Abstract
cABC I (chondroitinase ABC I) from Proteus vulgaris is a GalAG (galactosaminoglycan) depolymerizing lyase that cleaves its substrates at the glycosidic bond via β-elimination. cABC I cleaves a particularly broad range of GalAG substrates, including CS (chondroitin sulphate), DS (dermatan sulphate) and hyaluronic acid. We recently cloned and recombinantly expressed cABC I in Escherichia coli, and completed a preliminary biochemical characterization of the enzyme. In the present study, we have coupled site-directed mutagenesis of the recombinant cABC I with a structural model of the enzyme–substrate complex in order to investigate in detail the roles of active site amino acids in the catalytic action of the enzyme. The putative catalytic residues His-501, Tyr-508, Arg-560 and Glu-653 were probed systematically via mutagenesis. Assessment of these mutants in kinetic and end-point assays provided direct evidence on the catalytic roles of these active-site residues. The crystal structure of the native enzyme provided a framework for molecular docking of representative CS and DS substrates. This enabled us to construct recombinant enzyme–substrate structural complexes. These studies together provided structural insights into the effects of the mutations on the catalytic mechanism of cABC I and the differences in its processing of CS and DS substrates. All His-501 mutants were essentially inactive and thereby implicating this amino acid to play the critical role of proton abstraction during catalysis. The kinetic data for Glu-653 mutants indicated that it is involved in a hydrogen bonding network in the active site. The proximity of Tyr-508 to the glycosidic oxygen of the substrate at the site of cleavage suggested its potential role in protonating the leaving group. Arg-560 was proximal to the uronic acid C-5 proton, suggesting its possible role in the stabilization of the carbanion intermediate formed during catalysis.
Keywords: active site, chondroitin sulphate (CS), chondroitinase ABC (cABC), dermatan sulphate, mode of action, mutagenesis
Abbreviations: C4S, chondroitin-4-sulphate; C6S, chondroitin-6-sulphate; cABC, chondroitinase ABC; cAC, chondroitinase AC; cB, chondroitinase B; CE, capillary electrophoresis; CS, chondroitin sulphate; DS, dermatan sulphate; GAG, glycosaminoglycan; GalAG, galactosaminoglycan; GalNAc, N-acetyl-D-galactosamine; GlcA, β-D-glucuronic acid; HAL, hyaluronate lyase; IdoA, α-L-iduronic acid
INTRODUCTION
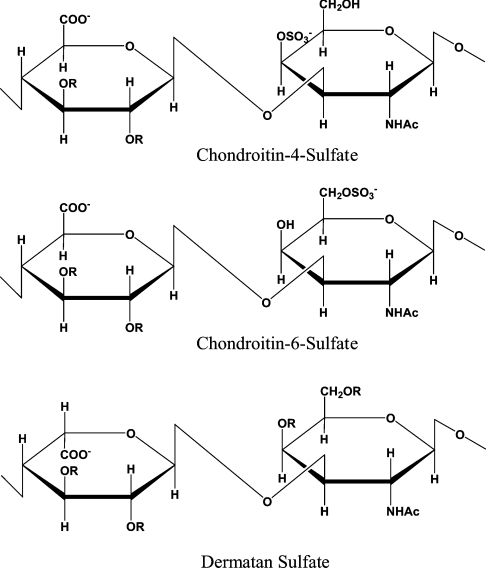
GalAGs (galactosaminoglycans) are one of the four classes of structurally complex linear polysaccharides called GAGs (glycosaminoglycans) [1]. GalAGs are composed of disaccharide repeat units of uronic acid [IdoA (α-L-iduronic acid) or GlcA (β-D-glucuronic acid)] (1→3) linked to GalNAc (N-acetyl-D-galactosamine). These basic disaccharide units (Figure 1) are linearly associated via β-(1→4) linkages to form polymers of CS (chondroitin sulphate) or DS (dermatan sulphate). The uronic acids in CS are exclusively GlcA; with DS, epimerization at the C-5 position of the uronic acid moiety during biosynthesis results in a mixture of IdoA and GlcA epimers. Biosynthesis of CS and DS also involves sulphation of the sugar backbone at various positions, which generates diversity in their oligosaccharide sequences. CS is commonly O-sulphated at the C-4 of the galactosamine [C4S (chondroitin-4-sulphate) or CSA] or the C-6 of the galactosamine [C6S (chondroitin-6-sulphate) or CSC]. Other rare modifications in CS, such as 2-O-or 3-O-sulphation of the GlcA moiety, have also been reported [2,3]. In the case of DS, O-sulphation at C-4 of GalNAc is a common modification; O-sulphation at C-6 of GalNAc and C-2 of the IdoA also occur, but with less frequency.
Figure 1. GalAG disaccharide chemical structures.
GAGs are polymers of repeated disaccharide units consisting of an uronic acid and a hexosamine. In the case of GalAGs, the hexosamine moiety is an N-acetyl-D-galactosamine (R=SO3− or H).
Due to their ubiquitous presence on the cell surface and in the extracellular matrix, GalAGs interact with a wide variety of proteins such as growth factors, chemokines, lipoproteins and enzymes in the extracellular environment. These interactions play critical roles in modulating the function of the protein [4,5]. DS is known to bind with thrombin [6] and activated protein C [7] to influence anticoagulation; collagen [8], fibronectin [9–11] and tenascin-X [12] to stabilize the extracellular matrix; transforming growth factor-β [13,14] to regulate growth; and hepatocyte growth factor/scatter factor [15,16] to spur cellular proliferation and organogenesis. It is becoming increasingly evident that there is sequence-specificity in GalAG–protein interactions in terms of the precise modifications in the chemical structure of GalAGs that bind with high affinity to a given protein [17,18]. Manipulation of GAG chemical structure has been shown to promote anti-tumour activities, inhibiting angiogenesis and tumour metastasis [19]. Modification of CS-containing proteoglycans has been observed in a variety of human cancers including those of the colon [20,21], blood [21] and larynx [22]. Defined GalAG oligosaccharides are also being developed as therapeutics for blood coagulation disorders [23,24]. Thus the characterization of structure–function relationships of GalAGs is important for understanding their biological roles.
The structural characterization of complex acidic polysaccharides, like GAGs, is a challenging task. Due to their complex non-template-based biosynthesis, it has been difficult to develop methodologies to obtain sufficient material containing pure GAG oligosaccharides. Furthermore, the chemical heterogeneity and highly acidic nature of GAGs have complicated their analysis. Despite these challenges, significant advances have been made in the development of methodologies for sequencing biologically important GAGs [25]. These techniques involve the precise depolymerization of GAGs at specific linkages and analysis of the resulting fragments using sensitive analytical methods such as MS and CE (capillary electrophoresis) [26].
Various microorganisms express GAG-depolymerizing polysaccharide lyases. Mechanistically, these lyases degrade their substrates via a β-elimination reaction that generates products with an unsaturated 4,5-bond on the uronic acid at the site of cleavage. Extensive biochemical characterization of the activity and substrate specificity of some of these enzymes, such as the heparinases from Flavobacterium heparinum (now known as Pedobacter heparinus) have successfully enabled their utilization as tools for the structural characterization of HSGAGs (heparin and heparan sulphate GAGs) [25,27].
Having extensively characterized the heparinases, we focused our efforts on the cloning, expression and characterization of the GalAG-depolymerizing enzymes. The GalAG lyases include cAC (chondroitinase AC) (EC 4.2.2.5) and cB (chondroitinase B) from P. heparinus, and cABC I (chondroitinase ABC I) and cABC II (EC 4.2.2.4) from Proteus vulgaris. cAC shows activity against C4S and C6S. cB cleaves DS as its sole substrate. cABC I and cABC II process a variety of substrates including C4S, C6S, DS and hyaluronan. Particularly striking is the ability of these broad substrate specificity enzymes (cABC I and II) to process GalAGs containing either of the uronic acid epimers. We have already cloned and recombinantly expressed cB and cAC from P. heparinus [28]. Unlike the heparinases, there is abundant structural information available on the chondroitinases. Co-crystal structures of cB with the disaccharide reaction products and cAC with different CS and DS substrates have been solved [29,30]. These co-crystal structures provided a framework to design site-directed mutagenesis experiments to obtain a detailed understanding of the role of the active site residues and bivalent ions (such as Ca2+ in the case of cB) in the catalytic mechanism and substrate specificity of these enzymes [30,31,35].
The biological significance of cABC has been demonstrated in studies that have utilized this enzyme for functional recovery through nerve regeneration following spinal cord injury, thus offering a potential treatment strategy for an unmet medical condition [32]. We recently cloned and recombinantly expressed cABC I from P. vulgaris [33]. We also completed a preliminary biochemical characterization of this enzyme [33], which involved mutating to alanine the four structurally conserved amino acid residues (His-501, Tyr-508, Arg-560 and Glu-653) that were identified in the cABC I crystal structure studies [34]. Our observation that all four mutants were inactive towards both CS and DS substrates suggested that these residues could potentially be part of the enzyme's catalytic machinery. In the present study, we systematically engineered conservative and non-conservative site-directed mutants of the putative catalytic amino acids and analysed the activity of these mutants using both short-term kinetic assays and overnight end-point product formation assays. Additionally, we modelled structural complexes of cABC I with CS and DS substrates, providing a framework to guide our mutagenesis studies and to further develop a chemical context for enzyme activity. These structural complexes were constructed by molecular docking of substrates into the proposed active site of the enzyme, modelled using the cABC I crystal structure [34].
EXPERIMENTAL
Materials
Porcine intestinal mucosa DS (average molecular mass 35000 Da) and shark cartilage C6S (average molecular mass 50000 Da) were purchased from Sigma. C4S (super special grade, average molecular mass 50000 Da) was purchased from Seikagaku/Associates of Cape Cod (Falmouth, MA, U.S.A.). Oligonucleotides were purchased from Invitrogen (Carlsbad, CA, U.S.A.). The QuikChange site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA, U.S.A.). All other materials are from common sources or are as given below.
Subcloning and site-directed mutagenesis of cABC I
Genomic DNA was isolated from cultures of P. vulgaris (A.T.C.C. 6896) using a Qiagen DNeasy purification kit. Subcloning procedures were as previously described [28,33]. The QuikChange site-directed mutagenesis kit was used to produce mutants of cABC I, as previously described [33]. Primer sequences for all studies are presented in Table 1. The plasmids were prepared using a miniprep kit (Qiagen, Chatsworth, CA, U.S.A.). Each clone was sequenced to confirm the presence of the desired mutation. Plasmid DNA was used to transform BL21 (DE3) E. coli.
Table 1. Summary of primer sequences for site-directed mutagenesis studies.
Mutation codons are indicated in bold; bases modified in order to create the desired point mutations are underlined.
| Mutant | Primer pair sequences |
|---|---|
| T154A | 5′-ACTGGCTGGCGTGCTGTGGGAGTCTCT-3′ |
| 5′-AGAGACTCCCACAGCACGCCAGCCAGT-3′ | |
| V309I | 5′-GGAACGCAAGGCAGACATCTGATCACTGATAAACAAATC-3′ |
| 5′-GATTTGTTTATCAGTGATCAGATGTCTGCCTTGCGTTCC-3′ | |
| P322L | 5′-CAACCAGAGAATCTTAACTCTCAAGATAAACAACTATTTG-3′ |
| 5′-CAAATAGTTGTTTATCTTGAGAGTTAAGATTCTCTGGTTG-3′ | |
| P694Q | 5′-GGTTGGGATTGGAATAGAATGCAAGGGGCAACCACT-3′ |
| 5′-AGTGGTTGCCCCTTGCATTCTATTCCAATCCCAACC-3′ | |
| H501A | 5′-TGATGGTACAGCATGGCGAGCTGAAGGCAACTATCCGGGCTA-3′ |
| 5′-TAGCCCGGATAGTTGCCTTCAGCTCGCCATGCTGTACCATCA-3′ | |
| Y508A | 5′-GGCAACTATCCGGGCGCCTCTTTCCCAGCC-3′ |
| 5′-GGCTGGGAAAGAGGCGCCCGGATAGTTGCC-3′ | |
| R560A | 5′-CCGCTTGCAGGAGCACACCCTTTTAACTCACCTTCG-3′ |
| 5′-CGAAGGTGAGTTAAAAGGGTGTGCTCCTGCAAGCGG-3′ | |
| E653A | 5′-CACCAATGTTTGGTCATCTGCAATTTATAACAAAGATAACCGT-3′ |
| 5′-ACGGTTATCTTTGTTATAAATTGCAGATGACCAAACATTGGTG-3′ | |
| R500A | 5′-CCTGATGGTACAGCATGGGCACATGAAGGCAACTATCCGGGC-3′ |
| 5′-GCCCGGATAGTTGCCTTCATGTGCCCATGCTGTACCATCAGG-3′ | |
| H501K | 5′-GGTACAGCATGGCGAAAGGAAGGCAACTATCCGGGC-3′ |
| 5′-GCCCGGATAGTTGCCTTCCTTTCGCCATGCTGTACC-3′ | |
| H501R | 5′-ACAGCATGGCGACGTGAAGGCAACTATCCGGGC-3′ |
| 5′-GCCCGGATAGTTGCCTTCACGTCGCCATGCTGT-3′ | |
| Y508F | 5′-AACTATCCGGGCTTCTCTTTCCCAGCC-3′ |
| 5′-GGCTGGGAAAGAGAAGCCCGGATAGTT-3′ | |
| E653D | 5′-CAATGTTTGGTCATCTGATATTTATAACAAAGATAACCGTTATGG-3′ |
| 5′-CCATAACGGTTATCTTTGTTATAAATATCAGATGACCAAACATTG-3′ | |
| E653Q | 5′-CAATGTTTGGTCATCTCAAATTTATAACAAAGATAACCGTTATGG-3′ |
| 5′-CCATAACGGTTATCTTTGTTATAAATTTGAGATGACCAAACATTG-3′ |
Recombinant expression and protein purification of cABC I and mutants
Recombinant cABC I and the site-directed mutants were expressed and purified as previously described [28,33]. The purity of the enzymes was assessed by SDS/PAGE using a precast Invitrogen NuPAGE 12% (w/v) Bis/Tris gel and Simply Blue SafeStain. Protein concentration was measured using the Bio-Rad Laboratories (Hercules, CA, U.S.A.) Bradford assay kit.
CE
To study the activities and product profiles of the proteins on each substrate (C6S, DS and C4S), digests of 100 μg/ml substrate, 50 mM Tris/HCl, 50 mM sodium acetate (pH 8.0) with 1 μg of recombinant cABC I or the site-directed mutants were placed at 37 °C for 14 h. The digests were analysed using CE as previously described [28,35].
CD
CD spectra were recorded at 25 °C on an Aviv 202 CD spectrophotometer using Quartz cuvettes with an optical path length of 0.1 cm. Scans were collected between 300 and 195 nm with a 1.0 nm bandwidth and a scan rate of 1 nm/min. Three scans were averaged for each protein. For melting experiments, spectra were collected at 5 °C intervals from 5 to 75 °C. Recombinantly expressed proteins were concentrated and buffer-exchanged into 50 mM sodium phosphate (pH 7.5) using Centricon 10 filters (Millipore, Watertown, MA, U.S.A.). All spectra were collected using a protein concentration of 0.2 mg/ml. The buffer contribution was subtracted for all spectra. The signal was normalized to molar ellipticity, [θ]m, in degrees·cm2·dmol−1.
Kinetic analysis
Recombinant proteins were concentrated and buffer-exchanged into 50 mM Tris/HCl and 50 mM sodium acetate (pH 8.0). In order to evaluate the activity of cABC I and mutants in a semi-high throughput manner, the kinetic analysis was adapted to a 96-well plate format. The studies were performed in a quartz 96-well plate at 37 °C using a Molecular Devices Spectramax 190. The assay was initiated by adding 0.2–6 μg of enzyme to 249 μl of a solution containing different concentrations of GalAG substrates (C4S, C6S and DS) in 50 mM Tris/HCl and 50 mM sodium acetate (pH 8.0), total reaction volume of 250 μl. Each well contained different substrate concentrations (0.1–5 mg/ml). Product formation was monitored by measuring the absorbance A at 232 nm every 2–3 s. Evaluation of the kinetic data was based on the initial reaction rate and was performed as previously described [33].
Docking of CS and DS substrates in the active site of cABC I
The SARF2 program [36] was used to determine the CA (Cα) atoms in the cABC I and cAC structures that gave an optimal rmsd (root mean-squared deviation) upon superimposition. Superposition of the 452 CA atoms identified by the SARF2 program in the cAC co-crystal structures (with C4S and DS) and the cABC I crystal structure [rmsd of 2.2 Å (1 Å=10−10 m or 0.1 nm)] provided the initial location and orientation of the CS and DS substrates relative to the putative active site of cABC I. The co-ordinates of C4S (GlcA-GalNAc4S)2 and DS (IdoA-GalNAc4S)2 tetrasaccharides were available from the two cAC co-crystal structures [30]. In the case of the DS tetrasaccharide, the C-5 proton was facing away from the putative active site residues (His-501, Tyr-508 and Arg-560). Thus this substrate was further reorientated to ensure that the C-5 proton was accessible for abstraction by the active site amino acids.
The starting structural models of the enzyme–substrate complexes were subject to energy minimization. The AMBER force field modified for carbohydrates was further modified to include O-sulphate and sulphamate groups [37,38]. This modified AMBER force field was used to assign the potentials for both the enzyme as well as the tetrasaccharide substrates. A subset of the enzyme co-ordinates around the active site groove was defined to include all of the putative active site amino acids and several additional amino acids that could potentially be involved in the catalytic activity. The enzyme–substrate complex was subject to minimization first without charges and then with charges using 500 steps of steepest descent and 500 steps of conjugate-gradient methods. Most of the protein was fixed and only the amino acids that were a part of the active site subset were allowed to move during the minimization. The final rms derivatives of the energies were below 0.1. The ring conformation of the monosaccharides was not distorted by the minimization procedure. The Viewer and Discover modules of InsightII (Release 2000.1; Accelrys, San Diego, CA, U.S.A.) were used for the orientation of the substrate and energy minimization respectively.
RESULTS AND DISCUSSION
cABC I characterization
The gene for cABC I was cloned from P. vulgaris genomic DNA without its putative leader sequence. The PCR product was subcloned into pET28a and expressed in E. coli with an N-terminal His6 tag. As reported in our previous study [33], there were sequence discrepancies between our clone and the reported protein sequence of the enzyme [34,39,40]. Thus it was necessary to perform site-directed mutagenesis to make our protein sequence consistent with the published sequence [34]. The catalytic activity of our cloned cABC I was previously characterized with various GalAG substrates (including C4S, C6S and DS) using a semi-high throughput procedure [33]. This procedure enabled us to obtain multiple initial velocity measurements between one substrate–enzyme pair within 1 min. Multiple runs for each enzyme–substrate pair could also be performed in a short period of time to establish a better confidence in the data generated. Km and Vmax information could be generated from these data almost immediately. The data suggest robust activity on all three substrates; however, there is a clear preference for the chondroitin substrates (Tables 2 and 3) [33]. The catalytic efficiency is about 3-fold higher for CS compared with DS.
Table 2. Kinetic analysis of cABC I and mutants with C6S as substrate.
Values are the means±S.D. for at least three experiments. NA, not available.
| Enzyme | Km (μM) | Kcat (min−1) | Kcat/Km (μM−1·min−1) |
|---|---|---|---|
| cABC I* | 1.2±0.6 | 37000±6500 | 31000 |
| H501A or K or R | NA‡ | NA | NA |
| Y508A | NA | NA | NA |
| Y508F | 36.4±0.8 | 31 | 0.85 |
| R560A | NA | NA | NA |
| E653A or D | NA | NA | NA |
| E653Q | 6.1±0.4 | 1600±300 | 260 |
| R500A | 19.9±1.0 | 410±50 | 21 |
* Values are consistent with previously reported data [33].
Table 3. Kinetic analysis of cABC I and mutants with DS as substrate.
Values are the means±S.D. for at least three experiments. NA, not available.
| Enzyme | Km (μM) | Kcat (min−1) | Kcat/Km (μM−1·min−1) |
|---|---|---|---|
| cABC I* | 2.5±0.5 | 27000±2500 | 11000 |
| H501A or K or R | NA* | NA | NA |
| Y508A | NA | NA | NA |
| Y508F | 48.9±0.6 | 110±40 | 2.2 |
| R560A | NA | NA | NA |
| E653A or D | NA | NA | NA |
| E653Q | 4.2±0.5 | 5200±400 | 1200 |
| R500A | 35.7±0.5 | 160±40 | 4.5 |
* Values are consistent with previously reported data [33].
Site-directed mutagenesis studies
Building on our preliminary biochemical characterization [33], we sought to establish unambiguously the roles of the four putative catalytic amino acids His-501, Tyr-508, Arg-560 and Glu-653. Our model of the enzyme–substrate structural complex (see below) was used as a framework to systematically design additional conservative mutants. In order to evaluate the role of His-501 in cABC I's activity (substrate binding versus direct contribution to catalysis), the histidine residue was conservatively mutated to lysine and then to an arginine residue, retaining this position's basic character. Y508F (Tyr508→Phe) was constructed to evaluate the importance of the hydrophobic group in positioning the substrate, compared with the hydroxy group that could potentially protonate the glycosidic bond after cleavage. Based on our enzyme–substrate structural complex, Glu-653 was proposed to be involved in a hydrogen bonding network with neighbouring His-501 and arginine. Thus two different Glu-653 mutants were designed: E653D to study the effect of side-chain length and E653Q to study the effect of acidic versus amide group on the hydrogen bonding network in the active site.
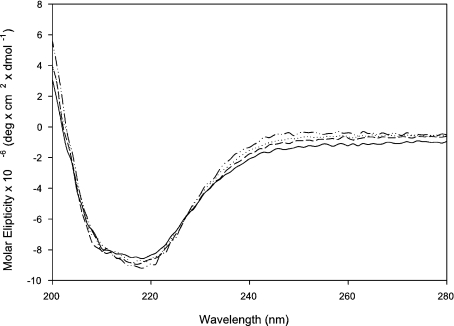
These mutants were analysed for their activity against GalAG substrates by scanning for product formation (as measured using A232 detection) against C6S and DS (Tables 2 and 3). Mutants with detectable activities were further characterized through kinetics assays. CE studies allowed for an inspection of an end-point product profile analysis following an exhaustive 14–18 h digestion with C6S, DS or C4S as substrate. CD data on all mutants was also collected and heat-denaturing experiments performed to ensure that mutagenesis did not compromise the enzyme structure (Figure 2). All mutants displayed melting transitions of approx. 45 °C.
Figure 2. CD spectra of recombinant enzymes.
The recombinant cABC I (—-) and the mutants H501K (····), H501R (–––) and E653D (–··–) were concentrated and buffer-exchanged into 50 mM sodium phosphate buffer (pH 8.0). Proteins were analysed in a 1 mm path length quartz cell. The slight deviations in spectral intensity can be attributed to errors inherent in protein quantification.
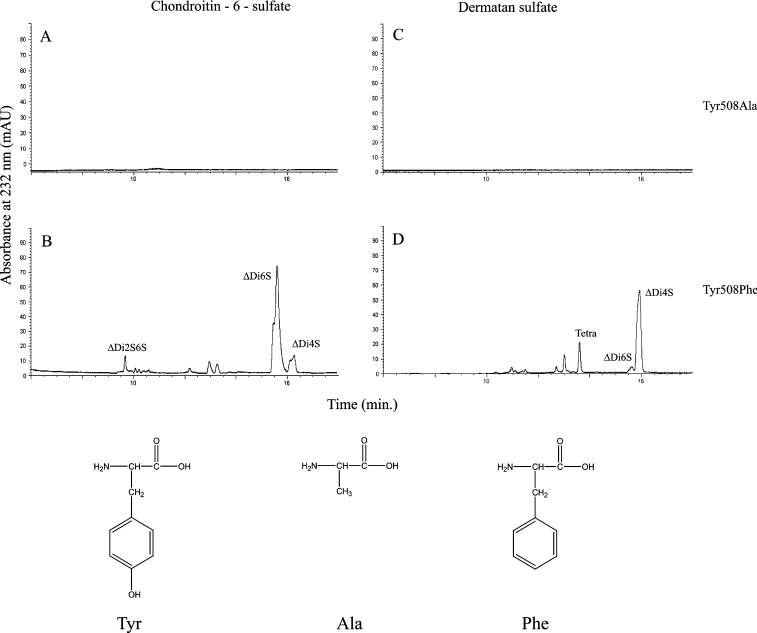
All His-501 mutations (H501A, H501K and H501R) showed no activity against C6S and DS substrates while scanning for product formation. Additionally, the His-501 mutants did not produce any products in CE assays, suggesting that His-501 is absolutely critical for cABC I activity. Though Y508A proved inactive against C6S and DS while scanning for product formation and was unable to degrade C6S, DS and C4S in exhaustive digestions, Y508F was able to process GalAG substrates. In an exhaustive digestion, product profile analysis revealed electrophoretograms virtually indistinguishable from those produced with recombinant cABC I. Y508F processes C6S (Figure 3) with both diminished binding (Km of 36.4 μM compared with 1.2 μM for wild-type recombinant cABC I) and markedly reduced catalytic-centre activity (kcat of 31 min−1 compared with 37000 min−1 for recombinant cABC I). Y508F acts on DS in similar fashion (Figure 3), with a Km of 48.9 μM and a kcat of 110 min−1 (compared with a Km of 2.5 μM and a kcat of 27000 min−1 for recombinant cABC I). Therefore the tyrosine to phenylalanine mutation results in an enzyme with a much higher Km and a greatly reduced kcat, suggesting that this residue possibly plays an important role in substrate positioning and turnover. Unlike His-501, the Y508F mutant does not seem to affect a critical step in GalAG degradation since it still produces products in an end-point analysis.
Figure 3. CE analysis of cABC I Tyr-508 mutants.
Product profiles for (A) Tyr508Ala acting on C6S, (B) Tyr508Phe acting on C6S, (C) Tyr508Ala acting on DS and (D) Tyr508Phe acting on DS. Depicted below are the relevant amino acids, which illustrate the nature of the chemical groups involved and the relative protrusion of each side chain into the catalytic pocket. Abbreviations: ΔDi2S,6S, ΔU2S-GalNAc6S (where ΔU2S is 2-O-sulphated uronic acid with a Δ4,5 unsaturated bond); ΔDi4S, ΔU-GalNAc4S (where ΔU is uronic acid with a Δ4,5 unsaturated bond and GalNAc4S is 4-O-sulphated N-acetylgalactosamine); ΔDi6S, ΔU-GalNAc6S (where GalNAc6S is 6-O-sulphated N-acetylgalactosamine; mAU, milli-absorbance units; Tetra, tetrasaccharide.
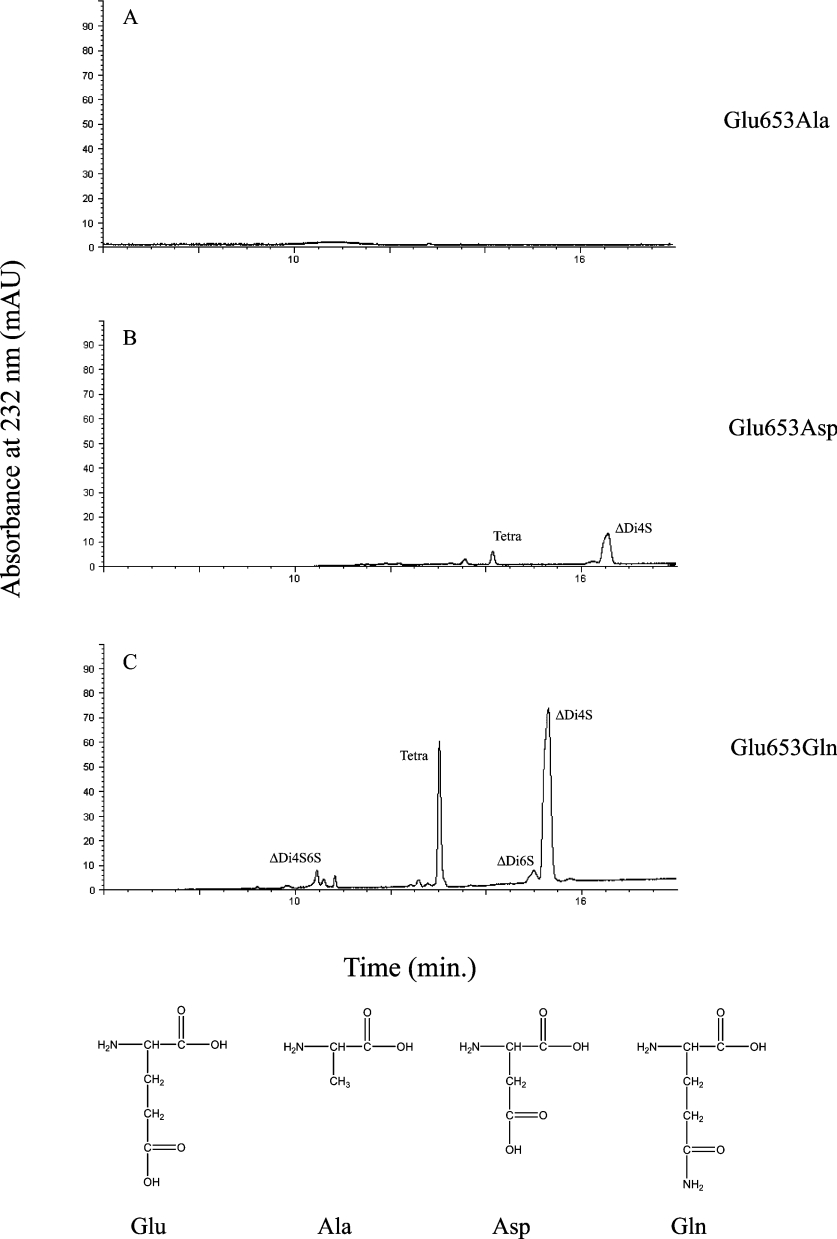
The other mutants were active against both C6S and DS, though with far less processing efficiency than recombinant cABC I (Tables 2 and 3). Although the E653D mutant proved to be catalytically inactive against both C6S and DS in kinetics assays, on overnight digestion this mutant was able to produce products on these substrates as well as on C4S. E653Q maintained some level of activity against both C6S and DS, with a slight increase in Km for both substrates, and a greater than 20-fold reduction in kcat for C6S and a 5-fold reduction in kcat for DS (Figure 4). The results suggest that while this residue does not play a major role in substrate positioning, its major role is probably in affecting the catalytic turnover of the enzyme. Against C6S, R500A remained active, but with a 17-fold increase in Km and a 1500-fold reduction in catalytic efficiency. With DS, R500A showed similar losses, with a 14-fold increase in Km and a greater than 2000-fold decrease in catalytic efficiency. The results of the kinetic analysis of the mutants were interpreted using the modelled enzyme–substrate structural complexes to understand the roles of the amino acid in the catalytic mechanism of cABC I.
Figure 4. CE analysis of cABC I Glu-653 mutants.
Products of DS degradation following digestion by (A) E653A, (B) E653D and (C) E653Q. Depicted below are the relevant amino acids. For each side chain, the relative length of protrusion into the catalytic pocket and the potential to participate in hydrogen bonding determine the end-product profiles. mAU, milli-absorbance units.
Enzyme–substrate structural complex: putative roles for active site amino acids
The structure of cABC I contains three domains, namely an N-terminal β-domain with a jellyroll fold, the catalytic α-helix domain [incomplete toroid (α/α)5-fold] and a C-terminal antiparallel β-sheet domain. The structural fold of cABC I, comprising the catalytic α-helix domain and the C-terminal β-sheet domain, is very similar to that of cAC and bacterial HALs (hyaluronate lyases) [41]. To obtain a clearer picture of the active site and positioning of the substrate within cABC I, its structure was superimposed on the enzyme–substrate co-crystal structures of the structurally related cAC [30] and HAL [41]. This superimposition aligned most of the C-terminal β-sheet domains. However, the α-helix domain did not align very well, since the cleft formed by the N- and C-terminal regions of this domain was more open in cABC I when compared with the closed grooves found in cAC and HAL. Consistent with the earlier structural alignment [34], the four structurally conserved amino acids His-501, Tyr-508, Arg-560 and Glu-653 appear to be involved in the catalytic activity of cABC I.
The superimposition of the cAC co-crystal structures with their CS and DS substrates [30,42] and cABC I provided a guiding framework for modelling the cABC I–substrate structural complexes. Using the information from the co-crystal structures as a starting point, the representative low-energy enzyme–substrate complexes were obtained by systematic energy minimization methods where the active site amino acids and the substrates were allowed to move freely. First, unfavourable steric contacts were removed by an initial energy minimization to obtain good starting structures for the enzyme–substrate complexes. These structures were further subjected to total energy minimization to obtain local minimum energy structural complexes.
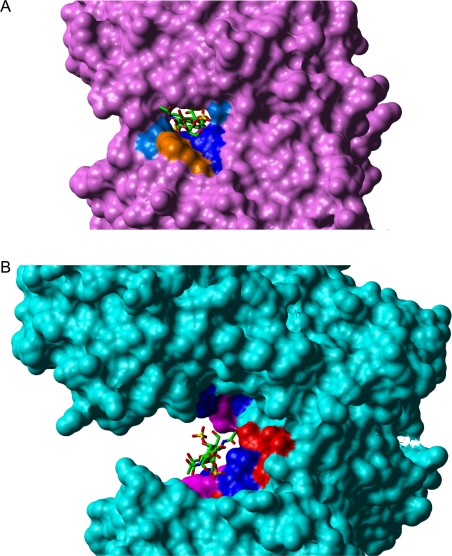
In the case of cABC I, the active site groove is more open, and there are no tryptophan residues to provide the necessary constraints on the orientation by means of ring-stacking interactions. These differences in the topology of the active sites of cAC and cABC I could perhaps account for the broader substrate specificity of cABC I (Figure 5). These differences are outlined in further detail in the following sections, which discuss the interactions of the active site amino acids of cABC I with C4S and DS substrates.
Figure 5. Structural comparison of cAC and cABC I.
Grasp rendering of (A) cAC (pink) and (B) cABC I (blue) structural complexes with DS. Note that the active site groove of cAC is more closed compared with that of cABC I. This narrower groove and the presence of Trp-127 and Trp-427 (both orange) in cAC locks the dermatan substrate in an orientation that allows binding to the active site but does not allow cleavage. On the other hand, the wider active site of cABC I provides room for the dermatan substrate to reorient during catalysis.
Interactions with C4S substrate
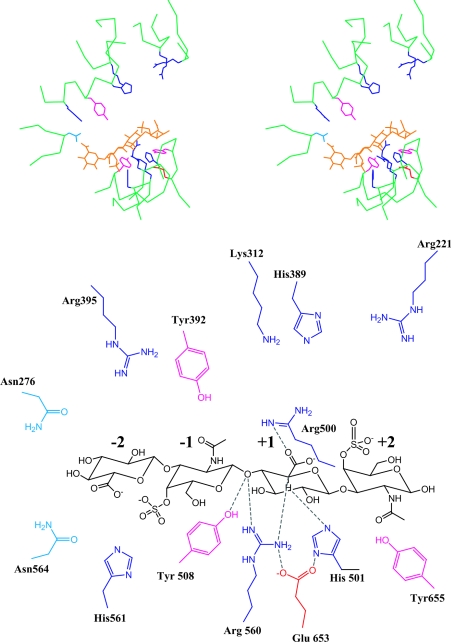
The position and orientation of C4S relative to the active site of cABC I was obtained using the cAC co-crystal structure with the C4S substrate as a starting point (Figure 6). From the proximity of the amino acids towards the C-5 proton of GlcA in the +1 subsite, His-501 is positioned more favourably to abstract this proton compared with Tyr-508. The close proximity between the glycosidic oxygen of the −1, +1 glycosidic bond and Tyr-508 suggests that it is more liable to play a role in protonating the leaving group. Arg-560 is also proximal to the glycosidic oxygen of the scissile bond and its equivalent cAC Arg-288 has been implicated to play a role in protonation of the leaving group [30]. However, the R288A mutant in cAC did not completely abolish its activity [30]. Glu-653 does not seem to be involved directly in catalysis, but it is at a hydrogen bonding distance with His-501 and Arg-560 and thus probably plays a critical role in positioning these residues for catalysis. The hydrogen bonding interaction of the analogous cAC Glu-371 with the corresponding histidine and arginine residues has been observed in cAC and a similar role for Glu-371 has been proposed [30]. Another important step in the β-eliminative cleavage of lyases is the neutralization of the charge on the carboxy group of the uronic acid to facilitate the abstraction of the proton. Based on our structural complex, Arg-500 is positioned to interact with the carboxylate group of the uronic acid at the +1 subsite. This role for Arg-500 has also been implicated in the earlier cABC I crystal structure study [34]. The stabilization of the carbanion intermediate could be achieved by potential interactions of either the protonated base His-501 or Arg-500 or Arg-560, since all of these groups are positioned to interact with the C-5 carbanion on the uronic acid formed after abstraction of the α-proton.
Figure 6. cABC I and the C4S substrate.
Shown above is a stereoview of C4S substrate in the active site. The oligosaccharide is coloured in orange. The basic amino acids (histidine, arginine and lysine in dark blue), acidic amino acids (aspartic and glutamine in red), and tyrosine (in pink) are also shown. Shown below is a detailed schematic of the different amino acids in the active site (numbered according to the crystal structure) and their proximity to the oligosaccharide. The cleavable bond is between the −1 and +1 sites, and the monosaccharides on the non-reducing side of the cleavable bond are numbered −2, −3, etc. and those on the reducing side are numbered +2, +3, etc.
Interactions with DS substrate
The initial positioning of the DS substrate based on its relative orientation in the cAC active site was such that the C-5 proton of the IdoA in the +1 subsite was facing away from the putative catalytic amino acids. In the cAC active site, the DS substrate is locked in this orientation due to the structural constraints imposed by the more closed active site groove and the two tryptophan amino acids. These observations are consistent with the inability of cAC to cleave DS. Rather, it has been observed that DS binds to the active site of cAC and inhibits its activity towards chondroitin substrates [30].
The chemical differences between IdoA in DS and GlcA in C4S provide further insights into differences in the positioning of these substrates within the active site. Whereas in GlcA the C-5 proton and the glycosidic oxygen of the cleavable bond are in the cis-orientation, in IdoA these two atoms are in the trans-orientation. Since the active site amino acids involved in the proton abstraction (from C-5 of uronic acid) and donation (to the glycosidic oxygen of the cleaved bond) are on the same face of the active site groove, the DS substrate would need to reorient in order for these amino acids to cleave it. Thus there are salient differences between the mechanism governing DS cleavage and the mechanism cABC I employs for C4S degradation.
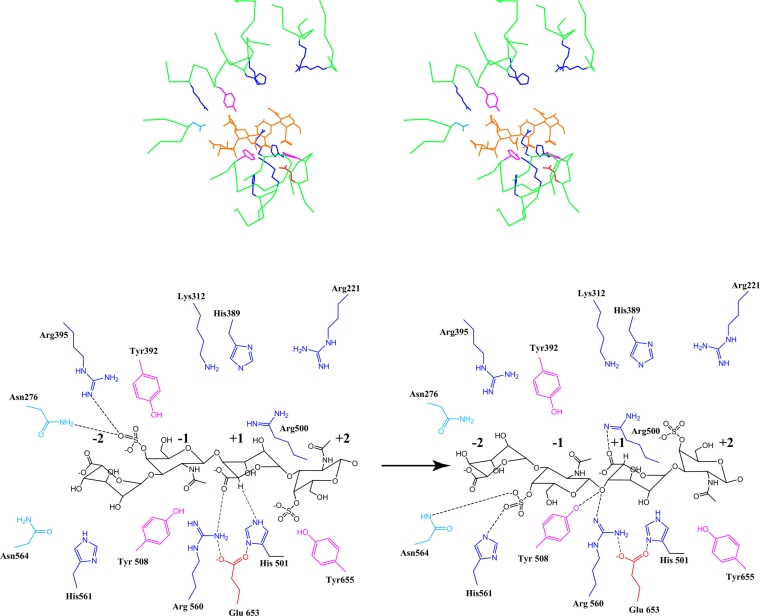
Based on our model of the cABC I–DS structural complex, His-501 is still the best candidate to serve as the general base that abstracts the C-5 proton from the IdoA in the +1 subsite (Figure 7). The positioning of the C-5 proton such that it would be abstracted by His-501 results in the carboxylate group of the IdoA being proximal to Arg-560. Thus Arg-560 could potentially play a role in neutralizing the charge on this group to make the C-5 proton more labile (Figure 7). It is possible that the dermatan substrate reorients itself after proton abstraction for the lone pair of glycosidic oxygen to face the active site Tyr-508 or Arg-560. The fact that Arg-560 and Arg-500 are on the opposite sides of the substrate provide further flexibility for interaction with substrate during the proton abstraction and donation processes (Figure 7).
Figure 7. cABC I and the DS substrate.
Shown above is a stereoview of DS in the active site. The colour scheme is analogous to Figure 6. Shown below is a detailed schematic of the interaction between various active site amino acids and the dermatan substrate orientated optimally for proton abstraction (left) and proton donation (right). The numbering of monosaccharides (−2, −1, +1 and +2) is described in Figure 6. Note that the two schematics are shown for clarity. It is possible that there is a reorientation of the substrate during catalysis. However, this cannot be determined conclusively.
Implications for the mode of action of cABC I
The results from the site-directed mutagenesis of the active site amino acids are consistent with the interactions observed in the theoretical enzyme–substrate structural complexes. Coupled analysis of both the kinetics experiments and the investigations of product profiles by CE strongly support the notion that His-501 is absolutely critical for the activity of cABC I on C6S, DS and C4S. From this we suggest that His-501 plays a central role in catalysis, the abstraction of the C-5 proton from the uronic acid moiety. The proximity of Tyr-508 to the C-5 proton and the glycosidic oxygen in the cABC I–chondroitin structural complex suggested its potential role in protonating the leaving group. However, the Y508F mutant did produce products against both C6S and DS in the overnight digestion. This mutant retains the hydrophobic group of tyrosine but abolishes the proton donating hydroxy group. Thus the role of Tyr-508 in proton donation is less conclusive and it can potentially be compensated by water molecules in the active site, albeit at reduced catalytic efficiency consistent with the kinetics data. This also suggests that the aromatic ring of Tyr-508 is likely to play a role in the positioning of the substrate.
The role of Glu-653 in forming the hydrogen bonding network with His-501 and Arg-560 is confirmed by the kinetic data and overnight product formation of E653D and E653Q mutants. Indeed, the relatively unchanged Km between cABC I and E653Q for both C6S (Table 2) and DS (Table 3) suggests that this residue is not important for directly binding the substrate. Furthermore, the diminished protrusion of the aspartic acid hydrogen bonding side chain as compared with the glutamic acid and glutamine side chains is consistent with the relative order of activities (Figure 4).
The proximity of Arg-560 to the C-5 atom and the glycosidic oxygen suggests that it may play a role in proton donation and/or stabilization of the carbanion intermediate following proton abstraction. It is more likely to play the latter role since proton donation can be compensated via neighbouring water molecules, and further, the Tyr-508 is better positioned to protonate the leaving group based on our model. Arg-560 could potentially play an additional role in neutralizing the carboxylate of the IdoA in DS based on its proximity to this group in the modelled cABC I–dermatan structural complex. Our structural model of the cABC I–CS complex and the earlier crystal structure studies [30,34] suggest that Arg-500 is likely to neutralize the charge on the carboxylate group of GlcA in CS. However, the R500A mutant does not conclusively justify this role since this mutant retains its activity towards chondroitin and dermatan substrates.
In addition to the catalytic amino acids already described, the cABC I active site contains several other basic residues. While some of these have structural analogues in the cAC and HAL co-crystal structures, there are many others that are unique to cABC I. The highly basic nature of the active site could potentially be involved in accommodating a wide variety of substrates including C4S, C6S, DS and HA, which have different charge distributions due to their differences in sulphation pattern. His-561 and Asn-564 are positioned to interact with the 4-O-sulphate group of the GalNAc4S at the −1 subsite (present in both C4S and DS). Furthermore, several additional basic amino acids, including His-388, Arg-395, Arg-105 and Lys-312 are located on the upper side of the active site cleft towards the N-terminal β-sheet domain (Figure 5). These basic amino acids could play a potential role in governing the specificity of cABC I towards different substrates.
The catalytic machinery of cABC I
The present study provides a first step towards identifying and defining the roles of the active site amino acids in substrate recognition, positioning and processing functions of cABC I. cABC I, like other GalAG lyases, cleaves its substrate at the glycosidic linkage between the hexosamine and uronic acid via a β-elimination mechanism. The fundamental steps involved in this mechanism are proton acceptance and donation [43]: (i) the abstraction of the C-5 proton on the uronic acid moiety, causing the formation of a double bond between C-4 and C-5, (ii) the stabilization of the carbanion intermediate, and (iii) the protonation of the anomeric oxygen, breaking the glycosidic bond. By coupling kinetic analysis of site-directed mutants of the active site amino acids with the construction of theoretical enzyme–substrate structural complexes to interpret the effects of the mutants, the detailed roles of the four active site amino acids have been outlined.
His-501 is an absolutely critical residue, as its mutants (Ala, Lys and Arg) proved null against all GalAG substrates. This residue is probably involved in proton abstraction from the uronic acid moiety of the GalAG. Although our initial studies with Y508A proved consistent with its acting as a general base, our Y508F mutant eliminated the likelihood of this function. Y508F generated products on overnight digestion with all GalAG substrates, making it unlikely that this residue modulates such a central role in catalysis. Our results suggest that Tyr-508 plays a role in the positioning of substrate via hydrogen bonding with the glycosidic bond of the GalAG. Similarly, the role of Glu-653 with other catalytic residues via hydrogen bonding is confirmed by our mutagenesis studies. Arg-560 seems to play an important part in cABC I's activity, perhaps in stabilizing the enolate intermediate through its positive charge. It also may act as the general acid in the reaction.
We also performed mutagenesis on Arg-500, since it was previously suggested that Arg-500 would be able to interact with the carboxylate group of either of the epimers of uronic acid, thus enabling cABC I's most remarkable ability to process both CS and DS [34]. Through its side chain flexibility, Arg-500's guanidinium group was thought to aid in charge neutralization of either of the uronic acid configurations. However, our results indicate that Arg-500 is not crucial for the processing of GalAG substrates, since the R500A mutant did generate products in both product profile studies and kinetics assays. It may be that a number of residues are important in this charge neutralization process.
Based on the enzyme–substrate structural complexes, we obtained insights into the differences in processing of CS and DS by cABC I. Our structural models suggest that the catalytic residues in cABC I are positioned to cleave chondroitin substrates more favourably than dermatan substrates. This is consistent with the kinetics of the wild-type enzyme, which shows increased catalytic efficiency towards chondroitin versus dermatan substrates. The enzyme–substrate structural models also revealed several other amino acids in the active site that are likely to play a role in substrate positioning and specificity. We are currently studying the roles of these amino acids in the substrate specificity and processing of cABC I. In summary, engineering cABC enzymes via site-directed mutagenesis would provide a repertoire of mutants with fine-tuned substrate specificities. These mutants not only facilitate the structural characterization of GalAGs, but they can also be directly utilized in physiological scenarios such as nerve regeneration after spinal cord injury, thus expanding the scope of treatment strategies.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) grant GM57073. We thank the Multiuser Facility for the study of complex macromolecular systems (The Massachusetts Institute of Technology; NSF-0070319 and NIH-GM68762) for access to instrumentation. V.P. was the recipient of the National Institutes of Health Biotechnology training grant (5-T32-GM08334) and a DuPont Fellowship (Massachusetts Institute of Technology). C.J.B. was funded through the NIH/NIEHS training grant in environmental toxicology (5-T32-ES0720).
References
- 1.Ernst S., Langer R., Cooney C. L., Sasisekharan R. Enzymatic degradation of glycosaminoglycans. Crit. Rev. Biochem. Mol. Biol. 1995;30:387–444. doi: 10.3109/10409239509083490. [DOI] [PubMed] [Google Scholar]
- 2.Nadanaka S., Sugahara K. The unusual tetrasaccharide sequence GlcA β1-3GalNAc(4-sulfate)β1-4GlcA(2-sulfate)β1-3GalNAc(6-sulfate) found in the hexasaccharides prepared by testicular hyaluronidase digestion of shark cartilage chondroitin sulfate D. Glycobiology. 1997;7:253–263. doi: 10.1093/glycob/7.2.253. [DOI] [PubMed] [Google Scholar]
- 3.Sugahara K., Tanaka Y., Yamada S., Seno N., Kitagawa H., Haslam S. M., Morris H. R., Dell A. Novel sulfated oligosaccharides containing 3-O-sulfated glucuronic acid from king crab cartilage chondroitin sulfate K: unexpected degradation by chondroitinase ABC. J. Biol. Chem. 1996;271:26745–26754. doi: 10.1074/jbc.271.43.26745. [DOI] [PubMed] [Google Scholar]
- 4.Trowbridge J. M., Gallo R. L. Dermatan sulfate: new functions from an old glycosaminoglycan. Glycobiology. 2002;12:117R–125R. doi: 10.1093/glycob/cwf066. [DOI] [PubMed] [Google Scholar]
- 5.Sugahara K., Mikami T., Uyama T., Mizuguchi S., Nomura K., Kitagawa H. Recent advances in the structural biology of chondroitin sulfate and dermatan sulfate. Curr. Opin. Struct. Biol. 2003;13:612–620. doi: 10.1016/j.sbi.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 6.Liaw P. C., Austin R. C., Fredenburgh J. C., Stafford A. R., Weitz J. I. Comparison of heparin- and dermatan-sulfate-mediated catalysis of thrombin inactivation by heparin cofactor II. J. Biol. Chem. 1999;274:27597–27604. doi: 10.1074/jbc.274.39.27597. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez J. A., Petaja J., Griffin J. H. Dermatan sulfate and LMW heparin enhance the anticoagulant action of activated protein C. Thromb. Haemostasis. 1999;82:1462–1468. [PubMed] [Google Scholar]
- 8.Iozzo R. V. The family of the small leucine-rich proteoglycans: key regulators of matrix assembly and cellular growth. Crit. Rev. Biochem. Mol. Biol. 1997;32:141–174. doi: 10.3109/10409239709108551. [DOI] [PubMed] [Google Scholar]
- 9.Tumova S., Woods A., Couchman J. R. Heparan sulfate chains from glypican and syndecans bind the Hep III domain of fibronectin similarly despite minor structural differences. J. Biol. Chem. 2000;275:9410–9417. doi: 10.1074/jbc.275.13.9410. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt G., Robenek H., Harrach B., Glossl J., Nolte V., Hormann H., Richter H., Kresse H. Interaction of small dermatan sulfate proteoglycan from fibroblasts with fibronectin. J. Cell Biol. 1987;104:1683–1691. doi: 10.1083/jcb.104.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker A., Gallagher J. T. Structural domains of heparan sulphate for specific recognition of the C-terminal heparin-binding domain of human plasma fibronectin (HEPII) Biochem. J. 1996;317:871–877. doi: 10.1042/bj3170871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elefteriou F., Exposito J. Y., Garrone R., Lethias C. Binding of tenascin-X to decorin. FEBS Lett. 2001;495:44–47. doi: 10.1016/s0014-5793(01)02361-4. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi Y., Mann D. M., Ruoslahti E. Negative regulation of transforming growth factor-β by the proteoglycan decorin. Nature (London) 1990;346:281–284. doi: 10.1038/346281a0. [DOI] [PubMed] [Google Scholar]
- 14.Hildebrand A., Romaris M., Rasmussen L. M., Heinegard D., Twardzik D. R., Border W. A., Ruoslahti E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor β. Biochem. J. 1994;302:527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon M., Deakin J. A., Mizuno K., Nakamura T., Gallagher J. T. Interaction of hepatocyte growth factor with heparan sulfate: elucidation of the major heparan sulfate structural determinants. J. Biol. Chem. 1994;269:11216–11223. [PubMed] [Google Scholar]
- 16.Lyon M., Deakin J. A., Rahmoune H., Fernig D. G., Nakamura T., Gallagher J. T. Hepatocyte growth factor/scatter factor binds with high affinity to dermatan sulfate. J. Biol. Chem. 1998;273:271–278. doi: 10.1074/jbc.273.1.271. [DOI] [PubMed] [Google Scholar]
- 17.Mascellani G., Liverani L., Bianchini P., Parma B., Torri G., Bisio A., Guerrini M., Casu B. Structure and contribution to the heparin cofactor II-mediated inhibition of thrombin of naturally oversulphated sequences of dermatan sulphate. Biochem. J. 1993;296:639–648. doi: 10.1042/bj2960639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maimone M. M., Tollefsen D. M. Structure of a dermatan sulfate hexasaccharide that binds to heparin cofactor II with high affinity. J. Biol. Chem. 1990;265:18263–18271. [PubMed] [Google Scholar]
- 19.Denholm E. M., Lin Y. Q., Silver P. J. Anti-tumor activities of chondroitinase AC and chondroitinase B: inhibition of angiogenesis, proliferation and invasion. Eur. J. Pharmacol. 2001;416:213–221. doi: 10.1016/s0014-2999(01)00884-6. [DOI] [PubMed] [Google Scholar]
- 20.Iozzo R. V., Cohen I. Altered proteoglycan gene expression and the tumor stroma. Experientia. 1993;49:447–455. doi: 10.1007/BF01923588. [DOI] [PubMed] [Google Scholar]
- 21.Makatsori E., Lamari F. N., Theocharis A. D., Anagnostides S., Hjerpe A., Tsegenidis T., Karamanos N. K. Large matrix proteoglycans, versican and perlecan, are expressed and secreted by human leukemic monocytes. Anticancer Res. 2003;23:3303–3309. [PubMed] [Google Scholar]
- 22.Papadas T. A., Stylianou M., Mastronikolis N. S., Papageorgakopoulou N., Skandalis S., Goumas P., Theocharis D. A., Vynios D. H. Alterations in the content and composition of glycosaminoglycans in human laryngeal carcinoma. Acta Otolaryngol. 2002;122:330–337. doi: 10.1080/000164802753648268. [DOI] [PubMed] [Google Scholar]
- 23.Vicente C. P., Zancan P., Peixoto L. L., Alves-Sa R., Araujo F. S., Mourao P. A., Pavao M. S. Unbalanced effects of dermatan sulfates with different sulfation patterns on coagulation, thrombosis and bleeding. Thromb. Haemostasis. 2001;86:1215–1220. [PubMed] [Google Scholar]
- 24.Gandra M., Cavalcante M., Pavao M. Anticoagulant sulfated glycosaminoglycans in the tissues of the primitive chordate Styela plicata (Tunicata) Glycobiology. 2000;10:1333–1340. doi: 10.1093/glycob/10.12.1333. [DOI] [PubMed] [Google Scholar]
- 25.Venkataraman G., Shriver Z., Raman R., Sasisekharan R. Sequencing complex polysaccharides. Science. 1999;286:537–542. doi: 10.1126/science.286.5439.537. [DOI] [PubMed] [Google Scholar]
- 26.Rhomberg A. J., Ernst S., Sasisekharan R., Biemann K. Mass spectrometric and capillary electrophoretic investigation of the enyzmatic degradation of heparin-like glycosaminoglycans. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4176–4181. doi: 10.1073/pnas.95.8.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst S., Rhomberg A. J., Biemann K., Sasisekharan R. Direct evidence for a predominantly exolytic processive mechanism for depolymerization of heparin-like glycosaminoglycans by heparinase I. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pojasek K., Shriver Z., Kiley P., Venkataraman G., Sasisekharan R. Recombinant expression, purification, and kinetic characterization of chondroitinase AC and chondroitinase B from Flavobacterium heparinum. Biochem. Biophys. Res. Commun. 2001;286:343–351. doi: 10.1006/bbrc.2001.5380. [DOI] [PubMed] [Google Scholar]
- 29.Huang W., Matte A., Li Y., Kim Y. S., Linhardt R. J., Su H., Cygler M. Crystal structure of chondroitinase B from Flavobacterium heparinum and its complex with a disaccaride product at 1.7 Å resolution. J. Mol. Biol. 1999;294:1257–1269. doi: 10.1006/jmbi.1999.3292. [DOI] [PubMed] [Google Scholar]
- 30.Huang W., Boju L., Tkalec L., Su H., Yang H. O., Gunay N. S., Linhardt R. J., Kim Y. S., Matte A., Cygler M. Active site of chondroitin AC lyase revealed by the structure of enzyme–oligosaccharide complexes and mutagenesis. Biochemistry. 2001;40:2359–2372. doi: 10.1021/bi0024254. [DOI] [PubMed] [Google Scholar]
- 31.Michel G., Pojasek K., Li Y., Sulea T., Linhardt R. J., Raman R., Prabhakar V., Sasisekharan R., Cygler M. The structure of chondroitin B lyase complexed with glycosaminoglycan oligosaccharides unravels a calcium-dependent catalytic machinery. J. Biol. Chem. 2004;279:32882–32896. doi: 10.1074/jbc.M403421200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradbury E. J., Moon L. D., Popat R. J., King V. R., Bennett G. S., Patel P. N., Fawcett J. W., McMahon S. B. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature (London) 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 33.Prabhakar V., Capila I., Bosques C. J., Pojasek K., Sasisekharan R. Chondroitinase ABC I from Proteus vulgaris: cloning, recombinant expression, and active site identification. Biochem. J. 2005;386:103–112. doi: 10.1042/BJ20041222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W., Lunin V. V., Li Y., Suzuki S., Sugiura N., Miyazono H., Cygler M. Crystal structure of Proteus vulgaris chondroitin sulfate ABC lyase I at 1.9 Å resolution. J. Mol. Biol. 2003;328:623–634. doi: 10.1016/s0022-2836(03)00345-0. [DOI] [PubMed] [Google Scholar]
- 35.Pojasek K., Raman R., Kiley P., Venkataraman G., Sasisekharan R. Biochemical characterization of the chondroitinase B active site. J. Biol. Chem. 2002;277:31179–31186. doi: 10.1074/jbc.M201552200. [DOI] [PubMed] [Google Scholar]
- 36.Alexandrov N. N., Fischer D. Analysis of topological and nontopological structural similarities in the PDB: new examples with old structures. Proteins. 1996;25:354–365. doi: 10.1002/(SICI)1097-0134(199607)25:3<354::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 37.Homans S. W. A molecular mechanical force field for the conformational analysis of oligosaccharides: comparison of theoretical and crystal structures of Man α1-3Man β1-4GlcNAc. Biochemistry. 1990;29:9110–9118. doi: 10.1021/bi00491a003. [DOI] [PubMed] [Google Scholar]
- 38.Ernst S., Venkataraman G., Sasisekharan V., Cooney C. L., Sasisekharan R. Pyranose ring flexibility: mapping of physical data for iduronate in continuous conformational space. J. Am. Chem. Soc. 1998;120:2099–2107. [Google Scholar]
- 39.Hamai A., Hashimoto N., Mochizuki H., Kato F., Makiguchi Y., Horie K., Suzuki S. Two distinct chondroitin sulfate ABC lyases: an endoeliminase yielding tetrasaccharides and an exoeliminase preferentially acting on oligosaccharides. J. Biol. Chem. 1997;272:9123–9130. doi: 10.1074/jbc.272.14.9123. [DOI] [PubMed] [Google Scholar]
- 40.Sato N., Shimada M., Nakajima H., Oda H., Kimura S. Cloning and expression in Escherichia coli of the gene encoding the Proteus vulgaris chondroitin ABC lyase. Appl. Microbiol. Biotechnol. 1994;41:39–46. doi: 10.1007/BF00166079. [DOI] [PubMed] [Google Scholar]
- 41.Jedrzejas M. J., Mello L. V., de Groot B. L., Li S. Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase: structures of complexes with the substrate. J. Biol. Chem. 2002;277:28287–28297. doi: 10.1074/jbc.M112009200. [DOI] [PubMed] [Google Scholar]
- 42.Lunin V. V., Li Y., Linhardt R. J., Miyazono H., Kyogashima M., Kaneko T., Bell A. W., Cygler M. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: an enzyme–substrate complex defines the catalytic mechanism. J. Mol. Biol. 2004;337:367–386. doi: 10.1016/j.jmb.2003.12.071. [DOI] [PubMed] [Google Scholar]
- 43.Jedrzejas M. J. Structural and functional comparison of polysaccharide-degrading enzymes. Crit. Rev. Biochem. Mol. Biol. 2000;35:221–251. doi: 10.1080/10409230091169195. [DOI] [PubMed] [Google Scholar]