Abstract
The efficiency and fidelity of nucleotide incorporation by high-fidelity replicative DNA polymerases (Pols) are governed by the geometric constraints imposed upon the nascent base pair by the active site. Consequently, these polymerases can efficiently and accurately replicate through the template bases which are isosteric to natural DNA bases but which lack the ability to engage in Watson-Crick (W-C) hydrogen bonding. DNA synthesis by Polη, a low-fidelity polymerase able to replicate through DNA lesions, however, is inhibited in the presence of such an analog, suggesting a dependence of this polymerase upon W-C hydrogen bonding. Here we examine whether human Polκ, which differs from Polη in having a higher fidelity and which, unlike Polη, is inhibited at inserting nucleotides opposite DNA lesions, shows less of a dependence upon W-C hydrogen bonding than does Polη. We find that an isosteric thymidine analog is replicated with low efficiency by Polκ, whereas a nucleobase analog lacking minor-groove H bonding potential is replicated with high efficiency. These observations suggest that both Polη and Polκ rely on W-C hydrogen bonding for localizing the nascent base pair in the active site for the polymerization reaction to occur, thus overcoming these enzymes' low geometric selectivity.
Classical DNA polymerases (Pols) replicate DNA with a high fidelity and are unable to replicate through DNA-distorting lesions. From studies with nonpolar, isosteric analogs, which lack the ability to form canonical Watson-Crick (W-C) hydrogen bonds, it has been concluded that W-C H bonding is not the determining factor for the efficiency and accuracy of nucleotide incorporation in high-fidelity polymerases such as T7 and Escherichia coli Pol I (20, 23, 24); rather, it is apparently the geometric fit within the active site of the incoming nucleoside triphosphate with the templating nucleotide that governs polymerase efficiency and accuracy (3, 5, 13, 15).
Members of the Y family of DNA polymerases replicate DNA with a low fidelity, and unlike the classical polymerases, they are able to replicate through DNA lesions (28, 29). Humans have four Y family Pols: η, ι, κ, and Rev1. Of these, Rev1 is a highly specialized polymerase which predominantly incorporates a C opposite template G and also opposite an abasic site (8, 26), and genetic studies of Saccharomyces cerevisiae have suggested that a major role of Rev1 is to act as an assembly factor in Polζ-dependent lesion bypass (28). Although not as extreme in its nucleotide insertion specificity as Rev1, Polι incorporates nucleotides opposite the four different template bases with very different efficiencies and fidelities, and it incorporates nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines (6, 11, 34, 39, 46). From the ternary crystal structure of Polι, with a templating A and an incoming dTTP, it has been determined that unlike all other DNA polymerases, which impose Watson-Crick base pairing in their active site, Polι uses Hoogsteen base pairing for DNA synthesis (25).
In contrast to the specificity of Rev1 and Polι for nucleotide incorporation opposite template bases, all other known DNA polymerases, including the high-fidelity replicative/repair DNA polymerases, as well as the two other members of the Y family, Polη and Polκ (10, 12, 41), form the four possible correct base pairs with nearly equivalent catalytic efficiencies. Pols η and κ, however, differ from the high-fidelity polymerases in their low fidelity of nucleotide incorporation and in their ability to replicate through DNA lesions. For example, both yeast and human Polη misincorporate nucleotides with a frequency of ∼10−2 to 10−3, and both of these enzymes replicate through a cis-syn TT dimer by inserting two A’s opposite the two T’s of the dimer with the same efficiency and fidelity as opposite the undamaged T’s (12, 40, 41, 43). The structure of yeast Polη modeled with a templating TT dimer and an incoming dATP has indicated that it can accommodate both of the residues of the dimer in its active site (35); in this respect, Polη differs from other polymerases, including the other Y family polymerases, as they can accommodate only the templating residue in their active site, while the next 5′ template base and the rest of the unpaired template are pushed out of the active site at a 90o angle (28).
One possible consequence of the proficient ability of Polη to hold two templating residues in its active site instead of one is that its active site may not be as closely juxtaposed to the templating base and the incoming deoxynucleoside triphosphate (dNTP) as occurs in the high-fidelity polymerases, which exercise a high degree of geometric selection because of the tight fit of the correct base pair in their active sites (3, 5, 14, 15). We have shown previously that Polη differs from high-fidelity replication/repair polymerases in its inability to replicate non-hydrogen-bonding nucleotide analogs. We hypothesized that this enzyme, lacking a tight geometric constraint, may rely instead upon Watson-Crick hydrogen bonding for the efficiency and fidelity of DNA synthesis (37).
Polκ differs from Polη in several respects, including its higher fidelity and its inability to incorporate nucleotides opposite DNA lesions. Polκ exhibits the highest fidelity among the Y family polymerases, as it misincorporates nucleotides with a frequency of ∼10−3 to 10−4 opposite all four template bases (10). Also, in contrast to Polη, Polκ is unable to replicate through a cis-syn TT dimer, which results primarily from its inability to incorporate nucleotides opposite the 3′ T of the dimer (10, 38). Polκ is also highly inefficient at incorporating nucleotides opposite various other lesions, such as a (6-4) TT photoproduct, an N2-acetyl-aminofluorene adduct, and others (9, 10, 17, 33, 38). For some DNA lesions, however, Polκ can promote the extension reaction wherein, following the incorporation of a nucleotide opposite the lesion site by another DNA polymerase, it performs the further extension of the primer terminus (7, 38, 42).
The higher fidelity and the inability to incorporate nucleotides opposite DNA lesions could result if the Polκ active site were more geometrically constrained than that of Polη. The structure of the Polκ apoenzyme and the modeling of Polκ in a ternary complex with DNA and an incoming nucleotide have suggested this to be the case, as Polκ's active site appears to be more constrained in the vicinity of the templating base than that of Polη (36). Since the more enclosed active site of Polκ could provide for a higher degree of geometric selection than would be possible for a polymerase with a more open active site, the dependence of Polκ upon other factors such as Watson-Crick or minor-groove hydrogen bonding may be lessened.
Here we examine the effects of difluorotoluene (F), which is identical in shape, size, and conformation to thymine but lacks the ability to form W-C hydrogen bonds with adenine (A) (30, 31), on DNA synthesis by human Polκ (hPolκ). Interestingly, we find that F is highly inhibitory to DNA synthesis by Polκ, regardless of whether it is used as a templating residue or as an incoming nucleotide. This is consistent with the notion that W-C hydrogen bonding makes a paramount contribution to DNA synthesis by Polκ. The implications of this observation are discussed.
MATERIALS AND METHODS
Purification of DNA polymerases.
S. cerevisiae strain BJ5464 was transformed with plasmid pPol42, which carries the gene encoding wild-type hPolκ fused in frame with glutathione S-transferase. Cells were grown and hPolκ was purified as described previously (10). Cleavage of the glutathione S-transferase tag from Polκ resulted in a seven-amino-acid peptide leader sequence attached to the N terminus of the polymerase. The protein concentration was determined by a Bio-Rad protein assay (Bio-Rad) and by UV absorbance at 280 nm under denaturing conditions (8 M urea), using the molar extinction coefficient calculated from the amino acid composition. The purified hPolκ was stored in 5-μl aliquots at −80°C. Taq was purchased from Gene Choice, Inc.
Nucleotides and DNA substrates.
5′-dNTPs (100 mM) were purchased from Roche Diagnostics and stored in aliquots at −20°C. Synthetic oliogodeoxynucleotide template and primers were used to prepare the nondamaged DNA substrates. The 3-deazaguanine (3DG)- and F-containing DNA substrates were synthesized and purified as described previously (37, 45). For the 3DG-containing substrates, the following template/primer pairs were used. The incoming nucleotide (P0) template/primer was 5′-CTGCGACTGCTGCGTCTGCGGTGC-3′/5′-GCACCGCAGACGCAGCA. For the P0 substrate, the bold and underlined nucleotide is the templating base, and either a dGTP or a 3DG nucleoside triphosphate (d3DGTP) was used as the incoming dNTP. The templating residue (T0) template/primer was 5′-CTGCGACTXCTGCGTCTGCGGTGC-3′/5′-GCACCGCAGACGCAG. For the T0 substrate, the bold and underlined nucleotide represents a G or a 3DG. The 2,4-difluorotoluene-containing template sequence was 5′-ACTGXTCTCCCTATAGTGAGTCGTATTA-3′, where the bold and underlined X represents either an A, a T, or an F. The standing-start and running-start primer sequences were 5′-TAATACGACTCACTATAGGGAGA-3′ and 5′-TAATACGACTCACTATAG-3′, respectively. For the polymerase assays, 32P-5′-end-labeled primers (1.0 μM) were annealed to templates (1.5 μM) in 50 mM Tris HCl (pH 7.5) and 100 mM NaCl by being heated to 95°C for 2 min, followed by slow cooling to room temperature. The resulting DNA substrates were used to perform DNA synthesis reactions and to determine the steady-state kinetics parameters.
DNA polymerase assays.
The standard DNA polymerase reaction mixture contained 25 mM Tris HCl (pH 7.5), 5 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 5 mM MgCl2, 10 nM DNA, 1 nM polymerase, and dNTPs. Reactions were carried out at 23°C for 3DG or at 30°C for F, and reaction mixtures were subsequently quenched with 95% formamide loading buffer. Quenched reaction products were heat denatured at 95°C for 3 min, and product formation was determined by 15% polyacrylamide gel electrophoresis (8 M urea) and quantified by PhosphorImager analysis (Molecular Dynamics).
For the steady-state kinetic analyses of the 3DG-containing substrates, standard DNA polymerase reaction conditions were used, with the addition of a single dNTP complementary to the templating base (e.g., dCTP opposite G and 3DG, dATP opposite T, etc.) in a concentration range appropriate for Km determinations. Deoxynucleotide incorporation was measured at multiple dNTP concentrations for a single time point, and the steady-state kinetic parameters kcat and Km were determined by the best fit to the Michaelis-Menten equation (Sigma Plot 7.0).
For the steady-state kinetic analyses of the 2,4-difluorotoluene-containing substrates, standard DNA polymerase reaction conditions were used, with the addition of a single dNTP complementary to the templating base (e.g., dATP opposite T and F, dFTP opposite A, or dTTP opposite A) in a concentration range appropriate for Km determinations. Deoxynucleotide incorporation was measured at multiple time points for each respective dNTP concentration, and the observed rate of nucleotide incorporation was determined by linear regression. Values of the steady-state parameters kcat and Km for nucleotide incorporation were then determined by the best fit to the Michaelis-Menten equation (Sigma Plot 7.0).
RESULTS
DNA synthesis past a template F residue by Polκ.
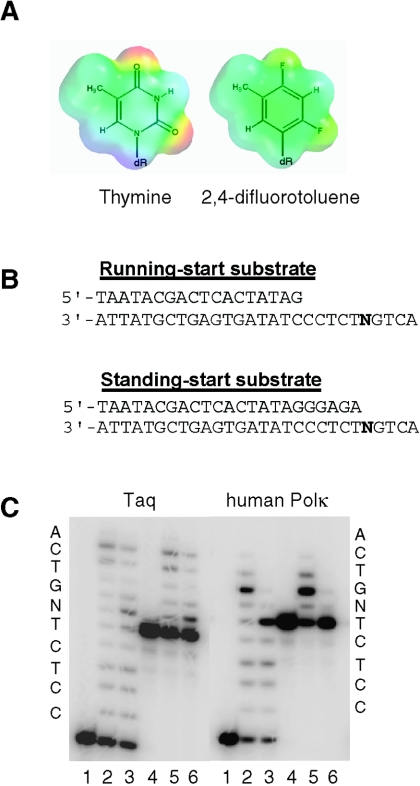
To determine whether human Polκ could synthesize DNA in the absence of Watson-Crick hydrogen bonds, we examined its ability to polymerize nucleotides past a template F, which is a nonpolar, isosteric analog of thymine. As F lacks the Watson-Crick O2 and O4 hydrogen bond acceptors and N-H3 donor of thymine (Fig. 1A), it is unable to participate in W-C hydrogen bonding (31). Since W-C hydrogen bonding is not needed for nucleotide incorporation by the Thermus aquaticus (Taq) DNA polymerase (23), we compared the abilities of Taq and Polκ to synthesize DNA on substrates containing an F residue at either the sixth or the first templating position of the DNA substrate (Fig. 1B). As expected, Taq was able to incorporate a nucleotide opposite the template F residue (Fig. 1C, left panel, lanes 3 and 6) and could synthesize DNA past it, but it was somewhat hindered in extending from the nucleotide incorporated opposite F, as evidenced by the presence of a stall site opposite the lesion. Polκ, on the other hand, was unable to synthesize DNA when F was present at either of the template positions (Fig. 1C, right panel, lanes 3 and 6). Also, in the running-start assay, a strong stall site was observed immediately before the F residue, indicating that Polκ is strongly inhibited at incorporating a nucleotide opposite the F residue.
FIG. 1.
(A) Overlay of the electrostatic potential and chemical structure of thymine (left) and the nonpolar, isosteric nucleobase analog, 2,4-difluorotoluene (right). (B) Running-start and standing-start DNA substrates. The 28-mer templates have either a T or an F at position 24, shown as a bold N. (C) Running-start and standing-start assays with Taq and human Polκ. Lanes 1 to 3 are running-start experiments, and lanes 4 to 6 are standing-start experiments. Lanes 1 and 4 contain no protein, lanes 2 and 5 contain a template T residue, and lanes 3 and 6 contain a template F residue. The stalling of synthesis by Polκ 1 to 2 nucleotides prior to the end of the DNA template is a feature that is characteristic of this polymerase (10).
Steady-state kinetics of nucleotide incorporation by Polκ.
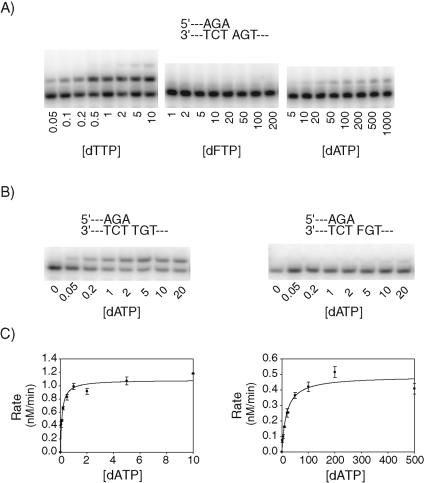
Next, we quantified the effects of an F residue on the efficiency and accuracy of nucleotide incorporation by Polκ, by using steady-state kinetic assays on DNA substrates that contained an F residue at the position of the incoming nucleotide or the templating base. From the rate of nucleotide incorporation, graphed as a function of [dNTP], and the subsequent best fit to the Michaelis-Menten equation, the steady-state kinetic parameters kcat and Km and the efficiency (kcat/Km) of nucleotide incorporation with respect to F substitution were determined. When F was used as the incoming dNTP opposite template A (i.e., a dFTP · A base pair), nucleotide incorporation was below detectable limits (Fig. 2A). Based upon the estimated minimal detectable levels of incorporation, the dFTP incorporation efficiencies were determined to be ≤1 × 10−5. For comparison, the misincorporation of an A opposite template A occurred with an efficiency of 4 × 10−3 (Table 1). We then determined the efficiency of nucleotide incorporation when F is the templating base (Fig. 2B). Polκ incorporates an A opposite template T with an efficiency of 1.7 (Fig. 2C, left panel; Table 1), and it misincorporates a T opposite a template T with an efficiency of 3.4 × 10−4 (Table 1). When F occupies the position of the templating base, dATP is incorporated with an efficiency of 4.5 × 10−3 (Fig. 2C, right panel; Table 1), suggesting that disrupting the Watson-Crick hydrogen bond interactions with the incoming dNTP results in a block which is almost as severe as the block for mispair formation. Thus, the substitution of an F for a T, whether at the templating position or the incoming nucleotide position, elicits a large reduction in the efficiency and fidelity of nucleotide incorporation by Polκ.
FIG. 2.
Effects of F on DNA synthesis by hPolκ. (A) dTTP, dFTP, or dATP incorporation opposite template A. (B) Incorporation of dATP opposite a template T or F. (C) Rates of dATP incorporation opposite a template T (left) and a template F (right), graphed as a function of [dATP]. The solid line represents the best fit to the Michaelis-Menten equation. Steady-state kinetic parameters are listed in Table 1. A portion of the DNA substrates used is shown in panel B.
TABLE 1.
Steady-state kinetic parameters for nucleotide incorporation by hPolκ with 2,4-difluorotoluene-containing DNA substrates
| DNA substrate | dNTP | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) | Relative efficiency |
|---|---|---|---|---|---|
| 5′-AGA | dTTP | 0.19 ± 0.008 | 0.16 ± 0.03 | 1.2 | 1 |
| 3′-TCTA | dFTP | NDa | ND | ≤1 × 10−5 | ≤8 × 10−6 |
| dATP | 0.036 ± 0.002 | 8.6 ± 3 | 4.2 × 10−3 | 4 × 10−3 | |
| 5′-AGA | dATP | 0.20 ± 0.006 | 0.12 ± 0.02 | 1.7 | 1 |
| 3′-TCTT | dTTP | 0.086 ± 0.004 | 250 ± 30 | 3.4 × 10−4 | 2 × 10−4 |
| 5′-AGA | dATP | 0.095 ± 0.005 | 21 ± 4 | 4.5 × 10−3 | 3 × 10−3 |
| 3′-TCTF | dTTP | 0.11 ± 0.01 | 630 ± 140 | 1.7 × 10−4 | 1 × 10−4 |
ND, no nucleotide incorporation detected. Nucleotide incorporation efficiencies are based upon the estimated minimal detectable levels of incorporation.
DNA synthesis by Polκ on substrates containing a 3DG substitution.
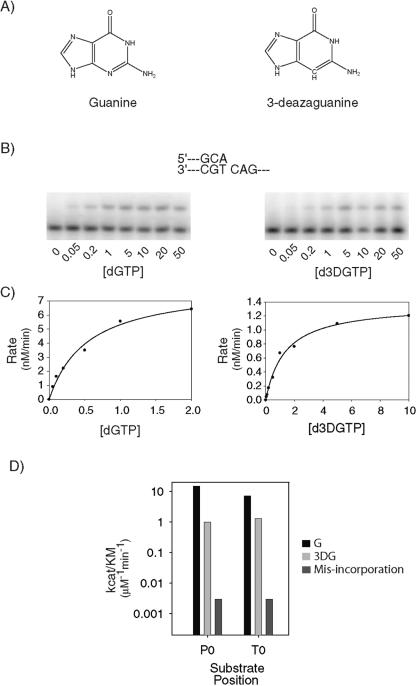
In addition to the geometric constraints imposed by a tight active site, DNA polymerases can check for the correctness of a base pair by participating in specific hydrogen bonding interactions with the minor-groove hydrogen bond acceptors, N3 for purines and O2 for pyrimidines, in DNA (2, 4, 18, 19, 21, 22, 32, 45). Because of the absence of the O2 hydrogen bond acceptor in F, we considered the possibility that the reduction in the efficiency and fidelity of nucleotide incorporation that occurs when F is used as a templating residue or as an incoming nucleotide originates from a possible dependence of Polκ on minor-groove hydrogen bonding interactions with both the templating base and the incoming nucleotide. To probe for such effects, we used 3DG, which is a base analog of guanine (G) but differs from G in having a carbon instead of a nitrogen at position 3 of the base (Fig. 3A). Because of the absence of an N3 hydrogen bond acceptor, the 3DG analog cannot participate in minor-groove hydrogen bonding interactions with the polymerase.
FIG. 3.
Effects of 3DG on DNA synthesis by hPolκ. (A) Chemical structures of guanine (left) and 3DG (right). (B) Incorporation of dGTP or d3DGTP opposite template C. A portion of the DNA substrate used is shown (top). (C) Rates of dGTP and d3DGTP incorporation opposite a template C, graphed as a function of [dNTP]. The solid line represents the best fit to the Michaelis-Menten equation. Steady-state kinetic parameters are listed in Table 2. (D) Bar graph showing the effect of 3DG substitution on the efficiency (kcat/Km) of correct nucleotide incorporation when a G (black) or 3DG (light gray) is used as the incoming nucleotide (P0) opposite template C or as the templating nucleotide (T0) for the incoming C. For comparison, the efficiencies of misincorporation of a C (P0) opposite a template C and of a G opposite template G (T0) are shown (dark gray).
We examined the effects of a 3DG substitution for G at the position of the incoming nucleotide, referred to as P0, or at the templating position, referred to as T0. Compared to the efficiency of G incorporation opposite template C, Polκ incorporated 3DG with an ∼15-fold reduction in efficiency (Fig. 3B, C, and D; Table 2). By contrast, Polκ misincorporated a C opposite template C with an ∼5,000-fold reduction in efficiency (Table 2; Fig. 3D). The substitution of 3DG for G at the templating position led to only an approximately fivefold reduction in the efficiency of C incorporation, while the efficiency of G misincorporation opposite template G was reduced by ∼2,500-fold (Table 2; Fig. 3D). Thus, the ablation of any possible minor-groove hydrogen bonding interactions with the P0 and T0 positions in DNA confers only a small impairment in the efficiency of correct nucleotide incorporation by Polκ. The results therefore indicate strongly that the large reduction in the efficiency and fidelity of nucleotide incorporation that occurs with analog F must derive from a different molecular origin.
TABLE 2.
Steady-state kinetic parameters for nucleotide incorporation by hPolκ with 3DG-containing DNA substrates
| DNA substratea | dNTP/template | kcat (min−1) | Km (μM) | kcat/Km (μM−1 min−1) | Relative efficiency |
|---|---|---|---|---|---|
| P0-G | G/C | 8.2 ± 0.6 | 0.54 ± 0.09 | 15 | 1 |
| C/C | 0.52 ± 0.01 | 150 ± 10 | 3.4 × 10−3 | 2 × 10−4 | |
| P0-3DG | 3DG/C | 1.4 ± 0.05 | 1.4 ± 0.2 | 1.0 | 0.07 |
| T0-G | C/G | 4.1 ± 0.4 | 0.57 ± 0.2 | 7.2 | 1 |
| G/G | 1.2 ± 0.07 | 400 ± 60 | 3.0 × 10−3 | 4 × 10−4 | |
| T0-3DG | C/3DG | 2.3 ± 0.14 | 1.7 ± 0.2 | 1.3 | 0.18 |
| G/3DG | 0.35 ± 0.008 | 150 ± 10 | 2.3 × 10−3 | 3 × 10−4 |
P0-G and P0-3DG refer, respectively, to the incoming G and 3DG nucleotides opposite template C. T0-G and T0-3DG refer, respectively, to the templates G and 3DG.
DISCUSSION
Based upon thermal denaturation studies of DNA alone, the base pairing free energy differences (ΔΔG°) between correct Watson-Crick base pairs and mismatched base pairs are as high as 4.0 kcal/mol (1, 16). At the terminus of DNA, where DNA synthesis occurs, however, selectivities are considerably smaller (27). Thus, these free energy differences are far too small to account for the millionfold stringency manifested by replicative DNA polymerases. Furthermore, the replacement of normal nucleotides with the nucleotide analogs which impair Watson-Crick hydrogen bonding has very little effect on the efficiency and fidelity of nucleotide incorporation by DNA polymerases such as E. coli Klenow fragment and T7 (20, 23, 24). From such analyses, it has been inferred that W-C hydrogen bonding is not needed for base pair synthesis by the high-fidelity polymerases.
In the high-fidelity DNA polymerases, the long α helices of the fingers domain close tightly on the incoming nucleotide and the templating residue, and the resulting snug active site may provide for a high degree of geometric selectivity that these enzymes apparently possess (2, 4, 18, 19). Consequently, any need for W-C hydrogen bonding is minimized. By contrast, the active site of Y family polymerases is much more open and sterically less constrained around the nascent base pair, and this is primarily due to the fingers domain being very small and stubby (28). As a consequence, the Y family polymerases are not as sensitive to the geometric distortions conferred upon DNA by the presence of lesions.
In a previous study, we showed that the substitution of an F for a T at the templating site or as an incoming nucleotide is highly inhibitory to DNA synthesis by yeast Polη (37). Here we provide evidence that DNA synthesis by Polκ is severely impaired when F is used either as a templating residue or as an incoming nucleotide. From these and other observations reported here, we hypothesize that DNA synthesis by Polκ is strongly dependent upon W-C hydrogen bonding, and in this respect, it closely resembles Polη. We suggest that, in a relatively roomy active site, the analog does not fill the available space and is therefore not fixed in the correct location for proper alignment of the triphosphate group. In contrast to this, thymine would be constrained by optimum hydrogen bonding geometry to a fixed location, thus aligning the reactive triphosphate for phosphodiester bond formation. This would also be consistent with the poor activity of mismatched pairs, which, although hydrogen bonded, would be fixed in an unfavorable position for base pair synthesis.
In making this hypothesis, we considered a number of other possible reasons for the poor activity of the analog F with Polκ. First is the size of the F · A base pair, which is likely larger than the natural T · A pair by a small amount (approximately 0.3 Å), due to the lack of a hydrogen bonding contraction. However, since the enzyme can replicate through lesions and form mispairs with greater efficiency, this seems an unlikely explanation for the present findings. Second is the aforementioned lack of a minor-groove H bond acceptor in F (31); however, our data with 3DG show clearly that this lack has only a very small kinetic effect, both in the template and in the incoming nucleotide. Since the chief chemical difference between F and T is the strong electrostatic difference along the Watson-Crick edge, we arrived at the lack of Watson-Crick hydrogen bonding as the best available explanation.
The observation that Polκ shows the same high degree of apparent dependence upon W-C hydrogen bonding as Polη may seem surprising, in view of the very considerable differences that exist in their fidelities and damage bypass abilities. Whereas Polη misincorporates nucleotides with a frequency of ∼10−2 to 10−3 and is able to proficiently replicate through cyclobutane pyrimidine dimers as well as through many other sorts of DNA lesions, Polκ displays a fidelity of ∼10−3 to 10−4 and is unable to replicate through cyclobutane pyrimidine dimers and other DNA lesions, which primarily results from its inability to incorporate nucleotides opposite the lesion site. As the lower fidelity and the more proficient ability of Polη to incorporate nucleotides opposite lesions such as cyclobutane pyrimidine dimers (and others) could be ascribed to this polymerase having a more open active site, for Polκ, the higher fidelity and the inability to incorporate nucleotides opposite lesion sites could suggest a more constrained active site. Consequently, Polκ's active site could have provided for a higher degree of geometric selection than that of Polη, thereby lessening the dependence of Polκ upon W-C hydrogen bonding. Our finding that a loss of W-C hydrogen bonding is apparently as detrimental to DNA synthesis by Polκ as it is to DNA synthesis by Polη, however, is more in line with the view that, overall, the active sites of both of these polymerases share common structural features which do not allow for a great deal of geometric selectivity. That, in turn, puts a strong premium upon W-C hydrogen bonding for the proper positioning of the nascent base pair in the active site, so that the subsequent induced fit conformational change (44) and the chemical reaction of phosphodiester bond formation may occur. We presume then that the different fidelities and damage bypass abilities of Polη and Polκ arise from subtle structural differences in their active sites and not from a large global change that could have allowed for a significantly higher degree of geometric selectivity in one polymerase than in the other.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences grant ES012411 and by grant GM072705 from NIGMS.
REFERENCES
- 1.Aboul-ela, F., D. Koh, I. Tinoco, Jr., and F. H. Martin. 1985. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A, C, G, T). Nucleic Acids Res. 13:4811-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doublie, S., S. Tabor, A. M. Long, C. C. Richardson, and T. Ellenberger. 1998. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature 391:251-258. [DOI] [PubMed] [Google Scholar]
- 3.Echols, H., and M. F. Goodman. 1991. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 60:477-511. [DOI] [PubMed] [Google Scholar]
- 4.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a Pol α family DNA polymerase. Cell 105:657-667. [DOI] [PubMed] [Google Scholar]
- 5.Goodman, M. F. 1997. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. USA 94:10493-10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska, L., L. Prakash, and S. Prakash. 2002. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl. Acad. Sci. USA 99:16000-16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haracska, L., S. Prakash, and L. Prakash. 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 277:15546-15551. [DOI] [PubMed] [Google Scholar]
- 9.Haracska, L., I. Unk, R. E. Johnson, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2002. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell. Biol. 22:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, R. E., S. Prakash, and L. Prakash. 2000. The human DINB1 gene encodes the DNA polymerase Polθ. Proc. Natl. Acad. Sci. USA 97:3838-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 13.Kool, E. T. 2002. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 71:191-219. [DOI] [PubMed] [Google Scholar]
- 14.Kool, E. T. 2001. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 30:1-22. [DOI] [PubMed] [Google Scholar]
- 15.Kool, E. T. 1998. Replication of non-hydrogen bonded bases by DNA polymerases: a mechanism for steric matching. Biopolymers 48:3-17. [DOI] [PubMed] [Google Scholar]
- 16.Law, S. M., R. Eritja, M. F. Goodman, and K. J. Breslauer. 1996. Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry 35:12329-12337. [DOI] [PubMed] [Google Scholar]
- 17.Levine, R. L., H. Miller, A. Grollman, E. Ohashi, H. Ohmori, C. Masutani, F. Hanaoka, and M. Moriya. 2001. Translesion DNA synthesis catalyzed by human Pol η and Pol κ across 1,N6-ethenodeoxyadenosine. J. Biol. Chem. 276:18717-18721. [DOI] [PubMed] [Google Scholar]
- 18.Li, Y., S. Korolev, and G. Waksman. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17:7514-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Y., V. Mitaxov, and G. Waksman. 1999. Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation. Proc. Natl. Acad. Sci. USA 96:9491-9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales, J. C., and E. T. Kool. 1998. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat. Struct. Biol. 5:950-954. [DOI] [PubMed] [Google Scholar]
- 21.Morales, J. C., and E. T. Kool. 2000. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry 39:12979-12988. [DOI] [PubMed] [Google Scholar]
- 22.Morales, J. C., and E. T. Kool. 1999. Minor groove interactions between polymerase and DNA: more essential to replication than Watson-Crick hydrogen bonds? J. Am. Chem. Soc. 121:2323-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morales, J. C., and E. T. Kool. 2000. Varied molecular interactions at the active sites of several DNA polymerases: nonpolar nucleotide isosteres as probes. J. Am. Chem. Soc. 122:1001-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran, S., R. X.-F. Ren, and E. T. Kool. 1997. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl. Acad. Sci. USA 94:10506-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nair, D. T., R. E. Johnson, S. Prakash, L. Prakash, and A. K. Aggarwal. 2004. Replication by human DNA polymerase ι occurs via Hoogsteen base-pairing. Nature 430:377-380. [DOI] [PubMed] [Google Scholar]
- 26.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382:729-731. [DOI] [PubMed] [Google Scholar]
- 27.Petruska, J., M. F. Goodman, M. S. Boosalis, L. C. Sowers, C. Cheong, and I. Tinoco, Jr. 1988. Comparison between DNA melting thermodynamics and DNA polymerase fidelity. Proc. Natl. Acad. Sci. USA 85:6252-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 29.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 30.Schweitzer, B. A., and E. T. Kool. 1994. Aromatic nonpolar nucleosides as hydrophobic isosteres of pyrimidine and purine nucleosides. J. Org. Chem. 59:7238-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweitzer, B. A., and E. T. Kool. 1995. Hydrophobic, non-hydrogen-bonding bases and base pairs in DNA. J. Am. Chem. Soc. 117:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spratt, T. E. 2001. Identification of hydrogen bonds betweeen Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry 40:2647-2652. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, N., E. Ohashi, K. Hayashi, H. Ohmori, A. P. Grollman, and S. Shibutani. 2001. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry 40:15176-15183. [DOI] [PubMed] [Google Scholar]
- 34.Tissier, A., J. P. McDonald, E. G. Frank, and R. Woodgate. 2000. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 14:1642-1650. [PMC free article] [PubMed] [Google Scholar]
- 35.Trincao, J., R. E. Johnson, C. R. Escalante, S. Prakash, L. Prakash, and A. K. Aggarwal. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8:417-426. [DOI] [PubMed] [Google Scholar]
- 36.Uljon, S. N., R. E. Johnson, T. A. Edwards, S. Prakash, L. Prakash, and A. K. Aggarwal. 2004. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure 12:1395-1404. [DOI] [PubMed] [Google Scholar]
- 37.Washington, M. T., S. A. Helquist, E. T. Kool, L. Prakash, and S. Prakash. 2003. Requirement of Watson-Crick hydrogen bonding for DNA synthesis by yeast DNA polymerase η. Mol. Cell. Biol. 23:5107-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2002. Human DINB1-encoded DNA polymerase κ is a promiscuous extender of mispaired primer termini. Proc. Natl. Acad. Sci. USA 99:1910-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2004. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 24:936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274:36835-36838. [DOI] [PubMed] [Google Scholar]
- 42.Washington, M. T., I. G. Minko, R. E. Johnson, W. T. Wolfle, T. M. Harris, R. S. Lloyd, S. Prakash, and L. Prakash. 2004. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 24:5687-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Washington, M. T., L. Prakash, and S. Prakash. 2003. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Natl. Acad. Sci. USA 100:12093-12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Washington, M. T., L. Prakash, and S. Prakash. 2001. Yeast DNA polymerase η utilizes an induced fit mechanism of nucleotide incorporation. Cell 107:917-927. [DOI] [PubMed] [Google Scholar]
- 45.Washington, M. T., W. T. Wolfle, T. E. Spratt, L. Prakash, and S. Prakash. 2003. Yeast DNA polymerase η makes functional contacts with the DNA minor groove only at the incoming nucleoside triphosphate. Proc. Natl. Acad. Sci. USA 100:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, Y., F. Yuan, X. Wu, and Z. Wang. 2000. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase ι. Mol. Cell. Biol. 20:7099-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]