Abstract
Transcriptional control is a major mechanism for regulating gene expression. The complex machinery required to effect this control is still emerging from functional and evolutionary analysis of genomic architecture. In addition to the promoter, many other regulatory elements are required for spatiotemporally and quantitatively correct gene expression. Enhancer and repressor elements may reside in introns or up- and downstream of the transcription unit. For some genes with highly complex expression patterns—often those that function as key developmental control genes—the cis-regulatory domain can extend long distances outside the transcription unit. Some of the earliest hints of this came from disease-associated chromosomal breaks positioned well outside the relevant gene. With the availability of wide-ranging genome sequence comparisons, strong conservation of many noncoding regions became obvious. Functional studies have shown many of these conserved sites to be transcriptional regulatory elements that sometimes reside inside unrelated neighboring genes. Such sequence-conserved elements generally harbor sites for tissue-specific DNA-binding proteins. Developmentally variable chromatin conformation can control protein access to these sites and can regulate transcription. Disruption of these finely tuned mechanisms can cause disease. Some regulatory element mutations will be associated with phenotypes distinct from any identified for coding-region mutations.
Introduction
Thanks to the completion of the annotated human sequence and to the huge progress in many other genome programs, >1,500 genes have been linked with specific clinical phenotypes (Valle 2004). In most reported cases, disease-associated mutations alter the protein coding sequence of the gene in some way. However, there are potentially many different mechanisms that can disrupt normal gene function and can lead to pathological states. Growing insight into genomic organization (Gaffney and Keightley 2004) and into the multiple levels of transcriptional regulation (Levine and Tjian 2003) have revealed ways in which gene function may be disturbed. In addition, heroic detective work to understand some unusual human disease mutations has uncovered novel insights into the mechanisms of gene regulation.
Functional Gene Domains Extend beyond the Transcription Unit
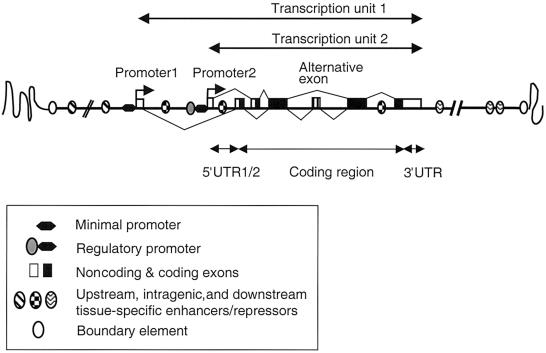
The majority of protein-coding genes are organized into multiple exons, which must be spliced to produce the mRNA that is translated into protein (fig. 1). A 5′ promoter element, contiguous with the transcription start site (the minimal or core promoter), is required to assemble the protein complex necessary for RNA synthesis (Levine and Tjian 2003). For many genes, the region immediately upstream of the minimal promoter contains sufficient transcription factor binding sites to direct correct expression of the gene—these are termed “regulatory promoters.” However, many genes also require multiple cis-acting distant genomic elements for spatiotemporally correct expression (Howard and Davidson 2004). These are often defined as enhancers, although some will be repressors or insulators; they can be located upstream, within introns, or downstream of the “transcription unit,” which comprises the transcribed exons and introns from the promoter to the polyadenylation site (fig. 1). Additional complexities include the possible presence of multiple alternative promoters and exons. The genomic regions harboring regulatory elements can stretch as much as 1 Mb in either direction from the transcription unit (Pfeifer et al. 1999; Kimura-Yoshida et al. 2004). Some or all of these elements may reside within the introns of neighboring gene(s), often with function unrelated to the regulated gene (Kleinjan et al. 2001; Lettice et al. 2002).
Figure 1.
Schematic representation of a theoretical gene locus, highlighting various cis elements that contribute to the regulation of gene expression. Exons are indicated by rectangular boxes, with the protein-coding portions in black. Complexity of gene output can be achieved through use of alternative promoters and/or exons. Multiple cis-regulatory elements, indicated by ovals, control the quantitative and spatiotemporal specific expression. These elements may be at considerable distances from the promoter, either upstream or downstream, and are sometimes within or beyond an adjacent gene. The chromatin structure of the locus is determined by a combination of the activities of these cis elements and the wider chromosomal and nuclear environment. In some loci, the outermost cis elements may carry some boundary activity, isolating the specific chromatin structure of the gene domain from that of adjacent chromosomal segments.
Cis-acting regulators can only fulfill their role if the chromatin structure in the region is appropriate. Some of the cis elements contribute to the organization of that structure. Chromatin exists in what is probably a structural continuum between the closed heterochromatic and open euchromatic state. Constitutive heterochromatin is found at centromeres and telomeres and in regions containing repetitive DNA. Facultative heterochromatin represents transiently silenced euchromatin. For high-level transcription, chromatin needs to be in an open (euchromatic) conformation (Elgin and Grewal 2003; Vermaak et al. 2003). A great deal of work is emerging that defines the machinery required for modulation of chromatin organization, with many protein components—and, recently, RNA components—still being identified (Elgin and Grewal 2003; Grewal and Rice 2004; Schramke and Allshire 2004). As with transcription factors, both general and tissue-specific components are required for this process. Many factors that affect chromatin and histone structure have been identified as regulating transcriptional control through acetylation, methylation, and ubiquitination of histone tails (Cho et al. 2004; Egger et al. 2004). The combination of specific histone modifications at a locus is usually referred to as the “histone code” (Turner 2002). Little is known about the factors that determine higher-order chromatin structure, such as the formation of hetero- and euchromatin.
Chromosomal Rearrangements Can Lead to Aberrant Gene Transcription through Different Mechanisms
Transcriptional control can be disrupted by one or both of the following mechanisms: (1) disturbing the interactions of the promoter and transcription unit with its cis-acting regulators, either by mutation or by physical dissociation of the transcribed gene from its full set of regulators (examples below and in table 1); and/or (2) alteration of local or global regulation of chromatin structure (Kleinjan and van Heyningen 1998; Jiang et al. 2003; Saveliev et al. 2003; Tufarelli et al. 2003). Chromosomal rearrangements can act through both these mechanisms. Because they are readily visualized through cytogenetic analysis, they were the first to be implicated in Drosophila position-effect variegation (PEV), in which mosaic patterns of gene expression were observed when genes that normally resided in euchromatic domains were transposed close to a heterochromatic region (Wallrath and Elgin 1995). Subsequently, germline chromosomal rearrangements were identified in some human diseases in which the implicated gene had already been firmly established but the phenotype-associated breakpoints were found to lie significantly outside the transcription unit. By analogy with Drosophila PEV mutations, such extragenic rearrangements were initially described as “position effects.” In recent years, an increasing number of such disease-related position-effect cases have come to light and are reviewed here. Extensive study of some of these cases has highlighted the importance of long-range transcriptional control in the affected genes, and, in some cases, such study has been instrumental in identifying distant cis-regulatory elements. In most cases, the genetic defect can be explained as a disruption of normal cis regulation of transcription, without clear evidence of altered chromatin organization, although mechanisms are difficult to assess in human disease, in which access to the affected tissues is often impossible. No variegation effects have been reported in humans, although the phenomenon has been observed in transgenic mice in which incomplete functional gene domains became inserted into heterochromatic genomic regions (Milot et al. 1996; Festenstein et al. 1999). Nonetheless, there are human disease states for which altered chromatin states, associated with chromosomal rearrangement, are implicated in the pathological mechanism (e.g., facioscapulohumeral dystrophy [FSHD] [see below]). Malignancy-associated somatic changes associated with altered chromatin conformation are not discussed here.
Table 1.
Position-Effect Genes in Human Diseases[Note]
| Gene | Gene Function | Domains/Motifs | Disease | Distance ofFurthest Breakpointa(kb) | 3′ or 5′ Side | Reference |
| PAX6 | TF | Paired box and homeodomain | Aniridia | 125 | 3′ | Kleinjan et al. 2001 |
| TWIST | TF | … | Saethre-Chotzen syndrome | 260 | 3′ | Cai et al. 2003 |
| POU3F4 | TF | POU homeodomain | X-linked deafness | 900 | 5′ | de Kok et al. 1996 |
| PITX2 | TF | Homeodomain | Rieger syndrome | 90 | 5′ | Trembath et al. 2004 |
| GLI3 | TF | Zinc finger | Greig cephalopolysyndactyly syndrome | 10 | 3′ | Wild et al. 1997 |
| MAF | TF | bZIP | Cataract, ocular anterior segment dysgenesis, and coloboma | 1,000 | 5′ | Jamieson et al. 2002 |
| FOXC1 | TF | Forkhead | Glaucoma/autosomal dominant iridogoniodysgenesis | 25/1,200 | 5′ | Davies et al. 1999 |
| FOXC2 | TF | Forkhead | Lymphedema distichiasis | 120 | 3′ | Fang et al. 2000 |
| FOXL2 | TF | Forkhead | Blepharophimosis-ptosis-epicanthus inversus syndrome | 170 | 5′ | Crisponi et al. 2004 |
| SOX9 | TF | HMG box | Campomelic dysplasia | 850 | 5′ | Bagheri-Fam et al. 2001; Pop et al. 2004 |
| SRY | TF | HMG box | Sex reversal | 3 | 5′/3′ | McElreavy et al. 1992 |
| SIX3 | TF | Homeodomain | Holoprosencephaly (HPE2) | <200 | 5′ | Wallis et al. 1999 |
| SHH | Signaling | … | Holoprosencephaly (HPE3) | 265 | 5′ | Roessler et al. 1997 |
| SHH | Signaling | … | Preaxial polydactyly | 1,000 | 5′ | Lettice et al. 2003 |
| SHFM1 | TF | DLX5/DLX6? | Split-hand/split-foot malformation | ∼450 | 5′/3′ | Crackower et al. 1996 |
| FSHD | ?? | … | Facioscapulohumeral dystrophy | 100 | 3′ | Gabellini et al. 2002; Jiang et al. 2003; Masny et al. 2004 |
| HBB | Oxygen carrier | Globin | γβ-Thalassemia | 50 | 5′ | Kioussis et al. 1983 |
| HBA | Oxygen carrier | Globin | α-Thalassemia | 18 | 3′ | Tufarelli et al. 2003 |
| Hoxd complex | TF | Homeodomain | Mesomelic dysplasia and vertebral defects | 60 | 3′ | Spitz et al. 2002 |
| LCT | Enzyme | Lactase | Lactase persistence | 15/20 | 5′ | Enattah et al. 2002 |
Note.— TF = transcription factor.
In the case of 3′ breakpoints, the distance refers to the distance from the breakpoint to the 3′ end of the gene or complex.
Phenotypes Associated with Regulatory Mutants
In humans, disturbance of long-range control by chromosomal rearrangement or deletion is most readily identified when the phenotype is a loss-of-function change very similar to those caused by point mutations within the coding region of the gene. However, it is important to realize that sequence changes (mutations) or localized deletion of specific elements may also to lead to novel phenotypes through partial, tissue-specific loss or gain of expression. Some examples have been identified, initially through mouse models (Lettice et al. 2003) or through detailed mapping and sequencing studies around a strong candidate gene (Enattah et al. 2002). Validation for the unpredictable effects of noncoding-region variation is now emerging (Knight 2003).
Disturbance of Long-Range Regulation at Human Disease Loci
Aniridia and PAX6: The Essential Role of a 200-kb Downstream Regulatory Domain
Aniridia (absence of the iris [MIM 106210]) and related eye anomalies are caused mainly by haploinsufficiency of the paired box/homeodomain gene, PAX6, at chromosome 11p13 (van Heyningen and Williamson 2002). A good model is provided by the Small eye (Sey) mouse (Hill et al. 1991). A number of aniridia cases have been described with no identifiable mutation in the transcription unit. Instead, chromosomal rearrangements disrupt the region downstream of the PAX6 transcription unit (Fantes et al. 1995; Lauderdale et al. 2000; Kleinjan et al. 2001; Crolla and van Heyningen 2002) (fig. 2A). Detailed mapping of the breakpoints placed them downstream of PAX6, with the furthest located 125 kb beyond the final exon (Fantes et al. 1995; Kleinjan et al. 2001, 2002). In silico transcript analysis of this region revealed the presence of a ubiquitously expressed neighboring gene, ELP4, which encodes a transcriptional elongation protein that is transcribed from the opposite strand (i.e., tail-to-tail with PAX6). All the human aniridia breakpoints map within the large (>100 kb) final intron of ELP4, presumably disrupting function of that allele, but transgenic YAC rescue of a mouse Sey deletion mutant showed that haploinsufficiency for Elp4 does not contribute to the eye phenotype (Kleinjan et al. 2002). Complete rescue of the heterozygous eye phenotype—and of the homozygous lethality, caused by a mouse Pax6 null mutation—is achieved with a 420-kb human YAC ending at YA (but not with one 80 kb shorter, ending at YB [fig. 2A]) (Schedl et al. 1996; Kleinjan et al. 2001). Essential regulatory elements were identified in Pax6-expressing cells between YA and YB by the presence of DNase I hypersensitive sites (HSs)—regions of chromatin open for regulatory factor binding. Transgenic reporter studies in mice revealed tissue-specific enhancer function for several of these elements (Kleinjan et al. 2001; Griffin et al. 2002). The cis-regulatory role of the distant elements was neatly illustrated in mouse-human somatic cell hybrids capable of expressing Pax6, in which human PAX6 was only expressed when a normal human chromosome 11 was retained, but not when harboring patient chromosomes with the PAX6 transcription unit intact but with distant downstream regulatory elements deleted (Lauderdale et al. 2000).
Figure 2.
Details of position-effect cases caused by disruption of long-range gene control. In all cases, the affected gene(s) are shown in red, and other genes are shown in purple or blue. Filled boxes indicate individual exons, and hashed boxes represent full genes. L-shaped black arrows indicate the direction of transcription. A, Human PAX6 locus. The loss of a set of DNase I HSs downstream from one allele causes aniridia. The HSs are located within introns of the adjacent ubiquitously expressed ELP4 gene. Some documented aniridia-associated breakpoints are denoted by blue arrows. The downstream end of the correcting YAC transgene (YA) and the noncorrecting one (YB) are shown in green. Both upstream YAC ends are ∼200 kb 5′ of the PAX6 promoters. Isolated HSs have been shown to act as tissue-specific enhancers for lens and retinal expression. B, The human POU3F4 deafness locus. The microdeletion of an 8-kb region located 900 kb upstream of the gene contains a conserved noncoding sequence, the loss of which leads to congenital deafness. The mouse slf inversion breakpoint X leaves the neural tube enhancer (nt) intact. C, Mouse/human upstream SHH region. A complex hotspot for limb abnormalities is found 1 Mb upstream of SHH, within the introns of LMBR1. The region contains a conserved noncoding element that is capable of functioning as an enhancer that drives SHH expression in the limb bud in both an anterior and posterior zone, as well as a repressor element that silences the anterior expression. The Sasquatch insertion disrupts the anterior repression function, whereas the acheiropodia deletion is thought to disrupt positive enhancer activity. D, Human FSHD region. Deletion of an integral number of D4Z4 repeats from the tip of the long arm of chromosome 4 to below a threshold of 10 repeats results in FSHD. A contentious model suggests that a multiprotein repressor complex fails to bind adequately to the deleted allele, which leads to derepression of several genes in the region proximal to the repeat array and causes the phenotype. E, Human α-globin locus (HBA). Deletion of the polyadenylation signal from the ubiquitously expressed LUC7L gene on the opposite strand leads to transcription of an antisense RNA that runs through the HBA2 gene, resulting in silencing and methylation of the HBA2 promoter. Open ovals indicate unmethylated CpG islands; the gray oval depicts the methylated CpG island. F, Mouse Hoxd cluster. A GCR regulates expression of multiple consecutive Hoxd genes in a tissue-specific manner. In the distal limb, the GCR also regulates the expression of Lnp, Evx2, and Hoxd13–10, whereas in the CNS it controls Lnp and Evx2. G, Mouse IL4/IL13 region. A conserved noncoding element (CNE) located between IL4 and IL13 controls expression of both genes, as well as IL5, but does not influence expression of the KIF3a and RAD50 genes. H, Human β-globin locus (HBB). Deletion of a large genomic region upstream of the human β-globin genes, including the LCR, results in reduced DNase I sensitivity and histone acetylation levels across the locus, which causes loss of globin expression. The β-globin locus is embedded within a region that contains numerous OR genes.
TWIST, POU3F4, PITX2, and GLI3
A number of developmental regulators with well-established haploinsufficiency phenotypes have been documented to give rise to similar loss-of-function phenotypes through disruption of long-range control: TWIST, a bHLH transcription factor; POU3F4 (previously, Brn4), with a POU domain and homeobox; PITX2, a paired-type homeodomain gene; and GLI3, a zinc finger gene homologous to Drosophila, cubitus interruptus (ci), which is a DNA-binding component of the sonic hedgehog signaling pathway. Loss-of-function mutations in TWIST lead to Saethre-Chotzen syndrome, an autosomal dominant form of craniosynostosis (Chun et al. 2002). By use of real-time PCR to analyze the allele dosage by “walking” across the critical region, translocation or inversion breakpoints, located at least 260 kb downstream of the TWIST gene, were found in two patients, which is suggestive of long-range disruption of gene function (Cai et al. 2003).
The POU3F4 gene (fig. 2B) is involved in the pathogenesis of the most common form of X-linked deafness, deafness type 3 (MIM 304400). Clinical features include fixation of the stapes and a widening of the internal auditory canal, which allows entry of cerebrospinal fluid into the inner ear. In addition to a spectrum of missense and truncating mutations, a complex duplication/inversion case involving distant upstream regions of POU3F4, as well as several genomic deletion cases, suggested upstream cis control of the gene (de Kok et al. 1995). The deletions observed in multiple independent patient cases overlap in a small region ∼900 kb upstream of the gene (de Kok et al. 1996). The smallest of these deletions comprises an 8-kb fragment containing a 2-kb sequence that is 80% conserved between mouse and human (Cremers and Cremers 2004). In addition, the mouse mutant sex-linked fidget (slf), generated in a random mutagenesis screen, was found to have developmental malformations of the inner ear that result in hearing loss and vestibular dysfunction and was defined by molecular analysis to result from a regulatory mutation of Pou3f4/Brn4 (Phippard et al. 2000). A large X-linked inversion was identified, with one breakpoint (“X” in fig. 2B) near, but not in, the Pou3f4 transcription unit. Expression in the embryonic inner ear, but not in the neural tube, was abolished, as two neural tube enhancers (“nt” in fig. 2B) within 6 kb of the promoter (Heydemann et al. 2001) were not disrupted.
Rieger syndrome type 1 (MIM 180500) is an autosomal dominant disorder characterized by dental hypoplasia and malformation of the umbilicus and anterior segment of the eye. The locus maps to 4q25, and mutations in PITX2, a paired-related bicoid-type homeobox gene encoding multiple isoforms, have been identified in individuals with Rieger syndrome (Alward et al. 1998). In addition to deletions and mutations in the gene itself, a translocation breakpoint 90 kb upstream of the gene was found in one patient (Flomen et al. 1998). Two further translocation breakpoints mapping 15–90 kb upstream of PITX2 have recently been identified (Trembath et al. 2004). The presence of a complex regulatory region around PITX2 is not surprising in light of its complex expression pattern, fulfilling diverse roles throughout development, not just in the eye but also in tooth, pituitary, heart, and laterality determination.
The zinc finger gene GLI3 on chromosome 7p13 is involved in the embryonal development of the brain, the limbs, and the skull and is an important effector of the hedgehog signaling network. Greig cephalopolysyndactyly syndrome (MIM 175700) is caused by haploinsufficiency of GLI3 on 7p13 (Wild et al. 1997). A probable position effect that results from a translocation has been described in a patient with a breakpoint 10 kb downstream from the last exon of GLI3 (Wild et al. 1997). Two dominant Gli3 mutant alleles in the mouse, the extra toes (Xt and XtJ) phenotypes, are caused by mutations in Gli3 itself (Vortkamp et al. 1992). The weak recessive Xt allele, anterior digit deformity (add), is caused by a transgene integration, combined with the deletion of an 80-kb region ∼40 kb upstream of Gli3 (van der Hoeven et al. 1993).
Lens Developmental Regulator MAF
Awareness of the possibility of long-range effects, coupled with biological insight into what makes a good candidate gene, can alert us to question whether an observed translocation is exerting its effect by haploinsufficiency of the disrupted gene. The chromosomal rearrangement, which apparently affects one gene, may, in fact, dissociate remote control elements from a more meaningful candidate gene located some distance away. Breakpoint analysis of a t(5;16) translocation in a family with cataracts and ocular anterior segment anomalies indicated that the translocation occurred in an intron of the WWOX gene, within a common fragile-site region on chromosome 16 (Paige et al. 2000). However, since WWOX is a widely expressed putative tumor suppressor gene and since the phenotype was observed in both balanced and unbalanced forms of the translocation, WWOX seemed an unlikely candidate to cause an eye phenotype. On the basis of its expression pattern and known involvement in eye development, misregulation of the bZIP transcription factor gene MAF (v-maf musculoaponeurotic fibrosarcoma oncogene homolog [avian]), located ∼1 Mb telomeric of the breakpoint, was considered a more likely candidate. Subsequent identification of a MAF (MIM 177075) missense mutation in another family with lens and iris anomalies confirmed the role of MAF in lens development (Jamieson et al. 2002). The observation that balanced and unbalanced forms of the t(5,16) translocation give rise to similar phenotypes suggests that the translocation leads to dominant misregulation of MAF, perhaps through sequences on the der(5) chromosome. Identification of a novel Maf missense mutation in the Ofl (opaque flecks in lens) mouse mutant confirms the dominant phenotype hypothesis for the missense mutation, since heterozygous Maf-null mice have no phenotype, and it provides a model for exploring Maf gene function in more detail (Lyon et al. 2003).
Forkhead Genes and Eye Development
A number of long-range regulatory disruptions are associated with genes of the forkhead/winged helix group of transcription factors, indicating the complex regulatory requirements of this gene family. Forkhead genes are involved in a diverse range of developmental pathways, with a significant number involved in eye development (Lehmann et al. 2003). FOXC1 (previously, FKHL7) lies in a cluster of forkhead genes on chromosome 6p25. Mutations cause ocular malformations with iris hypoplasia and glaucoma (iridogoniodysgenesis type 1 [IRID1] [MIM 601631]). The phenotypes are very similar, whether a segment is duplicated or deleted, which suggests that precise gene dosage is critical for normal eye development (Nishimura et al. 2001; Lehmann et al. 2003). In addition to intragenic point mutations and dosage effects for the FOXC1 gene in individuals with IRID1, a balanced translocation that mapped 25 kb from the gene was found in a case with primary congenital glaucoma (Nishimura et al. 1998). A further patient, with glaucoma and autosomal dominant iris anomaly, was shown to carry an interstitial deletion of 6p24-p25, where the proximal breakpoint was estimated to lie 1,200 kb from the FOXC1 locus (Davies et al. 1999). Despite the large distance, this patient could well represent another FOXC1 position-effect case. However, the situation is not clear-cut, because other possible candidate genes, notably the transcription factor TFAP2A, are located in the deletion interval.
A closely related forkhead gene, FOXC2, maps to chromosome 16q24 (Fang et al. 2000). Inactivating mutations have been identified in patients with lymphedema distichiasis (LD [MIM 153400]), an autosomal dominant disorder characterized by lymphedema that affects the limbs and by double rows of eyelashes (distichiasis). Other complications may include cardiac defects, cleft palate, extradural cysts, and photophobia, which highlight the pleiotropic functions of FOXC2 during development. A translocation t(Y;16)(q12;q24.3) with the breakpoint mapping ∼120 kb 3′ of the FOXC2 gene was reported in a patient with neonatal lymphedema. The translocation did not appear to interrupt a gene on chromosome 16, nor were any candidate genes found on the Y chromosome. This made a clear case for a position effect on FOXC2, once multiple inactivating mutations had been identified in other individuals with this phenotype. Interestingly, another forkhead gene, FOXL1, also maps between FOXC2 and the breakpoint and thus could also be inactivated and have phenotypic effects in the translocation patient (Fang et al. 2000). Because many intragenic FOXC2 mutations have been documented in LD individuals (Finegold et al. 2001; Brice et al. 2002), disruption of FOXC2 and not FOXL1 is ascribed to this chromosomal rearrangement.
FOXL2 is the forkhead family member in which translocation breakpoints some distance upstream of the gene cause phenotypes indistinguishable from loss-of-function mutations within that gene. Coding-region mutations in FOXL2 lead to blepharophimosis-ptosis-epicanthus inversus syndrome (MIM 110100), an eyelid and forehead dysmorphology in both sexes that is often associated with gonadal dysgenesis and premature ovarian failure in women (Crisponi et al. 2001). The breakpoints all map ∼170 kb 5′ of FOXL2, in intron 6 of the ubiquitously expressed MRPS22 gene (De Baere et al. 2001, 2003; Crisponi et al. 2004). Sequence comparisons between human and mouse reveal the presence of three highly conserved segments beyond the furthest breakpoint, in introns 6, 11, and 12 of MRPS22. Interestingly, polled intersex syndrome is a genetic syndrome in goats that combines a craniofacial defect resulting in polledness (absence of horns), female infertility, and XX sex reversal. It is caused by an 11.7-kb deletion in the homologous FOXL2 region of the goat genome, encompassing the conserved sequence in intron 11 of MRPS22 (Pailhoux et al. 2001). These elements are strong candidates to function as distant cis-regulatory elements affecting FOXL2 expression.
SOX9 Campomelic Dysplasia (CD) and Sex Determination
The HMG box gene SOX9 was identified as the gene responsible for autosomal sex reversal and CD (MIM 114290); coding-region mutations for this gene have been defined, and a high frequency of chromosomal translocations that do not disrupt the transcribed gene have been detected (Foster et al. 1994; Wagner et al. 1994). SOX9 is therefore one of the oldest examples of a human gene in which long-range regulation has been implicated and studied. The phenotypes observed are variable but generally severe, so that vertical transmission of mutations is rarely observed. In addition to the variable osteochondrodysplasia, XY sex reversal is found in about two-thirds of karyotypic male cases. All the rearrangements are found from 50 kb to 950 kb upstream of SOX9 (Pfeifer et al. 1999). The phenotypes of the breakpoint cases are generally similar to, although less severe than, the loss-of-function intragenic mutations. This, combined with the absence of any validated ORFs in the 1-Mb region upstream, suggests that the chromosomal rearrangements remove one or more cis-regulatory elements.
As in other cases, the availability of mouse models and comparative sequence analysis has aided the dissection of regulatory function in the SOX9 region. Heterozygous knockout of Sox9 in the mouse recapitulates the CD phenotype, except for the sex reversal (Kist et al. 2002). The involvement of long-range gene control is supported by the fact that mice transgenic for human SOX9-spanning YACs showed transgene expression patterns similar to those in endogenous Sox9 (except in gonads), but only when the YAC transgene contained a 350-kb sequence upstream of SOX9 and not with a truncated YAC that contained only 75 kb of 5′ flanking sequence (Wunderle et al. 1998). Comparative sequence analysis between human and Fugu rubripes revealed five conserved elements, E1–E5, in the 290-kb region 5′ of human SOX9 and three further elements, E6–E8, 3′ of SOX9 (Bagheri-Fam et al. 2001). On the basis of the expression pattern in the YAC transgenics, elements E3–E5 are candidate enhancers for SOX9 expression in limb and vertebral column. Of 10 CD translocation breakpoints analyzed so far, 8 separate these elements from SOX9, with 2 other breakpoint cases that suggest additional, more-distal enhancers (Pop et al. 2004).
A dominant insertional mutation in the mouse, named “Odsex” (Ods [ocular degeneration with sex reversal]), has been shown to dysregulate Sox9 expression. In a transgenic experiment, a tyrosinase minigene driven by the dopachrome tautomerase (Dct) promoter was randomly inserted ∼1 Mb upstream of Sox9, additionally causing a 134-kb deletion around the insertion site. In contrast to the male-to-female sex reversal associated with human SOX9 loss-of-function changes, Ods mice show female-to-male sex reversal, as well as microphthalmia with pigmentation defects and cataracts. The XX sex-reversal phenotype is accompanied by misexpression of Sox9 in the XX gonad, where Sox9 is usually repressed. Initially, it was proposed that the Ods deletion had removed gonad-specific long-range regulatory element(s) that would normally mediate the female-specific repression of Sox9, thus resulting in up-regulation of Sox9 and the consequent male development (Bishop et al. 2000). However, more-recent experiments have shown that the 134-kb deletion alone is insufficient to cause the sex reversal (Qin et al. 2004). A double-gene–targeting strategy was used to recreate the Ods insertional deletion and the tyrosinase insertion—but this time driven by its own promoter—no eye or sex-reversal phenotype was observed. This suggests that long-range interaction occurs specifically between the Dct promoter and Sox9 in the Ods mutant. Indeed, the Ods eye phenotype was recapitulated in transgenic mice with a Dct-Sox9 minigene cassette, and a temporal misexpression of Sox9 under the control of the Dct promoter was demonstrated. In the eyes of Ods mice, the inserted Dct “promoter” element seems to act as a long-range activator of the Sox9 promoter over a distance of 980 kb. The mechanism behind the sex-reversal phenotype is more complex. No sex reversal was seen in Dct-Sox9 transgenics, which suggests that, in the gonads of Ods mice, the Dct promoter interacts with Sox9 by an indirect mechanism, possibly involving endogenous gonad-specific Sox9 enhancers and chromosomal conformation changes as a result of the deletion (Qin et al. 2004). Furthermore, the XX sex-reversal phenotype, driven by the original Ods transgene, is highly background dependent, and a strong modifier locus has been identified (Qin et al. 2003).
Holoprosencephaly (HPE) Is Associated with Long-Range Regulator Effects for SIX3 and SHH
HPE is a complex brain malformation that results from incomplete cleavage of the prosencephalon and that affects both the forebrain and the face. The etiology of HPE is very heterogeneous, with at least 12 different genetic loci implicated. One of these, HPE2 (MIM 157170), was mapped to chromosome 2p21. Subsequent analysis of SIX3, an atypical homeobox gene, as a strong candidate gene within the critical interval revealed four different heterozygous point mutations that are predicted to interfere with SIX3 transcriptional competence. Two further patients with HPE were shown to carry translocation breakpoints outside the SIX3 transcribed region; these were located by pulsed-field gel electrophoresis at 10–200 kb upstream of the transcription initiation point (Wallis et al. 1999).
HPE3 (MIM 142945) was defined as another HPE locus and was associated with chromosome 7q36 deletions, and eventually point mutations were identified within the candidate gene sonic hedgehog (SHH). Phenotypic expressivity can vary, even within families—from a single cerebral ventricle and cyclopia (lethal) to clinically unaffected carriers, perhaps with a single central incisor (Belloni et al. 1996; Roessler et al. 1996). Interestingly, whereas HPE3 in humans is caused by haploinsufficiency of SHH, in the mouse both alleles need to be inactivated to produce a similar phenotype, which indicates a more critical role for correct SHH dosage in humans (Chiang et al. 1996). As for SIX3, balanced translocations with breakpoints up to 265 kb 5′ of the SHH transcription start site (Belloni et al. 1996; Roessler et al. 1996, 1997) were documented in individuals with HPE. A further familial translocation, 315 kb upstream of the SHH start, did not produce a detectable HPE phenotype in the seven individuals examined, thus limiting the region in which to look for the putative control element(s) (Roessler et al. 1997).
Distant Regulators for SHH Expression Are Also Involved in Limb Development
As mentioned, the phenotype caused by a regulatory mutation can be very different from that caused by coding-region mutations, because expression may only be affected in a subset of expressing tissues. The involvement of SHH in preaxial polydactyly (PPD [MIM 174500]) fits such a scenario. In addition to its many functions in brain and neural development, SHH plays a key role in defining the limb anterior-posterior axis. Normally, Shh is transiently expressed in the posterior part of the mouse limb bud and sets up a morphogen gradient from this zone of polarizing activity (ZPA). PPD is associated with mirror-image duplication of the hand/foot. One form is seen in the mouse mutant Sasquatch (Ssq), which arose during transgenic studies as a result of random insertion of a reporter cassette 1 Mb upstream of the Shh gene (Sharpe et al. 1999) (fig. 2C). None of the other independent transgene insertion mice showed this phenotype, which suggests that the Ssq phenotype is caused by the insertion of the transgene into that specific site, which was revealed by molecular analysis as intron 5 of the limb region 1 gene (Lmbr1) and which lies beyond the adjacent testis- and ovary-specific gene Rnf32. Homozygous transgenic mice have a more severe limb phenotype than heterozygotes. Genetic analysis of two other mouse limb mutants, Hemimelia extra-toes (Hx) and Hammertoe (Hm), also mapped to Lmbr1 and the expression of this gene was found to be altered in Hx embryos at the relevant time in limb formation (Clark et al. 2000). However, the transgenic Ssq mice with PPD, as well as Hx and Hm mice with the disease, all showed ectopic expression of Shh in an anterior, as well as the normal posterior, region of the limb bud (Sharpe et al. 1999; Lettice et al. 2002; Hill et al. 2003). This explains the mirror-image duplication phenotype and suggests that the correct control of Shh expression in the limb bud is disrupted by the transgene insertion (Lettice et al. 2002). To strengthen this conclusion, a cis-trans test was performed, which showed that when the Ssq transgene insertion (or the Hx mutant) is recombined to work in cis with a functionally null Shh allele, the PPD phenotype is abolished (Lettice et al. 2002; Sagai et al. 2004). These results strongly suggested the presence of a limb regulatory element for Shh expression 1 Mb upstream of the gene. By use of phylogenetic sequence comparison, an evolutionarily conserved sequence that brackets the Ssq insertion site was identified, and analysis in transgenic mice revealed reporter expression in the posterior margin of the limb buds, which implies that the isolated regulatory element, now designated as the “ZRS” (ZPA regulatory sequence), can function as a limb-specific enhancer (Lettice et al. 2003). The endogenous ZRS is also predicted to harbor repressor function in the anterior limb bud, since its disturbance leads to ectopic Shh expression there.
A chromosomal breakpoint in a translocation-associated case of human PPD was mapped within the homologous intron of LMBR1 (Lettice et al. 2002). PPD is a relatively frequent human phenotype, which had been tightly linked in many families to the LMBR1 gene. Interestingly, in patients with the severe human recessive limb anomaly acheiropodia (bilateral congenital amputations of the upper and lower extremities and aplasia of the hands and feet), a deletion that apparently removes exon 4 of LMBR1 was identified, and, initially, it was suggested that LMBR1 itself plays a major role in the disorder (Ianakiev et al. 2001). However, in light of the evidence cited above, it seems more likely that the involvement of LMBR1 is merely due to it harboring regulatory elements that control Shh expression in the limb.
To assess whether mutations in the ZRS could account for the PPD phenotype in some affected families without translocations or deletions, the element was sequenced in a large number of affected and unaffected family members and controls. Amazingly, single point mutations in the ZRS were found in four families with PPD; these mutations were observed in all of the affected and none of the unaffected individuals in these families (Lettice et al. 2003). All four point mutations were in different parts of the ZRS. Analysis of the ZRS in the Hx mouse and an N-ethyl-N-nitrosourea–induced mutant, M100081, identified two more independent point mutations (Sagai et al. 2004). As mentioned, the ZRS is thought to have a dual function in the regulation of SHH: (1) driving the initiation of expression in the limb bud and (2) restricting this expression to the posterior margin. The Ssq insertion and the Hx, M100081, and PPD point mutations affect the latter (but not the former) activity of the ZRS, which leads to the observed anterior ectopic SHH expression.
This illustrates the remarkable fact that point mutations in a regulatory element at a distance of 1 Mb from the gene promoter can have a detrimental effect on development. The scattered positioning of the mutations within the element suggests that different transcription-factor binding sites may be disrupted, with similar outcomes. An interesting link with development and evolution is that the ZRS is recognizably conserved in all vertebrates with some sort of locomotory appendage (limbs, wings, or fins), but it is absent in snakes and a limbless newt (Lettice et al. 2003; Sagai et al. 2004).
Split-Hand/Split-Foot Malformation
To underscore our current ignorance of long-range regulatory mechanisms, we touch on another phenotype: split-hand/split-foot malformation type 1 (SHFM1 [MIM 183600]), a heterogeneous limb developmental disorder characterized by missing digits, fusion of remaining digits, and a deep median cleft in the hands and feet—a phenotype that is almost certainly caused by a long-range regulatory anomaly. The SHFM1 critical region has been mapped to 7q21.3-q22.1, on the basis of chromosomal rearrangements (Scherer et al. 1994; Crackower et al. 1996; Ignatius et al. 1996). Three genes, DSS1 and the distalless homeobox genes DLX5 and DLX6, are located in this region, but none of these genes is directly interrupted by the translocations and deletions. The absence of patients with intragenic coding-region mutations has so far prevented the clear designation of the SHFM1 gene(s) in humans. Targeted inactivation of either Dlx5 or Dlx6 alone does not produce a limb phenotype in mouse (Robledo et al. 2002). However, the targeted double inactivation of Dlx5 and Dlx6 causes bilateral ectrodactyly with a severe defect of the central ray of the hind limbs, a malformation typical of SHFM1. Dss1 expression in these mice is normal. This raises the possibility that SHFM1 is due to the disruption of a shared regulatory element for the two DLX genes and possibly DSS1 (Merlo et al. 2002).
Aberrant Gene Expression in Human Disease through Altered Chromatin Structure
FSHD
FSHD (MIM 158900) is a neuromuscular disorder affecting predominantly the facial and shoulder girdle muscles. It is inherited as an autosomal dominant trait, but ∼10%–30% of the mutations are de novo, and about half of these are somatic mosaics. The molecular rearrangement associated with FSHD is the deletion of an integral number of 3.3-kb tandem repeats from the subtelomeric region of the long arm of chromosome 4 (Wijmenga et al. 1992). Unaffected individuals carry between 11 and 150 copies of this repeat (fig. 2D), named D4Z4, whereas patients carry ⩽10 copies on one of their chromosomes (van Deutekom et al. 1993). In general, the lower the number of D4Z4 repeats, the greater the severity of the phenotype and the younger the age at onset. The D4Z4 repeat contains internal VNTR-type repeats and a putative ORF (DUX4), but no protein-coding transcripts have been identified from the repeat sequence, despite intense efforts. FSHD is therefore thought to result from some form of cis interaction, between the shortened 4q35 repeat region and more-centromeric genes in the 4q35 region. This is supported by the fact that an almost identical repeat array on chromosome 10q26, with further homology on both sides of the repeat, does not cause disease when shortened (Bakker et al. 1995). A number of genes or putative genes have been identified in the region centromeric to the D4Z4 repeats on chromosome 4q35: FRG2, DUX4C, FRG1, and ANT1. ANT1, the most centromeric of these, located ∼5 Mb from the proximal repeat boundary, is considered the most likely candidate because it encodes an adenine nucleotide translocator that has been implicated in myopathy and is predominantly expressed in heart and skeletal muscle (Gabellini et al. 2002). Interestingly, individuals with deletion of the entire array, including D4Z4, FRG2, DUX4C, and FRG1, show no phenotypic consequences (Tupler et al. 1996). This suggests that the loss of repeats results in a gain-of-function mutation, but much debate continues over the mechanism. Recently, it was claimed that the D4Z4 repeats bind a multiprotein repressor complex that negatively regulates expression of genes in the 4q35 region, possibly through promoting a cis-spreading heterochromatinization of the region. In this model, D4Z4 deletions would lower the number of binding sites for the repressor complex below a critical threshold, thus allowing local decondensation of chromatin and the consequent derepression (up-regulation) of the genes in the region (Gabellini et al. 2002). In support of this position-effect hypothesis, one study found that all the genes in the region were up-regulated in muscle from patients with FSHD, and a protein complex that contained a subunit with known repressor activity was isolated and was shown to bind specifically to a site in the D4Z4 repeat (Gabellini et al. 2002). However, another recent study found no difference in expression levels of ANT1 and FRG1 between muscle tissue of patients with FSHD and unaffected individuals. Furthermore, histone H4 acetylation levels over the various gene promoters suggested that the region adopts a nonexpressed euchromatinlike structure, both in control individuals and in patients with FSHD (Jiang et al. 2003). Instead, a model was proposed in which a short array of D4Z4 repeats forms a long-distance loop that interacts directly with an as-yet unknown gene or genes on 4q35 to increase expression, whereas longer arrays of D4Z4 repeats form intra-array loops and thus sequester the array (Jiang et al. 2003). Recent work furthermore suggests that the 4q telomere always localizes at the nuclear periphery, requiring lamin A/C to do so. Although this specific localization is observed in both normal and D4Z4-deleted alleles, FSHD is proposed to arise from improper interactions with transcription factors or chromatin modifiers at the nuclear envelope (Masny et al. 2004).
α-Thalassemia and Antisense RNA
A completely different type of mechanism was shown to be responsible for a recently reported case of α-thalassemia (MIM 141800) (Tufarelli et al. 2003) (fig. 2E). Thalassemias result from an imbalance in the levels of the α- and β-globin chains that constitute the oxygen-carrying hemoglobin tetramer in human red blood cells. In general, this is caused by mutation or deletion of one or more of the globin genes or, as in the case of the Spanish and Dutch thalassemias, by the deletion of the β-globin locus control region. The α-globin gene is normally transcribed from four major gene copies (HBA1 and HBA2, on each chromosome 16), under the control of the 5′ HS-40 region. In the recently reported case of α-thalassemia, an 18-kb deletion, encompassing the HBA1 and HBQ1 genes, was identified. The HBA2 gene and HS-40 remained completely intact. One copy of the HBA1 gene was deleted in this family, but the severity of the phenotype suggested that expression from the intact HBA2 gene on the deleted chromosome could also be affected. The absence of HBA2 expression from the abnormal chromosome was confirmed in an experiment that used permissive cell hybrids. Subsequently, a 2-kb region, including the HBA2 CpG island, was found to be densely methylated in all tissues of this individual, whereas under normal circumstances the α-globin CpG islands always remain unmethylated, even in nonexpressing tissues. Further analysis of this mutation showed that silencing and methylation of the HBA2 promoter were strongly correlated with the presence of antisense RNA transcripts derived from the truncated neighboring LUC7L gene on the opposite strand (fig. 2E). In addition to the HBA1 and HBQ genes, the 18-kb deletion had removed the final three exons of the LUC7L gene, including its polyadenylation signal, which caused the RNA polymerase to read through into the HBA2 gene and promoter. The silencing and methylation of the HBA2 promoter, induced by the aberrant read-through transcription of LUC7L, was confined to the deleted chromosome, which indicates that it occurs through a cis-acting mechanism (Tufarelli et al. 2003).
This case presents a very different mutational mechanism that can result in turning off expression at a neighboring locus. Although it does not involve disruption of long-range gene control required for normal expression of the gene, it does represent a mechanism that could easily cause problems at many other gene loci, particularly in gene-dense regions of the genome. Other examples are likely to come to light when a highly expressed truncated gene lacking a polyA signal as a result of deletion or translocation prevents expression of an intact, adjacent disease gene (on the opposite strand) by an aberrant read-through mechanism (fig. 2E).
Exploring Gene Regulation through Animal Models
The cases described above highlight genes in which long-range transcriptional control has been identified through the analysis of patients with genetic malformations. There are, however, many more genes for which long-range gene regulation undoubtedly plays an important role but for which no position-effect–type mechanism of disease has so far been shown (e.g., Di Leone et al. 2000; Hadchouel et al. 2003). For some of these genes, long-range regulatory control has been explored through gene manipulation—sometimes accidentally, in mouse gene knockouts with unexpected outcomes (Olson et al. 1996). Associated human (or mouse) mutant phenotypes may exist but will not be readily identified if the phenotype is not caused by haploinsufficiency. In other cases, the disruption may have only a subtle effect or may give rise to a different phenotype from the one shown in the case of complete gene inactivation and therefore would not readily be associated with the gene of interest. Below, we describe three gene loci for which studies in mouse and sheep have contributed to our understanding of long-range gene control.
Hoxd Cluster
An interacting gene complex in which long-range control has been studied extensively in the mouse is the Hoxd cluster. The locus contains a number of genes, including the Hoxd complex and the Evx2 and lunapark (Lnp) genes, that are involved in patterning of the body axis in the lumbosacral region, as well as in limb development and the correct formation of the digits (Spitz et al. 2003). Elegant studies of the cluster, with its characteristic colinearity in spatiotemporal expression of the genes correlating with gene order along the chromosome, have led to the notion that the ancestral role of the cluster was the specification of morphogenesis along the main body axis and that, later in evolution, the Hoxd genes were co-opted to function in the development of novel structures, such as the limbs (Spitz et al. 2001). Consistent with this scenario, it was shown that the regulatory controls for the ancestral and colinear expression are located within the Hoxd cluster, whereas the co-opted expression domains depend on enhancers located at remote positions outside the cluster (Spitz et al. 2001). During limb development, Hoxd10–13 and the neighboring genes Evx2 and Lnp (fig. 2F) are coexpressed, with very similar profiles, in digits (Spitz et al. 2003), which suggests the possibility that a single enhancer may control digit expression for all these genes (van der Hoeven et al. 1996). A search for such an enhancer through BAC transgenics and sequence comparisons led to the identification of a region far upstream of the locus (fig. 2F) that controls tissue-specific expression in multiple tissues of a contiguous set of genes in the locus. This region, termed the “global control region” (GCR), is proposed to create a widespread regulatory landscape, sharing its enhancing activity over a defined number of genes in a tissue-specific manner (Spitz et al. 2003). Digit activity of the GCR spreads over six genes that cover at least 240 kb, whereas CNS activity is limited to the Lnp and Evx2 genes. A direct demonstration of the role of the GCR was provided by analysis of a semidominant mouse limb mutant, Ulnaless (Herault et al. 1997; Peichel et al. 1997). Ulnaless carries a paracentric inversion with one breakpoint in the Lnp gene and the other breakpoint 770 kb more telomeric; this inverts the Evx2 and the Hoxd cluster and moves it ∼700 kb from the remains of Lnp. When Ulnaless was bred against a targeted allele with a deletion of the region from Evx2 to Hoxd11, expression of Evx2 and Hoxd13 was shown to be lost in the distal limb (Spitz et al. 2003). This suggests that the inversion either has moved the limb-expressed genes out of reach of the GCR, or it has introduced some insulating activity between the genes and the GCR. Evolutionary sequence comparison of the human GCR region revealed very strong sequence conservation over several kb in mouse, as well as in two shorter regions in the teleost fish F. rubripes. The latter has no digits or external genitalia and lacks the Hoxd1, Hoxd8, and Hoxd13 genes, although Hoxd12, Evx2, and Lnp are present (Spitz et al. 2003). Although no human cases of chromosomal disruptions between the GCR and the HOXD genes have been described, a putative regulatory change has been reported at the other (telomeric) end of the cluster. A patient suffering from mesomelic dysplasia (shortening of the forearms and forelegs) with vertebral defects was found to carry a balanced translocation breakpoint 56 kb telomeric of the HOXD1 gene (Spitz et al. 2002) near MTX2.
Limb Deformity
Recent reassessment of a much-studied mouse recessive mutant, limb deformity (ld), has revealed that the original assignment of the phenotype to loss of function of the disrupted formin gene (Fmn) was incorrect. Three ld alleles were clearly shown, some time ago, to truncate Fmn (Maas et al. 1990; Wang et al. 1997). However, the out-of-frame deletion of exon 10 in the large Fmn gene fails to produce limb deformity, although a larger deletion from exon 10 to exon 24 does recapitulate the ld phenotype. Two previously uncharacterized ld alleles have now been shown to disrupt the neighboring gene gremlin (Grem1), and Grem1-targeted knockout mice are allelic with ld (Michos et al. 2004; Zuniga et al. 2004). Gremlin is a cystine knot protein that functions as a bone morphogenic protein (BMP) antagonist and that is itself regulated by SHH. Further investigation reveals that the region from exon 19 to exon 23 of Fmn is the site of a GCR required for GREM1 expression in the limb and that this region is responsive to SHH signaling. This is therefore an example in which, for over a decade, the wrong gene was implicated in a developmental anomaly because several alleles caused disruption of the gene that houses a major regulatory element for the real culprit, Grem1. The alleles that disrupt Grem1 directly have a more severe homozygous phenotype than the Fmn-disrupting alleles. The Grem1−/− mice have complete renal agenesis and lung septation defects, in addition to the limb anomaly. It will be interesting to see whether GREM1 mutations will be found in appropriate human limb/kidney phenotypes, possibly only in populations with frequent consanguinity. This is also a salutary example of how readily we can be misled into associating disease pathology with the wrong gene when mutations disrupt a neighboring, functionally unrelated gene that harbors cis elements controlling the real culprit.
Callipyge (CLPG) Mutation in Sheep
A striking example of the effect of a single-nucleotide substitution in a putative long-range regulatory element is provided by the CLPG mutation in the imprinted region DLK1–GTL2 of sheep (Georges et al. 2003). The callipyge (CLPG [“beautiful buttocks”]) phenotype is characterized by hindquarter muscle overgrowth that only affects heterozygotes with paternal inheritance of the CLPG mutation (Cockett et al. 1996). The complicated mechanism behind CLPG, termed “polar overdominance,” is thought to be a combination of a direct long-range cis-regulatory effect on transcription of the genes in the region and a transinteraction with a tightly linked transacting repressor. The CLPG region contains two paternally expressed protein-encoding genes, DLK1 and PEG11, and two maternally expressed noncoding genes, GTL2 and MEG8. All four genes show a postnatal increase in muscle-specific expression, when linked in cis to the CLPG mutation, while maintaining their imprinted status. By use of DNA from the mutants and from the mosaic founder individual, the CLPG mutation was identified as a single-base substitution in a region of strong sequence conservation 33 kb upstream of the GTL2 gene (Freking et al. 2002). In the current model, the paternally expressed DLK1 or PEG11 is proposed to encode an effector for muscle growth, whereas either of the maternally expressed GTL2 and MEG8 noncoding RNAs could act as a repressor. Only when the CLPG regulatory mutant allele is paternally inherited would there be an overexpression of the effector without the concomitant up-regulation of the repressor (Georges et al. 2003).
Identifying Novel Distant Regulatory Elements
Even quite recently, tissue-specific regulatory elements were generally identified by promoter/enhancer deletion studies in transgenic mice (Pfeffer et al. 2002) or by DNase I hypersensitivity mapping in expressing tissues (Elgin 1988; Kleinjan et al. 2001). The latter technique is based on the observation that local disruptions of the regular nucleosomal array create preferential targets for DNase I at low concentrations of the enzyme. Binding of transcription factors to the DNA at cis-regulatory elements is thought to cause the removal or dispersal of a nucleosome. Thus DNase I HSs are often found at active cis-regulatory elements. Genomic regions close to the promoter are sometimes further explored by footprinting and by electrophoretic mobility-shift assays, which seek to identify regulatory protein-binding DNA regions (Knight 2003). With the explosion of genomic sequences available for many model organisms, multiple sequence comparisons between evolutionarily diverged species have revealed many highly conserved genomic regions outside coding domains (Dubchak and Frazer 2003). The conservation of these sequences is taken to imply functional significance, most likely in gene regulation (Hardison 2000; Pennacchio and Rubin 2001). Indeed, a high proportion of regulatory elements that were previously validated—for example, by transgenic reporter studies—were found to be highly conserved, often across several vertebrate classes. Conversely, sequence conservation is now often the starting point for functional studies to reveal regulatory properties in novel transgenic and gene-targeting studies (Williams et al. 1998; Griffin et al. 2002; Nobrega et al. 2003; Uchikawa et al. 2003; Kimura-Yoshida et al. 2004; Kleinjan et al. 2004).
A conserved element was identified by cross-species sequence comparison in the intergenic space between the Il4 and Il13 cytokine genes (Loots et al. 2000). Targeted deletion in mice revealed it to be a coordinate enhancer for Il4 and Il13, as well as for the more distant Il5 gene; this deletion influences gene expression in TH2 cells and leaves expression in mast cells unaltered. Two other genes in the region—Kif3a, upstream of Il4, and Rad50, located between Il13 and Il5—were shown to be unaffected by the enhancer (Loots et al. 2000; Mohrs et al. 2001) (fig. 2G).
In most cases, functional analysis of conserved elements has been through the generation of transient or permanent transgenic reporter animals, which are produced to show that the reporter expression pattern mirrors a subset of the expression spectrum shown by the endogenous gene (Kleinjan et al. 2001; Griffin et al. 2002; Nobrega et al. 2003). Some of these analyses have now become quite systematic: large flanking regions around genes that are known to require complex spatiotemporal expression for full function have been screened. Selection of a distant species for comparison can help focus on the major functionally conserved elements, as shown by the comparison of mouse and chick Sox2 (Uchikawa et al. 2003). Use of the more compact F. rubripes genomic sequence for comparison means that a larger region of the genome can be surveyed, which is useful for genes like Otx2 and others with a very large regulatory domain (Kimura-Yoshida et al. 2004). This new capacity to identify regulatory elements more readily through evolutionary sequence comparison will allow the development of methods to identify and validate functionally significant variants and pathologic mutations in these regions.
Long-Range Regulation: Common Theme or Selected Few?
It is intriguing to note that most genes in which disturbance of long-range control has been observed are key developmental regulators. The examples above mainly involve tissue-restricted transcription factors that directly or indirectly bind to DNA. The remainder are developmental signaling molecules such as SHH. All of these genes play multiple recurrent roles in the determination, patterning, and differentiation of many tissues and organs—they are frequently reused at different stages of development (even within a single tissue), sometimes in collaboration with different partner proteins. For example, PAX6 and SOX2 are implicated in early lens determination and also later differentiation (Treisman and Lang 2002), and both are also differentially expressed in the retina and the brain (van Heyningen and Williamson 2002). To fulfill these multiple roles, they require complex spatiotemporal expression control, which evolves through phylogenetic diversification (see below).
From a regulatory viewpoint, genes can be grouped broadly into three classes: (1) “Housekeeping” genes, which are required for the functioning of most or all cells and therefore generally ubiquitously expressed; these genes usually have promoters that are active in all cells without the need for specific enhancer elements. (2) Tissue-specific genes, which play a specific role in the particular function of the differentiated cell-type; these genes are usually regulated through one or a few specific enhancers. (3) Developmental regulator genes, which function in specific tissues at defined time-points in development—sometimes at critically defined levels—and have to be strictly inactive in all other tissues and time-points; to achieve such sophisticated expression profiles, these genes require multiple enhancer elements that all need to be fitted into the cis region surrounding the gene. Despite the common use of terminology, which suggests some sort of engineered design in the structure of gene loci with respect to transcription and other biological processes, the acquisition and loss of regulatory elements does not occur through conscious design but rather as a result of evolutionary tinkering and selection (Duboule and Wilkins 1998; Carroll et al. 2001). The redeployment of developmental regulatory genes in novel tissues and pathways has become a recognized feature in the evolution of greater complexity in higher organisms. To a large extent, this depends on the chance appearance of a new combination of sequences with regulatory activity in the vicinity of the appropriate promoter. As long as the element can establish interaction with the promoter, does not interfere with the existing regulatory control in a disadvantageous manner, and presents some kind of evolutionary advantage itself, the new cis element may become fixed. The further appearance of elements synergizing with existing ones to achieve the optimal expression level more reliably will similarly confer selective advantage and retention. Interestingly, in some cases in which there is evidence of the redeployment of regulatory factors in more recent evolutionary adaptations, a correlation often exists between the more proximal location of enhancers involved in the ancestral function of the gene and the more distal position of regulatory elements required for more recently co-opted expression in novel tissues, as described above for the Hoxd cluster.
Evolution of Regulatory Control
Gain and Loss of Elements
The increase in complexity from yeast to mammals is not adequately reflected in expanded gene numbers. The emerging view is that the growth in evolutionary complexity is predominantly brought about by increasing regulatory intricacy (Carroll et al. 2001; Levine and Tjian 2003) and that a significant component of this is through alterations in cis regulation (Wray et al. 2003; Howard and Davidson 2004). New cis-regulatory elements can arise by several mechanisms, including random sequence mutation and genomic insertions (for example, through retrotransposition) (Han et al. 2004). These elements can bring, from elsewhere in the genome, functionally active sequences with regulatory capacity novel to the host gene. On the other hand, transposon insertion can also be highly deleterious and can cause disease (Druker and Whitelaw 2004). Another frequently observed mechanism for acquiring novel function is through gene duplication followed by divergence, not only of coding regions but even more readily in regulatory domains. Gene duplications may arise locally by aberrant recombination or replication (Hurles 2004) and may encompass one or a few adjacent genes. Occasionally, chromosome or genomewide duplications take place, generally followed by loss of many of the duplicated segments, as in teleost fishes (Robinson-Rechavi et al. 2001). There is ample evidence of all these types of events in most organisms. Phenotypic changes can also arise through loss of cis elements associated with a particular gene, as exemplified by the pelvic reduction seen in threespine sticklebacks (Shapiro et al. 2004).
Genomic Organization
Some developmental regulator genes with a network of multiple elements to control their intricate spatiotemporal expression pattern have a relatively simple genomic organization: they reside in regions that have been termed “gene deserts” (Nobrega et al. 2003). The regulatory domains of DACH (Nobrega et al. 2003; Boffelli et al. 2004) and SOX9 (Pfeifer et al. 1999) apparently cover 1 Mb or more of genomic sequence, with no other recognizable genes in the vicinity. For many genes, the situation is much more complex. A strange consequence of the complex genomic movements and rearrangements associated with the ongoing evolution of regulatory functions is that cis elements for one gene are quite often embedded within another nearby gene, generally within its introns. The neighboring genes usually fulfill very different functions—for example, highly tissue-specific genes may have regulatory elements embedded in ubiquitously or differently expressed genes (Bennani-Baiti et al. 1998; Kleinjan et al. 2002; Lettice et al. 2002; Zuniga et al. 2004). This type of functional link between otherwise apparently unrelated genes can provide a mechanism underlying conservation of synteny across classes (Kleinjan et al. 2002; Santagati et al. 2003).
Mechanisms of Long-Range Regulation
Structural Versus Functional Domains: Hypotheses
In the context of such complexity and diversity of genomic organization, it is difficult to develop a unified model for regulatory function. To explain how genes maintain their independence, the concept of “structural gene domains” has been widely discussed for many years. However, this model is largely based on the analysis of a small number of intensely studied gene loci. In the structural domain model, genes enjoy functional autonomy through physical separation from neighboring domains by specific sequences (Dillon and Sabbattini 2000). Adjacent structural domains would be distinguished by differences in their local chromatin structure, with transcriptionally active domains displaying an “open” chromatin structure, whereas neighboring inactive domains would have “closed” compacted chromatin. In this model, an important role is played by the boundary, or insulator, elements—sequences that function specifically to prevent the spread of active or inactive chromatin from one domain into the next. Strong support for the concept of structural domains was provided by the intensely studied relationship between chromatin structure and gene activation at loci such as the chicken β-globin locus (Prioleau et al. 1999). These studies have, indeed, led to the identification of such insulator elements at the borders of domains. Insulator elements are defined as sequences with the capacity to block enhancer-promoter interaction when placed between the two element types in certain assays; they can also insulate a flanking reporter cassette from repressive chromatin at the site of integration in transgenic assays (Bell et al. 2001; Burgess-Beusse et al. 2002). However, the finding that independently regulated loci can partially or completely overlap and that their cis-regulatory elements can be found within or beyond neighboring unrelated genes puts the universal applicability of the structural domain model into question.
Direct evidence for the structural domain model is already somewhat less convincing in the human β-globin locus, and it is clear that it does not apply to the α-globin locus. The α- and β-globin domains probably arose by duplication of a single ancestral gene. In contrast to the β-globin gene cluster, the α-globin gene is located in a large region of “open” chromatin that contains a number of housekeeping genes (Flint et al. 2001). In common with some of the disease genes described above, the main critical regulatory element of the α-globin locus, the −40-kb enhancer, is located within an intron of a neighboring, ubiquitously expressed housekeeping gene (Vyas et al. 1995). In the case of PAX6, a number of regulatory elements are spread throughout at least three introns of the neighboring ELP4 gene (Kleinjan et al. 2001). In the human growth hormone cluster, elements required for tissue-specific expression of the pituitary-specific GHN gene and the placenta-specific CSL, CSA, GHV, and CSB genes are located beyond a B-cell–specific gene (CD79B), within introns of the muscle-specific SCN4A gene (Bennani-Baiti et al. 1998). In this case, this genomic organization has led to the fortuitous activation of CD79B in the pituitary, where it has no known function, as a result of a bystander effect (Cajiao et al. 2004). This challenges the common assumption that specific high-level expression in a particular tissue implies a function for the gene in that tissue. Additional examples are provided by other cases described above. These observations provide compelling arguments against a fundamental requirement for the physical isolation of a gene and its regulatory sequences. Rather it has been proposed that specificity of enhancer-promoter interactions (fig. 3) is the key to maintaining functional autonomy of adjacent genes (Dillon and Sabbattini 2000). Viewed in the light of evolution, transcriptional control elements and chromatin structure at a locus will have coevolved under positive and negative selection to form the gene domains we observe today. In instances such as the β-globin locus, this will have led to domains that are structurally isolated from their surroundings, whereas, in many other cases, including α-globin, a large, integrated gene region without distinct boundaries has emerged. Although the concept of a gene as a physical entity, with a distinct map position on the chromosome, is still useful in many instances, a new model has been proposed that is based on the old definition of the gene as a “unit of inheritance” (Dillon 2003). Here, genes are defined in functional terms as “functional expression modules” that encompass both the transcribed regions and their cis-regulatory control systems and that function appropriately in different cells types (fig. 3) within the context of the local chromatin architecture.
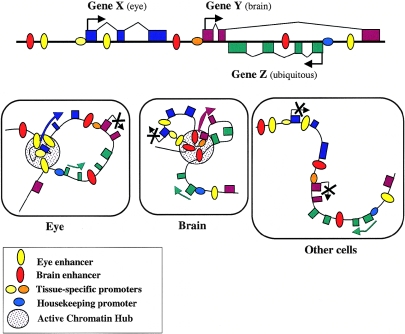
Figure 3.
Model for the coexistence of physically overlapping but independently regulated “functional gene expression modules” in the same genomic region. A hypothetical region containing two tissue-specific genes and one housekeeping gene. Gene X (blue exons) is expressed in eye tissue, gene Y (purple exons) is expressed in brain, and gene Z (green exons) is ubiquitously expressed. Transcriptional activity depends on the formation of an ACH that encompasses tissue-specific cis-acting elements with bound transcription factor complexes and selective interaction with the relevant gene promoter. Formation of an ACH provides a high local concentration of transcription factors and positive chromatin-modifying enzymes. The housekeeping promoter is active in all cells and does not rely on tissue-specific ACH formation.
Setting Up Models
Participation of a promoter is essential for transcription. Spatiotemporal and quantitative regulation of gene expression is brought about through the influence of cis-regulatory elements, such as enhancers and repressors. Since the first demonstration of the existence of distant enhancers, one of the main questions has been how these elements communicate regulatory activity to their target promoters over such large spans of intervening DNA. This question has been studied most extensively at the β-globin locus. Spurred by the original observations of γβ-thalassemias in patients with deletions of a distant upstream regulatory region (the locus control region [LCR]), but with intact promoters and globin transcription units (fig. 2H) (Kioussis et al. 1983; Driscoll et al. 1989), the globin locus been used extensively to explore how the LCR may work to drive high-level expression of the globin genes in erythroid cells (Fraser and Grosveld 1998). The model that is most commonly encountered in the context of enhancer-promoter interactions is the “looping” model; in this model, transcription factors bound at the enhancer make direct contact with the promoter and/or with factors bound at the promoter, while the intervening DNA loops out (Bulger and Groudine 1999). In the favored “random collision” model, both the enhancer and the promoter initially move around at random until they encounter each other, whereupon contact is established, which leads to activation of transcription. Other models differ from this mainly in the way the enhancer-bound factors establish contact with the promoter. In the “tracking” model, the initially formed enhancer-bound complex actively scans along the DNA in search of a promoter. In the “linking” model, a chain of proteins/complexes extends from the enhancer along the chromatin fiber to the promoter, where it subsequently mediates gene activation. Combinations of the tracking and looping models have also been proposed, in which the complex tracking along the DNA remains attached to the enhancer, dragging it along to create a loop. In another model, the distant enhancer provides a nucleation site from which a signal, distinct from the enhancer-bound factors, is transmitted along the chromatin fiber to the promoter. The existence of RNA transcripts that originate from active enhancers at some loci is intriguing and suggests a possible role for transcription itself in chromatin modification of actively transcribed regions (Gribnau et al. 2000; Kim and Dean 2004).
Visualizing Enhancer-Promoter Interaction
Two novel techniques provide strong support for a mechanism of long-range interaction that involves close contact between the enhancer and the promoter, as in the looping model. Chromosome conformation capture (3C) takes native chromatin from appropriate gene-expressing tissue and uses formaldehyde to crosslink adjacent proteins and DNA (Tolhuis et al. 2002). The DNA fiber is then chopped up by restriction-enzyme digestion, followed by religation under dilute conditions, such that crosslinked restriction fragments will become preferentially ligated to each other (intramolecular reaction), compared with ligation of random fragments. PCRs with one fixed primer (e.g., near the promoter) and a selection of second primers from potential enhancers and from intervening fragments throughout the locus are used to detect specific chromatin interactions. The relative abundance of PCR products obtained from distinct primer combinations is considered proportional to the interaction frequency between the chromatin segments they represent. With the use of this information, models for cis interactions can be developed. In a second assay—RNA-TRAP (tagging and recovery of associated proteins)—a clever combination of in situ hybridization and chromatin immunoprecipitation is used, again in expressing cells. An intron-specific probe for the gene of interest, which has been labeled with digoxygenin, is hybridized to specific, nascent pre-mRNA transcripts in formaldehyde-fixed nuclei. Next, antidigoxygenin antibodies conjugated to horseradish peroxidase are introduced to target peroxidase activity to the site of transcription. This activity produces free radicals that catalyze the covalent binding of the biotin tag to nearby proteins. Chromatin fragments tagged with biotin are then purified on streptavidin-agarose columns, and the relative enrichment of a specific DNA fragment (detected by PCR) indicates that the tagged sequence was in the vicinity of the nascent mRNA and, by extrapolation, of the promoter (Carter et al. 2002).
3-D Clustering of Active Cis Elements
Both techniques were applied to the endogenous mouse β-globin locus, which is organizationally very similar to the human locus shown in figure 2H. The murine cluster, which spans ∼130 kb, consists of the LCR with six DNase I HSs (5′→3′: HS6–HS1) and the four structural globin loci, each with its own promoter (ɛγ and βh1 expressed at embryonic stages and the adult βmajor and βminor genes). There is an additional pair of HSs at around −60 kb upstream of ɛγ and another site, 3′ HS1, downstream of βminor. The whole cluster is embedded in a large cluster of olfactory receptor (OR) genes (Bulger et al. 1999).
Detailed analysis of the RNA-TRAP and 3C data has not only shown a close interaction between the LCR and the globin gene promoters, but the data also suggest that, in β-globin-expressing cells, the whole cluster adopts a distinct conformation, with all HSs (including the active βmajor and βminor promoters) being clustered together. The inactive ɛγ and βh1 globin genes, as well as the silent OR genes, loop out and do not participate in this clustering. In nonexpressing brain cells, no specific close interactions are observed: the locus seems to remain in a linear conformation (Tolhuis et al. 2002). To describe the spatial clustering of regulatory elements and active promoters, the term “active chromatin hub” (ACH) was coined. The successful setting up of an ACH was proposed to be the key event required for the establishment of a “functional expression module” (de Laat and Grosveld 2003). The result of ACH formation is a high-density clustering of regulatory elements, their cognate-binding factors, associated coactivators, and chromatin modifiers, which sets up a suitable local environment to generate precisely the required expression level. Depending on the nature of its sequence and the bound recognition factors, promoters and other regulatory elements may or may not compete for interaction with the ACH. So far, the use of the 3C and RNA-TRAP techniques has focused on the β-globin locus, and much work remains to confirm and validate the ACH model at other loci. However, the model provides a compelling explanation for how overlapping genes can be independently regulated and expressed. It is well known that some enhancers display strong specificity in their preference for certain promoter types over others. Conversely, this could be interpreted as a differential affinity of the promoters of adjacent genes for a specific ACH. Elegant experiments in Drosophila, for instance, have shown that the AE1 enhancer from the Drosophila Antennapedia gene complex (ANT-C) and the IAB5 enhancer from the Bithorax complex (BX-C) preferentially activate TATA-containing promoters when located equidistant from a TATA-containing and a TATA-less promoter (Ohtsuki et al. 1998). Promoter affinity may be specifically enhanced by the presence of a so-called promoter-proximal tethering element. Such a novel type of element has been implicated in the regulation of the sex combs reducedgene (Scr) in the Drosophila ANT-C. A second enhancer in this complex, the T1 enhancer, bypasses its neighboring fushi tarazu gene (ftz) and interacts with the distant Scr promoter to activate expression in posterior head segments. The tethering element is a 450-bp promoter-proximal sequence that is essential for T1-Scr interactions (Calhoun et al. 2002). Apart from promoter selectivity (see also Hochheimer and Tjian 2003), distance between enhancer and promoter on the linear DNA fiber must play a role, since regulatory elements need to be in cis to the gene they activate (barring some poorly understood phenomena such as transvection). The linear distance between enhancer and promoter is very large in some cases (e.g., SHH and the limb enhancer ZRS); nevertheless, the distance is presumably not unlimited. On the other hand, in some multigene loci where competition for interaction with a shared enhancer plays a role, as in the globin gene and Hox clusters, distance is a distinct factor, with proximal genes more likely to be activated than more distant ones (Dillon et al. 1997).
Facilitators of Long-Range Interactions
Although the 3C (and, to some extent, the RNA-TRAP) assays show the clustering of cis-regulatory elements and promoters in nuclear space in an ACH, these techniques do not reveal the mechanism by which an ACH is set up in the first place. Whether the loops are formed through “random collision,” “looping and tracking,” or some other mechanism remains an open question. Theoretical calculations suggest that a process that depends on random collision alone would be highly inefficient when distances of more than a few kilobases are involved (Rippe 2001). A class of proteins implicated in promoting contact between promoters and distal enhancers is that of “facilitator proteins,” exemplified by the Nipped-B and Chip proteins originally identified in Drosophila (Dorsett 1999). CHIP, a LIM-domain–binding protein, was shown to facilitate the activation of multiple genes by interacting with diverse DNA binding proteins—for example, by promoting the cooperative binding of homeobox factors to DNA. Nipped-B, on the basis of its homology with yeast sister chromatid cohesion protein SCC2, was suggested to play a dual role. The more ancient function of facilitating transinteractions between sequences on sister chromatids has been adapted to include an additional role in long-range transcriptional regulation (Rollins et al. 2004). Recently, the human homologue of Nipped-B, NIPBL, was identified as the gene mutated in individuals with Cornelia de Lange syndrome (CDLS [MIM 122470]) (Krantz et al. 2004; Tonkin et al. 2004). CDLS is a multiple malformation disorder that is characterized by facial dysmorphology, mental retardation, growth delay, and limb reduction. In keeping with the putative general role of NIPBL in facilitating long-range regulation of multiple genes, dysregulation of several developmental control genes may underlie the diverse phenotypic anomalies seen in CDLS.
Regulatory Mutations and Variation in Human Disease
This review aimed to emphasize that, although disease-associated genetic changes are commonly thought of as affecting the coding regions of genes, some may exert their effect through abnormal gene expression that results from mutations that disturb the normal processes of control. Transcriptional regulation is exerted through interactions between cis-regulatory elements associated with a gene and through the chromatin structure of the gene locus. Disruption of either of these may lead to disease. The PAX6 and SHH position-effect cases are good examples of the disruption of cis-regulatory elements; FSHD cases and the α-thalassemia case described by Tufarelli et al. (2003) may be caused by aberrant long- or short-range chromatin structure. Chromatin changes are certainly known to be associated with diseases of imprinting (Walter and Paulsen 2003) and also, in many cases, with malignancy (Cho et al. 2004; Egger et al. 2004). Cytological or molecular detection of these changes is a component of molecular diagnostic technology, particularly in cancer. Advances in this type of technology may lead to improved diagnostic capacity in other disease-associated regulatory changes. The exciting prospect of modulating gene expression through the therapeutic alteration of epigenetic state is actively being explored (Egger et al. 2004). Other diseases that exert their effect through altered regulation of chromatin conformation are those associated with mutations in genes with a role in chromatin modification (see the recent review by Bickmore and van der Maarel [2003]).
The most obvious cases of transcriptional misregulation as the cause of genetic disease are associated with visible chromosomal rearrangements. However, as illustrated by many cases cited above, interpretation of what gene is implicated can be difficult, since the disruption may lie entirely in one gene while affecting expression of a nearby gene. Furthermore, the phenotypes observed may be very different from those that may be elicited by coding-region changes in the causative gene. An even more daunting task is the identification of small deletions or mutations in cis elements, as exemplified by the case of PPD and sonic hedgehog in which a single-nucleotide substitution located 1 Mb from the causative gene produces a severe genetic defect (Lettice et al. 2003). An emerging area of research aims to address this task through the implementation of mutation-screening technology by genomic array hybridization, which may identify cryptic disease-associated deletions (Shaw-Smith et al. 2004). To facilitate the detection of alterations that specifically involve cis-regulatory regions, arrays carrying the potentially relevant genomic elements can be produced after their identification in multispecies sequence comparisons (Dubchak and Frazer 2003).
By far the largest category of as-yet poorly explored regulatory variation will involve subtle sequence changes in regulatory elements, which may alter the expression pattern or level of the cognate gene, sometimes without immediate or clearly recognizable effect. There is already good evidence from several studies that, in cases in which alleles can be distinguished, there is high-frequency heritable variation in the level of expression from different alleles, as a result of polymorphisms at cis-regulatory sites. Such variation may represent a significant proportion of disease-associated QTLs. Data supporting this notion have been gathered in mouse strains (Cowles et al. 2002), as well as in humans (Yan et al. 2002; Knight 2004; Pastinen et al. 2004). Elegant evidence for evolutionary change that arises through this pathway comes from Drosophila studies (Wittkopp et al. 2004). Examples of regulatory QTLs associated with human variations are beginning to be elucidated. One of the first to be identified was the ancient polymorphism for lactase persistence (MIM 223100) (Enattah et al. 2002), which allows a high proportion of adults in European populations to digest milk. Two single-nucleotide variants in cis-regulatory elements, located 13.9 kb and 22 kb upstream of the lactase gene promoter, are strongly correlated with continued high-level expression of lactase (Olds and Sibley 2003).
In this postgenome era, we are just beginning to understand how long-range organization of the genome is related to function. The detailed analysis of human genetic malformations has proved to be invaluable—first, in highlighting the occurrence and scope of long-distance transcriptional regulation in the genome, and second, in the elucidation of some of the plethora of different mechanisms for long-range gene control. It seems likely that complex transcriptional control is essential for a substantial subset of all genes. The genes and diseases discussed here form a small sample of that group. They have shown us, however, that—despite the ever-increasing technical advances in high-throughput techniques—the laborious identification of the disease loci and regulatory mechanisms involved in currently “unsolved” human disorders remains a huge but rewarding task.
Electronic-Database Information
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for aniridia, deafness type 3, Rieger syndrome type 1, Greig cephalopolysyndactyly syndrome, MAF, LD, blepharophimosis-ptosis-epicanthus inversus syndrome, CD, HPE2, HPE3, PPD, SHFM1, FSHD, α-thalassemia, CDLS, and lactase expression)
References
- Alward WL, Semina EV, Kalenak JW, Heon E, Sheth BP, Stone EM, Murray JC (1998) Autosomal dominant iris hypoplasia is caused by a mutation in the Rieger syndrome (RIEG/PITX2) gene. Am J Ophthalmol 125:98–100 10.1016/S0002-9394(99)80242-6 [DOI] [PubMed] [Google Scholar]
- Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D (2001) Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics 78:73–82 10.1006/geno.2001.6648 [DOI] [PubMed] [Google Scholar]
- Bakker E, Wijmenga C, Vossen RH, Padberg GW, Hewitt J, van der WM, Rasmussen K, Frants RR (1995) The FSHD-linked locus D4F104S1 (p13E-11) on 4q35 has a homologue on 10qter. Muscle Nerve 2:S39–S44 [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G (2001) Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291:447–450 10.1126/science.291.5503.447 [DOI] [PubMed] [Google Scholar]
- Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW (1996) Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet 14:353–356 10.1038/ng1196-353 [DOI] [PubMed] [Google Scholar]
- Bennani-Baiti IM, Cooke NE, Liebhaber SA (1998) Physical linkage of the human growth hormone gene cluster and the CD79B (Igβ/B29) gene. Genomics 48:258–264 10.1006/geno.1997.5171 [DOI] [PubMed] [Google Scholar]
- Bickmore WA, van der Maarel SM (2003) Perturbations of chromatin structure in human genetic disease: recent advances. Hum Mol Genet 12:R207–R213 10.1093/hmg/ddg260 [DOI] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA (2000) A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet 26:490–494 10.1038/82652 [DOI] [PubMed] [Google Scholar]
- Boffelli D, Nobrega MA, Rubin EM (2004) Comparative genomics at the vertebrate extremes. Nat Rev Genet 5:456–465 10.1038/nrg1350 [DOI] [PubMed] [Google Scholar]
- Brice G, Mansour S, Bell R, Collin JR, Child AH, Brady AF, Sarfarazi M, Burnand KG, Jeffery S, Mortimer P, Murday VA (2002) Analysis of the phenotypic abnormalities in lymphoedema-distichiasis syndrome in 74 patients with FOXC2 mutations or linkage to 16q24. J Med Genet 39:478–483 10.1136/jmg.39.7.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M (1999) Looping versus linking: toward a model for long-distance gene activation. Genes Dev 13:2465–2477 10.1101/gad.13.19.2465 [DOI] [PubMed] [Google Scholar]
- Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, Felsenfeld G (1999) Conservation of sequence and structure flanking the mouse and human β-globin loci: the β-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci USA 96:5129–5134 10.1073/pnas.96.9.5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess-Beusse B, Farrell C, Gaszner M, Litt M, Mutskov V, Recillas-Targa F, Simpson M, West A, Felsenfeld G (2002) The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci USA Suppl 99:16433–16437 10.1073/pnas.162342499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Goodman BK, Patel AS, Mulliken JB, Van Maldergem L, Hoganson GE, Paznekas WA, Ben Neriah Z, Sheffer R, Cunningham ML, Daentl DL, Jabs EW (2003) Increased risk for developmental delay in Saethre-Chotzen syndrome is associated with TWIST deletions: an improved strategy for TWIST mutation screening. Hum Genet 114:68–76 10.1007/s00439-003-1012-7 [DOI] [PubMed] [Google Scholar]
- Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA (2004) Bystander gene activation by a locus control region. EMBO J 23:3854–3863 10.1038/sj.emboj.7600365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VC, Stathopoulos A, Levine M (2002) Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex. Proc Natl Acad Sci USA 99:9243–9247 10.1073/pnas.142291299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD (2001) From DNA to diversity: molecular genetics and the evolution of animal design. Blackwell, Malden, MA [Google Scholar]
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P (2002) Long-range chromatin regulatory interactions in vivo. Nat Genet 32:623–626 10.1038/ng1051 [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383:407–413 10.1038/383407a0 [DOI] [PubMed] [Google Scholar]
- Cho KS, Elizondo LI, Boerkoel CF (2004) Advances in chromatin remodeling and human disease. Curr Opin Genet Dev 14:308–315 10.1016/j.gde.2004.04.015 [DOI] [PubMed] [Google Scholar]
- Chun K, Teebi AS, Jung JH, Kennedy S, Laframboise R, Meschino WS, Nakabayashi K, Scherer SW, Ray PN, Teshima I (2002) Genetic analysis of patients with the Saethre-Chotzen phenotype. Am J Med Genet 110:136–143 10.1002/ajmg.10400 [DOI] [PubMed] [Google Scholar]
- Clark RM, Marker PC, Kingsley DM (2000) A novel candidate gene for mouse and human preaxial polydactyly with altered expression in limbs of Hemimelic extra-toes mutant mice. Genomics 67:19–27 10.1006/geno.2000.6225 [DOI] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M (1996) Polar overdominance at the ovine callipyge locus. Science 273:236–238 [DOI] [PubMed] [Google Scholar]
- Cowles CR, Hirschhorn JN, Altshuler D, Lander ES (2002) Detection of regulatory variation in mouse genes. Nat Genet 32:432–437 10.1038/ng992 [DOI] [PubMed] [Google Scholar]
- Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, Cobben JM, Hudgins L, Evans JP, Tsui LC (1996) Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet 5:571–579 10.1093/hmg/5.5.571 [DOI] [PubMed] [Google Scholar]
- Cremers FP, Cremers CW (2004) POU3F4 and mixed deafness with temporal bone defects (DFN3). In: Epstein CJ, Erickson RP, Wynshaw-Boris A (eds) Inborn errors of development: the molecular basis of clinical disorders of morphogenesis. Oxford University Press, New York [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27:159–166 10.1038/84781 [DOI] [PubMed] [Google Scholar]
- Crisponi L, Uda M, Deiana M, Loi A, Nagaraja R, Chiappe F, Schlessinger D, Cao A, Pilia G (2004) FOXL2 inactivation by a translocation 171 kb away: analysis of 500 kb of chromosome 3 for candidate long-range regulatory sequences. Genomics 83:757–764 10.1016/j.ygeno.2003.11.010 [DOI] [PubMed] [Google Scholar]
- Crolla JA, van Heyningen V (2002) Frequent chromosome aberrations revealed by molecular cytogenetic studies in patients with aniridia. Am J Hum Genet 71:1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AF, Mirza G, Flinter F, Ragoussis J (1999) An interstitial deletion of 6p24-p25 proximal to the FKHL7 locus and including AP-2α that affects anterior eye chamber development. J Med Genet 36:708–710 [PMC free article] [PubMed] [Google Scholar]
- De Baere E, Beysen D, Oley C, Lorenz B, Cocquet J, De Sutter P, Devriendt K, Dixon M, Fellous M, Fryns JP, Garza A, Jonsrud C, Koivisto PA, Krause A, Leroy BP, Meire F, Plomp A, Van Maldergem L, De Paepe A, Veitia R, Messiaen L (2003) FOXL2 and BPES: mutational hotspots, phenotypic variability, and revision of the genotype-phenotype correlation. Am J Hum Genet 72:478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Baere E, Dixon MJ, Small KW, Jabs EW, Leroy BP, Devriendt K, et al (2001) Spectrum of FOXL2 gene mutations in blepharophimosis-ptosis-epicanthus inversus (BPES) families demonstrates a genotype—phenotype correlation. Hum Mol Genet 10:1591–1600 10.1093/hmg/10.15.1591 [DOI] [PubMed] [Google Scholar]
- de Kok Y, Merkx GF, van der Maarel SM, Huber I, Malcolm S, Ropers HH, Cremers FP (1995) A duplication/paracentric inversion associated with familial X-linked deafness (DFN3) suggests the presence of a regulatory element more than 400 kb upstream of the POU3F4 gene. Hum Mol Genet 4:2145–2150 [DOI] [PubMed] [Google Scholar]
- de Kok Y, Vossenaar ER, Cremers CW, Dahl N, Laporte J, Hu LJ, Lacombe D, Fischel-Ghodsian N, Friedman RA, Parnes LS, Thorpe P, Bitner-Glindzicz M, Pander HJ, Heilbronner H, Graveline J, den Dunnen JT, Brunner HG, Ropers HH, Cremers FP (1996) Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum Mol Genet 5:1229–1235 10.1093/hmg/5.9.1229 [DOI] [PubMed] [Google Scholar]
- de Laat W, Grosveld F (2003) Spatial organization of gene expression: the active chromatin hub. Chromosome Res 11:447–459 10.1023/A:1024922626726 [DOI] [PubMed] [Google Scholar]
- Di Leone RJ, Marcus GA, Johnson MD, Kingsley DM (2000) Efficient studies of long-distance Bmp5 gene regulation using bacterial artificial chromosomes. Proc Natl Acad Sci USA 97:1612–1617 10.1073/pnas.97.4.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N (2003) Gene autonomy: positions, please. Nature 425:457 10.1038/425457a [DOI] [PubMed] [Google Scholar]
- Dillon N, Sabbattini P (2000) Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays 22:657–665 [DOI] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F (1997) The effect of distance on long-range chromatin interactions. Mol Cell 1:131–139 10.1016/S1097-2765(00)80014-3 [DOI] [PubMed] [Google Scholar]
- Dorsett D (1999) Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev 9:505–514 10.1016/S0959-437X(99)00002-7 [DOI] [PubMed] [Google Scholar]
- Driscoll MC, Dobkin CS, Alter BP (1989) γδβ-thalassemia due to a de novo mutation deleting the 5′ β-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci USA 86:7470–7474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker R, Whitelaw E (2004) Retrotransposon-derived elements in the mammalian genome: a potential source of disease. J Inherit Metab Dis 27:319–330 10.1023/B:BOLI.0000031096.81518.66 [DOI] [PubMed] [Google Scholar]
- Dubchak I, Frazer K (2003) Multi-species sequence comparison: the next frontier in genome annotation. Genome Biol 4:122 10.1186/gb-2003-4-12-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Wilkins AS (1998) The evolution of “bricolage.” Trends Genet 14:54–59 [DOI] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463 10.1038/nature02625 [DOI] [PubMed] [Google Scholar]
- Elgin SC (1988) The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem 263:19259–19262 [PubMed] [Google Scholar]
- Elgin SC, Grewal SI (2003) Heterochromatin: silence is golden. Curr Biol 13:R895–R898 10.1016/j.cub.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I (2002) Identification of a variant associated with adult-type hypolactasia. Nat Genet 30:233–237 10.1038/ng826 [DOI] [PubMed] [Google Scholar]
- Fang J, Dagenais SL, Erickson RP, Arlt MF, Glynn MW, Gorski JL, Seaver LH, Glover TW (2000) Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet 67:1382–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Redeker B, Breen M, Boyle S, Brown J, Fletcher J, Jones S, Bickmore W, Fukushima Y, Mannens M, Danes S, van Heyningen V, Hanson I (1995) Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype. Hum Mol Genet 4:415–422 [DOI] [PubMed] [Google Scholar]
- Festenstein R, Sharghi-Namini S, Fox M, Roderick K, Tolaini M, Norton T, Saveliev A, Kioussis D, Singh P (1999) Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat Genet 23:457–461 10.1038/70579 [DOI] [PubMed] [Google Scholar]
- Finegold DN, Kimak MA, Lawrence EC, Levinson KL, Cherniske EM, Pober BR, Dunlap JW, Ferrell RE (2001) Truncating mutations in FOXC2 cause multiple lymphedema syndromes. Hum Mol Genet 10:1185–1189 10.1093/hmg/10.11.1185 [DOI] [PubMed] [Google Scholar]
- Flint J, Tufarelli C, Peden J, Clark K, Daniels RJ, Hardison R, Miller W, Philipsen S, Tan-Un KC, McMorrow T, Frampton J, Alter BP, Frischauf AM, Higgs DR (2001) Comparative genome analysis delimits a chromosomal domain and identifies key regulatory elements in the α globin cluster. Hum Mol Genet 10:371–382 10.1093/hmg/10.4.371 [DOI] [PubMed] [Google Scholar]
- Flomen RH, Vatcheva R, Gorman PA, Baptista PR, Groet J, Barisic I, Ligutic I, Nizetic D (1998) Construction and analysis of a sequence-ready map in 4q25: Rieger syndrome can be caused by haploinsufficiency of RIEG, but also by chromosome breaks approximately 90 kb upstream of this gene. Genomics 47:409–413 10.1006/geno.1997.5127 [DOI] [PubMed] [Google Scholar]
- Foster JW, Dominguez-Steglich MA, Guioli S, Kowk G, Weller PA, Stevanovic M, Weissenbach J, Mansour S, Young ID, Goodfellow PN (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525–530 [DOI] [PubMed] [Google Scholar]
- Fraser P, Grosveld F (1998) Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol 10:361–365 10.1016/S0955-0674(98)80012-4 [DOI] [PubMed] [Google Scholar]
- Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, Jirtle RL, Smith TP (2002) Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res 12:1496–1506 10.1101/gr.571002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110:339–348 10.1016/S0092-8674(02)00826-7 [DOI] [PubMed] [Google Scholar]
- Gaffney DJ, Keightley PD (2004) Unexpected conserved non-coding DNA blocks in mammals. Trends Genet 20:332–337 10.1016/j.tig.2004.06.011 [DOI] [PubMed] [Google Scholar]
- Georges M, Charlier C, Cockett N (2003) The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet 19:248–252 10.1016/S0168-9525(03)00082-9 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Rice JC (2004) Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol 16:230–238 10.1016/j.ceb.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell 5:377–386 10.1016/S1097-2765(00)80432-3 [DOI] [PubMed] [Google Scholar]
- Griffin C, Kleinjan DA, Doe B, van Heyningen V (2002) New 3′ elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech Dev 112:89–100 10.1016/S0925-4773(01)00646-3 [DOI] [PubMed] [Google Scholar]
- Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Rigby PW, Buckingham M (2003) Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development 130:3415–3426 10.1242/dev.00552 [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD (2004) Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429:268–274 10.1038/nature02536 [DOI] [PubMed] [Google Scholar]
- Hardison RC (2000) Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet 16:369–372 10.1016/S0168-9525(00)02081-3 [DOI] [PubMed] [Google Scholar]
- Herault Y, Fraudeau N, Zakany J, Duboule D (1997) Ulnaless (Ul), a regulatory mutation inducing both loss-of-function and gain-of-function of posterior Hoxd genes. Development 124:3493–3500 [DOI] [PubMed] [Google Scholar]
- Heydemann A, Nguyen LC, Crenshaw EB III (2001) Regulatory regions from the Brn4 promoter direct LACZ expression to the developing forebrain and neural tube. Brain Res Dev Brain Res 128:83–90 10.1016/S0165-3806(01)00137-7 [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BM, Ton CCT, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V (1991) Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354:522–525 10.1038/354522a0 [DOI] [PubMed] [Google Scholar]
- Hill RE, Heaney SJ, Lettice LA (2003) Sonic hedgehog: restricted expression and limb dysmorphologies. J Anat 202:13–20 10.1046/j.1469-7580.2003.00148.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheimer A, Tjian R (2003) Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev 17:1309–1320 10.1101/gad.1099903 [DOI] [PubMed] [Google Scholar]
- Howard ML, Davidson EH (2004) cis-Regulatory control circuits in development. Dev Biol 271:109–118 10.1016/j.ydbio.2004.03.031 [DOI] [PubMed] [Google Scholar]
- Hurles M (2004) Gene duplication: the genomic trade in spare parts. PLoS Biol 2:E206 10.1371/journal.pbio.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianakiev P, van Baren MJ, Daly MJ, Toledo SP, Cavalcanti MG, Neto JC, Silveira EL, Freire-Maia A, Heutink P, Kilpatrick MW, Tsipouras P (2001) Acheiropodia is caused by a genomic deletion in C7orf2, the human orthologue of the Lmbr1 gene. Am J Hum Genet 68:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius J, Knuutila S, Scherer SW, Trask B, Kere J (1996) Split hand/split foot malformation, deafness, and mental retardation with a complex cytogenetic rearrangement involving 7q21.3. J Med Genet 33:507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson RV, Perveen R, Kerr B, Carette M, Yardley J, Heon E, Wirth MG, van Heyningen V, Donnai D, Munier F, Black GC (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11:33–42 10.1093/hmg/11.1.33 [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang F, Van Overveld PG, Vedanarayanan V, van der MS, Ehrlich M (2003) Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum Mol Genet 12:2909–2921 10.1093/hmg/ddg323 [DOI] [PubMed] [Google Scholar]
- Kim A, Dean A (2004) Developmental stage differences in chromatin subdomains of the β-globin locus. Proc Natl Acad Sci USA 101:7028–7033 10.1073/pnas.0307985101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I (2004) Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development 131:57–71 10.1242/dev.00877 [DOI] [PubMed] [Google Scholar]
- Kioussis D, Vanin E, deLange T, Flavell RA, Grosveld FG (1983) β-globin gene inactivation by DNA translocation in γβ-thalassaemia. Nature 306:662–666 [DOI] [PubMed] [Google Scholar]
- Kist R, Schrewe H, Balling R, Scherer G (2002) Conditional inactivation of Sox9: a mouse model for campomelic dysplasia. Genesis 32:121–123 10.1002/gene.10050 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Childs AJ, van Heyningen V (2004) Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev Biol 265:462–477 10.1016/j.ydbio.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Elgar G, van Heyningen V (2002) Characterization of a novel gene adjacent to PAX6, revealing synteny conservation with functional significance. Mamm Genome 13:102–107 10.1007/s00335-001-3058-y [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, Seawright A, Schedl A, Quinlan RA, Danes S, van Heyningen V (2001) Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum Mol Genet 10:2049–2059 10.1093/hmg/10.19.2049 [DOI] [PubMed] [Google Scholar]
- Kleinjan DJ, van Heyningen V (1998) Position effect in human genetic disease. Hum Mol Genet 7:1611–1618 10.1093/hmg/7.10.1611 [DOI] [PubMed] [Google Scholar]
- Knight JC (2003) Functional implications of genetic variation in non-coding DNA for disease susceptibility and gene regulation. Clin Sci 104:493–501 10.1042/CS20020304 [DOI] [PubMed] [Google Scholar]
- Knight JC (2004) Allele-specific gene expression uncovered. Trends Genet 20:113–116 10.1016/j.tig.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635 10.1038/ng1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale JD, Wilensky JS, Oliver ER, Walton DS, Glaser T (2000) 3′ deletions cause aniridia by preventing PAX6 gene expression. Proc Natl Acad Sci USA 97:13755–13759 10.1073/pnas.240398797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS (2003) Fox’s in development and disease. Trends Genet 19:339–344 10.1016/S0168-9525(03)00111-2 [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735 10.1093/hmg/ddg180 [DOI] [PubMed] [Google Scholar]
- Lettice LA, Horikoshi T, Heaney SJ, van Baren MJ, van der Linde HC, Breedveld GJ, Joosse M, Akarsu N, Oostra BA, Endo N, Shibata M, Suzuki M, Takahashi E, Shinka T, Nakahori Y, Ayusawa D, Nakabayashi K, Scherer SW, Heutink P, Hill RE, Noji S (2002) Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc Natl Acad Sci USA 99:7548–7553 10.1073/pnas.112212199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R (2003) Transcription regulation and animal diversity. Nature 424:147–151 10.1038/nature01763 [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA (2000) Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136–140 10.1126/science.288.5463.136 [DOI] [PubMed] [Google Scholar]
- Lyon MF, Jamieson RV, Perveen R, Glenister PH, Griffiths R, Boyd Y, Glimcher LH, Favor J, Munier FL, Black GC (2003) A dominant mutation within the DNA-binding domain of the bZIP transcription factor Maf causes murine cataract and results in selective alteration in DNA binding. Hum Mol Genet 12:585–594 10.1093/hmg/12.6.585 [DOI] [PubMed] [Google Scholar]
- Maas RL, Zeller R, Woychik RP, Vogt TF, Leder P (1990) Disruption of formin-encoding transcripts in two mutant limb deformity alleles. Nature 346:853–855 10.1038/346853a0 [DOI] [PubMed] [Google Scholar]
- Masny PS, Bengtsson U, Chung SA, Martin JH, Van Engelen B, van der Maarel SM, Winokur ST (2004) Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet 13:1857–1871 10.1093/hmg/ddh205 [DOI] [PubMed] [Google Scholar]
- McElreavy K, Vilain E, Abbas N, Costa JM, Souleyreau N, Kucheria K, Boucekkine C, Thibaud E, Brauner R, Flamant F, Fellous M (1992) XY sex reversal associated with a deletion 5′ to the SRY “HMG box” in the testis-determining region. Proc Natl Acad Sci USA 89:11016–11020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo GR, Paleari L, Mantero S, Genova F, Beverdam A, Palmisano GL, Barbieri O, Levi G (2002) Mouse model of split hand/foot malformation type I. Genesis 33:97–101 10.1002/gene.10098 [DOI] [PubMed] [Google Scholar]
- Michos O, Panman L, Vintersten K, Beier K, Zeller R, Zuniga A (2004) Gremlin-mediated BMP antagonism induces the epithelial-mesenchymal feedback signaling controlling metanephric kidney and limb organogenesis. Development 131:3401–3410 [DOI] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de BE, Langeveld A, Tan-Un K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P (1996) Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell 87:105–114 [DOI] [PubMed] [Google Scholar]
- Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM (2001) Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat Immunol 2:842–847 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, Kawase K, Kanis AB, Patil SR, Stone EM, Sheffield VC (2001) A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet 68:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147 [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN (1998) Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev 12:547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olds LC, Sibley E (2003) Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 12:2333–2340 [DOI] [PubMed] [Google Scholar]
- Olson EN, Arnold HH, Rigby PW, Wold BJ (1996) Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1–4 [DOI] [PubMed] [Google Scholar]
- Paige AJ, Taylor KJ, Stewart A, Sgouros JG, Gabra H, Sellar GC, Smyth JF, Porteous DJ, Watson JE (2000) A 700-kb physical map of a region of 16q23.2 homozygously deleted in multiple cancers and spanning the common fragile site FRA16D. Cancer Res 60:1690–1697 [PubMed] [Google Scholar]
- Pailhoux E, Vigier B, Chaffaux S, Servel N, Taourit S, Furet JP, Fellous M, Grosclaude F, Cribiu EP, Cotinot C, Vaiman D (2001) A 11.7-kb deletion triggers intersexuality and polledness in goats. Nat Genet 29:453–458 [DOI] [PubMed] [Google Scholar]
- Pastinen T, Sladek R, Gurd S, Sammak A, Ge B, Lepage P, Lavergne K, Villeneuve A, Gaudin T, Brandstrom H, Beck A, Verner A, Kingsley J, Harmsen E, Labuda D, Morgan K, Vohl MC, Naumova AK, Sinnett D, Hudson TJ (2004) A survey of genetic and epigenetic variation affecting human gene expression. Physiol Genomics 16:184–193 [DOI] [PubMed] [Google Scholar]
- Peichel CL, Prabhakaran B, Vogt TF (1997) The mouse Ulnaless mutation deregulates posterior HoxD gene expression and alters appendicular patterning. Development 124:3481–3492 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM (2001) Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet 2:100–109 [DOI] [PubMed] [Google Scholar]
- Pfeffer PL, Payer B, Reim G, di Magliano MP, Busslinger M (2002) The activation and maintenance of Pax2 expression at the mid-hindbrain boundary is controlled by separate enhancers. Development 129:307–318 [DOI] [PubMed] [Google Scholar]
- Pfeifer D, Kist R, Dewar K, Devon K, Lander ES, Birren B, Korniszewski L, Back E, Scherer G (1999) Campomelic dysplasia translocation breakpoints are scattered over 1 Mb proximal to SOX9: evidence for an extended control region. Am J Hum Genet 65:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippard D, Boyd Y, Reed V, Fisher G, Masson WK, Evans EP, Saunders JC, Crenshaw EB, III (2000) The sex-linked fidget mutation abolishes Brn4/Pou3f4 gene expression in the embryonic inner ear. Hum Mol Genet 9:79–85 [DOI] [PubMed] [Google Scholar]
- Pop R, Conz C, Lindenberg KS, Blesson S, Schmalenberger B, Briault S, Pfeifer D, Scherer G (2004) Screening of the 1 Mb SOX9 5′ control region by array CGH identifies a large deletion in a case of campomelic dysplasia with XY sex reversal. J Med Genet 41:e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioleau MN, Nony P, Simpson M, Felsenfeld G (1999) An insulator element and condensed chromatin region separate the chicken β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J 18:4035–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE (2004) Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet 13:1213–1218 [DOI] [PubMed] [Google Scholar]
- Qin Y, Poirier C, Truong C, Schumacher A, Agoulnik AI, Bishop CE (2003) A major locus on mouse chromosome 18 controls XX sex reversal in Odd Sex (Ods) mice. Hum Mol Genet 12:509–515 [DOI] [PubMed] [Google Scholar]
- Rippe K (2001) Making contacts on a nucleic acid polymer. Trends Biochem Sci 26:733–740 [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Bardet PL, Zelus D, Hughes S, Laudet V (2001) Euteleost fish genomes are characterized by expansion of gene families. Genome Res 11:781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T (2002) The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev 16:1089–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M (1996) Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet 14:357–360 [DOI] [PubMed] [Google Scholar]
- Roessler E, Ward DE, Gaudenz K, Belloni E, Scherer SW, Donnai D, Siegel-Bartelt J, Tsui LC, Muenke M (1997) Cytogenetic rearrangements involving the loss of the Sonic Hedgehog gene at 7q36 cause holoprosencephaly. Hum Genet 100:172–181 [DOI] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D (2004) Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24:3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T, Masuya H, Tamura M, Shimizu K, Yada Y, Wakana S, Gondo Y, Noda T, Shiroishi T (2004) Phylogenetic conservation of a limb-specific, cis-acting regulator of Sonic hedgehog ( Shh). Mamm Genome 15:23–34 [DOI] [PubMed] [Google Scholar]
- Santagati F, Abe K, Schmidt V, Schmitt-John T, Suzuki M, Yamamura K, Imai K (2003) Identification of cis-regulatory elements in the mouse Pax9/Nkx2-9 genomic region: implication for evolutionary conserved synteny. Genetics 165:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R (2003) DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature 422:909–913 [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND (1996) Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell 86:71–82 [DOI] [PubMed] [Google Scholar]
- Scherer SW, Poorkaj P, Massa H, Soder S, Allen T, Nunes M, Geshuri D, Wong E, Belloni E, Little S (1994) Physical mapping of the split hand/split foot locus on chromosome 7 and implication in syndromic ectrodactyly. Hum Mol Genet 3:1345–1354 [DOI] [PubMed] [Google Scholar]
- Schramke V, Allshire R (2004) Those interfering little RNAs! Silencing and eliminating chromatin. Curr Opin Genet Dev 14:174–180 [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM (2004) Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428:717–723 [DOI] [PubMed] [Google Scholar]
- Sharpe J, Lettice L, Hecksher-Sorensen J, Fox M, Hill R, Krumlauf R (1999) Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr Biol 9:97–100 [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, Colleaux L, Bobrow M, Carter NP (2004) Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet 41:241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D (2003) A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113:405–417 [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Peichel C, Vogt TF, Duboule D, Zakany J (2001) Large scale transgenic and cluster deletion analysis of the HoxD complex separate an ancestral regulatory module from evolutionary innovations. Genes Dev 15:2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Montavon T, Monso-Hinard C, Morris M, Ventruto ML, Antonarakis S, Ventruto V, Duboule D (2002) A t(2;8) balanced translocation with breakpoints near the human HOXD complex causes mesomelic dysplasia and vertebral defects. Genomics 79:493–498 [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W (2002) Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 10:1453–1465 [DOI] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641 [DOI] [PubMed] [Google Scholar]
- Treisman J, Lang R (2002) Development and evolution of the eye: Fondation des Treilles, September, 2001. Mech Dev 112:3–8 [DOI] [PubMed] [Google Scholar]
- Trembath DG, Semina EV, Jones DH, Patil SR, Qian Q, Amendt BA, Russo AF, Murray JC (2004) Analysis of two translocation breakpoints and identification of a negative regulatory element in patients with Rieger’s syndrome. Birth Defects Res Part A Clin Mol Teratol 70:82–91 [DOI] [PubMed] [Google Scholar]
- Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR (2003) Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34:157–165 [DOI] [PubMed] [Google Scholar]
- Tupler R, Berardinelli A, Barbierato L, Frants R, Hewitt JE, Lanzi G, Maraschio P, Tiepolo L (1996) Monosomy of distal 4q does not cause facioscapulohumeral muscular dystrophy. J Med Genet 33:366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2002) Cellular memory and the histone code. Cell 111:285–291 [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H (2003) Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4:509–519 [DOI] [PubMed] [Google Scholar]
- Valle D (2004) Genetics, individuality, and medicine in the 21st century. Am J Hum Genet 74:374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven F, Schimmang T, Vortkamp A, Ruther U (1993) Molecular linkage of the morphogenetic mutation add and the zinc finger gene Gli3. Mamm Genome 4:276–277 [DOI] [PubMed] [Google Scholar]
- van der Hoeven F, Zakany J, Duboule D (1996) Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell 85:1025–1035 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2:2037–2042 [DOI] [PubMed] [Google Scholar]
- van Heyningen V, Williamson KA (2002) PAX6 in sensory development. Hum Mol Genet 11:1161–1167 [DOI] [PubMed] [Google Scholar]
- Vermaak D, Ahmad K, Henikoff S (2003) Maintenance of chromatin states: an open-and-shut case. Curr Opin Cell Biol 15:266–274 [DOI] [PubMed] [Google Scholar]
- Vortkamp A, Franz T, Gessler M, Grzeschik KH (1992) Deletion of GLI3 supports the homology of the human Greig cephalopolysyndactyly syndrome (GCPS) and the mouse mutant extra toes (Xt). Mamm Genome 3:461–463 [DOI] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Picketts DJ, Higgs DR (1995) Conservation of position and sequence of a novel, widely expressed gene containing the major human α-globin regulatory element. Genomics 29:679–689 [DOI] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell 79:1111–1120 [DOI] [PubMed] [Google Scholar]
- Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M (1999) Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet 22:196–198 [DOI] [PubMed] [Google Scholar]
- Wallrath LL, Elgin SC (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9:1263–1277 [DOI] [PubMed] [Google Scholar]
- Walter J, Paulsen M (2003) Imprinting and disease. Semin Cell Dev Biol 14:101–110 [DOI] [PubMed] [Google Scholar]
- Wang CC, Chan DC, Leder P (1997) The mouse formin (Fmn) gene: genomic structure, novel exons, and genetic mapping. Genomics 39:303–311 [DOI] [PubMed] [Google Scholar]
- Wijmenga C, Hewitt JE, Sandkuijl LA, Clark LN, Wright TJ, Dauwerse HG, Gruter AM, Hofker MH, Moerer P, Williamson R (1992) Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet 2:26–30 [DOI] [PubMed] [Google Scholar]
- Wild A, Kalff-Suske M, Vortkamp A, Bornholdt D, Konig R, Grzeschik KH (1997) Point mutations in human GLI3 cause Greig syndrome. Hum Mol Genet 6:1979–1984 [DOI] [PubMed] [Google Scholar]
- Williams SC, Altmann CR, Chow RL, Hemmati-Brivanlou A, Lang RA (1998) A highly conserved lens transcriptional control element from the Pax-6 gene. Mech Dev 73:225–229 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG (2004) Evolutionary changes in cis and trans gene regulation. Nature 430:85–88 [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA (2003) The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20:1377–1419 [DOI] [PubMed] [Google Scholar]
- Wunderle VM, Critcher R, Hastie N, Goodfellow PN, Schedl A (1998) Deletion of long-range regulatory elements upstream of SOX9 causes campomelic dysplasia. Proc Natl Acad Sci USA 95:10649–10654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW (2002) Allelic variation in human gene expression. Science 297:1143 [DOI] [PubMed] [Google Scholar]
- Zuniga A, Michos O, Spitz F, Haramis AP, Panman L, Galli A, Vintersten K, Klasen C, Mansfield W, Kuc S, Duboule D, Dono R, Zeller R (2004) Mouse limb deformity mutations disrupt a global control region within the large regulatory landscape required for Gremlin expression. Genes Dev 18:1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]