Abstract
In a recent study, we provided evidence that strong promoter-bound transcriptional activators result in higher levels of splicing and 3′-end cleavage of nascent pre-mRNA than do weak promoter-bound activators and that this effect of strong activators requires the carboxyl-terminal domain (CTD) of RNA polymerase II (pol II). In the present study, we have investigated the mechanism of activator- and CTD-mediated stimulation of pre-mRNA processing. Affinity chromatography experiments reveal that two factors previously implicated in the coupling of transcription and pre-mRNA processing, PSF and p54nrb/NonO, preferentially bind a strong rather than a weak activation domain. Elevated expression in human 293 cells of PSF bypasses the requirement for a strong activator to promote efficient splicing and 3′-end cleavage. Truncation of the pol II CTD, which consists of 52 repeats of the consensus heptapeptide sequence YSPTSPS, to 15 heptapeptide repeats prevents PSF-dependent stimulation of splicing and 3′-end cleavage. Moreover, PSF and p54nrb/NonO bind in vitro to the wild-type CTD but not to the truncated 15-repeat CTD, and domains in PSF that are required for binding to activators and to the CTD are also important for the stimulation of pre-mRNA processing. Interestingly, activator- and CTD-dependent stimulation of splicing mediated by PSF appears to primarily affect the removal of first introns. Collectively, these results suggest that the recruitment of PSF to activated promoters and the pol II CTD provides a mechanism by which transcription and pre-mRNA processing are coordinated within the cell.
The processing of precursor mRNA (pre-mRNA) to mRNA in the nucleus of eukaryotic cells occurs by a series of coordinated steps that include the addition of a 5′-end m7G cap, splicing, and 3′-end formation. In most cases, 3′-end formation involves an endonucleolytic cleavage reaction downstream of an AAUAAA signal sequence, followed by the addition of a poly(A) tail (11, 68). Each of these processing steps can occur independently of one another in vitro, although in vivo, increasing evidence indicates that there is extensive communication between factors that participate in each of the processing reactions (28, 35, 41, 52). In some cases, one step can modulate the efficiency of another. Currently, little is known about the specific mechanisms by which different steps in the maturation of mRNA regulate each other.
Transcription is initiated within the context of a multisubunit holoenzyme complex that contains general transcription factors and RNA polymerase II (pol II) (26, 37). Sequence-specific DNA binding transcriptional activators play an important role in the recruitment of the pol II holoenzyme complex to promoter regions (53). RNA pol II contains a unique C-terminal domain (CTD) that consists in mammalian cells of 52 repeats of the heptapeptide consensus sequence YSPTSPS (2, 12). The CTD is important for the response of certain genes to promoter-bound activators (1, 25, 40, 60), although activators can also stimulate transcription via interactions involving other transcription factors, which may be direct or indirect, i.e., mediated by coactivators (6, 39). During transcription elongation, the CTD is important for efficient pre-mRNA processing, as transcription of pre-mRNAs with pol II containing a truncated CTD has been shown to result in reduced levels of 5′-end cap formation, splicing, and 3′-end cleavage (10, 43, 44). The different roles of the pol II CTD in transcription and pre-mRNA processing are in part controlled by the action of different kinases that phosphorylate the CTD on serines located at positions 2 and 5 in the heptapeptide consensus sequence (4, 49). The initiating form of pol II contains a hypophosphorylated CTD (pol IIa), whereas the elongating form of pol II contains a CTD that is hyperphosphorylated (pol IIo). This transition in phosphorylation status between pol IIa and pol IIo is thought to result in the recruitment of some processing factors to the CTD. For example, some splicing factors have been detected in association with pol IIo but not pol IIa (34, 47, 65), and pol IIo can specifically stimulate splicing in vitro (29). In other studies, however, certain pre-mRNA processing factors have been detected in association with pol IIa-containing holoenzyme complexes (14, 21, 54). These findings are of particular interest given the increasing number of observations indicating that the type of promoter or promoter-bound activator used to drive transcription can influence the efficiency of specific pre-mRNA processing steps in the corresponding transcribed RNAs.
Previous studies have shown that tethering different activators or swapping promoters upstream of a reporter gene can influence the level of inclusion of an alternatively spliced exon located in the middle of the transcribed reporter pre-mRNA (3, 13, 30, 48). It is evident that naturally occurring alternative promoters may also affect alternative splicing levels, since large-scale cDNA and expressed sequence tag sequencing efforts have revealed numerous examples of transcripts apparently initiated from different transcription start sites that have different spliced exon configurations (73). Recently, it was shown that the strength of a promoter-bound activator can also influence the efficiency of constitutive splicing and 3′-end cleavage of different reporter pre-mRNAs (55). This activator-dependent increase in pre-mRNA processing efficiency was found to be independent of the overall levels of the reporter transcript generated and to require the pol II CTD. These studies, together with numerous observations of interactions between transcription initiation and pre-mRNA processing components (18, 24, 36, 46), suggest that steps occurring at an early stage in the transcription cycle can be important for determining the efficiency of pre-mRNA processing steps, as well as for the final exon composition of mRNA.
The polypyrimidine tract binding protein-associated splicing factor (PSF) (50) and a closely related protein, p54nrb/NonO (17, 70), have been detected in association with transcription initiation (holoenzyme) and elongation complexes and with the pol II CTD (21, 31). PSF and p54nrb/NonO belong to a larger family of nucleic acid binding proteins that share a highly conserved inner core region that includes two canonical RNA recognition motifs (RRMs), as well as a domain that functions in the formation of homo- and heterodimers between members of the PSF and p54nrb/NonO-related family of proteins (33). PSF and p54nrb/NonO are thought to associate through this domain and have been identified as partner proteins in several reports (51, 62, 74). Both proteins have been detected in or proximal to nuclear “speckle” domains, which are enriched in numerous splicing factors as well as in pol IIo (20, 23). PSF is important for efficient pre-mRNA splicing and splicing complex formation in vitro (27, 50), whereas the function of p54nrb/NonO is less clear. Since PSF and p54nrb/NonO have been identified in association with factors and complexes that operate in different processes, including pre-mRNA processing and transcription, it is possible that they have multiple roles in the cell (61). In this regard, they are similar to members of the abundant class of heteronuclear ribonucleoproteins (RNPs), many of which have been assigned different functions associated with nucleic acid metabolism (19).
In the present study, we provide evidence that PSF specifically functions in mediating transcriptional activator- and CTD-dependent stimulation of constitutive pre-mRNA processing reactions. We further show that this function of PSF may be directed primarily at promoting more efficient splicing of 5′-end proximal introns, whereas its effect on 3′-end cleavage may be more indirect. The results provide new insights into how the transcriptional and pre-mRNA processing machineries are physically coupled and further suggest a mechanism by which the levels of transcription and pre-mRNA processing are coordinated in the cell.
MATERIALS AND METHODS
Antibodies.
Antisera used in this study include rabbit polyclonal antibodies specific for PSF and p54nrb/NonO (42), anti-phosphorylated CTD, anti-FLAG (M5; Sigma), anti-Myc (9E10; Sigma), anti-U1-70K (Cappel), anti-E2F1 (KH20 and KH95; Santa Cruz Biotechnology), anti-p62 (gift of Jack Greenblatt), Y12 (38), anti-SRm160 (B1C8) (8), anti-SR (MAb104) (57), and anti-hypophosphorylated pol II (8WG16) (63).
Cell lines.
Human embryonic kidney 293 (HEK293) cells (American Type Culture Collection) were used for all transient-transfection experiments involving analyses of reporter pre-mRNAs.
Plasmids.
Plasmids for expression of Gal4 fusion proteins containing transcriptional activation domains, including Gal4-SW6 (pSGSW6), Gal4-TAT (pHKGTat1-48), Gal4-FBP, Gal4-VP16 (pSGVPΔ490), and Gal4-p53 (pGalMp53), and expression of VA RNA (pSP-VA1) have been described previously (7). Plasmids for bacterial expression of glutathione S-transferase (GST)-SW6 (amino acids 410 to 490), GST-VP16 (410-490), GST-CTD 1-15, GST-CTD 1-15mt, GST-CTD wt, and pol II 1-15 were from gifts from David Bentley, and GST-p53 (amino acids 1 to 73) was a gift from Jack Greenblatt. Other RNA pol II expression plasmids used in this study, as well as the plasmids for the pre-mRNA reporters pGal5HIV2dsx(+/− intron)(+/− ESE [exonic splicing enhancer]) and pSVEDaGal5HIV2 and the plasmids used to generate RNase protection probes for these reporters, have been described previously (55).
The expression plasmid for Myc-PSF was constructed by digestion of hemagglutinin (HA)-PSF (42) with SalI and XhoI to release the PSF cDNA, filling in of the cohesive ends using avian myeloblastosis virus reverse transcriptase (Roche), and insertion into a pCMV-Myc vector (gift of Alan Cochrane, University of Toronto) after digestion with HindIII. The expression vector for FLAG-p54 was constructed by PCR amplification from HA-p54nrb/NonO (74) with the following primers: 5′-GCGGGATCCAATGCAGAGTAATAAAAC-3′ (which includes a BamHI restriction site upstream of the start codon) and 5′-CCGCTCGAGTTAGTATCGGCGACGTTTG-3′ (which includes a XhoI site downstream of the stop codon). The amplicon was digested with BamHI and XhoI, and the resulting product was ligated into vector pcDNA3-FLAG (modified from pcDNA3) after digestion with these enzymes.
Plasmids for expression of the Myc-tagged PSF deletion mutants ΔRGG, ΔPQ, and Δ+/− were created by PCR with Phusion enzyme (Finnzymes) using the Myc-PSF plasmid as a template and primer pairs designed to amplify around this plasmid, leaving the desired region deleted. The resulting PCR products were then ligated to recircularize the plasmid. The plasmids for expression of Myc-tagged mutants ΔN, ΔP, ΔRRM1, ΔRRM2, and ΔRRM1 and ΔRRM2 were generated by amplification of fragments of PSF upstream and downstream of the deleted region, using a green fluorescent protein-PSF (gift of James Patton) expression vector as a template (all primer sequences can be obtained from the authors upon request). The fragment upstream of the deletion region contained an NsiI restriction site, and the fragment downstream of the deletion region contained an SpeI site; after digestion with these enzymes, the fragments were ligated into the pCMV-Myc vector predigested with PstI and SpeI, in a triple-ligation reaction.
Bacterial protein expression and affinity chromatography.
The GST fusion proteins GST-SW6, GST-VP16, and GST-p53 and GST alone were bacterially expressed essentially as described previously (59). One milliliter of each bacterial lysate was pelleted and resuspended in sample buffer (140 mM Tris HCl, pH 6.8, 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.02% bromophenol blue), and the protein concentration was determined by comparison with known concentrations of purified GST by polyacrylamide gel electrophoresis and Coomassie blue staining. The remaining sample was sonicated (3 min, power 8, duty cycle 40% on a Branson Sonifier) and rocked for 10 min at 4°C after addition of Triton X-100 to 1%. Samples were centrifuged at 22,000 × g to collect the supernatant. Glutathione-Sepharose resin was added to the samples to result in a bound GST fusion protein concentration of 4 mg/ml and was rocked for 1 h at 4°C. The resin was washed three times in lysis buffer and then once with 50 ml of 20 mM HEPES, pH 7.9, 100 mM NaCl, 1 mM dithiothreitol (DTT), 0.1 mM EDTA, and 50% glycerol. Samples were stored in this buffer as a 1:5 (resin volume-to-buffer volume) resuspension at −20°C.
One milliliter of the GST and GST fusion protein resins was added to a disposable chromatography column (Bio-Rad), prerinsed in “high-salt” buffer (20 mM HEPES, pH 7.9, 1 M NaCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol). Columns were washed once with high-salt buffer and then once with “low-salt” buffer (20 mM HEPES, pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol), before the addition of HeLa nuclear extract (at 10 mg/ml). Prior to affinity chromatography, the HeLa nuclear extract was pretreated by incubation with 1.75 mM ATP, 6 mM creatine phosphate, 0.05% Nonidet P-40, 2 mM MgCl2 for 15 min at 30°C. Where indicated, the HeLa nuclear extract was also preincubated with RNase (Roche; 50 μg/ml) and 1,000 U DNase. Two milliliters of pretreated HeLa nuclear extract was added to each column, the columns were then washed with 1.75 ml of low-salt buffer, and bound proteins were eluted with 1.75 ml of high-salt buffer; 150-μl fractions were collected, and the protein concentrations of the fractions were determined. The four fractions containing the majority of the eluted protein were pooled, snap-frozen in liquid nitrogen, and stored at −80°C.
For the GST pull-down assays shown in Fig. 5, HEK293 cells were transfected with 10 μg of the expression plasmids for wild-type and mutant PSF proteins. Forty-eight hours after transfection, the cells were harvested for extract preparation, as described previously (16). The extracts were incubated for 2 h at 4°C with 100 μl glutathione-Sepharose beads precoupled to GST-VP16, GST-SW6, and GST (at 4 mg/ml of resin). The beads were washed three times with “low-salt” buffer (20 mM HEPES [pH 7.9], 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT, Complete Mini protease inhibitor cocktail tablet [Roche]) and then eluted by incubation on ice for 20 min in a “high-salt” buffer (same as the low-salt buffer except with 1 M KCl). The eluates were separated on a 10% SDS-polyacrylamide gel and analyzed by immunoblotting with anti-Myc antibody (9E10; Sigma).
FIG. 5.
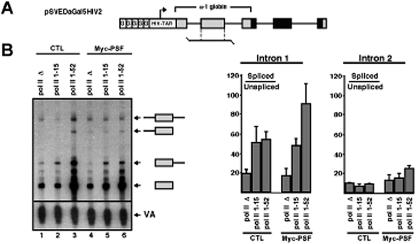
PSF preferentially stimulates splicing of the first intron in a human α-1 globin pre-mRNA reporter. (A) pSVEDaGal5HIV2 pre-mRNA reporter used to analyze splicing (48). This reporter contains five Gal4 DNA binding sites upstream of an HIV-2 promoter, human α-1 globin gene exons (shaded gray), and fibronectin gene exons (shaded black). Levels of splicing of introns 1 and 2 of the α-1 globin gene were analyzed by RNase protection assays with the probe indicated. (B) HEK293 cells were transfected with the pSVEDaGal5HIV2 pre-mRNA reporter, after transient transfection of the cells with expression plasmids for Gal4-VP16 (lanes 1 to 6); Myc-PSF (lanes 4 to 6); an empty expression vector (CTR, lanes 1 to 3); and α-amanitin-resistant RNA pol II-Δ (lanes 1 and 4), pol II 1-15 (lanes 2 and 5), and pol II 1-52 (lanes 3 and 6). The transfected cells were grown in the presence of α-amanitin after transfection of the VP16, PSF, and pol II expression vectors and prior to transfection of the pSVEDaGal5HIV2 pre-mRNA reporter, as described in Fig. 3. A representative RNase protection analysis of the splicing levels of α-1 globin introns 1 and 2 is shown. The identity of the unspliced and spliced RNAs corresponding to processing of each of the two α-1 globin introns is indicated (see also panel A), and quantification of the splicing levels is shown in the adjacent bar graph, with standard deviations indicated (corresponding to two separate experiments).
Tandem mass spectrometry.
Affinity chromatography eluates were concentrated by centrifugation in a Speed-Vac instrument, until the volume of each sample was approximately 20 μl. Tandem mass spectrometry was performed on the samples as described previously (9).
Immunoblotting and silver staining.
Thirty micrograms of protein lysates or 5 to 40 μg of the affinity chromatography elution fractions was analyzed on 7.5% SDS-polyacrylamide gels and detected by silver staining and immunoblotting (using chemiluminescence detection), per standard protocols.
Transfections and RNA analysis.
Human HEK293 cells grown in 15-cm plates were transfected with 5 μg of reporter and 10 μg of expression plasmids, as previously described (55, 56). Cells were harvested 48 h posttransfection, and RNA was isolated by RNase protection assays, as described previously (71). In experiments employing vectors for expression of α-amanitin-resistant forms of RNA pol II, transfections were performed as described previously (56). RNase protection products were analyzed in 6% denaturing polyacrylamide gels, detected by autoradiography, and quantified using a Bio-Rad phosphorimager.
RESULTS
Identification of factors that bind preferentially to strong rather than weak transcriptional activators.
In order to identify factors that potentially mediate the ability of strong transcriptional activators to stimulate splicing and 3′-end cleavage, we used mass spectrometry to identify HeLa nuclear extract proteins which preferentially associate with strong (i.e., VP16 and p53) rather than weak (i.e., SW6 [69]) activation domains. GST fusion proteins containing the activation domains of VP16 (amino acids 410 to 490), p53 (amino acids 1 to 73), and SW6 (a mutant version of the VP16 activation domain containing four Phe-to-Ala alterations) were immobilized on glutathione S-Sepharose columns and used to affinity-purify proteins in HeLa nuclear extract. Proteins eluted from each column were identified by tandem mass spectrometry.
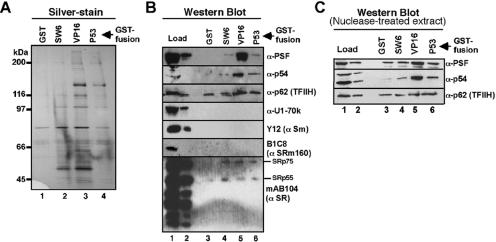
Analysis of the fractions eluted from each column by SDS-polyacrylamide gel electrophoresis and silver staining revealed several polypeptides that are specifically enriched by the VP16 and p53 activation domains, compared to the SW6 domain, or GST alone (Fig. 1A, compare lanes 3 and 4 with lanes 1 and 2). Mass spectrometric analysis of each fraction identified with high confidence over 300 different protein species in total, the majority of which bound to VP16 and p53 columns, compared to the SW6 and GST columns (data not shown). A subset of the proteins identified by the highest number of individually detected peptides in the VP16 and/or p53 column eluate, compared to the SW6 and GST eluates, are shown in Table 1. Among the proteins are PSF and p54nrb/NonO. PSF and p54nrb/NonO belong to a family of proteins that share close homology within a 246-amino-acid central region that contains two RRMs and a homo-/heterodimerization domain (17, 33, 50, 70). These proteins have been shown to bind each other as well as to single- and double-stranded nucleic acids. Both proteins have also been implicated in splicing and transcription (61) (see Discussion). We and others have recently shown that PSF and p54nrb/NonO are components of an RNA pol II holoenzyme complex and bind to the CTD of pol II (21, 31). Taken together with the results described above suggesting that PSF might bind preferentially to the VP16 and p53 activation domains and that p54nrb/NonO might bind preferentially to the VP16 activation domain and previous work demonstrating that the CTD is important for the stimulation of splicing and 3′-end cleavage of nascent transcripts, it was of interest to determine whether PSF or p54nrb/NonO is important for mediating the stimulatory effects of a strong activator and the CTD on pre-mRNA processing.
FIG. 1.
Affinity chromatography of factors associating with transcriptional activation domains of different strengths. GST fusion proteins containing the activation domains of the strong transcriptional activators VP16 (410 to 490) and p53 (1 to 73) or the weak transcriptional activator SW6 (amino acids 410 to 490) (7) were immobilized on glutathione-Sepharose columns and used to affinity purify proteins from HeLa nuclear extract. Proteins eluted from the GST activation domains and GST alone were separated by SDS-polyacrylamide gel electrophoresis and detected by silver staining (A) and by immunoblotting with antisera specific to proteins detected by mass spectrometry analysis as being enriched in the GST-VP16 and GST-p53 eluates, compared to the GST-SW6 and GST eluates (B; see Results). HeLa nuclear extract was treated extensively with both RNases and DNases prior to affinity chromatography, and proteins eluted from each column were analyzed by immunoblotting with the antisera indicated (C). Eluted proteins analyzed in lanes 3 to 6 of panels B and C represent the total material recovered after affinity chromatography with 1 mg of HeLa nuclear extract per column; 100 μg and 10 μg of total HeLa nuclear extract, respectively, were separated in lanes 1 and 2. See Materials and Methods for additional experimental details.
TABLE 1.
Selected HeLa nuclear extract proteins identified by tandem mass spectrometry in the elution fractions from affinity columns containing immobilized GST-transcriptional activation domain fusions
| Protein | Accession no. | No. of peptide hitsa
|
|||
|---|---|---|---|---|---|
| GST | SW6 | VP16 | p53 | ||
| LRPPRC: 130-kDa leucine-rich protein (LRP 130) (GP130) (leucine-rich PPR motif-containing protein) | P42704 | 0 | 0 | 18 | 6 |
| PSF: splicing factor, proline and glutamine rich (polypyrimidine tract-binding protein-associated splicing factor) (PTB-associated splicing factor) | P23246 | 0 | 0 | 16 | 2 |
| p54nrb/NonO: 54-kDa nuclear RNA and DNA binding protein (p54nrb) (55-kDa nuclear protein) (NMT55) (non-POU-domain-containing octamer-binding protein) | Q15233 | 1 | 1 | 11 | 1 |
| FUS: RNA binding protein FUS (oncogene FUS) (oncogene TLS) (translocated in liposarcoma protein) (POMp75) (75-kDa DNA-pairing protein) | P35637 | 2 | 0 | 2 | 6 |
| hnRNP A2/B1: heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2/hnRNP B1) | P22626 | 2 | 1 | 4 | 1 |
| DC50 (DC23) (hypothetical protein) | Q9GZT3 | 0 | 1 | 5 | 2 |
| PC4: activated RNA polymerase II transcriptional coactivator p15 (PC4) (p14) | P53999 | 0 | 1 | 4 | 1 |
| p68: probable RNA-dependent helicase p68 (DEAD-box protein p68) (DEAD-box protein 5) | P17844 | 0 | 0 | 5 | 2 |
Number of independently identified peptides in each elution fraction.
PSF and p54nrb/NonO associate preferentially with strong activation domains.
To further assess whether PSF and p54nrb/NonO associate with strong rather than weak activation domains, the elution fractions from the affinity chromatography experiments in Fig. 1A were analyzed by immunoblotting using polyclonal antisera specific for these two proteins (Fig. 1B). This analysis confirmed that both PSF and p54nrb/NonO are preferentially bound by the VP16 activation domain compared to the SW6 activation domain or GST alone (compare lane 5 with lanes 3 and 4). A modest increase in binding of these proteins to the p53 activation domain, compared to the SW6 domain and GST alone, was also apparent (compare lane 6 with lanes 3 and 4). To assess the specificity of our affinity chromatography procedure, we also analyzed the elution fractions with an antibody specific for the p62 subunit of the basal transcription factor TFIIH, which is known to bind preferentially to both VP16 and p53, compared to weak activators (7). Consistent with these previous results, we observe that p62 binds more strongly to the VP16 activation domain, and at a level slightly increased over that of the p53 activation, compared to the SW6 activation domain and GST alone (Fig. 1B, compare lanes 5 and 6 with lanes 3 and 4; data not shown).
Antisera specific for the abundant splicing components U1 snRNP-70K protein, snRNP Sm proteins, and SRm160 splicing coactivator did not detect these factors in any of the elution fractions, although they were easily detected in the load fractions (Fig. 1B, compare lanes 1 and 2 with lanes 3 to 6). Moreover, the majority of abundant SR family proteins detected by the monoclonal antibody MAb104 also were not detected at appreciable levels in the elution fractions; very low levels of the SR family proteins SRp55 and SRp75 are equally apparent in all of the fractions (Fig. 1B, lanes 3 to 6). Extensive pretreatment of the HeLa nuclear extract with both RNases and DNases (refer to Materials and Methods) did not substantially alter the level of binding of PSF, p54nrb/NonO, and p62 to the strong versus weak activation domains (Fig. 1C, lanes 3 to 6). These results of the affinity chromatography experiments, together with the mass spectrometry data, provide evidence that the splicing and transcription-associated proteins PSF and p54nrb/NonO preferentially and specifically associate with the strong VP16 activation domain, compared to the weak SW6 activation domain, and that both proteins bind with lower affinity to the p53 activation domain, at a level that may be only slightly higher than their binding to the SW6 activation domain.
PSF, but not p54nrb/NonO, is rate limiting for transcription activator-dependent stimulation of splicing and 3′-end cleavage of reporter pre-mRNAs.
Given that PSF and p54nrb/NonO preferentially associated with the activation domain of VP16, compared to the SW6 activation domain, we hypothesized that one or both of these proteins might mediate strong activator-dependent stimulation of pre-mRNA processing. Such a role would be expected if PSF and/or p54nrb/NonO is normally rate limiting for splicing and 3′-end formation in vivo. A strong activator such as VP16 might then function to increase the local concentration of PSF and p54nrb/NonO such that these factors are available to facilitate processing of increased levels of nascent pre-mRNA resulting from activated transcription. In this model, a strong activator would function to coordinate increased requirements for processing factors with increased transcript levels. To initially test this idea, we overexpressed PSF and p54nrb/NonO and asked whether increased levels of either protein can function to bypass VP16-dependent stimulation of splicing and 3′-end cleavage. Expression plasmids under the control of the cytomegalovirus promoter and containing the activation domains of SW6 or VP16 fused to the DNA binding domain (DBD) of Gal4 (Gal-SW6 and Gal-VP16, respectively) (7), or Gal4 with no activation domain (N.A.), were cotransfected into HEK293 cells with the pre-mRNA reporter plasmid Gal5HIV2dsxΔESE (55), along with either a control “empty” plasmid (CTL), or expression plasmids (also under the control of the cytomegalovirus promoter) for Myc epitope-tagged PSF (Myc-PSF), FLAG-epitope-tagged p54nrb/NonO (FLAG-p54), or both of these expression plasmids combined. The Gal5HIV2dsxΔESE pre-mRNA reporter consists of five Gal4-DBD recognition sites upstream of the human immunodeficiency virus type 2 (HIV-2) promoter, sequences derived from the exon 3-intron-exon 4 region of the Drosophila melanogaster doublesex (dsx) gene, and the simian virus 40 late polyadenylation signal (Fig. 2A). This pre-mRNA is accurately spliced in vitro and in vivo in mammalian systems and is responsive to the insertion of mammalian ESEs in exon 4 (45, 72). Transcription activated by the expressed Gal4 fusion proteins used in this study is accurately initiated and is dependent on the presence of the five Gal4 DNA binding sites in the promoter, since none of the Gal4 fusion proteins activated transcription from a version of the Gal5HIV2dsxΔESE containing an adenovirus major late promoter and lacking Gal4 DNA binding sites (55). RNA recovered from the transfected cells was analyzed by RNase protection assays using probes designed specifically for monitoring splicing and 3′-end cleavage efficiency (Fig. 2A) (45, 71).
FIG. 2.
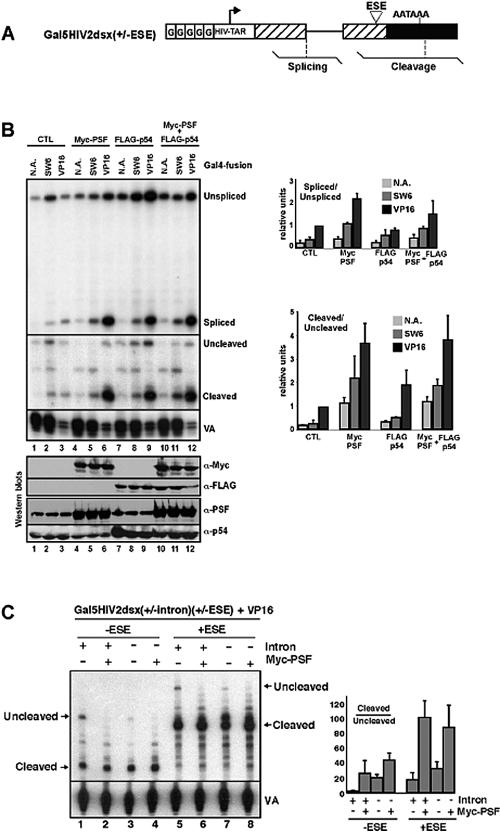
Increasing PSF expression stimulates splicing and 3′-end cleavage and can bypass the requirement for a strong activator to promote these pre-mRNA processing steps. (A) The Gal5HIV2dsx reporter plasmid (55) used for analyzing splicing and 3′-end cleavage efficiency in transient-transfection analyses, with RNase protection probes indicated. (B) HEK293 cells were transfected with the Gal5HIV2dsxΔESE (i.e., lacking an ESE) pre-mRNA reporter, together with an expression plasmid for Myc-tagged PSF (Myc-PSF), FLAG-tagged p54nrb/NonO (FLAG-p54), both Myc-PSF and FLAG-p54 together, or a control empty vector. Cotransfected with each of the above was an expression plasmid for fusion proteins containing the Gal4 DBD fused to amino acids 410 to 490 of SW6 (Gal4-SW6), amino acids 410 to 490 of VP16 (Gal4-VP16), or Gal4-DBD alone (N.A.), as indicated. As an internal control for transfection efficiency and RNA recovery, the plasmid pSPVA was also included in each transfection. This plasmid expresses an RNA polymerase III virus-associated transcript, VA. Cells were harvested posttransfection, and the lysates were analyzed by immunoblotting with antisera specific for PSF and p54nrb/NonO and with epitope tag-specific antibodies (anti-Myc or anti-FLAG), as indicated in the lower panels. RNase protection analysis of RNAs recovered from the transfected cells is shown in the upper panels. The panels show RNase protection performed with the probes for analyzing splicing, 3′-end cleavage, and the VA RNA. One-tenth less RNA was analyzed for the VP16 samples (lanes 3, 6, 9, and 12) than for the other samples in order to facilitate direct comparisons within the same gel (typically, VP16 activation resulted in 10-fold-more activation of the Gal5HIV2dsxΔESE reporter than SW6). Quantification of splicing and 3′-end cleavage levels from the RNase protectionassays is shown in the adjacent bar graphs and represents three separate analyses. (C) PSF stimulates 3′-end cleavage in the absence of splicing. Plasmids containing versions of the Gal5HIV2dsx reporter with or without an intron and with or without an ESE (consisting of six GAA repeats) in the second exon were cotransfected into HEK293 cells, with or without an expression plasmid for Myc-tagged PSF, as indicated. RNA was analyzed by RNase protection using probes for monitoring 3′-end cleavage, as described for panel B.
Immunoblotting of the transfected cell lysates with antibodies specific for PSF or p54nrb/NonO indicated that these proteins were overexpressed approximately fivefold over the levels of the endogenous proteins (Fig. 2B, bottom panels; compare lanes 4 to 6 and 10 to 12 with lanes 1 to 3 and 7 to 9 for PSF expression levels and lanes 7 to 12 with lanes 1 to 6 for p54nrb/NonO expression levels). Independent of the activator used to drive transcription, increased expression of PSF resulted in approximately a twofold-higher level of splicing (measured as the ratio of spliced to unspliced RNA) of the Gal5HIV2dsxΔESE pre-mRNA compared to the N.A. control (Fig. 2B, compare lanes 4 to 6 with lanes 1 to 3, and adjacent bar graph). Also independent of the activator used to drive transcription, increased expression of PSF resulted in at least a threefold-higher level of 3′-end cleavage (measured as the ratio of cleaved to uncleaved RNA). In contrast, increased expression of p54nrb/NonO did not significantly affect the ratios of spliced to unspliced or cleaved to uncleaved RNAs (Fig. 2B, compare lanes 7 and 8 with lanes 1 to 3, and adjacent bar graph). In some experiments, an increased total level of RNA was observed upon increased expression of p54nrb/NonO, although this effect was less pronounced in repeat experiments (data not shown). Simultaneous overexpression of PSF and p54nrb/NonO resulted in levels of splicing and 3′-end cleavage that were comparable to those when PSF was overexpressed alone (compare lanes 10 to 12 with 1 to 3 and 4 to 6, Fig. 2B). Notably, increased expression of PSF was sufficient to increase the level of splicing and 3′-end cleavage of SW6-activated transcripts to levels that are comparable to those when transcription is activated with VP16 at endogenous levels of PSF (Fig. 2B, compare lanes 5 and 3). These results demonstrate that the endogenous level of PSF, but not p54nrb/NonO, is normally rate limiting for splicing and 3′-end cleavage of the Gal5HIV2dsx reporter pre-mRNA and that activation of transcription with a strong activator such as VP16 can partially bypass this rate-limiting step.
PSF stimulates 3′-end cleavage in the absence of splicing.
We next assessed the extent to which the stimulation of 3′-end cleavage by PSF is a consequence of splicing of an adjacent intron, as opposed to a more direct effect on 3′-end processing that might occur in the absence of splicing. In this experiment we also asked to what extent a purine-rich ESE, which can promote both splicing and 3′-end cleavage of the Gal5HIV2dsx pre-mRNA reporter (45), influences the PSF-dependent stimulation of these processing activities. The Myc-PSF expression plasmid, or a control plasmid, was cotransfected into HEK293 cells with the Gal4-VP16 expression plasmid, and versions of the Gal5HIV2dsx pre-mRNA reporter with or without an intron and with or without an ESE consisting of six GAA repeats inserted into the second exon (Fig. 2A).
Overexpression of PSF resulted in an increase in the level of 3′-end cleavage, even in the absence of an intron (Fig. 2C). In the absence of an intron, PSF stimulated cleavage by approximately twofold (compare lanes 4 and 8 with lanes 3 and 7, respectively, and adjacent bar graph), whereas in the presence of an intron, overexpression of PSF resulted in a further increase in 3′-end cleavage levels (compare lanes 2 and 6 with lanes 1 and 5, respectively; refer to adjacent bar graph). The ESE resulted in higher levels of 3′-end cleavage, even in the absence of an intron (compare lanes 5 to 8 with 1 to 4), although it did not significantly influence the relative levels of PSF-dependent stimulation of 3′-end cleavage (compare lanes 1 and 2 with 5 and 6 and lanes 3 and 4 with 7 and 8). Thus, PSF can stimulate 3′-end cleavage in the absence of splicing, although its 3′-end cleavage-stimulatory activity is augmented by the presence of an intron. Consistent with previous evidence supporting a role for PSF in splicing in vitro (27, 50) and extensive evidence indicating that splicing components assembled adjacent to a cleavage/polyadenylation signal can stimulate 3′-end cleavage (28, 41, 52), the data in Fig. 2 provide evidence that PSF-dependent stimulation of 3′-end cleavage results in part from its role in promoting splicing, as well as from a more direct effect on 3′-end processing.
PSF-dependent stimulation of pre-mRNA processing requires the RNA pol II CTD.
To further assess whether PSF is involved in mediating transcription activator-dependent stimulation of splicing and 3′-end formation, we next asked whether its ability to stimulate pre-mRNA processing requires the RNA pol II CTD. This might be expected since we found previously that stimulation of splicing and 3′-end cleavage by VP16, over the levels of splicing and 3′-end cleavage obtained when transcription is driven by SW6, requires the CTD (55), and both PSF and p54nrb/NonO can bind directly to this structure (21). Myc-PSF, FLAG-p54nrb/NonO, or both proteins were overexpressed in HEK293 cells cotransfected with the Gal5HIV2dsxΔESE reporter and plasmids for expression of α-amanitin-resistant derivatives of human pol II with either no CTD (pol II-Δ), a truncated CTD containing heptapeptide repeats 1 to 15 (pol II 1-15), or a wild-type CTD containing heptapeptide repeats 1 to 52 (pol II 1-52) (Fig. 3A). In order to study the effects of overexpression of PSF and p54nrb/NonO when the pre-mRNA is transcribed by the plasmid-encoded pol II derivatives, cells were initially transfected with the expression plasmids for Myc-PSF and FLAG-p54nrb/NonO, activator proteins, and wild-type and mutant pol II large subunits and then grown in the presence of α-amanitin, which results in the degradation of endogenous pol II. The cells were subsequently transfected with the Gal5HIV2dsxΔESE pre-mRNA reporter. RNA recovered from the transfected cells was analyzed by RNase protection assays using probes to monitor splicing and 3′-end cleavage levels, as in Fig. 2. As before, PSF and p54nrb/NonO were expressed at comparable levels (approximately fivefold) over the levels of the endogenous proteins (bottom panels, Fig. 3A).
FIG. 3.
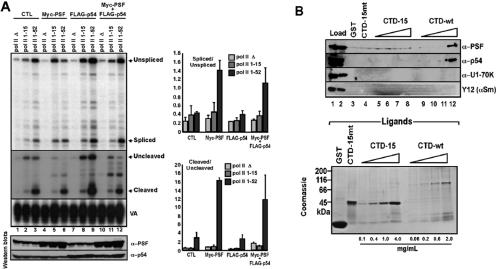
Stimulation of splicing and 3′-end cleavage by PSF requires the RNA pol II CTD. (A) HEK293 cells were transfected with expression plasmids for Gal4-VP16 (lanes 1 to 12), Myc-PSF (lanes 4 to 6), FLAG-p54 (lanes 7 to 9), and Myc-PSF and FLAG-p54 together (lanes 10 to 12), as well as with expression plasmids for α-amanitin-resistant versions of RNA pol II with no CTD (pol II Δ) (lanes 4, 7, and 10), with a CTD consisting of 15 heptapeptide repeats (pol II 1-15) (lanes 5, 8, and 11), or with a wild-type CTD (pol II 1-52) (lanes 6, 9, and 12). HEK293 cells were also transfected in parallel with these pol II expression plasmids but without expression plasmids for Myc-PSF or FLAG-p54 (CTL, lanes 1 to 3). After selection with α-amanitin the cells were subsequently transfected with the Gal5HIVdsxΔESE pre-mRNA reporter plasmid (lanes 1 to 12). Splicing and 3′-end cleavage levels of the Gal4-VP16-activated Gal5HIVdsxΔESE transcripts were determined by RNase protection assays as described in Fig. 2. Splicing, 3′-end cleavage, and VA RNase protection analyses are shown in the upper panels, as indicated. The lower panels show immunoblot analyses assaying the expression levels of PSF and p54nrb/NonO, as indicated in Fig. 2B. The adjacent bar graphs show the relative ratios of spliced/unspliced and cleaved/uncleaved RNAs, determined from three separate analyses. (B) GST alone and different concentrations (as indicated) of immobilized GST fusion proteins containing either the full-length human CTD (CTD-wt), a truncated CTD consisting of 15 heptapeptide repeats (CTD-15), or 15 repeats of the mutant heptapeptide sequence YSPTAPS (CTD-15mt) were used to affinity purify proteins from HeLa nuclear extract. Proteins eluted from each column were analyzed by immunoblotting with antisera specific for PSF, p54nrb/NonO, U1-70kDa, and snRNP Sm proteins, as indicated (upper panel). Samples of the immobilized GST fusion proteins were analyzed on a Coomassie blue-stained SDS-polyacrylamide gel (lower panel).
Consistent with the results in Fig. 2, PSF stimulated splicing (by approximately threefold) and 3′-end cleavage (by fivefold) when the pre-mRNA was transcribed with pol II containing a wild-type CTD (Fig. 3A, compare lanes 4 to 6 with 1 to 3, and refer to the adjacent bar graph). However, PSF did not stimulate splicing when the pre-mRNA was transcribed with pol II 1-15 or pol II-Δ. As before, overexpression of p54nrb/NonO did not affect splicing or 3′-end cleavage levels (compare lanes 7 to 9 with 1 to 3), whereas coexpression of PSF and p54nrb/NonO resulted in similar levels of splicing and 3′-end cleavage as when PSF was expressed alone (compare lanes 10 to 12 with 1 to 3 and adjacent bar graphs). As observed when PSF is overexpressed alone, only pol II with a full-length CTD, and not pol II 1-15 or pol II-Δ, supported the stimulation of splicing and 3′-end cleavage when PSF and p54nrb/NonO were coexpressed. These results demonstrate that increased levels of PSF, but not of p54nrb/NonO, stimulate splicing and 3′-end cleavage via a mechanism that is dependent strongly on the presence of the pol II CTD. Although significant levels of the reporter pre-mRNA were transcribed by pol II 1-15, a CTD consisting of 15 heptapeptide repeats is not sufficient to mediate PSF-dependent stimulation of splicing and 3′-end cleavage. Since in our previous study (55) it was found that the CTD was required specifically for a VP16-dependent increase in splicing, and not for levels of splicing obtained when transcription was driven by SW6, the results described above, together with the data in Fig. 1, are consistent with a role for PSF in mediating VP16- and CTD-dependent stimulation of splicing.
PSF and p54nrb/NonO associate with wild-type pol II CTD but not a truncated CTD consisting of 15 heptapeptide repeats.
In our previous study, we demonstrated that PSF and p54nrb/NonO bind efficiently to both phosphorylated and nonphosphorylated forms of the pol II CTD (21). To assess whether the inability of pol II 1-15 to mediate PSF-dependent stimulation of splicing and 3′-end formation might be a consequence of its inability to bind to a pol II CTD consisting of 15 heptapeptide repeats, we performed affinity chromatography of HeLa nuclear extract with GST fusion proteins containing either a wild-type unphosphorylated CTD or a 15-repeat CTD (Fig. 3B). For controls, affinity chromatography was performed with GST alone and with a GST fusion containing 15 repeats of the mutant heptapeptide sequence in which all serines at position 5 in the heptapeptide consensus sequence are replaced with alanine residues (YSPTAPS; CTD-15mt). Proteins bound to the columns were immunoblotted with antisera specific for PSF and p54nrb/NonO, as well as with antisera specific for the abundant spliceosomal snRNP U1-70K and Sm proteins.
As expected, PSF and p54nrb/NonO both bound to the wild-type CTD and binding was observed only at the highest concentration of ligand (2.0 mg/ml) (compare lane 12 to lane 8, Fig. 3B). This binding appears to be quite specific, since neither U1-70K nor Sm proteins are detected in the same elution fractions, although they are easily detected in the load fractions (Fig. 3B). In contrast, PSF and p54nrb/NonO did not bind to the 15-repeat CTD, even when the highest concentration of this ligand was used (4.0 mg/ml) for affinity chromatography. As the concentration of individual heptapeptide repeat present on the CTD 1-15 column at 4.0 mg/ml is in excess over the concentration of repeat on the wild-type CTD column at 2.0 mg/ml, the absence of any binding of PSF and p54nrb/NonO to the CTD 1-15 ligand is not a consequence of any difference in the relative concentrations of heptapeptide repeats between the affinity columns but is most likely due to the shorter length of the CTD. PSF and p54nrb/NonO also did not bind to either GST alone or CTD-15mt proteins. These data indicate that binding of PSF and p54nrb/NonO to the pol II CTD is quite specific among abundant splicing factors and requires a minimum number of consecutive heptapeptide repeats greater than 15. The results provide a mechanistic explanation for the inability of increased levels of PSF to promote splicing and 3′-end cleavage when pre-mRNA is transcribed with pol II containing a 15-repeat CTD. Even though this mutant version of pol II can support moderate levels of VP16-dependent transcription (i.e., in comparison with wild-type pol II and pol II-Δ; compare lanes 2 to lanes 1 and 3, Fig. 3A), it does not mediate a PSF-dependent increase in splicing and 3′-end cleavage, most likely because it contains an insufficient number of CTD heptapeptide repeats to support the binding of this factor.
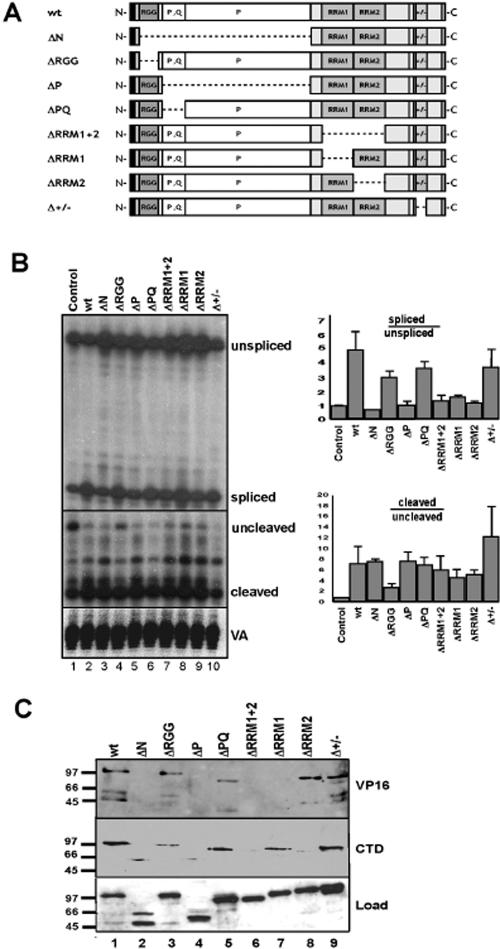
Domains of PSF required for stimulation of pre-mRNA processing and association with transcription components.
In order to investigate in further detail the mechanism by which PSF functions in cotranscriptional splicing and 3′-end cleavage, we next asked which domains in the protein are critical for these processing activities, as well as for binding to the VP16 activation domain and to the CTD (Fig. 4). A series of Myc epitope-tagged deletion derivatives of PSF were constructed which specifically remove the RGG box (ΔRGG, residues 9 to 27), the Pro/Glu-rich domain (ΔPQ, residues 34 to 113), RRM1 (ΔRRM1, residues 298 to 365), RRM2 (ΔRRM2, residues 375 to 429), and a basic/acidic-rich domain (Δ+/−, residues 575 to 611) (Fig. 4A). Also constructed were more extensive deletions which remove the entire N-terminal half of PSF (ΔN, residues 9 to 264), a proline-rich region (ΔP, residues 26 to 267), and both RRMs together (ΔRRM1 + 2, residues 298 to 429). Expression plasmids for each of these deletion constructs were transfected into HEK293 cells with the expression plasmid for Gal4-VP16 and the Gal5HIV2dsxΔESE pre-mRNA reporter (Fig. 4B). RNA isolated from the transfected cells was analyzed by RNase protection as described for Fig. 2. Lysates prepared from the cells were analyzed by immunoblotting with anti-Myc antibody to monitor expression of the wild-type and mutant PSF proteins. Comparable expression levels were observed between the proteins in this and several repeat transfection experiments (Fig. 4C, lower panel; data not shown). Moreover, all of the proteins were localized in the nucleus of the transfected cells, as detected by immunofluorescence localization with the anti-Myc epitope antibody (data not shown). This is consistent with the preservation of two previously mapped nuclear localization signal sequences in all of the deletion constructs (Fig. 4A) (20).
FIG. 4.
Domains of PSF required for stimulation of splicing, 3′-end cleavage, and binding to VP16 and the pol II CTD. (A) Diagram showing the domain organization of PSF and the regions removed in different mutant PSF expression plasmids (see Results for amino acid residues deleted in each construct). (B) Wild-type and mutant PSF expression vectors were cotransfected into HEK293 cells with the Gal5HIV2dsxDESE pre-mRNA reporter, and RNA recovered from the transfected cells was analyzed using RNase protection assays as described in Fig. 2. The results of representative RNase protection assays for analyzing splicing and 3′-end cleavage levels and VA RNA levels are shown, and bar graphs quantifying the data from three separate analyses are shown on the right. (C) GST fusion proteins containing the VP16 activation domain and wild-type pol II CTD were incubated in extracts prepared from HEK293 cells expressing each of the different mutant Myc epitope-tagged PSF proteins shown in panel A. After extensive washing of the beads, bound proteins were eluted and analyzed by immunoblotting using anti-Myc antibody. Levels of each of the wild-type and mutant proteins are shown in the total extract (Load) and elution fractions, as indicated.
Deletion of the RGG box and Pro/Glu- and basic/acidic-rich domains resulted in only a slight decrease in splicing-stimulatory activity compared to wild-type PSF (Fig. 4B, compare lanes 2, 4, 6,and 10; see adjacent bar graph). In contrast, the ΔN and ΔP deletion mutants, as well as each of the RRM deletion mutants (single and double), did not stimulate splicing activity. A related although somewhat different domain dependence for 3′-end cleavage stimulation was observed. Deletion of either RRM reduced slightly the levels of 3′-end cleavage stimulation, whereas deletion of the RGG box resulted in a more pronounced reduction in 3′-end cleavage levels (Fig. 4B, middle panel, compare lane 4 with lanes 1 to 3 and 5 to 10; see adjacent bar graph). A possible explanation for these results is that deletion of domains, such as the two RRMs, required for the stimulation of splicing may also influence indirectly 3′-end cleavage levels, since splicing and 3′-end cleavage are coupled processes (see the introduction), whereas the RGG box may be important for interactions that stimulate 3′-end cleavage more directly. However, removal of this domain in the context of a more extensive deletion (ΔN) restored 3′-end cleavage-stimulatory activity, perhaps indicating that regions of the protein exposed as a consequence of altered folding or interactions following deletion of the N-terminal half of the protein can spuriously stimulate 3′-end cleavage activity. It is also possible that the PSF domain requirements for 3′-end cleavage-stimulatory activity are less specific than they are for splicing stimulation.
We next assessed whether the loss of splicing-stimulatory activity for the PSF deletion mutants described above might be a consequence of their loss of association with the VP16 activation domain (which was used to drive transcription of the reporter in Fig. 4B) and/or with the pol II CTD. Lysates from transfected HEK293 cells expressing wild-type and mutant PSF proteins were chromatographed over immobilized GST-VP16 activation domain and GST-CTD fusion proteins, as described in Fig. 1 and 3, and the bound proteins were then eluted and analyzed by immunoblotting using the anti-Myc epitope antibody. In virtually all cases, the pattern of binding of the mutant proteins reflected their ability to stimulate splicing. For example, the ΔN, ΔP, and ΔRRM1 + 2 PSF mutants did not bind to the VP16 activation domain or the CTD, whereas the ΔRGG, ΔPQ and Δ+/− PSF mutants did bind to these proteins. Interestingly, deletion of RRM1, but not RRM2, prevented binding to VP16, whereas deletion of RRM2, but not RRM1, prevented binding to the CTD. This indicates that the loss of splicing stimulation resulting from the individual deletion of RRM1 and RRM2 may result from different effects on the association of PSF with the transcriptional machinery; RRM1 may play a more important role in binding to activators, whereas RRM2 may play a more important role in binding to the CTD (see Discussion below). These results thus suggest that PSF-dependent stimulation of cotranscriptional splicing involves separate and nonredundant roles for the two highly conserved RRMs in the protein, which may be different from their presumed functions in binding RNA.
Cotranscriptional stimulation of splicing by PSF preferentially affects the first intron of the human α-1 globin gene.
In our previous study (55), we demonstrated that the strong activators VP16 and p53, compared to the weak activators SW6, Tat, and FBP, preferentially stimulate the splicing of the first of two introns at the 5′ end of a human α-1 globin gene reporter (pSVEDaGal5HIV2) (55). This suggested that activator-dependent stimulation of splicing might primarily affect 5′-end-most introns. If PSF is acting via an activator-dependent pathway, then it might also be expected to preferentially stimulate splicing of the 5′-end-most intron of the pSVEDaGal5HIV2 pre-mRNA reporter. To test this, we assayed splicing of introns 1 and 2 of the pSVEDaGal5HIV2 reporter pre-mRNA (Fig. 5A), when transcribed with pol II with or without a full-length CTD, or with pol II containing a 15-heptapeptide-repeat CTD. In each case, transcription was driven by Gal4-VP16. In the absence of PSF overexpression, splicing of intron 1 of the pSVEDaGal5HIV2 pre-mRNA reporter was stimulated by two- to threefold when transcribed by wild-type pol II compared to pol II lacking a CTD (compare lane 3 to lane 1, Fig. 5B and adjacent bar graph). Interestingly, pol II with the 15-repeat CTD resulted in levels of splicing comparable to those observed for wild-type pol II (lanes 2 and 3, Fig. 5B), in contrast to the situation observed for the Gal5HIV2dsxΔESE pre-mRNA reporter, which was not spliced efficiently when transcribed with pol II 1-15. In the absence of PSF overexpression, splicing of intron 2 of the pSVEDaGal5HIV2 was relatively inefficient and was not significantly affected by the length of the CTD (lanes 1 to 3 and adjacent bar graph, Fig. 5B). These observations are consistent with our previous results demonstrating that different introns have different CTD length requirements for efficient splicing (56) and suggest that the CTD, like activators, may preferentially affect constitutive splicing of 5′-end-most introns (55) (see Discussion).
Upon overexpression of PSF, splicing of pSVEDaGal5HIV2 pre-mRNA intron 1 was stimulated by approximately twofold when transcription was driven by wild-type pol II (compare lane 6 to lanes 4 and 5 and refer to bar graphs, Fig. 5B). PSF also resulted in a modest stimulation of splicing of pSVEDaGal5HIV2 pre-mRNA intron 2 when transcribed by wild-type pol II, although the level of splicing stimulation was lower than the level of stimulation of splicing of intron 1 (lanes 4 to 6 and bar graphs, Fig. 5B). Consistent with the previous results obtained with the Gal5HIV2dsx pre-mRNA reporter, for both human α-1 globin introns, stimulation of splicing by PSF was strongly dependent on the presence of a full-length CTD, since transcription with pol II 1-15 or pol II-Δ did not support a PSF-dependent increase in splicing activity (lanes 1, 2, 4, and 5). Taken together with the results described above demonstrating interactions between PSF and both activators and the CTD, the results with the pSVEDaGal5HIV2 reporter also support a role for PSF in mediating strong activator-dependent stimulation of splicing activity through interactions that require the CTD and suggest that this mechanism may preferentially act on 5′-end-most introns.
DISCUSSION
This study addresses the factor requirements and mechanism responsible for the stimulation of pre-mRNA processing by a strong promoter-bound transcription activator and by the RNA polymerase II CTD. Evidence was obtained that PSF mediates the ability of a strong activator such as VP16 to promote efficient cotranscriptional splicing and 3′-end cleavage. Several observations support this role for PSF. First, PSF, and its partner protein p54nrb/NonO, preferentially bound to the VP16 activation domain rather than the weak activation domain SW6. Second, elevated expression levels in vivo of PSF (but not p54nrb/NonO) bypassed the requirement for VP16 to facilitate efficient splicing and 3′-end cleavage; this suggests that PSF is normally present at limiting concentrations for the processing of the two different reporter pre-mRNAs analyzed in this study and that a strong activator can function to overcome this rate-limiting step. Third, PSF, like strong activators, required the pol II CTD to stimulate splicing and 3′-end cleavage; transcription with a truncated CTD consisting of 15 heptapeptide repeats, which unlike the wild-type CTD did not support binding of PSF or p54nrb/NonO, resulted in the loss of both activator- and PSF-dependent stimulation of pre-mRNA processing. Fourth, domains in PSF that were critical for the stimulation of splicing were also important for its association with a strong activator and the CTD. Finally, increased levels of expression of PSF, like strong activators, appeared to preferentially stimulate the splicing of the first intron of a multi-intron substrate. These results thus provide insights into how the transcription machinery can positively influence pre-mRNA processing levels and suggest a mechanism for how transcription and pre-mRNA processing levels are coordinated in the cell.
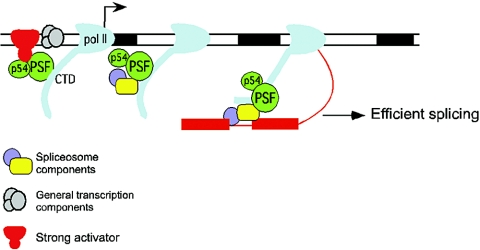
Taken together, our results suggest a model (Fig. 6) in which a promoter-bound activator facilitates splicing, and more indirectly 3′-end cleavage, by the recruitment of PSF to promoters. Initially, a promoter-bound activator binds PSF and p54nrb/NonO, either directly or in the context of a holoenzyme complex containing the initiating form of pol II. PSF and p54nrb/NonO associate with the CTD of the initiating pol II and travel with pol II as it synthesizes nascent pre-mRNA. The association of PSF and p54nrb/NonO subsequently facilitates the assembly of splicing complexes and splicing on the first intron to emerge. A strong activator bound to a promoter thus functions to increase the local concentration of PSF, such that efficient splicing results. Such a mechanism could afford the ability to coordinate transcription and pre-mRNA processing levels. For example, it is interesting that the preferential increase in splicing of first introns might be sufficient to establish a more active “processing state” on the nascent pre-mRNA. The assembly of splicing complexes on first introns may facilitate subsequent splicing of downstream introns by communication across adjacent splice sites, as envisaged by the exon definition model of Berget (5). Moreover, the assembly of splicing components on nascent RNA, at least in vitro, has been shown to reciprocally stimulate transcription elongation (22), such that efficient processing of the nascent RNA promoted by activators and PSF may also ensure subsequent increased production of full-length transcripts.
FIG. 6.
A model for the role of PSF in activator- and pol II CTD-dependent stimulation of pre-mRNA processing. Strong transcriptional activators bound to a promoter recruit PSF and p54nrb/NonO, possibly within the context of a pol II holoenzyme complex. PSF and/or p54nrb/NonO associates with the pol II CTD, and this association is maintained, at least initially, during transcription elongation and facilitates the assembly of splicing complexes on the nascent RNA, thereby resulting in the stimulation of splicing. This mechanism may apply specifically to the stimulation of splicing of first introns. Such a mechanism could in turn facilitate the increased processing of downstream introns (i.e., via exon-definition-type interactions) as well as 3′ processing of a proximal 3′-end cleavage and polyadenylation signal.
We have also obtained evidence that activator- and CTD-dependent stimulation of pre-mRNA processing via PSF may operate in the context of different promoters and in the context of activation of transcription of endogenous transcripts. For example, the CTD was required for PSF-dependent stimulation of splicing and 3′-end cleavage of the dsx reporter pre-mRNA used in the present study, when transcribed from the adenovirus major late promoter (E. Rosonina and B. J. Blencowe, unpublished). Moreover, increased expression of the cell cycle-regulated activator E2F1 can function to increase specifically the efficiency of splicing of intron 1, but not of introns 2 and 3, of endogenous transcripts from the human TP73 gene (H. Siu and B. J. Blencowe, unpublished). However, establishing a role for PSF in this activity was hampered by the difficulty in reducing its level efficiently in vivo using small interfering RNA-mediated knockdown. Nevertheless, these experiments suggest that activator- and CTD-dependent pre-mRNA processing, via a mechanism involving PSF, may play a more general role in the coordination of transcription and pre-mRNA processing levels. The results also indicate that PSF can function via interactions with different components of the transcription machinery, including activators and the pol II CTD, although they do not distinguish whether these interactions are coordinated or act independently (see below).
Our results add to a growing body of evidence indicating that transcriptional activators and coactivators can modulate pre-mRNA processing levels of target genes. Altering promoter architecture, either by swapping promoters or by tethering different activators to the promoters of transfected reporter genes, has been shown to modulate the levels of inclusion of alternative exons and to promote the constitutive splicing levels of 5′-end-most introns (3, 13, 48, 55). Two different mechanisms, which are not mutually exclusive, were proposed to explain these effects. In one model, activators increase the rate of elongation of pol II, which kinetically favors the simultaneous exposure of distal splice sites located upstream and downstream of an alternative exon (35). If the distal splice sites are stronger than the splice sites flanking the alternative exon, then their simultaneous exposure results in skipping of the alternative exon. Consistent with this model, conditions which reduce the elongation rate of pol II, including drug treatments and a pol II mutation that slows processivity, allow sufficient time for factors to associate with the relatively weak splice sites flanking the alternative exons, prior to exposure of the downstream strong 3′ splice site, thereby favoring inclusion of the alternative exon (15, 30). The extent to which such a mechanism operates in vivo in the context of endogenous transcripts is not known.
In the second model, which is supported by the results in the present study, binding of activators to promoters facilitates, either directly or indirectly, the recruitment of one or more rate-limiting pre-mRNA processing factors, which are then made available to the nascent RNA to facilitate processing. Previous studies have provided support for a role for transcriptional activators and coactivators in modulating the levels of processing of nascent RNA (see the introduction). For example, SR family proteins, which contain a domain rich in alternative arginine and serine residues (RS domain) and function in constitutive and alternative splicing, were found to associate with an RS-rich “loop” region in the epidermodysplasia verruciformis-associated human papillomavirus E2 protein (36). Activation of transcription of a pre-mRNA reporter containing a promoter with E2 binding sites using a mutant version of the epidermodysplasia verruciformis-associated human papillomavirus E2 protein lacking the RS loop region resulted in the reduced efficiency of splicing of the resulting RNA. Similarly, the PCG-1 transcriptional coactivator, which is involved in the regulation of genes involved in adaptive thermogenesis, contains an RS domain that interacts with SR family proteins and has been shown to affect the alternative splicing of pre-mRNAs transcribed from PCG-1-responsive pre-mRNA reporters (46). Other studies have also revealed functional associations between SR proteins and SR-related splicing components with transcriptional activator or coactivator proteins (18, 24).
PSF and p54nrb/NonO are known to associate with each other as well as with both single- and double-stranded nucleic acids. Both proteins are thought to be multifunctional since they have been identified in complexes that operate in different cellular processes, including splicing and transcription (61) (see the introduction). As mentioned above, PSF has been shown to be important for splicing in vitro. Its depletion from nuclear extracts reduces the efficiency of splicing of exogenously added pre-mRNAs, and activity can be restored by addition of recombinant protein (27, 50). PSF was originally identified as a factor that associates with the polypyrimidine tract binding protein, which has important roles in the regulation of alternative splicing (67). More recently, PSF and p54nrb/NonO were found to bind to the conserved stem 1b in U5 snRNA (51). Moreover, PSF and p54nrb/NonO have been shown to associate with a large multisubunit complex containing splicing and transcription factors, and p54nrb/NonO has been found to cross-link to a 5′-splice site oligonucleotide in this complex (31). Evidence was also presented in this study that both proteins associate with elongating transcription complexes assembled on biotinylated templates in vitro (31). Additional associations with the transcription machinery have been reported in studies implicating PSF and p54nrb/NonO in the negative regulation of nuclear receptor-activated transcription, by association with corepressor complexes (42, 64).
Our recent work demonstrated that PSF and p54nrb/NonO can bind directly to the pol II CTD and facilitate binding of RNA to this structure (21). This suggested a role for these proteins in tethering nascent RNP complexes to the CTD in order to facilitate their assembly into functional splicing complexes. Taken together with the results in the present study, a picture emerges in which PSF and p54nrb/NonO forge multiple interactions with the transcription machinery. PSF may associate initially with activators and/or coregulators, then subsequently with the pol II CTD, and ultimately with nascent pre-mRNA processing complexes (Fig. 6). However, as discussed above, these interactions need not be interdependent. Based on the analysis of the function of different domains in PSF in the present study, it is possible that the two highly conserved RRM domains support a tethering function for PSF. RRM1 appears to be more important for the association of PSF with activators, whereas RRM2 is more important for its association with the CTD. It is possible therefore that, following its initial recruitment by activators via RRM1, PSF binds to the CTD via RRM2. Following promoter clearance and loss of the association with the promoter-bound activator, RRM1 may become available for subsequent interactions with nascent RNP complexes. The mechanism by which PSF associates with activators and the CTD via its RRMs is not clear, although previous evidence indicating that the CTD can bind RNA (21, 58) suggests that RNA may play a role in stabilizing this interaction. Growing evidence also indicates a role for RRMs and flanking amino acid residues in the formation of protein-protein interactions (32), and it is therefore interesting to consider that these domains in PSF may function in protein-protein as well as protein-RNA interactions. Such a role for the RRMs would be consistent with the previously proposed roles for multifunctional RNA binding proteins such as PSF and p54nrb/NonO in chaperonin-type activities (21, 66), whereby their activity in facilitating cotranscription pre-mRNA processing could arise from their ability to confer altered RNA/protein folding conformations that favor the assembly of pre-mRNA processing complexes. Our identification in the present study of PSF as a rate-limiting factor for mediating activator- and CTD-dependent stimulation of pre-mRNA processing provides new insights into one possible mechanism by which transcription and pre-mRNA processing levels are coordinated within the cell.
Acknowledgments
We thank David Bentley, Jack Greenblatt, Alberto Kornblihtt, Jim Patton, and Andrew Philips for providing clones, antisera, and cell lines.
E.R. and J.Y.Y.I. were the recipients of National Cancer Institute of Canada (NCIC) and University of Toronto Open Studentships, respectively. This work was supported by an NCIC operating grant and Premier's Research Excellence Award to B.J.B.
REFERENCES
- 1.Allison, L. A., and C. J. Ingles. 1989. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc. Natl. Acad. Sci. USA 86:2794-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, L. A., M. Moyle, M. Shales, and C. J. Ingles. 1985. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell 42:599-610. [DOI] [PubMed] [Google Scholar]
- 3.Auboeuf, D., A. Honig, S. M. Berget, and B. W. O'Malley. 2002. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416-419. [DOI] [PubMed] [Google Scholar]
- 4.Bensaude, O., F. Bonnet, C. Casse, M. F. Dubois, V. T. Nguyen, and B. Palancade. 1999. Regulated phosphorylation of the RNA polymerase II C-terminal domain (CTD). Biochem. Cell Biol. 77:249-255. [PubMed] [Google Scholar]
- 5.Berget, S. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 6.Berk, A. J. 1999. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 11:330-335. [DOI] [PubMed] [Google Scholar]
- 7.Blau, J., H. Xiao, S. McCracken, P. O'Hare, J. Greenblatt, and D. Bentley. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16:2044-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blencowe, B. J., J. A. Nickerson, R. Issner, S. Penman, and P. A. Sharp. 1994. Association of nuclear matrix antigens with exon-containing splicing complexes. J. Cell Biol. 127:593-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cagney, G., and A. Emili. 2002. De novo peptide sequencing and quantitative profiling of complex protein mixtures using mass-coded abundance tagging. Nat. Biotechnol. 20:163-170. [DOI] [PubMed] [Google Scholar]
- 10.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 12.Corden, J. L., D. L. Cadena, J. M. Ahearn, Jr., and M. E. Dahmus. 1985. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc. Natl. Acad. Sci. USA 82:7934-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer, P., C. G. Pesce, F. E. Baralle, and A. R. Kornblihtt. 1997. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 15.de la Mata, M., C. R. Alonso, S. Kadener, J. P. Fededa, M. Blaustein, F. Pelisch, P. Cramer, D. Bentley, and A. R. Kornblihtt. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12:525-532. [DOI] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. D. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, B., D. S. Horowitz, R. Kobayashi, and A. R. Krainer. 1993. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein. Nucleic Acids Res. 21:4085-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowhan, D. H., E. P. Hong, D. Auboeuf, A. P. Dennis, M. M. Wilson, S. M. Berget, and B. W. O'Malley. 2005. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell 17:429-439. [DOI] [PubMed] [Google Scholar]
- 19.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 20.Dye, B. T., and J. G. Patton. 2001. An RNA recognition motif (RRM) is required for the localization of PTB-associated splicing factor (PSF) to subnuclear speckles. Exp. Cell Res. 263:131-144. [DOI] [PubMed] [Google Scholar]
- 21.Emili, A., M. Shales, S. McCracken, W. Xie, P. W. Tucker, R. Kobayashi, B. J. Blencowe, and C. J. Ingles. 2002. Splicing and transcription-associated proteins PSF and p54nrb/nonO bind to the RNA polymerase II CTD. RNA 8:1102-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong, Y. W., and Q. Zhou. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414:929-933. [DOI] [PubMed] [Google Scholar]
- 23.Fox, A. H., Y. W. Lam, A. K. Leung, C. E. Lyon, J. Andersen, M. Mann, and A. I. Lamond. 2002. Paraspeckles. A novel nuclear domain. Curr. Biol. 12:13-25. [DOI] [PubMed] [Google Scholar]
- 24.Ge, H., Y. Si, and A. P. Wolffe. 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell 2:751-759. [DOI] [PubMed] [Google Scholar]
- 25.Gerber, H. P., M. Hagmann, K. Seipel, O. Georgiev, M. A. West, Y. Litingtung, W. Schaffner, and J. L. Corden. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374:660-662. [DOI] [PubMed] [Google Scholar]
- 26.Gill, G. 2001. Regulation of the initiation of eukaryotic transcription. Essays Biochem. 37:33-43. [DOI] [PubMed] [Google Scholar]
- 27.Gozani, O., J. G. Patton, and R. Reed. 1994. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 13:3356-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 29.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kadener, S., J. P. Fededa, M. Rosbash, and A. R. Kornblihtt. 2002. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. USA 99:8185-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kameoka, S., P. Duque, and M. M. Konarska. 2004. p54(nrb) associates with the 5′ splice site within large transcription/splicing complexes. EMBO J. 23:1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiesler, E., F. Miralles, A. K. Ostlund Farrants, and N. Visa. 2003. The Hrp65 self-interaction is mediated by an evolutionarily conserved domain and is required for nuclear import of Hrp65 isoforms that lack a nuclear localization signal. J. Cell Sci. 116:3949-3956. [DOI] [PubMed] [Google Scholar]
- 34.Kim, E., L. Du, D. Bregman, and S. Warren. 1997. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J. Cell Biol. 136:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornblihtt, A. R., M. de la Mata, J. P. Fededa, M. J. Munoz, and G. Nogues. 2004. Multiple links between transcription and splicing. RNA 10:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, M. C., B. H. Teh, and W. Y. Tarn. 1999. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 274:11832-11841. [DOI] [PubMed] [Google Scholar]
- 37.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 38.Lerner, E. A., M. R. Lerner, L. A. Janeway, and J. A. Steitz. 1981. Monoclonal antibodies to nucleic acid containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA 78:2737-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis, B. A., and D. Reinberg. 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116:3667-3675. [DOI] [PubMed] [Google Scholar]
- 40.Makela, T. P., J. D. Parvin, J. Kim, L. J. Huber, P. A. Sharp, and R. A. Weinberg. 1995. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc. Natl. Acad. Sci. USA 92:5174-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 42.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997b. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 45.McCracken, S., M. Lambermon, and B. J. Blencowe. 2002. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22:148-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307-316. [DOI] [PubMed] [Google Scholar]
- 47.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nogues, G., S. Kadener, P. Cramer, D. Bentley, and A. Kornblihtt. 2002. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 277:43110-43114. [DOI] [PubMed] [Google Scholar]
- 49.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 270:3859-3870. [DOI] [PubMed] [Google Scholar]
- 50.Patton, J. G., E. B. Porro, J. Galceran, P. Tempst, and B. Nadal-Ginard. 1993. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7:393-406. [DOI] [PubMed] [Google Scholar]
- 51.Peng, R., B. T. Dye, I. Perez, D. C. Barnard, A. B. Thompson, and J. G. Patton. 2002. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 8:1334-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 53.Ranish, J. A., and S. Hahn. 1996. Transcription: basal factors and activation. Curr. Opin. Genet. Dev. 6:151-158. [DOI] [PubMed] [Google Scholar]
- 54.Robert, F., M. Blanchette, O. Maes, B. Chabot, and B. Coulombe. 2002. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 277:9302-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosonina, E., M. A. Bakowski, S. McCracken, and B. J. Blencowe. 2003. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 278:43034-43040. [DOI] [PubMed] [Google Scholar]
- 56.Rosonina, E., and B. J. Blencowe. 2004. Analysis of the requirement for RNA polymerase II CTD heptapeptide repeats in pre-mRNA splicing and 3′-end cleavage. RNA 10:581-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roth, M. B., C. Murphy, and J. G. Gall. 1990. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J. Cell Biol. 111:2217-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan, K., K. G. Murthy, S. Kaneko, and J. L. Manley. 2002. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 22:1684-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Scafe, C., D. Chao, J. Lopes, J. P. Hirsch, S. Henry, and R. A. Young. 1990. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature 347:491-494. [DOI] [PubMed] [Google Scholar]
- 61.Shav-Tal, Y., and D. Zipori. 2002. PSF and p54(nrb)/NonO-multi-functional nuclear proteins. FEBS Lett. 531:109-114. [DOI] [PubMed] [Google Scholar]
- 62.Straub, T., P. Grue, A. Uhse, M. Lisby, B. R. Knudsen, T. O. Tange, O. Westergaard, and F. Boege. 1998. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J. Biol. Chem. 273:26261-26264. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, N. E., D. B. Aronson, and R. R. Burgess. 1990. Purification of eukaryotic RNA polymerase II by immunoaffinity chromatography. Elution of active enzyme with protein stabilizing agents from a polyol-responsive monoclonal antibody. J. Biol. Chem. 265:7069-7077. [PubMed] [Google Scholar]
- 64.Urban, R. J., Y. Bodenburg, A. Kurosky, T. G. Wood, and S. Gasic. 2000. Polypyrimidine tract-binding protein-associated splicing factor is a negative regulator of transcriptional activity of the porcine p450scc insulin-like growth factor response element. Mol. Endocrinol. 14:774-782. [DOI] [PubMed] [Google Scholar]
- 65.Vincent, M., P. Lauriault, M. Dubois, S. Lavoie, O. Bensaude, and B. Chabot. 1996. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 24:4649-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Virbasius, C. M., S. Wagner, and M. R. Green. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4:219-228. [DOI] [PubMed] [Google Scholar]
- 67.Wagner, E. J., and M. A. Garcia-Blanco. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wahle, E., and W. Keller. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21:247-250. [PubMed] [Google Scholar]
- 69.Walker, S., R. Greaves, and P. O'Hare. 1993. Transcriptional activation by the acidic domain of Vmw65 requires the integrity of the domain and involves additional determinants distinct from those necessary for TFIIB binding. Mol. Cell. Biol 13:5233-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, Y. S., J. H. Hanke, L. Carayannopoulos, C. M. Craft, J. D. Capra, and P. W. Tucker. 1993. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 13:5593-5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yankulov, K., J. Blau, T. Purton, S. Roberts, and D. L. Bentley. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749-759. [DOI] [PubMed] [Google Scholar]
- 72.Yeakley, J. M., J. P. Morfin, M. G. Rosenfeld, and X. D. Fu. 1996. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc. Natl. Acad. Sci. USA 93:7582-7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zavolan, M., S. Kondo, C. Schonbach, J. Adachi, D. A. Hume, Y. Hayashizaki, and T. Gaasterland. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13:1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, W. W., L. X. Zhang, R. K. Busch, J. Farres, and H. Busch. 1993. Purification and characterization of a DNA-binding heterodimer of 52 and 100 kDa from HeLa cells. Biochem. J. 290:267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]