Abstract
Copper-containing nitrite reductases catalyze the reduction of nitrite to nitric oxide (NO), a key step in denitrification that results in the loss of terrestrial nitrogen to the atmosphere. They are found in a wide variety of denitrifying bacteria and fungi of different physiology from a range of soil and aquatic ecosystems. Structural analysis of potential intermediates in the catalytic cycle is an important goal in understanding enzyme mechanism. Using “crystal harvesting” and substrate-soaking techniques, we have determined atomic resolution structures of four forms of the green Cu-nitrite reductase, from the soil bacterium Achromobacter cycloclastes. These structures are the resting state of the enzyme at 0.9 Å, two species exhibiting different conformations of nitrite bound at the catalytic type 2 Cu, one of which is stable and also has NO present, at 1.10 Å and 1.15 Å, and a stable form with the product NO bound side-on to the catalytic type 2 Cu, at 1.12 Å resolution. These structures provide incisive insights into the initial binding of substrate, its repositioning before catalysis, bond breakage (O–NO), and the formation of a stable NO adduct.
Keywords: catalysis, denitrification, enzyme mechanism, nitrite and nitric oxide binding, crystal structures
Denitrification is the process by which some microorganisms couple respiratory ATP synthesis to the reduction of nitrate or nitrite to dinitrogen via the gaseous N-oxides NO and N2O. Denitrification forms an important step in the global nitrogen cycle. It has agronomic, environmental, and medical impacts (1–3). The individual reactions of denitrification are catalyzed by distinct reductases that variously contain Mo, Fe, Cu, or heme centers. The chemistry is that of small-molecule binding and activation and coupled electron–proton transfer in one or two electron reductions steps. The reduction of nitrite to NO is catalyzed by nitrite reductase (NiR). Bacteria employ one of two different forms of NiR, containing either heme cd1- or Cu in their prosthetic group (2, 3). Cu-nitrite reductases are divided into two subclasses based on their colors, green and blue, and their redox partners, pseudoazurin and azurin, respectively. Both subclasses exhibit a trimeric structure and constitute a highly conserved family of proteins that catalyze the chemical conversion of nitrite (NO–2) to NO, the first committed step in the denitrification process of the geobiological nitrogen cycle.
Each monomer of CuNiR has two types of Cu centers. A type 1 Cu ion (T1Cu) with (His)2-Cys-Met ligation is located within each monomer and is responsible for giving rise to the color of the enzyme (green or blue). It has been assigned a role in electron delivery to the catalytic type 2 center located at the interface of adjacent monomers. The type 2 Cu ion (T2Cu) has (His)3-H2O (or OH–) ligation with one of the His residues being provided by an adjacent monomer. Mutagenesis studies have shown that the active site pocket also has conserved aspartyl (Asp-98) and histidinyl (His-255) residues that are essential for enzymatic activity (4–7) and an isoleucine (Ile-257) residue that is important for optimum positioning of nitrite before catalysis (8). The residues His-255 and Ile-257 are provided by the adjacent monomer.
X-ray crystal structures have been determined for the green NiRs from Achromobacter cycloclastes (AcNiR) (9, 10) and Alcaligenes faecalis NiR (4, 11–13) and the blue Alcaligenes xylosoxidans (AxNiR) (14, 15), for which an atomic resolution structure has been reported recently (5). For the green nitrite reductases, resolution of the structure has steadily improved with the highest resolution being 1.4 Å. The nature of substrate binding in CuNiRs has been the focus of a number of biochemical, spectroscopic (electron nuclear double resonance/x-ray absorption fine structure), and crystallographic studies (10, 13, 14, 16–19). These studies have been in general agreement that nitrite binds to the T2Cu ion via O in an asymmetric bidentate fashion. Recently, Tocheva et al. (20) have noted a face-on interaction of the nitrite with Cu in their most recent 1.4-Å resolution structure compared with previous observations of bent O coordination. An examination of the Protein Data Bank (21) reveals a significant variation in the position and orientation of nitrite in the nitrite-bound CuNiRs. This variation raises an interesting possibility that some of these may represent different stages of binding/activation relevant to intermediates of the catalytic cycle. Using “crystal harvesting” and substrate-soaking techniques, we have determined atomic resolution structures of four forms of AcNiR, including two with nitrite bound (in two conformations) and one with the product NO bound. These structures are reported together with the structure of the resting state of the enzyme at 0.9 Å, the highest resolution obtained for any Cu protein. Our structures provide direct visualization of the initial binding of substrate, its reorientation before catalysis, and product formation.
Experimental Procedures
Growth of the organism and protein purification are described in Supporting Text, which is published as supporting information on the PNAS web site. Crystals were grown using the vapor diffusion hanging drop method with a protein concentration of 10 mg/ml. The reservoir solution consisted of 1.6 M ammonium sulfate and 0.1 M sodium acetate (pH 4.75). Crystals grew overnight in space group P213 and were soaked in a cryoprotectant solution comprising 3.5 M sodium malonate (pH 5.0) before flash-cooling in the cryostream. X-ray data were collected at 100 K on a Quantum 4 charge-coupled device detector (Area Detector Systems Corporation, Poway, CA) by using a 0.87-Å wavelength on station 9.6 and on a MAR225 charge-coupled device detector by using a 1.07-Å wavelength on station 10.1 at the Synchrotron Radiation Source, Daresbury Laboratory. A total of 20 crystals were examined. The data resolution was assessed according to the completeness in the outermost resolution shell. The data were indexed, scaled, and merged by using hkl2000 (22).
The resting-state structure was solved by using molrep (23) with AcNiR (PDB ID code 1NIF) as the search model. One monomer was located in the crystallographic asymmetric unit. The initial model was refined by the maximum-likelihood method implemented in refmac5 (24) as part of the ccp4i program suite (25) and rebuilt interactively by using 2Fobs – Fcalc and Fobs – Fcalc electron density maps in the programs o (26) and coot (27). Five percent of the data were set aside from refinement to calculate a free R factor. At no point in the refinement were restraints applied to the Cu–ligand distances or bond angles. Occupancies of the T2Cu and substrate/product atoms were estimated from the fitness of their B factors with those of the T2Cu ligands, and identification and modeling of substrate was aided by difference electron density analysis. A bulk solvent correction was applied, and solvent molecules were added by using arp/warp (28). The final model was then used as the starting structure for analyzing subsequent data from harvested crystals. The data collection and refinement statistics are summarized in Table 1 (and see Table 2, which is published as supporting information on the PNAS web site).
Table 1. Crystallographic data collection, processing, and refinement statistics.
| Resting enzyme | Nitrite-soaked | Endogenous NO | Endogenous nitrite + NO | |
|---|---|---|---|---|
| Resolution, Å | 20-0.90 | 30-1.10 | 50-1.12 | 17-1.15 |
| (last shell) | (0.93-0.90) | (1.13-1.10) | (1.16-1.12) | (1.19-1.15) |
| Completeness (%) | 98.1 (98.5) | 98.2 (93.3) | 97.8 (88.0) | 98.9 (97.7) |
| Redundancy | 5.2 (3.0) | 4.4 (3.6) | 4.4 (2.6) | 3.1 (2.3) |
| Unique reflections | 208,276 | 113,799 | 108,076 | 103,981 |
| Rmerge (%) | 5.9 (55.2) | 7.1 (60.9) | 5.4 (40.4) | 5.7 (42.1) |
| Wilson B factor (Å2) | 5.7 | 9.7 | 11.2 | 7.2 |
| Overall B factor (Å2) | 7.0 | 11.5 | 12.0 | 9.1 |
| R factor (Rfree) (%) | 11.7 (13.4) | 11.9 (14.5) | 12.7 (16.3) | 11.3 (14.3) |
| ML positional ESU, Å | 0.011 | 0.018 | 0.022 | 0.020 |
ML, maximum likelihood; ESU, estimated standard uncertainty.
Results and Discussion
The work presented here arises from our initial interest in determining an atomic resolution structure of a green nitrite reductase, for which pseudoazurin is the redox partner, with the aim of making a detailed comparison with the blue Cu-nitrite reductase, for which we had published the atomic resolution (1.04 Å) structure (5) and for which azurin is the redox partner. Surprisingly, in our first structure (1.55-Å resolution; data not shown) of the “as-isolated” AcNiR we found that the T2Cu site had NO bound in a side-on manner, similar to that reported recently for crystals of reduced Alcaligenes faecalis NiR exposed to NO under anaerobic conditions (20). Because we had not added exogenous NO, and because the crystals or protein had not been reduced, we decided to look at further crystals from the same bulk culture. We discovered that in some cases we had NO bound to the catalytic T2Cu and in others we had the water molecule bound to T2Cu as expected for the resting state. This observation provided the motivation to harvest crystals and collect very-high-resolution data sets from >20 crystals, some of these represented resting state, some represented NO bound, and some revealed both nitrite and NO bound to T2Cu. Here, only the highest-resolution structures for each category are presented. In addition, data were collected from nitrite-soaked crystals. We also purified the protein using bulk crystallization, and crystals grown from this purification were also used for additional data collection. Analysis of these data has provided atomic resolution structures representing the resting-state enzyme, the enzyme with nitrite bound in two different orientations, which we suggest represent different intermediate stages of substrate binding/reorganization of the catalytic center, and a NO-bound state. In one of the nitrite-bound structures, NO is also present at the T2Cu site in a fraction of the molecules, where it occupies a position that is virtually the same as that of the equivalent atoms of the nitrite ligand. We suggest these represent structures before bond breakage and product formation, respectively.
Structure of the Resting-State Enzyme at 0.9-Å Resolution. Two data sets collected were found to represent the resting state of the enzyme. The higher-resolution structure, determined at 0.9 Å, is the highest resolution reported for any Cu protein. It has a very low overall Wilson B factor of 5.7 Å2, indicating a well ordered structure. Recently, we had determined the structure of the blue Cu-nitrite reductase AxNiR at 1.04-Å resolution (5). The near doubling of the data (number of unique observations) compared with the atomic resolution structure of AxNiR enables the identification of dual conformations of key residues with significantly increased confidence. Moreover, this in principle would also allow a detailed study of the anisotropic behavior [through full matrix refinement (not yet done)] of residues in the putative channels and pockets involved in electron and proton delivery, as well as substrate guidance to the active site.
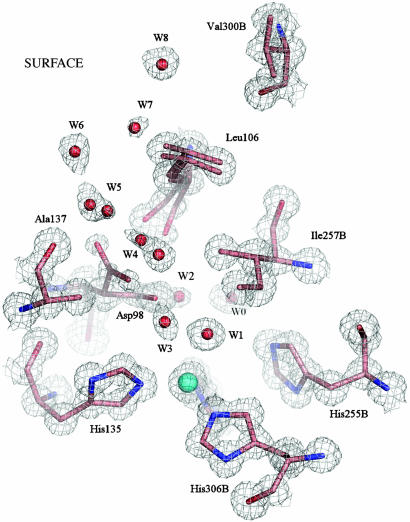
The catalytic T2Cu site is located in the interdomain cleft ≈12 Å from the protein surface (Fig. 1). A hydrophobic channel ≈6 Å wide, formed by residues belonging to adjacent monomers, provides the likely route of substrate entry. Two of these residues, Ala-137 and Leu-106, are shown in Fig. 1. The Asp-98 residue lies at the bottom of the hydrophobic channel. Two positions for the side chain of Asp-98 are clearly visible in the electron density map. To our knowledge, this is the first time that two conformations of Asp-98 have been observed in any structure of NiR, and its significance from the catalytic viewpoint becomes apparent (see below). The main orientation (80%), which will be referred to as the “proximal” position of Asp, is in a favorable position to form H-bonds with Cu ligand W1 at 2.8 Å and with water molecule W0 at 3.2 Å. W0 is also H-bonded to the NE2 atom of His-255 at 2.8 Å and is part of the water network that has previously been identified as belonging to the proton delivery channel (the anisotropy of the T2Cu site and water network comprising the proton delivery channel is shown in Fig. 5, which is published as supporting information on the PNAS web site) (5, 6, 15). In the second position, Asp-98 points into the substrate entry pocket and henceforth will be referred to as the “gatekeeper” position in analogy with the equivalent residue in carbonic anhydrase (18). The positions adopted by several water molecules and the Leu-106 side chain are influenced by this change in orientation of Asp-98. The occupancies of these water molecules and the Asp-98/Leu-106 residues are well correlated. Full details of the active site water network are given in Supporting Text.
Fig. 1.
The T2Cu site of the resting state enzyme at 0.9 Å resolution is shown with 2Fobs – Fcalc electron density contoured at 1.2σ. The site is well ordered, with isotropic B factors of 5.3 Å2 for the Cu2+ atom and an average of 5.4 Å2 for the NE2 atoms of the three ligating histidine ligands (100, 135, 306B), coordinated at 1.99 Å. A water molecule, W1, is the fourth Cu ligand, at 2.2 Å with a B factor of 5.9 Å2. The Ile257B residue from the second monomer provides a steric constraint (W1 to Ile257B is ≈3.4 Å) on the substrate binding site (8). A well ordered network of water molecules, W3–W8, leads from the substrate-binding site to the protein surface along the probable route of nitrite entry. W2, W4, and W5 have partial or dual occupancy, depending on the orientation of the Asp-98 side chain, either toward the substrate-binding pocket (proximal orientation) or into the substrate-entry channel (gatekeeper orientation). In the resting enzyme, the gatekeeper orientation has lower occupancy, as suggested by the weaker electron density for this position of the Asp-98 side chain. The Leu-106 residue also exhibits a dual conformation, corresponding to the different positions of Asp-98, W4 and W5. Water molecule W0 belongs to the proton-transfer water network.
Structure of the Nitrite-Soaked Complex at 1.10-Å Resolution. A number of crystal structures in the resolution range 1.1–1.4 Å were obtained from crystals soaked with nitrite for different durations. These structures revealed that despite increased exposure to substrate, the occupancy of nitrite remained at ≈50%. The following describes the active site refined at 1.1-Å resolution.
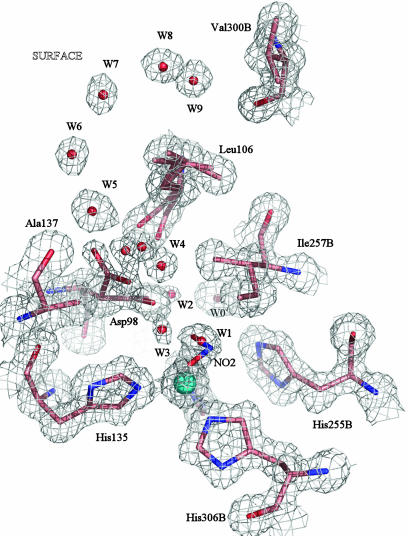
A nitrite molecule is coordinated as the fourth ligand to the T2Cu in 40% of the molecules (Fig. 2). The dual conformation of the Asp-98 ligand is more evident in the electron density of this complex than in the resting state structure shown in Fig. 1. Steric considerations suggest that the fraction of molecules with nitrite bound to the T2Cu correspond to the orientation of the Asp-98 side chain toward the substrate entry pocket, in the gatekeeper position, giving a separation of ≈3.4 Å between the O2(nitrite) and OD2(Asp-98) atoms (Fig. 2). The alternative, proximal conformation, with Asp-98 pointing into the proton channel, would result in a much closer contact between the side-chain oxygen atom and the O2(nitrite), falling in the range 1.9–2.4 Å, depending on the crystal examined. Therefore, this second conformation of Asp-98 most likely corresponds to the fraction of the molecules with water W1 at the T2Cu in place of nitrite with an Asp-98(OD2)–W1 distance of 2.8 Å.
Fig. 2.
The T2Cu site of the nitrite-soaked enzyme at 1.10 Å resolution, shown with 2Fobs – Fcalc electron density contoured at 1.2σ. Either a water or nitrite molecule occupies the substrate-binding site, with the nitrite OD2/OD1 atoms at 1.98/2.19 Å and the N atom at 2.15 Å or a water molecule, W1 at 2.0 Å from T2Cu. The B factors of the Cu atom and nitrite ligand are 10.1 Å2 and 16.0 Å2, respectively, compared with the B factors for ligating N(His), which average 10.0 Å2. The Cu occupancy of the protein was 90%. The nitrite is oriented so that the O1 and O2 atoms form H-bonds with water W2 at 2.4 Å and with water W3, at 2.6 Å. W2 is 2.4 Å from the conserved proton channel water molecule, W0. The substrate-entry water network of the resting-state enzyme is conserved in this structure, while the gatekeeper conformation of the Asp-98 is more pronounced in the electron density map.
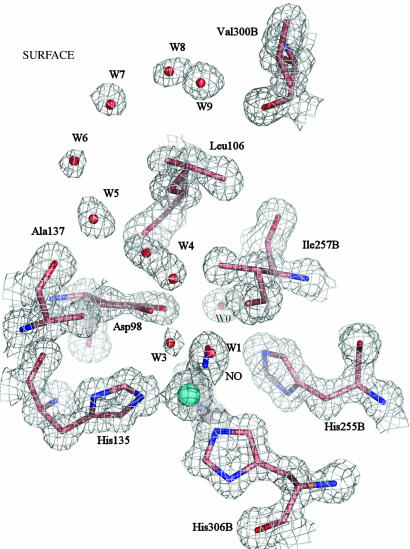
Structure of the Endogenously Bound NO Complex at 1.12-Å Resolution. The T2Cu sites are fully occupied in this structure with 65% of them having NO bound and 35% with a water molecule, W1, as the fourth ligand (Fig. 3). The NO is bound in a side-on manner,§ with the Cu–NNO and Cu–ONO bond lengths of 2.2 Å and an N–O bond length of 1.4 Å. Comparison with the catalytic site structure of the resting-state enzyme (Fig. 1) shows that the Cu atom and its ligands occupy similar positions, implying that the Cu atom is oxidized in the NO complex. A single conformation for the Leu-106 residue is present in this structure, while the electron density (Fig. 3) shows that the Asp-98 side chain only exhibits the proximal position, where it forms an H-bond with the N atom of the NO ligand, at 2.6–2.9 Å. This small spread (0.3 Å) of contact distances arises because the electron density for the Asp-98 side chain is modeled with a “fan-like” spread of positions rotated through 20° into the proton channel.
Fig. 3.
The T2Cu site of the enzyme with endogenously bound NO at 1.12 Å resolution, shown with 2Fobs – Fcalc electron density contoured at 1.2σ. The Cu atom is coordinated by either water or NO. The B factors of the Cu, NNO, and ONO atoms are 10.2, 10.9, and 12.6 Å2, respectively. The close correspondence of these B values indicates the correctness of the atomic assignments of the diatomic molecule. In molecules with water bound to T2Cu in place of NO, W1 is at 2.2 Å from the metal, with a B factor of 10.2 Å2. The proton channel water, W0, is indirectly linked to the NO via H-bonding through Asp-98: the W0—OD1(Asp-98) distance is 3.0–3.17 Å. The direct separation between the NO and W0 is 3.8 Å. The N atom of NO also has an H-bond to W3, at 2.6 Å. The His-135 is coordinated at 2.04 Å with the other histidine ligands at 2.0 Å; the average N(His) B factor is 9.8 Å2. The Asp-98 side chain is oriented toward the proton delivery channel, where it is H-bonded to the N atom of NO. The gatekeeper orientation is absent in this structure. Correspondingly, there is only a single conformation of Leu-106 and a single chain of water molecules in the substrate-entry channel. W4 refers to two distinct water molecules separated by 2.3 Å and not a single water with dual occupancy.
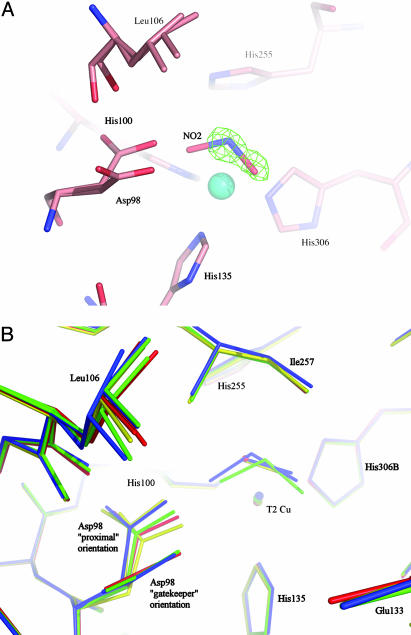
Structure of Endogenously Bound Nitrite and NO in the Same Crystal at 1.15-Å Resolution. In one of the crystals, solved at 1.15-Å resolution, the structure revealed the presence of both nitrite (0.5) and NO (0.3) at the active site with 80% T2Cu occupancy (Fig. 4). This model is supported by the examination of difference density maps and by superposition with the atomic resolution NO-bound and nitrite-bound structures (see Fig. 4A). This is the only structure in which the T2Cu occupancy matches that of the substrate/product species, and we suggest that in this crystal the two bound ligand species are in equilibrium. The proximal and gatekeeper orientations of the Asp-98 are again observed in this structure. The occupancy of the gatekeeper conformation of Asp-98 has been modeled as equivalent to that of bound nitrite. The NO is coordinated to the T2Cu in the “side-on” manner, with bond lengths to NNO and ONO of 1.97 and 2.05 Å, respectively. The T2Cu–N(nitrite) distance is 2.11 Å, with the OD1 and OD2 nitrite atoms coordinated at 2.12 and 2.14 Å, respectively. Comparison with the nitrite-soaked structure (Fig. 4B) shows that the endogenously bound nitrite has a different orientation relative to the T2Cu and the Asp-98. The “endogenous nitrite” ligand is coordinated in an almost “side-on” symmetrical orientation, with its three atoms within 0.03 Å of the T2Cu, while the “soaked-in nitrite” is more asymmetrically bound, by 0.2 Å, to the T2Cu. These differences in conformation could arise from different protonation states of Asp-98 at the pH used for crystallization, due to actual pH varying from crystal to crystal.
Fig. 4.
Comparison of T2Cu site in resting, nitrite-bound, NO bound, or mixed (NO and NO2) forms. (A) The catalytic T2 Cu site of AcNiR with endogenously bound nitrite and NO in the same crystal, with the NO removed. The nitrite ligand has been modeled in the 2Fobs – Fcalc electron density map, while the presence of the NO ligand is evident in the resulting Fobs – Fcalc difference electron density map, contoured at 3.2σ. (B) Comparison of the T2Cu site of the enzyme in the resting state (red), nitrite soaked (green), endogenously bound NO (yellow), and endogenously bound nitrite and NO trapped in the same crystal (blue). A perspective view is shown from the surface of the protein into the substrate-entry channel. For clarity, all water molecules except the bound water in the resting state have been omitted. The flexibility of the Leu-106 residue and the nearly 90° flip of the Asp-98 side chain, between the gatekeeper orientation (pointing out of the page) and the proximal orientation, are clearly illustrated.
In both structures where nitrite is present (Fig. 4), the presence of the substrate results in a lengthening of the coordination distances of the three histidine ligands by an average of 0.05 Å compared with the resting-state enzyme. The Cu atom is also shifted toward the substrate-binding pocket by 0.1 Å relative to the nitrite-soaked structure and by 0.3 Å relative to the resting-state enzyme. These positional changes at the active site suggest that the T2Cu atom is predominantly in the reduced state.¶ Furthermore, in view of the presence of both nitrite (50%) and the NO product (30%) in the same crystal, we are persuaded that these data represent the most probable structure before and during bond breakage.
Substrate Guidance and Product Formation. Our observation of the gatekeeper conformation of Asp-98 is highly significant and is consistent with its role within nitrite reductase for substrate guidance. The occupancy of this conformation and its close correspondence with the occupancy of nitrite at the T2Cu site is strongly suggestive of its role in “hand-shaking” the substrate when it comes down the substrate entry pocket (Leu-106/Ala-137) and guiding it to the T2Cu. Likewise, the observation of primarily the proximal conformation of Asp-98 in correspondence with the occupancy of product (NO) and ligated water argues strongly that this conformation of Asp is appropriately poised for proton abstraction, leading to bond-cleavage and formation of the product, and for product release (see below).
The anisotropic behavior of structural elements around the catalytic Cu site was discussed previously (5) when we had determined the structure of AxNiR in the resting state. The thermal ellipsoids for the three Cu-ligating histidines were found to be spherical, whereas the thermal ellipsoids of His-249 (His-255 in AcNiR) and Asp-92 (Asp-98 in AcNiR) deviate significantly from sphericity. We argued that the anisotropy of Asp-92 would be diminished when nitrite is anchored in position at the T2Cu by the Asp-92 side chain, and this is indeed the case in both the nitrite-bound and NO-bound structures of AcNiR (see Fig. 5). Atomic resolution structure of the resting state of AcNiR and the recently determined atomic resolution structure of the blue AxNiR allows an insight into the overall charge distribution from these highly accurate structures. Despite a major difference in the overall charge distribution, both enzymes reveal that the surface at the site of substrate entry is negatively charged (see Fig. 6, which is published as supporting information on the PNAS web site), suggesting that the substrate enters the pocket as HNO2.
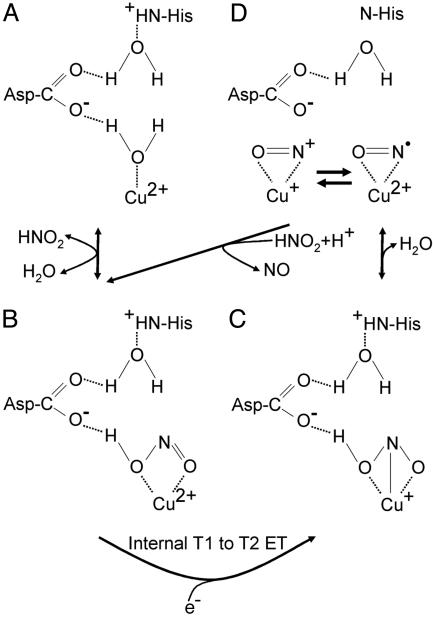
Structure-Based Enzyme Mechanism for Cu-Containing Nitrite Reductases. The steps in the mechanism of nitrite reduction by CuNiRs involves the binding of HNO2 to the oxidized enzyme, reduction and dehydration of bound intermediates, followed by the release of the product NO to reform the oxidized enzyme (30). Even though the origin of the nitrite and NO-bound adducts harvested via high-throughput crystal structure determination is unclear, it is evident that they represent substrate/product adducts of AcNiR that have remained essentially stable (product in Scheme 1D) because of the curtailment of enzyme turnover under conditions used for the isolation of the enzyme. The resting enzyme (Scheme 1 A) has a water molecule bound to the oxidized T2Cu site that is also H-bonded to a negatively charged Asp-98 residue (Fig. 1). The His-255 residue is protonated and H-bonded to Asp-98 via a water molecule. Nitrite-soaked crystals reveal that protonated, uncharged nitrite binds to the T2Cu ion (Fig. 2) via O-coordination displacing the bound water (intermediate in Scheme 1B). A number of structures for these substrate-soaked crystals were determined, mostly to atomic resolution. The binding of nitrite results in an increased midpoint potential of the T2Cu that facilitates the otherwise unfavorable electron transfer (ET) from the type 1 site (31, 32). On reduction of the T2Cu site, the proton is transferred from the Asp to the nitrite to form the intermediate Cu+NOOH (intermediate in scheme 1C). In the widely accepted mechanism, the imidazolium group of His-255 donates the second proton that facilitates cleavage of the N–ON bond to form NO, which dissociates to leave water bound to the oxidized T2Cu ion.
Scheme 1.
Our finding that stable species of AcNiR can be isolated, which have nitrite (Fig. 2), NO (Fig. 3), or both (Fig. 4A) bound to the T2Cu ion, requires refinement of this mechanistic scheme, and we hypothesize that two conditions are required to confer stability. First, nitrite forms a stable complex when the T2Cu ion that it is bound to is reduced. Although nitrite has a very low affinity for reduced NiR, it has a high affinity for NiR-T1CuoxT2Cuox and binding is reversible. In contrast to the isolated enzyme where pulse radiolysis studies have shown the T1Cu and T2Cu sites to be in rapid equilibrium (1), we propose that during turnover the internal ET from the T1Cu center to the T2Cu center is irreversible. This difference arises from the reduction of the nitrite-bound T2Cu ion by internal ET from the T1Cu site resulting in a center with sufficiently high redox potential so as to prevent reverse ET to the T1Cu site. Although it may appear counterintuitive that nitrite would not be directly reduced at such a site, the structures of reduced wild-type Alcaligenes faecalis NiR (13) and those of the reduced Asp-98 to Asn-98 and His-255 to Asn-255 mutants with bound nitrite have been determined (33). In the former, low temperatures were used to trap the state,∥ and in the latter, perturbation of the H-bonding network results in isolating the substrate bound to the reduced form of the enzyme. These findings emphasize the importance of protonation events in controlling both catalysis and internal ET (34). Secondly, we propose that the stability of the AcNiR Cu-nitrosyl could be accounted for if nitrite were required to displace the bound NO from the active site in an ordered mechanism. If the Cu-nitrosyl species and stable nitrite-bound species were in equilibrium, as suggested by the equivalence of the sum of bound nitrite and NO to the occupancy of Cu in the T2Cu site, then the species that we have structurally characterized (Fig. 4A) represent trapped intermediates of the catalytic cycle.
Key structural changes in the positioning of the essential residue Asp-98 are revealed for the first time in these structures. In the nitrite-bound derivative, the Asp-98 side chain assumes a dual conformation, linking the proton access channel to the substrate entry pocket (Fig. 2). This second conformation is absent in the NO-bound structures (Fig. 3). In the atomic resolution resting-state structure, where a water molecule is bound to the T2Cu, the Asp-98 again adopts a dual conformation (Fig. 1). The two positions of the Asp-98 side chain are mirrored by two distinct orientations of the Leu-106 residue, which is one of several hydrophobic residues that form the active-site cleft. A clearly defined and well conserved water network from the Cu atom to the surface is seen in the structures, including several water molecules with multiple conformations, depending on the orientation of the Asp-98 side chain and the presence or absence of substrate. In none of the nitrite-bound structures examined did we observe full occupancy of either substrate or product at the active site. However, the very high resolution of these structures has allowed us to model the two conformations of the Asp-98 and to link them quantitatively with the variable occupancies of Leu-106, the substrate, water molecules, and the T2Cu atom. The 1.15-Å structure (Fig. 4) shows 50% occupancy of nitrite and 30% NO. This nitrite-bound form represents the second intermediate where Cu has become reduced and nitrite does not exchange. This intermediate (Scheme 1C) is committed to the forward reaction because the increase in redox potential of the T2Cu consequent on nitrite binding prevents the rereduction of T1Cu. The NO complex seen in this structure represents the first stage of product formation and is the species in Scheme 1D. The trapping of this intermediate in the crystal strongly suggests that nitrite is required for turnover, generating the species in Scheme 1B, arguing that a displacement mechanism is required to regenerate a T2Cu2+ site. We propose that under conditions of limiting reductant, nitrite dissociates to form free oxidized AcNiR.
Conclusions
Atomic resolution structures of the resting-state, substrate-bound, and product-bound Cu-nitrite reductase have allowed structure-based elucidation of this kinetically intractable enzyme and provide a paradigm shift in our understanding of the catalytic cycle. Our structural data show that the Asp-98 residue has multiple functional roles, namely that of substrate guidance, proton abstraction, and product formation/release. For substrate-guidance, several residues, including Leu-106 and Ala-137, work in concert with Asp-98. Thus, the use of high-throughput structure determination and crystal harvesting, in combination with high-resolution structural information, opens up an avenue for trapping and defining different reaction intermediates of a catalytic cycle.
Supplementary Material
Acknowledgments
We thank Dr. Mark Ellis, Dr. Mike Hough, and Mr. Kostas Paraskevopoulos for their help and interest in this work. This work was supported by Biotechnology and Biological Sciences Research Council Biomolecular Sciences Program Grant 719/B14224 (to S.S.H.). We thank the Council for the Central Laboratory of the Research Councils and the Biotechnology and Biological Sciences Research Council for provision of facilities at Daresbury Laboratory and the John Innes Centre, respectively.
Author contributions: R.R.E. and S.S.H. designed research; S.V.A. performed research; R.R.E. contributed new reagents/analytic tools; S.V.A. and R.W.S. analyzed data; and R.W.S., G.S., R.R.E., and S.S.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NiR, nitrite reductase; AcNiR, Achromobacter cycloclastes NiR; AxNiR, Alcaligenes xylosoxidans NiR; ET, electron transfer; T1Cu, type 1 Cu; T2Cu, type 2 Cu.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2BW4 (resting state), 2BW5 (NO), 2BWD (NO+NO2), and 2BWI (NO2 soaked)].
Footnotes
Even though the side-on bonding of NO is also observed here and the position of individual atoms is reversed, the unrestrained interatomic distance is 1.4 Å instead of 1.67 Å (see supplementary material in ref. 20). In the absence of constraints imposed by partial occupancy of water molecule, this interatomic distance refines at 1.2 Å but with obvious disparity between the B values of the atoms of NO.
In the atomic resolution structure of superoxide dismutase, a 0.8-Å movement of Cu is observed on reduction when Cu changes from penta- to tri-coordination (29). Here, as the ligand is bound and Cu is penta-coordinate, a smaller movement of Cu is expected in going from Cu2+ to Cu1+.
In our case, isolation of the intermediate at room temperature presumably arises from T2Cu being reduced once nitrite is bound while T1Cu remains oxidized, and the lack of exogenous nitrite, as in the experiments of ref. 13, prevents turnover.
References
- 1.Eady, R. R. & Hasnain, S. S. (2003) Compr. Coord. Chem. II 8, 759–786. [Google Scholar]
- 2.Suzuki, S., Kataoka, K. & Yamaguchi, K. (2000) Acc. Chem. Res. 33, 728–735. [DOI] [PubMed] [Google Scholar]
- 3.Zumft, W. G. (1997) Microbiol. Mol. Biol. Rev. 61, 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulanger, M. J., Kukimoto, M., Nishiyama, M., Horinouchi, S. & Murphy, M. E. P. (2000) J. Biol. Chem. 275, 23957–23964. [DOI] [PubMed] [Google Scholar]
- 5.Ellis, M. J., Dodd, F. E., Sawyers, G., Eady, R. R. & Hasnain, S. S. (2003) J. Mol. Biol. 328, 429–438. [DOI] [PubMed] [Google Scholar]
- 6.Ellis, M. J., Prudencio, M., Dodd, F. E., Strange, R. W., Sawyers, G., Eady, R. R. & Hasnain, S. S. (2002) J. Mol. Biol. 316, 51–64. [DOI] [PubMed] [Google Scholar]
- 7.Prudencio, M., Eady, R. R. & Sawyers, G. (2001) Biochem. J. 353, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boulanger, M. J. & Murphy, M. E. P. (2003) Protein Sci. 12, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godden, J. W., Turley, S., Teller, D. C., Adman, E. T., Liu, M. Y., Payne, W. J. & LeGall, J. (1991) Science 253, 438–442. [DOI] [PubMed] [Google Scholar]
- 10.Adman, E. T., Godden, J. E. & Turley, S. (1995) J. Biol. Chem. 270, 27458–27474. [DOI] [PubMed] [Google Scholar]
- 11.Kukimoto, M., Nishiyama, M., Murphy, M. E. P., Turley, S., Adman, E. T., Horinuchi, S. & Beppu, T. (1994) Biochemistry 33, 5246–5252. [DOI] [PubMed] [Google Scholar]
- 12.Murphy, M. E. P., Turley, S., Kukimoto, M., Nishiyama, M., Horinouchi, S., Sasaki, H., Tanoruki, M. & Adman, E. T. (1995) Biochemistry 34, 12107–12117. [DOI] [PubMed] [Google Scholar]
- 13.Murphy, M. E. P., Turley, S. & Adman, E. T. (1997) J. Biol. Chem. 272, 28455–28460. [DOI] [PubMed] [Google Scholar]
- 14.Dodd, F. E., Hasnain, S. S., Abraham, Z. H. L., Eady, R. R. & Smith, B. E. (1997) Acta Crystallogr. D 53, 406–418. [DOI] [PubMed] [Google Scholar]
- 15.Dodd, F. E., Van Beeumen, J., Eady, R. R. & Hasnain, S. S. (1998) J. Mol. Biol. 282, 369–382. [DOI] [PubMed] [Google Scholar]
- 16.Howes, B. D., Abraham, Z. H. L., Lowe, D. J., Brüser, T., Eady, R. R. & Smith, B. E. (1994) Biochemistry 33, 3171–3177. [DOI] [PubMed] [Google Scholar]
- 17.Abraham, Z. H. L., Smith, B. E., Howes, B. D., Lowe, D. & Eady, R. R. (1997) Biochem. J. 324, 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strange, R. W., Dodd, F. E., Abraham, Z. H. L., Grössmann, J. G., Brüser, T., Eady, R. R., Smith, B. E. & Hasnain, S. S. (1995) Nat. Struct. Biol. 2, 287–292. [DOI] [PubMed] [Google Scholar]
- 19.Barrett, M. L., Harris, R. L., Antonyuk, S., Hough, M. A., Ellis, M. J., Sawers, G., Eady, R. R. & Hasnain, S. S. (2004) Biochemistry 43, 16311–16319. [DOI] [PubMed] [Google Scholar]
- 20.Tocheva, E. I., Rosell, F. I., Mauk, A. G. & Murphy, M. E. P. (2004) Science 304, 867–870. [DOI] [PubMed] [Google Scholar]
- 21.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 23.Vagin, A. A. & Teplyakov, A. (1997) J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- 24.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 25.Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. (2003) Acta Crystallogr. D 59, 1131–1137. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt, G. J. & Jones, T. A. (1996) Acta Crystallogr. D 52, 829–832. [DOI] [PubMed] [Google Scholar]
- 27.Emsley, P. & Cowtan, K. (2004) Acta Crystallogr. B 20, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 28.Lamzin, V. S. & Wilson, K. S. (1993) Acta Crystallogr. D 49, 129–147. [DOI] [PubMed] [Google Scholar]
- 29.Hough, M. & Hasnain, S. S. (2003) Structure 11, 937–946. [DOI] [PubMed] [Google Scholar]
- 30.Averill, B. A. (1996) Chem. Rev. 96, 2951–2964. [DOI] [PubMed] [Google Scholar]
- 31.Pinho, D., Besson, S., Brondino, C. D., de Castro, B. & Moura, I. (2004) Eur. J. Biochem. 271, 2361–2369. [DOI] [PubMed] [Google Scholar]
- 32.Hough, M. A., Ellis, M. J., Antonyuk, S., Strange, R. W., Sawyers, G., Eady, R. R. & Hasnain, S. S. (2005) J. Mol. Biol., in press. [DOI] [PubMed]
- 33.Boulanger, M. J. & Murphy, M. E. P. (2001) Biochemistry 40, 9132–9141. [DOI] [PubMed] [Google Scholar]
- 34.Kataoka, K., Furusawa, H., Takagi, K., Yamaguchi, K. & Suzuki, S. (2002) J. Biochem. 127, 345–350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.