Abstract
Background
SERPINI1 is a protein-coding gene, which has been reported to be related to malignancies, and the encoding protein is a secreted protein. Nevertheless, the specific effect of SERPINI1 on Hepatocellular carcinoma (HCC) remains unclear.
Methods
The expression level of SERPINI1 in cancers was detected by the Gene Expression Omnibus (GEO) database, the Gene Expression Profiling Interactive Analysis (GEPIA) database and the collected serum of HCC patients. The receiver operating characteristic (ROC) curve and area under curve (AUC) were used to evaluate the diagnostic effectiveness of serum SERPINI1 and the combination of AFP and SERPINI1 for HCC. The Kaplan-Meier (KM) survival was used to evaluate the prognostic capacity of SERPINI1 for HCC in GEPIA database. Furthermore, the correlations between clinicopathological characteristics and the level of serum SERPINI1 were analyzed. Besides, we detected the expression of SERPINI1 in HepG2 by qPCR and western blot, and confirmed the biological function of SERPINI1 through MTT, EdU, wound healing and transwell invasion assay.
Results
The results indicated that the level of SERPINI1 was significantly increased in tissue and serum of HCC patients. ROC analysis displayed that SERPINI1 had a significantly diagnostic value for HCC, the combination of AFP and SERPINI1 gained the higher specificity and sensitivity. The KM survival curves indicated that patients with SERPINI1 overexpression had worse overall survival. Furthermore, we found the positive correlations between serum SERPINI1 level and some clinicopathological characteristics, such as tumor size, differentiation degrees and so on. In addition, in vitro experiments revealed that SERPINI1 could promote the proliferation and invasion of HCC.
Conclusions
Taken together, our study demonstrates that SERPINI1, which is highly expressed in HCC and closely related to cell proliferation and invasion, may serve as a novel biomarker for diagnosis and prognosis of HCC.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12935-025-03716-y.
Keywords: SERPINI1, Hepatocellular carcinoma, Biomarker, Diagnosis
Introduction
Globally, liver cancer poses a serious threat to human health, which has an increasing incidence and mortality. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for approximately 90% of all cases [1]. In recent years, great progresses have been made in HCC therapy, but the prognosis of HCC patients still remains poor, because of high rate of recurrence and being diagnosed at an advanced stage. Over the past years, magnetic resonance imaging (MRI), computer tomography (CT), ultrasound examinations, and the histological pathological examination were the important diagnostic tools of HCC [2]. However, their application has been limited because of insensitivity to small tumors, invasiveness, and expensive. Currently, more and more tumor markers for HCC have been reported in different articles, but only a few of them have been validated and successfully used in clinical practice. Alpha fetoprotein (AFP) and prothrombin induced by vitamin K absence or antagonist-II (PIVKA-II) are the most widely used biomarkers for HCC diagnosis clinically [3]. Nevertheless, the sensitivity and specificity of which are unsatisfactory, especially for patients with early-stage HCC. Therefore, identification of novel molecular markers is desperately needed for improving the diagnosis and prognosis of HCC.
Bioinformatic analysis is more likely to overcome the shortcomings of known markers, because it allows researchers to comprehensive analyze data from a larger number of clinical samples from differently independent studies deeply, with these data, researchers can find many promising biomarkers and update understanding to diseases [4–6]. In recent years, additional biomarkers for HCC have been proposed, including cytokeratin 19, Golgi protein 73, and so on. It has been reported that the expression of cytokeratin 19 was associated with the poor differentiated grade, metastasis and microvascular invasion, nevertheless, which has not been considered enough for clinical application as indicator for HCC diagnosis. Golgi protein 73 has been considered as a biomarker for HCC, but its levels in patients with hepatic parenchymal tumor may also be increased. In this study, through bioinformatic analysis, we find a potential diagnostic and prognostic marker for HCC. Serpin peptidase inhibitor, clade I, member 1 (SERPINI1) is a secreted protease which can inhibit tissue type plasminogen activator and plasmin [7]. Although study remains less, the SERPINI1 gene has been reported to be related to malignancies. For instance, it has been reported that SERPINI1, S100 calcium binding proteins A8/A9, and growth differentiation factor 15 may be potential biomarkers for the diagnosis of colorectal cancer (CRC) [8]. SERPINI1 also can regulate the progress of ovarian cancer and the epithelial–mesenchymal transition (EMT) in an orthotopic implantation model of colorectal cancer [9]. In addition, SERPINI1 was reported to be abnormally down-regulated in gastric cancer [10]. All these studies suggested that SERPINI1 may be carcinogenic or anticancer in different cancers. However, so far, the expression and biological functions of SERPINI1 in HCC have rarely been reported. SERPINI1 may be a promising biomarker compared to other candidates.
Here, we performed an integrative analysis with clinical features and survival data of HCC in public databases and the collected serum samples to ascertain the value of SERPINI1 in HCC. We found that the high expression of SERPINI1 was correlated with the tumor size, differentiation degrees, stage, vascular invasion and lymph node metastasis. The up-regulated SERPINI1 can promote the proliferation, migration and invasion of HCC. Thus, our research identified the potential role of SERPINI1, which may be a novel diagnostic and prognostic biomarker in HCC.
Materials and methods
Patients and specimens
A total of 291 serum samples were acquired from 153 patients diagnosed with HCC and 138 healthy controls (HC) between October 2021 and June 2023 at Tianjin Medical University Cancer Institute and Hospital (Tianjin, China). Participators, who have taken anticoagulant medications such as vitamin K or warfarin within 6 months, and those with thrombotic or hemorrhagic diseases, were ruled out. Venous blood was obtained from the participants before treatment, centrifuged 20 min at 3000 rpm, transferred the supernatant to EP tube, and frozen to − 80 °C quickly, avoiding thawing and freezing cycles. Clinicopathological feature data, including gender, age, tumor number and size, stage, differentiation degree, vascular invasion, lymph node metastasis, distant metastasis, and complications were obtained.
Quantification of SERPINI1 levels in serum using enzyme-linked immunosorbent assays (ELISAs)
According to the manufacturer’s instructions, we detected the levels of serum SERPINI1 in the participants using the ELISA kit (USCN Life Sciences, USA) [8]. Briefly, 100 µl serum samples, standard substances and diluent (blank) were added into 96 well plates, separately. The plates were placed quietly 1 h at 37 °C. Then, add 100µL Detection Reagent A, incubate at 37 °C for 1 h. After three washes with PBST, added 100 µl enzyme-labeled antibody and incubated 30 min at 37 °C. Subsequently, washed with PBST three more times, added 90 µl substrate solution and 50 µl stop solution to each well. The absorbance of samples, standard substances and blank wells at 450 nm were measured by microplate reader (Thermo, Walsham, Massachusetts, USA).
Quantification of AFP levels in serum using electrochemiluminescence immunoassays
Using the Roche Cobas E601 electrochemical immunoluminescence analyzer (Roche Diagnostics, Mannheim, Germany) equipped with Roche dedicated reagents, we detected the levels of AFP in serum, according with the manufacturer’s instructions.
Cell culture and antibodies
The human hepatocellular cells HepG2, the human hepatic cells L02 and human umbilical vein endothelial cells (HUVECs) were obtained from the cell resource center of Shanghai (Shanghai, China). We cultivated the HepG2 in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Camarillo, CA, USA) supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml) and 10% fetal bovine serum (FBS, Gibco, Gaithersburg, MD, USA). We cultivated the L02 and HUVECs in Roswell Park Memorial Institute 1640 Medium (Gibco, Grand Island, NY, USA) supplemented with penicillin (100 U/ml), streptomycin (100 µg/ml) and 10% FBS. All cells were cultivated in an atmosphere containing 5% CO2 at 37 °C. The antibodies for SERPINI (Affinity, AF4065), E-Cadherin (Affinity, AF0131), Vimentin (Affinity, AF7013), MMP9 (Affinity, AF5228), GAPDH (Cell Signaling Technology, 5174), goat anti-mouse IgG (Abcam, ab97023), goat anti-rabbit IgG (Abcam, ab97051) and neuroserpin (Aipudi Biological Technology, China) were purchased.
Transfection of small interfering RNA (siRNA) and plasmids
We purchased the siRNA targeting the sequence of SERPINI1 and the overexpressed plasmids of SERPINI1 from GenePharma Biological Technology (Shanghai, China), to further analyze the function of gene knockdown or overexpression. When the cells were cultured to 50% confluence, they were transfected with the siRNAs or the overexpressed plasmids using Lipofectamine 3000 (Life Invitrogen Technology, NY, USA) and Opti-MEM (Gibco, Gaithersburg, MD, USA) following the manufacturer’s instructions. After culturing for 48 hours, the total RNA was extracted and analyzed by real-time PCR to ascertain the knockdown or overexpressed efficiency. The sequences of siRNA targeting SERPINI1 were as follows: SERPINI1 si-1 sense 5’-GCUGUGCUGUAUCCUCAAGUUTT-3’ and antisense 5’-AACUUGAGGAUACAGCACAGCTT-3’; SERPINI1 si-2 sense 5’-GGUUCACAGUGGAACAGGAAATT-3’ and antisense 5’-UUUCCUGUUCCACUGUGAACCTT-3’; SERPINI1 si-3 sense 5’-UCCUUCCUAGAGGUUAAUGAATT-3’ and antisense 5’-UUCAUUAACCUCUAGGAAGGATT − 3’. The experiments were performed triplicately.
RNA extraction and quantitative real-time polymerase chain reaction (qPCR)
According to the manufacturer’s protocol of the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), we extracted the total RNA from cells. Firstly, the total RNA was processed with DNase I (Takara, Dalian, China). Then, the cDNA was reverse transcriptase synthesized using the First-strand cDNA synthesize kit (#1622, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions with random hexamer primers. Furthermore, the synthesized cDNA templates were amplified with SYBR Green master kit (Takara, Dalian, China), and the expression level of GAPDH was used as the internal control. The relative expression levels were calculated by comparing the normalized cycle threshold (Ct) [11]. The sequences of primers were as follows: SERPINI1 (forward): 5’-TAGCCGTGGCCAACTACATC-3’, (reverse): 5’-GGCAGCATCAAAATCCCTTG-3’; GAPDH (forward): 5’-ACCCAGAAGACTGTGGATGG-3’, (reverse): 5’-TTCAGCTCAGGGATGACCTT-3’. The experiments were performed triplicately.
Western blot analysis
We extracted the total proteins using cell lysis buffer radioimmunoprecipitation assay (RIPA, Cell Signal Technology, USA) with 1% phenylmethylsulfonyl fluoride (PMSF) and 1% phosphatase inhibitor. The BCATM Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA) was used to detect the protein concentrations, and the BSA served as standard. Along with 4 × loading buffer, ddH2O, and moderate amounts of samples were boiled for 4 min and then subjected to SDS-PAGE. After electrophoresis, the separated proteins were blotted onto PVDF membranes with 300 mA constant current at 4 °C for 3 h. Sequentially, the PVDF membranes were washed in TBST, blocked with 5% non-fat milk, and incubated with the primary antibodies, including SERPINI1 (1:1000), E-Cadherin (1:1000), Vinmentin (1:1000) MMP9 (1:1000) and GAPDH (1:100000) overnight at 4 °C. The levels of GAPDH were severed as control. Next day, the membranes were incubated with secondary antibodies at room temperature for 1 h. Finally, we detected the signal by chemiluminescence (ECL) system (Merck Millipore, Darmstadt, Germany) [11]. The experiments were performed triplicately.
Cell proliferation assay
Cell proliferation ability was detected by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazo-lium bromide (MTT) (Sigma-Aldrich, St. Louis, MO, USA), according to the manufacturer’s instructions. Briefly, cells were cultured in 96-well culture plats (5 × 103 cells/well), attached for 24 h, and then transfected with different siRNAs or plasmids. Approximately after 48 h, the MTT (5 mg/ml) was added into the cells at 37℃ for 4 h. Absorbance at 490 nm was detected with the plate reader. In addition, we assessed cell proliferation using the Cell-Light EdU Apollo567 In Vitro Kit (C10310-1, RIBOBIO) in accordance with the instruction. The experiments were performed triplicately.
Wound healing assay
cells (1 × 106 cells/well) were seeded into 6-well plates and cultivated in DMEM with 10% FBS. When a monolayer of cells was formed, we scratched the cells using a 200-µl pipette tip. Then the cells were washed with PBS, and cultivated in DMEM without FBS for 48 h. Under a light microscope, the wound width was observed and photographed at 0 h and 48 h after scratching. The experiments were performed triplicately.
Matrigel transwell invasion assay
1 × 105 cells were seeded on top of 8 μm transwell inserts (BD Biosciences, Ontario, Canada) with 0.1% FBS and pre-coated with 1 mg/mL Matrigel (Becton, Dickinson and Company, Ontario, Canada). 10% FBS was used as chemoattractant. Next day, we transfected the cells with siRNAs or plasmids, separately. After 24 h, cells that had spanned the transwell inserts were fixed and stained using 0.1% crystal violet stain (Solarbio Life Sciences, Beijing, China) for 20 min. The experiments were performed triplicately.
Statistical analysis
The area under the curve (AUC), specificity, sensitivity and cut-off values were obtained through ROC analysis. The combined ROC analysis of SERPINI1 and AFP was performed by the predicted probability in binary regression. Z-test was performed to compare the AUCs by Medcalc. The Mann-Whitney U test and the non-parametric Spearman’s correlation test were performed to verify the relationship between biochemical parameters and the level of serum SERPINI1. The differences between groups were evaluated by one-way analysis of variance (ANOVA) or the student’s t-test. P-value < 0.05 was considered significant difference statistically. The statistical analyses were performed by SPSS 20.0 (IBM, Armonk, NY, USA).
Results
Identification of differentially expressed genes in HCC datasets
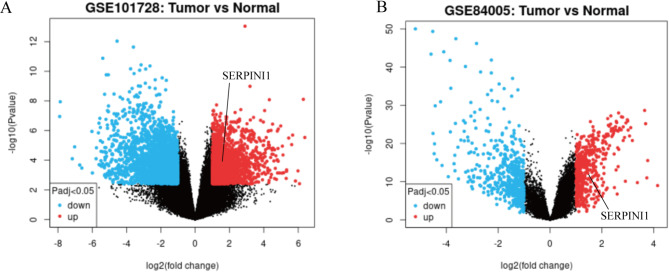
We screened the differentially expressed genes in HCC from GSE101728 and GSE84005 in the Gene Expression Omnibus (GEO) database. The GSE101728 contained the profile of RNA expression in 7 pairs of HCC and matched adjacent tumor-free tissues, and the GSE84005 was based on RNA expression profile data collected from tumor and adjacent normal tissues of thirty-eight HCC patients. Both sets of data were taken from human-derived liver cancer and adjacent tissue specimens, and a larger clinical sample size makes the results more credible. 5993 differentially expressed genes, including long non-coding RNAs and mRNAs, were identified in HCC datasets GSE101725, of which 2637 genes were up-regulated and 3356 genes were down-regulated, compared with the paracancerous tissue (Fig. 1A, Supplementary Table 1). Meanwhile, as shown in Fig. 1B and Supplementary Tables 2, 1032 differentially expressed genes were identified in GSE84005, of which 500 genes were up-regulated and 532 genes were down-regulated. A lot of overlapping up-regulated genes were obtained from the two GSE datasets, such as CDCA2, AKR1B10, CCL20 and so on. According to previous studies, CDCA2 has been identified as a diagnostic and prognostic marker for HCC. Serum AKR1B10 may serve as an indicator of unfavorable survival of HCC. The chemokine CCL20 can assist AFP in serological diagnosis of hepatocellular carcinoma [12–14]. A rarely reported gene, SERPINI1, caught our attention. Although it has been reported that SERPINI1 can play an important role in the regulation of synaptic plasticity and axonal growth [15], the function of which in HCC still remains elusive. Therefore, we focus on the research of SERPINI1 in HCC.
Fig. 1.
The differentially expressed genes in hepatocellular carcinoma (HCC) datasets. (A-B) Volcano plot showed the gene expression in GSE101728 (A) and GSE84005 (B) datasets, the red/blue marker represent the up-regulated/down-regulated genes based on the criteria of adjust p-value < 0.05 and|log2FoldChange| >1
The diagnostic and prognostic value of SERPINI1 in HCC patients
By examining the expression of SERPINI1 in different kinds of cancers, we found that the expression level of SERPINI1 is increased obviously in HCC in the GEPIA database (Fig. 2A, B). Furthermore, we analyzed the protein level of SERPINI1 in serum samples by ELISA, which were gathered from the patients with HCC and healthy control. The results also indicated that the level of SERPINI1 was elevated in HCC (Fig. 2C). To further explore the value of SERPINI1 in HCC as a promising diagnostic molecular marker, we plotted the ROC curves of SERPINI1 and AFP in participants. As shown in Fig. 2D; Table 1, both SERPINI1 and AFP showed significantly diagnostic value in distinguishing HCC from HC. The AUCs of the combined markers were remarkable bigger than SERPINI1 or AFP alone. The sensitivity and the specificity of the two combined markers were also elevated, obviously. In addition, we analyzed the influence of SERPINI1 on the prognosis of HCC in GEPIA database by Kaplan-Meier (KM) survival, and the results suggested that the increased level of SERPINI1 was related to a poor prognosis in HCC patients. These results indicated that SERPINI1 has excellent diagnostic and prognostic value in HCC.
Fig. 2.
The diagnostic and prognostic value of SERPINI1 in HCC. (A) SERPINI1 expression in various tumors in GEPIA dataset. The red name represents upregulation in the cancer, the green name represents downregulation in the cancer, the black name represents differences without statistically significance. (B) GEPIA dataset showed that the expression of SERPINI1 in HCC. The tumor number = 369, the normal number = 160. (C) The levels of SERPINI1 in serum of patients with HCC (number = 153) and healthy controls (HC) (number = 138) were detected by ELISA. (D) ROC of SERPINI1, AFP, and SERPINI1 + AFP were plotted to distinguish HCC from HC. (E) Overall survival analysis of SERPINI1 high (number = 181) and low (number = 181) expression in HCC in GEPIA dataset. *p-value < 0.05
Table 1.
Performances of SERPINI1 and AFP for the diagnosis of HCC patients
| HCC vs. HC | Sensitivity | Specificity | Cut-off | AUC | z-test | p-value |
|---|---|---|---|---|---|---|
| SERPINI1 | 0.680 | 0.855 | 3.27 (ng/ml) | 0.842* | 5.196 | < 0.01 |
| AFP | 0.686 | 0.957 | 91.20 (ng/ml) | 0.844* | 4.368 | < 0.01 |
| SERPINI1 + AFP | 0.771 | 0.978 | 0.933 |
*p < 0.01 in comparison with SERPINI1 + AFP
Correlations between the levels of SERPINI1 and clinical pathological characteristics of HCC
To investigate the clinical significance of SERPINI1 expression in HCC, we summarized the relevance between the levels of serum SERPINI1 and clinical pathological characteristics in HCCs patients (Table 2). The results revealed that the expression of SERPINI1 was significantly related to tumor size and differentiation degrees (both p < 0.05). The levels of serum SERPINI1 were correlated with the tumor stage and vascular invasion positively (both p < 0.01). Serum SERPINI1 levels in HCC patients with lymph node metastasis were elevated significantly (p < 0.05). Nevertheless, there was no significant relationship between the levels of SERPINI1 and age, gender, tumor numbers, distant metastasis, or complications.
Table 2.
Correlation of serum SERPINI1 levels with clinicopathological parameters in HCC
| Characteristics | Number (%) | Serum SERPINI1 levels | p-value | ||
|---|---|---|---|---|---|
| Low levels ≤ 4.00 ng/ml (n = 76) Number (%) |
High levels > 4.00 ng/ml (n = 77) Number (%) |
||||
| Age (years) | ≤ 60 | 69 (45.10) | 37 (24.18) | 32 (20.92) | 0.571 |
| > 60 | 84 (54.90) | 39 (25.49) | 45 (29.41) | ||
| Gender | Male | 112 (73.20) | 55 (35.95) | 57 (37.25) | 0.394 |
| Female | 41 (26.80) | 21 (13.73) | 20 (13.07) | ||
| Tumor size | ≤ 5 cm | 115 (75.16) | 58 (37.91) | 57 (37.25) | 0.014* |
| > 5 cm | 38 (24.84) | 18 (11.76) | 20 (13.07) | ||
| Tumor number | Single | 60 (39.22) | 36 (23.53) | 24 (15.69) | 0.126 |
| Multiple | 93 (60.78) | 40 (26.14) | 53 (34.64) | ||
| Differentiation degrees | Well | 43 (28.10) | 27 (17.65) | 16 (10.46) | 0.016* |
| Moderate | 75 (49.02) | 36 (23.53) | 39 (25.49) | ||
| Poor | 35 (22.88) | 13 (8.50) | 22 (14.38) | ||
| Tumor stage | 1 | 52 (33.99) | 37 (24.18) | 15 (9.80) | < 0.01* |
| 2 | 68 (44.44) | 29 (18.95) | 39 (25.49) | ||
| 3 | 29 (18.95) | 10 (6.54) | 19 (12.42) | ||
| 4 | 4 (2.62) | 0 (0.00) | 4 (2.61) | ||
| Vascular invasion | Yes | 93 (60.78) | 31 (20.26) | 62 (40.52) | < 0.01* |
| No | 60 (39.22) | 45 (29.41) | 15 (9.80) | ||
| Lymph node metastasis | Yes | 61 (39.87) | 24 (15.69) | 37 (24.18) | 0.016* |
| No | 92 (60.13) | 52 (33.99) | 40 (26.14) | ||
| Distant metastasis | Yes | 71 (46.41) | 30 (19.61) | 41 (26.80) | 0.095 |
| No | 82 (53.59) | 46 (30.07) | 36 (23.53) | ||
| Complications | Yes | 73 (47.71) | 33 (21.57) | 40 (26.14) | 0.275 |
| No | 80 (52.29) | 43 (28.10) | 37 (24.18) | ||
Validation of the biological function of SERPINI1 in HCC
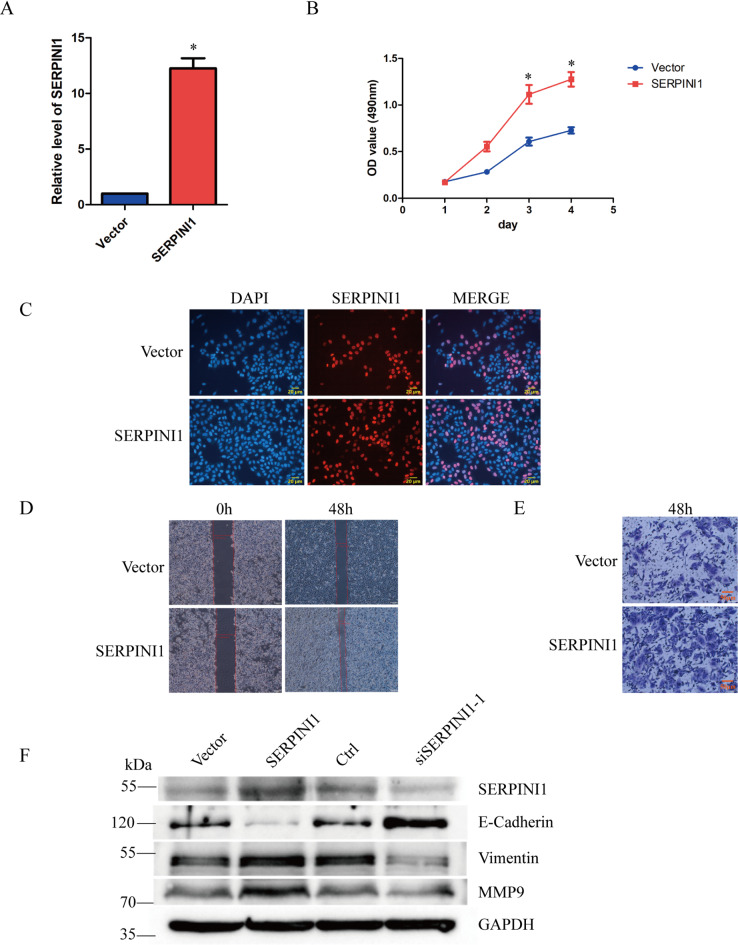
To further corroborate the function of SERPINI1 in HCC, we first detected the expression of SERPINI1 in HepG2, L02 and HUVEC cells. Total RNA and protein were extracted respectively, and the results of qPCR and western blot suggested that the levels of SERPINI1 were higher in HepG2 than in L02 and HUVEC cells (Fig. 3A, B). Next, HepG2 cells were selected to be the main experimental cell line. The transfection effectiveness of SERPINI1 siRNA was validated through qPCR (Fig. 3C). MTT and EdU assay showed that knockdown of SERPINI1 significantly reduced the cell proliferation ability (Fig. 3D, E). The wound healing and transwell assays showed that depletion of SERPINI1 dramatically inhibited both cell migration and invasion capacity (Fig. 3F, G). In addition, we overexpressed SERPINI1 in HepG2 cells with transfection of plasmid, qPCR showed the transfection effect of SERPINI1 overexpressed plasmid in HepG2 cells (Fig. 4A). The results of MTT and EdU assay demonstrated that overexpression of SERPINI1 dramatically promoted the cell proliferation (Fig. 4B, C), and the results of wound healing and transwell assays showed that the overexpressed SERPINI1 significantly increased the cell invasion and migration ability, compared with the vector group (Fig. 4D, E). In order to assess the potential therapeutic value of targeting SERPINI1 in HCC, we investigated the alteration in HepG2 treated with recombinant neuroserpin as a positive control. The wound healing and transwell assays indicated that the neuroserpin promoted cell migration and invasion capacity, obviously (Supplementary Fig. 1A, B). Furthermore, western blot revealed that knockdown of SERPINI1 increased the expression of E-cadherin, but suppressed the expression of vimentin and MMP9, obviously. Overexpression of SERPINI1 significantly decreased the level of E-cadherin, but increased the level of vimentin and MMP9 (Fig. 4F). These results demonstrated that SERPINI1 may regulate the proliferation, invasion and migration of HepG2 through EMT.
Fig. 3.
Depletion of SERPINI1 inhibits the proliferation, migration and invasion of HCC. (A) The expression of SERPINI1 in HUVEC, LO2 and HepG2 cells were detected by qPCR. (B) The expression of SERPINI1 in HUVEC, LO2 and HepG2 cells were detected by western blot. (C) The transfection efficiency of siSERPINI1 were measured in HepG2 by qPCR. (D-E) The effect of SERPINI1 knockdown on cell proliferation was examined by MTT assay (D) and Cell-Light EdU assay (E). (F-G) The migration and invasion ability of HepG2 were detected by wound healing assay (F) and transwell assay (G) when SERPINI1 was knockdown. The experiments were performed triplicately. *p-value < 0.05
Fig. 4.
Overexpression of SERPINI1 promotes the proliferation, migration and invasion of HCC. (A) The transfection efficiency of SERPINI1 were measured in HepG2 by qPCR. (B-C) The effect of SERPINI1 overexpression on cell proliferation was detected by MTT assay (B) and Cell-Light EdU assay (C). (D-E) The migration and invasion ability of HepG2 were detected by wound healing assay (D) and transwell assay (E) when SERPINI1 was overexpressed. (F) The expression levels of SERPINI1, E-Cadherin, Vinmentin and MMP9 were detected by western blot in HepG2, when SERPINI1 was knockdown or overexpressed. The experiments were performed triplicately. *p-value < 0.05
Discussion
In most patients with HCC, they are diagnosed at an advanced stage and the best time for treatment has been missed. It has been reported that excellent biomarkers could help diagnose of early-stage HCC and predict prognosis accurately. In this study, we identified the diagnostic and prognostic value of SERPINI1 in HCC, and the biological function of which in HCC for the first time.
Currently, identification of novel serum biomarkers for the diagnosis of early-stage HCC still is an important goal. Because of the finite study data or diagnostic capability, only a small part biomarker candidates have been applied to clinical practice. For example, there is still only AFP serving as the most commonly used serological biomarker for HCC patients so far, though its unsatisfactory specificity and sensitivity [7]. Glypican-3 (GPC3) has been suggested to be a serum marker for HCC, but the sensitivity of which was 55%, and the specificity was 58% in a meta-analysis [16, 17]. Apart from the markers mentioned above, circulating nucleic acid markers, such as cell-free DNA (cfDNA), are emerging gradually as biomarkers for HCC according to the liquid biopsy [18]. It has been reported that circulating tumor DNA (ctDNA), including 5- hydroxymethylcytosine (5hmC), methylation and mutation, based on their epigenetic or genomic variations, can serve as promising candidates for early-stage HCC and monitor the minimal residual disease [19–21]. Some other liquid biopsy markers, such as cell-free mitochondrial DNA (mtDNA), cell-free virus DNA, cfRNAs and extracellular vesicle (EV), have all been paid attention for their potential role in HCC [22–24]. All these reports have revealed a new non-invasive way for diagnosis of HCC by examination of circulating DNA, RNA or protein. In this study, we found that the levels of SERPINI1 in HCC patients were dramatically higher than that in healthy controls. The ROC analysis showed that the AUCs of the combined SERPINI1 and AFP were larger than one alone in distinguishing HCC from HC. Meanwhile, the sensitivity and specificity of the two combined markers were also better than one alone. In addition, that high expression of SERPINI1 was related to poor prognosis of HCC. All these findings suggested that SERPINI1 may serve as a potential diagnostic and prognostic biomarker for HCC.
During the process of cancer progress including invasion and metastasis, malignant epithelial cells lost their abilities of cell–cell adhesion and cell polarity, and induced their transition to the mesenchymal phenotype [25, 26]. In this process, the expression of EMT-associated genes, including E-cadherin, vimentin, MMP9, Slug, β-catenin, Snail, TWIST, and ZEB1 were changed diversely. Among these altered genes, the suppression of E-cadherin was thought to be one of the most important alterations during EMT [27–29]. In our study, in order to corroborate the function of SERPINI1 on the molecular and phenotypic changes of HCC, we first conformed the elevated level of SERPINI1 in HepG2. Furthermore, we examined the effect of SERPINI1 on the proliferation, invasion, and migration of HCC in vitro. We found that the malignant phenotype of HepG2 was suppressed when the expression of SERPINI1 was knocked down, and vice versa, thus indicating an oncogenic role of SERPINI1 in HCC. At the same time, knockdown of SERPINI1 increased the expression of E-cadherin, but suppressed the expression of vimentin and MMP9, and vice versa, indicating that SERPINI1 may exert the oncogenic effect through the EMT pathway. However, the molecular mechanisms underlying SERPINI1’s regulation of EMT proteins is still not elucidated, which will be our future work. A secretome study reported that a great deal of SERPINI1 were secreted to the supernatants of colon cancer cells. The protein of SERPINI1 was extracted and further analyzed functionally in their study [8]. Some secreted proteases, such as the ADAMTS family, have been reported to have vital roles in the invasion of CRC [30]. It has been reported that anile hepatic stellate cells can secrete a kind of protein profile, which promoting dysplastic or steatotic hepatocytes occurred malignant transformation by activating Wnt or hedgehog signaling pathways in the progression of NASH to cancer [31]. Therefore, it was important to analyze secreted SERPINI1 protein. Our study provides a new idea for the molecular function of SERPINI1 and which can be further investigated.
Although our study shows a comprehensive analysis of the biological function of SERPINI1 in HCC, there are still some inadequacies. The prognostic analysis of SERPINI1 in HCC patients were conducted only reliance on in vitro data and public datasets, which need further studies in larger clinical cohorts. The downstream regulatory molecules and related signaling pathways of SERPINI1 need to be explored in vivo experiments, which will be the direction of our future study.
Conclusions
In summary, this study showed that the significantly diagnostic and prognostic value of SERPINI1 in HCC, revealed that the elevated expression of SERPINI1 was associated with the tumor size, stage, differentiation degrees, vascular invasion and lymph node metastasis, and uncovered that SERPINI1 can promote the proliferation, migration and invasion of HCC. These findings suggest that SERPINI1 may be a novel serum biomarker for patients with HCC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- HCC
Hepatocellular carcinoma
- CT
Computer tomography
- MRI
Magnetic resonance imaging
- AFP
Alpha fetoprotein
- PIVKA-II
Prothrombin induced by vitamin K absence or antagonist-II
- SERPINI1
Serpin peptidase inhibitor, clade I, member 1
- CRC
Solorectal cancer
- EMT
Epithelial–mesenchymal transition
- HC
Healthy controls
- ELISAs
Enzyme-linked immunosorbent assays
- siRNA
Small interfering RNA
- qPCR
Quantitative real-time polymerase chain reaction
- RIPA
Radioimmunoprecipitation assay
- PMSF
Phenylmethylsulfonyl fluoride
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazo-lium bromide
- AUC
Area under the curve
- GEO
Gene Expression Omnibus
- KM survival
Kaplan-Meier survival
- GPC3
Glypican-3
- cfDNA
Cell-free DNA
- ctDNA
Circulating tumor DNA
- 5hmC
5- hydroxymethylcytosine
- mtDNA
Mitochondrial DNA
- EV
Extracellular vesicle
Author contributions
Y-WH, G-YW and E-WH contributed to the study design, analysis of the data, and first draft of the manuscript. Y-WH, S-TY, RZ and Y-YL collected samples and participated in the experiments. Y-WH, MZ, Y-GL and LR conceptualized and supervised this study, revised of the manuscript, and gave final approval of the version to be published. All authors confirmed that they had full access to all the data in the study and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 82372894), the Scientific Research Project of Tianjin Education Commission (No. 2020KJ129, 2020KJ141), and the Tianjin Health Commission, Tianjin Administration of Traditional Chinese Medicine, Integrated Chinese and Western medicine research program (No. 2023088).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All procedures performed in this study that involved participants were conformed to the ethical standards of the 1964 Helsinki Declaration and approved by the Research Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. All participants were over 18 years of age and signed informed consent forms voluntarily.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yawei Han, Gaoyv Wang and Erwei Han contributed equally to this work.
Contributor Information
Meng Zhao, Email: zhaomengtj@yeah.net.
Yueguo Li, Email: kesaqitj@126.com.
Li Ren, Email: liren@tmu.edu.cn.
References
- 1.Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7(1):6. [DOI] [PubMed] [Google Scholar]
- 2.Singal AG, Kanwal F, Llovet JM. Global trends in hepatocellular carcinoma epidemiology: implications for screening, prevention and therapy. Nat Rev Clin Oncol. 2023;20(12):864–84. [DOI] [PubMed] [Google Scholar]
- 3.Feng H, Li B, Li Z, Wei Q, Ren L. PIVKA-II serves as a potential biomarker that complements AFP for the diagnosis of hepatocellular carcinoma. BMC Cancer. 2021;21(1):401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dijk EL, Naquin D, Gorrichon K, Jaszczyszyn Y, Ouazahrou R, Thermes C, Hernandez C. Genomics in the long-read sequencing era. Trends Genet. 2023;39(9):649–71. [DOI] [PubMed] [Google Scholar]
- 5.Jiang P, Sinha S, Aldape K, Hannenhalli S, Sahinalp C, Ruppin E. Big data in basic and translational cancer research. Nat Rev Cancer. 2022;22(11):625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cortes-Ciriano I, Gulhan DC, Lee JJ, Melloni GEM, Park PJ. Computational analysis of cancer genome sequencing data. Nat Rev Genet. 2022;23(5):298–314. [DOI] [PubMed] [Google Scholar]
- 7.Zattoni M, Mearelli M, Vanni S, Colini Baldeschi A, Tran TH, Ferracin C, Catania M, Moda F, Di Fede G, Giaccone G, et al. Serpin signatures in prion and Alzheimer’s diseases. Mol Neurobiol. 2022;59(6):3778–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barderas R, Mendes M, Torres S, Bartolome RA, Lopez-Lucendo M, Villar-Vazquez R, Pelaez-Garcia A, Fuente E, Bonilla F, Casal JI. In-depth characterization of the secretome of colorectal cancer metastatic cells identifies key proteins in cell adhesion, migration, and invasion. Mol Cell Proteom. 2013;12(6):1602–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuda Y, Miura K, Yamane J, Shima H, Fujibuchi W, Ishida K, Fujishima F, Ohnuma S, Sasaki H, Nagao M, et al. SERPINI1 regulates epithelial-mesenchymal transition in an orthotopic implantation model of colorectal cancer. Cancer Sci. 2016;107(5):619–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamanaka S, Olaru AV, An F, Luvsanjav D, Jin Z, Agarwal R, Tomuleasa C, Popescu I, Alexandrescu S, Dima S, et al. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig Liver Dis. 2012;44(7):589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Zhang MX, Meng XX, Zhu J, Wang JJ, He YF, Li YH, Zhao SC, Shi ZM, Zheng LN, et al. Targeting GPR65 alleviates hepatic inflammation and fibrosis by suppressing the JNK and NF-kappaB pathways. Mil Med Res. 2023;10(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu Z, Zhang Y, Shao S, Liu Q, Li Y, Du X, Zhang K, Zhang M, Yuan H, Yuan Q, et al. Identification of CDCA2 as a diagnostic and prognostic marker for hepatocellular carcinoma. Front Oncol. 2021;11:755814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie C, Ye X, Zeng L, Zeng X, Cao D. Serum AKR1B10 as an indicator of unfavorable survival of hepatocellular carcinoma. J Gastroenterol. 2023;58(10):1030–42. [DOI] [PubMed] [Google Scholar]
- 14.Vucur M, Ghallab A, Schneider AT, Adili A, Cheng M, Castoldi M, Singer MT, Buttner V, Keysberg LS, Kusgens L, et al. Sublethal necroptosis signaling promotes inflammation and liver cancer. Immunity. 2023;56(7):1578–e15951578. [DOI] [PubMed] [Google Scholar]
- 15.Noto R, Randazzo L, Raccosta S, Caccia S, Moriconi C, Miranda E, Martorana V, Manno M. The stability and activity of human neuroserpin are modulated by a salt Bridge that stabilises the reactive centre loop. Sci Rep. 2015;5:13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Liu X, Lei Y, Wang G, Liu M. Glypican-3: A novel and promising target for the treatment of hepatocellular carcinoma. Front Oncol. 2022;12:824208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu D, Su C, Sun L, Gao Y, Li Y. Performance of serum glypican 3 in diagnosis of hepatocellular carcinoma: A meta-analysis. Ann Hepatol. 2019;18(1):58–67. [DOI] [PubMed] [Google Scholar]
- 18.Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19(10):670–81. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Huang A, Wang YP, Yin Y, Fu PY, Zhang X, Zhou J. Circulating tumor DNA correlates with microvascular invasion and predicts tumor recurrence of hepatocellular carcinoma. Ann Transl Med. 2020;8(5):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards VD, Roberts LR, Kisiel JB, Reddy KR, Lidgard GP, Johnson SC, et al. A novel Blood-Based panel of methylated DNA and protein markers for detection of Early-Stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2021;19(12):2597–e26052594. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Abou-Alfa GK, Zheng B, Liu JF, Bai J, Du LT, Qian YS, Fan R, Liu XL, Wu L, et al. Genome-scale profiling of Circulating cell-free DNA signatures for early detection of hepatocellular carcinoma in cirrhotic patients. Cell Res. 2021;31(5):589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Zhou K, Guo S, Wang Y, Ji X, Yuan Q, Su L, Guo X, Gu X, Xing J. NGS-based accurate and efficient detection of Circulating cell-free mitochondrial DNA in cancer patients. Mol Ther Nucleic Acids. 2021;23:657–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, Yang P, Wang JJ, Smalley M, Chen PJ, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun. 2020;11(1):4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Y, Jiang W, Wang Y, Zhao M, Li Y, Ren L. Serum long non-coding RNA SCARNA10 serves as a potential diagnostic biomarker for hepatocellular carcinoma. BMC Cancer. 2022;22(1):431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JXT, Tan WR, Low ZS, Lee JQ, Chua D, Yeo WDC, See B, Vos MIG, Yasuda T, Nomura S, et al. YWHAG deficiency disrupts the EMT-Associated network to induce oxidative cell death and prevent metastasis. Adv Sci (Weinh). 2023;10(31):e2301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. [DOI] [PubMed] [Google Scholar]
- 27.Xu D, Wang Y, Wu J, Lin S, Chen Y, Zheng J. Identification and clinical validation of EMT-associated prognostic features based on hepatocellular carcinoma. Cancer Cell Int. 2021;21(1):621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karpinski P, Rosales I, Laczmanski L, Kowalik A, Wenson S, Hoang MP. Expression of genes associated with Epithelial-Mesenchymal transition in Merkel cell Polyomavirus-Negative Merkel cell carcinoma. Lab Invest. 2023;103(8):100177. [DOI] [PubMed] [Google Scholar]
- 29.Sun XY, Li HZ, Xie DF, Gao SS, Huang X, Guan H, Bai CJ, Zhou PK. LPAR5 confers radioresistance to cancer cells associated with EMT activation via the ERK/Snail pathway. J Transl Med. 2022;20(1):456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelwick R, Desanlis I, Wheeler GN, Edwards DR. The ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs) family. Genome Biol. 2015;16(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang L, Ma Y, Xie L, Yang YY, Jin C, Chen H, Zhou Y, Song GQ, Ding J, et al. Secretome of senescent hepatic stellate cells favors malignant transformation from nonalcoholic steatohepatitis-fibrotic progression to hepatocellular carcinoma. Theranostics. 2023;13(13):4430–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.