Abstract
Six pairs of transcription profiles between glycopeptide-intermediate S. aureus (GISA [or vancomycin-intermediate S. aureus; VISA]) and glycopeptide-susceptible S. aureus (vancomycin-susceptible S. aureus [VSSA], including glycopeptide-susceptible isogenic mutants from VISA) strains were compared using a microarray. Ninety-two open reading frames which were or tended to be increased in transcription in VISA in at least five out of six array combination pairs were evaluated for their effects on glycopeptide susceptibility by introducing these genes one by one into VSSA strain N315 to construct an overexpression library. By screening the library, 17 genes including 8 novel genes were identified as associated with glycopeptide resistance since their experimental overexpression reduced vancomycin and/or teicoplanin susceptibility of N315. The raised MICs of vancomycin and teicoplanin were 1.25 to 3.0 and 1.5 to 6.0 mg/liter, respectively, as compared to 1.0 mg/liter of N315. Three of these genes, namely graF, msrA2, and mgrA, also raised the oxacillin MIC from 8.0 mg/liter for N315 to 64 to ∼128 mg/liter when they were overexpressed in N315. Their contribution to vancomycin and beta-lactam resistance was further supported by gene knockout and trans-complementation assay. By using a plasmid-based promoter-green fluorescent protein gene (gfp) transcriptional fusion system, graF promoter-activated cells were purified, and subsequent susceptibility tests and Northern blot analysis demonstrated that the cells with up-regulated activity of graF promoter showed reduced susceptibility to vancomycin, teicoplanin, and oxacillin. In addition, cell morphology studies showed that graF and msrA2 overexpression increased cell wall thickness of N315 by factors of 23.91 and 22.27%, respectively, accompanied by glycopeptide MIC increments of 3- to 6-fold, when they were overexpressed in N315. Moreover, extended experiments and analyses indicate that many of the genes identified above are related to the cell wall biosynthetic pathway, including active nutrient transport systems. We propose that the genes which raise glycopeptide resistance in S. aureus function toward altering the cell wall metabolic pathway.
The glycopeptide vancomycin has been used successfully for the treatment of serious infections caused by multidrug-resistant Staphylococcus aureus for several decades, and some isolates were reported to be susceptible only to this antibiotic (30, 36). However, with the worldwide emergence of glycopeptide-intermediate S. aureus (GISA [or vancomycin-intermediate S. aureus; VISA]) strains with vancomycin MICs of 8 or 16 mg/liter (5, 17), the likelihood of this resistance reaching global magnitude causes great concern about the possibility of losing the last treatment option for multidrug-resistant staphylococcus infection. Several factors have been reported to be involved in the mechanism of glycopeptide resistance in VISA, including thickened cell wall (7, 11), accumulation of surplus cell wall material (48), reduced peptidoglycan cross-linking (4, 12), inactivation of penicillin-binding protein D (PBP4) (9, 47), a decrease in the level of the muropeptide amidation (4, 7), and/or other cell wall alterations, such as increased glycan chain length (23). The phenomenon of thickened cell wall was discovered in our early study of the first VISA strain, Mu50 (11). Subsequent investigation of the isogenic cells with different cell wall thickness prepared from a single colony of Mu50 cells demonstrated that cell wall thickening is the major factor contributing to glycopeptide resistance in Mu50 (7). Recently we found that 16 VISA clinical isolates collected from seven countries all showed a distinct thickened cell wall compared to glycopeptide-susceptible S. aureus strains (6, 21, 41). An immanent link between cell wall thickness and the level of glycopeptide resistance was also demonstrated in a comparison study using triple sets of 16 VISA strains, their glycopeptide-susceptible derivatives (vancomycin-resistant S. aureus [VRSA]-P) produced by passage on drug-free medium, and phonotically glycopeptide-resistant revertants (VRSA-PR) selected from VRSA-P on vancomycin plates (5, 6). Contrary to the phenotypic mechanism, the genetic mechanism of VISA is far from being clear. Different VISA strains have been studied by many researchers, and altered expression of genes such as pbp2 (46), pbpD (9, 47), sigB (2, 11), ddh (3, 34), tcaA (33), and vraSR (26) were reported. Several sets of up- and down-regulated genes associated with vancomycin resistance, including regulators, were also reported (26, 35, 54). However, until now none of the genes was altered without exception in VISA strains and none could be attributed to VISA strains specifically. Thus there is a need for additional knowledge on the genetic mechanism(s) of glycopeptide resistance acquisition in VISA and for defining genetic alteration(s) resulting in phenotypic change, such as cell wall thickening. Recently, in an effort to efficiently discover genes involved in the mechanism of glycopeptide resistance in S. aureus, a DNA microarray which consists of PCR products covering the whole genome of N315 was used to identify open reading frames (ORFs) that are expressed differently between VISA and VSSA strains. In this report, we describe our findings from transcriptional profiling studies on genes that are differentially expressed as well as genetic studies on genes that are increased in transcription in glycopeptide-resistant strains.
MATERIALS AND METHODS
Bacterial strains.
The sources and relevant characteristics of the bacterial strains used in this study are listed in Table 1. Glycopeptide-susceptible isogenic derivatives (Mu50-P and MI-P) from clinical VISA strains Mu50 and MI and phenotypically glycopeptide-resistant revertants (Mu50-PR and MI-PR) selected from Mu50-P and MI-P were generated in vitro and characterized in our previous study along with their original strains, Mu50 and MI (6, 7). Hetero-VISA strain Mu3 and VISA strain Mu50 share an identical pulsed-field gel electrophoresis banding pattern, and are supposed to have originated from the same ancestor of clonotype II-A represented by VSSA strain N315 (18). All strains were grown in brain heart infusion (BHI) broth (Difco, Detroit, MI) in the absence of antibiotics at 37°C with aeration unless indicated otherwise. For each experiment, an overnight culture was diluted 100-fold in prewarmed fresh BHI broth and further incubated with aeration to ensure exponential growth condition before sampling. Cell growth was monitored by measuring the optical density of the culture at 578 nm (OD578) with a spectrophotometer (Pharmacia LKB Biotechnology, Inc., Uppsala, Sweden).
TABLE 1.
Phenotypic characteristics of strains used in transcriptional profile comparison
| Straina | Source | Antibiotic MIC (mg/liter)b
|
Cell wall thickness in nm (mean ± SD)c | |||
|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | Oxacillin | Imipenem | |||
| Mu50 | Clinical isolate | 8 | 13 | 512 | 26 | 35.02 ± 4.01 |
| Mu50-P | In vitro mutant | 2 | 7 | 512 | 58 | 24.45 ± 7.80 |
| Mu50-PR | In vitro mutant | 7 | 16 | 512 | 29 | 34.19 ± 4.96 |
| MI | Clinical isolate | 9 | 24 | 512 | 55 | 32.03 ± 3.60 |
| MI-P | In vitro mutant | 2 | 2 | 256 | 43 | 24.13 ± 5.54 |
| MI-PR | In vitro mutant | 7 | 9 | 512 | 64 | 32.03 ± 6.75 |
| Mu3 | Clinical isolate | 2 | 17 | 1,024 | 64 | 26.53 ± 4.80 |
| N315 | Clinical isolate | 1 | 1 | 8 | 1 | 21.46 ± 2.25 |
Mu50-P and MI-P are vancomycin-susceptible derivatives obtained from VISA strains Mu50 and MI by passaging them in drug-free medium for 35 and 84 days, respectively. Mu50-PR and MI-PR are vancomycin-resistant derivatives of Mu50-P and MI-P, respectively, obtained by one-step selection with vancomycin at 4 mg/liter (6).
The MIC determination was performed by the agar dilution method (6).
Cell wall thickness was measured from a transmission electron microscopy photograph (6).
Susceptibility tests.
The MIC determination was performed by both agar dilution and E-test, and population analysis was performed as described previously (16). For gene overexpression library screening, an antibiotic gradient gel assay was adopted by swabbing a 0.5 McFarland standard suspension of overnight cultures along an antibiotic gradient in rectangular agar plates.
Transmission electron microscopy.
Preparation for and examination of S. aureus cells by transmission electron microscopy were performed as described previously (7). Morphometric evaluation of cell wall thickness was performed using photographic images at a final magnification of ×30,000.
DNA microarray preparation, hybridization, and data analysis.
Two glass DNA microarray systems were used in this study. One consisted of 2,166 PCR products covering the whole genome of N315, prepared as described previously (25). This array was designed to cover the whole genome of N315 via selection of 2,166 out of 35,000 clones that were used in the N315 whole-genome sequencing project (27). One advantage of this array was that it covers many intergenic regions; however, some genes which are small in size were not cloned individually and some spots contained more than one gene, resulting in an inability to evaluate all genes accurately in the transcription profile. Therefore, another version of the microarray was developed. The new version consists of 2,472 PCR products from the N315 genome that represent all N315 ORFs. ORFs having at least 70% similarity in nucleotide sequence within the N315 genome were defined as the homologue ORF set (HOS), and only one of them was retained for preparing array probes. As a result, 123 ORFs belonging to HOS were eliminated from original 2,595 ORFs. The full length of the ORF (if <1.5 kb) or about a 1.5-kb fragment of ORFs that were greater than 1.5 kb in size was PCR amplified and cloned into the pCR2.1-TOPO vector to establish an N315 ORF library in Escherichia coli. PCR products amplified from the N315 ORF library with the universal primer set for pCR2.1-TOPO were used to prepare a glass DNA microarray as described previously (25). RNA preparation and the subsequent entire microarray procedure except for mRNA labeling were also performed as described previously (25). Targeting RNA was labeled with Cy3 or Cy5 dyes by using SuperScript indirect cDNA labeling system (Invitrogen Life Technologies, Carlsbad, California). A total of six paired competitive hybridizations (see Table S1 in the supplemental material) were performed using two different versions of microarrays. For each version, experiments were performed with different lots of RNA in duplicated experiment sets. Data analysis was performed by introducing a total RNA approach (13, 25), and the results were given by the average signal intensity ratio of VISA versus VSSA for each array target.
Construction of pYT3-based gene overexpression library.
A DNA fragment containing the entire gene (putative promoter, ORF, and terminator) was obtained by PCR amplification with genomic DNA of strain Mu50 as a template and cloned downstream of the tetL gene of shuttle vector pYT3 (11). Genes that were mutated in Mu50 compared to N315 were cloned from both Mu50 and N315 (see Table S1 in the supplemental material). The resultant plasmid was introduced into VSSA strain N315 by electroporation to generate a gene overexpression library.
DNA isolation, recombinant DNA techniques, and bacterial electroporation.
Plasmid DNA was isolated from E. coli using the Quantum Prep miniprep kit (Bio-Rad) according to the manufacturer's instructions. S. aureus DNA isolations were performed as described previously (52). Restriction endonuclease digestion, ligation reactions, and DNA cloning were carried out in accordance with the manufacturers' instructions. Electroporation of S. aureus was performed with a Bio-Rad Gene Pulser with a pulse controller as described previously (28).
Construction of mutants by allelic exchange.
To generate a gene-null knockout mutant, the 822-bp DNA fragment containing the cat gene was amplified from plasmid pRIT5 (38) with forward and reverse primers. The PCR product was digested with HindIII and SalI then ligated into the cut site of pUC118, which had been cut previously with HindIII and SalI enzyme to generate a chloramphenicol-resistant cassette vector. After that, about 1.5 kb of each junction (L- and R-junctions) of the target gene were cloned upstream and downstream of the cat gene cassette, respectively. The insertion was subcloned into tetracycline-resistant temperature-sensitive E. coli-S. aureus shuttle vector pYT3, which was then introduced into N315 by electroporation to yield chloramphenicol- and tetracycline-resistant transformants. The transformants with vector integration into chromosome DNA (single crossover) were selected by growing on 43°C and verified by PCR with primer sets located at pYT3 vector specifically and beyond the L-junction and/or R-junction on the chromosome DNA. For selection of allelic replacement mutants, the resulting single-crossover mutants were cultured with BHI broth containing 10 mg/liter chloramphenicol at 30°C overnight, and their serial dilutions were plated on BHI agar plates containing 10 mg/liter chloramphenicol. Following overnight incubation, the colonies that grew were replicated onto plates containing 10 mg/liter tetracycline, to select for the colonies that are chloramphenicol resistant and tetracycline susceptible. The allelic replacement mutants (representing possible double-crossover events) were verified by PCR. Finally, the resultant mutants were verified by sequencing if necessary.
Construction of a plasmid based promoter-green fluorescent protein gene (gfp) transcriptional fusion system and fluorescence-activated cell sorting.
The reporter plasmid pRIT5dS-graFp-gfp containing a fusion of the graF promoter region with gfp (a gene coding for GFP) was constructed by cloning the genes on promoterless multiclone site of E. coli-S. aureus shuttle vector pRIT5dS. pRIT5dS was a derivative of pRIT5 with a deletion of spa promoter upstream of the multiclone site. The vector pRIT5 (Pharmacia Biotech, Piscataway, N.J.,) was cut with NdeI and HindIII and self-ligated to generate pRIT5dS. A 734-bp fragment encompassing the gfp gene was PCR amplified with the following primers containing restriction sites (underlined): 5′-AAAAAGGATCC(BamHI)ATGGCTAGCAAAGGAGAAGA-3′ and 5′-AAAAACTGCAC (PstI)CTAGTTAGTCAATCGATGTT-3′, using the vector pQB125/50-fPA (Wako Pure Chemical Industries, Ltd., Osaka, Japan), which harbors the gfp gene as a template. This fragment was first cloned into pRIT5dS, followed by cloning of the graF promoter just upstream of gfp gene to generate pRIT5dS-graFp-gfp. A graF promoter fragment (graFp) extending 151 bp upstream of initial codon was PCR amplified from S. aureus strain Mu50 with primers containing restriction sites (5′-AAAAAGAATTC(EcoRI)TATGTTCACCTCAAAATCAT-3′ and 5′-AAAAAGGATCC(BamHI)TTCCTCCCAAATTGGATAAT-3′). Based on our pre-experiment, six nucleotides at the 3′ end of this fragment were replaced by BamHI restriction site for efficient GFP expression. Restriction analysis and DNA sequencing confirmed the orientation and authenticity of the promoter-reporter gene constructs. The reporter plasmid pRIT5dS-graFp-gfp, containing a fusion of the graF promoter region with gfp (a gene coding for GFP), was introduced into N315 by electroporation, and the culture of the resultant transformant was subjected to cell sorting. Fluorescence levels for the reporter were determined by flow cytometry with a fluorescence-activated cell sorter (FACScan; Becton Dickinson, Paramus, NJ), and fluorescence-activated cells were sorted by automated cell sorter (FACSAria; Becton Dickinson, Paramus, NJ). Appropriate settings for flow cytometry were determined by analyzing known mixtures of GFP-fluorescent and nonfluorescent S. aureus cells using the FL-1 channel. Cells from stationary-phase cultures were then analyzed using identical flow cytometry settings, and the data were analyzed using CellQuest (version 3.3; Becton Dickinson, NJ) software. For fluorescence-activated cell sorting, cells were gated by size based on log-scale forward and side scatter. Samples were run at low sample differential, resulting in a sample flow rate of 6,000 events/s, and cells exhibiting high fluorescence were collected. To isolate the populations exhibiting high fluorescence, the gate was set for collecting 1 in 105 cells. The cells were directly sorted on BHI agar plates, and colonies were harvested after overnight incubation at 37°C.
Northern blot analysis.
Seven micrograms of total RNA was denatured by incubation at 65°C for 15 min after mixing with formamide-bromophenol dye, followed by a quick chill on ice. Subsequent procedures except for probe preparation were performed as described elsewhere (25). The probe was prepared by labeling 50 ng of graF-specific (within ORF) PCR product using Ready-To-Go DNA labeling beads (−dCTP) (Amersham Biosciences, Piscataway, NJ), following removal of unincorporated nucleotides using Centri-Sep spin columns (Applied Biosystems, Foster City, CA).
RESULTS
Identification of genes differentially expressed between VISA and VSSA.
Our overall objective was to identify ORFs that exhibit change commonly in VISA in mRNA abundance in relation to glycopeptide-resistant phenotype. Two approaches were chosen to identify the genes that are associated with the glycopeptide-resistant phenotype. In experiment 1, the transcriptional profiles of two genetically distinct VISA strains (Mu50 and MI) and their isogenic derivatives (Mu50-P, Mu50-PR, MI-P, and MI-PR) that have different levels of glycopeptide resistance were analyzed (Table 1), along with Mu3 and N315. By comparing the transcriptional profiles of the VSSA with that of their respective isogenic or similar genetic background VISA strains, 185 to 265 genes representing approximately 7.13 to 10.20% of the entire N315 ORFs were identified as differentially expressed when a cutoff value of a twofold change was set (Table 2). It was also found that genes with increased transcription were greater in number than repressed ones in VISA compared to VSSA.
TABLE 2.
Number of differentially expressed genes in vancomycin-resistant strains versus vancomycin-susceptible strains
| Combination pair in microarray (VISA vs VSSA or h-VISA) | No. of genes altered ≥2-fold
|
P valuea | ||
|---|---|---|---|---|
| Up | Down | Total | ||
| Mu50 vs N315 | 111 | 91 | 202 | 0.075 |
| Mu50 vs Mu3 | 94 | 92 | 186 | 0.441 |
| Mu50 vs Mu50-P | 102 | 83 | 185 | 0.077 |
| Mu50-PR vs Mu50-P | 210 | 38 | 248 | <0.001 |
| MI vs MI-P | 220 | 45 | 265 | <0.001 |
| MI-PR vs MI-P | 174 | 52 | 226 | <0.001 |
Welch's approximate t test was performed for the difference between the number of up- and down-regulated genes against the total ORF number of 2,594 in N315.
Evaluation of genes commonly increased in gene expression in VISA.
The approach used in experiment 1 provided useful information for identifying genes associated with glycopeptide resistance in S. aureus. In the following approach, genes commonly increased in gene expression in VISA were selected to be cloned individually into E. coli-S. aureus shuttle vector pYT3 to construct gene overexpression library. To evaluate as many of the genes possible that would probably give higher expression in VISA, genes that had higher transcription levels in VISA compared to VSSA in at least five out of six combination pairs were selected. Some genes, even if the transcriptional levels were increased less than twofold, were also selected. Table S1 in the supplemental material shows detailed information for the selected genes along with microarray results. Ninety-two genes selected were found to be distributed to all functional categories assigned to the N315 genome previously (27). The recombinant vectors were introduced into VSSA strain N315 to develop gene overexpression transformants. After that, the transformants were examined for their susceptibility changes to glycopeptide and beta-lactam antibiotics by antibiotic gradient gel assay. Three to five transformants for each gene overexpression were examined, and those having decreased susceptibility without exception in all transformants tested (transformants which were introduced the same vector) were pooled for further study. Transformants showing an increased level of glycopeptide resistance compared to that of controls (parent strain N315 and the transformant harboring control vector only) in the above screening were further tested for their antibiotic susceptibility by MIC determination. Finally, 17 genes which reduced the susceptibility of N315 to glycopeptides by their overexpression were identified. The 17 genes included eight novel genes that we designated as the glycopeptide resistance-associated gene (gra). MICs of vancomycin, teicoplanin, oxacillin, imipenem, and ceftizoxime for these gene transformants are given in Table 3. The raised vancomycin MICs were 1.25 to 3.0 mg/liter, and the teicoplanin MICs were 1.5 to 6.0 mg/liter as compared to the MIC of 1.0 mg/liter for N315 by E-test. Three of them, namely graF, msrA2, and mgrA, also raised the oxacillin MIC from 8.0 mg/liter for N315 to 64 to ∼128 mg/liter. In addition, graR, mrsR, graD, graE, malR, sgtB, sigB, rsbU, and murZ raised oxacillin and/or imipenem susceptibility to a slight extent while they reduced glycopeptide susceptibility by its overexpression in N315.
TABLE 3.
MIC profile of overexpressed transformants of N315a
| Strains | Geneb
|
MIC (mg/liter)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vancomycin
|
Teicoplanin
|
Oxacillin
|
Imipenem
|
Ceftizoxime (agar) | |||||||
| IDd | Name | E-test | Agar | E-test | Agar | E-test | Agar | E-test | Agar | ||
| N315 | 1 | 0.5 | 1 | 1 | 8 | 8 | 0.5 | 1 | 4 | ||
| N315(pYT3) | 1 | 0.5 | 1 | 1 | 7 | 8 | 0.38 | 1 | 4 | ||
| N315(pYT3-graR) | SA0614 | graR | 1.75 | 1 | 1 | 1 | 1.5 | 0.5 | 0.125 | 1 | 4 |
| N315(pYT3-graS) | SA0615 | graS | 1.25 | 1 | 1 | 1 | 8 | 8 | 0.5 | 1 | 4 |
| N315(pYT3-graA) | SA0639 | graA | 2 | 1 | 2 | 2 | 8 | 16 | 0.75 | 1.5 | 4 |
| N315(pYT3-mgrA) | SA0641 | mgrA | 1.5 | 1 | 1.5 | 1 | 128 | 128 | 8 | 8 | 8, >512c |
| N315(pYT3-graB) | SA0743 | graB | 1.25 | 1 | 1 | 1 | 7 | 8 | 0.5 | 1 | 4 |
| N315(pYT3-msrA2) | SA1194 | msrA2 | 3 | 2 | 3 | 4 | 64 | 64 | 1.5 | 4 | 32 |
| N315(pYT3-msrR) | SA1195 | msrR | 1 | 1 | 2 | 2 | 4 | 4 | 0.094 | 0.125 | 4 |
| N315(pYT3-graC) | SAS044 | graC | 1.5 | 0.5 | 2 | 2 | 7 | 8 | 0.38 | 1 | 4 |
| N315(pYT3-lysC) | SA1225 | lysC | 1.75 | 1 | 1 | 2 | 8 | 8 | 0.5 | 0.5 | 4 |
| N315(pYT3-graD) | SA1318 | graD | 1.75 | 1 | 2 | 2 | 6 | 4 | 0.5 | 0.5 | 4 |
| N315(pYT3-graE) | SA1337 | graE | 1.5 | 1 | 1 | 1 | 3 | 4 | 0.25 | 0.13 | 4 |
| N315(pYT3-malR) | SA1339 | malR | 2 | 1 | 2 | 1 | 4 | 4 | 0.38 | 1 | 4 |
| N315(pYT3-sgtB) | SA1691 | sgtB | 2 | 1 | 1 | 2 | 4 | 8 | 0.25 | 0.5 | 4 |
| N315(pYT3-sigB) | SA1869 | sigB | 1.75 | 1 | 2 | 2 | 6 | 8 | 0.5 | 1 | 4 |
| N315(pYT3-rsbU) | SA1872 | rsbU | 1.75 | 1 | 2 | 2 | 4 | 4 | 0.38 | 1 | 4 |
| N315(pYT3-murZ) | SA1926 | murZ | 2 | 1 | 3 | 2 | 6 | 8 | 0.38 | 1 | 8 |
| N315(pYT3-graF) | SAS030 | graF | 3 | 2 | 6 | 4 | 128 | 32 | 2 | 1 | 16 |
MICs were determined by both E-test and agar dilution methods.
Mu50 genes were introduced into N315 cells by cloning on E. coli-S. aureus shuttle vector pYT3. ID, identification number.
Eagle phenotype, which is characteristic in its resistance to high drug concentrations and sensitivity to low drug concentrations (24).
Assigned gene numbers in the N315 genome (27).
Characterization of genes which raised the glycopeptide resistance level when overexpressed in N315.
Fourteen out of 17 genes identified as associated with glycopeptide resistance comprised two broad functional categories: category I, cell envelope and cellular processes, including the signal sensor; and category III, the information pathway, including the regulator. Nine genes belong to category III and five to category I. Two out of the remaining three belong to the intermediary metabolism category and one to another function category (Table 4).
TABLE 4.
Genes identified as associated with glycopeptide resistance in S. aureus
| Gene | Categorya | Function and/or product | Reference |
|---|---|---|---|
| graS | I | 2-component sensor histidine kinase | This study |
| graA | I | ABC-type transport system involved in cytochrome bd biosynthesis | This study |
| graD | I | Similar to LMP group of surface-lacated membrane protein | This study |
| sgtB | I | Similar to penicillin-binding protein 1A/1B (monofunctional glycosyltransferase) | 27 |
| murZ | I | UDP-N-acetylglucosamine 1-carboxylvinyl transferase 2 | 27 |
| graC | II | 4-Oxalocrotonate tautomerase | This study |
| lysC | II | Aspartokinase II | 56 |
| graR | III | Two-component response regulator | This study |
| mgrA | III | Transcriptional regulator (MarR family) | 31 |
| msrA2 | III | Peptide methionine sulfoxide reductase | 42 |
| msrR | III | Peptide methionine sulfoxide reductase regulator MsrR | 42 |
| graE | III | Transcription regulator AraC/XylS family homolog | This study |
| malR | III | Maltose operon transcriptional repressor | 8 |
| sigB | III | Sigma factor B | 50 |
| rsbU | III | Sigma B regulation protein RsbU | 50 |
| graF | III | Hypothetical protein (contains bipartite nuclear targeting sequence) | This study |
| graB | IV | Similar to staphylocoagulase precursor (cell-bound form of staphylocoagulase) | This study |
Category I, cell envelope and cellular processes (including sensor); II, intermediary metabolism; III, information pathways (including regulator); IV, other function.
graR and graS have highest homology to bceR and bceS, which encode a two-component regulatory system in Bacillus subtilis. The bceRS genes are located immediately upstream of bceAB genes (encoding the ABC transporter system) and control their expression by sensing extracellular bacitracin, affecting bacitracin susceptibility in B. subtilis (39). Similarly, graR and graS were located immediately upstream of ABC transporter genes vraF and vraG, which were found to be increased in gene expression more than threefold in VISA and were identified as associated with vancomycin resistance in our previous study (26). Interestingly, Mu50 graR carried a nucleotide substitution resulting in replacement of Ser197 by Asn197 compared to VSSA strain N315 and hetero-VISA strain Mu3. The role of this missense mutation remains unclear. However, while overexpression of Mu50 graR caused a decrease in vancomycin susceptibility and increase in beta-lactam susceptibility (Table 3), no such effect was observed when Mu3 graR was overexpressed in N315 (data not shown), indicating that this mutation affects the physical function of this gene.
mgrA has been found to negatively regulate autolysis genes and affects expression of many genes, including sarA, agr, cap8, hla, spa, and norA (19, 31, 53). In the present study, we found that glycopeptide and beta-lactam susceptibility were affected by this gene. This gene raised the MIC of oxacillin more than fourfold and caused a slight decrease of glycopeptide susceptibility when it was overexpressed in N315 (Table 3).
sigB was reported to have an effect on glycopeptide and beta-lactam resistance in S. aureus (1, 37, 50), and rsbU was known as a positive regulator of sigB gene. Accordingly, in our overexpression experiment, they slightly increased the level of resistance to glycopeptides, but not to beta-lactams, when they were overexpressed in N315.
graE belongs to one of the other most common positive regulator family, ArcC/XylS, having three common main regulatory functions: carbon metabolism, stress response, and pathogenesis. graE was found to have some identity to aarP gene (hit score 60, E value, 6e−9) that was identified in Providencia stuartii as a transcriptional activator of the 2′-N-acetyltransferase (32). The amino acid identity of GraE to AarP suggested a possible functional similarity as well.
The malRA operon is known to be essential for maltose-maltotriose utilization in S. xylosus, and was found to be highly expressed in five out of six VISA versus VSSA combination pairs (see Table S1 in the supplemental material). In a subsequent overexpression of malA and malR in N315 individually, malR raised the MIC of vancomycin and teicoplanin (Table 2).
graF is a small gene of 277 bp (inclusive of putative promoter, transcription termination signal, and 135-bp ORF) and is not similar to any gene by BLAST search against available database. In a search for an ortholog against genomic sequences of 166 microorganism species, no ortholog was found other than 10 staphylococcal species sequences. Moreover, domain search revealed that this gene has a bipartite nuclear targeting sequence occurring frequently in nuclear proteins but rarely in cytoplasmic protein in eukaryotic cell, suggesting that this gene is involved in the information pathway. msrA2, which encodes methionine sulfoxide reductase, was identified as being induced by cell wall-active antibiotics (42).
graF and msrA2 are the most influential genes in decreasing glycopeptide susceptibility among the genes identified as associated with glycopeptide resistance in the present study. The increments of vancomycin, teicoplanin, and oxacillin MICs for graF-overexpressed transformants were 3-, 6-, and 16-fold, and those for msrA2 transformants were 3-, 3-, and 6-fold, respectively. Their effects on drug resistance were also verified by gene-null knockout and trans-complementation assay. Table 5 shows the antibiotic susceptibility in MICs for graF and msrA2 overexpression, knockout, and complement strains, respectively. Knockout strains of graF and msrA2 did not cause any change in susceptibility to vancomycin and teicoplanin, while a slight increase in oxacillin susceptibility was observed in the graF knockout mutant but not in the msrA2 knockout mutant. By introducing the same plasmid that was used in overexpression into the respective gene knockout mutants, the level of glycopeptide susceptibility was complemented to the same level of gene-overexpressed transformants. Oxacillin resistance was also complemented, reaching a level as high as gene-overexpressed transformants in the graF knockout mutant, but it dropped remarkably in msrA2 knockout mutants in all three separate trials when the same plasmid used in gene overexpression was introduced. The reason for the noncomplemented phenotype of oxacillin resistance in the msrA2 knockout mutant remains to be clarified.
TABLE 5.
MICs of vancomycin, teicoplanin, and oxacillin for N315 derivativesa
| Strain | Description | MIC (mg/liter)
|
||
|---|---|---|---|---|
| Vancomycin | Teicoplanin | Oxacillin | ||
| N315 | N315 strain | 1 | 1 | 8 |
| N315(pTY3) | N315 harboring pYT3 | 1 | 1 | 8 |
| N315(pTY3-graF) | graF overexpression | 3 | 6 | 128 |
| N315-ΔgraF | graF knockout | 1 | 1 | 6 |
| N315-ΔgraF(pTY3-graF) | graF complementation | 3 | 4 | 128 |
| N315(pTY3-msrA2) | msrA2 overexpression | 3 | 3 | 64 |
| N315-ΔmsrA2 | msrA2 knockout | 1 | 1 | 8 |
| N315-ΔmsrA2(pTY3-msrA2) | msrA2 complementation | 3 | 2 | 0.25 |
| N315(pRIT5dS-graFp-gfp) | N315 harboring graF promoter-gfp fusion | 1 | 1 | 8 |
| N315(pRIT5dS-graFp-gfp)St | GFP fluorescence-activated strain from N315(pRIT5dS-graFp-gfp) | 2 | 3 | 32 |
MIC was determined by E-test on BHI agar for glycopeptides and MH agar for oxacillin.
MsrR shares high sequence identity and similarity to Psr, the PBP5 synthesis repressor of E. faecalis and E. hirae, and with LytR of B. subtilis. All belong to the LytR-CpasA-Psr family of cell envelope-related transcriptional regulators. msrR was strongly expressed in glycopeptide-resistant strains compared to glycopeptide-susceptible strains (see Table S1 in the supplemental material), and a slight decrease in glycopeptide susceptibility was observed when it was overexpressed in N315 (Table 3). This result is consistent with recent reports that inactivation of msrR leads to increased teicoplanin susceptibility, and transcription of this gene was inducted by cell wall-related antibiotics including vancomycin and oxacillin (43, 54).
In addition, five cell wall-related genes raised the level of vancomycin and/or teicoplanin resistance, which included graA, graB, graD, sgtB, and murZ (Table 3). graA, encoding a heterodimeric ATP-binding cassette (ABC) transporter, is similar to the cydD gene of E. coli. CydD is required for proper assembly of the E. coli cytochrome bd-type quinol oxidase. In E. coli, cytochrome bd may perform two physiological roles: contributing to energy conservation under microaerobiosis and protecting the cell from oxidative stress (14). GraD was found to have a repeated sequence element found in the LMP group of surface-located membrane proteins of Mycoplasma hominis. The number of repeats in the protein affects the tendency of cells to aggregate spontaneously. In general, the physiological roles of this protein group in bacteria are far from being fully understood. However, it has been known that aggregation may be an important factor in colonization. Non-aggregating microorganisms might easily be dispersed, whereas aggregation might provide a better chance to avoid an antibody response since some of the epitopes may be buried (20). sgtB and murZ, genes involved in cell wall biosynthesis, were found to have an increasing effect on glycopeptide resistance when they were overexpressed (Table 3). Besides these genes, which are categorized into informational pathway or cell envelope and cellular processes, some genes belonging to other functional categories were also identified as associated with glycopeptide resistance. They are graC, encoding 4-oxalocrotonate tautomerase (4-OT), lysC, encoding aspartokinase II, and graB, encoding staphylocoagulase precursor, the cell-bound form of staphylocoagulase. 4-OT is one of the enzymes involved in the Krebs cycle, and aspartokinase II catalyzes the first specific step in the biosynthesis of l-lysine, l-threonine, and l-methionine. lysC is the start gene of dap operon that consists of an eight-gene transcription unit, and this operon is involved in the biosynthesis of lysine that is an essential amino acid in cell wall biosynthesis (56).
graF and msrA2 overexpression and cell wall thickness.
We previously reported that a thickened cell wall is the phenotypic determinant of glycopeptide resistance in S. aureus (5, 6). As graF and msrA2 caused the most decrease in glycopeptide susceptibility when overexpressed, we measured cell wall thickness of the strains introduced with these two genes. Cell wall thickness was evaluated by measuring 130 cells for each strain along with that of N315(pYT3) control. Mathematical calculation showed that cell wall thicknesses of N315(pYT3-graF) and N315(pYT3-msrA2) were 26.07 ± 4.81 and 25.82 ± 4.47 nm, respectively. These values correspond to cell wall thickness increments of 23.91 and 22.27%, respectively, and are significantly different (P < 0.01) when compared to that of N315(pTY3) (21.04 ± 3.55 nm). The cell wall thickness of the transformants introduced with the genes other than graF and msrA2 remains to be determined.
graF promoter activity and glycopeptide resistance.
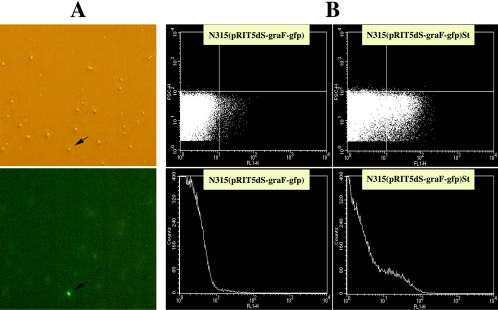
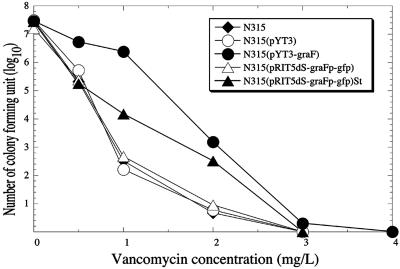
A bacterial culture is a complex matrix of a heterogeneous population with regard to its phenotype, especially in antibiotic susceptibility. Detailed analysis of single populations can provide insight into the mechanisms and rates of resistance factor acquisition and can estimate the capacity of a pathogenic species for developing and/or increasing levels of resistance. Based on evidence that (i) transcription of graF was increased in glycopeptide-resistant S. aureus compared to that of their glycopeptide-susceptible counterpart and (ii) overexpression of graF in N315 caused a decreased susceptibility to glycopeptide and/or beta-lactam antibiotics, we employed an approach to isolate a single population which gives higher graF expression from the parent N315 to see whether the isolated cultures have decreased susceptibility to glycopeptide and beta-lactam antibiotics. In this experiment, we first constructed a plasmid-based promoter-green fluorescent protein gene (gfp) transcriptional fusion system. After introducing the recombinant plasmid pRIT5dS-graFp-gfp into the N315 strain, the cells with high expression of the graF promoter were then purified by sorting the fluorescence-activated cells. Figure 1 shows results of fluorescence-activated cell sorter analysis for the representative cells collected in comparison with that before sorted. The population of fluorescence-activated cells was enriched 24.85 times after sorting [N315(pRIT5dS-graFp-gfp)St], and the sorted cells showed decreased susceptibility to vancomycin, teicoplanin, and oxacillin (Table 5 and Fig. 2). In order to verify the above observation, Northern hybridization was performed with RNA from the cells which are fluorescence activated and that of the original (before sorted), along with the graF-overexpressed, graF knockout, graF trans-complemented, and control cells. As expected, the cells that had decreased susceptibility to glycopeptides and beta-lactams showed strong expression of the graF gene compared to that of susceptible strains (Fig. 3). In addition, 20 vancomycin-resistant colonies generated from N315(pRIT5dS-graFp-gfp) by one-step selection on BHI agar containing 1 to 2 mg/liter vancomycin showed a fluorescence-activated pattern similar to N315(pRIT5dS-graFp-gfp)St in FACS analysis without any exception (data not shown).
FIG. 1.
Flow cytometric analysis of N315(pRIT5dS-graFp-gfp) and N315(pRIT5dS-graFp-gfp)St. The graF promoter-green fluorescent protein gene (gfp) transcriptional fusion plasmid pRIT5dS-graFp-gfp was introduced into N315 to generate N315(pRIT5dS-graFp-gfp), and fluorescence-activated cells were sorted by automated cell sorter to establish N315(pRIT5dS-graFp-gfp)St. (A) Representative field of microscopy (above) and fluorescent microscopy (below) for N315(pRIT5dS-graFp-gfp) cells. An arrow indicates a fluorescence-activated cell. (B) Flow cytometric profiles of N315(pRIT5dS-graFp-gfp) and N315(pRIT5dS-graFp-gfp)St cells after overnight culture in MH broth. Note that N315(pRIT5dS-graFp-gfp)St showed an increased number of fluorescence-activated cells compared to N315(pRIT5dS-graFp-gfp).
FIG. 2.
Analysis of vancomycin-resistant subpopulations of graF-relevant strains. The analysis was performed by spreading serially diluted cultures onto BHI agar plates containing vancomycin with a 1-mg/liter increment. The plates were then incubated at 37°C for 48 h before the number of CFU was counted.
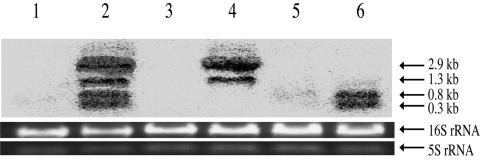
FIG. 3.
Northern blot analysis for graF-relevant derivatives of N315. Mid-log-phase cultures of bacteria were harvested to extract total RNA. Seven micrograms of total RNA was loaded per line, and the transcripts were detected with a graF-specific DNA probe. Electrophoresis images of 16S and 5S rRNA are shown as an internal control. Strains: 1, N315(pYT3), N315 harboring vector pYT3; 2, N315(pYT3-graF), graF-overexpressed transformant by pYT3-graF vector introduction; 3, N315-ΔgraF, graF-null mutant from N315; 4, N315-ΔgraF(pYT3-graF), N315-ΔgraF complemented with graF by pYT3-graF vector introduction; 5, N315(pRIT5dS-graFp-gfp), transformant of N315 generated by pRIT5dS-graFp-gfp introduction; 6, N315(pRIT5dS-graF-gfp)St, fluorescence-activated cell isolated from N315(pRIT5dS-graF-gfp). Four bands different in size correspond to transcripts beginning from vector pYT3-graF telL and the graF promoter and two other graF promoters on the chromosome, respectively.
DISCUSSION
Thickened cell wall was identified as a common feature in clinical VISA (6) and was found to directly contribute to glycopeptide resistance by serving as an obstacle to prevent vancomycin from reaching the target site (5). By cell wall thickening, cells would decrease the vancomycin concentration in the media or infected human tissue due to the increased number of vancomycin-binding targets in the cell wall, which affinity traps more vancomycin and decreases the vancomycin diffusion constant by having a “clogging effect” against the penetration of vancomycin molecules (L. Cui et al., submitted for publication). In contrast to the obvious correlation between cell wall thickness and level of glycopeptide resistance, the genetic mechanism of cell wall thickening remains unclear. One purpose of this study is to provide some new insight on this subject. Genes identified in this study as associated with glycopeptide-resistance (Table 4) were found to increase the level of glycopeptide resistance when they were overexpressed in glycopeptide-susceptible strain N315 (Tables 3 and 5). As expected, a subsequent morphological study found that msrA2 and graF, which raised the level of glycopeptide resistance, caused significant thickening of cell wall. The extent of cell wall thickness presented in these two gene transformants [N315(pYT3-graF) and N315(pYT3-msrA2)] is consistent with that found in our previous study, where regression analysis was performed between cell wall thickness and glycopeptide MICs with 48 S. aureus strains that have various levels of glycopeptide susceptibility (6). sigB and its positive regulator gene, rsbU, were also identified in this study as associated with glycopeptide resistance since their overexpression in the glycopeptide-susceptible strain raised the level of glycopeptide resistance. It was also reported that sigB caused thickening of cell wall when overexpressed in S. aureus (37). Hence, it is likely that the genetic mechanism of decreased glycopeptide susceptibility in S. aureus is, at least partially, directly associated with cell wall thickening.
Thickened cell wall is presumably due to the accumulation of an increased amount of peptidoglycan. Theoretically, there are two different ways to thicken the cell wall (peptidoglycan layers increment). One way is to produce excess amounts of peptidoglycan, which is employed by Mu50, and another is to reduce peptidoglycan turnover (15). In the present study, some genes related to cell wall biosynthesis were identified as associated with glycopeptide resistance, including murA, murZ, sgtB, and gcaD. These genes code for enzymes directly involved in the later step of the cell wall peptidoglycan (or its precursor) synthetic pathway (see Table S1 in the supplemental material). GlmS, a key enzyme in controlling cell wall synthesis in S. aureus (22), was reported to be activated in VISA strain Mu50 (7). In addition, several genes which are thought to be related to nutrient supplementation for cell wall biosynthesis were also increased in gene expression, including the dap operon (lysC, asd, dapA, dapD), proP, SA1987 (glycine betaine transporter opuD homolog), and opp-2B (see Table S1 in the supplemental material). Enzymes coded by the dap operon are key to the conversion of l-aspartate to l-lysine, which is an important component of the cell wall peptidoglycan (55, 56). Activation of the transport system for glycine betaine (SA1987) and proline betaine (proP) is supposed to increase the glycine pool (51), which is an immediate amino acid for cell wall peptidoglycan biosynthesis. opp-2B is related to oligopeptide metabolism. These corresponding genes are all involved in either generating the building blocks for cell-wall synthesis or the biosynthesis itself. Taken altogether, these findings agree with the view that an accelerated peptidoglycan synthesis occurs in glycopeptide-resistant S. aureus strains. We have previously reported that glycopeptide-resistant S. aureus strain Mu50 has activated cell wall biosynthesis pathways, including enhanced incorporation of N-acetylglucosamine (GlcNAc) into the cell wall, increased pool size of the cytoplasmic murein monomer precursor (UDP-N-acetylmuramyl-pentapeptide), increased cell wall turnover, increased production of PBP2 and 2′ (PBP2′), and an activated “GlmS pathway,” which digresses from the Embden-Meyerhof pathway at the Fru-6-P step to generate GlcN-6-P (7, 11, 12).
The malRA operon is known to be essential for maltose-maltotriose utilization in Staphylococcus xylosus and was found to be highly expressed in VISA (see Table S1 in the supplemental material). In a subsequent overexpression of malA and malR in N315 individually, malR raised the MIC of vancomycin and teicoplanin (Table 3). A study carried out with Staphylococcus xylosus on maltose-maltotriose found that chromosomal inactivation of malR led to reduced maltose utilization, and the indelibility of the maltose transport system needs its full positive expression (8). As the end product of the malRA operon is glucose, which is one of the major carbon resources for cell wall biosynthesis, an activated malRA operon would satisfy the need for cell wall biosynthesis. This is in accordance with our previous observation, where we found that increased uptake of glucose is one of the characteristics of glycopeptide-resistant strain Mu50 (7). In addition, the nas/nar operon, encoding the nitrite/nitrate assimilation system, was found to be repressed in the glycopeptide-resistant N315(pYT3-graF) strain compared to that of its isogenic control N315(pYT3) strain (L. Cui et al., in preparation). Nitrate (NO3−) is an important nitrogen source for many bacteria. Bacteria can use nitrate and nitrite (NO2−) as the sole nitrogen source during aerobic growth. Nitrate is converted through nitrite to ammonium by assimilatory nitrate and nitrite reductases. The resulting ammonium is then incorporated into amino acids through the action of glutamine synthetase and glutamate synthase (29). The decreased transcription of all genes that constitute the nas/nar operon, together with increased GlmS activity, indicates depletion of glutamine in the glycopeptide-resistant strain. GlmS requires glutamine as the ammonium source to convert Fru-6-P into GlcN-6-P. It has been reported that the amidation enzyme of glutamate residues of murein monomer precursors requires glutamine as the NH4+ donor (49). If the intracellular glutamine pool gets low, the activity of the amidation enzyme would be dampened, leaving more murein monomers nonamidated at the iso-glutamate residue (7). We previously reported that increased production of nonamidated muropeptides was one of the reasons causing reduction of peptidoglycan cross-linking in cell wall, which contributed positively to glycopeptide resistance in S. aureus (7).
It appears that a single genetic or biochemical change is unlikely to account for the glycopeptide resistance phenotype in VISA isolates observed to date (5, 10, 33, 35). Genes increased in transcription in glycopeptide-resistant strains were found to be distributed into many functional classes (see Table S1 in the supplemental material), and the genes identified to be associated with glycopeptide resistance are not directly related to each other in biological function nor physical location (Table 4). This suggests that multiple genes and pathway alteration may be needed to give rise to the diverse changes, which would then together constitute glycopeptide resistance. In addition, among the newly identified genes which were assigned to be associated with glycopeptide resistance in this study, 9 out of 17 genes belonged to regulator group or its related category, indicating a complex matrix of resistance mechanisms in glycopeptide resistance. In the present study, we employed Mu50 and MI as VISA parent strains and their respective glycopeptide-susceptible derivatives (Mu50-P and MI-P) and in vitro-developed glycopeptide-resistant (phenotypic) revertants (Mu50-PR and MI-PR) (6) to assess genomic changes in gene transcription (the transcriptome). These isogenic strain sets would be ideal tools for exaggerating the changes associated with the glycopeptide-resistant phenotype. However, changes that might convert a fully vancomycin-susceptible strain—i.e., from N315 with a vancomycin MIC of 1 mg/liter to a pre-VISA condition (as in Mu50-P and MI-P, vancomycin MIC of 2 mg/liter)—would have been missed in this study. Unlike the pre-VISA strain, the fully vancomycin-susceptible strain could not become a VISA strain by one-step selection (L. Cui et al., in preparation). Taking into account the above issue, it is reasonable to consider a stepwise process and multiple genetic alterations in achieving VISA status in S. aureus.
VISA resists glycopeptides by producing a thick cell wall. To achieve thickened cell wall, it has to accumulate multiple genetic alterations (mutations) that activate pathways for cell wall biosynthesis, increase uptake of nutrients, and change the flow of metabolites toward the generation of cell wall building blocks. In regard to nucleotide mutation, only a few studies have been performed. Schaaff et al. (45) hypothesized that an elevated mutation frequency was more likely in mutator strains defective in DNA mismatch repair (mutS knockout strains), especially when grown in the presence of vancomycin. Using a restriction-negative derivative of NCTC 8325, they developed a mutS knockout mutant. Both were subjected to a stepwise vancomycin selection procedure. Vancomycin resistance appeared much more quickly in the mutator background than in the wild type. However, whether this hypothesis can be applied to clinical isolates remains to be proven, since no difference in nucleotide sequence of mutS had been found in VISA strain Mu50 compared to VSSA strain N315 (40). Various agr point mutations were noted in several clinical VISA and hetero-VISA isolates, and a defective agr function has also been suggested to be associated with an increased probability of glycopeptide resistance formation upon vancomycin pressure (44). In our current study, 10 genes that had a mutation in Mu50 compared to N315 and increased gene expression in VISA strains were selected for overexpression by cloning them from both N315 and Mu50 DNA (see Table S1 in the supplemental material). Surprisingly, only one gene, graR, that carried a nucleotide substitution in Mu50 compared to N315 was found to cause a decrease in vancomycin susceptibility when it was overexpressed, while N315 graR did not, suggesting that missense mutation of this gene somehow affects the pathway toward achievement of resistance, even though the precise role of this mutation remains to be revealed. The extent of the effect on the resistance was also slight. Therefore, other mutations or combinations of mutations are necessary besides the alteration of genes mentioned above for VSSA cells to achieve VISA status.
Supplementary Material
Acknowledgments
We thank Tsai-Ling Lauderdale for her critical reading and discussion and Maria Kapi, Makoto Kuroda, and Fumihiko Takeuchi for their technical assistance in microarray preparation and assay.
This work was supported by a grant from Daiichi Pharmaceutical Co. Ltd., a Grant-in-Aid for 21st Century COE Research, and a Grant-in-Aid for Scientific Research on Priority Areas (13226114) from The Ministry of Education, Science, Sports, Culture and Technology of Japan.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bischoff, M., and B. Berger-Bachi. 2001. Teicoplanin stress-selected mutations increasing σB activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bachi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. [DOI] [PubMed] [Google Scholar]
- 3.Boyle-Vavra, S., B. L. M. de Jonge, C. C. Ebert, and R. S. Daum. 1997. Cloning of the Staphylococcus aureus ddh gene encoding NAD+-dependent d-lactate dehydrogenase and insertional inactivation in a glycopeptide-resistant isolate. J. Bacteriol. 179:6756-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes occurs in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui, L., and K. Hiramatsu. 2003. Vancomycin-resistant Staphylococcus aureus, p. 187-212. In A. C. Fluit and F. J. Schmitz (ed.), MRSA: current perspectives. Caister Academic Press, Norfolk, England.
- 6.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M.-N. Kim, M.-C. Ploy, N. El Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egeter, O., and R. Bruckner. 1995. Characterization of a genetic locus essential for maltose-maltotriose utilization in Staphylococcus xylosus. J. Bacteriol. 177:2408-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finan, J. E., G. L. Archer, M. J. Pucci, and M. W. Climo. 2001. Role of penicillin-binding protein 4 in expression of vancomycin resistance among clinical isolates of oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:3070-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gemmell, C. G. 2004. Glycopeptide resistance in Staphylococcus aureus: is it a real threat? J. Infect. Chemother. 10:69-75. [DOI] [PubMed] [Google Scholar]
- 11.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 12.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 13.Hill, A., E. Brown, M. Whitley, G. Tucker-Kellogg, C. Hunter, and D. Slonim. 21. November 2001, posting date. Evaluation of normalization procedures for oligonucleotide array data based on spiked cRNA controls. Genome Biol. 2:RESEARCH0055.1-0055.13. [Online.] doi: 10.1186/gb-2001-2-12-research0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill, S., S. Viollet, A. T. Smith, and C. Anthony. 1990. Roles for enteric d-type cytochrome oxidase in N2 fixation and microaerobiosis. J. Bacteriol. 172:2071-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 16.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 17.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. (Letter.) [DOI] [PubMed] [Google Scholar]
- 18.Hiramatsu, K., T. Ito, and H. Hanaki. 1999. Mechanisms of methicillin and vancomycin resistance in Staphylococcus aureus, p. 211-242. In R. G. Finch and R. J. Williams (ed.), Bailliere's clinical infectious diseases, vol. 5. Bailliere Tindall, London, United Kingdom. [Google Scholar]
- 19.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, L., S. Ladefoged, S. Birkelund, and G. Christiansen. 1995. Selection of Mycoplasma hominis PG21 deletion mutants by cultivation in the presence of monoclonal antibody 552. Infect. Immun. 63:3336-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, M.-N., C. H. Pai, J. H. Woo, J. S. Ryu, and K. Hiramatsu. 2000. Vancomycin-intermediate Staphylococcus aureus in Korea. J. Clin. Microbiol. 38:3879-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komatsuzawa, H., T. Fujiwara, H. Nishi, S. Yamada, M. Ohara, N. McCallum, B. Berger-Bachi, and M. Sugai. 2004. The gate controlling cell wall synthesis in Staphylococcus aureus. Mol. Microbiol. 53:1221-1231. [DOI] [PubMed] [Google Scholar]
- 23.Komatsuzawa, H., K. Ohta, S. Yamada, K. Ehlert, H. Labischinski, J. Kajimura, T. Fujiwara, and M. Sugai. 2002. Increased glycan chain length distribution and decreased susceptibility to moenomycin in a vancomycin-resistant Staphylococcus aureus mutant. Antimicrob. Agents Chemother. 46:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, N., K. Kuwahara-Arai, H. Kuroda-Murakami, E. Tateda-Suzuki, and K. Hiramatsu. 2001. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob. Agents Chemother. 45:815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 26.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 27.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 28.Kuwahara-Arai, K., N. Kondo, S. Hori, E. Tateda-Suzuki, and K. Hiramatsu. 1996. Suppression of methicillin resistance in a mecA-containing pre-methicillin-resistant Staphylococcus aureus strain is caused by the mecI-mediated repression of PBP 2′ production. Antimicrob. Agents Chemother. 40:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, J., B. Goldman, and V. Stewart. 1994. The nasFEDCBA operon for nitrate and nitrite assimilation in Klebsiella pneumoniae M5al. J. Bacteriol. 176:2551-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linares, J. 2001. The VISA/GISA problem: therapeutic implications. Clin. Microbiol. Infect. 7(Suppl. 4):8-15. [DOI] [PubMed] [Google Scholar]
- 31.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macinga, D. R., M. Parojcic, and P. N. Rather. 1995. Identification and analysis of aarP, a transcriptional activator of the 2′-N-acetyltransferase in Providencia stuartii. J. Bacteriol. 177:3407-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bächi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milewski, W. M., S. Boyle-Vavra, B. Moreira, C. C. Ebert, and R. S. Daum. 1996. Overproduction of a 37-kilodalton cytoplasmic protein homologous to NAD+-linked d-lactate dehydrogenase associated with vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, M. R., F. Perdreau-Remington, and H. F. Chambers. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morikawa, K., A. Maruyama, Y. Inose, M. Higashide, H. Hayashi, and T. Ohta. 2001. Overexpression of sigma factor, varsigma (B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem. Biophys. Res. Commun. 288:385-389. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson, B., L. Abrahmsen, and M. Uhlen. 1985. Immobilization and purification of enzymes with staphylococcal protein A gene fusion vectors. EMBO J. 4:1075-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohki, R., Giyanto, K. Tateno, W. Masuyama, S. Moriya, K. Kobayashi, and N. Ogasawara. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135-1144. [DOI] [PubMed] [Google Scholar]
- 40.Ohta, T., H. Hirakawa, K. Morikawa, A. Maruyama, Y. Inose, A. Yamashita, K. Oshima, M. Kuroda, M. Hattori, K. Hiramatsu, S. Kuhara, and H. Hayashi. 2004. Nucleotide substitutions in Staphylococcus aureus strains, Mu50, Mu3, and N315. DNA Res. 11:51-56. [DOI] [PubMed] [Google Scholar]
- 41.Oliveira, G. A., A. M. Aquila, R. A. Masiero, C. Levy, S. G. Gomes, L. Cui, K. Hiramatsu, and E. M. Mamizuka. 2001. Isolation in Brazil of nosocomial Staphylococcus aureus with reduced susceptibility to vancomycin. Infect. Control Hosp. Epidemiol. 22:443-448. [DOI] [PubMed] [Google Scholar]
- 42.Pechous, R., N. Ledala, B. J. Wilkinson, and R. K. Jayaswal. 2004. Regulation of the expression of cell wall stress stimulon member gene msrA1 in methicillin-susceptible or -resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 48:3057-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi, J., M. Bischoff, A. Wada, and B. Berger-Bächi. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47:2558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaaff, F., A. Reipert, and G. Bierbaum. 2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlaes, D. M., J. H. Shlaes, S. Vincent, L. Etter, P. D. Fey, and R. V. Goering. 1993. Teicoplanin-resistant Staphylococcus aureus expresses a novel membrane protein and increases expression of penicillin-binding protein 2 complex. Antimicrob. Agents Chemother. 37:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 48.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siewert, G., and J. Strominger. 1968. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J. Biol. Chem. 243:783-790. [PubMed] [Google Scholar]
- 50.Singh, V. K., J. L. Schmidt, R. K. Jayaswal, and B. J. Wilkinson. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int. J. Antimicrob. Agents 21:256-261. [DOI] [PubMed] [Google Scholar]
- 51.Smith, L. T., J.-A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki, E., K. Hiramatsu, and T. Yokota. 1992. Survey of methicillin-resistant clinical strains of coagulase-negative staphylococci for mecA gene distribution. Antimicrob. Agents Chemother. 36:429-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truong-Bolduc, Q. C., X. Zhang, and D. C. Hooper. 2003. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J. Bacteriol. 185:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Utaida, S., P. Dunman, D. Macapagal, E. Murphy, S. Projan, V. Singh, R. Jayaswal, and B. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 55.Velasco, A., J. Leguina, and A. Lazcano. 2002. Molecular evolution of the lysine biosynthetic pathways. J Mol. Evol 55:445-459. [DOI] [PubMed] [Google Scholar]
- 56.Wiltshire, M. D., and S. J. Foster. 2001. Identification and analysis of Staphylococcus aureus components expressed by a model system of growth in serum. Infect. Immun. 69:5198-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.