).
In early heart embryos, P30–1× GFP (57 kDa, similar in size to 2× sGFP, 54 kDa) moves throughout the whole embryo (Fig. 3A; compare with Fig. 2C), and P30–2× GFP (84 kDa, similar in size to 3× sGFP, 81 kDa) shows restricted movement around the SAM (Fig. 3B; compare with Fig. 2D). All P30–GFP fusions form puncta in the cell wall; such targeted localization is functionally diagnostic for PD specific labeling (Fig. 3 A, arrow, and K for high magnification view; also see Fig. 8 A and D, which is published as supporting information on the PNAS web site). At the early torpedo stage, all P30–GFP fusions show restricted movement to the cotyledons but appear in all cells in the hypocotyl (Fig. 3 D and E; compare with Fig. 2 I and J). Differential movement to the root occurs so that only P30–1× GFP moves into the root (Fig. 3D, arrowhead; compare with Fig. 2I, arrowhead), whereas P30–2× GFP does not (Fig. 3E, arrowhead; compare with Fig. 2 J, arrowhead). The lack of movement of P30–2× GFP to the root is more evident than that shown for 3× sGFP in Fig. 2 J, because embryos were viewed at a slightly later stage. At this early torpedo stage, embryos are ≈1.4-fold larger than late heart embryos because of elongation of cotyledons and hypocotyls.
At the midtorpedo stage, the movement pattern of the two constructs remains unchanged: there is a lack of movement to the cotyledons for both constructs and lack of movement to the root for P30–2× GFP (Fig. 3 G and H; compare with Fig. 2 O and P). Both constructs exhibit movement within the hypocotyl subdomain.
These data might imply that P30 cannot function to gate PD during embryogenesis; however, this is not the case. P30 fusions exclusively target cell walls in a punctate pattern diagnostic for P30 function, and are excluded from nuclei (unlike sGFPs). P30–1× GFP freely moves down to the root tip so that all of the cells of the root are intensely labeled. In contrast, similarly sized 2× sGFP moves less extensively, exhibiting a gradient in GFP signal intensity with much weaker fluorescence toward the root tip (compare Figs. 3G and 2O, arrowheads; also compare boxes b and c in Fig. 4K). In addition, both P30 fusions move more extensively into the midregion of the hypocotyl than do their similarly sized sGFP counterparts (see box a in Fig. 4K). These data show that P30 is functional in gating PD within subdomains. Thus, lack of movement of either construct into the cotyledons, and lack of movement of P30–2× GFP into the root, implies that there are boundaries between symplastic subdomains that cannot be overcome by P30. These data provide additional support for the formation of symplastic subdomains during embryogenesis. Fig. 9, which is published as supporting information on the PNAS web site, presents the images shown in Fig. 3 A–I without background red autofluorescence.
Localization of TMV P30-GFP in Embryos. Higher magnification observation of the movement patterns of P30 fusion constructs reveals that P30–1× GFP (like 2× sGFP) moves several cell layers up into the cotyledon from the SAM, but does not advance further (Fig. 3J, arrow). Thus, P30–1× GFP (like 2× sGFP) freely moves within the shoot apex symplastic subdomain including the SAM, but not into the symplastic subdomain of cotyledon. Although the movement extent of P30–GFP fusions between subdomains is similar to its equivalently sized sGFPs, its subcellular localization is clearly distinct; 1×, 2×, and 3× sGFP localize to nuclei as well as the cytoplasm (for example, see Fig. 4 A and F, arrowheads), whereas P30 fusions localize to cell walls (for example, see Fig. 4 B and G) as puncta in higher magnification (Fig. 3K).
Surprisingly, P30-GFP fusions preferentially localize to different cell walls in different cell types (Fig. 3 K and L). P30–GFPs targets puncta evenly on four sides of cell walls within (and close to) the SAM (box a in Fig. 3L), but preferably targets the basal and apical ends of cell walls in hypocotyls where puncta are less discernible because of their higher density (box b in Fig. 3L). Such differential targeting may reflect that cell-to-cell transport in/near the SAM occurs in all directions, whereas cell-to-cell transport in the hypocotyl is likely directional in either an acropetal or basipetal manner. In transgenic tobacco leaves, P30–1× GFP forms discrete puncta uniformly distributed around all cell walls of epidermal cells, but targets only apical and basal cell walls in trichomes where P30–1× GFP moves in only one direction (compare boxes a and b in Fig. 3L with Fig. 8 A and D).
Detailed Examination of the Root Symplastic Subdomain. To examine the boundary between the hypocotyl and the root in detail, the movement patterns of sGFP after expression from two different promoters in two different tissues were compared. After expression at the base of the hypocotyl driven by the STM promoter (Fig. 2 Q and R, arrows), neither 3× sGFP nor P30–2× GFP moves down into the root (Fig. 4 A, B, F, and G). Upon expression at the root apical meristem (RAM) in the MSG2 line (15), 2× sGFP moves freely toward the root tip but does not move upward into the hypocotyl (Fig. 4 C and H). Here, when 2× sGFP was expressed at the bottom of the hypocotyl by the STM promoter, it moves down toward the root tip (Fig. 2O and 4K). The data with the MSG2 line could be interpreted to suggest either selective/directional blockage of movement from the root to the hypocotyl or preferential movement downward to the root tip within the root subdomain, or both. However, the present data showing movement of 2× sGFP but not 3× sGFP down from the hypocotyl into the root suggests that PD aperture is regulated at the hypocotyl–root junction. These two sets of data are complementary and strongly support the existence of the root subdomain for cell-to-cell transport.
To determine the cell types that comprise the root symplastic subdomain, we compared the movement extent of GFPs to GUS marker lines for the root cap (Fig. 4E) and the quiescent center (Fig. 4J). These marker lines suggest that the symplastic subdomain of the root includes the central root cap and at least part of the RAM.
Discussion
In Arabidopsis embryos, all cells are interconnected by PD and integrated into a single symplast, the domain of common cytoplasm that is bounded by the plasma membranes of connected cells (reviewed in ref. 13). As the embryo develops, the functional aperture of PD is down-regulated. Embryos fail to grow to normal plants when this down-regulation is perturbed, showing that the regulation of PD is significant in embryo development (14). Previous studies showed free movement of 1× sGFP throughout the entire embryo, and limited movement of 2× sGFP near meristematic regions (15), consistent with the general notion that cells with less-determined fate in/near meristems and sink tissues, have more dilated PD (27, 28). Endogenous proteins such as transcriptional factors also move in the SAM (reviewed in ref. 10).
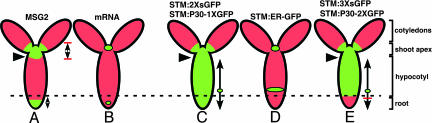
Here we provide evidence for four subdomains of symplastic transport in the Arabidopsis embryo (Figs. 5 and 6B). First, the shoot apex subdomain is indicated by the movement of 2× sGFP and P30–1× GFP (and 3× sGFP and P30–2× GFP) from the site of their initial synthesis, the SAM, under the control of the STM promoter (Fig. 5 C–E). The shoot apex subdomain is flanked by the cotyledons above and the hypocotyl below, indicated by the lack of movement of tracers greater than 2× sGFP into the cotyledons (Fig. 5 C and E), and the lack of movement from the SAM down into the hypocotyl in MSG2 (Fig. 5A, arrowhead). Second, the cotyledon subdomain is indicated consistently by the failure of all constructs larger than 2× sGFP to move into the cotyledons (Fig. 5 A, C, and E). Third, the hypocotyl subdomain is revealed by the movement of 2× sGFP and P30–1× GFP, and 3× sGFP and P30–2× GFP. The upper boundary of the hypocotyl subdomain is at the base of the shoot apex subdomain, indicated by the lack of movement of 2× sGFP into this domain from the SAM in MSG2 (Fig. 5A, arrowhead); and the lower boundary of this domain is indicated by the lack of movement of 3× sGFP and P30–2× GFP into the root (Fig. 5E, dotted line). The cells along the length of the hypocotyl exhibit a gradient in PD aperture. For example, cells in the mid hypocotyl are less open for transport of 2× sGFP than 1× sGFP, and even less for 3× sGFP (box a Fig. 4K). Fourth, the root subdomain is indicated by the movement of 2× sGFP and P30–1× GFP into this domain, and the lack of movement into this domain of 3× sGFP and P30–2× GFP (Fig. 5 C and E, dotted line). Results with the MSG2 line complement these data and provide additional support for the root symplastic subdomain; 2× sGFP (in MSG2) does not move into the hypocotyl from its site of synthesis in the RAM (Fig. 5A).
Fig. 5.
Summary of cell-to-cell transport of symplastic tracers in torpedo embryos. Red represents autofluorescence and green indicates the presence of GFP in cells. Small green circles represent the site of GFP synthesis, and arrows indicate the direction/extent of the cell-to-cell movement of each sGFP from the site of synthesis. Arrowheads mark the boundary between symplastic subdomains of the shoot apex and hypocotyl, and the dotted line shows the boundary between hypocotyls and the root. The MSG2 line expresses 2× sGFP in the SAM and RAM.
Fig. 6.
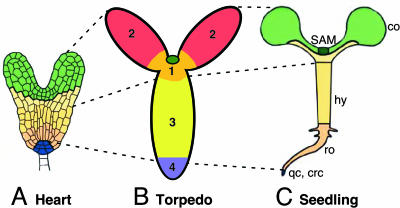
Axial body patterns of Arabidopsis embryos and seedlings. Colors in A and C represent the corresponding clonal regions in early embryos and seedlings. (B) Four subdomains of symplastic transport form along the apical–basal body axis. 1, shoot apex including the meristem (darker green circle) and adjacent cells (orange); 2, cotyledons; 3, hypocotyl; 4, root; co, cotyledons; hy, hypocotyl; ro, root; crc, central root cap; qc, quiescent center. A and C were adapted with permission from ref. 5.
It is critical to note that these symplastic subdomains could not have been uncovered without the correlative observation of distinctly localized boundaries between each of the subdomains where PD aperture is reduced. Otherwise, one would expect to observe a gradient of diffusion of different sized GFP tracers across these boundaries. The presence of boundaries between symplastic subdomains is significantly reinforced by data with P30 fused to sGFP. Thus, the results presented reveal that the P30 protein of TMV is unable to overcome boundaries between symplastic subdomains observed for sGFPs. The data suggest that this behavior is not due to lack of P30 function, because P30 localizes to embryonic PD just as it localizes to PD after germination, and P30-GFP fusion proteins move more freely within the hypocotyl (to the midregion) and within the root (to the tip) than their similarly sized sGFP counterparts. Thus, P30 both targets to PD and gates PD to allow more extensive movement than the diffusion of non-P30 containing proteins within symplastic subdomains.
Finally, the fate maps of early embryos and seedlings are well established (reviewed in refs. 4 and 5). Here, we found that symplastic subdomains are established along the body axis by the midtorpedo stage. These subdomains can be extrapolated to regions of the early embryo (and seedling) defined by gene expression profiles and clonal analyses (Fig. 6). Future studies should address the exact timing of the formation of the symplastic subdomains by analyses of the movement patterns of GFP tracers after crossing to Arabidopsis lines expressing various cell type-specific markers.
Supplementary Material
Acknowledgments
We gratefully acknowledge Kathy Barton (Carnegie Institution of Washington, Stanford, CA) for the STM promoter and ProSTM:GUS transgenic Arabidopsis, Shmuel Wolf and Ayelet Omid (Hebrew University, Jerusalem) for ProCaMV 35S:TMV P30–1× GFP transgenic tobacco, Ben Scheres (Utrecht University, Utrecht, The Netherlands) for QC 25 line, and Steve Ruzin and Denise Schichnes (Center for Biological Imaging, University of California, Berkeley) for invaluable advice on microscopy. This work was supported by National Institutes of Health Grant GM 45244.
Author contributions: I.K. designed research; I.K., K.K., and E.C. performed research; I.K. analyzed data; and I.K. and P.C.Z. wrote the paper.
Abbreviations: TMV, tobacco mosaic virus; GUS, β-glucuronidase; sGFP, soluble GFP; SAM, shoot apical meristem; ER, endoplasmic reticulum; ER-GFP, ER-tethered GFP; PD, plasmodesmata.
Footnotes
Fernandez, A. G., Long, J., Joy, R. E. & Barton, M. K. (2000) 11th International Confrerence on Arabidopsis Research, June 24–28, 2000, Madison, WI, abstr.
References
- 1.Scheres, B., Wolkenfelt, H., Willemsen, V., Terlouw, M., Lawson, E., Dean, C. & Weisbeek, P. (1994) Development (Cambridge, U.K.) 120, 2475-2487. [Google Scholar]
- 2.Poethig, R., Coe, E. & Johri, M. (1986) Dev. Biol. 117, 392-404. [Google Scholar]
- 3.Saulsberry, A., Martin, P. R., O'Brien, T., Sieburth, L. E. & Pickett, F. B. (2002) Development (Cambridge, U.K.) 129, 3403-3410. [DOI] [PubMed] [Google Scholar]
- 4.Berleth, T. & Chatfield, S. (2002) in The Arabidopsis Book, eds. Somerville, C. & Meyerowitz, E. (Am. Soc. Plant. Biol., Rockville, MD), pp. 1-22.
- 5.Laux, T., Wurschum, T. & Breuninger, H. (2004) Plant Cell 16, Suppl., S190-S202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zambryski, P. (2004) J. Cell Biol. 164, 165-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oparka, K. J. (2004) Trends Plant Sci. 9, 33-41. [DOI] [PubMed] [Google Scholar]
- 8.Lucas, W. J. & Lee, J. Y. (2004) Nat. Rev. Mol. Cell. Biol. 5, 712-726. [DOI] [PubMed] [Google Scholar]
- 9.Heinlein, M. & Epel, B. L. (2004) Int. Rev. Cytol. 235, 93-164. [DOI] [PubMed] [Google Scholar]
- 10.Kim, J. Y. (2005) Curr. Opin. Plant Biol. 8, 45-52. [DOI] [PubMed] [Google Scholar]
- 11.Citovsky, V. (1999) Philos. Trans. R. Soc. London B 54, 637-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansfield, S. G. & Briarty, L. G. (1991) Can. J. Bot. 69, 461-476. [Google Scholar]
- 13.McLean, B. G., Hempel, F. D. & Zambryski, P. C. (1997) Plant Cell 9, 1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, I., Hempel, F. D., Sha, K., Pfluger, J. & Zambryski, P. C. (2002) Development (Cambridge, U.K.) 129, 1261-1272. [DOI] [PubMed] [Google Scholar]
- 15.Kim, I., Cho, E., Crawford, K., Hempel, F. D. & Zambryski, P. C. (2005) Proc. Natl. Acad. Sci. USA 102, 2227-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long, J. A. & Barton, M. K. (1998) Development (Cambridge, U.K.) 125, 3027-3035. [DOI] [PubMed] [Google Scholar]
- 17.Crawford, K. M. & Zambryski, P. C. (2000) Curr. Biol. 10, 1032-1040. [DOI] [PubMed] [Google Scholar]
- 18.Carrington, J. C. & Freed, D. D. (1990) J. Virol. 61, 1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 20.Oparka, K. & Read, N. (1994) Plant Cell Biol. 139, 27-50 [Google Scholar]
- 21.Mantis, J. & Tague, B. W. (2000) Plant Mol. Biol. Rep. 18, 319-330. [Google Scholar]
- 22.Goldberg, R. B., De Paiva, G. & Yadegari, R. (1994) Science 266, 605-614. [DOI] [PubMed] [Google Scholar]
- 23.Kleinjan, D. A. & van Heyningen, V. (2005) Am. J. Hum. Genet. 76, 8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf, S., Deom, C. M., Beachy, R. N. & Lucas, W. J. (1989) Science 246, 377-379. [DOI] [PubMed] [Google Scholar]
- 25.Oparka, K. J., Prior, D. A., Santa Cruz, S., Padgett, H. S. & Beachy, R. N. (1997) Plant J. 12, 781-789. [DOI] [PubMed] [Google Scholar]
- 26.Hamann, T., Mayer, U. & Jurgens, G. (1999) Development (Cambridge, U.K.) 126, 1387-1395. [DOI] [PubMed] [Google Scholar]
- 27.Crawford, K. M. & Zambryski, P. C. (2001) Plant Physiol. 125, 1802-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oparka, K. J., Roberts, A. G., Boevink, P., Santa Cruz, S., Roberts, I., Pradel, K. S., Imlau, A., Kotlizky, G., Sauer, N. & Epel, B. (1999) Cell 97, 743-754. [DOI] [PubMed] [Google Scholar]
- 29.Sabatini, S., Heidstra, R., Wildwater, M. & Scheres, B. (2003) Genes Dev. 17, 354-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.