Abstract
Drosophila N-cadherin is required for the formation of precise patterns of connections in the fly brain. Alternative splicing is predicted to give rise to 12 N-cadherin isoforms. We identified an N-cadherin allele, N-cad18Astop, that eliminates the six isoforms containing alternative exon 18A and demonstrate that it strongly disrupts the connections of R7 photoreceptor neurons. During the first half of pupal development, N-cadherin is required for R7 growth cones to terminate within a temporary target layer in the medulla. N-cadherin isoforms containing exon 18B are sufficient for this initial targeting. By contrast, 18A isoforms are preferentially expressed in R7 during the second half of pupal development and are necessary for R7 to terminate in the appropriate synaptic layer in the medulla neuropil. Transgene rescue experiments suggest that differences in isoform expression, rather than biochemical differences between isoforms, underlie the 18A isoform requirement in R7 neurons.
Keywords: synapse, visual system, alternative splicing, synaptic specificity
Selection of appropriate synaptic partners is a crucial step in the formation of neuronal connections (1). However, the cellular recognition mechanisms underlying the formation of specific synaptic connections remain poorly understood. Several large families of related cell-surface molecules, for example, cadherins (2, 3), neurexins (4), and Down syndrome cell adhesion molecules (5, 6), have attracted considerable interest as potential cell surface “tags” that could promote specific recognition events leading to precise targeting.
Classical cadherins are homophilic cell adhesion molecules with characteristic extracellular sequence motifs (the cadherin repeats) and a conserved cytoplasmic domain that mediates interactions with intracellular ligands (the catenins). Differences in cadherin expression can mediate cell sorting in vitro (7, 8) and, in at least some cases, in vivo (9, 10). In the nervous system, several classical cadherins are expressed in distinct patterns (11, 12) and localize at synapses from early stages of synaptogenesis (13-15). These properties (early synaptic localization, molecular diversity, selective adhesion, and regional expression) support the proposal that cadherin mediated cell-cell recognition is important for synaptic specificity (16). N-cadherin, one of three classical cadherins in Drosophila, is necessary for the assembly of the embryonic CNS (17), for axonal targeting and dendritic patterning in the adult olfactory system (18, 19), and for targeting of photoreceptor neurons (R cells) (20-22).
The experimental accessibility of R7 targeting makes it an attractive model for exploring the role of N-cadherin in synaptic specificity. Three classes of R cells, R1-R6, R7, and R8 form synaptic connections in distinct regions of the brain. R1-R6 neurons synapse in the lamina ganglion that lies directly beneath the retina, whereas R7 and R8 project through the lamina and form synaptic connections in the medulla. Within the medulla, R8 and R7 neurons terminate in two distinct layers, M3 and M6, respectively. R7 neurons that lack N-cadherin target to the M3 rather than the M6 layer (20). R7 targeting within the medulla involves at least two distinct steps (23): R7 growth cones initially project to a temporary target layer and only reach their adult target layer, M6, after reorganization of processes within the medulla neuropil during a later phase of pupal development. N-cadherin is necessary for targeting to the temporary layer (ref. 23; also see Results and Discussion). Whether N-cadherin also contributes to the second phase of R7 layer selection is not known.
Although N-cadherin is essential for R7 targeting, its widespread expression in R cells and the target region makes it unlikely that homophilic N-cadherin interactions alone are sufficient for the specification of R7 target choice. However, the recent finding that alternative splicing generates 12 N-cadherin isoforms (23) raises the intriguing notion that these could contribute to differential cell recognition leading to synaptic specificity. If so, isoform-specific mutants affecting only some N-cadherin functions should exist. In this study, we describe the identification and characterization of one such mutant that reduces N-cadherin diversity from 12 to 6 isoforms. This mutant allele disrupts a subset of N-cadherin functions as assessed in several different developmental contexts. We demonstrate that different subsets of N-cadherin isoforms act at early and late stages of R7 targeting.
Materials and Methods
Genetics. Approximately 50,000 mosaic flies generated with GMR-FLP carrying an ethylmethane sulfonate-mutagenized (24) FRT40 chromosome were behaviorally screened for mutations affecting R7 function by using a UV/visual light choice test (20). F1 mutant flies that selected visible over UV light at least two of three times were retested as F2 populations. Behavior mutants missing R7 cells were discarded. The remaining lines were histologically examined for targeting phenotypes by using mosaic analysis with a repressible cell marker (MARCM) (25) with PANR7GAL4 upstream activating sequence (UAS) N-sybGFP (20). N-cad alleles were identified by complementation tests with N-cadM19.
Homozygous mutant clones were positively labeled by using MARCM (25), negatively labeled by loss of red fluorescent protein (RFP) expression, or both. For quantification of R7 targeting defects, developmental analyses, and rescue experiments, GMR-FLP MARCM clones were marked with Tubulin-Gal4, UAS N-synaptobrevin GFP (N-sybGFP). Tubulin-GAL4 is expressed throughout pupal development and is R7-specific in the medulla in this context, because GMR-FLP induces mitotic recombination in R7, but not R8, precursors. In the rescue experiments Tubulin-Gal4 also drives expression of UAS N-cad transgenes in addition to UAS N-sybGFP.
ey-FLP (26, 27) and ey3.5-FLP (I. Salecker, personal communication) were used to induce mitotic recombination in both the developing retina and in optic lobe precursors or only in the retina, respectively. Olfactory receptor neuron targeting was assessed in eyFLP MARCM clones with OR22aGAL4,UAS mCD8GFP or OR46aGAL4,UAS N-sybGFP (18). N-cadherin protein expression was examined in mosaic animals with the following genotype: ey3.5FLP;GMR-myr-mRFP FRT40/N-cad FRT40. In these animals, mutant retinal cells were marked by the absence of RFP. To combine this method with the MARCM technique, GMR-myr-mRFP was recombined onto Tubulin-Gal80 FRT40.
Histology. Fixation, Ab staining, and confocal imaging were essentially as described in ref. 20. Eye discs were fixed for 30 min; other samples were fixed for 90 min. The following Abs were used: mAb 24B10 at (1:50) (28), mAb nc82 (1:10) (29), rabbit anti-GFP (1:2,000) (Molecular Probes), rabbit anti-RFP (1:5,000) (Clontech), Cy5-labeled anti-horseradish peroxidase Ab (1:100) (Cappel, St. Louis), and rat anti-N-cadherin mAb NcadEx8 (1:20) (17). Secondary Abs were from Molecular Probes (Alexa Fluor 488 goat anti-rabbit and goat anti-rat, both at concentrations of 1:500) and from The Jackson Laboratory (Cy3 goat anti-mouse, 1:300; Cy5 goat anti-rat, 1:100).
Immunoblotting. Third-instar larval eye-brain complexes or adult flies were homogenized in SDS/PAGE sample buffer and analyzed by Western blotting with a rat mAb against the cytoplasmic domain of N-cadherin [NcadIn (17), 1:100]. Actin [rabbit anti-actin (Sigma), 1:1,000] served as a loading control.
Molecular Biology and Transgenes. For sequencing of mutant alleles, N-cadherin exons were amplified by PCR from genomic DNA. All N-cadherin transgenes were derived from the same N-cadherin cDNA (17) with alternative exons 7B, 13A, and 18A (referred to in the text as N-cad 18A). N-cad 7B13A18B (referred to in the text as N-cad18B) was obtained by replacing part of N-cad 18A with a fragment of an RT-PCR product containing exon 18B. N-cad 18B and N-cad 18A were cloned into pUAST to result in pUAST 18B and pUAST 18A, respectively. Except for a number of polymorphisms already present in N-cad 18A (17), sequencing of N-cad 18B did not reveal any mutations. Transgenic flies were generated by using standard procedures. N-cad 18B and N-cad 18A #2 are insertions of pUAST 18B and pUAST 18A. N-cad 18A #1 is an insertion described in ref. 17.
Results and Discussion
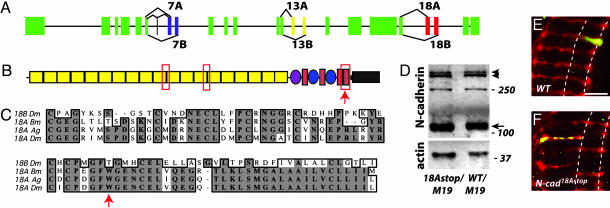
N-cad18Astop Is an Isoform-Specific N-cadherin Allele with a Strong R7-Targeting Phenotype. In a screen for R7-targeting mutants on chromosome 2L, we identified 16 alleles of N-cadherin (see Materials and Methods). Recently, the N-cadherin locus has been shown to give rise to 12 different isoforms [see the FlyBase Drosophila genome database (23)]. This diversity is the result of the selective splicing of two alternative versions of exon 7, exon 13, and exon 18. These pairs of alternative exons, which encode portions of the extracellular domain and part of the predicted transmembrane domain, share sequence similarity and are conserved between different insect orders (23) (Fig. 1 A-C). In addition, exon 7A can be modified by inclusion of a small exon 5′ to exon 7A. By sequencing the alternative exons of the new N-cad alleles, we identified one, N-cad18Astop, with a point mutation leading to a stop codon within alternative exon 18A (Fig. 1C) but no other changes in the coding region. Hence, all full-length N-cadherin in N-cad18Astop contains exon 18B; translation of all transcripts with exon 18A would be truncated before the transmembrane domain. That exon 18B-containing isoforms are expressed in N-cad18Astop is supported by Western blots showing similar levels of full-length N-cadherin in third-instar eye-brain complexes from N-cad18Astop and wild-type larvae (Fig. 1D). In addition, immunohistological studies indicate that 18A-containing isoforms are not expressed in N-cad18Astop (see Fig. 6).
Fig. 1.
N-cad18Astop is an isoform-specific N-cadherin allele. (A) N-cadherin gene structure [see the FlyBase Drosophila genome database (23)]. Constant exons are indicated in green, and alternatively spliced exons 7A/7B, 13A/13B, and 18A/18B are indicated in blue, yellow, and red, respectively. In addition, a small exon encoding 4 aa can be spliced into transcripts just 5′ to alternative exon 7A. (B) Domain structure of N-cadherin protein (31), with alternative regions marked with the red box outline. The exact number of cadherin repeats is unclear (17, 31). Yellow rectangles, cadherin repeats; purple oval, domain found in invertebrate classical cadherins and recently also identified in some nonmammalian vertebrate classical cadherins (32); red filled rectangles, EGF repeats; blue ovals, laminin G domains; thick black line, transmembrane domain; gray rectangle, cytoplasmic tail. (B and C) Arrows indicate the location of the nonsense mutation in N-cad18Astop. (C) The 18A alternative exon of Drosophila melanogaster (Dm) shares a high degree of sequence conservation with the mosquito Anopheles gambiae (Ag), and the silkworm Bombyx mori (Bm). Exons 18B (D. melanogaster sequence shown as example) are also conserved between Drosophila, mosquito, and silkworm. (D) N-cad18Astop larvae express full-length N-cadherin. An immunoblot of protein extracts prepared from eye-brain complexes of N-cad18Astop/N-cadM19 and N-cad wild-type/N-cadM19 third-instar larvae is shown. N-cadherin was detected with an Ab against the cytoplasmic domain, and anti-actin staining was used as a loading control. N-cadherin is proteolytically processed (17). The major C-terminal fragment (arrow) and full-length N-cadherin (arrowheads) (17) are indicated. Because homozygous protein-null animals do not survive to third instar, it remains unclear whether the 250-kDa species is an additional N-cadherin breakdown product or a background cross-reactive band. (E and F) N-cad18Astop R7 mutants terminate in the R8 recipient layer, M3, instead of the slightly deeper M6 layer in the medulla neuropil. Single-cell R7 clones were visualized in the adult with MARCM. The approximate positions of the M3 and M6 layers are indicated by dashed lines. (E) Wild-type (FRT40). (Scale bar, 10 μm.) (F) N-cad18Astop. Most N-cad18Astop R7 mutants terminate in the M3 layer (for quantification, see Results and Discussion). In addition, the synaptic marker synaptobrevin-GFP within R7 frequently is no longer restricted to the terminus but is distributed in a punctate pattern along the length of the R7 axon within the medulla neuropil.
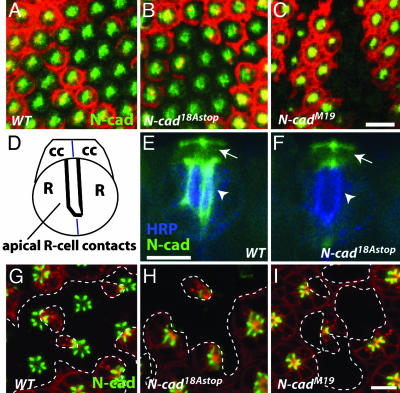
Fig. 6.
N-cadherin isoform expression changes during pupal development. Anti-N-cadherin staining (green) was examined in retinal clones [marked by loss of RFP (red)] homozygous for the indicated mutations. (A-C) Horizontal confocal sections through the apical region of early pupal eye discs (≈20%APF) show N-cadherin staining in wild-type (A) and N-cad18AStop (B), but not in N-cadM19 (C) clones. (Scale bars, 10 μm.) (D) Schematic of R cells (R) and cone cells (cc) in midpupal retina (side view). (E and F) At this stage (≈45% APF) N-cadherin is concentrated at apical R cell junctions (now oriented perpendicular to the eye surface along the developing rhabdomere as in D) (arrow-heads). N-cadherin also is localized to cone cells with strong staining at cone cell contacts (arrow). Comparison of N-cad18AStop (F) to wild-type (FRT40) control (E) clones shows a marked reduction of N-cadherin on R cells but not on cone cells. Anti-horseradish peroxidase staining (blue) in (E and F) labels all R cells. (G-I) Horizontal confocal sections of older eye discs (≈45% APF) at the R cell level shows N-cadherin enriched at the apical contacts of neighboring R cells. R8 cells are in a different focal plane and are not shown. Apical N-cadherin staining in N-cad18AStop R cells (H) is strongly reduced, indicating a loss of 18B isoform expression at this stage in development. (I) Control N-cadM19 clones also show no apical N-cadherin. Clone boundaries are marked by dashed lines. (Scale bar, 5 μm.).
Single N-cad18Astop mutant R7 cells generated by MARCM (25) frequently target to the M3 instead of the M6 layer in the medulla neuropil (Fig. 1 E and F), as described for N-cad-null mutants in ref. 20. Quantification of the R7-targeting defects of N-cad18Astop and a strong loss-of-function allele of N-cad, N-cadM19 (17, 19, 20), showed a similar frequency of R7 mistargeting. We observed that 100% (n = 288 R7 cells from seven brains) and 84% (n = 569 R7 cells from 12 brains) of the R7 terminals were in the R8 layer in N-cadM19 and N-cad18Astop, respectively, which indicates that exon 18A-containing isoforms are required for R7 targeting.
In contrast to N-cad-null mutants, which die during late embryogenesis, N-cad18Astop animals survived to late pupal stages. This result suggested that N-cadherin 18A isoforms might be required for only some N-cadherin functions, for example, the targeting of only subsets of neurons or for distinct steps in the targeting of a particular neuron. To further explore this possibility, we examined N-cad18Astop phenotypes at different stages of R7 targeting and in a number of other neuronal and nonneuronal cell types.
Alternative Exon 18A Is Required for a Late Step of R7 Targeting. Recent studies indicate that R7 targeting in the medulla involves early targeting to a temporary layer and later progression to the final target layer (23). N-cadherin is required for targeting to the temporary target layer (ref. 23 and this study). To assess when N-cad 18A-containing isoforms were required, we analyzed the R7 phenotypes of N-cad18Astop at 35% and 48% of development after pupal formation (APF) (Fig. 2). The later time point is just before the relocation of R7 terminals from their temporary to their final target layer [at ≈50% APF (23)]. All wild-type R7 growth cones were in the correct layer at both stages (Fig. 2 A). At ≈35% APF, 61% (n = 215 R7 cells from seven brains) of the N-cadM19 mutant growth cones were observed in the appropriate target layer. By 48% APF, this fraction had decreased to 38% (n = 106 R7 cells from four brains) (Fig. 2C). At both time points, the remaining R7 growth cones terminated in the R8 recipient layer or between the R7 and R8 recipient layers (Fig. 2 B and C). By contrast, all N-cad18Astop mutant R7 growth cones remained in the correct layer at both 35% (n = 172 R7 cells from six brains) and 48% (n = 168 R7 cells from five brains) (Fig. 2D). In the adult, most N-cad18Astop and all N-cadM19 R7 terminals were found in the R8 recipient layer, M3 (see above). These data indicate that N-cadherin is required for targeting and stabilization of R7 growth cones in the correct medulla layer during the first half of pupal development and that 18B-containing isoforms are sufficient at these early steps. Furthermore, N-cadherin 18A-containing isoforms appear to be only necessary for interaction of R7 neurons with their adult, but not with their temporary, target layer in the medulla.
Fig. 2.
N-cadherin 18A isoforms are required at a late step in R7 targeting. Targeting of N-cad mutant R7 growth cones (green) was examined at ≈35% and 48% APF. Single-mutant R7 neurons were generated by using the MARCM method with GMR-FLP to promote mitotic recombination and with labeled cells marked by Tubulin-Gal4 expression of N-syb-GFP (green). All R cells (red) were labeled with mAb 24B10. (A) Wild type. At 35% APF, all R7 terminals reside in the correct layer. (Scale bar, 10 μm.) (B and C) N-cadM19 clones. At 35% APF (B) and 48% APF (C), some R7 growth cones were seen at intermediate positions between the two layers, presumably indicating an ongoing process of axon retraction (arrows in B and C). The number of mistargeted R7 growth cones increased between 35% and 48% APF (from 39% to 62%; see Results and Discussion). (D) N-cad18AStop clones. In contrast to N-cadM19, homozygous N-cad18AStop R7 mutants remained in the correct target layer at 35% (data not shown) and 48%.
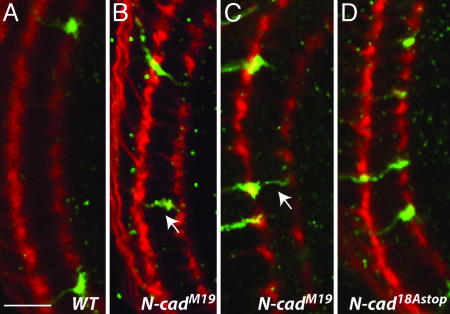
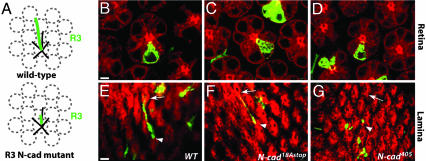
N-Cadherin 18A Isoforms Are Required for Late Stages of R1-R6 Targeting. In the lamina, each R1-R6 axon makes a short lateral extension to a single target, following a route that is specific to each R cell subtype (Fig. 3A). We compared the targeting of single N-cad18Astop, N-cad-null, and wild-type R1-R6 cells at 42% APF by using the MARCM technique (Fig. 3 B-G). Whereas wild-type axons invariably extended to the appropriate targets in the lamina (100% extension, n = 52), all R cell axons homozygous for a null N-cad allele, N-cad405 (20, 21), extended aberrantly (64%, n = 34 of 53) or did not extend at all (36%, n = 19 of 53). In contrast, many N-cad18Astop R1-R6 axons (69%, n = 73 of 106) extended normally, and all but four of these extensions were to their correct targets (95%). Only 7% (n = 8 of 106) of N-cad18Astop axons failed to extend. The remaining 24% (n = 25 of 106) extended aberrantly, forming thin, branched filopodia that nonetheless followed a normal trajectory. A similar pattern of axon extension was seen for all R1-R6 subtypes. Although the R1-R6 phenotype at 42% was mild, the phenotype in the adult was similar to N-cad405 (data not shown). Hence, as described for R7, N-cadherin 18A isoforms are required selectively at a late stage of R1-R6 targeting.
Fig. 3.
Initial targeting of R1-R6 axons to the correct cartridge within the pupal lamina is only mildly affected by N-cad18AStop.(A) Schematic of R1-R6 targeting in the developing lamina. R1-R6 (black lines; except R3, green line) from one ommatidium extend to different lamina cartridges (gray dashed circles) within the plane of the lamina. Cells homozygous for strong N-cad loss-of-function mutants, such as N-cadM19 and N-cad405, frequently fail to extend (illustrated for R3). (B-G) Examples of single-mutant R3 cells generated by MARCM at 42% APF (30). Retina (B-D) and lamina (E-G) views are shown. (B and E) Wild-type (FRT40) control. (C and F) N-cad18Astop. (Scale bars, 3 μm.) (D and G) N-cad405.(E) In wild-type, all R3 axons elaborate a normal projection, as indicated, extending from the arrowhead to the arrow. (F) Most (11 of 19) N-cad18Astop R3 neurons show a normal projection. (G) No normal projections were seen for N-cad405axons.
N-Cadherin 18B Isoforms Are Sufficient for Olfactory Receptor Neuron Targeting and Cone Cell Patterning. We also examined N-cad 18A isoform requirements in cone cells, a nonneuronal cell type in the eye, and in olfactory receptor neurons. In the olfactory system, N-cadherin is required for the formation of glomeruli (18), discreet bulbous structures in the antennal lobe in which olfactory receptor neurons synapse with their targets (Fig. 4). We examined two olfactory receptor neuron subclasses with strong N-cadherin-null phenotypes, Or22a and Or46a, in adult antennal lobes. In both cases, olfactory receptor neurons homozygous for N-cad18Astop appeared to innervate glomeruli in a fashion similar to the wild type (Fig. 4 C-H). Similarly, although N-cadherin plays a crucial role in regulating the arrangement of cone cells in midpupal retinas (10) (A.N. and S.L.Z, unpublished data) loss of N-cad 18A isoforms did not alter cone cell patterns (Fig. 5). These results further indicate that loss of 18A isoforms disrupts only a subset of N-cadherin functions.
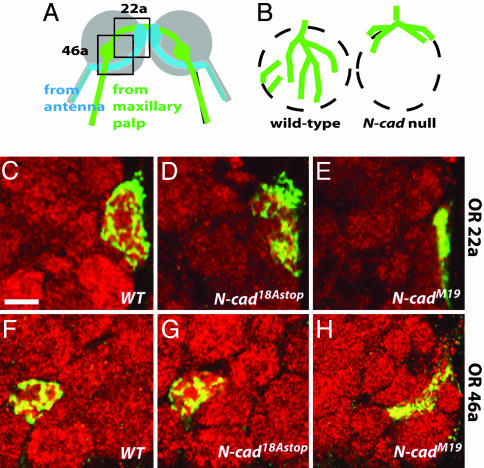
Fig. 4.
N-Cadherin 18A isoforms are not essential for glomerulus formation in the olfactory system. (A) Schematic representation of the projections of two classes of olfactory receptor neurons terminating in glomeruli in the adult antennal lobe. OR46a-expressing (green) and OR22a-expressing (blue) neurons from the maxillary palp and the antenna, respectively, target to specific glomeruli in the antennal lobe. The gray circle indicates the antennal lobe, and the green and blue blobs are glomeruli. In addition to forming connections ipsilaterally, axons extend branches to glomeruli in the contralateral lobe. Boxed areas indicate approximate regions shown in C-H.(B) N-cad olfactory receptor neuron phenotype (18) illustrated for a single glomerulus. Olfactory receptor neurons lacking N-cadherin (green lines) target to the correct region of the antennal lobe but remain largely on the antennal lobe surface and fail to elaborate glomeruli (dashed circles). (C-H) GFP-labeled (green) MARCM clones of adult OR22a-expressing (C-E) and OR46a-expressing (F-H) neurons homozygous for the indicated mutations. The antennal lobe was stained with mAb nc82 (red). Wild-type (FRT40) (C and F), N-cad18AStop (D and G), and N-cadM19 (E and H) phenotypes are shown. Whereas N-cadM19 fibers remain at the surface of the antennal lobe, the N-cad18Astop fibers innervate glomeruli in a fashion similar to wild type. (Scale bar, 10 μm.)
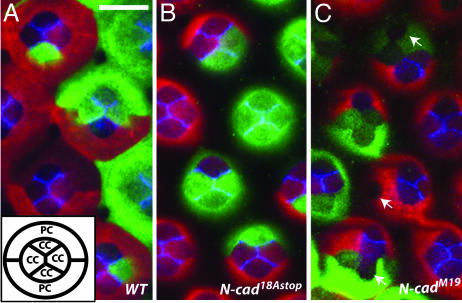
Fig. 5.
Cone cell patterning in midpupal retinas does not require 18A-containing N-cadherin isoforms. (A-C) N-cad phenotypes in clones in midpupal retinas (40-50% APF). (A) (Inset) The arrangement of cone cells (cc) and primary pigment cells (pc) as seen from above. Cells homozygous for a wild-type control (A), N-cad18Astop (B), or N-cadM19 (C) were labeled with GFP (green) using MARCM whereas all other retinal cells expressed RFP (red). Within a single confocal section GFP and RFP staining were occasionally difficult to see in some cone cells. Retinas also were stained with an N-cadherin Ab (blue). (A) N-Cadherin is expressed in cone cells but not pigment cells and localizes to regions of contact between cone cells. (Scale bar, 10 μm.) (C) When one or more cone cells in a cluster lacks N-cadherin their precise packing is disrupted. Frequently, N-cadherin-positive and -negative cone cells separate from each other (arrows). (A and B) In contrast, cone cell arrangements and N-Cadherin staining in clones of N-cad18Astop (B) were indistinguishable from wild type (A).
N-Cadherin Isoform Expression Changes During Midpupal Development. To assess the expression of N-cadherin isoforms containing alternative exons 18A and 18B, we examined N-cadherin protein expression in mosaic animals containing retinal tissue homozygous for N-cadM19 or N-cad18Astop alleles and wild-type cells. Loss of anti-N-cadherin staining in N-cad18Astop clones would indicate selective expression of 18A isoforms in the corresponding wild-type cells. Conversely, staining within clones would indicate expression of 18B isoforms solely or in combination with 18A isoforms. In third-instar larval (data not shown) and early pupal eye discs (≈20% APF) anti-N-cadherin staining in N-cad18Astop clones was similar to wild type (Fig. 6 A and B); no staining was seen in N-cadM19 mutant clones, which provided a control for Ab specificity (Fig. 6C). In contrast, in older eyes (≈45% APF), in which N-cadherin was concentrated at R cell junctions (Fig. 6 D, E, and G), staining was not detected in N-cad18Astop R cell clones (Fig. 6 F and H). Because an 18B-containing isoform expressed from a transgene at this stage of development in a null mutant clone was detected at R cell contacts (data not shown), we think it is unlikely that the absence of N-cadherin protein in N-cad18Astop R cell clones is due to a failure of 18B isoforms to localize to R cell junctions. Because of the strong and widespread N-cadherin expression in the medulla, we were unable to directly assess N-cadherin protein expression in R7 and R8 growth cones (data not shown). In the lamina, N-cadherin staining on R1-R6 growth cones homozygous for N-cad18Astop was detectable in third-instar larvae as well as at 45% APF. The residual staining at 45% APF probably reflects perdurance of N-cadherin 18B. This interpretation would be consistent with the weak N-cad18Astop phenotype in R1-R6 targeting seen at 42% APF (Fig. 3).
In summary, R cells initially express 18B alone or in combination with 18A but switch to express 18A isoforms predominantly at ≈45% APF. In contrast to R cells, N-cadherin expression in N-cad18Astop cone cells was indistinguishable from the wild type. Hence, there are differences in isoform expression between different cell types (cone cells and R cells) at the same developmental stage, and there are temporal changes in isoform expression within a cell type.
Expression of 18A- and 18B-Encoding N-Cadherin Transgenes Rescues the Targeting Phenotypes in Single N-Cad-Null Mutant R7 Neurons. We next examined whether individual 18A or 18B isoforms were able to functionally replace endogenous N-cadherin. We used three transgenic lines, two encoding N-cad 18A (#1 and #2) and one encoding N-cad 18B. Western blot and immunofluorescence analyses indicated that N-cad 18A #1 is expressed at much higher levels than N-cad 18B, which in turn was more strongly expressed than N-cad 18A #2 (Fig. 7D and data not shown). Wild-type R7 cells expressing an N-cad 18A or 18B transgene showed correct target layer selection (data not shown). We compared the targeting phenotypes of single N-cad-null mutant R7 neurons expressing these transgenes to that of single N-cad-null mutant R7 cells (Fig. 7). Expression of N-cad 18A #1, N-cad 18B, or N-cad 18A #2 increased the percentage of mutant R7 cells with correct target layer selection from 0% to 95%, 78%, or 65%, respectively. The differences in the fraction of mutant R7 neurons rescued correlated with the levels of protein expression driven by these different transgenes. Hence, the requirement for 18A isoforms in R7 target layer selection does not reflect intrinsic differences in biochemical function. Both 18A and 18B isoforms also rescue R1-R6 targeting defects; expression of N-cad 18B increased the proportion of normal R1-R6 targeting from 0% to 81% (46 of 57 R1-R6 projections scored). Rescue was observed for all R1-R6 subtypes and was similar to results with N-cad 18A (30).
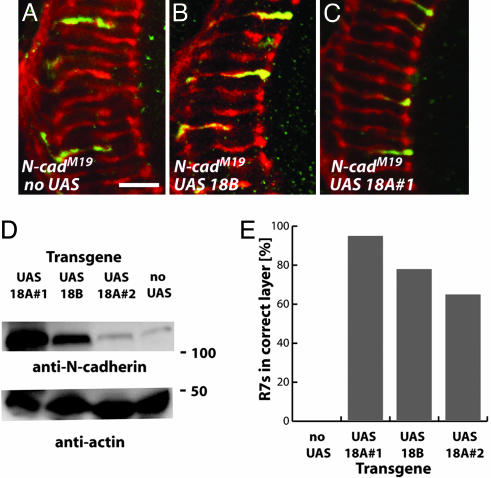
Fig. 7.
N-cadherin 18A- and 18B-containing isoforms rescue R7 layer-specific targeting defects. The MARCM technique with a Tubulin-GAL4 driver was used to express UAS N-cadherin transgenes containing the 18A or 18B exons in single N-cadherin-null mutant R7 cells throughout pupal development. R7 targeting was examined at the adult stage. (A-C) Confocal images of adult R7 clones homozygous for N-cadM19 (A) or N-cadM19 expressing UAS N-cad 18B (B), or UAS N-cad 18A#1 (C). (Scale bar, 10 μm.) (D) Immunoblot analyses of adult flies expressing the indicated UAS N-cadherin constructs from a constitutive promoter (hsGAL4). N-cadherin was detected with an Ab against the cytoplasmic domain. Only the main C-terminal fragment (compare with Fig. 1) is shown. The band in the no UAS lane is endogenous N-cadherin. Anti-actin staining served as a loading control. (E) Summary of rescue experiments. Both 18A and 18B transgenes can restore correct layer selection to a substantial fraction of mutant R7 cells. Correct targeting of R7 cells of the indicated genotypes: N-cadM19 without rescue construct, 1 of 288; N-cadM19 with UAS N-cad 18A #1, 598 of 628; N-cadM19 with UAS N-cad 18B, 716 of 917; and N-cadM19with UAS N-cad 18A #2, 210 of 323. The R7 targeting of a second strong loss-of-function mutant, N-cad405, was also rescued by 18A and 18B transgenes (data not shown).
Conclusion
Alternative splicing at the N-cadherin locus potentially generates 12 different protein isoforms. Here we report a specific requirement for isoforms containing alternative exon 18A for the appropriate target layer selection of R7 growth cones. We demonstrate that it is differences in expression rather than in biochemical properties between isoforms that underlies the requirement of exon 18A-containing isoforms for R7 targeting. Isoforms containing alternative exon 18A may be important for aspects of R7 synapse formation or function. Addressing these possibilities awaits the development of more incisive tools. Because the N-cad18Astop allele is homozygous lethal, it remains possible that biochemical differences between isoforms are important in other developmental or physiological contexts.
Acknowledgments
We thank members of the S.L.Z. laboratory for comments on the manuscript; Chi-Hon Lee and Akira Chiba for communicating results before publication; Sahael Stapleton for sequencing some of the N-cadherin alleles; and Iris Salecker (National Institute for Medical Research, London), Tadashi Uemura (Kyoto University, Kyoto), the Bloomington Stock Center (Indiana University, Bloomington), and the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City) for reagents. This work was supported by postdoctoral fellowships from the European Molecular Biology Organization (to A.N.), the Human Frontiers Science Program (to A.N.), and the Burroughs-Wellcome Fund for Biomedical Research (to T.H.); a predoctoral fellowship from the National Institutes of Health (to L.-V.T.N.); and the Stanford Medical Sciences Training Program (to S.P.). T.R.C. is a Sloan Fellow, a Searle Scholar, and a recipient of a Burroughs-Wellcome Career Development Award, and work in his laboratory was supported by National Institutes of Health Grant R01 EYO 15231-01A1. S.L.Z. is an investigator of the Howard Hughes Medical Institute.
Author contributions: A.N., T.H., and S.L.Z. designed research; A.N., L.-V.T.N., S.P., and T.R.C. performed research; A.N., L.-V.T.N., S.P., T.R.C., and S.L.Z. analyzed data; and A.N., T.R.C., and S.L.Z. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MARCM, mosaic analysis with a repressible cell marker; R cell, photoreceptor neuron; RFP, red fluorescent protein; UAS, upstream activating sequence.
References
- 1.Tessier-Lavigne, M. & Goodman, C. S. (1996) Science 274, 1123-1133. [DOI] [PubMed] [Google Scholar]
- 2.Fannon, A. M. & Colman, D. R. (1996) Neuron 17, 423-434. [DOI] [PubMed] [Google Scholar]
- 3.Uemura, T. (1998) Cell 93, 1095-1098. [DOI] [PubMed] [Google Scholar]
- 4.Missler, M. & Sudhof, T. C. (1998) Trends Genet. 14, 20-26. [DOI] [PubMed] [Google Scholar]
- 5.Schmucker, D., Clemens, J. C., Shu, H., Worby, C. A., Xiao, J., Muda, M., Dixon, J. E. & Zipursky, S. L. (2000) Cell 101, 671-684. [DOI] [PubMed] [Google Scholar]
- 6.Wojtowicz, W. M., Flanagan, J. J., Millard, S. S., Zipursky, S. L. & Clemens, J. C. (2004) Cell 118, 619-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg, M. S. & Takeichi, M. (1994) Proc. Natl. Acad. Sci. USA 91, 206-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foty, R. A. & Steinberg, M. S. (2005) Dev. Biol. 278, 255-263. [DOI] [PubMed] [Google Scholar]
- 9.Price, S. R., De Marco Garcia, N. V., Ranscht, B. & Jessell, T. M. (2002) Cell 109, 205-216. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi, T. & Carthew, R. W. (2004) Nature 431, 647-652. [DOI] [PubMed] [Google Scholar]
- 11.Yamagata, M., Herman, J. P. & Sanes, J. R. (1995) J. Neurosci. 15, 4556-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miskevich, F., Zhu, Y., Ranscht, B. & Sanes, J. R. (1998) Mol. Cell. Neurosci. 12, 240-255. [DOI] [PubMed] [Google Scholar]
- 13.Benson, D. L. & Tanaka, H. (1998) J. Neurosci. 18, 6892-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jontes, J. D., Emond, M. R. & Smith, S. J. (2004) J. Neurosci. 24, 9027-9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uchida, N., Honjo, Y., Johnson, K. R., Wheelock, M. J. & Takeichi, M. (1996) J. Cell Biol. 135, 767-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro, L. & Colman, D. R. (1999) Neuron 23, 427-430. [DOI] [PubMed] [Google Scholar]
- 17.Iwai, Y., Usui, T., Hirano, S., Steward, R., Takeichi, M. & Uemura, T. (1997) Neuron 19, 77-89. [DOI] [PubMed] [Google Scholar]
- 18.Hummel, T. & Zipursky, S. L. (2004) Neuron 42, 77-88. [DOI] [PubMed] [Google Scholar]
- 19.Zhu, H. & Luo, L. (2004) Neuron 42, 63-75. [DOI] [PubMed] [Google Scholar]
- 20.Lee, C. H., Herman, T., Clandinin, T. R., Lee, R. & Zipursky, S. L. (2001) Neuron 30, 437-450. [DOI] [PubMed] [Google Scholar]
- 21.Clandinin, T. R., Lee, C. H., Herman, T., Lee, R. C., Yang, A. Y., Ovasapyan, S. & Zipursky, S. L. (2001) Neuron 32, 237-248. [DOI] [PubMed] [Google Scholar]
- 22.Iwai, Y., Hirota, Y., Ozaki, K., Okano, H., Takeichi, M. & Uemura, T. (2002) Mol. Cell. Neurosci. 19, 375-388. [DOI] [PubMed] [Google Scholar]
- 23.Ting, C. Y., Yonekura, S., Chung, P., Hsu, S. N., Robertson, H. M., Chiba, A. & Lee, C. H. (2005) Development (Cambridge, U.K.) 132, 953-963. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner, M. (1989) Drosophila: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 25.Lee, T. & Luo, L. (2001) Trends Neurosci. 24, 251-254. [DOI] [PubMed] [Google Scholar]
- 26.Newsome, T. P., Asling, B. & Dickson, B. J. (2000) Development (Cambridge, U.K.) 127, 851-860. [DOI] [PubMed] [Google Scholar]
- 27.Stowers, R. S. & Schwarz, T. L. (1999) Genetics 152, 1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zipursky, S. L., Venkatesh, T. R., Teplow, D. B. & Benzer, S. (1984) Cell 36, 15-26. [DOI] [PubMed] [Google Scholar]
- 29.Stortkuhl, K. F., Hofbauer, A., Keller, V., Gendre, N. & Stocker, R. F. (1994) Cell Tissue Res. 275, 27-38. [DOI] [PubMed] [Google Scholar]
- 30.Prakash, S., Caldwell, J. C., Eberl, D. F. & Clandinin, T. R. (2005) Nat. Neurosci. 8, 443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill, E., Broadbent, I. D., Chothia, C. & Pettitt, J. (2001) J. Mol. Biol. 305, 1011-1024. [DOI] [PubMed] [Google Scholar]
- 32.Tanabe, K., Takeichi, M. & Nakagawa, S. (2004) Dev. Dyn. 229, 899-906. [DOI] [PubMed] [Google Scholar]