Abstract
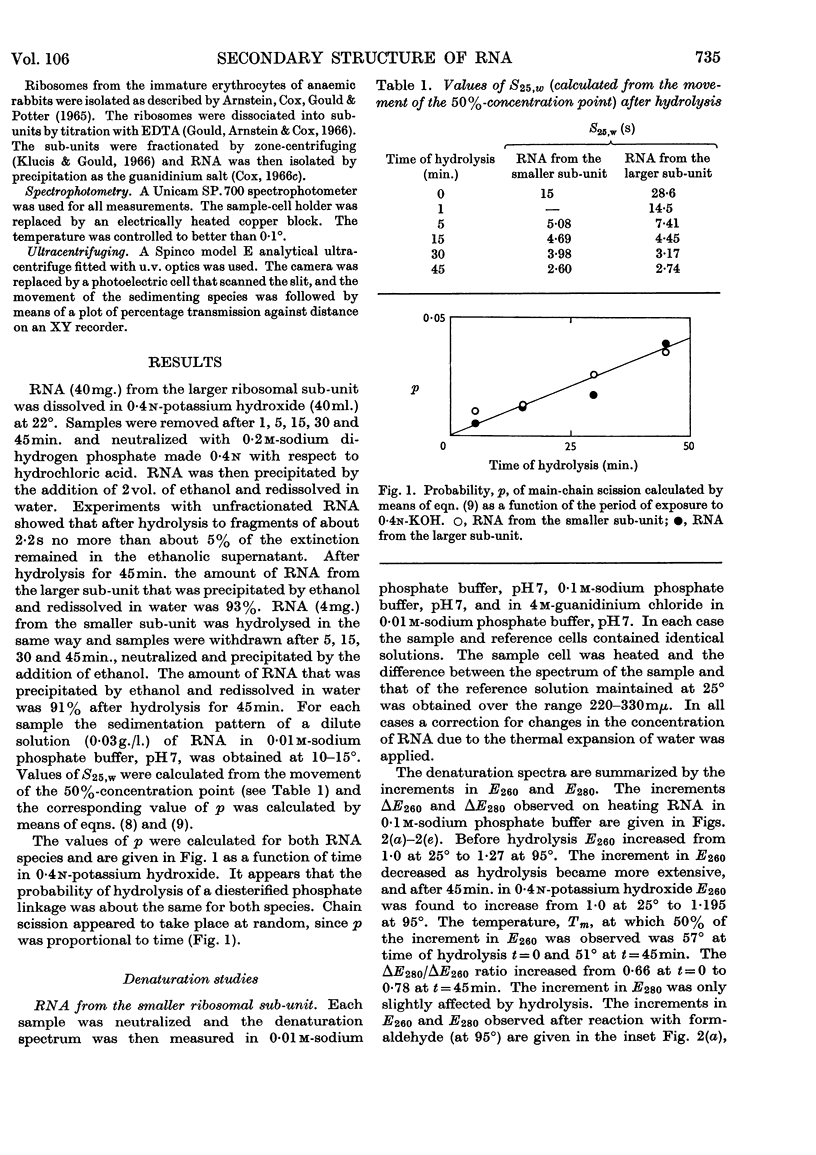
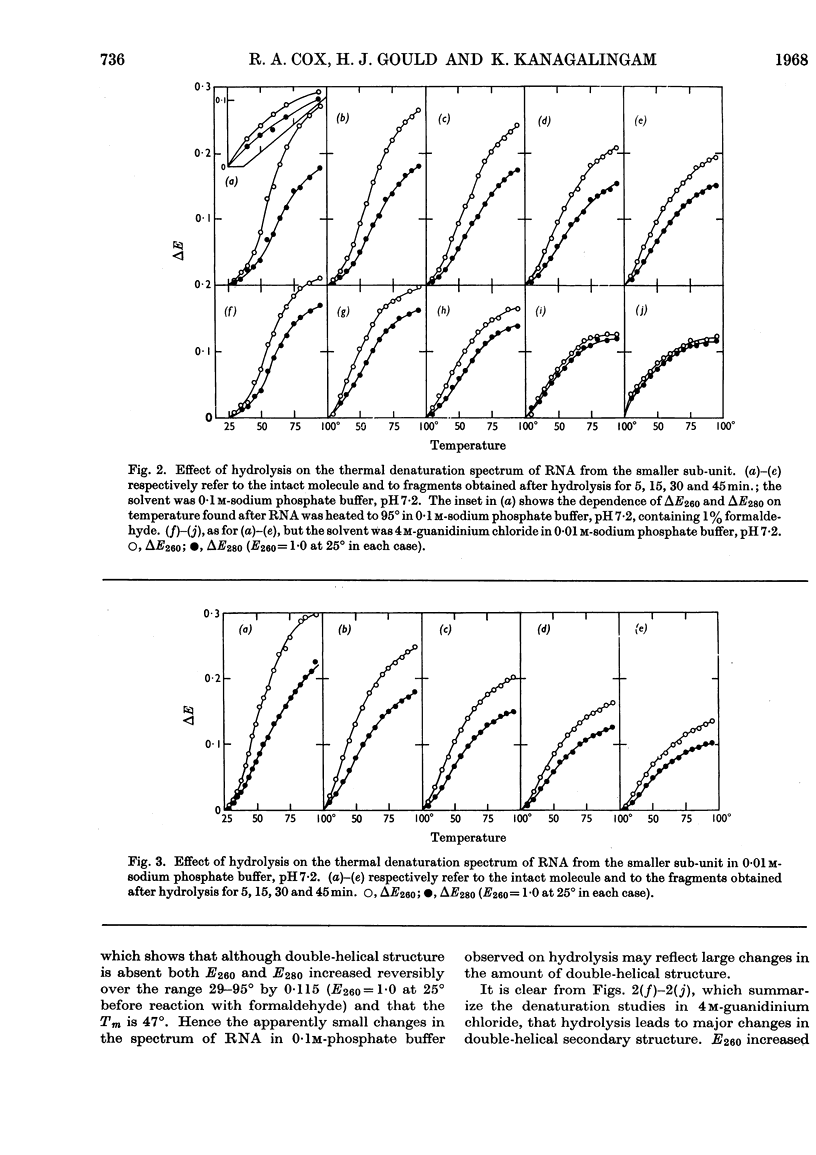
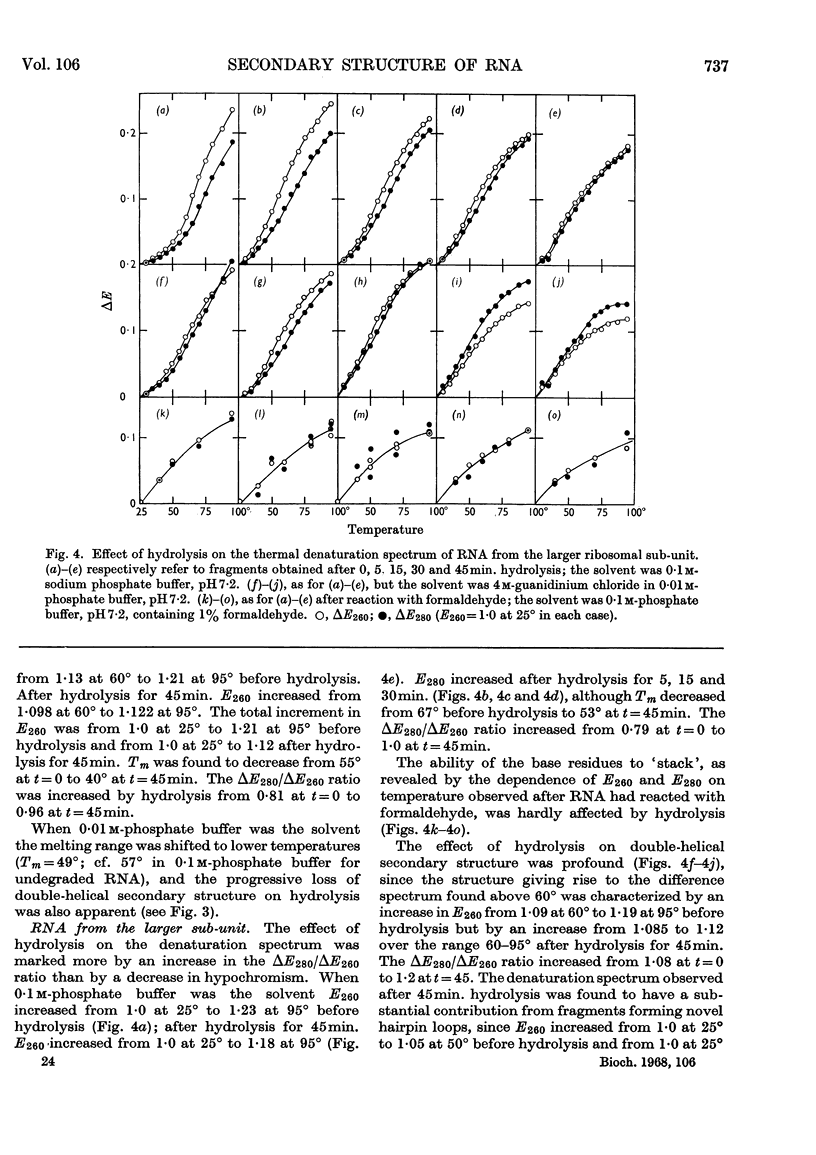
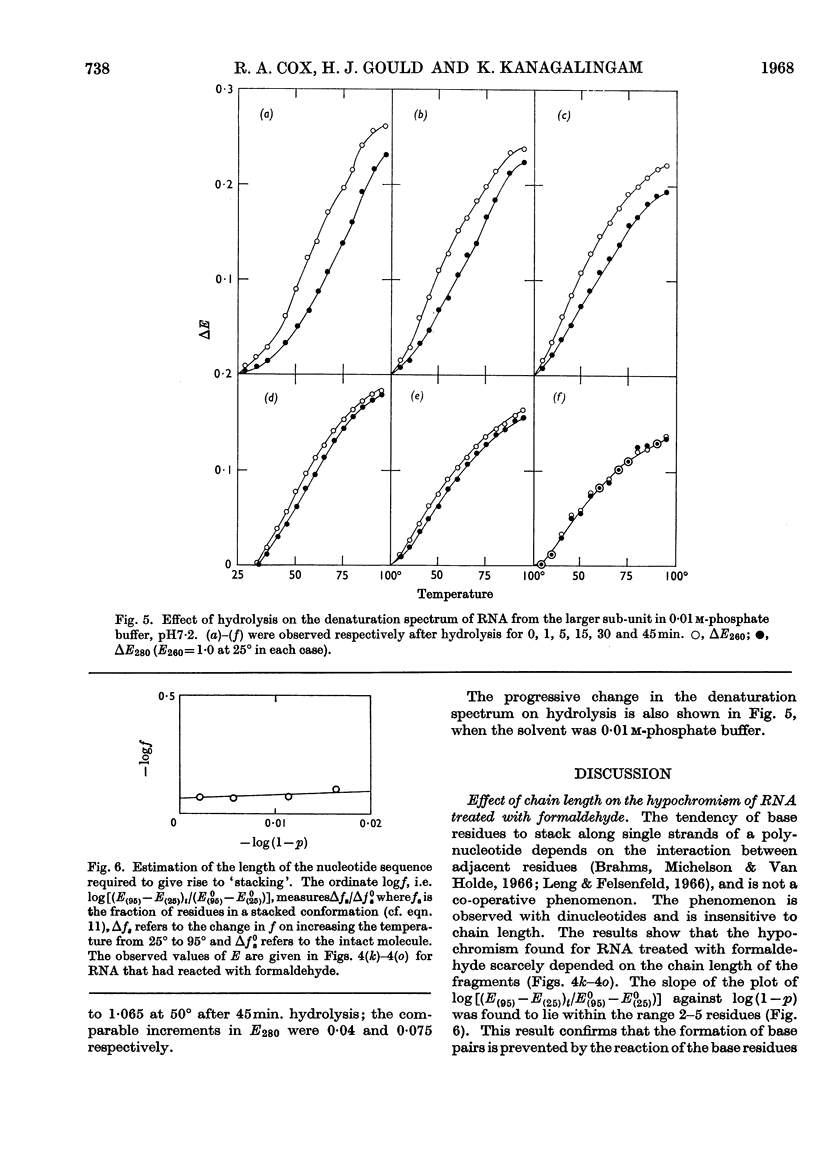
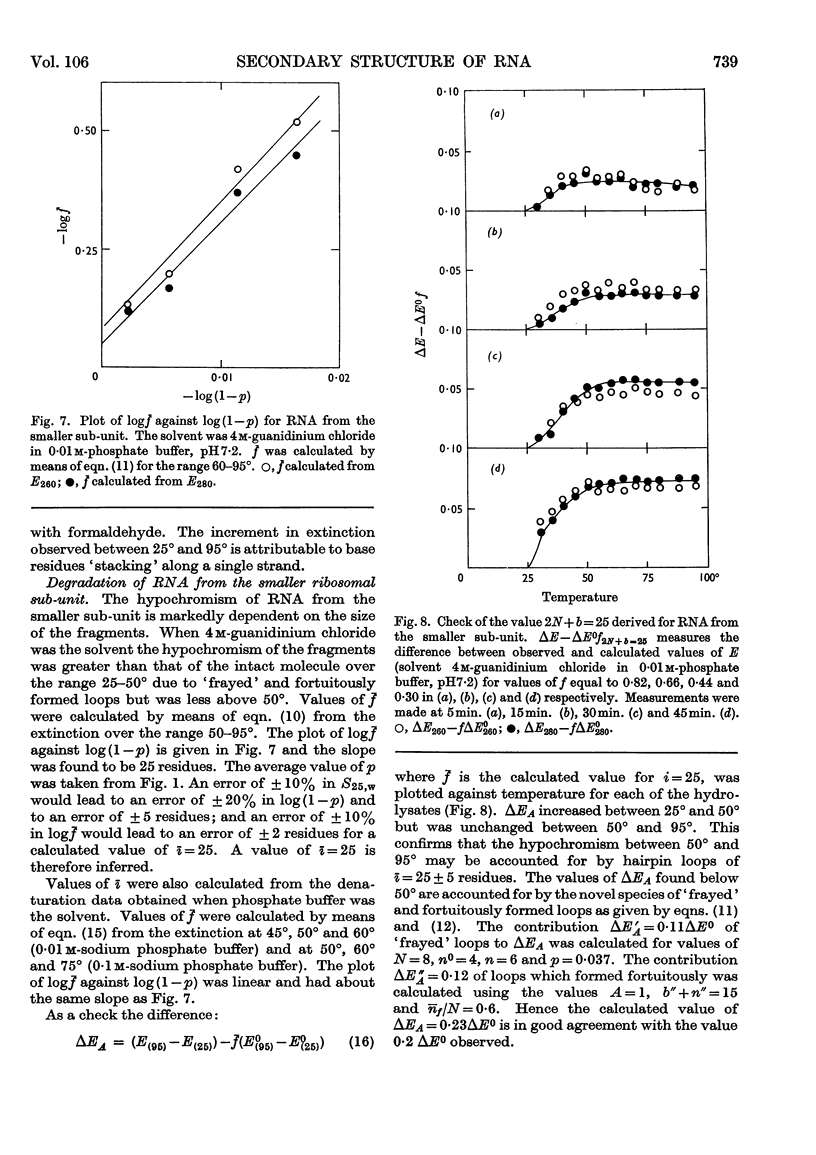
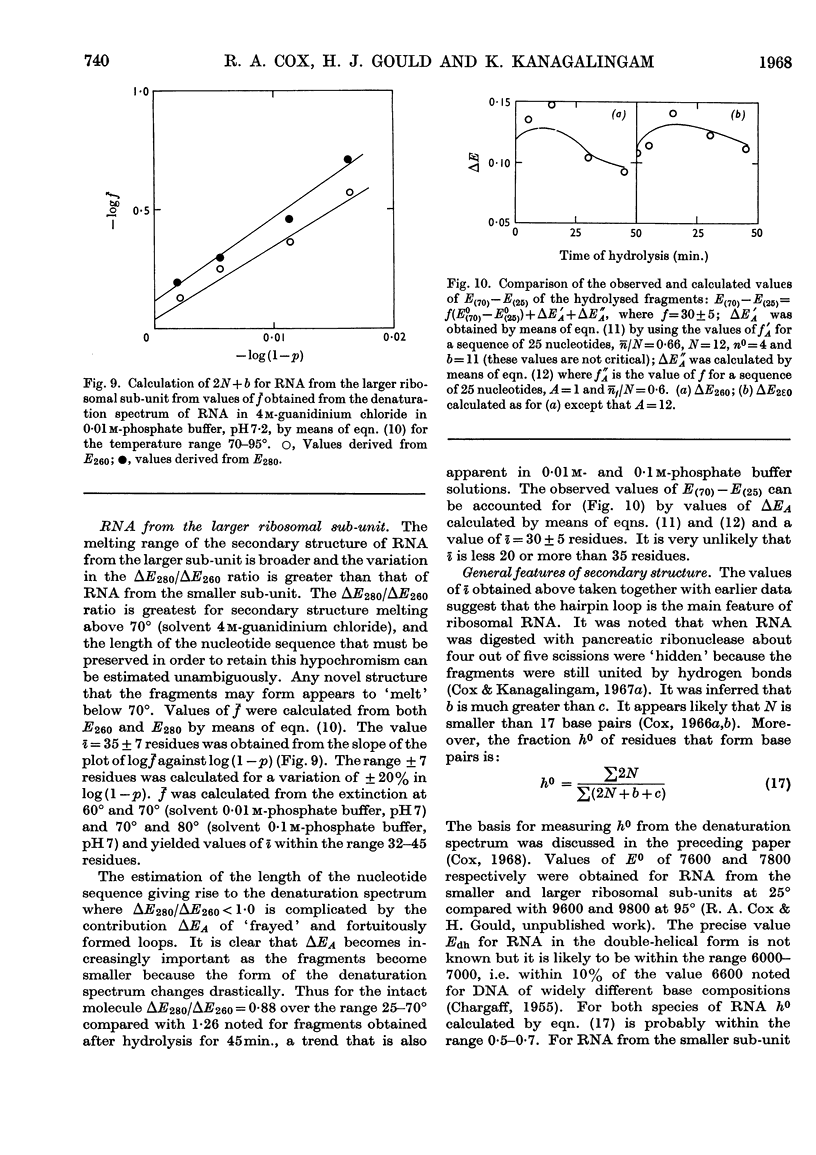
1. RNA isolated from the sub-units of rabbit reticulocyte ribosomes was hydrolysed by 0·4n-potassium hydroxide at 20°. The probability of main-chain scission was calculated from the number-average chain length, which was obtained from S25,w in 0·01m-phosphate buffer. 2. The fraction, f, of the original secondary structure that the fragments re-formed at neutral pH in 4m-guanidinium chloride, as well as in 0·01m- and 0·1m-phosphate buffer, was derived from changes in extinction over the range 220–310mμ on thermal denaturation. 3. The secondary structure of RNA is regarded as an assembly of hairpin loops each of 2N+b residues on average, where N is the number of base-paired residues and b is the number of unpaired residues. 4. If chain scission takes place at random then 2N+b=logf/log(1–p). 5. For RNA from the smaller sub-unit 2N+b was estimated as 25±5 residues, compared with 30±5 residues for the less stable species and 35±5 residues for the more stable species of hairpin loop of RNA from the larger sub-unit.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnstein H. R., Cox R. A., Gould H., Potter H. A comparison of methods for the isolation and fractionation of reticulocyte ribosomes. Biochem J. 1965 Aug;96(2):500–506. doi: 10.1042/bj0960500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms J., Michelson A. M., Van Holde K. E. Adenylate oligomers in single- and double-strand conformation. J Mol Biol. 1966 Feb;15(2):467–488. doi: 10.1016/s0022-2836(66)80122-5. [DOI] [PubMed] [Google Scholar]
- Cox R. A. A possible method for characterizing the secondary structure of ribonucleic acids. Biochem J. 1966 Jul;100(1):146–168. doi: 10.1042/bj1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A. Hydrolysis of polynucleotides and the characterization of their secondary structure. A theoretical study. Biochem J. 1968 Feb;106(3):725–731. doi: 10.1042/bj1060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Kanagalingam K. A spectrophotometric study of the secondary structure of ribonucleic acid based on a method for diminishing single-stranded base-'stacking' without affecting multi-helical structures. Biochem J. 1967 Jun;103(3):749–758. doi: 10.1042/bj1030749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A., Kanagalingam K. A study of the hydrolysis of unfractionated reticulocyte ribosomal ribonucleic acid by pancreatic ribonuclease and its relevance to secondary structure. Biochem J. 1967 May;103(2):431–452. doi: 10.1042/bj1030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R. A. The secondary structure of ribosomal ribonucleic acid in solution. Biochem J. 1966 Mar;98(3):841–857. doi: 10.1042/bj0980841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty P., Boedtker H., Fresco J. R., Haselkorn R., Litt M. SECONDARY STRUCTURE IN RIBONUCLEIC ACIDS. Proc Natl Acad Sci U S A. 1959 Apr;45(4):482–499. doi: 10.1073/pnas.45.4.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FASMAN G. D., LINDBLOW C., GROSSMAN L. THE HELICAL CONFORMATIONS OF POLYCYTIDYLIC ACID: STUDIES ON THE FORCES INVOLVED. Biochemistry. 1964 Aug;3:1015–1021. doi: 10.1021/bi00896a002. [DOI] [PubMed] [Google Scholar]
- FELSENFELD G., CANTONI G. L. USE OF THERMAL DENATURATION STUDIES TO INVESTIGATE THE BASE SEQUENCE OF YEAST SERINE SRNA. Proc Natl Acad Sci U S A. 1964 May;51:818–826. doi: 10.1073/pnas.51.5.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould H. J., Arnstein H. R., Cox R. A. The dissociation of reticulocyte polysomes into subunits and the location of messenger RNA. J Mol Biol. 1966 Feb;15(2):600–618. doi: 10.1016/s0022-2836(66)80130-4. [DOI] [PubMed] [Google Scholar]
- KALTREIDER H. B., SCOTT J. F. Concerning the alkaline hydrolysis of soluble ribonucleic acid. Biochim Biophys Acta. 1962 Mar 5;55:379–381. doi: 10.1016/0006-3002(62)90793-x. [DOI] [PubMed] [Google Scholar]
- Klucis E. S., Gould H. J. Zonal ultracentrifuge for the separation of ribosomal subunits. Science. 1966 Apr 15;152(3720):378–378. doi: 10.1126/science.152.3720.378. [DOI] [PubMed] [Google Scholar]
- Leng M., Felsenfeld G. A study of polyadenylic acid at neutral pH. J Mol Biol. 1966 Feb;15(2):455–466. doi: 10.1016/s0022-2836(66)80121-3. [DOI] [PubMed] [Google Scholar]
- Stevens C. L., Rosenfeld A. The secondary structure of polyadenylic acid Inferences from its reaction with formaldehyde. Biochemistry. 1966 Aug;5(8):2714–2721. doi: 10.1021/bi00872a031. [DOI] [PubMed] [Google Scholar]
- THOMAS C. A., Jr, BERNS K. I. The utility of formaldehyde in stabilizing polynucleotide chains from bacteriophage DNA. J Mol Biol. 1962 Apr;4:309–312. doi: 10.1016/s0022-2836(62)80008-4. [DOI] [PubMed] [Google Scholar]
- TIMASHEFF S. N., WITZ J., LUZZATI V. The structure of high molecular weight ribonucleic acid in solution. A smallangle x-ray scattering study. Biophys J. 1961 Sep;1:525–537. doi: 10.1016/s0006-3495(61)86906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


