Abstract
We purified the KH-type splicing regulatory protein (KSRP) as a protein interacting with the 3′-untranslated region (3′-UTR) of the human inducible nitric oxide (iNOS) mRNA. Immunodepletion of KSRP enhanced iNOS 3′-UTR RNA stability in in vitro-degradation assays. In DLD-1 cells overexpressing KSRP cytokine-induced iNOS expression was markedly reduced. In accordance, downregulation of KSRP expression increases iNOS expression by stabilizing iNOS mRNA. Co-immunoprecipitations showed interaction of KSRP with the exosome and tristetraprolin (TTP). To analyze the role of KSRP binding to the 3′-UTR we studied iNOS expression in DLD-1 cells overexpressing a non-binding mutant of KSRP. In these cells, iNOS expression was increased. Mapping of the binding site revealed KSRP interacting with the most 3′-located AU-rich element (ARE) of the human iNOS mRNA. This sequence is also the target for HuR, an iNOS mRNA stabilizing protein. We were able to demonstrate that KSRP and HuR compete for this binding site, and that intracellular binding to the iNOS mRNA was reduced for KSRP and enhanced for HuR after cytokine treatment. Finally, a complex interplay of KSRP with TTP and HuR seems to be essential for iNOS mRNA stabilization after cytokine stimulation.
INTRODUCTION
Gene expression is controlled by transcriptional and post-transcriptional mechanisms. A central part of the post-transcriptional modulation of gene expression is mediated by regulation of mRNA stability. The stability varies considerably between mRNAs and can be modulated by extracellular stimuli (1–3). A tight control of mRNA stability permits rapid changes in the level of mRNAs and provides a mechanism for prompt termination of protein production. The rate of mRNA decay is determined by cis-acting sequences within the mRNA, which are recognized by trans-acting factors. Dysregulation of the expression of RNA-bps or mutation of important cis-acting binding sequences and thereby dysregulation of mRNA stability has been associated with different human diseases like chronic inflammatory diseases (4,5), α-thalassemia (6), cancer (7–10) and Alzheimer's disease (11).
AU-rich elements (AREs) are critical cis-acting elements in the 3′-untranslated regions (3′-UTRs) of many inherently unstable mRNAs that code for cytokines, transcription factors and proto-oncogenes and are targets for trans-acting proteins regulating mRNA stability and translation (12,13). A number of ARE binding proteins (ARE-bps) have been identified that can interact with AU- and U-rich regions. These include the ELAV protein family members (most important HuR) (14), the ARE/poly-(U)-binding/degradation factor 1 (AUF-1, also named hnRNP D) (15), the heteronuclear ribonucleoprotein A1 (hnRNP A1) (16), the KH-type splicing regulatory protein (KSRP) (17), tristetraprolin (TTP) (18), the T cell-restricted intracellular antigen (TIA)-1 and the TIA-related protein (TIAR) (19,20).
The mechanism by which AREs mediate mRNA decay is not clear. In yeast, 3′- to 5′-mRNA degradation is mediated by the exosome, a multisubunit particle. Chen et al. (17) have purified and characterized the human exosome by mass spectrometry (MS) and found its composition to be similar to its yeast counterpart. Using a cell-free RNA decay system, these authors also demonstrate that the mammalian exosome is required for degradation of ARE-containing RNAs. However, the mammalian exosome does not seem to recognize the ARE-containing RNAs on its own but requires certain ARE-bps, like KSRP or TTP. These ARE-bps interact with the exosome and recruit it to unstable RNAs, thereby promoting their rapid degradation (17). By siRNA-mediated depletion of KSRP in HeLa cells and by creating a dominant negative KSRP isoform (by deletion of the most carboxyterminal located RNA binding domain, KH4) Gherzi et al. (21) showed that KSRP is essential for ARE-RNA decay in vivo.
The expression of inducible nitric oxide synthase (iNOS) can be induced in different types of cells and tissues following exposure to immunologic and inflammatory stimuli such as cytokines or lipopolysaccharide (LPS) (22). Once synthesized, the enzyme is active for hours to days (23) and generates large amounts of NO that can have beneficial effects, such as anti-microbial, anti-atherogenic or anti-apoptotic actions (24). However, inappropriate iNOS induction can have detrimental consequences, such as cellular injury in arthritis or colitis and in septic shock (22,25). The expression of human iNOS is controlled in large part by post-transcriptional mechanisms (22,26). In human epithelial A549, AKN or DLD-1 cells, nuclear run-on and transfection experiments revealed a significant basal activity of the human iNOS promoter that was only enhanced 2- to 5-fold by cytokines (27,28). In contrast, only after cytokine induction a significant iNOS mRNA expression could be detected.
Sequence analysis of the human iNOS mRNA reveals the presence of five AREs in the 3′-UTR. In transfection experiments with human A549 or DLD-1 cells, the 3′-UTR of the human iNOS mRNA destabilized the mRNA of a heterologous reporter gene (28).
Gel retardation experiments showed interaction of the ARE-bp HuR with the human iNOS 3′-UTR (28). HuR is known to stabilize ARE-containing mRNAs (29). Stable overexpression of HuR in human DLD-1 cells resulted in an upregulation of cytokine-induced iNOS expression. Accordingly, downregulation of HuR expression resulted in a reduction of cytokine-induced iNOS expression (28).
In recent studies, we showed that in addition to HuR TTP is essentially involved in the post-transcriptional regulation of human iNOS expression (30). In contrast to its published destabilizing effect, overexpression of TTP in DLD-1 cells resulted in a marked increase in iNOS expression, without any TTP binding to the human iNOS mRNA.
UV-crosslinking experiments with cellular extracts from DLD-1 cells and a 32P-radiolabeled probe of the iNOS 3′-UTR revealed that several proteins formed RNA–protein complexes in this sequence (31). Therefore, it seems very likely that in addition to HuR other RNA-bps are involved in the post-transcriptional regulation of human iNOS expression.
In the current study, we identified KSRP as a key regulator of human iNOS expression. KSRP destabilizes iNOS mRNA by binding to its 3′-UTR and interaction with the exosome. For cytokine induction of iNOS expression a complex interaction of the iNOS mRNA 3′-UTR and the RNA-bps KSRP, TTP and HuR seems to be essential.
MATERIALS AND METHODS
Materials
Trypsin-, glutamine-, and pyruvate-solutions, phenylmethylsulfonyl fluoride, leupeptin, aprotinin, agarose, isopropyl-β-d-thiogalactopyranoside, tRNA, BSA, actinomycin D, streptavidin-coated agarose beads, horseradish-peroxidase-coupled anti-goat and anti-mouse IgG were purchased from Sigma, Deisenhofen, Germany. Isotopes were obtained from NEN/Dupont, Köln, Germany. Restriction enzymes, Taq polymerase, Klenow DNA polymerase, dNTPs, pGEX2T and glutathione-agarose affinity beads were purchased from Amersham-Pharmacia, Freiburg, Germany. RNase A, RNase T1, DNase I, T3 and T7 RNA polymerase and the biotin RNA labeling mix were obtained from Roche Diagnostics, Mannheim, Germany. The mMESSAGE mMACHINE® T7 Kit was obtained from Ambion, Huntingdon, UK. The QuantiTect Probe RT–PCR Kit, the RNAeasy Mini Kit and polyclonal anti-HisTag-antibodies were from Qiagen, Hilden, Germany. All oligonucleotides and dual-labeled probes were from MWG-Biotech, Ebersberg, Germany. The MessageMuter™ shRNA Production Kit was from Epicentre/Biozym, Oldendorf, Germany. Human interferon-γ (IFN-γ), interleukin-1β (IL-1β), and tumor necrosis factor-α (TNF-α) were obtained from Strathmann, Hannover, Germany. Fetal calf serum (FCS) and DMEM were purchased from PAN-Systems, Nürnberg, Germany. G418 was purchased from Calbiochem, Bad Soden, Germany. pcDNA3 was purchased from Invitrogen, Groningen, The Netherlands. pCR-Script was from Stratagene, Heidelberg, Germany. The Bradford reagent mix for determination of protein concentration was obtained from BioRad, Munich, Germany. Protein A, G and protein A/G plus agarose beads were purchased from Santa Cruz Biotechnology, Heidelberg, Germany. The polyclonal anti-TTP antibody was a kind gift from Dr William Rigby (Department of Medicine, Dartmouth Medical School, Lebanon, USA), the polyclonal anti-KSRP antibody was a kind gift from Dr Ching-Yi Chen (Department of Biochemistry and Molecular Genetics, University of Alabama, Birmingham, USA), the polyclonal anti-PM-Scl 100-antibody was a kind gift from Dr G. Pruijn (Department of Biochemistry, University of Nijmegen, The Netherlands) and the monoclonal anti-KSRP antibody, the procaryotic (pET15b-KSRP) and eucaryotic (pcDNA3.1-His-KSRP) expression vectors for a His-tagged KSRP protein were a kind gift from Dr Douglas L. Black (Howard Hughes Medical Institute at UCLA, Los Angeles, USA).
Cell culture, cytokine treatment and RNA isolation
Human epithelial colon carcinoma DLD-1 cells (ATCC, #CCL-221) were used for this study. These cells had been shown to be inducible for human iNOS expression by incubation with cytokines (32). The cells were grown in DMEM with 10% inactivated fetal bovine serum, 2 mM l-glutamine, penicillin and streptomycin. Eighteen hours before cytokine induction, the cells were washed with phosphate-buffered saline (PBS) and incubated with DMEM containing 2 mM l-glutamine in the absence of serum and phenol red. iNOS expression in DLD-1 cells was induced with a cytokine mixture (CM) containing 100 U/ml IFN-γ, 50 U/ml IL-1β and 10 ng/ml TNF-α for the corresponding time periods depending on the experiment. Later, supernatant of the cells (300 µl) was used to measure by the Griess reaction or the Sievers Nitric Oxide Analyzer (ADInstruments, Spechbach, Germany), and the cells were processed for RNA isolation by guanidinium thiocyanate/phenol/chloroform extraction or for protein extraction as described below (33,34).
Quantitative RT–PCR (qRT–PCR)
One-Step RT–PCR was performed with the QuantiTect RT–PCR Kit (Qiagen, Hilden, Germany) in 25 µl reactions in a 96-well spectrofluorometric thermal cycler (iCycler, BioRad, München, Germany). RNA was isolated as described above. For real-time qRT–PCR (30 min 50°C, 15 min 95°C and 40 cycles of 15 s 94°C, 60 s 60°C), the oligonucleotides listed below served as sense and antisense primers and Taqman hybridization probes.
iNOS: sense, TGCAGACACGTGCGTTACTCC; antisense, GGTAGCCAGCATAGCGGATG; probe, TGGCAAGCACGACTTCCGGGTG. GAPDH: sense, CCCATGTTCGTCATGGGTGT; antisense, TGGTCATGAGTCCTTCCACGATA; probe, CTGCACCACCAACTGCTTAGCACCC. KSRP: sense, TGCAGCAAGCCTGTGAGATG; antisense, TCCGTACTCATTCCGGTCCC; probe, TGGACATCCTCCGGGAACGTGACC. Luciferase: sense, AAAAAGTTGCGCGGAGGAG; antisense, TTTTTCTTGCGTCGAGTTTTCC; probe, TGTGTTTGTGGACGAAGTACCGAAAGGTCTTAC.
Taqman hybridization probes were double labeled with 6-carboxyfluorescein (FAM) as reporter fluorophore and carboxytetramethyl-rhodamine (TAMRA) as quencher. All primers and dual-labeled probes (5′-FAM, 3′-TAMRA) were from MWG-Biotech, Ebersberg, Germany. Fluorescence was monitored at each 60°C step.
Each experimental reaction was performed in triplicate. All primer/probes sets had efficiencies of 100% (±10%).
To calculate the relative expression of iNOS- or KSRP mRNA in DLD-1 cells the 2[−ΔΔC(T)] method (35) was used. According to this method the C(T) values for iNOS- or KSRP mRNA expression in each sample were normalized to the C(T) values of GAPDH mRNA in the same sample. Then the values of untreated cell samples were set 100% and the percentage of iNOS- or KSRP expression was calculated.
Actinomycin D experiments
To analyze the effects of experimental interventions on iNOS mRNA stability, cells were incubated as indicated and iNOS expression was induced by cytokines for 6 h. Then, 10 µM actinomycin D was added and RNAs were prepared from 0 to 18 h thereafter. Relative iNOS- and GAPDH mRNA amounts were determined by qRT–PCR and iNOS mRNA was normalized to GAPDH mRNA. The relative amount of iNOS mRNA at 0 h and actinomycin D was set to 100%. Curve fittings of the resulting actinomycin D time curves were performed by non-linear regression using Graphpad Prism 3.0 (GraphPad Software, San Diego, USA).
Western blot experiments
To study the protein expression in DLD-1 cells total cell protein was fractionated into nuclear and cytoplasmic extracts as described (36). To study the expression of human KSRP, TTP and PM-Scl 100 cytoplasmic extracts (10–50 µg protein) were separated on SDS–PAGE and transferred to nitrocellulose membrane by semi-dry electroblotting. All further steps were performed as described (28). For detection of KSRP a polyclonal anti-KSRP antibody (21) or a monoclonal anti-KSRP antibody (37) was used. For the detection of TTP a polyclonal anti-TTP antibody (38) was used. For detection of PM-Scl 100 a polyclonal anti-PM-Scl 100-antibody (39) was used. The immunoreactive proteins on the blots were visualized by the enhanced chemiluminescence detection system (ECL, Amersham).
Streptavidin-biotin RNA-affinity chromatography
To generate biotinylated-RNA sense probes for the affinity purification 0.5–1 µg of the linearized plasmids pCR-iNOS 3′-UTR or pCR-iNOS 3′-UTR-non-AU (28) were in vitro transcribed using the biotin RNA labeling mix (Roche Diagnostics, Mannheim, Germany) as described by the manufacturer. Purification of the proteins was performed as described below. Protein extracts from CM-induced DLD-1 cells were first precleared by incubating 200 mg of cytoplasmic proteins with 3 ml of a 50% slurry pre-blocked streptavidin-coated agarose beads, for 3 h at 4°C in 50 ml binding buffer [10 mM HEPES (pH 7.6), 3 mM MgCl2, 5 mM EDTA, 2 mM DTT, 5% Glycerol, 0.5% NP-40, 3 mg/ml Heparin and 0.5 mg/ml Yeast RNA] supplemented with 40 mM KCl and RNasin (0.3 U/μl, final). Beads were pelleted and the supernatant was incubated with 600 µg biotinylated-RNA probe as described above for 2 h. The binding mixture was then incubated with 3 ml of the 50% slurry pre-blocked streptavidin-coated agarose beads for 2 h. Beads were pelleted and washed three times in binding buffer with 40 mM KCl and twice in binding buffer with 300 mM KCl. Isolated proteins were eluted from the beads by incubating with elution buffer [10 mM HEPES (pH 7.6), 3 mM MgCl2, 5 mM EDTA, 2 mM DTT, 0.2% glycerol and 2 M KCl] for 20 min at 4°C. To identify the proteins eluates were separated on SDS–polyacrylamide gels and the proteins were stained with colloidal Coomassie blue. The stained protein bands were cut out and the identity of the proteins was determined by peptide mass fingerprinting (Toplab, München, Germany).
Purification of GST-KSRP, GST-HuR and His-tagged KSRP proteins
Purified GST, GST-KSRP or GST-HuR fusion proteins were prepared using the plasmids pGEX2T, pGEX2T-KSRP (40), or pGEX2T-HuR as described previously (28). The yield of the purification procedure was determined by comparison to a BSA standard on Coomassie blue-stained SDS–PAGE.
For purification of His-tagged KSRP the plasmid pET15b-His-KSRP and the Ni-NTA Agarose from Qiagen (Hilden, Germany) were used as described by the manufacturer.
UV-crosslinking experiments
cDNAs encoding for subfragments of the human iNOS 3′-UTR have been described previously (28). To generate radiolabeled iNOS 3′-UTR sense probes for RNA binding experiments, 0.5–1 µg of DNA (linearized plasmids or PCR fragments) were in vitro transcribed as described above. Radiolabeled transcripts were analyzed by urea-denaturing electrophoresis in order to estimate the yield and the specific activity. Incorporated radioactivity in transcripts was usually >80% and the specific activity ranged from 0.2–0.5 µCi/pmol.
UV-crosslinking experiments were performed as described previously (30,41).
Establishment of cell lines expressing sense or antisense KSRP mRNA or a dominant negative KSRPdKH4 protein
The plasmid pcDNA3-KSRPas was generated by digesting the plasmid pcDNA3-His-KSRP (42) with EcoRI and XbaI and incubating with Klenow DNA polymerase to generate blunt ends. The fragment containing the KSRP cDNA was isolated and cloned into pcDNA3 restricted with EcoRI and XbaI and treated with Klenow enzyme. Plasmids containing the KSRP cDNA in antisense orientation were selected. To generate the plasmid pcDNA3-KSRPdKH4 pcDNA-His-KSRP was digested with Eco81I and XbaI and treated with Klenow enzyme. The obtained linearized blunt ended plasmid was religated. DNA sequences of all clones were controlled using the dideoxy chain termination method with a sequencing kit from Pharmacia.
To generate DLD-1 cells overexpressing a sense or antisense KSRP cDNA or a dominant negative KSRPdKH4 protein, cells plated on a 10 cm plate were transfected with 5 µg of pcDNA-His-KSRP, pcDNA-KSRPas or pcDNA-KSRPdKH4 with FuGene according to the manufacturer's recommendations. Pools of stable transfectants form one plate (usually >30) were selected with G418 (1 mg/ml). As a control, DLD-1 cells stably transfected with the pcDNA3 vector were also generated. The stable transfected cells were characterized for the expression of His-tagged KSRP or His-tagged KSRPdKH4 protein by western blots using a monoclonal anti-HisTag-antibody and by PCR analyses using primers specifically amplifying the integrated plasmid sequences. To test for antisense KSRP RNA expression PCRs with specific primers were performed.
Downregulation of KSRP expression by RNA interference
shRNAs were produced in vitro using chemically synthesized DNA oligonucleotide templates (MWG-Biotech, Ebersberg, Germany) and the MessageMuter™ shRNA Production Kit (Epicentre/Biozym, Oldendorf, Germany) as described by the manufacturer. Transcription templates were designed such that they contained T7 promoter sequences at the 3′ end. The target sequence for KSRP based on sequences within the KSRP coding region (GAUCAACCGGAGAGCAAGA) (21). As controls shRNAs with target sequences within the GL2-luciferase coding region (CGUACGCGGAAUACUUCGA) (43) were synthesized.
Immunoprecipitation
For immunoprecipitation, cell extracts were digested for 30 min at 30°C with RNase A (40 µg) and RNase T1 (100 000 U). Then these extracts were preincubated with protein A-agarose beads or protein G-agarose beads (Santa Cruz Biotechnology, Heidelberg, Germany) in RIPA buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 50 mM NaF, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM disodium glycerol phosphate, 10 nM okadaic acid, 2 mM EDTA, 10% glycerol, 1% NP-40 and 1/25 v/v complete EDTA-free protease inhibitor cocktail] for 1 h at 4°C. These precleared extracts were incubated with polyclonal or monoclonal antibodies overnight at 4°C in RIPA buffer. Subsequently protein–antibody complexes were captured by incubating with protein A or G-agarose beads for 5 h at 4°C in RIPA buffer. Beads were washed three times with RIPA buffer and co-immunoprecipitated proteins were analyzed by western blotting.
Immunoprecipitation-qRT–PCR assay
For determination of intracellular protein–RNA interactions DLD-1 cells were incubated for 4 h with or without the cytokine mixture. Cells were lyzed in a buffer containing 10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.1% NP-40, 50 mM NaF, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM disodium glycerol phosphate, 10 nM okadaic acid, 0.2% VRC, 100 U/ml RNasin and 1/25 v/v complete EDTA-free protease inhibitor cocktail. Cell lysates (1.5 mg) were preincubated with 20 µl protein A/G plus agarose beads (Santa Cruz Biotechnology, Heidelberg, Germany) in 500 µl NT2 buffer [50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40, 50 mM NaF, 10 mM Na3VO4, 10 mM sodium pyrophosphate, 50 mM disodium glycerol phosphate, 10 nM okadaic acid, 1 mM DTT, 2 mM EDTA, 0.2% VRC, 100 U/ml RNasin and 1/25 v/v complete EDTA-free protease inhibitor cocktail] for 30 min at 4°C. After a centrifugation at 1200 g for 5 min at 4°C the supernatant was incubated with 50 µl protein A/G plus agarose beads precoated with the specific antibody in 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40, 0.5 µg/µl tRNA and 0.5 mg/ml Heparin for 16 h at 4°C for 3 h at 4°C. Subsequently the beads were washed four times in 1 ml NT2 buffer and then digested with 0.5 mg/ml Proteinase K in NT2 buffer containing 0.1% SDS for 15 min at 55°C. Then 1 ng/sample of luciferase RNA transcribed in vitro was added to normalize for subsequent purification steps and RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) as described by the manufacturer. The amount of iNOS mRNA bound by KSRP or HuR was determined by qRT–PCR using the primers and probes described above. The C(T)-values for iNOS mRNA were normalized to the C(T)-values of luciferase mRNA.
In vitro decay assay
To generate sense 5′-capped and 3′-polyadenylated radiolabeled iNOS 3′-UTR or non-AU-RNAs for the in vitro decay assay the plasmids pCR-iNOS 3′-UTR-polyA and pCR-iNOS 3′-UTR-non-AU-polyA (28) were used. These plasmids contain the human iNOS 3′-UTR sequence or a human 3′-UTR sequence with deleted AREs cloned in front of a poly A100 tail. pCR-iNOS 3′-UTR-polyA or pCR-iNOS 3′-UTR-non-AU DNA was restricted with Eco ICRI and purified by phenol extraction and ethanol precipitation. The linearized DNA (0.5 µg) was transcribed in vitro using the mMESSAGE mMACHINE® T7 Kit (Ambion Ltd Huntingdon, UK) as described by the manufacturer.
The in vitro decay assay was performed as described (44). In brief, 32P-labeled 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNA was incubated with cytoplasmic extracts from DLD-1 cells (15 µg protein per assay) in 25 µl buffer containing 100 mM KCH3COOH, 2 mM Mg(CH3COOH)2, 10 mM Tris–HCl (pH 7.6), 2 mM DTT, 10 mM creatine phosphate, 1 µg creatine phosphokinase, 1 mM ATP, 0.4 mM GTP and 0.1 mM spermine. Reactions were incubated at 37°C for 0–30 min and stopped with 100 µl stop buffer [400 mM NaCl, 25 mM Tris–HCl (pH 7.6), 0.1% SDS] containing 100 000 c.p.m. of an in vitro transcribed β-actin RNA to normalize for subsequent purification steps. RNAs were phenol/chloroform extracted, ethanol precipitated and analyzed on denaturating urea 5% polyacrylamide gels (running buffer 1× TBE).
Statistics
Data represent means ± SEM. Statistical differences were determined by factorial analysis of variance followed by Fisher's protected least-significant-difference (PLSD) test for comparison of multiple means.
RESULTS
Purification and identification of iNOS 3′-UTR RNA-bps
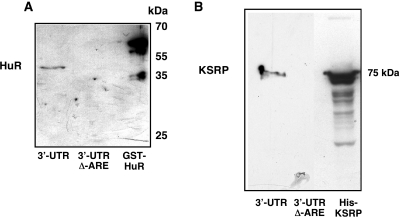
In order to purify proteins binding to the 3′-UTR of the human iNOS mRNA we performed affinity chromatographies using biotinylated iNOS 3′-UTR RNA (45,46) with (3′-UTR) or without (3′-UTR Δ-ARE) the AU-rich sequences and extracts from cytokine-induced DLD-1 cells. After several washing steps the RNA-bps were eluted by incubation with 2 M KCl. Proofing the applicability of this method we found by western blot that HuR known to interact with the iNOS 3′-UTR (28) was purified (Figure 1A).
Figure 1.
Purification of RNA-bps interacting with the 3′-UTR of the human iNOS mRNA. To purify proteins binding to the 3′-UTR of the human iNOS mRNA affinity chromatographies using biotinylated iNOS 3′-UTR RNA were performed (45,46) as described in Materials and Methods. DLD-1 cells were preincubated for 18 h in medium without FCS and phenol red. Then cells were incubated with the cytokine mixture for 6 h and protein extracts were isolated. These extracts were incubated with biotinylated iNOS 3′-UTR RNA (3′-UTR) or biotinylated iNOS 3′-UTR RNA without the ARE-sequences (3′-UTR Δ–ARE) and streptavidine-agarose beads. After several washing and centrifugation steps the RNA-bps were eluted by 2 M KCl. (A) Proofing the applicability of this method western blots using a specific anti-HuR antibody were performed, since HuR is known to interact with the human iNOS 3′-UTR (28). As a positive control bacterial expressed GST-HuR fusion protein was also loaded on the SDS gel. One representative blot of three different experiments were shown. (B) The eluates were tested for the presence of KSRP by western blots using a specific anti-KSRP antibody. As a positive control bacterial expressed His-KSRP fusion protein was also loaded on the SDS gel. One representative blot of three different experiments were shown.
To identify additional proteins binding to the human iNOS 3′-UTR the eluates were separated on SDS–polyacrylamide gels (see Supplementary Figures) and the proteins were stained with colloidal Coomassie blue. The stained protein bands were cut out and the identity of the proteins was determined by peptide mass fingerprinting (Toplab, München, Germany).
This analysis resulted in the identification of the KH-type splicing regulatory protein (KSRP, Figure 1B), the polyadenylate-binding protein 1 (PABP), (see Supplementary Figures) and the heteronuclear ribonucleoprotein E1 (hnRNP E1), (see Supplementary Figures) as proteins interacting with the human iNOS mRNA 3′-UTR.
Immunodepletion of KSRP enhances iNOS 3′-UTR RNA stability in in vitro decay assays
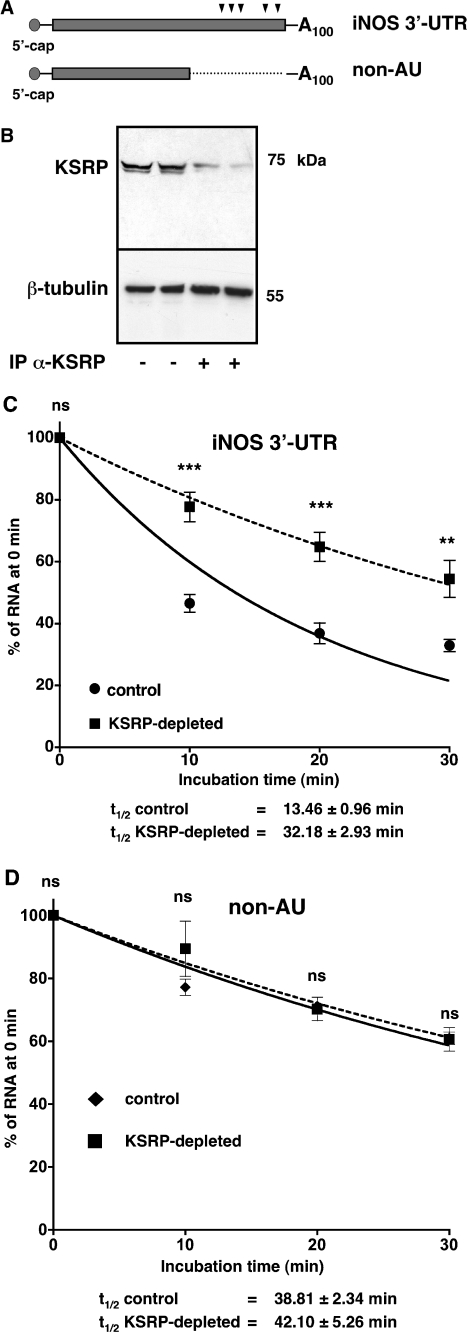
KSRP has been described as a protein needed for exosome-mediated degradation of ARE-containing mRNAs (17,21). Therefore, we analyzed the role of KSRP in human iNOS expression. First, we tested the effect of KSRP immunodepletion on iNOS 3′-UTR RNA stability in an in vitro degradation assay using transcripts containing the AREs (iNOS 3′-UTR) or with deleted AREs (non-AU). Radiolabeled 5′-capped iNOS 3′-UTR- or non-AU-RNAs containing a 100 nt poly-A-tail (see Figure 2A) were generated. These radiolabeled transcripts were incubated with cytoplasmic extracts from DLD-1 cells treated with cytokine mixture (control). To analyze the influence of KSRP on the stability of the iNOS 3′-UTR RNA the extracts were KSRP-depleted using a specific polyclonal anti-KSRP antibody (KSRP-depl, see Figure 2B). As shown in Figure 2C and D, immunodepletion of KSRP enhanced the stability of the iNOS 3′-UTR RNA (iNOS 3′-UTR) in the degradation experiments (t1/2control: 13.46 ± 0.96 min; t1/2KSRP-depleted: 32.18 ± 2.93 min) but had no influence on the degradation of the human iNOS 3′-UTR RNA with deleted AREs (non-AU; t1/2control: 38.81 ± 2.34 min; t1/2KSRP-depleted: 42.10 ± 5.26 min).
Figure 2.
Depletion of KSRP enhances iNOS 3′-UTR RNA stability in in vitro degradation assays. DLD-1 cells were preincubated for 18 h in medium without FCS and phenol red. Then the cells were incubated with the CM for 6 h and cytoplasmic proteins were isolated. Parts of these extracts were immunodepleted of KSRP by using a polyclonal anti-KSRP antibody (KSRP-depl). The extracts were incubated with radiolabeled 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNA with (iNOS 3′-UTR) or without (non-AU) the AREs for different time periods from 0 to 30 min. Then the degradation of the RNA in the different samples was stopped by adding SDS. To normalize the subsequent purification steps in vitro transcribed radiolabeled β-actin RNA fragments were added. After phenol extraction and ethanol precipitation the material was separated on denaturing urea polyacrylamide gels. (A) Scheme of the human 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNAs used for in vitro decay experiments. The positions of AU-repeats are indicated by arrowheads. (B) The degree of KSRP depletion of two different immunodepletion reactions (IP α-KSRP) were analyzed by western blotting using specific anti-KSRP- and anti-β-tubulin antibodies. For comparison the corresponding untreated samples were analyzed. The position of KSRP and β-tubulin is indicated. (C) Summary of densitometric analyses of eight different in vitro decay assays using 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNAs containing the AREs (iNOS 3′-UTR). Data points (means ± SEM) represent relative iNOS 3′-UTR RNA levels (100% = 0 min incubation; circles: undepleted extracts from CM-treated cells; squares: KSRP-depleted extracts from CM-treated cells; **P < 0.01; ***P < 0.001; ns = not significant versus undepleted extract). (D) Summary of densitometric analyses of five different in vitro decay assays using 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNAs without the AREs (non-AU). Data points (means ± SEM) represent relative non-AU iNOS 3′-UTR RNA levels (100% = 0 min incubation; diamonds: undepleted extracts from CM-treated cells; squares: KSRP-depleted extracts from CM-treated cells; ns = not significant versus undepleted extract).
KSRP regulates human iNOS expression by destabilization of the mRNA
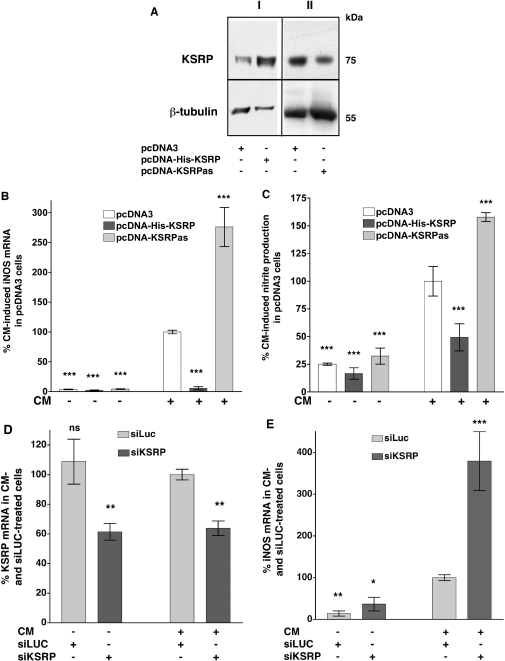
To determine whether KSRP plays a role in the regulation of iNOS gene expression in intact cells, we generated pools of stably transfected DLD-1 cells, which constitutively express a His-tagged KSRP protein (pcDNA-His-KSRP) or an antisense KSRP RNA (pcDNA-KSRPas) under the control of a CMV promoter (pcDNA3 expression plasmid). These cell pools were analyzed for stable integration of the DNA (data not shown) and KSRP expression (see Figure 3A). As a control, DLD-1 cells stably transfected with the pcDNA3 vector were generated as well (pcDNA3). Overexpression of a His-tagged KSRP protein resulted in nearly complete inhibition of cytokine-induced iNOS mRNA expression (Figure 3B). Consistent with these results, reduction of KSRP expression by the antisense approach markedly enhanced cytokine-induced iNOS mRNA expression (Figure 3B). Also cytokine-induced iNOS-dependent NO-production was decreased by KSRP overexpression in DLD-1-pcDNA-His-KSRP cells and increased by KSRP downregulation in DLD-1-pcDNA-KSRPas cells (see Figure 3C).
Figure 3.
Modulation of KSRP expression alters cytokine-induced iNOS mRNA expression and iNOS-dependent NO-production. Plasmid constructs allowing high level expression of sense (pcDNA-His-KSRP) or antisense (pcDNA-KSRPas) KSRP cDNA were stably transfected into DLD-1 cells. Cells transfected with the pcDNA3 vector backbone (pcDNA3) were used as controls. For analysis of iNOS expression pools of stable transfected cell were preincubated for 18 h in medium without FCS and phenol red. Then cells were incubated with (CM) or without (Co) the cytokine mixture for 6 h, RNA was isolated and iNOS and GAPDH mRNA expression was analyzed. To determine iNOS-mediated NO-production cells were incubated for 24 h with or without CM and the supernatant of the cells was analyzed for nitrite content. As another approach to downregulate KSRP expression the RNA interference technique was used. DLD-1 cells were transfected with siRNA directed against luciferase (control, siLUC) or KSRP (siKSRP). After 24 h the transfected cells were preincubated for 18 h in medium without FCS and phenol red. Then the cells were incubated with or without CM for 6 h, RNA was isolated and iNOS, KSRP and GAPDH mRNA expression was analyzed by real-time RT–PCR. (A) Western blots using specific anti-KSRP- and anti-β-tubulin antibodies and extracts from the stable transfected DLD-1 cell pools. The blots are representative of four other blots showing similar results. The positions of KSRP and β-tubulin are indicated. (B) A summary of 10 qRT–PCR analyses is shown using RNAs form DLD-1-pcDNA3 (pcDNA3), DLD-1-pcDNA-His-KSRP (pcDNA-His-KSRP) or DLD-1-pcDNA3-KSRPas (pcDNA-KSRPas) cells. Data (means ± SEM) represent relative iNOS mRNA levels (***P < 0.001 versus CM-treated pcDNA3 cells). (C) A summary of 12 nitrite analyses is shown using supernatants from DLD-1-pcDNA3 (pcDNA3), DLD-1-pcDNA-His-KSRP (pcDNA-His-KSRP) or DLD-1-pcDNA3-KSRPas (pcDNA-KSRPas) cells. Data (means ± SEM) represent relative nitrite levels (***P < 0.001 versus CM-treated pcDNA3 cells). (D) A summary of six qRT–PCR analyses is shown using RNAs from DLD-1 cells transfected with luciferase siRNAs (siLUC) or KSRP siRNAs (siKSRP) and incubated with or without cytokine mixture (CM). Data (means ± SEM) represent relative KSRP mRNA levels (**P < 0,01 versus CM- and siLUC-treated cells). (E) A summary of six qRT–PCR analyses is shown using the same RNAs as in (D) but analyzing iNOS mRNA expression. Data (means ± SEM) represent relative iNOS mRNA levels (*P < 0.05; **P < 0.01; ***P < 0.001 versus CM- and siLUC-treated cells).
To support these results of KSRP-mediated reduction of iNOS expression we aimed to downregulate KSRP expression using the RNA interference technique. Therefore, we generated siRNAs containing a hairpin structure (shRNAs) by in vitro transcription (see Material and Methods) directed against the KSRP coding region (siKSRP). These shRNAs were transfected into DLD-1 cells. After transfection for 40 h, the cells were incubated in the presence or absence of the CM and the KSRP- and iNOS mRNA expression was measured by real-time RT–PCR. As a control, DLD-1 cells were transfected with shRNAs directed against the GL2-luciferase coding region (siLuc). As shown in Figure 3D, KSRP mRNA expression was reduced in DLD-1 cells transfected with the anti-KSRP shRNAs (siKSRP) compared to cells transfected with the siLuc control shRNAs. Analysis of iNOS mRNA expression in the same RNAs (see Figure 3E) showed that downregulation of KSRP expression by siKSRP markedly enhanced iNOS mRNA expression. Also in A549/8 cells siRNA-mediated downregulation of KSRP expression resulted in an enhancement of cytokine-induced iNOS mRNA expression (data not shown).
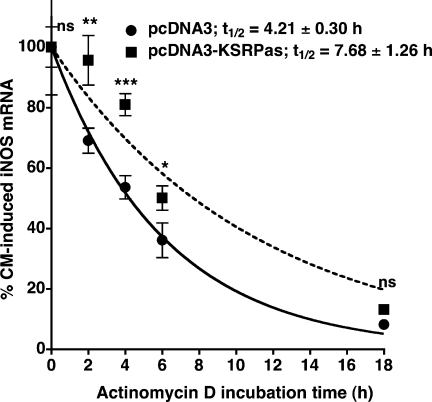
To test whether the effect of KSRP on iNOS expression resulted from KSRP-mediated destabilization of the human iNOS mRNA, we performed actinomycin D experiments. DLD-1-pcDNA3 and -pcDNA-KSRPas cells were incubated with CM for 6 h. Then actinomycin D (10 µg/ml) was added to stop transcription and RNA was isolated after 0, 2, 4, 6 and 18 h. Expression of iNOS mRNA in comparison to GAPDH was determined by qRT–PCR. As shown in Figure 4 compared to pcDNA3 cells (t1/2 = 4.21 ± 0.30 h), downregulation of KSRP expression enhanced iNOS mRNA stability (t1/2 = 7.68 ± 1.26 h).
Figure 4.
Effect of downregulation of KSRP expression on human iNOS mRNA stability. DLD-1-pcDNA3 (filled circles) and pcDNA-KSRPas (filled squares) cells were preincubated for 18 h in medium without FCS and phenol red. Subsequently cells were incubated with a cytokine mixture for 6 h. Then 10 µg/ml actinomycin D was added and RNAs were prepared after 0 to 18 h. iNOS mRNA and GAPDH mRNA concentrations were determined by qRT–PCR and iNOS mRNA was normalized to GAPDH mRNA. A summary of four qRT–PCR analyses is shown. Data (means ± SEM) represent relative iNOS mRNA levels in DLD-1-pcDNA3- (filled circles) and DLD-1-pcDNA-KSRPas- (filled squares) cells (*P < 0.05; **P < 0,01; ***P < 0.001; ns = not significant versus 0 h actinomycin D). Curve fitting was performed using Graphpad Prism for Macintosh.
In summary, KSRP regulates human iNOS expression in DLD-1 cells by destabilizing the human iNOS mRNA.
Cytokine incubation does not change KSRP expression in DLD-1 cells
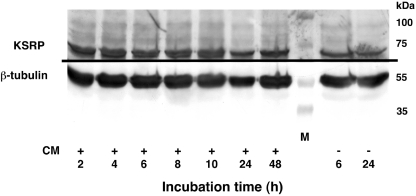
In order to get an insight in the mechanism of KSRP-mediated destabilization of iNOS mRNA we studied the expression of KSRP in DLD-1 cells. Cells were incubated with or without CM for 2 to 18 h and RNA and protein were isolated. Analysis of the KSRP mRNA expression showed no influence of cytokine induction on KSRP mRNA expression (data not shown). Also, as shown in Figure 5 cytokine treatment did not change KSRP protein expression.
Figure 5.
Cytokine incubation does not change KSRP expression in human DLD-1 cells. DLD-1 cells were preincubated for 18 h in medium without FCS and phenol red. Then the cells were incubated with or without a CM. Cytoplasmic protein extracts were prepared after the time periods indicated. Western blots were performed using specific anti-KSRP- and anti-β-tubulin antibodies and cytoplasmic extracts from DLD-1 cells. This blot is representative of three other blots showing similar results.
KSRP interacts with proteins involved in the exosomal degradation of ARE-containing mRNAs
The data above showed post-transcriptional regulation of cytokine-induced iNOS expression by KSRP without cytokine-mediated changes in KSRP expression. Therefore, it is likely that KSRP interacts with other proteins important for the stability of iNOS mRNA. KSRP has been shown to be an essential component of the degradation machinery of ARE-containing mRNAs by interaction with the exosome complex (17,21). Consequently, we tested the interaction of KSRP and the exosomal component PM-Scl 100 performing co-immunoprecipitation assays. In these experiments cell extracts were used which have been treated with RNase to exclude that binding to the same RNA resulted in coprecipitation by the specific antibodies. As shown for other human cell lines, also in DLD-1 cells these co-immunoprecipitation experiments showed a clear protein–protein interaction of KSRP with the exosomal component PM-Scl 100 (data not shown). However, these interactions were not changed by cytokine treatment (data not shown).
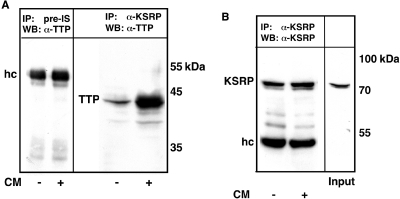
In addition to KSRP the RNA-bp TTP has been shown to interact with the exosome and to recruit this complex to ARE-containing mRNAs (17). By overexpression of TTP or downregulation of TTP expression we showed that TTP, which did not bind to the human iNOS mRNA, enhanced human iNOS expression (30). Therefore we analyzed the interaction of KSRP and TTP in human DLD-1 cells. Cellular extracts from CM- or untreated DLD-1 cells were pretreated with RNase and then incubated with monoclonal anti-KSRP antibodies. The immunoprecipitated material was analyzed by western blotting using a polyclonal anti-TTP antibody. As shown in Figure 6A, KSRP displayed a protein–protein interaction with TTP, which was enhanced by cytokine treatment. Analysis of the immunoprecipitated material for KSRP (Figure 6B) showed equal amounts of precipitated KSRP.
Figure 6.
KSRP interacts with TTP in human DLD-1 cells. DLD-1 cells were preincubated for 18 h in medium without FCS and phenol red. Then the cells were incubated with (CM) or without (Co) a cytokine mixture for 6 h. Extracts were prepared as described in Material and Methods. (A) Cell lysates were subjected to immunoprecipitation using monoclonal anti-KSRP antibodies (IP: α-KSRP) or a rabbit pre-immune serum (IP: Pre-IS). Coprecipitation of TTP was analyzed by western blots using polyclonal anti-TTP antibodies (WB: α-TTP). The positions of immunoglobulin heavy chains (hc) and TTP are indicated. One representative out of four co-immunoprecipitation analyses is shown. (B) To normalize for precipitated KSRP protein the amount of KSRP protein was analyzed performing western blots using monoclonal anti-KSRP antibodies (WB: α-KSRP). Also the KSRP expression in a control extract used for immunoprecipitation (Input, 10%) was analyzed. The positions of KSRP and immunoglobulin heavy chains (hc) are indicated. One representative blot out of four is shown.
Overexpression of a mutant KSRP protein, which is not able to bind to RNA, increases cytokine-induced iNOS expression in DLD-1 cells
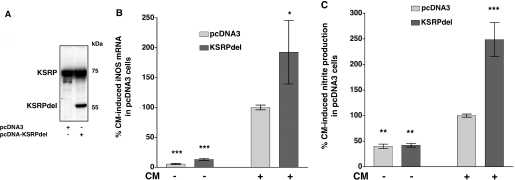
Since TTP mediates its effects on iNOS mRNA without binding to it, we aimed to analyze the biological role of KSRP binding to the iNOS 3′-UTR. Gherzi et al. (21) showed that deletion of the most carboxyterminal RNA binding domain (KH4) from KSRP generated a dominant negative KSRP isoform that was able to interact with the exosome but did not bind to ARE-containing RNAs. To analyze the effects of this mutant KSRP protein on cytokine-induced iNOS expression, we generated DLD-1 cell pools expressing this dominant negative KSRP isoform (see Figure 7A).
Figure 7.
Overexpression of a KSRP mutant which is not able to bind to RNA enhances iNOS expression. Plasmids allowing high level expression of a mutant KSRP protein not able to bind to RNA (KSRPdel) were stably transfected into DLD-1 cells. Cells transfected with the pcDNA3 vector backbone (pcDNA3) were used as controls. For analysis of iNOS expression pools of stable transfected cell were preincubated for 18 h in medium without FCS and phenol red. Then cells were incubated with (CM) or without (Co) the cytokine mixture for 6 h, RNA was isolated and iNOS and GAPDH mRNA expression was analyzed by real-time RT–PCR. To determine iNOS-mediated NO-production cells were incubated for 24 h with or without CM and the supernatant of the cells was analyzed for nitrite content. (A) Pooled populations of pcDNA3- or pcDNA-KSRPdel cells were analyzed for KSRP expression by western blots using specific anti-KSRP antibodies. One representative blot out of three is shown. (B) A summary of 5 qRT–PCR analyses is shown using RNAs from DLD-1-pcDNA3 (pcDNA3) or DLD-1-pcDNA3-KSRPdel (KSRPdel) cells. Data (means ± SEM) represent relative iNOS mRNA levels (*P < 0.05; ***P < 0.001 versus CM-treated pcDNA3 cells). (C) A summary of 5 nitrite analyses is shown using supernatants from DLD-1-pcDNA3 (pcDNA3) or DLD-1-pcDNA-KSRPdel (KSRPdel) cells. Columns (means ± SEM) represent relative nitrite levels (**P < 0.01; ***P < 0.01 versus CM-treated pcDNA3 cells).
Indicating the importance of KSRP binding, overexpression of this non-binding mutant isoform in DLD-1 cells (KSRPdel) resulted in an enhanced cytokine-induced iNOS mRNA expression and iNOS-dependent NO-production (see Figure 7B and C).
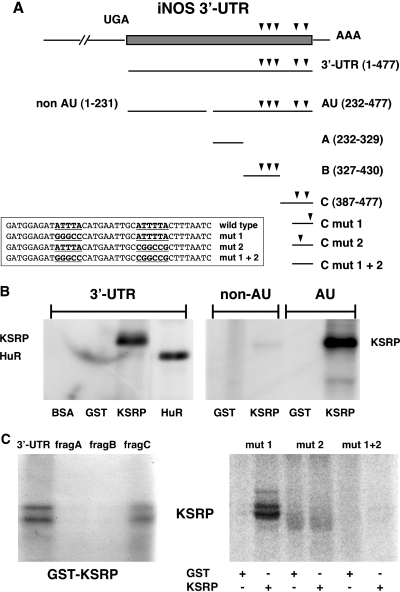
KSRP binds to the most 3′-located ARE in the 3′-UTR of the human iNOS mRNA
To characterize the binding site of KSRP to the human iNOS 3′-UTR (for sequence informations see GenBank L09210), recombinant GST-KSRP fusion protein was incubated with 32P-labeled transcripts comprising different nucleotide regions (see Figure 8A) and KSRP/RNA complex formation was assayed by UV-crosslinking experiments. Performing RNA gel retardation experiments instead of the UV-crosslinking method showed the same results (data not shown). We detected complex formation between recombinant GST-KSRP protein (or GST-HuR protein as control) and the whole 3′-UTR transcript. This binding activity was not observed with the GST protein (Figure 8B, left panel). To localize the KSRP binding site within the iNOS 3′-UTR, the region was first dissected into two subfragments, one without AU-repeats (non-AU) and the other containing these putative regulatory elements (AU). Only the subfragment containing the AU-repeats interacted with the KSRP protein (Figure 8B, right panel). Subsequently, the AU-fragment was dissected into three subfragments: subfragment A (232–329, no AU-repeats), subfragment B (327–428, three AUUUA-motifs), and subfragment C (387–477, one AUUUA and an AUUUUA element). As shown in Figure 8C (left panel), significant KSRP binding was found only for subfragment C. Therefore, as described for HuR (28), the KSRP binding site seems to be located at positions 439–477, with one or both of the AU-repeats being involved in the complex formation. As shown in Figure 8C, (right panel) binding activity of KSRP to a transcript comprising positions 443–477 was not affected when the AU-rich element at positions 449–453 was mutated to a GC-rich element (mut 1). In contrast, mutation of the second AU-element at positions 464–469 (mut 2) reduced complex formation considerably. The double mutant (mut 1/2) showed virtually no binding activity of KSRP.
Figure 8.
Analysis of the KSRP binding site in the human iNOS 3′-UTR RNA. Purified BSA, glutathione-S-transferase (GST), and GST-KSRP or GST-HuR fusion proteins were incubated with different radiolabeled RNAs generated by in vitro transcription using the different iNOS 3′-UTR fragments shown in Figure A. After binding, proteins were UV crosslinked to the RNA and the complexes were digested with RNase. RNA–protein complexes were separated on SDS–polyacrylamide gels. (A) Structure of the human iNOS 3′-UTR mRNA and fragments used in RNA binding studies. Scheme of the human iNOS 3′-UTR mRNA (477 nt) and transcripts used in RNA binding studies. The initial UGA nucleotide sequence (−3 to −1) corresponds to the translation termination codon. AUUUA and AUUUUA repeats are indicated by arrowheads. The sequences of different mutations in fragment C are shown. (B) KSRP binds to the AU-fragment of the human iNOS 3′-UTR. 32P-radiolabeled RNA transcripts [3′-UTR; non-AU; AU; see (A)] were incubated with BSA, GST, GST-KSRP fusion protein (KSRP) or GST-HuR fusion protein (HuR). The positions of RNA–protein complexes are indicated. (C) KSRP binds to the most 3′-located ARE in the human iNOS 3′-UTR. 32P-radiolabeled 3′-UTR, subfragment A (232–319, frag A), subfragment B (317–420; frag B) or subfragment C (387–477; frag C) were incubated with GST-KSRP (GST-KSRP) protein. The positions of RNA–protein complexes are indicated (left side). 32P-radiolabeled RNA transcripts (5′-ARE mutated: mut 1; 3′-ARE mutated: mut 2; both AREs mutated: mut 1 + 2) were incubated with either GST or GST-KSRP fusion protein (KSRP). The positions of RNA–protein complexes are indicated (right side).
The binding of HuR and KSRP to the human iNOS 3′-UTR sequence is mutually exclusive
As the analyses described above showed that KSRP binds to the same iNOS 3′-UTR sequences as HuR (28), we aimed to analyze whether KSRP and HuR compete for this binding sites. As shown in Figure 9, UV-crosslinking experiments revealed that enhanced amounts of HuR were able to replace KSRP from these binding sites and vice versa.
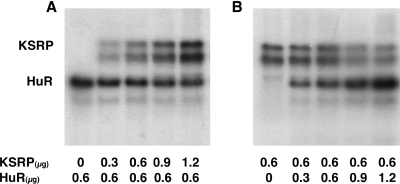
Figure 9.
Binding of KSRP and HuR to the 3′-UTR of the human iNOS mRNA is mutually exclusive. 32P-radiolabeled in vitro transcribed RNAs containing the sequence of subfragment C (387–477) were incubated in parallel with fixed amounts (0.6 µg) of GST-HuR or GST-KSRP and increasing amounts of GST-KSRP or GST-HuR (0 to 1.2 µg), respectively. After binding, proteins were UV crosslinked to the RNA and the complexes were digested with RNase. RNA–protein complexes were separated on SDS–polyacrylamide gels. The positions of RNA–protein complexes are indicated. (A) Radiolabeled fragment C-RNAs were incubated with 0.6 µg GST-HuR (0.6 µg) and 0 to 1.2 µg GST-KSRP. (B) Radiolabeled fragment C-RNAs were incubated with 0.6 µg GST-KSRP (0.6 µg) and 0–1.2 µg GST-HuR.
Cytokine incubation reduces intracellular binding of KSRP and enhances intracellular binding of HuR to the human iNOS mRNA
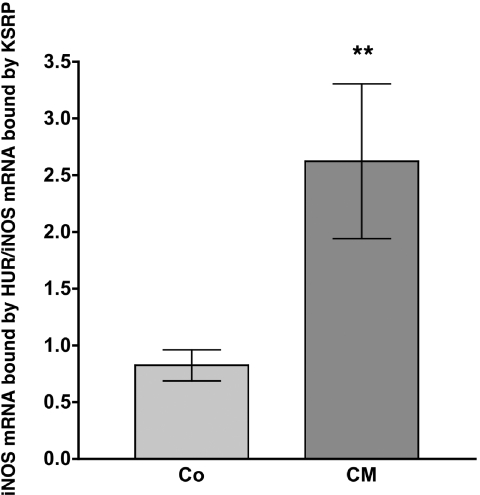
To verify a role for this competitive binding of KSRP and HuR to the iNOS mRNA in the regulation of iNOS expression in intact cells, we tested intracellular binding of these proteins to the iNOS mRNA by immunoprecipitation-qRT–PCR assays (see Figure 10). These analyses showed a cytokine-induced increase of the ratio of HuR-bound iNOS mRNA to KSRP-bound iNOS mRNA, representing enhanced binding of HuR and reduced binding of KSRP to the human iNOS mRNA after cytokine incubation.
Figure 10.
Cytokine incubation reduces intracellular binding of KSRP and enhances intracellular binding of HuR to the iNOS mRNA. DLD-1 cells were incubated for 4 h with or without CM. Cells were lyzed and RNA bound by KSRP or HuR protein was immunoprecipitated by specific antibodies. Immunoprecipitation with IgG was used as negative control. To normalize for the subsequent RNA purification steps 1 ng/sample of luciferase RNA transcribed in vitro was added before the RNA was isolated from immunoprecipitated proteins. The amount of iNOS mRNA bound by KSRP or HuR was determined by qRT–PCR using the luciferase RNA as normalization control. The values of the IgG controls were subtracted and the values for the iNOS mRNA bound by HuR were divided by the values for the iNOS mRNA bound by KSRP. A summary of 12 immunoprecipitation-qRT–PCR analyses is shown. Columns (means ± SEM) represent relative iNOS mRNA levels bound by HuR divided by the relative iNOS mRNA levels bound by KSRP (**P < 0.01 versus untreated DLD-1 cells).
Since KSRP destabilizes while HuR stabilizes iNOS mRNA (28), these changes in RNA binding of both proteins contribute to the stabilization of iNOS mRNA after cytokine induction.
DISCUSSION
Modulation of mRNA stability is an important control mechanism for regulation of human iNOS expression (22,26,30).
The rate of mRNA decay is determined by cis-acting sequences within the mRNA, which are recognized by trans-acting factors. The best-characterized cis-acting sequences responsible for mRNA decay in mammalian cells are the AREs present within the 3′-UTRs of short-lived mRNAs, whose expression has to be regulated exactly (12,47). These AREs are involved in deadenylation and subsequent degradation of mRNAs (48,49) and have also been described to stimulate 5′-decapping (50). Five such AREs are present in the 3′-UTR of the human iNOS mRNA. Furthermore, transfection experiments showed destabilization of the mRNA of a heterologous reporter gene by the human iNOS 3′-UTR (28).
More than 15 proteins are known to bind to AREs (11). Only few of them have been shown to regulate mRNA stability. These include the ARE/poly-(U)-binding/degradation factor 1 (AUF-1) (15,51), the embryonic lethal abnormal vision (ELAV) proteins (also named Hu proteins), especially HuR (29), the KSRP, (17,21) and the TTP family of zinc-finger RNA-bps (52,53). HuR has been shown to be involved in the regulation of human iNOS expression (28).
In an attempt to isolate proteins interacting with the 3′-UTR of the human iNOS mRNA, we performed RNA-affinity purifications of iNOS 3′-UTR RNA-bps (Figure 1). This method resulted in the identification of KSRP, PABP and hnRNP E1 as RNA-bps interacting with the human iNOS mRNA.
Besides regulating the translation of different mRNAs (54), hnRNP E1 has also been shown to regulate mRNA stability of the human renin (55) and collagen types I and III mRNAs (56). PABP is known to bind to the poly-A-tail of mRNAs and seems to be important for different levels of post-transcriptional control (57). However recent data suggest that PABP also binds specifically to ARE-containing mRNAs (46,58,59).
KSRP has been described as a protein essential for exosome-mediated degradation of ARE-containing mRNAs (17,21). As recent data from our laboratory (30) indicated that KSRP is involved in the post-transcriptional regulation of human iNOS expression, we analyzed the role of KSRP in human iNOS expression in more detail.
Using an in vitro degradation assay (Figure 2), we were able to show that immunodepletion of KSRP of extracts from DLD-1 cells resulted in enhancement of the stability of a 5′-capped and 3′-polyadenylated iNOS 3′-UTR RNA. Deletion of the AREs (non-AU) resulted in an enhancement of the stability of this RNA in these in vitro decay assays. Most important is that, without the AREs the depletion of KSRP has no effect on the stability of the 3′-UTR RNA. In the limitations of such in vitro decay assays this confirms an important role of KSRP in the regulation of iNOS expression at the level of mRNA stability.
To analyze the influence of KSRP on iNOS expression in intact cells, we generated stably transfected DLD-1 cell lines with enhanced or reduced KSRP expression. Results obtained with these cell lines as well as results generated using a RNA interference approach in DLD-1 and A549/8 cells confirmed the negative regulatory effect of KSRP on iNOS expression (see Figure 3). Additionally, we observed downregulation of TNF-α mRNA expression in DLD-1 cells overexpressing KSRP (data not shown). Actinomycin D experiments revealed a reduced decay of iNOS mRNA in cells with diminished KSRP expression, confirming that its effect occurs at the level of mRNA stability (Figure 4). In addition, cotransfection experiments showed no influence of KSRP on the cytokine-induced activity of the human iNOS promoter (see Supplementary Figures). Therefore, in accordance with its proposed destabilizing activity (17,21), KSRP is an important factor for the degradation of ARE-containing mRNAs in DLD-1 cells.
The results reported above indicate a direct negative effect of KSRP on human iNOS expression. Since induction of iNOS expression in DLD-1 cells depends on a complex CM we analyzed the effects of CM-incubation on endogenous KSRP expression. As shown in Figure 5 CM-incubation of DLD-1 cells did not change endogenous KSRP protein expression. Additionally, no clear evidence for modified KSRP localization was found (data not shown). Also analyses of KSRP phosphorylation revealed no changes in tyrosine or serine phosphorylation after cytokine treatment (data not shown). Therefore, the enhancement in iNOS mRNA expression measured after CM-incubation does not seem to correlate with changed KSRP expression, post-translational modifications or localization. In summary, these data imply that an interplay of different proteins in a complex network could be necessary to explain changes in iNOS mRNA expression after cytokine stimulation.
For example, KSRP has been described to recruit the exosome to ARE-containing mRNAs and thereby enabling exosome-mediated degradation of these RNAs (17,21). The exosome is a complex of 10 different proteins important for 3′– > 5′ degradation of ARE-containing mRNAs in mammalian cells (50–52). Therefore, we analyzed the interaction of KSRP with the exosome in DLD-1 cells by co-immunoprecipitation experiments. To exclude that binding of different RNA-bps to the same RNA resulted in a RNA-dependent co-immunoprecepitation we always pretreated the extracts with RNases. These analyses showed that KSRP displayed a protein–protein interaction with the exosome in DLD-1 cells (data not shown) as shown before for other human cell lines. However, this interaction was not modified by cytokine incubation of DLD-1 cells (data not shown). Therefore, KSRP seems to destabilize iNOS mRNA in interplay with the exosome independent of cytokine stimulation. This implies that for stabilization of iNOS mRNA after cytokine treatment, other factors must be involved that prevent KSRP-mediated decay.
In addition to KSRP, TTP has been shown to interact with the exosome and to recruit this nuclease complex to ARE-containing mRNAs (17). As shown in Figure 6, KSRP also interacts with TTP in DLD-1 cells. This interaction was enhanced by cytokine incubation, most likely due to increased TTP expression under these conditions. Interestingly, we have found that overexpression of TTP increases cytokine-induced iNOS expression in DLD-1 cells without any TTP binding to the human iNOS mRNA (30).
To verify that KSRP binding to the human iNOS mRNA is necessary for its regulatory action, we analyzed the effect of a non-binding KSRP mutant lacking the most carboxyterminal RNA binding domain KH4, (21), (Figure 7A). Loss of binding activity was tested in UV-crosslinking experiments using the iNOS 3′-UTR (data not shown). As shown in Figure 7B and C overexpression of this KSRP isoform (KSRPdel) in DLD-1 cells resulted in an enhancement of cytokine-induced iNOS mRNA expression and iNOS-dependent NO-production. Therefore, KSRP binding to the 3′-UTR of the human iNOS mRNA is essential for the negative regulation of iNOS mRNA stability by this RNA-bp.
Mapping of the exact binding sequence revealed the most 3′-located AU-rich element as binding site for KSRP (Figure 8). In former experiments we have shown that the ELAV protein HuR binds to the same ARE in the human iNOS mRNA (28). In in vitro binding assays (see Figure 9) using fixed amounts of KSRP and increasing amounts of HuR (or vice versa) a mutually exclusive binding of KSRP or HuR to the human iNOS 3′-UTR sequence was observed.
Analysis of the affinity of KSRP binding to the iNOS mRNA in in vitro assays using externally added iNOS 3′-UTR RNA (RNA-affinity chromatography or immunoprecipitation/UV-crosslinking) revealed no differences between untreated and cytokine-stimulated cells (data not shown). Therefore, enhanced iNOS mRNA expression after cytokine induction cannot be explained by changes in the affinity of KSRP binding to the human iNOS mRNA.
In contrast to this in vitro approaches, analysis of the relative intracellular binding of KSRP and HuR to the endogenous iNOS mRNA (see Figure 10) revealed a clear cytokine-induced change of the ratio of HuR and KSRP-bound to the iNOS mRNA. Whereas in intact DLD-1 cells KSRP binding was reduced, the binding of HuR was enhanced by cytokine incubation. Therefore, the enhanced expression of iNOS mRNA after cytokine stimulation can be explained by the shift from degradation mediated by KSRP to enhanced stabilization mediated by HuR resulting from these changes in RNA binding.
In summary, our data suggest the following mechanism of iNOS mRNA stabilization after cytokine stimulation: In untreated DLD-1 cells KSRP binds to the human iNOS mRNA 3′-UTR and recruits the exosome to the mRNA, thus enabling mRNA degradation. After cytokine treatment TTP expression and TTP/KSRP interaction are enhanced. In consequence, since TTP does not bind to the iNOS mRNA this is supposed to result in a dislodgement of the KSRP/exosome complex from the iNOS mRNA. As HuR and KSRP compete for the same binding site in the iNOS 3′-UTR sequence, this dislodgement of KSRP enhances HuR binding to the iNOS mRNA. This interaction results in an increase in iNOS mRNA stability and thus enhanced iNOS expression.
Finally complex interactions in a RNA-bp network seem to be required for regulation of human iNOS expression. In this network, KSRP appears to be a key regulator. It connects actions of other RNA-bps like TTP and HuR essential for iNOS mRNA stabilization. Subtle changes within this system may be responsible for the marked induction of iNOS expression after cytokine stimulation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr W. F. C. Rigby for providing the polyclonal anti-TTP antibody, Dr C. Y. Chen for providing the polyclonal anti-KSRP antibody, Dr G. J. M. Pruijn for providing the polyclonal anti-PM-Scl 100-antibody, Dr D. L. Black for providing the monoclonal anti-KSRP antibody and the expression plasmids pET15b-His-KSRP and pcDNA3.1-His-KSRP and Dr M. Bros for providing the pGl3-T7-Basic plasmid. This work was supported by Grant 8312–8338 62 61/322a,b from the Innovation Foundation of the State of Rhineland-Palatinate and by the Collaborative Research Center SFB 553 (Project A7 to HK). Funding to pay the Open Access publication charges for this article was provided by the Collaborative Research Center SFB 553 from the Deutsche Forschungsgemeinschaft (Bonn, Germany).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ross J. Control of messenger RNA stability in higher eukaryotes. Trends Genet. 1996;12:171–175. doi: 10.1016/0168-9525(96)10016-0. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz C.J., Gao M., Jones C.L., Wilusz J., Peltz S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- 3.Wilusz C.J., Wormington M., Peltz S.W. The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell. Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 4.Kontoyiannis D., Pasparakis M., Pizarro T.T., Cominelli F., Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 5.Di Marco S., Hel Z., Lachance C., Furneaux H., Radzioch D. Polymorphism in the 3′-untranslated region of TNFalpha mRNA impairs binding of the post-transcriptional regulatory protein HuR to TNFalpha mRNA. Nucleic Acids Res. 2001;29:863–871. doi: 10.1093/nar/29.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales J., Russell J.E., Liebhaber S.A. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 7.Jeon S., Lambert P.F. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross R.A., Lazarova D.L., Manley G.T., Smitt P.S., Spengler B.A., Posner J.B., Biedler J.L. HuD, a neuronal-specific RNA-binding protein, is a potential regulator of MYCN expression in human neuroblastoma cells. Eur. J. Cancer. 1997;33:2071–2074. doi: 10.1016/s0959-8049(97)00331-6. [DOI] [PubMed] [Google Scholar]
- 9.Dixon D.A., Tolley N.D., King P.H., Nabors L.B., McIntyre T.M., Zimmerman G.A., Prescott S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouble A., Grazide S., Meggetto F., Mercier P., Delsol G., Morello D. A new player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice. Cancer Res. 2002;62:1489–1495. [PubMed] [Google Scholar]
- 11.Hollams E.M., Giles K.M., Thomson A.M., Leedman P.J. mRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- 12.Chen C.Y., Shyu A.B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 13.Bevilacqua A., Ceriani M.C., Capaccioli S., Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 14.Good P.J. A conserved family of elav-like genes in vertebrates. Proc. Natl Acad. Sci. USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W., Wagner B.J., Ehrenman K., Schaefer A.W., DeMaria C.T., Crater D., DeHaven K., Long L., Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton B.J., Nagy E., Malter J.S., Arrick B.A., Rigby W.F. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 1993;268:8881–8887. [PubMed] [Google Scholar]
- 17.Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 18.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 19.Dember L.M., Kim N.D., Liu K.Q., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- 20.Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- 21.Gherzi R., Lee K.Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C.Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Kleinert H., Pautz A., Linker K., Schwarz P.M. Regulation of the expression of inducible nitric oxide synthase. Eur. J. Pharmacol. 2004;500:255–266. doi: 10.1016/j.ejphar.2004.07.030. [DOI] [PubMed] [Google Scholar]
- 23.MacMicking J., Xie Q.W., Nathan C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 24.Bogdan C. Nitric oxide and the immune response. Nature Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 25.Kröncke K.D., Fehsel K., Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clin. Exp. Immunol. 1998;113:147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Förstermann U., Li H., Schwarz P.M., Kleinert H. Signal transduction by reactive oxygen and nitrogen species. In: Forman H.J., Fukuto J., Torres M., editors. Dordrecht: Kluwer Academic Publishers; 2003. pp. 119–154. NO Synthesis and NOS Regulation. [Google Scholar]
- 27.de Vera M.E., Shapiro R.A., Nüssler A.K., Mudgett J.S., Simmons R.L., Morris S.M., Jr, Billiar T.R., Geller D.A. Transcriptional regulation of human inducible nitric oxide synthase (NOS2) gene by cytokines: initial analysis of the human NOS2 promoter. Proc. Natl Acad. Sci. USA. 1996;93:1054–1059. doi: 10.1073/pnas.93.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Pascual F., Hausding M., Ihrig-Biedert I., Furneaux H., Levy A.P., Förstermann U., Kleinert H. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J. Biol. Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. [DOI] [PubMed] [Google Scholar]
- 29.Brennan C.M., Steitz J.A. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fechir M., Linker K., Pautz A., Hubrich T., Förstermann U., Rodriguez-Pascual F., Kleinert H. Tristetraprolin regulates the expression of the human inducible nitric oxide synthase gene. Mol. Pharmacol. 2005;67:2148–2161. doi: 10.1124/mol.104.008763. [DOI] [PubMed] [Google Scholar]
- 31.Fechir M., Schneid C., Linker K., Förstermann U., Kleinert H. Complex regulation of human iNOS-expression by RNA binding proteins. Naunyn-Schmiedeb. Arch. Pharmacol. 2003;367:R41. [Google Scholar]
- 32.Kleinert H., Euchenhofer C., Fritz G., Ihrig-Biedert I., Förstermann U. Involvement of protein kinases in the induction of NO synthase II in human DLD-1 cells. Br. J. Pharmacol. 1998;123:1716–1722. doi: 10.1038/sj.bjp.0701782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 34.Kleinert H., Euchenhofer C., Ihrig Biedert I., Forstermann U. Glucocorticoids inhibit the induction of nitric oxide synthase II by down-regulating cytokine-induced activity of transcription factor nuclear factor-κB. Mol. Pharmacol. 1996;49:15–21. [PubMed] [Google Scholar]
- 35.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg M.E., Ziff E.B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 37.Hall M.P., Huang S., Black D.L. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brooks S.A., Connolly J.E., Diegel R.J., Fava R.A., Rigby W.F. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 2002;46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
- 39.Brouwer R., Allmang C., Raijmakers R., van Aarssen Y., Egberts W.V., Petfalski E., van Venrooij W.J., Tollervey D., Pruijn G.J. Three novel components of the human exosome. J. Biol. Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- 40.Lellek H., Kirsten R., Diehl I., Apostel F., Buck F., Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 2000;275:19848–19856. doi: 10.1074/jbc.M001786200. [DOI] [PubMed] [Google Scholar]
- 41.Wilson G.M., Brewer G. Identification and characterization of proteins binding A + U-rich elements. Methods. 1999;17:74–83. doi: 10.1006/meth.1998.0709. [DOI] [PubMed] [Google Scholar]
- 42.Min H., Turck C.W., Nikolic J.M., Black D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 43.Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 44.Chen C.Y., Gherzi R., Andersen J.S., Gaietta G., Jurchott K., Royer H.D., Mann M., Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 45.Grosset C., Chen C.Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A.B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 46.Bollig F., Winzen R., Gaestel M., Kostka S., Resch K., Holtmann H. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 2003;301:665–670. doi: 10.1016/s0006-291x(03)00015-9. [DOI] [PubMed] [Google Scholar]
- 47.Bakheet T., Frevel M., Williams B.R., Greer W., Khabar K.S. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shyu A.B., Belasco J.G., Greenberg M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 49.Xu N., Chen C.Y., Shyu A.B. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol. Cell. Biol. 1997;17:4611–4621. doi: 10.1128/mcb.17.8.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao M., Wilusz C.J., Peltz S.W., Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blaxall B.C., Pende A., Wu S.C., Port J.D. Correlation between intrinsic mRNA stability and the affinity of AUF1 (hnRNP D) and HuR for A+U-rich mRNAs. Mol. Cell. Biochem. 2002;232:1–11. doi: 10.1023/a:1014819016552. [DOI] [PubMed] [Google Scholar]
- 52.Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoecklin G., Ming X.F., Looser R., Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostareck-Lederer A., Ostareck D.H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., Hentze M.W. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morris B.J., Adams D.J., Beveridge D.J., van der Weyden L., Mangs H., Leedman P.J. cAMP controls human renin mRNA stability via specific RNA-binding proteins. Acta. Physiol. Scand. 2004;181:369–373. doi: 10.1111/j.1365-201X.2004.01307.x. [DOI] [PubMed] [Google Scholar]
- 56.Thiele B.J., Doller A., Kahne T., Pregla R., Hetzer R., Regitz-Zagrosek V. RNA-binding proteins heterogeneous nuclear ribonucleoprotein A1, E1, and K are involved in post-transcriptional control of collagen I and III synthesis. Circ. Res. 2004;95:1058–1066. doi: 10.1161/01.RES.0000149166.33833.08. [DOI] [PubMed] [Google Scholar]
- 57.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dean J.L., Sully G., Clark A.R., Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilization. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Sladic R.T., Lagnado C.A., Bagley C.J., Goodall G.J. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 2004;271:450–457. doi: 10.1046/j.1432-1033.2003.03945.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.