Abstract
IRE1-alpha is an integral membrane protein of the endoplasmic reticulum (ER) that is a key sensor in the cellular transcriptional response to stress in the ER. Upon induction of ER stress, IRE1-alpha is activated, resulting in the synthesis of the active form of the transcription factor XBP1 via IRE1-mediated splicing of its mRNA. In this report, we have examined the role of IRE1-alpha and XBP1 in activation of the hepatitis B virus S promoter by ER stress. Cotransfection experiments revealed that overexpression of either IRE1-alpha or XBP1 activated this promoter. Conversely, cotransfected dominant-negative IRE1-alpha or small interfering RNA directed against XBP1 decreased the activation of the S promoter by ER stress, confirming an important role for the IRE1-alpha/XBP1 signaling pathway in activation of the S promoter. However, XBP1 does not bind directly to the S promoter; rather, a novel S promoter-binding complex that does not contain XBP1 is induced in cells undergoing ER stress in an XBP1-dependent manner. This complex, as well as transcriptional activation of the S promoter, is induced by ER stress in hepatocytes but not in fibroblasts, despite the presence of active XBP1 in the latter. Thus, the hepatitis B virus S promoter responds to a novel, cell type-restricted transcriptional pathway downstream of IRE1-alpha and XBP1.
Eucaryotic cells have evolved sophisticated mechanisms to sense stress in various intracellular compartments and respond appropriately by modulating nuclear gene expression. Within the endoplasmic reticulum (ER), stress is induced by the presence of large amounts of unfolded or misfolded proteins, which leads to signal transduction to both the cytosol and the nucleus (reviewed in references 9, 27, 34, and 61). In the yeast Saccharomyces cerevisiae, the proximal sensor of ER stress is the transmembrane ER protein Ire1p (2, 28). Ire1p is both a kinase and an endoribonuclease that is active only in the presence of ER stress (3, 43). Upon activation, Ire1p becomes competent to cleave the Hac1 mRNA to initiate its splicing via an unconventional pathway, resulting in a sequence substitution at the 3′ end of the open reading frame (3). The unspliced mRNA is not translated, while the spliced mRNA is translated into the transcription factor Hac1p that binds to and activates the promoters of the genes coding for various ER chaperones as well as other selected proteins, such as lipid biosynthetic enzymes (15, 29, 31, 37).
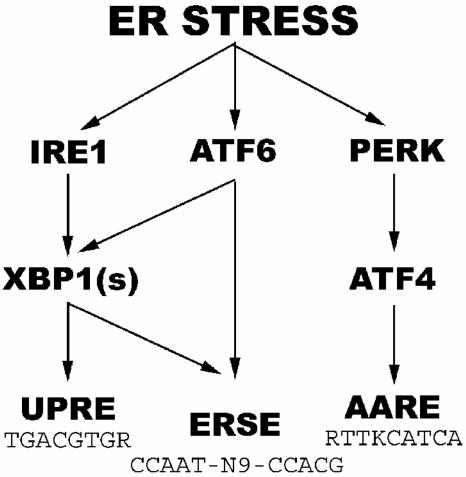
Metazoans, such as helminths and mammals, have preserved a homologous pathway wherein the downstream factor is the basic leucine zipper (b-ZIP) transcription factor called XBP1 (59) (Fig. 1). As in the yeast, metazoan IRE1-alpha activated by ER stress splices the mRNA for XBP1, resulting in the translation of the active form of this protein, called XBP1(s) (1, 41, 45, 49, 59). XBP1(s) activates the promoters of many genes, including those coding for proteins necessary for degradation of ER proteins, by binding to so-called unfolded protein response elements (UPRE) with the consensus sequence TGACGTGG/A (18, 58). In addition, metazoans have developed other pathways to sense and act on ER stress. The transmembrane ER protein PERK (also called PEK) has a similar ER luminal domain as IRE1 and thus also senses ER stress (11, 22, 42). The cytosolic domain is a kinase that phosphorylates the translation initiation factor eIF2-alpha and thereby decreases global translation (11, 22, 42). The phosphorylation of eIF2-alpha also paradoxically increases the translation of ATF4, a b-ZIP transcription factor that activates the transcription of genes coding for amino acid synthetic enzymes, antioxidant proteins, and certain regulatory proteins, such as CHOP and GADD34, via binding to the amino acid response element (AARE) with the consensus sequence RTTKCATCA (10, 12, 25, 26, 32, 47) (Fig. 1). Finally, upon ER stress the transmembrane ER protein ATF6-alpha is transported to the Golgi apparatus and cleaved by intramembranous proteases to release its N terminus, which is imported into the nucleus and functions as a b-ZIP transcription factor to activate promoters with ER stress response elements (ERSE; consensus sequence, CCAAT-N9-CCACG) (13, 20, 36, 40, 55, 57) (Fig. 1). Notably, ATF6-alpha recognizes the CCACG sequence, but its binding depends on the simultaneous binding of the ubiquitous transcription factor NF-Y to the CCAAT box at the other end of the ERSE (20, 60). Genes with ERSE include those coding for GRP78 (BiP) and other ER chaperones (57). Broadly speaking, it appears that the ATF6-alpha and PERK pathways function rapidly to decrease the amount of unfolded ER proteins by increasing folding and decreasing load, respectively, while XBP1(s) functions on a longer-term basis to increase both the folding and degradation of ER proteins (58).
FIG. 1.
Simplified diagram of ER stress pathways in metazoan cells.
Regulation of gene expression by ER stress in metazoans is complicated by extensive cross talk among the three pathways described above. For example, at least under certain conditions ATF6-alpha activates the transcription of xbp1 via an ERSE in its promoter (19), while XBP1(s) in conjunction with NF-Y also binds to ERSE and thus induces its own transcription as well as that of ER chaperones (59) (Fig. 1). In addition, some genes such as CHOP contain both ERSE and AARE and hence can be activated by all three b-ZIP factors (25, 32). Finally, ATF6-alpha and XBP1(s) can heterodimerize with each other and probably other b-ZIP transcription factors (30). In addition, as-yet-unknown pathways are likely involved. For example, ER stress does not activate transcription of an ERSE-containing reporter gene in cells doubly negative for ATF6-alpha and XBP1, as expected, yet the endogenous grp78 gene in these cells is still activated by ER stress, albeit to a slightly lower level than wild-type cells (18). Thus, despite recent rapid progress on our understanding of the mammalian ER stress response, much remains to be learned.
We have previously described the activation of the hepatitis B virus (HBV) S promoter by ER stress (52). The S promoter gives rise to the major envelope protein of HBV (small surface protein) and, thus, its activity is important in the viral life cycle (56). In further characterizing the S promoter sequences that respond to ER stress, we have found that it does not contain elements that resemble ERSE, UPRE, or AARE, nor does it bind to ATF6-alpha, XBP1(s), or ATF4. Rather, a novel factor in hepatoma cells binds to CT-rich elements in the promoter, in cooperation with NF-Y binding to a CCAAT element. This factor is induced by the IRE1-XBP1 pathway and appears to be absent in fibroblasts, despite the presence of XBP1 splicing in the latter cells. Thus, we have uncovered a previously unknown, cell type-restricted pathway of ER stress that is downstream of IRE1-alpha.
MATERIALS AND METHODS
Plasmids.
The reporter plasmids pS1CAT, pS1M2mutCAT, pS1Z1+2mutCAT (52), and pSV2CAT (6), as well as the expression plasmids pcDNA-IRE1, pcDNA/ATF6α, pcDNA-ATF6 (373), pcDNA/XBP1-s, pcDNA/XBP1-u (59, 60), pCHOPCAT, pcDNA/rATF4, and pATF4-HA (25), have been described previously in the cited references. The plasmids pcDNA-IRE1-ΔC (59), pgrp78(−457)CAT (21), and p5xATF6GL3 (50) were obtained from Kazunori Imaizumi (Nara Institute of Science and Technology), Amy Lee (University of Southern California), and Ron Prywes (Columbia University), respectively. The plasmids pGL3-control and pRL-CMV were purchased from Promega. The plasmid pS1-Luc contains the S promoter inserted between the BglII and HindIII sites of the plasmid pGL3-control (Promega), upstream of the firefly luciferase gene. The S promoter fragment was synthesized by using BglII and HindIII restriction enzymes to digest the PCR-amplified product from the plasmid pHBV1.3 (51), using the primers 5′-ATATAGATCTAACAATTCCTCCTCCTGCCTCCA and 5′-ATATAAGCTTCTGCAGAGTTTGGTGGAAGGCAG; its sequence in the final plasmid was verified at the UCSF Biomolecular Resource Facility.
Cell transfection and protein assays.
All cultured cell lines were grown at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin-streptomycin, under an atmosphere of 7% CO2. Cells growing in 60-mm plates were changed into medium without antibiotics, transfected with 3 μg of plasmid DNA using 6 μl of FuGENE6 (Roche), and harvested at 24 h after transfection. Where indicated, cells were treated for the last 17 h before harvesting with tunicamycin (50 μg/ml, diluted from a stock solution of 5 mg/ml in dimethyl sulfoxide) or the equivalent amount of dimethyl sulfoxide.
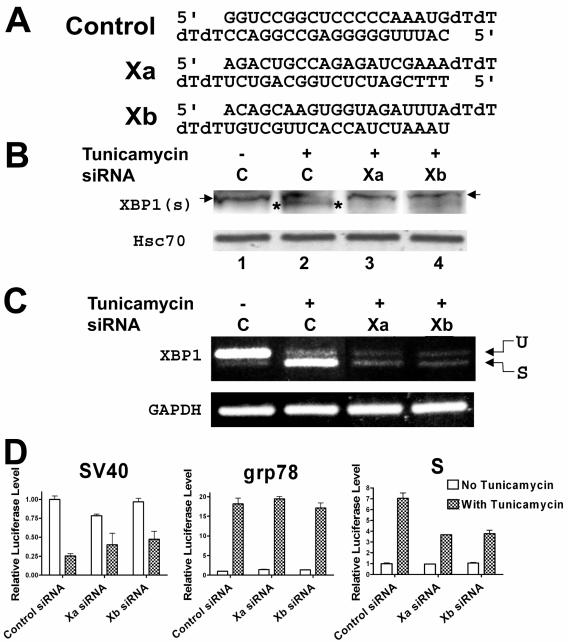
For experiments involving transfection of small interfering RNA (siRNA), the X-treme GENE siRNA transfection reagent (Roche) was used, according to the manufacturer's recommended protocol. Briefly, HuH7 cells grown in 150-cm dishes at 80% confluence were trypsinized, and 1/40 of the cells was placed into 1 ml of medium, mixed with 0.4 μg of the siRNA premixed with 5 μl of the transfection reagent, and immediately seeded into 12-well plates. After 24 h, the medium was changed and each well of cells was similarly transfected with 0.2 μg of siRNA and 0.4 μg of a plasmid premixed with 5 μl of the transfection reagent. The medium was replaced after 7 h, and either tunicamycin or dimethyl sulfoxide was added, as above. After 17 h, the cells were harvested for reporter assays. For protein or RNA quantitation, the same procedure was used but the amount of cells and reagents were doubled. The sequences of the siRNAs are shown below in Fig. 4A. The control siRNA was originally targeted to the nuclear protein CA150 but was found to be inactive (48). The two siRNAs directed against XBP1 were designed with the assistance of the Dharmacon siDESIGN Center (http://design.dharmacon.com/default.aspx), using GenBank accession number NM_005080, nucleotides 49 to 834. The Xa siRNA targets nucleotides 302 to 320, while the Xb siRNA targets nucleotides 345 to 563.
FIG. 4.
A. Sequences of the control siRNA (48) and the two siRNAs targeted against XBP1. B. Western blot of XBP1(s) after transfection with siRNAs against XBP1 or nonspecific siRNA. Proteins from HuH7 cells transfected with control siRNA (C) or two different XBP1 siRNAs (Xa and Xb, respectively) were separated on an sodium dodecyl sulfate-polyacrylamide gel and blotted for XBP1(s), as detailed in Materials and Methods. Note that there is a nonspecific band (marked with arrows) immediately above the XBP1(s) band (marked with asterisks). The same blot was stripped and reprobed for cellular Hsc70 to confirm the presence of similar amounts of proteins in each lane. A duplicate experiment yielded similar results. C. RT-PCR for xbp1 mRNA in HuH7 cells transfected with control siRNA (C) or XBP1 siRNAs (Xa and Xb). The top band represents unspliced xbp1 mRNA (U), while the bottom band represents spliced xbp1 mRNA (S). RT-PCR for GAPDH mRNA was performed on the same RNA samples to confirm that similar amounts of cDNA were present in each sample. A duplicate experiment revealed similar results. D. Effects of siRNAs targeting XBP1 on activation of the grp78 and S promoters by tunicamycin. HuH7 cells were cotransfected with a firefly luciferase reporter plasmid driven by the SV40, grp78, or S promoter and a Renilla luciferase reporter plasmid driven by the cytomegalovirus promoter, as well as siRNA Xa or Xb directed against XBP1, or a control siRNA, as detailed in Materials and Methods. One set of cells was treated with dimethyl sulfoxide (control), while another was treated with tunicamycin. The firefly luciferase activity in each extract was normalized to the Renilla luciferase activity, and the normalized activity in the cells transfected with control siRNA and treated with dimethyl sulfoxide was set as 1. Note that tunicamycin decreases expression from the SV40 promoter, possibly because this promoter is more sensitive to the nonspecific effects of tunicamycin than the cytomegalovirus promoter used to drive the expression of the Renilla luciferase used for normalization.
Chloramphenicol acetyltransferase (CAT) reporter activity in extracts of transfected cells was quantitated by the thin-layer chromatography method, using [14C]chloramphenicol (6). Cell extracts were diluted when necessary so that the activity would remain within the linear range of the assay (<50% acetylation of chloramphenicol).
For luciferase reporter assays, cells were harvested by scraping with a rubber policeman in 1 ml of phosphate-buffered saline. Each cell pellet was resuspended and lysed in 100 μl of passive lysis buffer (Promega) with vortexing and pipetting up and down and was cleared by centrifugation at 14,000 rpm for 10 min at 4°C. Firefly luciferase and Renilla luciferase activities were measured with 10 μl of cell extract in the dual-luciferase reporter assay system (Promega).
To measure XBP1(s) protein levels, cells grown in 12-well plates were lysed in 100 μl of reducing sodium dodecyl sulfate buffer. Twenty-microliter aliquots of samples were loaded on sodium dodecyl sulfate-polyacrylamide gels and subjected to electrophoresis. The samples were transferred to a polyvinylidene fluoride membrane (Millipore) and probed with a rabbit polyclonal antibody to XBP1 (59) at a dilution of 1/500. The secondary antibody was peroxidase-labeled goat anti-rabbit antibody from the Supersignal West Femto maximum sensitivity substrate kit (Pierce) diluted 1/1,000. To assess whether equivalent amounts of lysates were present in each lane, the blots were stripped with reducing sodium dodecyl sulfate buffer at 50°C for 30 min and reprobed with Hsc70 (K-19) goat polyclonal antibody (Santa Cruz Biotechnology) at a dilution of 1/2,000. The secondary antibody was peroxidase-labeled donkey anti-goat antibody (Santa Cruz Biotechnology) diluted 1/50,000. Supersignal West Femto maximum sensitivity substrate (Pierce) was used to develop all the blots.
Preparation of nuclear extracts and transcription factors.
All cell lines were cultured as above in 150-mm plates. Primary mouse embryo fibroblasts (MEFs) from C57BL6×157sv mice, derived as described by Flores et al. (4), were provided by Eric Huang and Guangwei Wei. Where indicated, the cells were treated for 12 h before harvesting with tunicamycin (50 μg/ml) or the equivalent amount of solvent (dimethyl sulfoxide). Nuclear extracts were prepared as described by Keller et al. (16). Briefly, cells were washed in phosphate-buffered saline and resuspended in five volumes of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethane sulfonyl fluoride), incubated on ice for 10 min, centrifuged at 250 × g for 5 min, and resuspended in three volumes of buffer A containing 0.05% Nonidet P-40. Nuclei were then released by using a Dounce homogenizer and pelleted by centrifugation at 250 × g for 10 min. After resuspension in 1 ml of buffer A with 0.05% Nonidet P-40, the nuclei were homogenized again, pelleted by centrifugation at 250 × g for 5 min, resuspended in 500 μl buffer C (5 mM HEPES at pH 7.9, 26% glycerol, 1.5 mM MgCl2, 300 mM NaCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethane sulfonyl fluoride), and incubated on ice for 30 min. After incubation, the lysed nuclei were centrifuged at 24,000 × g for 30 min at 4°C, and the supernatant was snap-frozen in liquid nitrogen and stored at −80°C until use. Protein concentration was measured with the Bio-Rad protein assay using bovine serum albumin as the standard.
For preparation of nuclear extracts from mouse livers, 3-month-old male B6D2F1 mice were injected intraperitoneally with either 0.4 ml of 50-μg/ml tunicamycin in 150 mM glucose or 0.4 ml of 150 mM glucose alone (62). After 24 h, the mice were euthanized by carbon dioxide narcosis, and the livers were harvested. All animal work was preapproved by the San Francisco Veterans Affairs Medical Center Animal Studies Subcommittee. Approximately 1 g of liver was homogenized in 2 M sucrose, 25 mM HEPES (pH 7.9), 25 mM KCl, 1 mM EDTA, 10% glycerol, 0.15 mM spermine, 0.5 mM spermidine, 2 μg/ml aprotinin, and 0.5 mM phenylmethane sulfonyl fluoride, and the nuclei were isolated by sucrose density gradient centrifugation (5, 7). The resulting nuclei were resuspended in five volumes of lysis buffer (10 mM HEPES [pH 7.5], 100 mM KCl, 3 mM MgCl2, 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 2 μg/ml aprotinin, and 0.5 mM phenylmethane sulfonyl fluoride) and lysed by adding 3 M ammonium sulfate (pH 7.9) dropwise to a final concentration of 0.4 M. After gentle shaking for 30 min at 4°C, the mixture was centrifuged at 10,000 × g for 1 h. The supernatant was collected, and solid ammonium sulfate was added to a final concentration of 0.3 g/ml. After sitting on ice for 30 min, the mixture was centrifuged at 10,000 × g for 30 min. The pellet was dissolved in 500 μl of dialysis buffer (25 mM HEPES [pH 7.6], 40 mM KCl, 0.1 mM EDTA, 10% [vol/vol] glycerol, 1 mM dithiothreitol, 2 μg/ml aprotinin, and 0.5 mM phenylmethane sulfonyl fluoride) and dialyzed for 24 h at 4°C against 500 ml of dialysis buffer, with two changes.
For in vitro translation of transcription factors, 1 μg of each plasmid was used in 50 μl of the TnT T7 quick-coupled transcription/translation system (Promega) for 90 min at 30°C, using the manufacturer's recommended protocol. Recombinant NF-Y trimer was reconstituted from bacterially expressed subunits as previously described (60).
EMSA.
Double-stranded synthetic oligonucleotides were used for an electrophoretic mobility shift assay (EMSA). The sequences of the wild-type S promoter fragment and various mutants are shown below in Fig. 5A. The S promoter CCAAT element sequence used is 5′-CCTCCACCAATCGGCAGT. The grp78 promoter ERSE1 sequence containing the composite NF-Y/ATF6 site is 5′-GGAGGGCCTTCACCAATCGGCGGCCTCCACGACGGGGCTGG (59). The human major histocompatibility (MHC) DRA promoter sequence containing the XBP1 site is 5′-CCTAGCAACAGATGCGTCATCTCAAAA (33). The wild-type and mutant 2X-c/EBP-ATF4 sequences are 5′-GATCCGGTTGCCAAACATTGCATCATCCA and 5′-GATCCGGTTGCCAAACGCGTCGGTCACCA, respectively (25). The oligonucleotides were radiolabeled using the Klenow fragment of DNA polymerase I and [α-32P]dCTP (Amersham) or T4 polynucleotide kinase and [γ-32P]ATP (Perkin-Elmer/NEN). Labeled oligonucleotides were purified by centrifugation through G50 Sephadex columns (Amersham) (38).
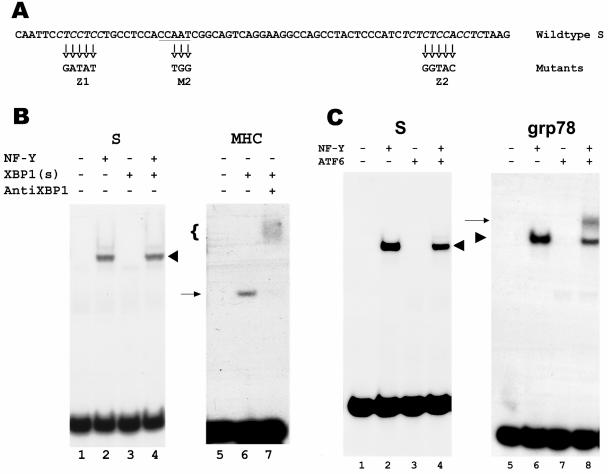
FIG. 5.
Lack of binding of XBP1(s) and ATF6-alpha to the HBV S promoter. A. Sequence of the S promoter, with the CCAAT element (underlined) and the CT-rich elements (italics) (52). The mutated sequences in the M2 and Z1+Z2 mutants are shown below. B. EMSA of the S promoter, using in vitro-synthesized XBP1(s) in the absence or presence of NF-Y (lanes 3 and 4, respectively). The shifted band resulting from binding by NF-Y is marked with a chevron. As a negative control, luciferase synthesized in vitro from the plasmid luciferase T7 control DNA (Promega) was substituted for XBP1(s) (lanes 1 and 2). As a positive control, the MHC DRA promoter X box was used to bind XBP1(s) (lane 6). The shifted band is marked with an arrow, and the presence of XBP1(s) in this band was confirmed by supershifting with antibodies to XBP1 (lane 7, marked with a bracket). C. EMSA of the S promoter, using in vitro-synthesized ATF6-alpha in the absence or presence of NF-Y (lanes 3 and 4, respectively). The shifted band resulting from binding by NF-Y is marked with a chevron. As a negative control, in vitro-synthesized luciferase was substituted for ATF6-alpha (lanes 1 and 2). As a positive control, the grp78 promoter ERSE was used to bind NF-Y alone, ATF6-alpha alone, or both (lanes 6, 7, and 8, respectively). The band resulting from binding by NF-Y alone is marked with a chevron, and the band resulting from binding by both NF-Y and ATF6-alpha is marked with an arrow.
For each EMSA tube, 5 μg of nuclear extract from cultured cells or mouse liver, 10 fmol of recombinant NF-Y, and/or 2 μl of in vitro-translated proteins was mixed with 2 μl 5× binding buffer [50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM dithiothreitol, 250 mM NaCl, 250 μg/ml poly(dI-dC) (Roche), 4% glycerol] in a total volume of 10 μl. The mixture was incubated at room temperature for 10 min, and 1 μl of the radiolabeled oligonucleotide probe (0.1 to 0.3 pmol; ∼10,000 to 30,000 cpm) was added and incubated at room temperature for another 20 min. Samples were loaded onto 4% polyacrylamide gels and separated by electrophoresis at 4°C at 200 V for 2 to ∼3 h in 45 mM Tris-borate and 1 mM EDTA, pH 8.4. In selected experiments, the samples were preincubated with 2 μg of antibodies at 37°C for 20 min prior to incubation with binding buffer. Antibodies against NF-Y (B-subunit specific) and XBP1 were purchased from Biodesign and Santa Cruz Biotechnology, Inc., respectively.
RNA assays.
For detection of xbp1 mRNA with reverse transcription-PCR (RT-PCR), total RNA was extracted from cultured cells with TRIzol reagent (Invitrogen), and cDNA was synthesized from 5 μg of RNA with the SuperScript first-strand synthesis system for RT-PCR (Invitrogen), using random hexamers as primers. An aliquot (1/250) of the reaction product was used for PCR-mediated amplification of the xbp1 mRNA, using cloned Pfu DNA polymerase (Stratagene) and the primer pairs 5′-CCTTGTAGTTGAGAACCAGG and 5′-GGGGCTTGGTATATATGTGG for human RNA (44) or 5′-CCTTGTGGTTGAGAACCAGG and 5′-GAGGCTTGGTGTATACATGG for murine RNA; these primers span the intron spliced out by IRE1-alpha. As internal control, primers specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were used; their sequences are 5′-GGGCTCTCCAGAACATCATC and 5′-CCATGTGGGCCATGAGGTCC. The products were separated by electrophoresis on a 1% agarose gel and visualized with ethidium bromide staining.
For detection of HBV pre-S1 and S mRNA, total RNA was extracted from transfected cells as above and 5 μg of each RNA sample was used in primer extension assays, using primers specific for the HBV pre-S1 and S mRNAs, as described previously (14).
RESULTS
Activation of HBV S promoter by IRE1-alpha and XBP1.
We have previously shown that ER stress activates the HBV S promoter, either situated endogenously in the viral genome or when placed upstream of a reporter gene (52). Figure 2A demonstrates a typical result with a CAT reporter plasmid in HuH7 hepatoma cells. To determine if ATF4, the b-ZIP factor which is activated by the ER stress pathway downstream of PERK (Fig. 1), may be involved in transducing the stress signal to the S promoter, we cotransfected into HuH7 hepatoma cells a plasmid expressing ATF4 with a plasmid containing the CAT reporter gene driven by the S promoter. As seen in Fig. 2B, ATF4 failed to activate the S promoter or the simian virus 40 (SV40) promoter (a negative control), while it was capable of activating the CHOP promoter, as expected. Therefore, the PERK-ATF4 pathway does not appear to be involved.
FIG. 2.
Effect of tunicamycin, ATF4, ATF6-alpha, IRE1-alpha, and XBP1(s) on the S promoter. HuH7 cells were cotransfected with a reporter plasmid containing the CAT gene under the control of the S promoter and plasmids expressing the indicated protein (B, C, D, and E), and the CAT activity was normalized to that in cells cotransfected with the empty vector instead of the expression plasmid. A. Cells were transfected with the reporter plasmid only, and one set of cells was treated with tunicamycin for 17 h before harvesting. The CAT activity in these cells was normalized to that in cells treated with dimethyl sulfoxide. As a positive control, the CAT gene under the control of the grp78 promoter (A, C, D, and E) or of the CHOP promoter (B) was used, while as a negative control the CAT gene under the control of the SV40 enhancer/early promoter was used. The results shown in this and subsequent figures represent the mean + standard error of the mean of three experiments.
Cotransfection of ATF6-alpha, another transcription factor induced by ER stress (Fig. 1), activated both the S and grp78 promoters but not the SV40 promoter (Fig. 2C). However, because ATF6-alpha activates the xbp1 promoter (19) and, because overexpressed ATF6-alpha can bind to and activate XBP1-responsive DNA elements (UPRE) in a nonphysiological manner (19), this result does not necessarily indicate a role for endogenous ATF6-alpha in the activation of the S promoter but may merely reflect a role for XBP1 (see Discussion). Thus, to determine if the IRE1-XBP1 pathway may be involved in transducing the stress signal to the S promoter, we coexpressed IRE1-alpha with the S promoter reporter plasmid. As seen in Fig. 2D, cotransfection of IRE1-alpha resulted in activation of the S promoter. As expected, IRE1-alpha also activated the grp78 promoter but not the SV40 early promoter (Fig. 2D). The b-ZIP transcription factor XBP1(s) is downstream of IRE1-alpha (Fig. 1). Therefore, if this pathway can activate the S promoter, forced expression of XBP1(s) from a cotransfected plasmid should also activate the S promoter. This indeed turned out to be the case (Fig. 2E), providing further support for involvement of the IRE1-XBP1 pathway in the activation of the S promoter by ER stress.
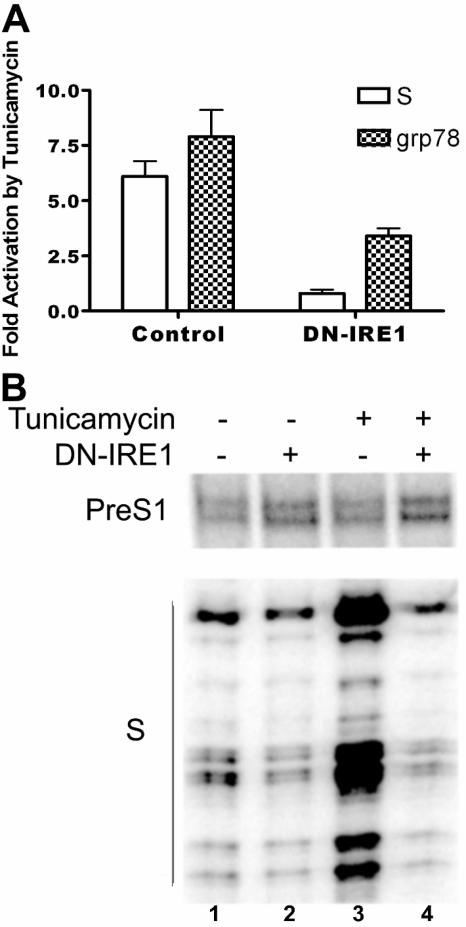
To further confirm the role of IRE1-alpha in activating the S promoter, we used a dominant-negative form of IRE1-alpha (59). Cotransfection of a plasmid expressing this protein resulted in a partial block of the activation of the grp78 promoter by tunicamycin (Fig. 3A). This result is in concert with published data and possibly reflects the fact that the grp78 promoter can be activated by both the IRE1-alpha and the ATF6 pathways (59). Cotransfected dominant-negative IRE1-alpha also decreased the activation of the S promoter by tunicamycin (Fig. 3A) and even to a greater extent than for the grp78 promoter, supporting the inference that the IRE1-alpha pathway is important for activation of the S promoter by ER stress. Dominant-negative IRE1-alpha similarly blocked the activation of the endogenous S promoter in the entire HBV genome by tunicamycin (Fig. 3B, lane 4; compare with lane 2), while the non-stress-inducible pre-S1 promoter was neither activated by tunicamycin nor repressed by the dominant-negative IRE1-alpha (Fig. 3B, lanes 2 to 4). Taken together, these results confirm an important role for IRE1-alpha in the activation of the S promoter by ER stress.
FIG. 3.
Effects of dominant-negative IRE1-alpha on activation of the S and grp78 promoters by tunicamycin. A. Effects on reporter plasmids. HuH7 cells were cotransfected with a CAT reporter plasmid and either an empty vector (left side, control) or a plasmid expressing dominant-negative IRE1-alpha (right side, DN-IRE1). One set of cells was treated with tunicamycin, and another was treated with dimethyl sulfoxide. The CAT activity in cells treated with tunicamycin was normalized to that in cells treated with dimethyl sulfoxide. The white bars represent cells transfected with the CAT reporter plasmid driven by the S promoter, while the hatched bars represent cells transfected with the CAT reporter plasmid driven by the grp78 promoter. B. Effects on endogenous HBV promoters in the context of the entire viral genome. HuH7 cells were cotransfected with pHBV1.3, which contains a longer-than-full-length clone of HBV genomic DNA that gives rise to all viral mRNAs (53), and either an empty vector or a plasmid expressing dominant-negative IRE1-alpha (DN-IRE1). After treatment with tunicamycin or dimethyl sulfoxide for 16 h, total RNA was harvested and the amount of HBV pre-S1 or S mRNA was measured with primer extension. Note that the dominant-negative IRE1-alpha has a small negative effect on the S promoter even without tunicamycin treatment, since the small amount of large surface protein synthesized during transient transfection appears to activate a low degree of ER stress.
To confirm the role of XBP1 in activating the S promoter, we used two different siRNAs directed against XBP1 (arbitrarily named Xa and Xb; sequences are shown in Fig. 4A). We first used Western blotting against XBP1(s) to assess the efficacy of these siRNAs. As seen in Fig. 4B, the induction of XBP1(s) by tunicamycin appeared to be significantly decreased by either siRNA compared to a control siRNA that has no known cellular targets (48) (compare the band flanked by asterisks in lane 2 with the corresponding locations in lanes 3 and 4). However, this antibody recognized a cross-reacting band (Fig. 4B) that migrated slightly above the XBP1(s) band, rendering it difficult to see the latter clearly. Thus, we also used RT-PCR to examine the amount of xbp1 mRNA in these cells. This assay showed a significant decrease in xbp1 mRNA, especially of the spliced variant, caused by the Xa and Xb siRNAs in cells treated with tunicamycin (Fig. 4C, compare lanes 3 and 4 with lane 2). Taken together, these data show that Xa and Xb have the ability to knock down XBP1(s) expression in transfected cells. As seen in Fig. 4D, these siRNAs also significantly decreased activation of the HBV S promoter by tunicamycin, compared with the control siRNA. In contrast, the XBP1 siRNAs had no significant effect on either the grp78 promoter or SV40 promoter (Fig. 4D). It is not clear why the effect of the dominant-negative IRE1-alpha appears to be stronger than that of the siRNAs against XBP1 in blocking the activation by tunicamycin of either the grp78 or S promoter (compare Fig. 3A with 4D), but this difference may reflect incomplete silencing of XBP1(s) (note a residual amount of spliced xbp1 mRNA in Fig. 4C, lanes 3 and 4) and/or side effects of the dominant-negative IRE1-alpha protein (see Discussion).
Lack of XBP1 binding site on S promoter.
One possible mechanism for the IRE1/XBP1 pathway to activate the S promoter would be for XBP1(s) to bind directly to this promoter. However, inspection of the minimal S promoter revealed no UPRE or ERSE (compare Fig. 1 and 5A), although there is a CCAAT element that we have previously shown to bind NF-Y (23, 24) (Fig. 5A). To test directly whether XBP1(s) can bind to the S promoter, EMSA was performed with radiolabeled S promoter and XBP1(s) synthesized in vitro. As seen in Fig. 5B, lane 3, no shifted band was detected. When NF-Y was added to the mixture, a single shifted band was present (Fig. 5B, lane 4); this band reflected the binding of NF-Y alone, since NF-Y without XBP1(s) gave rise to a band of identical mobility (Fig. 5B, lane 2). The XBP1(s) used in this assay was active, as it was able to bind to the X box of the MHC DRA promoter (Fig. 5B, lane 6), and the resulting band was supershifted by an antibody to XBP1(s) (Fig. 5B, lane 7).
Similarly, we were not able to demonstrate binding of ATF6-alpha to the S promoter, either in the absence or presence of NF-Y (Fig. 5C, lanes 3 and 4, respectively), while ATF6-alpha was able to bind the grp78 ERSE, albeit only in cooperation with NF-Y (Fig. 5C, lane 8), consistent with previously published data (60). As expected from the inability of cotransfected ATF4 to activate the S promoter, ATF4 also did not bind this promoter (data not shown).
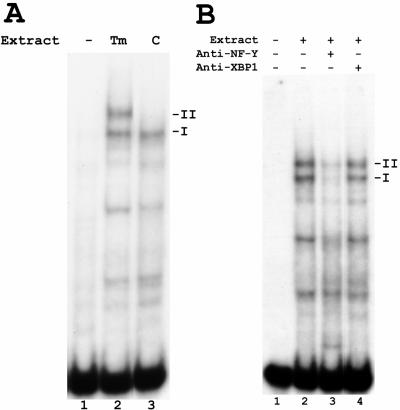
Induction of novel factor Z by ER stress.
Previous work demonstrated that the CCAAT element in the S promoter is important not only for its basal activity but also for activation by ER stress (23, 24, 52). The trimeric transcription factor NF-Y binds to the CCAAT element in the S promoter, and this binding can be readily demonstrated using EMSA with nuclear extracts from cultured cells, such as the human hepatoma cell line HuH7 (24) (Fig. 6A, lane 3, complex I). Interestingly, when nuclear extracts from tunicamycin-treated HuH7 cells were used, an additional band of slower mobility was observed (Fig. 6A, lane 2, complex II). Treatment with thapsigargin, a different inducer of ER stress, also resulted in the appearance of this band (data not shown). This ER stress-induced complex II contained NF-Y, as its intensity was markedly diminished by a blocking antibody against NF-Y, similar to the constitutive complex I (Fig. 6B, lane 3). However, it did not contain XBP1(s), as the supershifting antibody against XBP1 had no effect on its mobility (Fig. 6B, lane 4), further confirming that XBP1(s) does not bind to the S promoter.
FIG. 6.
Induction of new protein-S promoter complex by tunicamycin. A. EMSA of the S promoter, using nuclear extracts from tunicamycin-treated (Tm) or dimethyl sulfoxide-treated control (C) HuH7 cells. Control extracts gave rise to only one shifted band (I), corresponding to binding by NF-Y, while tunicamycin-treated extracts gave rise to an additional, slower-mobility band (II). B. EMSA of the S promoter using nuclear extracts from tunicamycin-treated (Tm) HuH7 cells. A blocking antibody against NF-Y reduced the intensity of both complexes I and II (lane 3, compare with lane 2), while a supershifting antibody against XBP1 had no effect (lane 4).
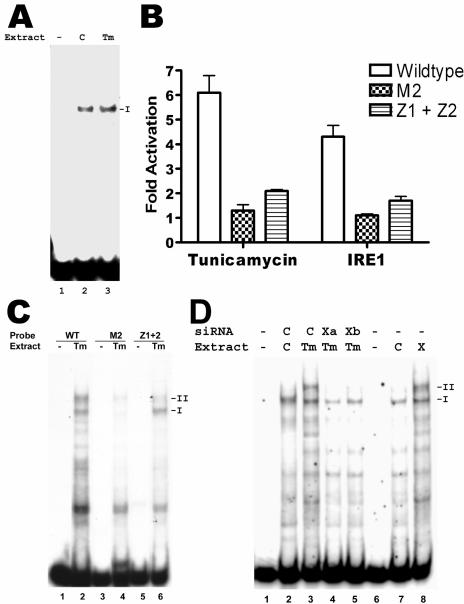
Complex II may have resulted from the binding of the CCAAT element by a modified form of NF-Y found only in ER-stressed cells. However, EMSA using the CCAAT element alone resulted only in complex I formation, using extracts from either untreated or tunicamycin-treated cells (Fig. 7A, lanes 2 and 3, respectively). This result suggested that complex II comprised NF-Y bound to the CCAAT element and another factor bound to a different sequence in the S promoter (similar to the formation of the upper band by NF-Y and ATF6-alpha binding to adjacent sequences of ERSE in Fig. 5C, lane 8). Indeed, the CCAAT element of the S promoter is necessary but not sufficient for activation by ER stress. Two CT-rich elements flanking the CCAAT element of the S promoter (Fig. 5A, Z1 and Z2 sites) are also important (52), as confirmed by the inability of tunicamycin and IRE1-alpha to activate the S promoter with these elements mutated (Fig. 7B, Z1+Z2), just as the S promoter with the CCAAT element mutated cannot be activated (Fig. 7B, M2). Thus, the CT-rich elements may bind the tunicamycin-induced factor. To test this hypothesis, we performed EMSA with mutated S promoter sequences. As shown in Fig. 7C, lane 6, the S promoter with mutated CT-rich elements formed almost no complex II when incubated with nuclear extracts from tunicamycin-treated cells, although complex I, corresponding to binding to NF-Y alone, was still formed by this mutant. This result suggested that the CT-rich elements are involved in the formation of complex II. In contrast, the S promoter mutant with a mutated CCAAT element showed defects in the formation of both complexes I and II (Fig. 7C, lane 4), in agreement with our results that both complexes depend on the CCAAT element binding to NF-Y. This result is also in agreement with the essential role for this element in both basal and stress-activated transcription.
FIG. 7.
Dependence of complex II formation on both CCAAT element and CT-rich elements of the S promoter, as well as XBP1(s). A. EMSA of CCAAT element of the S promoter, using nuclear extracts from tunicamycin-treated (Tm) or dimethyl sulfoxide-treated control (C) HuH7 cells. Both extracts gave rise to only one shifted band (I), corresponding to binding by NF-Y. B. Activation of wild-type or mutant S promoters by tunicamycin and IRE1-alpha. On the left side, CAT reporter plasmids driven by the wild-type S promoter, or mutated S promoters with lesions in the CCAAT element (M2) or the CT-rich elements (Z1+Z2), were transfected into HuH7 cells, and one set of cells was treated with tunicamycin for 17 h before harvesting. The CAT activity in these cells was normalized to that in cells treated with dimethyl sulfoxide. On the right side, HuH7 cells were cotransfected with the indicated S promoter reporter plasmid and the plasmid expressing IRE1-alpha, and the CAT activity was normalized to that in cells cotransfected with the empty vector. C. EMSA of wild-type or mutated S promoters, using extracts from tunicamycin-treated HuH7 cells. Mutation of the CCAAT element (M2) prevented formation of both complexes I and II, while mutation of the CT-rich elements (Z1+Z2) prevented formation of only complex II. D. Lanes 1 to 5: EMSA of wild-type S promoter, using extracts from HuH7 cells treated with tunicamycin (Tm; lanes 3, 4, and 5) or dimethyl sulfoxide (C; lane 2) and transfected with two different siRNAs against XBP1 (Xa and Xb; lanes 4 and 5, respectively) or a control siRNA (C; lanes 2 and 3). Lanes 6 to 8: EMSA of wild-type S promoter, using extracts from HuH7 cells transfected with a plasmid expressing XBP1(s) (X; lane 8) or the empty vector (C; lane 7). No extracts were used in lanes 1 and 6.
Thus, ER stress appears to induce a new, unidentified factor which binds in conjunction with NF-Y to the S promoter to form complex II. For convenience, we shall refer to this factor as factor Z. Our data showing that the activation of the S promoter is downstream of XBP1(s) (Fig. 4) predict that the induction of factor Z should be also dependent on XBP1(s). Indeed, transfection of HuH7 cells with a plasmid expressing XBP1(s) induced the formation of complex II without treatment with tunicamycin (Fig. 7D, lane 8). Conversely, treatment with either siRNA against XBP1 blocked the formation of complex II, even in the presence of tunicamycin (Fig. 7D, lanes 4 and 5). In contrast, a control siRNA did not prevent the formation of complex II (Fig. 7D, lane 3). Taken together, these data strongly support the inference that activation of factor Z by XBP1(s) is required for activation of the S promoter by ER stress.
Cell type restriction of S promoter activation by ER stress.
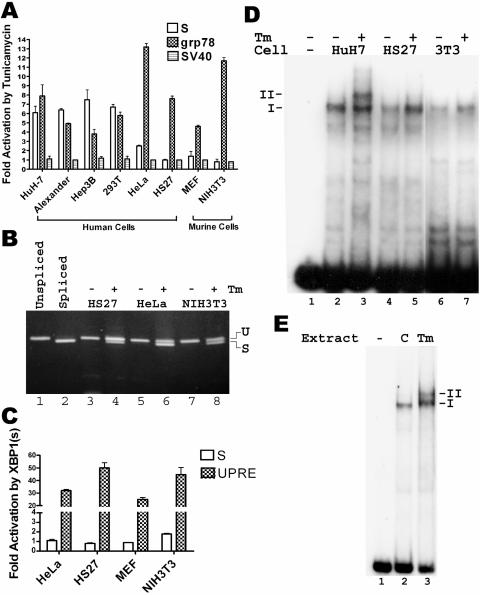
The most definitive test to confirm the importance of the IRE1/XBP1 pathway in activating the S promoter would be to use cells deficient in either IRE1-alpha or XBP1. Both ire1α−/− and xbp1−/− mice die in utero, but MEFs from these mice have been described in the literature (35, 46). However, preliminary experiments showed that the S promoter was not activated even in wild-type MEFs (Fig. 8A), even though the grp78 promoter showed the expected activation (Fig. 8A). Further analysis of different cell lines revealed marked variation in their ability to activate the S promoter in response to tunicamycin, regardless of whether they were of human or murine origin, and the variation did not correlate with the activation of the grp78 promoter (Fig. 8A), which was readily detected in all cells. Specifically, fibroblasts of either human or murine origin (HS27, MEF, and NIH 3T3) failed to reveal activation of the S promoter. The method of ER stress induction did not make a difference, as the HBV large surface protein also failed to induce the S promoter in HS27 cells (data not shown), unlike the robust induction in HuH7 cells (52). Similarly, HeLa human cervical carcinoma cells showed at best a weak activation of the S promoter, but two other human hepatoma cell lines and 293T human embryonic kidney cells showed similar activation as HuH7 cells (Fig. 8A). The difference could not be explained by the lack of xbp1 mRNA splicing in fibroblasts and HeLa cells, as the spliced mRNA could be easily detected after tunicamycin treatment of these cells (Fig. 8B, lanes 4, 6, and 8), similar to the situation in Huh7 cells (data not shown).
FIG. 8.
Cell type-restricted activation of S promoter and induction of complex II by tunicamycin. A. Effect of tunicamycin on the S, grp78, and SV40 promoters in various cell lines. The experiment was performed as described for Fig. 2A. HuH7, Alexander, and Hep3B are human hepatoma cell lines, 293T is a human embryonic kidney cell line, HeLa is a human cervical carcinoma cell line, HS27 is a human fibroblast cell line, MEFs are primary murine embryo fibroblasts, and NIH 3T3 is a murine fibroblast cell line. B. Splicing of xbp1 mRNA in response to tunicamycin in various cell lines. The indicated cell lines were treated with dimethyl sulfoxide or tunicamycin (Tm) for 8 h (- and +, respectively), and RT-PCR was performed on total RNA. The upper band corresponds to the unspliced mRNA, while the lower band corresponds to the spliced mRNA, as demonstrated by their comigration with PCR products from the respective cDNAs (lanes 1 and 2, respectively). C. Effect of XBP1(s) on the S and UPRE-containing promoters in various cell lines. The experiment was performed as for Fig. 2E, except that firefly luciferase was used as the reporter, and the firefly luciferase activity in each cell extract was normalized to the Renilla luciferase activity expressed from a cotransfected plasmid (pRL-CMV) under the control of the cytomegalovirus immediate-early promoter. The UPRE reporter plasmid (p5XATF6GL3) contains five copies of the UPRE upstream of a minimal SV40 early promoter (50). D. EMSA of the S promoter, using nuclear extracts from various cells treated with dimethyl sulfoxide or tunicamycin (Tm) (- and +, respectively). 3T3, NIH 3T3 cells. E. EMSA of the S promoter, using nuclear extracts from livers of mice treated with dimethyl sulfoxide (C) or tunicamycin (Tm).
The above result suggested that factor Z could not be induced by XBP1(s) in fibroblasts. This inference is supported by two lines of evidence. First, XBP1(s) expressed from a transfected plasmid failed to activate the S promoter in fibroblasts and HeLa cells, even though it was able to activate a UPRE-driven reporter plasmid in these cells (Fig. 8C). Second, an EMSA using nuclear extracts from tunicamycin-treated fibroblasts failed to show induction of complex II, despite the presence of complex I (Fig. 8D, lanes 5 and 7). Complex I in HS27 cells nevertheless contained NF-Y, as revealed by a supershift induced by specific antibodies (data not shown). Finally, complex II was induced by tunicamycin in the mouse liver (Fig. 8E), ruling out any species differences. Thus, the pathway from XBP1(s) to factor Z operates only in certain cell types in both human beings and mice.
DISCUSSION
It is now well established that ER stress in metazoans activates gene transcription via three different, albeit interconnected, pathways: the IRE1/XBP1 pathway, the PERK/ATF4 pathway, and the ATF6 pathway (reviewed in references 9, 27, and 61). Here, we have presented data showing that either cotransfected IRE1-alpha or XBP1(s) activated the S promoter and that dominant-negative IRE1-alpha and siRNAs against XBP1 significantly decreased this activation. These data suggest that the IRE1/XBP1 pathway is important for ER stress activation of the S promoter. Yet, XBP1(s) does not bind directly to the S promoter. Rather, XBP1(s) appears to activate the expression of a factor, here called factor Z, distinct from XBP1(s), ATF4, and ATF6-alpha. Factor Z binds to CT-rich elements in the S promoter in cooperation with the ubiquitous factor NF-Y, which binds to the CCAAT element. The induction of factor Z, similarly to the activation of the S promoter by ER stress, is dependent on XBP1(s). Thus, the activation of the S promoter depends on a previously unknown pathway downstream of IRE1/XBP1. Indeed, XBP1(s) has recently been found not only to increase expression of genes coding for proteins involved in the secretory pathway but also to cause increases in organelle biogenesis, protein synthesis, and cell size via unknown mechanisms (39). It is possible that HBV may have parasitized one of these pathways for regulating its gene expression.
Our results do not point to any role for the PERK/ATF4 pathway in activation of the S promoter by ER stress, but they cannot rigorously rule out the possibility that ATF6-alpha may be involved. This uncertainty results from the facts that ATF6-alpha and XBP1(s) bind to similar DNA sequences, particularly when overexpressed, and that these proteins can form heterodimers (19, 30). Nevertheless, the ability of dominant-negative IRE1-alpha to block completely activation of the S promoter by tunicamycin is an indication that ATF6-alpha alone cannot activate the S promoter. This result is in contrast to that for the grp78 promoter, whose activation was only partially blocked by dominant-negative IRE1-alpha, since this promoter is activated directly by ATF6-alpha.
The siRNAs against XBP1 showed a weaker blocking effect against either the grp78 or S promoter than dominant-negative IRE1-alpha (compare Fig. 3A and 4D). One possible explanation for this difference is that the siRNAs are not completely efficient in shutting down XBP1(s) expression (Fig. 4C). Another, non-mutually exclusive possibility is that dominant-negative IRE1-alpha may have effects other than shutting down the IRE1/XBP1 pathway. For example, there may be an unidentified homolog of IRE1-alpha that can heterodimerize with IRE1-alpha and splice another transcription factor that functions in a parallel pathway as XBP1(s) to activate both the grp78 and S promoters. In possible support of this scenario, previous publications have shown that dominant-negative IRE1-alpha partially blocks activation of the grp78 promoter by tunicamycin (59), yet activation of this promoter by tunicamycin in ire1α−/− MEFs is entirely unimpaired (54).
Although it is likely that XBP1(s) activates the synthesis of factor Z at the transcriptional level, we cannot exclude the possibilities that factor Z activity is induced at the translational or even posttranslational level via an indirect mechanism downstream of XBP1(s). Inhibitors such as actinomycin D (to stop transcription) or cycloheximide (to stop translation) cannot be used to distinguish among these possibilities, as the tunicamycin-induced transcription and translation of XBP1(s) itself would be blocked by these agents. Furthermore, it is even formally possible that NF-Y rather than factor Z is the target of the IRE1/XBP1 pathway, for example, by modifying NF-Y to allow it to stabilize the binding of factor Z to the S promoter.
Perhaps the most unexpected and interesting feature of the induction of factor Z is that it appears to be cell type restricted. Factor Z is induced by ER stress in human embryonic kidney cells and both human and murine liver-derived cells, but its binding activity cannot be induced in HeLa cells and human and murine fibroblasts. This feature is unlike the previously characterized ER stress pathways, which all appear to be ubiquitous. However, it is notable that expression of IRE1-beta, a homolog of IRE1-alpha of uncertain function, is largely restricted to intestinal cells (49), and Kondo et al. (17) recently showed that the b-ZIP transcription factor OASIS appears to be specifically activated by ER stress in astrocytes to increase grp78 expression. Thus, it is likely that certain cell types have developed specialized ER stress pathways to deal with specific stresses unique to those cells. It should be noted that XBP1(s) appears to be essential for liver development, as xbp1 knockout mice die in utero because of liver failure (35). It is possible that the presence of factor Z in liver cells explains at least in part the specific need for XBP1(s) in the developing liver.
The significance of factor Z for the HBV life cycle is currently unknown. However, we have previously demonstrated that the HBV large surface protein activates ER stress when markedly overexpressed (52). More recent results indicate that even lower amounts of large surface protein, expressed from the viral genome, can activate ER stress under certain circumstances (Z. M. Huang and T. S. B. Yen, unpublished observations). Thus, it is possible that transcription of the HBV S promoter in the infected hepatocyte partially depends on factor Z, secondary to ER stress induced by the large surface protein and/or other factors. The presence of factor Z in hepatocytes and not in certain other cell types may thereby partially explain the preferential replication of HBV in hepatocytes. In this regard, it is worth noting that the kidney cell line 293T supports the activation of the S promoter by ER stress (Fig. 8A), and HBV S gene expression is as high in the kidney of transgenic mice as in the liver (8). Thus, our results not only provide new information on the ER stress response in mammalian cells but also may point to new targets for small molecules that block HBV transcription and hence replication. The future isolation and characterization of factor Z will allow the detailed analysis of this apparently novel stress-induced factor in terms of its regulation, role in HBV gene expression, and importance for hepatocyte function and development in normal and diseased conditions.
Acknowledgments
We thank K. C. Lim for assistance with animal care and handling, Kazunori Imaizumi, Amy Lee, and Ron Prywes for plasmids, Eric Huang and Guangwei Wei for MEFs, and Jim Ou for critical reading of the manuscript.
This work was supported by NIH grant R01CA55578 and a VA Merit Review Award (to T.S.B.Y.).
REFERENCES
- 1.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 2.Cox, J. S., C. E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197-1206. [DOI] [PubMed] [Google Scholar]
- 3.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 4.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 5.Ghoshal, K., S. Majumder, Q. Zhu, J. Hunzeker, J. Datta, M. Shah, J. F. Sheridan, and S. T. Jacob. 2001. Influenza virus infection induces metallothionein gene expression in the mouse liver and lung by overlapping but distinct molecular mechanisms. Mol. Cell. Biol. 21:8301-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorman, C. 1985. High efficiency gene transfer into mammalian cells. In D. M. Glover (ed.), DNA cloning, vol. 2. IRL Press, Oxford, England.
- 7.Gorski, K., M. Carneiro, and U. Schibler. 1986. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell 47:767-776. [DOI] [PubMed] [Google Scholar]
- 8.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 10.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 11.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 12.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 13.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Z. M., and T. S. Yen. 1993. Dysregulated surface gene expression from disrupted hepatitis B virus genomes. J. Virol. 67:7032-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawahara, T., H. Yanagi, T. Yura, and K. Mori. 1997. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8:1845-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, E. T., C. Chang, and W. B. Ershler. 1996. Inhibition of NFκB activity through maintenance of IκBα levels contributes to dihydrotestosterone-mediated repression of the interleukin-6 promoter. J. Biol. Chem. 271:26267-26275. [DOI] [PubMed] [Google Scholar]
- 17.Kondo, S., T. Murakami, K. Tatsumi, M. Ogata, S. Kanemoto, K. Otori, K. Iseki, A. Wanaka, and K. Imaizumi. 2005. OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat. Cell Biol. 7:186-194. [DOI] [PubMed] [Google Scholar]
- 18.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, M., P. Baumeister, B. Roy, T. Phan, D. Foti, S. Luo, and A. S. Lee. 2000. ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 20:5096-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, W. W., S. Alexandre, X. Cao, and A. S. Lee. 1993. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin. J. Biol. Chem. 268:12003-12009. [PubMed] [Google Scholar]
- 22.Liu, C. Y., M. Schroder, and R. J. Kaufman. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275:24881-24885. [DOI] [PubMed] [Google Scholar]
- 23.Lu, C. C., M. Chen, J. H. Ou, and T. S. Yen. 1995. Key role of a CCAAT element in regulating hepatitis B virus surface protein expression. Virology 206:1155-1158. [DOI] [PubMed] [Google Scholar]
- 24.Lu, C. C., and T. S. Yen. 1996. Activation of the hepatitis B virus S promoter by transcription factor NF-Y via a CCAAT element. Virology 225:387-394. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Y., J. W. Brewer, J. A. Diehl, and L. M. Hendershot. 2002. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318:1351-1365. [DOI] [PubMed] [Google Scholar]
- 26.Ma, Y., and L. M. Hendershot. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278:34864-34873. [DOI] [PubMed] [Google Scholar]
- 27.Ma, Y., and L. M. Hendershot. 2001. The unfolding tale of the unfolded protein response. Cell 107:827-830. [DOI] [PubMed] [Google Scholar]
- 28.Mori, K., W. Ma, M. J. Gething, and J. Sambrook. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:743-756. [DOI] [PubMed] [Google Scholar]
- 29.Mori, K., N. Ogawa, T. Kawahara, H. Yanagi, and T. Yura. 2000. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 97:4660-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman, J. R., and A. E. Keating. 2003. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097-2101. [DOI] [PubMed] [Google Scholar]
- 31.Ng, D. T., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada, T., H. Yoshida, R. Akazawa, M. Negishi, and K. Mori. 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366:585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono, S. J., H. C. Liou, R. Davidon, J. L. Strominger, and L. H. Glimcher. 1991. Human X-box-binding protein 1 is required for the transcription of a subset of human class II major histocompatibility genes and forms a heterodimer with c-fos. Proc. Natl. Acad. Sci. USA 88:4309-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-355. [DOI] [PubMed] [Google Scholar]
- 35.Reimold, A. M., A. Etkin, I. Clauss, A. Perkins, D. S. Friend, J. Zhang, H. F. Horton, A. Scott, S. H. Orkin, M. C. Byrne, M. J. Grusby, and L. H. Glimcher. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152-157. [PMC free article] [PubMed] [Google Scholar]
- 36.Roy, B., and A. S. Lee. 1999. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 27:1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruegsegger, U., J. H. Leber, and P. Walter. 2001. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107:103-114. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Shaffer, A. L., M. Shapiro-Shelef, N. N. Iwakoshi, A. H. Lee, S. B. Qian, H. Zhao, X. Yu, L. Yang, B. K. Tan, A. Rosenwald, E. M. Hurt, E. Petroulakis, N. Sonenberg, J. W. Yewdell, K. Calame, L. H. Glimcher, and L. M. Staudt. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81-93. [DOI] [PubMed] [Google Scholar]
- 40.Shen, J., X. Chen, L. Hendershot, and R. Prywes. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3:99-111. [DOI] [PubMed] [Google Scholar]
- 41.Shen, X., R. E. Ellis, K. Lee, C. Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D. M. Kurnit, K. Mori, and R. J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107:893-903. [DOI] [PubMed] [Google Scholar]
- 42.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidrauski, C., and P. Walter. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031-1039. [DOI] [PubMed] [Google Scholar]
- 44.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279:17158-17164. [DOI] [PubMed] [Google Scholar]
- 45.Tirasophon, W., A. A. Welihinda, and R. J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urano, F., X. Wang, A. Bertolotti, Y. Zhang, P. Chung, H. P. Harding, and D. Ron. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664-666. [DOI] [PubMed] [Google Scholar]
- 47.Vattem, K. M., and R. C. Wek. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA 101:11269-11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner, E. J., and M. A. Garcia-Blanco. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10:943-949. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X. Z., H. P. Harding, Y. Zhang, E. M. Jolicoeur, M. Kuroda, and D. Ron. 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17:5708-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y., J. Shen, N. Arenzana, W. Tirasophon, R. J. Kaufman, and R. Prywes. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275:27013-27020. [DOI] [PubMed] [Google Scholar]
- 51.Xu, Z., V. Bruss, and T. S. Yen. 1997. Formation of intracellular particles by hepatitis B virus large surface protein. J. Virol. 71:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, Z., G. Jensen, and T. S. Yen. 1997. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J. Virol. 71:7387-7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu, Z., T. S. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto, K., H. Yoshida, K. Kokame, R. J. Kaufman, and K. Mori. 2004. Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. (Tokyo) 136:343-350. [DOI] [PubMed] [Google Scholar]
- 55.Ye, J., R. B. Rawson, R. Komuro, X. Chen, U. P. Dave, R. Prywes, M. S. Brown, and J. L. Goldstein. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355-1364. [DOI] [PubMed] [Google Scholar]
- 56.Yen, T. S. 1993. Regulation of hepatitis B virus gene expression. Semin. Virol. 4:33-42. [Google Scholar]
- 57.Yoshida, H., K. Haze, H. Yanagi, T. Yura, and K. Mori. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273:33741-33749. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida, H., T. Matsui, N. Hosokawa, R. J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4:265-271. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, K., and R. J. Kaufman. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279:25935-25938. [DOI] [PubMed] [Google Scholar]
- 62.Zinszner, H., M. Kuroda, X. Wang, N. Batchvarova, R. T. Lightfoot, H. Remotti, J. L. Stevens, and D. Ron. 1998. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12:982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]