Abstract
We recently found that many residues in enzyme active sites can be computationally predicted by the optimization of scoring functions based on substrate binding affinity, subject to constraints on the geometry of catalytic residues and protein stability. Here, we explore the generality of this surprising observation. First, the impact of hydrogen-bonding networks necessary for catalysis on the accuracy of sequence optimization is assessed; incorporation of these networks, where relevant, into the set of catalytic constraints is found to be essential. Next, the impact of multiple substrate selectivity on sequence optimization is probed by carrying out independent calculations for complexes of deoxyribonucleoside kinases with various cognate ligands, revealing how simultaneous selection pressures determined active-site sequences of these enzymes. Including previous calculations on simpler enzymes, computational sequence optimization correctly predicts 76% of all active-site residues tested (86% correct, with 93% similar, for naturally conserved residues). In these studies, the ligand is fixed in its native conformation. To assess the applicability of these methods to de novo active-site design, the effect of small ligand motions around the native pose is also examined. Robustness of sequence accuracy for topologically similar poses is demonstrated for selected kinases, but not for a model peptidase. Based on these observations, we introduce the notion of the designability of an enzyme active site, a metric that may be used to guide the search for protein scaffolds suitable for the introduction of de novo activity for a desired chemical reaction.
Since it was first observed that all known natural proteins adopt one of only ≈1,000 distinct protein folds (1), researchers working in the field of computational protein design have endeavored to understand the basis of this biophysical phenomenon and apply the resultant knowledge to the construction of unnatural proteins. Motivated by the observation that certain folds are associated with more natural sequences than others, the principle of fold designability (the number of unique sequences compatible with the backbone structure of the fold) was introduced (1). To place the problem on a mathematical foundation, an analogy has been drawn between sequence space and phase space in statistical physics, where the backbone structure represents a macroscopic state of the system and distinct sequences are microscopic states whose occupation of the macroscopic state is associated with an energy, the folding free energy of the protein. In this language, a fold's designability is correlated with its sequence entropy (2).
Recently, we explored the extension of the concept of equilibrium in sequence space from protein cores to the functional surface residues of proteins, specifically ligand binding sites and enzyme active sites (3). The findings of this work strongly suggested that the enzyme-substrate binding affinity, subject to constraints on the total protein energy and conformations of catalytic residues, should take the place of the folding free energy as the scoring function in active-site sequence space. Here, we further explore this concept by studying active sites that evolved under more complex selection pressures. In particular, we focus on enzymes whose active-site residues are involved in long-range hydrogen-bonding networks essential for catalysis and those capable of accommodating multiple substrates of qualitatively different structures. The impact of multiple substrate selectivity on sequence optimization is probed by carrying out independent calculations for complexes of selected enzymes with various cognate ligands.
In addition, we endeavor to extend the conceptual foundation for theoretical active-site design by asking: what is the macroscopic state associated with the active site that corresponds to the structure of the protein fold in core designability? In the most natural formulation, the macroscopic state is specified jointly by the active-site backbone trace and ligand conformation. By analogy to the manner in which core sequences are optimized for ensembles of related backbone traces (2), we explore the effects of moving the ligand from its native conformation on predicted sequence distributions. Implications of these results for de novo active-site design and the notion of active-site designability are analyzed.
Materials and Methods
Active-site sequence optimization was carried out by using an iterative three-step algorithm as described (3), consisting of side-chain conformational optimization, calculation of substrate binding affinity, and selection of the residue type/conformation with the highest binding affinity that satisfied auxiliary constraints. The starting point for these calculations was an active site where all residues involved in critical contacts to the substrate were mutated to alanine or random identities. These steps were iterated self-consistently until convergence in the ligand binding affinity was achieved. Unlike the optimization algorithms (e.g., Monte Carlo and dead-end elimination) commonly used in protein design (4), our approach has been developed for the constrained optimization problem of maximizing binding affinity under restrictions on the folding free energy and catalytic residues. The features of this algorithm essential for its accuracy in predicting enzyme active-site sequences are attention to the physicochemical details of side-chain conformations and enzyme-substrate binding.
Conformations are optimized for a fixed amino acid identity by using the all-atom OPLS force field and an implicit solvent model consisting of the surface-generalized Born model of polar solvation and a nonpolar free energy estimator (5, 6). The sampling of single side-chain conformations was accomplished primarily by using a highly detailed (10-deg resolution) rotamer library constructed from a database of 297 proteins. Computational expense was mitigated by prescreening rotamers by using only hard sphere overlap before energy evaluations (7, 8).
Enzyme-substrate binding affinities are computed by using an extensively developed semiempirical potential function, glidescore, that is sensitive to the delicate balance of interactions in enzyme active sites. glidescore consists of a lipophilic-lipophilic contact term and a hydrogen-bonding term separated into weighted components based on donor and acceptor charge, a Coulomb and van der Waals interaction term that reduces charges and van der Waals interaction energies from their gas-phase values, and a solvation model based on the computational introduction of explicit waters and empirical scoring terms that measure the exposure of various groups to the explicit waters (9). Nonnative ligand poses used to assess active-site designability were generated by redocking the ligand with the glide docking algorithm (8) and retaining low-energy excitations of the native pose. Unlike other methods for docking ligands to the 3D structure of a protein, glide docking approximates a complete search of the conformational, orientational, and positional space of the docked ligand for qualitatively superior binding mode predictions (10).
Geometric constraints on catalytic residues were generally applied as filters on the rank-ordered sequence lists produced by self-consistent optimization (3). In the case of β-lactamase, however, they were applied at each step of optimization as was the constraint on the total protein energy.
Results
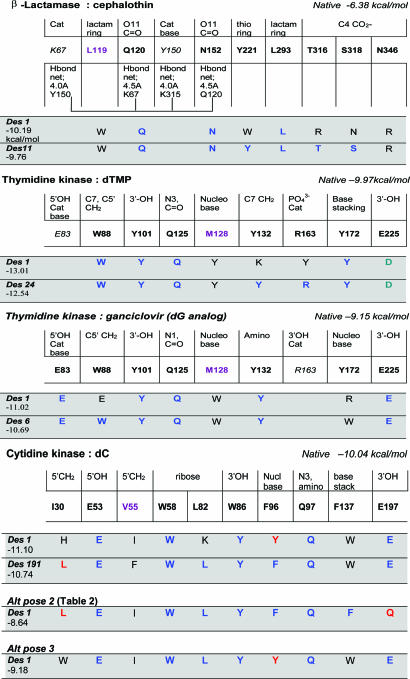
The active sites of the enzymes studied (Fig. 1; see also Supporting Text and Figs. 5–13, which are published as supporting information on the PNAS web site) were undoubtedly subject to multiple complex selective pressures during the course of natural evolution and therefore represent stringent tests of the generality of sequence optimization algorithms. β-Lactamases, serine hydrolases that bind and cleave penicillin-type antibiotics, are believed to have evolved rapidly from the so-called penicillin-binding proteins, carboxypeptidases, which, being capable of binding but not hydrolyzing antibiotics, are the primary targets of β-lactams (11). Mutagenesis data have suggested that a hydrogen-bond network connecting residues 67, 120, 150, 152, and 315 in the Enterobacter cloacae P99 β-lactamase may be important for hydrolysis of its cognate antibiotic cephalothin (12). Such a network has not been identified in the ancestral carboxypeptidases, for which our sequence optimization algorithm predicted many active-site residues correctly without hydrogen-bonding constraints (3). As a case study of the impact of catalytic hydrogen-bonding networks on sequence prediction accuracy, side chains of eight residues involved in contacts with cephalothin were subjected to sequence optimization, under the constraint that the hydrogen-bonding network is not disrupted (Fig. 1). Tighter constraints were applied to those side chains close to the general base Tyr-150.
Fig. 1.
Comparison of native and computationally optimized active-site sequences. For each enzyme–substrate complex, residues forming essential contacts with the substrate or in the catalytic (Cat) mechanism are listed (bold type indicates computationally repredicted; italics indicates catalytic conformationally optimized under constraints with fixed identity; purple indicates functionally promiscuous or displaying high variability in MSAs). Complementary moieties on the substrate are listed above the native residues. Computationally predicted active-site sequences are listed in the gray bars. The first sequence is that displaying the highest binding affinity while satisfying all geometric constraints. The second sequence is that displaying the highest sequence identity to the native active site within the top 0.5 kcal/mol of ranked sequences (designed number corresponds to rank in calculated sequence list). Blue amino acids are identical to native; red amino acids are isosteric to the native and engage in the same mode of interaction with the substrate (e.g., Tyr vs. Phe, Gln vs. Glu); green amino acids are the same type as native and engage in the same mode of interaction (e.g., Asp vs. Glu, Lys vs. Arg); black amino acids are none of the above. Native energy corresponds to binding affinity of native sequence/structure after side-chain conformational optimization. Constraints are: β-lactamase, H-bond network; TK:dTMP, Arg-163 within 3.6 Å of PO42–; TK:ganciclovir, Glu-83 within 3.5 Å of 5′OH, Arg-163 within 4.5 Å of PO42– (TK:dT is the same except residue 163's identity is unrestricted, see Fig. 10); CK, residue 53 (acid/base) within 3.5 Å of 5′OH.
The family of deoxyribonucleoside kinase enzymes represents a model case study of multiple substrate selectivity. Thymidine kinase (TK) catalyzes the transfer of the γ-phosphate from ATP to the 5′ hydroxyl of deoxythymidine (dT) to form dT-5′-monophosphate (dTMP) (13). We used herpes simplex virus TK, a multisubstrate enzyme that phosphorylates guanosine, cytosine, and dTMP as well as thymidine, to probe the effects of substrate selectivity on sequence optimization. Optimizations were carried out on the complexes of TK with thymidine, dTMP, and ganciclovir (guanosine analog, used in the absence of a crystal structure of the dG complex). Glu-83 serves as the general base in accepting the 5′ OH proton in phosphate ester formation. Arg-163 stabilizes the phosphate and facilitates the second esterification reaction. Constraints were imposed on the distances of these residues to their interaction partners on the ligand; more relaxed constraints were applied to Arg-163 for the thymidine and ganciclovir substrates because they lack a phosphate group. The Glu-83 constraint was dropped for dTMP because the 5′OH of this substrate is phosphorylated. The human cytidine kinase (CK) enzyme, which primarily phosphorylates deoxycytidine but also deoxyguanosine and deoxyadenosine (14), was also subjected to sequence optimization (with the cytidine ligand), with the general base Glu-53 constrained.
Accuracy of Active-Site Sequence and Structure Optimization. Data pertaining to optimized active-site sequences, those within the top 0.5 kcal/mol of the rank-ordered sequence lists (filtered for geometric constraints on catalysis) that are most similar to the native sequence, are presented in Table 1. Fig. 1 displays both the top-ranked sequences and those most similar to the native, along with the geometric constraints imposed in each case. For each complex, a sequence with at least 50% sequence identity to the native active site is found within the top 0.5 kcal/mol (and usually within the top 10–15 sequences), extending the generality of our previous study carried out with a more limited test set of receptor proteins and enzymes subject to simpler selection pressures. Moreover, at almost every residue position, the native amino acid is one of the three most frequently found in the optimized sequence lists (Fig. 6). Structural accuracy is somewhat lower for polar and charged side chains compared with nonpolar side chains, but most errors are close to the limits of crystallographic accuracy (Fig. 7). Conversely, the sequence prediction accuracy for nonpolar side chains (67% for all complexes studied) is lower than that for polar (83%) and charged (75%) side chains (Fig. 8), suggesting that nonpolar contacts are more promiscuous, and that some of the nonpolar side chains included in sequence prediction lists may not be naturally optimized, a conclusion supported by the higher amino acid variability at these positions observed in multiple sequence alignments (MSAs) (3). It is interesting to note that Trp is selected at six of the nine positions for which predictions are neither correct nor similar (including β-gal from ref. 3); this bulky and inflexible residue may interfere with multisubstrate selectivity or substrate motions that occur during catalysis (see below). Excluding residues that display high variability in MSAs (e.g., Met-128 in TK, Val-55 in CK, and Leu-119 in P99 β-lactamase; Fig. 9) or have additional functional roles (3), 86% of active-site residues in enzymes tested to date are predicted correctly (93% similar) in a sequence within the top 0.5 kcal/mol.
Table 1. Active site sequence design results for various enzyme families.
| No. of residues
|
|||||
|---|---|---|---|---|---|
| Enzyme | Substrate | Predicted† | Correct/similar | Mean correct | rmsds of correct, Å |
| Streptomyces R61 DD-peptidase* | Glycyl-l-α-amino-ε-pimelyl-d-Ala-d-Ala | 7 (6) | 6/6 | 3.96 | 0.55 |
| E. cloacae P99 β-lactamase | Cephalothin | 8 (7) | 6/6 | 2.77 | 0.77 |
| HSV-1 TK | dTMP | 8 (7) | 6/7 | 3.95 | 0.62 |
| Deoxythymidine | 8 (7) | 4/6 | 2.71 | 0.6 | |
| Ganciclovir (deoxyguanosine analog) | 8 (7) | 6/6 | 3.55 | 0.70 | |
| Human CK | Deoxycytidine | 10 (9) | 7/8 | 5.50 | 1.05 |
| E. coli thymidylate synthase* | dUMP | 6 | 6/6 | 3.81 | 1.02‡ |
| Penicillium β-galactosidase* | α-Galactose | 10 (8) | 6/7 | 5.73 | 0.62 |
| Total | 49 | 37 (76%)/40 (82%) | |||
Correct is the number of residues predicted correctly in the sequence from the top 0.5 kcal/mol of ranked sequences (binding affinity + constraints) bearing the highest sequence identity to native. Similar includes residues isosteric or functionally identical to native amino acid. Mean correct is the average number of residues matching the native sequence within the top 1 kcal/mol of ranked sequences. HSV, herpes simplex virus. Italics designate alternate substrates for TK.
Previously reported (3).
Excluding residues with auxiliary functions or high variability in MSAs, 86% (93%) of residue predictions are correct (similar).
Excluding Arg-21, which was predicted correctly despite omission of nearby crystallographic water.
We attribute the success of these results to the use of energy functions and solvation models capable of accurately modeling active sites and search algorithms capable of sampling them. Instead of the popular dead-end elimination algorithms, which can sample only pairwise-decomposable potentials that necessarily compromise the treatment of solvation (15), we use a side-chain conformation prediction algorithm (3) that can sample a potential incorporating a realistic treatment of solvation effects (the surface-generalized Born continuum model). Our findings reinforce the importance of deploying continuum solvation models in the design of protein surfaces (16). The treatments of electrostatics and solvation in our algorithm appear to be sufficiently accurate for the purposes of high-resolution enzyme active-site design. In such applications, the introduction of a Trp penalty [analogous to the Met penalty used in protein core design (17)] might effectively diminish bias toward this residue, especially given the small energy difference often observed between Trp and the native residues (e.g., 0.27 kcal/mol for Leu/Trp-119 in β-lactamase).
Incorporating Nonlocal Catalytic Selection Pressures into Active-Site Sequence Optimization. For the nucleoside kinase family and the enzymes examined in our previous study (3), the highest-affinity sequence was retained at each step of the self-consistent algorithm, irrespective of whether it satisfied the geometric constraints; the final rank-ordered sequence list was later filtered for those sequences that satisfied the constraints. For β-lactamase, it was found that the interconnected geometry of the hydrogen-bonding network rendered such an approach inadequate for producing native-like sequences; rather it was necessary to accept the highest-affinity sequence that satisfied all constraints on the network distances at each self-consistent step. Six of eight residues in β-lactamase optimized by using this modified approach matched the native sequence, with discrepancies at positions 119 and 346. In MSA profiles, however, position 119 displays particularly high variability (Fig. 9); omitting this position, accuracy is comparable to that achieved for the evolutionarily related DD-peptidase, where all of the residue contacts to the peptide substrate were predicted correctly (3). It is noteworthy that the imposition of fixed (not annealed) constraints on a binding affinity-based scoring function is capable of reproducing most of the essential active-site residues that have undergone nonlocal selection for catalytic efficiency.
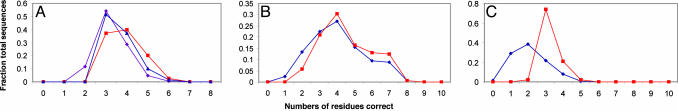
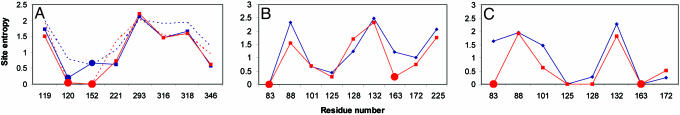
Fig. 2 displays the similarity of predicted sequence distributions to the native sequence, and Fig. 3 displays site (sequence) entropies of active-site residues for several of the complexes studied. Site entropies, defined as Si =–∑(a = 1... 20) [f(ia) ln f(ia)], where the sum is over all amino acid types and f(ia) is the frequency of amino acid a at position i, represent a quantitative measure of the variability of the amino acid identity at that site (18). For β-lactamase, the effects on site entropies of narrowing the binding energy window (dotted traces in Fig. 2 represent 2 kcal/mol) and imposing geometric constraints during optimization are similar for some residues (residue 152) but not others (residue 120). Applying constraints only as a filter (Fig. 2, blue dots) is clearly less effective for this enzyme. In effect, constraints decrease the size of the sampling space and therefore may render the search for optimal sequences simpler for a given number of amino acids. Zou and Saven (19) noted the importance of constrained sequence optimization in protein design. In the language of statistical mechanics, one may define the evolutionary temperature of a protein as the negative derivative of the energy with respect to the sequence entropy (T = –dE/dS). Shakhnovich et al. (20) have noted that the evolutionary temperature can be interpreted as a measure of the stringency of the selection pressure. In these terms, the constraints imposed on sequence optimization in enzyme active sites correspond to an increase in evolutionary temperature compared with noncatalytic binding sites.
Fig. 2.
Similarity of predicted sequence distributions to the native sequence and the effect of catalytic constraints. Purple traces (♦) correspond to sequence distributions drawn from binding affinity windows of +2 kcal/mol (relative to the highest affinity sequence), blue traces (▴) correspond to sequence distributions drawn from binding affinity windows of 1 kcal/mol, and red traces (▪) correspond to sequence distributions drawn from binding affinity windows of +1 kcal/mol with catalytic constraints. (A) TK-ganciclovir. (B) CK-cytidine native pose. (C) CK–cytidine nonnative pose 1 (see Table 2).
Fig. 3.
Sequence (site) entropies {Si = – ∑(a = 1... 20) [f(ia) ln f(ia)], sum over all amino acids a at site I} for residues in active sites of four representative enzyme:substrate complexes and the effect of catalytic constraints. Constrained residues are depicted as heavy dots (red trace). (A) P99 β-lactamase. Constraints are: hydrogen bond network, hydrogen-bonding atoms of residues 120 and 152 within 4.5 Å, residue 152 and Lys-65 within 4.0 Å, and Lys-65 and Lys-315 within 4.0 Å of Tyr 150. Dotted traces show residue entropies using a +2 kcal/mol rather than +1 kcal/mol window in binding affinities. Medium blue dots indicate that constraints were imposed during sequence optimization rather than as a filter after optimization; the red trace is constrained during optimization and also subsequently filtered. (B) TK complexed to dTMP. Residue 83 was assigned as the catalytic Glu, but residue 163 was selected freely. Constraints are: Glu-83 O-γ within 4.5 Å of 5′OH and residue 163 hydrogen-bonding atom within 3.6 Å of phosphate O–.(C) TK complexed to ganciclovir (deoxyguanosine analog). Residue 83 was selected freely but residue 163 was fixed to Arg because of the lack of a phosphate moiety in the ligand. Constraints are: Glu-83 O-γ within 3.5 Å of 5′OH and residue 163 hydrogen-bonding atom within 4.5 Å of phosphate O–.
Research in computational enzyme core redesign has revealed that core residues with high site entropies may constitute ideal “hot spots” for experimental sampling in combinatorial mutagenesis experiments (18). In contrast to core mutagenesis, the effects of which on active-site structure are difficult to predict, the computational active-site mutagenesis carried out here is of high structural resolution; therefore, a natural application of these site entropies would be in a combined computational side-chain selection/ligand docking algorithm where entropies are proportional to the relative amount of sampling that should be devoted to each residue.
Sequence Optimization Vis-à-Vis Multiple Substrate Selectivity. The TK enzyme was used as a model for the effects of multiple substrate selectivity on sequence optimization, in part because its different substrates display noticeably different pose geometries. In particular, the additional phosphate in dTMP causes its 3′OH to move closer to residues 101 and 225, optimizing hydrogen-bond distances to these residues compared with the thymidine complex. To explore the effects of the phosphate group in the nucleoside substrates of TK on sequence optimization, residue 163 was included in the design list of the thymidine complex but not of the ganciclovir complex. Conversely, the catalytic residue 83 was freely selected in the ganciclovir complex, but constrained to Glu in the other two structures. Interestingly, although the active site appears to be most thoroughly optimized for the dTMP substrate, the combined sequence profiles for dTMP and ganciclovir (Fig. 1), but neither profile in isolation is capable of reproducing the native sequence at all essential active-site residue positions, excluding Met-128, which is poorly conserved in MSAs and often replaced by Phe, an isostere of the frequently predicted Tyr (Fig. 10). In particular, the erroneous prediction of Trp-172 in the ganciclovir complex is corrected with Tyr in the dTMP complex, supporting the hypothesis that omission of multisubstrate selectivity may be responsible for the observed computational bias toward Trp prediction (omission of substrate motions, which are examined below, may also play a role). In the case of CK, where all active-site residues were optimized with the primary substrate cytidine, discrepancies occur only at residues 30, 55, and 137, all hydrophobic contacts, which as discussed above are likely more indiscriminate.
For the CK:cytidine complex, narrowing the window of binding affinities for acceptable sequences (e.g., from 2 to 1 kcal/mol) had a greater effect on the accuracy of predicted sequence distributions than catalytic constraints (Fig. 2). In this case, the catalytic residues in the most tightly binding sequences generally adopted suitable geometries spontaneously, without the need for additional filters, indicating that the active-site backbone is preengineered to produce a correlation between binding affinity and catalytic geometry. For TK, the imposition of catalytic constraints had a varying effect depending on the substrate. As an example, we compared the effect of imposing the requirement in TK that Arg-163 remain within 3.5 A of the 5′OH, to stabilize the product of phosphorylation, in the complexes of the enzyme with both thymidine and the guanosine analog ganciclovir. Although both of these ligands are devoid of a phosphate group, imposing this constraint on the ganciclovir complex has a noticeable effect on sequence similarity to native, whereas there is no effect for thymidine (Fig. 11).
The interplay between multiple substrate selectivity and site entropy is considered in Fig. 3 for TK:dTMP and TK:ganciclovir. Comparing the site entropy profiles of these two complexes reveals features not apparent from the sequence similarity distributions alone. The introduction of catalytic constraints roughly equalizes the site entropies at residue 88 (which forms an essential hydrophobic contact to the nucleoside base in the native enzyme) and residue 101, but has opposite effects at residue 128. The nonlocal effects of catalytic constraints on sequence optimization are thus seen to vary with substrate structure in a manner that is difficult to predict by simple inspection alone.
Designability of Enzyme Active Sites. Having established the ability of high-resolution sequence optimization to repredict active-site residues given the native ligand pose, we applied these methods to a preliminary investigation of the issue of active-site designability. A protein structure's designability refers to the number of sequences with which it is energetically compatible (1). Recently, it has been shown that the designability of a protein fold is correlated with its topological properties (21), and moreover that assessment of designabililty requires the consideration of not just a single structure, but families of topologically related structures, those whose geometries may differ slightly, but whose connectivity of backbone contacts is the same (22). In an enzyme active site, the relevant topology is the connectivity of contacts between the backbone and the ligand. Numerous reports have demonstrated that the volume of sequence space compatible with families of designable protein core structures is highly restricted to a region around the native sequence (23). An analogous feature is expected to be particularly important for enzyme active sites, where sequence dissimilarity is likely to render the assumption of a fixed ligand pose invalid, decreasing the number of sequences compatible with the pose. For the notion of designability to be applicable to active sites, it is therefore essential that families of geometrically related substrate poses are related in sequence space, i.e., that sequence prediction accuracy is robust to small substrate motions.
Perturbations of the ligand conformation around the native pose were thus examined for CK and the R61 DD-peptidase. Because the amino acid residues subjected to sequence selection were fixed to include only those forming contacts to the native ligand pose, care was taken to restrict alternate poses so that their “contact shells” were composed of the same residues as the native pose. In the case of the DD-peptidase, it was additionally necessary to ensure that four hydrogen bonds between the peptide substrate and backbone atoms were maintained. Nonnative poses were generated by redocking the ligand into the native active-site sequence by using the glide docking algorithm, which successfully repredicted native ligand poses within 0.75-Å rms deviation (rmsd) for the majority of enzyme:substrate complexes studied. Poses for sequence optimization were chosen from those that displayed binding affinities to the native active-site sequence (“starting affinity”) between 2 and 3 kcal/mol lower than that of the native pose, such that the native sequence did not constitute a tight-binding sequence for the pose (to avoid biasing the results toward the native.) Because we were interested in examining enzyme designability, acceptable poses were further restricted to those that satisfied distance constraints placed on catalytic residues. For CK, this amounted to enforcing the constraint that the 3′O of the deoxyribose was within 3.5 Å of a Glu-53 ε-O. Three cytidine poses were chosen to explore the effects of differences in both the topology of contacts between the ligand and active site (pose 1) and small perturbations in the geometry of distinct portions of the ligand (poses 2 and 3). CK pose 1 displayed a high total rmsd to the native pose but differed primarily in a 180° rotation of the nucleobase roughly superimposing C2 ( O) with C6 in the native pose. Pose 2 was close to the native pose over the nucleobase atoms and differed primarily in the sugar, while pose 3 displayed substantial deviations over both base and sugar.
O) with C6 in the native pose. Pose 2 was close to the native pose over the nucleobase atoms and differed primarily in the sugar, while pose 3 displayed substantial deviations over both base and sugar.
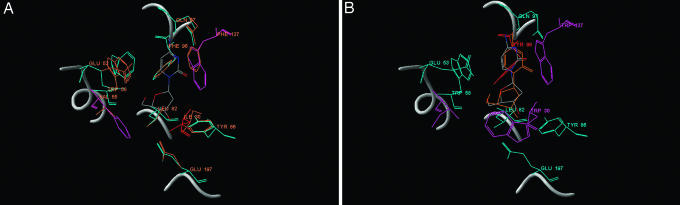
The results in Table 2 show that the most highly ranked sequences for the nonnative CK poses 2 and 3 bear a high level of similarity to the native active-site sequence, comparable with that of the native pose. However, the binding affinities for these sequence-pose combinations are not as favorable as those corresponding to the native pose. This feature of sequence similarity associated with modest energetic dissimilarity was previously noted in the context of sequence optimization for ensembles of near-native core backbone conformations (23). It strongly suggests that designability is a meaningful concept in active-site sequence space. Structures of the optimized active sites corresponding to the native pose and pose 3 are compared in Fig. 4 (see Fig. 12 for other poses).
Table 2. Designability of enzyme active sites: Sequence optimization for nonnative ligand poses.
| Total rmsd, Å
|
rmsd by region, Å
|
Starting/best affinity, kcal/mol
|
No. correct/predicted
|
|||
|---|---|---|---|---|---|---|
| Ligand pose | Base | Sugar | Constraints, <3.0 Å | |||
| CK native | -10.04/-10.74 | 7/10 | ||||
| CK 1 | 1.66 | 2.00 | 0.47 | E53-3′ OH | -7.85/-9.72 | 4/10 |
| CK 2 | 0.85 | 0.35 | 1.15 | E53-3′ OH | -7.83/-8.64 | 7/10 |
| CK 3 | 1.16 | 1.35 | 0.92 | E53-3′ OH | -7.26/-9.18 | 6/10 |
| C term | N term | |||||
| Peptidase native | N327, T301, S62 backbone H bonds | -10.02/-12.12 | 6/7 | |||
| Peptidase 1 | 0.98 | 0.95 | 1.00 | -8.22/-10.85 | 3/7 | |
C term, atoms C-terminal of the first pimely CH2; N term, all remaining atoms. Best affinity is that produced by sequence optimization.
Fig. 4.
Comparison of predicted active-site sequences/geometries for human deoxycytidine kinase bound to deoxycytidine in native (A) and nonnative (B) poses. Crystallographic conformations of side chains involved in binding the ligand or in catalysis (Glu-53) are shown in orange; predicted side-chain conformations at these positions from the most similar high-affinity optimized sequences (CK-designed sequences in Fig. 1) are shown in blue where residue identities match the native sequence, are shown in red where the predicted residue is isosteric to the native residue, and are shown in purple where the predicted residue is not isosteric. (A) The predicted sequence differs from the native sequence at positions 30 (Leu in place of Ile), 55 (Phe/Val), and 137 (Trp/Phe). All predicted amino acids interact with the ligand in the same mode as the native residues. (B) The nonnative substrate pose corresponds to pose 3 in Table 2 (crystallographic pose shown in orange). Native side chains and all hydrogen atoms are omitted for clarity.
Interestingly, the best binding affinity of the topologically distinct pose/sequence combination for CK was more favorable than that corresponding to the pose with the lowest rmsd. However, examination of the sequence distributions generated for these poses reveals substantial differences. Indeed, the mean number of residues predicted correctly for pose 1 is considerably lower than that for pose 3, which has a roughly comparable rmsd. The effect of catalytic constraints on sequence distributions sheds further light on the designabilities of these poses. We compared the effects of the requirement that residue 53 be capable of acid-base catalysis and remain in proximity of the sugar 5′OH in the sequence-optimized structures for the native pose, a topologically distinct pose, and a topologically similar but geometrically perturbed pose (Fig. 2). The catalytic constraint has a significantly greater effect on the topologically distinct pose than on the similar poses; binding affinity does not correlate well with optimal catalytic geometry for the former. A similar distinction is observed in the respective sequence entropy profiles (Fig. 13). These observations indicate that a complete definition of active-site designability should take into account the comparative effects of catalytic constraints on the sequences compatible with distinct poses.
In contrast to the CK:cytidine complex, a small (<1-Å rmsd) displacement of the DD-peptidase substrate from the native pose, which was roughly homogeneous throughout the peptide, resulted in qualitatively different optimized sequences. Moreover, as with topologically distinct poses of CK, the imposition of catalytic constraints (in this case on the hydrolytic triad) had a substantially greater effect in shifting the sequence distribution toward more residues correct for the nonnative pose than for the native pose (Fig. 10). Because the DD-peptidase is capable of accommodating various peptide substrates, this result suggests that active-site designability is not necessarily correlated with multisubstrate selectivity and constitutes a novel metric that cannot be assessed on the basis of native structure alone.
Discussion
Given that efficient enzymes must be capable of substrate binding, transition-state stabilization, and product release, it may appear surprising that sequence optimization for a substrate binding affinity-based scoring function is capable of accurately reproducing many native active-site residues. The scoring function used here incorporates the former two effects in terms of ligand affinity and catalytic constraints, but does not directly address product release. Indeed, native sequences seldom score highest and are often 1–2 kcal/mol behind the best-predicted sequences, a feature that may be important for reversible binding. Moreover, for the majority of enzymes studied, structural differences between reactant and transition state are localized near catalytic residues, whose identities were often fixed. Ultimately, it is likely that attention to properties of electronic structure, for example, via mixed quantum/molecular mechanics methods (24), will be required for full generality without heuristic assumptions regarding catalytic residues.
Nonetheless, the success of the optimization algorithm we have used herein indicates that this simple approach captures essential features of natural enzyme evolution. In this regard, it is important to note the distinction between catalytic perfection and active-site sequence optimization, even for an ideal scoring function. In contrast to catalytic perfection, which describes the absolute extent to which the enzyme is capable of accelerating a reaction, the extent of sequence equilibrium for a given active-site shape (3) is a relative quantity that presupposes a fixed backbone structure. Because much of the backbone trace of an active site evolves before the introduction of catalytic function (25), the maximum catalytic activity of an enzyme may be fundamentally limited even if active-site sequence equilibrium is achieved.
Designability refers to the sequence degeneracy associated with a particular substrate pose/chemical reaction, which is again a function of the backbone trace of the active site. Indications that certain native active-site ligand-backbone topologies are designable bode well for the de novo design of enzyme active sites, just as the designability of native protein core topologies underscored the feasibility of designing novel protein folds (23, 26). The designability of an active site may be particularly important for the design of catalytically robust enzymes, especially given the manner in which the density of sequence states compatible with a reaction can be considerably diminished by chemical constraints. Recent attempts at computational de novo enzyme design showed that enzymatic activity could be imparted to arbitrarily chosen catalytically inert scaffolds by side-chain optimization, but the activities were orders of magnitude below those of natural enzymes catalyzing the same reactions (27, 28). In choosing scaffolds for de novo enzyme design efforts, an effective strategy might be to find a scaffold that displays a maximum sequence designability given the geometric constraints necessary for catalysis. In addition, designability may be a useful tool for screening ligand poses with different topologies when attempting to determine the optimal sequence for an active site given only the ligand structure and the backbone geometry. Relaxation of auxiliary constraints on ligand conformation and catalytic residues should extend the domain of applicability of the present algorithm from sequence optimization and designability to the fully de novo design of functional enzymes.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health Postdoctoral Fellowship (to R.C.) and National Institutes of Health Grant GM52018 (to R.A.F.).
Author contributions: R.C. and R.A.F. designed research; R.C. performed research; R.C., A.M.K., and R.A.F. analyzed data; and R.C. wrote the paper.
Abbreviations: TK, thymidine kinase; dT, deoxythymidine; dTMP, dT-5′-monophosphate; CK, cytidine kinase; MSA, multiple sequence alignment; rmsd, rms deviation.
References
- 1.Li, H., Helling, R., Tang, C. & Wingreen, N. (1996) Science 273, 666–669. [DOI] [PubMed] [Google Scholar]
- 2.Taverna, D. M. & Goldstein, R. A. (2000) Biopolymers 53, 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Chakrabarti, R., Klibanov, A. M. & Friesner, R. A. (2005) Proc. Natl. Acad. Sci. USA 102, 10153–10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voigt, C. A., Gordon, D. B. & Mayo, S. L. (2000) J. Mol. Biol. 299, 789–803. [DOI] [PubMed] [Google Scholar]
- 5.Jorgensen, W. L. & Tirado-Rives, J. T. (1988) J. Am. Chem. Soc. 110, 1657–1666. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, A., Rapp, C. S. & Friesner, R. A. (1998) J. Phys. Chem. B 102, 10983–10990. [Google Scholar]
- 7.Jacobson, M. P., Kaminski, G. A., Rapp, C. S. & Friesner, R. A. (2002) J. Phys. Chem. B 106, 11673–11680. [Google Scholar]
- 8.Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J., Honig, B., Shaw, D. E. & Friesner, R. A. (2004) Proteins 55, 351–367. [DOI] [PubMed] [Google Scholar]
- 9.Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., et al. (2004) J. Med. Chem. 47, 1739–1749. [DOI] [PubMed] [Google Scholar]
- 10.Perola, E., Walters, W. P. & Charifson, P. S. (2004) Proteins 56, 235–249. [DOI] [PubMed] [Google Scholar]
- 11.Matagne, A., Dubus, A., Galleni, M. & Frere, J.-M. (1999) Nat. Prod. Rep. 16, 1–19. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg, S. D., Iannuccilli, W., Nguyen, T., Ju, J. & Cornish, V. W. (2003) Protein Sci. 12, 1633–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wild, K., Bohner, T., Folkers, G. & Schulz, G. E. (1997) Protein Sci. 6, 2097–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabini, E., Ort, S., Monnerjahn, C., Konrad, M. & Lavie, A. (2003) Nat. Struct. Biol. 10, 513–519. [DOI] [PubMed] [Google Scholar]
- 15.Desjarlais, J. R. & Clarke, N. D. (1998) Curr. Opin. Struct. Biol. 8, 471–476. [DOI] [PubMed] [Google Scholar]
- 16.Pokala, N. & Handel, T. M. (2005) J. Mol. Biol. 347, 203–227. [DOI] [PubMed] [Google Scholar]
- 17.Mooers, B. H. M., Datta, D., Baase, W. A., Zollars, E. S., Mayo, S. L. & Matthews, B. W. (2003) J. Mol. Biol. 332, 741–756. [DOI] [PubMed] [Google Scholar]
- 18.Voigt, C. A., Mayo, S. L., Arnold, F. H. & Wang, Z.-G. (2001) Proc. Natl. Acad. Sci. USA 98, 3778–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou, J. & Saven, J. G. (2000) J. Mol. Biol. 296, 281–294. [DOI] [PubMed] [Google Scholar]
- 20.Tiana, G., Shakhnovich, B. E., Dokholyan, N. V. & Shakhnovich, E. I. (2004) Proc. Natl. Acad. Sci. USA 101, 2846–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koehl, P. & Levitt, M. (2002) Proc. Natl. Acad. Sci. USA 99, 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson, S. M., England, J. L., Desjarlais, J. R. & Pande, V. S. (2002) Protein Sci. 11, 2804–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhlman, B. & Baker, D. (2000) Proc. Natl. Acad. Sci. USA 98, 10383–10388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gherman, B. F., Goldberg, S. D., Cornish, V. W. & Friesner, R. A. (2004) J. Am. Chem. Soc. 126, 7652–7664. [DOI] [PubMed] [Google Scholar]
- 25.Todd, A. E., Orengo, C. A. & Thornton, J. M. (1999) Curr. Opin. Chem. Biol. 3, 548–556. [DOI] [PubMed] [Google Scholar]
- 26.Kuhlman, B., Dantas, G., Ireton, G. C., Varani, G., Stoddard, B. L. & Baker, D. (2003) Science 302, 1364–1368. [DOI] [PubMed] [Google Scholar]
- 27.Bolon, D. N. & Mayo, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 14274–14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dwyer, M. A., Looger, L. L. & Hellinga, H. W. (2004) Science 304, 1965–1971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.