Abstract
R2 elements are non-long terminal repeat retrotransposons that specifically insert into 28S rRNA genes of many animal groups. These elements encode a single protein with reverse transcriptase and endonuclease activities as well as specific DNA and RNA binding properties. In this report, gel shift experiments were conducted to investigate the stoichiometry of the DNA, RNA, and protein components of the integration reaction. The enzymatic functions associated with each of the protein complexes were also determined, and DNase I digests were used to footprint the protein onto the target DNA. Additionally, a short polypeptide containing the N-terminal putative DNA-binding motifs was footprinted on the DNA target site. These combined findings revealed that one protein subunit binds the R2 RNA template and the DNA 10 to 40 bp upstream of the insertion site. This subunit cleaves the first DNA strand and uses that cleavage to prime reverse transcription of the R2 RNA transcript. Another protein subunit(s) uses the N-terminal DNA binding motifs to bind to the 18 bp of target DNA downstream of the insertion site and is responsible for cleavage of the second DNA strand. A complete model for the R2 integration reaction is presented, which with minor modifications is adaptable to other non-LTR retrotransposons.
While originally viewed as the unique property of retroviruses, the reverse transcription of RNA templates is now known to be a mechanism used by many eukaryotic mobile elements. One class of elements, frequently referred to as the LTR retrotransposons because they contain long terminal repeats, utilize the same replication mechanism as retroviruses (reviewed in reference 33). Reverse transcription of the RNA template is usually primed by the 3′ end of a tRNA annealed to the template. Full-length first and second DNA strands are made from the RNA template by the polymerase using the terminal repeats to jump from one end of the template to the other. The linear DNA product generated by reverse transcription is then inserted into chromosomal sites by an integrase.
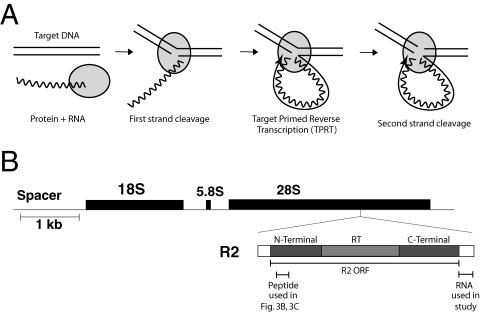
A second class of elements, usually referred to as the non-LTR retrotransposons because they lack terminal repeats, uses a different mechanism of integration. In this mechanism the chromosomal DNA target site is cleaved by an endonuclease, and the 3′ end generated by this cleavage is used to prime the reverse transcription directly onto the DNA target (Fig. 1A). This target-primed reverse transcription, or TPRT mechanism, has been most comprehensively documented for the R2 element of Bombyx mori (22), but in vitro and in vivo assays involving other elements are consistent with the basic features of the TPRT model (7, 11, 25, 32). One side effect of not requiring precise terminal repeats in any step of the reaction is that the reverse transcriptase of non-LTR retrotransposons is able to reverse transcribe other cellular RNA templates. Thus, the TPRT mechanism has been shown to generate short interspersed nuclear element (e.g., Alu) insertions as well as processed pseudogenes (12, 13, 17, 34).
FIG. 1.
R2 elements and the TPRT mechanism. (A) Diagram of the R2 TPRT mechanism. RNA corresponding to the 3′-UTR RNA of the R2 element and the R2 protein form a specific RNP complex. The RNP complex binds the target DNA and cleaves the first (bottom) strand. The 3′ DNA end generated by this nick is used to prime first-strand cDNA. Second (top)-strand cleavage occurs after reverse transcription. (B) R2 elements insert into a specific site within the 28S rRNA gene. R2 elements encode a 120-kDa protein containing an N-terminal domain with cysteine-histidine zinc fingers and c-Myb DNA binding motifs, a central reverse transcriptase domain (RT), and a C-terminal domain with a restriction-like-endonuclease domain and putative nucleic acid binding motif(s). Indicated below the R2 diagram is the 3′-UTR sequence used as R2 RNA in this study and the N-terminal peptide used for Fig. 3B and C.
The TPRT mechanism of insertion used by non-LTR retrotransposons may have originated with the mobile group II introns of bacteria. The TPRT mechanism utilized by group II introns differs from that of the non-LTR retrotransposons in that insertion is initiated by the RNA template reverse splicing into the chromosomal DNA target site. However, similar to the non-LTR mechanism, the DNA strand of the target site which is used for TPRT is cleaved by an element-encoded endonuclease (40). It has also been suggested that the TPRT mechanism of non-LTR retrotransposons may have originated with telomerase (26). The ability of telomerase to reverse transcribe a short RNA sequence onto a chromosome end shows striking similarity to TPRT.
Many questions remain concerning the mechanism of a complete TPRT reaction. How do the proteins interact with the target DNA? Does the element reverse transcriptase make the second DNA strand? R2 remains one of the most attractive model systems for studying this mechanism, because the enzyme encoded by this element is highly specific both for the DNA target site and for the RNA that is used for reverse transcription. The element inserts into a unique sequence of the 28S rRNA genes (Fig. 1B). All the initial steps of the TPRT reaction can occur in vitro with bacterially expressed protein that specifically binds the 60-bp target site (9) and efficiently reverse transcribes only R2 RNA (21). In this report we present evidence that a complete TPRT complex involves two R2 protein subunits. The first subunit binds upstream of the cleavage site and is responsible for the initial cleavage and reverse transcription step, while the second subunit binds downstream and is responsible for second-strand cleavage.
MATERIALS AND METHODS
Protein purification and nucleic acid preparation.
R2 protein of Bombyx mori was purified and stored as described previously (9). The DNA sequence corresponding to codons 89 to 229 of the R2 open reading frame (ORF) were PCR amplified with primers 5′-GGGAATTCCATATGCGAACAGGCGATAACCCGACTGTGCGAGGTTCC-3′ and 5′-CGCGGATCCTTAGCTAGGCTCGGCCGAGCAC-3′. The sense primer NdeI site and antisense primer BamHI site were used to clone the fragment into the expression vector pET28a (Novagen). The expression construct was transformed into BL21(DE3)-codon-plus bacteria (Stratagene) for expression. Cells were grown in 200 ml of LB at 37° on a shaker until an A600 of 0.6, and the cells were cooled, induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside, and grown at room temperature until an A600 of 1.2. The cells were spun down and suspended in 3 ml of loading buffer (50 mM HEPES [pH 7.5], 300 mM NaCl, 0.05 mM ZnCl2, 0.2% Triton 100, 1 mM beta-mercaptoethanol, 10% glycerol, 5 mM imidizole). Fifty micrograms of lysozyme/milliliter, 20 units DNase I, and 5 mM MgCl2 were added to the resuspended cells, and the cells were incubated at 37°C for 10 min and then at 22°C for 10 min. Cells were put on ice and then lysed further by sonication. The lysate was cleared twice by centrifugation at 13,000 × g for 15 min at 4°C. The peptide was affinity purified from the final soluble fraction on a 150-μl bed volume of Talon resin (Clonetech). The column was washed with a series of increasingly stringent 1-ml washes (loading buffer, 0.5 M NaCl, 15 mM imidazole, 30 mM imidazole, and 60 mM imidazole). The peptide was eluted with 0.3 ml loading buffer with 150 mM imidazole. The eluate was adjusted to 0.5× elution buffer, 50% glycerol, and 2 mM dithiothreitol and stored at −20°C.
R2 3′ untranslated region (UTR) RNA was made by in vitro transcription as described previously (9). 32P-labeled 3′-UTR RNA was made by treating the RNA with calf intestinal phosphatase (Gibco BRL), and the reaction was stopped by heat denaturation (95°C), and then 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase (Fermentas).
The 184-bp target DNA substrate was generated by PCR using primers that annealed to sites approximately 90 bp to either side of the R2 target site (9). The 60-bp and 100-bp DNA substrates were made by annealing complementary oligonucleotides. The 60-bp substrate spanned from 42 bases upstream of the R2 insertion site to 18 bases downstream on the insertion site. The 100-bp substrates spanned either from 50 bp upstream to 50 bp downstream (used to footprint the bottom strand) or from 70 bp upstream to 30 bp downstream (used to footprint the top strand).
The DNA substrates were 5′ end labeled on either the top or bottom strand by treating 20 pmol of the appropriate primer with 70 μCi γ-ATP (Perkin-Elmer/Life-Science, 6,000 mCi/mMol) and 10 units T4 polynucleotide kinase (Fermentas) for 1 h at 37°C in 30-μl reactions. The reactions were terminated by heating to 65°C for 15 min. The labeled primer was then annealed to a complementary primer (60-bp and 100-bp substrates) or used in a PCR with a pairing primer (184-bp substrate). The DNA substrates were then gel purified and eluted as previously described (9). The final pellet was dissolved in 80 μl 10 mM Tris-HCl (pH 8)-0.5 mM EDTA to a concentration of 60 fmol/μl. The nonspecific competitor poly(dIdC) was added to 25 μg/ml, and the solution was stored at −20°C.
R2 reactions and gel electrophoresis mobility shift assay (EMSA).
Unless otherwise noted, all binding, cleavage, and TPRT reactions were 13 μl and contained ∼80 fmol labeled substrate DNA, 40 fmol R2 protein, 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.1 mg/ml bovine serum albumin, 0.01% Triton X-100, and 10 to 12% glycerol. In addition, either 1.2 pmol of R2 3′-UTR RNA or 1 μg of RNase A was present. TPRT reactions contained 25 μM of each deoxynucleoside triphosphate (dNTP). Reactions were assembled and allowed to preincubate at 25°C for 5 min to allow the RNA to bind to the R2 protein or to allow the RNase A to digest any contaminating RNA. DNA binding was started by the addition of substrate DNA and continued at 37°C for 30 min. The reactions were chilled on ice prior to loading onto 5% native (1× Tris-borate-EDTA) polyacrylamide gels. Gels (20 cm by 20 cm) were run for 1 h at 350 V in a cold room (4°C). Gels were dried and exposed to a phosphorimager screen. Gels were exposed wet (at 4°C) to X-ray film in cases where the complexes were to be isolated for further analysis.
Analysis of complexes.
To determine what steps in the TPRT reaction had occurred with the various protein-DNA complexes, bands from EMSA gels were eluted by crushing gel pieces and soaking them in a solution containing 0.3 M sodium acetate (pH 5.2), 1 mM EDTA, and 0.1% sodium dodecyl sulfate. The DNA was ethanol precipitated, redissolved in 85% formamide, 1× Tris-borate-EDTA, and 50 ng sheared calf thymus DNA, and analyzed on a denaturing 6% acrylamide gel.
For DNase I footprints, a fivefold scaleup of a typical reaction with regard to DNA, RNA, and protein amounts was carried out in a 35-μl-volume reaction. To prevent DNA cleavage, the single-amino-acid mutation D966A protein, which is completely deficient in endonuclease activity (EN−), was used (37). Twenty-five μM of dCTP was present in the binding reaction. One unit of DNase I (Promega) was added and incubated for 2 min at room temperature. The reaction was stopped on ice, and the reaction product was directly loaded onto an EMSA gel to separate the bound DNA complexes. Isolated EMSA complexes were analyzed on a denaturing 6.5% polyacrylamide sequencing gel.
In the sequential addition reactions, the first subunit, either mutant or wild type, was added to the threshold where all of the DNA substrate had been bound. The D996A mutation was used for the EN− protein, and the D628Y mutation was used for the reverse transcriptase-deficient protein (37). After an initial incubation period, the second subunit was added at a concentration five times higher than that of the first protein and allowed to incubate for a second period. The reactions were analyzed both by denaturing gels and by EMSA gels.
RESULTS
Protein-DNA complexes in the presence and absence of RNA.
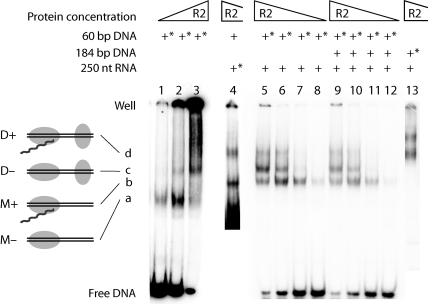
Figure 2 shows DNA EMSA with full-length R2 protein conducted in the presence or absence of RNA. For maximum resolution, the DNA target in these assays was only 60 bp, extending from 42 bp upstream to 18 bp downstream of the 28S rRNA gene insertion site. The RNA corresponded to the 250-nucleotide (nt) 3′-UTR of the R2 element and is the minimum RNA that is specifically bound and efficiently utilized by the R2 protein in a TPRT reaction (21). All incubations were conducted in the absence of DNA cleavage, either by removing Mg2+ from the assay or by the use of an endonuclease mutant protein (EN−) that lacks the ability to cleave DNA but maintains both reverse transcriptase and DNA binding activities (9, 37).
FIG. 2.
EMSA of the R2 protein and target DNA in the presence and absence of RNA. The triangles above the lanes represent the relative protein concentrations between the various lanes (12 to 360 fmol/reaction). The DNA and RNA substrates used for a given reaction are indicated above each lane. An asterisk indicates that the substrate was 32P labeled. The R2 protein was allowed to bind to substrate DNA at 37°C for 15 min (lanes 4 to 13) or 30 min (lanes 1 to 3). DNA cleavage was prevented by the presence of EDTA (lanes 1 to 3) or by using an endonuclease-mutant R2 protein (lanes 4 to 13). Shifted complexes, a, b, c, and d, in the remainder of this report are referred to as follows: M−, protein monomer without RNA; M+, monomer with RNA; D−, protein dimer without RNA; and D+, dimer with RNA. The structures of each complex based on the footprint studies shown in Fig. 3 are diagramed to the left of the gel.
In the absence of RNA, a single complex was formed at low protein-to-DNA ratios (Fig. 2A, lane 1, labeled a). At higher protein-to-DNA ratios, much of the DNA was shifted into a complex that remained trapped within the well of the gel (lanes 2 and 3); however, a small fraction of the protein could be seen migrating as a second distinct complex, labeled c. The matrix of protein and DNA stuck in the wells appears to involve specific protein-DNA interactions, because it gives rise to specific footprints and its formation is not inhibited by nonspecific competitor DNA (9). The amount of material stuck in the wells was reduced in Fig. 2 by conducting the electrophoresis after a 30-min preincubation at 37°C in the absence of Mg2+ (9).
In the presence of RNA, three distinct migrating complexes were formed (lanes 5 to 8). One of these complexes migrated at the same position as complex c formed in the absence of RNA. To determine if the other two complexes (labeled b and d) contained RNA, the incubation conditions in lane 5 were repeated, but instead of labeling the DNA target, the RNA template was labeled (lane 4). Comparison of lanes 4 and 5 confirmed that the middle complex did not contain RNA and was thus the same as complex c formed in the absence of RNA, while complexes b and d did contain RNA.
The formation of complexes c and d at higher protein concentrations could be the result of the association of more protein subunits in the complex and/or the involvement of additional DNA or RNA substrates. The possibility of multiple DNA substrates was a concern, because the R2 protein has the ability to form a DNA/protein matrix in the absence of RNA (Fig. 2, lane 3). Also, as will be described in Discussion, the R2 protein has similarities to type IIs restriction enzymes. These restriction enzymes can form protein dimers when monomers are bound to separate DNA binding sites. In order to address whether multiple DNA substrates were present in the complexes observed in the presence of RNA, the R2 protein was incubated with an equal molar mixture of two different-size DNA substrates: the labeled 60-bp DNA and an unlabeled 184-bp substrate also containing the R2 recognition site (Fig. 2, lanes 9 to 12). Previous work has shown that the 60-bp and 184-bp DNA targets are bound equally well by the R2 protein (9). For reference, the migration position of the R2 complexes formed on a labeled 184-bp DNA substrate is shown in lane 13. No new complexes migrating at novel positions appeared in the incubations containing both DNA substrates (compare lanes 5 to 8 and lanes 9 to 12), suggesting that there was only one DNA molecule present in complexes b, c, and d.
Experiments were also conducted to address whether multiple RNA substrates were present in the R2 protein complexes. In an incubation with the labeled 250-nt RNA template, the mobilities of the shifted complexes were unaffected by the presence of a second unlabeled RNA 500 nt in length (data not shown). Therefore, complexes b and d seen in Fig. 2 also contained a single RNA substrate.
These results suggest that the higher-order complexes formed at increasing ratios of R2 protein to its DNA target were a result of additional protein subunits binding to a single DNA target and RNA template. In an earlier study, we showed that the complexes, which formed at low protein ratios in the presence or absence of RNA, had the same number of protein subunits (9). Several independent experiments have suggested that these complexes are monomers. First, in the absence of RNA the stoichiometry of R2 protein subunits to bottom-strand cleavage in a single-round reaction corresponds to that of a monomer (36). Second, the protein in the absence of RNA and DNA sediments as a monomer (36). Third, UV cross-linking of the R2 protein to the DNA target is consistent with a single protein subunit in complexes a and b (J. Ye and T. H. Eickbush, unpublished data). For the remainder of this report, we will refer to complexes a and b as the monomer complexes, M+ or M−, depending upon whether they contain RNA. The slower-migrating complexes formed at higher protein ratios (c and d) represent a protein multimer bound to DNA. We will refer to these structures as the dimer complexes, D+ or D−, because of similarities to type IIs restriction enzymes (see Discussion); however, we have no direct evidence that they contain only two protein subunits.
The bipartite binding of the R2 protein to the DNA target.
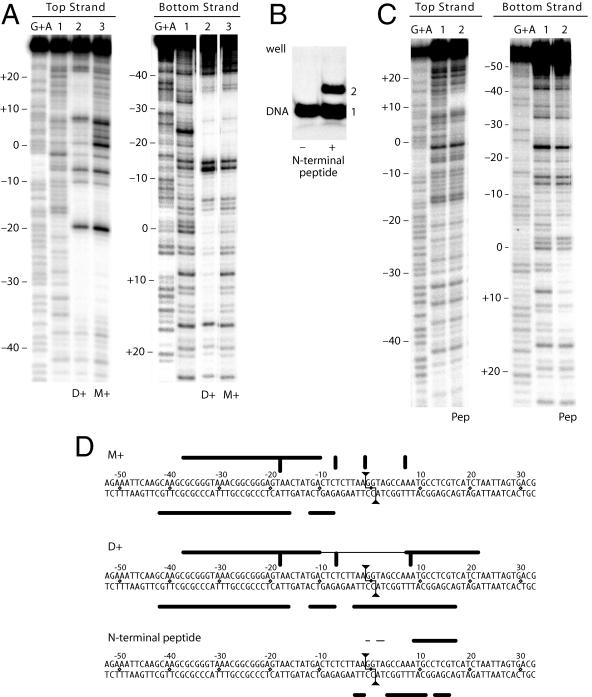
Figure 3A shows the DNase I footprint of excised M+ and D+ complexes of the R2 protein bound to its target DNA. A diagram of this footprint on the DNA sequence is shown in Fig. 3D. The EN− protein has again been used in order to monitor the footprint before cleavage. In the case of the M+ complex, the footprint of the top strand (Fig. 3A, lane 3) extended from −36 to −10 with respect to the cleavage/integration site, and there was a series of hypersensitive sites at −18, −7, +1, and +7. The footprint of the bottom strand (lane 3) was also upstream of the cleavage site from −42 to −7.
FIG. 3.
Bipartite binding of the R2 protein revealed by DNase I footprints. (A) Comparison of the DNase I footprints for the monomer (M+, lane 3) and the dimer (D+, lane 2). The target DNA was 5′end labeled on either the top (left panel) or bottom strand (right panel). The endonuclease mutant was used in order to observe the complexes prior to cleavage. Lane 1, DNase I pattern of naked DNA; lanes labeled G+A, guanosine-plus-adenine ladders. Numbers to the left of the footprint correspond to base positions relative to the R2 cleavage dyad (see panel D). (B) Electrophoresis mobility shift assay in the presence (+) and absence (−) of a 140-amino-acid N-terminal peptide of the R2 protein containing the zinc-finger and c-Myb motifs. Free DNA and bound DNA are labeled 1 and 2, respectively. (C) DNase I footprint of the shifted N-terminal peptide bound to DNA (lane 2) compared to free DNA (lane 1) in each panel. (D) Summary of the R2 protein footprints. The 2-bp staggered cleavage by R2 is indicated by the lines ending in triangles. Nucleotide positions are numbered relative to the dyad cleavage site, with negative numbers corresponding to upstream sequences (relative to transcription of the 28S gene; see Fig. 1B). Thick horizontal lines represent areas of greatest protection by the R2 protein, while thinner horizontal lines indicate weaker protection. Thick vertical lines are DNase I-hypersensitive sites.
The footprint of the D+ complex (Fig. 3A, lanes 2) remained the same as that of the M+ complex upstream of the cleavage site but now also extended downstream of this site. The additional protection of the top strand extended from +7 to +22, while the hypersensitive site at −1 was reduced and the hypersensitive site at +7 was shifted further from the cleavage site. The additional protection of the bottom strand extended from −5 to +17. Thus, binding of the R2 protein to the DNA target was bipartite, with interactions predominately far upstream of the cleavage sites in the M+ complex and both upstream and downstream in the D+ complex. The region surrounding the cleavage site on the top strand remained accessible to DNase I in both complexes. The cleavage region on the bottom strand was accessible to DNase I in the M+ complex but partially protected in the D+ complex.
The single protein encoded by R2 elements from diverse arthropods contains three conserved domains (5). As shown in Fig. 1B, these domains are an N-terminal domain with two putative DNA binding domains (a cysteine-histidine zinc finger and a c-Myb domain), a central reverse transcriptase domain, and a C-terminal domain that contains an endonuclease domain and a cysteine-histidine motif (37). We have attempted to express the three domains of R2 separately. To date the reverse transcriptase and C-terminal domains have not been obtained in a soluble form suitable for either enzymatic or binding assays. However, a 120-amino-acid peptide containing the putative DNA-binding motifs of the N-terminal domain was soluble and readily bound to the target DNA (Fig. 3B). The DNase I footprint of the shifted protein-DNA complex is shown in Fig. 3C, and the area of protection is diagrammed on the target sequence in Fig. 3D. The N-terminal peptide protected the top strand of the target site from +8 to +17 and more weakly from −1 to +3. The footprint of the bottom strand extended from −4 to +16 with gaps from 0 to +2 and at +11. Comparison of the DNase I footprints of this N-terminal peptide with that of the total R2 protein indicated that the N-terminal peptide accounted for most, if not all, of the additional footprint observed in the D+ complex compared to the M+ complex.
These footprint studies suggested that the downstream DNA binding found in the D+ complex was a result of the N-terminal domain of the second protein subunit. Because the DNA sequence and size of the upstream binding site have no similarity to those of the downstream site, and the N-terminal peptide has no affinity for the former, we propose that the major upstream DNA binding is conducted by the C-terminal domain of the protein. Because upstream binding is centered 25 to 30 bp from the cleavage site, this C-terminal domain presumably contains distinct subdomains for specific DNA binding and endonuclease activity.
Endonuclease activities associated with the M+ and D+ complexes.
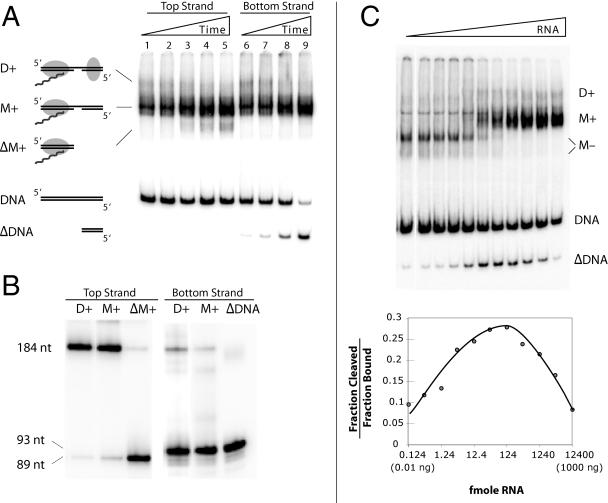
The mobility shift assay shown in Fig. 4A shows the time course of a cleavage reaction in the presence of R2 RNA and the wild-type (i.e., EN+) R2 protein. The 184-bp DNA substrate was 5′ end labeled on either the top (lanes 1 to 5) or the bottom (lanes 6 to 9) strand. Individual bands from the gel shifts were excised, and the DNA was run on a denaturing gel to determine the extent of top and bottom strand cleavage (Fig. 4B). At the protein concentration used in this assay, high levels of the M+ complex and low levels of the D+ complex formed immediately. Both complexes showed nearly complete bottom-strand cleavage (Fig. 4B). Over the 30 min of incubation, the D+ complex decreased in abundance and there was an increase in level of a complex (labeled ΔM+) migrating faster than M+ when the top strand was labeled and faster than the substrate DNA (labeled ΔDNA) when the bottom DNA strand was labeled. These late-appearing bands were correlated with top-strand cleavage (Fig. 4B). These findings are consistent with our original observation that supercoiled DNA containing the target site is rapidly nicked, while double-stranded cleavage occurred slowly over a 30-min reaction (22). The results here indicate that after top-strand cleavage, the R2 protein remained associated with the upstream DNA sequences but released the downstream sequences.
FIG. 4.
Endonuclease activities associated with the M+ and D+ complexes. (A) Time course of R2 binding and cleavage as revealed by EMSA. The 184-bp DNA substrate was 5′ end labeled on either the top (lanes 1 to 5) or bottom strand (lanes 6 to 8). The individual time points were 1, 3, 12, 20, and 30 min at 37°C; the 20-min time point was omitted in the bottom-strand assay. The M+ and D+ complexes are diagramed as in Fig. 2 except that DNA cleavage is indicated by a gap in the DNA. After top-strand cleavage, the R2 protein remains bound to the upstream DNA (ΔM+) but releases the downstream DNA (ΔDNA). (B) The cleavage state of the 184-bp DNA in the various complexes shown in panel A excised and run on a denaturing gel. Equivalent amounts of radioactive counts were loaded onto each lane of the gel. (C) Effects of 3′-UTR RNA concentration on DNA cleavage. Assays were conducted with the target DNA 5′ end labeled on either the top or bottom strand. Each reaction had 12 fmol R2 protein, 20 fmol 184-bp 5′ end-labeled DNA, 8 ng poly(dIdC), and 0.124 to 12400 fmol 3′-UTR RNA. Half of each assay was analyzed by EMSA to determine the fraction of DNA bound by the protein, and the second half was analyzed by denaturing gel electrophoresis to determine the fraction of the DNA cleaved. The EMSA gel (top panel) is from the bottom strand-labeled assay, and the graph (bottom panel) is from the top strand-labeled assay. It should be noted that the M− complex contained two conformations, a predominant slower-migrating form in which the target DNA was cleaved on the bottom strand and a faster-migrating noncleaved form (see reference 9 and Fig. 3 and 7).
Because the first R2 subunit binds RNA while the second subunit does not, we next investigated the degree to which the RNA concentration could be used to manipulate the level of top-strand cleavage. Figure 4C shows an experiment in which the amount of protein and DNA target was held constant while the amount of the 3′-UTR RNA was varied over a wide range. DNA cleavage was allowed for 30 min before electrophoresis (top panel). The level of top-strand cleavage relative to the amount of bound DNA was plotted in the bottom panel. At low RNA concentrations (0.124 fmol/reaction), most of the DNA complexes were in the M− form or the protein/DNA network extending to the top of the gel, and only low levels of top-strand cleavage were observed. Top-strand cleavage (bottom panel, also observed as the ΔDNA band in the top panel) peaked at intermediate RNA concentrations (approximately 124 fmol/reaction), which were also the concentrations at which the ratios of D+ to M+ complexes were the highest. At high RNA concentrations (12,400 fmol/reaction), complex formation was shifted away from the D+ complex towards the M+ complex. This shift away from the D+ complex resulted in lower levels of top-strand cleavage.
The results of Fig. 4 suggested that bottom-strand cleavage occurred under all conditions of R2 binding, while top-strand cleavage efficiently occurred only under conditions where D+ complexes were formed. After both DNA strands were cleaved, the downstream DNA sequences were released from the R2 protein complex.
Reverse transcriptase activity associated with the M+ and D+ complexes.
To determine which protein complex was associated with reverse transcription of the RNA template, TPRT reactions were conducted as a function of protein concentration. These TPRT reactions differ from the previous cleavage reactions only by the addition of dNTPs. Shown in Fig. 5A are the results of a series of TPRT reactions with the top or bottom DNA strand end labeled and the products separated on either an EMSA or a denaturing gel. The graph plots four activities: the fraction of DNA at each protein concentration that was bound by protein, cleaved on the bottom strand, cleaved on the top strand, or had undergone TPRT. Protein binding, closely followed by bottom-strand cleavage, rapidly increased with R2 protein concentration. TPRT and top-strand cleavage were less efficient but also increased with protein concentration.
FIG. 5.
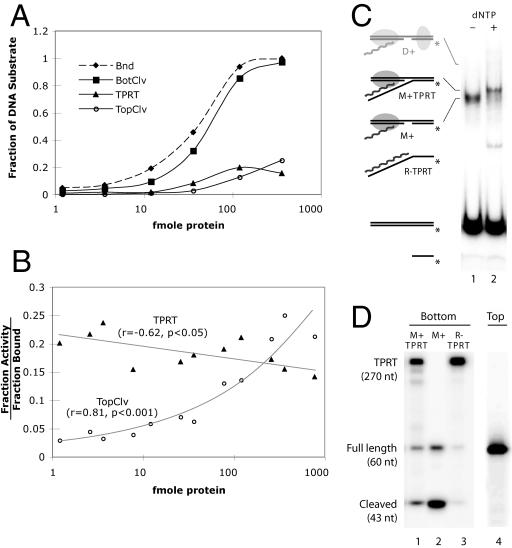
Reverse transcriptase activities associated with the M+ and D+ complexes. (A) Effect of protein concentration on the TPRT reaction. The y axis represents the fraction of the total DNA that was bound by protein, cleaved on the top or bottom strand, or had undergone TPRT. A value of 1 indicates 100% of the DNA in the reaction. The x axis is a logarithmic scale of protein concentration (fmol in each reaction). Each reaction was a standard TPRT reaction (see Material and Methods) with the 184-bp DNA substrate, the 250-nt R2 RNA, and R2 protein in the range of 0.12 to 360 fmol. The data are derived from two separate reactions (bottom or top strand end labeled) with reaction products analyzed on both EMSA and denaturing polyacrylamide gels. In the labeled bottom-strand reaction, a ∼340-nt product represented the DNA that had undergone TPRT, while the combined DNA corresponding to the ∼340-nt product and the 93-bp product represented the total level of bottom-strand cleavage. The level of top-strand cleavage was determined in the labeled top-strand reaction by the accumulation of an 89-nt product. (B) Effect of protein concentration on TPRT and top-strand cleavage. The y axis is the fraction of DNA that had undergone TPRT (triangles) or top-strand (circles) cleavage divided by the amount of DNA bound at each protein concentration. The x axis is the same as in panel A. The data presented are the combined results of two independent experiments. (C) EMSA analysis of a TPRT reaction at a low protein concentration. Reactions were preincubated for 30 min (lane 1) to reduce the presence of dimers, then 25 μM dNTPs were added and the incubation continued for 15 min (lane 2). Reactions were standard TPRT assays except that the amount of protein was only 4 fmol. Complexes that have undergone TPRT are marked (M+TPRT). R−TPRT is an artifact resulting from denaturation of the bottom 17-bp downstream DNA after TPRT. (D) Levels of TPRT and top-strand cleavage in the complexes in panel C. Lanes 1 to 3, complexes excised from the EMSA gel and the DNA analyzed on a denaturing polyacrylamide gel. Lane 4, a separate reaction similar to that in panel C but with top strand labeled to monitor top-strand cleavage.
Because both TPRT and top-strand cleavage occurred with lower efficiency, in Fig. 5B the levels of these two reactions at each protein concentration were normalized to the total level of shifted complexes seen on the EMSA gel. The level of TPRT relative to the amount of DNA bound by protein was at its highest at low protein concentrations and remained the same or declined slightly as the concentration of R2 protein increased in the reaction. The ability of TPRT to occur at similar efficiencies at low and high protein concentrations strongly suggested that the TPRT reaction could occur within both the M+ and the D+ complexes. Top-strand cleavage, on the other hand, was minimal at low concentrations and increased in proportion to the amount of protein added to the reaction. Thus, top-strand cleavage does appear to require the formation of the D+ complex.
As a second means to show that TPRT could occur within a M+ complex, the TPRT reactions in Fig. 5C were conducted at low protein and high RNA concentrations to reduce the level of D+ formation. To determine the extent of cleavage and TPRT, the complexes were excised from the native gel in Fig. 5C, and the DNA was run on a denaturing gel (Fig. 5D). After preincubation of the protein, DNA, and RNA, only the M+ complex was present (lane 1). Fifteen minutes after the addition of dNTPs (lane 2), much of the M+ complex was shifted to a somewhat slower-migrating band (M+TPRT). As shown on the denaturing gel (Fig. 5D, lane 1), the bottom strand of this slower-migrating complex had increased in length to ∼270 nt, indicating that it had undergone TPRT (17 nt of downstream DNA plus 250 nt of cDNA derived from the R2 RNA). Also generated during the reaction was a faster-migrating band (R−TPRT), which had also undergone TPRT (Fig. 5D, lane 3). This TPRT product was the result of the melting of the short downstream duplex (only 17 bp) and its release from the protein complex, not because of top-strand cleavage. If incubations identical to that in Fig. 5C were conducted with the top strand labeled, no cleavage of the top strand was detected (Fig. 5D, lane 4), suggesting that few or no D+ complexes had formed during the assay. These findings add further support to the suggestion that TPRT can efficiently occur within an M+ complex, and thus, it is the first R2 subunit that supplies the catalytic activity for reverse transcription.
Sequential addition of R2 subunits in a TPRT reaction.
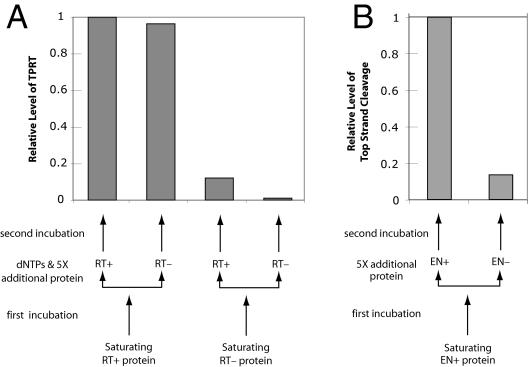
As a final test of whether the first or second subunit is responsible for the reverse transcription and top-strand cleavage, we took advantage of the ability of the R2 monomer to remain associated with the DNA substrate after nicking (36). This tight association allows the formation of R2 heterodimers through the sequential addition of wild-type and mutant R2 proteins. In the first series of experiments (Fig. 6A), a near-saturating amount of either wild-type (RT+) or a reverse transcriptase-deficient mutant (RT−) protein (37) was incubated at 37°C with substrate DNA. The RT− protein has been shown to have normal DNA binding and nicking activity (37; S. Christensen, unpublished data). This first incubation period was designed to allow binding and first-strand nicking and thus “fix” the first subunit. A fivefold excess of the “second” subunit, either RT+ or RT−, was then added to the reaction along with the dNTPs during a second incubation. The reactions were assayed for TPRT activity at the end of the second incubation period. If the RT+ protein was positioned as the first subunit, then TPRT products were seen at similar levels whether the second subunit was RT+ or RT−. If the RT− protein was positioned as the first subunit, then only low levels of TPRT products were observed, again independently of whether the second subunit was RT+ or RT−. These findings confirm that the first subunit provides the reverse transcriptase activity for TPRT.
FIG. 6.
Sequential addition of protein subunits to the DNA target site. (A) TPRT assays. Near-saturating levels (140 fmol) of wild-type (RT+) or reverse transcriptase-deficient protein (RT−) were added to the reaction and allowed to bind the bottom strand-labeled DNA substrate (60 fmol) for 17 min at 37°C. The reaction was split in two, and 5× more protein (350 fmol), either RT− or RT+, was added along with 25 μM dNTPs (final concentration) for an additional incubation at 37°C for 17 min. The bar graph reports the relative level of TPRT attained in each reaction. (B) Second-strand cleavage. Similar to panel A, wild-type R2 protein (EN+) was added to the reaction and allowed to bind the top strand-labeled DNA substrate. The reaction mixture was split in two, and 5× more protein, either EN+ or EN−, was added before the second incubation.
The second series of experiments was designed to determine which protein subunit was responsible for top-strand cleavage using wild type (EN+) and endonuclease mutant (EN−) proteins (37) (see Materials and Methods). Because the first R2 subunit can be displaced from the target DNA prior to bottom-strand cleavage (data not shown), the only viable sequential addition experiment employed the EN+ protein as the first subunit bound to the target DNA, followed by either EN+ or EN− as the second subunit (Fig. 6B). When the EN+ protein was used as the second subunit, then top-strand cleavage readily occurred; however, when the EN− protein was positioned as the second subunit, the level of top-strand cleavage was six- to sevenfold lower. This experiment confirmed our previous findings (Fig. 4) that top-strand cleavage requires the protein dimer and further suggests that it is the endonuclease domain of the second subunit that supplies the catalytic activity for top-strand cleavage.
DISCUSSION
Experiments in this report suggest that the R2 protein forms two distinct complexes with the target DNA. At low concentrations, the protein binds upstream of the target site and cleaves the bottom strand. TPRT occurs if R2 RNA and dNTPs are present. At higher concentrations, the protein also binds downstream of the target site, and in addition to bottom-strand cleavage and TPRT, top-strand cleavage occurs. We have previously shown that the stoichiometry of protein subunits to the level of bottom-strand cleavage in a single-round reaction is consistent with that of a monomer (36), that the protein sediments as a monomer (36), and that a single protein subunit cross-linked to the DNA target can comigrate with the complex capable of bottom-strand cleavage (J. Ye and T. Eickbush, unpublished data). Thus, we suggest that the R2 complex at low protein concentrations is a monomer, while the complex formed at higher protein concentrations is a dimer (although a higher-order complex is possible). The ability of the R2 reverse transcriptase to function as a monomer is similar to those of the reverse transcriptases from murine leukemia virus or the Ty3 LTR retrotransposon, which appear to function as monomers (28, 30), but differs from that of the reverse transcriptase from the human immunodeficiency virus, which appears to function as a dimer (18).
The R2 endonuclease has an active site with sequence similarity to those of certain restriction endonucleases (37). Restriction endonucleases employ two active sites in opposite orientation to cleave both DNA strands (19, 29). This is usually conducted by homodimers recognizing a palindromic recognition site or by monomers dimerizing after each binds to separate sites. The characterized type IIs restriction endonucleases, e.g., FokI, MboII, and MlyI, have DNA binding domains which recognize the target DNA at positions distant from the site of cleavage, while the catalytic domains cannot be easily footprinted (20, 39). These restriction enzymes are nickases as monomers and make double-stranded cleavages as dimers (2, 4, 31). R2 shares these traits: R2 subunits bind asymmetric DNA sites at a distance from the cleavage sites, the cleavage sites are weakly footprinted, and R2 is a nickase as a monomer and cleaves double-stranded DNA as a dimer. These properties are ideal for the TPRT reaction, since it allows the endonuclease catalytic domain to cleave the integration site and then move out to allow the reverse transcriptase to gain access, all while the protein remains bound to the target DNA.
There is little to no sequence similarity between the large upstream and smaller downstream binding regions of the target DNA (Fig. 3D); thus, these regions are likely contacted by separate DNA binding domains of the R2 protein. The footprint of the N-terminal peptide establishes the likely protein-DNA interactions responsible for downstream-DNA binding. While we have not identified the domain that contacts the upstream DNA, the most likely candidate is the C-terminal domain. Part of this DNA binding domain may be the highly conserved cysteine-histidine motif (C-X3-C-X7-H-X4-C), a motif separated from the active site residues of the endonuclease subdomain (5, 37).
The shift in equilibrium between the two binding states of the R2 protein appears to be brought about by R2 RNA. High concentrations of RNA reduced the formation of the D+ complex and top-strand cleavage (Fig. 4C). The RNA may directly bind the N-terminal domain of the protein or induce a conformational change that sequesters the N-terminal domain. In the absence of R2 RNA, both DNA binding domains are presumably accessible and can bind the upstream and downstream sites of two separate DNA molecules. Such cross-links can explain the matrix of R2 protein and DNA seen on mobility shift assays conducted in the absence of RNA (Fig. 2) (9).
Model of the R2 TPRT reaction.
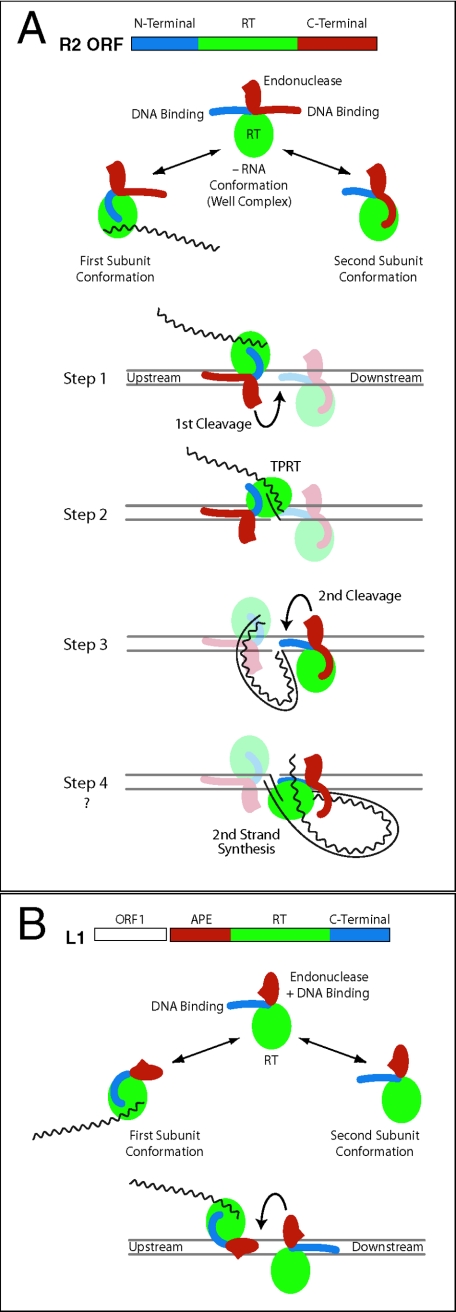
Based on these findings, we present an updated model for R2 retrotransposition (Fig. 7A). The three domains of the R2 protein, N-terminal DNA binding, C-terminal endonuclease, and central reverse transcriptase domains, are drawn as separate modules. The C-terminal domain is assumed to have separate DNA binding and endonuclease submodules. The complete complex is postulated to be a dimer, because a monomer subunit can catalyze the first half of essentially a symmetric integration reaction. The two subunits of the dimer are in different conformations and destined to perform different catalytic roles. The primary determinant for one subunit appears to be the binding of element RNA, while the determinant for the second subunit appears to be its interaction with the RNA-bound subunit on the DNA target. In each conformation, the unused DNA binding domain is sequestered.
FIG.7.
Model of a complete R2 integration reaction. (A) The R2 ORF is divided into three domains: N-terminal DNA binding (blue), reverse transcriptase (green), and C terminal (red) (5). The C-terminal domain has been subdivided into DNA-binding and endonuclease subdomains. The DNA cleavage domain (red oval with spike) is on a flexible linker, similar to the type IIs restriction endonucleases. In the context of a dimer, each R2 subunit exposes only one DNA binding domain, while the other domain is sequestered. The subunit with bound 3′-UTR RNA exposes the C-terminal DNA-binding domain and binds upstream of the cleavage/integration site. The subunit lacking RNA exposes the N-terminal DNA binding domain and binds in opposite orientation downstream of the site. While it is likely that protein-protein interactions play a role in dimer formation, such interactions have not been depicted. Step 1 (first-strand cleavage). The EN domain of the upstream subunit cleaves the bottom strand. Step 2 (first-strand synthesis). The upstream subunit undergoes a conformational change, placing its reverse transcriptase over the bottom strand cut, which can then catalyze TPRT. Step 3 (second-strand cleavage). The EN domain of the downstream subunit cleaves the top strand. Step 4 (second-strand synthesis). The upstream subunit undergoes a conformational change, placing its RT over the top-strand cut, which can then catalyze second-strand synthesis. This last step does not occur in our in vitro reaction. (B) Model of L1 integration. L1 elements encode two ORFs (27). The second ORF contains a N-terminal AP endonuclease domain (red), a reverse transcriptase domain (green), and a C-terminal domain, which is postulated in the model to be DNA binding (blue). As in the R2 model, the active complex is assumed to be a dimer, with each subunit in opposite orientation conducting one-half of the reactions. One subunit binds RNA, binds the DNA target by means of the APE domain, cleaves the first strand, and conducts TPRT. The second subunit binds by means of the C-terminal domain, cleaves the second strand, and conducts second-strand DNA synthesis. No evidence yet exists for the L1 C-terminal domain binding DNA.
The complete integration reaction is postulated to occur in a symmetric manner involving four steps. The first subunit binds the element RNA, contacts the upstream region of the target DNA, and nicks the bottom strand (step 1). A conformational change in this first subunit then positions the reverse transcriptase domain next to the nicked site for TPRT (step 2). Evidence for this shift is the expansion of the footprint of the M+ complex over the cleavage site after bottom-strand cleavage (9). In step 3, the second subunit, which lacks element RNA and binds to the target DNA downstream of the insertion site via its N-terminal domain, is responsible for performing top-strand cleavage. We assume that the R2 endonuclease is similar to that of type IIs restriction enzymes in requiring oppositely oriented active sites to achieve double-stranded cleavage. The second subunit would thus be in the correct orientation to conduct second-strand synthesis (step 4). This last step is the only step that has not been seen in our in vitro reactions.
While not observed in vitro, R2 reverse transcriptase seems to have the catalytic potential to conduct the fourth step of a TPRT reaction. The protein can efficiently utilize single-stranded DNA templates in polymerization reactions and has the unusual ability to displace RNA strands that are annealed to these DNA templates (3; A. Bibillo and T. Eickbush, unpublished data). Failure to observe second-strand synthesis is thus likely the result of difficulty in initiating this step. One possible source of this inefficiency is the dissociation of the second protein subunit after second-strand cleavage (Fig. 4A). Perhaps in vivo second-strand synthesis can occur because various chromatin components help stabilize the R2 complex. The R2 protein appears able to cope with chromatin structures, since all the initial steps of the TPRT reaction can occur when the target site is assembled into a nucleosome core particle (38). It is interesting that the 5′ ends of R2 insertions in most insects are highly variable (5). Thus, even in vivo there is an imprecise, and potentially inefficient, mechanism for initiating second-strand synthesis.
Relationship to other non-LTR retrotransposons.
A number of other non-LTR retrotransposons, many of which are site specific, appear to share the same domain structure as the R2 ORF, i.e., N-terminal DNA binding motifs and a C-terminal restriction-like endonuclease (6, 23). It is likely that these elements conduct the TPRT reaction in a manner similar to that of R2.
Many other non-LTR retrotransposons contain an apurinic-apyrimidinic-endonuclease (APE) located N terminal to the reverse transcriptase domain (23, 27). Most current TPRT models proposed for these elements assume that the element-encoded reverse transcriptase synthesizes the second DNA strand (14, 15, 27). Because the APE domains of these elements have also been characterized as nickases (1, 8, 10, 14), the basic features of the R2 model may be common to the APE-encoding elements: namely, two subunits asymmetrically bound to target DNA through independent binding domains, with one subunit responsible for the first-strand nick and reverse transcriptase activity and the other subunit responsible for second-strand cleavage and second-strand synthesis.
The ORF structure of the best-characterized APE-containing element, the L1 element of mammals, is shown in Fig. 7B (27). ORF2 contains a centrally located reverse transcriptase domain, an N-terminal APE domain, and a C-terminal domain. The C-terminal domain contains a cysteine-histidine motif similar to that of R2, and mutagenesis experiments have shown the C-terminal domain is critical in L1 retrotransposition (25). We propose that the first subunit binds the target site by means of the APE domain and nicks the DNA. The finding that DNA cleavage specificity by isolated APE domains is usually highest for the strand used to prime reverse transcription (1, 8, 14) supports the model in which the APE domain is primarily associated with the binding of the first subunit. The second subunit is proposed to bind downstream of the DNA target by the C-terminal domain and cleaves the second DNA strand. One of the subunits would conduct the TPRT reactions, while the second would be responsible for second-strand synthesis.
This general model of L1 integration differs from R2 integration in that binding of the first subunit, via the APE domain, is directly at the cleavage site (10, 35); hence, the reverse transcriptase might have to access the nicked DNA by means of the other DNA groove or through the partial melting of the cleaved DNA, perhaps by the ORF1 protein (15, 16, 24, 27). Also unlike R2, L1 integration generates large target-site duplications, possibly because the bound APE of the first subunit forces DNA cleavage by the second subunit to occur well downstream of first-strand cleavage (27). This model for L1 integration is highly speculative but would be strongly supported if the C-terminal domain of an APE-containing element was found to bind downstream of the insertion site.
Acknowledgments
We thank Danna Eickbush and William Burke for their insightful comments on the manuscript.
This work was supported by National Institutes of Health grant GM42790 to T.H.E.
REFERENCES
- 1.Anzai, T., H. Takahashi, and H. Fujiwara. 2001. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)n by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol. Cell. Biol. 21:100-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bath, A. J., S. E. Milsom, N. A. Gormley, and S. E. Halford. 2002. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 277:4024-4033. [DOI] [PubMed] [Google Scholar]
- 3.Bibillo, A., and T. H. Eickbush. 2002. The reverse transcriptase of the R2 non-LTR retrotransposon: continuous synthesis of cDNA on non-continuous RNA templates. J. Mol. Biol. 316:459-473. [DOI] [PubMed] [Google Scholar]
- 4.Bitinaite, J., D. A. Wah, A. K. Aggarwal, and I. Schildkraut. 1998. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA 95:10570-10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, W. D., H. S. Malik, J. P. Jones, and T. H. Eickbush. 1999. The domain structure and retrotransposition mechanism of R2 elements are conserved throughout arthropods. Mol. Biol. Evol. 16:502-511. [DOI] [PubMed] [Google Scholar]
- 6.Burke, W. D., D. Singh, and T. H. Eickbush. 2003. R5 retrotransposons insert into a family of infrequently transcribed 28S rRNA genes of planaria. Mol. Biol. Evol. 20:1260-1270. [DOI] [PubMed] [Google Scholar]
- 7.Chambeyron, S., A. Bucheton, and I. Busseau. 2002. Tandem UAA repeats at the 3′-end of the transcript are essential for the precise initiation of reverse transcription of the I factor in Drosophila melanogaster. J. Biol. Chem. 277:17877-17882. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S., G. Pont-Kingdom, and D. Carroll. 2000. Target specificity of the endonuclease from the Xenopus laevis non-long terminal repeat retroransposon, Tx1L. Mol. Cell. Biol. 20:1219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, S., and T. H. Eickbush. 2004. Footprint of the retrotransposon R2Bm protein on its target site before and after cleavage. J. Mol. Biol. 336:1035-1045. [DOI] [PubMed] [Google Scholar]
- 10.Cost, G. J., and J. D. Boeke. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081-18093. [DOI] [PubMed] [Google Scholar]
- 11.Cost, G. J., Q. Feng, A. Jacquier, and J. D. Boeke. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 13.Esnault, C., J. Maestre, and T. Heidmann. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24:363-367. [DOI] [PubMed] [Google Scholar]
- 14.Feng, Q., G. Schumann, and J. D. Boeke. 1998. Retrotransposon R1Bm endonuclease cleaves the target sequence. Proc. Natl. Acad. Sci. USA 95:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, N., S. LutzPrigge, and J. V. Moran. 2002. Genomic deletions created upon LINE-1 retrotransposition. Cell 110:315-325. [DOI] [PubMed] [Google Scholar]
- 16.Hohjoh, H., and M. F. Singer. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16:6034-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajikawa, M., and N. Okada. 2002. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell 111:433-444. [DOI] [PubMed] [Google Scholar]
- 18.Kohlstaedt, L. A., J. Wang, J. M. Friedmen, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 19.Kovall, R. A., and B. W. Matthews. 1999. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 3:578-583. [DOI] [PubMed] [Google Scholar]
- 20.Li, L., L. P. Wu, and S. Chandrasegaran. 1992. Functional domains in Fok I restriction endonuclease. Proc. Natl. Acad. Sci. USA 89:4275-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luan, D. D., and T. H. Eickbush. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15:3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 23.Malik, H. S., W. D. Burke, and T. H. Eickbush. 1999. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 16:793-805. [DOI] [PubMed] [Google Scholar]
- 24.Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, T. M., and T. R. Cech. 1998. Reversing time: origin of telomerase. Cell 92:587-590. [DOI] [PubMed] [Google Scholar]
- 27.Ostertag, E. M., and H. H. Kazazian, Jr. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35:501-538. [DOI] [PubMed] [Google Scholar]
- 28.Pandey, P. K., N. Kaushik, T. T. Talele, P. N. Yadav, and V. N. Pandey. 2001. Insertion of a peptide from MuLV RT into the connection subdomain of HIV-RT results in a functionally active chimeric enzyme in monomeric conformation. Mol. Cell Biochem. 225:135-144. [DOI] [PubMed] [Google Scholar]
- 29.Pingoud, A., and A. Jeltsch. 2001. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 29:3705-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rausch, J. W., M. K. Bona-Le Grice, M. H. Nymark-McMahon, J. T. Miller, and S. F. J. Le Grice. 2000. Interaction of p55 reverse transcriptase from Saccharomyces cerevisiae retrotransposon Ty3 with conformationally distinct nucleic acid duplexes. J. Biol. Chem. 275:13879-13887. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J., M. Bibikova, F. G. Whitby, A. R. Reddy, S. Chandrasegaran, and D. Carroll. 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc-finger DNA-recognition domains. Nucleic Acids Res. 28:3361-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, H., and H. Fujiwara. 2002. Transplantation of target site specificity by swapping the endonuclease domains of two LINEs. EMBO J. 21:408-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voytas, D. F., and J. D. Boeke. 2002. Ty1 and Ty5 of Sacharomyces cerevisiae, p. 631-662. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 34.Wei, W., N. Gilbert, S.-L. Ooi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weichenrieder, O., K. Repanas, and A. Perrakis. 2004. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure 12:975-986. [DOI] [PubMed] [Google Scholar]
- 36.Yang, J., and T. H. Eickbush. 1998. RNA-induced changes in the activity of the endonuclease encoded by the R2 retrotransposable element. Mol. Cell. Biol. 18:3455-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, J., H. S. Malik, and T. H. Eickbush. 1999. Identification of the endonuclease domain encoded by R2 and other site-specific, non-long terminal repeat retrotransposable elements. Proc. Natl. Acad. Sci. USA 96:7847-7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye, J., Z. Yang, J. J. Hayes, and T. H. Eickbush. 2002. R2 retrotransposition on assembled nucleosomes depends on the translational position of the target site. EMBO J. 21:6853-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yonezawa, A., and Y. Sugiura. 1994. DNA binding mode of class-IIS restriction endonuclease FokI revealed by DNA footprinting analysis. Biochim. Biophys. Acta 1219:369-379. [DOI] [PubMed] [Google Scholar]
- 40.Zimmerly, S., H. Guo, P. S. Perlman, and A. M. Lambowitz. 1995. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell 82:545-554. [DOI] [PubMed] [Google Scholar]