Abstract
Perturbations of the p53 pathway are associated with more aggressive and therapeutically refractory tumors. However, molecular assessment of p53 status, by using sequence analysis and immunohistochemistry, are incomplete assessors of p53 functional effects. We posited that the transcriptional fingerprint is a more definitive downstream indicator of p53 function. Herein, we analyzed transcript profiles of 251 p53-sequenced primary breast tumors and identified a clinically embedded 32-gene expression signature that distinguishes p53-mutant and wild-type tumors of different histologies and outperforms sequence-based assessments of p53 in predicting prognosis and therapeutic response. Moreover, the p53 signature identified a subset of aggressive tumors absent of sequence mutations in p53 yet exhibiting expression characteristics consistent with p53 deficiency because of attenuated p53 transcript levels. Our results show the primary importance of p53 functional status in predicting clinical breast cancer behavior.
Keywords: microarray, expression analysis, tumor profiling, class prediction
The p53 tumor suppressor is a critical regulator of tissue homeostasis, and its inactivation at the gene or protein level confers cellular properties conducive for oncogenesis and cancer progression. Mutations in p53 occur in >50% of human cancers (1, 2), and the mutational status of p53 is prognostic in many malignancies (3). In breast cancer, p53 mutations are associated with worse overall and disease-free survival, independent of other risk factors (4), and have been implicated in resistance to anticancer therapies (5-11). These observations, however, have been inconsistent (12, 13), owing, in part, to the variable accuracy of the methods to ascertain p53 status, variation in disease severity attributable to the different forms of p53 mutation, and studies of insufficient size (8, 11, 14). Further confounding the association between p53 status and patient risk is the growing number of alternative molecular mechanisms (e.g., MDM2) that compromise p53 function.
In this study, we explored the possibility that a gene-expression signature, derived from differences between p53 mutant (mt) and wild-type (wt) breast tumors, could provide a more accurate measure of the functional configuration of p53, thereby improving its prognostic utility. Using oligonucleotide microarrays covering >30,000 genes, we analyzed the global transcript levels of 251 primary invasive breast tumors for which we have detailed information on p53 status, as determined by cDNA sequencing (6) and pursued a validation strategy of intersecting alternative array data sets. We found that, in most cases, tumors with mt and wt p53 can readily be distinguished by their expression profiles and that a 32-gene p53 signature is consistently associated with patient survival in different patient subsets, independent of other risk factors, and is a superior prognostic and predictive indicator, compared with p53 mutation status alone.
Methods
Patients and Specimens. Frozen tissue was collected from 315 consecutively presented primary breast cancers representing 65% of all those resected in Uppsala County, Sweden, from January 1, 1987 to December 31, 1989 (6). Of these tissues, 251 were comprised predominantly of diseased tissue, were sequenced for p53 (6), and yielded sufficient RNA for array analysis. Clinicopathological variables measured at diagnosis were obtained from patient records and are described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. This microarray study was approved by the ethical committee at the Karolinska Institute, Stockholm, Sweden.
Expression Profiling. Total RNA was extracted from samples by using RNEasy Mini kit (Qiagen, Hilden, Germany) and evaluated on a 2100 Bioanalyzer (Agilent Technologies). In vitro transcription products were prepared from 2-5 μg of total RNA, hybridized to the Affymetrix U133 A and B arrays and washed and scanned according to the manufacturer's instructions.
Microarray Data Processing. Raw data were normalized by using the global mean method. Probe-set signal values were natural log transformed and scaled by adjusting the mean intensity to a target signal value of log 500. Samples with suboptimal average signal intensities (i.e., scaling factors >3.5) or GAPDH 3′/5′ ratios >3.5 were relabeled and rehybridized on new arrays. If visible artifacts were observed, the same cRNA was rehybridized on new chips.
Class Prediction. For gene selection, we fit a linear model to the expression data with expression level as the response and p53 status, estrogen-receptor (ER) status, and grade status as the predictor variables. As an initial filter, we excluded genes with a P value for model fit >0.001 and ranked genes in decreasing order of the absolute value of the p53 status coefficient. For class prediction, we evaluated several supervised learning methods, including diagonal linear discriminant analysis (15), k nearest neighbors (16), and support vector machines (17), as described in Supporting Materials and Methods.
Data Analysis. For all hierarchical cluster analyses, log expression values of each gene were mean centered, and genes and tumors were clustered by using Pearson correlation and average linkage (cluster and treeview software, http://rana.lbl.gov/EisenSoftware.htm).
The Kaplan-Meier estimate was used to compute survival curves, and the P value of the likelihood-ratio test was used to assess statistical significance of the hazard ratios. All patients with contralateral or bilateral cancers were omitted, and patients who died of their cancer 10 years after diagnosis were systematically censored.
For association tests, the χ2 test was used, unless the number of events was <5 in any category, in which case Fisher's exact test was used.
Cox regression was used to confirm the prognostic significance of the p53 classifier in multivariate analyses. The initial model, comprising all conventional predictors, and p53 mutation status and the p53 signature as competing measures of p53 activity, was simplified by using a stepwise model-selection procedure based on the Akaike information criterion. Remaining predictors were assessed by likelihood-ratio test.
Independent Datasets. The Sørlie et al. (18) and Chen et al. (19) data and clinical annotations were obtained from the Stanford microarray database by using filtering parameters as described by the authors. The Ma et al. (20) “whole tumor” data set was downloaded from the Gene Expression Omnibus with accession no. GSE1379, and each array was mean centered. The van't Veer et al. (21) data and survival annotation were accessed through the Rosetta Inpharmatics publications archive. All image clone IDs or GenBank accession nos. of array probes were mapped to UniGene build no. 167.
Results
P53 Mutant and WT Tumors Are Molecularly Distinct. Transcript profiles of 251 primary breast tumors were assessed by using Affymetrix U133 oligonucleotide microarrays. Previously, cDNA sequence analysis revealed that 58 of these tumors had p53 mutations resulting in protein-level changes, whereas the remaining 193 tumors were p53 wt (6). By unsupervised hierarchical cluster analysis, we found that p53 mt and wt tumors are distinguished by pervasive molecular differences. With the top 2,000 most variably expressed genes (selected independent of p53 status), >80% of the p53 mt tumors clustered into one branch and >70% of the p53 wts into the other (P = 5.6 × 10-13; see Fig. 5, which is published as supporting information on the PNAS web site). Importantly, this separation remained highly significant (P < 2 × 10-12) across a range of gene panels from the top 5,000 genes with highest variance to the top 125 (see Table 1, which is published as supporting information on the PNAS web site). This separation was most heavily influenced by three predominant gene clusters comprising genes involved in immune response, proliferation, and estrogen response (Fig. 5). Univariate analysis by statistical analysis of microarrays (SAM) (22) identified 6,545 Affymetrix probe sets representing ≈5,290 distinct genes whose expression patterns distinguished p53 mt and wt tumors with a false discovery rate (q value) <1% and d score (modified t statistic) >2.0 (see Table 2, which is published as supporting information on the PNAS web site), further illuminating the extensive nature of the molecular variation underlying p53 status. Topping the list of genes most highly expressed in p53 mt tumors were those with roles in cell cycle and proliferation, consistent with the observation that wt p53 has a negative regulatory effect on cell-cycle genes. The genes more highly expressed in the p53 wt tumors included uncharacterized genes, signaling molecules and transcription factors, transcriptional targets of p53, and estrogen-inducible genes.
The p53 status was also correlated with two other clinical parameters, ER status and tumor grade (Fig. 5). Within the p53 mt-rich cluster, we observed 89% of ER-negative tumors (P = 1.9 × 10-10), 79% of grade III tumors (P = 3.8 × 10-11), and only 14% of grade I tumors (P = 2.5 × 10-7). The finding that p53 mutant tumors are correlated with ER negativity and grade III status is consistent with previous reports that p53 mutations associate with ER negativity and high tumor grade (23).
A Gene Expression Classifier Predicts p53 Status in Independent Breast and Liver Cancer Data Sets. We considered the possibility that the differential expression observed between p53 mt and wt tumors might, to some extent, reflect changes in the operational configuration of the p53 pathway. We reasoned that some p53 wt tumors would be p53 deficient through mechanisms other than p53 mutation, such as MDM2 amplification or p14/ARF deletion and, thus, possess expression profiles more akin to p53 mt tumors with dysfunctional p53. To explore this possibility, we fitted a multivariate linear regression model (i.e., linear modelfit) (24) that allowed us to rank genes by their correlation with p53 status, while controlling for histologic grade and ER status. As a result, many cell-cycle genes correlated with p53 status by univariate analysis were no longer well associated (see Fig. 6, which is published as supporting information on the PNAS web site), suggesting that the transcriptional profiles of most cell-cycle genes are more related to histologic grade than to p53 status.
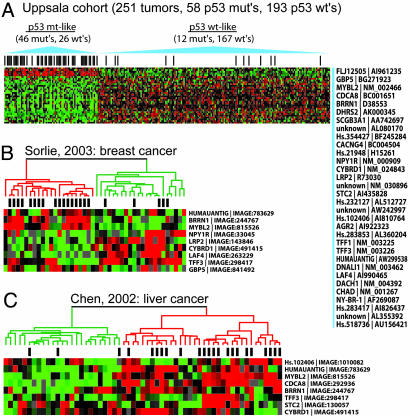
For class discrimination, we evaluated several linear learning methods including: diagonal linear discriminant analysis (DLDA) (15), k-nearest neighbors (kNN) (16), and support vector machines (SVM) (17). In each case, the optimal gene classifier was obtained by leave-one-out cross validation, where the linear model-fit procedure was iteratively applied to all samples minus the left-out sample. The resulting prediction accuracies were highly similar, ranging from 84.9% to 85.7% (see Supporting Materials and Methods). Interestingly, 20 tumors were consistently “misclassified” by all three methods (8 wt and 12 mt), indicating a surprising degree of concordance among misclassified tumors. DLDA showed the highest sensitivity for detecting p53 mutants (i.e., 79% sensitivity compared with 53% for both kNN and SVM) and was therefore selected for further analysis. By DLDA, the optimal classifier was comprised of 32 genes, whereby 26 of the wt tumors were misclassified as mutant-like, and 12 mutants were misclassified as wt-like (Fig. 1A).
Fig. 1.
The p53 signature is associated with p53 status in independent data sets. Clustergrams are oriented as outlined in Fig. 5. (A) Expression profiles of the Uppsala tumors segregated by the 32-gene signature. Unigene symbols and GenBank IDs are listed to the right. (B) P53 mt and wt breast tumors from Sørlie et al. (18) were clustered by using a nine-gene subset of the p53 signature. (C) P53 mt and wt liver tumors (predicted by immunohistochemistry) from Chen et al. (19) were clustered by using an eight-gene subset of the p53 signature. Green dendrogram branches denote tumors with the wt-like configuration; red branches indicate those with mt-like profiles. Probe IMAGE clone IDs from the original studies are listed. Black bars denote mt p53 status.
To evaluate the performance of the classifier genes (referred to hereafter as the p53 signature genes) as a clinical discriminator of p53 status, we accessed two publicly available cDNA microarray data sets where p53 mutational status was known: a breast cancer study by Sørlie et al. (18) and a liver cancer study by Chen et al. (19). In the Sørlie data set, 69 breast tumors had been sequenced for p53 mutations. Of our p53 signature genes, 28 mapped to established UniGene IDs, and more than half of these 28 genes were represented on the Sørlie et al. microarray. However, only nine were found to correspond to cDNA probes having expression measurements present in >50% of tumors, where the tumors possessed measurements for >50% of genes (resulting in a subset of 44 well sampled tumors). Because the classification rules could not be directly applied, we used this 9-gene subset of the p53 signature to hierarchically cluster the tumors in an unsupervised manner. Fig. 1B shows a significant separation of p53 mt and wt tumors: 77% of mutants clustered into one branch, and 77% of wts clustered into the other (P = 0.0003). By Monte Carlo simulations, we estimated the probability that a randomly selected nine-gene subset could cluster the samples with equivalent or better significance was P = 0.008, thus reaffirming the robust discriminative power of the p53 signature genes.
In the Chen et al. liver cancer data set (38), p53 protein levels had been ascertained by immunohistochemistry (IHC). Eight of our signature genes could be mapped to all 59 tumors assayed for p53, with each gene having data present in >90% of all tumors and where each tumor contained data for >50% of the genes. We observed that even this eight-gene subset was able to cluster the liver cancers into two primary clusters significantly correlated with p53 levels: 87% of the IHC-positive (predicted mts) in one cluster, and 61% of the predicted wts in the other (P = 0.00035) (Fig. 1C). Again, the probability of this clustering occurring by random chance was P = 0.009 by Monte Carlo P value estimation. Taken together, these observations suggest that the genes comprising the p53 signature are robust in their ability to classify not only breast tumors but also liver cancers according to their p53 mutational status and, therefore, may have generalizable utility in predicting p53 status in a range of cancer types.
Transcript Analysis of p53 Pathway Genes Corroborates Tumor Classifications. We hypothesized that the p53 expression signature may better reflect the relative intactness of p53 function in the tumor than sequence mutation status alone, implying that p53 sequence-wt tumors “misclassified” as mt-like may, in fact, be p53 deficient by other means. First, we considered the possibility that p53 deficiency could result from reduced p53 transcript levels. We compared the transcript levels of p53 among the different tumor classes (Fig. 2). We observed that the overall expression level of p53 was significantly reduced in the 26 wt tumors with mt-like signatures (referred to henceforth as the “26 mt-like” tumors), compared with the remaining 167 wt tumors classified as wt-like (P = 1.8 × 10-4), strongly suggesting that reduced p53 transcripts can result in biological consequences in vivo.
Fig. 2.
Transcript levels of p53 and its transcriptional targets are consistent with classification results. Expression levels of p53-pathway-relevant genes were examined in different tumor subgroups. The four tumor subgroups are defined as follows: (i) p53 mt tumors classified as mt-like (n = 46), (ii) p53 wt tumors classified as mt-like (n = 26), (iii) p53 wt tumors classified as wt-like (n = 167), and (iv) p53 mt tumors classified as wt-like (n = 12). Differences in transcript levels were determine by t test and are shown in a summary table to the right; P values >0.05 are shown in gray.
We further hypothesized that known transcriptional targets of p53 would show altered transcription in p53-deficient tumors. Indeed, a number of p53 target genes demonstrated expression patterns consistent with a mutant p53 status (Fig. 2). The TP53-inducible genes TP53INP1, SEMA3B, PMAIP1 (NOXA), FDXR, CCNG1, and LRDD, which all contain functional p53-binding sites in their promoters, showed significantly lower expression in the 26 mt-like tumors, compared with the other wt (all at P < 0.05). In a consistent manner, all but one of these genes were also significantly reduced in the p53 mt tumors, compared with all wt tumors. Furthermore, in all but two cases, these genes showed significantly higher expression in the set of 12 sequence-mt tumors classified as wt-like when compared with the other mts, suggesting that the p53 mutations in these 12 tumors may have a more benign effect, with respect to p53 functionality. CHEK1 and CHEK2 are both upstream effectors of p53 function known to be transcriptionally repressed by p53. Significantly, their mRNA levels were elevated in both the p53 mt and p53 mt-like classes. Again, the 12 mts classified as wt-like showed a reversed pattern, i.e., displaying significantly lower expression of these genes, compared with the other 46 p53 mutants. Together, these observations suggest that the “misclassified” tumors more correctly reflect the active/inactive status of the p53 pathway and are consistent with the notion that reduced p53 levels in breast tumors result in downstream transcriptional changes similar to those found in p53 mutations.
Of note, the canonical marker of p53 activity, CDKN1A (p21/WAF1), was only moderately higher in p53 wt tumors, compared with those with sequence mutations (P = 0.02), and not significantly lower in the 26 mt-like tumors, compared with the other wts (P = 0.09; data not shown). Furthermore, the known p53-inducible genes PERP, BAX, and SFN (14-3-3 sigma) were, paradoxically, all expressed at higher levels in the p53 mutants and the 26 mt-like tumors rather than the expected lower levels (Fig. 2). These observations may reflect cross-talk among different transcriptional regulators in the consensus of primary tissues, as compared with dynamic changes in single cell lines. For example, the p53 target genes p21 and BAX are also directly regulated by the breast cancer oncogene, c-Myc, in a manner independent of, and antagonistic to, p53 (25, 26). The regulation of p53 target genes by alternative transcriptional modifiers acting independently of p53 or in the context of p53 deficiency (e.g., PERP, BAX, and SFN) may have implications for p53 tumor-suppressor activity.
We next asked whether the mutational spectrum of p53 in our tumors could explain the different functional consequences, as measured by the expression profiles. Of the 46 p53 mt tumors correctly classified as mts, 43% (20 of 46) possessed “severe” mutations, defined as insertions (n = 2), deletions (n = 11), and stop codons (n = 7) resulting in frame shifts and truncations, whereas in the 12 p53 mutants classified as wt-like by the expression signature, only 1 contained a severe mutation, a 3-bp insertion in the DNA-binding domain, resulting in the in-frame addition of a glycine residue. Notably, this difference was statistically significant at P = 0.02. Using the IARC TP53 mutation database (ITMD) (27), we cross-compared the missense point mutations (mpms) in each tumor group with the ITMD's index of 418 mutants previously analyzed for dominant-negative function. Only 1 of the 11 mpms among the 12 wt-like mutants had been demonstrated previously to have dominant-negative activity, compared with 12 of 27 within the mt-like group (P = 0.039). Together, these data suggest that, at the sequence level, the 12 p53 mutants classified as wt-like may, in fact, represent p53 mutant forms that have less biological effect.
The p53 Signature Predicts Outcome Better Than p53 Mutation Status Alone. We next asked whether the p53 signature could predict disease-specific survival in the patients of the Uppsala cohort. The classifier separated patients into low and high risk groups with a much higher statistical significance than the sequence-based p53 status alone (P = 0.0006 versus P = 0.01, respectively) (Fig. 3 A and B). More interestingly, when the classifier was tested on the subset of women with wt p53 by sequence, we again observed a significant separation of patients by survival (P = 0.02; Fig. 3C), indicating that women with p53 sequence-wt tumors, yet exhibiting the mt-like expression signature, have a greater likelihood of dying from their cancer. Fig. 3D shows that the survival curve for this tumor type is highly similar to that of p53 mt tumors classified as mt-like (blue and green curves, respectively), whereas the 12 individuals with p53 mt tumors classified as wt-like do not have significantly unique outcomes.
Fig. 3.
The p53 classifier has greater prognostic significance than p53 mutation status alone. Kaplan-Meier survival plots for disease-specific survival are shown for patients classified according to p53 mutation status (A and E), the p53 classifier (B, C, and F), or both (D). All patients were assessed in A, B, and D. Only the patients with p53 wt tumors were assessed in C. Sixty-seven ER+, hormone-treated (TAM) patients were assessed in E and F.
To further test the clinical utility of the p53 signature, we analyzed its prognostic performance on therapy-specific treatment groups. In a subpopulation of the Uppsala cohort consisting of 67 ER+ patients who received only adjuvant hormonal therapy after surgery, the signature was a significant predictor of disease-specific survival (P = 0.05), whereas p53 mutation status alone was not (P = 0.4) (Fig. 3 E and F). Importantly, by multivariate Cox regression analysis, the p53 classifier remained significantly associated with survival in the hormone-treated group (P = 0.02), the complete cohort (P = 0.02), and the p53 wt group (P = 0.002), even when controlling for the classical predictors (ER and progesterone receptor) and prognostic factors (lymph node status, Elston grade, tumor size, and patient age), whereas the p53 mutation status, as determined by sequencing, did not. This demonstrates that the expression classifier is more directly prognostic of patient survival than is p53 mutation status alone.
The p53 Signature Predicts Outcome in Independent Therapy-Specific Data Sets. We next assessed the prognostic capability of the p53 signature genes in therapy-specific cohorts by using independent microarray data sets from the public domain (Fig. 4; and see Fig. 7, which is published as supporting information on the PNAS web site). First, we evaluated whether the signature genes were prognostic of tumor recurrence in the Ma et al. (20) data set of 60 breast tumors derived from patients treated with postoperative radiation and adjuvant tamoxifen monotherapy. In this cohort, patients with and without recurrent disease were matched with respect to tumor grade and tumor node metastasis stage. Twenty-two of the p53 signature genes mapped to 27 probes on the Ma et al. spotted oligonucleotide array. Hierarchical cluster analysis with these genes revealed two to three primary tumor clusters with expression profiles that resembled the mt-like and wt-like configurations (Fig. 4A). Using these tumor clusters to define patient survival groups, we analyzed disease-free survival (DFS) by the Kaplan-Meier estimate. As shown in Fig. 4 B and C, the clusters were significantly associated with tumor recurrence [P = 0.01 (two clusters, C1 and C2) and P = 0.005 (three clusters, C1, C2, and C3)]. Thus, concordant results in two independent studies suggest that functional p53 deficiency, as assessed by an expression readout, is predictive of outcome to hormonal therapy.
Fig. 4.
The p53 signature predicts survival in independent clinically diverse data sets. (A) Tumor dendrogram from clustering 60 tumors and 22 genes (27 probes) from Ma et al. (20). (B and C) Patient subgroups determined by the primary tumor branches (C1-C4) were analyzed for correlations with DFS. (D) Tumor dendrogram from clustering 76 tumors and 9 genes from Sørlie et al. (18). Patient subgroups defined by the primary tumor branches (C1 and C2) were analyzed for correlations with disease-specific survival (DSS) (E) and DFS (F). (G) Tumor dendrogram from clustering 97 tumors and 21 genes (25 probes) from van't Veer et al. (21). (H and I) Primary tumor clusters (C1-C5) defined patient subgroups for DFS analysis. Red branches denote tumors with the p53 mt-like signature; black branches identify those with the wt-like signature. Black triangles indicate patients who relapsed within 5 years. See Fig. 7 for gene heat maps and probe IDs.
To examine the prognostic performance of the p53 signature genes in patients treated with systemic chemotherapy, we used the Sørlie et al. cDNA microarray data set. The majority of patients (>80%) in the Sørlie study received weekly doxorubicin or 5FU and mitomycin and were comprised mostly of late-stage patients (10, 11). Here, the nine-gene partial signature that could distinguish mt and wt tumors with 77% accuracy, was used to hierarchically cluster 76 well sampled tumors with corresponding treatment and survival data (Fig. 4D). Again, we observed the tumors cluster into two primary branches with expression patterns characteristic of the wt-like and mt-like configurations. Survival analysis resulted in a highly significant difference in outcome between patients with mt-like and wt-like tumors [P = 7.5 × 10-5 (disease-specific survival) and P = 5.0 × 10-5 (DFS)]; Fig. 4 E and F) despite the small number of genes used. Notably, Fig. 4E predicts a remarkable 5-year 90% survival rate for the 31 p53 wt-like patients, compared with a 35% probability of 5-year survival for the 44 p53 mt-like patients.
Next, we tested the performance of the signature genes on a set of 97 early stage tumors (T1/T2, N0), from patients <55 years of age at diagnosis and treated by radiotherapy alone (21). From our 32-gene signature, we were able to map 25 probes corresponding to 21 signature genes to all 97 tumors with outcome information. Unsupervised clustering revealed two primary and four secondary tumor clusters (Fig. 4G) that could significantly discern patients based on time to distant metastasis within a 5-year period [Fig. 4 H and I; P = 0.0006 (two clusters, C1 and C2) and P = 0.001 (four clusters, C1, C2, C3, and C4)]. Notably, of the 24 tumors in cluster 1 (C1) that bear the molecular configuration of p53 mt-like tumors, 75% belonged to patients who developed a distant metastasis within 5 years, compared with 26% of 34 patients with tumors comprising C4 (which most closely resemble the p53 wt-like signature). These findings indicate that the p53 signature is also prognostic of recurrence in early stage, locally treated breast cancer.
The p53 Signature Genes Are Not Canonical p53 Targets. To gain some mechanistic insights, we examined the functional annotations of the signature genes for clues to explain their correlations with p53 status and patient outcome. We found that none of the signature genes are known transcriptional targets of p53, nor have they been previously implicated in the p53 pathway. Moreover, promoter analysis revealed no evidence of p53-binding sites. Of the characterized genes, a number are associated with cell growth and proliferation (MYBL2, TFF1, BRRN1, CHAD, SCGB3A1, DACH, and CDCA8), transcription (LAF4, NY-BR-1, DACH, and MYBL2), ion transport (CACNG4, CY-BRD1, and LRP2), and breast cancer biology (SCGB3A1, TFF1, STC2, NY-BR-1, and AGR2). Interestingly, MYBL2, which was transcriptionally up-regulated in the p53 mt-like tumors, is a growth-promoting transcription factor structurally related to the c-MYB oncogene. MYBL2 maps to a chromosomal region frequently amplified in breast cancer (20q13) and has previously been reported to be overexpressed in breast cancer cell lines and sporadic ovarian carcinomas (28, 29). SCGB3A1 (HIN1), which we observed to be down-regulated in the p53 mt-like tumors, is a putative tumor-suppressor gene that can inhibit breast cancer cell growth when overexpressed and has been found to be transcriptionally silenced by promoter hypermethylation in early stages of breast tumorigenesis (30). Thus, some of the p53 signature genes may contribute mechanistically to the poor prognosis associated with the p53 mt-like tumors.
Discussion
Breast cancers are characterized by multiple genetic alterations that, together, comprise the genotype that dictates tumor behavior. It is therefore reasonable that the compilation of genetic changes is a better indicator of clinical behavior than a single gene. Herein, we show that an expression signature, deduced from differences in the molecular configurations of p53 wt and mt tumors, predicts for p53 functional inactivation in primary breast cancers and provides a more accurate and useful measure of p53 clinical functionality than p53 mutation status alone. We show that, in independent data sets of both breast and liver cancers and regardless of other clinical features, subsets of the p53 signature can predict p53 status with significant accuracy. As a predictor of disease-specific survival, we found that the signature significantly outperformed p53 mutation status in a large patient cohort with heterogeneous treatment. Importantly, the p53 signature could significantly distinguish patients having more or less benefit from specific systemic adjuvant therapies and locoregional radiotherapy. Recently, Ma et al. identified by microarray analysis two genes (HOXB13 and IL17RB) whose expression ratio was predictive of tamoxifen response. Notably, we found that these genes were also predictive of disease-specific survival in the 67 Uppsala patients treated with tamoxifen monotherapy (P < 0.01; data not shown). However, these genes were not prognostic of recurrence in the van't Veer data set, nor were the van't Veer 70 genes prognostic of recurrence in the Ma data set (20), suggesting that tumor stage and/or therapeutic context is an important determinant of the prognostic capacity of some genes. In contrast, we demonstrate that the p53 signature genes are robustly prognostic of survival and recurrence in both early and late stage disease and in different therapeutic settings.
Although the p53 pathway may be compromised at some level in most human cancers, our analysis of genes involved in the p53 pathway suggests that the p53 expression signature defines some operational configuration of this pathway in breast tumors (more so than p53 mutation status alone) that impacts patient survival and therapeutic response. Recent evidence suggests that tumor sensitivity to some anti-cancer agents may depend largely on the relative intactness of p53-dependent mechanisms of apoptosis (7, 8, 10, 11) and that taxols (microtubule stabilizers), in particular, may have greater efficacy against p53-mt breast tumors than anthracycline-based (genotoxic) compounds (9). Whether the p53 classifier genes identified here are involved in some aspect of this p53 function or will have robust clinically utility as a predictor of therapeutic response warrants further investigation.
Other studies have elucidated gene expression signatures prognostic of breast cancer outcomes (21, 31). Although a 21-gene subset of our p53 signature could significantly distinguish patients with recurrent and nonrecurrent disease in the van't Veer study (21), none of these genes were found to overlap with the 231 genes identified as prognostic discriminators in the van't Veer set; and only one of the classifier genes, MYBL2, was found in the Sotiriou 485 survival-correlated genes (31). Similarly, none of the p53 signature genes were found in the top 25 relapse-associated genes reported by Ma et al. Thus, the p53 signature genes identified here represent a previously unrecognized prognostic cassette.
In cancer, it is clear that not all p53 mutations have equal effects; some simply confer loss of function, whereas others have a dominant-negative effect (such as transdominant suppression of wt p53 or oncogenic gain of function), whereas still others show only a partial loss of function where, for example, only a fraction of p53 target genes are dysregulated (32, 33). For these reasons, no single molecular assessment of p53 status appears to provide an absolute indication of the complete p53 function. Although the p53 classification method developed here seeks to categorize all tumors as either p53-deficient or not, it is likely that intermediate types exist with partial p53 functionality, distinguished by expression patterns that fall between those of the predominant mt-like and wt-like classes. Further investigation will be required to resolve the biological and clinical implications of such intermediate tumor classes.
Supplementary Material
Acknowledgments
We thank A. Ramasamy and K. R. Govindarajan for developing the database for cross-study analyses and other analytical tools used herein and Professor Hans Nordgren for Elston-grading of the tumor material. This study was supported by the Singapore Agency for Science Technology and Research and by grants from the Swedish Cancer Society, the Stockholm Cancer Society, and the King Gustav Fifth Jubilee Fund.
Abbreviations: DFS, disease-free survival; ER, estrogen receptor; mt, mutant; wt, wild type.
References
- 1.Hollstein, M., Sidransky, D., Vogelstein, B. & Harris, C. C. (1991) Science 253, 49-53. [DOI] [PubMed] [Google Scholar]
- 2.Levine, A. J., Momand, J. & Finlay, C. A. (1991) Nature 351, 453-456. [DOI] [PubMed] [Google Scholar]
- 3.Peller, S. (1998) Semin. Cancer Biol. 8, 379-387. [DOI] [PubMed] [Google Scholar]
- 4.Pharoah, P. D., Day, N. E. & Caldas, C. (1999) Br. J. Cancer 80, 1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe, S. W., Bodis, S., McClatchey, A., Remington, L., Ruley, H. E., Fisher, D. E., Housman, D. E. & Jacks, T. (1994) Science 266, 807-810. [DOI] [PubMed] [Google Scholar]
- 6.Bergh, J., Norberg, T., Sjogren, S., Lindgren, A. & Holmberg, L. (1995) Nat. Med. 1, 1029-1034. [DOI] [PubMed] [Google Scholar]
- 7.Aas, T., Borresen, A. L., Geisler, S., Smith-Sorensen, B., Johnsen, H., Varhaug, J. E., Akslen, L. A. & Lonning, P. E. (1996) Nat. Med. 2, 811-814. [DOI] [PubMed] [Google Scholar]
- 8.Berns, E. M., Foekens, J. A., Vossen, R., Look, M. P., Devilee, P., Henzen-Logmans, S. C., van Staveren, I. L., van Putten, W. L., Inganas, M., Meijer-van Gelder, M. E., et al. (2000) Cancer Res. 60, 2155-2162. [PubMed] [Google Scholar]
- 9.Kandioler-Eckersberger, D., Ludwig, C., Rudas, M., Kappel, S., Janschek, E., Wenzel, C., Schlagbauer-Wadl, H., Mittlbock, M., Gnant, M., Steger, G. & Jakesz, R. (2000) Clin. Cancer Res. 6, 50-56. [PubMed] [Google Scholar]
- 10.Geisler, S., Borresen-Dale, A. L., Johnsen, H., Aas, T., Geisler, J., Akslen, L. A., Anker, G. & Lonning, P. E. (2003) Clin. Cancer Res. 9, 5582-5588. [PubMed] [Google Scholar]
- 11.Geisler, S., Lonning, P. E., Aas, T., Johnsen, H., Fluge, O., Haugen, D. F., Lillehaug, J. R., Akslen, L. A. & Borresen-Dale, A. L. (2001) Cancer Res. 61, 2505-2512. [PubMed] [Google Scholar]
- 12.Archer, S. G., Eliopoulos, A., Spandidos, D., Barnes, D., Ellis, I. O., Blamey, R. W., Nicholson, R. I. & Robertson, J. F. (1995) Br. J. Cancer 72, 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozan, S., Vincent-Salomon, A., Zafrani, B., Validire, P., De Cremoux, P., Bernoux, A., Nieruchalski, M., Fourquet, A., Clough, K., Dieras, V., et al. (1998) Int. J. Cancer 79, 27-33. [DOI] [PubMed] [Google Scholar]
- 14.Borresen-Dale, A. L. (2003) Hum. Mutat. 21, 292-300. [DOI] [PubMed] [Google Scholar]
- 15.Dudoit, S., Frilyand, J. & Speed, T. P. (2002) J. Am. Stat. Assoc. 97, 77-87. [Google Scholar]
- 16.Ripley, B. D. (1996) in Pattern Recognition and Neural Networks (Cambridge Univ. Press, Cambridge, U.K.), pp. 181-212.
- 17.Vapnik, V. (1995) The Nature of Statistical Learning Theory (Springer, New York).
- 18.Sørlie, T., Tibshirani, R., Parker, J., Hastie, T., Marron, J. S., Nobel, A., Deng, S., Johnsen, H., Pesich, R., Geisler, S., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8418-8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, X., Cheung, S. T., So, S., Fan, S. T., Barry, C., Higgins, J., Lai, K. M., Ji, J., Dudoit, S., Ng, I. O., et al. (2002) Mol. Biol. Cell 13, 1929-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma, X. J., Wang, Z., Ryan, P. D., Isakoff, S. J., Barmettler, A., Fuller, A., Muir, B., Mohapatra, G., Salunga, R., Tuggle, J. T., et al. (2004) Cancer Cell 5, 607-616. [DOI] [PubMed] [Google Scholar]
- 21.van 't Veer, L. J., Dai, H., van de Vijver, M. J., He, Y. D., Hart, A. A., Mao, M., Peterse, H. L., van der Kooy, K., Marton, M. J., Witteveen, A. T., et al. (2002) Nature 415, 530-536. [DOI] [PubMed] [Google Scholar]
- 22.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattoretti, G., Rilke, F., Andreola, S., D'Amato, L. & Delia, D. (1988) Int. J. Cancer 41, 178-183. [DOI] [PubMed] [Google Scholar]
- 24.Draper, N. R. & Smith, H. (1998) in Applied Regression Analysis (Wiley, New York), 3rd Ed, pp. 217-234.
- 25.Mitchell, K. O., Ricci, M. S., Miyashita, T., Dicker, D. T., Jin, Z., Reed, J. C. & El-Deiry, W. S. (2000) Cancer Res. 60, 6318-6325. [PubMed] [Google Scholar]
- 26.Ceballos, E., Delgado, M. D., Gutierrez, P., Richard, C., Muller, D., Eilers, M., Ehinger, M., Gullberg, U. & Leon, J. (2000) Oncogene 19, 2194-2204. [DOI] [PubMed] [Google Scholar]
- 27.Hainaut, P., Hernandez, T., Robinson, A., Rodriguez-Tome, P., Flores, T., Hollstein, M., Harris, C. C. & Montesano, R. (1998) Nucleic Acids Res. 26, 205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forozan, F., Mahlamaki, E. H., Monni, O., Chen, Y., Veldman, R., Jiang, Y., Gooden, G. C., Ethier, S. P., Kallioniemi, A. & Kallioniemi, O. P. (2000) Cancer Res. 60, 4519-4525. [PubMed] [Google Scholar]
- 29.Tanner, M. M., Grenman, S., Koul, A., Johannsson, O., Meltzer, P., Pejovic, T., Borg, A. & Isola, J. J. (2000) Clin. Cancer Res. 6, 1833-1839. [PubMed] [Google Scholar]
- 30.Krop, I. E., Sgroi, D., Porter, D. A., Lunetta, K. L., LeVangie, R., Seth, P., Kaelin, C. M., Rhei, E., Bosenberg, M., Schnitt, S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 9796-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sotiriou, C., Neo, S. Y., McShane, L. M., Korn, E. L., Long, P. M., Jazaeri, A., Martiat, P., Fox, S. B., Harris, A. L. & Liu, E. T. (2003) Proc. Natl. Acad. Sci. USA 100, 10393-10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein, C. B., Attiyeh, E. F., Hobson, D. A., Silver, A. L., Broach, J. R. & Levine, A. J. (1998) Oncogene 16, 2115-2122. [DOI] [PubMed] [Google Scholar]
- 33.Monti, P., Campomenosi, P., Ciribilli, Y., Iannone, R., Inga, A., Abbondandolo, A., Resnick, M. A. & Fronza, G. (2002) Oncogene 21, 1641-1648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.