Abstract
In rod-shaped bacteria, certain proteins are specifically localized to the cell poles. The nature of the positional information that leads to the proper localization of these proteins is unclear. In a screen for factors required for the localization of the Shigella sp. actin assembly protein IcsA to the bacterial pole, a mutant carrying a transposon insertion in mreB displayed altered targeting of IcsA. The phenotype of cells containing a transposon insertion in mreB was indistinguishable from that of cells containing a nonpolar mutation in mreB or that of wild-type cells treated with the MreB inhibitor A22. In cells lacking MreB, a green fluorescent protein (GFP) fusion to a cytoplasmic derivative of IcsA localized to multiple sites. Secreted full-length native IcsA was present in multiple faint patches on the surfaces of these cells in a pattern similar to that seen for the cytoplasmic IcsA-GFP fusion. EpsM, the polar Vibrio cholerae inner membrane protein, also localized to multiple sites in mreB cells and colocalized with IcsA, indicating that localization to multiple sites is not unique to IcsA. Our results are consistent with the requirement, either direct or indirect, for MreB in the restriction of certain polar material to defined sites within the cell and, in the absence of MreB, with the formation of ectopic sites containing polar material.
The poles of rod-shaped bacteria are unique both in terms of their contribution to many important cell functions and in terms of the composition of the cell envelope. The poles are the destination for proteins involved in motility, protein secretion, chemotaxis, development, cell division, chromosome segregation, virulence, and adhesion. Proteins involved in these processes and specifically localized to the pole include actin assembly proteins of Shigella spp. (IcsA [VirG]) and Listeria monocytogenes (ActA) (24, 32), components of the type II secretion apparatus of Vibrio cholerae (including the inner membrane protein EpsM) (49), components of the Dot/Icm secretion apparatus in Legionella pneumophila (13), components of the Escherichia coli and Caulobacter crescentus chemotaxis apparatuses (2, 38, 55), the cell cycle regulatory signal transduction proteins of C. crescentus (reviewed in reference 3), cell cycle proteins in Bacillus subtilis (4, 7, 18, 19, 40), and type IV pili in Pseudomonas aeruginosa (8). In addition, the cell division inhibitors MinC and MinD oscillate between the poles of E. coli cells and are stable at the poles of B. subtilis cells (reviewed in reference 20). Whereas it seems likely that multiple mechanisms for the polar localization of proteins exist (51), these mechanisms are at present incompletely understood.
In addition to the presence of the proteins listed above, the polar regions of the cell each possess a distinctive peptidoglycan and outer membrane (16, 17). Peptidoglycan is a rigid polymer that determines and maintains the shape of the cell. In E. coli cells, peptidoglycan is stable at the poles, with all newly synthesized peptidoglycan being incorporated into existing peptidoglycan along the lengths of the cells (16). Likewise, the protein content of the outer membranes that overlie the poles is relatively stable (17).
The proper localization of proteins to the pole requires coordination of cell shape determinants with positional information recognized by polar proteins (28). Cell shape is established by the controlled placement of peptidoglycan (59). Both the maintenance of inert peptidoglycan at the poles and the synthesis of preseptal and septal peptidoglycan at the midcell division site are dependent on the cell division protein FtsZ (14, 16), which forms the cytokinetic ring at midcell. Disruption of certain low-molecular-weight penicillin-binding proteins (LMW PBPs) leads to misshapen cells that have kinks, bends, split poles, and other morphological abnormalities (17). These morphological abnormalities contain positional information recognized by the polar proteins IcsA and EpsM (44), suggesting that cell shape is linked, at least in part, with the controlled placement of polar material. In a separate series of experiments, we have shown that proper placement of this same polar positional information is independent of FtsZ and cell division (27). These studies further demonstrated that the placement of this positional information is not strictly dependent on the concave shape of poles in wild-type cells or on kinks in the LMW PBP mutant cells (27). Thus, although shape is clearly linked to the placement of polar positional information, it is not the only or defining factor.
We performed a screen of a transposon mutant library in E. coli for nonessential factors involved in the polar localization of IcsA. As we describe herein, we isolated in our screen a mutant carrying a transposon insertion in mreB that displayed aberrant localization of an IcsA-green fluorescent protein (GFP) fusion. As previously described (57), cells that lack MreB are spherical in shape. In mreB cells, the IcsA-GFP fusion displays multiple fluorescent foci, which contrasts with the polar or bipolar fluorescence observed for the fusion in wild-type cells (12). We demonstrate that, in cells that lack MreB, a subgroup of polar proteins localizes to multiple sites, which is consistent with the presence of multiple positions that contain ectopic polar material.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
E. coli YK4104 (Nalr) (F− araD139 lacU169 rpsL thi pyrC46 gyrA thyA his flaD), a K-12 derivative that lacks flagella (26), was selected for use in the microscopic screen because the absence of flagellum-based motility facilitated the imaging of live cells. E. coli MC1000 ΔmreB::FRT-cat-FRT, which contains a nonpolar mutation in mreB, was a gift from K. Gerdes (34). E. coli 2443 (Strr) [thr-1 leuB6 Δ(gpt-proA)66 argE3 thi-1 rfbo8 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl51 rpsL31 kdgK51], which expresses intact O antigen serotype 8, was a gift from A. T. Maurelli (48). Because the E. coli outer membrane protease OmpT hydrolyzes IcsA that is localized to the outer membrane, ompT was inactivated by introducing the ompT::km allele from AD202 (1) into appropriate strains by P1L4 phage transduction (41). A22, a small molecule that inhibits MreB (23), was a gift of J. K. Wagner and Y. V. Brun and was used at 10 μg/ml as described previously (23).
Plasmids pBAD-IcsA1-104-GFP, pBAD-IcsA507-620-GFP, and pBAD-IcsAΔ507-729-GFP express hybrid proteins consisting of various segments of IcsA fused in frame to green fluorescent protein (12) under the control of the arabinose promoter of pBAD24 (25). Ptac-IcsA507-620-mCherry is pGZ119EH (35), with IcsA residues 507 through 620 fused in frame to a derivative of mCherry (50) that has been codon optimized for E. coli (gift of K. Marians and S. Sandler) under the control of the tac promoter. Complementation plasmids pACYC184-mreB and pACYC184-mreBCD were generated by cloning into the NcoI and EcoRI sites of pACYC184 (11) PCR products that contain mreB or mreBCD as well as 253 nucleotides upstream of the ATG start codon of mreB. Full-length IcsA was expressed from its own promoter in a pBR322 vector backbone (39). The GFP-EpsM expression vector was similar to that previously described by Scott et al. (49). It was generated by cloning gfp at the amino terminus of and in frame with epsM in pBAD24 (25), with the coding sequence for a six-residue linker (Leu-Cys-Leu-Gln-Gly-Glu) placed between the coding sequences of GFP and EpsM. The CheY-yellow fluorescent protein (YFP) expression vector pVS1 was a gift from V. Sourjik and H. Berg (55).
Except for the studies indicated below, bacteria were grown in Luria-Bertani (LB) broth and on LB agar plates. For the analysis of auxotrophy of the YK4101 pBAD-IcsA506-620-GFP transposon library, minimal media containing 0.2% glucose, M9 salts (6 g liter−1 Na2HPO4, 3 g liter−1 KH2PO4, 0.5 g liter−1 NaCl, 1 g liter−1 NH4Cl), 1.5% agar, 0.5 μg ml−1 thiamine, 150 μM thymine, 50 μM uracil, 0.1 mM CaCl2, 0.5 mM MgSO4, and 40 μg ml−1 histidine were used. CheY localization studies were performed in the 2443 strain background in tryptone broth medium (55), since in our MC1000 strain background or in other media, CheY did not localize to the pole. This was not surprising, as the expression of chemotaxis and flagellar proteins has been shown to be modulated by the presence or absence of mobile insertion sequence elements in many E. coli K-12 strains (5). Where appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 25 μg ml−1; tetracycline, 12.5 μg ml−1; and kanamycin, 50 μg ml−1.
Construction of a transposon mutant library of E. coli YK4104 carrying pBAD-IcsA507-620-GFP.
Transposon mutagenesis of E. coli YK4104 carrying pBAD-IcsA507-620-GFP was performed using Tn10 derivative number 105 (30). The transposon was introduced into cells via the delivery vehicle λNK1324 as described previously (30). In brief, 1 liter of an overnight culture of the recipient strain was concentrated 10-fold and was transduced with λNK1324 at a multiplicity of infection of between 0.1 and 1:1. The phage was allowed to absorb to the bacteria for 15 min at room temperature, which was followed by 15 min at 37°C. The cells were then centrifuged, washed with 1 ml of LB containing 50 mM sodium citrate, resuspended in the same medium, and incubated for 1 h at 37°C. The cells were then plated on selective media and incubated overnight at 39°C. Individual colonies were picked into 96-well plates, grown overnight, diluted 1:1 in LB containing 30% glycerol, and stored at −80°C.
The randomness of the library was investigated by determining the percentage of mutants that were auxotrophs. Random mutagenesis of the chromosome is accompanied by 1% of mutants displaying auxotrophy (31). In our library, 1.6% of mutants examined (n = 500) were unable to grow on minimal media containing glucose as the sole carbon source, in parallel with robust growth on LB, consistent with transposon insertion in E. coli YK4104 pBAD24-icsA507-620-gfp having occurred in a random fashion. Transposon insertion into pBAD24-icsA507-620-gfp occurred at a low frequency (0.4%) and therefore did not constitute a significant percentage of the library or contribute considerably to phenotypic false positives during subsequent screening.
Screen of transposon library of E. coli YK4104 pBAD-IcsA507-620-GFP for mutants with aberrant localization of IcsA507-620-GFP.
Individual mutants were inoculated in LB containing antibiotics and 0.2% glycerol in wells of 96-well plates and were grown overnight at 37°C. Overnight cultures were back diluted at a 1:20 ratio into fresh media lacking glycerol and were grown for 90 min at 37°C with agitation at approximately 450 to 500 rpm on a platform shaker (Lab-Line Instruments, Inc.). Expression of the IcsA507-620-GFP fusion protein was induced from the arabinose promoter by the addition of l-arabinose to a final concentration of 0.2% followed by incubation for 15 min at 37°C. Bacterial cultures were diluted 1:5 in fresh medium containing l-arabinose into wells of glass-bottomed 96-well plates and centrifuged at 700 × g for 10 min in a tabletop centrifuge. Microscopic determination of the localization pattern of the IcsA507-620-GFP fusion protein in each mutant screened was performed as described below.
Identification of the sites of transposon insertion.
The site at which the transposon had inserted in specific mutants was determined by arbitrary PCR and sequencing as described previously (45). To verify that the phenotype resulted from a single transposon insertion, the chloramphenicol-resistant locus was introduced back into the parent strain by P1L4 transduction (41), and the phenotype was confirmed for each of several transductants.
Expression of fusion proteins.
The imaging of protein localization in individual strains not part of the library screen was performed as follows. For expression of IcsA-GFP fusion proteins, GFP-EpsM, or GFP alone, l-arabinose was added to final concentrations of 0.2% to exponential phase cultures in LB medium, and growth was continued for either 10 min at 37°C or 40 to 50 min at 25°C, essentially as described previously (12). For expression of CheY-YFP, l-arabinose was added to final concentrations of 0.005% to back dilutions in tryptone broth medium of overnight cultures and growth was continued for 4 h at 33°C, as described previously (55). For expression of IcsA507-620-mCherry, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to final concentrations of 1 mM to exponential-phase cultures in LB medium, and growth was continued for either 10 min at 37°C or 40 to 50 min at 25°C.
Microscopy.
Imaging of full-length secreted IcsA on the surfaces of intact fixed cells was performed by indirect immunofluorescence using an antibody to IcsA as described previously (24). Imaging of GFP and YFP protein fusions in live cells was performed as follows. Five microliters of culture was placed on agarose-coated microscope slides and left to sit at room temperature for 10 min to allow solidification of the agarose, which immobilizes the cells. Fluorescence and phase or differential interference contrast (DIC) microscopy was performed using a 100× oil immersion objective on a Nikon TE300 or Nikon Eclipse TE2000 microscope with Chroma Technology filters. Images were captured digitally using a black-and-white Photometrics Sensys or CoolSNAP HQ (Photometrics, Roper Scientific) charge-coupled-device camera and IP Lab (Scanalytics, Billerica, Mass.) software.
For imaging by confocal microscopy, bacteria were centrifuged onto cover glasses and fixed as described previously (24). Bacterial membranes were labeled by growth in the presence of 1 μg ml−1 of the membrane dye FM4-64 (Molecular Probes) for 1 h at 37°C prior to harvesting and fixation. Microscopy was performed using a Bio-Rad Radiance 2000 microscope and Image-Pro Plus software (Media Cybernetics, Silver Spring, Md.). Color figures were assembled by separately capturing signals with each of the appropriate filter sets and digitally pseudocoloring the images using Adobe Photoshop software.
Tabulation of numbers of foci.
The number of GFP foci per cell was determined for live cells by visualizing multiple focal planes at the microscope. All foci that were detectable by eye were counted. The number of foci in 60 bacteria chosen randomly under phase imaging from each of three independent experiments was tabulated. Individual cells not expressing IcsA-GFP fusion proteins were not tabulated.
RESULTS
Identification of mreBCD in a screen for transposon insertion mutations leading to altered localization of the polar Shigella protein IcsA.
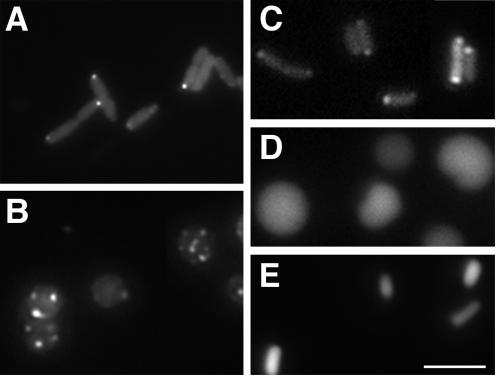
The Shigella actin assembly protein IcsA localizes to the Shigella pole (24). In an effort to identify nonessential proteins required for the polar localization of IcsA, we conducted a screen of transposon insertion mutants for mutations that lead to altered localization of IcsA. IcsA residues 507 to 620 contain one of two IcsA sequences that are able to localize a GFP fusion to the pole within the bacterial cytoplasm (12). Although IcsA is present only in Shigella, when full-length IcsA or a polar IcsA-GFP fusion is introduced into other members of the Enterobacteriaceae family, it also localizes to the poles (12), indicating that the mechanism of polar localization of IcsA is conserved. For this reason and because the distribution of IcsA507-620-GFP can be easily determined in living cells by microscopy, we performed our screen in an E. coli strain that carries an arabinose-inducible IcsA507-620-GFP (pBAD24-icsA507-620-gfp [12]). To facilitate the microscopy of live cells, we used a strain that is nonflagellated (strain YK4104 [26]). In this strain, IcsA507-620-GFP formed polar foci (Fig. 1A), as has been described previously for other E. coli strains (12, 27, 44). A library of approximately 30,000 individual transposon insertions was generated in YK4104 pBAD24-icsA507-620-gfp, and single colonies were stored individually in wells of 96-well plates.
FIG. 1.
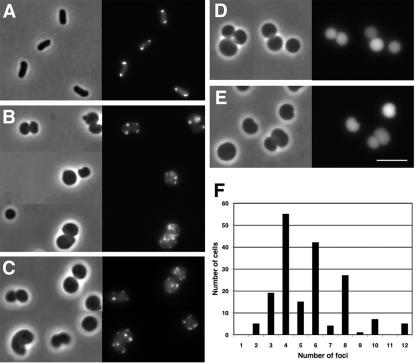
Localization of a cytoplasmic derivative of IcsA to multiple sites in cells that lack MreB. Fluorescence and bright-field microscopy of live E. coli cells expressing translational fusions of cytoplasmic derivatives of IcsA to GFP. Fluorescence images (right panels) and phase images (left panels) of the same microscopic fields are shown. (A) Rod-shaped strain YK4104 expressing IcsA507-620-GFP, which contains a polar targeting sequence of IcsA. (B) Spherically shaped Tn10 library mutant YK4104 mreB::Tn10 expressing IcsA507-620-GFP. (C) Nonpolar mreB mutant MC1000 ΔmreB::FRT-cat-FRT expressing IcsA507-620-GFP. (D) MC1000 ΔmreB::FRT-cat-FRT expressing IcsAΔ507-730-GFP, which lacks polar targeting sequences of IcsA. (E) MC1000 ΔmreB::FRT-cat-FRT expressing GFP alone. (F) Distribution of the numbers of IcsA507-620-GFP foci per MC1000 ΔmreB::FRT-cat-FRT cell (n = 180). Size bar, 5 μm.
Approximately 7,000 mutants were screened by fluorescence microscopy for altered localization of IcsA507-620-GFP. About 0.3 percent of the mutants showed filamentous growth, a mixed population of rods and filaments, minicells and filaments, or chains; in each of these, IcsA-GFP foci were present in a stepwise fashion along the lengths of the filaments at sites corresponding to or near potential cell division sites, as we have observed previously in cells in which cell division is blocked (27, 44). One mutant displayed an IcsA-GFP localization pattern that differed dramatically from that which we have observed previously. The cells of this mutant were rounded and contained multiple fluorescent IcsA-GFP foci (Fig. 1B).
To verify that the phenotype of the round mutant was due to a single Tn10 insertion into the chromosome of E. coli YK4104, the mutation was moved into the parental background by P1L4 transduction (41). Each of several transductants displayed morphologies and targeting patterns of IcsA507-620-GFP identical to those of the original mutant (data not shown). Sequencing of the DNA flanking the Tn10 transposon in this strain revealed that the transposon was inserted within mreB, 323 nucleotides downstream of its translational start codon.
MreB is a highly conserved homolog of eukaryotic actin that forms a helical filament within the bacterial cytoplasm (29, 34, 56) and is important for determining cylindrical cell shape in E. coli, B. subtilis, and C. crescentus (14, 36, 57). In the absence of MreB, cells display an aberrant spherical shape (22, 29, 34, 36, 52, 57) and defective chromosome segregation (23, 34, 54). In C. crescentus, alteration of the level of MreB has also been shown to disrupt the polar localization of certain dynamic cell cycle regulatory proteins (22). The molecular mechanism by which MreB controls cell width is unknown.
Requirement of mreBCD to complement the IcsA localization phenotype as well as the shape phenotype of mreB cells.
In E. coli, mreB is organized upstream of and in an operon with mreC and mreD. To assess the effect of disruption of mreB on IcsA localization independent of polar effects on the transcription of mreC and mreD, we examined IcsA localization in a strain in which mreB is disrupted by a chloramphenicol acetyltransferase (cat) cassette designed to provide read-through transcription of downstream genes (strain MC1000 ΔmreB::FRT-cat-FRT; gift of K. Gerdes). In this strain, the localization pattern of IcsA507-620-GFP was indistinguishable from that of the transposon insertion mutant (Fig. 1C). Moreover, the localization of the IcsA-GFP fusion was unaltered in a derivative of MC1000 ΔmreB::FRT-cat-FRT in which the cat gene had been removed by expression of Flp recombinase (data not shown). Except where otherwise noted, the MC1000 ΔmreB::FRT-cat-FRT strain was used for all subsequent studies.
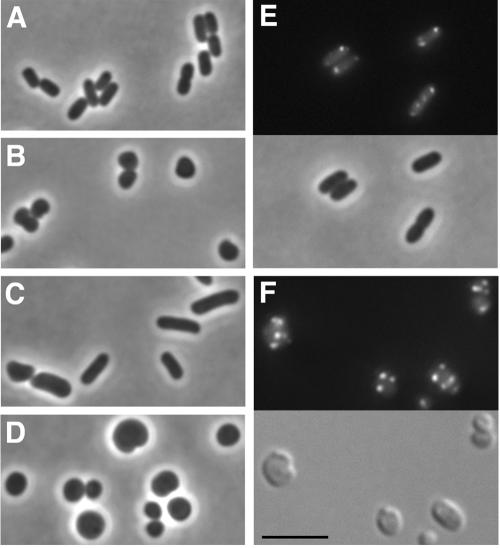
Investigators have previously observed that expression in trans of all three genes of the mreB operon (mreBCD) is required to complement the shape defect of strains carrying deletions in mreB, even when the deletion is in frame (E. coli [34]) or otherwise provides read-through transcription of downstream genes (B. subtilis [29]). We observed a similar requirement for all three genes to complement the shape defect of either mreB::Tn10 cells or ΔmreB::FRT-cat-FRT cells (Fig. 2A through D and data not shown). mreB cells in which mreBCD was expressed in trans were rod shaped, albeit slightly wider in diameter and longer than wild-type cells (Fig. 2, compare C to A). In contrast, mreB cells in which only mreB was expressed in trans were round and indistinguishable in shape from mreB mutant cells (Fig. 2, compare D to B). We then examined whether the partial rescue of the cell shape defect upon expression of mreBCD in trans led to the rescue of the IcsA localization phenotype. In mreB cells in which mreBCD was expressed in trans, IcsA507-620-GFP formed discrete fluorescent foci at the cell poles, in a manner similar to its pattern of localization in wild-type cells (Fig. 2E), indicating that the localization of IcsA was rescued by the expression of mreBCD in trans.
FIG. 2.
Requirement of mreBCD for partial complementation of morphological and IcsA localization phenotypes of mreB cells. Bright-field (panels A to D and E, bottom), DIC (panel F, bottom), and fluorescence (panels E and F, top) microscopy of live E. coli mreB cells, with or without complementation with mreB or mreBCD in trans. (A) Rod-shaped wild-type strain. (B) Spherically shaped MC1000 ΔmreB::FRT-cat-FRT. (C) MC1000 ΔmreB::FRT-cat-FRT expressing mreBCD in trans. (D) MC1000 ΔmreB::FRT-cat-FRT expressing mreB in trans. (E) MC1000 ΔmreB::FRT-cat-FRT expressing mreBCD and IcsA507-620-GFP. (F) MC1000 incubated with the MreB inhibitor A22 for 2 h and expressing IcsA507-620-GFP. Size bar, 5 μm.
IcsA localization following specific inhibition of MreB with A22.
Data published while this work was under review indicate that mreB is essential in E. coli (33), a fact which suggests that the mreB mutant strains used for many of the experiments presented here likely contain a mutation(s) that suppresses the viability defect that results from the disruption of mreB. To determine whether the localization phenotype we observed was due to the mutation in mreB rather than to a suppressor mutation, we examined the localization of IcsA-GFP in wild-type cells treated with A22, a small molecule that specifically inhibits MreB (23). After 1 h of incubation of exponential-phase wild-type cells (MC1000) with A22, many cells appeared rounded, and by 2 h of incubation with A22, the cells were morphologically indistinguishable from mreB mutant cells (Fig. 2F, bottom panel). Upon expression of IcsA-GFP in the A22-treated cells, multiple fluorescent foci were visible within individual cells, indistinguishable from the appearance of IcsA-GFP in mreB mutant cells (Fig. 2F, top panel; compare to Fig. 1B and C). We conclude that the IcsA localization phenotype observed in mreB mutant cells is due to the mutation in our mreB and not to a suppressor mutation.
Specificity of the IcsA localization pattern in mreB cells.
We have shown previously that localization of the IcsA507-620-GFP fusion protein is specific and not a result of the formation of inclusion bodies or protein aggregates (12, 27, 44). To assess the specificity of the targeting patterns we observed here, we examined the distribution in mreB cells of IcsAΔ507-729-GFP, which lacks residues 507 to 620 and therefore targets significantly less efficiently than IcsA507-620 in wild-type cells (12, 44). Similar to the pattern seen previously for this fusion in other strains, in the mreB strain, expression of IcsAΔ507-729-GFP led to diffuse fluorescence in the majority of cells (Fig. 1D), with foci present in only a small minority of cells. In addition, the expression of GFP alone led to diffuse fluorescence (Fig. 1E). These distribution patterns were similar in all strain backgrounds used in this study and are consistent with the localization of IcsA507-620-GFP being specific rather than a result of nonspecific aggregation or inclusion body formation. Consistent with this conclusion is the observation that when inclusion body formation is induced in E. coli, most cells that contain inclusion bodies contain only one (10), whereas in the vast majority of mreB cells expressing IcsA507-620-GFP in the present study, multiple fluorescent foci were observed. Thus, although we cannot definitively eliminate the possibility that the observed fluorescent foci represent inclusion bodies, our results indicate that in the absence of MreB, positional information recognized by IcsA may be present at multiple sites in the cell. Since IcsA507-620-GFP is cytoplasmic (12), these data indicate that, in cells lacking MreB, multiple sites that contain polar positional information recognized by IcsA may be present at the cytoplasmic face of the inner membrane. Thus, MreB may play a role in modulating the organization or number of polelike sites per cell.
Confocal microscopy of IcsA localization in mreB.
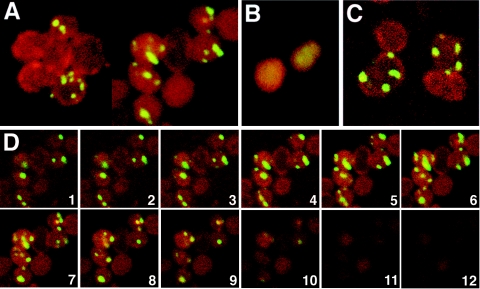
As viewed by standard fluorescent microscopy, IcsA-GFP foci appeared to be distributed in multiple focal planes of mreB cells. To better characterize the three-dimensional distribution of these foci, we imaged ΔmreB::FRT-cat-FRT cells expressing IcsA507-620-GFP by confocal microscopy. In essentially every cell that visibly expressed the fusion protein, multiple fluorescent foci were present (Fig. 3A). In individual z planes, with images taken at 0.3-μm intervals through the cell, the foci were generally at the cell periphery, adjacent to the membrane, although a few foci appeared to be more centrally located (Fig. 3C and D). However, no distinct pattern of distribution of the fluorescent foci could be ascertained. The expression of IcsAΔ507-729-GFP, which lacks polar localization residues 507 to 620, led to a fluorescent signal that was diffuse in the cell cytoplasm (Fig. 3B).
FIG. 3.
Confocal microscopic analysis of IcsA localization to multiple sites in mreB cells. Shown are fluorescence confocal microscopic images of mreB cells expressing IcsA507-620-GFP or IcsAΔ507-730-GFP (green) and stained with the membrane dye FM4-64 (red). (A) Stacks of z series images of MC1000 ΔmreB::FRT-cat-FRT cells expressing IcsA507-620-GFP, taken at 0.3-μm intervals. (B) Stacks of z series images of MC1000 ΔmreB::FRT-cat-FRT cells expressing IcsAΔ507-730-GFP, which lacks polar targeting sequences, taken at 0.3-μm intervals. (C) Single z plane image of MC1000 ΔmreB::FRT-cat-FRT cells expressing IcsA507-620-GFP. (D) Images from a single z series, taken at 0.3-μm intervals, from the lower half through to the top of a cluster of MC1000 ΔmreB::FRT-cat-FRT cells expressing IcsA507-620-GFP.
Quantification of the number of IcsA507-620-GFP foci per mreB cell.
As indicated by the micrographs described above, the number of IcsA507-620-GFP foci per mreB cell was much greater than the number normally present in wild-type cells. Whereas the mean number of foci per wild-type cell is 1.5 (27), the mean number of foci per mreB cell was 5 and the median was 4 (Fig. 1F), excluding cells in which the GFP signal was below the level of detection. The number of foci in a single mreB cell ranged from 2 to 12. Of note is the fact that a significant majority of cells (78% ± 13%) contained an even number of foci, whereas a minority (22% ± 13%) contained an odd number of foci (Fig. 1F).
On the surfaces of mreB cells, IcsA is present in multiple faint patches.
Whereas the distribution of the IcsA-GFP fusion was consistent with polar positional information being present at multiple sites within the cytoplasms of mreB cells, we wished to examine whether these sites might resemble cell poles in all layers of the cell envelope. We therefore tested whether full-length IcsA, which is normally translocated to the cell surface, showed a pattern of distribution similar to that of the IcsA-GFP derivatives. In wild-type cells, full-length IcsA is secreted at the pole (9), and in the presence of a complete lipopolysaccharide (LPS), is retained at the pole (46, 48). In the absence of a complete LPS, diffusion in the outer membrane is altered such that IcsA becomes delocalized from the pole, leading to a more uniform distribution of the protein on the bacterial surface (46-48). Such is the case in E. coli K-12 strains (48), which express a truncated O antigen (37). Since, in these experiments, we wished to examine the localization of IcsA on the surfaces of mreB cells, we introduced the mreB::Tn10 mutation into E. coli strain 2443, which expresses an intact LPS of the O8 serotype (42, 48). In addition, we introduced into this strain a disruption of the gene encoding the outer membrane protease OmpT, since OmpT cleaves IcsA, thereby removing it from the cell surface (43). The cellular morphology and IcsA507-620-GFP localization phenotype of 2443 mreB ompT cells were identical to those of the other mreB strains used in this study (data not shown).
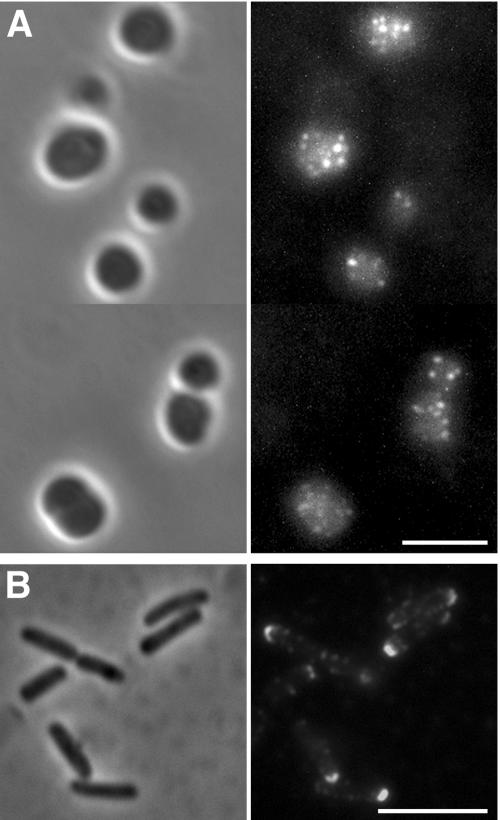
On intact 2443 mreB ompT cells, full-length IcsA was detected in multiple patches that appeared to be distributed over the entire cell surface (Fig. 4A). This distribution is distinctly similar to what we observed for IcsA-GFP foci in the cytoplasm (Fig. 1A and B). In contrast, on cells of the rod-shaped isogenic MreB+ strain (2443 ompT), full-length IcsA formed a cap on the cell pole (Fig. 4B) as has been described previously (44, 47). The fluorescent signal detected from IcsA on the surface of mreB cells was markedly weaker than that detected from IcsA on the surface of wild-type cells, requiring digital enhancement before it could be readily visualized. However, the raw pixel value of the foci was consistently 1.5- to 1.7-fold greater than the background on the slide and 1.2- to 1.4-fold greater than the diffuse signal within the cells. Along with the observation that cytoplasmic IcsA localizes to multiple sites in mreB cells, these data are consistent with the presence in these cells of multiple polelike sites that not only contain polar positional information but are at least partially competent for the secretion of polar outer membrane proteins.
FIG. 4.
Distribution of full-length secreted IcsA on the surface of mreB cells. Shown are results from immunofluorescence (right panels) and bright-field (left panels) microscopy of intact cells expressing full-length secreted IcsA. Derivatives of E. coli strain 2443, which contains an intact LPS, are depicted. Detection of surface IcsA was by indirect immunofluorescence using antibody to the extracellular domain of IcsA. (A) 2443 mreB::Tn10 ompT expressing IcsA. Because the signal was weak, the images were adjusted to enhance the intensity of the fluorescent signal and its contrast with the background signal. (B) 2443 ompT expressing IcsA. Size bars, 5 μm.
Other polar proteins and polar localization sequences localize to multiple sites in mreB cells.
Like IcsA residues 506 to 620, IcsA residues 1 to 104 efficiently localize a GFP fusion to the pole, likely by recognizing a distinct structure or epitope (12). As described above, our screen was performed using a GFP fusion to IcsA507-620. We reasoned that if the sites at which IcsA506-620-GFP is localizing in mreB cells contain positional information normally present at poles, then IcsA1-104-GFP would be predicted to display a distribution similar to that of IcsA506-620-GFP. Indeed, in mreB cells, IcsA1-104-GFP formed multiple foci in a pattern that was indistinguishable from that of IcsA507-620-GFP (data not shown).
Many proteins other than IcsA localize to the poles of rod-shaped bacteria. To assess whether the role of MreB in positioning of IcsA is unique to IcsA or common among polar proteins, we examined the localization in mreB cells of the V. cholerae type II secretion protein EpsM and the E. coli chemotaxis protein CheY, each of which localizes to the poles of wild-type E. coli cells (49, 55). EpsM is an inner membrane protein, and a translational fusion of GFP to the extreme amino terminus of EpsM localizes properly in both V. cholerae and E. coli (49). CheY is a cytoplasmic protein, and a translational fusion of GFP (or a GFP derivative) to the extreme carboxy terminus of CheY is functional and localizes properly (55).
In mreB cells expressing GFP-EpsM, multiple fluorescent foci were present (Fig. 5B). The number of foci per cell ranged from 1 to 10. As reported previously (49), in wild-type cells, EpsM-GFP localized to distinct foci at the poles (Fig. 5A). Thus, both the number and distribution of fluorescent foci were remarkably similar to those described above for each of the IcsA-GFP fusions. These results are consistent with the presence in mreB cells of multiple sites that contain positional information recognized by at least a subgroup of polar proteins.
FIG. 5.
Localization of EpsM, but not CheY, to multiple sites in cells that lack MreB. Shown is fluorescence microscopy of live E. coli cells. (A) Wild-type E. coli expressing GFP-EpsM. (B) MC1000 ΔmreB::FRT-cat-FRT expressing GFP-EpsM. (C) E. coli 2443 expressing CheY-YFP. (D) E. coli 2443 mreB::Tn10 expressing CheY-YFP. (E) E. coli 2443 mreB::Tn10 expressing mreBCD and CheY-YFP. Size bar, 5 μm.
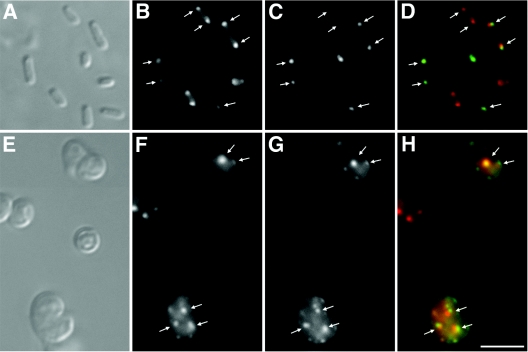
The pole to which IcsA and EpsM preferentially localize is the old pole (24, 49), with, at least in the case of IcsA, a percentage of cells also showing localization to the new pole (27). We examined whether, when expressed in the same cell, IcsA and EpsM would colocalize to the same pole. We expressed in wild-type cells the GFP-EpsM fusion protein and a fusion of IcsA507-620-mCherry. In the majority of cells, the GFP and mCherry foci colocalized (Fig. 6A through D), indicating that IcsA and EpsM target the same pole. Since mCherry is a monomer (50), the colocalization of the two fluorophores does not result from dimerization between mCherry and GFP. The colocalization of IcsA507-620-mCherry with EpsM-GFP also indicates that the two protein fusions do not interfere with each other's localization, suggesting that the epitopes they recognize at the pole are distinct.
FIG. 6.
Simultaneous localization of IcsA and EpsM in wild-type cells and in cells that lack MreB. DIC (A and E) and fluorescence (B through D and F through H) microscopy of live E. coli cells expressing both IcsA507-620-mCherry and GFP-EpsM. Panels A through D show MC1000. Panels E through H show MC1000 ΔmreB. Panels B and F show IcsA507-620-mCherry, panels C and G show GFP-EpsM, and panels D and H show overlays of IcsA507-620-mCherry signal (red) and GFP-EpsM signal (green). Arrows indicate sites of colocalization of the IcsA507-620-mCherry signal and the GFP-EpsM signal. Size bar, 5 μm.
We then tested whether the sites to which EpsM localizes in the mreB cells are the same as those to which IcsA localizes. As in the wild-type cells, IcsA507-620-mCherry fluorescent foci colocalized with GFP-EpsM fluorescent foci (Fig. 6E through H). The colocalization of IcsA507-620-mCherry with GFP-EpsM is consistent with localization of IcsA and EpsM in mreB cells not being a result of nonspecific aggregation or inclusion body formation, although we cannot absolutely exclude the possibility that the two proteins aggregate at the same sites. A minority of cells either expressed only one of the two fluorophores or expressed no fluorophore. However, in those cells that expressed both fluorophores, the number of GFP foci was similar to the number of mCherry foci (Fig. 6E through H), consistent with the possibility that individual mreB cells have a distinct number of polelike sites.
In contrast to IcsA-GFP and GFP-EpsM, a YFP fusion to the polar chemotaxis protein CheY displayed a diffuse fluorescent signal in mreB cells (Fig. 5D), whereas under the same growth conditions, CheY-YFP localized to the poles of wild-type cells (Fig. 5C). When mreBCD was expressed in trans, the localization of CheY to the poles was not rescued, although the rod-shaped morphology of the cells was largely restored (Fig. 5E). Therefore, the delocalization of CheY in the mreB cells could be due either to a mutation outside of the mreBCD locus or to incomplete complementation by the expression of mreBCD in trans. Thus, we were not able to definitively determine whether the positional information recognized by CheY at the poles of wild-type cells is present in distinct sites within mreB cells.
DISCUSSION
Our data indicate that E. coli cells that lack the cytoskeletal protein MreB have multiple sites that contain polar material. Two distinct polar targeting sequences of the Shigella protein IcsA localize to multiple sites within the cytoplasms of mreB cells. The polar V. cholerae protein EpsM also localizes to multiple sites within these cells and, when expressed in cells that also express IcsA, colocalizes with IcsA. These data indicate that this pattern of localization is not unique to IcsA but rather common to at least a subgroup of polar proteins. Moreover, they are consistent with a model in which, in the absence of MreB, polar material recognized by these proteins is misplaced and present in multiple sites within the cell. The accumulation of these polypeptides at these sites is independent of the engagement of the secretion apparatus and independent of insertion into the membrane, since one of the IcsA sequences that displays this pattern (IcsA507-620-GFP) lacks both a signal peptide and membrane-spanning domains. Localization of IcsA does not appear to be determined simply by convex shape, since in cells that have been filamented, IcsA will localize at sites of potential poles along the lengths of the filaments (27). Of note is the fact that other investigators have observed a similar distribution of IcsA in cells lacking MreB (L. Rothfield, personal communication). Our results indicate that MreB is required, either directly or indirectly, for the restriction of certain polar material to defined sites within the cell and that, in the absence of MreB, ectopic sites that contain polar material arise.
These data are consistent with recent results pertaining to the role of MreB in the polarity of the developmentally asymmetric organism C. crescentus, which forms a stalk at the older of its two poles. Gitai et al. have shown that upon depletion of MreB, each of four polar proteins is delocalized from the poles and appears either in puncta around the cell perimeter or diffusely in the cell cytoplasm (22). In addition, Wagner et al. have demonstrated that, following depletion and repletion of MreB, cell stalks form at multiple sites around the cell (58). Thus, the role of MreB in localizing polar material appears to be conserved.
We observed that the number of IcsA-GFP foci per cell was even in almost 80% of cells (Fig. 1F). Moreover, when IcsA507-620-mCherry was coexpressed with GFP-EpsM, mCherry foci colocalized with GFP foci, and the number of mCherry foci in a cell was generally the same as the number of GFP foci. The quantity of foci per cell and the predominance of even numbers of foci in individual cells are strikingly similar to what has been observed previously for chromosomes by Kruse et al. (34). These investigators observed that cells with depletions of MreB segregated their chromosomes in pairs and contained increased numbers of chromosomes compared to wild-type cells. More recent data indicate that, in C. crescentus, MreB is required for the segregation of the chromosome origins (23). Taken together, these findings are consistent with the possibility that MreB may play a role in the organization of polar positional information and chromosome segregation in a parallel or linked manner.
The presence of multiple sites to which these polar proteins localize in the absence of MreB could represent a defect in the consolidation of polar material at the two normal polar sites, the unregulated recruitment of polar material to multiple ectopic sites, or a splitting of existing polar material by abnormal new synthesis. MreB, a homolog of eukaryotic actin, polymerizes into filaments that extend the lengths of rod-shaped cells and appear as helices (21, 29, 34). During most of the cell cycle, these helical filaments extend the length of the cell, but in predivisional cells of C. crescentus, they form a tight spiral at midcell (21, 22). Within the filaments, MreB monomers are oriented in a head-to-tail manner (56) such that, like actin filaments, MreB filaments have inherent polarity. Moreover, in B. subtilis, MreB filaments have been shown to display dynamic movement from midcell towards each of the poles and along helical tracts at rates that approximate that of polymerization from the barbed ends of actin filaments (15). In theory, this could be explained by the occurrence of active polymerization at filament ends situated at midcell in association with depolymerization at filament ends situated at the cell poles. To our knowledge, whether a similar directional movement of MreB filaments along helical tracts occurs in other organisms has not yet been tested, but given the conservation of both the MreB protein sequence and the helical distribution of the filaments across all gram-negative and gram-positive bacterial organisms examined to date, it seems likely that its dynamic behavior will be similar in these organisms.
Both our data and previously published data are consistent with a recently proposed model in which material required for establishing polarity might be actively moved along the MreB helix to the two ends of the cell (28), analogous to the way actin is known to mediate the transport of organelles in eukaryotic cells (53). In such a model, proteins translated anywhere within the cell cytoplasm would directly or indirectly bind the MreB filament, and directional movement of the filament subunits would transport the newly translated protein cargo to the poles, where it would dissociate from the filament and be incorporated into the polar site. It is unclear whether the molecules theoretically transported by the MreB filaments would merely establish, either directly or indirectly, a localized polar structure that then directly recruits other molecules to the pole by an MreB-independent mechanism or whether the majority of polar molecules would use the MreB transport system to get to the pole.
Certain data are consistent with MreB serving as a scaffold for peptidoglycan synthesis required for cell elongation and possibly for the coordination of the switch in peptidoglycan synthesis from cell elongation to septum formation that occurs at the time of cell division. In C. crescentus, the positioning of the peptidoglycan-biosynthetic enzyme that is required for cell elongation, PBP2 (6), is dependent on MreB (21). Moreover, the positioning of MreB in spirals at midcell during cell division is dependent on FtsZ (21), and the overexpression of FtsQAZ in mreB cells suppresses the lethality of mutations in mreBCD (33). In contrast, the positional information required for the proper localization of polar proteins such as IcsA to the pole is independent of FtsZ and cell division (27). Therefore, these data are consistent with a model in which the perturbations of polar positional information that we observed in cells that lack MreB are an indirect result of the absence of MreB. The absence of an interaction between IcsA polar localization sequences and MreB in two hybrid analyses (T. Nilsen and M. B. Goldberg, unpublished data), while not eliminating the possibility of a direct interaction, is also consistent with the role of MreB in the polar localization of IcsA being indirect.
Other than MreB, proteins that are important for the development of polarity in bacteria include the LMW PBPs. E. coli cells that lack multiple LMW PBPs are misshapen, with bends, kinks, and branches (17). We have previously shown that, in these LMW PBP mutants, the sites of morphological abnormalities represent ectopic poles, in that Shigella IcsA and V. cholerae EpsM efficiently localize to them (44). The ectopic poles of the LMW PBP mutants permit the efficient translocation of IcsA to the surface (44), suggesting that, at the sites of morphological abnormalities in the LMW PBP mutants, each layer of the cell envelope functions as a pole. How LMW PBP activity, PBP2 activity, and the possible scaffold role of MreB are coordinated in the determination of cell shape is incompletely understood.
Based on our data, we are not able to distinguish whether the effect of the mreB mutation is a defect in the consolidation of polar material at the two normal polar sites or the unregulated recruitment of polar material to multiple ectopic sites. However, two aspects of the data suggest that the altered placement of polar material in these cells is irregular and is not a coordinated misdirection of normal cellular processes. First, the spacing between foci was irregular, even as visualized in single z planes of confocal series (Fig. 3C). Second, the translocation of IcsA into the outer membrane appeared to be inefficient. If specific proteins or structures are required for the translocation of IcsA or other secreted polar proteins into the outer membrane, then these proteins or structures would necessarily need to all be present at the same site, i.e., in wild-type cells, at the pole, and, in mreB cells, at the site to which the protein is targeted in the cytoplasm. The weak signal we observed from secretion-competent IcsA on the surfaces of mreB cells (Fig. 4) suggests that some protein or structure required for its efficient translocation to the outer membrane is inconsistently present at the sites to which the protein initially localizes. This is distinctly different from ectopic poles present in cells that lack multiple peptidoglycan-modifying enzymes, where IcsA is efficiently translocated to the surface (44). The defect in the mreB cells could result from the uncoordinated delivery of components of a translocation machinery to different sites, rather than to the same sites, around the sphere. Alternatively, it could result from pleiotropic effects of the mreB mutation on the cell envelope. Determination of the mechanism by which MreB regulates placement of polar positional information at the ends of rod-shaped cells and of whether this is an indirect effect of a role of MreB in peptidoglycan synthesis requires further investigation.
Acknowledgments
We thank K. Gerdes, A. T. Maurelli, and H. C. Berg for providing strains and plasmids used in this study. We are particularly grateful to K. Marians and S. Sandler for providing the E. coli codon-optimized mCherry gene prior to their publication of it.
This work was supported by grant AI035817 (to M.B.G.) from the National Institutes of Health, a postdoctoral fellowship from the Research Council of Norway (to T.N.), and a Tufts University School of Medicine Harold Williams Scholarship (to A.W.Y.).
REFERENCES
- 1.Akiyama, Y., and K. Ito. 1990. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem. Biophys. Res. Commun. 167:711-715. [DOI] [PubMed] [Google Scholar]
- 2.Alley, M. R., J. R. Maddock, and L. Shapiro. 1992. Polar localization of a bacterial chemoreceptor. Genes Dev. 6:825-836. [DOI] [PubMed] [Google Scholar]
- 3.Ausmees, N., and C. Jacobs-Wagner. 2003. Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu. Rev. Microbiol. 57:225-247. [DOI] [PubMed] [Google Scholar]
- 4.Autret, S., and J. Errington. 2003. A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis. Mol. Microbiol. 47:159-169. [DOI] [PubMed] [Google Scholar]
- 5.Barker, C. S., B. M. Pruss, and P. Matsumura. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532-536. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, J. M. 2000. Localization of the histidine kinase PilS to the poles of Pseudomonas aeruginosa and identification of a localization domain. Mol. Microbiol. 36:153-162. [DOI] [PubMed] [Google Scholar]
- 9.Brandon, L. D., N. Goehring, A. Janakiraman, A. W. Yan, T. Wu, J. Beckwith, and M. B. Goldberg. 2003. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 50:45-60. [DOI] [PubMed] [Google Scholar]
- 10.Carrio, M. M., J. L. Corchero, and A. Villaverde. 1998. Dynamics of in vivo protein aggregation: building inclusion bodies in recombinant bacteria. FEMS Microbiol. Lett. 169:9-15. [DOI] [PubMed] [Google Scholar]
- 11.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles, M., M. Perez, J. H. Kobil, and M. B. Goldberg. 2001. Polar targeting of Shigella virulence factor IcsA in Enterobacteriacae [sic] and Vibrio. Proc. Natl. Acad. Sci. USA 98:9871-9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2003. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 14.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 15.Defeu Soufo, H. J., and P. L. Graumann. 2004. Dynamic movement of actin-like proteins within bacterial cells. EMBO Rep. 5:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Pedro, M. A., K. D. Young, J. V. Holtje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards, D. H., and J. Errington. 1997. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol. Microbiol. 24:905-915. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, D. H., H. B. Thomaides, and J. Errington. 2000. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 19:2719-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 22.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg, M. B., O. Barzu, C. Parsot, and P. J. Sansonetti. 1993. Unipolar localization and ATPase activity of IcsA, a Shigella flexneri protein involved in intracellular movement. J. Bacteriol. 175:2189-2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homma, M., K. Kutsukake, and T. Iino. 1985. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J. Bacteriol. 163:464-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janakiraman, A., and M. B. Goldberg. 2004. Evidence for polar positional information independent of cell division and nucleoid occlusion. Proc. Natl. Acad. Sci. USA 101:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janakiraman, A., and M. B. Goldberg. 2004. Recent advances on the development of bacterial poles. Trends Microbiol. 12:518-525. [DOI] [PubMed] [Google Scholar]
- 29.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 30.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 31.Kleckner, N., R. K. Chan, B. K. Tye, and D. Botstein. 1975. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J. Mol. Biol. 97:561-575. [DOI] [PubMed] [Google Scholar]
- 32.Kocks, C., R. Hellio, P. Gounon, H. Ohayon, and P. Cossart. 1993. Polarized distribution of Listeria monocytogenes surface protein ActA at the site of directional actin assembly. J. Cell Sci. 105:699-710. [DOI] [PubMed] [Google Scholar]
- 33.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 34.Kruse, T., J. Moller-Jensen, A. Lobner-Olesen, and K. Gerdes. 2003. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 22:5283-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin, P. A., P. S. Margolis, P. Setlow, R. Losick, and D. Sun. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174:6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, D., and P. R. Reeves. 1994. Escherichia coli K12 regains its O antigen. Microbiology 140:49-57. [DOI] [PubMed] [Google Scholar]
- 38.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 39.Magdalena, J., and M. B. Goldberg. 2002. Quantification of Shigella IcsA required for bacterial actin polymerization. Cell Motil. Cytoskelet. 51:187-196. [DOI] [PubMed] [Google Scholar]
- 40.Marston, A. L., H. B. Thomaides, D. H. Edwards, M. E. Sharpe, and J. Errington. 1998. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 12:3419-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maurelli, A. T., and R. Curtiss III. 1984. Bacteriophage Mu d1(Apr lac) generates vir-lac operon fusions in Shigella flexneri 2a. Infect. Immun. 45:642-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meier, U., and H. Mayer. 1985. Genetic location of genes encoding enterobacterial common antigen. J. Bacteriol. 163:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakata, N., T. Tobe, I. Fukuda, T. Suzuki, K. Komatsu, M. Yoshikawa, and C. Sasakawa. 1993. The absence of a surface protease, OmpT, determines the intercellular spreading ability of Shigella: the relationship between the ompT and kcpA loci. Mol. Microbiol. 9:459-468. [DOI] [PubMed] [Google Scholar]
- 44.Nilsen, T., A. S. Ghosh, M. B. Goldberg, and K. D. Young. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 46.Robbins, J. R., D. Monack, S. J. McCallum, A. Vegas, E. Pham, M. B. Goldberg, and J. A. Theriot. 2001. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol. Microbiol. 41:861-872. [DOI] [PubMed] [Google Scholar]
- 47.Sandlin, R. C., K. A. Lampel, S. P. Keasler, M. B. Goldberg, A. L. Stolzer, and A. T. Maurelli. 1995. Avirulence of rough mutants of Shigella flexneri: requirement of O antigen for correct unipolar localization of IcsA in the bacterial outer membrane. Infect. Immun. 63:229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandlin, R. C., and A. T. Maurelli. 1999. Establishment of unipolar localization of IcsA in Shigella flexneri 2a is not dependent on virulence plasmid determinants. Infect. Immun. 67:350-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scott, M. E., Z. Y. Dossani, and M. Sandkvist. 2001. Directed polar secretion of protease from single cells of Vibrio cholerae via the type II secretion pathway. Proc. Natl. Acad. Sci. USA 98:13978-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 51.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 52.Shih, Y. L., T. Le, and L. Rothfield. 2003. Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc. Natl. Acad. Sci. USA 100:7865-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simon, V. R., and L. A. Pon. 1996. Actin-based organelle movement. Experientia 52:1117-1122. [DOI] [PubMed] [Google Scholar]
- 54.Soufo, H. J., and P. L. Graumann. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916-1920. [DOI] [PubMed] [Google Scholar]
- 55.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 56.van den Ent, F., L. A. Amos, and J. Lowe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 57.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner, J., S. Setayeshgar, and Y. V. Brun. 2005. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J. Bacteriol. 187:544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, K. D. 2003. Bacterial shape. Mol. Microbiol. 49:571-580. [DOI] [PubMed] [Google Scholar]