Abstract
The use of matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry for the analysis of carbohydrates and glycoconjugates is a well‐established technique and this review is the 12th update of the original article published in 1999 and brings coverage of the literature to the end of 2022. As with previous review, this review also includes a few papers that describe methods appropriate to analysis by MALDI, such as sample preparation, even though the ionization method is not MALDI. The review follows the same format as previous reviews. It is divided into three sections: (1) general aspects such as theory of the MALDI process, matrices, derivatization, MALDI imaging, fragmentation, quantification and the use of computer software for structural identification. (2) Applications to various structural types such as oligo‐ and polysaccharides, glycoproteins, glycolipids, glycosides and biopharmaceuticals, and (3) other general areas such as medicine, industrial processes, natural products and glycan synthesis where MALDI is extensively used. Much of the material relating to applications is presented in tabular form. MALDI is still an ideal technique for carbohydrate analysis, particularly in its ability to produce single ions from each analyte and advancements in the technique and range of applications show little sign of diminishing.
Keywords: carbohydrates, glycolipids, glycoproteins, MALDI, synthesis, natural products
Abbreviations
- p (as in Galp)
pyranose form of sugar
- 2‐AB
2‐aminobenzamide
- 2VP
butyl‐terminated poly(2‐vinylpyridine
- A2F
core‐fucosylated biantennary, N‐glycan
- AA
aminoacridine
- AC
aminocinnoline‐3‐carboxamide
- ACE2
angiotensin converting enzyme 2
- ADCC
antibody‐dependent cellular cytotoxicity
- AEAB
2‐amino(N‐aminoethyl)benzamide
- AEC
anion‐exchange chromatography
- AETMA
(2‐aminoethyl)trimethylammonium chloride hydrochloride
- AGE
advanced glycation end products
- AGP
alpha‐1‐acid glycoprotein
- Ala
alanine
- ALG
mannosyltransferase (gene)
- AlgL
alginate lyase
- AMAC
aminoacridone
- AP
aminopyridine, or atmospheric pressure, or 1‐(2‐aminoethyl)piperazine
- APBA
3‐aminophenylboronic acid
- APCI
atmospheric pressure chemical ionization
- apoC
apolipoprotein C
- APP
amyloid‐β precursor protein
- APTS
8‐aminopyrene‐1,3,6‐trisulphonic acid
- AQ
aminoquinoline
- AQC
6‐aminoquinolyl‐N‐hydroxysuccinimidyl carbamate
- AraN
aminoarabinose
- Arg
arginine
- Asn
asparagine
- Asp
aspartic acid
- ATCC
American Type Culture Collection (bacteria)
- ATD
arrival time distribution
- ATP
adenosine triphosphate
- ATR
attenuated total reflection
- AuNPs
gold nanoparticles
- BCG
Bacillus Calmette–Guérin (vaccine)
- BDA
bovine serum albumin
- BNDM
1,10‐binaphthyl‐2,20‐diamine
- BOA
O‐benzylhydroxylamine
- BODIPY
boron‐dipyrromethene(4,4‐difluoro‐4‐bora‐3a,4a‐diaza‐s‐indacene)
- BSH
benzenesulfonyl hydrazine
- CA
caffeic acid
- CAMLG
calcium modulating ligand (gene)
- CBM
carbohydrate binding module
- CD
cyclodextrin
- CDG
congenital disorders of glycosylation
- CE
capillary electrophoresis
- Cer
ceramide
- CFTR
cystic fibrosis transmembrane conductance regulator
- CHCA
α‐cyano‐4‐hydroxycinnamic acid
- Chit42
endochitinase 42
- CHO
Chinese hamster ovary
- CI
chemical ionization
- CID
collision‐induced dissociation
- Cit
citric acid
- ClCCA
4‐chloro‐α‐cyanocinnamic acid
- CMBT
5‐chloro‐2‐mercaptobenzothiazole
- CNF
carbon fiber
- CNS
central nervous system
- COG6
component of oligomeric Golgi complex 6 (gene)
- COPD
chronic obstructive pulmonary disease
- CORA
Cellular O‐Glycome Reporter/Amplification
- CoV
coronavirus
- COV
covalent organic framework
- CPH
1‐(4‐cyanophenyl)‐4‐piperidinyl hydrazide
- CPMP
carboxy‐1‐phenyl‐3‐methyl‐5‐pyrazolone
- CRC
colorectal cancer
- CRISPR
clustered regularly interspaced short palindromic repeats
- CRM
cross‐reacting material
- CSDB
Carbohydrate Structure Database
- CSF
cerebrospinal fluid
- CTA
2‑cyano‐3‐(2‐thienyl)acrylic acid
- CTD
charge‐transfer dissociation
- CuACC
Copper‐catalysed 1,3‐dipolar azide‐alkyne cycloaddition
- CV
coefficient of variation
- CZE
capillary zone electrophoresis
- Da
Dalton
- DABP
3,4‐diaminobenzophenone
- DAN
1,5‐diaminonaphthalene
- DBA
4‐(dimethylamino)phenylboronic acid
- DBD
dielectric barrier discharge
- DCLK1
doublecortin like kinase 1
- DC‐SIGN
dendritic cell‐specific ICAM3‐grabbing nonintegrin
- DCTB
2‐[4‐tert‐butylphenyl‐2‐methylprop‐2‐enylidene]‐malonitrile
- DESI
desorption electrospray ionization
- DHA (or DHAP)
2,5‐dihydroxyacetophenone
- DHB
dihydroxybenzoic acid (2,5‐isomer unless otherwise stated)
- DIUTHAME
desorption ionization using through‐hole alumina membrane
- DMA
dimethylamine
- DMABA
4‐dimethylaminobenzaldehyde
- DMAPA
N,N‐dimethylamino‐p‐phenylenediamine
- DMCA
3,4‐dimethoxycinnamic acid
- DMDT
N,N‐dimethylpropylenetriamine
- DMEN
N,N‐dimethylenediamine
- DMHA
N,O‐dimethylhydroxylamine
- DMPA
3‐(dimethylamino)‐1‐propylamine
- DMSO
dimethylsulfoxide
- DMT‐MM
4‐(4,6‐dimethoxy‐1,2,3‐triazil‐2‐yl)‐4‐methylmorpholinium chloride
- DNA
deoxyribonucleic acid
- DOSG+
derivatization of sialylated glycopeptides plus
- DP
degree of polymerization
- DSPE
1,2‐distearoyl‐sn‐glycero‐3‐phosphoethanolamine
- DTT
1,4‐dithiothreitol
- EAD
electron‐activated dissociation
- ECD
electron‐capture dissociation
- EDC
1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimide
- EDD
electron detachment dissociation
- EDMA
ethylene glycol dimethacrylate
- EDTA
ethylenediamine tetra‐acetic acid
- EED
electronic excitation dissociation
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- EI
electron ionization (impact)
- EIEIO
electron‐impact excitation of ions from organics
- EMBL
European Molecular Biology Laboratory
- Endo
endoglycosidase
- EPO
erythropoietin
- EPS
exopolysaccharide
- ER
endoplasmic reticulum
- ESI
electrospray ionization
- EThcD
electron‐transfer/higher‐energy collision dissociation
- EtN
ethanolamine
- f (as in Galf)
furanose form of sugar
- FAB
fast atom bombardment
- FAIMS
high‐field asymmetric waveform ion mobility spectrometry
- Fc
fragment (crystallisable) region of IgG
- FFPE
formalin‐fixed and paraffin‐embedded
- FLAT
fast lipid analysis technique
- FLR
fluorescence
- FRET
fluorescence resonance energy transfer
- Fru
fructose
- FT
Fourier‐transfer
- Fuc
fucose
- FUT
fucosyltransferase
- FWHM
full width at half maximum
- GADS
Glycopeptide Abundance Distribution Spectra
- GAG
glycosaminoglycan
- Gal
galactose
- GalA
galacturonic acid
- GALAXY
Glycoanalysis by the Three Axes of MS and Chromatography
- GalN
galactosamine
- GalNAc
N‐acetylgalactosamine
- GAQ
glucosylated aminoquinoline
- GC/MS
combined gas chromatography/mass spectrometry
- GDP
guanosine diphosphate
- GLC
gas‐liquid chromatography
- Glc
glucose
- GlcA
glucuronic acid
- GlcNAc
N‐acetyl glucosamine
- GLP
glucagon‐like peptide
- Glu
glutamine
- GM3
ganglioside (αNeu5Ac‐(2→3)‐β‐d‐Galp‐(1→4)‐β‐d‐Glcp‐(1→1)Cer)
- GO
graphene oxide
- GOS
galactooligosaccharide
- GSL
glycosphingolipid
- HA
hyaluronic acid
- HABA
2‐(4‐hydroxyphenylazo)benzoic acid
- Hb
haemoglobin
- HBA
3‐hydrazinobenzoic acid
- HCD
higher‐energy collisional dissociation
- HCQ
hydroxychloroquine
- HDX
hydrogen/deuterium exchange
- HEK
human embryonic kidney
- HeLa
Henrietta Lacks cancer cell line
- Hex
hexose
- HexCer
hexosylceramide
- HexNAc
N‐acetylhexosamine
- HF
high field
- HILIC
hydrophilic interaction liquid chromatography
- HIV
human immunodeficiency virus
- HOBt
1‐hydroxybenzotriazole
- HOIL‐1
heme‐oxidized IRP2 ubiquitin ligase 1
- HPA
hydroxypyridine‐2‐carboxylic acid
- HPAEC
high performance anion exchange chromatography
- HPLC
high‐performance liquid chromatography
- HQ
2‐hydrazinoquinoline
- HRP
horseradish peroxidase
- HSA
human serum albumin
- IC
ion chromatography
- ICR
ion cyclotron resonance
- IDA
iminodiacetic acid
- IFMALDI
intensity‐fading matrix‐assisted laser desorption ionization
- IGF
insulin‐like growth factor
- IgG(M)
immunoglobulin G(M)
- IM
ion mobility
- IMAC
immobilized metal affinity chromatography
- INLIGHT
Individuality Normalization when Labeling with Isotopic Glycan Hydrazide Tags
- IR
infrared
- IRP2
iron regulatory protein 2
- ISD
in‐source decay
- IT
ion trap
- ITO
indium‐tin oxide
- IUPAC
International Union of Pure and Applied Chemistry
- KCP
keratinocyte‐associated protein
- KDO
3‐deoxy‐D‐manno‐oct‐2‐ulosonic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KLH
keyhole limpet antigen
- Ko
glycero‐d‐talo‐oct‐2‐ulosonic acid
- L
linear (as in L‐TOF)
- Lac
lactose
- LAESI
laser ablation electrospray ionization
- LALDI‐MS
label‐assisted laser desorption/ionization mass spectrometry
- LC
liquid chromatography
- LDI
laser desorption/ionization
- LIF
laser‐induced fluorescence
- LINUCS
Linear Notation for Unique Description of Carbohydrate Sequences
- LNT
lacto‐N‐triaose
- LOD
limit of detection
- LODES
logically derived sequence
- LOQ
limit of quantification
- LOS
lipooligosaccharides
- LPMO
lytic polysaccharide monooxygenase
- LPS
lipopolysaccharide
- LTQ
linear trap quadrupole
- MALDESI
Combined MALDI and ESI
- MALDI
matrix‐assisted laser desorption/ionization
- Man
mannose
- ManNAc
N‐acetylmannosamine
- MBA
methylbenzylamine
- MBT
2‐mercaptobenzothiazole
- MCR
mobile colistin resistance
- MEKC
micellar electrokinetic chromatography
- Met
methionine
- MFSD1
major facilitator superfamily domain containing 1 (protein‐coding gene)
- MGAT
mannosyl‐glycoprotein‐2‐beta‐N‐acetylglucosaminyltransferase
- MIRAGE
minimum information required for a glycomics experiment
- MOF
metal‐organic framework
- MOGS
mannosyl‐oligosaccharide glucosidase
- MPI
mannose phosphate isomerase
- MPyCA
2‐mercaptopyridine‐3‐carboxylic acid
- MRI
magnetic resonance imaging
- MS
mass spectrometry
- MSn
successive MS fragmentation n times
- MSI
mass spectrometry imaging
- MUC
mucin
- MurNAc
N‐acetylmuraminic acid
- MW
molecular weight
- NAH
1‐naphthaleneacethydrazide
- NAO
neoagarooligosaccharide
- NAPA
(silicon) nanopost arrays
- NAT
natural
- NCBI
National Center for Biotechnology Information
- NEDC
N‐(1‐naphthyl) ethylenediamine dihydrochloride
- NETD
negative electron transfer dissociation
- Neu5Ac
N‐acetylneuraminic acid
- Neu5Gc
N‐glycolylneuraminic acid
- NIMS
nanostructure‑initiator mass spectrometry
- NK
natural killer
- NMCR
nonmobile colistin resistance
- NMR
nuclear magnetic resonance
- NSI
nanoelectrospray
- P4HZD
(4‐hydrazidebutyl)triphenylphosphonium bromide
- P2VP
butyl‐terminated poly(2‐vinylpyridine
- PAD
pulsed amperometric detection
- PAGE
polyacrylamide gel electrophoresis
- PAMAM
poly(amidoamine)
- PAN
polyacrylonitrile
- PAPAN
2‐phenyl‐3‐(p‐aminophenyl)acrylonitrile
- PC
phosphorylcholine
- PEG
polyethylene glycol
- Pen
pentose
- PET
polyethylene terephthalate
- PEtN
phosphatidylethanolamine
- PGC
porous graphitic carbon
- PMM
phosphomannomutase
- PMP
1‐phenyl‐3‐methyl‐5‐pyrazolone
- pNA
para‐nitroaniline
- PNGase
peptide‐N‐glycosidase
- PSA
prostate‐specific antigen
- PSD
postsource decay
- PSSE
poly‐synchronous surface extraction
- PTM
posttranslational modification
- PVDF
polyvinylidene fluoride
- PVK
N‐vinylcarbazole
- PYAB
2‐amino‐N‐(prop‐2‐yn‐1‐yl)benzamide
- PyAOP
(7‐azabenzotriazol‐1‐yloxy)tripyrrolidinophosphonium hexafluorophosphate
- Q
quadrupole
- R
reflectron (as in R‐TOF)
- RBC
red blood cells
- RBD
receptor‐binding domain
- REMPI
resonance enhanced two‐photon ionization
- RF
radio frequency
- Rha
rhamnose
- RNase
ribonuclease
- RP
reversed phase
- RSD
relative standard deviation
- SA
sinapinic acid
- SALDI
surface‐assisted laser desorption/ionization
- SALSA
sialic acid linkage‐specific alkylamidation
- SARS
severe acute respiratory syndrome
- SDC
sodium deoxycholate
- s‐DHB
super DHB (DHB plus 2‐hydroxy‐5‐methoxybenzoic acid)
- SDS
sodium dodecyl sulfate
- SEC
size‐exclusion chromatography
- Ser
serine
- SETs
surface energy traps
- SICRIT
soft ionization by chemical reaction in transfer
- SIL
stable isotope label
- SIMS
secondary ion mass spectrometry
- SK3
small conductance calcium‐activated potassium channel 3
- SLC
solute carrier
- SLGO
single‐layer graphene oxide
- SLIM
structures for lossless ion manipulation
- SNFG
symbolic nomenclature for glycans
- SPE
solid‐phase extraction
- TAG
Toolbox Accelerating Glycomics
- TEA
trimethylamine
- TFA
trifluoroacetic acid
- THAP
2,4,6‐trihydroxyacetophenone
- Thr
threonine
- TIMS
trapped ion mobility spectrometry
- TLC
thin‐layer chromatography
- TLR
toll‐like receptor
- TMS
trimethylsilyl
- Tn
Thomsen Friesenreich (antigen)
- TOF
time‐of‐flight
- TSG
N‐(3‐triethoxysilylpropyl)gluconamide
- TWIMS
travelling wave ion mobility spectrometry
- Tyr
tyrosine
- UDP
uridine diphosphate
- UltraGIG
Ultrafast Glycoprotein Immobilization for Glycan extraction
- UPLC
ultra‐performance liquid chromatography
- UV
ultraviolet
- VPA
vinylphosphonic acid
- VPBA
4‐vinylbenzeneboronic acid
- WAX
weak anion exchange
- Xyl (or X)
xylose
- YAG
yttrium aluminium garnet
- YLF
yttrium lithium fluoride
- ZIC
zwitterionic
1. INTRODUCTION
This review is a continuation of the 11 earlier ones in this series (Harvey, 1999, 2006, 2008, 2009, 2011, 2012, 2015, 2017, 2018, 2021, 2023) on the application of matrix‐assisted laser desorption/ionization (MALDI) mass spectrometry to the analysis of carbohydrates and glycoconjugates. It is intended to bring the coverage of the literature to the end of 2022 and includes papers with cover dates of 2021 and 2022 (as well as a few papers that were missed in earlier reviews). Papers published on preprint servers are not included because these have not been peer reviewed. Also excluded are uncorrected proofs and other versions of papers that are not fully published; these will be included in later reviews when the final versions are available. In addition, the review does not cover papers that simply report the mass of glycoproteins and those concerned with nucleotides and nucleosides. It does, however include papers describing methods for carbohydrate analysis that are relevant to MALDI analysis, even though MALDI has not been used as the analytical technique. Most applications of MALDI analysis are reported in tables with the main text being restricted to reports of analytical methods. Some papers are difficult to classify; for example, a paper on MALDI imaging of cancer biomarkers might be listed under imaging or medical applications. For reviews, the number of cited references is include to give the reader some idea of the extent of coverage.
2. GENERAL
Several books and review articles directly concerned with, or including MALDI analysis of carbohydrates and glycoconjugates, have been published during the review period. Those of a general nature are listed in Table 1; those concerned with specific carbohydrate types are listed in the appropriate sections.
Table 1.
Books and general reviews on the analysis of carbohydrates with specific reference to matrix‐assisted laser desorption/ionization analysis.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Mass spectrometry in metabolomics | General review of mass spectrometers and applications to biomarkers, drug development, nutrition, toxicology, and forensic science | 53 | Amoresano and Pucci (2022) |
| Carbohydrate analysis by mass spectrometry | General review of different types of mass spectrometry | ‐ | Chizhov (2022) |
| Glycosylation: Methods and Protocols (Book) | Several sections: Analytical and Bioinformatics, glycoengineering, glycan networks and biomarkers. Several chapters covered in this review | ‐ | Davy (2022) |
| The value of coupling thin‐layer chromatography to mass spectrometry in lipid research (glycolipids also included) | Emphases the importance of separating components of mixtures to prevent phenomena such as ion suppression | 73 | Engel and Schiller (2021) |
| The Art of Carbohydrate Analysis (Book) | General coverage with protocols | ‐ | Gerwig (2021g) |
| Analytical techniques to study carbohydrates | Short overview of different methods including hydrolysis, separation techniques, (TLC, SEC, HPLC, PGC, anion/cation exchange chromatography, high pH, AEC), glycan labelling (with protocol), permethylation, GLC. | 102 | Gerwig (2021c) |
| Analysis of carbohydrates by mass spectrometry | Short general review with emphasis on N‐ and O‐linked glycans | 76 | Gerwig (2021b) |
| Mass spectrometry‐based techniques to elucidate the sugar code | Instrumentation, sugar types (milk sugars, N‐ and O‐glycans, GAGs, glycopeptides) | 655 | Grabarics et al. (2022) |
| Tools for mammalian glycoscience research | Primer, glycan structure and analysis, synthesis, glycan‐protein interactions, mention of MALDI imaging but not much else on MALDI. | 172 | Griffin and Hsieh‐Wilson (2022) |
| Recent advances in mass spectrometry‐based structural elucidation techniques | General review with sections on proteins and lipids as well as glycans | 173 | Ma (2022) |
| An overview of biological applications and fundamentals of new inlet and vacuum ionization technologies | Covers ESI, laserspray, vacuum laserspray, vacuum MALDI and applications | 153 | Trimpin et al. (2021) |
| Essentials of Glycobiology, Fourth edition | Main Glycobiology textbook | ‐ | Varki et al. (2022) |
| Mass spectrometry for structural elucidation and sequencing of carbohydrates | Methods for monosaccharide identification, linkage, sequence determination, applications | 168 | Wang, Zhao, Nie, et al. (2021) |
| Mass spectrometry as a crucial analytical basis for omics sciences | General review with a small section on glycomics | 175 | Zaikin and Borisov (2021) |
3. THEORY
Fewer papers on the theory of the MALDI process have been published than in previous years. However, the ionization mechanism of UV‐MALDI using 2,5‐dihydroxybenzoic acid (DHB, 1) as the matrix has been studied with two separate temperature‐dependent experiments. First, the angular resolved intensity and velocity distributions of neutrals desorbed from a solid sample of DHB with a UV laser (355 nm) were measured using a rotating quadrupole mass spectrometer. Second, the desorbed neutrals, at an angle normal to the surface, and the desorbed ions were simultaneously detected for each laser shot using a quadrupole mass spectrometer and a time‐of‐flight (TOF) mass spectrometer, respectively. Both experiments were conducted at initial temperatures of 100 and 300oK and the measurements were used to calculate the initial temperature dependence of the ion‐to‐neutral ratio. The results closely agreed with the predictions of the temperature‐dependent ion‐to neutral ratio using the thermal model, indicating that thermally induced proton transfer is the dominant reaction that generates initial ions from DHB in UV‐MALDI (Lin, Dyakov, et al., 2021).

The matrix α‐cyano‐4‐hydroxycinnamic acid (CHCA, 2) is able to protonate some compounds and form alkali metal adducts from others. Lou, Miley, et al. (2021) have provided evidence that the matrix can exist in two interconverting forms; the alkali metal (e.g., Na) adduct of the acid ([[CHCA]Na]+) or a protonated alkali metal salt ([[CHCA‐H+Na]H]+) with each version able to produce the appropriate MALDI ion.
The dynamics initiated by both chirped picosecond and femtosecond laser pulses have been investigated and three‐dimensional (3D) momentum images of desorbed ions from DHB have been obtained for the first time. The two different pulses produced a striking difference between the processes initiated by each one. The lack of initial momentum in ions produced by femtosecond pulses suggested a suppression of plume formation, which could be exploited to increase the sensitivity of the ionization process (Stewart et al., 2022).
4. INSTRUMENTATION
Murray (2021) has reviewed lasers used for MALDI over the 35 years that the technique has been used. The original lasers were UV fixed‐wavelength nitrogen and Nd:YAG lasers, but over the years, several additional types of laser have been introduced with wavelengths ranging from the IR to the UV and pulse widths ranging from nanoseconds to femtoseconds. Wavelength tuneable lasers have been employed in both the IR and UV ranges, and repetition rates have increased from tens of Hz to tens of kHz as MALDI has been used for mass spectrometry imaging. Dual‐pulse configurations have been implemented with two lasers directed at the target or with a second laser generating ions in the plume of desorbed material. These techniques are described in more detail in the section on MALDI imaging.
5. METHODS
A review on “Recent advances in combinations of TLC with MALDI and other desorption/ionization mass‐spectrometry techniques” with 82 references (Borisov et al., 2021) covers recent advances in the combined techniques.
5.1. Calibration
Butyl‐terminated poly(2‐vinylpyridine) (P2VP, 3), C4H9(C7H7N)nH, has been reported to be an excellent external and internal mass calibrant for positive‐ion MALDI‐MS covering the range m/z 450–4500 with ion spacings of 105.0578 mass units ([M + H]+ ions). It was found suitable to calibrate a TOF mass spectrometer in linear and reflector mode, an ion mobility‐quadrupole‐time‐of‐flight (IM‐Q‐TOF) mass spectrometer, and an Fourier‐transfer ion cyclotron resonance (FT‐ICR) instrument (Gross, 2021).

5.2. Ion mobility mass spectrometry
A review with 60 references on the application of ion mobility to glycomics covering free and permethylated N‐ and O‐linked glycans, glycosaminoglycans (GAGs) and glycolipids has been published in the book “New Developments in Mass Spectrometry No. 11” (Struwe, 2021). Ion mobility collision cross sections (singly‐, doubly‐, and triply‐protonated ions) and liquid chromatography retention times from 71 pyridylaminated N ‑linked oligosaccharides have been published (Manabe et al., 2022).
Mookherjee et al. (2021) have shown that although the MS2 and MS3 spectra of Gal‐GlcNAc and Fuc‐GlcNAc in different linkages (4–9) are very similar, some differences can be observed in their ion mobility spectra. In nitrogen, although the arrival‐time distributions for the [M – H2O]+ ion from the β1→3‐ and β1→4‐ linkage isomers of Gal‐GlcNAc (4,5) were virtually identical, the β1→6‐isomer (6) gave two semi‐resolved peaks, clearly providing differentiation (Figure 1). Separations of the corresponding ion from Fuc‐GlcNAc (9) was even more pronounced. The ions formed by further loss of galactose or fucose (m/z 204) from the β1→6‐isomers (6, 9) also gave a different ATD from the others showing that the ions formed from the different isomers had different gas‐phase structures that retained some of the original linkage information, a phenomenon termed linkage memory.
Figure 1.

ATDs (N2) of (A) m/z 366 ([M – H2O]+) from the three isomers (4–6) of Gal‐GlcNAc, (B) The corresponding ATDs from the isomers (7–9) from Fuc‐GlcNAc. (C) m/z 204 ([M – H2O ‐ Gal]+) from the three isomers (4–6) of Gal‐GlcNAc, (D) The corresponding ATDs from the isomers (7–9) from Fuc‐GlcNAc. From (Mookherjee et al., 2021), with permission from the American Chemical Society. The glycan symbols have been changed to the “Oxford” system to conform with those used in the rest of the review for consistency.

Symbols for the monosaccharides used in this review are shown below. These symbols from the so‐called “Oxford” system (Harvey et al., 2009) are used in preference to those from the more commonly used “Symbol Nomenclature for Glycans” (SNFG) system (Neelamegham et al., 2019; Varki et al., 2015) because they overcome some of the problems and inconsistencies inherent with the SNFG system.



Isomer separation is a major application of ion mobility and is of particular relevance to glycomics. One application where ion mobility has had an impact is in the separation of sialic acid isomers. In one method that used a Waters travelling wave (TWIMS) instrument, it was found that ion mobility could successfully distinguish between α2→3‐ and α2→6‐linked sialic acids in complex N‐glycans by separation of the fragment ions Neu5Ac‐Gal‐GlcNAc (18, 19) respectively following collision‐induced dissociation (CID) (Feng et al., 2021). Using the method, the authors demonstrated aberrant sialylation of haptoglobin in hepatocellular carcinoma where the ratios of α2→3‐ to α2→6‐ sialylation of seven N‐glycopeptides were found to be significantly altered (p < 0.01) in cancer (n = 27) compared with healthy controls (n = 27). Quantification was also possible with good linearity (R 2 = 0.99) with a dynamic range of two orders of magnitude and high reproducibility (coefficient of variation [CV] < 10%, n = 3).

Early instruments with ion mobility cells did not possess sufficient resolution to separate many isomers but two recent instruments, the Waters cyclic‐TWIMS mass spectrometer (Giles et al., 2019) and the instrument based on lossless ion manipulation (SLIM) technology (Deng et al., 2017) produce considerably improved resolutions. Using these instruments, separation of many glycan isomers has been possible. Thus, monogalactosylated biantennary isomers (20, 21) have been separated to base line with a Waters cyclic TWIMS mass spectrometer after two circuits of the cyclic mobility cell (Oganesyan et al., 2022). Further cycles produced partial separation of various conformers. The concomitant separation of conformers or anomers somewhat complicates the picture and sometimes requires separate experiments to distinguish between the two. Confirmation of nonisomeric separations was provided in the work by Oganesyan et al. by the observation of multiple peaks with the aglycosylated biantennary glycan (22) after eight cycles and after three cycles for the fully galactosylated glycan (23). Neither of these glycans should contain isomers.

Isomers of the core‐fucosylated analogues of glycans 20 and 21 (glycans 25 and 24) have been resolved to baseline with a SLIM device (60 M flight path giving an estimated resolution of about 5000), (Figure 2). Two peaks were resolved for each isomer, probably attributed to anomers (Dyukova et al., 2021).
Figure 2.

Arrival time distributions for the biantennary glycans 24 and 25 ([M + 2Na]2+ ions) recorded with the SLIM device. From Dyukova et al. (2021) with permission from the Royal Society of Chemistry.

Ion mobility is proving to be a great asset to glycan analysis (Struwe, 2021). In addition to its ability to separate isomers (Gao, Li, et al., 2021; Mastellone et al., 2022), as discussed above, it provides the ability to measure collisional cross sections which are relatively instrument independent and provide an alternative to the glucose units that are familiar to most glycobiologists. In addition, it provides a method for cleaning spectra by removing extraneous ions from noisy backgrounds (Harvey, Crispin, et al., 2015), particularly when these ions are multiply charged. The technique is also invaluable for removing contaminating ions from MS/MS spectra (Harvey et al., 2016). It is expected that ion mobility will be increasingly used for glycan analysis in the coming years.
6. MATRICES
Reviews and general articles relating to MALDI matrices are listed in Table 2.
Table 2.
Reviews and general articles on matrices.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Recent progress in the matrix for analysis of low molecular weight compounds using matrix assisted laser desorption ionization time‐of‐flight mass spectrometry | Comprehensive review. Discusses each type of matrix. In Chinese | 89 | Chen, Gao, et al. (2022) |
| Inorganic matrices assisted laser desorption/ionization mass spectrometry for metabolic analysis in biofluids | General coverage with several references to glycan analysis | 89 | Ding et al. (2022) |
| Recent advancements of carbon dots in analytical techniques | General chapter on carbon dots. Short section on MALDI | 82 | Gedda et al. (2022) |
| Diverse applications of ionic liquids: A comprehensive review | General review with short section on use of ionic liquids as MALDI matrices | 258 | Kaur et al. (2022) |
| Graphene oxide derivatives and their nanohybrid structures for MALDI analysis of small molecules | Applications mainly to amino acids, peptides, monosaccharides and small oligosaccharides | 104 | Kim, Kwon et al. (2021) |
| Nanostructured substrates as matrices for surface assisted laser desorption/ionization mass spectrometry: A progress report from material research to biomedical applications | General review including references to carbohydrates | 178 | Ma, Li, Li, et al. (2021) |
| Interfacial assembly of functional mesoporous nanomatrices for laser desorption/ionization mass spectrometry | Summarises recent advances in the fabrication strategies, properties and MALDI‐MS mechanisms of optical heterostructures based on mesoporous nanomaterials | 308 | Ma, Xie, et al. (2022) |
| MALDI Matrices for the analysis of low molecular weight compounds: Rational design, challenges and perspectives | Classic matrices, binary, hybrid and nanomaterial‐based matrices, reactive matrices, negative ion matrices | 126 | Qiao and Lissel (2021) |
The development of new matrices continues with much of the emphasis on those for low‐mass compounds that give ions in the same region as many organic matrices. These new matrices enable molecules such as small as monosaccharides to be examined.
6.1. Simple organic matrices
2‑Cyano‐3‐(2‐thienyl)acrylic acid (CTA, 26) has been reported as a new matrix for a wide variety of analytes such as peptides, lipids, polyethylene glycol (PEG), carbohydrates (β‐cyclodextrin [β‐CD, 27], maltotriose [28], sugammadex [29], and lactose [30]) and glycosides (Yerra et al., 2021). Signal strengths were reported to be higher than those produced by common matrices such as DHB although peptides gave similar signals with this matrix and CHCA. As with DHB, carbohydrates gave [M + Na]+ and [M + K]+ ions.





Sinapinic acid (SA), the most widely used matrix for proteins and glycoproteins, exists as two isomers: E‐ (31) and Z‐SA (32). It has long been known that Z‐cinnamic acid outperforms the E‐acids when acting as a MALDI matrix. Using ESI, MS/MS and titration experiments, and a variety of carbohydrates, De León et al. (2022) have shown that the Z‐isomer forms stronger gas‐phase complexes with the carbohydrates than the E‐isomer, thus explaining the phenomenon. Over time, the Z‐form isomerizes to E‐SA accounting for the reduction in signal strength of analytes with aged matrix samples.

Dealkaline lignin (complex branched polymer formed mainly from p‐coumaryl alcohol (33), coniferyl alcohol (34) and sinapyl alcohol (35) has been found to be a good matrix for several types of small molecule including oligosaccharides, glycosides, esters, vitamins, amino acids, hydroxyl‐acids, and fatty acids in both positive and negative ion modes. Linear quantitative results were obtained with excellent correlation with parallel high‐performance liquid chromatographic (HPLC) analyses. The performance of lignin as a matrix was said to be due to its superior optical property and abundant conjugated structure (Zhao, Wang, Liu, et al., 2021).

6.2. Binary and mixed matrices
Urakami and Hinou (2022c) have developed a mixed matrix of 1,5‐diaminonaphthalene (DAN, 36)/DHB/Na (2:10:1) and have use this to examine small glycopeptides directly. The matrix gave a more homogeneous target than DHB and promoted in‐source (ISD) fragmentation such that the glycans were released as 0,2A and 2,4A fragments from the reducing end (see Scheme 1). Further fragmentation in the TOF/TOF instrument yielded mainly glycosidic cleavage ions. Applications were to ovomucoid and egg white but some of the reported structures, high‐mannose glycans in particular, deviate from those established from known biosynthetic pathways.
Scheme 1.

Method for naming fragment ions as devised by Domon and Costello (1988). Fragments with the charge at the nonreducing end of the molecule are designated with the letters A (cross‐ring), B and C (glycosidic) with the following subscript number indicating the position of cleavage. Corresponding ions from the reducing end are designated X, Y, and X. For the cross‐ring ions, the bonds that are cleaved are indicated by superscript numbers preceeding the lertters.

6.3. Ionic liquid matrices
Ionic liquid matrices present a homogeneous surface to the laser beam, thus eliminating the concept of “sweet spots” and a review with 61 references on their use for quantification of small molecules, including carbohydrates has been published by Kobylis et al. (2021). However, little is known about their properties. In one of the latest of a series of papers investigating ionicity (Kobylis et al., 2022; MacFarlane et al., 2009) have studied four truly liquid matrices (see Kobylis et al., 2019), namely CHCA/trimethylamine (TEA), ferulic acid (37)/TEA, 2‐(4‐hydroxyphenylazo)benzoic acid (HABA, 38)/(α‐methylbenzylamine (α‐MBA, 39), and 2,5‐DHB/α‐MBA), The results, particularly as shown by a Walden plot (Molar conductivity against viscosity) showed that HABA/α‐MBA was the best ionic matrix. The ionicity of the other matrices was reduced because of intermolecular interactions. It was concluded that although the tested matrices differed in iconicity, this made no difference to their auto‐ionization properties.

Urakami and Hinou (2022b) have analysed N‐glycans from ribonuclease B (RNase B) with the ionic liquid matrix DHB‐aniline‐Na and observed both molecular ([M + Na]+) and 0,2A and 2,4A in‐source cleavage ions from the high‐mannose glycans. Formation of peptide fragment ions were of minor relative abundance. Further LIFT fragmentation was used to characterise the glycans.
6.4. Carbon‐based matrices
Carbon fiber (CNF), prepared by carbonization of electrospun polyacrylonitrile (PAN) fibers, has proved to be an excellent matrix for small molecules, especially carbohydrates such as glucose (11), sorbitol (40), mannitol (41) and sucrose (42) (Chae et al., 2021). The matrix exhibited a high salt tolerance and high sensitivity in both positive ([M + Na]+ ions) and negative ([M – H]‐ ions) ionization modes. A linear response for sucrose was recorded over the range 0–500 pmol allowing quantitation. Other compounds that were successfully analysed included amino acids and synthetic polymers such as PEG.


Nitrogen and boron codoped carbon nanofiber has also been reported as a good matrix for a range of compounds such as carbohydrates, amino acids, and polymers. This matrix showed high signal to noise ratio, excellent salt‐tolerance and homogeneous ion distribution and was reported to be superior to CHCA and to a C nanofiber matrix acting as a control (Zhao, Wang, Zhao, et al., 2021).
Highly curved onion‐like carbon nanoparticles have been synthesized from soot collected on a glass slide from the centre of a candle flame. The particles had a large surface area and good hydrophilicity. They exhibited superior performance for the detection of xylose (Xyl, 15), glucose (11), maltose (43) monohydrate, and raffinose (44) pentahydrate in positive‐ion mode with low background noise, a homogeneous target, excellent reproducibility, good salt‐tolerance and high sensitivity compared to traditional matrices such as CHCA. Using the matrix, the authors developed a quantitative assay for glucose in rat serum (Zhao, Zhao, et al., 2022).


6.5. Nanoparticles and related substances
As these compounds, mainly consisting of metal and metal oxides, lack the organic structure of traditional matrices, they are useful for examination of small molecules. “Green metal” nanoparticles have been prepared from the leaves of Cudrania tricuspidata and silver nitrate and used as a MALDI matrix for various small molecules (MW<500 Da) such as glucose, lysine, sucrose (42) and glutamic acid (Sharma, Rejeeth, et al., 2021). A low detection limit (4–20 nmol) was reported with peaks of higher intensity than those obtained using conventional CHCA. Background noise was low. By using the matrix, the authors were able to detect 13 low molecular weight metabolites in human healthy serum samples and another distinct 18 low molecular weight compounds in pancreatic cancer serum samples.
Zhao, Ma, et al. (2022) have prepared sandwich‐like gold nanoparticles@mesoporous silica nanocomposite@silver nanoparticles (Au@MSN@Ag) by a layer‐by‐layer super‐assembly strategy as a novel matrix for the quantitative detection and enrichment of small biomolecules. The sandwich‐like nanospheres were said to form a unique plasma resonant cavity that effectively absorbed the laser energy, while the homogeneous mesoporous structure of the nanoparticles could lock the analyte. Compared to traditional matrices, Au@MSN@Ag produced a low background, a wide application range, high sensitivity, good high salt and protein tolerance, and good stability. As an example of its performance, the detection limit of glucose was 5 fmol, and showed a good linear relationship in the range of 1−750 μg/mL. Gold nanoparticles coated with 2‐mercaptopyridine‐3‐carboxylic acid (MPyCA, 45) has also proved to be an effective matrix for small molecules, including glucose and has been reported to give stronger signals from this compound than when ionized by CHCA (Kakuta et al., 2022).

Palladium nanoparticles decorated thiol‐functionalized metal organic framework (MOF) nanocomposite (UiO‐66‐(SH)2@Pd NPs) has been synthesised as a matrix for analysis of di‐, tri‐ and tetra‐saccharides. The ionization efficiency was significantly improved over that of conventional matrices owning to the synergistic effect of MOF and Pd nanoparticles. By combining laser desorption‐LIFT‐TOF/TOF, 24 oligosaccharide isomers including disaccharides, trisaccharides and tetrasaccharides, were effectively differentiated. In addition, the relative quantification curves for isomeric oligosaccharides were established with good linear correlations. The method was successfully applied to the identification and quantification of sucrose and maltose in three batches of Asian and American ginseng respectively (Luo, Zhao, et al., 2022).
Among metal oxides, Fe3O4 nanoparticles have been reported as excellent matrices for a number of small molecules such as d,l‐pyroglutamic acid, d,l‐aspartic acid, l‐proline, l‐phenylalanine, sucrose, raffinose, and the triglycerides tripalmitin and triolein in positive ion mode (Zhao, Xu, Gong, et al., 2021). The matrix increased the MS peak strength and reduced the background noise compared with conventional matrices. The relative standard deviations in in‐spot and spot‐to‐spot repeatability were less than 3.2% and 6.0%, respectively and the linear correlation coefficients between MS peak intensity and concentrations were no less than 0.997 in the concentration range of 0.05–1.0 mg/mL.
TiO2 Nanoparticles have been reported to be a promising matrix for a variety of lipids including LacCer (47) (Peng, Zhang, et al., 2021). To prepare the target, the sample was mixed with the matrix solution in ethanol and NaCl was added if needed. The mixture was added to the target and allowed to evaporate. Strong signals were produced in both positive and negative ion modes with few interfering signals. P25 Titania, another TiO2 product has been shown to provide better ionization of small metabolites than either DHB or CHCA (Chen, Zhang, Wu, et al., 2022). Ten peaks were observed from a standard metabolite mixture consisting of glutamine acid, methionine, histidine, phenylalanine, taurine, aspartic acid, mannitol, and glucose whereas only two and five peaks were observed from DHB and CHCA respectively. Furthermore, the two matrices showed abundant matrix‐related peaks in the metabolite region. The method was used to examine metabolic patterns in membranous nephropathy. The material (Ti3C2(OH) x ), synthesised from the new two‐dimensional material MXene, has also shown excellent properties as a matrix for small molecules such as mono‐ and disaccharides and amino acids. Furthermore, the material showed good storage properties and was stable for at least 8 months (Li, Ma, et al., 2022).

Of a series of MOFs synthesised by Ma, Yang, et al. (2022) the maltose‐functional MOF MIL‐101‐maltose has proved to be the best. Glucose was included in five test compounds and the matrix provided ultrahigh ionization efficiency, free of matrix background, uniform crystallization, good dispersibility, a short analysis time, strong salt tolerance (500 mM NaCl), and satisfactory reproducibility. The matrix was used for serum glucose determination and successfully identified diabetic patients from healthy controls.
6.6. Matrices for negative ion mode
Neutral compounds tend not to produce ions in negative ion mode with many traditional matrices although compounds such as norharmane (49) are effective. Acidic compounds such as carboxylic acids perform better but a number of more specialised matrices have been introduced. Among these are the deprotonating matrices 4‐dimethylaminobenzaldehyde (DMABA, 50), N,N‐dimethylamino‐p‐phenylenediamine (DMAPA, 51), and 3‐aminoquinoline (3‐AQ, 52) (Krivosheina et al., 2021) which give limits of detection in the low ng/mL range with DMABA producing the strongest signals from acids and a number of neutral compounds.

A disadvantage of metal‐containing nanoparticles is the unwanted appearance of metal adducts in positive ion mode. To overcome this disadvantage Tang et al. (2022) have investigated a bismuth oxide‐graphene oxide (Bi2O3‐GO) semiconductor nanomaterial for analysis of small molecules. The matrix was characterized using conventional methods and its performance for the detection of small molecules was compared with traditional matrices (e.g., CHCA, DHB, 9‐aminoacridine [9‐AA, 53] and graphene oxide [GO]). The results showed that the negative ion spectra of small molecules were free of matrix‐related interferences, and possessed good signal intensity and repeatability. Application of Bi2O3@GO to the quantitative determination of glucose in human serum and soft drinks confirmed that the hybrid matrix could also be applied to complex samples. Conclusions drawn from the experimental results, computational chemistry calculations, and previous studies, suggesting that interfacial photogenerated thermal electron transfer and capture are key processes in the LDI mechanism. Other matrices for negative ion work (Veličković, Sharma, et al., 2022) are discussed in the section on MALDI imaging.

6.7. Matrices for dual‐polarity investigations
Most matrices for MALDI‐TOF MS of small‐molecules are only suitable for either positive or negative ion mode and, with the exception of carbon‐based nanomaterials, are not suitable for operation in dual‐ion mode. To achieve this property, two materials, poly N‐vinylcarbazole (PVK, 54) and single‐layer graphene oxide (SLGO), have recently been combined to provide both positive‐ and negative‐ion‐mode spectra of amino acids, nucleic acid bases, environmental endocrine disruptors, antibiotics, and various small molecules such as sugars (Chen, Wang, Luo, et al., 2022). The lone‐pair electrons on the nitrogen atom of PVK can serve as a Lewis base with strong electron‐donation effects, which is favourable for production of negative ion spectra. The surface of SLGO, which contains many oxygen atoms in carboxyl and hydroxyl groups that act as Lewis acids provides favourable protonation sites for positive ion mode detection. The PVK/SLGO combined matrix was compared with PVK, SLGO, and the commercially available matrices 9‐AA and CHCA where the tested analytes were shown to give strong signals in both ion modes with the new matrix. Limits of detection ranged from 0.1 to 0.0001 and 0.01 to 0.0001 mg/mL in the positive and negative ion modes, respectively.

6.8. Combined matrices and derivatization agents
These compounds are used to form derivatives at the reducing end of the analyte molecules, sometimes directly on the MALDI plate before analysis. 2‐Phenyl‐3‐(p‐aminophenyl)acrylonitrile (PAPAN, 55, Scheme 2) has been developed as one of these matrices (Ling et al., 2019). It forms a Schiff base (57) with the carbohydrates and the derivatives have been claimed to show increased ionization efficiency and reproducibility than DHB. Sample preparation involved mixing the acidified sample and PAPAN and heating at 60oC for 1 h and depositing the mixture directly onto the MALDI plate. The matrix was used to investigate maltooligosaccharides from beer (Ling, Jiang, et al., 2021).
Scheme 2.

Derivatization of glucose with PAPAN (55, Schiff base) and 2‐HQ (56, hydrazone).
2‐Hydrazinoquinoline (2‐HQ, 56), forming a hydrazone derivative (58, Scheme 2), has also been used as a dual‐mode matrix. Samples were reacted with 2‐HQ in methanol containing 5% acetic acid for 10 min at 35oC, following which the solution was deposited onto the MALDI target and allowed to air‐dry. Use of the resulting glycan hydrazones were claimed to provide an enhancement in detection sensitivity of 10 and 100 fold over that provided by 3‐AQ or DHB respectively. The matrix worked in both positive and negative ion modes (neutral glycans as Cl‐ adducts) (Lin, Xiao, et al., 2021). 4‐Hydrazinoquinazoline (59), also introduced by the same research group (Ling, Yu, et al., 2021) and used in a similar fashion, was claimed to give a 100‐fold increase in sensitivity for maltoheptaose and a 30 fold improvement for the triantennary N‐glycan (60) compared with conventional matrices such as DHB. The matrix also formed homogeneous crystals and, thus, showed good shot‐to‐shot reproducibility. It was successfully applied to the analysis of N‐glycans released from ovalbumin, bovine fetuin and human serum.

A mixture of 3‐AQ (52) and CHCA has been used to provide on‐target derivatization of various carbohydrates in an attempt to improve sensitivity (Wang, Zhao, Nie, et al., 2022). CHCA and 3‐AQ were mixed with ammonium dihydrogen phosphate and the carbohydrate (maltooligosaccharides and cyclodextrins), were deposited onto the MALDI plate and heated at 60oC for 1 h. MALDI‐TOF/TOF spectra were recorded and the sugars appeared as phosphate adducts in negative ion mode. Improved detection limits were achieved and the 3‐AQ derivatized glycans gave informative fragmentation spectra with A‐type cross‐ring cleavage ions providing useful linkage information.
Another combination of derivatization reagent and matrix is O‐benzylhydroxylamine (BOA, 61) mixed with DHB and a small amount of a sodium salt (Barada & Hinou, 2022). Derivatization suppressed in‐ and post‐source fragments from the reducing end of the glycans and was reported to give excellent results from both O‐ and N‐linked glycans. MALDI targets were prepared simply by mixing the sample and reagents with sodium bicarbonate and spotting onto AnchorChipTM 400/384 TF plates.

Other new matrices are described in the section on MALDI imaging.
6.9. Matrix‐free methods
The absence of a matrix overcomes the problem of matrix ions masking ions produced by low molecular weight glycans. Hauser et al. (2021) have developed a technique, which they refer to as “label‐assisted laser desorption/ionization mass spectrometry” (LALDI‐MS) that dispenses with the traditional matrix. Sugars were tagged at the reducing terminal with pyrene‐based reagents (62 – 66, Scheme 3), which behaves in a similar way to the matrix by absorbing the laser energy. The labels were designed to avoid the laser‐induced loss of ketene inherent in earlier pyrene tags (Yoneda et al., 2016). In this way, only the labelled compounds in a mixture were detected. The method was demonstrated by detecting lactose (30) and extending it to its detection directly in cow's milk.
Scheme 3.

Pyrene derivatives for LALDI‐MS.
Electrochemical deposition of silver from silver trifluoroacetate at 10 V for 15 min has produced a surface that showed intense surface‐assisted laser desorption/ionization (SALDI)‐MS signals for standards from various classes of compounds including sugars, lipids, fatty acids and cyclitols at a concentration of 1 nmol/spot, with values of the signal‐to‐noise ratio greater than 50. The values of the limit of detection were 0.71 μM for adonitol (67), 2.08 μM for glucose and 0.39 μM for palmitic acid per spot (Arendowski et al., 2022). Using a through‐hole alumina membrane as an ionization‐assisting substrate, Fukuoka et al. (2021) have successfully analysed a series of mannosylerythritol biosurfactants (68) with molecular weights below about 750 Da.

New matrices relevant to MALDI imaging are covered in Section 7.2.1.
7. MALDI IMAGING
MALDI imaging is possibly the fastest growing area in the use of MALDI ionization. New methods are constantly being developed with greater sensitivity and resolution. Many applications now involve enzymatic digestion of samples by spraying enzymes onto the material to be imaged and there are many new matrices being developed for specific purposes. Several reviews have been published over the 2‐year period covered by this review: These are summarized in Table 3.
Table 3.
Reviews and general articles on matrix‐assisted laser desorption/ionization imaging.
| Subject | Contents | Citations | References |
|---|---|---|---|
| Imaging of the human brain | Imaging in different disease states (cancer, Alzheimer's, epilepsy, etc.) | 140 | Ajith et al. (2021) |
| Mass spectrometry imaging for spatial chemical profiling of vegetative parts of plants | General review. Different types of imaging (MALDI, DESI, SIMS, LAESI). Applications–disease, etc. | 163 | Ajith et al. (2022) |
| MALDI Mass spectrometry imaging and glycomics | Discussion of glycan types: N‐glycans, GSLs, GAGs, glycosides, free glycans | 117 | Blaschke and Drake (2022) |
| Sample preparation of biological tissues for MALDI‐MSI | Embedding, storage, sectioning, FFPE samples, washing (lipids, glycans proteins) enzyme digestion (proteins, glycans), derivatization, matrix selection | 101 | Cillero‐Pastor and Cuypers (2022) |
| Exploring natural products through mass spectrometry imaging | Concentrates on recent progress with plants and microorganisms | 133 | Dong and Aharoni (2022) |
| Imaging mass spectrometry | General review with sections on different compound types, including N‐glycans | 171 | Drake et al. (2021) |
| Applications of stable isotopes in MALDI imaging | Application to measurements of UDP‐glucose and glucose phosphates in bovine lens | 111 | Grey et al. (2021) |
| On‐tissue chemical derivatization in mass spectrometry imaging | Covers ionization techniques. On‐tissue derivatization of various functional groups, reagent deposition, applications to glycomics, lipidomics and proteomics. Table of reagents | 151 | Harkin et al. (2022) |
| Mass spectrometry imaging of the brain glycome | Contains tables listing deglycosylation methods and MALDI matrices used for brain studies | 190 | Hasan et al. (2021) |
| Mass spectrometry imaging for direct visualization of components in plant tissues | General review, ionization methods, matrices, applications to compound type | 115 | Hu, Han, et al. (2021) |
| Recent advances in surface‑assisted laser desorption/ionization mass spectrometry and its imaging for small molecules | Discussion of different types of substrate and applications | 132 | Huang, Ouyang, et al. (2022) |
| Advanced applications of mass spectrometry imaging technology in quality control and safety assessments of traditional Chinese medicines | Covers topics such as sample preparation, matrix selection and various applications | 123 | Jiang, Zhang, et al. (2022) |
| An introduction to MALDI ionization mechanisms for users of mass spectrometry imaging | Covers laser ablation, plume pressure, temperature and velocity, laser spot size, ionization “lucky survivors”, thermal and non‐thermal ionization, metal surfaces, secondary ionization, matrix and analyte suppression | 76 | Knochenmuss (2021) |
| Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research | Methods (instrumentation, matrices, matrix deposition, quantification), applications (identification of disease, biomarkers, drug distribution) | 142 | Lee, Yeoh, et al. (2021) |
| Mass spectrometry imaging of small molecules. Methods and protocols | Book | ‐ | Lee (2022) |
| Matrix‐assisted laser desorption/ionization mass spectrometry imaging for in situ analysis of endogenous small molecules in biological samples | General review, matrices with extensive table of matrices for various compounds, matrix coating methods, instrumentation, applications | 192 | Liu, Pan, et al. (2022) |
| Surface‐assisted laser desorption/ionization mass spectrometry imaging: A review | Definition of SALDI. Mechanisms. Strategies for SALDI imaging. Applications | 274 | Müller et al. (2022) |
| MALDI Mass spectrometry imaging in lipidomics (and glycolipidomics) | Sample preparation, MALDI matrices and application, identification of lipids by accurate mass measurements, MS/MS, ion mobility. Applications (cancer research, brain injury, liver disease), MALDI‐2, single cell analysis, use of stable isotopes | 171 | Mutuku and Ellis (2022) |
| Cell and tissue imaging by TOF‐SIMS and MALDI‐TOF: An overview for biological and pharmaceutical analysis | General review, methods, applications to cancer, toxicology, drug detection, combination with other methods | 262 | Noun et al. (2022) |
| Mass spectrometry‐based lipid analysis and imaging | General article on methods | 189 | Pathmasiri et al. (2021) |
| MALDI mass spectrometry imaging: The metabolomics visualization | Brief general review with applications to glycolipids, | 48 | Salviati et al. (2022) |
| Unravelling the local complexity of biological environments by MALDI mass spectrometry imaging | Reviews MALDI imaging for a wide range of compounds | 114 | Sgobba et al. (2021) |
| Introduction to spatial mapping of biomolecules by imaging mass spectrometry | Book, General coverage with chapters on methods and different compound types | ‐ | Shrestha (2021a) |
| Imaging mass spectrometry: Glycans | Brief general coverage | 29 | Shrestha (2021c) |
| Imaging mass spectrometry: Gangliosides in brain tissue | Book chapter, brief coverage | 28 | Shrestha (2021b) |
| Instrumentation for MALDI‐MSI – Part I. Ionization sources and design | Vacuum, intermediate and atmospheric pressure sources, special resolution, modes of illumination, postionization, MALDI‐2, MALDESI | 70 | Soltwisch (2022) |
| Quantitative mass spectrometry imaging of biological systems | Topics such as matrix effects on imaging, quant. of small molecules in tissues, addn. of standards, proteins | 96 | Unsihuay et al. (2021) |
| Research progress of derivatization methods in MALDI mass spectrometry imaging | Derivatives for various functional groups. Linkage‐specific sialic acid derivatization. In Chinese | 95 | Wang, Zhang, and Guo (2021) |
| Recent developments of novel matrices and on‐tissue chemical derivatization reagents for MALDI‐MSI | General review covering different compound types | 94 | Zhou et al. (2021) |
| Advances in MALDI mass spectrometry imaging single cell and tissues | General review on methods. Small section on N‐glycoproteomes | 214 | Zhu, Xu, et al. (2022) |
7.1. Methods
Current matrix deposition methods face technical problems related to the inhomogeneous distribution of crystals and the low analyte extraction and cocrystallization efficiency prompting several investigators to develop techniques that are more efficient. In the approach adopted by Li, Wu, et al. (2022), an integrated matrix sublimation device with synchronous solvent nebulization has been developed. In operation, droplets of solvents were directly introduced into the chamber of the sublimator by using a miniaturized ultrasonic nebulizer unit and, at the same time, the matrix (DHB) was sublimed. Both synchronous and asynchronous modes of solvent nebulization and matrix sublimation were systematically investigated, but the synchronous technique was found to give the best results. Imaging of both protein (from 2,5‐dihydroxyacetophenone [DHA, 69]) and small metabolites (e.g., sulfatide [48] in negative mode) was achieved in mouse brain tissue sections with clearly improved performance compared with those of conventional spray and sublimation methods. Luo, Song, Mao, et al. (2022) have overcome some of the problems related to matrix deposition by developing an automated heated sprayer system which they claim produces a more even matrix deposition and increases sensitivity by twofold to fivefold. To reduce analyte movement within the sample caused by matrix application, Nambiar et al. (2021) have developed a freeze‐spot method for matrix (DHB) application whereupon the matrix solution freezes on contact with the sample and the solvent dissipates by sublimation. The method was found to be particularly useful for small sample sections.

Three methods for matrix application have been evaluated by Deng, He, et al. (2021) for imaging of potato glycoalkaloids such as α‐solanine (70). Each method has advantages and disadvantages. Sublimation reduces analyte diffusion because there is no solvent sprayed directly onto the tissue. The main advantages are the small matrix crystals and the homogenous matrix layer that is formed. However, the sensitivity of the method is usually lower than matrix application by spraying. Airbrush spraying is relatively fast and simple but tends to generate matrix crystals that are too large for high spatial resolution imaging. The third method was a “two‐step matrix application” technique (Shimma et al., 2013), which combined matrix sublimation and airbrushing. By comparing these methods, ionization efficiencies were ranked according to the average ion signal intensity of four glycoalkaloids as follows: sublimation < airbrushing < sublimation & airbrushing resulting in the combination of sublimation and airbrushing being chosen as the matrix deposition method of choice.

The size and distribution of matrix crystals deposited on the surface of a tissue section are critical for satisfactory imaging. Xie, Wu, et al. (2021) have achieved uniform distribution and a restricted size of matrix crystals by use of a homemade matrix sublimation device with a subzero controllable crystallization temperature, giving homogeneous matrix crystals with diameters <0.2 μm. The method was applied to endogenous and exogenous components in the tissues of strawberries, kidneys and mussels. Good reproducibility was achieved, and the quality of the ion images was significantly improved compared with the use of more traditional methods. Compounds such as pelargonidin‐3,5‐diglucoside (71), a previously undetected compound, were found in strawberries at −15oC illustrating the power of the technique.

Rather than spraying the matrix on top of the sample, Xu, Deng, Ye, et al. (2021) have used prepared slides containing a single layer of the matrix graphene oxide film on indium‐tin oxide (ITO) slides, onto which the sample was placed, Using rat brain slices they imaged 60 kinds of lipids including HexCer (46), phospholipids, cyclic adenosine monophosphate, inosine, and cholesterol. The slides could be stored for over a month and their use avoided problems such as sprayer nozzles becoming blocked.
Lipids, including HexCer, have been imaged in mouse brain in both positive and negative modes using a dual polarity approach (Müller, Verdin, et al., 2021) on alternate pixels. Gold nanoparticles were used as the matrix in both polarities with an FT‐ICR instrument. Images from six accumulated laser shots were acquired from each pixel at a repetition rate of 60 Hz with Kendrick mass defect filtering used to aid lipid identification. Approximately 200 lipid species were identified. Blanc et al. (2021) have emphasised the advantages of using administered substrates incorporating the stable isotopes 12C and 13C for deconvolution of metabolic pathways and have also used the Kendrick mass defect method to analyse the data. Applications included a study of cancer metastases in mouse brain.
With the latest instrumental developments, where pixel sizes in the micrometre range can be obtained, investigations are becoming increasingly focused on single cell analysis. Traditional methods of matrix application at this scale can be problematical because of imperfections or inhomogeneities in the matrix layer. A solution is to use premanufactured, homogeneous ionization‐assisting devices such as a matrix‐free imaging technique called Desorption Ionization Using Through‐Hole Alumina Membrane (DIUTHAME) in which a premanufactured nanostructured membrane is deposited on top of a tissue section rather than by use of a spray coating of an organic matrix. By use of this method, Müller, Bhandari, et al. (2021) acquired spectra at atmospheric pressure and, compared to MALDI MSI, DIUTHAME MS images displayed higher signal homogeneities, higher contrast and reduced background signals, while signal intensities were reduced by about one order of magnitude. DIUTHAME membranes used on tissue sections thicker than 50 μm, were successful for mammal, insect and plant tissue with a high lateral resolution down to 5 μm.
Problems exist in the application of MALDI imaging to adipose tissues arising from poor matrix distribution and crystallization caused by excess liquid lipids on the tissue surface. The problem particularly affects lipid‐rich white adipose tissue. Wang, Sun, Kunzke, et al. (2022) have developed a simple and low‐cost preparation step which they refer to as “filter paper application” It consists of placing a filter paper onto the tissue before matrix application to remove the layer of excess liquid lipids. Thirty seconds was found to be optimal and the method resulted in a higher number of detected m/z species, including nucleotides, carbohydrates, and amino acids, and higher ion intensities than before the filter paper application.
McEwen et al. (2022) have developed a new liquid tissue sampling method which they call “poly‐synchronous surface extraction” (PSSE) that uses an omniphobic substrate patterned with hydrophilic surface energy traps (SETs) which, when wet with a solvent, form a dense microdroplet array. When in contact with a tissue sample, each microdroplet extracts analytes from the tissue surface, which can be analyzed by MALDI‐IMS. The method was used to examine glycosides, such as pelargonidin‐3‐O‐glucoside (see 71), in slices of a strawberry (Fragaria × ananassa) and the method was shown to produce similar results to direct analysis and demonstrated the potential of the method to increase the speed of ambient MS tissue imaging techniques by decreasing the number of steps required for sample preparation.
As a method for increasing confidence of compound analysis, Rensner and Lee (2022) have used hydrogen/deuterium exchange (HDX) to provide information on the number of exchangeable hydrogen atoms for up to 17 labile hydrogens. HDX efficiency of 73%−85% were achieved by introducing D2O vapour into a heated MALDI source in combination with a deuterium labelled matrix (DHA). The D2O vapour was introduced directly into the ion funnel of an Orbitrap mass spectrometer by bubbling a stream of nitrogen through D2O. Complications arose because of the presence of 13C isotope peaks which needed a resolution of 280,000 for separation; higher than that of the Orbitrap. This problem was overcome by subtracting the contribution of the 13C isotope calculated from the number of carbon atoms in the compound's molecular formula. Applications were to the study of metabolites in sections of the fronds from Lemna minor (duckweed).
Dreisbach et al. (2021) have interfaced an autofocusing atmospheric pressure AP‐SMALDI AF high‐resolution MALDI imaging ion source to a Q Exactive HF Orbitrap mass spectrometer to obtain 3D images of cardiac glycosides produced by wounded leaves from the plant Asclepias curassavica. The ion source incorporated a diode‐pumped solid‐state laser operating at 343‐nm wavelength and at 100 Hz, irradiating the sample at 35° relative to the transfer capillary axis of the mass spectrometer. This system enabled the authors to keep the desorption/ionisation laser focus, fluence and ablation spot size constant across sample height differences by adjusting the sample stage position according to the sample height profile for each measurement spot. The instrument was operated at a resolution of 240,000 (at m/z 200) over a mass range of m/z 250 to 1000. The results showed an increased latex flow rate towards the point of leaf damage leading to an accumulation of defence substances in the affected area.
7.1.1. Sample preparation
A report on optimization of sample preparation protocols for MALDI imaging of single cells has concentrated on washing, drying, chemical fixation, and matrix coating steps (Bien et al., 2021). Incubation of cells with formalin for about 5 min after isotonic washing and drying, resulted in a robust protocol that largely preserved not only cell morphologies, but also the molecular integrities of amine group‐containing cell membrane phospholipids. The method was demonstrated with four model cell lines, cultured directly on ITO‐coated glass slides. Transmission (t‐)mode MALDI‐2 gave a pixel size of 2 μm.
7.2. Matrices
Angerer et al. (2022), using an atmospheric pressure (AP‐) MALDI ion source coupled to an Orbitrap Elite mass spectrometer have evaluated six MALDI matrices and several protocols for analysis of lipids and glycolipids in mouse brain sections. Of the matrices CHCA, norharmane, DHB, 2,6‐dihydroxyacetophenone DHAP (72), 2,4,6‐trihydroxyacetophenone (THAP, (73), and DAN (36), the largest number of lipids were detected with CHCA and THAP, while THAP and DAN provided the best signal intensities. In negative‐ion mode, DAN showed the best lipid coverage and DHAP gave the best results for gangliosides. One hundred fifty‐five lipids were detected in positive ion mode with THAP and 137 in negative‐ion mode with DAN. The spatial resolution achievable with DAN was 10 μm and the overall results show that the performance of AP‐MALDI is comparable to that of vacuum MALDI.

Treu and Römpp (2021) have advocated the use of cluster ions from common matrices as calibration standards for imaging experiments. DHB, for example, can form clusters with added NH4 + or added alkali metal ions of the type [aM + X+‐bH2O]+ (where X = added ion) with masses up to m/z 1378.19427. In negative ion mode, ions of the type [aM –bH + (b‐1)X–cH2O]‐ or [aM–bH–H + (b‐1)X]‐ can be formed. CHCA, sinapinic acid, trans‐2‐[4‐tert‐butylphenyl‐2‐methylprop‐2‐enylidene]‐malonitrile (DCTB, 74), 4‐nitroaniline (pNA, 75), 1,5‐DAN and norharmane all formed both positive and negative ion clusters but THAP and 9‐AA worked best in negative ion mode.

7.2.1. New matrices
Several new matrices for MALDI imaging have been introduced during the review period.
7.2.1.1. Organic matrices
Hydralazine (76) has been found to be a versatile and universal matrix for MALDI imaging of a wide range of endogenous compounds between 50.0 and 20,000.0 Da including glucosylceramides, galactosylceramides, sulfatides and gangliosides in both positive and negative ion modes. To improve its performance the matrix was doped with NH4OH or trifluoroacetic acid (TFA), resulting in superior performance for imaging biologically relevant compounds in the negative and positive‐ion modes, respectively. Compared with conventional matrices such as DHB, CHCA, and 9‐AA), hydralazine provided higher sensitivity, broader molecular coverage, and improved signal intensities and was applied successfully for the visualization of tissue‐specific distributions and changes of small molecules, lipids, and proteins in murine kidney and liver sections (Tang, Gordon, et al., 2021).

Gold nanoparticles (AuNPs) modified TiO2 nanospheres modified with gallic acid (77) to give TiO2@GA nanospheres have been used as a surface‐assisted (SALDI) substrate for imaging, They were sprayed onto ITO glass slides using a gas‐assisted electric sprayer and compared with matrices such as DHB, 2‐mercaptobenzothiazole (MBT, 78), DAN, DHA, and 9‐AA. The nanospheres provide higher detection sensitivity, lower background interference, dual‐polarity detection and enhanced ionization efficiency of various endogenous molecules. Animal tissues (mouse brain, kidney, and liver) yielded mainly neutral lipids but plant tissues such as potato tubers additionally enabled glycoalkaloids to be mapped (Sun, Tang, et al., 2022).

Another new matrix for small molecules consists of yolk‐shell Ni/NiO nanoparticles anchored onto nitrogen‐doped graphene (Ni/NiO/N‐Gr) and capable of analysing molecules in both ion modes (Zhao, Li, et al., 2022). The matrix showed the superior behaviour for the analysis of various small molecular metabolites such as carbohydrates amino acids, spermidine (79), creatinine (80), hippuric acid (81), dopamine (82), and ascorbic acid (83) with high sensitivity and excellent salt tolerance compared to the traditional CHCA and control substances (Ni/N‐Gr and NiO/N‐Gr). The matrix gave accurate quantitation of blood glucose in mice with a linearity concentration range of 0.2–7.5 mM and qualitative detection of various endogenous small molecular metabolites in murine serum and urine. Excellent spatial distribution of lipids in imaging the hippocampus region of mice brain was obtained.

4‐Aminocinnoline‐3‐carboxamide (4‐AC, 84) has been developed as a new dual‐polarity matrix and compared with traditionally matrices such as DHB and 9‐AA. It was reported to exhibit superior performance in UV absorption at 355 nm, better ion yields, low background interference and vacuum stability than the more traditional matrices. It was used to map many types of compound in mouse brain in a transgenic mouse model of Alzheimer's disease. Ninety‐three metabolites were shown to exhibit different levels of regional changes compared to the age‐matched controls (Chen, Hu, et al., 2022).

Several glycosylated matrices have been synthesised by combining glucose with common MALDI matrices such as 3‐AQ, 6‐AQ, and DAN. Compared with their parent matrices, the glycosylated matrices exhibited remarkably improved sensitivity and higher signal reproducibility in detecting small metabolites. Glucosylated 6‐AQ (6‐GAQ, 85) exhibited the best performance with a detection limit for citric acid in the low fmol range. The matrix was used to image metabolites from mouse kidney sections, and showed higher sensitivity and lower background noise than the commonly used matrices. More importantly, this matrix could selectively detect hydrophilic metabolites, especially the hydrophilic lipids in the mouse kidney (Ma, Zhao, et al., 2022).

7.2.1.2. Nanoparticles and quantum dots
To overcome problems such as low‐mass matrix peaks, various inorganic nanomaterials, such as gold nanoparticles, and metal oxides such as TiO2 have proved to be successful. TiO2, in particular has been preferred because of its favourable UV absorbing property, high chemical stability, and facile surface modification properties. Sun, Zhang, Tang, et al. (2022) have utilized this latter property to combine TiO2 submicron particles with various DHB isomers and have found that 3,4‐DHB (86)–TiO2 provides superior performance than the conventional matrices such as DHB or CHCA. The matrix exhibited low background noise and high detection sensitivity for the visualization of spatial distribution patterns of secondary metabolites such as flavonoids in the roots of the differently aged medicinal herb Scutellaria baicalensis Georgi (Chinese skullcap).

Nitrogen‐doped quantum dots, with their electron‐rich sites, promoted deprotonation and formation of negative ion spectra from compounds such as amino acids, carbohydrates and fatty acids. The matrices were highly salt‐tolerant and produced reproducible spectra as demonstrated by imaging of low molecular mass species in rat brain tissue (Jin, Liu, et al., 2022). Plasmonic Gold Nanoshell (SiO2@Au) is another type of new matrix that is stated to outperform that of conventional matrices and to be appropriate for a wide range of molecules such as carbohydrates, amino acids, peptides, drugs, nucleosides and dyestuffs (Du, Chen, et al., 2022). It has been used to image strawberry tissues at a pixel size of 100 μm without the presence of imaging artefacts and for mapping the lipid distribution within the whole‐body tissues of zebrafish (Danio rerio), honeybees (Apis cerana), and mouse brain tissues in a spatially resolved manner at pixel sizes of 55, 30, and 50 μm, respectively.
7.3. Surface‐assisted laser desorption/ionization mass spectrometry (SALDI‐MS)
SALDI‐MS, Because of its low background, has been successfully applied in the analysis of various small molecules. A new substrate, gold nanoparticles/thiol‐β‐cyclodextrin‐functionalized TiO2 nanowires (AuNPs/SH‐β‐CD‐TiO2 NWs) have been prepared on ITO glass slides and their performance compared with that of conventional organic matrices such as DHB (Wang & Li, 2022). The new substrate showed superior performances on detection sensitivity, repeatability and analyte coverage of various small molecules, such as carbohydrates, fatty acids, and bile acids in negative‐and positive ion mode and was used to profile several natural products in spearmint leaves and potato tubers. Magnetron‐sputtered niobium nanoparticles (a monoisotopic metal) has also been used as an alternative to expensive noble metals and found to work particularly well with GalCer and phospholipids (Pleskunov et al., 2022).
7.4. Nanopost arrays
MALDI using traditional matrices is relatively ineffective at ionizing neutral lipids and glycolipids, particularly in the presence of phospholipids. A recent innovation that improves the situation is to use silicon nanopost arrays (NAPA). Fincher et al. (2021) have produced NAPA wafers from low‐resistivity p‐type silicon wafers using UV projection photolithography followed by deep reactive ion etching to give arrays with a final dimensions of 1100 nm in height, 150 nm in diameter, and with a periodicity of 350 nm. Use of these arrays was then combined with trapped ion mobility imaging mass spectrometry (TIMS IMS) for examination of intact rat brain and kidney tissue which were placed directly on the arrays. The method provided enhanced ionization efficiency for neutral lipid species and provided complementary coverage to MALDI imaging. It enabled imaging of neutral lipid species at 20 μm spatial resolution and increased molecular coverage greater than twofold as the result of separation of molecular species, such as triglycerides, cholesteryl esters, HexCers and phospholipids, into distinct mobility‐m/z bands using gas‐phase ion mobility separations. In addition, the method allowed for the separation of isomeric species, including mobility resolved isomers of Cer(d42:2) (m/z 686.585).
Dufresne et al. (2021) have developed a precoated substrate that enables high spatial resolution of phospholipids, neutral lipids and glycolipids in positive ion mode as metal cation adducts. The substrates were constructed by depositing a layer of CHCA and potassium salts onto silicon nanopost arrays before tissue mounting. The matrix/salt precoated NAPA substrate was shown to significantly enhance all detected lipid signals allowing lipids to be detected at lower laser energies than could be obtained with bare NAPA. The method enabled ion images to be generated at 10 μm spatial resolution from samples such as rat retinal tissue. Signal intensity increases of at least 5.8 ± 0.1‐fold for phospholipids and 2.0 ± 0.1‐fold for neutral lipids compared to bare NAPA were obtained.
TiO2 Nanopillar arrays have been developed by Yamada et al. (2021) and shown to be effective at ionizing small amino acids, sugars, pesticides, peptides, and proteins with molecular weights of up to 24,000. A substrate with a lower surface density exhibited more intense signals for the detection of small (∼1.2 kDa) analytes as the result of more effective heat confinement. Wetting behaviour was another factor facilitating better performance for smaller molecules at lower surface density. On the other hand, the homogeneous adsorption of target molecules onto the nanopillared surface was thought to be a dominant factor for the detection of the larger proteins.
7.5. MALDI‐2 and related methods
With pixel sizes in the 5–20 μm range, the number of ions produced by each pixel is small resulting in low sensitivity. Detection of the sample molecules can also be compromised by ion suppression effects caused by ready ionization of major and easily ionisable constituents such as phosphatidylcholine (87). One way to alleviate the problem is to use a second laser to produce post‐ionization, a technique known as MALDI‐2, capable of boosting sensitivity by 2–3 orders of magnitude and well described in the protocol published by Dreisewerd et al. (2022). The authors state that two prerequisites are required for exploiting the MALDI‐2 effect, namely the use of ion sources that are operated under elevated pressure (a few mbar of nitrogen buffer gas) and the use of a pulsed UV post‐ionization laser. The wavelength of this laser should fall below the two‐photon threshold of the utilized MALDI matrix which, for aromatic matrices, such as, for example, DHB, CHCA or DHAP, is around 310 nm. Frequency‐quadrupled Q‐switched Nd:YAG lasers, which emit at 266 nm are suitable. Experiments with two pulse widths (28 ps and 6 ns at a wavelength of 266 nm) for the ionizing laser support a resonance enhanced two‐photon ionization (REMPI) of neutral matrix molecules desorbed by the MALDI laser from DHB or DHA (Potthoff et al., 2022). In a modification, known as transmission (t‐) mode MALDI, the target is illuminated from the back. Pixel sizes in the 1–2 μm range have been achieved and the method has been used, in combination with an optical microscope to image lipids and glycolipids in single cells and intercellular matrices at a pixel size of 2 μm (Bien et al., 2022).

Postionization has also been implemented in an IR‐atmospheric ion source with improvements designed to overcome some of the disadvantages associated with AP sources (Schneemann et al., 2022). Ambient MS imaging comes with the advantage that visualizing biomolecules from tissues involves no or minimal sample preparation but it suffers from a pronounced bias towards either polar or nonpolar analytes. The improvements to the source devised by the authors involves use of an in‐capillary dielectric barrier discharge (DBD) module for postionization of neutrals desorbed by the IR‐MALDI) MSI source. The device was found to enhance the signal intensities of nonpolar compounds by up to 104 compared to IR‐MALDI, without affecting transmission of IR‐MALDI ions. It was used to study mouse tissue and Danaus plexippus caterpillar tissue sections, visualizing the distribution of glycolipids, sterols, fatty acids, monoglycerides, and diglycerides that are not detected in IR‐MALDI MSI experiments and allowed mapping of nonpolar analytes with pixel resolutions down to 20 μm.
A related postionization technique involves irradiating the MALDI target with a series of nsec‐long UV laser pulses of 349 nm wavelength on a pixel‐by‐pixel basis, analogous to a classical MALDI‐MSI experiment. To induce secondary ionization in the MALDI plume, this material was irradiated by three RF‐Krypton discharge lamps operated at 13.560 MHz. Pulses of light were synchronized with the MALDI laser and the ion source was operated at about 10 mbar of N2 and dopant vapour (e.g., acetone) was introduced via a capillary system. Under these conditions, samples reacted with the dopant gas, and residual water vapour to give chemical ionization (CI)‐type reactions that were dissimilar to those seen in MALDI‐2. The technique was used to image lipids and glycolipids from animal tissues and it was reported that signal intensities could be boosted by up to 2–3 orders of magnitude (Bookmeyer et al., 2022).
Another technique involves plasma‐based postionization after the MALDI ion source using a commercially available “soft ionization by chemical reaction in transfer” (SICRIT) system interfaced to a trapped ion mobility mass spectrometer (Michael et al., 2022). The instrument worked particularly well for lipids, including glycosphingolipids and the ion mobility function was invaluable for separating isomers that were not resolved in the m/z dimension.
7.6. Formalin‐fixed and paraffin‐embedded (FFPE) samples
Applications in this area are particularly relevant to clinical studies. Sections of retina analyzed by imaging‐MS are typically fresh‐frozen. However, paraformaldehyde fixation facilitates the preservation of tissue morphology by forming methylene bridge crosslinks between formaldehyde and amine/thiols in biomolecules and would possibly be a better method for sample preparation for imaging. Consequently, Kotnala et al. (2021) have compared the molecular identity of lipids and glycolipids generated by MALDI‐IMS and LC–MS/MS for fixed and fresh‐frozen retina tissues in both positive and negative ion modes. More lipid signals were observed in fixed compared with fresh‐frozen retina. More potassium adducts were observed in fresh‐frozen tissues than in fixed tissues because the fixation process caused displacement of potassium adducts to protonated and sodiated species in ion positive ion mode. LC–MS/MS analysis showed an overall decrease in lipid signals due to fixation, particularly with glycerophospholipids and glycerolipids. However, the method largely conserved the signals from most sphingolipids and cholesteryl esters.
7.6.1. Use of enzymes
For studies of compounds such as large polymers and N‐linked glycans from glycoproteins, methods, usually enzymatic, are needed to render them suitable for mass spectrometric analysis. Several investigators have used such methods for analysis of various compounds in FFPE tissues.
7.6.1.1. Release of N‐linked glycans from glycoproteins with peptide‐N‐glycosidase F (PNGase F)
Unstandardized and uncontrolled incubation steps in the N‐glycan release step often cause significant delocalization of released N‐glycans, resulting in the inability to link given N‐glycan composition to a specific microanatomical region of the tissue. Veličković, Sharma, et al. (2022) have investigated this problem and have optimized the incubation step by use of methods to maintain constant relative humidity in the incubation chamber. They tested saturated solutions of various salts and showed that the best performance was achieved using a saturated solution of KNO3 that maintained an 89% relative humidity, Under these conditions, near maximal sensitivity was achieved with only minimal ion delocalization. The method was demonstrated at a 35 μm spatial resolution with a kidney nephrectomy tissue section. Another digestion device that controls humidity, this time by cyclic ventilation and heating of the slide holder and the chamber lid has been developed by Fülöp et al. (2022). The device was designed to enable controlled micro‐condensation on the slide and to stabilize and monitor the digestion process. It was used to study sagittal mouse brain sections and xenografted human U87 glioblastoma cells in CD1 nu/nu mouse brain.
Although many methods have been developed for examination of FFPE soft tissues, problems arise for hard tissues such as cartilage‐bone, tooth and whole mouse body. For example, there can be loss of morphology during the heat‐induced epitope retrieval step on commercially available conductive ITO slides. To overcome the problem, Lee, Briggs et al. (2021) have taken conductive ITO slides precoated with gelatin and chromium potassium sulfate dodecahydrate to improve the adherence of FFPE human osteoarthritic cartilage‐bone tissue sections for monitoring N‐glycans. Tissues were sprayed with PNGase F and incubated for at least 2 h at 37oC. A peptide calibration standard was added followed by the CHCA matrix, which was sprayed on with the same system as was used for the enzyme. Scanning was conducted with a TOF/TOF instrument. Use of the gelatin‐coated ITO slides resulted in overall higher N‐glycan signal intensity not only for FFPE osteoarthritic cartilage‐bone tissue but also for FFPE hard‐boiled egg white used as a quality control.
Pace et al. (2022) have reported the first use of the hybrid technique, IR‐MALDESI for imaging N‐glycans in FFPE samples. IR Ionization has the advantage of minimizing losses of sialic acids and avoids the necessity for derivatization. The method was demonstrated with FFPE embedded human prostate tissue, which was analyzed in negative ionization mode after pneumatic application of PNGase F to cleave the glycans. The mass spectrometer was an Orbitrap Exploris 240 with a resolving power of 240,000 (FWHM at 200 m/z). Fifty‐three N‐linked glycans were identified; more than 60% contained sialic acid residues.
Amidation with aniline of sialylated N‐glycans in FFPE tissue samples from human laryngeal cancer patients has been reported to provide increased detection sensitivity. After dewaxing, the sialic acids were amidated, the N‐glycans were released by spraying a solution of PNGase F and incubating at 37oC for 12 h and the glycans were examined by MALDI‐TOF after spraying with CHCA. Identification was by database matching (Zhang, Shi, et al., 2022).
7.6.1.2. Use of other enzymes
Quantification of hyaluronic acid (HA, 88) in human skin sections has been achieved following incubation with hyaluronidase (H1136). The enzyme was sprayed onto the tissue with a TM sprayer and the tissue was incubated at 37oC for 18 h. For MALDI analyses, the matrix was DHB/DMA, which was also sprayed onto the tissue. HA was detected in each skin section in negative ion mode by targeting its specific digestion fragment (6‐mer, 88, n = 3) at m/z 1180.2900 ([M−3H+2Na]− ion). The method was said to be better than existing methods based on Raman imaging or use of a biotinylated HA‐binding protein and was used to measure the HA concentration in the epidermis, upper dermis, and lower dermis following treatment with a cosmetic formulation (Legouffe et al., 2022).

7.6.1.3. Use of several enzymes
Serial treatment of FFPE tissue sections with several enzymes has been used to study constituents from the extracellular matrix from aortic valve sections (Clift, Drake, et al., 2021). For example, chondroitin sulfates were imaged after treatment with a chondroitinase, after which, treatment with PNGase F allowed N‐glycans to be studied. Peptides were then imaged by treatment of the tissues with elastase. Imaging was performed with an FT‐ICR instrument with CHCA as the matrix. A protocol for the method, aimed at fibrosis research has been published (Clift, Mehta, et al., 2021).
Denti et al. (2022) have developed a multiomics approach for visualizing lipids, N‐glycans and tryptic peptides on a single slide. The slides were first heated for 1 h at 60°C followed by two washes for 8 min each with toluene. The 9‐AA matrix was sprayed onto the surface with a TM‐Sprayer and Phosphorus Red, used as a calibration standard, was then spotted onto the slide, which was analysed by MALDI‐TOF. Next, the 9‐AA was removed and rehydration was performed with consecutive washes in 100% ethanol (1 × 3 min), 70% ethanol (1 × 3 min), and H2O (2 × 2 min). A citric acid antigen retrieval step was performed in a bath of citrate buffer (pH 5.9, 10 mM) at 97°C for 45 min before washes in H2O (20 min). PNGase F from Elizabethkingia meningoseptica was deposited using an iMatrixSpray and the slide was incubated overnight in a humidity chamber at 42°C. Finally, CHCA in 70% acetonitrile solution was sprayed using a TM‐Sprayer and MALDI‐TOF was again performed. The CHCA was then removed from the slides and rehydration was performed as above. Trypsin deposition (20 ng/μL) was performed using an iMatrixSpray and the slides were incubated in a humidity chamber overnight at 40°C. Finally, a solution of CHCA in 70% acetonitrile with 0.1% trifluoroacetic acid was applied with a TM‐Sprayer and the slide was examined by MALDI‐TOF. The method was applied to murine brain and renal carcinoma tissue providing complementary information that characterized different histological regions.
PNGase F/Endo F3 Glycan release, combined with neuraminidase digestion have provided a means to improve sensitivity and provide more information on chain branching, Enzymes were sprayed onto deparaffinised tissue slices and incubated for 2 h at 37oC (DelaCourt et al., 2022).
7.7. On‐tissue derivatization
On‐tissue derivatization has been used by several investigators to improve detection of specific compounds. The topic has been reviewed: “On‐tissue chemical derivatization reagents for matrix‐assisted laser desorption/ionization mass spectrometry imaging” (82 references) (Merdas et al., 2021). This is a general review but contains a small section on carbohydrates.
A new method for analysis of steroid glycosides has been reported and used to improve the study of cardiac‐glycoside sequestration in D. plexippus (Dreisbach et al., 2022). The method involved derivatization of the 19‐oxo group of 89 with Girard's T reagent (Scheme 4) which gave an improvement of at least an order of magnitude over the use of underivatized samples.
Scheme 4.

Derivatization of calotropin/calactin with Girard's T reagent (90).
Han, Zhao, et al. (2022) have developed an on‐tissue derivatization method, to image and quantify the aldose and ketose isomers of monosaccharides in biological tissues. The new derivatization reagent, 1‐naphthaleneacethydrazide (NAH, 92) was synthesized and was shown to significantly enhance the sensitivity of detection of the monosaccharides. In addition, the NAH‐derivatized aldose and ketose monosaccharides gave isomer‐specific diagnostic ions upon fragmentation (see Figure 3). Specifically, aldose carbohydrates, illustrated by glucose, gave m/z 265 and 143, whereas the keto‐monosaccharides, illustrated by fructose, gave m/z 295 and 119. For derivative formation, a solution of the reagent (0.5 mg/mL in acetonitrile/acetic acid (7:3 [v:v]), was sprayed onto the tissue and incubated for 2 h at 60oC. MALDI used 1,5‐DAN as the matrix. A quantitative method was also developed and applied to tissues from strawberry, carrot, mulberry, and burdock.
Figure 3.

Formation of aldose‐ and ketose‐specific ions from the monosaccharides glucose and fructose following derivatization with NAH. From Han, Zhao, et al. (2022) with permission from Elsevier.

4‐(Dimethylamino)phenylboronic acid (DBA, 93), applied with a TM sprayer, has been used to derivatize cis‐diol metabolites, including several carbohydrates, in cryo‐sectioned tissues from maize. The presence of the derivative improved the signal from the target compounds and identification was facilitated by use of the 10B/11B isotope pattern (Forsman et al., 2021).

The use of multiple derivatization agents in parallel increases metabolite coverage even further but produces large and complex datasets that can be challenging to analyze. To address this problem, Larson et al. (2022) have developed “Metaspace,” for annotation of results from multiple derivatization experiments. Maize roots were used as a model system to obtain MSI data sets after parallel chemical derivatization with four different reagents, Girard's T (90, Scheme 4) and P (94) for carbonyl groups, coniferyl aldehyde (95) for primary amines, and 2‐picolylamine (96) for carboxylic acids. Using this method, 631 unique metabolites were identified compared with 256 from the underivatized data set. Analysis time was also shorter. An additional feature is a method to remove false derivatized annotations, which can clean 5%−25% of these annotations from the derivatized data.
Protocols for various methods for MALDI imaging are listed in Table 4 and applications of MALDI imaging are listed in Table 5.
Table 4.
Protocols for methods relating to matrix‐assisted laser desorption/ionization imaging.
| Subject | References |
|---|---|
| Array‐based N‐glycan profiling of cells in culture | Angel, Mehta, et al. (2021) |
| Preparing ductal epithelial organoids for high‐spatial‐resolution molecular profiling | Bakker et al. (2022) |
| MALDI‐2 and t‐MALDI‐2 mass spectrometry imaging | Dreisewerd et al. (2022) |
| Matrix‐assisted laser desorption/ionization mass spectrometry imaging of glycogen in situ | Hawkinson and Sun (2022) |
| Ambient mass spectrometry imaging of small molecules from cells and tissues | Kim, Lim, et al. (2022) |
| Enhancing metabolite coverage for matrix‐assisted laser desorption/ionization mass spectrometry imaging through multiple on‐tissue chemical derivatizations | O'Neill, Dueñas, et al. (2022) |
| Single‐cell metabolomics with rapid determination of chemical formulas from isotopic fine structures | Samarah, Vertes, and Anderton (2022) |
| Mass spectrometry imaging of biological tissues by laser desorption ionization from silicon nanopost arrays | Samarah and Vertes (2022) |
| Sample preparation for imaging mass spectrometry | Shrestha (2021d) |
| MALDI Methods used in imaging, sample preparation, etc. | Shrestha (2021e) |
| Regional N‐glycan and lipid analysis from tissues using MALDI‐mass spectrometry imaging | Stanback et al. (2021) |
| MALDI Mass spectrometry imaging of lipids on free‐floating brain sections and immunohistochemically colocalized markers of neurodegeneration | Strnad et al. (2022) |
| TOF‐SIMS Imaging of biological tissue sections and structural determination using tandem MS | Van Nuffel and Brunelle (2022) |
| Optimization of multiple glycosidase and chemical stabilization strategies for N‐glycan isomer detection by mass spectrometry imaging in FFPE tissues | West, Lu, et al. (2021) |
Table 5.
Imaging.
| Target compound | MALDI | Notes | References |
|---|---|---|---|
| Acylated anthocyanins in rat jejunum membranes (e.g., cyanidin glucoside [97]) | TOF/TOF (DHB, ImagePrep sprayer) | Analysis of intestinal absorption of acylated anthocyanins in Sprague–Dawley rats | Hahm et al. (2021) |
| Alzheimer's brains (mouse and human) | PNGase F (TM sprayer), Q‐IM‐TOF (Waters Synapt G2‐Xs), (CHCA) | In situ spatial glycomic imaging. Changes in N‐linked glycosylation detected | Hawkinson, Clarke, et al. (2022) |
| Aminoglycoside and vancomycin antibiotics from mice and rabbit lung | FT‐ICR (DHB, spray) | Development of an optimized method for the detection and spatial distribution of aminoglycoside and vancomycin antibiotics | Wang, Dartois, et al. (2021) |
| Anthocyanins and carbohydrates from strawberries | TOF/TOF (DHB, TM sprayer) | Distribution of strawberry plant metabolites at different maturity stages | Wang, Yang, Chaurand, et al. (2021) |
| Arabidopsis thaliana leaves (glycolipids) | IR ablation atmospheric pressure photoionization, TOF (ice, sublimation) | Imaging at the single‐cell level | Hieta et al. (2021) |
| Arabinoxylans in developing wheat grain. | R‐TOF (DHB/DMA, nebulizing robot) | Spatial correlation of water distribution and fine structure of arabinoxylans | Fanuel et al. (2022) |
| Asperosaponin VI (glycoside, 98) in Dipsacus asperoides roots | TOF/TOF (DHB, TM sprayer) | Jasmonic acid biosynthesis and signalling shown to be associated with the biosynthesis of asperosaponin VI in D. asperoides | Xu, Hu, et al. (2022) |
| Carbohydrates, amino acids, monoamines in banana pulp | TOF/TOF (AuNPs) | Spatially resolved metabolomics reveals variety‐specific metabolic changes during postharvest senescence | Yin, Dong, et al. (2022) |
| Carbohydrates, amino acids, and various metabolites from Arctium lappa L. (burdock) roots | TOF/TOF (DHB.CHCA, spray) | Distribution of components in roots. Carbohydrates mainly in centre | Li, Qiu, et al. (2022) |
| Cellooligosaccharides in plant cells | TOF (DMA/DHB, nebulyser) | Real‐time imaging of enzymatic degradation of cellulose in pretreated maize internodes reveals different cell types have different profiles | Leroy et al. (2022) |
| Clausena lansium (Lour.) skeels | IT‐TOF (DHB/TFA, airbrush) | Visualizing the spatial distribution of metabolites | Tang, Zhao, et al. (2021) |
| Defensive cardiac glycosides in Asclepias curassavica | Orbitrap with 3D AP‐MALDI ion source, (DHB, pneumatic sprayer) | 3D‐Surface MALDI mass spectrometry imaging for visualising plant defensive cardiac glycosides | Dreisbach et al. (2021) |
| Disaccharide isomers in plant tissues | R‐TOF/TOF (NEDC, ImagePrep sprayer, ‐ve mode) | MALDI‐TOF/TOF tandem mass spectrometry imaging reveals nonuniform distribution of disaccharide isomers | Zhan et al. (2021) |
| Ellagitannins in Fragaria × ananassa (strawberry) | TOF/TOF (DAN, vapour deposition and spray), MS/MS, LC‐MS | Study of distribution in fruit | Enomoto (2021) |
| Fructans in stem and rhizome of Agave tequilana Weber var. azul | Q‐TOF (CHCA) | Higher DP fructans found toward the central section of the stem, lower DP fructans concentrated in the highly vascularized central core of rhizomes | Pérez‐López, et al. (2021) |
| Galactosylated glycerols and other defense‐related metabolites in Triticum spp. | AP‐MALDI, Orbitrap (DHB, spray) | For mapping the spatial distribution of defense‐related metabolites | Righetti et al. (2022) |
| Ginsenosides (e.g., 99) and other metabolites from Panax notoginseng | TOF/TOF (9‐AA, CHCA, DAN, TM sprayer) | Visualizing the distributions and spatiotemporal changes of metabolites | Sun, Ma, et al. (2021) |
| Ginsenosides in Panax notoginseng | Q‐TOF (DHB, CHCA [+ve], 9‐AA [‐ve] spray), MS/MS | For unveiling the transformations of ginsenosides during processing | Fan, Yang, Li, et al. (2022) |
| Glucose metabolites in bovine lens | FT‐ICR (9‐AA, DAN, DHB, NEDC, TM sprayer, ‐ve) | Development of method, NEDC matrix best | Zahraei et al. (2021) |
| Glucose in bovine lens cortex | FT‐ICR (NEDC, TM sprayer) | Mapping uptake, transport and metabolism | Zahraei et al. (2022) |
| Glycoalkaloids in potato tubers | QIT‐TOF (DHB, sublimation/spraying) | Distribution and changes of glycoalkaloids in potato tubers under different storage times | Deng, He, et al. (2021) |
| Glycogen and N‐glycans from human cancerous tissue (various) | PNGase F, IM‐Q‐TOF (CHCA, spray) | Imaging reveals heterogeneous glycogen stores in human normal and cancerous tissues | Young et al. (2022) |
| Glycosides and primary metabolites from bilberry (Vaccinium myrtillus) fruit | FT‐ICR (THAP, sublimation) | Determination of developmental distribution patterns | Dare et al. (2021) |
| Glycosides from Gliricidia sepium leaves | FT‐ICR (DHB, CHCA, MBT, nebulizer), LC | Optimization of imaging conditions and comparison with ESI | Pereira et al. (2022) |
| Glycosphingolipids from mouse retina | FT‐ICR (DHAP, sublimation, ‐ve) | Ganglioside GD3 synthase deletion shown to alter retinal structure and impair visual function in mice | Abreu et al. (2021) |
| Glycosphingolipids in ovarian cancer tissue | TOF/TOF (DAN, TM‐sprayer, +ve, ‐ve) | Glycosphingolipids shown to be mediators of cancer plasticity through independent signalling pathways | Cumin et al. (2022) |
| Glycosphingolipids in Gaucher disease mouse brain | TIMS‐TOF (DHB, sublimation) | Study of neuroinflammation in neuronopathic Gaucher disease | Boddupalli et al. (2022) |
| Lipids and glycolipids from postmortem human brain tissues | R‐TOF/TOF (DHB, +ve, DAN, ‐ve sublimation) | Regional lipid expression abnormalities shown to correspond to MRI‐defined white matter hyperintensities | Pinsky et al. (2021) |
| Lipids and glycolipids from rat brain | TOF/TOF (s‐DHB, DHB, CHCA, nebulizer) | To confirm Raman imaging study of posttraumatic stress injury | Chaichi et al. (2021) |
| Lipids and glycolipids from mouse brain | TOF/TOF (CHCA, spray) | Profiling changes in lipids over time under a high fat diet | Sighinolfi et al. (2021) |
| Lipids and sucrose in peanuts | TIMS‐TOF (DHB, TM‐sprayer) | Distribution of lipids | Wang, Chen, Liu, et al. (2022) |
| Lipids, including HexCer from human kidney | LTQ‐Orbitrap (DAN, ‐ve, sublimation) | High resolution imaging (10 μm) | Martín‐Saiz et al. (2021) |
| Lipids, including sulfatide, in cave‐dwelling fish | FT‐ICR (DHB/AgNPs, spray) | Study of lipid metabolic pathways underlying troglomorphic adaptations | Lam et al. (2022) |
| Lipids including sulfatide from mouse kidney | AP‐MALDI (Orbitrap), (DHB +ve, norharmane, ‐ve, spray) | In study of perfluorooctane sulfonate‐induced nephrotoxicity | Chen, Jiang, et al. (2022) |
| Maize tissue (metabolites) | LTQ‐Orbitrap (DHB, CHCA, DAN, TM sprayer) | Use of on‐tissue boronic acid derivatization for the analysis of vicina‐diol metabolites | Forsman et al. (2021) |
| Mannosylerythritol lipids in human skin cells | Ion trap | Studies of recovery effect on damaged skin cells | Kondo, Yasui, et al. (2022) |
| Metabolites such as gallotannins in Paeonia suffruticosa and Paeonia lactiflora roots | FT‐ICR (DHB and DHB‐Li, spray) | Spatial distribution of metabolites in roots | Li, Ge, et al. (2021) |
| Metabolites (e.g., rutin, 100) from Forsythia suspensa | TOF/TOF (DAN, spray, ‐ve ion) | Spatial distribution of functional metabolites at different harvest stages | Jing et al. (2022) |
| Metabolites (various) in mouse kidney | FT‐ICR (9‐AA, DAN, NEDC [preferred]) | Use of stable isotopes to monitor metabolic activity | Wang, Xing, et al. (2022) |
| Metabolites (various) in rat kidney | Orbitrap, TOF/TOF (DAN, TM sprayer) | Identification of tissue‐specific metabolic reprogramming | Wang, Fu, et al. (2021) |
| Metabolites in a transgenic mouse model of Alzheimer's disease | TOF/TOF (4‐AC, spray) | Use of 4‐AC as a new matrix | Chen, Hu, et al. (2022) |
| Metabolites from plant roots | FT‐ICR (NEDC, spray) | Elucidating drought‐tolerance mechanisms in plant roots | Honeker et al. (2022) |
| Metabolites (trehalose) from Sphagnum (peat moss) | FT‐ICR (DHB/CHCA (+ve), NEDC (‐ve), TM sprayer) | Novel metabolic interactions and environmental conditions shown to mediate the boreal peat moss‐cyanobacteria mutualism | Carrell et al. (2022) |
| Metabolites in continuously cropped Salvia miltiorrhiza Bge | TOF/TOF (1,5‐DAN, BNDM, TM sprayer) | Visualization of the spatial distribution and alteration of metabolite profiles | Sun, Cui, et al. (2022) |
| N‐Glycans from aortic valve tissue | PNGase F, FT‐ICR (CHCA, TM sprayer), N‐glycans | Spatial N‐glycomics of the human aortic valve in development and pediatric endstage congenital aortic valve stenosis | Angel, Drake, et al. (2021) |
| N‐Glycans in colorectal cancer | PNGase F, TOF/TOF (CHCA), N‐glycans (amide derivatization) | Cancer cells found to have higher levels of sialylation and high‐mannose glycans, together with less fucosylation and branching | Boyaval et al. (2021) |
| N‐Glycans in endometrial cancer tissue (FFPE preparation) | PNGase F (spray), R‐TOF/TOF (CHCA), N‐glycans | Detection of altered N‐linked glycosylation in endometrial cancer | Mittal et al. (2021) |
| N‐Glycans from alcoholic FFPE mouse liver | PNGase F, TOF/TOF (DHB, airbrush), N‐glycans, sialic acid benzylation | Investigation of benzylamidation of sialic acids. | Saito et al. (2021) |
| N‐Glycans from pancreatic cancer tissue | PNGase F or endo F3, FT‐ICR, Q‐TOF (CHCA, TM sprayer), amidation of sialic acids | Imaging of N‐glycans, high‐mannose, bi‐, tri‐, tetra‐antennary complex. Increased sialylation in cancer tissue | McDowell et al. (2021) |
| N‐Glycans from striatal neuroinflammation in the rodent brain | PNGase F, Q‐TOF (CHCA, spray) | Neuroinflammation caused a significant decrease in the abundance of sialylated and core fucosylated structures and an increase in high‐mannose glycans | Rebelo et al. (2021) |
| N‐Glycans from mouse brain | PNGase F, Q‐IM‐TOF, FT‐ICR (CHCA, spray) | Brain glycogen shown to serve as a critical glucosamine cache required for protein glycosylation | Sun, Young, et al. (2021) |
| N‐Glycans from 15 types of cancer tissue | PNGase F, TOF/TOF (CHCA, TM sprayer), high‐mannose N‐glycans | Re‐evaluation of previous data and re‐examination of tissues to evaluate contribution of high‐mannose N‐glycans to cancer | Chatterjee et al. (2021) |
| N Glycans from canine glioma | PNGase F, TOF (DHB) | Identification of biantennary glycan on haptoglobin | Malaker et al. (2022) |
| N‐Glycans in soybean root nodules | PNGase H, F, (FT‐ICR (CHCA, TM‐sprayer) | Spatial mapping provides insights into legume‐rhizobia symbiosis | Veličković et al. (2022) |
| N‑glycans in human knee osteoarthritis tissue | PNGase F, R‐TOF/TOF (CHCA, spray) | Identification of complex‑type N‑glycans as putative cartilage degradation markers | Lee, Briggs, et al. (2022) |
| Oligosaccharides from maize kernels | Orbitrap (THAP, spray) | Imaging following in situ enzymatic treatment | Granborg et al. (2022) |
| Oligosaccharides from FFPE slides from pancreatic ductal adenocarcinoma | FT‐ICR (9‐AA, spray), free glycans | Native glycan fragments shown to be independent prognostic factors of cancer | Sun, Trajkovic‑Arsic, et al. (2021) |
| Oligosaccharides, fatty acids, polyphenols from Pisum sativum seed coats | Q‐TOF (DHB, THAP) | Use of electronically driven micromanipulation and MALDI for analysis of seed coat layers in study of seed dormancy | Krejčí et al. (2022) |
| Planteose (101) in the parasitic weed Orobanche minor | iMScope TRIO (DHB, sublimation) | Study of the involvement of the enzyme in planteose hydrolysis during seed germination | Okazawa et al. (2022) |
| Polysaccharides in soybean root nodules | Laser desorption ionization from silicon nanopost arrays and MALDI, LTQ‐Orbitrap (HABA, CHCA, 9‐AA, DHB, nebulizer and airbrush) | Characterization of number and weight average molar mass, polydispersity, and oligomer size distributions across the tissue section. Comparison with MALDI | Samarah et al. (2021) |
| Prostate tissue | PNGase F, Q‐TOF (CHCA, spray), N‐glycans | Direct N‐glycan profiling | Blaschke, Hartig, et al. (2021) |
| Small molecules, including sugars, in brain tissues | TOF/TOF (ZnO nanoparticles, TM sprayer) | Use of zinc oxide nanoparticles as matrix | Chen, Laviolette, et al. (2021) |
| Steroidal glycosides in Allium macrostemon Bge. and A. chinense G. Don | TOF (DHB, TM sprayer) | Structural identification and structure/activity relationships | Duan et al. (2022) |
| Sucrose metabolite from Vitis vinifera L. (grapevine) infected with Plasmopara viticola | FT‐ICR (DHB, CHCA, 9‐AA [9‐AA poor]) | Investigation of first moments of pathogen interaction | Maia et al. (2022) |
| Sugar phosphates and other metabolites from FFPE renal cancer tissue | FT‐ICR (9‐AA, spray), ‐ve mode | Identification of prognostic pathways and metabolites for renal cell carcinomas | Erlmeier et al. (2022) |
| Sulfatides in murine kidney | TOF (9‐AA [‐ve], CHCA [+ve] sublimation) | Identification of sulfatide with ceramide composed of phytosphingosine (t18:0) and 2‐hydroxy FAs in renal intercalated cells | Nakashima et al. (2022) |
| Sulfatides in rat brain | TOF (9‐AA, DHB, electrosprayed) | Imaging revealed sulfatide depletion in brain tissues of rats exposed in real air with high fine particulate matter | Diao et al. (2022) |
| Various endogenous compounds (mono‐, di‐saccharides, glycosides from Wolfberry fruit (Lycium barbarum L.) | QIT‐TOF (DHB [+ve], 9‐AA [‐ve] sublimation) | Visualizing the spatial distribution of endogenous molecules at different development stages | Zhao, Zhang, Shi, et al. (2021) |
| Various from food | Orbitrap (DHB, pneumatic sprayer) | Demonstration of MALDI imaging for ingredients, contaminants and additives in processed food | Kokesch‐Himmelreich et al. (2022) |





8. DERIVATIVES
Although reducing terminal derivatization is normally associated with the attachment of fluorescent derivatives for HPLC work, such derivatization methods can also attach moieties such as those containing a charge, that can enhance mass spectral performance. Several other derivatization methods such as permethylation and linkage‐specific derivatization of sialic acids are also used. Several reviews have appeared; these are listed in Table 6.
Table 6.
General reviews on derivatives.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Recent advancements in glycoproteomic studies: Glycopeptide enrichment and derivatization | Brief; derivatization of glycopeptides, permethylation and derivatization of sialic acids | 106 | Pujić and Perreault (2022) |
| Targeting out of range biomolecules: Chemical labeling strategies for qualitative and quantitative MALDI MS‐based detection | Includes reductive amination reactions of carbohydrates | 75 | Sejalon‐Cipolla et al. (2021) |
| Chemical derivatization for mass spectrometric analysis of metabolite isomers | In Chinese, table and references in English | 70 | Wang, Li and Abliz (2021) |
| Derivatization of carbohydrates for analysis by liquid chromatography and capillary electrophoresis | Derivatization for various detection systems. Brief mention of MALDI | 66 | Yu, Dalman, et al. (2021) |
| Options of the main derivatization approaches for analytical ESI and MALDI mass spectrometry | Extensive review with large table showing derivative structures | 410 | Zaikin and Borisov (2022) |
8.1. Reducing terminal derivatives
8.1.1. Reducing terminal derivatives prepared by reductive amination
Reaction of the aldehyde group of the open chain form of reducing carbohydrates with an amine yields a Schiff base, which can be stabilized by reduction. Two protocols have been published for attachment of 2‐aminobenzamide (anthranilamide (2‐AB) 102): “Profiling of N‐linked oligosaccharides of a glycoprotein by UPLC‐FLR‐ESI‐MS after derivatization with fluorescent anthranilamide” (Butré, Largy, & Delobel, 2021), and “Profiling, relative quantification, and identification of sialylated N‐linked oligosaccharides by UPLC‐FLR‐ESI/MS after derivatization with fluorescent anthranilamide” (Butré, Largy, Cantais, & Delobel, 2021). The 2‐AB reagent is used in excess and needs to be removed before mass spectral analysis. A method using a monolithic disc‐packed spin column has been reported (Yu, Dalman, et al., 2021). MonoSpin amide and MonoSpin‐NH2 columns showed the same efficiency as conventional solid‐phase extraction methods in the removal of the 2‐AB reagent and the recovery of the labelled glycans (Yui et al., 2022).

MALDI‐TOF/TOF has been used to characterize 2‐aminoacridone (2‐AMAC, 103) derivatives of chitooligosaccharides in a study whereby they were encapsulated in alginate nanospheres to enhance bioavailability and antiliver fibrotic effects (Liu, Li, et al., 2022).
8.1.2. Hydrazides
Benzenesulfonyl hydrazine (BSH, 104) derivatives, whose identity was shown by MALDI‐TOF‐MS of their per‐methylated derivatives, have been shown to be suitable for glycan separation by 2D‐HPLC. Furthermore, the underivatized glycans could be recovered by heating at 70oC for 30 min. (Wang, Gao, et al., 2022).

8.1.3. Reducing‐terminal derivatives prepared by other methods
Reducing sugars have been shown to react with N,O‐dimethylhydroxylamine hydrochloride (DMHA, 105) to form the substituted glycosylamine (106, shown for glucose) (Norberg et al., 2022). The derivatives gave good RP‐HPLC performance with a single peak for each sugar and enabled many isomers to be separated. The MALDI and ESI spectra were also reported to be excellent. Furthermore, as above, the free carbohydrates could be recovered quantitatively following mild acid hydrolysis with HCl or acetic acid.

1‐Phenyl‐3‐methyl‐5‐pyrazolone (PMP) has long been used as a derivatization reagent for carbohydrates. To produce high sensitivity, this reagent has now been modified (107, Scheme 5) to incorporate two carboxylic acids (CPMP) into the derivatized carbohydrate (108). The resulting derivatives gave exceptionally strong signals in negative ion mode. For derivatized disaccharides, the limits of detection (LODs) and limits of quantification (LOQ) ranged from 3.90 to 8.67 and 12.99 to 28.92 ng L−1, respectively (Ma, Chen, et al., 2022). Although analysis was by ESI, this derivative should work equally well by MALDI.
Scheme 5.

Formation of CPMP derivatives (108, shown for glucose).
8.1.4. Direct derivatization of PNGase F‐released glycans
When glycoproteins are incubated with PNGase F, the N‐glycans are released as glycosylamines, which slowly hydrolyse to the native glycans. Some investigators have developed methods for derivatizing the glycosylamines directly and thus, are able to save time. One such reaction is with 6‐aminoquinolyl‐N‐hydroxysuccinimidyl carbamate (AQC, 110) as shown in Scheme 6 (Wu, Zhang, Li, et al., 2022). The reaction is rapid and labelling was achieved in 20 min directly from the PNGase F incubation mixture.
Scheme 6.

Derivatization of the reducing terminal GlcNAc residue of N‐glycans with AQC (110).
8.1.5. Derivative removal
A method for removal of a wide range of common florescent tags from glycans has been reported by Zhang, Wang, Li, et al. (2022). Glycans were incubated with 1% Oxone (trade name for 2KHSO5·KHSO4·K2SO4) containing 0.1% TFA for 0.5 h after which, the reaction mixture was purified with a RP‐C18 SPE cartridge. Unfortunately, reactions were not quantitative. Several products were generally produced as illustrated for glucose derivatized with 2‐amino(N‐aminoethyl) benzamide (AEAB (Scheme 7). Yields varied depending on the glycan and the derivative with glucose and lactose performing badly with only 31% and 27% of the required product for the AEAB derivative respectively, and 53% and 63% for the 2‐AB and 2‐aminobenzoic acid (2‐AA, 115) derivatives respectively. N‐Acetylamino‐sugars, on the other hand gave high yields (e.g., 75% and 86% for GlcNAc [117] and GalNAc [118] respectively for the 2‐AA derivative. For aniline derivatives, the yield was 100% of the released glycan. 2‐aminopyridine (2‐AP, 116) derivatives, however, failed to give a reaction, attributed to oxidation of the pyridine.
Scheme 7.

Derivative removal with Oxone.

8.2. Derivatives of other sites
8.2.1. Hydroxyl groups—Permethylation
Permethylation is possibly the oldest derivatization technique for carbohydrates and was originally used for gas‐liquid chromatographic (GLC) and combined GC/MS work. It featured prominently in a method for linkage analysis that is still used today. A tutorial on the technique has been published by Black et al. (2021). Later, it was used for fast‐atom bombardment (FAB) mass spectrometry and it still finds uses for improving sensitivity for MALDI analysis. Many examples can be found in Tables 12, 13, 15, 20, 21, 22, 33, 36, 39, 40, 43 and 50. Several methods of preparation have been published; an up‐to‐date one can be found in the paper by Cho, Banazadeh, et al. (2021).
Table 12.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of carbohydrate polymers from plants, animals and algae.
| Species or glycan source | Carbohydrate | Methodsa | Notes | References |
|---|---|---|---|---|
| Agave angustifolia Haw | Fructans | R‐TOF (DHB) | Identification and evaluation of the fermentation of acetylated agave fructans with Saccharomyces boulardii as a probiotic | Buitrago‐Arias et al. (2021) |
| Agave tequilana Weber var. azul | Fructans | TOF, IMS‐Q‐TOF, (CHCA) imaging | Localization and composition in stem and rhizome | Pérez‐López et al. (2021) |
| Agavin (fructan) from Agave tequilana Weber Var. Blue (commercial) | Fructan | TOF (DHB) | Study of the effect of dietary agavin supplementation in blood parameters and antioxidant enzymes of juvenile Nile tilapia (Oreochromis niloticus) under stress conditions | Flores‐Méndez et al. (2022) |
| Alhagi pseudalhagi, (camel thorn) | Hetero‐polysaccharide (14 residues) | L‐TOF/TOF (DHB) | Structural elucidation and osteogenic activity | Ye, Li, et al. (2021) |
| Allium sativum (garlic) | Oligosaccharide with Fru, Glc, GalA, Gal, Man, Ara, Rha | TOF | Preparation and structural characterization | Jiang, Ran, et al. (2022) |
| Allium schoenoprasum | Major polysaccharide with Ara, Gal, Glc, Fru (ratio 1:2:2:5) | R‐TOF/TOF (CMBT) | Purification and structural characterization | Zhang, Zheng, et al. (2021) |
| Apium graveolens (celery) | Rhamnogalacturonan‑II | TOF (DHB), ESI, GC/MS | Structural characterization | Barnes et al. (2021) |
| Arabidopsis thaliana | Xyloglucans | TOF/TOF (CMBT/DHB) | Study of the effect of O‐acetylation levels of cell wall xyloglucan on sensitivity to aluminium | Wu, Tao, et al. (2022) |
| Aspergillus flavus and A. fumigatus | Chito‐oligosaccharides | TOF/TOF (DHB) | For study on inhibitory activity and mechanism of chitosan oligosaccharides on Aspergillus flavus and A. fumigatus | Ke et al. (2022) |
| Astragalus arbusculinus gum | Carbohydrate with Glc, pinitol, and Ara (relative molar ratio of 4:1) | TOF (CHCA) | Isolation, characterization, and antioxidant activity | Ahmadi, Rezadoost, et al. (2022) |
| Avena sativa (oat, bran) | β ‐Glucan | TOF (DHB) | Structural studies of water‐insoluble β‐glucan and its effect on improving lipid metabolism in mice fed high‐fat diet | Yu, Wang, et al. (2021) |
| Birch and beech wood | Xylo‐oligosaccharides | TOF/TOF (DHB) | Hydrolysis of xylans catalyzed by xylanase from Bacillus subtilis | Wei et al. (2021) |
| Bletilla formosana | Glucomannans | R‐TOF/TOF | Structural determination of two glucomannans and their protective effect on inflammation via inhibiting NF‐κB pathway | Gu (2022) |
| Chicken jejunum | Oligosaccharides | TOF (DHB), (per‐Me) | Qualitative and quantitative profiles of jejunal oligosaccharides in broiler chickens receiving different dietary levels of fiber, protein and exogenous enzymes | Lin and Olukosi (2021) |
| Coreopsis tinctoria (Kunlun chrysanthemum flower tea) | Oligosaccharides | L‐TOF/TOF | Structural elucidation of three novel oligosaccharides and their bioactivities (hyperglycemia and neuroinflammation) | Yu, Chen, et al. (2021) |
| Crassostrea hongkongensis (oyster) | Polysaccharide (α‐(1→4) d‐linked Glc backbone and (→4,6)‐α‐d‐Glc‐(1→) branches every 4.7 residues | TOF/TOF | Oyster polysaccharides shown to ameliorate intestinal mucositis and improve metabolism in 5‐fluorouracil‐treated S180 tumour‐bearing mice | Baxa et al. (2020) |
| Crataegus azarolus (yellow hawthorn) fruit | Polysaccharides | TOF (DHB) | β‐(1 → 4)‐Linked glucose and mannose residues with monosaccharide branches of α‐(1 → 6) galactose and O‐acetyl substituents. | Bensaci et al. (2022) |
| Cremastra appendiculata (medicinal plant) | Mannoglucan | TOF, HPLC, NMR, GC/MS, IR | Structural characterization | Zhang, Bi, et al. (2021) |
| Cyamopsis tetragonolobus (guar) | Oligosaccharides | MALDI‐TOF | Characterization of resultant oligosaccharides from guar galactomannan upon depolymerization by nonspecific enzymes | Shobha et al. (2022) |
| Cynara cardunculus var. scolymus (artichoke) | Pectic oligosaccharides | TOF | Characterisation and virtual screening of prebiotic properties using in silico colonic fermentation | Sabater, Blanco‐Doval, Margolles, et al. (2021) |
| Digesta and excreta from broiler chicken | Arabinoxylo‐oligosaccharides | TOF/TOF (DHB) | Dietary endo‐xylanase shown to alter arabinoxylan utilization | Kouzounis et al. (2022) |
| Evodia lepta (Spreng) Merr. | Oligosaccharides | TOF (1‐(4‐cyanophenyl)‐4‐piperidinyl hydrazide (CPH) derivative) | Characterization, antioxidant and antitumor activities. Use of different extraction methods. Microwave‐assisted extraction best. | Xiong, Liang, et al. (2022) |
| Glycine max (soybean) | Polysaccharides | L‐TOF (DHB) | Study of the effect of microwave‐assisted acid extraction on the physicochemical properties and structure of soy hull polysaccharides | Cai, Zhang, et al. (2022) |
| Glycine max (soybean) | Polysaccharides | TOF (DHB) | Chemical composition and sugar spectroscopy of polysaccharides obtained by microwave‐assisted salt extraction | Li, Zhang, Cheng, et al. (2022) |
| Hermetia illucens (black soldier flies) | Chitosan | R‐TOF/TOF (CHCA, DHB) | Structure and enzymatic hydrolysis | Lee, Kim, Nam, et al. (2022) |
| Hordeum vulgare (spring barley) | Fructans | TOF/TOF (DHB) | Genome‐wide association study reveals the genetic complexity of fructan accumulation patterns in barley grain | Matros et al. (2021) |
| Hordeum vulgare (spring barley) | Fructose | IMS‐TOF (DHB), MS/MS, GC/MS | Structural determination and immunomodulatory properties | Lemieszek et al. (2022) |
| Hordeum vulgare (barley) | BF‐1 (Mixture of (arabinoxylan, yeast‐derived β‐glucan, barley‐derived β‐glucan, and type II arabinogalactan) | TOF/TOF (DHB), LC‐MS/MS | Structural identification of active moiety in antitumor metastatic polysaccharide purified from fermented barley | Son et al. (2022) |
| Horse gut (feces and stomach contents) | Cellulose | TOF (per‐Me) | Oxidation of cellulose by lytic polysaccharide monooxygenases | Liu, Yu, et al. (2022) |
| Inula helenium L. | Inulin (fructan) | R‐TOF (DHB) | Optimization of methods for inulin extraction | Ahmadi, Farimani, et al. (2022) |
| Lignosus rhinocerotis (Cooke) Ryvarden (fungus) | β‐Glucans | R‐TOF/TOF (CHCA) | Structural determination and effect on intestinal mucosal wound healing | Veeraperumal et al. (2021) |
| Malus domestica (apple, pomace) | Xyloglucans | TOF (DHB/DMA) | Analysis of xyloglucans for potential developments in industrial applications | Chen, Mac‐Béar, et al. (2022) |
| Nyctanthesarbor‐tristis leaves | Xylo‐oligosaccharides | R‐TOF (DHB), SEC, HPAEC, GC/MS | Production and identification of bioactive oligosaccharides by a combination of enzymatic, HPAEC and MALDI‐TOF‐MS techniques | Ali, Mukherjee, et al. (2021) |
| Oryza sativa (rice) | Oligosaccharides | R‐TOF/TOF (DHB, CMBT) | Poaceae‐specific cell wall‐derived oligosaccharides shown to activate plant immunity via OsCERK1 during Magnaporthe oryzae infection | Yang, Liu, et al. (2021) |
| Oryza sativa (rice), cell walls | Xylans | TOF (DHB) | Organically‐bound silicon shown to enhance resistance to enzymatic degradation | Pu et al. (2021) |
| Oryza sativa (rice) | Hydroxycinnamic acid‐modified xylan | TOF/TOF (DHB), (procainamide derivative) | Hydroxycinnamic acid‐modified xylan side chains and their cross‐linking products in rice cell walls are shown to be reduced in the xylosyl arabinosyl substitution of xylan 1 mutant | Feijao et al. (2022) |
| Panax quinquefolius, L (ginseng) | Polysaccharides | TOF/TOF (DHB) | Structural analysis of red ginseng polysaccharides | Jin, Oh, et al. (2021) |
| Pennisetum glaucum (pearl millet) | Oligosaccharide (Glc, Gal) | TOF/TOF (DHB), GC/MS, TLC, FTIR | Characterization and evaluation of their prebiotic potential | Mondal et al. (2022) |
| Pinctada fucata (pearl oyster), shells | Sulfated polysaccharide | TOF/TOF (DHB) | Sulfated polysaccharide shown to improve scopolamine‐induced memory impairment | Yamagami et al. (2021) |
| Polygonatum cyrtonema | Fructan and galactan | TOF/TOF (DHB) | Structures and their utilization by probiotic bacteria | Zhang, Chen, Luo, et al. (2021) |
| Polygonatum cyrtonema Hua | Fructo‐oligosaccharide | TOF (DHB) | Structural characterization and treatment of LPS‐induced peritonitis in mice | He et al. (2021) |
| Polygonatum odoratum (Mill.) Druce | Cell wall polysaccharides | TOF/TOF (DHB) | Structure and biological activities of cell wall polysaccharides in the rhizome, stem, and leaf | Li, Hsiung, et al. (2022) |
| Poplar sawdust (genus Populus) | Cello‐oligosaccharides | TOF (DHB) | Production of high‐yield short‐chain oligomers from cellulose via selective hydrolysis in molten salt hydrates | Ma, Lin, et al. (2022) |
| Rhizobium radiobacter ATCC 1333 | Cyclic β‐1,2‐glucans | TOF (DHB) | Isolation and use for increasing solubility of curcumin by complexation | Wu, Zhang, Gao, et al. (2022) |
| Rosaceae family: Apple and sweet cherry | Xyloglucans | TOF/TOF (DHB/DMA) | Comparison of cell wall chemical evolution during the development of fruits | Lahaye et al. (2021) |
| Sabia parviflora | α‐Glucoside | TOF | Isolation, structure identification and hepatoprotective activity | Zhang, Li, et al. (2021) |
| Sepioteuthis lessoniana (squid) | Sulfated chitosan | R‐TOF (CHCA) | Sulfated chitosan converted to low molecular weight form with gamma radiation. Antituberculosis activity | Ramachandran et al. (2022) |
| Solanum lycopersicum (tomato) | Hemicellulose | Cutinase and endo‐1‐4‐ß‐d‐glucanase, TOF (DHB/DMA) | Investigation of cutin polymer matrix structure during fruit development | Reynoud et al. (2022) |
| Taraxacum kok‐saghyz Rodin | Inulin | R‐TOF (DHB) | Optimization of extraction by response surface methodology | Chen, Wang, Dong, et al. (2022) |
| Tichocarpus crinitus (red alga) | Kappa/beta‐carrageenan | R‐TOF/TOF (DHB), MS/MS, LC‐MS | Structural determination. Potential inhibitor of HIV‐1 | Yermak et al. (2021) |
| Ulva sp. | Oligo‐ and polysaccharides | TOF | Effect on human skin fibroblasts | Fournière et al. (2021) |
| Ulva fasciata (green seaweed) | Ulvan, a water‐soluble polysaccharide (‐4‐β‐d‐GlcpA‐(1→4)‐α‐l‐Rhap‐(1→)‐ repeat | L‐TOF/TOF (DHB, ‐ve) | Ulvan shown to consist of rhamnose, rhamnose‐3‐sulfate, xylose and numerous uronic acid residues and to induce resistance in wheat against Zymoseptoria tritici without major alteration of leaf metabolome | de Borba et al. (2021) |
| Vaccinium macrocarpon (cranberry) | Oligosaccharides and proanthocyanidins | R‐TOF (DHB) | Proanthocyanidin‐enriched cranberry extract shown to induce resilient bacterial community dynamics in a gnotobiotic mouse model | Neto et al. (2021) |
| Vaccinium sect. Cyanococcus (bulberry) | Xyloglucan and pectin | R‐TOF/TOF (DHB) | Structure and composition of glycans that bind anthocyanins during fruit puree processing | Hotchkiss et al. (2021) |
| Wolffiella repanda (duckweed) | Rhamno‐galacturonan‑II | TOF (DHB), ESI, GC/MS | Structural characterization | Barnes et al. (2021) |
| Wood (oak, hornbeam, walnut) | Various oligosaccharides | TOF (DHB) | Destructive behaviour of wood by the white‐rot fungus Fomes fomentarius | Bari et al. (2021) |
| Commercial | Hyaluronic acid (low molecular weight) | TOF/TOF (DHB) | For study of rheological properties of hyaluronic acid diluted solutions as components of cosmetics | Saitarly et al. (2022) |
| Commercial | Chitin and chitosan | TOF (DHB) | Development of prediction models for adsorption properties of chitin and chitosan for micropollutants | Cho, Lim, et al. (2021) |
| Commercial | Chitosan | TOF (CHCA [peptides], DHB [glycans]) | Structural characterization by MALDI | Jung, Lee, et al. (2021) |
Format (not all items present): Depolymerization method, MALDI method (matrix), compounds run (derivative), other methods.
Table 13.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of carbohydrate polymers from lower organisms.
| Species | Carbohydrate | Techniquesa | Notes | References |
|---|---|---|---|---|
| Auricularia auricula‐judae | Black fungus polysaccharide (β‐glucan) | TOF/TOF (DHB) | Investigation of anti‐hepatoma activity | Cai, Zhou, et al. (2021) |
| Bacillus amyloliquefaciens WX‐1 | Konjac glucomannan | TOF (DHB) | Production, characterization, and prebiotic activity | Wan, Wei, et al. (2021) |
| Colaconema formosanum | Mannose | TOF | Investigation of bioactive compounds for industrial use | Lee, Huang, et al. (2022) |
| Enterobacter soli | Exopolysaccharides | TOF (DHB, +ve, ‐ve) | Shown to be effective at removing chromium from industrial effluent | Kailasam et al. (2022) |
| Ganoderma lucidum | β‐d‐glucan | TOF/TOF (DHB/TFA) | Microwave‐assisted degradation and the structural and immunoregulatory properties of oligosaccharide fractions | Qin, Ma, et al. (2022) |
| Lactobacillus paraplantarum KM1 | Exopolysaccharides (EPS) | TOF (DHB) | Purification and characterization of novel exopolysaccharides produced from L. paraplantarum KM1 isolated from human milk and its cytotoxicity | Sharma, Sharma, et al. (2021) |
| Mucilaginibacter sp. ERMR7:07 (glacier bacterium) | Exopolysaccharides | TOF/TOF (DHB), GC/MS, NMR, UV, FTIR | Production, characterisation, and applications | Kumar et al. (2022) |
| Mycobacterium bovis (BCG) | Polysaccharides | TOF (per‐Me) | Structural chacterization | Luo, Song, Chang, et al. (2022) |
| Nostoc commune | Mannose | TOF | Investigation of bioactive compounds for industrial use | Lee, Huang, et al. (2022) |
| Paenibacillus polymyxa A 26 | Exopolysaccharides | TOF (DHB) | Silica particles shown to trigger the EPS production of harsh environment isolates of growth‐promoting rhizobacteria and increase their ability to enhance wheat biomass in drought‐stressed soils | Fetsiukh et al. (2021) |
| Pseudomonas aeruginosa | Pel polysaccharide | R‐TOF (DHB), GC/MS, NMR | Structural determination (dimeric repeat of α−1,4 linked GalN and GalNAc) | Le Mauff et al. (2022) |
| P. aeruginosa | Malto‐oligosaccharides | TOF/TOF (DHB) | Trehalose and α‐glucan shown to mediate distinct abiotic stress responses | Woodcock et al. (2021) |
| Saccharomyces cerevisiae CNCM I‐3856 | Hetero‐polysaccharides | R‐TOF/TOF (DHB), (per‐Me), GC/MS | Structural characterization and anti‐adhesive properties against E. coli associated with Crohn's disease | Sivignon et al. (2021) |
| Sarcodia suae | Mannose | TOF | Investigation of bioactive compounds for industrial use | Lee, Huang, et al. (2022) |
| Sargassum muticum | Heterofucoidans | TOF | Use of acetone precipitation to extract heterofucoidans from autohydrolysis extracts | Acevedo‑García et al. (2021) |
| Sphaerotilus montanus (sheath‐forming bacterium) | Sheath‐forming polysaccharide | R‐TOF (2,3‐DHB) | Structural characterization using thiopeptidoglycan lyase which recognizes the 1→4 linkage between α‐d‐GalN and β‐d‐GlcA | Kashiwabara et al. (2021) |
| Spirulina platensis | Oligosaccharide | TOF/TOF (DHB) | Structural characterization and its effect on the faecal microbiota in vitro | Cai, Yi, et al. (2022) |
| Usnea sp. (lichen) | Amide‑containing β‑glucan | R‐TOF/TOF (DHB) | Development of method to release the glucan | Fernandes et al. (2021) |
| Xanthomonas pathogens | Xyloglucans | TOF/TOF | Investigation of xyloglucan processing machinery and its role in the transcriptional activation of virulence factors | Vieira et al. (2021) |
| Zygnematophyceae (green algae), several species | Xyloglucan | TOF/TOF (DHB) | Ancient origin of fucosylated xyloglucan discovered in charophycean green algae | Mikkelsen et al. (2021) |
Format (not all items present): MALDI method (matrix), (derivative), other methods.
Table 15.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for the characterization of carbohydrates from milk and milk products.
| Source | Methodsa | Notes | References |
|---|---|---|---|
| Human milk | R‐TOF/TOF (CHCA), (per‐Me) | Identification of new (Gal)3 and Fuc‐(Gal)3‐containing 6‐antennae (see text) | Hanisch and Kunz (2021) |
| Human milk | TOF, (DHB) (linkage‐specific amidation with d5‐aniline, Girard's P), LC‐MS | Identification of novel α2,3‐linked di‐/tri‐sialylated oligosaccharide isomers | Jin, Lu, et al. (2022) |
| Human colostrum and mature milk | PNGase F, TOF (DHB), (linkage‐specific amidation) | A preliminary study on isomer‐specific quantification of sialylated N‐glycans released from whey glycoproteins | Jin, Li, et al. (2021) |
| Human, cow, goat, sheep, and camel milk | R‐TOF/TOF (DHB), (per‐Me) | Identification and absolute quantification of milk oligosaccharides in different species | Shi, Han, et al. (2021) |
| Human milk | FT‐ICR, LTQ (MS/MS), (DHB) | Antibiofilm activity against multidrug resistant and susceptible isolates of Acinetobacter baumannii | Spicer et al. (2021) |
Format (not all items present): MALDI method (matrix), (derivative), other methods.
Table 20.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of N‐glycans from specific glycoproteins.
| Glycoprotein | Methodsa | Notes | References |
|---|---|---|---|
| 4‐1BB receptor (human) | PNGase F, TOF/TOF (per‐Me) | Structural determination and demonstration that N‐glycosylation facilitates 4‐1BB membrane localization by avoiding its multimerization | Sun, Kim, et al. (2022) |
| Alkaline phosphatase from Neurospora crassa | PNGase F, TOF (per‐Me), GC/MS | Characterization the N. crassa DFG‐5 α−1,6‐mannanase and demonstration of binding to the α−1,6‐mannose backbone of an N‐linked galactomannan found on cell wall glycoproteins. | Patel et al. (2022) |
| Alpha‐1‐acid glycoprotein (AGP) | PNGase F, TOF/TOF (DHB), (per‐Me) | Different glycoforms of AGP shown to contribute to its functional alterations in platelets and neutrophils | Sumanth et al. (2021) |
| Apo‐H (beta‐2‐glycoprotein) | PNGase F, TOF (s‐DHB), (Et esters) | Structural characterization (bi‐, tri‐antennary complex) | Javeed et al. (2021) |
| Alpha fetoprotein | PNGase F, MALDI (DHB), (Girard's P) | In development of a dual‐modal ratiometric immunoassay for diagnosis of hepatocellular carcinoma | Li, Pang, et al. (2022) |
| Asialofetuin | PNGase F, TOF, (+ve, ‐ve), (per‐Me) | Use of a novel lamprey antibody to characterize 3‐O‐sulfation | McKitrick et al. (2021) |
| Bilirubin oxidase | PNGase F, endo‐F1, TOF/TOF (DHB), (Bz oximes, Glycoblotting) | Effects on direct electron transfer‐type bioelectrocatalysis (high mannose) | Suzuki, Itoh, et al. (2022) |
| Bovine fetuin | PNGase F, TOF (CHCA) (amidation with aniline) | Control experiment on aniline derivatization of sialic acids for increased sensitivity for tissue imaging | Zhang, Shi, et al. (2022) |
| Bovine lactoferrin | PNGase F TOF (DHB), (Et ester, Girard's P) | Detection of N‐glycan changes of bovine lactoferrin at different stages of lactation | Jia et al. (2021) |
| Dynactin‑associated protein | PNGase F, Spiral‐TOF (CHCA), (3‐AQ) | Glycosylation of T/S cluster region (anomalous behaviour in SDS‐PAGE analysis) | Yin, Konishi, et al. (2022) |
| Erythropoietin | FT‐ICR (DHAP, Cl‐CCA, CHCA, SA) glycoprotein | Glycoform analysis of intact erythropoietin | Lippold et al. (2021) |
| Glycoprotein from Abelmoschus esculentus L. Moench (okra) | PNGase A, TOF (DHB) | Structural determination and antioxidant activity | Zhao, Xu, et al. (2021) |
| H11 protein from Haemonchus contortus | PNGase F, TOF/TOF (DHB), (per‐Me) | H11‐induced immunoprotection shown to be predominantly linked to N‐glycan moieties during infection | Wang, Liu, et al. (2022) |
| Human ACE2/IgG1‐Fc domain (ACE2‐Fc) | PNGase F, R‐TOF/TOF (DHB), (reduction, per‐Me) | SARS‐CoV‐2 spike protein variant binding affinity | Matthews et al. (2022) |
| IgG | PNGase F, IT‐TOF (DHB), (per‐Me) | Cytokines in the immune microenvironment shown to change the glycosylation of IgG by regulating intracellular glycosyltransferases | Cao, Song, et al. (2022) |
| IgG | PNGase F, R‐TOF/TOF (DHB), (Per‐Me and Me ester) | Enzyme ST6Gal1 shown not to be required for IgG sialylation | Oswald et al. (2022) |
| IgG, (surface variable domain) | PNGase F, TOF/TOF (s‐DHB), (Et ester) | Surface IgG glycosylation shown to affect autoantigen binding and acts as threshold for human autoreactive B cell activation | Kissel et al. (2022) |
| IgG (monoclonal against aflatoxin B1) | PNGase F, TOF (DHB) | As part of detailed structural analysis | Xing et al. (2022) |
| IgG from rhesus macaque | PNGase F, TOF/TOF (DHB), (2‐AA, Et ester/amide, linkage‐specific) | Alteration of N‑glycome during infection with the human parasitic filarial nematode Brugia malayi | Petralia, Santha, et al. (2022) |
| IgG1 and FcgRIIIa (158F and 158V allotypes) | PNGase F, R‐TOF (s‐DHB), (Et esters, linkage‐specific) | Study of the role of N‐glycosylation in FcgRIIIa interaction with IgG | Van Coillie et al. (2022) |
| Invertase glycoforms from Saccharomyces cerevisiae | PNGase F, R‐TOF/TOF (DHB) | To obtain optimum glycosylation for use as synthetic enzymes for methyl β‐d‐fructofuranoside | Andjelković et al. (2021) |
| Lactoperoxidase | EndoBI‐1, TOF (DHB), (2‐AA) | Model compound for evaluation of immobilized bifidobacterial endo‐ß‐N‐acetylglucosaminidase to generate bioactive compounds for food industry | Pekdemir et al. (2022) |
| Palivizumab (SynagisR), (monoclonal antibody) | PNGase F, R‐TOF/TOF (SA), (2‐AB), UHPLC, database structural interpretation | Structural determination mainly by HPLC (Man5GlcNAc2, biantennary complex) | Sran et al. (2022) |
| Prolyl‐alanyl‐specific endoprotease endopro from Aspergillus niger | PNGase F, FT‐ICR (s‐DHB), (2‐AA) | Structural determination and functional proteoform characterization (high‐mannose, site analysis) | van Schaick et al. (2021) |
| Prostate‐specific antigen (PSA) | Trypsin, FT‐ICR (s‐DHB), (sialic acid amidation) | Glycopeptide profiling of PSA from seminal plasma by MALDI‐MS | Wang, Kałuża, et al. (2021) |
| SARS‐CoV‐2, Spike protein S1 subunit RBD (Arg319‐Phe541) | PNGase F, TOF/TOF (DHB), (per‐Me), MS/MS | Site‐specific analysis (bi‐, tri‐antennary complex) | Antonopoulos et al. (2021) |
| SARS‐CoV‑2 Spike glycoprotein (from HEK293 and baculovirus‐insect cells) | PNGase F, TOF/TOF (DHB) | Site‐specific analysis (high‐mannose, bi‐, tri‐antennary complex) | Wang, Wu, et al. (2021) |
| SARS‐CoV‑2 nucleocapsid protein in HEK293 cells | PNGase F, TOF/TOF (DHB), per‐Me | Structural identification, (high‐mannose, bi‐antennary complex) | Supekar et al. (2021) |
| SARS‐CoV‑2 spike glycoprotein in CHO and HEK293 cells | PNGase F, TOF/TOF (di‐Me amide/amide derivs) | Use of a linkage‐specific sialic acid labeling strategy to define site‐specific glycosylation patterns | Wang, Wang, et al. (2021) |
| SARS‐CoV‐2 spike protein | PNGase F, QIT‐TOF (DHB), (per‐Me) | Study of the effect of N‐glycosylation of SARS‐CoV‐2 spike protein on the virus interaction with the host cell ACE2 receptor | Huang, Tan, et al. (2021) |
| Shark‐derived IgG new antigen receptor | PNGase F, TOF/TOF (DHB), BlotGlyco method | Identification of N‐glycans (high‐mannose, hybrid, di‐, tri‐ and tetra‐antennary complex) and production of monoclonal IgG from CHO cells | Enatsu et al. (2021) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), (derivative), other methods.
Table 21.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of N‐glycans from intact organisms, tissues or glycoprotein mixtures.
| Source | Methodsa | Notes | References |
|---|---|---|---|
| Abelmoschus esculentus L. Moench (okra) | PNGase A, TOF (DHB) | Structural characterisation and antioxidant activity of a novel N‐linked glycoprotein | Zhao, Xu, et al. (2021) |
| Amniotic membrane (human) | PNGase F R‐TOF/TOF (DHB), (per‐Me) | Glycoproteins in amniotic membrane shown to contain bisected complex N‐glycans | Chen, Zhang, Zhang, et al. (2022) |
| Arabidopsis thaliana | PNGase F, TOF | Changes of protein N‐glycosylation in the growth of A. thaliana and effects of enzymatic deglycosylation on root development (in Chinese) | Wang, Yang, Zhao, et al. (2021) |
| B3GAT1 and mCherry A549 cells | PNGase F, R‐TOF/TOF (DHB), (per‐Me) | Inhibition of sialyltransferase prevents infection by influenza respiratory viruses | Trimarco et al. (2022) |
| Bombus terrestris, (bumblebee, queen) | PNGase F, QIT‐TOF (DHB) | Hex5HexNAc3dHex2, Hex3HexNAc3dHex2, and Hex4HexNAc3Pen1 identified in study of the role of GAGs in aged rats | Ahn et al. (2021) |
| Brugia malayi (filarial nematode) | PNGase F, TOF/TOF (DHB), (2‐AA), exoglycosidase | Identification of high‐mannose glycans and complex glycans with GlcA and phosphatidylchloline | Petralia, van Diepen, et al. (2022) |
| Caveolin‑1 knockout mouse serum | PNGase F, QIT‐TOF (per‐Me) | Caveolin‐1 (protein) shown to influence N‐glycosylation | Chen, Wang, Wu, et al. (2022) |
| Chlorella vulgaris UTEX395 | PNGase A, TOF/TOF | Structural analysis of secretome and N‐glycosylation, Man3GlcNAc2Pen3(Me)2 or 3 | Choi et al. (2021) |
| CHO Cell lines (CHO‐K1, CHO‐S, and CHO‐Pro5) | PNGase F, R‐TOF/TOF | Structural determination. Differences in sialylation and fucosylation | Wang, Wang, Wu, et al. (2022) |
| EXT1 k.d. cells (ER membranes) | PNGase F, TOF/TOF (per‐Me) | Alternative glycosylation shown to control endoplasmic reticulum (ER) dynamics and tubular extension in mammalian cells | Kerselidou et al. (2021) |
| Extracellular vesicles secreted from Plasmodium falciparum‐infected red blood cells | PNGase F, TOF/TOF (s‐DHB), (per‐Me) | Sialylated N‐glycans shown to mediate monocyte uptake of the extracellular vesicles | Pilo et al. (2022) |
| Fish cell lines (6 species) | PNGase F, R‐TOF/TOF, (Me ester) | High‐mannose, hybrid, bi‐, tri‐antennary complex glycans. For predicting cellular receptors to the nervous necrosis virus | Gye and Nishizawa (2022) |
| Funaria hygrometrica (moss) | PNGase A, TOF (DHB), LC‐MS/MS, GC‐MS, (reduced) | Identification of plant‐like glycans (Man3GlcNAc2Fuc1Xyl1) and glycans with 2,6‐di‐Me‐Man at 6‐antenna. Also methylated glycans from other mosses. (see text) | Stenitzer, Mócsai, Zechmeister, et al. (2022) |
| Haemonchus (parasitic nematode) | PNGase A, F, TOF/TOF (DHB), (per‐Me) | Structural characterization, high‐mannose, paucimannosidic | Wang, Gao, et al. (2021) |
| HCT116 CRC cell line | PNGase F, TOF/TOF (s‐DHB), (Et ester) | Experiments to determine best lectin for detecting core fucose | Rubén et al. (2021) |
| HepG2 Cells | PNGase F, TOF/TOF (DHB), (aoWR derivatives), exoglycosidase | To evaluate effect of swainsonine on N‐glycosylation and its contribution to toxicosis in livestock grazing swainsonine‐producing plants | Morikawa et al. (2022) |
| HepG2 Cells | PNGase F, R‐TOF/TOF (DHB), (per‐Me) | Study of the origin of cytoplasmic GDP‐fucose used for glycan assembly | Sosicka et al. (2022) |
| HepG2 cells (plasma glycoproteins) | PNGase F, TOF/TOF (DHB), (2‐AB), HPLC, LC‐MS | DNA Hypomethylation shown to upregulate expression of the MGAT3 gene and lead to changes in N‐glycosylation of secreted glycoproteins | Klasić et al. (2022) |
| HL‐60 promyelocytes | PNGase F, TOF/TOF (DHB), (per‐Me) | In study of inhibition of O‐glycan biosynthesis using the hexosamine analog Ac5GalNTGc | Wang, del Solar, et al. (2021) |
| HL‐60 cells | PNGase F, TOF/TOF (DHB/DMA), (Et ester, p‐toluidine) | Demonstration of altered sialidase expression in human myeloid cells undergoing apoptosis and differentiation | Hyun et al. (2022) |
| Human brain | R‐TOF, LC/MS, CID | Bisecting Lewis X in hybrid‐type glycans identified (see text) | Helm et al. (2021) |
| Human cervicovaginal fluid | PNGase F, TOF/TOF (DABP), (per‐Me) | Structural characterization. Found to reflect microbial community, immune activity, and pregnancy status | Wu, Grassi, et al. (2022) |
| Human dermal endothelial cells | PNGase F, TOF/TOF (DHB), (per‐Me) | Sialoglycans on lymphatic endothelial cells shown to augment interactions with Siglec‐1 (CD169) of lymph node macrophages | D'Addio et al. (2021) |
| Human erythrocytes | PNGase F, TOF/TOF (CMBT), (per‐Me | Structural determination, high‐mannose, hybrid, bi‐, tri‐antennary complex, bisects, N‐acetyl‐lactosamine extensions | Bua et al. (2021) |
| Human serum and cerebrospinal fluid | PNGase F, R‐TOF/TOF, (aoWR), exoglycosidase (2‐AB) | Detection of novel low‐molecular‐weight blood group‐specific glycans in serum and cerebrospinal fluid | Furukawa et al. (2021) |
| Human umbellar vein endothelial cells | PNGase F, TOF (per‐Me) | To assess adhesion of cells to PET woven fabrics used in medicine. No significant change, unlike O‐glycans | Hu, Sheng, et al. (2022) |
| Leptinotarsa decemlineata (Colorado potato beetle), peritrophic membrane | PNGase A, FT‐ICR (DABP), (per‐Me) | Study of changes to peritrophic membrane in mannosidase‐Ia silenced insects. Accumulation of high‐mannose glycans | Liu, De Schutter, et al. (2022) |
| Madin‐Darby canine kidney (MDCK) cells and humanized MDCK cell line | PNGase F, TOF/TOF (DHB), (per‐Me) | Analysis of sialylated and sulfated glycans in humanized cell line | Byrd‑Leotis et al. (2022) |
| MDCK sialic acid knockout cells | PNGase F, QIT‐TOF (DHB), (linkage‐specific amidation, 2‐AA) | Investigation of influenza A virus agnostic receptor tropism with terminal sialic acid knockout cells | Kamiki et al. (2022) |
| Middle silk gland of silkworm (Bombyx mori) | Hydrazinolysis, R‐TOF (DHB), (2‐AP) | Temporal analysis of N‐acetylglucosamine extension of N‐glycans | Kajiura et al. (2022) |
| Mouse liver | PNGase F, TOF (DHB), (per‐Me) | Nuclear receptors (farnesoid X receptor and small heterodimer partner) found to regulate protein N‐glycan modifications in the liver | Mathur et al. (2021) |
| Mouse serum | PNGase F, TOF/TOF (DHB), (per‐Me) | Identification of glycans separated by molecular matrix electrophoresis | Liu, Liu, Li, et al. (2021) |
| Mouse serum | PNGase F, TOF/TOF (CHCA), (Me ester, BOA) | Study of anxiety‐related behaviors in single versus group‐housed male mice | Abou‐Elnaga et al. (2021) |
| Mouse brain tissue | PNGase F, TOF/TOF (DHB), (per‐Me) | Brain glycoproteins shown to exhibit diminished glycan complexity compared to other tissues | Williams et al. (2022) |
| Mouse peritoneal macrophage sub‐populations | PNGase F, TOF/TOF (DHB), (per‐Me), MS/MS | Resident and elicited murine macrophages shown to differ in expression of their glycomes and glycan‐binding proteins | Park, Chen, et al. (2021) |
| Mouse primary mesangial cell | PNGase F, FT‐ICR (CHCA, TM sprayer on glass slides) | Increased sialylation in lupus | Sundararaj et al. (2021) |
| NK Cells | PNGase F, TOF | Glycoengineering of NK cells with glycan ligands of CD22 and selectins for B‐Cell lymphoma therapy | Hong et al. (2021) |
| Oryzias latipes, (Japanese medaka, fish) | Hydrazine, TOF (DHB), (2‐AP) | Exposure of silver nanocolloids from environmental pollution shown to cause glycosylation disorders and embryonic deformities in medaka fish | Shimizu et al. (2021) |
| Oryzias latipes (Japanese medaka, fish) | Hydrazine, QIT‐TOF (DHB), (2‐AP) | Exposure of TiO2 nanoparticles from environmental pollution shown to cause glycosylation disorders and embryonic deformities in medaka fish | Horiuchi et al. (2021) |
| Pelagia noctiluca (jellyfish), mucus | PNGase F, R‐TOF/TOF (DHB), glycoblotting | High‐mannose. Use for accumulation of nanoparticles | Patwa et al. (2022) |
| Phaseolus lunatus beans | R‐TOF/TOF (DHB), (procainamide) | Investigation of the use of sodium hypochlorite to release Man9GlcNAc2 | Diaz et al. (2022) |
| Porcine bladder urothelial cells | PNGase F, FT‐ICR (DHB), (per‐Me) | Structural identification (high‐mannose, hybrid, di‐, tri‐, tetra‐antennary complex) | Wang, Bergström, et al. (2022) |
| Porcine endometrium | PNGase F, TOF/TOF (DHB) | Glycomics reveal that ST6GAL1‐mediated sialylation regulates uterine lumen closure during implantation | Han, Wang, et al. (2022) |
| Thorsmoerkia curvula gen. et spec. nov. (semi‑terrestrial microalga from Iceland | Pepsin, PNGase A, TOF (DHB), (per‐Me) | High‐mannose (Man2‐9) plus series with one deoxy‐hexose of one pentose. | Nicoletti et al. (2021) |
| Umbilical mesenchymal stem cells | Trypsin (release method not stated), QIT‐TOF (DHB), (per‐Me), database analysis | Proteomics and posttranslational modifications analysis during aging (Dubious high mannose structures) | Wang, Zhao, Chen, et al. (2022) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), (derivative), other methods.
Table 22.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of O‐glycans from specific glycopeptides.
| Glycoprotein and source | Methodsa | Notes | References |
|---|---|---|---|
| Vλ6 light chain mutant Wil, recombinant in Pichia pastoris | Trypsin, TOF/TOF (SA), glycopeptides | Effect of O‐glycosylation on amyloid fibril formation of the variable domain | Abe et al. (2021) |
| Apolipoprotein CIII | FT‐ICR (CHCA, SA) | Structural characterization | Demus et al. (2021) |
| Asprosin (from serum) | TOF/TOF (CHCA), (per‐Me) | Asprosin detection in clinical samples reveals serum/saliva correlation and indicates cartilage as source for serum asprosin | Morcos et al. (2022) |
| SARS‐CoV‑2 Receptor‐binding domain (RDB) | TOF (s‐DHB) | Structural and functional characterization of SARS‐CoV‑2 RBD domains produced in mammalian cells | Gstöttner, Zhang, et al. (2021) |
| Major peptide from the male ejaculatory duct of Rosophila melanogaster | R‐TOF/TOF (DHB) | Unspecified structure with one hexose, HexNAc and phosphoethanolamine | Sturm et al. (2021) |
| MUC2 from mouse small intestine and colon | β‐Elimination, TOF/TOF, (per‐Me) | The role of the mucin‐glycan foraging Ruminococcus gnavus in communication between the gut and the brain. Changes in sialylation detected | Coletto et al. (2022) |
| Mucins from murine submandibular glands | β‐Elimination, R‐TOF/TOF, QIT‐TOF (DHB), (per‐Me) | Study of the effect of aging on mucins | Kameyama, Tin, et al. (2021) |
| NOTCH1 EGF repeat fragments | L‐TOF/TOF (DHB), (glycoprotein) | Study of O‐GlcNAcylation of NOTCH1 | Tsukamoto et al. (2022) |
| Porcine gastric mucin | β‐Elimination, TOF (per‐Me) | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging | Wu, Crost, et al. (2021) |
| SARSCoV‑2 Spike glycoprotein (HEK293 and baculovirus‐insect cells) | β‐Elimination, TOF/TOF (DHB), (per‐Me) | Site‐specific analysis | Wang, Wu, et al. (2021) |
| SARS‐CoV‑2 nucleocapsid protein in HEK293 cells | β‐Elimination, TOF/TOF (DHB), (per‐Me) | Structural characterization | Supekar et al. (2021) |
| Thrombospondin‐1 | TOF, glycoprotein | O‐Fucosylation shown to stabilize the TSR3 motif in thrombospondin‐1 by interacting with nearby amino acids and protecting a disulfide bond | Berardinelli et al. (2022) |
| Various glycoproteins | β‐Elimination, TOF/TOF, (per‐Me) | To identify O‐glycans in new method for on‐line LC‐MS/MS identification of O‐glycosites by EThcD | Yang, Wang, et al. (2021) |
| Visgun (protein) from Drosophila melanogaster S2R+ cells | β‐Elimination, TOF/TOF (DHB) | Identification of Visgun as a Tc toxin receptor | Xu, Viswanatha, et al. (2022) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), compounds run (derivative).
Table 33.
Use of matrix‐assisted laser desorption/ionization mass spectrometry for the study of natural products.
| Source | Compound | Methods a | Notes | References |
|---|---|---|---|---|
| Carbohydrates | ||||
| Beta vulgaris L. (red beet) | Mono‐ and poly‐saccharides | R‐TOF/TOF (DHB) | Structural characterization | Hotchkiss et al. (2022) |
| Fucus vesiculosus | Fucoidan (240) | TOF/TOF (DHB (native), CHCA (per‐Me)) | In study of competitive inhibition of gastrointestinal norovirus binding | Hanisch, Aydogan, and Schroten (2021) |
| Sargassum horneri | Sulfated polysaccharides | TOF/TOF (DHB), MS/MS | Structural characteristics and immune‐enhancing activity | Kim, Hwang, et al. (2022) |
| Glycosides | ||||
| Agave marmorata Roezl | Flavonoids, phenolics, steroidal glycosides | R‐TOF/TOF (DHB) | Study of the micropropagation of seed‐derived clonal lines of this endangered plant and their compatibility with endophytes | Martinez‐Rodriguez et al. (2022) |
| Albizia julibrissin (Chinese medicinal herb) | Oleanane‐type glycoside (241) | R‐TOF/TOF | Structural determination and cytotoxic activity | Han et al. (2021) |
| Calicotome spinosa (Gorse) | Isoflavonoid glucosides (242) | MALDI | Extraction and identification of bioactive compounds | Mustafa et al. (2022) |
| Deschampsia antarctica | Various glycosides, e.g. orientin 2"‐O‐β‐arabino‐pyranoside (243) and isoswertiajaponin 2"‐O‐β‐arabinopyranoside | TOF/TOF (CHCA) | Chromatographic and mass spectrometric analysis of secondary metabolites of D. antarctica from Galindez Island, Argentine Islands | Ivannikov et al. (2022) |
| Holothuria poli (sea cucumber) | Triterpene glycosides (e.g. holothurin A, 244) | TOF (‐ve) | Characterization and investigation of anti‐proliferation in tumor cell lines | Mert‐Ozupek et al. (2022) |
| Holothuria scabra (viscera) | Hemolytic saponins (structures similar to holothurin A) | Q‐TOF (DHB/DMA) | Structural characterization, desulfation with microwaves to determine toxic fraction | Savarino et al. (2022) |
| Khaya ivorensis (African mahogany) | Quercetin‐7‐O‐hydroxybenzoic acid‐3‐O‐hexoside | TOF | Identification of phenolic compounds | Athomo et al. (2021) |
| Pinus pumila | Triterpene glycoside (245) | TOF | Structural characterization | Liu, Liu, Tao, et al. (2021) |
| Prosopis species | Glycosides and other compounds | TOF, TOF/TOF | Mainly review of various compounds | Picariello et al. (2022) |
| Quillaja saponaria (commercial) | Triterpenoid glycosides | TOF/TOF (DHB) | For development of low‐cost cage‐like particles to formulate veterinary vaccines | Lupi et al. (2022) |
| Scutellaria brevibracteata subsp. subvelutina | Iridoid glycosides (8, e.g., epiloganic acid (246), phenylethanoid glycoside (martynoside, 247) | TOF | Identification of secondary metabolites and their in vitro anti‐inflammatory activities | Erdoğan et al. (2021) |
| Glycolipids | ||||
| Natrinema halophilum sp. nov., Natrinema salinisoli sp. nov., Natrinema amylolyticum sp. nov. | Sulfated mannosyl glucosyl diethers | TOF/TOF (9‐AA) | Detection as part of description of species | Bao et al. (2022) |
| Haloterrigena alkaliphila sp.nov. | Sulfated mannosyl glucosyl diethers | TOF/TOF (9‐AA) | Detection as part of description of species | Bao et al. (2022) |
| Perilla frutescens (L.) | Glycoglycero‐lipids | TOF/TOF (DHB) | Structural characterization and anti‐inflammatory activities | Zi et al. (2021) |
| Quillaja lancifolia (Q. brasiliensis) | Quillaja saponins | TOF (DHB) | Investigation of nanoparticles formed from saponins | Cibulski et al. (2022) |
| Rohdea chinensis | Steroidal sapogenins (2) | Spiral‐TOF | Identification of two new steroidal sapogenins from rhizomes and their antifungal activity | Yao et al. (2022) |
| Streptomyces sp. | Tunicamycin | MALDI | Effect of acyl chain on activity | Price et al. (2021) |
| Glycopeptides, etc. | ||||
| Micromonospora chersina strain DSM 44154 | Lipoglyco‐depsipeptide Chersinamycin (antibiotic) | TOF/TOF | Discovery of six ramoplanin family gene clusters and the antibiotic | Morgan et al. (2021) |
| Bee pollen from rape (Brassica napus L.) | Reversibly glycosylated polypeptide‐2 | TOF, LC‐MS | Purification and characterization | Zhang, Sun, et al. (2021) |
| Major component of the venom of the ant Myrmecia gulosa. | O‐Linked glycopeptide (Mg7a) | L‐TOF/TOF (CHCA) | Identification and synthesis | Robinson et al. (2021) |
| Other compounds | ||||
| Actinokineospora spheciospongiae | Polyene macrolides (natamycin, luconsomycin, kineosporicin (248) | TOF | Sterol sponge mechanism of fungicidal action is shown to be conserved. | Guo, Zhang, et al. (2021) |
| Dolichos lablab L. hull | Pectin‐glucuronoxylan complex | L‐TOF/TOF (DHB) | Structural characterization | Liu, Tang, et al. (2022) |
| Haloprofundus salilacus sp. nov., H. halobius sp. nov. and H. salinisoli sp. nov.: | Various including sulfated mannosyl glucosyl diether, mannosyl glucosyl diether‐phosphatidic acid and sulfated mannosyl glucosyl diether‐phosphatidic acid | TOF/TOF (9‐AA, ‐ve) | Structural characterization | Li, Xin, et al. (2022) |
| Halosolutus amylolyticus gen. nov., sp. nov., H. halophilus sp. nov. and H. gelatinilyticus sp. nov. | Polar lipids and glycolipids | TOF | Description of species | Sun, Wang, et al. (2022) |
| Rice (Oryza sativa) straw | Soluble polysaccharides conjugated with p‐coumaric acid, ferulic acid, vanillic acid, and vanillin | R‐TOF/TOF | Structural characterization. Shown to alleviate ethanol fermentation stresses in Saccharomyces cerevisiae | Wang, Zheng, et al. (2022) |
| Rosa roxburghii | Ellagitannins | QIT‐TOF | Structural identification and suppression of poly(I:C)‑induced IL‑8 production in human keratinocytes | Takayama et al. (2021) |
| Tabernaemontana contorta Stapf. | Glyco‐cerebrosides (249) | TOF | Identification of new glycocerebrosides from the trunk and their antibacterial activity | Ebede et al. (2022) |
| Wood (oak, hornbeam, walnut) | Glycosylated conyferyl alcohols (250) | TOF (DHB) | Destructive behaviour of wood by the white‐rot fungus Fomes fomentarius | Bari et al. (2021) |
Format (not all items present): MALDI method (matrix), other methods.
Table 36.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of carbohydrate‐containing compounds in clinical studies.a
| Disease | Medium | Methodsb | Notes | References |
|---|---|---|---|---|
| Cancer | ||||
| Bladder cancer | Cancer cell line (150 μg) | PNGase F, TOF/TOF (3,4‐Di‐NH 2 ‐benzophenone) N‐, O‐glycans (per‐Me) | Identification of major differences in glycosylation and fucosyltransferase expression in low‐grade versus high‐grade bladder cancer cell lines | Ezeabikwa et al. (2021) |
| Bladder cancer | Cell culture | TOF/TOF (DHB), Free O‐glycans (per‐Me) | Characterization of the CD44 splicing code associated with bladder cancer invasion | Gaiteiro et al. (2022) |
| Bladder cancer | Urine (4 h collection) | PNGase F, TOF, N‐glycans | Use of lamprey immunity protein for early detection and recurrence monitoring by recognizing Neu5Gc‐modified uromodulin glycoprotein in urine | Teng et al. (2022) |
| Bladder cancer | FFPE tissue | PNGase F, TOF/TOF, N‐glycans (per‐Me) | High‐mannose H6‐9N2 and complex H6N5F1 increased in cancer. H5N3 (hybrid) and H4N3, H4N4 and H6N5F1S2 (complex) decreased (In Chinese) | Cheng, Sun, et al. (2022) |
| Brain tumors (secondary) | Metastatic tissue (4.31 mg of lipids) | TOF (DHB, ‐ve), GSLs | Preliminary analysis of the glycolipid profiles | Serb et al. (2022) |
| Breast cancer | Serum (6 μL) | PNGase F, FT‐ICR (sDHB), N‐glycans (Et ester) | Serum N‐glycan profiles shown to differ for various breast cancer subtypes | Vreeker et al. (2021) |
| Breast cancer | MCF7breast cancer cells | PNGase F, R‐TOF/TOF (DHB), N‐glycans | Characterization of paclitaxel resistance in breast cancer cells | Cao, Zhou, et al. (2021) |
| Breast cancer cells | Cancer cell line | β‐Elimination, R‐TOF/TOF (DHB), O‐glycans (reduced, per‐Me) | O‐Linked mucin‐type glycosylation shown to regulate the transcriptional programme downstream of the epidermal growth factor receptor (EGFR) | Tajadura‐Ortega et al. (2021) |
| Breast cancer cells | Breast cancer‐derived MCF‐7 cells (1 mg cell powder) | PNGase F, TOF/TOF (DHB), N‐ and O‐glycans (per‐Me) | Evaluation of the anticancer effect of violacein, phycocyanin and phycocyanobilin on apoptotic genes expression and glycan profiles | Hussein et al. (2021) |
| Breast cancer | Saliva (1 mL) | TOF/TOF, N‐glycans | Alternations of N‐glycans recognized by Phaseolus vulgaris leucoagglutinin in the saliva of patients with breast cancer | Yang, Ma, et al. (2021) |
| Breast cancer | Tissue (5 μM thick) | PNGase F, FT‐ICR (CHCA), imaging, N‐glycans | Clinical importance of high‐mannose, fucosylated, and complex N‐glycans in breast cancer metastasis | Ščupáková et al. (2021) |
| Breast cancer | Serum (1 μL) glycoproteins | PNGase F, (TM sprayer), (CHCA), N‐glycans | Differentiation between benign lesions or breast cancer in mammograms | Blaschke, Hill, et al. (2022) |
| Colon cancer | Serum (40 μL) and tissue (1 x 1 cm) | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me), LC‐MS/MS | Identification of differential N‐glycan compositions in the serum and tissue of colon cancer patients | Coura et al. (2021) |
| Colon carcinoma cell line (murine) | Murine cell line | PNGase F, L‐TOF/TOF (DHB), N‐glycans (2‐AB) | The solute carrier MFSD1 shown to decrease the activation status of β1 integrin and thus tumor metastasis | Roblek et al. (2022) |
| Colorectal cancer | Serum (10 μL) IgG | Trypsin, TOF/TOF (DHB), N‐glycopeptides | Revealing the changes of IgG subclass‐specific N‐glycosylation in colorectal cancer progression | Liu, Yu, et al. (2021) |
| Colorectal cancer | Serum (5 μL) | PNGase F, QIT‐TOF (s‐DHB) N‐glycans (reduced, Et ester) | Screening and diagnosis of colorectal cancer and advanced adenoma by Bionic Glycome method and machine learning | Pan, Zhang, Zhang, et al. (2021) |
| Colorectal cancer | Cell line | β‐Elimination, FT‐ICR (s‐DHB), O‐glycans (per‐Me) | As reference for development of automated method for O‐glycan profiling | Kotsias et al. (2021) |
| Colorectal cancer | FFPE tissue | Imaging, PNGase F, R‐TOF/TOF (CHCA, spray), (DMA derivatives) | Identification of high‐mannose N‐glycans as malignant progression markers in early‐stage colorectal cancer | Boyaval et al. (2022) |
| Colorectal cancer | Serum (10 μL) | PNGase F, TOF/TOF, N‐glycans (Me ester, Bz‐oxime) | Biomarker identification. (ratio of A1 to A2F biantennary glycans shown to be significant in detecting advanced cancer) | Takei et al. (2022) |
| Colorectal cancer (stage II) | FFPE tissue (6 μm sections) | PNGase F, TOF/TOF (CHCA, sprayer), N‐glycans (amide derivatization) | Cancer cells found to have higher levels of sialylation and high‐mannose glycans, less fucosylation and branching | Boyaval et al. (2021) |
| Endometrial cancer | FFPE tissue blocks | PNGase F (spray), R‐TOF/TOF (CHCA), N‐glycans | Detection of altered N‐linked glycosylation in endometrial cancer | Mittal et al. (2021) |
| Esophageal squamous cell carcinoma | Salivary (approx. 1 mL) glycoproteins | PNGase F, TOF/TOF (DHB), N‐glycans | Altered profiles in cancer patients. More complex, less fucosylation and high mannose | Shu et al. (2021) |
| Gastric cancer | Urinary (100 mL) exosomes | PNGase F, TOF, N‐glycans | Use of magnetic porous carbon‐dependent platform for the determination of N‐glycans from urine exosomes | Wu, Zhang, et al. (2021) |
| Glioblastoma | Tissue sections (5 or 10 μm sections) | Imaging, (9‐AA, TM sprayer), FT‐ICR, (‐ve ion), glycolipids | Discrimination between glioblastoma tumor cell subpopulations and different microvascular formations based on their lipid profiles | O'Neill, Liapis, et al. (2022) |
| Hepatocellular carcinoma | Tissue (imaging) | PNGase F, FT‐ICR (CHCA, TM sprayer), N‐glycans | N‐Glycosylation patterns shown to correlate with hepatocellular carcinoma genetic subtypes | DelaCourt et al. (2021) |
| Intrahepatic cholangio‐carcinoma | Human and rat tissue (approx. 25 mg) | TOF/TOF (DHB), GSLs (per‐Me) | Globo H shown to be a promising theranostic marker | Hung et al. (2022) |
| Invasive ductal carcinoma | FFPE tissue (10 μm sections) | PNGase F, TOF (s‐DHB), N‐glycans (Et ester, 2‐AA), LC‐MS/MS | Five N‐glycans (H5N2, H3N3F1, H6N2, H7N2, and H5N5F1) found to be significantly associated with invasive ductal carcinoma | Yaman, Kayili, et al. (2021) |
| Liver cancer | Hepatocellular carcinoma cells | TOF/TOF (DHB), glycosphingolipid | Ganglioside synthesis was increased in liver cancer | Su, Qin, et al. (2021) |
| Lung cancer | Serum haptoglobin | TOF, N‐glycans | Investigation of variation of fucose on biomarkers | Boonyapranai et al. (2021) |
| Lung cancer (A549) | Multicellular spheroids (14 μm sections) | R‐TOF (DHB, CHCA, SA, THAP, CA (255), DMCA (256), AQ, HCQ (258)), lipids and Glc‐Cer | Alterations of lipid metabolites in multicellular tumor spheroids in response to hydroxychloroquine revealed by imaging | Chen, Wang, et al. (2021) |
| Lung cancer | Cell culture | PNGase F, R‐TOF (DHB), N‐glycans (2‐AP) | Identification of distinct N‐glycosylation patterns on extracellular vesicles from small‐cell and non–small‐cell lung cancer cells | Kondo, Harada, et al. (2022) |
| Lung cancer | Tissue | MALDI imaging, FFPE sections | Use of high‐dimensionality reduction and clustering analysis and imaging to study metabolic heterogeneity | Conroy et al. (2022) |
| Melanoma | Cell line | TLC‐R‐TOF (9‐AA, ‐ve) Gangliosides and other lipids | Identification potential biomarkers in exosomes from melanoma cells with different metastatic potential | Lobasso et al. (2021) |
| Mucoepidermoid carcinoma | Salivary gland tissue | β ‐Elimination, TOF/TOF, QIT‐TOF (DHB), O‐glycans (per‐Me) | Characterization of tumor‐associated MUC1 | Isaka et al. (2021) |
| Neuroblastoma | Serum (5 μL) glycoproteins | PNGase F, QIT‐TOF (s‐DHB), N‐glycans (Et esters) | Identification of possible biomarkers for neuroblastoma | Qin et al. (2021) |
| Oral cancer | Tissue (100 μg protein) | PNGase F, TOF/TOF (DHB), (Fmoc, Me‐amide) | Minor differences found in the relative abundances of eight glycans in cancer patients | Wu, Liu, et al. (2022) |
| Ovarian cancer | Epithelial tissue, serum (5 μL) | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | N‐Glycome changes reflecting resistance to platinum‐based chemotherapy | Zahradnikova et al. (2021) |
| Ovarian cancer (epithelial) | Epithelial tissue (5 μm sections) | PNGase F, imaging, TOF (CHCA), (DMA, spray), (sialic acid derivatization), N‐glycans | Identification of biomarkers | Grzeski et al. (2022) |
| Pancreatic cancer | Tumor lysates (approx. 1 mg protein) | Hydrazine (gas‐phase), L‐TOF (DHB), N‐, O‐glycans (2‐AP) | Quantitative structural analysis of glycans expressed within tumors derived from pancreatic cancer patient‐derived xenograft mouse models | Hasehira et al. (2021) |
| Pancreatic cancer | Tumor tissue | PNGase F or endo F3, FT‐ICR, Q‐TOF (CHCA, TM sprayer), amidation of sialic acids, N‐glycans | Imaging of N‐glycans, high‐mannose, bi‐, tri‐, tetra‐antennary complex. Increased sialylation in cancer tissue | McDowell et al. (2021) |
| Pancreatic cancer | Cell line | β‐Elimination, FT‐ICR (s‐DHB), O‐glycans (per‐Me) | As reference for development of automated method for O‐glycan profiling | Kotsias et al. (2021) |
| Pancreatic cancer | Serum (25 μL) | Orbitrap (9‐AA) | Lipidomic profiling of human serum enables detection of pancreatic cancer | Wolrab et al. (2022) |
| Pancreatic cancer | Serum (6 μL) | PNGase F, FT‐ICR (s‐DHB), N‐glycans (Et ester) | Longitudinal changes of serum protein N‐glycan levels may support earlier detection of pancreatic cancer in high‐risk individuals | Levink et al. (2022) |
| Pancreatic ductal adenocarcinoma | FFPE samples (3 μm sections) | FT‐ICR (9‐AA, spray), free glycans | Native glycan fragments shown to be independent prognostic factors of cancer | Sun, Trajkovic‐Arsic, et al. (2021) |
| Papillary thyroid cancer | Plasma | PNGase F TOF/TOF (DHB), N‐glycans, (Et ester) | To distinguish benign and malignant thyroid nodules and to identify lymph node metastasis | Zhang, Reiding, et al. (2021) |
| Papillary thyroid cancer | Serum (5 μL) | PNGase F, TOF (s‐DHB), N‐glycans (Et ester) | Serum linkage‐specific sialylation changes shown to be potential biomarkers for monitoring and predicting the recurrence of papillary thyroid cancer following thyroidectomy | Cao, Zhang, et al. (2022) |
| Papillary thyroid microcarcinoma | Serum (10 μL) | PNGase F, TOF/TOF (DHB), (Et‐ester/lactone), N‐glycans | Use of nomograms for diagnosis of papillary thyroid microcarcinoma and prediction of lymph node metastasis | Zhang, Cao, et al. (2022) |
| Prostate cancer | Tissue (4 μm sections) | PNGase F (imaging, TM sprayer) (CHCA), N‐glycans | Investigation of N‐glycans as potential biomarkers of prostate cancer. (Higher high‐mannose, tri‐ and tetra‐antennary complex) | Conroy et al. (2021) |
| Pseudomyxoma peritoneil (mucinous adenocarcinoma) | FFPE tissue sections | PNGase F, R‐TOF/TOF, N‐glycans (Et esters) | Detection of altered linkage pattern of N‐glycan sialic acids | Nummela et al. (2021) |
| Renal cell carcinoma | Plasma (25 μL), urine (2 mL) and tissue (25 mg) | Orbitrap (9‐AA, ‐ve), sulfatides | Identification of altered profiles of sulfatides and sphingomyelins in patients with renal cell carcinoma | Jirásko et al. (2022) |
| Thyroid cancer | Plasma (70 μL blood) IgG | PNGase F, TOF (s‐DHB), N‐glycans | Diagnostic potential of plasma IgG N‐glycans in discriminating thyroid cancer from benign thyroid nodules | Zhang, Wu, et al. (2021) |
| Various (15 types) | Tissues | PNGase F, TOF/TOF (CHCA, TM sprayer), high‐mannose N‐glycans | Re‐evaluation of previous data and re‐examination of tissues to evaluate contribution of high‐mannose N‐glycans to cancer | Chatterjee et al. (2021) |
| Various (4 types in mice) | Cancer cells (2–3 x 106 cells) | PNGase F, R‐TOF/TOF (DHB), (Me esters, aoWR derivatives, “Glycoblotting” method) | Investigation of the role of the glycocalyx of tumor cell‐derived exosomes in organotropic cancer metastasis | Koide et al. (2022) |
| Congenital disorders of glycosylation (CDG) | ||||
| ALG2‐CDG | Serum (10 μL) glycopeptides | PNGase F, R‐TOF/TOF (CMBT), N‐glycans (per‐Me) | Characterization of ALG2‐CDG in Argentinean patients with a new genetic variant in homozygosis | Papazoglu et al. (2021) |
| ALG12‐CDG | Serum (10 μL) transferrin | Trypsin, MALDI | Absence of N‐glycans on Asn611 | Hiraide et al. (2021) |
| ALG12‐CDG | Serum (10 μL) glycoproteins | PNGase F, R‐TOF/TOF (DHB), N‐glycans | A novel homozygous mutation in the human ALG12 gene found to produce an aberrant profile of high‐mannose N‐glycans in patient's serum (Man5‐7GlcNAc2 up, Man8‐9GlcNAc2 down) | Ziburová et al. (2021) |
| CAMLG‐CDG | Serum (125‐150 μL) transferrin | R‐TOF/TOF (DHB) | Identification of a novel CDG linked to defective membrane trafficking | Wilson, Durin, et al. (2022) |
| CDG with Golgi homeostasis disruption | Serum (5 μL) apoC‐III | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | ApoC‐III glycosylation used to diagnose disease when transferrin glycosylation appeared normal | Raynor, Vincent‐Delorme, et al. (2021) |
| COG6‐CD | Serum (10 μL) | TOF, N‐, O‐glycans (per‐Me) | Case study. badly disrupted glycosylation, under processed glycans | Cirnigliaro et al. (2022) |
| DPM2 deficient CDG | Serum transferrin | TOF, N‐glycans | Expanding the clinical and metabolic phenotype | Radenkovic et al. (2021) |
| GM2 Gangliosidoses | Glycoproteins | R‐TOF/TOF (DHB), N‐glycans | Increased phosphorylation of HexM shown to improve lysosomal uptake and potential for managing GM2 gangliosidoses | Benzie et al. (2022) |
| HNF1a Variant and liver adenomatosis | Serum (10 μL) and serum glycoproteins (100 μL) | PNGase F, TOF/TOF (CMBT), N‐glycans (per‐Me) | Detection of highly sialylated complex glycans (two Neu5Ac per antenna) | Sturiale et al. (2021) |
| Leukocyte adhesion deficiency II | HEK293T and HepG2 cells | TOF, (DHB), N‐ (2‐AB) and O‐glycans (per‐Me), desialylated | Identification and investigation of salvage pathway. Cαaused by mutations in the SLC35C1 gene encoding Golgi GDP‐fucose transporter | Skurska et al. (2022) |
| MAN1B1‐CDG | Serum (5 μL) | PNGase F, Endo H, R‐TOF/TOF (DHB) N‐glycans (per‐Me) | Identification of disease in three individuals | Sakhi et al. (2021) |
| MAN1B1‐CDG | Serum transferrin | Trypsin, TOF (DHB), N‐glycopeptides | No change in glycosylation observed after disulfiram treatment | Kemme et al. (2021) |
| MAN1B1‐CDG | Serum transferrin | TOF (DHB), N‐glycans, HPLC | Siblings with MAN1B1‐CDG showing novel biochemical profiles | Okamoto et al. (2021) |
| MOGS‐CDG | IgG | MALDI, N‐glycans | Epilepsy and movement disorders in the oldest‐known MOGS‐CDG patient | Lo Barco et al. (2021) |
| MOGS‐CDG | Urine | R‐TOF/TOF (DHB), (per‐Me), HPLC (2‐AB), oligosaccharides | Clinical, biochemical and genetic characteristics of the disease | Shimada et al. (2022) |
| MPI‐CDG | Serum (20 μL) and serum transferrin | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | Variation of the serum N‐glycosylation during the pregnancy of a MPI‐CDG patient. Glycosylation improved | Lebredonchel et al. (2021) |
| PMM2‐CDG (Zebrafish embryo model) | Tissue imaging | PNGase F, Q‐TOF, N‐glycans | Protease‐dependent defects in N‐cadherin processing shown to drive PMM2‐CDG pathogenesis | Klaver et al. (2021) |
| Slc35a1 Deficiency | Platelets | PNGase F, β‐elimination, TOF, N‐, O‐glycans (per‐Me) | Slc35a1 Deficiency shown to cause thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver | Ma, Li, Kondo, et al. (2021) |
| SLC35A2‐CDG | Serum transferrin | TOF | Identification of novel variant | Quelhas et al. (2021) |
| SLC37A4‐CDG | Serum glycoproteins | PNGase F, TOF, N‐glycans (per‐Me) | Mutation in SLC37A4 shown to cause a dominantly inherited CDG characterized by liver dysfunction | Ng et al. (2021) |
| SLC37A4‐CDG | Serum glycoproteins | Endo‐H, PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | High‐mannose and hybrid glycans. Abnormally high Man5GlcNAc | Raynor, Haouari, et al. (2021) |
| Mild variant of leukocyte adhesion deficiency type II (SLC35C1‐CDG) | Serum N‐glycoproteins | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | Study of the response of two children to oral fucose therapy | Tahata et al. (2022) |
| Various | Serum transferrin and apoCIII | TOF (DHB), glycoproteins | Profiling of apoCIII to monitor changes in O‐glycosylation | Wada and Okamoto (2021) |
| SLC37A4‐CDG | Serum transferrin | PNGase F, TOF (THAP, ‐ve), N‐glycans | Second patient with novel variant | Wilson et al. (2021) |
| SLC39A8‑CDG | Serum (10 μL) transferrin | PNGase F, R‐TOF/TOF, N‐glycans (per‐Me) | Glycan profile showed small decrease in galactosylation | Bonaventura et al. (2021) |
| Other | ||||
| Aging heart | Tissue (20 μg protein) | PNGase F, TOF/TOF, N‐glycans (per‐Me) | Proposal that changes in the heart glycoproteome likely contribute to the age‐related functional decline of the cardiovascular system. | Franzka et al. (2021) |
| Alzheimer's disease | Plasma (3 μL) and CSF (100 μL) | L‐TOF/TOF, glycoproteins | Distinct patterns of apolipoprotein C‐I, C‐II, and C‐III isoforms shown to be associated with markers of Alzheimer's disease | Hu, Meuret, et al. (2021) |
| Alzheimer's disease | Mouse brain slices | R‐TOF/TOF (NEDC, ‐ve), sulfatide | Adult‐onset CNS myelin sulfatide deficiency shown to cause Alzheimer's disease‐like neuroinflammation and cognitive impairment | Qiu et al. (2021) |
| Alzheimer's disease | Serum (5 μL) and brain (20‐100 mg) | TOF/TOF, N‐glycans (per‐Me) | N‐Glycome profiling in Alzheimer's disease and Alzheimer's disease‐related dementias | Yu, Huo, et al. (2021) |
| Anti‐neutrophil cytoplasmic antibody‐associated vasculitis | Serum (50 μL) | R‐TOF/TOF (9‐AA), sulfatide | Serum sulfatide levels identified as a biomarker | Harada et al. (2022) |
| Anti‐PLA2R1–associated membranous nephropathy | Serum IgG4 | PNGase F, TOF N‐glycans (sialic acid derivatization) | Altered glycosylation of IgG4 shown to promote lectin complement pathway activation in anti‐PLA2R1–associated membranous nephropathy | Haddad et al. (2021) |
| Behcet's disease | Serum (50 μL) glycoproteins | R‐TOF/TOF (DHB), N‐glycans | Isomer‐specific monitoring (PGC chromatography) of sialylated N‐glycans reveals association of α2,3‐linked sialic acid with Behcet's disease | Seo et al. (2021) |
| Carotid atherosclerosis in patients with rheumatoid arthritis | Serum sulfatides | TOF/TOF (‐ve) | Serum sulfatide level proposed as a predictor (biomarker) for the progression of accelerated atherosclerosis in rheumatoid arthritis cases. | Li, Yin, et al. (2022) |
| Chronic obstructive pulmonary disease (COPD) lung transplant patients | Plasma IgG and IgG1‐3 | PNGase F, FT‐ICR (CHCA), N‐glycans | Pro‐inflammatory IgG1 N‐glycan signature shown to correlate with primary graft dysfunction onset in COPD patients | McQuiston et al. (2022) |
| Congenital aortic valve stenosis | Aortic valve tissue | PNGase F, FT‐ICR (CHCA, TM sprayer), N‐glycans | Spatial N‐glycomics of the human aortic valve in development and pediatric endstage congenital aortic valve stenosis | Angel, Drake, et al. (2021) |
| Covid‐19 | Plasma (20 μL) IgG1, total IgG2, and anti‐spike IgG, | Trypsin, R‐TOF/TOF (CHCA, ‐ve), glycopeptides | Differences in glycosylation (fucosylation and galactosylation), particularly in the anti‐spike IgG | Schwedler et al. (2022) |
| Covid‐19 | Glycated HSA hyperglycosylated IgG3 in serum | TOF (SA), glycated HSA | Patients recovering from Covid‐19 found to have increased levels of glycated HSA and IgG3 | Iles et al. (2022) |
| Crohn's disease | Serum | PNGase F, TOF (“carbon”), N‐glycans | Investigation of serum N‐glycan patterns for rapid and precise detection of Crohn's disease | Wu, Chen, et al. (2021) |
| Cystic fibrosis | Broncho‐alveolar lavage fluid | β‐Elimination, R‐TOF, (per‐Me) | Evidence of early increased sialylation of airway mucins and defective mucociliary clearance in CFTR‐deficient piglets | Caballero et al. (2021) |
| Diabetic nephropathy | Rat kidney | Orbitrap, TOF/TOF (DAN, TM sprayer) | Identification of tissue‐specific metabolic reprogramming | Wang, Fu, et al. (2021) |
| Endoplasmic reticulum (ER) stress (several disease states) | HeLa cells | PNGase F, Glycoblotting, R‐TOF/TOF (DHB), N‐glycans (Me ester, aoWR) | For quantitative evaluation of ER stress. Increased high‐mannose and ratio of sialylated and non‐sialylated glycans | Fujitani et al. (2021) |
| Fatty liver disease | Mouse plasma | PNGase F, TOF (DHB, THAP), N‐glycans (per‐Me) | Defective lipid droplet–lysosome interaction shown to cause fatty liver disease as evidenced by human mutations in TMEM199 and CCDC115 | Larsen, van den Boogert, et al. (2022) |
| Hereditary angioedema | Plasma (5 μL) | PNGase F, TOF/TOF (s‐DHB), N‐glycans (Et ester) | Validation of diagnostic and predictive N‐glycan biomarkers | Zhang, Wang, et al. (2021) |
| HIV | Plasma | TIMS‐TOF (norharmane, ‐ve), Lipid A | Variation in blood microbial LPS shown to contribute to immune reconstitution in response to suppressive antiretroviral therapy | Luo, Health, et al. (2022) |
| Invasive candidiasis | Serum | TOF/TOF, LTQ‐Orbitrap (DHB/pyridine) | Identification of fungal trehalose for the diagnosis of invasive candidiasis by mass spectrometry | Mery et al. (2022) |
| Isolated hyper‐prolactinaemia | Serum IgG (35 μg) | Trypsin, R‐TOF (Cl‐CHCA) glycopeptides | Altered immunoglobulin demonstrated in patients | Hirschberg et al. (2021) |
| Liver disease | Blood | PNGase F, FT‐ICR (CHCA), N‐glycans | For development of a comprehensive biomarker data model | Lyman et al. (2022) |
| Liver fibrosis | IgG from serum | PNGase F, FT/ICR (CHCA), N‐glycans | Development of biomarker | Scott et al. (2022) |
| Lupus nephritis | IgG (7 μg) from urine | PNGase F, R‐TOF/TOF (DHB), N‐glycans (per‐Me) | Presence of lupus nephritis indicated by aberrantly glycosylated IgG which elicits pathogenic signalling in podocytes | Bhargava et al. (2021) |
| Lyme disease | Serum (1 μL) IgG | Sialidase A, PNGase F, FT‐ICR, N‐glycans | Results show that during the acute phase of infection, IgG shifts its glycosylation profile to include structures that are not associated with the classic pro‐inflammatory IgG N‐glycan signature. | Haslund‐Gourley et al. (2022) |
| Meat allergy | Glycolipids and glycoproteins from rabbit erythrocytes | TOF/TOF | α‐Gal residues present on both glycolipids and glycoproteins contribute to immune response in meat‐allergic patients | Chakrapani, Fischer, et al. (2022) |
| Migraine | Serum IgG (10 μg) | Trypsin, L‐TOF (DHB), N‐glycopeptides | Bisected glycans increases but no change in fucosylation and sialylation | Xu, Wang, et al. (2022) |
| Nucleus pulposus (intervertebral discs) | Cell membranes | TOF/TOF (DHB). Identification by GlycoWorkBench | Enhancement of nucleus pulposus repair by glycoengineered (addition of unnatural sialic acids) adipose‐derived mesenchymal cells | Ying et al. (2022) |
| Parkinson's disease | Serum (5 μL) glycoproteins | PNGase F, TOF/TOF (DHB/DMA), N‐glycans (Et ester) | Increased abundance of glycans containing core fucose, sialic acid, and bisecting GlcNAc detected | Xu, Jin, et al. (2022) |
| Pediatric ulcerative colitis | Colonic aspirate (100 μg protein) | PNGase F, TOF/TOF (DHB), N‐glycans (Me‐amide derivs.) | Elevated colonic microbiota‐associated paucimannosidic and truncated N‐glycans | Li, Zhang, et al. (2021) |
| Pemphigus vulgaris (autoimmune disease after corticosteroid treatmennt) | Serum IgG (0.5 mg) | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | No evidence found for a correlation between the IgG N‐glycans profile in the active phase and in the remission phase of pemphigus | Petit et al. (2021) |
| Pemphigus | IgG | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | Changes in N‐glycan profile from IgG in patients treated with Rituximab (less galactosylation) | Font et al. (2022) |
| Plasma cell disorders | Serum | TOF, N‐glycans | MASS‐FIX (MALDI method) for the detection of monoclonal proteins and light chain N‐glycosylation in routine clinical practice: a cross‐sectional study of 6315 patients | Mellors et al. (2021) |
| Plasma cell disorders | Serum | PNGase F, TOF (DHB), N‐glycans (Girard's T) | Characterizing M‐protein light chain glycosylation | Miller et al. (2022) |
| Schizophrenia | Mouse brain regions (1 mg brain protein) | PNGase F, β‐elimination, TOF (DHB), N‐ and O‐glycans (per‐Me) | The schizophrenia‐associated variant in SLC39A8 found to alter protein glycosylation in mouse brain | Mealer et al. (2022) |
| Seasonal allergic rhinitis | Serum (50 μL) | PNGase F, R‐TOF/TOF‐MS/MS (s‐DHB) (Et ester) | Structural changes. Several triantennary glycans decreased, tetra‐antennary increased | Yaman, Avci, et al. (2021) |
| Sickle cell anaemia and malaria | Red blood cell ghosts | TOF/TOF, N‐glycans (per‐Me) | Patches of high mannose N‐glycans), expressed on diseased or oxidized RBC surfaces, shown to bind the mannose receptor (CD206) on phagocytes to mediate clearance. | Cao, Antonopoulos, et al. (2021) |
| Splenic function | Erythrocytes | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | Measurement of erythrocyte membrane high‐mannose glycans to assess splenic function | Cao, Mathur, et al. (2022) |
| Status epilepticus | Serum (10 μL) glycoproteins | PNGase F, R‐TOF/TOF (DHB), N‐glycans | N‐glycan profiling in pilocarpine induced status epilepticus in immature rats | Kapoor et al. (2022) |
| Systemic lupus erythematosus | Kidney FFPE tissue, | PNGase F R‐TOF/TOF, N‐glycans (DMA/NH2 amidation) | Increased production of high mannose glycans as a diagnostic and prognostic biomarker | Alves et al. (2021) |
| Vernal and atopic kerato‐conjunctivitis | Tears (8 μL) | PNGase F, TOF/TOF (CHCA), N‐glycans (per‐Me) | Over 150 high‐mannose, bi‐, tri‐ and tetra‐antennary complex glycans. Variations in bisected and fucosylated glycans detected. | Messina, Palmigiano, Tosto, et al. (2021) |
Human unless otherwise stated
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), compounds run (derivative), other methods.

Table 39.
Use of matrix‐assisted laser desorption/ionization analysis to monitor N‐ and O‐glycosylation in biopharmaceuticals and related materials.
| Biopharmaceutical and Expression System | Methodsa | Notes | References |
|---|---|---|---|
| Arabidopsis alg3 | TOF (2‐AP) | For production of N‐glycans lacking 3‐fucose and xylose substituents | Sariyatun et al. (2021) |
| Cell walls from Saccharomyces cerevisiae Δalg3 Δalg11 | PNGase F, TOF/TOF (s‐DHB, THAP), MS/MS, N‐glycans (per‐Me) | Production of galactosylated complex‑type N‑glycans in glycoengineered S. cerevisiae | Piirainen et al. (2022) |
| Colorectal cancer antigen produced in tomato fruits | Trypsin, PNGase A, TOF (DHB), N‐glycans | Immunotherapeutic effects | Park et al. (2022) |
| Erythropoietin (EPO) | PNGase F, R‐TOF/TOF (DHB), N‐glycans, (per‐Me) | Evaluation of erythropoietin biosimilars EpotinTM, Hemax® and JimaixinTM, Comparison with original alfa drug Eprex® | Capdevilleet al. (2021) |
| Erythropoietin (EPO) in Spodoptera frugiperda cells with multiple Mgat1 deletions | PNGase F, R‐TOF/TOF, N‐glycans | Production of a new insect cell line engineered to produce recombinant glycoproteins with cleavable N‐glycans | Mabashi‐Asazuma and Jarvis (2021) |
| Etanercept from CHO cells | PNGase F, TOF/TOF (DHB), N‐glycans (reduced) and O‐glycopeptides (per‐Me) | Production of an O‐glycovariant with enhanced potency | Biel et al. (2022) |
| Ectonucleotide pyrophosphatase phospodiesterase‐1 | PNGase F, TOF, N‐glycans (per‐Me) | Improvements to the pharmacodynamics and in vivo activity through protein and glycosylation engineering | Stabach et al. (2021) |
| Human tissue plasminogen activator, Reteplase fused to IgG Fc in Nicotiana benthamiana | PNGase F, MALDI | Reteplase Fc‐fusions produced in N. benthamiana shown to be able to dissolve blood clots ex vivo | Izadi et al. (2021) |
| Human acid α‑glucosidase in rice cells | PNGase A, R‐TOF/TOF (DHB), (2‐AP) | Production of recombinant human acid α‑glucosidase with mannosidic N‑glycans | Jung (2022) |
| IgG (anti‐CD20 antibody, with sequence similar to Rituximab) various cell lines | PNGase F, R‐TOF (Et ester, p‐toluidine amidation) | Study of the interplay of protein engineering and glycoengineering to fine‐tune antibody glycosylation and its impact on effector functions | Wang, Wang, Zhang, et al. (2022) |
| IgG From CHO cells | PNGase F, R‐TOF/TOF (DHB), per‐Me | Modulation of N‐glycan galactosylation and fucosylation in CHO cells by feeding with galactose and fucose | Prabhu et al. (2022) |
| Oryza sativa (rice) | TOF/TOF (DHB), N‐glycans (per‐Me) | Inactivation of the β (1, 2)‑xylosyltransferase and the α (1,3)‑fucosyltransferase gene by multiplex CRISPR/Cas9 strategy | Jung, Shin, et al. (2021) |
| Rituximab | PNGase F, TOF/TOF (SA), N‐glycans (2‐AB) | Comparison of glycoprofiles of Rituximab versions licensed for sale in India | Kaur, Shukla, et al. (2021) |
| Tobacco BY‐2 cells | PNGase F, FT‐ICR (DHB), N‐glycans (2‐AB) | Inactivation of N‐acetylglucosaminyl‐transferase I and α1,3‐fucosyltransferase genes in N. tabacum BY‐2 cells shown to give glycoproteins with highly homogeneous, high‐mannose N‐glycans | Herman et al. (2021) |
| Trastuzumab and cetuximab | PNGase F, TOF/TOF (DHB) | Remodelling by glycan cleavage with Endo S and attaching mannose‐6‐phosphate glycan ligands for targeted protein degradation | Zhang, Liu, et al. (2022) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), compounds run (derivative), other methods.
Table 40.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry to study general biochemistry.
| Study | Methodsa | References |
|---|---|---|
| Functional glycomics and anxiety‐related behaviors in single versus group‐housed C57BL/6 and DBA/2 male mice. Shows increase in sialylated N‐glycans | PNGase F, TOF/TOF (CHCA/Di‐Et‐ammonium salt), N‐glycans (Me ester, BOA derivs.) | Abou‐Elnaga et al. (2021) |
| Fungi hijack a ubiquitous plant apoplastic endoglucanase to release a ROS scavenging β‐glucan decasaccharide to subvert immune responses | R‐TOF (DHB) | Chandrasekar et al. (2022) |
| Analysis of the proteome and PTMomes of C2C12 myoblasts reveals that sialylation plays a role in the differentiation of skeletal muscle cells | PNGase F, TOF/TOF (DHB), N‐glycans (per‐Me) | Chen, Sun, et al. (2021) |
| Characterization of the noncovalent interactions between lysozyme and panaxadiol glycosides by intensity‐fading – matrix‐assisted laser desorption ionization – mass spectrometry (IFMALDI‐MS) | R‐TOF (DHB, SA) | Du, Du, et al. (2021) |
| In vitro fermentation of chitooligosaccharides and their effects on human fecal microbial community structure and metabolites | TOF | Ji, Chang, et al. (2021) |
| HOIL‐1 ubiquitin ligase activity shown to target unbranched glucosaccharides and is required to prevent polyglucosan accumulation | TOF (DHA, NH4Cit) of ubiquitin‐Glc7 | Kelsall et al. (2022) |
| Site‐selective chemoenzymatic modification on the core fucose of an antibody enhances its Fcγ receptor affinity and ADCC activity | TOF/TOF (DHB/DMA) | Li, Chong, et al. (2021) |
| Human gut Faecalibacterium prausnitzii shown to deploy a highly efficient conserved system to cross‐feed on β‐mannan‐derived oligosaccharides | TOF/TOF (DHB) | Lindstad et al. (2021) |
| Investigation of in vitro histone H3 glycosylation using H3 tail peptides. GlcNAcylation of histone H3 tail peptide in the presence of O‐GlcNAc transferase shown not to occur in vitro | R‐TOF/TOF (CHCA) | Merx et al. (2022) |
| Mechanistic studies and in vivo efficacy of an oxadiazole‐containing antibiotic. MALDI of Glc2‐diacyl glycerol (reduction of lipoteichoic acid synthesis) | TOF | Naclerio et al. (2022) |
| New insights into the molecular mechanism behind mannitol and erythritol fructosylation by β‑fructofuranosidase from Schwanniomyces occidentalis | R‐TOF/TOF (DHB) | Rodrigo‑Frutos et al. (2021) |
| Study on the origin of life; Investigation of the effect of proton irradiation on N‐glycosidic bond formation | Q‐Exactive | Saladino et al. (2021) |
| Effect of inhibitory mycobacterial cell wall lipids on survival of mycobacteria and their effect on the promotion of disease. | R‐TOF/TOF (DHB) | Weng et al. (2022) |
| Activation of regulatory T cells triggers specific changes in glycosylation associated with Siglec‐1‐dependent inflammatory responses | PNGase F, TOF (DHB), N‐glycans (per‐Me) | Wu, Murugesan, et al. (2021) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), compounds run (derivative).
Table 43.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for the characterization of carbohydrates from foods and drink.
| Compound | Methods1 | Notes | References |
|---|---|---|---|
| Noncovalent and covalent complexes between proteins and mono‐ or di‐glucoside anthocyanins | TOF/TOF (SA) | Effect of complexes on β‐lactoglobulin‐digestibility | Khalifa et al. (2022) |
| Glc2‐13 from Schisandra chinensis syrup | R‐TOF/TOF (DHB) | Synthesis and biological characterization | Kwak et al. (2022) |
| Pectin oligosaccharide | TOF (graphene oxide) | Effect of pectin oligosaccharide on quality control of quick‐frozen pumpkin puree | Li, Wang, et al. (2022) |
| Polysaccharides from Glycine max (soybean) | TOF (DHB) | Chemical composition and sugar spectroscopy of polysaccharides obtained by microwave‐assisted salt extraction | Li, Zhang, Chen, et al. (2022) |
| Maltooligosaccharides from beer | TOF (DHB and PAPAN) | Use of reactive matrix to form PAPAN derivatives (see text) | Ling, Jiang, et al. (2021) |
| β‐Mannans | TOF/TOF (DHB) | Human gut Faecalibacterium prausnitzii deploys a highly efficient conserved system to cross‐feed on β‐mannan‐derived oligosaccharides | Lindstad et al. (2021) |
| N‐Glycans | EndoBI‐1, rapifleX™ MALDI Tissuetyper™ (DHB) | Use of deglycosylated whey and chickpea protein matrices for enrichment by black mulberry polyphenols | Ozleyen et al. (2022) |
| Shiitake mushrooms | Q‐TOF (DHB) | Changes in the morphometric, textural, and aromatic characteristics of shiitake mushrooms during combined humid‐convective drying. Yield of polysaccharides, predominantly β‐glucans higher than with hot air | Subramaniama et al. (2021) |
| Metabolites and thymocytes from mice | PNGase F, R‐TOF/TOF (per‐Me) | Dietary glucosamine shown to overcome the defects in αβ‐T cell ontogeny caused by the loss of de novo hexosamine biosynthesis | Werlen et al. (2022) |
| Oligogalacturonide | TOF/TOF (2,5‐Di‐OH‐cinnamic acid (257) above) | Fungal polygalacturonase‐generated oligogalacturonide shown to restrain softening in ripening tomatoes | Yang, Lu, et al. (2022) |
| Shiitake mushrooms (Lentinula eddoes) | R‐TOF (DHB) | Analysis of glucan from chitin nanofibers prepared from Shiitake stipes | Zhang, Zhao, et al. (2022) |
| Pinot noir wines | TOF/TOF (DHB, ‐ve, +ve) | Isolation, characterization, and compositional analysis of polysaccharides | Zhu, Alcazar‐Magana, et al. (2022) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), (derivative).
Table 50.
Other methods for glycan and glycoconjugate analysis.
| Method | Compound type | Main technique | References |
|---|---|---|---|
| Capillary (gel) electrophoresis‐based methods for immunoglobulin glycosylation analysis | IgG Glycosylation | CE and CE‐MS | Cajic et al. (2021) |
| Exploiting pglB oligosaccharyltransferase‐positive and –negative Campylobacter jejuni and a multiprotease digestion strategy to identify novel sites modified by N‑linked protein glycosylation | Glycoproteins | LC‐FAIMS‐MS/MS | Cain et al. (2021) |
| A multidimensional mass spectrometry‐based workflow for de novo structural elucidation of oligosaccharides from polysaccharides | Polysaccharides | LC‐MS/MS, UHPLC‐QqQ‐TOF | Castillo et al. (2021) |
| Methods to improve quantitative glycoprotein coverage from bottom‐up LC‐MS data | Glycoproteins | LC‐MS | Chang and Zaia (2022) |
| Characterization of galacto‐oligosaccharides using high‐performance anion exchange chromatography‐tandem mass spectrometry | Galacto‐oligosaccharides | HPAEC‐MS/MS | Chen and Liu (2021) |
| High‐throughput analyses of glycans, glycosites, and intact glycopeptides using C4‐and C18/MAX‐tips and liquid handling system | Glycans, glycosites, and intact glycopeptides | Protocol, unspecified mass spectrometry | Chen, Clark, et al. (2021) |
| Resolving structural detail and occupancy of glycans on intact glycoproteins | N‐ and O‐glycans on glycoproteins | HPLC, LC‐MS, exoglycosidase | Chen, Wu, et al. (2021) |
| Mirror‐cutting‐based digestion strategy enables the in‐depth and accuracy characterization of N‑linked protein glycosylation | N‐Glycopeptides | LC‐MS/MS | Chen, Fang, et al. (2021) |
| Targeted N ‑glycan analysis with parallel reaction monitoring using a quadrupole‐Orbitrap hybrid mass spectrometer | N‐Glycans (per‐Me) | LC/MS/MS | Cho, Reyes, et al. (2022) |
| Desalting paper spay mass spectrometry for rapid detection of glycans and glycoconjugates | Glycans and glycoconjugates | Paper spray | Chiu et al. (2021) |
| In‐depth profiling of O‑glycan isomers in human cells using C18 nanoliquid chromatography‐mass spectrometry and glycogenomics | O‑Glycan isomers | LC/MS (2‐AB derivatives) | de Haan, Narimatsu, et al. (2022) |
| Data‐independent acquisition‐based mass spectrometry for quantitative analysis of intact N‐linked glycopeptides | N‐Glycopeptides | LC‐MS/MS | Dong et al. (2021) |
| Immobilized exoglycosidase matrix mediated solid phase glycan sequencing | N‐Glycans | CE, exoglycosidase digestion | Farsang et al. (2022) |
| Mesoporous graphitized carbon column for efficient isomeric separation of permethylated glycans | N‐Glycans | LC‐MS (LTQ Orbitrap) | Gautam et al. (2021) |
| Glycine additive shown to enhance sensitivity for N‐ and O‐glycan analysis with HILIC‐ESI‐MS | N‐ and O‐glycans | HILIC‐ESI‐MS | Guo, Nayak, et al. (2021) |
| Fast and ultrasensitive glycoform analysis by supercritical fluid chromatography−tandem mass spectrometry | N‐Glycans (per‐Ac) | SFC‐MS (main) also HPLC, MALDI‐TOF/TOF | Haga, Yamada, et al. (2022) |
| In‐source microdroplet derivatization using coaxial contained‐electrospray mass spectrometry for enhanced sensitivity in saccharide analysis | Oligosaccharides | ESI | Heiss and Badu‐Tawiah (2021) |
| Liquid chromatography−tandem mass spectrometry with online, in‐source droplet‐based phenylboronic acid derivatization for sensitive analysis of saccharides | Oligosaccharides | LC/MS | Heiss and Badu‐Tawiah (2022) |
| Fc glycosylation characterization of human immunoglobulins G using immunocapture and LC‐MS | N‐glycans from serum or plasma IgG, Fc | LC‐MS, protocol | Helali et al. (2021) |
| N‐glycan profiling of glycoproteins by hydrophilic interaction liquid chromatography with fluorescence and mass spectrometric detection | N‐Glycans | HILIC‐HPLC, LC‐MS/MS, video of protocol | Kayili and Salih (2021) |
| High‐throughput N‐glycan screening method for therapeutic antibodies using a microchip‐based DNA analyzer | N‐Glycans from monoclonal antibodies | CE‐MS of ANTS‐labelled N‐glycans | Kinoshita, Nakajima, et al. (2021) |
| Development of a novel, label‐free N‐glycan method using charged aerosol detection | N‐Glycans | HPLC without fluorescent derivatization | Knihtila et al. (2022) |
| Separation of glycoproteins using novel stationary phases modified with poly(ethylene glycol)‐conjugated boronic‐acid derivatives | Glycoproteins | HPLC | Kobayashi et al. (2022) |
| Characterization of protein glycoforms at intact level by Orbitrap mass spectrometry | Glycoproteins | LC‐MS (Orbitrap), protocol | Kristensen et al. (2021) |
| Capillary electrophoresis‐based N‐glycosylation analysis in the biomedical and biopharmaceutical fields | N‐Glycans | CE | Kun et al. (2021) |
| Separation and identification of permethylated glycan isomers by reversed phase nano‐LC‐NSI‐MSn | Oligosaccharides, N‐glycans | LC‐ESI‐MSn | Kurz et al. (2021) |
| Cross‐identification of N‐glycans by CE‐LIF using two capillary coatings and three labeling dyes | N‐glycans | CE‐LIF | Li, Wang, Guo, et al. (2022) |
| High sensitivity capillary electrophoresis with fluorescent detection for glycan mapping | Glucose oligomers, N‐glycans | CE with fluorescence | Liénard–Mayor et al. (2021) |
| Lab‐in‐droplet: From glycan sample treatment toward diagnostic screening of congenital disorders of glycosylation | N‐Glycans | CE with fluorescence | Liénard–Mayor et al. (2022) |
| High sensitivity acidic N‐glycan profiling with MS‐enhancing derivatization and mixed mode chromatography | N‐Glycans | LC/MS, charged derivatives (RapiFlour) | Liu, Wang, Lauber, et al. (2022) |
| Distinguishing carbohydrate isomers with rapid hydrogen/deuterium exchange‐mass spectrometry | Trisaccharides | ESI‐Orbitrap | Liyanage, Quintero, et al. (2021) |
| High‐sensitivity glycan profiling of blood‐derived IgG, plasma, and extracellular vesicle isolates with CZE‐MS | N‐Glycans | CZE‐ESI‐MS | Marie et al. (2021) |
| HILIC‐UPLC‐MS for high throughput and isomeric N‐glycan separation and characterization in CDGs and human diseases | N‐Glycans | HILIC‐UPLC‐ESI‐MS | Messina, Palmigiano, Esposito, et al. (2021) |
| Liquid chromatography and capillary electrophoresis in glycomic and glycoproteomic analysis | N‐Glycans | LC and CE | Molnarova et al. (2022) |
| Polysaccharide identification through oligosaccharide fingerprinting | Polysaccharides | HPLC‐QTOF | Nandita et al. (2021) |
| An improved method for galactosyl oligosaccharide characterization | Galactosyl oligosaccharides | HPAEC‐ESI‐MS | Patil and Rohrer (2021) |
| Lectin and liquid chromatography‐based methods for immunoglobulin glycosylation analysis | N‐ and O‐glycans | LC‐MS, Lectin chromatography, lectin microarrays | Petrović and Trbojević‐Akmačić (2021) |
| Nanoflow LC−MS method allowing in‐depth characterization of natural heterogeneity of complex bacterial lipopolysaccharides | Complex bacterial lipo‐polysaccharides | Nano‐LC‐MS | Pupo et al. (2021) |
| 2‐Dimensional ultrahigh performance liquid chromatography and methyl ester formation paired with tandem mass spectrometry for comprehensive serum N‐glycome characterization | N‐Glycans | Weak anion exchange (WAX) and HILIC chromatography‐MS/MS | Smith, Millán‐Martín, et al. (2021) |
| N‑glycomics of various tissue samples that may contain glycans with unknown or unexpected structures | N‐Glycans (2‐AP derivatives) | LC‐MS, exoglycosidase digestions. | Suzuki et al. (2021) |
| N ‑Glycan isomer differentiation by zero flow capillary electrophoresis coupled to mass spectrometry | N‐Glycans | Zero‐flow CE | Wagt et al. (2022) |
| Streamlined subclass‐specific absolute quantification of serum IgG glycopeptides using synthetic isotope‐labeled standards | Glycopeptides | Nano‐LC‐MS | Wang, Liu, Qu, et al. (2021) |
| Improving the sensitivity for quantifying heparan sulfate from biological samples | Heparan sulfate | LC‐MS/MS (AMAC label) | Wang, Dhurandhare, et al. (2021) |
| A LC‐MS/MS method to simultaneously profile 14 free monosaccharides in biofluids | Monosaccharides | LC‐MS | Wang, Zhang, Peng, et al. (2022) |
| Paired derivatization approach with H/D‐labeled hydroxylamine reagents for sensitive and accurate analysis of monosaccharides by liquid chromatography tandem mass spectrometry | Monosaccharides | LC‐MS/MS | Wang, Wang, Wu, Cai, et al. (2022) |
| High‐sensitivity glycoproteomic analysis of biological samples by CZE‐ESI‐MS ‐ Protocol | N‐Glycans | CE‐ESI‐MS | Wang and Lageveen‐Kammeijer (2022) |
| Carbon fiber paper spray ionization mass spectrometry. Said to give stronger spectra than MALDI for glycans | Glycans | Paper spray | Wang, Bai, et al. (2022) |
| A versatile strategy for high‐resolution separation of reducing glycan mixtures as hydrazones by two‐dimensional high‐performance liquid chromatography | Glycans (N‐glycans) (per‐Me) | HPLC, ESI‐MS/MS, MALDI | Wang, Gao, et al. (2022) |
| High‐throughput and high‐sensitivity N‐glycan profiling: A platform for biopharmaceutical development and disease biomarker discovery | N‐Glycans | HPLC | Xie, Mota, et al. (2021) |
| HPLC separation and preparative conditions for 8‐aminopyrene‐1,3,6‐trisulfonic acid‐labeled N‐glycans using a hydrophilic interaction column | N‐Glycans | HPLC | Yamamoto et al. (2022) |
| Glycan mapping of low‐molecular‐weight heparin using mass spectral correction based on chromatography fitting with “Glycomapping” software | Low‐molecular‐weight heparin | LC‐Q‐TOF | Yan et al. (2022) |
| Improved online LC‐MS/MS identification of O‐glycosites by EThcD fragmentation, chemoenzymatic reaction, and SPE enrichment | O‐Glycosites | EThcD Fragmentation, chemoenzymatic reaction, and SPE enrichment | Yang, Wang, et al. (2021) |
| In capillary labeling with APTS and online electrophoretic separation of N‐glycans from glycoproteins | N‐Glycans | CE | Yang, Mai, et al. (2022) |
| Capillary zone electrophoresis‐electrospray ionization tandem mass spectrometry for total analysis of chondroitin/dermatan sulfate oligosaccharides. ‐ Protocol | Chondroitin/dermatan sulfate oligosaccharides | CE‐ESI‐MS | Zamfir (2022) |
| Routine analysis of N‐glycans using liquid chromatography coupled to routine mass detection | N‐Glycans | LC‐MS, protocol | Zhang, Vimalraj, et al. (2021) |
| GlycoHybridSeq: Automated identification of N‑linked glycopeptides using electron transfer/high‐energy collision dissociation (EThcD) | N‐Glycans | Using electron transfer/high‐energy collision dissociation | Zhang, Zhu, et al. (2021) |
| Fractionation and characterization of sialyl linkage isomers of serum N‐glycans by CE‐MS | N‐Glycans | CE‐MS | Zhou, Song, et al. (2022) |
Reaction conditions for solid‐phase permethylation have been optimized in another recent paper (Guan, Zhang, et al., 2021). The authors concluded that 10/100 (v/v) water/DMSO solvent gave the best results with 100 μL of iodomethane and 200 mg of sodium hydroxide beads and an incubation time of 10 min at room temperature. The method was said to minimize side reactions and inhibit the removal of O‐acetylation from sialic acids.
8.2.2. Sialic acids
Sialic acids are generally unstable under MALDI conditions but they can be stabilized by ester (Powell & Harvey, 1996) or amide formation. Cheng, Shu, et al. (2022) have prepared four amides (with reagents 119–122) of the carboxylic group of sialylated N‐glycans (characterized by MALDI‐TOF MS) and have used them to separate linkage isomers of several N‐glycans by microfluidic capillary electrophoresis‐MS. DMDT (121) Was chosen as the most satisfactory of the four reagents. As well as separating the linkage isomers, the migration times also revealed the number of sialic acids. Using this method, 52 sialylated N‐glycans were quantified in human serum within 10 min.

Amide formation is a popular alternative to ester formation. Wang, Zhang, Gao, et al. (2022) have used monomethylamides to study N‐glycans in cases of multiple myeloma, and Li, Zhang, et al. (2021) have employed this reaction in a study of N‐glycans in pediatric ulcerative colitis. Derivatives were prepared by mixing the sample with methylamine and (7‐azabenzotriazol‐1‐yloxy)tris‐pyrrolidinophosphonium hexa‐fluorophoshate (PyAOP, 123) and allowing the mixture to incubate for 30 min at room temperature.

Jia et al. (2021) have used the same amides together with Girard's P derivatization to study changes in N‐glycans from bovine lactoferrin at different stages of lactation, and Cai, Ren, et al. (2022) have studied the urinary N‐glycome in diabetic kidney disease using similar methods. Rather than using PyAOP, Ret et al. (2022) have used the carboxylic acid activator 4‐(4,6‐dimethoxy‐1,2,3‐triazil‐2‐yl)‐4‐methylmorpholinium chloride (DMT‐MM, 124) to form methylamides from N‐glycans. Benzylamidation has also been used in this context (Saito et al., 2021); the authors claim higher sensitivity than is produced by use of other amides.

8.2.2.1. Linkage‐specific derivatization
Under certain conditions, such as by the reaction with methanol in the presence of DMT‐MM, α2→6‐linked sialic acids form methyl esters, whereas the α2→3‐linked acids form lactones, The 32 unit mass difference allows the linkage to be determined by mass measurement (Wheeler et al., 2009). Smith, Millán‐Martín, Mittermayr, et al. (2021) have used this method to study sialylation of human serum glycoproteins. Rather than formation of methyl esters, investigators now prefer ethyl esters, prepared by use of 1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimide (EDC, 125) as a carboxylic acid activator and 1‐hydroxybenzotriazole (HOBt, 126) as the catalyst (Aguedo et al., 2022; Cao, Zhang, et al., 2022; Van Coillie et al., 2022; Levink et al., 2022; Nummela et al., 2021; Pan, Zhang, Zhang, et al., 2021; Rubén et al., 2021; Xu, Jin, et al., 2022; Yaman, Kayili, et al., 2021; Zhang, Cao, et al., 2022; Zhang, Reiding, et al., 2021; Zhang, Wang, et al., 2021). Iso‐propyl alcohol has also been used for the α2→6‐esterification (Ohmi et al., 2021; Yang & Tian, 2022).

Several investigators have improved the original method, mainly by reacting the rather unstable lactone with a further amidation stage. An investigation of this latter reaction has confirmed that formation of the lactone is a prerequisite for amide formation and that the reaction involves direct amidation rather than prior hydrolysis of the lactone (Pongracz et al., 2021). The simplest reactions use the addition of ammonium hydroxide to convert the lactone from the α2→3‐linked acids to amides (Boyaval et al., 2022; Moran et al., 2022; Petralia, Santha, et al., 2022). Two publications have used p‐toluidine (127) as the second derivatization agent (Hyun et al., 2022; Wang, Wang, Zhang, et al., 2022). The α2→6 modification imparts a +28 amu tag whilst that of the p‐toluidine increases the molecular weight by 89 Da.

Amides have also been used in the first stage of the reaction as illustrated by the use of monomethyl‐ (Wang, Kałuża, et al., 2021) and dimethyl‐amides and ammonia (Alves et al., 2021; Zhu, Delbianco, et al., 2021). Ohmi et al. (2021), Yang and Tian (2022) have used iso‐propyl alcohol for the first esterification reaction and have followed it with methylamidation giving mass increases of 41.063 and 13.032 Da respectively, and the method has been adapted to allow it to be used with glycopeptides (Zhong, Huang, et al., 2021). The procedure also derivatized the COOH group of the peptide to give an approximately 4.6‐fold increase in signal intensity.
Many examples of these derivatization reactions employing use of several different alcohols for esterification and several amines for amidation, can be found in Tables 20, 21, 30, 36, 39 and 40. In a modification to allow α2→3‐linked acids to be specifically detected by electrophoresis (CE), the α2→6‐linked acids were amidated with methylamine and the α2→3‐linked acids were amidated with N,N‐dimethylethylenediamine (128) which could easily acquire a positive charge on the tertiary amine allowing it to separate from the isomer by CE. MALDI‐TOF was used to monitor the initial reaction with N‐glycans from fetuin (Cheng et al., 2021).
Table 30.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of glycosphingolipids.
| Source | Methodsa | Notes | References |
|---|---|---|---|
| Human serum and cerebrospinal fluid | Endoglucoceramidase, R‐TOF/TOF, Me‐ester | Detection of novel low‐molecular‐weight blood group‐specific glycans in serum and cerebrospinal fluid | Furukawa et al. (2021) |
| Mouse kidney | Endoglucoceramidase, R‐TOF/TOF (aoWR) | GM3 shown to prevent albuminuria and podocytopathy induced by anti‑nephrin antibody | Kawashima et al. (2022) |
| Brugia malayi (filarial nematode) | Endoglycoceramidase, TOF/TOF (DHB), (2‐AA), exoglycosidase | Identification of glycans with GlcA and phosphatidylchloline | Petralia, van Diepen, et al. (2022) |
| Human cervical cancer cells | R‐TOF/TOF (DHB) | GRASP55 protein shown to regulate intra‐Golgi localization of glycosylation enzymes to control glycosphingolipid biosynthesis | Pothukuchi et al. (2021) |
| Cervical and prostate cancer cells | TOF/TOF (DHB), intact GSLs | Golgi maturation‐dependent glycoenzyme recycling shown to control GSL biosynthesis and cell growth via the Golgi‐localised oncoprotein GOLPH3 | Rizzo et al. (2021) |
| Aspergillus fumigatus glycoinositol‐phosphoceramides | R‐TOF/TOF (ATT, ‐ve) | Characterization of a gene cluster involved in A. fumigatus zwitterionic glycosphingolipid synthesis | Seegers et al. (2022) |
Format (not all items present): Glycan cleavage, MALDI method (matrix), compounds studied, (derivative) other methods.

A high‐sensitivity method, termed derivatization of sialylated glycopeptides (DOSG), involves converting the α2→6‐ and α2→3‐linked acids in glycopeptides to iso‐propyl and methyl amides respectively. Carboxylic acid groups from amino acids are simultaneously converted into iso‐propyl amides (Zhong et al., 2021). An extension of this method, termed (DOSG+), which combines the linkage‐specific sialic acid derivatization with fixed charge derivatization has been developed (Zhong, Huang, et al., 2022). The sialic acids were reacted in a two‐stage process to convert the α2→6‐linked acids to iso‐propylamides and the α2→3‐linked acids to methyl amides. The acids were then reacted with sodium periodate to oxidize the side chain and the resulting aldehyde group was reacted with (2‐aminoethyl)trimethylammonium chloride hydrochloride (AETMA, 129) by reductive amination to introduce a positive charge. Using a model glycopeptide, sensitivity increases of about 30% were reported.

Jezková et al. (2022) have prepared alkyl esters of sialylated N‐glycans in a linkage‐specific fashion and then formed phenylhydrazone derivatives with phenylhydrazine (130). Under these conditions, the lactone ring was opened with the incorporation of a second phenylhydrazine moiety and phenylhydrazine was also added to the reducing terminal. Methyl rather than ethyl esterification was found to give the best results. The method was applied to the monitoring of sialylation in the serum of lung cancer patients.

8.2.2.1.1. Quantification
To avoid potential problems with quantification due to the fact that most of these methods convert the differently linked sialic acids into different compounds, Peng, Gu, et al. (2021) have used d0‐ and d3‐methylamide derivatization for the two stage reaction. Similar results were obtained with forward (d0‐ followed by d3) or backwards labelling. In a similar reaction, Jin, Li, et al. (2021) have used d0‐ and d6‐pyridine to label the α2→6‐ and α2→3‐linked sialic acids respectively. The N‐glycan solution was mixed with EDC, HOBt and d0‐aniline in DMSO and incubated at 60°C for 1 h. d5‐Aniline, together with additional EDC and HOBt in DMSO were then added and incubation was continued for a further hour. After purification, the glycans were derivatized with Girard's P reagent. Applications were to N‐glycans from glycoproteins in human colostrum and mature milk with a further study on human milk (Jin, Lu, et al., 2022).
8.3. Charged derivatives
Reagents with constitutive positive charges have been used to increase sensitivity in positive ion mode. Examples of the use of Girard's reagents T and P have been illustrated above and others are listed in Tables 15, 20, and 36. Another example is the imidazolium derivative (GITag, 131) which has been synthesized and used to derivatize glycans by reductive amination (Zhang, Ghirardello, et al., 2021). Gains in sensitivity of up to 600 times over that provided by 2‐AB labelling were claimed for derivatized GlcNAc and lactose (30). The derivatization reaction could be conducted directly on the MALDI target following N‐glycan release with PNGase F and the reaction was demonstrated by examination of glycans released from human serum.

Although originally designed for CE studies (4‐hydrazidebutyl)triphenylphosphonium bromide (P4HZD, 132), containing a permanent positive charge, has showed good MALDI performance. For example, the signals from derivatized maltodextrin were increased by an order of magnitude (Ma, Wang, et al., 2022). Larger oligomers were also detected when derivatized with this tag than were detected with the free sugars. Strong signals were also obtained from derivatized N‐glycans from therapeutic glycoproteins. Derivatization was said to increase the signal strength by more than the common labelling reagent, Girard's P, in both MALDI and ESI modes. Other examples of the use of Girard's P reagent are given in the section on linkage‐specific derivatization of sialic acids (Section Linkage‐specific derivatization).

Constituents of ionic liquids, attached to various positions of the glycan molecules, including the reducing terminal, provide another method for attaching a fixed charge for high sensitivity. The subject has recently been reviewed (Ghirardello et al., 2022) and highlights tags such as 133–136.

9. GLYCAN ARRAYS
A review with 138 references on glycan array technology has been published by Martinez et al. (2021). In a method to acquire N‐glycans for array construction Cao, Antonopoulos, et al., 2021) have used two‐dimensional hydrophilic interaction liquid chromatography and porous graphitized carbon chromatography to purify 31 N‐glycans from chicken ovalbumin. Purity of the glycans was estimated to be over 90% with identification by negative ion CID. The glycans were printed onto nitrocellulose‐coated glass slides and interrogated with wheat‐germ agglutinin, which mainly bound to hybrid‐type N‐glycans with a bisecting GlcNAc residue, compounds that were abundant in the released glycan mixture.
10. QUANTIFICATION
Reviews and general articles relating to quantitation of glycans and glycopeptides are listed in Table 7. “Methods to improve quantitative glycoprotein coverage from bottom‐up LC‐MS data” (Chang & Zaia, 2022) (136 references) is also of interest. Patabandige et al. (2022) have discussed quantitative clinical glycomics strategies and have concluded that there is no one best method and provide guidance on the best approach to take.
Table 7.
Reviews and general articles on glycan quantitation.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Quantitative characterization of O‐GalNAc glycosylation | Summarizes the most common quantitative strategies and discusses benefits and limitations of the various approaches | 51 | Čaval et al. (2021) |
| Recent advances in analytical approaches for glycan and glycopeptide quantitation | Glycan and glycopeptide quantitation. Methods for isotope labelling, software. | 208 | Delafield and Li (2021) |
| Recent advances in qualitative and quantitative analysis of polysaccharides in natural medicines: A critical review | Discusses general and mass spectrometric methods | 100 | Li, Zhang, Han, et al. (2022) |
| Qualitative and quantitative methods for N‐glycans in N‐glycomics | Book chapter. General coverage of glycan analysis with some common quantitative methods | 120 | Ren and Lu (2022) |
| Isotope labeling strategies of glycans for mass spectrometry‐based quantitative glycomics | Mainly discusses isotope‐labelled derivatives | 56 | Yun et al. (2021) |
10.1. N‐Glycopeptides
A 4‐plex method has been developed for quantification of glycopeptides. The reagents (DiLeuEN, Scheme 8) formed amides with the carboxylic acid groups and had the advantage of neutralizing the negative charge to improve sensitivity. The reporter ions allowed four samples to be quantified simultaneously (Li, Zhong, et al., 2022).
Scheme 8.

4‐Plex derivatives for glycopeptide quantification.
The lack of suitable standards for quantitation of glycopeptides from immunoglobulin G (IgG) has prompted the synthesis of fifteen such compounds (Wang, Liu, Qu, et al., 2021) by the attachment of 13C6‐fucose (Fuc, 14), which introduced a 6 Da mass shift, to the core region of unfucosylated glycopeptides using the enzyme FUT8. The reference material was used to measure IgG glycopeptides in colon cancer sera.
10.2. O‐Glycans
A 4‐plex method for O‐glycan quantification, using the same‐labelled leucine analogues (Scheme 9) as in the glycopeptide method described above, has been used for O‐glycan quatification (Li, Gu, et al., 2021). O‐glycan release using ammonium hydroxide and labelling to form the modified PMP derivatives (see Scheme 4) was achieved simultaneously by heating at 70oC for 24 h. Products were measured by MALDI‐MS or LC‐MS/MS. Applications were to core I O‐glycans from human serum.
Scheme 9.

4‐Plex derivatives for O‐glycan quantification.
11. FRAGMENTATION
Mechanisms leading to the formation of fragment ions in several carbohydrates continue to be a fruitful area for research with several methods for producing the ions being available. Nomenclature for the fragment ions follows that proposed by Domon and Costello (1988) (Scheme 1).
11.1. In‐source decay (ISD)
The [M + H]+ ion from α‐CD (as 27 but with six glucose rings) has been shown to exhibit two fragmentation pathways (Jang & Choi, 2021). After ring opening, the first pathway involves successive losses of glucose with the relative abundances of the fragments increasing as their mass decreases. The second series involved losses of OH and glucose units.
The disaccharide isomers gentiobiose (145), isomaltose (146), melibiose (147), lactose (30), maltose (43), cellobiose (148), and sucrose (42) have been ionized with a 349‐nm Nd:YLF UV laser from a graphene oxide matrix ([M + Na]+ ions) and shown to fragment in a manner that revealed differences between isomers (Lee, Kim, et al., 2021). Anomeric configurations of 1–6 and 1–4 linked isomers could be differentiated by comparing the peak intensity at m/z 267 with that at m/z 365. The α‐anomers (maltose [43], isomaltose [146], and melibiose [147] had m/z 267/m/z 365 ratios greater than 0.1, while those of the β‐anomers (cellobiose [148], lactose [30], and gentiobiose [145]) had ratios that were less than 0.1. Linkage isomers (1–4 and 1–6) were differentiated by the presence of a peak at m/z 275, which was only observed with 1–6 linked isomers such as gentiobiose, isomaltose, and melibiose.


Liew, Chen, and Ni (2022) have studied ISD in electrospray ion sources and noted 0.2%–3% dissociation of neutral glycans and more than 50% dissociation when sialic acid is present. Dissociation rose with increasing temperatures and products of the larger glycans, which were similar to those observed by CID, were smaller glycans, some of which occurred naturally. The authors point out that this property could have adverse effects on the apparent compositions of glycan mixtures.
11.2. CID and higher‐energy collisional dissociation (HCD)
Nguan, Tsai, and Ni (2022) have used quantum chemical calculations and experimental measurements to elucidate the fragmentation mechanisms of the [M + Na]+ ions from cellobiose and maltose. Four mechanisms were studied. Dehydration mainly occurred through the transfer of a hydrogen atom to O1 of the sugar at the reducing end, followed by a C1−O1 bond cleavage. Cross‐ring dissociation started with a ring‐opening reaction, which occurred through the transfer of a hydrogen atom from O1 to O5 (ring oxygen) of the sugar at the reducing end. The third route, generation of B1 and Y1 ions, occurred through the transfer of an H atom from O3 (cellobiose) or O2 (maltose) to O1 of the sugar at the nonreducing end, followed by a glycosidic bond cleavage. The fourth pathway, production of C1−Z1 fragmentation, had two mechanisms: (1) the transfer of a hydrogen atom from O3 or O2 to O4 of the sugar at the reducing end to generate C ions in the ring form and (2) the transfer of a hydrogen atom from O3 of the sugar at the reducing end to O5 of the sugar at the nonreducing end to produce C ions in the linear form.
Fragmentation of protonated β‐cyclodextrins (27) by CID and HCD, assisted by fragmentation of di‐ and tri‐methylated CD derivatives has shown initial glycosidic cleavage to open the ring (150, Scheme 10) followed by the elimination of glucose subunits and the subsequent release of water and formaldehyde moieties from the glucose monomer and dimer fragment ions (Bruni & Schürch, 2021).
Scheme 10.

Proposed mechanism for ring opening of cyclodextrin rings.
The resulting linear structure further decomposes in charge‐independent processes forming either a zwitterionic fragment from the boat conformation of the reducing‐terminal ring (Scheme 11), elimination of a 1,4‐anhydroglucose moiety (Scheme 12), or loss of a new macrocyclic structure and an oxonium ion (Scheme 13).
Scheme 11.

Proposed mechanism leading to elimination of a zwitterionic fragment from cyclodextrins.
Scheme 12.

Proposed loss of a 1,4‐anhydroglucose moiety during fragmentation of cyclodextrins.
Scheme 13.

Proposed mechanism for elimination of a new macrocyclic structure during fragmentation of cyclodextrins.
Fragmentation of [M + Na]+ ions from β‐CD has been studied by Rabus et al. (2021) using ion mobility and cryogenic IR. Electronic structure calculations were consistent with formation of a fragment with a 2‐ketone group as shown in Scheme 14. Other B‐type fragments were formed similarly. The structures of three other proposed fragments (160–162) are shown below.
Scheme 14.

Proposed mechanism for the formation of a fragment with a 2‐ketone group. From Rabus et al. (2021).
CID of the protonated ion from Lewis A trisaccharide (α‐l‐Fuc‐(1→4)‐[β‐d‐Gal‐(1→3)]‐d‐GlcNAc, 163) and its methyl glycoside has shown that fragmentation from the reducing end of the ion plays a key role in the fragmentation process. The main product of the fragmentation are Y‐type fragment ions and a combination of Y‐type fragmentation and the loss of water at the reducing end instead of Z‐type fragmentation as proposed earlier. It appears that fragmentation only occurs with the aid of the mobile proton added during ionization (Iwan & Grotemeyer, 2021).

A major fragmentation pathway for glycopeptides is glycosidic cleavage to give a B fragment with a conventional mechanism from the [M + H]+ ion from a monosaccharide giving m/z 204 (168) as shown in Scheme 15. MS3 experiments indicate that this ion fragments further to yield m/z 126 by loss of C2H6O3 but there appears to be no reasonable way in which this ion could be produced from the structure of m/z 204 shown in Scheme 15.
Scheme 15.

Formation of the ion at m/z 204 by the conventional mechanism. Shown for GlcNAc‐β‐1‐Asn + H+. From Guan and Bythell (2021).
Consequently, Guan and Bythell (2021) have investigated this fragmentation mechanism using hydrogen/deuterium exchange and energy calculations and have proposed that the reaction proceeds through a furanose form of the sugar as shown in Scheme 16.
Scheme 16.

New proposal for formation of the ion at m/z 204 (174). Shown for GlcNAc‐β‐1‐Asn + H+.
This ion is then proposed to decompose to m/z 126 by (178) the mechanism shown in Scheme 17.
Scheme 17.
Proposed mechanism for formation of the ion at m/z 126 (178).
Evidence supporting this proposed fragmentation mechanism has been supplied by IR action spectrometry (Rabus et al., 2022). This reaction, shown in Scheme 17, was found to occur irrespective of the glycosidic linkage stereochemistry (α or β) or the N‐acetylated hexose (GlcNAc or GalNAc). The authors comment that “Dissociation of the glycosidic and other bonds thus occur from the furanose isomer critically altering the reaction feasibility and product ion structures.”
Rumiantseva et al. (2022) have investigated 16O/18O and H/D exchange reactions in an attempt to gain more information on fragmentation reactions in general. Oxygen exchange was observed at the anomeric site of d‐glucose with a small amount occurring at the adjacent position as the result of aldose‐ketose reactions. The study showed that several of the cross‐ring fragment ions consisted of several species such as losses of C3 and C4 fragments from different parts of the molecule as shown for the C3 fragments in Scheme 18.
Scheme 18.

Formation of C3 fragments from d‐glucose. Atoms in the fragments are shown in red. From Rumiantseva et al. (2022).
Reasons for the differences in the CID spectra of the two glucose dimers, Glcα1→4‐Glc (maltose, 43) and Glcα1→6‐Glc (isomaltose, 146) as sodium adducts have been studied using high‐level quantum chemistry calculations (Nguan & Ni, 2022). These calculations revealed that, although the two disaccharides had similar dissociation mechanisms, energy differences between the lowest transition states of various dissociation channels led to different fragmentation patterns. The dissociation barriers for dehydration and glycosidic bond cleavage were similar for the two disaccharides, but the cross‐ring dissociation, which has the lowest dissociation barrier, exhibited differences. The cross‐ring dissociation barrier for α‐maltose was only slightly lower than those for dehydration and glycosidic bond cleavage. However, the corresponding barrier for α‐isomaltose was substantially lower. Furthermore, most of the α‐isomaltose conformers that led to dehydration also led to cross‐ring dissociation, resulting in suppression of dehydration by cross‐ring dissociation. The findings can explain the low branching ratios for dehydration and glycosidic bond cleavage observed in the CID spectrum of α‐isomaltose CID spectra.
11.2.1. CID of complexes
Differentiation of isomers has recently been facilitated by formation of complexes formed in the gas phase. For example, Chao and McLuckey (2021) have reacted deprotonated gangliosides with magnesium‐Terpy complex cations ([Mg(Terpy)2]2+, 183) to form magnesium complexes and have demonstrated isomeric differentiation between GD1a (184) and GD1b (185) as [GD1−H+Mg]+ ions. In addition, isomeric identification among GT1a, GT1b, and GT1c was also achieved. The method was applied to ganglioside profiling in a porcine brain extract where 34 gangliosides were profiled among only 20 precursor ion m/z values.



11.3. Electron‐transfer/higher‐energy collision dissociation (EThcD)
EThcD, triggered by HCD has been used to map N‐glycosylation on intact therapeutic antibodies (Li, Zhu, et al., 2022). The method was reported to provide higher quality spectra than use of EThcD alone and to differentiate between different N‐glycan classes such as high‐mannose, hybrid and complex.
11.4. Helium‐charge transfer dissociation (He‐CTD)
Using CTD with a modified ion trap instrument, Sasiene, Mendis, et al. (2021) have investigated the effect of Na/H exchange (sodium salt formation) on the fragmentation patterns of mannuronic acid oligomers (186). The conclusion was that the fewest possible number of Na/H exchanges will provide the most confident peak assignments and structural characterization.

A comparison of the low‐energy CID and He‐CTD spectra for the branched xyloglucan (X3G4, 187) has emphasised the superior results that can be obtained by the latter technique. The CID spectra of ions such as [M + Na]+ and [M + H + K]2+ contained numerous fragments produced by glycosidic cleavages but few cross‐ring cleavage ions and did not allow the 1→4 and 1→6 linkages of the glycan to be identified, He‐CTD, on the other hand, was able to identify the linkage. Different metal adducts (H+, Na+, K+, Ca2+, and Mg2+) were investigated but were found to have a negligible effect on the type of cross‐ring cleavages that were observed (Sasiene, Ropartz, et al., 2021).
11.5. LIFT fragmentation
Palladium nanoparticles decorated thiol‐functionalized metal organic framework nanocomposite (UiO‐66‐(SH)2@Pd NPs) have been prepared and shown to be an efficient MALDI matrix (Luo, Zhao, et al., 2022). By using this matrix combined with LIFT‐TOF/TOF, 24 oligosaccharide isomers including disaccharides, trisaccharides and tetrasaccharides, have been differentiated as shown in Scheme 19. Reducing and nonreducing disaccharides could be distinguished by the presence or absence of cross‐ring cleavage ions. Only B (m/z 185) and Y (m/z 203) fragment ions were observed in the MS/MS spectra of nonreducing sugars (trehalose and sucrose), whereas cross‐ring cleavage ions (m/z 305, 275, 245,143, and 113) were observed in the spectra of 10 reducing sugars to varying degrees. 1→4 and 1→6 linkage isomers produced m/z 305 whereas this ion was missing from the spectra of the 1→2 and 1→3 linkage isomers. The ion at m/z 275 appeared in the spectra of 1→6 and 1→3 linkage isomers but not in those from the 1→4 and 1→2 isomers. These and other diagnostic fragment ions are listed in Table 8.
Scheme 19.

Differentiation of disaccharides with different linkages using LIFT‐MS/MS. From Luo, Zhao, et al. (2022).
Table 8.
Diagnostic ions for differentiating linkage isomers from disaccharides (From Luo, Zhao, et al., 2022).
| Linkage position | Ions (m/z) | Relative ion abundance | |
|---|---|---|---|
| Present | Absent | ||
| 1→2 | 245, 203, 185, 143 | 305, 275 | α>15; β<5 or α>1>β |
| 1→3 | 275, 203, 185, 113 | 305 | α>1>β |
| 1→4 | 305, 245, 203, 185 | 275 | α>1>β |
| 1→6 | 305, 275, 203, 185 | ‐ | α>1>β |
11.6. Photofragmentation
A method for differentiation of disaccharide isomers using a combination of IR and UV photodissociation mass spectrometry with an FT‐ICR instrument has enabled ten disaccharides, chosen for differences in connectivity, configuration, and/or composition, to be resolved (Du, Zhang, et al., 2022). The disaccharides were complexed with 3,5‐diiodo‐l‐tyrosine (188) by ESI to add UV absorption properties and irradiated successively with light from a double‐beam laser. The IR laser produced mainly glycosidic B/Y and C/Z ions, whereas the UV laser produced other complementary fragments. Fragments were formed from both parts of the complexes and were, therefore, not assigned the Domon and Costello nomenclature. Major fragments involved losses of water or monosaccharide residues.

11.7. Multiple successive fragmentation (MS n )
Successive stages of fragmentation presents the analyst with multiple choices of which fragment ion to use for the next stage of fragmentation. This choice can become quite extensive after several stages. To simplify the analysis, Huang, Hsu, et al. (2021) and Ni et al. (2021) have developed a logically derived sequence (LODES) for galactose‐containing tri‐ and tetrasaccharides, but with the potential for it to be extended to other glycans. The method made use of ionic properties that differentiate between, for example, linear and branched compounds and their dissociation to disaccharides, which were compared to a database. The paper contains extensive figures explaining the recommended sequences. However, the success of the method for other glycans depends on the derived disaccharides being available in the database. The method has been extended to determine the structures of N‐glycan isomers and the paper contains a useful list of how the relative abundances of cross‐ring fragments relate to linkage (Liew, Yen, et al., 2021). The authors (Liew, Chan, et al., 2021) have also used the method to identify isomeric glycans derived from glycosphingolipids. In an application, Lin and Ni (2022) use the method to determine the structure of lichenin, a linear polymer with alternating β‐Glc‐(1 → 4)‐β‐Glc‐(1 → 4)‐β‐Glc‐(1 → 3)‐Glc, β‐Glc‐(1 → 4)‐β‐Glc‐(1 → 4)‐β‐Glc‐(1 → 4)‐β‐Glc‐(1→ 3)‐Glc repeats. A discussion as to what constitutes “good” (retaining much structural information) and “bad” fragment ions has also been published (Liew, Hsu, et al., 2022). The LODES/MS n method also features in a chapter in the reference work “Comprehensive Glycoscience” (Ni et al., 2021).
11.8. Electron ionization
A new electron‐activated dissociation (EAD) device has been developed and coupled to a Q‐TOF mass spectrometer (Baba et al., 2021). It features a new electron beam optics design allowing high electron currents up to the space‐charge limit of 0.5 μA in the reaction cell, and enables fast and efficient dissociation of various analytes ranging from singly charged small molecules to multiply protonated proteins. The tuneable electron energy provided access to different fragmentation regimes: electron‐capture dissociation (ECD), hot ECD, and electron‐impact (EI) excitation of ions from organics (EIEIO). The system was evaluated for several compound classes including intact proteins and glycopeptides. Application of hot ECD for the analysis of glycopeptides resulted in rich fragmentation with predominantly peptide backbone fragments; but with additional glycan fragments attributed to the ECD process.
11.9. Negative ion fragmentation
Negative ion fragmentation of N‐glycans produces more diagnostic ions than positive ion fragmentation (Hykollari et al., 2022) and is frequently conducted using phosphate adducts to stabilize the ions (Harvey, 2020). Ruf et al. (2022) have now shown that adduction with phosphate considerably enhances sensitivity for mono‐ and oligosaccharides and forms more hydrogen bonds with the sugars than Cl‐, another popular adduct.
11.10. Comparison of methods
Fragmentation spectra of high‐mannose glycans, predominantly Man5GlcNAc2 (189), as [M + Mg]2+ and [M + Na2]2+ ions, induced by CID, ECD or electronic excitation dissociation (EED), have been compared (Wong, Chen, Wu, et al., 2022). CID produced mainly glycosidic cleavages, although more cross‐ring fragments could be obtained at higher intensities when [M + Mg]2+ ions were fragmented. 0,2A3, 0,3A3, and 0,4A3 ions (cleavage of the core branching mannose) provided structural information on the 3 → 1 and 6 → 1 linkages of the mannoses. Some internal fragment ions, such as 2,4A5/Y3β, were also produced in high abundance. ECD produced fewer fragments compared to the other dissociation methods when either of the metal ions were used as charge carriers. Cross‐ring fragments were produced in relatively high abundance, with the charge mainly retained on the nonreducing end. EED produced extensive glycosidic and cross‐ring cleavages with either metal charge carrier. More structural‐specific fragments were produced when Na+ was used as the charge carrier and this metal also provided higher fragmentation efficiency. Of the 31 possible cross‐ring cleavages, 25 were found, thus providing extensive linkage information. Many fragment ions were produced by all three dissociation methods when Mg2+ was used as the charge carrier. Best results were obtained with CID of [M + Mg]2+ ions and EED of sodiated glycans.

These three dissociation methods have also been compared for structural characterization of doubly charged N‐glycopeptides. CID produced distinctively different positive ion mass spectra for glycopeptides adducted with different charge carriers (hydrogen, sodium, magnesium). Protonated species produced mainly glycosidic cleavages in high abundance. Glycopeptides adducted with magnesium formed more cross‐ring cleavages, whereas doubly sodiated species produced cleavages at both glycan and peptide moieties. The effect of charge carriers on fragmentation in ECD and EED was lower than that in CID. ECD produced mainly peptide backbone cleavages but few cleavages of the glycan, whereas EED of glycopeptides resulted in extensive fragmentation regardless of the charge carrier. However, magnesiated species gave more cross‐ring cleavages than other charge carriers (Wong, Chen, Zhang, et al., 2022).
12. COMPUTER ANALYSIS OF SPECTRA
Several reviews on computer applications are listed in Table 9.
Table 9.
Reviews and general articles on computer analysis of spectra.
| Subject | Contents | Citations | References |
|---|---|---|---|
| Glycoinformatics in the artificial intelligence era | General review, discusses early failures, describes how the field has benefitted from lessons learned from areas such as proteomics, and makes predictions on the future | 164 | Bojar and Lisacek (2022) |
| Recent advances in software tools for more generic and precise intact glycopeptide analysis | Includes table describing different software | 108 | Cao, Liu, et al. (2021) |
| Glycobioinformatics in deciphering the mammalian glycocode: Recent advances | Comprehensive list of software tools | 264 | Datta and Sukhija (2021) |
| Glycobioinformatics | Short overview. Discussion of various systems | 64 | Gerwig (2021a) |
| Artificial intelligence in the analysis of glycosylation data | Use to gain mechanistic insights into glycosylation machinery and to predict models of glycosylation | 78 | Li, Chiang, et al. (2022) |
| Book chapter ‐ Analytical software and databases in N‐glycoproteomics | Mainly glycopeptide identification. States that latest GPSeeker reports additional structure‐specific information of monosaccharide sequences unavailable from older systems such as Byonic and GPQuest. Additionally, search engines that support O‐glycosylation identification are also briefly introduced. | 74 | Qin and Tian (2022) |
A large amount of work has been devoted to designing software for analysing glycan spectra with the impression that lack of software is a serious problem for glycomics. Such attitudes must be viewed with caution because most of the software only gives pointers as to what a correct structure should be. Simply matching masses to structures in a database clearly is only a first step and obviously cannot assign a structure if it is not represented in the database and, of course, this method is incapable of identifying new structures. Also, taking a structure with the best fit out of several possibilities as the correct structure, cannot be accepted as good science; such putative structures must be confirmed with orthogonal techniques. In any case, a skilled analyst could probably identify a compound simply by looking at the spectrum and, in addition, spot the presence of additional compounds; something a software package would almost certainly miss.
Efforts to integrate various software packages for glycoinformatics have been summarised with emphasis on using consistent nomenclature across the various packages (Mariethoz et al., 2022) and various software packages have been compared with the identification of key variables that should guide future software developments and assist informatics decision‐making in glycoproteomics (Kawahara et al., 2021).
Several methods have been developed for converting glycan structures into computer‐readable formats. The major ones, International Union of Pure and Applied Chemistry (IUPAC), Linear Notation for Unique Description of Carbohydrate Sequences (LINUCS), Kyoto Encyclopedia of Genes and Genomes (KEGG), Chemical Function (KCF), GLYcan Data Exchange‐II (GLYDE‐II) and Glyco Connection Table (GlycoCT) have been summarised in a short review (Frey, 2022).
A glycosylation mapping tool, termed GlycoMaple, which visualizes and estimates glycan structures based on gene expression has been developed (Huang, Aoki, et al., 2021). Nine hundred and fifty genes involved in glycosylation and its regulation were selected and the expression profiles of these genes were mapped onto global glycan metabolic pathways to predict glycan structures. These structures were confirmed using glycomic analyses of N‐glycans in 40 knockout HEK293 cell lines. In addition, the glycan structures of 64 cell lines, 37 tissues, and primary colon tumor tissues were estimated and compared using publicly available databases. The authors point out that this is only a predictive tool for possible structures and that the structures of detected compounds must be confirmed by orthogonal methods. The code for GlycoMaple is available at https://glycosmos.org/glycomaples/index.
Zhou and Neelamegham (2022) have describes the development and usage of a package entitled “comparative Glycomics” (cGlyco) which is an open‐source program that can be used to compare data from multiple mass spectrometry runs. It has been used, for example, to compare differences in the metabolism of various human blood cell types and it may also be applied to different tissue types or to data collected from other mass spectrometers in the field.
There is currently much interest in increasing glycan data in bioinformatics databases such as ChEBI and PubChem, and connecting them to resources at the EMBL‐EBI and NCBI. Much material is available in databases such as GlyTouCan which contains glycans obtained primarily through batch upload from glycan repositories and individual laboratories and, as such, many glycan structures from such sources may not be fully defined. Databases like ChEBI and PubChem were designed to accommodate complete atomistic structures with well‐defined chemical linkages and, consequently, they cannot easily accommodate the structural ambiguity inherent in glycan databases. Therefore, there is a need to improve the organization of glycan data to enhance connectivity across the major NCBI, EMBL‐EBI and glycoscience databases. Navelkar et al. (2021) have developed a workflow in collaboration between GlyGen, ChEBI and PubChem to improve the connectivity of glycan data across these resources. GlyGen hosts a subset of glycans (∼29,000) from the GlyTouCan database and has submitted valuable glycan annotations to the PubChem database and integrated over 10,500 (including ambiguously defined) glycans into the ChEBI database. The current PubChem, ChEBI and GlyTouCan mappings can be downloaded from GlyGen (https://data.glygen.org).
12.1. Algorithms for analysing spectra
An addition to the Byonic software (Bern et al., 2012) that addresses the issues of finding glycopeptide spectra when they are a tiny fraction of the total spectra; assigning spectra with unanticipated glycans that are not in the initial glycan database; and finding, scoring, and labeling diagnostic peaks in tandem mass spectra has been developed (Roushan et al., 2021).
Claimed to be better than Byonic, the freely available package pGlyco3 (available at https://doi.org/10.1038/s41592-021-01306-0) is a “glycan‐first” application for analysing glycopeptides. It combines electron‐based (HCD, EDC) dissociation and provides site‐specific glycan localization. It is claimed to be 5‐40 times faster than other glycoproteomics search engines (Zeng, Cao, et al., 2021).
StrucGP is a new package for determination of N‐glycan structures from glycopeptides (Shen et al., 2021). It categorizes B and Y ions, produced by low‐energy MS/MS into three groups, core fragments, glycan subtype fragments and fragments from the antennae. Based on only these masses and LC data, it is claimed that complete structures can be determined. The method was tested satisfactorily with standard glycoproteins such as ovalbumin, fetuin, RNase B and IgG and then applied to mouse brain and other tissues. The detailed structures of 600 glycans were reported from mouse brain and 719 from five tissues. In another paper using StrucGP, 773 N‐glycans were identified in human seminal plasma (Xin, Xu, et al., 2022). Unfortunately, many of these structures do not conform to any reported biosynthetic pathways and the authors’ report that “It is worth to mention that the location of each branch structure (such as α−2,3 or α−2,6 mannose) can't actually be identified by StrucGP”. Thus, many of the reported structures are undoubtedly incorrect raising the question of how much reliance should be placed on the supposed identification using only computer algorithms such as this. It would appear that most “structures” can only be regarded as pointers to the correct structure and that these structures must be confirmed by orthogonal techniques as pointed out above. This conclusion is supported by statements in several other publications using computer algorithms such as “the structure with the highest score was taken to be correct.”, or the highest score within a given data set (Zhang, Peng, et al., 2022). Unfortunately, several publications have appeared in which StrucGP (Li, Zhao, et al., 2022; Xin, Xu, et al., 2022) and other computer software are the only methods used to “identify” N‐glycans with again, dubious structures being reported in many instances. Authors and reviewers of these papers must ensure that, at least, reported structures conform to the products of established biosynthetic pathways in the species being investigated. Thus, in publications where “identifications” have been made only by the use of computer algorithms and database matching with no follow‐up, results should be treated with caution. It is somewhat gratifying to see that at least one recent paper that used StrucGP to “identify” structures (of the Covid‐19 virus spike glycoprotein), that the authors only report the partial structures that the algorithm can correctly identify (Zhu, Chen, et al., 2022).
A software package called GlycanAnalyser has been developed for automatically interpreting the results of methods of glycan analysis that rely on HPLC separation of glycans and exoglycosidase digestion to determine the nonreducing terminal monosaccharide constituents. The paper describes a protocol for using the software and a table listing other software tools available to the glycobiologist (Walsh et al., 2022). The HPLC/exoglycosidase technique originated in Oxford in the 1990s and was developed in Dublin with the introduction of GlycoBase, a database containing retention data for many N‐linked glycans. Glycostore is a development of this work (Campbell et al., 2021). Its first release in October 2017) contained over 850 glycan entries accompanied by over 8500 retention times including data from HPLC, PGC interfaced with ESI‐MS/MS, and CE.
GAGrank is an algorithm that uses a bipartite graph model for sequencing GAGs from electron detachment dissociation (EDD) or negative electron transfer dissociation (NETD) tandem mass spectra. The process involves first assigning GAG product ions using the recently‐developed GAGfinder algorithm (Hogan et al., 2018) and secondly calculating every possible sequence for a given GAG composition. Sequences are given a higher ranking if they link to many important fragments. The system was optimized using ten training sequences and validated with three validation sequences. It was able to sequence isomeric mixtures using two mixtures at five different ratios (Hogan et al., 2021).
An algorithm for reconstruction of glycan structures, GlycoDeNovo, first reported in 2017 (Hong et al., 2017), has been upgraded to GlycoDeNovo2 with the inclusion of the calculation of glycan composition from precursor mass and the ability to calculate a p‐value from the predicted structures (Chen, Wei, et al., 2022).
One of the weaknesses of mass spectrometry for glycan analysis is the problem of identifying the nature of the constituent monosaccharides. An algorithm, termed HexNAcQuest has now been developed and is claimed to be able to differentiate GalNAc from GlcNAc with 97% accuracy (Li, Hou, et al., 2022). Essentially, the algorithm looks at the relative intensities of five fragment ions (HCD mode, Orbitrap) from oxonium ions derived from glycopeptides. Specifically, if the intensity of m/z 138 is much higher than that of m/z 144, the probability of GlcNAc is between 0.5 and 1, and if the ions at m/z 138 and 144 are of similar intensity, the corresponding probability will be between 0 and 0.5, indicating an O‐GalNAc modification.
The Toolbox Accelerating Glycomics (TAG) package for analysing MALDI spectra, was developed in 2020 (Miura et al., 2020) and consists of three units, “TAG List” which creates a glycan list that is used for database searching in “TAG Expression”; ‘TAG Expression’, which automatically annotates and quantifies glycan signals; and ‘TAG Pathway’ which maps the obtained expression information to biosynthetic pathways. This software has now been updated (Miura et al., 2022) to include some less common glycans such as those containing glucuronic acid and the linkage‐specific alkylamidation method (SALSA) (Hanamatsu et al., 2018, 2019; Nishikaze et al., 2017) for determining the linkage of sialic acids.
The web application “Glycoanalysis by the Three Axes of MS and Chromatography” (GALAXY) is a tool for assisting glycoprofiling by HPLC and MS data of 2‐AP‐derivatized glycans (Kato & Takahashi, 2009; Takahashi & Kato, 2003). A new version (3) has now appeared and includes new HPLC data on glucoronylated and sulfated glycans and an improved graphical user interface (Yagi et al., 2022).
12.2. Quantification
The “Individuality Normalization when Labeling with Isotopic Glycan Hydrazide Tags” (INLIGHT) strategy for glycan quantification uses hydrazide chemistry to derivatize the reducing end of N‐linked glycans and incorporates either a natural (NAT, 12C6) or a stable‐isotope label (SIL, 13C6) to enable relative quantification. GlycoHunter is software created in MATLAB that enables researchers to process quantitative glycomics data generated with but not limited to, INLIGHT. GlycoHunter accepts the commonly used data file formats imzML and mzXML and effectively identifies all peak pairs associated with NAT and SIL‐labeled N‐linked glycans using MS1 data. It also includes the ability to export the results for further analysis using Skyline or Excel. The software is available for no charge from the Web site https://glycohunter.wordpress.ncsu.edu/ (Kalmar et al., 2021). Ion mobility properties of INLIGHT derivatives of several N‐glycans released from commercial glycoproteins such as horseradish peroxidase (HRP) and fetuin have also been reported (Butler, Kalmar, et al., 2022).
gQuant, coded in Python, is another program for processing quantitative data from experiments using stable isotopes (Huang, Jiang, et al., 2021). In tests, reported quantitation ratios matched well with the experimental glycan mixture ratios ranging from 1:10 to 10:1. The application has a simple user interface and can easily be adapted by users for specific experimental designs, such as specific glycan databases or different derivatization types.
12.3. Databases
Five‐ reviews are relevant and are listed in Table 10.
Table 10.
Reviews on glycan databases.
| Subject | Contents | Citations | References |
|---|---|---|---|
| Analytical software and databases in N‐glycoproteomics | General overview of current analytical software and databases in N‐glycoproteomics | ‐ | Qin and Tian (2022) |
| Plant lipid databases | Overview of plant lipid databases focusing on nomenclature, structures as well as physical and chemical properties | 18 | Dörmann (2021) |
| Database search assisted N‐glycan structure identification | Concentrates on conventional glucose unit calculation, the virtual ladder approach and exoglycosidase glycan sequencing | 49 | Jarvas et al. (2021) |
| Databases and bioinformatic tools for glycobiology and glycoproteomics | Comprehensive review of different databases with comments on each | 117 | Li et al. (2020) |
| Glycosciences. De: Databases and tools to support research in glycomics and glycoproteomics | Overview of the individual databases and applications within Glycosciences. de, and their interconnections with each other and with external resources | 40 | Lütteke (2021) |
GlycoPOST is a database that accepts MS data from glycomics experiments and issues an accession number to provide traceability for reuse and reanalysis of the data. This system is based on the jPOST repository system (Okuda et al., 2017), a stable MS data repository for proteomics. The GlycoPOST system has been designed to make it easy to input various metadata such as experimental conditions and instrument settings (ion source, ion transfer optics, etc.) specific to glycomics. GlycoPOST is a part of the GlyCosmos portal (Yamada et al., 2020), which also includes UniCarb‐DR and GlyTouCan (Aoki‐Kinoshita et al., 2016) as associated repository systems. Because of this relationship between UniCarb‐DR and GlycoPOST, the authors have implemented a combined user registration system that handles user information for both repositories (Watanabe et al., 2021). Metadata should comply with the MIRAGE guidelines (York et al., 2014). Several of these guidelines, covering techniques such as mass spectrometry (Kolarich et al., 2013) and sample preparation (Struwe et al., 2016) have been published. The latest covers capillary electrophoresis (Lageveen‐Kammeijer, Rapp, et al., 2022).
The Carbohydrate Structure Database (CSDB), which has been in place for some 15 years, aims to incorporate the best features of other databases while avoiding their problems. A recent paper (Toukach & Shirkovskaya, 2022) summarizes other databases and outlines the main features of CSDB. The project features free access, annual data deposition and updates, search and correction of errors (including those in publications), and regular announcement of new services.
12.4. Tools for displaying structures
A useful discussion of various software tools for annotating and displaying glycan structures has been published in the book “Glycosylation,” part of the Methods in Molecular Biology series (Mariethoz et al., 2022). Unfortunately, there is heavy emphasis on the SNFG method (Varki et al., 2015) for drawing structures which, in this reviewer's opinion, is inferior to the so called “Oxford” system used in this review (Harvey et al., 2009b); which is more logical, is equally useful in black and white, and allows the reader to easily depict the structure of many newly discovered monosaccharide constituents without having to invent new symbols. This is because the Oxford system uses different shapes, not color, for the basic monosaccharides and shows modifications to these structures by additions such as a full fill for the presence of an N‐acetyl group, and the addition of one dot or two to show the presence of one (as in fucose) or two absent hydroxyl groups respectively. An additional advantage is that many new symbols drawn in this way can simply be “read” directly without the reader having to refer to a table of symbols, many of which do not follow a logical pattern. Linkage in the Oxford system is shown by the angle of the lines connecting the symbols. Although an advantage to the original SNFG system, which relied on the linkage being written on the bond, the Oxford linkage system is now recommended for use by the SNFG system. The colors used by the SNFG system for the monosaccharides have been incorporated into the Oxford system to make it more understandable for those not familiar with it.
12.5. Tools for annotating and displaying spectra
The software package Sweet‐SEQer (Serang et al., 2013) for annotation of tandem mass spectra, written in Python, has been modified and improved using C++. The new version, C‐SEQer, produces the same output but is claimed to do so in approximately 15‐fold less time than the Python version (Burgoyne & Smith, 2021).
A software package, termed Glycopeptide Abundance Distribution Spectra (GADS) has been developed for simplifying the visualization of glycopeptides for specific peptides (Remoroza et al., 2021). The presentation is in the form of a mass spectrum with glycopeptide peaks labelled with their glycan composition. The displayed mass (in place of m/z) of each peak is that of the glycan mass, and its abundance corresponds to its relative abundance in the electrospray MS1 spectrum. The method is illustrated with glycopeptides from several glycoproteins, including SARS‐CoV‐t spike protein. The software has been applied to human milk proteins (Remoroza et al., 2022) where two varieties of mass spectral libraries were generated. One contains GADS spectra, whereas the other contains tandem mass spectra of the underlying glycopeptides.
Mass Spectrum Peptide Annotation (MS_Piano) is software developed for annotation of peaks in CID spectra of peptides or N‐glycopeptides for given peptide sequences, charge states, and positional modifications. The program annotates each peak in high or low resolution spectra with possible product ion(s) and the mass difference between the measured and theoretical m/z values. Spectral quality is measured by two major parameters: the ratio between the sum of unannotated versus all peak intensities in the top 20 peaks, and the intensity of the highest unannotated peak. The software is freely available in .exe and .dll formats for the Windows operating system (Yang, Neta, et al., 2021).
To address the absence of a high‐throughput tool for visualization and molecular annotation of N‐glycans in MSI data, Veličković et al. (2021) have developed NGlycDB, a public database of N‐glycans based on METASPACE, an open‐source cloud engine for molecular annotation of MSI data to automatically annotate, visualize, analyze, and interpret high resolution mass spectrometry‐based spatial N‐glycomics data. Its applicability was demonstrated by analyzing MALDI‐MSI data from FFPE human kidney and murine lung tissue sections.
13. STUDIES ON SPECIFIC CARBOHYDRATE TYPES
13.1. Polysaccharides
Reviews and general articles are listed in Table 11.
Table 11.
reviews on the use of matrix‐assisted laser desorption/ionization for the analysis of polysaccharides.
| Subject | Notes | Citations | References |
|---|---|---|---|
| A comprehensive review on mutan (a mixed linkage of α‐1‐3 and α‐1‐6 glucans) from bacterial sources | General review (history, transferases, biosynthesis, analysis (little on MALDI), function) | 139 | Boddapati and Gummadi (2021) |
| Agar oligosaccharides: A review of preparation, structures, bioactivities and application | General review, brief MALDI references. Mainly biological activity | 60 | Chen, Fu, et al. (2021) |
| Monosaccharide composition analysis | Mainly GC/MS with protocols. Brief mention of MALDI | 26 | Gerwig (2021d) |
| Carrageenan oligosaccharides: A comprehensive review of preparation, isolation, purification, structure, biological activities and applications | Production of oligosaccharides, separation methods, structural identification (MALDI, NMR), biological activity | 186 | Guo, Wei, et al. (2021) |
| Recent advances in qualitative and quantitative analysis of polysaccharides in natural medicines: A critical review | Emphasis given to depolymerisation of polysaccharides to oligosaccharides and their subsequent analysis | 100 | Li, Zhang, Han, et al. (2022) |
| Enzymatic synthesis and characterization of different families of chitooligosaccharides and their bioactive properties | Includes use of MALDI for characterization of chitooligosaccharides | 87 | Miguez et al. (2021) |
| Date (Phoenix dactylifera L.) polysaccharides: A review on chemical structure and nutritional properties | Structure, extraction, identification and biological effects | 89 | Noorbakhsh and Khorasgani (2022) |
| The cell wall of hornworts and liverworts: Innovations in early land plant evolution? | Presents an overview on shared and divergent polysaccharide features between these two groups of bryophytes and vascular plants. | 175 | Pfeifer et al. (2022) |
| Exploiting the amazing diversity of natural source‐derived polysaccharides: Modern procedures of isolation, engineering, and optimization of antiviral activities | Concentrates on sulfated polysaccharides. Comparison of extraction techniques. Few references to MALDI | 230 | Ray et al. (2021) |
| Plants arabinogalactans: From structures to physico‐chemical and biological properties | Structure, occurrence in different plant parts, extraction, purification, properties. Some discussion of MALDI | 231 | Saeidy et al. (2021) |
| Alginate derived functional oligosaccharides: Recent developments, barriers, and future outlooks | Compound separation, purification, analysis and biological properties | 256 | Vasudevan et al. (2021) |
| Recent advances in marine algae oligosaccharides: Structure, analysis, and potential prebiotic activities | Brief review, structure and activity. MALDI analysis listed in table | 124 | Xie and Cheong (2022) |
| Recent research advances in polysaccharides from Undaria pinnatifida (edible seaweed): Isolation, structures, bioactivities, and applications | Contains large tables citing references to isolation, structural characterization and biological activity | 140 | Zeng et al. (2022) |
Two papers are highlighted. Using an FT‐ICR instrument in positive ion mode, Nicolardi et al. (2021) have recorded the MALDI spectra from super‐DHB (s‐DHB) of several large polysaccharides and confirmed their structure by observation of ISD fragments where both glycosiodic and cross‐ring fragments were observed. Polysaccharides included a 16‐mer chain with a [(→4)‐Rha‐α‐(1→3)‐Man‐β‐(1→) repeat ([M + Na]+, monoisotopic, m/z 9900.573 (calc. m/z 9900.543)), a 100‐mer linear polymannoside ([M + Na]+ major isotopic peak, m/z 16339.680 (calc. m/z 16339.397)) and a 151‐mer branched polymannoside (M + Na]+, average, m/z 24610.58)).
Depolymerization of plant polysaccharides with periodate at 121oC has been used to provide a unique MALDI‐TOF fingerprint profile of all investigated polysaccharides except xyloglycan (Pandeirada et al., 2022). The method was able to differentiate polysaccharides such as birch wood xylan vs wheat arabinoxylan vs rye arabinoxylan, and guar galactomannan vs locust bean galactomannan. Principal component analysis and hierarchical cluster analysis of the MALDI‐TOF MS data highlighted the structural heterogeneity of the polysaccharides.
Applications of MALDI to the analysis of polysaccharides in plants, animals and algae are listed in Table 12 and in lower organisms are in Table 13.
13.2. Milk sugars
Four reviews are relevant and are listed in Table 14.
Table 14.
Reviews and general articles on analysis of milk.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Oligosaccharides in human milk, achievements in analysis: A review | Short review | 40 | Belusko et al. (2022) |
| Human milk oligosaccharides: Structure and functions | Chapter from the 94th Nestlé Nutrition Institute workshop | 24 | Bode (2020) |
| Structural and functional aspects of milk oligosaccharides | Health benefits and analysis | ‐ | Debnath et al. (2022) |
| Evolution of milk oligosaccharides: Origin and selectivity of the ratio of milk oligosaccharides to lactose among mammals | Discusses structures of milk sugars in many different species | 182 | Urashima et al. (2022) |
Liou et al. (2021) have used carbon‐dioxide supercritical fluid chromatography (SFC), coupled with both evaporative light scattering detectors and UV‐vis detectors to separate 18 human milk glycans attached to an azidohexyl linker ((CH2)6‐N3). The authors were able to separate regioisomers and connectivity isomers which is a major limitation currently associated with carbohydrate analysis. The oligomers, with compositions ranging from disaccharides to hexasaccharides were well separated within 10 min.
A new class of milk sugars containing a (Gal)3 chain without (190) and with (191) an additional fucose residue has been identified in human milk (Hanisch & Kunz, 2021). The glycans are thought to be the first to be observed with branching on the 6‐arm of the terminal galactose of the core galactosyl‐lactose moiety.

Other publications on the application of MALDI MS to the analysis of milk products are listed in Table 15.
13.3. Glycoproteins
MALDI has had a considerable impact on the analysis of glycoproteins and their attached glycans. Because of the complexity of these compounds, several processes are involved in their analyses. Glycoproteins with few glycosylation sites and a limited number of glycans at each site can now be resolved with high‐resolution instruments, but this method only gives a composition for the glycans following subtraction of the protein mass. Although not involving MALDI‐MS, the review on high‐resolution native mass spectrometry by Tamara et al. (2022), contains much useful information on the mass spectrometry of intact glycoproteins. The type (mainly N‐ and O‐linked) and attachment sites of the glycans are usually determined by analysis of derived glycopeptides following enzymatic hydrolysis, typically with trypsin, but structural analyses of the glycans themselves are usually determined following their release from the protein by chemical or, more commonly, enzymatic means. Such glycan analyses involves their composition and the determination of linkage and branching patterns between the constituent monosaccharides. Finally, all the individual pieces of information are combined to give the complete structure. General articles and reviews on glycoproteins and their analysis are listed in Table 16.
Table 16.
Reviews and general articles on the analysis of glycoproteins.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Glycoproteomics | Glycoproteomic methods including sample selection; techniques for protein isolation, proteolytic digestion, glycopeptide enrichment and MS fragmentation | 449 | Bagdonaite et al. (2022) |
| Research progress in structure‐specific N‐glycoproteomics | Covers basic analytical procedures. In Chinese | 57 | Bi and Tian (2021) |
| The emerging role of cellular posttranslational modifications in modulating growth and productivity of recombinant Chinese hamster ovary cells | General review of glycosylation, phosphorylation and ubiquitination. | 287 | Bryan et al. (2021) |
| Site‐specific glycosylation of SARS‐CoV‐2: Big challenges in mass spectrometry analysis | N‐ and O‐glycans, Compares results from different analysis software. Little on MALDI | 105 | Campos et al. (2022) |
| Qualitative and quantitative analytical methods for intact glycopeptides | Book chapter – general overview | ‐ | Cao and Yang (2022) |
| Quantitative characterization of O‐GalNAc glycosylation | Short review several MS references but few that mention MALDI directly | 51 | Čaval et al. (2021) |
| Towards structure‐focused glycoproteomics | Covers literature for period 2018‐2020 | 254 | Chernykh et al. (2021) |
| Seeing the forest through the trees: Characterizing the glycoproteome | Emphasises the importance of studying intact glycoprotein | 102 | Critcher et al. (2022) |
| Developments and perspectives in high‐throughput protein glycomics: Enabling the analysis of thousands of samples | Summary of current high‐throughput methods and some applications | 92 | de Haan, Pučić‐Baković, et al. (2022) |
| Advances in mass spectrometry‐based glycomics—An update covering the period 2017–2021 | Glycan release, purification, derivatization, glycan separation, MS ionization, quantitation, bioinformatics | 211 | Donohoo et al. (2022) |
| Carbohydrate analysis of glycoconjugates | Short general review with protocols. Glycan release and analysis of glycopeptides | 110 | Gerwig (2021f) |
| Structural characterization of released glycans | N‐glycans (exoglycosidase digestion), O‐glycans, mucins, (with protocols) | 34 | Gerwig (2021e) |
| Analysis of sialic acids | General summary with protocols. Mass spectrometry but very brief on linkage‐specific methods. | 32 | Gerwig (2021h) |
| LC‐MS/MS in glycomics and lycoproteomics analyses | Derivatization including linkage‐specific methods. Separation methods. Software | 202 | Goli et al. (2021) |
| The glycosylation in SARS‐CoV‐2 and its receptor ACE2 | Comprehensive review, N‐ and O‐glycosylation | 477 | Gong et al. (2021) |
| Glycosylation analysis | Chapter in book on monoclonal antibodies. Biological effects and analysis | 152 | Gstöttner, Kaur, et al. (2021) |
| N‐Glycosylation of milk proteins: A review spanning 2010–2022 | Analysis protocol. Table of studies. Biological properties | 107 | Guan et al. (2022) |
| Glycomics and glycoproteomics: Approaches to address isomeric separation of glycans and glycopeptides | LC‐MS techniques, derivatization for sialic acid linkage. Table of methods | 150 | Gutierrez Reyes et al. (2021) |
| Advances in mass spectrometry‐based glycoproteomics: An update covering the period 2017‐2021 | Metabolic labelling, enrichment, derivatization, quantification, ion mobility, bioinformatics | 189 | Gutierrez‐Reyes et al. (2022) |
| Glycan nanostructures of human coronaviruses | Comprehensive review with sections on each type of virus | 119 | Guo, Lakshminarayanan, et al. (2021) |
| Mass spectrometry‐based methods for immunoglobulin G N‐glycosylation analysis | Very comprehensive review. Mass spec, instrumentation, sample preparation, fragmentation, MALDI, LC/MS, CE | 272 | Habazin et al. (2021) |
| Calculating glycoprotein similarities from mass spectrometric data | Reviews analytical and statistical methods for determining glycoprotein molecular similarities from glycoproteomics data. | 77 | Hackett and Zaia (2021) |
| Automation of immunoglobulin glycosylation analysis | Mainly automation of methods for sample preparation | 52 | Hendel et al. (2021) |
| Negative‐mode mass spectrometry in the analysis of invertebrate, fungal, and protist N‐glycans | Emphasises some of the advantages of using negative ion MS for structural identification. Mainly MALDI applications | 75 | Hykollari et al. (2022) |
| Recent progress of analytical methods of proteomics based on mass spectrometry | Identification and quantitation. In Chinese | 111 | Ji, Fu, et al. (2021) |
| A mass spectrometry‐based glycotype‐centric cellular glycomics is the more fruitful way forward to see the forest for the trees | General review with comments. Native glycans, sialic acid derivatization, MSn reactions | 120 | Khoo (2021) |
| Recent advances and trends in sample preparation and chemical modification for glycan analysis | Comprehensive review of glycan release, glycan enrichment, derivatization, use of stable isotopes | 189 | Kinoshita and Yamada (2021) |
| High sensitivity glycomics in biomedicine | Glycan sample preparation, clean‐up, analysis (CE‐MS, LC‐MS, PGC‐LC‐MS, ion mobility, MALDI), analysis of glycopeptides and glycoproteins | 191ü | Lageveen‐Kammeijer, Küster, et al. (2022) |
| Analysis of glycosylation of IgG using mass spectrometry and its application | Contains discussion of several mass spectrometry methods (in Chinese) | 146 | Lai, Zhou, et al. (2021) |
| Mass spectrometry‐based analysis of IgG glycosylation and its applications | Intact IgG, glycopeptides and released glycans | 132 | Liu, Sun, et al. (2022) |
| Analytical and biochemical perspectives of protein O‑GlcNAcylation | Research over 35 years. Protein characterization. MS methods (ionization and fragmentation), enzyme characterization | 680 | Ma, Wu, et al. (2021) |
| Protein glycosylation in extracellular vesicles: Structural characterization and biological functions | General review. Many MALDI papers listed in tabular form | 258 | Macedo‐da‐Silva et al. (2021) |
| The hitchhiker's guide to glycoproteomics | General review (glycopeptide preparation, purification, MS fragmentation) | 215 | Oliveira et al. (2021) |
| A perspective on the Protein Data Bank's impact on the field of glycobiology | Discusses under‐representation of structures containing glycans | 68 | Prestegard (2021) |
| SARS‐CoV‐2 | Combined with review on glycopeptide enrichment and derivatization | 106 | Pujić and Perreault (2022) |
| Progresses in mass spectrometry‐based plant and algae N‐glycomics and N‐glycoproteomics | Contains tables listing details of methods used and results | 90 | Qin, Qin, et al. (2022) |
| Qualitative and quantitative methods for N‐glycans in N‐glycomics | Covers ample pretreatment protocols N‐glycan release, purification and enrichment, separation, derivatization and some common qualitative and quantitative analysis strategies | 120 | Ren and Lu (2022) |
| N‐Glycoproteins in plant cell walls: A survey | Brief review covering structure of N‐glycans, overview of analytical methods, role of N‐glycans | 80 | San Clemente and Jamet (2022) |
| Quantitative methods for N‐glycosite containing peptides in N‐glycoproteomics | Book chapter – Reviews recently developed quantitative approaches in glycoproteomics | ‐ | Sun, Zhang and Lu, (2022) |
| High‐throughput glycomic methods | Comprehensive review. Historical overview. Discussion of various techniques including sample preparation for each one (HPLC, CE, MS, lectin microarrays), data processing, applications | 438 | Trbojević‐Akmačić et al. (2022) |
| mAbs N‐glycosylation: Implications for biotechnology and analytics | Small section on analytical methods | 181 | Wang, Liu and Voglmeir, et al. (2022) |
| Glycomics, glycoproteomics, and glycogenomics: An inter‐taxa evolutionary perspective | Discusses current glycomic, glycoproteomic, and glycogenomic methods to characterize protein glycosylation in less‐well‐studied eukaryotes | 126 | West, Malzl, et al. (2021) |
| The role of data‐independent acquisition for glycoproteomics | N‐ and O‐glycosylation, oxonium ions and limitations of the technique | 78 | Ye and Vakhrushev (2021) |
13.3.1. Isolation and concentration of glycoproteins and glycopeptides
Enrichment of glycoproteins, glycopeptides and purification of released glycans is an essential aspect of a successful structural analysis of these compounds and many methods have been devised; Table 17 lists a number of reviews. A few of the materials exhibit novel properties but most rely on established methods. Formation of boronate esters with compounds bearing cis‐diol groups is a popular method with the advantage that the boronate rings can easily be cleaved to release the free glycan. Other methods include hydrophilic attachment where many materials have been used. Of particular significance are zwitterionic materials, which are particularly useful for glycopeptides. Metal‐organic frameworks, with their high surface area, are also useful. Cotton has also been used, particularly for purification of N‐glycans and lectins provide a method for fractionating different types of these glycans. A protocol: “Enrichment of intact glycopeptides using strong anion exchange and electrostatic repulsion hydrophilic interaction chromatography” has been published (Bermudez & Pitteri, 2021).
Table 17.
Reviews on methods for glycoprotein and glycan enrichment.
| Subject | Notes | Citations | References |
|---|---|---|---|
| Recent strategies for using monolithic materials in glycoprotein and glycopeptide analysis | Use in chromatographic methods and for glycopeptide enrichment | 158 | Alla and Stine (2022) |
| Advances in enrichment methods for mass spectrometry‐based proteomics analysis of posttranslational modifications | Covers glycosylation and other PTMs such as phosphorylation and acetylation | 117 | Brandi et al. (2022) |
| Improving the study of protein glycosylation with new tools for glycopeptide enrichment | Materials used for glycan and glycopeptide enrichment classified by type | 53 | Chen, Dupard, et al. (2021) |
| Application of magnetic solid phase extraction in separation and enrichment of glycoproteins and glycopeptides | Covers categories of magnetic adsorbents and applications to human body fluids | 83 | Qi et al. (2021) |
| A review on recent advances in the enrichment of glycopeptides and glycoproteins by liquid chromatographic methods: 2016‐Present. | Nanoparticles, chromatographic methods, lectin affinity, metal‐organic frameworks | 128 | Kumari and Tetala (2022) |
| Recent progress and application of boronate affinity materials in bioanalysis | Uses in sample preparation and guidance for designing for specific requirements | 207 | Li, He, et al. (2021) |
| Advances in glycopeptide enrichment methods for the analysis of protein glycosylation over the past decade | Biological roles of glycosylation, analytical workflows, boronate affinity, O‐glycosylation | 143 | Li, Zhang, Xu, et al. (2022) |
| Methods for enrichment and assignment of N‐acetylglucosamine modification sites | Use of lectins, labelling methods, immunoprecipitation, MS analysis and software | 56 | Maynard and Chalkley (2021) |
| Recent advancements in glycoproteomic studies: Glycopeptide enrichment and derivatization | Briefly discusses different types of enrichment materials (combined with derivatization and SARS‐CoV‐2 glycosylation) | 106 | Pujić and Perreault (2022) |
| A guide to enrichment strategies for mass spectrometry–based glycoproteomics | Use of glycosidases, metal affinity chromatography, hydrophilic interaction chromatography, use of PGC and chemical coupling methods | 512 | Riley et al. (2021) |
| Simultaneous application of nanomaterials to separation of phosphorylated and glycosylated proteins | Book chapter, discusses methods such as immobilized metal affinity chromatography and metal oxide affinity chromatography | 27 | Sun, Deng and Shen (2021a) |
| Application of nanomaterials to separation of glycosylated proteins | Book chapter, discusses different carbohydrates and different chemical types (amino acids etc.) as functional groups, boronate affinity materials | 233 | Sun, Deng and Shen (2021b) |
| Selective enrichment methods for N‐glycosite containing peptides in N‐glycoproteomics | Current representative methods for glycoproteins/glycopeptides enrichment are summarized with discussion of advantages and limitations | 94 | Wang, Zhang and Lu (2022) |
| Advances in proteomic sample preparation and enrichment for phosphorylation and glycosylation analysis | Categorises enrichment methods by type of adsorbent | 134 | Xie, Feng, Zhang, et al. (2022) |
| Chemistry of magnetic covalent organic frameworks (MagCOFs): From synthesis to separation applications | General review with section on use for glycopeptide enrichment | 179 | Yadav et al. (2022) |
A method for sialoglycopeptide enrichment, but which modifies the glycan, involves periodate oxidation, coupling with an alkyn‐containing hydrazide and click chemistry was employed to link the derivatized glycopeptides to Dde‐Azide or PEG‐azide resin (Li, Huang, et al., 2022). After centrifugation to isolate the resin‐bound glycopeptides, the resin was removed by incubation with hydrazine. The derivatized glycopeptides could then be examined by MALDI‐TOF MS or LC/MS.
Cai, Ren, et al. (2022) have developed a method which they call Ultrafast Glycoprotein Immobilization for Glycan extraction (UltraGIG) in which proteins are captured with NHS‐activated agarose resin via amide linkages. Contaminating compounds, salts, and so forth could then easily be removed and the glycans recovered by enzymatic cleavage. The method was used to study urinary N‐glycans in patients with diabetic kidney disease.
Table 18 lists 88 of the other materials that have been reported for purification and isolation procedures during the review period.
Table 18.
Materials and methods used for the enrichment of carbohydrates, glycoproteins and glycopeptides.
| Method | MALDIa | Materials | References |
|---|---|---|---|
| Boronate‐based methods | |||
| Boronoisophthalic acid | L‐TOF (SA) | Glycoproteins, human milk | Ali, Hussain et al. (2021) |
| Phenylboronate functionalized magnetic nanoparticles | TOF/TOF (CHCA) | Low molecular weight glycoproteins | Dou et al. (2021) |
| Encapsulated magnetic nanoparticles with a polymer containing boronic acid groups | TOF | HRP Glycoprotein | Gharaghoushi et al. (2022) |
| Boronic acid‐functionalized mesoporous graphene−silica composites | TOF/TOF (DHB), LC/MS | N‐ and O‑linked glycopeptides (IgG, human serum) | Kong et al. (2021) |
| 6‐Aminopyridine‐3‐boronic acid functionalized magnetic nanoparticles | MEKC | Cis‐diol‐containing biomolecules (HRP, human urine). Compounds not specified | Li and Dong (2021) |
| Boronate‐immobilized cellulose nanofiber‐reinforced cellulose microspheres (pH‐dependent) | UV | Glycoproteins (ovalbumin) | Li, Qiao, et al. (2022) |
| Boronate affinity sorbents based on thiol‐functionalized polysiloxane‐polymethacrylate composite materials | TOF/TOF (CHCA) | Glycopeptides (HRP, human serum) | Mompó‐Roselló et al. (2021) |
| Boric acid‐functionalized metal–organic frameworks | TOF | Glycopeptides (HRP), serum of cervical cancer patients | Rao et al. (2022) |
| Boric acid imprinted magnetic nanoparticles | TOF | Glycoproteins (HRP, ovalbumin) | Wang, Duan, et al. (2021) |
| Boric acid–functionalized magnetic covalent organic framework | TOF (DHB) | N‐Glycopeptides (HRP, human saliva) | Wang, Liu, Yan, et al. (2021) |
| Covalent organic framework material rich in boronic acid sites | TOF (DHB) | Glycopeptides (HRP, human saliva and serum) | Xie, Yan, et al. (2022) |
| Electrochemical sensor with surface imprinted boric acid | Electrochemical | P‐Glycoproteins | Yang, Song, et al. (2022) |
| Hollow MnFe2O4@C@APBA nanospheres | TOF | Glycopeptides | Zhang, Jin, et al. (2021) |
| Hydrazide‐based methods | |||
| Chemical oxidation and reversible hydrazide chemistry | TOF/TOF (DHB), LC‐MS/MS | O‑GlcNAc Glycopeptides | Chen, Qin,et al. (2021) |
| Graphite and carbon‐based methods | |||
| Magnetic porous carbon‐dependent platform | TOF | N‐Glycans (ovalbumin, urinary exosomes) | Wu, Zhang, et al. (2021) |
| Carbohydrate‐functionalized materials | |||
| Bi‐amino acid functionalized biomimetic honeycomb chitosan membrane | TOF, LC‐MS/MS | N‐Glycopeptides (HRP), nasopharyngeal carcinoma serum | Fu et al. (2022) |
| Carrageenan functionalized magnetic carbon‐based framework | TOF (DHB) | N‐Glycopeptides from human saliva (IgG) | Jin, Zhu, et al. (2021) |
| Glycosyl imprinted mesoporous microspheres | LC/MS | Glycopeptide antibiotics | Tan et al. (2021) |
| Hydrophilic glucose functionalized quantum dots | TOF (DHB) | Glycopeptides (HRP, diabetic serum) | Xie, Feng, Fang, et al. (2022) |
| Absorbent cotton | LC‐MS/MS | Glycopeptides (mouse brain, seminal plasma) | Xin, You, et al. (2022) |
| Amino acid and peptide‐functionalized materials | |||
| Magnetic binary metal oxide composites with hydrophilic tripeptide | TOF | Glycopeptides (HRP) | Chu et al. (2022) |
| Amino acid–functionalized zinc sulphide quantum dots | R‐TOF (DHB) | N‐Glycopeptides (HRP, human saliva) | Feng et al. (2022) |
| Glutathione‐functionalized two‐dimensional cobalt sulphide nanosheets | R‐TOF/TOF (DHB/H3PO4) | N‐Glycopeptides (HRP, IgG, human serum) | Gao, Bai, et al. (2021) |
| Covalent organic frameworks with glutathione and cysteine (denoted as COF‐S@Au@GC) | TOF/TOF (DHB) | Glycopeptides (HRP), glycopeptides from serum exosomes | Hua et al. (2022) |
| Asparagine immobilized cellulose/polymer nanohybrid | TOF/TOF (DHB) | N‐Glycans (ovalbumin, IgG, human serum) | Sajid, Saleem, Jabeen, Saleem, et al. (2022) |
| O‐Phospho‐l‐serine‐poly(glycidyl methacrylate‐co‐ethylene dimethacrylate) microspheres | TOF/TOF (DHB) | N‐Glycopeptides (IgG, HeLa glycoproteins) and phosphopeptides | Tang, Yu, et al. (2021) |
| Dandelion‐like silica nanoparticles modified with l‐glutathione | TOF/TOF (DHB) | N‐Glycopeptides (IgG, human serum) | Tian et al. (2021) |
| Tannic acid and l‐cysteine functionalized magnetic composites | TOF/TOF (DHB) | N‐glycopeptides (HRP, human serum) | Wang, Xu, et al. (2022) |
| Glutathione‐functionalized magnetic thioether‐covalent organic frameworks | TOF | Glycopeptides (HRP, exosomes) | Xiong, Jia, et al. (2022) |
| β‑Amyloid peptide 1−42‐conjugated magnetic nanoparticles | LC‐MS/MS | Glycoproteins (egg white) | Zhen et al. (2021) |
| Hydrophilic arginine‐functionalized mesoporous polydopamine‐graphene oxide composites | R‐TOF (DHB) | Glycopeptides (IgG) | Zheng, Pu, et al. (2022) |
| Dipeptide‐based polymeric material | TOF | Glycoproteins (IgG, HRP) | Zheng, Zhang, et al. (2022) |
| Graphene functionalized with structurally complementary amino acids | TOF, LC‐MS/MS | N‐Glycopeptides (HRP, human saliva and serum) | Zhu, Wu, et al. (2021) |
| Metal‐organic frameworks | |||
| Melamine foam assisted in‐tip packed amine‐functionalized titanium metal–organic framework | TOF (CHCA, SA) | Glycopeptides (HRP, ovalbumin, IgG, human saliva) | Ali, Zhu, Wang, et al. (2021) |
| Metal‐organic framework (MF@PDA@UiO‐66‐NH2 composite) | TOF/TOF (CHCA) | Glycopeptides (HRP) | Ali, Zhu, Hussain, et al. (2021) |
| Gold nanoparticle‐glutathione functionalized metal‐organic frameworks | R‐TOF (DHB) | Glycopeptides (HRP, human saliva and serum) | Wu, Jin, et al. (2021) |
| Hydrophilic hollow zirconium organic frameworks | TOF | Glycopeptides (HRP) | He, Zheng, et al. (2022) |
| Zwitterionic dual‐functional metal‐organic framework nanocomposite.(In Chinese) | TOF (DHB) | Glycopeptides (HRP) | Li, Xie, et al. (2021) |
| Graphene oxide/chitosan foam incorporated with metal–organic framework | TOF/TOF (DHB) | Glycopeptides (HRP) | Liu, Gao, et al. (2022) |
| Bifunctional magnetic covalent organic framework | TOF (DHB) | Glycopeptides (IgG, rat liver) | Luo et al. (2021) |
| Fe3O4@SiO2@(ZreTi‐MOF)10‐NH2 Dual‐functionalized magnetic bimetallic metal‐organic framework composite | TOF/TOF (DHB), ESI‐MS/MS | Glycopeptides (IgG, human serum) | Pan, Zhang, Xiao, et al. (2021) |
| Magnetic dual‐hydrophilic metal organic framework | TOF (DHB) | N‐glycopeptides (HRP) | Su, Wang, et al. (2021) |
| Glutathione functionalized magnetic covalent organic frameworks | TOF (CHCA) | Glycopeptides (HRP) | Su et al. (2022) |
| Hydrophilic MOFs‐303‐functionalized magnetic probe | TOF (DHB) | Glycopeptides (HRP) | Wang, Wang, Li, et al. (2022) |
| Gold nanoparticle‐glutathione functionalized MOFs | TOF (DHB) | Glycopeptides (HRP) | Wu, Jin, et al. (2021) |
| Hydrophilic hierarchical porous metal‐organic frameworks | TOF (DHB), LC‐MS/MS | Glycopeptides (IgG) | Zhu, Gu, et al. (2022) |
| Lectins | |||
| α‐Mannose‐specific Burkholderia cenocepacia lectin A | LC‐MS/MS | C‑ and O‑mannosylated peptides | Hütte et al. (2022) |
| Zwitterionic materials | |||
| Zwitterionic polymer modified graphene oxide | TOF/TOF (DHB) | Glycopeptides from urine of healthy subjects and patients with lung adenocarcinoma | Bai et al. (2022) |
| Zwitterionic HILIC with exposed choline group | LC‐MS/MS | Sialoglycopeptides | Chen, Yen, et al. (2021) |
| ZIC‐cHILIC functionalized magnetic nanoparticle | L‐TOF/TOF (CHCA) | Glycopeptides (HRP, fetuin) | Pradita et al. (2021) |
| Zwitterionic carboxybetaine‐based hypercrosslinked polymers | TOF/TOF | Glycopeptides (IgG) | Sun, Xu, et al. (2022) |
| Zwitterionic sulfobetaine vinylimidazole‐based monoliths | TOF/TOF | Glycopeptides (IgG) | Wang, Sun, Wu, et al. (2022) |
| Zwitterionic microspheres (HILIC mode) | TOF (DHB), LC‐MS/MS | N‑Glycopeptides (IgG, human serum) | Wu, Tang, et al. (2021) |
| Zwitterionic‐HILIC (ZIC‐HILIC) nanosphere (Fe3O4‐CG) | TOF (DHB) | Glycopeptides (HRP, Alzheimer's disease patients’ serum) | Yi, Shao, et al. (2022) |
| Other methods | |||
| MXene cartridge (Ti3C2) | TOF/TOF (DHB) | N‐Glycans | Aguedo et al. (2022) |
| Mesoporous covalent organic framework microspheres | TOF/TOF | Glycopeptides (IgG) | Ba et al. (2022) |
| TiO2 | LC‐Q‐TOF | Glycopeptides, simultaneous enrichment, on‐line deglycosylation | Chen, Zhang, Dong, et al. (2021) |
| PAMAM dendrimer‐assisted 3‐carboxybenzoboroxole‐functionalized magnetic nanoparticles | TOF/TOF | Glycoproteins (HRP, human saliva) | Fan, Yang, Huang, et al. (2022) |
| Three hydrophilic poly(glycidyl methacrylate‐co‐ethylene glycol dimethacrylated macroporous adsorbent resins | TOF, LC‐MS | N‐Glycopeptides (IgG, human serum) | Gao, Tang, et al. (2021) |
| Highly crosslinking core–shell magnetic (Fe3O4) nanocomposites | SDS, UV | Glycoproteins (IgG) | Guo, Yao, et al. (2022) |
| Dual‐functional Ti(IV)‐IMAC material | TOF/TOF (DHB) | Glycopeptides (RNase B, mouse lung) | Huang, Liu, et al. (2021) |
| EDMA‐co‐VPBA‐co‐VPA) monolith) | TOF | Glycopeptides (HRP, human serum) | Huang, Zheng, et al. (2021) |
| Hydrophilic hydrogel with a 3D network structure (Zn2+/SAP) | TOF/TOF (DHB) | N‐glycopeptides (HRP) | Jin, Gao, et al. (2022) |
| Magnetic polyaniline nanomaterial (Fe3O4@PANI) | FT‐ICR (DHB) | N‐Glycopeptides (ovine fetuin, transferrin, haptoglobin) | Lai, Zhang, et al. (2021) |
| Nitrogen‐rich linear porous organic polymers | TOF/TOF (DHB), LC‐MS/MS | Glycopeptides (IgG) | Li, Xu, et al. (2022) |
| Titanium (IV) ion affinity chromatography materials | ESI‐Q‐TOF | O‐Glycopeptides (fetuin) | Li, Dong, et al. (2022) |
| Hydrophilic magnetic mesoporous silica microspheres | LC/MS | N‐Glycopeptides, N‐glycans (HRP, human serum) | Liu, Ma, He, et al. (2021) |
| HILIC HPLC, automated method | TOF/TOF (DHB), LC‐MS | N‐Glycopeptides (IgG, human serum) | Liu, Zhu, et al. (2021) |
| Cyclen‐containing hydrophilic polymeric monolithic materials | TOF/TOF, LC‐MS/MS | N‐Glycopeptides (IgG, human serum) | Ma, Tang, et al. (2021) |
| (Thio)urea and crown ether polymer | TOF (DHB) | Sialylated glycopeptides (bovine fetuin) | Mavliutova et al. (2021) |
| Iminodiacetic acid (IDA)‑generated mesoporous nanopolymer | TOF/TOF (DHB), LC‐MS/MS | Glycopeptides (HRP, IgG, human serum) | Sajid et al. (2021) |
| Methyl methacrylate/ethylene glycol dimethacrylate/1,2‐epoxy‐5‐hexene polymer plus cysteic acid | TOF/TOF (DHB), LC‐MS/MS | N‐glycopeptides (HRP, serum glycoproteins) | Sajid, Saleem, Jabeen, Najam‑ul‑Haq, et al. (2022) |
| Zirconium modified adenosine triphosphate functionalized monolith | CE‐LIF | N‐Glycans (RNase B) | Shao et al. (2021) |
| Al3+‐doped‐TiO2 monodisperse microspheres | TOF/TOF (DHB) | Glycopeptides (IgG, α‐casein, human serum, nonfat milk) | Sheng, Xue, et al. (2021) |
| Hydrophilic graphene oxide‐dopamine‐cationic cellulose composites | TOF/TOF (DHB) | Glycopeptides (IgG, human serum) | Sheng, Li, et al. (2021) |
| Nanoparticle biomolecular corona‐based enrichment | LC‐MS/MS | Glycoproteins (fibrinogen) | Trinh et al. (2022) |
| Dopamine/graphene oxide linked to trypsin for hydrolysis and enrichment | LC‐MS/MS | Glycopeptides | Wang, Zhang, Wei, et al. (2022) |
| Core‐shell microporous organic polymer‐coated silica microspheres | TOF, LC‐MS/MS | N‐Glycopeptides | Wang, Tang, et al. (2022) |
| Thiazolidine modified magnetic nanoparticles | TOF | Glycated peptides | Wu, Fei, et al. (2022) |
| Fluorescent molecular imprinted polymers | Fluorescence | Glycoproteins (ovalbumin) | Xie, Li, et al. (2022) |
| Strongly hydrophilic mesoporous silica (Fe3O4@mSiO2‐TSG) | R‐TOF/TOF | Glycopeptides (HRP) | Xu, Wu, et al. (2021) |
| Hydrophilic mesoporous channel coupled with metal oxide, Fe3O4@TiO 2@mSiO2 ‐TSG nanomaterial | TOF | Glycopeptides (HRP, IgG, salivary glycopeptides) | Xu, Wu, et al. (2022) |
| Polyoxometalate‐covalent organic framework conjugate | SDS‐PAGE | Glycoproteins, phosphoproteins | Xu, Cao, et al. (2022) |
| Bowl‐like mesoporous polydopamine | TOF/TOF (DHB) | Glycopeptides (HRP) | Yan et al. (2021) |
| Hydrophilic nano‐floral inter‐polymeric material | TOF (DHB) | Glycopeptides (HRP) | Yang, Gao, et al. (2022) |
| Bifunctional super‐hydrophilic mesoporous nanocomposite (mTiO2 @AuCG) | TOF | Glycopeptide (HRP), phosphopeptides | Yi, Fu, et al. (2022) |
| Immobilized metal ion affinity chromatography | LC‐MS/MS | O‑GalNAc glycopeptides (human serum) | Yue et al. (2021) |
Format (not all items present): MALDI method (matrix), other methods.
13.3.2. Problems encountered during sample preparation
Morgenstern et al. (2022) have commented on the fact that boronate methods of glycan enrichment often suffer from poor performance. On investigation, they found that the choice of buffer made a major difference to the method's performance. By eliminating amine‐containing buffers, glycan yields could be improved by as much as 10‐fold.
Despite its widespread use, hydrophilic enrichment methods are associated with several problems including the need for relatively large amounts of starting materials, potential introduction of chemical artefacts such as formylation when high concentrations of formic acid are used, and biasing or under‐sampling of specific classes of compound such as O‐linked glycopeptides. Izaham et al. (2021) have investigated these shortcomings for the study of bacterial glycoproteomes using three Burkholderia species (B. cenocepacia, B. Dolosa, and B. ubonensis), confirming that short aliphatic O‐linked glycopeptides are typically absent from hydrophilic interaction liquid chromatographic (HILIC) enrichments, yet are readily identified in whole proteome samples. Using high‐field asymmetric waveform ion mobility spectrometry (FAIMS) fractionation, they showed that at high compensation voltages, these compounds can be enriched from complex samples, providing an alternative method to HILIC enrichment.
Glycans and glycoproteins have been observed to develop an artefactual compound that produces a peak 28 mass units above that of the target compounds, which slowly increases in abundance when the samples were stored at −20oC. The peak was not observed in samples stored at room temperature, +4oC, −80oC, or −196oC. A corresponding reaction was not observed with acetic of trifluoroacetic solutions. The cause of the peak was traced to formylation of one of the hydroxyl groups on the glycan but it was not clear why the reaction was only observed at −20oC (Zhi et al., 2022).
The position of acetyl groups on the 7, 8 or 9 positions of sialic acids (192) influences the extent to which some pathogenic viruses bind. It has recently been reported (Oh et al., 2022) that acetyl groups can migrate between these positions in the presence of base making it important to control the conditions during sample preparation if determination of the position of such acetylation is important.

13.3.3. N‐glycans
13.3.3.1. Analysis of intact glycoproteins
The high resolution capabilities of FT‐ICR instruments are now sufficient to resolve glycoforms of many glycoproteins as shown in Figure 4 for erythropoietin (EPO), a glycoprotein with three N‐glycosylation sites occupied mainly by bi‐, tri‐, and tetra‐antennary complex glycans (Lippold et al., 2021). The MALDI‐FT‐ICR spectra were obtained from 2,5‐DHAP, a matrix which minimizes loss of sialic acid and the spectrum of the doubly charged ion before and after incubation with sialidase is shown in Figure 4A. Figure 4B shows the corresponding MALDI‐TOF spectrum illustrating the much poorer resolution.
Figure 4.

(A) MALDI‐FT‐ICR spectra of the doubly charged ion from EPO. (B) The spectrum of the glycans following desialylation. Spectra C and D are the corresponding MALDI‐TOF sprectra. From Lippold et al. (2021) with permission from Elsevier.
13.3.3.1.1. Use of mass spectrometry to detect glycosylation of proteins
Detection of possible glycosylation of glycoproteins can simply be made by measuring their mass before and after incubation with a suitable endoglucosidase such as PNFase F. Thus, for the soluble complement receptor 1 investigated by Wymann et al. (2021), the difference between the mass of the intact glycoprotein (169,251‐178,056 ± 50 Da) and the product of PNGase F digestion (148,165 ± 50 Da) was interpreted as showing the presence of N‐glycans. Similarly, laccase from Madurella mycetomatis gave a mass of 67.4 kDa, which reduced to 62.0 following deglycosylation with Endo H. The, result indicated 8.8% glycosylation (Tülek et al., 2021).
13.3.3.1.2. Detection of glycosylation sites and site occupancy
N‐Glycosylation occurs at asparagine (Asn) residues in a Asn‐Xxx‐Ser(Thr) motif where Xxx is any amino acid except proline. Not all sites are fully occupied and detection of glycosylation and its extent can be evaluated by the conversion of Asn to aspartic acid (Asp) giving a mass change of +1 Da following deglycosylation with PNGase F. An example of where this method has been used is the study by Dittner‐Moormann et al. (2021) on the transferrin biomarker for the congenital disorders of glycosylation (CDG) disease PMM2‐CDG where glycosylation with biantennary glycans is deficient. Transferrin has two N‐glycosylation sites and the results of PNGase F digestion of the glycoprotein from patients showed that the deficiency of biantennary glycosylation occurred equally at both sites.
13.3.3.2. Analysis of N‐glycans
Detailed structural analysis of the N‐linked glycans is most commonly performed following their release from the glycoprotein.
13.3.3.2.1. N‐Glycan release
Two types of method are commonly employed; chemical or enzymatic digestion.
13.3.3.2.1.1. Chemical release
Hydrazinolysis has been used extensively to remove both N‐ and O‐linked glycans in the past, but is now rarely used because of its associated hazards and the accompanying complete degradation of the protein. However, some other chemical procedures are still being investigated. Thus, for example, Diaz et al. (2022) have advocated the use of sodium hypochlorite for releasing Man9GlcNAc2 (193) from Phaseolus lunatus beans for use as a standard reference material. Some decomposition was observed at the reducing terminal GlcNAc residue, caused by the chlorination of intermediate imines.

Because of the expense of enzymatic release, chemical release, such as with ammonia, is used when large quantities of N‐glycans are required. It has been reported, however, that the high pH of this reaction can cause epimerization of the reducing‐terminal GlcNAc (117) residue to ManNAc (194) in relatively large quantities (Liew, Chen, Tsai, & Ni, 2022). CID spectra can be used to differentiate the two isomers: Thus, for N‐glycans with GlcNAc at the reducing end, the intensity of the fragment ion produced through dehydration (loss of neutral m = 18 Da) is smaller than the intensity (50%) of the fragment ion produced through glycosidic bond cleavage (loss of neutral m = 221). By contrast, for N‐glycans with ManNAc at the reducing end, the intensity of this fragment ion is larger than 80% of the intensity of the fragment ion produced through glycosidic bond cleavage.

13.3.3.2.1.2. Enzymatic release
Enzymatic release has now almost completely replaced chemical release and new enzymes are frequently being discovered. The most popular endoglycosidase for releasing N‐glycans is peptide N‐glycosidase F (PNGase F). However, this enzyme has some drawbacks. In particular, it operates optimally only in the neutral to slightly acidic pH range, suffers from steric inhibition, and its activity is severely compromised in the presence of reducing and denaturing substances. A new PNGase, isolated from the Gram‐negative bacterium Rudaea cellulosilytica (PNGase Rc) has been shown to overcome many of these disadvantages. It demonstrated broad substrate specificity for N‐glycan release from multiply occupied and natively folded proteins and is more tolerant to low pH conditions and to the presence of reagents such as urea and guanidinium chloride than PNGase F, making it suitable for use in hydrogen‐deuterium exchange experiments (Gramlich et al., 2022; Guo, Zhang, et al., 2022).
A recent study has compared the effects of detergents used to denature glycoproteins during release of N‐glycans with PNGase F (Kayili, Sakhta, et al., 2022). The released N‐glycans were labeled with procainamide (195), purified using cellulose‐containing solid‐phase extraction cartridges and analyzed by HPLC with fluorescence detection. The results showed that sodium dodecyl sulfate (SDS) and sodium deoxycholate (SDS + SDC) detergent combination provided the highest average fluorescence signal areas and intensities suggesting the most efficient release. It was also found that the average signal intensities of the detected N‐glycans were reduced when SDS and SDC were used with 1,4‐dithiothreitol (DTT) reducing agents. Profiles reflected the relative abundance of the released glycans rather than their compositions. A mixture of SDS, SDC and DTT produced a profile from human plasma consisting mainly of larger glycans (bi‐ and tri‐antennary) whereas the profile produced by SDS contained much more abundant low mass glycans (high‐mannose and degalactosylated biantennary). The authors also report the results of another relevant paper (Vilaj et al., 2020) in which it was reported that glycan profiles produced by PNGase F enzymes made by different manufacturers also differed.

Release of glycans with PNGase F followed by removal of the deglycosylated proteins by use of C18 cartridges is a popular method for N‐glycan analysis. However, it is not so applicable to large scale samples. Consequently, Wang, Peng, et al. (2022) have developed an alternative method in which the proteins are precipitated with acetone. The yield of N‐glycans was tested with the standard glycoprotein samples, bovine fetuin and human serum. Compared to the amounts of N‐glycans from the use of C18 cartridges, most of the sialylated N‐glycans from human serum were detected with higher abundance after acetone precipitation. However, C18 showed a slightly higher efficiency for protein removal. Using the unfiltered human serum, around 97.7% of the proteins were removed by acetone precipitation, while more than 99.9% of the proteins were removed by C18 cartridges.
13.3.3.2.2. Extraction and purification of released glycans
A comparative study of different methods for N‐glycan purification strategies, including filter‐aided sample preparation, de‐N‐glycosylated protein precipitation, and trypsin digestion followed by reversed phase‐based solid‐phase extraction (RP‐SPE) has concluded that the RP‐SPE method produced the best results (Guan, Zhang, Wang, et al., 2021). Glycans were permethylated using an optimized method (see Section 8.2.1.) and the method was used for examination of N‐glycans from the monoclonal antibodies trastuzumab and adalimumab.
Some methods, such as negative ion fragmentation, work best with neutral glycans and require a method for sialic acid removal. This can be accomplished enzymatically or by acid treatment. TFA is frequently used and has prompted Dong, Liu, et al. (2022) to study the reaction. It was found that, although most of the sialic acids were removed after heating at 75oC for 1 hour, it took 4 h for complete removal. Alternatively, when the concentration of TFA was raised to 5%, complete removal could be achieved in 1 h. However, the increased reaction times or TFA concentrations had adverse effects on peptides if the reactions were performed with glycopeptides.
13.3.3.2.3. Analysis of released glycans
Use of HPLC with exoglycosidase digestion of fluorescently labelled glycans has been a popular method for structural analysis of N‐glycans but suffers from the disadvantage that exoglycosidases are not available for identification of all structural features, one consequence of which is that new structures are difficult to analyse. Nevertheless, protocols continue to be published, such as that from McLeod et al. (2021). The article covers HPLC, LC‐MS, and CE, with glycans labelled with, for example, procainamide or 2‐AB. Products of exoglycosidase digestions can, of course, be monitored by mass spectrometry with the advantage that the measured mass leads directly to the composition of the glycan. MALDI is particularly useful because of its property of producing mainly single ions from each glycan. Techniques such as ESI tend to produce ions in different charge states, particularly for the larger glycans.
13.3.3.2.3.1. Methods to identify core‐fucosylated glycans
Abnormal expression of cell‐surface glycans with core fucosylation has been frequently observed in various cancers such as liver, colorectal, ovarian, prostate, and breast cancer, and has been associated with promotion of tumor growth, invasion, and metastasis. Consequently, there is much interest in methods that enable core from antenna fucosylation to be determined.
A method for achieving this distinction using low energy HCD claims to overcome problems caused by fucose migration leading to false positives (Chen, Shen, et al., 2022). The method involved observing the ratio of the Y1 + Fuc to the Y1 ions. If the ratio was greater than 0.1, the glycopeptide was considered to be core‐fucosylated, whereas, if the Y1F/Y1 ratio was less than 0.1, the glycopeptide was considered as solely antenna fucosylated. The method was tested with glycoproteins from human IgG (core fucosylation) and haptoglobin (antenna fucosylation). All 1026 core fucosylated glycopeptides from IgG were correctly identified whereas 156 of the 159 antenna‐fucosylated glycopeptides from haptoglobin were identified.
Another method has been developed by Tian, Wang, et al. (2022) with MALDI‐TOF being used to identify the final fucosylated glycopeptide. The enzyme Endo‐F3 from Elizabethkingia meningoseptica was used to cleave between the two GlcNAc residues of the core region of core‐fucosylated bi‐ and tri‐antennary glycans leaving Fuc‐GlcNAc attached to the protein. The, M‐endo‐F3 was used to attach the biotinylated probe 196 to the Fuc‐GlcNAc group effectively giving a biotinylated biantennary glycan attached to the protein. These labelled glycoproteins were cleaved with trypsin and the resulting fucosylated glycopeptides were captured with streptavidin beads. Cleavage with endo F3 left glycopeptides with attached Fuc‐GlcNAc which were identified by MALDI‐TOF and LC‐MS/MS.

13.3.3.3. Total methods for glycoprotein structure
A review with 91 references on sample preparation methods for N‐glycomics which covers glycan release, purification and enrichment, fluorescence labelling, permethylation and sialic acid derivatization has been published by Kayili, Atakay, et al. (2022). Protocols for various aspects of N‐glycan analysis are listed in Table 19.
Table 19.
Reviews and general articles on N‐glycan protocols.
| Subject | References |
|---|---|
| Site‐specific N‐glycosylation analysis of recombinant proteins by LC/MSE | Canis et al. (2021) |
| Analysis of intact glycoproteins by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry | Giménez et al. (2021) |
| Profiling of cellular glycoproteins and GSLs by Glycoblotting | Hanamatsu and Furukawa (2022) |
| Analysis of the biosynthesis and degradation of N‐glycan precursors in mammalian cells | Harada et al. (2021) |
| Analysis of monoclonal antibodies | Nmagu et al. (2021) |
An integrated on‐line deglycosylation, labeling and purification method for N‐glycan analysis is believed by the authors (Wu, Zhang, Li, et al., 2022) to be the first time that such a simplified method has been developed. The method consists of an on‐line immobilized enzyme reactor for PNGase F release of the N‐glycans, direct labeling of released N‐glycosylamines (see Section 8.1.4) with 6‐aminoquinolyl‐N‐hydroxysuccinimidyl carbamate (AQC) (Scheme 6) and purification of the derivatives on a microfluidic chip. The process could be completed in within about 30 min. Good reproducibility and stability were achieved with the relative standard deviation (RSD) less than 10%. Intermediate stages were monitored by MALDI‐TOF but the method itself was designed for HPLC monitoring.
Another integrated method that claims to reduce glycan release and labelling from 2 days to 2.5 h involves use of Stage Tips, prepared in pipette tips containing 3 mm of cotton wool. The glycoprotein (IgG) was added, followed by PNGase F and appropriate buffers and the tips were incubated for 1 h at 45oC. The released glycans were then labelled with procainamide by incubation at 70oC for 1 h. The Stage Tips were centrifuged and washed with aqueous acetonitrile and TFA and the labelled glycans were eluted with water, purified with solid‐phase extraction cartridges for analysis by MALDI with DHB. The tips were also used to produce glycopeptides by incubation with trypsin (Kayili, Ragoubi, et al., 2022).
A method for obtaining N‐glycans from human milk has involved acetone precipitation of the glycoproteins, removal of the glycans with PNGase F, methylation of the sialic acids and use of the glycoblotting technique (aoWR [197] labelling) and analysis by MALDI‐TOF/TOF from DHB. To enhance sensitivity, glycans were also permethylated for MALDI analysis. N‐Glycans from human milk were the normal range of high‐mannose, hybrid and bi‐, tri‐, and tetra‐antennary compounds. Bovine milk contained similar compounds but in different proportions such as less core fucosylation and more high‐mannose and bisected structures (Wang, Zhao, Tao, et al., 2021).

13.3.3.4. Identification of new N‐linked glycan structures
Several novel N‐glycan structures have been identified during the review period. These compounds are usually produced by “lower organisms” and those synthesised by Caenorhabditis elegans have been reviewed (Paschinger et al., 2021).
Methyl hexoses feature in several of these new structures. Thus, Man3GlcNAc2 with 3 pentoses, each carrying 0 or 1 Me group (e.g., 198) have been found in green algae (Chlorella species) (Choi et al., 2021).

Mosses have been shown to contain the normal plant paucimannosidic glycans such as 199 and some of them, such a Funaria hygrometrica and Plagiomnium undulatum contain methylated constituents such as 2,6‐dimethyl‐mannose comprising the 6‐antenna (200, 201) (Stenitzer & Altmann, 2022; Stenitzer, Mócsai, et al., 2022). Antennae with Lewis A termini (e.g., 202) are also common.


A new set of hybrid glycans with substituted bisecting GlcNAc residues (e.g., 203 and 204) has been detected in human brain (Helm et al., 2021).

High‐mannose glycans with additional substituents also feature in some species. Thus, Man3‐5GlcNAc2 with one additional (unidentified) pentose and Man2‐8GlcNAc2 with one deoxy‐hexose have been found in the semi‑terrestrial microalga Thorsmoerkia curvula gen. et spec. nov. (Trebouxiophyceae, Chlorophyta) from Iceland. Glycans bearing both substituents were not found (Nicoletti et al., 2021). High mannose glycans with an additional galactose residue has been found attached to invertase expressed in the industrial yeast Yarrowia lipolytica (Szymański et al., 2022).
Among other structures, N‐glycans with GlcA (205) and phosphorylcholine (206) substitutions have been found in N‐glycans from the filarial nematode Brugia malayi (Petralia, van Diepen, et al., 2022).

The N‐linked glycans from chloroviruses (family Phycodnaviridae) are considerably different from those of most other species and lack the normal trimannosyl chitobiose core. A recent review (Speciale, Notaro, et al., 2022) discusses their structure and biosynthesis. A new study on glycans from Paramecium bursaria chlorella virus MA‐1D by MALDI‐TOF/TOF‐MS (from DHB) and NMR has revealed three structures (207–209) that share several features with those of the other chloroviruses examined earlier except that they lack a distal xylose residue that was believed to be part of a conserved core structure for all the chloroviruses (Speciale, Di Lorenzo, et al., 2022). The authors believe that this result requires a reconsideration of the core structure for all chloroviruses.

Further applications on the use of MALDI MS to the analysis of N‐glycans in specific glycoproteins and tissues are listed in Tables 20 and 21 respectively.
13.3.4. O‐linked glycans
Analysis of O‐linked glycans has received much less attention than that of N‐glycans. Although generally smaller, they do not have the conserved core of N‐glycans and enzymatic release suffers from a lack of suitable enzymes. β‐Elimination is, thus, the preferred method for their release. Two recent reviews are of interest “Recent advances in demystifying O‐glycosylation in health and disease” (116 references) (Li, Guo, et al., 2022) and “Quantitative characterization of O‐GalNAc glycosylation” (51 references) (Čaval et al., 2021).
Recently, several O‐glycan‐specific endoproteases that can cleave the protein adjacent to the appended glycan have been described and used for the analysis of these compounds. To date, use of most of these enzymes suffer from problems such as inefficient cleavage of glycoproteins bearing sialylated O‐glycans, high selectivity for certain types of glycoproteins, or protein sequence bias. Vainauskas et al. (2022) have investigated a new immunomodulating metalloprotease from Pseudomonas aeruginosa using an array of synthetic peptides and their glycoforms. They showed that the enzyme has no specific residue preference and can tolerate most amino acids, except aspartic acid, at the position adjacent to the glycosylation site on the amino‐terminal side of the peptide. The enzyme was found not to cleave between two adjacent O‐glycosites. Glycopeptides with as few as two amino acids on either side of an O‐glycosite could be cleaved and the enzyme efficiently cleaved peptides and proteins carrying sialylated and asialylated O‐glycans.
A new method for identification of O‐GlcNAc‐modified proteins involves intracellular expression of a soluble GalNAc transferase, which then labels the GlcNAc residues with GalNAc. The resulting disaccharides can be detected by Wisteria japonica agglutinin, which is specific for this disaccharide, or by MALDI‐TOF MS (Abo et al., 2022).
13.3.4.1. Site analysis
Two protocols for site analysis of O‐glycans have been published: “Mapping O‐glycosylation sites using the O‐specific protease OpeRATOR and LC‐MS” (Nordgren et al., 2021) and “Site‐specific O‐glycosylation analysis by liquid chromatography–mass spectrometry with electron‐transfer/higher‐energy collisional dissociation” (Hashii & Suzuki, 2021).
13.3.4.2. Other studies
A protocol named Cellular O‐Glycome Reporter/Amplification (CORA) for studying mucin‐type O‐glycans of living cells has been developed by Kudelka et al. (2021). It involves incubation of cells with peracetylated Bn‐α‐GalNAc or N3‐Bn‐α‐GalNAc where cytosolic esterases generate the deacylated Bn‐α‐GalNAc or N3‐Bn‐α‐GalNAc. These glycans are transported into the Golgi apparatus and modified by native O‐glycosyltransferases which are secreted into the medium, purified, and then analyzed by MALDI‐TOF MS (from DHB) for Bn‐O‐glycans or derivatized with fluorescent tag 2‐amino‐N‐(prop‐2‐yn‐1‐yl)benzamide (PYAB, 210) followed by HPLC separation. The consequent amplification and secretion of the O‐glycome products was claimed to greatly facilitate their analysis and to aid functional studies.

Further applications of the use of MALDI for the analysis of O‐glycans in specific glycoproteins and tissues are listed in Tables 22 and 23 respectively.
Table 23.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of O‐glycans from intact organisms or tissues.
| Organism | Methodsa | Notes | References |
|---|---|---|---|
| Bovine submaxillary mucin | β‐Elimination, FT‐ICR (s‐DHB), (per‐Me) | As reference for development of automated method | Kotsias et al. (2021) |
| CHO Cell lines (CHO‐K1, CHO‐S, and CHO‐Pro5) | β‐Elimination, R‐TOF/TOF, (per‐Me) | Structural determination. Differences in sialylation and fucosylation | Wang, Wang, Wu, Lin, et al. (2022) |
| Human dermal endothelial cells | Glycans from cell medium, TOF/TOF (DHB), (per‐Me) | Sialoglycans on lymphatic endothelial cells shown to augment interactions with Siglec‐1 (CD169) of lymph node macrophages | D'Addio et al. (2021) |
| Human (serum and cerebrospinal fluid) | β‐Elimination, R‐TOF/TOF | Detection of novel low‐molecular‐weight blood group‐specific glycans in serum and cerebrospinal fluid | Furukawa et al. (2021) |
| Human umbellar vein endothelial cells | TOF (per‐Me) | To assess adhesion of cells to PET woven fabrics used in medicine. Profile changed, unlike N‐glycans | Hu, Sheng, et al. (2022) |
| Mouse brain | β‐Elimination, QIT‐TOF (DHB), amidation of sialic acids | Majority of α−2,6‐sialylated glycans in the adult mouse brain shown to exist in O‐glycans | Ohmi et al. (2021) |
| Mouse bain tissue | β‐Elimination, TOF/TOF (DHB), (per‐Me) | Brain glycoproteins shown to exhibit diminished glycan complexity compared to other tissues (most unbranched) | Williams et al. (2022) |
| Mouse serum | β‐Elimination, TOF/TOF (DHB), (per‐Me) | Identification of glycans separated by supported molecular matrix electrophoresis. α2,8‐Sialylated O‐glycans detected | Liu, Liu, Li, et al. (2021) |
| Mouse (submandibular gland) | β‐Elimination, R‐TOF/TOF, QIT‐TOF (DHB), (per‐Me) | Protein Bmi‐1 shown to regulate mucin levels and mucin O‐glycosylation | Kameyama, Nishijima, et al. (2021) |
| Mouse embryonic stem cell | Trypsin (TOF/TOF) | Identification of O‐GlcNAcylation of proteasome activator subunit 3 (Psme3) protein | Pecori et al. (2021) |
| Mouse peritoneal macrophage subpopulations | β‐Elimination, TOF/TOF (DHB), (per‐Me), MS/MS | Resident and elicited murine macrophages shown to differ in expression of their glycomes and glycan‐binding proteins | Park, Chen, et al. (2021) |
| Pelagia noctiluca (jellyfish), mucus | R‐TOF/TOF (DHB), glycoblotting | Use for accumulation of nanoparticles | Patwa et al. (2022) |
| Porcine bladder urothelial cells | β‐Elimination, FT‐ICR (DHB), (per‐Me) | Structural identification | Wang, Bergström, et al. (2022) |
| Pseudomonas aeruginosa | Non‐reductive β‐elimination, R‐TOF/TOF, (per‐Me) | Mucin glycans shown to signal through the sensor kinase RetS to inhibit virulence‐associated traits | Wang, Wheeler, et al. (2021) |
| Schmidtea mediterranea (flatworm) | β‐Elimination, TOF/TOF (s‐DHB), (per‐Me and free) | Structural determination of mucin‐type O‐glycans | Subramanian et al. (2022) |
| Tribolium castaneum (Insect) | β‐Elimination, FT‐ICR (s‐DHB), (per‐Me) | Pentasaccharide mucin‐type O‐glycans shown to be linked with pupation | Li, De Schutter, et al. (2022) |
Format (not all items present): Glycan release method and/or protease, MALDI method (matrix), (derivative), other methods.
13.4. Glycated proteins (nonenzymatic attachment of sugars)
Review: “Enrichment and analysis of glycated proteins” (Cho, Duong, et al., 2022), 126 references
d‐Glucose and d‐fructose react with amino groups in proteins to form Schiff bases that rearrange to more stable Amadori and Heyns products, respectively (Scheme 20). These compounds are difficult to differentiate by mass spectrometry because of their identical molecular masses and similar fragmentation patterns. However, it has now been shown that separation can be achieved by RP‐HPLC with phosphate‐buffered eluants providing the best separation (Schmutzler & Hoffmann, 2022).
Scheme 20.

Formation of products resulting from glycation of proteins.
Glycated hemoglobin (HbA1c) is used to monitor patients with diabetes but results can differ depending on the method used. Song, Xu, et al. (2022) have compared LC/MS and MALDI‐TOF to monitor this glycated protein and generally found good agreement between the two techniques. Variations were, however, found where variant hemoglobin was encountered.
Other applications of MALDI to the analysis if glycated proteins are listed in Table 24.
Table 24.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for the investigation of glycated proteins.
| Protein/amino acid and sugar | Methodsa | Notes | References |
|---|---|---|---|
| Alpha‐synuclein, methylglyoxal | TOF/TOF (DHA, NH4Cit) | Glycation shown to modulate alpha‐synuclein fibrillization kinetics | Farzadfard et al. (2022) |
| BSA, glucose | MALDI (DHB/SA) | Development of a benzothiazole‐phenothiazine conjugate‐based molecular probe for the differential detection of glycated albumin | Kumar et al. (2021) |
| BSA, glucose, fructose, methylglyoxal | TOF/TOF (SA) | In vitro chronic glycation shown to induce AGEs accumulation reducing insulin‐stimulated glucose uptake and increasing glucagon‐like peptide 1 (GLP1R) in adipocytes | Chilelli et al. (2021) |
| Hemoglobin, methylglyoxal | TOF | Methylglyoxal‐derived hemoglobin advanced glycation end products shown to induce apoptosis and oxidative stress in human umbilical vein endothelial cells | Lee, Samsuzzaman, et al. (2021) |
| HSA, d‐glucose | TOF | Effects of glycation in the binding of bioactive flavonoid 6‐hydroxyflavone by HSA | Sarmah et al. (2022) |
| HSA, d‐glucose | TOF/TOF (SA, CHCA, DHB) | Investigation of the effects of glycation on drug binding to HSA | Ghosh and Kishore (2022) |
| Murine cardiac proteins, fructose | TOF | Curcumin shown to prevent glycation of tricarboxylic acid cycle and cell respiration proteins in the hearts of mice fed with a high‐fructose diet | León‐García et al. (2022) |
| Myoglobin, glyoxal | TOF/TOF (CHCA) | Long‐term incubation of myoglobin with glyoxal shown to induce amyloid like aggregation of the heme protein | Banerjee (2021b) |
| Myoglobin, melibiose | MALDI | The melibiose‑derived glycation product shown to mimic a unique epitope present in human and animal tissues | Staniszewska et al. (2021) |
| Myoglobin, methylglyoxal | TOF/TOF (CHCA) | Role of advanced glycation end products in inducing protein structural alterations | Banerjee (2021a) |
Format (not all items present): MALDI method (matrix).
13.5. Peptidoglycans
Peptidoglycans are found in the cell walls of most bacteria and are composed of chains of GlcNAc‐β‐(1→4)‐MurNAc (where MurNAc = 214) cross‐linked by small peptides, commonly l‐Ala‐γ‐d‐Glu‐meso‐A2pm(or l‐Lys)‐d‐Ala‐. Their analysis usually involves hydrolysis to amino acids, peptides and amino sugars or enzymatic digestion of the glycan chain to muropeptides (disaccharide plus peptide).

Papers describing work with these compounds are listed in Table 25.
Table 25.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of bacterial peptidoglycans and muropeptides.
| Species | Peptidoglycan | Methods | Notes | References |
|---|---|---|---|---|
| Acinetobacter baumannii | Muropeptides | TOF | The bacterium is shown to be able to survive with an outer membrane lacking lipooligosaccharide due to structural support from elongasome peptidoglycan synthesis | Simpson et al. (2021) |
| Bacillus subtilis mreB mutants | Muropeptides | TOF | Magnesium shown to restore the rod shape of Bacillus subtilis mreB mutants through its inhibitory effect on peptidoglycan hydrolases | Tesson et al. (2022) |
| Vibrio cholerae | D‐Met‐and D‐Arg‐muropeptides | TOF/TOF | Binding of noncanonical peptidoglycan shown to control V. cholerae broad spectrum racemase activity | Espaillat et al. (2021) |
13.6. Glycosaminoglycans (GAGS)
Reviews and general articles on the analysis of glycosaminoglycans are listed in Table 26.
Table 26.
Reviews and general articles on the analysis of glycosaminoglycans.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Developments in mass spectrometry for glycosaminoglycan analysis | Covers sample preparation, composition analysis, sequencing (fragmentation), software and applications | 165 | Pepi, Sanderson, et al. (2021) |
| Insights into structure, affinity, specificity, and function of GAG‐protein interactions through the chemoenzymatic preparation of defined sulfated oligohyaluronans | Useful short section on analysis of GAGs by mass spectrometry | 32 | Schiller et al. (2021) |
| Analysis of the glycosaminoglycan chains of proteoglycans | Fairly comprehensive. Sample preparation, MS (mainly LC‐MS, NMR, hyphenated techniques). | 107 | Song et al. (2021) |
| State‐of‐the‐art glycosaminoglycan characterization | Comprehensive review covering structure, function, sample preparation, chromatographic and mass spectrometric analytical methods, ion mobility, IR spectroscopy | 352 | Zappe et al. (2022) |
A problem with analysis of these compounds by MALDI is loss of sulfate. A new method, reported Krüger et al. (2022) involves on‐target derivatization with 3‐hydrazinobenzoic acid (3‐HBA, 215) by heating at 70oC for 10 min. MALDI‐TOF/TOF was performed in negative ion mode. Disaccharides from dermatan (216) and chondroitin sulfates (217) were examined and MS/MS spectra allowed sulfation patterns to be resolved.



A detailed study of the photofragmentation of chondroitin sulfate isomers has shown promising results, particularly with differentiating isomers involving C‐5 uronic acid stereochemistry (Pepi, Leach, et al., 2021).
13.7. Glycolipids
Several types of glycolipid can be identified. They include lipopolysaccharides (LPS) found in the cell membranes of Gram negative bacteria, glycosphingolipids (GSLs) and a range of assorted structures, usually found in bacteria. Several general reviews are of interest. A review of recommendations for good practice in MS‐based lipidomics, while not providing a step‐by‐step protocol for best practice, nevertheless provides the reader with links to original publications concerning the state‐of‐the‐art practices in the field (Köfeler et al., 2021). MALDI methods, however, are specifically excluded because the authors state that the technique is overwhelmingly used for MALDI imaging which is beyond the scope of the review.
“Imaging lipids in biological samples with surface‐assisted laser desorption/ionization mass spectrometry” (SALDI‐MSI) with 169 references, is a concise review of work published during the last decade (Müller, De Pauw, et al., 2021) (169). The review describes the advantages of SALDI‐MSI for lipid analysis, such as the ability to perform analyses in both ionization modes with the same nanosubstrate, and the detection of lipids that exhibit low ionization efficiency in MALDI‐MS. The complementarity of SALDI and MALDI‐MSI is also discussed. The review contains a very comprehensive list of the use of SALDI in lipid and glycolipid analysis.
The third review “A new update of MALDI‐TOF mass spectrometry in lipid research” (361 references) (Engel et al., 2022), is a general review of lipids and glycolipids covering work over the past 10 years, with emphasis on glycerophospholipids. Particular attention is given to quantitative aspects of MALDI MS since this is widely considered as the most serious drawback of the technique. The choice of the MALDI matrix is shown to be crucial to be able to detect all lipid classes. MALDI imaging and the combination of MALDI with TLC are given special attention.
Other reviews are listed in Table 27.
Table 27.
Reviews on glycolipids.
| Subject | Notes | Citations | References |
|---|---|---|---|
| A journey from structure to function of bacterial lipopolysaccharides | General review. Structural identification of LPS. Comments on unusual monosaccharides | 284 | Di Lorenzo et al. (2022) |
| Lipopolysaccharide lipid A: A promising molecule for new immunity‐based therapies and antibiotics | Emphasises the dominance of MALDI as the best analytical method for lipid A with examples | 240 | Garcia‐Vello, Di Lorenzo, et al. (2022) |
| History of colistin resistance mechanisms in bacteria: Progress and challenges | Short section on MALDI analysis of lipid A | 110 | Hamel et al. (2021) |
| A comprehensive review on natural occurrence, synthesis and biological activities of glycolipids | Many types of glycolipid. Few references to MALDI | 318 | Jala et al. (2022) |
| Integrated mass spectrometry‐based multi‐omics for elucidating mechanisms of bacterial virulence | Mainly proteomics but section on lipid A | 276 | Man et al. (2021) |
| Solving the structural puzzle of the bacterial glycome | Short review, mass spectrometry, NMR, bioinformatics | 52 | Marchetti et al. (2021) |
| Structures and functions of the gut microbial lipidome | Covers several structural types such as glycoglycerolipids, sphingolopids, lipid A and steroidal glycolipids | 157 | Morozumi et al. (2022) |
| Modern techniques for separation, mass spectrometric detection, and characterization of glycolipids | Extraction and purification, TLC, CZE and ion mobility | 89 | Sarbu and Zamfir (2021) |
13.7.1. LPS and lipooligosaccharides (LOS)
These compounds are composed of lipid A, a glycolipid containing two glucosamine molecules attached to up to six fatty acyl chains and decorated with various groups such as phosphate (see structure 219 below for an example), a core region and usually a long carbohydrate chain consisting of repeat units. The term LOS is usually used for the smaller molecules. A protocol “Dissecting lipopolysaccharide composition and structure by GC‐MS and MALDI spectrometry” in Methods in Molecular Biology (Garcia‐Vello, Speciale, et al., 2022) describes methods for analysis of these compounds.
Lipid profiles as determined by MALDI‐MS in negative ion mode, combined with a machine‐learning algorithm have proved useful in discriminating between Escherichia coli, Shigella flexneri, and S. sonnei. The three species showed different profiles for cardiolipins (218) and bisphosphoryl lipid A with the Shigella species demonstrating higher mass peaks for lipid A (Pizzato et al., 2022).
Intact lipooligosaccharide from the deep‐sea marine bacterium Idiomarina zobellii KMM 231T, isolated at a depth of 4000 m (Kokoulin et al., 2022) has been analysed and shown to consist of only five sugar rings, two of which comprise the lipid A portion (219). The negative ion MALDI‐TOF spectrum (Figure 5) resolved three groups of peaks corresponding to tri‐, tetra‐, and penta‐acylated forms with acyl chains of different length. Deacylated LOS was studied by NMR and monosaccharide identification was performed by GC/MS. The lipid A portion of the molecule was isolated and each acylated form was studied by MALDI‐MS/MS.
Figure 5.

Negative ion MALDI‐TOF spectrum of the LOS from Idiomarina zobellii KMM 231T. From Kokoulin et al. (2022) with permission from MDPI).


13.7.1.1. Lipid A
Two detailed protocols for analysis of 4‐monophosphoryl lipid A by MALDI‐TOF have been published (Larrouy‐Maumus, 2021; Micoli et al., 2022). Most work was done in negative ion mode because of the anionic groups attached to the GlcN residues. Aissa et al. (2021) have studied the negative ion (deprotonated molecule) CID spectra of lipid A and their findings can be summarised as follows: (i) cleavage of the C‐3 primary fatty acid to leave an epoxide group attached to the reducing sugar (Scheme 21); (ii) cleavage of the C‐3’ primary fatty acid (as an acid) which generates a cyclic phosphate connected to the nonreducing sugar (Scheme 22); (iii) cleavage of the C‐2’ secondary fatty acid which is observed to occur both in acid and ketene forms; (iv) the C‐2 and C‐2’ primary fatty acids are eliminated as an amide and ketene, respectively; (v) the 0,2A2 cross‐ring fragment from the reducing terminal ring contains a four‐membered ring (oxetanose, Scheme 23); (vi) the 0,4A2 ion is formed from this ion; and (vii) formations of H2PO4 ‐ and PO3 ‐ ions are associated with the formation of sugar epoxide.
Scheme 21.

Formation of an epoxide by loss of the C3 primary acid group from the [M – H]‐ ion of lipid A.
Scheme 22.

Formation of a cyclic phosphate by loss of the C3’ primary acid group from the [M – H]‐ ion of lipid A.
Scheme 23.

Formation of an oxetanose ring.
Yang, Smith, Chandler, et al. (2022) have developed a tandem MS version of the earlier fast lipid analysis technique (FLAT) method termed FLAT n and used it to directly examine lipid A from a single bacterial colony of E. coli. Washed bacteria were deposited onto an indium tin oxide (ITO) slide, treated with citrate buffer and heated at 110oC for 30 min and examined with a timsTOF instrument. Detailed spectra of the lipid A, including cross‐ring fragments, were obtained. The method was developed into a direct‐from‐urine diagnostic for Gram‐negative pathogens (Yang, Smith, Sumner, et al., 2022).
The phosphate groups on lipid A are targeted by cationic antimicrobial peptides, such as polymyxins. However, resistance can develop because the bacteria are able to neutralize the negative charges by adding neutral groups such as aminoarabinose (AraN, 229) and ethanolamine (EtN). These modifications can be detected by MALDI‐TOF but only semiquantitatively. To overcome this disadvantage Sherman et al. (2022) have developed a GC/MS assay for the individual components of lipid A involving hydrolysis and derivatization as methyloxime/trimethylsilyl (TMS) derivatives. Using the method, increase in the abundance of AraN and EtN modifications were observed when resistant Enterobacter and E. coli strains were grown in the presence of colistin (polymyxin E). Because lipid A modifications serve as indicators of polymyxin resistance in Gram‐negative bacteria, this GC/MS method was claimed to provide an excellent method to monitor polymyxin resistance.

13.7.1.2. O‐Chain
A brief study of the fragmentation pattern of the O‐specific polysaccharide from Vibrio cholera O139 has been reported. In negative mode, the predominant fragmentation pathway was loss of neutral monosaccharide residues (Pančík, Pakanová, Mečárová, et al., 2022). A rapid method for measurement of the O‐chain by MALDI‐TOF directly from cells involves heating with HCl for 10 min at 90oC (Urakami & Hinou, 2022a) (paper in Japanese).
Table 28 lists work that has been reported with these compounds. Much is targeted in understanding mechanisms of antibiotic resistance and has resulted in the observation of increases phosphorylation of lipid A making it more acidic. As with lipid A, most of the MALDI work has been in negative ion mode with THAP and norharmane as the favoured matrices.
Table 28.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of glycolipids from Gram‐negative bacteria.
| Species | Type | Methodsa | Notes | References |
|---|---|---|---|---|
| Acinetobacter baumannii | Lipid A | R‐TOF/TOF (CMBT‐EDTA) | Colistin dependence in extensively drug‐resistant A. baumannii strain shown to be associated with ISAjo2 and ISAba13 insertions and multiple cellular responses | Chamoun et al. (2021) |
| A. baumannii | Lipid A | R‐TOF (CMBT) | Alteration of lipooligosaccharide structure during cold stress | Herrera et al. (2021) |
| A. baumannii | Lipid A | TOF (norharmane, ‐ve) | Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in clinical isolates with colistin–sulbactam combination therapy | Srisakul et al. (2022) |
| A. baumannii | Lipid A | TOF (DHB) | Phenotypic modification (PEtN incorporation and loss of a C12 acyl chain) of lipid A in clinical isolates | Kim, Yun, et al. (2022) |
| Aeromonas hydrophila | Lipid A | TOF (‐ve) | Investigation of colistin resistance | Liu, Xiao, et al. (2021) |
| Aeromonas salmonicida | Lipid A | TOF (‐ve) | Mobile colistin resistance enzyme MCR‐3 shown to facilitate bacterial evasion of host phagocytosis | Yin, Ling, et al. (2021) |
| Aeromonas veronii bv. sobria Strain K133 | LPS | Q‐TOF (THAP) | Structural characterization | Dworaczek et al. (2021) |
| Alcaligenes faecalis | LPS, Lipid A | TOF/TOF (THAP) | Complete structure characterization and chemical synthesis of its lipid A | Shimoyama et al. (2021) |
| Bacteroides thetaiotaomicron | LPS | TOF/TOF (DHB) | Structural characterization and immunological activity | Pither, Illiano, et al. (2022) |
| Cattle (rumen microbiome) | Lipid A | R‐TOF/TOF (THAP, ‐ve) | Lipid A acetylation pattern shown to differ between cows fed on different diets | Sarmikasoglou et al. (2021) |
| Caulobacter crescentus | Lipid A | TOF | lipid A shown to be conditionally dispensable in the absence of ferric uptake regulator fur and in the presence of anionic sphingolipids | Zik et al. (2022) |
| Echinicola pacifica KMM 6172T and E. vietnamensis KMM 6221T | Lipid A | TOF/TOF (THAP, DHB, ‐ve), MS/MS, GC/MS | Incorporation of GalA and modifications to acyl chains in response to survival in a marine environment | Pither, Mantova, et al. (2021) |
| Edwardsiella tarda PCM 1155 | O‐polysaccharide | TOF/TOF (DHB) | Structural determination shows the presence of unique β‐l‐RhapNAc3NAc derivative | Kaszowska et al. (2021) |
| Enterobacter cloacae | Lipid A | TOF (DHB) | Presence of 2‐hydroxymyristate on endotoxins shown to be associated with death in neonates with E. cloacae complex septic shock | Augusto et al. (2021) |
| E. cloacae | Lipid A | TOF (DHB, ‐ve) | Characterization of resistance mechanisms of E. cloacae complex co‐resistant to carbapenem and colistin | Liu, Fang, et al. (2021) |
| Enterobacter species | Lipid A | TOF (norharmane, ‐ve) | Development of a MALDI‐TOF assay for the rapid detection of colistin‐resistant enterobacter species | Smith, McElheny, et al. (2022) |
| Escherichia coli | Lipid A | TOF (DHB) | Investigation of colistin resistance | Wan, Xu, et al. (2021) |
| E. coli | Lipid A | TOF (ATT, DHB) | Diacylglycerol kinase A shown to be essential for polymyxin resistance provided by EptA, MCR‐1, and other lipid A phosphoethanolamine transferases | Purcell et al. (2022) |
| E. coli R1 and K12 | Entero‐bacterial common antigen | TOF/TOF (THAP, DHB) | Structural identification (repeats of →3)‐α‐d‐Fucp4NAc‐(1→4)‐β‐d‐ManpNAcA‐(1→4)‐α‐d‐GlcpNAc‐(1→ linear and cyclic forms | Gozdziewicz et al. (2021) |
| E. coli harbouring the mcr‐8 (nmcr‐2) mutants | Lipid A | TOF | Characterization of NMCR‐2, a new nonmobile colistin resistance enzyme: | Ullah et al. (2021) |
| E. coli BL21 carrying Ah762 (functional variant of MCR‐3) | Lipid A | TOF | The MCR‐3 inside linker appears as a facilitator of colistin resistance | Xu, Chen, et al. (2021) |
| E. coli | O25B Polysaccharide | Q‐TOF (DHB) | Development and characterization of an E. coli O25B bioconjugate vaccine | Kowarik et al. (2021) |
| E. coli NK5449. | Lipid A | TOF/TOF (DHB) | Study of genes in hospital wastewater breaking through the defence line of last‐resort antibiotics | Zhu, Shuai, et al. (2022) |
| Fusobacterium nucleatum ATCC 51191 | Lipid A | R‐TOF/TOF (THAP, ‐ve) | Structural characterization (and O‐antigen by NMR) | Garcia‐Vello et al. (2021) |
| Granulibacter bethesdensis | Lipid A | R‐TOF/TOF (THAP, ‐ve) | Bacterium shown to produce a penta‐acylated hypostimulatory glycero‐d‐talo‐oct‐2‐ulosonic acid–lipid A glycolipid (Ko‐lipid A) | Muszyński et al. (2021) |
| Herbaspirillum sp. Root189, isolated from the roots of Arabidopsis thaliana | O‐Antigen | FT‐ICR (s‐DHB) | LPS O‐antigen molecular and supramolecular modifications of plant root microbiota shown to be pivotal for host recognition | Vanacore et al. (2022) |
| Klebsiella pneumoniae | Lipid A | L‐TOF/TOF (CMBT/NH4‐Cit) | A K. pneumoniae DedA family membrane protein shown to be required for colistin resistance and for virulence in wax moth larvae | Tiwari et al. (2021) |
| K. pneumoniae | Lipid A | R‐TOF/TOF (DHB, –ve) | Pharmacodynamic and immunomodulatory effects of polymyxin B in combination with fosfomycin against KPC‐2‐producing K. pneumoniae | Sharma, Garcia, et al. (2022) |
| K. pneumoniae | LPS | TOF | Shown to induce host metabolic stress that promotes tolerance to pulmonary infection | Lung et al. (2022) |
| K. pneumonia and Acinetobacter baumannii | Lipid A | TOF (norharmane) | Benzimidazole isosteres of salicylanilides shown to be highly active colistin adjuvants. Changes to lipid A monitored by MALDI. | Li, Mattingly, et al. (2021) |
| Mycobacterium smegmatis | LOS | TOF/TOF (DHB) | Elimination of enzymes PknL and MSMEG_4242 in M. smegmatis shown to alter the character of the outer cell envelope | Báez‐Ramírez et al. (2021) |
| Neisseria gonorrhoeae | LOS/Lipid A | Q‐TOF (THAP/nitro‐cellulose) | Investigation of novel small molecules that increase the susceptibility of N. gonorrhoeae to cationic antimicrobial peptides by inhibiting lipid A phosphoethanolamine transferase | Mullally et al. (2022) |
| Pandoraea pulmonicola | Lipid A | TOF/TOF (THAP, DHB, ‐ve) | Chronic strain of the cystic fibrosis pathogen P. pulmonicola shown to express a heterogenous hypo‐acylated lipid A | Pither, McClean, et al. (2021) |
| Pseudoalteromonas nigrifaciens Sq02‐Rifr | LOS (230) | TOF/TOF (THAP), NMR | Complete structural characterization and study of its immunomodulatory activity | Di Guida et al. (2021) |
| Pseudomonas aeruginosa | Lipid A | TOF, (norharmane) | Detection of colistin resistance in Pseudomonas aeruginosa using the MALDIxin test on the routine MALDI Biotyper Sirius mass spectrometer | Jeannot et al. (2021) |
| P. aeruginosa | Lipid A | TOF (norharmane, ‐ve) | Loss of resistance‐nodulation‐division‐type multidrug efflux pumps shown to trigger iron starvation and lipid A modifications | Adamiak et al. (2021) |
| P. aeruginosa | Lipid A | R‐TOF (norharmane) | Genomic characterization of lytic bacteriophages targeting genetically diverse P. aeruginosa clinical isolates | Nordstrom et al. (2022) |
| Pseudomonas syringae pv. phaseolicola | Lipid A | TOF (norharmane) | Remodelling of lipid A in vitro | Gerster et al. (2022) |
| Rickettsia (4 species) | Lipid A | TOF (norharmane, ‐ve) | Structural characterization | Guillotte et al. (2021) |
| Salmonella enterica subsp. enterica serovar Liverpool | Lipid A | TIMS‐TOF (9‐AA, ‐ve) | Coculture with Acinetobacter johnsonii shown to enhance resistance to benzalkonium chloride disinfectant by triggering lipid A modifications to reduce net negative charge | Wilson, Fegan, et al. (2022) |
| Shigella sonnei | Lipid A | L‐TOF/TOF (s‐DHB, ‐ve) | Investigation of the contribution of O‑antigen and proteins to the immunogenicity of Shigella sonnei generalized modules for membrane antigens | Mancini et al. (2021) |
| S. sonnei | Entero‐bacterial common antigen | TOF/TOF (THAP, DHB) | Structural identification (repeats of →3)‐α‐d‐Fucp4NAc‐(1→4)‐β‐d‐ManpNAcA‐(1→4)‐α‐d‐GlcpNAc‐(1→ linear and cyclic forms | Gozdziewicz et al. (2021) |
| Yersinia pestis | Lipid A | TOF (norharmane, ‐ve) | Optimization of RG1‐VLP vaccine performance in mice with novel TLR4 agonists | Zacharia et al. (2021) |
| Zunongwangia profunda SM‑A87 | Lipid A | R‐TOF/TOF (THAP) | Structural characterization | Pither, Sun, et al. (2022) |
Format (not all items present): MALDI method (matrix), other methods.

13.7.2. Glycosphingolipids (GSLs)
These compounds consist of the amino alcohol, sphingosine (231) in which the amino group is amidated with a long chain saturated or unsaturated fatty acid giving ceramide (Cer) and the primary hydroxyl group is connected to an oligosaccharide chain (see structures 184 and 185). Du, Yu, et al. (2021) have published a protocol for analysis of glycosphingolipid glycans by lectin microarrays and MALDI‐TOF mass spectrometry. The review “Developments and applications of separation and microfluidics methods coupled to electrospray mass spectrometry in glycomics of nervous system gangliosides” (Sarbu, Ica, & Zamfir, 2021) is relevant and contains references to ion mobility and a few to MALDI. Other reviews are listed in Table 29.
Table 29.
Reviews and general articles on the analysis of glycosphingolipids (GSLs).
| Subject | Comments | Citations | References |
|---|---|---|---|
| Recent progress in O‐acetylated gangliosides analysis and functions in cancer | Short discussion on analysis of acylated GSLs by MALDI | 88 | Groux‐Degroote et al. (2021) |
| Recognition and avoidance of ion source‐generated artefacts in lipidomics analysis | Fragments generated by ISD. Mainly glycerolipids but some glycosphingolipids mentioned | 98 | Hu, Luo, et al. (2022) |
| Glycolipids being viewed in vivo or in vitro | Review of analytical methods. Small section on MALDI | 35 | Yilmaz (2021) |

13.7.2.1. Analysis of intact molecules
A simple method for separating GSLs from other lipids, including phospholipids and cholesterol, using zirconium dioxide (zirconia, ZrO2) has been developed (Nagasawa et al., 2022). The lipid mixture consisting of GSLs, cholesterol and phospholipids was loaded onto a ZrO2 column where cholesterol did not bind. The column was eluted with DHB in methanol when GSLs but not phospholipids were recovered; leaving the phospholipids bound to the ZrO2 particles. This method worked well for GSLs such as triglycosylceramides, tetraglycosylceramides and some pentaglycosylceramides, sulfatide (48) and GM3 (232) located in the lower phase of a Folch's partition, where significant amounts of phospholipids, cholesterol and neutral lipids were found along with GSLs.

A method involving AP‐MALDI interfaced to an Orbitrap mass spectrometer with a THAP matrix spiked with lithium salts has given improved detection of lipids, particularly HexCer from enveloped viruses (Tran, Monreal, et al., 2021). Use of the method resulted in the identification of over 130 lipids from influenza A virions.
Analysis of sphingo‐ and glycosphingo‐lipids in complex mixtures is greatly facilitated by using basic hydrolysis to remove contaminating glycero‐ and phospho‐lipids. KOH is traditionally used for this purpose leading to the lipids being detected as potassium adducts. Tran, Wan, et al. (2021) have reported that LiOH hydrolysis gives better detection of ceramides and glycoceramides and results in Li adducts of the lipids in the resulting MALDI spectrum. They have, consequently, developed a method using LiOH and have found that THAP provides the best signals. The method was applied to sphingolipid detection from a high‐fat‐induced obesity mouse model.
Use of thin‐layer chromatography (TLC) plates with or without blotting onto hydrophilic polyvinylidene fluoride (PVDF) membranes is frequently used in work with these compounds. A problem can arise from background peaks in the MALDI spectrum that often mask those from the sample. Matsushita et al. (2021) have addressed this problem by pre‐washing the plates with 1,2‐dichloroethane before development and found that the background peaks could successfully be removed.
Positive ion spectra of gangliosides are often weak because of the presence of acidic groups. As with N‐glycans, this situation has been reversed by derivatization to block the negative charge sites. Liu, Yang, Li, et al. (2022) have used 1,1‐dimethylethylenediamine (DMEN, 119 above) as the derivatizing reagent which, not only formed amides with the carboxylic acid groups, but also provided a site that was easy to protonate for high sensitivity, The detection of gangliosides was reported to be improved by at least four fold. By using DMEN derivatization, 45 glycosphingolipids were identified from human plasma, including 30 gangliosides and 15 neutral glycosphingolipids.
13.7.2.2. Analysis of released glycans
Because of the heterogeneity of the ceramide, some investigators remove this part of the molecule to study the glycan portion. Endoglycoceramidase I is a suitable enzyme and was employed by Furukawa et al. (2021) in a study of blood group‐specific antigens in serum/plasma and cerebrospinal fluid (CSF). The results suggest that blood group‐specific antigens are predominantly present on GSLs and lipoproteins rather than on glycoproteins. Petralia, van Diepen, et al. (2022) have used the enzyme in a study of the glycome from the filarial nematode B. malayi where it was shown that the GSLs contained both unusual glucuronic acid and phosphorylcholine (PC).
Other applications are listed in Table 30.
13.7.3. Bacterial glycolipids
A number of diverse structures are included under this heading and relevant papers are listed in Table 31.
Table 31.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for examination of glycolipids from bacteria, plants and similar organisms.
| Source | Glycolipid | Methods a | Notes | References |
|---|---|---|---|---|
| Bifidobacterium animalis subsp. lactis BPL1 | Lipoteichoic acid (233) | R‐TOF/TOF (CHCA) | Shown to reduce fat deposition via the IGF‐1 pathway | Balaguer et al. (2022) |
| Castor oil | Mannosylerythritol lipids (e.g., 234) | TOF/TOF | Structural characterization | Beck et al. (2021) |
| Mycobacterium abscessus | Glycopeptidolipids | L‐TOF (s‐DHB) | An improved method for rapid detection of M. abscessus complex based on species‐specific lipid fingerprint by routine MALDI‐TOF | Khor et al. (2021) |
| Apilactobacillus kosoi 10HT, Lactiplantibacillus plantarum JCM1149T and Lacticaseibacillus rhamnosus GG | Lipoteichoic acid anchor region | R‐TOF/TOF (DHB) | Investigation of the role of lipoteichoic acid from the genus Apilactobacillus in inducing a strong IgA response | Matsuzaki et al. (2022) |
| E. coli | Membrane protein integrase glycolipid (235) | TOF | Summary of discovery, structure, synthesis and biological activity | Fujikawa et al. (2019) |
| Halorientalis salina sp. nov., H. marina sp. nov., and H. litorea sp. nov. | Sulfated mannosyl glucosyl diethers | TOF (‐ve ion) | Results indicated three novel species. | Wang, Sun, Wu, Zheng et al. (2022) |
| Lactococcus cremoris 3107 | Cell‐wall polysaccharide | TOF/TOF (DHB) | Investigation of mechanism for lactococcal phage TP901‐1 infection. Glycan modification not implicated | Ruiz‐Cruz et al. (2022) |
| Mycobacterium abscessus | Glycopeptidolipid | QIT‐TOF (DHB), MS/MS | Glycopeptidolipid glycosylation shown to control surface properties and pathogenicity | Daher et al. (2022) |
| Mycobacterium bovis BCG | Trehalose dimycolate (236) | TOF | The liposome of trehalose dimycolate extracted from M. bovis BCG shown to induce antitumor immunity via the activation of dendritic cells and CD8+ T cells | Shiga et al. (2021) |
| Pseudomonas aeruginosa | Rhamonolipids (e.g., 237) | TOF/TOF (CHCA) | The rhamnolipids shown to promote methane hydrates formation in fixed bed silica gel medium | Arora et al. (2021) |
| P. aeruginosa | Rhamonolipids | R‐TOF/TOF | Characterization and cytotoxicity of rhamnolipids against breast cancer MDA‐MB‐231 cell line | Mishra et al. (2021) |
Format (not all items present): MALDI method (matrix), other methods.





13.7.4. Glycosides ‐ Natural Products
Although identification of natural products, mainly glycosides from plants, is a very active field, most mass spectrometric work appears to be conducted with ESI and LC‐MS/MS techniques. However, MALDI features in several publications and relevant reviews relating to the technique are summarised in Table 32.
Table 32.
Reviews and general articles on the analysis miscellaneous natural products.
| Subject | Notes | Citations | References |
|---|---|---|---|
| Current knowledge of intestinal absorption of anthocyanins | Short overview including MALDI imaging | 59 | Hahm et al. (2022) |
| Chemical constituents and chemical analysis of Ginkgo biloba leaf, extract, and phytopharmaceuticals | Extensive list of flavonoids and glycosides, discussion of analytical methods and quantification | 122 | Liu, Wang, Zhang, et al. (2021) |
| Extraction, purification, structural characteristics and biological properties of the polysaccharides from Codonopsis pilosula | Includes table listing use of MALDI to investigate structures of the glycans | 136 | Luan et al. (2021) |
| Application of MS‐based metabolomic approaches in analysis of starfish and sea cucumber bioactive compounds | Contains table with details of analysis of starfish polar steroids and sea cucumber triterpene glycosides | 183 | Popov et al. (2022) |
| Holothuria triterpene glycosides: A comprehensive guide for their structure elucidation and critical appraisal of reported compounds | Comprehensive review. Table of reported structures. Mass spectral fragmentation | 93 | Puspitasari et al. (2022) |
| Bioactive polysaccharides and oligosaccharides from garlic (Allium sativum L.): Production, physicochemical and biological properties, and structure–function relationships | Comprehensive review. Occurrence, production and extraction of sugars. Modifications such as sulfation. Characterization (MALDI). Metabolism, biological activity | 433 | Qiu et al. (2022) |
| Separation procedures for complicated mixtures of sea cucumber triterpene glycosides | With several references to MALDI analysis and MALDI imaging | 89 | Silchenko et al. (2022) |
A method claiming to provide improved coverage of plant metabolites involves examination of powder derived from dried plant fragments rather than the products of liquid extraction (Islam et al., 2022). Ground plant powder was fixed to a metal plate using double‐sided adhesive tape and interrogated directly with the laser in an FT‐ICR instrument. No matrix was required, various compounds in the powder were presumed to provide this function. The method required a smaller amount of sample (~200 μg) compared with traditional methods. By employing the powder method using Centella asiatica leaves, a higher number of reproducible molecular formulae (>5000) and metabolites (>650) were obtained than with the conventional methods. Flavonoids, phenolic acids, xanthones, lipids, carbohydrates, terpenoids and alkaloids were all detected from leaves, stems and roots of the plant.
Yamagaki et al. (2022) have shown that the postsource decay (PSD) fragmentation spectra of the [M + Na]+ ions of 4′‐O‐galloylpaeoniflorin (238) and 4‐O‐galloylalbiflorin (239) are different even though they are positional isomers. In particular, 4’‐galloylpaeoniflorin tended to eliminate a galloyl group to produce major ions whereas 4‐O‐galloylalbiflorin eliminated the sugar residue.

Investigations of enzyme‐assisted extraction of various compounds from plant material, with the aim of maximizing the quality of the extracts, have been made by Rafińska et al. (2022). Pectinase was shown to be particularly efficient in obtaining high quality extracts with a low content of interfering compounds using Medicago sativa L. as a test plant. The types of compounds that were investigated included carbohydrates, flavonoids and phenolic acids.
Other publications are listed in Table 33.









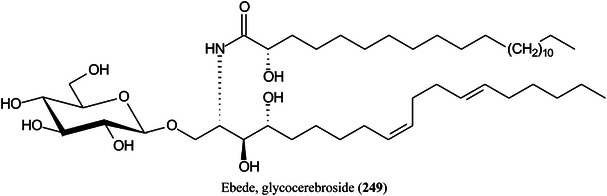

14. USE OF MALDI MASS SPECTROMETRY IN OTHER FIELDS
14.1. Enzymes
Another area where MALDI finds use is in monitoring the products of enzymatic digestions. One group of enzymes that has received considerable attention (see Table 34) are lytic polysaccharide monooxygenases (LPMOs) and a review on tools for assessing their activity on cellulose (includes HPLC, MALDI‐TOF, LC‐MS, NMR) has been published by Calderaro et al. (2021).
Table 34.
Use of matrix‐assisted laser desorption/ionization to study the products of enzymes' action on carbohydrates.
| Enzyme | Source | Methods a | Notes | References |
|---|---|---|---|---|
| Glycosyltransferases | ||||
| Acholetin phosphorylase | Acholeplasma laidlawii | MALDI, glycans | Shown to synthesise poly‐β‐1,3‐GlcNAc, completing the suite of β‑linked GlcNAc polysaccharides | Macdonald et al. (2022) |
| Archaeal β‐1,4‐N‐acetylglucosaminyl‐transferase (Agl24), | Sulfolobus acidocaldarius | TOF (DHB/TFA), glycolipids | Investigation of enzyme responsible for synthesis of N‐glycan chitobiose core in archaea | Meyer, Adam, et al. (2022) |
| Cycloisomalto‐oligosaccharide glucanotransferase | Thermo‐anaerobacter thermocopriae | TOF/TOF (DHB), glycans | Carbohydrate‐binding module of the enzyme shown to improves its cyclodextran production | Hong et al. (2022) |
| Enzyme from blr2358 gene | Bradyrhizobium diazoefficiens USDA110 | TOF/TOF (2,3‐DHB), glycans | Identification of gene involved in exopolysaccharide biosynthesis | Xu, Ruan, et al. (2021) |
| Fucosyl transferase FUT8 | Recombinant in HEK293F cells | PNGase F, TOF/TOF (DHB), N‐glycans | Investigation as to which modifications affect enzyme's ability to fucosylated core of N‐glycans | Zhang, Yang, et al. (2021) |
| Fucosyl transferase FUT8 | Human FUT8 | Glycans from egg yolk, TOF/TOF (DHB) | Investigation of factors influencing core fucosylation | García‐García et al. (2021) |
| Galacturonosyl‐transferases | Arabidopsis thaliana | TOF, glycans | Multiple Arabidopsis Gal‐transferases shown to synthesize polymeric homogalacturonan by oligosaccharide acceptor‐dependent or de novo synthesis | Engle et al. (2022) |
| α‐1,3‐Glucosyl‐transferase | Pneumococcus serotype 8C | TOF, glycans | Biochemical characterization and synthetic application | Wang, Sun, et al. (2021) |
| Glycosyl‐transferases, SdgB and SdgA, | Recombinant in E. coli | TOF/TOF (DHB) glycopeptides | SdgB but not SdgA transferred GlcNAc to staphylococcal adhesive proteins | Kim, Baek, et al. (2021) |
| Glycosyltransferase GT43 family | Phyllostachys edulis (bamboo) | TOF, glycans | Identification and expression analysis of enzymes reveal their potential function in xylan biosynthesis during rapid growth | Li, Wang, Yang, et al. (2021) |
| Glycosyltransferase family 61 | Conifers | R‐TOF/TOF, glycans | Study of xylan substitutions in conifers | Zhong, Phillips, et al. (2022) |
| Human exostosin‐like 3 | Human | TOF (DHB), glycans | Investigation into its mechanism of action for heparin sulfate synthesis | Wilson, Dendooven, et al. (2022) |
| Inverting S/O‐HexNAc‐transferase | Streptomyces venezuelae ATCC 15439 | TOF, glycopeptides | First experimental evidence of S‐linked glycosylation in actinobacteria | Sharma, Ahlawat, et al. (2022) |
| IRX10 Xylan synthases | Rice (Oryza sativa) | TOF/TOF (DHB), glycans (procainamide derivatives) | Identification of a xylan‐rich nanodomain at pit borders of xylem vessels | Wang, Yang, Wen, et al. (2022) |
| β‑KDO transferase KpsS | Recombinant | MALDI | Characterization of the initiating enzyme in the biosynthesis of the lipid acceptor for E. coli polysialic acid | Lanz et al. (2021) |
| Mannan synthase | Arabidopsis thaliana | R‐TOF (DHB/DMA) | The TaCslA12 gene expressed in the wheat grain endosperm shown to synthesize wheat‐like mannan when expressed in yeast and Arabidopsis | Verhertbruggen et al. (2021) |
| Protein S‐glycosyl‐transferases (thuS) | Bacillus thuringiensis serovar andalousiensis BGSC 4AW1 | TOF, glycoprotein, glycopeptides | Structural and mechanistic investigations of protein S‐glycosyltransferases | Fujinami et al. (2021) |
| RG‐1 Rhamnosyl‐transferases | Arabidopsis thaliana | TOF (DHB), glycans (2‐AB) | Investigation of the role of the enzyme in the biosynthesis of rhamnogalacturonan I in plants | Amos et al, (2022) |
| Trans‐sialidase | Trypanosoma congolense | R‐TOF/TOF (CHCA), glycopeptides | N‐Glycosylation shown to modulate enzymatic activity | Rosenau et al. (2022) |
| UTP‐glucose‐1‐phosphate uridylyltransferase YngB | Bacillus subtili | R‐TOF (DHB/s‐DHB), R‐TOF/TOF‐MS/MS, glycolipids | Enzyme shown to contribute to wall teichoic acid glucosylation and glycolipid formation during anaerobic growth | Wu, Rismondo, et al. (2022) |
| Xylan arabinosyl‐transferases (OsXATs) | Various grasses | R‐TOF (DHB/HIQ), glycans | Results show that that multiple OsXATs are involved in 3‐O‐arabinosylation of xylan | Zhong, Cui, et al. (2021) |
| Xylan arabinosyl‐transferases | Grasses recombinant in Arabidopsis thaliana | R‐TOF/TOF, glycans | Identification of enzymes catalyzing addition of 2‐O‐xylosyl residue onto arabinosyl side chains of xylan in grass species | Zhong, Lee, et al. (2022) |
| Xyloglucan xylosyltransferase | Arabidopsis thaliana and Oryza sativa (rice) in HEK293 cells | TOF (DHB), glycans | A single xyloglucan xylosyltransferase shown to be sufficient for generation of the XXXG xylosylation pattern of xyloglucan | Zhong, Phillips, et al. (2021) |
| Xyloglucan xylosyltransferase 1 | Arabidopsis thaliana. | TOF (DHB), glycans | Enzyme shown to display promiscuity toward donor substrates during in vitro reactions | Ehrlich et al. (2021) |
| Glycosidases | ||||
| Chitosanase | Bacillus toyonensis CCT 7899 | TOF/TOF (DHB), glycans | Purification and functional oligosaccharide production | Dantas et al. (2022) |
| β‐N‐Acetylhexosaminidase | Cateni‐bacterium mitsuokai | TOF/TOF, glycan (LNT2 glycan, 251) | Biochemical characterization of a form suitable for the synthesis of lacto‐N‐triose II | Liu, Ma, Shi, et al. (2021) |
| α‐Agarases | Colwellia echini A3T | TOF (DHB), glycans | A novel auxiliary agarolytic pathway shown to expand the metabolic versatility in the agar‐degrading marine bacterium C echini A3T | Pathiraja et al. (2021) |
| β‐Agarase | Cellulophga sp. J9‐3 | TOF/TOF, glycans | Purification and biochemical characterization of β‐agarase produced by marine microorganism (In Korean) | Kim, Kim, et al. (2021) |
| Alginate lyases (AlgLs) | Pseudomonas aeruginosa (Pae‐AlgL) and Azotobacter vinelandii (Avi‐AlgL) | TOF/TOF, glycans | Investigation of mannuronate preference | Zeng, Li, et al. (2021) |
| α‐Amylase (Amyrel) | Diptera muscomorpha) (Drosophila) | R‐TOF/TOF, glycans | A novel glucose‐forming α‐amylase with 4‐α‐glucanotransferase activity | Feller et al. (2021) |
| α‐l‐Arabino‐furanosidase, rCsAbf62A | Cellulomonas sp. B6 | TOF (DHB), glycans | Investigation of the xylan degradation system | Garrido et al. (2022) |
| Chitinase | Enterobacter cloacae subsp. cloacae (EcChi2) | TOF/TOF (DHB), glycans | Catalytic efficiency of multi‐domain transglycosylating chitinase is influenced by polycystic kidney disease domains | Mallakuntla and Podile (2021) |
| Chitinase | Streptococcus macrosporeus VTCC 940003 | TOF/TOF (DHB), glycans | Chito‐oligosaccharide production by the enzyme and their inhibition activities on Botrytis cinerea | Anh et al. (2021) |
| Chitosanase | Bacillus amyloliquefaciens | TOF, glycans and protein | Identification of a new class of chitosanase from B. amyloliquefaciens for the generation of chitooligosaccharides | Bhuvanachandra et al. (2021) |
| Chitosanase | Streptomyces niveus | R‐TOF/TOF (DHB, +ve. –ve), glycans | Expression and biochemical characterization of enzyme suitable for preparation of chitobiose | Chen, Cheng, et al. (2021) |
| Difructose dianhydride I synthase/hydrolase | Bifidobacterium dentium | TOF (5‐Me‐DHB), glycans (free and OAc) | Identification of a novel glycoside hydrolase family | Kashima et al. (2021) |
| Endo‐chitosanase, AqCoA | Aquabacterium sp. A7‐Y | TOF/TOF (DHB), glycans | Use for synthesis of active chitooligosaccharides and their application in fungal disease protection | Wang, Li, Liu, et al. (2021) |
| Endoglucanase RfGH5_4 | Recombinant from Ruminococcus flavefaciens FD‐1 v3 | R‐TOF (DHB), glycans | Use for recycling lignocellulosic plant biomasses | Gavande et al. (2022) |
| Exo‐β‐1,3‐glucanase | Moose rumen | TOF/TOF, glycans | Enzyme shows a structural framework similar to yeast exo‐β‐1,3‐glucanases | Kalyani et al. (2021) |
| α‐l‐Fucosidases | Human, Lactobacillus casei and Bacteroides fragilis. | R‐TOF/TOF (DHB/DMA), N‐glycans | Comparative studies on the substrate specificity and defucosylation activity of three α‐l‐fucosidases using synthetic fucosylated glycopeptides and glycoproteins as substrates | Prabhu et al. (2021) |
| β‐1,3‐Galactosidase (WceF) | Pantoea stewartii | MALDI, glycans | WceF shown to be a glycan biofilm‐modifying enzyme with a bacteriophage tailspike‐like fold | Irmscher et al. (2021) |
| β‐Galactosidase | Arion lusitanicus and Arion vulgaris (A0A0B7AQJ9), from Sf9 cells | TOF/TOF (CHCA) glycans | Identification, characterization, and expression | Thoma et al. (2022) |
| 1,3‐β‐Glucanases | Penicillium sumatraense | R‐TOF/TOF (DHB), glycans | Investigation into why the enzyme digests food but not endogenous glycans | Scafati et al. (2022) |
| β‐1,3‐Glucanase Gns6 | Oryza sativa (rice) | TOF/TOF (DHB), glycans | Enzyme shown to possess antifungal activity against Magnaporthe oryzae | Wang, Liu, Wang, et al. (2021) |
| GH13 α ‐glucosidase | Weissella cibaria | TOF/TOF (DHB), glycans | Enzyme shown to uncommonly act on short‐chain maltooligosaccharides | Wangpaiboon, Laohawuttichai, et al. (2021) |
| Endo‐glucanase 16 (EG16) | Various | TOF (DHB), glycans | Demonstrates conservation of enzyme activity across highly divergent plant lineages | Behar et al. (2021) |
| β‑1,3‐Glucanase (MoGluB) | Magnaporthe oryzae | TOF/TOF (DHB), glycans | Functional characterization and biocontrol of M. oryzae | Wang, Zhao, Wang, et al. (2021) |
| GH10 endo‐xylanase rCsXyn10A | Cellulomonas sp. B6 | TOF (DHB), glycans | Investigation of the xylan degradation system | Garrido et al. (2022) |
| GH18 chitinase | Recombinant (E. coli BL21(DE3)star cells | R‐TOF (DHB) | Auxiliary active site mutations shown to enhance the glycosynthase activity for polymerization of chitooligosaccharides | Alsina et al. (2021) |
| β‐1,3‐Glucanase (thermostable) | Trichoderma harzianum in Pichia pastoris | R‐TOF/TOF, glycans | Expression and use in oligoglucoside hydrolysis | Gao, Yan, et al. (2021) |
| Family 55 β‑1,3‑glucanase, AcGluA | Archangium sp strain AC19 | TOF, glycans | Heterologous expression and characterization | Wang, Li, Dong, et al. (2021) |
| Glucomannanase | Paenibacillus polymyxa | TOF/TOF, glycans (konjac glucomannan) | Identification and characterization | Li, Jiang, et al. (2021) |
| Glycoside hydrolyses (family 39) | Bifidobacterium longum subsp. longum | TOF (DHB), glycans | Mechanism of cooperative degradation of gum arabic arabinogalactan protein by B. longum surface enzymes | Sasaki et al. (2022) |
| Glycoside hydrolyses | Ustilago maydis | TOF (DHB/DMA), glycans | Identification of glycoside hydrolases and carbohydrate oxidases directed toward components of the fungal cell wall | Reyre et al. (2022) |
| β‐Hexosaminidases | Nicotiana benthamiana | R‐TOF/TOF (‐ve), glycans | β‐Hexosaminidases along the secretory pathway of N. benthamiana shown to have distinct specificities toward engineered helminth N‐glycans on recombinant glycoproteins | Alvisi et al. (2021) |
| Laccase | Madurella mycetomatis | Endo H, TOF, glycoprotein | Enzyme immobilized in silica‐coated ZIF‐8 nanocomposites for environmentally friendly cotton bleaching process | Tülek et al. (2021) |
| β‐Mannanase TrMan5A variants | Trichoderma reesei | R‐TOF/TOF (DHB) | Transglycosylation activity and enzyme synergy for synthesis of allyl glycosides from galactomannan | Butler, Birgersson, et al. (2022) |
| Pectate lyase AnPL9 | Aspergillus nidulans in Pichia pastoris | TOF/TOF, glycans | First report of a fungal pectate lyase belonging to the PL9 family. | Suzuki, Morishima, et al. (2022) |
| Pectinase | Streptomyces hydrogenans YAM1 | TOF (DHB), glycans | Investigation of antioxidant and anticancer activities of unsaturated oligo‐galacturonic acids | Abari et al. (2021) |
| PL17 Oligoalginate lyase | Zobellia galactanivorans DsijT | TOF, glycan | Structure–function analysis | Jouanneau et al. (2021) |
| α‐Rhamnosidases | Lactobacillus plantarum WCFS1 | TOF/TOF (DHB), glycosides | Production and role in deglycosylation of dietary flavonoids naringin and rutin | Ferreira‐Lazarte, et al. (2021) |
| Xyloglucanase MtXgh74 | Recombinant strain Pichia pastoris GS115 | TOF, glycans | Strategic aromatic residues in the catalytic cleft shown to modify thermostability, mode of enzyme action, and viscosity reduction ability | Berezina et al. (2021) |
| Xyloglucanase | Rhizomucor miehei CAU432 in Pichia pastoris | TOF/TOF (DHB), glycans | High level expression for production of xyloglucan oligosaccharides and its application in yoghurt | Wang, Li, Miao, et al. (2021) |
| GH74 xyloglucanase | Paenibacillus sp. | TOF (DHB/DMA), glycans | Mode of action on tamarind seed xyloglucan | Chen, Ropartz, et al. (2022) |
| Xyloglucanase B | Rhizomucor miehei exptessed in Pichia pastoris. | TOF/TOF, glycans | High‐level expression and its application in the preparation of partially hydrolysed apple pomace xyloglucan | Wang, Li, et al. (2022) |
| Xyloglucanase GH74 | Thielavia terrestris | TOF/TOF (DHB), glycans | Comparison of the roles of GH74 xyloglucanase and its CBM‐deleted variant in the degradation of xyloglucan‐rich biomass | Wang, Chen, Zhang, et al. (2022) |
| β‐Xylosidases GH8, GH39, and GH52 | Bacillus halodurans C‐125 | TOF/TOF, glycans | Substrate specificities toward substituted xylooligosaccharides | Teramoto et al. (2021) |
| α‑Xylosidase | Aspergillus oryzae | TOF/TOF (DHB), glycans | Characterization of an extracellular α‑xylosidase involved in xyloglucan degradation | Matsuzawa et al. (2022) |
| α‐Xylosidase 1 | Arabidopsis thaliana | TOF (s‐DHB), glycans | Cell wall modifications by the enzyme shown to be required for control of seed and fruit size | Di Marzo et al. (2022) |
| Yeast GH30 xylanase | Sugiyamaella lignohabitans | TOF/TOF (DHB), glycans | Enzyme shown to be a glucuronoxylanase with auxiliary xylobiohydrolase activity | Šuchová et al. (2022) |
| Other enzymes acting on sugars | ||||
| Acetyl xylan esterase GELP7 | Arabidopsis thaliana | TOF, glycans | Overexpression shown to improve saccharification efficiency | Rastogi et al. (2022) |
| Bifunctional feruloyl and acetyl xylan esterase | Metagenomes from beaver droppings and moose rumen | R‐TOF (DHB), glycans | Biochemical characterization and crystal structure | Hameleers et al. (2021) |
| Carbohydrate esterase family 16 | Arabidopsis thaliana | TOF/TOF (DHB), glycans | Enzyme shown to contain fungal hemicellulose acetyl esterases with varying specificity | Venegas et al. (2022) |
| Chondroitin sulfate/dermatan sulfate 4‐O‐endosulfatase | Commercial from squid cartilage | R‐TOF (HABA/TMG 2, (252), ‐ve), glycans | Investigation of mode of action | Wang, Przybylski, et al. (2021) |
| Galactan precursor transporter | Mycobacterium smegmatis | R‐TOF/TOF (DHB), glycans | An ATP‐binding cassette transporter Wzm–Wzt shown to catalyze translocation of lipid‐linked galactan across the plasma membrane in mycobacteria | Savková et al. (2021) |
| Heterologous invertase | Expressed in Yarrowia lipolytica | PNGase F, Endo H, TOF/TOF (DHB), HPLC, glycans, exoglycosidases | Study of the influence of Y. lipolytica glycosylation on the biochemical properties and oligomerization of heterologous invertase | Szymański et al. (2022) |
| Human Gb3/CD77 synthase | Human | TOF/TOF (norharmane), GSLs | One (Asn121) of the two N‐glycans on human Gb3/CD77 synthase shown to be expendable | Mikolajczyk et al. (2021) |
| Lytic polysaccharide monooxygenase | Eupenicillium parvum 4‐14 | TOF/TOF (DHB), glycans | Identification of a highly xyloglucan active enzyme that shows boosting effect on hydrolysis of complex lignocellulosic substrates | Shi, Chen, et al. (2021) |
| Lytic polysaccharide monooxygenases | Coptotermes gestroi (termite) | TOF/TOF (DHB), glycans | Shown not to be involved in lignocellulose digestion but might play a role in termite development | Cairo et al. (2021) |
| Lytic polysaccharide monooxygenases | Pleurotus ostreatus | TOF/TOF (DHB), glycans | Enhanced konjac glucomannan hydrolysis and generation of prebiotic oligosaccharides | Li, Sun, et al. (2021) |
| Lytic polysaccharide monooxygenases (cellulose‑active) | Cellulomonas species | TOF/TOF (DHB), glycans | Identification and characterization of four lytic polysaccharide monooxygenases | Li, Solhi, et al. (2021) |
| Lytic polysaccharide monooxygenase TgAA11 | Trichoderma guizhouense NJAU 4742 | TOF/TOF (DHB/TFA), glycans | Functional characterization of a novel copper‐dependent enzyme in the oxidative degradation of chitin | Ma, Liu, et al. (2021) |
| Lytic polysaccharide monooxygenase | Podosphaera xanthii | TOF/TOF glycans | The enzyme from the cucurbit powdery mildew pathogen P., xanthii contributes to the suppression of chitin‐triggered immunity | Polonio et al. (2021) |
| Lytic polysaccharide monooxygenases | Aspergillus oryzae | TOF, glycans | Comparison of C4‐oxidizing and C1/C4‐oxidizing AA9 LPMOs in substrate adsorption, H2O2‐driven activity and synergy with cellulase on celluloses of different crystallinity | Chen, Zhang, Long, et al. (2021) |
| Lytic polysaccharide monooxygenases | Aphanomyces astaci (fungus) | TOF/TOF, glycans | Enzyme identified as a chitin‐specific virulence factors in “crayfish plague” | Sabbadin et al. (2021) |
| Lytic polysaccharide monooxygenase | Aspergillus fumigatus (AfAA11B) | TOF, glycans, HPAEC‐PAD | Enzyme shown to have a preference for soluble substrates and absence of monooxygenase activity | Rieder et al. (2021) |
| Lytic polysaccharide monooxygenase | Cellvibrio japonicus | TOF/TOF (DHB) | C‐type cytochrome shown to initiate reduction of bacterial LPMOs | Branch et al. (2021) |
| Lytic polysaccharide monooxygenases | Ceriporiopsis subvermispora | TOF/TOF (DHB), glycans | Identification of two C1‐oxidizing monooxygenases and demonstration of enhancement of the saccharification of wheat straw | Long et al. (2021) |
| Lytic polysaccharide monooxygenases | Sordaria brevicollis | TOF/TOF (DHB), glycans | Two C1‑oxidizing AA9 lytic polysaccharide monooxygenases differ in thermostability, activity, and synergy with cellulase | Zhang, Chen, Long, et al. (2021) |
| Lytic polysaccharide monooxygenase | Aspergillus fumigatus | TOF/TOF (DHB), glycans | Characterization of enzyme which shows functional variation among family AA11 fungal LPMOs | Støpamo et al. (2021) |
| Lytic polysaccharide monooxygenase | Irpex lacteus 254 | TOF/TOF (DHB), glycans | Investigation of lignin degradation via Fenton reaction | Li, Zhao, et al. (2021) |
| Lytic polysaccharide monooxygenase | Ceriporiopsis subvermispora | TOF/TOF (DHB), glycans | Functional and structural characterizations | Nguyen et al. (2022) |
| Lytic polysaccharide monooxygenase | Thermoascus aurantiacus | TLC. TOF/TOF (CMBT/DHB) | Purification, structural characterization and identification of its C1‐ and C4‐oxidized reaction products | Yu et al. (2022) |
| Lytic polysaccharide monooxygenases | Thielavia terrestris, TtAA9F and TtAA9G, in Trichoderma reesei | TOF/TOF (CMBT, DHB), glycans | For development of a high‑throughput gluco‑oligosaccharide oxidase‑based HRP colorimetric method for assaying LPMO activity | Wu, Tian, et al. (2022) |
| Lytic polysaccharide monooxygenases | Thermothielavioides terrestris | R‐TOF/TOF (DHB), glycans | Comparison of six LPMOs | Tõlgo et al. (2022) |
| Lytic polysaccharide monooxygenase | Natrialbaceae archaeon | TOF/TOF (DHB), glycans | Characterization and application for chitin biodegradation | Li, Liu, et al. (2022) |
| Lytic polysaccharide monooxygenase cMPO2 | Compost | TOF/TOF (DHB), glycans | Structural and functional study for the oxidative degradation of cellulose | Ma, Li, et al. (2022) |
| Lytic polysaccharide monooxygenase, SscLPMO10B | Streptomyces scabies | TOF/TOF (DHB), glycans from cellulose | Apparent monooxygenase activity observed in reactions without exogenously added H2O2 reflects a peroxygenase reaction | Stepnov et al. (2022) |
| Lytic polysaccharide monooxygenase PpAA10 | Pseudomonas putida W619 recombinant in E. coli | TOF/TOF (DHB), glycans | Activity and substrate specificity: An ATR FTIR‐based sensitive assay using attenuated total reflection‐FT‐ICR | Serra et al. (2022) |
| Lytic polysaccharide monooxygenase AA15 | Tribolium castaneum and Locusta migratoria | TOF/TOF, glycans | Enzyme shown to be required for efficient chitinous cuticle turnover during insect molting | Qu et al. (2022) |
| Lytic polysaccharide monooxygenase AA9 | Aspergillus nidulans | TOF/TOF, glycans | Deletion of AA9 LPMO shown to impact secretome and growth on lignocellulose | Terrasan et al. (2022) |
| Lytic polysaccharide monooxygenase | Cellulomonas flavigena | TOF/TOF (DHB), glycans | Chitin‐active LPMOs shown to be rare in Cellulomonas species | Li, Goddard‐Borger, et al. (2022) |
| Lytic polysaccharide monooxygenase | Chaetomium thermophilum | TOF, glycans | Oxidation properties and synergism (In Chinese) | Xia, Liu, et al. (2022) |
| Lytic polysaccharide monooxygenase | Aspergillus fumigatus | TOF, glycans | Light shown to boost the activity of enzyme | Velasco et al. (2022) |
| Sucrose‐6‐phosphate hydrolase | Lactobacillus gasseri | TOF (DHB), glycans | Crystal structure and potential applications in fructan production and the food industry | de Lima et al. (2021) |
| Xylan O‐acetyltransferase 1 (XOAT1) | Recombinant and engineered | TOF (DHB), glycans (2‐AB) | Redesign for controlled functionalization of acetylated xylan for cell‐free polymer biosynthesis | Wang, Bharadwaj, et al. (2021) |
Format (not all items present): MALDI method (matrix), compounds studied (derivative) other methods.

A multiplexed nanostructure‑initiator mass spectrometry (NIMS) assay has been described for simultaneously detecting glycosyl hydrolase and lignin modifying enzyme activities (Ing, et al., 2021). [U]‐13C glucose and [U]‐13C cellobiose were used as internal standards with detection by MALDI‐TOF MS.
Other work on enzymes is summarized in Table 34.
14.2. Medical applications
MALDI has been used extensively in medical research and is involved in the identification of biomarkers, tracking changes in glycosylation in various disease states, particularly cancer, identification of congenital disorders of glycosylation CDG), and patient monitoring. Reviews on these and other topics are listed in Table 35.
Table 35.
Reviews and general articles on the application of matrix‐assisted laser desorption/ionization to disease.
| Subject | Comments | Citations | References |
|---|---|---|---|
| General | |||
| Glycan imaging mass spectrometry: Progress in developing clinical diagnostic assays for tissues, biofluids, and cells | General review: Covers instrumentation, FFPE samples, sample preparation | 120 | Blaschke, McDowell, et al. (2021) |
| Advances in understanding N‐glycosylation structure, function, and regulation in health and disease | Short review. Structure and function of N‐glycans. N‐Glycosylation in disease states | 104 | Esmail and Manolson (2021) |
| Clinical applications of mass spectrometry | Covers clinical applications published between 2015 and 2020 and includes glycoproteome and glycome profiling, potential biomarker and drug target discovery and characterization of therapeutic glycoproteins | 268 | Fang and Lu (2022) |
| Mass spectrometry imaging spatial tissue analysis toward personalized medicine | Several references to the use of MALDI for glycan analysis | 105 | Gonçalves et al. (2022) |
| Immunoglobulin A glycosylation and its role in disease | Mainly biology, very little on MALDI | 249 | Hansen et al. (2021) |
| Enhancing precision medicine through clinical mass spectrometry platform | General review. Emphasis on MALDI. imaging | 100 | Hristova and Svinarov (2022) |
| Mass spectrometric biosensing: A powerful approach for multiplexed analysis of clinical biomolecules | Use of compound‐specific mass tags. Much use of MALDI analysis | 140 | Hu, Liu, et al. (2021) |
| Importance of evaluating protein glycosylation in pluripotent stem cell‑derived cardiomyocytes for research and clinical applications | Contains table listing published methods where MALDI used for glycan detection | 125 | Kelly et al. (2021) |
| Progress of proteomics‐driven precision medicine: From a glycosylation view | Glycoproteomics of cancer | 78 | Liang, Fu, et al. (2022) |
| Lipids and glycolipids as biomarkers of mycobacterial infections | Emphasised advantages of MALDI in providing rapid diagnosis | 124 | Liu and Larrouy‐Maumus (2022) |
| Quantitative clinical glycomics strategies: A guide for selecting the best analysis approach | Compares performance of different methods (e.g. MALDI, LC‐MS) of released glycans and glycopeptides | 111 | Patabandige et al. (2022) |
| MALDI Mass spectrometry imaging in the clinical landscape | Use of MALDI for disease detection, disease subtyping, disease outcome prediction | 188 | Schwamborn (2022) |
| Proteomic and glyco(proteo)mic tools in the profiling of cardiac progenitors and pluripotent stem cell derived cardiomyocytes: Accelerating translation into therapy | Short section on glycomic and proteomic analysis and table listing methods for analysis of cardiac progenitor cells | 170 | Sebastião et al. (2021) |
| Glycomic technology and its application in disease marker mining | Concentrates on methods based on MALDI (In Chinese) | Shifang and Jianxin (2022) | |
| MALDI‐TOF Mass spectrometry technology as a tool for the rapid diagnosis of antimicrobial resistance in bacteria | Identification of bacteria with small section on MS of lipid A | 80 | Yoon and Jeong (2021) |
| Cancer | |||
| Lipid (and glycolipid) biomarkers for breast cancer diagnostics | Mainly lipids. A few glycolipids included | 89 | Bibi et al. (2022) |
| Glycosylation changes in prostate cancer progression | General review with references to MALDI analysis | 110 | Butler and Huang (2021) |
| Importance of glycosphingolipids on cellular and molecular mechanisms associated with epithelial‐to‐mesenchymal transition in cancer | Discusses individual compound types with section on analytical methods | 139 | Cumin et al. (2021) |
| Glycoproteogenomics: Setting the course for next‐generation cancer neoantigen discovery for cancer vaccines | Discusses cancer vaccines, glycosylation in cancer and glycomics and glycoproteomic methods | 206 | Ferreira et al. (2021) |
| Mass spectrometry‐based glycoproteomics and prostate cancer | Few direct references to MALDI | 76 | Gabriele et al. (2021) |
| Cancer glycomics offers potential biomarkers and therapeutic targets in the framework of predictive, preventive and personalized (3P) medicine | N‐ and O‐glycans. Different analytical methods – mass spectrometry, lectins, immunological, fluorescence imaging | 170 | Guo, Jia, et al. (2022) |
| Glycosylation in cancer: Its application as a biomarker and recent advances of analytical techniques | Brief review, biochemistry, analysis and use for biomarker discovery | 131 | Haga and Ueda (2022) |
| Urinary glycan biomarkers in prostate cancer | Detection of biomarkers and MALDI methods for N‐glycan analysis | 118 | Hatakeyama et al. (2021) |
| The repertoire of glycan alterations and glycoproteins in human cancers | Tables listing use of MALDI in clinical studies | 277 | Kori et al. (2021) |
| Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research | Methods (instrumentation, matrices, matrix deposition, quantification), applications (identification of disease, biomarkers, drug distribution) | 142 | Lee, Yeoh, et al. (2021) |
| Glycosphingolipids in human embryonic stem cells and breast cancer stem cells, and potential cancer therapy strategies based on their structures and functions | MALDI and other analytical methods. GSLs as potential biomarkers for breast cancer | 125 | Liang (2022) |
| Blood‐based protein biomarkers in bladder urothelial tumors | Protein and glycoprotein biomarkers | 246 | López‐Cortés et al. (2021) |
| Mass spectrometry in the lipid study of cancer | Mainly concentrates on changes in lipid metabolism | 165 | Nabi et al. (2021) |
| Glycosylation alterations in cancer cells, prognostic value of glycan biomarkers and their potential as novel therapeutic targets in breast cancer | Glycans as biomarkers and use for development of targeted therapies | 118 | Peric et al. (2022) |
| Glycosylation and its research progress in endometrial cancer | Role of glycosylation, characterization of cancer biomarkers, chemotherapy | 173 | Pu et al. (2022) |
| Mass spectrometry: A powerful method for monitoring various types of leukemia, especially MALDI‐TOF in leukemia's proteomics studies | Mainly concentrates on proteins but a few references to glycoproteins and N‐glycans | 130 | Ramandi et al. (2022) |
| Mass spectrometry imaging in gynecological cancers: The best is yet to come | Different types of cancer, problems with imaging, types of imaging. | 98 | Pietkiewicz et al. (2022) |
| Separation based characterization methods for the N‐glycosylation analysis of prostate‐specific antigen | Covers MS techniques, including MALDI, LC/MS and CE/MS, | 90 | Reider et al. (2021) |
| Recent advances in mass spectrometry‐based glycomic and glycoproteomic studies of pancreatic diseases | Subjects include diabetes and cancer biomarkers | 157 | Tabang et al. (2021) |
| Causal link between immunoglobulin G glycosylation and cancer: A potential glycobiomarker for early tumor detection | Reports promising novel biomarkers for noninvasive‐cancer diagnosis, Only few MALDI references | 102 | Wang, Huang, et al. (2021) |
| MS imaging of multicellular tumor spheroids and organoids as an emerging tool for personalized medicine and drug discovery | Ionization methods, sample preparation, data analysis, quantification. | 116 | Wang and Hummon (2021) |
| Glycomic‐based biomarkers for ovarian cancer: Advances and challenges | Concentrates on N‐glycans. Short section on instrumentation | 103 | Wanyama and Blanchard (2021) |
| MALDI‐TOF/MS Analysis of noninvasive human urine and saliva samples for the identification of new cancer biomarkers | Mainly concentrates on proteins but some references to glycoproteins and glycopeptides | 89 | Zambonin and Aresta (2022) |
| Aging | |||
| Glycosylation and aging | Discusses various changes in the aging process; little on MALDI | 267 | Cindrić et al. (2021) |
| Immunoglobulin G glycans – Biomarkers and molecular effectors of aging | Contains table listing methods (many MALDI) used for examination of changes to IgG glycosylation. | 173 | Krištić et al. (2022) |
| Glycosylation biomarkers associated with age‐related diseases and current methods for glycan analysis | Discusses various disease types with tables of application. Also analytical methods | 223 | Paton et al. (2021) |
| Neurological disease | |||
| What can N‐glycomics and N‐glycoproteomics of cerebrospinal fluid tell us about Alzheimer's disease? | Discusses methods used for analysis of N‐glycans | 112 | Gaunitz et al. (2021) |
| Glycomic and glycoproteomic techniques in neurodegenerative disorders and neurotrauma: Towards personalized markers | Methods (enrichment, ionization, chromatography‐MS, software), applications to different diseases | 377 | Kobeissy et al. (2022) |
| MS‐based glycomics: An analytical tool to assess nervous system diseases | First section deals with techniques – MALDI, LC‐MS (permethylation), CE‐MS, ion mobility, fragmentation. Second section on applications (biomarkers) – Alzheimer's disease, Parkinson's disease, CNS disease, traumatic brain injury | 311 | Peng et al. (2022) |
| Role and therapeutic implications of protein glycosylation in neuroinflammation | Types of glycosylation, results of injury to CNS, neuroinflammation, glycodysregulations | 127 | Rebelo et al. (2022) |
| Gangliosides as biomarkers of human brain diseases: Trends in discovery and characterization by high‐performance mass spectrometry | Discusses the use of MS in different diseases. | 214 | Sarbu et al. (2022) |
| MALDI imaging mass spectrometry: An emerging tool in neurology | Discusses applications to different diseases and summarises in a table | 85 | Schnackenberg et al. (2022) |
| Other diseases | |||
| Congenital disorders of glycosylation | Contains table of different diseases but with no associated references. Also incorrect N‐glycan structures. | 24 | Mendes et al. (2022) |
| Mass spectrometry‐based N‐glycosylation analysis in kidney disease | Sample preparation, general workflows, N‐glycosylation in specific diseases, table of applications with many MALDI references. | 122 | Ren, Bian, and Cai (2022) |
| Diagnostics of lysosomal storage diseases by mass spectrometry | Concentrates on the identification of biomarkers for various diseases | 59 | Pančík, Pakanová, Květoň, et al. (2022) |
| Recent advances and potential future applications of MALDI‐TOF mass spectrometry for identification of helminths | Causative agents of major neglected tropical diseases. Several references to the use of MALDI for glycan and glycoprotein identification. | 52 | Sy et al. (2022) |
| Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders | Comprehensive review covering various glycoproteins with notes on analytical methods | 379 | Ugonotti et al. (2021) |
| Glycosylation in autoimmune diseases | Discusses various diseases such as multiple sclerosis and rheumatoid arthritis. Little on MALDI | 86 | Ząbczyńska et al. (2021) |
Since 2018 (MALDI‐TOF‐MS), termed MASS‐FIX, has replaced serum immunofixation for the detection and isotyping of serum monoclonal protein at Mayo Clinic Rochester campus. It offers the advantages of rapid throughput, high sensitivity and specificity for the detection of monoclonal protein, and ability to differentiate therapeutic monoclonal antibodies. It can easily identify light chain N‐glycosylation which has diagnostic implications, as it is more common in some disorders than others (Mellors, Dasari, et al., 2021; Kohlhagen et al., 2021). It has been used to detect the novel antibody drug conjugate Belantamab mafodotin (Mellors, Kohlhagen, et al., 2021). Since its introduction, MASS‐FIX has been used extensively. Google Scholar lists over 100 papers although few mention glycans.
14.2.1. Cancer
The application of MALDI with imaging MS in cancer research, with particular emphasis on the sample preparation step, has been discussed by Buszewska‐Forajta et al. (2022). Several protocols based on cryosections and FFPE tissue were compared, taking into account the measured metabolites of potential diagnostic importance for a given type of cancer. The importance of the sample collection and storage, pretreatment protocols were emphasised and it was noted that proper preservation of tissue material should start during collection. Use of an appropriate quenching method will stop the reactive enzymatic autolysis and tissue degradation. The choice of the MALDI matrix and its method of application is also critical.
Much work has been aimed at the discovery of biomarkers. Many examples are listed in Table 36. N‐Glycosylation often appears significantly different in cancer patients as exemplified by a study of stage II and III colon cancer in serum and tissues. N‐glycosylation was generally decreased in serum whereas high‐mannose, hypogalactosylated, and tetra‐antennary glycans were overexpressed in tumor tissues (Coura et al., 2021). The quantities of multiantennary glycans were also elevated in some reports (Takei et al., 2022) as were α2→3‐linked sialic acids (Boyaval et al., 2022). Fucosylation levels have also been reported to vary. With respect to the methods that are generally used for MALDI analysis; permethylation is popular as is the preparation of linkage‐specific derivatives for sialic acids.
14.2.2. Congenital disorders of glycosylation (CDG)
These disorders are comparatively rare but have attracted much use of MALDI mass spectrometry for their diagnosis. The diseases affect various aspects of the glycosylation of proteins and lipids and are usually diagnosed by observations of the glycosylation of serum transferrin, a glycoprotein with two N‐linked glycosylation sites that are normally occupied by sialylated biantennary glycans. However, recently three cases have been detected where serum transferrin glycosylation appeared normal. Diagnosis was achieved by observation of abnormally glycosylated apolipoprotein C‐III (Raynor, Vincent‐Delorme, et al., 2021).
14.2.3. Biomarkers for other diseases
Some unusually large glycolipids (e.g., 253 and 254) have been detected by negative ion MALDI‐TOF in human breath of tuberculosis patients and have been proposed as biomarkers for the disease (Mosquera‐Restrepo et al., 2022).


Using a solid‐phase glycoblotting technique and MALDI‐TOFMS‐based quantitative glycomics, Otaki et al. (2022) have mapped N‐glycosylation patterns of 16 mouse organs/tissues, serum, and serum‐derived exosomes from Slc:ddY mice. Data are presented mainly as heat maps in the main paper but full quantitative data are presented in the Supplementary information. A preliminary examination showed that machine learning analysis of the mouse lung N‐glycome data set enables differentiation of lungs from different mouse strains such as the outbred mouse Slc:ddY, inbred mouse DBA/2Crslc, and systemic lupus erythematosus model mouse MRL‐lpr/lpr emphasising the usefulness of a similar human organ/tissue glycome database for understanding the importance of the N‐glycome‐for identification of disease‐specific biomarkers.
Other applications are listed in Table 36.
14.3. Biopharmaceuticals
Relevant general reviews are listed in Table 37.
Table 37.
Reviews and general articles on the analysis of biopharmaceuticals.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Glycoproteomics technologies in glycobiotechnology | Short sections on different techniques such as MALDI and ion mobility | 92 | Alagesan et al. (2021) |
| Characterization of glycosylation in monoclonal antibodies and its importance in therapeutic antibody development | Summarizes analytical techniques for monitoring glycosylation and effects of glycosylation on function | 93 | Kaur (2021) |
| Biopharmaceutical quality control with mass spectrometry | Recent advances, including post‐translation modifications and structural characterization | 87 | Liu and Schulz (2021) |
| State‐of‐the‐art glycomics technologies in glycobiotechnology | General review of different methods: CE, MALDI, LC‐MS, ion mobility | 217 | Pralow et al. (2021) |
| N‐Glycosylation of monoclonal antibody therapeutics: A comprehensive review on significance and characterization | Structure, function and analysis | 180 | Shrivastava et al. (2022) |
14.3.1. Monoclonal antibodies
Much of the work being undertaken with biopharmaceuticals is concerned with the production of monoclonal antibodies in species other than human and where glycosylation is invariably different. In some cases, such as when terminal α‐galactose residues or glycolylneuraminic acids are added, these glycans can be antigenic prompting the development of methods for their detection and great scope for genetic engineering to “humanize” the glycosylation in such nonhuman species.
Martín et al. (2021) have investigated the effect of storage on IgG Fc N‐glycosylation in the commonly analyzed biofluids, serum and plasma. Stability was tested by incubating samples from three healthy donors for up to 2 weeks at 50°C compared with storage at −80°C for 2 weeks. All tested IgG glycosylation features, namely sialylation, galactosylation, bisection, and fucosylation remained unchanged up to room temperature as well as during multiple freeze−thaw cycles and exposure to light. Only when subjected to 37°C or 50°C for 2 weeks, did galactosylation and sialylation subtly change.
14.3.2. Vaccines
Effective vaccines against pathological bacteria can be prepared by linking expressed O‐antigen polysaccharides with specific carrier proteins. These polysaccharides are typically polydisperse, and the carrier proteins can have multiple glycosylation sites. Consequently, the resultant recombinant glycoconjugate vaccines frequently have a high structural heterogeneity, making their characterization difficult. Nicolardi et al. (2022) have addressed this problem using three glycoconjugate vaccine candidates, obtained from the bioconjugation of the O‐antigen polysaccharides from E. coli serotypes O2, O6A, and O25B with the genetically detoxified exotoxin A from Pseudomonas aeruginosa. They used MALDI‐ISD FT‐ICR MS to analyse protein and glycan ISD fragment ions which were selectively detected using DAN and s‐DHB respectively. MS/MS analysis of O‐antigen ISD fragments enabled detection of specific O‐repeats and fragments from the ends of the protein chain provided identification. The rapid method required only minute sample amounts and avoided the use of chemical derivatization.
Vaccine development involving conjugation of glycans to proteins such as BSA is another area where MALDI has played a major part in monitoring products and estimating the number of sugars that are attached to the protein. Relevant work is listed in Table 38. A protocol: “Oligosaccharide antigen conjugation to carrier proteins to formulate glycoconjugate vaccines employing dicarboxylic acid linkers” has been published (Smith & Guo, 2021) and the review with 199 references: “Cross‐reacting material (CRM197) as a carrier protein for carbohydrate conjugate vaccines targeted at bacterial and fungal pathogens” (Khatuntseva & Nifantiev, 2022) is also relevant.
Table 38.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry to study carbohydrate‐protein conjugates.
| Sugar | Protein | Methods a | Notes | References |
|---|---|---|---|---|
| Azido‐gluconolactone (259) | Gly‐His‐Tagged proteins | TOF | Synthesis of N‐terminally modified proteins | Brune et al. (2021) |
| Methylated rhamnan oligosaccharide | HSA | TOF | For development of vaccine against Pseudomonas aeruginosa | Cairns et al. (2022) |
| Oligomannose, from hepta‐ and nona‐high‐mannose glycans | BSA | TOF/TOF (DHA) | As specific HIV‐1‐neutralizing antibodies | Cattin et al. (2022) |
| Globo H analogs | CRM197 | TOF (SA) | Chemoenzymatic synthesis for immunogenicity evaluation | Chen, Lin, et al. (2022) |
| Lipoteichoic acid and PS‐II polysaccharide from Clostridium difficile | HSA | TOF (SA) | To develop a vaccine against the dental pathogen Streptococcus mutans | Cox et al. (2021) |
| Glucosinolates | BSA | TOF | Use of the myrosinase‐glucosinolate system to generate neoglycoproteins | Cutolo et al. (2022) |
| Biantennary and high‐mannose N‐glycans | Bacteriophage Qβ nanoparticles and BSA | TOF/TOF (SA) | Synthesis and immunological study reveals dominant antibody responses to the conserved chitobiose core | Donahue et al. (2022) |
| α‐d‐Rha4NFo (260)‐containing oligosaccharides | BSA | TOF | As potential vaccine against Brucellosis infection | Duncombe et al. (2022) |
| Saponin adjuvants and the Tn antigen plus linker (261) | BSA | TOF/TOF (SA) | Design, synthesis, and initial immunological evaluation as self‐adjuvanting glycoconjugate cancer vaccine | Fuentes, Aguinagalde, et al. (2021) |
| Pyruvylated‐human‐type complex N‐glycans | HiLyte Fluor 750‐conjugated HSA | TOF | In vivo imaging of fluorescent albumin modified with pyruvylated‐human‐type complex oligosaccharide reveals sialylation‐like biodistribution and kinetics | Fukuhara et al. (2022) |
| Tetravalent glycodendrons (αGal, βGal and/or αFuc) | BSA | TOF/TOF | Prepared by click chemistry, as ligands for bacterial lectins | Goyard et al. (2022) |
| Rha4NFo(1 → 2)Rha4NFo from Brucella sp. | BSA, CRM197 | TOF/TOF (SA) | Synthesis and immunogenicity | Hao et al. (2021) |
| C‐3‐Substituted N,N’‐diacetyl‐lactosamine glycomimetics (262) | HSA | TOF | Chemoenzymatic synthesis for inhibition of cancer‐related galectin‐3 | Heine et al. (2021) |
| 3‐O‐Methyl‐d‐rhamnose oligosaccharide (263) | HSA | TOF | Synthesis and immunogenicity of a methyl rhamnan pentasaccharide conjugate from Pseudomonas aeruginosa A‑band polysaccharide | Jamshidi et al. (2022) |
| Several Lewisa six‐aminohexyl glycoside (e.g., 264) | BSA | TOF (SA) | Anti‐Lea monoclonal antibody SPM 522 recognizes an extended Lea epitope | Jegatheeswaran et al. (2022) |
| β‐1,2‐Mannans | HSA | TOF | As potential antifungal vaccines | Liao, Pan, et al. (2022) |
| Trisaccharides (265) related to Bacillus anthracis | KLH, BSA | TOF | Potential vaccine development. Induced immune response in mice | Liao, Zhuo, et al. (2022) |
| Man5GlcNAc2 | BSA, CRM197 | TOF/TOF (DHB) | Synthesis and immunological evaluation as HIV‑1 vaccine candidates | Liu, Huo, et al. (2022) |
| α‐Gal‐containing oligosaccharides derived from Leishmania major (266) | BSA | TOF (SA) | Reversed immunoglycomics used to identify α‐galactosyl‐bearing glycotopes specific for L. major infection | Montoya et al. (2021) |
| Synthetic β ‐Galf ‐containing glycans | BSA | TOF (SA) | For specific recognition of β ‐galactofuranose‐containing glycans of synthetic neoglycoproteins by sera of chronic Chagas disease patients | Montoya et al. (2022) |
| O‐Antigen from E. coli O25B (267) | CRM197 | TOF/TOF (SA) | For vaccine against E. coli O25B | Naini et al. (2022) |
| Pentasaccharide repeating unit of LPS derived from virulent E. coli O1 | BSA | TOF | Synthesis of pentasaccharide and identification of a glycotope candidate of avian pathogenic E. coli O1 | Nishi et al. (2021) |
| Amarogentin (glycoside) (268) | BSA, HSA | TOF | For development of competitive immune‐chromatographic assay | Nuntawong et al. (2021) |
| Structurally rigid TnThr mimic (Gal analogue) | BSA | TOF/TOF | As template for molecularly imprinted polymers. A promising tool for cancer diagnostics | Palladino et al. (2022) |
| Lactose and 3‐ and 2′ ‐fucosyl lactose | Erythrina cristagalli lectin, Aleuria aurantia lectin, and Ulex europaeus agglutinin‐I | TOF/TOF (CHCA) | Synthesis of photoactivable oligosaccharide derivatives from 1,2‐cyclic carbamate building blocks and study of their interaction with carbohydrate‐binding proteins | Podvalnyy et al. (2021) |
| Streptococcus pneumoniae serotype 14 capsular polysaccharide (269) | Adenoviral type 3 dodecahedron | Q‐TOF (SA) | Investigation of the use of adenovirus dodecahedron as a carrier for glycoconjugate vaccines | Prasanna et al. (2021) |
| Type K9 capsular polysaccharide of Acinetobacter baumannii | BSA, chicken ovalbumin and snail hemocyanin [KLH]) | L‐TOF/TOF (DHB) | Determination of immune response | Rudenko et al. (2022) |
| High‐mannose and complex N‐glycans | CRM197 | TOF (SA) | Immunogenicity evaluation of N‑glycans recognized by HIV broadly neutralizing antibodies | Shivatare et al. (2021) |
| Various | Phage pVIII protein | TOF (SA) | For construction of multivalent liquid glycan array | Sojitra et al. (2021) |
| Lipid A analog CRX‐527 | Synthetic peptides | TOF, FT‐ICR (DHB) | Shown to enhance vaccination efficacy and tumor control | Tondini et al. (2022) |
| Mannose dendrimers | CRM197 | TOF | For development of site‐specific multifunctionalization of CRM197 by disulfide rebridging for conjugate vaccine development | Trattnig et al. (2022) |
| Phenolic glycolipids from Mycobacterium leprae, | BSA | TOF/TOF | For development of diagnostic tests for leprosy | van Dijk et al. (2021) |
| Galf‐β1→3‐Man‐α‐, Gal‐α1→3‐Galf‐β1→3‐Man‐α‐ and Gal‐α1→6‐Gal‐α1→3‐Galf‐β1→3‐Man‐α‐ | BSA | TOF | For monitoring of New‐World tegumentary leishmaniasis using synthetic type‐2 glycoinositolphospholipid‐based neoglycoproteins | Viana et al. (2022) |
| Group A streptococcal trisaccharide | Fn, Fn2, rsScpA193 or CRM197 | TOF (SA) | Investigation of best potential carrier protein for glycoconjugate vaccine development | Wang, Zhao, Zhao, et al. (2021) |
| GalCer | Receptor‐binding domain (RBD) of SARS‐Cov‐2 | TOF/TOF (SA) | Shown to induce potent immunity against SARS‐CoV‑2 and its variants of concern | Wang, Wen, et al. (2022) |
| d‐Glycero‐β‑d‐mannoheptose phosphate | HSA | L‐TOF (SA) | For production of molecular probes | Williams et al. (2021) |
| 9NHAc‐GD2 | BSA | TOF (SA) | To overcome the hydrolytic instability of O‐acetylated‐GD2 for anticancer conjugate vaccine development | Wu, Ye, et al. (2021) |
| GalNAc | CRM197 | L‐TOF (SA) | Development of a GalNAc‐tyrosine‐specific monoclonal antibody and detection of tyrosine O ‑GalNAcylation | Xia, Bellomo, et al. (2022) |
| GM3 Glycan | BSA | TOF | Synthesis and evaluation of liposomal anti‐GM3 cancer vaccine candidates | Yin, Lu, et al. (2021) |
| Tn antigen | HSA, CRM197 | TOF/TOF (DHB) | As part of Tn‐based three‐component cancer vaccine | Yang, Luo, et al. (2022) |
| Tetrasaccharide haptens from Vibrio vulnificus MO6‐24 and BO62316 (270) | CRM197 or BSA | IT‐TOF (SA) | Total synthesis of glycans and immunological evaluation of their protein conjugates | Zhang, Wang, Meng, et al. (2022) |
Format: MALDI method (matrix).
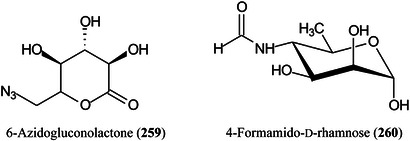










Other work in the area of biopharmaceuticals is summarized in Table 39.
14.4. General Biochemistry
Applications to general biochemistry are listed in Table 40.
14.5. Industrial applications
Two reviews are of interest: “Algal glycobiotechnology: Omics approaches for strain improvement” (Sirohi et al., 2021), (68 references). Contains a table of metabolic studies listing analysis of N‐ and O‐glycans by MALDI; and “Comprehensive approach of methods for microstructural analysis and analytical tools in lignocellulosic biomass assessment” (Rodrigues et al., 2022), 118 references.
Several applications can be found listed in Table 12 (Polysaccharides). Others are in Table 41.
Table 41.
Industrial and other applications.
| Method/notes | Methodsa | References |
|---|---|---|
| Bioactive compounds (mainly glycosides) in waste by‐products from olive oil production: Applications and structural characterization by mass spectrometry techniques | Many methods (review) | Abbattista et al. (2021) |
| Saccharification of cellulose‐containing raw materials using Aspergillus niger | TOF | Budenkova et al. (2021) |
| A simple procedure to obtain a medium‐size oligogalacturonic acids fraction from orange peel and apple pomace wastes | Q‐TOF (DCTB) | Cano et al. (2021) |
| Manufacturing of hemicellulosic oligosaccharides from fast‐growing Paulownia wood via autohydrolysis: Microwave versus conventional heating | TOF/TOF (DHB/TFA) | del Río et al. (2022) |
| Fast saccharide mapping method for quality consistency evaluation of commercial xylooligosaccharides collected in China | TOF/TOF (DHB) | Deng, Chen, et al. (2021) |
| Strategy for recycling miscellaneous waste carbohydrates from high‐fructose syrup production by Pichia pastoris fermentation | TOF/TOF | Gao, Duan, et al. (2021) |
| Fiber‐degrading enzymes released oligosaccharides in the upper gastrointestinal tract in wheat‐fed broilers to increase animal growth | TOF/TOF (DHB) | Kouzounis et al. (2021) |
| Compositional analysis of commercial galactooligosaccharide product NeoGOS‐P70 | TOF/TOF (DHB) | Park, Eom, et al. (2021) |
| Cyttaria hariotii E. Fisch. as a promising source of pullulan and Mn(II)‐pullulan complexes for Mn‐deficiency remediation in winter cereals | TOF/TOF | Ramos‐Sánchez et al. (2021) |
| Antioxidant neoagarooligosaccharides (NAOs) and dietary fiber production from red algae Gracilariopsis lemaneiformis using an enzyme assisted one‐step process | TOF/TOF (DHB) | Song, Liu, et al. (2022) |
| Chitosan grafting via one‐enzyme double catalysis: An effective approach for improving performance of wool | TOF/TOF (DHB) | Wang, Zhang, et al. (2021) |
| Novel two‐step process in cellulose depolymerization: Hematite‐mediated photocatalysis by lytic polysaccharide monooxygenase and Fenton reaction | TOF (DHB) | Wang, Kao, et al. (2022) |
| Environmentally friendly chitosan adhesives for plywood bonding | L‐TOF (DHB) | Xi et al. (2022) |
| Efficient and green production of manno‐oligosaccharides from Gleditsia microphylla galactomannans using CO2 and solid acid in subcritical water | TOF (DHB) | Xu, Han, et al. (2022) |
| Novel immunological and mass spectrometry methods for comprehensive analysis of recalcitrant oligosaccharides in ammonia fiber expansion pretreated corn stover. Presence of methylated uronic acids | TOF, GC/MS | Xue et al. (2022) |
| Efficient removal of bacterial endotoxin and related risks in tailwater by dielectric barrier discharge plasma | TOF | Zhang, Wang, Zhou, et al. (2022) |
Format (not all items present): MALDI method (matrix).
14.6. Food and Drink
Table 42 lists some reviews and general articles and applications are listed in Table 43.
Table 42.
Reviews and general articles on the analysis of food and drink.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Recent advances in the knowledge of wine oligosaccharides | Summary of work published in the last 10 years. Origins of oligosaccharides, isolation, structure determination and dependence on grape origin | 120 | Apolinar‐Valiente et al. (2021) |
| Progress in the pretreatment and analysis of carbohydrates in food: An update since 2013 | Sample preparation, analytical methods (LC, LC‐MS, MALDI, SEC, HPAEC, GC, CE, SFC) | 112 | Jie et al. (2021) |
| Biomolecular profiling by MALDI‐TOF mass spectrometry in food and beverage analyses | Analysis categorized by food type (milk and milk products, edible oils and fats, wine, beer, other foods). | 104 | Šebela (2022) |
| Recent trends in the analysis of honey constituents | Discusses various compound types (phenols, carbohydrates, amino acids and proteins, vitamins, lipids, minerals and organic acids) | 120 | Valverde et al. (2022) |
| The practice of application and features of the control of oligosaccharides in the production of specialized food products | Contains references to analysis of milk oligosaccharides by MALDI (In Russian) | 59 | Yurova and Ananyeva (2022) |
14.7. Carbohydrate synthesis
Relevant reviews are listed in Table 44.
Table 44.
Reviews and general articles containing applications of matrix‐assisted laser desorption/ionization to carbohydrate synthesis.
| Subject | Citations | References |
|---|---|---|
| Synthesis of cello‐oligosaccharides by depolymerization of cellulose | 97 | Chen, Shrotri, et al. (2021) |
| Chemical synthesis of cell wall constituents of Mycobacterium tuberculosis | 347 | Holzheimer et al. (2021) |
| Glucan phosphorylase‐catalyzed enzymatic synthesis of unnatural oligosaccharides and polysaccharides using nonnative substrates | 77 | Kadokawa (2022) |
| Ring‐opening of cyclodextrins: An efficient route to pure maltohexa‐, hepta‐, and octa‐oses | 59 | Pélingre et al. (2021) |
| Discovery of semi‐ and fully‐synthetic carbohydrate vaccines against bacterial infections using a medicinal chemistry approach | 208 | Seeberger (2021) |
| Carbohydrate‐based macromolecular biomaterials | 667 | Su, Feng, et al. (2021) |
| Chemical synthesis of polysaccharides | 53 | Wang, Yang, Zhu, et al. (2022) |
As reported in the previous review, a large number of papers were found with the experimental details for MALDI measurements detailed in the Methods section of the paper but with no subsequent indication of its use; for example, all individual products were examined by ESI‐MS or Atmospheric pressure chemical ionization (APCI)‐MS (sometimes both) but with no details of these techniques in the “Methods” section. In several other cases, spectra were clearly acquired by ESI with the unfortunately named “MALDI‐Synapt” instrument. Many false positives were produced by computer searches for MALDI and carbohydrate names when this instrument was employed. In one publication, spectra were labelled as MALDI‐TOF spectra when, clearly, they had been obtained by ESI with this instrument. In another case, ESI spectra were said to be acquired with a MALDI‐TOF/TOF Ultraflex instrument, attributed to the wrong manufacturer and referred to as MALDI‐TOF spectra in the text. In yet another publication, samples were injected directly into a MALDI‐TOF mass spectrometer. Many papers omit to report the matrix even though the type of compound used is important to enable the analyte to “fly.” One paper reported the matrix as MeCN, H2O, TFA (50/49.5/0.5, v/v/v). Clearly, greater care needs to be taken with the description of methods and better reviewing is required. Needless to say, most of these papers are not cited in this review.
Synthesis of N‐glycans is hampered by the limited availability of functional glycoenzymes, many of which are membrane proteins that fail to express in heterologous hosts. Jaroentomeechai et al. (2022) have devised a method converting membrane‐bound glycosyltransferases into water soluble enzymes, which are expressed at high levels in the cytoplasm of living cells with retention of biological activity. Ninety eight difficult‐to‐express enzymes, predominantly of human origin, were produced and used to remodel both free and protein‐linked glycans including those found on the monoclonal antibody therapeutic trastuzumab.
14.7.1. Synthesis of multivalent carbohydrates, dendrimers and glycoclusters
Two reviews are relevant: “Cu(I)‐catalyzed click chemistry in glycoscience and their diverse applications” (1331 references) (Agrahari, Bose, et al., 2021) which mainly discusses synthesis of glycodendrimers, and “Review of photoresponsive and glycoside dendrimers in biomaterials and sensors applications” (87 references) (Rajasekar et al., 2022).
Applications are listed in Table 45 with the largest compounds analysed being those shown in 271 and 280. Compound 271 had 36 acetylated galactose residues and a molecular formula of C768H876Cl32N148O372, giving a calculated molecular mass of 19,240.4212. The mass found by MALDI‐TOF was 19,300 approx. (Agrahari, Jaiswal, et al., 2021). Compound 280 gave a mass of 98,900 with DHB as the matrix.
Table 45.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for studies on glycodendrimers and glycoclusters.
| Scaffold | Sugara | Methodsb | Notes | References |
|---|---|---|---|---|
| Cyclen (1,4,7,10‐tetra‐azacyclododecane) (271) | Galactose OAc (12, 36) | TOF (DHB, CHCA) | CuAAC mediated synthesis | Agrahari, Jaiswal, et al. (2021) |
| Porphyrin (272) | Mannose (4) | TOF | As fluorescent sensors for Cu(II) ions. Synthesis by click chemistry | Agrahari et al. (2022) |
| Tris, tetrakis and hexakis‐(4‐(sulfanylmethyl) phenylacetic acid) benzene (273) | Fucose (3, 4, 6) | TOF/TOF (DHB, CHCA) | Used for targeting β ‑propeller lectins from lung pathogens. Show promising anti‐adhesive properties | Duca et al. (2022) |
| PAMAM (274) | β‐Cyclodextrin (2, 4) | TOF (DHB) | Synthesis by copper(I)‐catalyzed alkyne–azide cycloaddition click chemistry under microwave irradiation with yields up to 99%. As potential drug carriers | González‐Méndez et al. (2022) |
| Tetravalent benzene (275) | 1→2‐Di‐pseudo‐mannose (12) | TOF (DHB) | For DC‐SIGN targeting | Goti et al. (2022) |
| Linear (276) | Mannose (32) | TOF | Synthetic glycomacromolecules with defined valency | Hartweg et al. (2021) |
| Di‐COOH benzene plus two solanesol groups (277) | Malto‐oligosaccharides (2) | R‐TOF/TOF (DHB) | For construction of star‐shaped molecules | Isono et al. (2022) |
| Poly‐(propyleneimine) | Maltose (43), mannose (6) | R‐TOF/TOF (DHB) | Synthesis of nanoparticles for directed immunomodulation | Jatczak‐Pawlik et al. (2021) |
| PAMAM | Mannose (~110) | TOF | For study of the adherence of Escherichia coli 83972 on α‐biphenyl mannoside‐presenting polydimethylsiloxane surfaces | Liu, Liang, et al. (2021) |
| Di‐amide, tetra‐alcohol | Mannose (6, 12) | MALDI | Synthesis by photoinitiated thiol‐ene click protocol for efficient inhibition of gram‐negative bacteria | Mahadevegowda et al. (2021) |
| Peptide (278) | Galactose (4, 8) | TOF | Synthesis and evaluation as inhibitors of the adhesion of Candida albicans | Martin, Masterson, et al. (2021) |
| Cyclic decapeptide | Galactose (12, 32) | TOF | As inhibitors of the adhesion of fungal pathogen Candida albicans to human buccal epithelial cells | Martin, Goyard, et al. (2021) |
| Carbosilane (279) | GlcNAc (6) | TOF/TOF (DHB, CHCA) | Synthesis of dendritic maleimide‐thiol adducts carrying pendant glycosides as high‐affinity ligands for wheat germ agglutinin | Matsushita, Toda, et al. (2022) |
| PAMAM | GlcNAc and large glycans on peptides (generation 6, 231 glycopeptides) | TOF | MALDI‐TOF to study mass of released glycopeptides with S. aureus V8 protease | Matsushita, Hinou, et al. (2022) |
| Carbosilane | Galactose, glucose (4‐32) | R‐TOF/TOF (DHB) | For anticancer drug delivery: Synthetic route, characterization, and biological effect of glycodendrimer‐doxorubicin complexes | Müllerová et al. (2022) |
| Pentaerythritol‐peptides | Fucose (9) | TOF (DCTB) | Fucodendropeptides shown to induce changes in cells of the immune system in food allergic patients via DC‐SIGN receptor | Palomares et al. (2022) |
| Fullerene | Mannose (10) | TOF (DCTB), ESI | As EBOLA virus inhibitors | Ramos‐Soriano et al. (2022) |
| Dipentaerythritol | Galactose (24) | L‐TOF | For hepatocyte‐specific targeting and intracellular drug delivery for the treatment of liver disorders | Sharma, Porterfield et al. (2021) |
| PAMAM (280) | Galactose β1‑4 fucose (10, 24, 48, 117) | TOF (IAA, DHB) | Preparation of nanoparticles to study lectins in Caenorhabditis elegans | VanKoten et al. (2021) |
| Tris(2‐aminoethyl)amine | Sulfated mono‐, di‐ and tri‐saccharides (6) | MALDI | Synthesis of sulfoglycodendrimer therapeutics for HIV‐1 and SARS‐CoV‐2 | Wells et al. (2021) |
| Perylene bisimide | Iminosugars (6) | MALDI | As multivalent glucosidase inhibitors | Yang, Li, et al. (2022) |
Number of sugar residues in parentheses
Format (not all items present): MALDI method (matrix). “MALDI” used when instrument not specified.










14.7.2. Other synthesised compounds
Many other types of glycan or glyca,‐containing compounds have been synthesised. These are listed in Table 46.
Table 46.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry for monitoring products of synthetic reactions.
| Carbohydrate | Methodsa | Synthetic methods and/or comments | References |
|---|---|---|---|
| Monosaccharides | |||
| Many derivatized and amino acid conjugates | TOF | Synthesis of chiral acidic amino acids as tethers for intramolecular glycosylation | Fukushima et al. (2021) |
| Acetylated of d‐glucosamine 3‐O‐sulfate | TOF | For studies on lysosomal degradation of 3‐O‐sulfate containing heparan sulfate by arylsulfatase G | Kowalewski et al. (2021) |
| Oxidized trehalose | TOF | Synthesis and application as a hydrophilic anti‐crease finishing reagent for cotton fabric | Lou, Yuan, et al. (2021) |
| α‐d‐Ribofuranose derivatives | TOF | Synthesis, in vivo and in silico analgesic and anti‐inflammatory properties | Spriha et al. (2021) |
| Functionalized 4‐acetylthio‐butyl glucopyranosides | R‐TOF (Dithranol) | For studies of carbohydrate‐carbohydrate‐interactions by atomic force microscopy | Thimm et al. (2022) |
| Oligosaccharides | |||
| (GlcNAc)5 | TOF | For studies on the impact of HILIC amino‑based column equilibration conditions on the analysis of chitooligosaccharides | Abla et al. (2022) |
| N,N‐Diacetyllactosamine | TOF (6‐ATT) | Enzymatic synthesis. Mouse β1α4‐GalT1 wild‐type and mutant Y286L found to perform best for transferring β1→4‐Gal and β1→4‐GalNAc residues | Cao, Li, et al. (2022) |
| Maltose phosphate | QIT‐TOF (DHB) | To study polysaccharide storage in Chlamydiae | Colpaert et al. (2021) |
| Agaro‐oligosaccharides | L‐TOF (DHB) | Synthesis by microwave assisted hydrothermal hydrolysis | Dan et al. (2022) |
| Branched oligoglucosides | TOF | One‐step production using mutant endo‐β−1→3‐glucanase | Gao, Xu, et al. (2021) |
| Gellan oligosaccharides | TOF | Synthesis by irradiation treatment and acid hydrolysis of gellan gum | Gao, Li, et al. (2022) |
| Lewis b hexasaccharide thioglycoside donor | TOF | Synthesis and use towards an extended mucin core Tn heptasaccharide structure and a photoreactive biotinylated serine linked hexasaccharide | Hollinger et al. (2022) |
| Fe(III)‐Rhamnoxylan | R‐TOF (DHB) | Synthesis of a novel high spin Fe(iii) octahedral complex | Hayat et al. (2022) |
| Galβ1–3GlcNAc | TOF | In study identifying N‐glycans with one reducing‐terminal GlcNAc at the reducing terminus | Huang, Seino, et al. (2022) |
| Fluorogenic biantennary dextrins | R‐TOF/TOF (DHB) | For study of the mechanism of action of glycogen debranching enzyme | Ikeda et al. (2022) |
| β‐l‐Arabinofuranosyl‐l‐arabinofuranosides | TOF | Towards the substrate specificity evaluation of β‐l‐arabinofuranosidase | Ishiwata et al. (2022) |
| Mixed linkage trisaccharide derivatives | R‐TOF/TOF (ATT) | As endo‐β‐glucanase inhibitors | Jain et al. (2021) |
| Chitosan | TOF (DHB, CHCA) | By hydrolysis of high molecular weight chitosan. Antibacterial activity | Lee, Park, et al. (2022) |
| Short‐chain glucan oligomers | TOF/TOF | Production and separation from corn stover in an unacidified LiBr molten salt hydrate via pre‐extraction of hemicellulose | Liu, Zhou, et al. (2022) |
| 6, 6’‐Carboxy trehalose and ring opened | R‐TOF/TOF (DHB) | For improving the antiwrinkle and hydrophilicity performance of cotton fabric via crosslinking cellulose | Lou, Wang, et al. (2021) |
| Cyclic α‐nigerosyl‐(1→6)‐nigerose (Ac and Me derivatives) | TOF | Derivatives to achieve complete protection | Matsuoka et al. (2022) |
| (LacNAc)3 plus 13C‐labels | TOF/TOF (DHB) | To study protein binding epitopes by NMR | Moure et al. (2021) |
| Keratan sulfate oligosaccharides | TOF/TOF | Use of blockwise synthesis | Ozaki et al. (2022) |
| Hexasaccharide from pneumococcal serotype 3 capsular polysaccharide | TOF | For studies of ligand binding of pneumococcal serotype 3 capsular polysaccharide‐specific protective antibodies | Ozdilek et al. (2021) |
| Methacryl‐10,2,20,3,30,4,6,60‐hepta‐O‐acetyl‐d‐maltose | TOF | For polymer synthesis | Palodkar et al. (2022) |
| Alginate oligosaccharides | TOF | Alginate oligosaccharides shown to maintain activities of lysosomes under low pH condition | Park, Nguyen, et al. (2021) |
| Long‐chain isomaltooligosaccharide | TOF/TOF (DHB) | Synthesis from maltodextrin with a novel glucosyltransferase derived from Thermoanaerobacter thermocopriae | Park, Park, et al. (2021) |
| β(1→4)‐GlcNAc oligosaccharides | TOF/TOF (DHB) | Size‐controlled synthesis using an endo‐glycosynthase | Rousseau et al. (2021) |
| Modified artichoke pectin and pectic ligosaccharides. | TOF | Optimisation of an enzymatic method using artificial neural network tools | Sabater, Blanco‐Doval, Montilla, et al. (2021) |
| Oligogalactofuranosides | TOF/TOF | Automated glycan assembly reveals the influence of protecting groups on oligosaccharide stability | Sabbavarapu and Seeberger (2021) |
| Alkyl β‐celluloside | TOF/TOF (DHB) | Investigation of parallel versus antiparallel molecular arrangements in crystalline assemblies | Serizawa et al. (2021) |
| α‐Glucosidase inhibitory oligosaccharides | TOF/TOF (DHB) | Preparation, structure and α‐glucosidase inhibitory by enzymatic hydrolysis from Annona squamosa polysaccharide | Sun, Sun, et al. (2022) |
| α(1→6)‐d‐Mannans and α(1→5)‐d‐arabinans | TOF | Synthesis and nitric oxide‐inducing activities | Suwanwong et al. (2022) |
| Xylβ1→2Manβ | TOF (DHB) | As the core fragment of plant N‐glycans | Tsygankova et al. (2022) |
| Chitooligosaccharides | TOF (DHB) | Use of plant chitinase mutants as the catalysts with sugar oxazoline derivatives | Umemoto et al. (2022) |
| Branched β−1→3‐glucan oligosaccharide | TOF/TOF | Synthesis by fermentation of β−1,3‐glucan producing fungi and Trichoderma harzianum capable of secreting endo‐β−1,3‐glucanase with Sclerotium rolfsii and Schizophyllum commune | Wu, Yang, et al. (2021) |
| Mannooligosaccharide | TOF (DHB) | Production from Gleditsia microphylla galactomannan using acetic acid and FeCl2 | Xu, Han, et al. (2021) |
| Curdlan oligosaccharides | TOF/TOF (DHB) | Synthesis from curdlan by hydrolysis with HCl | Xu, Wang, et al. (2021) |
| Gellan gum oligosaccharides | TOF/TOF (DHB) | Synthesis from gellan by hydrolysis with HCl | Xu, Wang, et al. (2021) |
| Xanthan gum oligosaccharides | TOF/TOF (DHB) | Synthesis by treatment of xanthan gum with H2O2 for 5 days | Xu, Wang, et al. (2021) |
| Pullulan oligosaccharides | TOF/TOF (DHB) | Synthesis by hydrolysis of pullulan with pullulanase | Xu, Wang, et al. (2021) |
| Xylooligosaccharides | TOF/TOF (DHB) | Production from Camellia oleifera Abel fruit shell using a shell‐based solid acid catalyst | Xu, Zhang, et al. (2022) |
| Polysialic acid | TOF/TOF (ATT) | For development of photothermal therapy of neuroblastoma | Xu, Zhao, et al. (2022) |
| Homogalacturonan | TOF/TOF (DHB) | For study of binding togalectin‐3 | Zheng et al. (2021) |
| Azide‐modified disaccharide oxazolines | TOF/TOF (DHB) | As enzyme substrates for single‐step Fc glycan‐mediated antibody‐drug conjugation | Zhang, Ou, et al. (2022) |
| Polysaccharides | |||
| Bacterial cellulose nanocrystals | TOF/TOF (DHB) | Use of lytic polysaccharide monooxygenases and cellulases | Buruaga‐Ramiro et al. (2022) |
| Chitosan (from crab shells). | L‐TOF/TOF (Dithranol) | For preparation of pH‐sensitive nanoparticles loaded with dolutegravir as milk and food admixture for paediatric anti‐HIV therapy | Dharshini et al. (2021) |
| Fluorinated cellodextrins | TOF | Chemoenzymatic synthesis and identification of a new allomorph for cellulose‐like materials | de Andrade et al. (2021) |
| Quaternized and sulfated xylan‐derivatives | TOF/TOF (s‐DHB) | With enhanced microbiological and antioxidant properties over natural xylans | Fröhlich et al. (2022) |
| Oligocellulose | TOF/TOF (DHB) | Oligocellulose production from acid hydrolysis: A revisit | Jiang et al. (2021) |
| Oxidized inulin | L‐TOF (CHCA/TFA) | Use for synthesis of oxidized inulin cross‐linked collagen‐ZrO2 hybrid scaffolds for tissue engineering applications | Kalirajan et al. (2022) |
| Terminally carboxylated cellulose oligomers | TOF | For synthesis of organic−inorganic hybrid hydrogels | Sugiura et al. (2022) |
| Cellulose | TOF/TOF (DHB) | Solvent‐assisted fractionation of oligomeric cellulose and reversible transformation of cellulose II and IV | Zhang, Jiang, et al. (2021) |
| Reducing end thiol‐modified nanocellulose | TOF/TOF (DHB) | 13% In two steps. For binding studies | Zhong, Zajki‐Zechmeister, et al. (2021) |
| Reducing‐end thiol‐modified cellulose | R‐TOF/TOF (DHB) | Use of cellodextrin phosphorylase from Clostridium stercorarium | Zhong, Nidetzky, (2022) |
| Chitooligosaccharides | |||
| Sulfated chitosans | TOF/TOF (CHCA) | Extracted from marine waste. For evaluation of antibacterial, teratogenicity and antibiofilm effect against microorganisms | Gomathy et al. (2021) |
| Chitooligosaccharides | TOF/TOF (DHB) | Synthesis and anti‐inflammatory activity on VitD3‐induced human THP‐1 monocytes | Jitprasertwong et al. (2021) |
| Chitooligosaccharides | R‐TOF/TOF (DHB) | Production and characterization by the fungal chitinase Chit42 immobilized on magnetic nanoparticles and chitosan beads | Kidibule et al. (2021) |
| Chitooligosaccharides | TOF/TOF (DHB) | Production of structurally defined chito‐oligosaccharides with a single N‑acetylation at their reducing end using a new chitinase from Paenibacillus pabuli | Li, Wang, Chang, et al. (2021) |
| Chitooligosaccharides | TOF/TOF | Synthesis by hydrolysis of chitosan (to model chitooligosaccharides found in seawater) Action in diabetic mice. | You et al. (2022) |
| Chitosan (commercial) | L‐TOF/TOF (DHB, CHCA, SA) | For determination of chitosan characteristics in electrolyte solutions | Lupa et al. (2022) |
| Chitooligosaccharides | TOF | For preparation of chitooligosaccharide ‐polyphenol conjugates | Mittal et al. (2022) |
| Protected precursors of chitin oligosaccharides | TOF | Synthesis by electrochemical polyglycosylation of thioglycosides | Rahman et al. (2022) |
| Oligomeric chitin | TOF (DHB) | Efficient production of oligomeric chitin with narrow distributions of degree of polymerization using sonication‐assisted phosphoric acid hydrolysis | Zhang, Mao, et al. (2022) |
| Glycosaminoglycans and related compounds | |||
| Non‐glycosaminoglycan‐type heparin‐analogue trisaccharides | R‐TOF (DHB) | Synthesis and cell growth inhibitory activity | Lisztes et al. (2021) |
| N ‐linked glycans | |||
| BODIPY‐labelled Neu5Ac‐CMP | TOF/TOF (9‐AA) | Development of BODIPY labelled sialic acids as sialyltransferase substrates for direct detection of terminal galactose on N‐ and O‐linked glycans | Abukar et al. (2021) |
| High‐mannose‐Asn‐Fmoc | TOF/TOF (DHB) | For development of array for recognition of oligomannose isomers by glycan‐binding proteins involved in innate and adaptive immunity | Gao Stavenhagen et al. (2021) |
| Sulfated N‐glycans | TOF | Site‐selective sulfation of N‐glycans by human GlcNAc‐6‐O‐sulfotransferase 1 and chemoenzymatic synthesis of sulfated antibody glycoforms | Huang, Li, et al. (2022) |
| Decamannoside | MALDI | Binding evaluation of pradimicins for oligomannose motifs from fungal mannans | Nakagawa et al. (2021) |
| High‐mannose glycans | TOF | One‐pot glycosylation strategy assisted by ion mobility−mass spectrometry analysis | Ponnapalli et al. (2022) |
| Paucimannosidic glycans | TOF | To determine the minimal glycan recognition epitope for Mannitou IgM | Robakiewicz et al. (2021) |
| N‐Glycans from the parasite Schistosoma mansoni | TOF | Chemoenzymatic synthesis and examination of importance of epitope presentation on DC‐SIGN recognition | Srivastava et al. (2021) |
| O ‐linked glycans | |||
| Sulfated and nonsulfated core 2 O‑GalNAc glycans | TOF (DHB) | Chemoenzymatic synthesis | Xu, Deng, Zhang, et al. (2021) |
| Glycopeptides/glycoproteins | |||
| GM1 Glycolipid plus dodecapeptide | TOF | Ceramide structure shown to dictate glycosphingolipid nanodomain assembly and function | Arumugam et al. (2021) |
| MUC1 Glycopeptides | TOF (CHCA) | For calorimetric analysis of the interplay between synthetic Tn antigen‐presenting MUC1 glycopeptides and human macrophage galactose‐type lectin | Beckwith et al. (2021) |
| Glycocins | TOF/TOF (CHCA) | Development of SELECT‐GLYCOCIN: A recombinant microbial system for expression and high‐throughput screening of glycocins | Choudhary and Rao (2021) |
| Fluorine‐substituted MUC1 glycopeptide | R‐TOF/TOF (DHB, SA) | As a self‐adjuvanting antitumor nanoliposomal vaccine | Dong, Cheng, et al. (2022) |
| Fluorescently labelled glycopeptide (biantennary glycan) | TOF | In vivo imaging reveals sialylation‐like biodistribution and kinetics | Fukuhara et al. (2022) |
| IgG ((Fucα1, 6)GlcNAc‐rituximab or GlcNAc‐rituximab) by transglycosylation with endo‐S2 | PNGase F, TOF (2‐AA) | For development of synthetic nanobodies as tools to distinguish IgG Fc glycoforms | Kao et al. (2022) |
| N‐Glycoproteins | TOF (SA) | Synthesis using a combination of genetic code expansion and chemoselective ligation techniques (click chemistry) | Hyun et al. (2021) |
| Evasin‐3 | TOF | Use of 2,20‐dipyridyl disulfide‐mediated thiazolidine ring‐opening reaction | Katayama and Nagata (2021) |
| Insulin‐like androgenic gland factor from crayfish | TOF | Chemical synthesis and functional evaluation | Katayama et al. (2022) |
| D‐α‐Galp‐l‐Ser/l‐Thr‐l‐Ala‐l‐Ala | TOF (DHB), FAB | As precursors of new glycopeptide antibiotics | Khodair et al. (2022) |
| Glycopeptide with biantennary glycan from trasnsferrin | R‐TOF/TOF (CHCA) | Screening for glycan‐specific aptamers using the glycosylated peptide as a scaffold | Li, Ma, et al. (2021) |
| N‐Glycoproteins | PNGase F, TOF, glycans (per‐Me) | Design of a new bacmid for customized protein glycosylation pathway engineering in the baculovirus‐insect cell system | Maghodia et al. (2021) |
| C‑Mannosyl tryptophan | MALDI | For quantification of serum C‑mannosyl tryptophan by novel assay to evaluate renal function and vascular complications in patients with type 2 diabetes | Morita et al. (2021) |
| O‐Glycopeptides (mucins) | TOF | Use of gene engineered cells | Nason et al. (2021) |
| O‐Glycopeptide (Mg7a). Major component of the venom of the ant Myrmecia gulosa | L‐TOF/TOF (CHCA) | Synthesis by solid‐phase peptide synthesis, combined with diselenide–selenoester ligation‐deselenization chemistry | Robinson et al. (2021) |
| SARS‐CoV‑2 homogeneous O ‑linked glycopeptides | TOF | Chemoenzymatic synthesis. For exploring their inhibition functions | Rong et al. (2022) |
| D‑Fructose‑derived Heyns peptides | R‐TOF/TOF (CHCA) | Synthesis utilizing Nα‑Fmoc‑Lysin‐[Nε‑(2‑deoxy‑D‑glucos‑2‑yl),Nε‑Boc]‑OH as building block | Schmutzler et al. (2021) |
| Clusterin glycopeptides | TOF | For development of a selective reaction monitoring approach using structure‐defined synthetic glycopeptides for validating glycopeptide biomarkers | Shiratori et al. (2022) |
| Amyloid‐β precursor protein with GalNAc at Tyr681 | TOF (CHCA) | Tyrosine O‑GalNAc shown to alter the conformation and proteolytic susceptibility of APP model glycopeptides | Singh et al. (2021) |
| GalAc plus nonapeptide | TOF | As an antibiotic nano‐adjuvant to inhibit Pseudomonas aeruginosa biofilm and enhance antibacterial activity | Song, Zhang, et al. (2022) |
| [2H3]‐Methylamide labelled glycopeptides | R‐TOF/TOF | For quantitative method for measuring N‐glycopeptides | Sun, Ji et al. (2021) |
| N‐Glycopeptides with 13C‐fucose | R‐TOF/TOF (DHB) | For LC/MS quantitation of serum IgG glycopeptides | Wang, Liu, Qu, et al. (2021) |
| Carbohydrates from bacteria | |||
| Lactic acid bacteria exopolysaccharides | TOF | Identification of binding sites for oligosaccharide repeats from lactic acid bacteria exopolysaccharides on bovine β‑lactoglobulin identified by NMR | Birch et al. (2021) |
| Lipid A mimetics | TOF (6‐ATT, ‐ve) | Synthesis based on an unnatural disaccharide scaffold as potent TLR4 agonists for prospective immunotherapeutics and adjuvants | Strobl et al. (2022) |
| Oligosaccharides derived from the capsular polysaccharide of Streptococcus pneumoniae serotypes 6A and 6B | TOF | Synthesis and immunological studies | Mettu et al. (2022) |
| Carbohydrates from fungi | |||
| Cordyceps militaris glycans | TOF/TOF | Total synthesis via stereoselective orthogonal one‐pot glycosylation and α ‑glycosylation strategies | Ma, Jiang, et al. (2022) |
| Glycosphingolipids and related glycans | |||
| Deuterium‐labelled acyl‐globotriaosylceramide | TOF (CHCA), FAB | Synthesis by transesterification of N3‐lyso GM3 with 2H35−18:0 acid p‐nitrophenyl ester for potential imaging of subcellular localization of GB3 using nanoSIMS | Aly and El Azab (2021) |
| 6‐NH2‐α‐GalCer. | FT‐ICR (DHB, CHCA) | For construction of an antitumor vaccine of MUC1 glycopeptide and α‐GalCer via a gold‐nanoparticle delivery system. | Liu, Wang, Yu, et al. (2021) |
| Fluorescently labelled lacto‐series ganglioside | TOF (CHCA) | For single molecule imaging | Takahashi et al. (2022) |
| Other glycolipids | |||
| 2‐O‐Ac‐3,4,6‐tri‐O‐Ac‐α‐d‐Glcp‐(1→6)−1‐O‐(2‐oleoyl‐1‐stearoyl‐sn‐glycero‐3‐phosphonate)−2,3,4,5‐tetra‐O‐Ac‐d‐myo‐inositol | MALDI | Glycosylphosphatidylinositol oligosaccharide intermediate | Guerrero et al. (2021) |
| Poly‐amido‐saccharides containing myristoyl, palmitoyl, or stearoyl terminal chains | TOF | As water‐soluble biosurfactants | Sockett et al. (2022) |
| Glycosides and related compounds | |||
| Propargyl‐(oligo)‐mannopyranoside | TOF | Intermediate in the synthesis of mannose‐based surfactants | Argudo, Spitzer, Ibarboure, et al. (2022) |
| 6’’‐O‐Lauroyl‐1‐kestose and 6’’’‐O‐lauroylnystose | TOF (DHB) | Regioselective synthesis by sequential enzymatic reactions of transfructosylation and acylation | Campos‐Valdez et al. (2022) |
| Pixatimod (PG545), a sulfated oligosaccharide‐steroid conjugate | Spiral‐TOF (DHB, ‐ve) | Development of improved synthetic routes | Chhabra et al. (2021) |
| 2 and 6‑S‑Hexyl‑β‑d‐glucopyranose S‑linked maleimide | TOF (DHB) | For study of a UDP‑glucose, cereblon‑dependent proinsulin degrader | Cho, Miyagawa et al. (2022) |
| Modified QS‐21 glycoside | MALDI | Replacing the rhamnose‐xylose moiety with simpler terminal disaccharide units attenuates adjuvant activity in truncated saponin variants | Fuentes, Ruiz‐de‐Angulo, et al. (2021) |
| Rutin polyglucoside | TOF (DHB) | Addition of (Glc)n catalyzed by a cyclodextrin glucanotransferase to increase solubility | González‐Alfonso et al. (2021) |
| Glycosylated polyene macrolides labelled with 3,6‐di‐2‐pyridyl‐1,2,4,5‐tetrazine | TOF | To investigate antifungal action by sterol sponge mechanism | Guo, Zhang, et al. (2021) |
| β‐C‐glycoside‐2‐aminoundecanes | R‐TOF (DHB) | From glucose, lactose, and maltose. For use in personal care and cleaning products | Jackson et al. (2021) |
| Glucosyl‐α‐(1→6)‐mangiferin | TOF | For enhancement of water solubility and antioxidant capacities of mangiferin | Lee, Kim, Moon, et al. (2022) |
| Flavonoid glycosides, oroxins C and D from the seeds of Oroxylum indium | MALDI | Concise synthesis and antidiabetic activity | Li, Wang, Tong, et al. (2021) |
| 2‐Deoxyiminosugar C‐glycosides | TOF/TOF (DHB, DCTB) | Stereocontrolled synthesis and evaluation as glycosidase inhibitors | Lumbroso et al. (2021) |
| Fisetin‐ 4’‐O‐α‐d‐glucopyranoside | TOF (DHB) | Synthesised with dextransucrase from Leuconostoc mesenteroides | Moon et al. (2022) |
| Transglycosylated mogrosides (steroid glycosides) from Siraitia grosvenorii, (Luo Han Guo fruit) | TOF (DHB), LC/MS | High‐yield synthesis improves the flavor profile of monk fruit extract sweeteners | Muñoz‐Labrador et al. (2021) |
| Galactooligosaccharide‐ modified mogrosides | TOF (DHB) | As new sweeteners | Muñoz‐Labrador et al. (2022) |
| Maltoheptaose‐palmitate ester | TOF/TOF (DHB) | Synthesis of a sugar ester having excellent emulsifying properties | Nguyen et al. (2021) |
| Acetoglucose‐substituted methacrylate | TOF | Intermediate in the synthesis of carbohydrate‐based block copolymer micelles for photodynamic therapy | Park, Jung, et al. (2021) |
| Bis‐glucosides | TOF/TOF (DCTP) | Synthesis and characterization of a small library of bis‐glucosides | Patry et al. (2021) |
| Thio‐ether functionalized glycolipids | TOF/TOF (dithranol, CHCA) | Synthesis and use to reveal a potent activator of SK3 channel with vasorelaxation effect | Sevrain et al. (2021) |
| Schaftoside | TOF | Total chemical synthesis in 11 steps (8.83% yield) | Shang et al. (2021) |
| Triterpene glycoside from Eupentacta fraudatrix | TOF | In study of structure‐activity relationships of holothuroid's triterpene glycosides | Zelepuga et al. (2021) |
| Cyclodextrins (CDs) | |||
| 6I–O‐Allyl‐γ‐CD | TOF | For synthesis of γ‐CD poly (glycidyl‐co‐ethylene dimethacrylate) for host‐guest interactions for extracting antibiotics | Belenguer‐Sapiña et al. (2021) |
| β‐CD + (PhCHO)7 | TOF/TOF | For preparation of glycopeptide dendrimers: | Bi et al. (2022) |
| Betulinic acid‐CD conjugates | TOF/TOF | For inhibition of influenza infection | Chen, Wang, Ma, et al. (2022) |
| Heptakis‐6‐octanethio‐β‐CD (CD‐C8) and CD‐C12 | TOF (DCTB) | Amphiphilic CD‐based nanoparticulate vaccines shown to trigger T‐cell immune responses | Geisshüsler et al. (2022) |
| Heptavalent glycyrrhetinic acid β‐CD conjugates | TOF/TOF (CHCA) | Synthesis, characterization, and anti‐influenza activity | Liang, Ma, et al. (2022) |
| BODIPY‐modified β‐CD | TOF | For use in fluorescence sensing of glutathione thiyl radical | Liu, Dai, et al. (2022) |
| Oligopeptide‐decorated amphiphilic CD nanomagnet intermediates | TOF/TOF | For selective amyloid beta recognition and fishing | Mazzaglia et al. (2022) |
| γ‐CD–Fullerene | TOF | Synthesis of amphiphilic γ‐CD–fullerene complexes with photodynamic activity | Miki et al. (2022) |
| Peptide/BODIPY‐modified per‐O‐methyl‐β‐CDs | TOF | For FRET‐based in‐cell detection of highly selective supramolecular complexes of meso‐tetraarylporphyrin | Nakagami et al. (2021) |
| β‐CD‐Fluvastatin conjugates | TOF/TOF (DHB) | Synthesis and biological evaluation as potential therapeutics for neuronal disorders such as Alzheimer's and Niemann Pick type C disease | Nicolosi et al. (2021) |
| Lactose‐appended hydroxypropyl‐β‐CD | TOF | Shown to lower cholesterol accumulation and alleviate motor dysfunction in Niemann−Pick type C disease model mice | Nishida et al. (2022) |
| CD‐oligocaprolactone derivatives | TOF/TOF (DHB, CHCA) | Synthesis and structural characterization by MALDI‐MS/MS | Peptu et al. (2022) |
| Sulfur‐bridged β‐CD dimers | TOF | For enantiodifferentiating photocyclodimerization of 2‐anthracenecarboxylate. Protocol | Wei et al. (2022) |
| α‐, β‐, and γ‐Ureido‐CDs | TOF | Synthesis of upper critical solution temperature‐type responsive cyclodextrins | Zhu, Liu, et al. (2021) |
| Rotaxanes | |||
| Pillar[5]arene‐based polycationic glyco[2]rotaxanes | R‐TOF/TOF (DCTB) | As Pseudomonas aeruginosa antibiofilm agents | El Dine et al. (2021) |
| Polyfluorene/permodified CD polyrotaxanes | TOF/TOF (DCTB) | Synthesis, photophysics and production of Langmuir films | El Haitami et al. (2021) |
| Polymers | |||
| Branched glycopolymer–pyropheophorbide‐a conjugate | IT‐TOF (DHB, CHCA) | For cancer chemotherapy | Duan et al. (2021) |
| Sulfur‐linked sugar polymers | R‐TOF/TOF | Chemoenzymatic synthesis. As heparanase inhibitors | He, Zhang, et al. (2022) |
| Sucrose‐1,6‐hexamethylene diisocyanate polymer | TOF/TOF (DHB) | For synthesis of novel polyurethane networks | Lakatos et al. (2022) |
| Glycosylated poly(ethylene oxide)–poly(propylene oxide) | TOF | For preparation of glucosylated polymeric micelles to actively target breast cancer | Lecot et al. (2021) |
| Polymers from d‐ and l‐xylose | TOF | Synthesis with control of crystallinity and stereocomplexation | McGuire, Bowles, et al. (2021) |
| Maltotriose‐b‐poly(N‐n‐propylglycine | TOF (CHCA) | For construction of permeable polymer vesicles | Okuno et al. (2021) |
| Fructo‐oligosaccharides/inulin polymers | TOF (DHB) | For preparation of nanodisks | Ravula and Ramamoorthy (2021) |
| Antibiotics and other drugs | |||
| Teicoplanin derivative | R‐TOF (DHB) | Synthesis of the first dimeric derivatives | Bereczki et al. (2022) |
| Vancomycin derivative | TOF | Synthesis of novel semisynthetic antibiotics active against Staphylococcus aureus biofilms and cells in late stationary growth phase | Vimberg et al. (2021) |
| Microarrays, Nanoparticles | |||
| Trehalose‐based nanoparticles | L‐TOF/TOF (DHB, DHAP) | For organ‐selective gene delivery | Carbajo‐Gordillo et al. (2021) |
| β‐CD‐poly (β‐amino ester) nanoparticles | TOF/TOF (DHB) | Nanoparticles for high loading and sustained release of histone deacetylase inhibitors | Chaudhuri et al. (2021) |
| Controlled density glycodendron microarrays | TOF | For studying carbohydrate–lectin interactions | Di Maio et al. (2021) |
| Doublecortin like kinase 1 (DCLK1) antibody functionalized folic acid conjugated hesperetin encapsulated chitosan nanoparticles | TOF/TOF | For targeting colon cancer stem cells | Lazer et al. (2022) |
| N‐Acetylgalactosamine‐decorated nanoliposomes | TOF/TOF | For targeted delivery of paclitaxel to hepatocellular carcinoma | Li, Yu, et al. (2021) |
| Glycans linked to glycerophosphate | QIT‐TOF (THAP) | Synthesis of noncovalent microarrays from synthetic amino‐terminating glycans | Li, Palma, et al. (2021) |
| Sialylated solid lipid microparticles | TOF | As inhibitors of influenza A virus infection | Richard et al. (2022) |
| TLR7 Ligands on gold nanoparticles | TOF/TOF | The resulting glyco‐nanoadjuvants shown to affect their immunostimulatory activities | Shinchi et al. (2021) |
| Manα1→6Glc‐nanoparticles | TOF/TOF | For study of the effect of linker length for conjugating a synthetic small‐molecule TLR7 ligand to glyco‐nanoparticles on immunostimulatory effects | Shinchi et al. (2022) |
| Xylan microparticles | TOF (DHB) | Enzymatic synthesis of xylan microparticles with tunable morphologies | Smith, Curry, et al. (2022) |
| Gold nanoparticles with lactose | TOF/TOF (DHB) | For investigations of affinity labelling for target protein analysis | Suto et al. (2021) |
| Sugar‐coated hierarchical platinum nanostructures | TOF/TOF (DHB) | Method to support and heterogenize nanoparticles | Woitassek et al. (2022) |
| Peptide‐CD nanoparticles | TOF/TOF | Effect and mechanism of action on hepatoma of nanoparticles loaded with tyroserleutide | Wu, Hua, et al. (2021) |
| Miscellaneous | |||
| Mono‐, di‐, and trivalent α‐d‐mannopyranosyl conjugates | MALDI (CHCA) | On an aromatic scaffold. Synthesis and hemagglutination inhibitory properties | Al‐Mughaid and Khazaaleh (2021) |
| Almost‐linear mannose polysaccharides linked to an oleic or ricinoleic acid | TOF/TOF (DHB) | Design and self‐assembly of sugar‐based amphiphiles: Spherical to cylindrical micelles. Use of click chemistry | Argudo, Spitzer, Jerome, et al. (2022) |
| Liposomes displaying glycan ligands | TOF | Increasing phagocytosis of micoglia by targeting CD33 with product | Bhattacherjee et al. (2021) |
| Triazolylisatins glycoconjugates | TOF/TOF | Use of click reaction of propargylisatins with some azido‐sugars | Bogdanov et al. (2021) |
| Glucosylated 5‐hydroxymethyl‐pyrimidines | TOF | As epigenetic DNA bases regulating transcription and restriction cleavage | Chakrapani, Ruiz‐Larrabeiti, et al. (2022) |
| Galactose‐modified multifunctional nanoprobe | TOF | For cancer therapy based on nitric oxide prodrug delivery and release | Dang et al. (2021) |
| S‐Linked sugar‐nucleotide analogues | R‐TOF (DHB) | Synthesis of potential glycosyl transferase inhibitors by thio‐click reactions | Debreczeni et al. (2021) |
| Porphyrin glycoconjugates | TOF (CHCA) | Synthesis and evaluation of porphyrin glycoconjugates varying in linker length and preliminary effects on the photodynamic inactivation of Mycobacterium smegmatis | Dixon et al. (2021) |
| DSPE‐PEG (2000)‐GalNAc | R‐TOF/TOF | For construction of liposome‐GalNAc nanoparticles for hepatocellular carcinoma chemotherapy | Farinha et al. (2021) |
| Glycan‐oligonucleotide conjugates. | TOF (3‐HPA) | For preparation of glycan chips for on‐chip biosynthesis of cancer‐associated complex glycans | Heo et al. (2021) |
| Glycocholic acid‐chitosan oligosaccharide conjugate | TOF | For oral administration of chemotherapeutic drugs | Liu, Han, et al. (2022) |
| Lactose‐functionalized dimeric camptothecin | TOF | For targeted and fluorescence imaging‐guided chemo‐photodynamic therapy | Ma, Shi, et al. (2022) |
| Guanosine diphosphate l‐fucose | TOF/TOF (9‐AA) | In vitro synthesis using multi‐enzyme cascades | Mahour et al. (2021) |
| N,N‐Bis(hexyl α‐d‐acetylmannosyl) acrylamide | TOF (DHB) | Synthesised as a by‐product of the monomer | Miyagawa et al. (2021) |
| Sugar‐polyolefin conjugates | MALDI | For synthesis of stable thermotropic 3D and 2D double gyroid nanostructures with sub‐2‐nm feature size | Nowak et al. (2021) |
| Oxazolidine boronate sugar complexes | TOF | In construction of fluorescent sensor array for quantitative determination of saccharides | Pushina et al. (2021) |
| Carbohydrate‐attached fullerene derivative | TOF | For selective localization in ordered carbohydrate‐block‐poly(3‐hexylthiophene) nanodomains | Sakai‐Otsuka et al. (2021) |
| Mannosylated‐calix[4]arene | TOF | Dynamic self‐assembly into micelles for the delivery of hydrophobic drugs | Sreedevi et al. (2021) |
| Cholesterol‑undecanoate‐glucose conjugate | TOF | For the treatment of cerebral malaria | Tian, Zheng, et al. (2022) |
| α‑Dystroglycan mucin type core m1 (glyco)peptide library | TOF/TOF (DHB) | Exploring the in situ pairing of human galectins toward synthetic O‑mannosylated core M1 glycopeptides of α‑dystroglycan | Villones et al. (2022) |
| 64Cu‐Containing carbohydrate fluorescence biomarker | TOF/TOF | Fluorescence marker for improved surgical precision | Wang, Hansen, et al. (2021) |
| Cyanidin‐3‐O‐glucoside and β‐lactoglobulin conjugate | TOF | Effect of ultrasound on binding interaction and functional properties | Wang, Wang, Luo, et al. (2022) |
| Glucose−lipopeptide conjugates | TOF | Conjugates reveal the role of glucose modification position in complexation and the potential of malignant melanoma therapy | Zhao, Zhang, Li, et al. (2021) |
| Disaccharide oxazolines carrying four or six azide tags | R‐TOF (DHB) | Chemoenzymatic method for glycan‐mediated site‐specific labeling and conjugation of antibodies | Zhang, Ou, et al. (2021) |
Format (not all items present): MALDI method (matrix), “MALDI” is used when the instrument is not specified.
A number of other papers, listed in Table 47, report more general methods and a few report work on synthetic mechanisms (Table 48).
Table 47.
Use of matrix‐assisted laser desorption/ionization‐mass spectrometry to study methods for general synthesis.
| Method/notes | Methodsa | References |
|---|---|---|
| Bifidobacterial β‐galactosidase‐mediated production of galacto‐oligosaccharides | TOF (DHB) | Ambrogi et al. (2021) |
| Facile synthesis of per(6‐O‐tertbutyldimethylsilyl)‐α‐, β‐, and γ‐cyclodextrin as protected intermediates for the functionalization of the secondary face of the macrocycles | TOF | Benkovics et al. (2021) |
| Site‐selective attachment of polymer chains to glycoproteins by sodium periodate oxidation of glycans (shown for HRP) | TOF/TOF | Bi, Xiong, et al. (2021) |
| MALDI mass spectrometry monitoring of cyclodextrin‐oligolactide derivatives synthesis | TOF, TOF/TOF (CHCA, DHB) | Blaj et al. (2021) |
| Diisobutyl aluminum hydride promoted debenzylation of α‐cyclodextrin | MALDI | Bols and Friis (2022) |
| Synthesis of poly‐β‐1,4‐glucan derivatives by use of promiscuous glycosyltransferase | TOF | Bulmer et al. (2021) |
| Conformation‐controlled hydrogen‐bond‐mediated aglycone delivery method for α‑xylosylation | TOF/TOF | Cai, Bian, et al. (2021) |
| A mild glycosylation protocol with glycosyl 1‐methylimidazole‐2‐carboxylates as donors | MALDI | Chen, Tang, et al. (2021) |
| High‐quality palladium on carbon catalysts for hydrogenolysis—use with serotype A decasaccharide | TOF | Crawford et al. (2021) |
| Endo‐M mediated chemoenzymatic approach enables reversible glycopeptide labeling for O‐GlcNAcylation analysis | TOF | Chen, Tang, et al. (2022) |
| Enhanced binding and reduced immunogenicity of glycoconjugates prepared via solid‐state photoactivation of aliphatic diazirine carbohydrates | TOF | Congdon and Gildersleeve (2021) |
| Solid‐phase synthesis of polysaccharides from unprotected glucose catalyzed by Hβ zeolites | TOF/TOF (DHB) | Dong, Xiao, et al. (2022) |
| Controlled depolymerization of cellulose by photoelectrochemical bioreactor using a lytic polysaccharide monooxygenase | TOF | Gao, Zhang, et al. (2022) |
| Phosphine‐mediated three‐component bioconjugation of amino‐ and azidosaccharides in ionic liquids | MALDI | Hall et al. (2022) |
| Synthesis of unnatural cyclodextrins with cyclodextrin glucanotransferase | MALDI | Larsen, Ferreira et al. (2022) |
| Insights into the synergistic effect of catalyst acidity and solvent basicity for effective production of pentose from glucose | TOF | Li, Lin, et al. (2022) |
| Iterative synthesis of 2‐deoxyoligosaccharides enabled by stereoselective visible‐light‐promoted glycosylation | TOF/TOF | Liu, Wang, Guo, et al. (2022) |
| In vitro glycosylation of membrane proteins using N‑glycosyltransferase | TOF (CHCA) | Liyanage, Harris, et al. (2021) |
| Sustainable polyesters via direct functionalization of lignocellulosic sugars | TOF | Manker et al. (2022) |
| Ring‐opening copolymerization of a d‑xylose anhydrosugar oxetane to produce polymers from sugars and cyclic anhydrides | TOF | McGuire, Clark, et al. (2021) |
| Precursors of iminosugars with 7‐membered rings | Q‐TOF/TOF | Osuch‐Kwiatkowska and Jarosz (2022) |
| β‐1,3‐Glucan synthesis, novel supramolecular self‐assembly, characterization and application | TOF | Pylkkänen et al. (2022) |
| Design, synthesis, and characterization of stapled oligosaccharides | TOF | Ricardo et al. (2022) |
| Solid‐phase synthesis of glucosyl glycopeptides from synthesised amino acid derivatives: Optimization of the synthetic route | TOF | Rodríguez et al. (2021) |
| Phosphorylase‐catalyzed synthesis and self‐assembled structures of cellulose oligomers in the presence of protein denaturants | TOF (DHB) | Sakurai et al. (2022) |
| Enzyme‐catalyzed propagation of cello‐oligosaccharide chains from bifunctional oligomeric primers for the preparation of block co‐oligomers and their crystalline assemblies | L‐TOF (DHB) | Sugiura et al. (2021) |
| Catalytic, regioselective sulfonylation of carbohydrates with dibutyltin oxide under solvent‐free conditions | MALDI | Traboni et al. (2021) |
| Introducing hyaluronic acid into supramolecular polymers and hydrogels | TOF (CHCA, DCTB) | Varela‐Aramburu et al. (2021) |
| Synthesis and characterization of regioselectively functionalized mono‐sulfated and ‐phosphorylated anionic poly‐amido‐saccharides | R‐TOF (aminoacridine) | Varghese et al. (2022) |
| Per‐glycosylation of the surface‐accessible lysines: One‐pot aqueous route to stabilized proteins with native activity | TOF (SA) | Walther et al. (2021) |
| Chemoenzymatic modular assembly of O‐GalNAc glycans for functional glycomics | R‐TOF/TOF | Wang, Chen, et al. (2021) |
| Synergistic enzyme cocktail between levansucrase and inulosucrase for levan‐type fructooligosaccharide synthesis | TOF | Wangpaiboon, Klaewkla, et al. (2021) |
| Facile preparation of polysaccharide−polypeptide conjugates via a biphasic solution ring‐opening polymerization | TOF | Yang, Liu, et al. (2022) |
| Merging reagent modulation and remote anchimeric assistance for glycosylation: Highly stereoselective synthesis of α‐glycans up to a 30‐mer | TOF/TOF | Zhang, He, et al. (2021) |
| Production of water‐soluble sugar from cellulose and corn stover via molten salt hydrate impregnation and separation | TOF | Zhou, Liu, et al. (2022) |
| Automated assembly of starch and glycogen polysaccharides | TOF | Zhu, Delbiabco, et al. (2021) |
Format (not all items present): MALDI method (matrix). “MALDI” used when instrument not specified.
Table 48.
Use of matrix‐assisted laser desorption/ionization‐ mass spectrometry to study carbohydrate reactions.
| Reaction | Methods | References |
|---|---|---|
| Regioselective reductive ring‐opening reactions of 4,6‑O‑halobenzylidene acetals of glucopyranosides | R‐TOF/TOF (DHB) | Mezö et al. (2021) |
| VaporSPOT: Parallel synthesis of oligosaccharides on membranes | TOF/TOF | Tsouka et al. (2022) |
| A study of the diisobutylaluminum hydride‐promoted selective debenzylation of α‐CD protected with two different benzyl groups | TOF | Yousefi et al. (2022) |
15. MISCELLANEOUS STUDIES
A method for detection of ricin B by MALDI using lactosylated Fe3O4 magnetic nanoparticles has been developed and used to detect ricin B spiked into corn starch (Kandasamy et al., 2021). The nanoparticles were prepared by attaching lactose to the surface of aminated nanoparticles through the Maillard reaction and enrichment of ricin B took about 1 h by incubating the nanoparticles with samples under shaking at room temperature, followed by magnetic isolation. The limit of detection toward ricin B was about 3 nM.
The use of fluorescently labelled glycans has been advocated as a convenient method for the study of microbial degradation of glycans such as those pertaining to the gut microbiome. Acetylated galactoglucomannan from Norwegian spruce wood and acetylated (arabino) glucuronoxylan from Norwegian birch wood were used in the study with 2‐AB as the fluorescent label, chosen for its similar size to the monosaccharide constituents of the target sugars (Leivers et al., 2022). Monitoring of the labelling reaction was performed by MALDI and HPLC.
16. OTHER METHODS FOR GLYCAN AND GLYCOCONJUGATE ANALYSIS
As mentioned in the Introduction, this review also includes methods for the analysis of carbohydrates other than those based on MALDI analysis. Most of these methods could, however, be adapted for MALDI analysis. Several relevant reviews have been published as listed in Table 49.
Table 49.
Reviews and general articles relating to methods other than matrix‐assisted laser desorption/ionization.
| Subject | Comments | Citations | References |
|---|---|---|---|
| Capillary electrophoresis‐mass spectrometry of carbohydrates and glycoconjugates | MS interfacing, oligosaccharides and glycoconjugates | 101 | do Lago et al. (2021) |
| Reversed‐phase and hydrophobic interaction chromatography of carbohydrates and glycoconjugates | Comprehensive review. Some references to MALDI | 333 | El Rassi (2021b) |
| Carbohydrate analysis by modern liquid phase separation techniques, second edition | Book | ‐ | El Rassi (2021a) |
| Capillary electrophoresis and links with MALDI: Advances in N‐glycomics and glycoproteomics | Coupling of CE with MS and applications to glycan analysis | 66 | Makrydaki et al. (2021) |
| Advances in ion chromatography‐mass spectrometry (IC‐MS) for improved separation and analysis of carbohydrates | Mainly O‐glycans | 43 | Rumachik et al. (2021) |
Addition of a dopant to the gas flow after separation in an LC/MS system has been shown to improve the signal strength in negative ion mode (Madunić et al., 2021). Isopropanol‐enriched gas was shown to greatly improve the detection of both N‐and O‐glycans and their MS/MS mass spectra, particularly for the early eluting species.
Monoclonal antibodies radiolabelled with positron emitting radionuclides incorporated by use of bifunctional chelators, are widely used in nuclear imaging. Three methods for assessment of the average functionalisation and heterogeneity of the conjugated mAbs, radiometric and photometric titrations, MALDI‐TOF‐MS and UPLC/ESI‐TOF MS, have shown that all three gave comparable results. MALDI/TOF MS provided equivalent results to those obtained by the radio‐ and photo‐metric titrations although investigation of the heterogeneity of the conjugates was challenging and inaccurate, whereas UPLC/ESI‐TOF gave good peak resolution but was unable to discriminate between different smaller conjugates (Feiner et al., 2021).
Meyer, Montero, et al. (2022) have compared four common chromatographic methods (SFC, HILIC, RP‐LC, and GC) with detection by triple quadrupole mass spectrometer for analysis of monosaccharides. They showed that GC and RP‐LC, with suitable derivatization, are superior to the other two methods in terms of separation performance. Overall, RP‐LC–MS in MRM mode after derivatization with PMP gave the best separation, sensitivity and repeatability.
Wang, Wang, Wu, et al. (2022) have investigated several hydroxylamine reagents for analysis of monosaccharides and found that O‐(4‐methoxybenzyl)‐hydroxylamine hydrochloride gave the best results. Twelve monosaccharides were readily detected although not all were fully separated by HPLC. The d3 analogue of the derivatization reagent was also synthesised and used for quantitative studies.
Other methods are listed in Table 50.
17. REPORT OF RETRACTIONS
The paper by Zhao et al. (2014), reported in the review (Harvey, 2018); has been retracted. The paper “Discrimination of urinary exosomes from microvesicles by lipidomics using thin layer liquid chromatography (TLC) coupled with MALDI‐TOF mass spectrometry” by Singhto et al. (2022) has also been retracted because the mass tolerance was too large and the ion mode was at times inappropriate for the lipid species analysed.
18. CONCLUSIONS
MALDI continues to be a major technique for the analysis of carbohydrates and glycoconjugates with its advantage of speed and production of mainly singly charged ions. Thus, unlike ESI, the technique allows profiles of mixtures to be reproduced accurately. Although the rapid year‐by‐year increase in the number of publications reported in earlier reviews appears to have slowed somewhat, largely as the result of more analyses being conducted by LC‐MS, new methods and applications continue to appear. Growth areas are in applications to clinical practice for biomarker discovery, and particularly MALDI imaging with many new methods and matrices being reported. The incorporation of ion mobility into glycan assays, particularly for isomer separation is another growing area, and the use of linkage‐specific sialic acid derivatization, now appears in a large number of publications. The next review in this series (period from 2023 to 2024) will probably see a continuation of the trends reported above, particularly in the areas of MALDI imaging and incorporation of ion mobility into glycan analysis.
AUTHOR CONTRIBUTIONS
David J Harvey: Conceptualization; Project administration; Writing—original draft; Writing—review & editing.
CONFLICT OF INTEREST STATEMENT
The author declares no conflict of interest.
Harvey DJ. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2021‐2022. Mass Spectrom Rev. 2025;44:213‐453. 10.1002/mas.21873
REFERENCES
- Abari AH, Rourani HA, Ghasemi SM, Kim H, Kim YG. 2021. Investigation of antioxidant and anticancer activities of unsaturated oligo‑galacturonic acids produced by pectinase of Streptomyces hydrogenans YAM1. Sci. Rep. 11:8491. 10.1038/s41598-021-87804-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbattista R, Ventura G, Calvano CD, Cataldi TRI, Losito I. 2021. Bioactive compounds in waste by‐products from olive oil production: Applications and structural characterization by mass spectrometry techniques. Foods, 10:1236. 10.3390/foods10061236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y, Shibata H, Oyama K, Ueda T. 2021. Effect of O‐glycosylation on amyloid fibril formation of the variable domain in the Vλ6 light chain mutant Wil. Int. J. Biol. Macromol. 166:342–351. 10.1016/j.ijbiomac.2020.10.194 [DOI] [PubMed] [Google Scholar]
- Abla M, Ladavière C, Trombotto S. 2022. Impact of HILIC amino‑based column equilibration conditions on the analysis of chitooligosaccharides. Chromatographia, 85:55–63. 10.1007/s10337-021-04109-9 [DOI] [Google Scholar]
- Abo H, Kume M, Pecori F, Miura T, Matsumoto N, Nishihara S, Yamamoto K. 2022. Disaccharide‐tag for highly sensitive identification of O‐GlcNAc‐modified proteins in mammalian cells. PlosOne, 17:e0267804. 10.1371/journal.pone.0267804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou‐Elnaga AF, Batiha GE, Potter CT, Rehan IF. 2021. Functional glycomics and anxiety‐related behaviors in single versus group‐housed C57BL/6 and DBA/2 male mice. Adv. Anim. Vet. Sci. 9:1532–1546. 10.17582/journal.aavs/2021/9.10.1532.1546 [DOI] [Google Scholar]
- Abreu CA, Teixeira‐Pinheiro LC, Lani‐Louzada R, da Silva‐Junior AJ, Vasques JF, Gubert F, Nascimento‐dos‐Santos G, Mohana‐Borges R, de Souza Matos E, Pimentel‐Coelho PM, Santiago MF, Mendez‐Otero R. 2021. GD3 synthase deletion alters retinal structure and impairs visual function in mice. J. Neurochem. 158:694–709. 10.1111/jnc.15443 [DOI] [PubMed] [Google Scholar]
- Abukar T, Rahmani S, Thompson NK, Antonescu CN, Wakarchuk WW. 2021. Development of BODIPY labelled sialic acids as sialyltransferase substrates for direct detection of terminal galactose on N‐ and O‐linked glycans. Carbohydr. Res. 500:108249. 10.1016/j.carres.2021.108249 [DOI] [PubMed] [Google Scholar]
- Acevedo‑García V, Flórez‑Fernández N, López‑García M, Vilariño JML, Domínguez H, Torres MD. 2021. Acetone precipitation of heterofucoidans from Sargassum muticum autohydrolysis extracts. Waste Biomass Valori., 12:867–877. 10.1007/s12649-020-01044-y [DOI] [Google Scholar]
- Adamiak JW, Jhawar V, Bonifay V, Chandler CE, Leus IV, Ernst RK, Schweizer HP, Zgurskaya HI. 2021. Loss of RND‐type multidrug efflux pumps triggers iron starvation and lipid A modifications in Pseudomonas aeruginosa . Antimicrob. Agents Chemother. 65:e00592‐00521. 10.1128/aac.00592-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrahari AK, Bose P, Jaiswal MK, Rajkhowa S, Singh AS, Hotha S, Mishra N, Tiwari VK. 2021. Cu(I)‐catalyzed click chemistry in glycoscience and their diverse applications. Chem. Rev. 121:7638–7956. 10.1021/acs.chemrev.0c00920 [DOI] [PubMed] [Google Scholar]
- Agrahari AK, Jaiswal MK, Yadav MS, Tiwari VK. 2021. CuAAC mediated synthesis of cyclen cored glycodendrimers of high sugar tethers at low generation. Carbohydr. Res. 508:108403. 10.1016/j.carres.2021.108403 [DOI] [PubMed] [Google Scholar]
- Agrahari AK, Kumar S, Pandey MD, Rajkhowa S, Jaiswal MK, Tiwari VK. 2022. Click chemistry‐inspired synthesis of porphyrin hybrid glycodendrimers as fluorescent sensor for Cu(II) ions. ChemistrySelect, 7:e202202273. 10.1002/slct.202202273 [DOI] [Google Scholar]
- Aguedo J, Pakanova Z, Lorencova L, Nemcovic M, Kasak P, Barath M, Farkas P, Tkac J. 2022. MXene as a novel cartridge for N‐glycan enrichment. Anal. Chim. Acta, 1234:340512. 10.1016/j.aca.2022.340512 [DOI] [PubMed] [Google Scholar]
- Ahmadi E, Farimani MM, Rezadoost H. 2022. Optimization of inulin extraction from Inula helenium L. using response surface methodology followed by its MALDI‐TOF and TLC‐FLD‐based characterization . J. Med. Plants, 21:43–55. http://jmp.ir/article-1-3303-en.html [Google Scholar]
- Ahmadi E, Rezadoost H, Farimani MM. 2022. Isolation, characterization, and antioxidant activity of neutral carbohydrates from Astragalus arbusculinus gum. S. Afr. J. Bot., 146:669–675. 10.1016/j.sajb.2021.12.006 [DOI] [Google Scholar]
- Ahn MY, Yoon HJ, Hwang JS, Jin JM, Park KK. 2021. The role of noble bumblebee (Bombus terrestris) queen glycosaminoglycan in aged rat and gene expression profile based on DNA microarray. Toxicol. Res. 37:85–98. 10.1007/s43188-020-00065-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aissa I, Kilár A, Dörnyei Á. 2021. Study on the CID fragmentation pathways of deprotonated 4’‐monophosphoryl lipid A. Molecules, 26:5961. 10.3390/molecules26195961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajith A, Sthanikam Y, Banerjee S. 2021. Chemical analysis of the human brain by imaging mass spectrometry. Analyst, 146:5451–5473. 10.1039/D1AN01109J [DOI] [PubMed] [Google Scholar]
- Ajith A, Milnes PJ, Johnson GN, Lockyer NP. 2022. Mass spectrometry imaging for spatial chemical profiling of vegetative parts of plants. Plants, 11:1234. 10.3390/plants11091234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Mughaid H, Khazaaleh M. 2021. α‐d‐Mannoside ligands with a valency ranging from one to three: Synthesis and hemagglutination inhibitory properties. Carbohydr. Res. 508:108396. 10.1016/j.carres.2021.108396 [DOI] [PubMed] [Google Scholar]
- Alagesan K, Hoffmann M, Rapp E, Kolarich D. 2021. Glycoproteomics technologies in glycobiotechnology. Adv. Biochem. Eng. Biotechnol. 175:413–434. 10.1007/10_2020_144 [DOI] [PubMed] [Google Scholar]
- Ali I, Mukherjee S, Jana S, Khawas S, Ray B, Ray S. 2021. Production and identification of intricate bioactive oligosaccharides from Nyctanthesarbor‐tristis leaves by a combination of enzymatic, HPAEC and MALDI‐TOF‐MS techniques. Ind. J. Chem. B, 60B:1471–1477. [Google Scholar]
- Ali MM, Hussain D, Tang Y, Sun X, Shen Z, Zhang F, Du Z. 2021. Boronoisophthalic acid as a novel affinity ligand for the selective capture and release of glycoproteins near physiological pH. Talanta, 225:121896. 10.1016/j.talanta.2020.121896 [DOI] [PubMed] [Google Scholar]
- Ali MM, Zhu Z, Hussain D, Shen Z, He Y, Du Z. 2021. Flexible and hierarchical metal‐organic framework composite as solid‐phase media for facile affinity‐tip fabrication to selectively enrich glycopeptides and phosphopeptides. Talanta, 233:122576. 10.1016/j.talanta.2021.122576 [DOI] [PubMed] [Google Scholar]
- Ali MM, Zhu Z, Wang M, Hussain D, Gao X, Wang J, Du Z. 2021. Melamine foam assisted in‐tip packed amine‐functionalized titanium metal‐organic framework for the selective enrichment of endogenous glycopeptides. J. Chromatogr., A, 1636:461711. 10.1016/j.chroma.2020.461711 [DOI] [PubMed] [Google Scholar]
- Alla AJ, Stine KJ. 2022. Recent strategies for using monolithic materials in glycoprotein and glycopeptide analysis. Separations, 9:44. 10.3390/separations9020044 [DOI] [Google Scholar]
- Alsina C, Sancho‐Vaello E, Aranda‐Martínez A, Faijes M, Planas A. 2021. Auxiliary active site mutations enhance the glycosynthase activity of a GH18 chitinase for polymerization of chitooligosaccharides. Carbohydr. Polym. 252:117121. 10.1016/j.carbpol.2020.117121 [DOI] [PubMed] [Google Scholar]
- Alves I, Santos‐Pereira B, Dalebout H, Santos S, Vicente MM, Campar A, Thepaut M, Fieschi F, Strahl S, Boyaval F, Vizcaíno R, Silva R, Holst‐Bernal S, Vasconcelos C, Santos L, Wuhrer M, Marinho A, Heijs B, Pinho SS. 2021. Protein mannosylation as a diagnostic and prognostic biomarker of lupus nephritis: An unusual glycan neoepitope in systemic lupus erythematosus. Arthritis Rheumatol., 73:2069–2077. 10.1002/art.41768 [DOI] [PubMed] [Google Scholar]
- Alvisi N, van Noort K, Dwiani S, Geschiere N, Sukarta O, Varossieau K, Nguyen D‐L, Strasser R, Hokke CH, Schots A, Wilbers RHP. 2021. β‐Hexosaminidases along the secretory pathway of Nicotiana benthamiana have distinct specificities toward engineered helminth N‐glycans on recombinant glycoproteins. Front. Plant Sci. 12:638454. 10.3389/fpls.2021.638454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly MRE, El Azab IH. 2021. Synthesis of a deuterium‐labeled globotriaosylceramide probe for potential imaging of subcellular localization of Gb3 using nanoSIMS. Russ. J. Org, Chem. 57:1719–1724. 10.1134/S1070428021100213 [DOI] [Google Scholar]
- Ambrogi V, Bottacini F, Sharry JM, van Breen J, O'Keeffe E, Walsh D, Schoemaker B, Cao L, Kuipers B, Lindner C, Jimeno ML, Doyagüez EG, Hernandez‐Hernandez O, Moreno FJ, Schoterman M, van Sinderen D. 2021. Bifidobacterial β‐galactosidase‐mediated production of galacto‐oligosaccharides: Structural and preliminary functional assessments. Front. Microbiol. 12:750635. 10.3389/fmicb.2021.750635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoresano A, Pucci P. 2022. Mass spectrometry in metabolomics. In: Troisi J, Editor, Metabolomics Perspectives. From Theory to Practical Application. London, San Diego, Cambridge MA, Oxford, UK: Elsevier. p 109–147. 10.1016/B978-0-323-85062-9.00004-0 [DOI] [Google Scholar]
- Amos RA, Atmodjo MA, Huang C, Gao Z, Venkat A, Taujale R, Kannan N, Moremen KW, Mohnen D. 2022. Polymerization of the backbone of the pectic polysaccharide rhamnogalacturonan I. Nat. Plants, 8:1289–1303. 10.1038/s41477-022-01270-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andjelković U, Gudelj I, Klarić T, Hinneburg H, Vinković M, Wittine K, Dovezenski N, Vikić‐Topić D, Lauc G, Vujčić Z, Josić D. 2021. Increased yield of enzymatic synthesis by chromatographic selection of different N‐glycoforms of yeast invertase. Electrophoresis, 42:2626–2636. 10.1002/elps.202000092 [DOI] [PubMed] [Google Scholar]
- Angel PM, Drake RR, Park Y, Clift CL, West C, Berkhiser S, Hardiman G, Mehta AS, Bichell DP, Su YR. 2021. Spatial N‐glycomics of the human aortic valve in development and pediatric endstage congenital aortic valve stenosis. J. Mol. Cell. Cardiol., 154:6–20. 10.1016/j.yjmcc.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel PM, Mehta AS, Drake RR. 2021. Array‐based N‐glycan profiling of cells in culture. Method. Mol. Biol. 2271:331–342. 10.1007/978-1-0716-1241-5_23 [DOI] [PubMed] [Google Scholar]
- Angerer TB, Bour J, Biagi J‐L, Moskovets E, Frache G. 2022. Evaluation of 6 MALDI‐matrices for 10 μm lipid imaging and on‐tissue MSn with AP‐MALDI‐Orbitrap. J. Am. Soc. Mass Spectrom. 33:760–771. 10.1021/jasms.1c00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anh TTV, Uyen NQ, Hop DV, Dommes J, Versali M‐F, Van Hoang V. 2021. Chito‐oligosaccharide production by chitinase of Streptococcus macrosporeus VTCC 940003 and their inhibition activities on Botrytis cinerea . Eur. J. Plant Pathol., 161:185–193. 10.1007/s10658-021-02313-9 [DOI] [Google Scholar]
- Antonopoulos A, Broome S, Sharov V, Ziegenfuss C, Easton RL, Panico M, Dell A, Morris HR, Haslam SM. 2021. Site‐specific characterization of SARS‐CoV‐2 spike glycoprotein receptor‐binding domain. Glycobiology, 31:181–187. 10.1093/glycob/cwaa085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki‐Kinoshita K, Agravat S, Aoki NP, Arpinar S, Cummings RD, Fujita A, Fujita N, Hart GM, Haslam SM, Kawasaki T, Matsubara M, Moreman KW, Okuda S, Pierce M, Ranzinger R, Shikanai T, Shinmachi D, Solovieva E, Suzuki Y, Tsuchiya S, Yamada I, York WS, Zaia J, Narimatsu H. 2016. GlyTouCan 1.0—The international glycan structure repository. Nucleic Acids Res., 44:D1237–D1242. 10.1093/nar/gkv1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolinar‐Valiente R, Williams P, Doco T. 2021. Recent advances in the knowledge of wine oligosaccharides. Food Chem., 342:128330. 10.1016/j.foodchem.2020.128330 [DOI] [PubMed] [Google Scholar]
- Arendowski A, Sagandykova G, Mametov R, Rafińska K, Pryshchepa O, Pomastowski P. 2022. Nanostructured layer of silver for detection of small biomolecules in surface‐assisted laser desorption ionization mass spectrometry. Mater. Adv. 15:4076. 10.3390/ma15124076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argudo PG, Spitzer L, Ibarboure E, Jerome F, Cramail H, Lecommandoux S. 2022. Mannose‐based surfactant as biofunctional nanoemulsion stabilizer. Colloid Surface B, 220:112877. 10.1016/j.colsurfb.2022.112877 [DOI] [PubMed] [Google Scholar]
- Argudo PG, Spitzer L, Jerome F, Cramail H, Camacho L, Lecommandoux S. 2022. Design and self‐assembly of sugar‐based amphiphiles: Spherical to cylindrical micelles. Langmuir, 38:7535–7544. 10.1021/acs.langmuir.2c00579 [DOI] [PubMed] [Google Scholar]
- Arora A, Cameotra SS, Balomajumder C, Kumar R, Singh AK, Santhakumari B, Kumar P, Laik S. 2021. Rhamonolipids produced by Pseudomonas aeruginosa promotes methane hydrates formation in fixed bed silica gel medium. Mar. Geophys. Res. 42:5. 10.1007/s11001-020-09426-6 [DOI] [Google Scholar]
- Arumugam S, Schmieder S, Pezeshkian W, Becken U, Wunder C, Chinnapen D, Ipsen JH, Kenworthy AK, Lencer W, Mayor S, Johannes L. 2021. Ceramide structure dictates glycosphingolipid nanodomain assembly and function. Nat. Commun. 12:3675. 10.1038/s41467-021-23961-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athomo ABB, Anris SPE, Tchiama RS, Eyma F, Arnaudguilhem C, Charrier B. 2021. Identification of phenolic compounds from K. ivorensis by selected chromatographic and spectrometric techniques. J. Renew. Mater., 9:35–48. 10.32604/jrm.2021.013626 [DOI] [Google Scholar]
- Augusto LA, Bourgeois‐Nicolaos N, Breton A, Barreault S, Alonso EH, Gera S, Faraut‐Derouin V, Semaan N, De Luca D, Chaby R, Doucet‐Populaire F, Tissières P. 2021. Presence of 2‐hydroxymyristate on endotoxins is associated with death in neonates with Enterobacter cloacae complex septic shock. iScience, 24:102916. 10.1016/j.isci.2021.102916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba S, Luo B, Li Z, He J, Lan F, Wu Y. 2022. Mesoporous covalent organic framework microspheres with dual‐phase separation strategy for high‐purity glycopeptide enrichment. J. Chromatogr., A, 1684:463575. 10.1016/j.chroma.2022.463575 [DOI] [PubMed] [Google Scholar]
- Baba T, Ryumin P, Duchoslav E, Chen K, Chelur A, Loyd B, Chernushevich I. 2021. Dissociation of biomolecules by an intense low‐energy electron beam in a high sensitivity time‐of‐flight mass spectrometer. J. Am. Soc. Mass Spectrom. 32:1964–1975. 10.1021/jasms.0c00425 [DOI] [PubMed] [Google Scholar]
- Báez‐Ramírez E, Querales L, Aranaga CA, López G, Guerrero E, Kremer L, Carrère‐Kremer S, Viljoen A, Daff M, Laval F, Cole ST, Benjak A, Alzari P, André‐Leroux G, Jacobs WR, Vilcheze C, Takiff HE. 2021. Elimination of PknL and MSMEG_4242 in Mycobacterium smegmatis alters the character of the outer cell envelope and selects for mutations in Lsr2. Cell Surf., 7:100060. 10.1016/j.tcsw.2021.100060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I, Malaker SA, Polasky DA, Riley NM, Schjoldager K, Vakhrushev SY, Halim A, Aoki‐Kinoshita KF, Nesvizhskii AI, Bertozzi CR, Wandall HH, Parker BL, Thaysen‐Andersen M, Scott NE. 2022. Glycoproteomics. Nat. Rev. 2:48. 10.1038/s43586-022-00128-4 [DOI] [Google Scholar]
- Bai H, Zhang B, Cheng X, Liu J, Wang X, Qin W, Zhang M. 2022. Synthesis of zwitterionic polymer modified graphene oxide for hydrophilic enrichment of N‐glycopeptides from urine of healthy subjects and patients with lung adenocarcinoma. Talanta, 237:122938. 10.1016/j.talanta.2021.122938 [DOI] [PubMed] [Google Scholar]
- Bakker B, Vaes RDW, Aberle MR, Welbers T, Hankemeier T, Rensen SS, Damink SWMO, Heeren RMA. 2022. Preparing ductal epithelial organoids for high‐spatial‐resolution molecular profiling using mass spectrometry imaging. Nat. Protoc. 17:962–979. 10.1038/s41596-021-00661-8 [DOI] [PubMed] [Google Scholar]
- Balaguer F, Enrique M, Llopis S, Barrena M, Navarro V, Álvarez B, Chenoll E, Ramón D, Tortajada M, Martorell P. 2022. Lipoteichoic acid from Bifidobacterium animalis subsp. lactis BPL1: A novel postbiotic that reduces fat deposition via IGF‐1 pathway. Microb. Biotechnol. 15:805–816. 10.1111/1751-7915.13769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. 2021a. Biophysical and mass spectrometry based characterization of methylglyoxal‐modified myoglobin: Role of advanced glycation end products in inducing protein structural alterations. Int. J. Biol. Macromol. 193:2165–2172. 10.1016/j.ijbiomac.2021.11.047 [DOI] [PubMed] [Google Scholar]
- Banerjee S. 2021b. Long‐term incubation of myoglobin with glyoxal induces amyloid like aggregation of the heme protein: Implications of advanced glycation end products in protein conformational disorders. J. Mol. Liquids, 326:115256. 10.1016/j.molliq.2020.115256 [DOI] [Google Scholar]
- Bao C‐X, Li S‐Y, Xin Y‐J, Hou J, Cui H‐L. 2022. Natrinema halophilum sp. nov., Natrinema salinisoli sp. nov., Natrinema amylolyticum sp. nov. and Haloterrigena alkaliphila sp. nov., four extremely halophilic archaea isolated from salt mine, saline soil and salt lake. Int. J. Systemat. Evolut. Microbiol., 72:005385. 10.1099/ijsem.0.005385 [DOI] [PubMed] [Google Scholar]
- Barada E, Hinou H. 2022. BOA/DHB/Na: An efficient UV‐MALDI matrix for high‐sensitivity and auto‐tagging glycomics. Int. J. Mol. Sci. 23:12510. 10.3390/ijms232012510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari E, Pizzi A, Schmidt O, Amirou S, Tajick‐Ghanbary MA, Humar M. 2021. Differentiation of fungal destructive behaviour of wood by the white‐rot fungus Fomes fomentarius by MALDI‐TOF mass spectrometry. J. Renew. Mater. 9:381–397. 10.32604/jrm.2021.015288 [DOI] [Google Scholar]
- Barnes WJ, Koj S, Black IM, Archer‑Hartmann SA, Azadi P, Urbanowicz BR, Peña MJ, O'Neill MA. 2021. Protocols for isolating and characterizing polysaccharides from plant cell walls: a case study using rhamnogalacturonan‑II. Biotechnol. Biofuels, 14:142. 10.1186/s13068-021-01992-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxa U, Weintraub A, Seckler R. 2020. Self‐competitive inhibition of the bacteriophage P22 tailspike endorhamnosidase by O‑antigen oligosaccharides. Biochemistry, 59:4845–4855. 10.1021/acs.biochem.0c00872 [DOI] [PubMed] [Google Scholar]
- Beck A, Haitz F, Thier I, Siems K, Jakupovic S, Rupp S, Zibek S. 2021. Novel mannosylerythritol lipid biosurfactant structures from castor oil revealed by advanced structure analysis. J. Ind. Microbiol. Biotechnol. 48:kuab042. 10.1093/jimb/kuab042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith DM, FitzGerald FG, Benavente MCR, Mercer ER, Ludwig A‐K, Michalak M, Kaltner H, Kopitz Jr., Gabius H‐J, Cudic M. 2021. Calorimetric analysis of the interplay between synthetic Tn antigen‐presenting MUC1 glycopeptides and human macrophage galactose‐type lectin. Biochemistry, 60:547–558. 10.1021/acs.biochem.0c00942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar H, Tamura K, Wagner ER, Cosgrove DJ, Brumer H. 2021. Conservation of endo‐glucanase 16 (EG16) activity across highly divergent plant lineages. Biochem. J. 478:3063–3078. 10.1042/bcj20210341 [DOI] [PubMed] [Google Scholar]
- Belenguer‐Sapiña C, Pellicer‐Castell E, Chali SP, Ravoo BJ, Amorós P, Simó‐Alfonso EF, Mauri‐Aucejo AR. 2021. Host‐guest interactions for extracting antibiotics with a γ‐cyclodextrin poly (glycidyl‐co‐ethylene dimethacrylate) hybrid sorbent. Talanta, 232:122478. 10.1016/j.talanta.2021.122478 [DOI] [PubMed] [Google Scholar]
- Belusko A, Aumeistere L, Ciprovica I. 2022. Oligosaccharides in human milk, achievements in analysis: A review. Food Sci., 37:100–105. 10.22616/rrd.28.2022.015 [DOI] [Google Scholar]
- Benkovics G, Malanga M, Cutrone G, Béni S, Vargas‐Berenguel A, Casas‐Solvas JM. 2021. Facile synthesis of per(6‐O‐tertbutyldimethylsilyl)‐α‐, β‐, and γ‐cyclodextrin as protected intermediates for the functionalization of the secondary face of the macrocycles. Nat. Protoc. 16:965–987. 10.1038/s41596-020-00443-8 [DOI] [PubMed] [Google Scholar]
- Bensaci N, Abdi A, Aziza HB, Aouadi S. 2022. Characterization and biological evaluation of Crataegus azarolus fruit polysaccharides. J. Mol. Struct. 1270:133889. 10.1016/j.molstruc.2022.133889 [DOI] [Google Scholar]
- Benzie G, Bouma K, Battellino T, Cooper S, Hemming R, Kammouni W, Liu L, Do C, Khajehpour M, Perreault H, Kornfeld S, Triggs‐Raine B, Mark BL. 2022. Increased phosphorylation of HexM improves lysosomal uptake and potential for managing GM2 gangliosidoses. BBA Adv. 2:100032. 10.1016/j.bbadva.2021.100032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berardinelli SJ, Eletsky A, Valero‐González J, Ito A, Manjunath R, Hurtado‐Guerrero R, Prestegard JH, Woods RJ, Haltiwanger RS. 2022. O‐Fucosylation stabilizes the TSR3 motif in thrombospondin‐1 by interacting with nearby amino acids and protecting a disulfide bond. J. Biol. Chem., 298:102047. 10.1016/j.jbc.2022.102047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereczki I, Szűcs Z, Batta G, Nagy TM, Ostorházi E, Kövér KE, Borbás A, Herczegh P. 2022. The first dimeric derivatives of the glycopeptide antibiotic teicoplanin. Pharmaceuticals, 15:77. 10.3390/ph15010077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezina OV, Rykov SV, Polyakova AK, Bozdaganyan ME, Sidochenko AV, Baudrexl M, Schwarz WH, Zverlov VV, Yarotsky SV. 2021. Strategic aromatic residues in the catalytic cleft of the xyloglucanase MtXgh74 modifying thermostability, mode of enzyme action, and viscosity reduction ability. Appl. Microbiol. Biotechnol. 105:1461–1476. 10.1007/s00253-021-11106-3 [DOI] [PubMed] [Google Scholar]
- Bermudez A, Pitteri SJ. 2021. Enrichment of intact glycopeptides using strong anion exchange and electrostatic repulsion hydrophilic interaction chromatography. Method. Mol. Biol. 2271:107–120. 10.1007/978-1-0716-1241-5_8 [DOI] [PubMed] [Google Scholar]
- Bern M, Kil YJ, Bechker C. 2012. Byonic: Advanced peptide and protein identification software. Curr, Protoc, Bioinformatics, 40:13.20.11–13.20.14. 10.1002/0471250953.bi1320s40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava R, Lehoux S, Maeda K, Tsokos MG, Krishfield S, Ellezian L, Pollak M, Stillman IE, Cummings RD, Tsokos GC. 2021. Aberrantly glycosylated IgG elicits pathogenic signaling in podocytes and signifies lupus nephritis. JCI Insight, 6:e147789. 10.1172/jci.insight.147789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacherjee A, Daskhan GC, Bains A, Watson AES, Eskandari‐Sedighi G, St. Laurent CD, Voronova A, Macauley MS. 2021. Increasing phagocytosis of micoglia by targeting CD33 with liposomes displaying glycan ligands. J. Control Release, 338:680–693. 10.1016/j.jconrel.2021.09.010 [DOI] [PubMed] [Google Scholar]
- Bhuvanachandra B, Sivaramakrishna D, Alim S, Preethiba G, Rambabu S, Swamy MJ, Podile AR. 2021. New class of chitosanase from Bacillus amyloliquefaciens for the generation of chitooligosaccharides. J. Agric. Food Chem., 69:78–87. 10.1021/acs.jafc.0c05078 [DOI] [PubMed] [Google Scholar]
- Bi F, Zhang J, Wei Z, Yu D, Zheng S, Wang J, Li H, Hua Z, Zhang H, Yang G. 2022. Dynamic glycopeptide dendrimers: Synthesis and their controllable self‐assembly into varied glyco‐nanostructures for the biomimicry of glycans. Biomacromolecules, 23:128–139. 10.1021/acs.biomac.1c01137 [DOI] [PubMed] [Google Scholar]
- Bi M, Tian Z‐X. 2021. Research progress in structure‐specific N‐glycoproteomics. J. Chinese Mass Spectrom. Soc. 42:897–913. 10.7538/zpxb.2021.0122 [DOI] [Google Scholar]
- Bi X, Xiong W, He Ja, Ma S, Zhang J, Fang Y, Wu Y. 2021. Site‐selective and biocompatible growth of polymers from glycan moieties of glycoproteins and living cells. Biomacromolecules, 22:4237–4243. 10.1021/acs.biomac.1c00792 [DOI] [PubMed] [Google Scholar]
- Bibi N, Yamin M, Awan AT, Ahmad K, Khattak R. 2022. Lipid biomarkers for breast cancer diagnostics. In: Malik SS, Masood N, Editors., Breast Cancer: From Bench to Personalized Medicine. Singapore: Springer Nature. p 235–262. 10.1007/978-981-19-0197-3_11 [DOI] [Google Scholar]
- Biel TG, Faison T, Matthews AM, Zou G, Ortega‐Rodriguez U, Pegues MA, Azer N, Gomez F, Johnson S, Rogstad S, Chen K, Xie H, Agarabi C, Rao VA, Ju T. 2022. An etanercept O‐glycovariant with enhanced potency. Mol. Therap. Meth. Clin. Develop. 25:124–135. 10.1016/j.omtm.2022.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien T, Bessler S, Dreisewerd K, Soltwisch J. 2021. Transmission‐mode MALDI mass spectrometry imaging of single cells: Optimizing sample preparation protocols. Anal. Chem. 93:4513–4520. 10.1021/acs.analchem.0c04905 [DOI] [PubMed] [Google Scholar]
- Bien T, Koerfer K, Schwenzfeier J, Dreisewerd K, Soltwisch J. 2022. Mass spectrometry imaging to explore molecular heterogeneity in cell culture. Proc. Natl. Acad. Sci., USA, 119:e2114365119. 10.1073/pnas.2114365119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch J, Khan S, Madsen M, Kjeldsen C, Møller MS, Stender EGP, Peters GHJ, Duus JØ, Kragelund BB, Svensson B. 2021. Binding sites for oligosaccharide repeats from lactic acid bacteria exopolysaccharides on bovine β‑lactoglobulin identified by NMR spectroscopy. ACS Omega, 6:9039–9052. 10.1021/acsomega.1c00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black I, Heiss C, Carlson RW, Azadi P. 2021. Linkage analysis of oligosaccharides and polysaccharides: A tutorial. Method. Mol. Biol. 2271:249–271. 10.1007/978-1-0716-1241-5_18 [DOI] [PubMed] [Google Scholar]
- Blaj D‐A, Balan‐Porcarasu M, Petre BA, Harabagiu V, Peptu C. 2021. MALDI mass spectrometry monitoring of cyclodextrin‐oligolactide derivatives synthesis. Polymer, 233:124186. 10.1016/j.polymer.2021.124186 [DOI] [Google Scholar]
- Blanc L, Ferraro GB, Tuck M, Prideaux B, Dartois V, Jain RK, Desbenoit N. 2021. Kendrick mass defect variation to decipher isotopic labeling in brain metastases studied by mass spectrometry imaging. Anal. Chem. 93:16314–16319. 10.1021/acs.analchem.1c03916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke CRK, Hartig JP, Grimsley G, Liu L, Semmes OJ, Wu JD, Ippolito JE, Hughes‐Halbert C, Nyalwidhe JO, Drake RR. 2021. Direct N‐glycosylation profiling of urine and prostatic fluid glycoproteins and extracellular vesicles. Front. Chem. 9:734280. 10.3389/fchem.2021.734280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke CRK, McDowell CT, Black AP, Mehta AS, Angel PM, Drake RR. 2021. Glycan imaging mass spectrometry: Progress in developing clinical diagnostic assays for tissues, biofluids, and cells. Clinics Lab. Med., 41:247–266. 10.1016/j.cll.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke CRK, Drake RR. 2022. MALDI Mass spectrometry imaging and glycomics. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 207–233. 10.1039/9781839165191-00207 [DOI] [Google Scholar]
- Blaschke CRK, Hill EG, Mehta AS, Angel PM, Laronga C, Drake RR. 2022. Integrating age, BMI, and serum N‑glycans detected by MALDI mass spectrometry to classify suspicious mammogram findings as benign lesions or breast cancer. Sci. Rep. 12:20801. 10.1038/s41598-022-25401-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddapati S, Gummadi SN. 2021. A comprehensive review on mutan (a mixed linkage of α‐1‐3 and α‐1‐6 glucans) from bacterial sources. Biotechnol. Genetic. Eng. Rev. 37:208–237. 10.1080/02648725.2021.2003072 [DOI] [PubMed] [Google Scholar]
- Boddupalli CS, Nair S, Belinsky G, Gans J, Teeple E, Nguyen T‐H, Mehta S, Guo L, Kramer ML, Ruan J, Wang H, Davison M, Kumar D, Vidyadhara DJ, Zhang B, Klinger K, Mistry PK. 2022. Neuroinflammation in neuronopathic Gaucher disease: Role of microglia and NK cells, biomarkers, and response to substrate reduction therapy. eLife, 11:e79830. 10.7554/eLife.79830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. 2020. Human milk oligosaccharides: Structure and functions. In: Ogra PL, Walker WA, Lönnerdal B, Editors., Milk, Mucosal Immunity and the Microbiome: Impact on the Neonate, 94th Nestlé Nutrition Institute workshop, Lausanne, September 2019. Basel: Karger. p 115–123. 10.1159/000505339 [DOI] [PubMed]
- Bogdanov AV, Andreeva OV, Belenok MG, Voloshina AD, Enikeeva KI, Samorodov AV, Mironov VF. 2021. Synthesis of triazolylisatins glycoconjugates and some ammonium hydrazones on their basis. Russ. J. Gen. Chem. 91:1282–1291. 10.1134/S1070363221070045 [DOI] [Google Scholar]
- Bojar D, Lisacek F. 2022. Glycoinformatics in the artificial intelligence era Chem. Rev. 122:15971–15988. 10.1021/acs.chemrev.2c00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bols M, Friis V. 2022. Taming of the DIBAL promoted debenzylation of α‐cyclodextrin. Kinetics, substituent effects and efficient synthesis of Lings tetrol. Chem. Eur. J. 28:e202200564. 10.1002/chem.202200564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventura E, Barone R, Sturiale L, Pasquariello R, Alessandrì MG, Pinto AM, Renieri A, Panteghini C, Garavaglia B, Cioni G, Battini R. 2021. Clinical, molecular and glycophenotype insights in SLC39A8‑CDG. Orphanet J. Rare Dis., 16:307. 10.1186/s13023-021-01941-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookmeyer C, Röhling U, Dreisewerd K, Soltwisch J. 2022. Single‐photon‐induced post‐ionization to boost ion yields in MALDI mass spectrometry imaging. Angew. Chem. Int. Ed. 61:e202202165. 10.1002/anie.202202165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyapranai K, Ounjaijean S, Kulprachakarn K, Potpromanee L, Chen MC‐M, Tsai H‐Y, Chen ST. 2021. Haptoglobin polymorphisms and fucosylation change: Possible influence of variation on the identified lung cancer‐relevant biomarkers. Curr. Proteomics, 18:380–389. 10.2174/1570164617666200817115006 [DOI] [Google Scholar]
- Borisov R, Kanateva A, Zhilyaev D. 2021. Recent advances in combinations of TLC with MALDI and other desorption/ionization mass‐spectrometry techniques. Front. Chem. 9:771801. 10.3389/fchem.2021.771801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaval F, van Zeijl R, Dalebout H, Holst S, van Pelt G, Fariña‐Sarasqueta A, Mesker W, Tollenaar R, Morreau H, Wuhrer M, Heijs B. 2021. N‐Glycomic signature of stage II colorectal cancer and its association with the tumor microenvironment. Mol. Cell. Proteomics, 20:100057. 10.1074/mcp.ra120.002215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyaval F, Dalebout H, Van Zeijl R, Wang W, Sarasqueta AF, Lageveen‐Kammeijer GSM, Boonstra JJ, McDonnell LA, Wuhrer M, Morreau H, Heijs B. 2022. High‐mannose N‐glycans as malignant progression markers in early‐stage colorectal cancer. Cancers, 14:1552. 10.3390/cancers14061552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch J, Rajagopal BS, Paradisi A, Yates N, Lindley PJ, Smith J, Hollingsworth K, Turnbull WB, Henrissat B, Parkin A, Berry A, Hemsworth GR. 2021. C‐type cytochrome‐initiated reduction of bacterial lytic polysaccharide monooxygenases. Biochem. J. 478:2927–2944. 10.1042/BCJ20210376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandi J, Noberini R, Bonaldi T, Cecconi D. 2022. Advances in enrichment methods for mass spectrometry‐based proteomics analysis of post‐translational modifications. J. Chromatogr., A, 1678:463352. 10.1016/j.chroma.2022.463352 [DOI] [PubMed] [Google Scholar]
- Brune KD, Liekniņa I, Sutov G, Morris AR, Jovicevic D, Kalniņš G, Kazāks A, Kluga R, Kastaljana S, Zajakina A, Jansons J, Skrastiņa D, Spunde K, Cohen AA, Bjorkman PJ, Morris HR, Suna E, Tārs K. 2021. N‐Terminal modification of Gly‐His‐tagged proteins with azidogluconolactone. ChemBioChem, 22:3199–3207. 10.1002/cbic.202100381 [DOI] [PubMed] [Google Scholar]
- Bruni PS, Schürch S. 2021. Fragmentation mechanisms of protonated cyclodextrins in tandem mass spectrometry. Carbohydr. Res. 504:108316. 10.1016/j.carres.2021.108316 [DOI] [PubMed] [Google Scholar]
- Bryan L, Clynes M, Meleady P. 2021. The emerging role of cellular post‐translational modifications in modulating growth and productivity of recombinant Chinese hamster ovary cells. Biotechnol. Adv. 49:107757. 10.1016/j.biotechadv.2021.107757 [DOI] [PubMed] [Google Scholar]
- Bua RO, Messina A, Sturiale L, Barone R, Garozzo D, Palmigiano A. 2021. N‐Glycomics of human erythrocytes. Int. J. Mol. Sci. 22:8063. 10.3390/ijms22158063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budenkova E, Sukhikh S, Ivanova S, Babich O, Dolganyuk V, Michaud P, Kriger O. 2021. Improvement of enzymatic saccharification of cellulose‐containing raw materials using Aspergillus niger . Processes, 9:1360. 10.3390/pr9081360 [DOI] [Google Scholar]
- Buitrago‐Arias C, Londoño‐Moreno A, Avila‐Reyes SV, Arenas‐Ocampo ML, Alamilla‐Beltran L, Jimenez‐Aparicio AR, Camacho‐Diaz BH. 2021. Evaluation of the fermentation of acetylated agave fructans (agavins), with Saccharomyces boulardii as a probiotic. Rev. Mex. Ing. Quím. 20:Poly2533. 10.24275/rmiq/Poly2533 [DOI] [Google Scholar]
- Bulmer GS, Mattey AP, Parmeggiani F, Williams R, Ledru H, Marchesi A, Seibt LS, Both P, Huang K, Galan MC, Flitsch SL, Green AP, van Munster JM. 2021. A promiscuous glycosyltransferase generates poly‐β‐1,4‐glucan derivatives that facilitate mass spectrometry‐based detection of cellulolytic enzymes. Org. Biomol. Chem. 19:5529–5533. 10.1039/d1ob00971k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne C, Smith R. 2021. C‐SEQer: An open‐source de novo glycan identification tool in C++. J. Proteome Res., 20:4068–4074. 10.1021/acs.jproteome.1c00379 [DOI] [PubMed] [Google Scholar]
- Buruaga‐Ramiro C, Fernández‐Gándara N, Cabañas‐Romero LV, Valenzuela SV, Pastor FIJ, Diaz P, Martinez J. 2022. Lytic polysaccharide monooxygenases and cellulases on the production of bacterial cellulose nanocrystals. Eur. Polymer J. 163:110939. 10.1016/j.eurpolymj.2021.110939 [DOI] [Google Scholar]
- Buszewska‐Forajta M, Rafińska K, Buszewski B. 2022. Tissue sample preparations for preclinical research determined by molecular imaging mass spectrometry using matrix‐assisted laser desorption/ionization. J. Sep. Sci., 45:1345–1361. 10.1002/jssc.202100578 [DOI] [PubMed] [Google Scholar]
- Butler KE, Kalmar JG, Muddiman DC, Baker ES. 2022. Utilizing liquid chromatography, ion mobility spectrometry, and mass spectrometry to assess INLIGHT™ derivatized N‐linked glycans in biological samples. Anal. Bioanal. Chem. 414:623–637. 10.1007/s00216-021-03570-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SJ, Birgersson S, Wiemann M, Arcos‐Hernandez M, Stålbrand H. 2022. Transglycosylation by β‐mannanase TrMan5A variants and enzyme synergy for synthesis of allyl glycosides from galactomannan. Proc. Biochem. 112:154–166. 10.1016/j.procbio.2021.11.028 [DOI] [Google Scholar]
- Butler W, Huang J. 2021. Glycosylation changes in prostate cancer progression. Front. Oncol. 11:809170. 10.3389/fonc.2021.809170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butré CI, Largy E, Cantais F, Delobel A. 2021. Profiling, relative quantification, and identification of sialylated N‐linked oligosaccharides by UPLC‐FLR‐ESI/MS after derivatization with fluorescent anthranilamide. Method. Mol. Biol. 2271:237–247. 10.1007/978-1-0716-1241-5_17 [DOI] [PubMed] [Google Scholar]
- Butré CI, Largy E, Delobel A. 2021. Profiling of N‐linked oligosaccharides of a glycoprotein by UPLC‐FLR‐ESI‐MS after derivatization with fluorescent anthranilamide. Method Mol. Biol. 2271:179–188. 10.1007/978-1-0716-1241-5_13 [DOI] [PubMed] [Google Scholar]
- Byrd‑Leotis L, Jia N, Matsumoto Y, Lu D, Kawaoka Y, Steinhauer DA, Cummings RD. 2022. Sialylated and sulfated N‑glycans in MDCK and engineered MDCK cells for influenza virus studies. Sci. Rep. 12:12757. 10.1038/s41598-022-16605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero I, Ringot‐Destrez B, Si‐Tahar M, Barbry P, Guillon A, Lantier I, Berri M, Chevaleyre C, Fleurot I, Barc C, Ramphal R, Pons N, Paquet A, Lebrigand K, Baron C, Bähr A, Klymiuk N, Léonard R, Robbe‐Masselot C. 2021. Evidence of early increased sialylation of airway mucins and defective mucociliary clearance in CFTR‐deficient piglets. J. Cyst. Fibros. 20:173–182. 10.1016/j.jcf.2020.09.009 [DOI] [PubMed] [Google Scholar]
- Cai B, Yi X, Han Q, Pan J, Chen H, Sun H, Wan P. 2022. Structural characterization of oligosaccharide from Spirulina platensis and its effect on the faecal microbiota in vitro . Food Sci. Hum. Wellness, 11:109–118. 10.1016/j.fshw.2021.07.012 [DOI] [Google Scholar]
- Cai D, Bian Y, Wu S, Ding K. 2021. Conformation‐controlled hydrogen‐bond‐mediated aglycone delivery method for α‑xylosylation. J. Org. Chem., 86:9945–9960. 10.1021/acs.joc.1c00187 [DOI] [PubMed] [Google Scholar]
- Cai L, Zhou S, Wang Y, Xu X, Zhang L, Cai Z. 2021. New insights into the anti‐hepatoma mechanism of triple‐helix β‐glucan by metabolomics profiling. Carbohydr. Polym. 269:118289. 10.1016/j.carbpol.2021.118289 [DOI] [PubMed] [Google Scholar]
- Cai W, Zhang H, Chen X, Yan S, Yang L, Song H, Li J, Liu J, Yu H, Liu H, Zhu D. 2022. Effect of microwave‐assisted acid extraction on the physicochemical properties and structure of soy hull polysaccharides. Int. J. Food Sci. Technol. 57:6744–6754. 10.1111/ijfs.16034 [DOI] [Google Scholar]
- Cai Y, Ren W, Wang H, Bian Q. 2022. In‐depth profiling of urinary N‐glycome in diabetic kidney disease by ultrafast glycoprotein immobilization for glycan extraction (UltraGIG). Anal. Chim. Acta, 1221:340144. 10.1016/j.aca.2022.340144 [DOI] [PubMed] [Google Scholar]
- Cain JA, Dale AL, Cordwell SJ. 2021. Exploiting pglB oligosaccharyltransferase‐positive and ‐negative Campylobacter jejuni and a multiprotease digestion strategy to identify novel sites modified by N‑linked protein glycosylation. J. Proteome Res., 20:4995–5009. 10.1021/acs.jproteome.1c00482 [DOI] [PubMed] [Google Scholar]
- Cairns CM, St. Michael F, Jamshidi M, van Faassen H, Yang Q, Henry KA, Hussack G, Sauvageau J, Vinogradov EV, Cox AD. 2022. Structural characterization and evaluation of an epitope at the tip of the A‑band rhamnan polysaccharide of Pseudomonas aeruginosa . ACS Infect. Dis. 8:1336–1346. 10.1021/acsinfecdis.2c00183 [DOI] [PubMed] [Google Scholar]
- Cairo JPLF, Cannella D, Oliveira LC, Gonçalves TA, Rubio MV, Terrasan CRF, Tramontina R, Mofatto LS, Carazzolle MF, Garcia W, Felby C, Damasio A, Walton PH, Squina F. 2021. On the roles of AA15 lytic polysaccharide monooxygenases derived from the termite Coptotermes gestroi . J. Inorg. Biochem. 216:111316. 10.1016/j.jinorgbio.2020.111316 [DOI] [PubMed] [Google Scholar]
- Cajic S, Hennig R, Burock R, Rapp E. 2021. Capillary (Gel) electrophoresis‐based methods for immunoglobulin (G) glycosylation analysis. In: Pezer M, Editor, Antibody Glycosylation. Cham, Switzerland: Springer Nature. p 137–172. 10.1007/978-3-030-76912-3_4 [DOI] [PubMed] [Google Scholar]
- Calderaro F, Bevers LE, van den Berg MA. 2021. Oxidative power: Tools for assessing LPMO activity on cellulose. Biomolecules, 11:1098. 10.3390/biom11081098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MP, Zhao S, Abrahams JL, Nguyen‐Khuong T, Rudd PM. 2021. GlycoStore: A platform for H/UPLC and capillary electrophoresis glycan data. Method. Mol. Biol. 2370:25–40. 10.1007/978-1-0716-1685-7_2 [DOI] [PubMed] [Google Scholar]
- Campos‐Valdez AR, Casas‐Godoy L, Sandoval G, Hernández L, Sassaki GL, de Menezes LRA, Campos‐Terán J, Reyes‐Duarte D, Arrizon J. 2022. Regioselective synthesis of 6”‐O‐lauroyl‐1‐kestose and 6”’‐O‐lauroylnystose by sequential enzymatic reactions of transfructosylation and acylation. Biocatal. Biotransfor. 40:133–143. 10.1080/10242422.2021.1952192 [DOI] [Google Scholar]
- Campos D, Girgis M, Sanda M. 2022. Site‐specific glycosylation of SARS‐CoV‐2: Big challenges in mass spectrometry analysis. Proteomics, 22:2100322. 10.1002/pmic.202100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canis K, Garénaux E, Boe J‐F. 2021. Site‐specific N‐glycosylation analysis of recombinant proteins by LC/MSE . Method. Mol. Biol. 2271:133–154. 10.1007/978-1-0716-1241-5_10 [DOI] [PubMed] [Google Scholar]
- Cano ME, García‐Martín A, Ladero M, Lesur D, Pilard S, Kovensky J. 2021. A simple procedure to obtain a medium‐size oligogalacturonic acids fraction from orange peel and apple pomace wastes. Food Chem., 346:128909. 10.1016/j.foodchem.2020.128909 [DOI] [PubMed] [Google Scholar]
- Cao C, Yu L, Yan J, Fu D, Yuan J, Liang X. 2021. Purification of natural neutral N‐glycans by using two‐dimensional hydrophilic interaction liquid chromatography x porous graphitized carbon chromatography for glycan‐microarray assay. Talanta, 221:121382. 10.1016/j.talanta.2020.121382 [DOI] [PubMed] [Google Scholar]
- Cao H, Antonopoulos A, Henderson S, Wassall H, Brewin J, Masson A, Shepherd J, Konieczny G, Patel B, Williams M‐L, Davie A, Forrester MA, Hall L, Minter B, Tampakis D, Moss M, Lennon C, Pickford W, Erwig L, Robertson B, Dell A, Brown GD, Wilson HM, Rees DC, Haslam SM, Rowe JA, Barker RN, Vickers MA. 2021. Red blood cell mannoses as phagocytic ligands mediating both sickle cell anaemia and malaria resistance. Nat. Commun. 12:1792. 10.1038/s41467-021-21814-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Mathur A, Robertson C, Antonopoulos A, Henderson S, Girard L‐P, Wong JH, Davie A, Wright S, Brewin J, Rees DC, Dell A, Haslam SM, Vickers MA. 2022. Measurement of erythrocyte membrane mannoses to assess splenic function. Br. J. Haematol. 198:155–164. 10.1111/bjh.18164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Zhou Y, Li X, Lin S, Tan Z, Guan F. 2021. Integrating transcriptomics, proteomics, glycomics and glycoproteomics to characterize paclitaxel resistance in breast cancer cells. J. Proteomics, 243:104266. 10.1016/j.jprot.2021.104266 [DOI] [PubMed] [Google Scholar]
- Cao R, Li J‐X, Chen H, Cao C, Zheng F, Huang K, Chen Y‐R, Flitsch SL, Liu L, Voglmeir J. 2022. Complete shift in glycosyl donor specificity in mammalian, but not C. elegans β1,4‐GalT1 Y286L mutants, enables the synthesis of N,N‐diacetyllactosamine. ChemCatChem, 14:e202101699. 10.1002/cctc.202101699 [DOI] [Google Scholar]
- Cao W, Liu M, Kong S, Wu M, Zhang Y, Yan P. 2021. Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol. Cell. Proteomics, 20:100060. 10.1074/mcp.r120.002090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Yang P. 2022. Qualitative and quantitative analytical methods for intact glycopeptides. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. Boca Raton: CRC Press. p 91–130. 10.1201/9781003185833-4 [DOI] [Google Scholar]
- Cao Y, Song Z, Guo Z, Zhao X, Gong Y, Zhao K, Qu C, Huang Y, Li Y, Gao Y, Zhang J, Guo X. 2022. Cytokines in the immune microenvironment change the glycosylation of IgG by regulating intracellular glycosyltransferases. Front. Immunol. 12:724379. 10.3389/fimmu.2021.724379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Zhang Z, Liu R, Wu M, Li Z, Xu X, Liu Z. 2022. Serum linkage‐specific sialylation changes are potential biomarkers for monitoring and predicting the recurrence of papillary thyroid cancer following thyroidectomy. Front. Endocrinol. 13:858325. 10.3389/fendo.2022.858325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdeville P, Martin L, Cholet S, Damont A, Audran M, Ericsson M, Fenaille F, Marchand A. 2021. Evaluation of erythropoietin biosimilars EpotinTM, Hemax®and JimaixinTM by electrophoretic methods used for doping control analysis and specific N‐glycan analysis revealed structural differences from original epoetin alfa drug Eprex®. J. Pharmaceut. Biomed. Anal. 194:113750. 10.1016/j.jpba.2020.113750 [DOI] [PubMed] [Google Scholar]
- Carbajo‐Gordillo AI, González‐Cuesta M, Blanco JLJ, Benito JM, Santana‐Armas ML, Carmona T, Di Giorgio C, Przybylski C, Mellet CO, de Ilarduya CT, Mendicuti F, Fernández JMG. 2021. Trifaceted Mickey Mouse amphiphiles for programmable self‐assembly, DNA complexation and organ‐selective gene delivery. Chem. Eur. J. 27:9429–9438. 10.1002/chem.202100832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell AA, Veličković D, Lawrence TJ, Bowen BP, Louie KB, Carper DL, Chu RK, Mitchell HD, Orr G, Markillie LM, Jawdy SS, Grimwood J, Shaw AJ, Schmutz J, Northen TR, Anderton CR, Pelletier DA, Weston DJ. 2022. Novel metabolic interactions and environmental conditions mediate the boreal peatmoss‐cyanobacteria mutualism. ISME J. 16:1074–1085. 10.1038/s41396-021-01136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo JJ, Galermo AG, Amicucci MJ, Nandita E, Couture G, Bacalzo N, Chen Y, Lebrilla CB. 2021. A multidimensional mass spectrometry‐based workflow for de novo structural elucidation of oligosaccharides from polysaccharides. J. Am. Soc. Mass Spectrom., 32:2175–2185. 10.1021/jasms.1c00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin M, Bruxelle J‐F, Ng K, Blaukopf M, Pantophlet R, Kosma P. 2022. Synthetic neoglycoconjugates of hepta‐ and nonamannoside ligands for eliciting oligomannose‐specific HIV‐1‐neutralizing antibodies. ChemBioChem, 23:e202200061. 10.1002/cbic.202200061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čaval T, de Haan N, Konstantinidi A, Vakhrushev SY. 2021. Quantitative characterization of O‐GalNAc glycosylation. Curr. Opin. Struct. Biol. 68:135–141. 10.1016/j.sbi.2020.12.010 [DOI] [PubMed] [Google Scholar]
- Chae A, Lee G, Koh D‐Y, Yang C‐M, Lee S, Kim Y‐K. 2021. Polyacrylonitrile‐based carbon nanofibers as a matrix for laser desorption/ionization time‐of‐flight mass spectrometric analysis of small molecules under both positive and negative ionization modes. Anal. Bioanal. Chem. 413:1193–1202. 10.1007/s00216-020-03083-9 [DOI] [PubMed] [Google Scholar]
- Chaichi A, Hasan SMA, Mehta N, Donnarumma F, Ebenezer P, Murray KK, Francis J, Gartia MR. 2021. Label‐free lipidome study of paraventricular thalamic nucleus (PVT) of rat brain with posttraumatic stress injury by Raman imaging. Analyst, 146:170–183. 10.1039/D0AN01615B [DOI] [PubMed] [Google Scholar]
- Chakrapani A, Ruiz‐Larrabeiti O, Pohl R, Svoboda M, Krásný L, Hocek M. 2022. Glucosylated 5‐hydroxymethylpyrimidines as epigenetic DNA bases regulating transcription and restriction cleavage. Chem. Eur. J. 28:e202200911. 10.1002/chem.202200911 [DOI] [PubMed] [Google Scholar]
- Chakrapani N, Fischer J, Swiontek K, Codreanu‐Morel F, Hannachi F, Morisset M, Mugemana C, Bulaev D, Blank S, Bindslev‐Jensen C, Biedermann T, Ollert M, Hilger C. 2022. α‐Gal present on both glycolipids and glycoproteins contributes to immune response in meat‐allergic patients. J. Allergy Clin. Immunol. 150:396–405. 10.1016/j.jaci.2022.02.030 [DOI] [PubMed] [Google Scholar]
- Chamoun S, Welander J, Martis‐Thiele M‐M, Ntzouni M, Claesson C, Vikström E, Turkina MV. 2021. Colistin dependence in extensively drug‐resistant Acinetobacter baumannii strain is associated with ISAjo2 and ISAba13 insertions and multiple cellular responses. Int. J. Mol. Sci. 22:576. 10.3390/ijms22020576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar B, Wanke A, Wawra S, Saake P, Mahdi L, Charura N, Neidert M, Poschmann G, Malisic M, Thiele M, Stühler K, Dama M, Pauly M, Zuccaro A. 2022. Fungi hijack a ubiquitous plant apoplastic endoglucanase to release a ROS scavenging β‐glucan decasaccharide to subvert immune responses. Plant Cell, 34:2765–2784. 10.1093/plcell/koac114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Zaia J. 2022. Methods to improve quantitative glycoprotein coverage from bottom‐up LC‐MS data. Mass Spectrom. Rev. 41:922–937. 10.1002/mas.21692 [DOI] [PubMed] [Google Scholar]
- Chao H‐C, McLuckey SA. 2021. Manipulation of ion types via gas‐phase ion/ion chemistry for the structural characterization of the glycan moiety on gangliosides. Anal. Chem. 93:15752–15760. 10.1021/acs.analchem.1c03876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Ugonotti J, Lee LY, Everest‐Dass A, Kawahara R, Thaysen‐Andersen M. 2021. Trends in oligomannosylation and α1,2‐mannosidase expression in human cancers. Oncotarget, 12:2188–2205. 10.18632/oncotarget.28064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Fowler MJ, Baker C, Stopka SA, Regan MS, Sablatura L, Broughton CW, Knight BE, Stabenfeldt SE, Agar NYR, Sirianni RW. 2021. β‑Cyclodextrin‐poly (β‐amino ester) nanoparticles are a generalizable strategy for high loading and sustained release of HDAC inhibitors. ACS Appl. Mater. Interfaces, 13:20960–20973. 10.1021/acsami.0c22587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C‐Y, Lin Y‐W, Wang S‐W, Lin Y‐C, Cheng Y‐Y, Ren C‐T, Wong C‐H, Wu C‐Y. 2022. Synthesis of azido‐globo H analogs for immunogenicity evaluation. ACS Cent. Sci. 8:77–85. 10.1021/acscentsci.1c01277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Laviolette SR, Whitehead SN, Renaud JB, Yeung KK‐C. 2021. Imaging of neurotransmitters and small molecules in brain tissues using laser desorption/ionization mass spectrometry assisted with zinc oxide nanoparticles. J. Am. Soc. Mass Spectrom. 32:1065–1079. 10.1021/jasms.1c00021 [DOI] [PubMed] [Google Scholar]
- Chen C, Zhang X, Dong X, Zhou H, Li X, Liang X. 2021. TiO2 Simultaneous enrichment, on‐line deglycosylation, and sequential analysis of glyco‐ and phosphopeptides. Front. Chem. 9:703176. 10.3389/fchem.2021.703176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhang N, Wu Y, Yang C, Xie Q, Deng C, Sun N. 2022. Investigation of urinary exosome metabolic patterns in membranous nephropathy by titania‐assisted intact exosome mass spectrometry. Small Sci., 2:2100118. 10.1002/smsc.202100118 [DOI] [Google Scholar]
- Chen J, Tang Y, Yu B. 2021. A mild glycosylation protocol with glycosyl 1‐methylimidazole‐2‐carboxylates as donors. Eur. J. Org. Chem. 4333–4344. 10.1002/ejoc.202100677 [DOI] [Google Scholar]
- Chen J, Wang W, Dong Y, Zhang J, Hu K, Ma Q. 2022. Optimization of extraction of inulin from Taraxacum kok‐saghyz Rodin by response surface methodology and its MALDI‐TOF MS analysis. Sci. Technol. Food Indust. 43:205–212. 10.13386/j.issn1002-0306.2021040173 [DOI] [Google Scholar]
- Chen K, Zhang X, Long L, Ding S. 2021. Comparison of C4‐oxidizing and C1/C4‐oxidizing AA9 LPMOs in substrate adsorption, H2O2‐driven activity and synergy with cellulase on celluloses of different crystallinity. Carbohydr. Polym. 269:118305. 10.1016/j.carbpol.2021.118305 [DOI] [PubMed] [Google Scholar]
- Chen M, Dupard SJ, McClung CM, Ruse CI, Ganatra MB, Vainauskas S, Taron CH, Samuelson JC. 2021. Improving the study of protein glycosylation with new tools for glycopeptide enrichment. In: Raghav A, Ahmad J, Editors, Fundamentals of Glycosylation, London, IntechOpen, p. 447–534. 10.5772/intechopen.97339 [DOI]
- Chen M, Mac‐Béar J, Ropartz D, Lahaye M. 2022. Biorefinery of apple pomace: New insights into xyloglucan building blocks. Carbohydr. Polym. 290:119526. 10.1016/j.carbpol.2022.119526 [DOI] [PubMed] [Google Scholar]
- Chen M, Ropartz D, Mac‐Béar J, Bonnin E, Lahaye M. 2022. New insight into the mode of action of a GH74 xyloglucanase on tamarind seed xyloglucan: Action pattern and cleavage site. Carbohydr. Res. 521:108661. 10.1016/j.carres.2022.108661 [DOI] [PubMed] [Google Scholar]
- Chen P, Shrotri A, Fukuoka A. 2021. Synthesis of cello‐oligosaccharides by depolymerization of cellulose: A review. Appl. Catal. A, Gen. 621:118177. 10.1016/j.apcata.2021.118177 [DOI] [Google Scholar]
- Chen Q, Zhang Y, Zhang K, Liu J, Pan H, Wang X, Li S, Hu D, Lin Z, Zhao Y, Hou G, Guan F, Li H, Liu S, Ren Y. 2022. Profiling the bisecting N‐acetylglucosamine modification in amniotic membrane via mass spectrometry. Genomics Proteomics Bioinform., 20:648–656. 10.1016/j.gpb.2021.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S‐Y, Clark DJ, Zhang H. 2021. High‐throughput analyses of glycans, glycosites, and intact glycopeptides using C4‐and C18/MAX‐tips and liquid handling system. Curr. Protoc., 1:e186. 10.1002/cpz1.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wu D, Robinson CV, Struwe WB. 2021. Native mass spectrometry meets glycomics: Resolving structural detail and occupancy of glycans on intact glycoproteins. Anal. Chem. 93:10435–10443. 10.1021/acs.analchem.1c01460 [DOI] [PubMed] [Google Scholar]
- Chen T, Cheng G, Jiao S, Ren L, Zhao C, Wei J, Han J, Pei M, Du Y, Li J‐J. 2021. Expression and biochemical characterization of a novel marine chitosanase from Streptomyces niveus suitable for preparation of chitobiose. Mar. Drugs, 19:300. 10.3390/md19060300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X‐Y, Gao B‐X, Zhou H‐Y. 2022. Recent progress in matrix for analysis of low molecular weight compounds using matrix assisted laser desorption ionization time‐of‐flight mass spectrometry. Chin. J. Anal. Chem. 50:12–24. 10.19756/j.issn.0253-3820.211012 [DOI] [Google Scholar]
- Chen X, Fu X, Huang L, Xu J, Gao X. 2021. Agar oligosaccharides: A review of preparation, structures, bioactivities and application. Carbohydr. Polym. 265:118076. 10.1016/j.carbpol.2021.118076 [DOI] [PubMed] [Google Scholar]
- Chen X, Sun Y, Zhang T, Roepstorff P, Yang F. 2021. Comprehensive analysis of the proteome and PTMomes of C2C12 myoblasts reveals that sialylation plays a role in the differentiation of skeletal muscle cells. J. Proteome Res. 20:222–235. 10.1021/acs.jproteome.0c00353 [DOI] [PubMed] [Google Scholar]
- Chen X, Wang L, Wu Y, Zhang H, Dong W, Yu X, Huang C, Li Y, Wang S, Zhang J. 2022. Caveolin‑1 knockout mice have altered serum N‑glycan profile and sialyltransferase tissue expression. J. Physiol. Biochem., 78:73–83. 10.1007/s13105-021-00840-x [DOI] [PubMed] [Google Scholar]
- Chen X, Wang Y, Luo Y, Gao Z, Han T, Zhou H. 2022. Composite PVK/SLGO as matrix for MALDI‐TOF MS detection of small molecules in dual‐ion mode. ACS Omega, 7:39028–39038. 10.1021/acsomega.2c04772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y‐J, Yen T‐C, Lin Y‐H, Chen Y‐L, Khoo K‐H, Chen Y‐J. 2021. ZIC‐cHILIC‐based stagetip for simultaneous glycopeptide enrichment and fractionation toward large‐scale N‑sialoglycoproteomics. Anal. Chem. 93:15931–15940. 10.1021/acs.analchem.1c03224 [DOI] [PubMed] [Google Scholar]
- Chen Y, Fang Z, Zhou J, Qin H, Ye M. 2021. Mirror‐cutting‐based digestion strategy enables the in‐depth and accuracy characterization of N‑linked protein glycosylation. J. Proteome Res. 20:4948–4958. 10.1021/acs.jproteome.1c00333 [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu Y. 2021. Characterization of galacto‐oligosaccharides using high‐performance anion exchange chromatography‐tandem mass spectrometry. J. Sep. Sci. 44:2221–2233. 10.1002/jssc.202100064 [DOI] [PubMed] [Google Scholar]
- Chen Y, Qin H, Yue X, Zhou J, Liu L, Nie Y, Ye M. 2021. Highly efficient enrichment of O‑GlcNAc glycopeptides based on chemical oxidation and reversible hydrazide chemistry. Anal. Chem. 93:16618–16627. 10.1021/acs.analchem.1c04031 [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang T, Xie P, Song Y, Wang J, Cai Z. 2021. Mass spectrometry imaging revealed alterations of lipid metabolites in multicellular tumor spheroids in response to hydroxychloroquine. Anal. Chim. Acta, 1184:339011. 10.1016/j.aca.2021.339011 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hu D, Zhao L, Tang W, Li B. 2022. Unraveling metabolic alterations in transgenic mouse model of Alzheimer's disease using MALDI MS imaging with 4‐aminocinnoline‐3‐carboxamide matrix. Anal. Chim. Acta, 1192:339337. 10.1016/j.aca.2021.339337 [DOI] [PubMed] [Google Scholar]
- Chen Y, Jiang L, Zhang R, Shi Z, Xie C, Hong Y, Wang J, Cai Z. 2022. Spatially revealed perfluorooctane sulfonate‐induced nephrotoxicity in mouse kidney using atmospheric pressure MALDI mass spectrometry imaging. Sci. Total Environ. 838:156380. 10.1016/j.scitotenv.2022.156380 [DOI] [PubMed] [Google Scholar]
- Chen Y, Tang F, Qin H, Yue X, Nie Y, Huang W, Ye M. 2022. Endo‐M mediated chemoenzymatic approach enables reversible glycopeptide labeling for O‐GlcNAcylation analysis. Angew. Chem. Int. Ed. 61:e202117849. 10.1002/anie.202117849 [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang X, Ma X, Liang S, Gao Q, Tretyakova EV, Zhang Y, Zhou D, Xiao S. 2022. Facial synthesis and bioevaluation of well‐defined OEGylated betulinic acid‐cyclodextrin conjugates for inhibition of influenza infection. Molecules, 27:1163. 10.3390/molecules27041163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Shen J, Dong W, Li P, Xin M, Liu D, Jia L, Zhu B, Li W, Sun S. 2022. Recognition of core‐fucosylated glycopeptides based on the Y1+Fuc/Y1 ratio in low‐energy HCD spectra. Anal. Chem. 94:17349–17353. 10.1021/acs.analchem.2c03182 [DOI] [PubMed] [Google Scholar]
- Chen Z, Wei J, Tang Y, Lin C, Costello CE, Hong P. 2022. GlycoDeNovo2: An improved MS/MS‐based de novo glycan topology reconstruction algorithm. J. Am. Soc. Mass Spectrom. 33:436–445. 10.1021/jasms.1c00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Shu H, Peng Y, Feng X, Yan G, Zhang L, Yao J, Bao H, Lu H. 2021. Specific analysis of α‑2,3‐sialylated N‑glycan linkage isomers by microchip capillary electrophoresis−mass spectrometry. Anal. Chem. 93:5537–5546. 10.1021/acs.analchem.1c00064 [DOI] [PubMed] [Google Scholar]
- Cheng M, Shu H, Yang M, Yan G, Zhang L, Wang L, Wang W, Lu H. 2022. Fast discrimination of sialylated N‑glycan linkage isomers with one‐step derivatization by microfluidic capillary electrophoresis−mass spectrometry. Anal. Chem. 94:4666–4676. 10.1021/acs.analchem.1c04760 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Sun C, Qin Y, Shi S, Li Y, Fan Q, Yang G, Gao X. 2022. In situ enzymatic hydrolysis and analysis of bladder cancer N‐glycans based on FFPE tissue sections. Prog. Biochem. Biophys, 49:2001–2014. 10.16476/j.pibb.2021.0393 [DOI] [Google Scholar]
- Chernykh A, Kawahara R, Thaysen‐Andersen M. 2021. Towards structure‐focused glycoproteomics. Biochem. Soc. Trans. 49:161–186. 10.1042/bst20200222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra M, Wimmer N, He QQ, Ferro V. 2021. Development of improved synthetic routes to pixatimod (PG545), a sulfated oligosaccharide‐steroid conjugate. Bioconj. Chem. 32:2420–2431. 10.1021/acs.bioconjchem.1c00453 [DOI] [PubMed] [Google Scholar]
- Chilelli N‐C, Faggian A, Favaretto F, Milan G, Compagnin C, Dassie F, Bettini S, Roverso M, Seraglia R, Lapolla A, Vettor R. 2021. In vitro chronic glycation induces AGEs accumulation reducing insulin‐stimulated glucose uptake and increasing GLP1R in adipocytes. Am. J. Physiol. Endocrinol. Metab., 320:E976–E988. 10.1152/ajpendo.00156.2020 [DOI] [PubMed] [Google Scholar]
- Chiu K‐Y, Wang Q, Gunawardena HP, Held M, Faik A, Chen H. 2021. Desalting paper spay mass spectrometry (DPS‐MS) for rapid detection of glycans and glycoconjugates. Int. J. Mass Spectrom. 469:116688. 10.1016/j.ijms.2021.116688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhov AO. 2022. Carbohydrate analysis by mass spectrometry In: Nollet LML, Winkler R, Editors., Mass Spectrometry in Food Analysis. New York: CRC Press. p 85–108. 10.1201/9781003091226-8 [DOI] [Google Scholar]
- Cho BG, Banazadeh A, Peng W, Zhao J, Goli M, Gautam S, Hussein A, Mechref Y. 2021. Determination of isomeric glycan structures by permethylation and liquid chromatography‐mass spectrometry (LC‐MS). Method. Mol. Biol. 2271:281–301. 10.1007/978-1-0716-1241-5_20 [DOI] [PubMed] [Google Scholar]
- Cho BG, Reyes CDG, Goli M, Gautam S, Banazadeh A, Mechref Y. 2022. Targeted N‑glycan analysis with parallel reaction monitoring using a quadrupole‐Orbitrap hybrid mass spectrometer. Anal. Chem. 94:15215–15222. 10.1021/acs.analchem.2c01975 [DOI] [PubMed] [Google Scholar]
- Cho C‐W, Lim C‐R, Cho B‐G, Mun S‐B, Choi J‐W, Zhao Y, Kim S, Yun Y‐S. 2021. Development of prediction models for adsorption properties of chitin and chitosan for micropollutants. Chem. Eng. J. 426:131341. 10.1016/j.cej.2021.131341 [DOI] [Google Scholar]
- Cho J, Miyagawa A, Yamaguchi K, Abe W, Tsugawa Y, Yamamura H, Imai T. 2022. UDP‑glucose, cereblon‑dependent proinsulin degrader. Sci. Rep. 12:14568. 10.1038/s41598-022-18902-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Duong V‐A, Mok J‐H, Joo M, Park J‐M, Lee H. 2022. Enrichment and analysis of glycated proteins. Rev. Anal. Chem. 41:83–97. 10.1515/revac-2022-0036 [DOI] [Google Scholar]
- Choi J, Shin J‐H, An HJ, Oh MJ, Kim S‐R. 2021. Analysis of secretome and N‐glycosylation of Chlorella species. Algal Res., 59:102466. 10.1016/j.algal.2021.102466 [DOI] [Google Scholar]
- Choudhary P, Rao A. 2021. SELECT‐GLYCOCIN: A recombinant microbial system for expression and high‐throughput screening of glycocins. Glycoconj. J. 38:233–250. 10.1007/s10719-020-09960-w [DOI] [PubMed] [Google Scholar]
- Chu H, Zheng H, Sun N, Deng C. 2022. Simultaneous analysis of cellular glycoproteome and phosphoproteome in cervical carcinoma by one‐pot specific enrichment. Anal. Chim. Acta, 1195:338693. 10.1016/j.aca.2021.338693 [DOI] [PubMed] [Google Scholar]
- Cibulski S, de Souza TA, Raimundo JP, Nascimento YM, Abreu LS, Suarez N, Miraballes I, Roehe PM, de Araújo DAM, Tavares JF, da Silva MS, Silveira F. 2022. ISCOM‑Matrices nanoformulation using the raw aqueous extract of Quillaja lancifolia (Q. brasiliensis). BioNanoScience, 12:1166–1171. 10.1007/s12668-022-01023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillero‐Pastor B, Cuypers E. 2022. Sample preparation of biological tissues for MALDI‐MSI. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 87–104. 10.1039/9781839165191-00087 [DOI] [Google Scholar]
- Cindrić A, Krištić J, Kavur MM, Pezer M. 2021. Glycosylation and aging. Adv. Exp. Med. Biol. 1325:341–373. 10.1007/978-3-030-70115-4_17 [DOI] [PubMed] [Google Scholar]
- Cirnigliaro L, Bianchi P, Sturiale L, Garozzo D, Mangili G, Keldermans L, Rizzo R, Matthijs G, Fiumara A, Jaeken J, Barone R. 2022. COG6‐CDG: Novel variants and novel malformation. Birth Defects Res., 114:165–174. 10.1002/bdr2.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift CL, Drake RR, Mehta A, Angel PM. 2021. Multiplexed imaging mass spectrometry of the extracellular matrix using serial enzyme digests from formalin‐fixed paraffin‐embedded tissue sections. Anal. Bioanal. Chem. 413:2709–2719. 10.1007/s00216-020-03047-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift CL, Mehta A, Drake RR, Angel PM. 2021. Multiplexed imaging mass spectrometry of histological staining, N‐glycan and extracellular matrix from one tissue section: A tool for fibrosis research. Method. Mol. Biol. 2350:313–329. 10.1007/978-1-0716-1593-5_20 [DOI] [PubMed] [Google Scholar]
- Coletto E, Latousakis D, Pontifex MG, Crost EH, Vaux L, Santamarina EP, Goldson A, Brion A, Hajihosseini MK, Vauzour D, Savva GM, Juge N. 2022. The role of the mucin‐glycan foraging Ruminococcus gnavus in the communication between the gut and the brain. Gut Microb., 14:e2073784. 10.1080/19490976.2022.2073784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert M, Kadouche D, Ducatez M, Pillonel T, Kebbi‐Beghdadi C, Cenci U, Huang B, Chabi M, Maes E, Coddeville B, Couderc L, Touzet H, Bray F, Tirtiaux C, Ball S, Greub G, Colleoni C. 2021. Conservation of the glycogen metabolism pathway underlines a pivotal function of storage polysaccharides in Chlamydiae . Commun. Biol. 4:296. 10.1038/s42003-021-01794-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon MD, Gildersleeve JC. 2021. Enhanced binding and reduced immunogenicity of glycoconjugates prepared via solid‐state photoactivation of aliphatic diazirine carbohydrates. Bioconj. Chem. 32:133–142. 10.1021/acs.bioconjchem.0c00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy LR, Stanback AE, Young LEA, Clarke HA, Austin GL, Liu J, Allison DB, Sun RC. 2021. In situ analysis of N‐linked glycans as potential biomarkers of clinical course in human prostate cancer. Mol. Cancer Res., 19:1727–1738. 10.1158/1541-7786.mcr-20-0967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conroy LR, Chang JE, Sun Q, Clarke HA, Buoncristiani MD, Young LEA, McDonald RJ, Liu J, Gentry MS, Allison DB, Sun RC. 2022. High‐dimensionality reduction clustering of complex carbohydrates to study lung cancer metabolic heterogeneity. Adv. Cancer Res. 154:227–251. 10.1016/bs.acr.2022.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coura MdMA, Barbosa EA, Brand GD, Bloch C Jr., de Sousa JB. 2021. Identification of differential N‐glycan compositions in the serum and tissue of colon cancer patients by mass spectrometry. Biology, 10:343. 10.3390/biology10040343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, St. Michael F, Aubry A, Strong PCR, Hayes AC, Logan SM. 2021. Comparison of polysaccharide glycoconjugates as candidate vaccines to combat Clostridiodes (Clostridium) difficile . Glycoconj. J. 38:493–508. 10.1007/s10719-020-09937-9 [DOI] [PubMed] [Google Scholar]
- Crawford CJ, Qiao Y, Liu Y, Huang D, Yan W, Seeberger PH, Oscarson S, Chen S. 2021. Defining the qualities of high‐quality palladium on carbon catalysts for hydrogenolysis. Org. Process Res. Dev. 25:1573–1578. 10.1021/acs.oprd.0c00536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critcher M, Hassan AA, Huang ML. 2022. Seeing the forest through the trees: Characterizing the glycoproteome. Trends Biochem. Sci. 47:492–505. 10.1016/j.tibs.2022.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumin C, Huang Y‐L, Everest‐Dass A, Jacob F. 2021. Deciphering the importance of glycosphingolipids on cellular and molecular mechanisms associated with epithelial‐to‐mesenchymal transition in cancer. Biomolecules, 11:62. 10.3390/biom11010062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumin C, Huang Y‐L, Rossdam C, Ruoff F, Céspedes SP, Liang C‐Y, Lombardo FC, Coelho R, Rimmer N, Konantz M, López MN, Alam S, Schmidt A, Calabrese D, Fedier A, Vlajnic T, von Itzstein M, Templin M, Buettner FFR, Everest‐Dass A, Heinzelmann‐Schwarz V, Jacob F. 2022. Glycosphingolipids are mediators of cancer plasticity through independent signaling pathways. Cell Rep., 40:111181. 10.1016/j.celrep.2022.111181 [DOI] [PubMed] [Google Scholar]
- Cutolo G, Didak B, Tomas J, Roubinet B, Lafite P, Nehmé R, Schuler M, Landemarre L, Tatibouët A. 2022. The myrosinase‐glucosinolate system to generate neoglycoproteins: A case study targeting mannose binding lectins. Carbohydr. Res. 516:108562. 10.1016/j.carres.2022.108562 [DOI] [PubMed] [Google Scholar]
- D'Addio M, Frey J, Tacconi C, Commerford CD, Halin C, Detmar M, Cummings RD, Otto VI. 2021. Sialoglycans on lymphatic endothelial cells augment interactions with Siglec‐1 (CD169) of lymph node macrophages. FASEB J., 35:e22017. 10.1096/fj.202100300r [DOI] [PubMed] [Google Scholar]
- Daher W, Leclercq L‐D, Johansen MD, Hamela C, Karam J, Trivelli X, Nigou J, Guérardel Y, Kremer L. 2022. Glycopeptidolipid glycosylation controls surface properties and pathogenicity in Mycobacterium abscessus . Cell Chem. Biol. 29:910–924. 10.1016/j.chembiol.2022.03.008 [DOI] [PubMed] [Google Scholar]
- Dan M, Shen J, Zhao G, Wang D. 2022. Complete conversion of agarose into water soluble agaro‐oligosaccharides by microwave assisted hydrothermal hydrolysis. Food Chem., 395:133622. 10.1016/j.foodchem.2022.133622 [DOI] [PubMed] [Google Scholar]
- Dang Y, Ruan L, Tian Y, Xu Z, Zhang W. 2021. Nitric oxide prodrug delivery and release monitoring based on a galactose‐modified multifunctional nanoprobe. Anal. Chem. 93:7625–7634. 10.1021/acs.analchem.1c00287 [DOI] [PubMed] [Google Scholar]
- Dantas JMdM, de Araújo NK, da Silva NS, Torres‐Rêgo M, Furtado AA, de Assis CF, Araújo RM, Teixeira JA, Ferreira LdS, Fernandes‐Pedrosa MdF, dos Santos ES. 2022. Purification of chitosanases produced by Bacillus toyonensis CCT 7899 and functional oligosaccharides production. Prep. Biochem. Biotechnol. 52:443–451. 10.1080/10826068.2021.1961273 [DOI] [PubMed] [Google Scholar]
- Dare AP, Günther CS, Grey AC, Guo G, Demarais NJ, Cordiner S, McGhie TK, Boldingh H, Hunt M, Deng C, Karppinen K, Jaakola L, Espley RV. 2021. Resolving the developmental distribution patterns of polyphenols and related primary metabolites in bilberry (Vaccinium myrtillus) fruit. Food Chem., 374:131703. 10.1016/j.foodchem.2021.131703 [DOI] [PubMed] [Google Scholar]
- Datta AK, Sukhija N. 2021. Glycobioinformatics in deciphering the mammalian glycocode: Recent advances. In: Banerjee DK, Editor, Glycome: The Hidden Code in Biology. Hauppauge, NY: Nova Science Publishers. p 323–375. [Google Scholar]
- Davy GP, Editor 2022. Glycosylation. New York: Humana Press (part of Springer Nature). [Google Scholar]
- de Andrade P, Muñoz‐García JC, Pergolizzi G, Gabrielli V, Nepogodiev SA, Iuga D, Fábián L, Nigmatullin R, Johns MA, Harniman R, Eichhorn SJ, Angulo J, Khimyak YZ, Field RA. 2021. Chemoenzymatic synthesis of fluorinated cellodextrins identifies a new allomorph for cellulose‐like materials. Chem. Eur. J. 27:1374–1382. 10.1002/chem.202003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Borba MC, Velho AC, Grondard AM, Baltenweck R, Magnin‐Robert M, Randoux B, Holvoet M, Hilbert J‐L, Flahaut C, Reignault P, Hugueney P, Stadnik MJ, Siah A. 2021. The algal polysaccharide ulvan induces resistance in wheat against Zymoseptoria tritici without major alteration of leaf metabolome. Front. Plant Sci. 12:703712. 10.3389/fpls.2021.703712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan N, Narimatsu Y, Aasted MKM, Larsen ISB, Marinova IN, Dabelsteen S, Vakhrushev SY, Wandall HH. 2022. In‐depth profiling of O‑glycan isomers in human cells using C18 nanoliquid chromatography−mass spectrometry and glycogenomics. Anal. Chem. 94:4343–4351. 10.1021/acs.analchem.1c05068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan N, Pučić‐Baković M, Novokmet M, Falck D, Lageveen‐Kammeijer G, Razdorov G, Vučković F, Trbojević‐Akmacic I, Gornik O, Hanić M, Wuhrer M, Lauc G, The Human Glycome Project 2022. Developments and perspectives in high‐throughput protein glycomics: Enabling the analysis of thousands of samples. Glycobiology, 32:651–663. 10.1093/glycob/cwac026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De León TS, Salum ML, Matsushita Y, Fukushima K, Monge ME, Erra‐Balsells R. 2022. ESI‐MS reveals preferential complex formation of carbohydrates with Z‐sinapinic acid compared with the E‐isomer. New J. Chem. 46:18563–18574. 10.1039/D2NJ02789E [DOI] [Google Scholar]
- de Lima MZT, de Almeida LR, Mera AM, Bernardes A, Garcia W, Muniz JRC. 2021. Crystal structure of a sucrose‐6‐phosphate hydrolase from Lactobacillus gasseri with potential applications in fructan production and the food industry. J. Agric. Food Chem., 69:10223–10234. 10.1021/acs.jafc.1c03901 [DOI] [PubMed] [Google Scholar]
- Debnath A, Patil S, Bansal V. 2022. Structural and functional aspects of milk oligosaccharides. In: Goyal MR, Veena N, Mishra SK, Editors., Functional Dairy Ingredients and Nutraceuticals. New York: Apple Academic Press. [Google Scholar]
- Debreczeni N, Bege M, Borbás A. 2021. Synthesis of potential glycosyl transferase inhibitors by thio‐click reactions. Eur. J. Org. Chem. 6743–6747. 10.1002/ejoc.202101220 [DOI] [Google Scholar]
- del Río PG, Pérez‐Pérez A, Garrote G, Gullón B. 2022. Manufacturing of hemicellulosic oligosaccharides from fast‐growing Paulownia wood via autohydrolysis: Microwave versus conventional heating. Ind. Crops Prod. 187:115313. 10.1016/j.indcrop.2022.115313 [DOI] [Google Scholar]
- DelaCourt A, Black A, Angel P, Drake R, Hoshida Y, Singal A, Lewin D, Taouli B, Lewis S, Schwarz M, Fiel MI, Mehta AS. 2021. N‐Glycosylation patterns correlate with hepatocellular carcinoma genetic subtypes. Mol. Cancer Res. 19:1868–1877. 10.1158/1541-7786.mcr-21-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelaCourt AT, Liang H, Drake RR, Angel PM, Mehta AS. 2022. Novel combined enzymatic approach to analyze nonsialylated N‑linked glycans through MALDI imaging mass spectrometry. J. Proteome Res. 21:1930–1938. 10.1021/acs.jproteome.2c00193 [DOI] [PubMed] [Google Scholar]
- Delafield DG, Li L. 2021. Recent advances in analytical approaches for glycan and glycopeptide quantitation. Mol. Cell. Proteomics, 20:100054. 10.1074/mcp.r120.002095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demus D, Naber A, Dotz V, Jansen BC, Bladergroen MR, Nouta J, Sijbrands EJG, Van Hoek M, Nicolardi S, Wuhrer M. 2021. Large‐scale analysis of apolipoprotein CIII glycosylation by ultrahigh resolution mass spectrometry. Front. Chem. 9:678883. 10.3389/fchem.2021.678883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Webb IK, Garimella SVB, Hamid AM, Zheng X, Norheim RV, Prost SA, Anderson GA, Sandoval JA, Baker ES, Ibrahim YM, Smith RD. 2017. Serpentine ultralong path with extended routing (SUPER) high resolution traveling wave ion mobility‐MS using structures for lossless ion manipulations. Anal. Chem. 89:4628–4634. 10.1021/acs.analchem.7b00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Chen C, Chen L, Han B, Li S, Zhao J. 2021. Fast saccharide mapping method for quality consistency evaluation of commercial xylooligosaccharides collected in China. J. Pharmaceut. Anal. 11:284–291. 10.1016/j.jpha.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, He M, Feng F, Feng X, Zhang Y, Zhang F. 2021. The distribution and changes of glycoalkaloids in potato tubers under different storage time based on MALDI‐TOF mass spectrometry imaging. Talanta, 221:121453. 10.1016/j.talanta.2020.121453 [DOI] [PubMed] [Google Scholar]
- Denti V, Capitoli G, Piga I, Clerici F, Pagani L, Criscuolo L, Bindi G, Principi L, Chinello C, Paglia G, Magni F, Smith A. 2022. Spatial multiomics of lipids, N‑glycans, and tryptic peptides on a single FFPE tissue section. J. Proteome Res., 21:2798–2809. 10.1021/acs.jproteome.2c00601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharshini KP, Fang H, Ramya Devi D, Yang J‐X, Luo R‐H, Zheng Y‐T, Brzeziński M, Vedha Hari BN. 2021. pH‐Sensitive chitosan nanoparticles loaded with dolutegravir as milk and food admixture for paediatric anti‐HIV therapy. Carbohydr. Polym. 256:117440. 10.1016/j.carbpol.2020.117440 [DOI] [PubMed] [Google Scholar]
- Di Guida R, Casillo A, Stellavato A, Di Meo C, Kawai S, Kawamoto J, Ogawa T, Kurihara T, Schiraldi C, Corsaro MM. 2021. Complete lipooligosaccharide structure from Pseudoalteromonas nigrifaciens Sq02‐Rifr and study of its immunomodulatory activity. Mar. Drugs, 19:646. 10.3390/md19110646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo F, Duda KA, Lanzetta R, Silipo A, De Castro C, Antonio M. 2022. A journey from structure to function of bacterial lipopolysaccharides. Chem. Rev. 122:15767–15821. 10.1021/acs.chemrev.0c01321 [DOI] [PubMed] [Google Scholar]
- Di Maio A, Cioce A, Achilli S, Thépaut M, Vivès C, Fieschi F, Rojo J, Reichardt N‐C. 2021. Controlled density glycodendron microarrays for studying carbohydrate‐lectin interactions. Org. Biomol. Chem. 19:7357–7362. 10.1039/d1ob00872b [DOI] [PubMed] [Google Scholar]
- Di Marzo M, Viana VE, Banfi C, Cassina V, Corti R, Herrera‐Ubaldo H, Babolin N, Guazzotti A, Kiegle E, Gregis V, de Folter S, Sampedro J, Mantegazza F, Colombo L, Ezquer I. 2022. Cell wall modifications by α‐XYLOSIDASE1 are required for control of seed and fruit size in Arabidopsis J. Exp. Bot. 73:1499–1515. 10.1093/jxb/erab514 [DOI] [PubMed] [Google Scholar]
- Diao X, Xie C, Xie G, Song Y, Liang Y, Li R, Dong C, Zhu L, Wang J, Cai Z. 2022. Mass spectrometry imaging revealed sulfatides depletion in brain tissues of rats exposed in real air with high fine particulate matter. Environ. Sci. Technol. Lett. 9:856–862. 10.1021/acs.estlett.2c00615 [DOI] [Google Scholar]
- Diaz JMM, Peel SR, Spencer DIR, Hendel JL. 2022. Extraction and purification of a high mannose type oligosaccharide from Phaseolus lunatus beans by oxidative release with sodium hypochlorite. Carbohydr. Res. 517:108583. 10.1016/j.carres.2022.108583 [DOI] [PubMed] [Google Scholar]
- Ding Y, Pei C, Shu W, Wan J. 2022. Inorganic matrices assisted laser desorption/ionization mass spectrometry for metabolic analysis in biofluids. Chem. Asian J. 17:e202101310. 10.1002/asia.202101310 [DOI] [PubMed] [Google Scholar]
- Dittner‐Moormann S, Lourenco CM, Reunert J, Nishinakamura R, Tanaka SS, Werner C, Debus V, Zimmer K‐P, Wetzel G, Naim HY, Wada Y, Rust S, Marquardt T. 2021. TRAPγ‐CDG shows asymmetric glycosylation and an effect on processing of proteins required in higher organisms. J. Med. Genet. 58:213–216. 10.1136/jmedgenet-2019-106279 [DOI] [PubMed] [Google Scholar]
- Dixon CF, Nottingham AN, Lozano AF, Sizemore JA, Russell LA, Valiton C, Newell KL, Babin D, Bridges WT, Parris MR, Shchirov DV, Snyder NL, Ruppel JV. 2021. Synthesis and evaluation of porphyrin glycoconjugates varying in linker length: Preliminary effects on the photodynamic inactivation of Mycobacterium smegmatis . RSC Adv., 11:7037–7042. 10.1039/D0RA10793J [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Lago CL, Daniel D, Lopes FS, Cieslarova Z. 2021. Capillary electrophoresis‐mass spectrometry of carbohydrates and glycoconjugates. In: El Rassi Z, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 443–484. 10.1016/B978-0-12-821447-3.00016-0 [DOI] [Google Scholar]
- Domon B, Costello CE. 1988. A systematic nomenclature for carbohydrate fragmentations in FAB‐MS/MS spectra of glycoconjugates. Glycoconj. J. 5:397–409. 10.1007/BF01049915 [DOI] [Google Scholar]
- Donahue TC, Zong G, O'Brien NA, Ou C, Gildersleeve JC, Wang L‐X. 2022. Synthesis and immunological study of N‑glycan‐bacteriophage Qβ conjugates reveal dominant antibody responses to the conserved chitobiose core. Bioconj. Chem., 33:1350–1362. 10.1021/acs.bioconjchem.2c00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Xiao Y, Zhang M, Gong P, Ye Y, Peng S, Wang K, Fan M. 2022. Solid‐phase synthesis of polysaccharides from unprotected glucose catalyzed by Hβ zeolites. ACS Sustain. Chem. Eng. 10:9707–9718. 10.1021/acssuschemeng.1c08583 [DOI] [Google Scholar]
- Dong M, Lih T‐SM, Ao M, Hu Y, Chen S‐Y, Eguez RV, Zhang H. 2021. Data‐independent acquisition‐based mass spectrometry (DIA‐MS) for quantitative analysis of intact N‑linked glycopeptides. Anal. Chem. 93:13774–13782. 10.1021/acs.analchem.1c01659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Cheng S, Wang Y, Gao H, Zhang Y, Zhu T, Yu P, Meng X. 2022. A self‐adjuvanting anti‐tumor nanoliposomal vaccine based on fluorine‐substituted MUC1 glycopeptide. Chem. Commun. 58:8642–8645. 10.1039/D2CC02143A [DOI] [PubMed] [Google Scholar]
- Dong W, Liu H, Chen Z, Chen L, Jia L, Shen J, Zhu B, Li P, Fan D, Sun S. 2022. De‐sialylation of glycopeptides by acid treatment: Enhancing sialic acid removal without reducing the identification. Anal. Meth. 14:2913–2919. 10.1039/D2AY00949H [DOI] [PubMed] [Google Scholar]
- Dong Y, Aharoni A. 2022. Image to insight: Exploring natural products through mass spectrometry imaging. Nat. Prod. Rep. 39:1510–1530. 10.1039/d2np00011c [DOI] [PubMed] [Google Scholar]
- Donohoo KB, Wang J, Goli M, Yu A, Peng W, Hakim MA, Mechref Y. 2022. Advances in mass spectrometry‐based glycomics‐An update covering the period 2017‐2021. Electrophoresis, 43:119–142. 10.1002/elps.202100199 [DOI] [PubMed] [Google Scholar]
- Dörmann P. 2021. Plant lipid databases. Method. Mol. Biol. 2295:441–454. 10.1007/978-1-0716-1362-7_25 [DOI] [PubMed] [Google Scholar]
- Dou P, Xiang Y, Liang L, Liu Z. 2021. Preparation of multi‐functional magnetic nanoparticles for harvestying low‐molecular‐weight glycoproteins. Chin. J. Chromatogr. 39:1102–1110. 10.3724/sp.j.1123.2021.07019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake RR, Scott DA, Angel PM. 2021. Imaging mass spectrometry. In: Ross BD, Gambhir SS, Editors., Molecular Imaging; Principles and Practice, Volumn 1, Second edition. London, UK, San Diego, CA; Cambridge, MA, Oxford, UK: Elsevier (Academic Press). p 303–323. 10.1016/B978-0-12-816386-3.00017-X [DOI] [Google Scholar]
- Dreisbach D, Petschenka G, Spengler B, Bhandari DR. 2021. 3D‐surface MALDI mass spectrometry imaging for visualising plant defensive cardiac glycosides in Asclepias curassavica . Anal. Bioanal. Chem. 413:2125–2134. 10.1007/s00216-021-03177-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisbach D, Heiles S, Bhandari DR, Petschenka G, Spengler B. 2022. Molecular networking and on‐tissue chemical derivatization for enhanced identification and visualization of steroid glycosides by MALDI mass spectrometry imaging. Anal. Chem. 94:15971–15979. 10.1021/acs.analchem.2c02694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisewerd K, Bien T, Soltwisch J. 2022. MALDI‐2 and t‐MALDI‐2 mass spectrometry imaging. Method . Mol. Biol. 2437:21–40. 10.1007/978-1-0716-2030-4_2 [DOI] [PubMed] [Google Scholar]
- Du H, Yu H, Yang F, Li Z. 2021. Comprehensive analysis of glycosphingolipid glycans by lectin microarrays and MALDI‐TOF mass spectrometry. Nat. Protoc. 16:3470–3491. 10.1038/s41596-021-00544-y [DOI] [PubMed] [Google Scholar]
- Du M, Chen D, Chen Y, Huang Y, Ma L, Xie Q, Xu Y, Zhu X, Chen Z, Yin Z, Xu H, Wu X. 2022. Plasmonic gold nanoshell‐assisted laser desorption/ionization mass spectrometry for small‐biomolecule analysis and tissue imaging. ACS Appl. Nano Mater. 5:9633–9645. 10.1021/acsanm.2c01850 [DOI] [Google Scholar]
- Du M, Zhang K, Jiao L, Xu Y, Kong X. 2022. Differentiation of disaccharide isomers via a combination of IR and UV photodissociation mass spectrometry. Rapid Commun. Mass Spectrom., 36:e9218. 10.1002/rcm.9218 [DOI] [PubMed] [Google Scholar]
- Du Y, Du Y, Cui M, Liu Z. 2021. Characterization of the noncovalent interactions between lysozyme and panaxadiol glycosides by intensity‐fading‐matrix‐assisted laser desorption ionization‐mass spectrometry (IF‐MALDI‐MS). Anal. Letts., 54:2387–2394. 10.1080/00032719.2020.1867995 [DOI] [Google Scholar]
- Duan S, Li X, Yao Z, Zhang X, Yao X, Yang J, Qin Z. 2022. Visual authentication of steroidal saponins in Allium macrostemon Bge. and Allium chinense G. Don using MALDI‐TOF imaging mass spectrometry and their structure activity relationship. Arabian J. Chem., 15:104138. 10.1016/j.arabjc.2022.104138 [DOI] [Google Scholar]
- Duan Z, Luo Q, Dai X, Li X, Gu L, Zhu H, Tian X, Zhang H, Gong Q, Gu Z, Luo K. 2021. Synergistic therapy of a naturally inspired glycopolymer‐based biomimetic nanomedicine harnessing tumor genomic instability. Adv. Mater. 33:2104594. 10.1002/adma.202104594 [DOI] [PubMed] [Google Scholar]
- Duca M, Haksar D, van Neer J, Thies‐Weesie DME, Martínez‐Alarcón D, de Cock H, Varrot A, Pieters RJ. 2022. Multivalent fucosides targeting β‑propeller lectins from lung pathogens with promising anti‐adhesive properties. ACS Chem. Biol. 17:3515–3526. 10.1021/acschembio.2c00708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne M, Fincher JA, Patterson NH, Schey KL, Norris JL, Caprioli RM, Spraggins JM. 2021. α‑Cyano‐4‐hydroxycinnamic acid and tri‐potassium citrate salt pre‐coated silicon nanopost array provides enhanced lipid detection for high spatial resolution MALDI imaging mass spectrometry. Anal. Chem. 93:12243–12249. 10.1021/acs.analchem.1c01560 [DOI] [PubMed] [Google Scholar]
- Duncombe L, Howells L, Haughey A, Taylor AV, Kaveh D, Gôrbilek SE, Dell A, Hitchen PG, Haslam SM, Mandal SS, Ganesh NV, Bundle DR, McGiven J. 2022. The tip of Brucella O‐polysaccharide is a potent epitope in response to brucellosis infection and enables short synthetic antigens to be superior diagnostic reagents. Microorganisms, 10:708. 10.3390/microorganisms10040708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworaczek K, Kurzylewska M, Laban M, Drzewiecka D, Pękala‐Safińska A, Turska‐Szewczuk A. 2021. Structural studies of the lipopolysaccharide of Aeromonas veronii bv. sobria strain K133 which represents new provisional serogroup PGO1 prevailing among mesophilic aeromonads on Polish fish farms. Int. J. Mol. Sci., 22:4272. 10.3390/ijms22084272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyukova I, Faleh AB, Warnke S, Yalovenko N, Yatsyna V, Bansal P, Rizzo TR. 2021. A new approach for identifying positional isomers of glycans cleaved from monoclonal antibodies. Analyst, 146:4789–4795. 10.1039/D1AN00780G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebede GR, Mbing JN, Nama AB, Shehla N, Rahman A, Pegnyemb DE, Ndongo JT, Choudhary MI. 2022. New glycocerebrosides from the trunk of Tabernaemontana contorta Stapf. (Apocynaceae) and their antibacterial activity. Biochem. System. Ecol. 101:104396. 10.1016/j.bse.2022.104396 [DOI] [Google Scholar]
- Ehrlich JJ, Weerts RM, Shome S, Culbertson AT, Honzatko RB, Jernigan RL, Zabotina OA. 2021. Xyloglucan xylosyltransferase 1 displays promiscuity toward donor substrates during in vitro reactions. Plant Cell Physiol., 62:1890–1901. 10.1093/pcp/pcab114 [DOI] [PubMed] [Google Scholar]
- El Dine TM, Jimmidi R, Diaconu A, Fransolet M, Michiels C, De Winter J, Gillon E, Imberty A, Coenye T, Vincent SP. 2021. Pillar[5]arene‐based polycationic glyco[2]rotaxanes designed as Pseudomonas aeruginosa antibiofilm agents. J. Med. Chem. 64:14728–14744. 10.1021/acs.jmedchem.1c01241 [DOI] [PubMed] [Google Scholar]
- El Haitami A, Resmerita A‐M, Fichet O, Cantin S, Aubert P‐H, Farcas A. 2021. Synthesis, photophysics, and Langmuir films of polyfluorene/permodified cyclodextrin polyrotaxanes. Langmuir, 37:11406–11413. 10.1021/acs.langmuir.1c02014 [DOI] [PubMed] [Google Scholar]
- El Rassi Z. 2021a. Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. 10.1016/B978-0-12-821447-3.09995-9 [DOI] [Google Scholar]
- El Rassi Z. 2021b. Reversed‐phase and hydrophobic interaction chromatography of carbohydrates and glycoconjugates. In: El Rassi Z, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 35–124. 10.1016/B978-0-12-821447-3.00017-2 [DOI] [Google Scholar]
- Enatsu H, Okamoto N, Nomura Y, Onitsuka M, Yamano‐Adachi N, Koga Y, Omasa T. 2021. Production of monoclonal shark‐derived immunoglobulin new antigen receptor antibodies using Chinese hamster ovary cell expression system. J. Biosci. Bioeng. 132:302–309. 10.1016/j.jbiosc.2021.04.015 [DOI] [PubMed] [Google Scholar]
- Engel KM, Schiller J. 2021. The value of coupling thin‐layer chromatography to mass spectrometry in lipid research ‐ a review. J. Chromatogr., B, 1185:123001. 10.1016/j.jchromb.2021.123001 [DOI] [PubMed] [Google Scholar]
- Engel KM, Prabutzki P, Leopold J, Nimptsch A, Lemmnitzer K, Vos DRN, Hopf C, Schiller J. 2022. A new update of MALDI‐TOF mass spectrometry in lipid research. Prog. Lipid Res. 86:101145. 10.1016/j.plipres.2021.101145 [DOI] [PubMed] [Google Scholar]
- Engle KA, Amos RA, Yang J‐Y, Glushka J, Atmodjo MA, Tan L, Huang C, Moremen KW, Mohnen D. 2022. Multiple Arabidopsis galacturonosyltransferases synthesize polymeric homogalacturonan by oligosaccharide acceptor‐dependent or de novo synthesis. Plant J. 109:1441–1456. 10.1111/tpj.15640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto H. 2021. Unique distribution of ellagitannins in ripe strawberry fruit revealed by mass spectrometry imaging. Curr. Res. Food Sci. 4:821–828. 10.1016/j.crfs.2021.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdoğan M, Konya R, Özhan Y, Sipahi H, Çinbilgel I, Masullo M, Piacente S, Kırmızıbekmez H. 2021. Secondary metabolites from Scutellaria brevibracteata subsp. subvelutina and their in vitro anti‐inflammatory activities. S. Afr. J. Bot., 139:12–18. 10.1016/j.sajb.2021.01.028 [DOI] [Google Scholar]
- Erlmeier F, Sun N, Shen J, Feuchtinger A, Buck A, Prade VM, Kunzke T, Schraml P, Moch H, Autenrieth M, Weichert W, Hartmann A, Walch A. 2022. MALDI Mass spectrometry imaging‐prognostic pathways and metabolites for renal cell carcinomas. Cancers 14:1763. 10.3390/cancers14071763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmail S, Manolson MF. 2021. Advances in understanding N‐glycosylation structure, function, and regulation in health and disease. Eur. J. Cell Biol., 100:151186. 10.1016/j.ejcb.2021.151186 [DOI] [PubMed] [Google Scholar]
- Espaillat A, Carrasco‐López C, Bernardo‐García N, Rojas‐Altuve A, Klett J, Morreale A, Hermoso JA, Cava F. 2021. Binding of non‐canonical peptidoglycan controls Vibrio cholerae broad spectrum racemase activity. Comput. Struct. Biotechnol. J. 19:1119–1126. 10.1016/j.csbj.2021.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeabikwa B, Mondal N, Antonopoulos A, Haslam SM, Matsumoto Y, Martin‐Caraballo M, Lehoux S, Mandalasi M, Ishaque A, Heimburg‐Molinaro J, Cummings RD, Nyame AK. 2021. Major differences in glycosylation and fucosyltransferase expression in low‐grade versus high‐grade bladder cancer cell lines. Glycobiology, 31:1444–1463. 10.1093/glycob/cwab083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Yang Y, Li L, Fan L, Wang Z, Yang L. 2022. Mass spectrometry‐based profiling and imaging strategy, a fit‐for‐purpose tool for unveiling the transformations of ginsenosides in Panax notoginseng during processing. Phytomedicine, 103:154223. 10.1016/j.phymed.2022.154223 [DOI] [PubMed] [Google Scholar]
- Fan Y, Yang Y, Huang Y, Cai K, Qiao Y. 2022. Polyamidoamine dendrimer‐assisted 3‐carboxybenzoboroxole‐functionalized magnetic nanoparticles for highly efficient capture of trace cis‐diol‐containing biomacromolecules New J. Chem. 46:9889–9896. 10.1039/D2NJ01242A [DOI] [Google Scholar]
- Fang C, Lu H. 2022. Clinical applications. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. London: Taylor and Francis. p 167–235. 10.1201/9781003185833-6 [DOI] [Google Scholar]
- Fanuel M, Grélard F, Foucat L, Alvarado C, Arnaud B, Chateigner‐Boutin A‐L, Saulnier L, Legland D, Rogniaux H. 2022. Spatial correlation of water distribution and fine structure of arabinoxylans in the developing wheat grain. Carbohydr. Polym. 294:119738. 10.1016/j.carbpol.2022.119738 [DOI] [PubMed] [Google Scholar]
- Farinha D, Migawa M, Sarmento‐Ribeiro A, Faneca H. 2021. A combined antitumor strategy mediated by a new targeted nanosystem to hepatocellular carcinoma. Int. J. Nanomed. 16:3385–3405. 10.2147/IJN.S302288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsang R, Kovács N, Szigeti M, Jankovics H, Vonderviszt F, Guttman A. 2022. Immobilized exoglycosidase matrix mediated solid phase glycan sequencing. Anal. Chim. Acta, 1215:339906. 10.1016/j.aca.2022.339906 [DOI] [PubMed] [Google Scholar]
- Farzadfard A, König A, Petersen SV, Nielsen J, Vasili E, Dominguez‐Meijide A, Buell AK, Outeiro TF, Otzen DE. 2022. Glycation modulates alpha‐synuclein fibrillization kinetics: A sweet spot for inhibition. J. Biol. Chem. 298:101848. 10.1016/j.jbc.2022.101848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijao C, Morreel K, Anders N, Tryfona T, Busse‐Wicher M, Kotake T, Boerjan W, Dupree P. 2022. Hydroxycinnamic acid‐modified xylan side chains and their cross‐linking products in rice cell walls are reduced in the Xylosyl arabinosyl substitution of xylan 1 mutant. Plant J. 109:1152–1167. 10.1111/tpj.15620 [DOI] [PubMed] [Google Scholar]
- Feiner IVJ, Longo B, Gómez‐Vallejo V, Calvo J, Chomet M, Vugts DJ, Windhorst AD, Padro D, Zanda M, Rejc L, Llop J. 2021. Comparison of analytical methods for antibody conjugates with application in nuclear imaging—Report from the trenches. Nucl. Med. Biol. 102–103:24–33. 10.1016/j.nucmedbio.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Feller G, Bonneau M, Da Lage J‐L. 2021. Amyrel, a novel glucose‐forming α‐amylase from Drosophila with 4‐α‐glucanotransferase activity by disproportionation and hydrolysis of maltooligosaccharides. Glycobiology, 31:1134–1144. 10.1093/glycob/cwab036 [DOI] [PubMed] [Google Scholar]
- Feng Q, Xie Z, Liang H, Zhang Z, Yan Y, Ding C‐F. 2022. Hydrophilic, dual amino acid‐functionalized zinc sulfide quantum dot for specific identification of N‐glycopeptides from biological samples. Rapid Commun. Mass Spectrom., 36:e9405. 10.1002/rcm.9405 [DOI] [PubMed] [Google Scholar]
- Feng X, Shu H, Zhang S, Peng Y, Zhang L, Cao X, Wei L, Lu H. 2021. Relative quantification of N‑glycopeptide sialic acid linkage isomers by ion mobility mass spectrometry. Anal. Chem. 93:15617–15625. 10.1021/acs.analchem.1c02803 [DOI] [PubMed] [Google Scholar]
- Fernandes RF, Alves GAS, Gonçalves RV, Temperini MLA. 2021. A methodology to identify the releasing of the amide‑containing β‑glucan from the Usnea lichen: A spectroscopic study. J. Polym. Environ. 29:3105–3115. 10.1007/s10924-021-02104-7 [DOI] [Google Scholar]
- Ferreira‐Lazarte A, Plaza‐Vinuesa L, de las Rivas B, Villamiel M, Muñoz R, Moreno FJ. 2021. Production of α‐rhamnosidases from Lactobacillus plantarum WCFS1 and their role in deglycosylation of dietary flavonoids naringin and rutin. Int. J. Biol. Macromol., 193:1093–1102. 10.1016/j.ijbiomac.2021.11.053 [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Relvas‐Santos M, Peixoto A, Silva AMN, Santos LL. 2021. Glycoproteogenomics: Setting the course for next‐generation cancer neoantigen discovery for cancer vaccines. Genomics Proteomics Bioinform. 19:25–43. 10.1016/j.gpb.2021.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetsiukh A, Conrad J, Bergquist J, Timmusk S. 2021. Silica particles trigger the exopolysaccharide production of harsh environment isolates of growth‐promoting rhizobacteria and increase their ability to enhance wheat biomass in drought‐stressed soils. Int. J. Mol. Sci. 22:6201. 10.3390/ijms22126201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher JA, Djambazova KV, Klein DR, Dufresne M, Migas LG, Van de Plas R, Caprioli RM, Spraggins JM. 2021. Molecular mapping of neutral lipids using silicon nanopost arrays and TIMS imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 32:2519–2527. 10.1021/jasms.1c00159 [DOI] [PubMed] [Google Scholar]
- Flores‐Méndez LC, Lizárraga‐Velázquez CE, Sánchez‐Gutiérrez EY, Arrizon J, Leyva‐López N, Hernández C. 2022. Study of the effect of dietary agavin supplementation in blood parameters and antioxidant enzymes of juvenile Nile tilapia (Oreochromis niloticus) under stress conditions. Fishes, 7:340. 10.3390/fishes7060340 [DOI] [Google Scholar]
- Font G, Walet‐Balieu M‐L, Petit M, Burel C, Maho‐Vaillant M, Hébert V, Chan P, Fréret M, Boyer O, Joly P, Calbo S, Bardor M, Golinski M‐L. 2022. IgG N‐glycosylation from patients with pemphigus treated with rituximab. Biomedicines, 10:1774. 10.3390/biomedicines10081774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman TT, Dueñas ME, Lee YJ. 2021. On‐tissue boronic acid derivatization for the analysis of vicinal diol metabolites in maize with MALDI‐MS imaging. J. Mass Spectrom. 56:e4709. 10.1002/jms.4709 [DOI] [PubMed] [Google Scholar]
- Fournière M, Bedoux G, Lebonvallet N, Leschiera R, Le Goff‐Pain C, Bourgougnon N, Latire T. 2021. Poly‐ and oligosaccharide Ulva sp. Fractions from enzyme‐assisted extraction modulate the metabolism of extracellular matrix in human skin fibroblasts: Potential in anti‐aging dermo‐cosmetic applications. Mar. Drugs, 19:156. 10.3390/md19030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzka P, Krüger L, Schurig MK, Olecka M, Hoffmann S, Blanchard V, Hübner CA. 2021. Altered glycosylation in the aging heart. Front. Mol. Biosci. 8:673044. 10.3389/fmolb.2021.673044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey LJ. 2022. Informatics ecosystems to advance the biology of glycans. Method. Mol. Biol. 2303:655–673. 10.1007/978-1-0716-1398-6_50 [DOI] [PubMed] [Google Scholar]
- Fröhlich AC, Bazzo GC, Stulzer HK, Parize AL. 2022. Synthesis and physico‐chemical characterization of quaternized and sulfated xylan‐derivates with enhanced microbiological and antioxidant properties. Biocatal. Agric. Biotechnol. 43:102416. 10.1016/j.bcab.2022.102416 [DOI] [Google Scholar]
- Fu M, Wang B, Yi L, Jin X, Yan Y, Ding C‐F. 2022. Bi‐amino acid functionalized biomimetic honeycomb chitosan membrane as a multifunctional hydrophilic probe for specific capture of N‐linked glycopeptides in nasopharyngeal carcinoma's disease patient's serum. J. Sep. Sci., 45:1580–1589. 10.1002/jssc.202100993 [DOI] [PubMed] [Google Scholar]
- Fuentes R, Aguinagalde L, Sacristána N, Fernández‐Tejada A. 2021. Design, synthesis, and initial immunological evaluation of glycoconjugates based on saponin adjuvants and the Tn antigen. Chem. Commun. 57:11382–11385. 10.1039/D1CC04459A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes R, Ruiz‐de‐Angulo A, Sacristán N, Navo CD, Jiménez‐Osés G, Anguita J, Fernández‐Tejada A. 2021. Replacing the rhamnose‐xylose moiety of QS‐21 with simpler terminal disaccharide units attenuates adjuvant activity in truncated saponin variants. Chem. Eur. J. 27:4731–4737. 10.1002/chem.202004705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa K, Nishiyama K‐i, Shimamoto K. 2019. Enzyme‐like glycolipids MPIase involved in membrane protein integration of E. coli . Trends Glycosci. Glycotechnol. 31:E151–E158. 10.4052/tigg.1705.1E [DOI] [Google Scholar]
- Fujinami D, de Gonzalo CVG, Biswas S, Hao Y, Wang H, Garg N, Lukk T, Nair SK, van der Donk WA. 2021. Structural and mechanistic investigations of protein S‐glycosyltransferases. Cell Chem. Biol. 28:1740–1749. 10.1016/j.chembiol.2021.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani N, Ariki S, Hasegawa Y, Uehara Y, Saito A, Takahashi M. 2021. Integrated structural analysis of N‑glycans and free oligosaccharides allows for a quantitative evaluation of ER stress. Biochemistry, 60:1708–1721. 10.1021/acs.biochem.0c00969 [DOI] [PubMed] [Google Scholar]
- Fukuhara R, Ogura A, Yoshinaga S, Fukunaga T, Kinoshita T, Sumiyoshi W, Higuchi Y, Tanaka K, Takegawa K. 2022. In vivo imaging of fluorescent albumin modified with pyruvylated‐human‐type complex oligosaccharide reveals sialylation‐like biodistribution and kinetics. Bioorg. Med. Chem., 70:116943. 10.1016/j.bmc.2022.116943 [DOI] [PubMed] [Google Scholar]
- Fukuoka T, Nakamura S, Morita T, Ohmura T, Kotani M, Naito Y, Sato H. 2021. Surface‐assisted laser desorption/ionization mass spectrometry analysis of the glycolipid biosurfactants, mannosylerythritol lipids, using an ionization‐assisting substrate. J. Oleo Sci. 70:1175–1179. 10.5650/jos.ess21084 [DOI] [PubMed] [Google Scholar]
- Fukushima K, Kikuma T, Takeda Y. 2021. Chiral acidic amino acids as tethers for intramolecular glycosylation. J. Carbohydr. Chem. 40:283–307. 10.1080/07328303.2021.2015364 [DOI] [Google Scholar]
- Fülöp A, Marsching C, Barka F, Ucal Y, Pfänder P, Opitz CA, Barka G, Hopf C. 2022. Device‐controlled microcondensation for spatially confined on‐tissue digests in MALDI imaging of N‐glycans. Pharmaceuticals, 15:1356. 10.3390/ph15111356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa J‐i, Hanamatsu H, Yokota I, Hirayama M, Ando T, Kobayashi H, Ohnishi S, Miura N, Okada K, Sakai S, Yuyama K, Igarashi Y, Ito M, Shinohara Y, Sakamoto N. 2021. Comprehensive glycomic approach reveals novel low‐molecular‐weight blood group‐specific glycans in serum and cerebrospinal fluid. J. Proteome Res. 20:2812–2822. 10.1021/acs.jproteome.1c00056 [DOI] [PubMed] [Google Scholar]
- Gabriele C, Prestagiacomo LE, Cuda G, Gaspari M. 2021. Mass spectrometry‐based glycoproteomics and prostate cancer. Int. J. Mol. Sci. 22:5222. 10.3390/ijms22105222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteiro C, Soares J, Relvas‐Santos M, Peixoto A, Ferreira D, Paulo P, Brandão A, Fernandes E, Azevedo R, Palmeira C, Freitas R, Miranda A, Osório H, Prieto J, Lima L, Silva AMN, Santos LL, Ferreira JA. 2022. Glycoproteogenomics characterizes the CD44 splicing code associated with bladder cancer invasion. Theranostics, 12:3150–3177. 10.7150/thno.67409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Stavenhagen K, Eckmair B, McKitrick TR, Mehta AY, Matsumoto Y, McQuillan AM, Hanes MS, Eris D, Baker KJ, Jia N, Wei M, Heimburg‐Molinaro J, Ernst B, Cummings RD. 2021. Differential recognition of oligomannose isomers by glycan‐binding proteins involved in innate and adaptive immunity. Sci. Adv. 7:eabf6834. 10.1126/sciadv.abf6834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M‐J, Yan J‐J, Zhao Y, Zhu L, Yang G‐S, Zhan X‐B. 2021. Expression of a thermostable β−1,3‐glucanase from Trichoderma harzianum in Pichia pastoris and use in oligoglucosides hydrolysis. Proc. Biochem. 107:74–82. 10.1016/j.procbio.2021.05.010 [DOI] [Google Scholar]
- Gao M, Duan F, Liu L, Hu X, Zhu L, Jiang Y, Zhan X. 2021. An innovative strategy of recycling miscellaneous waste carbohydrates from high‐fructose syrup production for Pichia pastoris fermentation. J. Clean. Prod. 326:129404. 10.1016/j.jclepro.2021.129404 [DOI] [Google Scholar]
- Gao M, Xu Y, Yang G, Jin S, Hu X, Jiang Y, Zhu L, Li Z, Zhan X. 2021. One‐step production of functional branched oligoglucosides with coupled fermentation of Pichia pastoris GS115 and Sclerotium rolfsii WSH‐G01. Bioresource Technol., 335:125286. 10.1016/j.biortech.2021.125286 [DOI] [PubMed] [Google Scholar]
- Gao M, Li H, Yang T, Li Z, Hu X, Wang Z, Jiang Y, Zhu L, Zhan X. 2022. Production of prebiotic gellan oligosaccharides based on the irradiation treatment and acid hydrolysis of gellan gum. Carbohydr. Polym. 279:119007. 10.1016/j.carbpol.2021.119007 [DOI] [PubMed] [Google Scholar]
- Gao W, Bai Y, Liu H. 2021. Glutathione‐functionalized two‐dimensional cobalt sulfide nanosheets for rapid and highly efficient enrichment of N‐glycopeptides. Microchim. Acta, 188:274. 10.1007/s00604-021-04909-8 [DOI] [PubMed] [Google Scholar]
- Gao W, Zhang H, Li T, Ju J, Zhou H, Zong X, Yin H. 2022. Controlled depolymerization of cellulose by photoelectrochemical bioreactor using a lytic polysaccharide monooxygenase. Biochem. Eng. J. 187:108597. 10.1016/j.bej.2022.108597 [DOI] [Google Scholar]
- Gao Z, Li L, Chen W, Ma Z, Li Y, Gao Y, Ding C‐F, Zhao X, Pan Y. 2021. Distinguishment of glycan isomers by trapped ion mobility spectrometry. Anal. Chem. 93:9209–9217. 10.1021/acs.analchem.1c01461 [DOI] [PubMed] [Google Scholar]
- Gao Z, Tang R, Ma S, Jia S, Zhang S, Gong B, Ou J. 2021. Design and construction of a hydrophilic coating on macroporous adsorbent resins for enrichment of glycopeptides. Anal. Methods‐UK 13:4515–4527. 10.1039/D1AY01276B [DOI] [PubMed] [Google Scholar]
- García‐García A, Serna S, Yang Z, Delso I, Taleb V, Hicks T, Artschwager R, Vakhrushev SY, Clausen H, Angulo J., Corzana F, Reichardt NC, Hurtado‐Guerrero R. 2021. FUT8‐Directed core fucosylation of N‑glycans is regulated by the glycan structure and protein environment. ACS Catal., 11:9052–9065. 10.1021/acscatal.1c01698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Vello P, Di Lorenzo F, Lamprinaki D, Notaro A, Speciale I, Molinaro A, Juge N, De Castro C. 2021. Structure of the O‐antigen and the lipid A from the lipopolysaccharide of Fusobacterium nucleatum ATCC 51191. ChemBioChem, 22:1252–1260. 10.1002/cbic.202000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Vello P, Di Lorenzo F, Zucchetta D, Zamyatina A, De Castro C, Molinaro A. 2022. Lipopolysaccharide lipid A: A promising molecule for new immunity‐based therapies and antibiotics. Pharmacol. Therap. 230:107970. 10.1016/j.pharmthera.2021.107970 [DOI] [PubMed] [Google Scholar]
- Garcia‐Vello P, Speciale I, Di Lorenzo F, Molinaro A, De Castro C. 2022. Dissecting lipopolysaccharide composition and structure by GC‐MS and MALDI spectrometry. Method. Mol. Biol. 2548:181–209. 10.1007/978-1-0716-2581-1_12 [DOI] [PubMed] [Google Scholar]
- Garrido MM, Piccinni FE, Landoni M, Peña MJ, Topalian J, Couto A, Wirth SA, Urbanowicz BR, Campos E. 2022. Insights into the xylan degradation system of Cellulomonas sp. B6: Biochemical characterization of rCsXyn10A and rCsAbf62A. Appl. Microbiol. Biotechnol. 106:5035–5049. 10.1007/s00253-022-12061-3 [DOI] [PubMed] [Google Scholar]
- Gaunitz S, Tjernberg LO, Schedin‐Weiss S. 2021. What can N‐glycomics and N‐glycoproteomics of cerebrospinal fluid tell us about Alzheimer disease? Biomolecules, 11:858. 10.3390/biom11060858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam S, Banazadeh A, Cho BG, Goli M, Zhong J, Mechref Y. 2021. Mesoporous graphitized carbon column for efficient isomeric separation of permethylated glycans. Anal. Chem. 93:5061–5070. 10.1021/acs.analchem.0c04395 [DOI] [PubMed] [Google Scholar]
- Gavande PV, Nath P, Kumar K, Ahmed N, Fontes CMGA, Goyal A. 2022. Highly efficient, processive and multifunctional recombinant endoglucanase RfGH5_4 from Ruminococcus flavefaciens FD‐1 v3 for recycling lignocellulosic plant biomasses. Int. J. Biol. Macromol. 209:801–813. 10.1016/j.ijbiomac.2022.04.059 [DOI] [PubMed] [Google Scholar]
- Gedda G, Tiruveedhi VLNBG, Ganesh G, Suribabu J. 2022. Recent advancements of carbon dots in analytical techniques. In: Kailasa SK, Hussain CM, Editors., Carbon Dots in Analytical Chemistry. Amsterdam, Oxford, Cambridge: Elsevier. p 137–147. 10.1016/B978-0-323-98350-1.00017-7 [DOI] [Google Scholar]
- Geisshüsler S, Schineis P, Langer L, Wäckerle‐Men Y, Leroux J‐C, Halin C, Vogel‐Kindgen S, Johansen P, Gander B. 2022. Amphiphilic cyclodextrin‐based nanoparticulate vaccines can trigger T‐cell immune responses. Adv. Nanobiomed. Res. 2:2100082. 10.1002/anbr.202100082 [DOI] [Google Scholar]
- Gerster T, Wröbel M, Hofstaedter CE, Schwudke D, Ernst RK, Ranf S, Gisch N. 2022. Remodeling of lipid A in Pseudomonas syringae pv. phaseolicola in vitro . Int. J. Mol. Sci., 23:1996. 10.3390/ijms23041996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwig GJ. 2021a. Glycobioinformatics. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 297–312. 10.1007/978-3-030-77791-3_13 [DOI] [Google Scholar]
- Gerwig GJ. 2021b. Analysis of carbohydrates by mass spectrometry. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 253–271. 10.1007/978-3-030-77791-3_11 [DOI] [Google Scholar]
- Gerwig GJ. 2021c. Analytical techniques to study carbohydrates. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 89–126. 10.1007/978-3-030-77791-3_5 [DOI] [Google Scholar]
- Gerwig GJ. 2021d. Monosaccharide composition analysis. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 127–156. 10.1007/978-3-030-77791-3_6 [DOI] [Google Scholar]
- Gerwig GJ. 2021e. Structural characterization of released glycans. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Narure. p 211–228. 10.1007/978-3-030-77791-3_8 [DOI] [Google Scholar]
- Gerwig GJ. 2021f. Carbohydrate analysis of glycoconjugates. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 157–209. 10.1007/978-3-030-77791-3_7 [DOI] [Google Scholar]
- Gerwig GJ. 2021g. The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. 10.1007/978-3-030-77791-3 [DOI] [Google Scholar]
- Gerwig GJ. 2021h. Analysis of sialic acids. In: Gerwig GJ, Editor, The Art of Carbohydrate Analysis. Cham, Switzerland: Springer Nature. p 229–243. 10.1007/978-3-030-77791-3_9 [DOI] [Google Scholar]
- Gharaghoushi SM, Nezhati MN, Baharvand H, Mohammadian T, Panahi HA. 2022. Encapsulated magnetic nanoparticles with a polymer containing boronic acid groups for separation and enrichment of horseradish peroxidase glycoprotein. Int. J. Polym. Mater. Polym. Biomater., 71:946–958. 10.1080/00914037.2021.1931208 [DOI] [Google Scholar]
- Ghirardello M, Zhang Y‐Y, Voglmeir J, Galan MC. 2022. Recent applications of ionic liquid‐based tags in glycoscience. Carbohydr. Res. 520:108643. 10.1016/j.carres.2022.108643 [DOI] [PubMed] [Google Scholar]
- Ghosh R, Kishore N. 2022. Mechanistic physicochemical insights into glycation and drug binding by serum albumin: Implications in diabetic conditions. Biochimie, 193:16–37. 10.1016/j.biochi.2021.10.008 [DOI] [PubMed] [Google Scholar]
- Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, Langridge D, Green M. 2019. A cyclic ion mobility‐mass spectrometry system. Anal. Chem. 91:8564–8573. 10.1021/acs.analchem.9b01838 [DOI] [PubMed] [Google Scholar]
- Giménez E, Mancera‐Arteu M, Benavente F, Sanz‐Nebot V. 2021. Analysis of intact glycoproteins by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry. Method. Mol. Biol. 2271:47–66. 10.1007/978-1-0716-1241-5_3 [DOI] [PubMed] [Google Scholar]
- Goli M, Yu A, Cho BG, Gautam S, Wang J, Gutierrez‐Reyes CD, Jiang P, Peng W, Mechref Y. 2021. LC‐MS/MS in glycomics and glycoproteomics analyses. In: El Rassi X, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 391–441. 10.1016/B978-0-12-821447-3.00005-6 [DOI] [Google Scholar]
- Gomathy V, Manigandan V, Vignesh N, Thabitha A, Saravanan R. 2021. Evaluation of antibacterial, teratogenicity and antibiofilm effect of sulfated chitosans extracted from marine waste against microorganism. J. Bioact. Compat. Pol. 36: 249–258. 10.1177/08839115211014225 [DOI] [Google Scholar]
- Gonçalves JPL, Bollwein C, Schwamborn K. 2022. Mass spectrometry imaging spatial tissue analysis toward personalized medicine. Life, 12:1037. 10.3390/life12071037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Qin S, Dai L, Tian Z. 2021. The glycosylation in SARS‐CoV‐2 and its receptor ACE2. Sig. Transduct. Target. Therap. 6:396. 10.1038/s41392-021-00809-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Alfonso JL, Poveda A, Arribas M, Hirose Y, Fernández‐Lobato M, Ballesteros AO, Jiménez‐Barbero J., Plou FJ. 2021. Polyglucosylation of rutin catalyzed by cyclodextrin glucanotransferase from Geobacillus sp.: Optimization and chemical characterization of products. Ind. Eng. Chem. Res. 60:18651–18659. 10.1021/acs.iecr.1c03070 [DOI] [Google Scholar]
- González‐Méndez I, Loera‐Loera E, Sorroza‐Martínez K, Vonlanthen M, Cuétara‐Guadarrama F, Bernad‐Bernad MJ, Rivera E, Gracia‐Mora J. 2022. Synthesis of β‐cyclodextrin‐decorated dendritic compounds based on EDTA core: A new class of PAMAM endrimer analogs. Pharmaceutics, 14:2363. 10.3390/pharmaceutics14112363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goti G, Colombo C, Achilli S, Vivès C, Thépaut M, Luczkowiak J, Labiod N, Delgado R, Fieschi F, Bernardi A. 2022. Precision glycodendrimers for DC‐SIGN targeting. Eur. J. Org. Chem. 35–43. 10.1002/ejoc.202200113 [DOI] [Google Scholar]
- Goyard D, Roubinet B, Vena F, Landemarre L, Renaudet O. 2022. Homo‐ and heterovalent neoglycoproteins as ligands for bacterial lectins. ChemPlusChem, 87:e202100481. 10.1002/cplu.202100481 [DOI] [PubMed] [Google Scholar]
- Gozdziewicz TK, Maciejewska A, Tsybulska A, Lugowski C, Lukasiewicz J. 2021. A new look at the enterobacterial common antigen forms obtained during rough lipopolysaccharides purification. Int. J. Mol. Sci. 22:701. 10.3390/ijms22020701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarics M, Lettow M, Kirschbaum C, Greis K, Manz C, Pagel K. 2022. Mass spectrometry‐based techniques to elucidate the sugar code. Chem. Rev. 122:7840–7908. 10.1021/acs.chemrev.1c00380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramlich M, Maier S, Kaiser PD, Traenkle B, Wagner TR, Voglmeir J, Stoll D, Rothbauer U, Zeck A. 2022. A novel PNGase Rc for improved protein N‑deglycosylation in bioanalytics and hydrogen−deuterium exchange coupled with mass spectrometry epitope mapping under challenging conditions. Anal. Chem. 94:9863–9871. 10.1021/acs.analchem.2c01748 [DOI] [PubMed] [Google Scholar]
- Granborg JR, Kaasgaard SG, Janfelt C. 2022. Mass spectrometry imaging of oligosaccharides following in situ enzymatic treatment of maize kernels. Carbohydr. Polym., 275:118693. 10.1016/j.carbpol.2021.118693 [DOI] [PubMed] [Google Scholar]
- Grey AC, Tang M, Zahraei A, Guo G, Demarais NJ. 2021. Applications of stable isotopes in MALDI imaging: Current approaches and an eye on the future. Anal. Bioanal. Chem. 413:2637–2653. 10.1007/s00216-021-03189-8 [DOI] [PubMed] [Google Scholar]
- Griffin ME, Hsieh‐Wilson LC. 2022. Tools for mammalian glycoscience research. Cell, 185:2657–2677. 10.1016/j.cell.2022.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JH. 2021. Poly(2‐vinylpyridine) as a reference compound for mass calibration in positive‐ion matrix‐assisted laser desorption/ionization‐mass spectrometry on different instrumental platforms. Eur. J. Mass Spectrom. 27:191–204. 10.1177/14690667211055701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groux‐Degroote S, Cavdarli S, Yamakawa N, Guérardel Y, Delannoy P. 2021. Recent progress in O‐acetylated gangliosides analysis and functions in cancer. Carbohydr. Chem. 45:537–552. 10.1039/1465-1963 [DOI] [Google Scholar]
- Grzeski M, Taube ET, Braicu EI, Sehouli J, Blanchard V, Klein O. 2022. In situ N‐glycosylation signatures of epithelial ovarian cancer tissue as defined by MALDI mass spectrometry imaging. Cancers, 14:1021. 10.3390/cancers14041021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gstöttner C, Kaur H, Wuhrer M. 2021. Glycosylation analysis. In: Kaur H, Reusch D, Editors., Monoclonal Antibodies; Physicochemical Analysis. London, UK; San Diego, CA; Cambridge, MA; Oxford, UK: Elsevier, Academic Press. p 65–92. 10.1016/B978-0-12-822318-5.00002-8 [DOI] [Google Scholar]
- Gstöttner C, Zhang T, Resemann A, Ruben S, Pengelley S, Suckau D, Welsink T, Wuhrer M, Domínguez‐Vega E. 2021. Structural and functional characterization of SARS‐CoV‑2 RBD domains produced in mammalian cells. Anal. Chem. 93:6839–6847. 10.1021/acs.analchem.1c00893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Liu Y, Zhen L, Zhao T, Luo L, Zhang J, Deng T, Wu M, Cheng G, Hu J. 2022. The structures of two glucomannans from Bletilla formosana and their protective effect on inflammation via inhibiting NF‐κB pathway. Carbohydr. Polymers. 292:119694. 10.1016/j.carbpol.2022.119694 [DOI] [PubMed] [Google Scholar]
- Guan B, Zhang Z, Chai Y, Amantai X, Chen X, Cao X, Yue X. 2022. N‐Glycosylation of milk proteins: A review spanning 2010‐2022. Trends Food Sci. Technol., 128:1–21. 10.1016/j.tifs.2022.07.017 [DOI] [Google Scholar]
- Guan S, Bythell BJ. 2021. Evidence of gas‐phase pyranose‐to‐furanose isomerization in protonated peptidoglycans. Phys. Chem. Chem. Phys. 23:23256–23266. 10.1039/D1CP03842G [DOI] [PubMed] [Google Scholar]
- Guan Y, Zhang M, Wang J, Schlüter H. 2021. Comparative analysis of different N‑glycan preparation approaches and development of optimized solid‐phase permethylation using mass spectrometry. J. Proteome Res., 20:2914–2922. 10.1021/acs.jproteome.1c00135 [DOI] [PubMed] [Google Scholar]
- Guerrero PA, Murakami Y, Malik A, Seeberger PH, Kinoshita T, Silva DV. 2021. Rescue of glycosylphosphatidylinositol‐anchored protein biosynthesis using synthetic glycosylphosphatidylinositol oligosaccharides. ACS Chem. Biol. 16:2297–2306. 10.1021/acschembio.1c00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillotte ML, Chandler CE, Verhoeve VI, Gillespie JJ, Driscoll TP, Rahman MS, Ernst RK, Azad AF. 2021. Lipid A structural divergence in Rickettsia pathogens. mSphere, 6:e00184–00121. 10.1128/mSphere.00184-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Nayak S, Mao Y, Li N. 2021. Glycine additive enhances sensitivity for N‐ and O‐glycan analysis with hydrophilic interaction chromatography‐electrospray ionization‐mass spectrometry. Anal. Biochem. 635:114447. 10.1016/j.ab.2021.114447 [DOI] [PubMed] [Google Scholar]
- Guo R‐R, Zhang T‐C, Lambert TOT, Wang T, Voglmeir J, Rand KD, Liu L. 2022. PNGase H + variant from Rudaea cellulosilytica with improved deglycosylation efficiency for rapid analysis of eukaryotic N‐glycans and hydrogen deuterium exchange mass spectrometry analysis of glycoproteins. Rapid Commun. Mass Spectrom., 36:e9376. 10.1002/rcm.9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Lakshminarayanan H, Rodriguez‐Palacios A, Salata RA, Xu K, Draz MS. 2021. Glycan nanostructures of human coronaviruses. Int. J. Nanomed. 16:4813–4830. 10.2147/IJN.S302516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Zhang J, Li X, Xiao E, Lange JD, Rienstra CM, Burke MD, Mitchell DA. 2021. Sterol sponge mechanism is conserved for glycosylated polyene macrolides. ACS Cent. Sci. 7:781–791. 10.1021/acscentsci.1c00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Jia W, Yang J, Zhan X. 2022. Cancer glycomics offers potential biomarkers and therapeutic targets in the framework of 3P medicine. Front. Endocrinol. 13:970489. 10.3389/fendo.2022.970489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Wei Y, Zhang Y, Xu Y, Zheng L, Zhu B, Yao Z. 2021. Carrageenan oligosaccharides: A comprehensive review of preparation, isolation, purification, structure, biological activities and applications. Algal Res., 61:102593. 10.1016/j.algal.2021.102593 [DOI] [Google Scholar]
- Guo ZY, Yao QH, Zheng WH, Jiao Rw, Zhang C, Zhang L, Ye TX, Chen X. 2022. Highly crosslinking core‐shell magnetic nanocomposites based catalyst and heat free polymerization for isolation of glycoprotein. Anal. Bioanal. Chem. 414:6393–6402. 10.1007/s00216-022-04202-4 [DOI] [PubMed] [Google Scholar]
- Gutierrez‐Reyes CD, Jiang P, Atashi M, Bennett A, Yu A, Peng W, Zhong J, Mechref Y. 2022. Advances in mass spectrometry‐based glycoproteomics: An update covering the period 2017‐2021. Electrophoresis, 43:370–387. 10.1002/elps.202100188 [DOI] [PubMed] [Google Scholar]
- Gutierrez Reyes CD, Jiang P, Donohoo K, Atashi M, Mechref YS. 2021. Glycomics and glycoproteomics: Approaches to address isomeric separation of glycans and glycopeptides. J. Sep. Sci. 44:403–425. 10.1002/jssc.202000878 [DOI] [PubMed] [Google Scholar]
- Gye HJ, Nishizawa T. 2022. Analysis of sialylated N‐linked glycans on fish cell lines permissive to nervous necrosis virus for predicting cellular receptors of the virus. Aquaculture, 555:738198. 10.1016/j.aquaculture.2022.738198 [DOI] [Google Scholar]
- Habazin S, Štambuk J, Šimunović J, Keser T, Razdorov G, Novokmet M. 2021. Mass spectrometry‐based methods for immunoglobulin G N‐glycosylation analysis. In: Pezer M, Editor, Antibody Glycosylation. Cham, Switzerland: Springer Nature. p 73–135. 10.1007/978-3-030-76912-3_3 [DOI] [PubMed] [Google Scholar]
- Hackett WE, Zaia J. 2021. Calculating glycoprotein similarities from mass spectrometric data. Mol. Cell. Proteomics, 20:100028. 10.1074/mcp.R120.002223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad G, Lorenzen JM, Ma H, de Haan N, Seeger H, Zaghrini C, Brandt S, Kölling M, Wegmann U, Kiss B, Pál G, Gál P, Wüthrich RP, Wuhrer M, Beck LH, Salant DJ, Lambeau G, Kistler AD. 2021. Altered glycosylation of IgG4 promotes lectin complement pathway activation in anti‐PLA2R1‐associated membranous nephropathy. J. Clin. Invest. 131:e140453. 10.1172/jci140453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga Y, Ueda K. 2022. Glycosylation in cancer: Its application as a biomarker and recent advances of analytical techniques. Glycoconj. J. 39:303–313. 10.1007/s10719-022-10043-1 [DOI] [PubMed] [Google Scholar]
- Haga Y, Yamada M, Fujii R, Saichi N, Yokokawa T, Hama T, Hayakawa Y, Ueda K. 2022. Fast and ultrasensitive glycoform analysis by supercritical fluid chromatography−tandem mass spectrometry. Anal. Chem. 94:15948–15955. 10.1021/acs.analchem.2c01721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm TH, Tanaka M, Nguyen H‐N, Tsutsumi A, Aizawa K, Matsui T. 2021. Matrix‐assisted laser desorption/ionization mass spectrometry‐guided visualization analysis of intestinal absorption of acylated anthocyanins in Sprague‐Dawley rats. Food Chem., 334:127586. 10.1016/j.foodchem.2020.127586 [DOI] [PubMed] [Google Scholar]
- Hahm TH, Tanaka M, Matsui T. 2022. Current knowledge on intestinal absorption of anthocyanins. J. Agric. Food Chem. 70:2501–2509. 10.1021/acs.jafc.1c08207 [DOI] [PubMed] [Google Scholar]
- Hall YD, Uzoewulu CP, Nizam ZM, Ishizawa S, El‐Shaffey HM, Ohata J. 2022. Phosphine‐mediated three‐component bioconjugation of amino‐ and azidosaccharides in ionic liquids. Chem. Commun. 58:10568. 10.1039/D2CC04013A [DOI] [PubMed] [Google Scholar]
- Hamel M, Rolain J‐M, Baron SA. 2021. The history of colistin resistance mechanisms in bacteria: Progress and challenges. Microorganisms, 9:442. 10.3390/microorganisms9020442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameleers L, Penttinen L, Ikonen M, Jaillot L, Fauré R, Terrapon N, Deuss PJ, Hakulinen N, Master ER, Jurak E. 2021. Polysaccharide utilization loci‑driven enzyme discovery reveals BD‑FAE: A bifunctional feruloyl and acetyl xylan esterase active on complex natural xylans. Biotechnol. Biofuels, 14:127. 10.1186/s13068-021-01976-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Wang F, Yue Y, Tan X, Tian M, Miao Y, Zhao S, Dong W, Yu M. 2022. Glycomics reveal that ST6GAL1‐mediated sialylation regulates uterine lumen closure during implantation. Cell Prolif., 55:e13169. 10.1111/cpr.13169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Qian Y, Wang X, Zhang Q, Cui J, Tu P, Liang H. 2021. Oleanane‐type saponins and prosapogenins from Albizia julibrissin and their cytotoxic activities. Phytochemistry, 185:112674. 10.1016/j.phytochem.2021.112674 [DOI] [PubMed] [Google Scholar]
- Han Y, Zhao Y, Chen P, Wang L, Hu Q, Wang X, Sun C. 2022. On‐tissue derivatization for isomer‐specific mass spectrometry imaging and relative quantification of monosaccharides in biological tissues. Anal. Chim. Acta, 1225:340241. 10.1016/j.aca.2022.340241 [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Nishikaze T, Miura N, Piao J, Okada K, Sekiya S, Iwamoto S, Sakamoto N, Tanaka K, Furukawa J‐i. 2018. Sialic acid linkage‐specific derivatization of glycosphingolipid glycans by ring‐opening aminolysis of lactones. Anal. Chem. 90:13193–13199 [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Nishikaze T, Tsumoto H, Ogawa K, Kobayashi T, Yokota I, Morikawa K, Suda G, Sho T, Nakai M, Miura N, Higashino K, Sekiya S, Iwamoto S, Miura Y, Furukawa J‐i, Tanaka K, Sakamoto N. 2019. Comparative glycomic analysis of sialyl linkage isomers by sialic acid linkage‐specific alkylamidation in combination with stable isotope labeling of α2,3‐linked sialic acid residues. Anal. Chem. 91:13343–13348. 10.1021/acs.analchem.9b03617 [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Furukawa J‐i. 2022. Comprehensive cellular glycan profiling of glycoproteins and glycosphingolipids by glycoblotting and BEP methods. Method. Mol. Biol. 2556:1–18. 10.1007/978-1-0716-2635-1_1 [DOI] [PubMed] [Google Scholar]
- Hanisch F‐G, Aydogan C, Schroten H. 2021. Fucoidan and derived oligo‐fucoses: Structural features with relevance in competitive inhibition of gastrointestinal norovirus binding. Mar. Drugs, 19:591. 10.3390/md19110591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch F‐G, Kunz C. 2021. Novel class of human milk oligosaccharides based on 6′‐galactosyllactose containing N‑acetylglucosamine branches extended by oligogalactoses. J. Proteome Res. 20:3865–3874. 10.1021/acs.jproteome.1c00154 [DOI] [PubMed] [Google Scholar]
- Hansen AL, Reily C, Novak J, Renfrow MB. 2021. Immunoglobulin A glycosylation and its role in disease. In: Pezer M, Editor, Antibody Glycosylation. Cham, Switzerland: Springer Nature. p 433–477. 10.1007/978-3-030-76912-3_14 [DOI] [PubMed] [Google Scholar]
- Hao T, Wang H, Liu J, Zhang Q, Liu Y, Zhao L, Zhu T, Yu P, Guo N, Meng X. 2021. Synthesis and immunogenicity of Brucella monovalent neoglycoconjugate. Carbohydr. Res. 499:108196. 10.1016/j.carres.2020.108196 [DOI] [PubMed] [Google Scholar]
- Harada M, Nakajima T, Yamada Y, Aomura D, Yamaguchi A, Sonoda K, Tanaka N, Hashimoto K, Kamijo Y. 2022. Serum sulfatide levels as a biomarker of active glomerular lesion in patients with anti‐neutrophil cytoplasmic antibody‐associated vasculitis: A single center pilot study. J. Clin. Med. 1:762. 10.3390/jcm11030762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Nakajima K, Li S, Suzuki T, Taniguchi N. 2021. Protocol for analyzing the biosynthesis and degradation of N‐glycan precursors in mammalian cells. STAR Protoc., 2:100316. 10.1016/j.xpro.2021.100316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin C, Smith KW, Cruickshank FL, Mackay CL, Flinders B, Heeren RMA, Moore T, Brockbank S, Cobice DF. 2022. On‐tissue chemical derivatization in mass spectrometry imaging. Mass Spectrom. Rev. 41:662–694. 10.1002/mas.21680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweg M, Jiang Y, Yilmaz G, Jarvis CM, Nguyen HV‐T, Primo GA, Monaco A, Beyer VP, Chen KK, Mohapatra S, Axelrod S, Gómez‐Bombarelli R, Kiessling LL, Becer CR, Johnson JA. 2021. Synthetic glycomacromolecules of defined valency, absolute configuration, and topology distinguish between human lectins. JACS Au, 1:1621–1630. 10.1021/jacsau.1c00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ. 1999. Matrix‐assisted laser desorption/ionization mass spectrometry of carbohydrates. Mass Spectrom. Rev. 18:349–451. [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2006. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update covering the period 1999‐2000. Mass Spectrom. Rev. 25:595–662. 10.1002/mas.20080 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2008. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update covering the period 2001‐2002. Mass Spectrom. Rev. 27:125–201. 10.1002/mas.20157 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2009. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2003‐2004. Mass Spectrom. Rev. 28:273–361. 10.1002/mas.20192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. 2009. Proposal for a standard system for drawing structural diagrams of N‐ and O‐linked carbohydrates and related compounds. Proteomics, 9:3796–3801. 10.1002/pmic.200900096 [DOI] [PubMed] [Google Scholar]; Correction, Proteomics, 9:5002. 10.1002/pmic.200990087 [DOI] [Google Scholar]
- Harvey DJ. 2011. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for the period 2005‐2006. Mass Spectrom. Rev. 30:1–100. 10.1002/mas.20265 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2012. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2007‐2008. Mass Spectrom. Rev. 31:183–311. 10.1002/mas.20333 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2015. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2009‐2010. Mass Spectrom. Rev. 34:268–422. 10.1002/mas.21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ, Crispin M, Bonomelli C, Scrivens JH. 2015. Ion mobility mass spectrometry for ion recovery and clean‐up of MS and MS/MS spectra obtained from low abundance viral samples. J. Am. Soc. Mass Spectrom., 26:1754–1767. 10.1007/s13361-015-1163-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ, Scarff C, Edgeworth M, Crispin M, Scrivens J. 2016. Travelling‐wave ion mobility mass spectrometry and negative ion fragmentation of hybrid and complex N‐glycans. J. Mass Spectrom. 51:1064–1079. 10.1002/jms.3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey DJ. 2017. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2011–2012. Mass Spectrom. Rev. 36:255–422. 10.1002/mas.21471 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2018. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2013‐2014. Mass Spectrom. Rev. 37:353–491. 10.1002/mas.21530 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2020. Negative ion mass spectrometry for the analysis of N‐linked glycans. Mass Spectrom. Rev. 39:586–679. 10.1002/mas.21622 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2021. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2015‐2016. Mass Spectrom. Rev. 40:408–565. 10.1002/mas.21651 [DOI] [PubMed] [Google Scholar]
- Harvey DJ. 2023. Analysis of carbohydrates and glycoconjugates by matrix‐assisted laser desorption/ionization mass spectrometry: An update for 2017‐2018. Mass Spectrom. Rev. 42:227–431. 10.1002/mas.21721 [DOI] [PubMed] [Google Scholar]
- Hasan M, Mimi A, Mamun A, Islam A, Waliullah ASM, Nabi M, Tamannaa Z, Kahyo T, Setou M. 2021. Mass spectrometry imaging for glycome in the brain. Front. Neuroanat. 15:711955. 10.3389/fnana.2021.711955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasehira K, Furuta T, Shimomura O, Asada M, Oda T, Tateno H. 2021. Quantitative structural analysis of glycans expressed within tumors derived from pancreatic cancer patient‐derived xenograft mouse models. Biochem. Biophys. Res. Commun. 534:310–316. 10.1016/j.bbrc.2020.11.087 [DOI] [PubMed] [Google Scholar]
- Hashii N, Suzuki J. 2021. Site‐specific O‐glycosylation analysis by liquid chromatography–mass spectrometry with electron‐transfer/higher‐energy collisional dissociation. Method. Mol. Biol. 2271:169–178. 10.1007/978-1-0716-1241-5_12 [DOI] [PubMed] [Google Scholar]
- Haslund‐Gourley BS, Grauzam S, Mehta AS, Wigdahl B, Comunale MA. 2022. Acute lyme disease IgG N‐linked glycans contrast the canonical inflammatory signature. Front. Immunol. 13:949118. 10.3389/fimmu.2022.949118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Yoneyama T, Tobisawa Y, Yamamoto H, Ohyama C. 2021. Narrative review of urinary glycan biomarkers in prostate cancer. Transl. Androl. Urol. 10:1850–1864. 10.21037/tau-20-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser JR, Bergström ET, Kulak AN, Warriner SL, Thomas‐Oates J, Bon RS. 2021. Pyrene tags for the detection of carbohydrates by label‐assisted laser desorption/ionisation mass spectrometry. ChemBioChem, 22:1430–1439. 10.1002/cbic.202000721 [DOI] [PubMed] [Google Scholar]
- Hawkinson TR, Clarke HA, Young LEA, Conroy LR, Markussen KH, Kerch KM, Johnson LA, Nelson PT, Wang C, Allison DB, Gentry MS, Sun RC. 2022. In situ spatial glycomic imaging of mouse and human Alzheimer's disease brains. Alzheimer's Dement., 18:1721–1735. 10.1002/alz.12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkinson TR, Sun RC. 2022. Matrix‐assisted laser desorption/ionization mass spectrometry imaging of glycogen in situ . Method. Mol. Biol. 2437:215–228. 10.1007/978-1-0716-2030-4_15 [DOI] [PubMed] [Google Scholar]
- Hayat A, Iqbal MS, Ahmad N, Alruwaili NK, Atta‐Ur‐Rehman. 2022. Fe(III)‐rhamnoxylan ‐ A novel high spin Fe(iii) octahedral complex having versatile physical and biological properties. Polymers, 14:4290. 10.3390/polym14204290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Yan B, Yao C, Chen X, Li L, Wu Y, Song Z, Song S, Zhang Z, Luo P. 2021. Oligosaccharides from Polygonatum cyrtonema Hua: Structural characterization and treatment of LPS‐induced peritonitis in mice. Carbohydr. Polym. 255:117392. 10.1016/j.carbpol.2020.117392 [DOI] [PubMed] [Google Scholar]
- He P, Zhang X, Xia K, Green DE, Baytas S, Xu Y, Pham T, Liu J, Zhang F, Almond A, Linhardt RJ, DeAngelis PL. 2022. Chemoenzymatic synthesis of sulfur‐linked sugar polymers as heparanase inhibitors. Nat. Commun. 13:7438. 10.1038/s41467-022-34788-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zheng Q, Huang H, Ji Y, Lin Z. 2022. Synergistic synthesis of hydrophilic hollow zirconium organic frameworks for simultaneous recognition and capture of phosphorylated and glycosylated peptides. Anal. Chim. Acta, 1198:339552. 10.1016/j.aca.2022.339552 [DOI] [PubMed] [Google Scholar]
- Heine V, Hovorková M, Vlachová M, Filipová M, Bumba L, Janoušková O, Hubálek M, Cvačka J, Petrásková L, Pelantová H, Křen V, Elling L, Bojarová P. 2021. Immunoprotective neo‐glycoproteins: Chemoenzymatic synthesis of multivalent glycomimetics for inhibition of cancer‐related galectin‐3. Eur. J. Med. Chem. 220:113500. 10.1016/j.ejmech.2021.113500 [DOI] [PubMed] [Google Scholar]
- Heiss DR, Badu‐Tawiah AK. 2021. In‐source microdroplet derivatization using coaxial contained‐electrospray mass spectrometry for enhanced sensitivity in saccharide analysis. Anal. Chem. 93:16779–16786. 10.1021/acs.analchem.1c02897 [DOI] [PubMed] [Google Scholar]
- Heiss DR, Badu‐Tawiah AK. 2022. Liquid chromatography‐tandem mass spectrometry with online, in‐source droplet‐based phenylboronic acid derivatization for sensitive analysis of saccharides. Anal. Chem. 94:14071–14078. 10.1021/acs.analchem.2c03736 [DOI] [PubMed] [Google Scholar]
- Helali Y, Sharma S, Vandeput M, Welba D, Van Antwerpen P, Marchant A, Delporte C. 2021. Fc Glycosylation characterization of human immunoglobulins G using immunocapture and LC‐MS. Method. Mol. Biol. 2271:57–71. 10.1007/978-1-0716-1241-5_4 [DOI] [PubMed] [Google Scholar]
- Helm J, Grünwald‐Gruber C, Thader A, Urteil J, Führer J, Stenitzer D, Maresch D, Neumann L, Pabst M, Altmann F. 2021. Bisecting Lewis X in hybrid‐type N‑glycans of human brain revealed by deep structural glycomics. Anal. Chem. 93:15175–15182. 10.1021/acs.analchem.1c03793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel JL, Gardner RA, Spencer DIR. 2021. Automation of immunoglobulin glycosylation analysis. In: Pezer M, Editor, Antibody Glycosylation. Cham Switzerland: Springer Nature. p 173–204. 10.1007/978-3-030-76912-3_5 [DOI] [PubMed] [Google Scholar]
- Heo HR, Joo KI, Seo JH, Kim CS, Cha HJ. 2021. Glycan chip based on structure‐switchable DNA linker for on‐chip biosynthesis of cancer‐associated complex glycans. Nat. Commun. 12:1395. 10.1038/s41467-021-21538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman X, Far J, Courtoy A, Bouhon L, Quinton L, De Pauw E, Chaumont F, Navarre C. 2021. Inactivation of N‐acetylglucosaminyltransferase I and α1,3‐fucosyltransferase genes in Nicotiana tabacum BY‐2 cells results in glycoproteins with highly homogeneous, high‐mannose N‐glycans. Front. Plant Sci. 12:634023. 10.3389/fpls.2021.634023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Voss BJ, Trent MS. 2021. Homeoviscous adaptation of the Acinetobacter baumannii outer membrane: Alteration of lipooligosaccharide structure during cold stress. mBio, 12:e01295–01221. 10.1128/mbio.01295-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieta J‐P, Sipari N, Räikkönen H, Keinänen M, Kostiainen R. 2021. Mass spectrometry imaging of Arabidopsis thaliana leaves at the single‐cell level by infrared laser ablation atmospheric pressure photoionization (LAAPPI). J. Am. Soc. Mass Spectrom., 32:2895–2903. 10.1021/jasms.1c00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraide T, Wada Y, Matsubayashi T, Kadoya M, Masunaga Y, Ohkubo Y, Nakashima M, Okamoto N, Ogata T, Saitsu H. 2021. Novel ALG12 variants and hydronephrosis in siblings with impaired N‐glycosylation. Brain Develop., 43:945–951. 10.1016/j.braindev.2021.05.013 [DOI] [PubMed] [Google Scholar]
- Hirschberg D, Ekman B, Wahlberg J, Landberg E. 2021. Altered immunoglobulin G glycosylation in patients with isolated hyperprolactinaemia. PLoS One, 16:e0247805. 10.1371/journal.pone.0247805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan JD, Klein JA, Wu J, Chopra P, Boons G‐J, Carvalho L, Lin C, Zaia J. 2018. Software for peak finding and elemental composition assignment for glycosaminoglycan tandem mass spectra. Mol. Cell. Proteomics, 17:1448–1456. 10.1074/mcp.RA118.000590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan JD, Wu J, Klein JA, Lin C, Carvalho L, Zaia J. 2021. GAGrank: Software for glycosaminoglycan sequence ranking using a bipartite graph model. Mol. Cell. Proteomics, 20:100093. 10.1016/j.mcpro.2021.100093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinger M, Bonaccorsi F, Cheallaigh AN, Oscarson S. 2022. Synthesis of a Lewis b hexasaccharide thioglycoside donor and its use towards an extended mucin core Tn heptasaccharide structure and a photoreactive biotinylated serine linked hexasaccharide. Org. Biomol. Chem. 20:4431–4440. 10.1039/d2ob00477a [DOI] [PubMed] [Google Scholar]
- Holzheimer M, Buter J, Minnaard AJ. 2021. Chemical synthesis of cell wall constituents of Mycobacterium tuberculosis . Chem. Rev. 1231:9554–9643. 10.1021/acs.chemrev.1c00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeker LK, Hildebrand GA, Fudyma JD, Daber LE, Hoyt D, Flowers SE, Gil‐Loaiza J, Kübert A, Bamberger I, Anderton CR, Cliff J, Leichty S, AminiTabrizi R, Kreuzwieser J, Shi L, Bai X, Velickovic D, Dippold MA, Ladd SN, Werner C, Meredith LK, Tfaily MM. 2022. Elucidating drought‐tolerance mechanisms in plant roots through 1H NMR metabolomics in parallel with MALDI‐MS, and nanoSIMS imaging techniques. Environ. Sci. Technol. 56:2021–2032. 10.1021/acs.est.1c06772 [DOI] [PubMed] [Google Scholar]
- Hong P, Sun H, Sha L, Pu Y, Khatri K, Yu X, Tang Y, Lin C. 2017. GlycoDeNovo ‐ an efficient algorithm for accurate de novo glycan topology reconstruction from tandem mass spectra. J. Am. Soc. Mass Spectrom., 28:2288–2301. 10.1007/s13361-017-1760-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S‐J, Park B‐R, Lee H‐N, Jang DE, Kang H‐J, Ameer K, Kim S‐J, Kim Y‐M. 2022. Carbohydrate‐binding module of cycloisomaltooligosaccharide glucanotransferase from Thermoanaerobacter thermocopriae improves its cyclodextran production. Enzyme Microb. Technol. 157:110023. 10.1016/j.enzmictec.2022.110023 [DOI] [PubMed] [Google Scholar]
- Hong S, Yu C, Wang P, Shi Y, Cao W, Cheng B, Chapla DG, Ma Y, Li J, Rodrigues E, Narimatsu Y, Yates JR III, Chen X, Clausen H, Moremen KW, Macauley MS, Paulson JC, Wu P. 2021. Glycoengineering of NK cells with glycan ligands of CD22 and selectins for B‐cell lymphoma therapy. Angew. Chem. Int. Ed. 60:3603–3610. 10.1002/anie.202005934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi R, Ozawa M, Tomii T, Kashiwada S, Miyanishi N. 2021. Structural analysis of N‐glycans in medaka gut exposed to silver and titanium dioxide nanoparticles. Environ. Sci. Pollut. Res. 28:58799–58806. 10.1007/s11356-021-14773-x [DOI] [PubMed] [Google Scholar]
- Hotchkiss, Jr. AT , Chau HK, Strahan GD, Nünez A, Simon S, White AK, Dieng S, Heuberger ER, Yadav MP, Hirsch J. 2022. Structural characterization of red beet fiber and pectin. Food Hydrocolloid., 129:107549. 10.1016/j.foodhyd.2022.107549 [DOI] [Google Scholar]
- Hotchkiss, Jr. AT , Chau HK, Strahan GD, Nuñez A, Simon S, White AK, Dieng S, Heuberger ER, Yadav MP, Hirsch J. 2021. Structure and composition of blueberry fiber pectin and xyloglucan that bind anthocyanins during fruit puree processing. Food Hydrocolloid. 116:106572. 10.1016/j.foodhyd.2020.106572 [DOI] [Google Scholar]
- Hristova J, Svinarov D. 2022. Enhancing precision medicine through clinical mass spectrometry platform. Biotechnol. Biotec. Eq. 36:106–116. 10.1080/13102818.2022.2053342 [DOI] [Google Scholar]
- Hu C, Luo W, Xu J, Han X. 2022. Recognition and avoidance of ion source‐generated artifacts in lipidomics analysis. Mass Spectrom. Rev. 41:15–31. 10.1002/mas.21659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu F, Chen Y, Shangguan G, Ju H. 2021. Mass spectrometric biosensing: A powerful approach for multiplexed analysis of clinical biomolecules. ACS Sensors, 6:3517–3535. 10.1021/acssensors.1c01394 [DOI] [PubMed] [Google Scholar]
- Hu W, Han Y, Sheng Y, Wang Y, Pan Q, Nie H. 2021. Mass spectrometry imaging for direct visualization of components in plants tissues. J. Sep. Sci. 44:3462–3476. 10.1002/jssc.202100138 [DOI] [PubMed] [Google Scholar]
- Hu X, Sheng J, Guan G, Ju T, Smith DF, Wang L. 2022. Morphology of biomaterials affect O‐glycosylation of HUVECs. J Funct. Biomater. 13:235. 10.3390/jfb13040235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Meuret C, Martinez A, Yassine HN, Nedelkov D. 2021. Distinct patterns of apolipoprotein C‐I, C‐II, and C‐III isoforms are associated with markers of Alzheimer's disease. J. Lipid Res. 62:100014. 10.1194/jlr.ra120000919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Feng Q, Xie Z, Mao H, Zhou Y, Yan Y, Ding C‐F. 2022. Post‐synthesis of covalent organic frameworks with dual‐hydrophilic groups for specific capture of serum exosomes. J. Chromatogr., A, 1679:463406. 10.1016/j.chroma.2022.463406 [DOI] [PubMed] [Google Scholar]
- Huang C, Tan Z, Zhao K, Zou W, Wang H, Gao H, Sun S, Bu D, Chai W, Li Y. 2021. The effect of N‐glycosylation of SARS‐CoV‐2 spike protein on the virus interaction with the host cell ACE2 receptor. iScience, 24:103272. 10.1016/j.isci.2021.103272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Seino J, Fujihira H, Sato K, Fujinawa R, Sumer‐Bayraktar Z, Ishii N, Matsuo I, Nakaya S, Suzuki T. 2022. Occurrence of free N‐glycans with a single GlcNAc at the reducing termini in animal sera. Glycobiology, 32:314–332. 10.1093/glycob/cwab124 [DOI] [PubMed] [Google Scholar]
- Huang H, Zheng Q, He Y, Zhong C, Tian W, Zhang S, Lin J, Lin Z. 2021. Facile synthesis of bifunctional polymer monolithic column for tunable and specific capture of glycoproteins and phosphoproteins. J. Chromatogr., A, 1651:462329. 10.1016/j.chroma.2021.462329 [DOI] [PubMed] [Google Scholar]
- Huang H, Ouyang D, Lin ZA. 2022. Recent advances in surface‑assisted laser desorption/ionization mass spectrometry and its imaging for small molecules. J. Anal. Test. 6:217–234. 10.1007/s41664-022-00211-5 [DOI] [Google Scholar]
- Huang J, Jiang B, Liu M, Yang P, Cao W. 2021. gQuant, an automated tool for quantitative glycomic data analysis. Front. Chem. 9:707738. 10.3389/fchem.2021.707738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu X, Wang D, Cui Y, Shi X, Dong J, Ye M, Li L. 2021. Dual‐functional Ti(IV)‐IMAC material enables simultaneous enrichment and separation of diverse glycopeptides and phosphopeptides. Anal. Chem. 93:8568–8576. 10.1021/acs.analchem.1c01324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Li C, Zong G, Prabhu SK, Chapla DG, Moremen KW, Wang L‐X. 2022. Site‐selective sulfation of N‐glycans by human GlcNAc‐6‐O‐sulfotransferase 1 (CHST2) and chemoenzymatic synthesis of sulfated antibody glycoforms. Bioorg. Chem. 128:106070. 10.1016/j.bioorg.2022.106070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S‐P, Hsu HC, Liew CY, Tsai S‐T, Ni C‐K. 2021. Logically derived sequence tandem mass spectrometry for structural determination of galactose oligosaccharides. Glycoconj. J. 38:177–189. 10.1007/s10719-020-09915-1 [DOI] [PubMed] [Google Scholar]
- Huang Y‐F, Aoki K, Akase S, Ishihara M, Liu Y‐S, Yang G, Kizuka Y, Mizumoto S, Tiemeyer M, Gao X‐D, Aoki‐Kinoshita KF, Fujita M. 2021. Global mapping of glycosylation pathways in human‐derived cells. Dev. Cell, 56:1195–1209. 10.1016/j.devcel.2021.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T‐H, Hung J‐T, Wu C‐E, Huang Y, Lee C‐W, Yeh C‐T, Chung Y‐H, Lo F‐Y, Lai L‐C, Tung JK, Yu J, Yeh C‐N, Yu AL. 2022. Globo H is a promising theranostic marker for intrahepatic cholangiocarcinoma. Hepatol. Commun. 6:194–208. 10.1002/hep4.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein NA, Ebied SA, Saleh MM. 2021. Evaluation of the anticancer effect of violacein, phycocyanin and phycocyanobilin on apoptotic genes expression and glycan profiles in breast cancer cells. Int. J. Cancer Biomed, Res., 5:81–97. 10.21608/jcbr.2021.46268.1079 [DOI] [Google Scholar]
- Hütte HJ, Tiemann B, Shcherbakova A, Grote V, Hoffmann M, Povolo L, Lommel M, Strahl S, Vakhrushev SY, Rapp E, Buettner FFR, Halim A, Imberty A, Bakker H. 2022. A bacterial mannose binding lectin as a tool for the enrichment of C‑ and O‑mannosylated peptides. Anal. Chem. 94:7329–7338. 10.1021/acs.analchem.2c00742 [DOI] [PubMed] [Google Scholar]
- Hykollari A, Paschinger K, Wilson IBH. 2022. Negative‐mode mass spectrometry in the analysis of invertebrate, fungal, and protist N‐glycans. Mass Spectrom. Rev. 41:945–963. 10.1002/mas.21693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun JY, Kim S, Lee C‐H, Lee HS, Shin I. 2021. Efficient preparation and bioactivity evaluation of glycan‐defined glycoproteins. ACS Chem. Biol. 16:1930–1940. 10.1021/acschembio.0c00629 [DOI] [PubMed] [Google Scholar]
- Hyun SW, Feng C, Liu A, Lillehoj EP, Trotta R, Kingsbury TJ, Passaniti A, Lugkey KN, Chauhan S, Cipollo JF, Luzina IG, Atamas SP, Cross AS, Goldblum SE. 2022. Altered sialidase expression in human myeloid cells undergoing apoptosis and differentiation. Sci. Rep. 12:14173. 10.1038/s41598-022-18448-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda A, Makino Y, Matsubara H. 2022. Glycogen debranching pathway deduced from substrate specificity of glycogen debranching enzyme. Glycoconj. J. 39:345–355. 10.1007/s10719-022-10046-y [DOI] [PubMed] [Google Scholar]
- Iles RK, Iles JK, Lacey J, Gardiner A, Zmuidinaite R. 2022. Direct detection of glycated human serum albumin and hyperglycosylated IgG3 in serum, by MALDI‐ToF mass spectrometry, as a predictor of COVID‐19 severity. Diagnostics, 12:2521. 10.3390/diagnostics12102521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ing N, Deng K, Chen Y, Aulitto M, Gin JW, Pham TLM, Petzold CJ, Singer SW, Bowen B, Sale KL, Simmons BA, Singh AK, Adams PD, Northen TR. 2021. A multiplexed nanostructure‑initiator mass spectrometry (NIMS) assay for simultaneously detecting glycosyl hydrolase and lignin modifying enzyme activities. Sci. Rep. 11:11803. 10.1038/s41598-021-91181-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irmscher T, Roske Y, Gayk I, Dunsing V, Chiantia S, Heinemann U, Barbirz S. 2021. Pantoea stewartii WceF is a glycan biofilm‐modifying enzyme with a bacteriophage tailspike‐like fold. J. Biol. Chem., 296:100286. 10.1016/j.jbc.2021.100286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaka E, Sugiura T, Hashimoto K, Kikuta K, Anazawa U, Nomura T, Kameyama A. 2021. Characterization of tumor‑associated MUC1 and its glycans expressed in mucoepidermoid carcinoma. Oncology Lett. 22:702. 10.3892/ol.2021.12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwata A, Fujita K, Fushinobu S, Tanaka K, Ito Y. 2022. Synthesis of naturally occurring β‐l‐arabinofuranosyl‐l‐arabinofuranoside structures towards the substrate specificity evaluation of β‐l‐arabinofuranosidase. Bioorg. Med. Chem. 68:116849. 10.1016/j.bmc.2022.116849 [DOI] [PubMed] [Google Scholar]
- Islam S, Alam R, Kim S. 2022. Improved coverage of plant metabolites using powder laser desorption/ionization coupled with Fourier‐transform ion cyclotron mass spectrometry. Food Chem., 373:131541. 10.1016/j.foodchem.2021.131541 [DOI] [PubMed] [Google Scholar]
- Isono T, Komaki R, Kawakami N, Chen K, Chen H‐L, Lee C, Suzuki K, Ree BJ, Mamiya H, Yamamoto T, Borsali R, Tajima K, Satoh T. 2022. Tailored solid‐state carbohydrate nanostructures based on star‐shaped discrete block co‐oligomers. Biomacromolecules, 23:3978–3989. 10.1021/acs.biomac.2c00813 [DOI] [PubMed] [Google Scholar]
- Ivannikov R, Anishchenko V, Kuzema P, Stavinskaya O, Laguta I, Poronnik O, Parnikoza I. 2022. Chromatographic and mass spectrometric analysis of secondary metabolites of Deschampsia antarctica from Galindez Island, Argentine Islands. Polish Polar Res., 43:341–362. 10.24425/ppr.2022.140369 [DOI] [Google Scholar]
- Iwan V, Grotemeyer J. 2021. Elucidating the fragmentation mechanism of protonated Lewis A trisaccharide using MSn CID. Eur. J. Mass Spectrom. 27:256–265. 10.1177/14690667211069033 [DOI] [PubMed] [Google Scholar]
- Izadi S, Javaran MJ, Monfared SR, Castilho A. 2021. Reteplase Fc‐fusions produced in N. benthamiana are able to dissolve blood clots ex vivo . PLoS One, 16:e0260796. 10.1371/journal.pone.0260796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaham ARA, Ang C‐S, Nie S, Bird LE, Williamson NA, Scott NE. 2021. What are we missing by using hydrophilic enrichment? Improving bacterial glycoproteome coverage using total proteome and FAIMS analyses. J. Proteome Res. 20:599–612. 10.1021/acs.jproteome.0c00565 [DOI] [PubMed] [Google Scholar]
- Jackson MA, Evans KO, Price NPJ, Blackburn JA, Ward CJ, Ray KJ, Vermillion KE. 2021. New family of surfactants from biobased materials. ACS Sustain. Chem. Eng. 9:13842–13850. 10.1021/acssuschemeng.1c04703 [DOI] [Google Scholar]
- Jain N, Tamura K, Déjean G, Van Petegem F, Brumer H. 2021. Orthogonal active‐site labels for mixed‐linkage endo‐β‐glucanases. ACS Chem. Biol. 16:1968–1984. 10.1021/acschembio.1c00063 [DOI] [PubMed] [Google Scholar]
- Jala RCR, Vudhgiri S, Kumar CG. 2022. A comprehensive review on natural occurrence, synthesis and biological activities of glycolipids. Carbohydr. Res. 516:108556. 10.1016/j.carres.2022.108556 [DOI] [PubMed] [Google Scholar]
- Jamshidi MP, Cairns C, Chong S, St. Michael F, Vinogradov EV, Cox AD, Sauvageau J. 2022. Synthesis and immunogenicity of a methyl rhamnan pentasaccharide conjugate from Pseudomonas aeruginosa A‑band polysaccharide. ACS Infect. Dis. 8:1347–1355. 10.1021/acsinfecdis.2c00184 [DOI] [PubMed] [Google Scholar]
- Jang S, Choi S‐S. 2021. Characterization of the fragmentation behaviors of protonated α‐cyclodextrin generated by electrospray ionization. Rapid Commun. Mass Spectrom., 35:e8967. 10.1002/rcm.8967 [DOI] [PubMed] [Google Scholar]
- Jaroentomeechai T, Kwon YH, Liu Y, Young O, Bhawal R, Wilson JD, Li M, Chapla DG, Moremen KW, Jewett MC, Mizrachi D, DeLisa MP. 2022. A universal glycoenzyme biosynthesis pipeline that enables efficient cell‐free remodeling of glycans. Nat. Commun. 13:6325. 10.1038/s41467-022-34029-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvas G, Szigeti M, Campbell MP, Guttman A. 2021. Database search assisted N‐glycan structure identification. In: El Rassi Z, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 843–858. 10.1016/B978-0-12-821447-3.00010-X [DOI] [Google Scholar]
- Jatczak‐Pawlik I, Gorzkiewicz M, Studzian M, Zinke R, Appelhans D, Klajnert‐Maculewicz B, Pułaski Ł. 2021. Nanoparticles for directed immunomodulation: Mannose‐functionalized glycodendrimers induce interleukin‑8 in myeloid cell lines. Biomacromolecules, 22:3396–3407. 10.1021/acs.biomac.1c00476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javeed R, Hussain D, Jabeen F, Sajid MS, Fatima B, Ashiq MN, Najam‐ul‐Haq M. 2021. Apo‐H (beta‐2‐glycoprotein) intact N‐glycan analysis by MALDI‐TOF‐MS using sialic acid derivatization. Anal. Bioanal. Chem. 413:7441–7449. 10.1007/s00216-021-03701-0 [DOI] [PubMed] [Google Scholar]
- Jeannot K, Hagart K, Dortet L, Kostrzewa M, Filloux A, Plesiat P, Larrouy‐Maumus G. 2021. Detection of colistin resistance in Pseudomonas aeruginosa using the MALDIxin test on the routine MALDI Biotyper Sirius mass spectrometer. Front. Microbiol. 12:725383. 10.3389/fmicb.2021.725383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegatheeswaran S, Guillemineau M, Giovane R, Borrillo L, Liao L, Kuir D, Auzanneau F‐I. 2022. Anti‐Lea monoclonal antibody SPM 522 recognizes an extended Lea epitope. Bioorg. Med. Chem. 56:116628. 10.1016/j.bmc.2022.116628 [DOI] [PubMed] [Google Scholar]
- Jezková P, Skřičková J, Wimmer G, Jr., Zelinková J, Zdráhal Z, Lattová E. 2022. Differentiation of sialyl linkages using a combination of alkyl esterification and phenylhydrazine derivatization: Application for N‑glycan profiling in the sera of patients with lung cancer. Anal. Chem. 94:6736–6744. 10.1021/acs.analchem.2c00105 [DOI] [PubMed] [Google Scholar]
- Ji M‐C, Fu B, Zhang Y‐J. 2021. Recent progress of analytical methods of proteomics based on mass spectrometry. J. Chinese Mass Spectrom. Soc. 42:862–877. 10.7538/zpxb.2021.0091 [DOI] [Google Scholar]
- Ji X‐G, Chang K‐L, Chen M, Zhu L‐L, Osman A, Yin H, Zhao L‐M. 2021. In vitro fermentation of chitooligosaccharides and their effects on human fecal microbial community structure and metabolites. LWT ‐ Food Sci. Technol., 144:111224. 10.1016/j.lwt.2021.111224 [DOI] [Google Scholar]
- Jia Y, Lu Y, Wang X, Yang Y, Zou M, Liu J, Jin W, Wang X, Pang G, Huang L, Wang Z. 2021. Mass spectrometry based quantitative and qualitative analyses reveal N‐glycan changes of bovine lactoferrin at different stages of lactation. LWT ‐ Food Sci. Technol., 147:111626. 10.1016/j.lwt.2021.111626 [DOI] [Google Scholar]
- Jiang F, Zhang X, Hwang W, Nishiyama Y, Briber RM, Wang H. 2021. Oligocellulose from acid hydrolysis: A revisit. Appl. Surf. Sci. 537:147783. 10.1016/j.apsusc.2020.147783 [DOI] [Google Scholar]
- Jiang H, Zhang Y, Liu Z, Wang X, He J, Jin H. 2022. Advanced applications of mass spectrometry imaging technology in quality control and safety assessments of traditional Chinese medicines. J. Ethnopharmacol. 284:114760. 10.1016/j.jep.2021.114760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Ran Y, Liu N, Huang X. 2022. Preparation and structural characterization of water‐soluble polysaccharide from garlic skin. Food Sci., 43:57–65. 10.7506/spkx1002-6630-20210323-289 [DOI] [Google Scholar]
- Jie L, Yuan Z, Yu Z, Xue‐song F. 2021. Progress in the pretreatment and analysis of carbohydrates in food: An update since 2013. J. Chromatogr., A, 1655:462496. 10.1016/j.chroma.2021.462496 [DOI] [PubMed] [Google Scholar]
- Jin H, Gao W, Liu R, Yang J, Zhang S, Han R, Lin J, Zhang S, Yu J, Tang K. 2022. A novel hydrophilic hydrogel with a 3D network structure for the highly efficient enrichment of N‐glycopeptides. Analyst, 147:2425–2432. 10.1039/D2AN00516F [DOI] [PubMed] [Google Scholar]
- Jin W, Li C, Zou M, Lu Y, Wei M, Nan L, Jia Y, Wang C, Huang L, Wang Z. 2021. A preliminary study on isomer‐specific quantification of sialylated N‐glycans released from whey glycoproteins in human colostrum and mature milk using a glycoqueuing strategy. Food Chem., 339:127866. 10.1016/j.foodchem.2020.127866 [DOI] [PubMed] [Google Scholar]
- Jin W, Lu Y, Li C, Zou M, Chen Q, Nan L, Wei M, Wang C, Huang L, Wang Z. 2022. Improved glycoqueuing strategy reveals novel α2,3‐linked di‐/tri‐sialylated oligosaccharide isomers in human milk. J. Agric. Food Chem. 70:13996–14004. 10.1021/acs.jafc.2c04499 [DOI] [PubMed] [Google Scholar]
- Jin X, Zhu C, Wu J, Yan Y, Ding C‐F, Tang K, Zhang D. 2021. Hydrophilic carrageenan functionalized magnetic carbon‐based framework linked by silane coupling agent for the enrichment of N‐glycopeptides from human saliva. J. Sep. Sci. 44:143–2152. 10.1002/jssc.202001216 [DOI] [PubMed] [Google Scholar]
- Jin YR, Oh MJ, Yuk HJ, An HJ, Kim DS. 2021. Novel analysis procedure for red ginseng polysaccharides by matrix‐assisted laser desorption/ionization time‐of‐flight/time‐of‐flight mass spectrometry. J. Ginseng Res. 45:539–545. 10.1016/j.jgr.2021.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Liu M, Huang X, Zhang X, Qu Z, Zhu J‐J, Min Q. 2022. Top‐down rational engineering of heteroatom‐doped graphene quantum dots for laser desorption/ionization mass spectrometry detection and imaging of small biomolecules. Anal. Chem. 94:7609–7618. 10.1021/acs.analchem.2c00802 [DOI] [PubMed] [Google Scholar]
- Jing F, Wang L, Yang M, Wu C, Li J, Shi L, Feng S, Li F. 2022. Visualizing the spatial distribution of functional metabolites in Forsythia suspensa at different harvest stages by MALDI mass spectrometry imaging. Fitoterapia, 162:105285. 10.1016/j.fitote.2022.105285 [DOI] [PubMed] [Google Scholar]
- Jirásko R, Idkowiak J, Wolrab D, Kvasnička A, Friedecký D, Polański K, Študentová H, Študent V, Melichar B, Holčapek M. 2022. Altered plasma, urine, and tissue profiles of sulfatides and sphingomyelins in patients with renal cell carcinoma. Cancers, 14:4622. 10.3390/cancers14194622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jitprasertwong P, Khamphio M, Petsrichuang P, Eijsink VGH, Poolsri W, Muanprasat C, Rangnoi K, Yamabhai M. 2021. Anti‐inflammatory activity of soluble chitooligosaccharides (CHOS) on VitD3‐induced human THP‐1 monocytes. PLoS One, 16:e0246381. 10.1371/journal.pone.0246381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanneau D, Klau LJ, Larocque R, Jaffrennou A, Duval G, Le Duff N, Roret T, Jeudy A, Aachmann FL, Czjzek M, Thomas F. 2021. Structure‐function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT . Glycobiology, 31:1364–1377. 10.1093/glycob/cwab058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JW, Shin JH, Lee WK, Begum H, Min CH, Jang MH, Oh HB, Yang MS, Kim SR. 2021. Inactivation of the β (1,2)‑xylosyltransferase and the α (1,3)‑fucosyltransferase gene in rice (Oryza sativa) by multiplex CRISPR/Cas9 strategy. Plant Cell Rep., 40:1025–1035. 10.1007/s00299-021-02667-8 [DOI] [PubMed] [Google Scholar]
- Jung JW. 2022. Production of recombinant human acid α‑glucosidase with mannosidic N‑glycans in α‑mannosidase I mutant rice cell suspension culture. Plant Biotechnol. Rep. 16:333–342. 10.1007/s11816-022-00757-x [DOI] [Google Scholar]
- Jung SH, Lee YH, Jeong DY, Jeong YR, Jung BO, Park JK. 2021. Matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry analysis for molecular characterization of chitosan. J. Chitin Chitosan, 26:76–82. 10.17642/jcc.26.2.5 [DOI] [Google Scholar]
- Kadokawa J‐i. 2022. Glucan phosphorylase‐catalyzed enzymatic synthesis of unnatural oligosaccharides and polysaccharides using nonnative substrates. Polymer J. 54:413–426. 10.1038/s41428-021-00584-x [DOI] [Google Scholar]
- Kailasam S, Arumugam S, Balaji K, Kanth SV. 2022. Adsorption of chromium by exopolysaccharides extracted from lignolytic phosphate solubilizing bacteria. Int. J. Biol. Macromol. 206:788–798. 10.1016/j.ijbiomac.2022.03.047 [DOI] [PubMed] [Google Scholar]
- Kajiura H, Eguchi T, Uchino K, Tatematsu K‐i, Tamura T, Sezutsu H, Fujiyama K. 2022. Temporal analysis of N‐acetylglucosamine extension of N‐glycans in the middle silk gland of silkworm Bombyx mori . J. Biosci. Bioeng. 133:533–540. 10.1016/j.jbiosc.2022.03.001 [DOI] [PubMed] [Google Scholar]
- Kakuta T, Manyuan N, Kawasaki H. 2022. UV‐Absorbing ligand capped gold nanoparticles for the SALDI‐MS analysis of small molecules. Mass Spectrom. (Tokyo), 11:Ao107. 10.5702/massspectrometry.A0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalirajan C, Behera H, Selvaraj V, Palanisamy T. 2022. In vitro probing of oxidized inulin cross‐linked collagen‐ZrO2 hybrid scaffolds for tissue engineering applications. Carbohydr. Polym., 289:119458. 10.1016/j.carbpol.2022.119458 [DOI] [PubMed] [Google Scholar]
- Kalmar JG, Garrard KP, Muddiman DC. 2021. GlycoHunter: An open‐source software for the detection and relative quantification of INLIGHT‐labeled N‑linked glycans. J. Proteome Res. 20:1855–1863. 10.1021/acs.jproteome.0c00840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyani DC, Reichenbach T, Aspeborg H, Divne C. 2021. A homodimeric bacterial exo‐β‐1,3‐glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo‐β‐1,3‐glucanases. Enzyme Microb. Technol. 143:109723. 10.1016/j.enzmictec.2020.109723 [DOI] [PubMed] [Google Scholar]
- Kameyama A, Nishijima R, Yamakoshi K. 2021. Bmi‐1 regulates mucin levels and mucin O‐glycosylation in the submandibular gland of mice. PLoS One, 16:e0245607. 10.1371/journal.pone.0245607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kameyama A, Tin WWT, Nishijima R, Yamakoshi K. 2021. Alteration of mucins in the submandibular gland during aging in mice. Arch. Oral Biol. 121:104967. 10.1016/j.archoralbio.2020.104967 [DOI] [PubMed] [Google Scholar]
- Kamiki H, Murakami S, Nishikaze T, Hiono T, Igarashi M, Furuse Y, Matsugo H, Ishida H, Katayama M, Sekine W, Muraki Y, Takahashi M, Takenaka‐Uema A, Horimotoa T. 2022. Influenza A virus agnostic receptor tropism revealed using a novel biological system with terminal sialic acid knockout cells. J. Virol. 96:e0041622. 10.1128/jvi.00416-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K, Selvaprakash K, Chen Y‐C. 2021. Functional magnetic nanoparticle‐based affinity probe for MALDI mass spectrometric detection of ricin B. Microchim. Acta, 188:339. 10.1007/s00604-021-04991-y [DOI] [PubMed] [Google Scholar]
- Kao KS, Gupta A, Zong G, Li C, Kerschbaumer I, Borghi S, Achkar JM, Bournazos S, Wang L‐X, Ravetch JV. 2022. Synthetic nanobodies as tools to distinguish IgG Fc glycoforms. Proc. Natl. Acad. Sci., USA, 119:e2212658119. 10.1073/pnas.2212658119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Nemcovic M, Folbergrova J, Kala D, Svoboda J, Otahal J, Brnoliakova Z. 2022. N‐Glycans profiling in pilocarpine induced status epilepticus in immature rats. Eur. Pharm. J. 69(2):1–4. 10.2478/afpuc-2022-0011 [DOI] [Google Scholar]
- Kashima T, (鹿), Okumura K, (奥), Ishiwata A, (石) , Kaieda M, (海), Terada T, (寺), Arakawa T, (荒), Yamada C, (山), Shimizu K, (清), Tanaka K, (田), Kitaoka M, (北), Ito Y, (伊), Fujita K, (藤), Fushinobu S, (伏). 2021. Identification of difructose dianhydride I synthase/hydrolase from an oral bacterium establishes a novel glycoside hydrolase family. J. Biol. Chem. 297:101324. 10.1016/j.jbc.2021.101324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabara D, Kondo K, Usami R, Kan D, Kawamura I, Kawasaki Y, Sato M, Nittami T, Suzuki I, Katahira M, Takeda M. 2021. Structural determination of the sheath‐forming polysaccharide of Sphaerotilus montanus using thiopeptidoglycan lyase which recognizes the 1,4 linkage between α‐d‐GalN and β‐d‐GlcA. Int. J. Biol. Macromol. 183:992–1001. 10.1016/j.ijbiomac.2021.05.001 [DOI] [PubMed] [Google Scholar]
- Kaszowska M, Górska S, Knirel Y, Kalinchuk N, Gamian A, Katzenellenbogen E. 2021. Structural analysis of Edwardsiella tarda PCM 1155 O‐polysaccharide revealed the presence of unique β‐l‐RhapNAc3NAc derivative. Carbohydr. Res. 509:108423. 10.1016/j.carres.2021.108423 [DOI] [PubMed] [Google Scholar]
- Katayama H, Nagata K. 2021. Application of 2,20‐dipyridyl disulfide‐mediated thiazolidine ring‐opening reaction to glycoprotein synthesis: Total chemical synthesis of evasin‐3. J. Peptide Sci. 27:e3290. 10.1002/psc.3290 [DOI] [PubMed] [Google Scholar]
- Katayama H, Toyota K, Tanaka H, Ohira T. 2022. Chemical synthesis and functional evaluation of the crayfish insulin‐like androgenic gland factor. Bioorg. Chem. 122:105738. 10.1016/j.bioorg.2022.105738 [DOI] [PubMed] [Google Scholar]
- Kato K, Takahashi N. 2009. GALAXY database and pyridylaminated oligosaccharide library. In: Taniguchi N, Suzuki A, Ito Y, Narimatsu H, Kawasaki T, Hase S, Editors., Experimental Glycoscience. Tokyo: Springer. p 413–416. 10.1007/978-4-431-77922-3_100 [DOI] [Google Scholar]
- Kaur G, Kumar H, Singla M. 2022. Diverse applications of ionic liquids: A comprehensive review. J. Mol. Liquids, 351:118556. 10.1016/j.molliq.2022.118556 [DOI] [Google Scholar]
- Kaur H. 2021. Characterization of glycosylation in monoclonal antibodies and its importance in therapeutic antibody development. Crit. Rev. Biotechnol. 41:300–315. 10.1080/07388551.2020.1869684 [DOI] [PubMed] [Google Scholar]
- Kaur T, Shukla BN, Yadav VK, Kulkarni MJ, Rao A. 2021. Comparison of glycoprofiles of rituximab versions licensed for sale in India and an analytical approach for quality assessment. J. Proteomics, 244:104267. 10.1016/j.jprot.2021.104267 [DOI] [PubMed] [Google Scholar]
- Kawahara R, Chernykh A, Alagesan K, Bern M, Cao W, Chalkley RJ, Cheng K, Choo MS, Edwards N, Goldman R, Hoffmann M, Hu Y, Huang Y, Kim JY, Kletter D, Liquet B, Liu M, Mechref Y, Meng B, Neelamegham S, Nguyen‐Khuong T, Nilsson J, Pap A, Park GW, Parker BL, Pegg CL, Penninger JM, Phung TK, Pioch M, Rapp E, Sakalli E, Sanda M, Schulz BL, Scott NE, Sofronov G, Stadlmann J, Vakhrushev SY, Woo CM, Wu H‐Y, Yang P, Ying W, Zhang H, Zhang Y, Zhao J, Zaia J, Haslam SM, Palmisano G, Yoo JS, Larson G, Khoo K‐H, Medzihradszky KF, Kolarich D, Packer NH, Thaysen‐Andersen M. 2021. Community evaluation of glycoproteomics informatics solutions reveals high‐performance search strategies for serum glycopeptide analysis. Nat. Methods 18:1304–1316. 10.1038/s41592-021-01309-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima N, Naito S, Hanamatsu H, Nagane M, Takeuchi Y, Furukawa J‐i, Iwasaki N, Yamashita T, Nakayama K‐i. 2022. Glycosphingolipid GM3 prevents albuminuria and podocytopathy induced by anti‑nephrin antibody. Sci. Rep. 12:16058. 10.1038/s41598-022-20265-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayili HM, Salih B. 2021. N‐glycan profiling of glycoproteins by hydrophilic interaction liquid chromatography with fluorescence and mass spectrometric detection. J. Vis. Exp. 175:e62751. 10.3791/62751 [DOI] [PubMed] [Google Scholar]
- Kayili HM, Atakay M, Hayatu A, Salih B. 2022. Sample preparation methods for N‐glycomics. Adv. Sample Prep., 4:100042. 10.1016/j.sampre.2022.100042 [DOI] [Google Scholar]
- Kayili HM, Ragoubi ZME, Salih B. 2022. An integrated stage‐tip‐based glycomic and glycoproteomic approach for simple and rapid N‐glycosylation profiling of glycoproteins. Biomed. Chromatogr. 36:e5503. 10.1002/bmc.5503 [DOI] [PubMed] [Google Scholar]
- Kayili HM, Sakhta R, Salİh B. 2022. Comparison of denaturing agent effects in enzymatic N‐glycan release for human plasma N‐glycan analysis. Turk. J. Chem. 46:15. 10.55730/1300-0527.3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Ding B, Zhang M, Dong T, Fu Y, Lv Q, Ding W, Wang X. 2022. Study on inhibitory activity and mechanism of chitosan oligosaccharides on Aspergillus flavus and Aspergillus fumigatus . Carbohydr. Polym. 275:118673. 10.1016/j.carbpol.2021.118673 [DOI] [PubMed] [Google Scholar]
- Kelly MI, Albahrani M, Castro C, Poon E, Yan B, Littrell J, Waas M, Boheler KR, Gundry RL. 2021. Importance of evaluating protein glycosylation in pluripotent stem cell‑derived cardiomyocytes for research and clinical applications. Pflüg. Archiv Eur. J. Physiol. 473:1041–1059. 10.1007/s00424-021-02554-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, McCrory EH, Xu Y, Scudamore CL, Nanda SK, Mancebo‐Gamella P, Wood NT, Knebel A, Matthews SJ, Cohen P. 2022. HOIL‐1 ubiquitin ligase activity targets unbranched glucosaccharides and is required to prevent polyglucosan accumulation. EMBO J., 41:e109700. 10.15252/embj.2021109700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemme L, Grüneberg M, Reunert J, Rust S, Park J, Westermann C, Wada Y, Schwartz O, Marquardt T. 2021. Translational balancing questioned: Unaltered glycosylation during disulfiram treatment in mannosyl‐oligosaccharide alpha‐1,2‐mannnosidase‐congenital disorders of glycosylation MAN1B1‐CDG). JIMD Rep., 60:42–55. 10.1002/jmd2.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerselidou D, Dohai BS, Nelson DR, Daakour S, De Cock N, Hassoun ZAO, Kim D‐K, Olivet J, El Assal DC, Jaiswal A, Alzahmi A, Saha D, Pain C, Matthijssens F, Lemaitre P, Herfs M, Chapuis J, Ghesquiere B, Vertommen D, Kriechbaumer V, Knoops K, Lopez‐Iglesias C, van Zandvoort M, Lambert J‐C, Hanson J, Desmet C, Thiry M, Lauersen KJ, Vidal M, Van Vlierberghe P, Dequiedt F, Salehi‐Ashtiani K, Twizere J‐C. 2021. Alternative glycosylation controls endoplasmic reticulum dynamics and tubular extension in mammalian cells. Sci. Adv. 7:eabe8349. 10.1126/sciadv.abe8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa I, Lorenzo JM, Bangar SP, Morsy OM, Nawaz A, Walayat N, Sobhy R. 2022. Effect of the non‐covalent and covalent interactions between proteins and mono‐ or di‐glucoside anthocyanins on β‐lactoglobulin‐digestibility. Food Hydrocolloid., 133:107952. 10.1016/j.foodhyd.2022.107952 [DOI] [Google Scholar]
- Khatuntseva EA, Nifantiev NE. 2022. Cross reacting material (CRM197) as a carrier protein for carbohydrate conjugate vaccines targeted at bacterial and fungal pathogens. Int. J. Biol. Macromol. 218:775–798. 10.1016/j.ijbiomac.2022.07.137 [DOI] [PubMed] [Google Scholar]
- Khodair AI, Kassab SE, Kheder NA, Fahim AM. 2022. Synthesis of novel d‐α‐galactopyranosyl‐l‐seryl/l‐threonyl‐l‐alanyl‐l‐alanine as useful precursors of new glycopeptide antibiotics with computational calculations studies. Carbohydr. Res. 514:108546. 10.1016/j.carres.2022.108546 [DOI] [PubMed] [Google Scholar]
- Khoo K‐H. 2021. A mass spectrometry‐based glycotope‐centric cellular glycomics is the more fruitful way forward to see the forest for the trees. Biochem. Soc. Trans. 49:55–69. 10.1042/bst20190861 [DOI] [PubMed] [Google Scholar]
- Khor MJ, Broda A, Kostrzewa M, Drobniewski F, Larrouy‐Maumus G. 2021. An improved method for rapid detection of Mycobacterium abscessus complex based on species‐specific lipid fingerprint by routine MALDI‐TOF. Front. Chem. 9:715890. 10.3389/fchem.2021.715890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidibule PE, Costa J, Atrei A, Plou FJ, Fernandez‐Lobato M, Pogni R. 2021. Production and characterization of chitooligosaccharides by the fungal chitinase Chit42 immobilized on magnetic nanoparticles and chitosan beads: Selectivity, specificity and improved operational utility. RSC Adv., 11:5529–5536. 10.1039/D0RA10409D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D‐G, Baek I, Lee Y, Kim H, Kim JY, Bang G, Kim S, Yoon HJ, Han BW, Suh SW, Kim HS. 2021. Structural basis for SdgB‐ and SdgA‐mediated glycosylation of staphylococcal adhesive proteins. Acta Chromatogr., D77:1460–1474. 10.1107/s2059798321010068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Kim J‐H, Chi W‐J. 2021. Purification and biochemical characterization of β‐agarase produced by marine microorganism Cellulophga sp. J9‐3. Microbiol. Biotechnol. Lett, 49:329–336. 10.48022/mbl.2105.05009 [DOI] [Google Scholar]
- Kim JY, Lim H, Moon DW. 2022. Ambient mass spectrometry imaging of small molecules from cells and tissues. Method. Mol. Biol. 2437:41–59. 10.1007/978-1-0716-2030-4_3 [DOI] [PubMed] [Google Scholar]
- Kim S‐H, Yun S, Park W. 2022. Constitutive phenotypic modification of lipid A in clinical Acinetobacter baumannii isolates. Microbiol. Spectrum, 10:e0129522. 10.1128/spectrum.01295-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S‐W, Kwon S, Kim Y‐K. 2021. Graphene oxide derivatives and their nanohybrid structures for laser desorption/ionization time‐of‐flight mass spectrometry analysis of small molecules. Nanomaterials, 11:288. 10.3390/nano11020288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y‐S, Hwang J, Lee SG, Jo HY, Oh MJ, Liyanage NM, Je J‐G, An HJ, Jeon Y‐J. 2022. Structural characteristics of sulfated polysaccharides from Sargassum horneri and immune‐enhancing activity of polysaccharides combined with lactic acid bacteria. Food Funct., 13:8214–8227. 10.1039/D1FO03946F [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Nakajima K, Yamamoto S, Suzuki S. 2021. High‐throughput N‐glycan screening method for therapeutic antibodies using a microchip‐based DNA analyzer: A promising methodology for monitoring monoclonal antibody N‐glycosylation. Anal. Bioanal. Chem. 413:4727–4738. 10.1007/s00216-021-03434-0 [DOI] [PubMed] [Google Scholar]
- Kinoshita M, Yamada K. 2021. Recent advances and trends in sample preparation and chemical modification for glycan analysis. J. Pharmaceut. Biomed. Anal. 207:114424. 10.1016/j.jpba.2021.114424 [DOI] [PubMed] [Google Scholar]
- Kissel T, Ge C, Hafkenscheid L, Kwekkeboom JC, Slot LM, Cavallari M, He Y, van Schie KA, Vergroesen RD, Kampstra ASB, Reijm S, Stoeken‐Rijsbergen G, Koeleman C, Voortman LM, Heitman LH, Xu B, Pruijn GJM, Wuhrer M, Rispens T, Huizinga TWJ, Scherer HU, Reth M, Holmdahl R, Toes REM. 2022. Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation. Sci. Adv. 8:eabm1759. 10.1126/sciadv.abm1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasić M, Krištić J, Korać P, Horvat T, Markulin D, Vojta A, Reiding KR, Wuhrer M, Lauc G, Zoldoš V. 2022. DNA hypomethylation upregulates expression of the MGAT3 gene in HepG2 cells and leads to changes in N‐glycosylation of secreted glycoproteins. Sci. Rep. 6:24363. 10.1038/srep24363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaver EJ, Dukes‐Rimsky L, Kumar B, Xia Z‐J, Dang T, Lehrman MA, Angel P, Drake RR, Freeze HH, Steet R, Flanagan‐Steet H. 2021. Protease‐dependent defects in N‐cadherin processing drive PMM2‐CDG pathogenesis. JCI Insight, 6:e153474. 10.1172/jci.insight.153474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knihtila R, Chemmalil L, Sawant P, Bhavsar S, Kuang J, Shao C, Atsma J, Li Z, Ding J. 2022. Development of a novel, label‐free N‐glycan method using charged aerosol detection. J. Chromatogr., B, 1212:123502. 10.1016/j.jchromb.2022.123502 [DOI] [PubMed] [Google Scholar]
- Knochenmuss R. 2021. An introduction to MALDI ionization mechanisms for users of mass spectrometry imaging. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 3–19. 10.1039/9781839165191-00001 [DOI] [Google Scholar]
- Kobayashi H, Masuda Y, Takaya H, Kubo T, Otsuka K. 2022. Separation of glycoproteins based on sugar chains using novel stationary phases modified with poly(ethylene glycol)‐conjugated boronic‐acid derivatives. Anal. Chem. 94:6882–6892. 10.1021/acs.analchem.2c01002 [DOI] [PubMed] [Google Scholar]
- Kobeissy F, Kobaisi A, Peng W, Barsa C, Goli M, Sibahi A, El Hayek S, Abdelhady S, Haidar MA, Sabra M, Orešič M, Logroscino G, Mondello S, Eid AH, Mechref Y. 2022. Glycomic and glycoproteomic techniques in neurodegenerative disorders and neurotrauma: Towards personalized markers. Cells, 11:581. 10.3390/cells11030581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobylis P, Lis H, Stepnowski P, Caban M. 2019. Spectroscopic verification of ionic matrices for MALDI analysis. J. Mol. Liquids, 284:328–342. 10.1016/j.molliq.2019.03.137 [DOI] [Google Scholar]
- Kobylis P, Stepnowski P, Caban M. 2021. Review of the applicability of ionic liquid matrices for the quantification of small molecules by MALDI MS. Microchem. J. 164:105983. 10.1016/j.microc.2021.105983 [DOI] [Google Scholar]
- Kobylis P, Kasprzyk M, Nowacki A, Caban M. 2022. An investigation of the ionicity of selected ionic liquid matrices used for matrix‐assisted laser desorption/ionization. J. Mol. Liquids, 349:118106. 10.1016/j.molliq.2021.118106 [DOI] [Google Scholar]
- Köfeler HC, Ahrends R, Baker ES, Ekroos K, Han X, Hoffmann N, Holčapek M, Wenk MR, Liebisch G. 2021. Recommendations for good practice in MS‐based lipidomics. J. Lipid Res. 62:100138. 10.1016/j.jlr.2021.100138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhagen M, Dasari S, Willrich M, Hetrick M, Netzel B, Dispenzieri A, Murray DL. 2021. Automation and validation of a MALDI‐TOF MS (Mass‐Fix) replacement of immunofixation electrophoresis in the clinical lab. Clin. Chem. Lab. Med. 59:155–163. 10.1515/cclm-2020-0581 [DOI] [PubMed] [Google Scholar]
- Koide R, Hirane N, Kambe D, Yokoi Y, Otaki M, Nishimura S‐I. 2022. Antiadhesive nanosome elicits role of glycocalyx of tumor cell‐derived exosomes in the organotropic cancer metastasis. Biomaterials, 280:121314. 10.1016/j.biomaterials.2021.121314 [DOI] [PubMed] [Google Scholar]
- Kokesch‐Himmelreich J, Wittek O, Race AM, Rakete S, Schlicht C, Busch U, Römpp A. 2022. MALDI mass spectrometry imaging: From constituents in fresh food to ingredients, contaminants and additives in processed food. Food Chem., 385:132529. 10.1016/j.foodchem.2022.132529 [DOI] [PubMed] [Google Scholar]
- Kokoulin MS, Dmitrenok PS, Romanenko LA. 2022. Structure of the lipooligosaccharide from the deep‐sea marine bacterium Idiomarina zobellii KMM 231T, isolated at a depth of 4000 meters. Mar. Drugs, 20:700. 10.3390/md20110700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarich D, Rapp E, Struwe WB, Haslam SM, Zaia J, McBride R, Agravat S, Campbell MP, Kato M, Ranzinger R, Kettner C, York WS. 2013. The minimum information required for a glycomics experiment (MIRAGE) project: Improving the standards for reporting mass‐spectrometry‐based glycoanalytic data. Mol. Cell. Proteomics, 12:991–995. 10.1074/mcp.o112.026492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Harada Y, Nakano M, Suzuki T, Fukushige T, Hanzawa K, Yagi H, Takagi K, Mizuno K, Miyamoto Y, Taniguchi N, Kato K, Kanekura T, Dohmae N, Machida K, Maruyama I, Inoue H. 2022. Identification of distinct N‐glycosylation patterns on extracellular vesicles from small‐cell and non‐small‐cell lung cancer cells. J. Biol. Chem., 298:101950. 10.1016/j.jbc.2022.101950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Yasui C, Banno T, Asakura K, Fukuoka T, Ushimaru K, Koga M, Minamikawa H, Saika A, Morita T, Takahashi D, Toshima K. 2022. Self‐assembling properties and recovery effects on damaged skin cells of chemically synthesized mannosylerythritol lipids. ChemBioChem, 23:e202100631. 10.1002/cbic.202100631 [DOI] [PubMed] [Google Scholar]
- Kong S, Zhang Q, Yang L, Huang Y, Liu M, Yan G, Zhao H, Wu M, Zhang X, Yang P, Cao W. 2021. Effective enrichment strategy using boronic acid‐functionalized mesoporous graphene‐silica composites for intact N‐ and O‑linked glycopeptide analysis in human serum. Anal. Chem. 93:6682–6691. 10.1021/acs.analchem.0c05482 [DOI] [PubMed] [Google Scholar]
- Kori M, Aydin B, Gulfidan G, Beklen H, Kelesoglu N, Iscan AC, Turanli B, Erzik C, Karademir B, Arga KY. 2021. The repertoire of glycan alterations and glycoproteins in human cancers. OMICS J. Integrat. Biol. 25:139–168. 10.1089/omi.2020.0210 [DOI] [PubMed] [Google Scholar]
- Kotnala A, Anderson DMG, Patterson NH, Cantrell LS, Messinger JD, Curcio CA, Schey KL. 2021. Tissue fixation effects on human retinal lipid analysis by MALDI imaging and LC‐MS/MS technologies. J. Mass Spectrom. 56:e4798. 10.1002/jms.4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsias M, Madunić K, Nicolardi S, Kozak RP, Gardner RA, Jansen BC, Spencer DIR, Wuhrer M. 2021. A semi‐automated, high throughput approach for O‐glycosylation profiling of in vitro established cancer cell lines by MALDI‐FT‐ICR MS. Glycoconj. J. 38:747–756. 10.1007/s10719-021-10003-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzounis D, Hageman JA, Soares N, Michiels J, Schols HA. 2021. Impact of xylanase and glucanase on oligosaccharide formation, carbohydrate fermentation patterns, and nutrientutilization in the gastrointestinal tract of broilers. Animals, 11:1285. 10.3390/ani11051285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzounis D, Jonathan MC, Soares N, Kabel MA, Schols HA. 2022. In vivo formation of arabinoxylo‐oligosaccharides by dietary endo‐xylanase alters arabinoxylan utilization in broilers. Carbohydr. Polym., 291:119527. 10.1016/j.carbpol.2022.119527 [DOI] [PubMed] [Google Scholar]
- Kowalewski B, Lange H, Galle S, Dierks T, Lübke T, Damme M. 2021. Decoding the consecutive lysosomal degradation of 3‐O‐sulfate containing heparan sulfate by Arylsulfatase G (ARSG). Biochem. J. 478:3221–3237. 10.1042/bcj20210415 [DOI] [PubMed] [Google Scholar]
- Kowarik M, Wetter M, Haeuptle MA, Braun M, Steffen M, Kemmler S, Ravenscroft N, De Benedetto G, Zuppiger M, Sirena D, Cescutti P, Wacker M. 2021. The development and characterization of an E. coli O25B bioconjugate vaccine. Glycoconj. J. 38:421–435. 10.1007/s10719-021-09985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejčí P, Cechová MZ, Nádvorníková J, Barták P, Kobrlová L, Balarynová J, Smýkal P, Bednář P. 2022. Combination of electronically driven micromanipulation with laser desorption ionization mass spectrometry—The unique tool for analysis of seed coat layers and revealing the mystery of seed dormancy. Talanta, 242:123303. 10.1016/j.talanta.2022.123303 [DOI] [PubMed] [Google Scholar]
- Kristensen DB, Sloth TM, Orgaard M, Jensen PF. 2021. Characterization of protein glycoforms at intact level by orbitrap mass spectrometry. Method. Mol. Biol. 2271:23–45. 10.1007/978-1-0716-1241-5_2 [DOI] [PubMed] [Google Scholar]
- Krištić J, Lauc G, Pezer M. 2022. Immunoglobulin G glycans ‐ Biomarkers and molecular effectors of aging. Clin. Chim. Acta, 535:30–45. 10.1016/j.cca.2022.08.006 [DOI] [PubMed] [Google Scholar]
- Krivosheina MS, Borisov RS, Zhilyaev DI, Matveeva MD, Zaikin VG. 2021. New suitable deprotonating matrices for the analysis of carboxylic acids and some acidic compounds by matrix‐assisted laser desorption/ionization mass spectrometry in negative ion mode. Rapid Commun. Mass Spectrom., 35:e8954. 10.1002/rcm.8954 [DOI] [PubMed] [Google Scholar]
- Krüger L, Biskup K, Karampelas V, Ludwig A, Kasper A‐S, Poller WC, Blanchard V. 2022. Straightforward analysis of sulfated glycosaminoglycans by MALDI‐TOF mass spectrometry from biological samples. Biology, 11:506. 10.3390/biology11040506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka MR, Li Z, Chernova TA, Smith DF, Song X, Cummings RD, Ju T. 2021. Cellular O‐glycome reporter/amplification (CORA): Analytical and preparative tools to study mucin‐type O‐glycans of living cells. Curr. Protoc., 1:e142. 10.1002/cpz1.142 [DOI] [PubMed] [Google Scholar]
- Kumar A, Datta LP, Samanta S, Arora H, Govindaraju T. 2021. Benzothiazole‐phenothiazine conjugate based molecular probe for the differential detection of glycated albumin. Israel J. Chem. 61:222–230. 10.1002/ijch.202000098 [DOI] [Google Scholar]
- Kumar A, Mukhia S, Kumar R. 2022. Production, characterisation, and application of exopolysaccharide extracted from a glacier bacterium Mucilaginibacter sp. ERMR7:07. Proc. Biochem. 113:27–36. 10.1016/j.procbio.2021.12.018 [DOI] [Google Scholar]
- Kumari M, Tetala KKR. 2022. A review on recent advances in the enrichment of glycopeptides and glycoproteins by liquid chromatographic methods: 2016‐Present. Electrophoresis, 43:388–402. 10.1002/elps.202100172 [DOI] [PubMed] [Google Scholar]
- Kun R, Jóna E, Guttman A. 2021. Capillary electrophoresis‐based N‐glycosylation analysis in the biomedical and biopharmaceutical fields. Adv. Exp. Med. Biol. 1336:129–137. 10.1007/978-3-030-77252-9_7 [DOI] [PubMed] [Google Scholar]
- Kurz S, Sheikh MO, Lu S, Wells L, Tiemeyer M. 2021. Separation and identification of permethylated glycan isomers by reversed phase nanoLC‐NSI‐MSn . Mol. Cell. Proteomics, 20:100045. 10.1074/mcp.ra120.002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S‐H, Kim H, Lee S, Lim J, Pal K, Chung B, Kang D‐H, Kim D. 2022. Synthesis and biological characterization of low‐calorie Schisandra chinensis syrup. Food Sci. Biotechnol. 31:857–865. 10.1007/s10068-022-01061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lageveen‐Kammeijer GSM, Rapp E, Chang D, Rudd PM, Kettner C, Zaia J. 2022. The minimum information required for a glycomics experiment (MIRAGE): Reporting guidelines for capillary electrophoresis. Glycobiology, 32:580–587. 10.1093/glycob/cwac021 [DOI] [PubMed] [Google Scholar]
- Lageveen‐Kammeijer GSM, Küster B, Reusch D, Wuhrer M. 2022. High sensitivity glycomics in biomedicine. Mass Spectrom. Rev. 41:1014–1039. 10.1002/mas.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye M, Tabi W, Le Bot L, Delaire M, Orsel M, Campoy JA, Garcia JQ, Le Gall S. 2021. Comparison of cell wall chemical evolution during the development of fruits of two contrasting quality from two members of the Rosaceae family: Apple and sweet cherry. Plant Physiol. Biochem. 168:93–104. 10.1016/j.plaphy.2021.10.002 [DOI] [PubMed] [Google Scholar]
- Lai Z‐Z, Zhou J‐Y, Li Z‐L. 2021. Analysis of glycosylation of immunoglobulin G using mass spectrometry and its application. J. Chinese Mass Spectrom. Soc., 42:878–896. 10.7538/zpxb.2021.0119 [DOI] [Google Scholar]
- Lai Z, Zhang M, Zhou J, Chen T, Li D, Shen X, Liu J, Zhou J, Li Z. 2021. Fe3O4@PANI: A magnetic polyaniline nanomaterial for highly efficient and handy enrichment of intact N‐glycopeptides. Analyst, 146:4261–4267. 10.1039/D1AN00580D [DOI] [PubMed] [Google Scholar]
- Lakatos C, Kordován MÁ, Czifrák K, Nagy L, Vadkerti B, Daróczi L, Zsuga M, Kéki S. 2022. Synthesis of sucrose‐HDI cooligomers: New polyols for novel polyurethane networks. Int. J. Mol. Sci. 23:1444. 10.3390/ijms23031444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SM, Li J, Sun H, Mao W, Lu Z, Zhao Q, Han C, Gong X, Jiang B, Chua GH, Zhao Z, Meng F, Shui G. 2022. Quantitative lipidomics and spatial MS‐imaging uncovered neurological and systemic lipid metabolic pathways underlying troglomorphic adaptations in cave‐dwelling fish. Mol. Biol. Evol. 39:msac050. 10.1093/molbev/msac050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz ND, Ming SA, Thon V, Veeramachineni VM, Azurmendi HF, Vann WF. 2021. Characterization of the β‑KDO transferase KpsS, the initiating enzyme in the biosynthesis of the lipid acceptor for Escherichia coli polysialic acid. Biochemistry, 60:2044–2054. 10.1021/acs.biochem.1c00088 [DOI] [PubMed] [Google Scholar]
- Larrouy‐Maumus G. 2021. Shotgun bacterial lipid A analysis using routine MALDI‐TOF mass spectrometry. Method. Mol. Biol. 2306:275–283. 10.1007/978-1-0716-1410-5_18 [DOI] [PubMed] [Google Scholar]
- Larsen D, Ferreira M, Tilloy S, Monflier E, Beeren SR. 2022. Unnatural cyclodextrins can be accessed from enzyme‐mediated dynamic combinatorial libraries. Chem. Commun. 58:2287–2290. 10.1039/D1CC06452E [DOI] [PubMed] [Google Scholar]
- Larsen LE, van den Boogert MAW, Rios‐Ocampo WA, Jansen JC, Conlon D, Chong PLE, Levels JHM, Eilers RE, Sachdev VV, Zelcer N, Raabe T, He M, Hand NJ, Drenth JPH, Rader DJ, Stroes ESG, Lefeber DJ, Jonker JW, Holleboom AG. 2022. Defective lipid droplet‐lysosome interaction causes fatty liver disease as evidenced by human mutations in TMEM199 and CCDC115 . Cell. Mol. Gastroenterol. Hepatol. 13:583–597. 10.1016/j.jcmgh.2021.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson EA, Forsman TT, Stuart L, Alexandrov T, Lee YJ. 2022. Rapid and automatic annotation of multiple on‐tissue chemical modifications in mass spectrometry imaging with metaspace. Anal. Chem. 94:8983–8991. 10.1021/acs.analchem.2c00979 [DOI] [PubMed] [Google Scholar]
- Lazer LM, Kesavan Y, Gor R, Ramachandran I, Pathak S, Narayan S, Anbalagan M, Ramalingam S. 2022. Targeting colon cancer stem cells using novel doublecortin like kinase 1 antibody functionalized folic acid conjugated hesperetin encapsulated chitosan nanoparticles. Colloid Surface B, 217:112612. 10.1016/j.colsurfb.2022.112612 [DOI] [PubMed] [Google Scholar]
- Le Mauff F, Razvi E, Reichhardt C, Sivarajah P, Parsek MR, Howell L, Sheppard DC. 2022. The Pel polysaccharide is predominantly composed of a dimeric repeat of α‐1,4 linked galactosamine and N‐acetylgalactosamine. Commun. Biol. 5:502. 10.1038/s42003-022-03453-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebredonchel E, Duvet S, Douillard C, Foulquier F, Klein A. 2021. Variation of the serum N‐glycosylation during the pregnancy of a MPI‐CDG patient. JIMD Rep., 62:22–29. 10.1002/jmd2.12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecot N, Glisoni R, Oddone N, Benech J, Fernández M, Gambini JP, Cabral P, Sosnik A. 2021. Glucosylated polymeric micelles actively target a breast cancer model. Adv. Therap. 4:2000010. 10.1002/adtp.202000010 [DOI] [Google Scholar]
- Lee D, Kim Y, Jalaludin I, Nguyen H‐Q, Kim M, Seo J, Jang K‐S, Kim J. 2021. MALDI‐MS analysis of disaccharide isomers using graphene oxide as MALDI matrix. Food Chem., 342:128356. 10.1016/j.foodchem.2020.128356 [DOI] [PubMed] [Google Scholar]
- Lee JH, Samsuzzaman M, Park MG, Park SJ, Kim SY. 2021. Methylglyoxal‐derived hemoglobin advanced glycation end products induce apoptosis and oxidative stress in human umbilical vein endothelial cells. Int. J. Biol. Macromol. 187:409–421. 10.1016/j.ijbiomac.2021.07.058 [DOI] [PubMed] [Google Scholar]
- Lee JY, Kim H, Moon Y, Kwak S, Kang CG, Park C, Jo J, Kim SW, Pal K, Kang DH, Kim D. 2022. Enhancement of the water solubility and antioxidant capacities of mangiferin by transglucosylation using a cyclodextrin glycosyltransferase. Enzyme Microb. Technol. 159:110065. 10.1016/j.enzmictec.2022.110065 [DOI] [PubMed] [Google Scholar]
- Lee M‐C, Huang C‐Y, Lai C‐L, Yeh H‐Y, Huang J, Lung WQC, Lee P‐T, Nan F‐H. 2022. Colaconema formosanum, Sarcodia suae, and Nostoc commune as fermentation substrates for bioactive substance production. Fermentation, 8:343. 10.3390/fermentation8070343 [DOI] [Google Scholar]
- Lee PY, Yeoh Y, Omar N, Pung Y‐F, Lim LC, Low TY. 2021. Molecular tissue profiling by MALDI imaging: Recent progress and applications in cancer research. Crit. Rev. Clin. Lab. Sci. 58:513–529. 10.1080/10408363.2021.1942781 [DOI] [PubMed] [Google Scholar]
- Lee Y‐J. 2022. Mass Spectrometry Imaging of Small Molecules. New York, Heidelberg, Dordrecht: Springer Nature. 10.1007/978-1-4939-1357-2 [DOI] [Google Scholar]
- Lee Y‐R, Briggs MT, Kuliwaba JS, Anderson PH, Condina MR, Hoffmann P. 2021. Gelatin‐coated indium tin oxide slides improve human cartilage‐bone tissue adherence and N‐glycan signal intensity for mass spectrometry imaging. Anal. Bioanal. Chem. 413:2675–2682. 10.1007/s00216-020-02986-x [DOI] [PubMed] [Google Scholar]
- Lee YH, Kim SC, Nam KD, Kim TH, Jung BO, Park Y‐I, Synytsya A, Park JK. 2022. Chitosan isolated from black soldier flies Hermetia illucens: Structure and enzymatic hydrolysis. Proc. Biochem. 118:171–181. 10.1016/j.procbio.2022.04.020 [DOI] [Google Scholar]
- Lee YH, Park SY, Hwang YJ, Park JK. 2022. Molecular weight determination of chitosan with antibacterial activity using matrix‐assisted laser desorption/ionization‐time of flight mass spectrometry analysis. Macromol. Res. 30:90–98. 10.1007/s13233-022-0013-0 [DOI] [Google Scholar]
- Lee YR, Briggs MT, Young C, Condina MR, Kuliwaba JS, Anderson PH, Hoffmann P. 2022. Mass spectrometry imaging spatially identifies complex‑type N‑glycans as putative cartilage degradation markers in human knee osteoarthritis tissue. Anal. Bioanal. Chem. 414:7597–7607. 10.1007/s00216-022-04289-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legouffe R, Jeanneton O, Gaudin M, Tomezyk A, Gerstenberg A, Dumas M, Heusèle C, Bonnel D, Stauber J, Schnebert S. 2022. Hyaluronic acid detection and relative quantification by mass spectrometry imaging in human skin tissues. Anal. Bioanal. Chem. 414:5781–5791. 10.1007/s00216-022-04139-8 [DOI] [PubMed] [Google Scholar]
- Leivers S, Lagos L, Garbers P, La Rosa SL, Westereng B. 2022. Technical pipeline for screening microbial communities as a function of substrate specificity through fluorescent labelling. Commun. Biol. 5:444. 10.1038/s42003-022-03383-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieszek MK, Komaniecka I, Chojnacki M, Choma A, Rzeski W. 2022. Immunomodulatory properties of polysaccharide‐rich young green barley (Hordeum vulgare) extract and its structural characterization. Molecules, 27:1742. 10.3390/molecules27051742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León‐García MC, Silva‐Gaona OG, Hernández‐Ortiz M, Vargas‐Ortiz K, Ramírez‐Emiliano J, Garay‐Sevilla ME, Encarnación‐Guevara S, Pérez‐Vázquez V. 2022. Curcumin prevents the glycation of tricarboxylic acid cycle and cell respiration proteins in the heart of mice fed with a high‐fructose diet. Curr. Pharmaceut. Des. 28:1769–1778. 10.2174/1381612828666220331160501 [DOI] [PubMed] [Google Scholar]
- Leroy A, Devaux M‐F, Fanuel M, Chauvet H, Durand S, Alvarado C, Habrant A, Sandt C, Rogniaux H, Paës G, Guillon F. 2022. Real‐time imaging of enzymatic degradation of pretreated maize internodes reveals different cell types have different profiles. Bioresource Technol., 353:127140. 10.1016/j.biortech.2022.127140 [DOI] [PubMed] [Google Scholar]
- Levink IJM, Klatte DCF, Hanna‐Sawires RG, Vreeker GCM, Ibrahim IS, van der Burgt YEM, Overbeek KA, Koopmann BDM, Cahen DL, Fuhler GM, Wuhrer M, Bonsing BA, Tollenaar RAEM, Vleggaar FP, Vasen HFA, van Leerdam ME, Bruno MJ, Mesker WE. 2022. Longitudinal changes of serum protein N‐glycan levels for earlier detection of pancreatic cancer in high‐risk individuals. Pancreatology, 22:497–506. 10.1016/j.pan.2022.03.021 [DOI] [PubMed] [Google Scholar]
- Li B, Ge J, Liu W, Hu D, Li P. 2021. Unveiling spatial metabolome of Paeonia suffruticosa and Paeonia lactiflora roots using MALDI MS imaging. New Phytol., 231:892–902. 10.1111/nph.17393 [DOI] [PubMed] [Google Scholar]
- Li C, Chong G, Zong G, Knorr DA, Bournazos S, Aytenfisu AH, Henry GK, Ravetch JV, MacKerell AD Jr., Wang L‐X. 2021. Site‐selective chemoenzymatic modification on the core fucose of an antibody enhances its Fcγ receptor affinity and ADCC activity. J. Am. Chem. Soc. 143:7828–7838. 10.1021/jacs.1c03174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Palma AS, Zhang P, Zhang Y, Gao C, Silva LM, Li Z, Trovão F, Weishaupt M, Seeberger PH, Likhosherstov LM, Piskarev V, Yu J, Westerlind U, Chai W. 2021. Noncovalent microarrays from synthetic amino‐terminating glycans: Implications in expanding glycan microarray diversity and platform comparison. Glycobiology, 31:931–946. 10.1093/glycob/cwab037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Dong S. 2021. 6‐Aminopyridine‐3‐boronic acid functionalized magnetic nanoparticles for highly efficient enrichment of cis‐diol‐containing biomolecules. Anal. Meth., 13:2331–2337. 10.1039/D1AY00414J [DOI] [PubMed] [Google Scholar]
- Li D, Xie G, Xie P, Zhu L, Cai Z. 2021. Synthesis of zwitterionic dual‐functional metal‐organic framework nanocomposite with ultra‐hydrophilicity for selective enrichment of glycopeptides. Chin. J. Chromatogr. 39:205–210. 10.3724/sp.j.1123.2020.11006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Sun X, Yu W, Shi C, Zhang X, Yu H, Ma F. 2021. Enhanced konjac glucomannan hydrolysis by lytic polysaccharide monooxygenases and generating prebiotic oligosaccharides. Carbohydr. Polym. 253:117241. 10.1016/j.carbpol.2020.117241 [DOI] [PubMed] [Google Scholar]
- Li F, Zhao H, Shao R, Zhang X, Yu H. 2021. Enhanced Fenton reaction for xenobiotic compounds and lignin degradation fueled by quinone redox cycling by lytic polysaccharide monooxygenases. J. Agric. Food Chem. 69:7104–7114. 10.1021/acs.jafc.1c01684 [DOI] [PubMed] [Google Scholar]
- Li F, Liu Y, Liu Y, Li Y, Yu H. 2022. Heterologous expression and characterization of a novel lytic polysaccharide monooxygenase from Natrialbaceae archaeon and its application for chitin biodegradation. Bioresource Technol., 354:127174. 10.1016/j.biortech.2022.127174 [DOI] [PubMed] [Google Scholar]
- Li G, Wang G, Tong Y, Zhu J, Yun T, Ye X, Li F, Yuan S, Liu Q. 2021. Concise synthesis and antidiabetic activity of natural flavonoid glycosides, oroxins C and D, isolated from the seeds of Oroxylum indium . J. Chem. Res. 45:68–75. 10.1177/1747519820927966 [DOI] [Google Scholar]
- Li H, He H, Liu Z. 2021. Recent progress and application of boronate affinity materials in bioanalysis. Trends Anal. Chem. 140:116271. 10.1016/j.trac.2021.116271 [DOI] [Google Scholar]
- Li H, Mattingly AE, Jania LA, Smith R, Melander RJ, Ernst RK, Koller BH, Melander C. 2021. Benzimidazole isosteres of salicylanilides are highly active colistin adjuvants. ACS Infect. Dis. 7:3303–3313. 10.1021/acsinfecdis.1c00463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang X, Chen R, Cheng K, Ning Z, Li J, Twine S, Stintzi A, Mack D, Figeys D. 2021. Elevated colonic microbiota‐associated paucimannosidic and truncated N‐glycans in pediatric ulcerative colitis. J. Proteomics, 249:104369. 10.1016/j.jprot.2021.104369 [DOI] [PubMed] [Google Scholar]
- Li H, Chiang AWT, Lewis NE. 2022. Artificial intelligence in the analysis of glycosylation data. Biotechnol. Adv. 60:108008. 10.1016/j.biotechadv.2022.108008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ma C, Gao M, Li Y, Xie L, Zhao D, Zhang R, Zhang G, Li W, Rong R, Kong B. 2022. Ti3C2(OH)x‑assisted LDI‑TOF‑MS for the rapid analysis of natural small molecules. Anal. Bioanal. Chem. 414:8447–8461. 10.1007/s00216-022-04382-z [DOI] [PubMed] [Google Scholar]
- Li H, Wu R, Hu Q, Chen X, Chan T‐WD. 2022. A matrix sublimation device with an integrated solvent nebulizer for MALDI‐MSI. J. Am. Soc. Mass Spectrom. 33:11–16. 10.1021/jasms.1c00335 [DOI] [PubMed] [Google Scholar]
- Li J, Solhi L, Goddard‑Borger ED, Mathieu Y, Wakarchuk WW, Withers SG, Brumer H. 2021. Four cellulose‑active lytic polysaccharide monooxygenases from Cellulomonas species. Biotechnol. Biofuels, 14:29. 10.1186/s13068-020-01860-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang D, Chang S‐C, Liang P‐H, Srivastava V, Guu S‐Y, Shie J‐J, Khoo K‐H, Bulone V, Hsieh YSY. 2021. Production of structurally defined chito‐oligosaccharides with a single N‑acetylation at their reducing end using a newly discovered chitinase from Paenibacillus pabuli . J. Agric. Food Chem. 69:3371–3379. 10.1021/acs.jafc.0c06804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Dong X, Cui Y, Li S, Chen C, Zhang X, Li X, Liang X, Zhu Y. 2022. Simultaneous enrichment and sequential separation of O‐linked glycopeptides and phosphopeptides with immobilized titanium (IV) ion affinity chromatography materials. J. Chromatogr., A, 1681:463462. 10.1016/j.chroma.2022.463462 [DOI] [PubMed] [Google Scholar]
- Li J, Goddard‐Borger ED, Raji O, Saxena H, Solhi L, Mathieu Y, Master ER, Wakarchuk WW, Brumer H. 2022. Chitin‐active lytic polysaccharide monooxygenases are rare in Cellulomonas species. Appl. Environ. Microbiol. 88:1–15. 10.1128/aem.00968-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo B, Zhang W, Yue S, Huang S, Gao S, Ma J, Cipollo JF, Yang S. 2022. Recent advances in demystifying O‐glycosylation in health and disease. Proteomics, 22:2200156. 10.1002/pmic.202200156 [DOI] [PubMed] [Google Scholar]
- Li J, Hsiung S‐Y, Kao M‐R, Xing X, Chang S‐C, Wang D, Hsieh P‐Y, Liang P‐H, Zhu Z, Cheng T‐JR, Shie J‐J, Liou J‐P, Abbott DW, Kwon SW, Hsieh YSY. 2022. Structural compositions and biological activities of cell wall polysaccharides in the rhizome, stem, and leaf of Polygonatum odoratum (Mill.) Druce. Carbohydr. Res. 521:108662. 10.1016/j.carres.2022.108662 [DOI] [PubMed] [Google Scholar]
- Li J, Yin L, Qi X, Huang Y. 2022. Serum sulfatide as a biomarker of carotid atherosclerosis in patients with rheumatoid arthritis. Clin. Chim. Acta, 534:6–13. 10.1016/j.cca.2022.06.030 [DOI] [PubMed] [Google Scholar]
- Li J, Zhang J, Xu M, Yang Z, Yue S, Zhou W, Gui C, Zhang H, Li S, Wang PG, Yang S. 2022. Advances in glycopeptide enrichment methods for the analysis of protein glycosylation over the past decade. J. Sep. Sci. 45:3169–3186. 10.1002/jssc.202200292 [DOI] [PubMed] [Google Scholar]
- Li J, Zhao T, Li J, Shen J, Jia L, Zhu B, Dang L, Ma C, Liu D, Mu F, Hu L, Sun S. 2022. Precision N‐glycoproteomics reveals elevated LacdiNAc as a novel signature of intrahepatic cholangiocarcinoma. Mol. Oncol. 16:2135–2152. 10.1002/1878-0261.13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Jiang C, Tan H, Li J, Xu Y, Tang D, Zhao X, Liu Q, Li J, Yin H. 2021. Identification and characterization of a novel glucomannanase from Paenibacillus polymyxa . 3 Biotech, 11:129. 10.1007/s13205-021-02676-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L‐F, Zhang Q‐W, Han Q‐B. 2022. Recent advances in qualitative and quantitative analysis of polysaccharides in natural medicines: A critical review. J. Pharmaceut. Biomed. Anal. 220:115016. 10.1016/j.jpba.2022.115016 [DOI] [PubMed] [Google Scholar]
- Li L, Qiu Z, Jiang M, Zhang B, Chen Q, Zhang C, Zheng Z, Qiao X. 2022. Visualizing the spatial distribution of Arctium lappa L. root components by MALDI‐TOF mass spectrometry imaging. Foods, 11:3957. 10.3390/foods11243957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhang H, Chen X, Yan S, Yang L, Song H, Li J, Liu J, Yu H, Liu H, Zhu D. 2022. Chemical composition and sugar spectroscopy of soy hull polysaccharides obtained by microwave‐assisted salt extraction. J. Food Process. Preserv. 46:e16869. 10.1111/jfpp.16869 [DOI] [Google Scholar]
- Li M, Gu T‐j, Lin X, Li L. 2021. DiLeuPMP: A multiplexed isobaric labeling method for quantitative analysis of O‑glycans. Anal. Chem. 93:9845–9852. 10.1021/acs.analchem.1c01433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Huang J, Ma M, Shi X, Li L. 2022. Selective enrichment of sialylglycopeptides enabled by click chemistry and dynamic covalent exchange. Anal. Chem. 94:6681–6688. 10.1021/acs.analchem.1c05158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhong X, Feng Y, Li L. 2022. Novel isobaric tagging reagent enabled multiplex quantitative glycoproteomics via electron‐transfer/higher‐energy collisional dissociation (EThcD) mass spectrometry. J. Am. Soc. Mass Spectrom. 33:1874–1882. 10.1021/jasms.2c00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhu W, Zheng H, Zhang J. 2022. Efficient HCD‐pd‐EThcD approach for N‐glycan mapping of therapeutic antibodies at intact glycopeptide level. Anal. Chim. Acta, 1189:339232. 10.1016/j.aca.2021.339232 [DOI] [PubMed] [Google Scholar]
- Li P, Pang J, Xu S, He H, Ma Y, Liu Z. 2022. A glycoform‐resolved dual‐modal ratiometric immunoassay improves the diagnostic precision for hepatocellular carcinoma. Angew. Chem. Int. Ed., 61:e202113528. 10.1002/anie.202113528 [DOI] [PubMed] [Google Scholar]
- Li P, Wang L, Guo R, Feng H, Ji Y, Lim SY, Ng BH, Laserna AKC, Khan S, Chen S‐M, Li SFY. 2022. Cross‐identification of N‐glycans by CE‐LIF using two capillary coatings and three labeling dyes. Talanta, 239:123061. 10.1016/j.talanta.2021.123061 [DOI] [PubMed] [Google Scholar]
- Li R, Lin Q, Ren J, Guo Z, Wang Y, Yang X, Wang X. 2022. Insights into the synergistic effect of catalyst acidity and solvent basicity for effective production of pentose from glucose. Chem. Eng. J. 442:136224. 10.1016/j.cej.2022.136224 [DOI] [Google Scholar]
- Li S, Qiao L, Liang C, Zhao L, Du K. 2022. Boronate‐immobilized cellulose nanofiber‐reinforced cellulose microspheres for pH‐dependent adsorption of glycoproteins. Carbohydr. Polym. 298:120068. 10.1016/j.carbpol.2022.120068 [DOI] [PubMed] [Google Scholar]
- Li SY, Xin YJ, Bao CX, Hou J, Cui HL. 2022. Haloprofundus salilacus sp. nov., Haloprofundus halobius sp. nov. and Haloprofundus salinisoli sp. nov.: Three extremely halophilic archaea isolated from salt lake and saline soil. Extremophiles, 26:6. 10.1007/s00792-021-01255-8 [DOI] [PubMed] [Google Scholar]
- Li T, Yu P, Chen Y, Sun B, Dong P, Zhu T, Meng X. 2021. N‐Acetylgalactosamine‐decorated nanoliposomes for targeted delivery of paclitaxel to hepatocellular carcinoma. Eur. J. Med. Chem. 222:113605. 10.1016/j.ejmech.2021.113605 [DOI] [PubMed] [Google Scholar]
- Li W, Ma Y, Guo Z, Xing R, Liu Z. 2021. Efficient screening of glycan‐specific aptamers using a glycosylated peptide as a scaffold. Anal. Chem. 93:956–963. 10.1021/acs.analchem.0c03675 [DOI] [PubMed] [Google Scholar]
- Li W, De Schutter K, Van Damme EJM, Smagghe G. 2022. Developmental O‐glycan profile analysis shows pentasaccharide mucin‐type O‐glycans are linked with pupation of Tribolium castaneum . Arch. Insect Biochem. Physiol. 109:e21852. 10.1002/arch.21852 [DOI] [PubMed] [Google Scholar]
- Li W, Hou C, Li Y, Wu C, Ma J. 2022. HexNAcQuest: A tool to distinguish O‑GlcNAc and O‑GalNAc. J. Am. Soc. Mass Spectrom., 33:2008–2012. 10.1021/jasms.2c00172 [DOI] [PubMed] [Google Scholar]
- Li W, Wang H, Yang D, Liu J, Wu J, Ge Y. 2022. Effect of pectin oligosaccharide on quality control of quick‐frozen pumpkin puree. Int. J. Food Sci. Technol., 57:1061–1073. 10.1111/ijfs.15469 [DOI] [Google Scholar]
- Li X, Xu Z, Hong X, Zhang Y, Zou X. 2020. Databases and bioinformatic tools for glycobiology and glycoproteomics. Int. J. Mol. Sci. 21:6727. 10.3390/ijms21186727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu J, Li X, Ma S, Wei Y, Ou J. 2022. One‐step fabrication of nitrogen‐rich linear porous organic polymer‐based micron‐sized sphere for selective enrichment of glycopeptides. Anal. Chim. Acta, 1215:339988. 10.1016/j.aca.2022.339988 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang X, Yang K, Zhu C, Yuan T, Wang J, Li Y, Gao Z. 2021. Identification and expression analysis of the glycosyltransferase GT43 family members in bamboo reveal their potential function in xylan biosynthesis during rapid growth. BMC Genomics, 22:867. 10.1186/s12864-021-08192-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Ma X, Li M, Yi Y, Gao Q, Zhang Y, Zhang L, Zhou D, Xiao S. 2022. Novel β‐cyclodextrin‐based heptavalent glycyrrhetinic acid conjugates: Synthesis, characterization, and anti‐influenza activity. Front. Chem. 10:836955. 10.3389/fchem.2022.836955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Fu B, Zhang Y, Lu H. 2022. Progress of proteomics‐driven precision medicine: From a glycosylation view. Rapid Commun. Mass Spectrom., 36:e9288. 10.1002/rcm.9288 [DOI] [PubMed] [Google Scholar]
- Liang YJ. 2022. Glycosphingolipids in human embryonic stem cells and breast cancer stem cells, and potential cancer therapy strategies based on their structures and functions. Glycoconj. J. 39:177–195. 10.1007/s10719-021-10032-w [DOI] [PubMed] [Google Scholar]
- Liao J, Pan B, Zhuo X, Liao G, Gao Y, Yao Z, Wang L, Wu Q, Pan W, Jiao B, Zhao Q. 2022. β‐1,2‐Mannan‐based glycoconjugates as potential antifungal vaccines. Chinese Chem. Lett. 33:4345–4349. 10.1016/j.cclet.2021.12.065 [DOI] [Google Scholar]
- Liao J, Zhuo X, Pan B, Zou Y, Chai X, Wu Q, Yu S, Pan W, Zhao Q. 2022. Synthesis and preliminary immunologic properties of di‐/trisaccharide‐conjugates related to Bacillus anthracis . Bioorg. Med. Chem. Lett., 76:128986. 10.1016/j.bmcl.2022.128986 [DOI] [PubMed] [Google Scholar]
- Liénard–Mayor T, Yang B, Tran NT, Bruneel A, Guttman A, Taverna M, Mai TD. 2021. High sensitivity capillary electrophoresis with fluorescent detection for glycan mapping. J. Chromatogr., A, 1657:462593. 10.1016/j.chroma.2021.462593 [DOI] [PubMed] [Google Scholar]
- Liénard–Mayor T, Bricteux C, Bendali A, Tran N‐T, Bruneel A, Taverna M, Mai TD. 2022. Lab‐in‐droplet: From glycan sample treatment toward diagnostic screening of congenital disorders of glycosylation. Anal. Chim. Acta, 1221:340150. 10.1016/j.aca.2022.340150 [DOI] [PubMed] [Google Scholar]
- Liew CY, Chan C‐K, Huang S‐P, Cheng Y‐T, Tsai S‐T, Hsu HC, Wang C‐C, Ni C‐K. 2021. De novo structural determination of oligosaccharide isomers in glycosphingolipids using logically derived sequence tandem mass spectrometry. Analyst, 146:7345–7357. 10.1039/D1AN01448J [DOI] [PubMed] [Google Scholar]
- Liew CY, Yen C‐C, Chen J‐L, Tsai S‐T, Pawar S, Wu C‐Y, Ni C‐K. 2021. Structural identification of N‐glycan isomers using logically derived sequence tandem mass spectrometry. Commun. Chem. 4:92. 10.1038/s42004-021-00532-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CY, Chen J‐L, Ni C‐K. 2022. Electrospray ionization in‐source decay of N‐glycans and the effects on N‐glycan structural identification. Rapid Commun. Mass Spectrom., 36:e9352. 10.1002/rcm.9352 [DOI] [PubMed] [Google Scholar]
- Liew CY, Chen J‐L, Tsai S‐T, Ni C‐K. 2022. Identification of side‐reaction products generated during the ammonia‐catalyzed release of N‐glycans. Carbohydr. Res. 522:108686. 10.1016/j.carres.2022.108686 [DOI] [PubMed] [Google Scholar]
- Liew CY, Hsu HC, Nguan H‐S, Huang Y‐C, Zhong Y‐Q, Hung S‐C, Ni C‐K. 2022. The good, the bad, and the ugly memories of carbohydrate fragments in collision‐induced dissociation. J. Am. Soc. Mass Spectrom. 33:1891–1903. 10.1021/jasms.2c00180 [DOI] [PubMed] [Google Scholar]
- Lin H‐Y, Dyakov YA, Lee YT, Ni C‐K. 2021. Temperature dependence of desorbed ions and neutrals and ionization mechanism of matrix‐assisted laser desorption/ionization. J. Am. Soc. Mass Spectrom. 32:95–105. 10.1021/jasms.0c00101 [DOI] [PubMed] [Google Scholar]
- Lin H‐Y, Ni C‐K. 2022. Structural determination of polysaccharides lichenin using logically derived sequence tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 33:335–346. 10.1021/jasms.1c00325 [DOI] [PubMed] [Google Scholar]
- Lin X, Xiao C, Ling L, Guo L, Guo X. 2021. A dual‐mode reactive matrix for sensitive and quantitative analysis of carbohydrates by MALDI‐TOF MS. Talanta, 235:122792. 10.1016/j.talanta.2021.122792 [DOI] [PubMed] [Google Scholar]
- Lin Y, Olukosi OA. 2021. Qualitative and quantitative profiles of jejunal oligosaccharides and cecal short‐chain fatty acids in broiler chickens receiving different dietary levels of fiber, protein and exogenous enzymes. J. Sci. Food Agric. 101:5190–5201. 10.1002/jsfa.11165 [DOI] [PubMed] [Google Scholar]
- Lindstad LJ, Lo G, Leivers S, Lu Z, Michalak L, Pereira GV, Røhr ÅK, Martens EC, McKee LS, Louis P, Duncan SH, Westereng B, Pope PB, La Rosa SL. 2021. Human gut Faecalibacterium prausnitzii deploys a highly efficient conserved system to cross‐feed on β‐mannan‐derived oligosaccharides. mBio, 12:e03628‐03620. 10.1128/mbio.03628-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Xiao C, Ma Y, Jiang L, Wang S, Guo L, Jiang S, Guo X. 2019. 2‐Phenyl‐3‑(p‑aminophenyl) acrylonitrile: A reactive matrix for sensitive and selective analysis of glycans by MALDI‐MS. Anal. Chem. 91:8801–8807. 10.1021/acs.analchem.9b01434 [DOI] [PubMed] [Google Scholar]
- Ling L, Jiang L, Chen Q, Zhao B, Li Y, Guo X. 2021. Rapid and accurate profiling of oligosaccharides in beer by using a reactive matrix via MALDI‐TOF MS. Food Chem., 340:128208. 10.1016/j.foodchem.2020.128208 [DOI] [PubMed] [Google Scholar]
- Ling L, Yu S, Ding C. 2021. 4‐Hydrazinoquinazoline acting as a reactive matrix for the rapid and sensitive analysis of neutral and sialylated glycans using MALDI MS. Analyst, 146:6840–6845. 10.1039/D1AN01452H [DOI] [PubMed] [Google Scholar]
- Liou S‐W, Fang J‐L, Lin H‐W, Tsai T‐W, Huang H‐H, Liang C‐Y, Yang C‐R, Wei G‐T, Yu C‐C. 2021. Effective separation of human milk glycosides using carbon dioxide supercritical fluid chromatography. Chem. Asian J. 16:492–497. 10.1002/asia.202001404 [DOI] [PubMed] [Google Scholar]
- Lippold S, Thavarajah R, Reusch D, Wuhrer M, Nicolardi S. 2021. Glycoform analysis of intact erythropoietin by MALDI FT‐ICR mass spectrometry. Anal. Chim. Acta, 1185:339084. 10.1016/j.aca.2021.339084 [DOI] [PubMed] [Google Scholar]
- Lisztes E, Mező E, Demeter F, Horváth L, Bősze S, Tóth BI, Borbás A, Herczeg M. 2021. Synthesis and cell growth inhibitory activity of six non‐glycosaminoglycan‐type heparin‐analogue trisaccharides. ChemMedChem, 16:1467–1476. 10.1002/cmdc.202000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C‐C, Huo C‐X, Zhai C, Zheng X‐J, Xiong D‐C, Ye X‐S. 2022. Synthesis and immunological evaluation of pentamannose‐based HIV‑1 vaccine candidates. Bioconj. Chem. 33:807–820. 10.1021/acs.bioconjchem.2c00079 [DOI] [PubMed] [Google Scholar]
- Liu D, Liu G, Li Y, Wang Y, Zheng Y, Sha S, Li W, Kameyama A, Dong W. 2021. Rapid glycosylation analysis of mouse serum glycoproteins separated by supported molecular matrix electrophoresis. J. Proteomics, 234:104098. 10.1016/j.jprot.2020.104098 [DOI] [PubMed] [Google Scholar]
- Liu D, De Schutter K, Far J, Staes A, Dewettinck K, Quinton L, Gevaert K, Smagghe G. 2022. RNAi of mannosidase‐Ia in the Colorado potato beetle and changes in the midgut and peritrophic membrane. Pest Manag. Sci. 78:5071–5079. 10.1002/ps.7145 [DOI] [PubMed] [Google Scholar]
- Liu D, Tang W, Huang X‐J, Hu J‐L, Wang J‐Q, Yin J‐Y, Nie S‐P, Xie M‐Y. 2022. Structural characteristic of pectin‐glucuronoxylan complex from Dolichos lablab L. hull. Carbohydr. Polym. 298:120023. 10.1016/j.carbpol.2022.120023 [DOI] [PubMed] [Google Scholar]
- Liu H, Pan Y, Xiong C, Han J, Wang X, Chen J, Nie Z. 2022. Matrix‐assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) for in situ analysis of endogenous small molecules in biological samples. Trends Anal. Chem., 157:116809. 10.1016/j.trac.2022.116809 [DOI] [Google Scholar]
- Liu J, Liang M, Bi X, Cao S, Zhang C, Zhu Z. 2021. Study of the adherence of Escherichia coli 83972 on α‐biphenyl mannoside‐presenting PDMS surfaces. Colloid Interface Sci. Commun. 45:100507. 10.1016/j.colcom.2021.100507 [DOI] [Google Scholar]
- Liu J, Xiao G, Zhou W, Yang J, Wang Y, Wu Y, Cheng X, Sun Z. 2021. Various novel colistin resistance mechanisms interact to facilitate adaptation of Aeromonas hydrophila to complex colistin environments. Antimicrob. Agents Chemother. 65:e00071–00021. 10.1128/aac.00071-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K‐M, Wang P‐Y, Guo Z‐Y, Xiong D‐C, Qin X‐J, Liu M, Liu M, Xue W‐Y, Ye X‐S. 2022. Iterative synthesis of 2‐deoxyoligosaccharides enabled by stereoselective visible‐light‐promoted glycosylation. Angew. Chem. Int. Ed., 61:e202114726. 10.1002/anie.202114726 [DOI] [PubMed] [Google Scholar]
- Liu K, Liu H‐Y, Tao X, Li Z‐J, Si C‐L, Yu H‐Y, Yan X‐N, Nie S, Wang J‐H, Cong R‐Z, Wei R, Wang S‐Y. 2021. A new triterpene glycoside from Pinus pumila . Chem. Nat. Compds., 57:115–119. 10.1007/s10600-021-03294-1 [DOI] [Google Scholar]
- Liu L, Wang Y, Zhang J, Wang S. 2021. Advances in the chemical constituents and chemical analysis of Ginkgo biloba leaf, extract, and phytopharmaceuticals. J. Pharmaceut. Biomed. Anal. 193:113704. 10.1016/j.jpba.2020.113704 [DOI] [PubMed] [Google Scholar]
- Liu L, Zhu B, Fang Z, Zhang N, Qin H, Guo Z, Liang X, Yao Z, Ye M. 2021. Automated intact glycopeptide enrichment method facilitating highly reproducible analysis of serum site‐specific N‑glycoproteome. Anal. Chem. 93:7473–7480. 10.1021/acs.analchem.1c00645 [DOI] [PubMed] [Google Scholar]
- Liu N, Yu W, Guo X, Chen J, Xia D, Yu J, Li D. 2022. Oxidative cleavage of cellulose in the horse gut. Microb. Cell Factories, 21:38. 10.1186/s12934-022-01767-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li H, Li R, Geng Y, Gong J, Xu H, Xu Z, Shi J. 2022. Nanoencapsulation of chitooligosaccharides enhances its oral bioavailability and anti‐liver fibrotic effects. Food Res. Int. 157:111471. 10.1016/j.foodres.2022.111471 [DOI] [PubMed] [Google Scholar]
- Liu Q, Zhou L, Xie X, Fan D, Ouyang X, Fan W, Qiu X. 2022. Enhanced production and separation of short‐chain glucan oligomers from corn stover in an unacidified LiBr molten salt hydrate via pre‐extraction of hemicellulose. Green Chem., 24:8812–8819. 10.1039/D2GC03396H [DOI] [Google Scholar]
- Liu R, Gao W, Yang J, Zhang S, Wang C, Lin J, Zhang S, Yu J, Tang K. 2022. A novel graphene oxide/chitosan foam incorporated with metal‐organic framework stationary phase for simultaneous enrichment of glycopeptide and phosphopeptide with high efficiency. Anal. Bioanal. Chem. 414:2251–2263. 10.1007/s00216-021-03861-z [DOI] [PubMed] [Google Scholar]
- Liu S, Fang R, Zhang Y, Chen L, Huang N, Yu K, Zhou C, Cao J, Zhou T. 2021. Characterization of resistance mechanisms of Enterobacter cloacae complex coresistant to carbapenem and colistin. BMC Microbiol., 21:208. 10.1186/s12866-021-02250-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Schulz BL. 2021. Biopharmaceutical quality control with mass spectrometry. Bioanalysis, 13:1275–1291. 10.4155/bio-2021-0123 [DOI] [PubMed] [Google Scholar]
- Liu S, Yu Y, Liu Y, Lin J, Fu Y, Cheng L, Liu X. 2021. Revealing the changes of IgG subclass‐specific N‐glycosylation in colorectal cancer progression by high‐throughput assay. Proteomics Clin. Appl. 15:2000022. 10.1002/prca.202000022 [DOI] [PubMed] [Google Scholar]
- Liu W, Han Y, Xin X, Chen L, Liu Y, Liu C, Zhang X, Jin M, Jin J, Gao Z, Huang W. 2022. Biomimetic and temporal‑controlled nanocarriers with ileum transporter targeting for achieving oral administration of chemotherapeutic drugs. J. Nanobiotechnol. 20:281. 10.1186/s12951-022-01460-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Z, Li Z, Zhang Y, Lu H. 2022. Mass spectrometry‐based analysis of IgG glycosylation and its applications. Int. J. Mass Spectrom. 474:116799. 10.1016/j.ijms.2022.116799 [DOI] [Google Scholar]
- Liu X, Wang Q, Lauber MA. 2022. High sensitivity acidic N‐glycan profiling with MS‐enhancing derivatization and mixed mode chromatography. J. Chromatogr., B, 1191:123120. 10.1016/j.jchromb.2022.123120 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ma J, Shi R, Li T, Yan Q, Jiang Z, Yang S. 2021. Biochemical characterization of a β‐N‐acetylhexosaminidase from Catenibacterium mitsuokai suitable for the synthesis of lacto‐N‐triose II. Proc. Biochem. 102:360–368. 10.1016/j.procbio.2021.01.013 [DOI] [Google Scholar]
- Liu Y, Ma W, He Y, Chen Z, Lin Z. 2021. Facile synthesis of hydrophilic magnetic mesoporous silica microspheres for selective enrichment of glycopeptides and glycans. Anal. Lett. 54:966–978. 10.1080/00032719.2020.1789161 [DOI] [Google Scholar]
- Liu Y, Wang Z, Yu F, Li M, Zhu H, Wang K, Meng M, Zhao W. 2021. The adjuvant of α‐galactosylceramide presented by gold nanoparticles enhances antitumor immune responses of MUC1 antigen‐based tumor vaccines. Int. J. Nanomed. 16:403–420. 10.2147/ijn.s273883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Larrouy‐Maumus G. 2022. Lipids and glycolipids as biomarkers of mycobacterial infections. In: Fatima Z, Canaan S, Editors., Biology of Mycobacterial Lipids London, San Diego, Cambridge, Oxford: Elsevier. p 83–104. 10.1016/B978-0-323-91948-7.00014-2 [DOI] [Google Scholar]
- Liu Y, Yang L, Li H, Liu J, Tian R. 2022. Derivatization strategy for sensitive identification of neutral and acidic glycosphingolipids using RPLC‐MS. Int. J. Mass Spectrom. 482:116937. 10.1016/j.ijms.2022.116937 [DOI] [Google Scholar]
- Liu Z, Dai X, Xu Q, Sun X, Liu Y. 2022. Fluorescence sensing of glutathione thiyl radical by BODIPY modified β‐ cyclodextrin. Chinese J. Chem. 40:493–499. 10.1002/cjoc.202100662 [DOI] [Google Scholar]
- Liyanage LA, Harris MS, Cook GA. 2021. In vitro glycosylation of membrane proteins using N‑glycosyltransferase. ACS Omega, 6:12133–12142. 10.1021/acsomega.1c00835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage OT, Quintero AV, Hatvany JB, Gallagher ES. 2021. Distinguishing carbohydrate isomers with rapid hydrogen/deuterium exchange‐mass spectrometry. J. Am. Soc. Mass Spectrom. 32:152–156. 10.1021/jasms.0c00314 [DOI] [PubMed] [Google Scholar]
- Lo Barco T, Osanni E, Bordugo A, Rodella G, Iascone M, Tenconi R, Barone R, Bernardina BD, Cantalupo G. 2021. Epilepsy and movement disorders in CDG: Report on the oldest‐known MOGS‐CDG patient. Am. J. Med. Genet. A, 185A:219–222. 10.1002/ajmg.a.61916 [DOI] [PubMed] [Google Scholar]
- Lobasso S, Tanzarella P, Mannavola F, Tucci M, Silvestris F, Felici C, Ingrosso C, Corcelli A, Lopalco P. 2021. A lipidomic approach to identify potential biomarkers in exosomes from melanoma cells with different metastatic potential. Front. Physiol. 12:748895. 10.3389/fphys.2021.748895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L, Sun L, Ding D, Chen K, Lin Q, Ding S. 2021. Two C1‐oxidizing lytic polysaccharide monooxygenases from Ceriporiopsis subvermispora enhance the saccharification of wheat straw by a commercial cellulase cocktail. Proc. Biochem. 110:243–250. 10.1016/j.procbio.2021.08.013 [DOI] [Google Scholar]
- López‐Cortés R, Gómez BB, Vázquez‐Estévez S, Pérez‐Fentes D, Núñez C. 2021. Blood‐based protein biomarkers in bladder urothelial tumors. J. Proteomics, 247:104329. 10.1016/j.jprot.2021.104329 [DOI] [PubMed] [Google Scholar]
- Lou J, Wang D, Yuan J, Xu J, Fan X. 2021. Improving the anti‐wrinkle and hydrophilicity performance of cotton fabric via crosslinking cellulose with carboxylated polyaldehyde trehalose. Cellulose, 28:5135–5149. 10.1007/s10570-021-03831-9 [DOI] [Google Scholar]
- Lou J, Yuan J, Wang Q, Fan X. 2021. Synthesis and application of oxidized trehalose as a hydrophilic anti‐crease finishing reagent for cotton fabric. Fibers Polym. 22:1016–1024. 10.1007/s12221-021-9661-5 [DOI] [Google Scholar]
- Lou X, Miley G, Van Dongen JLJ. 2021. Dual roles of [CHCA + Na/K/Cs]+ as a cation adduct or a protonated salt for analyte ionization in matrix‐assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom., 35:e9111. 10.1002/rcm.9111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan F, Ji Y, Peng L, Liu Q, Cao H, Yang Y, He X, Zeng N. 2021. Extraction, purification, structural characteristics and biological properties of the polysaccharides from Codonopsis pilosula: A review. Carbohydr. Polym. 261:117863. 10.1016/j.carbpol.2021.117863 [DOI] [PubMed] [Google Scholar]
- Lumbroso A, Berthonneau C, Beaudet I, Quintard J‐P, Planchat A, García‐Moreno MI, Mellet CO, Le Grognec E. 2021. A versatile stereocontrolled synthesis of 2‐deoxyiminosugar C‐glycosides and their evaluation as glycosidase inhibitors. Org. Biomol. Chem. 19:1083–1099. 10.1039/D0OB02249G [DOI] [PubMed] [Google Scholar]
- Lung TWF, Charytonowicz D, Beaumont KG, Shah SS, Sridhar SH, Gorrie CL, Mu A, Hofstaedter CE, Varisco D, McConville TH, Drikic M, Fowler B, Urso A, Shi W, Fucich D, Annavajhala MK, Khan IN, Oussenko I, Francoeur N, Smith ML, Stockwell BR, Lewis IA, Hachani A, Baskota SU, Uhlemann A‐C, Ahn D, Ernst RK, Howden BP, Sebra R, Prince A. 2022. Klebsiella pneumoniae induces host metabolic stress that promotes tolerance to pulmonary infection. Cell Metab., 34:761–774. 10.1016/j.cmet.2022.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Yan S, Zhang Y, Zhou J, Lan F, Wu Y. 2021. Bifunctional magnetic covalent organic framework for simultaneous enrichment of phosphopeptides and glycopeptides. Anal. Chim. Acta, 1177:338761. 10.1016/j.aca.2021.338761 [DOI] [PubMed] [Google Scholar]
- Luo L, Song X, Chang X, Huang S, Zhou Y, Yang S, Zhu Y, Zhang L, Wu Y, Zhang J, Zhou Z, Wu M. 2022. Detailed structural analysis of the immunoregulatory polysaccharides from the Mycobacterium bovis BCG. Molecules, 27:5691. 10.3390/molecules27175691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Song C, Mao J, Peng Z, Sun S, Zhang Y, Yu A, Zhang W, Zhao W, Ouyang G. 2022. Developing a noncontact heating matrix spraying apparatus with controllable matrix film formation for MALDI mass spectrometry imaging. Anal. Chem. 94:12136–12143. 10.1021/acs.analchem.2c02192 [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhao X, Gao Z, Wang H, Liu Y, Guo C, Pan Y. 2022. Pd nanoparticles decorated thiol‐functionalized MOF as an efficient matrix for differentiation and quantitation of oligosaccharide isomers by laser desorption/ionization mass spectrometry. Anal. Chim. Acta, 1202:339665. 10.1016/j.aca.2022.339665 [DOI] [PubMed] [Google Scholar]
- Luo Z, Health SL, Li M, Yang H, Wu Y, Collins M, Deeks SG, Martin JN, Scott A, Jiang W. 2022. Variation in blood microbial lipopolysaccharide (LPS) contributes to immune reconstitution in response to suppressive antiretroviral therapy in HIV. eBioMedicine, 80:104037. 10.1016/j.ebiom.2022.104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupa D, Płaziński W, Michna A, Wasilewska M, Pomastowski P, Gołębiowski A, Buszewski B, Adamczyk Z. 2022. Chitosan characteristics in electrolyte solutions: Combined molecular dynamics modeling and slender body hydrodynamics. Carbohydr. Polym. 292:119676. 10.1016/j.carbpol.2022.119676 [DOI] [PubMed] [Google Scholar]
- Lupi GA, Valtierra FXS, Cabrera G, Spinelli R, Siano ÁS, González V, Osuna A, Oresti GM, Marcipar I. 2022. Development of low‐cost cage‐like particles to formulate veterinary vaccines. Vet. Immunol. Immunopathol. 251:110460. 10.1016/j.vetimm.2022.110460 [DOI] [PubMed] [Google Scholar]
- Lütteke T. 2021. Glycosciences.De: Databases and tools to support research in glycomics and glycoproteomics. In: Barchi JJ,Jr., Editor, Comprehensive Glycoscience (Second edition). Amsterdam, Kidlington UK, Cambridge MA: Elsevier. p 432–438. 10.1016/b978-0-12-819475-1.00020-1 [DOI] [Google Scholar]
- Lyman DF, Bell A, Black A, Dingerdissen H, Cauley E, Gogate N, Liu D, Joseph A, Kahsay R, Crichton DJ, Mehta A, Mazumder R. 2022. Modeling and integration of N‐glycan biomarkers in a comprehensive biomarker data model. Glycobiology, 32:855–870. 10.1093/glycob/cwac046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Tang R, Wang Y, Ma S, Tang S, Zhang J, Ou J. 2021. One‐step preparation of cyclen‐containing hydrophilic polymeric monolithic materials via epoxy‐amine ring‐opening reaction and their application in enrichment of N‐glycopeptides. Talanta, 225:122049. 10.1016/j.talanta.2020.122049 [DOI] [PubMed] [Google Scholar]
- Ma C, Xie L, Wang X, Liang K, Kong B. 2022. Interfacial assembly of functional mesoporous nanomatrices for laser desorption/ionization mass spectrometry. Nano Today, 42:101365. 10.1016/j.nantod.2021.101365 [DOI] [Google Scholar]
- Ma G, Zhao X, Guo M, Liu Y, Shi K, Guo C, Pan Y. 2022. 6‐Glycosylaminoquinoline‐assisted LDI MS for detection and imaging of small molecules with enhanced detection selectivity and sensitivity. Anal. Chim. Acta, 1201:339620. 10.1016/j.aca.2022.339620 [DOI] [PubMed] [Google Scholar]
- Ma J, Wu C, Hart GW. 2021. Analytical and biochemical perspectives of protein O‑GlcNAcylation. Chem. Rev. 121:1513–1581. 10.1021/acs.chemrev.0c00884 [DOI] [PubMed] [Google Scholar]
- Ma K, Shi J, Pei Y, Pei Z. 2022. A carrier‐free supramolecular nanoprodrug based on lactose‐functionalized dimeric camptothecin via self‐assembly in water for targeted and fluorescence imaging‐guided chemo‐photodynamic therapy. J. Colloid Interf. Sci., 609:353–363. 10.1016/j.jcis.2021.12.002 [DOI] [PubMed] [Google Scholar]
- Ma L, Liu Z, Kong Z, Wang M, Li T, Zhu H, Wan Q, Liu D, Shen Q. 2021. Functional characterization of a novel copper‐dependent lytic polysaccharide monooxygenase TgAA11 from Trichoderma guizhouense NJAU 4742 in the oxidative degradation of chitin. Carbohydr. Polym. 258:117708. 10.1016/j.carbpol.2021.117708 [DOI] [PubMed] [Google Scholar]
- Ma L, Li G, Xu H, Liu Z, Wan Q, Liu D, Shen Q. 2022. Structural and functional study of a novel lytic polysaccharide monooxygenase cPMO2 from compost sample in the oxidative degradation of cellulose. Chem. Eng. J. 433:134509. 10.1016/j.cej.2022.134509 [DOI] [Google Scholar]
- Ma Q, Lin J, Guan M, Liang H, Liu Q. 2022. Enhanced cello‐oligosaccharides production from cellulose hydrolysis in molten salt hydrate over lignin‐based hyper‐cross‐linked polymer (LHCP) adsorption. Appl. Catal. A, Gen. 644:118808. 10.1016/j.apcata.2022.118808 [DOI] [Google Scholar]
- Ma Q, Wang W, Yang X, Chen Y, Liu Y, Chen H, Zhao Y. 2022. Development and application of a sensitive phosphonium‐hydrazide oligosaccharide labelling reagent in capillary electrophoresis‐electrospray ionization‐mass spectrometry. J. Chromatogr., A, 1680:463409. 10.1016/j.chroma.2022.463409 [DOI] [PubMed] [Google Scholar]
- Ma S, Chen F, Zhang M, Yuan H, Ouyang G, Zhao W, Zhang S, Zhao Y. 2022. Carboxyl‐based CPMP tag for ultrasensitive analysis of disaccharides by negative tandem mass spectrometry. Anal. Chem. 94:9557–9563. 10.1021/acs.analchem.2c00287 [DOI] [PubMed] [Google Scholar]
- Ma W, Li J, Li X, Bai Y, Liu H. 2021. Nanostructured substrates as matrices for surface assisted laser desorption/ionization mass spectrometry: A progress report from material research to biomedical applications. Small Meth. 5:2100762. 10.1002/smtd.202100762 [DOI] [PubMed] [Google Scholar]
- Ma W, Yang B, Li J, Liu M, Li X, Liu H. 2022. Maltose‑functional metal‐organic framework assisted laser desorption/ionization mass spectrometry for small biomolecule determination. Microchim. Acta, 189:253. 10.1007/s00604-022-05338-x [DOI] [PubMed] [Google Scholar]
- Ma X, Li Y, Kondo Y, Shi H, Han J, Jiang Y, Bai X, Archer‐Hartmann SA, Azadi P, Ruan C, Fu J, Xia L. 2021. Slc35a1 deficiency causes thrombocytopenia due to impaired megakaryocytopoiesis and excessive platelet clearance in the liver. Haematologica, 106:759–769. 10.3324/haematol.2019.225987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. 2022. Recent advances in mass spectrometry‐based structural elucidation techniques. Molecules, 27:6466. 10.3390/molecules27196466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Jiang Q, Wang X, Xiao G. 2022. Total synthesis of Cordyceps militaris glycans via stereoselective orthogonal one‐pot glycosylation and α‑glycosylation strategies. Org. Lett. 24:7950–7954. 10.1021/acs.orglett.2c03081 [DOI] [PubMed] [Google Scholar]
- Mabashi‐Asazuma H, Jarvis DL. 2021. A new insect cell line engineered to produce recombinant glycoproteins with cleavable N‐glycans. J. Biol. Chem. 298:101454. 10.1016/j.jbc.2021.101454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald SS, Pereira JH, Liu F, Tegl G, DeGiovanni A, Wardman JF, Deutsch S, Yoshikuni Y, Adams PD, Withers SG. 2022. A synthetic gene library yields a previously unknown glycoside phosphorylase that degrades and assembles poly‐β‐1,3‐GlcNAc, completing the suite of β‑linked GlcNAc polysaccharides. ACS Cent. Sci. 8:430–440. 10.1021/acscentsci.1c01570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo‐da‐Silva J, Santiago VF, Rosa‐Fernandes L, Marinho CRF, Palmisano G. 2021. Protein glycosylation in extracellular vesicles: Structural characterization and biological functions. Molec. Immunol. 135:226–246. 10.1016/j.molimm.2021.04.017 [DOI] [PubMed] [Google Scholar]
- MacFarlane DR, Forsyth M, Izgorodina EI, Abbott AP, Annat G, Fraser K. 2009. On the concept of ionicity in ionic liquids. Phys. Chem. Chem. Phys. 11:4962–4967. 10.1039/B900201D [DOI] [PubMed] [Google Scholar]
- Madunić K, Wagt S, Zhang T, Wuhrer M, Lageveen‐Kammeijer GSM. 2021. Dopant‐enriched nitrogen gas for enhanced electrospray ionization of released glycans in negative ion mode. Anal. Chem. 93:6919–6923. 10.1021/acs.analchem.1c00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghodia AB, Geisler C, Jarvis DL. 2021. A new bacmid for customized protein glycosylation pathway engineering in the baculovirus‐insect cell system. ACS Chem. Biol. 16:1941–1950. 10.1021/acschembio.0c00974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevegowda SH, Ruan L, Zhang J, Hou S, Raju C, Chan‐Park MB. 2021. Synthesis of dimeric and tetrameric trithiomannoside clusters through convenient photoinitiated thiol‐ene click protocol for efficient inhibition of Gram‐negative bacteria. J. Carbohydr. Chem. 40:83–96. 10.1080/07328303.2021.1928154 [DOI] [Google Scholar]
- Mahour R, Marichal‐Gallardo PA, Rexer TFT, Reichl U. 2021. Multi‐enzyme cascades for the in vitro synthesis of guanosine diphosphate l‐fucose. ChemCatChem, 13:1981–1989. 10.1002/cctc.202001854 [DOI] [Google Scholar]
- Maia M, McCann A, Malherbe C, Far J, Cunha J, Eiras‐Dias J, Cordeiro C, Eppe G, Quinton L, Figueiredo A, De Pauw E, Silva MS. 2022. Grapevine leaf MALDI‐MS imaging reveals the localisation of a putatively identified sucrose metabolite associated to Plasmopara viticola development. Front. Plant Sci. 13:1012636. 10.3389/fpls.2022.1012636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrydaki E, Kotidis P, Polizzi KM, Kontoravdi C. 2021. Hitting the sweet spot with capillary electrophoresis: Advances in N‐glycomics and glycoproteomics. Curr. Opin. Biotechnol. 71:182–190. 10.1016/j.copbio.2021.07.013 [DOI] [PubMed] [Google Scholar]
- Malaker SA, Quanico J, Raffo‐Romero A, Kobeissy F, Aboulouard S, Tierny D, Bertozzi CR, Fournier I, Salzet M. 2022. On‐tissue spatially resolved glycoproteomics guided by N‐glycan imaging reveal global dysregulation of canine glioma glycoproteomic landscape. Cell Chem. Biol. 29:30–42. 10.1016/j.chembiol.2021.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallakuntla MK, Podile AR. 2021. Catalytic efficiency of a multi‐domain transglycosylating chitinase from Enterobacter cloacae subsp. cloacae (EcChi2) is influenced by polycystic kidney disease domains. Enzyme Microb. Technol. 143:109702. 10.1016/j.enzmictec.2020.109702 [DOI] [PubMed] [Google Scholar]
- Man L, Klare WP, Dale AL, Cain JA, Cordwell SJ. 2021. Integrated mass spectrometry‐based multi‐omics for elucidating mechanisms of bacterial virulence. Biochem. Soc. Trans. 49:1905–1926. 10.1042/bst20191088 [DOI] [PubMed] [Google Scholar]
- Manabe N, Ohno S, Matsumoto K, Kawase T, Hirose K, Masuda K, Yamaguchi Y. 2022. A data set of ion mobility collision cross sections and liquid chromatography retention times from 71 pyridylaminated N‑linked oligosaccharides. J. Am. Soc. Mass Spectrom. 33:1772–1783. 10.1021/jasms.2c00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini F, Gasperini G, Rossi O, Aruta MG, Raso MM, Alfini R, Biagini M, Necchi F, Micoli F. 2021. Dissecting the contribution of O‑antigen and proteins to the immunogenicity of Shigella sonnei generalized modules for membrane antigens (GMMA). Sci. Rep. 11:906. 10.1038/s41598-020-80421-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manker LP, Dick GR, Demongeot A, Hedou MA, Rayroud C, Rambert T, Jones MJ, Sulaeva I, Vieli M, Leterrier Y, Potthast A, Maréchal F, Michaud V, Klok H‐A, Luterbacher JS. 2022. Sustainable polyesters via direct functionalization of lignocellulosic sugars. Nat. Chem. 14:976–984. 10.1038/s41557-022-00974-5 [DOI] [PubMed] [Google Scholar]
- Marchetti R, Forgione RE, Fabregat FN, Di Carluccio C, Molinaro A, Silipo A. 2021. Solving the structural puzzle of bacterial glycome. Curr. Opin. Struct. Biol. 68:74–83. 10.1016/j.sbi.2020.12.003 [DOI] [PubMed] [Google Scholar]
- Marie A‐L, Ray S, Lu S, Jones J, Ghiran I, Ivanov AR. 2021. High‐sensitivity glycan profiling of blood‐derived immunoglobulin G, plasma, and extracellular vesicle isolates with capillary zone electrophoresis‐mass spectrometry. Anal. Chem. 93:1991–2002. 10.1021/acs.analchem.0c03102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariethoz J, Alocci D, Karlsson NG, Packer NH, Lisacek F. 2022. An interactive view of glycosylation. Method Mol. Biol. 2370:41–65. 10.1007/978-1-0716-1685-7_3 [DOI] [PubMed] [Google Scholar]
- Martín‐Saiz L, Mosteiro L, Solano‐Iturri JD, Rueda Y, Martín‐Allende J, Imaz I, Olano I, Ochoa B, Fresnedo O, Fernández JA, Larrinaga G. 2021. High‐resolution human kidney molecular histology by imaging mass spectrometry of lipids. Anal. Chem. 93:9364–9372. 10.1021/acs.analchem.1c00649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Goyard D, Margalit A, Doherty K, Renaudet O, Kavanagh K, Velasco‐Torrijos T. 2021. Multivalent presentations of glycomimetic inhibitor of the adhesion of fungal pathogen Candida albicans to human buccal epithelial cells. Bioconj. Chem., 32:971–982. 10.1021/acs.bioconjchem.1c00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H, Masterson H, Kavanagh K, Velasco‐Torrijos T. 2021. The synthesis and evaluation of multivalent glycopeptoids as inhibitors of the adhesion of Candida albicans . Pathogens, 10:572. 10.3390/pathogens10050572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín MA, Wuhrer M, Falck D. 2021. Serum and plasma immunoglobulin G Fc N‑glycosylation is stable during storage. J. Proteome Res. 20:2935–2941. 10.1021/acs.jproteome.1c00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Rodriguez A, Beltran‐Garcia C, Valdez‐Salas B, Santacruz‐Ruvalcaba F, Di Mascio P, Beltran‐Garcia MJ. 2022. Micropropagation of seed‐derived clonal lines of the endangered Agave marmorata Roezl and their compatibility with endophytes. Biology, 11:1423. 10.3390/biology11101423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JER, Thomas B, Flitsch SL. 2021. Glycan array technology. Adv. Biochem. Eng. Biotechnol. 175:435–456. 10.1007/10_2019_112 [DOI] [PubMed] [Google Scholar]
- Mastellone J, Kabir KMM, Donald WA. 2022. Separation of disaccharide epimers, anomers and connectivity isomers by high resolution differential ion mobility mass spectrometry. Anal. Chim. Acta, 1206:339783. 10.1016/j.aca.2022.339783 [DOI] [PubMed] [Google Scholar]
- Mathur B, Shajahan A, Arif W, Chen Q, Hand NJ, Abramowitz LK, Schoonjans K, Rader DJ, Kalsotra A, Hanover JA, Azadi P, Anakk S. 2021. Nuclear receptors FXR and SHP regulate protein N‐glycan modifications in the liver. Sci. Adv. 7:eabf4865. 10.1126/sciadv.abf4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matros A, Houston K, Tucker MR, Schreiber M, Berger B, Aubert MK, Wilkinson LG, Witzel K, Waugh R, Seiffert U, Burton RA. 2021. Genome‐wide association study reveals the genetic complexity of fructan accumulation patterns in barley grain. J. Exp. Bot. 72:2383–2402. 10.1093/jxb/erab002 [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Endo D, Adachi R, Koyama T, Matsushita T, Hatano K. 2022. Chemical modification of CNN 1. Complete protection of CNN. Tetrahedron Lett., 103:153986. 10.1016/j.tetlet.2022.153986 [DOI] [Google Scholar]
- Matsushita S, Hasegawa T, Hiraoka M, Hayashi A, Suzuki Y. 2021. TLC‐based MS imaging analysis of glycosphingolipids and glycerin fatty acid esters after 1,2‐dichloroethane washing. Anal. Sci. 37:1491–1495. 10.2116/analsci.21c009 [DOI] [PubMed] [Google Scholar]
- Matsushita T, Hinou H, Nishimura S‐I. 2022. Generation 6 polyamidoamine dendrimer provides an ideal nanoparticular platform for enzyme‐assisted synthesis of glycopeptides having bulky and complex glycans. Chem. Lett. 51:1044–1048. 10.1246/cl.220344 [DOI] [Google Scholar]
- Matsushita T, Toda N, Koyama T, Hatano K, Matsuoka K. 2022. Dendritic maleimide‐thiol adducts carrying pendant glycosides as high‐affinity ligands. Bioorg. Chem. 128:106061. 10.1016/j.bioorg.2022.106061 [DOI] [PubMed] [Google Scholar]
- Matsuzaki C, Shiraishi T, Chiou T‐Y, Nakashima Y, Higashimura Y, Yokota S‐i, Yamamoto K, Takahashif T. 2022. Role of lipoteichoic acid from the genus Apilactobacillus in inducing a strong IgA response. Appl. Environ. Microbiol. 88:e00190–00122. 10.1128/aem.00190-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T, Watanabe M, Nakamichi Y, Kameyama A, Kojima N, Yaoi K. 2022. Characterization of an extracellular α‑xylosidase involved in xyloglucan degradation in Aspergillus oryzae . Appl. Microbiol. Biotechnol. 106:675–687. 10.1007/s00253-021-11744-7 [DOI] [PubMed] [Google Scholar]
- Matthews AM, Biel TG, Ortega‐Rodriguez U, Falkowski VM, Bush X, Faison T, Xie H, Agarabi C, Rao VA, Ju T. 2022. SARS‐CoV‐2 spike protein variant binding affinity to an angiotensin‐converting enzyme 2 fusion glycoproteins. PLoS One, 17:e0278294. 10.1371/journal.pone.0278294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavliutova L, Verduci E, Sellergren B. 2021. Combinatorial design of a sialic acid imprinted binding site exploring a dual ion receptor approach. RSC Adv., 11:34329–34337. 10.1039/d1ra06962d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard JC, Chalkley RJ. 2021. Methods for enrichment and assignment of N‐acetylglucosamine modification sites. Mol. Cell. Proteomics, 20:100031. 10.1074/mcp.r120.002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzaglia A, Di Natale G, Tosto R, Scala A, Sortino G, Piperno A, Casaletto MP, Riminucci A, Giuffrida ML, Mineo PG, Villari V, Micali N, Pappalardo G. 2022. KLVFF oligopeptide‐decorated amphiphilic cyclodextrin nanomagnets for selective amyloid beta recognition and fishing. J. Colloid Interf. Sci., 613:814–826. 10.1016/j.jcis.2022.01.051 [DOI] [PubMed] [Google Scholar]
- McDowell CT, Klamer Z, Hall J, West CA, Wisniewski L, Powers TW, Angel PM, Mehta AS, Lewin DN, Haab BB, Drake RR. 2021. Imaging mass spectrometry and lectin analysis of N‐linked glycans in carbohydrate antigen‐defined pancreatic cancer tissues. Mol. Cell. Proteomics, 20:100012. 10.1074/mcp.ra120.002256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen RAH, Hermann M, Metwally H, Donovan K, Liu C, Le Blanc JCY, Covey TR, Richard O. 2022. Discontinuously dewetting solvent arrays: Droplet formation and poly‐synchronous surface extraction for mass spectrometry imaging applications. Anal. Chem. 94:7219–7228. 10.1021/acs.analchem.2c00161 [DOI] [PubMed] [Google Scholar]
- McGuire TM, Bowles J, Deane E, Farrar EHE, Grayson MN, Buchard A. 2021. Control of crystallinity and stereocomplexation of synthetic carbohydrate polymers from d‐ and l‐xylose. Angew. Chem. Int. Ed., 60:4524–4528. 10.1002/anie.202013562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TM, Clark EF, Buchard A. 2021. Polymers from sugars and cyclic anhydrides: Ring‐opening copolymerization of a d‑xylose anhydrosugar oxetane. Macromolecules, 54:5094–5105. 10.1021/acs.macromol.1c00365 [DOI] [Google Scholar]
- McKitrick TR, Bernard SM, Noll AJ, Collins BC, Goth CK, McQuillan AM, Heimburg‐Molinaro J, Herrin BR, Wilson IA, Cooper MD, Cummings RD. 2021. Novel lamprey antibody recognizes terminal sulfated galactose epitopes on mammalian glycoproteins. Commun. Biol. 4:674. 10.1038/s42003-021-02199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod E, Magnelli P, Shi Z. 2021. Use of exoglycosidases for the structural characterization of glycans. Method. Mol. Biol. 2271:273–280. 10.1007/978-1-0716-1241-5_19 [DOI] [PubMed] [Google Scholar]
- McQuiston A, Scott D, Nord D, Langerude L, Pelaez A, Machuca T, Mehta A, Drake RR, Christie JD, Angel P, Atkinson C. 2022. Pro‐inflammatory IgG1 N‐glycan signature correlates with primary graft dysfunction onset in COPD patients. Transplant. Immunol. 71:101491. 10.1016/j.trim.2021.101491 [DOI] [PubMed] [Google Scholar]
- Mealer RG, Williams SE, Noel M, Yang B, D'Souza AK, Nakata T, Graham DB, Creasey EA, Cetinbas M, Sadreyev RI, Scolnick EM, Woo CM, Smoller JW, Xavier RJ, Cummings RD. 2022. The schizophrenia‐associated variant in SLC39A8 alters protein glycosylation in the mouse brain. Mol. Psychiatry, 27:1405–1415. 10.1038/s41380-022-01490-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors PW, Dasari S, Kohlhagen MC, Kourelis T, Go RS, Muchtar E, Gertz MA, Kumar SK, Buadi FK, Willrich MAV, Lust JA, Kapoor P, Lacy MQ, Dingli D, Hwa Y, Fonder A, Hobbs M, Hayman S, Warsame R, Leung NR, Lin Y, Gonsalves W, Siddiqui M, Kyle RA, Rajkumar SV, Murray DL, Dispenzieri A. 2021. MASS‐FIX for the detection of monoclonal proteins and light chain N‐glycosylation in routine clinical practice: A cross‐sectional study of 6315 patients. Blood Cancer J. 11:110. 10.1038/s41408-021-00496-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors PW, Kohlhagen MC, Dasari S, Willrich MAV, Gertz MA, Kumar SK, Lacy MQ, Murray DL, Dispenzieri A. 2021. Belantamab mafodotin detection by MASS‐FIX and immunofixation. Clin. Chem. Lab. Med. 59:e430–e433. 10.1515/cclm-2021-0326 [DOI] [PubMed] [Google Scholar]
- Mendes AR, Quelhas D, Correia J, Coelho MP, Bandeira A, Martins E. 2022. Congenital disorders of glycosylation. Birth Growth Med. J. 31:38–54. 10.25753/BirthGrowthMJ.v31.i1.26341 [DOI] [Google Scholar]
- Merdas M, Lagarrigue M, Vanbellingen Q, Umbdenstock T, Da Violante G, Pineau C. 2021. On‐tissue chemical derivatization reagents for matrix‐assisted laser desorption/ionization mass spectrometry imaging. J. Mass Spectrom. 56:e4731. 10.1002/jms.4731 [DOI] [PubMed] [Google Scholar]
- Mert‐Ozupek N, Basbinar Y, Uysal‐Kilic T, Koz O, Ellidokuz H, Cavas L. 2022. Semi‐purified saponins of Holothuria poli associated antiproliferation in tumor cell lines. Nutr. Cancer, 74:1511–1518. 10.1080/01635581.2021.1952630 [DOI] [PubMed] [Google Scholar]
- Merx J, Hintzen JCJ, Proietti G, Elferink H, Wang Y, Porzberg MRB, Sondag D, Bilgin N, Park J, Mecinović J, Boltje TJ. 2022. Investigation of in vitro histone H3 glycosylation using H3 tail peptides. Sci. Rep. 12:19251. 10.1038/s41598-022-21883-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mery A, Jawhara S, François N, Cornu M, Poissy J, Martinez‐Esparza M, Poulain D, Sendid B, Guerardel Y. 2022. Identification of fungal trehalose for the diagnosis of invasive candidiasis by mass spectrometry. Biochim. Biophys, Acta ‐ Gen. Sub. 1866:130083. 10.1016/j.bbagen.2022.130083 [DOI] [PubMed] [Google Scholar]
- Messina A, Palmigiano A, Esposito F, Fiumara A, Bordugo A, Barone R, Sturiale L, Jaeken J, Garozzo D. 2021. HILIC‐UPLC‐MS for high throughput and isomeric N‐glycan separation and characterization in Congenital Disorders Glycosylation and human diseases. Glycoconj. J. 38:201–211. 10.1007/s10719-020-09947-7 [DOI] [PubMed] [Google Scholar]
- Messina A, Palmigiano A, Tosto C, Romeo DA, Sturiale L, Garozzo D, Leonardi A. 2021. Tear N‐glycomics in vernal and atopic keratoconjunctivitis. Allergy, 76:2500–2509. 10.1111/all.14775 [DOI] [PubMed] [Google Scholar]
- Mettu R, Lih Y‐H, Vulupala HR, Chen C‐Y, Hsu M‐H, Lo H‐J, Liao K‐S, Cheng Y‐Y, Chiu C‐H, Wu C‐Y. 2022. Synthetic library of oligosaccharides derived from the capsular polysaccharide of Streptococcus pneumoniae Serotypes 6A and 6B and their immunological studies. ACS Infect. Dis. 8:626–634. 10.1021/acsinfecdis.1c00646 [DOI] [PubMed] [Google Scholar]
- Meyer BH, Adam PS, Wagstaff BA, Kolyfetis GE, Probst AJ, Albers SV, Dorfmueller HC. 2022. Agl24 is an ancient archaeal homolog of the eukaryotic N‐glycan chitobiose synthesis enzymes. eLife, 11:e67448. 10.7554/eLife.67448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Montero L, Meckelmann SW, Schmitz OJ. 2022. Comparative study for analysis of carbohydrates in biological samples. Anal. Bioanal. Chem. 414:2117–2130. 10.1007/s00216-021-03845-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezö E, Herczeg M, Demeter F, Bereczki I, Csávás M, Borbás A. 2021. Systematic study of regioselective reductive ring‐opening reactions of 4,6‑O‑halobenzylidene acetals of glucopyranosides. J. Org. Chem. 86:12973–12987. 10.1021/acs.joc.1c01667 [DOI] [PubMed] [Google Scholar]
- Michael JA, Mutuku SM, Ucur B, Sarretto T, Maccarone AT, Niehaus M, Trevitt AJ, Ellis SR. 2022. Mass spectrometry imaging of lipids using MALDI coupled with plasma‐based post‐ionization on a trapped ion mobility mass spectrometer. Anal. Chem. 94:17494–17503. 10.1021/acs.analchem.2c03745 [DOI] [PubMed] [Google Scholar]
- Micoli F, Alfini R, Giannelli C. 2022. Methods for assessment of OMV/GMMA quality and stability. Method. Mol. Biol. 2414:227–279. 10.1007/978-1-0716-1900-1_14 [DOI] [PubMed] [Google Scholar]
- Miguez N, Kidibule P, Santos‐Moriano P, Ballesteros AO, Fernandez‐Lobato M, Plou FJ. 2021. Enzymatic synthesis and characterization of different families of chitooligosaccharides and their bioactive properties. Appl. Sci. 11:3212. 10.3390/app11073212 [DOI] [Google Scholar]
- Miki K, Zhang ZD, Kaneko K, Kakiuchi Y, Kojima K, Enomoto A, Oe M, Nogita K, Murata Y, Harada H, Ohe K. 2022. Amphiphilic γ‐cyclodextrin‐fullerene complexes with photodynamic activity. Mater. Adv. 3:312–317. 10.1039/D1MA00743B [DOI] [Google Scholar]
- Mikkelsen MD, Harholt J, Westereng B, Domozych D, Fry SC, Johansen IE, Fangel JU, Łężyk M, Feng T, Nancke L, Mikkelsen JD, Willats WGT, Ulvskov P. 2021. Ancient origin of fucosylated xyloglucan in charophycean green algae. Commun. Biol. 4:754. 10.1038/s42003-021-02277-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolajczyk K, Bereznicka A, Szymczak‐Kulus K, Haczkiewicz‐Lesniak K, Szulc B, Olczak M, Rossowska J, Majorczyk E, Kapczynska K, Bovin N, Lisowska M, Kaczmarek R, Miazek A, Czerwinski M. 2021. Missing the sweet spot: One of the two N‐glycans on human Gb3/CD77 synthase is expendable. Glycobiology, 31:1145–1162. 10.1093/glycob/cwab041 [DOI] [PubMed] [Google Scholar]
- Miller ID, Kohlhagen MC, Ladwig PM, Dasari S, Kumar S, Dispenzieri A, Willrich MAV, Murray DL. 2022. Characterizing M‐protein light chain glycosylation via mass spectrometry. Clin. Biochem. 109–110:11–16. 10.1016/j.clinbiochem.2022.09.004 [DOI] [PubMed] [Google Scholar]
- Mishra N, Rana K, Seelam SD, Kumar R, Pandey V, Salimath BP, Agsar D. 2021. Characterization and cytotoxicity of Pseudomonas mediated rhamnolipids against breast cancer MDA‐MB‐231 cell line. Front. Bioeng. Biotechnol. 9:761266. 10.3389/fbioe.2021.761266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal A, Singh A, Zhang B, Visessanguan W, Benjakul S. 2022. Chitooligosaccharide conjugates prepared using several phenolic compounds via ascorbic acid/H2O2 free radical grafting: Characteristics, antioxidant, antidiabetic, and antimicrobial activities. Foods, 11:920. 10.3390/foods11070920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P, Briggs M, Klingler‐Hoffmann M, Kaur G, Packer NH, Oehler MK, Hoffmann P. 2021. Altered N‐linked glycosylation in endometrial cancer. Anal. Bioanal. Chem. 413:2721–2733. 10.1007/s00216-020-03039-z [DOI] [PubMed] [Google Scholar]
- Miura N, Hanamatsu H, Yokota I, Okada K, Furukawa J‐I, Shinohara Y. 2020. Toolbox accelerating glycomics (TAG): glycan annotation from MALDI‐TOF MS spectra and mapping expression variation to biosynthetic pathways. Biomolecules, 10:1383. 10.3390/biom10101383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura N, Hanamatsu H, Yokota I, Akasaka‐Manya K, Manya H, Endo T, Shinohara Y, Furukawa J‐I. 2022. Toolbox accelerating glycomics (TAG): Improving large‐scale serum glycomics and refinement to identify SALSA‐modified and rare glycans. Int. J. Mol. Sci. 23:13097. 10.3390/ijms232113097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa A, Ohno S, Yamamura H. 2021. N,N‐Bis(hexyl‐d‐acetylmannosyl) acrylamide. Molbank, 2021:M1255. 10.3390/M1255 [DOI] [Google Scholar]
- Molnarova K, Cokrtova K, Tomnikova A, Krizek T, Kozlik P. 2022. Liquid chromatography and capillary electrophoresis in glycomic and glycoproteomic analysis. Monatsh. Chem. 153:659–686. 10.1007/s00706-022-02938-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompó‐Roselló Ó, Vergara‐Barberán M, Lerma‐García MJ, Simó‐Alfonso EF, Herrero‐Martínez JM. 2021. Boronate affinity sorbents based on thiol‐functionalized polysiloxane‐polymethacrylate composite materials in syringe format for selective extraction of glycopeptides. Microchem. J. 164:106018. 10.1016/j.microc.2021.106018 [DOI] [Google Scholar]
- Mondal S, Balasubramanian A, Biswas P, Agrawal S, Ghosh S, Dey S. 2022. Characterization of pearl millet oligosaccharides and evaluation of their prebiotic potential. Bioact. Carbohydr. Diet. Fibre, 28:100324. 10.1016/j.bcdf.2022.100324 [DOI] [Google Scholar]
- Montoya AL, Austin VM, Portillo S, Vinales I, Ashmus RA, Estevao I, Jankuru SR, Alraey Y, Al‐Salem WS, Acosta‐Serrano Al, Almeida IC, Michael K. 2021. Reversed immunoglycomics identifies α‑galactosyl‐bearing glycotopes specific for Leishmania major infection. JACS Au, 1:1275–1287. 10.1021/jacsau.1c00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya AL, Gil ER, Heydemann EL, Estevao IL, Luna BE, Ellis CC, Jankuru SR, de Noya BA, Noya O, Zago MP, Almeida IC, Michael K. 2022. Specific recognition of β‐galactofuranose‐containing glycans of synthetic neoglycoproteins by sera of chronic Chagas disease patients. Molecules, 27:411. 10.3390/molecules27020411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mookherjee A, Uppal SS, Murphree TA, Guttman M. 2021. Linkage memory in underivatized protonated carbohydrates. J. Am. Soc. Mass Spectrom. 32:581–589. 10.1021/jasms.0c00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon Y, Kim H, Kang CG, Park C, Kim SW, Kim D. 2022. Biochemical characterization of synthesized fisetin glucoside by dextransucrase from Leuconostoc mesenteroides NRRL B‐1299CB4 with enhanced water solubility. Enzyme Microb. Technol. 161:110111. 10.1016/j.enzmictec.2022.110111 [DOI] [PubMed] [Google Scholar]
- Moran AB, Gardner RA, Wuhrer M, Lageveen‐Kammeijer GSM, Spencer DIR. 2022. Sialic acid derivatization of fluorescently labeled N‑glycans allows linkage differentiation by reversed‐phase liquid chromatography‐fluorescence detection‐mass spectrometry. Anal. Chem. 94:6639–6648. 10.1021/acs.analchem.1c02610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcos YAT, Lütke S, Tenbieg A, Hanisch FG, Pryymachuk G, Piekarek N, Hoffmann T, Keller T, Janoschek R, Niehoff A, Zaucke F, Dötsch J, Hucklenbruch‑Rother E, Sengle G. 2022. Sensitive asprosin detection in clinical samples reveals serum/saliva correlation and indicates cartilage as source for serum asprosin. Sci. Rep. 12:1340. 10.1038/s41598-022-05060-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan KT, Zheng J, McCafferty DG. 2021. Discovery of six ramoplanin family gene clusters and the lipoglycodepsipeptide chersinamycin. ChemBioChem, 22:176–185. 10.1002/cbic.202000555 [DOI] [PubMed] [Google Scholar]
- Morgenstern D, Wolf‐Levy H, Tickotsky‐Moskovitz N, Cooper I, Buchman AS, Bennett DA, Beeri MS, Levin Y. 2022. Optimized glycopeptide enrichment method−it is all about the sauce. Anal. Chem. 94:10308–10313. 10.1021/acs.analchem.2c00524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa C, Sugiura K, Kondo K, Yamamoto Y, Kojima Y, Ozawa Y, Yoshioka H, Miura N, Piao J, Okada K, Hanamatsu H, Tsuda M, Tanaka S, Furukawa J‐i, Yasuro S. 2022. Evaluation of the context of downstream N‐ and free N‐glycomic alterations induced by swainsonine in HepG2 cells. Biochim. Biophys, Acta Gen. Sub. 1866:130168. 10.1016/j.bbagen.2022.130168 [DOI] [PubMed] [Google Scholar]
- Morita S, Inai Y, Minakata S, Kishimoto S, Manabe S, Iwahashi N, Ino K, Ito Y, Akamizu T, Ihara Y. 2021. Quantification of serum C‑mannosyl tryptophan by novel assay to evaluate renal function and vascular complications in patients with type 2 diabetes. Sci. Rep. 11:1946. 10.1038/s41598-021-81479-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozumi S, Ueda M, Okahashi N, Arita M. 2022. Structures and functions of the gut microbial lipidome. Biochim. Biophys. Acta ‐ Mol. Cell Biol.Lipids, 1867:159110. 10.1016/j.bbalip.2021.159110 [DOI] [PubMed] [Google Scholar]
- Mosquera‐Restrepo SF, Zuberogoïtia S, Gouxette L, Layre E, Gilleron M, Stella A, Rengel D, Burlet‐Schiltz O, Caro AC, Garcia LF, Segura C, Jaramillo CAP, Rojas M, Nigou J. 2022. A Mycobacterium tuberculosis fingerprint in human breath allows tuberculosis detection. Nat. Commun. 13:7751. 10.1038/s41467-022-35453-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure MJ, Gimeno A, Delgado S, Diercks T, Boons G‐J, Jiménez‐Barbero J, Ard A. 2021. Selective 13C‐labels on repeating glycan oligomers to reveal protein binding epitopes through NMR: Polylactosamine binding to galectins. Angew. Chem. Int. Ed., 60:18777–18782. 10.1002/anie.202106056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally C, Stubbs KA, Thai VC, Anandan A, Bartley S, Scanlon MJ, Jarvis GA, John CM, Lim KYL, Sullivan CM, Sarkar‐Tyson M, Vrielink A, Kahler CM. 2022. Novel small molecules that increase the susceptibility of Neisseria gonorrhoeae to cationic antimicrobial peptides by inhibiting lipid A phosphoethanolamine transferase. J. Antimicrob. Chemother., 77:2441–2447. 10.1093/jac/dkac204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MA, Bhandari DR, Spengler B. 2021. Matrix‐free high‐resolution atmospheric‐pressure SALDI mass spectrometry imaging of biological samples using nanostructured DIUTHAME membranes. Metabolites, 11:624. 10.3390/metabo11090624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller WH, De Pauw E, Far J, Malherbe C, Eppe G. 2021. Imaging lipids in biological samples with surface‐assisted laser desorption/ionization mass spectrometry: A concise review of the last decade. Prog. Lipid Res. 83:101114. 10.1016/j.plipres.2021.101114 [DOI] [PubMed] [Google Scholar]
- Müller WH, Verdin A, Kune C, Far J, De Pauw E, Malherbe C, Eppe G. 2021. Dual‐polarity SALDI FT‐ICR MS imaging and Kendrick mass defect data filtering for lipid analysis. Anal. Bioanal. Chem. 413:2821–2830. 10.1007/s00216-020-03020-w [DOI] [PubMed] [Google Scholar]
- Müller WH, Verdin A, De Pauw E, Malherbe C, Eppe G. 2022. Surface‐assisted laser desorption/ionization mass spectrometry imaging: A review. Mass Spectrom. Rev. 41:373–420. 10.1002/mas.21670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllerová M, Maciel D, Nunes N, Wrobel D, Stofik M, Štástná LC, Krupková A, Cuří̌nová P, Nováková K, Božík M, Malý M, Malý J, Rodrigues J, Strašák T. 2022. Carbosilane glycodendrimers for anticancer drug delivery: Synthetic route, characterization, and biological effect of glycodendrimer‐doxorubicin complexes. Biomacromolecules, 23:276–290. 10.1021/acs.biomac.1c01264 [DOI] [PubMed] [Google Scholar]
- Muñoz‐Labrador A, Azcarate S, Lebrón‐Aguilar R, Quintanilla‐López JE, Galindo‐Iranzo P, Kolida S, Methven L, Rastall RA, Moreno FJ, Hernandez‐Hernandez O. 2021. High‐yield synthesis of transglycosylated mogrosides improves the flavor profile of monk fruit extract sweeteners. J. Agric. Food Chem. 69:1011–1019. 10.1021/acs.jafc.0c07267 [DOI] [PubMed] [Google Scholar]
- Muñoz‐Labrador A, Lebrón‐Aguilar R, Quintanilla‐López JE, Galindo‐Iranzo P, Azcarate SM, Kolida S, Kachrimanidou V, Garcia‐Cañas V, Methven L, Rastall RA, Moreno FJ, Hernandez‐Hernandez O. 2022. Prebiotic potential of a new sweetener based on galactooligosaccharides and modified mogrosides. J. Agric. Food Chem. 70:9048–9056. 10.1021/acs.jafc.2c01363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray KK. 2021. Lasers for matrix‐assisted laser desorption ionization. J. Mass Spectrom., 56:e4664. 10.1002/jms.4664 [DOI] [PubMed] [Google Scholar]
- Mustafa HH, Elahmar MAI, Hameed RT, Alsultan M, Nesseef L, Swiegers GF. 2022. Extraction and identification of effective compounds from natural plants. J. Compos. Sci. 6:149. 10.3390/jcs6050149 [DOI] [Google Scholar]
- Muszyński A, Zarember KA, Heiss C, Shiloach J, Berg LJ, Audley J, Kozyr A, Greenberg DE, Holland SM, Malech HL, Azadi P, Carlson RW, Gallin JI. 2021. Granulibacter bethesdensis, a pathogen from patients with chronic granulomatous disease, produces a penta‐acylated hypostimulatory glycero‐d‐talo‐oct‐2‐ulosonic acid‐lipid A glycolipid (Ko‐lipid A). Int. J. Mol. Sci., 22:3303. 10.3390/ijms22073303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutuku SM, Ellis SR. 2022. MALDI Mass spectrometry imaging in lipidomics. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 157–187. 10.1039/9781839165191-00157 [DOI] [Google Scholar]
- Nabi M, Mamun A, Islam A, Hasan M, Waliullah ASM, Tamannaa Z, Sato T, Kahyo T, Setou M. 2021. Mass spectrometry in the lipid study of cancer. Expert Rev. Proteomics, 18:201–219. 10.1080/14789450.2021.1912602 [DOI] [PubMed] [Google Scholar]
- Naclerio GA, Abutaleb NS, Onyedibe KI, Karanja C, Eldesouky HE, Liang H‐W, Dieterly A, Aryal UK, Lyle T, Seleem MN, Sintim HO. 2022. Mechanistic studies and in vivo efficacy of an oxadiazole‐containing antibiotic. J. Med. Chem., 65:6612–6630. 10.1021/acs.jmedchem.1c02034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa H, Miyazaki S, Kyogashima M. 2022. Simple separation of glycosphingolipids in the lower phase of a Folch's partition from crude lipid fractions using zirconium dioxide. Glycoconj. J. 39:789–795. 10.1007/s10719-022-10080-w [DOI] [PubMed] [Google Scholar]
- Naini A, Bartetzko MP, Sanapala SR, Broecker F, Wirtz V, Lisboa MP, Parameswarappa SG, Knopp D, Przygodda J, Hakelberg M, Pan R, Patel A, Chorro L, Illenberger A, Ponce C, Kodali S, Lypowy J, Anderson AS, Donald RGK, von Bonin A, Pereira CL. 2022. Semisynthetic glycoconjugate vaccine candidates against Escherichia coli O25B induce functional IgG antibodies in mice. JACS Au, 2:2135–2151. 10.1021/jacsau.2c00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami A, Mao Q, Gouhier G, Arima H, Kitagishi H. 2021. FRET‐Based in‐cell detection of highly selective supramolecular complexes of meso‐tetraarylporphyrin with peptide/BODIPY‐modified per‐O‐methyl‐β‐cyclodextrins. ChemBioChem, 22:3190–3198. 10.1002/cbic.202100380 [DOI] [PubMed] [Google Scholar]
- Nakagawa Y, Yamaji F, Miyanishi W, Ojika M, Igarashi Y, Ito Y. 2021. Binding evaluation of pradimicins for oligomannose motifs from fungal mannans. Bull. Chem. Soc. Jpn. 94:732–754. 10.1246/bcsj.20200305 [DOI] [Google Scholar]
- Nakashima K, Hirahara Y, Koike T, Tanaka S, Gamo K, Oe S, Hayashi S, Seki‐Omura R, Nakano Y, Ohe C, Yoshida T, Kataoka Y, Tsuda M, Yamashita T, Honke K, Kitada M. 2022. Sulfatide with ceramide composed of phytosphingosine (t18:0) and 2‐hydroxy FAs in renal intercalated cells. J. Lipid Res. 63:100210. 10.1016/j.jlr.2022.100210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar S, Kahn N, Gummer JPA. 2021. Matrix‐assisted laser desorption ionization mass spectrometry imaging by freeze‐spot deposition of the matrix. J. Am. Soc. Mass Spectrom. 32:1829–1836. 10.1021/jasms.1c00063 [DOI] [PubMed] [Google Scholar]
- Nandita E, Bacalzo, Jr. NP , Ranque CL, Amicucci MJ, Galermo A, Lebrilla CB. 2021. Polysaccharide identification through oligosaccharide fingerprinting. Carbohydr. Polym. 257:117570. 10.1016/j.carbpol.2020.117570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nason R, Büll C, Konstantinidi A, Sun L, Ye Z, Halim A, Du W, Sørensen DM, Durbesson F, Furukawa S, Mandel U, Joshi HJ, Dworkin LA, Hansen L, David L, Iverson TM, Bensing BA, Sullam PM, Varki A, de Vries E, de Haan CAM, Vincentelli R, Henrissat B, Vakhrushev SY, Clausen H, Narimatsu Y. 2021. Display of the human mucinome with defined O‐glycans by gene engineered cells. Nat. Commun. 12:4070. 10.1038/s41467-021-24366-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navelkar R, Owen G, Mutherkrishnan V, Thiessen P, Cheng T, Bolton E, Edwards N, Tiemeyer M, Campbell MP, Martin M, Vora J, Kahsay R, Mazumder R. 2021. Enhancing the interoperability of glycan data flow between ChEBI, PubChem and GlyGen. Glycobiology, 31:1510–1519. 10.1093/glycob/cwab078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S, Aoki‐Kinoshita K, Bolton E, Frank M, Lisacek F, Lütteke T, O'Boyle N, Packer NH, Stanley P, Toukach P, Varki A, Woods RJ, Group TSD. 2019. Updates to the symbol nomenclature for glycans guidelines. Glycobiology, 29:620–624. 10.1093/glycob/cwz045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto CC, Mortzfeld BM, Turbitt JR, Bhattara SK, Yeliseyev V, DiBenedetto N, Bry L, Bucci V. 2021. Proanthocyanidin enriched cranberry extract induces resilient bacterial community dynamics in a gnotobiotic mouse model. Microb. Cell, 8:131–142. 10.15698/mic2021.06.752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, Sosicka P, Fenaille F, Harroche A, Vuillaumier‐Barrot S, Porterfield M, Xia Z‐J, Wagner S, Bamshad MJ, Vergnes‐Boiteux M‐C, Cholet S, Dalton S, Dell A, Dupré T, Fiore M, Haslam SM, Huguenin Y, Kumagai T, Kulik M, McGoogan K, Michot C, Nickerson DA, Pascreau T, Borgel D, Raymond K, Warad D, University of Washington Center for Mendelian Genomics (UW‐CMG), Flanagan‐Steet H, Steet R, Tiemeyer M, Seta N, Bruneel A, Freeze HH. 2021. A mutation in SLC37A4 causes a dominantly inherited congenital disorder of glycosylation characterized by liver dysfunction. Am. J. Hum. Genet., 108:1040–1052. 10.1016/j.ajhg.2021.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguan H‐S, Ni C‐K. 2022. Collision‐induced dissociation of α‑isomaltose and α‑maltose. J. Phys. Chem. A, 126:8799–8808. 10.1021/acs.jpca.2c04278 [DOI] [PubMed] [Google Scholar]
- Nguan H‐S, Tsai S‐T, Ni C‐K. 2022. Collision‐induced dissociation of cellobiose and maltose. J. Phys. Chem. A, 126:1486–1495. 10.1021/acs.jpca.1c10046 [DOI] [PubMed] [Google Scholar]
- Nguyen H, Kondo K, Yagi Y, Iseki Y, Okuoka N, Watanabe T, Mikami B, Nagata T, Katahira M. 2022. Functional and structural characterizations of lytic polysaccharide monooxygenase, which cooperates synergistically with cellulases, from Ceriporiopsis subvermispora . ACS Sustain. Chem. Eng. 10:923–934. 10.1021/acssuschemeng.1c06810 [DOI] [Google Scholar]
- Nguyen PC, Nguyen MTT, Kim J‐H, Hong S‐T, Kim H‐L, Park J‐T. 2021. A novel maltoheptaose‐based sugar ester having excellent emulsifying properties and optimization of its lipase‐catalyzed synthesis. Food Chem., 352:129358. 10.1016/j.foodchem.2021.129358 [DOI] [PubMed] [Google Scholar]
- Ni C‐K, Hsu HC, Liew CY, Huang S‐P, Tsai S‐T. 2021. Modern mass spectrometry techniques for oligosaccharide structure determination: Logically derived sequence tandem mass spectrometry for automatic oligosaccharide structural determination. In: Barchi JJ, Jr., Editor, Comprehensive Glycoscience; From Chemistry to Systems Biology, Second edition. Amsterdam: Elsevier. p 309–339. 10.1016/b978-0-12-409547-2.14903-0 [DOI] [Google Scholar]
- Nicolardi S, Joseph AA, Zhu Q, Shen Z, Pardo‐Vargas A, Chiodo F, Molinaro A, Silipo A, van der Burgt YEM, Yu B, Seeberger PH, Wuhrer M. 2021. Analysis of synthetic monodisperse polysaccharides by wide mass range ultrahigh‐resolution MALDI mass spectrometry. Anal. Chem. 93:4666–4675. 10.1021/acs.analchem.1c00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolardi S, Danuser R, Dotz V, Domínguez‐Vega E, Kaabi AA, Beurret M, Anish C, Wuhrer M. 2022. Glycan and protein analysis of glycoengineered bacterial E. coli vaccines by MALDI‐in‐source decay FT‐ICR mass spectrometry. Anal. Chem., 94:4979–4987. 10.1021/acs.analchem.1c04690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti C, Procházková L, Nedbalová L, Mócsai R, Altmann F, Holzinger A, Remias D. 2021. Thorsmoerkia curvula gen. et spec. nov. (Trebouxiophyceae, Chlorophyta), a semi‑terrestrial microalga from Iceland exhibits high levels of unsaturated fatty acids. J. Appl. Phycol., 33:3671–3682. 10.1007/s10811-021-02577-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolosi M, Bellia F, Giuffrida ML, Zimbone S, Oliveri V, Vecchio G. 2021. Synthesis and biological evaluation of novel β‐cyclodextrin‐fluvastatin conjugates. Res. Chem. 3:100230. 10.1016/j.rechem.2021.100230 [DOI] [Google Scholar]
- Nishi N, Seki K, Takahashi D, Toshima K. 2021. Synthesis of a pentasaccharide repeating unit of lipopolysaccharide derived from virulent E. coli O1 and identification of a glycotope candidate of avian pathogenic E. coli O1. Angew. Chem. Int. Ed. 60:1789–1796. 10.1002/anie.202013729 [DOI] [PubMed] [Google Scholar]
- Nishida T, Yokoyama R, Kubohira Y, Maeda Y, Takeo T, Nakagata N, Takagi H, Ishikura K, Yanagihara K, Misumi S, Kishimoto N, Ishitsuka Y, Kondo Y, Irie T, Soga M, Era T, Onodera R, Higashi T, Motoyama K. 2022. Lactose‐appended hydroxypropyl‐β‐cyclodextrin lowers cholesterol accumulation and alleviates motor dysfunction in Niemann−Pick type C disease model mice. ACS Appl. Bio Mater., 5:2377–2388. 10.1021/acsabm.2c00233 [DOI] [PubMed] [Google Scholar]
- Nishikaze T, Tsumoto H, Sekiya S, Iwamoto S, Miura Y, Tanaka K. 2017. Differentiation of sialyl linkage isomers by one‐pot sialic acid derivatization for mass spectrometry‐based glycan profiling. Anal. Chem. 89:2353–2360. 10.1021/acs.analchem.6b04150 [DOI] [PubMed] [Google Scholar]
- Nmagu D, Singh SK, Lee KH. 2021. Creation of monoclonal antibody expressing CHO cell lines grown with sodium butyrate and characterization of resulting antibody glycosylation. Method. Enzymol. 660:267–295. 10.1016/bs.mie.2021.06.039 [DOI] [PubMed] [Google Scholar]
- Noorbakhsh H, Khorasgani MR. 2022. Date (Phoenix dactylifera L.) polysaccharides: A review on chemical structure and nutritional properties. J. Food Meas. Charact., 16:3240–3250. 10.1007/s11694-022-01425-y [DOI] [Google Scholar]
- Norberg T, Johansson G, Kallin E. 2022. Derivatization of sugars with N,O‐dimethylhydroxylamine. Efficient RP‐HPLC separation of sugar mixtures. Carbohydr. Res. 520:108635. 10.1016/j.carres.2022.108635 [DOI] [PubMed] [Google Scholar]
- Nordgren M, Nägeli A, Nyhlén H, Sjögren J. 2021. Mapping O‐glycosylation sites using OpeRATOR and LC‐MS. Method. Mol. Biol. 2271:155–167. 10.1007/978-1-0716-1241-5_11 [DOI] [PubMed] [Google Scholar]
- Nordstrom HR, Evans DR, Finney AG, Westbrook KJ, Zamora PF, Hofstaedter CE, Yassin MH, Pradhan A, Iovleva A, Ernst RK, Bomberger JM, Shields RK, Doi Y, Van Tyne D. 2022. Genomic characterization of lytic bacteriophages targeting genetically diverse Pseudomonas aeruginosa clinical isolates. iScience, 25:104372. 10.1016/j.isci.2022.104372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noun M, Akoumeh R, Abbas I. 2022. Cell and tissue imaging by TOF‐SIMS and MALDI‐TOF: An overview for biological and pharmaceutical analysis. Microsc. Microanal. 28:1–26. 10.1017/s1431927621013593 [DOI] [PubMed] [Google Scholar]
- Nowak SR, Lachmayr KK, Yager KG, Sita LR. 2021. Stable thermotropic 3D and 2D double gyroid nanostructures with sub‐2‐nm feature size from scalable sugar‐polyolefin conjugates. Angew. Chem. Int. Ed. 60:8710–8716. 10.1002/anie.202016384 [DOI] [PubMed] [Google Scholar]
- Nummela P, Heiskanen A, Kytölä S, Haglund C, Lepistö A, Satomaa T, Ristimäki A. 2021. Altered linkage pattern of N‐glycan sialic acids in Pseudomyxoma peritonei . Glycobiology, 31:211–222. 10.1093/glycob/cwaa079 [DOI] [PubMed] [Google Scholar]
- Nuntawong P, Lohseethong K, Juengwatanatrakul T, Yusakul G, Putalun W, Tanaka H, Sakamoto S, Morimoto S. 2021. Competitive immunochromatographic test strips for the rapid semi‐quantitative analysis of the biologically active bitter glycoside, amarogentin. J. Immunoass. Immunochem. 42:48–61. 10.1080/15321819.2020.1819308 [DOI] [PubMed] [Google Scholar]
- O'Neill KC, Dueñas ME, Larson E, Forsman TT, Lee Y‐J. 2022. Enhancing metabolite coverage for matrix‐assisted laser desorption/ionization mass spectrometry imaging through multiple on‐tissue chemical derivatizations. Method. Mol. Biol. 2437:197–213. 10.1007/978-1-0716-2030-4_14 [DOI] [PubMed] [Google Scholar]
- O'Neill KC, Liapis E, Harris BT, Perlin DS, Carter CL. 2022. Mass spectrometry imaging discriminates glioblastoma tumor cell subpopulations and different microvascular formations based on their lipid profiles. Sci. Rep. 12:17069. 10.1038/s41598-022-22093-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan I, Hajduk J, Harrison JA, Marchand A, Czar MF, Renato Z. 2022. Exploring gas‐phase MS methodologies for structural elucidation of branched N‑glycan isomers. Anal. Chem. 94:10531–10539. 10.1021/acs.analchem.2c02019 [DOI] [PubMed] [Google Scholar]
- Oh L, Ji Y, Li W, Varki A, Chen X, Wang L‐P. 2022. O‑Acetyl migration within the sialic acid side chain: A mechanistic study using the ab initio nanoreactor. Biochemistry, 61:2007–2013. 10.1021/acs.biochem.2c00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi Y, Nishikaze T, Kitaura Y, Ito T, Yamamoto S, Sugiyama F, Matsuyama M, Takahashi Y, Takeda A, Kawahara T, Okajima T, Furukawa K, Furukawa K. 2021. Majority of alpha2,6‐sialylated glycans in the adult mouse brain exist in O‐glycans: SALSA‐MS analysis for knockout mice of alpha2,6‐sialyltransferase genes. Glycobiology, 31:557–570. 10.1093/glycob/cwaa105 [DOI] [PubMed] [Google Scholar]
- Okamoto N, Ohto T, Enokizono T, Wada Y, Kohmoto T, Imoto I, Haga Y, Seino J, Suzuki T. 2021. Siblings with MAN1B1‐CDG showing novel biochemical profiles. Cells, 10:3117. 10.3390/cells10113117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa A, Baba A, Okano H, Tokunaga T, Nakaue T, Ogawa T, Shimma S, Sugimoto Y, Ohta D. 2022. Involvement of α‐galactosidase OmAGAL2 in planteose hydrolysis during seed germination of Orobanche minor . J. Exp. Bot. 73:1992–2004. 10.1093/jxb/erab527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M, Takami T, Kobayashi D, Araki N, Yoshizawa AC, Tabata T, Sugiyama N, Goto S, Ishihama Y. 2017. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res., 45:D1107–D1111. 10.1093/nar/gkw1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Nishimura T, Sasaki Y, Akiyoshi K. 2021. Thermoresponsive carbohydrate‑b‑polypeptoid polymer vesicles with selective solute permeability and permeable factors for solutes. Biomacromolecules, 22:3099–3106. 10.1021/acs.biomac.1c00530 [DOI] [PubMed] [Google Scholar]
- Oliveira T, Thaysen‐Andersen M, Packer NH, Kolarich D. 2021. The Hitchhiker's guide to glycoproteomics. Biochem. Soc. Trans. 49:1643–1662. 10.1042/bst20200879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuch‐Kwiatkowska A, Jarosz S. 2022. Synthesis of the precursors of iminosugars with 7‐membered ring. Carbohydr. Res. 518:108584. 10.1016/j.carres.2022.108584 [DOI] [PubMed] [Google Scholar]
- Oswald DM, Lehoux SD, Zhou JY, Glendenning LM, Cummings RD, Cobb BA. 2022. ST6Gal1 in plasma is dispensable for IgG sialylation. Glycobiology, 32:803–813. 10.1093/glycob/cwac039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaki M, Hirane N, Natsume‑Kitatani Y, Itoh MN, Shindo M, Kurebayashi Y, Nishimura SI. 2022. Mouse tissue glycome atlas 2022 highlights inter‑organ variation in major N‑glycan profiles. Sci. Rep. 12:17804. 10.1038/s41598-022-21758-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki H, Asano T, Tanaka H‐N, Komura N, Ando H, Ishida H, Imamura A. 2022. Blockwise synthesis of polylactosamine fragments and keratan sulfate oligosaccharides comprised of dimeric Galβ(1 → 4)GlcNAc6Sβ. Carbohydr. Res. 512:108502. 10.1016/j.carres.2022.108502 [DOI] [PubMed] [Google Scholar]
- Ozdilek A, Huang J, Babb R, Paschall AV, Middleton DR, Duke JA, Pirofski L‐a, Mousa JJ, Avcia FY. 2021. A structural model for the ligand binding of pneumococcal serotype 3 capsular polysaccharide‐specific protective antibodies. mBio, 12:e00800–e00821. 10.1128/mbio.00800-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozleyen A, Cinar ZO, Karav S, Bayraktar A, Arslan A, Kayili HM, Salih B, Tumer TB. 2022. Biofortified whey/deglycosylated whey and chickpea protein matrices: Functional enrichment by black mulberry polyphenols. Plant Foods Hum. Nutr. 77:51–61. 10.1007/s11130-021-00943-2 [DOI] [PubMed] [Google Scholar]
- Pace CL, Angel PM, Drake RR, Muddiman DC. 2022. Mass spectrometry imaging of N‑linked glycans in a formalin‐fixed paraffin‐embedded human prostate by infrared matrix‐assisted laser desorption electrospray ionization. J. Proteome Res. 21:243–249. 10.1021/acs.jproteome.1c00822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino P, Papi F, Minunni M, Nativi C, Scarano S. 2022. Structurally constrained MUC1‐Tn mimetic antigen as template for molecularly imprinted polymers (MIPs): A promising tool for cancer diagnostics. ChemPlusChem, 87:e202200068. 10.1002/cplu.202200068 [DOI] [PubMed] [Google Scholar]
- Palodkar KK, Rao NNM, Iyer S, Puttalingaiah RT, Sadhu V, Aminabhavi TM, Reddy KR, Sainath AVS. 2022. Maltose‐based methacrylated polymer architectures and their biocompatibility. Mater. Today Chem. 23:100669. 10.1016/j.mtchem.2021.100669 [DOI] [Google Scholar]
- Palomares F, Gomez F, de la Fuente MC, Perez‐Sanchez N, Torres MJ, Mayorga C, Rojo J, Ramos‐Soriano J. 2022. Fucodendropeptides induce changes in cells of the immune system in food allergic patients via DC‐SIGN receptor. Carbohydr. Res. 517:108580. 10.1016/j.carres.2022.108580 [DOI] [PubMed] [Google Scholar]
- Pan Y, Zhang C, Xiao R, Zhang L, Zhang W. 2021. Dual‐functionalized magnetic bimetallic metal‐organic framework composite for highly specific enrichments of phosphopeptides and glycopeptides. Anal. Chim. Acta, 1158:338412. 10.1016/j.aca.2021.338412 [DOI] [PubMed] [Google Scholar]
- Pan Y, Zhang L, Zhang R, Han J, Qin W, Gu Y, Sha J, Xu X, Feng Y, Ren Z, Dai J, Huang B, Ren S, Gu J. 2021. Screening and diagnosis of colorectal cancer and advanced adenoma by Bionic Glycome method and machine learning. Am. J. Cancer Res. 11:3002–3020 [PMC free article] [PubMed] [Google Scholar]
- Pančík F, Pakanová Z, Květoň F, Baráth P. 2022. Diagnostics of lysosomal storage diseases by mass spectrometry: A review. Chem. Papers, 76:3995–4004. 10.1007/s11696-022-02153-9 [DOI] [Google Scholar]
- Pančík F, Pakanová Z, Mečárová J, Čížová A, Bystrický S, Kozmon S, Baráth P. 2022. Fragmentation analysis of O‐specific polysaccharide from bacteria Vibrio cholerae O139 by MALDI‐TOF and LC/ESI‐MS/MS. Eur. J. Mass Spectrom., 28:47–55. 10.1177/14690667221099119 [DOI] [PubMed] [Google Scholar]
- Pandeirada CO, Hageman JA, Janssen H‐G, Westphal Y, Schols HA. 2022. Identification of plant polysaccharides by MALDI‐TOF MS fingerprinting after periodate oxidation and thermal hydrolysis. Carbohydr. Polym. 292:119685. 10.1016/j.carbpol.2022.119685 [DOI] [PubMed] [Google Scholar]
- Papazoglu GM, Cubilla M, Pereyra M, de Kremer RD, Pérez B, Sturiale L, Asteggiano CG. 2021. Mass spectrometry glycophenotype characterization of ALG2‐CDG in Argentinean patients with a new genetic variant in homozygosis. Glycoconj. J. 38:191–200. 10.1007/s10719-021-09976-w [DOI] [PubMed] [Google Scholar]
- Park B‐R, Park JY, Lee SH, Hong S‐J, Jeong JH, Choi J‐H, Park S‐Y, Park CS, Lee H‐N, Kim Y‐M. 2021. Synthesis of improved long‐chain isomaltooligosaccharide, using a novel glucosyltransferase derived from Thermoanaerobacter thermocopriae, with maltodextrin. Enzyme Microb. Technol. 147:109788. 10.1016/j.enzmictec.2021.109788 [DOI] [PubMed] [Google Scholar]
- Park DD, Chen J, Kudelka MR, Jia N, Haller CA, Kosaraju R, Premji AM, Galizzi M, Nairn AV, Moremen KW, Cummings RD, Chaikof EL. 2021. Resident and elicited murine macrophages differ in expression of their glycomes and glycan‐binding proteins. Cell Chem. Biol. 28:567–582. 10.1016/j.chembiol.2020.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H‐R, Eom DH, Kim JH, Shin J‐C, Shin M‐S, Shin K‐S. 2021. Composition analysis and oral administered effects on dextran sulfate sodium‐induced colitis of galactooligosaccharides bioconverted by Bacillus circulans . Carbohydr. Polym. 270:118389. 10.1016/j.carbpol.2021.118389 [DOI] [PubMed] [Google Scholar]
- Park JM, Jung CY, Jang W‐D, Jaung JY. 2021. Silicon tetrapyrazinoporphyrazine derivatives‐incorporated carbohydrate‐based block copolymer micelles for photodynamic therapy. ACS Appl. Bio Mater. 4:1988–2000. 10.1021/acsabm.0c00256 [DOI] [PubMed] [Google Scholar]
- Park RM, Nguyen NHT, Lee SM, Kim YH, Min J. 2021. Alginate oligosaccharides can maintain activities of lysosomes under low pH condition. Sci. Rep. 11:11504. 10.1038/s41598-021-91175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Ji KY, Park SY, Kim HM, Ma SH, Do JH, Kang H, Kang HS, Oh DB, Shim JS, Joung YH. 2022. Immunotherapeutic effects of recombinant colorectal cancer antigen produced in tomato fruits. Sci. Rep. 12:9723. 10.1038/s41598-022-13839-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K, Yan S, Pohl NLB, Wilson IBH. 2021. Glycobiology of Caenorhabditis elegans . In: Taniguchi N, Editor, Comprehensive Glycoscience (Second edition). Amsterdam, Kidlington UK, Cambridge MA: Elsevier. p 36–54. 10.1016/b978-0-12-819475-1.00071-7 [DOI] [Google Scholar]
- Patabandige MW, Pfeifer LD, Nguyen HT, Desaire H. 2022. Quantitative clinical glycomics strategies: A guide for selecting the best analysis approach. Mass Spectrom. Rev. 41:901–921. 10.1002/mas.21688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Tung SK, Porfirio S, Sonon R, Azadi P, Free SJ. 2022. Extracellular targeting of Neurospora crassa cell wall and secreted glycoproteins by DFG‐5. Fungal Genet. Biol. 160:103686. 10.1016/j.fgb.2022.103686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathiraja D, Christiansen L, Park B, Schultz‐Johansen M, Bang G, Stougaard P, Choi I‐G. 2021. A novel auxiliary agarolytic pathway expands metabolic versatility in the agar‐degrading marine bacterium Colwellia echini A3T . Appl. Environ. Microbiol. 87:e00230–00221. 10.1128/aem.00230-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathmasiri KC, Nguyen TTA, Khamidova N, Cologna SM. 2021. Mass spectrometry‐based lipid analysis and imaging. Curr. Top. Memb. 88:315–357. 10.1016/bs.ctm.2021.10.005 [DOI] [PubMed] [Google Scholar]
- Patil S, Rohrer J. 2021. An improved method for galactosyl oligosaccharide characterization. J. Chromatogr., B, 1184:122967. 10.1016/j.jchromb.2021.122967 [DOI] [PubMed] [Google Scholar]
- Paton B, Suarez M, Herrero P, Canela N. 2021. Glycosylation biomarkers associated with age‐related diseases and current methods for glycan analysis. Int. J. Mol. Sci. 22:5788. 10.3390/ijms22115788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patry S, Robitzer M, Jean‐Pierre H. 2021. Synthesis and characterization of a small library of bisglucosides: Influence of the nature of the diol/diphenol used in O‐glucosylation. Carbohydr. Res. 500:108217. 10.1016/j.carres.2020.108217 [DOI] [PubMed] [Google Scholar]
- Patwa A, Thiéry A, Lombard F, Lilley MKS, Boisset C, Bramard J‐F, Bottero J‐Y, Barthélémy P. 2022. Accumulation of nanoparticles in “jellyfish” mucus: A bio‐inspired route to decontamination of nanowaste. Sci. Rep. 5:11387. 10.1038/srep11387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecori F, Kondo N, Ogura C, Miura T, Kume M, Minamijima Y, Yamamoto K, Nishihara S. 2021. Site‐specific O‐GlcNAcylation of Psme3 maintains mouse stem cell pluripotency by impairing P‐body homeostasis. Cell Rep., 36:109361. 10.1016/j.celrep.2021.109361 [DOI] [PubMed] [Google Scholar]
- Pekdemir B, Duman H, Arslan A, Kaplan M, Karyelioğlu M, Özer T, Kayılı HM, Salih B, Henrick BM, Duar RM, Karav S. 2022. Immobilization of a bifidobacterial endo‐ß‐N‐acetylglucosaminidase to generate bioactive compounds for food industry. Front. Bioeng. Biotechnol. 10:922423. 10.3389/fbioe.2022.922423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pélingre M, Teki DS‐EK, El‐Abid J, Chagnault V, Kovensky J, Bonnet V. 2021. Ring‐opening of cyclodextrins: An efficient route to pure maltohexa‐, hepta‐, and octaoses. Organics, 2:287–305. 10.3390/org2030015 [DOI] [Google Scholar]
- Peng C, Zhang Q, Liu J‐a, Wang Z‐p, Zhao Z‐w, Kang N, Chen Y, Huo Q. 2021. Study on titanium dioxide nanoparticles as MALDI MS matrix for the determination of lipids in the brain. Green Proc. Synth. 10:700–710. 10.1515/gps-2021-0056 [DOI] [Google Scholar]
- Peng W, Kobeissy F, Mondello S, Barsa C, Mechref Y. 2022. MS‐based glycomics: An analytical tool to assess nervous system diseases. Front. Neurosci. 16:1000179. 10.3389/fnins.2022.1000179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Gu B, Sun Z, Li Y, Zhang Y, Lu H. 2021. Linkage‐selective derivatization for glycosylation site‐ and glycoform‐specific characterization of sialic acid isomers using mass spectrometry. Chem. Commun. 57:9590–9593. 10.1039/D1CC04142H [DOI] [PubMed] [Google Scholar]
- Pepi LE, Leach, III FE , Klein DR, Brodbelt JS, Amster IJ. 2021. Investigation of the experimental parameters of ultraviolet photodissociation for the structural characterization of chondroitin sulfate glycosaminoglycan isomers. J. Am. Soc. Mass Spectrom. 32:1759–1770. 10.1021/jasms.1c00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepi LE, Sanderson P, Stickney M, Amster IJ. 2021. Developments in mass spectrometry for glycosaminoglycan analysis: A review. Mol. Cell. Proteomics, 20:100025. 10.1074/mcp.R120.002267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peptu C, Blaj D‐A, Balan‐Porcarasu M, Rydz J. 2022. Cyclodextrin‐oligocaprolactone derivatives‐synthesis and advanced structural characterization by MALDI mass spectrometry. Polymers, 14:1436. 10.3390/polym14071436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira ACH, Auer AC, Biedel L, de Almeida CM, Romão W, Endringer DC. 2022. Analysis of Gliricidia sepium leaves by MALDI mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 33:573–583. 10.1021/jasms.1c00367 [DOI] [PubMed] [Google Scholar]
- Pérez‐López AV, Simpson J, Clench MR, Gomez‐Vargas AD, Ordaz‐Ortiz JJ. 2021. Localization and composition of fructans in stem and rhizome of Agave tequilana Weber var. azul. Front. Plant Sci. 11:608850. 10.3389/fpls.2020.608850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric L, Vukadin S, Petrovic A, Kuna L, Puseljic N, Sikora R, Rozac K, Vcev A, Smolic M. 2022. Glycosylation alterations in cancer cells, prognostic value of glycan biomarkers and their potential as novel therapeutic targets in breast cancer. Biomedicines, 10:3265. 10.3390/biomedicines10123265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit M, Walet‐Balieu M‐L, Schapman D, Golinski M‐L, Burel C, Barray M, Drouot L, Maho‐Vaillant M, Hébert V, Boyer O, Bardor M, Joly P, Calbo S. 2021. Longitudinal pathogenic properties and N‐glycosylation profile of antibodies from patients with pemphigus after corticosteroid treatment. Biomedicines, 9:1411. 10.3390/biomedicines9101411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia LMC, Santha E, Behrens AJ, Nguyen DL, Ganatra MB, Taron CH, Khatri V, Kalyanasundaram R, van Diepen A, Hokke CH, Foster JM. 2022. Alteration of rhesus macaque serum N‑glycome during infection with the human parasitic filarial nematode Brugia malayi . Sci. Rep. 12:15763. 10.1038/s41598-022-19964-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia LMC, van Diepen A, Lokker LA, Nguyen DL, Sartono E, Khatri V, Kalyanasundaram R, Taron CH, Foster JM, Hokke CH. 2022. Mass spectrometric and glycan microarray‐based characterization of the filarial nematode Brugia malayi glycome reveals anionic and zwitterionic glycan antigens. Mol. Cell. Proteomics, 21:100201. 10.1016/j.mcpro.2022.100201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrović T, Trbojević‐Akmačić I. 2021. Lectin and liquid chromatography‐based methods for immunoglobulin (G) glycosylation analysis. In: Pezer M, Editor, Antibody Glycosylation. Cham Switzerland: Springer Nature. p 29–72. 10.1007/978-3-030-76912-3_2 [DOI] [PubMed] [Google Scholar]
- Pfeifer L, Mueller K‐K, Classen B. 2022. The cell wall of hornworts and liverworts: Innovations in early land plant evolution? J. Exp. Bot. 73:4454–4472. 10.1093/jxb/erac157 [DOI] [PubMed] [Google Scholar]
- Picariello G, Sciammaro LP, Puppo MC, Mamone G. 2022. Omic sciences for analysis of different Prosopis species. In: Puppo MC, Felker P, Editors., Prosopis as a Heat Tolerant Nitrogen Fixing Desert Food Legume. Prospects for Economic Development in Arid Lands. Santiago del Estero, Argentina, Vista, California: Elsevier. p 263–273. 10.1016/B978-0-12-823320-7.00007-9 [DOI] [Google Scholar]
- Pietkiewicz D, Plewa S, Zaborowski M, Garrett TJ, Matuszewska E, Kokot ZJ, Matysiak J. 2022. Mass spectrometry imaging in gynecological cancers: The best is yet to come. Cancer Cell Int., 22:414. 10.1186/s12935-022-02832-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piirainen MA, Salminen H, Frey AD. 2022. Production of galactosylated complex‑type N‑glycans in glycoengineered Saccharomyces cerevisiae . Appl. Microbiol. Biotechnol. 106:301–315. 10.1007/s00253-021-11727-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilo HBA, Khilji SK, Lühle J, Biskup K, Gal BL, Goldian IR, Alfandari D, Revach O‐Y, Kiper E, Morandi MI, Rotkopf R, Porat Z, Blanchard V, Seeberger PH, Regev‐Rudzki N, Moscovitz O. 2022. Sialylated N‐glycans mediate monocyte uptake of extracellular vesicles secreted from Plasmodium falciparum‐infected red blood cells. J. Extracellular Biol. 1:33. 10.1002/jex2.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky W, Harris A, Roseborough AD, Wang W, Khan AR, Jurcic K, Yeung KK‐C, Pasternak SH, Whitehead SN. 2021. Regional lipid expression abnormalities identified using MALDI IMS correspond to MRI‐defined white matter hyperintensities within post‐mortem human brain tissues. Anal. Chem. 93:2652–2659. 10.1021/acs.analchem.0c05017 [DOI] [PubMed] [Google Scholar]
- Pither MD, Mantova G, Scaglione E, Pagliuca C, Colicchio R, Vitiello M, Chernikov OV, Hua K‐F, Kokoulin MS, Silipo A, Salvatore P, Molinaro A, Di Lorenzo F. 2021. The unusual lipid A structure and immunoinhibitory activity of LPS from marine bacteria Echinicola pacifica KMM 6172T and Echinicola vietnamensis KMM 6221T . Microorganisms, 9:2552. 10.3390/microorganisms9122552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pither MD, McClean S, Silipo A, Molinaro A, Di Lorenzo F. 2021. A chronic strain of the cystic fibrosis pathogen Pandoraea pulmonicola expresses a heterogenous hypo‐acylated lipid A. Glycoconj. J. 38:135–144. 10.1007/s10719-020-09954-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pither MD, Illiano A, Pagliuca C, Jacobson A, Mantova G, Stornaiuolo A, Colicchio R, Vitiello M, Pinto G, Silipo A, Fischbach MA, Salvatore P, Amoresano A, Molinaro A, Di Lorenzo F. 2022. Bacteroides thetaiotaomicron rough‐type lipopolysaccharide: The chemical structure and the immunological activity. Carbohydr. Polym. 297:120040. 10.1016/j.carbpol.2022.120040 [DOI] [PubMed] [Google Scholar]
- Pither MD, Sun ML, Speciale I, Silipo A, Zhang YZ, Molinaro A, Di Lorenzo F. 2022. Structural determination of the lipid A from the deep‑sea bacterium Zunongwangia profunda SM‑A87: A small‑scale approach. Glycoconj. J. 39:565–578. 10.1007/s10719-022-10076-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzato J, Tang W, Bernabeu S, Bonnin RA, Bille E, Farfour E, Guillard T, Barraud O, Cattoir V, Plouzeau C, Corvec S, Shahrezaei V, Dortet L, Larrouy‐Maumus G. 2022. Discrimination of Escherichia coli, Shigella flexneri, and Shigella sonnei using lipid profiling by MALDI‐TOF mass spectrometry paired with machine learning. Microbiol. Open, 11:e1313. 10.1002/mbo3.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleskunov P, Prysiazhnyi V, Nikitin D, Košutová T, Cieslar M, Gordeev I, Krtouš Z, Ali‐Ogly S, Šomvársky J, Protsak M, Biliak K, Kishenina K, Bednařík A, Dopita M, Preisler J, Choukourov A. 2022. Magnetron‐sputtered niobium nanoparticles for molecular imaging of brain tissues through surface‐assisted laser desorption/ionization mass spectrometry. Anal. Chem. 94:12865–12875. 10.1021/acsanm.2c02734 [DOI] [Google Scholar]
- Podvalnyy NM, Chesnov S, Nanni P, Gut M, Holland JP, Hennet T. 2021. Synthesis of photoactivable oligosaccharide derivatives from 1,2‐cyclic carbamate building blocks and study of their interaction with carbohydrate‐binding proteins. Carbohydr. Res. 508:108399. 10.1016/j.carres.2021.108399 [DOI] [PubMed] [Google Scholar]
- Polonio Á, Fernández‐Ortuño D, de Vicente A, Pérez‐García A. 2021. A haustorial‐expressed lytic polysaccharide monooxygenase from the cucurbit powdery mildew pathogen Podosphaera xanthii contributes to the suppression of chitin‐triggered immunity. Mol. Plant Pathol. 22:580–601. 10.1111/mpp.13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongracz T, Verhoeven A, Wuhrer M, de Haan N. 2021. The structure and role of lactone intermediates in linkage‐specific sialic acid derivatization reactions. Glycoconj. J. 38:157–166. 10.1007/s10719-020-09971-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnapalli KK, Ho Y‐C, Tseng M‐C, Vasamsetti BVS, Shie J‐J. 2022. One‐pot glycosylation strategy assisted by ion mobility−mass spectrometry analysis toward the synthesis of N‑linked oligosaccharides. J. Org. Chem. 87:5339–5357. 10.1021/acs.joc.2c00184 [DOI] [PubMed] [Google Scholar]
- Popov RS, Ivanchina NV, Dmitrenok PS. 2022. Application of MS‐based metabolomic approaches in analysis of starfish and sea cucumber bioactive compounds. Mar. Drugs, 20:320. 10.3390/md20050320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothukuchi P, Agliarulo I, Pirozzi M, Rizzo R, Russo D, Turacchio G, Nüchel J, Yang J‐S, Gehin C, Capolupo L, Hernandez‐Corbacho MJ, Biswas A, Vanacore G, Dathan N, Nitta T, Henklein P, Thattai M, Inokuchi J‐I, Hsu VW, Plomann M, Obeid LM, Hannun YA, Luini A, D'Angelo G, Parashuraman S. 2021. GRASP55 regulates intra‐Golgi localization of glycosylation enzymes to control glycosphingolipid biosynthesis. EMBO J. 40:e107766. 10.15252/embj.2021107766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff A, Minte O, Dreisewerd K, Soltwisch J. 2022. Effect of the laser pulse width in MALDI‐2: A comparative study of picosecond versus nanosecond wide pulses for laser postionization. J. Am. Soc. Mass Spectrom. 33:315–321. 10.1021/jasms.1c00308 [DOI] [PubMed] [Google Scholar]
- Powell AK, Harvey DJ. 1996. Stabilisation of sialic acids in N‐linked oligosaccharides and gangliosides for analysis by positive ion matrix‐assisted laser desorption‐ionization mass spectrometry. Rapid Commun. Mass Spectrom., 10:1027–1032. [DOI] [PubMed] [Google Scholar]
- Prabhu A, Shanmugam D, Gadgil M. 2022. Engineering nucleotide sugar synthesis pathways for independent and simultaneous modulation of N‐glycan galactosylation and fucosylation in CHO cells. Metab. Eng. 74:61–71. 10.1016/j.ymben.2022.09.003 [DOI] [PubMed] [Google Scholar]
- Prabhu SK, Li C, Zong G, Zhang R, Wang L‐X. 2021. Comparative studies on the substrate specificity and defucosylation activity of three α‐l‐fucosidases using synthetic fucosylated glycopeptides and glycoproteins as substrates. Bioorg. Med. Chem. 42:116243. 10.1016/j.bmc.2021.116243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradita T, Chen Y‐J, Mernie EG, Bendulo SN, Chen Y‐J. 2021. ZIC‐cHILIC functionalized magnetic nanoparticle for rapid and sensitive glycopeptide enrichment from <1 μL serum. Nanomaterials, 11:2159. 10.3390/nano11092159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralow A, Cajic S, Alagesan K, Kolarich D, Rapp E. 2021. State‐of‐the‐art glycomics technologies in glycobiotechnology. Adv. Biochem. Eng. Biotechnol. 175:379–412. 10.1007/10_2020_143 [DOI] [PubMed] [Google Scholar]
- Prasanna M, Podsiadla‐Bialoskorska M, Mielecki D, Ruffier N, Fateh A, Lambert A, Fanuel M, Camberlein E, Szolajska E, Grandjean C. 2021. On the use of adenovirus dodecahedron as a carrier for glycoconjugate vaccines. Glycoconj. J. 38:437–446. 10.1007/s10719-021-09999-3 [DOI] [PubMed] [Google Scholar]
- Prestegard JH. 2021. A perspective on the PDB's impact on the field of glycobiology. J. Biol. Chem., 296:100556. 10.1016/j.jbc.2021.100556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NPJ, Jackson MA, Hartman TM, Brändén G, Ek M, Koch AA, Kennedy PD. 2021. Branched chain lipid metabolism as a determinant of the N‑acyl variation of Streptomyces natural products. ACS Chem. Biol. 16:116–124. 10.1021/acschembio.0c00799 [DOI] [PubMed] [Google Scholar]
- Pu C, Biyuan, Xu K , Zhao Y. 2022. Glycosylation and its research progress in endometrial cancer. Clinical Transl. Oncol. 24:1865–1880. 10.1007/s12094-022-02858-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J, Wang L, Zhang W, Ma J, Zhang X, Putnis CV. 2021. Organically‐bound silicon enhances resistance to enzymatic degradation and nanomechanical properties of rice plant cell walls. Carbohydr. Polym. 266:118057. 10.1016/j.carbpol.2021.118057 [DOI] [PubMed] [Google Scholar]
- Pujić I, Perreault H. 2022. Recent advancements in glycoproteomic studies: Glycopeptide enrichment and derivatization, characterization of glycosylation in SARS CoV2, and interacting glycoproteins. Mass Spectrom. Rev. 41:488–507. 10.1002/mas.21679 [DOI] [PubMed] [Google Scholar]
- Pupo E, van der Ley P, Meiring HD. 2021. Nanoflow LC−MS method allowing in‐depth characterization of natural heterogeneity of complex bacterial lipopolysaccharides. Anal. Chem. 93:15832–15839. 10.1021/acs.analchem.1c01043 [DOI] [PubMed] [Google Scholar]
- Purcell AB, Voss BJ, Trent MS. 2022. Diacylglycerol kinase A is essential for polymyxin resistance provided by EptA, MCR‐1, and other lipid A phosphoethanolamine transferases. J. Bacteriol. 204:e00498–00421. 10.1128/jb.00498-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushina M, Penavic A, Farshbaf S, Anzenbacher, Jr. P 2021. Fluorescent sensor array for quantitative determination of saccharides. ACS Sensors, 6:4001–4008. 10.1021/acssensors.1c01371 [DOI] [PubMed] [Google Scholar]
- Puspitasari YE, De Bruyne T, Foubert K, Aulanni'am A, Pieters L, Hermans N, Tuenter E. 2022. Holothuria triterpene glycosides: A comprehensive guide for their structure elucidation and critical appraisal of reported compounds. Phytochem. Rev. 21:1315–1358. 10.1007/s11101-021-09783-z [DOI] [Google Scholar]
- Pylkkänen R, Mohammadi P, Liljeström V, Płaziński W, Beaune G, Timonen JVI, Penttilä M. 2022. β‐1,3‐Glucan synthesis, novel supramolecular self‐assembly, characterization and application. Nanoscale, 14:15533–15541. 10.1039/D2NR02731C [DOI] [PubMed] [Google Scholar]
- Qi H, Jiang L, Jia Q. 2021. Application of magnetic solid phase extraction in separation and enrichment of glycoproteins and glycopeptides. Chinese Chem. Lett. 32:2629–2636. 10.1016/j.cclet.2021.01.037 [DOI] [Google Scholar]
- Qiao Z, Lissel F. 2021. MALDI Matrices for the analysis of low molecular weight compounds: Rational design, challenges and perspectives. Chem. Asian J. 16:868–878. 10.1002/asia.202100044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Qin S, Tian Z. 2022. Progresses in mass spectrometry‐based plant N‐glycomics and N‐glycoproteomics. Int. J. Mass Spectrom., 481:116917. 10.1016/j.ijms.2022.116917 [DOI] [Google Scholar]
- Qin S, Tian Z. 2022. Analytical software and databases in N‐glycoproteomics. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. London: Taylor and Francis. p 131–166. 10.1201/9781003185833-5 [DOI] [Google Scholar]
- Qin W, Pei H, Li X, Li J, Yao X, Zhang R. 2021. Serum protein N‐glycosylation signatures of neuroblastoma. Front. Oncol. 11:603417. 10.3389/fonc.2021.603417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Ma G, Liu L, Feng J, Zhou S, Han W, Zhou J, Liu Y, Zhang J. 2022. Microwave‐assisted degradation of β‐d‐glucan from Ganoderma lucidum and the structural and immunoregulatory properties of oligosaccharide fractions. Int. J. Biol. Macromol. 229:1197–1211. 10.1016/j.ijbiomac.2022.08.128 [DOI] [PubMed] [Google Scholar]
- Qiu S, Palavicini JP, Wang J, Gonzalez NS, He S, Dustin E, Zou C, Ding L, Bhattacharjee A, Van Skike CE, Galvan V, Dupree JL, Han X. 2021. Adult‐onset CNS myelin sulfatide deficiency is sufficient to cause Alzheimer's disease‐like neuroinflammation and cognitive impairment. Mol. Neurodegen. 16:64. 10.1186/s13024-021-00488-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Qiao Y, Zhang B, Sun‐Waterhouse D, Zheng Z. 2022. Bioactive polysaccharides and oligosaccharides from garlic (Allium sativum L.): Production, physicochemical and biological properties, and structure‐function relationships. Comp. Rev. Food Sci. Food Safety, 21:3033–3095. 10.1111/1541-4337.12972 [DOI] [PubMed] [Google Scholar]
- Qu M, Guo X, Tian S, Yang Q, Kim M, Mun S, Noh MY, Kramer KJ, Muthukrishnan S, Arakane Y. 2022. AA15 lytic polysaccharide monooxygenase is required for efficient chitinous cuticle turnover during insect molting. Commun. Biol. 5:518. 10.1038/s42003-022-03469-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quelhas D, Correia J, Jaeken J, Azevedo L, Lopes‐Marques M, Bandeira A, Keldermans L, Matthijs G, Sturiale L, Martins E. 2021. SLC35A2‐CDG: Novel variant and review. Mol. Genet. Metab. Rep. 26:100717. 10.1016/j.ymgmr.2021.100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus JM, Pellegrinelli RP, Khodr AHA, Bythell BJ, Rizzo TR, Carrascosa E. 2021. Unravelling the structures of sodiated β‐cyclodextrin and its fragments. Phys. Chem. Chem. Phys. 23:13714–13723. 10.1039/D1CP01058A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus JM, Guan S, Schultz LM, Abutokaikah MT, Maître P, Bythell BJ. 2022. Protonated α‑N‑acetyl galactose glycopeptide dissociation chemistry. J. Am. Soc. Mass Spectrom., 33:1745–1752. 10.1021/jasms.2c00155 [DOI] [PubMed] [Google Scholar]
- Radenkovic S, Fitzpatrick‐Schmidt T, Byeon SK, Madugundu AK, Saraswat M, Lichty A, Wong SYW, McGee S, Kubiak K, Ligezka A, Ranatunga W, Zhang Y, Wood T, Friez MJ, Clarkson K, Pandey A, Jones JR, Morava E. 2021. Expanding the clinical and metabolic phenotype of DPM2 deficient congenital disorders of glycosylation. Mol. Genet. Metab. 132:27–37. 10.1016/j.ymgme.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafińska K, Wrona O, Krakowska‐Sieprawska A, Walczak‐Skierska J, Kiełbasa A, Rafiński Z, Pomastowski P, Kolankowski M, Buszewski B. 2022. Enzyme‐assisted extraction of plant material ‐ New functional aspects of the process on an example of Medicago sativa L. Ind. Crops Prod. 187:115424. 10.1016/j.indcrop.2022.115424 [DOI] [Google Scholar]
- Rahman MA, Kuroda K, Endo H, Sasaki N, Hamada T, Sakai H, Nokami T. 2022. Synthesis of protected precursors of chitin oligosaccharides by electrochemical polyglycosylation of thioglycosides. Beilstein J. Org. Chem. 18:1133–1139. 10.3762/bjoc.18.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekar M, Agash SGS, Rajasekar K. 2022. Review of photoresponsive and glycoside dendrimers in biomaterials and sensors applications. RSC Adv., 12:35123–35150. 10.1039/D2RA06563K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Narasimman V, Rajesh P. 2022. Low molecular weight sulfated chitosan isolation, characterization and anti‐tuberculosis activity derived from Sepioteuthis lessoniana . Int. J. Biol. Macromol., 206:29–39. 10.1016/j.ijbiomac.2022.02.121 [DOI] [PubMed] [Google Scholar]
- Ramandi NF, Faranoush M, Ghassempour A, Aboul‐Enein HY. 2022. Mass spectrometry: A powerful method for monitoring various type of leukemia, especially MALDI‐TOF in leukemia's proteomics studies. Crit. Rev. Anal. Chem. 52:1259–1286. 10.1080/10408347.2021.1871844 [DOI] [PubMed] [Google Scholar]
- Ramos‐Sánchez MC, Martín‐Gil J, Buzón‐Durán L, Martín‐Ramos P. 2021. Cyttaria hariotii E. Fisch. as a promising source of pullulan and Mn(II)‐pullulan complexes for Mn‐deficiency remediation in winter cereals. Nat. Prod. Res. 35:6158–6162. 10.1080/14786419.2020.1831493 [DOI] [PubMed] [Google Scholar]
- Ramos‐Soriano J, Illescas BM, Pérez‐Sánchez A, Sánchez‐Bento R, Lasala F, Rojo J, Delgado R, Martín N. 2022. Topological and multivalent effects in glycofullerene oligomers as EBOLA virus inhibitors. Int. J. Mol. Sci. 23:5083. 10.3390/ijms23095083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D, Wang B, Zhong H, Yan Y, Ding C‐F. 2022. Construction of boric acid‐functionalized metal‐organic frameworks for glycopeptide recognition in the serum of cervical cancer patients. Rapid Commun. Mass Spectrom., 36:e9314. 10.1002/rcm.9314 [DOI] [PubMed] [Google Scholar]
- Rastogi L, Chaudhari AA, Sharma R, Pawar PAM. 2022. Arabidopsis GELP7 functions as a plasma membrane‑localized acetyl xylan esterase, and its overexpression improves saccharification efficiency. Plant Mol. Biol. 109:781–797. 10.1007/s11103-022-01275-8 [DOI] [PubMed] [Google Scholar]
- Ravula T, Ramamoorthy A. 2021. Synthesis, characterization, and nanodisc formation of non‐ionic polymers. Angew. Chem. Int. Ed. 60:16885–16888. 10.1002/anie.202101950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B, Schütz M, Mukherjee S, Jana S, Ray S, Marschall M. 2021. Exploiting the amazing diversity of natural source‐derived polysaccharides: Modern procedures of isolation, engineering, and optimization of antiviral activities. Polymers, 13:136. 10.3390/polym13010136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynor A, Haouari W, Ng BG, Cholet S, Harroche A, Raulet‐Bussian C, Lounis‐Ouaras S, Vuillaumier‐Barrot S, Pascreau T, Borgel D, Freeze HH, Fenaille F, Bruneel A. 2021. SLC37A4‐CDG: New biochemical insights for an emerging congenital disorder of glycosylation with major coagulopathy. Clin. Chim. Acta, 521:104–106. 10.1016/j.cca.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynor A, Vincent‐Delorme C, Alaix A‐S, Cholet S, Dupré T, Vuillaumier‐Barrot S, Fenaille F, Besmond C, Bruneel A. 2021. Normal transferrin patterns in congenital disorders of glycosylation with Golgi homeostasis disruption: Apolipoprotein C‐III at the rescue! Clin. Chim. Acta, 519:285–290. 10.1016/j.cca.2021.05.016 [DOI] [PubMed] [Google Scholar]
- Rebelo AL, Gubinelli F, Roost P, Jan C, Brouillet E, Van Camp N, Drake RR, Saldova R, Pandit A. 2021. Complete spatial characterisation of N‐glycosylation upon striatal neuroinflammation in the rodent brain. J. Neuroinflamm. 18:116. 10.1186/s12974-021-02163-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AL, Chevalier MT, Russo L, Pandit A. 2022. Role and therapeutic implications of protein glycosylation in neuroinflammation. Trends Mol. Med. 28:270–289. 10.1016/j.molmed.2022.01.004 [DOI] [PubMed] [Google Scholar]
- Reider B, Jarvas G, Krenkova J, Guttman A. 2021. Separation based characterization methods for the N‐glycosylation analysis of prostate‐specific antigen. J. Pharmaceut. Biomed. Anal., 194:113797. 10.1016/j.jpba.2020.113797 [DOI] [PubMed] [Google Scholar]
- Remoroza CA, Burke MC, Liu Y, Mirokhin YA, Tchekhovskoi DV, Yang X, Stein SE. 2021. Representing and comparing site‐specific glycan abundance distributions of glycoproteins. J. Proteome Res. 20:4475–4486. 10.1021/acs.jproteome.1c00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remoroza CA, Burke MC, Yang X, Sheetlin S, Mirokhin Y, Markey SP, Tchekhovskoi DV, Stein SE. 2022. Mass spectral library methods for analysis of site‐specific N‑glycosylation: Application to human milk proteins. J. Proteome Res. 21:2421–2434. 10.1021/acs.jproteome.2c00286 [DOI] [PubMed] [Google Scholar]
- Ren S, Lu H. 2022. Qualitative and quantitative methods for N‐glycans in N‐glycomics. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. London: Taylor and Francis. p 55–89. 10.1201/9781003185833-3 [DOI] [Google Scholar]
- Ren W, Bian Q, Cai Y. 2022. Mass spectrometry‐based N‐glycosylation analysis in kidney disease. Front. Mol. Biosci. 9:976298. 10.3389/fmolb.2022.976298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensner JJ, Lee YJ. 2022. Efficient hydrogen−deuterium exchange in matrix‐assisted laser desorption/ionization mass spectrometry imaging for confident metabolite identification. Anal. Chem. 94:11129–11133. 10.1021/acs.analchem.2c00978 [DOI] [PubMed] [Google Scholar]
- Ret D, Stefenatti L, Gentile A, Rohrhofer J, Knaus S, Untersmayr E. 2022. DMTMM‐mediated methylamidation for MALDI mass spectrometry analysis of N‐glycans with structurally conserved sialic acid residues in biological fluids “via direttissima”. Talanta, 242:123326. 10.1016/j.talanta.2022.123326 [DOI] [PubMed] [Google Scholar]
- Reynoud N, Geneix N, Petit J, D'Orlando A, Fanuel M, Marion D, Rothan C, Lahaye M, Bakan B. 2022. The cutin polymer matrix undergoes a fine architectural tuning from early tomato fruit development to ripening. Plant Physiol., 190:1821–1840. 10.1093/plphys/kiac392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyre J‐L, Grisel S, Haon M, Navarro D, Ropartz D, Le Gall S, Record E, Sciara G, Tranquet O, Berrin J‐G, Bissaro B. 2022. The maize pathogen Ustilago maydis secretes glycoside hydrolases and carbohydrate oxidases directed toward components of the fungal cell wall. Appl. Environ. Microbiol. 88:e0158122. 10.1128/aem.01581-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo MG, Reuber EE, Yao L, Danglad‐Flores J, Delbianco M, Seeberger PH. 2022. Design, synthesis, and characterization of stapled oligosaccharides. J. Am. Chem. Soc. 144:18429–18434. 10.1021/jacs.2c06882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard E, Traversier A, Julien T, Rosa‐Calatrava M, Putaux J‐L, Jeacomine I, Samain E. 2022. Biotechnological production of sialylated solid lipid microparticles as inhibitors of influenza A virus infection. Glycobiology, 32:949–961. 10.1093/glycob/cwac054 [DOI] [PubMed] [Google Scholar]
- Rieder L, Petrović D, Väljamäe P, Eijsink VGH, Sørlie M. 2021. Kinetic characterization of a putatively chitin‐active LPMO reveals a preference for soluble substrates and absence of monooxygenase activity. ACS Catal. 11:11685–11695. 10.1021/acscatal.1c03344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Righetti L, Gottwald S, Tortorella S, Spengler B, Bhandari DR. 2022. Mass spectrometry imaging disclosed spatial distribution of defense‐related metabolites in Triticum spp. Metabolites, 12:48. 10.3390/metabo12010048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley NM, Bertozzi CR, Pitteri SJ. 2021. A pragmatic guide to enrichment strategies for mass spectrometry‐based glycoproteomics. Mol. Cell. Proteomics, 20:100029. 10.1074/mcp.r120.002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo R, Russo D, Kurokawa K, Sahu P, Lombardi B, Supino D, Zhukovsky MA, Vocat A, Pothukuchi P, Kunnathully V, Capolupo L, Boncompain G, Vitagliano C, Marino FZ, Aquino G, Montariello D, Henklein P, Mandrich L, Botti G, Clausen H, Mandel U, Yamaji T, Hanada K, Budillon A, Perez F, Parashuraman S, Hannun YA, Nakano A, Corda D, D'Angelo G, Luini A. 2021. Golgi maturation‐dependent glycoenzyme recycling controls glycosphingolipid biosynthesis and cell growth via GOLPH3. EMBO J., 40:e107238. 10.15252/embj.2020107238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robakiewicz S, Bridot C, Serna S, Gimeno A, Echeverria B, Delgado S, de Ruyck J, Semwal S, Charro D, Dansercoer A, Verstraete K, Azkargorta M, van Noort K, Wilbers RHP, Savvides SN, Abrescia NGA, Arda A, Reichardt NC, Jiménez‐Barbero J, Bouckaert J. 2021. Minimal epitope for Mannitou IgM on paucimannose‐carrying glycoproteins. Glycobiology, 31:1005–1017. 10.1093/glycob/cwab027 [DOI] [PubMed] [Google Scholar]
- Robinson SD, Kambanis L, Clayton D, Hinneburg H, Corcilius L, Mueller A, Walker AA, Keramidas A, Kulkarni SS, Jones A, Vetter I, Thaysen‐Andersen M, Payne RJ, King GF, Undheim EAB. 2021. A pain‐causing and paralytic ant venom glycopeptide. iScience, 24:103175. 10.1016/j.isci.2021.103175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roblek M, Bicher J, van Gogh M, György A, Seeböck R, Szulc B, Damme M, Olczak M, Borsig L, Siekhaus DE. 2022. The solute carrier MFSD1 decreases the activation status of β1 integrin and thus tumor metastasis. Front. Oncol. 12:777634. 10.3389/fonc.2022.777634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo‑Frutos D, Jiménez‑Ortega E, Piedrabuena D, Ramírez‑Escudero M, Míguez N, Plou FJ, Sanz‑Aparicio J, Fernández‑Lobato M. 2021. New insights into the molecular mechanism behind mannitol and erythritol fructosylation by β‑fructofuranosidase from Schwanniomyces occidentalis . Sci. Rep. 11:7158. 10.1038/s41598-021-86568-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RCLB, Rodrigues BG, Canettieri EV, Martinez EA, Palladino F, Wisniewski, Jr. A , Rodrigues, Jr. D 2022. Comprehensive approach of methods for microstructural analysis and analytical tools in lignocellulosic biomass assessment—A review. Bioresource Technol., 348:126627. 10.1016/j.biortech.2021.126627 [DOI] [PubMed] [Google Scholar]
- Rodríguez V, Román J, Insuasty‐Cepeda D, Fierro‐Medina R, Rivera‐Monroy Z, García‐Castañeda J. 2021. Synthesis of glucosyl amino acid derivatives for obtaining N‐glucopeptides via SPPS: Optimization of the synthetic route. ChemistrySelect, 6:4083–4088. 10.1002/slct.202100063 [DOI] [Google Scholar]
- Rong Y, Wang X, Mao W, Yuan F, Chen M, Wang S, Wang PG, Wu Z, He Y, Kong Y. 2022. Chemoenzymatic synthesis of SARS‐CoV‑2 homogeneous O‑linked glycopeptides for exploring their inhibition functions. ACS Infect. Dis. 8:2198–2206. 10.1021/acsinfecdis.2c00383 [DOI] [PubMed] [Google Scholar]
- Rosenau J, Grothaus IL, Yang Y, Kumar ND, Ciacchi LC, Kelm S, Waespy M. 2022. N‐Glycosylation modulates enzymatic activity of Trypanosoma congolense trans‐sialidase. J. Biol. Chem. 298:102403. 10.1016/j.jbc.2022.102403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roushan A, Wilson GM, Kletter D, Sen KI, Tang W, Kil YJ, Carlson E, Bern M. 2021. Peak filtering, peak annotation, and wildcard search for glycoproteomics. Mol. Cell. Proteomics, 20:100011. 10.1074/mcp.ra120.002260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau A, Armand S, Cottaz S, Fort S. 2021. Size‐controlled synthesis of β(1→4)‐GlcNAc oligosaccharides using an endo‐glycosynthase. Chem. Eur. J. 27:17637–17646. 10.1002/chem.202103212 [DOI] [PubMed] [Google Scholar]
- Rubén L‐C, Laura M‐R, Almudena F‐B, Emilio GM. 2021. Glycan array analysis of Pholiota squarrosa lectin and other fucose‐oriented lectins. Glycobiology, 31:459–476. 10.1093/glycob/cwaa093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko N, Karatovskaya A, Zamyatina A, Shepelyakovskaya A, Semushina S, Brovko F, Shpirt A, Torgov V, Kolotyrkina N, Zinin A, Kasimova A, Perepelov A, Shneider M, Knirel Y. 2022. Immune response to conjugates of fragments of the type K9 capsular polysaccharide of Acinetobacter baumannii with carrier proteins. Microbiol. Spectrum, 10:e01674‐22. 10.1128/spectrum.01674-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf A, Kanawati B, Schmitt‐Kopplin P. 2022. Dihydrogen phosphate anion boosts the detection of sugars in electrospray ionization mass spectrometry: A combined experimental and computational investigation. Rapid Commun. Mass Spectrom., 36:e9283. 10.1002/rcm.9283 [DOI] [PubMed] [Google Scholar]
- Ruiz‐Cruz S, Garzon AE, Kelleher P, Bottacini F, Breum SØ, Neve H, Heller KJ, Vogensen FK, Palussière S, Courtin P, Chapot‐Chartier M‐P, Vinogradov E, Sadovskaya I, Mahony J, van Sinderen D. 2022. Host genetic requirements for DNA release of lactococcal phage TP901‐1. Microb. Biotechnol. 15:2875–2889. 10.1111/1751-7915.14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumachik N, Tian T, Hou Y, Saini C, Cheng J, Pohl C, Liu Y. 2021. Towards a more complete glycome: Advances in ion chromatography‐mass spectrometry (IC‐MS) for improved separation and analysis of carbohydrates. J. Chromatogr., B, 1175:122719. 10.1016/j.jchromb.2021.122719 [DOI] [PubMed] [Google Scholar]
- Rumiantseva L, Osipenko S, Zharikov A, Kireev A, Nikolaev EN, Kostyukevich Y. 2022. Analysis of 16O/18O and H/D exchange reactions between carbohydrates and heavy water using high‐resolution mass spectrometry. Int. J. Mol. Sci. 23:3585. 10.3390/ijms23073585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater C, Blanco‐Doval A, Margolles A, Corzo N, Montilla A. 2021. Artichoke pectic oligosaccharide characterisation and virtual screening of prebiotic properties using in silico colonic fermentation. Carbohydr. Polym. 255:117367. 10.1016/j.carbpol.2020.117367 [DOI] [PubMed] [Google Scholar]
- Sabater C, Blanco‐Doval A, Montilla A, Corzo N. 2021. Optimisation of an enzymatic method to obtain modified artichoke pectin and pectic oligosaccharides using artificial neural network tools. In silico and in vitro assessment of the antioxidant activity. Food Hydrocolloid. 110:106161. 10.1016/j.foodhyd.2020.106161 [DOI] [Google Scholar]
- Sabbadin F, Henrissat B, Bruce NC, McQueen‐Mason SJ. 2021. Lytic polysaccharide monooxygenases as chitin‐specific virulence factors in crayfish plague. Biomolecules, 11:1180. 10.3390/biom11081180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbavarapu NM, Seeberger PH. 2021. Automated glycan assembly of oligogalactofuranosides reveals the influence of protecting groups on oligosaccharide stability. J. Org. Chem. 86:7280–7287. 10.1021/acs.joc.1c00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeidy S, Petera B, Pierre G, Fenoradosoa TA, Djomdi D, Michaud P, Delattre C. 2021. Plants arabinogalactans: From structures to physico‐chemical and biological properties. Biotechnol. Adv. 53:107771. 10.1016/j.biotechadv.2021.107771 [DOI] [PubMed] [Google Scholar]
- Saitarly S, Pushkarev Y, Nesterkina M, Öztürk S, Salih B, Kravchenko I. 2022. Rheological properties of hyaluronic acid diluted solutions as components of cosmetics. Biointerface Res. Appl. Chem. 12:1907–1915. 10.33263/BRIAC122.19071915 [DOI] [Google Scholar]
- Saito T, Watanabe A, Nakano M, Matsuo K. 2021. MALDI‐TOF mass spectrometry imaging for N‐glycans on FFPE tissue sections of mouse NASH liver through sialic acid benzylamidation. Glycoconj. J. 38:167–175. 10.1007/s10719-021-09984-w [DOI] [PubMed] [Google Scholar]
- Sajid MS, Saleem S, Jabeen F, Fatima B, Zulfikar M, Ashiq MN, Ressom HW, Pukala TL, Najam‑ul‑Haq M. 2021. Iminodiacetic acid (IDA)‑generated mesoporous nanopolymer: A template to relate surface area, hydrophilicity, and glycopeptides enrichment. Microchim. Acta, 188:417. 10.1007/s00604-021-05074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid MS, Saleem MN, Jabeen F, Saleem S, Iqbal S, Habib S, Ashiq MN, Ressom HW, Najam‐ul‐Haq M. 2022. Human serum N‐glycome profiling via the newly developed asparagine immobilized cellulose/polymer nanohybrid. J. Sep. Sci., 45:4236–4244. 10.1002/jssc.202200179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajid MS, Saleem S, Jabeen F, Najam‑ul‑Haq M, Ressom HW. 2022. Terpolymeric platform with enhanced hydrophilicity via cysteic acid for serum intact glycopeptide analysis. Microchim. Acta, 189:277. 10.1007/s00604-022-05343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai‐Otsuka Y, Ogawa Y, Satoh T, Chen W‐C, Borsali R. 2021. Carbohydrate‐attached fullerene derivative for selective localization in ordered carbohydrate‐block‐poly(3‐hexylthiophene) nanodomains. Carbohydr. Polym. 255:117528. 10.1016/j.carbpol.2020.117528 [DOI] [PubMed] [Google Scholar]
- Sakhi S, Cholet S, Wehbi S, Isidor B, Cogne B, Vuillaumier‐Barrot S, Dupré T, Detleft T, Schmitt E, Leheup B, Bonnet C, Feillet F, Muti C, Fenaille F, Bruneel A. 2021. MAN1B1‐CDG: Three new individuals and associated biochemical profiles. Mol. Genet. Metab. Rep. 28:100775. 10.1016/j.ymgmr.2021.100775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai Y, Sawada T, Serizawa T. 2022. Phosphorylase‐catalyzed synthesis and self‐assembled structures of cellulose oligomers in the presence of protein denaturants. Polymer J. 54:561–569. 10.1038/s41428-021-00592-x [DOI] [Google Scholar]
- Saladino R, Bizzarri BM, Botta L, Šponer J, Šponer JE, Georgelin T, Jaber M, Rigaud B, Kapralov M, Timoshenko GN, Rozanov A, Krasavin E, Timperio AM, Di Mauro E. 2021. Proton irradiation: A key to the challenge of N‐glycosidic bond formation in a prebiotic context. Sci. Rep. 7:14709. 10.1038/s41598-017-15392-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviati E, Sommella E, Campiglia P. 2022. MALDI‐mass spectrometry imaging: The metabolomic visualization. In: Troisi J, Editor, Metabolomics Perspectives. From Theory to Practical Application. London; San Diego; Oxford, UK, Cambridge, MA: Elsevier Academic Press. p 535–551. 10.1016/B978-0-323-85062-9.00015-5 [DOI] [Google Scholar]
- Samarah LZ, Tran TH, Stacey G, Vertes A. 2021. Mass spectrometry imaging of bio‐oligomer polydispersity in plant tissues by laser desorption ionization from silicon nanopost arrays. Angew. Chem. Int. Ed., 60:9071–9077. 10.1002/anie.202015251 [DOI] [PubMed] [Google Scholar]
- Samarah LZ, Vertes A. 2022. Mass spectrometry imaging of biological tissues by laser desorption ionization from silicon nanopost arrays. Method. Mol. Biol. 2437:89–98. 10.1007/978-1-0716-2030-4_6 [DOI] [PubMed] [Google Scholar]
- Samarah LZ, Vertes A, Anderton CR. 2022. Single‐cell metabolomics with rapid determination of chemical formulas from isotopic fine structures. Method. Mol. Biol. 2437:61–75. 10.1007/978-1-0716-2030-4_4 [DOI] [PubMed] [Google Scholar]
- San Clemente H, Jamet E. 2022. N‐Glycoproteins in plant cell walls: A survey. Plants, 11:3204. 10.3390/plants11233204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbu M, Ica R, Zamfir AD. 2021. Developments and applications of separation and microfluidics methods coupled to electrospray mass spectrometry in glycomics of nervous system gangliosides. Electrophoresis, 42:429–449. 10.1002/elps.202000236 [DOI] [PubMed] [Google Scholar]
- Sarbu M, Zamfir AD. 2021. Modern techniques for separation, mass spectrometric detection, and characterization of glycolipids. In: El Rassi Z, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 485–527. 10.1016/B978-0-12-821447-3.00006-8 [DOI] [Google Scholar]
- Sarbu M, Ica R, Zamfir AD. 2022. Gangliosides as biomarkers of human brain diseases: Trends in discovery and characterization by high‐performance mass spectrometry. Int. J. Mol. Sci. 23:693. 10.3390/ijms23020693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyatun R, Kajiura H, Limkul J, Misaki R, Fujiyama K. 2021. Analysis of N‐glycan profile of Arabidopsis alg3 cell culture. Plant Biotechnol., 38:463–467. 10.5511/plantbiotechnology.21.1025a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah S, Dhurua S, Belwal VK, Lyndem S, Jana M, Roy AS. 2022. Combined spectroscopic and computational approaches for the recognition of bioactive flavonoid 6‐hydroxyflavone by human serum albumin: Effects of non‐enzymatic glycation in the binding. J. Mol. Liquids, 346:118288. 10.1016/j.molliq.2021.118288 [DOI] [Google Scholar]
- Sarmikasoglou E, Vinyard JR, Khan MS, Jiranantasak T, Ravelo A, Lobo RR, Fan P, Jeong KC, Tuanyok A, Faciola A. 2021. Ruminal lipid A analysis by matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry. Polysaccharides, 2:817–824. 10.3390/polysaccharides2040049 [DOI] [Google Scholar]
- Sasaki Y, Komeno M, Ishiwata A, Horigome A, Odamaki T, Xiao J‐Z, Tanaka K, Ito Y, Kitahara K, Ashida H, Fujita K. 2022. Mechanism of cooperative degradation of gum arabic arabinogalactan protein by Bifidobacterium longum surface enzymes. Appl. Environ. Microbiol. 88:e02187–02121. 10.1128/aem.02187-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasiene ZJ, Mendis PM, Ropartz D, Rogniaux H, Jackson GP. 2021. The influence of Na/H exchange on the charge transfer dissociation (CTD) spectra of mannuronic acid oligomers. Int. J. Mass Spectrom. 468:116634. 10.1016/j.ijms.2021.116634 [DOI] [Google Scholar]
- Sasiene ZJ, Ropartz D, Rogniaux H, Jackson GP. 2021. Charge transfer dissociation of a branched glycan with alkali and alkaline earth metal adducts. J. Mass Spectrom. 56:e4774. 10.1002/jms.4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino P, Colson E, Caulier G, Eeckhaut I, Flammang P, Gerbaux P. 2022. Microwave‐assisted desulfation of the hemolytic saponins extracted from Holothuria scabra viscera. Molecules, 27:537. 10.3390/molecules27020537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savková K, Huszár S, Baráth P, Pakanová Z, Kozmon S, Vancová M, Tesařová M, Blaško J, Kaliňák M, Singh V, Korduláková J, Mikušová K. 2021. An ABC transporter Wzm‐Wzt catalyzes translocation of lipid‐linked galactan across the plasma membrane in mycobacteria. Proc. Natl. Acad. Sci. USA, 118:e2023663118. 10.1073/pnas.2023663118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafati V, Troilo F, Ponziani S, Giovannoni M, Scortica A, Pontiggia D, Angelucci F, Di Matteo A, Mattei B, Benedetti M. 2022. Characterization of two 1,3‑β‑glucan‑modifying enzymes from Penicillium sumatraense reveals new insights into 1,3‑β‑glucan metabolism of fungal saprotrophs. Biotechnol. Biofuels Bioprod., 15:138. 10.1186/s13068-022-02233-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller J, Lemmnitzer K, Dürig J‐N, Rademann J. 2021. Insights into structure, affinity, specificity, and function of GAG‐protein interactions through the chemoenzymatic preparation of defined sulfated oligohyaluronans. Biol. Chem. 402:1375–1384. 10.1515/hsz-2021-0165 [DOI] [PubMed] [Google Scholar]
- Schmutzler S, Knappe D, Marx A, Hoffmann R. 2021. Solid‑phase synthesis of d‐fructose‐derived Heyns peptides utilizing Nα‑Fmoc‐Lysin[Nε‐(2‐deoxy‐d‐glucos‐2‐yl),Nε‐Boc]‐OH as building block. Amino Acids, 53:881–891. 10.1007/s00726-021-02989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutzler S, Hoffmann R. 2022. Chromatographic separation of glycated peptide isomers derived from glucose and fructose. Anal. Bioanal. Chem. 414:6801–6812. 10.1007/s00216-022-04243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg LK, Thorn DA, Barnette D, Jones EE. 2022. MALDI imaging mass spectrometry: An emerging tool in neurology. Meta. Brain Dis. 37:105–121. 10.1007/s11011-021-00797-2 [DOI] [PubMed] [Google Scholar]
- Schneemann J, Schäfer K‐C, Spengler B, Heiles S. 2022. IR‐MALDI mass spectrometry imaging with plasma post‐ionization of nonpolar metabolites. Anal. Chem. 94:16086–16094. 10.1021/acs.analchem.2c03247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn K. 2022. MALDI mass spectrometry imaging in the clinical landscape. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 408–431. 10.1039/9781839165191-00408 [DOI] [Google Scholar]
- Schwedler C, Grzeski M, Kappert K, Rust J, Heymann G, Hoppe B, Blanchard V. 2022. Coronavirus disease 2019‐related alterations of total and anti‐spike IgG glycosylation in relation to age and anti‐spike IgG titer. Front. Microbiol. 13:775186. 10.3389/fmicb.2022.775186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, Wang M, Grauzam S, Pippin S, Black A, Angel PM, Drake RR, Castellino S, Kono Y, Rockey DC, Mehta AS. 2022. GlycoFibroTyper: A novel method for the glycan analysis of IgG and the development of a biomarker signature of liver fibrosis. Front. Immunol. 13:797460. 10.3389/fimmu.2022.797460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ščupáková K, Adelaja OT, Balluff B, Ayyappan V, Tressler CM, Jenkinson NM, Claes BSR, Bowman AP, Cimino‐Mathews AM, White MJ, Argani P, Heeren RMA, Glunde K. 2021. Clinical importance of high‐mannose, fucosylated, and complex N‐glycans in breast cancer metastasis. JCI Insight, 6:146945. 10.1172/jci.insight.146945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastião MJ, Marcos‐Silva L, Gomes‐Alves P, Alves PM. 2021. Proteomic and glyco(proteo)mic tools in the profiling of cardiac progenitors and pluripotent stem cell derived cardiomyocytes: Accelerating translation into therapy. Biotechnol. Adv. 49:107755. 10.1016/j.biotechadv.2021.107755 [DOI] [PubMed] [Google Scholar]
- Šebela M. 2022. Biomolecular profiling by MALDI‐TOF mass spectrometry in food and beverage analyses. Int. J. Mol. Sci. 23:13631. 10.3390/ijms232113631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberger PH. 2021. Discovery of semi‐ and fully‐synthetic carbohydrate vaccines against bacterial infections using a medicinal chemistry approach. Chem. Rev. 121:3598–3626. 10.1021/acs.chemrev.0c01210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegers C, Roth IR, Zarnovican P, Buettner FFR, Routier FH. 2022. Characterization of a gene cluster involved in Aspergillus fumigatus zwitterionic glycosphingolipid synthesis. Glycobiology, 32:814–824. 10.1093/glycob/cwac036 [DOI] [PubMed] [Google Scholar]
- Sejalon‐Cipolla M, Bruyat P, Bregant S, Malgorn C, Devel L, Subra G, Cantel S. 2021. Targeting out of range biomolecules: Chemical labeling strategies for qualitative and quantitative MALDI MS‐based detection. Trends Anal. Chem. 143:116399. 10.1016/j.trac.2021.116399 [DOI] [Google Scholar]
- Seo N, Lee H, Oh MJ, Kim GH, Lee SG, Ahn JK, Cha H‐S, Kim KH, Kim J, An HJ. 2021. Isomer‐specific monitoring of sialylated N‐glycans reveals association of α2,3‐linked sialic acid epitope with Behcet's disease. Front. Mol. Biosci. 8:778851. 10.3389/fmolb.2021.778851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serang O, Froehlich JW, Muntel J, McDowell G, Steen H, Lee RS, Steen JA. 2013. SweetSEQer, Simple de novo filtering and annotation of glycoconjugate mass spectra. Mol. Cell. Proteomics, 12:1735–1740. 10.1074/mcp.O112.025940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serb A‐F, Novaconi C, Georgescu M, Puiu M, Dema A, Onulov R, Sisu E. 2022. Preliminary analysis of the glycolipid profile in secondary brain tumors. BioMed Res. Int. 2022:4293172,. 10.1155/2022/4293172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa T, Tanaka S, Sawada T. 2021. Control of parallel versus antiparallel molecular arrangements in crystalline assemblies of alkyl β‐cellulosides. J. Colloid Interf. Sci., 601:505–516. 10.1016/j.jcis.2021.05.117 [DOI] [PubMed] [Google Scholar]
- Serra I, Piccinini D, Paradisi A, Ciano L, Bellei M, Bortolotti CA, Battistuzzi G, Sola M, Walton PH, Di Rocco G. 2022. Activity and substrate specificity of lytic polysaccharide monooxygenases: An ATR FTIR‐based sensitive assay tested on a novel species from Pseudomonas putida . Protein Sci., 31:591–601. 10.1002/pro.4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrain CM, Fontaine D, Bauduin A, Guéguinou M, Zhang BL, Chantôme A, Mahéo K, Pasqualin C, Maupoil V, Couthon H, Vandier C, Jaffrès P‐A. 2021. Thio‐ether functionalized glycolipid amphiphilic compounds reveal a potent activator of SK3 channel with vasorelaxation effect. Org. Biomol. Chem. 19:2753–2766. 10.1039/d1ob00021g [DOI] [PubMed] [Google Scholar]
- Sgobba E, Daguerre Y, Giampà M. 2021. Unravel the local complexity of biological environments by MALDI mass spectrometry imaging. Int. J. Mol. Sci. 22:12393. 10.3390/ijms222212393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C, Zhao C, Nie P, Liu J, Du Y. 2021. Effective total synthesis of schaftoside. Tetrahedron, 91:132216. 10.1016/j.tet.2021.132216 [DOI] [Google Scholar]
- Shao H, Lai L, Xu D, Crommen J, Wang Q, Jiang Z. 2021. Development of zirconium modified adenosine triphosphate functionalized monolith for specific enrichment of N‐glycans. J. Chromatogr., A, 1644:462090. 10.1016/j.chroma.2021.462090 [DOI] [PubMed] [Google Scholar]
- Sharma A, Rejeeth C, Vivek R, Babu VN, Ding X. 2021. Novel green silver nanoparticles as matrix in the detection of small molecules using matrix‐assisted laser desorption ionization mass spectrometry (MALDI‐MS). J. Pharmaceut Innovat. 16:715–725. 10.1007/s12247-020-09486-6 [DOI] [Google Scholar]
- Sharma K, Sharma N, Handa S, Pathania S. 2021. Purification and characterization of novel exopolysaccharides produced from Lactobacillus paraplantarum KM1 isolated from human milk and its cytotoxicity. J. Genetic Eng. Biotechnol., 18:56. 10.1186/s43141-020-00063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Porterfield JE, An H‐T, Jimenez AS, Lee S, Kannan S, Sharma A, Kannan RM. 2021. Rationally designed galactose dendrimer for hepatocyte‐specific targeting and intracellular drug delivery for the treatment of liver disorders. Biomacromolecules, 22:3574–3589. 10.1021/acs.biomac.1c00649 [DOI] [PubMed] [Google Scholar]
- Sharma R, Garcia E, Diep JK, Lee VH, Minhaj F, Jermain B, Ellis‐Grosse EJ, Abboud CS, Rao GG. 2022. Pharmacodynamic and immunomodulatory effects of polymyxin B in combination with fosfomycin against KPC‐2‐producing Klebsiella pneumoniae . Int. J. Antimicrob. Agents, 59:106566. 10.1016/j.ijantimicag.2022.106566 [DOI] [PubMed] [Google Scholar]
- Sharma Y, Ahlawat S, Rao A. 2022. Biochemical characterization of an inverting S/O‐HexNAc‐transferase and evidence of S‐linked glycosylation in Actinobacteria. Glycobiology, 32:148–161. 10.1093/glycob/cwab089 [DOI] [PubMed] [Google Scholar]
- Shen J, Jia L, Dang L, Su Y, Zhang J, Xu Y, Zhu B, Chen Z, Wu J, Lan R, Hao Z, Ma C, Zhao T, Gao N, Bai J, Zhi Y, Li J, Zhang J, Sun S. 2021. StrucGP: De novo structural sequencing of site‐specific N‐glycan on glycoproteins using a modularization strategy. Nat. Meth. 18:921–929. 10.1038/s41592-021-01209-0 [DOI] [PubMed] [Google Scholar]
- Sheng Q, Li J, Chen Y, Liang X, Lan M. 2021. Hydrophilic graphene oxide‐dopamine‐cationic cellulose composites and their applications in N‐glycopeptides enrichment. Talanta, 226:122112. 10.1016/j.talanta.2021.122112 [DOI] [PubMed] [Google Scholar]
- Sheng Q, Xue C, Zhou Y, Li J, Yuan H, Ke Y, Lan M. 2021. Synthesis of Al3+‐doping‐TiO2 monodisperse microspheres and their application for phosphopeptides and glycopeptides enrichment. Talanta, 223:121715. 10.1016/j.talanta.2020.121715 [DOI] [PubMed] [Google Scholar]
- Sherman ME, Smith RD, Gardner FM, Goodlett DR, Ernst RK. 2022. A sensitive GC‐MS method for quantitation of lipid A backbone components and terminal phosphate modifications. J. Am. Soc. Mass Spectrom. 33:2301–2309. 10.1021/jasms.2c00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen K, Long L, Ding S. 2021. A highly xyloglucan active lytic polysaccharide monooxygenase EpLPMO9A from Eupenicillium parvum 4‐14 shows boosting effect on hydrolysis of complex lignocellulosic substrates. Int. J. Biol. Macromol., 167:202–213. 10.1016/j.ijbiomac.2020.11.177 [DOI] [PubMed] [Google Scholar]
- Shi Y, Han B, Zhang L, Zhou P. 2021. Comprehensive identification and absolute quantification of milk oligosaccharides in different species. J. Agric. Food Chem., 69:15585–15597. 10.1021/acs.jafc.1c05872 [DOI] [PubMed] [Google Scholar]
- Shifang R, Jianxin G. 2022. Glycomic technology and its application in disease marker mining. Chin. J. Lab. Med. 45:318–322. 10.3760/cma.j.cn114452-20220208-00056 [DOI] [Google Scholar]
- Shiga M, Miyazaki J, Tanuma K, Nagumo Y, Yoshino T, Kandori S, Negoro H, Kojima T, Tanaka R, Okiyama N, Fujisawa Y, Watanabe M, Yamasaki S, Kiyohara H, Watanabe M, Sato T, Tahara H, Nishiyama H, Yano I. 2021. The liposome of trehalose dimycolate extracted from M. bovis BCG induces antitumor immunity via the activation of dendritic cells and CD8+ T cells. Cancer Immunol. Immunother., 70:2529–2543. 10.1007/s00262-021-02870-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S, Ng BG, White AL, Nickander KK, Turgeon C, Liedtke KL, Lam CT, Font‐Montgomery E, Lourenco CM, He M, Peck DS, Umana LA, Uhles CL, Haynes D, Wheeler PG, Bamshad MJ, Nickerson DA, Cushing T, Gates R, Gomez‐Ospina N, Byers HM, UW Center for Mendelian Genomics, Scalco FB, Martinez NN, Sachdev R, Smith L, Poduri A, Malone S, Harris RV, Scheffer IE, Rosenzweig SD, Adams DR, Gahl WA, Malicdan MCV, Raymond KM, Freeze HH, Wolfe LA. 2022. Clinical, biochemical and genetic characteristics of MOGS‐CDG: A rare congenital disorder of glycosylation. J. Med. Genet. 59:1104–1115. 10.1136/jmedgenet-2021-108177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Kotajima D, Fukao K, Mogi F, Horiuchi R, Kataoka C, Kagami Y, Fujita M, Miyanishi N, Kashiwada S. 2021. Exposure of silver nanocolloids causes glycosylation disorders and embryonic deformities in medaka. Toxicol, Appl, Pharmacol., 430:115714. 10.1016/j.taap.2021.115714 [DOI] [PubMed] [Google Scholar]
- Shimma S, Takashima Y, Hashimoto J, Yonemori K, Tamura K, Hamada A. 2013. Alternative two‐step matrix application method for imaging mass spectrometry to avoid tissue shrinkage and improve ionization efficiency. J. Mass Spectrom. 48:1285–1290. 10.1002/jms.3288 [DOI] [PubMed] [Google Scholar]
- Shimoyama A, Di Lorenzo F, Yamaura H, Mizote K, Palmigiano A, Pither MD, Speciale I, Uto T, Masui S, Sturiale L, Garozzo D, Hosomi K, Shibata N, Kabayama K, Fujimoto Y, Silipo A, Kunisawa J, Kiyono H, Molinaro A. 2021. Lipopolysaccharide from gut‐associated lymphoid‐tissue‐resident Alcaligenes faecalis: Complete structure determination and chemical synthesis of its lipid A. Angew . Chem. Int. Ed. 60:10023–10031. 10.1002/anie.202012374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinchi H, Yuki M, Yamauchi T, Niimura M, Wakao M, Cottam HB, Hayashi T, Carson DA, Moroishi T, Suda Y. 2021. Glyco‐nanoadjuvants: sugar structures on carriers of a small molecule TLR7 ligand affect their immunostimulatory activities. ACS Appl. Bio Mater. 4:2732–2741. 10.1021/acsabm.0c01639 [DOI] [PubMed] [Google Scholar]
- Shinchi H, Komaki F, Yuki M, Ohara H, Hayakawa N, Wakao M, Cottam HB, Hayashi T, Carson DA, Moroishi T, Suda Y. 2022. Glyco‐nanoadjuvants: impact of linker length for conjugating a synthetic small‐molecule TLR7 ligand to glyco‐nanoparticles on immunostimulatory effects. ACS Chem. Biol. 17:957–968. 10.1021/acschembio.2c00108 [DOI] [PubMed] [Google Scholar]
- Shiratori K, Yokoi Y, Wakui H, Hirane N, Otaki M, Hinou H, Yoneyama T, Hatakeyama S, Kimura S, Ohyama C, Nishimura S‐I. 2022. Selective reaction monitoring approach using structure‐defined synthetic glycopeptides for validating glycopeptide biomarkers predetermined by bottom‐up glycoproteomics. RSC Adv., 12:21385. 10.1039/d2ra02903k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivatare SS, Cheng T‐JR, Cheng Y‐Y, Shivatare VS, Tsai T‐I, Chuang H‐Y, Wu C‐Y, Wong C‐H. 2021. Immunogenicity evaluation of N‑glycans recognized by HIV broadly neutralizing antibodies. ACS Chem. Biol. 16:2016–2025. 10.1021/acschembio.1c00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobha MS, Harish Prashanth KVH, Srinivasa PC, Tharanathan RN. 2022. Characterization of resultant oligosaccharides from guar galactomannan upon depolymerization by non‐specific enzymes. Trends Carbohydr. Res. 14:15–22 [Google Scholar]
- Shrestha B. 2021a. Introduction to spatial mapping of biomolecules by imaging mass spectrometry. Amsterdam, Oxford UK, Cambridge, MA, USA: Elsevier; [Google Scholar]
- Shrestha B. 2021b. Imaging mass spectrometry: Gangliosides in brain tissue, Introduction to spatial mapping of biomolecules by imaging mass spectrometry. Amsterdam, Oxford UK, Cambridge, MA, USA: Elsevier. p 245–254 [Google Scholar]
- Shrestha B. 2021c. Imaging mass spectrometry: Glycans, introduction to spatial mapping of biomolecules by imaging mass spectrometry. Amsterdam, Oxford UK, Cambridge, MA, USA: Elsevier. p 203–210 [Google Scholar]
- Shrestha B. 2021d. Sample preparation for imaging mass spectrometry, Introduction to spatial mapping of biomolecules by imaging mass spectrometry. Amsterdam, Oxford UK, Cambridge, MS, USA: Elsevier. p 23–47. 10.1016/C2018-0-03962-6 [DOI] [Google Scholar]
- Shrestha B. 2021e. Matrix for matrix‐assisted laser desorption/ionization (MALDI), Introduction to spatial mapping of biomolecules by imaging mass spectrometry. Amsterdam, Oxford UK, Cambridge, MA, USA: Elsevier. p 61–67 [Google Scholar]
- Shrivastava A, Joshi S, Guttman A, Rathore AS. 2022. N‐Glycosylation of monoclonal antibody therapeutics: A comprehensive review on significance and characterization. Anal. Chim. Acta, 1209:339828. 10.1016/j.aca.2022.339828 [DOI] [PubMed] [Google Scholar]
- Shu J, Ma J, Ren X, Wang J, Wang Y, Zhang K, Yu H, Guo X, Li Z. 2021. The abnormal glycopatterns of salivary glycoproteins in esophageal squamous cell carcinoma patients. Front. Chem. 9:637730. 10.3389/fchem.2021.637730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sighinolfi G, Clark S, Blanc L, Cota D, Rhourri‑Frih B. 2021. Mass spectrometry imaging of mice brain lipid profile changes over time under high fat diet. Sci. Rep. 11:19664. 10.1038/s41598-021-97201-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silchenko AS, Avilov SA, Kalinin VI. 2022. Separation procedures for complicated mixtures of sea cucumber triterpene glycosides with isolation of individual glycosides, their comparison with HPLC/MS metabolomic approach, and biosynthetic interpretation of the obtained structural data. Stud. Nat. Prod. Chem. 72:103–146. 10.1016/B978-0-12-823944-5.00015-6 [DOI] [Google Scholar]
- Simpson BW, Nieckarz M, Pinedo V, McLean AB, Cava F, Trenta MS. 2021. Acinetobacter baumannii can survive with an outer membrane lacking lipooligosaccharide due to structural support from elongasome peptidoglycan synthesis. mBio, 12:e03099–03021. 10.1128/mbio.03099-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh Y, Ormaza D, Massetti A, Minond D, Cudic M. 2021. Tyrosine O‑GalNAc alters the conformation and proteolytic susceptibility of APP model glycopeptides. ACS Chem. Neurosci. 12:2974–2980. 10.1021/acschemneuro.1c00387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhto N, Vinaiphat A, Thongboonkerd V. 2022. Discrimination of urinary exosomes from microvesicles by lipidomics using thin layer liquid chromatography (TLC) coupled with MALDI‐TOF mass spectrometry. Sci. Rep. 9:13834. 10.1038/s41598-019-50195-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sirohi R, Joun J, Choi HI, Gaur VK, Sim SJ. 2021. Algal glycobiotechnology: Omics approaches for strain improvement. Microb. Cell Factories, 20:163. 10.1186/s12934-021-01656-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivignon A, Yu S‐Y, Ballet N, Vandekerckove P, Barnich N, Guerardel Y. 2021. Heteropolysaccharides from S. cerevisiae show anti‐adhesive properties against E. coli associated with Crohn's disease. Carbohydr. Polym. 271:118415. 10.1016/j.carbpol.2021.118415 [DOI] [PubMed] [Google Scholar]
- Skurska E, Szulc B, Maszczak‐Seneczko D, Wiktor M, Wiertelak W, Makowiecka A, Olczak M. 2022. Incorporation of fucose into glycans independent of the GDP‐fucose transporter SLC35C1 preferentially utilizes salvaged over de novo GDP‐fucose. J. Biol. Chem., 298:102206. 10.1016/j.jbc.2022.102206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BR, Guo Z. 2021. Oligosaccharide antigen conjugation to carrier proteins to formulate glycoconjugate vaccines. Method. Mol. Biol. 2183:305–312. 10.1007/978-1-0716-0795-4_15 [DOI] [PubMed] [Google Scholar]
- Smith J, Millán‐Martín S, Mittermayr S, Hilborne V, Davey G, Polom K, Roviello F, Bones J. 2021. 2‐Dimensional ultra‐high performance liquid chromatography and DMT‐MM derivatization paired with tandem mass spectrometry for comprehensive serum N‐glycome characterization. Anal. Chim. Acta, 1179:338840. 10.1016/j.aca.2021.338840 [DOI] [PubMed] [Google Scholar]
- Smith PJ, Curry TM, Yang J‐Y, Barnes WJ, Ziegler SJ, Mittal A, Moremen KW, York WS, Bomble YJ, Peña MJ, Urbanowicz BR. 2022. Enzymatic synthesis of xylan microparticles with tunable morphologies. ACS Mater., Au, 2:440–452. 10.1021/acsmaterialsau.2c00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, McElheny CL, Izac JR, Gardner FM, Chandler CE, Goodlett DR, Doi Y, Johnson K, Ernsta RK. 2022. A novel lipid‐based MALDI‐TOF assay for the rapid detection of colistin‐resistant enterobacter species. Microbiol. Spectrum, 10:e01445–01421. 10.1128/spectrum.01445-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sockett KA, Loffredo M, Korunes‐Miller J, Varghese M, Grinstaff MW. 2022. Synthesis and characterization of carbohydrate‐based biosurfactant mimetics. Carbohydr. Res. 522:108697. 10.1016/j.carres.2022.108697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sojitra M, Sarkar S, Maghera J, Rodrigues E, Carpenter EJ, Seth S, Vinals DF, Bennett NJ, Reddy R, Khalil A, Xue X, Bell MR, Zheng RB, Zhang P, Nycholat C, Bailey JJ, Ling C‐C, Lowary TL, Paulson JC, Macauley MS, Derda R. 2021. Genetically encoded multivalent liquid glycan array displayed on M13 bacteriophage. Nat. Chem. Biol. 17:806–816. 10.1038/s41589-021-00788-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltwisch J. 2022. Instrumentation for MALDI‐MSI ‐ Part IIonization sources and design. In: Siegel T, Editor, MALDI Mass Spectrometry Imaging: From Fundamentals to Spatial Omics. London: Royal Society of Chemistry. p 39–58. 10.1039/9781839165191-00039 [DOI] [Google Scholar]
- Son S‐U, Kim HW, Shin K‐S. 2022. Structural identification of active moiety in anti‐tumor metastatic polysaccharide purified from fermented barley by sequential enzymatic hydrolysis. Food Biosci. 59:101999. 10.1016/j.fbio.2022.101999 [DOI] [Google Scholar]
- Song T, Liu L, Tang Q, Xiang S, Wang B, Zhang S, Wang X, Chu Y, Luo D, Lin J. 2022. Antioxidant neoagarooligosaccharides (NAOs) and dietary fiber production from red algae Gracilariopsis lemaneiformis using enzyme assisted one‐step process. Food Hydrocolloid., 125:107382. 10.1016/j.foodhyd.2021.107382 [DOI] [Google Scholar]
- Song Y, Zhang F, Linhardt RJ. 2021. Analysis of the glycosaminoglycan chains of proteoglycans. J. Histochem. Cytochem. 69:121–135. 10.1369/0022155420937154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Xu A, Wang M, Ji L, Wang Q, Shi J, Zhao R, Fu W, Zhang R. 2022. Comparability of different methods of glycated hemoglobin measurement for samples of patients with variant and non‐variant hemoglobin. Clin. Chim. Acta, 533:168–174. 10.1016/j.cca.2022.06.024 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang J, Fei Y, Huang Z, Liu X, Li L‐L. 2022. Lipase‐activated glycopeptide nano‐assemblies as an antibiotic nano‐adjuvant to inhibit Pseudomonas aeruginosa biofilm and enhance antibacterial activity. Sci. China Chem. 65:2538–2547. 10.1007/s11426-022-1348-9 [DOI] [Google Scholar]
- Sosicka P, Ng BG, Pepi LE, Shajahan A, Wong M, Scott DA, Matsumoto K, Xia Z‐J, Lebrilla CB, Haltiwanger RS, Azadi P, Freeze HH. 2022. Origin of cytoplasmic GDP‐fucose determines its contribution to glycosylation reactions. J. Cell Biol. 221:e202205038. 10.1083/jcb.202205038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speciale I, Di Lorenzo F, Notaro A, Noel E, Agarkova I, Molinaro A, Van Etten JL, De Castro C. 2022. N‐glycans from Paramecium bursaria chlorella virus MA‐1D: Re‐evaluation of the oligosaccharide common core structure. Glycobiology, 32:260–273. 10.1093/glycob/cwab113 [DOI] [PubMed] [Google Scholar]
- Speciale I, Notaro A, Abergel C, Lanzetta R, Lowary TL, Molinaro A, Tonetti M, Van Etten JL, De Castro C. 2022. The astounding world of glycans from giant viruses. Chem. Rev. 122:15717–15766. 10.1021/acs.chemrev.2c00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer SK, Moore RE, Lu J, Guevara MA, Marshall DR, Manning SD, Damo SM, Townsend SD, Gaddy JA. 2021. Antibiofilm activity of human milk oligosaccharides against multidrug resistant and susceptible isolates of Acinetobacter baumannii . ACS Infect. Dis. 7:3254–3263. 10.1021/acsinfecdis.1c00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriha SE, Rahman FI, Rahman SMA. 2021. Synthesis, in vivo and in silico analgesic and anti‐inflammatory studies of α‐d‐ribofuranose derivatives. Saudi Pharmaceut. J. 29:981–991. 10.1016/j.jsps.2021.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sran KS, Sharma Y, Kaur T, Rao A. 2022. Post‑translational modifications and glycoprofiling of palivizumab by UHPLC‐RPLC/HILIC and mass spectrometry. J. Proteins Proteomics, 13:95–108. 10.1007/s42485-022-00086-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedevi P, Nair JB, Joseph MM, Murali VP, Suresh CH, Varma RL, Maiti KK. 2021. Dynamic self‐assembly of mannosylated‐calix[4]arene into micelles for the delivery of hydrophobic drugs. J. Control. Release, 339:284–296. 10.1016/j.jconrel.2021.09.038 [DOI] [PubMed] [Google Scholar]
- Srisakul S, Wannigama DL, Higgins PG, Hurst C, Abe S, Hongsing P, Saethang T, Luk‑in S, Liao T, Kueakulpattana N, Shein AMS, Gan L, Kupwiwat R, Tanasatitchai C, Wapeesittipan P, Phattharapornjaroen P, Badavath VN, Leelahavanichkul A, Chatsuwan T. 2022. Overcoming addition of phosphoethanolamine to lipid A mediated colistin resistance in Acinetobacter baumannii clinical isolates with colistin‐sulbactam combination therapy. Sci. Rep. 12:11390. 10.1038/s41598-022-15386-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AD, Unione L, Bunyatov M, Gagarinov IA, Delgado S, Abrescia NGA, Ard A, Boons G‐J. 2021. Chemoenzymatic synthesis of complex N‐glycans of the parasite S. mansoni to examine the importance of epitope presentation on DCSIGN recognition. Angew. Chem. Int. Ed., 60:19287–19296. 10.1002/anie.202105647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabach PR, Zimmerman K, Adame A, Kavanagh D, Saeui CT, Agatemor C, Gray S, Cao W, De La Cruz EM, Yarema KJ, Braddock DT. 2021. Improving the pharmacodynamics and in vivo activity of ENPP1‐Fc through protein and glycosylation engineering. Clin. Transl. Sci. 14:362–372. 10.1111/cts.12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanback AE, Conroy LR, Young LEA, Hawkinson TR, Markussen KH, Clarke HA, Allison DB, Sun RC. 2021. Regional N‐glycan and lipid analysis from tissues using MALDI‐mass spectrometry imaging. STAR Protoc., 2:100304. 10.1016/j.xpro.2021.100304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staniszewska M, Bronowicka‑Szydełko A, Gostomska‑Pampuch K, Szkudlarek J, Bartyś A, Bieg T, Gamian E, Kochman A, Picur B, Pietkiewicz J, Kuropka P, Szeja W, Wiśniewski J, Ziółkowski P, Gamian A. 2021. The melibiose‑derived glycation product mimics a unique epitope present in human and animal tissues. Sci. Rep. 11:2940. 10.1038/s41598-021-82585-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenitzer D, Altmann F. 2022. Protein glycosylation in bryophytes differs subtly from that in vascular plants. IntechOpen. 10.5772/intechopen.107035 [DOI] [Google Scholar]
- Stenitzer D, Mócsai R, Zechmeister H, Reski R, Decker EL, Altmann F. 2022. O‐Methylated N‐glycans distinguish mosses from vascular plants. Biomolecules, 12:136. 10.3390/biom12010136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepnov AA, Christensen IA, Forsberg Z, Aachmann FL, Courtade G, Eijsink VGH. 2022. The impact of reductants on the catalytic efficiency of a lytic polysaccharide monooxygenase and the special role of dehydroascorbic acid. FEBS Lett. 596:53–70. 10.1002/1873-3468.14246 [DOI] [PubMed] [Google Scholar]
- Stewart GA, Debrah D, Ranathunga Y, Olowolafe TA, Schlegel HB, Lee SK, Li W. 2022. Picosecond vs femtosecond: Are all laser desorption ionizations created equal ? J. Phys. Chem. C, 126:17135–17140. 10.1021/acs.jpcc.2c06573 [DOI] [Google Scholar]
- Støpamo FG, Røhr ÅK, Mekasha S, Petrović DM, Várnai A, Eijsink VGH. 2021. Characterization of a lytic polysaccharide monooxygenase from Aspergillus fumigatus shows functional variation among family AA11 fungal LPMOs. J. Biol. Chem. 297:101421. 10.1016/j.jbc.2021.101421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad Š, Strnadová V, Sýkora D, Cvaěka J, Maletínská L, Vrkoslav V. 2022. MALDI Mass spectrometry imaging of lipids on free‐floating brain sections and immunohistochemically colocalized markers of neurodegeneration. Method. Mol. Biol. 2437:229–239. 10.1007/978-1-0716-2030-4_16 [DOI] [PubMed] [Google Scholar]
- Strobl S, Hofbauer K, Heine H, Zamyatina A. 2022. Lipid A mimetics based on unnatural disaccharide scaffold as potent TLR4 agonists for prospective immunotherapeutics and adjuvants. Chem. Eur. J. 28:e202200547. 10.1002/chem.202200547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB, Agravat S, Aoki‐Kinoshita KF, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson NG, Khoo K‐H, Kolarich D, Liu Y, McBride R, Novotny MV, Packer NH, Paulson JC, Rapp E, Ranzinger R, Rudd PM, Smith DF, Tiemeyer M, Wells L, York WS, Zaia J, Kettner C. 2016. The minimum information required for a glycomics experiment (MIRAGE) project: Sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology, 26:907–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struwe WB. 2021. Glycomics and ion mobility. In: Ashcroft AE, Sobott F, Editors., Ion mobility‐Mass Spectrometry. Fundamentals and Applications. London: Royal Society of Chemistry. p 496–526. 10.1039/9781839162886-00496. [DOI] [Google Scholar]; doi: 10.1093/glycob/cww082. [DOI]
- Sturiale L, Nassogne M‐C, Palmigiano A, Messina A, Speciale I, Artuso R, Bertino G, Revencu N, Stephénne X, De Castro C, Matthijs G, Barone R, Jaeken J, Domenico G. 2021. Aberrant sialylation in a patient with a HNF1α variant and liver adenomatosis. iScience, 24:102323. 10.1016/j.isci.2021.102323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm S, Dowle A, Audsley N, Isaac RE. 2021. Mass spectrometric characterisation of the major peptides of the male ejaculatory duct, including a glycopeptide with an unusual zwitterionic glycosylation. J. Proteomics, 246:104307. 10.1016/j.jprot.2021.104307 [DOI] [PubMed] [Google Scholar]
- Su L, Feng Y, Wei K, Xu X, Liu R, Chen G. 2021. Carbohydrate‐based macromolecular biomaterials. Chem. Rev. 121:10950–11029. 10.1021/acs.chemrev.0c01338 [DOI] [PubMed] [Google Scholar]
- Su P, Wang Z, Li X, Li M, Li G, Gong Z, Song J, Yang Y. 2021. Fabrication of magnetic dual‐hydrophilic metal organic framework for highly efficient glycopeptide enrichment. Anal. Bioanal. Chem. 413:5267–5278. 10.1007/s00216-021-03535-w [DOI] [PubMed] [Google Scholar]
- Su P, Li M, Li X, Yuan X, Gong Z, Wu L, Song J, Yang Y. 2022. Glutathione functionalized magnetic covalent organic frameworks with dual‐hydrophilicity for highly efficient and selective enrichment of glycopeptides. J. Chromatogr., A, 1667:462869. 10.1016/j.chroma.2022.462869 [DOI] [PubMed] [Google Scholar]
- Su T, Qin X‐Y, Dohmae N, Wei F, Furutani Y, Kojima S, Yu W. 2021. Inhibition of ganglioside synthesis suppressed liver cancer cell proliferation through targeting kinetochore metaphase signaling. Metabolites, 11:167. 10.3390/metabo11030167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniama S, Wena X‐Y, Zhangb Z‐T, Jinga P. 2021. Changes in the morphometric, textural, and aromatic characteristics of shiitake mushrooms during combined humid‐convective drying. Drying Technol. 39:2206–2217. 10.1080/07373937.2020.1760878 [DOI] [Google Scholar]
- Subramanian SP, Lakshmanan V, Palakodeti D, Subramanian R. 2022. Glycomic and glycotranscriptomic profiling of mucin‐type O‐glycans in planarian Schmidtea mediterranea . Glycobiology, 32:36–49. 10.1093/glycob/cwab097 [DOI] [PubMed] [Google Scholar]
- Šuchová K, Chyba A, Hegyi Z, Rebroš M, Puchart V. 2022. Yeast GH30 xylanase from Sugiyamaella lignohabitans is a glucuronoxylanase with auxiliary xylobiohydrolase activity. Molecules, 27:751. 10.3390/molecules27030751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Sawada T, Tanaka H, Serizawa T. 2021. Enzyme‐catalyzed propagation of cello‐oligosaccharide chains from bifunctional oligomeric primers for the preparation of block co‐oligomers and their crystalline assemblies. Polymer J. 53:1133–1143. 10.1038/s41428-021-00513-y [DOI] [Google Scholar]
- Sugiura K, Saito M, Sawada T, Tanaka H, Serizawa T. 2022. Cellodextrin phosphorylase‐catalyzed single‐process production and superior mechanical properties of organic−inorganic hybrid hydrogels composed of surface‐carboxylated synthetic nanocelluloses and hydroxyapatite. ACS Sustain. Chem. Eng. 10:13484–13494. 10.1021/acssuschemeng.2c04349 [DOI] [Google Scholar]
- Sumanth MS, Jacob SP, Abhilasha KV, Manne BK, Basrur V, Lehoux S, Campbell RA, Yost CC, McIntyre TM, Cummings RD, Weyrich AS, Rondina MT, Marathe GK. 2021. Different glycoforms of alpha‐1‐acid glycoprotein contribute to its functional alterations in platelets and neutrophils. J. Leukocyte Biol. 109:915–930. 10.1002/jlb.3a0720-422r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Ma S, Li L, Wang D, Liu W, Liu F, Guo L, Wang X. 2021. Visualizing the distributions and spatiotemporal changes of metabolites in Panax notoginseng by MALDI mass spectrometry imaging. J. Ginseng Res., 45:726–733. 10.1016/j.jgr.2021.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Cui L, Zhou B, Wang X, Guo L, Liu W. 2022. Visualizing the spatial distribution and alteration of metabolites in continuously cropped Salvia miltiorrhiza Bge using MALDI‐MSI. J. Pharmaceut. Anal. 12:719–724. 10.1016/j.jpha.2021.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Xu D, Shen Y, Crommen J, Wang Q, Jiang Z. 2022. Photo‐assisted generation of versatile zwitterionic carboxybetaine‐based hypercrosslinked polymers for separation science. Chem. Eng. J. 431:133374. 10.1016/j.cej.2021.133374 [DOI] [Google Scholar]
- Sun N, Deng C, Shen X. 2021a. Simultaneous application of nanomaterials to separation of phosphorylated and glycosylated proteins. In: Sun N, Deng C, Shen X, Editors., Applications of Nanomaterials in Proteomics. Singapore: Springer. p 297–323. 10.1007/978-981-16-5816-7_5 [DOI] [Google Scholar]
- Sun N, Deng C, Shen X. 2021b. Application of nanomaterials to separation of glycosylated proteins. In: Sun N, Deng C, Shen X, Editors., Applications of Nanomaterials in Proteomics. Singapore: Springer. p 179–296. 10.1007/978-981-16-5816-7_4 [DOI] [Google Scholar]
- Sun N, Trajkovic‑Arsic M, Li F, Wu Y, Münch C, Kunzke T, Feuchtinger A, Steiger K, Schlitter AM, Weichert W, Esposito I, Siveke JT, Walch A. 2021. Native glycan fragments detected by MALDI mass spectrometry imaging are independent prognostic factors in pancreatic ductal adenocarcinoma. EJNMMI Res., 11:120. 10.1186/s13550-021-00862-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Kim AMJ, Murray AA, Lim S‐O. 2022. N‐Glycosylation facilitates 4‐1BB membrane localization by avoiding its multimerization. Cells, 11:162. 10.3390/cells11010162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Tang W, Li B. 2022. Gold‐TiO2@gallic acid nanospheres for enhanced surface‐assisted laser desorption/ionization mass spectrometry imaging. Appl. Mater. Today, 26:101336. 10.1016/j.apmt.2021.101336 [DOI] [Google Scholar]
- Sun R, Zhang Y, Tang W, Li B. 2022. Submicron 3,4‐dihydroxybenzoic acid‐TiO2 composite particles for enhanced MALDI MS imaging of secondary metabolites in the root of differently aged baical skullcap. Analyst, 147:3017–3024. 10.1039/D2AN00710J [DOI] [PubMed] [Google Scholar]
- Sun RC, Young LEA, Bruntz RC, Markussen KH, Zhou Z, Conroy LR, Hawkinson TR, Clarke HA, Stanback AE, Macedo JKA, Emanuelle S, Brewer MK, Rondon AL, Mestas A, Sanders WC, Mahalingan KK, Tang B, Chikwana VM, Segvich DM, Contreras CJ, Allenger EJ, Brainson CF, Johnson LA, Taylor RE, Armstrong DD, Shaffer R, Waechter CJ, Vander Kooi CW, DePaoli‐Roach AA, Roach PJ, Hurley TD, Drake RR, Gentry MS. 2021. Brain glycogen serves as a critical glucosamine cache required for protein glycosylation. Cell Metab., 33:1404–1417. 10.1016/j.cmet.2021.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y‐P, Wang B‐B, Zheng X‐W, Wu Z‐P, Hou J, Cui H‐L. 2022. Description of Halosolutus amylolyticus gen. nov., sp. nov., Halosolutus halophilus sp. nov. and Halosolutus gelatinilyticus sp. nov., and genome‐based taxonomy of genera Natribaculum and Halovarius . Int. J. Systemat. Evolut. Microbiol., 72:005598. 10.1099/ijsem.0.005598 [DOI] [PubMed] [Google Scholar]
- Sun Y‐Y, Sun H‐Q, Pan L‐C, Jia Y‐Q, Liu C‐Y, Wang H‐X, Liu X‐C, Zhu Z‐Y, Si C‐L. 2022. Preparation, structure and α‐glucosidase inhibitory of oligosaccharides by enzymatic hydrolysis from Annona squamosa polysaccharide. Ind. Crops Prod. 177:114468. 10.1016/j.indcrop.2021.114468 [DOI] [Google Scholar]
- Sun Z, Ji G, Wang G, Wei L, Zhang Y, Lu H. 2021. One step carboxyl group isotopic labeling for quantitative analysis of intact N‐glycopeptides by mass spectrometry. Chem. Commun. 57:4154–4157. 10.1039/D1CC00197C [DOI] [PubMed] [Google Scholar]
- Sun Z, Zhang Y, Lu H. 2022. Quantitative methods for N‐glycosite containing peptides in N‐glycoproteomics. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. Boca Raton: CRC Press. p 29–54. 10.1201/9781003185833-2 [DOI] [Google Scholar]
- Sundararaj K, Rodgers J, Angel P, Wolf B, Nowling TK. 2021. The role of neuraminidase in TLR4‐MAPK signalling and the release of cytokines by lupus serum‐stimulated mesangial cells. Immunology, 162:418–433. 10.1111/imm.13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar NT, Shajahan A, Gleinich AS, Rouhani DS, Heiss C, Chapla DG, Moremen KW, Azadi P. 2021. Variable posttranslational modifications of severe acute respiratory syndrome coronavirus 2 nucleocapsid protein. Glycobiology, 31:1080–1092. 10.1093/glycob/cwab044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto N, Kamoshita S, Hosoya S, Sakurai K. 2021. Exploration of the reactivity of multivalent electrophiles for affinity labeling: Sulfonyl fluoride as a highly efficient and selective label. Angew. Chem. Int. Ed. 60:17080–17087. 10.1002/anie.202104347 [DOI] [PubMed] [Google Scholar]
- Suwanwong N, Chatwichien J, Chainok K, Ruchirawat S, Boonyarattanakalin S. 2022. Discovery of nitric oxide‐inducing activities of synthetic LAM glycan motifs prepared by scalable rapid syntheses. Carbohydr. Polym. 296:119637. 10.1016/j.carbpol.2022.119637 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Morishima T, Handa A, Tsukagoshi H, Kato M, Shimizu M. 2022. Biochemical characterization of a pectate lyase AnPL9 from Aspergillus nidulans . Appl. Biochem. Biotechnol. 194:5627–5643. 10.1007/s12010-022-04036-x [DOI] [PubMed] [Google Scholar]
- Suzuki N, Abe T, Hanzawa K, Natsuka S. 2021. Toward robust N‑glycomics of various tissue samples that may contain glycans with unknown or unexpected structures. Sci. Rep. 11:6334. 10.1038/s41598-021-84668-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Itoh A, Kataoka K, Yamashita S, Kano K, Sowa K, Kitazumi Y, Shirai O. 2022. Effects of N‐linked glycans of bilirubin oxidase on direct electron transfer‐type bioelectrocatalysis. Bioelectrochemistry, 146:108141. 10.1016/j.bioelechem.2022.108141 [DOI] [PubMed] [Google Scholar]
- Sy I, Conrad L, Becker SL. 2022. Recent advances and potential future applications of MALDI‐TOF mass spectrometry for identification of helminths. Diagnostics, 12:3035. 10.3390/diagnostics12123035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymański K, Hapeta P, Moroz P, Wąsik B, Robak M, Lazar Z. 2022. The influence of Yarrowia lipolytica glycosylation on the biochemical properties and oligomerization of heterologous invertase. Sustainability, 14:7926. 10.3390/su14137926 [DOI] [Google Scholar]
- Tabang DN, Ford M, Li L. 2021. Recent advances in mass spectrometry‐based glycomic and glycoproteomic studies of pancreatic diseases. Front. Chem. 9:707387. 10.3389/fchem.2021.707387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahata S, Raymond K, Quade M, Barnes S, Boyer S, League S, Kumanovics A, Abraham R, Jacob E, Menon P, Morava E. 2022. Defining the mild variant of leukocyte adhesion deficiency type II (SLC35C1‐congenital disorder of glycosylation) and response to l‐fucose therapy: Insights from two new families and review of the literature. Am. J. Med. Genet. A, 188A:2005–2018. 10.1002/ajmg.a.62737 [DOI] [PubMed] [Google Scholar]
- Tajadura‐Ortega V, Gambardella G, Skinner A, Halim A, Van Coillie J, Schjoldager KT‐BG, Beatson R, Graham R, Achkova D, Taylor‐Papadimitriou J, Ciccarelli FD, Burchell JM. 2021. O‐Linked mucin‐type glycosylation regulates the transcriptional programme downstream of EGFR. Glycobiology, 31:200–210. 10.1093/glycob/cwaa075 [DOI] [PubMed] [Google Scholar]
- Takahashi M, Komura N, Yoshida Y, Yamaguchi E, Hasegawa A, Tanaka H‐N, Imamura A, Ishida H, Suzuki KGN, Ando H. 2022. Development of lacto‐series ganglioside fluorescent probe using late‐stage sialylation and behavior analysis with single‐molecule imaging. RSC Chem. Biol. 3:868–885. 10.1039/D2CB00083K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Kato K. 2003. GALAXY (Glycoanalysis by the Three Axes of MS and Chromatography): A web application that assists structural analyses of N‐glycans. Trends Glycosci. Glycotechnol. 15:235–251. 10.4052/tigg.15.235 [DOI] [Google Scholar]
- Takayama S, Kawanishi M, Yamauchi K, Tokumitsu D, Kojima H, Masutani T, Iddamalgoda A, Mitsunaga T, Tanaka H. 2021. Ellagitannins from Rosa roxburghii suppress poly(I:C)‑induced IL‑8 production in human keratinocytes. J. Nat. Med., 75:623–632. 10.1007/s11418-021-01509-x [DOI] [PubMed] [Google Scholar]
- Takei D, Harada K, Nouso K, Miyahara K, Dohi C, Matsushita H, Kinugasa H, Hiraoka S, Nishimura S‐I, Okada H. 2022. Clinical utility of a serum glycome analysis in patients with colorectal cancer. J. Gastroenterol. Hepatol. 37:727–733. 10.1111/jgh.15781 [DOI] [PubMed] [Google Scholar]
- Tamara S, den Boer MA, Heck AJR. 2022. High‐resolution native mass spectrometry. Chem. Rev. 122:7269–7326. 10.1021/acs.chemrev.1c00212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Deng F, Luo X, Pan X, Zhang L, Marina ML, Jiang Z. 2021. Glycosyl imprinted mesoporous microspheres for the determination of glycopeptide antibiotics using ultra‐high performance liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr., A, 1659:462630. 10.1016/j.chroma.2021.462630 [DOI] [PubMed] [Google Scholar]
- Tang R, Yu Y, Dong J, Yao Y, Ma S, Ou J, Ye M. 2021. Facile preparation of bifunctional adsorbents for efficiently enriching N‐glycopeptides and phosphopeptides. Anal. Chim. Acta, 1144:111–120. 10.1016/j.aca.2020.12.015 [DOI] [PubMed] [Google Scholar]
- Tang W, Gordon A, Wang F, Chen Y, Li B. 2021. Hydralazine as a versatile and universal matrix for high‐molecular coverage and dual‐polarity matrix‐assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 93:9083–9093. 10.1021/acs.analchem.1c00498 [DOI] [PubMed] [Google Scholar]
- Tang X, Zhao M, Chen Z, Huang J, Chen Y, Wang F, Wan K. 2021. Visualizing the spatial distribution of metabolites in Clausena lansium (Lour.) skeels using matrix‐assisted laser desorption/ionization mass spectrometry imaging. Phytochemistry, 192:112930. 10.1016/j.phytochem.2021.112930 [DOI] [PubMed] [Google Scholar]
- Tang X, Chen Z, Chen Y, Jiang X, Zhu F, Liu S, Wan K. 2022. Hybrid bismuth oxide‐graphine oxide nanomaterials improve the signal‐to‐noise response of small molecules analyzed by matrix assisted laser desorption ionization‐time‐of‐flight mass spectrometry. Talanta, 252:123768. 10.1016/j.talanta.2022.123768 [DOI] [PubMed] [Google Scholar]
- Teng H, Li Q, Gou M, Liu G, Cao X, Lu J, Han Y, Yu Y, Gao Z, Song X, Dong W, Pang Y. 2022. Lamprey immunity protein enables early detection and recurrence monitoring for bladder cancer through recognizing Neu5Gc‐modified uromodulin glycoprotein in urine. BBA ‐ Mol. Basis Dis. 1868:166493. 10.1016/j.bbadis.2022.166493 [DOI] [PubMed] [Google Scholar]
- Teramoto K, Tsutsui S, Sato T, Fujimoto Z, Kaneko S. 2021. Substrate specificities of GH8, GH39, and GH52 β‐xylosidases from Bacillus halodurans C‐125 toward substituted xylooligosaccharides. Appl. Biochem. Biotechnol. 193:1042–1055. 10.1007/s12010-020-03451-2 [DOI] [PubMed] [Google Scholar]
- Terrasan CRF, Rubio MV, Gerhardt JA, Cairo JPF, Contesini FJ, Zubieta MP, de Figueiredo FL, Valadares FL, Ribeiro CTL, Murakami MT, Franco TT, Davies GJ, Walton PH, Damasio A. 2022. Deletion of AA9 lytic polysaccharide monooxygenases impacts A. nidulans secretome and growth on lignocellulose. Microbiol. Spectrum, 10:e0212521. 10.1128/spectrum.02125-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson B, Dajkovic A, Keary R, Marlière C, Dupont‑Gillain CC, Carballido‑López R. 2022. Magnesium rescues the morphology of Bacillus subtilis mreB mutants through its inhibitory effect on peptidoglycan hydrolases. Sci. Rep. 12:1137. 10.1038/s41598-021-04294-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm JC, Beketow E, Thiem J. 2022. Studies of carbohydrate‐carbohydrate‐interactions by atomic force microscopy employing functionalized 4‐acetylthio‐butyl glucopyranoside. Carbohydr. Res. 521:108649. 10.1016/j.carres.2022.108649 [DOI] [PubMed] [Google Scholar]
- Thoma J, Stenitzer D, Grabherr R, Staudacher E. 2022. Identification, characterization, and expression of a β‐galactosidase from Arion species (Mollusca). Biomolecules, 12:1578. 10.3390/biom12111578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Tang R, Wang X, Zhou J, Li X, Ma S, Gong B, Ou J. 2021. Bioinspired dandelion‐like silica nanoparticles modified with l‐glutathione for highly efficient enrichment of N‐glycopeptides in biological samples. Anal. Chim. Acta, 1173:338694. 10.1016/j.aca.2021.338694 [DOI] [PubMed] [Google Scholar]
- Tian Y, Wang Y, Yin H, Luo Y, Wei F, Zhou H, Wen L. 2022. A sensitive and reversible labeling strategy enables global mapping of the core‐fucosylated glycoproteome on cell surfaces. Angew. Chem. Int. Ed. 61:e202206802. 10.1002/anie.202206802 [DOI] [PubMed] [Google Scholar]
- Tian Y, Zheng Z, Wang X, Liu S, Gu L, Mu J, Zheng X, Li Y, Shen S. 2022. Establishment and evaluation of glucose‑modified nanocomposite liposomes for the treatment of cerebral malaria. J. Nanobiotechnol. 20:318. 10.1186/s12951-022-01493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V, Panta PR, Billiot CE, Douglass MV, Herrera CM, Trent MS, Doerrler WT. 2021. A Klebsiella pneumoniae DedA family membrane protein is required for colistin resistance and for virulence in wax moth larvae. Sci. Rep. 11:24365. 10.1038/s41598-021-03834-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tõlgo M, Hegnar OA, Østby H, Várnai A, Vilaplana F, Eijsink VGH, Olssona L. 2022. Comparison of six lytic polysaccharide monooxygenases from Thermothielavioides terrestris shows that functional variation underlies the multiplicity of LPMO genes in filamentous fungi. Appl. Environ. Microbiol. 88:e00096–00022. 10.1128/aem.00096-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tondini E, Reintjens NRM, Castello G, Arakelian T, Isendoorn M, Camps M, Vree J, van der Marel GA, Filippov DV, Codee JDC, Ossendorp F. 2022. Lipid A analog CRX‐527 conjugated to synthetic peptides enhances vaccination efficacy and tumor control. npj Vaccines. 7:64. 10.1038/s41541-022-00484-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toukach PV, Shirkovskaya AI. 2022. Carbohydrate structure database and other glycan databases as an important element of glycoinformatics. J. Bioorg. Chem. 48:457–466. 10.1134/S1068162022030190 [DOI] [Google Scholar]
- Traboni S, Bedini E, Landolfi A, Vessella G, Iadonisi A. 2021. Catalytic, regioselective sulfonylation of carbohydrates with dibutyltin oxide under solvent‐free conditions. Catalysts, 11:202. 10.3390/catal11020202 [DOI] [Google Scholar]
- Tran A, Monreal IA, Moskovets E, Aguilar HC, Jones JW. 2021. Rapid detection of viral envelope lipids using lithium adducts and AP‐MALDI high‐resolution mass spectrometry. J. Am. Soc. Mass Spectrom. 32:2322–2333. 10.1021/jasms.1c00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran A, Wan L, Xu Z, Haro JM, Li B, Jones JW. 2021. Lithium hydroxide hydrolysis combined with MALDI TOF mass spectrometry for rapid sphingolipid detection. J. Am. Soc. Mass Spectrom. 32:289–300. 10.1021/jasms.0c00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trattnig N, Li Z, Bosman GP, Kosma P, Boons G‐J. 2022. Site‐specific multi‐functionalization of the carrier protein CRM197 by disulfide rebridging for conjugate vaccine development. ChemBioChem, 23:e202200408. 10.1002/cbic.202200408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trbojević‐Akmačić I, Lageveen‐Kammeijer GSM, Heijs B, Petrović T, Deriš H, Wuhrer M, Lauc G. 2022. High‐throughput glycomic methods. Chem. Rev. 122:15865–15913. 10.1021/acs.chemrev.1c01031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treu A, Römpp A. 2021. Matrix ions as internal standard for high mass accuracy matrix‐assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom., 35:e9110. 10.1002/rcm.9110 [DOI] [PubMed] [Google Scholar]
- Trimarco JD, Nelson SL, Chaparian RR, Wells AI, Murray NB, Azadi P, Coyne CB, Heaton NS. 2022. Cellular glycan modification by B3GAT1 broadly restricts influenza virus infection. Nat. Commun. 12:6456. 10.1038/s41467-022-34111-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimpin S, Marshall DD, Karki S, Madarshahian S, Hoang K, Meher AK, Pophristic M, Richards AL, Lietz CB, Fischer JL, Elia EA, Wang B, Pagnotti VS, Lutomski CA, El‐Baba TJ, Lu I‐C, Wager‐Miller J, Mackie K, McEwen CN, Inutan ED. 2021. An overview of biological applications and fundamentals of new inlet and vacuum ionization technologies. Rapid Commun. Mass Spectrom., 35(S1):e8829. 10.1002/rcm.8829 [DOI] [PubMed] [Google Scholar]
- Trinh DN, Gardner RA, Franciosi AN, McCarthy C, Keane MP, Soliman MG, O'Donnell JS, Meleady P, Spencer DIR, Monopoli MP. 2022. Nanoparticle biomolecular corona‐based enrichment of plasma glycoproteins for N‑glycan profiling and application in biomarker discovery. ACS Nano, 16:5463–5475. 10.1021/acsnano.1c09564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouka A, Dallabernardina P, Mende M, Sletten ET, Leichnitz S, Bienert K, Hoang KLM, Seeberger PH, Loeffler FF. 2022. VaporSPOT: Parallel synthesis of oligosaccharides on membranes. J. Am. Chem. Soc. 144:19832–19837. 10.1021/jacs.2c07285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Ogawa M, Yogi K, Tashima Y, Takeuchi H, Okajima T. 2022. Glycoproteomics of NOTCH1 EGF repeat fragments overexpressed with different glycosyltransferases in HEK293T cells reveals insights into O‐GlcNAcylation of NOTCH1. Glycobiology, 32:616–628. 10.1093/glycob/cwac015 [DOI] [PubMed] [Google Scholar]
- Tsygankova SV, Pazynina GV, Paramonov AS, Chizhov AO, Bovin NV. 2022. Synthesis of disaccharide Xylβ1‐2Manβ, the core fragment of plant N‐glycoproteins. Russ. J. Bioorg. Chem., 48:513–518. 10.1134/S1068162022030207 [DOI] [Google Scholar]
- Tülek A, Yıldırım D, Aydın D, Binay B. 2021. Highly‐stable Madurella mycetomatis laccase immobilized in silica‐coated ZIF‐8 nanocomposites for environmentally friendly cotton bleaching process. Colloid Surface B, 202:111672. 10.1016/j.colsurfb.2021.111672 [DOI] [PubMed] [Google Scholar]
- Ugonotti J, Chatterjee S, Thaysen‐Andersen M. 2021. Structural and functional diversity of neutrophil glycosylation in innate immunity and related disorders. Mol. Aspects Med. 79:100882. 10.1016/j.mam.2020.100882 [DOI] [PubMed] [Google Scholar]
- Ullah S, Ji K, Li J, Xu Y, Jiang C, Zhang H, Huang M, Feng Y. 2021. Characterization of NMCR‐2, a new non‐mobile colistin resistance enzyme: Implications for an MCR‐8 ancestor. Environ. Microbiol. 23:844–860. 10.1111/1462-2920.15171 [DOI] [PubMed] [Google Scholar]
- Umemoto N, Saito N, Noguchi M, Shoda S‐i, Ohnuma T, Watanabe T, Sakuda S, Fukamizo T. 2022. Plant chitinase mutants as the catalysts for chitooligosaccharide synthesis using the sugar oxazoline derivatives. J. Agric. Food Chem. 70:12897–12906. 10.1021/acs.jafc.2c04632 [DOI] [PubMed] [Google Scholar]
- Unsihuay D, Sanchez DM, Laskin J. 2021. Quantitative mass spectrometry imaging of biological systems. Annu. Rev. Phys. Chem. 72:307–329. 10.1146/annurev-physchem-061020-053416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakami S, Hinou H. 2022a. Development of O antigen rapid typing by MALDI‐TOF MS. J. Mass Spectrom. Soc. Jpn. 70:237–240. 10.5702/massspec.S22-60 [DOI] [Google Scholar]
- Urakami S, Hinou H. 2022b. Glycan‐selective MALDI in‐source decay analysis of intact glycoproteins. Anal. Sens. 2:e202100040. 10.1002/anse.202100040 [DOI] [Google Scholar]
- Urakami S, Hinou H. 2022c. Direct MALDI glycotyping of glycoproteins toward practical subtyping of biological samples. ACS Omega, 7:39280–39286. 10.1021/acsomega.2c05429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urashima T, Katayama T, Sakanaka M, Fukuda K, Messer M. 2022. Evolution of milk oligosaccharides: Origin and selectivity of the ratio of milk oligosaccharides to lactose among mammals. Biochim. Biophys. Acta ‐ Gen. Sub. 1866:130012. 10.1016/j.bbagen.2021.130012 [DOI] [PubMed] [Google Scholar]
- Vainauskas S, Guntz H, McLeod E, McClung C, Ruse C, Shi X, Taron CH. 2022. A broad‐specificity O‑glycoprotease that enables improved analysis of glycoproteins and glycopeptides containing intact complex O‑glycans. Anal. Chem. 94:1060–1069. 10.1021/acs.analchem.1c04055 [DOI] [PubMed] [Google Scholar]
- Valverde S, Ares AM, Elmore JS, Bernal J. 2022. Recent trends in the analysis of honey constituents. Food Chem., 387:132920. 10.1016/j.foodchem.2022.132920 [DOI] [PubMed] [Google Scholar]
- Van Coillie J, Schulz MA, Bentlage AEH, de Haan N, Ye Z, Geerdes DM, van Esch WJE, Hafkenscheid L, Miller RL, Narimatsu Y, Vakhrushev SY, Yang Z, Vidarsson G, Clausen H. 2022. Role of N‐glycosylation in FcgRIIIa interaction with IgG. Front. Immunol. 13:987151. 10.3389/fimmu.2022.987151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk JHM, van Hooij A, Groot LM, Geboers J, Moretti R, Verhard‐Seymonsbergen E, de Jong D, van der Marel GA, Corstjens PLAM, Codée JDC, Geluk A. 2021. Synthetic phenolic glycolipids for application in diagnostic tests for leprosy. ChemBioChem, 22:1487–1493. 10.1002/cbic.202000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nuffel S, Brunelle A. 2022. TOF‐SIMS Imaging of biological tissue sections and structural determination using tandem MS. Method. Mol. Biol. 2437:77–86. 10.1007/978-1-0716-2030-4_5 [DOI] [PubMed] [Google Scholar]
- van Schaick G, Domínguez‐Vega E, Gstöttner C, van den Berg‐Verleg JH, Schouten O, Akeroyd M, Olsthoorn MMA, Wuhrer M, Heck AJR, Abello N, Franc V. 2021. Native structural and functional proteoform characterization of the prolyl‐alanyl‐specific endoprotease endopro from Aspergillus niger . J. Proteome Res., 20:4875–4885. 10.1021/acs.jproteome.1c00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacore A, Vitiello G, Wanke A, Cavasso D, Clifton LA, Mahdi L, Campanero‐Rhodes MA, Solís D, Wuhrer M, Nicolardi S, Molinaro A, Marchetti R, Zuccaro A, Paduano L, Silipo A. 2022. Lipopolysaccharide O‐antigen molecular and supramolecular modifications of plant root microbiota are pivotal for host recognition. Carbohydr. Polym. 277:118839. 10.1016/j.carbpol.2021.118839 [DOI] [PubMed] [Google Scholar]
- VanKoten HW, Moore RS, Cloninger MJ. 2021. Nanoparticles to study lectins in Caenorhabditis elegans: Multivalent galactose β1‑4 fucose‐functionalized dendrimers provide protection from oxidative stress. Biomacromolecules, 22:4720–4729. 10.1021/acs.biomac.1c01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela‐Aramburu S, Su L, Mosquera J, Morgese G, Schoenmakers SMC, Cardinaels R, Palmans ARA, Meijer EW. 2021. Introducing hyaluronic acid into supramolecular polymers and hydrogels. Biomacromolecules, 22:4633–4641. 10.1021/acs.biomac.1c00927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese M, Haque F, Lu W, Grinstaff MW. 2022. Synthesis and characterization of regioselectively functionalized mono‐sulfated and ‐phosphorylated anionic poly‐amido‐saccharides. Biomacromolecules, 23:2075–2088. 10.1021/acs.biomac.2c00086 [DOI] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Aebi M, Packer NH, Seeberger PH, Esko JD, Stanley P, Hart G, Darvill A, Kinoshita T, Prestegard JJ, Schnaar RL, Freeze HH, Marth JD, Bertozzi CR, Etzler ME, Frank M, Vliegenthart JFG, Lütteke T, Perez S, Bolton E, Rudd P, Paulson J, Kanehisa M, Toukach P, Aoki‐Kinoshita KF, Dell A, Narimatsu H, York W, Taniguchi N, Kornfeld S. 2015. Symbol nomenclature for graphical representations of glycans. Glycobiology, 25:1323–1324. 10.1093/glycob/cwv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Prestegard JJ, Schnaar RL, Seeberger PH, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH. 2022. Essentials of Glycobiology, Fourth edition. New York: Cold Spring Harbor Laboratory Press. 10.1101/9781621824213 [DOI] [Google Scholar]
- Vasudevan UM, Lee OK, Lee EY. 2021. Alginate derived functional oligosaccharides: Recent developments, barriers, and future outlooks. Carbohydr. Polym. 267:118158. 10.1016/j.carbpol.2021.118158 [DOI] [PubMed] [Google Scholar]
- Veeraperumal S, Qiu H‐M, Tan C‐S, Ng S‐T, Zhang W, Tang S, Cheong K‐L, Liu Y. 2021. Restitution of epithelial cells during intestinal mucosal wound healing: The effect of a polysaccharide from the sclerotium of Lignosus rhinocerotis (Cooke) Ryvarden. J. Ethnopharmacol., 274:114024. 10.1016/j.jep.2021.114024 [DOI] [PubMed] [Google Scholar]
- Velasco J, Sepulchro AGV, Higasi PMR, Pellegrini VOA, Cannella D, de Oliveira LC, Polikarpov I, Segato F. 2022. Light boosts the activity of novel LPMO from Aspergillus fumigatus leading to oxidative cleavage of cellulose and hemicellulose. ACS Sustain Chem. Eng. 10:16969–16984. 10.1021/acssuschemeng.2c06281 [DOI] [Google Scholar]
- Veličković D, Liao Y‐C, Thibert S, Veličkovic M, Anderton C, Voglmeir J, Stacey G, Zhou M. 2022. Spatial mapping of plant N‐glycosylation cellular heterogeneity inside soybean root nodules provided insights into legume‐rhizobia symbiosis. Front. Plant Sci. 13:869281. 10.3389/fpls.2022.869281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veličković D, Bečejac T, Mamedov S, Sharma K, Ambalavanan N, Alexandrov T, Anderton CR. 2021. Rapid automated annotation and analysis of N‑glycan mass spectrometry imaging data sets using NGlycDB in METASPACE. Anal. Chem. 93:13421–13425. 10.1021/acs.analchem.1c02347 [DOI] [PubMed] [Google Scholar]
- Veličković D, Sharma K, Alexandrov T, Hodgin JB, Anderton CR. 2022. Controlled humidity levels for fine spatial detail information in enzyme‐assisted N‑glycan MALDI MSI. J. Am. Soc. Mass Spectrom. 33:1577–1580. 10.1021/jasms.2c00120 [DOI] [PubMed] [Google Scholar]
- Venegas FA, Koutaniemi S, Langeveld SMJ, Bellemare A, Chong S‐L, Dilokpimol A, Lowden MJ, Hilden KS, Leyva‐Illades JF, Mäkelä MR, Pham TTM, Peng M, Hancock MA, Zheng Y, Tsang A, Tenkanen M, Powlowski J, de Vries RP. 2022. Carbohydrate esterase family 16 contains fungal hemicellulose acetyl esterases (HAEs) with varying specificity. New Biotechnol., 70:28–38. 10.1016/j.nbt.2022.04.003 [DOI] [PubMed] [Google Scholar]
- Verhertbruggen Y, Bouder A, Vigouroux J, Alvarado C, Geairon A, Guillon F, Wilkinson MD, Stritt F, Pauly M, Lee MY, Mortimer JC, Scheller HV, Mitchell RAC, Voiniciuc C, Saulnier L, Chateigner‐Boutin A‐L. 2021. The TaCslA12 gene expressed in the wheat grain endosperm synthesizes wheat‐like mannan when expressed in yeast and Arabidopsis . Plant Sci., 302:110693. 10.1016/j.plantsci.2020.110693 [DOI] [PubMed] [Google Scholar]
- Viana SM, Montoya AL, Carvalho AM, de Mendonça BS, Portillo S, Olivas JJ, Karimi NH, Estevao IL, Ortega‐Rodriguez U, Carvalho EM, Dutra WO, Maldonaldo RA, Michael K, de Oliveira CI, Almeida IC. 2022. Serodiagnosis and therapeutic monitoring of New‐World tegumentary leishmaniasis using synthetic type‐2 glycoinositolphospholipid‐based neoglycoproteins. Emerg. Microbes Infect. 11:2147–2159. 10.1080/22221751.2022.2114852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira PS, Bonfim IM, Araujo EA, Melo RR, Lima AR, Fessel MR, Paixão DAA, Persinoti GF, Rocco SA, Lima TB, Pirolla RAS, Morais MAB, Correa JBL, Zanphorlin LM, Diogo JA, Lima EA, Grandis A, Buckeridge MS, Gozzo FC, Benedetti CE, Polikarpov I, Giuseppe PO, Murakami MT. 2021. Xyloglucan processing machinery in Xanthomonas pathogens and its role in the transcriptional activation of virulence factors. Nat. Commun. 12:4049. 10.1038/s41467-021-24277-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilaj M, Lauc G, Trbojević‐Akmačić I. 2020. Evaluation of different PNGase F enzymes in immunoglobulin G and total plasma N‐glycans analysis. Glycobiology. 10.1093/glycob/cwaa047 [DOI] [PubMed]
- Villones, Jr. LL , Ludwig AK, Kumeta H, Kikuchi S, Ochi R, Aizawa T, Nishimura SI, Gabius HJ, Hinou H. 2022. Exploring the in situ pairing of human galectins toward synthetic O‑mannosylated core M1 glycopeptides of α‑dystroglycan. Sci. Rep. 12:17800. 10.1038/s41598-022-22758-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimberg V, Zieglerova L, Mazumdar A, Szűcs Z, Borbás A, Herczegh P, Novotna GB. 2021. Two novel semisynthetic lipoglycopeptides active against Staphylococcus aureus biofilms and cells in late stationary growth phase. Pharmaceuticals, 14:1182. 10.3390/ph14111182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeker GCM, Vangangelt KMH, Bladergroen MR, Nicolardi S, Mesker WE, Wuhrer M, van der Burgt YEM, Tollenaar RAEM. 2021. Serum N‐glycan profiles differ for various breast cancer subtypes. Glycoconj. J. 38:387–395. 10.1007/s10719-021-10001-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Okamoto N. 2021. Apolipoprotein C‐III O‐glycoform profiling of 500 serum samples by matrix‐assisted laser desorption/ionization mass spectrometry for diagnosis of congenital disorders of glycosylation. J. Mass Spectrom. 56:e4597. 10.1002/jms.4597 [DOI] [PubMed] [Google Scholar]
- Wagt S, de Haan N, Wang W, Zhang T, Wuhrer M, Lageveen‐Kammeijer GSM. 2022. N‑Glycan isomer differentiation by zero flow capillary electrophoresis coupled to mass spectrometry. Anal. Chem. 94:12954–12959. 10.1021/acs.analchem.2c02840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh I, Zhao S, Wongtrakul‐Kish K, Choo M, Tay SJ, Taron CH, Rudd PM, Nguyen‐Khuong T. 2022. Glycoinformatics tools for comprehensive characterization of glycans enzymatically released from proteins. Method. Mol. Biol. 2370:3–23. 10.1007/978-1-0716-1685-7_1 [DOI] [PubMed] [Google Scholar]
- Walther R, Monge P, Pedersen AB, Benderoth A, Pedersen JN, Farzadfard A, Mandrup OA, Howard KA, Otzen DE, Zelikin AN. 2021. Per‐glycosylation of the surface‐accessible lysines: One‐pot aqueous route to stabilized proteins with native activity. ChemBioChem, 22:2478–2485. 10.1002/cbic.202100228 [DOI] [PubMed] [Google Scholar]
- Wan F, Xu L, Ruan Z, Luo Q. 2021. Genomic and transcriptomic analysis of colistin‐susceptible and colistin‐resistant isolates identify two‐component system EvgS/EvgA associated with colistin resistance in Escherichia coli . Infect. Drug Resist. 14:2437–2447. 10.2147/idr.s316963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X, Wei L, Zhang W, Lei Y, Kong Q, Zhang B. 2021. Production, characterization, and prebiotic activity of oligosaccharides from konjac glucomannan by Bacillus amyloliquefaciens WX‐1. J. Funct. Food, 88:104872. 10.1016/j.jff.2021.104872 [DOI] [Google Scholar]
- Wang B, Duan A, Xie S, Zhang J, Yuan L, Cao Q. 2021. The molecular imprinting of magnetic nanoparticles with boric acid affinity for the selective recognition and isolation of glycoproteins. RSC Adv., 11:25524–25529. 10.1039/D1RA00716E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Liu J, Yan Y, Ding C‐F, Tang K. 2021. Post‐synthesis of boric acid‐functionalized magnetic covalent organic framework as an affinity probe for the enrichment of N‐glycopeptides. Microchim. Acta, 188:336. 10.1007/s00604-021-04998-5 [DOI] [PubMed] [Google Scholar]
- Wang B, Chen K, Zhang P, Long L, Ding S. 2022. Comparison of the biochemical properties and roles in the xyloglucan‐rich biomass degradation of a GH74 xyloglucanase and its CBM‐deleted variant from Thielavia terrestris . Int. J. Mol. Sci., 23:5276. 10.3390/ijms23095276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Sun YP, Wu ZP, Zheng XW, Hou J, Cui HL. 2022. Halorientalis salina sp. nov., Halorientalis marina sp. nov., Halorientalis litorea sp. nov.: Three extremely halophilic archaea isolated from a salt lake and coarse sea salt. Extremophiles, 26:26. 10.1007/s00792-022-01275-y [DOI] [PubMed] [Google Scholar]
- Wang BX, Wheeler KM, Cady KC, Lehoux S, Cummings RD, Laub MT, Ribbeck K. 2021. Mucin glycans signal through the sensor kinase RetS to inhibit virulence‐associated traits in Pseudomonas aeruginosa . Curr. Biol. 31:90–102. 10.1016/j.cub.2020.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C‐Y, Bergström E, Southgate J, Thomas‐Oates J. 2022. Surface shave: Revealing the apical‐restricted uroglycome. J. Proteome Res. 21:360–374. 10.1021/acs.jproteome.1c00714 [DOI] [PubMed] [Google Scholar]
- Wang C, Gao W, Yan S, Zhu X‐Q, Suo X, Liu X, Gupta N, Hu M. 2021. N‐glycome and N‐glycoproteome of a hematophagous parasitic nematode Haemonchus . Comput. Struct. Biotechnol. J. 19:2486–2496. 10.1016/j.csbj.2021.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gao X, Gong G, Man L, Wei Q, Lan Y, Yang M, Han J, Jin W, Wei M, Huang L, Wang Z. 2022. A versatile strategy for high‐resolution separation of reducing glycan mixtures as hydrazones by two‐dimensional high‐performance liquid chromatography. J. Chromatogr., A, 1685:463599. 10.1016/j.chroma.2022.463599 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu L, Wang T, Liu X, Peng W, Srivastav RK, Zhu X‐Q, Gupta N, Gasser RB, Hu M. 2022. H11‐induced immunoprotection is predominantly linked to N‐glycan moieties during Haemonchus contortus infection. Front. Immunol. 13:1034820. 10.3389/fimmu.2022.1034820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tang R, Pan L, Wu W, Ma S, Wei Y, Ou J. 2022. Preparation of core‐shell microporous organic polymer‐coated silica microspheres for chromatographic separation and N‐glycopeptides. J. Sep. Sci. 45:1458–1468. 10.1002/jssc.202100466 [DOI] [PubMed] [Google Scholar]
- Wang C, Zhang C, Gao X, Lin JM. 2022. Isomer‑specific biomarker discovery in multiple myeloma with dual‑derivatized N‑glycans. Anal. Bioanal. Chem. 414:5617–5626. 10.1007/s00216-022-04010-w [DOI] [PubMed] [Google Scholar]
- Wang D, Kao M‐R, Li J, Sun P, Meng Q, Vyas A, Liang P‐H, Wang Y‐S, Hsieh YSY. 2022. Novel two‐step process in cellulose depolymerization: Hematite‐mediated photocatalysis by lytic polysaccharide monooxygenase and Fenton reaction. J. Agric. Food Chem. 70:9941–9947. 10.1021/acs.jafc.2c02445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao J, Zhao Y, Wang S, Feng S, Gu G. 2021. Immunogenicity assessment of different segments and domains of group A streptococcal C5a peptidase and their application potential as carrier protein for glycoconjugate vaccine development. Vaccines, 9:139. 10.3390/vaccines9020139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang Y, Lu H. 2022. Selective enrichment methods for N‐glycosite containing peptides in N‐glycoproteomics. In: Lu H, Editor, Mass Spectrometry‐Based Glycoproteomics and Its Clinic Application. London: Taylor and Francis. p 1–27. 10.1201/9781003185833-1 [DOI] [Google Scholar]
- Wang H‐T, Bharadwaj VS, Yang J‐Y, Curry TM, Moremen KW, Bomble YJ, Urbanowicz BR. 2021. Rational enzyme design for controlled functionalization of acetylated xylan for cell‐free polymer biosynthesis. Carbohydr. Polym. 273:118564. 10.1016/j.carbpol.2021.118564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sun C, Sun X, Zhang L, Zhao J, Liang M, Xiao M, Gu G. 2021. Biochemical characterization and synthetic application of α‐1,3‐glucosyltransferase from pneumococcus serotype 18C. ChemCatChem, 13:3350–3356. 10.1002/cctc.202100507 [DOI] [Google Scholar]
- Wang H, Yang H, Wen Z, Gao C, Gao Y, Tian Y, Xu Z, Liu X, Persson S, Zhang B, Zhou Y. 2022. Xylan‐based nanocompartments orchestrate plant vessel wall patterning. Nat. Plants, 8:295–306. 10.1038/s41477-022-01113-1 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang X, Peng Y, Pan B, Wang B, Peng DH, Guo W. 2022. A LC‐MS/MS method to simultaneously profile 14 free monosaccharides in biofluids. J. Chromatogr., B, 1192:123086. 10.1016/j.jchromb.2021.123086 [DOI] [PubMed] [Google Scholar]
- Wang J, Huang C, Zhou J, Zhao K, Li Y. 2021. Causal link between immunoglobulin G glycosylation and cancer: A potential glycobiomarker for early tumor detection. Cellular Immunol. 361:104282. 10.1016/j.cellimm.2021.104282 [DOI] [PubMed] [Google Scholar]
- Wang J, Yang E, Chaurand P, Raghavan V. 2021. Visualizing the distribution of strawberry plant metabolites at different maturity stages by MALDI‐TOF imaging mass spectrometry. Food Chem., 345:128838. 10.1016/j.foodchem.2020.128838 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao J, Nie S, Xie M, Li S. 2021. Mass spectrometry for structural elucidation and sequencing of carbohydrates. Trends Anal. Chem. 144:116436. 10.1016/j.trac.2021.116436 [DOI] [Google Scholar]
- Wang J, Peng W, Fowowe M, Daramola O, Mechref Y. 2022. An efficient and economical N‐glycome sample preparation using acetone precipitation. Metabolites, 12:1285. 10.3390/metabo12121285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang X, Li J, Xia Y, Gao M, Zhang X, Huang L‐H. 2022. A novel hydrophilic MOFs‐303‐functionalized magnetic probe for the highly efficient analysis of N‐linked glycopeptides. J. Mater. Chem. B, 10:2011–2018. 10.1039/D1TB02827H [DOI] [PubMed] [Google Scholar]
- Wang J, Wen Y, Zhou S‐H, Zhang H‐W, Peng X‐Q, Zhang R‐Y, Yin X‐G, Qiu H, Gong R, Yang G‐F, Guo J. 2022. Self‐adjuvanting lipoprotein conjugate αGalCer‐RBD induces potent immunity against SARS‐CoV‑2 and its variants of concern. J. Med. Chem., 65:2558–2570. 10.1021/acs.jmedchem.1c02000 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhao J, Nie S, Xie M, Li S. 2022. Rapid profiling strategy for oligosaccharides and polysaccharides by MALDI TOF mass spectrometry. Food Hydrocolloid., 124:107237. 10.1016/j.foodhyd.2021.107237 [DOI] [Google Scholar]
- Wang J, Zhao K, Chen L, Zhou J, Sun Q, Chen J, Su R, Li Y. 2022. Proteomics and post‐translational modifications analysis of umbilical mesenchymal stem cells aging. Anal. Biochem. 652:114770. 10.1016/j.ab.2022.114770 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang X, Luo F, Li Y. 2022. Effect of ultrasound on cyanidin‐3‐O‐glucoside and β‐lactoglobulin binding interaction and functional properties. Int. J. Food Sci. Technol. 57:7057–7065. 10.1111/ijfs.16037 [DOI] [Google Scholar]
- Wang L, Xing X, Zeng X, Jackson SR, TeSlaa T, Al‐Dalahmah O, Samarah LZ, Goodwin K, Yang L, McReynolds MR, Li X, Wolff JJ, Rabinowitz JD, Davidson SM. 2022. Spatially resolved isotope tracing reveals tissue metabolic activity. Nat. Meth. 19:223–230. 10.1038/s41592-021-01378-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N‐n, Li Y‐x, Miao M, Zhu C‐h, Yan Q‐j, Jiang Z‐q. 2021. High level expression of a xyloglucanase from Rhizomucor miehei in Pichia pastoris for production of xyloglucan oligosaccharides and its application in yoghurt. Int. J. Biol. Macromol., 190:845–852. 10.1016/j.ijbiomac.2021.09.035 [DOI] [PubMed] [Google Scholar]
- Wang N, Dartois V, Carter CL. 2021. An optimized method for the detection and spatial distribution of aminoglycoside and vancomycin antibiotics in tissue sections by mass spectrometry imaging. J. Mass Spectrom. 56:e4708. 10.1002/jms.4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N‐n, Li Y‐x, Zhu C‐h, Yan Q‐j, Shi R, Jiang Z‐q. 2022. High‐level expression of xyloglucanase B from Rhizomucor miehei and its application in the preparation of partially hydrolyzed apple pomace xyloglucan. Food Bioeng., 1:119–125. 10.1002/fbe2.12012 [DOI] [Google Scholar]
- Wang Q, Wang Y, Yang S, Lin C, Aliyu L, Chen Y, Parsons L, Tian Y, Jia H, Pekosz A, Betenbaugh MJ, Cipollo JF. 2021. A linkage‐specific sialic acid labeling strategy reveals different site‐specific glycosylation patterns in SARS‐CoV‐2 spike protein produced in CHO and HEK cell substrates. Front. Chem. 9:735558. 10.3389/fchem.2021.735558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun L, Wu H, Deng N, Zhao X, Zhou J, Zhang T, Han H, Jiang Z. 2022. Rapid fabrication of zwitterionic sulfobetaine vinylimidazole‐based monoliths via photoinitiated copolymerization for hydrophilic interaction chromatography. J. Pharmaceut. Anal., 12:783–790. 10.1016/j.jpha.2022.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Sun N, Kunzke T, Buck A, Shen J, Prade VM, Stöckl B, Wang J, Feuchtinger A, Walch A. 2022. A simple preparation step to remove excess liquid lipids in white adipose tissue enabling improved detection of metabolites via MALDI‑FTICR imaging MS. Histochem. Cell Biol. 157:595–605. 10.1007/s00418-022-02088-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang T, Wu WW, Lin C‐Y, Yang S, Yang G, Jankowska E, Hu Y, Shen R‐F, Betenbaugh MJ, Cipollo JF. 2022. Comprehensive N‐ and O‑glycoproteomic analysis of multiple chinese hamster ovary host cell lines. J. Proteome Res. 21:2341–2355. 10.1021/acs.jproteome.2c00207 [DOI] [PubMed] [Google Scholar]
- Wang Q, Wang T, Zhang R, Yang S, McFarland KS, Chung CY, Jia H, Wang LX, Cipollo JF, Betenbaugh MJ. 2022. The interplay of protein engineering and glycoengineering to fine‐tune antibody glycosylation and its impact on effector functions. Biotechnol. Bioeng. 119:102–117. 10.1002/bit.27953 [DOI] [PubMed] [Google Scholar]
- Wang S‐L, Wang Y, Wu L, Cai Y‐Y, Wang Z‐C, Alolga RN, Qi L‐W, Li B, Huang F‐Q. 2022. Paired derivatization approach with H/D‐labeled hydroxylamine reagents for sensitive and accurate analysis of monosaccharides by liquid chromatography tandem mass spectrometry. Anal. Chem. 94:3590–3599. 10.1021/acs.analchem.1c04924 [DOI] [PubMed] [Google Scholar]
- Wang S‐S, del Solar V, Yu X, Antonopoulos A, Friedman AE, Agarwal K, Garg M, Ahmed SM, Addhya A, Nasirikenari M, Lau JT, Dell A, Haslam SM, Sampathkumar S‐G, Neelamegham S. 2021. Efficient inhibition of O‐glycan biosynthesis using the hexosamine analog Ac5GalNTGc. Cell Chem. Biol. 28:699–710. 10.1016/j.chembiol.2021.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S‐S, Zhang Q, Guo Y‐l. 2021. Research progress of derivatization method in MALDI mass spectrometry imaging. J. Chinese Mass Spectrom. Soc., 42:995–1013. 10.7538/zpxb.2020.0074 [DOI] [Google Scholar]
- Wang S, Chen C, Gadi MR, Saikam V, Liu D, Zhu H, Bollag R, Liu K, Chen X, Wang F, Wang PG, Ling P, Guan W, Li L. 2021. Chemoenzymatic modular assembly of O‐GalNAc glycans for functional glycomics. Nat. Commun. 12:3573. 10.1038/s41467-021-23428-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu D, Qu J, Zhu H, Chen C, Gibbons C, Greenway H, Wang P, Bollag RJ, Liu K, Li L. 2021. Streamlined subclass‐specific absolute quantification of serum IgG glycopeptides using synthetic isotope‐labeled standards. Anal. Chem. 93:4449–4455. 10.1021/acs.analchem.0c04462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bai J, Wang K, Guo Y. 2022. Carbon fiber paper spray ionization mass spectrometry. Anal. Chim. Acta, 1232:340477. 10.1016/j.aca.2022.340477 [DOI] [PubMed] [Google Scholar]
- Wang S, Yang Y, Zhu Q, Lin G‐Q, Yu B. 2022. Chemical synthesis of polysaccharides. Curr. Opin. Chem. Biol. 69:102154. 10.1016/j.cbpa.2022.102154 [DOI] [PubMed] [Google Scholar]
- Wang T, Yang H, Zhao H, Voglmeir J, Liu L. 2021. Changes of protein N‐glycosylation in the growth of Arabidopsis thaliana and effects of enzymatic deglycosylation on root development. Chinese Bull. Bot. 56:262–274. 10.11983/CBB20163 [DOI] [Google Scholar]
- Wang T, Liu L, Voglmeir J. 2022. mAbs N‐glycosylation: Implications for biotechnology and analytics. Carbohydr. Res. 514:108541. 10.1016/j.carres.2022.108541 [DOI] [PubMed] [Google Scholar]
- Wang W, Hansen AE, Sun H, Fliedner FP, Kjaer A, Jensen AI, Andresen TL, Henriksen JR. 2021. Carbohydrate based biomarkers enable hybrid near infrared fluorescence and 64Cu based radio‐guidance for improved surgical precision. Nanotheranostics, 5:448–460. 10.7150/ntno.60295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kałuża A, Nouta J, Nicolardi S, Ferens‐Sieczkowska M, Wuhrer M, Lageveen‐Kammeijer GSM, de Haan N. 2021. High‐throughput glycopeptide profiling of prostate‐specific antigen from seminal plasma by MALDI‐MS. Talanta, 222:121495. 10.1016/j.talanta.2020.121495 [DOI] [PubMed] [Google Scholar]
- Wang W, Przybylski C, Cai X, Lopin‐Bon C, Jiao R, Shi L, Sugahara K, Neira JL, Daniel R, Li F. 2021. Investigation of action pattern of a novel chondroitin sulfate/dermatan sulfate 4‐O‐endosulfatase. Biochem. J. 478:281–298. 10.1042/bcj20200657 [DOI] [PubMed] [Google Scholar]
- Wang W, Lageveen‐Kammeijer GSM. 2022. High‐sensitivity glycoproteomic analysis of biological samples by CZE‐ESI‐MS. Method . Mol. Biol. 2530:143–162. 10.1007/978-1-0716-2493-7_10 [DOI] [PubMed] [Google Scholar]
- Wang X‐N, Li B. 2022. Monolithic gold nanoparticles/thiol‐β‐cyclodextrin‐functionalized TiO2 nanowires for enhanced SALDI MS detection and imaging of natural products. Anal. Chem. 94:952–959. 10.1021/acs.analchem.1c03764 [DOI] [PubMed] [Google Scholar]
- Wang X, Chen Y, Liu Y, Ouyang L, Yao R, Wang Z, Kang Y, Yan L, Huai D, Jiang H, Lei Y, Liao B. 2022. Visualizing the distribution of lipids in peanut seeds by MALDI mass spectrometric imaging. Foods, 11:3888. 10.3390/foods11233888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hummon AB. 2021. MS imaging of multicellular tumor spheroids and organoids as an emerging tool for personalized medicine and drug discovery. J. Biol. Chem. 297:101139. 10.1016/j.jbc.2021.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li D, Dong C, Zhao Y, Zhang L, Yang F, Ye X, Huang Y, Li Z, Cui Z. 2021. Heterologous expression and characterization of a novel glycoside hydrolase family 55 β‑1,3‑glucanase, AcGluA, from Archangium sp. strain AC19. Appl. Microbiol. Biotechnol. 105:6793–6803. 10.1007/s00253-021-11513-6 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li D, Liu M, Xia C, Fan Q, Li X, Lan Z, Shi G, Dong W, Li Z, Cui Z. 2021. Preparation of active chitooligosaccharides with a novel chitosanase AqCoA and their application in fungal disease protection. J. Agric. Food Chem., 69:3351–3361. 10.1021/acs.jafc.0c07802 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu M, Wang X, Zhong L, Shi G, Xu Y, Li Y, Li R, Huang Y, Ye X, Li Z, Cui Z. 2021. A novel β‐1,3‐glucanase Gns6 from rice possesses antifungal activity against Magnaporthe oryzae . J. Plant Physiol., 265:153493. 10.1016/j.jplph.2021.153493 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wu Z, Hu W, Hao P, Yang S. 2021. Impact of expressing cells on glycosylation and glycan of the SARSCoV‑2 spike glycoprotein. ACS Omega, 6:15988–15999. 10.1021/acsomega.1c01785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang N, Wang Q, Yu Y, Wang P. 2021. Chitosan grafting via one‐enzyme double catalysis: An effective approach for improving performance of wool. Carbohydr. Polym. 252:117157. 10.1016/j.carbpol.2020.117157 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao H, Tao J, Li M, Liu G, Dong W. 2021. A new method for purifying N‐glycans released from milk glycoprotein. J. Proteomics, 245:104283. 10.1016/j.jprot.2021.104283 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao Y, Wang X, Zhong L, Fan Q, Lan Z, Ye X, Huang Y, Li Z, Cui Z. 2021. Functional characterization of the novel laminaripentaose‐producing β‑1,3‐glucanase MoGluB and its biocontrol of Magnaporthe oryzae . J. Agric. Food Chem. 69:9571–9584. 10.1021/acs.jafc.1c03072 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu W, Xu H, Jia Q. 2022. Preparation of tannic acid and l‐cysteine functionalized magnetic composites for synergistic enrichment of N‐glycopeptides followed by mass spectrometric analysis. Anal. Meth. 14:3260–3269. 10.1039/d2ay01169g [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang X, Wei Z‐H, Jiao Y‐J, An D‐Y, Huang Y‐P, Liu Z‐S, Yan C. 2022. Dual‐function monolithic enzyme reactor based on dopamine/graphene oxide coating for simultaneous protein enzymatic hydrolysis and glycopeptide enrichment. J. Chromatogr., A, 1666:462848. 10.1016/j.chroma.2022.462848 [DOI] [PubMed] [Google Scholar]
- Wang Yz, Zheng J, Nawaz M, Yang F, Hu J, Gao M‐T. 2022. Oligosaccharide‐phenolic compound conjugates in soluble polysaccharides from rice straw alleviate ethanol fermentation stresses in Saccharomyces cerevisiae . Ind. Crops Prod. 181:114782. 10.1016/j.indcrop.2022.114782 [DOI] [Google Scholar]
- Wang Z, Dhurandhare VM, Mahung CA, Arnold K, Li J, Su G, Xu D, Maile R, Liu J. 2021. Improving the sensitivity for quantifying heparan sulfate from biological samples. Anal. Chem. 93:11191–11199. 10.1021/acs.analchem.1c01761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fu W, Huo M, He B, Liu Y, Tian L, Li W, Zhou Z, Wang B, Xia J, Chen Y, Wei J, Abliz Z. 2021. Spatial‐resolved metabolomics reveals tissue‐specific metabolic reprogramming in diabetic nephropathy by using mass spectrometry imaging. Acta Pharmaceut. Sinica B, 11:3665–3677. 10.1016/j.apsb.2021.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li, X. , Abliz, Z. 2021. Chemical derivatization for mass spectrometric analysis of metabolite isomers. Prog. Chem. 33:406–416. 10.7536/PC200555 [DOI] [Google Scholar]
- Wangpaiboon K, Klaewkla M, Charoenwongpaiboon T, Vongkusolkit N, Panpetch P, Kuttiyawong K, Visessanguan W, Pichyangkura R. 2021. Synergistic enzyme cocktail between levansucrase and inulosucrase for superb levan‐type fructooligosaccharide synthesis. Enzyme Microb. Technol. 154:109960. 10.1016/j.enzmictec.2021.109960 [DOI] [PubMed] [Google Scholar]
- Wangpaiboon K, Laohawuttichai P, Kim S‐Y, Mori T, Nakapong S, Pichyangkura R, Pongsawasdi P, Hakoshima T, Krusong K. 2021. A GH13 α‐glucosidase from Weissella cibaria uncommonly acts on short‐chain maltooligosaccharides. Acta Crystallogr. D Biol. Crystallogr., D77:1064–1076. 10.1107/s205979832100677x [DOI] [PubMed] [Google Scholar]
- Wanyama FM, Blanchard V. 2021. Glycomic‐based biomarkers for ovarian cancer: Advances and challenges. Diagnostics, 11:643. 10.3390/diagnostics11040643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Aoki‐Kinoshita KF, Ishihama Y, Okuda S. 2021. GlycoPOST realizes FAIR principles for glycomics mass spectrometry data. Nucleic Acids Res., 49:D1523–D1528. 10.1093/nar/gkaa1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Liu L, Bian Z, Wu Y. 2021. MALDI‐TOF‐MS analysis of hydrolysis products of beechwood and birchwood xylans catalyzed by xylanase from Bacillus subtilis . J. Food Biochem. 45:e13841. 10.1111/jfbc.13841 [DOI] [PubMed] [Google Scholar]
- Wei X, Ji J, Nie Y, Tang L, Rao M, Wang X, Wu W, Su D, Zhong Z, Yang C. 2022. Synthesis of cyclodextrin derivatives for enantiodifferentiating photocyclodimerization of 2‐anthracenecarboxylate. Nat. Protoc. 17:2494–2516. 10.1038/s41596-022-00722-6 [DOI] [PubMed] [Google Scholar]
- Wells L, Vierra C, Hardman J, Han Y, Dimas D, Gwarada‐Phillips LN, Blackeye R, Eggers DK, LaBranche CC, Král P, McReynolds KD. 2021. Sulfoglycodendrimer therapeutics for HIV‐1 and SARS‐CoV‐2. Adv. Therap. 4:2000210. 10.1002/adtp.202000210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Shepherd D, Liu Y, Krishnan N, Robertson BD, Platt N, Larrouy‐Maumus G, Platt FM. 2022. Inhibition of the Niemann‐Pick C1 protein is a conserved feature of multiple strains of pathogenic mycobacteria. Nat. Commun. 13:5320. 10.1038/s41467-022-32553-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werlen G, Li M‐L, Tottone L, da Silva‐Diz V, Su X, Herranz D, Jacinto E. 2022. Dietary glucosamine overcomes the defects in αβ‐T cell ontogeny caused by the loss of de novo hexosamine biosynthesis. Nat. Commun. 13:7404. 10.1038/s41467-022-35014-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- West CA, Lu X, Grimsley G, Norris‐Caneda K, Mehta AS, Angel PM, Drake RR. 2021. Optimization of multiple glycosidase and chemical stabilization strategies for N‐glycan isomer detection by mass spectrometry imaging in formalin‐fixed, paraffin‐embedded tissues. Method. Mol. Biol. 2271:303–316. 10.1007/978-1-0716-1241-5_21 [DOI] [PubMed] [Google Scholar]
- West CM, Malzl D, Hykollari A, Wilson IBH. 2021. Glycomics, glycoproteomics, and glycogenomics: An inter‐taxa evolutionary perspective. Mol. Cell. Proteomics, 20:100024. 10.1074/mcp.r120.002263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SF, Domann P, Harvey DJ. 2009. Derivatization of sialic acids for stabilization in matrix‐assisted laser desorption/ionization mass spectrometry and concomitant differentiation of α(2‐3) and α(2‐6) isomers. Rapid Commun. Mass Spectrom., 23:303–312. 10.1002/rcm.3867 [DOI] [PubMed] [Google Scholar]
- Williams D, Jamshidi MP, St. Michael F, Chisholm K, Cox A, Sauvageau J. 2021. d‐Glycero‐β‑d‐mannoheptose phosphate 7‑O‑modifications. J. Org. Chem. 86:2184–2199. 10.1021/acs.joc.0c02333 [DOI] [PubMed] [Google Scholar]
- Williams SE, Noel M, Lehoux S, Cetinbas M, Xavier RJ, Sadreyev RI, Scolnick EM, Smoller JW, Cummings RD, Mealer RG. 2022. Mammalian brain glycoproteins exhibit diminished glycan complexity compared to other tissues. Nat. Commun. 13:275. 10.1038/s41467-021-27781-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Fegan N, Turner MS. 2022. Co‐culture with Acinetobacter johnsonii enhances benzalkonium chloride resistance in Salmonella enterica via triggering lipid A modifications. Int. J. Food Microbiol., 381:109905. 10.1016/j.ijfoodmicro.2022.109905 [DOI] [PubMed] [Google Scholar]
- Wilson LFL, Dendooven T, Hardwick SW, Echevarría‐Poza A, Tryfona T, Krogh KBRM, Chirgadze DY, Luisi BF, Logan DT, Mani K, Dupree P. 2022. The structure of EXTL3 helps to explain the different roles of bi‐domain exostosins in heparan sulfate synthesis. Nat. Commun. 13:3314. 10.1038/s41467-022-31048-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MP, Quelhas D, Leão‐Teles E, Sturiale L, Rymen D, Keldermans L, Race V, Souche E, Rodrigues E, Campos T, Van Schaftingen E, Foulquier F, Garozzo D, Matthijs G, Jaeken J. 2021. SLC37A4‐CDG: Second patient. JIMD Rep., 58:122–128. 10.1002/jmd2.12195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MP, Durin Z, Unal Ö, Ng B, Marrecau T, Keldermans L, Souche E, Rymen D, Gündüz M, Köse G, Sturiale L, Garozzo D, Freeze H, Jaeken J, Foulquier F, Matthijs G. 2022. CAMLG‐CDG: A novel congenital disorder of glycosylation linked to defective membrane trafficking. Hum. Mol. Genet. 31:2571–2581. 10.1093/hmg/ddac055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woitassek D, Moya‐Cancino JG, Sun Y, Song Y, Woschko D, Roitsch S, Janiak C. 2022. Sweet, sugar‐coated hierarchical platinum nanostructures for easy support, heterogenization and separation. Chemistry, 4:1147–1160. 10.3390/chemistry4040078 [DOI] [Google Scholar]
- Wolrab D, Jirásko R, Cífková E, Höring M, Mei D, Chocholoušková M, Peterka O, Idkowiak J, Hrnčiarová T, Kuchař L, Ahrends R, Brumarová R, Friedecký D, Vivo‐Truyols G, Škrha P, Škrha J, Kučera R, Melichar B, Liebisch G, Burkhardt R, Wenk MR, Cazenave‐Gassiot A, Karásek P, Novotný I, Greplová K, Hrstka R, Holčapek M. 2022. Lipidomic profiling of human serum enables detection of pancreatic cancer. Nat. Commun. 13:124. 10.1038/s41467-021-27765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H‐TK, Chen X, Wu R, Wong Y‐LE, Hung Y‐LW, Chan T‐WD. 2022. Dissociation of mannose‐rich glycans using collision‐based and electron‐based ion activation methods. J. Am. Soc. Mass Spectrom. 33:803–812. 10.1021/jasms.1c00385 [DOI] [PubMed] [Google Scholar]
- Wong H‐TK, Chen X, Zhang S, Lui T‐Y, Hu D, Chan T‐WD. 2022. Tandem mass spectrometry for structural characterization of doubly‐charged N‑linked glycopeptides. J. Am. Soc. Mass Spectrom. 33:1458–1464. 10.1021/jasms.2c00143 [DOI] [PubMed] [Google Scholar]
- Woodcock SD, Syson K, Little RH, Ward D, Sifouna D, Brown JKM, Bornemann S, Malone JG. 2021. Trehalose and α‐glucan mediate distinct abiotic stress responses in Pseudomonas aeruginosa . PLoS Genet., 17:e1009524. 10.1371/journal.pgen.1009524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C‐C, Zhang H‐T, Gao Z‐X, Qu J‐J, Zhu L, Zhan X‐B. 2022. Enhanced solubility of curcumin by complexation with fermented cyclic β−1,2‐glucans. J. Pharmaceut. Biomed. Anal. 211:114613. 10.1016/j.jpba.2022.114613 [DOI] [PubMed] [Google Scholar]
- Wu C‐H, Rismondo J, Morgan RML, Shen Y, Loessner MJ, Larrouy‐Maumus G, Freemont PS, Gründling A. 2022. Bacillus subtilis YngB contributes to wall teichoic acid glucosylation and glycolipid formation during anaerobic growth. J. Biol. Chem., 296:100384. 10.1016/j.jbc.2021.100384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Murugesan G, Nagala M, McCraw A, Haslam SM, Dell A, Crocker PR. 2021. Activation of regulatory T cells triggers specific changes in glycosylation associated with Siglec‐1‐dependent inflammatory responses. Wellcome Open Res., 6:134. 10.12688/wellcomeopenres.16834.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Grassi P, MacIntyre DA, Molina BG, Sykes L, Kundu S, Hsiao CT, Khoo KH, Bennett PR, Dell A, Haslam SM. 2022. N‑Glycosylation of cervicovaginal fluid reflects microbial community, immune activity, and pregnancy status. Sci. Rep. 12:16948. 10.1038/s41598-022-20608-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Crost EH, Owen CD, van Bakel W, Gascueña AM, Latousakis D, Hicks T, Walpole S, Urbanowicz PA, Ndeh D, Monaco S, Salom LS, Griffiths R, Reynolds RS, Colvile A, Spencer DIR, Walsh M, Angulo J, Juge N. 2021. The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract. PLoS Biol., 19:e3001498. 10.1371/journal.pbio.3001498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Jin X, Zhu C, Yan Y, Ding C‐F, Tang K. 2021. Gold nanoparticle‐glutathione functionalized MOFs as hydrophilic materials for the selective enrichment of glycopeptides. Talanta, 228:122263. 10.1016/j.talanta.2021.122263 [DOI] [PubMed] [Google Scholar]
- Wu J, Yang Z, Yang X, Chen X, Zhang H, Zhan X. 2021. Synthesis of branched β‐1,3‐glucan oligosaccharide with narrow degree of polymerization by fungi co‐cultivation. Carbohydr. Polym. 273:118582. 10.1016/j.carbpol.2021.118582 [DOI] [PubMed] [Google Scholar]
- Wu L, Fei W, Liu Z, Zhang L, Fang C, Lu H. 2022. Specific and reversible enrichment of early‐stage glycated proteome based on thiazolidine chemistry and palladium‐mediated cleavage. Anal. Chem. 94:5213–5220. 10.1021/acs.analchem.1c03648 [DOI] [PubMed] [Google Scholar]
- Wu Q, Tao Y, Huang J, Liu YS, Yang XZ, Jing HK, Shen RF, Zhu XF. 2022. The MYB transcription factor MYB103 acts upstream of TRICHOME BIREFRINGENCE‐LIKE27 in regulating aluminum sensitivity by modulating the O‐acetylation level of cell wall xyloglucan in Arabidopsis thaliana . Plant J., 111:529–545. 10.1111/tpj.15837 [DOI] [PubMed] [Google Scholar]
- Wu S, Tian J, Xie N, Adnan M, Wang J, Liu G. 2022. A sensitive, accurate, and high‑throughput gluco‑oligosaccharide oxidase‑based HRP colorimetric method for assaying lytic polysaccharide monooxygenase activity. Biotechnol. Biofuels Bioprod. 15:15. 10.1186/s13068-022-02112-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Tang R, Pan L, Wang C, Zhang J, Ma S, Shen Y, Ou J. 2021. Fabrication of hydrophilic zwitterionic microspheres via inverse suspension polymerization for the enrichment of N‑glycopeptides. Microchim. Acta, 188:348. 10.1007/s00604-021-05010-w [DOI] [PubMed] [Google Scholar]
- Wu X, Hua Y, Wei T, Ma C, Wang Z, Zhang L, Wang J. 2021. Effect and mechanism of action in vitro of cyclodextrin derivative nanoparticles loaded with tyroserleutide on hepatoma. Nanotechnology, 32:285101. 10.1088/1361-6528/abf3f2 [DOI] [PubMed] [Google Scholar]
- Wu X, Ye J, DeLaitsch AT, Rashidijahanabad Z, Lang S, Kakeshpour T, Zhao Y, Ramadan S, Saavedra PV, Yuzbasiyan‐Gurkan V, Kavunja H, Cao H, Gildersleeve JC, Huang X. 2021. Chemoenzymatic synthesis of 9NHAc‐GD2 antigen to overcome the hydrolytic instability of O‐acetylated‐GD2 for anticancer conjugate vaccine development. Angew. Chem. Int. Ed. 60:24179–24188. 10.1002/anie.202108610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Chen Y, Chen H, Yang C, Shen X, Deng C, Sun N, Wu H. 2021. Probing serum N‐glycan patterns for rapid and precise detection of Crohn's disease. Chem. Commun. 57:11362–11365. 10.1039/D1CC04699C [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang N, Wu H, Sun N, Deng C. 2021. Magnetic porous carbon‐dependent platform for the determination of N‐glycans from urine exosomes. Microchim. Acta, 188:66. 10.1007/s00604-021-04728-x [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Shang Z, Liu X, Xu Y, Liu W. 2022. N‑Glycomic profiling reveals dysregulated glycans related to oral cancer using MALDI‑MS. Anal. Bioanal. Chem. 414:1881–1890. 10.1007/s00216-021-03822-6 [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang Y, Li W, Xu Y, Liu Y, Liu X, Xu Y, Liu W. 2022. Flowing on‐line preparation of deglycosylation, labeling and purification for N‐glycan analysis. Talanta, 249:123652. 10.1016/j.talanta.2022.123652 [DOI] [PubMed] [Google Scholar]
- Wymann S, Dai Y, Nair AG, Cao H, Powers GA, Schnell A, Martin‐Roussety G, Leong D, Simmonds J, Lieu KG, de Souza MJ, Mischnik M, Taylor S, Ow SY, Spycher M, Butcher RE, Pearse M, Zuercher AW, Morelli AB, Panousis C, Wilson MJ, Rowe T, Hardy MP. 2021. A novel soluble complement receptor 1 fragment with enhanced therapeutic potential. J. Biol. Chem. 296:100200. 10.1074/jbc.ra120.016127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi X, Pizzi A, Lei H, Zhang B, Chen X, Du G. 2022. Environmentally friendly chitosan adhesives for plywood bonding. Int. J. Adhes. Adhes. 112:103027. 10.1016/j.ijadhadh.2021.103027 [DOI] [Google Scholar]
- Xia D, Liu N, Guo X, Li D. 2022. The oxidation properties and synergism of polysaccharide monooxygenase from Chaetomium thermophilum . Mycosystema, 41:1068–1079. 10.13346/j.mycosystema.210363 [DOI] [Google Scholar]
- Xia L, Bellomo TR, Gibadullin R, Congdon MD, Edmondson EF, Li M, Wlodawer A, Li C, Temme JS, Patel P, Butcher D, Gildersleeve JC. 2022. Development of a GalNAc‐tyrosine‐specific monoclonal antibody and detection of tyrosine O‑GalNAcylation in numerous human tissues and cell lines. J. Am. Chem. Soc., 144:16410–16422. 10.1021/jacs.2c04477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Wu R, Hung YLW, Chen X, Chan T‐WD. 2021. Development of a matrix sublimation device with controllable crystallization temperature for MALDI mass spectrometry imaging. Anal. Chem. 93:6342–6347. 10.1021/acs.analchem.1c00260 [DOI] [PubMed] [Google Scholar]
- Xie X‐T, Cheong K‐L. 2022. Recent advances in marine algae oligosaccharides: Structure, analysis, and potential prebiotic activities. Crit. Rev. Food Sci. Nutr. 62:7703–7717. 10.1080/10408398.2021.1916736 [DOI] [PubMed] [Google Scholar]
- Xie X, Li J, Zhen X, Chen L, Yuan W, Feng Q, Liu X. 2022. Rational construction of fluorescent molecular imprinted polymers for highly efficient glycoprotein detection. Anal. Chim. Acta, 1209:339875. 10.1016/j.aca.2022.339875 [DOI] [PubMed] [Google Scholar]
- Xie Y, Mota LM, Bergin A, O'Flaherty R, Jones A, Morgan B, Butler M. 2021. High‐throughput and high‐sensitivity N‐glycan profiling: A platform for biopharmaceutical development and disease biomarker discovery. Anal. Biochem. 623:114205. 10.1016/j.ab.2021.114205 [DOI] [PubMed] [Google Scholar]
- Xie Z, Feng Q, Fang X, Dai X, Yan Y, Ding C‐F. 2022. One‐pot preparation of hydrophilic glucose functionalized quantum dots for diabetic serum glycopeptidome analysis. Microchem. J. 170:107397. 10.1016/j.microc.2022.107397 [DOI] [Google Scholar]
- Xie Z, Feng Q, Zhang S, Yan Y, Deng C, Ding C‐F. 2022. Advances in proteomics sample preparation and enrichment for phosphorylation and glycosylation analysis. Proteomics, 22:2200070. 10.1002/pmic.202200070 [DOI] [PubMed] [Google Scholar]
- Xie Z, Yan Y, Tang K, Ding C‐F. 2022. Post‐synthesis modification of covalent organic frameworks for ultrahigh enrichment of low‐abundance glycopeptides from human saliva and serum. Talanta, 236:122831. 10.1016/j.talanta.2021.122831 [DOI] [PubMed] [Google Scholar]
- Xin M, Xu Y, You S, Li C, Zhu B, Shen J, Chen Z, Shi W, Xue X, Shi J, Sun S. 2022. Precision structural interpretation of site‐specific N‐glycans in seminal plasma. J. Proteome Res. 21:1664–1674. 10.1021/acs.jproteome.2c00046 [DOI] [PubMed] [Google Scholar]
- Xin M, You S, Wu J, Xu Y, Li C, Zhu B, Shen J, Chen Z, Dang L, Dan W, Zhang X, Sun S. 2022. Evaluation of absorbent cotton for glycopeptide enrichment. Anal. Bioanal. Chem. 414:8245–8253. 10.1007/s00216-022-04353-4 [DOI] [PubMed] [Google Scholar]
- Xing C, Liu C, Kong Z, Wei K, Li P, Li G, Yuan J, Yan W. 2022. De novo assisted AFB1‐specific monoclonal antibody sequence assembly and comprehensive molecular characterization. Anal. Biochem. 656:114883. 10.1016/j.ab.2022.114883 [DOI] [PubMed] [Google Scholar]
- Xiong F, Jia J, Ma J, Jia Q. 2022. Glutathione‐functionalized magnetic thioether‐COFs for the simultaneous capture of urinary exosomes and enrichment of exosomal glycosylated and phosphorylated peptides. Nanoscale, 14:853–864. 10.1039/D1NR06587D [DOI] [PubMed] [Google Scholar]
- Xiong F, Liang H‐X, Zhang Z‐J, Mahmud T, Chan ASC, Li X, Lan W‐J. 2022. Characterization, antioxidant and antitumor activities of oligosaccharides isolated from Evodia lepta (Spreng) Merr. by different extraction methods. Antioxidants, 10:1842. 10.3390/antiox10111842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Ruan H, Cai W, Staehelin C, Dai W. 2021. Identification of an exopolysaccharide biosynthesis gene in Bradyrhizobium diazoefficiens USDA110 . Microorganisms, 9:2490. 10.3390/microorganisms9122490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang R, Zhang H, Wu J, Zhu L, Zhan X. 2021. In vitro assessment of prebiotic properties of oligosaccharides derived from four microbial polysaccharides. LWT ‐ Food Sci. Technol., 147:111544. 10.1016/j.lwt.2021.111544 [DOI] [PubMed] [Google Scholar]
- Xu J, Hu Z, He H, Ou X, Yang Y, Xiao C, Yang C, Li L, Jiang W, Zhou T. 2022. Transcriptome analysis reveals that jasmonic acid biosynthesis and signaling is associated with the biosynthesis of asperosaponin VI in Dipsacus asperoides . Front. Plant Sci. 13:1022075. 10.3389/fpls.2022.1022075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang Y, Han Y, Liu N, Liu Z, Guo H, Zou X, Zhang J. 2022. A preliminary study on change of serum immunoglobulin G glycosylation in patients with migraine. Front. Neurol. 13:860555. 10.3389/fneur.2022.860555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Jin H, Wu Z, Han Y, Chen J, Mao C, Hao P, Zhang X, Liu C‐F, Yang S. 2022. Mass spectrometry‐based analysis of serum N‑glycosylation changes in patients with Parkinson's disease. ACS Chem. Neurosci. 13:1719–1726. 10.1021/acschemneuro.2c00264 [DOI] [PubMed] [Google Scholar]
- Xu S, Zhao M, Gu Z, Lu H, Liu Z. 2022. Photothermal therapy of neuroblastoma via polysialic acid‐targeting nanomissiles. Small, 18:2201671. 10.1002/smll.202201671 [DOI] [PubMed] [Google Scholar]
- Xu W, Han M, Zhang W, Zhang F, Lei F, Wang K, Jiang J. 2021. Production of manno‐oligosaccharide from Gleditsia microphylla galactomannan using acetic acid and ferrous chloride. Food Chem., 346:128844. 10.1016/j.foodchem.2020.128844 [DOI] [PubMed] [Google Scholar]
- Xu W, Cao J‐F, Zhang X‐P, Shu Y, Wang J‐H. 2022. The concurrent enrichment of glycoproteins and phosphoproteins with polyoxometalate‐covalent organic framework conjugate as the adsorbent. J. Chromatogr., A, 1675:463183. 10.1016/j.chroma.2022.463183 [DOI] [PubMed] [Google Scholar]
- Xu W, Han M, Zhang W, Tang M, Zhang F, Jiang J. 2022. Efficient and green production of manno‐oligosaccharides from Gleditsia microphylla galactomannans using CO2 and solid acid in subcritical water. LWT ‐ Food Sci. Technol., 156:113019. 10.1016/j.lwt.2021.113019 [DOI] [Google Scholar]
- Xu W, Zhang W, Han M, Zhang F, Lei F, Cheng X, Ning R, Wang K, Ji L, Jiang J. 2022. Production of xylooligosaccharides from Camellia oleifera Abel fruit shell using a shell‐based solid acid catalyst. Bioresource Technol., 365:128173. 10.1016/j.biortech.2022.128173 [DOI] [PubMed] [Google Scholar]
- Xu Y, Chen H, Zhang H, Ullah S, Hou T, Feng Y. 2021. The MCR‐3 inside linker appears as a facilitator of colistin resistance. Cell Rep., 35:109135. 10.1016/j.celrep.2021.109135 [DOI] [PubMed] [Google Scholar]
- Xu Y, Deng Y, Ye R, Gong C, Liu Z, Zhao Y, Lu Y, Liu J, Xu X. 2021. MALDI‐MS imaging of lipids and small molecules in rat brain tissue based on graphene oxide film pre‐coated matrix. Int. J. Mass Spectrom. 464:116573. 10.1016/j.ijms.2021.116573 [DOI] [Google Scholar]
- Xu Y, Viswanatha R, Sitsel O, Roderer D, Zhao H, Ashwood C, Voelcker C, Tian S, Raunser S, Perrimon N, Dong M. 2022. CRISPR screens in Drosophila cells identify Vsg as a Tc toxin receptor. Nature, 610:349–355. 10.1038/s41586-022-05250-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Deng Y, Zhang Z, Ma W, Li W, Wen L, Li T. 2021. Diversity‐oriented chemoenzymatic synthesis of sulfated and nonsulfated core 2 O‑GalNAc glycans. J. Org. Chem. 86:10819–10828. 10.1021/acs.joc.1c01115 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wu Y, Deng Z, Long J, Sun N, Deng C. 2021. One‐step fabrication of strongly hydrophilic mesoporous silica for comprehensive analysis of serum glycopeptidome. Talanta, 234:122713. 10.1016/j.talanta.2021.122713 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wu Y, Hu X, Deng C, Sun N. 2022. Inherently hydrophilic mesoporous channel coupled with metal oxide for fishing endogenous salivary glycopeptides and phosphopeptides. Chinese Chem. Lett. 33:4695–4699. 10.1016/j.cclet.2021.12.069 [DOI] [Google Scholar]
- Xue S, Pattathil S, Sousa LdC, Ubanwa B, Dale B, Jones AD, Balan V. 2022. Understanding the structure and composition of recalcitrant oligosaccharides in hydrolysate using high‑throughput biotin‑based glycome profiling and mass spectrometry. Sci. Rep. 12:2521. 10.1038/s41598-022-06530-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P, Yadav M, Gaur R, Gupta R, Arora G, Srivastava A, Goswami A, Gawande MB, Sharma RK. 2022. Chemistry of magnetic covalent organic frameworks (MagCOFs): From synthesis to separation applications. Mater. Adv. 3:1432–1458. 10.1039/D1MA01060C [DOI] [Google Scholar]
- Yagi H, Amagasa E, Shiota M, Yamada I, Aoki‐Kinoshita KF, Kato K. 2022. GALAXY ver 3: Updated web application for glycosylation profiling based on 3D HPLC map. Glycobiology, 32:646–650. 10.1093/glycob/cwac025 [DOI] [PubMed] [Google Scholar]
- Yamada I, Shiota M, Shinmachi D, Ono T, Tsuchiya S, Hosoda M, Fujita A, Aoki NP, Watanabe Y, Fujita N, Angata K, Kaji H, Narimatsu H, Okuda S, Aoki‐Kinoshita KF. 2020. The GlyCosmos Portal: A unified and comprehensive web resource for the glycosciences. Nat. Meth., 17:649–650 [DOI] [PubMed] [Google Scholar]
- Yamada Y, Murase M, Yatsugi K, Mizoshita N. 2021. Laser desorption/ionization mass spectrometry using TiO2 nanopillar array substrates with tunable surface roughness and wettability. ACS Appl. Nano Mater., 4:13884–13895. 10.1021/acsanm.1c03222 [DOI] [Google Scholar]
- Yamagaki T, Sugahara K, Fujikawa K, Washida K. 2022. Fragmentation and ionization efficiency of positional and functional isomers of paeoniflorin derivatives in matrix‐assisted laser desorption/ionization time‐of‐flight mass spectrometry. Mass Spectrom. (Tokyo), 11:A0101. 10.5702/massspectrometry.a0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami H, Fuji T, Wako M, Hasegawa Y. 2021. Sulfated polysaccharide isolated from the nacre of pearl oyster improves scopolamine‐induced memory impairment. Antioxidants, 10:505. 10.3390/antiox10040505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Miyawaki N, Kawakami N, Kinoshita M, Suzuki S. 2022. HPLC separation and preparative conditions for 8‐aminopyrene‐1,3,6‐trisulfonic acid‐labeled N‐glycans using a hydrophilic interaction column. Bunseki Kagaku, 71:333–339. 10.2116/bunsekikagaku.71.333 [DOI] [Google Scholar]
- Yaman ME, Avci I, Atila NE, Atila A, Kayili HM, Salih B. 2021. Characterization of serum N‐glycome alterations in seasonal allergic rhinitis using MALDI‐TOF‐MS: A pilot study. J. Carbohydr. Chem., 40:269–282. 10.1080/07328303.2021.2009502 [DOI] [Google Scholar]
- Yaman ME, Kayili HM, Albayrak M, Kadioglu Y, Salih B. 2021. Differential N‐glycosylation profiling of formalin‐fixed paraffin‐embedded (FFPE) invasive ductal carcinoma tissues using MALDI‐TOF‐MS. Mol. Omics, 17:394–404. 10.1039/D0MO00150C [DOI] [PubMed] [Google Scholar]
- Yan N, Song F, Ouyang Y, Li D, Tian H, Yi L, Linhardt RJ, Zhang Z. 2022. Glycan mapping of low‐molecular‐weight heparin using mass spectral correction based on chromatography fitting with “Glycomapping” software. Anal. Chem. 94:13000–13009. 10.1021/acs.analchem.2c01579 [DOI] [PubMed] [Google Scholar]
- Yan Y, Han R, Hou Y, Zhang H, Yu J, Gao W, Xu L, Tang K. 2021. Bowl‐like mesoporous polydopamine with size exclusion for highly selective recognition of endogenous glycopeptides. Talanta, 233:122468. 10.1016/j.talanta.2021.122468 [DOI] [PubMed] [Google Scholar]
- Yang B, Mai TD, Tran NT, Taverna M. 2022. In capillary labeling and online electrophoretic separation of N‐glycans from glycoproteins J. Sep. Sci., 45:3594–3603. 10.1002/jssc.202200340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Liu R, Pang J, Ren B, Zhou H, Wang G, Wang E, Liu J. 2021. Poaceae‐specific cell wall‐derived oligosaccharides activate plant immunity via OsCERK1 during Magnaporthe oryzae infection in rice. Nat. Commun. 12:2178. 10.1038/s41467-021-22456-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Luo X, Lian Q, Gao L, Wang C, Qi X, Zhang R, Liu Z, Liao G. 2022. Fully synthetic Tn‐based three‐component cancer vaccine using covalently linked TLR4 ligand MPLA and iNKT cell agonist KRN‐7000 as built‐in adjuvant effectively protects mice from tumor development. Acta Pharmaceut. Sinica B, 12:4432–4445. 10.1016/j.apsb.2022.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Liu Z, Si W, Song Z, Yin L, Tang H. 2022. Facile preparation of polysaccharide−polypeptide conjugates via a biphasic solution ring‐opening polymerization. ACS Macro Lett., 11:663–668. 10.1021/acsmacrolett.2c00205 [DOI] [PubMed] [Google Scholar]
- Yang H, Smith RD, Chandler CE, Johnson JK, Jackson SN, Woods AS, Scott AJ, Goodlett DR, Ernst RK. 2022. Lipid A structural determination from a single colony. Anal. Chem. 94:7460–7465. 10.1021/acs.analchem.1c05394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Smith RD, Sumner KP, Goodlett DR, Johnson JK, Ernst RK. 2022. A matrix‐assisted laser desorption ionization‐time of flight mass spectrometry direct‐from‐urine‐specimen diagnostic for gram‐negative pathogens. Microbiol. Spectrum, 10:e0373022. 10.1128/spectrum.03730-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Song H, Suo Z, Li F, Jin Q, Zhu X, Chen Q. 2022. A molecularly imprinted electrochemical sensor based on surface imprinted polymerization and boric acid affinity for selective and sensitive detection of P‐glycoproteins. Anal. Chim. Acta, 1207:339797. 10.1016/j.aca.2022.339797 [DOI] [PubMed] [Google Scholar]
- Yang H, Tian Z. 2022. Sialic acid linkage‐specific quantitative N‐glycoproteomics using selective alkylamidation and multiplex TMT‐labeling. Anal. Chim. Acta, 1230:340391. 10.1016/j.aca.2022.340391 [DOI] [PubMed] [Google Scholar]
- Yang J‐X, Li J‐J, Yin F‐Q, Wang G‐Y, Wei W‐T, Li X‐L, Wang K‐R. 2022. Multivalent glucosidase inhibitors based on perylene bisimide and iminosugar conjugates. Eur. J. Med. Chem. 241:114621. 10.1016/j.ejmech.2022.114621 [DOI] [PubMed] [Google Scholar]
- Yang J, Ma T, Yu H, Yin M, Qiao Y, Niu L, Yang F, He J, Li Z. 2021. Alternations of N‐glycans recognized by Phaseolus vulgaris leucoagglutinin in the saliva of patients with breast cancer. Neoplasma, 68:994–1004. 10.4149/neo_2021_210301n264 [DOI] [PubMed] [Google Scholar]
- Yang J, Gao W, Liu R, Yu J, Wang C, Hu J, Tang K. 2022. One‐step synthesis of hydrophilic nano‐floral inter‐polymeric material for highly selective enrichment of N‐linked glycopeptides. Anal. Letts., 55:857–868. 10.1080/00032719.2021.1968888 [DOI] [Google Scholar]
- Yang S, Wang Y, Mann M, Wang Q, Tian E, Zhang L, Cipollo JF, Ten Hagen KG, Tabak LA. 2021. Improved online LC‐MS/MS identification of O‐glycosites by EThcD fragmentation, chemoenzymatic reaction, and SPE enrichment. Glycoconj. J. 38:145–156. 10.1007/s10719-020-09952-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Neta P, Mirokhin YA, Tchekhovskoi DV, Remoroza CA, Burke MC, Liang Y, Markey SP, Stein SE. 2021. MS_Piano: A software tool for annotating peaks in CID tandem mass spectra of peptides and N‑glycopeptides. J. Proteome Res. 20:4603–4609. 10.1021/acs.jproteome.1c00324 [DOI] [PubMed] [Google Scholar]
- Yang Y, Lu L, Sun D, Wang J, Wang N, Qiao L, Guo Q, Wang C. 2022. Fungus polygalacturonase‐generated oligogalacturonide restrains fruit softening in ripening tomato. J. Agric. Food Chem. 70:759–769. 10.1021/acs.jafc.1c04972 [DOI] [PubMed] [Google Scholar]
- Yao S‐F, Zhang H‐B, Cai Z‐S, Gao H, Wang D, Shang S‐B. 2022. Two new steroidal sapogenins from Rohdea chinensis (synonym Tupistra chinensis) rhizomes and their antifungal activity. J. Asian Nat. Prod. Res., 24:153–162. 10.1080/10286020.2021.1886088 [DOI] [PubMed] [Google Scholar]
- Ye Z, Li T, Qing D, Sun Y, Chen H, Yu Q, Yan C. 2021. Structural elucidation and osteogenic activity of a novel heteropolysaccharide from Alhagi pseudalhagi . Int. J. Biol. Macromol., 171:185–197. 10.1016/j.ijbiomac.2020.12.189 [DOI] [PubMed] [Google Scholar]
- Ye Z, Vakhrushev SY. 2021. The role of data‐independent acquisition for glycoproteomics. Mol. Cell. Proteomics, 20:100042. 10.1074/mcp.r120.002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yermak I, Anastyuk S, Kravchenko A, Helbert W, Glazunov V, Shulgin A, Spirin P, Prassolov V. 2021. New insights into the structure of kappa/beta‐carrageenan: A novel potential inhibitor of HIV‐1. Int. J. Mol. Sci., 22:12905. 10.3390/ijms222312905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerra NV, Dyaga B, Dadinaboyina SB, Pandeti S, Vaidya JR, Tabet J‐C, Thota JR. 2021. 2‑Cyano‐3‐(2‐thienyl)acrylic acid as a new MALDI matrix for the analysis of a broad spectrum of analytes. J. Am. Soc. Mass Spectrom., 32:387–393. 10.1021/jasms.0c00398 [DOI] [PubMed] [Google Scholar]
- Yi L, Fu M, Shao Y, Tang K, Yan Y, Ding C‐F. 2022. Bifunctional super‐hydrophilic mesoporous nanocomposite: A novel nanoprobe for investigation of glycosylation and phosphorylation in Alzheimer's disease. J. Chromatogr., A, 1676:463236. 10.1016/j.chroma.2022.463236 [DOI] [PubMed] [Google Scholar]
- Yi L, Shao Y, Fu M, Yan Y, Ding C‐F, Tang K. 2022. One‐step preparation of magnetic zwitterionic‐hydrophilic dual functional nanospheres for in‐depth glycopeptides analysis in Alzheimer's disease patients’ serum. J. Chromatogr., A, 1669:462929. 10.1016/j.chroma.2022.462929 [DOI] [PubMed] [Google Scholar]
- Yilmaz F. 2021. Glycolipids being viewed in vivo or in vitro . In: Demir K, Editor, Studies in Glycolipids. Cambridge: Cambridge Scholars Publishing. p 61–74. [Google Scholar]
- Yin W, Ling Z, Dong Y, Qiao L, Shen Y, Liu Z, Wu Y, Li W, Zhang R, Walsh TR, Dai C, Li J, Yang H, Liu D, Wang Y, Gao GF, Shen J. 2021. Mobile colistin resistance enzyme MCR‐3 facilitates bacterial evasion of host phagocytosis. Adv. Sci. 8:2101336. 10.1002/advs.202101336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X‐G, Lu J, Wang J, Zhang R‐Y, Wang X‐F, Liao C‐M, Liu X‐P, Liu Z, Guo J. 2021. Synthesis and evaluation of liposomal anti‐GM3 cancer vaccine candidates covalently and noncovalently adjuvanted by αGalCer. J. Med. Chem. 64:1951–1965. 10.1021/acs.jmedchem.0c01186 [DOI] [PubMed] [Google Scholar]
- Yin X, Konishi T, Horikawa K, Tanaka R, Togo Y, Noda T, Hosoi M, Tsuchida M, Kunoh T, Wada S, Nakamura T, Tsuda E, Sasaki R, Mizukami T, Hasegawa M. 2022. Structure and function of potential glycosylation sites of dynactin‑associated protein dynAP. Molec Biotechnol. 64:611–620. 10.1007/s12033-021-00435-3 [DOI] [PubMed] [Google Scholar]
- Yin Z, Dong T, Huang W, Du M, Chen D, Fernie AR, Yi G, Yan S. 2022. Spatially resolved metabolomics reveals variety‐specific metabolic changes in banana pulp during postharvest senescence. Food Chem. X, 15:100371. 10.1016/j.fochx.2022.100371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L, Liang C, Zhang Y, Wang J, Wang C, Xia K, Shi K, Yu C, Yang B, Xu H, Zhang Y, Shu J, Huang X, Xing H, Li F, Zhou X, Chen Q. 2022. Enhancement of nucleus pulposus repair by glycoengineered adipose‐derived mesenchymal cells. Biomaterials, 283:121463. 10.1016/j.biomaterials.2022.121463 [DOI] [PubMed] [Google Scholar]
- Yoneda K, Hu Y, Kita M, Kigoshi H. 2016. 6‐Amidopyrene as a label‐assisted laser desorption/ionization (LALDI) enhancing tag: Development of photoaffinity pyrene derivative. Sci. Rep. 5:17853. 10.1038/srep17853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon E‐J, Jeong SH. 2021. MALDI‐TOF Mass spectrometry technology as a tool for the rapid diagnosis of antimicrobial resistance in bacteria. Antibiotics, 10:982. 10.3390/antibiotics10080982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York WS, Agravat S, Aoki‐Kinoshita KF, McBride R, Campbell MP, Costello CE, Dell A, Feizi T, Haslam SM, Karlsson N, Khoo K‐H, Kolarich D, Liu Y, Novotny M, Packer NH, Paulson JC, Rapp E, Ranzinger R, Rudd PM, Smith DF, Struwe WB, Tiemeyer M, Wells L, Zaia J, Kettner C. 2014. MIRAGE: The minimum information required for a glycomics experiment. Glycobiology, 24:402–406. 10.1093/glycob/cwu018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Zhao M, Chen S, Jiang L, Gao S, Yin H, Zhao L. 2022. Effect of chitooligosaccharides with a specific degree of polymerization on multiple targets in T2DM mice. Bioresources Bioproc. 9:94. 10.1186/s40643-022-00579-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LEA, Conroy LR, Clarke HA, Hawkinson TR, Bolton KE, Sanders WC, Chang JE, Webb MB, Alilain WJ, Kooi CWV, Drake RR, Andres DA, Badgett TC, Wagner LM, Allison DB, Sun RC, Gentry MS. 2022. In situ mass spectrometry imaging reveals heterogeneous glycogen stores in human normal and cancerous tissues. EMBO Mol. Med., 14:e16029. 10.15252/emmm.202216029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi N‐A, Zimmermann ML, Bols M. 2022. A study of the DIBAL‐promoted selective debenzylation of α‐cyclodextrin protected with two different benzyl groups. Beilstein J. Org. Chem. 18:1553–1559. 10.3762/bjoc.18.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Huo Z, Yang J, Palma‐Gudiel H, Boyle PA, Schneider JA, Bennett DA, Zhao J. 2021. Human brain and blood N‐glycome profiling in Alzheimer's disease and Alzheimer's disease‐related dementias. Front. Aging Neurosci. 13:765259. 10.3389/fnagi.2021.765259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Chen W, Zhong J, Qing D, Yan C. 2021. Structural elucidation of three novel oligosaccharides from Kunlun Chrysanthemum flower tea and their bioactivities. Food Chem. Toxicol. 149:112032. 10.1016/j.fct.2021.112032 [DOI] [PubMed] [Google Scholar]
- Yu RB, Dalman NAV, Wuethrich A, Quirino JP. 2021. Derivatization of carbohydrates for analysis by liquid chromatography and capillary electrophoresis. In: El Rassi Z, Editor, Carbohydrate Analysis by Modern Liquid Phase Separation Techniques, Second edition. Amsterdam: Elsevier. p 1–33. 10.1016/B978-0-12-821447-3.00019-6 [DOI] [Google Scholar]
- Yu S, Wang J, Li Y, Wang X, Ren F, Wang X. 2021. Structural studies of water‐insoluble β‐glucan from oat bran and its effect on improving lipid metabolism in mice fed high‐fat diet. Nutrients, 13:3254. 10.3390/nu13093254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Mohsin I, Papageorgiou AC, Li D. 2022. Purification and structural characterization of the auxiliary activity 9 native lytic polysaccharide monooxygenase from Thermoascus aurantiacus and identification of its C1‐ and C4‐oxidized reaction products. Catalysts, 12:139. 10.3390/catal12020139 [DOI] [Google Scholar]
- Yue X, Qin H, Chen Y, Fang Z, Liu L, Zhu H, Liu X, Zhou J, Tian K, Qiao X, Ye M. 2021. Highly efficient enrichment of O‑GalNAc glycopeptides by using immobilized metal ion affinity chromatography. Anal. Chem. 93:7579–7587. 10.1021/acs.analchem.0c05236 [DOI] [PubMed] [Google Scholar]
- Yui Y, Ota S, Aoyama C, Kakuda M. 2022. Purification of 2‐aminobenzamide derivatized glycans using a monolithic solid‐phase extraction centrifugal column. Bunseki Kagaku, 71:365–368. 10.2116/bunsekikagaku.71.365 [DOI] [Google Scholar]
- Yun J, Jo J‐Y, Tuomivaara ST, Lim J‐M. 2021. Isotope labeling strategies of glycans for mass spectrometry‐based quantitative glycomics. Microchem. J. 170:106655. 10.1016/j.microc.2021.106655 [DOI] [Google Scholar]
- Yurova EA, Ananyeva NV. 2022. The practice of application and features of the control of oligosaccharides in the production of specialized food products. A review. Food Sys., 5:353–360. 10.21323/2618-9771-2022-5-4-353-360 [DOI] [Google Scholar]
- Ząbczyńska M, Link‐Lenczowski P, Pocheć E. 2021. Glycosylation in autoimmune diseases. Adv. Exp. Med. Biol. 1325:205–218. 10.1007/978-3-030-70115-4_10 [DOI] [PubMed] [Google Scholar]
- Zacharia A, Harberts E, Valencia SM, Myers B, Sanders C, Jain A, Larson NR, Middaugh CR, Picking WD, Difilippantonio S, Kirnbauer R, Roden RB, Pinto LA, Shoemaker RH, Ernst RK, Marshall JD. 2021. Optimization of RG1‐VLP vaccine performance in mice with novel TLR4 agonists. Vaccine, 39:292–302. 10.1016/j.vaccine.2020.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradnikova M, Ihnatova I, Lattova E, Uhrik L, Stuchlikova E, Nenutil R, Valik D, Nalezinska M, Chovanec J, Zdrahal Z, Vojtesek B, Hernychova L, Novotny MV. 2021. N‐Glycome changes reflecting resistance to platinum‐based chemotherapy in ovarian cancer. J. Proteomics, 230:103964. 10.1016/j.jprot.2020.103964 [DOI] [PubMed] [Google Scholar]
- Zahraei A, Guo G, Perwick RD, Donaldson PJ, Demarais NJ, Grey AC. 2021. Mapping glucose metabolites in the normal bovine lens: Evaluation and optimisation of a matrix‐assisted laser desorption/ionisation imaging mass spectrometry method. J. Mass Spectrom. 56:e4666. 10.1002/jms.4666 [DOI] [PubMed] [Google Scholar]
- Zahraei A, Guo G, Varnava KG, Demarais NJ, Donaldson PJ, Grey AC. 2022. Mapping glucose uptake, transport and metabolism in the bovine lens cortex. Front. Physiol. 13:901407. 10.3389/fphys.2022.901407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaikin VG, Borisov RS. 2021. Mass spectrometry as a crucial analytical basis for omics sciences. J. Anal. Chem. 76:1567–1587. 10.1134/S1061934821140094 [DOI] [Google Scholar]
- Zaikin VG, Borisov RS. 2022. Options of the main derivatization approaches for analytical ESI and MALDI mass spectrometry. Crit. Rev. Anal. Chem. 52:1287–1342. 10.1080/10408347.2021.1873100 [DOI] [PubMed] [Google Scholar]
- Zambonin C, Aresta A. 2022. MALDI‐TOF/MS Analysis of non‐invasive human urine and saliva samples for the identification of new cancer biomarkers. Molecules, 27:1925. 10.3390/molecules27061925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamfir AD. 2022. Capillary zone electrophoresis‐electrospray ionization tandem mass spectrometry for total analysis of chondroitin/dermatan sulfate oligosaccharides. Method. Mol. Biol. 2531:163–184. 10.1007/978-1-0716-2493-7_11 [DOI] [PubMed] [Google Scholar]
- Zappe A, Miller RL, Struwe WB, Pagel K. 2022. State‐of‐the‐art glycosaminoglycan characterization. Mass Spectrom. Rev. 41:1040–1071. 10.1002/mas.21737 [DOI] [PubMed] [Google Scholar]
- Zelepuga EA, Silchenko AS, Avilov SA, Kalinin VI. 2021. Structure‐activity relationships of holothuroid's triterpene glycosides and some in silico insights obtained by molecular dynamics study on the mechanisms of their membranolytic action. Mar. Drugs, 19:604. 10.3390/md19110604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Luan F, Hu J, Liu Y, Zhang X, Qin T, Zhang X, Liu R, Zeng N. 2022. Recent research advances in polysaccharides from Undaria pinnatifida: Isolation, structures, bioactivities, and applications. Int. J. Biol. Macromol. 206:325–354. 10.1016/j.ijbiomac.2022.02.138 [DOI] [PubMed] [Google Scholar]
- Zeng L, Li J, Cheng Y, Wang D, Gu J, Li F, Han W. 2021. Comparison of biochemical characteristics, action models, and enzymatic mechanisms of a novel exolytic and two endolytic lyases with mannuronate preference. Mar. Drugs, 19:706. 10.3390/md19120706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W‐F, Cao W‐Q, Liu M‐Q, He S‐M, Yang P‐Y. 2021. Precise, fast and comprehensive analysis of intact glycopeptides and modified glycans with pGlyco3. Nat. Meth. 18:1515–1523. 10.1038/s41592-021-01306-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Huang X, Xue J, Liu H, Xiong C, Wang J, Nie Z. 2021. MALDI‐TOF/TOF tandem mass spectrometry imaging reveals non‐uniform distribution of disaccharide isomers in plant tissues. Food Chem., 338:127984. 10.1016/j.foodchem.2020.127984 [DOI] [PubMed] [Google Scholar]
- Zhang C, Jin X, Wang L, Jin C, Han X, Ma W, Li X, Teng G. 2021. Hollow MnFe2O4@C@APBA nanospheres with size exclusion and pH response for efficient enrichment of endogenous glycopeptides. ACS Appl. Mater. Interfaces, 13:9714–9728. 10.1021/acsami.0c22221 [DOI] [PubMed] [Google Scholar]
- Zhang F, Zheng J, Li Z, Cai Z, Wang F, Yang D. 2021. Purification, characterization, and self‐assembly of the polysaccharide from Allium schoenoprasum . Foods, 10:1352. 10.3390/foods10061352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang T, Zhou J, Guo H, Qu G, Guo X, Jia H, Zhu L. 2022. Intrinsic mechanisms underlying the highly efficient removal of bacterial endotoxin and related risks in tailwater by dielectric barrier discharge plasma. Water Res., 226:119214. 10.1016/j.watres.2022.119214 [DOI] [PubMed] [Google Scholar]
- Zhang H, Shi X, Liu Y, Wang B, Xu M, Welham NV, Li L. 2022. On‑tissue amidation of sialic acid with aniline for sensitive imaging of sialylated N‑glycans from FFPE tissue sections via MALDI mass spectrometry. Anal. Bioanal. Chem. 414:5263–5274. 10.1007/s00216-022-03894-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang X, Meng Y, Yang X, Zhao Q, Gao J. 2022. Total synthesis of the tetrasaccharide haptens of Vibrio vulnificus MO6‐24 and BO62316 and immunological evaluation of their protein conjugates. J. Amer. Chem. Soc. Au, 2:97–108. 10.1021/jacsau.1c00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chen H, Luo L, Zhou Z, Wang Y, Gao T, Yang L, Peng T, Wu M. 2021. Structures of fructan and galactan from Polygonatum cyrtonema and their utilization by probiotic bacteria. Carbohydr. Polym. 267:118219. 10.1016/j.carbpol.2021.118219 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao K, Zhang K, Wang W, Xing J, Li Y. 2022. Influence of glucan on physicochemical and rheology properties of chitin nanofibers prepared from Shiitake stipes. Carbohydr. Polym. 294:119762. 10.1016/j.carbpol.2022.119762 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Sun T, Tuo X, Li Y, Yang H, Deng J. 2021. A novel reversibly glycosylated polypeptide‐2 of bee pollen from rape (Brassica napus L.): Purification and characterization. Protein Peptide Lett., 28:543–553. 10.2174/0929866527666201103161302 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang S, Li Z, Lasanajak Y, Li L, Song X. 2022. Regeneration of free reducing glycans from reductive amination‐tagged glycans by oxone. J. Org. Chem. 87:3736–3740. 10.1021/acs.joc.1c02709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Yang Q, Boruah BM, Zong G, Li C, Chapla D, Yang J‐Y, Moremen KW, Wang L‐X. 2021. Appropriate aglycone modification significantly expands the glycan substrate acceptability of α1,6‐fucosyltransferase (FUT8). Biochem. J. 478:1571–1583. 10.1042/bcj20210138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Zhu J, Lubman DM, Mechref Y, Tang H. 2021. GlycoHybridSeq: Automated identification of N‑linked glycopeptides using electron transfer/high‐energy collision dissociation (EThcD). J. Proteome Res. 20:3345–3352. 10.1021/acs.jproteome.1c00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Peng W, Huang Y, Gautam S, Wang J, Mechref Y, Tang H. 2022. A reciprocal best‐hit approach to characterize isomeric N‑glycans using tandem mass spectrometry. Anal. Chem. 94:10003–10010. 10.1021/acs.analchem.2c00229 [DOI] [PubMed] [Google Scholar]
- Zhang X‐X, Li J‐Q, Li M‐S, Chen Y, He H‐B, Liu S‐P, Cheng F, Liu C‐X, Zou K. 2021. Isolation, structure identification and hepatoprotective activity of a polysaccharide from Sabia parviflora . Bioorg. Med. Chem. Lett., 32:127719. 10.1016/j.bmcl.2020.127719 [DOI] [PubMed] [Google Scholar]
- Zhang X, Bi C, Shi H, Li X. 2021. Structural studies of a mannoglucan from Cremastra appendiculata (Orchidaceae) by chemical and enzymatic methods. Carbohydr. Polym. 272:118524. 10.1016/j.carbpol.2021.118524 [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen K, Long L, Ding S. 2021. Two C1‑oxidizing AA9 lytic polysaccharide monooxygenases from Sordaria brevicollis differ in thermostability, activity, and synergy with cellulase. Appl. Microbiol. Biotechnol. 105:8739–8759. 10.1007/s00253-021-11677-1 [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang F, Torres‐Luna C, Nishiyama Y, Briber RM, Wang H. 2021. Solvent‐assisted fractionation of oligomeric cellulose and reversible transformation of cellulose II and IV. ACS Biomater. Sci. Eng. 7:4792–4797. 10.1021/acsbiomaterials.1c00885 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ou C, Liu H, Prabhu SK, Li C, Yang Q, Wang L‐X. 2021. General and robust chemoenzymatic method for glycan‐mediated site‐specific labeling and conjugation of antibodies: Facile synthesis of homogeneous antibody−drug conjugates. ACS Chem. Biol. 16:2502–2514. 10.1021/acschembio.1c00597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Vimalraj V, Patel M. 2021. Routine analysis of N‐glycans using liquid chromatography coupled to routine mass detection. Method. Mol. Biol. 2271:205–219. 10.1007/978-1-0716-1241-5_15 [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu H, He J, Ou C, Donahue TC, Muthana MM, Su L, Wang L‐X. 2022. Site‐specific chemoenzymatic conjugation of high‐affinity M6P glycan ligands to antibodies for targeted protein degradation. ACS Chem. Biol. 17:3013–3023. 10.1021/acschembio.1c00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mao Y, Briber RM. 2022. Efficient production of oligomeric chitin with narrow distributions of degree of polymerization using sonication‐assisted phosphoric acid hydrolysis. Carbohydr. Polym. 276:118736. 10.1016/j.carbpol.2021.118736 [DOI] [PubMed] [Google Scholar]
- Zhang X, Ou C, Liu H, Wang L‐X. 2022. Synthesis and evaluation of three azide‐modified disaccharide oxazolines as enzyme substrates for single‐step Fc glycan‐mediated antibody‐drug conjugation. Bioconj. Chem. 33:1179–1191. 10.1021/acs.bioconjchem.2c00142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y‐Y, Ghirardello M, Wang T, Lu A‐M, Liu L, Voglmeir J, Galan MC. 2021. Imidazolium labelling permits the sensitive mass‐spectrometric detection of N‐glycosides directly from serum. Chem. Commun. 57:7003–7006. 10.1039/D1CC02100A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He H, Chen Z, Huang Y, Xiang G, Li P, Yang X, Lu G, Xiao G. 2021. Merging reagent modulation and remote anchimeric assistance for glycosylation: Highly stereoselective synthesis of α‐glycans up to a 30‐mer. Angew. Chem. Int. Ed. 60:12597–12606. 10.1002/anie.202103826 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Reiding KR, Wu J, Li Z, Xu X. 2021. Distinguishing benign and malignant thyroid nodules and identifying lymph node metastasis in papillary thyroid cancer by plasma N‐glycomics. Front. Endocrinol. 12:692910. 10.3389/fendo.2021.692910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang X, Gu J, Wu J, Cao Y, Xu Y, Li L, Guan K, Liu P, Yin J, Zhi Y, Zhang S. 2021. Validation of diagnostic and predictive biomarkers for hereditary angioedema via plasma N‐glycomics. Clin. Transl. Allergy, 11:e12090. 10.1002/clt2.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu J, Liu P, Kang L, Xu X. 2021. Diagnostic potential of plasma IgG N‐glycans in discriminating thyroid cancer from benign thyroid nodules and healthy controls. Front. Oncol. 11:658223. 10.3389/fonc.2021.658223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Cao Z, Liu R, Li Z, Wu J, Liu X, Wu M, Xu X, Liu Z. 2022. Nomograms based on serum N‐glycome for diagnosis of papillary thyroid microcarcinoma and prediction of lymph node metastasis. Curr. Oncol. 29:6018–6034. 10.3390/curroncol29090474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Ma C, Gao M, Li Y, Yang B, Li H, Zhang R, Hao M, Huang J, Liang K, Chen P, Xie L, Rong R, Kong B. 2022. Super‐assembled sandwich‐like Au@MSN@Ag nanomatrices for high‐throughput and efficient detection of small biomolecules. Nano Res., 15:2722–2733. 10.1007/s12274-021-3741-0 [DOI] [Google Scholar]
- Zhao H, Wang J, Zhao H, Liu Y, Li Y, Zhang R. 2021. One‐step synthesis of N, B‐codoped carbon nanofiber as a novel matrix for high‐throughput and efficient laser desorption/ionization mass spectrometry analysis. Microchem. J. 164:105966. 10.1016/j.microc.2021.105966 [DOI] [Google Scholar]
- Zhao H, Li Y, Zhao H, Zhao Z, Wang J, Zhang R. 2022. Yolk‐shell Ni/NiO anchored on N‐doped graphene synthesized as dual‐ion MALDI matrix for detecting and imaging bioactive small molecules. J. Colloid Interf. Sci., 613:285–296. 10.1016/j.jcis.2021.12.105 [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhao H, Wang J, Liu Y, Li Y, Zhang R. 2022. The local electric field effect of onion‐like carbon nanoparticles for improved laser desorption/ionization efficiency of saccharides. Colloid Surface B, 211:112321. 10.1016/j.colsurfb.2022.112321 [DOI] [PubMed] [Google Scholar]
- Zhao W‐H, Zhang Y‐D, Shi Y‐P. 2021. Visualizing the spatial distribution of endogenous molecules in wolfberry fruit at different development stages by matrix‐assisted laser desorption/ionization mass spectrometry imaging. Talanta, 234:122687. 10.1016/j.talanta.2021.122687 [DOI] [PubMed] [Google Scholar]
- Zhao W, Xu G, Chen Y, Yu Z, Li J, Yu H, Liao X. 2021. Glycan characterisation and antioxidant activity of a novel N‐linked glycoprotein from okra. Int. Food Res. J. 28:1119–1130. 10.47836/ifrj.28.6.03 [DOI] [Google Scholar]
- Zhao X, Wang H, Liu Y, Ou R, Liu Y, Li X, Pan Y. 2021. Lignin as a MALDI matrix for small molecules: A proof of concept. Analyst, 146:7573–7582. 10.1039/D1AN01632F [DOI] [PubMed] [Google Scholar]
- Zhao X, Zhang P, Li Y, Wu S, Li F, Wang Y, Liang S, He X, Zeng Y, Liu Z. 2021. Glucose‐lipopeptide conjugates reveal the role of glucose modification position in complexation and the potential of malignant melanoma therapy. J. Med. Chem., 64:11483–11495. 10.1021/acs.jmedchem.1c00805 [DOI] [PubMed] [Google Scholar]
- Zhao Y‐Z, Xu Y, Gong C, Ju Y‐R, Liu Z‐X, Xu X. 2021. Analysis of small molecule compounds by matrix‐assisted laser desorption ionization mass spectrometry with Fe3O4 nanoparticles as matrix. Chin. J. Anal. Chem. 49:103–112. 10.1016/S1872-2040(20)60074-3 [DOI] [Google Scholar]
- Zhao Y, Li Y, Ma H, Dong W, Zhou H. 2014. Modification of sialylation mediates the invasive properties and chemosensitivity of human hepatocellular carcinoma. Mol. Cell. Proteomics, 13:520–536. 10.1074/mcp.W119.001568 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhen Y, Chen L, Ma X, Ding G, Zhang D, Chen Q. 2021. β‑Amyloid peptide 1−42‐conjugated magnetic nanoparticles for the isolation and purification of glycoproteins in egg white. ACS Appl. Mater. Interfaces, 13:14028–14036. 10.1021/acsami.1c02356 [DOI] [PubMed] [Google Scholar]
- Zheng X, Zhang F, Zhao Y, Xiong Y, Zhang X, Shi Z, Qian S, Qin H, Qing G. 2022. Enrichment of IgG and HRP glycoprotein by dipeptide‐based polymeric material. Talanta, 241:123223. 10.1016/j.talanta.2022.123223 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Su J, Miller MC, Geng J, Xu X, Zhang T, Mayzel M, Zhou Y, Mayo KH, Tai G. 2021. Topsy‐turvy binding of negatively charged homogalacturonan oligosaccharides to galectin‐3. Glycobiology, 31:341–350. 10.1093/glycob/cwaa080 [DOI] [PubMed] [Google Scholar]
- Zheng Y, Pu C, Zhao H, Gu Q, Zhu T, Lan M. 2022. Hydrophilic arginine‐functionalized mesoporous polydopamine‐graphene oxide composites for glycopeptides analysis. J. Chromatogr., B, 1189:123049. 10.1016/j.jchromb.2021.123049 [DOI] [PubMed] [Google Scholar]
- Zhi Y, Jia L, Shen J, Li J, Chen Z, Zhu B, Hao Z, Xu Y, Sun S. 2022. Formylation: An undesirable modification on glycopeptides and glycans during storage in formic acid solution. Anal. Bioanal. Chem. 414:3311–3317. 10.1007/s00216-022-03989-6 [DOI] [PubMed] [Google Scholar]
- Zhong C, Zajki‐Zechmeister K, Nidetzky B. 2021. Reducing end thiol‐modified nanocellulose: Bottom‐up enzymatic synthesis and use for templated assembly of silver nanoparticles into biocidal composite material. Carbohydr. Polym. 260:117772. 10.1016/j.carbpol.2021.117772 [DOI] [PubMed] [Google Scholar]
- Zhong C, Nidetzky B. 2022. Precision synthesis of reducing‐end thiol‐modified cellulose enabled by enzyme selection. Polymer J. 54:551–560. 10.1038/s41428-021-00599-4 [DOI] [Google Scholar]
- Zhong J, Huang Y, Mechref Y. 2021. Derivatization of sialylated glycopeptides (DOSG) enabling site‐specific isomeric profiling using LC‐MS/MS. Anal. Chem. 93:5763–5772. 10.1021/acs.analchem.0c05149 [DOI] [PubMed] [Google Scholar]
- Zhong J, Huang Y, Jiang P, Mechref Y. 2022. Derivatization of sialylated glycopeptides plus based sialoglycopeptides enrichment using cation exchange media. Anal. Chim. Acta, 1233:340492. 10.1016/j.aca.2022.340492 [DOI] [PubMed] [Google Scholar]
- Zhong R, Cui D, Phillips DR, Sims NT, Ye ZH. 2021. Functional analysis of GT61 glycosyltransferases from grass species in xylan substitutions. Planta Med., 254:131. 10.1007/s00425-021-03794-y [DOI] [PubMed] [Google Scholar]
- Zhong R, Phillips DR, Ye Z‐H. 2021. A single xyloglucan xylosyltransferase is sufficient for generation of the XXXG xylosylation pattern of xyloglucan. Plant Cell Physiol., 62:1589–1602. 10.1093/pcp/pcab113 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Cui D, Phillips DR, Adams ER, Jeong H‐Y, Jung K‐H, Ye Z‐H. 2022. Identification of xylan arabinosyl 2‐O‐xylosyltransferases catalyzing the addition of 2‐O‐xylosyl residue onto arabinosyl side chains of xylan in grass species. Plant J., 112:193–206. 10.1111/tpj.15939 [DOI] [PubMed] [Google Scholar]
- Zhong R, Phillips DR, Ye ZH. 2022. Independent recruitment of glycosyltransferase family 61 members for xylan substitutions in conifers. Planta, 256:70. 10.1007/s00425-022-03989-x [DOI] [PubMed] [Google Scholar]
- Zhou L, Liu Q, Ma Q, Guan M, Ouyang X, Qiu X. 2022. Production of water‐soluble sugar from cellulose and corn stover via molten salt hydrate impregnation and separation. Cellulose, 29:879–891. 10.1007/s10570-021-04345-0 [DOI] [Google Scholar]
- Zhou Q, Fülöp A, Hopf C. 2021. Recent developments of novel matrices and on‐tissue chemical derivatization reagents for MALDI‐MSI. Anal. Bioanal. Chem. 413:2599–2617. 10.1007/s00216-020-03023-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Song W, Novotny MV, Jacobson SC. 2022. Fractionation and characterization of sialyl linkage isomers of serum N‐glycans by CE‐MS. J. Sep. Sci. 45:3348–3361. 10.1002/jssc.202200223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Neelamegham S. 2022. Comparative glycomics analysis of mass spectrometry data. Method. Mol. Biol. 2370:97–113. 10.1007/978-1-0716-1685-7_5 [DOI] [PubMed] [Google Scholar]
- Zhu B, Chen Z, Shen J, Xu Y, Lan R, Sun S. 2022. Structural‐ and site‐specific N‐glycosylation characterization of COVID‐19 virus spike with StrucGP. Anal. Chem. 94:12274–12279. 10.1021/acs.analchem.2c02265 [DOI] [PubMed] [Google Scholar]
- Zhu C, Wu J, Jin X, Yan Y, Ding C‐F, Tang K, Zhang Q. 2021. Graphene functionalized with structurally complementary amino acids for sensitive recognition of N‐linked glycopeptides. J. Chromatogr., A, 1655:462505. 10.1016/j.chroma.2021.462505 [DOI] [PubMed] [Google Scholar]
- Zhu D, Alcazar‐Magana A, Qian YP, Tao Y, Qian MC. 2022. Isolation, characterization, and compositional analysis of polysaccharides from Pinot Noir wines: An exploratory study. Molecules, 27:8330. 10.3390/molecules27238330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Liu K, Zheng S, Zhang X, Yan J, Li W, Zhang A. 2021. Upper critical solution temperature‐type responsive cyclodextrins with characteristic inclusion abilities. Chem. Eur. J. 27:10470–10476. 10.1002/chem.202101283 [DOI] [PubMed] [Google Scholar]
- Zhu L, Shuai X‐Y, Lin Z‐J, Sun Y‐J, Zhou Z‐C, Meng L‐X, Zhu Y‐G, Chen H. 2022. Landscape of genes in hospital wastewater breaking through the defense line of last‐resort antibiotics. Water Res., 209:117907. 10.1016/j.watres.2021.117907 [DOI] [PubMed] [Google Scholar]
- Zhu T, Gu Q, Liu Q, Zou X, Zhao H, Zhang Y, Pu C, Lan M. 2022. Nanostructure stable hydrophilic hierarchical porous metal‐organic frameworks for highly efficient enrichment of glycopeptides. Talanta, 240:123193. 10.1016/j.talanta.2021.123193 [DOI] [PubMed] [Google Scholar]
- Zhu X, Xu T, Peng C, Wu S. 2022. Advances in MALDI mass spectrometry imaging single cell and tissues. Front. Chem. 9:782432. 10.3389/fchem.2021.782432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Delbianco M, Seeberger PH. 2021. Automated assembly of starch and glycogen polysaccharides. J. Am. Chem. Soc. 143:9758–9768. 10.1021/jacs.1c02188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi Y, Yao M, Lu Z, Lu F, Bie X, Zhang C, Zhao H. 2021. Glycoglycerolipids from the leaves of Perilla frutescens (L.) Britton (Labiatae) and their anti‐inflammatory activities in lipopolysaccharide‐stimulated RAW264.7 cells. Phytochemistry, 184:112679. 10.1016/j.phytochem.2021.112679 [DOI] [PubMed] [Google Scholar]
- Ziburová J, Nemčovič M, Šesták S, Bellová J, Pakanová Z, Siváková B, Šalingová A, Šebová C, Ostrožlíková M, Lekka D‐E, Brucknerová J, Brucknerová I, Skokňová M, Mc Cullough A, Hrčková G, Hlavatá A, Bzdúch V, Mucha J, Baráth P. 2021. A novel homozygous mutation in the human ALG12 gene results in an aberrant profile of oligomannose N‐glycans in patient's serum. Am. J. Med. Genet. A, 185A:3494–3501. 10.1002/ajmg.a.62474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik JJ, Yoon SH, Guan Z, Skidmore GS, Gudoor RR, Davies KM, Deutschbauer AM, Goodlett DR, Klein EA, Ryan KR. 2022. Caulobacter lipid A is conditionally dispensable in the absence of fur and in the presence of anionic sphingolipids. Cell Rep., 39:110888. 10.1016/j.celrep.2022.110888 [DOI] [PMC free article] [PubMed] [Google Scholar]


