Abstract
Rpm2p, a protein subunit of yeast mitochondrial RNase P, has another function that is essential in cells lacking the wild-type mitochondrial genome. This function does not require the mitochondrial leader sequence and appears to affect transcription of nuclear genes. Rpm2p expressed as a fusion protein with green fluorescent protein localizes to the nucleus and activates transcription from promoters containing lexA-binding sites when fused to a heterologous DNA binding domain, lexA. The transcriptional activation region of Rpm2p contains two leucine zippers that are required for transcriptional activation and are conserved in the distantly related yeast Candida glabrata. The presence of a mitochondrial leader sequence does not prevent a portion of Rpm2p from locating to the nucleus, and several observations suggest that the nuclear location and transcriptional activation ability of Rpm2p are physiologically significant. The ability of RPM2 alleles to suppress tom40-3, a temperature-sensitive mutant of a component of the mitochondrial import apparatus, correlates with their ability to transactivate the reporter genes with lexA-binding sites. In cells lacking mitochondrial DNA, Rpm2p influences the levels of TOM40, TOM6, TOM20, TOM22, and TOM37 mRNAs, which encode components of the mitochondrial import apparatus, but not that of TOM70 mRNA. It also affects HSP60 and HSP10 mRNAs that encode essential mitochondrial chaperones. Rpm2p also increases the level of Tom40p, as well as Hsp60p, but not Atp2p, suggesting that some, but not all, nucleus-encoded mitochondrial components are affected.
In the yeast Saccharomyces cerevisiae, mitochondrial RNase P, a 5′ tRNA-processing enzyme, is coded in both mitochondrial and nuclear genomes. Mitochondrial DNA (mtDNA) codes for Rpm1r, the RNA subunit of mitochondrial RNase P, and nuclear DNA codes for the protein subunit, Rpm2p (18, 27, 45, 67). Moreover, Rpm2p is also required for maturation of the RNA subunit, Rpm1r (62), and separate domains of Rpm2p promote tRNA and Rpm1r maturation (63). Analysis of a mutant allele, rpm2-100, revealed that RPM2 has another mitochondrial function, in addition to the mitochondrial RNase P-related functions (64). Cells with rpm2-100 as their only source of Rpm2p have correctly processed mitochondrial tRNAs but are still respiration deficient. Pulse-chase analysis of mitochondrial translation revealed decreased rates of translation of COX1, COX2, and COX3 mRNAs. This decrease leads to low steady-state levels of Cox1p, Cox2p, and Cox3p, loss of visible spectra of aa3 cytochromes, and low cytochrome c oxidase activity in mutant mitochondria (64). Thus, Rpm2p has another role in mitochondrial biogenesis, in addition to its role as a subunit of mitochondrial RNase P.
Surprisingly, there is a synthetic lethal interaction between rpm2-100 and the loss of wild-type mtDNA (64). Cells with either the rpm2-100 mutation or the deletion of mtDNA grow on glucose. When both alterations occur in the same cell, there is no growth on any carbon source. This explains why a complete deletion of RPM2 is lethal (32). Loss of RNase P activity leads to loss of mtDNA, and loss of the second function requires maintenance of mtDNA for cell viability. Therefore, the RPM2 essential function is conditional and depends on the status of the mitochondrial genome.
A link between a non-RNase P function of Rpm2p and mitochondrial protein import was established when RPM2 was isolated as a high-copy-number suppressor of tom40-2, which encodes a temperature-sensitive component of the mitochondrial import channel (2, 32). The RNase P activity of Rpm2p is not required for suppression (32). This result is substantiated by the observation that rpm2-100 supports normal RNase P activity but does not suppress tom40-3 (64). Therefore, this non-RNase P function of Rpm2p could stem from a second role for Rpm2p within the mitochondria, or it could be the result of a function for Rpm2p elsewhere in the cell.
Here we present data that a portion of Rpm2p is nuclear. We also demonstrate that Rpm2p has a transcriptional activation domain and plays a role in defining the steady-state levels of mRNAs for some nucleus-encoded mitochondrial components. Furthermore, the ability of Rpm2p to act as a transcriptional activator in the nucleus strongly correlates with its ability to support a non-RNase P function of RPM2.
MATERIALS AND METHODS
Strains, media, and reagents.
Standard yeast manipulations were used (30). Yeast cells were transformed with plasmid DNA using a lithium acetate method (12). The plasmid shuffle protocol was performed as previously described (60). Rich medium included 1% Bacto Yeast Extract, 2% Bacto Peptone, and 2% glucose (YPD) or 3% glycerol and 2% ethanol instead of glucose. Synthetic complete (SD) medium lacking appropriate amino acids for plasmid retention contained 0.67% Bacto Nitrogen Base and 2% glucose. Solid medium for plates included 2% Bacto Agar. Synthetic complete medium containing 1 g/liter of 5-fluoroorotic acid (5-FOA) was used to select Ura− yeast segregants. Culture medium reagents were supplied by Fisher Scientific (Pittsburgh, PA) or Difco (Detroit, MI). The yeast strains used in this study were YML34.1 (MATa ade2-1 ade3Δ22 his3-11,15 leu2-3,11 trp1-1 ura3-1 can1-100 RPM2/Δrpm2::KanMX YEp352/RPM2) (41), KKY3.3 [MATa his3-Δ200 isp42::HIS3 leu2-3,112 ade2-101 suc2-Δ9 trp1-Δ901 ura3-52(pRS316/isp42-3)] (32), L40 [MATa ade2 his3Δ200 leu2-3,112 trp1-901 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ gal4 gal80] (26), BY4741 (MATa his3Δ leu2Δ lys2Δ met15Δ ura3Δ), and an isogenic haploid containing an ADA3 disruption (MATa his3Δ leu2Δ lys2Δ met15Δ ura3Δ ada3Δ) generated by the S. cerevisiae genome deletion project consortium and were obtained from Research Genetics.
Plasmid construction.
Standard procedures were used for the preparation and ligation of DNA fragments and recovery of plasmid DNA from Escherichia coli (55). Restriction and modification enzymes were used as recommended by the suppliers (New England Biolabs, Beverly, MA; MBI Fermentas, Vilnius, Lithuania). Plasmid DNA and DNA fragments were purified using QIAGEN kits (QIAGEN, Chatsworth, CA). To construct plasmids harboring lexA-RPM2, lexA-rpm2-ΔC, lexA-NLS-RPM2, and lexA-NLS-rpm2-ΔC, original constructs for yeast two-hybrid analysis, pAS2-RPM2 and pACT-rpm2-ΔC, were cut with XmaI and SalI. Fragments carrying RPM2 were gel purified and ligated into pLex-a and pLexN-a (a gift from Anne Vojtek) precut with SmaI and SalI. To construct plasmids carrying portions of N-terminally truncated RPM2 fused to lexA, PCR products were obtained with the following oligonucleotides: common (primes at ADH terminator of lexA-rpm2-ΔC), CCCCCGAGCTCATGCTATACCTGAGAAAG; 7 (amino acid 521), GCGGAATTCTTACTGCATCCAATCGGTG; 8 (amino acid 591), GCGGAATTCGCTGAGTTTATCAAGAAGAG; 9 (amino acid 644), GCGGAATTCAGCTATAATGGGCTAATATCA; 10 (amino acid 684), GCGGAATTCACTTACCCAATTTTGCAAAATG. The underlined sequences in the oligonucleotides indicate the coding sequences for either the ADH terminator (common) or RPM2 (7 to 10). PCR products were digested with EcoRI and SacI and ligated into pRS314/lexA-rpm2-ΔC precut with EcoRI and SacI. To construct MLS-lexA-RPM2, MLS-lexA-rpm2-100, MLS-lexA-rpm2-ΔC, and MLS-lexA-rpm2-ΔC(−20), a lexA coding sequence was obtained by PCR with the following oligonucleotides: forward, TCTACCGGCCGTCCGGGCGGAATGAAAGCGTTA; reverse, TAATGTACGGCCGAATTCCAGCCAGTCGCCGTT (underlined are coding sequences). PCR products were cut with EagI and ligated into an EagI site introduced downstream of the mitochondrial leader sequence in 314/RPM2, 314/rpm2-100, 314/rpm2-ΔC, and 314/rpm2-ΔC(−20), respectively. To construct the GFP-RPM2-containing plasmid, the parental vector pPS811 was modified to eliminate the NPL3 gene. A portion of the coding region of RPM2 (coding for amino acids 44 to 735) was inserted into a blunted BamHI site.
Real-time PCR.
Total RNA was isolated by hot-phenol extraction (34). Two micrograms of total RNA was used for cDNA synthesis with an iScript cDNA synthesis kit in a 40-μl reaction volume (Bio-Rad). One microliter of cDNA was then amplified in a total volume of 50 μl containing 1× SYBR green PCR mix (iQ SYBR green Supermix; Bio-Rad) and 300 nM gene-specific primers (sequences are available upon request). The thermal cycling conditions comprised an initial denaturation step of 95°C for 3 min and 40 cycles of 95°C for 30 s and 61°C for 45 s. All reactions were performed in triplicate. Normalization-quantification was performed using the comparative (ΔCT) method as described in reference 48. ATP2 was used as a reference gene because it was not affected by either SEF1 or RPM2. Briefly, the CT is the fractional cycle number for which the amount of amplified target reaches a fixed threshold. This amount is a constant depending on the primer set. The difference (ΔCT) between the CTs of the target gene and the reference gene depends on the RNA relative copy number between the target and the reference gene. The amount of target normalized to an endogenous reference, ATP2, and relative to a calibrator (XN,C), wild-type [rho+] cells, is given by the equation XN,C = 2−ΔΔCT, where ΔΔCT is the difference between the ΔCT of the sample and the ΔCT of the calibrator.
Western analysis.
Proteins were separated by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis using the buffer system of Laemmli, transferred to Immobilon-P membranes (Millipore, Bedford, MA), and treated with antibodies. The anti-Rpm2p antibodies were made against a peptide encoding amino acids 306 to 323 (QCB Inc., Hopkinton, MA) and were used at a 1:200 dilution. Antiserum (MD9) against Atp2p and antiserum (159-3) against Tom40p (a gift from M. G. Douglas) were used at a 1:1,000 dilution. Anti-yeast hsp60 antibodies were obtained from Stressgen Biotechnologies, Victoria, British Columbia, Canada. Protein concentrations were determined using a Bio-Rad Dc protein assay kit (Bio-Rad, Hercules, CA).
Microscopy.
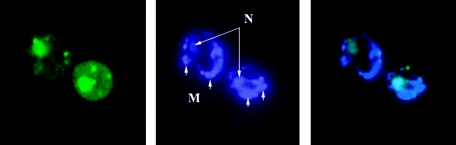
Yeast carrying GFP-RPM2 on a high-copy-number plasmid was grown in liquid glycerol-ethanol-containing synthetic medium lacking uracil. Expression of the GFP-RPM2 fusion gene was induced by addition of galactose (2% final concentration), and cells were harvested, fixed, and stained with 4′,6′-diamidino-2-phenylindole (DAPI). Fluorescent images were examined using an Axiovert 200 microscope (Zeiss). The photo in Fig. 1 is of strain YMW1 grown on raffinose and switched to galactose for 45 min. The same results were obtained with BY4741.
FIG. 1.
Microscopy. Yeast carrying GFP-RPM2 on a high-copy-number plasmid (left panel), stained with DAPI (middle panel), and merged (right panel). N, nuclei; M, mitochondria.
RESULTS
A portion of Rpm2p is nuclear.
Biochemical and genetic studies from our laboratory have clearly established that Rpm2p is located in mitochondria. Furthermore, Huh et al. (28) demonstrated that an Rpm2p-green fluorescent protein (GFP) fusion protein colocalizes with mitochondria, a result consistent with the known mitochondrial function of Rpm2p. However, Rpm2p has a second, essential function required in cells with dysfunctional mitochondria (64). We created a GFP-Rpm2 fusion protein, but unlike the construct of Huh et al. (28), we placed the GFP tag at the amino-terminal end by fusing the GFP coding sequence to the open reading frame 44 amino acids downstream from the initiator AUG. The Rpm2p used in this construct also lacks residues 736 to 1122, which are not required for growth by fermentation (32). Figure 1 shows the location of this fusion protein in cells grown in glycerol-ethanol-containing synthetic medium after GFP-RPM2 gene induction with galactose. There is no mitochondrial staining because this construct lacks the mitochondrial leader sequence and has GFP at the amino terminus. GFP-Rpm2p produced from this construct is clearly nuclear. In addition, there are foci of GFP-Rpm2p in the cytoplasm that do not appear to colocalize with any organelle.
Expression of Rpm2p as a lexA-Rpm2 fusion protein results in transcriptional activation of promoters containing lexA-binding sites.
Two observations suggested that the function of Rpm2p needed for growth on glucose might be linked to a role for this protein in nuclear transcription. First, we observed high backgrounds when Rpm2p, fused to the DNA binding domain of Gal4p, was used as bait in the yeast two-hybrid screen. This suggests that Rpm2p has endogenous transcriptional activation activity. Furthermore, similar to the behavior of Gal4-VP16, Gal4-E1A, Gal4-σ54, and Gal4-interleukin-1 fusion proteins, powerful transcriptional activators in yeast (9, 11, 59, 61), overexpression of the Gal4-Rpm2p fusion protein is toxic. Second, we found that multiple copies of SEF1, a gene encoding a putative transcription factor with a Zn(2)-Cys(2) binuclear cluster motif, suppresses RPM2 deletions and allows cells to grow by fermentation (23).
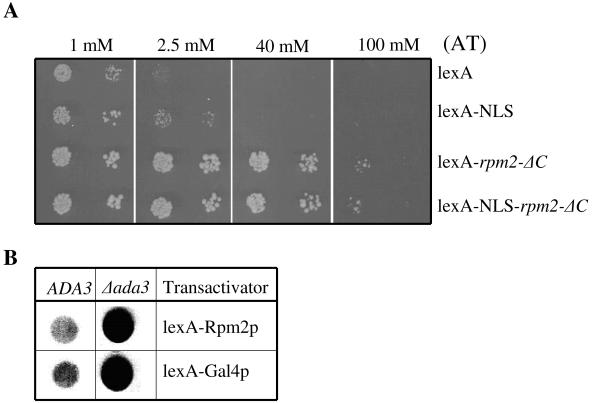
To test the idea that Rpm2p has transcriptional activity, we created fusion proteins. We employed two reporter genes, HIS3 and lacZ, that are under transcriptional control of the bacterial lexA operator. We made fusion constructs between the bacterial lexA protein and amino acids 44 to 735 of Rpm2p. In addition, we made constructs with the simian virus 40 (SV40) nuclear localization signal (NLS) inserted between the lexA and Rpm2p domains of the fusion proteins. The constructs were named lexA-NLS-rpm2-ΔC and lexA-rpm2-ΔC to reflect the presence or absence of the NLS, respectively. Like the GFP fusion protein described above, the Rpm2p used in these constructs lacks a portion of the amino-terminal mitochondrial leader sequence and residues 736 to 1202, which are not required for growth on glucose (32). A construct was made with lexA fused to the NLS alone to assay for transcriptional activation attributed to lexA in yeast. The different constructs were transformed into the reporter strain L40 (26) and transcriptional activation monitored by growth on medium lacking histidine and containing 3-aminotriazole (AT), a competitive inhibitor of the HIS3 gene product. Figure 2A shows that, in contrast to Gal4-RPM2 fusions, lexA-RPM2 fusions do not inhibit cell growth when expressed from either plasmid pLex-a or pLexN-a. Cells transformed with lexA alone grow on medium containing 1 mM AT, but growth ceases at higher concentrations of AT. This background activity is likely linked to the passive diffusion of lexA into the nucleus and a modest ability of lexA alone to activate transcription in yeast. Fusion of an NLS to lexA increased reporter activity somewhat above that seen with lexA alone, presumably due to higher nuclear accumulation of the lexA-NLS. In contrast, the lexA-RPM2 constructs showed robust reporter activity that was only inhibited by extremely high concentrations of AT. The reporter activity observed with the lexA-rpm2-ΔC fusions was not affected by the presence or absence of the SV40 NLS. These data confirm that Rpm2p contains information needed for nuclear localization and strongly suggest it plays a role in transcriptional activation.
FIG. 2.
Transcriptional activation by lexA-Rpm2p. (A) Complementation assay. Plasmids encoding lexA or lexA fusion proteins were introduced into the L40 yeast strain by selecting for Trp+ colonies. Ten to 15 individual colonies were combined and grown in Trp− liquid medium and plated on Trp− His− medium containing the indicated concentrations of AT, a competitive inhibitor of the HIS3 gene product. (B) Enhanced transcriptional activation by lexA-Rpm2p in an ADA3 deletion strain. Yeast strains expressing either lexA-Rpm2p or lexA-Gal4p activate the reporter lacZ on the indicator plates. Blue color intensity (appears black in the image shown) qualitatively reflects the amount of lacZ produced from the reporter construct.
Although there is a well-established correlation between the degree of AT resistance and the level of HIS3 mRNA (25), we also examined the expression of the gene for β-galactosidase, another reporter gene. In addition, to determine other requirements for transcription mediated by Rpm2p, we compared the level of transcriptional activation by lexA-Rpm2p in wild-type and ADA3 mutant yeast. ADA3 is a member of the histone acetyltransferase complex SAGA and is required for the integrity of SAGA (3). The Gcn5 histone acetyltransferase (8), along with SAGA/ADA complex proteins (Ada1p, Ada2p, and Ada3p), is required for some, but not all, yeast activator proteins (22, 54). Disruption of ADA3 leads to diminished transcription by Gcn4p (50) but enhanced transcription by Gal4p, Pdr1p, and Pdr3p (6, 42). Therefore, we asked whether the SAGA complex is associated with Rpm2p transactivation ability. To address this question, we transformed wild-type and ADA3 deletion strains with low-copy-number plasmids carrying either lexA-rpm2-ΔC or lexA-GAL4AD, together with the plasmid harboring lacZ, a reporter gene under control of the lexA operators. Transformants were grown on selective liquid medium, and the same amount of cells was spotted onto the indicator plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), a substrate of the lacZ gene product. In this in vivo assay, if the lexA fusion protein activates lacZ expression, cells will turn blue on the indicator plates. Figure 2B shows that lexA-Rpm2p does activate transcription of the lacZ reporter in both wild-type and ADA3 deletion strains. The loss of ADA3 leads to a substantial increase in expression of lacZ in cells producing either lexA-Rpm2 or lexA-Gal4 fusion protein. The latter has been reported previously (6). Therefore, ADA3, a specific transcriptional coactivator-corepressor protein represses transcriptional activation by Rpm2p.
lexA-Rpm2 fusion proteins provide the essential function of Rpm2p.
We wanted to determine whether the genes producing lexA-Rpm2 proteins that were located in the nucleus and supported transcriptional activation could also substitute for RPM2. Since lexA-rpm2-ΔC constructs are missing a portion of the Rpm2p mitochondrial leader, we predicted that they would not be imported and the cells would be deficient in respiration. Thus, these constructs could be useful in determining whether the essential function stems from a second role for Rpm2p in mitochondria or whether it is the result of a function elsewhere in the cell, for instance, in the nucleus, as suggested by the results in Fig. 1 and 2.
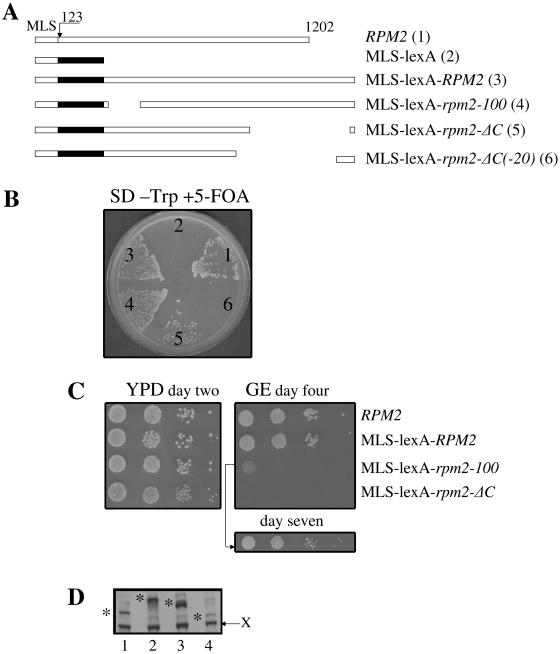
In addition to the C-terminally truncated lexA-rpm2-ΔC constructs used above, we created additional lexA-RPM2 constructs containing the full-length C terminus, with and without the heterologous SV40 NLS (Fig. 3A). We transformed cells containing a chromosomal deletion of RPM2 and a copy of RPM2 on a URA3 plasmid with a TRP1 plasmid containing either wild-type RPM2 or rpm2-ΔC, which supports growth on glucose but not RNase P activity (63), or different lexA-RPM2 constructs all lacking residues 1 to 44. The transformants were grown on 5-FOA to counterselect against the URA3-based plasmid. Only cells that lose the URA3-containing plasmid and have another source of functional Rpm2p can grow under these conditions. All of the strains containing either carboxyl-terminally truncated or full-length RPM2 formed colonies on 5-FOA plates (data not shown) and continued to grow when transferred to a fresh glucose plate. Figure 3B shows that lexA-RPM2 fusion genes coding either amino acids 44 to 735 (lexA-rpm2-ΔC) or amino acids 44 to 1202 (lexA-RPM2) grow comparably to those containing RPM2 sequences alone. The presence or absence of the SV40 NLS also does not affect growth.
FIG. 3.
Growth phenotypes of cells expressing lexA-Rpm2 fusion proteins. (A) Schematic diagram of various lex-Rpm2 fusion protein constructs. (B) Yeast expressing either wild-type RPM2 (strain 1), lexA-NLS-rpm2-ΔC (strain 2), lexA-rpm2-ΔC (strain 3), rpm2-ΔC (strain 4), lexA-RPM2 (strain 5), or lexA-NLS-RPM2 (strain 6) as its only source of RPM2 grows on glucose-containing medium (YPD). Only cells that express full-length Rpm2 protein with an intact mitochondrial leader sequence grow on nonfermentable carbon sources (glycerol and ethanol [GE]).
In contrast, cells containing lexA-RPM2 constructs that are missing the first 1 to 43 amino acids do not grow on nonfermentable carbon sources, indicating that mitochondrial RNase P activity is lost. Like lexA-RPM2-containing cells, cells with rpm2-ΔC do not grow on nonfermentable carbon sources (Fig. 3B). The product of the rpm2-ΔC allele is localized to mitochondria and supports RNase P function only when maintained on nonfermentable carbon sources (63). Since mitochondrial protein synthesis is required for the maintenance of the wild-type mtDNA (44), wild-type mtDNA is lost if cells are grown first on glucose (63). Thus, the ability of lexA-RPM2 to complement growth on fermentable carbon sources in an rpm2 null strain indicates that expression of RPM2, as a lexA-Rpm2 fusion protein, does not compromise its essential function and suggests that mitochondrial localization is not required for growth on glucose.
The transcriptional activation region of Rpm2 contains two conserved leucine zipper domains, both required for transcriptional activation.
Our primary structure analyses identified two putative leucine zipper domains at amino acid residues 527 to 555 and 696 to 717 in Rpm2p (Fig. 4A). Both leucine zippers are also present in a homolog from Candida glabrata, isolated in our laboratory by functional complementation of S. cerevisiae RPM2 (7; GenBank accession no. AF338039) (Fig. 3A). The second leucine zipper domain is included in the lexA-rpm2-ΔC construct used above, which terminates at position 735. We have shown previously that Rpm2p truncated at amino acid 735 supports growth on glucose, whereas a mutant form 20 residues shorter, truncated at amino acid 714, no longer supports growth on glucose (63). The latter truncation includes part of the putative leucine zipper domain. To examine whether the inability of this truncation to support growth on glucose correlates with its ability to act as a transcription activator, we truncated the lexA-RPM2 construct at residue 714 and assayed it for the ability to activate transcription from the HIS3 reporter gene. Figure 4B shows that the ability of lexA-rpm2-ΔC(−20) to activate HIS3 transcription is substantially diminished relative to lexA-rpm2-ΔC, but the fusion protein is made, albeit in a reduced amount (Fig. 4C). These data indicate that the ability of Rpm2p to support growth on glucose correlates with its ability to activate transcription and that both of these activities are lost in a construct where residues within the second leucine zipper domain are eliminated.
FIG. 4.
Deletion of amino acids 715 to 735 diminishes the ability of Rpm2p to activate transcription as a lexA fusion protein in the L40 strain. (A) Comparison of S. cerevisiae and C. glabrata Rpm2p in the region coding for the two putative leucine zippers. In the leucine zipper, every seventh amino acid is in boldface. (B) Growth of lexA-rpm2-ΔC-, lexA-rpm2-ΔC(−20)-, and lexA-GAL4-transformed cells on medium in the absence of histidine and in the presence of AT. (C) The top part shows a Western analysis of fusion proteins detected using antibodies against lexA protein. lexA-Rpm2.ΔCp, lexA-Rpm2.ΔC(−20), and lexA-Gal4p are in lanes 1 to 3, respectively. The bottom part shows a Western analysis of unknown yeast protein, marked with the letter X, which is recognized by lexA antibodies serving as a loading control. (D) Growth of lexA-rpm2-ΔC-, lexA-NLS-rpm2-ΔC-, lexA-rpm2-ΔC(−20)-, and lexA-NLS-rpm2-ΔC(−20)-transformed cells on histidine-lacking medium without AT or with 5.0 mM AT.
Since transcriptional activation using the lexA reporter system requires a fusion partner that provides both an NLS and transactivation activity, the construct truncated at position 714 may be defective in one or the other or both of these functions. To distinguish among these alternatives, we added a heterologous NLS to the lexA-rpm2-ΔC(−20) construct. Figure 4D shows that the ectopic NLS does not improve the ability of lexA-rpm2-ΔC(−20) to activate transcription from the HIS3 reporter gene. Thus, sequences between residues 715 and 735 of Rpm2p are necessary for transcriptional activation.
To determine the minimal domain for transcriptional activation, a set of N-terminal deletion mutant constructs, each designed to truncate Rpm2p by an average of 50 amino acids, fused to lexA were made and tested for their activities on the HIS3 reporter (Fig. 5A and B). However, this analysis was hampered by the apparent instability of many fusion proteins. Deletion of the amino-terminal 520 residues had no effect on transcriptional activation by Rpm2p (Fig. 5B). Further deletion to amino acid 591, which removes the first leucine zipper, resulted in diminished but detectable protein levels; however, transcriptional activation was totally lost. Deletion to amino acid 684 resulted in a stable fusion protein, which retains a portion of Rpm2p including the second leucine zipper. However, transcriptional activity of this mutant was equal to that observed for lexA alone. Therefore, together these data indicate that the region for transcriptional activation lies between amino acids 521 and 735 of Rpm2p and contains two putative leucine zippers.
FIG. 5.
(A) Schematic drawing of the deletion mutants and Western analysis showing the fusion proteins marked with asterisks. Lanes: 1, lexA-rpm2-ΔC; 2, lexA-rpm2521-735; 3, lexA-rpm2591-735; 4, lexA-rpm2644-735; 5, lexA-rpm2684-735. (B) Growth of lexA-rpm2 deletion mutants in the absence of histidine and in the presence of increasing amounts of AT. The numbering is the same as in panel A.
Rpm2p localizes lexA to both the nucleus and mitochondria.
It is clear that Rpm2p can reach the nucleus if a portion of the mitochondrial leader sequence is missing. However, if the nuclear location is physiologically meaningful, Rpm2p must localize to both the nucleus and the mitochondria. To determine whether some Rpm2p localizes to the nucleus when the mitochondrial leader sequence is intact, we inserted a lexA coding sequence downstream of the mitochondrial leader sequence in wild-type RPM2 and in the mutant alleles rpm2-100, rpm2-ΔC, and rpm2-ΔC(−20) (Fig. 6A). We used lexA fused to the Rpm2p mitochondrial leader sequence alone, MLS-lexA, as a control (Fig. 6A). We introduced these alleles on low-copy-number plasmids into cells carrying a wild-type RPM2 gene on a URA3-containing plasmid and then measured the ability of these cells to grow on medium containing 5-FOA. Figure 6B demonstrates that cells with alleles harboring coding regions of either full-length RPM2, rpm2-100, or rpm2-ΔC, but not rpm2-ΔC(−20), can grow in the absence of wild-type RPM2 on glucose. The latter has a truncated second leucine zipper.
FIG. 6.
(A) Schematic drawing of RPM2 genes containing lexA between the mitochondrial leader sequence and the mature protein. The constructs are described in the text. (B) Growth on SD medium lacking tryptophan and containing 5-FOA. Numbers correspond to those following the construct names in panel A. (C) Cells from plate B were diluted 10-fold subsequently four times and spotted on glucose (YPD)- and glycerol-ethanol (GE)-containing plates and incubated as indicated in the figure. (D) Western analysis showing the fusion proteins marked with asterisks. Lanes: 1, Rpm2p; 2, MLS-lexA-Rpm2p; 3, MLS-lexA-rpm2-100p; 4, MLS-lexA-rpm2-ΔCp. The migration of an unknown yeast protein which is recognized by lexA antibodies serves as a loading control (X).
Cells expressing these proteins from low-copy-number plasmids as their only source of RPM2 (Fig. 6D) grow on rich glucose-containing medium (YPD) (Fig. 6C). Rpm2-lexA protein supports mitochondrial protein synthesis since cells grow like the wild type on nonfermentable carbon sources (Fig. 6C). Cells expressing rpm2-100-lexA fusion protein grow poorly on glycerol-ethanol-containing medium (Fig. 6C). This is not a surprise because the rpm2-100 mutation itself gives the same phenotype (64). Cells expressing the C-terminal deletion mutation as an Rpm2-lexA fusion protein grow on glucose, but the cells lose their wild-type mitochondrial genome and are unable to grow by respiration, which is consistent with our published results (32, 63). Together, these results indicate that lexA does not interfere with any Rpm2p function(s).
To determine whether these fusion proteins function in the nucleus, we tested for the lexA operator-dependent activation of a HIS3 reporter gene in the L40 strain as described above. Figure 7 demonstrates that cells expressing either MLS-lexA-Rpm2 or MLS-lexA-Rpm2-ΔC protein can grow in the absence of histidine and in the presence of AT, a competitive inhibitor of the HIS3 gene product. Neither MLS-lexA-rpm2-100 nor MLS-rpm2-ΔC(−20) fusion protein can support growth in the absence of histidine. Thus, the rpm2-100-encoded mutant protein, which causes a translational defect of mitochondrion-encoded subunits of cytochrome c oxidase, loss of growth in the absence of the wild-type mitochondrial genome, and the ability to suppress the temperature-sensitive tom40-3 mutant (64), also abolished its ability to activate transcription as a lexA fusion protein. Unlike the rpm2 null mutation, the rpm2-100 mutation abrogates a specific function of RPM2, suggesting that the lack of transcriptional activation by rpm2-100-lexA fusion protein is due to the loss of a physiologically relevant function.
FIG. 7.
Growth of MLS-lexA (strain 1)-, MLS-lexA-RPM2 (strain 2)-, MLS-lexA-rpm2-100 (strain 3)-, MLS-lexA-rpm2-ΔC (strain 4)-, and MLS-rpm2-ΔC(−20) (strain 5)-transformed cells in the absence of histidine and the presence of AT.
Suppression of tom40-3 temperature-sensitive growth correlates with the efficiency of transcriptional activation.
Another very interesting observation from the previous experiment is that the C-terminally truncated protein has higher activity in transactivation compared to full-length Rpm2p when expressed as a lexA fusion protein. Cells expressing Rpm2-ΔC-lexA fusion protein can grow in the presence of 5 mM AT, while cells with Rpm2-lexA do not. The allele specificity of RPM2-lexA transcriptional activation suggested that this property could be utilized to determine other activities associated with RPM2 function. Multiple copies of RPM2, but not rpm2-100, can suppress a mutation in tom40-3, which encodes a temperature-sensitive component of the mitochondrial import channel (32, 64). Therefore, we asked whether the degree of suppression of a tom40-3 mutant by different Rpm2p alleles correlates with transcriptional activation by the same Rpm2p alleles expressed as fusion proteins with lexA. To address this question, we transformed tom40-3 mutant cells with RPM2 and rpm2-ΔC alleles on either low- or high-copy-number plasmids and determined their ability to suppress tom40-3 temperature-sensitive growth. Figure 8 demonstrates that the temperature-sensitive growth of tom40-3 mutant cells is suppressed by the rpm2-ΔC allele on a low-copy-number plasmid. In contrast, comparable suppression by wild-type RPM2 requires multiple copies. Thus, the ability of RPM2 to suppress the tom40-3 mutation correlates with its ability to support transcription.
FIG. 8.
Suppression of tom40-3 temperature-sensitive growth by various RPM2 constructs. Either a single-copy (cen) or a multicopy (2μm) plasmid coding for Rpm2p, rpm2-100p, or rpm2-ΔCp was introduced into cells carrying the tom40-3 temperature-sensitive allele. Transformants were spotted in 1:10 serial dilutions on YPD plates together with the wild-type strain (TOM40).
Effect of Rpm2p on nucleus-encoded mitochondrial components in ρo cells.
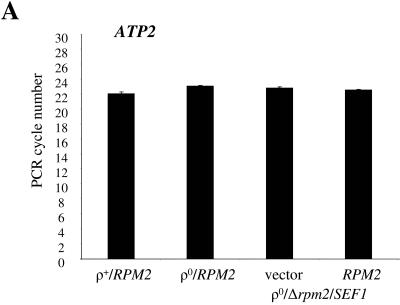
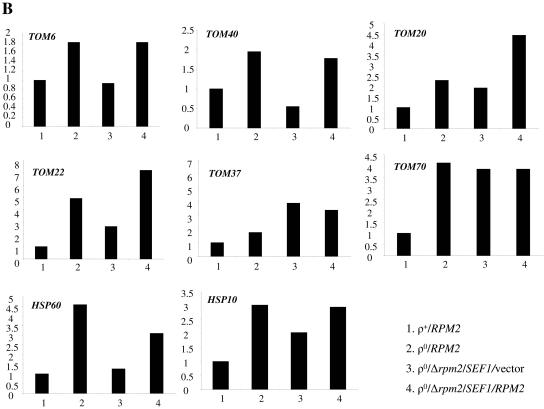
The fact that Rpm2p is clearly capable of localizing to both mitochondria and the nucleus favors a model where Rpm2p influences the synthesis of nucleus-encoded proteins involved in mitochondrial biogenesis and function. This model predicts that the levels of Rpm2p targets (directly or indirectly) will differ in the RPM2 and Δrpm2 backgrounds. To test this prediction, we started with a Δrpm2 strain that is viable because it carries the Δrpm2 multicopy suppressor SEF1 (23). SEF1 (suppressor of essential function) is a bypass suppressor of the essential function of RPM2 but not the RNase P function (23). Consequently, these cells lose their wild-type mitochondrial genomes. We transformed this strain with vector alone or with RPM2 and compared the steady-state levels of mRNAs coding for TOM complex components, the essential mitochondrial chaperones HSP10 and HSP60, and ATP2 mRNAs. In addition, we included wild-type strains with ([rho+]) and without ([rho0]) the wild-type mitochondrial genome as controls, since the loss of mtDNA itself induces changes in nuclear gene expression (10). Figure 9A demonstrates that the steady-state level of nucleus-encoded ATPase subunit β, ATP2, mRNAs is slightly reduced in cells without mtDNA compared to wild-type cells. However, this level remains comparable in the presence or absence of RPM2 and does not appear to be influenced by overexpression of SEF1. Therefore, we used the ATP2 mRNA as the normalization control in our real-time PCR experiments. After normalization to ATP2, we found that steady-state levels of mRNA encoding the multisubunit TOM complex components are increased in cells lacking mtDNA (compare strains 1 and 2 in Fig. 9B). These are the mRNAs encoding the core components Tom40p and Tom6p, as well as Tom20p-Tom22p and Tom37p-Tom70p, all components of two-receptor systems (2, 31, 32, 46, 49). In addition, mRNA levels of two essential mitochondrial chaperones, Hsp10p and Hsp60p, are higher in [rho0] cells. The increase of TOM6, TOM20, HSP60, and HSP10 mRNAs in [rho0] cells has been reported previously (66); however, the mechanism responsible for the observed changes is unknown. We demonstrate here first that the increase of TOM6, TOM40, and HSP60 mRNAs in [rho0] cells is RPM2 dependent and is not affected by overexpression of SEF1. Second, TOM20, TOM22, TOM37, and HSP10 mRNAs increase in [rho0] cells and also show an increase compared to [rho+] cells when there is overexpression of SEF1. Third, the levels of TOM20, TOM22, and HSP10 but not TOM37 further increase in cells overexpressing SEF1 and carrying RPM2 on a low-copy-number vector, suggesting an additive effect of both genes on certain TOM mRNAs. Fourth, expression of TOM70 mRNA is similar in all [rho0] cells (compare strains 2, 3, and 4 in Fig. 9B) and is independent of the overexpression of SEF1 and additional copies of RPM2. These results indicate that Rpm2p, as well as its bypass suppressor SEF1, can affect certain components of the TOM import machinery and essential mitochondrial chaperons in cells without mtDNA.
FIG. 9.
Steady-state levels of nucleus-encoded mRNAs. RNA was isolated from yeast cells as shown in the figure. Real-time PCR was carried out as described in Materials and Methods. (A) ATP2 threshold cycle. (B) Change (fold) of target mRNAs normalized with ATP2 relative to calibrator wild-type RNA.
Nucleus-encoded mitochondrial proteins Tom40p and Hsp60p, but not Atp2p, increase in the presence of RPM2.
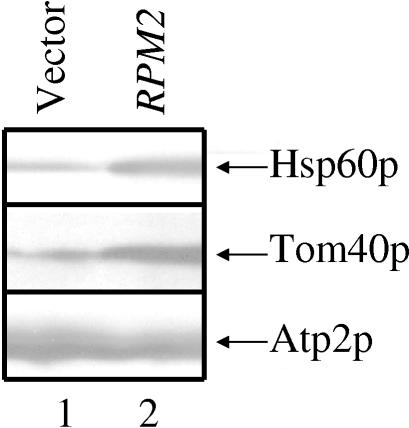
We performed Western analysis with proteins isolated from cells without mtDNA and either with or without RPM2 to determine whether protein levels change in the same way that the mRNA levels changed. Western analysis was performed on cell extracts from strains 3 and 4 (Fig. 9B). Figure 10 demonstrates a significant reduction in essential protein Tom40p (2) and Hsp60p (15) but not Atp2p (13) levels in cells lacking RPM2. Therefore, we conclude that there is a specific, rather than a general, effect of RPM2 on the steady-state level of mitochondrial proteins.
FIG. 10.
Steady-state levels of some mitochondrial proteins increase in the presence of RPM2. Total protein was prepared from SEF1/Δrpm2/vector (lane 1) and SEF1/Δrpm2/RPM2 (lane 2) cells and analyzed by Western blotting with antibodies that recognize nucleus-encoded mitochondrial proteins Hsp60p, Tom40p, and Atp2p.
DISCUSSION
The yeast S. cerevisiae Rpm2 protein was originally identified as a component of mitochondrial RNase P (45). We demonstrate here that Rpm2p also localizes to the nucleus and RPM2 constructs with a heterologous DNA-binding domain inserted downstream of the mitochondrial targeting signal support mitochondrial RNase P activity and activate transcription of a reporter gene in the nucleus. Therefore, Rpm2p has information for mitochondrial, as well as nuclear, localization. In addition, in cells with dysfunctional mitochondria, Rpm2p affects the levels of mRNAs encoding certain components of TOM complexes, as well as mitochondrial heat shock proteins.
Targeting and translocation of most nucleus-encoded mitochondrial proteins depend on N-terminal extensions referred to as mitochondrial leader sequences or presequences (45, 49). With a few exceptions, a presequence typically consists of about 15 to 40 amino acid residues, which is cleaved upon import into the organelle (5). The DNA sequence of RPM2 predicts a protein of 1,202 amino acids with a calculated molecular mass of 139,347 Da. Although we have never shown directly that the first 122 amino acids of Rpm2p are sufficient for directing a passenger protein into the organelle, it is clear that information for targeting is present in these first 122 amino acids. First, this peptide shares sequence features common to mitochondrial targeting signals such as the absence of acidic residues and the presence of basic residues and hydroxylated residues (49). Second, this sequence has a motif which is found in precursors that are cleaved in one step by mitochondrial processing peptidase (5). Third, the amino terminus of Rpm2p obtained by direct protein sequencing starts with amino acid 123 of the deduced protein sequence (45). Fourth, we show here that the lexA-Rpm2p fusion protein missing the first 43 amino acids provides the essential function but not the mitochondrial functions, suggesting that the non-RNase P function of Rpm2p is outside the mitochondria.
Studies with GFP revealed that amino acids 44 to 735 are sufficient to localize a GFP-Rpm2 fusion protein to the nucleus, which indicates that nuclear localization of the fusion protein is dependent on Rpm2p. Huh et al. (28) observed mitochondrial staining but not nuclear staining in their studies. We know from our own studies that the essential function phenotype is expressed even though Rpm2p protein cannot be detected by Western blot assays, suggesting that Rpm2p levels in nuclei are low. However, using another reporter gene assay we found that lexA-Rpm2 fusion protein missing the first 43 amino acids activates transcription of promoters containing lexA-binding sites, indicating that Rpm2p can be localized to the nucleus and act as a transcriptional activator. We also demonstrate here that Rpm2p localizes to both mitochondria and the nucleus in the presence of a mitochondrial leader sequence. Proteins larger than 60 kDa require active transport into the nucleus, but further work is necessary to understand the details of how nuclear localization is promoted.
Rpm2p, like many other transcriptional activators, contains two putative leucine zipper domains, at amino acid residues 527 to 555 and 696 to 717. A deletion mutation that eliminated two amino acids at the end of the second zipper domain abolished the essential function of Rpm2p without discernibly affecting protein stability (63). We demonstrate here that this domain is also critical for transcriptional activation. Furthermore, deletion mapping revealed that the region required for transcriptional activation lies between amino acids 521 and 735 of Rpm2p and requires both leucine zippers.
Unlike the rpm2 null mutation, another RPM2 mutation, rpm2-100, abrogates a specific function of RPM2. The function of RPM2 compromised by the rpm2-100 mutation makes yeast cells dependent on the wild-type mitochondrial genome for fermentative growth (64), indicating that Rpm2p is essential in cells with dysfunctional mitochondria. Other proteins required for respiratory growth also become essential for growth on fermentable carbon sources in the absence of wild-type mtDNA. These genes include AAC2, encoding the ADP/ATP translocase (36); PGS1, encoding the phosphatidylglycerol phosphate synthase (65); YME1, encoding the ATP-zinc-dependent mitochondrial protease (68); and genes encoding the α and β subunits of F1 ATPase (13). It is unclear why cells require wild-type mtDNA when these genes lose function, since the lack of function does not lead to an obvious common defect. Nonetheless, each of these genes can be tied, either directly or indirectly, to the mitochondrial import process (20, 35; for a review, see references 14 and 17). Indeed, there is direct evidence indicating that cells with defects in mitochondrial protein import depend on wild-type mtDNA for growth on fermentable carbon sources (20). Mutation of any of the six genes (TIM18, TIM54, TIM10, TIM9, TIM12, and TOM70) that function in the TIM22 pathway (transport inner membrane) is incompatible with loss of mtDNA (20, 33, 57). Thus, it is possible that mutations in these genes, as well as RPM2, may decrease the efficiency of mitochondrial import. If the efficiency is further reduced by the loss of the mitochondrial genome and the concurrent reduction in membrane potential, import efficiency may be reduced below the point necessary to maintain the organelle. This notion is supported by the results of Lefebvre-Balguerie et al. (37) indicating that F1-catalyzed hydrolysis of ATP is essential for maintaining an electrochemical mitochondrial membrane potential and when it is compromised, mitochondrial Hsp60 precursors accumulate.
The observation that multiple copies of RPM2 can suppress a mutation in a key component of the mitochondrial protein import channel (32) while rpm2-100 cannot (64) suggests a model relating a non-RNase P function to mitochondrial import. Although the mechanism of suppression of tom40-3 is unclear, we demonstrate here that increased levels of TOM6, TOM40, and HSP60 mRNAs depend on Rpm2p function in cells lacking wild-type mtDNA. In addition, the rpm2-100 mutation that causes a translational defect of mitochondrion-encoded subunits of cytochrome c oxidase and loss of growth in the absence of the wild-type mitochondrial genome (64) also abolishes the Rpm2p transcriptional activity of the Rpm2-lexA fusion protein. This suggests that the lack of transcriptional activation by rpm2-100-lexA protein is due to the loss of a physiologically relevant function. These observations lead to the model that Rpm2p affects transcription of nuclear genes and is required to maintain sufficient levels of nucleus-encoded components of the mitochondrial import machinery in cells with dysfunctional mitochondria. We found previously that reduced proteasome activity in pre4-2 and ump1-2 mutants allows growth in the absence of RPM2 (41). This is also consistent with the model because decreased degradation could also lead to increases in components of the mitochondrial import apparatus.
Changes in the functional state of mitochondria cause changes in nuclear gene expression (47). These changes are, for the most part, adaptive in that they represent cellular adjustments to altered mitochondrial states (for a review, see reference 10). The status of mtDNA affects nuclear gene expression in both yeast and mammalian cells (1, 4, 21, 24, 38, 47, 51, 52, 66). This important phenomenon, called retrograde regulation, was first discovered in the yeast S. cerevisiae (38, 47, 53). Rtg1p, Rtg2p, and Rtg3p (29, 56) are the key components of the retrograde signaling pathway required for the expression of some retrograde responsive genes (16, 21, 40). One function of retrograde signaling is to maintain glutamate levels in cells with dysfunctional mitochondria (21, 40). However, in the presence of glutamine, disruption of the RTG1-3 pathway does not prevent growth on either fermentable or nonfermentable carbon sources. Other genes that change expression in response to mitochondrial dysfunction do not appear to be under the control of the Rtg1-3 proteins. For example, a small number of retrograde-responsive genes that confer pleiotropic drug resistance are RTG independent and regulated by two closely related transcription factors, Pdr1p and Pdr3p (19, 24, 66). Other genes, however, might be regulated by additional RTG- and pleiotropic drug resistance-independent pathways involved in mitochondrion-to-nucleus signaling.
Four percent of yeast genes reproducibly alter transcription in yeast cells devoid of mtDNA when grown on glucose (66). Of these genes, 86% were induced between 1.5- and 6-fold while 14% were repressed. About one-fifth of the genes with elevated expression in cells lacking mtDNA encode proteins involved in mitochondrial biogenesis and function, indicating cellular accommodations to the mitochondrial defects. Previous work showed that TOM6, TOM20, HSP10, and HSP60 are upregulated in cells lacking mtDNA (66), and we show here that this increase in cells lacking mtDNA depends on RPM2. Moreover, the effects of SEF1 and RPM2 appear additive in upregulation of TOM20 and TOM22, while there is no effect of RPM2 on TOM37 and TOM70, although the latter mRNAs are induced in [rho0] cells. We believe that induction of TOM components and the essential chaperones in cells lacking mtDNA is most likely an adaptation to maintain efficient protein import upon reduction in membrane potential caused by the loss of mtDNA. The transcriptional induction of mitochondrial chaperones also occurs in mammalian cells after depletion of mtDNA (43), and overexpression of Hsp10 and Hsp60 proteins inhibits myocardial apoptosis in response to ischemic injury (39) and doxorubicin-induced cardiomyopathy (58).
Our observation that Rpm2p affects expression of nucleus-encoded mitochondrial proteins in cells with dysfunctional mitochondria and activates transcription if brought to the DNA strongly implicates Rpm2p in transcription. Since Rpm2p clearly has RNA binding activity in its role as a subunit of mitochondrial RNase P, it may be that RNA binding activity is also important to its role in transcription. There also may be a regulatory significance to mitochondrial biogenesis because Rpm2p is both mitochondrial and nuclear and because Rpm2p is required to maintain normal levels of essential mitochondrial chaperones in cells with dysfunctional mitochondria. In either case, it is clear that Rpm2p is a strong candidate for a regulatory protein intimately involved in mitochondrial biogenesis and critical to maintaining viability when cells lose their mitochondrial genome. Therefore, Rpm2p provides a vehicle to further our understanding of mitochondrial biogenesis and the complex regulatory networks necessary to the organization and maintenance of this critical and essential organelle.
Acknowledgments
We thank Pamela Silver for providing plasmid pPS811, Anne Vojtek for providing plasmids pLex-a and pLexN-a, Michael Douglas for providing a crude antiserum against Tom40p, and Hadi Falahatpisheh for help with the real-time PCR.
This work was supported by grant GM-27597 from the National Institutes of Health to N.C.M. and in part by the Center for Genetics and Molecular Medicine, University of Louisville, to V.S.
REFERENCES
- 1.Amuthan, G., G. Biswas, S.-Y. Zhang, A. Klein-Szanto, C. Vijayasarathy, and N. Avadhani. 2001. Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion. EMBO J. 20:1910-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, K. P., A. Schaniel, D. Vestweber, and G. Schatz. 1990. A yeast mitochondrial outer membrane protein essential for protein import and cell viability. Nature 348:605-609. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian, R., M. G. Pray-Grant, W. Selleck, P. A. Grant, and S. Tan. 2002. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277:7989-7995. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, G., O. A. Adebanjo, B. D. Freedman, K. Anandatheerthavarada, C. Vijayasarathy, M. Zaidi, M. Kotlikoff, and N. G. Avadhani. 1999. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 18:522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branda, S. S., and G. Isaya. 1995. Prediction and identification of new natural substrates of the yeast mitochondrial intermediate peptidase. J. Biol. Chem. 270:27366-27373. [DOI] [PubMed] [Google Scholar]
- 6.Brandl, C. J., A. M. Furlanetto, J. A. Martens, and K. S. Hamilton. 1993. Characterization of NGG1, a novel yeast gene required for glucose repression of GAL4p-regulated transcription. EMBO J. 12:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bristow, M. J., Jr. 2001. Ph.D. thesis. University of Louisville, Louisville, Ky.
- 8.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 9.Buryskova, M., M. Pospisek, A. Grothney, T. Simmet, and L. Burysek. 2004. Intracellular interleukin-1α functionally interacts with histone acetyltransferase complexes. J. Biol. Chem. 279:4017-4026. [DOI] [PubMed] [Google Scholar]
- 10.Butow, R. A., and N. G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Chen, B.-S., Z.-W. Sun, and M. Hampsey. 2001. A Gal4-σ54 hybrid protein that functions as a potent activator of RNA polymerase II transcription in yeast. J. Biol. Chem. 276:23881-23887. [DOI] [PubMed] [Google Scholar]
- 12.Chen, D. C., B. C. Yang, and T. T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X. J., and G. D. Clark-Walker. 1999. α and β subunits of F1-ATPase are required for survival of petite mutants in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:898-908. [DOI] [PubMed] [Google Scholar]
- 14.Chen, X. J., and G. D. Clark-Walker. 1999. The petite mutation in yeast: 50 years on. Int. Rev. Cytol. 194:197-238. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, M. Y., F.-U. Hartl, J. Martin, R. A. Pollock, F. Kalousek, W. Neupert, E. M. Hallberg, R. L. Hallberg, and A. L. Horwich. 1989. Mitochondrial heat-shock protein hsp60 is essential for assembly of proteins imported into yeast mitochondria. Nature 337:620-625. [DOI] [PubMed] [Google Scholar]
- 16.Chielstowska, A., and R. A. Butow. 1995. RTG genes in yeast in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J. Biol. Chem. 270:18141-18146. [DOI] [PubMed] [Google Scholar]
- 17.Contamine, V., and M. Picard. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64:281-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang, Y. L., and N. C. Martin. 1993. Yeast mitochondrial RNase P: sequence of the RPM2 gene and demonstration that its product is a protein subunit of the enzyme. J. Biol. Chem. 268:19791-19796. [PubMed] [Google Scholar]
- 19.Devaux, F., E. Carvajal, S. Moye-Rowley, and C. Jacq. 2001. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 515:25-28. [DOI] [PubMed] [Google Scholar]
- 20.Dunn, C. D., and R. E. Jensen. 2003. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins for viability of yeast cells lacking mitochondrial DNA. Genetics 165:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epstein, C. B., J. A. Waddle, W. Hale IV, V. Davé, J. Thornton, T. L. Macatee, H. R. Garner, and R. A. Butow. 2001. Genome-wide responses to mitochondrial dysfunctions. Mol. Biol. Cell 12:297-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of the Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 23.Groom, K. R., H. C. Heyman, M. C. Steffen, L. Hawkins, and N. C. Martin. 1998. Kluyveromyces lactis SEF1 and Saccharomyces cerevisiae homologue bypass the unknown essential function, but not the mitochondrial RNase P function, of the S. cerevisiae RPM2 gene. Yeast 14:77-87. [DOI] [PubMed] [Google Scholar]
- 24.Hallstrom, T. C., and W. S. Moye-Rowley. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347-37356. [DOI] [PubMed] [Google Scholar]
- 25.Hill, D. E., I. A. Hope, J. P. Macke, and K. Struhl. 1986. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science 234:451-457. [DOI] [PubMed] [Google Scholar]
- 26.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollingsworth, M. J., and N. C. Martin. 1986. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrial and nucleus-encoded components. Mol. Cell. Biol. 6:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huh, W.-K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 29.Jia, Y., B. Rothermel, J. Thornton, and R. A. Butow. 1997. A basic helix-loop-helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol. 17:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Kassenbrock, C. K., W. Cao, and M. G. Douglas. 1993. Genetic and biochemical characterization of ISP6, a small mitochondrial outer membrane protein associated with the protein translocation complex. EMBO J. 12:3023-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassenbrock, C. K., G.-J. Gao, K. R. Groom, P. Sulo, M. G. Douglas, and N. C. Martin. 1995. RPM2, independently of its mitochondrial RNase P function, suppresses an ISP42 mutant defective in mitochondrial import and is essential for normal growth. Mol. Cell. Biol. 15:4763-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerscher, O., N. B. Sepuri, and R. E. Jensen. 2000. Tim18p is a new component of the Tim54-Tim22p translocon in the mitochondrial inner membrane. Mol. Biol. Cell 11:103-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 35.Kominsky, D. J., M. P. Brownson, D. L. Updike, and P. E. Thorsness. 2002. Genetic and biochemical basis for viability of yeast lacking mitochondrial genomes. Genetics 162:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kováč, L., L. M. Lachowicz, and P. Slonimski. 1980. Biochemical genetics of oxidative phosphorylation. Science 158:1564-1567. [DOI] [PubMed] [Google Scholar]
- 37.Lefebvre-Balguerie, A., S. Duvezin-Caubet, M. F. Giraud, P. P. Slonimski, and J. P. Di Rago. 2003. F1-catalysed ATP hydrolysis is required for mitochondrial biogenesis in Saccharomyces cerevisiae growing under conditions where it cannot respire. Mol. Microbiol. 47:1329-1339. [DOI] [PubMed] [Google Scholar]
- 38.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 39.Lin, K. M., B. Lin, I. Y. Lian, R. Mestril, I. E. Scheffler, and W. H. Dillmann. 2001. Combined and individual mitochondrial HSP60 and HSP10 expression in cardiac myocytes protects mitochondrial function and prevents apoptotic death induced by stimulated ischemia-reoxygenation. Circulation 103:1787-1792. [DOI] [PubMed] [Google Scholar]
- 40.Liu, Z., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lutz, M. S., S. R. Ellis, and N. C. Martin. 2000. Proteasome mutants, pre4-2 and ump1-2, suppress the essential function but not the mitochondrial RNase P function of the Saccharomyces cerevisiae gene RPM2. Genetics 154:1013-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martens, J. A., J. Genereaux, A. Saleh, and C. J. Brandl. 1996. Transcriptional activation by yeast Pdr1p is inhibited by its association with Ngg1/Ada3p. J. Biol. Chem. 271:15884-15890. [DOI] [PubMed] [Google Scholar]
- 43.Martinus, R. D., G. P. Garth, T. L. Webster, P. Cartwright, D. J. Naylor, P. B. Hoj, and N. J. Hoogenraad. 1996. Selective induction of mitochondrial chaperones in response to loss of the mitochondrial genome. Eur. J. Biochem. 240:98-103. [DOI] [PubMed] [Google Scholar]
- 44.Meyers, A. M., L. K. Pape, and A. Tzagoloff. 1985. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 4:2087-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morales, M. J., Y. L. Dang, Y. C. Lou, P. Sulo, and N. C. Martin. 1992. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc. Natl. Acad. Sci. USA 89:9875-9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 47.Parikh, V. S., M. M. Morgan, R. Scott, L. S. Clements, and R. A. Butow. 1987. The mitochondrial genotype can influence nuclear gene expression in yeast. Science 235:576-580. [DOI] [PubMed] [Google Scholar]
- 48.Peinnequin, A., C. Mouret, O. Birot, A. Alonso, J. Mathieu, D. Clarencon, D. Agay, Y. Chancerelle, and E. Multon. 2004. Rat pro-inflammatory cytokine and cytokine related mRNA quantification by real-time polymerase chain reaction using SYBR green. BMC Immunol. 5:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfanner, N., and M. Meijer. 1997. The Tom and Tim machine. Curr. Biol. 7:100-103. [DOI] [PubMed] [Google Scholar]
- 50.Pina, B., S. Berger, G. A. Marcus, N. Silverman, J. Agapite, and L. Guarente. 1993. ADA3: a gene, identifies by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol. Cell. Biol. 13:5981-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poyton, R. O., and J. E. McEwen. 1996. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 65:563-607. [DOI] [PubMed] [Google Scholar]
- 52.Rochard, P., A. Rodier, F. Casas, I. Cassar-Malek, S. Marchal-Victorion, L. Daury, C. Wrutniak, and G. Cabello. 2000. Mitochondrial activity is involved in the regulation of myoblast differentiation through myogenin expression and activity of myogenic factors. J. Biol. Chem. 275:2733-2744. [DOI] [PubMed] [Google Scholar]
- 53.Rothermel, B. A., J. Thornton, and R. A. Butow. 1997. Rtg3p, a basic helix-loop-helix protein that functions in mitochondrial-induced changes in gene expression, contains independent activation domains. J. Biol. Chem. 272:19801-19807. [DOI] [PubMed] [Google Scholar]
- 54.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Sekito, T., J. Thornton, and R. A. Butow. 2000. Mitochondria-to-nucleus signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell 11:2103-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Senapin, S., X. J. Chen, and G. D. Clark-Walker. 2003. Transcription of TIM9, a new factor required for the petite-positive phenotype of Saccharomyces cerevisiae, is defective in spt7 mutants. Curr. Genet. 44:202-210. [DOI] [PubMed] [Google Scholar]
- 58.Shan, Y.-X., T.-J. Liu, H.-F. Su, A. Samsamshariat, R. Mestril, and P. H. Wang. 2003. Hsp10 and Hsp60 modulate Bcl-2 family and mitochondria apoptosis signaling induced by doxorubicin in cardiac muscle cells. J. Mol. Cell. Cardiol. 35:1135-1143. [DOI] [PubMed] [Google Scholar]
- 59.Shuen, M., N. Avvakumov, P. G. Walfish, C. J. Brandl, and J. S. Mymryk. 2002. The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J. Biol. Chem. 277:30844-30851. [DOI] [PubMed] [Google Scholar]
- 60.Sikorski, R. S., and J. D. Boeke. 1991. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 194:302-318. [DOI] [PubMed] [Google Scholar]
- 61.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stribinskis, V., G.-J. Gao, P. Sulo, Y. L. Dang, and N. C. Martin. 1996. Yeast mitochondrial RNase P RNA synthesis is altered in an RNase P protein subunit: insights into the biogenesis of a mitochondrial RNA-processing enzyme. Mol. Cell. Biol. 16:3429-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stribinskis, V., G.-J. Gao, P. Sulo, S. R. Ellis, and N. C. Martin. 2001. Rpm2p: separate domains promote tRNA and Rpm1r maturation in Saccharomyces cerevisiae mitochondria. Nucleic Acids Res. 29:3631-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stribinskis, V., G.-J. Gao, S. R. Ellis, and N. C. Martin. 2001. Rpm2p, the protein subunit of mitochondrial RNase P in Saccharomyces cerevisiae, also has a role in the translation of mitochondrially encoded subunits of cytochrome c oxidase. Genetics 158:573-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subik, J. 1974. A nuclear mutant of S. cerevisiae non-tolerating the cytoplasmic petite mutation. FEBS Lett. 42:309-312. [DOI] [PubMed] [Google Scholar]
- 66.Traven, A., J. M. S. Wong, D. Xu, M. Sopta, and J. C. Ingles. 2001. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276:4020-4027. [DOI] [PubMed] [Google Scholar]
- 67.Underbrink-Lyon, K., D. L. Miller, N. A. Ross, H. Fukuhara, and N. C. Martin. 1983. Characterization of a yeast mitochondrial locus necessary for tRNA biosynthesis. Mol. Gen. Genet. 191:512-518. [DOI] [PubMed] [Google Scholar]
- 68.Weber, E. R., R. S. Rooks, K. S. Shafer, J. W. Chase, and P. E. Thorsness. 1995. Mutations in the mitochondrial ATP synthase gamma subunit suppress a slow-growth phenotype of yme1 yeast lacking mitochondrial DNA. Genetics 140:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]