Abstract
Background
Natural language processing (NLP) and machine learning (ML) techniques may help harness unstructured free-text electronic health record (EHR) data to detect adverse drug events (ADEs) and thus improve pharmacovigilance. However, evidence of their real-world effectiveness remains unclear.
Objective
To summarise the evidence on the effectiveness of NLP/ML in detecting ADEs from unstructured EHR data and ultimately improve pharmacovigilance in comparison to other data sources.
Methods
A scoping review was conducted by searching six databases in July 2023. Studies leveraging NLP/ML to identify ADEs from EHR were included. Titles/abstracts were screened by two independent researchers as were full-text articles. Data extraction was conducted by one researcher and checked by another. A narrative synthesis summarises the research techniques, ADEs analysed, model performance and pharmacovigilance impacts.
Results
Seven studies met the inclusion criteria covering a wide range of ADEs and medications. The utilisation of rule-based NLP, statistical models, and deep learning approaches was observed. Natural language processing/ML techniques with unstructured data improved the detection of under-reported adverse events and safety signals. However, substantial variability was noted in the techniques and evaluation methods employed across the different studies and limitations exist in integrating the findings into practice.
Conclusions
Natural language processing (NLP) and machine learning (ML) have promising possibilities in extracting valuable insights with regard to pharmacovigilance from unstructured EHR data. These approaches have demonstrated proficiency in identifying specific adverse events and uncovering previously unknown safety signals that would not have been apparent through structured data alone. Nevertheless, challenges such as the absence of standardised methodologies and validation criteria obstruct the widespread adoption of NLP/ML for pharmacovigilance leveraging of unstructured EHR data.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40264-024-01505-6.
Key Points
| The studies included in this review demonstrated not only proficiency in identifying specific adverse events but also the ability to uncover previously unknown safety signals that would not have been apparent through structured data alone. |
| Unstructured text from EHRs could enrich pharmacovigilance programmes that have traditionally relied on other data sources. However, research into the ease of use, usability and effectiveness of using unstructured data from EHRs in comparison to traditional data sources is limited. |
| This review highlights great variability in methods used and validation techniques in obtaining data from unstructured notes in EHRs for pharmacovigilance, which may limit its wide-scale implementation. |
Introduction
Patient data on the now ubiquitous electronic health records (EHRs) have improved the quality and safety of healthcare globally [1, 2]. Digital patient data comprise structured data points (such as demographics, vital signs, prescriptions, and lab test results) and unstructured information (such as physician notes, progress notes, clinical notes, discharge summaries, patient narratives and imaging reports) [3, 4]. These comprehensive repositories of patient information can enhance the quality of care for individual patients through data analytics techniques that take advantage of patterns and trends derived from the records as a whole [5]. Unstructured data in EHRs accounts for over 80% of patient information, offering a valuable resource for gaining knowledge [6, 7], but it is rarely used [8] as it is much harder to utilise given the ambiguity of free text expressions [6, 9]. Furthermore, utilising one-for-all methods to extract the unstructured portion of EHRs could introduce errors such as perceived data distribution, complicating the process [10].
Progress in the field of natural language processing (NLP), machine learning (ML) and artificial intelligence (AI) enables automated extraction of key information from unstructured text. These advancements prove promising in various clinical applications, such as pharmacovigilance [11]. Pharmacovigilance involves detecting, evaluating, and preventing adverse drug events (ADEs) or reactions (ADRs). With the help of NLP techniques along with ML and AI, meaningful insights can be derived from unstructured clinical text to aid in this field [12]. Automation also makes it possible to efficiently analyse large amounts of textual information related to ADEs.
Adverse drug events pose a major threat to patient safety worldwide and can have severe consequences, including hospitalisation, disability, and even death [13]. Pharmacovigilance plays a crucial role in the continuous surveillance of drug safety following regulatory approval. Traditionally, pharmacovigilance relies heavily on healthcare professionals, pharmaceutical companies, clinical research organisations and more recently patients to report ADEs through a spontaneous reporting systems [14] such as the FDA Adverse Event Reporting System (FAERS). However, approximately 6 % of all ADEs are reported through spontaneous systems [15]. This significant underreporting hampers prompt and comprehensive identification of potentially important medication safety signals [16, 17]. Apart from incomplete information, spontaneous reporting systems suffer from reporting bias [18, 19].
The need to enhance pharmacovigilance capabilities at scale has led to a growing research focus on incorporating unstructured clinical text alongside structured EHR data [20]. Unstructured text offers valuable contextual details and may uncover suspected ADEs not specifically identified through coding processes, such as ICD-9 (International Classification of Diseases, 9th revision) [12, 21–25]. While the potential of using EHR data for pharmacovigilance has been acknowledged, there are still uncertainties regarding the value obtained from extracting information from unstructured clinical text compared to relying solely on structured data [9, 26–29]. There is currently no comprehensive synthesis of the effectiveness of utilising unstructured EHR data for pharmacovigilance purposes and patient safety outcomes in comparison to other data sources [30]. We aim to address this gap through a scoping review of the literature that assesses the performance and utility of using unstructured data in EHR data to identify adverse events as compared to other data sources.
Methods
This scoping review was conducted and reported according to PRISMA-ScR (Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews) checklist [31] (see checklist in Supplementary material). A predefined protocol was initially written to guide the process with any deviations or changes documented. The research question was developed using the PICO framework (Population, Intervention, Comparison, and Outcomes); the inclusion and exclusion criteria are presented in Table 1. Studies after 1990 were filtered for this review as there were key developments in AI methods for extracting and analysing EHRs after this date [32]. Due to resource constraints, only English-language studies were included.
Table 1.
Inclusion and exclusion criteria for studies on utilising ADEs/ADRs from unstructured EHR data
| Inclusion | Exclusion | |
|---|---|---|
| Participants | Patients of all age and demographic characteristics categories with reported adverse/side effects documented in unstructured text in EHRs/EMRs | Patients with no adverse/side effects documented in EHRs/EMRs |
| Intervention | Extraction and utilisation of unstructured data from EHRs/EMRs on pharmacovigilance practices (typically using NLP/ML methods) | Studies that primarily focused on structured data and studies not providing sufficient details on the impact of utilising unstructured data in pharmacovigilance practices |
| Comparator |
Studies that include a comparison group using one or more of the following as a comparator: Structured data from EHR/EMR: Studies that utilise structured data (well-defined, coded data) from EHR/EMR Spontaneous reporting systems (e.g., FDA Adverse Event Reporting System, MHRA Yellow Card Scheme) Clinical trials data SmPC Traditional pharmacovigilance methods |
Studies that do not have a comparison group or one of the comparators outlined in the inclusion criteria |
| Outcomes | Impact on identification of ADRs, including the completeness and accuracy of such reports. Secondary outcomes included the identification of safety signals, the timeliness of detecting safety signals, and any improvements in patient safety outcomes resulting from the utilisation of unstructured data in pharmacovigilance practices | Pure technological methods |
| Setting | Any healthcare setting from any geographical location | Non-healthcare settings |
| Study Design | Any type of research design. RCTs, non-randomised controlled studies, before-and-after studies, time series evaluations, cohort studies, case-control studies, and observational studies | Non-empirical research, opinion pieces, commentaries, letters to the editors, or editorials |
| Restrictions |
Published in or after 1990 In English language (or translation available) |
Published before 1990 In non-English language |
ADE adverse drug event, ADRs adverse drug reaction, EHRs electronic health records, EMR electronic medical record, MHRA Medicines & Healthcare products Regulatory Agency, NLP/ML natural language processing/machine learning, RCTs randomised control trials, SmPC Summary of Product Characteristics
Search Strategy
The following electronic databases were searched: Medline, Embase, PsycINFO, IEEE Xplore (Institute of Electrical and Electronics Engineers) and ACM Digital Library (Association of Computing Machinery) as well as Google Scholar (of which only the first 300 records were sifted) [33]. Additionally, the reference lists of selected studies were manually reviewed. The final search was completed on 18 July 2023. The full search strategy for all the databases is detailed in Supplementary material.
Screening
Title and abstract screening were conducted in Covidence [34], a web-based workflow platform for systematic reviews, by two reviewers independently. Full texts were retrieved for those that could not be excluded based on title and abstract review alone. Rigorous exclusion criteria were applied to eliminate papers that did not have a clear focus on NLP/ML or AI for pharmacovigilance, lacked relevant adverse event outcomes, or provided insufficient evidence, or did not have a comparator. Reasons for excluding selected full-text articles were noted.
Data Extraction
A customised data extraction form was created in Covidence. Details summarized in the extraction form included: study ID, paper title, authors, publication year, objectives, study design, methods, data source, population, interventions/exposures, outcomes, results/ conclusions, and how it was relevant to the review topic.
Quality parameters of the studies were extracted and discussed in lieu of formal methodological quality assessment (or risk of bias assessment)—as this is a scoping review. The quality parameters included research design, sample size, appropriateness of data sources, use of suitable NLP/ML techniques, evaluation metrics, comprehensive interpretation of results, reliability measures, and alignment with the review objective.
We also provided an overview of the NLP and ML techniques utilised in existing pharmacovigilance research for ADE and safety signal detection, as well as the specific adverse events analysed, outcome metrics used and overall findings. This scoping review aims to offer a comprehensive summary of the existing literature while underscoring its limitations and key gaps that should be addressed in future research on utilising NLP/ML for pharmacovigilance with EHR/EMR data.
Results
Selection of Studies
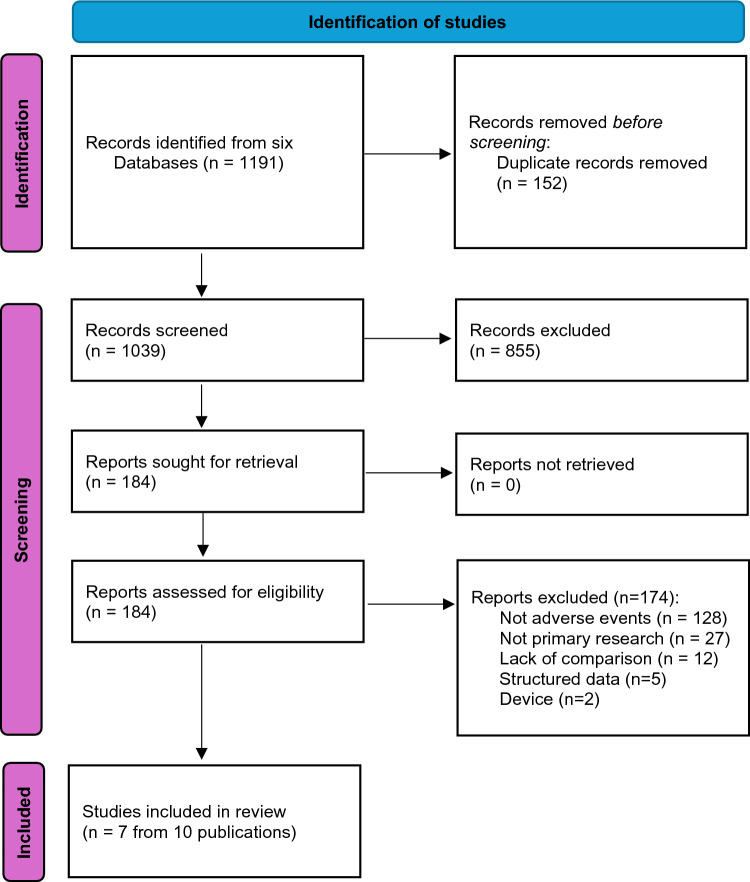
A total of 1191 references from the six databases were imported into Covidence after eliminating 152 duplicates. Of the 1039 remaining records screened based on titles and abstracts, 857 were excluded for being irrelevant to the review topic. Of the 184 articles reviewed for full-text screening, 174 were excluded based on the predetermined inclusion criteria (see Supplementary material). Ultimately, 7 studies from 10 publications (Table 2) spanning the publication years from 2013 to 2023 met all our inclusion criteria [35–44] (Fig. 1). For each study thereafter we cite all the related publications.
Table 2.
Studies included in this scoping review
| Author, year | Aim | Methods | Data source | Drug-ADE | Evaluation metrics used | Results | Conclusion |
|---|---|---|---|---|---|---|---|
| Cai 2016 [39] | Assess association between arthralgia and vedolizumab | Used NLP to extract mentions of arthralgias, then a logistic regression models to test the association between arthralgia and vedolizumab. The NLP approach was compared against ICD-9 coding information. Used unspecified NLP method for extraction | 367 IBD patients on vedolizumab and 1218 IBD patients on TNFi (control) in two US hospitals compared to ICD-9 codes | Vedolizumab—arthralgia |
Sensitivity, specificity, PPV, NPV PPV = 0.9, NPV = 0.88, Sensitivity = 0.83, Specificity = 0.93 |
The NLP approach had a higher accuracy for identifying arthralgias V/S ICD-9 codes: PPV 0.90 vs 0.78, NPV 0.88 vs 0.71. Using NLP, the prevalence of arthralgia was higher among IBD patients who received vedolizumab (77.1 %) compared to those who did not (49.1 %) | Confirmed increased arthralgia with vedolizumab. NLP was able to uncover AE-drug associations not visible through traditional coding |
| Geva 2020 [38] | Compare ADE rates determined from EHRs and administrative claims data among children treated with drugs for PH | Used NLP (Apache [cTAKES]) to extract textual mentions of medication and signs/symptoms that may represent ADE mentions from clinical notes, compared the results to EHR diagnostic codes and payor claims data. Used rule-based NLP tool for extraction | Two retrospective data sources including the EHR dataset from the Boston Children’s Hospital and the claims dataset from a national, private health plan in the USA. After data analysis and filtering, 286 patients that have used PH-targeted medication from EHR dataset and 253 patients that claimed for at least 1 PH-targeted drug from claims dataset. EHR dataset contains plain-text admission, discharge, consultation, progress, emergency department, procedure, and clinic notes | Drugs for pulmonary hypertension in children (sildenafil, tadalafil, bosentan, ambrisentan)—ADEs (anaemia, diarrhoea, oedema, headache, hearing loss, dizziness/hypotension, intracranial haemorrhage, priapism, rash/flushing, reflux, seizure, sinusitis, syncope/pre-syncope, thrombocytopenia/bleeding, transaminitis, visual changes (including ischaemic optic neuropathy) |
F1score, precision, recall. F1 = 0.78, precision = 0.69, recall = 0.9 |
cTAKES: F1 score on held out EHR data (38 notes for 12 patients) was 0.78. ADE rates differed between the EHR clinical notes and diagnostic codes. Of 40 potential ADEs examined, 6 (15 %) were identified significantly more frequently in the EHR clinical notes. An additional 13 potential ADEs were identified only in clinical notes but not in diagnostic codes. Only 1 potential ADE was identified significantly more frequently in the diagnostic codes. Some potential ADEs were similar in both data sources |
Analysis of clinical notes generally identifies more potential ADEs than diagnostic codes in either EHR or insurance claims datasets, but certain diagnoses are better represented in structured data |
| Green 2019 [35] also in Hazlehurst 2019 [40] | Enhance automated methods for opioid overdose identification and classification in EHRs | Built a rule-based NLP knowledge module based on the MediClass system to identify and classify the opioid overdose from clinical notes of EHR encounter records. They used terms or concepts related to the indicators of overdose and type of overdose as variables to enhance a code-based algorithm. Each NLP‐derived variable was tested using logistic regression analyses to determine whether or not its addition improved performance beyond that of each respective code‐based algorithm. Used rule-based NLP tool for extraction | 1006 (n = 627 used) records for the development dataset, 1696 (n = 710 used) records for the validation dataset, and 435 (n = 305 used) records for the portability dataset from two US health systems (ED, inpatient, outpatient, and telephone follow-up) compared to chart audit | Opioid—overdose |
Sensitivity, specificity, PPV, NPV Identification of the substance involved in ADR: PPV = 0.32, NPV = 0.89, Sensitivity = 0.24, specificity = 0.93 |
The NLP method performed well in identifying overdose (sensitivity = 0.80, specificity = 0.93), intentional overdose (sensitivity = 0.81, specificity = 0.98), and involvement of opioids (excluding heroin, sensitivity = 0.72, specificity = 0.96) and heroin (sensitivity = 0.84, specificity = 1.0). However, it performed poorly at identifying adverse drug reactions and overdose due to patient error and moderately at identifying substance abuse in opioid‐related unintentional overdose (sensitivity = 0.67, specificity = 0.96) | Accurately identified overdoses in EHR text and some types of overdoses. Performed well overall but poorly on some classifications. The NLP‐enhanced algorithms for suicides/suicide attempts and abuse‐related overdoses perform significantly better than code‐based algorithms and are appropriate for use in settings that have data and capacity to use NLP |
| Harpaz 2013 [37] | Develop a better signal detection strategy for potentially novel ADRs by requiring signalling from both AERS of the Food and Drug Administration and EHRs in both sources |
Used a rule-based NLP system—MedLEE to extract and normalize medications, diseases, and signs and symptoms. Temporal information corresponding to admission, discharge, and visit dates was also extracted. Laboratory test data directly available in structured form and based on internal New York Presbyterian Hospital (NYPH) codes were linked to each of the narratives, and, together with the data dimensions extracted from the narratives, formed the set of clinical variables used for statistical signal detection Used rule-based NLP tool for information extraction, and statistical analysis to estimate surrogate measures of association between specific drug-event (outcome) combinations |
Full dataset consisted of 7 years (2004–2010) of data, around 1.2 million narratives, and 178,000 patients from unstructured HER—clinical narratives—corresponding to discharge summaries, admission notes, and outpatient office visits compared to over 4 million FAERS reports | Any drug –rhabdomyolysis, acute pancreatitis, and QT prolongation |
Precision at K (signals selected), recall at k, F1and average precision. For the union of the established and plausible classes of ADRs: precision at K = 85, recall at k = 20, F1score = 30 |
The combined AERS and EHR system outperforms the AERS system for all values of K examined, and often by a large margin | Replicated signalling in AERS and EHRs can enhance signal detection under certain operating scenarios and objectives. This approach leads to improved accuracy of signal detection when the goal is to produce a highly selective ranked set of candidate ADRs. Such a system is not intended to replace, but rather augment, the portfolio of existing approaches |
| Lependu 2013 [43, 44] | Investigate adverse drug events in large volumes of free-text clinical notes | Used NLP pipelines to extract and normalize biomedical concepts such as drug, disease, device, and procedures. Uses ~5.6 million strings from existing terminologies; filters unambiguous terms that are predominantly noun phrases, uses the cleaned up lexicon for term recognition to tag or annotate the text; excludes negated terms or terms that apply to family and medical history; normalises all terms using the ontology hierarchies; and finally uses the time stamps of the note to produce a deidentified, temporally ordered patient–feature matrix. Compares to recalls or alerts or signals. Ontology based text processing workflow Extraction. Used rule-based NLP tool for extraction | Stanford Translational Research Integrated Database Environment, which spans 18 years of patient data from 1.8 million patients; it contains 19 million encounters, 35 million coded ICD-9, diagnoses, and >11 million unstructured clinical notes, which are a combination of pathology, radiology, and transcription reports | 12 distinct ADEs, 78 distinct drugs, 28 positive cases, and 165 negative cases |
Odds ratio, propensity score, ROC, AUROC Detecting 28 true positive associations from the single drug–adverse event reference set: AUROC (adjusted) = 0.804; a threshold of 1.0 on the lower bound of the 95 % CI, sensitivity = 0.39, Specificity = 0.975 |
Using the set of terms corresponding to the definition of the event of interest: sensitivity = 0.74, specificity = 0.96. For 28 true positive associations from the single drug–adverse event reference set: the overall performances of detecting associations between a single drug and its adverse event, with an AUROC of 75.3 % (unadjusted) and 80.4 % (adjusted). A threshold of 1.0 (a commonly used cut-off) on the lower bound of the 95 % CI of the adjusted ORs translates to 39 % sensitivity and 97.5 % specificity | Reproduced rofecoxib signal and other drug recalls or alerts. Provided methodology for mining clinical notes using NLP/terminologies to transform into structured data for pharmacovigilance. Analysis of textual clinical notes could detect adverse drug events 2 years before the official alert |
| Murphy 2023 [41] | Investigate drug-related AKI causes in ICU | Used keyword search to identify AKI and DAKI from clinical notes; and combined other data from the allergy module and diagnostic codes for an observational study on ICU data. Compares diagnosis codes (structured data), allergy module (semi-structured data), and clinical notes (unstructured data). Used keyword search for information retrieval | 8124 ICU admissions were included, with 542 (6.7 %) ICU admissions experiencing AKI stage 2 or 3 from ICU data from a hospital in the Netherlands | Nephrotoxic drugs –drug-induced acute kidney injury | NA (keyword search) | 18.8 % of AKI drug-related; all DAKI cases retrieved were documented in the clinical notes, none were retrieved by diagnostic codes | Highlighted limitations around identifying drug-associated AKI from unstructured documentation, informing the need for better NLP/ML to improve extraction, and reporting of ADEs |
| Wang 2017 and 2018 [36, 42] | Screen for ADEs combining FAERS and EMRs | Used NLP techniques (MedTagger—CRF based ML model) to extract treatment outcome information from unstructured EMR text and then standardize the extracted terms using UMLS, and adopted a case-crossover design with drug use as the exposure. Proposes methods combine FAERS and EMR data. Compares with SIDER and ADReCS. Used CRF-based machine learning model for information extraction, and a case-crossover design with drug use as the exposure for association analysis | Rheumatoid arthritis patients with 15,826 notes from a US medical centre | Disease-modifying antirheumatic drugs—ADEs |
Odds ratio, precision, recall, F1 score Precision = 0.1, Recall = 0.43, F1 = 0.156 |
For ADE signal detection of conventional DMARDs in rheumatoid arthritis patients: combining FAERS with EMR achieved better F1 scores in detecting ADEs. For instance. using ADReCS as gold standard, their approaches achieve 0.111 of F1 score on Methotrexate, and 0.153 of F1 score on leflunomide. Flexible mapping strategy helped improve recall, e.g., using SIDER as gold standard, 0.578 vs 0.376 of recall on methotrexate. Signals detected from EMR have considerably overlapped with signals detected from FAERS or ADE knowledge bases, implying the importance of EMR for pharmacovigilance | Recall was greatly increased when combining FAERS with EMR compared with FAERS alone and EMR alone. Combining data from clinical notes using NLP and FAERS data improved ADE detection |
ADE adverse drug event, AE adverse events, AERS adverse event reporting system, AKI acute kidney injury, CRF conditional random fields, cTAKES clinical Text Analysis Knowledge Extraction System, DAKI drug-associated acute kidney injury, DMARDS disease-modifying antirheumatic drugs, EHRs electronic health records, EMR electronic medical record, FAERS FDA Adverse Event Reporting System, FDA US Food and Drug Administration, HER human epidermal growth factor receptor, IBD inflammatory bowel disease, ICD-9 International Classification of Diseases 9th revision, ICU intensive care unit, NLP natural language processing, NPV negative predictive value, PH pulmonary hypertension, PPV positive predictive value, TNFi tumour necrosis factor inhibitor, UMLS Unified Medical Language System
Fig. 1.
Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) flow diagram for included studies
Study Sample Size
The sample sizes of the reviewed studies varied greatly, whilst some reported the number of patients (from 286 to 1.8 million) others reported the quantity of clinical notes (up to 11 million EHR notes). One study [41] focused specifically on ICU notes, four studies [35, 37, 38, 40, 43, 44] on emergency departments, inpatient departments or outpatient departments, whilst the remaining two studies [36, 39, 42] did not specify any particular note types. The outcomes examined revolved around the identification and documentation of specific adverse events including arthralgias, opioid overdoses, diarrhoea, hypoglycaemia, acute pancreatitis and acute kidney injury, among others. Comparisons of the data retrieved from unstructured notes to other data sources were mostly confined to comparisons with structured data [35, 38–41], with some comparisons to FAERS [36, 37, 42], claims data [35, 38, 40], and known signals [36, 37, 42–44].
NLP/ML Approaches
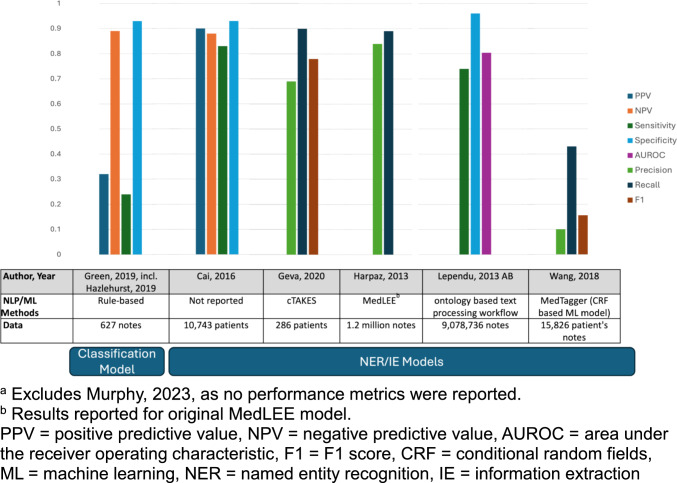
The studies listed in Table 2 cover a range of applications and methodologies of how NLP and ML are utilised for pharmacovigilance purposes and for evaluating patient safety outcomes. The NLP tasks covered in these reviewed studies include information extraction [36–39, 42–44], classification methods [35, 40] and information retrieval [41]. Two studies used information extraction to identify ADEs directly [37, 38], while the remaining three studies [36, 39, 42–44] used information extraction as a preliminary step for further analysis. An example showed that one study used extracted information to build logistic regression to assess the association between arthralgia and vedolizumab [35, 39, 40], another study used a temporally ordered patient feature matrix to investigate ADEs in EHR [43, 44], and another study used a case-crossover design to screen potential ADEs [36, 42]. For the classifier task, a rule-based classifier was used to identify opioid overdose [35, 40]. For the information retrieval task, one study [41] utilised keyword search to find documentation of drug-associated acute kidney injury (DAKI) in clinical notes. Clinical applications in these studies include detecting association between medications and their ADRs [36, 39, 42–44], identifying or classifying ADEs [35, 37, 38, 40], and investigating drug-related acute kidney injury [41]. Figure 2 provides an overview of the NLP/ML methods used and synthesizes the reported results of the best performing system for each study.
Fig. 2.
Overview of system performance for included studiesa
Overview of ADE Identification/Classification Performance
For identifying ADEs from clinical notes, three studies employed rule-based methods for ADE identification/classification tasks. Green et al (2019) [35, 40] leveraged NLP to build rule-based systems for identifying and classifying opioid-related overdoses using EHR data. By extracting information related to overdose indicators and types from clinical narratives, they demonstrated that NLP‐enhanced algorithms for suicides/suicide attempts and abuse‐related overdoses perform significantly better than code‐based algorithms and are appropriate for use in the setting that have data and capacity. Geva et al (2020) [38] used a NLP tool—Apache clinical Text Analysis Knowledge Extraction System (cTAKES)—to extract textual mentions of medication and signs/symptoms that may represent ADE mentions from clinical notes. In their experiments, using cTAKES to analyse clinical notes generally identified more potential ADEs than diagnostic codes in either EHR or insurance claims datasets. Similarly, Harpaz et al (2013) [37] used an NLP tool—MedLEE—to extract and normalise medications, diseases, and signs and symptoms from EHR notes. After combining this extracted information with the signal from adverse event reporting system (AERS), they found that this strategy enhanced signal detection under certain operating scenarios and objectives for potentially novel ADRs.
Overview of ADE-Drug Association Detection Model Performance
For detecting association between medications and their ADEs, these studies [36, 39, 41–44] first utilised NLP techniques to extract ADEs, and such ADE information was applied in different models for association analysis. For instance, Cai et al (2016) [39] employed NLP to extract mentions of arthralgias from clinical notes and subsequently used logistic regression models to assess the association between arthralgia and vedolizumab. Their NLP approach demonstrated higher accuracy compared to traditional ICD-9 coding methods, enabling more precise identification of adverse events. Lependu et al (2013) [43, 44] used NLP pipelines to extract and normalise biomedical concepts such as the drug, disease, device, and procedure, and used such information and the time stamps of the clinical notes to produce a deidentified, temporally ordered patient feature matrix, which was then used for detecting drug safety signals at scale. The overall performances of detecting associations between a single drug and its adverse event were reported [43, 44], with an area under the receiver operating characteristic (AUROC) curve of 75.3 % (unadjusted) and 80.4 % (adjusted). The unadjusted AUROC measures the raw association between drug and its ADEs; the adjusted AUROC is for cancelling the effect of potential confounders. Wang et al 2018 [36, 42] applied a ML-based NLP system (MedXN) [45] to extract and a Unified Medical Language System (UMLS) dictionary to normalise drugs and clinical outcomes from unstructured text, and then adopted case-crossover design to detect signals of association between drug and potential ADEs for patients with certain indications. They compared the extracted ADE from FAERS and Electronic Medical Record (EMR) against two ADE knowledge bases. Their finding showed that recall of ADE extraction greatly increased when combining FAERS with EMR compared with FAERS alone and EMR alone, especially for flexible mapping strategy. In addition, signals detected from EMR have considerably overlapped with signals detected from FAERS or ADE knowledge bases, implying the importance of EMR for pharmacovigilance.
Murphy et al (2023) [41] used keyword search to identify acute kidney injury (AKI) and drug-related AKI from ICU data. They highlighted limitations around identifying drug-associated AKI from unstructured documentation, informing on the need for better NLP/ML to improve the extraction and reporting of ADRs.
The comparisons with other data sources gave varying results but overall authors concluded that the addition of adverse events from unstructured data could improve pharmacovigilance. Lependu 2013 [43, 44] suggests that adverse events can be detected 2 years earlier with unstructured data. Others also indicate higher accuracy [37], higher recall [36, 38, 42], or new associations not visible by comparison sources [38, 39, 41]. However, some caution was recommended as certain adverse events are better represented in structured data [35, 38, 40]. For instance, in Green et al., the NLP performed well in identifying overdose and intentional opioid overdose but performed poorly at identifying overdose due to patient error [35, 40].
Discussion
In summary, this review evaluated the effectiveness of using NLP/ML to leverage unstructured EHR data to enhance pharmacovigilance. A total of 7 studies in 10 articles published between 2013 and 2023 were included, the majority of which were observational in design and conducted in healthcare settings to investigate a range of adverse drug events such as AKI, hypoglycaemia and arthralgias. Overall, these studies showed that NLP and ML approaches have potential for extracting clinically useful information on adverse events from unstructured EHR/EMR data. The same finding is also shown in prior research [46]. Various approaches in the fields of NLP and ML were utilised, including rule-based systems [34–44], statistical models [36, 37, 39, 42–44], and ML-based NLP tools [36, 42].
The overall performance of these models was reasonably high across most studies based on metrics such as sensitivity, specificity, F1 scores, and AUROC [39, 43, 44]. The NLP has facilitated the creation of unstructured datasets amenable to pharmacovigilance tasks including signal detection, risk identification, and detection of underreported adverse event-drug pairs [36, 42]. However, significant variability exists in techniques, data sources, evaluation methods, and pharmacovigilance outcomes examined. Only one study [36, 42] validated findings in external datasets or across multiple sites, which may limit the generalisability of the evidence. For future work of assessing ML/NLP studies in detecting ADEs from EHR notes, we refer readers to the prior study [47].
Performance varied across models and tasks, highlighting the need for continued research and validation on diverse clinical applications. Moreover, there is currently no widely accepted set of guidelines or criteria for reporting and critically evaluating NLP/ML research in analysing EHR notes. While a few well-known benchmark datasets [48] and shared tasks [49] exist for evaluating NLP/ML methods on detecting ADEs in EHR notes, they were not widely adopted in our included studies. As this emerging field continues to develop, it will be essential to establish standards for methodology and reporting to enhance the quality of studies and facilitate effective synthesis of evidence [50]. There is also no consensus on the best techniques for representative data sampling, addressing class imbalance, optimising models, and evaluating model performance. Three studies developed their own rule-based NLP methods [39, 41, 43, 44] for information extraction or information retrieval, while the remaining four studies leveraged existing tools, including three rule-based NLP tools [35, 37, 38, 40] and one ML-based tagger [36, 42]. For more detailed information on the effectiveness and characteristics of different NLP/ML methods for analysing EHR notes, we refer readers to a systematic review of NLP/ML methods applied to clinical notes [51, 52]. Additionally, studies focusing on information extraction and information retrieval [35, 36, 39, 40, 42–44] failed to compare their methods with widely used clinical NLP tools like cTAKES [53] and MetaMap [54] for analysing EHR data.
By leveraging the comprehensive information contained within EHRs, researchers can gain access to contextual documentation pertaining to medication usage patterns and their associated effects at scale [55]. Nevertheless, it is worth mentioning that NLP or ML in isolation cannot yield conclusive proof of causal connections between medications and adverse drug reactions. To establish such relationships, a comprehensive appraisal of all accessible evidence must be conducted [56]. Moving forward, the next crucial step involves assessing the real-world effectiveness of integrating information derived from unstructured data with other evidence obtained from EHRs and EMRs [57].
This review focuses on studies that assess the detection of specific adverse drug events from unstructured EHR data compared to other sources in order to improve patient safety. In contrast, previous reviews [58, 59] provide a general overview of using NLP for pharmacovigilance from diverse data sources or focus solely on technical issues.
This review synthesises evidence on the impact of unstructured EHR/EMR data on key pharmacovigilance metrics like adverse event detection, safety signals, risk identification, and documentation practices. Prior reviews in this field [46, 60] show a lack of such evidence.
Overall, there was a consensus that unstructured data have the potential to improve pharmacovigilance by being an adjunct to other sources. However, caution should be applied as variations in the performance of detecting adverse drug events were apparent according to adverse event type, and currently there is insufficient understanding of why such discrepancies exist.
Limitations
Certain limitations of this review must be acknowledged when interpreting the results. The exclusion of non-English studies could introduce potential language bias.
Most NLP/ML studies focused on simulated datasets or single research groups, limiting insights into real-world effectiveness and generalisability. Insufficient information regarding institutional data sources, patient demographics, model specifications, and evaluation methods in certain papers can also limit the analysis. Lastly, as this was a scoping review we did not conduct formal quality assessment of the studies. However, given the lack of a validated quality assessment tool for these types of studies, rigorous quality assessment with these studies would have been challenging.
Given the preliminary research stage in this area, caution should be exercised despite the potential of NLP/ML technologies for clinical use. Conducting well-designed validation studies and fostering interdisciplinary collaboration is crucial to establish effective frameworks for incorporating these tools into pharmacovigilance practice. While this review emphasised the current possibilities, it also emphasised substantial gaps that must be addressed through rigorous research and comparison to existing pharmacovigilance systems before widespread implementation.
Implications for Practice and Research
This review has significance for how future research will be undertaken and incorporated into pharmacovigilance practice. First, it re-affirms that NLP/ML systems cannot provide evidence of causality between drugs and adverse event outcomes [49]. Their importance comes from strengthening signal detection and structured/unstructured data processing to enable more proactive safety surveillance [61]. Second, collaboration between data scientists and clinical domain specialists is necessary to successfully design, develop, and use these systems [62]. Third, synthesising higher-quality evidence would be made possible by developing methodological standards and reporting criteria specifically for NLP/ML health research [63]. Finally, before considering large-scale clinical deployment of AI technologies, comprehensive multi-site prospective studies are essential to confirm utility across varied settings and populations [64], which help to optimise resources and prevent health disparities.
Overall, while this review reveals the promising potential of NLP and ML techniques for extracting insights on adverse drug events from unstructured EHR data, much work must be done to establish the best methods, determine the utility in detail, and incorporate these novel approaches into applied pharmacovigilance practice. The review summarises the available research progress while identifying the drawbacks, biases, and knowledge gaps that must be addressed in subsequent studies. The use of these instruments to enhance patient safety will depend on standardised guidelines, multidisciplinary cooperation, and cautious evaluation.
The integration of unstructured data and NLP/ML algorithms demonstrates the potential value of clinical data embedded in narrative notes for pharmacovigilance [65]. Nevertheless, the absence of standardised frameworks and varying systems for unstructured EHR/EMR data poses obstacles to implementation.
Recommendations for Future Research
To improve future research in the use of NLP or ML on methods on EHR data, we propose several key recommendations. First, the development and adherence to standardised reporting guidelines specific to NLP/ML studies in healthcare, such as those suggested by Stevens et al. [47], is crucial. These guidelines recommend reporting detailed descriptions of data sources, preprocessing steps, model architectures and rationale, and evaluation metrics. To provide a full assessment of a model’s performance, comprehensive evaluation metrics, including precision, recall, F1-score, and AUROC, should be consistently reported with confidence intervals and statistical significance tests [66]. Systematic comparisons between NLP/ML approaches and traditional pharmacovigilance methods, as well as established clinical NLP tools like cTAKES and MetaMap, are essential. These tools have been widely used in the clinical setting and are well established as baseline methods, which can be used as comparators for different datasets. Transparency and reproducibility can be enhanced by providing detailed documentation of cohort inclusion criteria, data collection and preprocessing, model development steps, and whenever possible, making code and de-identified datasets publicly available [67, 68]. Robust study designs should include multi-site investigations to assess generalisability, prospective validation in real-world clinical settings, and utilisation of larger, more diverse datasets to improve model performance and reduce bias [69, 70]. Finally, the development and utilisation of standardised benchmark datasets for evaluating NLP/ML models in pharmacovigilance would enable fair comparisons across different approaches and studies [49]. By following these recommendations, future research can produce more robust, comparable, and clinically relevant results, ultimately advancing the integration of NLP and ML techniques into pharmacovigilance practice.
Conclusion
This comprehensive scoping review indicates that utilising NLP and ML techniques has shown the potential to enhance pharmacovigilance by leveraging unstructured EHR/EMR data. These approaches have demonstrated proficiency in identifying specific adverse events and uncovering previously unknown safety signals that would not have been apparent through structured data alone. It is important to point out the significant differences in approach among various studies, and that the levels of validation and comparison to other data sources in terms of ease of use, utility, and effectiveness remain limited. To ensure reliability and effectiveness, practicality in healthcare settings, and definitive improvement in patient outcomes, it is essential to undertake further comprehensively planned studies involving multiple sites and comparisons.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
U.S. National Library of Medicine, R01LM011176, Graciela Gonzalez-Hernandez.
Conflicts of Interest
None declared.
Ethics Approval
Not applicable. Secondary analysis of publicly available literature.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
The data supporting the findings of this scoping review are derived from published studies and are publicly accessible. A comprehensive list of the studies included in the review, along with the excluded sources, can be found in the text and supplementary materials of this manuscript. For any additional inquiries regarding the data, please contact the corresponding author.
Code Availability
Not applicable.
Authors' Contributions
Study conception and design (SG). Development of search strategies (SG). Running of search strategies (MB). Screening (MB and SG/KO). Data extraction (MB/DX). Data synthesis (DX, KO and YW). The first draft of the manuscript was written by SG and authors (GG, DX, KO, YW) commented on previous versions of the manuscript. All authors read and approved the final manuscript (GG, DX, KO, YW, MB, SG).
References
- 1.Uslu A, Stausberg J. Value of the electronic medical record for hospital care: update from the literature. J Med Internet Res. 2021;23(12): e26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garets D, Davis M. Electronic medical records vs. electronic health records: yes, there is a difference. Policy white paper Chicago, HIMSS Analytics. 2006;1.
- 3.Knevel R, Liao KP. From real-world electronic health record data to real-world results using artificial intelligence. Ann Rheum Dis. 2023;82(3):306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrenstein V, Kharrazi H, Lehmann H, Taylor CO. Obtaining data from electronic health records. Tools and technologies for registry interoperability, registries for evaluating patient outcomes: A user’s guide, 3rd edn, Addendum 2 [Internet]: Agency for Healthcare Research and Quality (US); 2019.
- 5.Manca DP. Do electronic medical records improve quality of care? Yes. Can Fam Physician. 2015;61(10):846–7, 50–1. [PMC free article] [PubMed]
- 6.Kong HJ. Managing unstructured big data in healthcare system. Healthc Inf Res. 2019;25(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford E, Oswald M, Hassan L, Bozentko K, Nenadic G, Cassell J. Should free-text data in electronic medical records be shared for research? A citizens’ jury study in the UK. J Med Ethics. 2020;46(6):367–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarwar T, Seifollahi S, Chan J, Zhang X, Aksakalli V, Hudson I, et al. The secondary use of electronic health records for data mining: data characteristics and challenges. ACM Comput Surv (CSUR). 2022;55(2):1–40. [Google Scholar]
- 9.Tayefi M, Ngo P, Chomutare T, Dalianis H, Salvi E, Budrionis A, Godtliebsen F. Challenges and opportunities beyond structured data in analysis of electronic health records. Wiley Interdiscip Rev Comput Stat. 2021;13(6): e1549. [Google Scholar]
- 10.Holmes JH, Beinlich J, Boland MR, Bowles KH, Chen Y, Cook TS, et al. Why is the electronic health record so challenging for research and clinical care? Methods Inf Med. 2021;60(01/02):32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vora LK, Gholap AD, Jetha K, Thakur RRS, Solanki HK, Chavda VP. Artificial intelligence in pharmaceutical technology and drug delivery design. Pharmaceutics. 2023;15(7):1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilipiec P, Liwicki M, Bota A. Using machine learning for pharmacovigilance: a systematic review. Pharmaceutics. 2022;14(2):266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WH Organization. Global patient safety action plan 2021–2030: towards eliminating avoidable harm in health care. Geneva: World Health Organization; 2021. [Google Scholar]
- 14.Sawarkar A, Sharma R, Gautam V, Shramankar K, Dinodia N. Pharmacovigilance: present status and future perspectives. Pharma Innov J. 2019;8(8):84–92. [Google Scholar]
- 15.Adisa R, Omitogun TI. Awareness, knowledge, attitude and practice of adverse drug reaction reporting among health workers and patients in selected primary healthcare centres in Ibadan, southwestern Nigeria. BMC Health Serv Res. 2019;19(1):926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.García-Abeijon P, Costa C, Taracido M, Herdeiro MT, Torre C, Figueiras A. Factors associated with underreporting of adverse drug reactions by health care professionals: a systematic review update. Drug Saf. 2023;46(7):625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alomar M, Tawfiq AM, Hassan N, Palaian S. Post marketing surveillance of suspected adverse drug reactions through spontaneous reporting: current status, challenges and the future. Ther Adv Drug Saf. 2020;11:2042098620938595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mc Cord KA, Hemkens LG. Using electronic health records for clinical trials: where do we stand and where can we go? CMAJ. 2019;191(5):E128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen MR. Why error reporting systems should be voluntary. BMJ. 2000;320(7237):728–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haerian K, Salmasian H, Friedman C. Methods for identifying suicide or suicidal ideation in EHRs. AMIA Ann Sympos Proc AMIA Sympos. 2012;2012:1244–53. [PMC free article] [PubMed] [Google Scholar]
- 21.Wasylewicz A, van de Burgt B, Weterings A, Jessurun N, Korsten E, Egberts T, et al. Identifying adverse drug reactions from free-text electronic hospital health record notes. Br J Clin Pharmacol. 2022;88(3):1235–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kline A, Wang H, Li Y, Dennis S, Hutch M, Xu Z, et al. Multimodal machine learning in precision health: a scoping review. NPJ Digit Med. 2022;5(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorbello A, Haque SA, Hasan R, Jermyn R, Hussein A, Vega A, et al. Artificial intelligence-enabled software prototype to inform opioid pharmacovigilance from electronic health records: development and usability study. Jmir ai. 2023 Jan–Dec;2. [DOI] [PMC free article] [PubMed]
- 24.Mower J, Bernstam E, Xu H, Myneni S, Subramanian D, Cohen T. Improving pharmacovigilance signal detection from clinical notes with locality sensitive neural concept embeddings. AMIA Jt Summits Transl Sci Proc. 2022;2022:349–58. [PMC free article] [PubMed] [Google Scholar]
- 25.Hamid AAA, Rahim R, Teo SP. Pharmacovigilance and Its importance for primary health care professionals. Korean J Fam Med. 2022;43(5):290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malmasi S, Hosomura N, Chang LS, Brown CJ, Skentzos S, Turchin A. Extracting healthcare quality information from unstructured data. AMIA Annu Symp Proc. 2017;2017:1243–52. [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Koppel R, McGreevey JD 3rd, Craven CK, Schreiber R. Transitions from one electronic health record to another: challenges, pitfalls, and recommendations. Appl Clin Inf. 2020;11(5):742–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magoc T, Allen KS, McDonnell C, Russo JP, Cummins J, Vest JR, Harle CA. Generalizability and portability of natural language processing system to extract individual social risk factors. Int J Med Inf. 2023;177: 105115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis SE, Zabotka L, Desai RJ, Wang SV, Maro JC, Coughlin K, et al. Use of electronic health record data for drug safety signal identification: a scoping review. Drug Saf. 2023;46(8):725–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edrees H, Song W, Syrowatka A, Simona A, Amato MG, Bates DW. Intelligent telehealth in pharmacovigilance: a future perspective. Drug Saf. 2022;45(5):449–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tricco AC, Lillie E, Zarin W, O’Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. 2018;169(7):467–73. [DOI] [PubMed] [Google Scholar]
- 32.Evans RS. Electronic health records: then, now, and in the future. Yearb Med Inf. 2016;25(Suppl 1):S48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haddaway NR, Collins AM, Coughlin D, Kirk S. The role of google scholar in evidence reviews and its applicability to grey literature searching. PLoS ONE. 2015;10(9): e0138237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Covidence systematic review software, Veritas Health Innovation, Melbourne, Australia. [cited 2024 5th October]. Available from www.covidence.org. Accessed 5 Oct 2024.
- 35.Green CA, Perrin NA, Hazlehurst B, Janoff SL, DeVeaugh-Geiss A, Carrell DS, et al. Identifying and classifying opioid-related overdoses: a validation study. Pharmacoepidemiol Drug Saf. 2019;28(8):1127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L, Rastegar-Mojarad M, Liu S, Zhang H, Liu H. Discovering adverse drug events combining spontaneous reports with electronic medical records: a case study of conventional DMARDs and biologics for rheumatoid arthritis. AMIA Jt Summits Transl Sci Proc. 2017;2017:95–103. [PMC free article] [PubMed] [Google Scholar]
- 37.Harpaz R, Vilar S, Dumouchel W, Salmasian H, Haerian K, Shah NH, et al. Combing signals from spontaneous reports and electronic health records for detection of adverse drug reactions. J Am Med Inf Assoc JAMIA. 2013;20(3):413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geva A, Abman SH, Manzi SF, Ivy DD, Mullen MP, Griffin J, et al. Adverse drug event rates in pediatric pulmonary hypertension: a comparison of real-world data sources. J Am Med Inf Assoc JAMIA. 2020;27(2):294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai T, Kane-Wanger G, Bond A, Cagan A, Murphy SN, Ananthakrishnan A, Liao K. Natural language processing to rapidly identify potential signals for adverse events using electronic medical record data: example of arthralgias and vedolizumab. Arthritis Rheumatol. 2016;68(Supplement 10):2802–4. [Google Scholar]
- 40.Hazlehurst B, Green CA, Perrin NA, Brandes J, Carrell DS, Baer A, et al. Using natural language processing of clinical text to enhance identification of opioid-related overdoses in electronic health records data. Pharmacoepidemiol Drug Saf. 2019;28(8):1143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy RM, Dongelmans DA, Kom IY, Calixto I, Abu-Hanna A, Jager KJ, et al. Drug-related causes attributed to acute kidney injury and their documentation in intensive care patients. J Crit Care. 2023;75:154292. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Rastegar-Mojarad M, Ji Z, Liu S, Liu K, Moon S, et al. Detecting pharmacovigilance signals combining electronic medical records with spontaneous reports: a case study of conventional disease-modifying antirheumatic drugs for rheumatoid arthritis. Front Pharmacol. 2018;9(101548923):875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther. 2013;93(6):547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Ghebremariam YT, Cooke JP, Shah NH. Pharmacovigilance using Clinical Text. AMIA Jt Summits Transl Sci Proc. 2013;2013:109. [PMC free article] [PubMed] [Google Scholar]
- 45.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc. 2014;21(5):858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young IJB, Luz S, Lone N. A systematic review of natural language processing for classification tasks in the field of incident reporting and adverse event analysis. Int J Med Inf. 2019;132:103971. [DOI] [PubMed] [Google Scholar]
- 47.Stevens LM, Mortazavi BJ, Deo RC, Curtis L, Kao DP. Recommendations for reporting machine learning analyses in clinical research. Circ Cardiovasc Qual Outcomes. 2020;13(10): e006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henry S, Buchan K, Filannino M, Stubbs A, Uzuner O. 2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records. J Am Med Inf Assoc. 2020;27(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jagannatha A, Liu F, Liu W, Yu H. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 10). Drug Saf. 2019;42(1):99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bohr A, Memarzadeh K. The rise of artificial intelligence in healthcare applications. Int J Med Inf. 2019;132:103971. [Google Scholar]
- 51.Sim JA, Huang X, Horan MR, Stewart CM, Robison LL, Hudson MM, et al. Natural language processing with machine learning methods to analyze unstructured patient-reported outcomes derived from electronic health records: a systematic review. Artif Intell Med. 2023;146: 102701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inf Assoc. 2020;27(3):457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inf Assoc. 2010;17(5):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inf Assoc. 2010;17(3):229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Batko K, Ślęzak A. The use of big data analytics in healthcare. J Big Data. 2022;9(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalez-Hernandez G, Krallinger M, Muñoz M, Rodriguez-Esteban R, Uzuner Ö, Hirschman L. Challenges and opportunities for mining adverse drug reactions: perspectives from pharma, regulatory agencies, healthcare providers and consumers. Database. 2022. 10.1093/database/baac071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dang A. Real-world evidence: a primer. Pharmaceut Med. 2023;37(1):25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural language processing for ehr-based pharmacovigilance: a structured review. Drug Saf. 2017;40(11):1075–89. [DOI] [PubMed] [Google Scholar]
- 59.Wong A, Plasek JM, Montecalvo SP, Zhou L. Natural language processing and its implications for the future of medication safety: a narrative review of recent advances and challenges. Pharmacotherapy. 2018;38(8):822–41. [DOI] [PubMed] [Google Scholar]
- 60.Hauben M. The potential of artificial intelligence in pharmacovigilance. Clin Ther. 2021;43(2):372–9. [DOI] [PubMed] [Google Scholar]
- 61.Murali K, Kaur S, Prakash A, Medhi B. Artificial intelligence in pharmacovigilance: practical utility. Indian J Pharmacol. 2019;51(6):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bates DW, Levine D, Syrowatka A, Kuznetsova M, Craig KJT, Rui A, et al. The potential of artificial intelligence to improve patient safety: a scoping review. npj Digit Med. 2021;4(1):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arno A, Elliott J, Wallace B, Turner T, Thomas J. The views of health guideline developers on the use of automation in health evidence synthesis. Syst Rev. 2021;10(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Titler MG. The evidence for evidence-based practice implementation. In: Titler MG, Hughes RG, editors. Patient safety and quality: an evidence-based handbook for nurses. Rockville, MD: Agency for Healthcare Research and Quality; 2008. [PubMed] [Google Scholar]
- 65.Liang L, Hu J, Sun G, Hong N, Wu G, He Y, et al. Artificial intelligence-based pharmacovigilance in the setting of limitedresources. Drug Saf. 2022;45(5):511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hicks SA, Strümke I, Thambawita V, Hammou M, Riegler MA, Halvorsen P, Parasa S. On evaluation metrics for medical applications of artificial intelligence. Sci Rep. 2022;12(1):5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.National Academies of Sciences E, and Medicine; Policy and Global Affairs; Committee on Science, Engineering, Medicine, and Public Policy; Board on Research Data and Information; Division on Engineering and Physical Sciences; Committee on Applied and Theoretical Statistics; Board on Mathematical Sciences and Analytics; Division on Earth and Life Studies; Nuclear and Radiation Studies Board; Division of Behavioral and Social Sciences and Education; Committee on National Statistics; Board on Behavioral, Cognitive, and Sensory Sciences; Committee on Reproducibility and Replicability in Science. . Reproducibility and Replicability in Science. Washington (DC): National Academies Press (US); 2019 May 7. 6, Improving Reproducibility and Replicability; 2019. Available from https://www.ncbi.nlm.nih.gov/books/NBK547525/
- 68.O’Connor K, Golder S, Weissenbacher D, Klein AZ, Magge A, Gonzalez-Hernandez G. Methods and annotated data sets used to predict the gender and age of twitter users: scoping review. J Med Internet Res. 2024;26: e47923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Soltan AAS, Clifton DA. Machine learning generalizability across healthcare settings: insights from multi-site COVID-19 screening. NPJ Digit Med. 2022;5(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, Soltan AAS, Eyre DW, Clifton DA. Algorithmic fairness and bias mitigation for clinical machine learning with deep reinforcement learning. Nat Mach Intel. 2023;5(8):884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.