Abstract
Staufen1 is a component of transported ribonucleoprotein complexes. Genetic work in Drosophila has suggested that Staufen plays a role in the de-repression of translation of oskar mRNA following localization. To determine whether Staufen1 can play a similar role in mammals, we studied translation of transcripts in the presence or in the absence of Staufen1. Translationally repressed mRNAs were generated by fusing the structured human immunodeficiency virus type 1 trans-activating response (TAR) element to the 5′ end of a reporter transcript. In rabbit reticulocyte lysates and in mammalian cultured cells, the addition of Staufen1 resulted in the up-regulation of reporter activity when translation was driven by the TAR-bearing RNA. In contrast, Staufen1 had no effect on translation of efficiently translated mRNAs lacking an apparent structured 5′ end, suggesting that Staufen1-binding to the 5′ end is required for enhanced translation. Consistently, Staufen1 RNA-binding activity is necessary for this translational effect. In addition, similar up-regulation of translation was observed when Staufen1 was tethered to the 5′ end of mRNAs via other structured RNAs, the highest level of translational increase being obtained with the bona fide Staufen1-binding site of the Arf1 transcript. The expression of Staufen1 promoted polysomal loading of TAR-luciferase transcripts resulting in enhanced translation. Our results support a model in which the expression of Staufen1 and its interaction with the 5′ end of RNA and ribosomes facilitate translation initiation.
INTRODUCTION
Intracellular localization of mRNA is a universal phenomenon that is conserved in plants and animals (1–3). It has been shown to be important for cell motility (4), asymmetric cell division (5), axis formation during development (6), synaptic plasticity (7) and long-term potentiation (8). One general concept that emerges from studies on mRNA localization is the link that exists between localization and translation (3,9). mRNA is translationally silent during transport, but its translation becomes de-repressed following transport, resulting in localized protein synthesis. While several mechanisms to achieve translational repression during transport have been described, these largely involve the association of a repressor to the 3′-untranslated region (3′-UTR) (10–14) or the presence of an inhibitory RNA structure in the 5′-UTR to block translational initiation (15,16).
In contrast, there are few details known about the mechanism that controls the de-repression of translation following mRNA transport. Work in Drosophila has suggested that RNA-binding proteins can play a role here. For example, Staufen is a double-stranded RNA (dsRNA)-binding protein (17) that is required for the de-repression of oskar mRNA translation in oocytes (10,18). The mechanism underlying this function of Staufen is not known. In mammals, Staufen1 (Stau155) (19,20) is a component of RNA transport granules and ribosome-free RNA particles (21,22). It is believed to play important role(s) in the transport of RNA to dendrites (23–25) and because it is also associated with polysomes (19,20,26–28), it is likely to be involved in the translation of mRNAs.
To address the role of Staufen1 in translation, we used the well-studied model of translational repression involving the human immunodeficiency virus type 1 (HIV-1) trans-activating response region (TAR). The TAR RNA sequence is present at the 5′ end of all HIV-1 transcripts and forms a stable secondary RNA structure consisting of a stem–loop with a bulge. It inhibits translation by making the RNA cap structure inaccessible to translation initiation factors (29) and more generally by activating the interferon response pathway (15,30,31). In addition to the viral protein Tat (32), many cellular factors, including the dsRNA-dependent kinase (PKR) (33), the TAR-RNA-binding protein (TRBP) (34), the RNA helicase A (RHA) (35) and the La autoantigen (36), have been shown to interact with the RNA TAR structure. Some of these proteins also regulate translation of TAR-containing transcripts. PKR becomes activated when bound to TAR (15,31). Activated PKR phosphorylates the translation initiation factor 2α (eIF-2α) thus causing general inhibition of translation (37–40). In contrast, TRBP which was identified in a large-scale screen for cellular TAR interacting proteins (34) partially relieves the TAR-induced inhibition of translation through a PKR-independent pathway (41). Similarly, the La autoantigen stimulates translation of TAR-bearing transcripts through its ATP-dependent helicase activity that disrupts the TAR secondary structure (42,43).
In this report, we studied the role of Stau155 in translation in both rabbit reticulocyte lysates (RRLs) and in human embryonic kidney (HEK)293T cells. To generate translationally repressed transcripts, we fused dsRNA structures to the 5′ end of reporter transcripts. Our studies show that Stau155 stimulates translation of structure-repressed mRNA in a mechanism that requires Stau155-binding to the 5′ end of transcripts. Therefore, Staufen1 expression and binding are important factors to regulate translation of some RNAs and to contribute to the de-repression of translation following mRNA localization.
MATERIALS AND METHODS
Antibodies and reagents
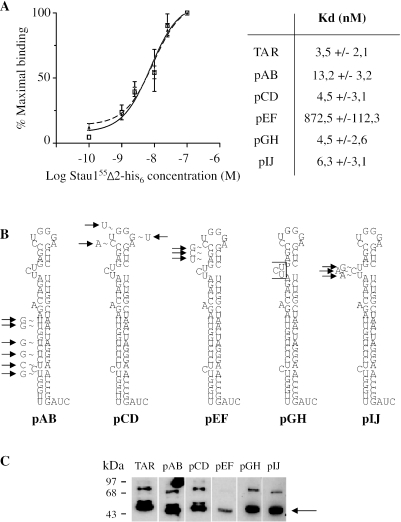
Mouse polyclonal anti-ribosomal L7/SPA protein antibodies were obtained from Novus Biologicals (Littleton, CO), mouse monoclonal anti-tubulin antibodies from ICN (Aurora, OH), rabbit polyclonal anti-calnexin antibodies from Stressgen Biotechnologies (Victoria, BC, Canada) and anti-mouse and anti-rabbit polyclonal antibodies conjugated to horseradish peroxidase from DakoCytomation (Mississauga, ON, Canada). To generate Stau155 monoclonal antibodies, bacterially expressed human GST-Stau155Δ2 was affinity purified on a glutathione Sepharose matrix (Amersham Biosciences Corp, Piscataway, NJ) and eluted from the column by cleavage with thrombin. Mice were injected with 10 µg of antigen per injection. Following serum conversion, spleens of positive mice were isolated and monoclonal antibodies (11C6) were prepared as described previously (44). G418 and 2-aminopurine (2-AP) were purchased from Sigma (Oakville, ON, Canada).
cDNA construction
Plasmids coding for Stau155Δ2-his6 (formerly named HFBDQ-his6), NEP-his6, Stau155-HA3 (20) and Stau155F-HA3 (26) were described previously. Plasmids containing mutations in TAR were generated from the p48 (pCD, pEF, pGH and pIJ) or p49 (pAB) plasmids (generous gifts from K. T. Jeang, NIH/NIAID Bethesda, MD) by ligation of double-stranded synthetic oligonucleotides (Table 1) within the TAR sequence. The p48 and p49 plasmids were first digested with SmaI/EcoRI and AvaI/EcoRI, respectively, treated with the Klenow fragment to blunt ends and ligated to remove the SacI restriction site in the multi-cloning site. Then, plasmids were digested with Sac1 in the TAR sequence and either BamH1 (pAB), BglII (pCD) or HindIII (pEF, pGH and pIJ), purified and ligated with oligonucleotides. La-his6 plasmid was obtained by subcloning the BamHI/HindIII fragment of La-pBS SK (a generous gift from K. T. Jeang) in frame with the his6 tag in the pQE31 expressing plasmid (Qiagen, Mississauga, ON, Canada). pcDNA3 RSV-Rluc plasmid was constructed by subcloning the HindIII–XbaI PCR fragment from pRluc-N1(h) (PerkinElmer Biosignal Inc, Montreal, QC, Canada) into the XbaI–HindIII sites of pcDNA3 RSV plasmid (45). The PCR was carried out with 2 U of Vent DNA polymerase (New England Biolabs, Pickering, ON, Canada) and the sense 5′-CTCACGCGTCTGCAG-3′ and antisense 5′-GGCTGATTATGCTCTAGATCG-3′ primers. The pcDNA3 RSV-TAR-Rluc plasmid was constructed by subcloning the TAR-containing HindIII fragment of pSp6TAR-CAT plasmid (a generous gift from E. Cohen, University of Montreal) into the HindIII site of pcDNA3 RSV-Rluc. The sh1 and sh2 plasmids were obtained by the PCR SHAGging strategy (46). Two rounds of PCR were carried out. A first PCR was carried out on the pGEM-U6 plasmid (a generous gift from G. Ferbeyre, University of Montreal) with 50 pmol of U6 sense 5′-GATTTAGGTGACACTATAG-3′ and sh1 5′-AAAAAAATAAGGATCA ACAGGCTTATACATCGGCTCAAGCTTCAACCAATGTATAAGCCTGTTGACCCTTACGGT GTTTCGTCCTTTCCACAA-3′ or sh2 5′-AAAAAAACACCTCCCACACACAGAC ATCGGCCCATCAAGCTTCACGGACCAATGTCTGTGTGTGGGAGGTGCGGTGTTTCGTCCTTTCCACAA-3′ antisense primers. PCR was carried out with 100 ng pGEM-U6 template, 5 U AmpliTaq DNA polymerase (Applied Biosystems), 4% dimethyl sulfoxide, 0.2 mM dNTPs and 1.5 mM MgCl2 (95°C for 3 min, 30 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 1 min, followed by a 10 min incubation at 72°C). PCR products were purified on low-melt agarose gel, and ethanol precipitated. They were then cloned as followed: using the first PCR SHAG products as template, a second PCR amplification was carried out with 0.2 µM of U6 sense 5′-ACAGAATTCGATTTAGGTGACACTATAG-3′ (underlined: the EcoRI restriction site) and sh1 antisense 5′-ACACTCGAGAAAAAAATAAGGATCAACAGGCTTA-3′(underlined: the XhoI restriction site) or sh2 antisense primers (5′-ACACTCGAGAAAAAAACACCTCCCACACACAGAC-3′ (underlined: the XhoI restriction site) at 95°C for 1 min, 58°C for 1 min, 72°C for 1 min for 35 cycles. The resulting PCR products were digested with EcoRI/XhoI and subcloned in pBluescript KS plasmid (Stratagene). The Stau155Δsh1-HA3 plasmid was PCR amplified from Stau155-HA3 plasmid with the all-around technique using the sense 5′-CTTACTCTCGGACGCAGTCCACCTA-3′ and antisense 5′-TAGGTGGACTGCGTCCGAGAGTAAG-3′ oligonucleotide primers. Similarly, Stau155KK-HA3 was PCR amplified from Stau155-HA3 plasmid using the sense 5′-AGCGCGGCGATTTCAAAGAAAAATGCC-3′ and antisense 5′-AATCGCCGCGCTTTTCCCTTCACC-3′ primers.
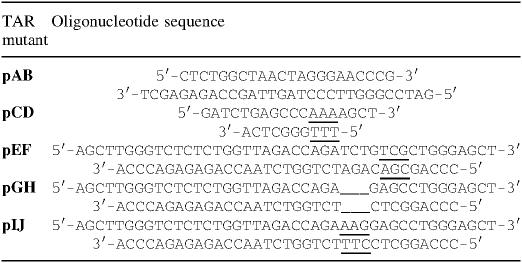
Table 1.
Oligonucleotides used to mutate the TAR sequence
The MS2bs-Rluc plasmid was obtained by subcloning the KpnI digestion fragment of the pSL-MS2 plasmid (a kind gift from P. Chartrand, University of Montreal) in the KpnI site of the pcDNA3 RSV-Rluc plasmid. The pcDNA3 RSV SBS-Rluc plasmid was obtained by PCR amplification of the Staufen1-binding site (SBS) from the pSport-Arf1 plasmid using the sense 5′-CAGCTCCGGAACCAGAAGTGAAC-3′ and antisense 5′-AGGACCCGGGAACACAGCGACTCCTGGAGG-3′ (underlined: the XmaI restriction site) at 95°C for 1 min, 57°C for 45 s, 72°C for 45 s for 35 cycles. PCR products were kinased and digested with XmaI restriction enzyme, purified and subcloned in the EcoRV/XmaI sites of pBluescript SK. pBluescript SK SBS plasmid was digested by HindIII/BamHI and the digestion fragment was subcloned in the HindIII/BamHI sites of the pcDNA3 RSV-Rluc plasmid.
Recombinant protein production and purification
Bacterially expressed Stau155Δ2-his6 and NEP-his6 were induced for 3 h with 1 mM isopropyl-β-d-thiogalactopyranoside and purified as described previously (20). Aliquots of purified proteins were stored at −80°C until use. Protein concentration was determined by the BioRad dye reagent and BSA as standard.
In vitro assays
The SpIII-10 CAT and pSp64TAR-CAT plasmids (generous gift from E. Cohen, University of Montreal) were linearized at the BamHI site, transcribed in vitro using the Sp6 RNA polymerase and m7GpppG CAP analog and used for in vitro translation in RRLs as described previously (42). One-fifth volume of CAT and one volume of TAR-CAT translation products were either loaded on gel and detected by autoradiography or quantified by enzyme-linked immunosorbent assay (ELISA) as indicated by the manufacturer (Roche Biochemicals, Laval, QC, Canada). For the in vitro ribosome pull-down assay, 250 ng of Stau155Δ2-his6, in the presence or in the absence of 50 ng of TAR-CAT RNA, were incubated with RRL for 1 h. The reaction mixture was diluted to 300 µl with ice-cold isotonic buffer (110 mM KOAc, 2 mM MgOAc, 10 mM HEPES–KOH, pH 7.3 and 2 mM DTT) and centrifuged in a Beckman SW 50.1C rotor for 45 min at 100 000 g at 4°C. The ribosome-enriched pellet was harvested and analysed by SDS–PAGE. The helicase assay was performed as described previously (43). Northwestern and filter binding assays were carried as described previously (20). For immunoprecipitation, transfected cells were lysed in 600 µl of lysis buffer (50 mM Tris–Cl, pH 7.5, 0.5% Triton X-100, EDTA 15 mM and DTT 1 mM). Cells extracts were pre-cleared with 60 µl of 50% v/v Protein A-Sepharose slurry (Roche) for 1 h and then incubated with 3 µl of anti-HA ascite fluid (12CA5) at 4°C for 2 h and with 150 µl of 50% v/v Protein A-Sepharose slurry at 4°C for 2 h.
RNA steady-state level
HEK293T cells were cultured in 12-well dishes and transfected with 100 ng of pcDNA3 RSV-Rluc or pcDNA3 RSV-TAR-Rluc in the absence or in the presence of increasing quantities of pcDNA3 RSV-Stau155-HA3 with FuGene6 transfection reagent (Roche). Twenty-four hours post-transfection, total RNA was extracted with Trizol reagent (Invitrogen Life Science, Burlington, ON, Canada) and treated with DNase I for 1 h at 37°C. Rluc, TAR-Rluc and GAPDH RNAs were reverse transcribed at 42°C for 17 min using 1 µg of total RNA, 5 pmol of Rluc (5′-CAGCACTCTCTCCACGAAGC-3′) and GAPDH (5′-CAAAAGTTGTCATGGATGACC-3′) antisense primers using the GENEAMP RNA PCR core kit (Applied Biosystems, Foster City, CA). Rluc (5′-GCAAGGTGTACGACCCCG-3′) and GAPDH (5′-CCTTCATTGACCTCAACTACAT-3′) sense primers (250 ng) were added for PCR amplification (95°C for 2 min, followed by 17 cycles at 94°C for 1 min, 54°C for 1 min and 72°C for 1 min). Resulting products were resolved on a 1.25% agarose gel. Similarly, 50 ng of CAT or TAR-CAT RNAs were incubated with RRL at 30°C for 0, 7.5, 15 or 30 min in the presence of 40 µg/µl of G418. RNA was extracted with Trizol reagent and reverse transcribed using 5 pmol of CAT antisense primer (5′-CCACTCATCGCAGTACTGTTG-3′). PCRs were carried out at 95°C for 2 min, followed by 14 cycles at 94°C for 1 min, 52.5°C for 1 min and 72°C for 1 min using 250 ng of CAT sense primer (5′-CCTATAACCAGACCGTTCAGC-3′) and antisense primer (5′-CCACTCATCGCAGTACTGTTG-3′). Resulting products were resolved on a 1.5% agarose gel.
Transfection and luciferase assays
HEK293T cells and PKR−/− MEF (a generous gift from A. Gatignol, McGill University) were cultured in DMEM (Invitrogen Life Science) supplemented with 10% Cosmic calf serum (Hyclone, Logan, UT), 5 µg/ml of penicillin–streptomycin and 2 mM l-Glutamine (Invitrogen Life Science). To test the role of Stau1 in translation, cells were transfected with FuGene6 (Roche Biochemicals) using 100 ng of pcDNA3 RSV-Rluc or pcDNA3 RSV-TAR-Rluc and increasing concentrations (0, 50, 100 and 500 ng) of plasmid coding for Stau155-HA3 or its mutants. Total amount of transfected DNA was adjusted to 600 ng with the pcDNA3 RSV plasmid. Cells were harvested 24 h post-transfection in 100 µl of lysis buffer (0.1 M Tris–Cl, pH 7.9, 0.5% NonidetP40 and 1 mM DTT) for 5 min on ice. An aliquot of 25 µl of the extracts and 5 µM Coelenterazine H (Molecular Probes, Burlington, ON, Canada) were used for Renilla reniformis luciferase assays. Luminescence was quantified with a Fusion α-FP (PerkinElmer-Canberra Packard BioScience) by measuring emitted light at 475–480 nm. To knockdown the expression of Stau1, HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen Life Science) using 700 ng of the silencing sh1 or the non-silencing sh2 plasmids. For translation assays, cells were re-transfected 24 h later using FuGene6 (Roche) and plasmid DNA as described above. Knockdown rescue was performed with 300 ng of plasmid coding for Stau155Δsh1-HA3.
Cell fractionation on sucrose gradients
Polyribosome profile was analysed as described previously (26). Briefly, transfected HEK293T cells were incubated for 20 min with cycloheximide (100 µg/ml), washed in cold phosphate-buffered saline and isotonic buffer and lysed in hypotonic buffer supplemented with cycloheximide (100 µg/ml). For the run-off experiments, cells were incubated for 30 min with sodium azide (25 mM) instead of cycloheximide. Cytoplasmic extracts (corresponding to ∼20 OD260) were centrifuged on a continuous 10–40% or 15–45% sucrose gradient containing 100 mM KCl, 10 mM KOAc, 2 mM MgOAc, 1 mM DTT and 5 mM HEPES–KOH, pH 7.3, with or without cycloheximide (100 µg/ml) in a SW41 rotor (Beckman) at 160 000 g for 150 min at 4°C. Fourteen fractions of ∼800 µl were recovered and the ribosomal profile was monitored at OD254 with a gradient fractionator (ISCO, Lincoln, USA). Aliquots containing 25 µl of each fraction were analysed by SDS–PAGE and western blotting. For RNA analysis, total RNA was extracted from a 250 µl aliquot of each fraction by adding an equal volume of denaturing buffer [7 M urea, 1% (w/v) SDS, 350 mM NaCl, 10 mM EDTA and 10 mM Tris–HCl, pH 7.5] followed by phenol–chloroform extraction and ethanol precipitation. RNA samples were incubated for 5 min at 65°C in RNA denaturing solution [66% (v/v) formamide, 8% (w/v) formaldehyde, 1× MOPS electrophoresis buffer] and slot-blotted onto nylon membrane using a HybriSlot apparatus (Gibco BRL). Membranes were hybridized with a random-prime 32P-labelled Rluc DNA fragment, exposed overnight and revealed by autoradiography.
RESULTS
Stau155 stimulates translation of inefficiently translated transcripts in RRL
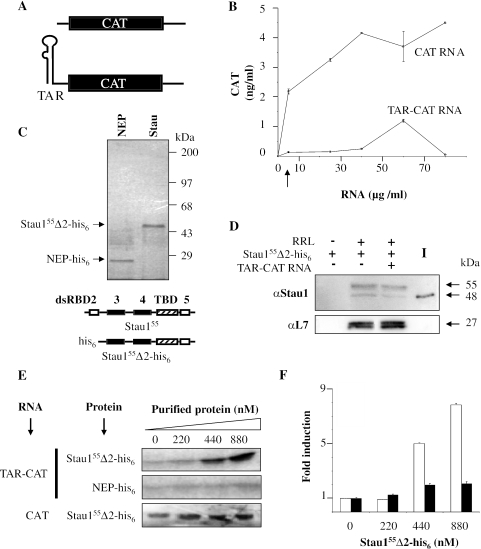
To determine whether Stau155 regulates translation, we first set-up an in vitro translation assay using RRLs. Translation efficiencies of capped chloramphenicol acetyl transferase (CAT) RNA and TAR-CAT RNA (Figure 1A) were compared. Similar to previous data (29,47), the translation of a transcript that contains TAR at the 5′ end is repressed in RRL (Figure 1B). Then, bacterially expressed and purified Stau155Δ2-his6 or NEP-his6 as control (Figure 1C) were tested for their capacity to associate with ribosomes as reported in cultured cells (26). Stau155Δ2-his6 was used instead of the full-length Stau155 because this protein is more soluble than full-length. RRLs were incubated in the presence or in the absence of 250 ng of recombinant Stau155Δ2-his6 and extracts were centrifuged at 100 000 g for 45 min to pellet ribosomes. Ribosomal pellets were separated by SDS–PAGE and analysed by western blotting using anti-Stau1 and anti-L7 ribosomal protein antibodies. After centrifugation, recombinant Stau155Δ2-his6 was found in the pellet with endogenous Stau1 and ribosomes (Figure 1D). The addition of TAR-CAT RNA had no effect on Staufen quantities in the ribosomal pellet, consistent with our previous demonstration that Stau1 interaction with ribosomes is RNA independent (26). When centrifuged in the absence of RRL, Stau155Δ2-his6 was not detected in the pellet (Figure 1D). These results show that purified Stau155Δ2-his6 associates with ribosomes in RRL in the presence or in the absence of added RNAs and that it is not over-represented as compared with endogenous Stau1 in this assay.
Figure 1.
Stau155 increases translation of TAR-bearing RNA in RRL. (A) Schematic representation of transcripts. RNAs coding for the CAT reporter protein are shown with or without a double-stranded TAR RNA structure at the 5′ end. (B) Translation of in vitro synthesized CAT and TAR-CAT transcripts in RRL. TAR structure at the 5′ end of the transcript represses translation. Vertical arrow, RNA concentration (4 µg/ml) subsequently used in the in vitro translation assays. (C) Bacterially expressed NEP-his6 (NEP) and Stau155Δ2-his6 (Stau) were purified on Ni-NTA columns, analysed by SDS–PAGE and Coomassie blue staining. Wild-type Stau155 and the truncated Stau155Δ2-his6 are schematically represented. Black boxes, full-length double-stranded RNA-binding domains (dsRBD) 3 and 4; white boxes, half size dsRBD 2 and 5; hatched box, the tubulin binding domain (TBD). (D) Bacterially expressed and purified Stau155Δ2-his6 was added (+) to RRL in the presence (+) or in the absence (−) of TAR-CAT RNA. Extracts were centrifuged and proteins in the pellet were analysed by SDS–PAGE. Proteins were revealed with anti-Stau1 and anti-ribosomal protein L7 antibodies. Endogenous Stau1 in RRL is visible at 55 kDa. I, input is 250 ng protein. (E) A representative experiment showing that Stau155Δ2-his6 increases translation of TAR-CAT RNA in RRL. CAT or TAR-CAT RNAs (4 µg/ml) were incubated in RRL with increasing concentrations of bacterially purified Stau155Δ2-his6 or NEP-his6, in the presence of [35S]methionine. Resulting proteins were analysed by SDS–PAGE and autoradiography. Note that one-fifth volume of CAT and one volume of TAR-CAT translation products were loaded on gel. (F) In this experiment, in vitro synthesized CAT was measured using ELISA (n = 4). Statistical analyses of TAR-CAT (open bars) and CAT (black bars) RNA translation in relation to the concentration of Stau155Δ2-his6. In the absence of Stau155Δ2-his6, a 17-fold repression of translation of the TAR-CAT RNA was observed as compared with translation of CAT RNA. Results are expressed as fold induction in CAT activity versus the concentration of the Stau155Δ2-his6 coding plasmid. To facilitate comparison, fold induction of the CAT activity in the absence of Stau155Δ2-his6 was defined as 1.
RRLs were then incubated with CAT or TAR-CAT transcripts (4 µg/ml). At this RNA concentration, the translation capacity of RRL was not saturated and a 17-fold repression of translation of the TAR-CAT RNA was observed as compared with translation of CAT RNA (Figure 1B). Increasing concentrations of recombinant Stau155Δ2-his6 or NEP-his6 were tested for their ability to stimulate translation. Stau155Δ2-his6 enhanced translation of the repressed TAR-CAT RNA in a dose-dependent manner reaching 15-fold (Figure 1E). This effect was specific since NEP-his6 had no effect on TAR-CAT RNA translation. In contrast, Stau155Δ2-his6 did not markedly affect translation efficiency of CAT RNA, showing a 2-fold increase (Figure 1E). A sensitive CAT ELISA was also used to quantify the in vitro synthesis of CAT. These assays demonstrated that Stau155Δ2-his6 increases translation of TAR-repressed transcripts by 10-fold and that of normally translated mRNAs by only 2-fold (Figure 1F).
Stau155 stimulates translation of TAR-containing transcripts in mammalian cells
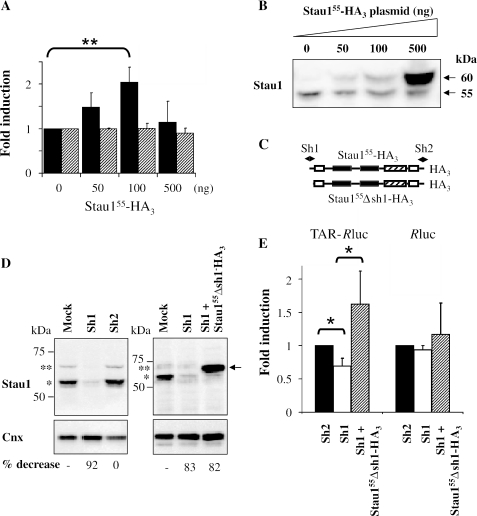
The effects of Staufen on translation efficiency were then measured in mammalian HEK293T cells. Cells were transfected with plasmids that expressed either Renilla reniformis luciferase (Rluc) or TAR-Rluc transcripts. The TAR-mediated translational repression was also observed in this context showing a 2-fold decrease in luciferase activity as compared with that of the Rluc transcript (Figure 2A). HEK293T cells were then co-transfected with increasing amounts of Stau155-HA3 expressor construct (Figure 2B) and either Rluc or TAR-Rluc plasmids. As shown in Figure 2A, Stau155-HA3 stimulated translation of TAR-Rluc RNA 2-fold when compared with TAR-Rluc RNA translation in the absence of transfected Stau155-HA3. In contrast, it had no effect on translation of Rluc transcript as indicated by a stable luciferase activity. Therefore, Stau155-HA3 rescued the inhibition imposed by the TAR sequence on the translation of TAR-Rluc transcripts. Importantly, the optimal amount of plasmid (100 ng) that enhances translation of TAR-Rluc mRNA results in Stau155-HA3 expression levels that are quantitatively close to that of endogenous Stau155, as shown in Figure 2B.
Figure 2.
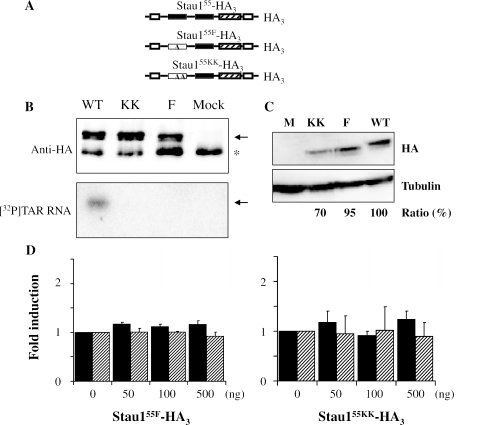
Stau155-HA3 increases translation of TAR-Rluc transcripts in HEK293T cells. (A) HEK293T cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of a plasmid coding for Stau155-HA3. Resulting luciferase activity was quantified 24 h post-transfection. In the absence of Stau155-HA3, a 2-fold repression of translation of the TAR-Rluc RNA was observed as compared with translation of Rluc RNA. Results are expressed as fold induction in luciferase activity versus the concentration of the Stau155-HA3 coding plasmid. To facilitate comparison, fold induction of the luciferase activity in the absence of Stau155-HA3 was defined as 1. Black bars, TAR-Rluc RNA; hatched bars, Rluc RNA. **P ≤ 0.01, n = 3. (B) HEK293T cells were transfected with different concentrations of a plasmid coding for Stau155-HA3. Western blot analyses showed that, for most dilutions, Stau155-HA3 (60 kDa) is not overexpressed as compared with endogenous Stau155 (55 kDa). (C) Schematic representation of Stau155Δsh1-HA3 and position of the sh1 and sh2 RNAs. Symbols are described in the legend of Figure 1. (D) HEK293T cells were transfected with plasmids expressing sh1 or shsh2 RNAs (left panel) or co-transfected with plasmids expressing sh1 RNA and Stau155Δsh1-HA3 (right panel). Proteins were analysed by SDS–PAGE and western blotting using anti-Stau1 and anti-calnexin (Cnx) antibodies. The percentage of down-regulation of endogenous Stau155 is indicated at the bottom of the figure. Position of endogenous Stau163 and of Stau155 is indicated by two and one asterisk, respectively. (E) HEK293T cells were transfected as described in (D) in the presence of either TAR-Rluc or Rluc expressors. Resulting luciferase activity was analysed as described in (B) (n = 3). Expression of sh1 decreased luciferase activity that is rescued by expression of Stau155Δsh1-HA3. *P ≤ 0.05.
In order to examine the effect of Staufen on translation, we then used RNAi to knockdown Staufen expression on HEK293T cells expressing the Rluc transcripts as above. Short hairpin (sh) RNA-expressing plasmids (Figure 2C) were used to knockdown endogenous expression of Stau1. sh1 was shown to result in a 92% decrease in Stau1 protein expression (Figure 2D). Therefore, sh1- (silencing) or control sh2- (non-silencing) expressing plasmids were co-transfected with plasmids that expressed either Rluc or TAR-Rluc. Sh1 caused a significant 30% decrease (P ≤ 0.05) in luciferase activity in cells expressing the TAR-Rluc transcript (Figure 2E). In contrast, sh1 had no effect on luciferase activity when the Rluc transcript was expressed. sh2 did not modulate luciferase activity when either RNA was expressed (Figure 2E). In an attempt to demonstrate that the effect of Staufen on translation was indeed due to Staufen, we performed an experiment in which sh1 was used to knockdown expression of endogenous Staufen, but co-expresssed Staufen in trans using a vector that does not have the sh1 target sequence (Stau155Δsh1-HA3, M. Luo and L. DesGroseillers, unpublished data) (Figure 2C and D). The results showed that the effects were specific to Staufen expression (Figure 2E). Taken together, the results obtained from three different approaches show that Stau155 modulates the expression of translationally repressed TAR-containing transcripts but has no effect on efficiently translated mRNAs.
Stau155-dependent translational stimulation is independent of PKR activity
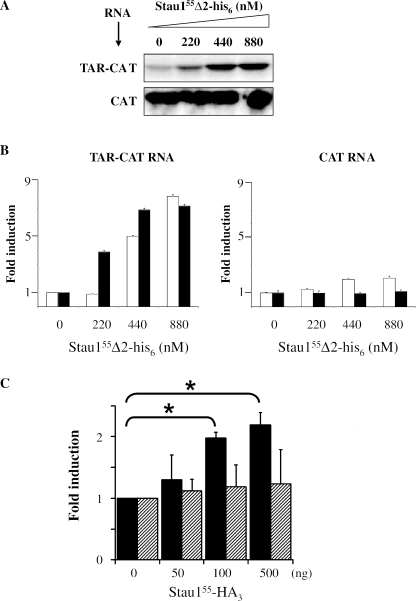
The ability of Stau155 to modulate the expression of TAR-repressed transcripts was distinguished from a competition with PKR for binding the dsRNA TAR structure by in vitro translation assay in the presence of 2-AP, a Ser/Thr kinase inhibitor (48). As shown before (49), 2-AP treatment of RRL slightly increased translation of the TAR-CAT transcript 2-fold likely by relieving a partial translational repression imposed by PKR. However, increasing concentrations of Stau155Δ2-his6 still stimulated translation of the TAR-containing transcript 8-fold. This shows that 2-AP did not prevent Stau155-dependent activation of translation (Figure 3A and B). In contrast, 2-AP prevented the 2-fold induction observed with Stau155Δ2-his6 on CAT RNA translation (Figure 3A and B), showing that, in this case, the increase in translation that is mediated by Staufen may be PKR dependent.
Figure 3.
Stau155-mediated increase of TAR-bearing RNA translation is independent of PKR. (A) In vitro protein synthesis in RRL was performed as described in Figure 1, except that PKR was inhibited by addition of the ser/thr kinase inhibitor 2-AP. The same volume of CAT and TAR-CAT translation products was loaded on gel. (B) Statistical analyses of TAR-CAT (left panel) and CAT (right panel) RNA translation in relation to the concentration of Stau155Δ2-his6. In vitro synthesized CAT was measured by ELISA (n = 4). Black bars, 2-AP; open bars, controls. (C) PKR−/− MEF cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of a plasmid coding for Stau155-HA3. Resulting luciferase activity was measured as described in Figure 2A. In the absence of Stau155-HA3, a 2-fold repression of translation of the TAR-Rluc RNA was observed as compared with translation of Rluc RNA. To facilitate comparison, fold induction of the luciferase activity in the absence of Stau155-HA3 was defined as 1. Black bars, TAR-Rluc RNA; hatched bars, Rluc RNA. *P ≤ 0.05, n = 3.
To confirm these results, we tested the effects of Stau155 in PKR-deficient cells (PKR−/−) (50). PKR−/− cells were co-transfected with plasmids coding for Stau155-HA3 and either Rluc or TAR-Rluc. As observed in HEK293T cells, Stau155-HA3 increased translation of TAR-Rluc mRNA by 2-fold, whereas it has no effect on Rluc transcripts (Figure 3C). Altogether, our results show that the Stau155-stimulating effect on TAR-bearing RNA translation is achieved through a PKR-independent pathway.
Stau155 binds to the TAR RNA structure
We next determined whether Stau155 binds the TAR RNA structure in vitro since it is the only difference between the Rluc and TAR-Rluc transcripts. Filter binding and Northwestern assays (20) were used to examine this. These analyses have allowed us to show that Stau155Δ2-his6 binds the TAR-RNA structure with high affinity with a Kd of 3.5 nM (Figure 4A). Several mutants that disrupt specific regions of the TAR structure (Figure 4B) (34) were tested to map the Stau155Δ2-his6 binding site. Although mutations in the lower stem, in the bulge or in the loop did not abolish binding of Stau155Δ2-his6, mutations in the upper stem between the bulge and the loop were critical (Figure 4A and C). As control, NEP-his6 did not bind TAR RNA (data not shown). Altogether, these results support a model in which Stau155Δ2-his6 binds to the TAR RNA structure and this may be important for the observed translational enhancement of RNAs that possess structured 5′ regions.
Figure 4.
Stau155Δ2-his6 binds the TAR-RNA structure in vitro. (A) Filter binding assays using bacterially expressed and column-purified Stau155Δ2-his6 and [32P]labelled TAR or bicoid 3′-UTR RNA (left panel). Open squares, TAR RNA; black triangles, bicoid 3′-UTR RNA. Similar experiments were performed with [32P]labelled TAR mutants, allowing to calculate Kd (right panel). (B) Schematic representation of different mutants already used to map TAR-binding proteins (34). Position and nature of substituted nucleotides are indicated. (C) Northwestern assays using bacterially expressed and column-purified Stau155Δ2-his6 and [32P]labelled TAR and TAR mutants. The arrow indicates the position of Stau155Δ2-his6. The upper band likely represents dimers.
Stau155 RNA-binding activity is required for TAR-induced translational activation
Since Stau155 binds the TAR RNA structure at the 5′ end of the TAR-Rluc transcript, we then tested whether Stau155 binding to RNA is required for the observed up-regulation of TAR-Rluc translation. Two mutants, Stau155F-HA3 (26) or Stau155KK-HA3 (C. Martel and L. DesGroseillers, manuscript submitted) (Figure 5A), were first tested for their capacity to bind the TAR RNA structure in vitro. Northwestern assays using immunoprecipitated proteins isolated from HEK293T cells showed that, in contrast to Stau155-HA3, both mutants were impaired in their RNA-binding activity (Figure 5B). Then, HEK293T cells were co-transfected with plasmids coding for either Stau155F-HA3 or Stau155KK-HA3 and with either TAR-Rluc or Rluc vectors. Western blot analysis showed that the levels of expression of the mutated proteins were quite similar to that of Stau155-HA3 (Figure 5C). However, these two mutants failed to stimulate the activity of either reporter (Figure 5D). In the same experiment, Stau155-HA3 stimulated translation of TAR-luciferase RNA 2-fold. These results suggest that Stau155 binding to RNA is critical for enhanced translation of TAR-containing transcripts.
Figure 5.
The Stau155 RNA-binding activity is required for TAR-bearing RNA translational regulation. (A) Schematic representation of wild-type Stau155-HA3 and of two mutants with disrupted RNA-binding activity. Symbols are described in the legend of Figure 1. (B) HEK293T cells were transfected with plasmids coding for either Stau155-HA3, Stau155F-HA3 or Stau155KK-HA3. Tagged proteins were immunoprecipitated with an anti-HA antibody and resulting proteins were analysed by SDS–PAGE. Western blotting (upper panel) using anti-Stau1 antibody and Northwestern blotting (lower panel) using [32P]labelled TAR RNA. Arrows indicate the position of Stau1-HA3. *Non-specific IgG bands. (C) HEK293T cells were transfected with plasmids coding for either Stau155-HA3, Stau155F-HA3 or Stau155KK-HA3 and proteins were analysed by SDS–PAGE and western blotting using anti-Stau1 and anti-tubulin antibodies. Similar amounts of proteins are expressed by the three plasmids. (D) HEK293T cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of plasmids coding for either Stau155F-HA3 (left panel) or Stau155KK-HA3 (right panel). Resulting luciferase activity was quantified as described in Figure 2B. In the absence of Stau155-HA3, a 2-fold repression of translation of the TAR-Rluc RNA was observed as compared with translation of Rluc RNA. To facilitate comparison, fold induction of the luciferase activity in the absence of Stau155-HA3 was defined as 1. Differences are not statistically significant (n = 3). Black bars, TAR-Rluc RNA; hatched bars, Rluc RNA.
The effects of Stau155 are at the level of translation
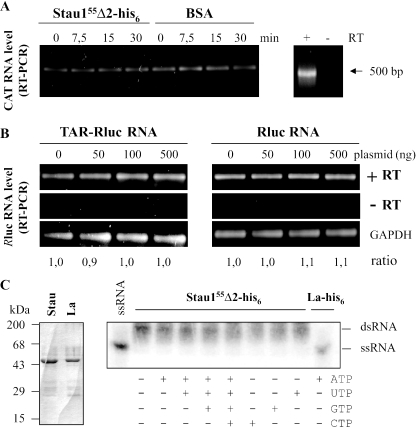
Three non-exclusive mechanisms may explain how Stau155 modulates translation of TAR-bearing transcripts: (i) increase in stability of TAR-containing RNA, (ii) unwinding activity of Stau1 on the TAR structure and/or (iii) direct binding to ribosomes or to translation factors and modulation of their activity. To distinguish between an effect on TAR-bearing mRNA itself and an effect on the translational machinery, we assessed the ability of Stau155 to act at the level of RNA metabolism. To address this possibility, TAR-CAT and CAT mRNAs were incubated in RRL in the presence or in the absence of Stau155Δ2-his6. An inhibitor of translation (G418) was added to prevent mRNA association/protection with translating ribosomes. Aliquots were taken at different time points and RT–PCR amplifications were carried out using primers in the CAT sequence. The presence of Stau155Δ2-his6, or that of BSA as control, did not affect TAR-CAT (Figure 6A) or CAT (data not shown) mRNA stability, indicating that the RNA is as stable as in control conditions.
Figure 6.
Stau155 mediated translational up-regulation does not involved RNA modification. (A) TAR-CAT RNA was incubated in RRL in the presence of 400 nM of bacterially expressed and purified Stau155Δ2-his6 or BSA for increasing periods of time. TAR-CAT RNA was then reverse transcribed and PCR amplified for 14 cycles to stay in the non-saturated part of the amplification curve. Resulting DNA was resolved on agarose gel. As control, the same experiment was performed in the absence of reverse transcriptase (right panel). (B) HEK293T cells were co-transfected with plasmids expressing either Rluc or TAR-Rluc transcripts and different concentrations of a plasmid coding for Stau155-HA3. Twenty-four hours post-transfection, RNA was isolated, reverse transcribed and PCR amplified. Resulting DNA was resolved on agarose gel. As control, the same experiment was performed in the absence of reverse transcriptase (−RT). RNA coding for GAPDH was RT–PCR and used to normalize the results. (C) Bacterially expressed and column-purified Stau155Δ2-his6 (Stau) and La-his6 (La) (left panel) were incubated with [32P]labelled double-stranded RNA in the presence of different combinations of ribonucleotides (right panel). RNA was resolved on agarose gel and revealed by autoradiography. While La-his6 displayed an helicase activity, Stau155Δ2-his6 was inactive in this assay.
Similarly, we tested whether the steady-state levels of TAR-Rluc and Rluc mRNAs in HEK293T cells varied with overexpression of Stau155-HA3. TAR-Rluc or Rluc expressing plasmids were co-transfected with increasing concentrations of Stau155-HA3 cDNAs. Twenty-four hours post-transfection, total RNA was extracted and semi-quantitative RT–PCR assays were performed using primers in the Rluc sequence. mRNA levels were normalized to endogenous gapdh mRNA. Expression of Stau155-HA3 did not affect the steady-state levels of TAR-Rluc and Rluc mRNAs (Figure 6B). Luciferase assays performed on the same cell extracts showed the expected increase in translation of TAR-Rluc RNA (data not shown).
Stau155 could potentially unwind the double-stranded TAR-RNA structure upon binding and facilitate translation. To test this possibility, recombinant Stau155Δ2-his6 or the RNA-binding protein La-his6 (43), used as a positive control (Figure 6C, left panel), were incubated in the presence of [32P]labelled dsRNA and different combinations of NTP. As shown in Figure 6C (right panel), a helicase activity was observed with the La autoantigen but could not be detected in the Stau155Δ2-his6 preparation. These results show that Stau155Δ2-his6 does not have helicase activity in vitro. These results suggest that Stau1 likely acts on the translational machinery to enhance structured RNA translation.
The Stau155 mutants that are unable to increase translation cofractionate with polysomes
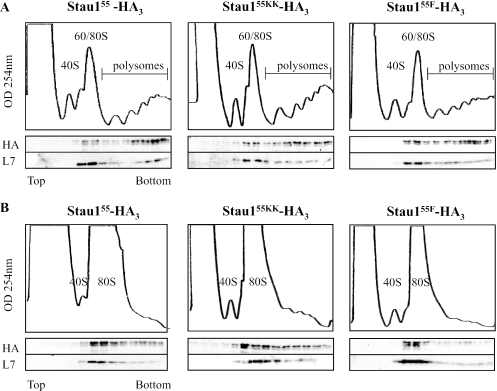
Since Stau155 associates with ribosomes (19,26), it might facilitate the positioning of the TAR-bearing transcript on the ribosomes and/or directly modulate ribosome activity. Therefore, one reason that Stau155 mutants failed to increase translation (Figure 5D) might be because the introduced mutations impaired their capacity to associate with ribosomes. To test this possibility, HEK293T cells were transfected with plasmids coding for Stau155-HA3 or Stau155-HA3 mutants and treated with cycloheximide. Cell extracts were centrifuged on a continuous 10–40% sucrose density gradient to separate free ribosomes from polysomes. As shown in Figure 7A, Stau155-HA3, Stau155F-HA3 and Stau155KK-HA3 were mainly found in fractions containing polysomes, showing that the mutations did not impair the capacity of the mutants to cosediment with ribosomes.
Figure 7.
Stau155 mutants unable to modulate TAR-Rluc translation are nevertheless associated with ribosomes. HEK293T cells were transfected with plasmids coding for Stau155-HA3, Stau155F-HA3 or Stau155KK-HA3. Cells were treated with either cyclohexemide (A) or sodium azide (B) for 20 and 30 min, respectively, before being lysed. Cell extracts were separated onto 10–40% sucrose gradients and proteins in each fraction were analysed by SDS–PAGE and western blotting using anti-HA and anti-ribosomal protein L7 antibodies. Positions of 40S, 60S ribosomal subunits, 80S ribosomes and polysomes are indicated as revealed by the OD254 scan of the gradients.
We next determined whether ribosomes that are associated with the Stau155 mutants have impaired translational capability, e.g. by forming non-translating polysomes. To explore this possibility, centrifugation experiments were repeated following treatment with sodium azide, a non-specific inhibitor of translation initiation (51). In this assay, mRNA-bound ribosomes are able to complete translation of the bound RNA but free ribosomes are unable to initiate translation. As a result, large polysomes disappear and the amounts of free ribosomes or ribosomal subunits increase. Following sodium azide treatment, large amounts of both Stau155-HA3 and Stau155-HA3 mutants were shifted from fractions containing heavy polysomes to fractions of monosomes and small polysomes (Figure 7B). Quantitation of the amounts of Stau155-HA3, Stau155F-HA3 and Stau155KK-HA3 in the presence or in the absence of sodium azide revealed a reduction of 54.0, 54.4 and 58.8%, respectively, in the association with heavy polysomes. These results demonstrate that the mutations did not impair the capacity of the mutated Stau155 proteins to bind actively translating polysomes. Although we do not exclude the possibility that the mutations prevent association with essential translation factors, these results suggest that Stau1 acts as a carrier to facilitate transport/positioning of target RNAs on translating ribosomes.
Stau155 overexpression causes a shift of TAR-containing transcripts to dense polysomes
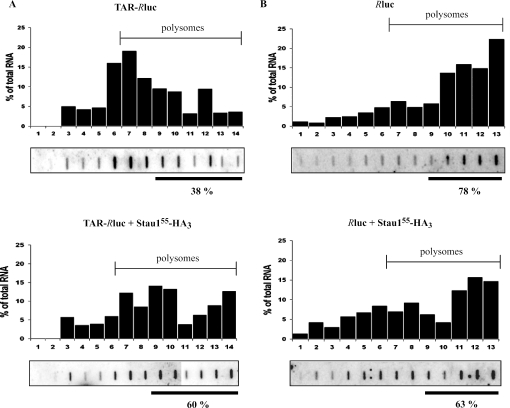
In order to determine how Stau1 was influencing the TAR-Rluc RNA profile in polysomes, cells were transfected with plasmids expressing TAR-Rluc or Rluc, in the absence or in the presence of plasmids coding for Stau155-HA3. In the absence of Stau155-HA3, the TAR-Rluc RNA was mostly found in fractions corresponding to 80S ribosomes and light polysomes that contain few ribosomes (Figure 8A). However, in the presence of Stau155-HA3, the TAR-Rluc RNA was shifted to heavy polysome fractions. In contrast, the Rluc RNA was found in heavy polysome fractions, in both the presence and absence of co-transfected Stau155-HA3 (Figure 8B). Interestingly, in the presence of Stau155-HA3, the percentage of TAR-Rluc RNA in heavy polysome fractions was roughly the same as that of Rluc RNA in the presence or in the absence of Stau155-HA3, consistent with the synthesis of similar amounts of Rluc protein (see above). These results suggest that Stau155 increases the probability to initiate TAR-Rluc RNA translation, thus increasing the number of attached ribosomes to form heavier polysomes and enhancing the global rate of translation of this transcript.
Figure 8.
In the presence of Stau155-HA3, TAR-Rluc transcripts are shifted to fractions containing heavy polysomes. HEK293T cells were transfected with plasmids expressing either TAR-Rluc (A) or Rluc (B) transcripts (top panels) or co-transfected with the same plasmids and plasmids coding for Stau155-HA3 (bottom panels). Twenty-four hours post-transfection, cell extracts were centrifuged onto 15–45% sucrose gradients and the position of ribosomal subunits, ribosomes and polysomes determined by OD254 (data not shown). Position of polysomes is indicated. RNA was isolated from each fraction and analysed by slot-blotting and hybridization using [32P]labelled luciferase cDNA. As control, the probe recognized a single band on a northern blot made with RNA isolated from Rluc-transfected HEK293T cells (data not shown). The percentage of RNA in each fraction in relation to the amount of total RNA is plotted above the hybridization blots. The percentage of RNA found in heavy polysomes is indicated below the blots. Similar results were obtained three times. The increase in the amounts of TAR-Rluc transcripts associated with heavy polysomes in the presence of Stau155-HA3 is statistically significant (P ≤ 0.05), whereas the small decrease in the amounts of Rluc transcripts associated with heavy polysomes in the presence of Stau155-HA3 is not statistically significant.
Stau1 increases translation of structure-repressed transcripts when tethered at the 5′ end
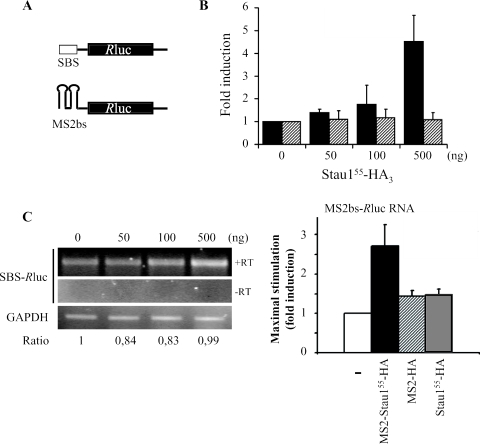
To establish a correlation between Stau1 binding to the 5′ end of transcripts and its ability to enhance translation, we substituted the TAR-RNA structure with other structured RNA sequences. The SBS that was recently identified by us (52) or two copies of the MS2 binding site (MS2bs) were fused to the 5′ end of the Rluc transcript (Figure 9A). First, HEK293T cells were co-transfected with a fixed amount of either Rluc or SBS-Rluc cDNA and increasing amounts of cDNA coding for Stau155-HA3. As shown in Figure 9B, Stau155-HA3 stimulated translation of SBS-Rluc RNA about 5-fold as compared with SBS-Rluc RNA translation in the absence of transfected Stau155-HA3. This effect was at the translational level because the SBS-Rluc transcript did not vary with Stau155-HA3 expression (Figure 9C). Similarly, cells were co-transfected with a fixed amount of the MS2bs-Rluc plasmid and increasing concentrations of MS2-Stau155-HA3 or MS2-HA3 as a negative control. MS2-Stau155-HA3 increased translation about 3-fold as compared with MS2-HA3 (Figure 9D) showing that the effect is specific to Staufen1. Importantly, co-transfection of Stau155-HA3 (instead of MS2-Stau155-HA3) did not increase translation of the MS2-Rluc transcript (Figure 9D) showing that Stau155 recruitment to the structured 5′ end of RNA is critical for increased translation of these RNAs.
Figure 9.
Binding of Stau155 to the 5′ end increases translation of structure-repressed transcripts. (A) Schematic representation of 5′-structure-repressed transcripts. RNAs coding for the Rluc reporter protein are shown with one copy of the SBS or two copies of the MS2-binding site (MS2bs) at the 5′ end. (B) HEK293T cells were co-transfected with plasmids expressing either Rluc or SBS-Rluc transcripts and different concentrations of a plasmid coding for Stau155-HA3. Resulting luciferase activity was quantified 24 h post-transfection. In the absence of Stau155-HA3, a 100-fold repression of translation of the SBS-Rluc RNA was observed as compared with translation of Rluc RNA. Results are expressed as luciferase activity versus concentration of the Stau155-HA3 coding plasmid. To facilitate comparison, the luciferase activity in the absence of Stau155-HA3 was defined as 1. P ≤ 0.01, n = 3. Black bars, SBS-Rluc RNA; hatched bars, Rluc RNA. (C) HEK293T cells were co-transfected with plasmids expressing the SBS-Rluc transcript and different concentrations of a plasmid coding for Stau155-HA3. Twenty-four hours post-transfection, RNA was isolated, reverse transcribed and PCR amplified. Resulting DNA was resolved on agarose gel. As control, the same experiment was performed in the absence of reverse transcriptase (−RT). RNA coding for GAPDH was RT–PCR and used to normalize the results. (D) HEK293T cells were co-transfected with plasmids expressing the MS2bs-Rluc transcript and different concentrations of plasmids coding for either MS2-Stau155-HA3, MS2-HA or Stau155-HA3. Resulting luciferase activity was quantified 24 h post-transfection. In the absence of MS2-Stau155-HA3, a 100-fold repression of translation of the MS2bs-Rluc RNA was observed as compared with translation of Rluc RNA. To facilitate comparison, the luciferase activity in the absence of expressor plasmids was defined as 1, n = 3.
DISCUSSION
In this paper, we show that Stau155 increases translation of mRNAs when bound to their 5′ end. Consequently, Stau155 is not a general regulator of translation but rather acts on specific mRNA targets, through direct binding to their 5′ end. Indeed, our data establish a significant correlation between the binding of Stau155 to the 5′ end of a transcript and its ability to enhance translation. Interestingly, a maximal increase of expression was observed when a natural SBS (52) was fused at the 5′ end of Rluc. The Stau155-mediated up-regulation of translation is of the same order of magnitude as those reported for TRBP on TAR-bearing RNAs (41) and for proteins of the exon-junction complex on MS2-containing RNAs (53). In both the RRL assays in vitro and following transfection of cultured cells, the ratio of recombinant to endogenous Stau155 proteins associated with ribosomes and necessary to observe an effect on translation of TAR-bearing mRNAs is quite similar. In both cases, the amount of recombinant Stau155 that is required for optimal stimulation of translation is about one-fifth the level of the endogenous Stau155 (Figures 1D and 2B).Therefore, it suggests that within a cell a slight variation in the expression of endogenous Stau155 may regulate the translation of Stau155-bound mRNAs. Physiologically relevant transcripts harbouring structured 5′ end are likely to be translationally repressed in the cells due to the translational block imposed by these structures (15,16). Therefore, their regulated interaction with Stau155 would allow a concerted expression of a specific set of Stau155-bound transcripts in response to cell's needs (see below).
Other RNA-binding proteins have been shown to bind the TAR RNA structure and affect translation of TAR-bearing transcripts. Since both TRBP (34,41) and La antoantigen (42) were shown to increase translation of TAR-bearing RNA, as does Stau155, an indirect mechanism of translational repression involving competition between these proteins for the TAR RNA structure can be excluded. Inhibition of their function through competition should rather repress translation of TAR-bearing transcripts. In contrast, PKR activation through RNA binding was shown to repress translation (15,40). Therefore, binding of Stau155 to the TAR RNA structure may mask the PKR binding site, preventing PKR activation and indirectly enhancing general translation. However, even if the Stau155-binding site on TAR partly overlaps that of PKR (33), we show that the Stau155-mediated increase in translation of TAR-bearing RNA is independent of PKR both in vitro and in cell cultures (Figure 3). Nevertheless, a mechanism of competition between Stau155 and PKR for binding RNAs or sequestration of PKR through direct binding to Stau155 may explain the slight increase in the translation of CAT transcripts in vitro. This small increase was not seen in cultured cells, however.
Stau155-mediated up-regulation of protein expression occurs at the translational level. Steady-state analyses of RNAs show that Stau155 does not protect TAR transcripts from degradation and does not increase transcription. Moreover, Stau155 does not unwind dsRNA through helicase activity at least in vitro. Therefore, although Stau155 RNA-binding activity is required, Stau155 has no observable effect on RNA metabolism. It was suggested that the secondary structure of TAR affects translation by preventing the accessibility of the cap structure (29). In this context, Stau155 may favour access of this highly structured RNA to the ribosomal and/or translational machinery. Binding of Stau155 to TAR-bearing transcripts may (i) facilitate transport and positioning of the transcripts on the ribosomes, (ii) destabilize the TAR RNA structure, allowing binding of eIF4E and/or (iii) facilitate interaction between co-factors, the TAR structure and the ribosomes. Therefore, our working hypothesis is that Stau155 first binds selected RNAs and facilitates their transport and positioning on the ribosomes through its capacity to associate with ribosomes. Then, Stau155- and/or Stau155-associated proteins may help to destabilize the 5′ end structure leading to better interaction with translation initiation factors, such as eIF4E and/or the translational machinery. This effect would allow more ribosomes to be bound to the transcript (Figure 8) and consequently should increase translation. Among putative co-factors, helicases are known to influence translation of mRNAs containing secondary structures at their 5′ ends, helping to disrupt the secondary structures and to translocate the RNA within the polysomes (42,54–57). Moreover, they are frequently associated with RNA-binding proteins (58,59). At least two RNA helicases, RHA and La autoantigen, were shown to bind the TAR RNA element and influence TAR-mediated transcription and translation, respectively (35,42). Interestingly, RHA was identified in two independent proteomic analyses of Stau1-containing complexes (27,28). While it would also be interesting to test if direct binding of Stau155 to ribosomes is required for its function on translation, all of the Stau1 mutations tested to date that prevent Stau155-ribosome association also impair Stau155 RNA-binding activity (26).
Alternatively, Stau155 through its observed association with the 60S ribosomal subunit (26) may directly modify ribosomal activity, favouring the joining of the 60S ribosomal subunit to the translation initiation complex and/or influencing translational elongation. Stau155 may also interact with chaperones that facilitate proper folding of nascent proteins thus increasing the translation elongation rate or it may interfere with the association of trans-acting protein repressors to 3′- or 5′-UTR of RNA. The former hypothesis is not likely since the same protein is translated from TAR-Rluc and Rluc transcripts. In Drosophila, Staufen protein was shown to be essential for the translational de-repression of oskar mRNA once properly localized in the oocyte (10,18). Translational repression of oskar mRNA depends on functional association of proteins that bind both its 3′- and 5′-UTR (10,60–62) and on protein involved in the stabilization of the nascent protein (63). Interestingly, the 5′-UTR of oskar mRNA contains an RNA sequence/element that is required for its translational de-repression when localized at the posterior pole (60).
What is the biological relevance of Staufen in translational regulation? Proteomic (25,27,28,64) and cell biology (21–24,65) experiments clearly establish that Stau1 is a component of the RNA transport machinery in several cell types. In neurons, Stau1 granules also contain RNAs and move in dendrites on microtubules (23,24), suggesting that Stau1 function might be linked to mRNA transport. This conclusion is supported by the observation that the down-regulation of Stau1 by siRNA reduces the amounts of RNA in dendrites of neurons (25). While the exact role of Staufen has yet to be defined, it is clear that Stau155 plays multiple role(s) in cells in addition to its role in RNA transport. Stau155 is associated with telomerase and/or telomeric RNA in the nucleolus (66,67), suggesting that it has nuclear function. In the cytoplasm, Stau155 regulates RNA stability (52) and it can also regulate translation of a subpopulation of transcripts (this paper). These functions may all be complementary to its putative role in RNA transport such that in both RNA transport granules and stress granules, translation is known to be repressed (68–70) and it must resume once the transcripts are localized or when physiological conditions are returned to normal. Stau155 is likely associated to RNA during transport and to translation following localization as demonstrated for Staufen in Drosophila (10). Since the 5′-SBS-bearing transcripts are translationally repressed, a 5-fold increase in their translation following Stau155 binding is functionally significant. Several Stau155-bound transcripts code for proteins involved in the cell metabolism and/or cell growth (52) making critical the regulation of their translation. The site of Stau155 binding on the RNA may determine the nature of the induced process. When bound downstream of a natural termination codon, Stau155 elicits RNA degradation (52) whereas its binding to the 5′-UTR of a transcript enhances translation but not RNA degradation. Similar observations were reported for the proteins of the exon-junction complex that can either induce non-sense mediated RNA decay or enhance translation (53). Stau155 post-translational modifications and/or its association with different co-factors may regulate these processes, in response to the cell's needs, for instance. These results indicate that Stau1 is a multifunctional protein. Analyses of Stau155-endogenous RNA ligands will contribute to elucidate these mechanisms.
Acknowledgments
The authors thank M. Fyfe for her help in the production of monoclonal antibody, L. Cournoyer for help in cell cultures and T.-S. Huang for bacterially expressed Stau1 preparation. The authors thank A. Gatignol (McGill University) for the PKR−/− cell line, E. Cohen (Université de Montréal) for the SpIII-10 CAT and pSp64TAR-CAT plasmids, K. T. Jeang (NIH/NIAID Bethesda, MD) for the p48, p49 and La-pBS SK plasmids, L. E. Maquat (Rochester University) for MS2-HA and MS2-Stau1-HA plasmids, P. Chartrand (Université de Montréal) for the pSL-MS2 plasmid and G. Ferbeyre (Université de Montréal) for the pGEM-U6 plasmid. This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) grant to L.D.G. and a Canadian Institute for Health Research (CIHR) grant to A.J.M. A.J.M. was supported by a CIHR New Investigator Award. K.B. and S.D.-B. were supported by studentships from NSERC and Fonds de la Recherche en Santé du Québec (FRSQ), respectively. Funding to pay the Open Access publication charges for this article was provided by Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kiebler M.A., DesGroseillers L. Molecular insights into mRNA transport and local translation in the mammalian nervous system. Neuron. 2000;25:19–28. doi: 10.1016/s0896-6273(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 2.Chartrand P., Singer R.H., Long R.M. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- 3.Johnstone O., Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 4.Kislauskis E.H., Zhu X., Singer R.H. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., Long R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 6.Li P., Yang X., Wasser M., Cai Y., Chia W. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 1997;90:437–447. doi: 10.1016/s0092-8674(00)80504-8. [DOI] [PubMed] [Google Scholar]
- 7.Steward O., Schuman E.M. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 8.Steward O., Worley P.F. A cellular mechanism for targeting newly synthesized mRNAs to synaptic sites on dendrites. Proc. Natl Acad. Sci. USA. 2001;98:7062–7068. doi: 10.1073/pnas.131146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartrand P., Meng X.H., Huttelmaier S., Donato D., Singer R.H. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 10.Kim-Ha J., Kerr K., Macdonald P.M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 11.Gavis E.R., Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369:315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 12.Rongo C., Gavis E.R., Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y.S., Richter J.D. Regulation of local mRNA translation. Curr. Opin. Cell Biol. 2004;16:308–313. doi: 10.1016/j.ceb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y.S., Carson J.H., Barbarese E., Richter J.D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edery I., Petryshyn R., Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 16.Goossen B., Hentze M.W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.St Johnston D., Beuchle D., Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 18.Micklem D.R., Adams J., Grunert S., St Johnston D. Distinct roles of two conserved Staufen domains in oskar mRNA localization and translation. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marion R.M., Fortes P., Beloso A., Dotti C., Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wickham L., Duchaine T., Luo M., Nabi I.R., DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallardo M., Deitinghoff A., Muller J., Goetze B., Macchi P., Peters C., Kiebler M.A. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl Acad. Sci. USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krichevsky A.M., Kosik K.S. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 23.Kiebler M.A., Hemraj I., Verkade P., Kohrmann M., Fortes P., Marion R.M., Ortin J., Dotti C.G. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C.G., Kiebler M.A. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 26.Luo M., Duchaine T.F., DesGroseillers L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem. J. 2002;365:817–824. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villace P., Marion R.M., Ortin J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brendel C., Rehbein M., Kreienkamp H.J., Buck F., Richter D., Kindler S. Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 2004;384:239–246. doi: 10.1042/BJ20040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkin N.T., Cohen E.A., Darveau A., Rosen C., Haseltine W., Sonenberg N. Mutational analysis of the 5′ non-coding region of human immunodeficiency virus type 1: effects of secondary structure on translation. EMBO J. 1988;7:2831–2837. doi: 10.1002/j.1460-2075.1988.tb03139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maitra R.K., McMillan N.A., Desai S., McSwiggen J., Hovanessian A.G., Sen G., Williams B.R., Silverman R.H. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 31.Sengupta D.N., Silverman R.H. Activation of interferon-regulated, dsRNA-dependent enzymes by human immunodeficiency virus-1 leader RNA. Nucleic Acids Res. 1989;17:969–978. doi: 10.1093/nar/17.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berkhout B., Silverman R.H., Jeang K.T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 33.Spanggord R.J., Vuyisich M., Beal P.A. Identification of binding sites for both dsRBMs of PKR on kinase-activating and kinase-inhibiting RNA ligands. Biochemistry. 2002;41:4511–4520. doi: 10.1021/bi0120594. [DOI] [PubMed] [Google Scholar]
- 34.Gatignol A., Buckler-White A., Berkhout B., Jeang K.T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 35.Fujii R., Okamoto M., Aratani S., Oishi T., Ohshima T., Taira K., Baba M., Fukamizu A., Nakajima T. A role of RNA Helicase A in cis-acting transactivation response element-mediated transcriptional regulation of human immunodeficiency virus type 1. J. Biol. Chem. 2001;276:5445–5451. doi: 10.1074/jbc.M006892200. [DOI] [PubMed] [Google Scholar]
- 36.Chang Y.N., Kenan D.J., Keene J.D., Gatignol A., Jeang K.T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J. Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Der S.D., Lau A.S. Involvement of the double-stranded-RNA-dependent kinase PKR in interferon expression and interferon-mediated antiviral activity. Proc. Natl Acad. Sci. USA. 1995;92:8841–8845. doi: 10.1073/pnas.92.19.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hershey J.W. Protein phosphorylation controls translation rates. J. Biol. Chem. 1989;264:20823–20826. [PubMed] [Google Scholar]
- 39.Hershey J.W. Translational control in mammalian cells. Annu. Rev. Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- 40.Lee S.B., Melkova Z., Yan W., Williams B.R., Hovanessian A.G., Esteban M. The interferon-induced double-stranded RNA-activated human p68 protein kinase potently inhibits protein synthesis in cultured cells. Virology. 1993;192:380–385. doi: 10.1006/viro.1993.1048. [DOI] [PubMed] [Google Scholar]
- 41.Dorin D., Bonnet M.C., Bannwarth S., Gatignol A., Meurs E.F., Vaquero C. The TAR RNA-binding protein, TRBP, stimulates the expression of TAR-containing RNAs in vitro and in vivo independently of its ability to inhibit the dsRNA-dependent kinase PKR. J. Biol. Chem. 2003;278:4440–4448. doi: 10.1074/jbc.M208954200. [DOI] [PubMed] [Google Scholar]
- 42.Svitkin Y.V., Pause A., Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J. Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huhn P., Pruijn G.J., van Venrooij W.J., Bachmann M. Characterization of the autoantigen La (SS-B) as a dsRNA unwinding enzyme. Nucleic Acids Res. 1997;25:410–416. doi: 10.1093/nar/25.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crine P., LeGrimellec C., Lemieux E., Labonte L., Fortin S., Blachier A., Aubry M. The production and characterization of a monoclonal antibody specific for the 94,000 dalton enkephalin-degrading peptidase from rabbit kidney brush border. Biochem. Biophys. Res. Commun. 1985;131:255–261. doi: 10.1016/0006-291x(85)91796-6. [DOI] [PubMed] [Google Scholar]
- 45.Duchaine T., Wang H.J., Luo M., Steinberg S.V., Nabi I.R., DesGroseillers L. A novel murine Staufen isoform modulates the RNA content of Staufen complexes. Mol. Cell. Biol. 2000;20:5592–5601. doi: 10.1128/mcb.20.15.5592-5601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paddison P.J., Caudy A.A., Sachidanandam R., Hannon G.J. Short hairpin activated gene silencing in mammalian cells. Methods Mol. Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- 47.Geballe A.P., Gray M.K. Variable inhibition of cell-free translation by HIV-1 transcript leader sequences. Nucleic Acids Res. 1992;20:4291–4297. doi: 10.1093/nar/20.16.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J.T., Schneider R.J. Adenovirus inhibition of cellular protein synthesis is prevented by the drug 2-aminopurine. Proc. Natl Acad. Sci. USA. 1990;87:7115–7119. doi: 10.1073/pnas.87.18.7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sengupta D.N., Berkhout B., Gatignol A., Zhou A.M., Silverman R.H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc. Natl Acad. Sci. USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y.L., Reis L.F., Pavlovic J., Aguzzi A., Schafer R., Kumar A., Williams B.R., Aguet M., Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feng Y., Absher D., Eberhart D.E., Brown V., Malter H.E., Warren S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y.K., Furic L., DesGroseillers L., Maquat L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 53.Nott A., Le Hir H., Moore M.J. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chuang R.Y., Weaver P.L., Liu Z., Chang T.H. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 55.Yoon H., Miller S.P., Pabich E.K., Donahue T.F. SSL1, a suppressor of a HIS4 5′-UTR stem–loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992;6:2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]
- 56.Gulyas K.D., Donahue T.F. SSL2, a suppressor of a stem–loop mutation in the HIS4 leader encodes the yeast homolog of human ERCC-3. Cell. 1992;69:1031–1042. doi: 10.1016/0092-8674(92)90621-i. [DOI] [PubMed] [Google Scholar]
- 57.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 58.Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yedavalli V.S., Neuveut C., Chi Y.H., Kleiman L., Jeang K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 60.Gunkel N., Yano T., Markussen F.H., Olsen L.C., Ephrussi A. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes Dev. 1998;12:1652–1664. doi: 10.1101/gad.12.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilhelm J.E., Hilton M., Amos Q., Henzel W.J. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J. Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yano T., De Quinto S.L., Matsui Y., Shevchenko A., Shevchenko A., Ephrussi A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- 63.Braat A.K., Yan N., Arn E., Harrison D., Macdonald P.M. Localization-dependent oskar protein accumulation; control after the initiation of translation. Dev. Cell. 2004;7:125–131. doi: 10.1016/j.devcel.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 64.Ohashi S., Koike K., Omori A., Ichinose S., Ohara S., Kobayashi S., Sato T.A., Anzai K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 2002;277:37804–37810. doi: 10.1074/jbc.M203608200. [DOI] [PubMed] [Google Scholar]
- 65.Thomas M.G., Tosar L.J., Loschi M., Pasquini J.M., Correale J., Kindler S., Boccaccio G.L. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell. 2005;16:405–420. doi: 10.1091/mbc.E04-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachand F., Triki I., Autexier C. Human telomerase RNA-protein interactions. Nucleic Acids Res. 2001;29:3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le S., Sternglanz R., Greider C.W. Identification of two RNA-binding proteins associated with human telomerase RNA. Mol. Biol. Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 69.Darnell R.B. RNA logic in time and space. Cell. 2002;110:545–550. doi: 10.1016/s0092-8674(02)00937-6. [DOI] [PubMed] [Google Scholar]
- 70.Wickens M., Goldstrohm A. Molecular biology. A place to die, a place to sleep. Science. 2003;300:753–755. doi: 10.1126/science.1084512. [DOI] [PubMed] [Google Scholar]