Abstract
We have developed a general profile for the proteins of the TetR family of repressors. The stretch that best defines the profile of this family is made up of 47 amino acid residues that correspond to the helix-turn-helix DNA binding motif and adjacent regions in the three-dimensional structures of TetR, QacR, CprB, and EthR, four family members for which the function and three-dimensional structure are known. We have detected a set of 2,353 nonredundant proteins belonging to this family by screening genome and protein databases with the TetR profile. Proteins of the TetR family have been found in 115 genera of gram-positive, α-, β-, and γ-proteobacteria, cyanobacteria, and archaea. The set of genes they regulate is known for 85 out of the 2,353 members of the family. These proteins are involved in the transcriptional control of multidrug efflux pumps, pathways for the biosynthesis of antibiotics, response to osmotic stress and toxic chemicals, control of catabolic pathways, differentiation processes, and pathogenicity. The regulatory network in which the family member is involved can be simple, as in TetR (i.e., TetR bound to the target operator represses tetA transcription and is released in the presence of tetracycline), or more complex, involving a series of regulatory cascades in which either the expression of the TetR family member is modulated by another regulator or the TetR family member triggers a cell response to react to environmental insults. Based on what has been learned from the cocrystals of TetR and QacR with their target operators and from their three-dimensional structures in the absence and in the presence of ligands, and based on multialignment analyses of the conserved stretch of 47 amino acids in the 2,353 TetR family members, two groups of residues have been identified. One group includes highly conserved positions involved in the proper orientation of the helix-turn-helix motif and hence seems to play a structural role. The other set of less conserved residues are involved in establishing contacts with the phosphate backbone and target bases in the operator. Information related to the TetR family of regulators has been updated in a database that can be accessed at www.bactregulators.org.
INTRODUCTION
Bacteria in the environment are exposed to variations in temperature and nutrient and water availability and the presence of toxic molecules that originate from their abiotic and biotic surroundings (including deleterious molecules that originate from their own metabolism). These changes can make their living conditions far from optimal. Survival in this unstable environment requires a wide range of rapid, adaptive responses which are triggered by regulatory proteins. These regulators respond to specific environmental and cellular signals that modulate transcription, translation, or some other event in gene expression, so that the physiological responses are modified appropriately (32, 52, 64, 104, 107, 145, 244, 311, 312, 326, 330, 379, 397, 409, 427).
In most cases, the adaptive responses are mediated by transcriptional regulators. Most microbial regulators involved in transcriptional control are two-domain proteins with a signal-receiving domain and a DNA-binding domain which transduces the signal (1, 18, 145, 152, 170, 207, 271, 292-294, 298, 303, 345, 369, 428, 431) (Table 1). In other cases, the sensing of signals that trigger a transcriptional process involves two proteins, as in two-component regulatory systems such as CzcR/CzcS; DcuS/DcuR; NifL/NifA; NtrB/NtrC; PhoP/PhoQ; and TodS/TodT (75, 139, 200, 206, 233, 234, 257, 307, 309, 316, 409, 423). One protein is usually a membrane-linked kinase that, upon sensing the appropriate signal, phosphorylates a DNA-binding protein that mediates transcription from its cognate promoter. Structural analyses have revealed that the helix-turn-helix (HTH) signature is the most recurrent DNA-binding motif in prokaryotic transcriptional factors, since almost 95% of all transcriptional factors described in prokaryotes use the HTH motif to bind their target DNA sequences (12, 19, 27, 41, 43, 104, 135, 136, 302, 335, 343).
TABLE 1.
Prokaryotic regulator families
| Family | Action | Some regulated functions | DBD motif | Position | Reference(s) |
|---|---|---|---|---|---|
| LysR | Activator/repressor | Carbon and nitrogen metabolism | HTH | N-terminal | 145, 342 |
| AraC/XylS | Activator | Carbon metabolism, stress response and pathogenesis | HTH | C-terminal | 109, 394 |
| TetR | Repressor | Biosynthesis of antibiotics, efflux pumps, osmotic stress, etc. | HTH | C-terminal | 9, 10, 11 |
| LuxR | Activator | Quorum sensing, biosynthesis and metabolism, etc. | HTH | C-terminal | 106, 298, 317 |
| LacI | Repressor | Carbon source utilization | HTH | N-terminal | 54, 420 |
| ArsR | Repressor | Metal resistance | HTH | Central | 49, 432 |
| IcIR | Repressor/activator | Carbon metabolism, efflux pumps | HTH | N-terminal | 265, 319, 321, 378 |
| MerR | Repressor | Resistance and detoxification | HTH | N-terminal | 144, 377 |
| AsnC | Activator/repressor | Amino acid biosynthesis | HTH | N-terminal | 103 |
| MarR | Activator/repressor | Multiple antibiotic resistance | HTH | Central | 4, 13, 352, 376 |
| NtrC (EBP) | Activator | Nitrogen assimilation, aromatic amino acid synthesis, flagella, catabolic pathways, phage response, etc. | HTH | C-terminal | 200, 257 |
| OmpR | Activator | Heavy metal and virulence (response regulator of a two-component system) | Winged helix | C-terminal | 237 |
| DeoR | Repressor | Sugar metabolism | HTH | N-terminal | 311, 405, 450 |
| Cold shock | Activator | Low-temperature resistance | RNA binding domain (CSD) | Variable | 42, 205, 344 |
| GntR | Repressor | General metabolism | HTH | N-terminal | 138, 318, 324 |
| Crp | Activator/repressor | Global responses, catabolite repression and anaerobiosis | HTH | C-terminal | 54, 110, 244 |
Prokaryotic transcriptional regulators are classified in families on the basis of sequence similarity and structural and functional criteria (49, 84, 106, 108, 120, 121, 138, 145, 146, 237, 274, 308, 313, 324, 342, 370, 377). Table 1 lists the most important families of microbial transcriptional regulators, the type of DNA binding motifs they exhibit, whether the members of the family are preferentially repressors or activators, and whether they show a dual action.
This review focuses on the TetR family, a family of transcriptional regulators that is well represented and widely distributed among bacteria with an HTH DNA-binding motif (210, 211, 246, 288).
Members of the TetR family of repressors are identified by a profile (see below) which can be easily used to recognize TetR family members in SWISS-PROT and TrEMBL and in all available proteins from prokaryotic genome sequences. After compiling data from protein and nucleic acid databases, the TetR family of regulators was found to include 2,353 nonredundant sequences (as of December 2004). The specific function regulated by members of the TetR family is known for only about 85 members (Table 2). These proteins control genes whose products are involved in multidrug resistance, enzymes implicated in different catabolic pathways, biosynthesis of antibiotics, osmotic stress, and pathogenicity of gram-negative and gram-positive bacteria (Table 2). The most relevant information on these proteins is collected in a database available at http://www.bactregulators.org (235). The database also supplies information for each member of the family, including identifiers, names, sequences, source, function, COG (clusters orthologous groups), position and orientation of the corresponding gene in the genome, and, when available, three-dimensional structures.
TABLE 2.
Specific functions regulated by members of the TetR family of repressors
| No. | SPTRa | Name | Organism | Function | Gb | Reference(s) |
|---|---|---|---|---|---|---|
| 1 | P34000 | AcrR | Escherichia coli | Represses the expression of the acrAB operon which confers multidrug resistance and probably also controls the gene micF | 1 | 102, 164, 223, 224, 314, 325, 425 |
| 2 | Q53901 | ActII | Streptomyces coelicolor | Located in the act cluster, which contains regulatory and antibiotic export genes | 1 | 50, 96 |
| 3 | Q9F8V9 | AmeR | Agrobacterium tumefaciens | Negatively regulates the ameABC operon, which encodes proteins similar to nodulation-cell division (RND)-type efflux systems | 1 | 301 |
| 4 | Q9RG61 | AmrR | Pseudomonas aeruginosa | Probably regulates amrAB genes encoding an efflux system involved in aminoglycoside impermeability phenotype in Pseudomonas aeruginosa | 1 | 423 |
| 5 | Q9KJC4 | ArpR | Pseudomonas putida S12 | Seems to be a repressor for the expression of the arpABC operon; ArpABC in Pseudomonas putida S12 is involved only in multidrug resistance and not in tolerance towards organic solvents. | 1 | 178 |
| 6 | Q6VV70 | BpeR | Burkholderia pseudomallel | Controls expression of the BpeAB-OprB efflux pump that extrudes gentamycin, streptomycin erythromycin, and acryflavine | 1 | 53 |
| 7 | P31676 | EnvR | E. coli K-12 | Regulates the acrEF efflux pump operon, which is relevant to multidrug resistance in E. coli. Its substrate specificity (antibiotics, basic dyes and detergents) is similar to that of AcrAB | 1 | 186 |
| 8 | P96222 | EthR | Mycobacterium tuberculosis | Ethionamide resistance | 1 | 28, 90, 111 |
| 9 | P72185 | HemR | Propionibacterium freudenreichii | Probably regulates hemX, which appears to be involved in heme transport | 1 | 137 |
| 10 | Q93QZ7 | HydR | Tn5398 from Clostridium difficile | Involved in erythromycin resistance | 1 | 93 |
| 11 | O68442 | IfeR | Agrobacterium tumefaciens 1D1609 | Seems to be a repressor that controls the expression of the putative ifeABR isoflavonoid efflux system | 1 | 296 |
| 12 | Q9ZGB7 | LanK | Streptomyces cyanogenus | Probably a landomycin A resistance regulator | 1 | 315, 424 |
| 13 | LfrR | Mycobacterium segmatis | Control of the lfrA gene whose end product confers resistance to fluoroquinolones, ethidium bromide, and acryflavine | 1 | 212 | |
| 14 | O34619 | LmrA | Bacillus subtilis | Probable repressor of the lincomycin-resistance operon | 1 | 197, 198, 260 |
| 15 | P39897 | MtrR | Neisseria gonorrhoeae | A transcriptional repressor that regulates transcription of the mtrCDE genes, which encode a multidrug efflux pump; MtrR acts directly or indirectly as a positive regulator of farAB gene expression | 1 | 72, 130, 131, 220, 221, 297, 333, 334, 339, 447, 448 |
| 16 | Q9F0Y2 | Pip | Streptomyces coelicolor | Pristinamycin I-induced regulator that controls mutidrug resistance genes | 1 | 99 |
| 17 | Q9F147 | PqrA | Streptomyces coelicolor | Probably the repressor of pqrB, which encodes an efflux pump conferring resistance to paraquat | 1 | 61 |
| 18 | P23217 | QacR | Staphylococcus aureus | Regulates the QacA multidrug efflux pump | 1 | 119, 120, 249, 299, 300, 332, 350, 351, 390 |
| 19 | O52558 | RifQ | Amycolatopsis mediterranei | Located in the rifamycin biosynthetic gene cluster and probably related to the adjacent gene that encodes a rifamycin efflux protein | 1 | 17 |
| 20 | Q9KIH5 | RmrR | Rhizobium etli plasmid B | Probably regulates the operon rmrAB related to a multidrug efflux pump involved in sensitivity to phytoalexins, flavonoids, and salicylic acids | 1 | 113 |
| 21 | Q9AMH9 | SimReg 2 | Streptomyces antibioticus | Included in the Streptomyces antibioticus simocyclinone biosynthetic gene cluster; probably regulates the putative export protein SimEX | 1 | 396 |
| 22 | Q8KLP4 | SmeT | Stenotrophomonas maltophilia | A repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF | 1 | 340, 452 |
| 23 | Q9R9T9 | SrpR | Pseudomonas putida | Probable regulator of the solvent resistance pump SrpABC of strain S12 | 1 | 163, 179, 180, 422 |
| 24 | P39885 | TcmR | Streptomyces glaucescens | A regulator of the tetracenomycin C resistance repressing the gene tcmA, which encodes an export pump | 1 | 126, 127 |
| 25 | P09164 | TetR | Escherichia coli | Controls the expression of tetracycline resistance mediated by the gene tetA, which encodes an efflux pump that acts as an antiporter by coupling the export of [MgTetracycline]+ out of the cell with the uptake of protons | 1 | 29, 30, 36, 38, 39, 142, 143, 148, 149, 150, 183, 189, 259, 286, 287, 288, 289, 393, 402, 430 |
| 26 | Q9AIU0 | TtgR | Pseudomonas putida | Regulates the TtgABC efflux pump mediating organic solvent tolerance and resistance to ampicillin, tetracycline, chloramphenicol, and nalidixic acid | 1 | 81, 391 |
| 27 | Q93PU7 | TtgW | Pseudomonas putida | ttgW is a pseudogene | 1 | 327, 329 |
| 28 | Q9RP98 | UrdK | Streptomyces fradiae Tu2717 | Probably regulates an urdamycinA efflux pump | 1 | 94 |
| 29 | Q9AJL5 | VarR | Streptomyces virginiae | Regulates transcription of varS, the virginiamycin S-specific transporter in a virginiamycin S-dependent manner | 1 | 263 |
| 30 | P96676 | YdeS | Bacillus subtilis | Similar to a regulator of antibiotic transport complexes in Streptomyces hygroscopicus | 1 | 34 |
| 31 | Q54189 | ArpA | Streptomyces griseus | Represses the expression of adpA; AdpA activates the expression of strR, and the StrR protein activates the expression of streptomycin biosynthetic genes. ArpA also controls morphogenesis | 2, 5, 8 | 157, 278, 282, 283, 285, 375, 412, 438 |
| No. | SPTRa | Name | Organism | Function | Gb | Reference(s) |
| 32 | Q93M20 | Aur1B | Streptomyces aureofaciens | Included in the Streptomyces aureofaciens auricin polyketide biosynthesis gene cluster | 2 | 275 |
| 33 | Q9LBV6 | BarA | Streptomyces virginiae | Probably involved in regulation of virginiamycin biosynthesis | 2 | 175, 262 |
| 34 | Q8KNI9 | CalR1 | Micromonospora echinospora | Included in the calicheamicin gene cluster | 2 | 2 |
| 35 | O66129 | CprB | Streptomyces coelicolor | CprB is involved in the control of actinorhodin and undecylprodigiosin biosynthesis and morphogenesis | 2 | 264, 284 |
| 36 | O24741 | FarA | Streptomyces lavendulae FRI-5 | IM-2-specific receptor; plays an important role in the regulation of secondary metabolism and the biosynthesis of the antibiotics showdomycin and minimycin in Streptomyces lavendulae; FarA acts as a negative transcriptional regulator for the biosynthesis of nucleoside antibiotics and blue pigment, switching on their expression in the presence of IM-2; also acts as a positive transcriptional regulator for the biosynthesis of d-cycloserine, switching off its expression in the presence of IM-2 | 2 | 184, 185, 413 |
| 37 | Q939Q2 | JadR* | Streptomyces venezuelae | Included in the cluster for the biosynthesis of the dideoxysugar component of jadomycin B | 2 | 416 |
| 38 | Q56153 | JadR2 | Streptomyces venezuelae | Represses the biosynthesis of jadomycin B and seems to control cellular pigmentation | 2 | 442, 443 |
| 39 | Q9ZN97 | MphB | Escherichia coli plasmid pTZ3721 | Repressor of antibiotic biosynthesis | 2 | 172, 272 |
| 40 | Q9XDF0 | NonG | Streptomyces griseus sbsp. griseus | Probably related to nonactin biosynthesis | 2 | 414 |
| 41 | Q9RF02 | PhlF | Pseudomonas fluorescens | A repressor of the phlABCD operon responsible for the biosynthesis of the antifungal 2,4-diacetylphloroglucinol (PHL) | 2 | 346 |
| 42 | Q9ZHP8 | TylQ | Streptomyces fradiae | Butyrolactone receptor TylQ is a potential regulator of production of the macrolide antibiotic tylosin | 2, 8 | 371 |
| 43 | Q8VQC6 | VanT | Vibrio anguillarum | Positively regulates serine metalloprotease, pigment and biofilm production | 2, 5 | 71 |
| 44 | Q9RPK9 | TarA | Streptomyces tendae | Hypothetical receptor of gamma-butyrolactone, which regulates nikkomycin synthesis | 2, 8 | 86 |
| 45 | Q9XCC7 | TylP | Streptomyces fradiae | Regulates tylosin production and morphological differentiation, and is probably a gamma-butyrolactone receptor | 2, 5, 8 | 25, 371, 372 |
| 46 | Q59213 | BM1P1 | Bacillus megaterium | Probably acts as positive regulatory protein involved in the expression of the P450BM-1 gene by interfering with the binding of the repressor protein, Bm3R1, to the regulatory regions of P450BM-1 | 3 | 358, 361, 362 |
| 47 | O68276 | Bm1P1 | Bacillus megaterium ATCC 14581 | Negatively affects basal-level expression of P450BM-1, a barbiturate-inducible P450 monooxygenase; cytochromes P450BM-3 and P450BM-1 catalyze the hydroxylation of fatty acids | 3 | 140, 214, 215, 295, 356, 358, 359, 362 |
| 48 | P43506 | Bm3R1 | Bacillus megaterium | A transcriptional repressor involved in the regulation of barbiturate-inducible proteins in Bacillus megaterium | 3 | 87, 88, 89, 140, 213, 214, 295, 356, 357, 358, 359, 361, 362 |
| 49 | Q9AJ68 | ButR | Streptomyces cinnamonensis | Putative transcriptional repressor of crotonyl-CoA reductase | 3 | 218, 219 |
| 50 | Q93TU7 | CampR | Rhodococcus sp. NCIMB 9784 | Probably regulates 6-oxocamphor hydrolase | 3 | 123 |
| 51 | Q51597 | CamR | Pseudomonas putida plasmid CAM | A negative regulator of the cytochrome P-450cam hydroxylase operon | 3 | 9, 10, 105 |
| 52 | O33453 | CymR | Pseudomonas putida | A repressor which controls expression of both the cym and cmt operons and is inducible by p-cumate but not p-cymene | 3 | 62, 82, 83, 279 |
| 53 | Q9RAJ1 | DhaR | Mycobacterium sp. GP1 | Appears to function as a repressor of dhaA expression, dhaA is an haloalkane dehalogenase gene included in the 1-clorobutane catabolic gene cluster | 3 | 306 |
| 54 | Q9RA03 | KstR | Rhodococcus erythropolis strain SQ1 | A repressor of kstD expression that encodes a 3-ketosteroid Δ-dehydrogenase protein involved in the degradation of steroid intermediates in phytosterol degradation | 3 | 404 |
| 55 | Q8VV87 | LexA-like | Terrabacter sp. strain DBF63 | Probably involved in degradation of dibenzofuran | 3 | 171 |
| 56 | AcnR | Corynebacterium glutamicum | Repressor of the acn gene encoding aconitrase and controlling the tricarboxylic acid cycle | 3 | 195 | |
| 57 | Q9FA56 | PaaR | Azoarcus evanssi | Probably regulates the paa genes, which are responsible for the aerobic phenylacetic acid catabolic pathway | 3 | 256 |
| 58 | Q9XDW2 | PsbI | Rhodopseudomonas palustris | Included in the cluster of genes participating in aerobic biodegradation of p-cumate | 3 | 310 |
| 59 | O85706 | ThlR | Clostridium acetobutylicum DSM 792 | Possibly acts as a transcriptional repressor of the thlRBC operon, which is involved in the biosynthesis of thiolase | 3 | 429 |
| 60 | Q59431 | UidR | E. coli | A repressor of the uidRABC (gusRABC) operon that comprises a beta-d-glucuronidase (uidA), a glucuronide permease (uidB) and a membrane-associated protein (uidC) | 3 | 40 |
| 61 | P22645 | YDH1 | Xanthobacter autotrophicus | Probably regulates the dhlA gene involved in 1,2-dichloroethane degradation | 3 | 162 |
| 62 | P17446 | BetI | Escherichia coli | A choline-sensing repressor of the bet regulon involved in osmotic stress | 4 | 8, 201, 202, 331 |
| 63 | Q8NLK1 | McbR | Corynebacterium glutamicum | In absence of l-methionine, represses the expression of six key enzymes for the biosynthesis of the sulfur- containing amino acids L-cysteine and L-methionine including sulfonate utilization and sulfite reduction | 4 | 322 |
| 64 | Q9EVJ6 | MphR | Escherichia coli | Represses the mph(A)-mrx-mphR(A) operon in the absence of erythromycin; erytromycin induces the synthesis of macrolide 2′-phosphotransferase I [Mph(A)], which inactivates erythromycin | 4 | 273 |
| 65 | Q9F9Z7 | PhaD | Pseudomonas oleovorans | Biosynthesis of medium-chain-length (MCL) poly-3-hydroxyalkanoates (PHAs) as intracellular storage material | 4 | 188, 451 |
| 66 | Q9ZF45 | Q9ZF45 | Lactococcus lactis | Regulates the operon purDEK, which encodes enzymes in the de novo pathway of purine nucleotides | 4 | 269 |
| 67 | P06969 | TtK | Escherichia coli | Co-transcribed with the dut (deoxyuridine triphosphatase) gene | 4 | 85, 415 |
| 68 | P32398 | Yhgd or YixD | Bacillus subtilis | Probably related to protoheme IX biosynthesis | 4 | 132 |
| 69 | Q9F6W0 | CasR | Rhizobium etlli | A repressor of the casA gene, which encodes the calmodulin-like protein calsymin involved in bacteroid development during symbiosis and in symbiotic nitrogen fixation | 5 | 433 |
| 70 | Q9RQQ0 | IcaR | Staphylococcus aureus | A repressor of the operon ica which is responsible for an intercellular polysaccharide compound that acts as the slime in biofilm formation | 5 | 70, 163 |
| 71 | Q8GLC6 | IcaR | Staphylococcus epidermidis | A repressor of the operon ica, which is reponsible for an intracellular polysaccharide compound that acts as the slime in biofilm formation | 5 | 60, 66, 67, 190, 456 |
| 72 | Q8KX64 | LitR | Vibrio fischeri | Important for the normal induction of luminescence, plays a positive role in modulating the ability to colonize juvenile squid, and may control the opacity/translucent phenotype of the colony | 5, 8 | 97 |
| 73 | P21308 | LuxR | Vibrio harveyi | Required for expression of the luxCDABEGH (luciferase) operon, responsible for bacterial luminescence | 5 | 23, 24, 51, 57, 167, 232, 240, 250, 251, 253, 254, 255, 355, 365, 380, 381, 382 |
| 74 | Q9ANS7 | LuxT | Vibrio harveyi | Activates the expression of LuxO, the phosphorelay protein that regulates luminescence in Vibrio harveyi | 5 | 216 |
| 75 | O50285 | OpaR | Vibrio parahaemolyticus | A transcriptional regulator that controls the opaque morphology in Vibrio parahaemolyticus colonies | 5 | 240, 355 |
| 76 | Q9XDV7 | Orf2 | Streptomyces griseus | Probably related to carbon-source-dependent differentiation in Streptomyces griseus | 5 | 398 |
| 77 | Q9L8G8 | SmcR | Vibrio vulnificus | Appears to play an important role in starvation adaptation and in the regulation of many growth phase-regulated genes, including some virulence factors (protease, hemolysin); ScmR represses motility, fimbria production, and biofilm production | 5 6 | 63, 242, 243, 355 |
| 78 | O30343 | HapR | Vibrio cholerae | A transcriptional regulator with a central role in control of the virulence of Vibrio cholerae, in a cell density-dependent way | 6 | 165, 194, 196, 240, 455 |
| 79 | Q8KU49 | Ef0113 | Enterococcus faecalis | Located in a pathogenicity island in vancomycin-resistant Enterococcus faecalis | 6 | 354 |
| 80 | Q63B57 | HlyIIR | Bacillus cereus | Regulates expression of hlyII whose gene product has haemolytic activity | 6 | 47 |
| 80 | O24739 | BarB | Streptomyces virginiae | Regulates virginiamycin biosynthesis | 7 | 182 |
| 81 | O86852 | ScbR | Streptomyces coelicolor | Acts as the cytoplasmic receptor that specifically binds SCB1 gamma-butarylactone and negatively regulates transcription of the scbA gene, responsible for gamma-butyrolactone SCB1 synthesis | 2, 7, 8 | 3, 385, 386 |
| 82 | Q9JN89 | MmfR | Streptomyces coelicolor plasmid SCP1 | Putative lactone-dependent transcriptional regulator | 8 | 437 |
| 83 | Q9S3L4 | AmtR | Corynebacterium glutamicum | Regulator of nitrogen control | 9 | 48,161 |
| 84 | Q9EX90 | PsrA | Pseudomonas putida | Involved in the regulatory cascade controlling rpoS gene regulation in response to cell density | 9 | 192 |
| 85 | P36656 | YjdC | Escherichia coli | Probably involved in copper tolerance | 10 | 101 |
Swiss-Prot and TrEMBL accession number.
1, regulation of efflux pumps and transporters involved in antibiotic resistance and tolerance to toxic compounds; 2, regulation of antibiotic biosynthesis; 3, regulation of catabolic pathways; 4, biosynthesis of products important for bacteria (e.g., osmoprotectants, nucleotides, amino acids, PHAs, protoheme); 5, regulation of differentiation (sporulation, mycelium formation), colony phenotype, biofilm formation; 6, regulation of genes involved in virulence; 7, regulation of butyrolactone synthesis; 8, butyrolactone or autoinducer receptors; 9, global regulation; 10, other.
DEFINING THE TetR FAMILY
TetR Family Profile
The TetR family is named after the member of this group that has been most completely characterized genetically and biochemically, the TetR protein (141, 148, 150, 168, 288, 395). This protein controls the expression of the tet genes, whose products confer resistance to tetracycline (150, 183, 209, 210, 337, 434, 435). Members of the TetR family exhibit a high degree of sequence similarity at the DNA binding domain (see below). Interpro (258) assigns proteins to the TetR family based on PROSITE signature PS01081 (364), PRINTS motif PR00455 (15, 16), and Pfam Hidden Markov Model (HMM) profile PF00440 (26, 27). To establish a single criterion defining the TetR family, we decided to develop a conventional profile, because conventional profiles are easy to manage and their sensitivity is equivalent to that of HMM profiles.
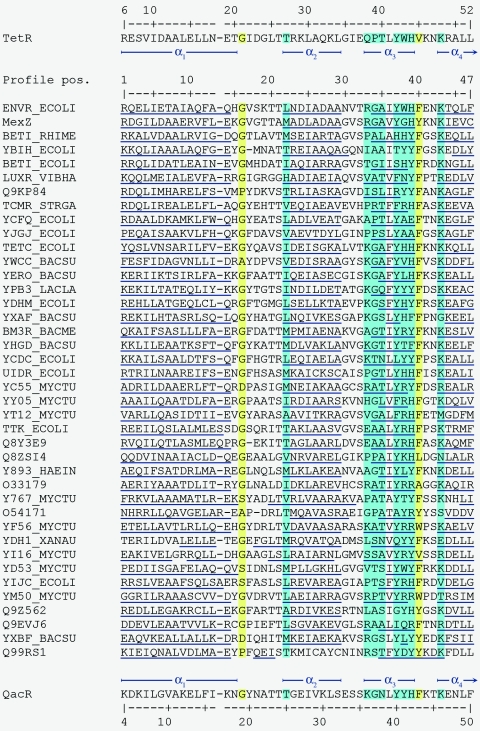
To develop the TetR family profile, we first selected a set of 120 sequences as belonging to the TetR family based on two criteria: a positive score for PROSITE signature PS01081, and a high score for PF00440 HMM. The 120 sequences were clustered into 42 groups using BLAST, and a representative sequence was selected and aligned for each cluster using CLUSTAL (http://clustalw.genome.ad.jp/). This revealed that the most conserved region corresponded to the HTH domain described in the TetR and QacR crystals (120, 150, 287, 288, 289, 349, 350, 351). The initial HTH motif was progressively extended until the global score of the multialignment diminished. Figure 1 shows the final alignment of the sequences. This conserved stretch corresponded in TetR and QacR crystals to the almost complete α-helix 1, the HTH domain formed by α-helices 2 and 3, and five residues of α-helix 4 that connect the DNA-interacting region with the core of the protein (see Fig. 2 for the three-dimensional structure of TetR).
FIG. 1.
Alignment of 42 members of the TetR family that exemplify the TetR family profile. The blue column indicates α-helix residues involved in DNA contacts in the crystal structure of TetR and QacR. The yellow column indicates turns. The most conserved residues are shaded. Abbreviations are as follows: BACME, Bacillus megaterium; BACSU, Bacillus subtilis; ECOLI, Escherichia coli; HAEIN, Haemophilus influenzae; LACLA, Lactobacillus lactis; MYCTU, Mycobacterium tuberculosis; RHIME, Rhizobium meliloti; STRGA, Streptomyces sp.; VIBHA, Vibrio haemophilus; XANAU, Xanthomonas sp.
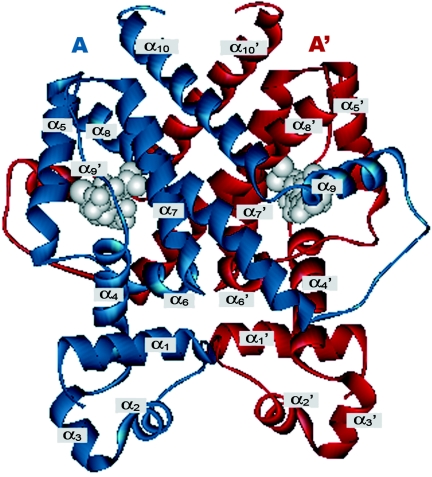
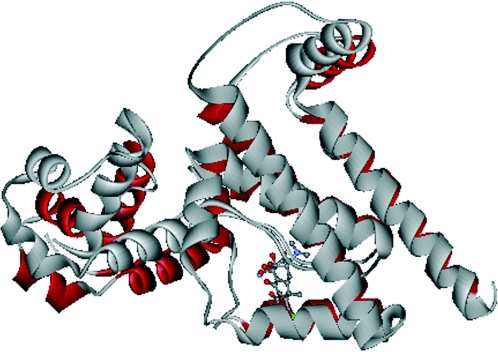
FIG. 2.
Ribbon diagram of a TetR homodimer. Monomers are shown in blue or red. Two tetracycline molecules, each bound to a monomer, are shown in grey. α-Helices 2 and 3 in the blue monomer and α2′ and α3′ in the red monomer constitute the shared HTH DNA binding domain. α-Helix 1 and part of helix α-4, together with α-helices 2 and 3, comprise the sequence that best defines the TetR family profile. (Adapted from Hinrichs et al. [150] with permission of the publisher.)
The final alignment shown in Fig. 1 was used as a seed for the construction of a conventional profile to detect TetR family members. The TetR profile was built using the pfmake program available at the Swiss Institute of Bioinformatics (http://npsa-pbil.ibcp.fr/cgi-bin/npsa__automat.pl?page=/NPSA/npsa__pfmake.html) (45, 46). The TetR profile was confronted against the 660,992 bacterial and archaeal proteins in the SWISS-PROT and TrEMBL databases (released December 2004) using the pfsearch program available at http://bioweb.pasteur.fr/seqanal/interfaces/pftools.html#pfsearch (46). The program, which proposes a tentative threshold Z-score of 8.5 to consider a protein a member of the TetR family, selected 2,357 proteins as putative members of the TetR family.
To verify the quality of this TetR profile for specificity (false positives) and sensitivity (false negatives), we implemented a new tool called Provalidator which uses Interpro, Swiss-Prot, Prodom, TIGRfam, CoGnitor, NCBI-RPS-BLAST, and PSI-BLAST resources (68, 128, 154, 323, 348, 387, 449). In the first step, we searched for false positives among the 2,353 proteins we assigned to the TetR family. Interpro assigned 2,315 proteins to the TetR family, and these 2,315 were considered true positives. The remaining 38 proteins were analyzed with other resources such as TIGRfam, Prodom, NCBI-RPS-BLAST and PSI-BLAST (128, 449). This allowed us to assign 34 proteins to the TetR family. Three of the false positives (Q89RN6, Q988I6, and Q6N8G8) that we found were protein members of the AraC/XylS family of transcription activators (109, 394). These proteins have two HTH motifs at the C-terminal end, typical of AraC/XylS family members (109, 229). These three proteins were identified as potential TetR members because one of its HTH is highly similar to the DNA-binding domain in TetR. The fourth false positive is a transposase (Q981E7).
Provalidator detected 15 false negatives (Q742Y2, Q8CJK3, Q73ZY1, Q6D1J7, Q8KU64, Q9A917, Q880T2, Q6D2Z4, Q885G7, Q8PC90, Q9A466, Q9S6C0, Q9ZH26, Q6A626, and Q8G822), which are proteins assigned to the TetR family by INTERPRO but whose Z-score was between 6.407 and 8.487. In summary, the TetR profile with a Z-score threshold of 8.5 identified proteins that were not detected by INTERPRO, and among the 660,992 proteins analyzed, only four false positives were found. These results indicate that the new algorithm is highly effective for the detection of members of the TetR family.
Identification of TetR Family Members in DNA and Protein Databases
Using the profile defined above for the TetR family, we searched for members of this family in the Swiss-Prot and TrEMBL databases and also searched the 196 complete and incomplete microbial genomes available in NCBI (Release December 2004). We detected 73 TetR proteins in Swiss-Prot, 2,277 in TrEMBL, and 2,410 in the translated open reading frame corresponding to 196 microbial genomes. To select nonredundant sequences the set of 4,758 TetR proteins was analyzed using the SEQUNIQ program developed in our laboratory (Molina-Henares et al., unpublished results). This program integrates the set of sequences available in nucleic acid and protein databases. We found 2,353 sequences in the TetR family that surpassed the threshold Z-score of 8.5. The HTH in 2,348 members of the family was located at the N-terminal end of the proteins.
Table 3 shows that members of the TetR family were detected in 144 microbial genomes belonging to 80 genera and 113 species of gram-positive and α-, β-, and γ-proteobacteria, cyanobacteria, and archaea, indicating wide taxonomic distribution. We have found that proteins of the TetR family are encoded both in chromosomes and in plasmids, and the mobility of the latter elements could be a source of the spread of genes in this family via horizontal transfer (147, 383), as is also the case with catabolic genes (77, 160, 236, 410, 426), antimicrobial resistance determinants (20, 100, 124), and 16S rRNA genes (347).
TABLE 3.
Distribution of TetR proteins in microbes
| Microorganism | Genome size (Mbp) | No. of members |
|---|---|---|
| Nocardia farcinica IFM 10152 | 6.21 | 151 |
| Streptomyces coelicolor A3(2) | 9.05 | 150 |
| Streptomyces avermitilis MA-4680 | 9.12 | 116 |
| Mycobacterium avium subsp. paratuberculosis k10 | 4.83 | 108 |
| Agrobacterium tumefaciens C58 | 11.35 | 61 |
| Bradyrhizobium japonicum USDA 110 | 9.11 | 59 |
| Mycobacterium bovis AF2122/97 | 4.35 | 51 |
| Mycobacterium tuberculosis CDC1551 | 4.40 | 51 |
| Mycobacterium tuberculosis H37Rv | 4.41 | 51 |
| Bacillus licheniformis ATCC 14580 | 8.44 | 48 |
| Mesorhizobium loti MAFF303099 | 7.60 | 47 |
| Rhodopseudomonas palustris CGA009 | 5.46 | 40 |
| Pseudomonas aeruginosa PAO1 | 6.26 | 38 |
| Bacillus cereus ATCC 10987 | 5.22 | 36 |
| Bacillus anthracis ‘Ames Ancestor’ | 5.50 | 32 |
| Bacillus anthracis A2012 | 5.37 | 32 |
| Bacillus anthracis Sterne | 5.23 | 31 |
| Bacillus anthracis Ames | 5.23 | 30 |
| Bacillus cereus ZK | 5.30 | 30 |
| Bacillus cereus ATCC 14579 | 5.43 | 29 |
| Bordetella bronchiseptica RB50 | 5.34 | 28 |
| Pseudomonas syringae pv. tomato DC3000 | 6.40 | 28 |
| Sinorhizobium meliloti 1021 | 6.69 | 28 |
| Bacillus thuringiensis serovar konkukian 97-27 | 5.24 | 27 |
| Clostridium acetobutylicum ATCC 824 | 4.13 | 27 |
| Caulobacter crescentus CB15 | 4.02 | 26 |
| Lactobacillus plantarum WCFS1 | 3.31 | 26 |
| Pseudomonas putida KT2440 | 6.18 | 25 |
| Burkholderia pseudomallei K96243 | 7.25 | 24 |
| Ralstonia solanacearum GMI1000 | 5.81 | 24 |
| Photobacterium profundum SS9 | 6.40 | 23 |
| Oceanobacillus iheyensis HTE831 | 3.63 | 22 |
| Bordetella parapertussis 12822 | 4.77 | 21 |
| Burkholderia mallei ATCC 23344 | 5.84 | 21 |
| Bacillus halodurans C-125 | 4.20 | 20 |
| Bacillus subtilis subsp. subtilis 168 | 4.21 | 20 |
| Acinetobacter sp. strain ADP1 | 3.60 | 19 |
| Bordetella pertussis Tohama I | 4.09 | 17 |
| Chromobacterium violaceum ATCC 12472 | 4.75 | 17 |
| Shewanella oneidensis MR-1 | 5.13 | 17 |
| Vibrio vulnificus CMCP6 | 5.13 | 17 |
| Escherichia coli CFT073 | 5.23 | 16 |
| Gloeobacter violaceus PCC 7421 | 4.66 | 16 |
| Methanosarcina acetivorans C2A | 5.75 | 16 |
| Streptococcus mutans UA159 | 2.03 | 16 |
| Vibrio parahaemolyticus RIMD 2210633 | 5.17 | 16 |
| Vibrio vulnificus YJ016 | 5.26 | 16 |
| Xanthomonas axonopodis pv. citri 306 | 5.18 | 16 |
| Corynebacterium glutamicum ATCC 13032 | 3.31 | 15 |
| Deinococcus radiodurans R1 | 3.28 | 15 |
| Erwinia carotovora subsp. atroseptica SCRI1043 | 5.06 | 15 |
| Xanthomonas campestris pv. campestris ATCC 33913 | 5.08 | 15 |
| Escherichia coli O157:H7 | 5.59 | 14 |
| Corynebacterium efficiens YS-314 | 3.15 | 13 |
| Escherichia coli O157:H7 EDL933 | 5.53 | 13 |
| Lactococcus lactis subsp. lactis I11403 | 2.37 | 13 |
| Leifsonia xyli subsp. xyli CTCB07 | 2.58 | 13 |
| Listeria monocytogenes 4b F2365 | 2.91 | 13 |
| Salmonella enterica subsp. enterica serovar Typhi CT18 | 5.13 | 13 |
| Salmonella typhimurium LT2 | 4.95 | 13 |
| Treponema denticola ATCC 35405 | 2.84 | 13 |
| Corynebacterium diphtheriae NCTC 13129 | 2.49 | 12 |
| Escherichia coli K12 | 4.64 | 12 |
| Listeria innocua Clip11262 | 3.01 | 12 |
| Listeria monocytogenes EGD-e | 2.94 | 12 |
| Salmonella enterica subsp. enterica serovar Typhi Ty2 | 4.79 | 12 |
| Shigella flexneri 2a 301 | 4.61 | 12 |
| Vibrio cholerae O1 biovar eltor N16961 | 4.03 | 12 |
| Nostoc sp. strain PCC 7120 | 7.21 | 11 |
| Enterococcus faecalis V583 | 3.22 | 10 |
| Mycobacterium leprae TN | 3.27 | 10 |
| Shigella flexneri 2a 2457T | 4.60 | 10 |
| Geobacter sulfurreducens PCA | 3.81 | 9 |
| Leptospira interrogans serovar Copenhageni Fiocruz L1-130 | 4.63 | 9 |
| Leptospira interrogans serovar Lai 56601 | 4.69 | 9 |
| Propionibacterium acnes KPA171202 | 2.56 | 9 |
| Staphylococcus aureus subsp. aureus MRSA252 | 2.90 | 9 |
| Shigella flexneri 2a 301 | 4.61 | 12 |
| Vibrio cholerae O1 biovar eltor N16961 | 4.03 | 12 |
| Nostoc sp. strain PCC 7120 | 7.21 | 11 |
| Enterococcus faecalis V583 | 3.22 | 10 |
| Mycobacterium leprae TN | 3.27 | 10 |
| Shigella flexneri 2a 2457T | 4.60 | 10 |
| Geobacter sulfurreducens PCA | 3.81 | 9 |
| Leptospira interrogans serovar Copenhageni Fiocruz L1-130 | 4.63 | 9 |
| Leptospira interrogans serovar Lai 56601 | 4.69 | 9 |
| Propionibacterium acnes KPA171202 | 2.56 | 9 |
| Staphylococcus aureus subsp. aureus MRSA252 | 2.90 | 9 |
| Bifidobacterium longum NCC2705 | 2.26 | 8 |
| Brucella melitensis 16M | 3.29 | 8 |
| Brucella suis 1330 | 3.32 | 8 |
| Photorhabdus luminescens subsp. laumondii TTO1 | 5.69 | 8 |
| Staphylococcus aureus subsp. aureus Mu50 | 2.90 | 8 |
| Symbiobacterium thermophilum IAM 14863 | 3.57 | 8 |
| Yersinia pestis biovar Medievalis 91001 | 4.80 | 8 |
| Yersinia pseudotuberculosis IP 32953 | 4.84 | 8 |
| Bacteroides fragilis YCH46 | 5.31 | 7 |
| Bacteroides thetaiotaomicron VPI-5482 | 6.26 | 7 |
| Bdellovibrio bacteriovorus HD100 | 3.78 | 7 |
| Desulfovibrio vulgaris subsp. vulgaris Hildenborough | 3.77 | 7 |
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | 2.17 | 7 |
| Staphylococcus aureus subsp. aureus MSSA476 | 2.80 | 7 |
| Staphylococcus aureus subsp. aureus MW2 | 2.82 | 7 |
| Staphylococcus aureus subsp. aureus N315 | 2.84 | 7 |
| Yersinia pestis CO92 | 4.83 | 7 |
| Desulfotalea psychrophila LSv54 | 3.66 | 6 |
| Lactobacillus johnsonii NCC 533 | 1.99 | 6 |
| Mannheimia succiniciproducens MBEL55E | 2.31 | 6 |
| Rhodopirellula baltica SH 1 | 7.15 | 6 |
| Streptococcus agalactiae NEM316 | 2.21 | 6 |
| Yersinia pestis KIM | 4.60 | 6 |
| Aquifex aeolicus VF5 | 1.59 | 5 |
| Methylococcus capsulatus Bath | 3.30 | 5 |
| Streptococcus agalactiae 2603V/R | 2.16 | 5 |
| Streptococcus pyogenes MGAS10394 | 1.90 | 5 |
| Streptococcus pyogenes MGAS8232 | 1.90 | 5 |
| Clostridium perfringens 13 | 3.09 | 4 |
| Methanococcus maripaludis S2 | 1.66 | 4 |
| Staphylococcus epidermidis ATCC 12228 | 2.50 | 4 |
| Streptococcus pyogenes M1 GAS | 1.85 | 4 |
| Streptococcus pyogenes MGAS315 | 1.90 | 4 |
| Streptococcus pyogenes SSI-1 | 1.89 | 4 |
| Thermus thermophilus HB27 | 2.13 | 4 |
| Clostridium tetani E88 | 2.80 | 3 |
| Haemophilus influenzae Rd KW20 | 1.83 | 3 |
| Methanosarcina mazei Go1 | 4.10 | 3 |
| Nitrosomonas europaea ATCC 19718 | 2.81 | 3 |
| Pasteurella multocida subsp. multocida Pm70 | 2.26 | 3 |
| Porphyromonas gingivalis W83 | 2.34 | 3 |
| Streptococcus pneumoniae R6 | 2.04 | 3 |
| Streptococcus pneumoniae TIGR4 | 2.16 | 3 |
| Synechocystis sp. strain PCC 6803 | 3.57 | 3 |
| Thermoanaerobacter tengcongensis | 2.69 | 3 |
| Wolinella succinogenes DSM 1740 | 2.11 | 3 |
| Haemophilus ducreyi 35000HP | 1.70 | 2 |
| Halobacterium salinarum NRC-1 | 2.57 | 2 |
| Legionella pneumophila str. Lens | 3.41 | 2 |
| Legionella pneumophila str. Paris | 3.64 | 2 |
| Legionella pneumophila subsp. pneumophila Philadelphia 1 | 3.40 | 2 |
| Methanothermobacter thermautotrophicus Delta H | 1.75 | 2 |
| Neisseria meningitidis MC58 | 2.27 | 2 |
| Neisseria meningitidis Z2491 | 2.18 | 2 |
| Thermotoga maritima MSB8 | 1.86 | 2 |
| Xylella fastidiosa 9a5c | 2.73 | 2 |
| Archaeoglobus fulgidus DSM 4304 | 2.18 | 1 |
| Campylobacter jejuni subsp. jejuni NCTC 11168 | 1.64 | 1 |
| Coxiella burnetii RSA 493 | 2.00 | 1 |
| Helicobacter hepaticus ATCC 51449 | 1.80 | 1 |
| Mycoplasma penetrans HF-2 | 1.36 | 1 |
| Picrophilus torridus DSM 9790 | 1.55 | 1 |
| Pyrococcus abyssi GE5 | 1.77 | 1 |
| Sulfolobus solfataricus P2 | 2.99 | 1 |
| Sulfolobus tokodaii 7 | 2.69 | 1 |
| Ureaplasma parvum serovar 3 ATCC 700970 | 0.75 | 1 |
We found that TetR family members are particularly abundant in microbes exposed to environmental changes, such as soil microorganisms (i.e., Nocardia, Streptomyces, Bradyrhizobium, Mesorhizobium, Pseudomonas, Bacillus, and Ralstonia spp.); plant and animal pathogens (i.e., Agrobacterium, Brucella, Escherichia coli, Bordetella, Mycobacterium, and Salmonella spp.), extremophiles (i.e., Deinococcus), and methanogenic bacteria such as Methanosarcina acetivorans. In contrast, TetR family members do not appear in intracellular pathogens such as chlamydias, mycoplasmas, and endosymbionts such as Buchera, in agreement with their life style in nonchanging environments (52). However, it should be noted that Dugan et al. (80) recently found that Chlamydia suis can acquire tetracycline resistance via horizontal gene transfer of genomic islands bearing the tet genes.
As a general collorarium, we can say that it seems that proteins of the TetR family are involved in the adaptation to complex and changing environments. This in turn correlates with the fact that many members of the TetR family are found among microbes with abundant extracytoplasmic function sigma factors (52, 227, 236, 277, 444).
PROTEINS WITH KNOWN THREE-DIMENSIONAL STRUCTURES
The high degree of primary sequence identity in the stretch that defines the HTH region of the TetR profile probably reflects a common three-dimensional structure in this domain in members of the family. This is supported by the almost identical three-dimensional structure of the HTH of TetR, QacR, CprB, and EthR, as deduced from the superimposition of these regions, and the high degree of sequence conservation in the alignment (79, 264, 349). As in other families of transcriptional regulators, no sequence conservation was found outside the HTH domain, which probably reflects differences in the kind of signal sensed by different regulators of the family, i.e., antibiotics with dissimilar structures, barbiturates, homoserine lactones, organic solvents, and choline (see Table 2). Nonetheless, some striking global structural conservation in the three-dimensional structure was found.
In addition, given that all members of the family whose function is known are repressors, they probably function in a similar way. Binding of an inducer molecule to the nonconserved domain of a TetR family member probably causes conformational changes in the conserved DNA-binding region that result in release of the repressor from the operator and thus allow transcription from the cognate promoter. To gain insights into the mechanisms of action of the TetR family members, we analyzed in detail the three-dimensional structure of the four members of the family, TetR, QacR, CprB, and EthR, whose crystal structures have been obtained (150, 264, 286-289, 349-351), in order to identify common and differential features of the TetR family members.
TetR Regulator
Tetracycline resistance and the role of the transcriptional regulator TetR.
Tetracyclines are among the most commonly used broad-spectrum antibiotics (209, 210). They act by binding to the small ribosomal subunit, thereby interrupting polypeptide chain elongation by an unknown mechanism. Many gram-negative bacteria have developed mechanisms of resistance against this antibiotic. The most frequent mechanism involves a membrane-associated protein (TetA) that exports the antibiotic out of the bacterial cell before it inhibits polypeptide elongation (169, 211, 389, 434, 435, 453).
Adjacent to tetA and divergently oriented is tetR (112), whose gene product tightly controls expression of both tetA and tetR (148, 150). The intergenic region between the tetR and tetA genes contains two identical operators separated by 11 bp. TetR binds to these operators and thus prevents transcription from both promoters (Fig. 3) and (288). In all TetR crystal structures elucidated to date (PDB identifiers: 2TCT; 2TRT; 1A6I; 1BJO; 1BJY; 1BJZ; 1ORK; and 1RP1), this repressor appears as a homodimer (29, 30, 159, 183, 287-289, 366). The TetR homodimer binds to the operator (Fig. 3). Each 15-bp operator shows an internal palindromic symmetry with an extra central base pair (Fig. 3A). The operator sequences overlap with promoters for tetA and tetR, thereby blocking the expression of both genes. When tetracycline complexed with Mg2+ binds to TetR (166, 384), a conformational change takes place that renders the TetR protein unable to bind DNA. As a consequence, TetR and TetA are expressed (286).
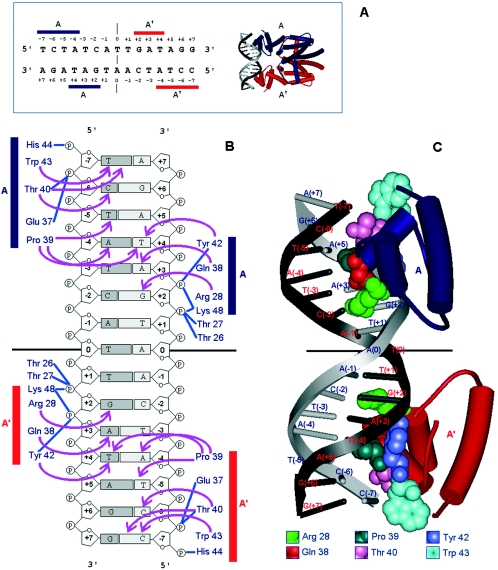
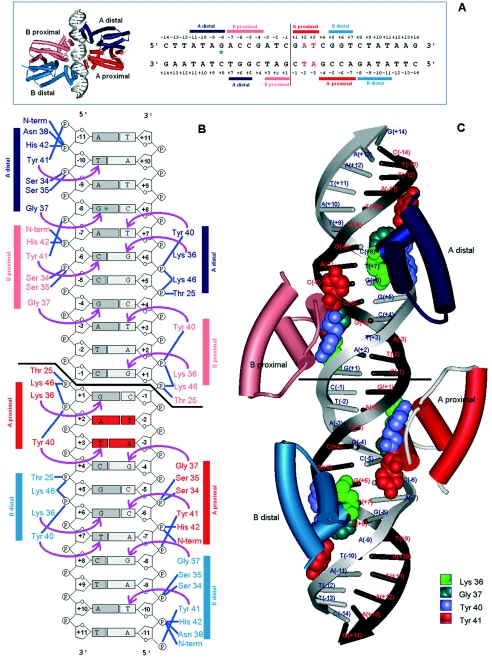
FIG. 3.
Binding of TetR to its operator site. A) tetR operator and contact regions. The tetR operator is a palindromic sequence. Horizontal bars show nucleotides contacted by each monomer of the TetR dimer. B) Interaction of TetR residues with specific nucleotides (arrows) and phosphate backbone (blue lines) in the operator region. The amino acids involved in DNA binding extend from residues 27 to 48. Contacts established with the operator were confirmed by footprint assays, by analysis of TetR mutants, and by crystallographic studies (29, 30, 159, 266, 366). C) Representation of each homodimer bound to the tet operator in a double-helix representation. (Adapted from Orth et al. [288] with permission of the publisher.)
The TetR homodimer is constituted by two identical monomers that fold into 10 α-helices with connecting turns and loops (Fig. 2). The three-dimensional structure of the TetR monomer is stabilized mainly by hydrophobic helix-to-helix contacts. The global structure of the TetR homodimer can be divided into two DNA-binding domains at the N-terminal end of each monomer, and a regulatory core domain involved in dimerization and ligand binding (150, 286-289). The DNA-binding domains are constituted by helices α1, α2, and α3 and their symmetric helices α1′, α2′, and α3′ (a prime denotes the second monomer). Helices α4 and α4′ connect these domains with the regulatory core domain composed of helices α5 to α10 and their symmetric counterparts α5′ and α10′ (150, 287, 289). The regulatory domain is responsible for dimerization and contains, for each monomer, a binding pocket that accommodates tetracycline in the presence of a divalent cation. Helices α5, α8, and α10 and their counterparts α5′, α8′, and α10′ constitute the scaffold of the core domain, and their structure is the most conserved in both TetR conformations (150, 287-289).
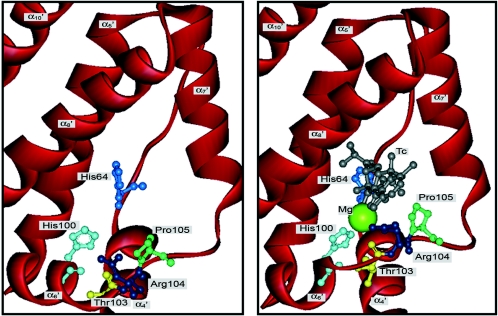
The tetracycline-binding pocket is identical in both monomers. The cavity to which the [TcMg]+ complex binds is depicted in Fig. 4 (286, 287, 289). The entrance of this cavity is controlled by α9′ and the C-terminal end of α8′ and the loop that connects both, while the exit is closed by loop 4-5 (287- 289). When [TcMg]+ enters the tunnel, its A ring makes contacts with loop 4-5, and the interaction with the effector triggers a cascade of conformational changes. The contacts that His100 and Thr103, both in α6, establish with the magnesium ion of the complex displace α6, which undergoes a conformational change in its C terminus to form a β-turn (Fig. 4). The 6-7 loop is also pushed near the inducer, so that Arg104 and Pro105 interact with tetracycline. Translation of α6 forces α4 to move in the same direction due to van der Waals contacts. His64 of α4, anchored to α5 and to tetracycline, acts as a pivot point, and α4 moves like a pendulum. As a consequence of the rotation of α4 and α4′, recognition helices α3 and α3′ move further apart, and the DNA contacts are disrupted (Fig. 5) (286, 287, 289). Tetracycline is impeded from freeing the binding cavity, and TetR cannot bind its target DNA again. It should be noted that residues outside the binding cavity can influence affinity for tetracycline, as revealed by Kamionka et al. (168), who isolated a double mutant (G96E, L205S) with reduced affinity for the antibiotic.
FIG. 4.
Representation of the TetR cavity involved in the binding of tetracycline. Left) In the absence of tetracycline. Right) In the presence of tetracycline. The green ball represents the Mg2+ ion. Specific interactions are not drawn for the sake of clarity but are described in the text. (Adapted from Orth et al. [287, 289] and Kisker et al. [183] with permission of the publishers.)
FIG. 5.
Docking of a TetR monomer without (grey) and with (red) tetracycline. Note the alterations induced in the α1-α3 region involved in binding to the target operators. The increase in distance between α3 and α3′ with tetracycline results in the inability of TetR to maintain the specific interactions shown in Fig. 3, and therefore the repressor is released. (Adapted from Orth et al. [288] with permission of the publisher.)
The on/off switch mechanism used by TetR to respond to specific signals may be used similarly in other TetR family members.
TetR DNA-binding domain: a symmetric TetR dimer binds a palindromic operator.
Cocrystallization of TetR with its operator DNA established that the TetR homodimer binds perpendicularly to the longitudinal DNA axis (Fig. 3A). Two adjacent DNA major groove regions covering a 6-base-pair area on both strands are involved in the almost perfect docking with the two TetR-interacting domains (Fig. 3A and 3B) (288). No water molecules were found at the TetR-DNA interface, where the crucial interactions are hydrophobic (288).
The interactions of each HTH domain with the operator DNA are summarized in Fig. 3A and 3B. The TetR monomer A binds the main strand from positions −4 to −7 while contacting the complementary strand from operator positions +4 to +2, and the symmetric monomer A' binds the main strand from positions +2 to +4 and the complementary strand from positions −4 to −7 (Fig. 3A and 3B).
Crystallographic analysis revealed that helix α3 (from Gln38 to His44) is the main element responsible for sequence-specific recognition, since all residues in this helix contribute to it, except for Leu41, which is part of the hydrophobic core stabilizing the α1, α2, and α3 helix bundle. Thr40 residue in monomer A establishes direct contacts with operator base pairs T(−7) and C(−6) in the main DNA strand (Fig. 3A and 3B). Trp43 interacts with T(−7) as well. Pro39 interacts with both strands at bases T(−5) and A(−4) of the main strand and T(+4) of the complementary strand. In the rest of the operator half site, the α3 helix of monomer A interacts with the complementary strand, Tyr42 contacting with T(+4) and Gln38 with A(+3). Helix α2 supplies an additional specific contact with the complementary strand, namely, Arg28 contacts G(+2).
Although the TetR DNA binding domain maintains its structure thanks to a hydrophobic core formed by residues from the α1, α2, and α3 bundle (288), interactions with DNA lead to changes in the TetR DNA binding domain. One such change is that α3 forms a 310-helical turn at the N-terminal end as a result of complex DNA contacts. The H-bonds between Arg28-G(+2), and Gln38-A(+3) increase the separation between base pairs 1 and 2 from 3.4 Å to 3.9 Å (288). The two phosphate groups accompanying the G at position +2 establish H-bonds with side chains of Thr26, Thr27, Tyr42, and Lys48, and with the amino groups of the main chain of Thr27 and Lys48 (Fig. 3B). These contacts draw DNA closer to TetR near G(+2). Although the DNA is kinked away from TetR at position +2 in both operator strands, bending toward TetR in the area corresponding to positions +3 to +6 compensates for the DNA deviation. Crystallographic studies revealed that Lys48 located in α4, outside the HTH motif, also established contacts with the target DNA region (Fig. 3B). This lysine is relatively well conserved among TetR family members, and we are tempted to suggest that this residue plays an equivalent role in other proteins of the TetR family.
QacR Regulator
Two QacR dimers bind the operator to repress the qacA multidrug transporter gene.
QacA confers resistance to monovalent and bivalent cationic lipophilic antiseptics and disinfectants such as quaternary ammonium compounds (hence the name Qac) (10, 11, 44, 239). The qac locus consists of the qacA gene and the divergently transcribed qacR gene, which are borne on a plasmid (119). In the absence of drug, the 188-residue QacR protein represses transcription of the qacA multidrug transporter gene by binding two nested palindromes located downstream from the qacA promoter and overlapping its transcription start site (119, 300). Therefore, QacR seems to repress transcription by hindering the transition of the RNA polymerase-promoter complex into a productively transcribing state rather than by blocking RNA polymerase binding.
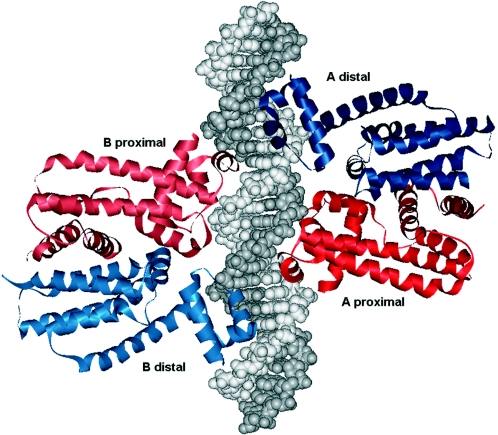
The three-dimensional structure of QacR (PDB identifiers 1JTX, 1JTG, 1JTY, 1JUM, 1JUP, 1JUS, and 1JTO) revealed that it is an all-helical protein which contains a DNA-binding HTH motif embedded within an N-terminal three-helix bundle and a second domain involved in drug binding and dimerization (350, 351). It should be noted that unlike TetR, two QacR dimers, rather than one, bind the operator site (339, 340) (Fig. 6).
FIG. 6.
Binding of QacR to its operator site. A) Interaction of QacR with the qac operator. B) Contacts established by residues at α-helix 3 of QacR homodimers A and B with specific nucleotides (arrows) and phosphate backbone (blue lines) in the synthetic operator used for QacR-DNA cocrystal (349, 350). C) Representation of the two QacR homodimers bound to the qac operator in a double-helix representation. (Adapted from Schumacher et al. [351] with permission of the publisher.)
The monomers of each dimer have been called proximal and distal to refer to their positions with respect to the center of symmetry of the palindromic operator (Fig. 6A and 6B). It was shown that the operator to which one dimer is bound is symmetric and partially overlaps that bound by the other dimer (351) (Fig. 6 and 7). The existence within the same fragment of DNA sequence of two overlapping partial palindromes with identical symmetric bases is therefore surprising (Fig. 6). In this sense the palindromic sequences recognized by QacR are equivalent to those described for the TetR interface except for the spacer sequence length, 3 bp for TetR versus 4 bp for QacR, supporting the hypothesis that interactions of other members of the family with their target sequences may be similar, independent of the number of dimers involved.
FIG. 7.
Ribbon representation of the two QacR homodimers bound to target DNA in a double-helix representation (A) and details of the contacts established by α-helix 3 of monomers of different homodimers when recognizing overlapping sites (B). (Adapted from Schumacher et al. [351] with permission of the publisher.)
The α3 helix of QacR A distal and B distal monomers establish the most extensive specific interactions with the operator (351). The Tyr41 residue of the A distal monomer (Fig. 6B) establishes hydrophobic contacts with base T(−10) of the DNA main strand as well as with the phosphate at position −11 in the main strand, while Tyr40 contacts T(+7) (Fig. 6B). In addition, tight docking with DNA is facilitated by specific hydrogen bonds between Lys36 and base G(+6) in the complementary strand, and between Gly37 and base G(−8) in the main strand. Gly37 is important in repression because nucleotide G(−8) is the transcription start site for the qacA gene. Monomers A and B proximal also establish a series of critical interactions. For instance, Tyr41 of B proximal contacts the C(−6) base in the main strand, whereas Tyr40 contacts base T(+3) and phosphate (+2) in the complementary strand (351). Gly37 in the A proximal monomer contacts G(−4) in the complementary strand, whereas Lys36 contacts G(+1) in the main strand. A number of residues in α2, loop α2-α3, α3 and the positive dipole of the α1 (N terminus) also interact with the phosphate backbone of both DNA strands (351).
Figure 6C shows how each dimer engages the DNA major groove in a face almost opposite to the other dimer, forming an angle between the two dimer axes of less than 180° (Fig. 7). Studies of QacR binding to DNA have indicated that the two dimers bind DNA cooperatively (120, 121, 351). Analysis of the three-dimensional structure suggested that such cooperativity does not arise from protein-protein interactions, as the closest approach of the dimers is 5.0 Å. Rather, binding cooperativity appears to be mediated through conversion of the DNA structure from a B-DNA conformation to the high-affinity undertwisted configuration observed in the crystal structure. Conversion of the DNA conformation is necessary because the optimal distance between each of the HTH motifs of the QacR dimer is 37 Å. This requires expansion of the 34-Å distance between successive major groove regions on one edge of the canonical B-DNA. It has been suggested that binding of the first QacR dimer forces this energetically unfavorable conformational change, which in turn produces an optimal DNA conformation for the easy binding of the second dimer (351). Experimental data reported by Grkovic et al. (121, 122) suggested that the two dimers must bind simultaneously and cooperatively to the operator in order to maintain the DNA deformation detected in the crystal.
Schumacher and Brennan (349) noticed that TetR and QacR achieve the same degree of specificity in DNA binding through different mechanisms. They noted that TetR, recruits Arg28, located outside its recognition helix, to make a base pair-specific contact (288), whereas QacR does not employ residues outside α3 to ensure DNA binding specificity. They also noted that TetR kinks its binding site and induces a 17° bend towards the protein to optimize the position of its HTH motifs for specific base interactions within each DNA half site; whereas QacR widens the major groove of the entire IR1 binding site smoothly and bends its DNA site by only 3°. These distinctions are reflected in the different HTH center-to-center distances observed in QacR (37 Å). Thus, an important lesson derived from comparisons of the QacR-DNA and TetR-DNA structures is that even structurally homologous proteins of the same family that share a similar function, i.e., repression, can utilize slightly different mechanisms of action.
QacR as a model for multidrug recognition.
QacR is released from the qacA operator by its interaction with a number of cationic lipophilic drugs such as rhodamine 6G, crystal violet, and ethidium (119). More recently, Grkovic et al. (122) showed that effector recognition of QacR can be extended to several bivalent cationic dyes and plant alkaloids. In spite of the existence of two binding pockets, only one drug molecule is bound by each homodimer, as determined by equilibrium dialysis studies and isothermal titration calorimetry for the QacR-R6G complex (350). The QacR crystal bound to different drugs revealed another remarkable finding: the presence of an expansive and multifaceted drug-binding pocket with a volume of 1,100 Å3, so that different drugs partially overlap different subpockets (349, 351). A similar cavity able to bind multiple drugs was reported by Yu et al. (445, 446) for the AcrB multidrug transporter.
Crystallographic studies by Schumacher et al. (350) and Murray et al. (261) have demonstrated that multidrug recognition mediated by the QacR dimer is a rather simple process that, contrary to expectations, does not require sophisticated molecular mechanisms. Indeed, the drug binding domain of QacR consists of six α-helices (PDB identifiers: 1JTX, 1JT6, 1JTY, 1JUP, 1JUS, 1JTO, 1RKW, and 1RPW). Entry to the mostly buried drug-binding pocket is through a small opening formed by the divergence of helices α6, α7, α8, and α8′. The stoichiometry of one drug molecule for two QacR subunits led to this asymmetric induction process, in which the drug-bound monomer undergoes a major structural change. Comparison of the drug-bound structure with the DNA-bound structure reveals that drug binding triggers a coil-to-helix transition of residues 89 to 93, which extends helix α5 by a turn. This transition removes the drug surrogates Tyr92 and Tyr93 from the hydrophobic core of the protein. Expulsion of these tyrosines also leads to the relocation of nearby helix α6 and its tethered DNA-binding domain. The result of this structural transition is a 9-Å translation and a 37° rotation of the DNA-binding domain, effectively rendering the QacR dimer unable to bind its target DNA.
Three-Dimensional Structure of CprB
The gram-positive bacterial genus Streptomyces uses γ-butyrolactones as autoregulators or microbial hormones, together with their specific receptors (γ-butyrolactone receptors), to control morphological differentiation, antibiotic production, or both (150, 151). The most representative of the γ-butyrolactone autoregulatory factors is 2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone, known as A-factor, which is essential for aerial mycelium formation, streptomycin production, streptomycin resistance, and yellow pigment production (133, 134, 155) in Streptomyces griseus. However, the A-factor receptor protein, known as ArpA, has proved to be difficult to purify. In contrast, the CprB protein from Streptomyces coelicolor A3(2), which is 30% identical to ArpA (284), has been purified and crystallized (264), although the ligand for CprB is still unknown. Nonetheless, CprB binds the same nucleotide sequence as does ArpA (375) and indeed CprB also serves as a negative regulator for both secondary metabolism and morphogenesis in S. coelicolor, as ArpA does in S. griseus (264, 284).
The CprB dimer is omega shaped, and the two subunits in the dimer are related by a pseudo-twofold axis. Each monomer of CprB is composed of 10 α-helices and has two domains: a DNA-binding domain (residues 1 to 52) and a regulatory domain (residues 77 to 215). The three-dimensional structure of CprB is essentially similar to that of QacR bound to DNA except for the lack of α10 (350, 351). In addition, the DNA-binding domains of the two proteins are very similar, so much so that the two DNA-binding domains can be superimposed with an rms deviation of 1.48 Å for 71 Cα atoms (264). Although no information on CprB-operator DNA is available, the high degree of sequence conservation allowed the authors to predict that the core of the DNA-binding domain is composed of Ile14, Ile15, Ala18, Phe22, Leu32, Ile35, Leu46, and Phe50.
It has been suggested that a CprB dimer binds to its target DNA as found in the TetR-DNA complex (150, 287, 288). This is because structure-based amino acid sequence alignment shows that at the amino acid sequence level the DNA-binding domains of CprB and TetR are highly identical. This suggests that there is an evolutionary relationship between the DNA-binding domains of the two proteins. The regulatory domain of CprB is composed of six α-helices (helices α5 to α10) (264), which can also be superimposed on the corresponding domain of TetR (286, 287, 289) (PDB code 1JT0).
EthR Structure
Ethionamide has been used for more than 30 years as a second-line chemotherapeutic treatment in tuberculosis patients who have developed resistance to first-line drugs such as isoniazid and rifampin. Activation of the prodrug ethionamide is regulated by the Baeyer-Villiger monooxygenase EthA and the TetR family repressor EthR, whose open reading frames are separated by 75 bp in the genome of Mycobacterium tuberculosis. EthR has been shown to repress transcription of the activator ethA gene by binding to the intergenic region and contributing to ethionamide resistance.
The expression of ethA is regulated by EthR in M. tuberculosis. Overexpression of ethR leads to ethionamide resistance, whereas chromosomal inactivation of ethR promotes ethionamide hypersensitivity (28). EthR was found to bind directly and specifically to DNA sequences corresponding to the ethRA intergenic region (28, 90). The large EthR operator, which comprises 55 bp in comparison with the 15-bp operators recognized by most other family members, is organized as a putative highly degenerated palindrome containing pairs of overlapping inverted and tandem repeat sequences (90). In the absence of DNA, EthR forms a homodimer in solution, and surface plasmon resonance measurements suggest that EthR octamerizes when bound to DNA (90).
The EthR monomer is an all-helical, two-domain molecule (79). The N-terminal domain comprises helices 1 to 3, with helices 2 and 3 forming the HTH DNA-binding motif seen in other TetR family protein structures. The larger C-terminal domain, which in QacR and TetR has been dubbed the drug-binding domain, consists of helices 4 to 9, and its function in EthR is unknown. The crystal structure revealed that the dimerization interface, a conserved structural feature among the TetR class of repressors, is primarily formed by helices 8 and 9 (288, 351).
One of the most striking features of the EthR structure is a narrow tunnel-like cavity formed by helices 4, 5, 7, and 8 that opens to the bottom of the molecule (79). The tunnel measures about 20 Å in length and is lined predominantly, albeit not exclusively, by aromatic residues, with helices 5 and 7 constituting the majority of side chains. The loop connecting helices 4 and 5 restricts the opening of the hydrophobic tunnel, and the electron density in this loop is only poorly defined, indicating a certain degree of structural flexibility in the loop. This cavity may serve as the binding site for an as yet unknown ligand.
Crystal structure of TetR family members with unknown functions.
New genomic/proteomic approaches are leading to the crystallization of a number of proteins, many of which have no assigned function. The following proteins of the TetR family have been crystallized: Cgl2612 of Corynebacterium glutamicum (pdb 1V7B); YbiH of Salmonella enterica serovar Typhimurium (pdb 1T33); YcdC of Escherichia coli (pdb 1PB6); and YfiR and YsiA from Bacillus subtilis (pdb 1RKT and 1VIO, respectively).
DNA-BINDING PREDICTIONS BASED ON TetR AND QacR CRYSTAL STRUCTURES
There is a perfect overlap of the DNA binding domains of QacR, TetR, CprB, and EthR, and no gaps were found in the α-helices involved in contacts with DNA in the multialignment of the 2,353 members of the TetR family in this domain. Based on these findings, we hypothesized that residues at the same position in the multialignment of all family members may play equivalent roles. This prompted us to analyze each amino acid in the multialignment within the DNA binding domain.
Relationship between Profile Positions and Structural Positioning
Analysis comparison of the cocrystal of QacR and TetR with their corresponding operators revealed that residues corresponding to positions 22, 33, 34, 35, 37, 38, 39, and 43 in the family multialignment are involved in interactions with target operator DNA (Fig. 1). We analyzed the occurrence of each amino acid at these positions in the multialignment of all members of the TetR family (Table 4).
TABLE 4.
Amino acid frequency at each of the positions critical for operator recognition by TetR family membersa
| Frequency at position:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22
|
33
|
34
|
35
|
37
|
38
|
39
|
43
|
||||||||
| AA | % | AA | % | AA | % | AA | % | AA | % | AA | % | AA | % | AA | % |
| V | 21.95 | K | 29.26 | G | 37.79 | T | 34.30 | Y | 74.16 | R | 22.75 | H | 44.50 | K | 77.25 |
| L | 20.60 | R | 20.07 | A | 18.93 | S | 20.87 | F | 8.05 | Y | 16.44 | Y | 33.83 | R | 10.07 |
| M | 18.66 | P | 10.27 | P | 12.55 | A | 19.6 | H | 4.63 | H | 13.29 | R | 4.90 | L | 2.82 |
| I | 17.65 | Q | 6.04 | S | 8.39 | L | 5.84 | L | 3.09 | N | 8.39 | F | 3.56 | I | 1.88 |
| T | 13.22 | V | 5.30 | R | 6.71 | N | 5.30 | T | 2.48 | K | 6.11 | A | 2.15 | M | 1.88 |
| F | 2.15 | A | 4.56 | T | 5.10 | G | 4.30 | S | 1.95 | W | 5.84 | N | 1.95 | V | 1.81 |
| H | 1.74 | T | 4.30 | Q | 3.56 | V | 2.28 | N | 1.88 | A | 5.64 | Q | 1.88 | T | 0.94 |
| A | 1.61 | L | 4.23 | M | 2.35 | M | 2.08 | R | 1.21 | L | 3.56 | E | 1.48 | G | 0.94 |
| Y | 1.28 | E | 3.36 | N | 1.07 | Q | 1.34 | Q | 0.6 | S | 3.56 | L | 1.34 | Q | 0.74 |
| S | 0.54 | I | 2.89 | K | 1.01 | Y | 1.28 | A | 0.47 | F | 2.89 | W | 1.21 | A | 0.47 |
| P | 0.40 | H | 2.89 | V | 0.81 | I | 1.14 | I | 0.47 | Q | 2.55 | S | 0.94 | H | 0.34 |
| N | 0.13 | S | 2.01 | D | 0.47 | P | 0.54 | G | 0.27 | T | 2.35 | T | 0.74 | F | 0.27 |
| E | 0.07 | N | 1.61 | L | 0.47 | R | 0.40 | M | 0.27 | E | 1.68 | V | 0.54 | P | 0.20 |
| G | 1.07 | E | 0.40 | E | 0.20 | V | 0.2 | V | 1.61 | C | 0.40 | S | 0.13 | ||
| D | 1.01 | F | 0.27 | H | 0.20 | K | 0.13 | G | 1.41 | K | 0.27 | Y | 0.07 | ||
| Y | 0.67 | I | 0.07 | K | 0.13 | C | 0.07 | D | 1.07 | I | 0.13 | E | 0.07 | ||
| M | 0.27 | H | 0.07 | D | 0.13 | P | 0.07 | I | 0.54 | D | 0.07 | N | 0.07 | ||
| C | 0.13 | C | 0.07 | C | 0.20 | G | 0.07 | ||||||||
| F | 0.07 | M | 0.07 | ||||||||||||
| P | 0.07 | ||||||||||||||
The amino acid (AA) frequency is expressed as a percentage and refers to the 2,353 TetR family members.
We found two types of position, one in which the residue was highly conserved and another in which the residue was poorly conserved, if at all. Positions 37, 39, and 43 were well conserved, whereas at positions 22, 33, 34, 35, and 38 the profile aligned different residues.
Tyr42 in TetR and Tyr40 in QacR corresponded to position 37 in the profile sequence displayed in Fig. 1, where a Tyr residue appeared in 74.16% of the aligned proteins (Table 4). The next most highly represented residues in this position are also aromatic amino acids: phenylalanine (8%) and histidine (4%) (Table 4). Tyr-42 in TetR and Tyr40 in QacR appear at the center of α-helix 3 and contact a thymine located at the center of the palindrome forming the operator and also contact a phosphate one position towards the center of the palindrome (Fig. 3B and 6B).
The residue at position 39 of the profile in the multialignment corresponds to His44 in TetR and His42 in QacR. In the corresponding cocrystals, these residues established contacts with the phosphate backbone (Fig. 3B and Fig. 6B). In the multiple sequence alignment of all family members, either histidine or tyrosine appears at position 39. We are tempted to propose that this residue is critical for interactions with the phosphate backbone.
A lysine-DNA phosphate interaction is shared at residues Lys48 in TetR and Lys46 in QacR, which correspond to position 43 in the multialignment and are located in the amino end of the α4 helix. A lysine residue is present in 77% of TetR proteins, and their interactions with DNA phosphates seem to be crucial to adjust the HTH domain to contact DNA (Fig. 3B and 6B). At position 22 of the profile (Thr27 in TetR and Thr25 in QacR), five residues are the most abundant (Val, Leu, Met, Ile, and Thr). Thr27 in TetR and Thr25 in QacR are involved in interactions with the phosphate backbone.
Thus, in the TetR family, the contacts established by the residue aligned at position 37 in α3 (tyrosine present in 74% of the cases) and 39 in α3 (His or Tyr present in 98% of the cases) and a residue at position 43 in α4 (Lys present in 77% of the cases) probably orient the HTH motif to interact with the DNA major groove and anchor the protein to the phosphate backbone.
Glycine at position 16, located at the end of α1, in the multialignment is highly conserved and is involved in changing the polypeptide direction in the TetR and QacR crystals to orient the HTH DNA binding domain properly.
Positions 33, 34, 35, and 38 of the profile align many different residues (Table 4). In TetR and QacR, the corresponding residues establish specific contacts with different DNA bases except Asn38 of QacR (position 35 in the multialignment), which contacts the phosphate backbone. Based on the high variability of these positions in the corresponding multiple alignment of the family, we are tempted to propose that these positions endow specificity to each protein so that it can recognize its operator through specific protein-DNA interactions.
SOME REGULATORS ARE PART OF COMPLEX REGULATORY CIRCUITS
Published data indicate that the specific function of 85 members of TetR family is known (Table 2). More information about each TetR protein is available at http://www.bactregulators.org (235). We have clustered the functions regulated by TetR family members into 10 groups (Table 2). The most frequent function performed by TetR family proteins is the regulation of efflux pumps and transporters involved in antibiotic resistance and tolerance to toxic chemicals. We have also observed that TetR family members often regulate their own synthesis, this feedback control ensures the transcriptional repressor level within optimal concentration limits (31, 73, 231, 338, 392). In this simple regulatory scheme, synthesis of the repressor and of the regulated protein(s) is derepressed in the presence of an inducer molecule.
However, TetR family proteins also participate in other types of regulatory networks that underlie complex processes, such as homeostasis in metabolism (biosynthesis of amino acids, nucleotides, protoheme, and reserve material), synthesis of osmoprotectants, quorum sensing, drug resistance, virulence, and processes related to growth phase-dependent differentiation (sporulation and biosynthesis of antibiotics) (Table 2) (www.bactregulators.org) (235).
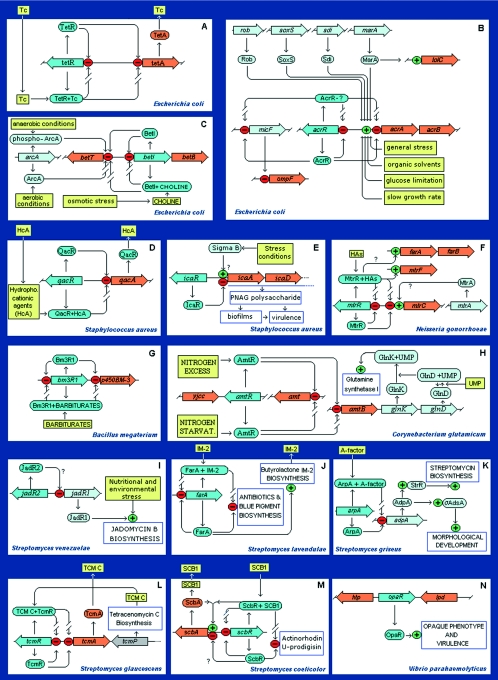
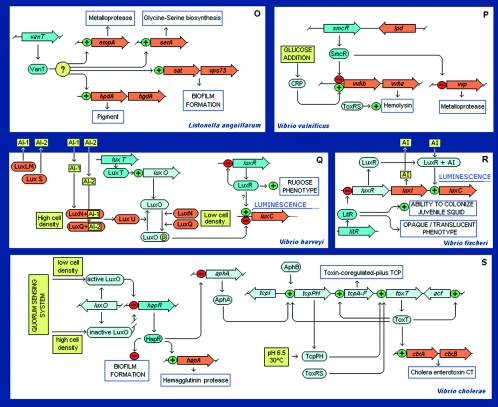
Figure 8 shows a series of schemes in which a TetR family member plays a role in complex circuits. Below, for the sake of brevity, we have analyzed only some representative sets of regulatory networks, including proteins involved in drug resistance (AcrR of E. coli and MtrR of Neisseria gonorrhoeae), biosynthesis of an osmoprotectant (BetI), a key protein involved in idiophase antibiotic production and differentiation in Streptomyces (ArpR), a protein involved in pathogenesis in Vibrio (HapR), and some proteins involved in quorum sensing.
FIG. 8.
Regulatory networks involving members of the TetR family. Although TetR and QacR cannot be considered part of a network, their type of control is shown because it is frequently found in members of the family. The following color code was used for complex networks: dark blue, TetR family member; orange, the gene directly regulated by the TetR family member; light blue, a regulator that modulates the expression of a TetR family member or which assists in the regulation of the gene under the control of a TetR family member; yellow boxes, signals and conditions influencing the system; open boxes, final results of the action of the system when the result is a scorable phenotype. References recommended for each circuit: panel A (29, 38, 286, 288, 388, 417, 418); panel B (4, 7, 13, 31, 35, 110, 176, 191, 230, 245, 270, 436); panel C (8, 91, 201-204, 222, 290, 331); panel D (119, 120, 122, 249, 261, 332, 350-351); panel E (5, 6, 66, 67, 116, 163); panel F (228, 238, 397, 333, 353, 408); panel G (87-89, 125, 140, 214, 215, 295, 360, 403); panel H (48, 161); panel I (78); panel J (56, 184, 185, 208, 413); panel K (133, 134, 158, 413); panel L (127); panel M (61, 266); panel N (240, 241, 345); panel O (248); panel P (59, 63, 242, 243, 355); panel Q (23, 24, 51, 107, 232, 255); panel R (97, 107, 117, 252, 341, 363); and panel S (107, 117, 181, 196, 217, 247).
AcrR Regulator Is the Local Specific Regulator of the acrAB Efflux Pump
Multiple antibiotic resistance in Escherichia coli has attracted recent attention, promoting the elucidation of a number of mechanisms that contribute to this phenomenon. One of these is the transport of diverse substrates out of the cell by the AcrAB-TolC efflux transporter, leading to a multiple antibiotic resistance (Mar) phenotype (267). The set of antibiotics to which AcrAB can confer resistance includes ampicillin, chloramphenicol, erythromycin, fluoroquinolones, β-lactams, novobiocin, tetracycline, tigecycline, and rifampin (151, 187, 223, 267, 268, 276).
AcrB is a large cytoplasmic membrane protein (224, 226, 445, 446) which associates with AcrA, a membrane fusion protein (281), and TolC, a protein that forms a channel for the extrusion of substrates into the medium (102, 193). The acrA and acrB genes form an operon (224) whose transcription is regulated by the acrR gene product. The acrR gene is divergently transcribed from the acrAB operon. Overexpression of AcrR represses the transcription of acrAB. This observation is consistent with the function of AcrR as a repressor for acrAB transcription. Evidence for this function has come also from gel shift mobility assays, which provided direct evidence for the binding of AcrR to the promoter region of acrAB. DNA sequencing (92) of certain isolates that overexpressed acrB mRNA revealed that the mutant strains had insertions that disrupted the acrR gene or point mutations that rendered a nonfunctional regulator, i.e., an amino acid substitution of cysteine for arginine at position 45 of AcrR. This biochemical and genetic evidence provides support for the regulatory role of AcrR.
MarA, SoxS, and Rob are related transcriptional activators of the AraC/XylS family (7, 112, 367) that activate acrAB expression, although they are not involved in the regulation of acrAB in response to general stress conditions (13, 14, 21, 35, 110, 224) because the acrAB operon can be activated in response to these stresses in genetic backgrounds lacking mar and sox (223-225). It was also found that general stress conditions increased the transcription of acrAB in the absence of functional AcrR, and these conditions, surprisingly, increased the transcription of acrR to a greater extent than that of acrAB. These results suggest the existence of a mar-sox-independent pathway to control acrAB expression in response to the general stress conditions. This transcriptional control of acrAB is also AcrR independent. Therefore, a major role of AcrR is to function as a specific secondary modulator to fine-tune the level of acrAB transcription and prevent unwanted overexpression of the efflux pump. This represents a novel mechanism for regulating gene expression in E. coli.
Mtr Circuit of Neisseria
The MtrCDE efflux pump of Neisseria gonorrhoeae provides gonococci with a mechanism to resist structurally diverse antimicrobial hydrophobic agents and antibiotic peptides that adopt β-sheet (protegenin 1) or two-helix (PC-8 and LC37) structures (130, 228, 238, 353). Mutations that render no expression or inactivation of mtrR, encoding a transcriptional repressor, resulted in high expression of the mtrCDE operon, concomitantly increasing resistance to hydrophobic agents (69, 130, 220, 221, 297, 353, 447). It was also found that strains of N. gonorrhoeae that display hypersusceptibility to hydrophobic agents often contained mutations in the mtrCDE efflux pump genes (406).
The mtrR gene is divergently transcribed with respect to the mtrCDE operon (Fig. 8F). The promoters of mtrR and mtrC overlap in their −35 boxes, and footprinting analysis showed that MtrR binds a 40-bp region within the −10 to −35 region of the mtrR promoter, which contains an inverted repeat (221). MtrR bound to its target site prevented expression from the efflux pump operon and its regulator (Fig. 8F). The expression of mtr genes is enhanced by the AraC/XylS member MtrA, although the mechanism of activation of this protein is unknown.
On the other hand, Veal and Shafer (407) have recently identified a gene that was designated mtrF, located downstream of the mtrR gene, that is predicted to encode a 56.1-kDa cytoplasmic membrane protein containing 12 transmembrane domains. Expression of mtrF was enhanced in a strain deficient in MtrR production, indicating that this gene, together with the closely linked mtrCDE operon, is subject to MtrR-dependent transcriptional control. Genetic evidence suggests that MtrF is also important in the expression of high-level detergent resistance by gonococci, and it was proposed that MtrF acts in conjunction with the MtrC-MtrD-MtrE efflux pump to confer high-level resistance to certain hydrophobic agents in gonococci. MtrR also controls the farAB operon, which encodes an efflux pump involved in resistance to long-chain fatty acids (Fig. 8F). The efflux pump FarAB uses MtrE as the outer membrane component (208).
BetI Controls the Choline-Glycine Betaine Pathway of E. coli
In Escherichia coli, glycine betaine serves as an osmoprotector in hyperosmotically stressed cells. This osmoprotector accumulates in large amounts in the cytoplasm, which allows cells to maintain appropriate osmotic strength and thus prevents dehydration. Glycine betaine is only one of several cellular osmolytes used by E. coli, but its accumulation allows this microbe to achieve its highest level of osmotic tolerance (199, 373). To accumulate glycine betaine, E. coli needs an external supply of this compound or its precursors choline and betaine aldehyde.
The osmoregulatory choline-glycine betaine pathway is encoded by the bet genes. The betA gene encodes choline dehydrogenase; betB encodes betaine aldehyde dehydrogenase; betT encodes a transport system for choline; and betI encodes a 21.8-kDa repressor protein involved in choline regulation of the bet genes (Fig. 8C). The bet genes are linked, with betT being transcribed divergently from the betIBA operon (203, 374). Primer extension analysis identified two partially overlapping promoters which were responsible for the divergent expression of the betT gene and betIBA operon. The transcripts are initiated 61 bp apart and are induced by osmotic stress, but for full expression choline is required in the growth medium (91, 202, 204). Because the ArcA protein represses the expression of bet genes in E. coli under anaerobiosis, the bet genes are expressed only under aerobic conditions. An arcA mutation caused complete derepression of the bet genes. A similar pattern for derepression by ArcA has been reported previously for other genes (sodA and arcA) which are directly regulated by ArcA (65).
Results from different laboratories suggest that choline regulation but not osmotic regulation of the bet promoters depended on BetI, a TetR family member. This was indicated by the requirement for choline, in addition to osmotic stress, for betT to be expressed in a mutant strain in which betI was supplied in trans. Furthermore, this choline effect was not seen in cells lacking betI. These findings indicate that betI encodes a repressor that reduced the expression of betT (331).
A chimeric BetI glutathione S-transferase fusion protein (BetI*) was purified, and gel mobility shift assays showed that BetI* formed a complex with a 41-bp DNA fragment carrying the intergenic betI promoter region. Footprinting revealed the presence of two sequences of dyad symmetry which probably constitute the BetI operator.
The Sinorhizobium meliloti bet genes have been cloned, and their involvement in response to osmotic stress has been analyzed (304, 305, 368).
ArpA Regulator from Streptomyces
Streptomycetes are filamentous, soil-living, gram-positive bacteria characterized for their ability to produce a wide variety of secondary metabolites, including antibiotics and biologically active substances, and for their complex morphological differentiation culminating in the formation of chains of spores (55). Secondary metabolite synthesis is sometimes called “physiological” differentiation because it occurs during the idiophase after the main period of rapid vegetative growth and assimilative metabolism (139, 153, 291).
The ultimate regulator of the mentioned processes in Streptomyces griseus is a homodimeric protein called A-factor receptor protein (ArpA) (155-158), which regulates the switch for physiological and morphological differentiation. The main biologically significant target of ArpA is the adpA gene. The AdpA protein in turn controls the expression of other genes. These genes include strR, which serves as a pathway-specific transcriptional activator for streptomycin biosynthetic genes (278); an open reading frame encoding a probable pathway-specific regulator for a polyketide compound (441); adsA, which encodes an extracytoplasmic function sigma factor of RNA polymerase essential for aerial mycelium formation (438); sgmA, which encodes a metalloendopeptidase probably involved in apoptosis of substrate hyphae during aerial mycelium development (174); ssgA, which encodes a small acidic protein essential for spore septum formation (437); amfR, essential for aerial hyphae formation (398, 440); and the sprT and sprU genes, which encode trypsin-like proteases (173).
In vitro, ArpA binds its target DNA site at the −10/−35 region, which is a 22-bp palindrome (5′-GG(T/C)CGGT(A/T)(T/C)G(T/G)-3′). Addition of γ-butyrolactone effector to the ArpA-DNA complex immediately releases ArpA from the DNA. A mutant strain deficient in ArpA or producing a mutant ArpA protein unable to bind to its target DNA overproduces streptomycin and forms aerial mycelia and spores earlier than the wild-type strain (282, 283). An amino acid replacement at Val-41 to Ala in ArpA in the HTH motif at the N-terminal portion of ArpA abolished DNA binding activity but not γ-butyrolactone binding activity, suggesting the involvement of this HTH in DNA binding. On the other hand, mutation of Trp-119 (Trp 119→Ala) generated a mutant unable to bind the γ-butyrolactone, resulting in a mutant protein that did not sense the presence of A-factor. These data suggest that ArpA consists of an HTH DNA-binding at the N-terminal end and an effector binding domain at the C-terminal end of the protein.
In the streptomycin biosynthetic gene cluster in S. griseus, StrR is the pathway-specific regulator that serves as a transcriptional activator for the other genes in the cluster (320). Expression of the strR gene was controlled by the AdpA protein, which binds the region upstream of the strR promoter and activates its transcription (311, 312). adpA knockout mutants produced no streptomycin, and overexpression of adpA caused the wild-type S. griseus strain to produce streptomycin at an earlier growth stage in a larger amount. This set of events explains how A-factor triggers streptomycin biosynthesis.
Disruption of the chromosomal adsA gene encoding σAdsA resulted in loss of aerial hypha formation but not streptomycin production, indicating that this sigma factor is involved in morphological development (401, 438).
Several receptor proteins for γ-butyrolactone-type autoregulators have been described in other species of Streptomyces. For example, CprA and CprB are involved in secondary metabolism and aerial mycelium formation in S. coelicolor A3(2) (284). The virginiae butanolyde receptor BarA is involved in virginiamycin biosynthesis in Streptomyces virginiae (280), and the IM-2 receptor, FarA is involved in blue pigment production in another Streptomyces strain (413).
FarA in Streptomyces lavendulae senses the concentration of γ-butyrolactone IM-2 and also transduces this signal, thus derepressing antibiotic and blue pigment biosynthesis. In addition, FarA also seems to be necessary for IM-2 biosynthesis (Fig. 8J).
The role of ScbR protein in the quorum-sensing circuit of Streptomyces coelicolor is similar to that of FarA in S. lavendulae. ScbR is also involved in three functions: positively regulating SCB1 synthesis (the γ-butyrolactone that acts as signal), receiving the signal, and transducing this signal, thereby derepressing the production of the antibiotics actinorhodin and U-prodigiosin (Fig. 8M).
HapR Regulates Virulence Genes in Vibrio cholerae
The development of cholera in humans is directly related to the production of two virulence factors: toxin-coregulated pilus (TCP), which mediates colonization, and cholera toxin (CT), responsible for the severe diarrhea, characteristic of this disease. The coordinated expression of TCP and CT occurs though a complex regulatory cascade (Fig. 8S) in which the regulators AphA and AphB synergistically activate tcpPH transcription (194). The TetR family protein HapR negatively regulates the expression of AphA and indirectly diminishes the production of TCP and CT.
HapR, in turn, is regulated by quorum-sensing signals that are sensed and transmitted by LuxO. The quorum-sensing apparatus in Vibrio cholerae is unusually complex and is composed of three parallel signaling systems (247). In contrast to other bacteria, in which high cell density triggers virulence gene expression, in Vibrio cholerae low cell density is the condition that activates the production of the pathogenic factors CT and TCP (455). At high cell density HapR positively regulates the expression of a hemagglutinin protease (Hap) that promotes detachment of Vibrio cholerae from the gastrointestinal epithelium (365) and exerts a negative effect on biofilm formation. Taking into account the regulatory functions of HapR and considering that some pathogenic biotypes lose HapR expression whereas others lose the aphA binding site, it appears that HapR expression is related to diminished toxicity and colonization capacity. These features offer potentially fruitful avenues of research to design drugs to modulate Vibrio cholerae pathogenicity.
Other Quorum-Sensing Circuits
Within the genus Vibrio, some TetR family proteins, i.e., LuxT, LuxR, and LitR, are involved in complex quorum-sensing circuits. In Vibrio harveyi, LuxT and LuxR participate in the circuit that regulates luminescence. This strain senses two autoinducers, AI-1 and AI-2 (58, 216). AI-1 is an acyl-homoserine lactone, as in other gram-negative bacteria. However, AI-2 is a novel autoinducer produced by many species that appears to be related to interspecies cell density detection. These two autoinducers use the same phosphorelay system to transduce the signal for bioluminescence regulation. Expression of luxR is in turn regulated by LitR (Fig. 8R).
BIOTECHNOLOGICAL APPLICATIONS AND FUTURE PROSPECTS
The Tet system is at present the most widely used system for conditional gene expression in eukaryotic cells (37). The system is based in the high affinity (10−9 M) of TetR for its operator, tetO, the favorable pharmacokinetics of tetracyclines (they diffuse through biological membranes), and their long record of safe clinical use. Cloning of the tetO elements adjacent to the TATA box of the target gene (114) was used successfully to control genes expressed by RNA polymerase I in Leishmania donovani and by RNA polymerase III in yeast and plant cells (76, 400). However, this system is not efficient in mammalian cells. For this reason, and based on the knowledge acquired about how TetR binds to its target operator, several chimeric versions of TetR fused to eukaryotic regulatory domains were constructed, such as the acidic activation domain (tTA) (22, 115, 403) and repression domains (tTS) (33, 336). Based on hybrid transregulators, transgenic mice able to produce diphtheria toxin or the regulated expression of Shiga toxin β to induce apoptosis in mammalian fibroblastic cells were obtained.
The tet system has also been used to study cancer and neurological disorders (95, 98, 419). In the future, advances to approach multifactorial biological processes like development and diseases are expected, which will be relevant for the treatment of complex diseases (37).
Solvent efflux pumps generally exhibit a broad substrate specificity, but some of them are highly specific and remove a certain number of chemicals. One such efflux pumps is the TtgABC pump of Pseudomonas putida DOT-T1E, which removes toluene, m-xylene, and propylbenzene as well as styrene and other aromatic hydrocarbons (327) in addition to several antibiotics. This efflux pump is under the control of the drug binding repressor TtgR, a member of the TetR family (391), and has applications in the safe development of solvent-resistant bacteria. Therefore, it is potentially suitable for the biotransformation of water-insoluble compounds into added-value products. One such system has been exploited to produce catechols from xylenes/toluenes in double-phase systems (328). Because these efflux systems are energy consuming, their expression in heterologous hosts has to be tightly controlled by their cognate repressor, which in turn has to be able to respond to the presence of the aromatic solvent. These types of pumps are presumed to be useful if transferred to a wide variety of bacteria that carry suitable biotransformation machineries.
As shown in Fig. 8, TetR family members are key players in multidrug resistance, virulence, and pathogenicity processes in certain bacterial pathogens. The development of drugs that bind irreversibly to the repressors and prevent their release from their cognate operators may be a strategy to fight these pathogens. Suitable screening procedures to search for these drugs can be envisaged based on current tools for gram-negative bacteria (74).
Acknowledgments
Work in our laboratories was supported by EC grants MIFRIEND QLK3-CT 2000-017 and BIOCARTE QLRT-2001-01923 and projects CICYT-2003 (BIO2003-00515) and Petri-NBT (PTR1995-0653 PO) to J.L.R. and grant 0021/2002 from the Human Frontier Science Program to M.T.G., K.W., and X.Z.
We thank W. Hindrich and S. Horinouchi for comments on the article. We thank Inés Abril and Carmen Lorente for secretarial assistance and Karen Shashok for checking the use of English in the manuscript. R. Tobes acknowledges support from Eduardo Pareja.
REFERENCES
- 1.Abo-Amer, A. E., J. Munn, K. Jackson, M. Aktas, P. Golby, D. J. Kelly, and S. C. Andrews. 2004. DNA interaction and phosphotransfer of the C4-dicarboxylate-responsive DcuS-DcuR two-component regulatory system from Escherichia coli. J. Bacteriol. 186:1879-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlert, J., E. Shepard, N. Lomovskaya, E. Zazopoulos, A. Staffa, B. O. Bachmann, K. Huang, L. Fonstein, A. Czisny, R. E. Whitwam, C. M. Farnet, and J. S. Thorson. 2002. The calicheamicin gene cluster and its iterative type I enediyne PKS. Science 297:1173-1176. [DOI] [PubMed] [Google Scholar]
- 3.Aigle, B., A. Wietzorrek, E. Takano, and M. J. Bibb. 2000. A single amino acid substitution in region 1.2 of the principal sigma factor of Streptomyces coelicolor A3(2) results in pleiotropic loss of antibiotic production. Mol. Microbiol. 37:995-1004. [DOI] [PubMed] [Google Scholar]
- 4.Alekshun, M. N., and S. B. Levy. 1999. Alteration of the repressor activity of MarR, the negative regulator of the Escherichia coli marRAB locus, by multiple chemicals in vitro. J. Bacteriol. 181:4669-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allignet, J., S. Aubert, K. G. Dyke, and N. El Solh. 2001. Staphylococcus caprae strains carry determinants known to be involved in pathogenicity: a gene encoding an autolysin-binding fibronectin and the ica operon involved in biofilm formation. Infect. Immun. 69:712-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allmeier, H., B. Cresnar, M. Greck, and R. Schmitt. 1992. Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111:11-120. [DOI] [PubMed] [Google Scholar]
- 7.Amabile-Cuevas, C. F., and B. Demple. 1991. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 19:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andresen, P. A., I. Kaasen, O. B. Styrvold, G. Boulnois, and A. R. Strøm. 1988. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J. Gen. Microbiol. 134:1737-1746. [DOI] [PubMed] [Google Scholar]
- 9.Aramaki, H., Y. Sagara, M. Hosoi, and T. Horiuchi. 1993. Evidence for autoregulation of camR, which encodes a repressor for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J. Bacteriol. 175:7828-7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aramaki, H., Y. Sagara, H. Kabata, N. Shimamoto, and T. Horiuchi. 1995. Purification and characterization of a cam repressor (CamR) for the cytochrome P-450cam hydroxylase operon on the Pseudomonas putida CAM plasmid. J. Bacteriol. 177:3120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aramaki, H., N. Yagi, and M. Suzuki. 1995. Residues important for the function of a multihelical DNA binding domain in the new transcription factor family of Cam and Tet repressors. Protein Eng. 8:1259-1266. [DOI] [PubMed] [Google Scholar]
- 12.Argos, P., M. Hanei, J. M. Wilson, and W. N. Kelley. 1983. A possible nucleotide-binding domain in the tertiary fold of phosphoribosyltransferases. J. Biol. Chem. 258:6450-6457. [PubMed] [Google Scholar]
- 13.Ariza, R. R., S. P. Cohen, N. Bachawat, S. B. Levy, and B. Demple. 1994. Repressor mutations in the marRAB operon that activate oxidative stress genes and multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 176:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariza, R. R., Z. Li, N. Ringstad, and B. Demple. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attwood, T. K., P. Bradley, D. R. Flower, A. Gaulton, N. Maudling, A. L. Mitchell, G. Moulton, A. Nordle, K. Paine, P. Taylor, A. Uddin, and C. Zygouri. 2003. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 31:400-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attwood, T. K., M. D. Croning, D. R. Flower, A. P. Lewis, J. E. Mabey, P. Scordis, J. N. Selley, and W. Wright. 2000. Prints-S: The database formerly known as Prints. Nucleic Acids Res. 28:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.August, P. R., L. Tang, Y. J. Yoon, S. Ning, R. Muller, T. W. Yu, M. Taylor, D. Hoffmann, C. G. Kim, X. Zhang, C. R. Hutchinson, and H. G. Floss. 1998. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 5:69-79. [DOI] [PubMed] [Google Scholar]
- 18.Babu, M. M., and S. A. Teichmann. 2003. Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res. 31:1234-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bairoch, A., and K. E. Rudd. 1995. PROSITE. Accession no. PDOC00874.
- 20.Balsalobre, L., M. J. Ferrándiz, J. Liñares, F. Tubau, and A. G. de la Campa. 2003. Viridans group Streptococci are donors in horizontal transfer of topoisomerase IV genes to Streptococcus pneumoniae. Antimicrob. Agents Chemother. 47:2072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron, U., M. Gossen, and H. Bujard. 1997. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 25:2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 24.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 25.Bate, N., A. R. Butler, A. R. Gandecha, and E. Cundliffe. 1999. Multiple regulatory genes in the tylosin biosynthetic cluster of Streptomyces fradiae. Chem. Biol. 6:617-624. [DOI] [PubMed] [Google Scholar]
- 26.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baulard, A. R., J. C. Betts, J. Ndong, S. Quan, R. A. McAdam, P. J. Brennan, C. Locht, and G. S. Besra. 2000. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275:28326-28331. [DOI] [PubMed] [Google Scholar]
- 29.Baumeister, R., V. Helbl, and W. Hillen. 1992. Contacts between Tet repressor and tet operator revealed by new recognition specificities of single amino acid replacement mutants. J. Mol. Biol. 226:1257-1270. [DOI] [PubMed] [Google Scholar]
- 30.Baumeister, R., G. Muller, B. Hecht, and W. Hillen. 1992. Functional roles of amino acid residues involved in forming the alpha-helix-turn-alpha-helix operator DNA binding motif of Tet repressor from Tn10. Proteins 14:168-177. [DOI] [PubMed] [Google Scholar]
- 31.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 32.Beinlich, K. L., R. Chuanchuen, and H. P. Schweizer. 2001. Contribution of multidrug efflux pumps to multiple antibiotic resistance in veterinary clinical isolates of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 198:129-134. [DOI] [PubMed] [Google Scholar]
- 33.Belli, G., E. Gari, L. Piedrafita, M. Aldea, and E. Herrero. 1998. An activator/repressor dual system allows right tetracycline regulated gene expression in budding yeast. Nucleic Acids Res. 26:942-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beloin, C., S. Ayora, R. Exley, L. Hirschbein, N. Ogasawara, Y. Kasahara, J. C. Alonso, and F. Le Hegarat. 1997. Characterization of an lrp-like (lrpC) gene from Bacillus subtilis. Mol. Gen. Genet. 256:63-71. [DOI] [PubMed] [Google Scholar]
- 35.Bennik, H. J. M., P. J. Pomposiello, D. F. Thorne, and B. Demple. 2000. Defining a rob regulon in Escherichia coli by using transposon mutagenesis. J. Bacteriol. 182:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berens, C., Altschmied, L., and W. Hillen. 1992. The role of the N terminus in Tet repressor for tet operator binding determined by a mutational analysis. J. Biol. Chem. 267:1945-1952. [PubMed] [Google Scholar]
- 37.Berens, C., and W. Hillen. 2003. Gene regulation by tetracycline. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 270:3109-3121. [DOI] [PubMed] [Google Scholar]
- 38.Berens, C., D. Schnappinger, and W. Hillen. 1997. The role of the variable region in Tet repressor for inducibility by tetracycline. J. Biol. Chem. 272:6936-6942. [DOI] [PubMed] [Google Scholar]
- 39.Biburger, M., C. Berens, T. Lederer, T. Krec, and W. Hillen. 1998. Intragenic suppressors of induction-deficient TetR mutants: localization and potential mechanism of action. J. Bacteriol. 180:737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blanco, C., P. Ritzenthaler, and M. Mata-Gilsinger. 1986. Negative dominant mutations of the uidR gene in Escherichia coli: genetic proof for a cooperative regulation of uidA expression. Genetics 112:173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branden, C., and J. Tooze. 1991. Introduction to protein structure. Garland Publishing Co., New York, N.Y.
- 42.Brandi, A., C. L. Pon, and C. O. Gualerzi. 1994. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie 76:1090-1098. [DOI] [PubMed] [Google Scholar]
- 43.Brennan, R. G., S. L. Roderick, Y. Takeda, and B. W. Matthews. 1990. Protein-DNA conformational changes in the crystal structure of a lambda-Cro-operator complex. Proc. Natl. Acad. Sci. USA 87:8165-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown, M. H., and R. A. Skurray. 2001. Staphylococcal multidrug efflux protein QacA. J. Mol. Biotechnol. 3:163-170. [PubMed] [Google Scholar]
- 45.Bucher, P., and A. Bairoch. 1994. A generalized profile syntax for biomolecular sequence motifs and its function in automatic sequence interpretation. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:53-61. [PubMed] [Google Scholar]
- 46.Bucher, P., K. Karplus, N. Moeri, and K. Hofmann. 1996. A flexible motif search technique based on generalized profiles. Comput. Chem. 20:3-24. [DOI] [PubMed] [Google Scholar]
- 47.Budarina, Z. I., D. V. Nikitin, N. Zenkin, M. Zakharova, E. Semenova, M. G. Shlyapnikov, E. A. Rodikova, S. Masyukova, O. Ogarkov, G. E. Baida, A. S. Solonin, and K. Severinov. 2004. A new Bacillus cereus, DNA-binding protein, HlyIIR, negatively regulates expression of B. cereus haemolysin II. Microbiology. 150:3691-3701. [DOI] [PubMed] [Google Scholar]
- 48.Burkovski, A. 2003. I do it my way: Regulation of ammonium uptake and ammonium assimilation in Corynebacterium glutamicum. Arch. Microbiol. 179:83-88. [DOI] [PubMed] [Google Scholar]
- 49.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 50.Caballero, J. L., F. Malpartida, and D. A. Hopwood. 1991. Transcriptional organization and regulation of an antibiotic export complex in the producing Streptomyces culture. Mol. Gen. Genet. 228:372-380. [DOI] [PubMed] [Google Scholar]
- 51.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-22166. [PubMed] [Google Scholar]
- 52.Cases, I., V. de Lorenzo, and C. A. Ouzounis. 2003. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 11:248-253. [DOI] [PubMed] [Google Scholar]
- 53.Chan, Y. Y., T. M. C. Tan, Y. M. Ong, and K. L. Chua. 2004. BpeAB-OprB, a multidrug efflux pump in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 48:1128-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandler, M. S. 1992. The gene encoding cAMP receptor protein is required for competence development in Haemophilus influenzae Rd. Proc. Natl. Acad. Sci. USA 89:1626-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chater, K. F. 1993. Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47:685-713. [DOI] [PubMed] [Google Scholar]
- 56.Chater, K. F., and C. J. Bruton. 1985. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 4:1893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterjee, J., C. M. Miyamoto, and E. A. Meighen. 1996. Autoregulation of luxR: the Vibrio harveyi lux-operon activator functions as a repressor. Mol. Microbiol. 20:415-425. [DOI] [PubMed] [Google Scholar]
- 58.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 59.Cheng, J. C., C. P. Shao, and L. I. Hor. 1996. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene 183:255-257. [DOI] [PubMed] [Google Scholar]
- 60.Cho, S. H., K. Naber, J. Hacker, and W. Ziebuhr. 2002. Detection of the icaADBC gene cluster and biofilm formation in Staphylococcus epidermidis isolates from catheter-related urinary tract infections. Int. J. Antimicrob. Agents. 19:570-575. [DOI] [PubMed] [Google Scholar]
- 61.Cho, Y. H., E. J. Kim, H. J. Chung, J. H. Choi, K. F. Chater, B. E. Ahn, J. H. Shin, and J. H. Roe. 2003. The pqrAB operon is responsible for paraquat resistance in Streptomyces coelicolor. J. Bacteriol. 185:6756-6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi, E. N., M. C. Cho, Y. Kim, C. K. Kim, and K. Lee. 2003. Expansion of growth substrate range in Pseudomonas putida F1 by mutations in both cymR and todS, which recruit a ring-fission hydrolase CmtE and induce the tod catabolic operon, respectively. Microbiology 149:795-805. [DOI] [PubMed] [Google Scholar]
- 63.Choi, H. K., N. Y. Park, D. I. Kim, H. J. Chung, S. Ryu, and S. H. Choi. 2002. Promoter analysis and regulatory characteristics of vvhBA encoding cytolytic hemolysin of Vibrio vulnificus. J. Biol. Chem. 277:47292-47299. [DOI] [PubMed] [Google Scholar]
- 64.Cohen, S. P., H. Hachler, and S. B. Levy. 1993. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J. Bacteriol. 175:1484-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Compan, I., and D. Touati. 1994. Anaerobic activation of arcA transcription in Escherichia coli: roles of Fnr and ArcA. Mol. Microbiol. 11:955-964. [DOI] [PubMed] [Google Scholar]
- 66.Conlon, K. M., H. Humphreys and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:171-177. [DOI] [PubMed] [Google Scholar]
- 67.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corpet, F., J. Gouzy, and D. Kahn. 1999. Recent improvements of the Prodom database of protein domain families. Nucleic Acids Res. 27:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cousin, S. L., Jr., W. L. H. Whittington, and M. C. Roberts. 2003. Acquired macrolide resistance genes and the 1 bp deletion in the mtrR promoter in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 51:131-133. [DOI] [PubMed] [Google Scholar]
- 70.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation.Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Croxatto, A., V. J. Chalker, J. Lauritz, J. Jass, A. Hardman, P. Williams, M. Camara, and D. L. Milton. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delahay, R. M., B. D. Robertson, J. T. Balthazar, W. M. Shafer, and C. A. Ison. 1997. Involvement of the gonococcal MtrE protein in the resistance of Neisseria gonorrhoeae to toxic hydrophobic agents. Microbiology 143:2127-2133. [DOI] [PubMed] [Google Scholar]
- 73.Delany, I., R. Ieva, C. Alaimo, R. Rappuoli, and V. Scarlato. 2003. The iron-responsive regulator Fur is transcriptionally autoregulated and not essential in Neisseria meningitidis. J. Bacteriol. 185:6032-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.de Lorenzo, V., and H. P. Schweizer. 2004. Molecular tools for genetic analysis of pseudomonads p. 317-350. In J. L. Ramos (ed.), Pseudomonas, vol. I. Kluwer, London, United Kingdom. [Google Scholar]
- 75.Derzelle, S., E. Turlin, E. Duchaud, S. Pages, F. Kunst, A. Givaudan, and A. Danchin. 2004. The PhoP-phoQ two-component regulatory system of Photorhabdus luminescens is essential for virulence in insects. J. Bacteriol. 186:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dingermann, T., U. Frank-Stoll, H. Wener, A. Wissmann, W. Hillen, M. Jacquet, and R. Marschalek. 1992. RNA polymerase III catalysed transcription can be regulated in Saccaharomyces cerevisiae by the bacterial tetracycline repressor-operator system. EMBO J. 11:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dogra, C., V. Raina, R. Pal, M. Suar, S. Lal, K. H. Gartemann, C. Holliger, J.R. van der Meer, and R. Lal. 2004. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J. Bacteriol. 186:2225-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doull, J. L., A. K. Singh, M. Hoare, and S. W. Ayer. 1994. Conditions for the production of jadomycin B by Streptomyces venezuelae ISP5230: effects of heat shock, ethanol treatment and phage infection. J. Ind. Microbiol. 13:120-125. [DOI] [PubMed] [Google Scholar]
- 79.Dover, L. G., P. E. Corsino, I. R. Daniels, S. L. Cocklin, V. Tatituri, G. S. Besra, and K. Fütterer. 2004. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 340:1095-1105. [DOI] [PubMed] [Google Scholar]
- 80.Dugan, J., D. D. Rockey, L. Jones, and A. A. Andersen. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duque, E., A. Segura, G. Mosqueda, and J. L. Ramos. 2001. Global and cognate regulators control the expression of the organic solvent efflux pumps TtgABC and TtgDEF of Pseudomonas putida. Mol. Microbiol. 39:1100-1106. [DOI] [PubMed] [Google Scholar]
- 82.Eaton, R. W. 1996. p-Cumate catabolic pathway in Pseudomonas putida Fl: cloning and characterization of DNA carrying the cmt operon. J. Bacteriol. 178:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eaton, R. W. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J. Bacteriol. 179:3171-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Egan, S. 2002. Growing repertoire of AraC/XylS activators. J. Bacteriol. 184:5529-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.El-Hajj, H. H., H. Zhang, and B. Weiss. 1988. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J. Bacteriol. 170:1069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Engel, P., L. L. Scharfenstein, J. M. Dyer, and J. W. Cary. 2001. Disruption of a gene encoding a putative gamma-butyrolactone-binding protein in Streptomyces tendae affects nikkomycin production. Appl. Microbiol. Biotechnol. 56:414-419. [DOI] [PubMed] [Google Scholar]
- 87.English, N., V. Hughes, and C. R. Wolf. 1994. Common pathways of cytochrome P450 gene regulation by peroxisome proliferators and barbiturates in Bacillus megaterium ATCC14581. J. Biol. Chem. 269:26836-26841. [PubMed] [Google Scholar]
- 88.English, N., V. Hughes, and C. R. Wolf. 1996. Induction of cytochrome P-450 BM-3 (CYP 102) by non-steroidal anti-inflammatory drugs in Bacillus megaterium. Biochem. J. 316:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.English, N., C. N. Palmer, W. L. Alworth, L. Kang, V. Hughes, and C. R. Wolf. 1997. Fatty acid signals in Bacillus megaterium are attenuated by cytochrome P-450-mediated hydroxylation. Biochem. J. 327:363-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engohang-Ndong, J., Baillat, D., Aumercier, M., Bellefontaine, F., Besra, G. S., Locht, C., and Baulard, A. R. 2004. EthR, a repressor of the TetR/CamR family implicated in ethionamide resistance in mycobacteria, octamerizes cooperatively on its operator. Mol. Microbiol. 51:175-188. [DOI] [PubMed] [Google Scholar]
- 91.Eshoo, M. W. 1988. lac fusion analysis of the bet genes of Escherichia coli: regulation by osmolarity, temperature, oxygen, choline, and glycine betaine. J. Bacteriol. 170:5208-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Everett, M., Y. F. Jin, V. Ricci, and L. J. V. Piddock. 1996. Contribution of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farrow, K. A., D. Lyras, and J. I. Rood. 2001. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 147:2717-2728. [DOI] [PubMed] [Google Scholar]
- 94.Faust, B., D. Hoffmeister, G. Weitnauer, L. Westrich, S. Haag, P. Schneider, H. Decker, E. Kunzel, J. Rohr, and A. Bechthold. 2000. Two new tailoring enzymes, a glycosyltransferase and an oxygenase, involved in biosynthesis of the angucycline antibiotic urdamycin A in. Streptomyces fradiae Tu2717. Microbiology. 146:147-154. [DOI] [PubMed] [Google Scholar]
- 95.Felscher, D. W., and J. M. Bishop. 1999. Reversible tumorogenesis by MYC in hematopoietic lineages. Mol. Cell 2:451-461. [DOI] [PubMed] [Google Scholar]
- 96.Fernández-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 97.Fidopiastis, P. M., C. M. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 98.Fischer, G. H., S.l. Wellen, D. Klimstra, J. M. Lenczowski, J. W. Tichelaar, M. J. Lizak, J. A. Whitsett, A. Korestsky, and H. E. Varmus. 2001. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Folcher, M., R. P. Morris, G. Dale, K. Salah-Bey-Hocini, P. H. Viollier, and C. J. Thompson. 2001. A transcriptional regulator of a pristinamycin resistance gene in Streptomyces coelicolor. J. Biol. Chem. 276:1479-1485. [DOI] [PubMed] [Google Scholar]
- 100.Fomenko, D. E., A. Z. Metlitskaya, J. Péduzzi, C. Goulard, G. S. Katrukha, L. V. Gening, S. Rebuffat, and I. A. Khmel. 2003. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 47:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fong, S. T., J. Camakaris, and B. T. Lee. 1995. Molecular genetics of a chromosomal locus involved in copper tolerance in Escherichia coli K-12. Mol. Microbiol. 15:1127-1137. [DOI] [PubMed] [Google Scholar]
- 102.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Friedberg, D., M. Midkiff, and J. M. Calvo. 2001. Global versus local regulatory roles for Lrp-related proteins: Haemophilus influenzae as a case study. J. Bacteriol. 183:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fromknecht, K., P. D. Vogel, and J. G. Wise. 2003. Combinatorial redesign of the DNA binding specificity of a prokaryotic helix-turn-helix repressor. J. Bacteriol. 185:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fujita, M., H. Aramaki, T. Horiuchi, and A. Amemura. 1993. Transcription of the cam operon and camR genes in Pseudomonas putida PpG1. J. Bacteriol. 175:6953-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 108.Gallegos, M. T., C. Michán, and J. L. Ramos. 1993. The XylS/AraC family of regulators. Nucleic Acids Res. 21:807-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. AraC/XylS of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gambino, L., S. J. Gracheck, and P. F. Miller. 1993. Overexpression of the MarA positive regulator is sufficient to confer multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 175:2888-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.George, A. M., and S. B. Levy. 1983. Gene in the major cotransduction gap of the Escherichia coli K-12 linkage map required for the expression of chromosomal resistance to tetracycline and other antibiotics. J. Bacteriol. 178:2507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.González-Pasayo, R., and E. Martínez-Romero. 2000. Multiresistance genes of Rhizobium etli CFN42. Mol. Plant-Microbe Interact. 13:572-577. [DOI] [PubMed] [Google Scholar]
- 114.Gossen, M., A. L. Bonin, and H. Bujard. 1993. Control of gene activity in higher eukaryotes cells by prokaryotic regulatory elements. Trends Biochem. Sci. 18:471-475. [DOI] [PubMed] [Google Scholar]
- 115.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 117.Gray, K. M., and J. R. Garey. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379-2387. [DOI] [PubMed] [Google Scholar]
- 118.Green, J., C. Scott, and J. R. Guest. 2001. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv. Microb. Physiol. 44:1-34. [DOI] [PubMed] [Google Scholar]
- 119.Grkovic, S., M. H. Brown, N. J. Roberts, I. T. Paulsen, and R. A. Skurray. 1998. QacR is a repressor protein that regulates expression of the Staphylococcus aureus multidrug efflux pump QacA. J. Biol. Chem. 273:18665-18673. [DOI] [PubMed] [Google Scholar]
- 120.Grkovic, S., M. H. Brown, M. A. Schumacher, R. G. Brennan, and R. A. Skurray. 2001. Staphylococcal QacR multidrug regulator binds a correctly spaced operator as a pair of dimers. J. Bacteriol. 183:7102-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Grkovic, S., M. H. Brown, and R. A. Skurray. 2001. Transcriptional regulation of multidrug efflux pumps in bacteria. Cell Dev. Biol. 12:225-237. [DOI] [PubMed] [Google Scholar]
- 122.Grkovic, S., K. M. Hardie, M. H. Brown, and R. A. Skurray. 2003. Interactions of the QacR multidrug-binding protein with structurally diverse ligands: implications for the evolution of the binding pocket. Biochemistry 42:15226-15236. [DOI] [PubMed] [Google Scholar]
- 123.Grogan, G., G. A. Roberts, D. Bougioukou, N. J. Turner, and S. L. Flitsch. 2001. The desymmetrization of bicyclic beta-diketones by enzymatic retro-claisen reaction. A new reaction of the crotonase superfamily. J. Biol. Chem. 276:12565-12572. [DOI] [PubMed] [Google Scholar]
- 124.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Guengerich, F. P. 1991. Reactions and significance of cytochrome P-450 enzymes. J. Biol. Chem. 266:10019-10022. [PubMed] [Google Scholar]
- 126.Guilfoile, P. G., and C. R. Hutchinson. 1992. Sequence and transcriptional analysis of the Streptomyces glaucescens tcmAR tetracenomycin C resistance and repressor gene loci. J. Bacteriol. 174:3651-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guilfoile, P. G., C. R. Hutchinson. 1992. The Streptomyces glaucescens TcmR protein represses transcription of the divergently oriented tcmR and tcmA genes by binding to an intergenic operator region. J. Bacteriol. 174:3659-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Haft, D. H., B. J. Loftus, D. L. Richardson, F. Yang, J. A. Eisen, I. T. Paulsen, and O. White. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29:41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hagen, K. D., and J. C. Meeks. 1999. Biochemical and genetic evidence for participation of DevR in a phosphorelay signal transduction pathway essential for heterocyst maturation in Nostoc punctiforme ATCC 29133. J. Bacteriol. 181:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hagman, K. E., W. Pan, B. G. Spratt, J. T. Balthazar, R. C. Judd, and W. M. Shafer. 1995. Resistance of Neisseria gonorrhoeae to antimicrobial agents is modulated by the mtrRCDE efflux system. Microbiology 141:611-622. [DOI] [PubMed] [Google Scholar]
- 131.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J. Bacteriol. 177:4162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hansson, M., and L. Hederstedt. 1992. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J. Bacteriol. 174:8081-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hara, O., and T. Beppu. 1982. Mutants blocked in streptomycin production in Streptomyces griseus—the role of A-factor. J. Antibiot. 35:349-358. [DOI] [PubMed] [Google Scholar]
- 134.Hara, O., and T. Beppu. 1982. Induction of streptomycin-inactivating enzyme by A-factor in Streptomyces griseus. J. Antibiot. 35:1208-1215. [DOI] [PubMed] [Google Scholar]
- 135.Harrison, S. C., and A. K. Aggarwal. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu. Rev. Biochem. 59:933-969. [DOI] [PubMed] [Google Scholar]
- 136.Harrison, S. C. 1991. A structural taxonomy of DNA-binding domains. Nature 353:715-719. [DOI] [PubMed] [Google Scholar]
- 137.Hashimoto, Y., M. Yamashita, and Y. Murooka. 1997. The Propionibacterium freudenreichii hemYHBXRL gene cluster, which encodes enzymes and a regulator involved in the biosynthetic pathway from glutamate to protoheme. Appl. Microbiol. Biotechnol. 47:385-392. [DOI] [PubMed] [Google Scholar]
- 138.Haydon, D. J., and J. R. Guest. 1991. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 63:291-295. [DOI] [PubMed] [Google Scholar]
- 139.Hayes, A., G. Hobbs, C. P. Smith, S. G. Oliver, and P. R. Butler. 1997. Environmental signals triggering methylenomycin production by Streptomyces coelicolor A3(2). J. Bacteriol. 179:5511-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He, J. S., Q. Liang, and A. J. Fulco. 1995. The molecular cloning and characterization of BM1P1 and BM1P2 proteins, putative positive transcription factors involved in barbiturate-mediated induction of the genes encoding cytochrome P450BM-1 of Bacillus megaterium. J. Biol. Chem. 270:18615-18625. [DOI] [PubMed] [Google Scholar]
- 141.Helbl, W., C. Berens, and W. Hillen. 1995. Proximity probing of Tet repressor to tet operator by dimenthylsulfate reveals protected and accessible functions for each recognized base-pair in the major groove. J. Mol. Biol. 245:538-548. [DOI] [PubMed] [Google Scholar]
- 142.Helbl, V., and W. Hillen. 1998. Stepwise selection of TetR variants recognizing tet operator 4C with high affinity and specificity. J. Mol. Biol. 276:313-318. [DOI] [PubMed] [Google Scholar]
- 143.Helbl, V., B. Tiebel, and W. Hillen. 1998. Stepwise selection of TetR variants recognizing tet operator 6C with high affinity and specificity. J. Mol. Biol. 276:319-324. [DOI] [PubMed] [Google Scholar]
- 144.Helmann, J. D., B. T. Ballard, and C. T. Walsh. 1990. The MerR metaloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science 247:946-948. [DOI] [PubMed] [Google Scholar]
- 145.Henikoff, S., G. W. Haughn, J. M. Calvo, and J. C. Wallace. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. USA 85:6602-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Henikoff, S., J. C. Wallace, and J. P. Brown. 1990. Finding protein similarities with nucleotide sequence databases. Methods Enzymol. 183:111-132. [DOI] [PubMed] [Google Scholar]
- 147.Heuer, H., R. Szczepanowski, S. Schneiker, A. Pühler. E.M. Top, and A. Schlüter. 2004. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1β group without any accessory genes. Microbiology 150:3591-3599. [DOI] [PubMed] [Google Scholar]
- 148.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 149.Hillen, W., K. Schollmeier, and C. Gatz. 1984. Control of expression of the Tn10-encoded tetracycline resistance operon. II. Interaction of RNA polymerase and Tet repressor with the tet operon regulatory region. J. Mol. Biol. 172:185-201. [DOI] [PubMed] [Google Scholar]
- 150.Hinrichs, W., C. Kisker, M. Düvel, A. Müller, K. Tovar, W. Hillen, and W. Saenger. 1994. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264:418-420. [DOI] [PubMed] [Google Scholar]
- 151.Hirata, T., A. Saito, K. Nishino, N. Tamura, and A. Yamaguchi. 2004. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob. Agents Chemother. 48:2179-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hitomi, M., H. Nishimura, Y. Tsujimoto, H. Matsui, and K. Watanabe. 2003. Identification of a helix-turn-helix motif of Bacillus thermoglucosidasius HrcA essential for binding to the CIRCE element and thermostability of the HrcA-CIRCE complex, indicating a role as a thermosensor. J. Bacteriol. 185:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hobbs, G., A. I. Obanye, J. Petty, J. C. Mason, E. Barratt, D. C. Gardner, F. Flett, C. P. Smith, P. Broda, and S. G. Oliver. 1992. An integrated approach to studying regulation of production of the antibiotic methylenomycin by Streptomyces coelicolor A3(2). J. Bacteriol. 174:1487-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The Prosite database, its status in 1999. Nucleic Acids Res. 27:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Horinouchi, S., and T. Beppu. 1990. Autoregulatory factors of secondary metabolism and morphogenesis in actinomycetes. Crit. Rev. Biotechnol. 10:191-204. [DOI] [PubMed] [Google Scholar]
- 156.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 157.Horinouchi, S., Y. Ohnishi, and D. K. Kang. 2001. The A-factor regulatory cascade and cAMP in the regulation of physiological and morphological development in Streptomyces griseus. J. Ind. Microbiol. Biotechnol. 27:177-182. [DOI] [PubMed] [Google Scholar]
- 158.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:2045-2057. [DOI] [PubMed] [Google Scholar]
- 159.Isackson, P. J., and K. P. Bertrand. 1985. Dominant negative mutations in the Tn10 Tet repressor: evidence for use of the conserved helix-turn-helix motif in DNA binding. Proc. Natl. Acad. Sci. USA 82:6226-6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Itoh, K., Y. Tashiro, K. Uobe, Y. Kamagata, K. Suyama, and H. Yamamoto. 2004. Root nodule Bradyrhizobium spp. harbor tfdAα and cadA, homologous with genes encoding 2,4-dichlorophenoxyacetic acid-degrading proteins. Appl. Environ. Microbiol. 70:2110-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jakoby, M., L. Nolden, J. Meier-Wagner, R. Kramer, and A. Burkovski. 2000. AmtR, a global repressor in the nitrogen regulation system of Corynebacterium glutamicum. Mol. Microbiol. 37:964-977. [DOI] [PubMed] [Google Scholar]
- 162.Janssen, D. B., F. Pries, J. van der Ploeg, B. Kazemier, P. Terpstra, and B. Witholt. 1989. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10 and expression and sequencing of the dhlA gene. J. Bacteriol. 171:6791-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jefferson, K. K., S. E. Cramton, F. Gotz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 164.Jellen-Ritter, A. S., and W. V. Kern. 2001. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob. Agents Chemother. 45:1467-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 166.Jogun, K. H., and J. J. Stezowski. 1976. Coordination and conformational aspects of oxytetracycline metal ion complexation. J. Am. Chem. Soc. 98:6018-6026. [DOI] [PubMed] [Google Scholar]
- 167.Johnston, T. C., R. B. Thompson, and T. O. Baldwin. 1986. Nucleotide sequence of the luxB gene of Vibrio harveyi and the complete amino acid sequence of the beta subunit of bacterial luciferase. J. Biol. Chem. 261:4805-4811. [PubMed] [Google Scholar]
- 168.Kamionka, A., J. Bogdanska-Urbaniak, O. Scholz, and W. Hillen. 2004. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 32:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kaneko, M., A. Yamaguchi, and T. Sawai. 1985. Energetics of tetracycline efflux system encoded by Tn10 in Escherichia coli. FEBS Lett. 193:893-899. [DOI] [PubMed] [Google Scholar]
- 170.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentiallly diverse roles in environmental signaling. J. Bacteriol. 186:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kasuga, K., H. Nojiri, H. Yamane, T. Kodama, and T. Omori. 1997. Cloning and characterization of genes involved in the degradation of dibenzofuran by Terrabacter sp. strain DBF63. J. Ferment. Bioeng. 84:387-399. [Google Scholar]
- 172.Katayama, J., and N. Noguchi. 1999. Nucleotide sequence of the gene cluster containing the mphB gene for macrolide 2′-phosphotransferase II. Biol. Pharm. Bull. 22:227-228. [DOI] [PubMed] [Google Scholar]
- 173.Kato, J.-Y., I. Miyahisa, M. Mashiko, Y. Ohnishi, and S. Horinouchi. 2004. A single target is sufficient to account for biological effects of the A-factor receptor protein of Streptomyces griseus. J. Bacteriol. 186:2206-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kato, J., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in acrial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6010-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kawachi, R., T. Akashi, Y. Kamitani, A. Sy, U. Wangchaisoonthorn, T. Nihira, and Y. Yamada. 2000. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36:302-313. [DOI] [PubMed] [Google Scholar]
- 176.Kawamura-Sato, K., K. Shibayama, T. Horii, Y. Iimuma, Y. Arakawa, and M. Ohta. 1999. Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol. Lett. 179:345-352. [DOI] [PubMed] [Google Scholar]
- 177.Kieboom, J., R. Bruinenberg, I. Keizer-Gunnink, and J. A. de Bont. 2001. Transposon mutations in the flagella biosynthetic pathway of the solvent-tolerant Pseudomonas putida S12 result in a decreased expression of solvent efflux genes. FEMS Microbiol. Lett. 198:117-122. [DOI] [PubMed] [Google Scholar]
- 178.Kieboom, J., and J. de Bont. 2001. Identification and molecular characterization of an efflux system involved in Pseudomonas putida S12 multidrug resistance. Microbiology 147:43-51. [DOI] [PubMed] [Google Scholar]
- 179.Kieboom, J., J. J. Dennis, J.A. de Bont, and G. J. Zylstra. 1998. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 273:85-91. [DOI] [PubMed] [Google Scholar]
- 180.Kieboom, J., J. J. Dennis, G. J. Zylstra, J.A. de Bont. 1998. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180:6769-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kim, Y. B., J. Okuda, C. Matsumoto, N. Takahashi, S. Hashimoto, and M. Nishibuchi. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37:1173-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kinoshita, H., H. Ipposhi, S. Okamoto, H. Nakano, T. Nihira, and Y. Yamada. 1997. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J. Bacteriol. 179:6986-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kisker, C., W. Hinrichs, K. Tovar, W. Hillen, and W. Saenger. 1995. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 247:260-280. [DOI] [PubMed] [Google Scholar]
- 184.Kitani, S., H. Kinoshita, T. Nihira, and Y. Yamada, Y. 1999. In vitro analysis of the butyrolactone autoregulator receptor protein (FarA) of Streptomyces lavendulae FRI-5 reveals that FarA acts as a DNA-binding transcriptional regulator that controls its own synthesis. J. Bacteriol. 181:5081-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kitani, S., Y. Yamada, and T. Nihira. 2001. Gene replacement analysis of the butyrolactone autoregulator receptor (FarA) reveals that FarA acts as a novel regulator in secondary metabolism of Streptomyces lavendulae FRI-5. J. Bacteriol. 183:4357-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Klein, J. R., B. Henrich, and R. Plapp. 1991. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol. Gen. Genet. 230:230-240. [DOI] [PubMed] [Google Scholar]
- 187.Klein, J. R., and R. Plapp. 1992. Locations of the envCD genes on the physical map of the Escherichia coli chromosome. J. Bacteriol. 174:3828-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Klinke, S., G. de Roo, B. Witholt, and B. Kessler. 2000. Role of phaD in accumulation of medium-chain-length poly(3-hydroxyalkanoates) in Pseudomonas oleovorans. Appl. Environ. Microbiol. 66:3705-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klock, G., B. Unger, C. Gatz, W. Hillen, J. Altenbuchner, K. Schmid, and R. Schmitt. 1985. Heterologous repressor-operator recognition among four classes of tetracycline resistance determinants. J. Bacteriol. 161:326-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kobayashi, K., N. Tsukagoshi, and R. Aono. 2001. Suppression of hypersensitivity of Escherichia coli acrB mutant to organic solvents by integrational activation of the acrEF operon with the IS1 or IS2 element. J. Bacteriol. 183:2646-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kojic, M., and V. Venturi, V. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 183:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 194.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 195.Krug, A., V. F. Wendisch, and M. Bott. 2005. Identification of AcnR, a TetR-type repressor of the acotinase gene acn in Corynebacterium glutamicum. J. Biol. Chem. 280:585-595. [DOI] [PubMed] [Google Scholar]
- 196.Krukonis, E. S., R. R. Yu, and V. J. Dirita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 197.Kumano, M., A. Tamakoshi, and K. Yamane. 1997. A 32 kb nucleotide sequence from the region of the lincomycin-resistance gene (22 degrees-25 degrees) of the Bacillus subtilis chromosome and identification of the site of the lin-2 mutation. Microbiology 143:2775-2782. [DOI] [PubMed] [Google Scholar]
- 198.Kumano, M., M. Fujita, K. Nakamura, M. Murata, R. Ohki, and K. Yamane. 2003. Lincomycin resistance mutations in two regions immediately downstream of the −10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 47:432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Kunin, C. M., T. H. Hua, W. L. Van Arsdale, and M. Villarejo. 1992. Growth of Escherichia coli in human urine: role of salt tolerance and accumulation of glycine betaine. J. Infect. Dis. 166:1311-1315. [DOI] [PubMed] [Google Scholar]
- 200.Kustu, S., A. K. North, and D. S. Weiss. 1991. Prokaryotic transcriptional enhancers and enhancer-binding proteins. Trends Biochem. Sci. 16:397-402. [DOI] [PubMed] [Google Scholar]
- 201.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strom. 1995. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 202.Lamark, T., T. P. Rokenes, J. McDougall, and A. R. Strom. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178(6):1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lamark, T., O. B. Styrvold, and A. R. Strøm. 1992. Efflux of choline and glycine betaine from osmoregulating cells of Escherichia coli. FEMS Microbiol. Lett. 96:149-154. [DOI] [PubMed] [Google Scholar]
- 204.Landfald, B., and A. R. Strom. 1986. Choline-glycine betaine pathway a high level of osmotic tolerance in Escherichia coli. J. Bacteriol. 165(3):849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.La Teana, A., A. Brandi, M. Falconi, R. Spurio, C. L. Pon, and C. O. Gualerzi. 1991. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. USA 88:10907-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lau, P. C. K., Y. Wang, A. Patel, D. Labbe, H. Bergeron, R. Brousseau, Y. Konishi, and M. Rawlings. 1997. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc. Natl. Acad. Sci. USA 94:1453-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lauble, H., Y. Georgalis, and U. Heinemann. 1989. Studies on the domain structure of the Salmonella typhimurium AraC protein. Eur. J. Biochem. 185:319-325. [DOI] [PubMed] [Google Scholar]
- 208.Lee, E. H., C. Rouquette-Loughlin, J. P. Folster, and W. M. Shafer. 2003. FarR regulates the farAB-encoded efflux pump of Neisseria gonorrhoeae via an MtrR regulatory mechanism. J. Bacteriol. 185:7145-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Levy, S. B. 1984. Resistance to tetracyclines p. 191-204. In L. E. Bryan (ed.), Antimicrobial drug resistance. Academic Press, New York, N.Y.
- 210.Levy, S. B. 1988. Tetracycline resistance determinants are widespread. ASM News. 54:418-421. [Google Scholar]
- 211.Levy, S. B., L. M. McMurry, T. M. Barbosa, V. Burdett, P. Courvalin, W. Hillen, M. C. Roberts, J. I. Rood, and D. E. Taylor. 1999. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 43:1523-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Li, X.-Z., L. Zhang, and H. Nikaido. 2004. Efflux pump-mediated intrinsic drug resistance in Mycobacterium smegmatis. Antimicrob. Agents Chemother. 48:2415-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liang, Q., L. Chen, and A. J. Fulco. 1998. In vivo roles of Bm3R1 repressor in the barbiturate-mediated induction of the cytochrome P450 genes (P450(BM-3) and P450(BM-)1) of Bacillus megaterium. Biochim. Biophys. Acta 1380:183-197. [DOI] [PubMed] [Google Scholar]
- 214.Liang, Q., and A. J. Fulco. 1995. Transcriptional regulation of the genes encoding cytochromes P450BM-1 and P450BM-3 in Bacillus megaterium by the binding of Bm3R1 repressor to Barbie box elements and operator sites. J. Biol. Chem. 270:18606-18614. [DOI] [PubMed] [Google Scholar]
- 215.Liang, Q., J. S. He, and A. J. Fulco, A. J. 1995. The role of Barbie box sequences as cis-acting elements involved in the barbiturate-mediated induction of cytochromes P450BM-1 and P450BM-3 in Bacillus megaterium. J. Biol. Chem. 270:4438-4450. [DOI] [PubMed] [Google Scholar]
- 216.Lin, Y. H., C. Miyamoto, and E. A. Meighen. 2000. Purification and characterization of a luxO promoter binding protein LuxT from Vibrio harveyi. Protein Expr. Purif. 20:87-94. [DOI] [PubMed] [Google Scholar]
- 217.Lin, Z., K. Kumagai, K. Baba, J. J. Mekalanos, and M. Nishibuchi. 1993. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J. Bacteriol. 175:3844-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Liu, H., and K. A. Reynolds. 1999. Role of crotonyl coenzyme A reductase in determining the ratio of polyketides monensin A and monensin B produced by Streptomyces cinnamonensis. J. Bacteriol. 181:6806-6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Liu, H., and K. A. Reynolds. 2001. Precursor supply for polyketide biosynthesis: the role of crotonyl-CoA reductase. Metab. Eng. 3:40-48. [DOI] [PubMed] [Google Scholar]
- 220.Lucas, C. E., K. E. Hagman, J. C. Levin, D. C. Stein, and W. M. Shafer. 1995. Importance of lipooligosaccharide structure in determining gonococcal resistance to hydrophobic antimicrobial agents resulting from the mtr efflux system. Mol. Microbiol. 16:1001-1009. [DOI] [PubMed] [Google Scholar]
- 221.Lucas, C. E., J. T. Balthazar, K. E. Hagman, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 179:4123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ly, A., J. Henderson, A. Lu, D. E. Culham, and J. M. Wood. 2004. Osmoregulatory systems of Escherichia coli: identification of betaine-carnitine-choline transporter family member BetU and distributions of betU and trkG among pathogenic and nonpathogenic isolates. J. Bacteriol. 186:296-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 224.Ma, D., M. Alberti, C. Lynch, N. G. Pon, H. Nikaido, and J. E. Hearst. 1995. Genes acrAB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45-55. [DOI] [PubMed] [Google Scholar]
- 225.Ma, D., D. N. Cook, M. Alberti, N. G. Pon, H. Nikaido, and J. E. Hearst. 1993. Molecular cloning and characterization of acrA and acrE genes of Eschericia coli. J. Bacteriol. 175:6299-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ma, D., D. N. Cook, J. Hearst, and H. N. Nikaido. 1994. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 2:489-493. [DOI] [PubMed] [Google Scholar]
- 227.Mahren, S., and W. Braun. 2003. The FecI extracytoplasmic-function sigma factor of Escherichia coli interacts with the β'subunit of RNA polymerase. J. Bacteriol. 185:1796-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128:321-330. [DOI] [PubMed] [Google Scholar]
- 229.Manzanera, M., S. Marqués, and J. L. Ramos. 2000. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 476:312-317. [DOI] [PubMed] [Google Scholar]
- 230.Marger, M. D., and M. H. Saier, Jr. 1993. A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18:13-20. [DOI] [PubMed] [Google Scholar]
- 231.Marqués, S., M. T. Gallegos, M. Manzanera, A. Holtel, K. N. Timmis, and J. L. Ramos. 1998. Activation and repression of transcription at the double tandem divergent promoters for the xylR and xylS genes of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 180:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Martin, M., R. Showalter, and M. Silverman. 1989. Identification of a locus controlling expression of luminescence genes in Vibrio harveyi. J. Bacteriol. 171:2406-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Martínez-Argudo, I., P. Salinas, R. Maldonado, and A. Contreras. 2002. Domain interactions on the ntr signal transduction pathway: two-hybrid analysis of mutant and truncated derivatives of histidine kinase NtrB. J. Bacteriol. 184:200-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Martínez-Argudo, I., and A. Contreras. 2002. PII T-Loop mutations affecting signal transduction to NtrB also abolish yeast two-hybrid interactions. J. Bacteriol. 184:3746-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Martinez-Bueno, M., Molina-Henares, A. J., Pareja, E., Ramos, J. L., Tobes, R. 2004. BacTregulators: a database of transcriptional regulators in bacteria and archaea. Bioinformatics 20:2787-2791. [DOI] [PubMed] [Google Scholar]
- 236.Martínez-Bueno, M., Tobes, R., Rey, M., and Ramos, J. L. 2002. Detection of multiple extracytoplasmic function (ECF) sigma factors in the genome of Pseudomonas putida KT2440 and their counterparts in Pseudomonas aeruginosa PA01. Environ. Microbiol. 4:842-855. [DOI] [PubMed] [Google Scholar]
- 237.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 238.Mavroidi, A., L. S. Tzouvelekis, K. P. Kyriakis, H. Avgerinou, M. Danilidou, and E. Tzelepi. 2001. Multidrug-resistant strains of Neisseria gonorrhoeae in Greece. Antimicrob. Agents Chemother. 45:2651-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mayer, K., G. Murphy, R. Tarchini, R. Wambutt, G. Volckaert, T. Pohl, A. Duesterhoeft, W. Stiekema, K. D. Entian, N. Terryn, K. Lemcke, D. Haase, C. R. Hall, A.M. van Dodeweerd, S. V. Tingey, H. W. Mewes, M. W. Bevan, and I. Bancroft. 2001. Conservation of microstructure between a sequenced region of the genome of rice and multiple segments of the genome of Arabidopsis thaliana. Genome Res. 11:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.McCarter, L. L. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1:51-57. [PubMed] [Google Scholar]
- 242.McDougald, D., S. A. Rice, and S. Kjelleberg. 2000. The marine pathogen Vibrio vulnificus encodes a putative homologue of the Vibrio harveyi regulatory gene, luxR: a genetic and phylogenetic comparison. Gene. 248:213-221. [DOI] [PubMed] [Google Scholar]
- 243.McDougald, D., S. A. Rice and S. Kjelleberg. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McKay, D. B., and T. A. Steitz. 1981. Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature 290:744-749. [DOI] [PubMed] [Google Scholar]
- 245.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 246.Méndez, B., C. Tachibana, and S. B. Levy. 1980. Heterogeneity of tetracycline resistance determinants. Plasmid 3:99-108. [DOI] [PubMed] [Google Scholar]
- 247.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 248.Milton, D. L., A. Hardman, M. Camara, S. R. Chhabra, B. W. Bycroft, G. S. Stewart, and P. William. 1997. Quorum sensing in Vibrio anguillarum: characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol. 179:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mitchell, B. A., I. T. Paulsen, M. H. Brown, and R. A. Skurray. 1999. Bioenergetics of the staphylococcal multidrug export protein QacA. Identification of distinct binding sites for monovalent and divalent cations. J. Biol. Chem. 274:3541-3548. [DOI] [PubMed] [Google Scholar]
- 250.Miyamoto, C. M., M. Boylan, A. F. Graham, and E. A. Meighen. 1988. Organization of the lux structural genes of Vibrio harveyi. Expression under the T7 bacteriophage promoter, mRNA analysis, and nucleotide sequence of the luxD gene. J. Biol. Chem. 263:13393-13399. [PubMed] [Google Scholar]
- 251.Miyamoto, C. M., J. Chatterjee, E. Swartzman, R. Szittner, and E. A. Meighen. 1996. The role of lux autoinducer in regulating luminescence in Vibrio harveyi; control of luxR expression. Mol. Microbiol. 19:767-775. [DOI] [PubMed] [Google Scholar]
- 252.Miyamoto, C. M., Y. H. Lin, and E. A. Meighen. 2000. Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol. Microbiol. 36:594-607. [DOI] [PubMed] [Google Scholar]
- 253.Miyamoto, C. M., E. A. Meighen, and A. F. Graham. 1990. Transcriptional regulation of lux genes transferred into Vibrio harveyi. J. Bacteriol. 172:2046-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Miyamoto, C. M., E. E. Smith, E. Swartzman, J. G. Cao, A. F. Graham, and E. A. Meighen. 1994. Proximal and distal sites bind LuxR independently and activate expression of the Vibrio harveyi lux operon. Mol. Microbiol. 14:255-262. [DOI] [PubMed] [Google Scholar]
- 255.Miyamoto, C. M., W. Sun, and E. A. Meighen. 1998. The LuxR regulator protein controls synthesis of polyhydroxybutyrate in Vibrio harveyi. Biochim. Biophys. Acta 1384:356-364. [DOI] [PubMed] [Google Scholar]
- 256.Mohamed, M. S., W. Ismail, J. Heider, and G. Fuchs. 2002. Aerobic metabolism of phenylacetic acids in Azoarcus evansii. Arch. Microbiol. 178:180-192. [DOI] [PubMed] [Google Scholar]
- 257.Morett, E., and L. Segovia. 1993. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 175:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Mulder, N. J., R. Apweiler, T. K. Attwood, A. Bairoch, D. Barrell, A. Bateman, D. Binns, M. Biswas, P. Bradley, P. Bork, P. Bucher, R. R. Copley, E. Courcelle, U. Das, R. Durbin, L. Falquet, W. Fleischmann, S. Griffiths-Jones, D. Haft, N. Harte, N. Hulo, D. Kahn, A. Kanapin, M. Krestyaninova, R. Lopez, I. Letunic, D. Lonsdale, V. Silventoinen, S. E. Orchard, M. Pagni, D. Peyruc, C. P. Ponting, J. D. Selengut, F. Servant, C. J. Sigrist, R. Vaughan, and E. M. Zdobnov. 2003. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 31:315-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Muller, G., B. Hecht, V. Helbl, W. Hinrichs, W. Saenger, and W. Hillen. 1995. Characterization of non-inducible Tet repressor mutants suggests conformational changes necessary for induction. Nat. Struct. Biol. 2:693-703. [DOI] [PubMed] [Google Scholar]
- 260.Murata, M., S. Ohno, M. Kumano, K. Yamane, and R. Ohki. 2003. Multidrug resistant phenotype of Bacillus subtilis spontaneous mutants isolated in the presence of puromycin and lincomycin. Can. J. Microbiol. 49:71-77. [DOI] [PubMed] [Google Scholar]
- 261.Murray, D. A., M. A. Schumacher, and R. G. Brennan. 2004. Crystal structures of QacR-diamidine complexes reveal additional multidrug-binding modes and a novel mechanism of drug charge neutralization. J. Biol. Chem. 279:14365-14371. [DOI] [PubMed] [Google Scholar]
- 262.Nakano, H., E. Takehara, T. Nihira, and Y. Yamada. 1998. Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J. Bacteriol. 180:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Namwat, W., C. K. Lee, H. Kinoshita, Y. Yamada, and T. Nihira. 2001. Identification of the varR gene as a transcriptional regulator of virginiamycin S resistance in Streptomyces virginiae. J. Bacteriol. 183:2025-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Natsume, R., Y. Ohnishi, T. Senda, and S. Horinouchi. 2004. Crystal structure of a γ-butyrolactone autoregulator receptor protein in Streptomyces coelicolor A3(2). J. Mol. Biol. 336:409-419. [DOI] [PubMed] [Google Scholar]
- 265.Neal, R. J., and K. F. Chater. 1987. Nucleotide sequence analysis reveals similarities between proteins determining methylenomycin A resistance in Streptomyces and tetracycline resistance in eubacteria. Gene 58:229-241. [DOI] [PubMed] [Google Scholar]
- 266.Neal, R. J., and K. F. Chater. 1991. Bidirectional promoter and terminator regions bracket mmr, a resistance gene embedded in the Streptomyces coelicolor A3(2) gene cluster encoding methylenomycin production. Gene 100:75-83. [DOI] [PubMed] [Google Scholar]
- 267.Nikaido, H. N. 1996. Multidrug efflux pumps of Gram-negative bacteria. J. Bacteriol. 178:5853-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nikaido, H. N. 1998. Antibiotic resistance caused by Gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27:532-541. [DOI] [PubMed] [Google Scholar]
- 269.Nilsson, D., and M. Kilstrup. 1998. Cloning and expression of the Lactococcus lactis purDEK genes, required for growth in milk. Appl. Environ. Microbiol. 64:4321-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nishino, K., and A. Yamaguchi. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nixon, B. T., C. W. Ronson, and F. M. Ausubel. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. USA 83:7850-7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Noguchi, N., J. Katayama, and M. Sasatsu. 2000. A transposon carrying the gene mphB for macrolide 2′-phosphotransferase II. FEMS Microbiol. Lett. 192:175-178. [DOI] [PubMed] [Google Scholar]
- 273.Noguchi, N., K. Takada, J. Katayama, A. Emura, and M. Sasatsu. 2000. Regulation of transcription of the mph(A) gene for macrolide 2′-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J. Bacteriol. 182:5052-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.North, A. K., K. E. Klose, K. M. Stedman, and S. Kustu. 1993. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J. Bacteriol. 175:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Novakova, R., J. Bistakova, D. Homerova, B. Rezuchova, and J. Kormanec. 2002. Cloning and characterization of a polyketide synthase gene cluster involved in biosynthesis of a proposed angucycline-like polyketide auricin in Streptomyces aureofaciens CCM 3239. Gene 297:197-208. [DOI] [PubMed] [Google Scholar]
- 276.Oethinger, M., I. Podglajen, W. V. Kern, and S. B. Levy. 1998. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob. Agents Chemother. 42:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ohki, R., K. Tateno, Y. Okada, H. Okajima, K. Asai, Y. Sadaie, M. Murata, and T. Aiso. 2003. A bacitracin-resistant Bacillus subtilis gene encodes a homologue of the membrane-spanning subunit of the Bacillus licheniformis ABC transporter. J. Bacteriol. 185:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptocyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 279.Ohta, Y., M. Maeda, and T. Kudo. 2001. Pseudomonas putida CE2010 can degrade biphenyl by a mosaic pathway encoded by the tod operon and cmtE, which are identical to those of P. putida F1 except for a single base difference in the operator-promoter region of the cmt operon. Microbiology 147:31-41. [DOI] [PubMed] [Google Scholar]
- 280.Okamoto, S., K. Nakamura, T. Nihira, and Y. Yamada. 1995. Virginiae butanolide binding protein from Streptomyces virginiae. Evidence that VbrA is not the virginiae butanolide binding protein and reidentification of the true binding protein. J. Biol. Chem. 270:12319-12326. [DOI] [PubMed] [Google Scholar]
- 281.Okusu, H., D. Ma, and H. N. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Onaka, H., N. Ando, T. Nihira, Y. Yamada, T. Beppu, and S. Horinouchi. 1995. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J. Bacteriol. 177:6083-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Onaka, H., and S. Horinouchi. 1997. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol. Microbiol. 24:991-1000. [DOI] [PubMed] [Google Scholar]
- 284.Onaka, H., T. Nakagawa, and S. Horinouchi. 1998. Involvement of two A-factor receptor homologues in Streptomyces coelicolor A3(2) in the regulation of secondary metabolism and morphogenesis. Mol. Microbiol. 28:743-753. [DOI] [PubMed] [Google Scholar]
- 285.Onaka, H., M. Sugiyama, and S. Horinouchi. 1997. A mutation at proline-115 in the A-factor receptor protein of Streptomyces griseus abolishes DNA-binding ability but not ligand-binding ability. J. Bacteriol. 179:2748-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Orth, P., F. Cordes, D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 1998. Conformational changes of the Tet repressor induced by tetracycline trapping. J. Mol. Biol. 279:439-447. [DOI] [PubMed] [Google Scholar]
- 287.Orth, P., W. Saenger, and W. Hinrichs. 1999. Tetracycline-chelated Mg2+ ion initiates helix unwinding in Tet repressor induction. Biochemistry 38:191-198. [DOI] [PubMed] [Google Scholar]
- 288.Orth, P., D. Schnappinger, W. Hillen, W. Saenger, and W. Hinrichs. 2000. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 7:215-219. [DOI] [PubMed] [Google Scholar]
- 289.Orth, P., D. Schnappinger, P.-E. Sum, G. A. Ellestad, W. Hillen, W. Saenger, and W. Hinrichs. 1999. Crystal structure of the Tet repressor in complex with a novel tetracycline, 9-(N,N-dimethylglycylamido)-6-demethyl-6-deoxy-tetracycline. J. Mol. Biol. 285:455-461. [DOI] [PubMed] [Google Scholar]
- 290.Osteras, M., E. Boncompagni, N. Vincent, M. C. Poggi, and D. Le Rudulier. 1998. Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. USA 95:11394-11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Otten, S. L., J. Ferguson, and C. R. Hutchinson. 1995. Regulation of daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J. Bacteriol. 177:1216-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Pabo, C. O., and M. Lewis. 1992. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature 298:443-447. [DOI] [PubMed] [Google Scholar]
- 293.Pabo, C. O., and R. T. Sauer. 1984. Protein-DNA recognition. Annu. Rev. Biochem. 53:293-321. [DOI] [PubMed] [Google Scholar]
- 294.Pabo, C. O., and R. T. Sauer. 1992. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Microbiol. 61:1053-1095. [DOI] [PubMed] [Google Scholar]
- 295.Palmer, C. N., E. Axen, V. Hughes, and C. R. Wolf. 1998. The repressor protein, Bm3R1, mediates an adaptive response to toxic fatty acids in Bacillus megaterium. J. Biol. Chem. 273(29):18109-18116. [DOI] [PubMed] [Google Scholar]
- 296.Palumbo, J. D., C. I. Kado, and D. A. Phillips. 1998. An isoflavonoid-inducible efflux pump in Agrobacterium tumefaciens is involved in competitive colonization of roots. J. Bacteriol. 180:3107-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope by the mtr system. Mol. Microbiol. 11:769-775. [DOI] [PubMed] [Google Scholar]
- 298.Pao, G. M., R. Tam, L. S. Lipschitz, and M. H. Saier, Jr. 1994. Response regulators: structure, function and evolution. Res. Microbiol. 145:356-362. [DOI] [PubMed] [Google Scholar]
- 299.Paulsen, I. T., M. H. Brown, S. J. Dunstan, and R. A. Skurray. 1995. Molecular characterization of the staphylococcal multidrug resistance export protein QacC. J. Bacteriol. 177:2827-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Paulsen, I. T., M. H. Brown, T. G. Littlejohn, B. A. Mitchell, and R. A. Skurray. 1996. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc. Natl. Acad. Sci. USA 93:3630-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Peng, W. T., and E. W. Nester. 2001. Characterization of a putative RND-type efflux system in Agrobacterium tumefaciens. Gene 270:245-252. [DOI] [PubMed] [Google Scholar]
- 302.Pérez-Rueda, E., and J. Collado-Vides. 2001. Common history at the origin of the position-function correlation in transcriptional regulators in archaea and bacteria. J. Mol. Evol. 53:172-179. [DOI] [PubMed] [Google Scholar]
- 303.Perron, K., O. Caille, C. Rossier, C. van Delden, J. L. Dumas, and T. Köhler. 2004. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 279:8761-8768. [DOI] [PubMed] [Google Scholar]
- 304.Pocard, J. A., T. Bernard, L. T. Smith, and D. Le Rudulier. 1989. Characterization of three choline transport activities in Rhizobium meliloti: modulation by choline and osmotic stress. J. Bacteriol. 171:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Pocard, J. A., N. Vincent, E. Boncompagni, L. T. Smith, M. C. Poggi, and D. Le Rudulier. 1997. Molecular characterization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34. Microbiology 143:1369-1379. [DOI] [PubMed] [Google Scholar]
- 306.Poelarends, G. J. , L. A. Kulakov, M. J. Larkin, J. E. van Hylckama Vlieg, and D. B. Janssen. 2000. Roles of horizontal gene transfer and gene integration in evolution of 1,3-dichloropropene- and 1,2-dibromoethane-degradative pathways. J. Bacteriol. 182:2191-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 308.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. E. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Puskas, L. G., M. Inui, Z. Kele, and H. Yukawa. 2000. Cloning of genes participating in aerobic biodegradation of p-cumate from Rhodopseudomonas palustris. DNA Seq. 11:9-20. [DOI] [PubMed] [Google Scholar]
- 311.Ramos, J. L., E. Duque, M. T. Gallegos, P. Godoy, M. I. Ramos-González, A. Rojas, W. Terán, and A. Segura. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 312.Ramos, J. L., M. T. Gallegos, S. Marqués, M. I. Ramos-González, M. Espinosa-Urgel, and A. Segura. 2001. Response of gram-negative bacteria to certain environmental stresses. Curr. Opin. Microbiol. 4:166-171. [DOI] [PubMed] [Google Scholar]
- 313.Ramos, J. L., F. Rojo, L. Zhou, and K. N. Timmis. 1990. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 18:2149-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Rand, J. D., S. G. Danby, D. L. Greenway, and R. R. England. 2002. Increased expression of the multidrug efflux genes acrAB occurs during slow growth of Escherichia coli. FEMS Microbiol. Lett. 207:91-95. [DOI] [PubMed] [Google Scholar]
- 315.Rebets, Y., B. Ostash, A. Luzhetskyy, D. Hoffmeister, A. Brana, C. Mendez, J. A. Salas, A. Bechthold, and V. Fedorenko. 2003. Production of landomycins in Streptomyces globisporus 1912 and S. cyanogenus S136 is regulated by genes encoding putative transcriptional activators. FEMS Microbiol. Lett. 222:149-153. [DOI] [PubMed] [Google Scholar]
- 316.Redenbach, M., K. Ikeda, M. Yamasaki, and H. Kinashi. 1998. Cloning and physical mapping of the EcoRI fragments of the giant linear plasmid SCP1. J. Bacteriol. 180:2796-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 318.Reizer, A., J. Deutscher, M. H. Saier, and J. Reizer. 1991. Analysis of the gluconate (gnt) operon of Bacillus subtilis. Mol. Microbiol. 5:1081-1089. [DOI] [PubMed] [Google Scholar]
- 319.Reizer, J. , I. T. Paulsen, A. Reizer, F. Titgemeyer, and M. H. Saier, Jr. 1996. Novel phosphotransferase system genes revealed by bacterial genome analysis: the complete complement of pts genes in Mycoplasma genitalium. Microb. Comp. Genomics 1:151-164. [DOI] [PubMed] [Google Scholar]
- 320.Retzlaff, L., and J. Distler. 1995. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 18:151-162. [DOI] [PubMed] [Google Scholar]
- 321.Reverchon, S., W. Nasser, and J. Robert-Baudouy. 1991. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol. Microbiol. 5:2203-2216. [DOI] [PubMed] [Google Scholar]
- 322.Rey, D. A., A. Puhler, and J. Kalinowski. 2003. The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 103:51-65. [DOI] [PubMed] [Google Scholar]
- 323.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 324.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 325.Rodionov, D. A., M. S. Gelfand, A. A. Mironov, and A. B. Rakhmaninova. 2001. Comparative approach to analysis of regulation in complete genomes: multidrug resistance systems in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol. 3:319-324. [PubMed] [Google Scholar]
- 326.Roessner, C. A., K. X. Huang, M. J. Warren, E. Raux, and A. I. Scott. 2002. Isolation and characterization of 14 additional genes specifying the anaerobic biosynthesis of cobalamin (vitamin B12) in. Propionibacterium freudenreichii (P. shermanii). Microbiology 148:1845-1853. [DOI] [PubMed] [Google Scholar]
- 327.Rojas, A., E. Duque, G. Mosqueda, G. Golden, A. Hurtado, J. L. Ramos, and A. Segura. 2001. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 183:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Rojas, A., Duque, E., Schmid, A., A. Hurtado, and J. L. Ramos. 2004. Biotransformation in double phase systems: physiological responses of Pseudomonas putida DOT T1E to a double phase made of aliphatic alcohols and biosynthesis of substituted catechols. Appl. Environ. Microbiol. 70:3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Rojas, A., A. Segura, M. E. Guazzaroni, W. Terán, A. Hurtado, M. T. Gallegos, and J. L. Ramos. 2003. In vivo and in vitro evidence that TtgV is the specific regulator of the TtgGHI multidrug and solvent efflux pump of Pseudomonas putida. J. Bacteriol. 185:4755-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Rojo, F. 1999. Repression of transcription initiation in bacteria. J. Bacteriol. 181:2987-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Røkenes, T. P., T. Lamark, and A. R. Strøm. 1996. DNA-binding properties of the BetI repressor protein of Escherichia coli: the inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 178:1663-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Rouch, D. A., D. S. Cram, D. DiBerardino, T. G. Littlejohn, and R. A. Skurray. 1990. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 4:2051-2062. [DOI] [PubMed] [Google Scholar]
- 333.Rouquette, C., J. B. Harmon, and W. M. Shafer. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33:651-658. [DOI] [PubMed] [Google Scholar]
- 334.Rouquette-Loughlin, C., I. Stojiljkovic, T. Hrobowski, J. T. Balthazar, and W. M. Shafer. 2002. Inducible, but not constitutive, resistance of gonococci to hydrophobic agents due to the MtrC-MtrD-MtrE efflux pump requires TonB-ExbB-ExbD proteins. Antimicrob. Agents Chemother. 46:561-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Rousseau, P., E. Guegen, G. Duval-Valentin, and M. Chandler. 2004. The helix-turn-helix motif of bacterial insertion sequence IS911 transposase is required for DNA binding. Nucleic Acids Res. 32:1335-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ryu, J. R., L. K. Olson, and D. N. Arnosti. 2001. Cell-type specificity of short-range transcriptional repressors. Proc. Natl. Acad. Sci. USA 98:12960-12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Saenger, W., P. Orth, C. Kisker, W. Hillen, and W. Hinrichs. 2000. The tetracycline repressor-A paradigm for a biological switch. Angew. Chem. Int. ed. Engl. 39:2042-2052. [DOI] [PubMed] [Google Scholar]
- 338.Sala, C., F. Forti, E. di Florio, F. Canneva, A. Milano, G. Riccardi, and D. Ghisotti. 2003. Mycobacterium tuberculosis FurA autoregulates its own expression. J. Bacteriol. 185:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Salah-Bey, K., and C. J. Thompson. 1995. Unusual regulatory mechanism for a Streptomyces multidrug resistance gene, ptr, involving three homologous protein-binding sites overlapping the promoter region. Mol. Microbiol. 17:1109-1119. [DOI] [PubMed] [Google Scholar]
- 340.Sánchez, P., A. Alonso, and J. L. Martínez. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob. Agents Chemother. 46:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Schauder, S., and B. L. Bassler. 2001. The languages of bacteria. Genes Dev. 15:1468-1480. [DOI] [PubMed] [Google Scholar]
- 342.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 343.Schevitz, R. W., Z. Otwinowski, A. Joachimiak, C. L Lawson, and P. B. Singler. 1985. The three-dimensional structure of trp repressor. Nature 317:782-786. [DOI] [PubMed] [Google Scholar]
- 344.Schindelin, H., M. A. Marahiel, and U. Heinemann. 1993. Universal nucleic acid-binding domain revealed by crystal structure of the B. subtilis major cold-shock protein. Nature 364:164-168. [DOI] [PubMed] [Google Scholar]
- 345.Schleif, R. 1987. The L-arabinose operon, p. 1473-1481. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Um-barger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 346.Schnider-Keel, U., A. Seematter, M. Maurhofer, C. Blumer, B. Duffy, C. Gigot-Bonnefoy, C. Reimmann, R. Notz, G. Defago, D. Haas, and C. Keel. 2000. Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin. J. Bacteriol. 182:1215-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Schouls, L. M., C. S. Schot, and J. A. Jacobs. 2003. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J. Bacteriol. 185:7241-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Schultz, J. , R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. Smart: A Web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Schumacher, M. A., and R. G. Brennan. 2002. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol. Microbiol. 45:885-893. [DOI] [PubMed] [Google Scholar]
- 350.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2001. Structural mechanisms of QacR induction and multidrug recognition. Science 294:2158-2163. [DOI] [PubMed] [Google Scholar]
- 351.Schumacher, M. A., M. C. Miller, S. Grkovic, M. H. Brown, R. A. Skurray, and R. G. Brennan. 2002. Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J. 21:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Shafer, W. M., J. T. Balthazar, K. E. Hagman, and S. A. Morse. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141:907-911. [DOI] [PubMed] [Google Scholar]
- 354.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 355.Shao, C. P., and L. I. Hor. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Shaw, G. C., and A. J. Fulco. 1992. Barbiturate-mediated regulation of expression of the cytochrome P450BM-3 gene of Bacillus megaterium by Bm3R1 protein. J. Biol. Chem. 267:5515-5526. [PubMed] [Google Scholar]
- 357.Shaw, G. C., and A. J. Fulco. 1993. Inhibition by barbiturates of the binding of Bm3R1 repressor to its operator site on the barbiturate-inducible cytochrome P450BM-3 gene of Bacillus megaterium. J. Biol. Chem. 268:2997-3004. [PubMed] [Google Scholar]
- 358.Shaw, G. C., Y. H. Hsueh, and H. S. Kao. 2000. The basal-level expression of the cytochrome P450(BM-1) gene is negatively affected by the bm1P1 gene of Bacillus megaterium. Curr. Microbiol. 40:47-50. [DOI] [PubMed] [Google Scholar]
- 359.Shaw, G. C., Y. H. Hsueh, C. C. Sung, Y. S. Chen, and C. H. Liu. 2000. Negative regulation of expression of the Bacillus megaterium bmlP1 gene by the bmlP1 3′ flanking region. J. Bacteriol. 182:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Shaw, G. C., H. S. Kao, C. C. Sung, and A. N. Chiang. 1997. Iron and salicylate induction of cytochrome P450BM-1 in Bacillus megaterium. Curr. Microbiol. 35:28-31. [DOI] [PubMed] [Google Scholar]
- 361.Shaw, G. C., C. C. Sung, C. H. Liu, and H. S. Kao. 1997. A 53-base-pair inverted repeat negatively regulates expression of the adjacent and divergently oriented cytochrome P450(BM-1) gene and its regulatory gene, bm1P1, in Bacillus megaterium. J. Bacteriol. 179:280-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Shaw, G. C., C. C. Sung, C. H. Liu, and C. H. Lin. 1998. Evidence against the Bm1P1 protein as a positive transcription factor for barbiturate-mediated induction of cytochrome P450BM-1 in bacillus megaterium. J. Biol. Chem. 273:7996-8002. [DOI] [PubMed] [Google Scholar]
- 363.Showalter, R. E., M. O. Martin, and M. R. Silverman. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Sigrist, C. J. , L. Cerutti, N. Hulo, A. Gattiker, L. Falquet, M. Pagni, A. Bairoch, and P. Bucher. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3:265-274. [DOI] [PubMed] [Google Scholar]
- 365.Silva, A. J. , K. Pham, and J. A. Benitez. 2003. Haemagglutinin/protease expression and mucin gel penetration in El Tor biotype Vibrio cholerae. Microbiology 149:1883-1891. [DOI] [PubMed] [Google Scholar]
- 366.Sizemore, C., A. Wissmann, U. Gülland, and W. Hillen. 1990. Quantitative analysis of Tn10 Tet repressor binding to a complete set of tet operator mutants. Nucleic Acids Res. 18:2875-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Skarstad, K., B. Thony, D. S. Hwang, and A. Kornberg. 1993. A novel binding protein of the origin of the Escherichia coli chromosome. J. Biol. Chem. 268:5365-5370. [PubMed] [Google Scholar]
- 368.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276:421-425. [DOI] [PubMed] [Google Scholar]
- 370.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie van Leeuwenhoek 66:23-36. [DOI] [PubMed] [Google Scholar]
- 371.Stratigopoulos, G., and E. Cundliffe. 2002. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ gene product. Chem. Biol. 9:71-78. [DOI] [PubMed] [Google Scholar]
- 372.Stratigopoulos, G., A. R. Gandecha, and E. Cundliffe. 2002. Regulation of tylosin production and morphological differentiation in Streptomyces fradiae by TylP, a deduced gamma-butyrolactone receptor. Mol. Microbiol. 45:735-744. [DOI] [PubMed] [Google Scholar]
- 373.Strom, A. R., P. Falkenberg, and B. Landfald. 1986. Genetics of osmoregulation in Escherichia coli: uptake and biosynthesis of organic osmolytes. FEMS Microbiol. Rev. 39:79-86. [Google Scholar]
- 374.Styrvold, O. B., P. Falkenberg, B. Landfald, M. W. Eshoo, T. Bjørnsen, and A. R. Strøm. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sugiyama, M., H. Onaka, T. Nakagawa, and S. Horinouchi. 1998. Site-directed mutagenesis of the A-factor receptor protein: Val-41 important for DNA-binding and Trp-119 important for ligand-binding. Gene 222:133-144. [DOI] [PubMed] [Google Scholar]
- 376.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436-446. [PMC free article] [PubMed] [Google Scholar]
- 377.Summers, A. O. 1992. Untwist and shout: a heavy metal-responsive transcriptional regulator. J. Bacteriol. 174:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Sunnarborg, A., D. Klumpp, T. Chung, and D. C. LaPorte. 1990. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J. Bacteriol. 172:2642-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Swartzman, E., C. Miyamoto, A. Graham, and E. Meighen. 1990. Delineation of the transcriptional boundaries of the lux operon of Vibrio harveyi demonstrates the presence of two new lux genes. J. Biol. Chem. 265:3513-3517. [PubMed] [Google Scholar]
- 381.Swartzman, E., and E. A. Meighen, E. A. 1993. Purification and characterization of a poly(dA-dT) lux-specific DNA-binding protein from Vibrio harveyi and identification as LuxR. J. Biol. Chem. 268:16706-16716. [PubMed] [Google Scholar]
- 382.Swartzman, E., M. Silverman, and E. A. Meighen. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 174:7490-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Szczepanowski, R., I. Krahn, B. Linke, A. Goesman, A. Pühler, and A. Schlüter. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613-3630. [DOI] [PubMed] [Google Scholar]
- 384.Takahashi, M., L. Altschmied, and W. Hillen. 1986. Kinetic and equilibrium characterization of the Tet-repressor-tetracycline complex by fluorescence measurements; evidence for divalent ion requirements and energy transfer. J. Mol. Biol. 187:341-348. [DOI] [PubMed] [Google Scholar]
- 385.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamad, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 386.Takano, E., T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada, and M. Bibb. 2000. Purification and structural determination of SCB1, a gamma-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275:11010-11016. [DOI] [PubMed] [Google Scholar]
- 387.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from. Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48:117-129. [DOI] [PubMed] [Google Scholar]
- 389.Tauch, A., A. Pühler, J. Kalinowski, and G. Thierbach. 2000. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid 44:285-291. [DOI] [PubMed] [Google Scholar]
- 390.Tennent, J. M., B. R. Lyon, M. T. Gillespie, J. W. May, and R. A. Skurray. 1985. Cloning and expression of Staphylococcus aureus plasmid-mediated quaternary ammonium resistance in Escherichia coli. Antimicrob. Agents Chemother. 27:79-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Terán, W., A. Felipe, A. Segura, A. Rojas, J. L. Ramos, and M. T. Gallegos. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 47:3067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Thieffry, D., A. M. Huerta, E. Perez-Rueda, and J. Collado-Vides. 1998. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli. Bioessays 20:433-440. [DOI] [PubMed] [Google Scholar]
- 393.Tiebel, B., K. Garke, and W. Hillen, W. 2000. Observing conformational and activity changes of tet repressor in vivo. Nat. Struct. Biol. 7:479-481. [DOI] [PubMed] [Google Scholar]
- 394.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30:318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Tovar, K., A. Ernst, and W. Hillen. 1988. Identification and nucleotide sequence of the class E tet regulatory elements and operator and inducer binding of the encoded purified Tet repressor. Mol. Gen. Genet. 215:76-80. [DOI] [PubMed] [Google Scholar]
- 396.Trefzer, A., S. Pelzer, J. Schimana, S. Stockert, C. Bihlmaier, H. P. Fiedler, K. Welzel, A. Vente, and A. Bechthold. 2002. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic. Antimicrob. Agents Chemother. 46:1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Tropel, D., and J. R. van der Meer. 2004. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68:474-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Ueda, K., K. Matsuda, H. Takano, and T. Beppu. 1999. A putative regulatory element for carbon-source-dependent differentiation in Streptomyces griseus. Microbiology 145:2265-2271. [DOI] [PubMed] [Google Scholar]
- 399.Ueda, K., H. Takano, M. Nishimoto, H. Inaba, and T. Beppu. 2005. Dual transcriptional control of amfTSBA, which regulates the onset of cellular differentiation in Streptomyces griseus. J. Bacteriol. 187:135-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Ulmasou, B., J. Capone, and W. Folk. 1997. Regulated expression of plant RNA genes by the prokaryotic tet and lac repressors. Plant Mol. Biol. 25:417-424. [DOI] [PubMed] [Google Scholar]
- 401.Umeyama, T., P. C. Lee, K. Ueda, and S. Horinouchi. 1999. An AfsK/AfsR system involved in the response of aerial mycelium formation to glucose in Streptomyces griseus. Microbiology 145:2281-2292. [DOI] [PubMed] [Google Scholar]
- 402.Unger, B., G. Klock, and W. Hillen. 1984. Nucleotide sequence of the repressor gene of the RA1 tetracycline resistance determinant: structural and functional comparison with three related Tet repressor genes. Nucleic Acids Res. 12:7693-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Urlinger, S., V. Helbl, J. Guthman, E. Pook, S. Grimm, and W. Hillen. 2000. The P65 domain from NF-KB is an efficient human activator in the tetracycline-regulatable gene expression system. Gene 247:103-110. [DOI] [PubMed] [Google Scholar]
- 404.van Der Geize, R., G. I. Hessels, R. van Gerwen, J. W. Vrijbloed, P. van Der Meijden, and L. Dijkhuizen. 2000. Targeted disruption of the kstD gene encoding a 3-ketosteroid delta(1)-dehydrogenase isoenzyme of Rhodococcus erythropolis strain SQ1. Appl. Environ. Microbiol. 66:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.van Rooijen, R. J. , and W. M. de Vos. 1990. Molecular cloning, transcriptional analysis and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J. Biol. Chem. 265:18499-18503. [PubMed] [Google Scholar]
- 406.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for chromosomically mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 184:5619-5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Veal, W. L., and W. M. Shafer. 2003. Identification of a cell envelope protein (MtrF) involved in hydrophobic antimicrobial resistance in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 51:27-32. [DOI] [PubMed] [Google Scholar]
- 408.Veal, W. L., A. Yellen, J. T. Balthazar, W. Pan, B. G. Spratt, and W. M. Shafer. 1998. Loss-of-function mutations in the mtr efflux system of Neisseria gonorrhoeae. Microbiology 144:621-627. [DOI] [PubMed] [Google Scholar]
- 409.Ventre, I., A. Filloux, and A. Lazdunski. 2004. Two-component signal transduction systems: a key to the adaptative potential of Pseudomonas aeruginosa p. 257-288. In J. L. Ramos (ed.), Pseudomonas, vol. II. Kluwer, London, England. [Google Scholar]
- 410.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Vujaklija, D., K. Ueda, S. K. Hong, T. Beppu, and S. Horinouchi. 1991. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol. Gen. Genet. 229:119-128. [DOI] [PubMed] [Google Scholar]
- 412.Vujaklija, D., S. Horinouchi, and T. Beppu. 1993. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 175:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Waki, M., T. Nihira, and Y. Yamada. 1997. Cloning and characterization of the gene (farA) encoding the receptor for an extracellular regulatory factor (IM-2) from Streptomyces sp. strain FRI-5. J. Bacteriol. 179:5131-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Walczak, R. J. , A. J. Woo, W. R. Strohl, and N. D. Priestley. 2000. Nonactin biosynthesis: the potential nonactin biosynthesis gene cluster contains type II polyketide synthase-like genes. FEMS Microbiol. Lett. 183:171-175. [DOI] [PubMed] [Google Scholar]
- 415.Wang, L., and B. Weiss. 1992. dcd (dCTP deaminase) gene of Escherichia coli: mapping, cloning, sequencing, and identification as a locus of suppressors of lethal dut (dUTPase) mutations. J. Bacteriol. 174:5647-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Wang, L., R. L. White, and L. C. Vining. 2002. Biosynthesis of the dideoxysugar component of jadomycin B: genes in the jad cluster of Streptomyces venezuelae ISP5230 for L-digitoxose assembly and transfer to the angucycline aglycone. Microbiology 148:1091-1103. [DOI] [PubMed] [Google Scholar]
- 417.Wasylewski, Z., P. Kaszycki, and M. Drwiega. 1996. A fluorescence study of Tn10-encoded tet repressor. J. Protein Chem. 15:45-58. [DOI] [PubMed] [Google Scholar]
- 418.Waters, S. H., P. Rogowsky, J. Grinsted, J. Altenbuchner, and R. Schmitt. 1983. The tetracycline resistance determinants of RP1 and Tn1721: nucleotide sequence analysis. Nucleic Acids Res. 11:6089-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Weber, W., R. Schoenmakers, M. Spielmann, M. D. El-Baba, M. Folcher, B. Keller, C. C. Weber, N. Link, P. van de Wetering, C. Heinzen, B. Jolivet, U. Sequin, D. Aubel, C. J. Thompson, and M. Fussenegger. 2003. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 31:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Weickert, M. J. , and S. Adhya. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869-15874. [PubMed] [Google Scholar]
- 421.Wen, L. P., and A. J. Fulco. 1987. Cloning of the gene encoding a catalytically self-sufficient cytochrome P-450 fatty acid monooxygenase induced by barbiturates in Bacillus megaterium and its functional expression and regulation in heterologous (Escherichia coli) and homologous (Bacillus megaterium) hosts. J. Biol. Chem. 262:6676-6682. [PubMed] [Google Scholar]
- 422.Wery, J. , B. Hidayat, J. Kieboom, and J. A. de Bont. 2001. An insertion sequence prepares Pseudomonas putida S12 for severe solvent stress. J. Biol. Chem. 276:5700-5706. [DOI] [PubMed] [Google Scholar]
- 423.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterizarion of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Westrich, L., S. Domann, B. Faust, D. Bedford, D. A. Hopwood, and A. Bechthold. 1999. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol. Lett. 170:381-387. [DOI] [PubMed] [Google Scholar]
- 425.White, D. G., J. D. Goldman, B. Demple, and S. B. Levy. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or rob in Escherichia coli. J. Bacteriol. 179:6122-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Wilson, M. S., J. B. Herrick, C. O. Jeon, D. E. Hinman, and E. L. Madsen. 2003. Horizontal transfer of phnAc dioxygenase genes within one of two phenotypically and genotypically distinctive naphthalene-degrading guilds from adjacent soil environments. Appl. Environ. Microbiol. 69:2172-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Winans, S. C., and B. L. Bassler. 2002. Mob psychology. J. Bacteriol. 184:873-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Wintjens, R., and M. Rooman. 1996. Structural classification of HTH DNA-binding domains and protein-DNA interaction modes. J. Mol. Biol. 262:294-313. [DOI] [PubMed] [Google Scholar]
- 429.Winzer, K., K. Lorenz, B. Zickner, and P. Durre. 2000. Differential regulation of two thiolase genes from Clostridium acetobutylicum DSM 792. J. Mol. Microbiol. Biotechnol. 2:531-541. [PubMed] [Google Scholar]
- 430.Wissmann, A., R. Baumeister, G. Muller, B. Hecht, V. Helbl, K. Pfleiderer, and W. Hillen. 1991. Amino acids determining operator binding specificity in the helix-turn-helix motif of Tn10 Tet repressor. EMBO J. 10:4145-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wösten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 432.Wu, J. , and B. P. Rosen. 1991. The ArsR protein is a trans-acting regulatory protein. Mol. Microbiol. 5:1331-1336. [DOI] [PubMed] [Google Scholar]
- 433.Xi, C., E. Schoeters, J. Vanderleyden, and J. Michiels. 2000. Symbiosis-specific expression of Rhizobium etli casA encoding a secreted calmodulin-related protein. Proc. Natl. Acad. Sci. USA 97:11114-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Yamaguchi, A., N. Ono, T. Akasaka, T. Noumi, and T. Sawai. 1999. Metal-tetracycline/H+ antiporter of Escherichia coli encoded by a transposon, Tn10. J. Biol. Chem. 265:15525-15530. [PubMed] [Google Scholar]
- 435.Yamaguchi, A., T. Udagawa, and T. Sawai. 1999. Transport of divalent cations with tetracycline as mediated by the transposon Tn10-encoded tetracycline resistance protein. J. Biol. Chem. 9:4809-4813. [PubMed] [Google Scholar]
- 436.Yamamoto, K., K. Yata, N. Fujita, and A. Ishihama. 2001. Novel mode of transcription regulation by SdiA, an Escherichia coli homologue of the quorum-sensing regulator. Mol. Microbiol. 41:1187-1198. [DOI] [PubMed] [Google Scholar]
- 437.Yamazaki, M., Y. Ikuto, A. Ohira, K. Chater, and H. Kinashi. 2003. Limited regions of homology between linear and circular plasmids encoding methylenomycin biosynthesis in two independently isolated streptomycetes. Microbiology 149:1351-1356. [DOI] [PubMed] [Google Scholar]
- 438.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 441.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]
- 442.Yang, K., L. Han, J. He, L. Wang, and L. C. Vining. 2001. A repressor-response regulator gene pair controlling jadomycin B production in. Streptomyces venezuelae ISP5230. Gene. 279:165-173. [DOI] [PubMed] [Google Scholar]
- 443.Yang, K., L. Han, and L. C. Vining. 1995. Regulation of jadomycin B production in Streptomyces venezuelae ISP5230: involvement of a repressor gene, jadR2. J. Bacteriol. 177:6111-61117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591-599. [DOI] [PubMed] [Google Scholar]
- 445.Yu, E. W., J. R. Aires, and H. Nikaido. 2003. AcrB multidrug efflux pump of Escherichia coli: composite substrate-binding cavity of exceptional flexibility generates its extremely wide substrate specificity. J. Bacteriol. 185:5657-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Yu, E. W., G. McDermott, H. I. Zgurskaya, H. Nikaido, D. E. Koshland, Jr. 2003. Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science 300:976-980. [DOI] [PubMed] [Google Scholar]
- 447.Zarantonelli, L., G. Borthagaray, E. H. Lee, and W. M. Shafer. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 43:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Zarantonelli, L., G. Borthagaray, E. H. Lee, E., W. Veal, and W. M. Shafer. 2001. Decreased susceptibility to azithromycin and erythromycin mediated by a novel mtr(R) promoter mutation in Neisseria gonorrhoeae. J. Antimicrob. Chemother. 47:651-654. [DOI] [PubMed] [Google Scholar]
- 449.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
- 450.Zeng, G., S. Ye, and T. J. Larson. 1996. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J. Bacteriol. 178:7080-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Zhang, G., X. Hang, P. Green, K. P. Ho, and G. Q. Chen. 2001. PCR cloning of type II polyhydroxyalkanoate biosynthesis genes from two Pseudomonas strains. FEMS Microbiol. Lett. 198:165-170. [DOI] [PubMed] [Google Scholar]
- 452.Zhang, L., X. Z. Li, and K. Poole. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Zhao, J. , and T. Aoki. 1992. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol. Immunol. 36:1051-1060. [DOI] [PubMed] [Google Scholar]
- 454.Zhou, L., L. Xiang-He, B. R. Bochner, and B. L. Wahner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Zhu, J. , M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]