Abstract
Pseudorabies virus (PRV) is a herpesvirus of swine, a member of the Alphaherpesvirinae subfamily, and the etiological agent of Aujeszky's disease. This review describes the contributions of PRV research to herpesvirus biology, neurobiology, and viral pathogenesis by focusing on (i) the molecular biology of PRV, (ii) model systems to study PRV pathogenesis and neurovirulence, (iii) PRV transsynaptic tracing of neuronal circuits, and (iv) veterinary aspects of pseudorabies disease. The structure of the enveloped infectious particle, the content of the viral DNA genome, and a step-by-step overview of the viral replication cycle are presented. PRV infection is initiated by binding to cellular receptors to allow penetration into the cell. After reaching the nucleus, the viral genome directs a regulated gene expression cascade that culminates with viral DNA replication and production of new virion constituents. Finally, progeny virions self-assemble and exit the host cells. Animal models and neuronal culture systems developed for the study of PRV pathogenesis and neurovirulence are discussed. PRV serves as a self-perpetuating transsynaptic tracer of neuronal circuitry, and we detail the original studies of PRV circuitry mapping, the biology underlying this application, and the development of the next generation of tracer viruses. The basic veterinary aspects of pseudorabies management and disease in swine are discussed. PRV infection progresses from acute infection of the respiratory epithelium to latent infection in the peripheral nervous system. Sporadic reactivation from latency can transmit PRV to new hosts. The successful management of PRV disease has relied on vaccination, prevention, and testing.
INTRODUCTION
Pseudorabies virus (PRV) is a pathogen of swine resulting in devastating disease and economic losses worldwide. PRV has been of interest to virologists and neurobiologists as well as those concerned about disease control in swine agriculture. This herpesvirus has served as a useful model organism for the study of herpesvirus biology. The virus has also been used as a “live” tracer of neuronal pathways, making use of its remarkable propensity to infect synaptically connected neurons. Finally, while efforts to eradicate PRV in the United States and Europe have shown great progress, it is still an endemic problem in many countries.
This review focuses on recent reports regarding the molecular biology of PRV, the use of laboratory animal models to study viral pathogenesis, the use of PRV as a neuronal tracer, and the agricultural impact of PRV. The broad coverage of this review targets not only virologists, but also those interested in neurobiology and veterinary medicine.
Virus Nomenclature
Pseudorabies virus is also known by its taxonomic name, suid herpesvirus 1, or by its original name, Aujeszky's disease virus. PRV is a swine herpesvirus of the Alphaherpesvirinae subfamily.
Herpesviridae and Alphaherpesvirinae
All herpesviruses have a double-stranded DNA genome, similar virion size (200 to 250 nm) and structure (capsid, tegument, and envelope), and undergo a latent phase in their life cycle (348). According to the International Committee on Taxonomy of Viruses (http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fr-fst-a.htm#H), most herpesviruses can be subdivided into three major subfamilies of herpesviruses based on their biological properties and genome content and organization: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. The recently discovered ictalurid herpesvirus 1, a channel catfish virus, represents the sole and founding member of a fourth subfamily (99). Additionally, there are dozens of herpesviruses awaiting classification.
The three major subfamilies differ in the cell type where latency is established and the length of their productive replication cycle (348). Alphaherpesviruses have the broadest host range, tend to replicate rapidly with cytopathic effects to produce viral particles in a matter of hours, and can establish latency in the sensory ganglia. Betaherpesviruses have the most restricted host range and the slowest rate of replication that is often accompanied by cell enlargement (cytomegalia), and establish latency in a number of tissues and cells, including secretory glands, kidneys, and lymphoreticular cells. Gammaherpesviruses infect lymphoblastoid cells and are usually specific for either T or B lymphocytes, establishing latency in lymphoid tissue.
Humans harbor three alphaherpesviruses: varicella-zoster virus (VZV) and herpes simplex virus type 1 (HSV-1) and type 2 (HSV-2). The monkey B virus can also be transmitted to humans with lethal consequences. Despite its significant homology to human alphaherpesviruses and its broad host range, PRV is not transmitted to humans. Anecdotal reports of rare human PRV infections have not been substantiated, and likely reflect the low cross-reactivity of antibodies against HSV-1 gB to PRV gB (344, 437). Owing to the significant homology between members of the Alphaherpesvirinae, information derived from the study of PRV provides a powerful opportunity for comparative molecular virology (118). Accordingly, research interests in PRV reflect more than the agricultural impact of the disease it causes. Other well-studied alphaherpesviruses are animal pathogens important to agriculture, including bovine herpesviruses (BHV-1 and BHV-5), equine herpesviruses (EHV-1 and EHV-4), ovine herpesvirus, and avian herpesviruses such as Marek's disease virus and infectious laryngotracheitis virus (ILTV) (348).
Based on molecular criteria and sequence analysis, the Alphaherpesvirinae subfamily can be subdivided further into the genera Simplexvirus (HSV-1), Varicellovirus (VZV), Marek's disease-like virus, and ILTV-like virus (265, 346, 348). PRV, and its closely related homologs BHV-1, EHV-1 and EHV-4, feline herpesvirus type 1, and canine herpesvirus type 1, are all members of the Varicellovirus genus.
MOLECULAR BIOLOGY OF PRV
A number of previous excellent reviews examine particular aspects of PRV molecular biology, with more recent reviews comprehensively describing specific features of the PRV replication cycle such as viral entry, virion morphogenesis, and viral egress (29, 121, 274, 276, 277).
Virion Structure
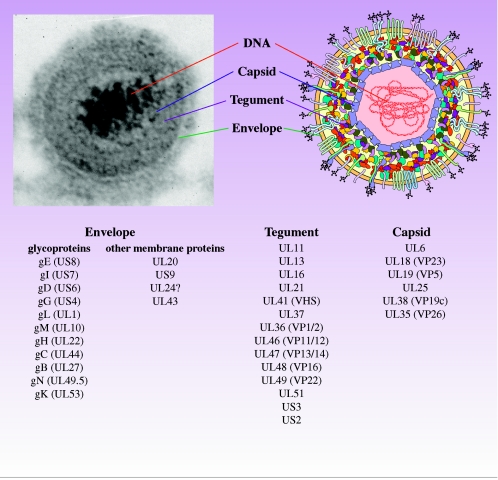
Membership in the Herpesviridae family was based historically on their unique virion architecture (348) and PRV virions resemble others in the family (29, 153, 274). The mature virion, or infectious viral particle, consists of four morphologically distinct structural components (Fig. 1): the central core contains the linear double-stranded DNA genome of the virus; the DNA is enclosed within a protective icosahedral capsid to form a nucleocapsid; the capsid is embedded in a protein matrix known as the tegument; finally, the tegument is surrounded by the envelope, a lipid membrane containing several viral glycoproteins. Nearly half of all the PRV gene products are structural components of the mature virion (Fig. 1; Table 1).
FIG. 1.
Structure of the PRV virion. PRV virions are composed of four structural elements. The double-stranded DNA genome is housed in an icosahedral capsid. The tegument is a collection of approximately 12 proteins organized into at least two layers, one which interacts with envelope proteins and one that is closely associated with the capsid. The envelope is a lipid bilayer infused with transmembrane proteins, many of which are modified by glycosylation. Listed are proteins thought to be components of the virion; however, not all proteins are represented in the cartoon.
TABLE 1.
PRV gene functions
| Gene | Size (kDa) | Common name | Proposed function(s)a | Structural role | Core |
|---|---|---|---|---|---|
| ORF1.2 | 35.3 | Unknown | Virion | No | |
| ORF1 | 21.8 | Unknown | Virion | No | |
| UL54 | 40.4 | ICP27 | Transcription modulation; cell-cell spread; RNA-binding protein | Nonstructural | Yes |
| UL53 | 33.8 | gK | Viral egress (secondary envelopment); glycoprotein K; type III membrane protein; gK/UL20 together inhibit glycoprotein-mediated membrane fusion | Virion (envelope) | No |
| UL52 | 103.3 | DNA replication; primase subunit of UL5/UL8/UL52 complex | Nonstructural | Yes | |
| UL51 | 25 | Viral egress (secondary envelopment); tegument protein, potentially palmytoilated | Virion (tegument) | Yes | |
| UL50 | 28.6 | dUTPase | dUTPase | Nonstructural | Yes |
| UL49.5 | 10.1 | gN | Immune evasion (TAP inhibitor); glycoprotein N; type I membrane protein; complexed with gM | Virion (envelope) | Yes |
| UL49 | 25.9 | VP22 | Interacts with C-terminal domains of gE & gM; tegument protein | Virion (tegument) | No |
| UL48 | 45.1 | VP16, α-TIF | Gene regulation (transactivator); viral egress (secondary envelopment); tegument protein | Virion (tegument) | No |
| UL47 | 80.4 | VP13/14 | Viral egress (secondary envelopment); tegument protein | Virion (tegument) | No |
| UL46 | 75.5 | VP11/12 | Unknown; tegument protein | Virion (tegument) | No |
| UL27 | 100.2 | gB | Viral entry (fusion); cell-cell spread; glycoprotein B; type I membrane protein | Virion (envelope) | Yes |
| UL28 | 78.9 | ICP18.5 | DNA cleavage and packaging; component of the UL15/UL28 terminase | Capsid precursor | Yes |
| UL29 | 125.3 | ICP8 | DNA replication and recombination; binds single stranded DNA | Nonstructural | Yes |
| UL30 | 115.3 | DNA replication; DNA polymerase subunit of UL30/UL42 holoenzyme | Nonstructural | Yes | |
| UL31 | 30.4 | Viral egress (nuclear egress); present only in primary enveloped virion; interacts with UL34 | Primary virion (tegument) | Yes | |
| UL32 | 51.6 | DNA packaging; efficient localization of capsids to replication compartments | Capsid precursor | Yes | |
| UL33 | 12.7 | DNA cleavage and packaging; associates with UL28 and UL15 | Nonstructural | Yes | |
| UL34 | 28.1 | Viral egress (nuclear egress); present only in primary enveloped virion; tail-anchored type II nuclear membrane protein; interacts with UL31 | Primary virion (envelope) | Yes | |
| UL35 | 11.5 | VP26 | Surface capsid protein | Virion (capsid) | Yes |
| UL36 | 324.4 | VP1/2 | Viral egress (capsid tegumentation); large tegument protein; interacts with UL37 and capsid | Virion (tegument) | Yes |
| UL37 | 98.2 | Viral egress (capsid tegumentation); interacts with UL36 | Virion (tegument) | Yes | |
| UL38 | 40 | VP19c | Minor capsid protein; UL38/UL18/UL18 triplex component | Virion (capsid) | Yes |
| UL39 | 91.1 | RR1 | Nucleotide synthesis; large subunit of ribonucleotide reductase | Nonstructural | Yes |
| UL40 | 34.4 | RR2 | Nucleotide synthesis; small subunit of ribonucleotide reductase | Nonstructural | No |
| UL41 | 40.1 | VHS | Gene regulation, RNAse, degrades host and viral mRNAs | Virion (tegument) | No |
| UL42 | 40.3 | DNA replication; polymerase accessory subunit of UL30/UL42 holoenzyme | Nonstructural | Yes | |
| UL43 | 38.1 | Inhibits glycoprotein-mediated membrane fusion; type III membrane protein | Virion (envelope) | No | |
| UL44 | 51.2 | gC | Viral entry (virion attachment); glycoprotein C; type I membrane protein; binds to heparan sulfate | Virion (envelope) | No |
| UL26.5 | 28.2 | pre-VP22a | Major scaffold protein; substrate for UL26 protease; capsid formation and maturation | Capsid precursor | Yes |
| UL26 | 54.6 | VP24 | Minor scaffold protein; capsid maturation protease | Capsid precursor | Yes |
| UL25 | 57.4 | Capsid-associated protein; required for capsid assembly | Virion (capsid) | Yes | |
| UL24 | 19.1 | Unknown; type III membrane protein | ? | Yes | |
| UL23 | 35 | TK | Nucleotide synthesis; thymidine kinase; selectively activates acyclovir | Nonstructural | No |
| UL22 | 71.9 | gH | Viral entry (fusion); cell-cell spread; glycoprotein H; type I membrane protein; complexed with gL | Virion (envelope) | Yes |
| UL21 | 55.2 | Unknown, capsid-associated tegument protein; interacts with UL16 | Virion (tegument) | Yes | |
| UL20 | 16.7 | Viral egress; type III membrane protein; required for gK processing; gK/UL20 together inhibit glycoprotein-mediated membrane fusion | ? | No | |
| UL19 | 146 | VP5 | Major capsid protein; forms hexons and pentons | Virion (capsid) | Yes |
| UL18 | 31.6 | VP23 | Minor capsid protein; UL38/UL18/UL18 triplex component | Virion (capsid) | Yes |
| UL17 | 64.2 | DNA cleavage and encapsidation | Virion (inner capsid) | Yes | |
| UL16 | 34.8 | Unknown; tegument protein; interacts with UL21 | Virion (tegument) | Yes | |
| UL15 | 79.1 | DNA cleavage/encapsidation; terminase subunit of the UL15/UL28 terminase; interacts with UL33, UL28 & UL6 | Capsid precursor | Yes | |
| UL14 | 17.9 | Unknown | ? | Yes | |
| UL13 | 41.1 | VP18.8 | Unknown; protein-serine/threonine kinase | Virion (tegument) | Yes |
| UL12 | 51.3 | AN | DNA recombination; alkaline exonuclease | ? | Yes |
| UL11 | 7 | Viral egress (secondary envelopment); membrane-associated tegument protein | Virion (tegument) | Yes | |
| UL10 | 41.5 | gM | Inhibits glycoprotein-mediated membrane fusion; glycoprotein M; type III membrane protein; C terminus interacts with tegument protein UL49; complexed with gN | Virion (envelope) | Yes |
| UL9 | 90.5 | OBP | Sequence-specific ori-binding protein, ATP-dependent helicase motif | Nonstructural | No |
| UL8.5 | 51 | OPBC | C-terminal domain of UL9 | ? | No |
| UL8 | 71.2 | DNA replication; part of UL5/UL8/UL52 helicase/primase complex | Nonstructural | Yes | |
| UL7 | 29 | Unknown | ? | Yes | |
| UL6 | 70.3 | Capsid protein; portal protein; docking site for terminase | Virion (capsid) | Yes | |
| UL5 | 92.1 | DNA replication; part of UL5/UL8/UL52 helicase/primase complex; helicase motif | Non-structural | Yes | |
| UL4 | 15.8 | Unknown | ? | No | |
| UL3.5 | 24 | Viral egress (secondary envelopment); membrane-associated protein | ? | No | |
| UL3 | 25.6 | Unknown | Nonstructural | No | |
| UL2 | 33 | UNG | DNA repair; Uracil-DNA glycosylase | Nonstructural | Yes |
| UL1 | 16.5 | gL | Viral entry (fusion); cell-cell spread; glycoprotein L; membrane anchored via complex with gH | Virion (envelope) | Yes |
| EP0 | 43.8 | ICP0 | Gene regulation (transactivator); early protein; ND10 structure modulation; contains RING finger motif | Virion | No |
| IE180 | 148.6 | ICP4 | Gene regulation (transactivator); immediate-early protein | Nonstructural | No |
| US1 | 39.6 | RSp40/ICP22 | Unknown; HSV-1 homolog (ICP22) acts as regulator of gene expression | ? | No |
| US3 (minor) | 42.9 | PK | Minor form of protein kinase (53-kDa mobility); inhibits apoptosis; mitochondrial targeting motif | ? | No |
| US3 (major) | 36.9 | PK | Viral egress (nuclear egress); inhibits apoptosis; major form of protein kinase (41-kDa mobility); found in both primary and secondary enveloped virions | Virion (tegument) | No |
| US4 | 53.7 | gG | Unknown; glycoprotein G (secreted) | Secreted | No |
| US6 | 44.3 | gD | Viral entry (cellular receptor binding protein); glycoprotein D; type I membrane protein | Virion (envelope) | No |
| US7 | 38.7 | gI | Cell-cell spread; glycoprotein I; type I membrane protein; complexed with gE | Virion (envelope) | No |
| US8 | 62.4 | gE | Cell-cell spread; glycoprotein E; type I membrane protein; complexed with gI; C-terminus interacts with UL49; protein sorting in axons | Virion (envelope) | No |
| US9 | 11.3 | 11K | Protein sorting in axons; type II tail-anchored membrane protein | Virion (envelope) | No |
| US2 | 27.7 | 28K | Tegument protein; membrane associated protein | Virion (tegument) | No |
Gene functions in italics rely primarily on studies of the HSV-1 homolog.
Genome and Gene Content
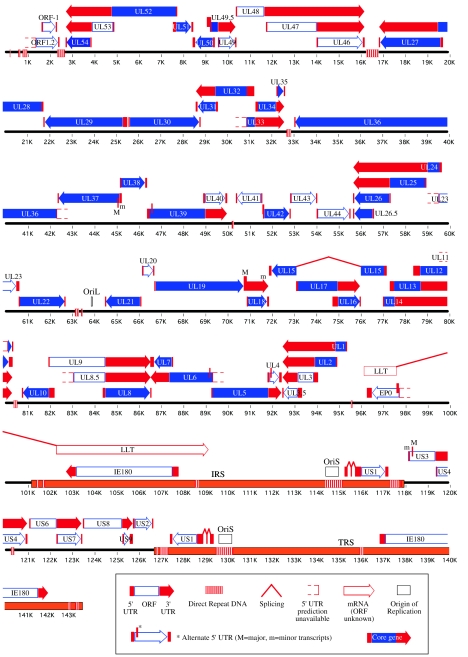
Alphaherpesvirus genomes have a partial colinear arrangement of genes encoding similar functions. Based on the overall arrangement of repeat sequences and unique regions, the herpesvirus genomes can be divided into six classes, designated by the letters A to F (348). The PRV genome, like that of VZV, belongs to the D class, characterized by two unique regions (UL and US), with the US region flanked by the internal and terminal repeat sequences (IRS and TRS, respectively) (Fig. 2) (29). The sequence and gene arrangement of the entire PRV genome are known and a map of the likely transcript organization, well supported by experimental data, has been established (Fig. 2) (219). Recombination between the inverted repeats can produce two possible isomers of the genome, with the US region in opposite orientation. Figure 2 presents only one of the isomers, though both isomers are infectious and found in equimolar amounts after infection (29).
FIG.2.
Linear map of the PRV genome: predicted gene and transcript organization. The PRV genome consists of a long and a short unique segment, named UL and US, respectively. The US region is flanked by the inverted repeats IRS and TRS. The predicted locations of core and accessory genes, transcripts, DNA repeats, splice sites, and the origin of replication are indicated. (Modified from reference 219.)
The functions of the 70 different genes identified in PRV are listed in Table 1. There are two copies of the genes encoding IE180 and US1 because of their location within the IRS and TRS. The major and minor forms of US3 are counted as separate genes because they show functional differences (414, 143). All PRV genes have homologs in one or more related alphaherpesviruses. Generally, gene names were derived from their location order within the UL or US region in accordance with the prototypical HSV-1, while protein nomenclature can vary widely (see Table 1). In this review we refer to gene names in italics and to protein names without. Commonly used protein names, mostly derived from studies of HSV-1 homologs, are also included: for example VP22 (viral protein 22), ICP35 (infected cell protein 35), and gB (glycoprotein B).
The genome of PRV is largely colinear with those of HSV-1 and other alphaherpesviruses, except for a large internal inversion in the UL region situated between UL46 and UL26.5 (32, 46, 110). PRV genes ORF1, ORF1.2, and UL3.5 are not found in HSV-1, while at least 16 HSV-1 genes are not found in PRV (219). Three origins of replication have been mapped in PRV: OriL, located in the UL region (Fig. 2), and two copies of OriS, each located in the inverted repeats (132, 440). The PRV origins of replication are structured as two inverted binding sites for the origin-binding protein UL9 (GTTCGCAC), separated by a 43-bp A-T-rich segment (76% A+T). This basic structure is found once in OriL, while three imperfect repeats of this basic arrangement constitute OriS. Likewise, HSV-1 contains two copies of oriS and one of oriL, consisting of palindromes of 45 bp and 144 bp, respectively, centered around an A-T-rich region of 18 bp (oriS) or 20 bp (oriL). The HSV-1 A-T regions are also flanked by one or two inverted binding sites for UL9 that vary in binding affinity (37, 241, 347).
Transcriptional architecture.
Many features of the PRV transcriptional architecture (Fig. 2) are conserved in the related alphaherpesviruses HSV-1 and BHV-1 (219). Many of the same genes form families of 3′-coterminal transcripts (266, 219, 331). The few genes found to be spliced in alphaherpesviruses are usually immediate-early genes or latency transcripts. PRV contains two known spliced transcripts (US1 and the Large Latency Transcript, LLT) and a putative spliced transcript (UL15), and all three homologs are spliced in HSV-1 (Fig. 2) (347, 219). Many of the core transcription elements are predicted to be shared between genes, with TATA boxes initiating divergent transcripts, or TATA boxes also functioning as polyadenylation signals for genes upstream. These features may be related to the high gene density and limited intergenic sequences found in alphaherpesvirus genomes, such as PRV. PRV also contains multiple short DNA repeat elements, often located between converging transcripts (Fig. 2), and these may serve to prevent transcription from one gene into an oppositely transcribed gene.
Core genes.
A set of 40 herpesvirus genes are conserved among all Alpha-, Beta-, and Gammaherpesvirinae (Fig. 2). These “core genes” encode proteins that perform steps fundamental to the replication of herpesviruses, in part, because of the common structure of nucleocapsids, the basic requirements for viral DNA replication and packaging, and the shared steps of entry into and egress from cells. Phylogenetic analysis of mammalian and avian herpesvirus genomes suggests that an ancestral virus contributed the 40 core genes to modern alpha-, beta-, and gammaherpesviruses (100). While many of the core genes show sequence conservation within the Herpesviridae, some share only the relative genome position and protein function. Table 1 notes the PRV core genes. Like those of other herpesviruses, all PRV core genes are found in the UL region (Fig. 2).
Capsid Proteins
Most of what is known about the PRV capsid is inferred from detailed studies of the prototypical HSV-1 virion. The major capsid protein (MCP or VP5) is encoded by UL19 and assembles into 162 capsomers (150 hexons and 12 pentons) arranged in a T = 16 icosahedral lattice (301). Both the structural arrangement and sequence of UL19 are well conserved in all herpesviruses, and the resulting capsids have a diameter of approximately 125 nm (348). The hexons, composed of six molecules of UL19 (VP5) and six molecules of UL35 (VP26), form the capsid edges and faces, while the 12 pentons comprise the vertices. Both hexons and pentons are connected in groups of three by triplexes, UL38 (VP19C)/UL18 (VP23)/UL18 (VP23) heterotrimers (43, 298, 408). Eleven of the pentons are pentamers of UL19 (VP5), while the twelfth vertex is a unique cylindrical portal made of 12 molecules of UL6 (300). Thus, each mature capsid contains 955 molecules of UL19 (VP5), 900 molecules of UL35 (VP26), 640 molecules of UL18 (VP23), 320 molecules of UL38 (VP19C), and 12 molecules of UL6 (portal protein). The portal's ring-like channel allows one copy of the viral DNA genome to be packaged into the preformed capsid (177).
Tegument Proteins
The tegument layer fills the space between capsids and envelope membranes of mature herpesvirus particles (Fig. 1; Table 1) (348). At least fourteen tegument proteins are of viral origin, but cellular actin is also incorporated into the tegument layer (108, 438). Tegument proteins play important roles during viral entry and virion morphogenesis (reviewed in reference 277). Following fusion of the viral envelope with the plasma membrane, the tegument proteins enter the cell with the capsid and assist with host-cell takeover. These events occur before any viral proteins are synthesized. For example, the UL48-encoded VP16 of PRV and HSV-1 both induce transcription of viral immediate-early genes (24, 57, 133). Another tegument protein, the product of the UL41 gene (PRV vhs) has RNase activity and degrades host mRNA in a manner similar to its HSV-1-encoded homolog (243).
Tegument complexity.
The origin and evolution of the tegument components remain enigmatic, as many of these proteins share almost no sequence homology between alpha-, beta-, and gammaherpesviruses, and most are dispensable for viral growth in cultured cells. The tegument is composed of at least two distinct structures: an inner layer that is tightly associated with capsid proteins and an outer layer that is asymmetrically organized, heterogeneous, and interacts with the cytoplasmic domains of viral membrane proteins. Examination of purified virions reveals variability in the amount of VP22-GFP incorporated into individual particles (107). Similar findings were reported for other tegument proteins (249). Cryoelectron microscopy and tomography revealed that the tegument of HSV-1 virions was pleimorphic, forming an asymmetric cap that occupied two thirds of the volume enclosed within the envelope (159). The innermost layer of tegument shows a more ordered icosahedral morphology, probably imparted by UL36 (VP1/2), an essential tegument protein that may associate with the major capsid protein UL19 (VP5), and UL37, a tegument protein found to interact with UL36 in PRV (136, 213, 249, 272, 452).
Envelope Proteins
Herpesvirus glycoproteins are found in almost all membranes of the infected cell as well as in the virion envelope. These membrane proteins function in virus entry, egress and cell-to-cell spread. They also modulate the immune response and promote syncytia formation. The genome of PRV encodes 16 membrane proteins (Table 1). Eleven membrane proteins are modified by N- or O-linked sugars designated as gB, gC, gD, gE, gG, gH, gI, gK, gL, gM, and gN (274). An additional four transmembrane proteins that are not glycosylated (UL20, UL43, US9, and possibly UL24) are found in the viral envelope. UL34 is a type II membrane protein found in the primary virion envelope but not in purified preparations of infectious virus (138, 217). During entry, the glycoproteins gC, gB, gD, gH, and gL are responsible for virion attachment to the host cell surface and the subsequent fusion of the viral envelope with plasma membrane. As surface constituents of virions and infected cells, glycoproteins represent dominant targets for the host's immune defense (278). A recent structural study of HSV-1 virions reported that the envelope contains 600 to 750 glycoprotein spikes. The spikes, of various length, spacing and angle of membrane emergence, were not distributed randomly, suggesting functional clustering (159).
Glycoprotein nomenclature.
The standard nomenclature of PRV and HSV envelope glycoproteins was adopted at the 18th International Herpesvirus Workshop in 1993. Most reports published before 1995 refer to PRV glycoproteins gB, gC, gD, gE, gG, and gI as gII, gIII, gp50, gI, gX, and gp63, respectively.
Glycoproteins and endocytosis.
Newly synthesized glycoproteins travel from the endoplasmic reticulum via the Golgi to the plasma membrane. However, several PRV envelope glycoproteins are subsequently internalized, either spontaneously or in response to binding of antigen-specific antibodies (125, 126, 304, 401, ). Antibody-mediated internalization of viral proteins from the cell surface may modulate the immune response and protect PRV-infected monocytes from efficient antibody-dependent, complement-mediated lysis (413). The contribution of antibody-independent, glycoprotein endocytosis to a successful viral life cycle is uncertain, but has been proposed to play a role in immune modulation, delivery of viral cell surface proteins to the intracellular compartment where viral envelopment takes place, and redirection of viral proteins to specific membrane surfaces (such as the apical, lateral, or basal surfaces of polarized cells) to facilitate cell to cell spread (reviewed in reference 49). The internalization of many alphaherpesvirus envelope proteins is mediated by tyrosine-based (YXXØ) or dileucine-based (LL) endocytosis motifs located in their cytoplasmic domain (305, 126, 94) (reviewed in reference 49). In addition, acidic clusters containing phosphorylation sites important in endocytosis of cell surface molecules can also occasionally be found. These motifs are known clathrin-mediated endocytosis motifs used by cellular receptors. Indeed, the internalization of PRV gB is mediated by the interaction between its endocytosis motif and the cellular clathrin-associated AP-2 adaptor complex (415).
Viral Replication Cycle
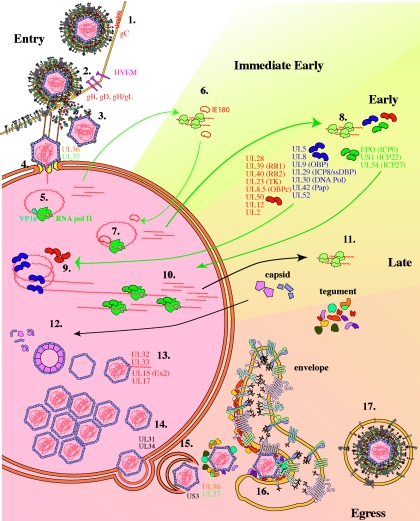
Figure 3 offers an overview of the productive PRV replication cycle.
FIG. 3.
Replication cycle of PRV. 1. Entry begins with attachment or binding of the virus particle to the cell surface. In PRV, this initial binding step is an interaction between gC in the virion envelope and heparan sulfate on the surface of the cell. 2. The next steps of entry require gD, gB, gH, and gL. In PRV, although gD is not essential for membrane fusion or cell-cell spread, gD interacts with the cellular herpesvirus entry mediator (HVEM) and is required for entry of extracellular virus (penetration). 3. After fusion of the virion envelope with the cell membrane, the capsid and tegument proteins are released into the cell. The viral tegument proteins begin takeover of the host cell protein synthesis machinery immediately after entering the cell. 4. The capsid and tightly bound inner tegument proteins are transported along microtubules to the cell nucleus. 5. The VP16 tegument protein localizes to the nucleus independent of the capsid and transactivates cellular RNA polymerase II transcription of the only immediate-early protein of PRV, the HSV ICP4 homolog IE180. 6. IE180 protein expressed in the cytoplasm is transported back to the nucleus. 7. There, it transactivates RNA polymerase II transcription of the early genes. 8. Early proteins fall into two main categories. The first category comprises 15 proteins involved in viral DNA synthesis. 9. Seven of these proteins (UL5, UL8, UL9/OBP, UL29/ssDNABP, UL30/DNA Pol, UL42/Pap, and UL52) (shown in blue) are essential for replication of the viral DNA. DNA replication occurs by a rolling-circle mechanism. 10. The second category comprises three proteins thought to act as transactivators of transcription (EP0, US1, and UL54). 11. Onset of DNA synthesis signals the start of the late stage of the PRV replication cycle and synthesis of true late proteins. 12. The capsid proteins are transported to the nucleus, where they assemble around a scaffold composed of the product of the UL26 and UL26.5 genes. 13. The mature capsid is composed of five proteins (UL19/VP5, UL18/VP23, UL25, UL38, and UL35). The product of the UL6 gene acts as a portal for insertion of the genomic DNAinto the capsid. UL32, UL33, UL15 (Ex2), and UL17 are all involved in cleavage and packaging of the viral DNA. 14. During primary envelopment, the fully assembled nucleocapsid buds out of the nucleus, temporarily entering the perinuclear space. This process involves the products of the UL31 and UL34 genes along with the US3 kinase. 15 and 16. The nucleocapsid (15) loses its primary envelope and (16) gains its final envelope by associating with tegument and envelope proteins and budding into the trans-Golgi apparatus. 17. The mature virus is brought to the cell surface within a sorting compartment/vesicle derived from the envelopment compartment.
Entry
The entry of herpesviruses virions into cells requires a cascade of events mediated by the viral glycoproteins (Fig. 3) (reviewed in reference 379). PRV virions first attach to cells by the interaction of gC with heparan sulfate proteoglycans in the extracellular matrix. PRV gD then binds to specific cellular receptors to stabilize the virion-cell interaction. Finally, PRV gB, gH and gL mediate the fusion of the viral envelope and the cellular plasma membrane to allow penetration of the viral capsid and tegument into the cell cytoplasm. Contrary to the adsorption step, membrane fusion is an energy- and temperature-dependent process (29). Tegument proteins in the outer layer (UL11, UL46, UL47, UL48 and UL49) quickly dissociate from the capsid following fusion of the viral envelope with the cell plasma membrane (154, 249), a process that may be regulated by phosphorylation (290). After fusion of HSV-1 virions, the capsids interact with dynein, a cellular microtubule-associated motor protein, for transport along microtubules from the cell periphery to the nuclear pore (113, 376). Similarly, intracellular PRV capsids frequently associate with microtubule-like structures shortly after infection (154, 156). One study identified PRV nucleocapsid antigens localized to a perinuclear structure thought to be the microtubule organizing center (200). A recent immunoelectron microscopy study in PRV found that the inner layer of tegument proteins (UL36, UL37, and US3) remained associated with the capsid during its transport across the cytoplasm to the nuclear pore (154). UL36 and UL37 also associate with capsids during retrograde axonal transport to the nucleus after virus entry in cultured chick neurons (249). After capsid docking at the nuclear pore, the PRV genomic DNA is released into the nucleus from what appear to be morphologically intact capsids (154, 156).
Herpesvirus entry mediators, cellular receptors for PRV entry.
Five cellular gD receptors, also known collectively as herpesvirus entry mediators, have been identified based on their capacity to allow entry of herpes simplex viruses into cells: HveA (TNFRSF14), HveB (PRR2, nectin 2), HveC (PRR1, nectin 1), HveD (PVR, CD55), and 3-O-sulfated heparan sulfate (reviewed in references 275, 377, and 378). Human HveB, HveC,and HevD, but not HveA or 3-O-sulfated heparan sulfate, could mediate entry of PRV into CHO cells, with HveC being the most effective (306). HveC encodes nectin 1, a protein important for cell adhesion, and the mammalian HveC homologs are found to be highly conserved: the porcine homolog shares 96% amino acid identity with the human homolog (284). Both human and porcine HveC can mediate the entry of HSV-1, HSV-2, PRV, and BHV-1 (88, 146, 284). HveC was also found as the primary receptor for HSV-1 infection in rat and mouse sensory neurons (337).
Regulation of viral glycoprotein-induced membrane fusion.
Transfected cells expressing PRV gB, gH, and gL can induce cell fusion in the absence of gD (221). In contrast, all four proteins are required for HSV1 glycoprotein-induced cell fusion (410). HSV-1 and PRV gD differ in glycosylation (PRV gD lacks the N-glycosylation sites found in HSV-1 and is instead only O-glycosylated [90, 321] and their role in cell-to-cell spread: gD is required for PRV penetration (Fig. 3-2) but not for cell-to-cell spread (316), while gD is required for both processes in HSV-1. The transfection-based assay serves as a model for membrane protein involvement in cell-cell spread. A number of PRV membrane proteins inhibit fusion when cotransfected with fusogenic glycoproteins: gM, UL43, or the combination of gK and UL20, all significantly reduce cell fusion (212, 221).
The fusion inhibition function of gM seems to be conserved among Herpesviridae. Like gM homologs in other herpesviruses, PRV gM forms a disulfide bond with the product of the UL49.5 gene, gN (2, 195, 231, 251, 238, 444). Yet, PRV gM is capable of inhibiting fusion in the absence of gN while the HSV-1, EHV-1, ILTV, BHV-1 and HHV8 homologs inhibit membrane fusion only when coexpressed with their UL49.5 homologs (94, 221, 223, 231). PRV gM inhibits fusion by internalizing viral glycoproteins from the plasma membrane and redirecting them to the Golgi apparatus (94). Likewise, the amount of cell surface viral glycoproteins is reduced when HSV-1 gK and UL20 are expressed together (11). PRV UL20 is required for the maturation of gK, and both proteins are required to inhibit fusion in the transfection based assay (111, 212). Inhibition of membrane fusion between infected and uninfected cells may promote efficient cell-cell spread of virus, while the internalization of fusogenic glycoproteins may direct them to sites of secondary envelopment.
Viral Transcription Activators and the Transcription Cascade
The transcription cascade is the temporally ordered sequence of RNA polymerase II-directed gene transcription that is a molecular hallmark of herpesvirus infection. The viral genes can be subdivided into at least three classes of successively expressed transcripts (reviewed in reference 29, 347). Viral transcription activators expressed during the first hour or two after infection propel the transcription cascade forward by activating transcription of the next set of viral genes. Herpesvirus immediate-early genes are synthesized directly following infection. Their appearance does not require new viral protein synthesis since their promoters are recognized by host transcription factors and RNA polymerase II. The early genes require the viral transactivators encoded by the immediate-early genes, and their transcription is sensitive to protein translation inhibitors such as cycloheximide. True late genes require viral DNA replication for efficient transcription and their expression is severely impaired in the presence of phosphonoacetic acid, an inhibitor of DNA replication. The mechanistic details of the switch from early to late gene transcription in PRV-infected cells remain unknown. Immediate-early gene products usually function either to activate the viral transcription cascade, to modulate host antiviral defense, or to exploit cell physiology required for productive viral infection. The early genes encode viral products required for DNA replication and nucleotide metabolism, while the late genes tend to encode proteins required for virion assembly and egress.
Earlier studies in rabbit kidney cells have described the general appearance kinetics of PRV-encoded transcripts and proteins (29). The length of time required to complete the PRV growth cycle varies according to cell types, and in the most commonly used cell types, viral progeny can be detected within 4 to 5 h. The immediate-early IE180 transcript is synthesized within 40 min of infection and the IE180 protein is synthesized up until 2.5 h postinfection. Early transcripts appear around 1 hour postinfection, with transcript levels peaking around 3 to 4 hours postinfection (early mRNAs) or later (early-late mRNAs). Early proteins are synthesized most abundantly between 1 and 4 hours postinfection, prior to and during the early stages of DNA replication. The products of early-late mRNAs appear around 1.5 hours postinfection but their synthesis peaks at later times (between 4 and 9 hours postinfection). Finally, late mRNAs are detected as early as 2.5 hours postinfection (when viral DNA synthesis starts), and their protein products are detectable around 3 hours postinfection, progressively accumulating to high levels thereafter.
IE180.
PRV encodes only one genuine immediate-early gene, IE180 (ICP4 homolog). In contrast, most herpesvirus genomes express several immediate-early proteins. For example, HSV-1 encodes at least five: RL2 (ICP0), UL54 (ICP27), RS1 (ICP4), US1 (ICP22), and US12 (ICP47). PRV does not encode an ICP47 (US12) homolog, because the PRV genome lacks the DNA corresponding to HSV-1 US10, US11, and US12. Though the PRV genome lacks the IRL and TRL repeats found in HSV-1, an ICP0 homolog called EP0 is located in the PRV UL region. Both EP0 and the ICP27 homolog encoded by UL54 are expressed with early kinetics in PRV (25, 76, 182). PRV US1 mRNA accumulates in the presence of cycloheximide and may be an immediate-early gene but no other analysis has been done (132). The related VZV contains three immediate-early proteins, ORF4 protein (ORF4), IE62 (ORF62) and IE63 (ORF63), homologs of ICP27 (UL54), ICP4 (RS1), and ICP22 (US1), respectively (91). VZV IE62 serves as the major immediate-early transactivator of viral genes during lytic infection, stimulating the transcription of all VZV promoters tested (91).
The IE180 transcript is the predominant PRV transcript detected in cycloheximide-treated rabbit kidney cells, appearing as early as 30 min postinfection (127, 185). As expected, the IE180 promoter drives expression of a reporter gene in the absence of any viral protein synthesis or infection (58, 232). Unlike some of the HSV-1 immediate-early genes (ICP0, ICP22, and ICP47), the IE180 transcript is not spliced in infected bovine kidney cells (79). The IE180 protein is 1,460 amino acids long and contains two ICP4-like domains (amino acids 493 to 669 and amino acids 1052 to 1366, respectively) (78). Though predicted to have a molecular mass of 153 kDa, the product of the IE180 gene migrates as a 180-kDa protein during sodium dodecyl sulfate-polyacrylamide gel electrophoresis, is reportedly phosphorylated (86), and accumulates in the nuclei of infected cells (391, 446). A nuclear localization signal was mapped to the positively charged region (amino acids 930 to 935) RRKRR (391).
Like HSV-1 ICP4, the PRV IE180 gene is present in two copies in the genome, located in the IRS and TRS repeats. The gene is essential for viral replication in tissue culture, as it is required for the efficient transcription of early (and possibly late) viral genes (reviewed in reference 29). When subjected to the nonpermissive temperature, the temperature-sensitive IE180 mutant tsG1 arrests the infection at the immediate-early stage, expressing only IE180 RNA and IE180 protein (185). Recombinant PRV deleted for both copies of IE180 fails to synthesize viral products (445). Since most of the IE180 gene overlaps with the oppositely transcribed large latency transcript (LLT) (76), deletions in IE180 delete a portion of LLT as well. Studies using a recombinant PRV with altered IE180 promoters determined that infection initiation in cultured cells depends on the induction of IE180 (152). Finally, cells expressing a dominant negative form of IE180 that strongly represses the IE180 promoter support PRV replication very poorly, but allow normal replication of HSV-1 (309).
The role of IE180 as a potent transcriptional activator has been well established. Like most typical cellular activators, IE180 contains a separate domain for DNA-binding and another for trans-activation (258, 441). A strong acidic activation domain maps to the N terminus (amino acids 1 to 34) of IE180 (258). In vivo, IE180 has been shown to activate gene expression from the following PRV promoters: US4 (gG), UL12 (AN), UL22 (gH), UL23 (thymidine kinase), and UL41 (vhs) (73, 312, 389). IE180 has a dose-dependent effect on the UL41 promoter, activating gene expression at low levels, but inhibiting it at higher doses (73). It is not known whether this mechanism reflects IE180 negative autoregulation, where expression of IE180 decreases gene expression from its own promoter (420). IE180 can also activate transcription from cellular promoters such as human beta-globin and topoisomerase I (157, 439) and viral promoters such as adenovirus 2 early genes (187, 442), simian virus 40 early genes (157), and human immunodeficiency virus long terminal repeat (449). Partially purified IE180 protein can activate transcription initiation in vitro from human promoters beta-globin and hsp70 (157, 439), and IE180 may aid the formation of a stable transcription preinitiation complex by enhancing TFIID binding (1).
Like its homolog in VZV (ORF62) and HSV-1 (ICP4), the IE180 DNA-binding activity is located in the first ICP4-like domain (441). IE180 has been shown to bind both single-stranded (86) and double-stranded DNA. Partially purified IE180 exhibits specific DNA-binding activity at the promoters of the adenovirus major late gene, human hsp70, PRV US4 (gG), and at the PRV LAP1 promoter (93, 312, 313). In DNA protection assays, IE180 protected sites located near the transcription initiation site as well as sites upstream of the core promoter. The high affinity binding sites (25 to 28 nt long) protected in herpesvirus promoters share only a 5′-ATCGT-3′ sequence (441), while the affinity sites in the adenovirus major late and human hsp70 promoters contained a near-identical 5′-CATCG-3′. The direct IE180 binding site at the PRV US4 promoter maps to a different sequence, the TEF-1 (transcription-enhancing factor 1) element, 5′-TGGAATGTG-3′ (312). However, we note that consensus TEF-1 elements are found only once in the PRV genome. It is clear that IE180, like ICP4, recognizes highly degenerate or nonconsensus DNA sequences.
As the first viral gene to be transcribed during infection, the IE180 promoter can direct expression of a reporter gene in the absence of any viral protein synthesis or infection (58, 232). The upstream region of the promoter contains numerous binding sites for cellular transcription factors: nine imperfect direct repeats (approximately 80 bp each) containing Oct-1 and NF-μE1 binding sites (232, 420). The core promoter itself contains a TATA, as well as Sp1 and CCAAT motifs (420). IFN-α treatment of Vero cells reduces gene expression from the IE180 promoter, and a negative regulation element was mapped to be within 90 bp upstream of the transcription initiation site (407). The negative autoregulation of IE180 transcription is probably direct, since the IE180 DNA-binding domain can bind the IE180 promoter (441). Mapping of the IE180 protein regions critical for negative autoregulation suggests that the DNA-binding domain is required (389). We notice two CATCGT elements flanking the IE180 transcription initiation, and suggest them as likely targets for IE180 binding.
Despite the effect of IE180 on cellular gene expression, stable IE180-expressing lines were established in both Vero and PK15 cells to complement IE180-deleted PRV, though the authors also noted that IE180 expression seemed to enhance or reactivate the production of endogenous retroviruses in PK15 (445). The effect of IE180 on global cellular gene transcription awaits further study, as its effects are unlikely to be confined to the beta-globin and hsp70 genes.
EP0.
As the name implies, the Early Protein 0 (EP0) gene is transcribed with early kinetics (76). The protein can be detected within 2 h after infection, and an additional slower migrating form appears later (310). The PRV EP0 has been detected within virions and shares the characteristics of a promiscuous transactivator (310). Recombinant EP0 can activate transcription initiation from synthetic TATA-based promoters in nuclear extracts (176). Expression of EP0 in vivo activates gene expression from PRV promoters, such as IE180, UL23 (thymidine kinase), and US4 (gG), as well as other viral promoters, such as VZV ORF29, HSV-1 UL23 (thymidine kinase), simian virus 40 early gene, and human immunodeficiency virus type 1 long terminal repeat (288, 424). However, EP0 has the opposite effect on the UL41 (vhs) promoter, reducing gene expression (73). Whether EP0 acts directly or indirectly to modulate transcription is not yet established. EP0 localizes to the nucleus following infection or transfection. Although no typical nuclear localization signal can be found in the EP0 protein sequence, deletion analysis suggests the existence of multiple nuclear localization signals within EP0 (423, 424).
Most of the EP0 gene overlaps with the oppositely transcribed large latency transcript (LLT) (76), so that deletions in EP0 inevitably delete part of the LLT as well. EP0 is dispensable for viral growth in cultured cells, but in its absence, viral titers and plaque size are reduced (18, 38). EP0-negative PRV mutants are attenuated in mice, swine and neonatal piglets (38, 81, 428). The lack of EP0 does not impair PRV in reaching and persisting in the trigeminal ganglia of swine after intranasal inoculation (80), but the amount of viral DNA harbored in trigeminal ganglia tissue is found to be reduced and dexamethasone is not effective in inducing the reactivation of infectious mutant virus.
There are two possible sources of the reactivation defect observed in EP0-negative infected animals: as the only gene known to be transcribed during latency, LLT seems the likely gene to be involved, but alternatively, the impaired reactivation could be due to the reduced replication of the mutant virus, a defect ascribed to the loss of EP0 function. Despite the reduced virulence and apparent defect in reactivation, an EP0 deletion mutant was able to elicit complete protective immunity in very young piglets (428), making the gene a potential target in PRV vaccine engineering.
EP0 is functionally homologous to the immediate-early ICP0 (RL2) gene of HSV-1, and they share little homology beyond a conserved C3HC4 RING finger domain, a zinc-binding motif thought to mediate protein-protein interactions (76). The PRV EP0 transactivation domains have been mapped to the N terminus and to the RING finger domain, and an intact RING domain is required for enhanced expression from PRV TK and IE180 promoters (423).
Expression of PRV EP0 or one of its homologs in related alphaherpesviruses (HSV-1 ICP0, VZV Vg61, BHV-1 BICP0, and EHV-1 Eg63) causes changes to ND10 structures and induces the colocalization of normally diffuse conjugated ubiquitin (314, 315). ND10 structures are repositories of transactivating factors, but their exact function remains uncertain (reviewed in reference 262). The growth defect of a PRV EP0 deletion mutant in cultured cells could be complemented by expression of the HSV-1 or VZV homolog (288). The conserved functions between alphaherpesvirus ICP0 homologs are likely to derive from the presence of the conserved RING finger domain. The RING finger region of HSV-1 ICP0 is essential for its regulation of gene expression, stimulation of lytic infection, enhancement of reactivation from quiescence, disruption of ND10 structures, induction of proteasome-dependent degradation of cellular proteins, and interaction with cyclin D3 (reviewed in reference 162). ICP0 also plays a role in blocking the antiviral effects of interferon in mice. It has recently been proposed that the multitude of functions demonstrated by HSV-1 ICP0 indirectly derive from its ability to serve as a component of the ubiquitin proteasome pathway (162). In this model, ICP0 does not regulate gene expression at the level of transcription, but rather at the level of protein stability (162).
Other regulators of gene expression: UL54, UL41, and UL48.
PRV UL54 and UL41 are likely to encode potent regulators of both viral and cellular gene expression. In HSV-1, UL54 encodes ICP27, a multifunctional RNA-binding protein that stimulates or inhibits transcription in a gene-specific manner, inhibits pre-mRNA splicing, modulates pre-mRNA polyadenylation and stability, and exports viral mRNAs into the cytoplasm (83, 166, 167, 223, 244, 267, 271, 336, 358). While HSV-1 UL54 (ICP27) is expressed as an immediate-early gene, PRV UL54 is expressed with early kinetics (182).
Like HSV-1, the PRV UL54 protein resides in the nucleus of infected cells and avidly binds poly(G) RNA (182). The predicted protein sequence shows a zinc-finger like motif of unknown importance at the C terminus and an N-terminal arginine-glycine rich stretch (amino acids 45 to 54) that resembles the RGG RNA-binding motif found in HSV-1 ICP27 (273). PRV deleted for UL54 shows reduced cell-cell spread, and this defect can be complemented by expression of the homologous proteins of HSV-1 (ICP27) or VZV (ORF4) (364). Furthermore, the absence of UL54 reduces viral replication and alters viral gene expression: gC (UL44) amounts were reduced, gK (UL53) was absent, and the levels of gB (UL27), gE (US8) and US9 were increased. Whether these changes can all be attributed to the loss UL54 is unclear: UL54, UL53, and UL52 are transcribed as 3′coterminal transcripts and a UL54 deletion is expected to alter the 3′ untranslated region of the UL53 and UL52 mRNAs (Fig. 2) (219). Indeed, deletion of UL54 reduces the mRNA levels of UL53 and UL52, encoding gK and a component of the viral replication machinery, respectively (364).
UL41 is conserved within alphaherpesviruses and encodes the vhs protein responsible for the virion host shut-off of cellular protein synthesis. Upon entry, the HSV-1 vhs protein present in the tegument induces the degradation of cellular (and viral) mRNAs (360, 235) by endoribonucleolytic cleavage of target RNAs (114). Subsequently, newly synthesized UL48 (VP16) binds to the vhs protein to inhibit its activity and allow the viral mRNAs to accumulate (239, 370). PRV UL41 lacks the VP16 binding site found in HSV-1, but has partially conserved the putative mRNA binding domain found in HSV-1 (6, 34). The PRV UL41 protein exhibits RNase activity, but is less active than HSV-1 vhs (114, 243, 359). As expected, deletion of PRV UL41 abrogates the degradation of host mRNAs and shows an early delay in viral growth (5). Unlike the early host protein shut-off observed with HSV-1, PRV infection results in a delayed shut-off similar to that seen in VZV, and requires de novo viral protein synthesis (5, 29, 30, 114, 185, 359). Like the VZV vhs homolog (ORF17), PRV UL41 is found in purified virion despite its lack of the VP16-binding site used for HSV-1 UL41 virion incorporation (243, 359). The PRV UL41 promoter can be transactivated by IE180 (73).
The tegument protein VP16 is encoded by UL48, a gene conserved among Alphaherpesvirinae (133). HSV-1 VP16 is known under many names (UL48, α-TIF, Vmw65, or ICP25), and possesses multiple functions during induction of viral gene expression and viral egress (57, 292). The transactivation properties of HSV-1 VP16 have been extensively studied (347). Like its homologs in other alphaherpesviruses (24, 286, 289), PRV VP16 enhances expression of viral immediate-early genes in newly infected host cells (133). Virions lacking the UL48 gene and UL48 protein failed to produce the normally abundant IE180 transcript upon infection, leading to delayed onset of replication, reduced titers, and small plaque size in cultured cells (133). Like many other tegument proteins, PRV UL48 also functions in virion morphogenesis and egress: UL48-negative PRV mutant accumulates unenveloped cytoplasmic capsids (133). In vivo, the UL48-negative PRV mutant exhibits reduced virulence and neuroinvasion after intranasal inoculation of mice (210). The late apparition of clinical signs and extended time to death correlates with, and seems to be explained by, a delayed neuroinvasion of both first-order and second-order neurons.
PRV also modulates the host translation machinery, though little is known about the mechanistic details (reviewed in reference 29). In vitro translation of infected cell mRNAs in rabbit reticulocyte lysates find that a significant proportion of cellular mRNAs fail to be translated and that some early viral mRNAs are translated poorly or not at all. Furthermore, the polysomal mRNA species isolated from infected cells represent only a subset of cytoplasmic mRNA species.
Host Transcript Changes during PRV Infection
DNA microarray technology has enabled investigators to examine the global modulation of cellular and gene transcription following internal and external stimuli, including infection by herpesviruses (54, 194, 291, 388, 400). A recent study compared the viral regulation of host cell gene expression during the productive infection by HSV-1 and PRV (334). Though the two viruses have distinct natural hosts and low DNA sequence homology, they display a high degree of similarity in their viral replication cycles, virion structures, gene organizations, and gene functions (29). Rat embryonic fibroblasts were used as a common permissive cell type for both viruses. While rats are not a natural host for either PRV or HSV-1, both viruses exhibit similar virulence and pathogenic effects in rodents as in their natural hosts, which may reflect common molecular interfaces of host and viral gene products during infection. Surprisingly, only 32% (498 out of 1,549) of cellular transcripts, representing diverse host functions, were similarly affected by viral infection of HSV-1 and PRV. Most of the alterations in cellular transcript levels occurred late in infection and were unlikely to derive from a general stress response, since more than a third of these late changes are virus-specific. Commonly affected genes included oxidative-stress response genes, heat shock genes, and genes involved in the phosphatidylinositol 3-kinase/Akt signaling pathway. Interferon- and interleukin-related genes were altered after HSV-1 but not PRV infection. Further comparison with array data from the transcriptional response of human cells to HSV-1 infection, find only 29 HSV-1-responsive genes shared by rat and human cells, and just 12 of those are similarly affected by PRV.
Gene Expression during Latency
After host survival of an acute infection, the herpesvirus genome resides in the nuclei of host cells for the remainder of the host's lifetime. Reactivation from latency allows spread to naive hosts and maintains the presence of the virus in the population (117). In pigs, neurons in the trigeminal ganglia are the primary site of PRV latency (161). PRV genomes in the trigeminal ganglia are transcriptionally active although only a small region of the genome is transcribed (77). These latency-associated transcripts (LATs) are transcribed from the strand opposite that encoding EP0 and IE180 in a region overlapping the IRS (76, 325, 326). LATs of multiple sizes can be detected in infected swine trigeminal ganglia (76, 77, 326). The largest is the 8.4-kb large latency transcript (LLT). It is possible that some of the smaller PRV LAT transcripts are stable introns spliced from the larger LLT as is the case for HSV-1 (124).
Transcription from the PRV LAT region is active during lytic infection of cultured mammalian (PK15 and MDBK) cells although a different set of transcripts is expressed (191). Two LLT promoters have been identified. The first latency-active promoter (LAP1) has a TATA box located 34 nucleotides upstream from the initiation site of the LLT (76). The LAP2 TATA sequence is 143 bp downstream. The roles of the two latency-active promoters appear to be similar to those described for HSV-1 (75). LAP1 is thought to be a neuron-specific promoter but is not required for LAT transcription in cultured cells (183, 192). LAP2 is active in both neuronal and nonneuronal cells (82, 390). The PRV LAT promoter (LAP1, LAP2, and upstream region) is sufficient to direct transgene expression in the trigeminal ganglia and other neuronal tissues of transgenic mice (392).
PRV IE180 is likely involved in the complicated, cell type-specific regulation of LAT transcription. IE180 binds oligonucleotide sequences corresponding to LAP1 and IE180 downregulates expression of a LAP1-driven reporter gene in mouse neuroblastoma Neuro-2a cells (313). However, IE180 downregulates transcription only in nonneuronal and not Neuro-2a cells when a larger region of the LAT promoter (LAP1, LAP2, and region upstream of LAP1) is used (390). The interaction of the immediate-early protein with the LAT promoter may be an initiating step in PRV reactivation from latency.
In contrast to the wealth of information regarding latent infection cycles of other alphaherpesviruses (193), relatively little is known about PRV gene expression during latency. Ongoing research, including the development of the mouse model of latent PRV infection and reactivation (discussed under Models for Reactivation from Latency) will undoubtedly identify the shared and unique aspects of the PRV latent infection cycle compared to other alphaherpesviruses.
DNA Replication
The structure of PRV DNA during replication is reviewed in reference 29), while the core functions of the herpesvirus DNA replication machinery is summarized in (347). Upon entry into the host nucleus, the linear viral DNA genomes assume a circular form and are quickly repaired of nicks and misincorporated ribonucleotides. Genome circularization most likely occurs by blunt end ligation of the free ends and does not require any viral protein synthesis. The circular genomes serve as the template for DNA synthesis, and the initial theta replication mechanism quickly switches towards a rolling-circle mechanism of DNA replication. The latter process produces replicated DNA in the form of long linear concatemeric genomes that serve as the substrate for genome encapsidation.
Serial passage of alphaherpesviruses at high multiplicities of infection can result in the establishment of a parasitic subpopulation of defective altered viral genomes that can be replicated and packaged into virion-like particles, but only in the presence of helper virus. The virion-like particles containing these genomes are called defective interfering particles (DIPs), as they can compete and interfere with the functional viral genome during DNA replication and encapsidation. Indeed PRV DIP genomes are found enriched for origins of replication and packaging signals (26, 28, 131, 174, 342, 343, 382, 440).
Herpesviruses encode many of the enzymes required for viral DNA replication. Seven HSV-1 proteins are required for origin-dependent synthesis of plasmid DNA: UL52, UL42, UL30, UL29, UL9, UL8, and UL5 (reviewed in reference 241). All seven genes are found conserved in PRV and are presumed to function similarly (Table 1). UL52, UL8, and UL5 are essential core genes encoding the subunits of the heterotrimeric primase-helicase complex, having been well studied in HSV-1, but not in PRV (reviewed in reference 241). UL30 and UL42 are essential genes conserved within all Herpesviridae, and encode the catalytic subunit (Pol) and polymerase-associated protein (Pap) of the viral DNA-dependent DNA polymerase holoenzyme, respectively (348). PRV UL30 possesses a DNA polymerase activity that could be stimulated by the addition of PRV UL42 in vitro, similar to what is seen for HSV-1 (35). The stimulation by UL42 was abrogated in a UL30 mutant missing the C-terminal 30 amino acids. Because of its potential as a target for antiviral drugs, HSV-1 UL30 has been extensively studied; antiherpetic drugs targeting UL30 include phosphonoacetic acid, foscarnet, and acyclovir.
PRV UL29 contains a conserved zinc-binding motif and a conserved DNA-binding region (443) and plays an essential role in viral genome replication (31). The protein is thought to bind the single-stranded DNA in unwound DNA and replication forks. Recombinant UL29 protein binds single-stranded DNA in a nonspecific and cooperative manner (443). Furthermore, the recombinant protein also physically interacts with the UL12 alkaline nuclease to stimulate its DNase activity, suggesting a possible role in viral recombination as well (179). A motif of the helicase type II superfamily is conserved among UL9 homologs, but little else is known about the PRV UL9 protein. HSV-1 UL9 initiates viral DNA replication by binding and unwinding viral origins of replication (oriS and oriL) (347). A separate transcript, encoded by PRV UL8.5 is translated into a protein of unknown function that corresponds to the C-terminal 470 amino acids of the UL9 protein (112). The UL8.5 protein is conserved with 47% identity in HSV-1 but no homolog has been described for other alphaherpesviruses, aside from the larger UL9 gene. HSV-1 UL8.5 has been designated OBPC and is capable of binding the HSV-1 origins of replication in vitro (19).
In addition to viral proteins, host proteins are likely required for PRV DNA replication. In HSV-1, cellular DNA polymerase alpha-primase, DNA ligase I, and topoisomerase II have all been proposed to participate in viral DNA synthesis (reviewed in reference 37). Host recombination proteins have also been suggested to play a role in HSV-1 DNA replication (reviewed in reference 434).
Nucleotide Metabolism
Herpesvirus genomes encode several enzymes involved in nucleotide metabolism. For example, both the HSV-1 and PRV genomes encode a dUTPase (UL50), a thymidine kinase (UL23), and a two-subunit ribonucleotide reductase (UL39/UL40). The PRV and HSV-1 genomes also encode a uracil DNA glycosylase (UL2) as well as an alkaline nuclease (UL12), which serve in viral DNA repair, recombination and DNA concatemer resolution (reviewed in reference 347). Because the corresponding host cell enzymes are virtually absent in nondividing and terminally differentiated cells, these viral gene products enable infection of resting cells (i.e., neurons). Indeed, these genes tend to be dispensable for viral replication in dividing cultured cells, but contribute to virulence in animal models (reviewed in reference 40).
PRV UL50 encodes a bona fide dUTPase that is not incorporated into virions (198). Host and viral dUTPases catalyze the hydrolysis of dUTP into dUMP and pyrophosphate. Reducing the amount of dUTP is predicted to decrease misincorporation of dUTP into viral DNA, while the new product, dUMP, can serve as a precursor for dTMP and dTTP synthesis. PRV UL50 is dispensable for replication in cultured cells, its absence only slightly delaying viral growth kinetics (198). However, the same UL50-negative PRV strain is attenuated when young pigs are inoculated intranasally (196). Prior infection with the UL50-negative strain conferred protective immunity against Aujeszky's disease, making UL50 a good deletion target for safe and potent live vaccines.
UL39 and UL40 encode the small subunit (RR1) and large subunit (RR2) of the viral ribonucleotide reductase, respectively (201). UL39 is conserved within Herpesviridae and contains blocks of highly conserved sequences that are also found within the large subunit of cellular ribonucleotide reductases (201). Ribonucleotide reductase catalyzes the reduction of ribonucleotides into deoxyribonucleotides, the substrates for DNA synthesis. As opposed to most cellular ribonucleotide reductases, the PRV-encoded enzyme is resistant to dTTP product feedback inhibition (reviewed in reference 29). PRV strains mutated for either UL39 or UL40 are able to replicate in cultured cells but are severely attenuated in pigs and mice (103, 104).
UL23 is only found within the Alphaherpesvirinae and Gammaherpesvirinae and encodes the viral thymidine kinase. Cells also contain a thymidine kinase, and the phosphorylation of deoxythymidine is a critical step in the synthesis pathway of dTTP, a substrate for DNA synthesis. Herpesvirus thymidine kinases have broader substrate specificity than their host counterparts, allowing the development of nucleoside analogs, such as acyclovir, that can be phosphorylated into antiviral compounds (347). PRV thymidine kinase possesses thymidine kinase activity in vitro though its substrate spectrum is much more limited than that of HSV-1 thymidine kinase (254; reviewed in reference 29). While PRV UL23 is not essential for viral growth in most cultured cells, UL23-negative PRV mutants prove to be highly attenuated in mice, rabbits and pigs, and confer protective immunity against PRV challenge in pigs (208, 268). A thymidine kinase defect is responsible for the attenuation of the Tatarov vaccine strain (245).
PRV UL12 encodes the alkaline nuclease, an endo-exonuclease with catalytic properties similar to that of the bacterial recombination DNase RecBCD (180, 181). A UL12 PRV insertion mutant shows a strong reduction of virulence in mice (104). The HSV-1 homolog is a nuclease involved in the processing of replication intermediates of viral genomic DNA (324).
PRV UL2 is predicted to encode a uracil-DNA glycosylase, an enzyme conserved within prokaryotes and eukaryotes (105). Uracil DNA glycosylases (UDG or UNG) serve to remove the uracil bases that can occur following DNA damage (347). Removal of the uracil base then allows DNA repair to proceed. PRV UL2 has not been studied yet. HSV-1 UL2 is not essential for viral replication in cultured cells, but plays a role in viral pathogenicity and reactivation from latency (329).
Capsid Formation
The alphaherpesvirus capsid assembly pathway is now fairly well understood thanks to a combination of in vivo and in vitro studies (299) (reviewed in references 177 and 383). Capsids assemble in the nucleus of the cell, and require the mature capsid constituents (UL38, UL35, UL25, UL19, UL18, and UL6), and two scaffolding proteins (UL26 and UL26.5) that participate in capsid formation but are not found in the mature virion. Like HSV-1, three types of capsids are found in PRV-infected cells, called A-, B-, and C-capsids (149, 333). The three differ in density and morphology because of the content inside their icosahedral shell: C-capsids resemble the capsids found in the infectious virion, and contain the viral genome DNA in densely coiled, liquid crystalline arrangement (42); B-capsids are mainly filled with VP22a, the cleaved form of the scaffold protein encoded by UL26.5 (298); and the inner core of A-capsids is devoid of protein and DNA, and is thought to represent an abortive form produced from failed attempts to package DNA (365). Pulse-chase experiments in EHV-1 showed that B-capsids could package DNA to mature into C-capsids and eventually into mature virions, while A-capsids could not (319). All three types of capsids are thought to arise from the maturation of a common precursor, called the procapsid (177).
Cyclooxygenases and capsid assembly
DNA microarray studies of cellular mRNA found cyclooxygenase-2 (COX-2) to be highly upregulated following infection of rat embryonic fibroblasts by PRV (334). Cyclooxygenases 1 and 2 function in the synthesis of prostaglandin, lipid-derived signaling molecules with multiple roles in inflammation and immune modulation (reviewed in reference 172). Further studies confirmed that the COX-2 protein levels were increased upon infection and that either the COX-1 or COX-2 isozyme was required for viral replication and morphogenesis (333). Studies with specific inhibitors of COX-1 or COX-2 show their function in viral replication to be partially redundant. Simultaneous inhibition of both isozymes resulted in a dramatic decrease of viral titers, accompanied by nuclear accumulation of a novel form of defective capsids. Like procapsids, the defective capsid integrity is cold sensitive, though they are distinctly polyhedral and resembled neither filled nor empty capsids.
DNA Encapsidation
PRV DNA encapsidation requires two linked events: cleavage of the replicated concatemeric DNA into monomeric units and packaging of the linear monomeric genomes into capsids (235, 236). DNA cleavage is dependent on capsid assembly and genetic analysis suggests the involvement of at least six PRV genes in DNA encapsidation (29). Herpesvirus genomes contain a highly conserved domain (pac1 and pac2) at each end of their linear genomes to direct the site-specific DNA cleavage and packaging (347). In PRV, the pac2 domain resides at the end of the UL region, while the pac1 domain is found both in the UL-proximal portion of the IRS and near the linear end of the TRS (169). The pac1 domain within the IRS does not function for concatemer cleavage and encapsidation (332).
Studies on the mechanism of DNA packaging in bacteriophages have provided considerable insight into the equivalent processes of the herpesviruses. It is likely that the basic mechanisms are conserved. For example, in most DNA viruses the two terminase subunits associate with the portal to form a powerful molecular motor to package DNA (70, 140). Herpesviruses encode a putative two-subunit terminase made up of UL15 and UL28 (3, 15, 16, 177), which can bind to the portal protein UL6 (432). In PRV, UL28 (ICP18.5) is required for DNA cleavage and encapsidation (282). In the absence of other viral proteins, PRV UL28 is distributed in the cytoplasm instead its normal nuclear location. Coexpression of HSV-1 UL15 and PRV UL28 is sufficient to target UL28 to the nucleus (229).
Other HSV-1 proteins involved in DNA cleavage and packaging include UL17, UL25, UL32, UL33, and the portal protein UL6 (347). PRV UL25 is found to be capsid-associated but has not yet been the subject of any mutagenesis study (200). The functions of the PRV genes UL6, UL32, and UL33 remain yet to be ascertained (229). Like HSV-1, PRV UL17 encodes an essential gene required for DNA cleavage and encapsidation (216, 357). PRV UL17 is a virion component and ultrastructural studies of infected cells strongly suggest that PRV UL17 is a located within the nucleocapsid, possibly associated with the viral DNA (216).
Egress
PRV nucleocapsids cross the nuclear envelope and participate in two separate envelopment events before infectious particles are released from a cell (reviewed in references 121, 275, and 276). The nuclear envelope consists of four major components: an inner nuclear membrane lined by a meshwork of intermediate filaments comprising the nuclear lamina, an outer nuclear membrane, and the two leaflets of the envelope are traversed by multiple nuclear pore complexes. Three proteins (US3, UL31, and UL34) allow the nucleocapsid to escape the egress barrier presented by the nuclear envelope. Once in the cytoplasm, direct binding of tegument proteins to the nucleocapsid, transport of the capsid to the site of secondary envelopment, and addition of more tegument and membrane proteins require the coordinated functions of multiple viral proteins.
Nuclear egress and primary envelopment.
The first step in egress is engagement of nucleocapsids with the inner nuclear membrane (Fig. 3). Primary envelopment occurs by budding of nucleocapsids through the inner nuclear membrane into the perinuclear space. This capsid with an envelope derived from the inner nuclear membrane is called the primary enveloped virion and the gene products of PRV UL31 and PRV UL34 contribute to its formation (138, 217). The UL34 protein is a type II, C-terminal anchored membrane protein, while the UL31 protein is a nuclear phosphoprotein. UL34 and UL31 colocalize in the nuclear envelope of infected cells and interact in a yeast two-hybrid assay (138). Nascent primary enveloped virions exit from the perinuclear space by fusion of the primary envelope with the outer nuclear membrane resulting in deenvelopment of the particles. Deenvelopment of the nascent virion results in entry of “naked” capsid structures into the cytoplasm, and requires the US3 protein kinase (215, 421).
(i) US3 promotes nuclear egress
PRV US3 is a serine/threonine protein kinase (328) found conserved in all alphaherpesviruses. The kinase substrate specificity is similar to HSV-1 US3, with the optimal consensus sequence defined as RRRX(S/T)Z, where X is any amino acid and Z is not an acidic amino acid (240). So far, only the ribosomal protein S6 has been identified as a substrate in vitro (202).
In the absence of US3, a striking accumulation of primary enveloped virions can be observed by electron microscopy within invaginated portions of the inner nuclear membrane of the nuclear envelope (421). These structures extend into the perinuclear space. The US3 protein likely plays a role in fusion of the primary envelope of nascent virions with the outer nuclear membrane of the nuclear envelope, thus enabling deenvelopment of primary virions and release of “naked” virions into the cytoplasmic compartment. However, US3 is dispensable for viral replication in cultured epithelial cells and the absence of US3 only mildly reduces the titer of extracellular particles (215). Thus, deenvelopment can still occur in the absence of US3, albeit less efficiently. It is currently unknown whether the impairment in deenvelopment observed in the absence of US3 protein reflects an important structural role for this protein in primary enveloped virions or whether it is due to effects of its kinase function.
The US3 kinase influences the nuclear membrane association of a UL34, a primary virion component critical for nuclear egress (215, 217). UL34 can still localize to inner and outer leaflets of the nuclear membrane in absence of US3 protein, but does so less efficiently. Metabolic labeling experiments found no difference in UL34 phosphorylation in the absence of US3, indicating that the viral kinase UL13 or a cellular kinase phosphorylates UL34 (215). US3 was also found to induce actin stress fiber disassembly in swine epithelial cells, a step that could be important viral egress (414). The use of kinase-inactive US3 mutants will answer whether these functions require a catalytically active US3 kinase.
Detection of striking perinuclear accumulation of nascent virions in the absence US3 was recapitulated and further characterized in ultrastructural studies (155). In PRV US3-null mutant-infected rabbit kidney cells analyzed by immunoelectron microscopy, PRV US3 protein was detectable in both primary and mature virions. This correlates with previous findings on PRV US3 distribution (215, 250). The absence of US3 protein does not affect virion incorporation of several tegument proteins (UL11, UL37, UL46, UL47, UL48, and UL49) and envelope glycoproteins (gB, gC, gD, gE, gI, gH, gK, or gM) (155), suggesting that PRV US3 protein is either retained during nuclear egress, or reacquired very early during tegumentation in the cytoplasm. Two recent studies have produced conflicting data regarding the presence of US3 in purified virions of the attenuated strain PRV-Bartha (155, 250).
Tegumentation and secondary envelopment
The next process during viral morphogenesis is the addition of tegument and secondary envelopment of viral particles in organelles of the secretory pathway (Fig. 3). While these two processes are closely linked, recent studies indicate that the addition of tegument proteins to the capsid occurs to some extent in an organized and stepwise fashion. The outer layer of the herpesvirus tegument is analogous to the matrix of some RNA viruses. Like matrix proteins, some of the outer tegument layer proteins interact with the inner side of the envelope and also with the inner tegument proteins attached to the capsid, joining these substructures together for the second, and final, envelopment process. The protein-protein interactions within the tegument and between tegument and envelope are likely to drive the secondary envelopment process and may be regulated by phosphorylation (290). Secondary envelopment is most commonly believed to occur in a compartment derived from the trans-Golgi network.
Tegument addition to cytoplasmic capsids is thought to be initiated by the direct interaction of the major capsid protein VP5 (UL19) and the UL36 tegument protein, which in turn can interact with another tegument protein, UL37 (136, 213, 452). This model is consistent with the observation that the inner tegument layer exhibits icosahedral symmetry (452).
Secondary envelopment of herpesviruses occurs on the cytoplasmic face of a specialized compartment within the infected cell derived from the trans-Golgi network (168). The interaction between tegument proteins and envelope could occur either via an interaction with the cytoplasmic domain of envelope proteins (135), or via direct membrane attachment following fatty acid modification of a tegument protein (252, 307). Interactions between the tegument and envelope proteins are likely to drive the secondary envelopment of herpesviruses: PRV VP22 interacts directly with the cytoplasmic domains of gM and gE (135). In HSV-1, VP16 interacts with gH (158), VP22 interacts with gD (84), and the UL13 protein kinase interacts with gE (302). However, there is undoubtedly much redundancy in the interactions between viral proteins required for secondary envelopment. Two of the most abundant tegument components are not absolutely required for secondary envelopment, UL46 (VP11/12) and UL49 (VP22) have each been deleted from the virus with only minor consequences to viral replication (109, 227). On the other hand, individual deletions of tegument proteins UL11, UL37, UL47, UL48, and UL51 have all resulted in accumulation of unenveloped cytoplasmic capsids, a finding that suggests a defect in secondary envelopment (133, 214, 218, 226, 227).
The UL20 membrane protein is a component of virions and plays a role in sorting mature enveloped virions from the site of secondary envelopment to be released from the cell (137). Another PRV protein implicated in the final envelopment of capsids is the product of UL3.5 (139).
L-particles.
Light or L-particles (so called because they are lighter than virions in Ficoll gradients) (387) are noninfectious structures produced during herpesvirus infection (reviewed in reference 384). L-particles contain tegument and envelope proteins but no capsid or genomic DNA. Most alphaherpesviruses, including HSV-1, EHV-1, PRV, and ILTV, produce L-particles during infection of cultured cells (153, 270, 387). L-particle production probably shares common assembly steps with normal tegument assembly and secondary envelope acquisition, and can still occur under conditions where capsids fail to leave the nucleus (341). L-particles fuse with cells, and may assist in the initiation of the infection cycle by delivering tegument and envelope proteins in addition to those brought by the infectious virus (95, 269). Support for their biological significance role comes from the finding that structures resembling L-particles are observed in epithelial cells and fibroblasts in the nasal mucosa of pigs after intranasal inoculation with PRV (4).
Functional redundancy and egress.
Many PRV genes can be deleted with little or no phenotype in cultured cells or in animal models of infection. PRV offers the possibility of testing mutants in the natural host. Despite the complications of working with infected animals, many studies confirm the phenotypes found in cultured cells or model animal infections. It is likely that some genes considered to be nonessential perform redundant functions. Recent studies using multiple deletions to probe for functional redundancy have found new or severe phenotypes not seen by the individual mutants (often called synthetic lethality).
The UL46, UL47, UL48, and UL49 gene cluster encodes outer layer tegument proteins dispensable for viral growth in tissue culture cells. Triple and quadruple deletion mutant were produced and compared to search for phenotypes hidden by functional redundancy (134). The quadruple deletion could still replicate in tissue culture cells, but formed very small plaques, replicated with delayed kinetics, and produced 100-fold fewer viruses. In this case, functional redundancy was difficult to define in cultured cells: UL47 and UL48 function independently during cell-cell spread and virus egress, while the simultaneous loss of UL46 and UL49 does not alter the replication of UL48-negative or UL47-negative PRV mutants.
The functional redundancy of structural components in virion maturation is exemplified by the finding that while gM or gE is dispensable for viral growth in cultured cells, a simultaneous deletion of gM and gE cytoplasmic domain produces a synthetic lethal defect in secondary envelopment (44, 45). Both proteins likely play a role in organizing structural virion components and recruiting them to sites of secondary envelopment. The cytoplasmic tails of both gE and gM interact with VP22 (135). In addition, PRV gM directs viral envelope proteins to the trans-Golgi apparatus in transfected cells (94). Thus, the synthetic lethality of the gM/gE double mutants may be attributed to a combined defect in tegument assembly and glycoprotein sequestration. Similar synthetic lethality results when PRV gM and UL11 or HSV-1 gE and gD are deleted simultaneously (123, 225).
Viral Antiapoptosis Genes
Viral infection of animal cells tends to generate proapoptotic signals that induce cell death and phagocytosis to limit virus replication and spread. Herpesviruses, like most large DNA viruses, encode proteins that interfere with this apoptotic response (reviewed in references 9 and 230). PRV, like HSV-1, is a cytocidal virus that induces rapid and extensive changes in cultured host cells. The cytopathic effects elicited by PRV infection are strain- and cell type-dependent and include inhibition of mitosis, formation of intranuclear inclusions, rounding up of cells, and, occasionally, formation of syncytia (reviewed in reference 29). In most cultured cells, the typical apoptotic effects, such as genomic DNA fragmentation or apoptotic-condensed nuclei, are not observed following infection. Neurons isolated from porcine trigeminal ganglia are more resistant to PRV-induced cell death than neurons isolated from the superior cervical ganglia and other porcine cell types. Trigeminal ganglia neurons survive productive PRV infection longer than other cell types, and this may extend the duration of infectious virus transmission during reactivation from latency (142). Several antiapoptosis genes have been implicated as modulators of apoptosis in HSV-1: UL54 (ICP27), US1 (ICP22), US3 (US3 PK), US5 (gJ), US6 (gD), RL2 (ICP34.5), and RL3 (LAT) (230). US3 is one of the most potent and best characterized antiapoptosis genes in the list.
US3 antiapoptosis activity
The US3 gene encodes a serine/threonine kinase that is conserved in Alphaherpesvirinae (265), and the US3 proteins from HSV-1, HSV-2, and PRV have all been shown to protect cells from apoptosis (143, 173, 190, 242, 308). HSV-1 US3 can counteract apoptosis triggered by either overexpression of pro-apoptotic genes of the Bcl-2 family (BAD, Bax, and Bid), or exogeneous inducers (osmotic shock and UV irradiation) (69, 190, 296, 303, 308). The antiapoptosis function of HSV-1 US3 is proposed to participate in viral immune evasion. US3 blocks the lysis of infected fibroblasts by cytotoxic T cells (68), a block attributed to the US3-dependent inhibition of caspase activation in the target cell. It is generally accepted that the US3 protein kinase prevents apoptosis by direct phosphorylation of target proteins, but the exact details remain unsolved. A recent study highlights the overlap between the substrates of protein kinase A, a cyclic AMP-dependent cellular kinase that can both inhibit and promote apoptosis, and HSV-1 US3, and found US3 expression to result in protein kinase A phosphorylation (33). Whether this signifies an activation of protein kinase A by US3, US3 mimicking protein kinase A antiapoptosis function, or something else is unknown.
PRV infection renders swine testicle cells resistant to apoptosis induced by staurosporine or sorbitol (143). However, cells infected with US3-negative PRV underwent apoptosis as measured by caspase-3 activation and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay for DNA fragmentation (143). US3-negative PRV also failed to prevent apoptosis in swine trigeminal ganglia neurons (142). Apoptosis was observed late in US3-null infected swine testicle cells (18-24 hours postinfection), suggesting that additional viral proteins may serve to inhibit apoptosis early in infection (143). So far, US3 is the only PRV gene found to have antiapoptosis activity (143). US3-negative PRV fails to suppress apoptotic, while US3 transfection experiments in the presence or absence of viral infection show US3 to be responsible for the antiapoptosis activity.
Two isoforms of US3 are detected during PRV infection (Table 1). The short isoform, a protein of 41 kDa, is more abundant (>95% of the US3 mRNA in infected cells) than the long isoform of 53 kDa (<5% of US3 mRNA) (418). The longer isoform possesses a functional mitochondrial localization sequence within the additional N-terminal 51 amino acids (55, 414). The localization of US3 seems to be dependent on the isoform examined, the cell type used, and whether US3 is expressed during viral infection or from a transfected plasmid (56, 414). The long isoform is able to restore antiapoptotic activity to the US3 deletion virus while the short isoform only partially compensates for the deletion of US3 (143). The short isoform of PRV US3 was also found to be as efficient as HSV-1 US3 at rescuing cells from Bax-induced apoptosis (308). Since the HSV-1 US3 kinase activity is required for its antiapoptotic activity (308), it is assumed that the same holds true for PRV, though this has yet to be formally tested. Finally, it is worth noting that US3 from the closely related BHV-1 does not seem to possess any antiapoptotic properties (393).
TAP Inhibition by UL49.5 (gN)
Herpesviruses cause lifelong infection of their hosts, and their persistence and repeated reactivation are facilitated by different immune evasion strategies, including inhibition of complement and antibody function, disruption of immune recognition of infected cells, and viral expression of cytokine-like and chemokine receptor-like molecules (reviewed in reference 97). In veterinary varicelloviruses (PRV, EHV-1, and BHV-1), the UL49.5 gene product is an inhibitor of TAP, the transporter associated with processing antigens into peptides for presentation by major histocompatibility complex (MHC) class I molecules at the cell surface (228). Inhibiting the presentation of virus-derived peptides by MHC class I molecules prevents the recognition and elimination of virus-infected cells by cytotoxic T lymphocytes. PRV UL49.5 encodes the virion envelope glycoprotein gN, a small O-glycosylated protein that forms a disulfide linked complex with gM (discussed above) and functions in viral immune evasion (197, 228). However, neither gM, nor any other viral proteins, was required for the gN-dependent TAP inhibition (228). A disruptive insertion in UL49.5 delayed virus entry and was unable to inhibit TAP (195, 228). A similar disruptive insertion in UL49.5 did not significantly alter the neurovirulence and neuroinvasiveness of the virus in mice after intranasal inoculation (260). UL49.5 is conserved within the Herpesviridae but is not glycosylated in all herpesviruses. In HSV-1, the TAP inhibition function is performed by ICP47 (UL41) (175, 448).
PRV AS A MODEL ORGANISM
PRV has proven to be a facile model system to investigate the many aspects of alphaherpesvirus biology, providing insights both into the basics of the viral life cycle and the more complex interactions with the host (reviewed in reference 118). Areas that benefited particularly from PRV research include the molecular mechanisms of virus attachment, entry, replication, assembly, intracellular trafficking and egress (reviewed in references 118 and 274). In addition, various facets of viral pathogenesis are under study, including the mechanisms of neuroinvasion, transneuronal spread, and host immune responses. The results from these studies have furthered our understanding of these complex phenomena and are bound to yield fascinating new insights.
For a number of reasons, PRV is an excellent choice for the study of alphaherpesvirus biology (reviewed in reference 118). PRV has a remarkably broad host range, causing a lethal infection in diverse animals, yet poses little to no danger to lab workers. PRV grows well and is easy to manipulate in the laboratory. Purified DNA is infectious and the techniques to replace and manipulate its genes are well established. Bacterial artificial chromosomes carrying the entire PRV genome have been constructed (226, 372). Indeed, some of the basic research on herpesvirus BAC technology was done using PRV (373). Animal experiments can be performed in the natural host, the pig, or in other favored model organisms such as rats, mice, and rabbits.
PRV Pathogenesis in Animal Models
The laboratory animal models of PRV pathogenesis have provided considerable insight into fundamental problems of herpes biology. These include the chicken embryo eye model (18), the rat eye model of infection (45, 222), the mouse skin flank model (51), and the mouse nasopharyngeal infection model (14).
Rat Eye Model: Genes Required for Neuroinvasion and Neurovirulence
In the rat eye model, virions are injected into the vitreous humor of the eye where retinal ganglion cells are the first to replicate the virus (64, 66). The infection spreads to retinorecipient structures of the brain primarily via the optic nerve and the third cranial nerve. This model has been applied successfully to delineate neural circuitry and to define the PRV gene products required for directional transport. The rat eye model led to the identification of three PRV genes required for anterograde spread of virus: US7 (gI), US8 (gE), and US9 (US9). Deletion of any of these genes abolished the anterograde, but not the retrograde, spread of PRV in synaptically connected neurons (47, 66, 184, 429, ). Moreover, additional studies demonstrated that gE and gI contribute to virulence, and that their function in virulence was distinct from their function in directional spread (402, 403, 447). PRV spread in this model is reviewed further in “Analyzing the Spread of PRV-Bartha in the Rat Eye Model” below and in reference 21.
Chicken Embryo Eye Model
An alternative model for the study of PRV neurovirulence and neuroinvasion uses chicken embryos (18). A viral inoculum is introduced through a window in the eggshell into the vitreous chamber of the eye at a developmental stage where retinal cells have developed synaptic connections with the optic tectum in the brain. At this stage of development, the eye is among the largest and most easily accessible tissues. The relatively large yet well-circumscribed target area of the vitreous eye chamber allows consistent, precise primary infections with little to no risk of nonspecific infection of other chick tissues. The innervation of the eye and eye structures are well known, facilitating analysis of spread.
Mouse Skin Flank Model: New Facets of PRV Pathogenesis
To study a primary PRV infection of an epithelial surface, Brittle et al. adapted a skin infection model well known in the HSV pathogenesis literature (51, 368, 426). In this model, shaved mouse skin flank is scarified and infected with a drop of viral inoculum, such that viral adsorption occurs on the underlying live epithelial tissue. A primary infection in epithelial tissue precedes virus entry into nerve endings of the peripheral nervous system, similar to the course of a natural herpesvirus infection. In permissive nonnatural hosts, such as the mouse, alphaherpesviruses can spread to the central nervous system. Thus, infection of epithelial tissue, virulence, and neuroinvasion of the peripheral and central nervous systems can all be examined in one animal.
Brittle et al. reported that virulent and attenuated virus strains exhibit distinct modes of neuroinvasion and lethality after inoculation (51). Mice infected with virulent PRV strains self-mutilated their flank regions in response to a virally induced, pruritic stimulus. In addition, these animals died rapidly with virtually no viral particles detectable in the brain by titering assays and immunohistochemical analyses. There were also no symptoms of central nervous system infection such as behavioral abnormalities. Mice infected with virulent PRV strain Becker, Kaplan, or NIA-3 died at approximately 77 h compared to attenuated vaccine strain Bartha-infected mice, which died at approximately 220 h. Mice infected with viruses lacking US9, gI, or gE died at roughly 101 h, 104 h, and 115 h, respectively. Interestingly, the longer-lived animals infected with the attenuated strains did not become pruritic or develop skin lesions on the infected flank region, yet developed profound ataxia and tremors associated with hindbrain lesions. In addition, immunohistochemical and titer analyses of specific brain regions found a preponderance of viral particles and infection in the hindbrain region at the later stages of infection (>96 h). An additional study with a UL54-null virus found a delay in symptom onset and a lengthened survival time following symptom appearance (364). Prior to death, the severity of pruritus and skin lesions of UL54-null and wild-type infected mice was indistinguishable.
Two Modes of PRV Lethality
It has long been assumed that nonnatural hosts of PRV died of classical herpesvirus encephalitis, defined by a massive necrotizing lesion of an expansive region of the brains. After all, PRV is highly neurotropic and the infected hosts suffered from neurological abnormalities. However, the severe pruritus and relatively shorter time to death by infection with wild-type strains of PRV, coupled with the barely detectable infection of the central nervous system, suggests that the fatal outcome of virulent infection is more a result of the host immune system response, or peripheral nervous system injury, rather than the result of fatal viral encephalitis. In comparison, the neurological symptoms, central nervous system viremia and long time to death seen after infection with attenuated strains, suggest that the cause of death may be quite different from the virulent strains, possibly a direct result of encephalitis, though no necrotizing brain tissue is observed (51).
Mouse Intranasal Infection Model
Over 60 years ago, Sabin used a murine model to study PRV neuroinvasion, and chose an intranasal route of infection to emulate the natural oronasal transmission of PRV in swine (355). Inoculation of the nasal cavity resulted in virally induced neuropathological lesions. The kinetics and locations of lesion appearance were consistent with a transneuronal spread of PRV from nasal epithelium to synaptically connected higher-order structures in the nervous system. Lesions were found in neurons of the olfactory system, parasympathetic system, sympathetic nervous system, and primary sensory neurons of the fifth cranial nerve.
The route of pseudorabies virus propagation in the mouse nervous system follows intranasal inoculation as described (14). The infection spreads through three neuronal pathways innervating the nasal cavity: (i) via anterograde spread in the trigeminal circuit, from trigeminal ganglia to spinal trigeminal nucleus, (ii) via retrograde spread in the sympathetic circuit, from superior cervical ganglion to sympathetic preganglionic neurons in the spinal cord's intermediolateral nucleus, and (iii) via retrograde spread in the parasympathetic circuit, from pterygopalatine ganglion to the superior salivatory nucleus. Infected mice exhibit distress symptoms (e.g., hyperactivity, facial pruritus, also known as “itch syndrome,” and hunched posture) prior to succumbing to infection within 3 days (14). Infected neurons could be identified by immunofluorescence with antibodies against viral antigens as well as by the activity of the lacZ reporter gene driven by the endogenous US4 (gG) promoter. The study further demonstrates that the relative neurovirulence and neuroinvasiveness of the wild-type strain and the US4-null lacZ reporter strain are similar (14).
As with the other animal models, a PRV US8 (gE)-null strain is severely attenuated in the mouse intranasal model, exhibiting reduced virulence and neuroinvasion (12). The infected mice survived longer and only the hunched posture syndrome was apparent. Infection of first-order neurons innervating the nasal mucosa still occurred in the absence of gE, but retrograde spread within the sympathetic circuit was reduced, while anterograde spread within the trigeminal circuit was completely abolished. Retrograde spread to second-order neurons in the parasympathetic circuit was not observed for US8 (gE)-null PRV; however, infection of the first-order neurons of this pathway could not be ascertained. The intranasal model has been used to systematically examine a collection of various gene deletions within PRV (12-14, 129, 210, 214, 260, 361; reviewed in reference 279).
Axonal Targeting, Transport, and Assembly of PRV in Cultured Neurons
Alphaherpesviruses maintain their presence in the host population by infecting and establishing a latent infection in the host peripheral nervous system. The cycle of infection, latency, and reactivation is critical to viral survival. The broad range of species permissive for PRV neuronal infection in vivo and ex vivo has made PRV invaluable for studying the molecular details of neuronal infection.
The study of PRV neuronal infection has enhanced our understanding of virus assembly and long distance transport in axons. The process of viral entry into neurons is thought to be similar to that found for most cultured cells (121). First, the viral envelope fuses with the cell membrane which, in the natural host setting, would be the axon terminal at the periphery of the peripheral nervous system. Next, the capsids (along with the inner tegument proteins) are transported retrogradely along the relatively great distance of the axon to the nucleus in the neuron cell soma. Live imaging studies of fluorescent capsids in neurons isolated from embryonic chick dorsal root ganglia have shown that the retrograde transport of capsids in axons is quite fast (1.17 ± 0.03 μm/second) (374). Similar live imaging studies have shown that the capsid remains associated with only a subset of tegument proteins, UL36 and UL37, but not VP13/14 (UL47), VP16 (UL48), or VP22 (UL49), during axonal transport in entry (249). The capsid transport in axons undoubtedly requires an association with molecular motors, most likely from the dynein family, since capsid movement by diffusion alone would be predicted to take years to reach the neuron soma (reviewed in reference 375).
Reactivation from latency involves triggering transcription, assembling mature virus, and transmitting infection from the cell body anterogradely through the axon back to epithelial cells at the periphery. Since capsids travel towards the nucleus after entry and towards the axon terminal during egress, a mechanism must exist to regulate the direction of capsid transport during infection. The direction of transport is presumably determined by the motor complex associated with the capsid: dynein would transport the capsid retrogradely, while kinesins would facilitate anterograde transport. Smith et al. have shown newly synthesized egressing green fluorescent protein (GFP)-labeled capsids pass monomeric red fluorescence protein-labeled capsids traveling in the opposite direction in the same axon (374). Thus, the modulation of transport direction, and hence motor association, occurs at the level of the individual capsid, rather than by global regulation of axonal motor transport.
Studies of HSV-1 point to a model in which egressing capsids are transported in axons separately from the viral envelope components rather than fully assembled virions (317, 285). In this model, assembly of mature particles occurs at a distal point along the axon, possibly at the axon terminal (reviewed in reference 406). Additional support for this model comes from studies of PRV infection of cultured rat motor neurons identifying viral proteins crucial to the anterograde transport of viral components. The envelope glycoprotein gE is required for efficient entry of glycoproteins, capsids and some tegument proteins into axons during viral egress (71). Meanwhile, the US9 envelope protein is required for sorting viral glycoproteins, but not capsids or tegument proteins into axons during viral egress (404). Thus, PRV capsids can be transported in axons independently of glycoproteins. However, entry of capsids into the axon is not sufficient to transmit infection to cells at the distal end of the axon.
To better study the neuron-to-cell spread of PRV, Ch'ng and Enquist recently developed a compartmented neuronal culture system wherein cell bodies and axon endings are separated by a physical barrier traversed only by the axons themselves (72). Additionally, the compartment containing the axonal endings is seeded with transformed epithelial cells to allow viral replication. In this model, the directional spread of PRV can be assessed by infecting the cells in one compartment and measuring the spread of virus through axons to cells in another compartment. In this system, gE, gI, and US9 were all required for transneuronal spread from the neuronal cell body compartment to the epithelial cells in the axon-end compartment. Electron microscopic analyses of PRV-infected neurons identified capsids that are transported within vesicles during anterograde axonal transport rather than directly associated with microtubules as in entry (72, 107). Since capsids can be transported independently of glycoproteins, the surrounding vesicle probably does not constitute the viral envelope. The origins and constituents of this transport vesicle remain unknown.
Models for Reactivation from Latency
Like other herpesviruses, in vitro reactivation of PRV can be induced in explants of trigeminal ganglia or tonsils, or in vivo in pigs or mice (52, 311, 396, 416). Reactivation of latent pseudorabies in pigs is induced by treatment with dexamethasone (reviewed in reference 52) or acetylcholine (395), drugs which influence the host immune and nervous systems, respectively. Reactivation from latency has also been studied using a mouse model. Because mice are normally unable to survive an acute infection of PRV, establishing a latent infection requires the passive transfer of high-titer neutralizing antibodies prior to viral inoculation (311). Subsequent stimuli can induce reactivation and virus production in trigeminal ganglia explants (311) or nasal cavity of live mice (396). Reactivation was observed in response to mild stress (restraint, cold and transportation), as well as acetylcholine and dexamethasone treatment (311, 394, 396, 397).
PRV AS A TRANSSYNAPTIC TRACER
While many viruses can infect the cells of the nervous system, only a handful have the unique ability to spread between synaptically connected chains of neurons: vesicular stomatitis virus (248), rabies virus (reviewed in reference 205), mouse hepatitis virus (21), betanodavirus (186), and alphaherpesviruses (reviewed in reference 121). Such neural spread requires that the virus must enter the neuron (the first-order neuron) and replicate. The encapsidated viral genome is then transported at or near sites of synaptic contact to a second order neuron where replication takes place again. This property of self-amplification allows the first order neuron to be as intensely labeled as the second and third order neurons. Tracing studies using PRV have been successfully employed in a number of different animal models: pigs (the natural host) (294), lambs and sheep (320), dogs (85), cats (101), chicken embryos (18), ferrets (36), and other rodents such as rats (reviewed in reference 121), mole rats (297), mice (51), gerbils (203), and hamsters (323). These three properties of pseudorabies virus—transsynaptic spread, self-amplification, and a broad host range—have allowed its use in an extensive number of neuroanatomical studies seeking to define the architecture of multisynaptic pathways (reviewed in reference 121).
Viral Tracers versus Nonviral Tracers
Conventional (nonviral) tracers that are transported in axons rely on antibody reactivity, radioactivity, enzymatic activity, or fluorescence to label the neurons taking them up (reviewed in reference 41). Commonly used conventional tracers include horseradish peroxidase alone or conjugated to wheat germ agglutinin, True Blue, Fast Blue, Fluoro-Gold, latex microspheres coated with fluorescing dyes, subunit B of cholera toxin (discussed below), the nontoxic fragment (C) of tetanus toxin, and dextran amines. While these tracers can travel within an axon to label distant cell bodies or dendrites, their use in multisynaptic neuronal circuit is limited by most tracers' inability to cross through the synapses from one neuron to the next. Some plant lectin-derived tracers, such as wheat germ agglutinin-horseradish peroxidase, can cross synapses (12, 170, 352), while others, such as Phaseolus vulgaris leucoagglutinin, cannot. Since propagation inevitably dilutes tracer concentrations and signal intensity, nonamplifiable tracers have limited utility in tracing higher order multisynaptic circuits.
In comparison, viral tracers are self-amplifying and do not decrease in signal intensity, whether assessed by the presence of viral antigens or marker genes. Finally, viral infections can proceed over several synapses to infect multiple segments of a neuronal circuit. PRV can replicate in all central nervous system neurons studied in permissive animals. As explained below, the multisynaptic tracing ability of PRV is heavily influenced by the architecture of the circuitry and the survival time of the infected host.
Transsynaptic Spread of PRV in the Nervous System
Electron microscopic analyses and tracing studies strongly support the view that PRV spreads in the nervous system primarily by direct cell-cell contact, rather than diffusion of virions through the extracellular space or spread via nonneuronal cells. Analysis of infected nervous tissue by electron microscopy reveals viral capsids and structural proteins localized at the synapses of infected nervous tissue (63, 67). However, the strongest evidence that PRV spreads through defined circuitry comes from carefully designed studies of virus transport within the nervous system. Much of this work has been done using the circuitry connecting the ventral musculature of the stomach to the brainstem and higher order structures of the central nervous system (Fig. 4). Three main points are as follows.
FIG. 4.
Stomach injection model. A. Sagittal view of the rat brain. S1, S2, and S3 refer to the levels of coronal sections depicted in panels B, C, D, and E. B. Innervation of smooth muscle of the ventral stomach. The area boxed in red is magnified to the right. Motor neurons from the dorsal motor nucleus of the vagus send projections through the vagus nerve to the ventral wall of the stomach. Sensory innervation of the ventral stomach through the left nodose ganglion is shown in pink. Neurons in the dorsal motor nucleus of the vagus exhibit anti-PRV immune reactivity 30 h after stomach injection of wild-type PRV-Becker (64, 338). PRV travels retrogradely to second-order neurons in the medial nucleus of the solitary tract between 50 and 60 h postinjection and to third-order neurons in the area postrema between 60 and 70 h postinjection. Labeling of neurons within the left nodose ganglia can be observed by 45 and 50 h postinjection. C. No cross talk with tongue or esophageal innervation after stomach injection. Injection of PRV-Becker into the ventrolateral musculature of the tongue (pathway shown in green) results in a very different pattern of infection from injection into the stomach (shown in blue) (64). After transport through the hypoglossal nerve, PRV immune reactivity can be seen in the hypoglossal nucleus (XII) 30 h postinjection. By about 52 h postinjection, PRV infection can be observed in second-order neurons in the spinal trigeminal nucleus (pars oralis and pars interpolaris) and the ventrolateral brainstem tegumentum and monoaminergic cell groups (not shown). Injection into the smooth muscle of the esophagus (shown in orange) produced labeling in the dorsal nucleus ambiguus. By 48 hours postinfection, labeling was detected in small bipolar neurons of the nucleus centralis of the medial NTS. The segregation of labeled structures following injection of stomach and esophagus is significant because the axons of these efferent circuits travel together in the vagus nerve, yet PRV infection is absent from the nucleus ambiguous following stomach injection and absent from the dorsal motor nucleus of the vagus followingesophageal injection. D. PRV requires an intact circuit for spread in the nervous system. In addition to surgical severance of the left vagus nerve which eliminates PRV transport to the left dorsal motor nucleus of the vagus, further proof that PRV neuronal spread requires intact, synaptically connected neurons is provided by tracing studies that span progressing developmental stages (339). PRV-Bartha immune reactivity in the central nervous system was examined 2.5 days after injection into the stomachs of newborn rats. Rats injected on postnatal day 1 (P1) exhibited PRV immune reactivity in the dorsal motor nucleus of the vagus, medial nucleus of the solitary tract, area postrema, and paraventricular nucleus of the hypothalamus by 2.5 days postinjection. No animals in the P1 group exhibited anti-PRV labeling in the central nucleus of the amygdala, lateral hypothalamic area, bed nucleus of the stria terminalis, insular cortex, or medial prefrontal cortex with the exception of one rat with six labeled neurons in the central nucleus of the amygdala. Rats injected at later developmental stages exhibit progressively more viral penetrance into the central nervous system; 2.5 days postinjection, P4 rats exhibit labeling of all structures observed in the P1 group plus extensive labeling in the central nucleus of the amygdala, lateral hypothalamic area, and bed nucleus of the stria terminalis. Only rats injected on P8 exhibit infection of neurons in the insular cortex and medial prefrontal cortex. E. Comparison of anterograde- and retrograde-defective alphaherpesviruses. In adult rats, stomach injection of PRV-Bartha results in retrograde-only transport of viral infection (pathway and PRV-immune reactive structures shown in orange) from the dorsal motor nucleus of the vagus to medial nucleus of the solitary tract to area postrema (340). Anti-PRV immunoreactivity 4 to 5 days postinjection does not change after elimination of anterograde transport by surgically severing the axons of pseudounipolar neurons projecting from the nodose ganglia (shown in pink; see panel B). HSV-H129 has a retrograde spread defect. HSV-H129 can spread retrogradelyin first-order neurons but only anterograde spread is observed in second-order pathways. Although HSV immune reactive structures appear similar to those infected by PRV after injection into the ventral stomach (HSV-H129, intact compare with PRV-Bartha), vagal deafferentation (illustrated by a red X on the sensory pathway from the nodose ganglia) eliminates infection of the medial nucleus of the solitary tract and area postrema (HSV-H129, vagal deafferentation). Abbreviations: AMBd, dorsal nucleus ambiguus; AP, area postrema; BNST, bed nucleus of the stria terminalis; CeA, central nucleus of the amygdala; DMV, dorsal motor nucleus of the vagus; IC, insular cortex; LHA, lateral hypothalamic area; mNST, medial nucleus of the solitary tract; mPFC, medial prefrontal cortex; nCen, nucleus centralis of the medial solitary nucleus; NG, nodose ganglion; PVN, paraventricular nucleus of the hypothalamus. (Figure modified from reference 386 with permission of the publisher.)
Circuit tracing by PRV faithfully reproduces tracing deduced by well-characterized, nonviral tracers.
After injection into the ventral stomach musculature, transport of the wild-type strain PRV-Becker to structures in the dorsal motor complex of the brainstem (Fig. 4B) was compared with transport of the beta subunit of cholera toxin, a compound used for both retrograde and anterograde tracing studies (7, 130). Here, the direction of virus spread is described in relation to the direction of impulse in a circuit, anterograde for spread from presynaptic to postsynaptic neuron, and retrograde for spread from postsynaptic to presynaptic neuron. Retrograde first-order neuronal transport to the dorsal motor nucleus of the vagus was seen for both PRV-infected and cholera toxin beta subunit-injected stomach (64). Cholera toxin beta subunit was also transported in an anterograde fashion from the stomach to label sensory afferents in the tractus solarius, something not seen in PRV-infected animals, presumably because the animals expired before the viral infection reached these distant structures.
Infection does not spread intraaxonally to synaptically unconnected but physically adjacent circuitry.
PRV injected into the tongue traveled through the hypoglossal nerve (XII) and reached the hypoglossal nucleus (Fig. 4C). No PRV antigens were ever detected in the adjacent dorsal motor nucleus of the vagus or nucleus of the solitary tract (NST) complex. Similarly, while injection of virus in the stomach can label the dorsal motor nucleus of the vagus and medial NST (mNST), injection of virus into the esophagus led to a pattern of labeling in the nearby nucleus ambiguous but not the dorsal motor nucleus of the vagus or the NST. This is notable because the axons that innervate the esophagus and ventral stomach travel together in the vagus nerve, yet virus labeling of neurons stays specific to the individual circuitries (64).
Spread in the nervous system requires an intact circuit and synaptic connections.
In the stomach infection model, severing the left vagus nerve eliminated virus transport to the left dorsal motor nucleus of the vagus, while transport through the right vagus nerve to the ipsilateral (right) dorsal motor nucleus of the vagus was unaffected (64).
The most compelling evidence that synaptic connections are required for PRV spread come from tracing studies to define the temporal stages of development of forebrain circuitry in the newborn rat (339). Ventral stomach injection of rats with PRV on postnatal day 1 (P1), P4, and P8 allowed a comparison of the circuitry connecting the dorsal motor nucleus of the vagus complex to higher ordered structures in the forebrain (Fig. 4D). While axonal projections from the NST to the medial prefrontal cortex and insular cortex are present early in development, PRV only spread from the dorsal motor nucleus of the vagus to these forebrain structures in older rats. The key assumption is that PRV is not transported to brain structures prior to the formation of synaptic connections. Although few studies have addressed the extent to which PRV transport requires a functional synapse in terms of neurotransmitter release and electrophysiology, recent reports have identified changes in PRV tracing after lesion-induced circuit reorganization (21, 178).
Containment of Neuronal Infection by Cytoarchitecture and Nonpermissive Cells
PRV is pantropic, that is, virions infect many different types of cells including neurons and epithelial cells. Yet, once introduced into a specific neuronal circuit, nonsynaptic spread to peripheral cells in the nervous system is severely limited. Recent studies have examined how and when nonsynaptic spread of PRV is limited in the brain, and whether PRV would be suitable for circuit tracing following intracerebral injection (reviewed in reference 8). Particular attention has been paid to the potential for mislabeling of neuronal circuits after intracranial injection of PRV. Direct PRV injection into brain ventricles, cerebral cavities filled with cerebrospinal fluid, showed that the neurons and the ependymal cells lining the ventricle and the caudal raphe could indeed be infected (74). Immunostaining for viral antigens shows that PRV can infect astrocytes and brain macrophages surrounding infected regions of the central nervous system. Temporally, infection of astrocytes follows neuronal infection, indicating spread is from neuron to astrocyte (64). However, since astrocytes are susceptible to PRV infection, but not permissive for viral replication, they do not contribute to trans-neuronal spread of the virus (63, 338). Viral infection does not spread from infected glial cells to nearby nonneuronal cells (405) or to nearby axons outside the circuitry being traced (64). Rather, infection of astroglia is thought to represent an effort of the local intrinsic and innate immune defense to contain the infection (62, 63, 338). When injected directly into brain tissue, PRV virions diffuse very little, producing a focal infection site. PRV could also infect neurons via fibers of passage, axons that traverse but do not synapse on cells at the injection site (74, 189). Though the potential for leakage into the cerebrospinal fluid exists, PRV performs well as a tracer following intracranial injection (62). Proper experimental design planning for tracing studies should minimize nonspecific labeling, while careful examination of ependymal cells should identify leaks into the cerebrospinal fluid.
Attenuated Strains Make Good Tracing Viruses
Attenuated virus strains possess mutations that reduce virulence. One of the best characterized attenuated PRV strains, PRV-Bartha was isolated after multiple passages of a virulent field isolate in cultured chicken cells and embryos (23). It was used as an effective vaccine against PRV in pigs (264). Though the complete sequence of the PRV-Bartha genome is unknown, molecular and genetic analyses have identified three independent mutations contributing to its reduced virulence: point mutations within UL21 (220), a signal sequence mutation in the UL44 (gC) gene (345), and a 3-kb deletion encompassing US8 (gE), US9 and a large portion of US7 (gI) and US2 (247, 281, 322). The attenuated mutants such as PRV-Bartha are favored for tracing studies because they penetrate further into neuronal circuits due to increased host survival time (reviewed in reference 116). In addition to their role in virulence, the membrane glycoproteins gE and gI and the membrane protein US9 are all required for efficient anterograde spread in the nervous system (47, 65, 184, 429).
Once introduced into the nervous system, PRV-Bartha spreads only in the retrograde direction in a circuit while wild-type strains, such as PRV-Becker and PRV-Kaplan, will spread in both the anterograde and retrograde directions (51). This monodirectional spread of Bartha clearly identifies the direction of neuronal circuits and often simplifies the analysis. Because PRV-Bartha can infect peripheral nervous system neurons projecting to central nervous system neurons and invade specific brain regions by retrograde transport, the strain has been widely exploited for defining the central nervous system circuits that modulate the autonomic and somatic peripheral outflows. A number of studies have used PRV-Bartha for circuit tracing following intracerebral injection as well (reviewed in reference 8).
Analyzing the Spread of PRV-Bartha in the Rat Eye Model
The retrograde-restricted spread phenotype of PRV-Bartha has been effectively characterized in rats and hamsters (66, 323, 369) by studying virus transport into the central nervous system following intraocular injection of PRV (Fig. 5) . In the rat eye model, the wild-type virus PRV-Becker infects the projection neurons of the retina, the retinal ganglion cells, and spreads by anterograde transport in axons through the optic nerve to infect second order neurons in the suprachiasmatic nucleus, the dorsal lateral geniculate nucleus, the ventral lateral geniculate nucleus, the intergeniculate leaflet, and superior colliculus (66, 287, 327). PRV can be detected in second-order neurons around 50 h postinoculation (64, 323), consistent with a direct path to these retinorecipient structures.
FIG. 5.
Rat eye infection model: comparison of wild-type (PRV-Becker) and attenuated (PRV-Bartha) neuronal spreads. A. Sagittal view of the rat brain. E1, E2, E3, and E4 refer to the level of coronal sections depicted in panels B and C. B. Spread of PRV-Becker (shown in red) after virus injection into the vitreous humor of the rat eye. PRV-Becker spreads from infected retinal ganglion cells (level E1) through the optic nerve to second-order neurons in the supracharismatic nucleus (level E2), dorsal and ventral aspects of the geniculate nuclei (dorsal aspect of the lateral geniculate nucleus and ventral aspect of the lateral geniculate nucleus) and intergeniculate nucleus (level E3), and the superior colliculus of thevisual cortex (level E4). Only first-order projections are shown. C. Transport of PRV-Bartha (shown in blue) is restricted to retrograde-only pathways. Although cells of the retinal ganglia (level E1) are infected with PRV-Bartha, this virus is restricted from anterograde spread through the optic nerve to retinorecipient neurons. Instead, retrograde spread of infection to first-order neurons in the ciliary ganglion leads to transport through the oculomotor nerve to second-order neurons in the Edinger-Westphal nucleus (level E4). Infection of neurons in the olivary pretectal nucleus, intergeniculate nucleus (level E3) and supracharismatic nucleus (level E2) is also by retrograde axonal transport of virus as shown (369). Abbreviations: CG, ciliary ganglion; dLGN, dorsal aspect of the lateral geniculate nucleus; EW, Edinger-Westphal nucleus; IGL, intergeniculate nucleus; OPN, olivary pretectal nucleus; SC, superior colliculus; SCN, suprachiasmatic nucleus; vLGN, ventral aspect of the lateral geniculate nucleus. (Figure modified from reference 386 with permission of the publisher.)
A more restricted set of structures are infected subsequent to intraocular PRV-Bartha injection, including the supracharismatic nucleus, the intergeniculate leaflet, the olivary pretectal nucleus, and the Edinger-Westphal nucleus, but never the superior colliculus or dorsal lateral geniculate nucleus. These results were not interpreted initially as a PRV-Bartha transport defect, but rather as an inability of PRV-Bartha to productively infect the subtypes of retinal ganglion cells leading to the restricted retinorecipient structures (66, 287). Additional studies suggested instead that PRV-Bartha was incapable of anterograde spread through chains of connected neurons (48, 120, 184). The apparent anterograde spread to the supracharismatic nucleus and intergeniculate leaflet by PRV-Bartha was actually found to occur by a different and retrograde route. Analysis of the pattern and temporal sequence of central infection of hamsters and rats with enhanced GFP-expressing derivatives of Bartha found that infection of the supracharismatic nucleus and intergeniculate leaflet occurs via the Edinger-Westphal nucleus by retrograde spread through the autonomic innervations of the eye (323, 369) (Fig. 5C). The bulk of the studies taken together clearly indicate that PRV-Bartha is incapable of spreading from presynaptic to postsynaptic neurons in a circuit.
Virulence and Neural Tracing
Though the exact time to death varies with the animal model and route of injection used, virulent wild-type strains induce dramatic symptoms and kill animals much faster than the attenuated strains. For example, PRV-Becker introduced into mice flank skin is usually fatal within three days (75 h), while PRV-Bartha causes death after 9 days (220 h) (51). Nearly a week of additional host survival time allows attenuated strain PRV-Bartha to infect distal neurons in midbrain and hindbrain following infection of the skin. A time course analysis of PRV-Bartha-infected animals shows that the viral infection spreads through efferent nerves innervating the skin, up the spinal cord, and into higher regions of the central nervous system before the animals die (51). A conservative estimate suggests that PRV-Bartha has spread to at least fourth-order neurons as early as 6 days after infection of the skin: sensory neuron in skin → interneuron in spinal cord → raphe magnus nucleus in brainstem → periaqueductal gray neurons in the midbrain.
In contrast, wild-type strain PRV-Becker follows a similar route of infection but the animals die before the infection spreads beyond the brain stem (51). Furthermore, PRV-Becker infection spreads by retrograde propagation in efferent nerves projecting to infected mouse flank. Infection with over a dozen PRV strains reveals an inverse relationship between a virulence (measured by time of death and appearance of pruritus) and ability to spread extensively within the nervous the system (51, 50). This inverse relationship is seen despite bidirectional and rapid spread of virulent strains (51). It is assumed that if there were a way to prolong the life of infected animals, wild-type viruses would label the central nervous system more extensively than attenuated strains. Despite considerable effort, no such treatments are known.
Recent Advances in Viral Tracing Techniques
The increasing popularity and acceptance of PRV for transneuronal tracing, has propelled the search for new tracing viruses and techniques to further enhance this powerful tool. Three promising advances have been examined in a recent review (119).
Dual tracing in circuitry analysis.
Dual tract tracing experiments have employed PRV-Bartha-derived mutants to determine whether different targets in the peripheral nervous system are under control of a common region of the central nervous system. One of the first studies to use this scheme sought to define the central autonomic neurons coordinating the cardiac and adrenal response during stress situations, such as fight-or-flight (189). Two antigenically distinct strains derived from PRV-Bartha were employed: one expressing the U244 (gC) allele of a virulent PRV strain (PRV-Kaplan), the other expressing bacterial β-galactosidase (product of lacZ). Injecting one virus into the stellate ganglion (the main sympathetic ganglion innervating the heart) and the other into the ipsilateral adrenal gland resulted in dual labeled neurons in the hypothalamus and brainstem. The dual tracing approach has been used in over a dozen collateralization studies (59, 144, 234, 351, 366; reviewed in references 15 and 119).
The bulk of the recent studies use isogenic strains of PRV-Bartha that express either the enhanced green fluorescence protein (EGFP) or β-galactosidase. Recently, Banfield et al. used two isogenic PRV-Bartha recombinants, expressing either green (EGFP) or red (monomeric red fluorescence protein) fluorescence protein (17). It is thought that for optimal dual labeling of central nervous system neurons, the two viral infections must reach the same neuron within hours of each other. Analysis of dual viral infection of cultured rat dorsal root ganglia found the percentage of double-labeled cells decreases from 100% to 27% when the second infection is delayed by a mere two hours (17). In both HSV-1 and PRV, this phenomenon of superinfection inhibition has been attributed, in part, to gD-mediated interference of viral entry (56, 145, 374). The situation is more complicated in sequential infection of neuronal circuitry in vivo. As the time of infection and the spread within two circuits by two different viruses may not be equivalent, a careful and empirical calibration of the relative time and dose of infection of the two viruses may be needed (61, 206). Clearly, the most serious limitation to the dual labeling method is the potential for false negatives. Nevertheless, the dual infection method remains a powerful approach to define collateralized pathways.
Electrophysiological recording of live neurons infected by a PRV tracer.
Infection with EGFP-expressing PRV-Bartha recombinants allows visualization of live infected neurons by fluorescence without any fixative staining. Recent studies have gone beyond the mere tracing of neuronal circuits to actually record the electrophysiological properties of infected neurons from live brain slices or retinal tissue of infected animals (98, 151, 188, 371, 422). The attenuated phenotype of the PRV strain used means reduced cytopathic effects and more physiologically intact neurons. Indeed, all five studies compared the electrophysiological properties of PRV-Bartha infected and uninfected neurons and found them to be indistinguishable. Of course, these results apply only to the particular stage of infection analyzed. Nevertheless, the potent combination of functional anatomy (tracing) and physiology (electrophysiological recording) opens the door to a much deeper understanding of neuronal circuit functions.
Conditional replication of PRV in specific neuronal populations.
A powerful extension of the classical viral tracing method has been introduced recently, relying on engineered viruses and hosts to make only a defined subpopulation of cells or neurons permissive for infection and viral propagation (106). The technique relies on Cre-LoxP: Cre is a site-specific recombinase able to recombine specific target DNA sequences, loxP, with high fidelity and without the need for cofactors (reviewed in reference 380). Briefly, transgenic mouse lines expressing Cre under the control of specific neuronal promoters are infected with a PRV-Bartha derivative, Ba2001, which is dependent on Cre-mediated recombination to allow expression of a reporter gene, GFP, and thymidine kinase. PRV thymidine kinase is not required for viral growth in mitotic cells, such as transformed tissue culture cells, but it is essential in nondividing cells, such as neurons. Thus, PRV Ba2001 will neither express GFP nor be replication-competent in neurons until the viral genome is rearranged following infection of a Cre-expressing neuron. Subsequent to the Cre recombination event, the altered Ba2001 virus will act like a normal transsynaptic retrograde tracer. De Falco and colleagues used Ba2001 and mice expressing Cre under the neuropeptide Y or leptin (ObRb) promoter to map the neural inputs to the neuropeptide Y-expressing cells and leptin-expressing cells that play a role in regulating feeding behavior (106).
Potential for monodirectional anterograde tracing with PRV.
The monodirectional, retrograde-only spread of PRV-Bartha in neuronal circuits has served to simplify the design and analysis of tracing studies. A monodirectional, anterograde-only PRV tracer would assuredly be equally useful and powerful. For example, just as the infection of peripheral organs with a retrograde-only viral tracer identified sympathetic and parasympathetic motor neuron circuits into the central nervous system, infection of peripheral organs with an anterograde-only tracer could identify the direct and relayed sensory input circuits into the central nervous system. No PRV strains with such a phenotype have been described yet. However, an HSV-1 strain, H129, has been reported to be incapable of, or severely impaired for, retrograde spread: H129 can spread in both direction within the axon of first order neurons (the neuronal cells at the primary site of infection), but transsynaptic spread occurs exclusively in an anterogradely manner to label second and higher order neurons (Fig. 4E) (340). A number of tracing studies have taken advantage of the new possibilities this strain offers (22, 141, 204, 340, 385, 451). The genetic basis for the retrograde spread of H129 is unknown. It is hoped that the mutation(s) will reside in genes conserved between HSV-1 and PRV and that a similar anterograde-restricted tracer can be engineered in PRV.
VETERINARY IMPACT OF PSEUDORABIES (AUJESZKY'S DISEASE)
Historical Discovery
As early as 1813, press reports from the United States of America point to sporadic outbreaks of animal disease consistent with PRV. The disease was first described as “mad itch” in cattle and was characterized by heavy itching (165). In 1902, a Hungarian veterinary surgeon, Aládar Aujeszky, provided the first scientific documentation that PRV is the etiological agent of the disease pseudorabies, or Aujeszky's disease (10; reviewed in references 160 and 437). Aujeszky chronicled his successful efforts to isolate PRV from an ox, dog, and cat. Inoculating the recovered virus into pigs and rabbits resulted in transmission of the disease. Examination of the disease syndrome in rabbits revealed that it was reminiscent, but distinguishable from rabies. Accordingly, the disease was assigned to pseudorabies. The viral nature of the causative agent of Aujeszky's disease was confirmed soon thereafter (362) and PRV was later identified as the cause of the mad itch (367).
Lethality of Pseudorabies Infection in Nonnative Hosts
In addition to infection of its natural swine host, PRV infects a broad range of vertebrates. These include cattle, sheep, dogs, cats, goats, chickens, raccoons, possums, skunks, rodents, rabbits, guinea pigs, and, rarely, horses (128, 160, 207, 263, 318, 435). Infection of carnivores, such as bears and wild felines, has also been reported to occur after consumption of raw PRV-infected meat (60, 150, 450). Infection of nonnative hosts with a wild-type PRV strain is uniformly lethal. While PRV does not infect humans, experimental studies in nonhuman primates indicate that rhesus monkeys and marmosets are susceptible to infection. Other higher-order primates, such as chimpanzees, are not susceptible to infection (118, 211).
Acute PRV Infection of Swine
Outbreaks of PRV in the United States increased substantially in the 1960s. This timing is coincident with a dramatic increase in the intensity of swine production and close-quarter confinement of large numbers of pigs in swine barns. The increased stresses imposed on the swine coupled with closer contact provided ideal conditions for spread of PRV (211). Primary viral replication occurs in the nasal and oropharyngeal mucosa (259). PRV is tropic for both respiratory and nervous system tissue of swine and viral particles enter sensory nerve endings innervating the infected mucosal epithelium. Morbidity and mortality associated with PRV infection varies with the age of the pig, overall health status of the animal, viral strain, and infectious dose. Younger swine are the most severely affected by PRV infection and typically exhibit symptoms of central nervous infection whereas older swine exhibit symptoms of respiratory disease (211).
For suckling piglets, the incubation period of PRV is typically 2 to 4 days. Initially, piglets are listless, febrile, and uninterested in nursing. Within 24 h of exhibiting these symptoms, the piglets will progressively develop signs of central nervous system infection including trembling, excessive salivation, incoordination, ataxia, and seizures. Infected piglets may sit on their haunches in a “dog-like” position because of hind limb paralysis, lay recumbent and paddle, or walk in circles. Once piglets develop central nervous system abnormalities, they die within 24 to 36 h. Mortality of suckling pigs with pseudorabies is extremely high, approaching 100%. The cause of death in piglets has historically been attributed to viral encephalitis. However, recent work suggests that a peripheral host immune response to viral infection may be a significant determining factor in the death of infected animals (51).
Weaned pigs, ages 3 to 9 weeks of age, tend to develop symptoms highly reminiscent of those described for suckling pigs. However, the mortality rate is much lower. Typically, 50% of infected 3- to 4-week-old animals die. Pigs 5 to 10 weeks of age become listless and anorectic, and exhibit temperatures of 41 to 42°C within 3 to 6 days of infection. Animals often develop respiratory signs such as sneezing, nasal discharge, a severe cough, and difficulty breathing. Pigs with respiratory illness often lose significant body weight, a condition that translates directly into economic loss for swine producers. Signs of infection typically resolve after 5 to 10 days, with most pigs making a rapid recovery upon resolution of the fever and anorexia. If carefully nursed through their illness and treated for secondary bacterial infections when necessary, mortality rarely exceeds 10% (211).
In adult swine, respiratory signs are the hallmark of PRV infection although, sporadically, adult animals may exhibit central nervous system abnormalities varying from mild muscle tremors to violent convulsions. Although morbidity is quite high (approaching 100% of infected animals), mortality is relatively low (1 to 2% of infected animals). Typically, clinical signs appear in 3 to 6 days. Symptoms include a febrile response (41 to 42°C), listless behavior, lack of interest in eating, and mild to severe respiratory signs. These animals will typically exhibit rhinitis evidenced by sneezing and nasal discharge. The respiratory illness may progress to pneumonia with a harsh cough and labored breathing. Sick animals will become emaciated and lose considerable body weight, resulting in financial losses for swine producers. The duration of clinical illness is 6 to 10 days, and infected animals typically recover rapidly (211).
Pregnant animals infected with PRV in the first trimester of pregnancy will usually reabsorb the fetuses in utero. If infection occurs within the second and third trimester of pregnancy, infection typically results in abortion, stillbirths, or weak piglets that die within 48 h of birth. PRV-induced infertility results from the virus crossing the placenta and infecting animals in utero. In fact, in a given litter, some pigs may be born normal, while others are weak and some are stillborn due to transplacental transmission of virus. Reproductive failure usually has a low incidence on an infected farm, occurring in 20% or less of pregnant animals (211).
PRV Latency in Swine
During an acute infection, viral particles replicate in the oropharyngeal mucosa then enter sensory nerve endings innervating the site of infection. Retrograde transport of virus particles occurs in the maxillary branch of the trigeminal nerve (cranial nerve V), the glossopharyngeal nerve (cranial nerve IX), and the olfactory nerve (cranial nerve I) (reviewed in reference 253). The term latent infection of swine is used to describe a long-term infection in which the PRV genome is quiescent and PRV virions cannot be recovered. The predominant sites of PRV latency are the trigeminal ganglia (originally described as gasserian ganglia) and the sacral ganglia in feral swine (discussed in Feral Swine: Reservoirs of Sexually Transmitted PRV in the United States). Similarly, the predominant sites of latent neuronal infection of the human alphaherpesviruses HSV-1 and HSV-2 are the trigeminal ganglia and the sacral ganglia (433). Latent PRV genomes can be detected in the other neural tissues such as the olfactory bulb and brain stem (355, 356, 354, 430). Each latently infected cell body of the trigeminal ganglia harbors approximately 30 copies of the viral genome (354). In addition, tonsillar lymph nodes are reported sites of latency (51, 52, 81, 356). However, the rate of PRV DNA detection in tonsils is often lower than that seen in trigeminal ganglia, and questions remain as to whether tonsils constitute a true site of latency rather than the site of low but persistent infection (reviewed in reference 253). The cell type harboring PRV within the tonsillar tissue remains unknown.
Reactivation and shedding of virus in latently infected animals frequently occurs after stressful experiences. Stressors include concomitant disease conditions, vehicular transport, poor animal husbandry, farrowing (i.e., giving birth to piglets), and treatment with immunosuppressive agents, e.g., corticosteroids (96, 353, 354, 399, 416, 436).
Modified Live Vaccines
The immunobiology of pseudorabies virus has been reviewed by Mettenleiter (278). Most PRV vaccines are attenuated modified live virus vaccines. The attenuation of modified live virus vaccines arises from their deletion of PRV UL23, the gene encoding the viral thymidine kinase. Thymidine kinase is critical for viral replication in nonmitotic tissues, such as neurons. Furthermore, attenuation is also mediated by deletion of gE (280).
Historically, attenuated modified live virus vaccine strains such as PRV-Bartha and PRV-Bucharest arose from extensive passaging of virulent field isolates in embryonic chicken cell cultures. For example, attenuated PRV-Bucharest was isolated after passaging a virulent strain over 800 times in chicken cells. Other methods for generation of attenuated strains include selection for drug resistance (23, 209).
In recent decades, advancements in understanding of PRV genetics and molecular biology permitted elucidation of essential PRV neurovirulence factors. PRV gE is an important swine neurovirulence factor (233, 246, 283). Quint et al. genetically engineered an attenuated “marker” vaccine strain (2.4N3A) by deleting the gE coding sequence from the genome of the highly virulent field strain NIA-3 (330). Deletion of gE from vaccine strains also makes it possible to serologically distinguish gE-null vaccine strain-infected animals from virulent field strain-infected animals.
By screening for 5-bromodeoxyuridine resistance in 2.4N3, a gE-null, thymidine kinase-null double knockout vaccine strain (PRV-Begonia) was isolated (419). Standardized vaccine efficacy tests demonstrate that Begonia exhibits the critical traits sought for a modified live virus vaccine (102). Vaccination with the Begonia strain imparts protective immunity against virulent field strain challenge and prevents or significantly diminishes excretion of challenge virus that could spread to other herd members. Vaccination also guards against the growth retardation associated with pseudorabies disease. The vaccine is safe for pigs of all ages, including pregnant females, other farm animals, including cattle and sheep, and pets such as dogs and cats (419).
gE/thymidine kinase double-knockout vaccine strains (gE null marker strains) are used preferentially worldwide. gG/thymidine kinase double-knockout vaccines (gG marker strains) are available as well. Preferential use of gE over gG marker strains is primarily due to the superiority of gE-targeted assays enzyme linked immunosorbant assays compared with gG-targeted diagnostic assays for differentiation of vaccinated and field strain infected animals (211, 363, 427, 431). Most regulatory organizations governing pseudorabies disease management in the United States and within the European Union require marking of pseudorabies vaccines by gE deletion (211, 295, 431).
DNA Vaccines
Globally, pork producers suffer substantial economic loss due to pseudorabies disease. Successful vaccination programs confer protection against disease, but are not sufficient to prevent PRV infection. Thus, successful protection of herds against PRV requires vaccination in conjunction with other measures including routine testing and selective culling of infected animals. The most efficacious virus-neutralizing antibodies target PRV gC and gD (89, 115 164, 256, 293, 425). Subunit vaccines consisting of gC or gD, as well as anti-idiotypic anti-gD antibodies elicit some degree of protective immune responses (257, 261, 409). A large portion of the neutralizing activity in convalescent swine sera is directed against gC (27). In addition, gC has been reported by Zuckerman et al. to be a primary target for porcine cytotoxic T cells (453). Taken together, these studies indicate PRV gC and gD represent the major targets for the host's immune system against infection.
DNA vaccines are DNA constructs encoding viral or bacterial immunogens. These constructs can be directly inoculated into animals and taken up by host cells, which then synthesize and process the encoded viral antigens for immune display by MHC class I molecules on their cellular surface (reviewed in reference 171, 410, 411). DNA vaccines, consisting of plasmids encoding PRV gC or gD have been tested for induction of a protective response against wild-type infections in swine (147). While vaccination with the gD plasmid offered no discernible protection, the results with the gC were more promising (147). Inoculation of swine with as little as 1 μg of gC-encoding plasmid DNA resulted in seroconversion of animals, the induction of a protective T-cell response, and full protection of animals against a challenge dose of a widely used vaccine challenge strain PRV-75V19 (6). However, the DNA vaccines provided only partial protection against the highly virulent PRV NIA-3 strain. Another promising result derived from this study was that intradermal inoculation of plasmid DNA resulted in a higher degree of seroconversion than introduction of plasmid DNA by an intramuscular route. This route of infection is particularly useful in that it facilitates efficient vaccination of a large herd of animals.
A multicomponent vaccine comprising the genes for gB, gC, gD, and gE was compared with commercial inactivated and modified live virus vaccines by Gerdts et al. The protection offered by the multicomponent vaccine exceeded that of the inactivated virus, but was less than the protective response elicited by the modified live virus (148).
Advantages of viral DNA vaccines include a relatively high degree of biological safety because infectious particles are not required for preparation of the vaccine, nor are infectious agent introduced into the animals. Additionally, proteins expressed from plasmid DNA constructs are incorporated into the antigen-processing machinery of both major histocompatibility complex class I and class II. Ultimately, this results in induction of not only effective humoral responses, as in the case for whole or subunit vaccines, but also in induction of protective cellular immune responses (87, 199, 255, 335, 398).
One additional advantage of a DNA vaccine is that the PRV UL49.5 gene product, gN, is not expressed in the vaccinated host's cells. PRV UL49.5 inactivates TAP (the transporter associated with antigen processing (228). TAP plays a critical role in translocating antigens from the cytoplasm into the endoplasmic reticulum of infected cells for presentation by major histocompatibility complex class I molecules. Major histocompatibility complex class I presentation of immunogens is essential for robust cytotoxic T cell response to the viral antigen target.
Diagnostic Tests for PRV
A number of assays can be performed in veterinary diagnostic laboratories to confirm a diagnosis of pseudorabies in the field. Such assays usually include viral isolation from infected tissues samples, immunofluorescence assays on tissue sections and nasal swabs, and serologic assays. Serologic assays consist of latex agglutination tests, serum neutralization assays, and enzyme-linked immunosorbent assays testing for a humoral response to gE. Enzyme-linked immunosorbent assays specifically identifying the presence of gE in an animal's blood are particularly useful because these distinguish an immune response to a field strain (gE+) infection from an immune response to a vaccine strain (gE−) (211).
Worldwide Distribution of Pseudorabies
Pseudorabies outbreaks occur in swine populations worldwide, resulting in substantial economic losses for affected countries. Many countries, such as Germany and the Netherlands, mounted successful agricultural campaigns to eradicate PRV from their swine populations. In 2001, Germany was listed as Aujeszky's disease-free. Eradication efforts included selective culling of PRV-positive herds, widespread vaccination programs with marker viruses such as gE-null vaccine strains, restricted importation of swine, and improved herd management practices to isolate swine from potential reservoirs of infection such as wild boar (295, 381, 417). In 2003, countries with documented cases of PRV (in alphabetical order) included Belarus, Brazil, Cuba, France, Hungary, Italy, Mexico, Panama, Poland, Portugal, Romania, Russia, Slovakia, Slovenia, Taiwan, and Ukraine. Pseudorabies outbreaks in Mexico are particularly troubling to U.S. state and federal agricultural officials, given Mexico's border with the United States. This underscores the importance of surveillance and monitoring of movement of animals across U.S. borders. Information regarding global pseudorabies outbreaks was provided by the World Organization for Animal Health/Office International des Epizooties (web site address: www.oie.int).
Economic Impact of PRV in the United States
Circa 1999, economic estimates indicated that the American pork production industry generates approximately US$30 billion in revenue annually. PRV has been the target of an effective nationwide eradication program. The annual cost of eradication efforts is approximately US$30 million. Of this amount, approximately US$17 million is spent vaccinating animals to protect against PRV, US$11 million in costs is attributable to government indemnity payments to swine producers for culling infected herds, and close to US$2 million is dedicated to disease surveillance. Reviewed information regarding pseudorabies and the status of pseudorabies eradication efforts in the United States is derived from the U.S. Department of Agriculture—Animal Plant Health Inspection Service (web site address, www.aphis.usda.gov).
Efforts to Eradicate PRV in the United States
In 1989, the U.S. Department of Agriculture implemented a program for eradicating PRV from herds in the United States. This eradication program is monitored and reported by individual states and includes the U.S. territories of Puerto Rico and U.S. Virgin Islands. According to information acquired from the U.S. Department of Agriculture-Animal Plant Health Inspection Service web site, five specific stages for eradication were outlined:
Stage I: Preparation and implementation of PRV programs under the auspices of state, federal, and industrial authorities.
Stage II: Surveillance, control, and culling of known infected PRV herds.
Stage III: Less than 1% of herds infected with PRV.
Stage IV: No new outbreaks reported within one calendar year and no herds testing positive for the presence of PRV infection.
Stage V: Two consecutive years at Stage IV.
The effectiveness of this program is readily apparent. As of April 2005, all states and U.S. territories are stage V.
Feral Swine: Reservoirs of Sexually Transmitted PRV in the United States
Studies on feral swine (Sus scrofa) populations in the United States have demonstrated that PRV is indigenous in these herds (92). In feral populations, PRV primarily spreads by a venereal route rather than a respiratory route typical in domestic herds (348a, 350). In contrast to the trigeminal ganglion latency of PRV in domestic swine, feral swine herds typically show latent infection of the sacral ganglia (349). To ensure the continued success of the U.S. eradication program, herd management practices require vigilant maintenance of physical barriers (e.g., double fences) between wild and domestic swine (350) (U.S. Department of Agriculture-Animal Plant Health Inspection Service web site). Viral strain differences may explain the difference in tropism, as a comparison study of virus strains originating from either domestic or feral pigs found the feral strains to be markedly attenuated (163).
OUTLOOK
As a distant relative of human alphaherpesviruses, PRV shares considerable positional and functional gene homology with HSV-1, HSV-2, and VZV, but little to no DNA sequence homology. Research on PRV has already provided considerable insight into basic biology and mechanisms of herpesvirus pathogenesis in general, highlighting both the conserved nature of viral replication strategies, and their unique adaptation to a particular ecological niche.
The pace of PRV research has accelerated in the past 20 years, with the genome sequence only recently completed. Recent investigations into the molecular biology of PRV have more than once yielded findings that challenged accepted dogma and led the way to a clearer revised picture of how alphaherpesviruses function, including the general mechanism of herpesvirus virion assembly, entry and egress, neuroinvasion, and pathogenesis. Ongoing efforts are directed to the systematic functional analysis of every gene by establishing and characterizing null mutants in tissue culture and animal models. For certain key genes, the studies are often extended to include more detailed studies using targeted genetics and cell biology. Clearly, the methodical investigation of PRV gene functions will continue into the near future. In addition to answering critical questions about the viral life cycle, neuronal spread and viral pathogenesis, the new findings are expected to lead to direct practical applications in diagnostic testing, vaccines and antiviral drug therapy. Future PRV research also promises significant contributions to the areas of comparative virology, neurobiology, and cellular biology.
The broad host range of PRV can be used to examine how host-pathogen interactions lead to radically different outcomes of PRV infection in the natural and nonnatural hosts. The strategies used by PRV to evade the intrinsic and innate host immune defenses in so many different animals have yet to be catalogued, and present an exciting and challenging new field of study.
The relatively modest number of genes encoded by the PRV genome simplifies the use of proteomics and genomics technology in comparative virology studies, and a first-generation PRV gene chip has been constructed (C. Hengartner and L. W. Enquist, personal communication). The molecular biology tools already available for PRV include bacterial artificial chromosomes of the viral genome, transposon mutagenesis, and epitope tagging. Neuronal culture systems, compartmented chamber culture systems, and live fluoroscopy imaging techniques have also been adapted for the study of virus intra-axonal transport, viral assembly, and transneuronal spread at the synapse. As described in the tracing section of this review, ever more complex and powerful viral vectors are being developed for mapping neuronal circuits.
Finally, as a veterinary pathogen, PRV is coming under control thanks to smart agricultural management, frequent testing, and extensive vaccination. The molecular aspects of the latency phase of the viral life cycle are not well understood, and progress in this area could serve to bring us closer to the goal of PRV eradication. However, research on the molecular biology and pathogenesis of the virus will undoubtedly continue to provide new insights into the biology of herpesviruses.
Acknowledgments
We acknowledge Lynn W. Enquist, E. Alex Flood, Toh Hean Ch'ng, Becket Feierbach, and Greg Smith for critical reading of the review, Gary Pickard for insightful comments on PRV tracing of neural circuitry, and Pat Card for invaluable suggestions regarding the tracing section and associated figures. The transmission electron micrograph of the extracellular PRV-Becker virion in Fig. 1 was produced by L. E. Pomeranz and L. W. Enquist.
L.E.P. was supported by NIH postdoctoral fellowship 1F32 NSO4 2967-01A1, A.E.R. was supported by NIH grant 1 K08 NS48150-01A1, and C.J.H. was supported by NIH grant 5 P01 CA87661.
REFERENCES
- 1.Abmayr, S. M., J. L. Workman, and R. G. Roeder. 1988. The pseudorabies immediate-early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 2:542-553. [DOI] [PubMed] [Google Scholar]
- 2.Adams, R., C. Cunningham, M. D. Davison, C. A. MacLean, and A. J. Davison. 1998. Characterization of the protein encoded by gene UL49A of herpes simplex virus type 1. J. Gen. Virol. 79:813-823. [DOI] [PubMed] [Google Scholar]
- 3.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aleman, N., M. I. Quiroga, M. Lopez-Pena, S. Vazquez, F. H. Guerrero, and J. M. Nieto. 2003. L-particle production during primary replication of pseudorabies virus in the nasal mucosa of swine. J. Virol. 77:5657-5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambagala, A. P., R. S. Gopinath, and S. Srikumaran. 2003. Inhibition of TAP by pseudorabies virus is independent of its vhs activity. Virus Res. 96:37-48. [DOI] [PubMed] [Google Scholar]
- 6.Andries, K., M. B. Pensaert, and J. Vandeputte. 1978. Effect of experimental infection with pseudorabies (Aujeszky's disease) virus on pigs with maternal immunity from vaccinated sows. Am. J. Vet. Res. 39:1282-1285. [PubMed] [Google Scholar]
- 7.Angelucci, A., F. Clasca, and M. Sur. 1996. Anterograde axonal tracing with the subunit B of cholera toxin: a highly sensitive immunohistochemical protocol for revealing fine axonal morphology in adult and neonatal brains. J. Neurosci. Methods 65:101-112. [DOI] [PubMed] [Google Scholar]
- 8.Aston-Jones, G., and J. P. Card. 2000. Use of pseudorabies virus to delineate multisynaptic circuits in brain: opportunities and limitations. J. Neurosci. Methods 103:51-61. [DOI] [PubMed] [Google Scholar]
- 9.Aubert, M., and J. A. Blaho. 2001. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 3:859-866. [DOI] [PubMed] [Google Scholar]
- 10.Aujeszky, A. 1902. Ueber eine neue Infektions-krankheit bei Haustieren. Zentralbl. Bakteriol. 32:353-357. (In German.) [Google Scholar]
- 11.Avitabile, E., G. Lombardi, T. Gianni, M. Capri, and G. Campadelli-Fiume. 2004. Coexpression of UL20p and gK inhibits cell-cell fusion mediated by herpes simplex virus glycoproteins gD, gH-gL, and wild-type gB or an endocytosis-defective gB mutant and downmodulates their cell surface expression. J. Virol. 78:8015-8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Babic, N., B. Klupp, A. Brack, T. C. Mettenleiter, G. Ugolini, and A. Flamand. 1996. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology 219:279-284. [DOI] [PubMed] [Google Scholar]
- 13.Babic, N., B. G. Klupp, B. Makoschey, A. Karger, A. Flamand, and T. C. Mettenleiter. 1996. Glycoprotein gH of pseudorabies virus is essential for penetration and propagation in cell culture and in the nervous system of mice. J. Gen. Virol. 77 (Pt 9):2277-2285. [DOI] [PubMed] [Google Scholar]
- 14.Babic, N., T. C. Mettenleiter, G. Ugolini, A. Flamand, and P. Coulon. 1994. Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology 204:616-625. [DOI] [PubMed] [Google Scholar]
- 15.Baines, J. D., C. Cunningham, D. Nalwanga, and A. Davison. 1997. The U(L)15 gene of herpes simplex virus type 1 contains within its second exon a novel open reading frame that is translated in frame with the U(L)15 gene product. J. Virol. 71:2666-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baines, J. D., A. P. Poon, J. Rovnak, and B. Roizman. 1994. The herpes simplex virus 1 UL15 gene encodes two proteins and is required for cleavage of genomic viral DNA. J. Virol. 68:8118-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banfield, B. W., J. D. Kaufman, J. A. Randall, and G. E. Pickard. 2003. Development of pseudorabies virus strains expressing red fluorescent proteins: new tools for multisynaptic labeling applications. J. Virol. 77:10106-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banfield, B. W., G. S. Yap, A. C. Knapp, and L. W. Enquist. 1998. A chicken embryo eye model for the analysis of alphaherpesvirus neuronal spread and virulence. J. Virol. 72:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baradaran, K., M. A. Hardwicke, C. E. Dabrowski, and P. A. Schaffer. 1996. Properties of the novel herpes simplex virus type 1 origin binding protein, OBPC. J. Virol. 70:5673-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bareyre, F. M., M. Kerschensteiner, O. Raineteau, T. C. Mettenleiter, O. Weinmann, and M. E. Schwab. 2004. The injured spinal cord spontaneously forms a new intraspinal circuit in adult rats. Nat. Neurosci. 7:269-277. [DOI] [PubMed] [Google Scholar]
- 21.Barnett, E. M., M. D. Cassell, and S. Perlman. 1993. Two neurotropic viruses, herpes simplex virus type 1 and mouse hepatitis virus, spread along different neural pathways from the main olfactory bulb. Neuroscience 57:1007-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnett, E. M., G. D. Evans, N. Sun, S. Perlman, and M. D. Cassell. 1995. Anterograde tracing of trigeminal afferent pathways from the murine tooth pulp to cortex using herpes simplex virus type 1. J. Neurosci. 15:2972-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartha, A. 1961. Experimental reduction of virulence of Aujeszky's disease virus. Mag. Allatorv. Lapja 16:42-45. (In Hungarian.) [Google Scholar]
- 24.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumeister, J., B. G. Klupp, and T. C. Mettenleiter. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Porat, T., J. M. Demarchi, and A. S. Kaplan. 1974. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology 61:29-37. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Porat, T., J. M. DeMarchi, B. Lomniczi, and A. S. Kaplan. 1986. Role of glycoproteins of pseudorabies virus in eliciting neutralizing antibodies. Virology 154:325-334. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Porat, T., and A. S. Kaplan. 1976. A comparison of two populations of defective, interfering pseudorabies virus particles. Virology 72:471-479. [DOI] [PubMed] [Google Scholar]
- 29.Ben-Porat, T., and A. S. Kaplan. 1985. Molecular biology of pseudorabies virus, p. 105-173. In B. Roizman (ed.), The herpesviruses, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 30.Ben-Porat, T., T. Rakusanova, and A. S. Kaplan. 1971. Early functions of the genome of herpesvirus. II. Inhibition of the formation of cell-specific polysomes. Virology 46:890-899. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Porat, T., R. A. Veach, and H. Hampl. 1983. Functions of the major nonstructural DNA binding protein of a herpesvirus (pseudorabies). Virology 124:411-424. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology 127:194-204. [DOI] [PubMed] [Google Scholar]
- 33.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 101:9411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berthomme, H., B. Jacquemont, and A. Epstein. 1993. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology 193:1028-1032. [DOI] [PubMed] [Google Scholar]
- 35.Berthomme, H., S. J. Monahan, D. S. Parris, B. Jacquemont, and A. L. Epstein. 1995. Cloning, sequencing, and functional characterization of the two subunits of the pseudorabies virus DNA polymerase holoenzyme: evidence for specificity of interaction. J. Virol. 69:2811-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Billig, I., J. P. Card, and B. J. Yates. 2003. Neurochemical phenotypes of MRF neurons influencing diaphragm and rectus abdominis activity. J. Appl. Physiol. 94:391-398. [DOI] [PubMed] [Google Scholar]
- 37.Boehmer, P. E., and I. R. Lehman. 1997. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 66:347-384. [DOI] [PubMed] [Google Scholar]
- 38.Boldogkoi, Z., A. Braun, and I. Fodor. 2000. Replication and virulence of early protein 0 and long latency transcript deficient mutants of the Aujeszky's disease (pseudorabies) virus. Microbes Infect. 2:1321-1328. [DOI] [PubMed] [Google Scholar]
- 39.Boldogkoi, Z., I. Medveczky, R. Glavits, A. Braun, and I. Fodor. 1996. In vivo studies on Aujeszky's disease virus mutants. Acta Microbiol. Immunol. Hung. 43:307-318. [PubMed] [Google Scholar]
- 40.Boldogkoi, Z., and A. Nogradi. 2003. Gene and cancer therapy-pseudorabies virus: a novel research and therapeutic tool? Curr. Gene Ther. 3:155-182. [DOI] [PubMed] [Google Scholar]
- 41.Boldogkoi, Z., A. Sik, A. Denes, A. Reichart, J. Toldi, I. Gerendai, K. J. Kovacs, and M. Palkovits. 2004. Novel tracing paradigms-genetically engineered herpesviruses as tools for mapping functional circuits within the CNS: present status and future prospects. Prog. Neurobiol. 72:417-445. [DOI] [PubMed] [Google Scholar]
- 42.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booy, F. P., B. L. Trus, W. W. Newcomb, J. C. Brown, J. F. Conway, and A. C. Steven. 1994. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus). Proc. Natl. Acad. Sci. USA 91:5652-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 47.Brideau, A. D., J. P. Card, and L. W. Enquist. 2000. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat nervous system. J. Virol. 74:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brideau, A. D., M. G. Eldridge, and L. W. Enquist. 2000. Directional transneuronal infection by pseudorabies virus is dependent on an acidic internalization motif in the Us9 cytoplasmic tail. J. Virol. 74:4549-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brideau, A. D., L. W. Enquist, and R. S. Tirabassi. 2000. The role of virion membrane protein endocytosis in the herpesvirus life cycle. J. Clin. Virol. 17:69-82. [DOI] [PubMed] [Google Scholar]
- 50.Brittle, E. E. 2005. Two modes of pseudorabies virus neuroinvasion and lethality in mice. Ph.D. Thesis. Princeton University, Princeton, N.J. [DOI] [PMC free article] [PubMed]
- 51.Brittle, E. E., A. E. Reynolds, and L. W. Enquist. 2004. Two modes of pseudorabies virus neuroinvasion and lethality in mice. J. Virol. 78:12951-12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brockmeier, S. L., K. M. Lager, and W. L. Mengeling. 1993. Comparison of in vivo reactivation, in vitro reactivation, and polymerase chain reaction for detection of latent pseudorabies virus infection in swine. J. Vet. Diagn. Investig. 5:505-509. [DOI] [PubMed] [Google Scholar]
- 53.Brown, T. T., Jr., K. O. Shin, and F. J. Fuller. 1995. Detection of pseudorabies viral DNA in tonsillar epithelial cells of latently infected pigs. Am. J. Vet. Res. 56:587-594. [PubMed] [Google Scholar]
- 54.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calton, C. M., J. A. Randall, M. W. Adkins, and B. W. Banfield. 2004. The pseudorabies virus serine/threonine kinase Us3 contains mitochondrial, nuclear and membrane localization signals. Virus Genes 29:131-145. [DOI] [PubMed] [Google Scholar]
- 56.Campadelli-Fiume, G., M. Arsenakis, F. Farabegoli, and B. Roizman. 1988. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J. Virol. 62:159-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 58.Campbell, M. E., and C. M. Preston. 1987. DNA sequences which regulate the expression of the pseudorabies virus major immediate early gene. Virology 157:307-316. [DOI] [PubMed] [Google Scholar]
- 59.Cano, G., J. P. Card, and A. F. Sved. 2004. Dual viral transneuronal tracing of central autonomic circuits involved in the innervation of the two kidneys in rat. J. Comp. Neurol 471:462-481. [DOI] [PubMed] [Google Scholar]
- 60.Capua, I., R. Fico, M. Banks, M. Tamba, and G. Calzetta. 1997. Isolation and characterisation of an Aujeszky's disease virus naturally infecting a wild boar (Sus scrofa). Vet. Microbiol. 55:141-146. [DOI] [PubMed] [Google Scholar]
- 61.Card, J. P., J. R. Dubin, M. E. Whealy, and L. W. Enquist. 1995. Influence of infectious dose upon productive replication and transynaptic passage of pseudorabies virus in rat central nervous system. J. Neurovirol. 1:349-358. [DOI] [PubMed] [Google Scholar]
- 62.Card, J. P., L. W. Enquist, and R. Y. Moore. 1999. Neuroinvasiveness of pseudorabies virus injected intracerebrally is dependent on viral concentration and terminal field density. J. Comp. Neurol. 407:438-452. [DOI] [PubMed] [Google Scholar]
- 63.Card, J. P., L. Rinaman, R. B. Lynn, B. H. Lee, R. P. Meade, R. R. Miselis, and L. W. Enquist. 1993. Pseudorabies virus infection of the rat central nervous system: ultrastructural characterization of viral replication, transport, and pathogenesis. J. Neurosci. 13:2515-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Card, J. P., L. Rinaman, J. S. Schwaber, R. R. Miselis, M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1990. Neurotropic properties of pseudorabies virus: uptake and transneuronal passage in the rat central nervous system. J. Neurosci. 10:1974-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Card, J. P., M. E. Whealy, A. K. Robbins, and L. W. Enquist. 1992. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J. Virol. 66:3032-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Card, J. P., M. E. Whealy, A. K. Robbins, R. Y. Moore, and L. W. Enquist. 1991. Two alpha-herpesvirus strains are transported differentially in the rodent visual system. Neuron 6:957-969. [DOI] [PubMed] [Google Scholar]
- 67.Carr, D. B., P. O'Donnell, J. P. Card, and S. R. Sesack. 1999. Dopamine terminals in the rat prefrontal cortex synapse on pyramidal cells that project to the nucleus accumbens. J. Neurosci. 19:11049-11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cartier, A., E. Broberg, T. Komai, M. Henriksson, and M. G. Masucci. 2003. The herpes simplex virus-1 Us3 protein kinase blocks CD8T cell lysis by preventing the cleavage of Bid by granzyme B. Cell Death Differ. 10:1320-1328. [DOI] [PubMed] [Google Scholar]
- 69.Cartier, A., T. Komai, and M. G. Masucci. 2003. The Us3 protein kinase of herpes simplex virus 1 blocks apoptosis and induces phosporylation of the Bcl-2 family member Bad. Exp. Cell Res. 291:242-250. [DOI] [PubMed] [Google Scholar]
- 70.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ch'ng, T. H., and L. W. Enquist. 2005. Efficient axonal localization of alphaherpesvirus structural proteins in cultured sympathetic neurons requires viral glycoprotein E. J. Virol. 79:8835-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ch'ng, T. H., and L. W. Enquist. Neuron-to-cell spread of pseudorabies virus in a compartmented neuronal culture system. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 73.Chang, Y. Y., H. W. Lin, M. L. Wong, and T. J. Chang. 2004. Regulation of the vhs gene promoter of pseudorabies virus by IE180 and EP0, and the requirement of a Sp1 Site for the promoter function. Virus Genes 28:247-258. [DOI] [PubMed] [Google Scholar]
- 74.Chen, S., M. Yang, R. R. Miselis, and G. Aston-Jones. 1999. Characterization of transsynaptic tracing with central application of pseudorabies virus. Brain Res. 838:171-183. [DOI] [PubMed] [Google Scholar]
- 75.Chen, X., M. C. Schmidt, W. F. Goins, and J. C. Glorioso. 1995. Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections. J. Virol. 69:7899-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheung, A. K. 1991. Cloning of the latency gene and the early protein 0 gene of pseudorabies virus. J. Virol. 65:5260-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheung, A. K. 1989. Detection of pseudorabies virus transcripts in trigeminal ganglia of latently infected swine. J. Virol. 63:2908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cheung, A. K. 1989. DNA nucleotide sequence analysis of the immediate-early gene of pseudorabies virus. Nucleic Acids Res. 17:4637-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheung, A. K. 1988. Fine mapping of the immediate-early gene of the Indiana-Funkhauser strain of pseudorabies virus. J. Virol. 62:4763-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheung, A. K. 1996. Latency characteristics of an EPO and LLT mutant of pseudorabies virus. J. Vet. Diagn. Investig. 8:112-115. [DOI] [PubMed] [Google Scholar]
- 81.Cheung, A. K., J. Fang, and R. D. Wesley. 1994. Characterization of a pseudorabies virus that is defective in the early protein 0 and latency genes. Am. J. Vet. Res. 55:1710-1716. [PubMed] [Google Scholar]
- 82.Cheung, A. K., and T. A. Smith. 1999. Analysis of the latency-associated transcript/UL1-3.5 gene cluster promoter complex of pseudorabies virus. Arch. Virol. 144:381-391. [DOI] [PubMed] [Google Scholar]
- 83.Cheung, P., K. S. Ellison, R. Verity, and J. R. Smiley. 2000. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated alpha-globin pre-mRNA in infected HeLa cells. J. Virol. 74:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 85.Chien, C. H., J. Y. Shieh, M. H. Liao, E. A. Ling, and C. Y. Wen. 1998. Neuronal connections between the auricular skin and the sympathetic pre- and postganglionic neurons of the dog as studied by using pseudorabies virus. Neurosci. Res. 30:169-175. [DOI] [PubMed] [Google Scholar]
- 86.Chlan, C. A., C. Coulter, and L. T. Feldman. 1987. Binding of the pseudorabies virus immediate-early protein to single-stranded DNA. J. Virol. 61:1855-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ciernik, I. F., J. A. Berzofsky, and D. P. Carbone. 1996. Induction of cytotoxic T lymphocytes and antitumor immunity with DNA vaccines expressing single T cell epitopes. J. Immunol. 156:2369-2375. [PubMed] [Google Scholar]
- 88.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coe, N. E., and W. L. Mengeling. 1990. Mapping and characterization of neutralizing epitopes of glycoproteins gIII and gp50 of the Indiana-Funkhauser strain of pseudorabies virus. Arch. Virol. 110:137-142. [DOI] [PubMed] [Google Scholar]
- 90.Cohen, G. H., D. Long, J. T. Matthews, M. May, and R. Eisenberg. 1983. Glycopeptides of the type-common glycoprotein gD of herpes simplex virus types 1 and 2. J. Virol. 46:679-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cohen, J. I., and S. E. Straus. 2001. Varicella-zoster virus and its replication, p. 2707-2730. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 92.Corn, J. L., D. E. Stallknecht, N. M. Mechlin, M. P. Luttrell, and J. R. Fischer. 2004. Persistence of pseudorabies virus in feral swine populations. J. Wildl. Dis. 40:307-310. [DOI] [PubMed] [Google Scholar]
- 93.Cromlish, W. A., S. M. Abmayr, J. L. Workman, M. Horikoshi, and R. G. Roeder. 1989. Transcriptionally active immediate-early protein of pseudorabies virus binds to specific sites on class II gene promoters. J. Virol. 63:1869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 95.Dargan, D. J., and J. H. Subak-Sharpe. 1997. The effect of herpes simplex virus type 1 L-particles on virus entry, replication, and the infectivity of naked herpesvirus DNA. Virology 239:378-388. [DOI] [PubMed] [Google Scholar]
- 96.Davies, E. B., and G. W. Beran. 1980. Spontaneous shedding of pseudorabies virus from a clinically recovered postparturient sow. J. Am. Vet. Med. Assoc. 176:1345-1347. [PubMed] [Google Scholar]
- 97.Davis-Poynter, N. J., and H. E. Farrell. 1996. Masters of deception: a review of herpesvirus immune evasion strategies. Immunol. Cell Biol. 74:513-522. [DOI] [PubMed] [Google Scholar]
- 98.Davis, S. F., K. W. Williams, W. Xu, N. R. Glatzer, and B. N. Smith. 2003. Selective enhancement of synaptic inhibition by hypocretin (orexin) in rat vagal motor neurons: implications for autonomic regulation. J. Neurosci. 23:3844-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davison, A. J. 1992. Channel catfish virus: a new type of herpesvirus. Virology 186:9-14. [DOI] [PubMed] [Google Scholar]
- 100.Davison, A. J. 2002. Evolution of the herpesviruses. Vet. Microbiol. 86:69-88. [DOI] [PubMed] [Google Scholar]
- 101.de Groat, W. C., I. Araki, M. A. Vizzard, M. Yoshiyama, N. Yoshimura, K. Sugaya, C. Tai, and J. R. Roppolo. 1998. Developmental and injury induced plasticity in the micturition reflex pathway. Behav. Brain Res. 92:127-140. [DOI] [PubMed] [Google Scholar]
- 102.de Leeuw, P. W., and J. T. van Oirschot. 1985. Vaccines against Aujeszky's disease: evaluation of their efficacy under standardized laboratory conditions. Vet. Q. 7:191-197. [DOI] [PubMed] [Google Scholar]
- 103.de Wind, N., A. Berns, A. Gielkens, and T. Kimman. 1993. Ribonucleotide reductase-deficient mutants of pseudorabies virus are avirulent for pigs and induce partial protective immunity. J. Gen. Virol. 74:351-359. [DOI] [PubMed] [Google Scholar]
- 104.de Wind, N., B. P. Peeters, A. Zuderveld, A. L. Gielkens, A. J. Berns, and T. G. Kimman. 1994. Mutagenesis and characterization of a 41-kilobase-pair region of the pseudorabies virus genome: transcription map, search for virulence genes, and comparison with homologs of herpes simplex virus type 1. Virology 200:784-790. [DOI] [PubMed] [Google Scholar]
- 105.Dean, H. J., and A. K. Cheung. 1993. A 3′ coterminal gene cluster in pseudorabies virus contains herpes simplex virus UL1, UL2, and UL3 gene homologs and a unique UL3.5 open reading frame. J. Virol. 67:5955-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DeFalco, J., M. Tomishima, H. Liu, C. Zhao, X. Cai, J. D. Marth, L. Enquist, and J. M. Friedman. 2001. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291:2608-2613. [DOI] [PubMed] [Google Scholar]
- 107.del Rio, T., T. H. Ch'ng, E. A. Flood, S. P. Gross, and L. W. Enquist. 2005. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J. Virol. 79:3903-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Del Rio, T., C. J. Decoste, and L. W. Enquist. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dézélée, S., F. Bras, P. Vende, B. Simonet, X. Nguyen, A. Flamand, and M. J. Masse. 1996. The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL 26.5 genes of herpes simplex virus type 1. Virus Res. 42:27-39. [DOI] [PubMed] [Google Scholar]
- 111.Dietz, P., B. G. Klupp, W. Fuchs, B. Kollner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dijkstra, J. M., W. Fuchs, T. C. Mettenleiter, and B. G. Klupp. 1997. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch. Virol. 142:17-35. [DOI] [PubMed] [Google Scholar]
- 113.Döhner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Elgadi, M. M., C. E. Hayes, and J. R. Smiley. 1999. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J. Virol. 73:7153-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eloit, M., D. Fargeaud, R. L'Haridon, and B. Toma. 1988. Identification of the pseudorabies virus glycoprotein gp50 as a major target of neutralizing antibodies. Arch. Virol. 99:45-56. [DOI] [PubMed] [Google Scholar]
- 116.Enquist, L. W. 2002. Exploiting circuit-specific spread of pseudorabies virus in the central nervous system: insights to pathogenesis and circuit tracers. J. Infect. Dis. 186(Suppl. 2):S209-214. [DOI] [PubMed] [Google Scholar]
- 117.Enquist, L. W. 1994. Infection of the mammalian nervous system by pseudorabies virus (PRV). Semin. Virol. 5:221-231. [Google Scholar]
- 118.Enquist, L. W. 1999. Life beyond eradication: veterinary viruses in basic science. Arch. Virol. Suppl. 15:87-109. [DOI] [PubMed] [Google Scholar]
- 119.Enquist, L. W., and J. P. Card. 2003. Recent advances in the use of neurotropic viruses for circuit analysis. Curr. Opin. Neurobiol. 13:603-606. [DOI] [PubMed] [Google Scholar]
- 120.Enquist, L. W., J. Dubin, M. E. Whealy, and J. P. Card. 1994. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J. Virol. 68:5275-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 122.Fabian, R. H., and J. D. Coulter. 1985. Transneuronal transport of lectins. Brain Res. 344:41-48. [DOI] [PubMed] [Google Scholar]
- 123.Farnsworth, A., K. Goldsmith, and D. C. Johnson. 2003. Herpes simplex virus glycoproteins gD and gE/gI serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J. Virol. 77:8481-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Farrell, M. J., A. T. Dobson, and L. T. Feldman. 1991. Herpes simplex virus latency-associated transcript is a stable intron. Proc. Natl. Acad. Sci. USA 88:790-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Favoreel, H. W., H. J. Nauwynck, and M. B. Pensaert. 1999. Role of the cytoplasmic tail of gE in antibody-induced redistribution of viral glycoproteins expressed on pseudorabies-virus-infected cells. Virology 259:141-147. [DOI] [PubMed] [Google Scholar]
- 126.Favoreel, H. W., G. Van Minnebruggen, H. J. Nauwynck, L. W. Enquist, and M. B. Pensaert. 2002. A tyrosine-based motif in the cytoplasmic tail of pseudorabies virus glycoprotein B is important for both antibody-induced internalization of viral glycoproteins and efficient cell-to-cell spread. J. Virol. 76:6845-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Feldman, L., F. J. Rixon, J. H. Jean, T. Ben-Porat, and A. S. Kaplan. 1979. Transcription of the genome of pseudorabies virus (a herpesvirus) is strictly controlled. Virology 97:316-327. [DOI] [PubMed] [Google Scholar]
- 128.Field, H. J., and T. J. Hill. 1974. The pathogenesis of pseudorabies in mice following peripheral inoculation. J. Gen. Virol. 23:145-157. [DOI] [PubMed] [Google Scholar]
- 129.Flamand, A., T. Bennardo, N. Babic, B. G. Klupp, and T. C. Mettenleiter. 2001. The absence of glycoprotein gL, but not gC or gK, severely impairs pseudorabies virus neuroinvasiveness. J. Virol. 75:11137-11145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fort, P., P. H. Luppi, and M. Jouvet. 1994. Afferents to the nucleus reticularis parvicellularis of the cat medulla oblongata: a tract-tracing study with cholera toxin B subunit. J. Comp. Neurol. 342:603-618. [DOI] [PubMed] [Google Scholar]
- 131.Frenkel, N., R. J. Jacob, R. W. Honess, G. S. Hayward, H. Locker, and B. Roizman. 1975. Anatomy of herpes simplex virus DNA. III. Characterization of defective DNA molecules and biological properties of virus populations containing them. J. Virol. 16:153-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fuchs, W., C. Ehrlich, B. G. Klupp, and T. C. Mettenleiter. 2000. Characterization of the replication origin (Ori(S)) and adjoining parts of the inverted repeat sequences of the pseudorabies virus genome. J. Gen. Virol. 81:1539-1543. [DOI] [PubMed] [Google Scholar]
- 133.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 141.Garner, J. A., and J. H. LaVail. 1999. Differential anterograde transport of HSV type 1 viral strains in the murine optic pathway. J. Neurovirol. 5:140-150. [DOI] [PubMed] [Google Scholar]
- 142.Geenen, K., H. W. Favoreel, and H. J. Nauwynck. 2005. Higher resistance of porcine trigeminal ganglion neurons towards pseudorabies virus-induced cell death compared with other porcine cell types in vitro. J. Gen. Virol. 86:1251-1260. [DOI] [PubMed] [Google Scholar]
- 143.Geenen, K., H. W. Favoreel, L. Olsen, L. W. Enquist, and H. J. Nauwynck. 2005. The pseudorabies virus US3 protein kinase possesses anti-apoptotic activity that protects cells from apoptosis during infection and after treatment with sorbitol or staurosporine. Virology 331:144-150. [DOI] [PubMed] [Google Scholar]
- 144.Geerling, J. C., T. C. Mettenleiter, and A. D. Loewy. 2003. Orexin neurons project to diverse sympathetic outflow systems. Neuroscience 122:541-550. [DOI] [PubMed] [Google Scholar]
- 145.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 146.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 147.Gerdts, V., A. Jöns, B. Makoschey, N. Visser, and T. C. Mettenleiter. 1997. Protection of pigs against Aujeszky's disease by DNA vaccination. J. Gen. Virol. 78:2139-2146. [DOI] [PubMed] [Google Scholar]
- 148.Gerdts, V., A. Jöns, and T. C. Mettenleiter. 1999. Potency of an experimental DNA vaccine against Aujeszky's disease in pigs. Vet. Microbiol. 66:1-13. [DOI] [PubMed] [Google Scholar]
- 149.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. 8. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Glass, C. M., R. G. McLean, J. B. Katz, D. S. Maehr, C. B. Cropp, L. J. Kirk, A. J. McKeirnan, and J. F. Evermann. 1994. Isolation of pseudorabies (Aujeszky's disease) virus from a Florida panther. J. Wildl. Dis. 30:180-184. [DOI] [PubMed] [Google Scholar]
- 151.Glatzer, N. R., C. P. Hasney, M. D. Bhaskaran, and B. N. Smith. 2003. Synaptic and morphologic properties in vitro of premotor rat nucleus tractus solitarius neurons labeled transneuronally from the stomach. J. Comp. Neurol. 464:525-539. [DOI] [PubMed] [Google Scholar]
- 152.Glazenburg, K. L., B. P. Peeters, J. M. Pol, A. L. Gielkens, and R. J. Moormann. 1995. Construction and properties of pseudorabies virus recombinants with altered control of immediate-early gene expression. J. Virol. 69:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Green, M. R., R. Treisman, and T. Maniatis. 1983. Transcriptional activation of cloned human beta-globin genes by viral immediate-early gene products. Cell 35:137-148. [DOI] [PubMed] [Google Scholar]
- 158.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of Herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 159.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 160.Gustafson, D. P. 1986. Pseudorabies, p. 209-223. In A. D. Leman, R. D. Glock, W. L. Mengeling, R. H. C. Penny, E. Scholl, and B. Straw (ed.), Diseases of swine, 6th ed. Iowa State University Press, Ames, Iowa.
- 161.Gutekunst, D. E., E. C. Pirtle, L. D. Miller, and W. C. Stewart. 1980. Isolation of pseudorabies virus from trigeminal ganglia of a latently infected sow. Am. J. Vet. Res. 41:1315-1316. [PubMed] [Google Scholar]
- 162.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hahn, E. C., G. R. Page, P. S. Hahn, K. D. Gillis, C. Romero, J. A. Annelli, and E. P. Gibbs. 1997. Mechanisms of transmission of Aujeszky's disease virus originating from feral swine in the USA. Vet. Microbiol. 55:123-130. [DOI] [PubMed] [Google Scholar]
- 164.Hampl, H., T. Ben-Porat, L. Ehrlicher, K. O. Habermehl, and A. S. Kaplan. 1984. Characterization of the envelope proteins of pseudorabies virus. J. Virol. 52:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hanson, R. P. 1954. The history of pseudorabies in the United States. J. Am. Vet. Med. Assoc. 124:259-261. [PubMed] [Google Scholar]
- 166.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Harley, C. A., A. Dasgupta, and D. W. Wilson. 2001. Characterization of herpes simplex virus-containing organelles by subcellular fractionation: role for organelle acidification in assembly of infectious particles. J. Virol. 75:1236-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Harper, L., J. Demarchi, and T. Ben-Porat. 1986. Sequence of the genome ends and of the junction between the ends in concatemeric DNA of pseudorabies virus. J. Virol. 60:1183-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Harrison, P. J., H. Hultborn, E. Jankowska, R. Katz, B. Storai, and D. Zytnicki. 1984. Labelling of interneurones by retrograde transsynaptic transport of horseradish peroxidase from motoneurones in rats and cats. Neurosci. Lett. 45:15-19. [DOI] [PubMed] [Google Scholar]
- 171.Hassett, D. E., and J. L. Whitton. 1996. DNA immunization. Trends Microbiol. 4:307-312. [DOI] [PubMed] [Google Scholar]
- 172.Hata, A. N., and R. M. Breyer. 2004. Pharmacology and signaling of prostaglandin receptors: multiple roles in inflammation and immune modulation. Pharmacol. Ther. 103:147-166. [DOI] [PubMed] [Google Scholar]
- 173.Hata, S., A. H. Koyama, H. Shiota, A. Adachi, F. Goshima, and Y. Nishiyama. 1999. Antiapoptotic activity of herpes simplex virus type 2: the role of US3 protein kinase gene. Microbes Infect. 1:601-607. [DOI] [PubMed] [Google Scholar]
- 174.Henry, B. E., W. W. Newcomb, and D. J. O'Callaghan. 1979. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology 92:495-506. [DOI] [PubMed] [Google Scholar]
- 175.Hill, A. B., B. C. Barnett, A. J. McMichael, and D. J. McGeoch. 1994. HLA class I molecules are not transported to the cell surface in cells infected with herpes simplex virus types 1 and 2. J. Immunol. 152:2736-2741. [PubMed] [Google Scholar]
- 176.Ho, T. Y., S. L. Wu, T. J. Chang, C. H. Hsiang, S. H. Chang, and C. Y. Hsiang. 1999. Pseudorabies virus early protein 0 trans-activates the TATA-associated promoter by stimulating the transcription initiation. Virus Res. 61:77-86. [DOI] [PubMed] [Google Scholar]
- 177.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 178.Horvath, S., E. Prandovszky, E. Pankotai, Z. Kis, T. Farkas, Z. Boldogkoi, K. Boda, Z. Janka, and J. Toldi. 2005. Use of a recombinant pseudorabies virus to analyze motor cortical reorganization after unilateral facial denervation. Cereb. Cortex 15:378-384. [DOI] [PubMed] [Google Scholar]
- 179.Hsiang, C. Y. 2002. Pseudorabies virus DNA-binding protein stimulates the exonuclease activity and regulates the processivity of pseudorabies virus DNase. Biochem. Biophys. Res. Commun. 293:1301-1308. [DOI] [PubMed] [Google Scholar]
- 180.Hsiang, C. Y., T. Y. Ho, and T. J. Chang. 1996. Identification of a pseudorabies virus UL12 (deoxyribonuclease) gene. Gene 177:109-113. [DOI] [PubMed] [Google Scholar]
- 181.Hsiang, C. Y., T. Y. Ho, C. H. Hsiang, and T. J. Chang. 1998. Recombinant pseudorabies virus DNase exhibits a RecBCD-like catalytic function. Biochem. J. 330:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Huang, C., and C. Y. Wu. 2004. Characterization and expression of the pseudorabies virus early gene UL54. J. Virol. Methods 119:129-136. [DOI] [PubMed] [Google Scholar]
- 183.Huang, C. J., M. K. Rice, G. B. Devi-Rao, and E. K. Wagner. 1994. The activity of the pseudorabies virus latency-associated transcript promoter is dependent on its genomic location in herpes simplex virus recombinants as well as on the type of cell infected. J. Virol. 68:1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Husak, P. J., T. Kuo, and L. W. Enquist. 2000. Pseudorabies virus membrane proteins gI and gE facilitate anterograde spread of infection in projection-specific neurons in the rat. J. Virol. 74:10975-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ihara, S., L. Feldman, S. Watanabe, and T. Ben-Porat. 1983. Characterization of the immediate-early functions of pseudorabies virus. Virology 131:437-454. [DOI] [PubMed] [Google Scholar]
- 186.Ikenaga, T., Y. Tatecho, T. Nakai, and K. Uematsu. 2002. Betanodavirus as a novel transneuronal tracer for fish. Neurosci. Lett. 331:55-59. [DOI] [PubMed] [Google Scholar]
- 187.Imperiale, M. J., L. T. Feldman, and J. R. Nevins. 1983. Activation of gene expression by adenovirus and herpesvirus regulatory genes acting in trans and by a cis-acting adenovirus enhancer element. Cell 35:127-136. [DOI] [PubMed] [Google Scholar]
- 188.Irnaten, M., R. A. Neff, J. Wang, A. D. Loewy, T. C. Mettenleiter, and D. Mendelowitz. 2001. Activity of cardiorespiratory networks revealed by transsynaptic virus expressing GFP. J. Neurophysiol. 85:435-438. [DOI] [PubMed] [Google Scholar]
- 189.Jansen, A. S., X. V. Nguyen, V. Karpitskiy, T. C. Mettenleiter, and A. D. Loewy. 1995. Central command neurons of the sympathetic nervous system: basis of the fight-or-flight response. Science 270:644-646. [DOI] [PubMed] [Google Scholar]
- 190.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 73:8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jin, L., and G. Scherba. 1999. Expression of the pseudorabies virus latency-associated transcript gene during productive infection of cultured cells. J. Virol. 73:9781-9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jin, L., W. M. Schnitzlein, and G. Scherba. 2000. Identification of the pseudorabies virus promoter required for latency-associated transcript gene expression in the natural host. J. Virol. 74:6333-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jones, J. O., and A. M. Arvin. 2003. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 77:1268-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jöns, A., V. Gerdts, E. Lange, V. Kaden, and T. C. Mettenleiter. 1997. Attenuation of dUTPase-deficient pseudorabies virus for the natural host. Vet. Microbiol. 56:47-54. [DOI] [PubMed] [Google Scholar]
- 197.Jöns, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jöns, A., and T. C. Mettenleiter. 1996. Identification and characterization of pseudorabies virus dUTPase. J. Virol. 70:1242-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Justewicz, D. M., M. J. Morin, H. L. Robinson, and R. G. Webster. 1995. Antibody-forming cell response to virus challenge in mice immunized with DNA encoding the influenza virus hemagglutinin. J. Virol. 69:7712-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kaliman, A. V., Z. Boldogkoi, and I. Fodor. 1994. Large and small subunits of the Aujeszky's disease virus ribonucleotide reductase: nucleotide sequence and putative structure. Biochim. Biophys. Acta 1219:151-156. [DOI] [PubMed] [Google Scholar]
- 202.Katan, M., M. J. McGarvey, W. S. Stevely, and D. P. Leader. 1986. The phosphorylation of ribosomal protein S6 by protein kinases from cells infected with pseudorabies virus. Biochem. J. 239:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Kaufman, G. D., M. J. Mustari, R. R. Miselis, and A. A. Perachio. 1996. Transneuronal pathways to the vestibulocerebellum. J. Comp. Neurol. 370:501-523. [DOI] [PubMed] [Google Scholar]
- 204.Kelly, R. M., and P. L. Strick. 2003. Cerebellar loops with motor cortex and prefrontal cortex of a nonhuman primate. J. Neurosci. 23:8432-8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kelly, R. M., and P. L. Strick. 2000. Rabies as a transneuronal tracer of circuits in the central nervous system. J. Neurosci. Methods 103:63-71. [DOI] [PubMed] [Google Scholar]
- 206.Kim, J. S., L. W. Enquist, and J. P. Card. 1999. Circuit-specific coinfection of neurons in the rat central nervous system with two pseudorabies virus recombinants. J. Virol. 73:9521-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kimman, T. G., G. J. Binkhorst, T. S. van den Ingh, J. M. Pol, A. L. Gielkens, and M. E. Roelvink. 1991. Aujeszky's disease in horses fulfils Koch's postulates. Vet. Rec. 128:103-106. [DOI] [PubMed] [Google Scholar]
- 208.Kit, S., M. Kit, and E. C. Pirtle. 1985. Attenuated properties of thymidine kinase-negative deletion mutant of pseudorabies virus. Am. J. Vet. Res. 46:1359-1367. [PubMed] [Google Scholar]
- 209.Kit, S., M. Sheppard, H. Ichimura, and M. Kit. 1987. Second-generation pseudorabies virus vaccine with deletions in thymidine kinase and glycoprotein genes. Am. J. Vet. Res. 48:780-793. [PubMed] [Google Scholar]
- 210.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kluge, J. P., G. W. Beran, H. T. Hill, and K. B. Platt. 1999. Pseudorabies (Aujeszky's disease), p. 233-46. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and T. J. Taylor (ed.), Diseases of swine, 8th ed. Iowa State University Press, Ames, Iowa.
- 212.Klupp, B. G., J. Altenschmidt, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2005. Identification and characterization of the pseudorabies virus UL43 protein. Virology 334:224-233. [DOI] [PubMed] [Google Scholar]
- 213.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Klupp, B. G., H. Granzow, R. Klopfleisch, W. Fuchs, M. Kopp, M. Lenk, and T. C. Mettenleiter. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 216.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. Identification, subviral localization and functional characterization of the pseudorabies virus UL17 protein. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 217.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Klupp, B. G., C. J. Hengartner, T. C. Mettenleiter, and L. W. Enquist. 2004. Complete, annotated sequence of the pseudorabies virus genome. J. Virol. 78:424-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Klupp, B. G., B. Lomniczi, N. Visser, W. Fuchs, and T. C. Mettenleiter. 1995. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology 212:466-473. [DOI] [PubMed] [Google Scholar]
- 221.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Knapp, A. C., P. J. Husak, and L. W. Enquist. 1997. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J. Virol. 71:5820-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Konig, P., K. Giesow, and G. M. Keil. 2002. Glycoprotein M of bovine herpesvirus 1 (BHV-1) is nonessential for replication in cell culture and is involved in inhibition of bovine respiratory syncytial virus F protein induced syncytium formation in recombinant BHV-1 infected cells. Vet. Microbiol. 86:37-49. [DOI] [PubMed] [Google Scholar]
- 225.Kopp, M., H. Granzow, W. Fuchs, B. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Koppers-Lalic, D., E. A. Reits, M. E. Ressing, A. D. Lipinska, R. Abele, J. Koch, M. Marcondes Rezende, P. Admiraal, D. van Leeuwen, K. Bienkowska-Szewczyk, T. C. Mettenleiter, F. A. Rijsewijk, R. Tampe, J. Neefjes, and E. J. Wiertz. 2005. Varicelloviruses avoid T cell recognition by UL49.5-mediated inactivation of the transporter associated with antigen processing. Proc. Natl. Acad. Sci. USA 102:5144-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Koslowski, K. M., P. R. Shaver, X. Y. Wang, D. J. Tenney, and N. E. Pederson. 1997. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J. Virol. 71:9118-9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Koyama, A. H., T. Fukumori, M. Fujita, H. Irie, and A. Adachi. 2000. Physiological significance of apoptosis in animal virus infection. Microbes Infect. 2:1111-1117. [DOI] [PubMed] [Google Scholar]
- 231.Koyano, S., E. C. Mar, F. R. Stamey, and N. Inoue. 2003. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J. Gen. Virol. 84:1485-1491. [DOI] [PubMed] [Google Scholar]
- 232.Kozmik, Z., L. Arnold, and V. Paces. 1991. Multiple sets of adjacent mu E1 and oct-1 binding sites upstream of the pseudorabies virus immediate-early gene promoter. Virology 182:239-249. [DOI] [PubMed] [Google Scholar]
- 233.Kritas, S. K., M. B. Pensaert, and T. C. Mettenleiter. 1994. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J. Gen. Virol. 75:2319-2327. [DOI] [PubMed] [Google Scholar]
- 234.Krout, K. E., T. C. Mettenleiter, and A. D. Loewy. 2003. Single CNS neurons link both central motor and cardiosympathetic systems: a double-virus tracing study. Neuroscience 118:853-866. [DOI] [PubMed] [Google Scholar]
- 235.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ladin, B. F., M. L. Blankenship, and T. Ben-Porat. 1980. Replication of herpesvirus DNA. V. Maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J. Virol. 33:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Ladin, B. F., S. Ihara, H. Hampl, and T. Ben-Porat. 1982. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology 116:544-561. [DOI] [PubMed] [Google Scholar]
- 238.Lake, C. M., S. J. Molesworth, and L. M. Hutt-Fletcher. 1998. The Epstein-Barr virus (EBV) gN homolog BLRF1 encodes a 15-kilodalton glycoprotein that cannot be authentically processed unless it is coexpressed with the EBV gM homolog BBRF3. J. Virol. 72:5559-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 240.Leader, D. P., A. D. Deana, F. Marchiori, F. C. Purves, and L. A. Pinna. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim. Biophys. Acta 1091:426-431. [DOI] [PubMed] [Google Scholar]
- 241.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 242.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lin, H. W., Y. Y. Chang, M. L. Wong, J. W. Lin, and T. J. Chang. 2004. Functional analysis of virion host shutoff protein of pseudorabies virus. Virology 324:412-418. [DOI] [PubMed] [Google Scholar]
- 244.Lindberg, A., and J. P. Kreivi. 2002. Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology 294:189-198. [DOI] [PubMed] [Google Scholar]
- 245.Lomniczi, B., A. S. Kaplan, and T. Ben-Porat. 1987. Multiple defects in the genome of pseudorabies virus can affect virulence without detectably affecting replication in cell culture. Virology 161:181-189. [DOI] [PubMed] [Google Scholar]
- 246.Lomniczi, B., S. Watanabe, T. Ben-Porat, and A. S. Kaplan. 1984. Genetic basis of the neurovirulence of pseudorabies virus. J. Virol. 52:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lomniczi, B., S. Watanabe, T. Ben-Porat, and A. S. Kaplan. 1987. Genome location and identification of functions defective in the Bartha vaccine strain of pseudorabies virus. J. Virol. 61:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lundh, B. 1990. Spread of vesicular stomatitis virus along the visual pathways after retinal infection in the mouse. Acta Neuropathol. (Berlin) 79:395-401. [DOI] [PubMed] [Google Scholar]
- 249.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Lyman, M. G., G. L. Demmin, and B. W. Banfield. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J. Virol. 77:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mach, M., B. Kropff, P. Dal Monte, and W. Britt. 2000. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 74:11881-11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 253.Maes, R. K., M. D. Sussman, A. Vilnis, and B. J. Thacker. 1997. Recent developments in latency and recombination of Aujeszky's disease (pseudorabies) virus. Vet. Microbiol. 55:13-27. [DOI] [PubMed] [Google Scholar]
- 254.Maga, G., A. Verri, L. Bonizzi, W. Ponti, G. Poli, A. Garbesi, D. Niccolai, S. Spadari, and F. Focher. 1993. Lack of stereospecificity of suid pseudorabies virus thymidine kinase. Biochem. J. 294:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Manickan, E., R. J. Rouse, Z. Yu, W. S. Wire, and B. T. Rouse. 1995. Genetic immunization against herpes simplex virus. Protection is mediated by CD4+ T lymphocytes. J. Immunol. 155:259-265. [PubMed] [Google Scholar]
- 256.Marchioli, C., R. J. Yancey, Jr., J. G. Timmins, L. E. Post, B. R. Young, and D. A. Povendo. 1988. Protection of mice and swine from pseudorabies virus-induced mortality by administration of pseudorabies virus-specific mouse monoclonal antibodies. Am. J. Vet. Res. 49:860-864. [PubMed] [Google Scholar]
- 257.Marchioli, C. C., R. J. Yancey, Jr., E. A. Petrovskis, J. G. Timmins, and L. E. Post. 1987. Evaluation of pseudorabies virus glycoprotein gp50 as a vaccine for Aujeszky's disease in mice and swine: expression by vaccinia virus and Chinese hamster ovary cells. J. Virol. 61:3977-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Martin, K. J., J. W. Lillie, and M. R. Green. 1990. Transcriptional activation by the pseudorabies virus immediate early protein. Genes Dev. 4:2376-2382. [DOI] [PubMed] [Google Scholar]
- 259.Masic, M., M. Ercegan, and M. Petrovic. 1965. The significance of the tonsils in the pathogenesis and diagnosis of Aujeszyk's disease in pigs. Zentralbl. Veterinarmed. B. 12:398-405. (In German.) [PubMed] [Google Scholar]
- 260.Masse, M. J., A. Jöns, J. M. Dijkstra, T. C. Mettenleiter, and A. Flamand. 1999. Glycoproteins gM and gN of pseudorabies virus are dispensable for viral penetration and propagation in the nervous systems of adult mice. J. Virol. 73:10503-10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Matsuda Tsuchida, A., S. Katayama, N. Okada, T. Okabe, and N. Sasaki. 1992. Protection from pseudorabies virus challenge in mice by a combination of purified gII, gIII and gVI antigens. J. Vet. Med. Sci. 54:447-452. [DOI] [PubMed] [Google Scholar]
- 262.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 263.McCracken, R. M., J. B. McFerran, and C. Dow. 1973. The neural spread of pseudorabies virus in calves. J. Gen. Virol. 20:17-28. [DOI] [PubMed] [Google Scholar]
- 264.McFerran, J. B., and C. Dow. 1975. Studies on immunisation of pigs with the Bartha strain of Aujeszky's disease virus. Res. Vet. Sci. 19:17-22. [PubMed] [Google Scholar]
- 265.McGeoch, D. J., and S. Cook. 1994. Molecular phylogeny of the alphaherpesvirinae subfamily and a proposed evolutionary timescale. J. Mol. Biol. 238:9-22. [DOI] [PubMed] [Google Scholar]
- 266.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 267.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly (A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 70:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.McGregor, S., B. C. Easterday, A. S. Kaplan, and T. Ben-Porat. 1985. Vaccination of swine with thymidine kinase-deficient mutants of pseudorabies virus. Am. J. Vet. Res. 46:1494-1497. [PubMed] [Google Scholar]
- 269.McLauchlan, J., C. Addison, M. C. Craigie, and F. J. Rixon. 1992. Noninfectious L-particles supply functions which can facilitate infection by HSV-1. Virology 190:682-688. [DOI] [PubMed] [Google Scholar]
- 270.McLauchlan, J., and F. J. Rixon. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J. Gen. Virol. 73:269-276. [DOI] [PubMed] [Google Scholar]
- 271.McMahan, L., and P. A. Schaffer. 1990. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to C-terminal regions and are required to modulate viral gene expression very early in infection. J. Virol. 64:3471-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 273.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis-state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 275.Mettenleiter, T. C. 2002. Brief overview on cellular virus receptors. Virus Res. 82:3-8. [DOI] [PubMed] [Google Scholar]
- 276.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 277.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Mettenleiter, T. C. 1996. Immunobiology of pseudorabies (Aujeszky's disease). Vet. Immunol. Immunopathol. 54:221-229. [DOI] [PubMed] [Google Scholar]
- 279.Mettenleiter, T. C. 2003. Pathogenesis of neurotropic herpesviruses: role of viral glycoproteins in neuroinvasion and transneuronal spread. Virus Res. 92:197-206. [DOI] [PubMed] [Google Scholar]
- 280.Mettenleiter, T. C., B. Lomniczi, L. Zsak, I. Medveczky, T. Ben-Porat, and A. S. Kaplan. 1989. Analysis of the factors that affect virulence of pseudorabies virus, p. 3-11. In J. T. van Oirschot (ed.), Vaccination and control of Aujeszky's disease. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 281.Mettenleiter, T. C., N. Lukacs, and H. J. Rziha. 1985. Pseudorabies virus avirulent strains fail to express a major glycoprotein. J. Virol. 56:307-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Mettenleiter, T. C., A. Saalmuller, and F. Weiland. 1993. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J. Virol. 67:1236-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Mettenleiter, T. C., L. Zsak, A. S. Kaplan, T. Ben-Porat, and B. Lomniczi. 1987. Role of a structural glycoprotein of pseudorabies in virus virulence. J. Virol. 61:4030-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Milne, R. S., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315-328. [DOI] [PubMed] [Google Scholar]
- 285.Miranda-Saksena, M., P. Armati, R. A. Boadle, D. J. Holland, and A. L. Cunningham. 2000. Anterograde transport of herpes simplex virus type 1 in cultured, dissociated human and rat dorsal root ganglion neurons. J. Virol. 74:1827-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Misra, V., A. C. Bratanich, D. Carpenter, and P. O'Hare. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J. Virol. 68:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Moore, R. Y., J. C. Speh, and J. P. Card. 1995. The retinohypothalamic tract originates from a distinct subset of retinal ganglion cells. J. Comp. Neurol. 352:351-366. [DOI] [PubMed] [Google Scholar]
- 288.Moriuchi, H., M. Moriuchi, H. Dean, A. K. Cheung, and J. I. Cohen. 1995. Pseudorabies virus EPO is functionally homologous to varicella-zoster virus ORF61 protein and herpes simplex virus type 1 ICPO. Virology 209:281-283. [DOI] [PubMed] [Google Scholar]
- 289.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 67:2739-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Morrison, E. E., Y. F. Wang, and D. M. Meredith. 1998. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72:7108-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Mukamoto, M., I. Watanabe, Y. Kobayashi, F. C. Icatlo, Jr., H. Ishii, and Y. Kodama. 1991. Immunogenicity in Aujeszky's disease virus structural glycoprotein gVI (gp50) in swine. Vet. Microbiol. 29:109-121. [DOI] [PubMed] [Google Scholar]
- 294.Mulder, W., J. Pol, T. Kimman, G. Kok, J. Priem, and B. Peeters. 1996. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J. Virol. 70:2191-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Müller, T., H. J. Bätza, H. Schlüter, F. J. Conraths, and T. C. Mettenleiter. 2003. Eradication of Aujeszky's disease in Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 50:207-213. [DOI] [PubMed] [Google Scholar]
- 296.Munger, J., A. V. Chee, and B. Roizman. 2001. The U(S)3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Negroni, J., N. C. Bennett, and H. M. Cooper. 2003. Organization of the circadian system in the subterranean mole rat, Cryptomys hottentotus (Bathyergidae). Brain Res. 967:48-62. [DOI] [PubMed] [Google Scholar]
- 298.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Newcomb, W. W., F. L. Homa, D. R. Thomsen, B. L. Trus, N. Cheng, A. Steven, F. Booy, and J. C. Brown. 1999. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J. Virol. 73:4239-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Newcomb, W. W., B. L. Trus, F. P. Booy, A. C. Steven, J. S. Wall, and J. C. Brown. 1993. Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol. 232:499-511. [DOI] [PubMed] [Google Scholar]
- 302.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 303.Nishiyama, Y., and T. Murata. 2002. Anti-apoptotic protein kinase of herpes simplex virus. Trends Microbiol. 10:105-107. [DOI] [PubMed] [Google Scholar]
- 304.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Nixdorf, R., B. G. Klupp, and T. C. Mettenleiter. 2001. Role of the cytoplasmic tails of pseudorabies virus glycoproteins B, E and M in intracellular localization and virion incorporation. J. Gen. Virol. 82:215-226. [DOI] [PubMed] [Google Scholar]
- 306.Nixdorf, R., J. Schmidt, A. Karger, and T. C. Mettenleiter. 1999. Infection of Chinese hamster ovary cells by pseudorabies virus. J. Virol. 73:8019-8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Nozawa, N., T. Daikoku, T. Koshizuka, Y. Yamauchi, T. Yoshikawa, and Y. Nishiyama. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77:3204-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Ogg, P. D., P. J. McDonell, B. J. Ryckman, C. M. Knudson, and R. J. Roller. 2004. The HSV-1 Us3 protein kinase is sufficient to block apoptosis induced by overexpression of a variety of Bcl-2 family members. Virology 319:212-224. [DOI] [PubMed] [Google Scholar]
- 309.Ono, E., S. Taharaguchi, S. Watanabe, H. Nikami, Y. Shimizu, and H. Kida. 1998. Suppression of pseudorabies virus replication by a mutant form of immediate-early protein IE180 repressing the viral gene transcription. Vet. Microbiol. 60:107-117. [DOI] [PubMed] [Google Scholar]
- 310.Ono, E., S. Watanabe, H. Nikami, T. Tasaki, and H. Kida. 1998. Pseudorabies virus (PRV) early protein 0 activates PRV gene transcription in combination with the immediate-early protein IE180 and enhances the infectivity of PRV genomic DNA. Vet. Microbiol. 63:99-107. [DOI] [PubMed] [Google Scholar]
- 311.Osorio, F. A., and D. L. Rock. 1992. A murine model of pseudorabies virus latency. Microb. Pathog. 12:39-46. [DOI] [PubMed] [Google Scholar]
- 312.Ou, C. J., M. L. Wong, and T. J. Chang. 2002. A TEF-1-element is required for activation of the promoter of pseudorabies virus glycoprotein X gene by IE180. Virus Genes 25:241-253. [DOI] [PubMed] [Google Scholar]
- 313.Ou, C. J., M. L. Wong, C. Huang, and T. J. Chang. 2002. Suppression of promoter activity of the LAT gene by IE180 of pseudorabies virus. Virus Genes 25:227-239. [DOI] [PubMed] [Google Scholar]
- 314.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Parkinson, J., and R. D. Everett. 2001. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 induce the formation of colocalizing, conjugated ubiquitin. J. Virol. 75:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Peeters, B., N. de Wind, M. Hooisma, F. Wagenaar, A. Gielkens, and R. Moormann. 1992. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J. Virol. 66:894-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Penfold, M. E., P. Armati, and A. L. Cunningham. 1994. Axonal transport of herpes simplex virions to epidermal cells: evidence for a specialized mode of virus transport and assembly. Proc. Natl. Acad. Sci. USA 91:6529-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Pensaert, M. B., and J. P. Kluge. 1989. Pseudorabies virus (Aujeszky's disease), p. 39-64. In M. B. Pensaert (ed.), Virus infections of porcines. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 319.Perdue, M. L., J. C. Cohen, C. C. Randall, and D. J. O'Callaghan. 1976. Biochemical studies of the maturation of herpesvirus nucleocapsid species. Virology 74:194-208. [PubMed] [Google Scholar]
- 320.Perez Fontan, J. J., and C. R. Velloff. 1997. Neuroanatomic organization of the parasympathetic bronchomotor system in developing sheep. Am. J. Physiol. 273:R121-33. [DOI] [PubMed] [Google Scholar]
- 321.Petrovskis, E. A., J. G. Timmins, M. A. Armentrout, C. C. Marchioli, R. J. Yancey, Jr., and L. E. Post. 1986. DNA sequence of the gene for pseudorabies virus gp50, a glycoprotein without N-linked glycosylation. J. Virol. 59:216-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Petrovskis, E. A., J. G. Timmins, T. M. Gierman, and L. E. Post. 1986. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 60:1166-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Pickard, G. E., C. A. Smeraski, C. C. Tomlinson, B. W. Banfield, J. Kaufman, C. L. Wilcox, L. W. Enquist, and P. J. Sollars. 2002. Intravitreal injection of the attenuated pseudorabies virus PRV Bartha results in infection of the hamster suprachiasmatic nucleus only by retrograde transsynaptic transport via autonomic circuits. J. Neurosci. 22:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Porter, I. M., and N. D. Stow. 2004. Virus particles produced by the herpes simplex virus type 1 alkaline nuclease null mutant ambUL12 contain abnormal genomes. J. Gen. Virol. 85:583-591. [DOI] [PubMed] [Google Scholar]
- 325.Priola, S. A., D. P. Gustafson, E. K. Wagner, and J. G. Stevens. 1990. A major portion of the latent pseudorabies virus genome is transcribed in trigeminal ganglia of pigs. J. Virol. 64:4755-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Priola, S. A., and J. G. Stevens. 1991. The 5′ and 3′ limits of transcription in the pseudorabies virus latency associated transcription unit. Virology 182:852-856. [DOI] [PubMed] [Google Scholar]
- 327.Provencio, I., H. M. Cooper, and R. G. Foster. 1998. Retinal projections in mice with inherited retinal degeneration: implications for circadian photoentrainment. J. Comp. Neurol. 395:417-439. [DOI] [PubMed] [Google Scholar]
- 328.Purves, F. C., A. D. Deana, F. Marchiori, D. P. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates [corrected] indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208-215. [DOI] [PubMed] [Google Scholar]
- 329.Pyles, R. B., and R. L. Thompson. 1994. Evidence that the herpes simplex virus type 1 uracil DNA glycosylase is required for efficient viral replication and latency in the murine nervous system. J. Virol. 68:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Quint, W., A. Gielkens, J. Van Oirschot, A. Berns, and H. T. Cuypers. 1987. Construction and characterization of deletion mutants of pseudorabies virus: a new generation of ‘live’ vaccines. J. Gen. Virol. 68:523-534. [DOI] [PubMed] [Google Scholar]
- 331.Rajcani, J., V. Andrea, and R. Ingeborg. 2004. Peculiarities of herpes simplex virus (HSV) transcription: an overview. Virus Genes 28:293-310. [DOI] [PubMed] [Google Scholar]
- 332.Rall, G. F., S. Kupershmidt, N. Sugg, R. A. Veach, and T. Ben-Porat. 1992. Functions of the sequences at the ends of the inverted repeats of pseudorabies virus. J. Virol. 66:1506-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Ray, N., M. E. Bisher, and L. W. Enquist. 2004. Cyclooxygenase-1 and -2 are required for production of infectious pseudorabies virus. J. Virol. 78:12964-12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ray, N., and L. W. Enquist. 2004. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 78:3489-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Raz, E., D. A. Carson, S. E. Parker, T. B. Parr, A. M. Abai, G. Aichinger, S. H. Gromkowski, M. Singh, D. Lew, M. A. Yankauckas, et al. 1994. Intradermal gene immunization: the possible role of DNA uptake in the induction of cellular immunity to viruses. Proc. Natl. Acad. Sci. USA 91:9519-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Richart, S. M., S. A. Simpson, C. Krummenacher, J. C. Whitbeck, L. I. Pizer, G. H. Cohen, R. J. Eisenberg, and C. L. Wilcox. 2003. Entry of herpes simplex virus type 1 into primary sensory neurons in vitro is mediated by Nectin-1/HveC. J. Virol. 77:3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Rinaman, L., J. P. Card, and L. W. Enquist. 1993. Spatiotemporal responses of astrocytes, ramified microglia, and brain macrophages to central neuronal infection with pseudorabies virus. J. Neurosci. 13:685-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Rinaman, L., P. Levitt, and J. P. Card. 2000. Progressive postnatal assembly of limbic-autonomic circuits revealed by central transneuronal transport of pseudorabies virus. J. Neurosci. 20:2731-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Rinaman, L., and G. Schwartz. 2004. Anterograde transneuronal viral tracing of central viscerosensory pathways in rats. J. Neurosci. 24:2782-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Rixon, F. J., C. Addison, and J. McLauchlan. 1992. Assembly of enveloped tegument structures (L particles) can occur independently of virion maturation in herpes simplex virus type 1-infected cells. J. Gen. Virol. 73:277-284. [DOI] [PubMed] [Google Scholar]
- 342.Rixon, F. J., and T. Ben-Porat. 1979. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology 97:151-163. [DOI] [PubMed] [Google Scholar]
- 343.Rixon, F. J., L. T. Feldman, and T. Ben-Porat. 1980. Expression of the genome of defective interfering pseudorabies virions in the presence or absence of helper functions provided by standard virus. J. Gen. Virol. 46:119-138. [DOI] [PubMed] [Google Scholar]
- 344.Robbins, A. K., D. J. Dorney, M. W. Wathen, M. E. Whealy, C. Gold, R. J. Watson, L. E. Holland, S. D. Weed, M. Levine, J. C. Glorioso, et al. 1987. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J. Virol. 61:2691-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Robbins, A. K., J. P. Ryan, M. E. Whealy, and L. W. Enquist. 1989. The gene encoding the gIII envelope protein of pseudorabies virus vaccine strain Bartha contains a mutation affecting protein localization. J. Virol. 63:250-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Roizman, B., R. C. Desrosiers, B. Fleckenstein, C. Lopez, A. C. Minson, and M. J. Studdert. 1992. The family Herpesviridae: an update. Arch. Virol. 123:425-449. [DOI] [PubMed] [Google Scholar]
- 347.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 348.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 348a.Romero, C. H., P. Meade, J. Santagata, K. Gillis, G. Lollis, E. C. Hahn, and E. P. Gibbs. 1997. Genital infection and transmission of pseudorabies virus in feral swine in Florida, USA. Vet. Microbiol. 55:131-139. [DOI] [PubMed]
- 349.Romero, C. H., P. N. Meade, B. L. Homer, J. E. Shultz, and G. Lollis. 2003. Potential sites of virus latency associated with indigenous pseudorabies viruses in feral swine. J. Wildl. Dis. 39:567-575. [DOI] [PubMed] [Google Scholar]
- 350.Romero, C. H., P. N. Meade, J. E. Shultz, H. Y. Chung, E. P. Gibbs, E. C. Hahn, and G. Lollis. 2001. Venereal transmission of pseudorabies viruses indigenous to feral swine. J. Wildl. Dis. 37:289-296. [DOI] [PubMed] [Google Scholar]
- 351.Rouzade-Dominguez, M. L., R. Miselis, and R. J. Valentino. 2003. Central representation of bladder and colon revealed by dual transsynaptic tracing in the rat: substrates for pelvic visceral coordination. Eur. J. Neurosci. 18:3311-3324. [DOI] [PubMed] [Google Scholar]
- 352.Ruda, M., and J. D. Coulter. 1982. Axonal and transneuronal transport of wheat germ agglutinin demonstrated by immunocytochemistry. Brain Res. 249:237-246. [DOI] [PubMed] [Google Scholar]
- 353.Rziha, H.-J., V. F. Ohlinger, D. Lüttiken, and N. Visser. 1989. Latency characteristics of pseudorabies vaccine virus, p. 79-85. In J. T. Van Oirschot (ed.), Vaccination and control of Aujeszky's disease. Kluwer Academic Publishers, Boston, Mass.
- 354.Rziha, H. J., T. C. Mettenleiter, V. Ohlinger, and G. Wittmann. 1986. Herpesvirus (pseudorabies virus) latency in swine: occurrence and physical state of viral DNA in neural tissues. Virology 155:600-613. [DOI] [PubMed] [Google Scholar]
- 355.Sabin, A. B. 1938. Progression of different nasally installed viruses along different nervous pathways in the same host. Proc. Soc. Exp. Biol. Med. 38:270-275. [Google Scholar]
- 356.Sabo, A., and J. Rajcani. 1976. Latent pseudorabies virus infection in pigs. Acta Virol. 20:208-214. [PubMed] [Google Scholar]
- 357.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 U(L)17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Sato, H., L. D. Callanan, L. Pesnicak, T. Krogmann, and J. I. Cohen. 2002. Varicella-zoster virus (VZV) ORF17 protein induces RNA cleavage and is critical for replication of VZV at 37 degrees C but not 33 degrees C. J. Virol. 76:11012-11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Schek, N., and S. L. Bachenheimer. 1985. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Schmidt, J., V. Gerdts, J. Beyer, B. G. Klupp, and T. C. Mettenleiter. 2001. Glycoprotein D-independent infectivity of pseudorabies virus results in an alteration of in vivo host range and correlates with mutations in glycoproteins B and H. J. Virol. 75:10054-10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Schmiedhofer, J. 1910. Beitraege zur Pathologie der Infektioesen Bulbaerparalyse (Aujeszkyschen Krankheit). Zeitschr. Infekt. Krkh. Haustiere 8:383-405. [Google Scholar]
- 363.Schmitt, B. J., F. A. Osorio, W. W. Stroup, and E. P. Gibbs. 1991. A comparison of differential diagnostic tests to detect antibodies to pseudorabies glycoproteins gX, gI, and gIII in naturally infected feral pigs. J. Vet. Diagn. Investig. 3:344-345. [DOI] [PubMed] [Google Scholar]
- 364.Schwartz, J. A., E. E. Brittle, A. E. Reynolds, L. W. Enquist, and S. J. Silverstein. Pseudorabies virus deleted of UL54 is attenuated in mice, but productively infects cells in culture. J. Virol., submitted for publication. [DOI] [PMC free article] [PubMed]
- 365.Sherman, G., and S. L. Bachenheimer. 1988. Characterization of intranuclear capsids made by ts morphogenic mutants of HSV-1. Virology 163:471-480. [DOI] [PubMed] [Google Scholar]
- 366.Shintani, T., A. R. Anker, I. Billig, J. P. Card, and B. J. Yates. 2003. Transneuronal tracing of neural pathways influencing both diaphragm and genioglossal muscle activity in the ferret. J. Appl. Physiol. 95:1453-1459. [DOI] [PubMed] [Google Scholar]
- 367.Shope, R. E. 1931. An experimental study of “mad itch” with especial reference to its relationship to pseudorabies. J. Exp. Med. 54:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Simmons, A., and A. A. Nash. 1984. Zosteriform spread of herpes simplex virus as a model of recrudescence and its use to investigate the role of immune cells in prevention of recurrent disease. J. Virol. 52:816-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Smeraski, C. A., P. J. Sollars, M. D. Ogilvie, L. W. Enquist, and G. E. Pickard. 2004. Suprachiasmatic nucleus input to autonomic circuits identified by retrograde transsynaptic transport of pseudorabies virus from the eye. J. Comp. Neurol. 471:298-313. [DOI] [PubMed] [Google Scholar]
- 370.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Smith, B. N., B. W. Banfield, C. A. Smeraski, C. L. Wilcox, F. E. Dudek, L. W. Enquist, and G. E. Pickard. 2000. Pseudorabies virus expressing enhanced green fluorescent protein: A tool for in vitro electrophysiological analysis of transsynaptically labeled neurons in identified central nervous system circuits. Proc. Natl. Acad. Sci. USA 97:9264-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 376.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Spear, P. G. 2001. A first step toward understanding membrane fusion induced by herpes simplex virus. Mol. Cell 8:2-4. [DOI] [PubMed] [Google Scholar]
- 378.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 379.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Stark, W. M., M. R. Boocock, and D. J. Sherratt. 1992. Catalysis by site-specific recombinases. Trends Genet. 8:432-439. [PubMed] [Google Scholar]
- 381.Stegeman, A., M. C. de Jong, A. van Nes, and A. Bouma. 1997. Dynamics of pseudorabies virus infections in vaccinated pig populations: a review. Vet. Q. 19:117-122. [DOI] [PubMed] [Google Scholar]
- 382.Stegmann, B., H. Zentgraf, A. Ott, and C. H. Schroder. 1978. Synthesis and packaging of herpes simplex virus DNA in the course of virus passages at high multiplicity. Intervirology 10:228-240. [DOI] [PubMed] [Google Scholar]
- 383.Steven, A. C., and P. G. Spear. 1997. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. Burnett, and R. L. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 384.Subak-Sharpe, J. H., and D. J. Dargan. 1998. HSV molecular biology: general aspects of herpes simplex virus molecular biology. Virus Genes 16:239-251. [DOI] [PubMed] [Google Scholar]
- 385.Sun, N., M. D. Cassell, and S. Perlman. 1996. Anterograde, transneuronal transport of herpes simplex virus type 1 strain H129 in the murine visual system. J. Virol. 70:5405-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Swanson, L. W. 1999 1998. Brain maps: structure of the rat brain, a laboratory guide with printed and electronic templates for data, models and schematics, 2nd revised ed. Elsevier Science, Amsterdam, The Netherlands.
- 387.Szilagyi, J. F., and C. Cunningham. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J. Gen. Virol. 72:661-668. [DOI] [PubMed] [Google Scholar]
- 388.Taddeo, B., A. Esclatine, and B. Roizman. 2002. The patterns of accumulation of cellular RNAs in cells infected with a wild-type and a mutant herpes simplex virus 1 lacking the virion host shutoff gene. Proc. Natl. Acad. Sci. USA 99:17031-17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Taharaguchi, S., H. Inoue, E. Ono, H. Kida, S. Yamada, and Y. Shimizu. 1994. Mapping of transcriptional regulatory domains of pseudorabies virus immediate-early protein. Arch. Virol. 137:289-302. [DOI] [PubMed] [Google Scholar]
- 390.Taharaguchi, S., T. Kobayashi, S. Yoshino, and E. Ono. 2002. Analysis of regulatory functions for the region located upstream from the latency-associated transcript (LAT) promoter of pseudorabies virus in cultured cells. Vet. Microbiol. 85:197-208. [DOI] [PubMed] [Google Scholar]
- 391.Taharaguchi, S., E. Ono, S. Yamada, Y. Shimizu, and H. Kida. 1995. Mapping of a functional region conferring nuclear localization of pseudorabies virus immediate-early protein. Arch. Virol. 140:1737-1746. [DOI] [PubMed] [Google Scholar]
- 392.Taharaguchi, S., S. Yoshino, K. Amagai, and E. Ono. 2003. The latency-associated transcript promoter of pseudorabies virus directs neuron-specific expression in trigeminal ganglia of transgenic mice. J. Gen. Virol. 84:2015-2022. [DOI] [PubMed] [Google Scholar]
- 393.Takashima, Y., H. Tamura, X. Xuan, and H. Otsuka. 1999. Identification of the US3 gene product of BHV-1 as a protein kinase and characterization of BHV-1 mutants of the US3 gene. Virus Res. 59:23-34. [DOI] [PubMed] [Google Scholar]
- 394.Tanaka, S., T. Imamura, M. Sakaguchi, and K. Mannen. 1998. Acetylcholine reactivates latent pseudorabies virus in mice. J. Virol. Methods 70:103-106. [DOI] [PubMed] [Google Scholar]
- 395.Tanaka, S., T. Imamura, M. Sakaguchi, K. Mannen, and K. Matsuo. 1996. Acetylcholine activates latent pseudorabies virus in pigs. Arch. Virol. 141:161-166. [DOI] [PubMed] [Google Scholar]
- 396.Tanaka, S., and K. Mannen. 2002. Activation of latent pseudorabies virus infection in mice treated with acetylcholine. Exp. Anim. 51:407-409. [DOI] [PubMed] [Google Scholar]
- 397.Tanaka, S., and K. Mannen. 2003. Effect of mild stress in mice latently infected pseudorabies virus. Exp. Anim. 52:383-386. [DOI] [PubMed] [Google Scholar]
- 398.Tang, D. C., M. DeVit, and S. A. Johnston. 1992. Genetic immunization is a simple method for eliciting an immune response. Nature 356:152-154. [DOI] [PubMed] [Google Scholar]
- 399.Thawley, D. G., R. F. Solorzano, and M. E. Johnson. 1984. Confirmation of pseudorabies virus infection, using virus recrudescence by dexamethasone treatment and in vitro lymphocyte stimulation. Am. J. Vet. Res. 45:981-983. [PubMed] [Google Scholar]
- 400.Thomas, J. T., S. T. Oh, S. S. Terhune, and L. A. Laimins. 2001. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J. Virol. 75:7564-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Tirabassi, R. S., and L. W. Enquist. 1998. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J. Virol. 72:4571-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Tirabassi, R. S., R. A. Townley, M. G. Eldridge, and L. W. Enquist. 1997. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J. Virol. 71:6455-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Tirabassi, R. S., R. A. Townley, M. G. Eldridge, and L. W. Enquist. 1998. Molecular mechanisms of neurotropic herpesvirus invasion and spread in the CNS. Neurosci. Biobehav Rev. 22:709-720. [DOI] [PubMed] [Google Scholar]
- 404.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Tomishima, M. J., and L. W. Enquist. 2002. In vivo egress of an alphaherpesvirus from axons. J. Virol. 76:8310-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Tomishima, M. J., G. A. Smith, and L. W. Enquist. 2001. Sorting and transport of alpha herpesviruses in axons. Traffic 2:429-436. [DOI] [PubMed] [Google Scholar]
- 407.Tonomura, N., E. Ono, Y. Shimizu, and H. Kida. 1996. Negative regulation of immediate-early gene expression of pseudorabies virus by interferon-alpha. Vet. Microbiol. 53:271-281. [DOI] [PubMed] [Google Scholar]
- 408.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Tsuda, T., T. Onodera, T. Sugimura, and Y. Murakami. 1992. Induction of protective immunity and neutralizing antibodies to pseudorabies virus by immunization of anti-idiotypic antibodies. Arch. Virol. 124:291-300. [DOI] [PubMed] [Google Scholar]
- 410.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Ulmer, J. B., J. J. Donnelly, S. E. Parker, G. H. Rhodes, P. L. Felgner, V. J. Dwarki, S. H. Gromkowski, R. R. Deck, C. M. DeWitt, A. Friedman, et al. 1993. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259:1745-1749. [DOI] [PubMed] [Google Scholar]
- 412.Ulmer, J. B., J. C. Sadoff, and M. A. Liu. 1996. DNA vaccines. Curr. Opin. Immunol. 8:531-536. [DOI] [PubMed] [Google Scholar]
- 413.Van de Walle, G. R., H. W. Favoreel, H. J. Nauwynck, and M. B. Pensaert. 2003. Antibody-induced internalization of viral glycoproteins and gE-gI Fc receptor activity protect pseudorabies virus-infected monocytes from efficient complement-mediated lysis. J. Gen. Virol. 84:939-948. [DOI] [PubMed] [Google Scholar]
- 414.Van Minnebruggen, G., H. W. Favoreel, L. Jacobs, and H. J. Nauwynck. 2003. Pseudorabies virus US3 protein kinase mediates actin stress fiber breakdown. J. Virol. 77:9074-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Van Minnebruggen, G., H. W. Favoreel, and H. J. Nauwynck. 2004. Internalization of pseudorabies virus glycoprotein B is mediated by an interaction between the YQRL motif in its cytoplasmic domain and the clathrin-associated AP-2 adaptor complex. J. Virol. 78:8852-8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.van Oirschot, J. T., and A. L. Gielkens. 1984. In vivo and in vitro reactivation of latent pseudorabies virus in pigs born to vaccinated sows. Am. J. Vet. Res. 45:567-571. [PubMed] [Google Scholar]
- 417.van Oirschot, J. T., H. J. Rziha, P. J. Moonen, J. M. Pol, and D. van Zaane. 1986. Differentiation of serum antibodies from pigs vaccinated or infected with Aujeszky's disease virus by a competitive enzyme immunoassay. J. Gen. Virol. 67:1179-1182. [DOI] [PubMed] [Google Scholar]
- 418.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747-1755. [DOI] [PubMed] [Google Scholar]
- 419.Visser, N., and D. Lütticken. 1989. Experiences with a gI-/TK-modified live pseudorabies virus vaccine: strain begonia, p. 37-44. In J. T. van Oirschot (ed.), Vaccination and control of Aujeszky's disease. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 420.Vlcek, C., Z. Kozmik, V. Paces, S. Schirm, and M. Schwyzer. 1990. Pseudorabies virus immediate-early gene overlaps with an oppositely oriented open reading frame: characterization of their promoter and enhancer regions. Virology 179:365-377. [DOI] [PubMed] [Google Scholar]
- 421.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 422.Wang, J., M. Irnaten, R. A. Neff, P. Venkatesan, C. Evans, A. D. Loewy, T. C. Mettenleiter, and D. Mendelowitz. 2001. Synaptic and neurotransmitter activation of cardiac vagal neurons in the nucleus ambiguus. Ann. N. Y. Acad. Sci. 940:237-246. [DOI] [PubMed] [Google Scholar]
- 423.Watanabe, S., E. Ono, Y. Shimizu, and H. Kida. 1996. Mapping of transregulatory domains of pseudorabies virus early protein 0 and identification of its dominant-negative mutant. Arch. Virol. 141:1001-1009. [DOI] [PubMed] [Google Scholar]
- 424.Watanabe, S., E. Ono, Y. Shimizu, and H. Kida. 1995. Pseudorabies virus early protein 0 transactivates the viral gene promoters. J. Gen. Virol. 76 (Pt 11):2881-2885. [DOI] [PubMed] [Google Scholar]
- 425.Wathen, M. W., and L. M. Wathen. 1984. Isolation, characterization, and physical mapping of a pseudorabies virus mutant containing antigenically altered gp50. J. Virol. 51:57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Weeks, B. S., R. S. Ramchandran, J. J. Hopkins, and H. M. Friedman. 2000. Herpes simplex virus type-1 and -2 pathogenesis is restricted by the epidermal basement membrane. Arch. Virol. 145:385-396. [DOI] [PubMed] [Google Scholar]
- 427.Weigel, R. M., W. F. Hall, G. Scherba, A. M. Siegel, E. C. Hahn, and J. R. Lehman. 1992. Evaluation of the sensitivity and specificity of two diagnostic tests for antibodies to pseudorabies virus glycoprotein X. J. Vet. Diagn. Investig. 4:238-244. [DOI] [PubMed] [Google Scholar]
- 428.Wesley, R. D., and A. K. Cheung. 1996. A pseudorabies virus mutant with deletions in the latency and early protein O genes: replication, virulence, and immunity in neonatal piglets. J. Vet. Diagn. Investig. 8:21-24. [DOI] [PubMed] [Google Scholar]
- 429.Whealy, M. E., J. P. Card, A. K. Robbins, J. R. Dubin, H. J. Rziha, and L. W. Enquist. 1993. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J. Virol. 67:3786-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Wheeler, J. G., and F. A. Osorio. 1991. Investigation of sites of pseudorabies virus latency, using polymerase chain reaction. Am. J. Vet. Res. 52:1799-1803. [PubMed] [Google Scholar]
- 431.White, A. K., J. Ciacci-Zanella, J. Galeota, S. Ele, and F. A. Osorio. 1996. Comparison of the abilities of serologic tests to detect pseudorabies-infected pigs during the latent phase of infection. Am. J. Vet. Res. 57:608-611. [PubMed] [Google Scholar]
- 432.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Whitley, R. J. 2001. Herpes simplex viruses, p. 2461-2509. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 434.Wilkinson, D. E., and S. K. Weller. 2003. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life 55:451-458. [DOI] [PubMed] [Google Scholar]
- 435.Wittmann, G., J. Jakubik, and R. Ahl. 1980. Multiplication and distribution of Aujeszky's disease (pseudorabies) virus in vaccinated and non-vaccinated pigs after intranasal infection. Arch. Virol. 66:227-240. [DOI] [PubMed] [Google Scholar]
- 436.Wittmann, G., V. Ohlinger, and H. J. Rziha. 1983. Occurrence and reactivation of latent Aujeszky's disease virus following challenge in previously vaccinated pigs. Arch. Virol. 75:29-41. [DOI] [PubMed] [Google Scholar]
- 437.Wittmann, G., and H.-J. Rziha. 1989. Aujeszky's disease (pseudorabies) in pigs, p. 230-325. In D. M. Knipe and P. M. Howley (ed.), Herpesvirus diseases of cattle, horses and pigs, vol. 9. Kluwer Academic Publishers, Boston, Mass. [Google Scholar]
- 438.Wong, M. L., and C. H. Chen. 1998. Evidence for the internal location of actin in the pseudorabies virion. Virus Res. 56:191-197. [DOI] [PubMed] [Google Scholar]
- 439.Wong, M. L., T. Y. Ho, J. H. Huang, C. Y. Hsiang, and T. J. Chang. 1997. Stimulation of type I DNA topoisomerase gene expression by pseudorabies virus. Arch. Virol. 142:2099-2105. [DOI] [PubMed] [Google Scholar]
- 440.Wu, C. A., L. Harper, and T. Ben-Porat. 1986. Molecular basis for interference of defective interfering particles of pseudorabies virus with replication of standard virus. J. Virol. 59:308-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Wu, C. L., and K. W. Wilcox. 1991. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J. Virol. 65:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Wu, L., and A. J. Berk. 1988. Transcriptional activation by the pseudorabies virus immediate early protein requires the TATA box element in the adenovirus 2 E1B promoter. Virology 167:318-322. [DOI] [PubMed] [Google Scholar]
- 443.Wu, S. L., C. Y. Hsiang, T. Y. Ho, and T. J. Chang. 1998. Identification, expression, and characterization of the pseudorabies virus DNA-binding protein gene and gene product. Virus Res. 56:1-9. [DOI] [PubMed] [Google Scholar]
- 444.Wu, S. X., X. P. Zhu, and G. J. Letchworth. 1998. Bovine herpesvirus 1 glycoprotein M forms a disulfide-linked heterodimer with the U(L)49.5 protein. J. Virol. 72:3029-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Yamada, S., and M. Shimizu. 1994. Isolation and characterization of mutants of pseudorabies virus with deletion in the immediate-early regulatory gene. Virology 199:366-375. [DOI] [PubMed] [Google Scholar]
- 446.Yamada, S., and M. Shimizu. 1994. Pseudorabies virus immediate-early regulatory protein IE180 expressed by recombinant baculovirus is functional. Virology 202:491-495. [DOI] [PubMed] [Google Scholar]
- 447.Yang, M., J. P. Card, R. S. Tirabassi, R. R. Miselis, and L. W. Enquist. 1999. Retrograde, transneuronal spread of pseudorabies virus in defined neuronal circuitry of the rat brain is facilitated by gE mutations that reduce virulence. J. Virol. 73:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.York, I. A., C. Roop, D. W. Andrews, S. R. Riddell, F. L. Graham, and D. C. Johnson. 1994. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell 77:525-535. [DOI] [PubMed] [Google Scholar]
- 449.Yuan, R., C. Bohan, F. C. Shiao, R. Robinson, H. J. Kaplan, and A. Srinivasan. 1989. Activation of HIV LTR-directed expression: analysis with pseudorabies virus immediate early gene. Virology 172:92-99. [DOI] [PubMed] [Google Scholar]
- 450.Zanin, E., I. Capua, C. Casaccia, A. Zuin, and A. Moresco. 1997. Isolation and characterization of Aujeszky's disease virus in captive brown bears from Italy. J. Wildl. Dis. 33:632-634. [DOI] [PubMed] [Google Scholar]
- 451.Zemanick, M. C., P. L. Strick, and R. D. Dix. 1991. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc. Natl. Acad. Sci. USA 88:8048-8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Zuckermann, F. A., L. Zsak, T. C. Mettenleiter, and T. Ben-Porat. 1990. Pseudorabies virus glycoprotein gIII is a major target antigen for murine and swine virus-specific cytotoxic T lymphocytes. J. Virol. 64:802-812. [DOI] [PMC free article] [PubMed] [Google Scholar]