Abstract
Matsumoto and colleagues recently identified PEX26 as the gene responsible for complementation group 8 of the peroxisome biogenesis disorders and showed that it encodes an integral peroxisomal membrane protein with a single C-terminal transmembrane domain and a cytosolic N-terminus that interacts with the PEX1/PEX6 heterodimer through direct binding to the latter. They proposed that PEX26 functions as the peroxisomal docking factor for the PEX1/PEX6 heterodimer. Here, we identify new PEX26 disease alleles, localize the PEX6-binding domain to the N-terminal half of the protein (aa 29–174), and show that, at the cellular level, PEX26 deficiency impairs peroxisomal import of both PTS1- and PTS2-targeted matrix proteins. Also, we find that PEX26 undergoes alternative splicing to produce several splice forms—including one, PEX26-Δex5, that maintains frame and encodes an isoform lacking the transmembrane domain of full-length PEX26 (PEX26-FL). Despite its cytosolic location, PEX26-Δex5 rescues peroxisome biogenesis in PEX26-deficient cells as efficiently as does PEX26-FL. To test our observation that a peroxisomal location is not required for PEX26 function, we made a chimeric protein (PEX26-Mito) with PEX26 as its N-terminus and the targeting segment of a mitochondrial outer membrane protein (OMP25) at its C-terminus. We found PEX26-Mito localized to the mitochondria and directed all detectable PEX6 and a fraction of PEX1 to this extraperoxisomal location; yet PEX26-Mito retains the full ability to rescue peroxisome biogenesis in PEX26-deficient cells. On the basis of these observations, we suggest that a peroxisomal localization of PEX26 and PEX6 is not required for their function and that the interaction of PEX6 with PEX1 is dynamic. This model predicts that, once activated in an extraperoxisomal location, PEX1 moves to the peroxisome and completes the function of the PEX1/6 heterodimer.
Introduction
Peroxisomes are ubiquitous, single-membrane–bound organelles that contain ⩾50 enzymes and participate in a variety of metabolic pathways. The importance of the peroxisome in development and metabolism is highlighted by inherited defects in peroxisome biogenesis in which most or all of peroxisomal metabolic functions are deficient (Wanders 2004a, 2004b). The clinical phenotype of the peroxisome-biogenesis disorders (PBDs) varies widely in severity and can be classified into two broad categories, the Zellweger spectrum and rhizomelic chondrodysplasia punctata (RCDP [MIM 215100]) (Gould et al. 2001). The Zellweger spectrum includes at least three overlapping phenotypes that represent segments of a phenotypic continuum, including Zellweger syndrome (ZS [MIM 214100]) at the severe end, neonatal adrenoleukodystrophy (NALD [MIM 202370]) as an intermediate form, and infantile Refsum disease (IRD [MIM 266510]) at the mild end of the spectrum. Dysmorphic features, neurological symptoms, hepatic disease, and skeletal abnormalities in varying degrees are common to all three (Gould et al. 2001). Reflecting the complexity of peroxisome assembly, somatic-cell-hybridization studies show that PBDs are genetically heterogeneous, with at least 13 human complementation groups (CGs) (Weller et al. 2003; Shimozawa et al. 2004).
At the cellular level, the PBDs can be divided into defects of peroxisomal membrane biogenesis and matrix protein import. Three CGs (9, 12, and 14) have blocks in peroxisome membrane biogenesis and lack identifiable peroxisomal structures entirely. The remaining 10 CGs (1, 2, 3, 4, 7, 8, 10, 11, 13, and K [Gifu]) display defects in peroxisome matrix protein import but retain the ability to form peroxisomal membranes (Sacksteder and Gould 2000; Shimozawa et al. 2004). Peroxisomal matrix proteins are synthesized on free cytosolic ribosomes and are targeted posttranslationally to the organelle by cytosolic receptors that bind to cis-acting sequences, the peroxisomal targeting signals (PTSs). The majority of matrix proteins are targeted by the PTS1, a C-terminal -SKL or conserved variant (Gould et al. 1988, 1989). A few matrix proteins are targeted by a different sequence, the PTS2, a degenerate sequence—(R/K)(L/V/I)X5(H/Q)(L/A)—located near the N-terminus (Osumi et al. 1991; Swinkels et al. 1991). PTS1 and PTS2 sequences are recognized by the cytosolic receptors PEX5 and PEX7, respectively. The receptor/matrix protein complexes assemble in the cytosol and shuttle to the peroxisome, where they dock to PEX14, an integral peroxisomal-membrane protein. Unloading and translocation of the cargo proteins into the peroxisomal matrix involves interactions with several other membrane peroxins (PEX13, PEX10, and PEX12) (Weller et al. 2003). The receptors are recycled back to the cytosol to participate in another round of import (Dodt and Gould 1996). Decreased steady-state levels of PEX5 in cell lines from patients with primary deficiencies of PEX1 or PEX6 suggest a role for these peroxins in stabilization and/or recycling of the former (Yahraus et al. 1996; Reuber et al. 1997; Collins et al. 2000). PEX1 and PEX6 are AAA ATPases that, in yeast, form heterodimeric complexes in an ATP-dependent manner (Faber et al. 1998a; Birschmann et al. 2005). AAA ATPases are involved in a diverse set of cellular processes, many involving protein unfolding, refolding, or the disassembly of protein complexes (Vale 2000; Lupas and Martin 2002). Although the precise roles of PEX1 and PEX6 in the peroxisome membrane protein import process are unknown, epistasis analyses utilizing the decreased level of PEX5 in PEX1 and PEX6-deficient cells indicated that they may act in the disassembly of the multimeric PEX5-PTS protein complex (preimplex) prior to or during the import process (Collins et al. 2000; Gould and Collins 2002).
Using expression cloning in Chinese hamster ovary (CHO) cells defective for peroxisome biogenesis, Matsumoto and colleagues (2003a) recently identified PEX26 as the gene responsible for CG8, the last unexplained human CG. In both human and mouse, PEX26 is a 305-aa protein with one transmembrane domain, located near the C-terminus of the protein (aa 252–269) and an N-terminal domain (NTD) exposed to the cytosol. Matsumoto and colleagues (2003a) showed that PEX26 binds PEX6 and proposed that it functions to recruit PEX1 and PEX6 to peroxisomal membranes. Interestingly, they also propose that PEX26 function is required for import of only catalase and PTS2-targeted proteins and is not required for import of classical PTS1-targeted proteins. To date, 12 pathologic PEX26 alleles have been described in studies of 12 patients with CG8, all with phenotypes in the Zellweger spectrum (Matsumoto et al. 2003a, 2003b; Steinberg et al. 2004).
Here, we identify all mutant PEX26 alleles in 10 CG8 probands, including five new PEX26 mutations, and quantify the peroxisomal import phenotype in CG8 cells of known genotype, showing that both PTS1- and PTS2-mediated import is impaired. We also determine the effect of PEX26 deficiency on PEX5 and PEX1 steady-state levels and map the PEX6-binding domain in PEX26. Additionally, we describe extensive alternative splicing of PEX26 in fibroblasts and various human tissues that creates a cytoplasmic, but nevertheless functional, PEX26 isoform. Extending this observation, we show that PEX26 mislocalized to the outer mitochondrial membrane retains its function in peroxisome biogenesis. Our findings extend the consequences of PEX26 deficiency to PTS1-mediated import and challenge the current model of PEX26 function as a docking factor for the PEX1-PEX6 protein complex at the peroxisomal membrane.
Material and Methods
PEX26 Amplification and Mutation Detection
We isolated genomic DNA and total RNA from transformed skin fibroblasts with the Puregene and Purescript kits (Gentra Systems), per the manufacturer's directions, and purchased additional human total cellular RNA from Ambion. We amplified PEX26 exons 1–6 from genomic DNA (250 ng) and total RNA (0.5–2 μg), using Taq polymerase (Invitrogen) or the OneStep Cycle kit (Qiagen), per the manufacturer's directions. Oligonucleotide sequences are shown in table 1 (primers 26-1–26-24). PCR products were purified from agarose gel (QIAquick Gel Extraction kit [Qiagen]) and were sequenced directly by automated fluorescent sequencing. We confirmed each mutation in amplicons from both genomic DNA and RNA.
Table 1.
Oligonucleotide Sequences[Note]
| Purpose and Oligonucleotide Number | Sequence |
| PEX26 exon amplification from genomic DNA: | |
| 26-1 | gaagctgtagcaccagtttg |
| 26-2 | caggattcttgcatccctg |
| 26-3 | cagcttttgcctcagatagg |
| 26-4 | caatctgggaaggacagaac |
| 26–5 | caaggtggaggtgggaatg |
| 26-6 | gatctggagggcaatagtag |
| 26-7 | gtggtcatgggaaatgaacc |
| 26-8 | tctaagctcgcaggagtttc |
| 26-9 | cttagaagcaagcacagagg |
| 26-10 | gacaaacacccactcccac |
| 26-11 | gtaattggtagtggagtctc |
| 26-12 | tggtcctgctttgtttccac |
| 26-13 | ctaatctcgcggagtgcac |
| 26-14 | agtagagatggggtttcacc |
| PEX26 RT-PCR: | |
| 26-15 | acagaaggcgatgggccg |
| 26-16 | cacttcaccaggctttttctg |
| 26-17 | actccgacgtctgaggac |
| 26-18 | tcttgccaccgatccatttc |
| 26-19 | ctctccctcttcgcccag |
| 26-20 | cacctcattggaggtgaag |
| 26-21 | cgaaccaacatcggtagtg |
| 26-22 | attcacacagcgaggcagc |
| 26-23 | ctgtggatgtgtcgctctg |
| 26-24 | ggtggtgagatttgatccag |
| ASO: | |
| 26-25 | ctcgttatgaagagc |
| 26-26 | ctcgttacgaagagc |
| 26-27 | gccgacctcctggt |
| 26-28 | gccgaccccctgg |
| 26-29 | gacctcctggtggt |
| 26-30 | gacctcccggtgg |
| 26-31 | aatggatcggtggca |
| 26-32 | aatggattggtggcaa |
| 26-33 | aagctaccccccaaa |
| 26-34 | aaagctactccccaaa |
| Mutagenesis: | |
| 26-35 | ggcggccgaccccctggtggtgcacc |
| 26-36 | ggtgcaccaccagggggtcggccgcc |
| 26-37 | ggcggccgacctcccggtggtgcacc |
| 26-38 | ggtgcaccaccgggaggtcggccgcc |
| Cloning: | |
| 26-39 | agcttggtaccgagctcggatccactagtaacggc |
| 26-40 | ccccctgaggggggctgcagaggtcgaagaatcgctcttcaggtcctcctcgctgatcagcttctgctccataacgagtccgggtcc |
| 26-41 | cccaagcttggtaccgagctcggatccactagtaacgg |
| 26-42 | cagggaggaaggggaagctttcattccaggtttggcttctgg |
| 26-43 | gccgcctcgagcaagagcgattcttcgacctctgcag |
| 26-44 | gctcgcgcggccgctcattccaggtttggcttctgggcctc |
| 26-45 | gccgcctcgagcccggcccgggcgccggccgtggac |
| 26-46 | gctcgcgcggccgctcataagcagcccagaggcagcagcac |
| 26-47 | gccgcctcgagcgctgtgctggatgtggtgggtgcc |
| 26-48 | gctcgcgcggccgctcatccaggctcttgcattttgctgtataaaag |
| 26-49 | gccgcctcgagcgccgtggcagaggaacccgcgggc |
| 26-50 | gctcgcgcggccgctcagtgaaattctgccaaggctccatattc |
| 26-51 | ccgcaagcttggactcgttatggagcagaagctg |
| 26-52 | cgcggaattcttccaggtttggcttctgggcctc |
| 26-53 | cgcggaattccatcgaggcgacggagagccgagtgg |
| 26-54 | ggaggcggccgctcagagctgctttcggtatctcacg |
| 26-55 | cccgtcgaccgcatgtccaaagaaaaagcgc |
| 26-56 | cccgcggccgctggtgctcgaggaatcaccgactttccgtttcttc |
Note.— Underlines indicate wild-type and mutant nucleotides, restriction sites, and the myc-tag and stop-codon, respectively.
Allele-Specific Oligonucleotide Hybridization (ASO)
We performed ASO analysis to determine the frequency of missense mutations M1T, L44P, L45P, R98W, and P117L in exons 2 and 3, as described elsewhere (Gärtner et al. 1992; Braverman et al. 1997). Sequences of the wild-type and mutant-specific probes are listed in table 1 (probes 26-25–26-34).
Cell Lines, Transfections, and Immunofluorescence
We analyzed PBD skin fibroblast cell lines assigned to CG8 by somatic complementation analysis in the collection of the Kennedy Krieger Institute, Baltimore. Cell lines are identified by a unique PBD number assigned to each proband and are used in all publications from our group. Cell lines PBD057, PBD111, PBD114, and PBD063 were also studied by Matsumoto and coworkers (2003b) (in which study they were designated GM11335, PDL35167, GM08771, and GM16865, respectively). Cell lines PBD059, PBD063, PBD057, and PBD114 from our study are also available from the Coriell cell repository (assigned numbers GM17398, GM16865, GM11335, and GM08771, respectively). Cell lines with null alleles for PEX1 (PBD009), PEX5 (PBD005), PEX6 (PBD106), and PEX3 (PBD400) have been reported elsewhere (Dodt et al. 1995; Yahraus et al. 1996; Reuber et al. 1997; South et al. 2000). All cell lines were transformed by transfection with a vector expressing SV40 large T-antigen, as described elsewhere (Dodt et al. 1995). Additionally, cell lines PBD009, PBD005, PBD106, and PBD400 were immortalized by stable transfection with a vector that constitutively expresses human telomerase (pBabePuro/hTER) (Hahn et al. 1999). We cultured the cells in Dulbecco’s modified Eagle medium high-glucose supplemented with 10% fetal bovine serum and 100 U penicillin and 100 μg streptomycin/ml (Gibco). We transfected the cells by electroporation, as described elsewhere (Chang et al. 1997), and processed the cells for immunofluorescence 48 h later (Warren et al. 1998). We used monoclonal anti-HA, mono- and polyclonal anti-myc (BD Bioscience and Santa Cruz Biotechnology), polyclonal anti-PEX14 (South et al. 2000), and anti-PMP70 (Shani et al. 1997) as primary antibodies, and we obtained fluorescent secondary antibodies from Vector Laboratories. Mitochondria were visualized using MitoTracker (Molecular Probes). Microscopy was performed on a Zeiss LSM510 Meta confocal microscope with the Carl Zeiss LSM510 Imaging Software.
Plasmids
We constructed the recombinant plasmids to express full-length human PEX26 cDNA and its splice variants by cloning RT-PCR amplicons from human control fibroblast RNA spanning exons 1–6 (probes 26-15–26-16) (TOPO TA Cloning [Invitrogen]). After sequence analysis, we transferred the inserts representing full-length (-FL), exon 5–less (-Δex5) and exon 2–less (-Δex2) PEX26 cDNA into pcDNA3 (Invitrogen), using either the KpnI and EcoRI restriction sites or the EcoRI restriction site only (PEX26-Δex2). To produce full-length PEX26 (PEX26-FL) and -Δex5 expression vectors with an N-terminal myc-tag, we modified the KpnI–Bsu36I fragment, using PCR with a reverse primer incorporating the myc-sequence (primers 26-39–26-40). To obtain the myc-tagged PEX26 expression vector lacking exon 5 and exon 6 (mycPEX26-Δex5+6), we inserted a stop codon (c.667GGC→TGA; G223X) following the sequence encoded by exon 4 in the cDNA. We PCR amplified a fragment of mycPEX26-Δex5 with oligonucleotide primers 26-41 and 26-42, the latter containing the -TGA stop codon, and cloned the HindIII fragment from this PCR product into the HindIII sites of mycPEX26-Δex5.
To mislocalize the myc-tagged N-terminus of PEX26 to the outer mitochondrial membrane, we PCR-amplified the corresponding segment of mycPEX26-FL with primers that incorporated HindIII and EcoRI restriction sites (primers 26-51–26-52). Using RT-PCR of rat liver RNA, we isolated the segment of rat Omp25 cDNA that encodes the transmembrane segment of the protein flanked by EcoRI and NotI restriction sites (probes 26-53–26-54). We ligated these fragments into pcDNA3 (HindIII and NotI) to create a recombinant vector expressing a chimeric protein, mycPEX26 (aa 2–222)–OMP25 (aa 170–206) (PEX26-Mito). We used the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene), per the manufacturer's directions, to introduce L44P (CTC→CCC) and L45P (CTG→CCG) into PEX26-FL cDNA expression vector and verified the mutations by sequencing (primers 26-35–26-38). For N-terminal myc-tagging of these clones, we used the PCR-modification strategy of the Kpn I-Bsu36 fragment that is described above (mycPEX26-L44P and -L45P). pcDNA3 with an N-terminal 3× HA epitope tag was created by inserting a PCR product coding for MTSGRIFYPYDVPDYAG YPYDVPD YA GSYPY-DVPDYALQCGR, flanked by BglII and XhoI-NotI-XbaI restriction sites into BamHI and XbaI sites of pcDNA3. To construct a plasmid with a 3× HA-tag and a 3× nuclear localization signal (HAnls) for N-terminal tagging of PEX26 fragments, we inserted a PCR amplicon containing the 3× nls signal (Jones et al. 1999, 2001) flanked by SalI and XhoI-NotI restriction sites, as a SalI–NotI fragment into XhoI and NotI sites of the pcDNA3–3× HA expression vector (probes 26-55–26-56). Using the new XhoI and the NotI restriction sites, we inserted PEX26 fragments obtained by PCR amplification with pcDNA3-PEX26-FL or PEX26-L44P/L45P as template for expression of PEX26 peptides corresponding to aa 2–222, aa 2-222-L44P, aa 2-222-L45P, aa 2–135, aa 2–163, aa 136–222, and aa 29–174 in frame with the HAnls signal at the N-terminus (probes 26-43–26-50). PCR-generated PEX26 fragments for expression of peptides corresponding to aa 2–174, aa 2–163, aa 29–174, and aa 69–174 were also inserted in frame with an N-terminal 3× myc-tag in pcDNA3 by use of XhoI and NotI restriction sites (Geisbrecht et al. 1998). pcDNA3-3× HAPEX6 (JM721) and pcDNA3-3× mycPEX6 (JM709) were constructed by inserting the SalI-NotI fragment from pTY3 (Yahraus et al. 1996) that contained the PEX6 ORF in the XhoI-NotI sites of the pcDNA3-3× HA (see above) and the pcDNA3-3× myc expression vector (Geisbrecht et al. 1998), respectively. The expression vector for pcDNA3-PEX1 with a C-terminal 3× HA-tag (pBER86) was made by inserting an ApaI-XhoI fragment that contained three repeats of the HA epitope tag (see above) between the ApaI-XhoI sites of pBER81 containing the PEX1 ORF (Reuber et al. 1997), resulting in an in-frame C-terminal fusion.
Oligonucleotides used for cloning and mutagenesis are listed in table 1. We sequenced all recombinant constructs to verify absence of PCR-introduced mutations. As reporters for peroxisomal-protein import, we used the following plasmids: pcDNA3-PAHXmyc (Mihalik et al. 1997), pcDNA3-NmycPECI (Geisbrecht and Gould 1999), pcDNA3-NmycHAOX1 (Liu et al. 1999), pcDNA3-NmycCatalase (Liu et al. 1999), and GFP-SKL.
Characterization of the Cellular Phenotype and Relative Rescue
We processed cells grown on cover slips for immunofluorescence and counted the numbers of cells expressing the matrix-protein import markers with variations of the PTS consensus sequence (see above). We placed each expressing cell into one of the following categories on the basis of the distribution of the marker protein: no import for cytoplasmic-only staining, partial import for cells with cytoplasmic and peroxisomal staining, and complete import when the marker protein was exclusively peroxisomal. In a subset of these experiments, the observer was blind to the cell line and to the transfected constructs, with results indistinguishable from the nonblind quantification. Additionally, recounting the same experiment in a blind fashion gave values differing by no more than 10%. To calculate percentages of partial and complete import, we scored at least 500 expressing cells in each experiment. To determine the complementing activity of PEX26 missense alleles L44P and L45P and PEX26 splice variants, we calculated the percentages of CG8 cells that displayed no import, partial import, and complete import of mycCAT or GFP-SKL after cotransfection with nonrecombinant pcDNA3, PEX26-FL, PEX26-L44P, PEX26-L45P, PEX26-Δex5, PEX26-Δex2, and PEX26-Mito expression vectors. Corrected rescue values were calculated by subtracting the percentage of import observed in cells transfected with nonrecombinant (empty) vector (typically with a range of 0%–10% for mycCAT/GFP-SKL in PBD059 cells) from the percentage of import observed in cells transfected with the PEX26-FL, PEX26 mutant, PEX26 splice variant, and PEX26-Mito expression vectors. Relative rescue values were obtained by dividing the corrected rescue values by the corrected rescue value determined for the cells transfected with the PEX26-FL expression vector.
Immunoblot Experiments
We used 15 μg of protein from whole-cell lysates from the indicated cells, electrophoresed in 7.5% or 10% Tris-HCl polyacrylamide gels (BioRad), and processed the gels for immunoblotting, following standard protocols. Mouse anti-PEX1 monoclonal antibody was purchased from BD Transduction Laboratories. Rabbit polyclonal anti-PEX5 antibody was described elsewhere (Dodt et al. 1995). Rabbit polyclonal anti-PEX13 antibody is directed against a C-terminal PEX13 peptide (South et al. 2000). We used anti-tubulin (Sigma) as a loading control. All results were obtained in duplicate in two experiments.
Antibodies directed against PEX26 were generated by immunization of rabbits with an N-terminal PEX26 peptide (NH2-MKSDSSTSAAPLRGLC-COOH) conjugated to keyhole limpet hemocyanin as a carrier protein (Covance Research Products). In vitro transcription and translation of full-length PEX26 was performed with the TnT T7 Coupled Reticulocyte Lysate System (Promega), per the manufacturer's directions.
Results
PEX26 Full-Length Transcripts and Orthologs
PEX26 maps to 16.9 Mb at 22q11.21, just centromeric of the region typically deleted in DiGeorge/velocardiofacial syndrome (McDermid and Morrow 2002), and has seven predicted exons distributed over 13 kb, with the translation start site located in exon 2. In our RT-PCR experiments, we routinely use a primer (26-15) corresponding to the 5′ end of exon 1, as predicted by the PEX26 cDNA with GenBank accession number NM_017929.
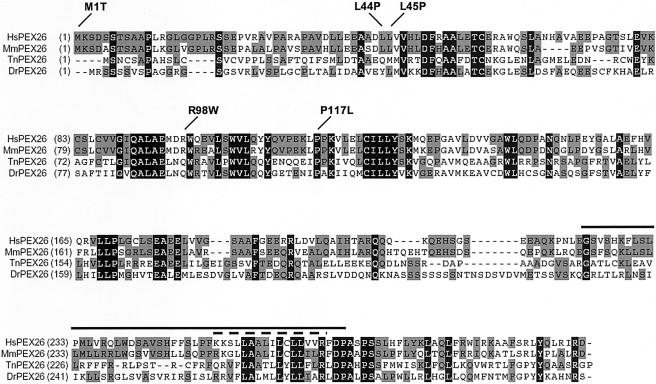
Using the human PEX26 amino acid sequence as a probe to screen public databases, we detected multiple vertebrate orthologs (Mus musculus, Rattus norvegicus, Gallus gallus, Danio rerio, Fugu rubripes, Tetraodon nigriviridis) but none in invertebrates. In D. rerio and T. nigriviridis, PEX26 orthologs show 30% and 32%, respectively, sequence identity with human PEX26 (fig. 1). Saccharomyces cerevisiae PEX15 resembles vertebrate PEX26 in topology (both are type II integral membrane proteins) and in binding of PEX6, but we found no detectable amino acid sequence similarity (12% overall identity between S. cerevisiae PEX15 and human PEX26, with no blocks of identical amino acids >2 residues).
Figure 1.
Sequence comparison of vertebrate PEX26 orthologs. Alignment of human (Hs), mouse (Mm), T. nigroviridis (Tn), and D. rerio (Dr) PEX26 amino acid sequence. HsPEX26 is 73%, 32%, and 31% identical to mouse, T. nigriviridis, and D. rerio PEX26, respectively. Amino acids identical across all four species are highlighted in black; those with a plurality are in gray. The positions of PEX26 missense mutations identified in this study are indicated. The solid line marks the segment of human PEX26 encoded by the alternatively spliced exon 5; the dashed line indicates the predicted transmembrane domain.
PEX26 Mutation Analysis
Using RT-PCR of patient fibroblast RNA with confirmation by amplification of the indicated exons in genomic DNA, we found nine PEX26 alleles that accounted for all 20 mutant PEX26 genes in 10 CG8 probands (table 2). Of these alleles, five are newly recognized (Matsumoto et al. 2003a, 2003b; Steinberg et al. 2004). One proband with a ZS phenotype (PBD059) is homozygous for a mutation in the splice-donor site of intron 2, c.230+1G→T, which leads to a frameshift at T77 and premature termination (T77fs139X) (see table 2). Four patients with an NALD or IRD phenotype (PBD111, PBD057, PBD161, and PBD065) are homozygous for a transition in exon 3, c.292C→T, which results in the previously reported missense mutation R98W. PBD063, with a ZS phenotype, is a compound heterozygote for R98W and an insertion of thymidine at position 254 in the cDNA (c.254insT; exon 3), which results in a frameshift and premature termination (C86fs114X). In PBD055, with an IRD phenotype, we detected three heterozygous mutations. In exon 3, a c.350C→T transition results in P117L; in exon 4, c.457C→G results in L153V; and, in exon 6, a single base-pair deletion (c.862delC) causes a frameshift at R288 with termination 79 aa downstream (R288fs366X). L153V is in cis with R288fs and is a known SNP (rs12484657). PBD110 is a compound heterozygote for L44P and R98W. PBD114 is a compound heterozygote for M1T and L45P. The first methionine downstream from M1 is M96, which is embedded in an acceptable Kozak consensus sequence (GCAGAAAUGG, compared with the consensus GCCPuCCAUGG), which suggests that translation could initiate at this position (Kozak and Harper 2000). PBD109 is a compound heterozygote for R98W and a complicated allele with a 122-bp duplication separated by an inserted thymidine (c.426_548dup122bpinsT) that maintains frame and predicts a protein in which aa 134–182 are tandemly duplicated, with substitution of glycine by valine at position 183 (A134_V182dup+G183V).
Table 2.
PEX26 Mutations in CG8 Patients
| Patient, Clinical Phenotype,and PEX26 Allele | PEX26 Mutation in cDNA | Comment |
| PBD059: | ||
| ZS: | ||
| T77fs139X homozygote | c.230+1G→T | This donor splice-site mutation results in the insertion of 173, 236, or 650 bp of intron 2, with the result of addition of a nonspecific sequence of 61 or 82 aa. For clarity, we designate the allele by the shortest insert. |
| PBD111: | ||
| NALD: | ||
| R98W homozygote | c.292C→Ta | Transition at CpG dinucleotide |
| PBD063: | ||
| IRD: | ||
| C86fs114X | c.254insT | Nonspecific sequence of 28 aa |
| R98W | c.292C→T | |
| PBD055: | ||
| IRD: | ||
| [L153V+R288fs366X] | [c.457C→G+c.862delC] | Nonspecific sequence of 78 aa; L153V and R288fs366C are in cis. |
| P117L | c.305C→Ta | |
| PBD057: | ||
| NALD: | ||
| R98W homozygote | c.292C→T | |
| PBD110: | ||
| NALD: | ||
| L44P | c.131T→Ca | |
| R98W | c.292C→T | |
| PBD114: | ||
| IRD: | ||
| M1T | c.2T→Ca | Downstream initiation at M96 theoretically possible |
| L45P | c.134T→Ca | |
| PBD109: | ||
| IRD: | ||
| R98W | c.292C→T | |
| A143_V182dup+G183V | c.426_548dup122bpinsT | Duplication within exon 4 maintains frame and predicts mutant PEX26p with duplication of aa 143–182 plus G183V |
| PBD161: | ||
| IRD: | ||
| R98W homozygote | c.292C→T | Clinical phenotype variant |
| PBD065: | ||
| NALD: | ||
| R98W homozygote | c.292C→T |
Frequencies of cDNA changes have been determined by ASO in 106 control chromosomes (see text).
Functional Consequence of PEX26 Missense Alleles L44P and L45P
Of these nine PEX26 alleles, four (M1T, L45P, R98W, and c.254insT) were shown elsewhere to be functionally significant by expression in a PEX26-deficient CHO cell line (Matsumoto et al. 2003b). We assumed functional significance for three nonsense or frameshift mutations that disrupted a large segment of PEX26 (c.230+1G→T, L153V+R288fs, and A143_V182dup+G183C). To test the functional consequences of PEX26-L44P and -L45P, we expressed them in transformed fibroblasts from a CG8 patient (PBD059) who was homozygous for a PEX26 null allele, and we compared their ability to restore matrix-protein import with wild-type PEX26 (relative rescue). In two separate experiments, PEX26-L44P had 1% and 8% relative-rescue activity, whereas PEX26-L45P had 0% relative-rescue activity in both trials. Thus, both these mutants severely reduce the function of PEX26.
Frequency of Mutant Alleles
To estimate the frequency of the five missense alleles (M1T, L44P, L45P, R98W, and P117L) in the general population, we amplified PEX26 exons 2 and 3 from genomic DNA of 53 individuals of mixed ethnicity, and we genotyped them by allele-specific oligonucleotide hybridization. We detected none of the mutations in these 106 control chromosomes and conclude all are relatively rare variants.
Cellular Phenotype of PEX26-Deficient Fibroblasts
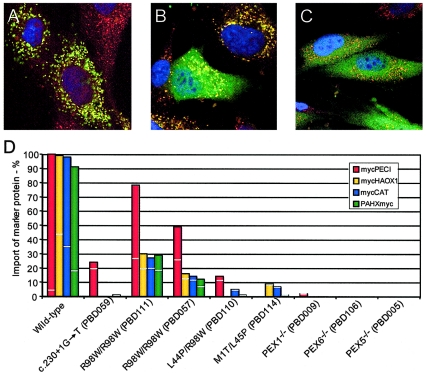
A previous study concluded that PTS1 import is normal in PEX26-deficient cells, a result that is surprising, given that the clinical phenotype of the patients is in the Zellweger spectrum rather than RCDP (Matsumoto et al. 2003b). To determine the extent and severity of the import defect for peroxisomal matrix enzymes in PEX26-deficient fibroblasts from individuals with known genotype, we made use of the differing import efficiency associated with variation in the PTS1 (Liu et al. 1999). We determined the matrix-protein import of a series of PTS1-targeted proteins ranging from the canonical -SKL targeted peroxisomal Δ3, Δ2 enoyl-CoA isomerase (PECI) (Geisbrecht and Gould 1999); the near-consensus -SKI targeted human peroxisomal glycolate peroxidase (HAOX1) (Jones et al. 2000); and the most atypical PTS1, -KANL, found in catalase (CAT) (Lazarow et al. 1982; Bell et al. 1986; Purdue and Lazarow 1996). We also tested PTS2 import by analyzing the distribution of phytanoyl-CoA hydroxylase (PAHX) (Mihalik et al. 1997). We compared the peroxisomal matrix import of these reporters in transformed fibroblasts of patients PBD059, PBD111, PBD057, PBD110, and PBD114 to that in cells from PEX1-, PEX5-, and PEX6-deficient patients and a control individual. As predicted by the degree of conservation of the PTS1, import for these markers varied with PECI>HAOX1>CAT (fig. 2). The cells of all PEX26-deficient patients had some residual import in this assay. PBD059, with a ZS phenotype (c.230+1G→T/c.230+1G→T), showed the most severe impairment, with residual import of mycPECI only. In contrast, cells from two R98W homozygotes (PBD111 and PBD057), both with an NALD phenotype, displayed the mildest import phenotype, with residual import for all marker proteins. Fibroblasts of PBD110 (L44P/R98W [NALD]) and PBD114 (M1T/L45P [IRD]) had minimal residual import for most marker proteins tested. Thus, there is a limited correlation between the cellular phenotype as measured in this assay and the clinical phenotype in these patients. Interestingly, cells from patients with null mutations in PEX1, PEX6, and PEX5 all had a more severe import phenotype than we observed in any of the PEX26-deficient cells, including the presumed null, PBD059, which had some residual import of the reporter with the most conserved PTS1 signal (PECI; 24% residual import). Our results differ from those of Matsumoto and coworkers, who interpreted their immunofluorescence data with an antibody directed against the canonical PTS1 (-SKL) as showing normal import of PTS1-targeted matrix proteins (Matsumoto et al. 2003a, 2003b). Our results showing a generalized defect in PTS1-mediated import are consistent with the Zellweger spectrum clinical phenotype of the patients with CG8.
Figure 2.
Cellular phenotype of cultured skin fibroblasts from PEX26-deficient patients. A–C, Example of merged images of transfected human fibroblasts showing complete (A), partial (B), or no (C) peroxisomal import of the indicated reporter. Indirect immunofluorescence staining of peroxisomal membranes was with anti-PEX14 antibody (Texas Red), and staining of a reporter peroxisomal matrix protein was with anti-myc antibody (FITC). Yellow indicates colocalization of the peroxisomal marker and the matrix-protein marker. D, Percentage of transfected cells displaying complete and partial import of the indicated matrix proteins. The vertical bars indicate the total percentage of cells showing import. The light horizontal line dividing each vertical bar indicates the fraction of cells scored as partial import (below the line) and complete import (above the line). The values represent the means of two experiments, except for (1) wild type, PBD059 and PBD111 transfected with mycPECI, and mycCAT (three experiments); and (2) PBD057 transfected with mycPECI, mycHAOX1, and PAHXmyc and PBD114 transfected with mycHAOX1, mycCAT, and PAHXmyc (one experiment); the value for PBD114 with mycPECI was not determined.
PEX26, PEX5, and PEX1 Steady-State Levels in PEX26-Deficient Fibroblasts
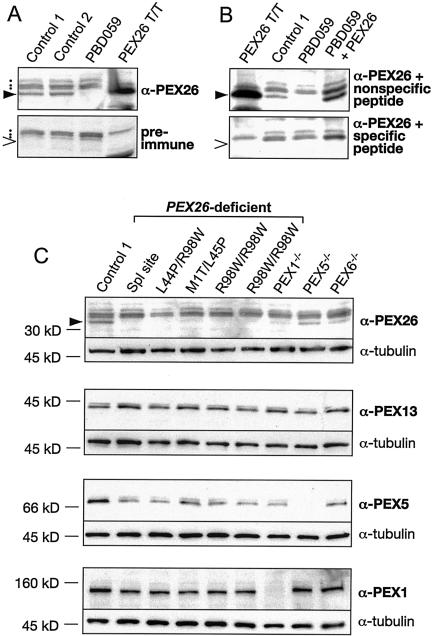
Immunoblot studies with anti-PEX26 antibody identified a protein of the size predicted for PEX26 (34 kD) in lysates of wild-type but not PBD059 cells. The antibody also detected a 34-kD protein produced in in vitro transcription and translation of a full-length PEX26 cDNA. Preincubation of the antibody with the specific but not with a nonspecific peptide blocked binding to the 34-kD protein (fig. 3). We conclude that our antibody specifically detects PEX26. A larger, cross-reacting protein is present in the immunoblots of wild-type and CG8 samples and is detected with either preimmune or immune serum (fig. 3).
Figure 3.
Immunoblot analysis of PEX26 and related peroxins in PBD fibroblasts. All panels show fibroblast lysates (15 μg/lane) or the products of in vitro transcribed and translated full-length PEX26 (PEX26T/T) electrophoresed in 7.5% or 10% polyacrylamide gels, under denaturing conditions, transferred to nylon filters, and detected with the indicated antibody or antiserum. Panels A and B show characterization of our anti-PEX26 N-terminal peptide (NH2-MKSDSSTSAAPLRGLC-COOH) antibody. A, Blots of lysates from control fibroblasts, a PEX26-deficient cell (PBD059, c.230+1G→T/c.230+1G→T), and PEX26T/T detected either with anti-PEX26 (top) or preimmune serum (bottom). The filled wedge (▸) indicates PEX26 (34 kDa), detected only in control fibroblasts and in the PEX26T/T products; the dots indicate nonspecific background bands present in fibroblast lysates; the unfilled wedge (>) indicates the expected location of PEX26 in the preimmune panel. B, Lysates from the indicated sources stained with anti-PEX26 antibody preincubated with either a nonspecific (unrelated) peptide (NH2-QKQEHSGSEEAQKPNC-COOH) (top) or with the specific N-terminal PEX26 peptide used as immunogen (NH2 -MKSDSSTSAAPLRGLC-COOH) (bottom). The lysate designated PBD059+PEX26 is from PEX26-null cells transfected with full-length PEX26. The filled wedge (▸) indicates PEX26 (top); the unfilled wedge (>) indicates expected location of PEX26 (bottom). Note that preincubation with the specific immunizing peptide but not with the unrelated nonspecific peptide completely blocks detection of PEX26. C, Immunoblots of lysates from cells of the indicated genotype stained with anti-PEX26 antiserum and anti-tubulin as a loading control (top panel). The second panel shows the same lysates stained with anti-PEX13 as a measure of the amount of peroxisome membrane in each sample, also with anti-tubulin as a loading control. The third panel shows the same lysate stained with anti-PEX5 and the bottom panel with anti-PEX1. In all panels, the PEX26 genotype is indicated above. The identity and genotypes of the cell lines with mutations in other PEX genes are as follows: PEX1−/−, PBD009 (c.2097insT/IVS18+1G→A) (Reuber et al. 1997); PEX5−/−, PBD005 (R390X/R390X) (Dodt et al. 1995); and PEX6−/−, PBD106 (c.del2398−2417insT/c.1962−1G→A).
Immunoblots of lysates of all CG8 cell lines tested showed no detectable PEX26, which indicates that the PEX26 mutations in these cell lines destabilize the protein product. Previous studies of monogenic defects in peroxisome biogenesis have shown that a primary defect in one component of the matrix-protein–import apparatus may result in a secondary deficiency of other components. For example, primary defects in PEX1 or PEX6 result in a marked secondary deficiency of PEX5 (Dodt and Gould 1996; Collins et al. 2000). Similarly, we found that, compared with wild type, PEX26 was reduced but detectable in lysates of reference PEX1-, PEX5-, and PEX6-deficient cell lines. In at least six CGs, including those caused by deficiency of PEX1, -5, and -6 (CG1, CG2, and CG4, respectively), peroxisomes are enlarged but show a 5-fold decrease in abundance (Chang et al. 1999). To exclude the possibility that the observed reduction in PEX26 level was secondary to a reduction of peroxisomal membrane, we examined the level of PEX13, an integral peroxisomal membrane protein, in this same set of cell lines. The levels of PEX13 were essentially the same as in control cells (fig. 3). Thus, the decrease in PEX26 seems to be a specific consequence of deficiency of PEX1, PEX5, or PEX6.
To determine if PEX26 deficiency affects the steady-state level of PEX5 or PEX1, we used total protein from whole-cell lysates from selected CG8-, CG1-, CG2-, and CG4-transformed fibroblasts for immunoblot experiments (fig. 3). As expected, we found no effect of PEX26, PEX5, or PEX6 deficiency on PEX1 levels. Unfortunately, neither we nor others have been able to develop an antibody to assess PEX6 levels. PEX5 levels were equally reduced in all CG8 cell lines and in PEX1- and PEX6-deficient cells.
Alternative Splicing of PEX26
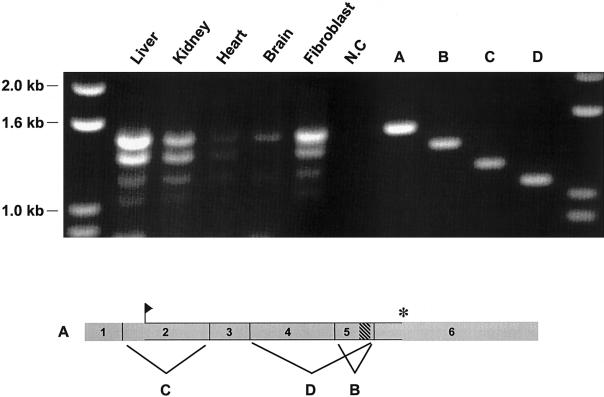
During the course of our RT-PCR experiments with fibroblast RNA, we detected three shorter amplified products in addition to the expected full-length product of 1,508 bp (PEX26-FL) (fig. 4). We cloned and sequenced these shorter cDNAs and found that they represented splice variants of PEX26. We found a similar pattern of these PEX26 splice forms in RNA from several tissues in relatively similar ratios (fig. 4). As compared with PEX26-FL, the next smaller product (1,361 bp) lacks exon 5 (PEX26-Δex5). The ORF of this splice form maintains frame and encodes a protein that is 49 aa shorter than full-length PEX26, lacking the 18-aa transmembrane domain of full-length PEX26 (fig. 4). The next shorter splice form (1,197 bp) results from skipping of exon 2 (PEX26-Δex2). Exon 2 encodes the N-terminus of PEX26, so this splice form would encode an N-terminal–truncated protein initiating at the next downstream methionine (M96). The next shorter splice form is 1,065 bp and lacks exons 4 and 5 (PEX26-Δex4+5). This splice form encodes a protein with the 124 N-terminal amino acids of PEX26, followed by a frameshift with an intact ORF, for 32 additional codons. In one RT-PCR experiment with heart mRNA, we also detected a faintly staining product that was 248 bp longer than PEX26-FL (data not shown). Cloning and sequencing revealed that intron 1 (248 bp) was retained in this product. Since the translation initiation site is located in exon 2, the complete ORF of PEX26-FL is intact in this splice form. A BLASTN search against the human EST database with use of PEX26 exon 1 plus exon 2 as a probe identified 19 ESTs containing exon 1 and exon 2 sequences. Intron 1 was retained in nine. In BLASTN searches with PEX26 cDNA exons 1–6, we found eight ESTs that contain or span the segment contributed by exon 5. Of these, two were the PEX26-Δex5 splice variant. We did not find a PEX26 EST corresponding to PEX26-Δex2.
Figure 4.
PEX26, alternatively spliced in fibroblasts and human tissues. Top panel, The products of RT-PCR amplification of PEX26 transcripts. RT-PCR products with total RNA from human liver, kidney, heart, and brain, by use of primers complementary to the 5′ and 3′ ends of full-length PEX26 transcript (probes 26-15 and 26-16, respectively), analyzed by agarose-gel electrophoresis and ethidium-bromide staining. The source of the RNA is indicated above the five lanes on the left. On the right, we cloned the RT-PCR products (1,508, 1,361, 1,197, and 1,065 bps) from fibroblast RNA, and the inserts representing full-length PEX26 cDNA (A), PEX26-Δex5 (B),PEX26-Δex2 (C), and PEX26-Δex4+5 (D) splice forms are shown as size references. Lower panel, Full-length PEX26 transcript, except for exon 7, which encodes 3′ untranslated sequence and is not included in the RT-PCR amplicons. The segments contributed by each exon are numbered and shown to scale. The flag shows the translation start site, the cross-hatched rectangle indicates the segment of exon 5 encoding the transmembrane domain, and the asterisk (*) indicates the translation stop site. The letters indicate the exon deleted from the similarly lettered splice form shown in the top panel.
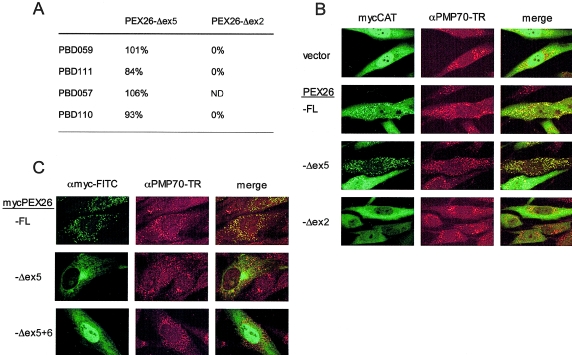
Complementing Activity and Subcellular Localization of PEX26 Isoforms
To examine the functional significance of the two splice variants that retain frame (PEX26-Δex5 and PEX26-Δex2), we expressed them in control fibroblasts and in PEX26-deficient fibroblasts with various PEX26 genotypes (PBD059, PBD111, PBD057, and PBD110). In control cells, neither PEX26 splice form had an effect on matrix-protein import (data not shown), nor did PEX26-Δex2 rescue-matrix–protein import in any of the PEX26-deficient cell lines. By contrast, the relative-rescue activity of PEX26-Δex5 was the same as PEX26-FL (fig. 5). This result was surprising, given that PEX26-Δex5 lacks the predicted transmembrane domain of PEX26, expected to be essential for its putative function as a docking protein for PEX1 and PEX6 (Matsumoto et al. 2003b).
Figure 5.
The PEX26-Δex5 splice form, which rescues PEX26-deficient cell lines without localizing to peroxisomal membranes. A, Relative-rescue activity of PEX26 isoforms for peroxisomal import of mycCAT. The numbers are the mean percent for import (partial plus complete) determined in three separate experiments for PBD059 (c.230+1G→T/c.230+1G→T), two trials for PBD111 (R98W/R98W) and PBD110 (L44P/R98W), and one trial for PBD057 (R98W/R98W). ND=not determined. B, Indirect immunofluorescence assays of the ability of PEX26 splice forms to rescue matrix import. We cotransfected PBD059 cells with mycCAT as reporter for peroxisomal import and either nonrecombinant pcDNA3 vector alone or recombinant pcDNA3 expressing the indicated PEX26 splice form. We localized peroxisomes, using a Texas Red–labeled αPMP70. C, Subcellular localization of the PEX26 isoforms. We transfected PBD059 cells with recombinant vectors expressing the indicated N-myc–tagged PEX26 isoform, and we localized the isoform (FITC) by indirect immunofluorescence and peroxisome membranes with anti-PM70 antibody (Texas Red [“TR”]). Images were made with a confocal laser scanning microscope (Zeiss). Colocalization is indicated by the yellow color in the merged images.
To determine the subcellular localization of the complementing PEX26 isoforms, we added N-terminal myc-tags to PEX26-FL and PEX26-Δex5. We also produced a myc-tagged PEX26 expression vector that lacked exon 5 and exon 6 (mycPEX26-Δex5+6, encoding PEX26 (aa 2–222). The relative-rescue activities for GFP-SKL import of mycPEX26-Δex5 and mycPEX26-Δex5+6, as compared with mycPEX26-FL, were 84%/77% and 65%/34% for PBD059/PBD110 cell lines. Thus, addition of the N-terminal myc-tag did not have a deleterious effect on the function of the PEX26 isoforms and, consistent with our results with PEX26-Δex5, PEX26-Δex5+6 encodes a partially functional protein. The subcellular localization of these various PEX26 isoforms is shown in figure 6. mycPEX26-FL is completely localized to peroxisomes, as indicated by a punctate staining pattern that colocalizes with PMP70. By contrast, the localization of mycPEX26-Δex5 and mycPEX26-Δex5+6 was similar and clearly different from that of mycPEX26-FL. Both show substantial localization to the cytosol, with some localization to the ER for mycPEX26-Δex5. These results suggest that a peroxisomal membrane location is not required for PEX26 function.
Figure 6.
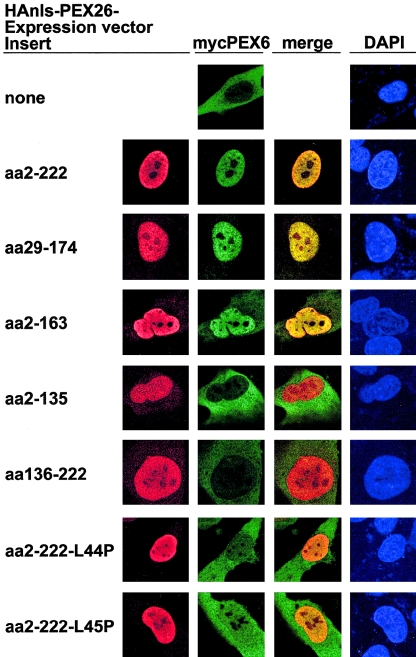
PEX26 segments aa 29–174 and 2–163, sufficient for PEX6 binding, as detected in a nuclear mislocalization assay. We transfected fibroblasts lacking peroxisomes due to an inherited deficiency of PEX3 (PBD400) with pcDNA3–3× mycPEX6 and either nonrecombinant pcDNA3 vector alone or recombinant pcDNA3 expressing N-terminal HA-tagged, nls-PEX26 fragments comprising the indicated segments of PEX26, and we analyzed them by indirect immunofluorescence with anti-HA (Texas Red), anti-myc (FITC) antibodies, and DAPI counterstaining, to show nuclei. The bottom two rows show expression of PEX26 segments sufficient to bind PEX6 but with the indicated missense mutations.
Mapping of PEX6 Binding in PEX26
Using Co-IP with epitope-tagged proteins and GST–pull down experiments, Matsumoto and colleagues (2003b) showed binding between full-length PEX26 and PEX6. We asked which part of PEX26 is essential for this interaction. To provide a discrete, easy-to-score subcellular location for these binding experiments, we mislocalized PEX26 fragments to the nucleus (HAnls-PEX26) (Sacksteder et al. 2000) in transformed fibroblasts from a PEX3-deficient patient (PBD400). These cells lack peroxisomes, as detected with antibodies to either peroxisome matrix or membrane proteins (South et al. 2000). We cotransfected PBD400 cells with myc-tagged PEX6 and either pcDNA3 or recombinant pcDNA3 carrying HAnls-PEX26 fragments (fig. 6). mycPEX6 was evenly distributed in the cytosol and was barely detectable in the nucleus when cotransfected with pcDNA3 vector alone. Cotransfection with HAnls-PEX26 (aa 2–222), which includes ∼90% of the cytoplasmic domain of PEX26, altered mycPEX6 localization to the nucleus. Using serial N- or C-terminal deletions of HAnls-PEX26, we identified aa 29–174 as a minimal fragment sufficient to localize mycPEX6 to the nucleus. HAnls-PEX26 (aa 2–163) had a variable effect on mycPEX6 localization, whereas two overlapping fragments, HAnls-PEX26 (aa 2–135) and HAnls-PEX26 (aa 136–222), showed no binding of mycPEX6. These results indicate that the segment of PEX26 required for interaction with PEX6 includes aa 29–163. Moreover, we found that HAnls-PEX26 (aa 2–222) with either the L44P or the L45P mutations produced a stable protein that did not direct mycPEX6 to the nucleus, which suggests that these mutations interfere with the binding of PEX6 to PEX26.
In line with these results, expression of full-length PEX26-L44P and PEX26-L45P does not result in the localization of mycPEX6 to peroxisomes that is observed with expression of wild-type PEX26-FL. We repeated this experiment with myc-tagged wild-type and mutant PEX26-FL (mycPEX26-FL, mycPEX26-L44P, and mycPEX26-L45P) and found that wild-type and mutant mycPEX26-FL are stable and localized on peroxisomal membranes, but only the wild-type protein directs HAPEX6 to peroxisomes (data not shown).
PEX26-Deletion Mutants and the Minimal Complementing Fragment
To determine whether the function of PEX26 necessary for peroxisome biogenesis can also be mapped to a smaller segment in the NTD, we constructed a series of deletion mutants with an N-terminal 3× myc-tag. We verified the expression of the deletion mutants in the cytosol of PBD059 cells, with indirect immunofluorescence staining against the myc-tag. The smallest fragment able to complement PBD059 cells comprised aa 2–174 (53% and 56% relative rescue of GFP-SKL import over PEX26-FL in two trials). PEX26 fragments comprising aa 2–163 or less had no complementing activity. Using N-terminal deletions, we found that a peptide comprising aa 29–174 retained 36% relative-rescue activity, whereas fragments with aa 69–174 or smaller did not complement PBD059 cells (data not shown). Thus, the functional domain of PEX26 corresponds to aa 29–174—or nearly exactly to the PEX6-binding domain.
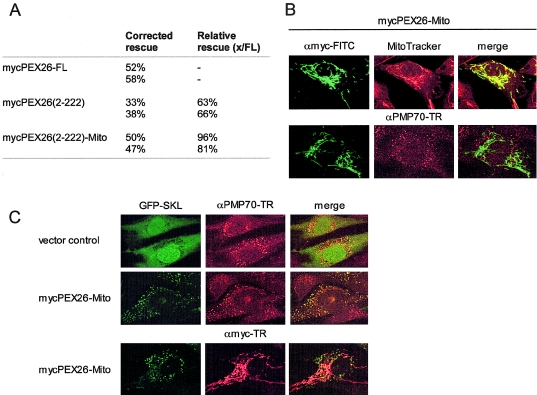
Expression of PEX26 on the Outer Mitochondrial Membrane
The results of our binding and splicing studies show that PEX26 does not require a peroxisomal location to rescue-matrix–protein import. To further test the function of extraperoxisomal PEX26, we elected to localize it to a discrete and remote site, the outer mitochondrial membrane. To do this, we expressed a chimeric protein consisting of an N-terminus of Nmyc-tagged PEX26 (aa 2–222) and a C-terminus of rat OMP25 (aa 170–206) (PEX26-Mito) in PBD059 cells. Targeting to the mitochondrial outer membrane with an Nout-Cin orientation for N-terminal chimeric proteins, with use of this fragment of rat OMP25, has been described elsewhere (Horie et al. 2002). We found that PEX26-Mito is localized exclusively on mitochondria (fig. 7), which is consistent with these earlier studies. Surprisingly, despite this ectopic location, expression of PEX26-Mito in PBD059 cells resulted in complete restoration of peroxisome matrix-protein import. Relative-rescue activity of PEX26-Mito compared with PEX26-FL was >80% and was even better than that of cytosolic mycPEX26-Δex5+6 (aa 2–222) in these experiments (63%–66%) (fig. 7).
Figure 7.
Expression of PEX26-Mito rescues the import defect of PBD059 cells. A, Relative rescue activity (partial plus complete) for a GFP-PTS1–tagged marker of mycPEX26 (aa 2–222) and PEX26-Mito (mycPEX26 [aa 2–222]–OMP25 [aa170–206]) in PBD059 cells. Values for two separate experiments are shown. Transfection of pcDNA3 vector alone as a control gave ∼10% import of GFP-SKL. B, PBD059 cells transfected with PEX26-Mito and analyzed by indirect immunofluorescence by use of anti-myc antibodies (FITC), anti-PMP70 antibodies (Texas Red), and Mito-Tracker (red) to confirm the mitochondrial localization of PEX26-Mito. C, PBD059 cells transfected with GFP-SKL and nonrecombinant pcDNA3 vector alone or recombinant pcDNA3 expressing PEX26-Mito, analyzed by indirect immunofluorescence staining with anti-PMP70 antibodies (red) or anti-myc antibodies (red) to show that peroxisomal import of GFP-SKL is rescued by PEX26-Mito expression.
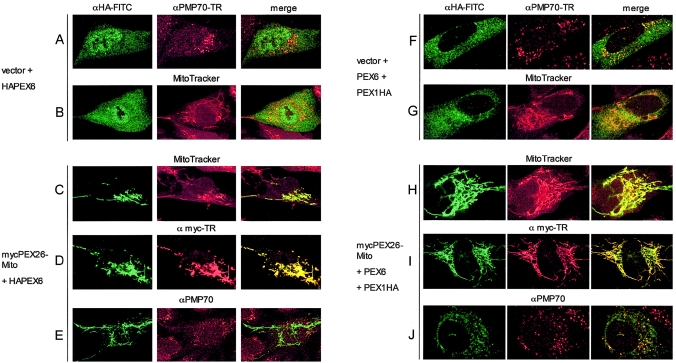
Localization of PEX6 and PEX1 in PEX26-Deficient Cells Expressing Functional PEX26-Mito
Previous work showed that PEX26 binds PEX6 directly and PEX1 indirectly through the PEX6 interaction (Matsumoto et al. 2003b). Our data show that localization to peroxisomes is not essential for PEX26 function and that the minimal PEX26 fragment necessary to complement CG8 cells retains the ability to bind PEX6. To understand how PEX26 functions in an extraperoxisomal location, we determined the localization of PEX6 and PEX1 in PBD059 cells complemented with PEX26-Mito (fig. 8). Colocalization of these AAA ATPases with PEX26-Mito would support the hypothesis that PEX26-PEX6-PEX1 interaction is essential for peroxisome matrix-protein import but would be surprising, because current models predict that the PEX1-PEX6 hetero-oligomer acts at the peroxisomal membrane; for example, in the disassembly of PEX5-PTS1 and PEX7-PTS2 complexes just prior to translocation of PTS1 and PTS2 proteins into the organelle (Gould and Collins 2002). In PBD059 cells cotransfected with empty vector and 3× HA-tagged PEX6 (JM721), we found that PEX6 localizes to the cytosol and does not accumulate on the membranes of either peroxisomes or mitochondria (fig. 8, A and B). Addition of PEX26-Mito results in a dramatic change, with complete colocalization of PEX6 with PEX26-Mito on the mitochondria (fig. 8, C and D). Under these conditions, we detected no colocalization of PEX6 with PMP70 on peroxisomal membranes (fig. 8, E).
Figure 8.
Expression of functional PEX26-Mito completely mislocalizes PEX6 and partially mislocalizes PEX1 to mitochondrial membranes. We analyzed PBD059 cells (c.230+1G→T/c.230+1G→T) by indirect immunofluorescence with anti-HA-FITC and anti-myc-Texas Red, as indicated. To mark peroxisomal or mitochondrial membranes, we used anti-PMP70 (Texas Red) or Mito-Tracker labeling (red), respectively. Yellow color in the merged column indicates colocalization. A–E, PBD059 cells were transfected with pcDNA3-HAPEX6 and either nonrecombinant pcDNA3 vector or recombinant pcDNA3 expressing mycPEX26-Mito (mycPEX26 [aa 2–222])–OMP25 [aa 170–206]). Panels A and B show a lack of colocalization of HAPEX6 with peroxisomes (A) or mitochondria (B) in cells cotransfected with pcDNA3 vector plus a recombinant vector expressing HAPEX6. By contrast, coexpression of mycPEX26-Mito redirects HAPEX6 to mitochondrial membranes (C), where it colocalizes with PEX26-Mito (D) but not with peroxisomes (E). In panels F–J, PBD059 cells were transfected with pcDNA3-PEX6 and pcDNA3-PEX1HA plus either nonrecombinant pcDNA3 (F and G) or recombinant pcDNA3 expressing mycPEX26-Mito (H–J). Panels F and G show a bimodal distribution of PEX1HA in the cytoplasm and on peroxisomes but no colocalization with mitochondria (G). Coexpression of mycPEX26-Mito redirects PEX1HA to mitochondrial membranes to a varying degree (H–J), with a fraction of PEX1 retaining a residual peroxisomal location (J). In multiple experiments, the amount of PEX1 on peroxisomes varied; in panel J, we show a typical ratio of peroxisomal to mitochondrial location of PEX1 from observations in three independent cotransfection experiments.
Interestingly, PBD059 cells transfected with PEX1-3× HA (pBER86)—untagged PEX6 and empty vector—show a bimodal distribution of PEX1, with most in the cytosol but some on peroxisomal membranes, as marked by PMP70 (fig. 8, F). This result suggests that a fraction of PEX1 localizes to the peroxisome membrane despite the lack of detectable PEX26 and PEX6 in this location. Addition of PEX26-Mito dramatically alters the localization of PEX1, with most now colocalizing with PEX26-Mito on the mitochondrial outer membrane (fig. 8, H and I). Interestingly, in contrast to PEX6, a fraction of PEX1 retains a residual peroxisomal localization in cells cotransfected with PEX26-Mito (fig. 8, J).
We tested 3× HA-PEX6 and PEX1-3× HA for their ability to complement PEX6- and PEX1-deficient cell lines, respectively. In two independent experiments, they maintained 52%–62% and 61%–77% relative-rescue activity, respectively, compared with the untagged proteins for import of GFP-SKL (data not shown). The localization of 3× HA-PEX6 and PEX1-3×HA with and without cotransfection of PEX26-Mito was confirmed in at least three independent experiments. Although the amount varied, we found some PEX1 on peroxisomes in all cotransfection experiments with PEX26-Mito, untagged PEX6, and tagged PEX1. Panel J of figure 8 shows a cell with a typical ratio of peroxisomal to mitochondrial location of PEX1.
These results suggest that peroxisomal localization of neither PEX26 nor PEX6 is essential for peroxisome matrix-protein import. By contrast, we detected some PEX1 on peroxisomal membranes in cotransfection experiments with PEX26-Mito and PEX6. Thus, a peroxisomal location of some fraction of PEX1 may be required for matrix-protein import. The observations raise the question of how the interaction of PEX26-PEX6—and possibly of PEX1—at an extraperoxisomal localization participates in peroxisome matrix-protein import.
Discussion
The recent identification of PEX26 by Matsumoto and colleagues completed the search for the genes responsible for the 12 recognized CGs for the Zellweger spectrum subgroup of the PBDs (Matsumoto et al. 2003a, 2003b) These investigators showed that PEX26 encodes a 305-aa, 34-kD integral peroxisomal membrane protein with a single C-terminal transmembrane domain near the C-terminus, a short (∼36 aa) intraperoxisomal C-terminal tail, and long (250 aa) N-terminus exposed to the cytosol. They also showed that PEX26 binds PEX6 directly and PEX1 indirectly, through the interaction with PEX6. The latter are AAA ATPases that have been proposed to act late in the peroxisomal matrix-protein import cycle (Collins et al. 2000; Vale 2000). Together, mutations in PEX1, PEX6, and PEX26 account for a large fraction (∼70%) of all patients with PBD (Gould and Valle 2000). In this work, we have identified 5 new PEX26 mutant alleles, which brings the total to 17. Of these, six are missense mutations in the N-terminal, cytosolic 60% of the protein, a segment with greater sequence identity across the recognized PEX26 vertebrate orthologs, as compared with the remaining C-terminal segment (e.g., human compared with Danio has 38% identity vs. 24% identity, respectively) (fig. 1). We find that none of these missense mutations are common in the general population, although R98W accounts for 14 (39%) of the mutant PEX26 genes in the total collection of 18 genotyped probands with CG8 (10 patients in our study and 7 in that of Matsumoto et al., with four overlaps, and 5 in that of Steinberg et al.). The high frequency of R98W may represent a founder effect, as has been described for certain alleles in other PBD CGs (Braverman et al. 1997), or recurrent mutations at a CpG dinucleotide in codon 98 (CGG→TGG). Haplotype studies will be necessary to distinguish between these possibilities.
In patient fibroblasts expressing endogenous PEX26, our immunoblot studies indicate that all of the missense mutations we tested are associated with a substantial reduction in steady-state levels of PEX26 (fig. 3). Thus, the major pathological consequence of these mutations in vivo appears to be destabilization of PEX26, a result that is similar to the consequences of disease-associated missense mutations at other loci (Brody et al. 1992). Additionally, our immunoblot studies suggest that cells with null mutations in PEX1 or PEX6 also appear to have some reduction in PEX26 that is not simply explained by alterations in the amount of peroxisomal membrane, as indicated by the similar levels of the integral peroxisome membrane protein PEX13 in all samples (fig. 3). The explanation for this observation is uncertain, but the finding is consistent with a model in which PEX26, PEX1, and PEX6 interact in a functional unit (see below).
As an additional aspect of the cellular phenotype, we also examined the steady-state levels of the PTS1 receptor PEX5 in the PEX26-deficient cells (fig. 3). Reduced abundance of PEX5 is characteristic of PEX1- and PEX6-deficient cell lines and has been reported elsewhere in a PEX26-deficient cell line (Dodt and Gould 1996) but is not observed in human cells with primary defects in PEX2, PEX7, PEX10, PEX12, or PEX16 (Dodt and Gould 1996; Yahraus et al. 1996; Sacksteder and Gould 2000). Reduced PEX5 steady-state levels are also found in yeast cells with primary defects in Pex1 and Pex6 (Collins et al. 2000). In our studies, we found a severe reduction in PEX5 in five different PEX26-deficient cell lines, independent of their genotype and similar in extent to that we observed in PEX1- and PEX6-deficient cells (fig. 3). This result also is consistent with a model in which PEX26, -1, and -6 work together as a functional unit whose activity influences the levels of PEX5. The levels of PEX1, by contrast, in the same set of cell lines, are normal or nearly so, and we are unable to measure PEX6 levels because of lack of an appropriate antibody.
Curiously, Matsumoto et al.—using indirect immunofluorescence staining for endogenous PTS1-targeted matrix proteins (detected with an anti-peptide antibody directly against the consensus PTS1, -SKL), catalase (targeted by an atypical PTS1, -KANL), and peroxisomal thiolase (a PTS2-targeted protein)—concluded that the cellular phenotype of PEX26-deficient cells was characterized by impaired peroxisomal import of catalase and PTS2-targeted proteins with normal import of classical PTS1-targeted proteins. Given that the clinical phenotype of primary deficiency of catalase involves either adult-onset oral ulcers or no symptoms at all (Wanders et al. 2001), this conclusion suggests that the clinical phenotype of PEX26-deficient patients would mainly reflect deficiency of PTS2-targeted proteins and, therefore, should resemble RCDP (caused by deficiency of the PTS2 receptor, PEX7) rather than the Zellweger spectrum, as described for patients with CG8 (see table 2). To address this discrepancy, we performed quantitative studies of the peroxisomal import of a set of four matrix proteins targeted by consensus (-SKL, PECI), near consensus (-SKI, HAOX1), or atypical (-KANL, catalase) PTS1-targeting signals plus a consensus PTS2-targeted protein, PAHX, in four CG8 cell lines with a variety of PEX26 genotypes. Although there is some residual import, our results show a clear and substantial defect in all three PTS1-targeted reporters, most severe for catalase but substantial for PECI and HAOX1 (14%–77% of control and 0%–30% of control, respectively, depending on cell line) (fig. 2). These results are consistent with the Zellweger spectrum clinical phenotype of PEX26-deficient patients and with a model of PEX26 function that involves interaction with PEX6 and PEX1, both of which are known to be necessary for the import of PTS1-targeted proteins. The reason(s) for the discrepancy between our results and those of Matsumoto et al. is unclear. It is possible that by using an antibody expected to detect any matrix protein with a classic (-SKL) PTS1-targeting signal, some or all of which may have a low level of residual import in PEX26-deficient cell lines, Matsumoto et al. may have underestimated the effect of PEX26 mutations on PTS-1–mediated import. Our studies also show a rough correlation of PEX26 genotype with cellular phenotype. The predicted null PBD059 (c.230+1G→T/c.230+1 G→T genotype) cells show the most severe defect in matrix-protein import and two cell lines homozygous for R98W showing the mildest defect, with some residual import for all four reporters. Moreover, there is a suggestion of correlation of genotype with clinical phenotype. PBD059 had ZS, the most severe clinical phenotype, whereas seven probands either homozygous or heterozygous for R98W had milder phenotypes, ranging from NALD to IRD (table 2).
At the mRNA level, we observed extensive alternative splicing of PEX26 transcripts in RNA isolated from fibroblasts and in multiple human tissues (fig. 4). The most informative of these alternatively spliced transcripts skips exon 5 and encodes a PEX26 isoform of 256 aa that lacks the transmembrane domain of the full-length protein (fig. 1). If, as previously suggested, the function of PEX26 is to recruit the PEX1/PEX6 heterodimer to peroxisomes, this splice form seemed likely to encode a functionally inactive form of PEX26. Surprisingly, our expression studies showed this isoform is functionally active despite being located in the cytosol (fig. 5). This result shows that a peroxisomal location is not required for PEX26 function and that the essential function(s) of PEX26 in peroxisome biogenesis involves more than recruitment of PEX6 to the organelle (see below).
Although we find no sequence similarity between vertebrate PEX26 and S. cerevisiae Pex15 (see above), they share a type II membrane protein topology, and both have been described as a PEX1/PEX6 anchoring site at the peroxisomal membrane (Birschmann et al. 2003; Matsumoto et al. 2003a). In an attempt to establish additional functional similarities between human PEX26 and S. cerevisiae Pex15, we asked if a cytosolic form of Pex15 is functional in peroxisome biogenesis. We tested two TMD-less constructs of PEX15 (aa 2–330 and aa 2–260). In contrast to our results with PEX26 in human cells and with full-length PEX15 (aa 2–283) in S. cerevisiae, we found that the transmembrane domain–less, cytosolic form of Pex15 did not rescue peroxisome matrix-protein import in a Δpex15 yeast strain (I.C., S.W., S.J.G, and D.V., unpublished results).
Estimating from our RT-PCR results, the ratio of PEX26-Δex5 transcript to full length is ∼1:2 and seemed similar in all the tissues we examined, with the possible exception of a lower amount in brain (fig. 4). In immunoblot experiments, however, we did not detect the shorter PEX26 isoform (predicted molecular weight of 28 vs. 34 kD for full-length PEX26). This suggests that either the shorter transcript is not translated with the same efficiency as for the full-length transcript or the shorter isoform of PEX26 is less stable. Additional studies determining the relative ratio of these transcripts and the amount of the short PEX26 isoform in various cells and tissues in response to metabolic and/or developmental perturbations will be required to determine potential physiologic consequences of this alternative splicing.
The other alternatively spliced PEX26 transcript we tested, PEX26-Δex2, lacks the normal translation start site and did not encode a functional PEX26 isoform. This result indicates that translation initiation at the most N-terminal internal methionine (Met96) either does not occur or produces an N-terminal–truncated protein that is unstable and/or nonfunctional. We favor the latter possibility. Met96 has an apparently acceptable Kozak consensus and might reasonably be expected to initiate translation. However, our functional studies (see below) and the N-terminal clustering of conserved sequence blocks and disease-producing missense mutations indicate that the N-terminus of PEX26 is essential for its function. These observations and conclusions contrast with those of Matsumoto et al., who report some residual activity for the PEX26-M1T allele expressed in PEX26-deficient CHO cells (ZP167) (Matsumoto et al. 2003b). The explanation for this difference is not clear, except possibly that the M1T transcript and/or its protein product behave differently in the CHO cell environment, as compared with our experiments in patient fibroblasts.
What do these results say about PEX26 function? First, our C- and N-terminal truncation studies of PEX26, mislocalized to the nucleus, delimit the PEX6-binding domain of PEX26 to the N-terminal half of the protein (residues 29–163). This result is consistent with the higher degree of sequence conservation among the vertebrate PEX26 orthologs in this segment of the protein and with the clustering of the disease-producing missense mutation to the same region. Moreover, our functional studies show that the minimal segment of PEX26 required to rescue peroxisome biogenesis in CG8 cells (residues 2–174) corresponds almost precisely to the minimal segment required for PEX6 binding. Additionally, we find that PEX26 undergoes alternative splicing, and, surprisingly, a splice form lacking the transmembrane (PEX26-Δex5) is fully functional, despite a cytosolic rather than peroxisomal location. The conclusion that PEX26 does not require peroxisomal location to restore biogenesis is substantiated by our studies with a chimeric protein (PEX26-Mito) comprising the N-terminal half of PEX26 and a C-terminal segment from a rat mitochondrial outer membrane protein that is exclusively (within the limits of detection by indirect immunofluorescence methods) localized to mitochondria. In this extraperoxisomal location, PEX26-Mito sequesters PEX6 and a substantial fraction of PEX1 (fig. 8). Despite the mislocalization of these peroxins, we find that peroxisome matrix-protein import is restored by the expression of PEX26-Mito in PEX26-deficient cells (fig. 7). Although the interaction of PEX26 with PEX6 is essential for matrix-protein import, our results indicate that neither protein requires a peroxisomal location to perform its function in peroxisome biogenesis.
Ultimately, understanding the role of PEX26 in peroxisome biogenesis will require better understanding of the functions of PEX1 and PEX6. Observations in yeast and humans suggest that these two, closely related, type II AAA ATPases (Vale 2000; Frickey and Lupas 2004) form a dynamic hetero-oligomeric complex whose subunit interactions depend on ATP binding and whose function depends on ATP hydrolysis (Faber et al. 1998b; Tamura et al. 1998; Kiel et al. 1999). For example, in the yeast Pichia pastoris, PEX1 and PEX6 coimmunoprecipitate only in the presence of ATP or ATPγS. In this regard, PEX1 and PEX6 appear to resemble a few AAA ATPases (e.g., VPS4 and katanin) that cycle between monomeric and oligomeric forms and contrast to other AAA ATPases (e.g., NSF, p97) that form stable two-tiered hexameric rings (Vale 2000). Consistent with the hypothesis that the interaction between PEX1 and PEX6 is dynamic, their subcellular localizations appear to be overlapping but not identical. Using epitope-tagged constructs with expression driven by a strong promoter in wild-type cells, we found that PEX6 appears to be exclusively cytosolic, whereas PEX1 is mainly cytosolic but with detectable signal also colocalizing with peroxisomes. Similar overlapping but nonidentical subcellular localizations were described for Pichia pastoris PEX1 and PEX6 (Faber et al. 1998b). Moreover, our results with PEX26-Mito showing full restoration of peroxisome matrix-protein import in PEX26-deficient cells despite mislocalization of PEX26, PEX6, and a fraction of PEX1 to the mitochondrial outer membrane predict that some protein is able to move from the PEX26-PEX6 complex on the mitochondrial outer membrane to the peroxisome. PEX1 is the most obvious candidate, given that its interaction with PEX6 appears to be dynamic and that we find PEX1 at both mitochondrial and peroxisomal locations. We also speculate that, in these mislocalization experiments, PEX1 cycles between a location on the peroxisomal membrane and the complex with PEX6 tethered to the mitochondrial outer membrane by PEX26-Mito. We also speculate that the interaction with PEX6 serves to “activate” PEX1, perhaps through some conformational change in a manner that allows it to complete its function for matrix-protein import after it relocates to the peroxisome membrane. This function could involve disassembly of the matrix-protein–receptor complex or release of the N-terminus of PEX5 from the peroxisome membrane (Costa-Rodrigues et al. 2004) or some other, as yet unappreciated, function. Consistent with this model, Shiozawa et al. (2004) have recently examined the structure of the NTD of PEX1 (aa 1–180). Using bioinformatics analyses, they noted that the NTD of PEX1 but not that of PEX6 resembles that of two other AAA ATPases, NSF and p97. Using crystallographic studies at a 2.05-Å resolution, they showed that the NTD of PEX1 folds independently as a globular domain, with striking similarities to the structure of the NTDs of NSF and p97. On the basis of similarity to the NTDs of NSF and p97, Shiozama et al. predict that the PEX1 NTD functions in substrate binding and/or as a site for the binding of a hypothetical adaptor protein. Our hypothesized “activation” of PEX1 might involve a conformation change in the NTD similar to those involved in the function of NSF or p97 (Brunger and DeLaBarre 2003; Shimozawa et al. 2004; Whiteheart and Matveeva 2004). A corollary of our model is that activated PEX1 has some mechanism for localizing to the peroxisome membrane independent of the PEX26-PEX6 complex, possibly by docking to a site on one of the integral peroxisomal membrane proteins or to a site on PEX5, which appears to insert into the peroxisomal membrane as it delivers peroxisomal matrix proteins to the organelle but leaves its N-terminus exposed (Gouveia et al. 2003; Oliveira et al. 2003; Costa-Rodrigues et al. 2004). Given that the release of unloaded PEX5 from the organelle requires ATP hydrolysis (Dodt and Gould 1996; Oliveira et al. 2003), some interaction with PEX1 at this stage seems likely.
Acknowledgments:
We acknowledge helpful discussions and the long-term interest in PBDs of Ann and Hugo Moser. S.W. was funded by German Bundesministerium fur Bildung und Forschung grant BMBF-LPD 9901/8-51. David Valle is an Investigator in the Howard Hughes Medical Institute. We thank Sandy Muscelli for assistance in manuscript preparation.
Electronic-Database Information
The accession number and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for PEX26 cDNA [accession number NM_017929])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for RCDP, ZS, NALD, and IRD)
References
- Bell GI, Najarian RC, Mullenbach GT, Hallewell RA (1986) cDNA sequence coding for human kidney catalase. Nucl Acids Res 14:5561–5562 [PMC free article] [PubMed] [Google Scholar]
- Birschmann I, Rosenkranz K, Erdmann R, Kunau WH (2005) Structural and functional analysis of the interaction of the AAA-peroxins Pex1p and Pex6p. FEBS J 272:47–58 [DOI] [PubMed] [Google Scholar]
- Birschmann I, Stroobants AK, van den Berg M, Schafer AI, Rosenkranz AR, Kunau WH, Tabak HF (2003) Pex15p of Saccharomyes cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6 to peroxisomal membranes. Mol Biol Cell 14:2226–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N, Steel G, Obie C, Moser A, Moser H, Gould SJ, Valle D (1997) Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet 15:369–376 [DOI] [PubMed] [Google Scholar]
- Brody LC, Mitchell GA, Obie C, Michaud J, Steel G, Fontaine G, Robert M-F, Kaiser-Kupfer MI, Valle D (1992) Ornithine-δ-aminotransferase mutations causing gyrate atrophy: allelic heterogeneity and functional consequences. J Biol Chem 267:3302–3307 [PubMed] [Google Scholar]
- Brunger AT, DeLaBarre B (2003) NSF and p97/VCP: similar at first, different at last. FEBS Lett 555:126–133 [DOI] [PubMed] [Google Scholar]
- Chang C-C, Lee W-H, Moser H, Valle D, Gould SJ (1997) Isolation of the human PEX12 gene, mutated in group 3 of the peroxisome biogenesis disorders. Nat Genet 15:385–388 [DOI] [PubMed] [Google Scholar]
- Chang C-C, South S, Warren D, Jones J, Moser AB, Moser HW, Gould SJ (1999) Metabolic control of peroxisome abundance. J Cell Sci 112:1579–1590 [DOI] [PubMed] [Google Scholar]
- Collins CS, Kalish JE, Morrell JC, Gould SJ (2000) The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol 20:7516–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Rodrigues J, Carvalho AF, Gouveia AM, Fransen M, Sa-Miranda C, Azevedo JE (2004) The N terminus of the peroxisomal cycling receptor, Pex5p, is required for redirecting the peroxisome-associated peroxin back to the cytosol. J Biol Chem 279:46573–46579 [DOI] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet 9:115–124 [DOI] [PubMed] [Google Scholar]
- Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Elgersma Y, Heyman JA, Koller A, Luers GH, Nuttley WM, Terlecky SR, Wenzel TJ, Subramani S (1998a) Use of Pichia pastoris as a model eukaryotic system: peroxisome biogenesis. Methods Mol Biol 103:121–147 [DOI] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S (1998b) Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol 18:936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T, Lupas AN (2004) Phylogenetic analysis of AAA proteins. J Struct Biol 146:2–10 [DOI] [PubMed] [Google Scholar]
- Gärtner J, Moser H, Valle D (1992) Mutations in the 70K peroxisomal membrane protein gene in Zellweger syndrome. Nat Genet 1:16–23 [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Collins CS, Reuber BE, Gould SJ (1998) Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA 95:8630–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht BV, Gould SJ (1999) The human PICD gene encodes a cytoplasmic and peroxisomal NADP+-dependent isocitrate dehydrogenase. J Biol Chem 274:30527–30533 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Collins CS (2002) Peroxisomal-protein import: is it really that complex? Nat Rev Mol Cell Biol 3:382–389 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S (1989) A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol 108:1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Subramani S (1988) Identification of peroxisomal targeting signals at the carboxy-terminus of four peroxisomal proteins. J Cell Biol 107:897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Raymond GV, Valle D (2001) The peroxisome biogenesis disorders. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 3181–3217 [Google Scholar]
- Gould SJ, Valle D (2000) Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet 16:340–344 [DOI] [PubMed] [Google Scholar]
- Gouveia AM, Guimarães CP, Oliveira ME, Reguenga C, Sá-Miranda C, Azevedo JE (2003) Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J Biol Chem 278:226–232 [DOI] [PubMed] [Google Scholar]
- Hahn SH, Lee EH, Jung JW, Hong CH, Yoon HR, Rinaldo P, Sims H, Gibson B, Strauss AW (1999) Very long chain acyl coenzyme A dehydrogenase deficiency in a 5 month old Korean boy: identification of a novel mutation. J Pediatr 135:250–253 [DOI] [PubMed] [Google Scholar]
- Horie C, Suzuki H, Sakaguchi M, Mihara K (2002) Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol Biol Cell 13:1615–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Morrell JC, Gould SJ (2000) Identification and characterization of HAOX1, HAOX2, and HAOX3, three human peroxisomal 2-hydroxy acid oxidases. J Biol Chem 275:12590–12597 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Multiple distinct targeting signals in integral peroxisomal membrane proteins. J Cell Biol 153:1141–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Nau K, Geraghty MT, Erdmann R, Gould SJ (1999) Identification of peroxisomal acyl-CoA thioesterases in yeast and humans. J Biol Chem 274:9216–9223 [DOI] [PubMed] [Google Scholar]
- Kiel JA, Hilbrands RE, van der Klei IJ, Rasmussen SW, Salomons FA, van der Heide M, Faber KN, Cregg JM, Veenhuis M (1999) Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast 15:1059–1078 [DOI] [PubMed] [Google Scholar]
- Kozak LP, Harper M-E (2000) Mitochondrial uncoupling proteins in energy expenditure. Annu Rev Nutr 20:339–363 [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Robbi M, Fujiki Y, Wong L (1982) Biogenesis of peroxisomal proteins in vivo and in vitro. Ann NY Acad Sci 386:285–300 [DOI] [PubMed] [Google Scholar]
- Liu Y, Björkman J, Urquhart A, Wanders RJA, Crane DI, Gould SJ (1999) PEX13 is mutated in complementation group 13 of peroxisome-biogenesis disorders. Am J Hum Genet 65:621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12:746–753 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Tamura S, Fujiki Y (2003a) The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 5:454–460 [DOI] [PubMed] [Google Scholar]
- Matsumoto N, Tamura S, Furuki S, Miyata N, Moser A, Shimozawa N, Moser HW, Suzuki Y, Kondo N, Fujiki Y (2003b) Mutations in novel peroxin gene PEX26 that cause peroxisome-biogenesis disorders of complementation group 8 provide a genotype-phenotype correlation. Am J Hum Genet 73:233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermid HE, Morrow BE (2002) Genomic disorders on 22q11. Am J Hum Genet 70:1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalik SJ, Morrell JC, Kim D, Sacksteder K, Watkins PA, Gould SJ (1997) Identification of PAHX, a Refsum-disease gene. Nat Genet 17:185–189 [DOI] [PubMed] [Google Scholar]
- Oliveira ME, Gouveia AM, Pinto RA, Sa-Miranda C, Azevedo JE (2003) The energetics of Pex5p-mediated peroxisomal protein import. J Biol Chem 278:39483–39488 [DOI] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T (1991) Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Comm 181:947–954 [DOI] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB (1996) Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol 134:849–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber BE, Collins CS, Germain-Lee E, Morrell JC, Ameritunga R, Moser HW, Valle D, Gould SJ (1997) Mutations in PEX1 are the most common cause of Zellweger syndrome, neonatal adrenoleukodystrophy and infantile Refsum disease. Nat Genet 17:445–448 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Gould SJ (2000) The genetics of peroxisome biogenesis. Annu Rev Genet 34:623–652 [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ (2000) PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol 148:931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani N, Jimenez-Sanchez G, Steel G, Dean M, Valle D (1997) Identification of a fourth half ABC transporter in the human peroxisomal membrane. Hum Mol Genet 6:1925–1931 [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Tsukamoto T, Nagase T, Takemoto Y, Koyama N, Suzuki Y, Komori M, Osumi T, Gootjes J, Wanders RJ, Kondo N (2004) Identification of a new complementation group of the peroxisome biogenesis disorders and PEX14 as a mutated gene. Hum Mutat 23:552–558 [DOI] [PubMed] [Google Scholar]
- Shiozawa K, Maita N, Tomii K, Seto A, Goda N, Akiyama Y, Shimizu T, Shirakawa M, Hiroaki H (2004) Structure of the N-terminal domain of PEX1 AAA-ATPase. J Biol Chem 279:50060–50068 [DOI] [PubMed] [Google Scholar]
- South ST, Sacksteder KA, Li X, Liu Y, Santos M, Gould SJ (2000) Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol 149:1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S, Chen L, Wei L, Moser A, Moser H, Cutting G, Braverman N (2004) The PEX gene screen: molecular diagnosis of peroxisome biogenesis disorders in the Zellweger syndrome spectrum. Mol Genet Metab 83:252–263 [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J 10:3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Shimozawa N, Suzuki Y, Tsukamoto T, Osumi T, Fujiki Y (1998) A cytoplasmic AAA family peroxin, Pex1p, interacts with Pex6p. Biochem Biophys Res Commun 245:883–886 [DOI] [PubMed] [Google Scholar]
- Vale RD (2000) AAA proteins: lords of the ring. J Cell Biol 150:F13–F19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ (2004a) Metabolic and molecular basis of peroxisomal disorders: a review. Am J Med Genet A 126:355–375 [DOI] [PubMed] [Google Scholar]
- ——— (2004b) Peroxisomes, lipid metabolism and peroxisomal disorders. Mol Genet Metab 83:16–27 [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Barth PG, Heymans HSA (2001) Single peroxisomal enzyme deficiencies. In: Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The metabolic and molecular bases of inherited disease. McGraw-Hill, New York, pp 3219–3256 [Google Scholar]
- Warren DS, Morrell JC, Moser HW, Valle D, Gould SJ (1998) Identification of PEX10, the gene defective in complementation group 7 of the peroxisome-biogenesis disorders. Am J Hum Genet 63:347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller S, Gould SJ, Valle D (2003) Peroxisome biogenesis disorders. Annu Rev Genomics Hum Genet 4:165–211 [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Matveeva EA (2004) Multiple binding proteins suggest diverse functions for the N-ethylmaleimide sensitive factor. J Struct Biol 146:32–43 [DOI] [PubMed] [Google Scholar]
- Yahraus T, Braverman N, Dodt G, Kalish JE, Morrell JC, Moser HW, Valle D, Gould SJ (1996) The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J 15:2914–2923 [PMC free article] [PubMed] [Google Scholar]