Abstract
In Saccharomyces cerevisiae, Mre11p/Rad50p/Xrs2p (MRX) complex plays a vital role in several nuclear processes including cellular response to DNA damage, telomere length maintenance, cell cycle checkpoint control and meiotic recombination. Telomeres are comprised of tandem repeats of G-rich DNA and are incorporated into non-nucleosomal chromatin. Although the structure of the yeast telomeric DNA is poorly understood, it has been suggested that the G-rich sequences can fold into G4 DNA, which has been shown to inhibit DNA synthesis by telomerase. However, little is known about the factors and mechanistic aspects of the generation of appropriate termini for DNA synthesis by telomerase. Here, we show that S.cerevisiae Mre11 protein (ScMre11p) possesses substantially higher binding affinity for G4 DNA, over single- or double-stranded DNA, and binding was inhibited by poly(dG) or porphyrin. Binding of ScMre11p to G4 DNA was most robust, compared with G2′ DNA and the resulting protein–DNA complexes were strikingly very resistant to dissociation by NaCl. Remarkably, binding of ScMre11p to G4 DNA and G-rich single-stranded DNA was accompanied by the endonucleolytic cleavage at sites flanking the array of G residues and G-quartets in Mn2+-dependent manner. Collectively, these results suggest that ScMre11p is likely to play a major role in generating appropriate substrates for DNA synthesis by telomerase and telomere-binding proteins. We discuss the implications of these findings with regard to telomere length maintenance by telomerase-dependent and independent mechanisms.
INTRODUCTION
Genetic and biochemical studies in Saccharomyces cerevisiae and humans indicate that Mre11p complex, consisting of Rad50, Mre11 and Xrs2/Nbs1 (MRX/N), plays a vital role in multiple nuclear processes including cellular response to DNA damage, telomere length maintenance, cell cycle checkpoint control and meiotic recombination [reviewed in Refs (1–5)]. Yeast strains lacking any one or all three members of the MRX complex display profound defects in these processes, thereby leading to genome instability and cell death (6–9). The MRX complex participates in the DNA double-strand break (DSB) repair via the homologous recombination and the non-homologous end-joining pathways (1–5).
Biochemical studies of the yeast and human MRX/N protein complexes have revealed that Mre11p possesses several enzymatic and non-enzymatic activities including DNA exo- and endonuclease activities, DNA strand annealing and activation of DNA checkpoint kinase, ATM (10–13). The nuclease activity of Mre11p is complex and is influenced in part by the type of DNA substrate (10,14–16). Mre11 by itself is an ATP-independent, Mn2+-dependent, 3′ to 5′ exo- and endonuclease, and cleaves hairpin DNA structures (14–17). The nuclease activities of Mre11p is stimulated by mismatched ends, duplex DNA with blunt ends and 3′ recessed ends, but inhibited by DNA with 3′ overhangs and delayed upon encountering an internal microhomology sequence (13,14,18–20). S.cerevisiae Mre11 protein (ScMre11p) is believed to be involved in the removal of Spo11p from meiotic DSBs (21). The biochemical activities of Mre11p are modulated by complex formation with Rad50 and Xrs2/Nbs1 (13–15,22).
The termini of most eukaryotic telomeres contain many tandem repeats of G-rich sequence, which form a single-stranded 3′ overhang (23,24). The presence of 3′ overhangs is conserved in eukaryotes, but the exact structure varies between species (23,24). The ability of G-rich genomic domains, the rDNA and telomeres, to fold into a variety of inter- or intramolecular four-stranded quadruplex structures has attracted considerable attention (25–27). A G-quartet is composed of a planar array of four guanine residues, where each guanine accepts and donates two Hoogsteen hydrogen bonds (25–27). The stacks of G-quartets in parallel orientation is termed G4 DNA and, as G2′ DNA when the two strands are in anti-parallel orientation (26). When the single-stranded 3′ end of telomere folds into a G4 DNA, it no longer acts as a primer for telomerase (28). However, little is known about the factors and the mechanism involved in generating the DNA termini for telomerase in yeast or any other organism.
One of the outstanding questions relating to ScMre11p is its biochemical role in telomere length maintenance. Here, we show that ScMre11p displays substantially higher binding affinity for G4 DNA and cleaves this DNA configuration as well as G-rich single-stranded DNA (ssDNA) in a structure- and sequence-specific manner. Consistent with our findings, genetic studies have implicated a role for S.cerevisiae MRE11 in the proper establishment of telomere end-structure and loading of telomere-binding proteins (29). Thus, ScMre11p might provide appropriate DNA termini for DNA synthesis by telomerase and telomere-binding proteins, thus uncovering a novel link between Mre11p and maintenance of telomere end structure in S.cerevisiae.
MATERIALS AND METHODS
Plasmids and DNA
The oligonucleotides (ODNs) used in this study are listed in Table 1. The lengths and sequences of oligonucleotides were selected according to the experimental needs, their resemblance to canonical telomeric arrays and their ability to form G4 DNA with appropriate conformational properties (25,26,30). DNA substrates were radiolabeled at the 5′ end. G4 DNA was prepared and purified by non-denaturing gel electrophoresis as described previously (30,31). G4 DNA isolated from polyacrylamide gels was devoid of ssDNA (data not shown). The following chemicals, reagents and materials were purchased from the indicated sources: DNA restriction and modifying enzymes were from New England Biolabs; TALON Metal Affinity Resin from Clonetech; Heparin-Agarose from Bio-Rad; oligonucleotides from Sigma and were purified by denaturing gel electrophoresis as described previously (30,31). Anti-His antibody was obtained from Qiagen GmbH, Germany. Other chemicals, reagents and materials were of the highest purity commercially available. DNA preparation and analysis, restriction digests, ligation, end-labeling, transformation, SDS–PAGE, protein estimation were performed according to standard techniques as described (31).
Table 1.
Oligonucleotides used in this study
| Name | Sequence | Bases |
|---|---|---|
| 4G3 | 5′-AATTCTGGGTGTGTGGGTGTGTGGGTGTGTGGGTGTGG-3′ | 38 |
| 4G3C | 5′-CCACACCCACACACCCACACACCCACACACCCAGAATT-3′ | 38 |
| 6G3 | 5′-AATTCTGGGTGTGTGGGTGTGTGGGTGTGTGGGTGTGTGGGTGTGTGGGTGTGG-3′ | 54 |
| 21G | 5′-GGGTTAGGGTTAGGGTTAGGG-3′ | 21 |
| 41SD | 5′-GCCGTGATCACCAATGCAGATTGACGAACCTTTGCCCACGT-3′ | 41 |
Expression and purification of ScMre11p
The pGBT9-ScMRE11 construct was a gift from Dr Wei Xiao (University of Saskatchewan), and S.cerevisiae MRE11 was sub-cloned into the expression vector pRSET B at EcoRI site. The resulting plasmid pRBMRE11 was introduced into Escherichia coli DH5α and the integrity of the S.cerevisiae MRE11 gene was confirmed by sequencing. ScMre11 protein was expressed in E.coli strain BL21 (DE3)pLysS (Novagen) grown in LB medium containing ampicillin (50 μg/ml) and chloramphenicol (34 μg/ml). Cells were grown until the OD600 reached 0.4, and then 0.4 mM isopropyl β-D-thiogalactoside (IPTG) was added to the medium, and further allowed to grow at 18°C to induce protein expression for 12 h. The cell pellet from 2.4 l culture was resuspended in buffer A [10 mM Tris–HCl (pH 8), 50 mM NaCl, 10% glycerol, 0.05% Triton X-100, 5 mM 2-mercaptoethanol and 1 mM phenyl methyl sulfonyl fluoride] supplemented with 0.2 mg/ml lysozyme for lysis. After incubation for 30 min, cells were disrupted by sonication. All steps of the following purification procedure were performed at 4°C. The cell lysate was centrifuged at 25 000 r.p.m. in a Beckman SW28 rotor. To the supernatant, Polymin P (pH 7.9) was added to a final concentration of 0.4% with continuous stirring over a period of 15 min. The precipitate was collected by centrifugation, and the pellet was resuspended in buffer B [10 mM Tris–HCl (pH 8), 10% glycerol, 0.5 M NaCl and 5 mM 2-mercaptoethanol]. Solid (NH4)2SO4 (0.56 g/ml) was added with continuous stirring; after 1 h, the precipitated protein was pelleted at 14 000 r.p.m. for 30 min in an SS34 rotor. The precipitate was resuspended in buffer B and dialyzed against the same buffer containing 150 mM NaCl. The protein solution was applied onto a Heparin Affi-gel column (5 ml bed volume) by gravity, which had been equilibrated with buffer C [10 mM Tris–HCl (pH 8), 10% glycerol, 150 mM NaCl and 5 mM 2-mercaptoethanol]. The column was washed with buffer C containing 275 mM NaCl until the absorbance at 280 nm fell to 0.01. The protein was eluted with a linear gradient of 275–750 mM NaCl in buffer C. Fractions containing ScMre11p were identified by SDS–PAGE and nuclease assays (13,15) were combined and dialyzed against buffer D [50 mM sodium phosphate (pH 7), 10% glycerol, 0.3 M NaCl and 5 mM 2-mercaptoethanol]. This highly enriched ScMre11p fraction was applied onto CO-NTA matrix column, which had been equilibrated with buffer D. The column was washed with buffer D containing 10 mM imidazole until the absorbance at 280 nm fell to 0.01. The bound protein was eluted with a linear gradient of 15–150 mM imidazole in buffer D. Fractions containing ScMre11p were identified based on its expected molecular mass (80 kDa) and were combined and dialyzed into 10 mM Tris–HCl (pH 7.5) buffer containing 300 mM NaCl and 10% glycerol, and stored at a concentration of 1.3 mg/ml at −80°C. The resulting 80 kDa protein, corresponding to the predicted size of ScMre11p, was >99% pure as judged by SDS–PAGE and western blotting with anti-Mre11 protein antibodies. Polyclonal antibodies for ScMre11p were produced in rabbits using the same excised and eluted from SDS–PAGE gels.
Electrophoretic mobility shift assays
The DNA-binding activity of ScMre11p was measured in a 20 μl reaction mixture containing 32P-labeled 1 nM single- (ODN 4G3, Table 1), double-stranded DNA (annealed using ODN 4G3 and 4G3C, Table 1) or 3 nM G4 DNA (formed by ODN 4G3, Table 1) in 20 mM Tris–HCl (pH 7.5), 0.1 mM DTT, 0.2 mg/ml bovine serum albumin (BSA) and indicated concentrations of ScMre11p. For the competition experiments, excess of unlabeled competitor was also included in the reaction mixture. After 10 min incubation at 37°C, reactions were terminated with the addition of 2 μl loading dye (0.1% each of bromophenol blue and xylene cyanol in 50% glycerol) and subjected to electrophoresis on a 6% polyacrylamide gel containing 10 mM KCl in 45 mM Tris-borate buffer (pH 8.3) containing 10 mM KCl and 1 mM EDTA at 10 V/cm for 4 h at 4°C. All assays were repeated a minimum of three times to ensure reproducibility of results. Gels were visualized by phosphorImaging and/or autoradiography, and quantified using UVI Band MAP software.
The binding constants were determined by mobility shift assays in polyacrylamide gels. Reactions were performed as described above. The intensity of bands corresponding to free DNA and Mre11–DNA complex in each lane was quantified using UVI-Band Map software. The data was analyzed by non-linear regression equation using the Graph pad Prism software.
Nitrocellulose filter binding assays
Quantitative determination of ScMre11p–DNA complexes was performed as described in (31). Nitrocellulose filters (pore size, 0.45 μm; Sartorius) were pretreated with 0.5 M KOH at 4°C for 30 min and then extensively washed with buffer [20 mM Tris–HCl (pH 7.5) containing 50 mM NaCl] prior to use. Typically, reaction mixtures (20 μl) contained 20 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.1 mM DTT, 0.2 mg/ml BSA, and a fixed amount of labeled DNA (single-stranded or G4 DNA) and increasing concentrations of ScMre11p. After 60 min incubation at 37°C, 10 μl was spotted on to a nitrocellulose filter (for determination of total radioactivity) and 10 μl aliquots were diluted with 0.8 ml cold buffer [20 mM Tris–HCl (pH 7.5) and 50 mM NaCl] applied directly to the filter under suction, and washed with 5 ml of same buffer. The filters were dried, and the bound radioactivity was quantified by liquid scintillation. The data were analyzed by non-linear regression equation using the Graph pad Prism software.
Nuclease assays
Nuclease assays were performed as described in (9–11). Briefly, the reaction mixtures (20 μl) contained 20 mM Tris–HCl (pH 7.5), 0.1 mM DTT, 5 mM MnCl2, 150 mM NaCl, 200 μg BSA/ml, 1 nM single-stranded (ODN 4G3, Table 1) or G4 DNA (formed by ODN 4G3) and indicated concentrations of ScMre11p. After incubation at 37°C for 1 h, the reactions were terminated by the addition of EDTA, SDS and proteinase K to a final concentration of 10 mM, 1% and 10 μg/ml respectively, and incubated for 15 min at 37°C. Samples were loaded onto a denaturing (7 M urea) 15% polyacrylamide gel and electrophoresed in 89 mM Tris-borate buffer (pH 8.3) at 250 V for 1 h. Gels were visualized by autoradiography. In immunodepletion experiments, His-tagged ScMre11p was incubated with anti-His antibody as suggested by the manufacturer, prior to incubation with G4 DNA as described above.
Determination of the cleavage sites
Reaction mixtures (50 μl) contained 1 nM of 32P-labeled G4 DNA (formed by ODN 4G3, Table 1) or its single-stranded constituent in 20 mM Tris–HCl (pH 7.5), 0.1 mM DTT, 5 mM MnCl2, 200 μg BSA/ml and indicated concentrations of ScMre11p. Samples were incubated at 37°C for 1 h. The reaction was terminated with the addition of proteinase K, SDS and EDTA to a final concentration of 10 μg/ml, 1% and 10 mM, respectively. The cleavage products were extracted with phenol:chloroform:butanol solution, dried and resuspended in 5 μl of formamide loading buffer. Samples were heated at 95°C for 5 min, cooled on ice and loaded onto a denaturing (7 M urea) 20% polyacrylamide gel and electrophoresed in 89 mM Tris-borate buffer (pH 8.3) at 1800 V for 75 min. Gels were visualized by phosphorImaging and/or autoradiography.
Circular dichroism
Circular dichroism (CD) measurements were performed as described previously (32). The ODN 4G3 or G4 DNA (formed by ODN 4G3) was diluted to a final concentration of 2 μM in 20 mM Tris–HCl (pH 8) and 1 mM EDTA. CD spectra were recorded on a Jasco-815 spectropolarimeter in the spectral region of 320–220 nm using 1 cm path length cuvette at 20°C.
RESULTS
Experimental rationale
Several lines of evidence have implicated a major role for the MRX complex in telomerase-mediated replication of telomeric DNA, and genetic studies have indicated a direct role for S.cerevisiae MRE11 in the proper establishment of telomere end-structure (29). However, the molecular mechanism by which ScMre11p generates appropriate DNA termini for telomerase is obscure. We have used two approaches to examine this hypothesis. First, we measured the binding of ScMre11p to ssDNA and G4 DNA, a configuration of DNA that is presumed to exist at telomeres (25,33). Second, we have examined the ability of ScMre11p to cleave G4 DNA, which has been shown to inhibit DNA synthesis by telomerase, and ssDNA to generate G-rich seed sequences.
Purification of ScMre11p
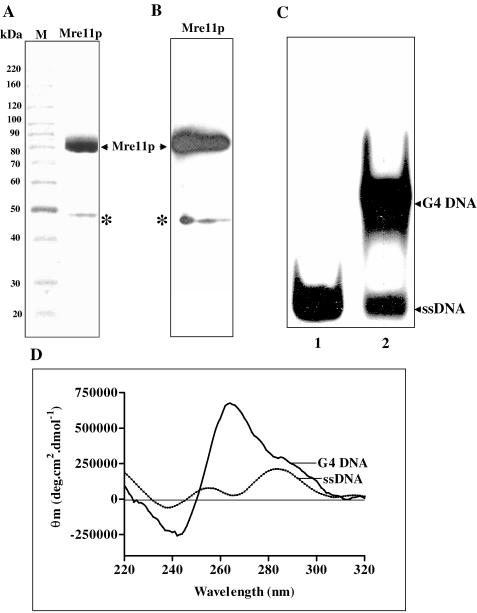
S.cerevisiae MRE11 was cloned and overexpressed in E.coli BL21 (DE3)pLysS host strain. ScMre11p was purified from cell-free lysates as described in Materials and Methods. The identity of ScMre11p was ascertained by 7.5% SDS–PAGE and visualized by Coomassie blue staining and western blotting with polyclonal antibodies against Mre11p (Figure 1A and B).
Figure 1.
Preparation of Mre11 protein and G4 DNA. (A) SDS–PAGE analysis of Mre11 protein: 15 μg of ScMre11p was analyzed by SDS–PAGE and visualized after staining with Coomassie blue. Lane M, molecular mass markers. (B) Western blot analysis of purified ScMre11p. An identical gel as shown in (A) was stained with anti-ScMre11p antibodies. The asterisks denote the truncated form of ScMre11p, which accounts to <1% in the final preparation. (C) Isolation and characterization of G4 DNA. Lane 1, ssDNA; and lane 2, mixture of single-stranded and G4 DNA. G4 DNA was isolated from the gel as described previously (30). (D) CD spectra of single-stranded and G4 DNA. Reactions were performed as described in Materials and Methods. Each spectra represent the best fit of four determinations. Buffer contributions were subtracted from the spectrum as appropriate. Subtractions and spectra were analyzed and plotted using software supplied with the spectrometer.
ScMre11p is a specific G4 DNA-binding protein
The telomeric DNA in S.cerevisiae and humans contain tracts of three guanines (23,24). Accordingly, we generated G4 DNA from oligonucleotides (Table 1, ODN 4G3 or 6G3) containing an array of three guanines, which also have been shown to fold into G4 DNA by chemical probing and spectroscopic techniques (32,34). G4 DNA was separated from single-stranded constituent by PAGE (Figure 1C). Circular dichroism (CD) has been used as a sensitive technique to ascertain the formation of G4 DNA. The CD spectrum with positive ellipticity maximum at ∼260 nm and negative ellipticity minimum at ∼240 nm is characteristic of G4 DNA (32). In addition, the crystal structure of G4 DNA containing four consecutive repeats of 3 G residues (4G3) has been shown to form an intramolecular G4 DNA (35). Consistent with previous findings (32), the CD spectra indicated that the DNA preparation was fully folded into a homogeneous population of G4 DNA (Figure 1D). For comparison, we monitored the spectra of its single-stranded constituent, which was devoid of the signals characteristic of G4 DNA.
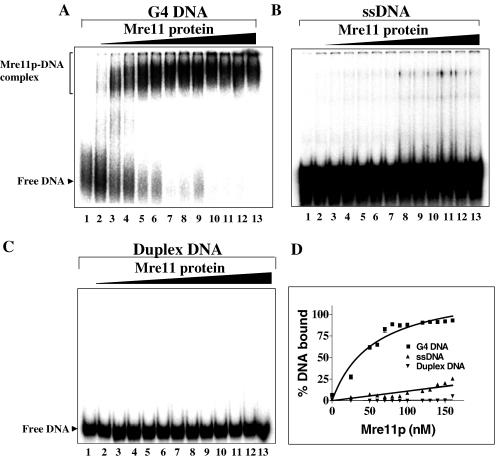
The ability of ScMre11p to bind G4 DNA was studied by gel mobility shift assay. Reactions were performed with increasing concentrations of ScMre11p and a fixed amount of 32P-labeled G4 DNA (formed by ODN 4G3, Table 1) or ssDNA in the absence of Mn2+. At lower concentrations, ScMre11p formed multiple species of protein–G4 DNA complexes as evidenced by smearing of the bands with slower mobility relative to free DNA (Figure 2A, lanes 2 and 3). However, the formation of a single species of protein–DNA complex progressively increased with increasing concentrations of ScMre11p (Figure 2A, lanes 5–13). In parallel, we examined the ability of ScMre11p to bind ssDNA (ODN 4G3, Table 1), and 38 bp duplex DNA (annealed using ODN 4G3 with its complementary strand ODN 4G3C, Table 1). ScMre11p was able to bind ssDNA to an extent of 20% albeit at higher concentrations (Figure 2B and D). However, ScMre11p failed to alter the mobility of duplex DNA at all the concentrations tested (Figure 2C).
Figure 2.
ScMre11 is a high affinity G4 DNA-binding protein. Reaction mixtures contained 3 nM of indicated radiolabeled DNA (lane 1) and ScMre11p at 25, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140 and 150 nM (lanes 2–13) in the absence of divalent metal ions. (A) 32P-labeled G4 DNA; (B) 32P-labeled ssDNA; (C) 32P-labeled duplex DNA (38 bp); and (D) graph showing quantitative comparison of ScMre11p binding to G4 DNA, G-rich single-stranded and duplex DNA. Samples were electrophoresed on native PAGE, and visualized by autoradiography, as described in Materials and Methods.
However, previous studies have shown binding of ScMre11p to duplex and ssDNA. Consequently, the binding assays were performed over a wide range of ScMre11p (0.1–7 μM). The extent of binding paralleled previously published results (10–13), and half-maximal binding to duplex and ssDNA relative to G4 DNA required 20-fold excess of ScMre11p (data not shown). Together, these results suggest that ScMre11p displays higher binding affinity for G4 DNA than to duplex DNA or ssDNA.
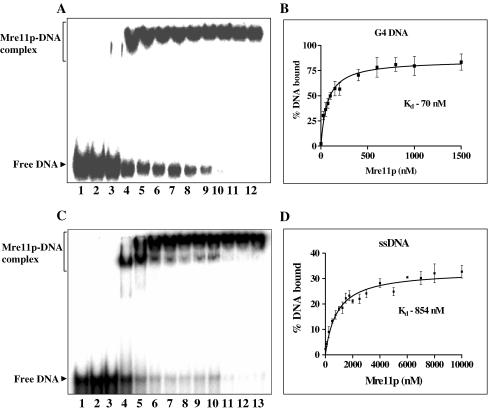
The binding constants for Mre11p binding to G4 DNA and its single-stranded constituent were determined by mobility shift assays (Figure 3A and C). The constants deduced from Mre11p concentration at which 50% DNA was bound, and represent mean values from six independent binding curves. This analysis yielded a Kd of 70 nM for G4 DNA and 854 nM for ssDNA, indicating that the affinity of ScMre11p for G4 DNA was ∼12-fold higher compared with ssDNA (Figure 3B and D). The calculated binding constants were validated by nitrocellulose filter binding assays. A fixed amount of 32P-labeled G4 DNA (formed by ODN 4G3, Table 1), or its monomeric constituent, was incubated with increasing concentrations of ScMre11p. Samples were analyzed as described in Materials and Methods. The binding constants determined by nitrocellulose filter binding assay suggested that the affinity of Mre11p to G4 DNA was 12- to 13-fold higher compared with ssDNA, which is consistent with that determined by mobility shift assays (data not shown).
Figure 3.
Determination of dissociation constants for ScMre11p–DNA complexes. Reactions were performed as described in the legend to Figure 2. The samples were separated by electrophoresis and analyzed as described in Materials and Methods. (A) 32P-labeled G4 DNA incubated with increasing concentrations of Mre11p. Reaction mixtures contained 1 nM of indicated radiolabeled DNA (lane 1) and ScMre11p at 25, 50, 75, 100, 150, 200, 250, 500, 750, 1000, 1500 nM (lanes 2–12) in the absence of divalent metal ions. (B) Graph showing binding of ScMre11p to G4 DNA. (C) 32P-labeled ssDNA incubated with increasing concentrations of Mre11p. Reaction mixtures contained 1 nM of indicated radiolabeled DNA (lane 1) and ScMre11p at 250, 500, 750, 1000, 2000, 3000, 4000, 5000, 6000, 7000, 8000, 10000 nM (lanes 2–13) in the absence of divalent metal ions. (D) Graph showing binding ScMre11p to ssDNA. Each data point in the graph is the average of six independent experiments. The results were analyzed by non-linear regression equation. The slope of the curves yielded the indicated Kd values.
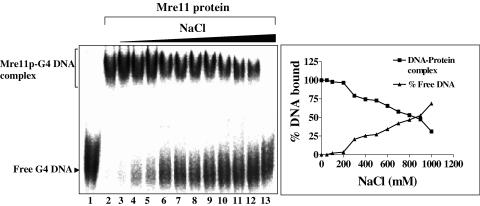
To further characterize the DNA-binding properties, the kinetics of binding of ScMre11p was measured with G4 DNA. The binding of ScMre11 to G4 DNA occurred rapidly with a t1/2 = 0.5 min (data not shown), which is comparable with the rates that have been reported for a variety of DNA-binding proteins in vitro (30,36). Binding was independent of temperature over a broad range (4–37°C) as well as nucleotide cofactors and metal ions (data not shown), indicating that ScMre11p by itself can rapidly access DNA. The G4 DNA–ScMre11p complexes were sensitive to proteinase K/SDS treatment (data not shown). The binding of ScMre11p to G4 DNA was extremely stable since >0.8 M NaCl was required to dissociate 50% of protein–DNA complexes (Figure 4).
Figure 4.
Effect of NaCl on ScMre11p–G4 DNA complex. Reaction mixtures contained 3 nM of 32P-labeled G4 DNA and 0.1 μM ScMre11p. After incubation for 10 min, NaCl was added to 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 and 1 M (lanes 3–13). Samples were electrophoresed on polyacrylamide gel, and this was followed by autoradiography, as described in Materials and Methods. In the right panel, the extent of dissociation of ScMre11p–G4 DNA complex is plotted versus varying amounts of NaCl.
Specificity of binding of ScMre11p to G4 DNA
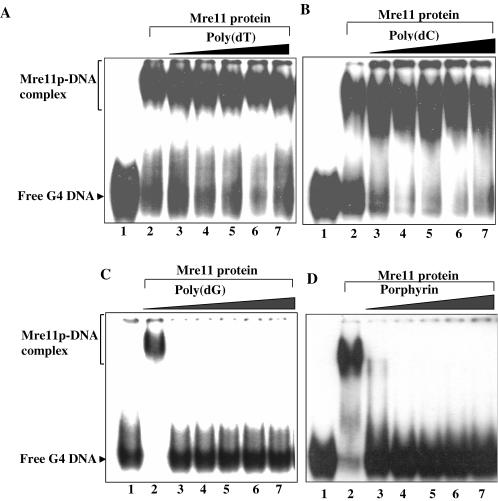
The higher affinity of ScMre11p for G4 DNA was ascertained in competition assays. The experiments consisted of mixing indicated amounts of the competitor with a constant amount of 32P-labeled G4 DNA (formed by ODN 4G3, Table 1). In accordance with the characteristics of specific protein–DNA interaction, the preference for G4 DNA was not diminished even at 17-fold excess of poly(dT) (Figure 5A), poly(dC) (Figure 5B) or poly(dA) (data not shown). In contrast, ∼3-fold excess of poly(dG), which spontaneously adopts G4 DNA configuration (33), competed efficiently for binding to ScMre11p (Figure 5C). It is possible that binding of poly(dG) to ScMre11p might be allosteric, inducing structural changes in ScMre11p that reduce its affinity. To ascertain this possibility, we used porphyrin, which is known to bind specifically to G4 DNA, stabilize its structure and influences protein activities on G4 DNA (37). As shown in Figure 5D, binding of ScMre11p to G4 DNA was inhibited at very low concentrations of porphyrin. These results support the notion that Mer11p recognizes distinct determinants in G4 DNA and inhibition by poly(dG) is via direct competition for DNA-binding by ScMre11p.
Figure 5.
ScMre11p binds preferentially to G4 DNA. Reactions were performed with 3 nM 32P-labeled G4 DNA and 0.1 μM ScMre11p as described in Materials and Methods. After incubation for 10 min, buffer or aliquots of indicated unlabeled competitors were added to the reaction mixture. Lane 2, no addition, and lanes 3–7, poly(dT) (A), poly(dC) (B) or poly(dG) (C) at 10, 20, 30, 40 and 50 nM; or porphyrin (D) at 1, 2, 3, 4 and 5 μM, respectively. Samples were analyzed as described in Materials and Methods.
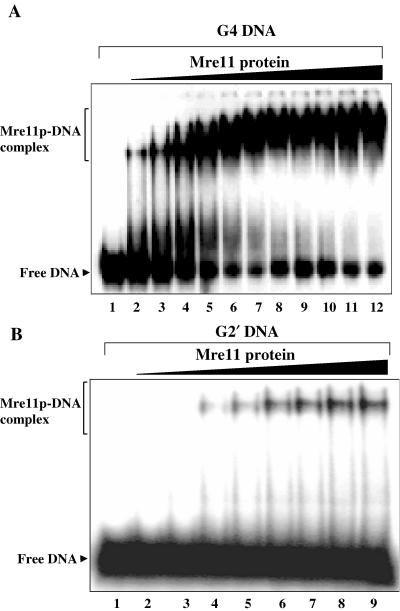
ScMre11p displays higher affinity for parallel G4 DNA
The foregoing observations revealed that ScMre11p was able to bind G4 DNA with high affinity. Since S.cerevisiae telomeric DNA sequence has been shown to form both parallel G4 DNA as well as antiparallel G2′ DNA in vitro (33), an important question is whether ScMre11p can bind to both G4 DNA as well as G2′ DNA. While ScMre11p was able to bind G4 DNA (formed by ODN 4G3; Table 1) at low amounts achieving plateau levels ∼150 nM (Figure 6A), the mobility of G2′ DNA (ODN 21G; Table 1) was reproducibly unchanged at equivalent concentrations. However, ScMre11p displayed binding to G2′ DNA in the μM range, but binding failed to exceed 20%, suggesting low-affinity interaction between ScMre11p and G2′ DNA (Figure 6B). Thus, specific binding of ScMre11p to G4 DNA suggests that it may be the natural substrate for Mre11 in vivo and that it exists in S.cerevisiae telomeric DNA.
Figure 6.
ScMre11p displays higher affinity for G4 DNA over G2′ DNA. (A) Reaction mixtures contained 3 nM 32P-labeled G4 DNA (lane 1) and ScMre11p at 50, 75, 100, 125, 150, 175, 200, 225, 250, 275 and 300 nM (lanes 2–12). (B) Reaction mixtures contained 3 nM 32P-labeled G2′ DNA (lane 1) and ScMre11p at 0.25, 0.5, 0.75, 1, 1.25, 1.5, 1.75 and 2 μM (lanes 2–9). Samples were analyzed as described in Materials and Methods.
Specific cleavage of G4 DNA and G-rich ssDNA by ScMre11p
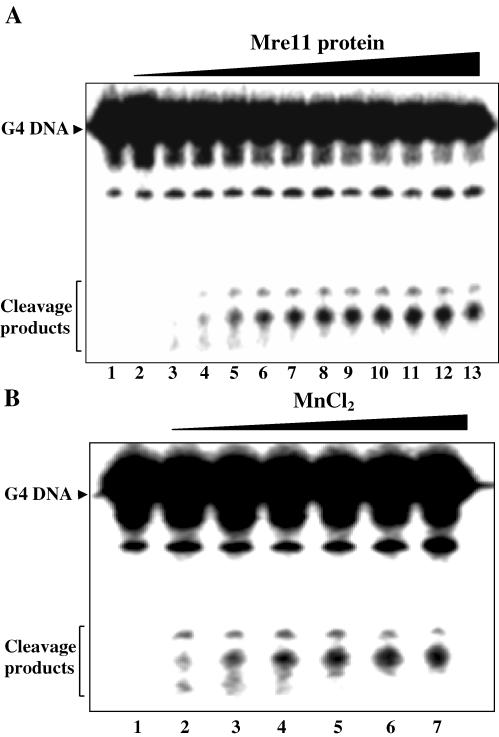
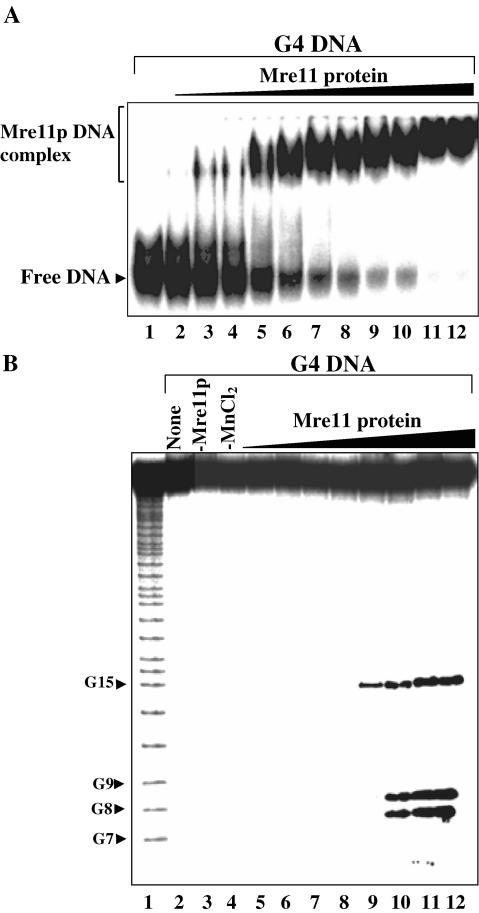
Since ScMre11p displayed high affinity for G4 DNA, this raised the possibility of whether ScMre11p possesses G4 DNA cleavage activity. To this end, 32P-labeled G4 DNA (formed by ODN 4G3; Table 1) was incubated with increasing concentrations of ScMre11p, and the products of the reaction were analyzed on denaturing polyacrylamide gels. The results show that ScMre11p generated two cleavage products, which increased linearly reaching plateau levels at ∼0.5 μM (Figure 7A). The cleavage increased with increasing concentrations of MnCl2, optimal being 3 mM (Figure 7B). But replacement of Mn2+ with 5 mM Mg2+ or Ca2+ failed to promote cleavage (data not shown), which distinguishes ScMre11p cleavage activity from the human GQN1, yeast KEM1/SEP1 or mXRN1 nuclease all of which require Mg2+ for their cleavage activity (38–40). Nuclease activity was significantly reduced at lower temperatures, abolished in the presence of porphyrin, and was neither enhanced nor diminished by rNTPs or dNTPs (data not shown).
Figure 7.
ScMre11p is a G4 DNA nuclease. (A) Reaction mixtures contained 1 nM 32P-labeled G4 DNA and ScMre11p at 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9 and 1 μM (lanes 3–13) in the absence (lane 2) or presence (lanes 3–13) of 5 mM Mn2+. Lane 2 contained 0.5 μM ScMre11p. (B) Cleavage of G4 DNA by ScMre11p in the absence or presence of increasing concentrations of Mn2+. Reaction mixtures contained 1 nM 32P-labeled G4 DNA and 0.5 μM ScMre11p in the absence (lane 1) or presence of 0.5, 1, 2, 3, 4 and 5 mM MnCl2 (lanes 2–7). Reaction products were analyzed on 15% PAGE in the presence of 7 M urea as described in Materials and Methods. Some G4 DNA preparations contained a small amount of ssDNA.
Then we sought to determine whether the endonuclease activity is intrinsic to ScMre11p or arises from a contaminating nuclease. To test this, His-tagged ScMre11p was incubated with anti-His antibody, and then incubated with G4 DNA under the same conditions as described above. In contrast to the control, endonuclease activity was not detectable in the immunodepleted reactions (data not shown). Together, these results suggest that G4 DNA endonuclease activity is intrinsic to ScMre11p.
Endonucleolytic cleavage of G-rich single-stranded and G4 DNA by ScMre11p
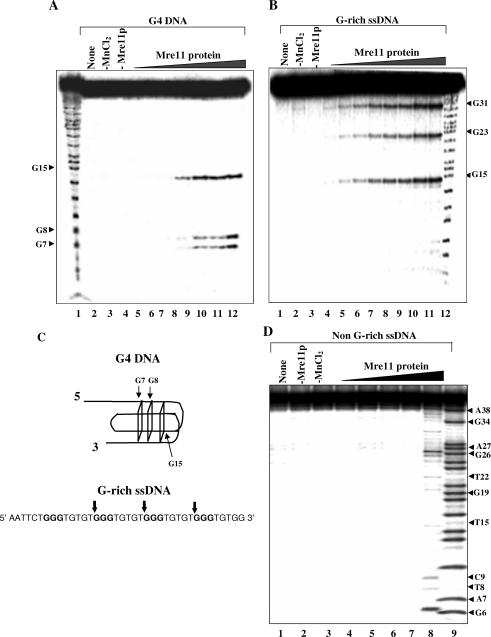
To identify the cleavage sites, the products of the cleavage reaction were separated by electrophoresis on sequencing gels and visualized by using autoradiography. The results are shown in Figure 8. ScMre11p cleaved G4 DNA (formed by ODN 4G3; Table 1) to generate three prominent bands and the extent of cleavage increased with increasing concentrations of ScMre11p (Figure 8A). In a similar experiment, we observed that 4G3 ODN was cleaved by Mre11p to generate three bands (Figure 8B). Identification of cleavage sites at the level of single-nucleotide resolution was made by comparison with a Maxam–Gilbert sequencing ladder using the same probe (4G3 ODN). Figure 8C summarizes the cleavage sites in G-rich single-stranded and G4 DNA. The cleavage sites in 4G3 ODN map onto the first G in the array of three G residues, while in G4 DNA they map within the vicinity of G-quartets. Together, these results indicate that cleavage of G-rich single-stranded and G4 DNA is due to the intrinsic endonuclease activity of Mre11p. For specificity controls, we incubated 32P-labeled non-G-rich ODN 41SD (Table 1) with concentrations of Mre11p as in Figure 8B. Analysis of reaction products suggested random cleavage of ODN by Mre11p at the highest concentration tested (lane 8, Figure 8D).
Figure 8.
Sequence- and structure-specific cleavage of G-rich single-stranded and G4 DNA by ScMre11p. (A) Reaction mixtures contained 1 nM 32P-labeled G4 DNA and ScMre11p at 0.1, 0.25, 0.5, 0.6, 0.7, 0.8, 0.9 and 1 μM (lanes 5–12) in the presence of 5 mM Mn2+. Lane 1, the G ladder. Controls performed in the absence of ScMre11p (lane 4), Mn2+ (lane 3) or no additions (lane 2) is indicated on top of each lane. (B) Reaction mixtures contained 1 nM 32P-labeled 38mer DNA and ScMre11p at 0.1, 0.25, 0.5, 0.6, 0.7, 0.8, 0.9 and 1 μM (lanes 4–11) in the presence of 5 mM Mn2+. Lane 12, G ladder. Controls performed in the absence of ScMre11p (lane 3), Mn2+ (lane 2) or no additions (lane 1) is indicated on top of each lane. Reaction products were analyzed on 20% PAGE in the presence of 7 M urea. (C) Summary of cleavage sites in G-rich single-stranded and G4 DNA by ScMre11p. The vertical arrows denote the position of cleavage. (D) Reaction mixtures contained 15 nM 32P-labeled non-G-rich ssDNA and ScMre11p at 0.1, 0.25, 0.5, 0.75 and 1 μM (lanes 4–8) in the presence of 5 mM Mn2+. Lane 9, the G+A ladder. Controls performed in the absence of ScMre11p (lane 2), Mn2+ (lane 3) or no additions (lane 1) is indicated on top of each lane.
We note that the extent of cleavage of G4 DNA that readily folds into a variety of non-B form DNA conformations (33), by Mre11p is less than that of B-DNA substrates (11). We speculate that this may be the result of non-B form DNA conformation of the substrate. It is possible that the cleavage of G4 DNA is exonucleolytic and the cleavage might be stalling at G-quartets. However, this possibility is unlikely because Mre11p nicks in the G4 DNA at two internal G-quartet sites (Figure 8C).
A caveat of the data presented above and the conclusions are based on the use of G4 DNA formed by ODN 4G3. To ascertain the generality of the phenomenon, we used an alternate DNA substrate. Monitoring the binding affinity of ScMre11p to G4 DNA formed by ODN 6G3 revealed a striking similarity to its binding to G4 DNA formed by ODN 4G3 (compare Figure 9A with Figure 2A). In addition, the efficiency and pattern of cleavage of G4 DNA formed by ODN 6G3 was similar to that of G4 DNA formed by ODN 4G3 (Figure 9B).
Figure 9.
ScMre11 displays higher affinity to G4 DNA formed by ODN 6G3 and cleaves it in a structure-specific manner. (A) Reaction mixtures contained 3 nM of indicated radiolabeled DNA (lane 1) and ScMre11p at 25, 50, 60, 70 80, 100, 110, 120, 130, 140 and 150 nM (lanes 2–12) in the absence of divalent metal ions. (B) Reactions were performed as described in the legend to Figure 8A.
DISCUSSION
In this study, we show that ScMre11p displays substantially higher binding affinity for G4 DNA, compared with single- or double-stranded DNA. Importantly, Mre11p cleaves G-rich single-stranded and G4 DNA in a sequence- and structure-specific manner. Our results are consistent with the genetic evidence implicating S.cerevisiae MRE11 in the establishment of proper telomere end structure (29). These observations suggest that ScMre11p might provide appropriate DNA termini for telomerase-mediated replication of telomeric DNA and loading of telomere-binding proteins.
Although elegant genetic and molecular biology approaches have led to the identification of components involved in telomere length maintenance, yet our understanding of how these proteins regulate telomere length is not clear. Several lines of evidence invoke MRX complex as the functional entity in regulation of telomere length. To begin to understand the mechanistic aspects of MRX function, we investigated the biochemical activities of Mre11p using DNA-binding and nuclease assays. We found that Mre11p displays higher binding affinity for G4 DNA, a configuration of DNA that is presumed to exist at telomeres (25,33), compared with single- or double-stranded DNA. Further analysis demonstrated that Mre11p binds G4 DNA with 12-fold higher binding affinity over ssDNA. Surprisingly, Mre11p remains stably bound even in the presence of non-physiological concentrations of NaCl. The physiological significance of its stable binding to G4 DNA remains to be elucidated. The observation that Mre11p binds G4 DNA with higher binding affinity led us to a second line of inquiry concerning its intrinsic nuclease activity. To address this question, we examined the ability of Mre11p to cleave G4 DNA in the presence of Mn2+. The endonucleolytic products generated by Mre11p with both single-stranded and G4 DNA contain G residue at the 5′ end. Thus, the ends generated by Mre11p could either directly serve as a template for telomerase or require further processing by other telomere-binding proteins.
In S.cerevisiae, both telomerase-dependent and telomerase-independent mechanisms co-exist to ensure proper telomere length. Inactivation of the components of telomerase triggers progressive telomere shortening culminating in genomic instability and cell death (41–43). In addition, telomerase-proficient cells but lacking any one or all three members of the MRX complex are non-viable and show severe defects in recombination and telomere length maintenance (6–8). When the telomeres reach a critical short length, a recombination process mediated by Rad52p, MRX complex and other components leads to telomere elongation, thus enabling few cells to regain wild-type growth potential (6–9,44). These survivors fall into two classes: type I cells that show amplification of the telomere-associated Y′ elements and have very short TG1–3 repeat tracts and type II cells that have long variable tracts of TG1–3 repeats and only modest Y′ element amplification (44,45). Type I-mediated recombination requires RAD51, RAD54, RAD57, CDC13 and the replicative polymerases, whereas type II survivors depend on RAD50, RAD59, MRE11, XRS2, TEL1 and MEC1 (6–9,46,47).
In Trypanosoma brucei, mouse and human cells, telomeric DNA adopts T-loop structures (48–50). Similarly, Taz1p of fission yeast catalyzes the formation of higher order structures and T-loops from model DNA substrates containing 3′ single-stranded telomeric overhangs (50). Although a similar structure has not been reported for S.cerevisiae, it is proposed that the formation of G-quartet and fold back structures may account for the repression of telomere proximal genes and stabilization of telomeres. S.cerevisiae strains lacking telomerase lose their telomeres gradually and eventually perish due to chromosome instability (23,24,50). It has been speculated that helicases disrupt G•G (Hoogsteen) base pairs in G4 DNA to generate 3′ TTAGGG overhangs, but the molecular mechanism of the generation of free-ends required for helicase(s) action is obscure (32,50). Genetic studies have shown that MRX complex is required for the generation of G-strand and loading of G-strand binding proteins (51,52). The most striking observation in this report is that Mre11p cleaves G4 DNA within the vicinity of G-quartets. In view of the fact that helicases and telomerase require free ends to serve substrates, these findings indicate that the endonucleolytic activity of Mre11p is likely to play a direct biological role in vivo.
A role for ScMre11p in the cleavage of G4 DNA can be visualized in the context of replication of telomerase-dependent telomeric DNA. The generation of suitable substrate for telomerase and telomere-binding proteins present a unique challenge to cells. Short direct DNA repeats comprise the underlying telomeric DNA and the strand that runs 5′ to 3′ towards the chromosome end is rich in guanines (G-tail). G-tails are presumed substrates for telomerase. Studies in S.cerevisiae have shown that telomeres gain long G-tails late in S phase after replication (50). The steps involved in elongation of telomeres include the generation of single-stranded TG1–3 tails, Cdc13p binding to this tail, recruitment of telomerase, Est2p-mediated lengthening of the TG1–3 tail, and C-strand synthesis (23,24,50). It has been speculated that Tel1p and MRX complex could act at any of these steps.
Careful inspection of the sites of telomerase addition revealed that healing at spontaneous breaks often takes place at naturally occurring G-rich seed sequences that resemble the G-rich repeats found at telomeres. This suggests that the sequence-specific telomere end-binding protein Cdc13, which is responsible for recruiting telomerase to chromosome ends (50,52,53), may similarly mediate the access of telomerase to DSBs. In strains of certain genetic backgrounds, the spectrum of chromosome healing events and telomere addition display a strong bias for sites that contain G-rich tracts, with essentially no healing at sites that lack a G-rich seed (50,54). Our data are consistent with a direct role for G-rich DNA endonuclease activity of ScMre11p in several nuclear processes including telomere length maintenance, cellular response to DNA damage and recombination.
Together, these results suggest that the nuclease activities of ScMre11p on G4 DNA and G-rich ssDNA are likely to be important for telomere replication and recombinational repair. Since the MRX complex is conserved, we speculate that a similar mechanistic role is likely to ensue for ScMre11p in vertebrate cells.
Acknowledgments
We thank the anonymous reviewers who meticulously reviewed the manuscript and provided us with extremely constructive comments. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Haber J.E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 2.Petrini J.H. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell. Biol. 2000;12:293–296. doi: 10.1016/s0955-0674(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 3.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours D., Jackson S.P. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nature Rev. Mol. Cell. Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- 5.Assenmacher N., Hopfner K.P. MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113:157–166. doi: 10.1007/s00412-004-0306-4. [DOI] [PubMed] [Google Scholar]
- 6.Kironmai K.M., Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 7.Boulton S.J., Jackson S.P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nugent C.I., Bosco G., Ross L.O., Evans S.K., Salinger A.P., Moore J.K., Haber J.E., Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr. Biol. 1998;8:657–662. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 9.Moreau S., Ferguson J.R., Symington L.S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paull T.T., Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol. Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 11.Trujillo K.M., Yuan S.-S.F., Lee Y.-H.P., Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J. Biol. Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 12.de Jager M., Dronkert M.L., Modesti M., Beerens C.E., Kanaar R., van Gent D.C. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 2001;29:1317–1325. doi: 10.1093/nar/29.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paull T.T., Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA. 2000;97:6409–6414. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paull T.T., Gellert M. Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 1999;13:1276–1288. doi: 10.1101/gad.13.10.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trujillo K.M., Sung P. DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50-Mre11 complex. J. Biol. Chem. 2001;276:35458–35464. doi: 10.1074/jbc.M105482200. [DOI] [PubMed] [Google Scholar]
- 16.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 17.Hopfner K.P., Karcher A., Shin D.S., Craig L., Arthur L.M., et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 18.de Jager M., van Noort J., van Gent D.C., Dekker C., Kanaar R., Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell. 2001;8:1129–1135. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 19.Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Jager M., Wyman C., van Gent D.C., Kanaar R. DNA end-binding specificity of human Rad50/Mre11 is influenced by ATP. Nucleic Acids Res. 2002;30:4425–4431. doi: 10.1093/nar/gkf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borde V., Lin W., Novikov E., Petrini J.H., Lichten M., Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- 22.Trujillo K.M., Roh D.H., Chen L., Van Komen S., Tomkinson A., Sung P. Yeast Xrs2 binds DNA and helps target Rad50 and Mre11 to DNA ends. J. Biol. Chem. 2003;278:48957–48964. doi: 10.1074/jbc.M309877200. [DOI] [PubMed] [Google Scholar]
- 23.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 24.Chakhparonian M., Wellinger R.J. Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet. 2003;19:439–446. doi: 10.1016/S0168-9525(03)00135-5. [DOI] [PubMed] [Google Scholar]
- 25.Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 26.Sen D., Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 27.Gellert M., Lipsett M.N., Davies D.R. Helix Formation by Guanylic Acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DMA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 29.Larrivee M., LeBel C., Wellinger R.J. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18:1391–1396. doi: 10.1101/gad.1199404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muniyappa K., Anuradha S., Byers B. Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol. 2000;20:1361–1369. doi: 10.1128/mcb.20.4.1361-1369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kironmai K.M., Muniyappa K., Friedman D.B., Hollingsworth N.M., Byers B. DNA-binding activities of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:1424–1435. doi: 10.1128/mcb.18.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giraldo R., Suzuki M., Chapman L., Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williamson J.R. G-quartet structures in telomeric DNA. Annu. Rev. Biophy. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 34.Giraldo R., Rhodes D. The yeast telomere-binding protein RAP1 binds to and promotes the formation of DNA quadruplexes in telomeric DNA. EMBO J. 1994;13:2411–2420. doi: 10.1002/j.1460-2075.1994.tb06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 36.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anantha N.V., Azam M., Sheardy R.D. Porphyrin binding to quadrupled T4G4. Biochemistry. 1998;37:2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- 38.Sun H., Yabuki A., Maizels N. A human nuclease specific for G4 DNA. Proc. Natl Acad. Sci. USA. 2001;98:12444–12449. doi: 10.1073/pnas.231479198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Z., Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 40.Bashkirov V.I., Scherthan H., Solinger J.A., Buerstedde J.M., Heyer W.D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundblad V., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 42.Hackett J.A., Feldser D.M., Greider C.W. Telomere dysfunction increases mutation rate and genomic instability. Cell. 2001;106:275–286. doi: 10.1016/s0092-8674(01)00457-3. [DOI] [PubMed] [Google Scholar]
- 43.Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 44.Teng S.C., Zakian V.A. Telomere–telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ritchie K.B., Petes T.D. The Mre11p/Rad50p/Xrs2p complex and the Tel1p function in a single pathway for telomere maintenance in yeast. Genetics. 2000;155:475–479. doi: 10.1093/genetics/155.1.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng S.C., Chang J., McCowan B., Zakian V.A. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 47.Stansel R.M., de Lange T., Griffith J.D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munoz-Jordan J.L., Cross G.A.M., de Lange T., Griffith J.D. t-loops at trypanosome telomeres. EMBO J. 2001;20:579–588. doi: 10.1093/emboj/20.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomaska L., Willcox S., Slezakova J., Nosek J., Griffith J.D. Taz1 binding to a fission yeast model telomere: formation of telomeric loops and higher order structures. J. Biol. Chem. 2004;279:50764–50772. doi: 10.1074/jbc.M409790200. [DOI] [PubMed] [Google Scholar]
- 50.Smogorzewska A., de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 51.Diede S.J., Gottschling D.E. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr. Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 52.Tsukamoto Y., Taggart A.K., Zakian V.A. The role of the Mre11-Rad50-Xrs2 complex in telomerase-mediated lengthening of Saccharomyces cerevisiae telomeres. Curr. Biol. 2001;11:1328–1335. doi: 10.1016/s0960-9822(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 53.Evans S.K., Bertuch A.A., Lundblad V. Telomeres and telomerase: at the end, it all comes together. Trends. Cell. Biol. 1999;9:329–331. doi: 10.1016/s0962-8924(99)01596-2. [DOI] [PubMed] [Google Scholar]
- 54.Pennock E., Buckley K., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]