Abstract
To probe the evolution and phylogeny of Listeria monocytogenes from defined host species and environments, L. monocytogenes isolates from human (n = 60) and animal (n = 30) listeriosis cases and food samples (n = 30) were randomly selected from a larger collection of isolates (n = 354) obtained in New York State between 1999 and 2001. Partial sequencing of four housekeeping genes (gap, prs, purM, and ribC), one stress response gene (sigB), and two virulence genes (actA and inlA) revealed between 11 (gap) and 33 (inlA) allelic types as well as 52 sequence types (unique combination of allelic types). actA, ribC, and purM demonstrated the highest levels of nucleotide diversity (π > 0.05). actA and inlA as well as prs and the hypervariable housekeeping genes ribC and purM showed evidence of horizontal gene transfer and recombination. actA and inlA also showed evidence of positive selection at specific amino acid sites. Maximum likelihood phylogenies for all seven genes confirmed that L. monocytogenes contains two deeply separated evolutionary lineages. Lineage I was found to be highly clonal, while lineage II showed greater diversity and evidence of horizontal gene transfer. Allelic types were exclusive to lineages, except for a single gap allele, and nucleotide distance within lineages was much lower than that between lineages, suggesting that genetic exchange between lineages is rare. Our data show that (i) L. monocytogenes is a highly diverse species with at least two distinct phylogenetic lineages differing in their evolutionary history and population structure and (ii) horizontal gene transfer as well as positive selection contributed to the evolution of L. monocytogenes.
Listeria monocytogenes is a facultative intracellular human food-borne and animal pathogen that may cause severe invasive disease in immunocompromised individuals (46). Clinical manifestations of invasive human listeriosis include meningitis, encephalitis, late-term spontaneous abortion, and septicemia. The vast majority of human listeriosis infections (99%) are thought to be food borne (28), and L. monocytogenes has been isolated from various raw and ready-to-eat foods (14, 57). In animals, L. monocytogenes has been associated with invasive infections of more than 40 species of mammals and birds. L. monocytogenes is able to survive and multiply outside mammalian hosts for extended periods of time and can endure environmental stress conditions that kill many other food-borne bacterial pathogens. Consequently, L. monocytogenes has been isolated from a variety of environmental sources, including surface water, soil, sewage, vegetation, and food-processing plants, and is often considered ubiquitous in nature (10). The ability of a pathogen to infect multiple host species and colonize diverse environments may play an important role in the transmission and pathogenesis of food-borne diseases (61). We thus selected L. monocytogenes as a model organism to study the evolution of food-borne and zoonotic pathogens with transmission dynamics that involve complex interactions between the pathogen, diverse environments, and multiple host species.
Traditionally, L. monocytogenes has been differentiated into 13 serotypes; only four of these serotypes (1/2a, 1/2b, 1/2c, and 4b), however, have been reported to cause a large majority of human listeriosis cases (reviewed in reference 58). Molecular subtyping methods (e.g., automated ribotyping and pulsed-field gel electrophoresis) allow for more sensitive discrimination of L. monocytogenes subtypes than phenotypic methods (e.g., serotyping) and have provided an initial understanding of the population structure of this pathogen (58). Data generated with most molecular subtyping methods have shown that L. monocytogenes isolates can be grouped into two major genetic divisions or lineages, termed lineages I and II (reviewed in reference 58). In addition, some studies have described the existence of a third, less common, L. monocytogenes lineage (lineage III), which appears to be predominantly associated with animals (19, 60). While a variety of different nomenclatures have been used to designate these L. monocytogenes lineages, we will refer to these lineages as I, II, and III following the designations previously used by our group (58, 60) and others (7, 55). Lineage I predominantly includes serotypes 1/2b, 3b, 3c, and 4b strains, while lineage II primarily includes serotypes 1/2a, 1/2c, and 3a (30). Interestingly, previous reports have shown that lineage I strains are significantly overrepresented among human clinical listeriosis cases compared to their prevalence among animal listeriosis cases and contaminated foods (15, 19, 34). On the other hand, lineage II strains show a significantly higher prevalence among food isolates and animal clinical cases than among human listeriosis cases (15, 19). In addition, lineage I isolates appear to have significantly greater pathogenic potential, as determined by their ability to spread to neighboring host cells in a cell culture plaque assay, than lineage II isolates (15, 34, 60). Thus, there is increasing consensus that lineage I strains may represent a human host-adapted lineage, while lineage II strains may represent an environmentally adapted lineage (58). While initial comparative genomics studies have shown considerable differences in genome content between lineage I and II isolates (8, 65), an improved understanding of the molecular phylogeny and evolution at the nucleotide level of these L. monocytogenes lineages is needed to provide a better understanding of the transmission, pathogenesis, and niche adaptation of this food-borne pathogen.
Most molecular epidemiology studies on L. monocytogenes have used DNA banding pattern-based subtyping methods (e.g., pulsed-field gel electrophoresis and ribotyping). However, these methods are difficult to standardize, and the data generated are not suitable for phylogenetic and evolutionary analyses (59). Multilocus sequence typing (MLST), on the other hand, represents a reliable high-throughput universal subtyping method that provides data appropriate for phylogenetic and evolutionary analyses. Similar to multilocus enzyme electrophoresis, MLST surveys several conserved loci, such as housekeeping genes, that diversify slowly and are not heavily influenced by evolutionary forces other than point mutations (i.e., positive selection and recombination) (26). Although preliminary L. monocytogenes MLST studies have been reported (4, 29, 42, 66), these studies used convenient and nonrepresentative sample sets, which were also usually small (<50 isolates, except for reference 42) and predominantly (except for reference 29) focused on development of subtyping methods. While Ward et al. (55) also recently reported a phylogenetic analysis of L. monocytogenes based on DNA sequence data for virulence genes located in the prfA virulence gene island, analysis of potentially horizontally transferred and positively selected genes in a single location does not provide for an accurate representation of the evolution of a particular group of organisms and allows conclusions on the evolution of only the specific virulence genes sequenced.
In order to overcome some of the limitations of previous MLST studies on L. monocytogenes, such as the lack of studies on a widely and randomly collected set of L. monocytogenes isolates (4, 29), we assembled a representative, geographically matched set of 120 L. monocytogenes isolates from humans and animals with clinical listeriosis as well as from foods. These isolates were characterized by partial sequencing of four housekeeping genes, one stress response gene, and two key L. monocytogenes virulence genes. Our data show that L. monocytogenes is a highly diverse species with at least two deeply separated evolutionary lineages, which have different population structures. We also showed that horizontal gene transfer as well as positive selection contributed to the evolution of L. monocytogenes present in the food continuum. This knowledge provides a critical framework to allow differentiation and characterization of L. monocytogenes strains that may have evolved to infect human hosts (i.e., “host-adapted strains”) and those that may have evolved both to survive under environmental stress conditions and to infect various hosts (i.e., “generalist strains”).
MATERIALS AND METHODS
L. monocytogenes isolates.
A total of 120 geographically and temporally matched L. monocytogenes isolates were selected from 354 human and animal clinical and food isolates collected between January 1999 and December 2001 in New York State as part of an ongoing study to probe the epidemiology and ecology of L. monocytogenes. These isolates were selected to represent the L. monocytogenes diversity associated with different sources in a given geographic region. Human isolates were obtained from the New York State Department of Health (NYSDOH) and the New York City Department of Health and Mental Hygiene (NYCDOHMH) as previously described (43). Food isolates were obtained from the New York State Department of Agriculture and Markets, which performs routine surveillance for L. monocytogenes in food products as previously described (43). Animal clinical isolates were obtained from the New York State Veterinary Diagnostic Laboratory. All isolates represent human and animal cases that occurred in New York State or foods that were collected from retail operations or processing plants located in New York State.
The 120 L. monocytogenes isolates used in this study were selected to include 60 isolates from human listeriosis cases (30 isolates from New York City residents [isolates obtained from the NYCDOHMH] and 30 isolates from residents of New York State excluding residents of New York City [isolates obtained from the NYSDOH]), 30 isolates from animal listeriosis cases, and 30 food isolates. Furthermore, the isolates were selected to evenly represent the 3-year time period from 1999 to 2001. More specifically, 40 isolates were selected to represent each year of this 3-year time period, so that the 40 isolates from a specific year included 10 human isolates from New York City, 10 human isolates from the remainder of New York State, 10 animal isolates, and 10 food isolates. All isolates were selected from the larger collection of 354 isolates using random-number tables; prior genetic characterization and lineage information for isolates were not used as factors in isolate selection. Human clinical isolates from New York City and the remainder of New York State (representing smaller cities and rural areas) were included to capture the L. monocytogenes diversity that might be associated with urban and rural populations; overrepresentation of human isolates should not affect the conclusions of our study, since no comparisons between diversity among different sources were performed.
All 120 L. monocytogenes isolates had previously been characterized by automated EcoRI ribotyping (41); EcoRI ribotype data were used to classify isolates into three genetic lineages (lineages I, II, and III) as previously described (60). Detailed information on these isolates is available in supplemental Table S1 (at http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary.txt) and in the PathogenTracker database (www.pathogentracker.net).
Selection of L. monocytogenes genes for DNA sequencing.
For all 120 L. monocytogenes isolates, partial DNA sequencing of seven genes (Table 1), including four housekeeping genes (gap, prs, purM, and ribC), two virulence genes (inlA and actA), and one stress response gene (sigB), was performed. Genes were also selected to represent distinct physical locations along the L. monocytogenes chromosome (Table 1) to sample the overall chromosomal diversity of L. monocytogenes (26). The virulence genes inlA and actA were selected because these genes are critical for the L. monocytogenes intracellular life cycle. inlA encodes internalin A (InlA), which is critical for L. monocytogenes ' ability to invade host cells that express E-cadherin, and actA encodes the ActA protein, which allows L. monocytogenes to spread to neighboring host cells during infection (reviewed in reference 40). Gene fragments between 561 and 771 bp were sequenced to allow sequencing with a single forward and reverse sequencing reaction according to standard MLST practices (26). The gene encoding the 16S RNA was not included in our sequencing approach, since it was previously shown to display very limited diversity within the L. monocytogenes species (5).
TABLE 1.
Description of L. monocytogenes genes sequenced
| Gene | Positiona | Length (in bp) | Gene product | Functional category |
|---|---|---|---|---|
| actA | 209.5 | 1,920 | Actin assembly inducer protein | Cell surface protein (virulence) |
| gap | 2532 | 1,011 | Glyceraldehyde 3-phosphate dehydrogenase | Energy pathway |
| inlA | 454.5 | 2,403 | Internalin A | Cell surface protein (virulence) |
| prs | 202.6 | 957 | Phosphoribosyl pyrophosphate synthetase | Translation, ribosomal structure, and biogenesis |
| purM | 1840 | 1,050 | Phosphoribosylaminoimidazole synthetase | Nucleotide and nucleic acid metabolism |
| ribC | 1358 | 945 | Riboflavin kinase and FADb synthase | Coenzyme and prosthetic group metabolism |
| sigB | 930.7 | 780 | Alternative sigma factor | Transcription initiation |
Relative position in kbp, as assigned in the full genome sequence for L. monocytogenes EGD-e (12).
FAD, flavin adenine dinucleotide.
PCR amplification and DNA sequencing.
PCR amplification of approximately 600- to 800-bp fragments of the genes detailed above was performed using Thermus aquaticus DNA polymerase, 1× PCR buffer, MgCl2 at a final concentration of 1.5 mM, and deoxynucleotide triphosphates at a total final concentration of 50 μM. Primers for PCR amplification and/or nucleotide sequencing and PCR amplification conditions are provided in supplemental Tables S2 and S3 (at http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary.txt). PCR products were purified using the Qiaquick PCR purification kit (QIAGEN, Inc., Valencia, CA). DNA concentrations of purified PCR products were estimated by comparing amplicon band intensities with band intensities of a DNA marker (pGEM; Promega, Madison, WI) with known DNA concentrations using LabImage software (Kapelan, Halle, Germany) or with a Bio-Rad fluorescent DNA quantification kit (Bio-Rad, Hercules, CA). DNA sequencing was performed by Cornell University's Bioresource Center (Ithaca, NY) or Macrogen, Inc. (Seoul, Korea) using PCR primers or internal sequencing primers (supplemental Table S2), Big Dye Terminator chemistry, and AmpliTaq-FS DNA polymerase. Sequencing reactions were run on either the ABI 3730xl or the ABI 3700 DNA analyzer. Nucleotide sequences were proofread and aligned with Seqman and Megalign (part of the DNAStar software package; Lasergene, Madison, WI), respectively. Multiple-sequence alignments were modified to include only amino acid-coding regions. Sequence data for all isolates are available through the PathogenTracker database (www.pathogentracker.net).
Descriptive analysis of sequence data.
Nucleotide diversity (π, average pairwise nucleotide difference/site; and k, average pairwise nucleotide difference/sequence), number of polymorphic sites, number of mutations, number of alleles, G+C content, Tajima's D test for neutrality, number of synonymous mutations (S), number of nonsynonymous mutations (N), and the dN/dS ratios (the number of nonsynonymous substitutions/nonsynonymous site [dN] to the number of synonymous substitutions/synonymous site [dS]) with a Jukes and Cantor correction were calculated using DnaSP version 3.99 (41). Allelic types, defined as a unique combination of polymorphisms within an individual gene, and sequence types (STs), defined by a unique combination of allelic types within the concatenated seven-gene data set, were also assigned using DnaSP.
Average uncorrected percent pairwise nucleotide differences were determined using the pairdiff procedure in PAUP* (50) and separate individual gene alignments containing all isolates, lineage I isolates only, and lineage II isolates only. Average uncorrected percent pairwise nucleotide differences were also determined for sigB sequences from other Listeria species, including Listeria innocua (n = 48), Listeria seeligeri (n = 48), and Listeria welshimeri (n = 48) isolates obtained from environmental samples collected throughout New York State in 2001 and 2002 (B. D. Sauders and M. Wiedmann, unpublished data).
Analysis for evidence of conflicting phylogeny.
A compatibility method, implemented in the RETICULATE program (18), was used to detect evidence of reticulate evolution. Analyses were conducted using concatenated nucleotide alignments of the seven gene fragments sequenced for all L. monocytogenes isolates as well as for lineage I and lineage II isolates only. RETICULATE assesses the level of phylogenetic cohesiveness for phylogenetically informative sites both within and between genes.
Sawyer's test for recombination was performed using the GENECONV program (44). GENECONV default settings were used, and analyses were performed using individual gene alignments with a single isolate representing each unique allelic type.
Phylogenetic analysis.
MODELTEST (37) was used to optimize parameters to infer maximum likelihood phylogenetic trees in PAUP* (50). The likelihood ratio test (LRT) was used to determine the most likely model of DNA substitution for each L. monocytogenes gene; this test compares nested models of DNA substitution using hierarchical hypothesis testing to determine which model best describes the observed sequence data for each gene. The LRT statistic was calculated by 2 × log Δ where Δ = L0 (null model data)/L1 (alternative model data), L0 is the likelihood estimate for the simple model, and L1 is the likelihood estimate for the model with more free parameters. The degrees of freedom were determined by the difference in the number of free parameters between the null and alternative models, and the test statistic was approximated to a χ2 distribution to determine statistical significance (37).
Due to intensive computational requirements, the molecular clock hypothesis was tested using a randomly selected subset (n = 30) of sequences. More specifically, a maximum likelihood tree using optimized parameters determined by MODELTEST was constructed both with and without the molecular clock constraint for each L. monocytogenes gene, and the LRT (with n − 2 degrees of freedom) was used to test the null hypothesis that all representative isolates in the data set evolved according to a molecular clock.
Due to the large size of our data set, a single isolate representing each unique ST (n = 52) was used to infer phylogenetic trees for individual L. monocytogenes genes. Maximum likelihood trees were generated with PAUP*. Heuristic searches were performed using equal weights for all sites, and the tree bisection-reconnection branch-swapping algorithm was employed. Confidence measures for maximum likelihood tree branch points were generated by performing 100 bootstrap replicates using the heuristic search algorithm described above and parameters defined by MODELTEST. Maximum likelihood trees generated for housekeeping genes and sigB were rooted with homologous gene sequences from Bacillus subtilis (http://genolist.pasteur.fr/SubtiList/). Sequences for the lineage III isolates (n = 3), which been have shown to be distantly related to lineages I and II (60), were used to root phylogenetic trees for actA and inlA, since these genes do not have a B. subtilis homologue.
Analysis for positive selection.
Genes with an average dN/dS ratio (ω) of >0.1, as estimated by DnaSP, were further tested for the presence of positive selection among amino acid sites using the Phylogenetic Analysis by Maximum Likelihood (PAML) software program version 3.13 (62). The LRT was used to compare null models 0, 1, and 7 to alternative models 3, 2, and 8, respectively. Comparing models 0 and 3 tests for variation in selective pressure along a sequence. Comparing model 1 to model 2 and model 7 to model 8 tests the hypothesis that a gene contains amino acid sites that are under positive selection. While models 1 and 2 use a discrete number of site classes for ω values (allowing for two and three distinct ω values, respectively), models 7 and 8 use a discrete approximation of a continuous distribution of ω values (allowing for 10 and 11 distinct ω values, respectively). Models 2 and 8 identify codons that fall into a category where ω is >1 and therefore may be under positive selection. In models 2 and 8, an empirical Bayesian approach is used to calculate the posterior probability that an amino acid site fits in each site class and sites with a high posterior probability of falling into the class of ω of >1 are considered to be under positive selection (1, 32, 64). Models 2 and 8 were run twice with initial ω estimates less than 1 and greater than 1. The best likelihood estimate from these two runs was used for the LRT.
RESULTS
Descriptive analysis of nucleotide sequence data.
A concatenated data set based on partial sequencing of 561- to 771-bp fragments (Table 2) of four housekeeping genes, one stress response gene, and two virulence genes revealed 506 polymorphic sites and 52 STs, while a concatenated data set that included the four housekeeping genes and the one stress response gene sequenced revealed only 341 polymorphic sites and 46 STs among the 120 isolates characterized.
TABLE 2.
Descriptive analysis of nucleotide sequence data
| Gene | Length (% of full ORFa) | No. of:
|
G + C content (%) | π/siteb | kc | Tajima's D valued | No. of mutationse:
|
dN/dSf | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymor- phisms | Muta- tions | Alleles | Syn. | Nonsyn. | |||||||
| All isolates (n = 120) | |||||||||||
| actA | 561 (29.2) | 93 | 99 | 30 | 38.50 | 0.0543 | 30.44 | 2.10 | 41 | 51 | 0.32 |
| gap | 569 (56.3) | 9 | 9 | 11 | 39.90 | 0.0037 | 2.11 | 0.62 | 6 | 3 | 0.04 |
| inlA | 771 (32.1) | 72 | 73 | 33 | 41.10 | 0.0240 | 17.38 | 0.88 | 42 | 31 | 0.11 |
| prs | 633 (66.1) | 46 | 46 | 12 | 40.80 | 0.0192 | 12.16 | 1.29 | 46 | 0 | |
| purM | 714 (68.0) | 146 | 162 | 29 | 42.00 | 0.0587 | 41.90 | 1.27 | 126 | 27 | 0.04 |
| ribC | 639 (67.6) | 102 | 106 | 24 | 39.50 | 0.0562 | 35.90 | 2.64 | 97 | 9 | 0.02 |
| sigB | 666 (85.4) | 38 | 38 | 14 | 38.60 | 0.0219 | 14.59 | 3.23 | 37 | 1 | 0.02 |
| Lineage I isolates (n = 69) | |||||||||||
| actA | 561 | 15 | 15 | 14 | 39.00 | 0.0061 | 3.39 | 0.25 | 6 | 9 | 0.97 |
| gap | 569 | 2 | 2 | 3 | 40.00 | 0.0010 | 0.56 | 0.56 | 2 | 0 | |
| inlA | 771 | 17 | 17 | 12 | 41.10 | 0.0046 | 3.37 | −0.14 | 6 | 11 | 0.32 |
| prs | 633 | 8 | 8 | 5 | 40.10 | 0.0021 | 1.30 | −0.56 | 8 | 0 | |
| purM | 714 | 8 | 9 | 12 | 42.40 | 0.0034 | 2.43 | 0.79 | 6 | 3 | 0.02 |
| ribC | 639 | 18 | 18 | 8 | 39.10 | 0.0062 | 3.99 | 0.19 | 15 | 3 | 0.06 |
| sigB | 666 | 9 | 9 | 8 | 38.60 | 0.0036 | 2.40 | 0.74 | 9 | 0 | |
| Lineage II isolates (n = 48) | |||||||||||
| actA | 561 | 16 | 16 | 13 | 37.90 | 0.0064 | 3.56 | −0.04 | 9 | 7 | 0.47 |
| gap | 569 | 9 | 9 | 8 | 39.80 | 0.0046 | 2.61 | 0.80 | 6 | 3 | 0.09 |
| inlA | 771 | 52 | 52 | 18 | 41.40 | 0.0218 | 16.60 | 1.45 | 34 | 18 | 0.12 |
| prs | 633 | 18 | 18 | 4 | 41.10 | 0.0027 | 1.73 | −1.82 | 18 | 0 | |
| purM | 714 | 122 | 132 | 15 | 41.60 | 0.0614 | 43.84 | 1.48 | 108 | 24 | 0.07 |
| ribC | 639 | 85 | 87 | 13 | 40.30 | 0.0359 | 22.94 | 0.61 | 80 | 7 | 0.01 |
| sigB | 666 | 27 | 27 | 4 | 38.50 | 0.0036 | 2.42 | −2.00 | 26 | 1 | 0.01 |
ORF, open reading frame.
Average pairwise nucleotide difference per site.
Average pairwise nucleotide differences per sequence.
Tajima's D values significantly different from 0 (indicating deviation from standard neutral model) are marked in bold type.
Syn., synonymous; Nonsyn., nonsynonymous.
No dN/dS ratio was determined when no nonsynonymous mutations were observed.
STs and allelic types for all isolates are available in supplemental Table S1 (at http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary.txt). All STs were exclusive to either lineage I or II. The 120 isolates represented a total of 39 unique EcoRI ribotypes, and multiple STs were thus found within a given EcoRI ribotype. Burst analysis (data not shown) further showed that the same ribotype could be found in multiple clonal complexes or could represent multiple singletons. The virulence genes actA and inlA along with purM and ribC were the most discriminatory loci; these four loci segregated for 30, 33, 29, and 24 unique alleles, respectively (Table 2). Housekeeping and stress response gene fragments contained between 9 and 146 polymorphic sites, while actA and inlA contained 93 and 72 polymorphic sites, respectively (Table 2). The greatest nucleotide diversity (π > 0.05) was observed for actA, ribC, and purM (Table 2). A total of three different inlA nonsense mutations (inlA alleles 3, 21, and 26; supplemental Table S1), which are predicted to lead to a truncated form of internalin A protein, were identified; these mutations were found in one human (allele 3) and eight food isolates (allele 3, n = 1; allele 26, n = 1; and allele 21, n = 6).
Descriptive analysis was also performed for nucleotide sequence data stratified by evolutionary lineage (I [n = 69] and II [n = 48]) (Table 2); sequence data for lineage III isolates were not analyzed separately due to the small number of isolates (n = 3). All L. monocytogenes isolates contained 433 binary parsimony informative sites along the 4,553 bases sequenced. Lineage I and II isolates contained 70 and 292 parsimony informative sites, respectively. Generally, alleles were unique to lineages. The only exception was a single gap allele (allele 1), which was found in 39 lineage I isolates as well as in a single lineage II isolate. However, this observation needs to be interpreted in the context of the overall nucleotide diversity of the genes studied here; gap showed by far the lowest level of diversity (π = 0.0037; number of polymorphisms = 9) of all genes sequenced. With the exception of sigB, nucleotide diversity was higher among lineage II isolates than among lineage I isolates; this difference in nucleotide diversity was particularly striking for purM (π = 0.0614 and 0.0034 for lineage II and I isolates, respectively) (Table 2). There were no apparent differences in G+C content between the lineage I and II sequences for each gene (Table 2).
Tajima's D test compares the population mutation rate, which is estimated from the number of segregating sites, to the average pairwise nucleotide distance to determine whether the observed frequency of segregating mutations agrees with the expected frequency under the standard neutral model (51). Tajima's D statistic for the data set containing all L. monocytogenes isolates yielded positive values for all genes and indicated that actA, ribC, and sigB deviate significantly (P < 0.05) from the standard neutral model. Within lineage I sequences, negative values for Tajima's D statistic were observed for two genes (inlA and prs) and Tajima's D statistic for none of the seven gene fragments deviated from the standard neutral model (P > 0.05). Within lineage II sequences, three genes (actA, prs, and sigB) yielded negative D statistic values (Table 2) and prs and sigB deviated significantly (P < 0.05) from the standard neutral model.
Models of evolution.
The DNA substitution models that best explain sequence evolution for individual genes were determined using MODELTEST and the LRT (37). Four genes (Table 3) followed an Hasegawa-Kishino-Yano model; this model allows for variable base frequencies and different substitution rates for transitions and transversions (16). Two genes followed the Tamura-Nei model; this model includes variable base and transition frequencies but equal transversion frequencies (52). actA followed a variation of the Kimura 1981 model, which allows variable base frequencies, equal transition frequencies, and variable transversion frequencies (two transversion rates are allowed in this model) (23, 24, 37). For all genes, inclusion of a gamma distribution (Table 3), which allows the rate of substitution to vary along a sequence (63), significantly improved the ability of the model to explain the sequence data. For gap and actA, addition of a parameter to define the proportion of invariate sites (Table 3) also significantly improved the ability of the model to explain the sequence data.
TABLE 3.
Molecular evolution parameters for the seven L. monocytogenes genes
| Gene | DNA substitution modela | ti/tvb | Alphac | Pinvard | −Ln L clocke | −Ln L no clockf | Test statistic for mol. clockg | Molecular clock conclusionh |
|---|---|---|---|---|---|---|---|---|
| actA | K81uf + G | Rate matrix | 0.2714 | −1282.03 | −1259.70 | 44.66 | Reject | |
| gap | HKY + I + G | 2.23 | 0.0002 | 0.8726 | −857.16 | −852.48 | 9.36 | Fail to reject |
| inlA | TRN + I + G | Rate matrix | 1.0079 | 0.7902 | −1803.98 | −1761.40 | 85.16 | Reject |
| prs | HKY + G | 3.01 | 0.0071 | −1097.63 | −1094.74 | 5.79 | Fail to reject | |
| purM | HKY + G | 1.34 | 0.1937 | −1902.35 | −1869.52 | 65.66 | Reject | |
| ribC | TRN + G | Rate matrix | 0.1433 | −1553.63 | −1518.86 | 69.54 | Reject | |
| sigB | HKY + G | Rate matrix | 0.1367 | −1140.37 | −1130.31 | 20.11 | Fail to reject |
As described by Posada and Crandall (37). Abbreviations: K81uf, Kimura 1981 model (23); G, gamma distribution; HKY, Hasegawa-Kishino-Yano model (16); I, proportion of invariate sites; TRN, Tamura-Nei model (52).
Ratio of transitions (ti) to transversions (tv).
Shape of the alpha parameter of the gamma distribution.
Pinvar, proportion of invariable sites; no value listed for genes that followed a model without an invariable site parameter.
−Ln L clock, −Ln likelihood score for null hypothesis that data set evolved according to a molecular clock.
−Ln L clock, −Ln likelihood estimate for the alternative hypothesis that data set did not evolve according to a molecular clock.
The test statistic for the molecular (mol.) clock was calculated as 2[(−Ln L clock) − (−Ln L no clock)].
Conclusion to reject or fail to reject the null hypothesis that data set follows a molecular clock.
A randomly selected subset (n = 30) of the full data set (n = 120) was used to evaluate the null hypothesis that a given gene evolved according to a molecular clock. Following a molecular clock implies that all branches in the sample set are the same distance from the ancestral node (tree root) and further suggests that all branches in the tree have evolved at the same rate (3). The null hypothesis that sequences evolve according to a molecular clock was rejected (P < 0.05) for virulence genes (actA and inlA) and the hypervariable housekeeping genes purM and ribC (29), while gap, prs, and sigB were shown to evolve according to a molecular clock (P > 0.05) (Table 3).
Phylogenetic compatibility within and between L. monocytogenes genes.
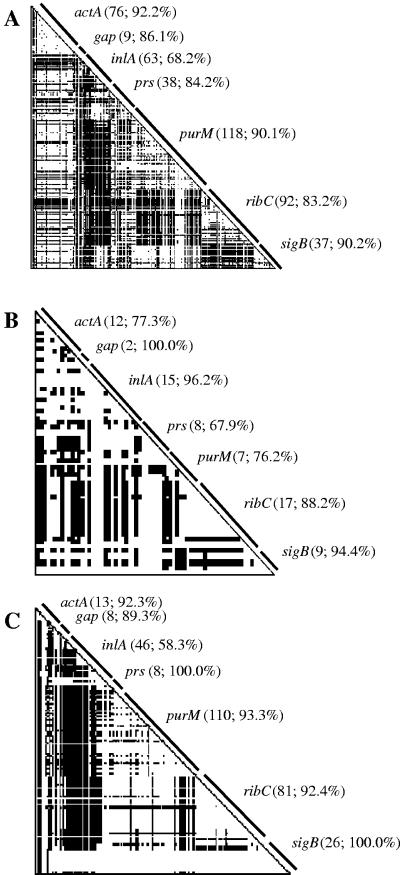
The fusion of sequence components from different sources to form a new hybrid sequence either by reciprocal recombination or by gene conversion is often referred to as reticulate evolution (18). The presence of reticulate evolution consequently means that different regions along a nucleotide alignment have different phylogenetic histories; thus, a single phylogenetic tree may fail to accurately describe the evolutionary history of a set of taxa (18). The RETICULATE software program (18) was used to construct compatibility matrices of all pairwise comparisons of binary parsimony informative sites within and between each of the seven gene fragments in order to probe for evidence of recombination. RETICULATE results can be displayed graphically, such that compatible pairs of sites are visualized as white cells, while incompatible pairs of sites are visualized as black cells. Overall compatibility contains two components including within-locus compatibility, represented by triangular regions, and between-locus compatibility, represented by rectangular regions (Fig. 1). All 120 L. monocytogenes isolates as well as lineage I and lineage II isolates only (Fig. 1) showed considerable within- and between-locus incompatibilities. The average within-locus compatibility was much higher than the average between-locus compatibility for all L. monocytogenes isolates as well as for lineage I and II isolates only (Table 4), indicating that polymorphisms in distinct chromosomal locations may deliver an incongruent phylogenetic signal. Although lineage II isolates showed relatively low compatibility between loci, sigB was highly compatible (97.2% to 100.0%) with all other loci with the exception of actA (53.8%). Interestingly, while most loci demonstrated similar levels of within-locus compatibility in lineage I and II isolates, the within-locus compatibility for inlA was notably higher for lineage I isolates than for lineage II isolates (Fig. 1).
FIG. 1.
Compatibility matrices for nucleotide polymorphisms within (A) all L. monocytogenes isolates, (B) lineage I isolates only, and (C) lineage II isolates only. Matrices contain all possible pairwise comparisons of binary parsimony informative sites that are phylogenetically compatible (white cells) or incompatible (black cells) within genes (triangular regions) or between genes (rectangular regions). For each gene, values in parentheses are the number of informative sites followed by the observed within-gene compatibility.
TABLE 4.
Summary of compatibility analyses for seven gene fragments sequenced
| Isolates includeda | Observed overall compatibility (%) | Neighbor similarity score (%)
|
P valueb | Mean compatibility (%)
|
||
|---|---|---|---|---|---|---|
| Observed | Mean random | Within genes | Between genes | |||
| All isolates | 71.72 | 78.72 | 67.44 | 0.0001 | 84.71 | 69.41 |
| Lineage I | 70.52 | 77.27 | 66.58 | 0.0001 | 83.36 | 68.36 |
| Lineage II | 72.60 | 87.68 | 70.17 | 0.0001 | 89.35 | 67.79 |
Compatibilities were analyzed separately for all L. monocytogenes isolates, lineage I isolates only, and lineage II isolates only.
P value comparing the observed neighbor similarity score to the mean random neighbor similarity score.
The observed overall compatibility for alignments containing all L. monocytogenes isolates as well as lineage I and II isolates only was similar (Table 4). The neighbor similarity score, or the fraction of adjacent neighboring sites with the same color (either compatible or incompatible), was higher than the observed overall compatibility for all L. monocytogenes isolates as well as for lineage I and II isolates only (Table 4). The neighbor similarity score also significantly (P < 0.0001 for all three data sets; Table 4) exceeded the neighbor similarity score based on 10,000 randomized matrices, indicating that the overall pattern of compatibility and incompatibility between sites is not random and that the order of sites along the nucleotide alignment increases clustering of compatible and incompatible sites. Thus, phylogenetic cohesiveness has not broken down completely due to recombination.
Horizontal gene transfer analysis.
Sawyer's test for recombination was used to identify segments of the aligned individual gene sequences for which a pair of sequences was sufficiently similar to imply a history of recombination. This method first eliminates monophyletic sites in a given alignment and proceeds to identify identical (concordant) stretches of polymorphisms or “fragments” between a given pair of sequences that are bracketed by two discordant polymorphisms or by one discordant polymorphism and the end of the alignment. Maximum condensed fragment (MCF) length scores were also calculated and compared to MCF scores obtained from 10,000 random permutations of the order of polymorphic sites. P values derived from comparing the observed MCF scores to the MCF scores from the permuted data set estimate the statistical significance of intragenic recombination in a given alignment. Significant global inner fragments indicate recombination events between ancestors of two sequences in the alignment. Significant global outer fragments suggest recombination events between ancestors outside of the alignment or indicate that recombination events have subsequently been obscured by mutation or recombination events (44, 45). MCF scores for inner and outer fragments showed statistically significant evidence (P < 0.05) for intragenic recombination in prs, purM, ribC, actA, and inlA. Multiple statistically significant (P < 0.05) global inner fragments were detected for actA, inlA, purM, and ribC, while only a single borderline significant (P = 0.0450) global inner fragment was detected for prs. actA was the only partial gene sequence with significant evidence for global outer recombination; only a single fragment with borderline significance (P = 0.04) was identified (Table 5).
TABLE 5.
Summary of recombination analysis using Sawyer's test
| Gene | Global inner recombinationa
|
Global outer recombinationb
|
Global inner events
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sim. P valuec | No. of fragmentsd | No. of eventse | Sim. P value | No. of fragments | No. of events | Within lineage
|
Between lineages
|
||||||
| I | II | III | I/II | I/III | II/III | Multiplef | |||||||
| actA | 0.0047* | 4 | 1 | 0.0409* | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| gap | 0.6786 | 0 | 0 | 0.2195 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| inlA | <0.0001* | 37 | 6 | 0.1520 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 0 | 3 |
| prs | 0.0450* | 1 | 1 | 0.0588 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| purM | <0.0001* | 50 | 7 | 1.0000 | 0 | 0 | 0 | 5 | 0 | 1 | 0 | 1 | 0 |
| ribC | <0.0001* | 45 | 7 | 0.0995 | 0 | 0 | 1 | 4 | 0 | 1 | 0 | 0 | 1 |
| sigB | 0.2128 | 0 | 0 | 1.0000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 137 | 22 | 1 | 1 | 1 | 10 | 0 | 4 | 0 | 3 | 4 | ||
Recombination events between ancestors of sequences in alignment.
Recombination events involving ancestor outside alignment or obscured by subsequent mutations.
Sim. P value, similar P value. P values indicating statistically significant (P < 0.05) evidence for recombination events are marked with asterisks.
Number of segments of alignment sufficiently similar to imply recombination.
Group of fragments linked to the same 5′ and/or 3′ breakpoints were classified as a single recombination event.
“Multiple” refers to global inner recombination events that involved significant fragments between and within lineages.
Because point mutations introduced after recombination may obscure the actual beginning and end points of a gene fragment involved in recombination, we defined an independent recombination event as a group of fragments that could be linked to the same 5′ or 3′ breakpoints. Using these criteria, the 137 global inner fragments initially identified by Sawyer's test (Table 5) could be grouped into 22 independent inner recombination events. While 21 inner recombination events involved lineage II isolates, only 9 and 4 recombination events involved lineage I and III isolates, respectively. While one event occurred within lineage I isolates, for the other events lineage I isolates appear to predominantly have served as donors in a conversion event. A total of 11 recombination events involved only isolates within a given lineage; only one of these events occurred in a virulence gene. On the other hand, of the 11 recombination events between lineages, six occurred in virulence genes. Four independent recombination events included global inner recombination fragments both within a given lineage and between lineages (three for inlA and one for ribC), possibly indicating ancestral recombination events before lineage divergence.
Gene trees.
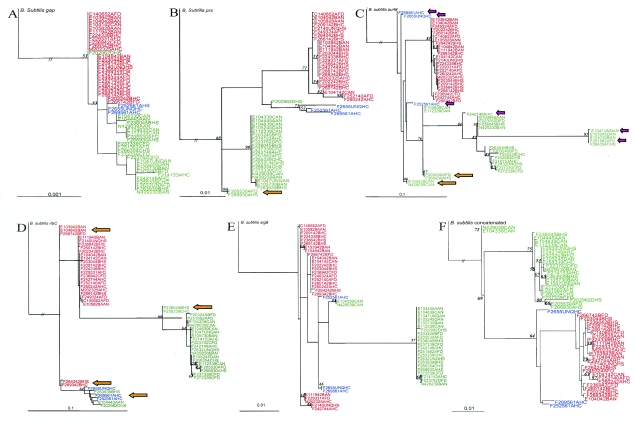
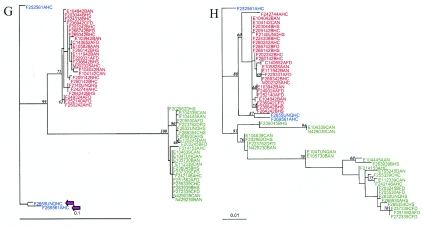
Phylogenetic trees were inferred by maximum likelihood methods from individual gene alignments containing a single isolate to represent each unique ST (n = 52). Lineage I (n = 26) and lineage II (n = 23) STs by and large represented two distinct evolutionary lineages in all gene trees (Fig. 2), which were generally strongly supported by bootstrap proportions (Fig. 2) and posterior probabilities (generated using Baysian methods; data not shown). Generally, for different gene trees, no consistent clustering of isolates and subtypes was observed within a given lineage, but some isolates grouped together in different trees (e.g., FSL NN-290 and FSL E1-043 in the gap and purM trees); none of these isolates showed apparent unique epidemiological aspects. While the vast majority of lineage I isolates formed tightly clustered monophyletic groups in all gene trees, lineage II isolates showed greater genetic diversity and often either represented polyphyletic groups (e.g., for sigB, inlA, ribC, and purM; Fig. 2) and/or contained sequence types that grouped with lineage I and III isolates (i.e., for prs and ribC; Fig. 2). While isolates generally grouped into the same phylogenetic group on the basis of all seven individual gene trees (Fig. 2), there were a few instances in which for one gene tree, a given isolate did not group within its predicted lineage. In general, only genes that showed significant evidence of multiple horizontal gene transfer events (i.e., ribC, purM, and inlA) yielded phylograms that did not show consistent clustering of lineage I and II isolates into distinct and well-separated clades; the specific gene sequences that did not cluster with their respective lineages or were located on unusually long branches generally had also been implicated in recombination events identified by Sawyer's test (Fig. 2). For example, while the ribC sequences for lineage I isolates FSL-F2-642 and FSL-F2-693 grouped with lineage II and III isolates, these isolates grouped with other lineage I isolates in all other gene trees. However, the ribC clade containing these two lineage I isolates was overall poorly supported by bootstrap data, and Sawyer's test found statistically significant evidence for intragenic recombination within ribC, including events involving the ribC sequences for isolates FSL-F2-642 and FSL-F2-693.
FIG. 2.
Phylograms inferred by maximum likelihood methods for (A) gap, (B) prs, (C) purM, (D) ribC, (E) sigB, and (F) a concatenated sequence containing gap, prs, and sigB (a neighbor-joining phylogram for this concatenated sequence that also includes L. innocua is available as supplemental Fig. 1 (at http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary.txt), as well as the virulence genes (G) actA and (H) inlA. Phylograms were constructed using one representative isolate for each sequence type. Taxon labels include the name (e.g., F2655, representing isolate FSL F2-655), ribotype (e.g., 44A represents DUP-1044A) and source (e.g., human isolate from NYSDOH [HS], human isolate from NYCDOH [HC], animal isolate [AN], and food isolate [FD]) for the representative isolate for each sequence type. Phylograms for gap, prs, purM, ribC, sigB, and the concatenated sequences for sigB, prs, and gap were rooted using Bacillus subtilis as an outgroup, while phylograms for actA and inlA were rooted using the three lineage III isolates in our data set as an outgroup. For some phylograms, the branch length of the outgroup was collapsed to best view the topology of the tree (collapsed branch length indicated by “//”). Maximum likelihood bootstrap support values (if >50) are shown as node labels. Genetic lineages assigned by EcoRI ribotyping are designated by different colors. Lineage I isolates (representing serotypes 1/2b [n = 32], 4b [n = 36], and 3b [n = 1]) are in red. Lineage II isolates (representing serotypes 1/2a [n = 43] and 1/2c [n = 4] as well one isolate that was untypeable [see supplemental Table 1]) are in green. Lineage III isolates (representing serotypes 4b [n = 1], 4a [n = 1] and one isolate that was untypeable [see supplemental Table 1]) are in blue. While lineage III isolates have previously been reported to predominantly represent serotypes 4a and 4c, recent data indicate that some lineage III isolates may also be serotype 4b (55). Large arrows indicate selected allelic types involved in recombination as identified by Sawyer's test (short purple and longer orange arrows indicate specific isolates and groups of isolates, respectively, involved in recombination); for inlA, allelic types involved in recombination were not identified due to the large number of recombination events in this gene.
Consistent with the evidence supporting reticulate evolution and inconsistent phylogenetic signals between gene sequences as well as multiple intragenic recombination events in actA, inlA, purM, and ribC, consensus trees inferred from concatenated alignments of either all seven L. monocytogenes loci or of the five housekeeping and stress response genes were inconsistent with topologies ascertained from the individual gene trees (not shown). More specifically, the presence of two distinct evolutionary lineages as observed in phylogenetic trees inferred from individual gene data sets was completely obscured when phylogenetic analysis was based on a five-gene or seven-gene concatenated data set. A phylogram based on a concatenated sequence for the three genes that followed the molecular clock hypothesis and showed no or limited evidence for horizontal gene transfer or diversifying selection (i.e., the housekeeping genes gap and prs and the stress response gene sigB) on the other hand allowed for construction of a phylogram consistent with the individual gene phylograms (Fig. 2F) and appears to provide a good representation of the phylogeny of L. monocytogenes.
Lineage III (n = 3) isolates were not only rare in the isolate set used in this study, but they also represent a rare L. monocytogenes clonal group in general (60). While lineage III isolates were genetically diverse and clustered differently in the individual gene trees, they broadly clustered with lineage I isolates (although on two separate and distinct clades) in the phylogram based on the concatenated gap, sigB, and prs sequences.
Nucleotide distances within and between lineages.
Average pairwise distances within and between lineage I and II isolates were calculated for each gene sequenced to provide a measure of genetic separation between these evolutionary lineages. With the exception of gap and purM, the pairwise distance between isolates representing lineages I and II was much greater than the pairwise distances observed within these lineages (Table 6). For all genes except sigB, lineage II isolates showed greater average pairwise distances than lineage I isolates (Table 6). The average pairwise nucleotide differences in sigB between L. innocua, L. seeligeri, and L. welshimeri isolates obtained from the natural environment were also determined to compare genetic distances between lineage I and II isolates to distances observed between different Listeria species (Table 7). The distance within L. innocua, L. seeligeri, and L. welshimeri isolates ranged from 0.32% to 1.33%, similar to sigB distances observed within both lineages I and II (0.36%), while distance between species ranged from 0.83% to 5.89%, consistent with the increased distance observed between L. monocytogenes lineages (2.19%).
TABLE 6.
Average percentage pairwise distances for each gene within and between L. monocytogenes lineage I and II isolates
| Gene | % Pairwise distance within:
|
% Pairwise distance between lineages | |
|---|---|---|---|
| Lineage I | Lineage II | ||
| actA | 0.60 | 0.63 | 5.33 |
| gap | 0.10 | 0.46 | 0.37 |
| inlA | 0.44 | 2.16 | 2.60 |
| prs | 0.21 | 0.27 | 1.85 |
| purM | 0.34 | 6.14 | 5.86 |
| ribC | 0.92 | 3.59 | 5.54 |
| sigB | 0.36 | 0.36 | 2.19 |
TABLE 7.
Average percentage pairwise distances for sigB in three Listeria speciesa
| Species | % Pairwise distance for sigB in species
|
||
|---|---|---|---|
| L. innocua | L. seeligeri | L. welshimeri | |
| L. innocua | 1.32 | ||
| L. seeligeri | 5.89 | 1.33 | |
| L. welshimeri | 3.99 | 0.83 | 0.32 |
A neighbor-joining sigB phylogram based on the unique sigB allelic types for the 120 L. monocytogenes isolates as well as the 48 L. innocua, 48 L. seeligeri, and 48 L. welshimeri isolates used to generate average percentage pairwise distances shown here is available (supplemental Fig. 2 at http://www.foodscience.cornell.edu/wiedmann/Nightingale%20Supplementary.txt).
Analysis for positive selection.
Selection at the amino acid level can be detected by comparing the number of nonsynonymous substitutions/nonsynonymous site (dN) to the number of synonymous substitutions/synonymous site (dS). While overall average dN/dS ratios (ω) of >1 provide strong evidence for diversifying or positive selection in a given gene, genes and gene fragments with an overall average ω of <1 may still contain specific amino acid sites that are under positive selection. A maximum likelihood approach that uses the LRT to compare nested models of heterogeneous selective pressure among amino acid sites was thus used to test genes with an overall ω of >0.1 (i.e., actA and inlA) to determine whether specific amino acid sites in these genes may be under positive selection. If the null models that do not include a category for sites with a ω of >1 (models 1 and 7) are rejected in favor of the alternative models 2 and 8, respectively, which include a category for sites with a ω of >1, then a gene is considered to be under positive selection. Statistically significant evidence (P < 0.05) for variation of selective pressure among sites (model 0 versus model 3) and positive selection was observed for the 3′ end of actA (Table 8). Models 2 and 8 categorized 12 amino acid sites into a site class where ω is >1; four of these sites showed statistically significant posterior probabilities (P > 0.95) for being under positive selection (Table 9). For inlA, significant evidence (P < 0.05) was observed for variation in selective pressure along the fragment sequenced, but the null hypothesis that all amino acid sites are under neutral or constrained selection could not be rejected (P > 0.05; Table 8). However, models 2 and 8 identified 10 amino acid sites that fit into a category where ω is >1 and a single codon (Table 9) showed statistically significant evidence for being under positive selection.
TABLE 8.
Summary of analysis for variable dN/dS ratios among sites
| Gene | Model code | pa | Ln Lb | Parameter estimate(s)c | Models testedd
|
||
|---|---|---|---|---|---|---|---|
| M0 vs M3 | M1 vs M2 | M7 vs M8 | |||||
| actA | 0 | 1 | −1432.5 | ω = 0.3478 | 17.86 | 3.52 | 7.42 |
| 1 | 1 | −1428.1 | p0 = 0.5749, p1 = 0.4251 | ||||
| 2 | 3 | −1426.3 | p0 = 0.0000, p1 = 0.2607, p2 = 0.7930; ω2 = 0.1018 | ||||
| 3 | 5 | −1423.5 | p0 = 0.0389, p1 = 0.8750, p2 = 0.0861; ω0 = 0.1975, ω1 = 0.1975, ω2 = 2.965 | ||||
| 7 | 2 | −1427.2 | p = 0.1546, q = 0.2794 | ||||
| 8 | 4 | −1423.5 | p0 = 0.9148, p1 = 0.0852; p = 24.630; q = 99.000; ω11 = 2.980 | ||||
| inlA | 0 | 1 | −1917.1 | ω = 0.1932 | 63.68 | 1.66 | 2.46 |
| 1 | 1 | −1886.1 | p0 = 0.8285, p1 = 0.1715 | ||||
| 2 | 3 | −1885.3 | p0 = 0.8316, p1 = 0.1477, p2 = 0.0207; ω2 = 2.496 | ||||
| 3 | 5 | −1885.2 | p0 = 0.8179, p1 = 0.1433, p2 = 0.0388; ω0 = 0.0000, ω1 = 0.7700, ω2 = 2.161 | ||||
| 7 | 2 | −1886.5 | p = 0.0024, q = 0.0102 | ||||
| 8 | 4 | −1885.3 | p0 = 0.9362, p1 = 0.0638; p = 0.0387, q = 0.4251; ω11 = 1.801 | ||||
p, number of free parameters.
Ln L, log likelihood estimate for a given model.
ω, dN/dS ratio; p0, proportion of sites with ω = 0; p1, proportion of sites with ω = 1; ω2, ω estimated from data; p and q are beta distribution parameters; ω11 = ω estimated from data for site class with ω >1.
Likelihood ratio test calculated as 2[ln L (null) − ln L (alternative)]; bold type indicates statistical significance at P < 0.05. M0 vs M3, model 0 versus model 3.
TABLE 9.
Positively selected amino acid sites in inlA and actAa
| Lineage | aa residue at inlA aa site 764b
|
aa residue at actA aa site:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 457b
|
461c
|
515b
|
522b
|
||||||||||||
| A | D | E | Kd | R | S | T | P | S | A | I | T | A | E | V | |
| I | 14e | 0 | 52e | 3 | 0 | 30 | 39e | 32 | 37 | 0 | 69e | 0 | 42e | 27e | 0 |
| II | 0 | 17e | 25 | 6 | 1 | 35e | 12 | 14 | 34 | 41e | 1 | 6e | 0 | 0 | 48e |
| III | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 0 |
Only amino acid (aa) sites that grouped in site class with ω > 1 with posterior probabilities of >0.95 are included.
Sites with posterior probability of >0.95.
Site with a posterior probability of >0.99.
Only isolates that carried nonsense mutations in inlA showed K at inlA aa site 764.
Significant (P < 0.05) association between a specific amino acid residue and an L. monocytogenes lineage.
Descriptive analyses of positively selected amino acid sites that were identified with posterior probabilities of >0.95 (Table 9) showed that four different amino acid residues occurred at site 764 in inlA, while two and three different amino acid residues occurred in one and three positively selected amino acid sites in actA, respectively. Statistical analyses indicated that the occurrence of specific amino acid residues at these sites was generally associated with lineage (P < 0.01 by chi-square test of independence); only amino acid residues at actA amino acid site 461 were not significantly lineage associated (P = 0.065). While two of the amino acid residues at inlA site 764 (A and D) showed exclusive associations with lineage, the other two amino acid residues at this site were found in multiple lineages. For actA, amino acid residues at two of the positively selected sites showed exclusive (site 522) or almost exclusive (site 515) associations with lineages, while specific amino acid residues at the other two sites were found across lineages (Table 9).
DISCUSSION
Partial nucleotide sequencing of multiple housekeeping genes, two virulence genes, and one stress response gene for 120 L. monocytogenes isolates representing defined sources throughout the food chain, including human and animal clinical listeriosis cases as well as contaminated food samples, provided a highly discriminatory subtyping method, which allowed us to differentiate L. monocytogenes isolates within previously defined EcoRI ribotypes. Phylogenetic analyses (i) confirmed that L. monocytogenes is a highly diverse bacterial species that contains two major, deeply separated evolutionary lineages (lineages I and II) as well as a less common lineage, lineage III, and (ii) showed that horizontal gene transfer as well as positive selection contributed to the evolution of L. monocytogenes virulence genes. While lineage I appears to be highly clonal, lineage II isolates show evidence of a greater history of horizontal gene transfer events in both housekeeping and virulence genes. These findings provide a framework for further studies on the evolution of host specificity and transmission characteristics of different L. monocytogenes strains and lineages. Although previous MLST studies have been conducted for L. monocytogenes (4, 29, 42, 66), these studies involved convenient retrospective and often small sets of isolates, which failed to adequately represent the genetic diversity of this bacterial species. In addition, these studies did not probe the contributions of both positive selection and recombination on the evolution of L. monocytogenes. The isolate set studied here is representative of the L. monocytogenes diversity among human, animal, and food isolates and thus allows for reliable estimation of evolutionary and population genetics parameters for L. monocytogenes associated with transmission through the food system.
Nucleotide sequencing of multiple loci provides a highly discriminatory method to differentiate L. monocytogenes.
Our study showed that partial sequencing of seven L. monocytogenes genes provides a highly discriminatory molecular subtyping method. Consistent with previous studies (4, 39, 66), inclusion of virulence genes in our DNA sequencing-based subtyping scheme increased the discriminatory power over an MLST scheme including only housekeeping genes. While traditional MLST schemes are based solely on housekeeping genes, which are not expected to experience strong selective pressure and are less likely to be affected by horizontal gene transfer (56), sequencing of virulence genes may also provide important information on the evolution of virulence characteristics of bacterial pathogens. For example, partial sequencing of inlA in our study uncovered three unique inlA nonsense mutations in 8 of 30 food isolates and 1 of 60 human clinical isolates, which are predicted to lead to a truncated internalin A (InlA) protein (K. K. Nightingale, K. Windham, K. E. Martin, M. Yeung, and M. Wiedmann, submitted for publication). Jonquieres et al. (20) previously discovered a similar inlA nonsense mutation that was shown to result in a truncated form of InlA, which is released into the medium rather than anchored to the bacterial cell membrane. Olier et al. (35, 36) further showed that L. monocytogenes isolates that harbor similar nonsense mutations in the 3′ end of inlA display a diminished capacity to invade human intestinal epithelial cells. The fact that alleles encoding a truncated InlA were predominantly associated with food isolates and rare among human clinical listeriosis isolates further supports that these isolates may show reduced human virulence and illustrates how virulence gene sequencing can aid in the identification and detection of L. monocytogenes strains that differ in virulence. These findings are also consistent with a recent study of 300 human clinical and 150 food isolates from France which identified, using Western blotting, truncated forms of InlA in 4% of human clinical isolates and 35% of food isolates (17).
L. monocytogenes is a very diverse species with at least two deeply separated, species-like evolutionary lineages.
Phylogenetic trees constructed for all seven genes supported the hypothesis that L. monocytogenes can be divided into two major evolutionary lineages, consistent with findings from previous studies using different molecular subtyping methods (29, 55, 60, 66). Lineage III isolates also were shown to group more closely to lineage I isolates than to lineage II isolates, consistent with a recent virulence gene-based phylogenetic analysis (55). Reconstruction of the phylogenetic history of a bacterial species may be complicated though by the fact that in addition to point mutations, horizontal gene transfer and subsequent recombination may contribute substantially to the divergence of clonal groups (38). For a given set of taxa, horizontal gene transfer can lead to conflicting phylogenies between trees based on different genes (6). Although we detected statistically significant evidence for multiple recombination events in actA, inlA, ribC, and purM, overall tree topologies ascertained from these genes still revealed clear separation of the two L. monocytogenes lineages, similar to findings for the genes that appear to have evolved primarily by point mutations (sigB, prs, and gap). While trees based on concatenated sequences that included genes that showed evidence for reticulate evolution and that did not follow a molecular clock did not group lineage I and II isolates into clear clades, a phylogenetic tree constructed from concatenated sequences of the three genes that followed a molecular clock and showed no (i.e., gap and sigB) or very limited (i.e., prs) indication for horizontal gene transfer supported the hypothesis that lineages I and II form two well-supported clades. Tajima's D test, which has been reported to have reasonable power to make inferences about population demographics (31, 47), also supported a population subdivision within L. monocytogenes. Specifically, a subdivided population tends to yield positive values for Tajima's D test statistic (31, 47). The facts that Tajima's D test statistic values derived from all L. monocytogenes isolates were positive for all genes and that the values for three genes (actA, ribC and sigB) were significantly higher than zero, the value expected under the standard neutral model, thus indicated a subdivided population. The observation that Tajima's D test statistic values derived from L. monocytogenes isolates stratified by lineage were generally close to or less than zero further supports the hypothesis that lineages I and II represent cohesive and separate populations, consistent with the tree topologies observed.
Our data reported here furthermore clearly indicate that L. monocytogenes lineages I and II represent distinct subpopulations. This hypothesis is supported by the observations that (i) horizontal gene transfer of core (nonvirulence) genes predominantly occurs between isolates that belong to the same lineage and (ii) average pairwise sigB gene distances between lineages are similar to those observed between Listeria spp. Specifically, our data not only showed limited horizontal gene transfer of housekeeping genes between lineages but also showed an almost complete absence of shared alleles between lineage I and II isolates, a strong indication for the presence of different species or subspecies in a set of taxa (13, 48, 49). Consistent with our observations, Salecado et al. (42) have previously reported MLST data that suggest that recombination should be rare between strains belonging to different L. monocytogenes genetic lineages. In addition, individual gene trees also support the hypothesis that lineages represent species or subspecies-like entities, as gene trees consistently grouped isolates into the same lineage and did not reveal consistent subgroups within lineages, which is consistent with a higher frequency of horizontal gene transfer within lineages than between lineages (25). The generally high level of genetic distance observed between lineages I and II thus appears to directly limit the amount of horizontal gene transfer between lineages. These findings are consistent with observations by Vulic et al. (54), who showed that as sequence divergence increases, the frequency of homologous genetic exchange decreases exponentially.
Using enteric bacterial taxa as a model, Wertz et al. (56) have shown that molecular evolution data may be used, under a core genome framework, to apply the biological species concept to bacteria. Lan and Reeves (25) defined core genes (e.g., housekeeping genes) as a set of genes shared within a bacterial species and proposed that there is generally no selective advantage to acquiring new core gene alleles by horizontal gene transfer. On the other hand, genes that permit adaptation of a bacterial species to a defined niche may be classified as auxiliary genes; these genes include virulence genes (such as actA and inlA in L. monocytogenes), antibiotic resistance genes, as well as genes encoding innovative metabolic functions, such as toxin genes (56). The core genome hypothesis thus entails an interspecies barrier for core gene recombination that does not hold true with respect to auxiliary genes. In the current study, gene conversion events for core genes (prs, purM, and ribC) occurred almost exclusively within the same L. monocytogenes evolutionary lineage, while gene conversion for auxiliary genes (actA and inlA) occurred more frequently between isolates from different lineages. Additionally, we observed a single instance of a lineage I isolate carrying a lineage II allele, indicating horizontal transfer of the full gene fragment sequenced. Interestingly, this putative horizontal gene transfer event occurred in gap, a gene for which the nucleotide distance between lineages I and II was much less than that observed between these two lineages for other loci studied. These data are consistent with a deep separation between L. monocytogenes lineages I and II, which appears to act as a barrier to the exchange of genetic information between these two lineages, even though exchange of genetic information between these lineages may occur rarely for nondivergent loci (e.g., gap) and more commonly for virulence genes.
Our conclusion that L. monocytogenes lineages I and II represent distinct evolutionary lineages is also supported by recent comparative genomics studies, which have revealed considerable differences in genome content between L. monocytogenes lineage I and II isolates (8, 66). For example, Doumith et al. (8) reported that, on the basis of full genome microarray data, approximately 8% of the sequences found in the lineage I strain CLIP 80459 were absent in the lineage II strain EGD-e; this level of genetic diversity resembles the 10% interspecies difference observed between the complete L. monocytogenes EGD-e and L. innocua genomes (12). Together with previous studies indicating that L. monocytogenes lineage I strains are more commonly associated with human disease outbreaks and cases, while lineage II strains are more commonly associated with animal clinical disease and environmental sources including foods (15, 19, 33), our results support the hypothesis that L. monocytogenes lineages I and II represent subspecies or species with distinct ecological preferences. Similar observations have been reported for other bacterial pathogens. For example, Neisseria meningitidis contains two genetically distinct evolutionary lineages, and most pathogenic strains belong to a few serotypes (2).
L. monocytogenes lineages I and II are characterized by distinct population structures and evolutionary histories.
Our data presented here support the hypothesis not only that L. monocytogenes lineages I and II represent distinct lineages but also that these lineages differ in their evolutionary histories and population structures. While lineage I isolates appear to be highly clonal with limited horizontal gene transfer, lineage II isolates show greater genetic diversity and evidence of a larger number of horizontal gene transfer events than lineage I isolates. The high degree of clonality observed for lineage I isolates may be indicative of a recent population bottleneck (29). Our observations on a large and diverse set of L. monocytogenes isolates are consistent with a previous study by Meinersmann et al. (29), who, based on a smaller set of isolates including many that were obtained from a single processing plant, found that lineage II strains showed a higher incidence of horizontal gene transfer than lineage I strains. Our findings are also consistent with previous studies, which have shown that different bacterial species can demonstrate considerable diversity of population structure from Neisseria, a species that shows almost free and rapid recombination to Salmonella, a species that appears to be highly clonal and largely unaffected by recombination (25). While it has historically been assumed that evolution of bacterial species predominantly occurs by point mutations and vertical transmission of genetic material, more recently horizontal gene transfer has been recognized to play an important role in the evolution of many bacterial species (48). Our study adds to an emerging theme that the balance of these two evolutionary forces may critically differ in subpopulations of bacterial pathogens that have adapted to distinct ecological niches and/or show distinct host and tissue specificities (27). Interestingly, the distributions of amino acid residues at positively selected amino acid sites in actA and inlA showed distinct patterns, with some residues showing exclusive or almost exclusive association with lineages, while others were found in multiple lineages, providing preliminary evidence for lineage-specific selection patterns at these sites.
Comparisons of the relative frequencies of L. monocytogenes lineages I and II in human listeriosis cases and in food products have consistently found that lineages I and II are over- and underrepresented, respectively, among isolates from human listeriosis cases (15, 55, 60). While Ward et al. (55) have proposed that it is unclear whether this overrepresentation of lineage I represents enhanced human virulence or unique ecological adaptations to food-associated environmental stress conditions, the enhanced cytopathogenicity of lineage I strains over lineage II strains (as determined by plaque assays; 14, 34, 60) supports the hypothesis that lineage I may represent a host-adapted lineage which maintained the ability to survive and multiply under food-associated stress conditions (7). Even though further comparative phenotypic and genomic analyses will be necessary to fully understand the ecology and transmission potential of L. monocytogenes lineages I and II, we propose that lineage I represents a highly clonal lineage which is adapted to food-borne transmission. Lineage II, on the other hand, appears to represent a “generalist” lineage, which is better adapted to survive and multiply in the environment while still maintaining the ability to cause human disease (7, 33). While some isolates in lineage II have reduced ability to cause human disease (e.g., through premature stop codon mutations in inlA), other isolates appear to have the ability to infect mammalian and particularly nonprimate mammalian hosts as indicated by their isolation from mammalian hosts with clinical listeriosis (15, 19). We hypothesize that elevated levels of genetic diversity and elevated recombination rates within lineage II isolates may be critical to maintain a greater ability to adapt to diverse host and/or environmental conditions. As previously noted (29), our findings are similar to those described for the species in the genus Mycobacterium, where the pathogenic mycobacteria (Mycobacterium tuberculosis, Mycobacterium leprae, and Mycobacterium avium subsp. paratuberculosis) appeared to have suffered a bottleneck as recently as 10 to 15 thousand years ago, while the environmental mycobacterial species are not clonal and maintain greater levels of horizontal gene transfer (11, 22). While differences in population structure between obligate intracellular pathogens and closely related environmental bacteria may not provide the strongest support for our hypothesis, limited other data are available on the difference in the population structure between closely related facultative pathogens and environmental microorganisms. For example, as pointed out by Feil and Spratt (9), even though pathogenic Escherichia coli bacteria show a clonal population structure, very little is known about the population structure of commensal or environmental E. coli.
Recombination and positive selection contribute to evolution of L. monocytogenes virulence genes.
As outlined above, our data indicate that horizontal gene transfer occurred within the virulence genes actA and inlA and that interlineage horizontal gene transfer was considerably more common for these virulence genes than for the core genes sequenced. In addition, actA and inlA showed higher dN/dS ratios than the core gene fragments sequenced. More in-depth analyses also showed that actA in particular showed significant evidence for positive selection among specific amino acid sites. Both horizontal gene transfer and positive selection thus appear to have contributed to the evolution of virulence genes in L. monocytogenes, although the relative contributions of these two forces may differ for different virulence genes. These findings add to an emerging body of evidence that both of these evolutionary forces critically contribute to the evolution of virulence factors in a variety of bacterial pathogens. For example, previous studies have shown that the evolution of the intimin gene in Escherichia coli O111 and the streptokinase gene in Streptococcus pyogenes also included recombination and positive selection as contributing factors (21, 53). Further studies on positive selection using full-length virulence gene sequences will be necessary though to fully define the contributions of positive selection to the evolution of L. monocytogenes lineages and their virulence characteristics.
Conclusions.
DNA sequencing of seven selected genes showed that L. monocytogenes isolates representing populations associated with human and animal clinical listeriosis and contaminated food represent two deeply separated species-like evolutionary lineages, which differ in their evolutionary history and population structure. These findings were also strongly supported by a core phylogeny based on three genes that follow a molecular clock; use of these genes to study the phylogeny of additional L. monocytogenes isolates will provide further insight into the evolution of this pathogen. In conjunction with other previous studies, our data suggest that lineage I strains may represent a predominantly host-adapted clonal group, while lineage II strains represent a genetically diverse L. monocytogenes evolutionary lineage with a more recombinatorial population structure. Lineage II appears to represent a “generalist” lineage, which not only survives well in diverse environments but also includes strains that have the ability to infect mammalian and possibly other hosts. While previous MLST-based studies have provided evidence for the importance of horizontal gene transfer in L. monocytogenes (29, 42), our study reported here provides for the first time specific evidence that positive selection of virulence genes also contributes critically to the evolution of L. monocytogenes, possibly facilitating fixation of recombination events.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R01GM63259 awarded to M. Wiedmann.
We are very grateful to Qi Sun (Computational Biology Service Unit, Cornell University) for his expertise in setting up the computing system to perform evolutionary analyses. We are also indebted to Esther Fortes and Alphina Ho for their expert technical support.
REFERENCES
- 1.Anisimova, M., R. Nielsen, and Z. Yang. 2003. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164:1229-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bart, A., C. Barnabe, M. Achtman, J. Dankert, A. van der Ende, and M. Tibayrenc. 2001. The population structure of Neisseria meningitidis serogroup A fits the predictions for clonality. Infect. Gen. Evol. 1:117-122. [DOI] [PubMed] [Google Scholar]
- 3.Bromham, L., and D. Penny. 2003. The modern molecular clock. Nat. Rev. Genet. 3:216-224. [DOI] [PubMed] [Google Scholar]
- 4.Cai, S., D. Y. Kabuki, A. Y. Kuaye, T. G. Cargioli, M. S. Chung, R. Nielsen, and M. Wiedmann. 2002. Rational design of DNA sequence-based strategies for subtyping Listeria monocytogenes. J. Clin. Microbiol. 40:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czajka, J., N. Bsat, M. Piani, W. Russ, K. Sultana, M. Wiedmann, R. Whitaker, and C. A. Batt. 1993. Differentiation of Listeria monocytogenes and Listeria innocua by 16S rRNA genes and intraspecies discrimination of Listeria monocytogenes strains by random amplified polymorphic DNA polymorphisms. Appl. Environ. Microbiol. 59:304-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daublin, V., N. A. Moran, and H. Ochman. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829-831. [DOI] [PubMed] [Google Scholar]
- 7.De Jesus, A. J., and R. C. Whiting. 2003. Thermal inactivation, growth, and survival studies of Listeria monocytogenes strains belonging to three distinct genotypic lineages. J. Food Prot. 66:1611-1617. [DOI] [PubMed] [Google Scholar]
- 8.Doumith, M., C. Cazalet, N. Simones, L. Frangeul, C. Jacquet, F. Kunst, P. Martin, P. Cossart, P. Glaser, and C. Buchrieser. 2004. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 72:1072-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 10.Fenlon, D. R. 1999. Listeria monocytogenes in the natural environment, p. 21-38. In E. T. Ryser and E. H. Marth (ed.), Listeria listeriosis and food safety, 2nd ed. Marcel Decker, Inc., New York, N.Y.
- 11.Frothingham, R. 1999. Evolutionary bottlenecks in the agents of tuberculosis, leprosy, and paratuberculosis. Med. Hypotheses 52:95-99. [DOI] [PubMed] [Google Scholar]
- 12.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusnoik, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couvelin, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simones, A. Tierrez, J. A. Vaszquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 13.Godoy, D., G. Randle, A. J. Simpson, D. M. Aanensen, T. L. Pitt, R. Kinoshita, and B. G. Spratt. 2003. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J. Clin. Microbiol. 41:2068-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gombas, D. E., Y. Chen, R. S. Clavero, and V. N. Scott. 2003. Survey of Listeria monocytogenes in ready-to-eat foods. J. Food Prot. 66:559-569. [DOI] [PubMed] [Google Scholar]
- 15.Gray, M. J., R. N. Zadoks, E. D. Fortes, B. Dogan, S. Cai, Y. Chen, V. N. Scott, D. E. Gombas, K. J. Boor, and M. Wiedmann. 2004. Food and human isolates of Listeria monocytogenes form distinct but overlapping populations. Appl. Environ. Microbiol. 70:5833-5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasegawa, M., H. Kishino, and T. Yano. 1985. Dating of the human-ape splitting by a molecular clock of mitchondrial DNA. J. Mol. Evol. 22:160-174. [DOI] [PubMed] [Google Scholar]
- 17.Jacquet, C., M. Doumith, J. I. Gordon, P. M. V. Martin, P. Cossart, and M. Lecuit. 2004. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J. Infect. Dis. 189:2094-2100. [DOI] [PubMed] [Google Scholar]
- 18.Jakobsen, I. B., and S. Easteal. 1996. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput. Appl. Biol. Sci. 12:291-295. [DOI] [PubMed] [Google Scholar]
- 19.Jeffers, G. T., J. L. Bruce, P. L. McDonough, J. Scarlet, K. J. Boor, and M. Wiedmann. 2001. Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147:1095-1104. [DOI] [PubMed] [Google Scholar]
- 20.Jonquieres, R., H. Biern, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalia, A., and D. E. Bessen. 2004. Natural selection and evolution of streptococcal virulence genes involved in tissue-specific adaptations. J. Bacteriol. 186:110-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur, V., T. S. Whittam, and J. M. Musser. 1994. Is Mycobacterium tuberculosis 15,000 years old? J. Infect. Dis. 17:1348-1349. [DOI] [PubMed] [Google Scholar]
- 23.Kimura, M. 1981. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci. USA 78:454-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura, M. 1980. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 25.Lan, R., and P. R. Reeves. 2001. When does a clone deserve a name? A perspective on bacterial species based on population genetics. Trends Microbiol. 9:419-424. [DOI] [PubMed] [Google Scholar]
- 26.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGregor, K. F., B. G. Spratt, A. Kalia, A. Bennett, N. Bilek, B. Beall, and D. E. Bessen. 2004. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J. Bacteriol. 186:4285-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCraig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meinersmann, R. J., R. W. Phillips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadon, C. A., D. L. Woodward, C. Young, F. G. Rodgers, and M. Wiedmann. 2001. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J. Clin. Microbiol. 39:2704-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen, R. 2001. Statistical tests of selective neutrality in the age of genomics. Heredity 86:641-647. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nightingale, K. K., Y. H. Schukken, C. R. Nightingale, E. D. Fortes, A. J. Ho, Z. Her, Y. T. Grohn, P. L. McDonough, and M. Wiedmann. 2004. Ecology and transmission of Listeria monocytogenes in ruminants and the farm environment. Appl. Environ. Microbiol. 70:4458-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norton, D. M., J. M. Scarlett, K. Horton, D. Sue, J. Thimothe, K. J. Boor, and M. Wiedmann. 2001. Characterization and pathogenic potential of Listeria monocytogenes isolates from the smoked fish industry. Appl. Environ. Microbiol. 67:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olier, M., F. Pierre, J. P. Lemaitre, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 36.Olier, M., F. Pierre, S. Rousseaux, J. P. Lamaitre, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes isolates carried asymptomatically by humans. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 38.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 39.Revazishvili, T., M. Kotetishvili, O. C. Stine, A. S. Kreger, J. G. Morris, Jr., and A. Sulakveldze. 2004. Comparative analysis of multilocus sequence typing and pulsed-field gel electrophoresis for characterizing Listeria monocytogenes strains isolated from environmental and clinical sources. J. Clin. Microbiol. 42:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, A. J., and M. Wiedmann. 2003. Pathogen, host and environmental factors contributing to the pathogenesis of listeriosis. Cell Mol. Life Sci. 60:904-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 42.Salcedo, C., L. Arreaza, B. Alcala, L. de la Fuente, J. A. Vazquez. 2003. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41:757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauders, B. D., K. Mangione, C. Vincent, J. Schermerhorn, C. M. Farchione, N. B. Dumas, D. Bopp, L. Kornstein, E. D. Fortes, K. Windham, and M. Wiedmann. 2004. Distribution of Listeria monocytogenes molecular subtypes among human and food isolates from New York State shows persistence of human disease-associated Listeria monocytogenes strains in retail environments. J. Food Prot. 7:1417-1428. [DOI] [PubMed] [Google Scholar]
- 44.Sawyer, S. A. 1999. GENECONV: a computer package for the statistical detection of gene conversion. Department of Mathematics, Washington University, St. Louis, Mo. [Online.] http://www.math.wustl.edu/∼sawyer.
- 45.Sawyer, S. A. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6:526-538. [DOI] [PubMed] [Google Scholar]
- 46.Schlech, W. F. 1998. Foodborne listeriosis. Clin. Infect. Dis. 31:770-775. [DOI] [PubMed] [Google Scholar]
- 47.Simonsen, K. L., G. A. Churchill, and C. F. Aquadro. 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141:413-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spratt, B. G., and M. C. Maiden. 1999. Bacterial population genetics, evolution, and epidemiology. Philos. Trans. R. Soc. Lond. B 354:701-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spratt, B. G. 2004. Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266:323-352. [DOI] [PubMed] [Google Scholar]
- 50.Swofford, D. 1997. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 51.Tajima, F. 1989. Statistical method for testing neutral mutation hypothesis by DNA polymorphism. Genetics 123:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 53.Tarr, C. L., and T. S. Whittam. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J. Bacteriol. 184:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vulic, M., F. Dionisio, F. Taddei, and M. Radman. 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. USA 94:9763-9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, T. J., L. Gorski, M. K. Borucki, R. E. Mandrell, J. Hutchins, and K. Pupedis. 2004. Intraspecific phylogeny and lineage group identification based on the prfA virulence gene cluster of Listeria monocytogenes. J. Bacteriol. 15:4994-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wertz, J. E., C. Goldstone, D. M. Gordon, and M. A. Riley. 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J. Evol. Biol. 16:1236-1248. [DOI] [PubMed] [Google Scholar]
- 57.Wiedmann, M. 2003. ADSA foundation scholar award—an integrated science-based approach to dairy food safety: Listeria monocytogenes as a model system. J. Dairy Sci. 86:1865-1875. [DOI] [PubMed] [Google Scholar]
- 58.Wiedmann, M. 2002. Molecular subtyping methods for Listeria monocytogenes. J. AOAC Int. 85:524-531. [PubMed] [Google Scholar]
- 59.Wiedmann, M. 2002. Subtyping technologies for bacterial foodborne pathogens. Nutr. Rev. 60:201-208. [DOI] [PubMed] [Google Scholar]
- 60.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woolhouse, M. E., L. H. Taylor, and D. T. Haydon. 2001. Population biology of the multihost pathogen. Science 292:1109-1112. [DOI] [PubMed] [Google Scholar]
- 62.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Z. 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11:367-372. [DOI] [PubMed] [Google Scholar]
- 64.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Bensen. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, W., B. M. Jayarao, and S. J. Knabel. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]