Abstract
Human immunodeficiency virus type 1 (HIV-1) evolved via cross-species transmission of simian immunodeficiency virus (SIVcpz) from chimpanzees (Pan troglodytes). Chimpanzees, like humans, are susceptible to infection by HIV-1. However, unlike humans, infected chimpanzees seldom develop immunodeficiency when infected with SIVcpz or HIV-1. SIVcpz and most strains of HIV-1 require the cell-surface receptor CC chemokine receptor 5 (CCR5) to infect specific leukocyte subsets, and, subsequent to infection, the level of CCR5 expression influences the amount of HIV-1 entry and the rate of HIV-1 replication. Evidence that variants in the 5′ cis-regulatory region of CCR5 (5′CCR5) affect disease progression in humans suggests that variation in CCR5 might also influence the response of chimpanzees to HIV-1/SIVcpz. To determine whether patterns of genetic variation at 5′CCR5 in chimpanzees are similar to those in humans, we analyzed patterns of DNA sequence variation in 37 wild-born chimpanzees (26 P. t. verus, 9 P. t. troglodytes, and 2 P. t. schweinfurthii), along with previously published 5′CCR5 data from 112 humans and 50 noncoding regions in the human and chimpanzee genomes. These analyses revealed that patterns of variation in 5′CCR5 differ dramatically between chimpanzees and humans. In chimpanzees, 5′CCR5 was less diverse than 80% of noncoding regions and was characterized by an excess of rare variants. In humans, 5′CCR5 was more diverse than 90% of noncoding regions and had an excess of common variants. Under a wide range of demographic histories, these patterns suggest that, whereas human 5′CCR5 has been subject to balancing selection, chimpanzee 5′CCR5 has been influenced by a selective sweep. This result suggests that chimpanzee 5′CCR5 might harbor or be linked to functional variants that influence chimpanzee resistance to disease caused by SIVcpz/HIV-1.
Introduction
The gene CC chemokine receptor 5 (CCR5 [MIM 601373]) encodes a cell-surface receptor that is exploited by the human immunodeficiency virus type 1 (HIV-1) to gain entry to several types of leukocytes (Alkhatib et al. 1996). Genetic variation in CCR5 has been associated with susceptibility to HIV-1 infection, possibly by affecting the rate at which HIV-1 enters a leukocyte (Lin et al. 2002; Ondoa et al. 2002). For example, homozygosity for a 32-bp deletion (i.e., CCR5-Δ32) of the ORF of CCR5 prevents the expression of functional CCR5 receptors on the cell surface and confers nearly complete resistance to infection by HIV-1 (Dean et al. 1996; Huang et al. 1996). Heterozygosity for CCR5-Δ32, as well as polymorphisms in the 5′ cis-regulatory region of CCR5 (5′CCR5), has also been associated with the rate at which individuals infected with HIV-1 progress to AIDS and death (Liu et al. 1996; Martin et al. 1998; Gonzalez et al. 1999; Cunningham et al. 2000; Mummidi et al. 2000). The mechanisms by which polymorphisms in 5′CCR5 may influence disease progression are unknown. However, different 5′CCR5 haplotypes have been associated with varied rates of CCR5 transcription (Mummidi et al. 2000). Furthermore, the expression level of CCR5 affects the amount of HIV-1 entry and may affect the postentry efficiency of HIV-1 replication in peripheral blood mononuclear cells (PBMCs) (Lin et al. 2002).
The HIV-1 pandemic originated via cross-species transmission of simian immunodeficiency virus (SIVcpz) from chimpanzee (Pan troglodytes), its natural reservoir (Gao et al. 1999). As with HIV-1, SIVcpz uses CCR5 to facilitate entry into leukocytes, but infection of chimpanzees in the wild by SIVcpz is infrequent and nonpathogenic. Chimpanzees are also susceptible to infection by HIV-1 (Corbet et al. 2000; Santiago et al. 2002), but, unlike humans, chimpanzees experimentally infected with HIV-1 rarely develop immunodeficiency or AIDS-related illnesses (O’Neil et al. 2000; Ondoa et al. 2002). Thus, chimpanzees are largely resistant to disease caused by SIVcpz/HIV-1. The reasons for this resistance are unknown, but it appears to result, in part, from the lesser capacity of chimpanzee PBMCs to support HIV-1 replication after cell entry (Ondoa et al. 2002), an effect similar to that observed in humans (Lin et al. 2002). These observations support the hypothesis that the 5′CCR5 region of chimpanzees harbors variants that influence the resistance of chimpanzees to disease caused by SIVcpz/HIV-1.
Population-genetics analyses of CCR5 in humans have revealed striking patterns of variation. For example, Stephens et al. (1998) showed that the CCR5-Δ32 allele has likely been driven to high frequency in European populations by positive natural selection, although probably not as a result of the fitness effects of HIV-1, which has entered human populations only recently (Stephens et al. 1998; Schliekelman et al. 2001; Bamshad et al. 2002; Galvani and Slatkin 2003; Mecsas et al. 2004). Similarly, Bamshad et al. (2002) showed that levels of genetic diversity in 5′CCR5 are among the highest in the human genome and have likely been maintained by balancing natural selection.
Information about the effects of natural selection on CCR5 in humans has been helpful in generating new hypotheses about the relationship between CCR5 variants and susceptibility to infectious disease (Bamshad and Wooding 2003). Evidence that balancing natural selection has maintained two haplotype clusters in 5′CCR5, for instance, has led to the hypothesis that the polymorphisms distinguishing these clusters may be especially good candidates for association studies (Bamshad et al. 2002). Such insights from population-genetics analyses in humans suggest that similar analyses in chimpanzees and other primates could be helpful in understanding the variable susceptibility of chimpanzees to infectious disease, especially HIV-1, as well as in understanding the differences between humans and chimpanzees. To explore inter- and intraspecific patterns of variation in 5′CCR5 and their implications for functional differences between humans and chimpanzees, we analyzed patterns of DNA sequence variation in 37 wild-born chimpanzees from each of three subspecies of chimpanzee (P. t. verus, P. t. troglodytes, and P. t. schweinfurthii) and compared them with patterns previously described in humans, along with patterns of variation in 50 noncoding regions of the chimpanzee genome.
Material and Methods
Population Samples
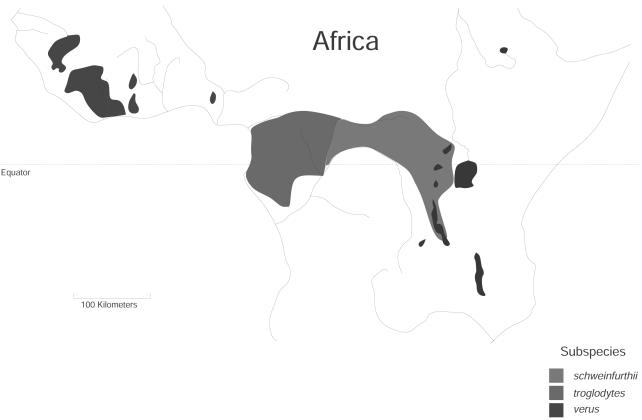
A 1,123-bp segment of the 5′ cis-regulatory region of CCR5 was sequenced in 37 unrelated, wild-born chimpanzees: 26 from West Africa (P. t. verus), 9 from Central Africa (P. t. troglodytes), and 2 from East Africa (P. t. schweinfurthii). The geographical distribution of these three subspecies is shown in figure 1. Three bonobos (Pan paniscus) and one gorilla (Gorilla gorilla gorilla) were also sequenced. These sequences were compared with previously reported sequences from 112 humans (Bamshad et al. 2002), including Africans (n=31), Asians (n=57), and Europeans (n=24) (see supplementary data [online only] for details). Nonhuman primate samples were imported in accordance with the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) under permit 99US013176/9. Informed consent was obtained from all human subjects.
Figure 1.
Geographical distributions of chimpanzee subspecies in Africa (redrawn from the article by Gagneux et al. [2001])
Patterns of diversity in the 5′CCR5 region in chimpanzees and humans were compared with patterns of diversity in two other data sets. First, a section of hypervariable sequence 1 (HV1) of the mtDNA control region was sequenced in all chimpanzee samples to confirm subspecies assignment and to allow comparison with other studies of diversity in mtDNA. Comparisons were also performed with data from 50 noncoding regions—each ∼500 bp in length—which were sequenced in 17 chimpanzees (6 P. t. verus, 5 P. t. troglodytes, 2 P. t. schweinfurthii, and 4 chimpanzees of unknown subspecies) and 30 humans (10 Africans, 10 Asians, and 10 Europeans) by Yu et al. (2002, 2003). The chimpanzee population examined by Yu et al. (2002, 2003) is somewhat smaller than that examined in our study, but it is similar in composition, containing all three chimpanzee subspecies.
Laboratory Methods
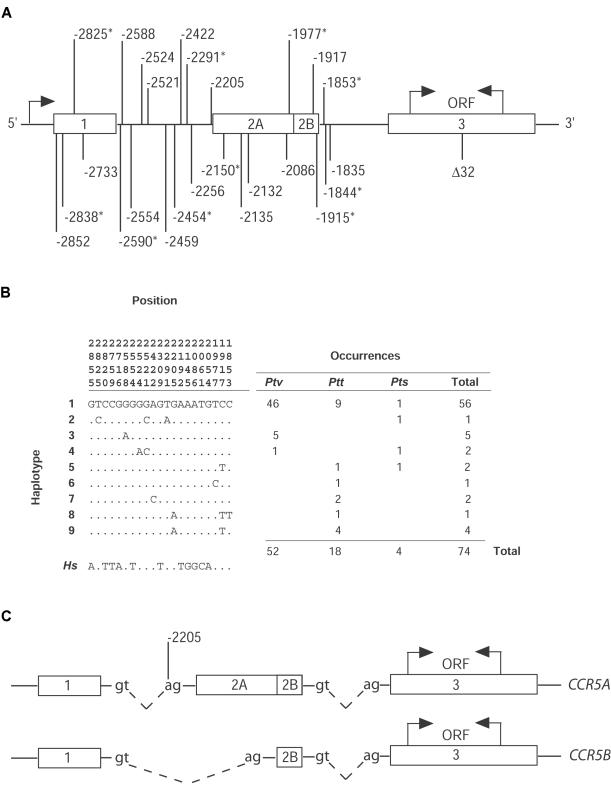
The region corresponding to the 5′ cis-regulatory region of human CCR5—which spans from −2867 to −1745 and for which −1 is the nucleotide immediately upstream of the translational start site at +1—was PCR amplified and sequenced on both strands (fig. 2A). This region includes the 3′ end of exon 1, intron 1, exons 2A and 2B, and the 5′ end of intron 2 (Mummidi et al. 2000). All of these exons are noncoding. PCR primer sequences were derived from the human genomic sequence (GenBank accession numbers AF031236 and AF031237). Mitochondrial HV1 of the control region was PCR amplified as described by Morin et al. (1994).
Figure 2.
CCR5 variation in chimpanzees and humans. A, Schematic drawing (not to scale) of CCR5, including exons (boxes) and introns. Polymorphisms found in the 5′ cis-regulatory region of CCR5, which spans from −2867 to −1745, are indicated and numbered. Genomic sequencing revealed 15 and 10 polymorphisms in humans (bottom) and chimpanzees (top), respectively. An excessive number of singletons is found in chimpanzees (asterisks [*]), whereas humans have more intermediate-frequency variants than expected under neutrality. B, 5′CCR5 haplotypes observed in chimpanzees. Positions are shown relative to the CCR5 translational start site (+1). Only positions that are variable in chimpanzees or that differ between the chimpanzee sequence and the human consensus sequence are shown. Columns at right indicate the number of times each haplotype was observed in P. t. verus (Ptv), P. t. troglodytes (Ptt), P. t. schweinfurthii (Pts), and the total sample. Hs = the human consensus sequence. C, Two of the CCR5 mRNA transcripts found in humans. An A→G mutation at −2205 in the splice-acceptor site of exon 2A eliminates the production of CCR5A in most chimpanzees. This site has reverted to A in some P. t. troglodytes, suggesting that they have regained the capability to make CCR5A.
Sequence-trace files were evaluated using the Phred, Phrap, and Consed programs (Ewing et al. 1998). Potential heterozygotes were identified using the PolyPhred program (Nickerson et al. 1997). Polymorphisms were verified by manual evaluation of the individual sequence traces. For most polymorphisms, it was possible to evaluate both the forward and the reverse sequences.
Statistical Analysis
Genetic diversity was measured using θW, a measure of genetic diversity that is based on the number of variable nucleotide positions in a sample (S) (Watterson 1975), and π, a measure of genetic diversity that is based on the mean pairwise difference (per nucleotide) between sequences (Tajima 1983).
Tests of evolutionary neutrality were performed using Tajima’s D statistic, which compares θW with π (Tajima 1989), and by use of Fu’s FS statistic (Fu 1997), which compares S with the number of haplotypes observed in a sample. Tajima’s D test was performed using the standard method, which is to simulate D values under the assumption that natural selection has been absent (Tajima 1989). To incorporate varying assumptions about population size change in human and chimpanzee populations, these simulations were also performed using the algorithm of Rogers (1995), as recently implemented by Wooding et al. (2004). This algorithm assumes that a population increased suddenly from an ancient population size (N1) to a larger population size (N0), t generations ago, at an infinite-sites mutation rate (μ).
Haplotypes were inferred using PHASE (version 2.02), and all haplotypes found had a high level of statistical support (Stephens and Donnelly 2003). Evolutionary relationships among haplotypes were inferred using the ARLEQUIN software package and were depicted by constructing rooted minimum-spanning (MS) networks. An orangutan was used as an outgroup species to determine the polarity of the character states. Networks constructed using parsimony and maximum-likelihood methods had similar topologies. Ambiguities in the MS network were drawn as reticulations.
Results and Discussion
Variation in the 5′ Cis-Regulatory Region of CCR5
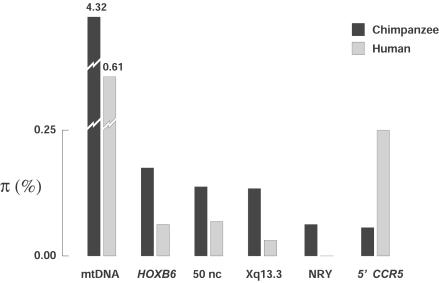
Ten variable nucleotide positions were identified in the 5′CCR5 region of chimpanzees (fig. 2A), and nucleotide diversity (π) in the 5′CCR5 region in chimpanzees was 0.00062±0.00053 (fig. 3). Most previously published studies have found higher levels of genetic diversity in chimpanzees than in humans (Deinard and Kidd 1999; Kaessmann et al. 2001; Stone et al. 2002; Fischer et al. 2004), but the estimated value of π for chimpanzee 5′CCR5 is lower than that of most other chimpanzee loci, as well as most human loci (fig. 3). Additionally, π was ∼4-fold lower in chimpanzees than 5′CCR5 in humans, in spite of the fact that the two major subspecies of chimpanzee (western and central) are estimated to have diverged from one another 430,000–650,000 years ago, on the basis of an analysis of DNA sequence data from nine unlinked, intergenic regions totaling ∼19,000 bp (Fischer et al. 2004). Ordinarily, such extended subdivision is expected to result in disproportionately high levels of genetic diversity. In contrast, the value of π in human 5′CCR5 (0.0023±0.0015) was substantially higher than the average π value in the 50 noncoding regions in humans (0.00088) (Bamshad et al. 2002).
Figure 3.
Nucleotide diversity in chimpanzees and humans. Each pair of bars represents a different locus/region. Data are from Stone et al. (2002) (for mtDNA), Deinard and Kidd (1999) (for HOXB6), Yu et al. (2002, 2003) (for 50 nc), Kaessmann et al. (1999, 2001) (for Xq13.3), Stone et al. (2002) (for NRY [noncoding region of the Y chromosome]), and from the present study (5′CCR5). The 5′CCR5 was the only region for which humans are more diverse than chimpanzees. In addition, whereas diversity at 5′CCR5 in chimpanzees was lower than that in the 50 noncoding (nc) regions (Yu et al. 2002, 2003), diversity at 5′CCR5 in humans was higher than that in the 50 noncoding regions.
Like the π values, θW values suggested that levels of diversity in 5′CCR5 are low in chimpanzees. The θW value in chimpanzees (0.18) was <95% of the values in the 50 noncoding regions in chimpanzees, whereas the θW value in humans (0.20) was >85% of the values in the homologous 50 regions in humans. And, as with π values, 5′CCR5 is the only gene region reported to date in which the θW value is lower in chimpanzees than in humans. Therefore, like the π values, the θW value suggests that 5′CCR5 diversity in chimpanzees is low, relative to both 5′CCR5 in humans and other loci in chimpanzees.
To determine whether the low π value in 5′CCR5 was the result of a sampling bias, we examined HV1 of the mitochondrial genome in the same animals in which we had resequenced 5′CCR5. The π value for HV1 in our sample, 0.0844, was slightly higher than the π value (0.075) that was estimated by Deinard and Kidd (1999) for HV1 sequences in a chimpanzee sample of similar size (n=41) and composition (i.e., with all three subspecies represented). If our sample were unusually homogeneous, the estimated value of π for HV1 would have been lower than Deinard and Kidd’s (1999) estimate of π—not higher, as was observed. This result suggests that the low level of diversity found in 5′CCR5 in our sample chimpanzees is not due to sampling bias.
Nine haplotypes were observed in the 5′CCR5 region of chimpanzees (fig. 2B), one of which accounted for >75% of sampled chromosomes. Other haplotypes ranged in frequency from 1.5% to 6.8%. A test using Fu’s FS statistic (Fu 1997)—which compares the observed number of haplotypes with the number expected, given S segregating sites—also yielded significant results (FS=-3.74; P<.025), suggesting that the number of haplotypes is greater than expected, given the observed θW. Together, these results indicate that not only is diversity in 5′CCR5 in chimpanzees lower than expected, but an excess of rare haplotypes is also present. Both of these patterns are consistent with the hypothesis that positive natural selection has acted to reduce diversity in this region.
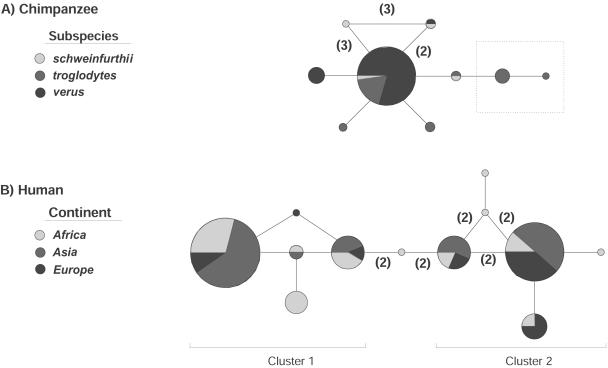
An MS network illustrating the mutational steps among chimpanzee 5′CCR5 haplotypes differed distinctly from that observed by Bamshad et al. (2002) for human 5′CCR5 (fig. 4). The network relating chimpanzee haplotypes was starlike, with one common haplotype and several rare haplotypes, whereas the network relating human haplotypes exhibited two major, divergent clusters, each of which contained two common haplotypes. Networks with such dissimilar topologies are often associated with different evolutionary histories. Starlike networks are often associated with population growth or positive natural selection, which cause reductions in genetic diversity and an excess of rare nucleotide variants. In contrast, networks with common but divergent haplotypes are more often associated with the effects of balancing natural selection or population decline, which cause increases in diversity and excesses of common nucleotide variants (Bamshad and Wooding 2003). Thus, the different haplotype networks observed in chimpanzees and humans could be the result of differences in population history, in selective pressure, or both.
Figure 4.
MS networks of haplotypes of 5′CCR5 in chimpanzees (A) and humans (B). Each circle represents a different haplotype, and the size of each circle is proportional to the relative frequency of the haplotype. For each haplotype, the extent of shading indicates the fraction of observations in Africans, Asians, Europeans, or different subspecies of chimpanzee. The lines between haplotypes correspond to one nucleotide substitution, except where the number of nucleotides is indicated in parentheses. Reticulations indicate ambiguous relationships.
An important distinction between the effects of population history and natural selection is that population history should affect all loci equally, whereas natural selection should affect only the local genomic region containing the target of selection. Thus, one way to distinguish the effects of natural selection on 5′CCR5 from those of population history is to compare the value of π for chimpanzee 5′CCR5 with the value of π for multiple genomic regions. A total of 50 noncoding regions, each ∼0.5 kb, were sequenced in humans (Yu et al. 2002) and chimpanzees (Yu et al. 2003). The value of π for chimpanzee 5′CCR5 was about half the overall diversity in chimpanzees (π=0.0013) and was lower than 40 of the 50 estimated values of π from individual regions in chimpanzees. In contrast, the estimated value of π at 5′CCR5 in humans was greater than 45 of the 50 estimated values of π from individual regions in humans. Together, our findings—(1) the relatively low level of diversity of 5′CCR5 in chimpanzees, relative to both human 5′CCR5 and the 50 noncoding regions in chimpanzee, and (2) the relatively high level of diversity in human 5′CCR5, relative to both chimpanzee 5′CCR5 and the 50 noncoding regions in humans—are consistent with the hypothesis that different selective pressures have acted on this locus in chimpanzees and humans. To investigate this possibility in more detail, we performed statistical tests of evolutionary neutrality.
Neutrality Tests
The presence of relatively low π and θW values at 5′CCR5 in chimpanzees is consistent with the hypothesis that either population growth or positive natural selection has acted to “sweep” old variants out of a population (Bamshad and Wooding 2003). However, low diversity levels can also occur when mutation rates or effective population sizes are low. To distinguish these alternatives, we used a test of Tajima’s D statistic (Tajima 1989), which exploits the fact that π and θW values are affected differently by natural selection and population history. For example, positive natural selection and population growth tend to produce excesses of young, low-frequency variants, compared with variants in a neutrally evolving region in a population of constant size. Because π is more sensitive to these effects, the value of π is reduced relative to the value of θW, resulting in negative D values. In contrast, balancing natural selection and population decrease tends to produce excesses of old, intermediate-frequency variants, increasing the value of π relative to θW and resulting in positive D values.
Because Tajima’s D statistic can be affected by both population history and natural selection, we performed these tests by using the method of Wooding et al. (2004). This method uses the standard approach, which is to generate the theoretical distribution of D by use of coalescent simulations (Tajima 1989). However, these simulations are performed for varying population histories, as described by Rogers and Harpending (1992) and Rogers (1995) (see also Hudson [1990, 2002]).
Several previous studies have reported that patterns of genetic variation in chimpanzees are consistent with the hypothesis that population sizes have been constant. However, more-recent evidence suggests that central and eastern chimpanzees, but not western chimpanzees, may have undergone growth (Gagneux et al. 1999; Goldberg and Ruvolo 1999; Kaessmann et al. 1999; Wooding and Rogers 2000; Fischer et al. 2004). In contrast, the preponderance of genetic evidence in humans suggests that human populations grew rapidly in the Upper Pleistocene (Harpending and Rogers 2000; Excoffier 2002; Tishkoff and Verrelli 2003). Because of uncertainty about the population history of chimpanzees and, to a lesser extent, humans, we performed tests of Tajima’s D statistic under varying assumptions about population history, to generate CIs.
CIs around the magnitude and timing of population expansion revealed that Tajima’s D statistic was significantly more negative (D=-1.73; P<.01) than expected under the assumption of constant population size or recent growth in chimpanzees (fig. 5). However, under the assumption of population decline or ancient population growth in chimpanzees (i.e., >375,000 years before the present [calculated from an effective population size of 25,000, 20 years per generation, and a time of expansion {τ} of 1.5 generations]), the estimate of D for 5′CCR5 in chimpanzees was not significantly different than expected. In humans, the opposite pattern was observed. Tajima’s D was significantly more positive than expected (D=0.82) under the assumption of ancient population growth but not under the assumption that human population size has been constant or has grown very recently (fig. 5). Importantly, when D values in both species differed significantly from expectation, D was significantly greater than expected in 5′CCR5 in humans but was significantly less than expected in chimpanzees. Given evidence that human populations increased in the Upper Pleistocene, whereas chimpanzee populations have likely remained nearly constant, the high value of Tajima’s D in humans is most consistent with the effects of balancing natural selection, whereas the low value of D in chimpanzees is better explained by recent positive selection.
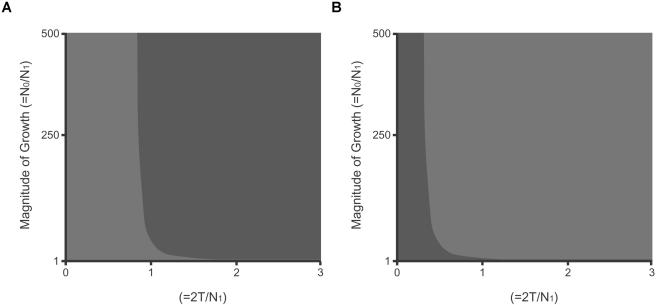
Figure 5.
CIs for tests of Tajima’s D, estimated for 5′CCR5 in chimpanzees (A) and humans (B). Each CI shows the demographic parameters for which neutrality could be rejected (red) or not rejected (blue) on the basis of Tajima’s D test. On the axes, N1 is the effective population size of the ancestral population, τ is the time of expansion (in generations), and N0 is the effective population size of the population after expansion. For example, if it is assumed that the chimpanzee population has a generation time of 20 years, that the ancient population size was 25,000, and that the population grew 250-fold 100,000 years ago, then τ=2(100,000/20)/25,000=0.4, and the hypothesis of neutrality cannot be rejected. For the negative value of Tajima’s D in chimpanzee 5′CCR5, the hypothesis of neutrality is rejected under constant population size or recent growth but not under ancient growth. In humans, the value of Tajima’s D for 5′CCR5 is positive, and neutrality cannot be rejected under either constant population size or recent population growth. Neutrality is rejected under older population growth.
Variation in population history between chimpanzee subspecies is also of potential importance. Evidence from earlier studies suggests that western chimpanzees (P. t. verus), in contrast to central chimpanzees (P. t. troglodytes), do not exhibit a high proportion of rare alleles, relative to the expectations of the standard neutral model; the mean value of Tajima’s D, estimated from intergenic DNA segments, is ∼0 (Yu et al. 2003; Fischer et al. 2004). This pattern is reversed in 5′CCR5. Two of the only three 5′CCR5 haplotypes in western chimpanzees were rare, and Tajima’s D for 5′CCR5 in western chimpanzees was more negative (D=-1.26; P<.075) than the value of D in central chimpanzees (D=-0.28; P<.25). Additionally, the value of π in 5′CCR5 of western chimpanzees (0.00022) was ∼4-fold lower than the value of π that was estimated from intergenic DNA segments in western chimpanzees (Fischer et al. 2004). This excess of rare alleles, albeit in a region of generally reduced nucleotide diversity, is further evidence that 5′CCR5 has been subject to positive selection. This is especially intriguing because, although naturally occurring infections have been found in both central and eastern chimpanzees, no western chimpanzee infected with SIVcpz has yet been reported (Santiago et al. 2002). Thus, evidence of a selective sweep on 5′CCR5 appears to be stronger in the only subspecies of chimpanzee that does not naturally harbor SIVcpz. This is consistent with the hypothesis that SIVcpz may have been a source of selective pressure on 5′CCR5.
To determine whether the value of Tajima’s D statistic for chimpanzee 5′CCR5 was lower than that for noncoding regions sampled from throughout the genome—as would be expected if positive selection has affected the region—we tested Tajima’s D in chimpanzees from pooled data from the 50 noncoding autosomal regions (∼0.5 kb each) sequenced by Yu et al. (2003). This test revealed that Tajima’s D among noncoding regions was negative (D=-1.2) but not significantly different from zero (P<.11). Other single loci assayed in chimpanzees have been found to have negative values for Tajima’s D as well. However, these values have not reached statistical significance, even when relatively large numbers of chromosomes have been examined (Morin et al. 1994; Deinard and Kidd 1999; Kaessmann et al. 2001; Stone et al. 2002). Thus, although there is a trend toward negative values of Tajima’s D in chimpanzees, the value of Tajima’s D for 5′CCR5 (D=-1.73) is more negative than estimates of the average value of Tajima’s D for individual loci examined to date. Despite evidence of a genomewide excess of rare alleles in chimpanzees, the excess of 5′CCR5 alleles is greater than in most regions of the chimpanzee genome examined to date.
The finding that the signature of a selective sweep in 5′CCR5 varies among chimpanzee subspecies raises questions as to when the modal 5′CCR5 haplotype originated, when the sweep may have occurred, and whether it was limited to or stronger in one subspecies of chimpanzee. The modal 5′CCR5 haplotype is common in all three chimpanzee subspecies, suggesting that it originated prior to the divergence of chimpanzee subspecies. Alternatively, it could have arisen in one of the chimpanzee subspecies and spread via migration to other subspecies, but there is little evidence of gene flow between subspecies of chimpanzee. Our results also suggest that the selective sweep occurred only in—or more recently in—western chimpanzees, even though 5′CCR5 diversity is reduced in all three chimpanzee subspecies. It is possible that the sweep occurred prior to the divergence of all chimpanzee subspecies, but, given that these subspecies diverged from one another ∼500,000 years ago, it is expected that the signature of a sweep would have been erased.
Possible Targets of a Selective Sweep
Collectively, patterns of population genetic variation in 5′CCR5 suggest that this region has been subjected to a selective sweep in chimpanzees. The biological significance of these findings remains to be explored. One reasonable hypothesis is that the modal 5′CCR5 haplotype in chimpanzees—or a variant in linkage disequilibrium that has yet to be identified—afforded some protection against disease caused by SIVcpz and/or other pathogens, such as poxviruses (Lalani et al. 1999). This hypothesis is motivated by the observation that the chimpanzee modal haplotype is most similar to the ancestral CCR5 human haplotype group A (HHA). Among human 5′CCR5 haplotypes tested to date, HHA exhibits the least promoter activity (Mummidi et al. 2000) and has been associated with delayed progression to AIDS in African Americans (Gonzalez et al. 1999). Thus, HHA may retain ancestral characteristics that confer some resistance to AIDS in humans.
The modal haplotype 5′CCR5 in chimpanzees is nearly fixed in western chimpanzees, and, despite the screening of several thousand western chimpanzees, none of these animals has been found to be infected with SIVcpz (Santiago et al. 2002). One explanation for the absence of SIVcpz in western chimpanzees is that the geographic separation of this subspecies from other chimpanzee subspecies predated the emergence of SIVcpz, so western chimpanzees simply may have not been exposed to SIVcpz. An alternative explanation that is consistent with our results is that adaptation has afforded some resistance to SIVcpz, possibly through evolution of the 5′CCR5 region.
The nature of the selective pressure is unknown, but one possible target might have been an A→G transition that abolishes the splice-acceptor site of exon 2A (fig. 2C). This substitution eliminates the production of CCR5A, an alternatively spliced transcript of CCR5, but does not eliminate the production of CCR5B (Mummidi et al. 2000). Whereas A is fixed at this site in humans and all other apes, this site is a G in >90% of chimpanzee chromosomes we tested, including the chimpanzee modal haplotype. In addition, the A variant appears to have been rederived in central chimpanzees from a haplotype bearing a G (fig. 3). If the G variant were a target of selection that afforded resistance to SIVcpz, we hypothesized that the A variant might contribute to disease susceptibility in the small percentage of chimpanzees who—when experimentally infected with HIV-1—develop immunodeficiency or AIDS (O’Neil et al. 2000).
To test the hypothesis that the A variant is more common in immunocompromised chimpanzees, we sequenced the 5′CCR5 region of six chimpanzees that became immunodeficient after infection with HIV-1. We found that none of these chimpanzees had the A variant. Indeed, each of these chimpanzees was instead homozygous for the modal chimpanzee haplotype. Thus, the inability to make the CCR5A isoform does not appear, in itself, to contribute to disease resistance in chimpanzees. However, the small size of our sample renders this test weak, and we argue that the A/G substitution merits particular attention in further efforts to understand HIV-1 disease and the differences in function, if any, between CCR5A and CCR5B.
Conclusions
Together, patterns of variation in 5′CCR5 in chimpanzees differ appreciably from those in humans. Whereas variation in 5′CCR5 in humans is dominated by the presence of two distinct haplotype clusters, variation in chimpanzees is dominated by a single haplotype. In addition, whereas levels of diversity at 5′CCR5 in chimpanzees are low, relative to those found in humans—as well as relative to those found in 50 noncoding regions of the chimpanzee genome—levels of diversity at 5′CCR5 in humans are high, relative to those found in 50 noncoding regions. Furthermore, whereas the value of Tajima’s D in humans is significantly greater than expected under reasonable assumptions about population history and is thus consistent with the presence of balancing natural selection, the value of D in chimpanzees is significantly less than expected under reasonable assumptions about population history and is thus consistent with the hypothesis that a recent selective sweep has occurred. Although the effects of population history cannot be ruled out as a partial explanation for our data, the disparate patterns of variation in humans and chimpanzees suggest that 5′CCR5 has evolved under appreciably different historical forces in these two species. The question of whether the differing patterns of variation at the 5′CCR5 locus in humans and chimpanzees can account for the disparate susceptibility of these two species to AIDS remains to be explored. More generally, these findings suggest that genetic variants that underlie differences between human and chimpanzee responses to HIV might be found by searching for signatures of selection at loci that influence the pathogenesis of AIDS. Such studies will be greatly facilitated by access to the complete chimpanzee genome sequence.
Supplementary Material
Acknowledgments
This research was supported by the National Institutes of Health (grants ES-12125, GM-59290, RR-00064, ES-10058, AI43279, and AI46326) and by the National Science Foundation (grants BCS-0073871, SBR-9514733, and SBR-9818215). We thank Alan Rogers and Jon Seger for helpful comments. Chimpanzee DNA samples were kindly provided by the Primate Foundation of Arizona, the Southwest Foundation for Biomedical Research, the New Iberia Primate Center, the Sunset Zoo, the Riverside Zoo, the Yerkes Primate Research Center, and the Jane Goodall Foundation, as well as by Harold McClure, Jean Wickings, and E. Zietkewicz.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human 5′CCR5 [accession numbers AF031236 and AF031237])
- PHASE, http://www.stat.washington.edu/stephens/software.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CCR5) [PubMed]
References
- Alkhatib G, Combadiere C, Broder CC, Fent Y, Kennedy PE, Murphy PM, Berger EA (1996) CC CKR5: a RANTES, MIP-1π, MIP-1α receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958 [DOI] [PubMed] [Google Scholar]
- Bamshad M, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Stone AC, Jorde LB, Ahuja SK, Weiss RB (2002) A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci USA 99:10539–10544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Wooding S (2003) Signatures of natural selection in the human genome. Nat Rev Genet 4:99–111 [DOI] [PubMed] [Google Scholar]
- Corbet S, Muller-Trutwin MC, Versmisse P, Delarue S, Ayouba A, Lewis J, Brunak S, Martin P, Brun-Vezinet F, Simon F, Barre-Sinoussi F, Mauclere P (2000) env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group n from the same geographic area. J Virol 74:529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham AL, Li S, Juarez J, Lynch G, Alali M, Naif H (2000) The level of HIV-1 infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol 68:311–317 [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study, O’Brien SJ (1996) Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856–1862 [DOI] [PubMed] [Google Scholar]
- Deinard A, Kidd K (1999) Evolution of a HOXB6 intergenic region within the great apes and humans. J Hum Evol 36:687–703 [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using PHRED. I. Accuracy assessment. Genome Res 8:175–185 [DOI] [PubMed] [Google Scholar]
- Excoffier L (2002) Human demographic history: refining the recent African origin model. Curr Opin Genet Dev 12:675–682 [DOI] [PubMed] [Google Scholar]
- Fischer A, Wiebe V, Paabo S, Przeworski M (2004) Evidence for a complex demographic history of chimpanzees. Mol Biol Evol 21:799–808 [DOI] [PubMed] [Google Scholar]
- Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Gonder MK, Goldberg TL, Morin PA (2001) Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos Trans R Soc Lond B Biol Sci 356:889–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P, Wills C, Gerloff U, Tautz D, Morin PA, Boesch C, Fruth B, Hohmann G, Ryder OA, Woodruff DS (1999) Mitochondrial sequences show diverse evolutionary histories of African homonoids. Proc Natl Acad Sci USA 96:5077–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani AP, Slatkin M (2003) Evaluating plague and smallpox as historical selective pressures for the CCR5-Δ-32 HIV-resistance allele. Proc Nat Acad Sci USA 100:15276–15279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH (1999) Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441 [DOI] [PubMed] [Google Scholar]
- Goldberg TL, Ruvolo M (1997) The geographic apportionment of mitochondrial genetic diversity in East African chimpanzees, Pan troglodytes schweinfurthii. Mol Biol Evol 14:976–984 [DOI] [PubMed] [Google Scholar]
- Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G, O’Connell P, Bowden DW, Freedman BI, Anderson SA, Walter EA, Evans JS, Stephan KT, Clark RA, Tyagi S, Ahuja SS, Dolan MJ, Ahuja SK (1999) Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA 96:12004–12009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending HC, Rogers A (2000) Genetic perspectives on human origins and differentiation. Annu Rev Genomics Hum Genet 1:361–385 [DOI] [PubMed] [Google Scholar]
- Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kuntsman K, Erikson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA (1996) The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med 2:1240–1243 [DOI] [PubMed] [Google Scholar]
- Hudson RR (1990) Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J (eds) Oxford series in evolutionary biology. Vol 7. Oxford University Press, Oxford, United Kingdom, pp 1–44 [Google Scholar]
- ——— (2002) Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Pääbo S (1999) Extensive nuclear DNA sequence diversity among chimpanzees. Science 286:1159–1162 [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Wiebe V, Weiss G, Pääbo S (2001) Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat Genet 27:155–156 [DOI] [PubMed] [Google Scholar]
- Lalani AS, Masters J, Zeng W, Barrett J, Pannu R, Everett H, Arendt CW, McFadden G (1999) Use of chemokine receptors by poxviruses. Science 286:1968–1971 [DOI] [PubMed] [Google Scholar]
- Lin Y, Mettling C, Portales P, Reynes J, Clot J, Corbeau P (2002) Cell surface CCR5 density determines the postentry efficiency of R5 HIV-1 infection. Proc Natl Acad Sci USA 99:15590–15595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR (1996) Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367–377 [DOI] [PubMed] [Google Scholar]
- Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S, Goedert JJ, O’Brien TR, Hilgartner MW, Vlahov D, O’Brien SJ, Carrington M (1998) Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907–1911 [DOI] [PubMed] [Google Scholar]
- Mecsas J, Franklin G, Kuziel WA, Brubaker RR, Falkof S, Mosier DE (2004) Evolutionary genetics: CCR5 mutation and plague protection. Nature 427:606 [DOI] [PubMed] [Google Scholar]
- Morin PA, Moore JJ, Chakraborty R, Jin L, Goodall J, Woodruff DS (1994) Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science 265:1193–1201 [DOI] [PubMed] [Google Scholar]
- Mummidi S, Bamshad M, Ahuja SS, Gonzalez E, Feuillet PM, Begum K, Galvis MC, Kostecki V, Valente AJ, Murthy KK, Haro L, Dolan MJ, Allan JS, Ahuja SK (2000) Evolution of human and non-primate CC chemokine receptor 5 gene and mRNA: potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem 275:18946–18961 [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single-nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondoa P, Davis D, Kestens L, Vereekcken C, Garcia Ribas S, Fransen K, Heeney J, van der Groen G (2002) In vitro susceptibility to infection with SIVcpz and HIV-1 is lower in chimpanzee than in human peripheral blood mononuclear cells. J Med Virol 67:301–311 [DOI] [PubMed] [Google Scholar]
- O’Neil SP, Novembre FJ, Hill AB, Suwyn C, Hart CE, Evans-Strickfaden T, Anderson DC, deRosayro J, Herndon JG, Saucier M, McClure HM (2000) Progressive infection in a subset of HIV-1–positive chimpanzees. J Infect Dis 182:1051–1062 [DOI] [PubMed] [Google Scholar]
- Rogers AR (1995) Genetic evidence for a Pleistocene population explosion. Evolution 49:608–615 [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending HC (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569 [DOI] [PubMed] [Google Scholar]
- Santiago ML, Rodenburg CM, Kamenya S, Bibollet-Ruche F, Gao F, Bailes E, Meleth S, Soong SJ, Kilby JM, Moldoveanu Z, Fahey B, Muller MN, Ayouba A, Nerrienet E, McClure HM, Heeney JL, Pusey AE, Collins DA, Boesch C, Wrangham RW, Goodall J, Sharp PM, Shaw GM, Hahn BH (2002) SIVcpz in wild chimpanzees. Science 295:465 [DOI] [PubMed] [Google Scholar]
- Schliekelman P, Gardner C, Slatkin M (2001) Natural selection and resistance to HIV. Nature 411:545–546 [DOI] [PubMed] [Google Scholar]
- Stephens JC, Reich DE, Goldstein DB, Shin HD, Smith MW, Carrington M, Winkler C, et al (1998) Dating the origin of the CCR5-Δ32 AIDS-resistance allele by the coalescence of haplotypes. Am J Hum Genet 62:1507–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P (2003) A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 73:1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone AC, Griffiths RC, Zegura SL, Hammer MF (2002) High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc Natl Acad Sci USA 99:43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F (1983) Evolutionary relationships of DNA sequences in finite populations. Genetics 105:437–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC (2003) Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 4:293–340 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim U, Bamshad M, Larsen J, Jorde L, Drayna D (2004) Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet 74:637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, Rogers A (2000) A Pleistocene population X-plosion? Hum Biol 72:693–695 [PubMed] [Google Scholar]
- Yu N, Chen FC, Ota S, Jorde LB, Pamilo P, Patthy L, Ramsay M, Jenkins T, Shyue SK, Li WH (2002) Larger genetic differences within Africans than between Africans and Eurasians. Genetics 161:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Jensen-Seaman MI, Chemnick L, Kidd JR, Deinard AS, Ryder O, Kidd KK, Li WH (2003) Low nucleotide diversity in chimpanzees and bonobos. Genetics 164:1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.