Abstract
Gene therapy for hemoglobinopathies requires efficient gene transfer into hematopoietic stem cells and high-level erythroid-specific gene expression. Toward this goal, we constructed a helper-dependent adenovirus vector carrying the β-globin locus control region (LCR) to drive green fluorescent protein (GFP) expression, whereby the LCR-GFP cassette is flanked by adeno-associated virus (AAV) inverted terminal repeats (Ad.LCR-β-GFP). This vector possesses the adenovirus type 35 fiber knob that allows efficient infection of hematopoietic cells. Transduction and vector integration studies were performed in MO7e cells, a growth factor-dependent CD34+ erythroleukemic cell line, and in cord blood-derived human CD34+ cells. Stable transduction of MO7e cells with Ad.LCR-β-GFP was more efficient and less subject to position effects and silencing than transduction with a vector that did not contain the β-globin LCR. Analysis of integration sites indicated that Ad.LCR-β-GFP integration in MO7e cells was not random but tethered to chromosome 11, specifically to the globin LCR. More than 10% of analyzed integration sites were within the chromosomal β-globin LCR. None of the Ad.LCR-β-GFP integrations occurred in exons. The integration pattern of a helper-dependent vector that contained X-chromosomal stuffer DNA was different from that of the β-globin LCR-containing vector. Infection of primary CD34+ cells with Ad.LCR-β-GFP did not affect the clonogenic capacity of CD34+ cells. Transduction of CD34+ cells with Ad.LCR-β-GFP resulted in vector integration and erythroid lineage-specific GFP expression.
Sickle cell disease is an autosomal recessive blood disorder characterized by the presence of abnormal adult hemoglobin in red blood cells. Early clinical observation and epidemiological studies indicated the existence of a fetal γ-hemoglobin, distinct from sickled adult hemoglobin, that could ameliorate the clinical course of patients with sickle cell disease (49). Current therapeutic approaches for sickle cell disease based on allogeneic bone marrow transplantation or pharmacological inducers of fetal hemoglobin synthesis are often ineffective or high risk and/or can be applied to only a small portion of patients (53). Our long-term goal is to develop a gene therapy for sickle cell anemia. In the case of sickle cell disease, high-level expression of the γ-globin gene can substitute for reduced or absent β-globin expression and ameliorate the clinical symptoms. For γ-globin gene therapy to be successful it is essential that the transferred gene be expressed in erythroid cells at relatively high levels. To achieve this, the β-globin locus control region (LCR) is thought to be needed (8). In the absence of the LCR, globin genes are subject to strong position effects when they are transferred into cultured erythroleukemic lines or CD34+ cells (11). A 22-kb β-globin LCR containing five DNaseI hypersensitive sites (HS) conferred erythroid-specific expression upon cis-linked genes in transgenic mice (12). Expression was independent of the position of integration and was as high as that of the endogenous mouse β-globin.
Viral gene transfer vectors for the treatment of sickle cell disease will need to efficiently deliver the LCR/globin cassette into human hematopoietic stem cells (HSCs) (as a lifelong source of erythrocytes) and also be able to integrate the transgene into the host genome. Current gene transfer vectors do not fulfill these requirements (for a review, see reference 52). Recombinant adeno-associated virus (AAV) or oncoretroviruses are inefficient at transducing quiescent human hematopoietic cells (27). The 22-kb β-globin LCR was also too large to be incorporated into retroviral vectors, and globin expression cassettes previously used in these vectors contained only “micro” or “nano” LCRs, had to be devoid of internal splice sites, and were sensitive to silencing and position effects (21, 40, 50).
Adenoviruses (Ads) have major advantages over other viral vectors, e.g., the production of high-titer virus preparations, the ability to efficiently infect nondividing cells, and a large cloning capacity. Helper-dependent (HD; also called “gutless”) Ad vectors, have all viral coding sequences deleted from the vector genome, which increases the capacity for the incorporation of heterologous DNA to ∼35 kb. (HD-Ad vector genomes are usually designed to be shorter than helper virus vector genomes. As a result, HD-Ad vector particles are less dense than helper vector particles and can be separated on equilibrium cesium gradients. In addition, small genome size should provide an advantage to the HD-Ad virus during successive cycles of replication [7].) Growth of HD vectors depends on coinfection of the producer cells with a helper virus, which provides all necessary Ad proteins in trans. Routinely, removal of helper virus from HD vector preparations is based on Cre-recombinase-mediated excision of the packaging signal during coinfection (17). Currently existing HD Ad vectors (based on Ad serotype 5 [Ad5]) are not suitable for stable gene transfer into HSCs due to their lack of tropism and inability to integrate. To address the lack of HSC tropism, a number of groups attempted to modify the capsids of Ad5 vectors (for a review, see reference 25). In this context, our group produced a chimeric Ad5-based vector containing Ad35 fibers that efficiently transduced human bone marrow cells with stem cell capacity (48). Integration of Ad vectors into the host genome can be stimulated by incorporating elements of integrating viruses, such as AAV (10, 23, 37-39) or oncoretroviruses (19, 30), into the Ad genome. Furthermore, transplantation of transposase- (59) or phage integrase-based (34) systems into Ad vectors is a promising means to mediate transgene integration. Our group focused on the development of Ad.AAV hybrid vectors devoid of all viral genes and containing AAV inverted terminal repeats (ITRs). The only requirements for random integration of recombinant AAV vectors are the ITRs and as-yet-unknown cellular factors (57). It is thought that AAV ITRs form specific secondary structures, which are prone to single- or double-strand breaks, and/or are substrates for cellular enzymes that mediate integration into the host genome. Sequencing of integration junctions has clearly established that recombinant AAV (rAAV) vectors efficiently integrate in vitro with an average frequency of 0.2 × 10−3 to 1.0 × 10−3 per infectious genome. In vivo, in mouse liver and muscle tissue, a large fraction of rAAV genomes can also persist as extrachromosomal vector DNA within the transduced cells in the form of monomeric circular and linear molecules (for a review, see reference 26).
A number of studies have demonstrated that the presence of two AAV ITRs in double-stranded DNA is sufficient to rescue a transgene cassette flanked by AAV ITRs and to mediate its integration (57, 58, 61). We showed that the incorporation of AAV ITRs into Ad vectors, which were devoid of all viral genes, mediated stable integration into the host genome (5, 23, 46).
In this study, we constructed capsid-modified, helper-dependent Ad.AAV vectors carrying the β-globin LCR to drive green fluorescent protein (GFP) expression. We analyzed GFP expression and vector integration in an erythroleukemia cell line and in primary human CD34+ cells.
MATERIALS AND METHODS
Ad vectors. (i) Ad.LCR vectors.
Corresponding shuttle plasmids were based on the cosmid vector pWE15 (Stratagene, La Jolla, CA). The ClaI site in pWE15 was deleted (pWE15c). pAd.AAV-3′HS1 contains the 5′ ITR (nucleotides 1 through 436) and 3′ ITR (nucleotides 35741 through 35938) of Ad5 and the 5′ ITR and 3′ ITR (nucleotides 3 through 145) of AAV type 2 (AAV2). To generate the Ad5 5′ ITR and packaging signal fragment, pHCA (47) was used as a template and the following primers were used: forward, 5′-GATCGATATCTAACTATAACGGTCCTAAGGTAGCGACATCATCAATAATATACCTTATTTTGG-3′, and reverse, 5′-GATCGATATCGTTTAAACGCGGCCGCATCGATGATCCTCGAGTTCGAAGATCGCTAGCCGCCAACTTTGACCCGGAACGCGGAAAACACC-3′. The PCR fragment was digested with EcoRV and inserted into the EcoRV site of pBluescript II KS(−) (pBS-Ad5ITRpsi). To PCR amplify the Ad5 3′ ITR, pHCA was used as a template, and the following primers were used: forward, 5′-GATCGTTTAAACGGTACCTAGGGATAACAGGGTAATGGCCGGCCCCTCAAATCGTCACTTCCGTTTTCCCACG-3′, and reverse, the same as 5′ end forward. The PCR fragment was digested with PmeI and EcoRV and inserted into the PmeI site of pBS-Ad5ITRpsi (pBS-Ad5). The 5′ ITR of AAV2 was cut from pΔE1.IRS.RSV.hAAT.ApoE (4) with XbaI and ClaI and inserted into pBS-Ad5 in NheI/BstBI (pBS-Ad-5′AAV). pWE15c was digested with EcoRI, blunted, and ligated with the EcoRV fragment from pBS-Ad-5′AAV (pWE15c-Ad5-5′AAV). A 6.7-kb BamHI fragment from pHCA was inserted into the BglII site of pWE15c-Ad5-AAV. The 3′HS1 fragment was obtained through PCR amplification by using a β-globin minilocus (12) (provided by F. Grosveld) as a template and the following primers: P1, (5′-GATCCTCGAGTAGGGATAACAGGGTAATCCAGGCTCCATTATTGATATAGTCATG-3′), and P2, (5′-GATCGAGCTCGGCCGGCCGCTAGCTTCGAAGGTGAAACCCTGTCTCTACTAAAACAC-3′). The 3′ ITR of AAV2 was inserted into the BstBI/NheI site of p3′HS1. Next, a 21.5-kb human β-globin LCR derived from the β-globin minilocous (SalI/ClaI fragment) was inserted in front of the 3′ HS1 into pAd.AAV-3′HS1 (XhoI/ClaI) (pAd.LCR). To generate pAd.LCR-β-GFP and pAd.LCR-CMV-GFP (Fig. 1A), the β-globin promoter-GFP-pA or CMV-GFP-pA cassette, respectively, was inserted into the ClaI site of pAd.LCR. The resulting plasmids were packaged into phages using Gigapack III Plus Packaging Extract (Stratagene, La Jolla, CA) and propagated. To generate the HD-Ad vectors Ad.LCR-β-GFP and Ad.LCR-CMV-GFP (Fig. 1A), C7.Cre cells (2), which express Cre recombinase, and Ad5/35 helper virus (47) were used. Ad5/35 helper virus is an E1-deleted Ad35 fiber-modified chimeric vector that contains a packaging signal flanked by loxP sites. For Ad.LCR-β-GFP and Ad.LCR-CMV-GFP rescue, viral genomes were released from the corresponding plasmids by I-CeuI digestion and transfected onto a 6-cm dish of C7-cre cells using a standard calcium phosphate method. At 12 to 16 h posttransfection, the cells were infected with 10 to 20 PFU per cell of Ad5/35 helper virus. Routinely, complete cytopathic effect was seen within 48 h. Infected cells were harvested and subjected to four cycles of freezing-thawing, and the crude viral lysates from five 6-cm dishes were used to infect five 10-cm plates of C7-cre cells, together with 10 to 20 PFU per cell of helper virus. When the cytopathic effect was complete, the crude viral lysate, prepared as above, was used to infect five 15-cm plates of C7-cre cells with helper virus supplementation. Virus DNA at this step was extracted from 400 μl of crude lysate and digested with HindIII to check the yield and intactness of recombinant viral genomes. For further amplification, virus was passaged to 10 15-cm plates and then to 40 15-cm plates of C7-cre cells. Lysate from the final step was subjected to cesium chloride separation and purification. Briefly, harvested cells were pelleted, resuspended in 18 ml phosphate-buffered saline (PBS) with 0.05% NP-40, freeze-thawed four times, and clarified by centrifugation at 2,000 rpm for 10 minutes. The supernatant was then supplemented with MgCl2 to 10 mM and treated with 100 μg/ml of DNase and 50 μg/ml of RNase A at 37°C for 30 min to digest unpackaged viral genomes and cellular nucleic acids. The lysate was then applied to a CsCl step gradient and centrifuged at 35,000 rpm at 14°C for 4 h. The band containing HD-Ad particles was subsequently mixed with 1.34 g/ml CsCl and centrifuged at 59,000 rpm for 12 h, followed by an additional centrifugation at 32,000 rpm for an additional 12 h at 4°C in a Beckman NVT90 rotor. The HD-Ad-containing band (Fig. 1B) was pulled from the gradient and dialyzed against 10 mM Tris-HCl, pH 7.5, containing 10 mM MgCl2, 150 mM NaCl, and 10% glycerol. The HD genome titer and helper contamination were analyzed by quantitative PCR or Southern blotting.
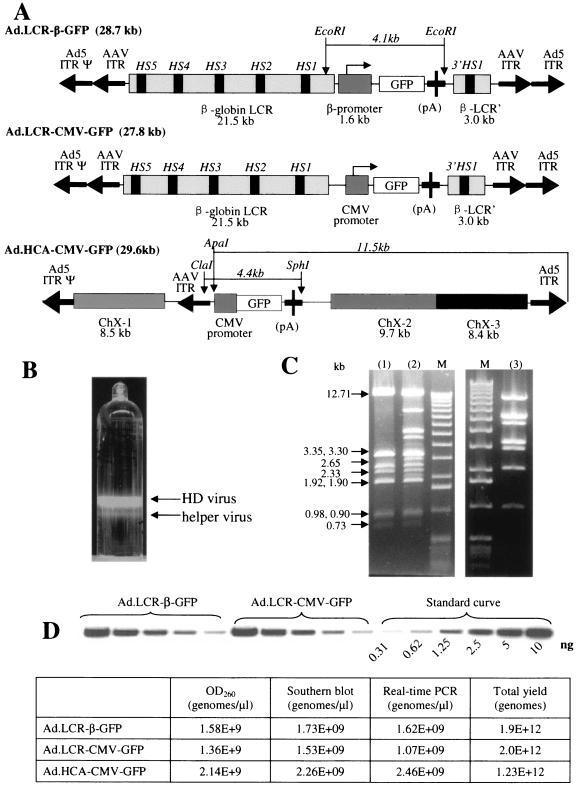
FIG. 1.
GFP expression in cell lines upon infection with helper-dependent Ad vectors. (A) Structure of helper-dependent Ad vectors. Ad.LCR-β-GFP contains the β-globin LCR, β-globin promoter, GFP gene, bPA polyadenylation signal (pA), and β-globin 3′ HS-1 region. This cassette is flanked by AAV ITRs. The GFP gene can be released from Ad.LCR-β-GFP as a 4.1-kb fragment by digestion with EcoRI. Ad.LCR-CMV-GFP has the same structure as Ad.LCR-β-GFP but contains the CMV promoter instead of the β-globin promoter. Ad.HCA-CMV-GFP contains the CMV-GFP expression cassette flanked by three fragments of stuffer DNA derived from human chromosome X (ChX-1 to ChX-3). Ad.HCA-CMV-GFP contains only one AAV ITR; a vector containing a second AAV ITR placed downstream of the GFP cassette was not stable and demonstrated rearrangements in or loss of the second AAV ITR. (B) Banded virus after second CsCl gradient. (C) Restriction analysis of viral DNA from banded virus particles. The figure shows a HindIII digest of Ad.LCR-β-GFP DNA isolated from the upper band (lane 1), DNA from the lower band a (mixture of helper and HD vector) (lane 2), and Ad5/35 helper virus DNA (lane 3). (D) Top, quantitative Southern blot analysis of vector titers: bottom, comparison of HD virus titer determined by measurement of optical density at 260 nm (OD260), quantitative Southern blotting, and real-time PCR.
(ii) Ad.HCA-CMV-GFP.
The Ad.HCA-CMV-GFP vector was generated earlier and described elsewhere (47). It contains an Ad5/35 capsid and a cytomegalovirus (CMV) promoter-GFP gene expression cassette flanked by stuffer DNA derived from human X-chromosomal (ChX) DNA (Fig. 1A). The stuffer DNA was selected to prevent homologous recombination with cellular sequences and is free of repeats, retrovirus elements, and known genes. The human sequence was disrupted into three parts and scrambled in orientation and order. The stuffer DNA sequences encompass the following X-chromosome regions: fragment 1, Chx 149334999→149326492; fragment 2, ChX 149308923→149299195; fragment 3, ChX 149294396→149285990).
Real-time PCR was used to quantify HD-vector genomes and helper contamination in each vector preparation. The plasmid pAd5GFP/F35 (48), with known concentration, was diluted serially and used for a standard curve. For quantification of HD-vector genomes, the following primers were used: GFP forward, 5′-TCGTGACCACCCTGACCTAC-3′; GFP reverse, 5′-GGTCTTGTAGTTGCCGTCGT-3′. For Ad5/35 helper contamination, the following primers were used: Ad helper forward, 5′-TGCAAGATACCCCTATCCTG-3′; Ad helper reverse, 5′-CCTGTTGCAGAGCGTTTGC-3′. Purified viruses were diluted 1:500 and 1:1,000 before PCR. The QuantiTect SYBR Green PCR kit (QIAGEN, Valencia, CA) was used. The following program was used on Lightcycler (Roche): 95°C, 20 minutes; 95°C, 15 s, 58°C, 20 s, 72°C, 17 s, 50 cycles; 95°C, 5 s, 60°C, 15 s; 40°C, 1 s. To measure HD and helper virus genomes in crude viral lysate, the cell pellet was freeze-thawed four times, centrifuged to remove cellular debris, and treated with 1 mg/ml DNase for 30 min at 37°C, followed by inactivation of DNase at 95°C for 20 min.
For quantitative Southern blotting, pAd5GFP/F35 was linearized with PacI and diluted serially, and a standard curve from 0 to 10 ng of plasmid DNA was used. A 10-μl aliquot of purified HD virus stock mixed with 190 μl PBS plus 2 × 105 293 cells (as a source of carrier DNA) was subjected to pronase digestion, subsequent phenol-chloroform purification, and ethanol precipitation. Serially diluted, extracted viral DNAs were run on an agarose gel together with the standard curve and analyzed by Southern blotting and phosphorimaging.
Cells.
Culture media (Dulbecco's modified Eagle's medium, Iscove’s modified Dulbecco’s medium and RPMI 1640) were purchased from GIBCO-BRL (Gaithersburg, MD). Fetal calf serum (FCS) was from HyClone (Logan, Utah). 293 cells (human embryonic kidney cells; Microbix, Toronto, Canada) and C7-cre cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% FCS, 2 mM glutamine, and 1× penicillin-streptomycin solution (Invitrogen, Carlsbad, Calif.). For C7-cre cells, 0.5 mg/ml G418 and 100 μg/ml hygromycin were added to maintain Cre, polymerase, and pTP expression (2). MO7e cells were maintained in RPMI 1640 medium containing 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 0.1 ng/ml of granulocyte-macrophage colony-stimulating factor (GM-CSF) (Immunex, Seattle, WA). Primary human CD34+ enriched cells were purified from cord blood by MiniMACS LS+ separation columns (Miltenyi Biotec, Auburn, Calif.) using the CD34+ progenitor cell isolation kit (StemCell Technologies, Vancouver, Canada) according to the manufacturer's protocol. Aliquots of cells were stored in liquid nitrogen. Sixteen hours before the experiment, CD34+ cells were recovered from frozen stocks and incubated overnight in Iscove’s modified Dulbecco’s medium supplemented with 20% FCS, 10−4 M β-mercaptoethanol, 100 μg/ml DNase I, 2 mM glutamine, 10 U/ml interleukin 3 (IL-3), 50 ng/ml stem cell factor, 50 ng/ml Flt3L, and 10 ng/ml thrombopoietin. The purity of CD34+ cell preparations was verified by flow cytometry and was consistently greater than 90%. For CFU assays, 3,000 infected CD34+ cells were cultivated with 1 ml MethoCult GF H4434 medium (StemCell Technologies) in a humidified atmosphere of 5% CO2 at 37°C in the presence of the following cytokines: IL-3, 50 U/ml; stem cell factor (SCF), 50 ng/ml; erythropoietin (Epo), 2 U/ml, granulocyte colony-stimulating factor, 6.36 ng/ml; GM-CSF, 50 ng/ml). After 14 days, the colonies were counted under an inverted microscopy. Etoposide was purchased from Sigma Chemical Corp. (St. Louis, MO). Z-VAD-FMK was purchased from Alexis Biochemicals Corp. (Lausen, Switzerland).
Cloning of transduced MO7e cells.
A total of 2 × 105 MO7e cells were infected with HD-Ad vector at indicated multiplicities of infection (MOI). Twenty-four hours postinfection, the cells were diluted and seeded onto 96-well plates at 0.5 cell/well. Two to 3 weeks later, when the number of cells in the colonies reached 2 × 103 to 3 × 103, the wells containing GFP-positive cells were counted and the percentage of GFP-positive cells in each given clone was estimated. Clones with more than 30% GFP-positive cells were sorted by fluorescence-activated cell sorter. The sorted cells were further expanded to ∼107 cells for Southern analysis.
Southern blot analysis of vector DNA.
Cellular DNA was isolated from transduced MO7e cell clones by a standard protocol and analyzed by Southern blotting as described elsewhere (47). The blots were hybridized with a CTP-labeled GFP probe. To equalize loading differences, filters were rehybridized with a probe specific for the genomic human α1-antitrypsin gene. To generate this 675-bp probe, pBS.hAAT (51) was digested with HindIII.
Isolation and analysis of vector-cellular-DNA junctions. (i) Inverse PCR.
Junctions in MO7e cell clones were analyzed by inverse PCR as described elsewhere (46). Briefly, 5 μg cellular DNA isolated from GFP-expressing clones was digested with HindIII and religated under conditions that promote intramolecular reaction. The ligation mixture was subjected to nested PCR (30 cycles each) using the Expand long Template PCR system (Roche, Mannheim, Germany) based on the manufacturer's protocol. For 5′ junctions, the following primer pairs were used: AAV-1, 5′-GATACCGTCGACCTCGATCTCAGGAAC-3′, and 5′HS5-a, 5′-CTCACCAAGCCAATGTTCCTCTCTATGTTG-3′; AAV-0, 5′-CGATCTCAGGAACCCCTAGTGATGGAGTTG-3′, and 5′HS5-b, 5′-CTATGTTGGCTCAAATGTCCTTGAACTTTCC-3′. For the 3′ junctions, the following primer pairs were used: AAV-1, 5′-GATACCGTCGACCTCGATCTCAGGAAC-3′, and 3′HS1-a, 5′-CATGCAGGTTTCTTCAGACTGACTACAAAGG-3′; AAV-0, 5′-CGATCTCAGGAACCCCTAGTGATGGAGTTG-3′, and 3′HS1-b, 5′-TTCAGACTGACTACAAAGGCTTCAGACTAGC-3′. PCR fragments obtained from different MO7e clones were cloned into pCR2.1-TA (Invitrogen, Carlsbad, CA) or pGEM-T Easy (Promega, Madison, WI), and junctions were sequenced using vector-specific primers.
The following primers were used to detect vector concatemers: P3, 5′-TGGTGAGGGATATCTGTCCTGATGTTGTCC-3′ (5′ end); P4, 5′-TGAAGCCTTTGTAGTCAGTCTGAAGAAACCTG-3′ (3′ end).
(ii) LAM-PCR.
Ligation-mediated PCR (LAM-PCR) analysis was conducted by the Clonal Analysis Core of the Fred Hutchinson Cancer Research Center. CD34+ derived GFP-positive single colonies (with 200 to 1,000 cells) were isolated from methylcellulose medium, flushed thoroughly with PBS, and digested with proteinase K, and genomic DNA was isolated. Reactions were conducted with 100 ng of genomic DNA. All amplifications were done with Advantage II Polymerase Mix and buffer system (BD Biosciences Clonetech, Palo Alto, CA), using the following cycling conditions: melting at 95°C for 20 s, annealing at 60°C for 45 s, and elongation at 68°C for 90 s. First, single-stranded copies of the virus-host junction region were made by 100 cycles of linear amplification, using a biotinylated ITR primer (biotin-5′-CCGTCGCTTACATGTGTTCC). The product was bound to streptavidin-coated magnetic M280 Dynabeads (Dynal Biotech, Oslo, Norway). Subsequent reactions and washes (three times with 200 μl 10 mM Tris, 10 mM NaCl, 0.01% Triton X-100 after every reaction) took place on this matrix. Matrix-bound DNA was resuspended in 20 μl random-priming reaction mixture containing 500 nM deoxynucleoside triphosphates, 100 ng/μl random hexamers, and 2 to 5 units Klenow enzyme (New England Biolabs, Beverly, MA) and manufacturer-supplied buffer at 1× and incubated at 37°C for 60 min. The double-stranded product was digested with Tsp509I (New England Biolabs, Beverly, MA). DNA that remained bound to the matrix was then ligated to a double-stranded anchor primer (AP), using Fast Link Ligase (Epicenter Technologies, Madison, Wis.). The component primers of the AP were GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG and AATTCCTAACTGCTGTGCCACTGAATTCAGATC, yielding a Tsp509I-compatible end. The resultant junction fragment, containing host DNA flanked by ITR and AP sequences, was eluted from the matrix in 5 μl 0.1 N NaOH and amplified by two rounds of nested PCR. Two microliters of eluate was amplified in a 50-μl PCR with ITR primer 1, GCCACACTTGCAACATCACAC, and AP primer 1, GACCCGGGAGATCTGAATTC. This reaction mixture was diluted 100-fold in water, and 2 μl template was amplified by 30 cycles of nested PCR using ITR primer 2, TTCCGCCACACTACTACGTC, and AP primer 2, GATCTGAATTCAGTGGCACAG. The amplified products were separated by electrophoresis on Novex 4 to 20% polyacrylamide Tris-borate-EDTA gels (Invitrogen, Carlsbad, CA), stained with ethidium bromide, and visualized by ultraviolet light fluorescence. For junction site identification, LAM-PCR products were separated on 2% agarose gels, and individual bands were excised and cloned into the TOPO-TA vector, pCR4 (Invitrogen, Carlsbad, CA). The cloned inserts were sequenced from flanking M13-F and M13-R primer sites in the vector, using Big Dye fluorescent dideoxynucleotide chemistry (Amersham) and a Prism 310 Genetic Analyzer (ABI). Sequences were screened for the presence of ITR and anchor-primer motifs, and the intervening sequence was compared to the human genome using the University of California—Santa Cruz BLAT service (http://genome.ucsc.edu/cgi-bin/hgBlat).
RESULTS
Vector production.
For globin gene therapy to be successful, it is essential that the transferred gene be expressed erythroid specifically at relatively high levels. To achieve this, we utilized elements of the human globin locus that have been shown to confer high-level, position-independent, erythroid-specific expression upon cis-linked genes in transgenic mice (12). These elements included the β-globin LCR encompassing the 5′ hypersensitivity regions HS5 to HS1 (21.5 kb), the β-globin promoter (1.6 kb), and the 3′ HS1 region (3.0 kb). The expression cassettes and HD-Ad vector genomes were assembled in a cosmid vector. Ad.LCR-β-GFP contains the globin LCR, the β-globin promoter, the 1.0-kb GFP gene, and the 3′ HS1 region (Fig. 1A). This cassette is flanked by AAV ITRs. The only Ad sequences in this vector were the Ad ITRs and packaging signal. To assess the properties of the vector, a series of control vectors were constructed. These included Ad.LCR-CMV-GFP, containing the CMV promoter instead of the β-globin promoter (Fig. 1A), and Ad.HCA-CMV-GFP, which contains stuffer DNA derived from human X-chromosomal DNA (see Materials and Methods and reference 47). The contiguous human sequence in Ad.HCA-CMV-GFP is disrupted into three parts (8.4, 8.5, and 9.7 kb in length) and scrambled in orientation and order (Fig. 1A).
All HD vectors were propagated using Ad5/35 helper virus and therefore possessed Ad5/35 capsids that allowed efficient transduction of hematopoietic cells (48). Analysis of HD vector genomes during propagation by restriction analysis demonstrated the absence of rearrangements in the vector genome (Fig. 1C). Genome titers and helper virus contamination of all vectors were analyzed by quantitative PCR. Yields were 1.9 × 1012, 2.0 × 1012, and 1.2 × 1012 genomes for Ad.LCR-β-GFP, Ad.LCR-CMV-GFP, and Ad.HCA-CMV-GFP, respectively. Genome titers obtained by quantitative PCR were confirmed by quantitative Southern blot analysis for viral DNA (Fig. 1D). Helper vector contamination in final vector preparations was less than 1% (based on quantitative PCR for vector genomes) or 0.05% (based on titering for helper vector PFU on 293 cells).
Transient GFP expression in MO7e cells.
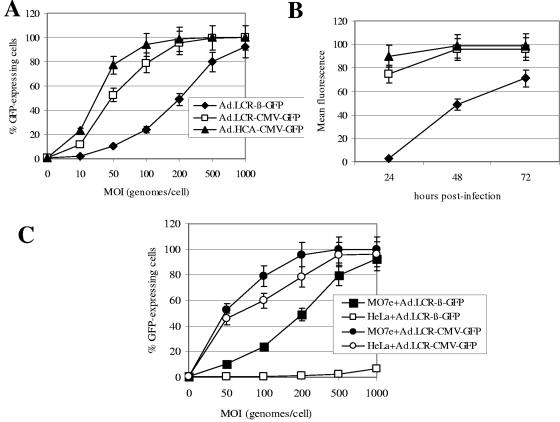
MO7e is a human, CD34-positive, c-kit-positive, growth factor-dependent erythroleukemic cell line (1). MO7e cells were infected with HD-Ad vectors at increasing MOIs, and GFP expression was analyzed 48 h later by flow cytometry (Fig. 2A). All vectors allowed efficient GFP expression in MO7e cells. GFP expression levels (mean fluorescence) of the Ad.LCR-β-GFP vector rose more slowly and were lower than those of CMV promoter-containing vectors (Ad.LCR-CMV-GFP and Ad.HCA-CMV-GFP) (Fig. 2B). There was significantly higher GFP expression from Ad.HCA-CMV-GFP than from Ad.LCR-CMV-GFP (P < 0.05), indicating that the globin LCR affects CMV promoter activity. To assess tissue specificity, MO7e cells and (nonerythroid) HeLa cells were infected with Ad.LCR-β-GFP and Ad.LCR-CMV-GFP, and GFP expression was analyzed 48 h later (Fig. 2C). This study demonstrated that β-globin promoter-driven GFP expression was selective for MO7e cells while the CMV promoter-containing vectors conferred high-level GFP expression on both cell lines.
FIG. 2.
Transient GFP expression upon HD-Ad vector transduction. (A) MO7e cells were infected with the indicated vectors at increasing MOIs. Forty-eight hours postinfection, the percentage of GFP-expressing cells was analyzed by flow cytometry. (B) MO7e cells were infected at an MOI of 200 genomes/cell, and GFP expression levels (mean fluorescence) were analyzed 24, 48, and 72 h postinfection. (C) MO7e cells and HeLa cells (as an example of nonerythroid cells) were infected with Ad.LCR-β-GFP and Ad.LCR-CMV-GFP at increasing MOIs, and 48 h postinfection, GFP expression was analyzed (n = 3). The error bars indicate standard deviations.
Analysis of GFP expression at the clonal level.
To discriminate transiently from stably transduced cells, we infected MO7e cells and obtained single clones by limiting dilution. Individual clones were expanded (for 2 to 3 weeks to >104 cells) and analyzed for GFP expression. Infection of MO7e cells with Ad.LCR-β-GFP and Ad.LCR-CMV-GFP at an MOI of 300 genomes/cell resulted in stable GFP expression in 18.6% and 16.6% of colonies, respectively, whereas transduction with Ad.HCA-CMV-GFP achieved stable GFP expression in 5.2% of the analyzed clones (Table 1). Interestingly, while 47% of all GFP-positive colonies transduced with Ad.LCR-β-GFP had more than 50% GFP-expressing cells per clone, all clones transduced with Ad.HCA-CMV-GFP had less than 30% GFP-expressing cells. Notably, GFP expression in more than 50% of cells in a given clone indicates vector integration during or prior to the first cell division. To study the reason for the observed differences in stable transduction between Ad.LCR-β-GFP and Ad.HCA-CMV-GFP, GFP-expressing cells from 20 Ad.LCR-β-GFP- and Ad.HCA-CMV-β-GFP-transduced clones were sorted and expanded. The number of vector copies present in each clone was analyzed by quantitative Southern blotting of genomic DNA digested with endonucleases that release the GFP gene (Fig. 1A) using a GFP-specific probe. The GFP cassette was present in all analyzed Ad.LCR-β-GFP- (Fig. 3A) and Ad.HCA-CMV-GFP-transduced clones (data not shown). The vector copy number ranged from 1 to 24 copies per cell for Ad.LCR-β-GFP-transduced cells and from 1 to 5 copies per cell for Ad.HCA-CMV-GFP-transduced clones. To assess whether the high copy number seen in most of the Ad.LCR-β-GFP-transduced clones was due to vector concatemerization, we performed PCR on genomic DNA using primers specific to a sequence located downstream of the AAV ITRs. Signals indicative of vector concatemerization (head to head, head to tail, and tail to tail) were observed in most Ad.LCR-β-GFP-transduced clones.
TABLE 1.
Analysis of GFP expression in Ad-transduced MO7e cell clonesa
| Ad | Parameter | Value | |||||
|---|---|---|---|---|---|---|---|
| Ad.LCR-β-GFP (n = 731 wells) | % GFP-positive cell per clone | >50 | 30-50 | 5-30 | <5 | 0 | |
| No. of wells | 65 | 17 | 30 | 24 | 595 | ||
| % Wells with GFP-positive clones | 8.89 | 2.32 | 4.1 | 3.28 | 81.4 | ||
| % Total GFP-positive colonies | 18.6 | ||||||
| Ad.LCR-CMV-GFP (n = 922 wells) | % GFP-positive cell per clone | >50 | 30-50 | 5-30 | <5 | 0 | |
| No. of wells | 26 | 16 | 63 | 48 | 769 | ||
| % Wells with GFP-positive clones | 2.82 | 1.73 | 6.83 | 5.2 | 83.4 | ||
| % Total GFP-positive colonies | 16.6 | ||||||
| Ad.HCA-CMV-GFP (n = 989 wells) | % GFP-positive cell per clone | >50 | 30-50 | 5-30 | <5 | 0 | |
| No. of wells | 0 | 0 | 13 | 38 | 938 | ||
| % Wells with GFP-positive clones | 0 | 0 | 1.31 | 3.84 | 94.8 | ||
| % Total GFP-positive colonies | 5.2 | ||||||
MO7e cells were infected with Ad.LCR-β-GFP, Ad.LCR-CMV-GFP, and Ad.HCA-CMV-GFP at an MOI of 300 genomes per cell. Twenty-four hours after infection, cells were plated onto 96-well plates with one cell per well (without sorting). Two weeks later, the plated cells were analyzed for GFP expression. The total number of colonies (n) and the number of colonies containing GFP-expressing cells were determined. Furthermore, the percentage of GFP-expressing cells per colony was estimated. The numbers of clones that contain more than 50%, 30 to 50%, 5 to 30%, or less than 5% GFP-expressing cells are shown.
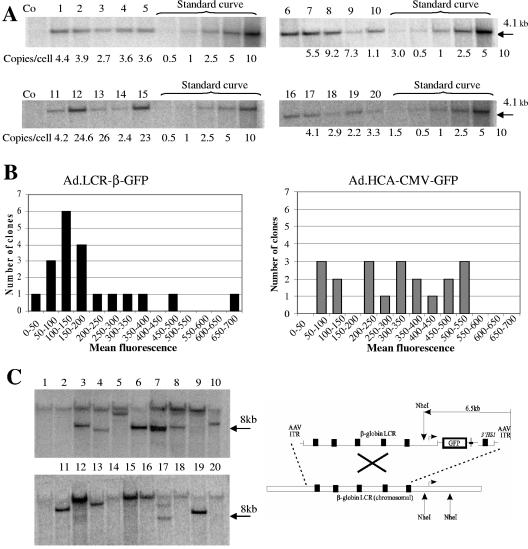
FIG. 3.
Clonal analysis of GFP expression and vector DNA in MO7e cells. (A) Analysis of integration copy numbers in GFP-positive clones transduced with Ad.LCR-β-GFP. GFP-expressing cells from 20 clones were sorted and expanded, and 10 μg of genomic DNA from each clone was analyzed by Southern blotting using a GFP-specific probe. EcoRI cuts twice within the vector sequence and releases a characteristic 4.1-kb fragment containing the GFP gene. The right side of the gel shows a standard curve for vector genomes using defined concentrations of plasmid DNA (pAd5GFP/F35) corresponding to 0.5, 1, 2.5, 5, and 10 vector copies per cell. Loading differences were equalized by rehybridizing the filters with a probe specific for the human α1-antitrypsin gene. The blots were analyzed using phosphorimaging, and the calculated number of vector copies per cell is shown for each clone. (B) Mean GFP fluorescence was measured by flow cytometry. The values were divided by the vector copies present in the given clone. The number of Ad.LCR-β-GFP and Ad.HCA-CMV-GFP clones that expressed GFP levels in the indicated range was plotted. (C) Analysis of integrated vectors in GFP-positive clones transduced with Ad.LCR-β-GFP. Ten micrograms of genomic DNA was digested with NheI, which cuts only once within the vector sequence. A GFP probe was used for hybridization. On the right is shown the localization of NheI sites in the vector and chromosomal globin LCR region. Theoretically, vector integration can result in a band that is larger than 6.5 kb (in the range of 7 to 9 kb).
In an attempt to evaluate whether the globin LCR offered protection from position effects and/or silencing of GFP expression upon vector integration, we measured the mean GFP fluorescence per vector copy in individual clones (Fig. 3B). GFP expression levels varied much less for Ad.LCR-β-GFP-transduced clones than for Ad.HCA-CMV-GFP-transduced clones, suggesting that the globin LCR offered protection from chromosomal position effects. Furthermore, during passaging over a period of 6 weeks, GFP expression levels remained constant in all Ad.LCR-β-GFP-transduced clones, whereas 2 out of 20 Ad.HCA-CMV-GFP-transduced clones lost GFP expression (GFP vector DNA was still present in these clones).
Taken together, GFP expression analysis at the clonal level indicated that stable transduction of MO7e cells with Ad.LCR-β-GFP is more efficient and less subject to position effects and silencing than transduction with the vector that does not contain the globin LCR.
Analysis of integrated vector DNA in MO7e clones by Southern blotting.
Southern blot analysis using an endonuclease that cuts only once in the vector DNA was used to assess the distribution of vector integration (Fig. 3C). NheI digestion of genomic DNA from Ad.LCR-β-GFP-transduced clones resulted in high-molecular-weight bands in the majority of clones, indicating vector integration. Interestingly, in 9 out of 20 clones, a prominent band with a size of 7 to 9 kb was detectable. Theoretically, this band could be the result of recombination between the 22-kb globin LCR within the vector and corresponding sequences within the cellular genome (Fig. 3C, right). Digestion of DNAs from Ad.HCA-CMV-GFP-transduced clones with ApaI demonstrated bands of different sizes, suggesting random integration (data not shown).
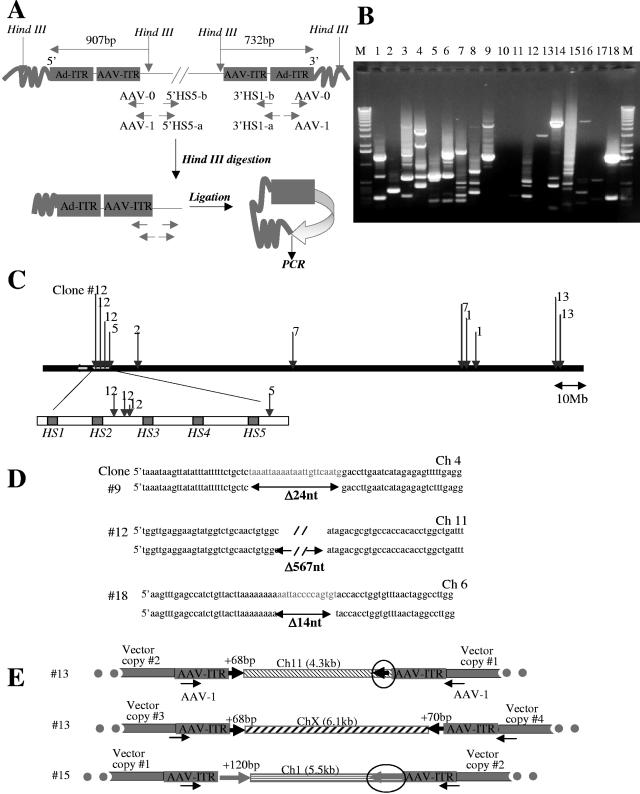
Because most of the clones had multiple integrants, we proceeded to analyze individual vector integration sites. Both 5′ and 3′ junctions between the vector and chromosomal DNA were analyzed using inverse PCR (Fig. 4A). Detailed data are shown for Ad.LCR-β-GFP-transduced clones (Fig. 4 and Table 2). The results for Ad.HCA-CMV-GFP-transduced clones are summarized in Table 3. The pattern of obtained PCR fragments in Ad.LCR-β-GFP-transduced clones corroborates the Southern blot data and demonstrates multiple integration sites (Fig. 4B). We analyzed a total of 33 independent integration sites (Table 2). Ten junctions contained AAV ITR sequences; 23 junctions were formed by the Ad ITR lacking various numbers of terminal nucleotides. Ten out of 33 integration junctions were found in genes, all of them in introns. None of the integrations occurred in oncogenes. Importantly, 11 out of 33 integrants were localized to chromosome 11, and 4 out of these 11 sites were within the globin LCR (within a sequence that was also present in the Ad.LCR-β-GFP genome) (Fig. 4C). For three integration sites, we were able to sequence both the 5′ and 3′ junctions of vector and chromosomal DNA (Fig. 4D). These limited data indicate that stretches of genomic DNA (14, 24, and 567 bp) were lost during vector integration, a phenomenon that is often observed during DNA integration via nonhomologous end joining (NHEJ) (24). In a number of clones with multiple integrants involving the AAV ITR, we also observed transposition of host genome sequences (Fig. 4E). For example, in clone 13, a short (68-bp) stretch of Ch11 host DNA that was adjacent to the 5′ end of vector DNA (vector copy no. 1 in Fig. 4E) appeared in three additional integration sites located on the same chromosome or on another chromosome, ChX. We speculate that this sequence (CCAGCCTGGACAAGAAGAGTGAAACTCCATCTCAAAAAAAAAAAAAAAAAAAAAAAAGGCGGGTGCGG) is prone to DNA breaks and that this might be involved in amplification and transposition of vector copies during DNA replication.
FIG. 4.
Analysis of integration junctions in MO7e clones by inverse PCR. (A) Schematic representation of inverse PCR. Five micrograms of genomic DNAs of 20 GFP-positive clones was digested with HindIII, relegated, and subjected to nested, inverse PCR with the indicated primers (see Materials and Methods). (B) 1% agarose gel showing inverse-PCR products from 18 clones. DNA bands were purified from the gel and cloned into pCR2.1 vector (Invitrogen) and transformed into the XL1-Blue strain, and junctions were sequenced. A sequence was considered to be from a genuine integration if it (i) contained both AAV ITR or Ad ITR sequence and the vector HindIII site, (ii) showed 95% or greater homology to the genomic sequence, and (iii) matched no more than one genomic locus with 95% or greater homology. (C) Schematic representation of integration sites found on chromosome 11. (D) Alignment of 5′ and 3′ junctions for three integration sites with corresponding genomic sequences found in GenBank (upper strand). Vector integration was associated with deletion of 24 bp, 567 bp, or 14 bp for the analyzed sites. (E) Schematic representation of transposition of genomic DNA fragments (circled) during vector integration (description in text).
TABLE 2.
Summary of Ad.LCR-β-GFP integration site analysis in MO7e clonesa
| Mo7e clone | 5′ or 3′ end | Ad or AAV ITR | XX (bp) | Integration (chromosome) | Integration (genome position) | Integrate into gene | Gene description |
|---|---|---|---|---|---|---|---|
| 1 | 5′ | Ad (Δ8 bp) | 32 | 11 | ←105,380,995 | − | |
| 3′ | Ad (Δ26 bp) | 11 | 104,906,474→ | − | |||
| 2 | 5′ | Ad (Δ6 bp) | 11 | 19,283,531→ | |||
| 3′ | Ad (Δ20 bp) | Repeat region? | |||||
| 3 | 5′ | Ad (Δ8 bp) | 1 | 3 | ←137,603,658 | STAG1 | Stromal antigen 1; intron |
| 4 | 3′ | Ad (Δ18 bp) | 8 | 106,809,443→ | ZFPM2 | Zinc finger protein, multitype 2; intron | |
| AAV | 15 | ←59,879,965 | − | ||||
| 5 | AAV | 11 | ←5,269,720* | − | |||
| 3′ | Ad (Δ20 bp) | Repeat region? | |||||
| 6 | 3′ | Ad (Δ9bp) | X | 43,412,862→ | MAOB | Monoamine oxidase B; intron | |
| Ad (Δ1bp) | 19 | X | ←15,570,024 | U2AF1L2 | U2 small nuclear ribonucleoprotein auxiliary; intron | ||
| 3′ | AAV | Repeat region | |||||
| 7 | 5′ | Ad | 10 | 11 | ←104,880,050 | − | |
| 5′ | Ad (Δ30 bp) | 11 | 59,277,649→ | − | |||
| 8 | |||||||
| 9 | 5′ | Ad (Δ12 bp) | 4⋆ | ←181,112,607 | − | ||
| 3′ | Ad (Δ8 bp) | 4 | 181,112,631→ | − | |||
| 10 | |||||||
| 11 | 5′ | Ad | 4 | 9 | 89,164,414→ | SECISBP2 | SECIS binding protein 2; intron |
| 12 | 5′ | Ad(Δ8 bp) | 11⋆ | 5,261,453*→ | − | ||
| Ad(Δ8 bp) | 14 | 11 | ←5,260,886* | − | |||
| 5′ | Ad(Δ205 bp) | 11 | ←5,261,972* | − | |||
| 13 | 5′ | Ad (Δ2bp) | 2 | 24,290,872→ | FLJ30851 | FLJ30851 protein; intron | |
| 3′ | AAV | 11 | ←125,680,575 | DCPS | mRNA-decapping enzyme; intron | ||
| AAV | 68 | 11 | 125,676,244→ | − | |||
| AAV | 68 | X | ←116,764,062 | − | |||
| AAV | 70 | X | 116,757,946→ | − | |||
| 14 | |||||||
| 15 | 5′ | Ad | 1 | ∼24,771,398→ | − | ||
| AAV | 1 | ←24,793,260 | − | ||||
| AAV | 120 | 1 | 24,787,753→ | − | |||
| 16 | |||||||
| 17 | AAV | 1 | 12 | ←52,955,718 | CBX5 | Chromobox homolog 5; intron | |
| 18 | 5′ | Ad (Δ10 bp) | 6⋆ | ←20,815,079 | CDKAL1 | CDK5 regulatory subunit-associated protein; intron | |
| 3′ | Ad(Δ8 bp) | 5 | 6 | 20,815,093→ | CDKAL1 | Intron | |
| 19 | 5′ | Ad (Δ8 bp) | 2 | 11,475,843→ | − | ||
| 20 | 3′ | Ad (Δ8 bp) | 3 | ←27,159,453 | − |
Localization of the globin LCR on chromosome 11, 5,270,125-5,248,599; localization of the 3′ HS1 on chromosome 11, 5,184,673-5,181,645; *, integration into the globin LCR; star, integration site for which both 5′ and 3′ ends were sequenced; XX, sequence was not homologous to the viral genome or chromosomal DNA; —, vector did not integrate into genes. Arrows indicate the orientation of the integrated vector. The USCS Genome Browser (http://genome.ucsc.edu/) was used to perform alignments of identified integration sites.
TABLE 3.
Summary of Ad.HCA-CMV-GFP integration site analysis in MO7e clonesa
| Clone | 5′ or 3′ | Ad ITR (bp) | XX (bp) | Integration (chromosome) | Integration (genome position) | Integrate into gene | Gene discription |
|---|---|---|---|---|---|---|---|
| 52/1 | 5′ | 6 | 17 | 27,353,505 | |||
| 15/1 | 3′ | 1 | 15 | 68,288,799 | − | − | |
| 18/2 | 3′ | 3 | X | 149,299,948 | CXorf6 | Chromosome X open reading frame 6; intron | |
| 22/5 | 3′ | 2 | 2 | 7,617,172 | − | − | |
| 40/5 | 3′ | 4 | 19 | 44,439,468 | − | − | |
| 41/1 | 3′ | 17 | 5 | 1 | 199,630,961 | ||
| 52/3 | 3′ | 1 | 6 | 3,262,660 | C6orf85 | Ion transporter protein; intron | |
| 65/3 | 3′ | 2 | 19 | 4,068,455 | MAP2k2 | Mitogen-activated protein kinase kinase 2; exon | |
| 69/1 | 3′ | 10 | 1 | 17 | 57,022,779 | ANAC | Alpha-NAC protein; exon |
| 69/7 | 3′ | 5 | 7 | 82,304,970 | − | − | |
| 73/2 | 3′ | 3 | 9 | 131,133,585 | NUP214 | Nucleoporin 214-kDa; intron | |
| 97/3 | 3′ | 10 | 6 | 140,127,347 | − | − | |
| 98/1 | 3′ | 2 | 1 | 54,402,564 | − | − | |
| 98/2 | 3′ | 20 | 4 | 149,457,240 | NR3C2 | Nuclear receptor subfamily 3 group C member 2; intron | |
| 98/3 | 3′ | 76 | 145 | 14 | 99,794,430 | YY1 | YY1 transcription factor; intron |
| 22/2 | 5′ | 630 | 10 | 67,881,795 | GRID1 | Glutamate receptor, ionotropic delta 1; intron | |
| 22/5 | 5′ | 630 | 17 | 49,687,702 | − | − | |
| 24/4 | 5′ | 630 | 10 | 125,243,823 | |||
| 52/1 | 5′ | 783 | 19 | 44,598,568 | PLEKHG2 | Pleckstrin homology domain containing, family G; intron | |
| 75-1/4 | 5′ | 630 | 21 | 33,339,549 | − | − | |
| 75-2/1 | 5′ | 630 | 146 | 1 | 205,929,122 | − | − |
| 75-2/2 | 5′ | 630 | 9 | 133,703,436 | VAV2 | Vav 2 oncogene; intron | |
| 75-2/3 | 5′ | 630 | 17 | 29,213,176 | ACCN1 | Neuronal amiloride-sensitive cation channel 1; intron |
Analysis of integration sites was conducted as described for Ad.LCR-β-GFP.
For Ad.HCA-CMV-GFP, we analyzed 24 integration sites (Table 3). Notably, Ad.HCA-CMV-GFP contains X-chromosomal stuffer DNA that does not form contiguous homology with cellular DNA. About half of the 5′ junctions involved AAV ITRs, while all 3′ junctions were formed by Ad ITR. Various numbers of 5′- and 3′-terminal Ad vector nucleotides were deleted when integration occurred via the Ad ITR. The integration sites were randomly distributed over the genome. None of the analyzed integration sites of this vector were in chromosome 11, and only one was in chromosome X. Ten out of 24 integrants were in genes; 2 out of these 10 junctions localized to exons. One integration occurred in a known oncogene (vav2).
In summary, Ad.LCR-β-GFP integration in MO7e cells does not appear to be random but is tethered to chromosome 11, specifically to the globin LCR. None of the Ad.LCR-β-GFP integration occurred in exons. The integration pattern of the vector that contains X-chromosomal stuffer DNA is clearly different from that of the LCR-containing vector. Integration is associated with deletions and rearrangements in genomic sequences.
Transduction and integration studies in human CD34+ cells.
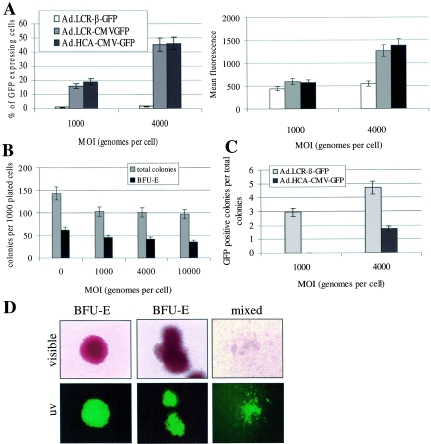
HSCs are the ultimate target for our vector, which aims for long-term correction of inherited blood disorders. Therefore, we performed transduction studies in CD34+ enriched, cord blood-derived cells, which are considered to contain HSCs. We have previously shown that first-generation Ad5/35 vectors efficiently transduce CD34+ cells, particularly subsets with stem cell capacity (48, 60), and that Ad5/35 transduction of CD34+ cells involves CD46 as a cellular receptor (9). Here, we demonstrate that HD Ad5/35 vectors allow efficient gene transfer into CD34+ cells. Forty-eight hours after infection with Ad.LCR-CMV-GFP or Ad.HCA-CMV-GFP at an MOI of 4,000 genomes per cell, more than 40% of CD34+ cells expressed GFP at high levels (Fig. 5A). As expected, the β-globin promoter in the context of the globin LCR has only low activity in CD34+ cells (36) and transient GFP expression from Ad.LCR-β-GFP is inefficient. Notably, from transduction studies in MO7e cells (Fig. 2A) and in CD34+ cells (Fig. 5A), the possibility that Ad.LCR-β-GFP and Ad.LCR-CMV-GFP have different infectivities cannot be excluded.
FIG. 5.
Analysis of GFP expression in transduced human cord blood-derived CD34+ cells. (A) Transient GFP expression in CD34+ cells. CD34+ cells were infected with Ad.LCR-β-GFP, Ad.LCR-CMV-GFP, and Ad.HCA-CMV-GFP at MOIs of 1,000 and 4,000 genomes per cell. The percentage (left) and mean fluorescence (right) of GFP-expressing cells were determined 48 h postinfection by flow cytometry. (B) CD34+ cells were mock infected or infected with Ad.LCR-CMV-GFP at the indicated MOIs. After infection, 3,000 mock-infected cells or 3,000 GFP-positive cells (after fluorescence-activated cell sorting) were plated for colony assays. The total numbers of colonies and of BFU-E colonies were determined 14 days after plating. (C) CD34+ cells were infected with Ad.LCR-β-GFP and Ad.HCA-CMV-GFP at MOIs of 1,000 and 4,000 genomes per cell. After infection, 3,000 cells were plated into semisolid medium without sorting, and 14 days later, the numbers of total colonies and GFP-positive colonies were determined. Shown is the percentage of GFP-expressing colonies. (D) Representative colonies derived from CD34+ cells after transduction with Ad.LCR-β-GFP at an MOI of 1,000 genomes per cell. Note that in mixed colonies, GFP expression is seen only in erythroid cells. The error bars indicate standard deviations.
To analyze stable transduction with our vectors, we performed colony assays with Ad-infected CD34+ cells in the presence of IL-3, SCF, Epo, granulocyte colony-stimulating factor, and GM-CSF. First, we demonstrated that infection of CD34+ cells with helper-dependent vectors did not affect the clonogenic capacity of CD34+ cells. The total number of colonies and the number of burst-forming units-erythroid (BFU-E) were comparable for mock-infected CD34+ cells and cells infected with Ad.LCR-β-GFP at MOIs ranging from 1,000 to 10,000 genomes per cell (Fig. 5B). Transduction of CD34+ cells with a first-generation Ad5/35 vector resulted in fewer colonies than with the helper-dependent vector, probably due to toxicity from expressed viral proteins (data not shown). Next, we analyzed GFP expression in colonies after transduction with Ad.LCR-β-GFP at an MOI of 4,000 genomes per cell. Fourteen days after being plated (without presorting), ∼5% of BFU-E were GFP positive (Fig. 5C). About 50% of the GFP-positive colonies were mixed colonies, indicating that the vector integrated into a pluripotent cell type. Notably, GFP expression in mixed colonies was restricted to primitive erythroid cells (Fig. 5D). In BFU-E, GFP expression was seen in all cells of a given colony, indicating that the vector integrated before the first cell division. Infection of CD34+ cells with Ad.LCR-CMV-GFP or Ad.HCA-CMV-GFP resulted in twofold fewer GFP-positive cells (Fig. 5C), so that GFP expression was not restricted to erythroid cells. It is unclear why transduction of CD34+ cells was less efficient with Ad.LCR-CMV-GFP than with Ad.LCR-β-GFP. We speculate that in contrast to the β-globin promoter, the CMV promoter is not active in all transduced cells.
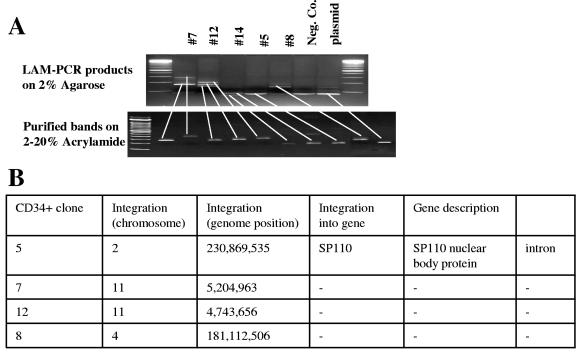
For integration analysis of CD34+ cells, we could not use the inverse-PCR method due to the limited amount of cellular DNA that can be obtained from colonies. We therefore employed LAM-PCR for integration site analysis (Fig. 6). We successfully mapped four integration sites for Ad.LCR-β-GFP in GFP-positive BFU-E colonies. Two of these sites were on chromosome 11, and the others were on chromosomes 2 and 4. Although very limited, these data indicate that the integration pattern of Ad.LCR-β-GFP in CD34+ cells is likely to be similar to that seen in MO7e cells.
FIG. 6.
LAM-PCR analysis of integration junctions in BFU-E colonies derived from CD34+ cells after transduction with Ad.LCR-β-GFP (MOI, 1,000 genomes per cell). (A) PCR fragments separated in 2% agarose (top). Selected bands were cut out and separated on polyacrylamide gels (bottom). Plasmid, pAd.AAV-LCR-β-GFP; negative control, DNA from untransduced BFU-E. (B) Summary of integration site analyses in four selected clones.
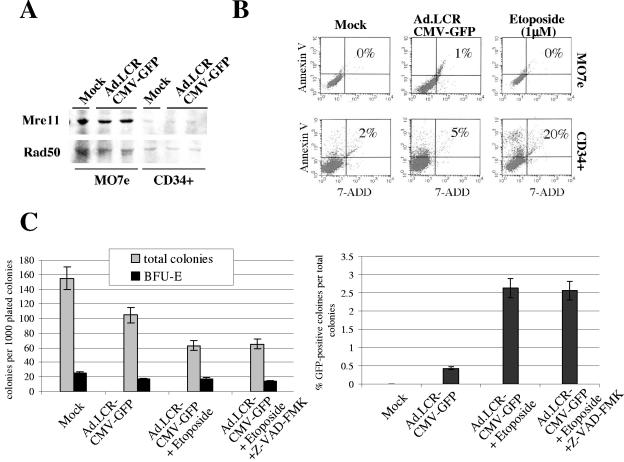
We also attempted to increase the frequency of stable transduction of CD34+ cells with our vectors. The results are shown for Ad.LCR-CMV-GFP transduction (Fig. 7). Similar data were obtained for the other HD Ad vectors. Increase of MOIs beyond 4,000 genomes per cell or increase of the cytokine concentration (to induce CD34+ cell division) during Ad infection did not lead to significantly higher numbers of GFP-positive colonies (data not shown). We hypothesized that vector integration involves NHEJ and the cellular DNA repair machinery and studied expression of the DNA repair proteins Mre11 and Rad50 in MO7e and primary CD34+ cells (Fig. 7A). While Mre11 and Rad50 were readily detectable by Western blot analysis in MO7e cells, expression of these proteins was markedly lower in CD34+ cells. Infection with Ad.LCR-CMV-GFP did not increase Mre11/Rad50 levels.
FIG. 7.
Induction of DNA breaks and DNA repair in CD34+ cells can increase stable transduction frequency with Ad.LCR vectors. (A) Western blot analysis of DNA repair enzymes Mre11 and Rad50 24 h after mock infection or Ad.LCR-CMV-GFP transduction (MOI, 1,000 genomes per cell) of CD34+ cells or MO7e cells. (B) Analysis of apoptosis (based on annexin V levels) and necrosis (based on 7-ADD levels) in CD34+ cells 34 h after mock infection, infection with Ad.LCR-CMV-GFP (MOI, 4,000 genomes per cell), or treatment with 1 μM etoposide. (C) Colony assays of transduced CD34+ cells. CD34+ cells were transduced with Ad.LCR-CMV-GFP in the presence or absence of 1 μM etoposide and 2 μM of the pancaspase inhibitor Z-VAD-FMK; 3,000 cells were plated in semisolid medium, and colonies were analyzed 14 days later. The error bars indicate standard deviations.
To stimulate DNA repair through NHEJ, we treated CD34+ cells with etoposide, which is known to induce double-strand DNA breaks. Induction of DNA damage in normal (p53+) cells can potentially induce apoptosis. To assess this, we measured levels of annexin V (a marker for apoptosis) and 7-ADD (a marker for necrosis) in CD34+ cells and MO7e cells (Fig. 7B). Etoposide treatment caused apoptosis in 20% of CD34+ cells but had no effect on MO7e cells. This study also showed that infection of CD34+ cells with Ad.LCR-CMV-GFP triggered apoptosis in only 5% of cells compared to 2% after mock infection. In agreement with the annexin V data, etoposide treatment of CD34+ cells also reduced the number of colonies in progenitor assays, and this effect could not be prevented by adding the pancaspase inhibitor Z-VAD-FMK (Fig. 7C). Despite toxicity, etoposide treatment increased the percentage of GFP-expressing colonies about fivefold.
In conclusion, infection of CD34+ cells with helper-dependent Ad.LCR vectors did not affect the clonogenic capacity of CD34+ cells. Transduction of CD34+ cells with Ad.LCR-β-GFP resulted in vector integration and erythroid cell-specific GFP expression.
The efficiency of stable transduction with Ad/LCR vectors can be increased by treatment of CD34+ cells with etoposide.
DISCUSSION
In this study, we show that incorporation of β-globin LCR elements into an HD Ad5/35.AAV vector allows stably erythroid-specific transgene expression in an HSC model cell line and in primary CD34+ derived human cells. Integration of LCR-containing Ad vectors (Ad.LCR) was more efficient and displayed a nonrandom distribution compared to a vector that contained stuffer DNA derived from the human X chromosome. A larger fraction of Ad.LCR integration occurred in the chromosomal β-globin LCR or in regions located on the same chromosome as the globin locus.
Ad.LCR: new features.
(i) Ad.LCR vectors possess Ad35 fiber knobs. Previous studies have shown that this allows targeting of human bone marrow-derived cells with potential stem cell capacity, including CD34+ C-Kit+, CD34− Lin−, CD38−, and a Hoechst-negative “side population” (48, 60), as well as NOD-SCID repopulating cells (33). Our finding that Ad.LCR vectors transduce pluripotent cells that are able to form mixed colonies in progenitor colony assays (Fig. 5E) provides further confirmation that Ad5/35 vectors are promising tools for HSC targeting. (ii) Ad.LCR contains AAV ITRs which are prone to single- or double-strand breaks and/or are substrates for cellular DNA repair enzymes and therefore stimulate vector integration (46). (iii) Most HD Ad vectors constructed so far contain nonfunctional stuffer DNA sequences that lack open reading frames or extended homology with the human genome, for example, X-chromosomal DNA (14, 17, 44). More than 90% of the Ad.LCR genome comprised globin LCR elements that fulfill a number of functions. First, the globin LCR elements in Ad.LCR-β-GFP allow erythroid-specific expression. While GFP expression was efficient in MO7e cells and BFU-E, only low GFP levels were detected in HeLa and undifferentiated CD34+ cells. Second, our data indicate that Ad.LCR is less subject to chromosomal position effects and silencing upon integration than the vector that does not contain the globin LCR. Notably, other gene transfer vectors that contained shorter globin LCR elements were not protected against position effects (21, 40, 46, 50). Finally, as discussed below, the LCR allows a certain degree of targeting of vector integration.
Integration frequency of Ad.LCR.
Integration frequencies for our HD vectors (Ad.LCR-CMV-GFP, Ad.LCR-β-GFP, and Ad.HCA-CMV-GFP) were higher than those reported for E1-deleted first-generation vectors, which were shown to integrate at frequencies of ∼10−3 to 10−5 into cultured mammalian cells (13, 15, 16). This could be due to the fact that HD vectors, in contrast to first-generation vectors, do not express viral gene products that can affect the expansion of stably transduced clones. Furthermore, it has been demonstrated that Ad E4 proteins (E4orf3 and E4orf6) can act to inhibit recombination and double-strand break repair (3), which might affect vector integration. More efficient integration of HD vectors versus first-generation vectors (1.8 × 10−3 versus 2 × 10−4) was also reported in a recent study by Hillgenberg et al. (14). Notably, in that study, stable transduction was assessed under G418 selection. This could have mimicked stress responses reported to affect the integration frequency and pattern (43). An important observation in our study was that the stable transduction frequency of Ad.LCR vectors in MO7e and CD34+ cells was higher than that of Ad.HCA, indicating that the presence of the globin LCR qualitatively and quantitatively influenced vector integration.
Mechanism of Ad.LCR integration.
All integrants contained the complete LCR/transgene sequence. Integration junctions contained AAV ITR and Ad ITR sequences. We speculate that Ad.LCR integration requires free vector DNA ends, resulting from single- or double-strand DNA breaks in the vector genome, and that the AAV and Ad ITRs are prone to such breaks due to their specific (secondary) structure. Notably, the termini of incoming Ad genomes are protected by the terminal protein (TP), which is thought to prevent efficient Ad integration (13, 14). Apparently, for Ad ITRs the terminal ITR nucleotides, together with the TP, must break off before integration. The latter speculation is supported by our finding that in all integrants that occurred via the Ad ITR, the terminal nucleotides of vector genomes were deleted. We further hypothesize that the free ends of linear Ad vector DNA are recognized as substrates for NHEJ, leading to integration of complete vector genomes at sites of double-strand breaks in chromosomal DNA, in a way similar to what has been described for double-stranded linear DNA (32) and rAAV (31). The following findings support this hypothesis: (i) a genotoxic drug such as etoposide that induces double-strand breaks increases Ad.LCR integration, (ii) increasing the Ad.LCR vector dose above a threshold level did not increase the frequency of stable transduction, (iii) vector integration is associated with deletions of chromosomal DNA, and (iv) vector integration was more efficient in MO7e cells that express higher levels of DNA repair proteins than in CD34+ cells.
Nonrandom integration pattern of Ad.LCR.
Analysis of Ad.LCR integration in MO7e cells demonstrated a nonrandom integration pattern, while Ad.HCA integrated randomly. Random integration of an HD Ad vector containing 27.4 kb of the human X chromosome was also reported by Hillgenberg et al. (14). Furthermore, first-generation Ad vectors (28), an Ad5-simian virus 40 vector (41), and wild-type Ad12 (62) have been shown to integrate randomly. Our integration studies of Ad.LCR in MO7e cells showed that 11 out of 33 junctions were localized to chromosome 11, and 4 out of these 11 sites were within the β-globin LCR. Nonrandom Ad.LCR integration of Ad.LCR is also supported by Southern analysis data (Fig. 3C), which show a prominent band in the range of 8 kb that could theoretically be the result of integration into the LCR. Tethering of Ad.LCR integration to the globin LCR is in agreement with our earlier study using a ΔAd.AAV vector containing a 5.3-kb β-globin LCR HS2/HS3 fragment, which showed that in two of five clones, integration occurred in the HS2/HS3 region of the chromosomal globin locus (46). We speculate that tethering of integration to the globin locus/chromosome 11 involves LCR-specific chromatin structures. The globin LCR present in the vector and chromosomal DNA contains DNase HS regions. HS regions are known to be associated with nonhistone proteins, to be prone to DNA breaks, and to be accessible to DNA repair enzymes (55). Furthermore, it is thought that transcription of distant genes occurs in discrete intranuclear foci called “transcription factories” (35). Recruitment of active genes into a particular transcription factory involves transcription factors and chromatin structures that are specific for a specific DNA region. Based on this, we speculate that the LCR carrying Ad vector colocalizes with the genomic β-globin LCR in a transcription factory and that this physical proximity, together with double-strand DNA breaks, allows more efficient NHEJ and preferential integration of our vector into the globin LCR or areas on chromosome 11 that are theoretically exposed on the same transcription factory. An interaction between chromatin elements in the incoming vector and chromosomal DNA is also supported by observations that distant HS sites in the globin locus interact, so that the intervening chromatin with inactive globin genes loops out (6, 35, 54).
The lower integration frequency and random pattern seen with Ad.HCA might therefore be due to a (“closed”) heterochromatin structure for X-chromosomal DNA present in the vector and/or the chromosome and/or their inability to be in physical proximity.
Existing vectors, including oncoretrovirus (56), lentivirus (45), and rAAV (42) vectors, have a predilection for integrating into genes, which bears the risk of activation of oncogenes. Tumorigenesis as a result of oncoretrovirus transduction has been demonstrated in mice (22) and became the focus of attention in a trial for ADA-SCID with murine oncoretrovirus vectors, in which patients developed leukemia (18). Therefore, targeting of vector integration is one of the major tasks in gene therapy. Homologous targeting with viral or plasmid vectors is generally a rare event in mammalian cells, with an average ratio of random to targeted insertion of around 1,000:1 (for a recent review, see reference 20). Our finding that >10% of integration events (4 out of 33) occurred in the globin LCR, and that this is due to the presence of the LCR in the vector, could form the basis for new targeting strategies. This, together with the erythroid-specific gene expression that is apparently not subject to silencing or position effects, makes Ad.LCR vectors promising tools for globin gene therapy. Our current efforts are aimed at increasing the frequency of Ad.LCR integration in primary human CD34+ cells by providing free terminal ends of vector DNA, by placing targeted double-strand breaks in chromosomal DNA using Zn finger proteins with nuclease activity (29), and/or by stimulation of the cellular DNA repair machinery.
Acknowledgments
We thank Daniel Stone for helpful discussions and for editing the manuscript.
This study was supported by NIH grants HL53750, HL-00-008,and R01 HLA078836 and a grant from the Doris Duke Charitable Foundation.
REFERENCES
- 1.Avanzi, G. C., P. Lista, B. Giovinazzo, R. Miniero, G. Saglio, G. Benetton, R. Coda, G. Cattoretti, and L. Pegoraro. 1988. Selective growth response to IL-3 of a human leukaemic cell line with megakaryoblastic features. Br. J. Haematol. 69:359-366. [DOI] [PubMed] [Google Scholar]
- 2.Barjot, C., D. Hartigan-O'Connor, G. Salvatori, J. M. Scott, and J. S. Chamberlain. 2002. Gutted adenoviral vector growth using E1/E2b/E3-deleted helper viruses. J. Gene Med. 4:480-489. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, C. A., D. M. Shayakhmetov, and A. Lieber. 2002. Restoration of a functional open reading frame by homologous recombination between two adenoviral vectors. Mol. Ther. 6:99-105. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, C. A., D. S. Steinwaerder, H. Stecher, D. M. Shayakhmetov, and A. Lieber. 2002. Rearrangements in adenoviral genomes mediated by inverted repeats. Methods Enzymol. 346:277-292. [DOI] [PubMed] [Google Scholar]
- 6.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 7.Chamberlain, J. S., C. Barjot, and J. Scott. 2003. Packaging cell lines for generating replication-defective and gutted adenoviral vectors. Methods Mol. Med. 76:153-166. [DOI] [PubMed] [Google Scholar]
- 8.Ellis, J., and D. Pannell. 2001. The beta-globin locus control region versus gene therapy vectors: a struggle for expression. Clin. Genet. 59:17-24. [DOI] [PubMed] [Google Scholar]
- 9.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 10.Goncalves, M. A., I. van der Velde, J. M. Janssen, B. T. Maassen, E. H. Heemskerk, D. J. Opstelten, S. Knaan-Shanzer, D. Valerio, and A. A. de Vries. 2002. Efficient generation and amplification of high-capacity adeno-associated virus/adenovirus hybrid vectors. J. Virol. 76:10734-10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grosveld, F., E. de Boer, N. Dillon, P. Fraser, J. Gribnau, E. Milot, T. Trimborn, and M. Wijgerde. 1998. The dynamics of globin gene expression and gene therapy vectors. Semin. Hematol. 35:105-111. [PubMed] [Google Scholar]
- 12.Grosveld, F., G. B. van Assendelft, D. R. Greaves, and G. Kollias. 1987. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell 51:975-985. [DOI] [PubMed] [Google Scholar]
- 13.Harui, A., S. Suzuki, S. Kochanek, and K. Mitani. 1999. Frequency and stability of chromosomal integration of adenovirus vectors. J. Virol. 73:6141-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hillgenberg, M., H. Tonnies, and M. Strauss. 2001. Chromosomal integration pattern of a helper-dependent minimal adenovirus vector with a selectable marker inserted into a 27.4-kilobase genomic stuffer. J. Virol. 75:9896-9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlsson, S., R. K. Humphries, Y. Gluzman, and A. W. Nienhuis. 1985. Transfer of genes into hematopoietic cells using recombinant DNA viruses. Proc. Natl. Acad. Sci. USA 82:158-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson, S., K. Van Doren, S. G. Schweiger, A. W. Nienhuis, and Y. Gluzman. 1986. Stable gene transfer and tissue-specific expression of a human globin gene using adenoviral vectors. EMBO J. 5:2377-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochanek, S., G. Schiedner, and C. Volpers. 2001. High-capacity ‘gutless’ adenoviral vectors. Curr. Opin. Mol. Ther. 3:454-463. [PubMed] [Google Scholar]
- 18.Kohn, D. B., M. Sadelain, and J. C. Glorioso. 2003. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat. Rev. Cancer 3:477-488. [DOI] [PubMed] [Google Scholar]
- 19.Kubo, S., and K. Mitani. 2003. A new hybrid system capable of efficient lentiviral vector production and stable gene transfer mediated by a single helper-dependent adenoviral vector. J. Virol. 77:2964-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanzov, V. A. 1999. Gene targeting for gene therapy: prospects. Mol. Genet. Metab. 68:276-282. [DOI] [PubMed] [Google Scholar]
- 21.Leboulch, P., G. M. Huang, R. K. Humphries, Y. H. Oh, C. J. Eaves, D. Y. Tuan, and I. M. London. 1994. Mutagenesis of retroviral vectors transducing human beta-globin gene and beta-globin locus control region derivatives results in stable transmission of an active transcriptional structure. EMBO J. 13:3065-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., J. Dullmann, B. Schiedlmeier, M. Schmidt, C. von Kalle, J. Meyer, M. Forster, C. Stocking, A. Wahlers, O. Frank, W. Ostertag, K. Kuhlcke, H. G. Eckert, B. Fehse, and C. Baum. 2002. Murine leukemia induced by retroviral gene marking. Science 296:497. [DOI] [PubMed] [Google Scholar]
- 23.Lieber, A., D. S. Steinwaerder, C. A. Carlson, and M. A. Kay. 1999. Integrating adenovirus-adeno-associated virus hybrid vectors devoid of all viral genes. J. Virol. 73:9314-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lieber, M. R., Y. Ma, U. Pannicke, and K. Schwarz. 2003. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev. Mol. Cell Biol. 4:712-720. [DOI] [PubMed] [Google Scholar]
- 25.Marini, F. C., III, Q. Yu, T. Wickham, I. Kovesdi, and M. Andreeff. 2000. Adenovirus as a gene therapy vector for hematopoietic cells. Cancer Gene Ther. 7:816-825. [DOI] [PubMed] [Google Scholar]
- 26.McCarty, D. M., S. M. Young, Jr., and R. J. Samulski. 2004. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu. Rev. Genet. 38:819-845. [DOI] [PubMed] [Google Scholar]
- 27.Medin, J. A., and S. Karlsson. 1997. Viral vectors for gene therapy of hematopoietic cells. Immunotechnology 3:3-19. [DOI] [PubMed] [Google Scholar]
- 28.Mitani, K., and S. Kubo. 2002. Adenovirus as an integrating vector. Curr. Gene Ther. 2:135-144. [DOI] [PubMed] [Google Scholar]
- 29.Moore, M., and C. Ullman. 2003. Recent developments in the engineering of zinc finger proteins. Brief. Funct. Genomic. Proteomic. 1:342-355. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, S. J., H. Chong, S. Bell, R. M. Diaz, and R. G. Vile. 2002. Novel integrating adenoviral/retroviral hybrid vector for gene therapy. Hum. Gene Ther. 13:745-760. [DOI] [PubMed] [Google Scholar]
- 31.Nahreini, P., S. H. Larsen, and A. Srivastava. 1992. Cloning and integration of DNA fragments in human cells via the inverted terminal repeats of the adeno-associated virus 2 genome. Gene 119:265-272. [DOI] [PubMed] [Google Scholar]
- 32.Nakai, H., E. Montini, S. Fuess, T. A. Storm, L. Meuse, M. Finegold, M. Grompe, and M. A. Kay. 2003. Helper-independent and AAV-ITR-independent chromosomal integration of double-stranded linear DNA vectors in mice. Mol. Ther. 7:101-111. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, M., S. Karlsson, and X. Fan. 2004. Functionally distinct subpopulations of cord blood CD34+ cells are transduced by adenoviral vectors with serotype 5 or 35 tropism. Mol. Ther. 9:377-388. [DOI] [PubMed] [Google Scholar]
- 34.Olivares, E. C., R. P. Hollis, T. W. Chalberg, L. Meuse, M. A. Kay, and M. P. Calos. 2002. Site-specific genomic integration produces therapeutic Factor IX levels in mice. Nat. Biotechnol. 20:1124-1128. [DOI] [PubMed] [Google Scholar]
- 35.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 36.Papayannopoulou, T., G. V. Priestley, A. Rohde, K. R. Peterson, and B. Nakamoto. 2000. Hemopoietic lineage commitment decisions: in vivo evidence from a transgenic mouse model harboring micro LCR-betapro-LacZ as a transgene. Blood 95:1274-1282. [PubMed] [Google Scholar]
- 37.Recchia, A., R. J. Parks, S. Lamartina, C. Toniatti, L. Pieroni, F. Palombo, G. Ciliberto, F. L. Graham, R. Cortese, N. La Monica, and S. Colloca. 1999. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc. Natl. Acad. Sci. USA 96:2615-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recchia, A., L. Perani, D. Sartori, C. Olgiati, and F. Mavilio. 2004. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 10:660-670. [DOI] [PubMed] [Google Scholar]
- 39.Recchia, A., L. Perani, D. Sartori, and F. Mavilio. 2003. Site-specific integration in human somatic cell DNA by Adeno/AAV hybrid vectors. Mol. Ther. 7:S199. [DOI] [PubMed] [Google Scholar]
- 40.Rivella, S., and M. Sadelain. 1998. Genetic treatment of severe hemoglobinopathies: the combat against transgene variegation and transgene silencing. Semin. Hematol. 35:112-125. [PubMed] [Google Scholar]
- 41.Romani, M., A. De Ambrosis, B. Alhadeff, M. Purrello, Y. Gluzman, and M. Siniscalco. 1990. Preferential integration of the Ad5/SV40 hybrid virus at the highly recombinogenic human chromosomal site 1p36. Gene 95:231-241. [DOI] [PubMed] [Google Scholar]
- 42.Russell, D. W. 2003. AAV loves an active genome. Nat. Genet. 34:241-242. [DOI] [PubMed] [Google Scholar]
- 43.Russell, D. W., I. E. Alexander, and A. D. Miller. 1995. DNA synthesis and topoisomerase inhibitors increase transduction by adeno-associated virus vectors. Proc. Natl. Acad. Sci. USA 92:5719-5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandig, V., R. Youil, A. J. Bett, L. L. Franlin, M. Oshima, D. Maione, F. Wang, M. L. Metzker, R. Savino, and C. T. Caskey. 2000. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. USA 97:1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 46.Shayakhmetov, D. M., C. A. Carlson, H. Stecher, Q. Li, G. Stamatoyannopoulos, and A. Lieber. 2002. A high-capacity, capsid-modified hybrid adenovirus/adeno-associated virus vector for stable transduction of human hematopoietic cells. J. Virol. 76:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shayakhmetov, D. M., Z. Y. Li, A. Gaggar, H. Gharwan, V. Ternovoi, V. Sandig, and A. Lieber. 2004. Genome size and structure determine efficiency of postinternalization steps and gene transfer of capsid-modified adenovirus vectors in a cell-type-specific manner. J. Virol. 78:10009-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatoyannopoulos, J. A. 1992. Future prospects for treatment of hemoglobinopathies. West J. Med. 157:631-636. [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatoyannopoulos, J. A., C. H. Clegg, and Q. Li. 1997. Sheltering of gamma-globin expression from position effects requires both an upstream locus control region and a regulatory element 3′ to the A gamma-globin gene. Mol. Cell. Biol. 17:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steinwaerder, D., and A. Lieber. 2000. Insulation from viral transcriptional regulatory elements improves inducible transgene expression from adenovirus vectors in vitro and in vivo. Gene Ther. 7:556-567. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, C. E., A. Ehrhardt, and M. A. Kay. 2003. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4:346-358. [DOI] [PubMed] [Google Scholar]
- 53.Tisdale, J., and M. Sadelain. 2001. Toward gene therapy for disorders of globin synthesis. Semin. Hematol. 38:382-392. [DOI] [PubMed] [Google Scholar]
- 54.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 55.Urnov, F. D. 2003. Chromatin remodeling as a guide to transcriptional regulatory networks in mammals. J. Cell Biochem. 88:684-694. [DOI] [PubMed] [Google Scholar]
- 56.Vijaya, S., D. L. Steffen, and H. L. Robinson. 1986. Acceptor sites for retroviral integrations map near DNase I-hypersensitive sites in chromatin. J. Virol. 60:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao, X., W. Xiao, J. Li, and R. J. Samulski. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71:941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71:9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yant, S. R., A. Ehrhardt, J. G. Mikkelsen, L. Meuse, T. Pham, and M. A. Kay. 2002. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat. Biotechnol. 20:999-1005. [DOI] [PubMed] [Google Scholar]
- 60.Yotnda, P., H. Onishi, H. E. Heslop, D. Shayakhmetov, A. Lieber, M. Brenner, and A. Davis. 2001. Efficient infection of primitive hematopoietic stem cells by modified adenovirus. Gene Ther. 8:930-937. [DOI] [PubMed] [Google Scholar]
- 61.Young, S. M., Jr., W. Xiao, and R. J. Samulski. 2000. Site-specific targeting of DNA plasmids to chromosome 19 using AAV cis and trans sequences. Methods Mol. Biol 133:111-126. [DOI] [PubMed] [Google Scholar]
- 62.Zeng, M., G. J. Cerniglia, S. L. Eck, and C. W. Stevens. 1997. High-efficiency stable gene transfer of adenovirus into mammalian cells using ionizing radiation. Hum. Gene Ther. 8:1025-1032. [DOI] [PubMed] [Google Scholar]