Abstract
In humans and in animal models, susceptibility to arthritis is under complex genetic control, reflecting influences on the immunological processes that initiate autoimmunity and on subsequent inflammatory mechanisms in the joints. The effector phases are conveniently modeled by the K/BxN serum transfer system, a robust model well suited for genetic analysis where arthritis is initiated by pathogenic Ig. Here, we mapped the genetic loci distinguishing the high-responder BALB/c vs. low-responder SJL strains. After computational modeling of potential breeding schemes, we adapted a stepwise selective breeding strategy, with a whole-genome scan performed on a limited number of animals. Several genomic regions proved significantly associated with high sensitivity to arthritis. One of these regions, on distal chr2, was centered on the interleukin 1 gene family. Quantitation of transcripts of the Il1a and Il1b candidate genes revealed a 10-fold greater induction of Il1b mRNA in BALB/c than in SJL splenocytes after injection of LPS, whereas Il1a showed much less difference. The differential activity of the Il1b gene was associated with a particular sequence haplotype of noncoding polymorphisms. The BALB/c haplotype was found in 75% of wild-derived strains but was rare among conventional inbred strains (4/33 tested, one of which is DBA/1, the prototype arthritis-susceptible strain) and was associated with vigorous Il1b responses in a panel of inbred strains. Inbred strains carrying this allele were far more responsive to serum-transferred arthritis, confirming its broad importance in controlling arthritis severity.
Keywords: genetics, autoimmunity, inflammation, Mouse Phenome Project
As might be expected for a complex disease that involves a number of cellular and molecular players, the genetic basis of arthritis has proven very complex and, despite significant efforts, remains even more uncharted than for many other multigenic diseases (reviewed in refs. 1-3). The importance of the genetic component of human rheumatoid arthritis (RA) has been well established by studies on familial aggregation, twins, and population segregation. The MHC, in particular the HLA-DR locus, is a major contributor to disease predisposition, accounting for one-third of the genetic effect (2). Although many non-MHC chromosomal regions exhibited suggestive linkage in several large-scale genetic studies worldwide, none has proven convincingly reproducible between cohorts (2).
In animal models, progress has also been hampered by the complexity of pathogenesis and of the underlying genetics. A strong influence of the MHC region was established for rats and mice (4, 5), but contributions from non-MHC genes have proven complex and more difficult to map (6). For rats, a major locus influencing the severity of pristane-induced arthritis localized to the Ncf1 gene, which controls oxidative burst (7). For mice, the C5 locus that encodes Complement Factor 5 was identified in both the collagen-induced arthritis (CIA) and K/BxN models as a key element for susceptibility and severity (8, 9), and the Ncf1 gene was recently found to affect CIA severity in mice as well (10). Yet, beyond these rare identified loci, it seems the surface has only been scratched, and there are many more arthritis modifier genes to identify (6, 11).
K/BxN T cell receptor transgenic mice spontaneously develop an autoimmune disease with many of the clinical, histological, and immunological features of RA in humans (12, 13). K/BxN arthritis is critically dependent on both T lymphocytes, at the apex of the autoimmune process, and B lymphocytes, which constitute an essential relay that produces arthritogenic Abs. K/BxN mice might be thought of as a generic model of inflammatory arthritis mediated by Ab and immune complex deposition in the joint. This patho-physiologic mechanism is a classic one for RA, but one which had fallen in disfavor until interest was recently rekindled by a number of observations, in particular the impact of B cell-based therapy (reviewed in ref. 14).
Transfer of serum from arthritic K/BxN mice into healthy animals provokes arthritis within days (13), providing a powerful experimental tool for analyzing effector-phase pathways, without the confounding influence of the immunological phase. The rapidity of the assay allows relatively facile genetic analysis, particularly because the disease does not completely depend on a single genetic background, in contrast to the situation with most other murine models of autoimmunity. Analysis of a panel of inbred mouse strains revealed a rich variability in the patterns of responses to K/BxN serum, ranging from complete resistance to explosive arthritis (8). An initial F2 analysis on a responder/nonresponder combination (C57BL/6 vs. NOD) highlighted a complex genetic determinism and revealed the critical role of the C5 locus. Indeed, the presence of the inactive variant of the C5 allele explains the complete nonresponsiveness of several inbred strains (8).
Here, we wished to explore the genetic basis for the very explosive arthritis induced in BALB/c mice by K/BxN serum, a rapidly progressing disease initiating within 1 day after serum injection. The SJL/J strain was chosen as a comparator, because preliminary data indicated that arthritis appears slowly after introduction of K/BxN serum, and the inflammation always remains mild (8). We mapped the intervals and genes responsible for aggressive arthritis with a selective breeding strategy, resulting in the identification of IL-1β as a key determinant of susceptibility.
Methods
Mice and Arthritis Induction. BALB/cJ, SJL/J, and NOD/LtJ mice were obtained from The Jackson Laboratory; all breedings and experimentation were performed in our facility (protocol 3024). In silico breedings were performed on s-plus (insightful; see Supporting Text, which is published as supporting information on the PNAS web site). Arthritis was induced as described (13) by i.p. injection of 10 μl/g of body weight serum from arthritic KRN×NOD mice at days 0 and 2 (except in Fig. 5b, 7.5 μl/g of weight). A clinical index was evaluated over time (0-3 points for each limb), except for the data of Fig. 5a, which reproduce a previous publication in which a 0-4 scale was used (0.5-1 point for each affected paw).
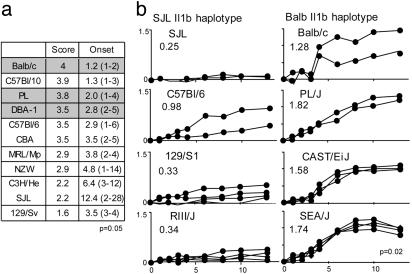
Fig. 5.
The BALB/c allele at IL-1b confers high responsiveness to arthritis induction. (a) Several strains that had been previously phenotyped for sensitivity to arthritis induction by K/BxN serum were genotyped for the -230 Il1b SNP. Strains with the BALB/c allele at Il1b allele are shaded. The values shown are the average maxAT (on a 0-4 scale) and the average (range) day of onset (phenotypic values reproduced from ref. 8); P value from Wilcoxon's rank sum test comparing scores in strains with either IL1b haplotype. (b) Additional mice carrying either the SJL or BALB/c alleles at Il1b (left and right columns, respectively) were challenged with K/BxN serum. Disease progression was measured by ankle thickening; the values in the boxes represent integrated ankle thickening (days 1-7); P value is from Student's t test comparing both groups.
Genetic Analysis. Genotyping with polymorphic fluorescent microsatellite markers was carried out as described (15). SNP genotyping was performed by allele-specific fluorogenic PCR, as detailed elsewhere (16). Genetic analyses were performed with qtdt, Ver. 2.4.3 (downloaded from www.sph.umich.edu/csg/abecasis/QTDT) and custom s-plus scripts (Supporting Text). For the latter, linkage peaks were identified by a combination of allele segregation and trait/marker regression, experiment-wise empirical P values being established by simulation tests (10,000 simulations of the pedigree with 100 neutral markers segregating randomly).
Quantitative PCR. Mice were challenged by i.p. injection of 50 μg of Escherichia coli LPS (Sigma L4524), RNA was isolated from frozen spleens, and quantitative real-time PCR was then performed with gene-specific fluorogenic assays (TaqMan, Applied Biosystems) for IL-1α and -1β transcripts; hypoxanthine guanine phosphoribosyl transferase (HPRT) was used as an internal standard, and arbitrary units of IL-1α and -1β mRNA were deduced as 2exp(Δ-ΔCT).
Results
Multiple Genetic Influences Underlie the Differential Susceptibility of BALB/c and SJL/J Mice to K/BxN Serum. The time-course data of Fig. 1a, representative of results from >10 experiments, illustrate the different responses of BALB/c and SJL/J mice to the injection of K/BxN serum. In most BALB/c recipients, whether male or female, inflammation of the joints was already clear on the first day after injection of serum from arthritic donors [onset: 1.24 ± 0.44 days; maximum ankle thickening (maxAT): 1.08 ± 0.13 mm] and was maximal by 4-6 days after injection. In SJL recipients, joint inflammation also occurred but in a much more sluggish fashion; the onset of visible swelling was delayed by several days and progressed more slowly, to a lower maximum value (onset, 5.0 ± 2.9; maxAT, 0.44 ± 0.24). The phenotype was intermediate in F1 animals (Fig. 1a). The timing of the responses of F2 animals covered the range of values bracketed by the parental phenotypes (not shown). Interestingly, maxAT was even higher in many of the F2s than in BALB/c mice, indicating that low-responder alleles at some loci may exist in the high-responder parental strain. Thus, a complex combination of loci seemed to control the aggressive arthritis of the BALB/c strain.
Fig. 1.
Whole-genome scan for linkage to arthritis severity in the pedigree. (a) Young adult BALB/cJ or SJL/J mice were challenged with serum from arthritis K/BxN donors, and the progression of arthritis was tracked as thickening of the ankle joint over time. (b) Marker/trait association score derived from combined information in different generations. Significance (-log10 of the P value) was derived from the frequency of observing by chance the same association metrics for unlinked markers segregating randomly through 10,000 simulated pedigrees of identical structure. (c) Selected logarithm of odds scores from the transmission disequilibrium test, for association to the different metrics of arthritis severity (onset, maxAT, or composite “severity index”). Light and dark shading denotes suggestive and significant association. Only the regions with suggestive or significant association are shown here; full data are in Table 2, which is published as supporting information on the PNAS web site.
Selective Breeding to Dissect the Genetic Basis of the BALB vs. SJL Difference. We opted to dissect the genetic elements that distinguish BALB/c from SJL mice by a series of selective crosses, in which the highest responders were selected at each generation for breeding. This classic approach has been used on occasion for quantitative trait loci (QTL) mapping in a number of genetic analyses (17, 18) and seemed of interest here because the arthritis induced by injection of K/BxN serum is reversible (13). The aim was to facilitate the genetic analysis by avoiding breeding and genotyping of a large F2 cohort; instead, genetic mapping information would be derived at each generation of the cross. The second aim was to generate “multicongenic” lines of mice that carry several genetic elements conferring high responsiveness to K/BxN serum amid a genome otherwise enriched for alleles from the low-responder background. In other words, rather than fixing a predefined genetic region as when generating conventional congenic lines, we would let the phenotypic selection drive the isolation of the responsible genetic regions. The breeding strategy was elaborated after computational simulations of the pedigrees and is detailed in Supporting Text and Fig. 6, which is published as supporting information on the PNAS web site. The actual breeding scheme is depicted in Fig. 7, which is published as supporting information on the PNAS web site.
A genome-wide scan was performed on a subset of animals throughout the pedigree. These animals included all 32 mice of the founder F2 cohort, the mice used for breeding at each generation up to H6, and a larger subset of mice of the H5 cohort, encompassing individuals that display more extreme phenotypes and were therefore likely to be more informative. The markers used were a combination of microsatellites and SNPs (105 markers altogether, with an average spacing of 18.3 Mb; Table 1, which is published as supporting information on the PNAS web site). In most cases, we used a composite “severity index” to represent disease aggressivity, which incorporated rate of onset, maximum clinical score, and maxAT, as a phenotypic metric.
Statistical methods are detailed in Supporting Text. The first test was designed to match the particular aspects of the pedigree, deriving combinatorial information from different generations in the cohort. Significance was estimated by comparison with a large number of randomly generated virtual pedigrees of identical structure, asking how frequent the phenotypic correlations and segregation patterns of markers in the true dataset would prove to be when a large number of neutral loci were simulated to segregate down an identical pedigree. Significant association (P < 0.001) was discerned from the combined information (Fig. 1b), primarily on Chr2 (50-160 Mb) and Chr6 (50-90 Mb). The analysis also highlighted loci where the BALB/c allele was negatively correlated with arthritis severity (not shown). This was the case on Chr5 and Chr1, quite close to a region that is positively associated with severity.
The second analysis used more conventional family-based tests, treating the entire pedigree as a group of families, and searching for linkage using qtdt software (19). Fig. 1c presents the regions with logarithm of odds scores >2 found on Chrs1, -2, -6, and -7, with a particularly robust association on Chr2. Similar association signals were observed for both time of onset and degree of inflammation.
Thus, the two analytical methods produced fairly concordant results, and the whole-genome screen identified prominent regions of significant linkage on Chr2 and Chr6, with the possibility of secondary regions on Chr1 and Chr7.
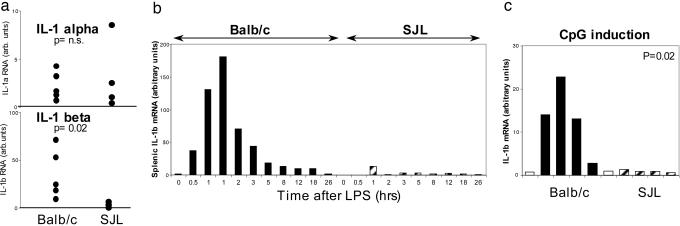
Il1b as a Candidate Gene for the Chr2 Region? Animal models of arthritis depend, to varying degrees, on one of the major inflammatory cytokines, TNFα and IL-1 (20). We previously showed that the K/BxN model requires IL-1, because mice deficient in the IL-1 receptor are refractory to disease (21). The two principal members of the IL-1 family, Il1a and Il1b, are encoded within 30 kb of each other, within the peak of the interval on Chr2 (129 Mb), and thus made immediate suspects for the major QTL on this chromosome. Hence, we measured the expression of both cytokines in BALB/c and SJL splenocytes (resting or after in vivo LPS challenge) by quantitative real-time PCR. There was a striking difference in levels of IL-1β transcripts that were induced up to 20-fold more in BALB/c than in SJL spleen cells, but the difference in IL-1α transcripts was less marked (Fig. 2a). Time-course experiments (Fig. 2b) indicated that the peak of IL-1β mRNA appeared at about the same time in both strains but was more pronounced in BALB/c than in SJL splenocytes (IL-1α mRNA followed the same kinetics). Because the induction of the IL-1α mRNA message did not show such divergence between the strains, the difference in IL-1β inducibility was unlikely to result from a variable sensitivity to LPS, as exists in the mutant C3H/HeJ mouse (22). This notion was confirmed in Fig. 2c by showing that IL-1β mRNA was also induced more strongly in BALB/c than in SJL mice after challenge with an independent inducer, a CpG dinucleotide-containing oligonucleotide that activates IL-1 transcription via the TLR9 receptor (23). This different response was specific to IL-1, because no such differential was seen in spleen IFNγ and TNFα mRNA levels in BALB/c vs. SJL after LPS challenge (data not shown).
Fig. 2.
The Il1b gene is differentially induced in BALB/c and SJL mice. (a) mRNA levels of the Il1a and Il1b candidate genes measured by quantitative real-time PCR in spleen RNA, from BALB/c, and SJL mice 6 h after i.p. injection of LPS; levels are standardized relative to hypoxanthine guanine phosphoribosyl transferase (HPRT) transcripts [units defined as 2exp(Δ-ΔCT)]. Each dot represents an individual mouse. (b) Time course of IL-1β transcripts after induction by i.p. LPS; each bar represents an individual mouse (data are representative of two similar experiments). (c) IL-1β mRNA levels (quantitative PCR) 6 h after injection of CpG oligonucleotide; white bars, untreated mice. P value from Student's t test.
These results suggested that the explosive responsiveness of BALB/c mice to K/BxN serum might be due, at least in part, to a much greater inducibility of the Il1b gene. To search for underlying sequence variation, we performed DNA sequence analysis on the Il1b locus of the BALB/c and SJL genomes. Recent findings indicate that genetic variation among inbred strains is dichotomous: some regions show very high divergence between any two strains, on the order of five SNPs per kb, whereas other areas have a far lower rate of sequence divergence (<0.05 SNP per kb) (24-26). This dichotomy is thought to reflect the mixed input from two main ancestral mouse genomes before the fixation of inbred mouse strains over the last century (27), such that any inbred strain is a mosaic of genetic material of domesticus and non-domesticus origin. We sequenced all Il1b exons and substantial stretches of promoter/intron sequences (Fig. 3a), focusing on the intronic regions conserved between human and mouse genomes. A number of differences were found, involving 39 of 4,011 bp sequenced, clearly in the range of high diversity. None of the polymorphisms alter the protein sequence, most belonging to flanking or intronic regions. The SJL sequence was always identical to, and BALB/c always different from, the database B6 sequence (in contrast, sequencing of a few segments of IL1a showed a far more intricate pattern of variation, with no simple dichotomy).
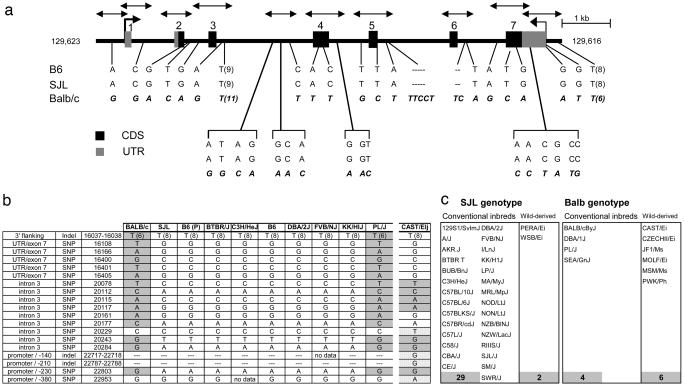
Fig. 3.
Sequence analysis of the IL-1β gene in inbred mouse strains. (a) Sequencing of the Il1b gene in three inbred strains. The intron-exon structure of the gene is schematized, with its position on mouse Chr2; double-pointed arrows indicating the PCR amplicons sequenced after amplification from tail DNA of Balb/cJ, SJL/J, and C57BL/6J mice. The bases at variable positions for each of the three strains are shown. (b) Additional sequencing in a broader panel of strains. Three of the Il1b amplicons shown in a were sequenced in DNA from a broader panel of inbred strains. White boxes, SJL haplotype; shaded, BALB/c haplotype; dotted, “private” variability of the non-domesticus strain CAST/EiJ; B6(P), reference sequence from public databases. (c) Genotyping for the -230 SNP of the IL-1b promoter, in the full panel of strains from the Mouse Phenome Project (plus DBA/1J and MRL/MpJ). The strains are separated into conventional and wild-derived inbreds.
The mosaic genomes of inbred mice, because of the patchwork nature of their genetic variation, offer great potential for QTL mapping (24-26). We thus asked whether the two haplotypes observed in the BALB/c vs. the SJL and B6 strains were conserved as a whole in other inbred strains, or whether strains carrying intragenic recombinant sequences might pinpoint the difference in inducibility. PCR amplicons located at each extremity and in the middle of the Il1b gene were sequenced in a set of eight inbred strains chosen for their distinct genealogies (27). The sequences partitioned cleanly into either BALB/c or SJL haplotypes; all had exactly the same sequence as either BALB/c or SJL at all positions sequenced (Fig. 3b). Only the wild-derived non-domesticus CAST/EiJ mouse (Mus musculus castaneus) had a more complex sequence. The SJL haplotype seemed more frequent, because only the PL/J strain carried the BALB/c haplotype. To better estimate the frequency of these haplotypes among strains, we determined the genotype of one diagnostic SNP (at -230 in the promoter region) in a broader panel of 42 strains constituting the reference panel of the Mouse Phenome Project (28). This analysis confirmed the skewed distribution of the Il1b haplotype (Fig. 3c). The BALB/c genotype was found in the majority of (6/8) “wild-derived” non-domesticus strains in the panel. In contrast, most conventional inbred strains carry the SJL haplotype; only four of them had the BALB/c haplotype: BALB/c, PL/J, SEA/J, and DBA/1. The identity of these exceptions is suggestive. DBA/1 is the prototypical high susceptibility strain in the CIA model, indicating that the present observations may apply to CIA too; SEA/GnJ derives from a BALB/cJ (27); PL/J is known to share similarities to wild-derived inbred strains in haplotype and disease susceptibility, notably West Nile Virus (29). Thus, the BALB/c haplotype at Il1b and the explosive transcriptional induction it seems to confer are quite frequent among wild-derived mice but appear to have been counterselected during fixation of the conventional inbred strains.
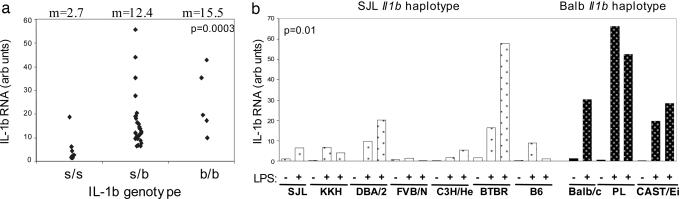
At this point, it was important to establish whether there was a direct relationship between the unusual Il1b haplotype of BALB/c mice, its high inducibility, and susceptibility to arthritis. The difference in Il1b gene transcription in BALB/c and SJL mice could theoretically be due to polymorphism at the locus itself or to differences in factors that regulate its expression, affecting up-stream events in the cascade of IL-1 induction. We tested whether IL-1β mRNA levels segregated with the Il1b locus in a set of F2 intercross mice. The mice were genotyped and challenged with LPS, and IL-1β responsiveness in spleen mRNA was measured by real-time PCR. A clear relationship was found (Fig. 4a), because mice with the BALB/c allele at Il1b showed higher levels of splenic IL-1β mRNA and serum cytokine than those with the SJL-derived allele. We extended this observation by testing the panel of eight inbred mouse strains whose Il1b gene had been partially sequenced (Fig. 4b). Here again, mice that shared the BALB/c allele (PL and CAST) showed higher IL-1β mRNA expression levels than did strains with the SJL allele (with the marked exception of BTBR). Thus, the strain difference in IL-1β expression is associated, at least in part, with the Il1b locus itself, in both BALB/c vs. SJL segregants and in a diverse panel of inbred strains.
Fig. 4.
Higher inducibility of the IL-1β gene in the BALB/c haplotype. (a) Il1b mRNA was measured by real-time PCR in spleen RNA 6 h after LPS in a cohort of (BALB/c × SJL) F2 mice genotyped for the -230 promoter region SNP. P values from a Kruskal-Wallis rank sum test. (b) A panel of inbred strains, carrying either the SJL or BALB/c allele at Il1b, were tested by LPS injection. Il1b transcripts were measured (spleen RNA, 4 h postLPS) in control (-) and LPS-injected (+) mice. P values are from Student's t test.
Finally, we asked whether the Il1b haplotype correlates with responsiveness to K/BxN arthritis in mouse strains in general. We had previously analyzed the susceptibility of several inbred strains, and Fig. 5a collates these data together with the newly determined Il1b genotype; quite clearly, the BALB/c allele at Il1b correlates with high disease susceptibility. This analysis was extended by comparing the course of arthritis in additional strains of mice after transfer of limiting doses of K/BxN serum (Fig. 5b). Strains with the BALB/c haplotype all showed high responsiveness, whereas strains with the SJL haplotype responded more weakly as a group. Thus, the analysis relying on inbred strain variability confirmed and extended the results of the intercross between BALB/c and SJL mice: the Il1b locus is associated with aggressive arthritis.
Discussion
The experiments described in this paper, initiated by in silico modeling of mouse genealogies, have resulted in the identification of Il1b as a gene whose variation strongly conditions mouse susceptibility to Ab-mediated arthritis. How worthwhile was the selective breeding approach in this endeavor? In one sense, it was successful; the major regions of significant linkage were detected, and <100 mice were genotyped, substantially less than would be required in conventional F2 crosses. But, quite clearly, the selection did not work as smoothly in breathing as in virtual mice (see Supporting Text). The rate of fixation of neutral markers largely matched the expected, but the association scores were significantly lower than had been hoped for, and it is only on the strongest QTL that significance could be established. Part of the discrepancy between the expected and the obtained can be attributed to uneven breeding and part to a higher phenotypic variability than had been modeled. Furthermore, the distribution of phenotypes at successive generations also indicated that more loci were at play than had been simulated, with a likely occurrence of dominant susceptibility loci from SJL. Quite clearly, the study did not exhaust all of the relevant polymorphism in the BALB/c vs. SJL combination but identified the major regions on Chr2 and Chr6.
As in previous studies on the K/BxN and other arthritis models (reviewed in refs. 6 and 11), the picture that emerged is one where a complex array of genetic polymorphisms determines the differential responsiveness of BALB/c and SJL mice, one that differs from previous results. We detected no sign of association with the region of the Ncf1 gene (10) or with the C5 locus (8). The region of association on Chr6 may be related to the Cia6 and Pgia19 loci identified in previous studies (30, 31); it may be relevant that a broad peak of association with CIA was detected on Chr6, suggesting the possibility of two independent QTL, compatible with the distribution observed here. Previous studies did not uncover a major effect of the Il1b locus, as observed herein. This discrepancy is readily explained by the strains used; the previously published experiments used a BALB/c vs. DBA/1 comparison, precisely the strains that shared the rare overinducible Il1b allele identified here (as discussed below, probably not by coincidence). The present results also differ from those of our previous analysis in a (B6x NOD)F2 intercross, which brought forth loci on Chr1 and proximal Chr2 (at C5) (8). The Chr1 QTL found in the two studies might be the same (our recent results point to a complex organization with two independent subregions, both of which are compatible with the QTL found in the BalbxSJL combination), but the C5 and Il1b loci are highlighted in only one of the two strain pairs. These different results illustrate the importance of testing multiple strain combinations (or out-bred crosses) to fully capture relevant genetic variation.
Definition of a susceptibility interval on Chr2 was followed by candidate gene analysis that resulted in the identification of a polymorphism in Il1b and consequently a difference in IL-1β mRNA induction as a major determinant of genetic susceptibility to K/BxN arthritis. We cannot exclude that other closely linked loci on Chr2 are involved as well, but Il1b had a major influence on the severity of arthritis in the backcross/intercross cohort and in inbred lines. This assignment fits well with the known role of IL-1 in this arthritis model; mice carrying knockout mutations in the IL-1 receptor locus or in the combined Il1a/Il1b loci were fully resistant to K/BxN arthritis (21, 32). An impact of quantitative variations in IL-1 expression on the unfolding of arthritis is also very consistent with the disease that developed when IL1-RA, the natural antagonist of IL-1 activity, was eliminated (ref. 33; interestingly, the phenotype was detected on the BALB/c but not on the C57BL/6 background) and with the protection against CIA afforded by overexpression of IL1-RA as a transgene (34). In fact, many interventions that modify IL-1 availability have had an impact on most mouse models of arthritis (reviewed in ref. 20).
Sequence analysis revealed variation at many positions of the BALB/c and SJL Il1b genes. These variations did not affect the protein sequence, occurring rather in promoter and intron sequences, and may thus influence mRNA levels, consistent with the very different inducibility of the Il1b haplotypes by ligands of Toll-like receptors. The time-course analysis suggested a difference not in mRNA stability but instead in transcriptional or splicing efficiency, because the timing of induction was essentially identical in the two strains. We had hoped that the analysis of inbred strains would allow us to determine the relevant variation by providing recombinant loci for SNP/phenotype correlation. This proved impossible, because the whole gene was transmitted en bloc in all conventional inbred strains sequenced, i.e., they all carried either the full BALB/c or the full SJL haplotype. The assignment of causal differences will require old-fashioned promoter dissection.
The BALB/c haplotype at the Il1b locus showed a strikingly imbalanced distribution among mouse strains; it was common in non-domesticus wild-derived mice but rare among the conventional inbreds, most of which were derived from mouse-fancier colonies of the late 1800s (27). Such a shift in haplotype distribution suggests that the BALB/c haplotype was for some reason subject to negative selective pressure during this domestication period. It is possible that the higher activity of the BALB/c Il1b locus is advantageous in wild mice, permitting them to respond to a particular set of pathogens encountered in the varied environments in which they reside, but that the activity of the Il1b locus also needs to be balanced at the population level by an allele that confers lower IL-1 responses. In the sheltered environment of pet or laboratory colonies, the lower IL-1 response might have been favored. Alternatively, this cytokine may somehow participate in the expression of behavioral traits, one set of phenotypes that most obviously distinguishes domesticated from wild mice and that must have been strongly selected; IL-1β has been implicated in murine behavioral traits in many studies (e.g., refs. 35 and 36).
Although variation at the Il1b locus has a strong effect on the IL-1β response, it is not the only determinant; even when homozygous for the BALB/c Il1b haplotype, mice of the (BALB/c × SJL), the H8 cohort did not recapitulate the typical BALB/c expression levels, suggesting that other elements from the SJL genome introduced in the pedigree may dampen the full expressivity of the Il1b haplotype. Similarly in inbred lines, whereas there was a correlation between the Il1b haplotype and mRNA inducibility, it was not absolute, and there were some clear exceptions (the high responsiveness of the SJL-like BTBR/J strain, for instance). Thus, other genes must influence the transcriptional activity conditioned by local variation at the Il1b locus. Genes encoding transcription or signaling factors that activate or silence the Il1b promoter/enhancer could modify the expression phenotype and may correspond to some of the other influences revealed in the genome scan.
The convergence of results from the genetic mapping and the inbred strain analyses demonstrate that genetic polymorphism at Il1b is an important determinant of arthritis severity in a broad sense, not merely when comparing the BALB/c and SJL inbred strains. What might be the relevance to arthritis in outbred humans? There is already evidence of suggestive association between RA severity and SNPs in genes encoding members of the IL-1 gene family (e.g., refs. 37-39). Although the human IL-1 family region obviously requires more profound analysis, these data do suggest that the role of IL1b in controlling arthritis development in mice may well have a parallel in humans. It will be of great interest to ascertain whether genetic variation in the human species harbors the same range of IL-1 expression levels found in mice, and how this diversity distributes among human populations and diseases.
Supplementary Material
Acknowledgments
We thank Q. M. Pham, T. Rogers, V. Bruklich, and J. Gile for help with the mice; G. Losyev for flow cytometry; the Arthritis Group for inspiring discussions; and S. Deveau (The Jackson Laboratory) for DNA. This work was supported by grants from the National Institutes of Health and by William T. Young Chair funds (to D.M. and C.B.), Joslin Diabetes Center's National Institute of Diabetes and Digestive and Kidney Diseases-funded Diabetes and Endocrinology Research Core, and the Mouse Phenome Project. K.O. received postdoctoral support from the Uehara Memorial Foundation and the National Arthritis Research Foundation.
Author contributions: K.O., J.R., D.M., and C.B. designed research; K.O., A.J., A.O.-L., P.D., M.R., W.B., M.B., and A.P. performed research; K.O., A.J., A.O.-L., P.D., M.R., W.B., M.B., A.P., M.L., D.M., and C.B. analyzed data; and K.O., D.M., and C.B. wrote the paper.
Abbreviations: RA, rheumatoid arthritis; CIA, collagen-induced arthritis; QTL, quantitative trait loci; maxAT, maximum ankle thickening.
References
- 1.Huizinga, T. W., Pisetsky, D. S. & Kimberly, R. P. (2004) Arthritis Rheum. 50, 2066-2071. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen, P. K. (2003) Clin. Immunol. 107, 1-9. [DOI] [PubMed] [Google Scholar]
- 3.Jirholt, J., Lindquist, A.-K. B. & Holmdahl, R. (2001) Arthritis Res. 3, 87-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wooley, P. H., Luthra, H. S., Stuart, J. M. & David, C. S. (1981) J. Exp. Med. 154, 688-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frankel, W. N. (1995) Trends Genet. 11, 471-477. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths, M. M. & Remmers, E. F. (2001) Immunol. Rev. 184, 172-183. [DOI] [PubMed] [Google Scholar]
- 7.Olofsson, P., Holmberg, J., Tordsson, J., Lu, S., Akerstrom, B. & Holmdahl, R. (2003) Nat. Genet. 33, 25-32. [DOI] [PubMed] [Google Scholar]
- 8.Ji, H., Gauguier, D., Ohmura, K., Gonzalez, A., Duchatelle, V., Danoy, P., Garchon, H. J., Degott, C., Lathrop, M., Benoist, C., et al. (2001) J. Exp. Med. 194, 321-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson, A. C., Sundler, M., Kjellen, P., Johannesson, M., Cook, A., Lindqvist, A. K., Nakken, B., Oldstad, A. I., Jonsson, R., Alarcon-Riquelme, M., et al. (2001) Eur. J. Immunol. 31, 1847-1856. [DOI] [PubMed] [Google Scholar]
- 10.Hultqvist, M., Olofsson, P., Holmberg, J., Backstrom, B. T., Tordsson, J. & Holmdahl, R. (2004) Proc. Natl. Acad. Sci. USA 101, 12646-12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmdahl, R. (2003) J Autoimmun. 21, 99-103. [DOI] [PubMed] [Google Scholar]
- 12.Kouskoff, V., Korganow, A.-S., Duchatelle, V., Degott, C., Benoist, C. & Mathis, D. (1996) Cell 87, 811-822. [DOI] [PubMed] [Google Scholar]
- 13.Korganow, A.-S., Ji, H., Mangialaio, S., Duchatelle, V., Pelanda, R., Martin, T., Degott, C., Kikutani, H., Rajewsky, K., Pasquali, J.-L., et al. (1999) Immunity 10, 451-461. [DOI] [PubMed] [Google Scholar]
- 14.Monach, P. A., Benoist, C. & Mathis, D. (2004) Adv. Immunol. 82, 217-248. [DOI] [PubMed] [Google Scholar]
- 15.Lefevre, C., Jobard, F., Caux, F., Bouadjar, B., Karaduman, A., Heilig, R., Lakhdar, H., Wollenberg, A., Verret, J. L., Weissenbach, J., et al. (2001) Am. J. Hum. Genet. 69, 1002-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zucchelli, S., Holler, P., Yamagata, T., Roy, M., Benoist, C. & Mathis, D. (2005) Immunity 22, 385-396. [DOI] [PubMed] [Google Scholar]
- 17.Garnett, I. & Falconer, D. S. (1975) Genet. Res. 25, 45-57. [DOI] [PubMed] [Google Scholar]
- 18.Keightley, P. D. & Bulfield, G. (1993) Genet. Res. 62, 195-203. [DOI] [PubMed] [Google Scholar]
- 19.Abecasis, G. R., Cardon, L. R. & Cookson, W. O. (2000) Am. J. Hum. Genet. 66, 279-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Berg, W. (2001) Arthritis Res. 3, 18-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji, H., Pettit, A., Ohmura, K., Ortiz-Lopez, A., Duchatelle, V., Degott, C., Gravallese, E. M., Mathis, D. & Benoist, C. (2002) J. Exp. Med. 196, 77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Van Huffel, C., Du, X., Birdwell, D., Alejos, E., Silva, M., Galanos, C., et al. (1998) Science 282, 2085-2088. [DOI] [PubMed] [Google Scholar]
- 23.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K., et al. (2000) Nature. 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 24.Wade, C. M., Kulbokas, E. J., 3rd, Kirby, A., Zody, M. C., Mullikin, J. C., Lander, E. S., Lindblad-Toh, K. & Daly, M. J. (2002) Nature 420, 574-578. [DOI] [PubMed] [Google Scholar]
- 25.Wiltshire, T., Pletcher, M. T., Batalov, S., Barnes, S. W., Tarantino, L. M., Cooke, M. P., Wu, H., Smylie, K., Santrosyan, A., Copeland, N. G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 3380-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yalcin, B., Fullerton, J., Miller, S., Keays, D. A., Brady, S., Bhomra, A., Jefferson, A., Volpi, E., Copley, R. R., Flint, J., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 9734-9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beck, J. A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J. T., Festing, M. F. & Fisher, E. M. (2000) Nat, Genet, 24, 23-25. [DOI] [PubMed] [Google Scholar]
- 28.Grubb, S. C., Churchill, G. A. & Bogue, M. A. (2004) Bioinformatics 20, 2857-2859. [DOI] [PubMed] [Google Scholar]
- 29.Mashimo, T., Lucas, M., Simon-Chazottes, D., Frenkiel, M. P., Montagutelli, X., Ceccaldi, P. E., Deubel, V., Guenet, J. L. & Despres, P. (2002) Proc. Natl. Acad. Sci. USA 99, 11311-11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, H. T., Jirholt, J., Svensson, L., Sundvall, M., Jansson, L., Pettersson, U. & Holmdahl, R. (1999) J. Immunol. 163, 2916-2921. [PubMed] [Google Scholar]
- 31.Adarichev, V. A., Valdez, J. C., Bardos, T., Finnegan, A., Mikecz, K. & Glant, T. T. (2003) J. Immunol. 170, 2283-2292. [DOI] [PubMed] [Google Scholar]
- 32.Choe, J. Y., Crain, B., Wu, S. R. & Corr, M. (2003) J. Exp. Med. 197, 537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horai, R., Saijo, S., Tanioka, H., Nakae, S., Sudo, K., Okahara, A., Ikuse, T., Asano, M. & Iwakura, Y. (2000) J. Exp. Med. 191, 313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer, G., Talabot-Ayer, D., Szalay-Quinodoz, L., Maret, M., Arend, W. P. & Gabay, C. (2003) Eur. J. Immunol. 33, 434-440. [DOI] [PubMed] [Google Scholar]
- 35.Merali, Z., Brennan, K., Brau, P. & Anisman, H. (2003) Psychopharmacology 165, 413-418. [DOI] [PubMed] [Google Scholar]
- 36.McCulley, M. C., Day, I. N. & Holmes, C. (2004) Am. J. Med. Genet. 124B, 50-53. [DOI] [PubMed] [Google Scholar]
- 37.Barrera, P., Faure, S., Prud'homme, J. F., Balsa, A., Migliorini, P., Chimenti, D., Radstake, T. R., Van de Putte, L. B., Pascual-Salcedo, D., Westhovens, R., et al. (2001) Clin. Exp. Rheumatol. 19, 709-714. [PubMed] [Google Scholar]
- 38.Buchs, N., Di Giovine, F. S., Silvestri, T., Vannier, E., Duff, G. W. & Miossec, P. (2001) Genes Immun. 2, 222-228. [DOI] [PubMed] [Google Scholar]
- 39.Genevay, S., Di Giovine, F. S., Perneger, T. V., Silvestri, T., Stingelin, S., Duff, G. & Guerne, P. A. (2002) Arthritis Rheum. 47, 303-309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.